Significance
Electrostatics have been thought to dominate interactions between a polybasic membrane anchoring motif and anionic lipids in the plasma membrane. Combining data from computational and imaging techniques, here we show that distinct conformational dynamics and interaction patterns of prenylated polybasic domains encode specificity for lipid acyl chain saturation. This previously unrecognized capacity to distinguish between acyl chain structures renders the anchor sensitive to the biophysical properties of the membrane, which are in turn determined by the acyl chain structure of the composite lipids. We propose that lipid acyl chain structure and dynamics participate in the regulation of the spatial distribution of prenylated polybasic membrane anchors found in many membrane-bound small GTPases, and possibly polybasic domains in other membrane associated proteins.
Keywords: KRAS, phosphatidylserine, nanoclusters, plasma membrane depolarization, acyl chains
Abstract
KRAS interacts with the inner leaflet of the plasma membrane (PM) using a hybrid anchor that comprises a lysine-rich polybasic domain (PBD) and a C-terminal farnesyl chain. Electrostatic interactions have been envisaged as the primary determinant of interactions between KRAS and membranes. Here, we integrated molecular dynamics (MD) simulations and superresolution spatial analysis in mammalian cells and systematically compared four equally charged KRAS anchors: the wild-type farnesyl hexa-lysine and engineered mutants comprising farnesyl hexa-arginine, geranylgeranyl hexa-lysine, and geranylgeranyl hexa-arginine. MD simulations show that these equally charged KRAS mutant anchors exhibit distinct interactions and packing patterns with different phosphatidylserine (PtdSer) species, indicating that prenylated PBD–bilayer interactions extend beyond electrostatics. Similar observations were apparent in intact cells, where each anchor exhibited binding specificities for PtdSer species with distinct acyl chain compositions. Acyl chain composition determined responsiveness of the spatial organization of different PtdSer species to diverse PM perturbations, including transmembrane potential, cholesterol depletion, and PM curvature. In consequence, the spatial organization and PM binding of each KRAS anchor precisely reflected the behavior of its preferred PtdSer ligand to these same PM perturbations. Taken together these results show that small GTPase PBD-prenyl anchors, such as that of KRAS, have the capacity to encode binding specificity for specific acyl chains as well as lipid headgroups, which allow differential responses to biophysical perturbations that may have biological and signaling consequences for the anchored GTPase.
KRAS4B (hereafter KRAS) is a lipid-anchored small GTPase that regulates multiple signaling pathways to control cell proliferation, survival, and migration (1, 2). KRAS is one of the most frequently mutated proteins in cancer, with mutations found in 98% of pancreatic tumors, 45% of colorectal tumors, and 31% of lung tumors (1, 2). KRAS signaling is mostly compartmentalized to the plasma membrane (PM) (3), where KRAS interacts with a specific set of lipids and undergoes spatial segregation to form nanometer-sized domains, termed nanoclusters (4, 5). Nanoclusters operate as transient platforms for KRAS signal transmission such that the extent of nanoclustering directly correlates with the efficiency of effector recruitment and MAPK signal output (6–8). KRAS effectors require synergistic binding with activated KRAS and specific lipids for efficient PM recruitment and activation (9), therefore concentrating a specific set of lipids within nanoclusters is essential to KRAS function (10, 11). In this context the KRAS C-terminal membrane anchor, which comprises a hexa-lysine polybasic domain (PBD) and a farnesylated, methylesterified cysteine residue (4, 5, 12, 13), selectively sorts the monovalent anionic phospholipid phosphatidylserine (PtdSer) into nanoclusters (7, 11, 14). In consequence, PtdSer levels in the PM modulate the extent of KRAS localization to, and nanoclustering on, the PM and hence regulate KRAS-dependent effector recruitment and signaling (7, 11, 14). Depletion of PtdSer compromises the proliferation of KRAS-driven cancer cell lines and KRAS oncogenicity in mouse xenograft models (15–20). Thus, PtdSer is a key structural component of KRAS signaling nanoclusters on the PM and plays important roles in KRAS function and pathology.
