Abstract
The COVID‐19 pandemic caused by SARS‐CoV‐2 is threating global health. Inhibiting interaction of the receptor‐binding domain of SARS‐CoV‐2 S protein (SRBD) and human ACE2 receptor is a promising treatment strategy. However, SARS‐CoV‐2 neutralizing antibodies are compromised by their risk of antibody‐dependent enhancement (ADE) and unfavorably large size for intranasal delivery. To avoid these limitations, we demonstrated an aptamer blocking strategy by engineering aptamers’ binding to the region on SRBD that directly mediates ACE2 receptor engagement, leading to block SARS‐CoV‐2 infection. With aptamer selection against SRBD and molecular docking, aptamer CoV2‐6 was identified and applied to prevent, compete with, and substitute ACE2 from binding to SRBD. CoV2‐6 was further shortened and engineered as a circular bivalent aptamer CoV2‐6C3 (cb‐CoV2‐6C3) to improve the stability, affinity, and inhibition efficacy. cb‐CoV2‐6C3 is stable in serum for more than 12 h and can be stored at room temperature for more than 14 days. Furthermore, cb‐CoV2‐6C3 binds to SRBD with high affinity (K d=0.13 nM) and blocks authentic SARS‐CoV‐2 virus with an IC50 of 0.42 nM.
Keywords: aptamers, neutralization therapy, SARS-CoV-2, viral infections
We propose an aptamer blocking strategy to inhibit SARS‐CoV‐2 infection. With the advantages of small size, rapid kinetics, high stability, sophisticated programmability and high security, our aptamers have great potential as prophylactic and therapeutic agents, which could greatly assist in the intervention of prevailing and emerging infectious diseases other than COVID‐19.
Introduction
Coronavirus disease 2019 (COVID‐19), caused by severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2), is having a deleterious impact on human lives and the global economy, highlighting the urgent search for effective therapeutic and prophylactic interventions. [1]
Because SARS‐CoV‐2 infection generally begins in the nasal cavity and gradually spreads to the lower respiratory tract, intranasal or pulmonary delivery of a viral inhibitor can have a direct and effective therapeutic benefit on the affected sites. [2] In addition, high concentrations of the viral inhibitor in the nose and respiratory system may provide prophylaxis, which is particularly useful for medical staff and others in frequent contact with infected individuals. Recently, several groups have identified SARS‐CoV‐2 neutralizing antibodies, which block the receptor‐binding domain of S (SRBD) from interacting with human angiotensin‐converting enzyme 2 (ACE2) receptor. [3] These neutralizing antibodies may serve as useful therapeutics and prophylactics to help control the COVID‐19 pandemic. Although some of neutralizing antibodies are in clinical trials as systemic treatments for COVID‐19, these antibodies are not ideal for intranasal or pulmonary delivery because of their large size (ca. 150 kDa). [4] Moreover, there is a potential for exacerbation of COVID‐19 through antibody‐dependent enhancement (ADE), which may lead to both increased virus infectivity and virulence. [5] Therefore, there is an urgent need to discover new neutralizing molecules with smaller sizes and enhanced safety to block virus infection of the host and to supplement existing treatment methods.
Inspired by neutralizing antibody, we developed an aptamer blocking strategy by engineering aptamers’ binding to the region on SRBD that mediates ACE2 receptor engagement, thereby inhibiting SARS‐CoV‐2 infection (Scheme 1). Compared to neutralizing antibodies, aptamers show several distinct advantages for SARS‐CoV‐2 treatment. The smaller sizes of aptamers (2–3 nm compared to >10 nm for an antibody) allow them to be directly delivered into the respiratory system through intranasal administration or nebulization. [6] Due to the chemical nature of nucleic acids, aptamers are highly thermal stable, [7] thus enabling them to be stored and transported at room temperature for several weeks, and to protect the environment and objects from viral contamination. Moreover, aptamer has no Fc fragment and therefore is less susceptible to the ADE effect and is expected to be applicable to a variety of species. And aptamers are emerging as highly programmable building blocks for the design of versatile molecular nanodevices with specific structures and functions. [8] In addition, the aptamer and neutralizing antibody can be used as a cocktail therapy, which is expected to resist mutations in the coronavirus and reduce the dosage of neutralizing antibody to decrease the probability of ADE. Therefore, aptamers with the ability to inhibit SARS‐CoV‐2 infection may provide an effective therapeutic and prophylactic solution to complement existing treatment and protection measures. Moreover, the selection and application of aptamers that block SARS‐CoV‐2 infection could also be expanded to infectious diseases other than COVID‐19 and serve as a technical reserve for rapid discovery of neutralizing molecules during possible future pandemics.
