Abstract
Plant fitness depends on the adequate morphological adjustment to the prevailing conditions of the environment. Therefore, plants sense environmental cues through their life cycle, including the presence of full darkness, light, or shade, the range of ambient temperatures, the direction of light and gravity vectors, and the presence of water and mineral nutrients (such as nitrate and phosphate) in the soil. The environmental information impinges on different aspects of the auxin system such as auxin synthesis, degradation, transport, perception, and downstream transcriptional regulation to modulate organ growth. Although a single environmental cue can affect several of these points, the relative impacts differ significantly among the various growth processes and cues. While stability in the generation of precise auxin gradients serves to guide the basic developmental pattern, dynamic changes in the auxin system fine-tune body shape to optimize the capture of environmental resources.
Given the role of auxin in the control of complex developmental programs that require stability, such as the organization of the shoot meristem (Vernoux and Pernisová 2020), we might expect homeostasis of auxin signaling status in response to environmental fluctuations. However, in this review we show that a wide array of environmental cues target auxin metabolism, perception, and/or transcriptional regulation to modify body shape in Arabidopsis thaliana. The cellular abundance of auxin depends on its metabolism (Casanova-Sáez et al. 2020) and transport (Schwechheimer et al. 2020) and is sensed by the coreceptors of the TRANSPORT INHIBITOR RESPONSE1/AUXIN SIGNALING F-BOX (TIR1/AFB) and the AUXIN RESISTANT/INDOLE-3 ACETIC ACID INDUCIBLE (Aux/IAA) families (Strader and Morffy 2020). The AUXIN RESPONSE FACTOR (ARF) family involves transcription factors that directly bind the promoters of auxin-responsive genes, such as SMALL AUXIN-UP RNA (SAUR) and Aux/IAA genes, to positively regulate their expression (except for negative ARFs). AUX/IAA proteins recruit transcriptional repressors to the regulatory region of auxin-responsive genes by binding to ARFs. Auxin binding to its coreceptors induces the proteasome-dependent degradation of Aux/IAA, releasing the activity of ARF transcription factors to induce the auxin-dependent expression profile.
Plants show remarkable plasticity (Casal et al. 2004). They rely on cues from the above- and below-ground environments to adjust their growth and developmental patterns to capture the available resources and use them efficiently. For instance, plants require light for photosynthesis. The occurrence of full darkness in the soil indicates that this resource is not available yet. For aerial tissues, the changes in spectral distribution and irradiance with respect to incident sunlight indicate the presence, nearness, and size of competitors for light, and ambient temperature weights the negative consequences of being shaded. Meanwhile, the direction of incident light indicates the location of impediments to reach light. In all these cases, the plant rapidly projects the foliage toward the places where the light resource is predicted to be more available. Plants also require water and mineral nutrients, and while gravity guides root growth toward the direction where these resources are more likely to be available, the occurrence of localized sources within the soil triggers changes in the root growth pattern that optimize their capture. Therefore, plants forage for light, water, and nutrients. In all these cases, auxin is between environmental sensing and the growth response.
FORAGING FOR LIGHT
Buried in the Darkness of the Soil
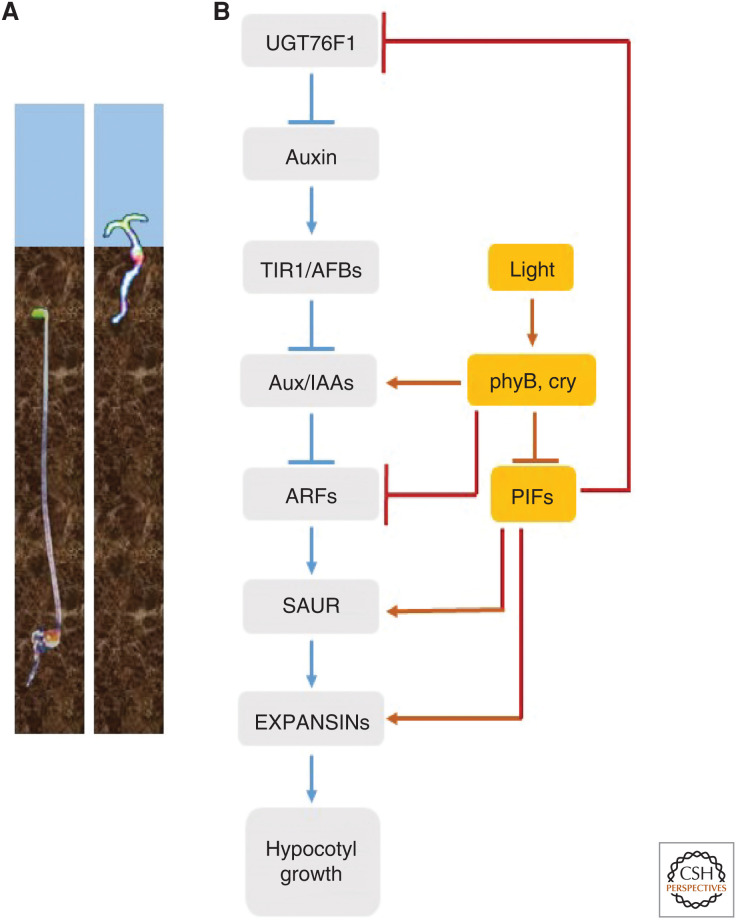
Seeds can germinate buried at certain deepness, where there is more water available than in the upper layers of the soil. There, the seedlings experience full darkness because light typically penetrates only a few millimeters into the soil. In response to this environment, the hypocotyl (A. thaliana seedlings) grows at a fast rate to minimize the time between germination and the emergence of the shoot to receive light for photosynthesis (i.e., the duration of a period when growth relies on seed reserves [heterotrophy]). Cotyledon expansion is rudimentary, which helps to save energy and avoids excessive attrition caused by soil roughness (Fig. 1). The upper section of the hypocotyl forms the plumular hook, which reduces the risk of apical meristem damage on its way through the soil. This growth pattern is called skotomorphogenesis (Arsovski et al. 2012) and the seedlings are described as etiolated. Auxin plays a fundamental role in skotomorphogenesis, and gain-of-function mutants producing stable Aux/IAA proteins that repress auxin-induced gene expression show a partially de-etiolated phenotype (Nagpal et al. 2000; Tian et al. 2002).
Figure 1.
The transition between skoto- and photomorphogenesis involves changes in the auxin system. (A) Etiolated (dark grown, left) and de-etiolated (light grown, right) seedling of Arabidopsis thaliana. (B) Light perceived by phytochromes (phy) and cryptochromes (cry) represses auxin levels and auxin signaling by direct interaction of these photosensory receptors with Aux/IAA and ARF proteins and by repressing the activity of PHYTOCHROME INTERACTING FACTORS (PIFs), which enhance auxin abundance and the expression of auxin targets.
Near the soil surface, the perception of UV-A/blue-light gradients by the plasma-membrane-associated kinases, phototropins (phot1, phot2; we follow the nomenclature established for photoreceptors by Quail et al. [1994], where the holoprotein containing the chromophore attached to the apoprotein is in lower case), induces hypocotyl bending toward the light (Liscum et al. 2020). This phototropic response guides the shoot to avoid physical barriers that might prevent successful emergence from the soil. Hypocotyl bending involves redistribution of growth-promoting auxin away from the illuminated side of the hypocotyl, and the formation of a gradient resulting from the regulation of auxin carriers (Fankhauser and Christie 2015). In darkness, phot1 interacts with PHYTOCHROME KINASE SUBSTRATE 4 (PKS4) and NON-PHOTOTROPIC HYPOCOTYL 3 (NPH3) at the plasma membrane. Light activation of phot1 leads to its dimerization and autophosphorylation, direct phosphorylation of PKS4, and indirect dephosphorylation of NPH3, which are considered important early events in downstream signaling (Liscum et al. 2020). phot1 also directly phosphorylates the multidrug resistance transporter ATP-BINDING-CASSETTE B19 (ABCB19), apparently reducing its polar auxin transport capacity to favor the formation of the horizontal gradient (Christie et al. 2011). The action of PIN-FORMED (PIN) efflux carriers (Schwechheimer et al. 2020) is important and PIN3 is laterally distributed in response to unilateral light (Ding et al. 2011). However, the exact mechanisms by which the phot1-PKS4-NPH3 complex establishes the auxin gradient remain to be elucidated (Fankhauser and Christie 2015).
As the uppermost regions of the shoot emerge from the soil, light activates photosensory receptors such as phytochromes (including phyA and phyB) (Burgie and Vierstra 2014) and cryptochromes (cry1 and cry2) (Wang and Lin 2020). Red light activates phyB, far-red light (proportionally more abundant in shaded environments) is the most effective wave band to activate phyA (Casal et al. 2014) and blue light activates cry1 and cry2. If the seedlings are in darkness, after synthesis, phytochromes remain in the cytosol in the inactive form. Light induces the migration of phyA and phyB to the nucleus (Kircher et al. 1999; Yamaguchi et al. 1999), where, together with cry1 and cry2, they negatively regulate auxin signaling (Fig. 1).