Electrostatics have long been considered to be the primary determinant of interactions between anionic lipids and the PBD of KRAS; however, in biological membranes these interactions are more complex. For example, KRAS nanoclusters are enriched with monovalent PtdSer, but not multivalent phosphoinositol 4,5-bisphosphate (PIP2) or phosphoinositol 3,4,5-trisphosphate (PIP3) (7, 11, 14, 21). Moreover, the KRAS anchor selectively binds and sorts mixed-chain PtdSer species comprising one saturated and one unsaturated acyl chain (16:0/18:1 PtdSer and 18:0/18:1 PtdSer), but not symmetric PtdSer species comprising two identical saturated or unsaturated acyl chains (di18:0 PtdSer, di18:1 PtdSer or di18:2 PtdSer) (11, 21). Molecular dynamics (MD) simulations reveal that the KRAS PBD samples diverse conformational states on bilayers (11), including a pseudohelical hairpin with only its center portion inserted into the bilayer core (11, 22). These findings suggest that the KRAS PBD anchor interacts with membranes in complex manners that extend beyond electrostatics. Here, we formally examine this hypothesis by integrating atomistic MD simulations with quantitative electron microscopy (EM)-spatial analysis of intact PM. We show that the KRAS prenyl group and PBD sequence synergistically contribute to the structure and lipid-binding pattern of the anchor on membranes. Different combinations of PBD sequence and prenyl chain can be engineered to preferentially interact with PtdSer species that have different acyl chain structures. When grafted onto KRAS the PtdSer acyl chain binding preferences of these anchors result in fundamentally different responses to multiple biophysical perturbations of PM properties, including transmembrane potential (ΔVm), cholesterol content, and membrane curvature. Together these results show that PBD prenyl anchors by recognizing phospholipid acyl chain structure link PM biophysics to small GTPase spatiotemporal organization and potentially biological function.
Results
Equally Charged KRAS PBD Anchors Favor Distinct Conformations and Sort Different PtdSer Species.
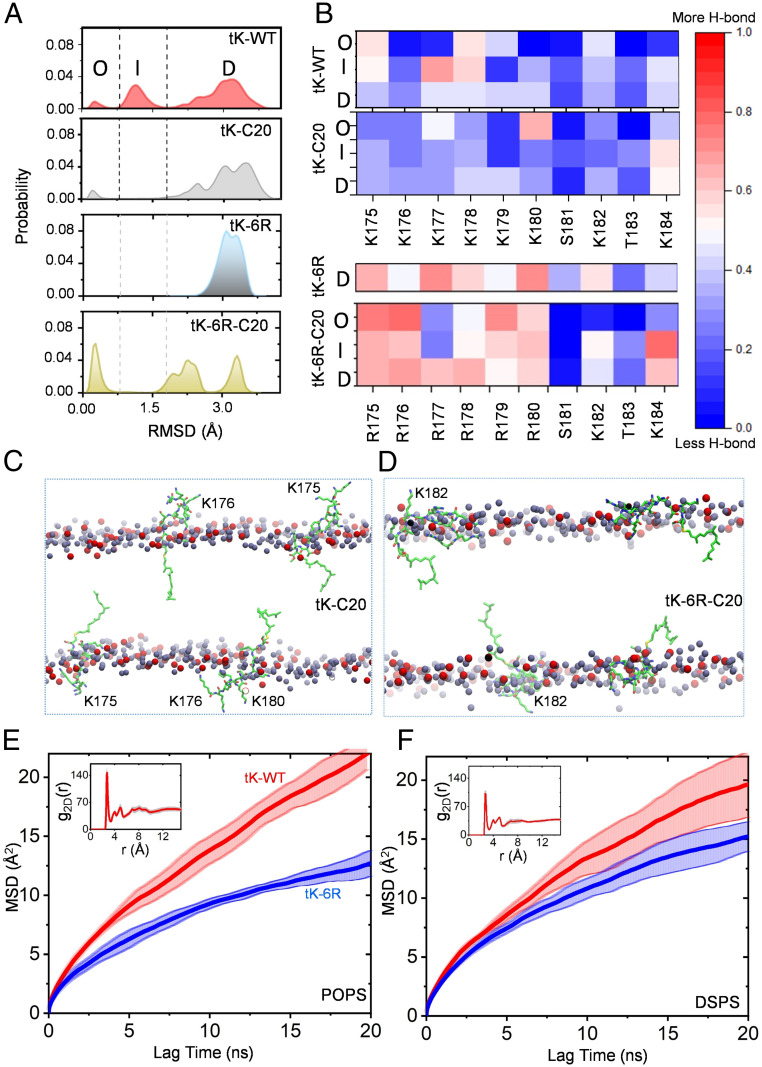
We previously used classical and metadynamics MD simulations to examine how the minimal KRAS C-terminal anchor, which comprises a hexa-lysine PBD and a farnesylated, methylesterified cysteine residue (termed tK-WT, illustrated in SI Appendix, Fig. S1) interacts with a model bilayer (11, 22). We showed that tK-WT samples ordered (O), intermediate (I), and disordered (D) conformations on a bilayer composed of 80% 16:0/18:1 PtdChol (POPC) and 20% 16:0/18:1 PtdSer (POPS; chemical structure shown in SI Appendix, Fig. S2). In this analysis, we used an ordered (semihelical) conformation as reference and defined O as the ensemble of conformers with rmsd < 0.8Å, D as those with rmsd > 1.8Å, and I as those with 0.8Å ≤ rmsd ≤ 1.8Å. We found that 64%, 29%, and 6% of the conformations sampled by the ∼1- to 1.5-μs-long simulations were in the D, I, and O state (11). In contrast, the geranylgeranylated counterpart of tK-WT, where farnesyl is replaced by geranylgeranyl (tK-C20, illustrated in SI Appendix, Fig. S1), almost exclusively adopted the D state (11). Here we extended these simulations out to 2.25 μs and found patterns for tK-WT and tK-C20 similar to those previously reported (Fig. 1A). Next, we compared two additional PBD anchors with six arginine residues in place of the lysine residues and either a farnesyl, termed tK-6R, or a geranylgeranyl, termed tK-6R-C20 (illustrated in SI Appendix, Fig. S1). All four PBD anchors therefore contain