Abstract
MID1 is an E3 ubiquitin ligase of the Tripartite Motif (TRIM) subfamily of RING-containing proteins, hence also known as TRIM18. MID1 is a microtubule-binding protein found in complex with the catalytic subunit of PP2A (PP2Ac) and its regulatory subunit alpha 4 (α4). To date, several substrates and interactors of MID1 have been described, providing evidence for the involvement of MID1 in a plethora of essential biological processes, especially during embryonic development. Mutations in the MID1 gene are responsible of the X-linked form of Opitz syndrome (XLOS), a multiple congenital disease characterised by defects in the development of midline structures during embryogenesis. Here, we review MID1-related physiological mechanisms as well as the pathological implication of the MID1 gene in XLOS and in other clinical conditions.
1. Introduction
The MID1 gene encodes an E3 ubiquitin ligase that belongs to the Tripartite Motif (TRIM) family, hence also the name TRIM18 for this gene (Reymond et al., 2001). The human MID1 gene is located on the short arm of the X chromosome very close to the pseudoautosomal boundary (Van den Veyver et al., 1998) and the study on its peculiar genomic location was concomitant with its identification as being responsible for a rare genetic disease, Opitz G/BBB Syndrome (OS) (Quaderi et al., 1997). OS is a congenital disorder characterised by defects in the development of embryonic midline structures and MID1, stemming from Midline-1, is reported mutated in patients with this disease (Opitz, 1987; Quaderi et al., 1997).
The human MID1 gene is located in the distal end of the X-specific region close to the pseudoautosomal region (PAR) and spans approximately 400 kb of the genome (Perry et al., 1998; Quaderi et al., 1997; Van den Veyver et al., 1998). Interestingly, its mouse orthologue, Mid1, is located on the same chromosome but spans the pseudoautosomal boundary with the first exons present only in the X-specific region while the 3′ exons contained in the PAR, hence both on the X and on the Y chromosome (Palmer et al., 1997). In both humans and rodents the gene is composed of 9 coding exons and upstream to the first coding exon, the MID1 gene presents alternative 5′ untranslated exons and its transcription is driven by at least five alternative promoters (Landry and Mager, 2002). This results in several MID1 transcript isoforms that involve not only the 5′ of the gene and several polyadenylation signals but also the coding region (Landry and Mager, 2002; Winter et al., 2004, 2007). The main features of the MID1 gene are summarised in Table 1. MID1 expression is complex and its regulation also includes the action of microRNAs. To date, the following have been found to regulate MID1 translation: miR-19, miR-340, miR-374 and miR-542 that bind the 3′-UTR in HEK293 cells (Unterbruner et al., 2018) and miR-135b in a lymphoblastoid cell line (Arigoni et al., 2013). We will also address MID1 regulation in specific context throughout the review.
Table 1.
Characteristics of the main isoform of the MID1 gene, transcript and protein in Homo sapiens, Mus musculus and Gallus gallus.
| Homo sapiens (a) | Mus musculus (b) | Gallus gallus (c) | |
|---|---|---|---|
| Gene symbol | MID1 | Mid1 | MID1 |
| NCBI Gene ID | 4281 | 17,318 | 373,920 |
| HGNC/MGI/CGNC | 7095 | 1,100,537 | 48,957 |
| OMIM | 300,552 | ||
| Chromosomal location | Xp22.2 | X F5 (79.19 cM) | 1 |
| Genome coordinates | chrX:10445309–10833682 | ChrX:169685199–169990798 | Chr1: 125930064–126095986 |
| GeneBank (gene) | NC_000023.11 | NC_000086.7 | NC_006088.5 |
| Ensembl transcript ID | ENSG00000101871 | ENSMUSG00000035299 | ENSGALG00000038504.2 |
| RefSeq (mRNA) | NM_000381.4 | NM_010797.3 | NM_204129.1 |
| RefSeq (protein) | NP_000372.1 | NP_034927.2 | NP_989460.1 |
| Gene length (kbp) | 665.9 | 305.6 | 185.92 |
| mRNA length (bp) | 6463 | 3974 | 2420 |
| Number of exons | 10 | 10 | 10 |
| Number of coding exons | 9 | 9 | 9 |
| CCDS code | CCDS14138.1 | CCDS41215.1 | |
| Uniprot name | E3 ubiquitin-protein ligase Midline-1 | E3 ubiquitin-protein ligase Midline-1 | Midline 1 |
| UniProtKB/Swiss-Prot ID | O15344–1 | O70583–1 | Q71R46–1 |
| Length of protein in AA | 667 | 680 | 667 |
| Mass (kDa) | 75.2 | 76.1 | 75.4 |
Data sources for Homo sapiens: https://www.ncbi.nlm.nih.gov/gene/4281; https://www.genecards.org/cgi-bin/carddisp.pl?gene=MID1; https://www.ensembl.org/Homo_sapiens/Gene/Summary?g=ENSG00000101871;r=X:10445310–10833654; https://omim.org/entry/300552?search=MID1&highlight=mid1; https://www.ncbi.nlm.nih.gov/CCDS/CcdsBrowse.cgi?REQUEST=CCDS&DATA=CCDS14138; https://www.uniprot.org/uniprot/O15344.
Data sources for Mus musculus: https://www.ncbi.nlm.nih.gov/gene/17318; https://www.ensembl.org/Mus_musculus/Gene/Summary?g=ENSMUSG00000035299;r=X:169685199–170005736; https://www.ncbi.nlm.nih.gov/CCDS/CcdsBrowse.cgi?REQUEST=CCDS&DATA=CCDS41215; https://www.uniprot.org/uniprot/O70583.
Data sources for Gallus gallus: https://www.ncbi.nlm.nih.gov/gene/373920; http://birdgenenames.org/cgnc/GeneReport?id=48957; https://www.uniprot.org/uniprot/Q71R46#sequences
Genomic coordinates based on GRCh38.p13
Genomic coordinates based on GRCm38.p6
Genomic coordinates based on GRCg6a
Despite more than 20 years of research on MID1 function, the pathogenesis of OS still remains unresolved. In addition, the involvement of MID1 in clinically relevant conditions also in adulthood has further highlighted the need of better understanding its biological function. Here, we will summarise the findings reported on MID1 referring to recent reviews for specific issues.
2. The MID1 gene product
The TRIM family members are characterised by the presence of an N-terminal module composed of 3 domains: a RING domain, one or two B-box domains and a coiled-coil region (Reymond et al., 2001). The family members are further classified with respect to their C-terminal domain composition (Short and Cox, 2006). In the case of MID1 and its close paralogues, the tripartite motif carries 2 B-boxes and is followed by a COS domain, a Fibronectin type III repeat (FN3), and a PRY-SPRY domain (Fig. 1) (Quaderi et al., 1997; Reymond et al., 2001; Short and Cox, 2006).
Fig. 1. MID1 protein domain structure and OS-associated mutations.
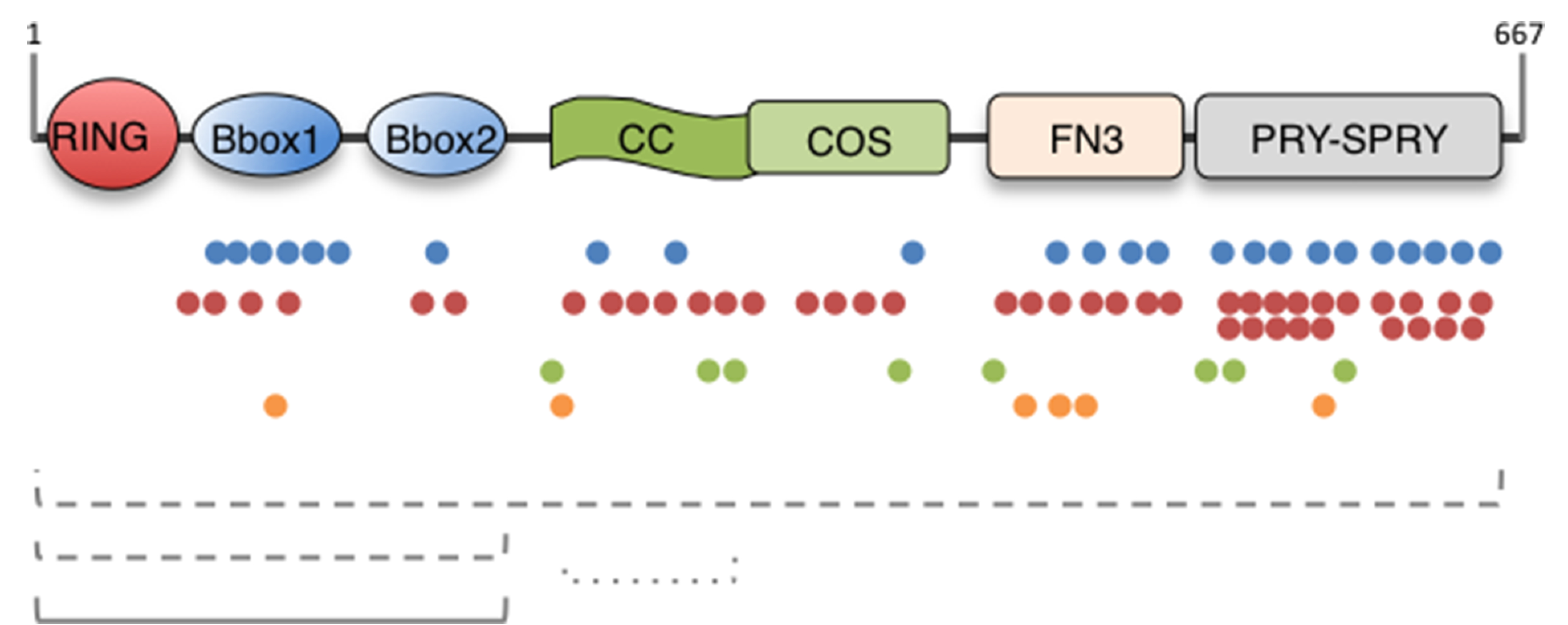
The domain composition of the MID1/TRIM18 protein is depicted. The MID1 protein is 667 residue-long and the limits of the single domains are following in brackets: RING (10–59), Really Interesting New Gene domain; B-Box (B1, 114–164; B2, 170–212), B-Box domain; CC (219–319), Coiled-coil; COS (320–380), C-terminal subgroup one signature; FN3 (382–472), Fibronectin type III repeat; PRY (483–528), domain associated with SPRY domains; SPRY (538–657), SPla and the RYanodine Receptor. Below the scheme, colour dots represent the different mutations reported so far in OS patients: blue dots, missense mutations; red dots, nonsense and truncating mutations; green dots, splice site mutations; orange dots, inframe indels. The dashed lines represent deletions and rearrangements; the continuous line represents duplications. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Despite the growing roles of the TRIM family members in several biological processes and medical conditions (Di Rienzo et al., 2020; Hatakeyama, 2017; van Gent et al., 2018), the biophysical structure of the TRIM proteins is still lacking, likely as a consequence of purification difficulties and propensity to aggregate. MID1 is no exception, although interesting structural details obtained by NMR spectroscopy are available for the TRIM proteins characteristic B-box domains. The first domain to be characterised was the B-box 1 with the discovery that its structure closely resembles the folds of the RING domain through the coordination of 2 zinc atoms (Massiah et al., 2006). Interestingly, even though there is minimal primary sequence similarity between B-box 1 and B-box 2 domains, the latter consists of a short α-helix and a structured loop of anti-parallel β-strands, which adopts a structure similar to MID1 B-box 1 and other RING structures (Massiah et al., 2007). In addition, when solved together, the two B-box domains pack against each other in a stable interaction reminiscent of the interaction of RING dimers (Tao et al., 2008). The other MID1 domain structurally characterised is the COS domain (Short and Cox, 2006) that follows the Coiled-coil region and that adopts a helix-loop-helix structure in which the N- and C-terminal ends are in close proximity (Wright et al., 2016). Although we are still far from a complete structure of the MID1 protein, these structural data have been helpful to define and interpret the localisation and biochemical activity of the MID1 protein as described in the following sections.
