ABSTRACT
Movement of the cell nucleus typically involves the cytoskeleton and either polymerization-based pushing forces or motor-based pulling forces. In the fission yeast Schizosaccharomyces pombe, nuclear movement and positioning are thought to depend on microtubule polymerization-based pushing forces. Here, we describe a novel, microtubule-independent, form of nuclear movement in fission yeast. Microtubule-independent nuclear movement is directed towards growing cell tips, and it is strongest when the nucleus is close to a growing cell tip, and weakest when the nucleus is far from that tip. Microtubule-independent nuclear movement requires actin cables but does not depend on actin polymerization-based pushing or myosin V-based pulling forces. The vesicle-associated membrane protein (VAMP)-associated proteins (VAPs) Scs2 and Scs22, which are critical for endoplasmic reticulum–plasma membrane contact sites in fission yeast, are also required for microtubule-independent nuclear movement. We also find that in cells in which microtubule-based pushing forces are present, disruption of actin cables leads to increased fluctuations in interphase nuclear positioning and subsequent altered septation. Our results suggest two non-exclusive mechanisms for microtubule-independent nuclear movement, which may help illuminate aspects of nuclear positioning in other cells.
KEY WORDS: Fission yeast, Schizosaccharomyces pombe, Nucleus, Nuclear movement, Actin, Endoplasmic reticulum
Summary: A new form of nuclear movement in fission yeast, termed microtubule-independent nuclear movement, that is dependent on actin cables and on proteins involved in ER–plasma membrane contacts is described.
INTRODUCTION
Movement and positioning of the cell nucleus depends on cell type, cell cycle stage, and cell migration and differentiation states (Bone and Starr, 2016; Dupin and Etienne-Manneville, 2011; Gundersen and Worman, 2013; Lele et al., 2018; Xiang, 2018). Precise nuclear positioning is important in cell division, cell migration, and development. In humans, mispositioning of the nucleus is associated with several different pathologies, including muscular dystrophy and cardiomyopathy, lissencephaly and premature ageing (Gundersen and Worman, 2013; Isermann and Lammerding, 2013). In the budding yeast Saccharomyces cerevisiae, nuclear positioning is required for the daughter cell to inherit a nucleus from the mother cell (Moore and Cooper, 2010; Pruyne et al., 2004), and in the cylindrically shaped fission yeast Schizosaccharomyces pombe, nuclear positioning is required to generate equal-sized daughter cells during cell division (Chang and Nurse, 1996; Daga and Chang, 2005; Tolić-Nørrelykke et al., 2005).
In organisms from humans to yeasts, nuclear movement generally involves proteins of the cytoskeleton and protein complexes associated with the nuclear envelope (NE) (Gundersen and Worman, 2013; Tapley and Starr, 2013). Both microtubules (MTs) and actin filaments have been implicated in generating forces for movement (Dupin and Etienne-Manneville, 2011; Reinsch and Gonczy, 1998); in some instances, intermediate filaments may also contribute (Dupin et al., 2011). In addition, in many cells, linkers of nucleoskeleton and cytoskeleton (LINC) protein complexes, which span the inner and outer nuclear membranes, help transmit forces from the cytoskeleton to the nucleus (Burke, 2019; Crisp et al., 2006; Padmakumar et al., 2005; Tapley and Starr, 2013).
MT-dependent nuclear movement can occur via multiple mechanisms. In some cases, the nucleus is closely associated with a microtubule organizing center [MTOC; e.g. the centrosome in animal cells, or the spindle pole body (SPB) in yeast], and movement of both the MTOC and the associated nucleus depends on dynamic changes in the MT network, driven variously by microtubule polymerization, depolymerization and/or cortex-localized MT motors (e.g. dynein). Examples of this include movement of the male pronucleus in Xenopus early development (Reinsch and Gonczy, 1998), the Drosophila oocyte nucleus during dorsal–ventral axis specification (Zhao et al., 2012), and fibroblast nuclei during cell migration (Levy and Holzbaur, 2008). In other cases, the nucleus itself moves along MTs, driven by motor proteins (e.g. kinesins or dyneins) associated with the NE. Examples include movement of the female pronucleus during Xenopus development (Reinsch and Gonczy, 1998), a variety of nuclear movements in Caenorhabditis elegans (Fridolfsson and Starr, 2010), and interkinetic nuclear migration of neuronal precursors during vertebrate brain development (Bertipaglia et al., 2018; Tsai et al., 2010).
Actin-dependent nuclear movement generally depends on forces that push the nucleus, as a result of actin polymerization, actin network flow and/or actomyosin cytoskeleton contractility. In some cases, these work in concert with microtubule-dependent forces. Actin-dependent nuclear movement is seen in migrating fibroblasts and astrocytes (Dupin et al., 2011; Gomes et al., 2005; Luxton et al., 2010), in certain types of neuroepithelia (Yanakieva et al., 2019), and in nurse cells in the Drosophila ovary (Huelsmann et al., 2013). Currently, there is little evidence for NE-associated motor proteins driving nuclear movement along actin filaments, at least in animal cells. However, in Arabidopsis thaliana, nuclear movement involves a plant-specific myosin, myosin XI-i (also known as Myo11G), which localizes to the NE and links the nucleus to actin filaments (Tamura et al., 2013).
In budding yeast, movement of the nucleus from the mother cell into the daughter bud neck involves MTs directly and actin filaments indirectly (Moore and Cooper, 2010; Pruyne et al., 2004). Initially, MTs emanating from one of the duplicated SPBs on the NE are guided along actin filaments into the bud neck, via association of MT plus-ends with the class V myosin Myo2. Subsequently, a combination of MT depolymerization at cortical MT contact sites in the daughter cell and cortical dynein-driven MT sliding provides force to pull the nucleus into the bud neck.
In fission yeast, the interphase nucleus is continuously maintained at the cell center as the cell grows by elongation at its tips. The position of the late-interphase/early-mitotic nucleus defines the future plane of cell division, and nuclear mispositioning leads to unequal segregation of cellular components and unequal-sized daughter cells (Chang and Nurse, 1996; Daga and Chang, 2005; Tolić-Nørrelykke et al., 2005). Because interphase growth in fission yeast is normally asymmetric (i.e. monopolar early in the cell cycle, and bipolar later in the cell cycle; Mitchison and Nurse, 1985), the precision of nuclear centering has suggested that an active mechanism positions the nucleus, and experiments using tubulin mutants, MT-depolymerizing drugs, cell centrifugation and optical tweezers have indicated that MTs are critical for nuclear centering (Daga et al., 2006; Hagan and Yanagida, 1997; Sawin et al., 2004; Toda et al., 1983; Tolić-Nørrelykke et al., 2005; Tran et al., 2001; Umesono et al., 1983). During interphase, fission yeast MTs exist as several independent antiparallel bundles that undergo repeated cycles of polymerization and depolymerization (Sawin and Tran, 2006), and these MTs can exert pushing forces on the nucleus during MT polymerization, leading to a model in which MT pushing forces drive nuclear positioning (Tran et al., 2001). Consistent with this model, the fission yeast SPB, which is associated with the nucleus, exhibits MT-dependent oscillations parallel to the long axis of the cell, and the NE similarly shows MT-dependent deformations (Tran et al., 2001).
Although the idea that MT-dependent forces position the fission yeast nucleus has been accepted for many years, the question of whether the nucleus can still move in a cell that lacks MTs has, surprisingly, never been investigated directly. In this work, using live-cell imaging and a combination of drug perturbations and mutants, we show that robust movement of the fission yeast nucleus persists even after MT depolymerization. This MT-independent nuclear movement is directed towards growing cell tips, and we further show that it depends on actin cables nucleated from growing tips, but not on myosin motors. We also find that MT-independent nuclear movement requires proper spatial organization of the endoplasmic reticulum (ER), which in yeast can link the NE with the plasma membrane. Overall, MT-independent movement of the fission yeast nucleus appears to involve mechanisms of force generation that are distinct from those previously described for nuclear movement. Finally, we show that in conjunction with MT-dependent pushing forces, the mechanisms underlying MT-independent nuclear movement contribute to accurate positioning of the nucleus at the cell center.
RESULTS
Unidirectional nuclear movement after microtubule depolymerization
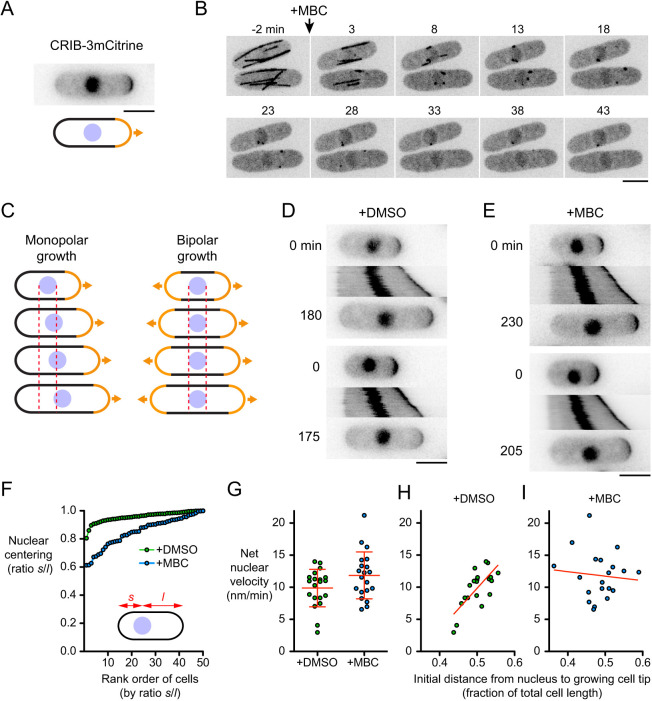
To investigate whether the fission yeast nucleus can move in the absence of MTs, we depolymerized MTs and imaged cells during interphase growth (cell elongation). We used a Cdc42/Rac interactive binding (CRIB)-3mCitrine fusion protein (Mutavchiev et al., 2016), which binds to the active (GTP-bound) form of Cdc42, as a reporter of both cell growth and nuclear position (Fig. 1A). CRIB–3mCitrine localizes to growing cell tips during interphase and to the septum during cell division; it is also present in the nucleus throughout the cell cycle, although this nuclear localization is artifactual and unrelated to Cdc42 function (Bendezú et al., 2015; Mutavchiev et al., 2016; Tatebe et al., 2005). We depolymerized MTs with the drug methyl benzimidazol-2-yl carbamate (MBC). Consistent with what has been shown in previous work (Sawin and Snaith, 2004), in cells expressing GFP–tubulin, nearly all MTs disappeared within 15 min after MBC addition (Fig. 1B). The only remaining MTs were short ‘stubs’ that would be unable to mediate pushing forces for MT-dependent nuclear centering, and even these stubs were barely visible after 20–30 min. We analyzed nuclear movement specifically in monopolar-growing cells (i.e. cells growing at only one tip). As shown in Fig. 1C, this is because in bipolar-growing cells, the nucleus would be expected to be in the cell center not only when there is an active mechanism positioning the nucleus to the cell center but also when there is no nuclear movement at all. By contrast, in monopolar-growing cells, an active nuclear positioning mechanism would result in unidirectional movement of the nucleus over time, in the same direction as growth of the growing cell tip.
Fig. 1.
Microtubules are not required for nuclear movement in fission yeast. (A) CRIB–3mCitrine as a reporter of growing cell tips and nuclear position. (B) Time points from movie showing kinetics of interphase microtubule (GFP–Atb2) depolymerization after treatment with 25 µg/ml methyl benzimidazol-2-yl carbamate (MBC). (C) Cartoon showing how nuclear movement is most easily assayed in monopolar-growing cells rather than bipolar-growing cells. Dashed lines indicate boundaries of nucleus at time zero. See text for details. (D,E) Timepoints and kymographs of nuclear movement from movies of DMSO-treated (D) and MBC-treated (E) cells. Time points (min) correspond to beginning and end of kymographs. Two examples are shown for each condition. (F) Nuclear centering in DMSO-treated and MBC-treated cells. Cells are ordered by ratio s/l, in which s and l are the shorter and longer distances, respectively, from the nucleus to cell tip (50 cells measured for each condition; see Materials and Methods). (G) Net nuclear velocity towards growing cell tips in monopolar-growing DMSO- and MBC-treated cells. Red lines show mean±s.d. (n=20 for each condition). Difference was not statistically significant (non-paired, two-tailed t-test, P=0.067). (H,I) Net nuclear velocity in DMSO-treated (H) and MBC-treated (I) cells, plotted against distance from nucleus to growing cell tip (as a fraction of total cell length) at beginning of movie. Data are from the same cells as in G. Red lines show linear regression (in H, r2=0.53, P=0.0003; in I, r2=0.009, P=0.69). Scale bars: 5 µm.
In control (DMSO-treated) monopolar-growing cells, the nucleus moved in the same direction as the growing cell tip in 100% of cells (100 out of 100 cells; Fig. 1D; Movie 1). Consistent with previous work (see Introduction), in these cells the nucleus was almost always near the cell center (Fig. 1F). Interestingly, in MBC-treated monopolar-growing cells, the nucleus also moved in the same direction as the growing cell tip, in 100% of cells (150 out of 150 cells) (Fig. 1E; Movie 1). We will refer to this movement as ‘MT-independent nuclear movement’ (MINM).
The net velocity of MINM (∼12 nm/min) was not significantly different from nuclear movement in DMSO-treated control cells (Fig. 1G). However, fidelity of nuclear centering in MBC-treated cells was significantly decreased compared to control cells (Fig. 1F). Although similarities in nuclear velocity would seem to be incompatible with differences in nuclear centering, these results were reconciled when we analyzed the relationship between nuclear velocity and the position of the nucleus at the start of imaging (Fig. 1H,I). In control cells, net nuclear velocity directed towards the growing cell tip was lowest when the nucleus was close to the growing cell tip (i.e. at the start of imaging) and highest when the nucleus was far from the growing tip (r2=0.53; P=0.0003), consistent with MTs providing a homeostatic nuclear-centering mechanism (Tran et al., 2001). By contrast, in MBC-treated cells, there was no specific relationship between nuclear velocity and the position of the nucleus at the start of imaging (r2=0.009; P=0.69), suggesting that MINM is largely independent of a centering mechanism. We also confirmed that MINM occurs in MBC-treated cells expressing reporters other than CRIB–3mCitrine (Fig. S1).
