Abstract
Electrical signaling in biology is typically associated with action potentials, transient spikes in membrane voltage that return to baseline. Hodgkin-Huxley and related conductance-based models of electrophysiology belong to a more general class of reaction-diffusion equations which could, in principle, support spontaneous emergence of patterns of membrane voltage which are stable in time but structured in space. Here we show theoretically and experimentally that homogeneous or nearly homogeneous tissues can undergo spontaneous spatial symmetry breaking through a purely electrophysiological mechanism, leading to formation of domains with different resting potentials separated by stable bioelectrical domain walls. Transitions from one resting potential to another can occur through long-range migration of these domain walls. We map bioelectrical domain wall motion using all-optical electrophysiology in an engineered cell line and in human induced pluripotent stem cell (iPSC)-derived myoblasts. Bioelectrical domain wall migration may occur during embryonic development and during physiological signaling processes in polarized tissues. These results demonstrate that nominally homogeneous tissues can undergo spontaneous bioelectrical symmetry breaking.
Introduction
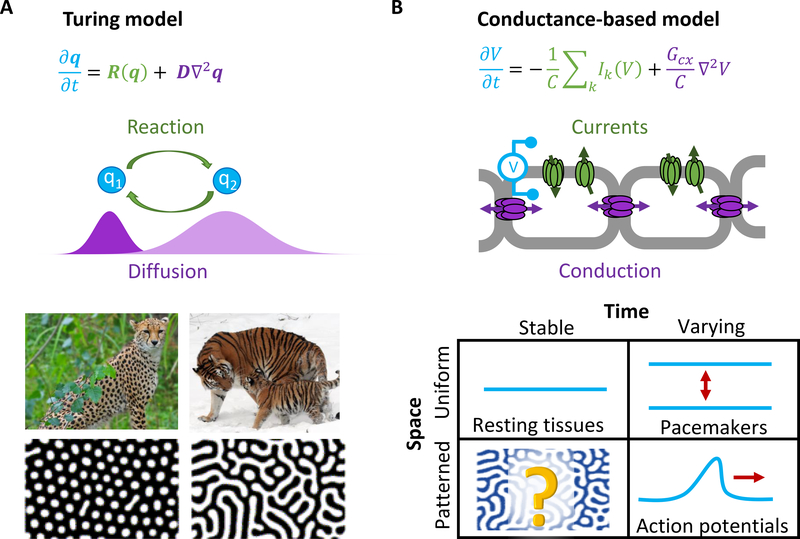
In 1952, Hodgkin and Huxley introduced a mathematical model of action potential propagation in the squid giant axon, based on nonlinear dynamics of electrically coupled ion channels.1 In the same year, Alan Turing proposed a model for biological pattern formation, based on diffusion and nonlinear reaction dynamics of chemical morphogens.2 These two seemingly unrelated models have an underlying mathematical kinship: both are nonlinear reaction-diffusion equations, first order in time and second order in space. Thus, from a mathematical perspective, one expects parallel classes of solutions. These solutions can be organized by whether they are uniform or patterned in space, and stable or varying in time (Fig. 1). All four combinations of spatial and temporal structure have been observed in chemical reaction-diffusion systems3, but only three of the four have been reported in nominally homogeneous systems governed by Hodgkin-Huxley-like conductance-based models. We thus sought to observe the fourth class of electrophysiological dynamics: spontaneous spatial symmetry breaking in a nominally homogeneous tissue to create patterns of membrane voltage that are static in time but that vary in space.
Figure 1. Biochemical and bioelectrical spontaneous pattern formation.
A) In chemical Turing patterns, a nonlinear chemical reaction coupled to diffusion leads to spontaneous formation of stable concentration patterns from homogeneous initial conditions, similar to ones seen in Nature. Here q = (q1, q2) is the vector of reagent concentrations, R(q) is the nonlinear relation between concentration and reaction rate, and D is the vector of diffusion coefficients. B) Conductance-based models have the same structure as the Turing equation. Here V is the membrane voltage, C is the membrane capacitance, Ik is the current through the kth ion channel, and Gcx is the connexin conductivity. The chart shows possible solutions to an initially homogeneous conductance-based model, classified by variation in space and time. Spontaneous patterns that vary in space but not in time are a little-explored possibility in electrophysiology. Images of natural and simulated patterns adapted from Wikipedia and Kondo et al3.
Spatial symmetry breaking might emerge during slow transitions in membrane potential, such as occur during embryonic development and in signaling processes in peripheral organs. While spontaneous pattern-forming processes in electrophysiology have been contemplated,4–8 unambiguous observations with clear mechanistic interpretations have been lacking. Part of the experimental challenge comes from the difficulty of spatially mapping membrane voltage. Patch clamp measurements of membrane potential probe the voltage at only discrete points in space, and are thus ill-suited to mapping spatial structure. Recent advances in voltage imaging facilitate spatially resolved measurements 9,10, and optogenetic stimulation offers the prospect to tune the electrophysiological state of a tissue and perhaps to drive it into a regime of spontaneous symmetry breaking.
Here, we explore these ideas experimentally in engineered cells expressing the inward-rectifying potassium channel Kir2.1 and the channelrhodopsin CheRiff (Fig. 2A,B). While this two-component cellular model is so simple as to appear almost trivial, we find that coupled ensembles of these cells show richly diverse transitional behaviors, including electrical bistability, bioelectrical domain walls, and noise-induced breakup into discrete electrical domains. We further show that similar dynamics occur in human induced pluripotent stem cell (iPSC)-derived myocytes during differentiation. Our results demonstrate bioelectrical pattern formation and domain wall motion as generic mechanisms by which tissues can switch from one membrane voltage to another.
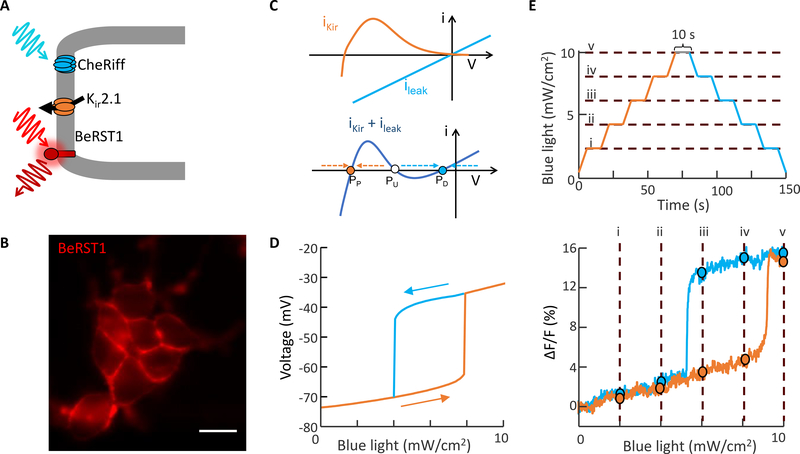
Figure 2. Electrophysiological bistability in an engineered cell line.
A) Bi-HEK cells expressed an inward-rectifier potassium channel, Kir2.1, and a light-gated ion channel, CheRiff. A far-red voltage-sensitive dye, BeRST1, reported membrane potential. B) Fluorescence image of bi-HEK cells labeled with BeRST1. Scale bar 20 μm. C) Expression of an inward rectifier potassium channel (e.g. Kir 2.1) and a non-selective leak conductance (e.g. channelrhodopsin) are sufficient, together, to produce electrical bistability. D) Numerical simulations showing hysteresis of steady-state membrane potential under ramped optogenetic stimulation. Simulations were based on measured I-V curves of bi-HEK cells (Methods). E) Top: optogenetic stimulus waveform. Epochs of constant illumination intensity have been indicated with Roman numerals. Bottom: Optical measurements of membrane voltage in a small cluster of bi-HEKs, exposed to the stimulus waveform above. Circles denote points where the optogenetic stimulus strength was held constant for 10 s.