One mechanism of regulation involves direct physical interaction of photosensory receptors with components of the auxin system. phyB and cry1 bind to Aux/IAA proteins favoring their dissociation from the TIR1 coreceptor and increasing their stability to negatively regulate auxin-induced gene expression (Tian et al. 2003; Xu et al. 2018). phyA might have a similar action (Colón-Carmona et al. 2000). Furthermore, the physical interaction of active phyB or cry1 with ARFs reduce the binding of these transcription factors to their target promoters (Fig. 1; Mao et al. 2020).
In darkness, the basic helix-loop-helix transcription factors PHYTOCHROME INTERACTING FACTOR (PIF) (Lee and Choi 2017; Pham et al. 2018) such as PIF3 and PIF4 bind the promoters of SAUR genes, promoting their expression in the hypocotyl and having the opposite effect in the cotyledons (Sun et al. 2016). SAUR proteins inhibit the activity of type 2C protein phosphatases belonging to the D clade (PP2C.D) that in turn inhibit plasma membrane proton-ATPases, favoring apoplast acidification (Spartz et al. 2014). Apoplast acidification enhances the activity of the wall-loosening protein expansins that promote growth (Cosgrove 2005). In response to light, active phyA and/or phyB bind PIFs and induce their phosphorylation and degradation in the proteasome (Lee and Choi 2017; Pham et al. 2018), whereas active cry1 and cry2 reduce the transcriptional activity of bound PIFs (Ma et al. 2016; Pedmale et al. 2016). Consequently, several SAUR genes are light-induced in cotyledons and/or repressed in hypocotyls, at least partially accounting for the differential growth responses of these organs to light (Sun et al. 2016). Compared to full darkness, light causes some reduction in auxin levels in the hypocotyl and the opposite response in the cotyledons (Sun et al. 2016). Indole-3-pyruvic acid can be converted to auxin by YUCCA (YUC) or glucosylated by the UDP-glycosyltransferase UGT76F1, which therefore reduces auxin levels (Casanova-Sáez et al. 2020; Chen et al. 2020). PIF4 binds to the UGT76F1 gene promoter to repress its activity and light enhances UGT76F1 to promote de-etiolation (Chen et al. 2020).
In summary, auxin signaling is elevated in darkness and, during de-etiolation, light reduces auxin levels and auxin sensitivity via direct physical interaction between the photosensory receptors and the core transcriptional machinery (Aux/IAAs and ARFs) and via the photosensory receptors–PIFs pathway directly affecting auxin-controlled or auxin metabolic genes.
Under Shade
“Sun plants” initiate shade-avoidance responses when exposed to the shade of neighboring vegetation or even to the threat of that possibility, anticipated by light reflected by potential competitors that still do not shade (Casal 2013; Ballaré and Pierik 2017). Shade-avoidance responses typically include the promotion of stem growth and petiole growth and the increased angle of insertion of the leaves to place the foliage at higher and better light strata of the canopy. Warm temperatures induce similar morphological responses (Quint et al. 2016; Casal and Balasubramanian 2019) that help cool down the tissues (Crawford et al. 2012) and enhance shade avoidance (Romero-Montepaone et al. 2020). The combination of shade and warm temperatures is predicted to be a complex scenario in terms of carbon balance due to low photosynthesis and high respiration.
Under sunlight conditions, the activity of the photosensory receptor phyB represses the action of the transcription factor PIF7 by cytoplasmic retention (Huang et al. 2018b) and that of PIF4 and PIF5 by facilitating their phosphorylation and degradation in the proteasome (Lorrain et al. 2008). The changes in spectral distribution (lowering of the red to far-red ratio) and overall irradiance caused by neighbor plants reduce the activity of phyB (Sellaro et al. 2019). Then, PIF7, PIF4, and PIF5 bind to the promoters of YUC genes to enhance auxin synthesis (Hornitschek et al. 2012; Li et al. 2012). The reduced blue light levels of shade lower cry1 and cry2 activity, enhancing these responses (de Wit et al. 2016; Pedmale et al. 2016). Warm temperatures enhance auxin levels by a mechanism that shows significant overlap with that of shade avoidance. Warm temperatures decrease the activity of phyB by accelerating the reversion of its active form to its inactive form (Jung et al. 2016; Legris et al. 2016), a mechanism that is different from the photochemical reaction that reduces phyB activity under shade. The latter in itself increases the activity of PIFs but temperature has additional points of action. At high temperature, an RNA hairpin within the 5′ untranslated region of PIF7 transcripts leads to enhanced translation, increasing PIF7 protein abundance (Chung et al. 2020). In addition, the EVENING COMPLEX, which contains ELF3, represses the expression of PIF4 and warm temperatures reduce the association of the transcription factor ELF3 to the promoter of PIF4 (Box et al. 2015). Warm temperature reduces the binding of the EVENING COMPLEX to DNA in vitro, suggesting that the complex acts as a direct thermosensor (Silva et al. 2020). Therefore, warm temperatures increase the expression and protein abundance of PIF4. In turn, PIF4 and PIF7 bind the promoters of YUC8, TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS (TAA1), and other genes to increase auxin levels (Franklin et al. 2011; Sun et al. 2012; Chung et al. 2020; Fiorucci et al. 2020).
Both neighbor cues and warm temperatures increase the levels of auxin in the cotyledons to promote the growth of the hypocotyl (Procko et al. 2014; Bellstaedt et al. 2019), and this activation appears to occur in the epidermis rather than in vascular tissues of the cotyledons (Kim et al. 2020). The sole induction of YUC3 expression in cotyledons is enough to trigger hypocotyl elongation in seedlings grown in the absence of neighbor signals or warm temperatures (Kohnen et al. 2016). Auxin travels via PINs and ABCBs from the cotyledons, where neighbor cues or warmth increase its levels, to the growing hypocotyl. Neighbor cues reorient PIN3 from the basal to the lateral side of the cell membrane to drive auxin flux to the epidermis (Keuskamp et al. 2010), which is the growth-limiting tissue of the organ. Neighbor cues reaching the hypocotyl help to conserve the additional auxin received from the cotyledons by lowering the expression of GH3.17, which is involved in a type of auxin conjugation that is followed by degradation (Procko et al. 2016).
The promotion of hypocotyl growth is not only about extra auxin synthesized in the cotyledons. Neighbor cues also affect the expression of genes involved in auxin conjugation, transport, perception, and signaling, many of which are targets of PIFs (Iglesias et al. 2018). Persistent shade reinforces the growth response but 1 day after the beginning of the environmental cue, auxin shows the same levels observed before the stimulation (Bou-Torrent et al. 2014; de Wit et al. 2015; Pucciariello et al. 2018). The hypocotyl growth response still requires auxin, but now a higher sensitivity to auxin mediates the shade effect (Pucciariello et al. 2018). Persistent shade reduces the expression of MIR393 and consequently increases the abundance of its targets, the auxin receptors (Pucciariello et al. 2018). In addition, persistent shade increases PIF4 abundance in the vascular tissues of the hypocotyl (Pucciariello et al. 2018) and there is evidence in favor of enhanced sensitivity to auxin mediated by PIF4 and PIF5 under shade (Hersch et al. 2014). Enhanced PIF4 abundance promotes the expression of its target genes IAA19 and IAA29, which in turn reduces the expression of the IAA17 gene to further accelerate growth (Pucciariello et al. 2018). Therefore, the shade-avoidance response initially involves increased auxin levels and subsequently a rewiring of auxin signaling that increases the sensitivity to the hormone. The promotion of YUC8 expression by warm temperature peaks at 4 hours and then decreases sharply (Sun et al. 2012; Huai et al. 2018). Then, enhanced sensitivity to auxin also plays a role in the response to warm temperatures, which increase HSP90 protein levels to stabilize the auxin coreceptor TIR1 (Wang et al. 2016).
Shade favors hypocotyl phototropic bending toward the site from which the seedlings receive more blue light perceived by phot1 and phot2, indicative of the position of a gap within the canopy (Goyal et al. 2016; Boccaccini et al. 2020). The reduced activities of phyB and cry1 under shade converge to enhance PIF4 abundance, respectively, by enhancing protein stability and gene expression. This establishes a strong auxin signaling status in the hypocotyl and a more pronounced auxin gradient in response to unilateral blue light to enhance light foraging (Goyal et al. 2016; Boccaccini et al. 2020).
Shade and warmth also cause leaf hyponasty. During this response, the lower face of the petiole grows faster than its upper face and the leaf adopts a more upright position. Adding a drop of auxin at the tip of the leaf lamina is enough to induce the hyponastic response. The neighbor cue perceived at the tip of the leaf lamina increases the activity of PIFs, locally enhancing the expression of YUC genes and the synthesis of auxin (Michaud et al. 2017; Pantazopoulou et al. 2017). In response to neighbors or warm temperatures, auxin produced in the lamina is transported to the petiole by the vasculature and distributed by PIN3 preferentially to the abaxial (fast growing) face of the petiole (Michaud et al. 2017; Pantazopoulou et al. 2017; Park et al. 2019). The epigenetic repressor ASYMMETRIC LEAVES 1 (AS1) provides leaf polarity information by repressing PIF4 binding to the PINOID (PID) gene promoter and hence PID expression more strongly in the adaxial than the abaxial face of the petiole (Park et al. 2019). PID is a protein kinase that phosphorylates the auxin efflux transporter PIN3 (Schwechheimer et al. 2020) and regulates its polarization to the outer membrane of abaxial petiole cells by vesicle-to-membrane trafficking (Park et al. 2019).
