Significance Statement
The traditional biomarkers currently used to monitor a kidney allograft for rejection are late markers of injury and they lack sensitivity and specificity. Allograft biopsies on the other hand, are invasive and costly. The authors describe the discovery and validation of two urinary exosomal mRNA multigene signatures for the diagnosis of acute T cell–mediated and antibody-mediated rejection and chronic, active antibody-mediated rejection in recipients of kidney transplant. Using a clinically validated platform for exosome isolation and analysis, they demonstrated the high stability of urinary exosomes and the reliability of this approach in monitoring patients for allograft rejection. One gene signature for all-cause rejection and another for discriminating T cell–mediated rejection from antibody-mediated rejection showed high predictive performances and offer clinicians the possibility of new tools for monitoring emergence of rejection in kidney allografts.
Keywords: kidney transplantation, urine exosome, mRNA, biomarker, rejection
Abstract
Background
Developing a noninvasive clinical test to accurately diagnose kidney allograft rejection is critical to improve allograft outcomes. Urinary exosomes, tiny vesicles released into the urine that carry parent cells’ proteins and nucleic acids, reflect the biologic function of the parent cells within the kidney, including immune cells. Their stability in urine makes them a potentially powerful tool for liquid biopsy and a noninvasive diagnostic biomarker for kidney-transplant rejection.
Methods
Using 192 of 220 urine samples with matched biopsy samples from 175 patients who underwent a clinically indicated kidney-transplant biopsy, we isolated urinary exosomal mRNAs and developed rejection signatures on the basis of differential gene expression. We used crossvalidation to assess the performance of the signatures on multiple data subsets.
Results
An exosomal mRNA signature discriminated between biopsy samples from patients with all-cause rejection and those with no rejection, yielding an area under the curve (AUC) of 0.93 (95% CI, 0.87 to 0.98), which is significantly better than the current standard of care (increase in eGFR AUC of 0.57; 95% CI, 0.49 to 0.65). The exosome-based signature’s negative predictive value was 93.3% and its positive predictive value was 86.2%. Using the same approach, we identified an additional gene signature that discriminated patients with T cell–mediated rejection from those with antibody-mediated rejection (with an AUC of 0.87; 95% CI, 0.76 to 0.97). This signature’s negative predictive value was 90.6% and its positive predictive value was 77.8%.
Conclusions
Our findings show that mRNA signatures derived from urinary exosomes represent a powerful and noninvasive tool to screen for kidney allograft rejection. This finding has the potential to assist clinicians in therapeutic decision making.
CKD is a major health concern in the Unites States and worldwide.1 Although patients with ESKD require either dialysis or transplantation to sustain their life, the latter remains the treatment of choice.2–4 However, long-term graft survival remains a major challenge, due mostly to acute and chronic rejection. Although the rate of acute rejection has decreased in the modern era of potent immunosuppression,5 recent reported incidence of acute rejections in literature ranges from 11% to 26%.6–9 During the first year after transplantation, the incidence of acute rejection is around 7.9%.10 This has been associated with poor long-term allograft survival.11 The implementation of the Banff classification in 1991 provided a valuable tool for histopathologic diagnosis of kidney-transplant injury and allowed for standardization when comparing biopsy sample results between different studies.12 Serum creatinine, eGFR and its increase (expressed as Δ eGFR), and urinary protein excretion are traditional biomarkers currently used to monitor the kidney allograft, but they lack desired sensitivity, specificity, and predictive ability.13 Kidney allograft biopsy specimens, with histopathologic evaluation, remain the gold standard in diagnosing acute rejection. However, there are limitations to their use because biopsies are invasive, costly, and can be associated with significant morbidity.14,15 Several biomarkers have been identified as potential noninvasive tools to diagnose early graft rejection, including CD3ɛ mRNA, IP-10 (CXCL10) mRNA, and 18S ribosomal RNA isolated from urine pellet, described in the CTOT04 study.8 Urinary CXCL9 mRNA was highly expressed in patients with acute rejection compared with patients without rejection in multicenter study which included 280 recipients of kidney transplants.16 Recently, donor-derived cellfree DNA (dd-cfDNA) has been introduced to the clinical practice as a novel biomarker for graft rejection after solid organ transplantation. Despite results showing good performances in discriminating active rejection from no-rejection status, dd-cfDNA using the currently defined threshold of 1% did not discriminate well between no rejection and lower grades of cellular rejection, such as acute cellular rejection (ACR) 1A.7
Exosomes are nanometer-sized vesicles (between 50 and 200 nm) released by cells to mediate cell-to-cell communication by delivering proteins and nucleic acids, such as mRNAs and microRNAs.17 Exosomes are released into the urine during fusion of the multivesicular body with the apical plasma cell membrane, or by direct budding of the plasma membrane.18 They carry the parent cells’ surface proteins and nucleic acids and, thus, reflect the biologic function of the parent cell. In the transplanted kidney, exosomes originate from glomerular podocytes, renal tubular cells, and from the uroepithelium.19 Exosomes shed into the urine and, therefore, represent an easily accessible, noninvasive window into ongoing pathologic processes within the kidney. We, and others, have recently shown that urinary exosomes are enriched with proteins derived from immune cells within the kidney transplant during rejection.20–22 Thus, urinary exosomes can provide investigators with a unique, concentrated sampling of membrane and cytosolic proteins during allograft rejection, and can further provide information regarding RNA derived from cells residing within the kidneys, including infiltrating lymphocytes.
Although there are no data on the association between urinary exosome–derived mRNA signatures and kidney allograft rejection, mRNA signatures from urinary cell pellets have been associated with active rejection.23,24 Compared with urinary mRNA isolated from whole urinary cells, urinary exosomal mRNA has shown a greater stability due to the encapsulation within membrane-bound vesicles.25 With recent techniques incorporating nanofiltration, affinity, microfluidics, and tangential flow fractionation, along with many others, it is possible to isolate exosome-enriched fractions.26 The RNA transcriptome can be efficiently profiled in urine exosomes, and this exosomal RNA has been shown to be a valuable source for biomarker discovery and integration of these gene signatures into clinical applications.27–29 Urinary exosome RNA diagnostic assays are being used today, and are even included in the National Comprehensive Cancer Network guidelines for early detection of prostate cancer.29–32
Our hypothesis was that the use of the urinary exosome mRNA gene signature could represent a rapid, noninvasive assay to diagnose acute rejection in kidney allografts. We included 192 urine samples collected from patients who received a renal transplant and were undergoing clinically indicated biopsies at three centers across the Unites States. We measured mRNA directly from urinary exosomes to identify a specific exosome RNA signature for kidney rejection. Whereas previous data identified a urinary cell mRNA signature,8,24 we report for the first time the development of a urinary exosome mRNA signature in recipients of kidney transplants undergoing T cell–mediated rejection (TCMR) and/or antibody-mediated rejection (ABMR).