Bistable membrane voltages
The lipid bilayer cell membrane behaves, electrically, as a parallel plate capacitor. Transmembrane protein channels can pass ionic currents which alter the intracellular charge, and hence the membrane voltage. In a single cell or a small isolated patch of tissue, the membrane voltage follows:
| [Eq. 1] |
where Cm is the membrane capacitance and I is the current through all ion channels (outward positive). Channel gating dynamics can impose a nonlinear and history-dependent relation between I and V which causes complex dynamics in excitable cells.
Resting potential in most polarized cells is set by an inward rectifier potassium channel, Kir. The current through the Kir channel is , where gK is the conductance (proportional to the number of channels in the membrane), and x∞(V) captures the voltage-dependent gating of the channel (shut at depolarized voltages, open at polarized voltages).11 The term (V − EK), where EK ∼ −90 mV is the potassium Nernst potential, accounts for the electrochemical driving force for ions to cross the membrane. The function IKir(V) crosses the x-axis at the potassium reversal potential. Inward rectification implies a drop in Kir current at more positive potentials. Together these attributes give Kir channels a non-monotonic I-V relationship (Fig. 2C).4,12 To a good approximation, the Kir conductance depends on present voltage only, not on history.
Cells typically have one or more leak conductances. We consider the simplest case: an Ohmic leak with reversal potential 0 mV and conductance gl, leading to a straight line I-V relation, Ileak = gl V. Leak conductances can be gated by external variables, e.g. by chemical ligands or mechanical forces. Below we use a non-selective cation-conducting channelrhodopsin, CheRiff, as a leak conductance where the value of gl is readily tunable via blue light 9.
The total current is the sum of the Kir and leak currents (Fig. 2C). When gl dominates, one has a single depolarized fixed point (I = 0) near 0 mV (PD). When gK dominates, one has a single polarized fixed point near −90 mV (PP). When gl and gK are approximately balanced, one has an N-shaped I-V curve which crosses the x-axis three times. This situation implies coexistence of stable fixed points PD and PP with an unstable fixed point (PU) in between, leading to overall bistability.13,14 From a dynamical systems perspective, this situation is analogous to the bistability observed in the famous E. coli Lac operon system.15,16
We genetically engineered a HEK293 cell line that stably expressed Kir2.1 and CheRiff (Methods).17,18 We call these bistable-HEK cells (bi-HEKs). Patch clamp measurements on small clusters (~50 μm diameter) of bi-HEKs revealed a non-monotonic I-V curve, which could be driven through two saddle-node bifurcations by light (Supplementary Fig. 1). We performed numerical simulations of a cell governed by Eq. 1, using the measured I-V curve (Methods). Under continuous variation in blue light the simulated membrane potential underwent sudden jumps at saddle node bifurcations where PU annihilated either PD or PP. The jumps occurred at different values of blue light in the polarizing and depolarizing directions, leading to hysteresis (Fig. 2D), i.e. within the hysteretic region, the membrane voltage was bistable.
We used a far-red voltage-sensitive dye, BeRST1 19, to report the membrane voltage in small clusters of bi-HEKs. Homogeneous blue illumination was slowly increased (0 to 10 mW/cm2 over 75 s) and then decreased using a piecewise-continuous waveform comprising linear ramps alternating with 10 s intervals of constant intensity (Fig. 2E, top). The 10 s periods of constant intensity were ~103-fold longer than the membrane electrical time constant (~10 ms), sufficient to ensure that the membrane voltage reached steady state. The optically recorded membrane voltage showed abrupt transitions and hysteresis, in close agreement with the numerical simulations (Fig. 2E, bottom). Furthermore, we did not detect drift in the membrane voltage during the periods of constant optogenetic drive, confirming that the dynamics were quasi-static. Thus, cells expressing Kir + leak exhibited a form of non-genetic electrophysiological memory: the steady-state membrane voltage was not uniquely specified by the ion channels alone. Rather, in the hysteretic regime the steady-state voltage depended on the history of ionic currents, which could in turn depend on the history of stimuli to the cell or, in principle, on the history of gene expression (e.g. whether the leak or the Kir channel was expressed first).4
Bioelectrical domains in extended tissues
In an extended tissue, neighboring cells can be coupled by gap junctions. When the voltage on a cell deviates from the mean of its neighbors, ionic currents flow through the gap junctions to minimize this deviation. The dynamics then become:
| [Eq. 2] |
where Gcxn is the sheet conductance due to gap junction channels. When the membrane potential is bistable (i.e. the ratio gl/gK is in the hysteretic portion of Fig. 2E), different regions of the tissue may sit at different resting potentials, PU and PD.20 A domain wall then emerges at the interface between these regions (Fig. 3A). In a homogeneous tissue, the domain wall is stationary only when the Kir and leak conductances are perfectly balanced, i.e. when the areas of the positive and negative portions of the I-V curve between PU and PD are equal.21 Otherwise the domain wall migrates to expand the territory of the stronger conductance (Supplementary Fig. 2A–C).
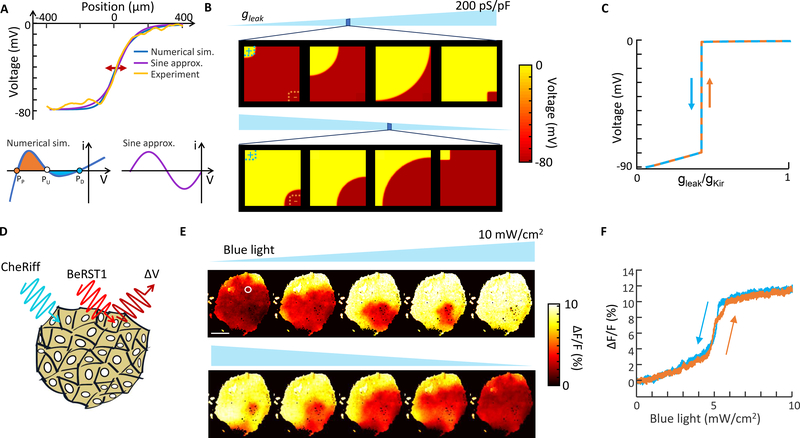
Figure 3. Bioelectric domain walls in an engineered cell line.
A) Top: Profile of a bioelectrical domain wall in one dimension, comparing numerical simulations based on a balanced Kir and leak current, an analytical approximation based on a sinusoidal I-V relation, and experimental data. Bottom: I-V curves based on a detailed biophysical model of Kir + leak (left) or a sinusoidal approximation (right). In a homogeneous tissue, the domain wall migrates in a direction set by the relative areas of the orange and blue shaded regions of the I-V curve, favoring the fixed point with the larger shaded region. B) Simulation of domain wall growth in a homogeneous tissue with two discrete defects to nucleate transitions (clamped at V = 0 on the top left, clamped at V = −90 mV on the bottom right). The transitions in the bulk tissue occurred over a narrow range of gleak. See Supplementary Movie S1. C) Simulation of membrane voltage in the tissue in (B) as a function of leak conductance. D) Confluent islands of bi-HEK cells were illuminated with uniform blue light to stimulate CheRiff, and with red light to elicit voltage-dependent fluorescence of BeRST1. E) Fluorescence images of an island of bi-HEKs under gradually increasing optogenetic stimulation. Scale bar 1 mm. See Supplementary Movies S2 and S3. White circle denotes region with voltage plotted in (F). F) Fluorescence as a function of optogenetic stimulus strength from the region circled in white in (E). Domain wall migration in the large island led to a step-like change in membrane potential without hysteresis.
To aid intuition, we introduce a simple analytical model of a bioelectrical domain wall. We approximate the I-V relation by a sinusoidal function, . While the sinusoidal approximation does not replicate the detailed shape of the I-V curve, it correctly captures the three zero-crossings between Ek and 0 mV (Fig. 3A). In one dimension, the steady-state domain wall profile () obeys
| [Eq. 3] |
Differential equations of this form appear in many contexts, perhaps most famously to describe the dynamics of a physical pendulum (with time replacing position as the independent variable).22 The domain wall profile is described by the separatrix solution which delineates the oscillatory from the rotatory solutions:
| [Eq. 4] |
The domain wall width scales as . The parameter A is a measure of the strength of the (non-gap junction) ionic currents, i.e. gK and gl in the more detailed biophysical model.
