Abstract
Purpose
Downregulation of MHC class I (MHC-I) is a common immune evasion strategy of many cancers. Similarly, two allogeneic clonal transmissible cancers have killed thousands of wild Tasmanian devils (Sarcophilus harrisii) and also modulate MHC-I expression to evade anti-cancer and allograft responses. IFNG treatment restores MHC-I expression on devil facial tumor (DFT) cells but is insufficient to control tumor growth. Transcriptional co-activator NLRC5 is a master regulator of MHC-I in humans and mice but its role in transmissible cancers remains unknown. In this study, we explored the regulation and role of MHC-I in these unique genetically mis-matched tumors.
Methods
We used transcriptome and flow cytometric analyses to determine how MHC-I shapes allogeneic and anti-tumor responses. Cell lines that overexpress NLRC5 to drive antigen presentation, and B2M-knockout cell lines incapable of presenting antigen on MHC-I were used to probe the role of MHC-I in rare cases of tumor regressions.
Results
Transcriptomic results suggest that NLRC5 plays a major role in MHC-I regulation in devils. NLRC5 was shown to drive the expression of many components of the antigen presentation pathway but did not upregulate PDL1. Serum from devils with tumor regressions showed strong binding to IFNG-treated and NLRC5 cell lines; antibody binding to IFNG-treated and NRLC5 transgenic tumor cells was diminished or absent following B2M knockout.
Conclusion
MHC-I could be identified as a target for anti-tumor and allogeneic immunity. Consequently, NLRC5 could be a promising target for immunotherapy and vaccines to protect devils from transmissible cancers and inform development of transplant and cancer therapies for humans.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00432-021-03601-x.
Keywords: Transmissible cancer, Devil facial tumor, Allograft, MHC-I, NLRC5, Immune evasion
Introduction
In 1996, a wild Tasmanian devil (Sarcophilus harrisii) was photographed with a large facial tumor. In subsequent years, similar devil facial tumors (DFTs) were recorded (Jones et al. 2004), and in 2006, it was confirmed that DFTs are clonally transmissible cancers that spread among devils through social interactions (Pearse and Swift 2006; Loh et al. 2006). In 2014, a second genetically independent transmissible devil facial tumor (DFT2) was discovered in southern Tasmania (Pye et al. 2016b). Despite the independent origin of the first devil facial tumor (DFT1) and DFT2, both clonal tumors arose from a Schwann cell lineage (Murchison et al. 2010; Patchett et al. 2020), suggesting devils could be prone to transmissible Schwann cell cancers. These lethal and unique tumors are simultaneously cancers, allografts, and infectious diseases, and have been the primary driver of an average 77% decline in devil populations across the island state of Tasmania (Lazenby et al. 2018).
The successful transmission and seeding of DFT cells from one devil to another as an allograft (Pearse and Swift 2006) reveals its ability to circumvent both allogeneic and anti-tumor immune responses. Genetic mismatches between host and tumor, particularly at the major histocompatibility complex (MHC) loci, should provide an allogeneic target for an immune response against DFTs. However, DFT1 cells generally express little or no MHC class I (MHC-I) on their surface (Siddle et al. 2013), an immune escape mechanism commonly observed in human cancers to avoid cytotoxic CD8+ T cell recognition (Yoshihama et al. 2016). Loss of MHC-I expression in DFT1 appears to be initiated by an impaired antigen presentation pathway. The clonal DFT1 cell lineage has a hemizygous mutation in beta-2-microglobulin (B2M) gene (Stammnitz et al. 2018) which encodes a light chain of the trimeric MHC-I complex. In addition to this, there is an epigenetic downregulation of B2M, and transporters associated with antigen processing TAP1 and TAP2 (Siddle et al. 2013), suggesting that immune evasion through reduced MHC-I expression has been a target of evolutionary selection pressure. Loss of MHC-I should lead to recognition and cytotoxic responses by natural killer (NK) cells. Devils have demonstrated NK-like activity in vitro (Brown et al. 2011) but the ongoing transmission of DFT1 cells suggests that NK cytotoxic response against DFT1 cells either do not occur or are ineffective. All DFT1 cell lines tested to date can upregulate MHC-I in response to interferon gamma (IFNG) treatment (Siddle et al. 2013). Rare cases of DFT1 regression have been reported in the wild (Pye et al. 2016a) and serum antibody responses of these devils are generally higher against cell lines treated with IFNG to upregulate MHC-I (Pye et al. 2016a, 2018). In contrast to DFT1 cells, DFT2 cells constitutively express MHC-I, but the most highly expressed alleles appear to be those shared by the DFT2 cells and the host devil (Caldwell et al. 2018). This further suggest a critical role of MHC-I in immune evasion of transmissible cancers such as DFTs.
Upregulation of MHC-I on DFT1 cells via treatment with IFNG has served as the foundation for a vaccine against devil facial tumor disease (DFTD), which is caused by DFT1 cells. However, there are caveats to using a pleiotropic cytokine such as IFNG. IFNG plays multiple roles in the innate and adaptive immune system and can function to drive either an anti-tumor or a pro-tumor response depending on the circumstances (He et al. 2005; Abiko et al. 2015; Folgiero et al. 2015; Lo et al. 2019; Song et al. 2019). While IFNG is well known for directing the immune response towards anti-tumor immunity, it causes the upregulation of programmed death ligand 1 (PDL1) on DFT cells (Flies et al. 2016) as well as non-classical, monomorphic MHC-I SAHA-UK (Caldwell et al. 2018). PDL1 and SAHA-UK molecules can be counterproductive to the cell-mediated immune response mediated by MHC-I recognition. Additionally, the inhibition of cell proliferation and increased DFT cell death associated with IFNG (Ong et al. 2019) constrain large-scale production of IFNG-treated DFT cells for whole cell vaccines.
NLRC5 (NLR caspase recruitment domain containing protein 5), a member of the NOD-like receptor (NLR) family, was first identified in 2010 as a regulator of MHC-I expression (Meissner et al. 2010). In subsequent years, several studies have independently shown the critical role of NLRC5 in constitutive and IFNG-induced MHC-I expression in humans and mice, although little is known about NLRC5 in other species (Staehli et al. 2012; Tong et al. 2012; Yao et al. 2012; Robbins et al. 2012; Biswas et al. 2012). NLRC5 acts as a co-activator with high specificity and contributes to MHC-I transcription by interacting with several other transcription factors to form a multi-protein complex called the enhanceosome (Gobin et al. 2001; Meissner et al. 2010, 2012; Neerincx et al. 2012). The enhanceosome activates the promoters of MHC-I genes and components of the antigen processing machinery such as B2M, immunoproteasome subunits PSMB8 (also known as LMP7) and PSMB9 (also known as LMP2), and TAP1 (Meissner et al. 2010; Yoshihama et al. 2016). Aside from MHC-I regulation, NLRC5 has been reported to be involved in innate immune responses as well as malignancy of certain cancers (Cui et al. 2010; Neerincx et al. 2010; Ranjan et al. 2015; Liu et al. 2016; Peng et al. 2016). Despite a potential central role of NLRC5 in immune evasion, studies of NLRC5 are limited and several hypothesized secondary roles of NLRC5 remain unexplored.
In this study, we take advantage of a unique natural experiment in which two independent clonal tumor cell lines have essentially been passaged through hundreds of free-living animals to assess the role of NLRC5 and MHC-I in immune evasion. The overexpression of NLRC5 in DFT1 and DFT2 cells induced the expression of B2M, MHC-I heavy chain SAHAI-01 and other functionally related genes. PDL1 and the non-classical MHC-I SAHA-UK which are upregulated by IFNG were not induced by NLRC5. MHC-I was constitutively expressed on the surface of DFT cells overexpressing NLRC5, which suggests that modulation of NLRC5 expression could be a potential substitute for IFNG to increase DFT cell immunogenicity. Additionally, MHC-I molecules on DFT cells were revealed to be an immunogenic target of allogeneic responses in wild devils.
Materials and methods
Cells and cell culture conditions
DFT1 cell line C5065 strain 3 (Pearse et al. 2012) (RRID:CVCL_LB79) and DFT2 cell lines RV (RRID:CVCL_LB80) and JV (RRID not available) were used in this study as indicated. DFT1 C5065 was provided by A-M Pearse and K. Swift of the Department of Primary Industries, Parks, Water and Environment (DPIPWE) (Hobart, TAS, Australia) and was previously established from DFT1 biopsies obtained under the approval of the Animal Ethics Committee of the Tasmanian Parks and Wildlife Service (permit numbers 33/2004–5 and 32/2005–6). DFT2 cell lines RV and JV were established from single cell suspensions obtained from tumor biopsies performed under the approval of the University of Tasmania Animal Ethics Committee (permit number A0012513) or under a Standard Operating Procedure approved by the General Manager, Natural and Cultural Heritage Division, Tasmanian Government DPIPWE. Cells were cultured at 35 °C with 5% CO2 in complete RPMI medium: RPMI 1640 medium with L-glutamine (Thermo Fisher Scientific, Waltham, MA, USA), 10% heat-inactivated fetal bovine serum (Bovogen Biologicals, Melbourne, VIC, Australia), 1% (v/v) Antibiotic–Antimycotic (100X) (Thermo Fisher Scientific), 10 mM HEPES (Thermo Fisher Scientific) and 50 μM 2-mercaptoethanol (Sigma-Aldrich, St. Louis, MO, USA).
RNA sequencing and analysis
Initial RNA sequencing was performed using DFT1 C5065 and DFT2 RV cells treated with and without 5 ng/mL recombinant devil IFNG (provided by Walter and Eliza Hall Institute (WEHI), Melbourne, VIC, Australia) for 24 h according to the previously described protocols (Patchett et al. 2018, 2020). For the remaining cell lines (Table 1, ID # 5–9), total RNA was extracted using the NucleoSpin® RNA plus kit (Macherey Nagel, Düren, Germany) per manufacturer’s instructions. Two replicates were prepared for each cell line. RNA sequencing was conducted at the Ramaciotti Centre for Genomics (Sydney, NSW, Australia) using the following methods. RNA integrity was assessed using Agilent TapeStation (Agilent Technologies, Santa Clara, CA, USA). All samples had RNA Integrity Number (RIN) scores of 10.0. mRNA libraries were prepared using the TruSeq Stranded mRNA Library Prep (Illumina Inc., San Diego, CA, USA). The libraries were sequenced on an Illumina NovaSeq 6000 platform (Illumina) with 100 base-pair single-end reads. The quality of the sequencing reads were analyzed using FastQC version 0.11.9 (Andrews 2010). Raw FASTQ files have been deposited to the European Nucleotide Archive (ENA) and are available at BioProject # PRJEB39847.
Table 1.