Methods
Patient and Sample Information
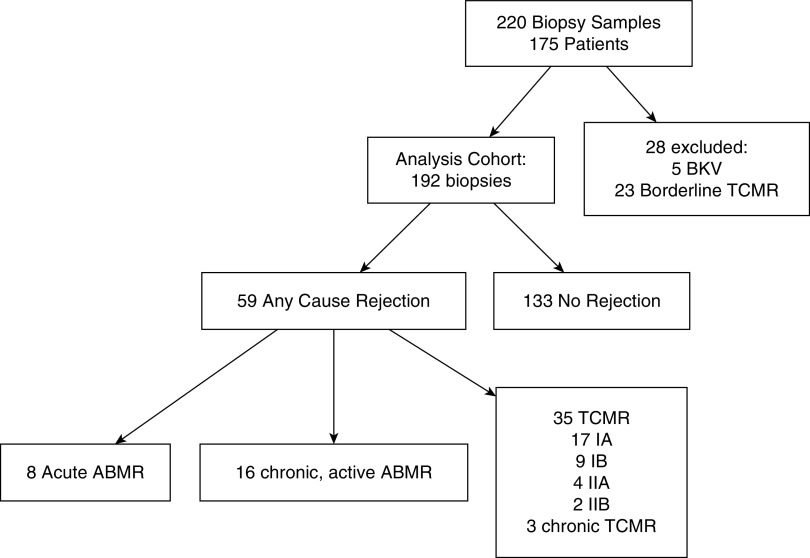
The study was approved by the institutional review board at each site, and the patients provided written informed consent in accord with the Declaration of Helsinki. We enrolled 175 recipients of kidney transplants at the time of a clinically indicated renal biopsy from three renal centers. A total of 220 urine samples were collected from patients with matched biopsy specimens for urinary exosomal mRNA profiling. Among the 175 patients, 44 had repeat biopsies, with 30 patients having two biopsies and seven patients having more than two biopsies. Demographic and clinical characteristics, and information on the donors, were collected from the medical chart. eGFR was calculated using the Modification of Diet in Renal Disease equation.33 We used the on-site pathologist’s renal transplant biopsy specimen report to define active rejection, in accordance with the Banff Working Groups’ criteria.34 We excluded from our primary analysis 23 samples that were diagnosed as borderline cell-mediated rejection, and five samples that were diagnosed with BK virus nephropathy. For our analyses, we integrated TCMR and acute and chronic active ABMR to form the rejection group, and we distinguished them from samples that were classified as having no rejection on the basis of biopsy specimen reports. Biopsy specimen reports with a diagnosis of mixed ABMR and TCMR were grouped with the TCMR subgroup, and those with mixed borderline TCMR and ABMR were grouped with the ABMR subgroup.
Detection of Donor-Specific Antibodies
The presence of anti-HLA antibodies was assessed by LABScreen Mixed (One Lambda Inc., Canoga Park, CA), analyzed on a Luminex platform. In the event of a positive assay, this was followed by LABScreen Single Antigen Class I/Class II (One Lambda Inc.). A normalized mean fluorescence intensity ≥3000 for class I or ≥1000 for class II is considered positive at our center.
Urinary Exosome Isolation, mRNA Extraction, and Gene-Expression Analysis
The second voided urine sample was collected on the morning of the biopsy, and whole urine samples were stored at −80°C. Three in-house controls were used, consisting of one-pooled male sample, one-pooled female sample, and one-pooled male and female sample. Samples were thawed and up to 20 ml urine was centrifuged to pellet cells and cellular debris at 2000 × g for 20 minutes before the extraction. Exosomes were isolated using a urine-exosome isolation kit, as described previously (Figure 1).29–31 RNA was eluted in 16 µl nuclease-free water, 14 µl of which was used in a 20 µl reverse-transcription reaction using the VILO cDNA synthesis kit (Thermo Fisher).
Figure 1.
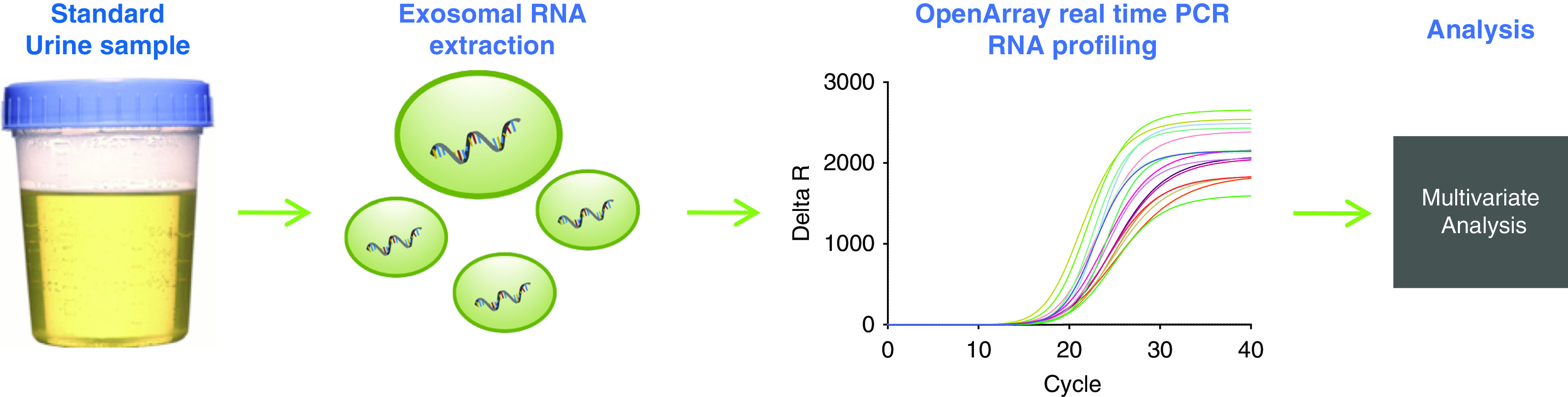
Simple collection protocol enables ease of use. Urine samples are collected in a standard urine cup at any time during the day. The exosomes are isolated from the urine sample, followed by an RNA extraction, and RT-qPCR analysis of the target genes. The relative quantities of each target gene are inputted into an algorithm to generate a single score from zero to one.
The first round of samples was analyzed using the TaqMan OpenArray Human Inflammation Panel (Thermo Fisher). This panel consists of 586 TaqMan assays for genes that have been studied as targets for a range of inflammatory diseases, and it includes 21 endogenous control assays. To prepare the samples for quantitative PCR (qPCR), 10 µl cDNA was split into two equal portions and preamplified with two pools of mixed primers, following the manufacturer’s directions. The preamplification reactions were mixed and diluted before mixing with TaqMan OpenArray Real-Time PCR Master Mix. Reaction mixes were loaded onto the OpenArray plates and the plates were run on the QuantStudio 12K Flex Real-Time PCR system (Thermo Fisher) using the preset protocol for this panel.
On the basis of the initial analysis, a subset of assays was identified and plated onto a custom TaqMan OpenArray panel. This panel consisted of 112 TaqMan assays. For this panel, 5 µl cDNA was preamplified with a pool of the 112 assays, using the manufacturer’s directions. The preamplification reactions were diluted before mixing samples with the TaqMan OpenArray Real-Time PCR Master Mix. Reaction mixes were loaded onto the OpenArray plates and the plates were run on the QuantStudio 12K Flex Real-Time PCR system (Thermo Fisher), using the preset protocol for this panel. Analysis of samples described here used the 112 TaqMan assays common to all samples.
Statistical Analyses
Genes with data missing from >20% of the samples were excluded from the analysis. Missing data were imputed using a nonparametric missing-value imputation.35 The Ct values from the OpenArray were normalized to PGK1. The Boruta36 algorithm was used to select genes that were most relevant for prediction.. A support vector machine (SVM) with a radial kernel was fit to the relevant features using a stratified repeated K-fold crossvalidation (K=10, repeats=10) to generate the rejection probabilities using the caret package.37 This approach gives a better indication of how well the model will perform on unseen data compared with just one train-test split in a hold-out method, which makes it highly dependent on how the data are split in test and train datasets. The pROC package was used to generate the receiver-operating-characteristic curves.38 Associations between clinical and demographic factors were computed using the t test for continuous variables and the Pearson chi-squared test for categoric variables. Area under the curve (AUC) comparison was performed using the DeLong test. Data reporting and analyses were conducted using R version 3.3. Two-tailed P values ≤0.05 were considered statistically significant. Sample size was calculated for a negative predictive value (NPV) and specificity of 90%, with a 10% width for the 95% CI at a prevalence of 20%.39 On the basis of this calculation, the required sample size was estimated to be 173 samples.