Shade and warmth also promote the elongation of the petiole. At least in the case of the shade-avoidance response, processes in the petiole itself perceive the neighbor cues, and auxin added to the lamina is not effective (Michaud et al. 2017; Pantazopoulou et al. 2017). In addition to effects on auxin levels, neighbor cues up-regulate the expression of TIR1/AFBs genes and enhance the accumulation of AFB1 protein in petioles with potential effects on auxin sensitivity (de Wit et al. 2015).
The environment is essentially dynamic across the diurnal cycle. Diurnal sensitivity to shade events depends on auxin signaling status and correlates with sensitivity to exogenously applied auxin (Sellaro et al. 2012). Furthermore, even under dense vegetation shade is often interrupted by light penetrating through gaps in the canopy. The occurrence of these interruptions depends on the interaction between the geometry of the canopy and solar angle, and therefore these events tend to repeat daily at a certain time in the photoperiod. phyB/phyA and the UV-B receptor UV-B RESISTANT 8 (UVR8), respectively, dominate the perception of afternoon and midday daily interruptions of shade (Sellaro et al. 2011; Moriconi et al. 2018). Light activation of phyB and phyA enhances the expression of ELONGATED HYPOCOTYL 5 (HY5) and the HY5 transcription factor represses the expression of auxin-induced genes (Sellaro et al. 2011). UV-B perceived by UVR8 reduces the abundance of PIF4 and PIF5 and the expression of auxin synthesis genes (Hayes et al. 2014). The sensitivity to light interruptions of shade also depends on auxin signaling status (Sellaro et al. 2011).
FORAGING FOR WATER AND NUTRIENTS
General Orientation to Explore the Soil
The direction of the gravity vector provides information that roots use to orient their growth in soil. Gravity sensing occurs in root columella cells, where the position of starch-accumulating amyloplasts depends on the direction of gravity (Nakamura et al. 2019). When the root grows in the direction of gravity, PIN3 evenly distributes auxin toward expanding cells in the root elongation zone. Disruption of root position causes PIN3 phosphorylation and relocation, increasing the flux of auxin toward the epidermal cells of the lower face of the root (Nakamura et al. 2019). In response to auxin, these cells reduce their rate of expansion causing root bending to reestablish the match with the direction of the gravity vector. This response is very rapid as it initiates in <30 sec after auxin reaches the growing cells (Fendrych et al. 2018). The action of auxin requires intracellular perception and the assembly of the TIR1/AFB–Aux/IAA coreceptor complex but it does not involve changes in transcription. Therefore, the canonical auxin receptor system appears to have a nontranscriptional output that is important for root positive gravitropism (Fendrych et al. 2018).
Taking Advantage of Water and Nitrate Patches
Although gravitropism provides a general sense of direction for the root, the soil is a heterogeneous environment. Therefore, the root can find previously unexplored areas where water and nutrients are more readily available. The root tip senses water gradients, biasing the radial position of the initiation of lateral roots when this critical resource for plant growth and development is available at soil–water microsites, an adaptive response termed hydropatterning (Bennett 2020). Lateral root development depends on auxin and ARF7, among other ARFs (Cavallari et al. 2020). Hydropatterning involves a change in the activity of ARF7 (Orosa-Puente et al. 2018). In fact, ARF7 is differentially SUMOylated at several lysine residues in the root area exposed to air, but not in the water exposed root section, by SMALL UBIQUITIN MODIFIER (SUMO) protein(s) that remain unidentified. Once SUMOylated, ARF7-SUMO is recruited and inhibited by IAA3, blocking the expression of LATERAL ORGAN BOUNDARIES 16 (LBD16) and other downstream genes required for lateral root development (Orosa-Puente et al. 2018).
Nitrogen is an essential macronutrient, vital for plant growth and productivity. In the soil, nitrogen is present in inorganic (e.g., nitrate, ammonium) or organic forms (e.g., amino acids, urea, etc.). In many species, including A. thaliana, nitrate is the main source of nitrogen for the plant (Crawford and Forde 2002). In soil with relatively low levels of nitrogen, the root takes advantage of rich spots by generating new branches and increasing the density of root hairs in response to the local nitrate stimulus. The plasma membrane protein NITRATE TRANSPORTER1.1 (NRT1.1) plays a key role in this branching response. NRT1.1 transports nitrate (with dual-affinity), senses nitrate (defined as a nitrate transceptor after these two functions), and facilitates auxin uptake (Krouk et al. 2010). Under low nitrate conditions, phosphorylated NRT1.1 transports auxin out of the lateral root apex preventing the accumulation of auxin required for the growth and consequent emergence of the lateral root (Bouguyon et al. 2015). When the root reaches a region containing high nitrate levels, auxin transport by NRT1.1 is inhibited, auxin accumulates, and the lateral root grows, exploring the place where nitrate is available (Bouguyon et al. 2015).
Nitrate can reduce primary root growth corrected by shoot growth (Linkohr et al. 2002). While ammonium is retained by the soil matrix, after rain, nitrate moves freely in solution toward lower strata. Compared to ammonium, nitrate triggers a rapid enhancement of root growth by increasing the number of cells that enter the root elongation zone (Ötvös et al. 2020), which would help foraging for this nutrient in deep soil strata. This response depends on the precise modulation of auxin transport routes between cortex and epidermis. The root adjusts to nitrogen sources by using a sensitive fine-tuning mechanism for auxin transport, mediated by PIN2 phosphorylation, trafficking, and localization (Ötvös et al. 2020).
Staying Superficial When There Is Not Enough Phosphate
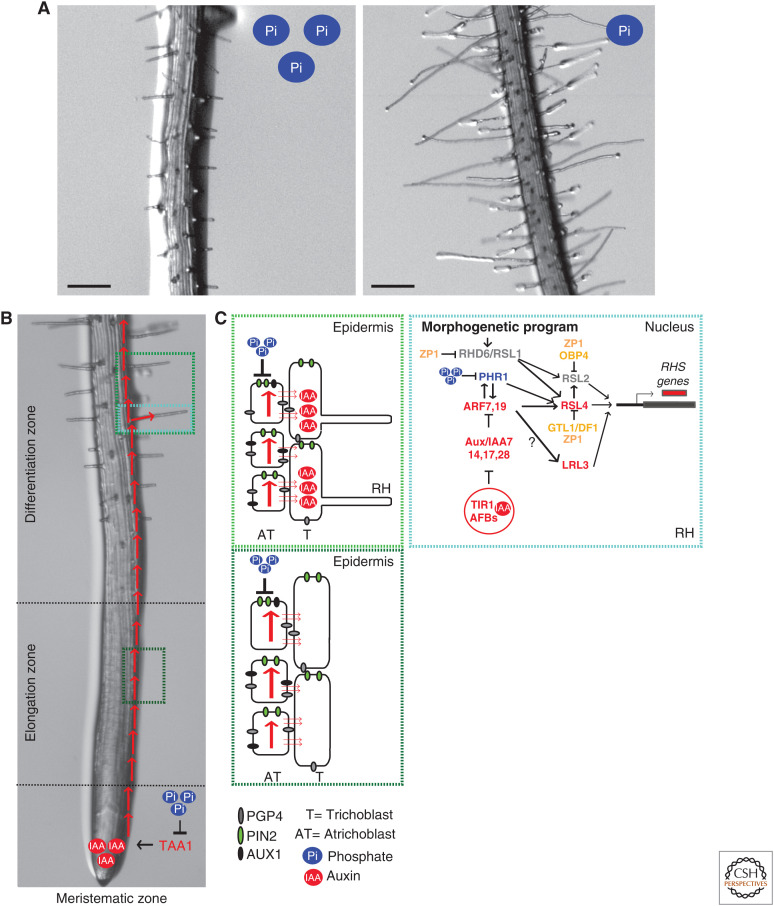
Phosphorus is an essential element for plant growth and development. Plants uptake phosphorus in the form of phosphate, which has low mobility in soil, is extremely reactive and usually precipitates with aluminum, iron, or calcium, making phosphorous one of the most frequently limiting nutrients. In response to low availability of phosphate, the growth of the primary root is restricted, root branching is more profuse, and the root hairs are longer (Fig. 2A). This pattern helps to explore the upper layers of the soils where plant detritus can release phosphorus.
Figure 2.
Low phosphate promotes hair root growth. (A) Detail of the root hairs of Arabidopsis thaliana seedlings grown at two different levels of phosphate (low and high). (B) Low phosphate increases auxin levels at the meristematic zone of the primary root and auxin travels from the meristematic zone through the lateral root cap and the epidermis to the differentiation zone. (C) This movement of auxin takes place via auxin carriers mostly in atrichoblasts to reach the root hairs (note negative regulation by phosphate). In the root hairs (RHs), auxin arriving from the meristematic zone is perceived to promote polar growth. Right-hand box shows the detail of nuclear events. Auxin signaling components (note negative regulation by phosphate) and auxin-responsive genes are in red, positive regulators of hair root growth are in gray, and negative regulators of hair root growth are in orange. (ZP1) ZINC FINGER PROTEIN 1 (Han et al. 2020), (GTL1/DF1) TRIHELIX TRANSCRIPTION FACTOR GT-2-LIKE 1 and its homolog (Shibata et al. 2018), (OBP4) OBF-BINDING PROTEIN 4 (Rymen et al. 2017), (RHS) ROOT HAIR SPECIFIC genes.