Fig. 3A compares the analytical approximation (Eq. 4) with numerical simulation using the complete expression for the Kir2.1 + leak I-V relation (Methods). The numerical and analytical domain wall profiles differ by at most 3.5 mV for a domain wall of height 77.5 mV.
Dual patch clamp measurements have shown that Gcxn is maximal at zero voltage difference between adjoining cells and decreases when the intercellular voltage exceeds approximately ±40 mV.23 Our simulations showed that in the domain walls the maximal nearest-neighbor voltage difference was < 1 mV, implying that the voltage dependence of Gcxn could safely be neglected. This effect may be necessary to include when the width of the domain wall approaches the size of a cell.
In two dimensions, simulations predicted that bioelectrical domains nucleated at defects (e.g. cells that expressed only leak or only Kir) and spread through the tissue (Fig. 3B, Supplementary Movie S1). In a tissue that was homogeneous but for an arbitrarily low density of nucleation points, the hysteresis vanished and the transition between depolarized and polarized states was abrupt (Fig. 3C). Thus the collective nature of the transition in an extended tissue was predicted to convert a gradual change in ionic currents into a highly sensitive phase change-like switch in membrane potential.
HEK cells endogenously express connexins 43 and 45 which mediate nearest-neighbor electrical coupling,24,25 so we reasoned that confluent monolayers of bi-HEK cells might support bioelectrical domain walls. We performed optogenetic stimulation and voltage imaging experiments in confluent islands of bi-HEKs with dimensions ~2 × 2 mm, corresponding to ~4×104 cells (Fig. 3D). Initially (in the absence of optogenetic stimulation) the tissue was homogeneously polarized. Illumination with dim blue light led to nucleation of depolarized domains near the tissue boundaries. We extracted the mean fluorescence intensity profile across the domain wall. When scaled to match the voltage axis, the profile agreed closely with the predictions of both the numerical simulation and the analytical approximation (Fig. 3A).
As the blue light further increased, the domain walls migrated across the tissue, until the whole tissue was depolarized (Fig. 3E, Supplementary Movies S2 and S3). The fluorescence intensity of most regions in the island showed step-like depolarization with little hysteresis, consistent with the theoretical predictions. Domain wall formation and migration were observed in 8 of 8 independently prepared and measured islands (Supplementary Fig. 3), though in some cases defects prevented depolarization of the entire island.
To test the stability of the domain walls, we applied a piecewise-continuous blue light waveform comprising linear ramps alternating with 10 s intervals of constant intensity, as in Fig. 2E. During each period of constant illumination the domain walls remained stationary. During each period of increasing or decreasing illumination, the domain walls advanced or retreated, respectively (Supplementary Fig. 4). The 10 s periods of domain wall stability were ~103-fold longer than the membrane relaxation time constant of ~10 ms, demonstrating the quasi-static nature of the electrical patterns. These experiments confirmed the existence of stable domain walls in a nominally homogeneous tissue, a hallmark of spontaneous spatial symmetry breaking.
We confirmed the role of gap junctions in domain wall migration by adding a gap junction blocker 2-aminoethyl diphenylborinate (2-APB, 50 μM) to island cultures of bi-HEKs. Before adding the gap junction blocker, a ramp of blue light caused the island to depolarize over a narrow range of blue light levels via domain wall migration, and the membrane potential showed little hysteresis. After adding the blocker, individual cells showed discrete hysteretic switching, each with its own transition points set by the cell-specific expression levels of Kir2.1 and CheRiff (Supplementary Fig. 5). Thus gap junctional coupling was necessary for the transition from zero-dimensional to two-dimensional behavior.
No tissue is perfectly uniform, so we explored via simulation tissues with cell-to-cell variations in expression of Kir or leak (Methods). Noisy ion channel expression introduced an effective friction for domain wall motion, stabilizing droplet-like domains of high or low voltage and broadening the transition in tissue-average voltage under a ramp in gl (Supplementary Fig. 6). Sufficiently strong heterogeneity led to stick/slip saltatory domain wall motion. The tissue-average voltage then showed Barkhausen-like fine-structure noise (Supplementary Fig. 7). Tissue heterogeneity also restored some degree of hysteresis in the tissue-average voltage, and, when strong enough, broke the tissue into discrete domains that switched independently. The predicted voltage dynamics of coupled cells expressing leak + Kir thus exhibited many of the features found in magnetization of a disordered soft ferromagnet.26
Signatures of noisy ion channel expression were observable in our experiments on island cultures. Due to regional variations in the depolarizing transition point, the voltage averaged over the whole island showed a broader transition than did local measurements (Supplementary Fig. 7). The island-average voltage showed Barkhausen-like fine-structure noise, and a small amount of hysteresis, indicative of stick-slip domain motion. Numerical simulations of islands with noisy gene expression recapitulated these effects (Supplementary Fig. 7). In cultures where transient transfection of Kir2.1 led to enhanced cell-to-cell variability in gK (Methods), we observed breakup into regions in which the voltage showed hysteretic zero-dimension-like behavior and regions which showed smooth and reversible depolarization, in concordance with simulations (Supplementary Fig. 7, Supplementary Movies S4 and S5).
Electrical bistability and hysteresis during myogenesis
Early embryonic tissue has a membrane voltage, Vm ~ 0 mV 27. During myogenesis, myoblast precursors polarize electrically, exit the cell cycle, and fuse into myocytes whose resting potential is ~−85 mV.28 Expression of Kir2.1 initiates this hyperpolarization.29 In mammals, myoblast precursors couple transiently via gap junctions during differentiation and prior to fusion.30 We thus hypothesized that bistability and bioelectric domain wall motion might occur during myogenesis.
We performed all-optical electrophysiology experiments in human induced pluripotent stem cell (hiPSC) derived myoblasts as they differentiated into myocyte fibers in vitro (Fig. 4A). HiPSC myoblasts were seeded at low density, lentivirally transduced to expresses CheRiff, and then allowed to proliferate to form a confluent monolayer (Methods; Fig. 4A). The cells were then differentiated into myocytes. After one week of differentiation, cells expressed stained positive for myogenin, PAX7, and myosin heavy chain, and adopted an elongated fiber-like morphology, indicative of differentiation toward mature myocytes (Fig. 4B). RNAseq measurements on matched samples showed a significant increase in Kir2.1 expression during the differentiation process (5.6-fold, p <0.001) and high expression of gap junction proteins Cx43 and Cx45 throughout (Methods). We performed voltage imaging under ramped wide-field optogenetic stimulation at two time-points during differentiation to test for signatures of electrical bistability in isolated cells and domain wall motion in confluent cultures.
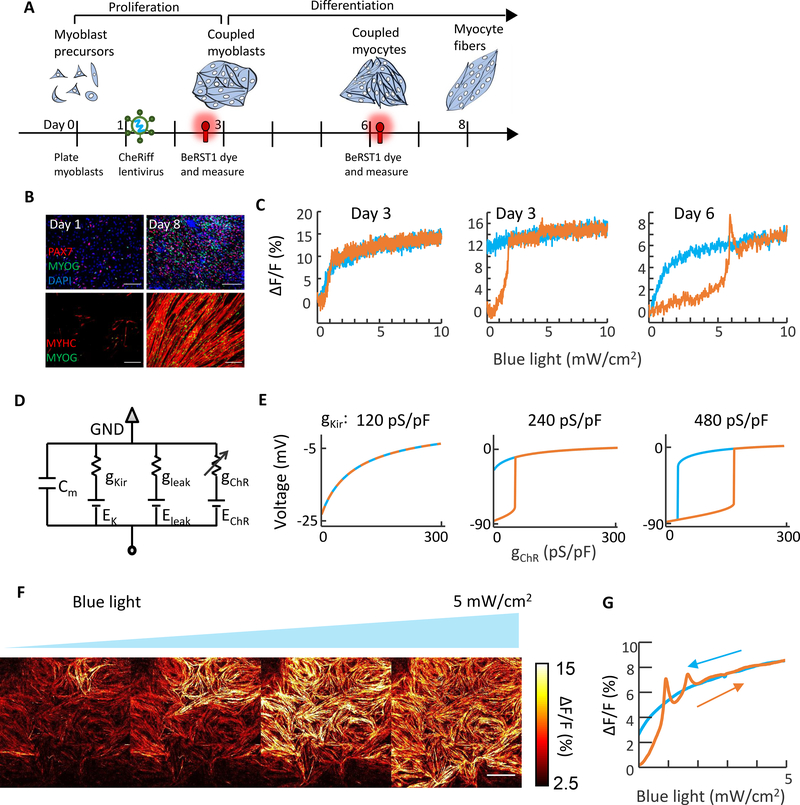
Figure 4: Bioelectric domain wall propagation in stem cell-derived myocytes.