Results
Patients’ Characteristics and Biopsy Specimens
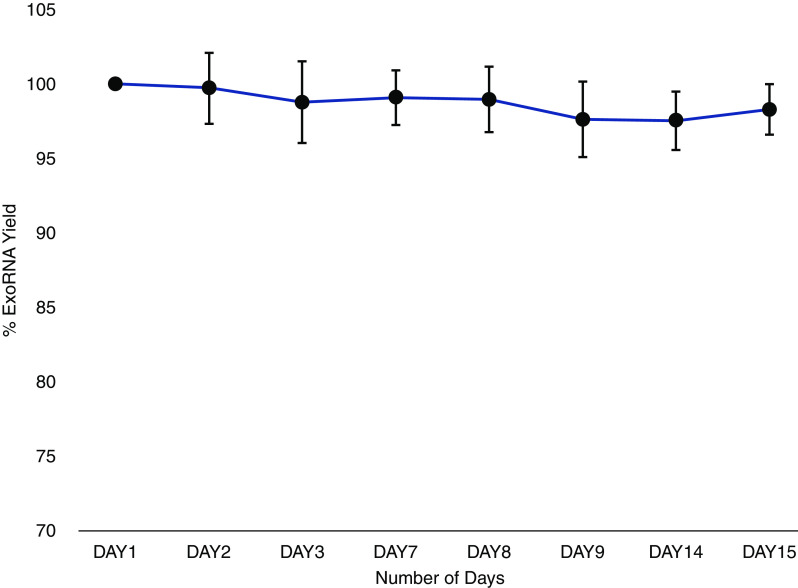
A total of 192 urine samples that have matched biopsy specimens were included. Exosomal mRNA showed excellent stability in urine stored at 4°C for 2 weeks. Figure 2 shows the expression of three targets from eight samples. The stability of mRNA is critical for developing clinically useful diagnostic tests because the samples can be safely cold-pack shipped from the patient’s residence to a central laboratory for analysis, where they can be either processed immediately or stored at +4°C for up to 2 weeks. Our study included matched urine samples for biopsy specimens showing TCMR (grades IA, IB, IIA, IIB) and acute active and chronic active ABMR subgroups of rejection, on the basis of the Banff classification, and we used the term “active rejection” to distinguish these samples from other biopsy specimens without rejection. There were 59 biopsy specimens with rejection, and 133 biopsy specimens without rejection (30.7% prevalence). Figure 3 shows the results for the 192 biopsy specimens that had matched urine samples. Table 1 shows the baseline characteristics of the study cohorts. The mean (interquartile range) age of patients with any-cause rejection was 51.0 (38.0–64.5) years, and 51.6 (40.8–65) years in patients without rejection. Median (interquartile range) eGFR levels were 32.85 (22.13–44.56) ml/min per 1.73 m2 in patients with any-cause rejection, and 37.89 (25.95–50.89) ml/min per 1.73 m2 in patients with no rejection. The any-cause rejection group included a higher proportion of patients with previous rejection episodes (P<0.001) and longer time since biopsy when compared with the group without rejection (P=0.02). The difference in the proportion of Black patients between the groups was NS (P=0.47). Among the any-cause rejection group, 59.3% of cases of rejection were due to active TCMR, and 40.7% were attributed to ABMR.
Figure 2.
Urinary exosomal RNA is stable over 2 weeks at 4°C. The urine samples were collected and stored at 4°C for up to 2 weeks. Exosomes were extracted at different time points, followed by RT-qPCR to analyze the yield and integrity of the RNA. The urinary exosome RNA (exoRNA) was stable over 2 weeks (average yield from three separate genes). The error bars represent the SD of the percentage of exosomal RNA yield across three different genes.
Figure 3.
CONSORT flow diagram and histologic diagnosis of enrolled patients. Of the 220 initially collected samples, 192 were further analyzed in this cohort.
Table 1.
Baseline characteristics of patients
| Characteristic | Clinical Cohort (n=192) | P Value | |
|---|---|---|---|
| No Rejection (n=133) | Any-Cause Rejection (n=59) | ||
| Age, yr | 51.6±15.1 | 51.0±16.2 | 0.80a |
| Female, % | 32.3 | 45.8 | |
| Race, % | |||
| White | 83.6 | 88.0 | 0.47b |
| Black | 16.4 | 22.0 | 0.47b |
| SCr at biopsy, mg/dl | 1.8 (1.5–2.6) | 2.2 (1.7–2.8) | 0.39a |
| eGFR ml/min per 1.73 m2 | 37.9 (25.6–50.9) | 32.9 (22.1–44.6) | 0.02a |
| Previous rejection, % | 15.2 | 42.4 | 8.23×10−07b |
| Deceased donor, % | 43.0 | 51.9 | 0.65b |
| Time to biopsy, d | 215 (46–1751) | 1250 (295–3063) | 0.02a |
| Thymoglobulin, % | 60.5 | 69.4 | 0.36b |
| UPCR | 0 (0–0.28) | 0 (0–1.51) | 0.27a |
| DSA, % | 18.5 | 44.2 | 0.002a |
| PRA, % | 2.0 (2.0–41.25) | 25.0 (2.0–61.25) | 0.02a |
Data presented as frequencies, mean±SD, and median (interquartile range). All demographic and clinical data are based on the day of biopsy, timed with the urine collection. SCr, serum creatinine; UPCR, urinary protein-creatinine ratio; PRA, panel-reactive antibody.
P values from t test.
P values from chi-squared test.
Identifying Any-Cause Rejection Signature from Urinary Exosomes
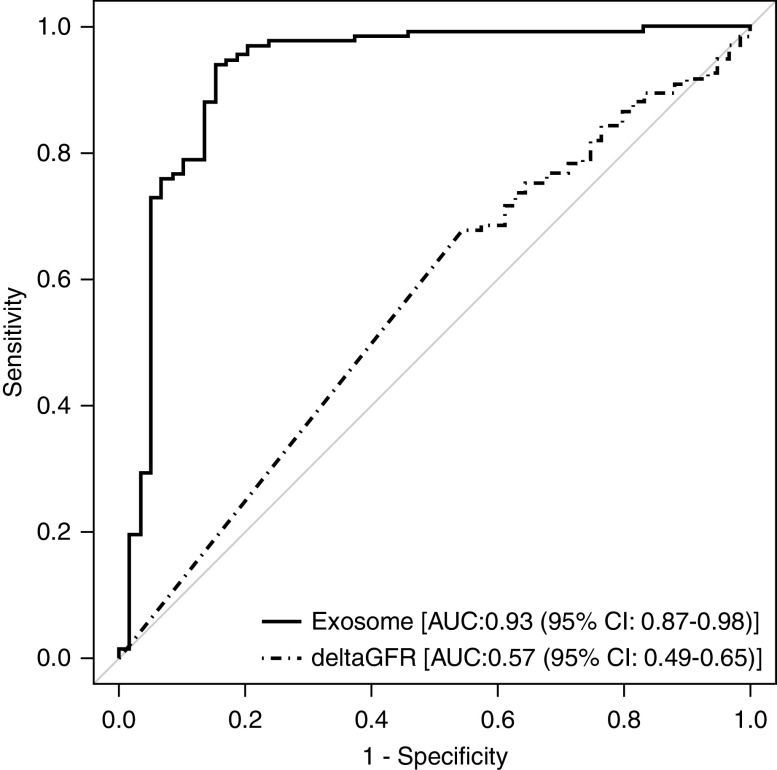
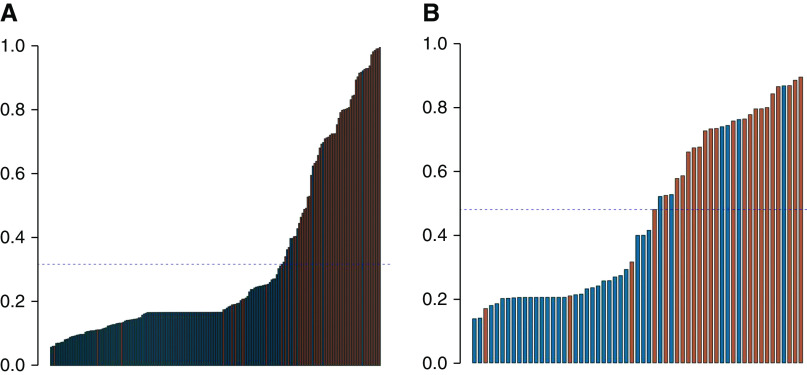
We compared mRNA from urinary exosomes collected from patients with biopsy sample–proven, any-cause rejection with urine samples from patients without rejection. To identify relevant genes in urinary exosomes that could predict any-cause rejection, we first analyzed the samples using the TaqMan OpenArray Human Inflammation Panel. This panel consists of 586 TaqMan assays for genes that have been studied as targets for a range of inflammatory diseases, and it includes 21 endogenous control assays. For subsequent analyses, a subset of 112 TaqMan assays was identified and plated onto a custom TaqMan OpenArray panel. Given the large number of investigated genes, we performed feature selection using Boruta to identify the relevant features. A repeated, stratified, K-fold classification