2.1. Association with the microtubules
MID1 is a microtubule-associated protein (Cainarca et al., 1999; Schweiger et al., 1999). This association occurs throughout the cell cycle phases, not only with interphase microtubules but also with the mitotic spindle and midbodies during cytokinesis (Cainarca et al., 1999; Gholkar et al., 2016; Zanchetta et al., 2017; Zanchetta and Meroni, 2019). Short and colleagues discovered a new domain, mentioned above, involved in microtubule binding and located between the coiled-coil and the FN3 domains: the COS-box that is to date a signature for microtubule-associated TRIM proteins (Fig. 1) (Short and Cox, 2006). More recently, structural data on the COS domain and further biochemical findings have revealed that both the coiled-coil and the adjacent COS domain are necessary for microtubule binding (Wright et al., 2016). MID1 is a phosphoprotein and its phosphorylation status can modulate microtubule binding (Liu et al., 2001). Initial findings showed that phosphorylation of MID1 promotes its association with microtubules; however, later studies using mutants in the putative serine residue (Ser96) undergoing phosphorylation indicated that MID1 phosphorylation reduces its association with microtubules (Aranda-Orgilles et al., 2008a). Whether this difference is due to diverse experimental conditions or to several phosphorylatable residues is presently unknown and this issue deserves further studies. MID1 phosphorylation status also regulates its own bi-directional transport on microtubules: both phosphatase inhibition and the simulation of a permanent phosphorylation of MID1 can block transport of MID1 on microtubules in a kinesin- and dynein-dependent process (Aranda-Orgilles et al., 2008a).
It is still not clear if MID1 association with microtubules affects their dynamics and stability. When MID1 microtubule localisation is impaired, e.g. when MID1 harbours mutations found in OS patients, microtubules distribution and dynamics are not affected (Cainarca et al., 1999; Schweiger et al., 1999). On the contrary, in some instances, expression of exogenous MID1 results in the protection of microtubules from depolymerising drugs (Schweiger et al., 1999). In addition, the cooperation of MID1 with one of its partners, MIG12 (MID1 interacting G-12 like protein, or MID1IP1), results in microtubule stabilisation (Berti et al., 2004). MIG12 is detected on microtubules if co-transfected together with MID1 and this interaction protects microtubules from depolymerisation. Moreover, antibodies against acetylated tubulin, marking stable tubulin, decorate the MID1-MIG12 bundles, suggesting that the complex is able to stabilise microtubules (Berti et al., 2004). The experiments above have been performed with overexpressed MID1 in Cos-7 cells, therefore the stabilising effect might be exacerbated by its sustained and strong expression. Physiologically, it is possible that microtubule stabilisation might occur only when needed, finely tuned and regulated. Interestingly, MID1-deprived HeLa cells display division defects, including cytokinetic arrest and delayed or aborted abscission, which induce cell binucleation or death (Gholkar et al., 2016). Moreover, in the same cell type a recently discovered partner of MID1, BRAF35 (BRCA2-Associated Factor 35), is involved in cytokinesis and both are localised at the midbody where stable microtubules are present (Zanchetta and Meroni, 2019; Zanchetta et al., 2017). These recent data suggest that MID1 can regulate microtubule dynamics in defined cell cycle phases. The MID1-dependent control of microtubule dynamics may also regulate other cellular processes, such as cell adhesion and cell migration, in which microtubules are involved (Demir et al., 2014; Latta and Golding, 2012).
2.2. E3 ubiquitin ligase activity
Ubiquitination is a post-translational modification that mainly targets a substrate for proteasome-dependent degradation, but can also modify its cellular localisation, modulate its activity or alter interaction with other proteins (Ciechanover, 2015; Hershko and Ciechanover, 1998; Schulman and Harper, 2009). Ubiquitin contains 7 lysines (K6, K11, K27, K29, K33, K48, K63) that can be employed to build chains with different topology. In addition, the N-terminal residue (Met1) can also be employed to generate linear chains (Huang and Zhang, 2020). Mono- or poly-ubiquitination determines the fate of the substrates (Kulathu and Komander, 2012). Linkage through K48 leads to proteasome-mediated degradation; K63 is involved in DNA repair, endocytosis and Nuclear Factor κ-light-chain-enhancer of activated B cells (NF-κB) signalling, whereas much less is known about the functioning of the other lysine linkages. Ubiquitin conjugation is catalysed by an enzymatic cascade, involving the E1 ubiquitin-activating enzyme, E2 ubiquitin-conjugating enzymes, and E3 ubiquitin ligases. E3 ubiquitin ligases are responsible for substrate recognition thus favouring ubiquitin transfer to it (Komander and Rape, 2012). On the basis of domain structures, E3 ubiquitin ligases can be classified into two main families: the homologous to E6-AP COOH Terminus (HECT) family and the RING-containing protein family (Huibregtse et al., 1995; Joazeiro and Weissman, 2000; Buetow and Huang, 2016).
MID1, like the other members of the TRIM family, belongs to the class of RING-containing E3 ubiquitin ligases (Meroni and Diez-Roux, 2005). Although the indication that MID1 is an E3 ubiquitin ligase was proposed by several authors for years (see below), only in 2011 MID1 enzymatic activity was biochemically characterised (Han et al., 2011; Napolitano et al., 2011). Bacterially produced recombinant MID1 shows auto-E3 ubiquitin ligase activity in vitro and, as expected, the RING domain is essential for this activity. MID1 is able to interact with several E2 conjugating enzymes thus possibly promoting several types of ubiquitin chains (Han et al., 2011; Napolitano et al., 2011). Although this is still a matter of debate in the TRIM family field, the B-boxes within the tripartite motif appear to have a role in the MID1 E3 ubiquitin ligase activity (Han et al., 2011). Indeed, some RING-less TRIM members, such as TRIM29 and TRIM16, have been shown to possess E3 ligase activity likely through their B-box domains (Bell et al., 2012; Dou et al., 2019) Acting as an E3 ubiquitin ligase, MID1 controls the stability and/or the activity of substrates, some of which are summarised in the next paragraphs.
2.3. MID1/α4/PP2A complex
One of the first discovered MID1 interactors was Alpha4 (α4), the mammalian homologue of the yeast protein TAP42 and a non-canonical subunit of protein phosphatase 2A (PP2A) (Di Como and Arndt, 1996; LeNoue-Newton et al., 2016). This interaction is mediated by the C-terminal domain of α4 (Liu et al., 2001) and the B-box1 domain of MID1 (Trockenbacher et al., 2001). α4 is diffusely distributed in both nucleus and cytoplasm, but it assumes a typical microtubular pattern in MID1-overexpressing Cos-7 cells (Liu et al., 2001). Indeed, Short and colleagues have proposed that MID1 tethers α4 to microtubules (Short et al., 2002). Conversely, high levels of α4 displace MID1 from microtubules, without affecting microtubular structure and dynamics (Liu et al., 2001). Studies performed in vitro demonstrated that a RING-B-box1 fragment is able to build poly-ubiquitin chains on α4 and that a form of this fragment harbouring the OS P151L mutation within B-box 1 cannot poly-ubiquitinate α4, although it is not clear if this is due to a lost interaction between MID1 and α4 (Wright et al., 2017). Nevertheless, no enrichment of ubiquitinated α4 and no changes in its level were observed upon MID1 overexpression in cells, leading to a first conclusion that α4 was not a substrate of MID1 E3 ubiquitin ligase activity. Instead, in presence of exogenous MID1 the level of the catalytic subunit of PP2A (PP2Ac) was decreased (Trockenbacher et al., 2001). Consistently, OS patients’ fibroblasts carrying MID1 mutations, show increased levels of PP2Ac, leading to a general hypo-phosphorylation of microtubule-associated proteins targeted by this phosphatase (Trockenbacher et al., 2001). Therefore, it was concluded that α4 serves as an adaptor protein promoting the formation of a ternary MID1/α4/PP2Ac complex. However, more detailed analyses in HEK293T cells have shown that α4 has one ubiquitin interacting motif (UIM) that permits α4 mono-ubiquitination, preventing the building of poly-ubiquitin chains on PP2Ac (LeNoue-Newton et al., 2011; McConnell et al., 2010).
Therefore, on one hand, some findings suggest that α4 promotes poly-ubiquitination of PP2Ac by scaffolding PP2Ac to MID1 (Trockenbacher et al., 2001); on the other hand, further studies show α4 to be a protective factor for PP2Ac degradation, through its UIM mono-ubiquitination (LeNoue-Newton et al., 2011; McConnell et al., 2010). The protective effect of α4 is not due to the fact that it can inhibit MID1 function or sequester MID1 from PP2Ac, since it is known that both MID1- and PP2Ac-binding domains are needed for α4 protective function. So, how can MID1 build poly-ubiquitin chains on PP2Ac? Several works show that MID1 mono-ubiquitinates α4, leading to its calpain-mediated cleavage, switching α4 activity from protective to degradative. In particular, the cleavage involves the C-terminal domain of α4, the one implicated in MID1 interaction (Watkins et al., 2012). Indeed, Han and colleagues show that the RING domain of MID1 mono-ubiquitinates a 45-amino acid polypeptide derived from the C-terminus of α4 (Han et al., 2011), further confirmed in Watkins analyses (Watkins et al., 2012). Taken together, all these results indicate that MID1 loss-of-function leads to a reduction of calpain-mediated cleavage of α4, increasing the protection of PP2Ac from degradation, explaining the hypo-phosphorylation of several Microtubule Associated Proteins (MAPs) as seen in OS fibroblasts (Trockenbacher et al., 2001). In this scenario, poly-ubiquitination of PP2Ac might be performed by (an)other E3 ubiquitin ligase(s), since the cleavage of α4 makes MID1 detach from MID1/α4/PP2Ac complex, leading to the hypothesis that cleaved α4 might redirect PP2Ac localisation promoting its polyubiquitination. In this way, only the microtubule pool of PP2Ac is subjected to proteasomal degradation.
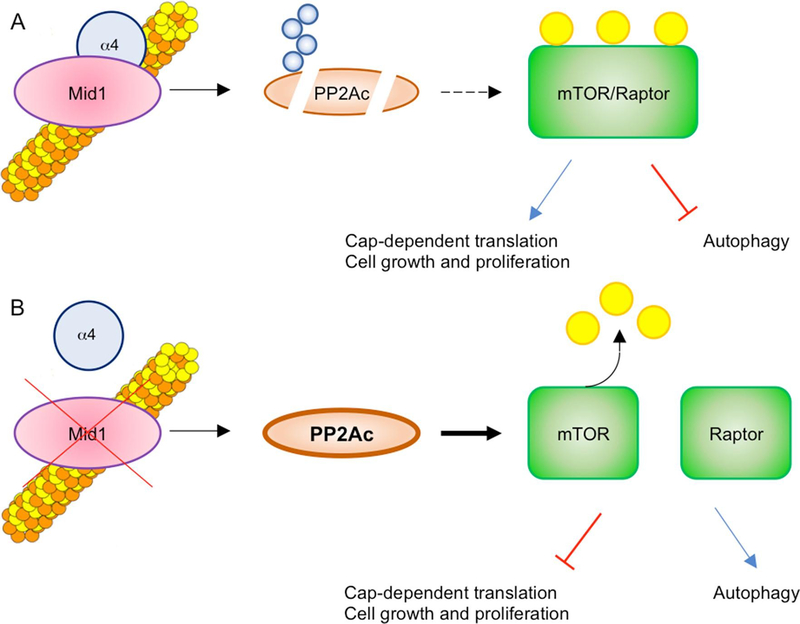
Given PP2A involvement in the regulation of several biological processes (Sontag, 2001), the MID1/α4/PP2Ac complex participates in the modulation of these activities mainly on microtubules. For instance, this complex has been shown to regulate mammalian Target Of Rapamycin Complex 1 (mTORC1) signalling in tumour cells (U2-OS, MCF-7, HCT-116) and in Mouse Embryonic Fibroblasts: increased PP2A levels, resulting from depletion of MID1 or α4, lead to disruption of the mTOR/Raptor complex and downregulation of mTORC1 signalling. In the same way, cells derived from OS patients show decreased mTORC1 formation, S6K1 phosphorylation, cell size, and cap-dependent translation, all of which is rescued by re-expression of wild-type MID1 (Liu et al., 2011). Further, MID1/α4/PP2A-dependent dysregulation of mTORC1 could influence additional pathways, especially those subjected to feedback inhibition by mTORC1, such as PI3K/AKT and Ras/ERK (Carracedo et al., 2008). mTORC1 signalling is implicated in cytoskeletal dynamics and intracellular transport as well as in cell migration (Huang and Fingar, 2014; Liu et al., 2011), so it is tempting to speculate that dysfunctions in this signalling pathway in OS might contribute to patients’ phenotype. Moreover, mTORC1 signalling is involved in processes like autophagy, protein synthesis, cell metabolism and cell growth and proliferation (Jhanwar-Uniyal et al., 2019) and given the role of MID1/α4/PP2Ac ternary complex in regulating mTORC1 complex association (Liu et al., 2011), MID1 absence might have an effect also in these processes (Fig. 2). Further, it was reported that MID1/α4/PP2Ac ternary complex is involved in TFEB (Transcriptional Factor EB) activity through FGF21 activation. FGF21, a fasting-induced hormone, induces calcium release from the ER, in fasting or starvation conditions, promoting DREAM (Downstream Regulatory Element Antagonist Modulator) shuttling into the nucleus. DREAM is a repressor of MID1 gene expression (see also “Role in embryonic development”) and, as consequence, PP2Ac accumulates, dephosphorylates TFEB allowing its translocation into the nucleus, where it activates target genes involved in lysosome biogenesis, autophagy and lipid metabolism (Chen et al., 2017).