A) Timeline for differentiation, viral transduction, and measurement. B) Immunocytochemistry staining of myocyte cultures during differentiation. Stains show PAX7, myogenin (MYOG), and myosin heavy chain (MYHC). Scale bars 200 μm. C) Optical measurements of membrane potential in individual isolated myoblasts at different times after differentiation. D) Simple electrical circuit model for myoblasts. The Kir channel was modeled as a nonlinear conductance, the leak was treated as Ohmic with Eleak = −20 mV, and the channelrhodopsin was treated as Ohmic with EChR = 0 mV. E) Simulations of optogenetically induced changes in membrane voltage at different values of gKir. All other parameters of the simulation were kept constant between the three panels. F) Bioelectrical domain wall migration in a monolayer of electrically coupled myocytes (measured 3 days after start of differentiation). Scale bar 1 mm. See Supplementary Movie S6. G) Optical measurements of membrane voltage as a function of optogenetic stimulus strength in the confluent culture. Depolarization-activated inward currents led to spikes atop the optogenetically induced depolarizations.
In myoblast precursors that had not yet reached confluence (day 3), we observed heterogeneous responses to ramped optogenetic stimulation: Cells showed either a smooth response with saturation-like behavior and little hysteresis (67%, 34 of 51, Fig. 4C left) or a step-wise depolarization, which did not reverse upon cessation of the optogenetic stimulus (33%, 17 of 51, Fig. 4C middle). In immature myocytes mechanically dissociated from a confluent culture (day 6, 3 days after start of differentiation), we observed sub-populations with behavior similar to day 3 (smooth depolarization with no hysteresis: 47%, 42 of 89; step-wise, irreversible depolarization: 29%, 26 of 89). We also observed a new sub-population comprising cells with closed hysteresis loops that resembled the bi-HEKs (24%, 21 of 89, Fig. 4C right, Supplementary Fig. 8, Methods).
These three seemingly disparate behaviors could all be explained by a simple model containing a leak, a channelrhodopsin, and Kir expression which increased on average between day 3 and 6 (Fig. 4D, S8). At the lowest Kir level, the I-V curve was monotonically increasing, so channelrhodopsin activation shifted a single stable voltage fixed point along the I = 0 axis. This led to a continuous and reversible change in voltage (Fig. 4E). At intermediate Kir level, the I-V curve was N-shaped and crossed the I=0 axis three times in the absence of channelrhodopsin activation. Blue light drove step-wise depolarization via a saddle node bifurcation. The depolarized state remained stable in the absence of optogenetic drive, leading to non-recovering depolarization. At the highest Kir level, the hysteresis curve shifted to the right and the cells repolarized in the absence of optogenetic drive. Thus, a simple model with one tuning parameter captured the three qualitatively distinct single-cell responses to channelrhodopsin activation.
In confluent monolayers at day 6, we used patterned optogenetic stimulation to excite a portion of the tissue. The evoked action potentials propagated beyond the stimulated region, confirming the presence of gap junctional electrical coupling (Supplementary Fig. 9). Under spatially homogeneous ramped optogenetic stimulation, we observed optogenetically induced domain wall propagation (Fig. 4F, Supplementary Movie S6). The presence of domain wall propagation was surprising, considering that only a minority (24%) of the isolated cells were bistable. Simulations showed that the due to strong electrotonic coupling, the global behavior of a tissue could be dominated by a minority of cells expressing Kir2.1 (Supplementary Fig. S6). As in the bi-HEKs, the whole-tissue average voltage showed little hysteresis as a function of optogenetic drive (Fig. 4G), consistent with depolarization via domain wall migration. These observations show that differentiating myoblasts exhibit electrical bistability when isolated and collective domain wall migration during an essential step of myogenesis.
In contrast to the bi-HEK cells, the myoblasts also supported propagation of regenerative action potential waves. These waves manifested as spikes in the whole-tissue fluorescence during a gradual optogenetic depolarization (Fig. 4G). The additional depolarizing drive associated with these spikes caused the waves to propagate rapidly across the tissue, without disruption from the defects which could pin the motion of domain walls.
Discussion
Quasi-static spatial variations in membrane potential are well known to arise in development (of animals31–33, plants34 and protists35), in wound healing36, and to persist in some mature tissues37. In most cases the ion channels responsible for these potentials are not known, but it is generally assumed that the membrane voltage in each patch of tissue is set by the locally expressed ion channels, i.e. that the bioelectrical patterns are ‘baked in’ to the tissue via conventional morphogen signaling pathways which govern ion channel gene expression. Our work shows that this need not be the case. Electrical instabilities can amplify minute (possibly undetectable) variations in ionic currents in an otherwise homogeneous tissue. Thus the conventional view that patterns of gene expression drive patterns of electrophysiology might, in some cases, be reversed. The pathways by which patterns of membrane voltage could affect patterns of gene expression remain a topic of much current research.33,38
In vitro, muscle cells must be aggregated to differentiate, a phenomenon called the “community effect”.39 Our results show that electrical coupling can mediate community effects, i.e. that the collective electrical dynamics of coupled cells can be strikingly different from the individuals, even if all cells are identical. Domain wall migration mediates polarization in extended tissues, whereas isolated cells or small clumps must polarize all at once. Consequently, under ramped Kir2.1 expression, an electrically coupled, extended tissue will polarize before an isolated cell or small patch, even if all other conditions are identical. Kir2.1 expression is required for the expression of the myogenic transcription factors Myogenin (MyoG) and Myocyte Enhancer Factor-2 (MEF2).29 Disruption of gap junction coupling is sufficient to disrupt myogenesis.40 Together, these observations suggest that bioelectric domain walls might play a functionally important role in myogenesis. This prediction merits further mechanistic tests in cultured myocytes and in vivo. It will be interesting to relate the bioelectrical response properties of developing muscle to the shifts that occur as myocytes fuse and gap junctional coupling diminishes during maturation.
Many combinations of ion channels can produce N-shaped I-V curves and thereby mediate electrical bistability. For instance, the combination of a K+-selective leak current and the steady-state window current of T-type CaV channels mediates plateau potentials in thalamocortical neurons.41,42 In this case, one would expect electrical bistability to be accompanied by bistability in intracellular Ca2+ concentration, which might then couple to downstream biochemical or genetic signaling pathways. The combination of a persistent voltage-gated sodium current and a K+-selective leak current can also drive bistability. Persistent NaV currents have been observed in striated cardiac and skeletal muscle and in many types of mammalian neurons.43 Persistent sodium and persistent L-type calcium currents contribute to sustained activation in a spinal cord injury model.44 Finally, persistent sodium currents are thought to play a role in propagating and amplifying the influence of distal synaptic inputs during dendritic integration.45 In principle, any of these bistable scenarios could produce bioelectrical domain walls in electrotonically extended systems, though we are not aware of any such direct observations.