model (K=10, repeats=10) with an SVM using a radial-basis-function kernel was used for classification. The stratification ensures there is a similar percentage of samples with rejection in each of the folds. This process is repeated ten times, with a different randomization in each repeat, to generate the final classification model. We used crossvalidation instead of a hold-out method because crossvalidation improves the generalizability of the gene signature by validating the performance on multiple train-test subsets of the data, and this results in a much more stable estimate of the performance. This allowed us to identify a multigene signature (CXCL11, CD74, IL32, STAT1, CXCL14, SERPINA1, B2M, C3, PYCARD, BMP7, TBP, NAMPT, IFNGR1, IRAK2, and IL18BP) that discriminated biopsy samples with any-cause rejection from those with no rejection. The AUC was 0.93 (95% CI, 0.87 to 0.98) (Figure 4). To compare the performance of this signature against current clinical practice, we also generated an AUC for the change (Δ) in eGFR (Δ eGFR) (Figure 4). The AUC for Δ eGFR for this set of patients was 0.57 (95% CI, 0.49 to 0.65), which was significantly inferior (P<0.001) to the performance of the multigene signature. We also derived a cut point to rule out any-cause rejection by optimizing Youden J (Figure 5A). This resulted in an NPV of 93.3% (95% CI, 87.7% to 96.4%) and a sensitivity of 84.7% (95% CI, 73.5% to 91.8%). The positive predictive value (PPV) for discriminating active rejection was 86.2% (95% CI, 75.1% to 92.8%) (Table 2).
Figure 4.
Exosome RNA signature significantly outperforms Δ eGFR in discriminating any-cause rejection. The receiver-operating-characteristic analysis and AUC is shown for the exosome RNA signature and compared with Δ eGFR. The fraction of true positive results (sensitivity) and the fraction of false positive results (1–specificity) for diagnosis of any-cause acute rejection are displayed on the y and x axis, respectively. The AUC for the RNA signature is 0.93 (95% CI, 0.87 to 0.98) and the AUC for Δ eGFR is 0.57 (95% CI, 0.49 to 0.65).
Figure 5.
Waterfall plot of the urinary exosome gene scores demonstrates the high NPV of the exosomal RNA signature. The blue dotted line represents the score cutoff for each of the gene signatures. (A) Discriminating any-cause rejection from no rejection (red bars denote samples that have clinical rejection). (B) Discriminating ABMR from TCMR (red bars denote samples with ABMR).
Table 2.
Gene signature discrimination performance characteristics for overall rejection
| Metric | Performance (95% CI), % |
|---|---|
| NPV | 93.3 (87.7 to 96.4) |
| Sensitivity | 84.7 (73.5 to 91.8) |
| Specificity | 94.0 (88.6 to 96.9) |
| PPV | 86.2 (75.1 to 92.8) |
We also analyzed the data on donor specific antibodies (DSAs) to assess whether the presence of DSAs against HLA class I or II was associated with an increased risk of ABMR and of any-cause rejection. As shown in Supplemental Figure 1, the presence of DSAs has an AUC (95% CI) of 0.72 (0.61–0.83) for ABMR and 0.64 (0.56–0.72) for any-cause rejection, which are significantly worse than our any-case rejection signature (P=0.03 and P=2.2 x 10-9 respectively). Combining Δ eGFR with DSA and urinary protein-creatinine ratio did not further improve the AUC (Supplemental Figure 1).
Discriminating TCMR from ABMR
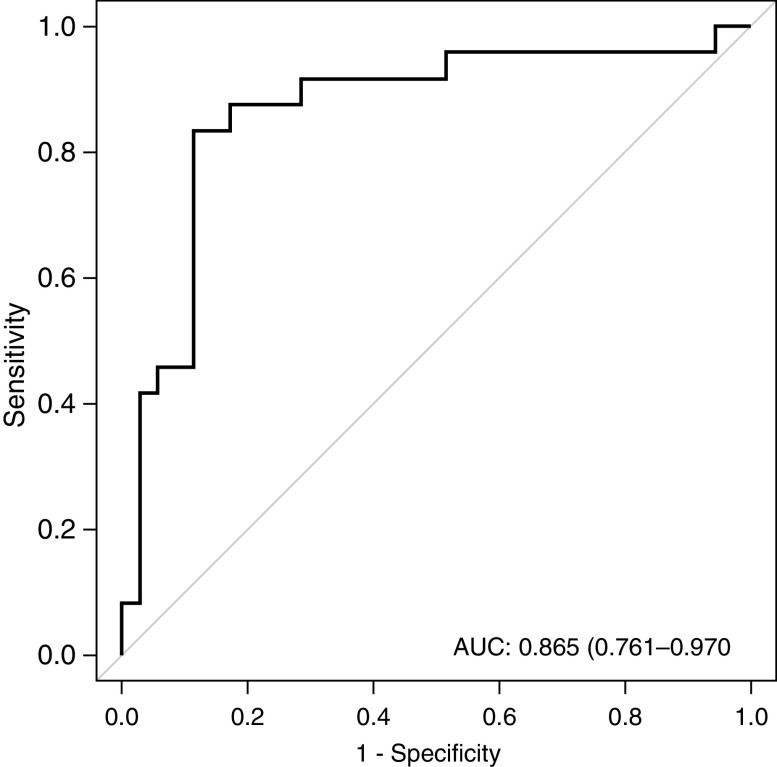
We also compared the TCMR samples with the ABMR samples to derive an additional signature to discriminate between these two forms of rejection. Applying the same optimization and classification approach used for any-cause rejection, we identified a multigene signature (CD74, C3, CXCL11, CD44, and IFNAR2) that could distinguish TCMR from ABMR. The AUC for this signature was 0.87 (95% CI, 0.76 to 0.97) (Figure 6). A cut point was derived to maximize the NPV and sensitivity to rule out ABMR (Figure 5B). Samples with a positive signature for all-cause rejection were analyzed for the second signature. If the second signature is negative, the patient has TCMR and ABMR is ruled out with an NPV of 90.6% (95% CI, 75.8% to 96.8%) and a PPV of 77.8% (95% CI, 59.2% to 89.4%). The sensitivity to discriminate TCMR from ABMR was 87.5% (95% CI, 69.0% to 95.7%) and the specificity was 82.9% (95% CI, 67.3% to 92.0%) (Table 3).
Figure 6.
Exosome RNA signature can accurately discriminate TCMR from ABMR. Receiver-operating-characteristic curve showing the fraction of true positive results (sensitivity) and the fraction of false positive results (1–specificity) for discriminating TCMR from ABMR (AUC, 0.87; 95% CI, 0.76 to 0.97).
Table 3.