Scheme 1.
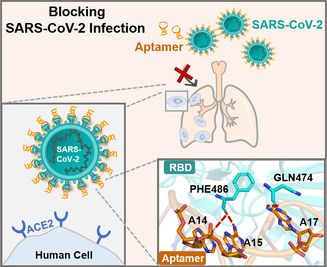
The mechanism of aptamer blocking strategy to inhibit SARS‐CoV‐2 infection.
Results and Discussion
Enrichment and Identification of Aptamers against SRBD
Since the SRBD obtained from different species have different glycosylation modifications which are difficult to ensure to be fully consistent with SARS‐CoV‐2 SRBD, [9] SARS‐CoV‐2 SRBD proteins expressed by baculovirus‐insect cells and HEK293 cells were chosen as targets to ensure generalizability of the obtained aptamers. Then we enriched ACE2‐competitive sequences and introduced molecular docking (MD) to predict the binding sites of candidate sequences, thus selecting those aptamers that have the same binding region on SRBD as ACE2 receptor.
Briefly, after 6 rounds of selection against SRBD expressed by HEK293 cells, baculovirus‐insect‐expressed SRBD was used as the selection target for 7 rounds, of which the last 5 rounds included ACE2 competition. The enrichment progress was monitored by flow cytometry. There was a clearly increasing trend of fluorescence in library binding against target (SRBD‐Ni‐beads) as selection progressed (Figure 1 A & S1). However, for the negative target (Ni‐beads), the fluorescence values of enrichment libraries were basically the same as that of the initial library (Figure 1 B). These results suggest that the DNA library was successfully enriched against SRBD. The 7th, 10th and 13th libraries were chosen for high‐throughput sequencing (Figure S2). Among the sequences with enrichment, we selected the 5 candidate sequences with the highest abundance among the 13 rounds (CoV2‐1 to CoV2‐5) and the sequences repeated in the three rounds of libraries (CoV2‐6 to CoV2‐8) for binding performance characterization (Figure 1 C & S3).
Figure 1.
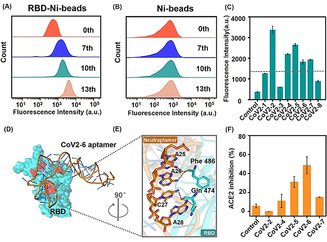
Aptamer CoV2‐6 identification and characterization. A,B) Flow cytometry to monitor the binding increment of enriched pools with RBD‐Ni‐beads (target beads) and Ni‐beads (control beads). C) Flow cytometry to investigate the binding performance of candidate sequences against SRBD. D) The results of molecular docking of overall structures of the CoV2‐6 aptamer (orange) and SRBD complex (SRBD is cyan and ACE2 binding amino acid residues are red), and E) the detailed analysis of the interface between CoV2‐6 aptamer and SRBD. F) The ACE2 inhibition efficiency of candidate sequences. SRBD was expressed by baculovirus‐insect cells.
Timing is critical in a pandemic: potent therapeutics are needed as quickly as possible. Because only aptamers with binding sites similar to that of ACE2 against SRBD hold promise to block viral infection, [10] to accelerate the selection of these aptamers, we applied molecular docking calculations to systematically model the minimized energy structures of the aptamer‐target complex and predict the possible binding sites. For the interaction of SRBD and ACE2, there is a “bridge” interaction model consisting of three cluster contact points, namely Gln 498, Thr 500, and Asn 501 at the N‐terminus, Lys 417 and Tyr 453 in the middle, and Phe 486 and Gln 474 at the C‐terminus of SRBD. [11] The MD results revealed that CoV2‐6 sequence embeds in the ACE2 binding sites, and sterically “covers” all binding amino acid residues on SRBD (Phe 486 and Gln 474) as the end of the ACE2‐SRBD “bridge” interaction model (Figure 1 D,E), the binding sites of the other candidate sequences are only close to the binding sites of ACE2 (Figure S4). This interaction model of CoV2‐6 sequence‐SRBD presumably leads to competition with ACE2 for SRBD binding.
To confirm this hypothesis, the inhibition between ACE2 and aptamer pre‐blocking SRBD was carried out to verify the accuracy of MD. Consistent with the predicted results, CoV2‐6 sequence has the highest ACE2 inhibitory effect among the candidate sequences (Figure 1 F). Thus, aptamer CoV2‐6 was chosen for further optimization. In this work, we developed a molecular docking‐based identification strategy which has great potential to obtain aptamers targeting new viral strains with inhibitory activity rapidly.