Auxin plays a key role in the response of root system architecture to phosphate availability (Nacry et al. 2005). PHOSPHATE RESPONSE 1 (PHR1) and related MYB transcription factors are crucial for adaptive responses to low phosphate. In the presence of phosphate, nuclear proteins that bear the SPX domain, also present in yeast phosphate sensors, reduce the interaction between PHR1 and its target promoters by direct physical interaction (Puga et al. 2014). Phosphate availability increases the concentration of inositol polyphosphate signaling molecules, which are the physiological ligands of the SPX-domain proteins, favoring their interaction with PHR1 and preventing adaptive responses to low phosphate (Wild et al. 2016). The specific connections between phosphate sensing and changes in auxin signaling remain to be fully elucidated, but genes involved in auxin synthesis, sensing, and downstream control of transcription are possible PHR1 targets involved root growth responses to phosphate starvation (Crombez et al. 2019). Furthermore, PHR1 is a direct target of ARF7 and ARF19 in the root, a regulation that is important for the formation of lateral root (Huang et al. 2018a).
Low levels of phosphate enhance the expression of the TAA1 gene at the primary root tip (Bhosale et al. 2018), auxin levels at the root apex and root hairs, and the expression of the DR5::VENUS reporter of auxin-induced transcriptional activity in the columella cells, quiescent center, stele, lateral root cap, and root hair zone (Fig. 2B; Nacry et al. 2005; Bhosale et al. 2018). The additional auxin synthesized at the root tip must reach the root hairs to promote their growth and the taa1 mutant displays a poor hair-root response to low phosphate (Bhosale et al. 2018).
Basipetal (shoot-ward) auxin movement via the lateral root cap and epidermal cells relies on the auxin efflux carrier PIN2, and the auxin influx carrier AUXIN RESISTANT 1 (AUX1) at the plasma membrane (Fig. 2C; Cho et al. 2007a; Ganguly et al. 2010; Bhosale et al. 2018). PIN2 shows higher abundance in nonhair cells (atrichoblast) than in hair cells (trichoblasts) of the root epidermis (Löfke et al. 2015) and AUX1 is only expressed in atrichoblast (Jones et al. 2009), suggesting that basipetal auxin transport though nonhair cells sustains root hair development (Lee and Cho 2006; Jones et al. 2009). When phosphate levels are normal, PIN2 accumulates in the vacuole in a process mediated by SORTING NEXIN (SNX)-containing endosomes (Lin et al. 2020). There, PIN2 is degraded, reducing auxin transport of auxin to the hair roots. Under phosphate starvation, PHR1 induces substantial remodeling of phospholipids, releasing phosphorus for other uses in the cell (Pant et al. 2015). The phosphatidic acid resulting from these lipid changes, binds to SNX1, preventing PIN2 accumulation and hence degradation in the vacuole, to favor root hair growth (Lin et al. 2020). Auxin can reach the trichoblast by diffusion from the atrichoblasts and this process could be accelerated by efflux carriers in atrichoblast plasma membranes adjacent to the trichoblast. The potential role of a symplastic pathway via plasmodesmata (Mellor et al. 2020) remains to be elucidated. The auxin efflux carrier ABCB4/P-GLYCOPROTEIN 1 (PGP4) can reduce the auxin stimulus via a local control (Santelia et al. 2005; Cho et al. 2007b).
Once in the trichoblasts, auxin sensed in situ by the TIR1/AFBs and Aux/IAAs coreceptors promotes root hair growth. Seedlings with root hair–specific TIR1 overexpression (Ganguly et al. 2010) have long root hairs, whereas tir1 afb1 afb3 mutants and axr1 mutants (affecting another of the SCFTIR1 complex components) have short root hairs (Lincoln et al. 1990; Pitts et al. 1998; Dharmasiri et al. 2005), restored by suppressors of the axr1 mutation (Cernac et al. 1997). Gain-of-function mutants of Aux/IAAs, such as IAA7 (axr2) (Lee and Cho 2006), IAA17 (axr3) (Knox et al. 2003), iaa28 (Rogg et al. 2001), and IAA14 (srl1) (Fukaki et al. 2002) all show reduced root hair growth (Fig. 2C). ARFs control the expansion of root hairs (Mangano et al. 2017; Bhosale et al. 2018; Schoenaers et al. 2018). ARF7 and ARF19 are the most abundantly expressed ARFs in root hair cells (Bargmann et al. 2013) and IAA14 is a repressor of both (Fukaki et al. 2002). ARF7 and ARF19 are direct targets of PHR1 (Crombez et al. 2019) and ARF7 and ARF19 enhance PHR1 expression (Huang et al. 2018a).
ARFs promote root hair growth by positively regulating the expression of several transcription factors such as ROOT HAIR DEFECTIVE-SIX-LIKE 2 (RSL2), RSL4 (Yi et al. 2010; Pires et al. 2013; Mangano et al. 2017), and LOTUS JAPONICUS RHL1-LIKE 3 (LRL3) (Karas et al. 2009; Tam et al. 2015), as well as other key components of polar growth such as the receptor-like kinase ERULUS (Schoenaers et al. 2018). RSL4 levels quantitatively relate to final root hair size (Datta et al. 2015). RSL1 and ROOT HAIR DEFECTIVE 6 (RHD6) are basic helix-loop-helix transcription factors (Menand et al. 2007; Pires et al. 2013) that act as master regulators, inducing the expression of their downstream basic helix-loop-helix transcription factors RSL2, RSL4, and LRL3 (Fig. 2C; Karas et al. 2009; Yi et al. 2010; Bruex et al. 2012).
The higher levels of auxin under low phosphate may mediate the early inhibition of primary root growth (Nacry et al. 2005), when the growth of epidermal cells is reduced (Sánchez-Calderón et al. 2005). However, low phosphate has a stronger effect on primary root growth at a later stage (Nacry et al. 2005), which might relate to a shift from indeterminate to determinate growth of the primary root under severely low phosphate (Sánchez-Calderón et al. 2005). In this case, there is a reduction in the number of cells in the meristem because of a lower rate of cell division plus accelerated cell differentiation, which could be caused by alterations in the activity and/or maintenance of the quiescent center (Sánchez-Calderón et al. 2005). These alterations in the quiescent center correlate with a reduction in the intensity of the DR5:GUS signal in the columella initials, the quiescent center, and the mature columella root cap (Sánchez-Calderón et al. 2005).
CONCLUDING REMARKS
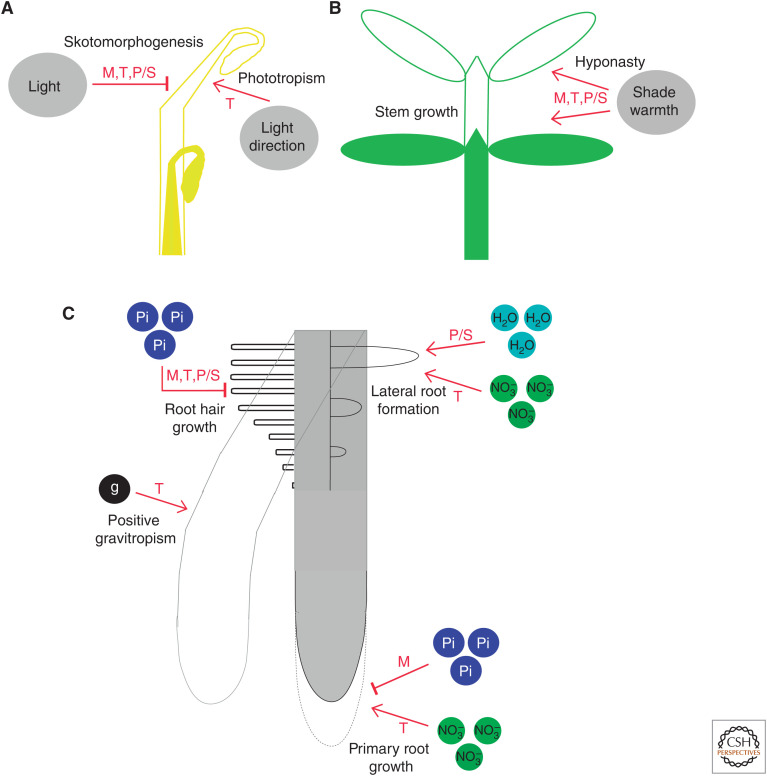
Auxin provides endogenous cues that are crucial for the organization of the plant body. Here we describe how external cues convey to the auxin system information about the environment that plants have to face. Environmental signaling converges on different aspects of the auxin system to reshape the plant body in a way that reduces the chances of stress (Fig. 3). For instance, light and temperature conditions affect the metabolic processes that control auxin levels in the shoot (Franklin et al. 2011; Hornitschek et al. 2012; Li et al. 2012; Sun et al. 2012; Procko et al. 2016; Chen et al. 2020; Chung et al. 2020; Fiorucci et al. 2020), and phosphate controls auxin levels in the root (Bhosale et al. 2018). The direction of light and gravity and the availability of nitrate or phosphate affect auxin transport and modify its distribution within the organ (Bouguyon et al. 2015; Fankhauser and Christie 2015; Nakamura et al. 2019; Lin et al. 2020; Ötvös et al. 2020). Light and temperature conditions can also affect the levels of TIR1/AFBs or Aux/IAA coreceptors (Tian et al. 2003; de Wit et al. 2015; Wang et al. 2016; Pucciariello et al. 2018; Xu et al. 2018). Light, water, and phosphate cues can affect the abundance and/or intrinsic activity of the ARF transcription factors (Orosa-Puente et al. 2018; Mao et al. 2020).