Collectively, these results indicate that nuclear movement in fission yeast does not depend exclusively on force generation by MTs; MT-independent mechanisms are also present. However, highly accurate centering of the nucleus requires MTs.
MINM requires actin cables
We hypothesized that MINM may depend on the actin cytoskeleton. During fission yeast interphase, actin filaments are present in two distinct structures – cortical actin patches and actin cables (Kovar et al., 2011; Mishra et al., 2014) (Fig. S2). Cortical actin patches are nucleated by the Arp2/3 complex, primarily at cell tips, and are involved in endocytosis. Actin cables, which contain multiple actin filaments, are nucleated by formin For3 at growing cell tips and are involved in intracellular transport. Because cortical actin patches are far away from the cell nucleus, they would not be expected to directly participate in MINM. By contrast, actin cables extend from growing cell tips into the cell interior and can easily contact the nucleus. We therefore investigated a role for actin cables in MINM.
To test whether MINM requires actin cables, we used for3Δ deletion mutants (see Materials and Methods). In for3Δ cells, interphase actin cables have been described as being either severely compromised or absent altogether, although polarized growth still occurs (Feierbach and Chang, 2001; Nakano et al., 2002). Using Lifeact–mCherry (Huang et al., 2012; Riedl et al., 2008), we observed a small number of residual actin cables in for3Δ cells, but these were shorter, fainter and fewer in number than in wild-type cells (Fig. S2). Residual actin cables may be due to low-level activity of the formin Cdc12, which is normally involved in cytokinesis (Burke et al., 2014). We analyzed monopolar-growing for3Δ cells as described above for wild-type cells (Fig. 2A,B,G; Movie 2). In DMSO-treated for3Δ cells, the nucleus moved in the same direction as the growing cell tip in 100% of cells (8 out of 8 cells; the number of cells was low because most untreated and DMSO-treated for3Δ cells showed premature bipolar growth; see Materials and Methods), By contrast, in MBC-treated for3Δ cells, the nucleus remained stationary in 91% of cells (75 out of 82 cells); the remaining 9% of cells showed slight movement, but this did not resemble MINM. These results indicate that For3 is required for MINM. Although for3Δ cells may contain a small number of actin cables, these are insufficient and/or unable to support MINM.
Fig. 2.
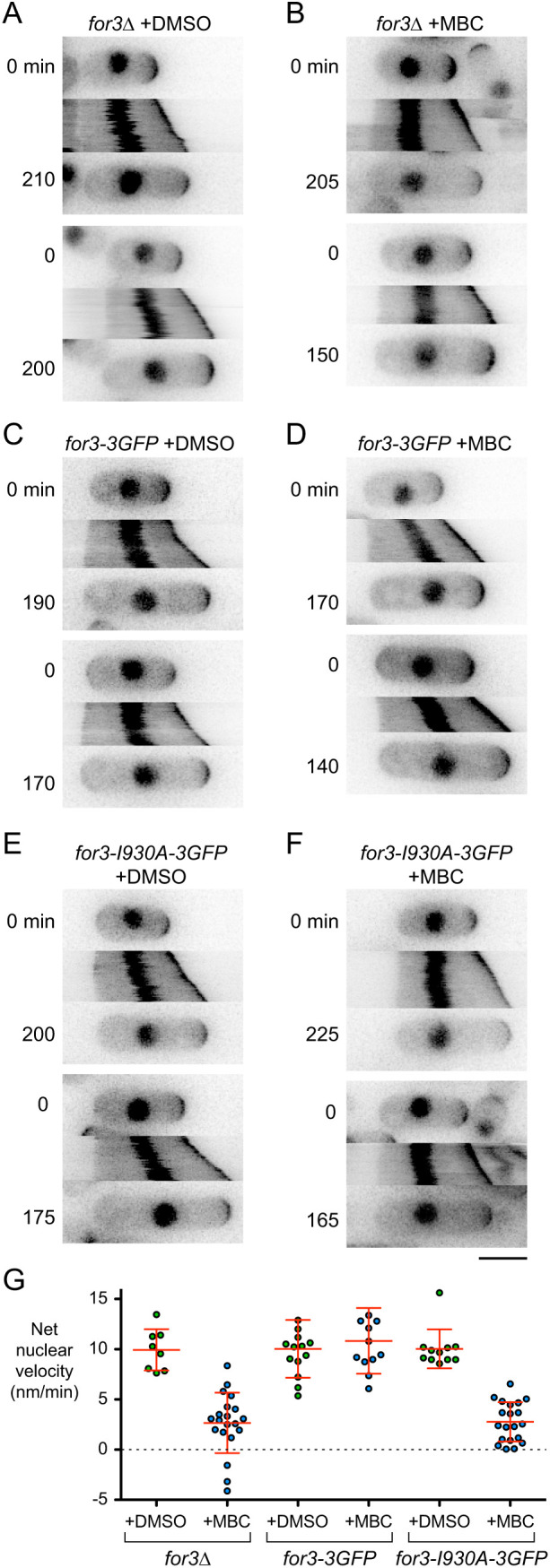
MINM requires actin cables. (A–F) Time points and kymographs of nuclear movement from movies of cells with the indicated genotypes and treatments. Time points (min) correspond to beginning and end of kymographs. Two examples are shown for each genotype and condition. (G) Net nuclear velocity towards growing cell tips in monopolar-growing cells for the genotypes and treatments indicated. Red lines show mean±s.d. For for3Δ and for3-I930A-3GFP cells, difference in net nuclear velocity in DMSO versus MBC was statistically significant (non-paired, two-tailed t-tests, P<0.0001). For for3-3GFP, difference was not statistically significant (non-paired, two-tailed t-test, P=0.521). Non-zero mean velocities for for3Δ+MBC and for for3-I930A-3GFP+MBC are likely an artifact of gradual swelling at cell non-growing tips, combined with the method used to determine net nuclear velocity (see Materials and Methods). Scale bar: 5 µm.
We next tested whether the role of For3 in MINM is specifically related to its role in actin nucleation. For3 contains several domains, with distinct functions and binding partners (Goode and Eck, 2007; Kovar, 2006; Martin and Chang, 2006; Martin et al., 2005, 2007). We analyzed for3-I930A mutants, which contain a single point mutation in the actin-binding region of the formin homology 2 (FH2) domain, directly involved in actin nucleation (Martin and Chang, 2006; Xu et al., 2004). We assayed nuclear movement in monopolar-growing for3-I930A-3GFP cells and for3-3GFP control cells after DMSO and MBC treatment [Fig. 2C–G; Movie 3; note that although both For3–3GFP and For3-I930A–3GFP localize to cell tips (Martin and Chang, 2006), they are too faint to be seen relative to CRIB–3mCitrine]. In DMSO- and MBC-treated for3-3GFP cells, the nucleus moved in the direction of the growing cell tip in 100% of cells (50 out of 50, and 60 out of 60 cells, respectively). DMSO-treated for3-I930A-3GFP cells showed similar behavior (11 out of 11 cells; the number of cells was low because, like for3Δ cells, for3-I930A-3GFP cells normally showed premature bipolar growth; see Materials and Methods). By contrast, in MBC-treated for3-I930A-3GFP cells the nucleus was stationary in 91% of cells (78 out of 86 cells); as with for3Δ cells, the remaining 9% of cells showed slight movement, but this did not resemble MINM. We conclude that For3-mediated actin cable nucleation is critical for MINM.
MINM depends on proximity of the nucleus to the growing cell tip
Because MINM is directed towards the growing cell tip and is dependent on actin cables, which are nucleated by For3 at the growing cell tip, we hypothesized that MINM may involve a mechanical connection between the nucleus and actin cables extending from the growing tip. If so, then MINM might be expected to be strongest when the nucleus is closest to the growing tip (i.e. where the local concentration of actin cables should be highest), and weakest when the nucleus is far away from the growing tip.
We tested this by treating cells with MBC and then centrifuging them to displace the nucleus randomly towards either cell tip, followed by imaging in the presence of MBC (Fig. 3A). When cells are centrifuged in the absence of MBC, the nucleus returns to the cell center within 20 min after centrifugation (Daga et al., 2006). Interestingly, we found that in the presence of MBC, the nucleus behaved very differently, and the type of behavior observed depended on whether centrifugation displaced the nucleus towards the growing or the non-growing cell tip. When displaced towards the growing cell tip, the nucleus continued to move in the direction of the growing tip (i.e. away from the cell center), and nuclear velocity was greatest when centrifugation displaced the nucleus very close to the growing tip (Fig. 3B,D; Movie 4). However, when displaced towards the non-growing tip, the nucleus showed essentially no movement (Fig. 3C,D; Movie 4). Intermediate displacements led to a range of nuclear velocities, all directed towards the growing tip. These results are consistent with a model in which MINM is most efficient when the nucleus is in a region of the cell containing a high local concentration of actin cables (see Discussion). Conversely, when the nucleus is ‘out of reach’ of actin cables, MINM cannot occur.
Fig. 3.
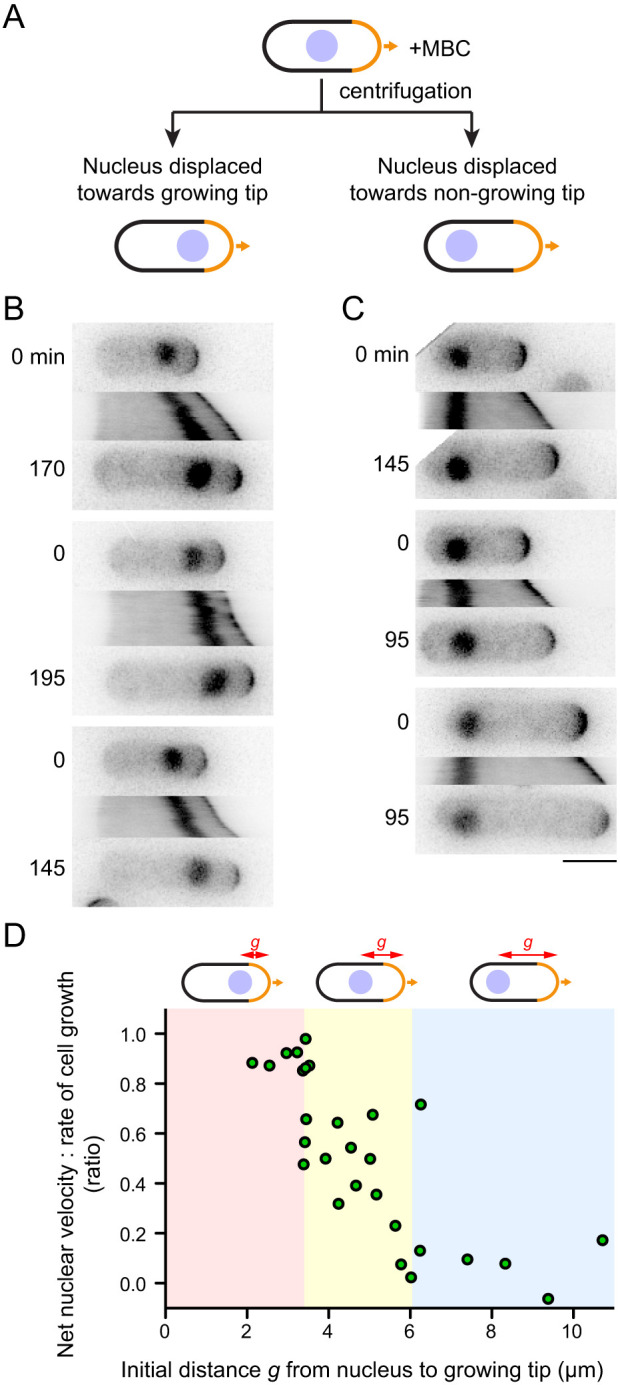
Velocity of MINM depends on distance of nucleus to growing cell tip. (A) Schematic of experiment to displace nucleus from cell center by MBC treatment and centrifugation, prior to imaging in presence of MBC. (B,C) Time points and kymographs of nuclear movement from movies of MBC-treated cells in which nucleus is displaced towards growing tip (B) or towards non-growing tip (C). Time points (min) correspond to beginning and end of kymographs. Three examples are shown for each condition. (D) Net nuclear velocity towards growing cell tips, normalized to rate of cell growth, in cells from these experiments, plotted against distance g from nucleus to growing cell tip at start of imaging. Colored regions of graph highlight different types of behavior. Scale bar: 5 µm.
MINM does not require class V myosins
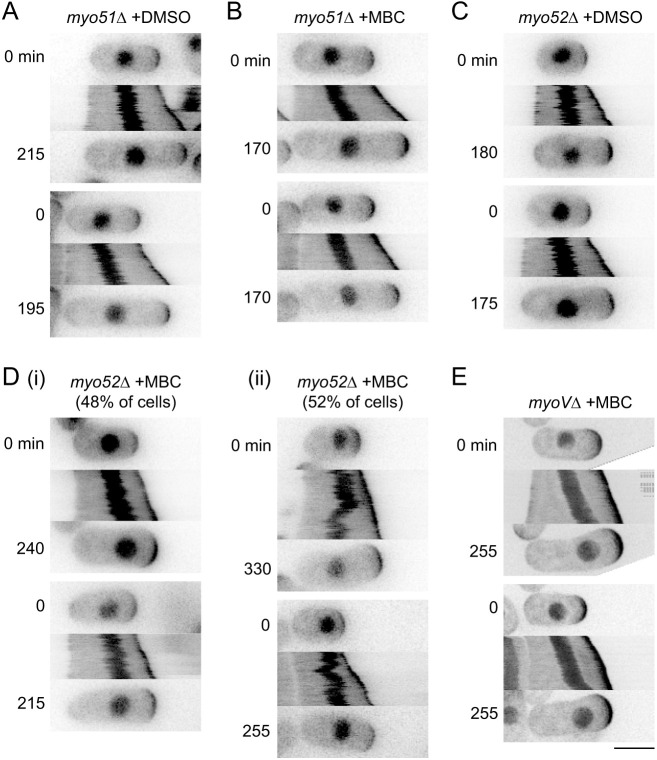
The importance of actin cables in MINM led us to ask whether myosin V motors provide force generation for MINM (see Introduction). Fission yeast contains five myosins: class I myosin Myo1; class II myosins Myo2 and Myp2; and class V myosins Myo51 and Myo52 (East and Mulvihill, 2011). Because Myo1 is associated with cortical actin patches, and Myo2 and Myp2 are associated with the contractile actomyosin ring during cytokinesis, they are unlikely to be involved in MINM and were not considered further. By contrast, class V myosins have broadly conserved roles in intracellular transport (Hammer and Sellers, 2011; Titus, 2018), and therefore we focused on Myo51 and Myo52 (Motegi et al., 2001; Win et al., 2001). Previous work has suggested that Myo51 and Myo52 have distinct functions in vivo. In vegetative cells, Myo51 makes minor contributions to interphase actin cable organization and assembly of the actomyosin ring (Huang et al., 2012; Lo Presti et al., 2012; Wang et al., 2014). Myo51 can also move actin filaments relative to each other in vitro (Tang et al., 2016), and, although it has no known natural cargoes, its motor domain can move an artificial cargo in vivo (Wang et al., 2014). Myo52 has a major role in actin-based transport of membrane-associated cargoes (Bendezú et al., 2012; Lo Presti and Martin, 2011; Mulvihill et al., 2006; Snaith et al., 2011), and myo52Δ deletion mutants have a stubby/spheroid appearance (Motegi et al., 2001; Win et al., 2001). Myo52 is also important for overall actin cable organization, as myo52Δ cells display curled and bundled actin cables (Lo Presti et al., 2012).