Aptamer Identification and Optimization
Generally, not all nucleotides are involved in target binding. Thus, a shorter sequence reduces molecular size, making the truncated sequence more available for synthesis and drug delivery. [12] The MD results suggested that the nucleotides on the two loops participated in the SRBD binding (Figure 2 A, bases in orange were predicted to interact with SRBD). Therefore, we removed some nucleotides that are not essential for hairpin formation, yielding a 46‐nt 2‐hairpin‐structured CoV2‐6C3 aptamer (Figure 2 A, 5′‐CGCAGCACCCAAGAACAAGGACTGCTTAGGATTGCGATAGGTTCGG −3′, 13.8 KDa). The binding capability and specificity of the truncated aptamer CoV2‐6C3 were evaluated. For RBD‐Ni‐beads, the fluorescence intensity increment of CoV2‐6C3 aptamer was similar to that of the full‐length CoV2‐6 aptamer, while for Ni‐beads, the fluorescence intensities of the aptamers were similar to that of the random sequence (Figure S5). In addition, the dissociation constant (K d) values of the CoV2‐6C3 and CoV2‐6 aptamers were determined. Against SRBD, the K d value of CoV2‐6C3 aptamer was 44.78±9.97 nM, about half that of the full‐length CoV2‐6 aptamer (Figure 2 B), indicating that CoV2‐6C3 aptamer has higher binding affinity after removing the unnecessary nucleotides for target binding. In addition to targeting baculovirus‐insect‐expressed SRBD, CoV2‐6C3 sequence also recognizes the HEK293‐expressed SRBD (Figure S6 & S7) and spike protein (Figure S8), but no to the SRBD or spike proteins of other viruses (Figure S10), indicating the ideal universality and selectivity of CoV2‐6C3 sequence. To further confirm the contribution of the loop region to target binding, the smaller loop of CoV2‐6C3 aptamer was deleted to obtain CoV2‐6C4 sequence. As speculated, CoV2‐6C4 sequence did not bind to SRBD (Figure S11). These results suggest that CoV2‐6C3 has the potential for efficient SARS‐CoV‐2 SRBD recognition.
Figure 2.
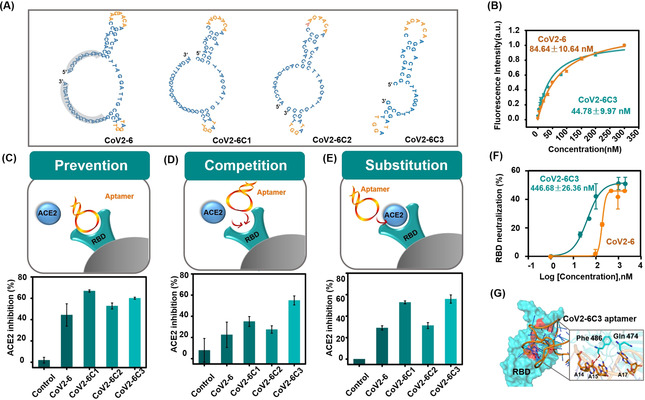
A) The secondary structures of CoV2‐6 aptamer and a series of truncated sequences simulated using mfold software; bases in orange were predicted to interact with SRBD, and bases with gray background were removed. B) Binding curves of CoV2‐6 (orange) and CoV2‐6C3 (green) against SRBD (expressed by baculovirus‐insect cells). Schematic illustration and ACE2 inhibition efficiency of candidate sequences on C) prevention, D) competition, and E) substitution of ACE2 binding to SRBD. F) Inhibition the interaction of SRBD (expressed by HEK293 cells) and ACE2 by CoV2‐6 (orange) and CoV2‐6C3 (green). G) Molecular dynamics simulated structure of CoV2‐6C3‐ SRBD complex.
Aptamer Characterization
To fully characterize the inhibitory effect, we investigated the inhibition efficiency of CoV2‐6C3 aptamer for prevention, competition, and substitution of ACE2 binding to SRBD protein, so as to simulate the effects of different scenarios during the process of SARS‐CoV‐2 infection. In the inhibition experiment, ACE2 was incubated with the aptamer pre‐binding SRBD, that was used to simulate the aptamer as COVID‐19 prevention reagents. Cov2‐6C and truncated sequences greatly inhibited the binding of SRBD and ACE2, and an obvious increase (44.34–66.84 %) was found in the ACE2 inhibition compared with the negative control group (1.91 %) (Figure 2 C). The competition experiment simulated the window period of SARS‐CoV‐2 infection, with SRBD, ACE2 and the aptamer mixed together at the same time, and the results showed that Cov2‐6C3 had the best inhibition efficiency up to 55.00 % (Figure 2 D). Similarly, CoV2‐6C3 aptamer replaced more than 55.91 % ACE2 bound to SRBD, indicating its potential for the treatment of infected patients (Figure 2 E). The half‐maximal inhibitory concentration (IC50) value of CoV2‐6C3 aptamer against SRBD is approximately 446.68 nM (Figure 2 F). These results showed the good versatility of the developed CoV2‐6C3 aptamer.
