Abstract
High Myopia (HM) is a common complex-trait eye disorder. There is essential evidence that genetic factors play a significant role in the development of nonsyndromic high myopia. Identification of susceptibility genes of high myopia will shed light on the pathophysiological mechanism underlying their genesis. This was a case control study examining the prospect of association of DLGAP1, EMILIN2 & MYOM1 genes on MYP2 locus in purely ethnic (Kashmiri) population representing a homogeneous cohort. Genomic DNA was extracted using phenol chloroform and salting out method. Extracted DNA was genotyped for polymorphic variations in MYOM1, EMILIN2 and DLGAP1 genes involving Sanger di-deoxy method. Allele frequencies were tested for Hardy-Weinberg disequilibrium in 224 cases and compared with 220 emmetropic controls. In DLGAP1, documented single nucleotide polymorphism (SNP); Pro517Pro was observed. A previously reported Asn451Asn SNP was observed in EMILIN2. MYOM1 showed five polymorphic variations; two in coding region (Gly333Gly & Gly341Ala) and three intronic (c.1022+23, G>A; c.3418+44 G>T & c.3418+65; C>G). All of the elucidated SNPs were having statistical significant role in increasing or decreasing the risk of disease. Although not statistically significant, a novel Glu507Lys SNP was observed in DLGAP1 (P>0.05). In silico predictions showed MYOM1 Gly341Ala to be benign & tolerated substitution while as DLGAP1 Glu507Lys to be possibly damaging substitution. The studied SNPs followed Over-Dominant, Recessive and Co-Dominant mode of inheritance with specific haplotypes associated with the disease. Our study reveals the involvement of MYP2 locus candidate gene polymorphism in the pathogenesis of HM.
Keywords: High myopia, MYP2 Locus, DLGAP1, EMILIN2, ethnic, MYOM1, novel, polymorphism
Introduction
Myopia is the multi-factorial ocular disorder with highest prevalence globally branded by spherical error of refraction (RE) and retinal defocus causing reduced visual insight. Approximately 60-80% of adult population in Taiwan, China, Korea, Japan and Singapore is hit by the disorder [1-6]. Pathological or high Myopia (RE>6D) is different from simply being “short sighted” [7]. HM is an advanced variant causing retinal detachment, lattice degeneration and chorio-retinal degeneration which are irreversible eye damages that cannot be corrected with glasses or contact lenses [7,8]. It can also lead to glaucoma [9] and incur a huge financial burden, even on developed countries [10]. The development and progression of HM is directly or indirectly influenced by various environmental factors including “near vision stimulus” in students and in some professions; educational status and time spent outdoors [11,12].
Genetic mapping involving successful efforts to determine disease genes rely on the use of functionally implicated candidate genes or probes, and the detection of significant cytogenetic findings of deletions, insertions, or translocations that consistently pair with the disease’s phenotype [13]. Genetic and environmental factors play a very important role in shaping refractive progress of an individual which is backed up by the fact that Myopia has a higher prevalence in developed Asian countries compared to Western world [14,15].
The influence of the genetic factors in the development of HM has also been demonstrated in Population-based association studies (or case-controlled studies) which have linked various genomic polymorphisms to HM phenotypes and pathogenesis. Recent multigenerational linkage studies have reported at least 23 Myopia susceptibility loci (MYP; MYP2 to MYP5) [16-19]. A genome-wide linkage analysis exposed a significant linkage of 18 centimorgan (cM) in MYP2 for non-syndromic autosomal dominant HM on 18 p11.31, which has been refined to 7.6 cM interval between markers D18S59 and D18S1138 by Haplotype analysis [16] and further tapered to an interval of 0.8 cM between markers D18S63 and D18S52 [17].
MYP2 locus harbor the genes involved in sclera formation and regulation, which are candidate genes for HM [20,21]. Numerous candidate genes for HM have been recognized within MYP2 which are involved in growth, maintenance and remodeling of sclera [20-22] which comprise Myomesin 1 (MYOM1), Elastin Microfibril Interfacer 2 (EMILIN2), Large Drosophila Homolog Associated Protein 1 (DLGAP1), Transforming Growth Β-Induced Factor (TGIF1), Lipin 2 (LPIN2), Myosin Regulatory Light Chain 2 (MRLC2), Myosin Regulatory Light Chain 3 (MRCL3), Clusterin-like 1 (CLUL1), and Zinc Finger Protein 161 Homolog (ZFP161) [20,21]. Definite role of MYP2 locus SNPs in the etiopathogenesis of HM has been established in different populations [23-25]. Yet, several studies have not shown any association between MYP2 locus SNPs and HM [20,21,26]. So, our study aimed to clarify this relationship with a case-control design keeping in view the ethnic purity of our population which may lead to effective therapies for severe forms of this potentially blinding eye disease.
Materials and methods
Study design
This is a Case-Control study conducted by the Department of Biotechnology, University of Kashmir and Department of Ophthalmology, Government Medical College (GMC) Srinagar and associated SMHS Hospital, Kashmir, India over a period of two years (2017-2019). The study has been approved by Ethical Committee of GMC Srinagar under ref no. 11A/ETH/GMC/ICM dated 22-02-2017 strictly adhering to the guidelines of the Declaration of Helsinki. Informed consent was obtained from the study subjects after explaining the nature and possible consequences of the study.
Study subjects
The study included cases with high Myopia (n=224) attending the Department of Ophthalmology, Govt. Medical College Srinagar and associated SMHS Hospital. Individuals with known ocular disease such as retinopathy, cataract or genetic disease associated with Myopia, such as Stickler or Marfan syndrome and any sort of genetic disorder were excluded from the study. Ophthalmic evaluation of each patient was done which included measuring visual acquity, keratometry, retinoscopy, slit lamp examination of the anterior segment, fundus examination and measurement of axial length. Controls (n=220) were randomly selected from a pool of healthy volunteers who visited the hospital for health check-up during the same period and enrolled in the study.
Sample collection and DNA extraction
05 ml of blood was collected from each patient and healthy control in EDTA vials; refrigerated at -80°C till further processing. Deoxyribonucleic acid (DNA) extraction of samples was carried out by standard procedures like phenol chloroform extraction and salting out. Extracted DNA was dissolved in tris-EDTA buffer for further use. The quality of DNA was checked on 2% agarose gel electrophoresis whereas purity and concentration was measured by using the NanoDrop 2000c Spectrophotometer (ThermoScientific, USA).
Polymerase chain reaction (PCR)
PCR was carried out in a total volume of 50 μl, using 50-100 ng genomic DNA, 2-6 pmole of each primer, 1× PCR buffer (Sigma Aldrich, USA) and 0.5 units of Taq DNA polymerase (Sigma Aldrich, USA). The PCR cycling conditions were as follows: one cycle of denaturation at 95°C for 5 min, 30 cycles of denaturation at 95°C for 45 s, annealing at t°C for 45 s, and extension at 72°C for 45 s, and one final 6 min extension cycle at 72°C, for amplifying different genes. All PCR products were verified on 2% agarose gel. The primer sequences, annealing temperatures and their corresponding amplicon size is shown in Table 1.
Table 1.