Figure 3.
Environmental cues affect plant growth by modifying auxin metabolisms (M), transport (T), perception (P), and/or signaling (S). (A) Etiolated shoot responses. (B) De-etiolated shoot responses. (C) Root responses.
Environmental cues introduce these changes by transcriptional and/or posttranscriptional regulation. Gene ontology terms related to auxin are often overrepresented in transcriptome responses to environmental cues. Shade (Kohnen et al. 2016) and warmth (Bellstaedt et al. 2019) provide clear examples, but we only partially understand the functional significance of these changes (Iglesias et al. 2018). Posttranscriptional regulation includes phosphorylation and relocalization (Nakamura et al. 2019; Ötvös et al. 2020), effects on turnover rates (Lin et al. 2020), and modified affinity (Bouguyon et al. 2015) of auxin carriers. Posttranscriptional regulation also includes the physical interaction of proteins carrying environmental information either with the Aux/IAAs to influence their interaction with the TIR1/ABFs coreceptors (Tian et al. 2003; Xu et al. 2018), with TIR1 to modify its stability (Wang et al. 2016) or with ARFs to prevent their interaction with the target promoters (Mao et al. 2020), and the SUMOylation of ARF transcription factors (Orosa-Puente et al. 2018). The list of growth processes and molecular mechanisms presented here does not pretend to be exhaustive. It simply illustrates that all aspects of the auxin system are under environmental control to substantially modify the patterns of plant growth and enhance fitness.
ACKNOWLEDGMENTS
Work at the Casal lab is supported by grants from Agencia Nacional de Promoción Científica y Tecnológica (ANPCYT, PICT-2018-01695), Universidad de Buenos Aires (20020170100505BA), and Consejo Nacional de Investigaciones Científicas y Técnicas (SNSF, SUIZ/17/04). Work at the Estevez lab is supported by grants from ICGEB (CRP/ARG16-03), ANPCYT (PICT2016-0132 and PICT2017-0066), and Fondo Nacional de Desarrollo Científico y Tecnológico (1200010) and Instituto Milenio iBio – Iniciativa Científica Milenio, MINECON.
Footnotes
Editors: Dolf Weijers, Karin Ljung, Mark Estelle, and Ottoline Leyser
Additional Perspectives on Auxin Signaling available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Arsovski AA, Galstyan A, Guseman JM, Nemhauser JL. 2012. Photomorphogenesis. Arab B 10: e0147. 10.1199/tab.0147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballaré CL, Pierik R. 2017. The shade-avoidance syndrome: multiple signals and ecological consequences. Plant Cell Environ 40: 2530–2543. 10.1111/pce.12914 [DOI] [PubMed] [Google Scholar]
- Bargmann BOR, Vanneste S, Krouk G, Nawy T, Efroni I, Shani E, Choe G, Friml J, Bergmann DC, Estelle M, et al. 2013. A map of cell type-specific auxin responses. Mol Syst Biol 9: 688. 10.1038/msb.2013.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellstaedt J, Trenner J, Lippmann R, Poeschl Y, Zhang X, Friml J, Quint M, Delker C. 2019. A mobile auxin signal connects temperature sensing in cotyledons with growth responses in hypocotyls. Plant Physiol 180: 757–766. 10.1104/pp.18.01377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Bennett M. 2020. Abiotic stress networks. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a040014 [DOI] [Google Scholar]
- Bhosale R, Giri J, Pandey BK, Giehl RFH, Hartmann A, Traini R, Truskina J, Leftley N, Hanlon M, Swarup K, et al. 2018. A mechanistic framework for auxin dependent Arabidopsis root hair elongation to low external phosphate. Nat Commun 9: 1409. 10.1038/s41467-018-03851-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boccaccini A, Legris M, Krahmer J, Allenbach Petrolati L, Goyal A, Ampudia CG, Vernoux T, Karayekov E, Casal JJ, Fankhauser C. 2020. Low blue light enhances phototropism by releasing cryptochrome 1-mediated inhibition of PIF4 expression. Plant Physiol 183: 1780–1793. 10.1104/pp.20.00243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouguyon E, Brun F, Meynard D, Kubeš M, Pervent M, Leran S, Lacombe B, Krouk G, Guiderdoni E, Zažímalová E, et al. 2015. Multiple mechanisms of nitrate sensing by Arabidopsis nitrate transceptor NRT1.1. Nat Plants 1: 15015. 10.1038/nplants.2015.15 [DOI] [PubMed] [Google Scholar]
- Bou-Torrent J, Galstyan A, Gallemí M, Cifuentes-Esquivel N, Molina-Contreras MJ, Salla-Martret M, Jikumaru Y, Yamaguchi S, Kamiya Y, Martínez-García JF. 2014. Plant proximity perception dynamically modulates hormone levels and sensitivity in Arabidopsis. J Exp Bot 65: 2937–2947. 10.1093/jxb/eru083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Box MS, Huang BE, Domijan M, Jaeger KE, Khattak AK, Yoo SJ, Sedivy EL, Jones DM, Hearn TJ, Webb AAR, et al. 2015. ELF3 controls thermoresponsive growth in Arabidopsis. Curr Biol 25: 194–199. 10.1016/j.cub.2014.10.076 [DOI] [PubMed] [Google Scholar]
- Bruex A, Kainkaryam RM, Wieckowski Y, Kang YH, Bernhardt C, Xia Y, Zheng X, Wang JY, Lee MM, Benfey P, et al. 2012. A gene regulatory network for root epidermis cell differentiation in Arabidopsis. PLoS Genet 8: e1002446. 10.1371/journal.pgen.1002446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgie ES, Vierstra RD. 2014. Phytochromes: an atomic perspective on photoactivation and signaling. Plant Cell 26: 4568–4583. 10.1105/tpc.114.131623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal JJ. 2013. Photoreceptor signaling networks in plant responses to shade. Annu Rev Plant Biol 64: 403–427. 10.1146/annurev-arplant-050312-120221 [DOI] [PubMed] [Google Scholar]
- Casal JJ, Balasubramanian S. 2019. Thermomorphogenesis. Annu Rev Plant Biol 70: 321–346. 10.1146/annurev-arplant-050718-095919 [DOI] [PubMed] [Google Scholar]
- Casal JJ, Fankhauser C, Coupland G, Blázquez MA. 2004. Signalling for developmental plasticity. Trends Plant Sci 9: 309–314. 10.1016/j.tplants.2004.04.007 [DOI] [PubMed] [Google Scholar]
- Casal JJ, Candia AN, Sellaro R. 2014. Light perception and signalling by phytochrome A. J Exp Bot 65: 2835–2845. 10.1093/jxb/ert379 [DOI] [PubMed] [Google Scholar]
- *.Casanova-Sáez R, Mateo-Bonmatí E, Ljung K. 2020. Auxin metabolism in plants. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a039867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Cavallari N, Artner C, Benkova E. 2020. Auxin-regulated lateral root organogenesis. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a039941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cernac A, Lincoln C, Lammer D, Estelle M, Schneider K, Lawson E, Dean C, Poethig S, Roberts K. 1997. The SAR1 gene of Arabidopsis acts downstream of the AXR1 gene in auxin response. Development 124: 1583–1591. [DOI] [PubMed] [Google Scholar]
- Chen L, Huang XX, Zhao SM, Xiao DW, Xiao LT, Tong JH, Wang WS, Li YJ, Ding Z, Hou BK. 2020. IPyA glucosylation mediates light and temperature signaling to regulate auxin-dependent hypocotyl elongation in Arabidopsis. Proc Natl Acad Sci 117: 6910–6917. 10.1073/pnas.2000172117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho M, Lee OR, Ganguly A, Cho HT. 2007a. Auxin-signaling: short and long. J Plant Biol 50: 79–89. 10.1007/BF03030615 [DOI] [Google Scholar]
- Cho M, Lee SH, Cho HT. 2007b. P-glycoprotein4 displays auxin efflux transporter-like action in Arabidopsis root hair cells and tobacco cells. Plant Cell 19: 3930–3943. 10.1105/tpc.107.054288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie JM, Yang H, Richter GL, Sullivan S, Thomson CE, Lin J, Titapiwatanakun B, Ennis M, Kaiserli E, Lee OR, et al. 2011. Phot1 inhibition of ABCB19 primes lateral auxin fluxes in the shoot apex required for phototropism. PLoS Biol 9: e1001076. 10.1371/journal.pbio.1001076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung BYW, Balcerowicz M, Di Antonio M, Jaeger KE, Geng F, Franaszek K, Marriott P, Brierley I, Firth AE, Wigge PA. 2020. An RNA thermoswitch regulates daytime growth in Arabidopsis. Nat Plants 6: 522–532. 10.1038/s41477-020-0633-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colón-Carmona A, Chen DL, Yeh KC, Abel S. 2000. Aux/IAA proteins are phosphorylated by phytochrome in vitro. Plant Physiol 124: 1728–1738. 10.1104/pp.124.4.1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ. 2005. Growth of the plant cell wall. Nat Rev Mol Cell Biol 6: 850–861. 10.1038/nrm1746 [DOI] [PubMed] [Google Scholar]
