Abstract
Coronavirus disease 2019 (COVID‐19), caused by severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2), continues to spread globally despite the worldwide implementation of preventive measures to combat the disease. Although most COVID‐19 cases are characterised by a mild, self‐limiting disease course, a considerable subset of patients develop a more severe condition, varying from pneumonia and acute respiratory distress syndrome (ARDS) to multi‐organ failure (MOF). Progression of COVID‐19 is thought to occur as a result of a complex interplay between multiple pathophysiological mechanisms, all of which may orchestrate SARS‐CoV‐2 infection and contribute to organ‐specific tissue damage. In this respect, dissecting currently available knowledge of COVID‐19 immunopathogenesis is crucially important, not only to improve our understanding of its pathophysiology but also to fuel the rationale of both novel and repurposed treatment modalities. Various immune‐mediated pathways during SARS‐CoV‐2 infection are relevant in this context, which relate to innate immunity, adaptive immunity, and autoimmunity. Pathological findings in tissue specimens of patients with COVID‐19 provide valuable information with regard to our understanding of pathophysiology as well as the development of evidence‐based treatment regimens. This review provides an updated overview of the main pathological changes observed in COVID‐19 within the most commonly affected organ systems, with special emphasis on immunopathology. Current management strategies for COVID‐19 include supportive care and the use of repurposed or symptomatic drugs, such as dexamethasone, remdesivir, and anticoagulants. Ultimately, prevention is key to combat COVID‐19, and this requires appropriate measures to attenuate its spread and, above all, the development and implementation of effective vaccines. © 2021 The Authors. The Journal of Pathology published by John Wiley & Sons, Ltd. on behalf of The Pathological Society of Great Britain and Ireland.
Keywords: coronavirus disease 2019 (COVID‐19), severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2), acute respiratory distress syndrome (ARDS), pathology, immunopathology, pathophysiology, diffuse alveolar damage (DAD), treatment, angiotensin‐converting enzyme 2 (ACE2), autoimmunity
Introduction
The outbreak of coronavirus disease 2019 (COVID‐19), caused by severe acute respiratory syndrome corona‐virus 2 (SARS‐CoV‐2), continues to spread globally despite the extensive implementation of preventive health measures. As of 24 January 2021, the pandemic has accumulated over 98 million confirmed cases, including more than 2.1 million deaths [1]. While most SARS‐CoV‐2 infections cause mild, self‐limiting upper respiratory complaints, a notable subset of patients require hospitalisation upon progression to severe disease, varying from lower respiratory tract infection to acute respiratory distress syndrome (ARDS) and multi‐organ failure (MOF) [2]. This is accompanied by abnormal laboratory parameters, including lymphopaenia and elevated levels of C‐reactive protein (CRP), D‐dimers, ferritin, and lactate dehydrogenase (LDH) [3]. Individuals particularly at risk for severe disease include the elderly, males, and those with comorbidities such as cardiovascular disease, diabetes, obesity, chronic respiratory disease, and immunocompromising conditions [4].
The complex pathophysiology of COVID‐19 seems to consist of multiple mechanisms, of which the immunopathological response to SARS‐CoV‐2 infection became a major area of interest. While a well‐regulated immune response is essential in controlling SARS‐CoV‐2 infection, a hyperinflammatory innate immune response along with an inadequate adaptive response may elicit extensive local and systemic tissue injury. In addition, recent reports demonstrate that SARS‐CoV‐2 infection may trigger autoimmune (adaptive immunity) and autoinflammatory (innate immunity) conditions, and several reactive autoantibodies have been found (see below). A profound understanding of the immune responses that lead to recovery instead of worsening disease is essential for developing effective treatment options for COVID‐19. To date, there are no specific treatment regimens to combat the disease, and current management revolves around supportive treatment and the use of (repurposed) drugs, such as dexamethasone, remdesivir, and anticoagulants. As for prevention of COVID‐19, several measures are held in place (e.g. social distancing, quarantine, wearing face masks, frequent handwashing) and emergency use authorisations have been issued for various vaccines, resulting in the implementation of large‐scale vaccination programmes. In this review, we elaborate on the various pathways and systems involved in the pathophysiology and immunopathogenesis of COVID‐19. Furthermore, we discuss the rationale and outcome of the use of several repurposed treatment modalities employed to combat SARS‐CoV‐2.
Proposed pathophysiological mechanisms underlying COVID‐19
COVID‐19 progression is considered to result from a complex interplay of multiple pathophysiological mechanisms, including (1) direct cytopathic effects of SARS‐CoV‐2; (2) angiotensin‐converting enzyme 2 (ACE2) downregulation with subsequent renin–angiotensin–aldosterone system (RAAS) disbalance and decreased inactivation of des‐Arg9‐bradykinin; (3) a dysregulated immune response featuring a ‘cytokine storm’; (4) coagulopathy associated with exocytosis of procoagulatory factors, thrombotic microangiopathy – probably caused by virus‐induced endothelial injury, complement activation, and cytokine effects – and uncontrolled localised and/or systemic ‘immunothrombosis’; and (5) autoimmunity [5, 6]. The relative contributions and interactions of these intertwined mechanisms to COVID‐19‐associated organ dysfunction remain undetermined, with the possibility of immunopathology being a hallmark of severe disease. Herein, we outline the various routes involved in the immunopathology of COVID‐19 based on a narrative review of the literature.
Immunopathology and immune defects
Innate immune response
The innate immune system provides the first line of immunological defence against SARS‐CoV‐2 infection. Following binding of the spike (S) protein to ACE2, SARS‐CoV‐2 may enter the cell either via endocytosis and cathepsin L (CTSL)‐dependent viral escape from the endosome – which is augmented by transmembrane serine protease 2 (TMPRSS2) – or by TMPRSS2‐dependent direct fusion of the viral envelope with the cell membrane (Figure 1) [7, 8, 9]. Unlike SARS‐CoV, cell entry of SARS‐CoV‐2 can also be facilitated by furin and neuropilin‐1 [10, 11]. Upon infection, viral pathogen‐associated molecular patterns (PAMPs) are sensed by endosomal pattern recognition receptors (PRRs), such as Toll‐like receptors (TLRs). This results in intracellular signalling cascades, which involve activation of transcription factors such as nuclear factor‐kappa B (NF‐κB) and interferon regulatory factors (IRFs) [12]. These events ultimately lead to the production of type I interferons (IFNs) and pro‐inflammatory cytokines (Figure 1).
Figure 1.
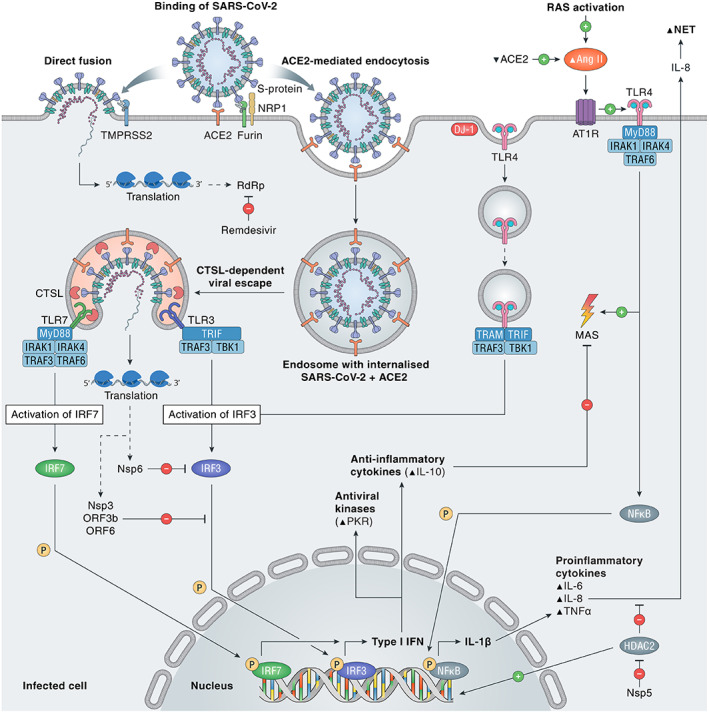
SARS‐CoV‐2 entry and immune activation. SARS‐CoV‐2 needs to bind to ACE2 to enter the cell, either by TMPRSS2‐dependent direct fusion of the viral envelope to the cell membrane or by TMPRSS2‐enhanced endocytosis. Furin and NRP1 can also facilitate viral entry. Viral escape by fusion to the endosomal membrane is CTSL‐dependent, as is activation of TLR7, which is essential in the recognition of single‐stranded RNA viruses and induction of type I IFN via IRF7. Activation of RAS by viral infection, via Ang II and AT1R, induces the TLR4/MyD88/NFκB pathway to increase pro‐inflammatory cytokines IL‐1β, IL‐6, IL‐8, and TNF‐α. After DJ‐1‐induced endocytosis of TLR4, type I IFN, antiviral kinases, and the anti‐inflammatory cytokine IL‐10 are activated. Nsp3 and ‐6 of SARS‐CoV‐2 inhibit IFN activation via the IRF3 pathway. Nsp5 blocks HDAC2, preventing it from decreasing IL‐8, a pro‐inflammatory cytokine with a role in NET formation. MAS is stimulated by the pro‐inflammatory TLR4/MyD88‐dependent pathway and inhibited by the anti‐inflammatory cytokine IL‐10. ACE2, angiotensin‐converting enzyme 2; Ang II, angiotensin II; AT1R, angiotensin II type 1 receptor; CTSL, cathepsin L; HDAC2, histone deacetylase 2; IL, interleukin‐1β/6/8/10; IRAK1/4, interleukin 1 receptor associated kinase 1/4; IRF3/7, interferon regulatory factor 3/7; MAS, macrophage activation syndrome; MyD88, myeloid differentiation primary response 88; NET, neutrophil extracellular trap; NFκB, nuclear factor kappa B; NRP1, neuropilin 1; Nsp3/5/6, nonstructural protein 3/5/6; ORF3b/6, open reading frame 3b/6; P, phosphate; PKR, double‐stranded (ds)RNA‐dependent protein kinase; RAS, renin–angiotensin system; RdRp, RNA‐dependent RNA polymerase; TBK1, TANK binding kinase 1; TLR, toll‐like receptor 3/4/7; TMPRSS2, transmembrane serine protease 2; TNFα, tumour necrosis factor α; TRAF6, tumour necrosis factor receptor‐associated factor 6; TRAM, TRIF‐related adaptor molecule; TRIF, Toll‐IL‐1 receptor domain‐containing adaptor inducing IFN‐β; type I IFN, type I interferon.
An adequate IFN response normally induces an antiviral immune state in infected cells that limits viral replication and induces apoptosis to protect the host from viral dissemination. However, multiple SARS‐CoV‐2 proteins [e.g. open reading frame 6 (ORF6) and ORF3b] have been shown to suppress antiviral type I IFN (IFN‐I) production and signalling [13, 14, 15]. The initial delay of the IFN‐I response is followed by unrestrained viral replication and dissemination in the infected host, thereby promoting an eventual increase of IFN‐I that can exacerbate hyperinflammation in the progression to severe disease [16]. Transcriptome profiling of respiratory cell types showed that SARS‐CoV‐2 infection elicited exceptionally low IFN levels, while inducing a robust pro‐inflammatory cytokine response [17]. In this study, there was also a significant induction of monocyte and neutrophil chemoattractants, in line with the commonly observed lung infiltration of macrophages and polymorphonuclear leukocytes in autopsied COVID‐19 cases [18, 19, 20]. Similar to other viral infections, inter‐individual variability in IFN responses may contribute to the heterogeneity of disease manifestations in COVID‐19. In a Dutch case series including four young male patients from two unrelated families admitted to the intensive care unit (ICU), rare genetic variants in the TLR7 gene were identified and associated with impaired IFN‐I and IFN‐II responses [21]. These findings may partially explain the discouraging results of the TLR7‐antagonising antimalarial drugs hydroxychloroquine and chloroquine [22, 23], whereas the TLR7‐agonist imiquimod may serve as suitable treatment option in some COVID‐19 patients [24]. Recently, two paired studies also reported specific underlying defects of IFN‐I signalling in life‐threatening COVID‐19, including inborn errors of TLR3‐ and IRF7‐dependent IFN‐I immunity [25], and the presence of neutralising autoantibodies against IFN‐I [26]. The latter study demonstrated that neutralising autoantibodies against IFN‐I were present in 10.2% of 987 patients with life‐threatening COVID‐19 pneumonia, ~15‐fold higher than the general population, and showed a male preponderance. These findings provide a rationale for the use of IFN‐based treatments in the early stage of COVID‐19 when only mild symptoms are present. Previously, some concerns were raised that conditions associated with IFN elevations could induce ACE2 expression – thereby promoting SARS‐CoV‐2 entry – since ACE2 was considered to be an interferon‐stimulated gene (ISG) [27]. However, a recent study discovered a novel truncated isoform of ACE2 – so‐called deltaACE2 – which was demonstrated to be an ISG as opposed to ACE2. DeltaACE2 neither acts as a receptor for SARS‐CoV‐2 nor acts as a carboxypeptidase for angiotensin II (Ang II) and des‐Arg9‐bradykinin [28]. Therefore, IFN‐induced deltaACE2 does not promote SARS‐CoV‐2 infection. Importantly, whereas the immune response is initially suppressed, an eventual overactivation of immune responses contributes to hyperinflammation and organ damage [16]. Hypercytokinaemia has been reported in severe COVID‐19 on several occasions. This condition is often referred to as a ‘cytokine storm’, being reminiscent of the macrophage activation syndrome (MAS) [29]. However, the role of a cytokine storm in COVID‐19 pathogenesis has been questioned recently, since the degree of pro‐inflammatory cytokinaemia in COVID‐19 has been shown to be profoundly less than in archetypical conditions associated with MAS [30].
Another key player in the innate immune response is the complement system, acting as a rapid immune surveillance system against invading pathogens, bridging innate and adaptive immunity [31]. In the case of COVID‐19, complement activation is overwhelming, which results in harmful acute and chronic inflammation, endothelial cell dysfunction, and intravascular coagulation [32]. Indeed, strong complement activation has been demonstrated in the systemic circulation [33, 34] as well as locally in various organs of COVID‐19 patients (see below) [34, 35, 36], providing a rationale for the use of complement inhibition as therapy in COVID‐19.
Finally, the innate immune system interacts with coagulation – a process known as ‘immunothrombosis’ – that is thought to be dysregulated in severe COVID‐19, leading to localised and/or systemic coagulopathy [37]. The detection of PAMPs and damage‐associated molecular patterns (DAMPs) by PRR‐expressing monocytes results in their enhanced expression of tissue factor (TF), which in turn activates the extrinsic pathway of coagulation [38]. In addition, activated neutrophils release neutrophil extracellular traps (NETs) – lattices composed of neutrophil‐derived DNA and acetylated histones – which trap and kill invading pathogens but may also induce a strong procoagulant response [38]. NETs can promote activation of the intrinsic coagulation pathway by activation of factor XII but can also bind TF to activate the extrinsic coagulation pathway [38]. NETs have been frequently demonstrated in COVID‐19, sometimes forming aggregates with platelets [39, 40], and they may be drivers of disease severity [41]. An overview of SARS‐CoV‐2 effects on the innate immune response is depicted in Figure 1.
Adaptive immune response
The adaptive immune system plays a pivotal role in SARS‐CoV‐2 clearance via activated cytotoxic T‐cells that destroy infected cells and through B‐cells that produce neutralising antibodies against virus‐specific antigens (Figure 2). A key feature of COVID‐19 is blood lymphopaenia, with reduced numbers of CD4+ T‐cells, CD8+ T‐cells, and B‐cells [42]. Lymphopaenia may be partially explained by an abnormal innate immune response featuring low IFN‐I, considering its essential role in the assembly of viral material for antigen presentation and the subsequent induction of adaptive immunity [43]. Other mechanisms that potentially contribute to COVID‐19‐associated lymphopaenia include direct SARS‐CoV‐2 infection of T‐cells [44], cytokine‐induced apoptosis and pyroptosis of lymphocytes [45], MAS‐related haemophagocytosis, lymphocyte sequestration in the lungs or other organs [46], reduced bone marrow haematopoiesis [47], and virus‐induced tissue damage of lymphatic organs [48, 49]. Notably, both spleen and lymph nodes showed the presence of SARS‐CoV‐2 [49], and pathological alterations (e.g. splenic white pulp atrophy and lymph node structure disruption) have been reported [48, 49], suggesting that direct SARS‐CoV‐2 cytotoxicity in lymphatic organs may impair the adaptive immune response in COVID‐19.
Figure 2.