Primers used for amplification of DLGAP1, EMILIN2 & MYOM1 gene and their annealing temperature and product size
| Gene | Exon | Primer sequence | Ta (°C) | Product size (bp) |
|---|---|---|---|---|
| MYOM1 | 4 | F; 5’ CATGAAGTTGTTTACACTTCAACTTAC 3’ | 63 | 260 |
| R; 5’ CTCAGTGTGATCACACAGCAT TGG 3’ | ||||
| 19 | F; 5’ TGCTTCTACACCTGCTTCTA CAG 3’ | 56 | 259 | |
| R; 5’ TTATATTCAGATAGCACACATTGA 3’ | ||||
| 29 | F; 5’ CCATTTCCTTTCAACCAGAAAGGG 3’ | 52 | 218 | |
| R; 5’ CATACATCTGCATG CCCTCCTGG 3’ | ||||
| EMILIN2 | 4 | F; 5’ TTGGTCAACAGATCAAGACATTGGACC 3’ | 66.7 | 300 |
| R; 5’ GAACGCTCCCCAGACGGTCTTCCAGAG 3’ | ||||
| DLGAP1 | 2 | F; 5’ GTCCACGGCATCCAAGCAGACCAC 3’ | 67.8 | 223 |
| R; 5’ TGTTTTCCTCAGGGACAGGCG 3’F | ||||
| 4 | F; 5’ CTGGAGTCGCAGGCCGTGGAAGCG 3’ | 67.8 | 300 | |
| R; 5’ ACATGGGTGGTATCTTGTTCCTGG 3’ |
DNA sequencing
PCR products were purified by sodium iodide method. All the Purified PCR products were sequenced, using the automated DNA sequencer ABI prism 310 (Applied Bio systems, USA) involving Sanger di deoxy method. DNA sequences of the amplicons were obtained in FASTA and PDF formats. The FASTA files were analyzed using ClustalX version 2 software (European Bioinformatics Institute, Cambridgeshire, UK) for sequence alignment and by ChromasPro version 1.49 beta2 software (Technelysiumpty Ltd, Australia) for the detailed inspection of the chromatograms individually.
Computational prediction tools
3D structure of protein in pdb format was predicted by an automated server (I-TASSER; zhang.bioinformatics.ku.edu/I-TASSER) [27]. Swiss PDB Viewer computed free energy of predicted 3D structures [28]. We also used Sorting Intolerant From Tolerant (SIFT) version 2, a program which predicts the tolerant and deleterious substitutions within a given sequence. Possible impact of an amino acid substitution on the structure and function of a human protein was predicted using PolyPhen-2 (Polymorphism Phenotyping version 2) [29].
Genetic association study and haplotyping
Adjusted odds ratios (ORs) were assessed using co-dominant, dominant, recessive and over-dominant inheritance models. The inheritance model with the lowest AIC (Akaike information criterion) is considered appropriate for the individual SNP data. Haplotype analysis for haplotypes with frequencies >1% was conducted using HAPSTAT 3.0 software and the risks were compared to the reference haplotype (Most common haplotype in control group). Haplotype frequencies were estimated from the genotyping data after stratification by gender and age.
Statistical analysis
For each polymorphism, the allelic and genotypic frequencies of cases and controls were compared by χ2-test with two degrees of freedom. The association of MYOM1, EMILIN2 & DLGAP1 genotypes with risk of disease was calculated by employing the logistic regression analysis. The relative risk was estimated by odds ratios (OR) and 95% confidence intervals (95% CI), P≤0.05 was considered as significant. Statistical analysis was done using SPSS 23.0 statistical package (SPSS Inc., Chicago IL, USA).
Results
Patient characteristics
Socio-demographic and clinicopathological parameters of cases and controls are revealed in Table 2. All cases and controls were matched as per their age, gender and smoking status. The calculated mean age of the high Myopia patients and control groups were 35.6±6.1 and 34.04±5.3 respectively. 61% (137 of 224) of patients were ≤30 of age and compared to controls where 59% (130 of 220) were ≤30 years old. Maximum number of cases and controls were passive smokers (62.5% vs 61.5%). 64.3% (144 of 224) of patients were having family history of high Myopia (Table 2).
Table 2.
Demographic and clinicopathological characteristics of Cases and controls enrolled for the study
| Characteristics | Cases N = 224 (%) | Controls N = 220 (%) | P Value |
|---|---|---|---|
| Age | |||
| ≤30 years | 137 (61.0) | 130 (59.0) | |
| >30 years | 87 (39.0) | 90 (41.0) | 0.3 |
| Gender | |||
| Male | 130 (58.0) | 140 (63.6) | |
| Female | 94 (42.0) | 80 (36.4) | 0.1 |
| Smoking Status | |||
| Non-smoker | 40 (17.8) | 45 (20.4) | |
| Passive smoker | 140 (62.5) | 135 (61.5) | 0.5 |
| Active smoker | 44 (19.7) | 40 (18.1) | |
| Occupation | |||
| Students | 80 (35.7) | 50 (22.7) | |
| Near workers | 100 (44.6) | 80 (36.3) | ≤0.05 |
| Others | 44 (19.7) | 90 (41.0) | |
| Family history | |||
| No | 80 (35.7) | - | - |
| Yes | 144 (64.3) | - | |
| Degree of Myopia | |||
| <-6 D | 124 (55.3) | - | - |
| ≥-6 D | 100 (44.7) | - |
D; Diopters.
Sequence analysis of MYP2 loci genes
This study detects sequence variations in MYOM1, EMILIN2 & DLGAP1 in a pure ethnic Kashmiri population. Mutational screening discovered a total of 8 polymorphic variations (05 in exons and 03 in introns) as shown in Table 3. MYOM1 showed five polymorphic variations; two in coding region and three intronic, EMILIN2 showed one polymorphic variation while as DLGAP1 showed two polymorphic variants (Table 3). MYOM1 Gly333Gly, MYOM1 c.1022+23, EMILIN2 Asn451Asn SNPs are significantly associated with the decreased risk while as MYOM1 Gly341Ala, MYOM1 c.3418+44, MYOM1 c.3418+65, and DLGAP1 Pro517Pro SNPs are associated with the increased risk of high myopia in study subjects (P≤0.05).
Table 3.
Variations detected in DLGAP1, EMILIN2 & MYOM1 genes of HM patients
| Gene | Wild nucleotide | SNP | rs number | Codon change (Amino acid change) | Codon Position |
|---|---|---|---|---|---|
| MYOM1 | |||||
| NM_003803 | G | G/A | rs2230162 | GGG to GGA (Gly>Gly) | Gly333Gly |
| NP_003794 | G | G/C | rs8099021 | GGA to GCA (Gly>Ala) | Gly341Ala |
| MYOM1 | |||||
| NM_003803.3 | G | G /A | rs17177479 | - | Intronic c.1022+23 G>A |
| G | G/T | rs55779127 | - | Intronic c.3418+44 G>T | |
| C | C/G | rs8096379 | - | Intronic c..3418+65 C>G | |
| EMILIN2 | |||||
| NM_032048 | T | T/C | rs3810067 | AAT to AAC (Asn>Asn) | Asn451Asn |
| NP_114437 | |||||
| DLGAP1 | |||||
| NM_004746 | G | G/A | Novel | GAG to AAG (Glu>Lys) | Glu507Lys |
| NP_004737 | G | G/A | rs3745051 | CCG to CCA (Pro>Pro) | Pro517Pro |
Glu; Glutamic acid, Lys; Lysine, Pro; Proline, Asn; Asparagine, Gly; Glysine, Ala; Alanine.