We analyzed MINM in monopolar-growing myo51Δ and myo52Δ single-deletion mutants and in a myo51Δ myo52Δ double mutant (here referred to as myoVΔ; Lo Presti et al., 2012). In DMSO-treated monopolar-growing myo51Δ and myo52Δ cells, nuclear movement was similar to that in wild-type cells, and in MBC-treated myo51Δ cells, MINM occurred as in wild-type cells (Fig. 4A–C; like for3 mutants, untreated and DMSO-treated myo52Δ and myoVΔ cells showed premature bipolar growth, and although enough monopolar-growing cells could be identified in DMSO-treated myo52Δ cells to serve as controls, this was not the case for myoVΔ cells; see Materials and Methods). Interestingly, in MBC-treated myo52Δ cells, nuclear movement was more complex (Fig. 4D; Movie 5). In approximately half of the cells (14 out of 29 cells), MINM occurred as in wild-type cells (Fig. 4Di), indicating that Myo52 is not required for MINM. However, in the remaining cells (15 out of 29 cells), the nucleus showed no movement for long periods and/or sudden random movements, including movement away from the growing tip (Fig. 4Dii). In MBC-treated myoVΔ cells, MINM occurred essentially as in wild-type cells in 100% of cells (35 out of 35 cells; Fig. 4E; Movie 6).
Fig. 4.
MINM does not require class V myosins Myo51 and Myo52. (A–E) Time points and kymographs of nuclear movement from movies of cells with the indicated genotypes and treatments. Di shows examples of normal nuclear movement in MBC-treated myo52Δ cells (48% of cells), while Dii shows examples of aberrant nuclear movement, including reversal of direction, in the same population (52% of cells). In E, ‘myoVΔ’ indicates double-mutant myo51Δ myo52Δ. Because essentially all DMSO-treated myoVΔ cells showed premature bipolar growth (see Materials and Methods), no DMSO-treated myoVΔ cells are shown. Time points (min) correspond to beginning and end of kymographs. Two examples are shown for each condition. Scale bar: 5 µm.
These results indicate that neither Myo51 nor Myo52 serves as a motor for MINM. Because MINM depends on actin cables, and actin cables are often disorganized in myo52Δ mutants (Lo Presti et al., 2012), we suspect that the aberrant nuclear movement seen in MBC-treated myo52Δ cells is due to transient changes in actin cable organization. It is not completely clear why the more complex nuclear movements seen in MBC-treated myo52Δ cells were not also observed in myoVΔ cells. Because myoVΔ cells may have slightly different actin cable organization compared to myo51Δ or myo52Δ single mutants (Lo Presti et al., 2012), we speculate that subtle differences in actin organization may be responsible for the observed differences in nuclear movement.
MINM requires VAPs Scs2 and Scs22
While the actin cytoskeleton represents one possible way to physically link the nucleus with the growing cell tip, another way to provide such a link could be via an endomembrane network involving the NE and the endoplasmic reticulum (ER). The NE is continuous with the ER, and in yeasts, much of the ER is cortical (cortER), which is closely apposed to the plasma membrane (PM) as a result of ER–PM membrane contact sites that tether cortER to the PM (Manford et al., 2012; Saheki and De Camilli, 2017; Scorrano et al., 2019; Stefan, 2018; West et al., 2011; Wu et al., 2018; Zhang et al., 2012). In both budding and fission yeast, cortER can cover up to 40% of the cytoplasmic face of the PM (Pichler et al., 2001; Pidoux and Armstrong, 1993; Schuck et al., 2009; Zhang et al., 2012). By contrast, in mammalian cells, which have a cortical actin cytoskeleton, less than 5% of the PM is covered by ER–PM contacts.
To investigate whether the NE–ER network contributes to MINM, we analyzed cells lacking the vesicle-associated membrane protein (VAMP)-associated proteins (VAPs) Scs2 and Scs22 (Manford et al., 2012; Murphy and Levine, 2016; Zhang et al., 2012). VAPs are conserved, ER-localized transmembrane proteins involved in lipid transfer, metabolism, signaling and membrane tethering at membrane contact sites, through interaction with a variety of protein partners on opposing membranes (Gatta and Levine, 2017; Murphy and Levine, 2016; Scorrano et al., 2019; Stefan, 2018, 2020). In fission yeast, Scs2 and Scs22 are particularly important for ER–PM contacts: in scs2Δ scs22Δ double-deletion mutants (herein referred to as scsΔ), the ER shows large-scale detachment from the PM throughout the cell (Ng et al., 2018; Zhang et al., 2012).
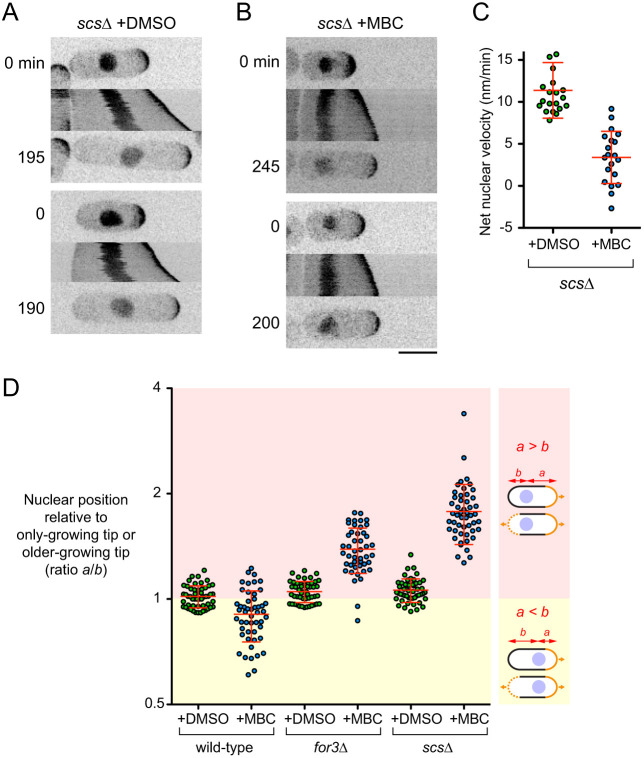
In DMSO-treated monopolar-growing scsΔ cells, the nucleus moved in the direction of the growing cell tip in 100% of cells (80 out of 80 cells) (Fig. 5A). However, after MBC treatment, there was essentially no nuclear movement in 92% of cells (59 out of 64 cells) (Fig. 5B,C; Movie 7); the remaining 8% of cells showed slight movement, but this did not resemble MINM. These results indicate that, like actin cables, fission yeast VAPs play a critical role in MINM.
Fig. 5.
MINM requires VAPs Scs2 and Scs22. (A,B) Time points and kymographs of nuclear movement from movies of DMSO-treated (A) and MBC-treated (B) double-mutant scs2Δ scs22Δ (scsΔ) cells. Time points (min) correspond to beginning and end of kymographs. Two examples of each condition are shown. (C) Net nuclear velocity towards growing cell tips in monopolar-growing DMSO- and MBC-treated scsΔ cells. The difference was statistically significant (non-paired, two-tailed t-test, P<0.0001). Non-zero mean velocities for scsΔ+MBC are likely an artifact of gradual swelling at cell non-growing tips, combined with the method used to determine net nuclear velocity (see Materials and Methods). (D) Nuclear position in late-interphase cells with indicated treatments and genotypes. Nuclear position was measured as ratio a/b, with a being the distance from the nucleus to the only-growing cell tip (for monopolar-growing cells; top cell in each diagram) or from the nucleus to the older-growing tip (for bipolar-growing cells; bottom cell in each diagram), and b being the distance from nucleus to the other cell tip (see Materials and Methods). For MBC treatments, only monopolar-growing cells were measured, because all cells were monopolar-growing. For DMSO treatments, a mixture of monopolar- and bipolar-growing cells were measured for wild-type and scsΔ, while essentially only bipolar-growing cells were measured for for3Δ, because for3Δ cells show premature bipolar growth (see Materials and Methods). In diagrams, solid orange lines indicate only-growing tips in monopolar-growing cells and older-growing tips in bipolar-growing cells; dashed orange lines indicate newer-growing tips in bipolar-growing cells. Yellow zone corresponds to cells with nuclei closer to only-growing or older-growing tips, while pink zone corresponds to cells with nuclei closer to non-growing or newer-growing tips. Note the log2 scale on Y-axis. For DMSO-treated cells, differences between wild-type and for3Δ and between wild-type and scsΔ were statistically significant (non-paired, two-tailed t-test, P=0.024 and 0.006, respectively). Red lines in graphs show mean±s.d. Scale bar: 5 µm.
Consistent with both actin cables and VAPs being required for MINM, we found that in MBC-treated for3Δ and scsΔ mutants, the nuclear position in late interphase (i.e. just prior to mitosis) was strongly biased towards the non-growing cell tip (Fig. 5D; i.e. in monopolar-growing cells; see Materials and Methods). By contrast, in MBC-treated wild-type cells, in which MINM can occur, nuclear position was biased towards the growing cell tip (Fig. 5D).
Actin cables can buffer MT-based pushing forces on the nucleus
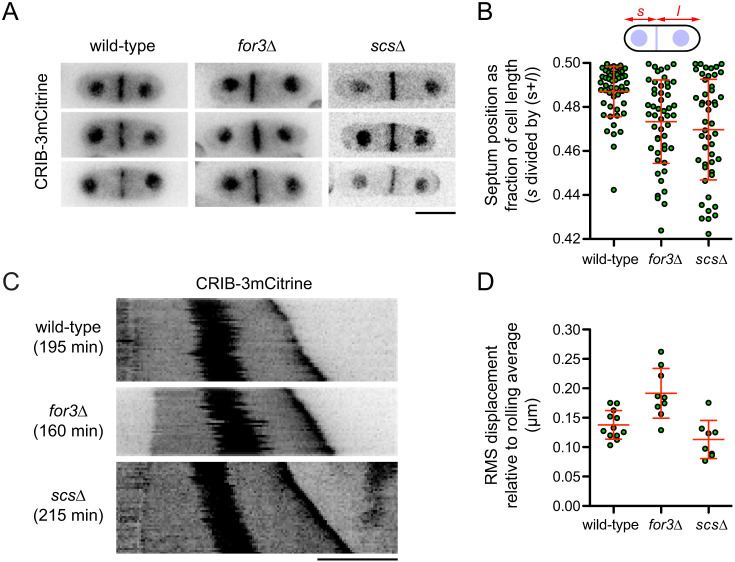
We next asked whether the mechanisms that drive MINM in MBC-treated cells also influence the behavior of the nucleus in unperturbed cells (i.e. in the presence of MTs). In control experiments measuring nuclear position, we noticed that, compared to DMSO-treated wild-type cells, the nucleus in DMSO-treated for3Δ and scsΔ cells showed a small but statistically significant bias towards the non-growing cell tip in monopolar-growing cells and/or the newer-growing cell tip in bipolar-growing cells (Fig. 5D; see Materials and Methods). This suggests that MINM-like mechanisms might also be present in untreated cells. If so, then for3Δ and scsΔ mutants might be expected to show an increased frequency of misplaced septa during normal cell division, because nuclear position in late interphase/early mitosis determines the subsequent plane of cell division (Daga and Chang, 2005). Consistent with this view, we found that while 78% of septa were positioned within 0.02 cell lengths of the cell center in untreated wild-type cells, this was the case for only 42% and 44% of septa in untreated for3Δ and scsΔ cells, respectively (Fig. 6A,B; Zhang et al., 2012).
Fig. 6.
Nucleus and septum positioning defects in for3Δ and scsΔ mutants. (A) CRIB–3mCitrine images showing septum positioning in untreated cells with the indicated genotypes. Three different cells are shown for each genotype. (B) Quantification of septum positioning from images as in A (n=50 cells each). In the diagram, s and l are the shorter and longer distances (respectively) from septum to cell tip. Differences between wild-type and for3Δ and between wild-type and scsΔ were statistically significant (Mann–Whitney U-test, P<0.0001 and P=0.0002, respectively). (C) Kymographs showing fluctuations in nuclear position in untreated cells with the indicated genotypes. Total time (min) for each kymograph is indicated. (D) Root-mean-square (RMS) displacement of nucleus relative to rolling average position over time, from experiments as in C but with higher time resolution (see Materials and Methods and Fig. S3). Each data point corresponds to an individual time-lapse movie. Difference between wild-type and for3Δ was statistically significant (non-paired, two-tailed t-test, P=0.0016). The difference between wild-type and scsΔ was not statistically significant (non-paired, two-tailed t-test, P=0.065). Red lines in graphs show mean±s.d. Scale bars: 5 µm.