To provide molecular level insight into the inhibition of CoV2‐6C3 aptamer against ACE2, we further used molecular dynamics simulations (MDS) to investigate the detailed aptamer‐SRBD interaction. The results indicated that SRBD and CoV2‐6C3 contain two consecutive binding interfaces, one of which is similar to that of the full‐length aptamer as the end of the “bridge” interaction be‐tween ACE2 and SRBD. Specifically, A14, A15, and A17 of the CoV2‐6C3 form a network (Figure 2 G). Moreover, sequences of mutations at the predicted binding sites exhibited significantly lower binding performance against SRBD compared to CoV2‐6C3 aptamer (Figure S12). In general, both the experimental and simulation results verified aptamer CoV2‐6C3 inhibition of the interaction of SRBD and ACE2, laying a solid foundation for applying the aptamer as a potential prophylactic and therapeutic agent against SARS‐CoV‐2.
Design and Characterization of Circularly Bivalent Aptamer
The programmability of aptamers enables them to be easily assembled into specific molecular devices to improve thermodynamic properties and stability. [13] To improve the binding affinity and stability, we constructed a circular bivalent aptamer (cb‐CoV2‐6C3, 38.9 kDa) by extending a flanking complementary sequence to form a tail‐to‐tail hybridization (Figure 3 A). Circular aptamers are resistant to exonuclease degradation, [14] helping them to retain sequence integrity, even after storage at room temperature for 14 days (Figure 3 B), incubation with 95 % human plasma for 12 hours (Figure 3 C) and cell media (containing 10 % FBS) for 48 hours, while the monovalent CoV2‐6C3 was easily degraded after incubation with cell media (containing 10 % FBS) for 6 hours (Figure S13). In addition, circular bivalent aptamer retained its SRBD binding ability after incubation in fresh human plasma for 12 hours (Figure 3 D). The enhanced biological stability of circular aptamer makes it highly desirable for applications in real biological samples without a temperature‐controlled supply chain.
Figure 3.
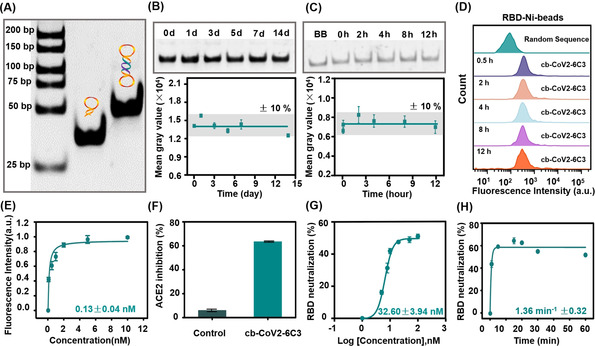
Formation and performance analysis of the circular aptamer (cb‐CoV2‐6C3). A) Formation of cb‐CoV2‐6C3, as determined by agarose gel electrophoresis. B,C) Stability analysis of the circular aptamer after storing at room temperature and incubated with 95 % human plasma. D) Flow cytometric analysis of cb‐CoV2‐6C3 binding to RBD‐Ni‐beads after incubation with 95 % human plasma for different times. E) Binding curve of cb‐CoV2‐6C3 against SRBD. F) ACE2 inhibition efficiency of cb‐CoV2‐6C3 on prevention for ACE2 binding to SRBD. G,H) Thermodynamic and kinetic curves of neutralization of SRBD by cb‐CoV2‐6C3. SRBD was expressed by baculovirus‐insect cells in (D–F) and by HEK293 cells in (G–H).