Devil facial tumor (DFT) cell lines and treatments
| ID # | Sample name | Parent cell line | Treatment |
|---|---|---|---|
| 1 | DFT1.WTa | DFT1 C5065 | Untreated |
| 2 | DFT1.WT + IFNG | DFT1 C5065 | 5 ng/mL IFNG, 24 h |
| 3 | DFT2.WTRVb | DFT2 RV | Untreated |
| 4 | DFT2.WTRV + IFNG | DFT2 RV | 5 ng/mL IFNG, 24 h |
| 5 | DFT1.BFP | DFT1 C5065 | Transfected with control vector pSBbi-BH |
| 6 | DFT1.NLRC5 | DFT1 C5065 | Transfected with NLRC5 vector pCO1 |
| 7 | DFT2.WT | DFT2 JV | Untreated |
| 8 | DFT2.BFP | DFT2 JV | Transfected with control vector pSBbi-BH |
| 9 | DFT2.NLRC5 | DFT2 JV | Transfected with NLRC5 vector pCO1 |
| 10 | DFT1.B2M−/− | DFT1 C5065 | Transfected with B2M targeting vector pAF217 |
| 11 | DFT1.B2M−/− + IFNG | DFT1 C5065 | Transfected with B2M targeting vector pAF217 and treated with 5 ng/mL IFNG for 24 h |
| 12 | DFT1.NLRC5.B2M−/− | DFT1 C5065 | Transfected with NLRC5 vector pCO1 and B2M targeting vector pAF218 |
The sequencing reads were mapped to the Tasmanian devil reference genome (GCA_902635505.1 mSarHar1.11) using Subread version 2.0.0 (Liao et al. 2013). Uniquely mapped reads were counted and assigned to genes using featureCounts (Liao et al. 2014). Differential expression analysis of gene counts was performed using statistical software RStudio (RStudio Team 2020) on R version 4.0.0 (R Core Team 2020). Firstly, genes with less than 100 aligned reads across all samples were filtered out to exclude lowly expressed genes. Gene counts were then normalized across samples by upper quartile normalization using edgeR (Robinson et al. 2009; Robinson and Oshlack 2010; Anders and Huber 2010) and EDASeq (Bullard et al. 2010; Risso et al. 2011). Normalized read counts were scaled by transcripts per kilobase million (TPM) to account for varied gene lengths. For differential expression analysis, gene expression of NLRC5-overexpressing cell lines (DFT1.NLRC5, DFT2.NLRC5) were compared against BFP-control cell lines (DFT1.BFP, DFT2.BFP) while IFNG-treated cells (DFT1.WT + IFNG, DFT2.WTRV + IFNG) were compared against the untreated wild-type (DFT1.WT, DFT2.WTRV), according to their respective tumor origin. Differential gene expression was calculated using the voom (Law et al. 2014) function in limma (Ritchie et al. 2015) with linear modelling and empirical Bayes moderation (Phipson et al. 2016) (Online Resource 1). Genes were defined as significantly differentially expressed by applying FDR < 0.05, and log2 fold change (FC) ≥ 2.0 (upregulated) or ≤ − 2.0 (downregulated) thresholds.
A bar plot of fold change in mRNA expression upon treatment was created from TPM values in GraphPad Prism version 5.03. Venn diagrams of differentially expressed genes were developed using Venny version 2.1 (Oliveros 2015). Heatmaps were created from log2TPM values using the ComplexHeatmap (Gu et al. 2016) package in R studio. For functional enrichment analysis, over-representation of gene ontology (GO) and Reactome pathways was analyzed on differentially expressed genes in R studio using functions enrichGO in ClusterProfiler (Yu et al. 2012) and enrichPathway in ReactomePA (Yu and He 2016), respectively. Significant GO terms and Reactome pathways were selected by applying the cut-offs p-value < 0.001, q-value < 0.05 and adjusted p-value < 0.05. P values were adjusted for multiple testing using Benjamini–Hochberg method.
Plasmid construction
The coding sequence for full length devil NLRC5 (ENSSHAT00000015489.1) was isolated from cDNA of devil lymph node mononuclear cells stimulated with recombinant devil IFNG (Flies et al. 2016) (10 ng/mL, 24 h). Devil NLRC5 was then cloned into plasmid pAF105 (detailed description of pAF105 plasmid construction available in Online Resource 2). For this study, devil NLRC5 was amplified from pAF105 with overlapping ends to the 5′ and 3′ SfiI sites of the Sleeping Beauty transposon plasmid pSBbi-BH (Kowarz et al. 2015) (a gift from Eric Kowarz; Addgene # 60,515, Cambridge, MA, USA) using Q5® Hotstart High-Fidelity 2X Master Mix (New England Biolabs (NEB), Ipswich, MA, USA) (see Online Resource 3 for primers and reaction conditions). The fragment was cloned into SfiI-digested (NEB) pSBbi-BH using NEBuilder® HiFi DNA Assembly Cloning Kit (NEB) and the assembled plasmid pCO1 was transformed into NEB® 5-alpha competent Escherichia coli (High Efficiency) (NEB) according to manufacturer’s instructions (see Online Resource 4 for plasmid maps). Positive clones were identified by colony PCR and the plasmid was purified using NucleoSpin® Plasmid EasyPure kit (Macherey–Nagel). The cloned devil NLRC5 transcript was verified by Sanger sequencing using Big Dye™ Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems (ABI), Foster City, CA, USA) and Agencourt® CleanSEQ® (Beckman Coulter, Brea, CA, USA) per manufacturer’s instructions. The sequences were analyzed on 3500xL Genetic Analyzer (ABI) (see Online Resource 5 for list of sequencing primers). For detailed step-by-step protocols for plasmid design and construction, reagent recipes, and generation of stable cell lines, see Bio-protocol # e3986 (Flies et al. 2020b).
Transfection and generation of stable cell lines
Stable cell lines of both DFT1 and DFT2 (C5065 and JV cell lines, respectively) overexpressing NLRC5 were prepared as follows. 5 × 105 cells were seeded in a six-well plate and incubated overnight to achieve 50–80% confluency on the day of transfection. As the vector constructed uses a Sleeping Beauty (SB) transposon system for gene transfer, co-transfection of an expression vector encoding an SB transposase enzyme pCMV(CAT)T7-SB100 (Mátés et al. 2009) (a gift from Zsuzsanna Izsvak; Addgene plasmid # 34,879) was needed to facilitate this process. Per 2.0 mL of culture volume, 2.0 μg of plasmid DNA (1.5 μg pCO1 + 0.5 μg pCMV(CAT)T7-SB100) was diluted in phosphate-buffered saline (PBS) to 100 μL and then added to 6.0 μg of polyethylenimine (PEI) (1 mg/mL, linear, 25 kDa; Polysciences, Warrington, FL, USA) diluted in PBS to 100 μL (3:1 ratio of PEI to DNA (w/w)). The DNA:PEI solution was mixed by gentle pipetting and incubated at room temperature for 15–20 min. The media on DFT cells were replaced with fresh complete RPMI medium and the transfection mix was added dropwise to the cells. The cells were incubated with the DNA:PEI solution overnight at 35 °C with 5% CO2. The next morning, media was replaced with fresh complete RPMI medium. 48 h post-transfection, the cells were observed for fluorescence through expression of reporter gene mTagBFP and were subjected to seven days of positive selection by adding 1 mg/mL hygromycin B (Sigma-Aldrich) in complete RPMI medium. Once selection was complete, the cells were maintained in 200 μg/mL hygromycin B in complete RPMI medium. pSBbi-BH was used as a control to account for the effects of the transfection and drug selection process.
Flow cytometric analysis of B2M expression
Cells were harvested and plated in a round-bottom 96-well plate (1 × 105 per well) and centrifuged at 500 g for 3 min at 4 °C to discard the medium. Cells were blocked with 50 μL of 1% normal goat serum (Thermo Fisher Scientific) in FACS buffer (PBS with 0.5% BSA, 0.02% sodium azide) for 10 min on ice. After blocking, 0.4 μL anti-devil B2M mouse antibody in supernatant (13-34-45, a gift from Hannah Siddle) (Siddle et al. 2013) diluted to a total of 50 μL in FACS buffer was added to the cells for 15 min on ice. The cells were washed with 150 µL FACS buffer and centrifuged at 500 g for 3 min at 4 °C. Goat anti-mouse IgG-Alexa Fluor 488 (Thermo Fisher Scientific) was diluted in FACS buffer to 4 µg/mL and 50 μL of the solution was incubated with the target cells in the dark for 30 min on ice. The cells were washed twice with FACS buffer to remove excess secondary antibody. Lastly, the cells were resuspended in 200 µL FACS buffer with propidium iodide (PI) (500 ng/mL) (Sigma-Aldrich) prior to analysis on BD FACSCanto™ II (BD Biosciences, Franklin Lakes, NJ, USA). As a positive control for surface B2M expression, DFT1 C5065 and DFT2 JV cells were stimulated with 5 ng/mL recombinant devil IFNG (Flies et al. 2016) for 24 h.
Generation of B2M CRISPR/Cas9 knockout cell lines (B2M−/−)
Two single guide RNAs (sgRNAs) targeting the first exon of devil B2M gene (ENSSHAG00000017005) were designed using a web-based CRISPR design tool CHOPCHOP (Labun et al. 2019) (Online Resource 6). Complementary oligonucleotides encoding each B2M sgRNA sequence were synthesized (Integrated DNA Technologies (IDT), Coralville, IA, USA), phosphorylated and annealed before cloning into lentiCRISPRv2 plasmid (Sanjana et al. 2014) (a gift from Feng Zhang; Addgene # 52,961) at BsmBI (NEB) restriction sites using T4 DNA ligase (NEB) (see Online Resource 7 for oligonucleotide sequences). The ligated plasmids pAF217 and pAF218 were then transformed into NEB® Stable Competent Escherichia coli (High Efficiency) (NEB). Single colonies were selected, and the plasmids were purified using ZymoPURE™ Plasmid Miniprep Kit (Zymo Research, Irvine, CA, USA). The sgRNA sequence in each plasmid was validated by Sanger sequencing according to the method described above (see Online Resource 5 for list of sequencing primers).
B2M targeting vectors pAF217 and pAF218 were each transfected into DFT1.WT and DFT1.NLRC5 cells to generate B2M knockout cell lines DFT1.B2M−/− and DFT1.NLRC5.B2M−/−. Transfection of cells were carried out as described above with the exception that 1.5 μg of plasmid was used instead of 2.0 μg. A day after transfection, the cells were subjected to positive selection by adding 100 μg/mL puromycin (InvivoGen, San Diego, CA, USA) for a week.
Post-drug selection, the cells were screened and sorted multiple rounds using a Beckman-Coulter MoFlo Astrios cell sorter to select DFT1.B2M−/− and DFT1.NLRC5.B2M−/− cells with negative B2M expression. DFT1.B2M−/− cells were treated with 10 ng/mL devil recombinant IFNG (Flies et al. 2016) for 24 h to stimulate surface B2M upregulation prior to analysis. For flow cytometry, cells were first harvested by centrifugation at 500 g for 3 min at 4 °C, and then blocked with 100 μL of 1% normal goat serum (Thermo Fisher Scientific) in complete RPMI medium for 10 min on ice. After blocking, the cells were incubated with 0.8 μL anti-devil B2M mouse antibody in supernatant (Siddle et al. 2013) diluted in complete RPMI to a total of 100 μL for 15 min on ice. The cells were washed with 2.0 mL complete RPMI and centrifuged at 500 g for 3 min at 4 °C. Next, the cells were incubated with 100 μL of 2 µg/mL goat anti-mouse IgG-Alexa Fluor 647 (Thermo Fisher Scientific) diluted in complete RPMI in the dark for 15 min on ice. The cells were washed with 2.0 mL of complete RPMI medium to remove excess secondary antibody. Lastly, the cells were resuspended to a concentration of 1 × 107 cells/mL in 200 ng/mL DAPI (Sigma-Aldrich) diluted in complete RPMI medium. B2M negative cells were selected and bulk-sorted using cell sorter Moflo Astrios EQ (Beckman Coulter).
After multiple rounds of sorting to establish a B2M negative population, genomic DNA of the cells was isolated and screened for mutations in the B2M gene by Sanger sequencing (see Online Resource 5 for sequencing primers). Indels (insertions or deletions) in the B2M gene were assessed using Inference of CRISPR Edits (ICE) analysis tool version 2.0 from Synthego (Synthego 2019) (Menlo Park, CA, USA) (Online Resource 6). B2M knockout cell lines: (i) DFT1.B2M−/− derived from DFT1 cells transfected with pAF217, and (ii) DFT1.NLRC5.B2M−/− derived from DFT1.NLRC5 transfected with pAF218 were selected for downstream analysis (see Table 1 for full list of cell lines).