To complement these experiments, we analyzed fluctuations in nuclear position in untreated interphase cells. Previously, Tran et al. have shown that MT-based pushing forces lead to fluctuations in nuclear position along the long axis of the cell (Tran et al., 2001), and we observed similar fluctuations in our experiments (compare for example, Fig. 1D vs 1E, or Fig. 2A vs 2B). We hypothesized that if the mechanisms that provide force generation for MINM in MBC-treated cells are also present in untreated cells, then these mechanisms might at least partially resist MT-based pushing forces on the nucleus. We therefore quantified the root-mean-square (RMS) displacement of the nucleus from its rolling-average position during growth of untreated wild-type, for3Δ and scsΔ cells (see Materials and Methods). Interestingly, and consistent with our hypothesis, the RMS displacement of the nucleus in for3Δ cells was on average 40–50% greater than in wild-type cells (Fig. 6C,D; Fig. S3). To further support these findings, we investigated MT organization in for3Δ cells, as it has been suggested that for3Δ mutants may have an altered MT distribution (Feierbach and Chang, 2001), and altered MT dynamics could have provided, at least in principle, an alternative explanation for increased fluctuations in nuclear position. However, we found no qualitative or quantitative differences in MT organization between for3Δ and wild-type cells (Fig. S4). Overall, we interpret these results as indicating that actin cables can normally dampen the transient pushing forces exerted on the nucleus by MTs, such that in the absence of actin cables, fluctuations are increased.
Unlike for3Δ mutants, scsΔ mutants did not show increased fluctuations in nuclear position compared to wild-type cells (Fig. 6C,D; Fig. S3). One possible reason for this may be that in scsΔ mutants, collapse of cortER into cytoplasm (Zhang et al., 2012) could itself dampen MT-based pushing forces on the nucleus (see also Discussion).
DISCUSSION
Nuclear movement in fission yeast is widely thought to depend on MTs (Dupin and Etienne-Manneville, 2011; Gundersen and Worman, 2013; Xiang, 2018), based on a strong body of evidence showing that MT-mediated pushing forces are critical for positioning the nucleus in the cell center (Daga et al., 2006; Sawin et al., 2004; Tolić-Nørrelykke et al., 2005; Tran et al., 2001). Here, we have shown that the fission yeast nucleus can move in the absence of MTs, and we have identified key features of microtubule-independent nuclear movement (MINM). MINM is observed in essentially all cells assayed (i.e. monopolar-growing cells) and is directed towards the growing cell tip. MINM does not exceed the rate of tip growth, and it is strongest when the nucleus is close to the growing tip. MINM requires actin cables but does not require class V myosin motor proteins, which are often employed for actin-based organelle transport (Hammer and Sellers, 2011; Titus, 2018). MINM also depends on the conserved VAP proteins Scs2 and Scs22, which maintain fission yeast cortER via ER–PM contacts (Zhang et al., 2012).
These features suggest that the mechanisms involved in MINM are fundamentally different from previously described mechanisms of nuclear movement (Dupin and Etienne-Manneville, 2011; Gundersen and Worman, 2013; Lele et al., 2018; Xiang, 2018). In nearly all cell types, nuclear movement is driven either by motor proteins acting on MTs or the actin cytoskeleton (‘pulling forces’) or by cytoskeletal polymerization itself (‘pushing forces’). By definition, MINM is independent of MTs. Our gene-deletion experiments rule out the only plausible actin-based motors, class V myosins, as force generators for MINM. We note that artificial tethering of class V myosin Myo52 to the nucleus has been shown to lead to nuclear translocation in fission yeast (Lo Presti et al., 2012); however, this is non-physiological and occurs at a rate of ∼65 nm/min, about six times faster than MINM. Actin-based pushing forces can also be ruled out as force generators for MINM, because actin cables in fission yeast are nucleated by formins specifically at the growing cell tip, and therefore any actin polymerization-based forces on the nucleus would be expected to oppose rather than support MINM.
Beyond these canonical mechanisms for nuclear movement, it has been shown in mouse oocytes that actomyosin-based vesicle movements generate a pressure gradient that can promote centering of the oocyte nucleus via a mechanism of active diffusion (Almonacid et al., 2015; Brangwynne et al., 2009). However, such a mechanism is unlikely to be involved in MINM, because of the small size of fission yeast relative to mammalian oocytes, as well as fundamental differences in actin organization between the two cell types. Moreover, actomyosin-based vesicle movements in the mouse oocyte depend on class V myosins (Almonacid et al., 2015). Another non-canonical mechanism of nuclear movement involving bulk flow is cytoplasmic streaming, which is generally restricted to very large cells or interconnected cell networks in which advective transport may be particularly beneficial (Goldstein and van de Meent, 2015). In animal cell embryos and large plant cells, cytoplasmic streaming is thought to be driven by molecular motors acting on the cytoskeleton (Quinlan, 2016; Goldstein and van de Meent, 2015; Kimura et al., 2017; Tominaga and Ito, 2015), while in multinucleate filamentous fungi, cytoplasmic streaming has been attributed to hydrostatic pressure gradients, with higher pressure at the center of the colony and lower pressure at hyphal tips (Lew, 2005, 2011; Xiang, 2018). However, there is no evidence, or apparent benefit, for cytoplasmic streaming in small single-celled fungi, in which small molecules can travel by diffusion. In addition, while cytoplasmic streaming in filamentous fungi is normally closely correlated with rapid hyphal growth [e.g. ∼13–25 µm/min in Neurospora crassa (Lew, 2005, 2011)], growth rates in fission yeast are much slower [∼0.02–0.04 µm/min at 25°C, depending on specific growth medium (Baumgärtner and Tolić-Nørrelykke, 2009; Mitchison and Nurse, 1985)], and cell tip growth can occur even when MINM cannot (e.g. in for3Δ and scsΔ mutants). MINM is thus unlikely to be due to cytoplasmic streaming.
While MINM appears to be different from known forms of nuclear movement, the specific mechanisms responsible for MINM force generation remain unclear. Below we discuss two non-exclusive, hypothetical models for this (Fig. 7). While both models are consistent with our results, both also contain speculative elements.
Fig. 7.
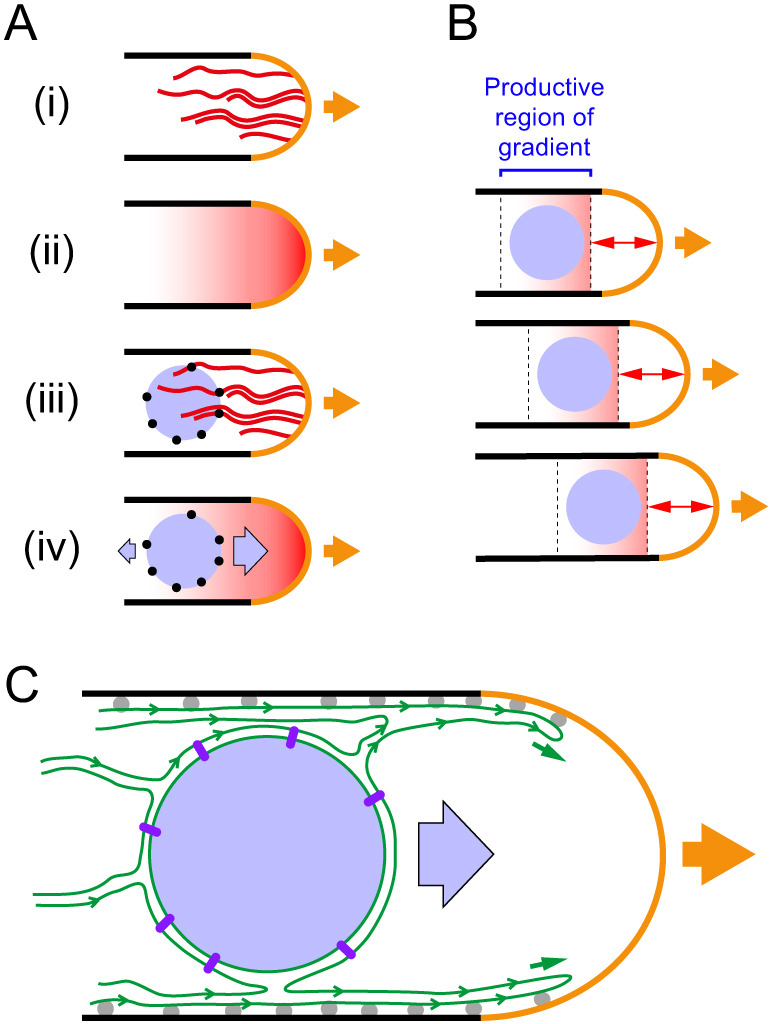
Candidate models for MINM force generation. In all diagrams, cells are drawn growing to the right (orange arrows). (A) Model for MINM based on hypothetical nuclear envelope-associated actin filament-binding proteins (NEAF-BPs) and an actin filament-concentration gradient. (i) Cables of dynamic actin filaments (red) are nucleated from the growing cell tip. (ii) Under steady-state conditions, there is an effective time-averaged gradient in the local concentration of actin filaments along long axis of cell. (iii) NEAF-BPs (black circles) on nucleus (light blue) interact with actin filaments. (iv) As a result of NEAF-BPs and the actin filament concentration gradient, diffusion-driven motion of the nucleus leads to net nuclear movement up the gradient, towards the growing cell tip. See main text for further details. (B) Illustration of how MINM could occur even if only a small portion of an actin filament concentration gradient is productive for diffusion-driven tip-directed movement. Panels show three successive time points during cell growth. Because actin filaments are nucleated from the cell tip and turn over as the tip grows, the productive region of the gradient maintains a constant distance from the growing tip (double-headed arrows) and thus moves relative to the deeper cell interior. According to this mechanism, and consistent with observations, the velocity of MINM would not exceed the rate of cell tip growth. (C) Model for MINM based on ER membrane dynamics. As the cell grows, ER (green) associated with cell cortex extends forward, as a result of ER-PM contact sites (gray balls). Net flow of ER membrane in direction of cell tip, which could be driven by several factors (see main text), is coupled to MINM via links from NE to nuclear interior (purple rods). Diagrams are schematic and not to scale.
Model 1
In the first model for MINM force generation, we hypothesize that dynamic, non-motor-based interactions between the nucleus and actin cables could harness the energy of thermal motion to generate directed nuclear movement (Fig. 7A,B). In this scenario, the nucleus would be physically linked to actin cables via NE-associated actin filament-binding proteins (we will refer to such hypothetical proteins as ‘NEAF-BPs’). Because actin cables are nucleated at the growing cell tip (by the formin For3) and contain dynamic filaments of varying length (Martin and Chang, 2006; Wang and Vavylonis, 2008), during steady-state growth (i.e. of monopolar cells) there should be a gradient in the local concentration of actin filaments along the long axis of the cell, with the highest filament concentration close to the growing tip. In the model, random fluctuations in nuclear position would lead to a net movement of nucleus in the direction of the growing cell tip, driven by an increased number of interactions of NEAF-BPs with actin filaments. Formally, this type of movement would be similar to haptotaxis of cells in an adhesion gradient (Carter, 1965; Petrie et al., 2009).
One possible objection to this model might be that actin cables may not always be evenly distributed in the region of the cell tip. However, compared to the slow speed of MINM (∼12 nm/min), the flexibility and rapid turnover of actin cables [estimated filament half-life ∼15–20 s (Tang et al., 2014)] may be sufficient to generate a reasonably smooth time-averaged gradient of binding sites. A second potential objection might be that as the nucleus gets very close to the growing cell tip, MINM could be opposed by pushing forces derived from actin polymerization itself; indeed, because actin nucleation occurs at the cell tip, such pushing forces would be greatest close to the tip. However, even if such forces were present, this would not necessarily invalidate the model. This is because as the cell tip (i.e. the site of actin nucleation) grows, the gradient of binding sites for NEAF-BPs moves with respect to the cell interior (i.e. in the direction of the cell tip) at the same rate as cell tip growth (Fig. 7B). Accordingly – and consistent with our observations – the mechanism we propose does not actually require the nucleus to be able to move all the way to the cell tip. Overall, as long as there is a productive region of the actin filament-concentration gradient within which directed movement of the nucleus can occur (i.e. a ‘sweet spot’), the model may be plausible.
Currently there are no clear candidates for the NEAF-BPs proposed here. At first glance, likely candidates might include KASH proteins, outer nuclear membrane proteins that interact with inner nuclear membrane SUN proteins to form LINC complexes, which connect the nucleus to the cytoskeleton in many cell types (Burke, 2019; Tapley and Starr, 2013). In metazoan cells, LINC complexes are present over most of the NE. However, in fission yeast, LINC proteins (i.e. the KASH proteins Kms1 and Kms2 and SUN protein Sad1) are restricted to the small region of the NE associated with the spindle pole body [i.e. in vegetatively growing cells (Hagan and Yanagida, 1995; Niwa et al., 2000; Walde and King, 2014)]. As this represents only a minute fraction of the nuclear surface, fission yeast LINC proteins are unlikely to fulfill the role suggested for NEAF-BPs. In addition, in preliminary experiments (Fig. S5A), we have found that MINM does not require KASH protein Kms1 (which, unlike Sad1, is nonessential for viability). We have also found (Fig. S5B) that MINM does not require inner nuclear membrane protein Ima1 (Hiraoka et al., 2011), the fission yeast homolog of mammalian Samp1, which supports LINC complex interactions with the nuclear interior (Borrego-Pinto et al., 2012).
In a mechanism for MINM involving NEAF-BPs and a gradient of actin filament concentration, the role of ER–PM contacts in MINM could be secondary or indirect. For example, loss of cortER (due to loss of VAPs Scs2 and Scs22) may lead to an increase in cytoplasmic ER (cytoER) sheets/cisternae or tubules, or abnormal cytoplasmic aggregation of ER, which could in turn lead to decreased diffusion of the nucleus, impaired NEAF-BP–actin interactions, or increased resistance against the weak forces driving MINM.
Model 2
In the second model for MINM force generation, we hypothesize that extension of cortER in the direction of the growing cell tip during cell growth may be a key driver of MINM (Fig. 7C). This model is motivated by our finding that Scs2 and Scs22, which are required for ER–PM contacts in fission yeast, are also required for MINM. Because the NE is continuous with cytoER and cortER, ER–PM contacts ultimately help link the NE to the PM. In the model, as the cell tip grows, ER–PM contacts would lead to extension of cortER that would in turn lead to movement of the NE and the nucleus (see below). By contrast, in scs2Δ scs22Δ mutants, in which ER–PM connectivity is lost and cortER is collapsed into the cytoplasm (Zhang et al., 2012), cell tip growth would not lead to movement of the NE and nucleus.