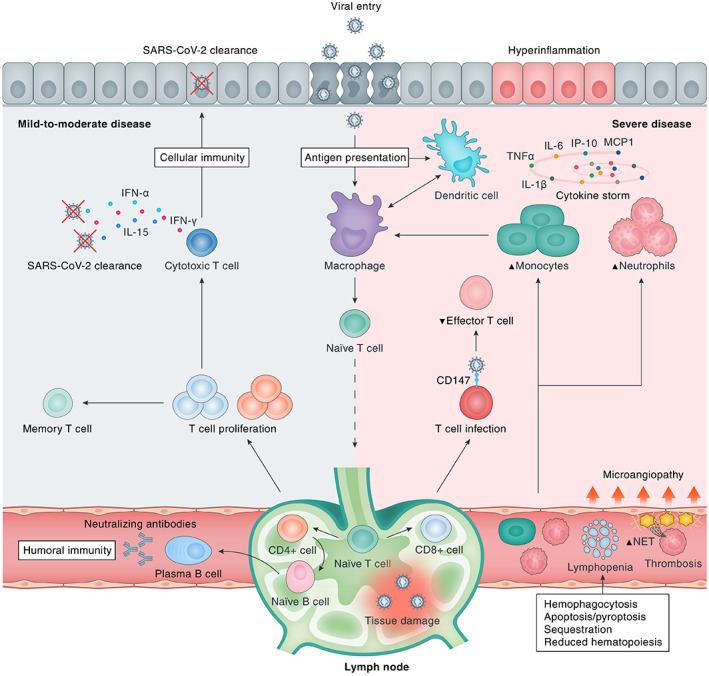
Immunological response to SARS‐CoV‐2 infection. Upon viral cell entry, SARS‐CoV‐2 antigens are processed by the innate immune system through antigen‐presenting cells (APCs), e.g. epithelial cells, macrophages, and/or dendritic cells. Subsequently, the adaptive immune system is activated by migration of APCs to the lymphoid system. Upon antigen recognition, T‐lymphocytes proliferate and differentiate into CD4+ and CD8+ T‐lymphocytes that are responsible for sequential events including cytokine production, activation of naïve B‐lymphocytes, and clearance of infected cells (CD8+ cytotoxic T‐lymphocytes). B‐lymphocytes proliferate and differentiate into plasma cells that produce large numbers of neutralising antibodies, representing humoral immunity. A bulk of cytokines is induced upon SARS‐CoV‐2 infection, most of which contribute to hyperinflammation as constituents of the ‘cytokine storm’ in severe disease (e.g. IL‐6, TNF‐α, IL‐1β, IP‐10, MCP‐1, CSFs, and IL‐17A), whereas others are particularly important for viral clearance (e.g. IL‐15, IFN‐α, IL‐12, IL‐21, and IFN‐γ) in mild‐to‐moderate disease. Severe COVID‐19 is marked by dysfunction of certain immune cells, with relatively increased abundances of neutrophils and monocytes and decreased levels of effector T‐lymphocytes. In addition, multiple downstream pathophysiological processes are activated, including an increased thrombogenic state [microangiopathy, formation of neutrophil extracellular traps (NETs)], haemophagocytosis, reduced haematopoiesis, and increased apoptosis/pyroptosis. CD147, cluster of differentiation 147; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2.
Loss of germinal centre formation in both spleen and lymph nodes provides a potential explanation for suboptimal humoral immunity, with the possibility of reinfection in certain individuals [50, 51]. Nevertheless, the vast majority of COVID‐19 patients with mild‐to‐moderate disease experience a robust adaptive immune response consisting of T‐cells (against S‐protein‐ and nucleoprotein/membrane protein‐derived antigens) and neutralising antibodies (against S‐protein‐derived antigens), which persists for months after primary infection [52, 53]. Overall, coordinated SARS‐CoV‐2‐specific adaptive immune responses are associated with milder disease and are therefore essential for optimally controlling viral infection [54].
Autoimmunity and autoinflammation
Recent research has highlighted the potential role of autoimmunity and autoinflammation in COVID‐19 (Figure 3). Dysregulation of TLRs has been discussed above. TLR7 recognises single‐stranded RNA of pathogens in order to initiate an immune response. To prevent the induction of an immune response against endogenous RNA, TLRs contain a self‐recognition mechanism, failure of which can lead to autoreactivity [55, 56]. During the course of the pandemic, cases of multisystem inflammatory syndrome in children (MIS‐C) have been repeatedly reported [57]. MIS‐C is an immunopathogenic illness consisting of systemic hyperinflammation and MOF, which may clinically resemble Kawasaki disease or toxic shock syndrome [58, 59]. While its pathogenesis in COVID‐19 is incompletely understood, MIS‐C represents an immune‐mediated post‐infectious process featuring autoantibodies, with several specificities likely being involved [60, 61]. Furthermore, a case report was published describing adult COVID‐19 patients with a disease state resembling MIS‐C, termed MIS in adults (MIS‐A) [62].
Figure 3.
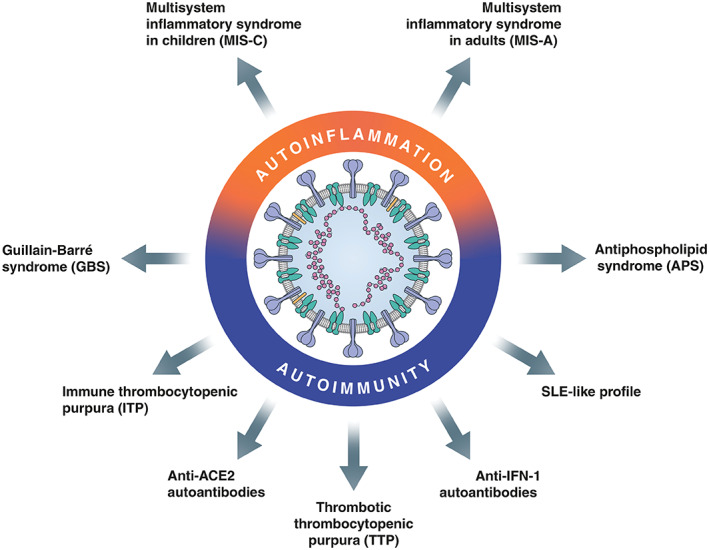
Autoimmunity and autoinflammation in COVID‐19. ACE2, angiotensin‐converting enzyme 2; IFN, interferon; SLE, systemic lupus erythematosus.
Autoimmune conditions that are potentially associated with COVID‐19 in selected cases comprise Guillain–Barré syndrome and its variants: immune thrombocytopenic purpura [63, 64]; antiphospholipid syndrome (APS), including its severe form catastrophic antiphospholipid syndrome (CAPS) [65, 66]; and thrombotic thrombocytopenic purpura (TTP) [67]. APS is an autoimmune disorder with formation of antiphospholipid antibodies (aPLs) causing an increased risk of (micro)vascular thrombosis [68]. Its severe variant CAPS is characterised by rapidly developing multi‐organ thrombotic injury [69], which may be linked to the hypercoagulability observed in severe COVID‐19. The presence of aPLs has been frequently reported in COVID‐19 patients [65, 70, 71, 72], although their association with thrombotic risk in this disease is still controversial [71, 73]. Interestingly, aPLs induce activation of pro‐inflammatory NF‐κB and increase the expression of TLR7, which may lead to overactivation of the pro‐inflammatory cytokine response [74]. Various viral infections have been shown to induce the production of autoantibodies against a disintegrin and metalloproteinase with a thrombospondin type 1 motif, member 13 (ADAMTS13) – a von Willebrand factor‐cleaving metalloprotease – leading to TTP, a condition that shares clinical similarities with COVID‐19, considering the frequent presence of endothelial cell injury and thrombotic microangiopathy [67, 75]. Furthermore, extrafollicular B‐cell responses in critically ill COVID‐19 patients have been associated with poor clinical outcomes [76]. Of note, the extrafollicular B‐cell signatures showed high similarity to those in autoimmune settings, especially systemic lupus erythematosus (SLE), suggesting that the antibodies were pathogenic due to autoreactivity [76]. Finally, a pre‐print study reported the presence of IgM autoantibodies against ACE2, which was strongly associated with severe clinical outcome in COVID‐19 patients [77]. The persistence of autoreactive antibodies may contribute to the post‐acute sequelae observed in a substantial proportion of symptomatic COVID‐19 patients – colloquially known as ‘long‐COVID’ [78, 79, 80]. These findings provide convincing evidence for the involvement of autoinflammation and autoimmunity in COVID‐19.
Host‐specific factors determining disease course
Inter‐individual differences in the adequacy of host defence mechanisms constitute a plausible explanation for the heterogeneity of disease course among infected individuals (Figure 4) [81]. Uncoordinated responses of adaptive immunity are associated with ageing and frequently precede disease control failure [54]. This can be explained by the concept of ‘immunosenescence’, which involves the age‐related decline in immune function with defects in both innate and adaptive immune responses, such as impaired pathogen recognition and chronic involution of the thymus [82, 83]. Other age‐related contributors include a chronic state of low‐grade systemic inflammation in the elderly (‘inflammaging’), an increased incidence of comorbidities in the elderly, and moderate to severe frailty [84]. Sex‐related variability in immune responses against SARS‐CoV‐2 may potentially underlie the increased disease vulnerability of males. In a study analysing sex differences in immune phenotypes, male patients showed a more robust innate immune response with higher plasma cytokine levels, whereas female patients had more robust T‐cell activation [85]. A poor T‐cell response against SARS‐CoV‐2 correlated with worse disease outcome in males, whereas higher levels of cytokines were associated with worsening of disease course in females, underscoring the potential rationale for sex‐specific treatment options. Apart from the aforementioned genetic variants in IFN‐related immunity [21, 25], additional undiscovered genetic immune variations may contribute to the variation in disease course, as well as pre‐existing T‐cell and B‐cell immune memory with antibody cross‐reactivity between seasonal coronaviruses and SARS‐CoV‐2 [86, 87, 88, 89].
Figure 4.
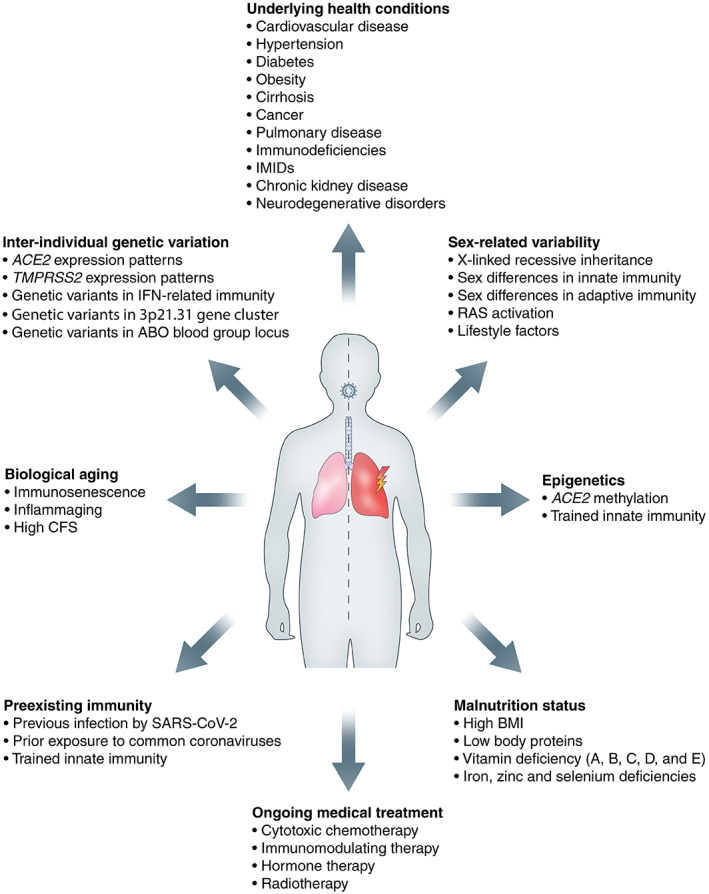
Host‐specific factors determining disease course. ACE2, angiotensin‐converting enzyme 2; BMI, body mass index; CFS, clinical frailty score; IFN, interferon; IMIDs, immune mediated inflammatory diseases; RAS, renin–angiotensin system; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2; TMPRSS2, transmembrane serine protease 2.
Individuals with multiple underlying morbidities or with severe single disease are at increased risk of developing severe COVID‐19, including those with cardiovascular disease, hypertension, diabetes, pulmonary disease, neurodegenerative disorders, immunodeficiencies, renal disease, liver damage, and obesity, among others [4]. Visceral fat, measured as abdominal adiposity, is associated with severe COVID‐19 disease [90]. Interestingly, a Dutch study on ICU‐admitted COVID‐19 patients found that serum leptin levels were increased in SARS‐CoV‐2‐infected patients compared with control patients with a similar body mass index (BMI) [91]. The authors hypothesised that this increase in leptin production, induced by SARS‐CoV‐2 infection of ACE2‐expressing adipocytes in visceral fat, led to activation of pulmonary leptin receptors resulting in enhanced local inflammation, thereby adding to the already existent chronic inflammatory state of obese patients. Treatment with resveratrol may be beneficial in this regard, considering its inhibitory effect on leptin secretion from adipocytes in addition to direct antiviral effects [92].
Furthermore, variations in ACE2 expression and activity among individuals are thought to affect the vulnerability to disease progression in COVID‐19. Paradoxically, increased membrane‐bound ACE2 may allow for more opportunities for SARS‐CoV‐2 to invade host cells, whereas downregulation of ACE2 (due to SARS‐CoV‐2‐induced endocytosis) precipitates tissue injury by (1) decreasing inactivation of des‐Arg9‐bradykinin, which potentiates the formation of pulmonary angioedema [93]; and (2) dysregulating the RAAS by causing a lack of conversion of Ang II into angiotensin(1–7) [94]. Interestingly, Ang II stimulates a disintegrin and metalloproteinase 17 (ADAM17), resulting in the release of cytokines [e.g. tumour necrosis factor alpha (TNF‐α)], thereby exacerbating inflammation, especially in individuals with pre‐existent chronically activated RAAS with high Ang II levels [95]. In addition, ADAM17 is a sheddase for ACE2, resulting in a soluble form of ACE2 (sACE2) [96]. Cardiovascular conditions and other COVID‐19 risk factors (e.g. male gender, older age, higher BMI) have been associated with increased levels of sACE2 [97, 98, 99]. A recent study showed a 97‐fold higher sACE2 activity in recovered SARS‐CoV‐2 patients compared with healthy controls, although its clinical significance is still a subject of debate [100]. Increased sACE2 may be deleterious by acting as an antigen (bound to SARS‐CoV‐2), thereby inducing autoimmunity against ACE2 [77, 101]. On the contrary, sACE2 may have beneficial effects as a competitive interceptor by preventing SARS‐CoV‐2 binding to membrane‐anchored ACE2 for host cell entry, a theory that has received experimental support from an in vitro study in which SARS‐CoV‐2 infection of human organoids could be inhibited by the administration of human recombinant sACE2 [102], in addition to favourable results from a single case study [103]. Finally, several other host factors are thought to determine COVID‐19 disease course, such as epigenetic mechanisms, malnutrition status, and ABO blood group status [104, 105, 106].
Pathology
COVID‐19 is a disease in which multiple organ systems can be involved. The main pathological alterations detected in the most commonly affected organ systems are discussed in the following paragraphs, with particular emphasis on immunopathology.
Respiratory system
SARS‐CoV‐2 primarily affects the respiratory system, with viral entry into upper and lower respiratory epithelial cells, causing influenza‐like clinical manifestations, such as fever, cough, and dyspnoea [107, 108]. The ensuing immune response is suppressed in some individuals, leading to unrestrained viral replication that facilitates progression to a pulmonary phase with pneumonia [109]. In late‐stage disease, hyperactivation of immune responses contributes to widespread hyperinflammation with the development of severe disease, including ARDS [109]. Histological examinations showed various patterns of diffuse alveolar damage (DAD) with interstitial T‐cell infiltration [40, 49, 110] (Figure 5). In some cases, an even more severe form of DAD could be observed, namely acute fibrinoid and organising pneumonia [111]. Typical features of DAD, with hyaline membrane formation and type II pneumocyte hyperplasia, were frequently encountered [110, 112]. Interestingly, abundant presence of SARS‐CoV‐2 has been demonstrated early in the disease course, whereas virus‐infected cells were only sporadically present at later stages of COVID‐19 [40, 46, 113]. The decrease in viral presence may be attributed to prompt immune activation, as demonstrated by extensive alveolar and interstitial inflammation [40] (Figure 6). In line with this, a recent autopsy study discovered two distinct immunopathological reaction patterns in the lungs of deceased COVID‐19 patients: in the first subgroup, patients died early after hospitalisation with high viral load and relatively limited pulmonary pathology, implying extra‐pulmonary factors as contributors to lethal outcome, while patients in the second subgroup had a longer disease course and low viral load, yet prominent DAD, which was associated with marked immune cell infiltration and local complement activation [46]. The disconnection between viral presence and pulmonary inflammation highlights the contribution of immunopathology in causing severe COVID‐19 [114].
Figure 5.