Fig. 2. MID1 regulates mTORC1 signalling through PP2A.
A) The microtubular pool of PP2Ac is the target of MID1 activity, leading to its proteasomal-dependent degradation upon poly-ubiquitination (Blue circles). Since mTOR/Raptor association is dependent on PP2A, the degradation of the latter leads to an increased formation of the mTOR/Raptor complex, activating its signalling pathway (phosphorylated form, yellow circles). B) MID1 loss-of-function reduces PP2Ac degradation, leading to de-phosphorylation and decreased association of mTOR and Raptor, causing a drop of active mTORC1 complex formation and signalling. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2.4. Association with mRNAs and translational control
MID1 acts not only on proteic substrates but is also part of ribonucleoprotein (RNP) complexes (Aranda-Orgilles et al., 2008b). In fact, the MID1/α4 complex is able to bind several mRNAs, specifically carrying purine-rich region and forming hairpin structures, also named MID1 association sequence (MIDAS) and whose consensus is [AT]GG\w (1,4)[AT]GG\w(1,4) [AT]GG\w(1,4) [AT]GG (Aranda-Orgilles et al., 2011, 2008b; Hettich et al., 2014). This RNA-protein interaction brings the transcripts in close proximity to ribosome- and translation-associated factors, namely elongation factor 1α (EF-1α-, receptor of activated protein kinase C1 (RACK1), Nucleophosmin (NPM), Annexin A2 (ANXA2) and several 40S ribosomal proteins, thus promoting protein translation (Aranda-Orgilles et al., 2008b; Hettich et al., 2014; Kohler et al., 2014; Krauss et al., 2013; Matthes et al., 2018b). The disruption of the MID1/α4 binding using an α4 mutated peptide is sufficient to abrogate the enhancing effect on the translation of mRNAs that are bound to the mRNP complex (Monteiro et al., 2018).
Among the MIDAS-harbouring transcripts, the 3-phosphoinositide dependent protein kinase-1 (PDPK-1) has been identified in association with the MID1-containing complex. PDPK-1 is a serine/threonine kinase involved in many cellular processes that plays a pivotal role, among others, in the mTOR signalling (Carneiro et al., 2015). Mutations in the MID1 gene have been demonstrated to reduce PDPK-1 protein translation efficiency, since a functional MID1 gene is able to rescue its synthesis (Aranda-Orgilles et al., 2011). Another example is represented by the β-secretase 1 (BACE1) sequence that encompasses several sites that fold like a MIDAS motif enabling the BACE1 transcript to bind the MID1-complex promoting its translation (Aranda-Orgilles et al., 2011).
However, neither the MID1 protein nor the other components of the multiprotein complex seem to contain any known RNA-binding domain (Aranda-Orgilles et al., 2008b), although newly identified RNA-interacting regions are expanding the number of putative RNA-binding proteins. Several members of the TRIM protein family, indeed, have recently emerged as direct RNA binding partners through the C-terminal domains (i.e. NHL or PRY/SPRY domains) (Williams et al., 2019). However, no evidence of a direct association between MID1 protein alone and RNA has been yet provided.
2.5. MID1 and the Sonic hedgehog pathway
The hedgehog family, comprising Sonic hedgehog (Shh), Indian hedgehog (Ihh) and Desert hedgehog (Dhh) is a group of signalling molecules involved in several biological processes ranging from cell proliferation, differentiation and survival to the establishment of the vertebrate body plan (Ingham and McMahon, 2001). Once produced, these signalling molecules bind to the membrane protein Patched (Ptch) thus activating another membrane protein, Smoothened (Smo). The signal is then transduced into the nucleus by Gli transcription factors, whose activity is modified by kinesin-like protein KIF7 and the suppressor of fused (SuFu) (Sasai et al., 2019).
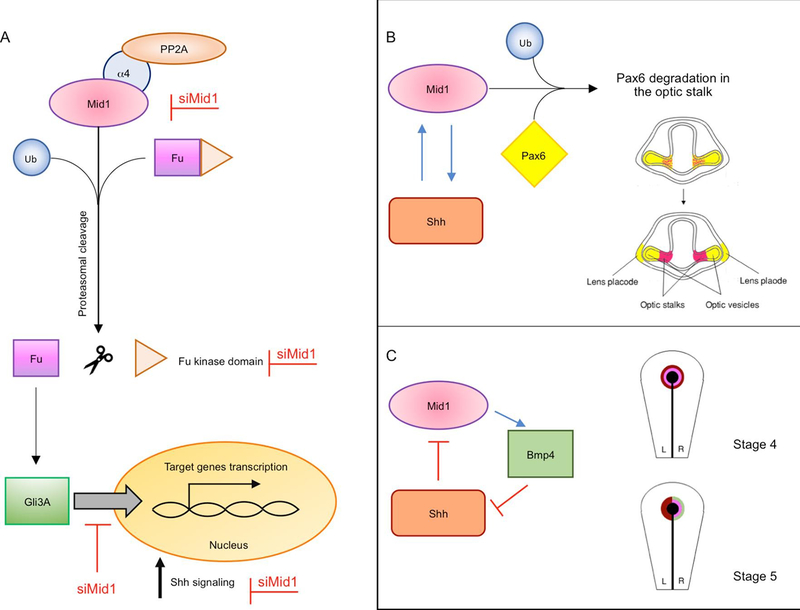
The findings above suggest that a tight link exists between MID1 and the mTORC1 signalling. Among the several networks in which mTORC1 is involved, it is observed that inhibition of mTORC1 activity upon rapamycin treatment reduces the translocation of the Sonic Hedgehog (Shh) pathway transcription factor GLI3 from the cytoplasm to the nucleus (Krauss et al., 2008). Likely, this involves the shuttling of the activator form of GLI3 (GLI3A), as the effect of this reduction in the nucleus leads to a decreased transcription of a Shh pathway target gene Cyclin D1. In HeLa cells, activation of PP2A via inhibition of its catalytic subunit degradation, mediated by MID1/α4 complex, leads to cytosolic retention of GLI3 and, as a consequence, a reduction of its function as a transcription activator (Krauss et al., 2008, 2009). In contrast, α4 overexpression significantly shifts GLI3 to the nucleus (Krauss et al., 2008). In the effort to explain how MID1/α4/PP2A axis regulates GLI3 localisation, it has been discovered that MID1 could interact with Fu, a component of the Shh pathway, promoting its ubiquitination (Schweiger et al., 2014). This ubiquitination involves K6-, K48- and K63 linkages but if this is a direct consequence of MID1 E3 ubiquitin ligase activity and if this modification leads to a proteasome-mediated cleavage of Fu producing a 90 kDa ΔN-terminal fragment is presently unclear. Though, a mimic of the putative ΔN-terminal fragment increases GLI3 nuclear localisation. If the cleavage of Fu is reduced, e.g. due to MID1 silencing, nuclear translocation of GLI3 decreases leading to a consequent reduction of the expression of SHH target genes (Schweiger et al., 2014) (Fig. 3).
Fig. 3. Model of Shh pathway regulated by MID1.
A) MID1 is involved in the proteasome-dependent cleavage of the Fu kinase domain, leading to GLI3A (activator) translocation into the nucleus. This process activates Shh target genes expression increasing Shh signalling. Silencing of MID1 (red writings) impairs Fu cleavage thus reducing Shh signalling. B) In Xenopus, an overlapping expression of mid1 (in pink) and pax6 (in yellow) is observed in the optic stalk, in early stages during the development of the visual system. mid1 is expressed within the forming optic stalk under the control of Shh. Here, mid1 regulates the ubiquitination and proteasomal degradation of Pax6 protein that is cleared from the optic stalk region, setting the border between the optic stalk and the retina via Mid1. C) In chicken development, both Shh (dark red) and cMid1 (pink) are initially expressed bilaterally in the Hensen’s node (black) until stage 5. cMid1 then induces the expression of Bmp4 (green) on the right side of the node. Bmp4 represses right-sided Shh expression, thus restricting Shh to the left side of the node. Left-sided Shh represses cMid1 expression on the left, restricting it to the right side of the node, together with Bmp4. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Indeed, several studies indicate that MID1 is involved in a reciprocal crosstalk with the Shh pathway. Although these data have been studied in cancer cell lines, the crosstalk between MID1 and the Shh pathway can be relevant also in development given the strong implication of the Shh pathway in ventral midline definition as also observed in the phenotypic signs of OS. Findings in Xenopus during development showed that ectopic Shh induces mid1 expression in the entire developing optic vesicle, but also in the prospective forebrain (Pfirrmann et al., 2016) (see also below) (Fig. 3). However, Shh can also suppress mid1 expression and mid1 can act upstream of Shh, due to the induction of bmp4 expression in the chicken Hensen’s node (Granata and Quaderi, 2003) (see also below). In this way, a feedback loop may exist, regulating the balance between mid1 expression with medium levels of Shh inducing mid1 expression, and lower levels suppressing mid1 expression (Fig. 3).
3. The role of MID1 in embryonic development
3.1. The MID1 gene and Opitz G/BBB syndrome
OS is a malformative disease characterised by defects in the development of embryonic midline structures and the MID1 gene is mutated in patients with the X-linked form of the disease, XLOS (Quaderi et al., 1997). Up to now, approximately one hundred mutations have been reported in the MID1 gene in association with OS (Cox et al., 2000; Fontanella et al., 2008; Li et al., 2016; Quaderi et al., 1997; Winter et al., 2016). They are distributed along the entire length of the gene and are represented by missense and nonsense mutations, splice site mutations, and indels, especially involving the 3′ of the gene. In some patients the MID1 gene is completely or partially deleted (Ferrentino et al., 2007) (Fig. 1). The presence of different kind of mutations involving the entire gene suggests loss-of-function (LOF) as the pathogenetic mechanism underlying this disease (Cox et al., 2000). It is interesting though that no point mutations have been found to date in the RING domain, suggesting possible different consequences of such alterations, if occurring.
XLOS, like several genetic developmental syndromes, is characterised by high variability of the clinical signs. In MID1-mutated patients, not only some of the signs might be absent but also if present their severity is variable. Variability is not dependent (or not completely dependent) on the type of mutations present as even affected members of the same family may show different clinical manifestations (Fontanella et al., 2008). The most frequently observed signs are: dysmorphic features, mainly represented by hypertelorism, often associated with other craniofacial features, such as cleft of lip and palate, frontal bossing, large nasal bridge, low-set ears, etc.; laryngo-tracheoesophageal abnormalities, ranging from tracheomalacia to esophageal clefts; and external genitalia abnormalities that, being an X-linked disease affecting mainly males, are predominantly represented by various-degree-hypospadias (Robin et al., 1996). In addition to the above clinical manifestations, XLOS MID1-mutated patients can present with cardiac abnormalities, mainly atrial-septal defects; anal defects, principally imperforate or ectopic anus; as well as other less frequent signs (Quaderi et al., 1997). XLOS also shows a neurological component presented as anatomical cerebellar vermis hypoplasia and agenesis or hypoplasia of the corpus callosum as well as cognitive and developmental delays (Fontanella et al., 2008). The prevalence of X-linked Opitz G/BBB syndrome ranges from 1:50,000 to 1:100,000 in males. The females are usually carriers of the mutation on one allele and they present minor manifestations, mainly ocular hypertelorism, likely as effect of X-inactivation mosaicism, although this issue would merit further investigations.
