Dear Editor,
Acute myeloid leukemia (AML) with t(8;21) is a heterogeneous disease and needs to be further stratified1–3. We previously reported that high-risk t(8;21) AML patients benefited from allogeneic hematopoietic stem cell transplantation (allo-HSCT)4, which implied that risk stratification could guide appropriate treatment selection for t(8;21) AML.
At present, KIT mutation is still the only widely accepted gene mutation with strong prognostic significance in t(8;21) AML5–10. Furthermore, RUNX1-RUNX1T1 transcript levels after treatment has been routinely tested to monitor minimal residual disease (MRD) and established as a powerful marker to predict relapse and guide treatment4,11–14. However, report on how to combine KIT mutation status with MRD levels to assess prognosis remains absent to date.
The current study included 287 t(8;21) AML patients who consecutively received treatment and achieved complete remission (CR) in our center from February 2009 to December 2019. The median age at diagnosis was 36 (range, 15–65) years. Information about patient treatment and samples availability before 2nd consolidation was shown in Fig. S1. The study was conducted in accordance with the Declaration of Helsinki and was approved by the Ethics Committee of the Peking University People’s Hospital. The cutoff date for the follow-up was October 31, 2020.
As we have previously reported4, induction chemotherapy was composed of 1–2 cycles of induction with the “3 + 7” regimen or the HAA regimen (homoharringtonine, cytarabine, and aclarubicin), and the post-remission therapy included intermediate-dose cytarabine-based chemotherapy (IDAC; 1–2 g/m2 every 12 h for 3 days; 2–4 cycles of cytarabine followed by 2–4 cycles of the “3 + 7”regimen), autologous-hematopoietic stem cell transplantation (auto-HSCT), or allogeneic-HSCT (allo-HSCT). After achieving CR, 162 patients received chemotherapy alone, 9 received chemotherapy followed by auto-HSCT, and 116 received chemotherapy followed by allo-HSCT (matched sibling donor, n = 38; haploidentical related donor, n = 72; matched unrelated donor, n = 6). The indications for the allo-HSCT were described in our previous studies14,15.
270 and 17 patients individually achieved CR after 1–2 and 3–4 cycles of induction, 80 patients (27.9%) experienced hematological relapse, and 250 patients (87.1%) were alive at the last follow-up. The median follow-up time was 28.5 (range, 3.3–109.0) months for the surviving patients. The 3-year cumulative incidence of relapse (CIR) and overall survival (OS) rate were 29.9% [95% confidence interval (CI), 21.6–38.6%] and 85.0% (95% CI, 79.5–89.1%), respectively.
Overall, 120 patients (41.8%) had KIT mutations (246 patients were screened at diagnosis and 41 screened after treatment with RUNX1-RUNX1T1 transcript levels higher than 5%). The mutations were categorized into the following six types: sole D816 (18.5%, n = 53; 38 D816V, 8 D816Y and 7 D816H), sole N822 (11.1%, n = 32; all were N822K), sole D820 (2.4%, n = 7; 5 D820G, 1 D820A, 1 D820Y), sole R815_D816delins (1.4%, n = 4), sole exon 8 delins (4.2%, n = 12, abbreviated as e8 thereafter) and compound mutations (4.2%, n = 12). The types of compound mutations were as follows: 5 D816 + D816, 1 D816 + I817, 1 D816 + D820, 1 D816 + N822, 2 D816 + e8, 1 D820 + N822 and 1 D820 + e8. Thus, all of the compound mutations contained D816 or/and D820 mutations.
First, patients were grouped according to their KIT mutation status. In our previous study, t(8;21) AML patients were categorized into D816/D820 mutation and N822/e8/WT groups and D816/D820 mutation was demonstrated to be an independent adverse prognostic factor for both relapse free survival and OS10. Here, patients with D816, compound mutations, 815_816delins and D820 mutations had similar 3-year CIR in the whole cohort and if censoring at the time of allo-HSCT [62.1% (95% CI, 48.1–73.3%) vs. 58.3% (95% CI, 31.4–77.8%) vs. 50.0% (95% CI, 5.8–84.5%) vs. 57.1% (95% CI, 5.0–90.0%), P = 0.60; censoring: P = 0.44, Fig. S2]. Because all patients in the above four groups had D816 or/and D820 mutations, they were merged and defined as KITD816/D820 (n = 76). In addition, because patients with e8 mutations had similar 3-year CIR to those with no mutation [8.3% (95% CI, 0–70.5%) vs. 19.9% (95% CI, 9.2–33.6%), P = 0.27; censoring: P = 0.42, Fig. S2], they were merged and defined as KITe8/WT. As a result, KITD816/D820 patients had significantly higher risk of relapse than both KITN822 and KITe8/WT patients [59.8% (95% CI, 47.6–70.0%), 22.6% (95% CI, 4.3–49.4%) and 19.0% (95% CI, 8.8–32.2%), P = 0.0025 and <0.0001; censoring: P = 0.0009 and <0.0001, Fig. S2]. CIR was not significantly different between KITN822 and KITe8/WT patients (P = 0.45, censoring: P = 0.19).