Gap junctions are necessary for proper formation of many tissues during development, including in heart, liver, skin, hair, cartilage, bone, and kidney,46,47 though the physiological roles of these gap junctions remain unclear. Our work suggests that gap junction-mediated bioelectrical domain wall motion may be an important feature in some of these systems. For instance, chondrocytes express Kir channels48, gap junctions49, and the ionotropic serotonin receptor 5-ht3a50 which is a nonselective cation channel electrically similar to channelrhodopsin. These conditions suggest that the ingredients are present for regulation of membrane potential via domain wall migration. Wounding in endothelial monolayers has been shown to induce slowly migrating zones of depolarization51, suggesting that these cells might also support bioelectrical domain walls. The phenomenon of spreading cortical depression may also reflect a form of electrical bistability, though the underlying mechanism is likely far more complex than the phenomena studied here.52 The presence of long-lived electrical bistability in tissues could provide a means to couple bioelectric patterns to biochemical and genetic signaling networks.4–6
The shape of the I-V curve in our experiments is qualitatively captured by a cubic (Fitzhugh Nagumo-type) nonlinearity.53,54 Models of this sort have been applied to describe similar dynamics (zero-dimensional hysteresis, domain nucleation, growth and disorder-driven breakup) in magnetic domain reversals in ferromagnets,55 in the spread of forest fires,56 in phase transitions,21 and in expanding species ranges with a strong Allee effect.57 Spontaneous spatial symmetry breaking and pattern formation are well established in neural field theories58 and in models of cardiac arrhythmias.59,60 In these systems, however, the membrane voltage varies with time, i.e. the systems are described by the lower right quadrant of Fig. 1B. Our work shows that the reaction-diffusion formalism can be applied to purely spatial symmetry breaking in electrophysiology.
In the Turing model, formation of quasi-periodic patterns requires interaction of two or more morphogens, often described as an activator and an inhibitor. Conductance-based models describe the dynamics of voltage, a scalar quantity. The spatial symmetry breaking studied in this report does not constitute a classical Turing pattern, in that voltage is only a one-dimensional state variable. As a result, the patterns of membrane voltage did not have a characteristic finite spatial frequency. To achieve a classical Turing-like pattern would require coupling of voltage to another diffusible species, e.g. Ca2+. It is not known whether classical Turing-like patterns of membrane voltage can be created via purely electrophysiological means.
Methods
Numerical modeling of electrically bistable cells and tissues
In the conductance-based model, the voltage dynamics are governed by the equation:
The gap junction sheet conductance is defined as GCxn = gCxn × l2, where gCxn is the gap junction conductance between adjacent cells, and l is the linear dimension of a cell. If gCxn is measured as an areal conductance (S/m2), then the units of GCxn are S (i.e. sheet conductance, which has no spatial dimension). If Cm is measured as an areal capacitance (F/m2), then the ratio GCxn/Cm has units of a diffusion coefficient (m2/s), making explicit the connection between the conductance-based model and the reaction-diffusion equation.
For convenience in simulations, units of space were 10 μm (corresponding to linear size of one cell), units of time were ms, and units of voltage were mV. We assumed the capacitance of a cell was 10 pF. Conductances were measured in nS/pF, ionic currents in pA/pF, and voltage in mV. Parameters used in all simulations are given in the Tables in the Model parameters section of the Methods.
Simulations were run in Matlab using custom software. Single-cell voltages in 0D were determined by finding fixed points of cell-autonomous current-voltage curves. Extended tissues were numerically simulated as two-dimensional grids of 100 × 100 cells with periodic boundary conditions and one grid-point per cell. Simulations and experiments thus had similar spatial scales. For simulations of nucleation events in homogenous tissues (Fig. 3B), 300 × 300 cell grids with no-flux boundary conditions were implemented to match experimental conditions. The discrete Laplacian was implemented using the matlab del2 function (with default spacing) and solutions were time-integrated using Euler’s method with 10 kHz sampling.
The inward rectifying potassium current from Kir2.1 was based on a model from ten Tusscher et al. 11 as: with reversal potential at EK = −90 mV. The parameter is a time-independent rectification factor that depends on voltage, with the following form:
The scaling factor x0 = 100 was introduced to make xK∞ of order unity between −90 and −60 mV. In our simulations the conductance magnitude gK was the only parameter varied to mimic changes in expression of Kir2.1. The variable leak was modeled as an Ohmic conductance with reversal potential 0 mV. For homogeneous tissues all cells had identical Kir2.1 and leak conductances.
To introduce disorder into the tissue, a fraction nK of the cells were randomly assigned to express Kir2.1 (all with conductance gK) while the remaining cells had no Kir2.1 expression (Supplementary Figs. 6, 7). Spatial correlations in Kir2.1 expression were introduced by assigning a random number to each cell, independently sampled from a uniform distribution on [0, 1]. The values where then smoothed with a two-dimensional Gaussian kernel of width d. A threshold was selected so that a fraction nK of the cells were above threshold. These cells were assigned to express Kir2.1 with conductance gK and cells below threshold did not express. The extent of the spatial correlations was tuned by varying d. In the simulation for Supplementary Figs. 7G,H, Kir2.1 and CheRiff expression were heterogeneous, and independent of each other. The distribution of CheRiff expression was calculated following the same procedure as for Kir2.1, using the same smoothing parameter, d, but different thresholds nK, and nChR.
Cells in simulated tissues were first initialized to their cell-autonomous resting potential, i.e. the resting potential in the absence of influences from the neighbors. Bistable cells were initialized to the fixed point which had the greater area under the curve between it and the unstable fixed point. Tissues were then time-evolved to generate an initial steady state voltage profile. To simulate bioelectrical dynamics under changes in parameters, conductances were gradually changed over timescales slower than any of the internal relaxation dynamics. For simulations of domain boundary velocities in homogeneous tissues with bistable I-V curves (Supplementary Fig. 2), the left half of the tissue was initialized in a depolarized state and the right half in a hyperpolarized state.
Bi-HEK cell generation and culture
Genetic constructs encoding the inward rectifying potassium channel Kir2.1 and the blue-shifted channelrhodopsin CheRiff were separately cloned into lentiviral expression backbones (FCK-CMV) and then co-expressed in HEK 293T cells along with the lentiviral packaging plasmid PsPAX2 (Addgene) and the envelope plasmid VsVg (Addgene) via polyethylenimine transfection (Sigma). Lentiviral particles were harvest at 36 and 72 hours post-transfection, and then concentrated 20-fold using the Lenti-X concentrator system (Takara).
For experiments where nominally homogeneous expression was the goal (Figs. 2, 3), HEK cells were incubated with both Kir2.1 and CheRiff lentiviral vectors for 48 hours prior to measurement, and then passaged and replated onto poly-D-lysine coated glass-bottom tissue culture dishes (MatTek). For patch clamp measurements (Supplementary Fig. 1), bi-HEKs were plated sparsely onto Matrigel coated dishes. For wide-field measurements (Fig. 3), adhesive islands were prepared by manually spotting poly-d-lysine onto MatTek plates. Bi-HEKs were plated onto these plates to create confluent patches of cells approximately 2 mm in diameter.
For experiments where disordered expression was the goal (Supplementary Fig. 7), Kir2.1 and CheRiff constructs were transiently expressed (using Mirus 293T) in HEK cells which were then grown to confluence for an additional 72 hours prior to measurement. High-disorder samples were not replated prior to measurement.
hiPSC myoblast and myocyte differentiation
HiPSC-derived myoblasts were differentiated into myocyte fibers according to an established serum-free differentiation protocol.61 Briefly, 3–4 weeks old primary myogenic cultures generated from wild-type hiPSCs were dissociated as described and myogenic progenitors (myoblasts) were replated at low density (35–40k/cm2) onto Matrigel (Corning, Cat#354277)-coated dishes in skeletal muscle growth media (SKGM-2, Lonza CC-3245) with 10 μM ROCK inhibitor.61 After 24 hours, medium was changed to SKGM-2 media without ROCK inhibitor and incubated with low-titer lentivirus encoding CheRiff-CFP. Myoblast cultures were proliferated for up to 72 hours, at which point cultures reached ~90% confluence. Cultures were then induced for myogenic differentiation with DMEM/F12 supplemented with 2% knock-out serum replacement (Invitrogen, Cat. # 10828028), 10 μM of the TGFβ inhibitor SB431542 (Tocris, Cat. # 1614), 1 μM Chiron (Tocris, Cat. # 4423), 0.2% Pen/Strep (Life Technologies, Cat. # 15140122) and 1x ITS (Life Technologies, Cat. # 41400045). 61 Following induction, medium was changed on days 1 and 2 and then was refreshed every other day for up to 10 days post-differentiation to generate mature and fused myocyte fibers. Samples were measured between 0 and 3 days after start of differentiation (i.e., after 3 and 6 days in culture).