We imagine that during cell tip growth (i.e. in wild-type cells), extension of cortER could be driven by diffusive spreading of newly synthesized ER membrane (Blom et al., 2011; Jacquemyn et al., 2017) into ER-free regions of the extending PM, followed by VAP-mediated ‘pinning’ of new cortER to the PM (Fig. 7C). We do not anticipate that actin cytoskeleton-based movements are directly involved in cortER extension, as it has previously been shown that neither For3 nor class V myosins are required for cortER extension during cell elongation [although they partially contribute to the ability of cortER to reach the extreme distal-most portion of the cell tip (Zhang et al., 2012)]. In budding yeast, the class V myosin Myo4 is important for cortER inheritance from mother cell to bud (Estrada et al., 2003), but this may be specifically related to helping cytoER tubules pass through a diffusion barrier at the bud neck (see, for example, Chao et al., 2014; Luedeke et al., 2005). No equivalent barrier would be expected in fission yeast, which does not form buds.
In order for cortER extension to contribute to MINM, we speculate that as cortER extends and new ER–PM contacts are made, dynamic changes in ER organization may lead to differential membrane tension [i.e. non-homogeneous in-plane tension (Pontes et al., 2017; Sens and Plastino, 2015)] within the cytoER network. Differential membrane tension should lead to flow of membrane components from lower-tension to higher-tension regions of the network (Dai and Sheetz, 1995). A net tension gradient within the ER network along the yeast-cell long axis (i.e. with higher tension closer to the growing tip) could therefore lead to movement of components of cytoER and of the NE, which itself is linked to the nuclear interior via association of chromatin with nuclear pores and inner nuclear membrane proteins (Matsuda et al., 2017; Schreiner et al., 2015). While intracellular membrane tension is more difficult to study than plasma membrane tension, there is evidence for different levels of tension in different intracellular membranes and, more recently, perhaps within the ER itself (Goujon et al., 2019; Pontes et al., 2017; Upadhyaya and Sheetz, 2004). One potential source of differential tension could be the dynamic ‘tug of war’ between sheets and tubules in the ER (Westrate et al., 2015), involving reticulon, REEP and atlastin family proteins (Wang and Rapoport, 2019; see also Rangamani et al., 2014). In this context it may be relevant that steady-state maintenance of the ER tubule network requires continuous input of energy, via atlastin-mediated GTP hydrolysis (Powers et al., 2017). Because most lipid synthesis occurs in the ER (Carman and Han, 2011; van Meer et al., 2008), another potential source of differential tension could be nonhomogeneous lipid synthesis within the ER network or, alternatively, dynamic local changes in ER lipid composition (Harayama and Riezman, 2018; Holthuis and Menon, 2014). Regardless of the source, resolution of differential tension via membrane flow might be expected to occur very rapidly (Keren et al., 2008); therefore, for the scenarios proposed here to be plausible, there must be a mechanism to sustain or regenerate differential tension in the longer term (see, for example, Lieber et al., 2015; Schweitzer et al., 2014). Given our finding that actin cables are required for MINM, it is possible that motor-independent interactions of ER with actin cables contribute to regulating differential tension and/or its consequences (Prinz et al., 2000). In this context, it is particularly interesting that in mammalian cells, immobilization of transmembrane proteins via interaction with the cortical actin cytoskeleton can strongly modulate tension-induced plasma membrane bulk flow (Cohen and Shi, 2020; Shi et al., 2018).
Finally, another possible way for cortER extension to contribute to MINM could be via ER lipid synthesis itself. ER synthesis during cell growth should scale with growth, in order to maintain constant ER density and organization, and if lipids are synthesized essentially homogeneously throughout the ER network during growth, then as the cortER expands unidirectionally in the direction of the growing cell tip, there should be a net flow of cytoER in the same direction. As with membrane flow derived from differential membrane tension, this may be capable of driving MINM. However, in this scenario, it is not immediately obvious how actin cables would contribute to MINM.
Currently, we do not have any evidence to suggest that the absence of MINM in for3Δ and scsΔ mutants is due to a single/interdependent defect, as we have found that upon MBC treatment, ER distribution in for3Δ cells is similar to wild-type cells (i.e. unlike ER distribution in scsΔ cells), and For3 localization in scsΔ cells is also similar to wild-type cells (Fig. S6).
Concluding remarks
Given previous work on nuclear positioning in fission yeast, MINM is both novel and unexpected. Because MINM is slow (∼12 nm/min), it may involve relatively weak forces, and our ability to detect it may depend on the highly polarized geometry of fission yeast growth. It is not yet known whether behaviors analogous to MINM exist in other types of cells. In this context, it is interesting that in the reference frame of the growing cell tip, MINM can potentially be viewed as maintenance of nuclear position relative to that tip. From this perspective, it is possible to imagine that in some cell types, a MINM-like mechanism could contribute to an apparent ‘fixed’ position of the nucleus, as part of a balance of forces. Accordingly, we note that lack of actin cables, which leads to loss of MINM when MTs are absent, leads to increased fluctuations of nuclear position when MTs are present.
Our characterization of MINM also highlights the fact that identifying the source(s) of force generation for intracellular movements is not always straightforward, especially when the associated movements are relatively subtle and/or the forces are unconventional. Recent examples of movement involving unconventional forces include the centering of the mouse oocyte nucleus by active diffusion, mentioned above (Almonacid et al., 2015; Brangwynne et al., 2009), and contraction of cytoskeletal filament networks in vitro via entropic forces (Braun et al., 2016; Hilitski et al., 2015; Lansky et al., 2015). How widespread these forces are in different cell types remains to be determined.
MATERIALS AND METHODS
Yeast strain construction and growth
Standard fission yeast techniques were used for strain construction, cell growth and genetic crosses (Ekwall and Thon, 2017; Petersen and Russell, 2016). Cells were grown as required in YE5S rich medium, using Difco yeast extract (Becton Dickinson), or in Edinburgh minimal medium containing sodium glutamate rather than ammonium chloride as nitrogen source (EMMG, also known as ‘pombe minimal glutamate’, or PMG; herein referred to as ‘minimal medium’). Solid medium used 2% Bacto agar (Becton Dickinson). For antibiotic selection, G418, nourseothricin and hygromycin were used at 100 µg/ml on YE5S plates. For genetic crosses, cells were mated on SPA plates and incubated for 3 days at 25°C. Crosses used either random spore analysis or tetrad dissection, and resulting strains were confirmed by colony PCR or fluorescence microscopy, depending on the genotype. A for3Δ deletion strain was constructed using PCR-based gene targeting (Bahler et al., 1998) and was confirmed using colony PCR. Myo52Δ and myo51Δ strains (Win et al., 2001) were obtained from Dan Mulvihill, University of Kent, UK. A myo51Δ myo52Δ strain (also known as myoVΔ) was constructed by deleting myo51 in a myo52Δ strain background. For3-3GFP and For3-I930A-3GFP strains (Martin and Chang, 2006) were obtained from Sophie Martin, University of Lausanne, Switzerland. A Lifeact-mCherry strain (Huang et al., 2012) was obtained from Mohan Balasubramanian, University of Warwick, UK. scs2Δ and scs22Δ strains (Zhang et al., 2012) and a GST–NLS–mCherry strain (Zhang and Oliferenko, 2014) were obtained from Snezhana Oliferenko, Francis Crick Institute, UK. An mCherry–Bgs4 strain (Cortés et al., 2005, 2015) was obtained from Juan Carlos Ribas, University of Salamanca, Spain. Strains and oligonucleotides used in this study are listed in Tables S1 and S2.
Drug treatments and centrifugal nuclear displacement
To depolymerize microtubules, methyl benzimidazol-2yl carbamate (MBC; Sigma) from a 2.5 mg/ml stock in DMSO was added to cultures at 25°C to a final concentration of 25 µg/ml MBC and 1% (v/v) DMSO (Sawin and Snaith, 2004). For analysis of nuclear movement, conditioned minimal medium containing MBC was added to cells and incubated for 15 min at 25°C to depolymerize microtubules prior to imaging (in movies, the ‘zero’ timepoint represents start of movie, not the time of MBC addition). In control experiments, DMSO alone was added to cultures to a final concentration of 1% (v/v).
In preliminary experiments to assess the role of actin cables in MINM, we attempted to use low concentrations of latrunculin A to specifically disrupt actin cables in wild-type cells (i.e. instead of using for3 mutants; Lo Presti and Martin, 2011). However, this was found to be unsuitable, as under our long-term imaging conditions, even low concentrations of latrunculin A (e.g. 2 µM) led to impaired cell polarity and cell growth (Mutavchiev et al., 2016).
Nuclear displacement was performed by first depolymerizing microtubules using 25 µg/ml MBC (as above) at 25°C for 15 min and then centrifuging cells for 3 min at 16,000 g in a microcentrifuge (Carazo-Salas and Nurse, 2006; Daga and Chang, 2005). After centrifugation, all subsequent steps to prepare cells for imaging (see below) were performed in MBC-containing media, to ensure that microtubules remained depolymerized.
Microscopy
To prepare cells for imaging, freshly-grown cells from YE5S solid medium were used to inoculate a 5 ml minimal medium liquid culture that was incubated overnight at 25°C to mid-log-phase density (0.25×107–1.0×107 cells/ml). Cells were then diluted in 25 ml minimal medium and incubated overnight at 25°C to mid-log-phase density (0.5×107–1.0×107 cells/ml). Approximately 400–600 µl of cell culture, depending on culture density, was then added to a 35 mm MatTek coverslip dish (Cat# P35G-0.170-14-C) that had been pretreated by coating with 20 µl of 1 mg/ml solution of soybean lectin (Sigma L1395) in deionized water, air-dried for 10 min and rinsed with deionized water. Cells were allowed to settle on the coverslip dish for 20 min at 25°C. Excess cells were removed using three to five washes of 1 ml conditioned minimal medium, which was prepared by growing a large volume of culture (using the same strain as that being imaged) to mid-log-phase density, centrifuging the culture (3488 g for 5 min; Heraeus Megafuge 40), and recovering the supernatant (Rupes et al., 1999; Sveiczer et al., 1996). After washing, 400 µl of conditioned minimal medium was finally added to the dish, and cells were then imaged. All imaging of cells was performed in conditioned minimal medium.
All imaging was performed at 25°C using temperature-controlled chambers. Nearly all time-lapse imaging used a 5 min interval between time points; the only exception to this was time-lapse imaging of fluctuations in nuclear position, which used a 1 min interval. The majority of live-cell imaging for cells expressing CRIB–3mCitrine (see below for exceptions) was performed on a Deltavision Elite system (Applied Precision) with solid-state illumination, UltimateFocus™, and environmental stage control. Imaging was performed using a 100×/1.40 NA UPLS Apo oil objective (Olympus) and acquired using a Cascade EMCCD camera (Photometrics). For each image, nine Z-sections were collected (100 ms each), with 0.6 µm spacing. Imaging of mCherry–atb2, GFP–atb2, Lifeact–mCherry, GST–NLS–mCherry, mCherry–Bgs4, GFP–ADEL, For3–3GFP (alone, without CRIB–3mCitrine) and of CRIB–3mCitrine in myo52Δ myo51Δ (myoVΔ) and scs2Δ scs22Δ (scsΔ) double-mutant strains (except for experiments measuring fluctuations in nuclear position in scsΔ) was performed on a customized spinning disc confocal microscope with a Nikon TE2000 microscope base, a modified Yokogawa CSU-10 unit (Visitech) and an iXon+ Du888 EMCCD camera (Andor). The microscope was equipped with a 100×/1.45 NA Plan Apo objective (Nikon), Optospin IV filter wheel (Cairn Research), and MS-2000 automated stage with CRISP autofocus (ASI), controlled using Metamorph software (Molecular Devices). For each image, 11 Z-sections were collected (100 ms each), with 0.6 µm spacing.
Under our imaging conditions, we found that in for3Δ, for3-I930A-GFP, myo52Δ, and myo51Δ myo52Δ mutants, most control cells (i.e. DMSO-treated cells) displayed premature bipolar growth. While this made it more difficult to identify monopolar-growing cells for analysis of nuclear movement under control conditions, in all cases except myo51Δ myo52Δ double mutants, sufficient numbers of cells could nevertheless be identified. To our knowledge, polarity growth patterns in myo52Δ and myo51Δ myo52Δ mutants have not previously been characterized. With regard to for3 mutants, the premature bipolar growth we observed is slightly different from the more complex pattern of polarized growth originally described for for3Δ cells, in which, after cell division, one daughter cell displays premature bipolar growth, while the other daughter cell shows only monopolar growth (Feierbach and Chang, 2001). While there is no immediately obvious explanation for these different observations, they may be due to differences in preparation of cells before and during imaging, as some live-cell imaging preparations can alter aspects of cell polarity in unintended ways, likely because of mild cell stress (Tay et al., 2018). The imaging protocol used in our work is specifically designed to minimize cell stress (Mutavchiev et al., 2016). In MBC-treated for3Δ, for3-I930A-GFP, myo52Δ, and myo51Δ myo52Δ cells, monopolar-growing cells were easily identified; we are currently investigating the basis for this different growth pattern.
For measurements of nuclear position in Fig. 1F, interphase (non-septating) cells were chosen for measurement from single time points in movies, avoiding cells that had immediately finished cytokinesis/septation. Both monopolar-growing and bipolar-growing cells were included in measurements. The distance from the center of nucleus to each of the two cell tips was measured, without regard for whether the tip was growing or not. Graph in Fig. 1F shows the ratio s/l, where s corresponds to the shorter of the two distances, and l corresponds to the longer of the two distances (i.e. the maximum possible value for s/l is 1).
For measurements of nuclear position in Fig. 5D, cells were chosen for measurement just prior to mitosis/cytokinesis in movies. Depending on the treatment and/or genotype, either monopolar-growing cells, bipolar-growing cells, or a combination of monopolar-growing and bipolar-growing cells were scored. For MBC-treated cells, all cells scored were monopolar-growing. For DMSO-treated wild-type and scsΔ cells, a mixture of monopolar-growing and bipolar-growing cells were scored, while for DMSO-treated for3Δ cells (most of which showed premature bipolar growth), essentially only bipolar cells were scored. For scoring, septating cells were first identified from movies, and then the movie was stepped back ∼3 frames until a single nucleus was present in the cell (i.e. before karyokinesis). The distance from the center of the nucleus to each of the two cell tips was then measured, taking into account movie data showing which cell tip was the growing tip (for monopolar-growing cells) or had initiated growth first (i.e. ‘older-growing tip’, for bipolar-growing cells). The distance from nucleus to the growing tip (for monopolar-growing cells) or older-growing tip (for bipolar-growing cells) was termed a, and the distance to the other tip was termed b. Graph in Fig. 5D shows the ratio a/b. Because this is a ratio, the Y-axis in Fig. 5D uses a log2 scale, and statistical tests were performed on log2-transformed data.