Gene signature discrimination performance characteristics discriminating TCMR from ABMR
| Metric | Performance (95% CI), % |
|---|---|
| NPV | 90.6 (75.8 to 96.8) |
| Sensitivity | 87.5 (69.0 to 95.7) |
| Specificity | 82.9 (67.3 to 92.0) |
| PPV | 77.8 (59.2 to 89.4) |
Borderline-Rejection Samples
We then applied our all-cause rejection signature to the urine samples in our cohort from patients diagnosed with borderline rejection. Follow-up information was available for 18 out of the 23 samples. One patient was excluded from the analysis for having recurrent glomerulopathy in addition to the histologic diagnosis of borderline rejection. Among the eight samples from patients with borderline rejection who showed a predicted negative signature of all-cause rejection, only two patients showed a decrease in eGFR of >30%, 12 months after biopsy. One of those two patients developed TCMR 1B and BK-virus nephritis less than a year later, and the other patient developed TCMR 1A on two subsequent biopsies, 1 and 3 months later. The other six patients remained stable with no change in eGFR (<5% change) or development of proteinuria, and they did not need any further intervention. Among the nine samples from patients with borderline rejection who showed a predicted positive signature, only three patients remained with stable eGFR (<5% change) and no proteinuria at 12 months postbiopsy. The other six patients had either a decrease in eGFR of >30% (two patients) and/or persistent proteinuria (five patients, urinary protein-creatinine ratio >0.9). It is important to note that borderline-rejection samples were not used in the development of the all-cause rejection signature, and the signature is only meant to diagnose ongoing rejection and not act as a predictor of rejection.
Discussion
In this study, we report a noninvasive test to detect active kidney-transplant rejection from urine samples of patients undergoing a for-cause biopsy, with diverse pathologic diagnosis including both ABMR and cellular-mediated rejection. We decided to exclude the borderline cellular-rejection samples because many studies have shown that borderline cellular rejection represents a heterogeneous group that ranges from insignificant inflammation to a clinically significant cellular rejection resulting in histologic consequences.40 Furthermore, the immunologic and clinical significance of borderline rejection is still a subject of debate in the transplant community.41 We used a clinically validated exosome isolation platform, which has been used for the ExoDx Prostate (Intelliscore) test and has been performed on >50,000 patients to date.29–31 This platform also enabled isolation and analysis of mRNA signatures in urinary exosomes to predict any-cause active rejection in patients with kidney transplant at the time of allograft biopsy. Compared with exosomal proteins, RNA is well protected inside the vesicle and can be stably assessed from urine samples even after prolonged storage, whereas urine samples for exosomal protein study can only be safely stored for about 12 hours,20 because proteins that are associated with the outer membrane are exposed to protease activity. Also, it is sometimes more challenging to robustly assess and discriminate exosome proteins from free proteins (or those sticking nonspecifically to the outside of the vesicle). RNA detection methods (such as RT-qPCR used here) is highly sensitive down to single-copy levels, and RT-PCR is a well-known method for clinical laboratories and is exceptionally well suited for diagnostic assays. We analyzed 112 target genes by TaqMan qPCR across all samples, using the OpenArray platform, and identified an exosomal mRNA signature that significantly correlates with active rejection. The mRNA signature was developed using an SVM classifier with a radial-basis-function kernel. An SVM classifier was chosen due to underlying nonlinearity of the data and its ability to better handle outliers. Stratification was chosen to ensure that all of the different folds had the same proportion of rejected samples to minimize any bias due to the underlying prevalence of rejection in the dataset. We also chose crossvalidation instead of a single training/test split to obtain a more accurate estimate of the performance of the model, which is not dependent on how the initial training/test split was generated. This potentially leads to lower variance in the performance of the classifier, leading to a better estimate of future performance on unseen data.
Both the any-cause rejection signature and the signature to distinguish between TCMR and ABMR demonstrated excellent correlation with histopathologic diagnosis. The any-cause rejection signature also demonstrated a higher performance over the current indicators of allograft function, such as eGFR, which showed an AUC of 0.57 discriminating active rejection from nonrejection status. All kidney transplant biopsies in our cohort were clinically indicated, and 30.7% of these biopsy specimens revealed active rejection. Considering that real-life active-rejection prevalence is consistently lower than it is in our study population,10 and that NPV and PPV are prevalence-dependent metrics (because NPV increases and PPV decreases when prevalence decreases), our signature is expected to show an even higher NPV if it holds up in a stable disease population. When we adjust the prevalence to 20%, the NPV will be 96.1%, and, when adjusted to the 7.9% first year incidence for any-cause rejection, NPV will be 98.6% and PPV will be 54.7%.
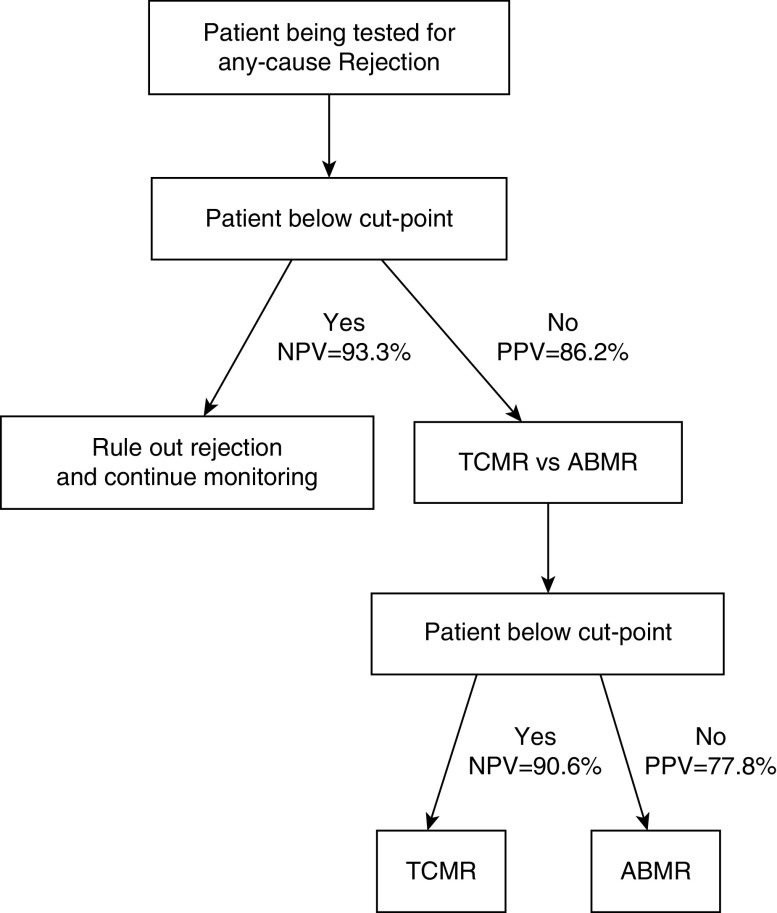
Therefore, the urinary exosome RNA assay for any-cause rejection can potentially be used to avoid unnecessary biopsies in patients with clinical suspicion of rejection. Whereas the high NPV suggests that only one in 15 patients would potentially miss a clinically indicated biopsy even at the very high (30.7%) prevalence in this cohort, the high PPV (86.2%) supports the possibility for patients with active rejection to be treated on time. The strong performances of urinary exosome gene signature in discriminating TCMR from ABMR status (AUC, 0.87; NPV, 90.6%) can help to refine the diagnosis by ruling out an ABMR and provide the clinicians with useful information for clinical management decisions (Figure 7).
Figure 7.
Schematic representation of the evaluation chart for recipients of kidney transplants. Serial monitoring of urinary exosome could be performed, and clinical management decisions made, according to the positivity or negativity of the signature.