an identical number of charged residues (SI Appendix, Fig. S1). MD simulation of these anchors on the same POPC/POPS bilayer showed that tK-6R exclusively adopted the D state whereas tK-6R-C20 adopted a similar distribution of O, I, and D states as the original tK-WT (Fig. 1A), although the I state here is more extended (1.5 Å ≤ rmsd ≤ 2.5 Å). Analysis of hydrogen bond interactions of each residue in each anchor, plotted as a heat map in Fig. 1B separately for the O, I, and D ensembles, shows that the hexa-arginine PBDs (tK-6R and tK-6R-C20) interact more extensively with POPS lipid headgroups than the hexa-lysine PBDs (tK and tK-C20). This is in part because the arginine guanidinium has a greater hydrogen bonding potential than the lysine ammonium group. However, it is interesting to observe that R177 of tK-6R-C20 in the O and I states does not interact strongly with lipids (Fig. 1B). Snapshots at the end of the simulation of tK-C20 and tK-6R-C20 are shown in Fig. 1 C and D, illustrating the rather dynamic organization of the peptides in the POPC/POPS bilayer. Importantly, there are significant differences beween the two peptides in terms of membrane adsorption and backbone organization, with tK-6R-C20 adopting a semiparallel orientation and its arginine side chains intercalating with lipids more extensively (Fig. 1D) than do the lysine side chains of tK-C20 (Fig. 1C). This is consistent with the observation from the hydrogen bond analysis (Fig. 1B). In sum the simulations show distinct conformational distributions and interaction patterns of equally charged prenylated PBD anchors on an anionic model bilayer.
Fig. 1.
The equally charged KRAS PBD anchors sample distinct conformational states on PtdSer bilayers. (A) Probability distribution of the rmsd of the Cα atoms of residues 177 to 182 was calculated for four equally charged KRAS PBD anchors: tK-WT (the original PBD), tK-C20, tK-6R, and tK-6R-C20 (sequences shown in SI Appendix, Fig. S1). The dotted lines demarcate the three distinct conformational groups of the peptides, O (<0.8 Å), D (>1.8 Å), and I, that lie in between these two (except for tK-6R-C20, where it spans the 1.5- to 2.5-Å range). (B) Heat maps of the frequency of hydrogen bonds between individual side chains with PtdSer separately for the KRAS PBD anchors in the O, I, and D conformational states. (C and D) The last snapshot from the simulation of tK-C20 (C) and tK-6K-C20 (D) in POPC/POPS (chemical structure illustrated in SI Appendix, Fig. S2) showing the dynamic structural organization of the four peptides per simulation (carbon in green, nitrogen in blue, and oxygen in red) in the bilayer. Only the phosphorus atoms of POPS (red) and POPC (dark gray) are shown; all other atoms including lipid acyl chains, water, and ions are omitted for clarity. Selected, transiently solvent-exposed residues of the tK peptides are labeled for reference. (E and F) MSD as a function of lag time for the Cα atom of the farnesylated cysteine of tK-WT (red) and tK-6R (blue) in a bilayer of 80% POPC and 20% POPS (E) or 20% DSPS (F); insets show the 2D radial pair distribution function, g2D(r), for the headgroup oxygen atoms of POPS (E) and DSPS (F) lipids around the side-chain nitrogen atoms of lysine and oxygen atoms of serine/threonine residues in tK-WT. These analyses were performed by dividing the last 300 ns of the simulation data into three blocks of 100 ns each so that the solid lines represent the mean and the shaded background the SD over the three blocks. Diffusion characteristics of POPS and DSPS lipids are shown in SI Appendix, Fig. S3.