Immunostaining and imaging
Human iPSC-derived myocyte cultures were fixed for 20 minutes in 4% formaldehyde. Cultures were rinsed three times in phosphate-buffered saline (PBS), followed by blocking buffer composed of PBS supplemented with 10% fetal bovine serum (FBS) and 0.1% Triton X-100. Primary antibodies were then diluted in blocking buffer and incubated overnight at 4 °C. Cultures were then washed three times with PBST (PBS supplemented with 0.5% Tween-20) and incubated with secondary antibodies conjugated with an AlexaFluor dye (Molecular probes) and DAPI (5 μg/mL) in blocking buffer for 2 h at room temperature. Cultures were washed with PBST followed by PBS, followed by imaging. Antibodies were: anti-PAX7 (Developmental Studies Hybridoma Bank, DSHB), anti-Myogenin (Santa Cruz, SC-576X) and embryonic anti-MyHC (DSHB, F1.652).
Transcriptomic profiling: Library preparation and sequencing
RNA was extracted from cells using Trizol (Invitrogen) or with the RNeasy Mini Kit (Qiagen). Libraries were prepared using Roche Kapa mRNA HyperPrep sample preparation kits from 100 ng of purified total RNA according to the manufacturer’s protocol. The finished dsDNA libraries were quantified by Qubit fluorometer, Agilent TapeStation 2200, and RT-qPCR using the Kapa Biosystems library quantification kit according to manufacturer’s protocols. Uniquely indexed libraries were pooled in equimolar ratios and sequenced on two Illumina NextSeq500 runs with single-end 75bp reads by the Dana-Farber Cancer Institute Molecular Biology Core Facilities.
Transcriptomic profiling: RNAseq Analysis
Sequenced reads were aligned to the UCSC hg19 reference genome assembly and gene counts were quantified using STAR (v2.5.1b)62. Differential gene expression testing was performed by DESeq2 (v1.10.1)63 and normalized read counts (FPKM) were calculated using cufflinks (v2.2.1)64. RNAseq analysis was performed using the VIPER snakemake pipeline.65
Patch clamp and all-optical electrophysiology
All electrophysiological measurements were performed in Tyrode’s solution, containing (in mM) 125 NaCl, 2 KCl, 2 CaCl2, 1 MgCl2, 10 HEPES, 30 glucose. The pH was adjusted to 7.3 with NaOH and the osmolality was adjusted to 305–310 mOsm with sucrose. Prior to measurements, 35-mm dishes were washed twice with 1 mL phosphate-buffered saline (PBS) to remove residual culture media, then filled with 2 mL Tyrode’s solution.
For patch clamp measurements, filamented glass micropipettes (WPI) were pulled to a resistance of 5–10 MΩ and filled with internal solution containing (in mM) 140 KCl, 1 MgCl2, 10 EGTA, 10 HEPES, 3 Mg-ATP, pH adjusted to 7.3 with KOH. The patch electrode was controlled via a low-noise patch clamp amplifier (A-M Systems, model 2400). Voltage traces were collected under I = 0 current clamp mode, and current traces were collected in voltage clamp mode. Blue light for optical stimulation (Coherent Obis 488 nm) was modulated using an acousto-optic tunable filter (Gooch and Housego GH18A series). Patch clamp measurements were performed on small clusters of cells (approximately 6 cells).
Spatially resolved optical electrophysiology measurements were performed using a home-built upright ultra-widefield microscope66 with a large field of view (4.6×4.6 mm2, with 2.25×2.25 μm2 pixel size) and high numerical aperture objective lens (Olympus MVPLAPO 2XC, NA 0.5). Fluorescence of BeRST1 was excited with a 639 nm laser (OptoEngine MLL-FN-639) at 100 mW/cm2, illuminating the sample from below at an oblique angle to minimize background autofluorescence. BeRST1 fluorescence was separated from scattered laser excitation via a dichroic beam splitter (Semrock Di01- R405/488/561/635-t3-60×85) and an emission filter (Semrock FF01-708/75-60-D). Images were collected at 100 Hz frame rate on a Hamamatsu Orca Flash 4.2 scientific CMOS camera. Optogenetic stimulation was performed by exciting CheRiff with a blue LED (Thorlabs M470L3) with a maximum intensity of at 10 mW/cm2.
Prior to measurement, cells were incubated with 1 μM BERST1 dye in phosphate buffered saline for 30 minutes in a tissue culture incubator. Samples were then washed and prepared in Tyrode’s solution immediately before imaging.
Data Analysis and Image Processing
Optical recordings of voltage-sensitive BeRST1 fluorescence were acquired for isolated cells, small clusters of cells (4–12 cells), and extended tissues (>2 mm linear size). Recordings were processed using custom MATLAB software. Briefly, to minimize uncorrelated shot-noise, movies were subjected to 4×4 binning, followed by pixel-by-pixel median filtering in the time domain (9 frame kernel). A background signal was calculated from a cell-free region of the field of view and subtracted from the region containing the cells. Mean sample images were generated by measuring the average fluorescence of the tissue prior to optogenetic stimulation. Functional recordings were divided pixel-wise by this baseline to generate movies of ΔF/F. Plots of voltage-dependent fluorescence were generated by averaging the time-lapse movies over the relevant region of interest (e.g. small clusters for 0D data; localized spots within extended cell culture islands for 2D local measurements; and over entire cell culture islands for 2D mean measurements).
Statistical Analysis
Statistical analysis was performed on optical electrophysiology recordings from immature myocyte precursors to assess the significance of population-level differences in electrophysiological phenotypes at different time points. Measurements were performed on populations of isolated cells taken at 3 and 6 days in culture (see hiPSC myoblast and myocyte differentiation methods above). Fluorescent recordings of voltage were acquired for 100 ms prior to illumination, during a 40 s ramp of blue light (increasing from 0 to 10 mW/cm2 for 20 s, then decreasing for 20 s), and then for 5 s after the blue light was turned off. Raw acquisitions were converted to ΔF/F for further analysis.
Electrophysiological phenotypes were parameterized by calculating the total endpoint hysteresis and the mean integral hysteresis for each identified cell (Supplementary Fig. 8). Endpoint hysteresis was defined as the difference in ΔF/F between the 5 s at the end of the acquisition and the 100 ms at the start of the acquisition. Integral hysteresis was defined as the difference between the mean ΔF/F over the decreasing phase of the blue light ramp and the mean ΔF/F during the increasing phase of the blue light ramp (each averaged over the corresponding full 20 s ramp).
In each sample, individual cells which responded to CheRiff stimulation were first manually identified via an overall mean ΔF/F image. 51 and 89 cells were identified in the day 3 and day 6 measurements, respectively. Endpoint hysteresis and mean hysteresis were then calculated for each identified cell, and a cell was sorted as hysteresis-positive if it demonstrated a value greater than 0.3 for either measure (Supplementary Fig. 8). Cells which showed endpoint hysteresis > 0.3 were sorted into the endpoint hysteresis cluster, regardless of their integral hysteresis value. Cells with integral hysteresis > 0.3 and endpoint hysteresis < 0.3 were sorted into the integral hysteresis cluster.