Most of the genes identified in our signatures have an established role in immune activation that can explain the rejection process. BMP7 (a protein of the TGFβ superfamily) and the proinflammatory cytokines CXCL14, B2M, and IL32 identified in our all-cause rejection signature have recently been shown to be significantly induced in a kidney transplant undergoing mixed cellular-mediated rejection and ABMR, as analyzed by single-cell analysis.42 Similarly, C3 and CXCL11, identified in both any-cause rejection and TCMR, have been found to be upregulated in gene-expression profiling in acute allograft rejection in multiple studies.43–46 Although some of the genes identified are not known to drive allograft rejection, such as NAMPT, they have been shown to play a role in inflammation and immune-cell activation.47 The PYCARD gene codes for the ASC protein, which is widely expressed in different cells, including B cells, monocytes, and mature T cells. It has been described as an adaptor protein that participates in the inflammasome assembly and has been reported to play a part in different autoimmune processes and in viral and bacterial infections.48,49 Interestingly, our signature that discriminates TCMR from ABMR showed some overlapping with the any-cause rejection signature. We note that three of these genes (CXCL11, CD74, and C3) are present in the all-cause rejection signature, and that two additional genes (IFNAR2 and CD44) can help distinguish ABMR from TCMR. All five of these genes which are part of the ABMR/TCMR signature are significantly overexpressed (P<0.05) in ABMR compared with TCMR.
CD74, the invariant chain of MHC class II, is strongly expressed in cells involved in the presentation of antigens like dendritic cells, B cells, and macrophages. It also plays a role in regulating protein trafficking; dendritic-cell migration; and T-lymphocyte homing, proliferation, and cytokine secretion.50 CD44, a cell surface glycoprotein involved in cell adhesion, migration, and homing,51,52 acts as a coreceptor to CD74.53 The IFNAR2 is a subunit of the INFA receptor coupled to the tyrosine protein kinase JAK1, and that binds to the latent form of the transcription factors STAT1 or STAT2, participating in amplifying the production of chemokines, such as CXCL10.54
Previous studies by Suthanthiran et al.8 have successfully used mRNA derived from urinary cell pellets to identify potential markers of acute rejection.24 Although cells in urine are often dead or dying, and RNA within dead cells is quickly subjected to degradation, the extreme stability of exosome RNA (stable in urine for 2 weeks) can enable a more robust diagnostic platform. A more recent biomarker that is currently used in the clinic is dd-cfDNA. Bloom et al.7 measured the plasma level of dd-cfDNA and used a threshold of 1% to discriminate patients with allograft rejection status from those without. Although they were able to show that dd-cfDNA discriminated an active rejection status with an AUC of 0.74 and an NPV of 84%, samples with grade-IA TCMR did not reach the 1% threshold.7 In a subsequent study, Huang et al.55 found that the same dd-cfDNA test has not been able to discriminate TCMR from nonrejection status, despite strong performance in ABMR. In contrast to these studies, our any-cause rejection signature is clearly able to discriminate TCMR from nonrejection, despite 42.9% of the cases being Banff IA. However, given the complexity of the molecular perturbations during kidney allograft rejection, it is becoming increasingly evident that the combination of multiple biomarkers can improve the performance of any individual test. In a recent publication, Sarwal et al.56 used a combination of genomic, metabolic, and proteomic urinary biomarkers in recipients of kidney transplants. Similarly, we envision that combining urinary exosomal transcriptomic and proteomic profiling can improve the performance of our current signature.
The strengths of this urinary exosome mRNA study is that we have (1) been able to unveil a combination of biomarkers involved in active allograft kidney rejection in a large number of patients and urine samples, (2) developed a signature that distinguishes TCMR from ABMR, and (3) showed high predictive performances in our results that will allow the clinical use of this test to rule out rejection in patients. Assuming a high prevalence of rejection in a cohort of patients with clinically indicated biopsies, and considering that urine samples are easy to collect and urinary exosomal mRNA is highly stable, with a good positive predictive value increasing with higher prevalence, our test can prove useful to also rule in rejection. This is particularly true in patients with high-risk biopsies, patients who live far from transplant centers, and in patients living through pandemics such as coronavirus disease 2019. This is important because we have shown that urine stored at 4°C maintains stable exosomal mRNA for up to 2 weeks, similar to what has been shown before.29
Integration of urinary exosomes into clinical practice has emerged in fields outside kidney transplantation. The world’s first exosome-based diagnostic assay (ExoDx Prostate IntelliScore test) was launched in 2016 and is a prostate cancer test which uses urinary exosome RNA. The ExoDx Prostate IntelliScore test has been validated in two independent, prospective, multicenter studies and in a large utility study, with a blinded control arm, showing the value of the exosome platform.29–31 This is the first study describing an exosome mRNA signature for any-cause kidney rejection, and we applied a similar approach as the prostate cancer signature on urinary exosomes.
There are a few limitations to this study. First, we have conducted a cross-sectional study and have not collected serial urine samples, which prevented us from testing how early the signature can predict rejection before the clinically indicated biopsy. Second, our study has not included an independent validation cohort. However, the crossvalidation technique we used has been shown to improve the variability and selection bias.
In this report, we show the high-performance characteristics of urinary exosome RNA to discriminate active rejection from no rejection and ABMR from TCMR. We did not directly compare how the exosome signature performs compared with a cell pellet RNA assay; however, the high stability of urinary exosomes supports the possibility of identifying the nucleic acid signature more accurately, even without complicated RNA preservatives in the urine-collection device. Our gene signatures offer the possibility to provide the clinician with a new tool to help differentiate causes of graft dysfunction in the kidney allograft. Further validating studies, with prospectively collected samples, would help confirm our results in larger patient cohorts, and evaluate the potentials of this assay for early detection of kidney allograft rejection.
Disclosures
J.R. Azzi, C. Coticchia, J. Hurley, J. Skog, and V. Tadigotla have intellectual properties related to this work. J.R. Azzi reports having intellectual properties and receiving royalties from Accrue Health Inc.; receiving research funding from the American Diabetes Association, American Heart Association, and Qatar Research Fund; being a scientific advisor for CareDx; and having intellectual properties in ExosomeDx. C. Coticchia, J. Hurley, J. Skog, and V. Tadigotla are employees of Exosome Diagnostics, a Bio-Techne brand. R.N. Formica reports being the president of the American Society of Transplantation; having consultancy agreements with Genentech Pharmaceutical and Veloxis Pharmaceuticals; being on a speakers bureau for Novartis Pharmaceuticals; being on the visiting committee for the Scientific Registry of Transplant Recipients (January 1, 2018); and being a member of the United Network for Organ Sharing/Organ Procurement and Transplantation Network Membership and Professional Standards Committee. J.F. Markmann reports having consultancy agreements with, ownership interest in, and being a scientific advisor for (or member of) eGenesis and QihanBio. L.V. Riella reports receiving research funding from Bristol Myers Squibb and Visterra, and being a scientific advisor for or member of CareDx. J. Skog reports having ownership interest in Bio-Techne and patents and inventions with Massachusetts General Hospital. A. Srivastava reports being on a speakers bureau for AstraZeneca; receiving honoraria from AstraZeneca and Horizon Therapeutics PLC; and having consultancy agreements with CVS Caremark and Tate & Latham (medicolegal consulting). A. Chandraker reports consultancy agreements with Mitobridge, Shire, Amgen, Immucor, Natera, and Allovir; receiving research funding from ReNu, CSL, Shire, Amgen, and Allovir; receiving honoraria from Hansa, Natera, and eGenesis; reports being a scientific advisor or membership with American Society of Transplantation as Development Chair, Transplant Therapeutics Consortium as Governance Committee and Past Chair, CEoT as Co-Chair, Transplant Metrics as Chair, and Scientific American as Editor for Nephrology. All remaining authors have nothing to disclose.