Flow cytometric analysis of serum antibody target
Serum samples of wild Tasmanian devils were obtained from previous studies as described (Pye et al. 2016a; Tovar et al. 2017). To induce surface expression of MHC-I, DFT cells were treated with 10 ng/mL devil recombinant IFNG (Flies et al. 2016) for 24 h prior to analysis. Cells were washed with cold FACS buffer and 1 × 105 cells per well were plated in a round-bottom 96-well plate. The cells were centrifuged at 500 g for 3 min at 4 °C to discard the medium. Serum samples (see Online Resource 8 for serum sample information) were thawed on ice and diluted 1:50 with FACS buffer. 50 μL of diluted serum was added to the cells and incubated for 1 h on ice. After incubation, the cells were washed twice with 200 μL FACS buffer. 50 μL of 10 μg/mL monoclonal mouse anti-devil IgG2b antibody (A4-D1-2-1, provided by WEHI) (Howson et al. 2014) in FACS buffer was added to the cells and incubated for 30 min on ice. The cells were washed twice with FACS buffer and then incubated with 50 µL of 4 μg/mL goat anti-mouse IgG-Alexa Fluor 488 (Thermo Fisher Scientific) in FACS buffer for 30 min on ice, protected from light. The cells were washed twice with ice-cold PBS (Thermo Fisher Scientific). After washing, the cells were stained with LIVE/DEAD™ Fixable Near-IR Dead Cell Stain (Thermo Fisher Scientific) per manufacturer’s instructions. For B2M surface expression analysis, the cells were stained as described in the protocol above. However, LIVE/DEAD™ Fixable Near-IR Dead Cell Stain (Thermo Fisher Scientific) was used instead of PI to determine cell viability. All cells were fixed with FACS fix (0.02% sodium azide, 1.0% glucose, 0.4% formaldehyde) prior to analysis on BD FACSCanto™ II (BD Biosciences).
Results
NLRC5 is upregulated in DFT1 and DFT2 cells treated with IFNG
IFNG has been shown to upregulate MHC-I (Siddle et al. 2013) and PDL1 (Flies et al. 2016) on DFT cells. To probe the mechanisms driving upregulation of these key immune proteins, we performed RNA-seq using mRNA extracted from IFNG-treated DFT1 cell line C5065 (DFT1.WT) and an IFNG-treated DFT2 cell line RV (DFT2.WTRV). Markers for Schwann cell differentiation, SRY-box 10 (SOX10) and neuroepithelial marker nestin (NES), that are expressed in both DFT1 and DFT2 cells (Patchett et al. 2020), were selected as internal gene controls. As expected, transcriptome analysis showed that B2M, MHC-I gene SAHAI-01, and PDL1 were strongly upregulated by IFNG. MHC-I transactivator NLRC5 was also upregulated upon IFNG treatment, more than a 100-fold in both DFT1.WT (275-fold) and DFT2.WTRV cells (124-fold) relative to untreated cells (Fig. 1).
Fig. 1.
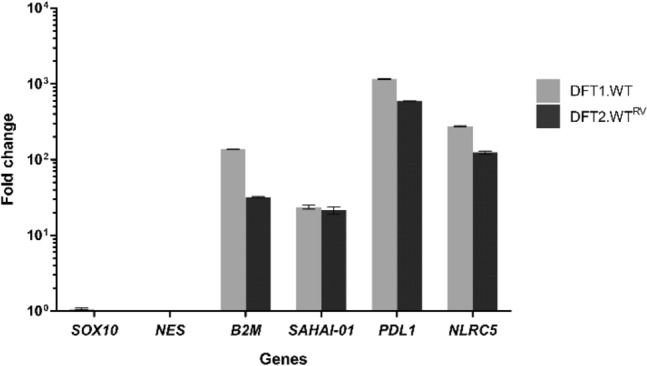
Upregulation of NLRC5 by IFNG in DFT1 and DFT2 cells. Fold change in mRNA expression (transcripts per kilobase million (TPM)) of B2M, MHC class I gene SAHAI-01, PDL1 and NLRC5 upon IFNG treatment in DFT1 C5065 cell line (DFT1.WT) and DFT2 RV cell line (DFT2.WTRV). SOX10 and NES were included as internal controls. Bars show the mean of N = 2 replicates per treatment. Error bars indicate standard deviation
NLRC5 upregulates MHC-I and antigen presentation genes but not PDL1 and non-classical MHC-I
To assess the role of NLRC5 in antigen processing and presentation, we developed an expression vector that stably upregulates NLRC5 in DFT cells. DFT1 cell line C5065 and DFT2 cell line JV were used for production of NLRC5-overexpressing DFT cells. Following drug selection to create stable cell lines, we performed RNA-seq on DFT1 and DFT2 cells stably transfected with BFP-control and NLRC5 vectors (see Table 1 for list of cell lines). Changes in the mRNA expression profile of DFT cells overexpressing NLRC5 relative to BFP-control cells were examined in parallel with changes observed in wild-type DFT cells following IFNG treatment (Fig. 2 and Online Resource 9). The transcriptome for IFNG-treated DFT2 cells was previously generated from the DFT2 RV cell line (DFT2.WTRV) (Patchett et al. 2020). Otherwise, all DFT2 results are from DFT2 JV.
Fig. 2.
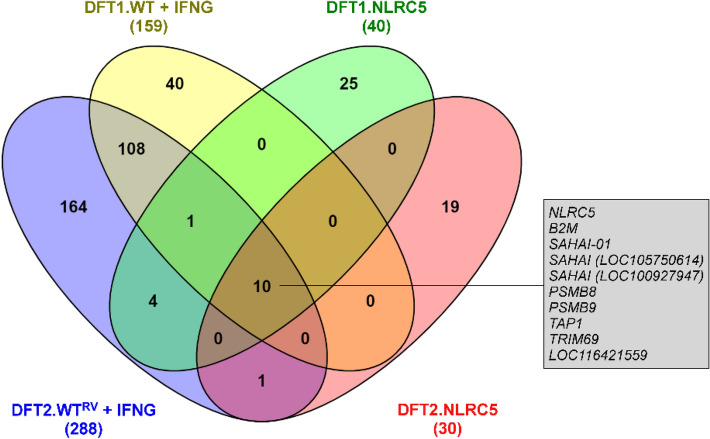
Venn diagram of genes significantly upregulated upon IFNG treatment and NLRC5 overexpression in DFT1 and DFT2 cells. Genes were defined as significantly upregulated when false discovery rate (FDR) < 0.05 and log2FC ≥ 2.0. Total number of genes upregulated for each treatment is indicated in parentheses under the sample name. The box shows genes upregulated in all four treatments: (i) IFNG-treated DFT1 cells (DFT1.WT + IFNG), (ii) IFNG-treated DFT2 cells (DFT2.WTRV + IFNG), (iii) NLRC5-overexpressing DFT1 cells (DFT1.NLRC5), and (iv) NLRC5-overexpressing DFT2 cells (DFT2.NLRC5). See Online Resource 1 for a full list of differentially expressed genes and Online Resource 10 for description of devil-specific genes (LOC symbols)
Differential expression analysis showed that 159 genes were upregulated by IFNG (DFT1.WT + IFNG) in contrast to 40 genes by NLRC5 (DFT1.NLRC5) in DFT1 cells (Fig. 2). In DFT2 cells, 288 genes were upregulated by IFNG (DFT2.WTRV + IFNG) and 30 genes by NLRC5 (DFT2.NLRC5) (Fig. 2). There were ten genes that were upregulated by both IFNG and NLRC5 in DFT1 and DFT2 cells. These shared genes were predominantly related to MHC-I antigen processing and presentation pathway which suggests a role of NLRC5 in IFNG-induced MHC-I expression.
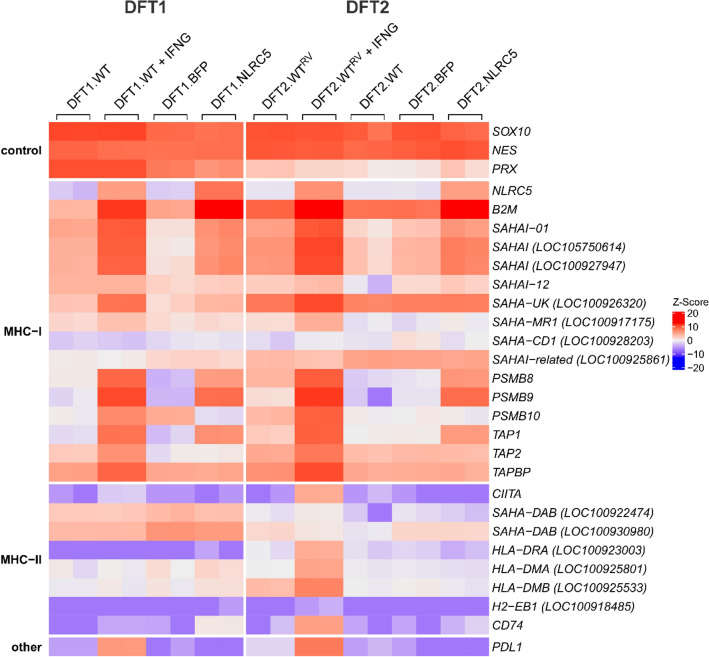
A heatmap was used to explore the expression profiles of genes associated with MHC-I and MHC-II antigen processing and presentation. In addition to SOX10 and NES, the myelin protein periaxin (PRX), a marker for DFT1 cells (Tovar et al. 2011), was included as an internal control. Overall, NLRC5 upregulated genes involved in MHC-I antigen presentation to a smaller magnitude than IFNG (Fig. 3). NLRC5 upregulated a subset of IFNG-induced MHC-I genes SAHAI-01, SAHAI (LOC105750614) and SAHAI (LOC100927947), and genes of the antigen processing machinery including B2M, PSMB8, PSMB9, and TAP1. In comparison, other IFNG-induced genes such as PSMB10, TAP2 and TAP binding protein (TAPBP) were not upregulated by NLRC5 in either DFT1.NLRC5 or DFT2.NLRC5 cells. MHC-I genes that were induced by IFNG but not NLRC5 include non-classical MHC-I genes SAHA-UK and SAHA-MR1, although the latter was only induced in DFT2 cells treated with IFNG. Additionally, PDL1 was upregulated by IFNG, but not NLRC5. Examination of the promoter elements immediately upstream of SAHA-UK and PDL1 did not identify the putative MHC-I-conserved SXY module (Cheng et al. 2012) necessary for NLRC5-mediated transcription in the devil genome. A putative interferon-stimulated response element (ISRE) for devil MHC-I genes was identified 127 bp upstream of the start codon of SAHA-UK (Online Resource 11).
Fig. 3.
Heatmap showing expression profiles of genes involved in MHC-I and MHC-II antigen processing and presentation pathways, and PDL1 in IFNG-treated, and NLRC5-overexpressing DFT1 and DFT2 cells. Log2TPM expression values were scaled across each gene (rows) and represented by Z-Score, with red and blue representing high and low relative expression, respectively. Replicates for each treatment (N = 2) are included in the heatmap. SAHAI encodes the Tasmanian devil MHC-I heavy chain gene. For genes with no official gene symbol (LOC symbols), alternative gene symbols were used according to the gene description on NCBI. See Online Resource 10 for corresponding NCBI gene symbols and description
NLRC5 did not consistently regulate MHC-II genes. However, the invariant chain associated with assembly of MHC-II complexes, CD74, was significantly upregulated in DFT1.NLRC5. Similarly, IFNG treatment on DFT1 cells only upregulated MHC-II transactivator CIITA. Strikingly, IFNG treatment on DFT2 cells induced several MHC-II genes such as HLA-DRA (LOC100923003), HLA-DMA (LOC100925801), HLA-DMB (LOC100925533), CD74 and CIITA.
NLRC5 primarily functions in MHC-I antigen processing and presentation but is not limited to immune-related functions
The majority of research into NLRC5 has been devoted to its role as a regulator of MHC-I expression. In addition, some studies have reported possible roles of NLRC5 in antiviral immunity, inflammation and cancer through modulation of various signaling pathways (Kuenzel et al. 2010; Cui et al. 2010; Benko et al. 2010; Neerincx et al. 2010; Davis et al. 2011; Wang et al. 2019). To identify additional biological functions of NLRC5 in DFT cells, over-representation analysis of gene ontology (GO) biological processes and Reactome pathways was performed using the list of differentially expressed genes between NLRC5-overexpressing DFT cells and BFP-controls (FDR < 0.05, log2FC ≥ 2.0 or ≤ − 2.0). Both analyses revealed significant up- and downregulation of genes associated with immune system processes and developmental processes in cells overexpressing NLRC5.