3.2. Expression during embryonic development
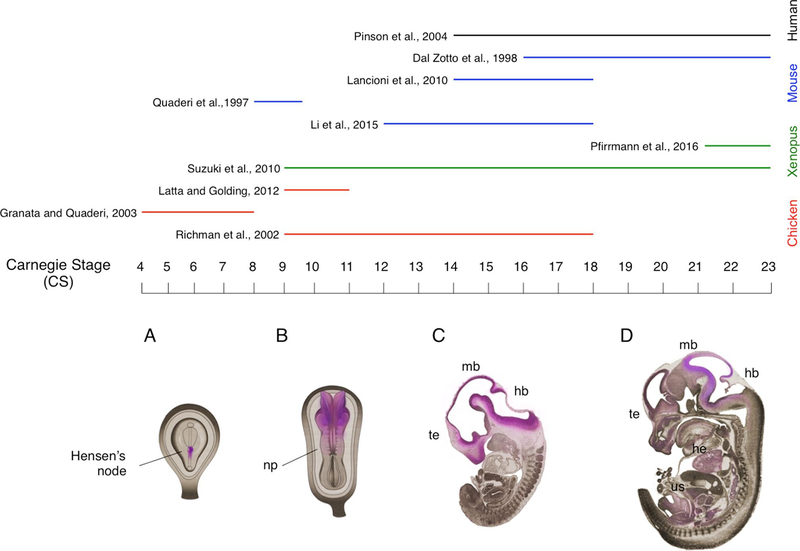
Being implicated in a congenital malformation disorder, MID1 is highly expressed during embryonic development where it exerts its main function. Embryogenesis is a long and complex process in which a series of remodelling events takes place in a sequential manner. The analysis of the physiological role and expression of MID1 in human tissues during development is an extremely difficult and controversial aspect, as such scientific knowledge regarding this issue is fairly restricted. Several model organisms recapitulate the same steps of organogenesis as in humans, thus representing the ideal alternative to human embryos for these studies providing also the experimental models to study the mechanisms. In general, the steps of organogenesis are conserved among different species. Several staging systems have been developed in order to describe the sequence of events that occurs during embryonic development: Hamburger-Hamilton (HH) for chick, Nieuwkoop-Faber (NF) for Xenopus, embryonic days (E) for mouse/rat. The Carnegie System (CS) provides a unified and standardised chronology of embryonic stages of the vertebrate embryos, based on the progressive morphological development. In line with that, the physiological expression profile and role of the orthologue genes of MID1 during embryogenesis have been explored at different time points during development in multiple animal models: mouse (Mid1) (Dal Zotto et al., 1998; Lancioni et al., 2010; Lu et al., 2013), chicken (cMid1) (Granata and Quaderi, 2003; Latta and Golding, 2012; Richman et al., 2002) and Xenopus (xMid1) (Pfirrmann et al., 2016; Suzuki et al., 2010), thus covering approximately all embryonic developmental phases (Fig. 4).
Fig. 4. Distribution of MID1 during embryonic development.
In the upper part, expression of MID1 in specific developmental stages is summarised. The coloured lines indicate the models used in the studies as indicated on the right-hand side. In the bottom part a schematic representation of Mid1 distribution at different stages of embryonic development is shown (pink shading); Carnegie stages (CS) are indicated. A) Mid1 distribution is restricted to the right side of the Hensen’s node; B) the cranial region of the neural plate (np) displays the strongest expression of Mid1 at CS9; C) Mid1 is mainly transcribed in the proliferating compartments of telencephalic vesicle (te), dorsal midbrain (mb) and hindbrain (hb); D) at late embryonic stages, high levels of Mid1 transcript are particularly described in the developing hindbrain and midbrain. Mid1 mRNA is also present in the heart (he) and in several organs of the urogenital system (us). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The earliest analysis of MID1 expression has been carried out in chicken embryos starting from gastrulation (Granata and Quaderi, 2003). At Carnegie stage (CS) 3 a weak expression of cMid1 in the anterior portion of the horseshoe-shaped domain is detected, which becomes stronger at CS4. Starting from that stage, cMid1 expression, which is first symmetrically detected in the ectoderm of Hensen’s node, is gradually restricted to the mesendoderm of the right side of the node. During neurulation, by CS8 cMid1 displays a weak bilateral symmetric pattern in the node and a strong signal is observed in the developing neural folds (Granata and Quaderi, 2003). The neural plate exhibits cMid1 expression especially in the cranial region at CS9 (Latta and Golding, 2012; Richman et al., 2002). Specific signal is detected in the neural epithelium of the forebrain, as well as in the head mesenchyme adjacent to midbrain and hindbrain, in the forming rhombomeres and in the presomitic mesoderm (Richman et al., 2002). In mouse and in Xenopus, at CS9 Mid1 is uniformly expressed at high level virtually in all embryonic tissues, except for the developing murine heart (Dal Zotto et al., 1998; Quaderi et al., 1997; Suzuki et al., 2010). Starting from stage CS10, in the late phase of neurulation, cMid1 mRNA shows a strong appearance in rhombomeres 2/3 in chicken embryos (Latta and Golding, 2012; Richman et al., 2002). At this stage xMid1 is expressed along the midline in the Xenopus neural tube, in the otic and optic vesicles and in the anlage of the cement gland (Suzuki et al., 2010). Within the prospective Central Nervous System (CNS), nearly the entire ventral neuroepithelium exhibits sensible expression of cMid1 transcript in chicken embryos at CS12, with notably strong levels within the diencephalic vesicle (Richman et al., 2002). Moreover, relatively high transcription of cMid1 at the base of the allantois is also observed at this stage (Richman et al., 2002).
At CS14 the strongest expression of Mid1 in murine developing CNS is described in the proliferating compartment of dorsal midbrain and hindbrain (Lancioni et al., 2010). Similar expression profile is found in human embryos at CS14, stage in which MID1 is transcribed particularly in the three primary brain vesicles of the developing CNS (forebrain, midbrain and hindbrain) (Pinson et al., 2004), but also in chicken embryos that display at this stage strong cMid1 expression in the craniofacial region, comprising frontal process of maxilla, the growing mandible and nasal mesenchyme (Richman et al., 2002). Furthermore, other developing structures present MID1 expression in different species between CS14 and CS15: the otic vesicle, the forming organs of the digestive system, the branchial arches, the mesonephros, the dorsal root ganglia, sclerotomes, anlages of limbs, the midgut (which will give rise to the tracheal region), the urogenital tract, the developing eyes and heart (Pinson et al., 2004; Richman et al., 2002). Up to CS17, murine and chicken embryos show a similar craniofacial expression as in the previous stages of development, with increasing level of Mid1 mRNA in the rostral part of the rising CNS, with particular regard to the proliferating telencephalic epithelium (Dal Zotto et al., 1998; Li et al., 2016; Richman et al., 2002).
As embryogenesis proceeds, human MID1 expression in the diencephalon is progressively reduced, while at CS18 a strong signal is detected in the telencephalic vesicles (Pinson et al., 2004). In addition, epithelia and mesenchyme of the respiratory and digestive developing organs (i.e the trachea and oesophagus), as well as the vertebra and the forming heart, even at lower extent, show appreciable expression of the transcript both in human and in chicken embryos (Pinson et al., 2004; Richman et al., 2002). Pattern of expression of human MID1 at CS19 is similar as at CS18, with even greater increase of signal in telencephalon and in metencephalon (especially in the cerebellar primordium), while it decreases in the mesencephalon (Pinson et al., 2004). High level of transcript is also observed in the epithelia of nasal and oral cavities, in the forming oesophagus and larynx, in the medial part of the tongue, in the spinal cord, in the dorsal root ganglia, in the neurosensory retina, in the metanephros and in the anal fold (Pinson et al., 2004). As in human, from the tailbud of Xenopus and E14.5 embryonic mouse (both corresponding to CS20) to the final embryonic stage (CS23), high expression level of Mid1 depicts the hindbrain and the midbrain, the developing kidney, the forming organs of the respiratory and the digestive system (e.g. lungs and stomach), the whole eye vesicle (mainly proliferating neurons of the neural retina and undifferentiated cells of the lens express Mid1) and the urethral epithelium (Dal Zotto et al., 1998; Pfirrmann et al., 2016; Suzuki et al., 2010); while only the interventricular septum shows transcription of MID1 gene within the defining human heart (Pinson et al., 2004).
The developmental expression of MID1 among different models is very similar, although not completely overlapping. For instance, while the chicken presents expression of the cMid1 mRNA in the whole developing heart (Richman et al., 2002), no heart expression is observed in murine embryos (Dal Zotto et al., 1998) and restriction of the transcript in the interventricular septum is detected in the forming human heart (Pinson et al., 2004). These observations could be due to biological variability in the species-specific expression of the gene, in accordance also with the fact that the MID1 gene maps within the X chromosome in a region characterised by high genomic instability, which is diversely regulated in distinct species (i.e. murine Mid1 gene spans the pseudoautosomal boundary, escaping X-inactivation; while in human MID1 is fully X-specific and subjected to X-inactivation), thus explaining divergence in gene expression (Dal Zotto et al., 1998). However, it noticeably emerges that the expression of MID1 transcript during embryogenesis correlates with structures and organs whose development is affected in OS.
3.3. Role in embryonic development
Even though the expression of MID1 transcript has been clearly detected in the structures and organs that are affected in OS patients, the first evidence of a direct involvement of MID1 gene in development has been provided through analysis in chicken embryos at early developing stages, assessing the implication of this E3 ubiquitin ligase in establishing the physiological/molecular asymmetry at the Hensen’s node (Granata and Quaderi, 2003). In fact, both ectopic overexpression of cMid1 in the physiologically Mid1-devoid left side and morpholino-induced knock-down of its expression on the right side of the node alter the proper asymmetrical expression of genes that are implicated in the definition of laterality early in chicken development (Granata and Quaderi, 2003). Importantly, cMid1 is able to modulate the levels of Shh in the perinodal region, thus assuming a pivotal role in controlling midline-lateral developmental progression with implication in the pathogenesis of OS (Granata and Quaderi, 2003) (Fig. 3C).
Furthermore, one of the features of the OS patients is the occurrence of neurological signs. Several studies conducted in mouse, Xenopus and chicken embryos highlight Mid1 physiological contribution to different neurodevelopmental events, starting from early migration of the Cranial Neural Crest (CNC) cells (Latta and Golding, 2012), through neural tube closure (Suzuki et al., 2010), to visual system and cerebellar development (Dierssen et al., 2012; Lancioni et al., 2010; Nakamura et al., 2017; Pfirrmann et al., 2016). Indeed, some of these processes are known to be defective in OS patients, in particular the morphogenesis of the cerebellum. According to a mouse model of the disease, depletion of Mid1 in the embryos causes anatomical brain abnormalities in the dorsal midbrain and cerebellar regions, i.e. hypoplasia of the anterior cerebellar vermis, the medial cerebellar region, caused by rostralisation of the midbrain/cerebellum boundary and downregulation of Fgf17, likely due to molecular defects occurring during midgestation (Fig. 5) (Lancioni et al., 2010). This defect has been shown to be associated with motor learning impairment (Lancioni et al., 2010). Similarly, another transgenic mouse line showing reduced level of Mid1 in cerebellum and hippocampus as consequence of the expression of a dominant active Downstream Regulatory Element Antagonistic Modulator (DREAM) mutant has been reported to exhibit a critical shortening of the rostro-caudal axis of the cerebellum, along with a severe delay in neuromotor development (Dierssen et al., 2012). Likewise, transgenic mice depleted for Rac1 and Rac3, that belong to the Rho family of small GTPases and are known to play important roles during embryogenesis show downregulation of Mid1 (Nakamura et al., 2017). Interestingly, axonogenesis, dendritogenesis, tangential and radial migrations of immature neurons are impaired in these transgenic mice, resulting in hypoplasia of the medial internal granule layer of the cerebellum. All these findings indicate a role of MID1 E3 ubiquitin ligase in contributing to cerebellar morphogenesis and function. In addition, the analysis of an independent Mid1 knock-out mouse line has revealed its essential role in axon development and precise neuronal projection pattern establishment in corpus callosum during embryogenesis (Lu et al., 2013). Indeed, Mid1 silencing in cultured neurons, isolated from cerebral cortex and hippocampus, accelerates axon growth and branch formation (Lu et al., 2013). This study suggests Mid1 as a negative regulator of axon growth that ensures axons precise patterning, which is crucial for proper circuit development.