Image analysis
Image processing and analysis was performed using ImageJ (NIH) and Metamorph (Molecular Devices) software. Movies are shown as maximum projection of Z-sections. Movie time points were registered along X- and Y-axes using the ‘StackReg’ plugin of ImageJ, using the ‘rigid body transformation’ option. Kymographs were generated by drawing an 11 pixel-wide line across the long axis of the cell and then using the ‘KymoResliceWide’ plugin of ImageJ. All kymographs have a 5 min interval between time points.
Nuclear positioning was determined by measuring the distance from the center of the nucleus to both cell tips, using the ‘Analyze’ function of ImageJ. Similarly, septum position was determined by measuring the distance from septum to both cell tips. The shorter distance was termed s, and the longer distance l, and the ratio s/(s+l) was plotted. Cell length was calculated using the ‘Analyze’ function of ImageJ. Net nuclear velocity was determined by measuring the distance from the center of the nucleus to the non-growing cell tip at the beginning and at the end of a movie sequence, and then dividing the distance moved by the duration of the movie. Cells were chosen randomly for quantification (for mutants defective in MINM, this meant that cells showing slight movements were also included in quantification). It is important to note that over long time periods, non-growing tips typically show a small amount of swelling and/or local shape changes that do not represent bona fide polarized growth. As a result, our measurement method can lead to apparent small but non-zero net nuclear velocities for nuclei that would otherwise be judged (e.g. by eye) to be not moving (see, for example, Fig. 2B,F,G). To avoid any unintended bias in quantification, we did not attempt to correct for these ‘apparent non-zero’ velocities, and the data are simply presented as measured.
Fluctuations in nuclear position were determined from movies using a custom in-house-written ImageJ plugin (doi: 10.5281/zenodo.3886623). Briefly, time-lapse movies (with 1-min time interval) are first checked for registration using the ImageJ StackReg plugin, to ensure that the cells to be analyzed are stationary in real space (i.e. apart from their growing cell tips). In the in-house-written plugin, a cut-out image of a user-selected yeast cell is then generated and rotated so that the long axis of the cell is oriented vertically. The cell nucleus is then identified by thresholding, and the centroid coordinates of the nucleus are then measured over time. Values for instantaneous nuclear position in a given cell are then ‘detrended’ by subtracting a running average of nuclear centroid position in the same cell, so that detrended values fluctuate around zero. The amplitude of fluctuations in a given cell is presented as the root-mean-square (RMS) displacement of the detrended data.
Numbers of microtubule bundles per cell in wild-type and for3Δ cells were counted manually from still images.
Numbers of replicate imaging experiments are described below; unless specified otherwise, each individual replicate imaging experiment typically involved imaging 10–15 fields of cells per condition. Experiments imaging MT depolymerization after MBC addition (Fig. 1B) were performed on two replicate occasions, with qualitatively similar results in each replicate. Experiments characterizing MINM and nuclear centering in wild-type cells (Fig. 1D–I) were performed on three replicate occasions, with similar results in each replicate. Quantification in Fig. 1F–I is from one representative replicate. Experiments involving for3Δ and for3-I930A-3GFP mutants (Fig. 2A,B,E,F,G) were performed on three replicate occasions, with similar results in each replicate. Experiments involving positive control for3-3GFP mutants (Fig. 2C,D,G) were performed on two replicate occasions, with similar results in each replicate. Experiments involving displacement of the nucleus by centrifugation (Fig. 3) were performed on three replicate occasions, and results were combined. Experiments involving myo51Δ mutants (Fig. 4A,B) were performed on two replicate occasions, with similar results in each replicate. Experiments involving myo52Δ mutants in DSMO (Fig. 4C) were performed on two replicate occasions, with similar results in each replicate. Experiments involving myo52Δ and myo51Δ myo52Δ (myoVΔ) mutants in MBC (Fig. 4D,E) were performed on three replicate occasions, with similar results in each replicate. Experiments involving scs2Δ scs22Δ (scsΔ) mutants (Fig. 5A,B,C) were performed on three replicate occasions, with similar results in each replicate. Net nuclear velocity was quantified from two of the three replicates. Experiments measuring nucleus and septum positioning in wild-type, for3Δ and scsΔ cells (Fig. 6A,C) were performed on three replicate occasions, with similar results in each replicate. Quantification from these experiments (Fig. 6B,D; Fig. S3) used two of the three replicates. Experiments characterizing MINM using GST–NLS–mCherry and mCherry–Bgs4 instead of CRIB–3mCitrine (Fig. S1) were performed on two replicate occasions, with similar results in each replicate. Experiments imaging actin organization (mCherry–Lifeact) in wild-type and for3Δ cells (Fig. S2) were performed on two replicate occasions, with similar results in each replicate. Experiments involving imaging MT bundles (mCherry–Atb2) in wild-type and for3Δ cells (Fig. S4) were performed on one occasion. Experiments involving kms1Δ and ima1Δ mutants (Fig. S5) were performed on one occasion. Experiments imaging GFP–ADEL in the presence of MBC in wild-type, for3Δ and scsΔ cells (Fig. S6A) were performed on one occasion, imaging 20 fields of per condition. Experiments imaging For3-3GFP alone (i.e. without CRIB-3mCitrine) in wild-type and scsΔ cells (Fig. S6B) were performed on one occasion, imaging 8 fields of cells in DMSO and 20 fields of cells in MBC.
Graphs were plotted using Graphpad Prism (GraphPad, San Diego, USA). Movies for presentation were assembled using ImageJ and QuickTime (Apple). For presentation purposes, most movies were rotated, using the ‘Rotate’ function (bicubic) of ImageJ. Figures were made using ImageJ, Photoshop (Adobe) and Illustrator (Adobe).
Supplementary Material
Acknowledgements
We thank Mohan Balasubramanian, Sophie Martin, Dan Mulvihill, Snezhana Oliferenko, Juan Carlos Ribas, and Dan Zhang for strains, and Adriana Dawes, Andrew Goryachev, Alex Mogilner, Snezhana Oliferenko and members of our laboratory for helpful discussions.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: S.A., K.E.S.; Methodology: S.A., K.E.S.; Software: D.A.K.; Investigation: S.A., Y.D.T.; Writing - original draft: S.A., K.E.S.; Writing - review & editing: S.A., Y.D.T., D.A.K., K.E.S.; Supervision: K.E.S.; Funding acquisition: K.E.S.
Funding
This work was supported by the Wellcome Trust (210659 to K.E.S.). S.A. was supported by a University of Edinburgh Principal's Career Development PhD Scholarship. The Wellcome Centre for Cell Biology is supported by core funding from the Wellcome Trust (203149). Open access funding provided by University of Edinburgh. Deposited in PMC for immediate release.
Supplementary information
Supplementary information available online at https://jcs.biologists.org/lookup/doi/10.1242/jcs.253021.supplemental
References
- Almonacid, M., Ahmed, W. W., Bussonnier, M., Mailly, P., Betz, T., Voituriez, R., Gov, N. S. and Verlhac, M.-H. (2015). Active diffusion positions the nucleus in mouse oocytes. Nat. Cell Biol. 17, 470-479. 10.1038/ncb3131 [DOI] [PubMed] [Google Scholar]
- Bahler, J., Wu, J.-Q., Longtine, M. S., Shah, N. G., McKenzie, A., III, Steever, A. B., Wach, A., Philippsen, P. and Pringle, J. R. (1998). Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14, 943-951. [DOI] [PubMed] [Google Scholar]
- Baumgärtner, S. and Tolić-Nørrelykke, I. M. (2009). Growth pattern of single fission yeast cells is bilinear and depends on temperature and DNA synthesis. Biophys. J. 96, 4336-4347. 10.1016/j.bpj.2009.02.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendezú, F. O., Vincenzetti, V. and Martin, S. G. (2012). Fission yeast Sec3 and Exo70 are transported on actin cables and localize the exocyst complex to cell poles. PLoS ONE 7, e40248. 10.1371/journal.pone.0040248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendezú, F. O., Vincenzetti, V., Vavylonis, D., Wyss, R., Vogel, H. and Martin, S. G. (2015). Spontaneous Cdc42 polarization independent of GDI-mediated extraction and actin-based trafficking. PLoS Biol. 13, e1002097. 10.1371/journal.pbio.1002097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertipaglia, C., Gonçalves, J. C. and Vallee, R. B. (2018). Nuclear migration in mammalian brain development. Semin. Cell Dev. Biol. 82, 57-66. 10.1016/j.semcdb.2017.11.033 [DOI] [PubMed] [Google Scholar]
- Blom, T., Somerharju, P. and Ikonen, E. (2011). Synthesis and biosynthetic trafficking of membrane lipids. Cold Spring Harb. Perspect. Biol. 3, a004713. 10.1101/cshperspect.a004713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bone, C. R. and Starr, D. A. (2016). Nuclear migration events throughout development. J. Cell Sci. 129, 1951-1961. 10.1242/jcs.179788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrego-Pinto, J., Jegou, T., Osorio, D. S., Aurade, F., Gorjanacz, M., Koch, B., Mattaj, I. W. and Gomes, E. R. (2012). Samp1 is a component of TAN lines and is required for nuclear movement. J. Cell Sci. 125, 1099-1105. 10.1242/jcs.087049 [DOI] [PubMed] [Google Scholar]
- Brangwynne, C. P., Koenderink, G. H., MacKintosh, F. C. and Weitz, D. A. (2009). Intracellular transport by active diffusion. Trends Cell Biol. 19, 423-427. 10.1016/j.tcb.2009.04.004 [DOI] [PubMed] [Google Scholar]
- Braun, M., Lansky, Z., Hilitski, F., Dogic, Z. and Diez, S. (2016). Entropic forces drive contraction of cytoskeletal networks. BioEssays 38, 474-481. 10.1002/bies.201500183 [DOI] [PubMed] [Google Scholar]
- Burke, B. (2019). Chain reaction: LINC complexes and nuclear positioning. F1000Res 8, 136. 10.12688/f1000research.16877.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke, T. A., Christensen, J. R., Barone, E., Suarez, C., Sirotkin, V. and Kovar, D. R. (2014). Homeostatic actin cytoskeleton networks are regulated by assembly factor competition for monomers. Curr. Biol. 24, 579-585. 10.1016/j.cub.2014.01.072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carazo-Salas, R. E. and Nurse, P. (2006). Self-organization of interphase microtubule arrays in fission yeast. Nat. Cell Biol. 8, 1102-1107. 10.1038/ncb1479 [DOI] [PubMed] [Google Scholar]
- Carman, G. M. and Han, G.-S. (2011). Regulation of phospholipid synthesis in the yeast Saccharomyces cerevisiae. Annu. Rev. Biochem. 80, 859-883. 10.1146/annurev-biochem-060409-092229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter, S. B. (1965). Principles of cell motility: the direction of cell movement and cancer invasion. Nature 208, 1183-1187. 10.1038/2081183a0 [DOI] [PubMed] [Google Scholar]
- Chang, F. and Nurse, P. (1996). How fission yeast fission in the middle. Cell 84, 191-194. 10.1016/S0092-8674(00)80973-3 [DOI] [PubMed] [Google Scholar]
- Chao, J. T., Wong, A. K. O., Tavassoli, S., Young, B. P., Chruscicki, A., Fang, N. N., Howe, L. A. J., Mayor, T., Foster, L. J. and Loewen, C. J. R. (2014). Polarization of the endoplasmic reticulum by ER-septin tethering. Cell 158, 620-632. 10.1016/j.cell.2014.06.033 [DOI] [PubMed] [Google Scholar]
- Cohen, A. E. and Shi, Z. (2020). Do cell membranes flow like honey or Jiggle like Jello? BioEssays 42, e1900142. 10.1002/bies.201900142 [DOI] [PubMed] [Google Scholar]
- Cortés, J. C. G., Carnero, E., Ishiguro, J., Sánchez, Y., Durán, A. and Ribas, J. C. (2005). The novel fission yeast (1,3)β-D-glucan synthase catalytic subunit Bgs4p is essential during both cytokinesis and polarized growth. J. Cell Sci. 118, 157-174. 10.1242/jcs.01585 [DOI] [PubMed] [Google Scholar]
- Cortes, J. C., Pujol, N., Sato, M., Pinar, M., Ramos, M., Moreno, B., Osumi, M., Ribas, J. C. and Perez, P. (2015). Cooperation between paxillin-like protein Pxl1 and glucan synthase Bgs1 is essential for actomyosin ring stability and septum formation in fission yeast. PLoS Genet. 11, e1005358. 10.1371/journal.pgen.1005358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisp, M., Liu, Q., Roux, K., Rattner, J. B., Shanahan, C., Burke, B., Stahl, P. D. and Hodzic, D. (2006). Coupling of the nucleus and cytoplasm: role of the LINC complex. J. Cell Biol. 172, 41-53. 10.1083/jcb.200509124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daga, R. R. and Chang, F. (2005). Dynamic positioning of the fission yeast cell division plane. Proc. Natl. Acad. Sci. USA 102, 8228-8232. 10.1073/pnas.0409021102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daga, R. R., Yonetani, A. and Chang, F. (2006). Asymmetric microtubule pushing forces in nuclear centering. Curr. Biol. 16, 1544-1550. 10.1016/j.cub.2006.06.026 [DOI] [PubMed] [Google Scholar]