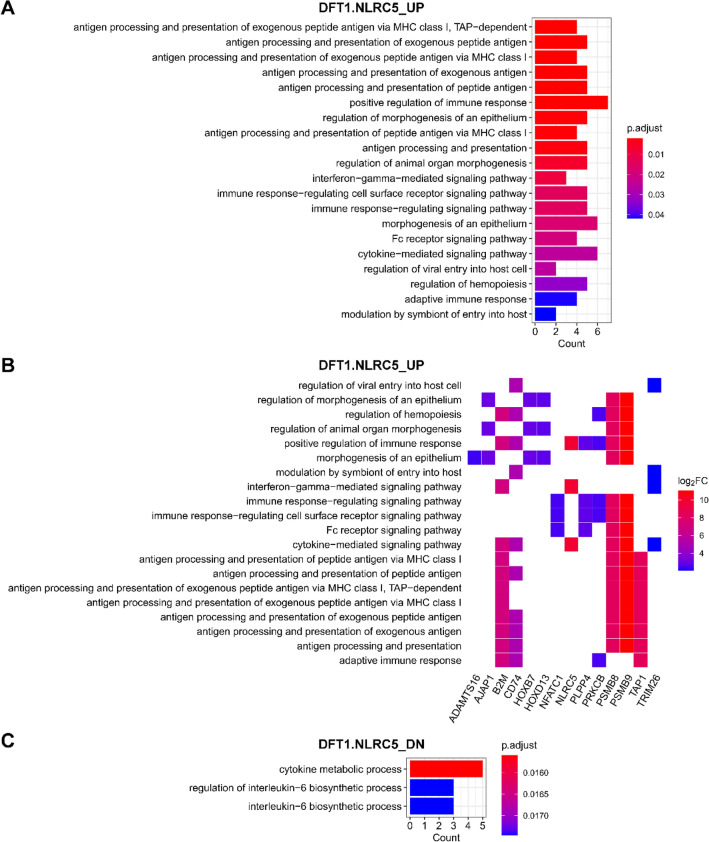
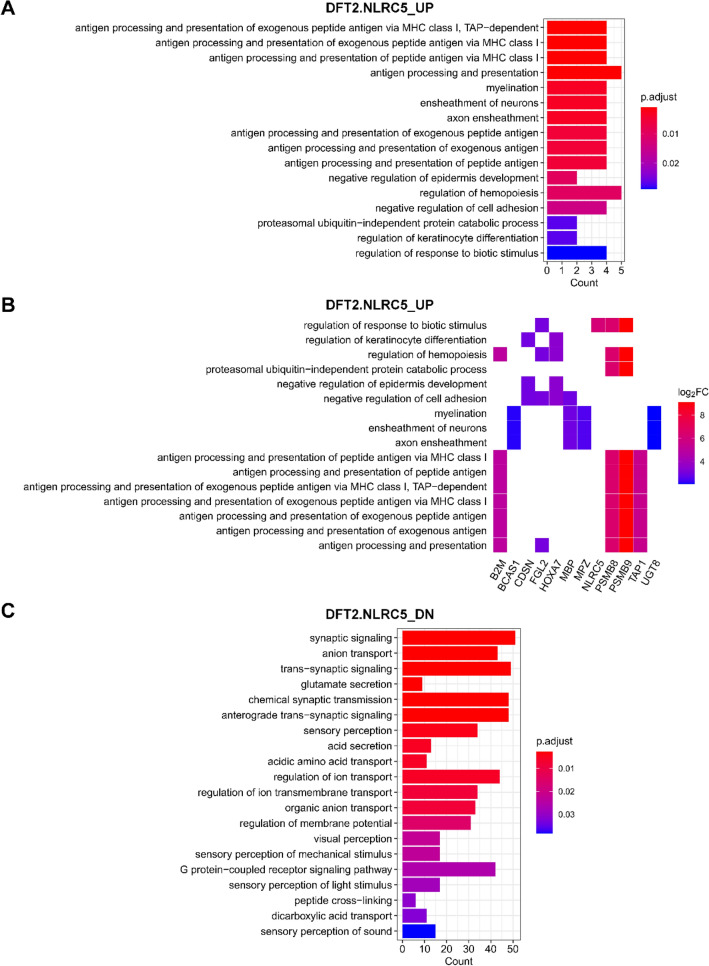
Among the list of genes upregulated in DFT1.NLRC5 and DFT2.NLRC5 cells, the most significantly associated GO biological process was antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent (Figs. 4a and 5a). Several additional immune-related processes were also associated with NLRC5 overexpression, particularly in DFT1 cells. Some of these included positive regulation of immune response, interferon-gamma-mediated signaling pathway, immune response-regulating cell surface receptor signaling pathway (Fig. 4a), and regulation of interleukin-6 biosynthetic process (Fig. 4c). In DFT1.NLRC5 and DFT2.NLRC5, GO terms related to development that were significantly over-represented included morphogenesis of an epithelium (Fig. 4a) and negative regulation of epidermis development (Fig. 5a), respectively.
Fig. 4.
GO biological processes that were enriched in DFT1 cells with NLRC5 overexpression. GO biological process terms associated with genes upregulated (UP) (a, b) and downregulated (DN) (c) in DFT1.NLRC5. b Heatplot of genes associated with each positively regulated GO term. The cut-offs p-value < 0.001 and adjusted p-value (p. adjust) < 0.05 were used to determine significant biological processes. P values were adjusted for multiple testing using Benjamini–Hochberg method. See also Online Resource 12 for full list of GO biological processes
Fig. 5.
GO biological processes that were enriched in DFT2 cells with NLRC5 overexpression. GO biological process terms associated with genes upregulated (UP) (a, b) and downregulated (DN) (c) in DFT2.NLRC5. b Heatplot of genes associated with each positively-regulated GO term. The cut-offs p-value < 0.001 and adjusted p-value (p. adjust) < 0.05 were used to determine significant biological processes. P values were adjusted for multiple testing using Benjamini–Hochberg method. See also Online Resource 13 for full list of GO biological processes
As DFT cells are of neuroendocrine origin, specifically of the Schwann cell lineage (Murchison et al. 2010; Patchett et al. 2020), a number of neural-related genes were targeted by NLRC5. In DFT2 cells, NLRC5 upregulated genes that are involved in myelination, which are usually expressed at low levels in DFT2 cells (Patchett et al. 2020) (Fig. 5a). These genes include brain enriched myelin associated protein 1 (BCAS1), myelin binding protein (MBP), myelin protein zero (MPZ) and UDP glycosyltransferase 8 (UGT8) (Fig. 5b). Furthermore, many of the downregulated genes in DFT2.NLRC5 were related to nervous system function, mainly pertaining to synaptic signaling and sensory perception (Fig. 5c).
Reactome pathway analysis revealed an enrichment of pathways that were consistent with those identified by GO analysis. This included enrichment of the ER-phagosome pathway and antigen processing-cross presentation in DFT1.NLRC5 (Table 2) and DFT2.NLRC5 (Table 3); signaling by the B cell receptor (BCR) in DFT1.NLRC5; and transmission across chemical synapses in DFT2.NLRC5 cells. Interestingly, nuclear factor of activated T cells 1 (NFATC1), protein kinase C beta (PRKCB), PSMB8 and PSMB9, associated with several GO immune-related processes in DFT1.NLRC5 (Fig. 4B), were enriched for the beta-catenin independent WNT signaling pathway (Table 2). Other enriched pathways included those involved in extracellular matrix organization such as collagen chain trimerization (Table 2) and assembly of collagen fibrils and other multimeric structures (Table 3).
Table 2.
Reactome pathways enriched in differentially expressed genes in DFT1.NLRC5
| Reactome ID | Pathway | Count | Term size | p value | p.adjust | Genes |
|---|---|---|---|---|---|---|
| Upregulated | ||||||
| R-HSA-1236974 | ER-Phagosome pathway | 4 | 74 | 4.69E-05 | 4.75E-03 | B2M, PSMB8, PSMB9, TAP1 |
| R-HSA-1168372 | Downstream signaling events of B Cell Receptor (BCR) | 4 | 80 | 6.37E-05 | 4.75E-03 | NFATC1, PRKCB, PSMB8, PSMB9 |
| R-HSA-1236975 | Antigen processing-Cross presentation | 4 | 81 | 6.69E-05 | 4.75E-03 | B2M, PSMB8, PSMB9, TAP1 |
| R-HSA-983705 | Signaling by the B Cell Receptor (BCR) | 4 | 104 | 1.77E-04 | 9.44E-03 | NFATC1, PRKCB, PSMB8, PSMB9 |
| R-HSA-3858494 | Beta-catenin independent WNT signaling | 4 | 129 | 4.06E-04 | 1.73E-02 | NFATC1, PRKCB, PSMB8, PSMB9 |
| R-HSA-1169091 | Activation of NF-kappaB in B cells | 3 | 64 | 7.19E-04 | 2.55E-02 | PRKCB, PSMB8, PSMB9 |
| Downregulated | ||||||
| R-HSA-216083 | Integrin cell surface interactions | 5 | 62 | 1.16E-05 | 4.04E-03 | CDH1, COL18A1, COL6A1, COL6A2, JAM2 |
| R-HSA-1251985 | Nuclear signaling by ERBB4 | 3 | 28 | 3.33E-04 | 4.93E-02 | EREG, GFAP, S100B |
| R-HSA-5173105 | O-linked glycosylation | 4 | 73 | 4.27E-04 | 4.93E-02 | ADAMTS7, B3GNT7, GALNT13, GALNT17 |
| R-HSA-913709 | O-linked glycosylation of mucins | 3 | 34 | 5.96E-04 | 4.93E-02 | B3GNT7, GALNT13, GALNT17 |
| R-HSA-8948216 | Collagen chain trimerization | 3 | 36 | 7.06E-04 | 4.93E-02 | COL18A1, COL6A1, COL6A2 |
Cut-offs p-value < 0.001 and p.adjust < 0.05 were used to display significant pathways. P values were adjusted (p.adjust) for multiple testing using Benjamini–Hochberg method. See also Online Resource 14 for full list of Reactome pathways
Table 3.
Reactome pathways enriched in differentially expressed genes in DFT2.NLRC5
| Reactome ID | Pathway | Count | Term size | p value | p.adjust | Genes |
|---|---|---|---|---|---|---|
| Upregulated | ||||||
| R-HSA-1236974 | ER-Phagosome pathway | 4 | 74 | 3.43E-06 | 3.80E-04 | B2M, PSMB8, PSMB9, TAP1 |
| R-HSA-1236975 | Antigen processing-Cross presentation | 4 | 81 | 4.93E-06 | 3.80E-04 | B2M, PSMB8, PSMB9, TAP1 |
| R-HSA-983169 | Class I MHC mediated antigen processing & presentation | 5 | 312 | 5.96E-05 | 3.06E-03 | B2M, PSMB8, PSMB9, TAP1, TRIM69 |
| R-HSA-983170 | Antigen Presentation: Folding, assembly and peptide loading of class I MHC | 2 | 18 | 3.31E-04 | 1.27E-02 | B2M, TAP1 |
| R-HSA-162909 | Host Interactions of HIV factors | 3 | 119 | 6.91E-04 | 1.36E-02 | B2M, PSMB8, PSMB9 |
| Downregulated | ||||||
| R-HSA-112316 | Neuronal System | 33 | 276 | 1.73E-06 | 1.28E-03 | see Online Resource 15 |
| R-HSA-1474228 | Degradation of the extracellular matrix | 16 | 97 | 1.82E-05 | 6.73E-03 | see Online Resource 15 |
| R-HSA-264642 | Acetylcholine Neurotransmitter Release Cycle | 5 | 10 | 5.87E-05 | 1.45E-02 | see Online Resource 15 |
| R-HSA-181429 | Serotonin Neurotransmitter Release Cycle | 5 | 12 | 1.70E-04 | 2.27E-02 | see Online Resource 15 |
| R-HSA-181430 | Norepinephrine Neurotransmitter Release Cycle | 5 | 12 | 1.70E-04 | 2.27E-02 | see Online Resource 15 |
| R-HSA-112315 | Transmission across Chemical Synapses | 21 | 179 | 1.84E-04 | 2.27E-02 | see Online Resource 15 |
| R-HSA-1474244 | Extracellular matrix organization | 24 | 224 | 2.65E-04 | 2.80E-02 | see Online Resource 15 |
| R-HSA-166658 | Complement cascade | 6 | 21 | 4.02E-04 | 3.38E-02 | see Online Resource 15 |
| R-HSA-1296072 | Voltage gated Potassium channels | 7 | 29 | 4.11E-04 | 3.38E-02 | see Online Resource 15 |
| R-HSA-2022090 | Assembly of collagen fibrils and other multimeric structures | 9 | 49 | 5.62E-04 | 4.16E-02 | see Online Resource 15 |
| R-HSA-210500 | Glutamate Neurotransmitter Release Cycle | 5 | 16 | 7.96E-04 | 4.91E-02 | see Online Resource 15 |
| R-HSA-212676 | Dopamine Neurotransmitter Release Cycle | 5 | 16 | 7.96E-04 | 4.91E-02 | see Online Resource 15 |
Cut-offs p-value < 0.001 and p.adjust < 0.05 were used to display significant pathways. P values were adjusted (p.adjust) for multiple testing using Benjamini–Hochberg method. See also Online Resource 15 for full list of Reactome pathways
NLRC5 induces MHC-I expression on the cell surface
To determine if NLRC5 is capable of regulating MHC-I expression at the protein level, surface MHC-I was analyzed by flow cytometry in DFT cells overexpressing NLRC5 using a monoclonal antibody against B2M (Siddle et al. 2013). The overexpression of NLRC5 induced upregulation of surface expression of B2M in both DFT1.NLRC5 (Fig. 6a) and DFT2.NLRC5 cells (Fig. 6b). The level of B2M expression was also comparable to wild-type DFT cells treated with IFNG.