To explore the physicochemical basis of the different interactions of these anchors with PtdSer, we compared simulations of tK-WT and tK-6R in bilayers of 80% POPC with either 20% mixed-chain POPS or 20% fully saturated di18:0 PtdSer (DSPS; chemical structure shown in SI Appendix, Fig. S2). The two-dimensional (2D) radial distribution function of PtdSer headgroups shows that POPS molecules clustered more extensively around tK-WT than did DSPS (Fig. 1 E and F, Insets), suggesting that tK-WT preferentially associates with POPS over DSPS. To examine how different PtdSer acyl chain structures might potentially impact the lateral dynamics of the anchor peptides, we compared the mean square displacement (MSD) of tK and tK-6R on the POPC/POPS and POPC/DSPS bilayers. As shown in Fig. 1 E and F main plots, based on the slopes of MSD, tK-6R diffused slower than tK on both the POPC/POPS and POPC/DSPS bilayers, in line with the more extensive hydrogen-bonding interactions of arginine compared to lysine (Fig. 1B). The difference in the rates of diffusion between tK and tK-6R was more pronounced in the presence of POPS than DSPS, consistent with the observed preferential clustering of POPS over DSPS around tK (Fig. 1 E and F, Insets). Radial distribution analyses (SI Appendix, Fig.S3 A and B) show no significant differences between the clustering of PtdSer around tK-C20 and tK-6R-C20, suggesting that the ability of arginine to form a multidentate hydrogen bond contributes to the higher number of hydrogen bonds in the poly-arginine peptides (Fig. 1B). Note that POPS and DSPS maintained similar diffusion characteristics in the simulations (SI Appendix, Fig. S3C); thus, the different diffusive behaviors of the tK and tK-6R anchors reflect different extents of lipid–anchor interactions, not different behaviors of the PtdSer species.
PtdSer Acyl Chain Binding Preferences Determine Spatial Distribution of KRAS Mutant Membrane Anchors.
We next compared how each PBD anchor interacts with PtdSer in intact cells by engineering the anchors onto the C terminus of green fluorescent protein (GFP)-tagged oncogenic mutant KRASG12V. For these experiments we used PSA3 cells, which are deleted for PtdSer synthase 1 (PSS1) and exhibit a ∼35% reduction in total PtdSer levels when grown in medium containing 10% dialyzed fetal bovine serum (DFBS) but which maintain wild-type PtdSer levels when supplemented with ethanolamine (Etn) (7, 11, 14, 21, 23, 24). To modulate the specific PtdSer content of the PM, we acutely add back synthetic PtdSer species to Etn-starved, PtdSer-depleted PSA3 cells (11, 21). DSPS, di18:1 PtdSer (DOPS), or POPS are all efficiently delivered structurally intact to the inner PM of PSA3 cells after acute addback as evidenced by Annexin V staining, immunogold labeling, fluorescence imaging, and lipidomics (11, 21, 24). PM sheets of PSA3 cells expressing the GFP-KRASG12V anchor variants were attached to EM grids and immunolabeled with 4.5-nm gold particles conjugated directly to anti-GFP antibody (SI Appendix, Fig. S4 A–E). The gold particle distribution was imaged using transmission EM (TEM) and the spatial distribution of the gold particles analyzed using univariate K-functions expressed as L(r)-r and plotted as a function of cluster radius, r (7, 8, 11, 25) (SI Appendix, Fig. S4F). L(r)-r values greater than 1 are outside the 99% CI for a random pattern and indicate statistically significant clustering. The peak L(r)-r value, termed Lmax, was used as a summary statistic, where larger Lmax values indicate more extensive nanoclustering. In addition, the number of gold nanoparticles per unit PM area quantifies the extent of PM binding of the ectopically expressed GFP-KRASG12V (7, 11, 14).