Next, patients were grouped according to MRD levels. The pretreatment baseline level of the RUNX1-RUNX1T1 transcript was 388% in our center4. We selected the median value at CR and after 1st consolidation, 4.0% (2-log reduction compared to baseline) and 0.4% (3-log reduction) as the individual cutoff value. In agreement with our previous reports4,10, 0.4% was selected as the cutoff value for the timepoint of after 2nd consolidation. Thus, patients with RUNX1-RUNX1T1 transcript levels higher and lower than the cutoff value were defined as high MRD levels and low MRD levels groups at individual timepoints. As shown in Fig. S3, patients with high MRD levels had significantly higher risk of relapse than those with low MRD levels at CR, after 1st consolidation and 2nd consolidation, respectively [CIR: 35.8% (95% CI, 24.2–47.6%) vs. 21.7% (95% CI, 9.6–36.9%), P = 0.0020; 38.1% (95% CI, 27.1–49.0%) vs. 17.4% (95% CI, 6.1–33.4%), P = 0.0001; 36.7% (95% CI, 23.7–49.8%) vs. 19.5% (95% CI, 8.4–34.0%), P = 0.0004].
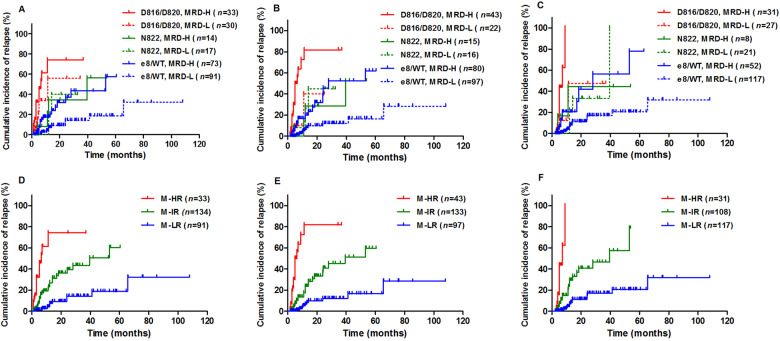
Then KIT mutation status and MRD levels were combined, and patients who received allo-HSCT were censored at the time of transplantation. As shown in Fig. 1, for KITD816/D820 patients, higher MRD levels at CR, after 1st consolidation and 2nd consolidation were significantly or tended to be significantly associated with an increased risk of relapse, respectively [74.2% (95% CI, 53.8–86.6%) vs. 55.9% (95% CI, 18.8–81.7%), P = 0.098; 81.8% (95% CI, 67.7–90.2%) vs. 40.0% (95% CI, 1.0–83.4%), P = 0.0048; 100.0% (95% CI, 100.0–100.0%) vs. 47.6% (95% CI, 9.7–79.0%), P = 0.0032]. Similarly for KITe8/WT patients, higher MRD levels at the three timepoints were significantly associated with an increased risk of relapse, respectively [43.4% (95% CI, 21.7–63.4%) vs. 14.3% (95% CI, 1.8–39.0%), P = 0.0008; 52.4% (95% CI, 29.7–70.9%) vs. 12.2% (95% CI, 1.4–35.7%), P < 0.0001; 56.5% (95% CI, 24.9–79.1%) vs. 17.0% (95% CI, 4.1–37.4%), P < 0.0001]. Whereas for KITN822 patients, MRD levels at all three timepoints had no impact on relapse [CIR: 34.5% (95% CI, 3.1–72.0%) vs. 40.0% (95% CI, 2.4–79.8%), P = 0.80; 28.6% (95% CI, 0.7–73.3%) vs. 45.0% (95% CI, 6.5–79.2%), P = 0.50; 44.4% (95% CI, 2.7–83.3%) vs. 33.3% (95% CI, 3.2–70.3%), P = 0.81].
Fig. 1. CIR with allo-HSCT patients censored at the time of transplantation and patients were categorized by the combination of KIT mutations and MRD status at different timepoints.
A, D At CR; B, E: after 1st consolidation; C, F: after 2nd consolidation. MRD-H and MRD-L represented <2-log reduction and ≥2-log reduction of RUNX1-RUNX1T1 at CR (A) and <3-log reduction and ≥3-log reduction of RUNX1-RUNX1T1 after 1st and 2nd consolidation (B, C), respectively.