Fig. 6.
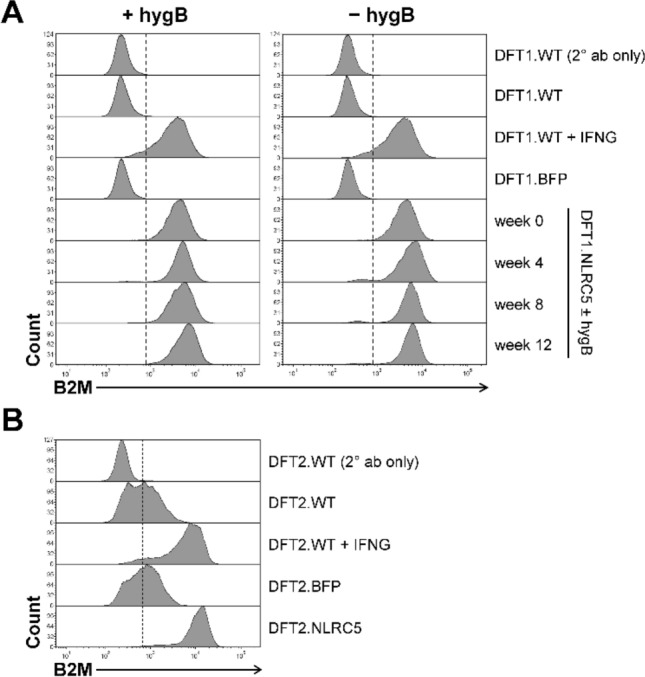
Upregulation of MHC-I following NLRC5 overexpression. Surface expression of B2M in DFT1.NLRC5 (a) and DFT2.NLRC5 (b). B2M expression in the NLRC5 cell lines were compared to wild-type (DFT.WT), BFP-control (DFT.BFP), and IFNG-treated (DFT.WT + IFNG) DFT cells. (a) Stable expression of B2M in DFT1.NLRC5 was assessed every 4 weeks for 12 weeks post-drug selection in the presence and absence of hygromycin B (hygB) selection pressure. Secondary antibody-only staining (DFT.WT (2° ab only)) was included as a control. The results shown are representative of N = 3 replicates/treatment
Next, we assessed the stability of NLRC5-induced MHC-I expression by examining the expression of B2M in long-term cultures. One-month post-drug selection, DFT1.NLRC5 cells cultured in the presence or absence of hygromycin B were stained for B2M every 4 weeks for a total of 12 weeks. As shown in Fig. 6a, MHC-I expression was stably maintained in DFT1.NLRC5 cells, with or without ongoing drug selection pressure throughout the 12-week culture thus, demonstrating the relative stability of the human EF1a promoter driving NLRC5 expression in long-term cell cultures. PDL1 was also not upregulated on the cell surface in NLRC5-overexpressing DFT cells compared to IFNG-treated DFT cells (Online Resource 16).
MHC-I is a predominant target of anti-DFT antibody responses
It was previously reported that the antibodies from devils infected with DFT1 were specific to MHC-I, as determined by incubating serum from these devils with IFNG-treated DFT cells (Pye et al. 2016a). Considering the diverse roles of IFNG, there could be other IFNG-induced antigens that can serve as targets for the anti-DFT antibody response.
To establish if MHC-I is the target of anti-DFT serum antibodies, surface MHC-I expression was first ablated by knocking out the hemizygous B2M allele (Stammnitz et al. 2018) in wild-type DFT1 cells (DFT1.WT) and NLRC5-overexpressing DFT1 cells (DFT1.NLRC5) using CRISPR/Cas9 technology. Gene disruption of B2M was confirmed by genomic DNA sequencing (Online Resource 6), and flow cytometry using a monoclonal anti-B2M antibody (Fig. 7). CRISPR/Cas9-mediated B2M knockout (B2M−/−) in DFT1 cells rendered the cells irreversibly deficient for surface expression of B2M despite IFNG and NLRC5 stimulation (DFT1.B2M−/− + IFNG and DFT1.NLRC5.B2M−/−). Due to the pivotal role of B2M in stability of MHC-I complex formation and surface presentation (Arce‐Gomez et al. 1978; Williams et al. 1989; Vitiello et al. 1990; Kozlowski et al. 1991; Boyd et al. 1992), absence of surface B2M is indicative of a lack of surface MHC-I expression.
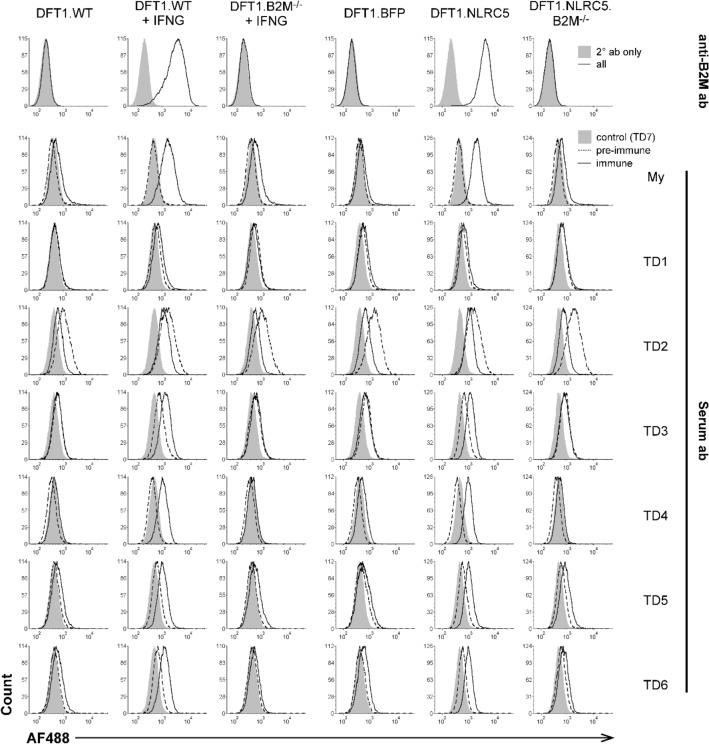
Fig. 7.
Flow cytometric analysis of serum antibody binding from devils with anti-DFT1 antibody response. Ablation of surface B2M in CRISPR/Cas9-mediated B2M knockout cells (B2M−/−) was confirmed using a monoclonal anti-B2M antibody (anti-B2M ab). Sera from six devils (TD1-TD6) with seroconversion (immune) following DFTD infection were tested against wild-type DFT1 (DFT1.WT), IFNG-treated DFT1 (DFT1.WT + IFNG), IFNG-treated B2M knockout DFT1 (DFT1.B2M−/− + IFNG), BFP-control (DFT1.BFP), DFT1 overexpressing NLRC5 (DFT1.NLRC5) and B2M knockout NLRC5-overexpressing DFT1 (DFT1.NLRC5.B2M−/−) cells. An immunized devil with induced tumor regression (My) was included as a positive control, meanwhile serum from a healthy devil (TD7) was included as a negative control as represented in the shaded grey area. Ab, antibody; AF488, Alexa Fluor 488
After surface MHC-I ablation was confirmed, serum from six wild devils (TD1–TD6) that demonstrated anti-DFT responses including natural DFT1 regressions (Pye et al. 2016a) was tested against B2M knockout cell lines DFT1.B2M−/− and DFT1.NLRC5.B2M−/−. Serum from a healthy devil (TD7) and an immunized devil with induced tumor regression (My) (Tovar et al. 2017) were used as negative and positive controls for antibody binding. All six sera from DFTD+ devils (TD1–TD6) showed weak to no binding to DFT1.WT and DFT1.BFP, which are inherently negative for surface MHC-I (Fig. 7). With forced expression of MHC-I using IFNG (DFT1.WT + IFNG) and NLRC5 (DFT1.NLRC5), a positive shift in antibody binding was observed. There was no apparent difference in the level of antibody binding between IFNG-treated and NLRC5-overexpressing DFT1 cells, suggesting a similarity between the antibody target(s) induced by IFNG and NLRC5. Following B2M knockout, antibody binding of all six sera was reduced in both IFNG-induced (DFT1.B2M−/− + IFNG) and NLRC5-induced B2M knockout DFT1 cells (DFT1.NLRC5.B2M−/−), suggesting that MHC-I is a target of DFT1-specific antibody responses in natural tumor regressions.
Discussion
Overexpression of NLRC5 in DFT cells has revealed a major and evolutionarily conserved role for NLRC5 in MHC-I antigen processing and presentation. Consistent with studies in human and mouse cell lines (Meissner et al. 2010; Yao et al. 2012; Biswas et al. 2012; Yoshihama et al. 2016; Rodriguez et al. 2016), NLRC5 induced the expression of classical MHC-I genes (SAHAI-01, SAHAI (LOC105750614), SAHAI (LOC100927947)), B2M, PSMB8, PSMB9 and TAP1 in both DFT1 and DFT2 cells. Despite the lack of increase in TAP2 expression, the selective upregulation of MHC-I and other functionally related genes by NLRC5 was sufficient to restore MHC-I molecules on the cell surface. Although the peptide transport function of TAP proteins typically involves the formation of TAP1 and TAP2 heterodimers, homodimerization of TAP proteins have been described (Lapinski et al. 2000; Antoniou et al. 2002). However, the functionality of TAP1 homodimers remains to be verified. The conservation of NLRC5 regulation of the MHC-I pathway across species highlights the important role of NLRC5 in MHC-I expression.
Previous studies have shown that sera from wild devils with anti-DFT immune response contained high titers of antibody that bound to IFNG-treated DFT1 cells. It was proposed that the primary antibody targets were MHC-I proteins on the tumor cell (Pye et al. 2016a). Additionally, some of these devils experienced tumor regression despite the lack of strong evidence for immune cell infiltration into tumors. Potential targets of these DFT-specific antibodies include: (i) non-MHC IFNG-induced cell surface antigens, (ii) tumor antigens complexed to MHC-I, or (iii) MHC-I molecules independent of the antigen. Because DFT1 and DFT2 are tumor cell lines that arose independently from single founder devils (Deakin et al. 2012; Murchison et al. 2012; Pye et al. 2016b), the different MHC alleles on the tumor compared to individual host devils may represent a source of antigens for an allogeneic response. The function of NLRC5 that is mainly restricted to MHC-I regulation compared with IFNG provided an opportunity to re-examine the antibody target(s) of serum antibodies from wild devils burdened with DFTs. A clear understanding of immunogenic targets of DFTs will provide direction for a more effective vaccine against DFTD.