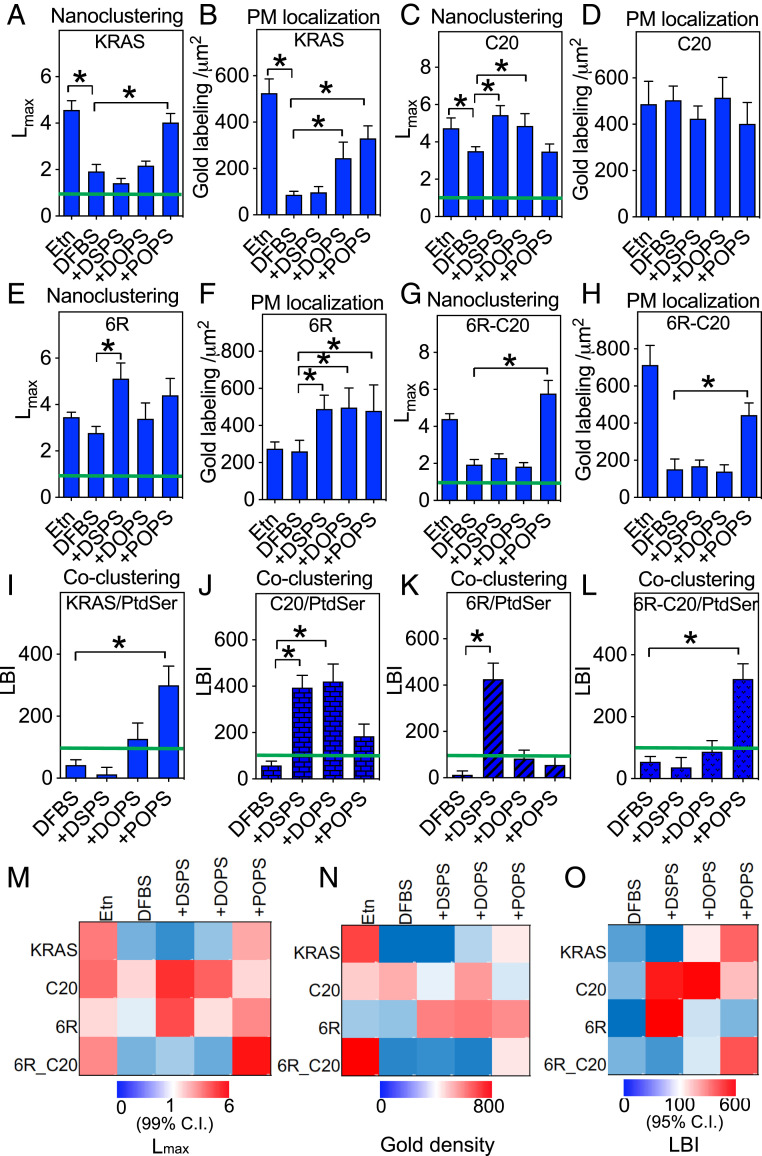
In control PSA3 cells with wild-type PtdSer levels, GFP-KRASG12V clustered and localized to the PM extensively (Fig. 2 A and B). Depleting endogenous PtdSer by Etn starvation significantly reduced PM localization and disrupted GFP-KRASG12V nanoclustering (Fig. 2 A and B). In PtdSer-depleted cells acute addback of only POPS, and neither DSPS nor DOPS (lipid structures shown in SI Appendix, Fig. S4), recovered the clustering of GFP-KRASG12V on the PM (Fig. 2A), whereas addback of POPS or DOPS did recover PM localization of GFP-KRASG12V (Fig. 2B). These data are consistent with our previous findings (11, 21). At wild-type PtdSer levels GFP-KRASG12V-C20 (anchor = a six-lysine PBD with geranylgeranyl) clustered on the PM (Fig. 2C) and localized to the PM (Fig. 2D) as efficiently as GFP-KRASG12V with a wild-type anchor. PtdSer depletion reduced the clustering of GFP-KRASG12V-C20 but not as extensively as for KRASG12V (Fig. 2C) and had no observable effect on the PM binding of GFP-KRASG12V-C20 (Fig. 2D). Acute addback of DSPS or DOPS, but not POPS, restored the nanoclustering of GFP-KRASG12V-C20 in PtdSer-depleted cells, (Fig. 2C), while PM localization of GFP-KRASG12V-C20 was unaffected by addback of any PtdSer species (Fig. 2D). PtdSer depletion had no effect on the clustering (Fig. 2E) or PM localization (Fig. 2F) of GFP-KRASG12V-6R (anchor = a six arginine PBD with farnesyl); however, addback of fully saturated DSPS, and not DOPS or POPS, enhanced GFP-KRASG12V-6R clustering (Fig. 2E) and addback of any PtdSer species elevated GFP-KRASG12V-6R PM binding (Fig. 2F). Strikingly, PtdSer depletion markedly disrupted the clustering (Fig. 2G) and PM localization (Fig. 2H) of GFP-KRASG12V-6R-C20 (anchor = a six arginine PBD with geranylgeranyl). Addback of only POPS, and not DSPS or DOPS, fully recovered both clustering (Fig. 2G) and PM localization (Fig. 2H) of GFP-KRASG12V-6R-C20. The nanoclustering response of GFP-KRASG12V-6R-C20 was therefore identical to the original GFP-KRASG12V in that only POPS supported nanoclustering but was more restrictive in the species of PtdSer that could also support PM binding. It is also worth noting, by comparing the nanoclustering values (Fig. 2 A, C, E, and G) with the PM localization values (Fig. 2 B, D, F, and H), that nanoclustering is independent of PM localization. This is consistent with previous studies (6, 8, 21, 26). The spatiotemporal organization data are summarized in heat maps of Lmax values (Fig. 2M) and gold labeling density values (Fig. 2N). Taken together, these data show that different PBD prenyl anchor combinations can discriminate between PtdSer species with different acyl chain compositions.