- Crawford NM, Forde BG. 2002. Molecular and developmental biology of inorganic nitrogen nutrition. Arab B 1: e0011. 10.1199/tab.0011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford AJ, McLachlan DH, Hetherington AM, Franklin KA. 2012. High temperature exposure increases plant cooling capacity. Curr Biol 22: R396–R397. 10.1016/j.cub.2012.03.044 [DOI] [PubMed] [Google Scholar]
- Crombez H, Motte H, Beeckman T. 2019. Tackling plant phosphate starvation by the roots. Dev Cell 48: 599–615. 10.1016/j.devcel.2019.01.002 [DOI] [PubMed] [Google Scholar]
- Datta S, Prescott H, Dolan L. 2015. Intensity of a pulse of RSL4 transcription factor synthesis determines Arabidopsis root hair cell size. Nat Plants 1: 15138. 10.1038/nplants.2015.138 [DOI] [PubMed] [Google Scholar]
- de Wit M, Ljung K, Fankhauser C. 2015. Contrasting growth responses in lamina and petiole during neighbor detection depend on differential auxin responsiveness rather than different auxin levels. New Phytol 3: 198–209. [DOI] [PubMed] [Google Scholar]
- de Wit M, Keuskamp DH, Bongers FJ, Martínez-Cerón C, Fankhauser C, Pierik R, Hornitschek P, Gommers CMM, Reinen E. 2016. Integration of phytochrome and cryptochrome signals determines plant growth during competition for light. Curr Biol 26: 3320–3326. 10.1016/j.cub.2016.10.031 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Estelle M. 2005. The F-box protein TIR1 is an auxin receptor. Nature 435: 441–445. 10.1038/nature03543 [DOI] [PubMed] [Google Scholar]
- Ding Z, Galván-Ampudia CS, Demarsy E, Łangowski Ł, Kleine-Vehn J, Fan Y, Morita MT, Tasaka M, Fankhauser C, Offringa R, et al. 2011. Light-mediated polarization of the PIN3 auxin transporter for the phototropic response in Arabidopsis. Nat Cell Biol 13: 447–452. 10.1038/ncb2208 [DOI] [PubMed] [Google Scholar]
- Fankhauser C, Christie JM. 2015. Plant phototropic growth. Curr Biol 25: R384–R389. 10.1016/j.cub.2015.03.020 [DOI] [PubMed] [Google Scholar]
- Fendrych M, Akhmanova M, Merrin J, Glanc M, Hagihara S, Takahashi K, Uchida N, Torii KU, Friml J. 2018. Rapid and reversible root growth inhibition by TIR1 auxin signalling. Nat Plants 4: 453–459. 10.1038/s41477-018-0190-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorucci AS, Galvão VC, Ince YÇ, Boccaccini A, Goyal A, Allenbach Petrolati L, Trevisan M, Fankhauser C. 2020. PHYTOCHROME INTERACTING FACTOR 7 is important for early responses to elevated temperature in Arabidopsis seedlings. New Phytol 226: 50–58. 10.1111/nph.16316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin KA, Lee SH, Patel D, Kumar SV, Spartz AK, Gu C, Ye S, Yu P, Breen G, Cohen JD, et al. 2011. PHYTOCHROME-INTERACTING FACTOR 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc Natl Acad Sci 108: 20231–20235. 10.1073/pnas.1110682108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukaki H, Tameda S, Masuda H, Tasaka M. 2002. Lateral root formation is blocked by a gain-of-function mutation in the SOLITARY-ROOT/IAA14 gene of Arabidopsis. Plant J 29: 153–168. 10.1046/j.0960-7412.2001.01201.x [DOI] [PubMed] [Google Scholar]
- Ganguly A, Lee SH, Cho M, Lee OR, Yoo H, Cho HT. 2010. Differential auxin-transporting activities of PIN-FORMED proteins in Arabidopsis root hair cells. Plant Physiol 153: 1046–1061. 10.1104/pp.110.156505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyal A, Karayekov E, Galvão VC, Ren H, Casal JJ, Fankhauser C. 2016. Shade promotes phototropism through phytochrome B-controlled auxin production. Curr Biol 26: 3280–3287. 10.1016/j.cub.2016.10.001 [DOI] [PubMed] [Google Scholar]
- Han G, Wei X, Dong X, Wang C, Sui N, Guo J, Yuan F, Gong Z, Li X, Zhang Y, et al. 2020. Arabidopsis ZINC FINGER PROTEIN1 acts downstream of GL2 to repress root hair initiation and elongation by directly suppressing bHLH genes. Plant Cell 32: 206–225. 10.1105/tpc.19.00226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes S, Velanis CN, Jenkins GI, Franklin KA. 2014. UV-B detected by the UVR8 photoreceptor antagonizes auxin signaling and plant shade avoidance. Proc Natl Acad Sci 111: 11894–11899. 10.1073/pnas.1403052111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hersch M, Lorrain S, de Wit M, Trevisan M, Ljung K, Bergmann S, Fankhauser C. 2014. Light intensity modulates the regulatory network of the shade avoidance response in Arabidopsis. Proc Natl Acad Sci 111: 6515–6520. 10.1073/pnas.1320355111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornitschek P, Kohnen MV, Lorrain S, Rougemont J, Ljung K, López-Vidriero I, Franco-Zorrilla JM, Solano R, Trevisan M, Pradervand S, et al. 2012. Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. Plant J 71: 699–711. 10.1111/j.1365-313X.2012.05033.x [DOI] [PubMed] [Google Scholar]
- Huai J, Zhang X, Li J, Ma T, Zha P, Jing Y, Lin R. 2018. SEUSS and PIF4 coordinately regulate light and temperature signaling pathways to control plant growth. Mol Plant 11: 928–942. 10.1016/j.molp.2018.04.005 [DOI] [PubMed] [Google Scholar]
- Huang KL, Ma GJ, Zhang ML, Xiong H, Wu H, Zhao CZ, Liu CS, Jia HX, Chen L, Kjorven JO, et al. 2018a. The ARF7 and ARF19 transcription factors positively regulate PHOSPHATE STARVATION RESPONSE1 in Arabidopsis roots. Plant Physiol 178: 413–427. 10.1104/pp.17.01713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Zhang Q, Jiang Y, Yang C, Wang Q, Li L. 2018b. Shade-induced nuclear localization of PIF7 is regulated by phosphorylation and 14-3-3 proteins in Arabidopsis. eLife 7: e31636. 10.7554/eLife.31636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias MJ, Sellaro R, Zurbriggen MD, Casal JJ. 2018. Multiple links between shade avoidance and auxin networks. J Exp Bot 69: 213–228. 10.1093/jxb/erx295 [DOI] [PubMed] [Google Scholar]
- Jones AR, Kramer EM, Knox K, Swarup R, Bennett MJ, Lazarus CM, Leyser HMO, Grierson CS. 2009. Auxin transport through non-hair cells sustains root-hair development. Nat Cell Biol 11: 78–84. 10.1038/ncb1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung JH, Domijan M, Klose C, Biswas S, Ezer D, Gao M, Box MS, Charoensawan V, Cortijo S, Locke JC, et al. 2016. Phytochromes function as thermosensors in Arabidopsis. Science 354: 886–889. 10.1126/science.aaf6005 [DOI] [PubMed] [Google Scholar]
- Karas B, Amyot L, Johansen C, Sato S, Tabata S, Kawaguchi M, Szczyglowski K. 2009. Conservation of lotus and Arabidopsis basic helix-loop-helix proteins reveals new players in root hair development. Plant Physiol 151: 1175–1185. 10.1104/pp.109.143867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keuskamp DH, Pollmann S, Voesenek LACJ, Peeters AJM, Pierik R. 2010. Auxin transport through PIN-FORMED 3 (PIN3) controls shade avoidance and fitness during competition. Proc Natl Acad Sci 107: 22740–22744. 10.1073/pnas.1013457108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Hwang G, Kim S, Thi TN, Kim H, Jeong J, Kim J, Kim J, Choi G, Oh E. 2020. The epidermis coordinates thermoresponsive growth through the phyB-PIF4-auxin pathway. Nat Commun 11: 1053. 10.1038/s41467-020-14905-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher S, Kozma-Bognar L, Kim L, Ádám E, Harter K, Schäfer E, Nagy F. 1999. Light-quality dependent nuclear import of the plant photoreceptors phytochrome A and B. Plant Cell 11: 1445–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox K, Grierson CS, Leyser O. 2003. AXR3 and SHY2 interact to regulate root hair development. Development 130: 5769–5777. 10.1242/dev.00659 [DOI] [PubMed] [Google Scholar]