Funding
This project was supported by American Heart Association award 13FTF17000018 (to J.R. Azzi) and National Institutes of Health (NIH) grant RO1 AI134842 (to J.R. Azzi). This project was also partially supported by Exosome Diagnostics, a Bio-Techne brand. A. Srivastava was supported by NIH Clinical Center grant F32DK11106.
Supplementary Material
Acknowledgments
The authors would like to thank the physician assistants at the Renal Transplant Division, Brigham and Women’s Hospital and the transplant coordinators at Yale School of Medicine and Massachusetts General Hospital for their logistic help in enrolling patients and collecting samples. In particular, the authors thank Michelle Kopp, Jill Lynch, Kaitlyn McGowan, Keri Foley, Jonathan Andrade, and Sheri Talbott. The authors also thank Ricarda Tomlin from Yale for the help.
Dr. Johan Skog and Dr. Jamil R. Azzi contributed to the conception and design; Dr. Anand Srivastava, Siawosh Eskandari, Dr. Albana B. Mihali, and Dr. Juliano Alhaddad contributed to data acquisition; Dr. James Hurley, Dr. Christine Coticchia, Dr. Vasisht Tadigotla, and Dr. Johan Skog performed assays and data analysis; Dr. Rania El Fekih and Dr. Jamil R. Azzi drafted the manuscript; all authors contributed to data interpretation and manuscript revisions, and all authors approved the final version of the manuscript.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
Supplemental Material
This article contains the following supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2020060850/-/DCSupplemental.
Supplemental Figure 1. Receiver-operating-characteristic (ROC) curves comparing performance of various clinical covariates.
References
- 1.Hill NR, Fatoba ST, Oke JL, Hirst JA, O’Callaghan CA, Lasserson DS, et al.: Global prevalence of chronic kidney disease – a systematic review and meta-analysis. PLoS One 11: e0158765, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mange KC, Joffe MM, Feldman HI: Effect of the use or nonuse of long-term dialysis on the subsequent survival of renal transplants from living donors. N Engl J Med 344: 726–731, 2001 [DOI] [PubMed] [Google Scholar]
- 3.Meier-Kriesche H-U, Kaplan B: Waiting time on dialysis as the strongest modifiable risk factor for renal transplant outcomes: A paired donor kidney analysis. Transplantation 74: 1377–1381, 2002 [DOI] [PubMed] [Google Scholar]
- 4.Gill JS, Tonelli M, Johnson N, Pereira BJG: Why do preemptive kidney transplant recipients have an allograft survival advantage? Transplantation 78: 873–879, 2004 [DOI] [PubMed] [Google Scholar]
- 5.Kaplan B, Meier-Kriesche HU: Renal transplantation: A half century of success and the long road ahead. J Am Soc Nephrol 15: 3270–3271, 2004 [DOI] [PubMed] [Google Scholar]
- 6.Sigdel TK, Archila FA, Constantin T, Prins SA, Liberto J, Damm I, et al.: Optimizing detection of kidney transplant injury by assessment of donor-derived cell-free DNA via massively multiplex PCR. J Clin Med 8: 19, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bloom RD, Bromberg JS, Poggio ED, Bunnapradist S, Langone AJ, Sood P, et al.; Circulating Donor-Derived Cell-Free DNA in Blood for Diagnosing Active Rejection in Kidney Transplant Recipients (DART) Study Investigators: Cell-free DNA and active rejection in kidney allografts. J Am Soc Nephrol 28: 2221–2232, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suthanthiran M, Schwartz JE, Ding R, Abecassis M, Dadhania D, Samstein B, et al.; Clinical Trials in Organ Transplantation 04 (CTOT-04) Study Investigators: Urinary-cell mRNA profile and acute cellular rejection in kidney allografts. N Engl J Med 369: 20–31, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christakoudi S, Runglall M, Mobillo P, Tsui T-L, Duff C, Domingo-Vila C, et al.: Development of a multivariable gene-expression signature targeting T-cell-mediated rejection in peripheral blood of kidney transplant recipients validated in cross-sectional and longitudinal samples. EBioMedicine 41: 571–583, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hart A, Smith JM, Skeans MA, Gustafson SK, Stewart DE, Cherikh WS, et al.: OPTN/SRTR 2015 annual data report: Kidney. Am J Transplant 17[Suppl 1]: 21–116, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Opelz G, Döhler B; Collaborative Transplant Study Report: Influence of time of rejection on long-term graft survival in renal transplantation. Transplantation 85: 661–666, 2008 [DOI] [PubMed] [Google Scholar]
- 12.Solez K, Axelsen RA, Benediktsson H, Burdick JF, Cohen AH, Colvin RB, et al.: International standardization of criteria for the histologic diagnosis of renal allograft rejection: The Banff working classification of kidney transplant pathology. Kidney Int 44: 411–422, 1993 [DOI] [PubMed] [Google Scholar]
- 13.Waikar SS, Betensky RA, Bonventre JV: Creatinine as the gold standard for kidney injury biomarker studies? Nephrol Dial Transplant 24: 3263–3265, 2009 [DOI] [PubMed] [Google Scholar]
- 14.Ferguson C, Winters S, Jackson S, McToal M, Low G: A retrospective analysis of complication and adequacy rates of ultrasound-guided native and transplant non-focal renal biopsies. Abdom Radiol (NY) 43: 2183–2189, 2018 [DOI] [PubMed] [Google Scholar]
- 15.Furness PN, Taub N; Convergence of European Renal Transplant Pathology Assessment Procedures (CERTPAP) Project: International variation in the interpretation of renal transplant biopsies: Report of the CERTPAP Project [published correction appears in Kidney Int 60: 2429, 2001]. Kidney Int 60: 1998–2012, 2001 [DOI] [PubMed] [Google Scholar]
- 16.Hricik DE, Nickerson P, Formica RN, Poggio ED, Rush D, Newell KA, et al.; CTOT-01 consortium: Multicenter validation of urinary CXCL9 as a risk-stratifying biomarker for kidney transplant injury. Am J Transplant 13: 2634–2644, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lai CP-K, Breakefield XO: Role of exosomes/microvesicles in the nervous system and use in emerging therapies. Front Physiol 3: 228, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gonzales PA, Pisitkun T, Hoffert JD, Tchapyjnikov D, Star RA, Kleta R, et al.: Large-scale proteomics and phosphoproteomics of urinary exosomes. J Am Soc Nephrol 20: 363–379, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pisitkun T, Shen R-F, Knepper MA: Identification and proteomic profiling of exosomes in human urine. Proc Natl Acad Sci U S A 101: 13368–13373, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sigdel TK, Ng YW, Lee S, Nicora CD, Qian W-J, Smith RD, et al.: Perturbations in the urinary exosome in transplant rejection. Front Med (Lausanne) 1: 57, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lim J-H, Lee C-H, Kim KY, Jung H-Y, Choi J-Y, Cho J-H, et al.: Novel urinary exosomal biomarkers of acute T cell-mediated rejection in kidney transplant recipients: A cross-sectional study. PLoS One 13: e0204204, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park J, Lin H-Y, Assaker JP, Jeong S, Huang C-H, Kurdi T, et al.: Integrated kidney exosome analysis for the detection of kidney transplant rejection. ACS Nano 11: 11041–11046, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Afaneh C, Muthukumar T, Lubetzky M, Ding R, Snopkowski C, Sharma VK, et al.: Urinary cell levels of mRNA for OX40, OX40L, PD-1, PD-L1, or PD-L2 and acute rejection of human renal allografts. Transplantation 90: 1381–1387, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muthukumar T, Dadhania D, Ding R, Snopkowski C, Naqvi R, Lee JB, et al.: Messenger RNA for FOXP3 in the urine of renal-allograft recipients. N Engl J Med 353: 2342–2351, 2005 [DOI] [PubMed] [Google Scholar]