Fig. 5. Mutations in Mid1 determine neuroanatomical defects during embryonic development.
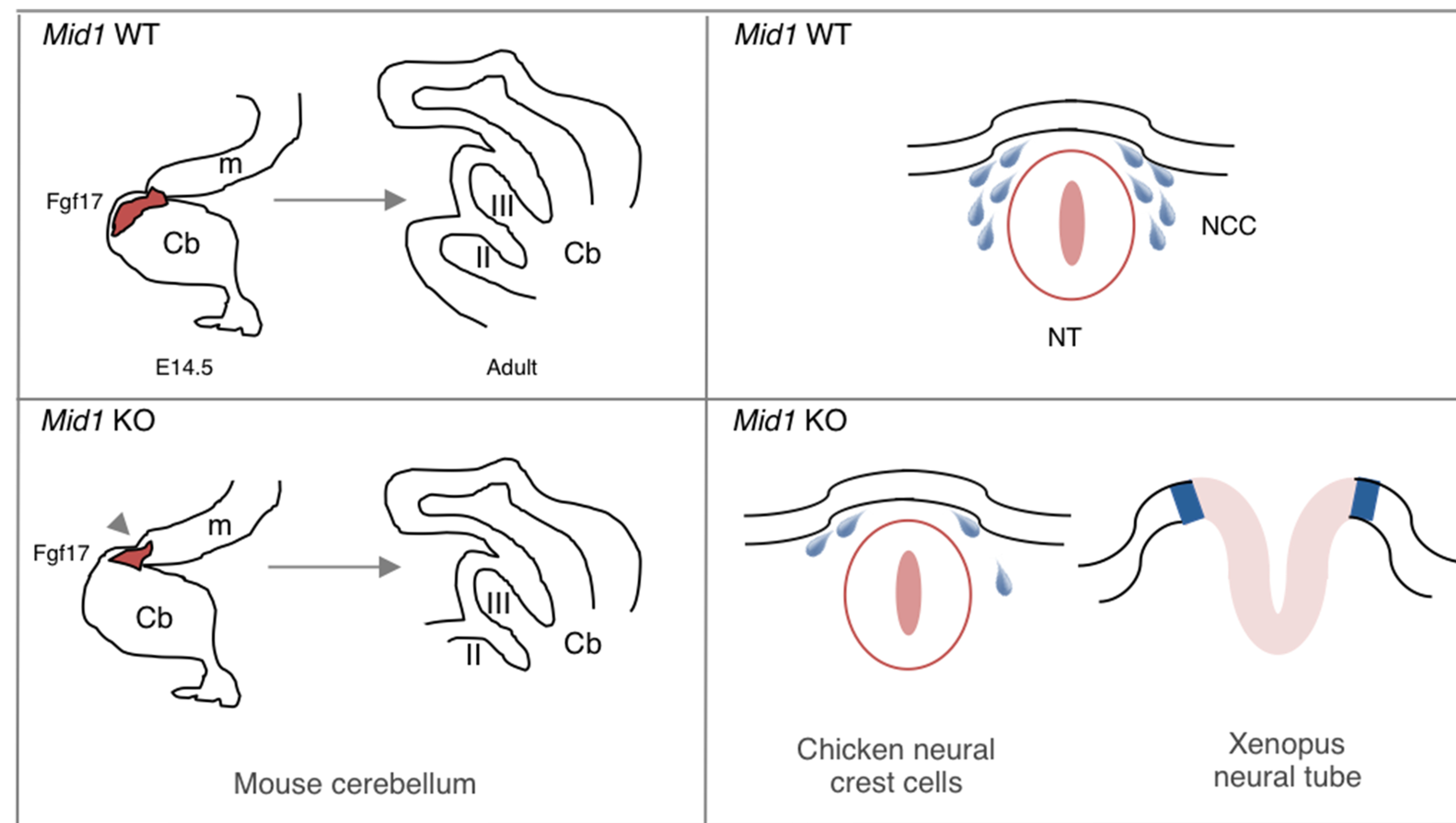
Left-hand side, mouse Mid1 knock-out causes a defective morphogenesis of the cerebellum (cb) and anterior midbrain (m) during development, including the rostralisation of the midbrain/hindbrain boundary (arrowhead) during midgestation (embryonic day 14.5, E14.5) and the consequent malformation of adult cerebellar most anterior lobes (lobes II and III). Right-hand side, Mid1 is implicated in proper neurulation event during embryonic development: defects in cranial neural crest cells (NCC) migration observed in chicken and in neural tube (NT) closure in Xenopus are associated with silencing of Mid1.
Evidence of Mid1 involvement in additional embryonic processes, albeit not directly associated with the occurrence of OS defects yet, has been provided. Following gastrulation, neurulation occurs; the neural plate bends and elevates, converging towards the dorsal midline and giving rise to the neural tube. To achieve a correct closure, this first rudiment of the CNS requires dynamic morphological changes of the neuroepithelial cells (De Pascalis and Etienne-Manneville, 2017; Leonard and Taneyhill, 2019). In Xenopus, the simultaneous depletion of xMid1 and of its close paralogue, xMid2, disrupts the proper microtubule organisation of midline neuroepithelial cells causing defective neural tube closure (Fig. 5) (Suzuki et al., 2010). Concomitant with the neural tube closure, a primary event is the migration of the CNC cells, destined among other fates to populate the branchial arches and the frontonasal process (Abramyan and Richman, 2018). Disruption of cMid1 function in chicken embryos results in reduction of CNC speed and subsequent deficit in cranial ganglia formation, especially the trigeminal one, through PP2Ac regulation (Fig. 5) (Latta and Golding, 2012).
In addition, silencing of xMid1 in Xenopus results in the enlargement of the eyes at tadpole stage, with aberrant retinal folds and loss of sharp boundaries of retinal layers (Pfirrmann et al., 2016). This is due to the lack of proteasomal degradation and consequential sustained level of Pax6, a transcription factor necessary for the specification of the eye prospective retinal field that later must be removed for the proper formation of the eyestalk and the differentiation of retinal precursor cells (Ashery-Padan et al., 2000; Pfirrmann et al., 2016; Plaza et al., 1995; Shaham et al., 2012).
In model organisms, the misregulation of Mid1 results in abnormalities comparable to those observed in OS patients. However, the phenotype seen in humans shows a wider range of defects that are often absent in other organisms. Nevertheless, when Mid1 functionality is compromised, distinct models commonly share the impairment in midline structures and laterality determination, corroborating the involvement of Mid1 in such developing processes.
4. The role of MID1 in adulthood
4.1. MID1 promotes allergic asthma
Following the first interest on the role of the MID1 gene in development driven by its involvement in XLOS, several subsequent findings indicate possible roles of the MID1 gene product also in adult life. In particular, mice sensitised and challenged with house dust mite (HDM) or rhinovirus infection, showing pathological signs of allergic asthma with airway hyper-reactivity and mucus production, upregulate Mid1 in bronchial cells at both mRNA and protein levels (Collison et al., 2013). The observed upregulation is Toll-Like Receptor 4 (TLR4)-dependent and can be recapitulated by administering recombinant Tumour necrosis factor–Related Apoptosis-Inducing Ligand (TRAIL) (Collison et al., 2013). This upregulation is associated with decreased PP2A activity and PP2Ac protein level and, conversely, silencing of Mid1 attenuates HDM-induced asthma signs. Therefore, Mid1 promotes allergic airway disease by limiting PP2A-mediated deactivation of NF-κB and p38 Mitogen-Activated Protein kinase (p38MAPK) among others (Collison et al., 2013; Foster et al., 2017). These results were confirmed by the observation that MID1 activates proinflammatory signalling in bronchial epithelial cells from human subjects. A similar mechanism is likely involved in a model of eosinophilic esophagitis and pulmonary fibrosis (Collison et al., 2019, 2015).
By considering the immune response, other evidence suggests that MID1 can control lytic granule exocytosis and polarisation as well as migration in mouse T cells. It is well known that microtubule cytoskeleton plays and important role in the secretion of lytic granules and thereby in cytotoxic lymphocytes (CTLs)-mediated target cell killing (Huse et al., 2008). Mid1 is strongly upregulated in murine CTLs, controlling T-cell receptor signalling, centrosome trafficking and exocytosis of lytic granules (Boding et al., 2014a). Another aspect described in murine T cells concerns Mid1 involvement in cell migration. Indeed, Mid1 localises to the uropod of migrating CTLs, modulating polarisation and migration (Boding et al., 2014b), probably by affecting microtubule dynamics known to be crucial in the in vivo migration of T-cells (Dong et al., 2013). Whether this function is present also in other cell types that use different migration mechanisms is an issue that deserves further studies.
4.2. Neurodegeneration and cancer
MID1 plays a role also in two highly relevant pathological areas, considering their social and clinical burden: neurodegeneration and cancer. We will briefly mention these issues as thoroughly reviewed elsewhere (Griesche et al., 2016; Winter et al., 2016).
Concerning the first area, neurodegeneration, among the transcripts that share the MIDAS motif is the family of CAG repeats-containing mRNAs. The trinucleotide CAG, which encodes for glutamine, represents unstable region that could vary in length, expand the number of the repetition of the codon within the gene and thus cause the synthesis of a toxic polyglutamine protein. Structurally, the RNAs with expanded CAG triplet can form hairpin structures, the stability of which increases with repeat-numbers (Gatchel and Zoghbi, 2005; Mirkin, 2007). Some of the CAG repeat mRNAs have been tested for their ability to associate with the MID1-complex. In particular, Huntingtin (HTT), Ataxin2 (ATXN2), Ataxin3 (ATXN3) and Ataxin7 (ATXN7) transcripts are all part of the MID1-containing mRNPs. Interestingly, both the strength of the interaction and the increase of protein translation are dependent on the CAG repeat size (Griesche et al., 2016; Krauss et al., 2013). Reducing MID1 protein level or masking RNA-MID1 association (i.e. by using the small molecule furamide able to inhibit the recruitment of MID1 to mutant HTT RNA) are sufficient to abrogate the interaction of those mRNAs with the MID1-containing complex and significantly reduce the protein levels, strongly corroborating the idea that MID1, within the mRNP complex, plays a crucial role in mediating the binding and the efficiency of the translation machinery with possible implications in neurodegeneration (Krauss et al., 2013; Matthes et al., 2018b). Interestingly, the mRNA of both APP, the amyloid precursor protein, and BACE1, the secretase responsible for the formation of the neurotoxic amyloid-β (Aβ) peptide, have been detected in association with MID1-containing complex (Matthes et al., 2018a). This interaction induces BACE1 and APP translation while, conversely, by interfering with the assembly of the mRNP complex both protein levels are reduced (Hettich et al., 2014; Matthes et al., 2018a). Interestingly, MID1 is overexpressed in post-mortem tissues from Alzheimer disease patients presenting with amyloid-β plaques and hyperphosphorylated Tau (Schweiger et al., 2017).
As true for several genes involved in developmental processes, some of the MID1-related networks are tightly controlled in somatic adult cells and when dysregulated can promote tumorigenesis. In the case of MID1, activation of the Shh pathway, as observed in some cell types, can be relevant in some cancer types, e.g. medulloblastoma (Kumar et al., 2019; Tamayo-Orrego and Charron, 2019). Another field where the role of MID1 might be relevant is prostate cancer. Indeed, MID1 can bind Androgen Receptor (AR) mRNA to induce its translation; AR transcript harbours a purine-rich region characterised by two trinucleotide repeats: the polyCAG and the polyGGY repeats, thereby depicting an additional type of mRNAs capable of interacting with MID1 complex (Demir et al., 2014; Kohler et al., 2014). Reciprocally, AR can control MID1 transcription. This crosstalk between MID1 and AR is important to regulate prostate cancer cells proliferation and consistently MID1 is over-expressed in prostate cancer cell lines showing aggressive phenotype (Kohler et al., 2014). Despite several reports on the role of MID1 in cancer, the precise role and types of tumours showing MID1 involvement still need to be further studied to evaluate the possibility of considering MID1 as a therapeutic target.