Since the circular bivalent aptamer has two recognition motifs, it was expected to have better binding ability than the monovalent aptamer. The K d value of the circular aptamer against SRBD was found to be 0.13 nM (Figure 3 E), indicating a 344‐fold higher binding affinity than that of monovalent CoV2‐6C3 aptamer (K d=44.78 nM). In addition, the circular aptamer can inhibit about 63.48 % SRBD binding to ACE2 (Figure 3 F). Moreover, the circular aptamer against SRBD was observed to have an IC50 value of ≈32.60 nM (Figure 3 G), which is much lower than that of the monovalent aptamer, indicating significantly improved inhibition ability. To fully characterize the neutralization performance, the inhibition kinetics of the circular aptamer was investigated. We observed rapid kinetics with a rate constant of 1.36 min−1 (Figure 3 H), 10‐fold faster than that of the reported neutralizing antibody. [15] Therefore, the circular bivalent aptamer obtained by facile enzymatic ligation, has improved stability and binding affinity over the monovalent aptamer, and comparable IC50 value and better neutralizing kinetic properties than those of reported neutralizing antibodies (Table S1).
Pseudotyped and Authentic SARS‐CoV‐2 Inhibition
The inhibition ability of the circular aptamer was further confirmed using pseudovirus neutralization assay, which is considered as a sensitive method to evaluate whether neutralizing agents specifically inhibit SARS‐CoV‐2 infection by influencing the interaction of S protein and ACE2. [16] The SARS‐CoV‐2 pseudovirus used here was packaged with S protein of SARS‐CoV‐2 as a surface capsid glycoprotein by a lentivirus packaging system, and the RNA genome containing GFP and luciferase gene driven by a CMV promoter. Therefore, the virus‐mediated cell entry can be conveniently determined via GFP or bioluminescence intensity (Figure S14). Varying concentrations of the circular aptamer were incubated with SARS‐CoV‐2 pseudovirus and then added to ACE2‐expressing 293T cells. The circular aptamer strongly blocked the pseudovirus infection with IC50 of 9.68 nM (Figure 4 A), while a control DNA sequence showed no neutralization activity (Figure S15). The fluorescence microscopy imaging also confirmed that some of the cells are almost completely uninfected by the aptamer neutralization, and the mean fluorescence intensity of even infected cells was much weaker than that of the group without treatment (Figure 4 B). Moreover, the circular aptamer displayed a high potency against authentic SARS‐CoV‐2 virus with an IC50 of 0.42 nM, and reduced the amount of viral RNA in the cells by 87.01 % compared to the sample without the aptamer (Figures 4 C). Immunofluorescent staining was used to further validate the effect of authentic virus neutralization, which indicated that the circular aptamer could significantly reduce the number of infected cells (Figures 4 D).
Figure 4.
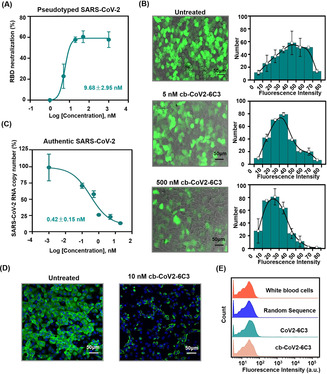
Pseudovirus and authentic SARS‐CoV‐2 inhibition by cb‐CoV2‐6C3. A) The inhibition of SARS‐CoV‐2 pseudovirus by cb‐CoV2‐6C3 assessed through IC50. B) Images of cb‐CoV2‐6C3 inhibition of SARS‐CoV‐2 pseudovirus infection of ACE2‐expressing 293T cells. C) Authentic SARS‐CoV‐2 neutralization curve of cb‐CoV2‐6C3 was determined via quantitative polymerase chain reaction (qPCR) of the viral genome in cellular RNA. D) Images of cb‐CoV2‐6C3 neutralization of authentic SARS‐CoV‐2 infection of Vero E6 cells. The cells were fixed and stained using an anti‐SARS‐CoV‐2 SRBD antibody for the virus and Hoechst dye for cell nucleus. E) Binding of cb‐CoV2‐6C3 and CoV2‐6C3 to white blood cells.
Previous trials of SARS‐CoV and other respiratory viruses suggested that neutralizing antibodies targeting SARS‐CoV‐2 could exacerbate COVID‐19 through ADE. [17] Fc receptor‐expressing phagocytic cell‐mediated virus internalization has been recognized as a main mechanism of ADE. The developed aptamers do not have Fc segments, so aptamers are theoretically not as susceptible to ADE as neutralizing antibodies. Taking white blood cells (WBCs) as the model phagocytic cells, there was no increase in fluorescence intensity for cb‐CoV2‐6C3 compared with pure white blood cells (Figure 4 E), suggesting that the cb‐CoV2‐6C3 aptamer does not bind to WBCs. In addition, cb‐CoV2‐6C3 showed no cell cytotoxicity (Figure S16). Overall, cb‐CoV2‐6C3 provides a safe and effective SARS‐CoV‐2 potential therapy, which is expected to supplement or combine with the existing COVID‐19 treatments.