Genotype and allele frequencies of MYP2 locus gene polymorphisms in cases and controls is shown in Table 4. In case of MYOM1 Gly333Gly (G>A; rs2230162) SNP, the frequency of variant genotype (GA+AA) in cases was 25% (56 of 224) as compared to controls (92 of 220; 42%) (P≤0.05). The partial electrophoretogram depicting change from G to A is shown in Figure 1A. Although the results suggest the possible association of homozygous variant state (AA) with disease phenotype but the combined effect of variant genotype (GA+AA) plays a protective role in predisposing an individual to risk of high Myopia. In case of MYOM1 Gly341Ala (G>C; rs8099021) SNP, homozygous wild genotype (GG) was absent in both cases and controls. The partial electrophoretogram depicting change from G to C is shown in Figure 1B. When variant genotype (GC+CC) was compared between cases and controls significance was not noted (P>0.05) however on comparing individual alleles (G vs C) statistical significance was noted (P≤0.05) with variant allele (C) conferring 2.3 times more risk of contracting high myopia. In case of MYOM1 c.1022+23 (G>A; rs17177479) SNP, the frequency of variant genotype (GA+AA) is almost double in controls when compared with cases (24% vs 13%) (P≤0.05). Figure 1C depicts the sequence variation from G to A. Although homozygous variant allele (AA) is present in 13% (28 of 224) of cases but the overall protective role of SNP is mediated by higher frequency of heterozygous genotype (GA) in controls (24%; 52 of 220). Among all studied SNPs, MYOM1 c.3418+44 (G>T; rs55779127) and MYOM1 c.3418+65 (C>G; rs8096379) SNPs in intronic region pose highest degree of risk to high Myopia as there is total absence of variant genotype (GT & TT; CC & GG) in controls and the effect is mediated by heterozygous genotypes (GT; 87%) being present only in cases (P≤0.05). Figure 1D, 1E shows the nucleotide change from G to T and C to T respectively. In case of, EMILIN2 Asn451Asn (T>C; rs3810067), heterozygotes (TC) are absent in both cases and controls. The nucleotide change from T to C is shown in Figure 2. The frequency of variant genotype (TC+CC) is higher in controls proving this SNP to be protective against disease risk and the effect is mediated by homozygotes (CC) only (P≤0.05). In case of DLGAP1 Pro517Pro (G>A; rs3745051) SNP, homozygous variant genotype (AA) was absent in both cases and controls and presence of heterozygous genotype (GA; 36%) in cases increases risk to high Myopia to a very high degree (P≤0.05). The nucleotide change from G to A is shown in Figure 3. Although a novel non-synonymous SNP, DLGAP1 Glu507Lys (G>A), was also found in our study but there was only presence of heterozygotes in cases as well as controls rendering this SNP statistically insignificant (P≥0.05; Table 4).
Table 4.
Genotype and allele frequencies of MYP2 locus gene polymorphisms in 224 cases and 220 controls
| Gene/variation | Genotype | Cases N = 224 (%) | Controls N = 220 (%) | OR (95% CI) | P value |
|---|---|---|---|---|---|
| MYOM1 Gly333Gly (G>A; rs2230162) | GG | 168 (75.0) | 128 (58.0) | Ref. (1.00) | |
| GA | 28 (12.5) | 92 (42.0) | 0.2 (0.14-0.37) | <0.0001 | |
| AA | 28 (12.5) | 00 (00.0) | 22.14 (2.9-38.6) | <0.0001 | |
| (GA+AA) | 56 (25.0) | 92 (42.0) | 0.4 (0.3-0.7) | <0.0001 | |
| G | 364 (81.0) | 348 (79.0) | Ref. (1.00) | ||
| A | 84 (19.0) | 92 (21.0) | 0.9 (0.6-1.2) | 0.2 | |
| MYOM1 Gly341Ala (G>C; rs8099021) | GG | 00 (00.0) | 00 (00.0) | Ref. (1.00) | |
| GC | 32 (14.0) | 68 (31.0) | 0.46 (0.02-7.6) | 0.3 | |
| CC | 192 (86.0) | 152 (69.0) | 1.2 (0.07-20.3) | 0.4 | |
| (GC+CC) | 224 (100.0) | 220 (100.0) | 1.01 (0.06-16.3) | 0.5 | |
| G | 32 (7.0) | 68 (16.0) | Ref. (1.00) | ||
| C | 416 (93.0) | 372 (84.0) | 2.3 (1.5-3.7) | <0.0001 | |
| MYOM1 c.1022+23 (G>A; rs17177479) | GG | 196 (87.0) | 168 (76.0) | Ref. (1.00) | |
| GA | 00 (00.0) | 52 (24.0) | 6.7 (1.8-45.1) | <0.0001 | |
| AA | 28 (13.0) | 00 (00.0) | 24.8 (3.3-184.2) | <0.0001 | |
| (GA+AA) | 28 (13.0) | 52 (24.0) | 0.46 (0.27-0.76) | <0.0001 | |
| G | 392 (87.0) | 398 (88.0) | Ref. (1.00) | ||
| A | 56 (13.0) | 52 (12.0) | 1.09 (0.7-1.6) | 0.3 | |
| MYOM1 c.3418+44 (G>T; rs55779127) | GG | 30 (13.0) | 220 (100.0) | Ref. (1.00) | |
| GT | 194 (87.0) | 00 (00.0) | 1390 (188-10280) | <0.0001 | |
| TT | 00 (00.0) | 00 (00.0) | 7.1 (0.4-11.6) | 0.12 | |
| (GT+TT) | 194 (87.0) | 00 (00.0) | 1390 (188-10280) | <0.0001 | |
| G | 254 (57.0) | 440 (100.0) | Ref. (1.00) | ||
| T | 194 (43.0) | 00 (00.0) | 33.7 (4.6-42.4) | <0.0001 | |
| MYOM1 c.3418+65 (C>G; rs8096379) | CC | 30 (13.0) | 220 (100.0) | Ref. (1.00) | |
| CG | 194 (87.0) | 00 (00.0) | 1390 (188-10280) | <0.0001 | |
| GG | 00 (0.0) | 00 (00.0) | 7.1 (0.4-11.6) | 0.12 | |
| (CG+GG) | 194 (87.0) | 00 (00.0) | 1390 (188-10280) | <0.0001 | |
| C | 254 (57.0) | 440 (100.0) | Ref. (1.00) | ||
| G | 194 (43.0) | 00 (00.0) | 33.7 (4.6-42.4) | <0.0001 | |
| EMILIN2 Asn451Asn (T>C; rs3810067) | TT | 186 (83.0) | 160 (72.0) | Ref. (1.00) | |
| TC | 38 (17.0) | 60 (28.0) | 0.54 (0.34-0.86) | 0.004 | |
| CC | 00 (0.0) | 00 (00.0) | 0.86 (0.05-13.8) | 0.4 | |
| (TC+CC) | 38 (17.0) | 60 (28.0) | 0.54 (0.34-0.86) | 0.004 | |
| T | 410 (91.0) | 380 (86.0) | Ref. (1.00) | ||
| C | 38 (9.0) | 28 (14.0) | 1.25 (0.75-2.09) | 0.2 | |
| DLGAP1 Glu507Lys (G>A; Novel) | GG | 00 (00.0) | 00 (00.0) | Ref. (1.00) | |
| GA | 224 (100.0) | 220 (100.0) | 1.02 (0.06-16.4) | 0.5 | |
| AA | 00 (00.0) | 00 (00.0) | 1.0 (0.01-50.3) | 0.5 | |
| (GA+AA) | 224 (100.0) | 220 (100.0) | 1.02 (0.06-16.4) | 0.5 | |
| G | 224 (50.0) | 220 (50.0) | Ref. (1.00) | ||
| A | 224 (50.0) | 220 (50.0) | 1.0 (0.7-1.3) | 0.5 | |
| DLGAP1 Pro517Pro (G>A; rs3745051) | GG | 144 (64.0) | 220 (100.0) | Ref. (1.00) | |
| GA | 80 (36.0) | 00 (00.0) | 124.3 (17.1-903.1) | <0.0001 | |
| AA | 00 (00.0) | 00 (00.0) | 1.5 (0.09-24.5) | 0.4 | |
| (GA+AA) | 80 (36.0) | 00 (00.0) | 124.3 (17.1-903.1) | <0.0001 | |
| G | 368 (82.0) | 440 (100.0) | Ref. (1.00) | ||
| A | 80 (18.0) | 00 (00.0) | 96.8 (13.4-698.9) | <0.0001 |
Figure 1.