Furthermore, the four groups, KITD816/D820 patients with low MRD levels, KITN822 patients with high MRD levels, KITN822 patients with low MRD levels and KITe8/WT patients with high MRD levels, had similar CIR at all three timepoints, respectively (P = 0.083, 0.94, and 0.94, Fig. 1A–C). Therefore, by considering KIT mutations and MRD status simultaneously, patients were recategorized into the following three groups: molecularly defined high-risk (M-HR; KITD816/D820 patients with high MRD levels), molecularly defined intermediate-risk (M-IR; KITD816/D820 patients with low MRD levels, KITN822 patients, KITe8/WT patients with high MRD levels) and molecularly defined low-risk (M-LR; KITe8/WT patients with low MRD levels) groups. As a result, M-HR, M-IR and M-LR patients had significantly different 3-year CIR at CR, after 1st consolidation and 2nd consolidation, respectively [74.2% (95% CI, 53.8–86.6%) vs. 43.4% (95% CI, 26.9–58.8%) vs. 14.3% (95% CI, 1.8–39.0%), 81.8% (95% CI, 67.7–90.2%) vs. 45.3% (95% CI, 27.3–61.7%) vs. 12.2% (95% CI, 1.4–35.7%), 100.0% (95% CI, 100.0–100.0%) vs. 46.6% (95% CI, 26.6–64.4%) vs. 17.0% (95% CI, 4.1–37.4%); all P < 0.0001, Fig. 1D–F]. Therefore, MRD levels could identify patients with better prognosis from KITD816/D820 and those with worse prognosis from KITe8/WT patients. It implied that KIT mutation and MRD levels had their unique prognostic roles and should be combined in order to better stratify t(8;21) AML.
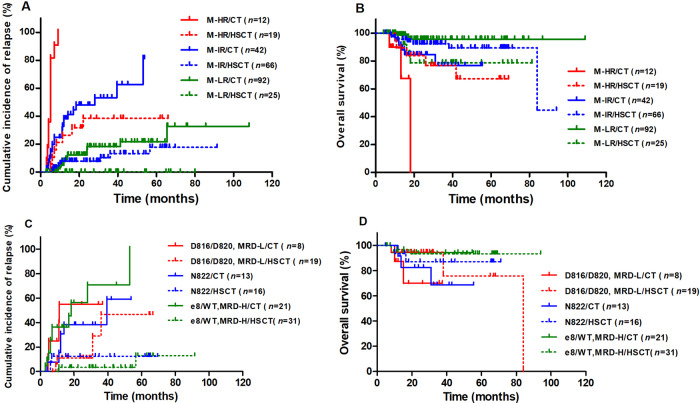
Because t(8;21) AML patients are evaluated whether to recommend to receive allo-HSCT after 2nd consolidation in our center4, we just compared the outcomes between patients with different molecularly defined risk at the timepoint of after 2nd consolidation (n = 256). As shown in Fig. 2A, B, for M-HR patients (n = 31, 12.1%), allo-HSCT had both significantly lower CIR and significantly higher OS than chemotherapy alone [CIR: 38.4% (95% CI, 12.9–63.9%) vs. 100.0% (95% CI, 100.0–100.0%), P < 0.0001; OS: 76.9% (95% CI, 49.0–90.8%) vs. 0% (95% CI, 0–0%), P = 0.035]; for M-IR patients (n = 108, 42.2%), allo-HSCT had significantly lower CIR than and similar OS to chemotherapy alone [CIR: 13.2% (95% CI, 1.2–39.5%) vs. 53.2% (95% CI, 35.4–68.1%), P < 0.0001; OS: 92.2% (95% CI, 82.3–96.7%) vs. 76.8% (95% CI, 52.0–89.9%), P = 0.11]; for M-LR patients (n = 117, 45.7%), allo-HSCT had significantly lower CIR than chemotherapy alone [CIR: 0% (95% CI, 0–0%) vs. 18.2% (95% CI, 4.7–38.7%), P = 0.025], whereas, the OS was significantly lower for allo-HSCT than that for chemotherapy alone [78.7% (95% CI, 56.1–90.5%) vs. 95.6% (95% CI, 86.9–98.6%), P = 0.011]. Comparisons were further made within M-IR groups (Fig. 2C, D), and allo-HSCT had significantly or tended to have significantly lower CIR than and had similar OS to chemotherapy alone for all three groups, KITD816/D820 patients with low MRD levels, KITN822 patients and KITe8/WT patients with high MRD levels. (CIR: P = 0.094, 0.060 and <0.0001; OS: P = 0.13, 0.37 and 0.84).
Fig. 2. CIR and OS of patients categorized by the molecularly defined risk and treatment modality.
A, C: CIR; B, D: OS. CT represented chemotherapy alone/auto-HSCT, HSCT represented allo-HSCT.