Conclusion
In conclusion, by exploiting the potential advantages of smaller size and minimum immunogenic risk, together with more rapid kinetics, stability and programmability over neutralizing antibodies (Table S1), we proposed an aptamer blocking strategy to inhibit SARS‐CoV‐2 infection. First, the developed aptamers are about 4–10‐fold smaller than a traditional antibody, and hence in an equal volume there is a larger number of aptamer molecules, increasing the concentration of a locally administrated drug or reducing side effects and dosage. The small size also makes aptamers potentially amenable to formulation in a gel or aerosol for nasal application, thus enabling direct delivery into the respiratory system and rapid neutralization kinetics. Second, ADE and immunogenicity are potential problems for SARS‐CoV‐2 neutralizing antibodies, but aptamers developed in this work and reported in previous researches have exhibited no ADE or little immune response. Third, the retention of aptamer integrity after more than 14 days at room temperature suggests that a temperature‐controlled transport chain will not be necessary. Although the inhibition efficiency of the aptamer does not reach 100 % and has not been tested in vivo for the time being, it is very promising to be improved through multiple aptamer‐based cocktail, multivalent recognition, specific topological structure formation. Overall, we proposed and demonstrated that an aptamer blocking strategy can be used to inhibit SARS‐CoV‐2 infection, suggesting a new direction for developing therapeutic agents against COVID‐19 and other emerging infectious diseases.
Conflict of interest
The authors declare no conflict of interest.
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supplementary
Acknowledgements
We thank the National Natural Science Foundation of China (22022409, 21735004, 21874089, 21705024), and the Program for Changjiang Scholars and Innovative Research Team in University (IRT13036).
M. Sun, S. Liu, X. Wei, S. Wan, M. Huang, T. Song, Y. Lu, X. Weng, Z. Lin, H. Chen, Y. Song, C. Yang, Angew. Chem. Int. Ed. 2021, 60, 10266.
Contributor Information
Prof. Honglin Chen, Email: hlchen@hku.hk.
Prof. Yanling Song, Email: ylsong@xmu.edu.cn.
Prof. Chaoyong Yang, Email: cyyang@xmu.edu.cn.
References
- 1.
- 1a. Wang C., Horby P. W., Hayden F. G., Gao G. F., Lancet 2020, 395, 470–473; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1b. Zhou P., Yang X. L., Wang X. G., Hu B., Zhang L., Zhang W., Si H. R., Zhu Y., Li B., Huang C. L., Chen H. D., Chen J., Luo Y., Guo H., Jiang R. D., Liu M. Q., Chen Y., Shen X. R., Wang X., Zheng X. S., Zhao K., Chen Q. J., Deng F., Liu L. L., Yan B., Zhan F. X., Wang Y. Y., Xiao G. F., Shi Z. L., Nature 2020, 579, 270–273; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1c. Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G. F., Tan W., N. Engl. J. Med. 2020, 382, 727–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hou Y. J., Okuda K., Edwards C. E., Martinez D. R., Asakura T., K. H. Dinnon 3rd , Kato T., Lee R. E., Yount B. L., Mascenik T. M., Chen G., Olivier K. N., Ghio A., Tse L. V., Leist S. R., Gralinski L. E., Schafer A., Dang H., Gilmore R., Nakano S., Sun L., Fulcher M. L., Livraghi-Butrico A., Nicely N. I., Cameron M., Cameron C., Kelvin D. J., de Silva A., Margolis D. M., Markmann A., Bartelt L., Zumwalt R., Martinez F. J., Salvatore S. P., Borczuk A., Tata P. R., Sontake V., Kimple A., Jaspers I., O'Neal W. K., Randell S. H., Boucher R. C., Baric R. S., Cell 2020, 182, 429–446 e414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.
- 3a. Ju B., Zhang Q., Ge J., Wang R., Sun J., Ge X., Yu J., Shan S., Zhou B., Song S., Tang X., Yu J., Lan J., Yuan J., Wang H., Zhao J., Zhang S., Wang Y., Shi X., Liu L., Zhao J., Wang X., Zhang Z., Zhang L., Nature 2020, 584, 115–119; [DOI] [PubMed] [Google Scholar]
- 3b. Cao Y., Su B., Guo X., Sun W., Deng Y., Bao L., Zhu Q., Zhang X., Zheng Y., Geng C., Chai X., He R., Li X., Lv Q., Zhu H., Deng W., Xu Y., Wang Y., Qiao L., Tan Y., Song L., Wang G., Du X., Gao N., Liu J., Xiao J., Su X. D., Du Z., Feng Y., Qin C., Qin C., Jin R., Xie X. S., Cell 2020, 182, 73–84 e16; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3c. Pinto D., Park Y. J., Beltramello M., Walls A. C., Tortorici M. A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A., Peter A., Guarino B., Spreafico R., Cameroni E., Case J. B., Chen R. E., Havenar-Daughton C., Snell G., Telenti A., Virgin H. W., Lanzavecchia A., Diamond M. S., Fink K., Veesler D., Corti D., Nature 2020, 583, 290–295. [DOI] [PubMed] [Google Scholar]
- 4.