- Kohnen MV, Schmid-Siegert E, Trevisan M, Petrolati LA, Sénéchal F, Müller-Moulé P, Maloof J, Xenarios I, Fankhauser C. 2016. Neighbor detection induces organ-specific transcriptomes, revealing patterns underlying hypocotyl-specific growth. Plant Cell 28: 2889–2904. 10.1105/tpc.16.00463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krouk G, Lacombe B, Bielach A, Perrine-Walker F, Malinska K, Mounier E, Hoyerova K, Tillard P, Leon S, Ljung K, et al. 2010. Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Dev Cell 18: 927–937. 10.1016/j.devcel.2010.05.008 [DOI] [PubMed] [Google Scholar]
- Lee SH, Cho HT. 2006. PINOID positively regulates auxin efflux in Arabidopsis root hair cells and tobacco cells. Plant Cell 18: 1604–1616. 10.1105/tpc.105.035972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee N, Choi G. 2017. Phytochrome-interacting factor from Arabidopsis to liverwort. Curr Opin Plant Biol 35: 54–60. 10.1016/j.pbi.2016.11.004 [DOI] [PubMed] [Google Scholar]
- Legris M, Klose C, Burgie E, Costigliolo Rojas C, Neme M, Hiltbrunner A, Wigge PA, Schäfer E, Vierstra RD, Casal JJ. 2016. Phytochrome B integrates light and temperature signals in Arabidopsis. Science 354: 897–900. 10.1126/science.aaf5656 [DOI] [PubMed] [Google Scholar]
- Li L, Ljung K, Breton G, Schmitz RJ, Pruneda-Paz J, Cowing-Zitron C, Cole BJ, Ivans LJ, Pedmale UV, Jung H-S, et al. 2012. Linking photoreceptor excitation to changes in plant architecture. Genes Dev 26: 785–790. 10.1101/gad.187849.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin DL, Yao HY, Jia LH, Tan JF, Xu ZH, Zheng WM, Xue HW. 2020. Phospholipase D-derived phosphatidic acid promotes root hair development under phosphorus deficiency by suppressing vacuolar degradation of PIN-FORMED2. New Phytol 226: 142–155. 10.1111/nph.16330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln C, Britton JH, Estelle M. 1990. Growth and development of the axr1 mutants of Arabidopsis. Plant Cell 2: 1071–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linkohr BI, Williamson LC, Fitter AH, Leyser HMO. 2002. Nitrate and phosphate availability and distribution have different effects on root system architecture of Arabidopsis. Plant J 29: 751–760. 10.1046/j.1365-313X.2002.01251.x [DOI] [PubMed] [Google Scholar]
- Liscum E, Nittler P, Koskie K. 2020. The continuing arc toward phototropic enlightenment. J Exp Bot 71: 1652–1658. 10.1093/jxb/eraa005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löfke C, Dünser K, Scheuring D, Kleine-Vehn J. 2015. Auxin regulates SNARE-dependent vacuolar morphology restricting cell size. eLife 4: e05868. 10.7554/eLife.05868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorrain S, Allen T, Duek PD, Whitelam GC, Fankhauser C. 2008. Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J 53: 312–323. 10.1111/j.1365-313X.2007.03341.x [DOI] [PubMed] [Google Scholar]
- Ma D, Li X, Guo Y, Chu J, Fang S, Yan C, Noel JP, Liu H. 2016. Cryptochrome 1 interacts with PIF4 to regulate high temperature-mediated hypocotyl elongation in response to blue light. Proc Natl Acad Sci 113: 224–229. 10.1073/pnas.1511437113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangano S, Denita-Juarez SP, Choi HS, Marzol E, Hwang Y, Ranocha P, Velasquez SM, Borassi C, Barberini ML, Aptekmann AA, et al. 2017. Molecular link between auxin and ROS-mediated polar growth. Proc Natl Acad Sci 114: 5289–5294. 10.1073/pnas.1701536114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Z, He S, Xu F, Wei X, Jiang L, Liu Y, Wang W, Li T, Xu P, Du S, et al. 2020. Photoexcited CRY1 and phyB interact directly with ARF6 and ARF8 to regulate their DNA-binding activity and auxin-induced hypocotyl elongation in Arabidopsis. New Phytol 225: 848–865. 10.1111/nph.16194 [DOI] [PubMed] [Google Scholar]
- Mellor NL, Voß U, Janes G, Bennett MJ, Wells DM, Band LR. 2020. Auxin fluxes through plasmodesmata modify root-tip auxin distribution. Development 147: dev181669. 10.1242/dev.181669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menand B, Yi K, Jouannic S, Hoffmann L, Ryan E, Linstead P, Schaefer DG, Dolan L. 2007. An ancient mechanism controls the development of cells with a rooting function in land plants. Science 316: 1477–1480. 10.1126/science.1142618 [DOI] [PubMed] [Google Scholar]
- Michaud O, Fiorucci A, Xenarios I, Fankhauser C. 2017. Local auxin production underlies a spatially restricted neighbor-detection response in Arabidopsis. Proc Natl Acad Sci 114: 7444–7449. 10.1073/pnas.1702276114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriconi V, Binkert M, Costigliolo C, Sellaro R, Ulm R, Casal JJ. 2018. Perception of sunflecks by the UV-B photoreceptor UV RESISTANCE LOCUS8. Plant Physiol 177: 75–81. 10.1104/pp.18.00048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nacry P, Canivenc G, Muller B, Azmi A, Van Onckelen H, Rossignol M, Doumas P. 2005. A role for auxin redistribution in the responses of the root system architecture to phosphate starvation in Arabidopsis. Plant Physiol 138: 2061–2074. 10.1104/pp.105.060061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagpal P, Walker LM, Young JC, Sonawala A, Timpte C, Estelle M, Reed JW. 2000. AXR2 encodes a member of the Aux/IAA protein family. Plant Physiol 123: 563–574. 10.1104/pp.123.2.563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M, Nishimura T, Morita MT. 2019. Bridging the gap between amyloplasts and directional auxin transport in plant gravitropism. Curr Opin Plant Biol 52: 54–60. 10.1016/j.pbi.2019.07.005 [DOI] [PubMed] [Google Scholar]
- Orosa-Puente B, Leftley N, von Wangenheim D, Banda J, Srivastava AK, Hill K, Truskina J, Bhosale R, Morris E, Srivastava M, et al. 2018. Root branching toward water involves posttranslational modification of transcription factor ARF7. Science 362: 1407–1410. 10.1126/science.aau3956 [DOI] [PubMed] [Google Scholar]
- Ötvös K, Marconi M, Vega A, Brien JO, Johnson A, Abualia R, Antonielli L, Montesinos JC, Zhang Y, Tan S, et al. 2020. Modulation of root growth by nutrient-defined fine-tuning of polar auxin transport. bioRxiv 2020.06.19.160994 [Google Scholar]
- Pant BD, Burgos A, Pant P, Cuadros-Inostroza A, Willmitzer L, Scheible W. 2015. The transcription factor PHR1 regulates lipid remodeling and triacylglycerol accumulation in Arabidopsis thaliana during phosphorus starvation. J Exp Bot 66: 1907–1918. 10.1093/jxb/eru535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantazopoulou CK, Bongers FJ, Küpers JJ, Reinen E, Das D, Evers JB, Anten NPR, Pierik R. 2017. Neighbor detection at the leaf tip adaptively regulates upward leaf movement through spatial auxin dynamics. Proc Natl Acad Sci 114: 7450–7455. 10.1073/pnas.1702275114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YJ, Lee HJ, Gil KE, Kim JY, Lee JH, Lee H, Cho HT, Vu LD, De Smet I, Park CM. 2019. Developmental programming of thermonastic leaf movement. Plant Physiol 180: 1185–1197. 10.1104/pp.19.00139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedmale UV, Huang SC, Zander M, Cole BJ, Hetzel J, Ljung K, Reis PAB, Sridevi P, Nito K, Nery JR, et al. 2016. Cryptochromes interact directly with PIFs to control plant growth in limiting blue light. Cell 164: 233–245. 10.1016/j.cell.2015.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham VN, Kathare PK, Huq E. 2018. Phytochromes and phytochrome interacting factors. Plant Physiol 176: 1025–1038. 10.1104/pp.17.01384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires ND, Yi K, Breuninger H, Catarino B, Menand B, Dolan L. 2013. Recruitment and remodeling of an ancient gene regulatory network during land plant evolution. Proc Natl Acad Sci 110: 9571–9576. 10.1073/pnas.1305457110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitts RJ, Cernac A, Estelle M. 1998. Auxin and ethylene promote root hair elongation in Arabidopsis. Plant J 16: 553–560. 10.1046/j.1365-313x.1998.00321.x [DOI] [PubMed] [Google Scholar]