- Dai, J. and Sheetz, M. P. (1995). Axon membrane flows from the growth cone to the cell body. Cell 83, 693-701. 10.1016/0092-8674(95)90182-5 [DOI] [PubMed] [Google Scholar]
- Dupin, I. and Etienne-Manneville, S. (2011). Nuclear positioning: mechanisms and functions. Int. J. Biochem. Cell Biol. 43, 1698-1707. 10.1016/j.biocel.2011.09.004 [DOI] [PubMed] [Google Scholar]
- Dupin, I., Sakamoto, Y. and Etienne-Manneville, S. (2011). Cytoplasmic intermediate filaments mediate actin-driven positioning of the nucleus. J. Cell Sci. 124, 865-872. 10.1242/jcs.076356 [DOI] [PubMed] [Google Scholar]
- East, D. A. and Mulvihill, D. P. (2011). Regulation and function of the fission yeast myosins. J. Cell Sci. 124, 1383-1390. 10.1242/jcs.078527 [DOI] [PubMed] [Google Scholar]
- Ekwall, K. and Thon, G. (2017). Spore analysis and tetrad dissection of Schizosaccharomyces pombe. Cold Spring Harb. Protoc. 2017, pdb.prot091710. 10.1101/pdb.prot091710 [DOI] [PubMed] [Google Scholar]
- Estrada, P., Kim, J., Coleman, J., Walker, L., Dunn, B., Takizawa, P., Novick, P. and Ferro-Novick, S. (2003). Myo4p and She3p are required for cortical ER inheritance in Saccharomyces cerevisiae. J. Cell Biol. 163, 1255-1266. 10.1083/jcb.200304030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feierbach, B. and Chang, F. (2001). Roles of the fission yeast formin for3p in cell polarity, actin cable formation and symmetric cell division. Curr. Biol. 11, 1656-1665. 10.1016/S0960-9822(01)00525-5 [DOI] [PubMed] [Google Scholar]
- Fridolfsson, H. N. and Starr, D. A. (2010). Kinesin-1 and dynein at the nuclear envelope mediate the bidirectional migrations of nuclei. J. Cell Biol. 191, 115-128. 10.1083/jcb.201004118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatta, A. T. and Levine, T. P. (2017). Piecing together the patchwork of contact sites. Trends Cell Biol. 27, 214-229. 10.1016/j.tcb.2016.08.010 [DOI] [PubMed] [Google Scholar]
- Goldstein, R. E. and van de Meent, J.-W. (2015). A physical perspective on cytoplasmic streaming. Interface Focus 5, 20150030. 10.1098/rsfs.2015.0030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes, E. R., Jani, S. and Gundersen, G. G. (2005). Nuclear movement regulated by Cdc42, MRCK, myosin, and actin flow establishes MTOC polarization in migrating cells. Cell 121, 451-463. 10.1016/j.cell.2005.02.022 [DOI] [PubMed] [Google Scholar]
- Goode, B. L. and Eck, M. J. (2007). Mechanism and function of formins in the control of actin assembly. Annu. Rev. Biochem. 76, 593-627. 10.1146/annurev.biochem.75.103004.142647 [DOI] [PubMed] [Google Scholar]
- Goujon, A., Colom, A., Straková, K., Mercier, V., Mahecic, D., Manley, S., Sakai, N., Roux, A. and Matile, S. (2019). Mechanosensitive fluorescent probes to image membrane tension in mitochondria, endoplasmic reticulum, and lysosomes. J. Am. Chem. Soc. 141, 3380-3384. 10.1021/jacs.8b13189 [DOI] [PubMed] [Google Scholar]
- Gundersen, G. G. and Worman, H. J. (2013). Nuclear positioning. Cell 152, 1376-1389. 10.1016/j.cell.2013.02.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan, I. and Yanagida, M. (1995). The product of the spindle formation gene sad1+ associates with the fission yeast spindle pole body and is essential for viability. J. Cell Biol. 129, 1033-1047. 10.1083/jcb.129.4.1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan, I. and Yanagida, M. (1997). Evidence for cell cycle-specific, spindle pole body-mediated, nuclear positioning in the fission yeast Schizosaccharomyces pombe. J. Cell Sci. 110, 1851-1866. [DOI] [PubMed] [Google Scholar]
- Hammer, J. A., III and Sellers, J. R. (2011). Walking to work: roles for class V myosins as cargo transporters. Nat. Rev. Mol. Cell Biol. 13, 13-26. 10.1038/nrm3248 [DOI] [PubMed] [Google Scholar]
- Harayama, T. and Riezman, H. (2018). Understanding the diversity of membrane lipid composition. Nat. Rev. Mol. Cell Biol. 19, 281-296. 10.1038/nrm.2017.138 [DOI] [PubMed] [Google Scholar]
- Hilitski, F., Ward, A. R., Cajamarca, L., Hagan, M. F., Grason, G. M. and Dogic, Z. (2015). Measuring cohesion between macromolecular filaments one pair at a time: depletion-induced microtubule bundling. Phys. Rev. Lett. 114, 138102. 10.1103/PhysRevLett.114.138102 [DOI] [PubMed] [Google Scholar]
- Hiraoka, Y., Maekawa, H., Asakawa, H., Chikashige, Y., Kojidani, T., Osakada, H., Matsuda, A. and Haraguchi, T. (2011). Inner nuclear membrane protein Ima1 is dispensable for intranuclear positioning of centromeres. Genes Cells 16, 1000-1011. 10.1111/j.1365-2443.2011.01544.x [DOI] [PubMed] [Google Scholar]
- Holthuis, J. C. M. and Menon, A. K. (2014). Lipid landscapes and pipelines in membrane homeostasis. Nature 510, 48-57. 10.1038/nature13474 [DOI] [PubMed] [Google Scholar]
- Huang, J., Huang, Y., Yu, H., Subramanian, D., Padmanabhan, A., Thadani, R., Tao, Y., Tang, X., Wedlich-Soldner, R. and Balasubramanian, M. K. (2012). Nonmedially assembled F-actin cables incorporate into the actomyosin ring in fission yeast. J. Cell Biol. 199, 831-847. 10.1083/jcb.201209044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsmann, S., Ylänne, J. and Brown, N. H. (2013). Filopodia-like actin cables position nuclei in association with perinuclear actin in Drosophila nurse cells. Dev. Cell 26, 604-615. 10.1016/j.devcel.2013.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isermann, P. and Lammerding, J. (2013). Nuclear mechanics and mechanotransduction in health and disease. Curr. Biol. 23, R1113-R1121. 10.1016/j.cub.2013.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquemyn, J., Cascalho, A. and Goodchild, R. E. (2017). The ins and outs of endoplasmic reticulum-controlled lipid biosynthesis. EMBO Rep. 18, 1905-1921. 10.15252/embr.201643426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren, K., Pincus, Z., Allen, G. M., Barnhart, E. L., Marriott, G., Mogilner, A. and Theriot, J. A. (2008). Mechanism of shape determination in motile cells. Nature 453, 475-480. 10.1038/nature06952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, K., Mamane, A., Sasaki, T., Sato, K., Takagi, J., Niwayama, R., Hufnagel, L., Shimamoto, Y., Joanny, J.-F., Uchida, S.et al. (2017). Endoplasmic-reticulum-mediated microtubule alignment governs cytoplasmic streaming. Nat. Cell Biol. 19, 399-406. 10.1038/ncb3490 [DOI] [PubMed] [Google Scholar]
- Kovar, D. R. (2006). Molecular details of formin-mediated actin assembly. Curr. Opin. Cell Biol. 18, 11-17. 10.1016/j.ceb.2005.12.011 [DOI] [PubMed] [Google Scholar]
- Kovar, D. R., Sirotkin, V. and Lord, M. (2011). Three's company: the fission yeast actin cytoskeleton. Trends Cell Biol. 21, 177-187. 10.1016/j.tcb.2010.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lansky, Z., Braun, M., Lüdecke, A., Schlierf, M., ten Wolde, P. R., Janson, M. E. and Diez, S. (2015). Diffusible crosslinkers generate directed forces in microtubule networks. Cell 160, 1159-1168. 10.1016/j.cell.2015.01.051 [DOI] [PubMed] [Google Scholar]
- Lele, T. P., Dickinson, R. B. and Gundersen, G. G. (2018). Mechanical principles of nuclear shaping and positioning. J. Cell Biol. 217, 3330-3342. 10.1083/jcb.201804052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy, J. R. and Holzbaur, E. L. F. (2008). Dynein drives nuclear rotation during forward progression of motile fibroblasts. J. Cell Sci. 121, 3187-3195. 10.1242/jcs.033878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew, R. R. (2005). Mass flow and pressure-driven hyphal extension in Neurospora crassa. Microbiology 151, 2685-2692. 10.1099/mic.0.27947-0 [DOI] [PubMed] [Google Scholar]
- Lew, R. R. (2011). How does a hypha grow? The biophysics of pressurized growth in fungi. Nat. Rev. Microbiol. 9, 509-518. 10.1038/nrmicro2591 [DOI] [PubMed] [Google Scholar]
- Lieber, A. D., Schweitzer, Y., Kozlov, M. M. and Keren, K. (2015). Front-to-rear membrane tension gradient in rapidly moving cells. Biophys. J. 108, 1599-1603. 10.1016/j.bpj.2015.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Presti, L. and Martin, S. G. (2011). Shaping fission yeast cells by rerouting actin-based transport on microtubules. Curr. Biol. 21, 2064-2069. 10.1016/j.cub.2011.10.033 [DOI] [PubMed] [Google Scholar]
- Lo Presti, L., Chang, F. and Martin, S. G. (2012). Myosin Vs organize actin cables in fission yeast. Mol. Biol. Cell 23, 4579-4591. 10.1091/mbc.e12-07-0499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luedeke, C., Frei, S. B., Sbalzarini, I., Schwarz, H., Spang, A. and Barral, Y. (2005). Septin-dependent compartmentalization of the endoplasmic reticulum during yeast polarized growth. J. Cell Biol. 169, 897-908. 10.1083/jcb.200412143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luxton, G. W. G., Gomes, E. R., Folker, E. S., Vintinner, E. and Gundersen, G. G. (2010). Linear arrays of nuclear envelope proteins harness retrograde actin flow for nuclear movement. Science 329, 956-959. 10.1126/science.1189072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manford, A. G., Stefan, C. J., Yuan, H. L., Macgurn, J. A. and Emr, S. D. (2012). ER-to-plasma membrane tethering proteins regulate cell signaling and ER morphology. Dev. Cell 23, 1129-1140. 10.1016/j.devcel.2012.11.004 [DOI] [PubMed] [Google Scholar]
- Martin, S. G. and Chang, F. (2006). Dynamics of the formin for3p in actin cable assembly. Curr. Biol. 16, 1161-1170. 10.1016/j.cub.2006.04.040 [DOI] [PubMed] [Google Scholar]
- Martin, S. G., McDonald, W. H., Yates, J. R., III and Chang, F. (2005). Tea4p links microtubule plus ends with the formin for3p in the establishment of cell polarity. Dev. Cell 8, 479-491. 10.1016/j.devcel.2005.02.008 [DOI] [PubMed] [Google Scholar]
- Martin, S. G., Rincón, S. A., Basu, R., Pérez, P. and Chang, F. (2007). Regulation of the formin for3p by cdc42p and bud6p. Mol. Biol. Cell 18, 4155-4167. 10.1091/mbc.e07-02-0094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda, A., Asakawa, H., Haraguchi, T. and Hiraoka, Y. (2017). Spatial organization of the Schizosaccharomyces pombe genome within the nucleus. Yeast 34, 55-66. 10.1002/yea.3217 [DOI] [PubMed] [Google Scholar]
- Mishra, M., Huang, J. and Balasubramanian, M. K. (2014). The yeast actin cytoskeleton. FEMS Microbiol. Rev. 38, 213-227. 10.1111/1574-6976.12064 [DOI] [PubMed] [Google Scholar]
- Mitchison, J. M. and Nurse, P. (1985). Growth in cell length in the fission yeast Schizosaccharomyces pombe. J. Cell Sci. 75, 357-376. [DOI] [PubMed] [Google Scholar]
- Moore, J. K. and Cooper, J. A. (2010). Coordinating mitosis with cell polarity: molecular motors at the cell cortex. Semin. Cell Dev. Biol. 21, 283-289. 10.1016/j.semcdb.2010.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motegi, F., Arai, R. and Mabuchi, I. (2001). Identification of two type V myosins in fission yeast, one of which functions in polarized cell growth and moves rapidly in the cell. Mol. Biol. Cell 12, 1367-1380. 10.1091/mbc.12.5.1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulvihill, D. P., Edwards, S. R. and Hyams, J. S. (2006). A critical role for the type V myosin, Myo52, in septum deposition and cell fission during cytokinesis in Schizosaccharomyces pombe. Cell Motil. Cytoskeleton 63, 149-161. 10.1002/cm.20113 [DOI] [PubMed] [Google Scholar]
- Murphy, S. E. and Levine, T. P. (2016). VAP, a versatile access point for the endoplasmic reticulum: review and analysis of FFAT-like motifs in the VAPome. Biochim. Biophys. Acta 1861, 952-961. 10.1016/j.bbalip.2016.02.009 [DOI] [PubMed] [Google Scholar]
- Mutavchiev, D. R., Leda, M. and Sawin, K. E. (2016). Remodeling of the fission yeast Cdc42 cell-polarity module via the Sty1 p38 stress-activated protein kinase pathway. Curr. Biol. 26, 2921-2928. 10.1016/j.cub.2016.08.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano, K., Imai, J., Arai, R., Toh, E. A., Matsui, Y. and Mabuchi, I. (2002). The small GTPase Rho3 and the diaphanous/formin For3 function in polarized cell growth in fission yeast. J. Cell Sci. 115, 4629-4639. 10.1242/jcs.00150 [DOI] [PubMed] [Google Scholar]
- Ng, A. Y. E., Ng, A. Q. E. and Zhang, D. (2018). ER-PM contacts restrict exocytic sites for polarized morphogenesis. Curr. Biol. 28, 146-153.e6. 10.1016/j.cub.2017.11.055 [DOI] [PubMed] [Google Scholar]
- Niwa, O., Shimanuki, M. and Miki, F. (2000). Telomere-led bouquet formation facilitates homologous chromosome pairing and restricts ectopic interaction in fission yeast meiosis. EMBO J. 19, 3831-3840. 10.1093/emboj/19.14.3831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmakumar, V. C., Libotte, T., Lu, W., Zaim, H., Abraham, S., Noegel, A. A., Gotzmann, J., Foisner, R. and Karakesisoglou, I. (2005). The inner nuclear membrane protein Sun1 mediates the anchorage of Nesprin-2 to the nuclear envelope. J. Cell Sci. 118, 3419-3430. 10.1242/jcs.02471 [DOI] [PubMed] [Google Scholar]
- Petersen, J. and Russell, P. (2016). Growth and the Environment of Schizosaccharomyces pombe. Cold Spring Harb. Protoc. 2016, pdb.top079764. 10.1101/pdb.top079764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrie, R. J., Doyle, A. D. and Yamada, K. M. (2009). Random versus directionally persistent cell migration. Nat. Rev. Mol. Cell Biol. 10, 538-549. 10.1038/nrm2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichler, H., Gaigg, B., Hrastnik, C., Achleitner, G., Kohlwein, S. D., Zellnig, G., Perktold, A. and Daum, G. (2001). A subfraction of the yeast endoplasmic reticulum associates with the plasma membrane and has a high capacity to synthesize lipids. Eur. J. Biochem. 268, 2351-2361. 10.1046/j.1432-1327.2001.02116.x [DOI] [PubMed] [Google Scholar]