In summary, combination of KIT mutation and MRD levels improved risk stratification and treatment guidance in t(8;21) AML. KITD816/D820 patients with <3-log reduction of RUNX1-RUNX1T1 transcript levels after 2nd consolidation had the poorest prognosis and benefited from allo-HSCT on both relapse and survival; KITe8/WT patients with ≥3-log reduction after 2nd consolidation had the best prognosis, and allo-HSCT decreased not only relapse but also survival; the remaining patients had the intermediate prognosis and allo-HSCT decreased relapse but had no significant effect on survival. Multicenter prospective studies are warranted to confirm the current results.
Supplementary information
Acknowledgements
This work was supported by the National Natural Science Foundation of China (81870125) and Key Program of the National Natural Science Foundation of China (81930004).
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41408-021-00461-z.
References
- 1.Byrd JC, et al. Pretreatment cytogenetic abnormalities are predictive of induction success, cumulative incidence of relapse, and overall survival in adult patients with de novo acute myeloid leukemia: results from Cancer and Leukemia Group B (CALGB 8461) Blood. 2002;100:4325–4336. doi: 10.1182/blood-2002-03-0772. [DOI] [PubMed] [Google Scholar]
- 2.Schlenk RF, et al. Individual patient data-based meta-analysis of patients aged 16 to 60 years with core binding factor acute myeloid leukemia: a survey of the German Acute Myeloid Leukemia Intergroup. J. Clin. Oncol. 2004;22:3741–3750. doi: 10.1200/JCO.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 3.Döhner H, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129:424–447. doi: 10.1182/blood-2016-08-733196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu HH, et al. MRD-directed risk stratification treatment may improve outcomes of t(8;21) AML in the first complete remission: results from the AML05 multicenter trial. Blood. 2013;121:4056–4062. doi: 10.1182/blood-2012-11-468348. [DOI] [PubMed] [Google Scholar]
- 5.Paschka P, et al. Adverse prognostic significance of KIT mutations in adult acute myeloid leukemia with inv(16) and t(8;21): a Cancer and Leukemia Group B Study. J. Clin. Oncol. 2006;24:3904–3911. doi: 10.1200/JCO.2006.06.9500. [DOI] [PubMed] [Google Scholar]
- 6.Duployez N, et al. Comprehensive mutational profiling of core binding factor acute myeloid leukemia. Blood. 2016;127:2451–2459. doi: 10.1182/blood-2015-12-688705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christen F, et al. Genomic landscape and clonal evolution of acute myeloid leukemia with t(8;21): an international study on 331 patients. Blood. 2019;133:1140–1151. doi: 10.1182/blood-2018-05-852822. [DOI] [PubMed] [Google Scholar]
- 8.Opatz S, et al. The clinical mutatome of core binding factor leukemia. Leukemia. 2020;34:1553–1562. doi: 10.1038/s41375-019-0697-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jahn N, et al. Genomic heterogeneity in core-binding factor acute myeloid leukemia and its clinical implication. Blood Adv. 2020;4:6342–6352. doi: 10.1182/bloodadvances.2020002673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qin YZ, et al. Heterogeneous prognosis among KIT mutation types in adult acute myeloid leukemia patients with t(8;21) Blood Cancer J. 2018;8:76. doi: 10.1038/s41408-018-0116-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yin JA, et al. Minimal residual disease monitoring by quantitative RT-PCR in core binding factor AML allows risk stratification and predicts relapse: results of the United Kingdom MRC AML-15 trial. Blood. 2012;120:2826–2835. doi: 10.1182/blood-2012-06-435669. [DOI] [PubMed] [Google Scholar]
- 12.Jourdan E, et al. Prospective evaluation of gene mutations and minimal residual disease in patients with core binding factor acute myeloid leukemia. Blood. 2013;121:2213–2223. doi: 10.1182/blood-2012-10-462879. [DOI] [PubMed] [Google Scholar]
- 13.Rücker FG, et al. Measurable residual disease monitoring in acute myeloid leukemia with t(8;21)(q22;q22.1): results from the AML Study Group. Blood. 2019;134:1608–1618. doi: 10.1182/blood.2019001425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qin YZ, et al. The dynamics of RUNX1-RUNX1T1 transcript levels after allogeneic hematopoietic stem cell transplantation predict relapse in patients with t(8;21) acute myeloid leukemia. J. Hematol. Oncol. 2017;10:44. doi: 10.1186/s13045-017-0414-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu L, et al. The consensus on indications, conditioning regimen, and donor selection of allogeneic hematopoietic cell transplantation for hematological diseases in China-recommendations from the Chinese Society of Hematology. J. Hematol. Oncol. 2018;11:33. doi: 10.1186/s13045-018-0564-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.