- 25.Miranda KC, Bond DT, McKee M, Skog J, Păunescu TG, Da Silva N, et al.: Nucleic acids within urinary exosomes/microvesicles are potential biomarkers for renal disease. Kidney Int 78: 191–199, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jia S, Zocco D, Samuels ML, Chou MF, Chammas R, Skog J, et al.: Emerging technologies in extracellular vesicle-based molecular diagnostics. Expert Rev Mol Diagn 14: 307–321, 2014 [DOI] [PubMed] [Google Scholar]
- 27.Yang F, Liao X, Tian Y, Li G: Exosome separation using microfluidic systems: Size-based, immunoaffinity-based and dynamic methodologies. Biotechnol J 12: 1600699, 2017 [DOI] [PubMed] [Google Scholar]
- 28.He L, Zhu D, Wang J, Wu X: A highly efficient method for isolating urinary exosomes. Int J Mol Med 43: 83–90, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McKiernan J, Donovan MJ, O’Neill V, Bentink S, Noerholm M, Belzer S, et al.: A novel urine exosome gene expression assay to predict high-grade prostate cancer at initial biopsy. JAMA Oncol 2: 882–889, 2016 [DOI] [PubMed] [Google Scholar]
- 30.McKiernan J, Donovan MJ, Margolis E, Partin A, Carter B, Brown G, et al.: A prospective adaptive utility trial to validate performance of a novel urine exosome gene expression assay to predict high-grade prostate cancer in patients with prostate-specific antigen 2-10ng/ml at initial biopsy. Eur Urol 74: 731–738, 2018 [DOI] [PubMed] [Google Scholar]
- 31.Tutrone R, Donovan MJ, Torkler P, Tadigotla V, McLain T, Noerholm M, et al.: Clinical utility of the exosome based ExoDx Prostate(IntelliScore) EPI test in men presenting for initial biopsy with a PSA 2-10 ng/mL. Prostate Cancer Prostatic Dis 23: 607–614, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carroll PR, Kellogg Parsons J, Andriole G, Bahnson RR, Castle EP, Catalona WJ, et al.: NCCN guidelines insights: Prostate cancer early detection, Version 2.2016. J Natl Compr Canc Netw 14: 509–519, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Levey AS, Stevens LA, Schmid CH, Zhang YL, Castro AF 3rd, Feldman HI, et al.: A new equation to estimate glomerular filtration rate [published correction appears in Ann Intern Med 155: 408, 2011]. Ann Intern Med 150: 604–612, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haas M, Loupy A, Lefaucheur C, Roufosse C, Glotz D, Nankivell BJ, et al.: The Banff 2017 Kidney Meeting Report: Revised diagnostic criteria for chronic active T cell-mediated rejection, antibody-mediated rejection, and prospects for integrative endpoints for next-generation clinical trials. Am J Transplant 18: 293–307, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stekhoven DJ, Bühlmann P: MissForest--non-parametric missing value imputation for mixed-type data. Bioinformatics 28: 112–118, 2012 [DOI] [PubMed] [Google Scholar]
- 36.Kursa MB, Rudnicki WR: Feature selection with the Boruta package. J Stat Softw 36: 1–13, 2010 [Google Scholar]
- 37.Kuhn M: Building predictive models in R using the caret package. J Stat Softw 28: 1–26, 2008. 27774042 [Google Scholar]
- 38.Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez J-C, et al.: pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics 12: 77, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Buderer NMF: Statistical methodology: I. Incorporating the prevalence of disease into the sample size calculation for sensitivity and specificity. Acad Emerg Med 3: 895–900, 1996 [DOI] [PubMed] [Google Scholar]
- 40.Nankivell BJ, Agrawal N, Sharma A, Taverniti A, P’Ng CH, Shingde M, et al.: The clinical and pathological significance of borderline T cell-mediated rejection. Am J Transplant 19: 1452–1463, 2019 [DOI] [PubMed] [Google Scholar]
- 41.Becker JU, Chang A, Nickeleit V, Randhawa P, Roufosse C: Banff borderline changes suspicious for acute T cell-mediated rejection: Where do we stand? Am J Transplant 16: 2654–2660, 2016 [DOI] [PubMed] [Google Scholar]
- 42.Wu H, Malone AF, Donnelly EL, Kirita Y, Uchimura K, Ramakrishnan SM, et al.: Single-cell transcriptomics of a human kidney allograft biopsy specimen defines a diverse inflammatory response. J Am Soc Nephrol 29: 2069–2080, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sreekumar R, Rasmussen DL, Wiesner RH, Charlton MR: Differential allograft gene expression in acute cellular rejection and recurrence of hepatitis C after liver transplantation. Liver Transpl 8: 814–821, 2002 [DOI] [PubMed] [Google Scholar]
- 44.Chen R, Sigdel TK, Li L, Kambham N, Dudley JT, Hsieh S-C, et al.: Differentially expressed RNA from public microarray data identifies serum protein biomarkers for cross-organ transplant rejection and other conditions. PLOS Comput Biol 6: e1000940, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spivey TL, Uccellini L, Ascierto ML, Zoppoli G, De Giorgi V, Delogu LG, et al.: Gene expression profiling in acute allograft rejection: Challenging the immunologic constant of rejection hypothesis. J Transl Med 9: 174, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Akalin E, Hendrix RC, Polavarapu RG, Pearson TC, Neylan JF, Larsen CP, et al.: Gene expression analysis in human renal allograft biopsy samples using high-density oligoarray technology. Transplantation 72: 948–953, 2001 [DOI] [PubMed] [Google Scholar]
- 47.Halvorsen B, Espeland MZ, Andersen GØ, Yndestad A, Sagen EL, Rashidi A, et al.: Increased expression of NAMPT in PBMC from patients with acute coronary syndrome and in inflammatory M1 macrophages. Atherosclerosis 243: 204–210, 2015 [DOI] [PubMed] [Google Scholar]
- 48.Martin BN, Wang C, Zhang CJ, Kang Z, Gulen MF, Zepp JA, et al.: T cell-intrinsic ASC critically promotes T(H)17-mediated experimental autoimmune encephalomyelitis. Nat Immunol 17: 583–592, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ichinohe T, Lee HK, Ogura Y, Flavell R, Iwasaki A: Inflammasome recognition of influenza virus is essential for adaptive immune responses. J Exp Med 206: 79–87, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Su H, Na N, Zhang X, Zhao Y: The biological function and significance of CD74 in immune diseases. Inflamm Res 66: 209–216, 2017 [DOI] [PubMed] [Google Scholar]
- 51.DeGrendele HC, Estess P, Picker LJ, Siegelman MH: CD44 and its ligand hyaluronate mediate rolling under physiologic flow: A novel lymphocyte-endothelial cell primary adhesion pathway. J Exp Med 183: 1119–1130, 1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ponta H, Sherman L, Herrlich PA: CD44: From adhesion molecules to signalling regulators. Nat Rev Mol Cell Biol 4: 33–45, 2003 [DOI] [PubMed] [Google Scholar]
- 53.Schröder B: The multifaceted roles of the invariant chain CD74--More than just a chaperone. Biochim Biophys Acta 1863[6 Pt A]: 1269–1281, 2016 [DOI] [PubMed] [Google Scholar]
- 54.Takaoka A, Yanai H: Interferon signalling network in innate defence. Cell Microbiol 8: 907–922, 2006 [DOI] [PubMed] [Google Scholar]
- 55.Huang E, Sethi S, Peng A, Najjar R, Mirocha J, Haas M, et al.: Early clinical experience using donor-derived cell-free dna to detect rejection in kidney transplant recipients. Am J Transplant 19: 1663–1670, 2019 [DOI] [PubMed] [Google Scholar]
- 56.Yang JYC, Sarwal RD, Sigdel TK, Damm I, Rosenbaum B, Liberto JM, et al.: A urine score for noninvasive accurate diagnosis and prediction of kidney transplant rejection. Sci Transl Med 12: eaba2501, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.