In this study, the MHC-I complex was identified as the predominant target of anti-DFT serum antibodies. The antibody binding intensity against NLRC5-overexpressing DFT cells was similar to IFNG-treated DFT cells, suggesting similar levels of target antigen expression. When MHC-I expression was ablated through B2M knockout, antibody binding was reduced to almost background levels despite IFNG and NLRC5 stimulation. This finding presents an option to exploit NLRC5 for induction of anti-DFT immunity, potentiated by humoral responses in Tasmanian devils. Although cellular immunity is likely a key mechanism for tumor rejection, B cells and antibodies can play eminent roles in transplant rejection (Schmitz et al. 2020) and anti-tumor immunity (Nelson 2010). B cells can promote rejection through antibody-dependent mechanisms that facilitate FcR-mediated phagocytosis by macrophages, antibody-dependent cellular cytotoxicity (ADCC) by NK cells, complement activation and antigen uptake by dendritic cells (reviewed in (Yuen et al. 2016)). Moreover, B cells can enhance immune surveillance and response through direct antigen presentation to T cells and production of immune-modulating molecules such as cytokines and chemokines (Yuen et al. 2016).
Caldwell et al. reported that the most highly expressed MHC alleles on DFT2 cells are those that matched host MHC alleles (Caldwell et al. 2018), which suggests that DFT cells may hide from host defenses or induce immunological tolerance via shared MHC alleles. If MHC-I is the major antibody target and potentially the overall immune system target, devils having the largest MHC mismatch with DFT cells will be the most likely to have strong MHC-I specific responses and reject DFTs, leading to natural selection in the wild. For example, previous studies have shown that some devils have no functional MHC-I allele at the UA loci and that these individuals can be homozygous at the UB and UC loci (Tovar et al. 2017). These individuals present a reduced MHC-peptide that would have the lowest probability of a match to the DFT MHC alleles that induce host DFT1 tolerance. However, selection for reduced genetic diversity in MHC alleles would be unfavorable for long-term conservation. A prophylactic vaccine would ideally be designed to assist in the preservation of the genetic diversity of wild devils (Flies et al. 2020c).
Although the MHC proteins themselves are likely a primary target of humoral and cellular immunity, MHC-I alleles generally differ by only a few amino acids (Caldwell et al. 2018; Gastaldello et al. 2020). Mutations in DFTs and somatic variation between host and tumor cells provide a rich source of additional antigenic targets for humoral and cellular immunity (Stammnitz et al. 2018). The reduction in antibody binding to B2M knockout cells suggests that these tumor antigens are unlikely to be the primary antibody targets, although binding of antibodies to peptide-MHC complexes and other B2M-associated proteins cannot be excluded. Besides classical MHC-I molecules, B2M has been found in association with several other membrane proteins that are MHC-I-like such as cluster of differentiation 1 (CD1), MHC class I-related (MR1), neonatal Fc receptor (FcRn), and homeostatic iron regulator (HFE) (Knowles and Bodmer 1982; Simister and Mostov 1989; Feder et al. 1996; Yamaguchi and Hashimoto 2002). However, none of these genes were upregulated upon NLRC5 and IFNG induction in DFT cells, which further suggest that the antibody target is restricted to classical MHC-I expression on tumor cells. Knocking out individual MHC alleles in DFT cells or overexpression of MHC alleles in alternative non-DFT cell lines could be used to disentangle the importance of specific alleles and investigate the potential for peptide-MHC complexes to be antibody targets.
Our results confirm that IFNG affects more immunoregulatory processes than NLRC5. However, the functional dichotomy of IFNG in cancer means that NLRC5 modulation could be an alternative to IFNG treatment for enhancing tumor cell immunogenicity in a range of species, including humans. Importantly, NLRC5 upregulated B2M on the surface of DFT cells to similar levels as IFNG, but it does not upregulate inhibitory molecules. The restoration of functional MHC-I molecules without concomitant upregulation of PDL1 and SAHA-UK has multiple advantages over IFNG for triggering effective cytotoxic responses against DFT cells. First, cells transfected with NLRC5 constitutively express MHC-I and therefore do not require culturing in IFNG, which can be problematic as IFNG can also reduce cell viability (Ong et al. 2019). Second, PDL1 negatively regulates T cell responses by inducing T cell anergy (Selenko-Gebauer et al. 2003) and apoptosis (Dong et al. 2002) while limiting T cell activity (Butte et al. 2007). Moreover, PDL1 promotes tumor growth and survival by stimulating cell proliferation (Ghebeh et al. 2007) and resistance to T cell killing (Iwai et al. 2002; Azuma et al. 2008). Third, the expression of monomorphic MHC-I SAHA-UK induced by IFNG would allow DFT cells to escape cytotoxic attack from both NK cells and CD8+ T cells (Kochan et al. 2013). Fourth, several other immune checkpoint protein receptor-ligand interactions were recently shown to be conserved in devils (Flies et al. 2017, 2020a), but we found no significant upregulation of these genes by NLRC5. The ability to improve tumor immunogenicity in the absence of inhibitory signals has positive implications for immunization and immunotherapeutic strategies. NLRC5 could evoke protective anti-tumor immunity against DFTs, similar to NLRC5-expressing B16-F10 melanoma cells in mice (Rodriguez et al. 2016).
The absence of a regulatory effect on SAHA-UK and PDL1 by NLRC5 in contrast to IFNG could be due to the composition of the promoter elements of these genes. The promoter of MHC class I genes consists of three conserved cis-regulatory elements: an NFκB-binding Enhancer A region, an interferon-stimulated response element (ISRE) and an SXY module (Van Den Elsen et al. 1998b, a). The SXY module is critical for NLRC5-mediated MHC-I transactivation as it serves as the binding site for the multi-protein complex formed between NLRC5 and various transcription factors (Meissner et al. 2012; Neerincx et al. 2012; Ludigs et al. 2015). An ISRE and SXY module is present within 200 base pairs of the start codon for all three classical devil MHC-I genes (Cheng et al. 2012). We identified an ISRE element in the SAHA-UK promoter region but were unable to identify an SXY module in this region. This could explain the upregulation of SAHA-UK upon IFNG stimulation but not in NLRC5-overexpressing DFT cells. Similarly, the SXY module was not identified in orthologues of SAHA-UK, which are Modo-UK in the grey short-tailed opossum (Belov et al. 2006) and Maeu-UK in the tammar wallaby (Siddle et al. 2009). The difference in regulation and therefore, pattern of expression of the UK gene in marsupials (Belov et al. 2006; Siddle et al. 2009; Cheng and Belov 2014) may reflect a separate function from classical MHC-I. The marsupial UK gene has been hypothesized to play a marsupial-specific role in conferring immune protection to vulnerable newborn marsupials during their pouch life (Siddle et al. 2009). SXY modules are typically not found in the promoter region of PDL1 (Lee et al. 2006) therefore, it is not expected for NLRC5 to be a regulator of PDL1. Rather, IFNG-mediated induction of PDL1 occurs via transcription factor interferon regulatory factor 1 (IRF-1) (Lee et al. 2006), which is induced by STAT1 (Loke and Allison 2003).
Beyond MHC-I regulation, NLRC5 expression in DFT1 cells displayed other beneficial immune-regulating functions, mainly via the non-canonical β-catenin-independent WNT signaling pathway. One of the downstream effectors that was upregulated by NLRC5 included PRKCB, an activator of NFκB in B cells (Saijo et al. 2002). NFκB is a family of pleiotropic transcription factors known to regulate several immune and inflammatory responses including cellular processes such as cell proliferation and apoptosis (Hayden and Ghosh 2004). In recent years, aberrations in NFκB signaling have been implicated in cancer development and progression (Xia et al. 2018; Verzella et al. 2020). The regulation of NFκB signaling by NLRC5 has been documented in several studies although the findings have been contradictory (Cui et al. 2010; Benko et al. 2010; Tong et al. 2012; Liu et al. 2017).
In summary, we have demonstrated the role of NLRC5 in MHC-I regulation of DFT cells thereby, displaying the functional conservation of NLRC5 across species. The finding that allogeneic MHC-I on DFT cells is a major antibody target in wild devils with anti-DFT response and natural DFT regression can help guide DFTD vaccine development and conservation management strategies. NLRC5-overexpressing DFT cells can be harnessed to elicit both cellular and humoral immunity against future and pre-existing DFT infections in wild devils using MHC-I as a target. Given the prevalence of altered MHC-I expression in cancer as a form of immune escape mechanism (Garrido et al. 1993; Hicklin et al. 1999; Campoli and Ferrone 2008), NLRC5 presents as a new target for providing an insight into the role of MHC-I in cancer as well as transplantation, and its manipulation for human cancer treatment and transplant tolerance.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors would like to thank Patrick Lennard, Peter Murphy, and Candida Wong for assistance in the lab and Terry Pinfold for assistance in flow cytometry. We thank Hannah Siddle for supplying the monoclonal antibody for B2M and for offering her expertise in devil MHC-I immunogenetics. We wish to thank G. Ralph for ongoing care of Tasmanian devils, the Bonorong Wildlife Sanctuary for providing access to Tasmanian devils, and R. Pye for providing care for devils and collecting blood samples. This work was supported by the Australian Research Council (ARC) DECRA grant # DE180100484 and ARC Discovery grant # DP180100520, University of Tasmania Foundation Dr. Eric Guiler Tasmanian Devil Research Grant through funds raised by the Save the Tasmanian Devil Appeal (2013, 2015, 2017).
Author contributions
ABL, ALP, ASF, CEBO and GMW designed the study. ALP, ASF, CEBO, GSL, JC, and JMD developed the technology. CEBO and JMD performed the experiments. ALP and CEBO performed bioinformatic analyses. CEBO created the figures. ALP, ASF, and CEBO analyzed the data. CEBO wrote the manuscript, and all authors edited the manuscript.
Funding
This work was supported by the Australian Research Council (ARC) DECRA grant # DE180100484 and ARC Discovery grant # DP180100520, University of Tasmania Foundation Dr. Eric Guiler Tasmanian Devil Research Grant through funds raised by the Save the Tasmanian Devil Appeal (2013, 2015, 2017).
Availability of data and material
RNA-seq datasets are available in the European Nucleotide Archive repository BioProject # PRJEB39847, # PRJEB28680 and # PRJNA416378. Other datasets supporting this article have been uploaded as part of the electronic supplementary material.
Declarations
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Ethics approval
All samples were obtained with ethics approval as indicated in the Material and Methods section.