5. Conclusions and future perspectives
MID1 is found within the cells in a ternary complex together with α4 and PP2Ac. This seems to be the central core of MID1 cellular function with implication in the regulation of the mTORC1 signalling pathway and of the Shh network. However, the MID1-α4-PP2Ac complex stoichiometry and dynamics still need to be further investigated. As explained above, only a restricted pool of PP2Ac is regulated by MID1 and, in turn, also the involved signalling pathways can be regulated in confined space and time within the cell. Moreover, it is still unclear if the translational control exerted by MID1 on selected mRNAs is dependent on α4 and PP2Ac and in such case, through which mechanism. The most important issue to unravel, however, is how MID1 alterations are pathogenetic during embryonic development leading to XLOS. Of the networks above, the role of the Shh pathway in the establishment of the midline during development has been long known and in fact mutations in the SHH gene cause holoprosencephaly, a complex human brain malformation associated with facial anomalies due to abnormal definition of the ventral midline (Dubourg et al., 2007). Additionally, mutations found in the GLI3 gene, another component of the Shh pathway, cause a range of phenotypes including Greig and Pallister-Hall syndromes, characterised by hypertelorism and polydactyly (Johnston et al., 2010; Vortkamp et al., 1991). Therefore, the existence of a crosstalk between Shh and MID1 is not surprising. MID1 loss-of-function mutations cause a phenotype that is somewhat opposite to holoprosencephaly suggesting that the reciprocal regulation between these two factors might be impaired in OS patients. Further, what is still unclear is whether the crosstalk between Shh and MID1 involves also mTORC1 signalling, since several works confirm the interrelationship between Shh pathway and mTORC1 signalling (Nanta et al., 2019). One can speculate the existence of a triple interplay involving MID1-α4-PP2Ac complex, mTORC1 and Shh pathway, in which mutations and/or alterations may interfere with the correct midline establishment during embryonic development.
In addition a feature not addressed in this review is MID1 ability to homo- and hetero-dimerise. Indeed, through its coiled-coil domain, MID1 is able to dimerise like all the members of the TRIM family (Cainarca et al., 1999; Short et al., 2002). As mentioned at the beginning, the structure of the entire MID1 protein is not solved, therefore at present, we cannot fully understand the conformation of MID1 homodimer and we do not appreciate the extent of homodimerisation necessity for microtubule association and binding to substrates (Short and Cox, 2006; Short et al., 2002). Within this topic, another issue that deserves further investigation is the functional relationship between MID1 and its close and interacting paralogue, MID2/TRIM1 (Buchner et al., 1999). Both bind α4 but whether they both participate in PP2A control and in the mTORC1 and Shh signalling is still unknown (Short et al., 2002). Functional redundancy of the two proteins has been shown in early chicken development (Granata et al., 2005) but other studies indicate that they can have overlapping but not coincident functions. For example, MID1 and MID2 interact with Astrin that is ubiquitinated only by MID2 (Gholkar et al., 2016; Zanchetta and Meroni, 2019); on the same line, despite several analyses, MID2 mutations are not associated to OS or similar pathologies but to an X-linked mental retardation syndrome characterised by a milder phenotype (Geetha et al., 2014). It would be interesting to investigate possible modulatory (and reciprocal?) functions of the two paralogues in the variable expressivity of these pathogenetic conditions in presence of different combination of mutated alleles.
In conclusion, there are still several open questions on the physiological functions of MID1 and further experimental research is required to get insights into MID1-related pathogenesis of developmental and adulthood clinical conditions.
Acknowledgements
This review and the corresponding Gene Wiki article are written as part of the Gene Wiki Review series–a series resulting from a collaboration between the journal GENE and the Gene Wiki Initiative. The Gene Wiki Initiative is supported by National Institutes of Health (GM089820). Additional support for Gene Wiki Reviews is provided by Elsevier, the publisher of GENE. This sentence has been indicated by the editor who invited for the review.
Work in Meroni’s lab is supported by PRIN2015-MIUR (Ministry of Education, University and Research), grant number 20152CB22L. The authors would like to apologise those colleagues whose work has not been cited due to space limitation.
The corresponding Gene Wiki entry for this review can be found here: https://en.wikipedia.org/wiki/MID1.
Abbreviations:
- ANXA2
Annexin A2
- APP
amyloid precursor protein
- AR
Androgen Receptor
- BACE1
β-secretase 1
- BRAF35
BRCA2-Associated Factor 35
- CNC
Cranial Neural Crest cells
- CNS
Central Nervous System
- COS
C-terminal subgroup One Signature
- CS
Carnegie System
- CTLs
cytotoxic lymphocytes
- DREAM
Downstream Regulatory Element Antagonist Regulator
- E
Embryonic days
- EF-1α
Elongation Factor 1α
- FGF21
Fibroblast Growth Factor 21
- FN3
Fibronectin type III repeat
- HECT
E6-AP COOH Terminus
- HH
Hamburger- Hamilton
- LOF
Loss-of-Function
- MAP
Microtubule Associated Protein
- MIDAS
MID1 Association Sequence
- MIG12
MID1 interacting G-12 like protein
- mTORC1
mammalian Target Of Rapamycin Complex 1
- NF-κB
Nuclear Factor κ-light-chain enhancer of activated B cells
- NF
Nieuwkoop- Faber
- NMR
Nuclear Magnetic Resonance
- NPM
Nucleophosmin
- OS
Opitz Syndrome
- p38MAPK
p38 Mitogen-Activated Protein kinase
- PAR
Pseudoautosomal region
- PDPK-1
3-Phosphoinositide Dependent Protein Kinase
- PP2A
Protein Phosphatase 2A
- PTCH
Patched
- RACK1
Receptor of Activated protein kinase C1
- RING
Really Interesting New Gene
- RNP
ribonucleoprotein
- SHH
Sonic Hedgehog
- SMO
Smoothened
- SPRY
SPla and the RYanodine receptor
- SUFU
Suppressor of Fused
- TFEB
Transcriptional Factor EB
- TLR4
Toll-Like Receptor 4
- TRAIL
Tumour necrosis factor–Related Apoptosis-Inducing Ligand
- TRIM
TRipartite Motif
- UIM
ubiquitin interacting motif
- XLOS
X-linked Opitz G/BBB Syndrome
Footnotes
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
The corresponding Gene Wiki entry for this review can be found here: https://en.wikipedia.org/wiki/MID1.
References
- Abramyan J, Richman JM, 2018. Craniofacial development: discoveries made in the chicken embryo. Int. J. Dev. Biol 62, 97–107. [DOI] [PubMed] [Google Scholar]
- Aranda-Orgilles B, Trockenbacher A, Winter J, Aigner J, Kohler A, Jastrzebska E, Stahl J, Muller EC, Otto A, Wanker EE, et al. , 2008b. The Opitz syndrome gene product MID1 assembles a microtubule-associated ribonucleoprotein complex. Hum. Genet 123, 163–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranda-Orgilles B, Aigner J, Kunath M, Lurz R, Schneider R, Schweiger S, 2008a. Active transport of the ubiquitin ligase MID1 along the microtubules is regulated by protein phosphatase 2A. PLoS ONE 3, e3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranda-Orgilles B, Rutschow D, Zeller R, Karagiannidis AI, Kohler A, Chen C, Wilson T, Krause S, Roepcke S, Lilley D, et al. , 2011. Protein phosphatase 2A (PP2A)-specific ubiquitin ligase MID1 is a sequence-dependent regulator of translation efficiency controlling 3-phosphoinositide-dependent protein kinase-1 (PDPK-1). J. Biol. Chem 286, 39945–39957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arigoni M, Barutello G, Riccardo F, Ercole E, Cantarella D, Orso F, Conti L, Lanzardo S, Taverna D, Merighi I, et al. , 2013. miR-135b coordinates progression of ErbB2-driven mammary carcinomas through suppression of MID1 and MTCH2. Am. J. Pathol 182, 2058–2070. [DOI] [PubMed] [Google Scholar]
- Ashery-Padan R, Marquardt T, Zhou X, Gruss P, 2000. Pax6 activity in the lens primordium is required for lens formation and for correct placement of a single retina in the eye. Genes. Dev 14, 2701–2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell JL, Malyukova A, Holien JK, Koach J, Parker MW, Kavallaris M, Marshall GM, Cheung BB, 2012. TRIM16 acts as an E3 ubiquitin ligase and can hetero-dimerize with other TRIM family members. PLoS ONE 7, e37470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berti C, Fontanella B, Ferrentino R, Meroni G, 2004. Mig12, a novel Opitz syndrome gene product partner, is expressed in the embryonic ventral midline and co-operates with Mid1 to bundle and stabilize microtubules. BMC Cell Biol 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boding L, Hansen AK, Nielsen MM, Meroni G, Braunstein TH, Woetmann A, Odum N, Bonefeld CM, Geisler C, 2014b. Midline 1 controls polarization and migration of murine cytotoxic T cells. Immun. Inflammation Dis 2, 262–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boding L, Hansen AK, Meroni G, Johansen BB, Braunstein TH, Bonefeld CM, Kongsbak M, Jensen BA, Woetmann A, Thomsen AR, et al. , 2014a. Midline 1 directs lytic granule exocytosis and cytotoxicity of mouse killer T cells. Eur. J. Immunol 44, 3109–3118. [DOI] [PubMed] [Google Scholar]
- Buchner G, Montini E, Andolfi G, Quaderi N, Cainarca S, Messali S, Bassi MT, Ballabio A, Meroni G, Franco B, 1999. MID2, a homologue of the Opitz syndrome gene MID1: similarities in subcellular localization and differences in expression during development. Hum. Mol. Genet 8, 1397–1407. [DOI] [PubMed] [Google Scholar]
- Buetow L, Huang DT, 2016. Structural insights into the catalysis and regulation of E3 ubiquitin ligases. Nat. Rev. Mol. Cell Biol 17, 626–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cainarca S, Messali S, Ballabio A, Meroni G, 1999. Functional characterization of the Opitz syndrome gene product (midin): evidence for homodimerization and association with microtubules throughout the cell cycle. Hum. Mol. Genet 8, 1387–1396. [DOI] [PubMed] [Google Scholar]
- Carneiro BA, Kaplan JB, Altman JK, Giles FJ, Platanias LC, 2015. Targeting mTOR signaling pathways and related negative feedback loops for the treatment of acute myeloid leukemia. Cancer Biol. Ther 16, 648–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carracedo A, Baselga J, Pandolfi PP, 2008. Deconstructing feedback-signaling networks to improve anticancer therapy with mTORC1 inhibitors. Cell Cycle 7, 3805–3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Wang K, Long A, Jia L, Zhang Y, Deng H, Li Y, Han J, Wang Y, 2017. Fasting-induced hormonal regulation of lysosomal function. Cell Res 27, 748–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A, 2015. The unravelling of the ubiquitin system. Nat. Rev. Mol. Cell Biol 16, 322–324. [DOI] [PubMed] [Google Scholar]
- Collison A, Hatchwell L, Verrills N, Wark PA, de Siqueira AP, Tooze M, Carpenter H, Don AS, Morris JC, Zimmermann N, et al. , 2013. The E3 ubiquitin ligase midline 1 promotes allergen and rhinovirus-induced asthma by inhibiting protein phosphatase 2A activity. Nat. Med 19, 232–237. [DOI] [PubMed] [Google Scholar]