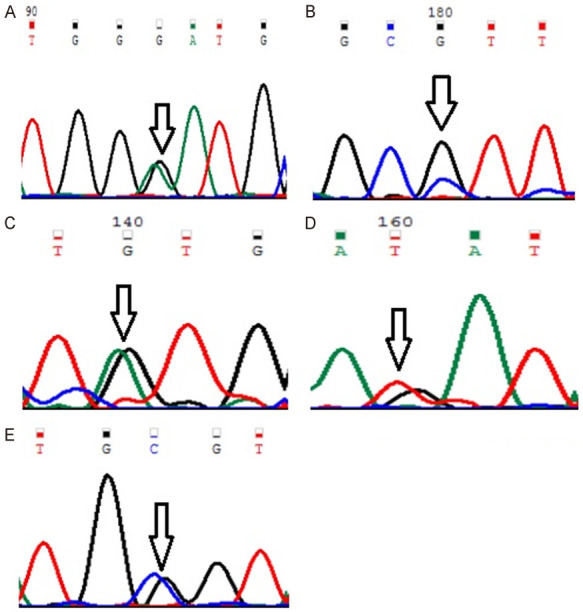
Representative partial chromatograms of affected samples showing sequence variations in MYOM1 (indicated by arrows). (A) Gly333Gly (G>A; rs2230162) (B) Gly341Ala (G>C; rs8099021) (C) c.1022+23 (G>A; rs17177479) (D) c.3418+44 (G>T; rs55779127) and (E) c.3418+65 (C>G; rs8096379).
Figure 2.
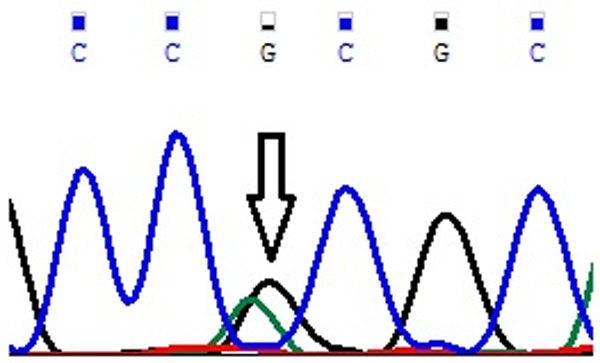
Representative partial chromatogram of affected samples showing sequence variations in EMILIN2 Asn451Asn (T>C; rs3810067) (indicated by arrows).
Figure 3.
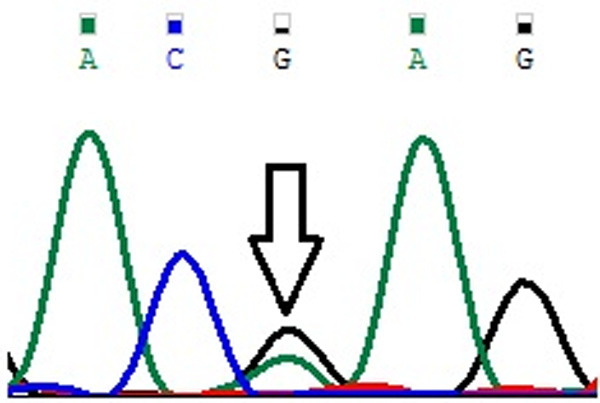
Representative partial chromatograms of affected samples showing sequence variations in DLGAP1 Pro517Pro (G>A; rs3745051) (indicated by arrows).
Stratification analysis of MYP2 loci gene variations and risk of High Myopia
To further assess the effect of MYOM1, EMILIN2 & DLGAP1 genotypes on disease risk with respect to various demographic and clinicopathological parameters of cases and controls, stratification analysis was carried out as shown in Tables 5, 6 and 7 respectively. MYOM1 Gly333Gly SNP showed a statistically significant association with age, gender, smoking status, occupation, family history and degree of myopia respectively with the frequency of variant genotype (GA+AA) higher in males, passive/active smokers, patients with no family history and degree of Myopia ≥-6 D (P≤0.05, Table 5). No significant association of any parameter with MYOM1 Gly341Ala SNP was found (P≥0.05; Table 5). MYOM1 c.1022+23 SNP showed statistical significance with occupation and family history with frequency of variant genotype higher in near workers and study subjects with no family history (P≤0.05, Table 6). Age, occupation, family history and degree of myopia were significantly associated with MYOM1 c.3418+44 SNP and the frequency of variant allele (GA+AA) was higher in study subjects with ≤30 years of age, family history of disease, ≥-6 D myopia and near workers (P≤0.05, Table 6). Statistical significance of MYOM1 c.3418+65 SNP was noted with age, occupation and family history of disease with frequency of variant allele (CG+GG) higher in study subjects with ≤30 years of age, family history of disease and in near workers (P≤0.05, Table 6). In case of EMILIN2 Asn451Asn SNP statistical significance was observed with smoking status, occupation, family history and degree of Myopia with the frequency of variant allele higher in passive smokers, near workers and in subjects with no family history and degree of Myopia ≥-6 D (P≤0.05, Table 7). DLGAP1 Pro517Pro SNP was significantly correlated with age, smoking status, family history and degree of Myopia with the frequency of variant allele (GA+AA) higher in study subjects with ≤30 years of age, family history of disease, <-6 D Myopia and non-smokers (P≤0.05, Table 7).
Table 5.
Association of MYOM1 gene alterations with demographic and clinicopathological variables in 444 subjects (224 cases and 220 controls)
| Parameters | MYOM1 Gly333Gly (G>A; rs2230162) | OR (95% CI) | P value | MYOM1 Gly341Ala (G>C; rs8099021) | OR (95% CI) | P value | ||
|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||
| GG | GA+AA | GG | GC+CC | |||||
| Age | ||||||||
| ≤30 years | 189 | 78 | Ref. (1.00) | 00 | 267 | Ref. (1.00) | 0.3 | |
| >30 years | 107 | 71 | 0.6 (0.4-0.9) | 0.01 | 00 | 177 | 1.5 (0.09-24.2) | |
| Gender | ||||||||
| Male | 163 | 107 | Ref. (1.00) | 00 | 270 | Ref. (1.00) | 0.4 | |
| Female | 133 | 41 | 2.1 (1.3-3.2) | 0.0002 | 00 | 174 | 1.5 (0.09-24.9) | |
| Smoking Status | ||||||||
| Non-Smoker | 70 | 15 | Ref. (1.00) | 00 | 85 | Ref. (1.00) | 0.2 | |
| Passive Smoker | 195 | 80 | 0.52 (0.28-0.96) | 0.01 | 00 | 275 | 0.3 (0.01-5.03) | 0.4 |
| Active Smoker | 31 | 53 | 0.12 (0.06-0.25) | <0.0001 | 00 | 84 | 1.01 (0.06-16.4) | |
| Occupation | ||||||||
| Student | 83 | 47 | Ref. (1.00) | 00 | 130 | Ref. (1.00) | ||
| Near Workers | 105 | 75 | 0.80 (0.49-1.26) | 0.1 | 00 | 180 | 0.7 (0.04-11.6) | 0.4 |
| Others | 108 | 26 | 2.3 (1.34-4.12) | 0.001 | 00 | 134 | 0.97 (0.06-15.6) | 0.5 |
| Family History | ||||||||
| No | 48 | 32 | Ref. (1.00) | 00 | 80 | Ref. (1.00) | 0.3 | |
| Yes | 120 | 24 | 3.3 (1.7-6.2) | <0.0001 | 00 | 144 | 0.5 (0.03-8.9) | |
| Degree of Myopia | ||||||||
| <-6 D | 100 | 24 | Ref. (1.00) | 00 | 124 | Ref. (1.00) | 0.4 | |
| ≥-6 D | 68 | 32 | 0.51 (0.27-0.94) | 0.01 | 00 | 100 | 1.2 (0.07-20.03) | |
D; Diopters.
Table 6.