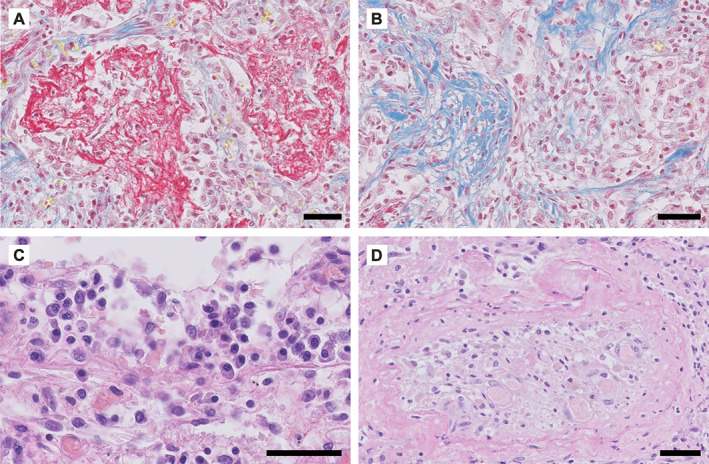
Representative examples of COVID‐19‐associated lung pathology. Panels A, B, and D show photomicrographs of the lung tissue of a 55‐year‐old male who died of COVID‐19 4–5 weeks after admission to the intensive care unit (ICU) of the University Medical Center Groningen. Panel C is from a 63‐year‐old male who died 2.5 weeks after admission to the ICU. (A) Alveolar spaces are filled with fibrin, stained in red. (B) Organising pneumonia is observed with fibrosis in blue; fibroblast proliferation can be observed on the right hand side. (C) Lymphoplasmocytic infiltration. (D) Occluded artery (thrombosis) with recanalization. (A, B) Martius scarlet blue; (C, D) H&E. Scale bar = 50 μm.
Figure 6.
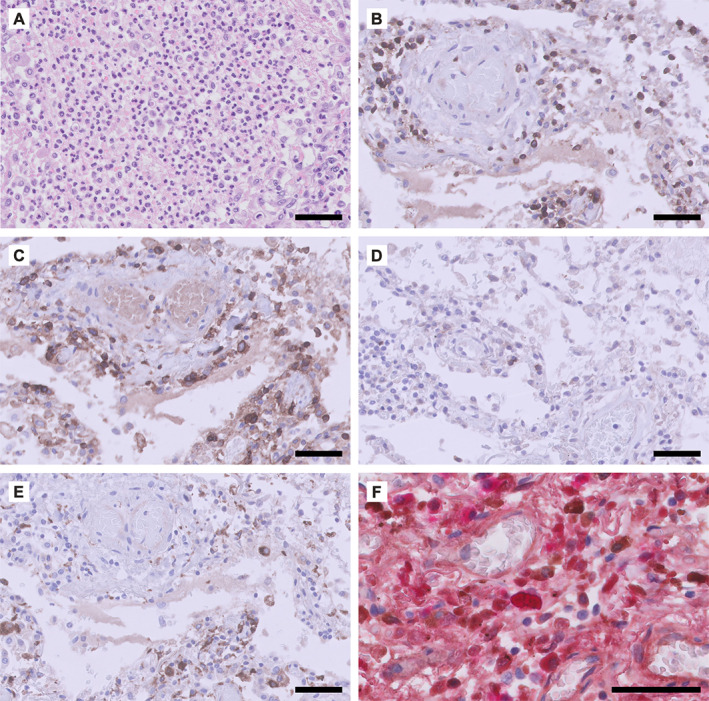
Phenotype of the inflammatory response observed in COVID‐19‐associated lung pathology. Panel A shows the lung tissue of a patient (male, 55 years) who died of COVID‐19 4–5 weeks after admission to the intensive care unit (ICU) of the University Medical Center Groningen. Panels B–F are from a male (63 years) who died 2.5 weeks after admission to the ICU. (A) Diffuse neutrophilic infiltrate in the alveolar spaces. Immunohistochemical staining for (B) CD3 (T‐cells), (C) CD4 (CD4+ T‐cells and macrophages), (D) CD8 (CD8+ T‐cells), (E) CD68 (macrophages), and (F) immunoglobulin kappa (brown) and lambda (red) light chains (double labelling). Scale bar = 50 μm.
Other main findings of fatal COVID‐19 pneumonia were vascular alterations, including the presence of thrombi and microthrombi, intussusceptive angiogenesis, and severe endothelial injury [115]. SARS‐CoV‐2‐mediated endothelial injury and endotheliitis can give rise to dysregulated local immunothrombosis, culminating in microvascular dysfunction and the formation of microthrombi [115, 116, 117]. In a recent post‐mortem study, neutrophil accumulation (Figure 6) was demonstrated in lung parenchyma and vasculature, with the formation of NETs, indicating a role for a maladaptive immune response in causing pulmonary coagulopathy [40, 118]. Furthermore, vigorous and dysregulated respiratory effort may also contribute to the progression of lung damage in COVID‐19, a phenomenon known as ‘patient self‐inflicted lung injury’ [119].
Cardiovascular system
Myocardial injury, reflected by increased blood troponin levels, has been reported frequently in COVID‐19 [120]. Pre‐existing cardiovascular disease is associated with poor prognosis [121], whereas COVID‐19 itself can induce cardiovascular complications, such as acute heart failure, arrhythmias, acute coronary syndrome, and myocarditis [120]. The latter has raised particular concern, with multiple case studies reporting on COVID‐19‐associated myocarditis [122, 123, 124] and studies demonstrating ongoing myocardial inflammation after recovery from COVID‐19 [125, 126]. Myocarditis may result from direct cardiotoxic effects of SARS‐CoV‐2 infection or from indirect effects, either via cytokine‐mediated cardiotoxicity or by triggering an autoimmune response against cardiac components [127]. In line with expression of ACE2 on different cell types in the human heart [128], SARS‐CoV‐2 has been detected in a variety of cells in myocardial tissue, including interstitial inflammatory cells [129, 130], cardiomyocytes [40, 124], and capillary endothelial cells [124]. These findings indicate that SARS‐CoV‐2 may directly infiltrate the heart via the bloodstream – consistent with the finding of cardiac endotheliitis [116] – or indirectly via immune cell transmigration. An autopsy study demonstrated that the presence of SARS‐CoV‐2 in myocardial tissue was not associated with a cardiac inflammatory response consistent with myocarditis [129]. Instead, COVID‐19‐associated myocarditis may rather result from secondary reactions to disease.
Cardiac inflammation has been described in several other reports [40, 131]. In a study analysing post‐mortem heart tissue from 20 COVID‐19 patients, epicarditis and endocarditis were present in all patients, whereas myocarditis was found in 55%, characterised by the presence of histiocytes, T‐cells, and neutrophils [40]. However, the presence of myocarditis showed no association with the type of myocardial injury and was inconsistent with clinical ECG observations and the presence of SARS‐CoV‐2 in the heart [40]. Another autopsy study including 21 patients who succumbed to COVID‐19 demonstrated that 18 patients had widespread interstitial macrophage infiltration without a clear association with myocardial injury, whereas multifocal lymphocytic myocarditis was found in three patients [131]. Besides cardiac inflammation, other forms of myocardial injury may be involved in COVID‐19, such as right ventricular strain injury secondary to lung pathology [131, 132], microvascular thrombi [40, 131], hypoxic injury [133], and pre‐existing injury compatible with underlying disease [40, 134].
Nervous system
Neurological manifestations have frequently been described in patients suffering from COVID‐19, ranging from anosmia, dysgeusia, headache, and ‘brain‐fog’ to severe conditions including ischaemic stroke and encephalopathy/encephalitis [135, 136]. Furthermore, cognitive deficits have been reported even after recovery, which is probably partly related to incident delirium in many cases [137]. The neurological manifestations may result from direct cytopathic effects of SARS‐CoV‐2 infection or from superimposed disease factors, including hypoxia‐induced damage [40, 138, 139], drug‐related changes, and harmful immune responses. The presence of SARS‐CoV‐2 has been demonstrated in the cerebrospinal fluid (CSF) [140, 141, 142] and post‐mortem brain tissues [138, 143, 144, 145] from COVID‐19 patients, indicating its neurotropic potential. The virus may enter the central nervous system (CNS) via the haematogenous pathway – involving immune cell transmigration or penetration of the blood–brain barrier – as well as via the neuronal pathway [146]. The identification of viral particles in the olfactory bulb [147], together with the common occurrence of anosmia [148], point to the olfactory pathway as a plausible route of neuroinvasion by SARS‐CoV‐2. On the other hand, ACE2 and TMPRSS2 have been found in microvillar cells, olfactory horizontal basal cells, and olfactory sustentacular cells but not in olfactory neuronal extensions. Therefore, the olfactory entry route into the CNS remains a conundrum [149]. Next to nervous and olfactory involvement, SARS‐CoV‐2 protein has recently been demonstrated intracellularly in the ocular tissues of a patient previously infected with this virus [150].
To date, the contribution of SARS‐CoV‐2 neuroinvasion to the pathogenesis of neurological manifestations in COVID‐19 remains undetermined. A study of SARS‐CoV in mice indicated that direct neuronal infection in vital brain regions (e.g. cardiorespiratory centres in the medulla oblongata) induces neuronal death and likely contributes to severe disease [151]. In human COVID‐19 autopsy studies, microglial activation and cytotoxic T‐cell infiltration were indeed most pronounced in the brainstem and cerebellum (Figure 7) [40, 145]. However, these studies also indicated that the role of neurotropism in causing COVID‐19‐related brain pathology may be rather limited. Autopsy series of COVID‐19 patients demonstrated the presence of SARS‐CoV‐2 in 53% of the brains investigated but showed no association with the severity of neuropathological alterations [145]. Instead, other factors are likely involved in the neuropathology observed in COVID‐19, such as an overstimulated immune response. An autopsy study of brain tissues from nine deceased COVID‐19 patients reported on extensive neuroinflammation, involving T‐cell infiltration and massive activation of microglia with formation of nodules, while SARS‐CoV‐2 RNA could not be detected [40]. This could indicate rapid viral clearance or widespread neuroinflammatory activation in response to serious SARS‐CoV‐2 infection elsewhere in the body. A disrupted blood–CSF barrier through SARS‐CoV‐2‐induced choroid plexus damage may also contribute to neuroinflammation by allowing entry of immune cells and cytokines [152]. An extensive inflammatory response was most notable in the olfactory bulbs – possibly leading to anosmia – as well as in the medulla oblongata, in which the primary respiratory control centre is located, and could therefore contribute to respiratory failure in COVID‐19 patients [40]. Another autopsy study found comparable neuroinflammatory findings [145]. Furthermore, the observation of neutrophilic plugs with NET formation in post‐mortem brain tissues indicates a role for immunothrombosis in thrombotic brain injury [40]. Finally, SARS‐CoV‐2 potentially acts as a trigger of neurodegenerative proteinopathies, such as Parkinson's disease and Alzheimer's disease, partly due to intracellular responses to inflammation that could lead to protein misfolding and aggregation [153, 154, 155]. Longitudinal studies should further clarify whether the COVID‐19 pandemic will lead to an increased burden of neurodegenerative and autoimmune conditions among COVID‐19 survivors.
Figure 7.
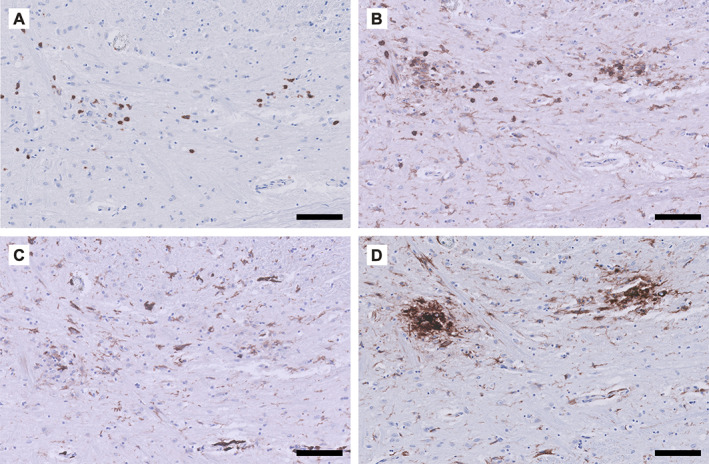
Phenotype of the inflammatory response observed in COVID‐19‐associated brain pathology. Detailed micrographs from the tegmental area of the medulla oblongata in a patient (male, 63 years) who died 2.5 weeks after admission to the ICU. Immunohistochemical staining for (A) CD3 (T‐cells), (B) CD45 (leukocytes including microglia), (C) CD163 (scavenger receptor, activated microglia), and (D) HLA‐DR (MHC class II, activated microglia). The T‐cell infiltrate (A) co‐localises with activated microglia (C and D). Scale bar = 100 μm.
Kidneys
Development of acute kidney injury (AKI) is common in COVID‐19 patients and is associated with poor clinical outcome [156]. COVID‐19 could affect renal tubular, glomerular, and vascular tissues, leading to conditions such as acute tubular injury, collapsing glomerulopathy, and thrombotic microangiopathy [157]. Potential mechanisms underlying AKI range from direct SARS‐CoV‐2 infection via ACE2, which is expressed in proximal tubular cells and glomerular epithelial cells [158], to indirect effects resulting from RAAS dysregulation, coagulopathy, haemodynamic instability, systemic inflammation, MOF, and iatrogenic injury [159]. A direct cytopathic effect of SARS‐CoV‐2 in the kidney is currently a subject of debate. Electron microscopy studies demonstrated viral particles within podocytes, proximal tubular epithelium, and endothelium [160, 161, 162], although careful interpretation is warranted as various cytoplasmic structures exhibit viral‐like morphology [163, 164]. Nevertheless, SARS‐CoV‐2 was detected in the renal parenchyma by alternative methodologies, which corroborated the concept of direct SARS‐CoV‐2 infection [40, 143, 162]. An autopsy study including 63 patients who had SARS‐CoV‐2 respiratory infection demonstrated an association between SARS‐CoV‐2 renal tropism with development of AKI and shorter patient survival time [165]. Interestingly, a characteristic post‐mortem finding in COVID‐19 patients was proximal tubular vacuolisation, which correlated with the presence of SARS‐CoV‐2‐like particles in renal tubular epithelial cells; this may therefore be indicative of a direct cytopathic effect of SARS‐CoV‐2 infection [40, 160, 162]. These findings are in contrast to multiple other studies in COVID‐19 patients with AKI that found no evidence of significant SARS‐CoV‐2 presence in renal tissue [157, 166, 167, 168].
Dysregulated immune responses in COVID‐19 probably play a role in the pathological tubular, glomerular, and vascular alterations in the kidney [157, 166]. AKI in COVID‐19 has been linked to sepsis‐associated AKI, partly due to the frequent observation of acute tubular necrosis, in which ischaemia and nephrotoxicity play a critical role [167]. Reports on COVID‐19‐associated collapsing glomerulopathy in primarily black patients suggested a role for cytokine‐mediated kidney disease in individuals with high‐risk apolipoprotein L1 genotypes with SARS‐CoV‐2 infection, as occurs with other forms of viral‐mediated collapsing glomerulopathy [166, 169, 170, 171]. Vascular pathology has been described sporadically in the kidneys of COVID‐19 patients and comprises vasculitis, thrombotic microangiopathy, and microthrombi [40, 157, 162], which may result from COVID‐19‐induced endothelial dysfunction, coagulopathy, and complement activation [157]. Figure 8 shows representative micrographs of the composition of the lymphocytic infiltrate present predominantly in the tubulointerstitial compartment.
Figure 8.
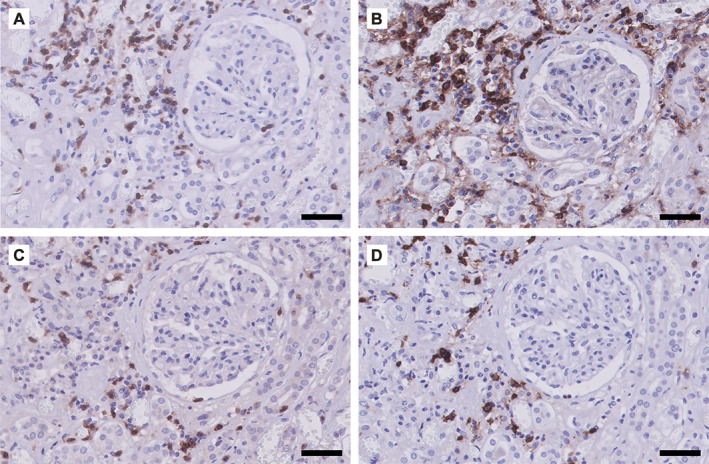
Phenotype of the inflammatory response observed in COVID‐19‐associated renal pathology. Micrographs of the renal cortical tissue from a patient (male, 65 years) who died of COVID‐19 5 weeks after appearance of the first symptoms. Immunohistochemical staining for (A) CD3 (T‐cells), (B) CD4 (CD4+ T‐cells and macrophages), (C) CD8 (CD8+ T‐cells), and (D) CD20 (B‐cells). The lymphocytic infiltrate is located mainly in the tubulointerstitium. Scale bar = 50 μm.