Model parameters
Default model parameters were as follows:
| Parameter | Explanation |
|---|---|
| Cm = 10 pF | Single cell capacitance |
| l = 10 μm | Size of an individual cell; discretization size of simulated tissues |
| EK = −90 mV | Potassium reversal potential |
| EChR = 0 mV | Channelrhodopsin reversal potential |
| Eleak = −20 mV | Additional leakage reversal potential. Used for myocyte models (Fig. 4) and for model of disorder-induced domain formation (Supplementary Fig. 7). |
|
GCxn = 2 nS/pF × l2 = 200 (nS/pF) × μm2 |
Strength of gap junction coupling between bi-HEK cells |
| nKir = 0 – 1 | Proportion of cells initialized to express Kir2.1 |
| nChR = 0.75 – 1 | Proportion of cells initialized to express CheRiff. Expression is uniform (nChR = 1) in all panels except for Supplementary Fig. 7H (in which nChR = 0.75). |
| d = 0.001 to 10 cells | Width of gaussian smoothing kernel used to introduce spatial correlations in Kir2.1 expression |
| gK = 0 – 500 pS/pF | Kir2.1 conductance. This conductance is multiplied by the inward rectifying function xK∞. |
| x0 = 100 | Scaling factor to set xK∞ to order unity between EK and the point of maximum outward current (Vmax ∼ −69 mV) |
| gChR, = 0 to 100 pS/pF | Conductance of the channelrhodopsin CheRiff. Varied continuously in simulations of blue light intensity ramps. |
| gleak = 5 pS/pF | Additional leak conductance added to myocyte model |
Model parameters for figure panels:
| Figure panel | Parameters |
|---|---|
| 2C | gChR, = 50 pS/pF, gK = 200 pS/pF |
| 2D | gChR, = 1 to 200 pS/pF, gK = 200 pS/pF |
| 3A |
imax = 0.54 pA/pF gChR = 50 pS/pF, gK = 200 pS/pF, nKir = 1 |
| 3B, top | gChR = 70 pS/pF, gK = 200 pS/pF, nKir = 1 |
| 3B, bottom | gChR = 60 pS/pF, gK = 200 pS/pF, nKir = 1 |
| 3C | gChR, = 1 to 200 pS/pF, gK = 200 pS/pF, nKir = 1 |
| 4E | gleak, = 100 pS/pF, gChR = 0.1 to 300 pS/pF (ramped), gK = 120, 240, 480 pS/pF (left, middle, and right panels) |
| S2B | gChR = 24.5 pS/pF, gK = 60, 72.5, 85, 97.5 pS/pF (top to bottom) |
| S2C | gChR = 22 pS/pF, nKir = 1 |
| S6B | gK = 1 to 500 pS/pF, gChR = 20 pS/pF, nKir = 1 |
| S6D,E | gK = 500 pS/pF, gChR = 20 pS/pF, d = 0.001, nKir = 0 to 1 |
| S6G,H | gK = 500 pS/pF, gChR = 20 pS/pF, d = 10, nKir = 0 to 1 |
| S7C,D | gK = 50 pS/pF, gleak = 20 pS/pF, Eleak = −20 mV, d = 3, nKir = 0.25, gChR= 0 to 250 pS/pF |
| S7G,H | gChR, = 1 to 300 pS/pF, gK = 600 pS/pF, nKir = 0.5, nChR = 0.75, d = 1.5 |
| S8A | gleak, = 100 pS/pF, gChR = 0.1 to 300 pS/pF (ramped), gK = 120, 240, 480 pS/pF (left, middle, and right panels) |
For simulations in S2 and S6, the channelrhodopsin and leak conductances were equivalent (i.e. there was a single ohmic leak conductance in the model with Eleak = EChR = 0 mV. For simulations of the three-conductance myocyte model in Figs. 4, S7 and S8, two separate Ohmic conductances were included (with Eleak = −20 mV, EChR = 0 mV).
Supplementary Material
Acknowledgments
We thank Urs Böhm, Amanda Klaeger, Juanita Mathews, and Michael Levin for helpful discussions. We thank Evan Miller for help providing the BeRST1 dye.
Funding: This work was supported by the Allen Discovery Center at Tufts University, the Vannevar Bush Fellowship Foundation, and the Howard Hughes Medical Institute. HMM was supported by the Department of Defense (DoD) through the National Defense Science & Engineering Graduate Fellowship (NDSEG) Program. GO was supported by the Howard Hughes Medical Institute Gilliam Fellowship.
Footnotes
Competing Interests: The authors declare no competing financial interests.
Data Availability
The plots of fluorescent voltage recordings in Figs. 2–4 are available as Source Data. All other data that support the plots within this paper and other findings of this study are available from the corresponding author upon reasonable request.
Code Availability
Custom-written code used for data analysis is available from the authors on request.
Material Availability
Lentiviral expression plasmids for CheRiff and Kir2.1 are available from the authors upon reasonable request.
References
- 1.Hodgkin AL & Huxley AF A quantitative description of membrane current and its application to conduction and excitation in nerve. J. Physiol. (Lond. ) 117, 500–544 (1952). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Turing AM The chemical basis of morphogenesis. Phil. Trans. R. Soc. Lond. B 237, 37–72 (1952). [Google Scholar]
- 3.Kondo S & Miura T Reaction-diffusion model as a framework for understanding biological pattern formation. Science 329, 1616–1620 (2010). [DOI] [PubMed] [Google Scholar]
- 4.Law R & Levin M Bioelectric memory: modeling resting potential bistability in amphibian embryos and mammalian cells. Theoretical Biology and Medical Modelling 12, 22 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pietak A & Levin M Bioelectric gene and reaction networks: computational modelling of genetic, biochemical and bioelectrical dynamics in pattern regulation. J. R. Soc. Interface 14, 20170425 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cervera J, Alcaraz A & Mafe S Bioelectrical signals and ion channels in the modeling of multicellular patterns and cancer biophysics. Sci. Rep 6, 20403 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brodsky M Turing-like patterns can arise from purely bioelectric mechanisms. bioRxiv, 336461 (2018). [Google Scholar]
- 8.Levin M Endogenous bioelectrical networks store non-genetic patterning information during development and regeneration. J. Physiol. (Lond. ) 592, 2295–2305 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hochbaum DR et al. All-optical electrophysiology in mammalian neurons using engineered microbial rhodopsins. Nat. Methods 11, 825–833 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adam Y et al. Voltage imaging and optogenetics reveal behaviour-dependent changes in hippocampal dynamics. Nature 569, 413 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.ten Tusscher KHWJ, Noble D, Noble PJ & Panfilov AV A model for human ventricular tissue. Am. J. Physiol. Heart Circ. Physiol 286, H1573–H1589 (2004). [DOI] [PubMed] [Google Scholar]
- 12.Hibino H et al. Inwardly rectifying potassium channels: their structure, function, and physiological roles. Physiol. Rev 90, 291–366 (2010). [DOI] [PubMed] [Google Scholar]
- 13.Cervera J, Alcaraz A & Mafe S Membrane potential bistability in nonexcitable cells as described by inward and outward voltage-gated ion channels. J. Phys. Chem. B 118, 12444–12450 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Gallaher J, Bier M & van Heukelom JS First order phase transition and hysteresis in a cell’s maintenance of the membrane potential—an essential role for the inward potassium rectifiers. BioSystems 101, 149–155 (2010). [DOI] [PubMed] [Google Scholar]
- 15.Ozbudak EM, Thattai M, Lim HN, Shraiman BI & Van Oudenaarden A Multistability in the lactose utilization network of Escherichia coli. Nature 427, 737 (2004). [DOI] [PubMed] [Google Scholar]
- 16.Novick A & Weiner M Enzyme Induction as an All-Or-None Phenomenon. Proc. Natl. Acad. Sci. U. S. A 43, 553–566 (1957). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McNamara HM, Zhang H, Werley CA & Cohen AE Optically controlled oscillators in an engineered bioelectric tissue. Phys. Rev. X 6, 031001 (2016). [Google Scholar]
- 18.McNamara HM et al. Geometry-dependent arrhythmias in electrically excitable tissues. Cell systems 7, 359–370. e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang YL, Walker AS & Miller EW A photostable silicon rhodamine platform for optical voltage sensing. J. Am. Chem. Soc 137, 10767–10776 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cervera J, Manzanares JA & Mafe S Electrical coupling in ensembles of nonexcitable cells: modeling the spatial map of single cell potentials. J. Phys. Chem. B 119, 2968–2978 (2015). [DOI] [PubMed] [Google Scholar]