Fig. 2.
KRAS PBD mutant anchors selectively sort distinct PtdSer species on intact cell PM. PSA3 cells ectopically expressing GFP-KRASG12V (A and B) or a GFP-KRASG12V anchor mutant (C–H) were grown in DFBS supplemented with 10 μM Etn (control) for 72 h to maintain wild-type PtdSer levels, or grown without Etn to deplete PM PtdSer levels (DFBS), or without Etn followed by acute addback of defined PtdSer species (DSPS, DOPS, or POPS at 10 μM for 1 h). Chemical structures of PtdSer species are illustrated in SI Appendix, Fig. S2. Intact PM sheets from the cells were attached to EM grids and immunolabeled with anti-GFP antibody conjugated to 4.5-nm gold particles. The gold distribution was imaged using TEM and analyzed using univariate K-functions plotted as L(r)-r, against cluster radius, r, in nanometers within randomly chosen 1-μm2 PM areas and standardized on the 99% CI for a random pattern (99% CI = green line). The peak value of the L(r)-r function, Lmax, was used as a summary statistic to quantify the extent of clustering. Sample EM images and calculations are shown and described in SI Appendix, Fig. S4. Graphs (A, C, E, and G) show mean Lmax values (± SEM) for each KRAS anchor calculated from 12 ≤ n ≤ 16 PM sheets selected from different cells. Significant differences from control Lmax values were assessed using bootstrap tests (*P < 0.05). In each case the mean number of gold particles per μm2 (± SEM) was calculated for the same set of PM sheets and used to estimate the extent of PM binding of each KRAS anchor (B, D, F, and H). Significant differences from control values were assessed using one-way ANOVAR (*P < 0.05). PSA3 cells coexpressing GFP-LactC2 with RFP-KRASG12V (I) or an RFP-KRASG12V anchor mutant (J–L) were grown and treated exactly as in A–H. Intact PM sheets from the cells were attached to EM grids and coimmunolabeled with anti-GFP antibody conjugated to 6-nm gold and anti-RFP antibody conjugated to 2-nm gold. The gold distributions were imaged using TEM and analyzed using bivariate K-functions plotted as Lbiv(r)-r, against cluster radius, r, in nanometers within randomly chosen 1-μm2 PM areas and standardized on the 95% CI for a random pattern (95% CI = green line). The area under the Lbiv(r)-r curve over the interval 10 < r < 110 nm, termed LBI, is used a summary statistic to evaluate the extent of colocalization. Sample EM images and calculations are illustrated in SI Appendix, Fig. S4. Graphs (I–L) show mean LBI values (± SEM) for each KRAS anchor calculated from 12 ≤ n ≤ 15 PM sheets selected from different cells. Significant differences from each control LBI value (DFBS) were assessed using bootstrap tests (*P < 0.05). Heat maps summarizing Lmax values from A, C, E, and G, gold labeling densities from B, D, F, and H, and LBI values from I–L, are shown in M–O, respectively.
KRAS Mutant Membrane Anchors Assemble Distinct PtdSer Species into Nanoclusters on the PM.