Gastrointestinal system
Gastrointestinal (GI) symptoms of COVID‐19 (e.g. anorexia, diarrhoea, abdominal pain) occur commonly and have been associated with more severe disease [172, 173]. Several lines of evidence support the GI tract as a potential site for active SARS‐CoV‐2 infection and replication. ACE2 was found to be abundantly present on the brush border of absorptive enterocytes in both the small and the large intestine, providing a prerequisite for SARS‐CoV‐2 infection and a potential route of faecal–oral transmission [158]. Furthermore, single‐cell transcriptomics revealed high co‐expression of ACE2 and TMPRSS2 in absorptive enterocytes along the entire GI tract [174]. Enterotropism of SARS‐CoV‐2 has been demonstrated in vitro by using human small intestinal organoids [175, 176, 177], in vivo in non‐human primate models [178, 179], and in human gastrointestinal tissue [180]. Apart from the intestinal epithelium, marked ACE2 expression has been observed within the intestinal endothelium and in smooth muscle cells of the intestinal muscular layers, suggesting a secondary, indirect possibility of intestinal transfection [108, 158]. Further evidence of viral GI infection is provided by prolonged presence of SARS‐CoV‐2 RNA in faecal samples of infected patients [181]. In some cases, RNA positivity persisted in faecal samples even after it vanished from respiratory tract samples [180, 181], which may be explained by the composition and functionality of the gut microbiota, which shows considerable inter‐individual diversity and diversifying responses of host immunity [182].
Previous autopsy studies revealed distinct histopathological features in GI tissues of patients with COVID‐19. Along different parts of the GI tract, interstitial oedema within the lamina propria and concurrent infiltration, mainly with lymphocytes, monocytes, and plasma cells, have been reported [180, 183]. Although the location and numbers of intestinal epithelial cells may have a natural appearance, they are sometimes accompanied by inflammatory infiltrates. Mucosal changes of the GI tract vary, from disruption of epithelial architecture, dilated and congested capillaries within the lamina propria and submucosa, and immune cell infiltration, to necrosis and shedding of the entire mucosa [184, 185].
Hepatic and biliary system
Patients with COVID‐19 are at risk of developing liver injury, manifested by increased liver transaminases with subtle hyperbilirubinaemia [186]. Pathological findings of liver injury have been reported most commonly in individuals with moderate‐to‐severe COVID‐19 [187]. Hepatic invasion by SARS‐CoV‐2 may be possible via constitutively expressed ACE2, mainly on cholangiocytes and, to a lesser extent, hepatocytes [188]. Thus, it has been hypothesised that SARS‐CoV‐2‐induced liver damage primarily results from biliary tract infection through cholangiocytes, with secondary injury to and compensatory proliferation of hepatocytes [108, 189]. In contrast to the endothelial linings of other organs, ACE2 expression of the hepatic sinusoidal endothelium is very limited [158]. Microscopically, pathological features of COVID‐19 in the liver include moderate macrovesicular steatosis, mild lobular and portal (mainly lymphocytic) infiltration, patchy hepatic necrosis, and both periportal and centrilobular sinusoidal dilatation [190, 191]. Apart from direct viral hepatic infection, liver damage may arise from immune‐mediated inflammation, drug toxicity, pre‐existing chronic liver disease, or concomitant hepatic infections [185, 192]. Finally, mild cases of COVID‐19 are not accompanied by signs of liver damage, and, if so, this injury is often transient and eventually shows spontaneous recovery [193].
Reproductive system and pregnancy
Concern has been raised about involvement of the reproductive system in COVID‐19, as this may affect fertility and poses a potential risk of sexual transmission. SARS‐CoV‐2 has been demonstrated in the semen of men with COVID‐19 in both the acute and the recovering stage of viral infection [194]. In line with this, ACE2 is highly expressed in the testis and epididymis – more specifically, in the spermatogonia, Leydig cells, and Sertoli cells [195, 196]. An autopsy study examining 12 testes from patients who succumbed to COVID‐19 showed significant damage to seminiferous tubules with ballooning of Sertoli cells, reduced numbers of Leydig cells, and a mild interstitial infiltration of predominantly T‐cells [197]. In this study, SARS‐CoV‐2 RNA was absent in 90% of patients, and no viral particles were demonstrated by electron microscopy, suggesting that viral presence had already been cleared by the immune response or that other factors were involved.
As for the female reproductive system, only few data are available on its association with SARS‐CoV‐2 infection. In a cohort of ten female patients with COVID‐19, no evidence of viral infection in vaginal fluids was reported [198], while another study did report on the presence of SARS‐CoV‐2 RNA in vaginal fluid [199]. Considering the well‐established functions of ACE2 within the female reproductive system and its ubiquitous expression in these organs, people have been concerned about the potential harm of COVID‐19 on female fertility and pregnancy‐related outcomes [200]. Although rare, transplacental transmission is possible, with subsequent significant placental inflammation and neonatal SARS‐CoV‐2 infection [201, 202]. Nevertheless, pregnancy outcomes in COVID‐19 are generally good [203].
Integumentary system
A diversity of dermatological manifestations have been described in patients suffering from COVID‐19. These include morbiliform rashes, pernio‐like acral lesions, urticaria, macular erythemas, vesicular eruptions, papulosquamous eruptions, and retiform purpura [204]. Many of these manifestations are non‐specific, although some may be secondary to COVID‐19‐related immune and inflammatory reactions [204]. Pernio (chilblain)‐like acral lesions in COVID‐19 – colloquially known as ‘COVID toes’ – typically present after the onset of COVID‐19 symptoms and are associated with a relatively mild disease course [204]. The underlying mechanism is thought to be primarily inflammatory with histopathological findings similar to those observed in idiopathic and autoimmune‐related pernio, including vacuolar interface damage with epidermal necrotic keratinocytes, perivascular and peri‐eccrine lymphocytic infiltration, and dermal oedema [204, 205, 206]. Interestingly, in the skin of COVID‐19 patients with pernio‐like lesions, SARS‐CoV‐2 was found in vascular endothelial cells and in epithelial cells of eccrine sweat glands [206], implying direct SARS‐CoV‐2 infection as a potential aetiopathogenic mechanism. In addition, a robust antiviral IFN response may be involved in causing pernio‐like lesions, thereby providing an explanation for the mild disease course observed in patients with pernio [207]. On the other hand, critically ill COVID‐19 patients may present with retiform purpura, livedo racemosa, and acral ischaemia, which are distinct from pernio with histopathological features of non‐inflammatory to pauci‐inflammatory thrombogenic vasculopathy [35, 204, 208]. Skin biopsies from COVID‐19 patients demonstrated the involvement of complement activation in the pathogenesis of cutaneous thrombosis, sometimes with co‐localisation of the SARS‐CoV‐2 S‐protein [35, 208].
Treatment
Current management strategies for COVID‐19 comprise the implementation of preventive (social) measures and vaccination to control viral dissemination, the implementation of supportive care when indicated, and the use of (repurposed) drugs. Considering that de novo development of specific anti‐SARS‐CoV‐2 drugs and vaccines takes time, clinical investigations have focused on drug repurposing. The efficacy and safety of many repurposed treatment modalities have been tested in preclinical studies and/or clinical trials, often with mixed results, and many trials are still ongoing.
During the course of the pandemic, progress has been made with regard to patient treatment. Successful treatment may depend on the correct timing of repurposed drugs, which requires an understanding of their mode of action, COVID‐19 pathophysiology, and disease stages. Drugs that inhibit viral entry and replication (i.e. antivirals) may hold more promise when applied early in the disease course, whereas immunomodulating therapy (including corticosteroids and anti‐cytokine antibodies) may have the greatest impact on the hyperinflammatory state in more advanced disease and can even be harmful during the initial stage, when an adequate immune reaction is indispensable for viral clearance. In keeping with this, a useful timeline was recently provided, demonstrating the potential contribution of each repurposed drug class to the treatment of the consecutive stages of COVID‐19 [93]. Immunomodulators have been widely used during the pandemic and will probably continue to play an important role. They are thought to act through several key mechanisms: (1) glucocorticoid‐mediated inhibition of pro‐inflammatory signals and activation of anti‐inflammatory signals; (2) inhibition of cytokines; (3) strengthening of antiviral defence by supplementation of IFN; (4) inhibition of the TLR‐signalling pathway; and (5) inhibition of the Janus kinase (JAK)/signal transducer and activator of transcription (STAT) pathway.
Based on the aforementioned pathophysiological and immunopathological considerations, we provide a detailed overview of currently advocated repurposed drug therapies with accompanying modes of action and clinical efficacy in Table 1. For an exhaustive list of experimental treatments, the reader is referred to the landscape document of the WHO [290]; the latest recommendations can be found in the living WHO guideline on drugs for COVID‐19 [291].
Table 1.
Overview of (repurposed) drugs for COVID‐19, based on the described pathophysiological and immunopathological mechanisms.
| Drug class | Drug names | Proposed mode of action | Clinical effectiveness | References |
|---|---|---|---|---|
| Antivirals | Remdesivir | Inhibition of RdRp | Reduces recovery time; halts progression to severe disease; no effect on survival | [209, 210] |
| Lopinavir/ritonavir | Inhibition of 3CLpro | Probably ineffective | [211] | |
| Ivermectin |
Inhibition of the IMP α/β receptor responsible for viral protein transmission into host cell nucleus Inhibition of LPS‐induced inflammation |
Inconclusive | [212, 213, 214, 215, 216, 217, 218, 219] | |
| Ribavirin | Inhibition of viral RNA synthesis/mRNA capping | Unrealistic dosing requirement | [220] | |
| Favipiravir | Inhibition of RdRp | Unrealistic dosing requirement | [221, 222, 223] | |
| Umifenovir | Impeding trimerization of SARS‐CoV‐2 S‐protein | Probably ineffective | [224] | |
| Zinc | Inhibition of RdRp | Uncertain | [225] | |
| Anticoagulants | LMWH (e.g. nadroparin) |
Potentiation of antithrombin‐mediated inhibition of coagulation factors Xa and IIa Conformational change of spike S1 receptor binding domain, possibly impeding binding to ACE2 |
Established | [226] |
| Unfractionated heparin |
Reducing viral entry by interacting with S‐protein Heparanase inhibition (associated with COVID‐19 severity) Neutralisation of chemokines and cytokines, and extracellular histones Interference of leukocyte trafficking through interaction with leukocyte surface ligands, preventing leukocyte attachment and extravasation Potentiation of antithrombin‐mediated inhibition of coagulation factors Xa and IIa |
[227, 228] | ||
| Immunomodulators – corticosteroids | Dexamethasone | Glucocorticoid‐mediated effects; inhibition of pro‐inflammatory signals and activation of anti‐inflammatory signals; mechanisms include lipocortin‐mediated PLA2 suppression (decreased arachidonic acid as precursor of prostaglandins, leukotrienes, and thromboxanes), COX inhibition (decreased prostaglandin synthesis), and inhibition of NF‐κB signalling, among others | Established | [229, 230, 231, 232] |
| Methylprednisolone | Similar to dexamethasone | Uncertain | [232, 233, 234] | |
| Hydrocortisone | Similar to dexamethasone | Uncertain | [235] | |
| Immunomodulators – biologicals | IFN β‐1a | Supplementation of interferon for antiviral defence |
Uncertain ACTT‐3 trial ongoing |
[236, 237] |
|
IL‐6R‐antagonists (e.g. tocilizumab) |
Inhibition of the pro‐inflammatory action of IL‐6 | Improves outcome in critical COVID‐19 | [238] | |
|
IL‐1R antagonists (e.g. anakinra) |
Inhibition of the pro‐inflammatory action of IL‐1 | Uncertain in severe disease; no improved outcomes in mild‐to‐moderate COVID‐19 | [239, 240] | |
|
TNF‐α inhibitors (e.g. adalimumab) |
Inhibition of the pro‐inflammatory cytokine TNF‐α | Uncertain | [241, 242, 243] | |
|
BTK inhibitors (e.g. ibrutinib) |
Inhibition of the TLR signalling pathway and thereby decreased cytokine production | Uncertain | [244] | |
|
JAK inhibitors (e.g. baricitinib, fedratinib) |
Inhibition of Janus kinase (JAK)/signal transducer and activator of transcription (STAT) pathway, a signalling cascade mediating cellular responses to multiple cytokines, growth factors, and other ligands (such as Ang II) after binding to their respective receptors Inhibition of upstream regulators of ACE2‐mediated endocytosis of SARS‐CoV‐2, including AAK1 and GAK |
Possible | [245, 246, 247] | |
| Calcineurin inhibitors (e.g. cyclosporine, tacrolimus) |
Inhibition of IL‐2 production and IL2‐R expression, leading to decreased T‐lymphocyte activation Potential antiviral activity by inhibiting viral replication, based on previous preclinical evidence for other coronaviruses (including SARS‐CoV) |
Uncertain | [248, 249, 250, 251, 252, 253] | |
| Complement inhibitors | Eculizumab | Terminal complement inhibitor that binds to C5 complement protein and blocks the generation of pro‐inflammatory C5a and the membrane attack complex (C5b‐9) | Uncertain | |
| Kinin–kallikrein pathway inhibitors | Lanadelumab | Inhibition of plasma kallikrein | Uncertain | |
| Icatibant | Selective antagonist of bradykinin receptor type 2 | Uncertain | [256] | |
| Angiotensin inhibitors | Angiotensin receptor blockers |
Prevention of Ang II binding to AT1R, thereby counteracting vasoconstriction, proliferation, fibrosis, thrombosis, and inflammation Prevention of ACE2 downregulation by endocytosis |
Uncertain | [108, 257, 258] |
| ACE inhibitors | Prevention of Ang II formation | Uncertain | [108, 257, 258] | |
| Recombinant human ACE2 (rhACE2) |
Binding of the viral spike protein and thereby neutralisation of SARS‐CoV‐2 Minimising COVID‐19‐associated organ damage as a result of RAAS hyperactivation/increased Ang II levels |
Uncertain | [103] | |
| SERPINs | C1 esterase inhibitor |
Inhibition of the complement cascade by binding to C1r and C1s Inhibition of the contact activation system (intrinsic coagulation and kinin–kallikrein pathways) by binding to FXIIa and kallikrein |
Uncertain |
[259] |
| Camostat mesylate | Inhibition of TMPRSS2 | Uncertain | [7] | |
| Nafamostat mesylate | Inhibition of various serine proteases: thrombin, FXII, TMPRSS2, and kallikrein | Uncertain | [260] | |
| Antimalarials | Hydroxychloroquine (HCQ)/chloroquine (CQ) | Inhibition of viral entry and endosome fusion/uncoating, reduction of cytokine production, and inhibition of platelet aggregation | Probably ineffective | [22, 23, 261, 262, 263, 264] |
| Blood‐derived products | Convalescent plasma | Neutralising antibodies from recuperated COVID‐19 patients | Probably ineffective | [265, 266, 267, 268] |
| Hyperimmune immunoglobulin | Neutralising SARS‐CoV‐2 viral antigens by administering antibodies from recuperated COVID‐19 patients with high antibody titres | Uncertain | [269] | |
| REGN‐COV2 | Cocktail of two neutralising antibodies against SARS‐CoV‐2 spike protein receptor binding domain | Uncertain | [270] | |
| Bamlanivimab | Anti‐spike neutralising IgG1 monoclonal antibody initially derived from a recovered COVID‐19 patient, intended for the treatment of mild to moderate COVID‐19 | Promising | [271, 272] | |
| Miscellaneous | Colchicine | Anti‐inflammation: inhibition of tubulin polymerisation, with effects on the inflammasome, cellular adhesion molecules, and inflammatory chemokines | Promising | [273] |
| Vitamin D |
Supports innate and adaptive immunity Inhibition of ADAM17 Counteracting NADPH oxidase activity resulting in decreased ROS production, thereby enhancing NO bioavailability Enhancing antioxidant enzymes that can scavenge free radicals Suppression of NF‐κB signalling and production of pro‐inflammatory cytokines |
Vitamin D deficiency associated with COVID‐19; effect of supplementation inconclusive | [274, 275] | |
| Azithromycin | Potential antiviral activity, based on evidence from other RNA viruses | Uncertain | [276, 277, 278, 279] | |
| Sirolimus | Inhibition of mTOR pathway, which plays a role in pro‐inflammatory T‐cell differentiation | Uncertain | [280] | |
| Resveratrol |
Reduction of leptin levels Suppression of Ang II Antioxidant effects Direct antiviral activity by inhibiting viral replication |
Uncertain | [92, 281, 282, 283] | |
| Heterologous vaccines | Influenza vaccine | Stimulation of trained innate immunity | Uncertain | [284, 285] |
| BCG | Stimulation of trained innate immunity | Uncertain; no significant effect in frail elderly | [286, 287, 288] | |
| Measles vaccine | Stimulation of trained innate immunity | Uncertain | [289] |
Vaccine development
The fight against COVID‐19 initiated a worldwide endeavour to develop a safe and effective vaccine. This has resulted in more than 230 vaccine candidates currently under development, 63 of which reached phase 1–3 clinical evaluation as of 26 January 2021 [292]. Several vaccines have already been authorised and used in vaccination programmes around the world. Currently, some of the most promising vaccine candidates include Pfizer–BioNTech (mRNA vaccine, efficacy 95.0%) [293], Moderna (mRNA vaccine, efficacy 94.1%) [294], Oxford–AstraZeneca (adenoviral vector vaccine, efficacy 70.4%) [295], Johnson and Johnson (adenoviral vector vaccine, efficacy 66%) [296], Novavax (protein subunit vaccine, efficacy 89.3%) [297], Sinopharm (inactivated vaccine, efficacy 79.3%) [298], Sinovac (inactivated vaccine, efficacy 50.4%) [299], and Sputnik V (adenoviral vector vaccine, efficacy 91.6%) [300]. However, it is important to note that the hitherto reported efficacy and safety results are based on interim analyses, as phase 3 trials for all of these candidates are still ongoing; more definite results and real‐world evidence are anxiously anticipated. Recently, concern has been raised about the emergence of three mutations of SARS‐CoV‐2 – the UK, South Africa, and Brazil variants – which may potentially hamper viral epitope recognition by induced antibodies. Other potential vaccine‐related limitations include insufficient immunogenicity (e.g. in the elderly), development of antibody‐dependent enhancement [301], failure of long‐lasting immunity, rapid vaccine development potentially compromising evaluation of long‐term effects, and vaccination hesitancy in the general population [302]. An extensive discussion on vaccine development in COVID‐19 is beyond the scope of this review and a comprehensive overview of current vaccine candidates can be found in the landscape document of the WHO [292]. Finally, some promising results were demonstrated with the use of heterologous vaccines, including influenza vaccine, bacillus Calmette‐Guérin (BCG) vaccine, and measles vaccines, which are incorporated in Table 1.