Association of intronic MYOM1 gene alterations with demographic and clinicopathological variables in 444 subjects (224 cases and 220 controls)
| Parameters | MYOM1 c.1022+23 (G>A; rs17177479) | OR (95% CI) | P value | MYOM1 c.3418+44 (G>T; rs55779127) | OR (95%CI) | P value | MYOM1 c.3418+65 (C>G; rs8096379) | OR (95%CI) | P value | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
||||||||||
| GG | GA+AA | GG | GT+TT | CC | CG+GG | |||||||
| Age | ||||||||||||
| ≤30 years | 213 | 54 | Ref. (1.00) | 140 | 127 | Ref. (1.00) | 140 | 127 | Ref. (1.00) | |||
| >30 years | 151 | 26 | 1.4 (0.88-2.4) | 0.06 | 110 | 67 | 1.5 (1.01-2.1) | 0.02 | 110 | 67 | 1.5 (1.01-2.1) | 0.02 |
| Gender | ||||||||||||
| Male | 220 | 50 | Ref. (1.00) | 152 | 118 | Ref. (1.00) | 150 | 120 | Ref. (1.00) | |||
| Female | 144 | 30 | 1.09 (0.66-1.7) | 0.3 | 98 | 76 | 1.0 (0.68-1.4) | 0.5 | 100 | 74 | 1.08 (0.7-1.5) | 0.3 |
| Smoking Status | ||||||||||||
| Non-Smoker | 70 | 15 | Ref. (1.00) | 65 | 20 | Ref. (1.00) | 60 | 25 | Ref. (1.00) | |||
| Passive Smoker | 230 | 45 | 1.09 (0.57-2.0) | 0.3 | 140 | 135 | 0.3 (0.2-0.5) | 0.001 | 145 | 130 | 0.4 (0.2-0.78) | 0.001 |
| Active Smoker | 64 | 20 | 0.6 (0.3-1.4) | 0.16 | 45 | 39 | 0.3 (0.2-0.7) | <0.001 | 45 | 39 | 0.5 (0.2-0.9) | 0.01 |
| Occupation | ||||||||||||
| Student | 115 | 15 | Ref. (1.00) | 55 | 75 | Ref. (1.00) | 55 | 75 | Ref. (1.00) | |||
| Near Workers | 135 | 45 | 0.4 (0.2-0.7) | 0.001 | 85 | 95 | 1.2 (0.7-1.9) | 0.2 | 85 | 95 | 1.2 (0.7-1.9) | 0.1 |
| Others | 114 | 20 | 0.7 (0.3-1.5) | 0.2 | 110 | 24 | 6.2 (3.5-10.7) | <0.001 | 110 | 24 | 6.2 (3.5-10.9) | <0.001 |
| Family History | ||||||||||||
| No | 60 | 20 | Ref. (1.00) | 20 | 60 | Ref. (1.00) | 20 | 60 | Ref. (1.00) | |||
| Yes | 136 | 08 | 5.6 (2.3-13.5) | <0.001 | 10 | 134 | 0.2 (0.09-0.5) | <0.001 | 10 | 134 | 0.2 (0.09-0.5) | <0.001 |
| Degree of Myopia | ||||||||||||
| <-6 D | 106 | 18 | Ref. (1.00) | 22 | 102 | Ref. (1.00) | 20 | 104 | Ref. (1.00) | |||
| ≥-6 D | 90 | 10 | 1.5 (0.6-3.4) | 0.15 | 08 | 92 | 0.4 (0.17-0.9) | 0.01 | 10 | 90 | 0.5 (0.2-1.2) | 0.09 |
D; Diopters.
Table 7.
Association of EMILIN2 and DLGAP1 gene alterations with demographic and clinicopathological variables in 444 subjects (224 cases and 220 controls)
| Parameters | EMILIN2 Asn451Asn (T>C; rs3810067) | OR (95% CI) | P value | DLGAP1 Pro517Pro (G>A; rs3745051) | OR (95%CI) | P value | ||
|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||
| TT | TC+CC | GG | GA+AA | |||||
| Age | ||||||||
| ≤30 years | 204 | 63 | Ref. (1.00) | 207 | 60 | Ref. (1.00) | ||
| >30 years | 142 | 35 | 1.2 (0.78-1.9) | 0.1 | 157 | 20 | 2.2 (1.3-3.9) | <0.001 |
| Gender | ||||||||
| Male | 215 | 55 | Ref. (1.00) | 220 | 50 | Ref. (1.00) | ||
| Female | 131 | 43 | 0.7 (0.5-1.2) | 0.1 | 144 | 30 | 0.86 (0.5-1.4) | 0.2 |
| Smoking Status | ||||||||
| Non-Smoker | 60 | 25 | Ref. (1.00) | 65 | 20 | Ref. (1.00) | ||
| Passive Smoker | 215 | 60 | 1.5 (0.8-2.5) | 0.07 | 235 | 40 | 1.8 (0.9-3.3) | 0.03 |
| Active Smoker | 70 | 13 | 2.2 (1.0-4.8) | 0.01 | 64 | 20 | 0.9 (0.4-2.0) | 0.4 |
| Occupation | ||||||||
| Student | 95 | 35 | Ref. (1.00) | 115 | 15 | Ref. (1.00) | ||
| Near Workers | 135 | 45 | 1.1 (0.6-1.8) | 0.3 | 140 | 40 | 0.4 (0.2-0.8) | 0.007 |
| Others | 116 | 18 | 2.3 (1.2-4.4) | 0.003 | 109 | 25 | 0.5 (0.2-1.1) | 0.06 |
| Family History | ||||||||
| No | 52 | 28 | Ref. (1.00) | 60 | 20 | Ref. (1.00) | ||
| Yes | 134 | 10 | 7.2 (3.2-15.9) | <0.001 | 84 | 60 | 0.4 (0.2-0.8) | 0.006 |
| Degree of Myopia | ||||||||
| <-6 D | 112 | 12 | Ref. (1.00) | 69 | 55 | Ref. (1.00) | ||
| ≥-6 D | 74 | 26 | 0.3 (0.1-0.6) | <0.001 | 75 | 25 | 2.3 (1.3-4.2) | 0.001 |
D; Diopters.
In silico prediction analysis
MYP2 locus genes were modeled by I-TASSER to obtain the PDB structures and predicted analysis (energy calculations) was done using PDB Viewer. In case of MYOM1, wild protein showed higher energy (-9702.442 kJ/mol) compared to mutant (-11496.317 kJ/mol) for codon Gly341Ala variation. PolyPhen Analysis for mutation MYOM1 Gly341Ala is predicted to be benign according to both HumVar and HumDiv datasets. Additionally SIFT analysis predicted the amino-acid substitution as tolerated with the score of 0.94. Provean score of 0.648 also predicts the mutation to be neutral.
Although statistically insignificant, the assessment of the I-TASSER protein structure showed higher energy for mutant DLGAP1 protein (20206.113 kJ/mol) compared to wild DLGAP1 protein (23265.684 kJ/mol). PolyPhen Analysis for mutation DLGAP1 Glu507Lys is predicted to be possibly damaging according to HumDiv dataset and benign according to HumVar dataset. SIFT analysis also predicts the amino acid substitution as damaging with the score of 0.01 and likewise, Provean score of 2.712 also predicts the mutation to be deleterious.
Genetic association studies and haplotyping
Various genetic association models have been applied on MYOM1, EMILIN2 and DLGAP1 gene polymorphisms; details of which are contained in Table 8. MYOM1 Gly333Gly, MYOM1 c.1022+23 and MYOM1 c.3418+44 SNPs follow Overdominant mode of inheritance. MYOM1 Gly341Ala and MYOM1 c.3418+65 SNPs follow Recessive mode while as EMILIN2 Asn451Asn SNP follows Co-dominant mode of inheritance.
Table 8.