Consent for publication
All authors consent to the publication of the manuscript in Journal of Cancer Research and Clinical Oncology.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Abiko K, Matsumura N, Hamanishi J, et al. IFN-γ from lymphocytes induces PD-L1 expression and promotes progression of ovarian cancer. Br J Cancer. 2015;112:1501–1509. doi: 10.1038/bjc.2015.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S (2010) FastQC: A quality control tool for high throughput sequence data. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- Antoniou AN, Ford S, Pilley ES, et al. Interactions formed by individually expressed TAP1 and TAP2 polypeptide subunits. Immunology. 2002;106:182–189. doi: 10.1046/j.1365-2567.2002.01415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arce-Gomez B, Jones EA, Barnstable CJ, et al. The genetic control of HLA-A and B antigens in somatic cell hybrids: requirement for β2 microglobulin. Tissue Antigens. 1978;11:96–112. doi: 10.1111/j.1399-0039.1978.tb01233.x. [DOI] [PubMed] [Google Scholar]
- Azuma T, Yao S, Zhu G, et al. B7–H1 is a ubiquitous antiapoptotic receptor on cancer cells. Blood. 2008;111:3635–3643. doi: 10.1182/blood-2007-11-123141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belov K, Deakin JE, Papenfuss AT, et al. Reconstructing an ancestral mammalian immune supercomplex from a marsupial major histocompatibility complex. PLoS Biol. 2006;4:e46. doi: 10.1371/journal.pbio.0040046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benko S, Magalhaes JG, Philpott DJ, Girardin SE. NLRC5 limits the activation of inflammatory pathways. J Immunol. 2010;185:1681–1691. doi: 10.4049/jimmunol.0903900. [DOI] [PubMed] [Google Scholar]
- Biswas A, Meissner TB, Kawai T, Kobayashi KS. Cutting edge: impaired MHC class I expression in mice deficient for NLRC5/class I transactivator. J Immunol. 2012;189:516–520. doi: 10.4049/jimmunol.1200064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd LF, Kozlowski S, Margulies DH. Solution binding of an antigenic peptide to a major histocompatibility complex class I molecule and the role of β2-microglobulin. Proc Natl Acad Sci U S A. 1992;89:2242–2246. doi: 10.1073/pnas.89.6.2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown GK, Kreiss A, Lyons AB, Woods GM. Natural killer cell mediated cytotoxic responses in the Tasmanian devil. PLoS ONE. 2011;6:e24475. doi: 10.1371/journal.pone.0024475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullard JH, Purdom E, Hansen KD, Dudoit S. Evaluation of statistical methods for normalization and differential expression in mRNA-Seq experiments. BMC Bioinformatics. 2010;11:94. doi: 10.1186/1471-2105-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butte MJ, Keir ME, Phamduy TB, et al. Programmed death-1 ligand 1 interacts specifically with the B7–1 costimulatory molecule to inhibit T cell responses. Immunity. 2007;27:111–122. doi: 10.1016/j.immuni.2007.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell A, Coleby R, Tovar C, et al. The newly-arisen Devil facial tumour disease 2 (DFT2) reveals a mechanism for the emergence of a contagious cancer. Elife. 2018;7:e35314. doi: 10.7554/eLife.35314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campoli M, Ferrone S. HLA antigen changes in malignant cells: Epigenetic mechanisms and biologic significance. Oncogene. 2008;27:5869–5885. doi: 10.1038/onc.2008.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Belov K. Characterisation of non-classical MHC class I genes in the Tasmanian devil (Sarcophilus harrisii) Immunogenetics. 2014;66:727–735. doi: 10.1007/s00251-014-0804-3. [DOI] [PubMed] [Google Scholar]
- Cheng Y, Stuart A, Morris K, et al. Antigen-presenting genes and genomic copy number variations in the Tasmanian devil MHC. BMC Genom. 2012;13:87. doi: 10.1186/1471-2164-13-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J, Zhu L, Xia X, et al. NLRC5 negatively regulates the NF-kappaB and type I interferon signaling pathways. Cell. 2010;141:483–496. doi: 10.1016/j.cell.2010.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis BK, Roberts RA, Huang MT, et al. Cutting edge: NLRC5-dependent activation of the inflammasome. J Immunol. 2011;186:1333–1337. doi: 10.4049/jimmunol.1003111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deakin JE, Bender HS, Pearse A-M, et al. Genomic restructuring in the Tasmanian devil facial tumour: chromosome painting and gene mapping provide clues to evolution of a transmissible tumour. PLoS Genet. 2012;8:e1002483. doi: 10.1371/journal.pgen.1002483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H, Strome SE, Salomao DR, et al. Tumor-associated B7–H1 promotes T-cell apoptosis: A potential mechanism of immune evasion. Nat Med. 2002;8:793–800. doi: 10.1038/nm730. [DOI] [PubMed] [Google Scholar]
- Feder JN, Gnirke A, Thomas W, et al. A novel MHC class I-like gene is mutated in patients with hereditary haemochromatosis. Nat Genet. 1996;13:399–408. doi: 10.1038/ng0896-399. [DOI] [PubMed] [Google Scholar]
- Flies AS, Lyons AB, Corcoran LM, et al. PD-L1 is not constitutively expressed on Tasmanian devil facial tumor cells but is strongly upregulated in response to IFN-γ and can be expressed in the tumor microenvironment. Front Immunol. 2016;7:581. doi: 10.3389/fimmu.2016.00581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flies AS, Blackburn NB, Lyons AB, et al. Comparative analysis of immune checkpoint molecules and their potential role in the transmissible tasmanian devil facial tumor disease. Front Immunol. 2017;8:513. doi: 10.3389/fimmu.2017.00513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flies AS, Flies EJ, Fox S, et al. An oral bait vaccination approach for the Tasmanian devil facial tumor diseases. Expert Rev Vaccines. 2020;19:1–10. doi: 10.1080/14760584.2020.1711058. [DOI] [PubMed] [Google Scholar]
- Flies AS, Darby JM, Lennard PR, et al. A novel system to map protein interactions reveals evolutionarily conserved immune evasion pathways on transmissible cancers. Sci Adv. 2020 doi: 10.1126/sciadv.aba5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flies AS, Darby JM, Murphy PR, et al (2020b) Generation and testing of fluorescent adaptable simple theranostic (FAST) proteins. Bio-protocol 10:e3696. 10.21769/BioProtoc.3696 [DOI] [PMC free article] [PubMed]
- Folgiero V, Cifaldi L, Pira GL, et al. TIM-3/Gal-9 interaction induces IFNγ-dependent IDO1 expression in acute myeloid leukemia blast cells. J Hematol Oncol. 2015;8:1–5. doi: 10.1186/s13045-015-0134-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido F, Cabrera T, Concha A, et al. Natural history of HLA expression during tumour development. Immunol Today. 1993;14:491–499. doi: 10.1016/0167-5699(93)90264-L. [DOI] [PubMed] [Google Scholar]
- Gastaldello A, Sh R, Bailey A, et al (2020) Passage of transmissible cancers in the Tasmanian devil is due to a dominant, shared peptide motif and a limited repertoire of MHC-I allotypes. bioRxiv. 10.1101/2020.07.03.184416
- Ghebeh H, Tulbah A, Mohammed S, et al. Expression of B7–H1 in breast cancer patients is strongly associated with high proliferative Ki-67-expressing tumor cells. Int J Cancer. 2007;121:751–758. doi: 10.1002/ijc.22703. [DOI] [PubMed] [Google Scholar]
- Gobin SJP, van Zutphen M, Westerheide SD, et al. The MHC-specific enhanceosome and its role in MHC class I and β2-microglobulin gene transactivation. J Immunol. 2001;167:5175–5184. doi: 10.4049/jimmunol.167.9.5175. [DOI] [PubMed] [Google Scholar]
- Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- Hayden MS, Ghosh S. Signaling to NF-κB. Genes Dev. 2004;18:2195–2224. doi: 10.1101/gad.1228704. [DOI] [PubMed] [Google Scholar]
- He YF, Wang XH, Zhang GM, et al. Sustained low-level expression of interferon-γ promotes tumor development: Potential insights in tumor prevention and tumor immunotherapy. Cancer Immunol Immunother. 2005;54:891–897. doi: 10.1007/s00262-004-0654-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicklin DJ, Marincola FM, Ferrone S. HLA class I antigen downregulation in human cancers: T-cell immunotherapy revives an old story. Mol Med Today. 1999;5:178–186. doi: 10.1016/S1357-4310(99)01451-3. [DOI] [PubMed] [Google Scholar]
- Howson LJ, Morris KM, Kobayashi T, et al. Identification of dendritic cells, B cell and T cell subsets in Tasmanian devil lymphoid tissue; evidence for poor immune cell infiltration into devil facial tumors. Anat Rec (Hoboken) 2014;297:925–938. doi: 10.1002/ar.22904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwai Y, Ishida M, Tanaka Y, et al. Involvement of PD-L1 on tumor cells in the escape from host immune system and tumor immunotherapy by PD-L1 blockade. Proc Natl Acad Sci U S A. 2002;99:12293–12297. doi: 10.1073/pnas.192461099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones ME, Paetkau D, Geffen E, Moritz C. Genetic diversity and population structure of Tasmanian devils, the largest marsupial carnivore. Mol Ecol. 2004;13:2197–2209. doi: 10.1111/j.1365-294X.2004.02239.x. [DOI] [PubMed] [Google Scholar]
- Knowles RW, Bodmer WF. A monoclonal antibody recognizing a human thymus leukemia-like antigen associated with β2-microglobulin. Eur J Immunol. 1982;12:676–681. doi: 10.1002/eji.1830120810. [DOI] [PubMed] [Google Scholar]
- Kochan G, Escors D, Breckpot K, Guerrero-Setas D. Role of non-classical MHC class I molecules in cancer immunosuppression. Oncoimmunology. 2013;2:e26491. doi: 10.4161/onci.26491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowarz E, Löscher D, Marschalek R. Optimized Sleeping Beauty transposons rapidly generate stable transgenic cell lines. Biotechnol J. 2015;10:647–653. doi: 10.1002/biot.201400821. [DOI] [PubMed] [Google Scholar]
- Kozlowski S, Takeshita T, Boehncke WH, et al. Excess β2 microglobulin promoting functional peptide association with purified soluble class I MHC molecules. Nature. 1991;349:74–77. doi: 10.1038/349074a0. [DOI] [PubMed] [Google Scholar]
- Kuenzel S, Till A, Winkler M, et al. The nucleotide-binding oligomerization domain-like receptor NLRC5 is involved in IFN-dependent antiviral immune responses. J Immunol. 2010;184:1990–2000. doi: 10.4049/jimmunol.0900557. [DOI] [PubMed] [Google Scholar]
- Labun K, Montague TG, Krause M, et al. CHOPCHOP v3: Expanding the CRISPR web toolbox beyond genome editing. Nucleic Acids Res. 2019;47:W171–W174. doi: 10.1093/nar/gkz365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapinski PE, Miller GG, Tampé R, Raghavan M. Pairing of the nucleotide binding domains of the transporter associated with antigen processing. J Biol Chem. 2000;275:6831–6840. doi: 10.1074/jbc.275.10.6831. [DOI] [PubMed] [Google Scholar]
- Law CW, Chen Y, Shi W, Smyth GK. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15:R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazenby BT, Tobler MW, Brown WE, et al. Density trends and demographic signals uncover the long-term impact of transmissible cancer in Tasmanian devils. J Appl Ecol. 2018;55:1368–1379. doi: 10.1111/1365-2664.13088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S-J, Jang B-C, Lee S-W, et al. Interferon regulatory factor-1 is prerequisite to the constitutive expression and IFN-γ-induced upregulation of B7–H1 (CD274) FEBS Lett. 2006;580:755–762. doi: 10.1016/j.febslet.2005.12.093. [DOI] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013;41:e108–e108. doi: 10.1093/nar/gkt214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- Liu X, Wu Y, Yang Y, et al. Role of NLRC5 in progression and reversal of hepatic fibrosis. Toxicol Appl Pharmacol. 2016;294:43–53. doi: 10.1016/j.taap.2016.01.012. [DOI] [PubMed] [Google Scholar]
- Liu Y, ru, Yan X, Yu H xia,, et al. NLRC5 promotes cell proliferation via regulating the NF-κB signaling pathway in Rheumatoid arthritis. Mol Immunol. 2017;91:24–34. doi: 10.1016/j.molimm.2017.08.024. [DOI] [PubMed] [Google Scholar]
- Lo UG, Pong RC, Yang D, et al. IFNG-induced IFIT5 promotes epithelial-to-mesenchymal transition in prostate cancer via miRNA processing. Cancer Res. 2019;79:1098–1112. doi: 10.1158/0008-5472.CAN-18-2207. [DOI] [PubMed] [Google Scholar]
- Loh R, Hayes D, Mahjoor A, et al. The immunohistochemical characterization of devil facial tumor disease (DFTD) in the tasmanian devil (Sarcophilus harrisii) Vet Pathol. 2006;43:896–903. doi: 10.1354/vp.43-6-896. [DOI] [PubMed] [Google Scholar]