We next assessed the capacity of different anchors to assemble distinct PtdSer species into nascent nanoclusters. To this end we evaluated the extent to which acute addback of defined PtdSer species to PtdSer-depleted cells induces coclustering of KRAS with LactC2, a PtdSer head group probe. Intact apical PM sheets of PSA3 cells coexpressing GFP-LactC2 and a red fluorescent protein (RFP)-tagged KRASG12V anchor mutant were attached to the EM grids and coimmunolabeled with anti-GFP and anti-RFP antibody coupled to 6-nm or 2-nm gold particles, respectively. The gold-labeled PM sheets were imaged using TEM (SI Appendix, Fig.S5 A and B) and colocalization between the two populations of gold particles quantified using bivariate K-functions expressed as Lbiv(r)-r and plotted as a function of cluster radius, r (7, 11) (SI Appendix, Fig. S5C). Lbiv(r)-r values greater than 1 are outside the 95% CI for random patterns and indicate statistically significant coclustering. As a summary statistic we integrated each bivariate K-function curve on the interval 10 < r < 110 to obtain the parameter l-function-bivariate-integrated (LBI). The upper 95% CI for LBI = 100, and thus LBI values >100 indicate significant coclustering of KRASG12V and LactC2.
Fig. 2 I–L show that PtdSer depletion abolishes colocalization between all KRASG12V anchor mutants and PtdSer as detected by the LactC2 probe. Addback of only POPS, but neither DSPS nor DOPS, restored colocalization between GFP-LactC2 and RFP-KRASG12V (Fig. 2I), indicating that only POPS is recruited into and enriched in KRASG12V nanoclusters (11, 21). By contrast, DSPS and DOPS, but not POPS, were recruited into and enriched in KRASG12V-C20 nanoclusters (Fig. 2J). Only DSPS was recruited into and enriched in KRASG12V-6R nanoclusters (Fig. 2K), and only POPS was recruited into and enriched in KRASG12V-6R-C20 nanoclusters (Fig. 2L). Fig. 2O summarizes these LactC2- KRASG12V colocalization data as a heat map of LBI values. Analyses of radial distribution functions in the tK-C20 and tK-6R-C20 simulations show no significant differences in terms of POPS clustering around the peptides despite the greater potential of tK-6R-C20 to form hydrogen bonds with POPS (Fig. 1 G and H). The EM-derived colocalization of POPS is also somewhat similar between KRASG12V and KRASG12V-6R-C20 (Fig. 2O). Together these findings show that equally charged PBD anchors can encode a high degree of binding specificity for distinct PtdSer species with defined acyl chain compositions.
Selective Sorting of Distinct PtdSer Species Mediates the Responsiveness of KRAS Nanoclustering to PM Depolarization.
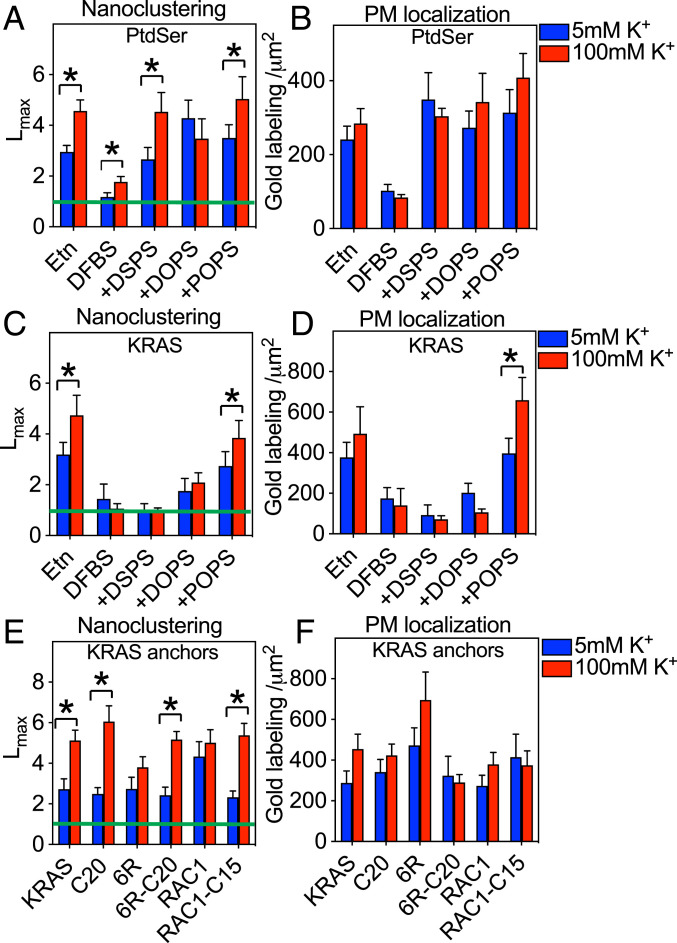
Previous work has shown that PM transmembrane potential, ΔVm, modulates PtdSer lateral distribution to regulate the extent of KRAS nanoclustering and signal output (7). To extend these observations we examined whether different PtdSer species are equivalently responsive to ΔVm. Control PSA3 cells expressing GFP-LactC2 were incubated in isotonic buffers containing 5mM K+ (polarized) or 100 mM K+ (depolarized) for 5 min and the PM spatial distribution of GFP-LactC2 was visualized by immunogold labeling and quantified by EM-spatial analysis. PM depolarization significantly elevated the Lmax of GFP-LactC2 (Fig. 3A), while not affecting gold labeling density (Fig. 3B), consistent