Conclusion
COVID‐19 progression is considered to result from a complex interplay of multiple pathophysiological mechanisms, among which the immunopathological response to SARS‐CoV‐2 infection is a key player in determining disease course, involving dysregulated innate and adaptive immunity as well as autoimmunity. Whereas the immune response is suppressed in the early stages of the disease, resulting in unrestrained viral replication and dissemination in infected host cells, an eventual increase of immune responses contributes to hyperinflammation and organ damage. Inter‐individual differences in failure of host defence mechanisms, along with other host‐specific factors, likely determine whether an individual progresses to severe disease. The multi‐faceted nature of the pathophysiology of COVID‐19, with its various disease stages, provides a basis for the use of various treatment regimens, ranging from antivirals early in disease when mild symptoms are present, to immunomodulating, antioxidant, and antithrombotic therapy in more advanced disease. Overall, COVID‐19 pathophysiology is complex with multiple interacting mechanisms being involved; approaching a single pathway would be insufficient for disease containment. Instead, a multilevel approach targeting multiple facets of the pathophysiology is required to tackle the COVID‐19 pandemic.
Ethics statement
Use of patient material was permitted according to regulations of the national ‘Code for Good Use of Patient Material’.
Abbreviations
- ACE2
angiotensin‐converting enzyme 2
- AKI
acute kidney injury
- Ang II
angiotensin II
- aPls
antiphospholipid antibodies
- APS
antiphospholipid syndrome
- ARDS
acute respiratory distress syndrome
- CAPS
catastrophic antiphospholipid syndrome
- COVID‐19
coronavirus disease 2019
- CRP
C‐reactive protein
- CSF
cerebrospinal fluid
- CTSL
cathepsin L
- DAD
diffuse alveolar damage
- DAMP
damage‐associated molecular pattern
- ICU
intensive care unit
- ISG
interferon‐stimulated gene
- LDH
lactate dehydrogenase
- MAS
macrophage activation syndrome
- MIS‐A
multisystem inflammatory syndrome in adults
- MIS‐C
multisystem inflammatory syndrome in children
- MOF
multi‐organ failure
- NET
neutrophil extracellular trap
- PAMP
pathogen‐associated molecular pattern
- PRR
pattern recognition receptor
- RAAS
renin–angiotensin–aldosterone system
- RAS
renin–angiotensin system
- sACE2
soluble ACE2
- SARS‐CoV‐2
severe acute respiratory syndrome coronavirus 2
- TF
tissue factor
- TMPRSS2
transmembrane serine protease 2
- TTP
thrombotic thrombocytopenic purpura
Author contributions statement
All the authors were involved in the conceptualisation of this review. LEvE, MB, ARB, AEA, AKO and HvG designed the outline of the review. LEvE, MB and HvG wrote the first draft. ARB and AKO wrote sections of the review. All the authors contributed to manuscript revision and approved the submitted final version.
No conflicts of interest were declared. This review was realised without funding.
References
- 1. World Health Organization . Weekly epidemiological update – 27 January 2021. [Accessed 31 January 2021]. Available from: https://www.who.int/publications/m/item/weekly-epidemiological-update-27-january-2021
- 2. Wu Z, McGoogan JM. Characteristics of and important lessons from the coronavirus disease 2019 (COVID‐19) outbreak in China: summary of a report of 72 314 cases from the Chinese Center for Disease Control and Prevention. JAMA 2020; 323 : 1239–1242. [DOI] [PubMed] [Google Scholar]
- 3. Velavan TP, Meyer CG. Mild versus severe COVID‐19: laboratory markers. Int J Infect Dis 2020; 95 : 304–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Williamson EJ, Walker AJ, Bhaskaran K, et al. Factors associated with COVID‐19‐related death using OpenSAFELY. Nature 2020; 584 : 430–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Binkhorst M, Offringa A, Hoeven J. COVID‐19: comprehensive synopsis of suggested pathophysiological mechanisms and repurposed drugs. Preprints 2020; 10.20944/preprints202007.0108.v1. [Not peer reviewed]. [DOI]
- 6. Ge X, Low B, Liang M, et al. Angiotensin II directly triggers endothelial exocytosis via protein kinase C‐dependent protein kinase D2 activation. J Pharmacol Sci 2007; 105 : 168–176. [DOI] [PubMed] [Google Scholar]
- 7. Hoffmann M, Kleine‐Weber H, Schroeder S, et al. SARS‐CoV‐2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020; 181 : 271–280.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Heurich A, Hofmann‐Winkler H, Gierer S, et al. TMPRSS2 and ADAM17 cleave ACE2 differentially and only proteolysis by TMPRSS2 augments entry driven by the severe acute respiratory syndrome coronavirus spike protein. J Virol 2014; 88 : 1293–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mahmoud IS, Jarrar YB, Alshaer W, et al. SARS‐CoV‐2 entry in host cells – multiple targets for treatment and prevention. Biochimie 2020; 175 : 93–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cantuti‐Castelvetri L, Ojha R, Pedro LD, et al. Neuropilin‐1 facilitates SARS‐CoV‐2 cell entry and infectivity. Science 2020; 370 : 856–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kielian M. Enhancing host cell infection by SARS‐CoV‐2. Science 2020; 370 : 765–766. [DOI] [PubMed] [Google Scholar]
- 12. Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell 2006; 124 : 783–801. [DOI] [PubMed] [Google Scholar]
- 13. Konno Y, Kimura I, Uriu K, et al. SARS‐CoV‐2 ORF3b is a potent interferon antagonist whose activity is increased by a naturally occurring elongation variant. Cell Rep 2020; 32 : 108185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Xia H, Cao Z, Xie X, et al. Evasion of type I interferon by SARS‐CoV‐2. Cell Rep 2020; 33 : 108234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lei X, Dong X, Ma R, et al. Activation and evasion of type I interferon responses by SARS‐CoV‐2. Nat Commun 2020; 11 : 3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tian W, Zhang N, Jin R, et al. Immune suppression in the early stage of COVID‐19 disease. Nat Commun 2020; 11 : 5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Blanco‐Melo D, Nilsson‐Payant BE, Liu WC, et al. Imbalanced host response to SARS‐CoV‐2 drives development of COVID‐19. Cell 2020; 181 : 1036–1045.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fox SE, Akmatbekov A, Harbert JL, et al. Pulmonary and cardiac pathology in African American patients with COVID‐19: an autopsy series from New Orleans. Lancet Respir Med 2020; 8 : 681–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Carsana L, Sonzogni A, Nasr A, et al. Pulmonary post‐mortem findings in a series of COVID‐19 cases from northern Italy: a two‐centre descriptive study. Lancet Infect Dis 2020; 20 : 1135–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Barnes BJ, Adrover JM, Baxter‐Stoltzfus A, et al. Targeting potential drivers of COVID‐19: neutrophil extracellular traps. J Exp Med 2020; 217 : e20200652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. van der Made CI, Simons A, Schuurs‐Hoeijmakers J, et al. Presence of genetic variants among young men with severe COVID‐19. JAMA 2020; 324 : 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Horby P, Mafham M, Linsell L, et al. Effect of hydroxychloroquine in hospitalized patients with Covid‐19. N Engl J Med 2020; 383 : 2030–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Abella BS, Jolkovsky EL, Biney BT, et al. Efficacy and safety of hydroxychloroquine vs placebo for pre‐exposure SARS‐CoV‐2 prophylaxis among health care workers: a randomized clinical trial. JAMA Intern Med 2021; 181 : 195–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Angelopoulou A, Alexandris N, Konstantinou E, et al. Imiquimod – a toll like receptor 7 agonist – is an ideal option for management of COVID 19. Environ Res 2020; 188 : 109858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang Q, Bastard P, Liu Z, et al. Inborn errors of type I IFN immunity in patients with life‐threatening COVID‐19. Science 2020; 370 : eabd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bastard P, Rosen LB, Zhang Q, et al. Auto‐antibodies against type I IFNs in patients with life‐threatening COVID‐19. Science 2020; 370 : eabd4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ziegler CGK, Allon SJ, Nyquist SK, et al. SARS‐CoV‐2 receptor ACE2 is an interferon‐stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell 2020; 181 : 1016–1035.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Onabajo OO, Banday AR, Stanifer ML, et al. Interferons and viruses induce a novel truncated ACE2 isoform and not the full‐length SARS‐CoV‐2 receptor. Nat Genet 2020; 52 : 1283–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mehta P, McAuley DF, Brown M, et al. COVID‐19: consider cytokine storm syndromes and immunosuppression. Lancet 2020; 395 : 1033–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Leisman DE, Ronner L, Pinotti R, et al. Cytokine elevation in severe and critical COVID‐19: a rapid systematic review, meta‐analysis, and comparison with other inflammatory syndromes. Lancet Respir Med 2020; 8 : 1233–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ricklin D, Hajishengallis G, Yang K, et al. Complement: a key system for immune surveillance and homeostasis. Nat Immunol 2010; 11 : 785–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Noris M, Benigni A, Remuzzi G. The case of complement activation in COVID‐19 multiorgan impact. Kidney Int 2020; 98 : 314–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Holter JC, Pischke SE, de Boer E, et al. Systemic complement activation is associated with respiratory failure in COVID‐19 hospitalized patients. Proc Natl Acad Sci U S A 2020; 117 : 25018–25025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gao T, Hu M, Zhang X, et al. Highly pathogenic coronavirus N protein aggravates lung injury by MASP‐2‐mediated complement over‐activation. medRxiv 2020. 10.1101/2020.03.29.20041962. [Not peer reviewed]. [DOI] [Google Scholar]
- 35. Magro C, Mulvey JJ, Berlin D, et al. Complement associated microvascular injury and thrombosis in the pathogenesis of severe COVID‐19 infection: a report of five cases. Transl Res 2020; 220 : 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Diao B, Wang C, Wang R, et al. Human kidney is a target for novel severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) infection. medRxiv 2020. 10.1101/2020.03.04.20031120. [Not peer reviewed]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nakazawa D, Ishizu A. Immunothrombosis in severe COVID‐19. EBioMedicine 2020; 59 : 102942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Engelmann B, Massberg S. Thrombosis as an intravascular effector of innate immunity. Nat Rev Immunol 2013; 13 : 34–45. [DOI] [PubMed] [Google Scholar]
- 39. Middleton EA, He XY, Denorme F, et al. Neutrophil extracellular traps contribute to immunothrombosis in COVID‐19 acute respiratory distress syndrome. Blood 2020; 136 : 1169–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schurink B, Roos E, Radonic T, et al. Viral presence and immunopathology in patients with lethal COVID‐19: a prospective autopsy cohort study. Lancet Microbe 2020; 1 : e290–e299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zuo Y, Yalavarthi S, Shi H, et al. Neutrophil extracellular traps in COVID‐19. JCI Insight 2020; 5 : e138999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhang X, Tan Y, Ling Y, et al. Viral and host factors related to the clinical outcome of COVID‐19. Nature 2020; 583 : 437–440. [DOI] [PubMed] [Google Scholar]
- 43. Kurche JS, Haluszczak C, McWilliams JA, et al. Type I IFN‐dependent T cell activation is mediated by IFN‐dependent dendritic cell OX40 ligand expression and is independent of T cell IFNR expression. J Immunol 2012; 188 : 585–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Helal MA, Shouman S, Abdelwaly A, et al. Molecular basis of the potential interaction of SARS‐CoV‐2 spike protein to CD147 in COVID‐19 associated‐lymphopenia. J Biomol Struct Dyn 2020. 10.1080/07391102.2020.1822208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Moon C. Fighting COVID‐19 exhausts T cells. Nat Rev Immunol 2020; 20 : 277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nienhold R, Ciani Y, Koelzer VH, et al. Two distinct immunopathological profiles in autopsy lungs of COVID‐19. Nat Commun 2020; 11 : 5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. van de Veerdonk FL, Janssen NAF, Grondman I, et al. A systems approach to inflammation identifies therapeutic targets in SARS‐CoV‐2 infection. medRxiv 2020. 10.1101/2020.05.23.20110916. [Not peer reviewed]. [DOI] [Google Scholar]
- 48. Liu Q, Shi Y, Cai J, et al. Pathological changes in the lungs and lymphatic organs of twelve COVID‐19 autopsy cases. Natl Sci Rev 2020; 7 : 1868–1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bradley BT, Maioli H, Johnston R, et al. Histopathology and ultrastructural findings of fatal COVID‐19 infections in Washington state: a case series. Lancet 2020; 396 : 320–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kaneko N, Kuo HH, Boucau J, et al. Loss of Bcl‐6‐expressing T follicular helper cells and germinal centers in COVID‐19. Cell 2020; 183 : 143–157.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tillett RL, Sevinsky JR, Hartley PD, et al. Genomic evidence for reinfection with SARS‐CoV‐2: a case study. Lancet Infect Dis 2021; 21 : 52–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wajnberg A, Amanat F, Firpo A, et al. Robust neutralizing antibodies to SARS‐CoV‐2 infection persist for months. Science 2020; 370 : 1227–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zuo J, Dowell A, Pearce H, et al. Robust SARS‐CoV‐2‐specific T‐cell immunity is maintained at 6 months following primary infection. bioRxiv 2020. 10.1101/2020.11.01.362319. [Not peer reviewed]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Rydyznski Moderbacher C, Ramirez SI, Dan JM, et al. Antigen‐specific adaptive immunity to SARS‐CoV‐2 in acute COVID‐19 and associations with age and disease severity. Cell 2020; 183 : 996–1012.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Duffy L, O'Reilly SC. Toll‐like receptors in the pathogenesis of autoimmune diseases: recent and emerging translational developments. Immunotargets Ther 2016; 5 : 69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Farrugia M, Baron B. The role of toll‐like receptors in autoimmune diseases through failure of the self‐recognition mechanism. Int J Inflam 2017; 2017 : 8391230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Abrams JY, Godfred‐Cato SE, Oster ME, et al. Multisystem inflammatory syndrome in children associated with severe acute respiratory syndrome coronavirus 2: a systematic review. J Pediatr 2020; 226 : 45–54.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Carter MJ, Shankar‐Hari M, Tibby SM. Paediatric inflammatory multisystem syndrome temporally‐associated with SARS‐CoV‐2 infection: an overview. Intensive Care Med 2020; 47 : 90–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Carter MJ, Fish M, Jennings A, et al. Peripheral immunophenotypes in children with multisystem inflammatory syndrome associated with SARS‐CoV‐2 infection. Nat Med 2020; 26 : 1701–1707. [DOI] [PubMed] [Google Scholar]
- 60. Gruber CN, Patel RS, Trachtman R, et al. Mapping systemic inflammation and antibody responses in multisystem inflammatory syndrome in children (MIS‐C). Cell 2020; 183 : 982–995.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Consiglio CR, Cotugno N, Sardh F, et al. The immunology of multisystem inflammatory syndrome in children with COVID‐19. Cell 2020; 183 : 968–981.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Morris SB, Schwartz N, Patel P, et al. Case Series of Multisystem Inflammatory Syndrome in Adults Associated with SARS‐CoV‐2 Infection – United Kingdom and United States, March–August 2020. MMWR Morb Mortal Wkly Rep 2020; 69 : 1450–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zulfiqar AA, Lorenzo‐Villalba N, Hassler P, et al. Immune thrombocytopenic purpura in a patient with Covid‐19. N Engl J Med 2020; 382 : e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Bomhof G, Mutsaers P, Leebeek FWG, et al. COVID‐19‐associated immune thrombocytopenia. Br J Haematol 2020; 190 : e61–e64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zhang Y, Xiao M, Zhang S, et al. Coagulopathy and antiphospholipid antibodies in patients with Covid‐19. N Engl J Med 2020; 382 : e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Cárdenas Suri H, Jimomila Bening D. Catastrophic antiphospholipid antibody syndrome and multiple organ dysfunctions in critically ill patients with COVID‐19. Expert Rev Respir Med 2020; 14 : 1071–1072. [DOI] [PubMed] [Google Scholar]
- 67. Hindilerden F, Yonal‐Hindilerden I, Akar E, et al. Covid‐19 associated autoimmune thrombotic thrombocytopenic purpura: report of a case. Thromb Res 2020; 195 : 136–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Miyakis S, Lockshin MD, Atsumi T, et al. International consensus statement on an update of the classification criteria for definite antiphospholipid syndrome (APS). J Thromb Haemost 2006; 4 : 295–306. [DOI] [PubMed] [Google Scholar]