Appropriate genetic association models for SNPs in MYOM1, EMILIN2 and DLGAP1 genes with response to HM phenotype (n = 444, adjusted by gender and age)
| Gene/variation | Model | Genotype | Case; n (%)n=224 | Control; n (%) n=220 | P Value | AIC |
|---|---|---|---|---|---|---|
| MYOM1 Gly333Gly (G>A; rs2230162) | Overdominant | G/G+A/A | 196 (87.0) | 128 (58.0) | ||
| G/A | 28 (13.0) | 92 (42.0) | <0.0001 | 1319.44 | ||
| MYOM1 Gly341Ala (G>C; rs8099021) | Recessive | G/G+G/C | 32 (14.0) | 68 (31.0) | ||
| C/C | 192 (86.0) | 152 (69.0) | <0.0001 | 1081.20 | ||
| MYOM1 c.1022+23 (G>A; rs17177479) | Overdominant | G/G+A/A | 224 (100.0) | 168 (76.0) | <0.0001 | 1196.02 |
| G/A | 00 (0.0) | 52 (24.0) | ||||
| MYOM1 c.3418+44 (G>T; rs55779127) | Overdominant | G/G+T/T | 30 (13.0) | 220 (100.0) | <0.0001 | 144488.4 |
| G/T | 194 (87.0) | 00 (0.0) | ||||
| MYOM1 c.3418+65 (C>G; rs8096379) | Recessive | C/C+C/G | 224 (100.0) | 220 (100.0) | ||
| G/G | 00 (0.0) | 00 (0.0) | <0.0001 | 44797.3 | ||
| EMILIN2 Asn451Asn (T>C; rs3810067) | Codominant | T/T | 186 (83.0) | 160 (73.0) | ||
| T/C | 38 (17.0) | 60 (27.0) | <0.0001 | 1087.4 | ||
| C/C | 00 (0.0) | 00 (0.0) | ||||
| DLGAP1 Pro517Pro (G>A; rs3745051) | Recessive | G/G+G/A | 224 (100.0) | 220 (100.0) | ||
| A/A | 00 (0.0) | 00 (0.0) | <0.001 | 514.14 |
The haplotype frequency estimation and its association with disease phenotype was done after adjustment by gender and age and the most common haplotype was taken as the reference group. Table 9 demonstrates MYOM1, EMILIN2 and DLGAP1 haplotype frequencies and its association with the disease.
Table 9.
MYOM1, EMILIN2, DLGAP1 Haplotype frequencies estimation and haplotype association with HM (adjusted by gender and age)
| Haplotype | Controls n=220 | Cases n=224 | Combined |
|---|---|---|---|
| MYOM1 | |||
| 00000 | 0.6209 | 0.4118 | 0.5083 |
| 00010 | 0.0000 | 0.0548 | 0.0273 |
| 00011 | 0.0045 | 0.3093 | 0.1649 |
| 00100 | 0.0109 | 0.0000 | 0.0058 |
| 00101 | 0.0000 | 0.0179 | 0.0082 |
| 01000 | 0.1545 | 0.0000 | 0.0874 |
| 01011 | 0.0000 | 0.0187 | 0.0000 |
| 10000 | 0.1018 | 0.0233 | 0.0640 |
| 10001 | 0.0000 | 0.0057 | 0.0025 |
| 10011 | 0.0000 | 0.0076 | 0.0034 |
| 10100 | 0.1073 | 0.0611 | 0.0841 |
| 10101 | 0.0000 | 0.0223 | 0.0121 |
| 10111 | 0.0000 | 0.0148 | 0.0070 |
| 11000 | 0.0000 | 0.0101 | 0.0000 |
| 11001 | 0.0000 | 0.0090 | 0.0045 |
| 11011 | 0.0000 | 0.0247 | 0.0162 |
| 11100 | 0.0000 | 0.0059 | 0.0025 |
| 11111 | 0.0000 | 0.0030 | 0.0020 |
| EMILIN2 | |||
| 0 | 0.1308 | 0.0957 | 0.1126 |
| 1 | 0.8692 | 0.9043 | 0.8874 |
| DLGAP1 | |||
| 00 | 0.5000 | 0.4128 | 0.4572 |
| 01 | 0.0000 | 0.0872 | 0.0428 |
| 10 | 0.5000 | 0.4128 | 0.4572 |
| 11 | 0.0000 | 0.0872 | 0.0428 |
Discussion
MYP2 is a candidate locus of the non-syndromic autosomal dominant HM, first identified by Young et al. who performed a genome-wide linkage investigation for MYP2; localized to chromosome 18p11.31 [30]. Candidate genes that map to MYP2 locus show expression in eye tissues and are vital for fundamental organization and preservation of connective tissue function [3]. The genes sheltering in this locus are also expressed in retina and impact the growth of sclera [31]. Experimental work indicates that neural control machinery is partly restricted to the retina itself, but how retinal signals directly regulate the growth of the outer coats of the eye is presently unknown [21].
Although some case control studies have been conducted relating MYP2 locus SNPs and HM, but tiny evidence has been collected from these studies owing mainly to insufficient sample size, lack of replica studies and heterogeneous nature of the populations studied. Previously MYP2 locus candidate genes like MYOM1, EMILIN2, TGIF, DLGAP1, CLUL1, LPIN2, MRCL3, MRLC2 and ZFP161 have been screened for polymorphic variations [21,26,32]. Population from Kashmir represents a homogeneous cohort of common ethnicity and provided an opportunity to revalidate the significance of MYP2 locus candidate gene variations (if any) for defining their relevance in the pathogenesis of the disease. Numerous studies indicate the association of MYP2 locus SNPs with HM which is in line with the results reported from our study [26,32]. In our study no association of DLGAP1 SNP with the risk of HM was found which is in coherence with previous studies [21].
MYOM1 is a structural constituent of cytoskeleton thought to integrate the thin and thick filaments and confer elasticity to the M-band of sarcomere in striated muscle [33]. In this study, two exonic (Gly333Gl; Gly341Ala) and 3 intronic (c.1022+23; c.3418+44; c.3418+65) polymorphic variations of MYOM1 gene were observed to be significantly associated with HM (P≤0.05, Table 4). MYOM1 intronic SNPs are reported to fall outside splice sites and outside promoter regions hence do not affect splicing [34].
EMILIN2 confers elasticity to the extracellular matrix [35]. Broadly expressed in connective tissues with cell adhesion promoting functions, it is abundant in blood vessels, skin, heart, lung, kidney, and cornea suggesting its central role in the process of elastogenesis in association with other extracellular matrix constituents [36]. EMILIN2 plays an important role in scleral wall elasticity seen in HM with elongated axial lengths [23]. Previously, EMILIN2 Asn451Asn SNP has not been associated with risk of HM [21] which is in contradiction with our study wherein we have found a significant association of disease with EMILIN2 Asn451Asn SNP.
DLGAP1 is a member of the PSD95 domain containing family of molecules that are collectively known as “chapsyns” for their function as channel associated proteins. It is known to be highly enriched in synaptosomal preparations of the brain, and is present in the post synaptic density [37]. Scavello et al. [21] have shown the expression of this gene in the retina of eye and have further proposed its role in regulating eye growth. The novel Glu507Lys SNP in DLGAP1 observed in our study group is apparently population specific and does not segregate with the disease phenotype (P=0.5) while as an additional documented Pro517Pro SNP appeared to associate significantly with the risk of HM (P<0.0001). Computational analysis and SIFT suggest DLGAP1 Glu507Lys polymorphism as damaging. DLGAP1 Glu507Lys SNP observed in our study must have a potentially significant implication among species because of the fact that sequences of different species like Homo sapiens, Oryctolaguscuniculus & Rattusnorvegicus were found to be completely conserved with respect to observed variation when aligned, whereas in Mus musculus G is replaced by A (G>A) which is a conserved variation in terms of protein coding, as GAA and GAG both code for same amino acid proving that the region has even been preferably conserved during evolution in terms of amino acid sequence [37]. None of the polymorphisms in DLGAP1 gene have been segregated with risk of HM as per previous studies [21].