- Collison AM, Sokulsky LA, Sherrill JD, Nightingale S, Hatchwell L, Talley NJ, Walker MM, Rothenberg ME, Mattes J, 2015. TNF-related apoptosis-inducing ligand (TRAIL) regulates midline-1, thymic stromal lymphopoietin, inflammation, and remodeling in experimental eosinophilic esophagitis. J. Allergy Clin. Immunol 136, 971–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collison AM, Li J, de Siqueira AP, Lv X, Toop HD, Morris JC, Starkey MR, Hansbro PM, Zhang J, Mattes J, 2019. TRAIL signals through the ubiquitin ligase MID1 to promote pulmonary fibrosis. BMC Pulmonary Med 19, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox TC, Allen LR, Cox LL, Hopwood B, Goodwin B, Haan E, Suthers GK, 2000. New mutations in MID1 provide support for loss of function as the cause of X-linked Opitz syndrome. Hum. Mol. Genet 9, 2553–2562. [DOI] [PubMed] [Google Scholar]
- Dal Zotto L, Quaderi NA, Elliott R, Lingerfelter PA, Carrel L, Valsecchi V, Montini E, Yen CH, Chapman V, Kalcheva I, et al. , 1998. The mouse Mid1 gene: implications for the pathogenesis of Opitz syndrome and the evolution of the mammalian pseudoautosomal region. Hum. Mol. Genet 7, 489–499. [DOI] [PubMed] [Google Scholar]
- De Pascalis C, Etienne-Manneville S, 2017. Single and collective cell migration: the mechanics of adhesions. Mol Biol Cell 28, 1833–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demir U, Koehler A, Schneider R, Schweiger S, Klocker H, 2014. Metformin antitumor effect via disruption of the MID1 translational regulator complex and AR downregulation in prostate cancer cells. BMC Cancer 14, 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Como CJ, Arndt KT, 1996. Nutrients, via the Tor proteins, stimulate the association of Tap42 with type 2A phosphatases. Genes Dev 10, 1904–1916. [DOI] [PubMed] [Google Scholar]
- Di Rienzo M, Romagnoli A, Antonioli M, Piacentini M, and Fimia GM (2020). TRIM proteins in autophagy: selective sensors in cell damage and innate immune responses. Cell Death Differ, Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dierssen M, Fedrizzi L, Gomez-Villafuertes R, de Lagran MM, Gutierrez-Adan A, Sahun I, Pintado B, Oliveros JC, Dopazo XM, Gonzalez P, et al. , 2012. Reduced Mid1 expression and delayed neuromotor development in daDREAM transgenic mice. Front. Mol. Neurosci 5, 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong B, Zhang SS, Gao W, Su H, Chen J, Jin F, Bhargava A, Chen X, Jorgensen L, Alberts AS, et al. , 2013. Mammalian diaphanous-related formin 1 regulates GSK3beta-dependent microtubule dynamics required for T cell migratory polarization. PLoS ONE 8, e80500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou Y, Xing J, Kong G, Wang G, Lou X, Xiao X, Vivier E, Li XC, Zhang Z, 2019. Identification of the E3 Ligase TRIM29 as a critical checkpoint regulator of NK cell functions. J. Immunol 203, 873–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubourg C, Bendavid C, Pasquier L, Henry C, Odent S, David V, 2007. Holoprosencephaly. Orphanet. J. Rare Dis 2, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrentino R, Bassi MT, Chitayat D, Tabolacci E, Meroni G, 2007. MID1 mutation screening in a large cohort of Opitz G/BBB syndrome patients: twenty-nine novel mutations identified. Hum. Mutat 28, 206–207. [DOI] [PubMed] [Google Scholar]
- Fontanella B, Russolillo G, Meroni G, 2008. MID1 mutations in patients with X-linked Opitz G/BBB syndrome. Hum. Mutat 29, 584–594. [DOI] [PubMed] [Google Scholar]
- Foster PS, Maltby S, Rosenberg HF, Tay HL, Hogan SP, Collison AM, Yang M, Kaiko GE, Hansbro PM, Kumar RK, et al. , 2017. Modeling TH 2 responses and airway inflammation to understand fundamental mechanisms regulating the pathogenesis of asthma. Immunol. Rev 278, 20–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatchel JR, Zoghbi HY, 2005. Diseases of unstable repeat expansion: mechanisms and common principles. Nat. Rev. Genet 6, 743–755. [DOI] [PubMed] [Google Scholar]
- Geetha TS, Michealraj KA, Kabra M, Kaur G, Juyal RC, Thelma BK, 2014. Targeted deep resequencing identifies MID2 mutation for X-linked intellectual disability with varied disease severity in a large kindred from India. Hum. Mutat 35, 41–44. [DOI] [PubMed] [Google Scholar]
- Gholkar AA, Senese S, Lo YC, Vides E, Contreras E, Hodara E, Capri J, Whitelegge JP, Torres JZ, 2016. The X-linked-intellectual-disability-associated ubiquitin ligase Mid2 interacts with astrin and regulates astrin levels to promote cell division. Cell Rep 14, 180–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granata A, Quaderi NA, 2003. The Opitz syndrome gene MID1 is essential for establishing asymmetric gene expression in Hensen's node. Dev. Biol 258, 397–405. [DOI] [PubMed] [Google Scholar]
- Granata A, Savery D, Hazan J, Cheung BM, Lumsden A, Quaderi NA, 2005. Evidence of functional redundancy between MID proteins: implications for the presentation of Opitz syndrome. Dev. Biol 277, 417–424. [DOI] [PubMed] [Google Scholar]
- Griesche N, Schilling J, Weber S, Rohm M, Pesch V, Matthes F, Auburger G, Krauss S, 2016. Regulation of mRNA translation by MID1: a common mechanism of expanded CAG repeat RNAs. Front. Cell. Neurosci 10, 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Du H, Massiah MA, 2011. Detection and characterization of the in vitro E3 ligase activity of the human MID1 protein. J. Mol. Biol 407, 505–520. [DOI] [PubMed] [Google Scholar]
- Hatakeyama S, 2017. TRIM family proteins: roles in autophagy, immunity, and carcinogenesis. Trends Biochem. Sci 42 (4), 297–311. [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A, 1998. The ubiquitin system. Annu. Rev. Biochem 67, 425–479. [DOI] [PubMed] [Google Scholar]
- Hettich MM, Matthes F, Ryan DP, Griesche N, Schroder S, Dorn S, Kraubeta S, Ehninger D, 2014. The anti-diabetic drug metformin reduces BACE1 protein level by interfering with the MID1 complex. PLoS ONE 9, e102420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K, Fingar DC, 2014. Growing knowledge of the mTOR signaling network. Semin. Cell Dev. Biol 36, 79–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Q, Zhang X, 2020. Emerging roles and research tools of atypical ubiquitination. Proteomics, e1900100. [DOI] [PubMed] [Google Scholar]
- Huibregtse JM, Scheffner M, Beaudenon S, Howley PM, 1995. A family of proteins structurally and functionally related to the E6-AP ubiquitin-protein ligase. Proc. Natl. Acad. Sci. U S A 92, 5249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huse M, Quann EJ, Davis MM, 2008. Shouts, whispers and the kiss of death: directional secretion in T cells. Nat. Immunol 9, 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham PW, McMahon AP, 2001. Hedgehog signaling in animal development: paradigms and principles. Genes Dev 15, 3059–3087. [DOI] [PubMed] [Google Scholar]
- Jhanwar-Uniyal M, Wainwright JV, Mohan AL, Tobias ME, Murali R, Gandhi CD, Schmidt MH, 2019. Diverse signaling mechanisms of mTOR complexes: mTORC1 and mTORC2 in forming a formidable relationship. Adv. Biol. Reg 72, 51–62. [DOI] [PubMed] [Google Scholar]
- Joazeiro CA, Weissman AM, 2000. RING finger proteins: mediators of ubiquitin ligase activity. Cell 102, 549–552. [DOI] [PubMed] [Google Scholar]
- Johnston JJ, Sapp JC, Turner JT, Amor D, Aftimos S, Aleck KA, Bocian M, Bodurtha JN, Cox GF, Curry CJ, et al. , 2010. Molecular analysis expands the spectrum of phenotypes associated with GLI3 mutations. Hum. Mutat 31, 1142–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler A, Demir U, Kickstein E, Krauss S, Aigner J, Aranda-Orgilles B, Karagiannidis AI, Achmuller C, Bu H, Wunderlich A, et al. , 2014. A hormone-dependent feedback-loop controls androgen receptor levels by limiting MID1, a novel translation enhancer and promoter of oncogenic signaling. Molecular Cancer 13, 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D, Rape M, 2012. The ubiquitin code. Annu. Rev. Biochem 81, 203–229. [DOI] [PubMed] [Google Scholar]
- Krauss S, Foerster J, Schneider R, Schweiger S, 2008. Protein phosphatase 2A and rapamycin regulate the nuclear localization and activity of the transcription factor GLI3. Cancer Res 68, 4658–4665. [DOI] [PubMed] [Google Scholar]
- Krauss S, So J, Hambrock M, Kohler A, Kunath M, Scharff C, Wessling M, Grzeschik KH, Schneider R, Schweiger S, 2009. Point mutations in GLI3 lead to misregulation of its subcellular localization. PLoS ONE 4, e7471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauss S, Griesche N, Jastrzebska E, Chen C, Rutschow D, Achmuller C, Dorn S, Boesch SM, Lalowski M, Wanker E, et al. , 2013. Translation of HTT mRNA with expanded CAG repeats is regulated by the MID1-PP2A protein complex. Nat. Commun 4, 1511. [DOI] [PubMed] [Google Scholar]
- Kulathu Y, Komander D, 2012. Atypical ubiquitylation - the unexplored world of polyubiquitin beyond Lys48 and Lys63 linkages. Nat. Rev. Mol. Cell Biol 13, 508–523. [DOI] [PubMed] [Google Scholar]
- Kumar R, Liu APY, and Northcott PA, 2019. Medulloblastoma Genomics in the Modern Molecular Era. Brain Pathol Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancioni A, Pizzo M, Fontanella B, Ferrentino R, Napolitano LM, De Leonibus E, Meroni G, 2010. Lack of Mid1, the mouse ortholog of the Opitz syndrome gene, causes abnormal development of the anterior cerebellar vermis. J. Neurosci 30, 2880–2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry JR, Mager DL, 2002. Widely spaced alternative promoters, conserved between human and rodent, control expression of the Opitz syndrome gene MID1. Genomics 80, 499–508. [PubMed] [Google Scholar]
- Latta EJ, Golding JP, 2012. Regulation of PP2A activity by Mid1 controls cranial neural crest speed and gangliogenesis. Mech. Dev 128, 560–576. [DOI] [PubMed] [Google Scholar]
- LeNoue-Newton ML, Wadzinski BE, Spiller BW, 2016. The three Type 2A protein phosphatases, PP2Ac, PP4c and PP6c, are differentially regulated by Alpha4. Biochem. Biophys. Res. Commun 475, 64–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeNoue-Newton M, Watkins GR, Zou P, Germane KL, McCorvey LR, Wadzinski BE, Spiller BW, 2011. The E3 ubiquitin ligase- and protein phosphatase 2A (PP2A)-binding domains of the Alpha4 protein are both required for Alpha4 to inhibit PP2A degradation. J. Biol. Chem 286, 17665–17671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard CE, Taneyhill LA, 2019. The road best traveled: Neural crest migration upon the extracellular matrix. Semin. Cell Dev. Biol Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Zhou T, Zou Y, 2016. Mid1/Mid2 expression in craniofacial development and a literature review of X-linked opitz syndrome. Mol. Genet. Genomic Med 4, 95–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu E, Knutzen CA, Krauss S, Schweiger S, Chiang GG, 2011. Control of mTORC1 signaling by the Opitz syndrome protein MID1. Proc. Natl. Acad. Sci. U S A 108, 8680–8685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Prickett TD, Elliott E, Meroni G, Brautigan DL, 2001. Phosphorylation and microtubule association of the Opitz syndrome protein mid-1 is regulated by protein phosphatase 2A via binding to the regulatory subunit alpha 4. Proc. Natl. Acad. Sci. U S A 98, 6650–6655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu T, Chen R, Cox TC, Moldrich RX, Kurniawan N, Tan G, Perry JK, Ashworth A, Bartlett PF, Xu L, et al. , 2013. X-linked microtubule-associated protein, Mid1, regulates axon development. Proc. Natl. Acad. Sci. U S A 110, 19131–19136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massiah MA, Simmons BN, Short KM, Cox TC, 2006. Solution structure of the RBCC/TRIM B-box1 domain of human MID1: B-box with a RING. J. Mol. Biol 358, 532–545. [DOI] [PubMed] [Google Scholar]