- 69. Cervera R, Rodríguez‐Pintó I, Espinosa G. The diagnosis and clinical management of the catastrophic antiphospholipid syndrome: a comprehensive review. J Autoimmun 2018; 92 : 1–11. [DOI] [PubMed] [Google Scholar]
- 70. Bowles L, Platton S, Yartey N, et al. Lupus anticoagulant and abnormal coagulation tests in patients with Covid‐19. N Engl J Med 2020; 383 : 288–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Reyes Gil M, Barouqa M, Szymanski J, et al. Assessment of lupus anticoagulant positivity in patients with coronavirus disease 2019 (COVID‐19). JAMA Netw Open 2020; 3 : e2017539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Zuo Y, Estes SK, Ali RA, et al. Prothrombotic autoantibodies in serum from patients hospitalized with COVID‐19. Sci Transl Med 2020; 12 : eabd3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Devreese KMJ, Linskens EA, Benoit D, et al. Antiphospholipid antibodies in patients with COVID‐19: a relevant observation? J Thromb Haemost 2020; 18 : 2191–2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Prinz N, Clemens N, Strand D, et al. Antiphospholipid antibodies induce translocation of TLR7 and TLR8 to the endosome in human monocytes and plasmacytoid dendritic cells. Blood 2011; 118 : 2322–2332. [DOI] [PubMed] [Google Scholar]
- 75. Lopes da Silva R. Viral‐associated thrombotic microangiopathies. Hematol Oncol Stem Cell Ther 2011; 4 : 51–59. [DOI] [PubMed] [Google Scholar]
- 76. Woodruff MC, Ramonell RP, Nguyen DC, et al. Extrafollicular B cell responses correlate with neutralizing antibodies and morbidity in COVID‐19. Nat Immunol 2020; 21 : 1506–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Casciola‐Rosen L, Thiemann DR, Andrade F, et al. IgM autoantibodies recognizing ACE2 are associated with severe COVID‐19. medRxiv 2020. 10.1101/2020.10.13.20211664. [Not peer reviewed]. [DOI] [Google Scholar]
- 78. Bhadelia N, Belkina AC, Olson A, et al. Distinct autoimmune antibody signatures between hospitalized acute COVID‐19 patients, SARS‐CoV‐2 convalescent individuals, and unexposed pre‐pandemic controls. medRxiv 2021. 10.1101/2021.01.21.21249176. [Not peer reviewed]. [DOI] [Google Scholar]
- 79. Woodruff MC, Ramonell RP, Lee FE‐H, et al. Clinically identifiable autoreactivity is common in severe SARS‐CoV‐2 infection. medRxiv 2020. 10.1101/2020.10.21.20216192. [Not peer reviewed]. [DOI] [Google Scholar]
- 80. Wang EY, Mao T, Klein J, et al. Diverse functional autoantibodies in patients with COVID‐19. medRxiv 2020; 10.1101/2020.12.10.20247205. [Not peer reviewed]. [DOI] [PubMed]
- 81. The Lancet Rheumatology . High‐stakes heterogeneity in COVID‐19. Lancet Rheumatol 2020; 2 : e577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Mueller AL, McNamara MS, Sinclair DA. Why does COVID‐19 disproportionately affect older people? Aging (Albany NY) 2020; 12 : 9959–9981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Zhavoronkov A. Geroprotective and senoremediative strategies to reduce the comorbidity, infection rates, severity, and lethality in gerophilic and gerolavic infections. Aging (Albany NY) 2020; 12 : 6492–6510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Hewitt J, Carter B, Vilches‐Moraga A, et al. The effect of frailty on survival in patients with COVID‐19 (COPE): a multicentre, European, observational cohort study. Lancet Public Health 2020; 5 : e444–e451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Takahashi T, Ellingson MK, Wong P, et al. Sex differences in immune responses that underlie COVID‐19 disease outcomes. Nature 2020; 588 : 315–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Mateus J, Grifoni A, Tarke A, et al. Selective and cross‐reactive SARS‐CoV‐2 T cell epitopes in unexposed humans. Science 2020; 370 : 89–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Nguyen‐Contant P, Embong AK, Kanagaiah P, et al. S protein‐reactive IgG and memory B cell production after human SARS‐CoV‐2 infection includes broad reactivity to the S2 subunit. MBio 2020; 11 : e01991‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Ng KW, Faulkner N, Cornish GH, et al. Preexisting and de novo humoral immunity to SARS‐CoV‐2 in humans. Science 2020; 370 : 1339–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Grifoni A, Weiskopf D, Ramirez SI, et al. Targets of T cell responses to SARS‐CoV‐2 coronavirus in humans with COVID‐19 disease and unexposed individuals. Cell 2020; 181 : 1489–1501.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. van Zelst CM, Janssen ML, Pouw N, et al. Analyses of abdominal adiposity and metabolic syndrome as risk factors for respiratory distress in COVID‐19. BMJ Open Respir Res 2020; 7 : e000792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. van der Voort PHJ, Moser J, Zandstra DF, et al. Leptin levels in SARS‐CoV‐2 infection related respiratory failure: a cross‐sectional study and a pathophysiological framework on the role of fat tissue. Heliyon 2020; 6 : e04696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Szkudelska K, Nogowski L, Szkudelski T. The inhibitory effect of resveratrol on leptin secretion from rat adipocytes. Eur J Clin Invest 2009; 39 : 899–905. [DOI] [PubMed] [Google Scholar]
- 93. van de Veerdonk FL, Netea MG, van Deuren M, et al. Kallikrein–kinin blockade in patients with COVID‐19 to prevent acute respiratory distress syndrome. Elife 2020; 9 : e57555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Hamming I, Cooper ME, Haagmans BL, et al. The emerging role of ACE2 in physiology and disease. J Pathol 2007; 212 : 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. de Queiroz TM, Lakkappa N, Lazartigues E. ADAM17‐mediated shedding of inflammatory cytokines in hypertension. Front Pharmacol 2020; 11 : 1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Lambert DW, Yarski M, Warner FJ, et al. Tumor necrosis factor‐alpha convertase (ADAM17) mediates regulated ectodomain shedding of the severe‐acute respiratory syndrome‐coronavirus (SARS‐CoV) receptor, angiotensin‐converting enzyme‐2 (ACE2). J Biol Chem 2005; 280 : 30113–30119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Narula S, Yusuf S, Chong M, et al. Plasma ACE2 and risk of death or cardiometabolic diseases: a case‐cohort analysis. Lancet 2020; 396 : 968–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Sama IE, Ravera A, Santema BT, et al. Circulating plasma concentrations of angiotensin‐converting enzyme 2 in men and women with heart failure and effects of renin–angiotensin–aldosterone inhibitors. Eur Heart J 2020; 41 : 1810–1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Swärd P, Edsfeldt A, Reepalu A, et al. Age and sex differences in soluble ACE2 may give insights for COVID‐19. Crit Care 2020; 24 : 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Patel SK, Juno JA, Lee WS, et al. Plasma ACE2 activity is persistently elevated following SARS‐CoV‐2 infection: implications for COVID‐19 pathogenesis and consequences. medRxiv 2020. 10.1101/2020.10.06.20207514. [Not peer reviewed]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. McMillan P, Uhal BD. COVID‐19 – a theory of autoimmunity to ACE‐2. MOJ Immunol 2020; 7 : 17–19. [PMC free article] [PubMed] [Google Scholar]
- 102. Monteil V, Kwon H, Prado P, et al. Inhibition of SARS‐CoV‐2 infections in engineered human tissues using clinical‐grade soluble human ACE2. Cell 2020; 181 : 905–913.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Zoufaly A, Poglitsch M, Aberle JH, et al. Human recombinant soluble ACE2 in severe COVID‐19. Lancet Respir Med; 8 : 1154–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Ellinghaus D, Degenhardt F, Bujanda L, et al. Genomewide association study of severe Covid‐19 with respiratory failure. N Engl J Med 2020; 383 : 1522–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Derosa L, Melenotte C, Griscelli F, et al. The immuno‐oncological challenge of COVID‐19. Nature Cancer 2020; 1 : 946–964. [DOI] [PubMed] [Google Scholar]
- 106. Briguglio M, Pregliasco FE, Lombardi G, et al. The malnutritional status of the host as a virulence factor for new coronavirus SARS‐CoV‐2. Front Med (Lausanne) 2020; 7 : 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Sungnak W, Huang N, Bécavin C, et al. SARS‐CoV‐2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat Med 2020; 26 : 681–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Bourgonje AR, Abdulle AE, Timens W, et al. Angiotensin‐converting enzyme 2 (ACE2), SARS‐CoV‐2 and the pathophysiology of coronavirus disease 2019 (COVID‐19). J Pathol 2020; 251 : 228–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Dos Santos WG. Natural history of COVID‐19 and current knowledge on treatment therapeutic options. Biomed Pharmacother 2020; 129 : 110493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Borczuk AC, Salvatore SP, Seshan SV, et al. COVID‐19 pulmonary pathology: a multi‐institutional autopsy cohort from Italy and New York City. Mod Pathol 2020; 33 : 2156–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Kory P, Kanne JP. SARS‐CoV‐2 organising pneumonia: ‘Has there been a widespread failure to identify and treat this prevalent condition in COVID‐19?’. BMJ Open Respir Res 2020; 7 : e000724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Polak SB, Van Gool IC, Cohen D, et al. A systematic review of pathological findings in COVID‐19: a pathophysiological timeline and possible mechanisms of disease progression. Mod Pathol 2020; 33 : 2128–2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Schaefer IM, Padera RF, Solomon IH, et al. In situ detection of SARS‐CoV‐2 in lungs and airways of patients with COVID‐19. Mod Pathol 2020; 33 : 2104–2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Dorward DA, Russell CD, Um IH, et al. Tissue‐specific immunopathology in fatal COVID‐19. Am J Respir Crit Care Med 2021; 203 : 192–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Ackermann M, Verleden SE, Kuehnel M, et al. Pulmonary vascular endothelialitis, thrombosis, and angiogenesis in Covid‐19. N Engl J Med 2020; 383 : 120–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Varga Z, Flammer AJ, Steiger P, et al. Endothelial cell infection and endotheliitis in COVID‐19. Lancet 2020; 395 : 1417–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. McGonagle D, O'Donnell JS, Sharif K, et al. Immune mechanisms of pulmonary intravascular coagulopathy in COVID‐19 pneumonia. Lancet Rheumatol 2020; 2 : e437–e445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Borczuk AC. Pulmonary pathology of COVID‐19: a review of autopsy studies. Curr Opin Pulm Med 2021; 27 : 184–192. [DOI] [PubMed] [Google Scholar]
- 119. Cruces P, Retamal J, Hurtado DE, et al. A physiological approach to understand the role of respiratory effort in the progression of lung injury in SARS‐CoV‐2 infection. Crit Care 2020; 24 : 494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Shi S, Qin M, Shen B, et al. Association of cardiac injury with mortality in hospitalized patients with COVID‐19 in Wuhan, China. JAMA Cardiol 2020; 5 : 802–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Inciardi RM, Adamo M, Lupi L, et al. Characteristics and outcomes of patients hospitalized for COVID‐19 and cardiac disease in Northern Italy. Eur Heart J 2020; 41 : 1821–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Hu H, Ma F, Wei X, et al. Coronavirus fulminant myocarditis treated with glucocorticoid and human immunoglobulin. Eur Heart J 2021; 42 : 206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Kesici S, Aykan HH, Orhan D, et al. Fulminant COVID‐19‐related myocarditis in an infant. Eur Heart J 2020; 41 : 3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Dolhnikoff M, Ferreira Ferranti J, de Almeida Monteiro RA, et al. SARS‐CoV‐2 in cardiac tissue of a child with COVID‐19‐related multisystem inflammatory syndrome. Lancet Child Adolesc Health 2020; 4 : 790–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Rajpal S, Tong MS, Borchers J, et al. Cardiovascular magnetic resonance findings in competitive athletes recovering from COVID‐19 infection. JAMA Cardiol 2020; 6 : 116–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Puntmann VO, Carerj ML, Wieters I, et al. Outcomes of cardiovascular magnetic resonance imaging in patients recently recovered from coronavirus disease 2019 (COVID‐19). JAMA Cardiol 2020; 5 : 1265–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Tschöpe C, Ammirati E, Bozkurt B, et al. Myocarditis and inflammatory cardiomyopathy: current evidence and future directions. Nat Rev Cardiol 2021; 18 169–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Nicin L, Abplanalp WT, Mellentin H, et al. Cell type‐specific expression of the putative SARS‐CoV‐2 receptor ACE2 in human hearts. Eur Heart J 2020; 41 : 1804–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Lindner D, Fitzek A, Bräuninger H, et al. Association of cardiac infection with SARS‐CoV‐2 in confirmed COVID‐19 autopsy cases. JAMA Cardiol 2020; 5 : 1281–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Tavazzi G, Pellegrini C, Maurelli M, et al. Myocardial localization of coronavirus in COVID‐19 cardiogenic shock. Eur J Heart Fail 2020; 22 : 911–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Basso C, Leone O, Rizzo S, et al. Pathological features of COVID‐19‐associated myocardial injury: a multicentre cardiovascular pathology study. Eur Heart J 2020; 41 : 3827–3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Creel‐Bulos C, Hockstein M, Amin N, et al. Acute cor pulmonale in critically ill patients with Covid‐19. N Engl J Med 2020; 382 : e70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Nan J, Jin YB, Myo Y, et al. Hypoxia in acute cardiac injury of coronavirus disease 2019: lesson learned from pathological studies. J Geriatr Cardiol 2020; 17 : 221–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Duarte‐Neto AN, de Almeida Monteiro RA, da Silva LFF, et al. Pulmonary and systemic involvement of COVID‐19 assessed by ultrasound‐guided minimally invasive autopsy. Histopathology 2020; 77 : 186–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Mao L, Jin H, Wang M, et al. Neurologic manifestations of hospitalized patients with coronavirus disease 2019 in Wuhan, China. JAMA Neurol 2020; 77 : 683–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Paterson RW, Brown RL, Benjamin L, et al. The emerging spectrum of COVID‐19 neurology: clinical, radiological and laboratory findings. Brain 2020; 143 : 3104–3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Hampshire A, Trender W, Chamberlain SR, et al. Cognitive deficits in people who have recovered from COVID‐19 relative to controls: an N=84,285 online study. medRxiv 2020. 10.1101/2020.10.20.20215863. [Not peer reviewed]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Solomon IH, Normandin E, Bhattacharyya S, et al. Neuropathological features of Covid‐19. N Engl J Med 2020; 383 : 989–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Kantonen J, Mahzabin S, Mäyränpää MI, et al. Neuropathologic features of four autopsied COVID‐19 patients. Brain Pathol 2020; 30 : 1012–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Moriguchi T, Harii N, Goto J, et al. A first case of meningitis/encephalitis associated with SARS‐Coronavirus‐2. Int J Infect Dis 2020; 94 : 55–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Huang YH, Jiang D, Huang JT. SARS‐CoV‐2 detected in cerebrospinal fluid by PCR in a case of COVID‐19 encephalitis. Brain Behav Immun 2020; 87 : 149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Virhammar J, Kumlien E, Fällmar D, et al. Acute necrotizing encephalopathy with SARS‐CoV‐2 RNA confirmed in cerebrospinal fluid. Neurology 2020; 95 : 445–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Puelles VG, Lütgehetmann M, Lindenmeyer MT, et al. Multiorgan and renal tropism of SARS‐CoV‐2. N Engl J Med 2020; 383 : 590–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Paniz‐Mondolfi A, Bryce C, Grimes Z, et al. Central nervous system involvement by severe acute respiratory syndrome coronavirus‐2 (SARS‐CoV‐2). J Med Virol 2020; 92 : 699–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Matschke J, Lütgehetmann M, Hagel C, et al. Neuropathology of patients with COVID‐19 in Germany: a post‐mortem case series. Lancet Neurol 2020; 19 : 919–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Iadecola C, Anrather J, Kamel H. Effects of COVID‐19 on the nervous system. Cell 2020; 183 : 16–27.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Morbini P, Benazzo M, Verga L, et al. Ultrastructural evidence of direct viral damage to the olfactory complex in patients testing positive for SARS‐CoV‐2. JAMA Otolaryngol Head Neck Surg 2020; 146 : 972–973. [DOI] [PubMed] [Google Scholar]