When stratified with respect to demographic and clinicopathological characteristics of HM patients, majority of SNPs were significantly associated with occupation or family history (P≤0.05). Relevance of genetic factors in HM has been substantiated by various twin and familial studies indicating correlations between refractive error in parents and siblings [38,39]. A detailed assessment of confounding effects and interactions between hereditary and environmental influences in HM by various researchers has shown that near work describes very little of the variance in refractive error compared to parental myopia [40]. In addition, near work exerted no confounding influence on the association between parent and child myopia, indicating that children do not become myopic by adopting parental reading habits. More importantly, there was no significant interaction between parental myopia and near work [45]. Reading has been weakly and equally associated with HM regardless of the number of myopic parents by various studies [41,42] which suggest that children inherit HM as a trait from parents. Multiple familial aggregation studies report a positive correlation between parental myopia susceptibility and myopia in their children, indicating heritable myopia [41-43]. Previous studies reported that children with a family history of myopia on an average had less hyperopia, deeper anterior chambers, and longer vitreous chambers even before becoming myopic [34,39]. In our study, most of MYP2 loci SNPs were significantly associated with smoking (P≤0.05). Cigarette smoke has around 60 cancer-causing compounds, including nitrosamines, aromatic amines and polycyclic aromatic hydrocarbons (PAH) which can form DNA adducts by metabolic activation leading to the mutations in genes and development of various diseases including HM [44]. As per earlier studies, genetic factors interact with cigarette smoke, signifying that diverse risk approximations relate to diverse genetic tendencies [45].
In this study, the screened MYP2 loci SNPs follow Overdominant, Recessive and Co-dominant mode of inheritance. An Over-dominant model assumes that heterozygote has the strongest impact on disease predisposition; Recessive model presumes the highest impact of homozygous mutant genotype while as Co-dominant model hypothesize that homozygous wild, heterozygous, homozygous mutant genotypes are associated with highest, intermediate and lowest risk respectively [46,47].
Conclusion
Our study supports the idea that the MYP2 locus candidate gene polymorphism contributes to the pathogenesis of HM. Since these SNPs appear to change the energy state of protein indicated by in silico analysis, a biological corroboration would be needed to elucidate the actual effect of these changes on the function of these proteins. The identification of MYP2 locus genes will not only provide insight into the molecular basis of this significant eye disease, but will also identify pathways that are involved in eye growth and development. In addition, this information may implicate other genes as possible myopia disease gene candidates. This effort may lead to effective therapies for the severe forms of this potentially blinding eye disease.
Acknowledgements
We are grateful to the patients who have willingly participated in the study. This work was financially supported by Department of Science and Technology, New Delhi, by grants to SR under Young Women Scientist scheme (Project No: -SR/WOS-A/LS-232/2007).
The study was performed with informed consent and following all the guidelines for experimental investigations required by the Institutional Review Board or Ethics Committee of which all authors are affiliated.
Disclosure of conflict of interest
None.
References
- 1.Hsu SL, Chang CH, Lai YH, Wen MH, Cheng KC, Ho CK. Refractive status of mountain aborigine School children in southern Taiwan. Kaohsiung J Med Sci. 2008;24:120–125. doi: 10.1016/S1607-551X(08)70139-6. [DOI] [PubMed] [Google Scholar]
- 2.Kim EC, Morgan IG, Kakizaki H, Kang S, Jee D. Prevalence and risk factors for refractive errors: Korean national health and nutrition examination survey 2008-2011. PLoS One. 2013;8:e80361. doi: 10.1371/journal.pone.0080361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen M, Wu A, Zhang L, Wang W, Chen X, Yu X, Wang K. The increasing prevalence of myopia and high myopia among high school students in Fenghua city, eastern China: a 15-year population-based survey. BMC Ophthalmol. 2018;18:159–170. doi: 10.1186/s12886-018-0829-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pan CW, Zheng YF, Anuar AR, Chew M, Gazzard G, Aung T, Cheng CY, Wong TY, Saw SM. Prevalence of refractive errors in a multiethnic Asian population: the Singapore epidemiology of eye disease study. Invest Ophthalmol Vis Sci. 2013;54:2590–2598. doi: 10.1167/iovs.13-11725. [DOI] [PubMed] [Google Scholar]
- 5.Sawada A, Tomidokoro A, Araie M, Iwase A, Yamamoto T Tajimi Study Group. Refractive errors in an elderly Japanese population: the Tajimi study. Ophthalmology. 2008;115:363–370. e3. doi: 10.1016/j.ophtha.2007.03.075. [DOI] [PubMed] [Google Scholar]
- 6.Yao Y, Shao J, Sun W, Zhu J, Hong Fu D, Guan H, Liu Q. Prevalence of blindness and causes of visual impairment among adults aged 50 years or above in southern Jiangsu Province of China. Pak J Med Sci. 2013;29:1203–1207. doi: 10.12669/pjms.295.3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Asaminew T, Gelaw Y, Bekele S, Solomon B. Retinal detachment in southwest ethiopia: a hospital based prospective study. PLoS One. 2013;8:e75693. doi: 10.1371/journal.pone.0075693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koh VT, Nah GK, Chang L, Yang AH, Lin ST, Ohno-Matsui K, Wong TY, Saw SM. Pathologic changes in highly myopic eyes of young males in Singapore. Ann Acad Med Singap. 2013;42:216–224. [PubMed] [Google Scholar]
- 9.Detry-Morel M. Is Myopia a risk factor for glaucoma? J Fr Ophtalmol. 2011;34:392–395. doi: 10.1016/j.jfo.2011.03.009. [DOI] [PubMed] [Google Scholar]
- 10.Javitt JC, Chiang YP. The socio economic aspects of laser refractive surgery. Arch Ophthal. 1994;112:1526–1530. doi: 10.1001/archopht.1994.01090240032022. [DOI] [PubMed] [Google Scholar]
- 11.Flitcroft DI. The complex interactions of retinal, optical and environmental factors in myopia aetiology. Prog Retin Eye Res. 2012;31:622–660. doi: 10.1016/j.preteyeres.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 12.Jones LA, Sinnott LT, Mutti DO, Mitchell GL, Moeschberger ML, Zadnik K. Parental history of myopia, sports and outdoor activities, and future myopia. Invest Ophthalmol Vis Sci. 2007;48:3524–3532. doi: 10.1167/iovs.06-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang WC, Yap MK, Yip SP. A review of current approaches to identifying human genes involved in myopia. Clin Exp Optom. 2008;91:4–22. doi: 10.1111/j.1444-0938.2007.00181.x. [DOI] [PubMed] [Google Scholar]
- 14.Zadnik K, Satariano WA, Mutti DO, Sholtz RI, Adams AJ. The effect of parental history of myopia on children’s eye size. J Am Med Assoc. 1994;271:1323–1327. [PubMed] [Google Scholar]
- 15.Goldschmidt E, Fledelius HC, Erlin Larsen F. Clinical features in high myopia. In: Fledelius HC, Alsbirk PH, Goldschmidt E, editors. Third International Conference on Myopia Copenhagen, 1980. Documenta Ophthalmologica Proceedings Series. 1981;28:987–1009. [Google Scholar]