- Pidoux, A. L. and Armstrong, J. (1993). The BiP protein and the endoplasmic reticulum of Schizosaccharomyces pombe: fate of the nuclear envelope during cell division. J. Cell Sci. 105, 1115-1120. [DOI] [PubMed] [Google Scholar]
- Pontes, B., Monzo, P. and Gauthier, N. C. (2017). Membrane tension: a challenging but universal physical parameter in cell biology. Semin. Cell Dev. Biol. 71, 30-41. 10.1016/j.semcdb.2017.08.030 [DOI] [PubMed] [Google Scholar]
- Powers, R. E., Wang, S., Liu, T. Y. and Rapoport, T. A. (2017). Reconstitution of the tubular endoplasmic reticulum network with purified components. Nature 543, 257-260. 10.1038/nature21387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz, W. A., Grzyb, L., Veenhuis, M., Kahana, J. A., Silver, P. A. and Rapoport, T. A. (2000). Mutants affecting the structure of the cortical endoplasmic reticulum in Saccharomyces cerevisiae. J. Cell Biol. 150, 461-474. 10.1083/jcb.150.3.461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruyne, D., Legesse-Miller, A., Gao, L., Dong, Y. and Bretscher, A. (2004). Mechanisms of polarized growth and organelle segregation in yeast. Annu. Rev. Cell Dev. Biol. 20, 559-591. 10.1146/annurev.cellbio.20.010403.103108 [DOI] [PubMed] [Google Scholar]
- Quinlan, M. E. (2016). Cytoplasmic streaming in the Drosophila oocyte. Annu. Rev. Cell Dev. Biol. 32, 173-195. 10.1146/annurev-cellbio-111315-125416 [DOI] [PubMed] [Google Scholar]
- Rangamani, P., Mandadap, K. K. and Oster, G. (2014). Protein-induced membrane curvature alters local membrane tension. Biophys. J. 107, 751-762. 10.1016/j.bpj.2014.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinsch, S. and Gonczy, P. (1998). Mechanisms of nuclear positioning. J. Cell Sci. 111, 2283-2295. [DOI] [PubMed] [Google Scholar]
- Riedl, J., Crevenna, A. H., Kessenbrock, K., Yu, J. H., Neukirchen, D., Bista, M., Bradke, F., Jenne, D., Holak, T. A., Werb, Z.et al. (2008). Lifeact: a versatile marker to visualize F-actin. Nat. Methods 5, 605-607. 10.1038/nmeth.1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupes, I., Jia, Z. and Young, P. G. (1999). Ssp1 promotes actin depolymerization and is involved in stress response and new end take-off control in fission yeast. Mol. Biol. Cell 10, 1495-1510. 10.1091/mbc.10.5.1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saheki, Y. and De Camilli, P. (2017). Endoplasmic reticulum-plasma membrane contact sites. Annu. Rev. Biochem. 86, 659-684. 10.1146/annurev-biochem-061516-044932 [DOI] [PubMed] [Google Scholar]
- Sawin, K. E. and Snaith, H. A. (2004). Role of microtubules and tea1p in establishment and maintenance of fission yeast cell polarity. J. Cell Sci. 117, 689-700. 10.1242/jcs.00925 [DOI] [PubMed] [Google Scholar]
- Sawin, K. E. and Tran, P. T. (2006). Cytoplasmic microtubule organization in fission yeast. Yeast 23, 1001-1014. 10.1002/yea.1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawin, K. E., Lourenco, P. C. C. and Snaith, H. A. (2004). Microtubule nucleation at non-spindle pole body microtubule-organizing centers requires fission yeast centrosomin-related protein mod20p. Curr. Biol. 14, 763-775. 10.1016/j.cub.2004.03.042 [DOI] [PubMed] [Google Scholar]
- Schreiner, S. M., Koo, P. K., Zhao, Y., Mochrie, S. G. J. and King, M. C. (2015). The tethering of chromatin to the nuclear envelope supports nuclear mechanics. Nat. Commun. 6, 7159. 10.1038/ncomms8159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck, S., Prinz, W. A., Thorn, K. S., Voss, C. and Walter, P. (2009). Membrane expansion alleviates endoplasmic reticulum stress independently of the unfolded protein response. J. Cell Biol. 187, 525-536. 10.1083/jcb.200907074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweitzer, Y., Lieber, A. D., Keren, K. and Kozlov, M. M. (2014). Theoretical analysis of membrane tension in moving cells. Biophys. J. 106, 84-92. 10.1016/j.bpj.2013.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scorrano, L., De Matteis, M. A., Emr, S., Giordano, F., Hajnóczky, G., Kornmann, B., Lackner, L. L., Levine, T. P., Pellegrini, L., Reinisch, K.et al. (2019). Coming together to define membrane contact sites. Nat. Commun. 10, 1287. 10.1038/s41467-019-09253-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sens, P. and Plastino, J. (2015). Membrane tension and cytoskeleton organization in cell motility. J. Phys. Condens. Matter 27, 273103. 10.1088/0953-8984/27/27/273103 [DOI] [PubMed] [Google Scholar]
- Shi, Z., Graber, Z. T., Baumgart, T., Stone, H. A. and Cohen, A. E. (2018). Cell membranes resist flow. Cell 175, 1769-1779.e13. 10.1016/j.cell.2018.09.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snaith, H. A., Thompson, J., Yates, J. R., III and Sawin, K. E. (2011). Characterization of Mug33 reveals complementary roles for actin cable-dependent transport and exocyst regulators in fission yeast exocytosis. J. Cell Sci. 124, 2187-2199. 10.1242/jcs.084038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefan, C. J. (2018). Building ER-PM contacts: keeping calm and ready on alarm. Curr. Opin. Cell Biol. 53, 1-8. 10.1016/j.ceb.2018.03.008 [DOI] [PubMed] [Google Scholar]
- Stefan, C. J. (2020). Endoplasmic reticulum-plasma membrane contacts: Principals of phosphoinositide and calcium signaling. Curr. Opin. Cell Biol. 63, 125-134. 10.1016/j.ceb.2020.01.010 [DOI] [PubMed] [Google Scholar]
- Sveiczer, A., Novak, B. and Mitchison, J. M. (1996). The size control of fission yeast revisited. J. Cell Sci. 109, 2947-2957. [DOI] [PubMed] [Google Scholar]
- Tamura, K., Iwabuchi, K., Fukao, Y., Kondo, M., Okamoto, K., Ueda, H., Nishimura, M. and Hara-Nishimura, I. (2013). Myosin XI-i links the nuclear membrane to the cytoskeleton to control nuclear movement and shape in Arabidopsis. Curr. Biol. 23, 1776-1781. 10.1016/j.cub.2013.07.035 [DOI] [PubMed] [Google Scholar]
- Tang, H., Laporte, D. and Vavylonis, D. (2014). Actin cable distribution and dynamics arising from cross-linking, motor pulling, and filament turnover. Mol. Biol. Cell 25, 3006-3016. 10.1091/mbc.e14-05-0965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, Q., Billington, N., Krementsova, E. B., Bookwalter, C. S., Lord, M. and Trybus, K. M. (2016). A single-headed fission yeast myosin V transports actin in a tropomyosin-dependent manner. J. Cell Biol. 214, 167-179. 10.1083/jcb.201511102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapley, E. C. and Starr, D. A. (2013). Connecting the nucleus to the cytoskeleton by SUN-KASH bridges across the nuclear envelope. Curr. Opin. Cell Biol. 25, 57-62. 10.1016/j.ceb.2012.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatebe, H., Shimada, K., Uzawa, S., Morigasaki, S. and Shiozaki, K. (2005). Wsh3/Tea4 is a novel cell-end factor essential for bipolar distribution of Tea1 and protects cell polarity under environmental stress in S. pombe. Curr. Biol. 15, 1006-1015. 10.1016/j.cub.2005.04.061 [DOI] [PubMed] [Google Scholar]
- Tay, Y. D., Leda, M., Goryachev, A. B. and Sawin, K. E. (2018). Local and global Cdc42 guanine nucleotide exchange factors for fission yeast cell polarity are coordinated by microtubules and the Tea1-Tea4-Pom1 axis. J. Cell Sci. 131, jcs216580. 10.1242/jcs.216580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titus, M. A. (2018). Myosin-driven intracellular transport. Cold Spring Harb. Perspect. Biol. 10, a021972. 10.1101/cshperspect.a021972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda, T., Umesono, K., Hirata, A. and Yanagida, M. (1983). Cold-sensitive nuclear division arrest mutants of the fission yeast Schizosaccharomyces pombe. J. Mol. Biol. 168, 251-270. 10.1016/S0022-2836(83)80017-5 [DOI] [PubMed] [Google Scholar]
- Tolić-Nørrelykke, I. M., Sacconi, L., Stringari, C., Raabe, I. and Pavone, F. S. (2005). Nuclear and division-plane positioning revealed by optical micromanipulation. Curr. Biol. 15, 1212-1216. 10.1016/j.cub.2005.05.052 [DOI] [PubMed] [Google Scholar]
- Tominaga, M. and Ito, K. (2015). The molecular mechanism and physiological role of cytoplasmic streaming. Curr. Opin. Plant Biol. 27, 104-110. 10.1016/j.pbi.2015.06.017 [DOI] [PubMed] [Google Scholar]
- Tran, P. T., Marsh, L., Doye, V., Inoué, S. and Chang, F. (2001). A mechanism for nuclear positioning in fission yeast based on microtubule pushing. J. Cell Biol. 153, 397-412. 10.1083/jcb.153.2.397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, J.-W., Lian, W.-N., Kemal, S., Kriegstein, A. R. and Vallee, R. B. (2010). Kinesin 3 and cytoplasmic dynein mediate interkinetic nuclear migration in neural stem cells. Nat. Neurosci. 13, 1463-1471. 10.1038/nn.2665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umesono, K., Toda, T., Hayashi, S. and Yanagida, M. (1983). Cell division cycle genes nda2 and nda3 of the fission yeast Schizosaccharomyces pombe control microtubular organization and sensitivity to anti-mitotic benzimidazole compounds. J. Mol. Biol. 168, 271-284. 10.1016/S0022-2836(83)80018-7 [DOI] [PubMed] [Google Scholar]
- Upadhyaya, A. and Sheetz, M. P. (2004). Tension in tubulovesicular networks of Golgi and endoplasmic reticulum membranes. Biophys. J. 86, 2923-2928. 10.1016/S0006-3495(04)74343-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Meer, G., Voelker, D. R. and Feigenson, G. W. (2008). Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 9, 112-124. 10.1038/nrm2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walde, S. and King, M. C. (2014). The KASH protein Kms2 coordinates mitotic remodeling of the spindle pole body. J. Cell Sci. 127, 3625-3640. 10.1242/jcs.154997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, N. and Rapoport, T. A. (2019). Reconstituting the reticular ER network - mechanistic implications and open questions. J. Cell Sci. 132, jcs227611. 10.1242/jcs.227611 [DOI] [PubMed] [Google Scholar]
- Wang, H. and Vavylonis, D. (2008). Model of For3p-mediated actin cable assembly in fission yeast. PLoS ONE 3, e4078. 10.1371/journal.pone.0004078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, N., Lo Presti, L., Zhu, Y.-H., Kang, M., Wu, Z., Martin, S. G. and Wu, J.-Q. (2014). The novel proteins Rng8 and Rng9 regulate the myosin-V Myo51 during fission yeast cytokinesis. J. Cell Biol. 205, 357-375. 10.1083/jcb.201308146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West, M., Zurek, N., Hoenger, A. and Voeltz, G. K. (2011). A 3D analysis of yeast ER structure reveals how ER domains are organized by membrane curvature. J. Cell Biol. 193, 333-346. 10.1083/jcb.201011039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westrate, L. M., Lee, J. E., Prinz, W. A. and Voeltz, G. K. (2015). Form follows function: the importance of endoplasmic reticulum shape. Annu. Rev. Biochem. 84, 791-811. 10.1146/annurev-biochem-072711-163501 [DOI] [PubMed] [Google Scholar]
- Win, T. Z., Gachet, Y., Mulvihill, D. P., May, K. M. and Hyams, J. S. (2001). Two type V myosins with non-overlapping functions in the fission yeast Schizosaccharomyces pombe: Myo52 is concerned with growth polarity and cytokinesis, Myo51 is a component of the cytokinetic actin ring. J. Cell Sci. 114, 69-79. [DOI] [PubMed] [Google Scholar]
- Wu, H., Carvalho, P. and Voeltz, G. K. (2018). Here, there, and everywhere: the importance of ER membrane contact sites. Science 361, eaan5835. 10.1126/science.aan5835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang, X. (2018). Nuclear movement in fungi. Semin. Cell Dev. Biol. 82, 3-16. 10.1016/j.semcdb.2017.10.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Y., Moseley, J. B., Sagot, I., Poy, F., Pellman, D., Goode, B. L. and Eck, M. J. (2004). Crystal structures of a Formin Homology-2 domain reveal a tethered dimer architecture. Cell 116, 711-723. 10.1016/S0092-8674(04)00210-7 [DOI] [PubMed] [Google Scholar]
- Yanakieva, I., Erzberger, A., Matejčić, M., Modes, C. D. and Norden, C. (2019). Cell and tissue morphology determine actin-dependent nuclear migration mechanisms in neuroepithelia. J. Cell Biol. 218, 3272-3289. 10.1083/jcb.201901077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, D. and Oliferenko, S. (2014). Tts1, the fission yeast homologue of the TMEM33 family, functions in NE remodeling during mitosis. Mol. Biol. Cell 25, 2970-2983. 10.1091/mbc.e13-12-0729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, D., Vjestica, A. and Oliferenko, S. (2012). Plasma membrane tethering of the cortical ER necessitates its finely reticulated architecture. Curr. Biol. 22, 2048-2052. 10.1016/j.cub.2012.08.047 [DOI] [PubMed] [Google Scholar]
- Zhao, T., Graham, O. S., Raposo, A. and St Johnston, D. (2012). Growing microtubules push the oocyte nucleus to polarize the Drosophila dorsal-ventral axis. Science 336, 999-1003. 10.1126/science.1219147 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.