- Loke P, Allison JP. PD-L1 and PD-L2 are differentially regulated by Th1 and Th2 cells. Proc Natl Acad Sci USA. 2003;100:5336–5341. doi: 10.1073/pnas.0931259100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludigs K, Seguín-Estévez Q, Lemeille S, et al. NLRC5 exclusively transactivates MHC class I and related genes through a distinctive SXY module. PLoS Genet. 2015;11:e1005088. doi: 10.1371/journal.pgen.1005088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mátés L, Chuah MKL, Belay E, et al. Molecular evolution of a novel hyperactive Sleeping Beauty transposase enables robust stable gene transfer in vertebrates. Nat Genet. 2009;41:753–761. doi: 10.1038/ng.343. [DOI] [PubMed] [Google Scholar]
- Meissner TB, Li A, Biswas A, et al. NLR family member NLRC5 is a transcriptional regulator of MHC class I genes. Proc Natl Acad Sci U S A. 2010;107:13794–13799. doi: 10.1073/pnas.1008684107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner TB, Liu Y-J, Lee K-H, et al. NLRC5 cooperates with the RFX transcription factor complex to induce MHC class I gene expression. J Immunol. 2012;188:4951–4958. doi: 10.4049/jimmunol.1103160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchison EP, Tovar C, Hsu A, et al. The Tasmanian devil transcriptome reveals Schwann cell origins of a clonally transmissible cancer. Science. 2010;327:84–87. doi: 10.1126/science.1180616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchison EP, Schulz-Trieglaff OB, Ning Z, et al. Genome sequencing and analysis of the Tasmanian devil and its transmissible cancer. Cell. 2012;148:780–791. doi: 10.1016/j.cell.2011.11.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neerincx A, Lautz K, Menning M, et al. A role for the human nucleotide-binding domain, leucine-rich repeat-containing family member NLRC5 in antiviral responses. J Biol Chem. 2010;285:26223–26232. doi: 10.1074/jbc.M110.109736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neerincx A, Rodriguez GM, Steimle V, Kufer TA. NLRC5 controls basal MHC class I gene expression in an MHC enhanceosome-dependent manner. J Immunol. 2012;188:4940–4950. doi: 10.4049/jimmunol.1103136. [DOI] [PubMed] [Google Scholar]
- Nelson BH. CD20 + B Cells: the other tumor-infiltrating lymphocytes. J Immunol. 2010;185:4977–4982. doi: 10.4049/jimmunol.1001323. [DOI] [PubMed] [Google Scholar]
- Oliveros JC (2015) Venny. An interactive tool for comparing lists with Venn’s diagrams. https://bioinfogp.cnb.csic.es/tools/venny/index.html
- Ong CEB, Lyons AB, Woods GM, Flies AS. Inducible IFN-γ expression for MHC-I upregulation in devil facial tumor cells. Front Immunol. 2019;9:3117. doi: 10.3389/fimmu.2018.03117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patchett AL, Wilson R, Charlesworth JC, et al (2018) Transcriptome and proteome profiling reveals stress-induced expression signatures of imiquimod-treated Tasmanian devil facial tumor disease (DFTD) cells. Oncotarget 9:15895–15914. 10.18632/oncotarget.24634 [DOI] [PMC free article] [PubMed]
- Patchett AL, Coorens THH, Darby J, et al. Two of a kind: transmissible Schwann cell cancers in the endangered Tasmanian devil (Sarcophilus harrisii) Cell Mol Life Sci. 2020;77:1847–1858. doi: 10.1007/s00018-019-03259-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearse A-M, Swift K. Allograft theory: Transmission of devil facial-tumour disease. Nature. 2006;439:549. doi: 10.1038/439549a. [DOI] [PubMed] [Google Scholar]
- Pearse A-M, Swift K, Hodson P, et al. Evolution in a transmissible cancer: a study of the chromosomal changes in devil facial tumor (DFT) as it spreads through the wild Tasmanian devil population. Cancer Genet. 2012;205:101–112. doi: 10.1016/j.cancergen.2011.12.001. [DOI] [PubMed] [Google Scholar]
- Peng Y, yun, He Y hua, Chen C,, et al. NLRC5 regulates cell proliferation, migration and invasion in hepatocellular carcinoma by targeting the Wnt/β-catenin signaling pathway. Cancer Lett. 2016;376:10–21. doi: 10.1016/j.canlet.2016.03.006. [DOI] [PubMed] [Google Scholar]
- Phipson B, Lee S, Majewski IJ, et al. Robust hyperparameter estimation protects against hypervariable genes and improves power to detect differential expression. Ann Appl Stat. 2016;10:946–963. doi: 10.1214/16-AOAS920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pye R, Hamede R, Siddle HV, et al. Demonstration of immune responses against devil facial tumour disease in wild Tasmanian devils. Biol Lett. 2016;12:20160553. doi: 10.1098/RSBL.2016.0553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pye RJ, Pemberton D, Tovar C, et al. A second transmissible cancer in Tasmanian devils. Proc Natl Acad Sci U S A. 2016;113:374–379. doi: 10.1073/pnas.1519691113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pye R, Patchett A, McLennan E, et al. Immunization strategies producing a humoral igg immune response against devil facial tumor disease in the majority of tasmanian devils destined for wild release. Front Immunol. 2018;9:259. doi: 10.3389/fimmu.2018.00259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2020) R: A Language and Environment for Statistical Computing
- Ranjan P, Singh N, Kumar A, et al. NLRC5 interacts with RIG-I to induce a robust antiviral response against influenza virus infection. Eur J Immunol. 2015;45:758–772. doi: 10.1002/eji.201344412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risso D, Schwartz K, Sherlock G, Dudoit S. GC-Content Normalization for RNA-Seq Data. BMC Bioinform. 2011;12:480. doi: 10.1186/1471-2105-12-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins GR, Truax AD, Davis BK, et al. Regulation of class I major histocompatibility complex (MHC) by nucleotide-binding domain, leucine-rich repeat-containing (NLR) proteins. J Biol Chem. 2012;287:24294–24303. doi: 10.1074/jbc.M112.364604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2009;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez GM, Bobbala D, Serrano D, et al. NLRC5 elicits antitumor immunity by enhancing processing and presentation of tumor antigens to CD8+ T lymphocytes. Oncoimmunology. 2016;5:e1151593. doi: 10.1080/2162402X.2016.1151593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RStudio Team (2020) RStudio: Integrated Development Environment for R
- Saijo K, Mecklenbräuker I, Santana A, et al. Protein kinase C β controls nuclear factor κB activation in B cells through selective regulation of the IκB kinase α. J Exp Med. 2002;195:1647–1652. doi: 10.1084/jem.20020408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz R, Fitch ZW, Schroder PM, et al. B cells in transplant tolerance and rejection: friends or foes? Transpl Int. 2020;33:30–40. doi: 10.1111/tri.13549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selenko-Gebauer N, Majdic O, Szekeres A, et al. B7–H1 (Programmed death-1 ligand) on dendritic cells is involved in the induction and maintenance of T cell anergy. J Immunol. 2003;170:3637–3644. doi: 10.4049/jimmunol.170.7.3637. [DOI] [PubMed] [Google Scholar]
- Siddle HV, Deakin JE, Coggill P, et al. MHC-linked and un-linked class I genes in the wallaby. BMC Genom. 2009;10:310. doi: 10.1186/1471-2164-10-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddle HV, Kreiss A, Tovar C, et al. Reversible epigenetic down-regulation of MHC molecules by devil facial tumour disease illustrates immune escape by a contagious cancer. Proc Natl Acad Sci U S A. 2013;110:5103–5108. doi: 10.1073/pnas.1219920110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simister NE, Mostov KE. An Fc receptor structurally related to MHC class I antigens. Nature. 1989;337:184–187. doi: 10.1038/337184a0. [DOI] [PubMed] [Google Scholar]
- Song M, Ping Y, Zhang K, et al. Low-dose IFNγ induces tumor cell stemness in tumor microenvironment of non–small cell lung cancer. Cancer Res. 2019;79:3737–3748. doi: 10.1158/0008-5472.CAN-19-0596. [DOI] [PubMed] [Google Scholar]
- Staehli F, Ludigs K, Heinz LX, et al. NLRC5 deficiency selectively impairs MHC class I- dependent lymphocyte killing by cytotoxic T cells. J Immunol. 2012;188:3820–3828. doi: 10.4049/jimmunol.1102671. [DOI] [PubMed] [Google Scholar]
- Stammnitz MR, Coorens THH, Gori KC, et al. The origins and vulnerabilities of two transmissible cancers in tasmanian devils. Cancer Cell. 2018;33:607–619.e15. doi: 10.1016/J.CCELL.2018.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Synthego (2019) Synthego performance analysis, ICE Analysis
- Tong Y, Cui J, Li Q, et al. Enhanced TLR-induced NF-κB signaling and type I interferon responses in NLRC5 deficient mice. Cell Res. 2012;22:822–835. doi: 10.1038/cr.2012.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tovar C, Obendorf D, Murchison EP, et al. Tumor-specific diagnostic marker for transmissible facial tumors of Tasmanian devils: immunohistochemistry studies. Vet Pathol. 2011;48:1195–1203. doi: 10.1177/0300985811400447. [DOI] [PubMed] [Google Scholar]
- Tovar C, Pye RJ, Kreiss A, et al. Regression of devil facial tumour disease following immunotherapy in immunised Tasmanian devils. Sci Rep. 2017;7:43827. doi: 10.1038/srep43827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Den Elsen PJ, Peijnenburg A, Van Eggermond MCJA, Gobin SJP. Shared regulatory elements in the promoters of MHC class I and class II genes. Immunol Today. 1998;19:308–312. doi: 10.1016/S0167-5699(98)01287-0. [DOI] [PubMed] [Google Scholar]
- Van Den Elsen PJ, Gobin SJP, Van Eggermond MCJA, Peijnenburg A. Regulation of MHC class I and II gene transcription: differences and similarities. Immunogenetics. 1998;48:208–221. doi: 10.1007/s002510050425. [DOI] [PubMed] [Google Scholar]
- Verzella D, Pescatore A, Capece D, et al. Life, death, and autophagy in cancer: NF-κB turns up everywhere. Cell Death Dis. 2020;11:1–14. doi: 10.1038/s41419-020-2399-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitiello A, Potter TA, Sherman LA. The role of beta 2-microglobulin in peptide binding by class I molecules. Science. 1990;80(250):1423–1426. doi: 10.1126/science.2124002. [DOI] [PubMed] [Google Scholar]
- Wang J, Quan L, Ru Y, Xia Q, et al. Emerging roles for NLRC5 in immune diseases. Front. Pharmacol. 2019;10:1352. doi: 10.3389/fphar.2019.01352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams DB, Barber BH, Flavell RA, Allen H. Role of beta 2-microglobulin in the intracellular transport and surface expression of murine class I histocompatibility molecules.[erratum appears in J Immunol 1989 Jul 15;143(2):761] J Immunol. 1989;142:2796–2806. [PubMed] [Google Scholar]
- Xia L, Tan S, Zhou Y, et al. Role of the NFκB-signaling pathway in cancer. Onco Targets Ther. 2018;11:2063–2073. doi: 10.2147/OTT.S161109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi H, Hashimoto K. Association of MR1 protein, an MHC class I-related molecule, with β2-microglobulin. Biochem Biophys Res Commun. 2002;290:722–729. doi: 10.1006/bbrc.2001.6277. [DOI] [PubMed] [Google Scholar]
- Yao Y, Wang Y, Chen F, et al. NLRC5 regulates MHC class I antigen presentation in host defense against intracellular pathogens. Cell Res. 2012;22:836–847. doi: 10.1038/cr.2012.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihama S, Roszik J, Downs I, et al. NLRC5/MHC class I transactivator is a target for immune evasion in cancer. Proc Natl Acad Sci U S A. 2016;113:5999–6004. doi: 10.1073/pnas.1602069113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, He QY. ReactomePA: An R/Bioconductor package for reactome pathway analysis and visualization. Mol Biosyst. 2016;12:477–479. doi: 10.1039/c5mb00663e. [DOI] [PubMed] [Google Scholar]
- Yu G, Wang LG, Han Y, He QY. ClusterProfiler: An R package for comparing biological themes among gene clusters. Omi A J Integr Biol. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen GJ, Demissie E, Pillai S. B lymphocytes and cancer: A love-hate relationship. Trends Cancer. 2016;2:747–757. doi: 10.1016/j.trecan.2016.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq datasets are available in the European Nucleotide Archive repository BioProject # PRJEB39847, # PRJEB28680 and # PRJNA416378. Other datasets supporting this article have been uploaded as part of the electronic supplementary material.