- 148. Mercante G, Ferreli F, De Virgilio A, et al. Prevalence of taste and smell dysfunction in coronavirus disease 2019. JAMA Otolaryngol Head Neck Surg 2020; 146 : 723–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Bilinska K, Jakubowska P, Von Bartheld CS, et al. Expression of the SARS‐CoV‐2 entry proteins, ACE2 and TMPRSS2, in cells of the olfactory epithelium: identification of cell types and trends with age. ACS Chem Nerosci 2020; 11 : 1555–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Yan Y, Diao B, Liu Y, et al. Severe acute respiratory syndrome coronavirus 2 nucleocapsid protein in the ocular tissues of a patient previously infected with coronavirus disease 2019. JAMA Ophthalmol 2020; 138 : 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Netland J, Meyerholz DK, Moore S, et al. Severe acute respiratory syndrome coronavirus infection causes neuronal death in the absence of encephalitis in mice transgenic for human ACE2. J Virol 2008; 82 : 7264–7275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Pellegrini L, Albecka A, Mallery DL, et al. SARS‐CoV‐2 infects the brain choroid plexus and disrupts the blood–CSF barrier in human brain organoids. Cell Stem Cell 2020; 27 : 951–961.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Sulzer D, Antonini A, Leta V, et al. COVID‐19 and possible links with Parkinson's disease and parkinsonism: from bench to bedside. NPJ Parkinson's Dis 2020; 6 : 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Lema Tomé CM, Tyson T, Rey NL, et al. Inflammation and α‐synuclein's prion‐like behavior in Parkinson's disease – is there a link? Mol Neurobiol 2013; 47 : 561–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Pavel A, Murray DK, Stoessl AJ. COVID‐19 and selective vulnerability to Parkinson's disease. Lancet Neurol 2020; 19 : 719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Cheng Y, Luo R, Wang K, et al. Kidney disease is associated with in‐hospital death of patients with COVID‐19. Kidney Int 2020; 97 : 829–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Sharma P, Uppal NN, Wanchoo R, et al. COVID‐19‐associated kidney injury: a case series of kidney biopsy findings. J Am Soc Nephrol 2020; 31 : 1948–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Hamming I, Timens W, Bulthuis ML, et al. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol 2004; 203 : 631–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Nadim MK, Forni LG, Mehta RL, et al. COVID‐19‐associated acute kidney injury: consensus report of the 25th Acute Disease Quality Initiative (ADQI) Workgroup. Nat Rev Nephrol 2020; 16 : 747–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Farkash EA, Wilson AM, Jentzen JM. Ultrastructural evidence for direct renal infection with SARS‐CoV‐2. J Am Soc Nephrol 2020; 31 : 1683–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Menter T, Haslbauer JD, Nienhold R, et al. Postmortem examination of COVID‐19 patients reveals diffuse alveolar damage with severe capillary congestion and variegated findings in lungs and other organs suggesting vascular dysfunction. Histopathology 2020; 77 : 198–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Su H, Yang M, Wan C, et al. Renal histopathological analysis of 26 postmortem findings of patients with COVID‐19 in China. Kidney Int 2020; 98 : 219–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Calomeni E, Satoskar A, Ayoub I, et al. Multivesicular bodies mimicking SARS‐CoV‐2 in patients without COVID‐19. Kidney Int 2020; 98 : 233–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Miller SE, Brealey JK. Visualization of putative coronavirus in kidney. Kidney Int 2020; 98 : 231–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Braun F, Lütgehetmann M, Pfefferle S, et al. SARS‐CoV‐2 renal tropism associates with acute kidney injury. Lancet 2020; 396 : 597–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Kudose S, Batal I, Santoriello D, et al. Kidney biopsy findings in patients with COVID‐19. J Am Soc Nephrol 2020; 31 : 1959–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Golmai P, Larsen CP, DeVita MV, et al. Histopathologic and ultrastructural findings in postmortem kidney biopsy material in 12 patients with AKI and COVID‐19. J Am Soc Nephrol 2020; 31 : 1944–1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Santoriello D, Khairallah P, Bomback AS, et al. Postmortem kidney pathology findings in patients with COVID‐19. J Am Soc Nephrol 2020; 31 : 2158–2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Peleg Y, Kudose S, D'Agati V, et al. Acute kidney injury due to collapsing glomerulopathy following COVID‐19 infection. Kidney Int Rep 2020; 5 : 940–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Larsen CP, Bourne TD, Wilson JD, et al. Collapsing glomerulopathy in a patient with COVID‐19. Kidney Int Rep 2020; 5 : 935–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Kissling S, Rotman S, Gerber C, et al. Collapsing glomerulopathy in a COVID‐19 patient. Kidney Int 2020; 98 : 228–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Jin X, Lian JS, Hu JH, et al. Epidemiological, clinical and virological characteristics of 74 cases of coronavirus‐infected disease 2019 (COVID‐19) with gastrointestinal symptoms. Gut 2020; 69 : 1002–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Cheung KS, Hung IFN, Chan PPY, et al. Gastrointestinal manifestations of SARS‐CoV‐2 infection and virus load in fecal samples from a Hong Kong cohort: systematic review and meta‐analysis. Gastroenterology 2020; 159 : 81–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Zhang H, Kang Z, Gong H, et al. Digestive system is a potential route of COVID‐19: an analysis of single‐cell coexpression pattern of key proteins in viral entry process. Gut 2020; 69 : 1010–1018. [Google Scholar]
- 175. Lamers MM, Beumer J, van der Vaart J, et al. SARS‐CoV‐2 productively infects human gut enterocytes. Science 2020; 369 : 50–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Zhou J, Li C, Liu X, et al. Infection of bat and human intestinal organoids by SARS‐CoV‐2. Nat Med 2020; 26 : 1077–1083. [DOI] [PubMed] [Google Scholar]
- 177. Zang R, Gomez Castro MF, McCune BT, et al. TMPRSS2 and TMPRSS4 promote SARS‐CoV‐2 infection of human small intestinal enterocytes. Sci Immunol 2020; 5 : eabc3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Sun SH, Chen Q, Gu HJ, et al. A mouse model of SARS‐CoV‐2 infection and pathogenesis. Cell Host Microbe 2020; 28 : 124–133.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Munster VJ, Feldmann F, Williamson BN, et al. Respiratory disease in rhesus macaques inoculated with SARS‐CoV‐2. Nature 2020; 585 : 268–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Xiao F, Tang M, Zheng X, et al. Evidence for gastrointestinal infection of SARS‐CoV‐2. Gastroenterology 2020; 158 : 1831–1833.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Wu Y, Guo C, Tang L, et al. Prolonged presence of SARS‐CoV‐2 viral RNA in faecal samples. Lancet Gastroenterol Hepatol 2020; 5 : 434–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Yang L, Tu L. Implications of gastrointestinal manifestations of COVID‐19. Lancet Gastroenterol Hepatol 2020; 5 : 629–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Su S, Shen J, Zhu L, et al. Involvement of digestive system in COVID‐19: manifestations, pathology, management and challenges. Therap Adv Gastroenterol 2020; 13 : 1756284820934626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Yao XH, Li TY, He ZC, et al. A pathological report of three COVID‐19 cases by minimal invasive autopsies. Zhonghua Bing Li Xue Za Zhi 2020; 49 : 411–417. [DOI] [PubMed] [Google Scholar]
- 185. Deshmukh V, Motwani R, Kumar A, et al. Histopathological observations in COVID‐19: a systematic review. J Clin Pathol 2021; 74 : 76–83. [DOI] [PubMed] [Google Scholar]
- 186. Mao R, Qiu Y, He JS, et al. Manifestations and prognosis of gastrointestinal and liver involvement in patients with COVID‐19: a systematic review and meta‐analysis. Lancet Gastroenterol Hepatol 2020; 5 : 667–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Li Y, Xiao SY. Hepatic involvement in COVID‐19 patients: pathology, pathogenesis, and clinical implications. J Med Virol 2020; 92 : 1491–1494. [DOI] [PubMed] [Google Scholar]
- 188. Chai X, Hu L, Zhang Y, et al. Specific ACE2 expression in cholangiocytes may cause liver damage after 2019‐nCoV infection. bioRxiv 2020. 10.1101/2020.02.03.931766. [Not peer reviewed]. [DOI] [Google Scholar]
- 189. Guan GW, Gao L, Wang JW, et al. [Exploring the mechanism of liver enzyme abnormalities in patients with novel coronavirus‐infected pneumonia]. Zhonghua Gan Zang Bing Za Zhi 2020; 28 : 100–106. [DOI] [PubMed] [Google Scholar]
- 190. Lagana SM, Kudose S, Iuga AC, et al. Hepatic pathology in patients dying of COVID‐19: a series of 40 cases including clinical, histologic, and virologic data. Mod Pathol 2020; 33 : 2147–2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Tian S, Xiong Y, Liu H, et al. Pathological study of the 2019 novel coronavirus disease (COVID‐19) through postmortem core biopsies. Mod Pathol 2020; 33 : 1007–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Gu J, Han B, Wang J. COVID‐19: gastrointestinal manifestations and potential fecal–oral transmission. Gastroenterology 2020; 158 : 1518–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Zhang C, Shi L, Wang FS. Liver injury in COVID‐19: management and challenges. Lancet Gastroenterol Hepatol 2020; 5 : 428–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Li D, Jin M, Bao P, et al. Clinical characteristics and results of semen tests among men with coronavirus disease 2019. JAMA Netw Open 2020; 3 : e208292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Wang Z, Xu X. scRNA‐seq profiling of human testes reveals the presence of the ACE2 receptor, a target for SARS‐CoV‐2 infection in spermatogonia, Leydig and Sertoli cells. Cell 2020; 9 : 920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. The Human Protein Atlas . Tissue expression of ACE2; epididymis. [Accessed 29 November 2020]. Available from: https://www.proteinatlas.org/ENSG00000130234-ACE2/tissue/epididymis
- 197. Yang M, Chen S, Huang B, et al. Pathological findings in the testes of COVID‐19 patients: clinical implications. Eur Urol Focus 2020; 6 : 1124–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Qiu L, Liu X, Xiao M, et al. SARS‐CoV‐2 is not detectable in the vaginal fluid of women with severe COVID‐19 infection. Clin Infect Dis 2020; 71 : 813–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Scorzolini L, Corpolongo A, Castilletti C, et al. Comment on the potential risks of sexual and vertical transmission of coronavirus disease‐19 infection 2. Clin Infect Dis 2020; 71 : 2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Jing Y, Run‐Qian L, Hao‐Ran W, et al. Potential influence of COVID‐19/ACE2 on the female reproductive system. Mol Hum Reprod 2020; 26 : 367–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Vivanti AJ, Vauloup‐Fellous C, Prevot S, et al. Transplacental transmission of SARS‐CoV‐2 infection. Nat Commun 2020; 11 : 3572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Fenizia C, Biasin M, Cetin I, et al. Analysis of SARS‐CoV‐2 vertical transmission during pregnancy. Nat Commun 2020; 11 : 5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Khalil A, Kalafat E, Benlioglu C, et al. SARS‐CoV‐2 infection in pregnancy: a systematic review and meta‐analysis of clinical features and pregnancy outcomes. EClinicalMedicine 2020; 25 : 100446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Freeman EE, McMahon DE, Lipoff JB, et al. The spectrum of COVID‐19‐associated dermatologic manifestations: an international registry of 716 patients from 31 countries. J Am Acad Dermatol 2020; 83 : 1118–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Kanitakis J, Lesort C, Danset M, et al. Chilblain‐like acral lesions during the COVID‐19 pandemic (“COVID toes”): histologic, immunofluorescence, and immunohistochemical study of 17 cases. J Am Acad Dermatol 2020; 83 : 870–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Colmenero I, Santonja C, Alonso‐Riaño M, et al. SARS‐CoV‐2 endothelial infection causes COVID‐19 chilblains: histopathological, immunohistochemical and ultrastructural study of seven paediatric cases. Br J Dermatol 2020; 183 : 729–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Damsky W, Peterson D, King B. When interferon tiptoes through COVID‐19: Pernio‐like lesions and their prognostic implications during SARS‐CoV‐2 infection. J Am Acad Dermatol 2020; 83 : e269–e270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Droesch C, Hoang M, DeSancho M, et al. Livedoid and purpuric skin eruptions associated with coagulopathy in severe COVID‐19. JAMA Dermatol 2020; 156 : 1022–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Beigel JH, Tomashek KM, Dodd LE, et al. Remdesivir for the treatment of Covid‐19 – final report. N Engl J Med 2020; 383 : 1813–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. Grein J, Ohmagari N, Shin D, et al. Compassionate use of remdesivir for patients with severe Covid‐19. N Engl J Med 2020; 382 : 2327–2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. RECOVERY Collaborative Group . Lopinavir–ritonavir in patients admitted to hospital with COVID‐19 (RECOVERY): a randomised, controlled, open‐label, platform trial. Lancet 2020; 396 : 1345–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Momekov G, Momekova D. Ivermectin as a potential COVID‐19 treatment from the pharmacokinetic point of view: antiviral levels are not likely attainable with known dosing regimens. Biotechnol Biotechnol Equip 2020; 34 : 469–474. [Google Scholar]
- 213. Caly L, Druce JD, Catton MG, et al. The FDA‐approved drug ivermectin inhibits the replication of SARS‐CoV‐2 in vitro . Antiviral Res 2020; 178 : 104787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214. Heidary F, Gharebaghi R. Ivermectin: a systematic review from antiviral effects to COVID‐19 complementary regimen. J Antibiot (Tokyo) 2020; 73 : 593–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Zhang X, Song Y, Ci X, et al. Ivermectin inhibits LPS‐induced production of inflammatory cytokines and improves LPS‐induced survival in mice. Inflamm Res 2008; 57 : 524–529. [DOI] [PubMed] [Google Scholar]
- 216. Bryant A, Lawrie TA, Dowswell T, et al. Ivermectin for prevention and treatment of COVID‐19 infection: a systematic review and meta‐analysis. OSF Preprints 2021; 10.31219/osf.io/k37ft. [Not peer reviewed]. [DOI] [PMC free article] [PubMed]
- 217. Kory P, Meduri G, Glesias J, et al. Review of the emerging evidence demonstrating the efficacy of ivermectin in the prophylaxis and treatment of COVID‐19. OSF Preprints 2021; 10.31219/osf.io/wx3zn. [Not peer reviewed]. [DOI] [PMC free article] [PubMed]
- 218. Hill A, Abdulamir A, Ahmed S, et al. Meta‐analysis of randomized trials of ivermectin to treat SARS‐CoV‐2 infection. Research Square Preprint 2021; 10.21203/rs.3.rs-148845/v1. [Not peer reviewed]. [DOI]
- 219. Castañeda‐Sabogal A, Chambergo‐Michilot D, Toro‐Huamanchumo CJ, et al. Outcomes of ivermectin in the treatment of COVID‐19: a systematic review and meta‐analysis. medRxiv 2021. 10.1101/2021.01.26.21250420. [Not peer reviewed]. [DOI] [Google Scholar]
- 220. Tong S, Su Y, Yu Y, et al. Ribavirin therapy for severe COVID‐19: a retrospective cohort study. Int J Antimicrob Agents 2020; 56 : 106114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221. Chen C, Zhang Y, Huang J, et al. Favipiravir versus Arbidol for COVID‐19: a randomized clinical trial. medRxiv 2020. 10.1101/2020.03.17.20037432. [Not peer reviewed]. [DOI] [Google Scholar]
- 222. Shannon A, Selisko B, Le N‐T‐T, et al. Rapid incorporation of Favipiravir by the fast and permissive viral RNA polymerase complex results in SARS‐CoV‐2 lethal mutagenesis. Nat Commun 2020; 11 : 4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223. Shannon A, Selisko B, Le N, et al. Favipiravir strikes the SARS‐CoV‐2 at its Achilles heel, the RNA polymerase. bioRxiv 2020. 10.1101/2020.05.15.098731. [Not peer reviewed]. [DOI] [Google Scholar]
- 224. Vankadari N. Arbidol: a potential antiviral drug for the treatment of SARS‐CoV‐2 by blocking trimerization of the spike glycoprotein. Int J Antimicrob Agents 2020; 56 : 105998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225. Vogel‐González M, Talló‐Parra M, Herrera‐Fernández V, et al. Low zinc levels at clinical admission associates with poor outcomes in COVID‐19. medRxiv 2020. 10.1101/2020.10.07.20208645. [Not peer reviewed]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226. Mycroft‐West C, Su D, Elli S, et al. The 2019 coronavirus (SARS‐CoV‐2) surface protein (spike) S1 receptor binding domain undergoes conformational change upon heparin binding. bioRxiv 2020. 10.1101/2020.02.29.971093. [Not peer reviewed]. [DOI] [Google Scholar]