- Procko C, Crenshaw CM, Ljung K, Noel JP, Chory J. 2014. Cotyledon-generated auxin is required for shade-induced hypocotyl growth in Brassica rapa. Plant Physiol 165: 1285–1301. 10.1104/pp.114.241844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Procko C, Burko Y, Jaillais Y, Ljung K, Long JA, Chory J. 2016. The epidermis coordinates auxin-induced stem growth in response to shade. Genes Dev 30: 1529–1541. 10.1101/gad.283234.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pucciariello O, Legris M, Rojas CC, Iglesias MJ, Hernando CE, Dezar C, Vazquez M, Yanovsky MJ, Finlayson SA, Prat S, et al. 2018. Rewiring of auxin signaling under persistent shade. Proc Natl Acad Sci 115: 5612–5617. 10.1073/pnas.1721110115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puga MI, Mateos I, Charukesi R, Wang Z, Franco-Zorrilla JM, De Lorenzo L, Irigoyen ML, Masiero S, Bustos R, Rodríguez J, et al. 2014. SPX1 is a phosphate-dependent inhibitor of phosphate starvation response 1 in Arabidopsis. Proc Natl Acad Sci 111: 14947–14952. 10.1073/pnas.1404654111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail PH, Briggs WR, Chory J, Hangarter RP, Harberd NP, Kendrick RE, Koornneef M, Parks B, Sharrock RA, Schäfer E, et al. 1994. Spotlight on phytochrome nomenclature. Plant Cell 6: 468–471. 10.2307/3869926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quint M, Delker C, Franklin KA, Wigge PA, Halliday KJ, van Zanten M. 2016. Molecular and genetic control of plant thermomorphogenesis. Nat Plants 2: 15190. 10.1038/nplants.2015.190 [DOI] [PubMed] [Google Scholar]
- Rogg LE, Lasswell J, Bartel B. 2001. A gain-of-function mutation in IAA28 suppresses lateral root development. Plant Cell 13: 465–480. 10.1105/tpc.13.3.465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Montepaone S, Poodts S, Fischbach P, Sellaro R, Zurbriggen MD, Casal JJ. 2020. Shade-avoidance responses become more aggressive in warm environments. Plant Cell Environ 43: 1625–1636. 10.1111/pce.13720 [DOI] [PubMed] [Google Scholar]
- Rymen B, Kawamura A, Schäfer S, Breuer C, Iwase A, Shibata M, Ikeda M, Mitsuda N, Koncz C, Ohme-Takagi M, et al. 2017. ABA suppresses root hair growth via the OBP4 transcriptional regulator. Plant Physiol 173: 1750–1762. 10.1104/pp.16.01945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Calderón L, López-Bucio J, Chacón-López A, Cruz-Ramírez A, Nieto-Jacobo F, Dubrovsky JG, Herrera-Estrella L. 2005. Phosphate starvation induces a determinate developmental program in the roots of Arabidopsis thaliana. Plant Cell Physiol 46: 174–184. 10.1093/pcp/pci011 [DOI] [PubMed] [Google Scholar]
- Santelia D, Vincenzetti V, Azzarello E, Bovet L, Fukao Y, Düchtig P, Mancuso S, Martinoia E, Geisler M. 2005. MDR-like ABC transporter AtPGP4 is involved in auxin-mediated lateral root and root hair development. FEBS Lett 579: 5399–5406. 10.1016/j.febslet.2005.08.061 [DOI] [PubMed] [Google Scholar]
- Schoenaers S, Balcerowicz D, Breen G, Hill K, Zdanio M, Mouille G, Holman TJ, Oh J, Wilson MH, Nikonorova N, et al. 2018. The auxin-regulated CrRLK1L kinase ERULUS controls cell wall composition during root hair tip growth. Curr Biol 28: 722–732.e6. 10.1016/j.cub.2018.01.050 [DOI] [PubMed] [Google Scholar]
- *.Schwechheimer C, Hammes U, Murphy A. 2020. Transport. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a039875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellaro R, Yanovsky MJ, Casal JJ. 2011. Repression of shade-avoidance reactions by sunfleck induction of HY5 expression in Arabidopsis. Plant J 68: 919–928. 10.1111/j.1365-313X.2011.04745.x [DOI] [PubMed] [Google Scholar]
- Sellaro R, Pacín M, Casal JJ. 2012. Diurnal dependence of growth responses to shade in Arabidopsis: role of hormone, clock, and light signaling. Mol Plant 5: 619–628. 10.1093/mp/ssr122 [DOI] [PubMed] [Google Scholar]
- Sellaro R, Smith RW, Legris M, Fleck C, Casal JJ. 2019. Phytochrome B dynamics departs from photoequilibrium in the field. Plant Cell Environ 42: 606–617. 10.1111/pce.13445 [DOI] [PubMed] [Google Scholar]
- Shibata M, Breuer C, Kawamura A, Clark NM, Rymen B, Braidwood L, Morohashi K, Busch W, Benfey PN, Sozzani R, et al. 2018. GTL1 and DF1 regulate root hair growth through transcriptional repression of ROOT HAIR DEFECTIVE 6-LIKE 4 in Arabidopsis. Dev 145: dev159707. 10.1242/dev.159707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva CS, Nayak A, Lai X, Hutin S, Hugouvieux V, Jung JH, López-Vidriero I, Franco-Zorrilla JM, Panigrahi KCS, Nanao MH, et al. 2020. Molecular mechanisms of evening complex activity in Arabidopsis. Proc Natl Acad Sci 117: 6901–6909. 10.1073/pnas.1920972117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spartz AK, Ren H, Park MY, Grandt KN, Lee SH, Murphy AS, Sussman MR, Overvoorde PJ, Gray WM. 2014. SAUR inhibition of PP2C-D phosphatases activates plasma membrane H+-ATPases to promote cell expansion in Arabidopsis. Plant Cell 26: 2129–2142. 10.1105/tpc.114.126037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Strader L, Morffy N. 2020. Response mechanism and structural aspects. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a039883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Qi L, Li Y, Chu J, Li C. 2012. PIF4-mediated activation of YUCCA8 expression integrates temperature into the auxin pathway in regulating Arabidopsis hypocotyl growth. PLoS Genet 8: e1002594. 10.1371/journal.pgen.1002594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun N, Wang J, Gao Z, Dong J, He H, Terzaghi W, Wei N, Deng XW, Chen H. 2016. Arabidopsis SAURs are critical for differential light regulation of the development of various organs. Proc Natl Acad Sci 113: 6071–6076. 10.1073/pnas.1604782113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam THY, Catarino B, Dolan L. 2015. Conserved regulatory mechanism controls the development of cells with rooting functions in land plants. Proc Natl Acad Sci 112: E3959–E3968. 10.1073/pnas.1416324112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Q, Uhlir NJ, Reed JW. 2002. Arabidopsis SHY2/IAA3 inhibits auxin-regulated gene expression. Plant Cell 14: 301–319. 10.1105/tpc.010283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Q, Nagpal P, Reed JW. 2003. Regulation of Arabidopsis SHY2/IAA3 protein turnover. Plant J 36: 643–651. 10.1046/j.1365-313X.2003.01909.x [DOI] [PubMed] [Google Scholar]
- *.Vernoux T, Pernisová M. 2020. Auxin in the shoot meristem. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a039925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Lin C. 2020. Mechanisms of cryptochrome-mediated photoresponses in plants. Annu Rev Plant Biol 71: 103–129. 10.1146/annurev-arplant-050718-100300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Zhang Y, Kieffer M, Yu H, Kepinski S, Estelle M. 2016. HSP90 regulates temperature-dependent seedling growth in Arabidopsis by stabilizing the auxin co-receptor F-box protein TIR1. Nat Commun 7: 10269. 10.1038/ncomms10269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild R, Gerasimaite R, Jung JY, Truffault V, Pavlovic I, Schmidt A, Saiardi A, Jacob Jessen H, Poirier Y, Hothorn M, et al. 2016. Control of eukaryotic phosphate homeostasis by inositol polyphosphate sensor domains. Science 352: 986–990. 10.1126/science.aad9858 [DOI] [PubMed] [Google Scholar]
- Xu F, He S, Zhang J, Mao Z, Wang W, Li T, Hua J, Du S, Xu P, Li L, et al. 2018. Photoactivated CRY1 and phyB interact directly with AUX/IAA proteins to inhibit auxin signaling in Arabidopsis. Mol Plant 11: 523–541. 10.1016/j.molp.2017.12.003 [DOI] [PubMed] [Google Scholar]
- Yamaguchi R, Nakamura M, Mochizuki N, Kay SA, Nagatani A. 1999. Light-dependent translocation of a phytochrome B-GFP fusion protein to the nucleus in transgenic Arabidopsis. J Cell Biol 145: 437–445. 10.1083/jcb.145.3.437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi K, Menand B, Bell E, Dolan L. 2010. A basic helix-loop-helix transcription factor controls cell growth and size in root hairs. Nat Genet 42: 264–267. 10.1038/ng.529 [DOI] [PubMed] [Google Scholar]