- 4a.B. E. Jones, P. L. Brown-Augsburger, K. S. Corbett, K. Westendorf, J. Davies, T. P. Cujec, C. M. Wiethoff, J. L. Blackbourne, B. A. Heinz, D. Foster, R. E. Higgs, D. Balasubramaniam, L. Wang, R. Bidshahri, L. Kraft, Y. Hwang, S. Zentelis, K. R. Jepson, R. Goya, M. A. Smith, D. W. Collins, S. J. Hinshaw, S. A. Tycho, D. Pellacani, P. Xiang, K. Muthuraman, S. Sobhanifar, M. H. Piper, F. J. Triana, J. Hendle, A. Pustilnik, A. C. Adams, S. J. Berens, R. S. Baric, D. R. Martinez, R. W. Cross, T. W. Geisbert, V. Borisevich, O. Abiona, H. M. Belli, M. de Vries, A. Mohamed, M. Dittmann, M. Samanovic, M. J. Mulligan, J. A. Goldsmith, C. L. Hsieh, N. V. Johnson, D. Wrapp, J. S. McLellan, B. C. Barnhart, B. S. Graham, J. R. Mascola, C. L. Hansen, E. Falconer, bioRxiv 2020, 10.1101/2020.09.30.318972; [DOI]
- 4b. Shi R., Shan C., Duan X., Chen Z., Liu P., Song J., Song T., Bi X., Han C., Wu L., Gao G., Hu X., Zhang Y., Tong Z., Huang W., Liu W. J., Wu G., Zhang B., Wang L., Qi J., Feng H., Wang F. S., Wang Q., Gao G. F., Yuan Z., Yan J., Nature 2020, 584, 120–124. [DOI] [PubMed] [Google Scholar]
- 5.
- 5a. DiLillo D. J., Tan G. S., Palese P., Ravetch J. V., Nat. Med. 2014, 20, 143–151; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5b. Klasse P. J., Adv. Biol. 2014, 2014, 157895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.
- 6a. Bing T., Shen L., Wang J., Wang L., Liu X., Zhang N., Xiao X., Shangguan D., Adv. Sci. 2019, 6, 1900143; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6b. Huang M., Yang J., Wang T., Song J., Xia J., Wu L., Wang W., Wu Q., Zhu Z., Song Y., Yang C., Angew. Chem. Int. Ed. 2020, 59, 4800–4805; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2020, 132, 4830–4835. [Google Scholar]
- 7.
- 7a. Yang Q., Deng Z., Wang D., He J., Zhang D., Tan Y., Peng T., Wang X. Q., Tan W., J. Am. Chem. Soc. 2020, 142, 2532–2540; [DOI] [PubMed] [Google Scholar]
- 7b. Li L., Jiang Y., Cui C., Yang Y., Zhang P., Stewart K., Pan X., Li X., Yang L., Qiu L., Tan W., J. Am. Chem. Soc. 2018, 140, 13335–13339; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7c. Shen L., Jia K., Bing T., Zhang Z., Zhen X., Liu X., Zhang N., Shangguan D., Anal. Chem. 2020, 92, 5370–5378. [DOI] [PubMed] [Google Scholar]
- 8.
- 8a. Zhang P., Gao D., An K., Shen Q., Wang C., Zhang Y., Pan X., Chen X., Lyv Y., Cui C., Liang T., Duan X., Liu J., Yang T., Hu X., Zhu J. J., Xu F., Tan W., Nat. Chem. 2020, 12, 381–390; [DOI] [PubMed] [Google Scholar]
- 8b. Fu X., Ke G., Peng F., Hu X., Li J., Shi Y., Kong G., Zhang X. B., Tan W., Nat. Commun. 2020, 11, 1518; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8c. Yang L., Sun H., Liu Y., Hou W., Yang Y., Cai R., Cui C., Zhang P., Pan X., Li X., Li L., Sumerlin B. S., Tan W., Angew. Chem. Int. Ed. 2018, 57, 17048–17052; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2018, 130, 17294–17298. [Google Scholar]
- 9.