- 21.Benguria R & Depassier M Speed of fronts of the reaction-diffusion equation. Phys. Rev. Lett 77, 1171 (1996). [DOI] [PubMed] [Google Scholar]
- 22.Cohen AE Nanoscale Mechanics. PhD Thesis, University of Cambridge, (2004). [Google Scholar]
- 23.Moreno A, Rook M, Fishman G & Spray DC Gap junction channels: distinct voltage-sensitive and-insensitive conductance states. Biophys. J 67, 113–119 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Butterweck A, Gergs U, Elfgang C, Willecke K & Traub O Immunochemical characterization of the gap junction protein connexin45 in mouse kidney and transfected human HeLa cells. J. Membr. Biol 141, 247–256 (1994). [DOI] [PubMed] [Google Scholar]
- 25.Langlois S, Cowan KN, Shao Q, Cowan BJ & Laird DW Caveolin-1 and-2 interact with connexin43 and regulate gap junctional intercellular communication in keratinocytes. Mol. Biol. Cell 19, 912–928 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barkhausen H Zwei mit Hilfe der neuen Verstärker entdeckte Erscheinungen. Phys.Z 20, 401 (1919). [Google Scholar]
- 27.Cross MH, Cross P & Brinster R Changes in membrane potential during mouse egg development. Dev. Biol 33, 412–416 (1973). [DOI] [PubMed] [Google Scholar]
- 28.Liu J et al. Role of an inward rectifier K current and of hyperpolarization in human myoblast fusion. J. Physiol. (Lond. ) 510, 467–476 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Konig S et al. Membrane hyperpolarization triggers myogenin and myocyte enhancer factor-2 expression during human myoblast differentiation. J. Biol. Chem 279, 28187–28196 (2004). [DOI] [PubMed] [Google Scholar]
- 30.Kalderon N, Epstein ML & Gilula NB Cell-to-cell communication and myogenesis. J. Cell Biol 75, 788–806 (1977). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Metcalf MM, Shi R & Borgens RB Endogenous ionic currents and voltages in amphibian embryos. J. Exp. Zool 268, 307–322 (1994). [Google Scholar]
- 32.Borgens RB, Rouleau MF & DeLanney LE A steady efflux of ionic current predicts hind limb development in the axolotl. J. Exp. Zool 228, 491–503 (1983). [DOI] [PubMed] [Google Scholar]
- 33.Whited JL & Levin M Bioelectrical controls of morphogenesis: from ancient mechanisms of cell coordination to biomedical opportunities. Curr. Opin. Genet. Dev 57, 61–69 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rinne PL & van der Schoot C Symplasmic fields in the tunica of the shoot apical meristem coordinate morphogenetic events. Development 125, 1477–1485 (1998). [DOI] [PubMed] [Google Scholar]
- 35.Achenbach F & Weisenseel M Ionic currents traverse the slime mould Physarum. Cell Biol. Int. Rep 5, 375–379 (1981). [DOI] [PubMed] [Google Scholar]
- 36.Zhao M Electrical fields in wound healing—an overriding signal that directs cell migration. Semin. Cell. Dev. Biol 20, 674–682 (2009). [DOI] [PubMed] [Google Scholar]
- 37.Borgens RB Endogenous ionic currents traverse intact and damaged bone. Science 225, 478–482 (1984). [DOI] [PubMed] [Google Scholar]
- 38.Zhou Y et al. Membrane potential modulates plasma membrane phospholipid dynamics and K-Ras signaling. Science 349, 873–876 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gurdon J, Tiller E, Roberts J & Kato K A community effect in muscle development. Current biology 3, 1–11 (1993). [DOI] [PubMed] [Google Scholar]
- 40.Proulx A, Merrifield PA & Naus CC Blocking gap junctional intercellular communication in myoblasts inhibits myogenin and MRF4 expression. Dev. Genet 20, 133–144 (1997). [DOI] [PubMed] [Google Scholar]
- 41.Crunelli V, Tóth TI, Cope DW, Blethyn K & Hughes SW The ‘window’T‐type calcium current in brain dynamics of different behavioural states. J. Physiol. (Lond. ) 562, 121–129 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hughes SW, Cope DW, Toth TI, Williams SR & Crunelli V All thalamocortical neurones possess a T-type Ca2 ‘window’current that enables the expression of bistability-mediated activities. J. Physiol. (Lond. ) 517, 805–815 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Crill WE Persistent sodium current in mammalian central neurons. Annu. Rev. Physiol. 58, 349–362 (1996). [DOI] [PubMed] [Google Scholar]
- 44.Li Y & Bennett DJ Persistent sodium and calcium currents cause plateau potentials in motoneurons of chronic spinal rats. J. Neurophysiol 90, 857–869 (2003). [DOI] [PubMed] [Google Scholar]
- 45.Schwindt PC & Crill WE Amplification of synaptic current by persistent sodium conductance in apical dendrite of neocortical neurons. J. Neurophysiol 74, 2220–2224 (1995). [DOI] [PubMed] [Google Scholar]
- 46.Constantin B & Cronier L Involvement of gap junctional communication in myogenesis. International Rev. Cytol 196, 1–65 (2000). [DOI] [PubMed] [Google Scholar]
- 47.Levin M Isolation and community: a review of the role of gap-junctional communication in embryonic patterning. J. Membr. Biol 185, 177–192 (2002). [DOI] [PubMed] [Google Scholar]
- 48.Mobasheri A et al. Potassium channels in articular chondrocytes. Channels 6, 416–425 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barrett-Jolley R, Lewis R, Fallman R & Mobasheri A The emerging chondrocyte channelome. Frontiers in physiology 1, 135 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tecott L, Shtrom S & Julius D Expression of a serotonin-gated ion channel in embryonic neural and nonneural tissues. Molecular and Cellular Neuroscience 6, 43–55 (1995). [DOI] [PubMed] [Google Scholar]
- 51.Chifflet S, Hernández JA & Grasso S A possible role for membrane depolarization in epithelial wound healing. American Journal of Physiology-Cell Physiology 288, C1420–C1430 (2005). [DOI] [PubMed] [Google Scholar]
- 52.Ayata C & Lauritzen M Spreading depression, spreading depolarizations, and the cerebral vasculature. Physiol. Rev 95, 953–993 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.FitzHugh R Impulses and physiological states in theoretical models of nerve membrane. Biophys. J 1, 445–466 (1961). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nagumo J, Arimoto S & Yoshizawa S An active pulse transmission line simulating nerve axon. Proceedings of the IRE 50, 2061–2070 (1962). [Google Scholar]
- 55.Onsager L Crystal statistics. I. A two-dimensional model with an order-disorder transition. Physical Review 65, 117 (1944). [Google Scholar]
- 56.Zeldovich Y & Frank-Kamenetskii D Theory of flame propagation. Zh.Fix.Khim 12, 100 (1938). [Google Scholar]
- 57.Allee WC, Park O, Emerson AE, Park T & Schmidt KP Principles of animal ecology (1949). [Google Scholar]
- 58.Coombes S Waves, bumps, and patterns in neural field theories. Biol. Cybern 93, 91–108 (2005). [DOI] [PubMed] [Google Scholar]
- 59.Shiferaw Y & Karma A Turing instability mediated by voltage and calcium diffusion in paced cardiac cells. Proc. Natl. Acad. Sci. U. S. A 103, 5670–5675 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Echebarria B & Karma A Instability and spatiotemporal dynamics of alternans in paced cardiac tissue. Phys. Rev. Lett 88, 208101 (2002). [DOI] [PubMed] [Google Scholar]
- 61.Chal J et al. Generation of human muscle fibers and satellite-like cells from human pluripotent stem cells in vitro. Nature protocols 11, 1833 (2016). [DOI] [PubMed] [Google Scholar]
- 62.Dobin A et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Love MI, Huber W & Anders S Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Trapnell C et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol 28, 511 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cornwell M et al. VIPER: Visualization Pipeline for RNA-seq, a Snakemake workflow for efficient and complete RNA-seq analysis. BMC Bioinformatics 19, 135 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Werley CA, Chien M & Cohen AE Ultrawidefield microscope for high-speed fluorescence imaging and targeted optogenetic stimulation. Biomedical optics express 8, 5794–5813 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.