- 227. Buijsers B, Yanginlar C, Maciej‐Hulme ML, et al. Beneficial non‐anticoagulant mechanisms underlying heparin treatment of COVID‐19 patients. EBioMedicine 2020; 59 : 102969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228. Gozzo L, Viale P, Longo L, et al. The potential role of heparin in patients with COVID‐19: beyond the anticoagulant effect. A review. Front Pharmacol 2020; 11 : 1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229. Horby P, Lim WS, Emberson JR, et al. Dexamethasone in hospitalized patients with Covid‐19 – preliminary report. N Engl J Med 2021; 384 : 693–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230. Tomazini BM, Maia IS, Cavalcanti AB, et al. Effect of dexamethasone on days alive and ventilator‐free in patients with moderate or severe acute respiratory distress syndrome and COVID‐19: the CoDEX randomized clinical trial. JAMA 2020; 324 : 1307–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231. Ramamoorthy S, Cidlowski JA. Corticosteroids: mechanisms of action in health and disease. Rheum Dis Clin North Am 2016; 42 : 15–31, vii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232. Sterne JAC, Murthy S, Diaz JV, et al. Association between administration of systemic corticosteroids and mortality among critically ill patients with COVID‐19: a meta‐analysis. JAMA 2020; 324 : 1330–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233. Wu C, Chen X, Cai Y, et al. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern Med 2020; 180 : 934–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234. Jeronimo CMP, Farias MEL, Val FFA, et al. Methylprednisolone as adjunctive therapy for patients hospitalized with COVID‐19 (Metcovid): a randomised, double‐blind, phase IIb, placebo‐controlled trial. Clin Infect Dis 2021; 72 : e373–e381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235. Angus DC, Derde L, Al‐Beidh F, et al. Effect of hydrocortisone on mortality and organ support in patients with severe COVID‐19: the REMAP‐CAP COVID‐19 corticosteroid domain randomized clinical trial. JAMA 2020; 324 : 1317–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236. Davoudi‐Monfared E, Rahmani H, Khalili H, et al. A randomized clinical trial of the efficacy and safety of interferon β‐1a in treatment of severe COVID‐19. Antimicrob Agents Chemother 2020; 64 : e01061–e01020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237. Monk PD, Marsden RJ, Tear VJ, et al. Safety and efficacy of inhaled nebulised interferon beta‐1a (SNG001) for treatment of SARS‐CoV‐2 infection: a randomised, double‐blind, placebo‐controlled, phase 2 trial. Lancet Respir Med 2021; 9 : 196–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238. Gordon AC, Mouncey PR, Al‐Beidh F, et al. Interleukin‐6 receptor antagonists in critically ill patients with Covid‐19 – preliminary report. medRxiv 2021. 10.1101/2021.01.07.21249390. [Not peer reviewed]. [DOI] [Google Scholar]
- 239. Huet T, Beaussier H, Voisin O, et al. Anakinra for severe forms of COVID‐19: a cohort study. Lancet Rheumatol 2020; 2 : e393–e400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240. CORIMUNO‐19 Collaborative group . Effect of anakinra versus usual care in adults in hospital with COVID‐19 and mild‐to‐moderate pneumonia (CORIMUNO‐ANA‐1): a randomised controlled trial. Lancet Respir Med 2021; 9 : 295–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241. Robinson PC, Richards D, Tanner HL, et al. Accumulating evidence suggests anti‐TNF therapy needs to be given trial priority in COVID‐19 treatment. Lancet Rheumatol 2020; 2 : e653–e655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242. Feldmann M, Maini RN, Woody JN, et al. Trials of anti‐tumour necrosis factor therapy for COVID‐19 are urgently needed. Lancet 2020; 395 : 1407–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243. Ungaro RC, Brenner EJ, Gearry RB, et al. Effect of IBD medications on COVID‐19 outcomes: results from an international registry. Gut 2021; 70 : 725–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244. Treon SP, Castillo JJ, Skarbnik AP, et al. The BTK inhibitor ibrutinib may protect against pulmonary injury in COVID‐19‐infected patients. Blood 2020; 135 : 1912–1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245. Richardson P, Griffin I, Tucker C, et al. Baricitinib as potential treatment for 2019‐nCoV acute respiratory disease. Lancet 2020; 395 : e30–e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246. Seif F, Aazami H, Khoshmirsafa M, et al. JAK inhibition as a new treatment strategy for patients with COVID‐19. Int Arch Allergy Immunol 2020; 181 : 467–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247. Stebbing J, Sánchez Nievas G, Falcone M, et al. JAK inhibition reduces SARS‐CoV‐2 liver infectivity and modulates inflammatory responses to reduce morbidity and mortality. Sci Adv 2021; 7 : eabe4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248. Cavagna L, Seminari E, Zanframundo G, et al. Calcineurin inhibitor‐based immunosuppression and COVID‐19: results from a multidisciplinary cohort of patients in northern Italy. Microorganisms 2020; 8 : 977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249. Kemmner S, Guba MO, Schönermarck U, et al. Cyclosporine as a preferred calcineurin inhibitor in renal allograft recipients with COVID‐19 infection. Kidney Int 2020; 98 : 507–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250. de Wilde AH, Zevenhoven‐Dobbe JC, van der Meer Y, et al. Cyclosporin A inhibits the replication of diverse coronaviruses. J Gen Virol 2011; 92(Pt 11): 2542–2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251. Tanaka Y, Sato Y, Sasaki T. Suppression of coronavirus replication by cyclophilin inhibitors. Viruses 2013; 5 : 1250–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252. Pfefferle S, Schöpf J, Kögl M, et al. The SARS‐coronavirus–host interactome: identification of cyclophilins as target for pan‐coronavirus inhibitors. PLoS Pathog 2011; 7 : e1002331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253. Carbajo‐Lozoya J, Müller MA, Kallies S, et al. Replication of human coronaviruses SARS‐CoV, HCoV‐NL63 and HCoV‐229E is inhibited by the drug FK506. Virus Res 2012; 165 : 112–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254. Laurence J, Mulvey JJ, Seshadri M, et al. Anti‐complement C5 therapy with eculizumab in three cases of critical COVID‐19. Clin Immunol 2020; 219 : 108555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255. Diurno F, Numis FG, Porta G, et al. Eculizumab treatment in patients with COVID‐19: preliminary results from real life ASL Napoli 2 Nord experience. Eur Rev Med Pharmacol Sci 2020; 24 : 4040–4047. [DOI] [PubMed] [Google Scholar]
- 256. van de Veerdonk FL, Kouijzer IJE, de Nooijer AH, et al. Outcomes associated with use of a kinin B2 receptor antagonist among patients with COVID‐19. JAMA Netw Open 2020; 3 : e2017708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257. Vaduganathan M, Vardeny O, Michel T, et al. Renin–angiotensin–aldosterone system inhibitors in patients with Covid‐19. N Engl J Med 2020; 382 : 1653–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258. Kalra A, Hawkins ES, Nowacki AS, et al. Angiotensin‐converting enzyme inhibitors versus angiotensin II receptor blockers: a comparison of outcomes in patients with COVID‐19. Circ Cardiovasc Qual Outcomes 2020; 13 : e007115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259. Urwyler P, Moser S, Charitos P, et al. Treatment of COVID‐19 with conestat alfa, a regulator of the complement, contact activation and kallikrein–kinin system. Front Immunol 2020; 11 : 2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260. Hoffmann M, Schroeder S, Kleine‐Weber H, et al. Nafamostat mesylate blocks activation of SARS‐CoV‐2: new treatment option for COVID‐19. Antimicrob Agents Chemother 2020; 64 : e00754–e00720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261. Schrezenmeier E, Dörner T. Mechanisms of action of hydroxychloroquine and chloroquine: implications for rheumatology. Nat Rev Rheumatol 2020; 16 : 155–166. [DOI] [PubMed] [Google Scholar]
- 262. Liu J, Cao R, Xu M, et al. Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS‐CoV‐2 infection in vitro . Cell Discov 2020; 6 : 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263. Lammers AJJ, Brohet RM, Theunissen REP, et al. Early hydroxychloroquine but not chloroquine use reduces ICU admission in COVID‐19 patients. Int J Infect Dis 2020; 101 : 283–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264. Self WH, Semler MW, Leither LM, et al. Effect of hydroxychloroquine on clinical status at 14 days in hospitalized patients with COVID‐19: a randomized clinical trial. JAMA 2020; 324 : 2165–2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265. Chai KL, Valk SJ, Piechotta V, et al. Convalescent plasma or hyperimmune immunoglobulin for people with COVID‐19: a living systematic review. Cochrane Database Syst Rev 2020; 7 : CD013600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266. Agarwal A, Mukherjee A, Kumar G, et al. Convalescent plasma in the management of moderate covid‐19 in adults in India: open label phase II multicentre randomised controlled trial (PLACID Trial). BMJ 2020; 371 : m3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267. Rubin R. Testing an old therapy against a new disease: convalescent plasma for COVID‐19. JAMA 2020; 323 : 2114–2117. [DOI] [PubMed] [Google Scholar]
- 268. Simonovich VA, Burgos Pratx LD, Scibona P, et al. A randomized trial of convalescent plasma in Covid‐19 severe pneumonia. N Engl J Med 2021; 384 : 619–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269. Nguyen AA, Habiballah SB, Platt CD, et al. Immunoglobulins in the treatment of COVID‐19 infection: proceed with caution! Clin Immunol 2020; 216 : 108459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270. Regeneron . REGN‐COV2 independent data monitoring committee recommends holding enrollment in hospitalized patients with high oxygen requirements and continuing enrollment in patients with low or no oxygen requirements. [Accessed 7 November 2020]. Available from: https://investor.regeneron.com/news-releases/news-release-details/regn-cov2-independent-data-monitoring-committee-recommends
- 271. Chen P, Nirula A, Heller B, et al. SARS‐CoV‐2 neutralizing antibody LY‐CoV555 in outpatients with Covid‐19. N Engl J Med 2020; 384 : 229–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272. Gottlieb RL, Nirula A, Chen P, et al. Effect of bamlanivimab as monotherapy or in combination with etesevimab on viral load in patients with mild to moderate COVID‐19: a randomized clinical trial. JAMA 2021: e210202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273. Tardif J‐C, Bouabdallaoui N, L'Allier PL, et al. Efficacy of colchicine in non‐hospitalized patients with COVID‐19. medRxiv 2021. 10.1101/2021.01.26.21250494. [Not peer reviewed]. [DOI] [Google Scholar]
- 274. Kim DH, Meza CA, Clarke H, et al. Vitamin D and endothelial function. Nutrients 2020; 12 : 575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275. Giannini S, Passeri G, Tripepi G, et al. Effectiveness of in‐hospital cholecalciferol use on clinical outcomes in comorbid COVID‐19 patients: a hypothesis‐generating study. Nutrients 2021; 13 : 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276. Oldenburg CE, Doan T. Azithromycin for severe COVID‐19. Lancet 2020; 396 : 936–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277. Furtado RHM, Berwanger O, Fonseca HA, et al. Azithromycin in addition to standard of care versus standard of care alone in the treatment of patients admitted to the hospital with severe COVID‐19 in Brazil (COALITION II): a randomised clinical trial. Lancet 2020; 396 : 959–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278. Retallack H, Di Lullo E, Arias C, et al. Zika virus cell tropism in the developing human brain and inhibition by azithromycin. Proc Natl Acad Sci U S A 2016; 113 : 14408–14413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279. Schögler A, Kopf BS, Edwards MR, et al. Novel antiviral properties of azithromycin in cystic fibrosis airway epithelial cells. Eur Respir J 2015; 45 : 428–439. [DOI] [PubMed] [Google Scholar]
- 280. Omarjee L, Janin A, Perrot F, et al. Targeting T‐cell senescence and cytokine storm with rapamycin to prevent severe progression in COVID‐19. Clin Immunol 2020; 216 : 108464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281. Jang IA, Kim EN, Lim JH, et al. Effects of resveratrol on the renin–angiotensin system in the aging kidney. Nutrients 2018; 10 : 1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282. Lian N, Zhang S, Huang J, et al. Resveratrol attenuates intermittent hypoxia‐induced lung injury by activating the Nrf2/ARE pathway. Lung 2020; 198 : 323–331. [DOI] [PubMed] [Google Scholar]
- 283. Ellen Ter BM, Dinesh Kumar N, Bouma EM, et al. Resveratrol and pterostilbene potently inhibit SARS‐CoV‐2 infection in vitro . bioRxiv 2020. 10.1101/2020.09.24.285940. [Not peer reviewed]. [DOI] [Google Scholar]
- 284. Marín‐Hernández D, Schwartz RE, Nixon DF. Epidemiological evidence for association between higher influenza vaccine uptake in the elderly and lower COVID‐19 deaths in Italy. J Med Virol 2021; 93 : 64–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285. Debisarun PA, Struycken P, Domínguez‐Andrés J, et al. The effect of influenza vaccination on trained immunity: impact on COVID‐19. medRxiv 2020. 10.1101/2020.10.14.20212498. [Not peer reviewed]. [DOI] [Google Scholar]
- 286. Giamarellos‐Bourboulis EJ, Tsilika M, Moorlag S, et al. Activate: randomized clinical trial of BCG vaccination against infection in the elderly. Cell 2020; 183 : 315–323.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287. Curtis N, Sparrow A, Ghebreyesus TA, et al. Considering BCG vaccination to reduce the impact of COVID‐19. Lancet 2020; 395 : 1545–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288. Nederlands Tijdschrift voor Geneeskunde (NTVG) . BCG voorkomt covid‐19 bij kwetsbare ouderen niet. [Accessed 31 January 2021]. Available from: https://www.ntvg.nl/artikelen/nieuws/bcg-voorkomt-covid-19-bij-kwetsbare-ouderen-niet/volledig
- 289. Ashford JW, Gold JE, Huenergardt MA, et al. MMR vaccination: a potential strategy to reduce severity and mortality of COVID‐19 illness. Am J Med 2021; 134 : 153–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290. World Health Organization . COVID 19 Landscape of experimental treatments. [Accessed 6 November 2020]. Available from: https://www.who.int/publications/i/item/covid‐19‐landscape‐of‐experimental‐treatments
- 291. Rochwerg B, Agoritsas T, Lamontagne F, et al. A living WHO guideline on drugs for covid‐19. BMJ 2020; 370 : m3379. [DOI] [PubMed] [Google Scholar]
- 292. World Health Organization . Draft landscape and tracker of COVID‐19 candidate vaccines. [Accessed 31 January 2021]. Available from: https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines
- 293. Polack FP, Thomas SJ, Kitchin N, et al. Safety and efficacy of the BNT162b2 mRNA Covid‐19 vaccine. N Engl J Med 2020; 383 : 2603–2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294. Baden LR, El Sahly HM, Essink B, et al. Efficacy and safety of the mRNA‐1273 SARS‐CoV‐2 vaccine. N Engl J Med 2021; 384 : 403–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295. Voysey M, Clemens SAC, Madhi SA, et al. Safety and efficacy of the ChAdOx1 nCoV‐19 vaccine (AZD1222) against SARS‐CoV‐2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet 2021; 397 : 99–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296. National Institutes of Health (NIH). Janssen Investigational COVID‐19 Vaccine: Interim Analysis of Phase 3 Clinical Data Released. [Accessed 4 February 2021]. Available from: https://www.nih.gov/news-events/news-releases/janssen-investigational-covid-19-vaccine-interim-analysis-phase-3-clinical-data-released
- 297. Novavax . Novavax COVID‐19 Vaccine Demonstrates 89.3% Efficacy in UK Phase 3 Trial. [Accessed 31 January 2021]. Available from: https://ir.novavax.com/news-releases/news-release-details/novavax-covid-19-vaccine-demonstrates-893-efficacy-uk-phase-3
- 298. Cable News Network (CNN) . China approves Sinopharm Covid‐19 vaccine, promises free shots for all citizens. [Accessed 4 February 2021]. Available from: https://edition.cnn.com/2020/12/30/asia/china-sinopharm-vaccine-efficacy-intl-hnk/index.html
- 299. BBC . Covid: What do we know about China's coronavirus vaccines? [Accessed 31 January 2021]. Available from: https://www.bbc.com/news/world-asia-china-55212787
- 300. Logunov DY, Dolzhikova IV, Shcheblyakov DV, et al. Safety and efficacy of an rAd26 and rAd5 vector‐based heterologous prime‐boost COVID‐19 vaccine: an interim analysis of a randomised controlled phase 3 trial in Russia. Lancet 2021. 397 : 671–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301. Lee WS, Wheatley AK, Kent SJ, et al. Antibody‐dependent enhancement and SARS‐CoV‐2 vaccines and therapies. Nat Microbiol 2020; 5 : 1185–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302. Marcec R, Majta M, Likic R. Will vaccination refusal prolong the war on SARS‐CoV‐2? Postgrad Med J 2021; 97 : 143–149. [DOI] [PMC free article] [PubMed] [Google Scholar]