- Massiah MA, Matts JA, Short KM, Simmons BN, Singireddy S, Yi Z, Cox TC, 2007. Solution structure of the MID1 B-box2 CHC(D/C)C(2)H(2) zinc-binding domain: insights into an evolutionarily conserved RING fold. J. Mol. Biol 369, 1–10. [DOI] [PubMed] [Google Scholar]
- Matthes F, Massari S, Bochicchio A, Schorpp K, Schilling J, Weber S, Offermann N, Desantis J, Wanker E, Carloni P, et al. , 2018b. Reducing mutant huntingtin protein expression in living cells by a newly identified RNA CAG binder. ACS Chem. Neurosci 9, 1399–1408. [DOI] [PubMed] [Google Scholar]
- Matthes F, Hettich MM, Schilling J, Flores-Dominguez D, Blank N, Wiglenda T, Buntru A, Wolf H, Weber S, Vorberg I, et al. , 2018a. Inhibition of the MID1 protein complex: a novel approach targeting APP protein synthesis. Cell Death Discovery 4, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell JL, Watkins GR, Soss SE, Franz HS, McCorvey LR, Spiller BW, Chazin WJ, Wadzinski BE, 2010. Alpha4 is a ubiquitin-binding protein that regulates protein serine/threonine phosphatase 2A ubiquitination. Biochemistry 49, 1713–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meroni G, Diez-Roux G, 2005. TRIM/RBCC, a novel class of 'single protein RING finger' E3 ubiquitin ligases. BioEssays 27, 1147–1157. [DOI] [PubMed] [Google Scholar]
- Mirkin SM, 2007. Expandable DNA repeats and human disease. Nature 447, 932–940. [DOI] [PubMed] [Google Scholar]
- Monteiro O, Chen C, Bingham R, Argyrou A, Buxton R, Pancevac Jonsson C, Jones E, Bridges A, Gatfield K, Krauss S, et al. , 2018. Pharmacological disruption of the MID1/alpha4 interaction reduces mutant Huntingtin levels in primary neuronal cultures. Neurosci. Lett 673, 44–50. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Ueyama T, Ninoyu Y, Sakaguchi H, Choijookhuu N, Hishikawa Y, Kiyonari H, Kohta M, Sakahara M, de Curtis I, et al. , 2017. Novel role of Rac-Mid1 signaling in medial cerebellar development. Development 144, 1863–1875. [DOI] [PubMed] [Google Scholar]
- Nanta R, Shrivastava A, Sharma J, Shankar S, Srivastava RK, 2019. Inhibition of sonic hedgehog and PI3K/Akt/mTOR pathways cooperate in suppressing survival, self-renewal and tumorigenic potential of glioblastoma-initiating cells. Mol. Cell. Biochem 454, 11–23. [DOI] [PubMed] [Google Scholar]
- Napolitano LM, Jaffray EG, Hay RT, Meroni G, 2011. Functional interactions between ubiquitin E2 enzymes and TRIM proteins. Biochem. J 434, 309–319. [DOI] [PubMed] [Google Scholar]
- Opitz JM, 1987. G syndrome (hypertelorism with esophageal abnormality and hypospadias, or hypospadias-dysphagia, or “Opitz-Frias” or “Opitz-G” syndrome)-perspective in 1987 and bibliography. Am. J. Med. Genet 28, 275–285. [DOI] [PubMed] [Google Scholar]
- Palmer S, Perry J, Kipling D, Ashworth A, 1997. A gene spans the pseudoautosomal boundary in mice. Proc. Natl. Acad. Sci. U S A 94, 12030–12035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry J, Feather S, Smith A, Palmer S, Ashworth A, 1998. The human FXY gene is located within Xp22.3: implications for evolution of the mammalian X chromosome. Hum. Mol. Genet 7, 299–305. [DOI] [PubMed] [Google Scholar]
- Pfirrmann T, Jandt E, Ranft S, Lokapally A, Neuhaus H, Perron M, Hollemann T, 2016. Hedgehog-dependent E3-ligase Midline1 regulates ubiquitin-mediated proteasomal degradation of Pax6 during visual system development. Proc. Natl. Acad. Sci. U S A 113, 10103–10108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinson L, Auge J, Audollent S, Mattei G, Etchevers H, Gigarel N, Razavi F, Lacombe D, Odent S, Le Merrer M, et al. , 2004. Embryonic expression of the human MID1 gene and its mutations in Opitz syndrome. J. Med. Genet 41, 381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaza S, Dozier C, Turque N, Saule S, 1995. Quail Pax-6 (Pax-QNR) mRNAs are expressed from two promoters used differentially during retina development and neuronal differentiation. Mol. Cell Biol 15, 3344–3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quaderi NA, Schweiger S, Gaudenz K, Franco B, Rugarli EI, Berger W, Feldman GJ, Volta M, Andolfi G, Gilgenkrantz S, et al. , 1997. Opitz G/BBB syndrome, a defect of midline development, is due to mutations in a new RING finger gene on Xp22. Nat. Genet 17, 285–291. [DOI] [PubMed] [Google Scholar]
- Reymond A, Meroni G, Fantozzi A, Merla G, Cairo S, Luzi L, Riganelli D, Zanaria E, Messali S, Cainarca S, et al. , 2001. The tripartite motif family identifies cell compartments. Embo. J 20, 2140–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richman JM, Fu KK, Cox LL, Sibbons JP, Cox TC, 2002. Isolation and characterisation of the chick orthologue of the Opitz syndrome gene, Mid1, supports a conserved role in vertebrate development. Int. J. Dev. Biol 46, 441–448. [PubMed] [Google Scholar]
- Robin NH, Opitz JM, Muenke M, 1996. Opitz G/BBB syndrome: clinical comparisons of families linked to Xp22 and 22q, and a review of the literature. Am. J. Med. Genet 62, 305–317. [DOI] [PubMed] [Google Scholar]
- Sasai N, Toriyama M, Kondo T, 2019. Hedgehog signal and genetic disorders. Front. Genet 10, 1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman BA, Harper JW, 2009. Ubiquitin-like protein activation by E1 enzymes: the apex for downstream signalling pathways. Nat. Rev. Mol. Cell Biol 10, 319–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweiger S, Foerster J, Lehmann T, Suckow V, Muller YA, Walter G, Davies T, Porter H, van Bokhoven H, Lunt PW, et al. , 1999. The Opitz syndrome gene product, MID1, associates with microtubules. Proc. Natl. Acad. Sci. U S A 96, 2794–2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweiger S, Dorn S, Fuchs M, Kohler A, Matthes F, Muller EC, Wanker E, Schneider R, Krauss S, 2014. The E3 ubiquitin ligase MID1 catalyzes ubiquitination and cleavage of Fu. J. Biol. Chem 289, 31805–31817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweiger S, Matthes F, Posey K, Kickstein E, Weber S, Hettich MM, Pfurtscheller S, Ehninger D, Schneider R, Krauss S, 2017. Resveratrol induces dephosphorylation of Tau by interfering with the MID1-PP2A complex. Sci. Rep 7, 13753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaham O, Menuchin Y, Farhy C, Ashery-Padan R, 2012. Pax6: a multi-level regulator of ocular development. Progress Retinal Eye Res 31, 351–376. [DOI] [PubMed] [Google Scholar]
- Short KM, Cox TC, 2006. Sub-classification of the rbcc/trim superfamily reveals a novel motif necessary for microtubule binding. J. Biol. Chem 281, 8970–8980. [DOI] [PubMed] [Google Scholar]
- Short KM, Hopwood B, Yi Z, Cox TC, 2002. MID1 and MID2 homo- and hetero-dimerise to tether the rapamycin- sensitive PP2A regulatory subunit, Alpha 4, to microtubules: implications for the clinical variability of X-linked Opitz GBBB syndrome and other developmental disorders. BMC Cell Biol 3, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sontag E, 2001. Protein phosphatase 2A: the Trojan Horse of cellular signaling. Cell Signal 13, 7–16. [DOI] [PubMed] [Google Scholar]
- Suzuki M, Hara Y, Takagi C, Yamamoto TS, Ueno N, 2010. MID1 and MID2 are required for Xenopus neural tube closure through the regulation of microtubule organization. Development 137, 2329–2339. [DOI] [PubMed] [Google Scholar]
- Tamayo-Orrego L, Charron F, 2019. Recent advances in SHH medulloblastoma progression: tumor suppressor mechanisms and the tumor microenvironment. F1000Research 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao H, Simmons BN, Singireddy S, Jakkidi M, Short KM, Cox TC, Massiah MA, 2008. Structure of the MID1 tandem B-boxes reveals an interaction reminiscent of intermolecular ring heterodimers. Biochemistry 47, 2450–2457. [DOI] [PubMed] [Google Scholar]
- Trockenbacher A, Suckow V, Foerster J, Winter J, Krauss S, Ropers HH, Schneider R, Schweiger S, 2001. MID1, mutated in Opitz syndrome, encodes an ubiquitin ligase that targets phosphatase 2A for degradation. Nat. Genet 29, 287–294. [DOI] [PubMed] [Google Scholar]
- Unterbruner K, Matthes F, Schilling J, Nalavade R, Weber S, Winter J, Krauss S, 2018. MicroRNAs miR-19, miR-340, miR-374 and miR-542 regulate MID1 protein expression. PLoS ONE 13, e0190437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Veyver IB, Cormier TA, Jurecic V, Baldini A, Zoghbi HY, 1998. Characterization and physical mapping in human and mouse of a novel RING finger gene in Xp22. Genomics 51, 251–261. [DOI] [PubMed] [Google Scholar]
- van Gent M, Sparrer KMJ, Gack MU, 2018. TRIM proteins and their roles in antiviral host defenses. Ann. Rev. Virol 5, 385–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vortkamp A, Gessler M, Grzeschik KH, 1991. GLI3 zinc-finger gene interrupted by translocations in Greig syndrome families. Nature 352, 539–540. [DOI] [PubMed] [Google Scholar]
- Watkins GR, Wang N, Mazalouskas MD, Gomez RJ, Guthrie CR, Kraemer BC, Schweiger S, Spiller BW, Wadzinski BE, 2012. Monoubiquitination promotes calpain cleavage of the protein phosphatase 2A (PP2A) regulatory subunit alpha4, altering PP2A stability and microtubule-associated protein phosphorylation. J. Biol. Chem 287, 24207–24215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams FP, Haubrich K, Perez-Borrajero C, Hennig J, 2019. Emerging RNA-binding roles in the TRIM family of ubiquitin ligases. Biol. Chem 400, 1443–1464. [DOI] [PubMed] [Google Scholar]
- Winter J, Lehmann T, Krauss S, Trockenbacher A, Kijas Z, Foerster J, Suckow V, Yaspo ML, Kulozik A, Kalscheuer V, et al. , 2004. Regulation of the MID1 protein function is fine-tuned by a complex pattern of alternative splicing. Hum. Genet 114, 541–552. [DOI] [PubMed] [Google Scholar]
- Winter J, Kunath M, Roepcke S, Krause S, Schneider R, Schweiger S, 2007. Alternative polyadenylation signals and promoters act in concert to control tissue-specific expression of the Opitz Syndrome gene MID1. BMC Mol. Biol 8, 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter J, Basilicata MF, Stemmler MP, Krauss S, 2016. The MID1 protein is a central player during development and in disease. Front. Biosci 21, 664–682. [DOI] [PubMed] [Google Scholar]
- Wright KM, Du H, Dagnachew M, Massiah MA, 2016. Solution structure of the microtubule-targeting COS domain of MID1. FEBS J 283, 3089–3102. [DOI] [PubMed] [Google Scholar]
- Wright KM, Du H, Massiah MA, 2017. Structural and functional observations of the P151L MID1 mutation reveal alpha4 plays a significant role in X-linked Opitz Syndrome. FEBS J 284, 2183–2193. [DOI] [PubMed] [Google Scholar]
- Zanchetta ME, Meroni G, 2019. Emerging Roles of the TRIM E3 Ubiquitin Ligases MID1 and MID2 in Cytokinesis. Front. Physiol 10, 274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanchetta ME, Napolitano LMR, Maddalo D, Meroni G, 2017. The E3 ubiquitin ligase MID1/TRIM18 promotes atypical ubiquitination of the BRCA2-associated factor 35, BRAF35. Biochim. Biophys. Acta, Mol. Cell. Res 1864, 1844–1854. [DOI] [PubMed] [Google Scholar]