- 9a. Watanabe Y., Allen J. D., Wrapp D., McLellan J. S., Cris-pin M. J. S., Science 2020, 369, 330–333; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9b.Y. Zhang, W. Zhao, Y. Mao, S. Wang, Y. Zhong, T. Su, M. Gong, X. Lu, J. Cheng, H. J. Yang, bioRxiv 2020, 10.1101/2020.03.28.013276. [DOI]
- 10.
- 10a. Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X., Nature 2020, 581, 215–220; [DOI] [PubMed] [Google Scholar]
- 10b. Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T. S., Herrler G., Wu N. H., Nitsche A., Muller M. A., Drosten C., Pohlmann S., Cell 2020, 181, 271–280 e278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. J. S., Science 2020, 367, 1444–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.
- 12a. Wu W., Zhang T., Han D., Fan H., Zhu G., Ding X., Wu C., You M., Qiu L., Li J., Zhang L., Lian X., Hu R., Mu Y., Zhou J., Tan W., Chem. Sci. 2018, 9, 3050–3055; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12b. Song Y., Song J., Wei X., Huang M., Sun M., Zhu L., Lin B., Shen H., Zhu Z., Yang C., Anal. Chem. 2020, 92, 9895–9900. [DOI] [PubMed] [Google Scholar]
- 13.
- 13a. Mao Y., Gu J., Chang D., Wang L., Yao L., Ma Q., Luo Z., Qu H., Li Y., Zheng L., Nucleic Acids Res. 2020, 48, 10680–10690; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13b. Liu M., Yin Q., Chang Y., Zhang Q., Brennan J. D., Li Y., Angew. Chem. Int. Ed. 2019, 58, 8013–8017; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2019, 131, 8097–8101; [Google Scholar]
- 13c. Yang Y., Sun X., Xu J., Cui C., Safari Yazd H., Pan X., Zhu Y., Chen X., Li X., Li J., Tan W., ACS Nano 2020, 14, 9562–9571; [DOI] [PubMed] [Google Scholar]
- 13d. Xue C., Zhang S., Yu X., Hu S., Lu Y., Wu Z. S., Angew. Chem. Int. Ed. 2020, 59, 17540 Angew. Chem. 2020, 132, 17693–17700. [DOI] [PubMed] [Google Scholar]
- 14.
- 14a. Kuai H., Zhao Z., Mo L., Liu H., Hu X., Fu T., Zhang X., Tan W., J. Am. Chem. Soc. 2017, 139, 9128–9131; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14b. Li X., Yang Y., Zhao H., Zhu T., Yang Z., Xu H., Fu Y., Lin F., Pan X., Li L., Cui C., Hong M., Yang L., Wang K. K., Tan W., J. Am. Chem. Soc. 2020, 142, 3862–3872. [DOI] [PubMed] [Google Scholar]
- 15. Wrapp D., De Vlieger D., Corbett K. S., Torres G. M., Wang N., Van Breedam W., Roose K., van Schie L., Team V.-C. C.-R., Hoffmann M., Pohlmann S., Graham B. S., Callewaert N., Schepens B., Saelens X., McLellan J. S., Cell 2020, 181, 1004–1015 e1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yang J., Wang W., Chen Z., Lu S., Yang F., Bi Z., Bao L., Mo F., Li X., Huang Y., Hong W., Yang Y., Zhao Y., Ye F., Lin S., Deng W., Chen H., Lei H., Zhang Z., Luo M., Gao H., Zheng Y., Gong Y., Jiang X., Xu Y., Lv Q., Li D., Wang M., Li F., Wang S., Wang G., Yu P., Qu Y., Yang L., Deng H., Tong A., Li J., Wang Z., Yang J., Shen G., Zhao Z., Li Y., Luo J., Liu H., Yu W., Yang M., Xu J., Wang J., Li H., Wang H., Kuang D., Lin P., Hu Z., Guo W., Cheng W., He Y., Song X., Chen C., Xue Z., Yao S., Chen L., Ma X., Chen S., Gou M., Huang W., Wang Y., Fan C., Tian Z., Shi M., Wang F. S., Dai L., Wu M., Li G., Wang G., Peng Y., Qian Z., Huang C., Lau J. Y., Yang Z., Wei Y., Cen X., Peng X., Qin C., Zhang K., Lu G., Wei X., Nature 2020, 586, 572–577. [DOI] [PubMed] [Google Scholar]
- 17. Lee W. S., Wheatley A. K., Kent S. J., DeKosky B. J., Nat. Microbiol. 2020, 5, 1185–1191. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supplementary