- 16.Young TL, Ronan SM, Drahozal LA, Wildenberg SC, Alvear AB, Oetting WS, Atwood LD, Wilkin DJ, King RA. Evidence that a locus for familial high myopia maps to chromosome 18p. Am J Hum Genet. 1997;63:109–119. doi: 10.1086/301907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Young TL, Atwood LD, Ronan SM, Dewan AT, Alvear AB, Peterson J, Holleschau A, King RA. Further refinement of the MYP2 locus for autosomal dominant high myopia by linkage disequilibrium analysis. Ophthalmic Genet. 2001;22:69–75. doi: 10.1076/opge.22.2.69.2233. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Q, Guo X, Xiao X, Jia X, Li S, Hejtmanciket JF. Novel locus for X linked recessive high myopia maps to Xq23-q25 but outside MYP1. J Med Genet. 2006;43:e20. doi: 10.1136/jmg.2005.037853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Verhoeven VJ. Genome wide meta-analyses of multi-ancestry cohorts identify multiple new susceptibility loci for refractive error and Myopia. Nat Genet. 2013;45:314–318. doi: 10.1038/ng.2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamane T, Mok J, Oka A, Okada E, Nishizaki R, Meguro A, Yonemoto J, Kulski JK, Ohno S, Inoko H, Mizuki N. Lack of association with high myopia and the MYP2 locus in the Japanese population by high resolution microsatellite analysis on chromosome 18. Clin Ophthalmol. 2007;1:311–316. [PMC free article] [PubMed] [Google Scholar]
- 21.Scavello GS, Paluru PC, Zhou J, White PS, Rappaport EF, Young TL. Genomic structure and organization of the high grade Myopia-2 locus (MYP2) critical region: mutation screening of 9 positional candidate genes. Mol Vis. 2005;11:97–110. [PubMed] [Google Scholar]
- 22.Kusakari T, Sato T, Tokoro T. Visual deprivation stimulates the exchange of the fibrous sclera into the cartilaginous sclera in chicks. Exp Eye Res. 2001;73:533–546. doi: 10.1006/exer.2001.1064. [DOI] [PubMed] [Google Scholar]
- 23.Mutti DO, Cooper ME, O’Brien S, Jones LA, Marazita ML, Murray JC, Zadnik K. Candidate gene and locus analysis of myopia. Mol Vis. 2007;13:1012–1019. [PMC free article] [PubMed] [Google Scholar]
- 24.Metlapally R, Michaelides M, Bulusu A, Li YJ, Schwartz M, Rosenberg T, Hunt DM, Moore AT, Züchner S, Rickman CB, Young TL. Evaluation of the X-linked high-grade Myopia locus (MYP1) with cone dysfunction and color vision deficiencies. Invest Ophthalmol Vis Sci. 2009;50:1552–1558. doi: 10.1167/iovs.08-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen JH, Chen H, Huang S, Lin J, Zheng Y, Xie M, Lin W, Pang CP, Zhang M. Endo-phenotyping reveals differential phenotype-genotype correlations between myopia-associated polymorphisms and eye biometric parameters. Mol Vis. 2012;18:765–778. [PMC free article] [PubMed] [Google Scholar]
- 26.Heath S, Robledo R, Beggs W, Feola G, Parodo C, Rinaldi A, Contu L, Dana D, Stambolian D, Siniscalco M. A novel approach to search for identity by descent in small samples of patient and controls from the same Mendelian breeding unit: a pilot study on myopia. Hum Hered. 2001;52:183–90. doi: 10.1159/000053375. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y. Template-based modeling and free modeling by I-TASSER in CASP7. Proteins. 2007;8:108–117. doi: 10.1002/prot.21702. [DOI] [PubMed] [Google Scholar]
- 28.Comeau SR, Gatchell DW, Vajda S, Camacho CJ. ClusPro: an automated docking and discrimination method for the prediction of protein complexes. Bioinformatics. 2004;20:45–50. doi: 10.1093/bioinformatics/btg371. [DOI] [PubMed] [Google Scholar]
- 29.Choi Y, Sims GE, Murphy S, Miller JR, Chan AP. Predicting the functional effect of amino acid substitutions and indels. PLoS One. 2012;7:e46688. doi: 10.1371/journal.pone.0046688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamane T, Mok J, Oka A, Okada E, Nishizaki R, Meguro A, Yonemoto J, Kulski JK, Ohno S, Inoko H, Mizuki N. Lack of association with high myopia and the MYP2 locus in the Japanese population by high resolution microsatellite analysis on chromosome 18. Clin Ophthalmol. 2007;1:311–316. [PMC free article] [PubMed] [Google Scholar]
- 31.Wallman J. Retinal control of eye growth and refraction. Prog Retin Eye Res. 1993;12:134–153. [Google Scholar]
- 32.Paget S, Julia S, Vitezica ZG, Soler V, Malecaze F, Calvas P. Linkage analysis of high myopia susceptibility locus in 26 families. Mol Vis. 2008;14:2566–2574. [PMC free article] [PubMed] [Google Scholar]
- 33.Wang K. Sarcomere-associated cytoskeletal lattices in striated muscle. Review and hypothesis. Cell Muscle Motil. 1985;6:315–369. doi: 10.1007/978-1-4757-4723-2_10. [DOI] [PubMed] [Google Scholar]
- 34.Terri LY. The molecular genetics of human myopia: an update. Optom Vis Sci. 2009;86:E8–E22. doi: 10.1097/OPX.0b013e3181940655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Doliana R, Bot S, Mungiguerra G, Canton A, Cilli SP, Colombatti A. Isolation and characterization of EMILIN-2, a new component of the growing EMILINs family and a member of the EMI domain-containing superfamily. Biol Chem. 2001;276:12003–12011. doi: 10.1074/jbc.M011591200. [DOI] [PubMed] [Google Scholar]
- 36.Colombatti A. The elastin associated glycoprotein gp115. Synthesis and secretion by chick cells in culture. J Biol Chem. 1988;263:17534–17540. [PubMed] [Google Scholar]
- 37.Andreas HR, Hanne BR, Asli S. The DLGAP family: neuronal expression, function and role in brain disorders. Mol Brain. 2017;10:43–49. doi: 10.1186/s13041-017-0324-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Teikari JM, O’Donnell J, Kaprio J, Koskenvuo M. Impact of heredity in myopia. Hum Hered. 1991;41:151–156. doi: 10.1159/000153994. [DOI] [PubMed] [Google Scholar]
- 39.Wojciechowski R. Nature and nurture: the complex genetics of Myopia and refractive error. Clin Genet. 2011;79:301–320. doi: 10.1111/j.1399-0004.2010.01592.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mutti DO, Mitchell GL, Moeschberger ML, Jones LA, Zadnik K. Parental myopia, near work, school achievement, and children’s refractive error. Invest. Ophthalmol Vis Sci. 2002;43:3633–3640. [PubMed] [Google Scholar]
- 41.Goss DA, Jackson TW. Clinical findings before the onset of myopia in youth: 4. parental history of myopia. Optom Vis Sci. 1996;73:279–282. doi: 10.1097/00006324-199604000-00010. [DOI] [PubMed] [Google Scholar]
- 42.Pacella R, McLellan J, Grice K, Del Bono EA, Wiggs JL, Gwiazda JE. Role of genetic factors in the etiology of juvenile-onset myopia based on a longitudinal study of refractive error. Optom Vis Sci. 1999;76:381–386. doi: 10.1097/00006324-199906000-00017. [DOI] [PubMed] [Google Scholar]
- 43.Zadnik K. The Glenn A. Fry Award Lecture. Myopia development in childhood. Optom Vis Sci. 1995;74:603–608. [PubMed] [Google Scholar]
- 44.Schaal C, Chellappan SP. Nicotine-mediated cell proliferation and tumor progression in smoking-related cancers. Mol Cancer Res. 2014;12:14–23. doi: 10.1158/1541-7786.MCR-13-0541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Raimondi S, Botteri E, Iodice S, Lowenfels AB, Maisonneuve P. Gene-smoking interaction on colorectal adenoma and cancer risk: review and meta-analysis. Mutat Res. 2009;670:6–14. doi: 10.1016/j.mrfmmm.2009.06.013. [DOI] [PubMed] [Google Scholar]
- 46.Thakkinstian A, McElduff P, D’Este C, Duffy D, Attia J. A method for meta-analysis of molecular association studies. Stat Med. 2005;24:1291–1306. doi: 10.1002/sim.2010. [DOI] [PubMed] [Google Scholar]
- 47.Attia J, Thakkinstian A, D’Este C. Meta-analyses of molecular association studies: methodologic lessons for genetic epidemiology. J Clin Epidemiol. 2003;56:297–303. doi: 10.1016/s0895-4356(03)00011-8. [DOI] [PubMed] [Google Scholar]