with previous observations (7). PtdSer depletion of GFP-Lact2–expressing PSA3 cells significantly reduced the gold-labeling density and GFP-Lact2 Lmax values under both polarized and depolarized conditions (Fig. 3 A and B); however, a numerically small, but significant, increase in GFP-Lact2 Lmax values was still evident on PM depolarization (Fig. 3A). This is expected since the remaining PtdSer in the PM of the PtdSer-depleted PSA3 cells is still sensitive to ΔVm. Acutely adding back any one of the three synthetic PtdSer species to the PtdSer-depleted cells effectively restored GFP-LactC2 to the PM (Fig. 3B), consistent with previous studies (11, 21, 24). However, addback of only DSPS or POPS, but not DOPS, restored the full responsiveness of GFP-LactC2 clustering to ΔVm (Fig. 3A). We interpret this result to mean that the lateral distribution of DSPS and POPS, but not DOPS, is sensitive to ΔVm. Thus, not all PtdSer species respond equally to changing PM transmembrane potential. We next tested whether the PtdSer acyl chain binding specificity of different PBD anchors could confer differential ΔVm responsiveness to KRASG12V. PM depolarization of PSA3 cells significantly enhanced the nanoclustering of GFP-KRASG12V at wild-type PtdSer levels, but the ΔVm responsiveness of GFP-KRASG12V remaining on the PM after PtdSer depletion was abolished (Fig. 3 C and D). Acute addback of mixed-chain POPS, but not DSPS or DOPS, recovered GFP-KRASG12V nanoclustering and effectively restored the responsiveness of GFP-KRASG12V to ΔVm (Fig. 3 C and D). The PM localization of GFP-KRASG12V was also further elevated upon PM depolarization in the POPS supplemented cells (Fig. 3D). PM depolarization also elevated the nanoclustering of GFP-KRASG12V-C20 and GFP-KRASG12V-6R-C20, but not that of GFP-KRASG12V-6R (Fig. 3E). Changing PM voltage did not affect the PM localization of any of the KRASG12V PBD mutants (Fig. 3F). Thus, the ΔVm responsiveness of each of the synthetic PBD anchors in the context of KRASG12V exactly matches the ΔVm responsiveness of the PtdSer species to which the anchor binds. One seemingly discrepant result here is the apparant insensitivity of KRASG12V-6R to ΔVm, despite selectively sorting fully saturated DSPS, which is sensitive to ΔVm. This may simply reflect the very low levels of fully saturated PtdSer species (<1%) present in mammalian cell membranes.
Fig. 3.
Different PtdSer species selectively mediate the responses of KRAS to PM depolarization. PSA3 cells ectopically expressing GFP-LactC2 were grown in 10 μM Etn to maintain wild-type level of PtdSer, or depleted of endogenous PtdSer without (DFBS) or with acute addback of defined PtdSer species exactly as described in Fig. 2. Cells were then incubated in isotonic Hepes buffers containing 5 mM [K+] (polarized) or 100 mM [K+] (depolarized) for 5 min before preparation of PM sheets followed by anti-GFP immunogold labeling and EM univariate K-function analysis as described in Fig. 2. A and B show mean Lmax values (± SEM) and gold labeling densities (± SEM), respectively, of immunogold-labeled GFP-LactC2 calculated from 12 ≤ n ≤ 15 PM sheets selected from different cells. The depolarization experiment in A and B was repeated with PSA3 cells ectopically expressing GFP-KRASG12V. C and D show mean Lmax values (± SEM) and gold labeling densities (± SEM), respectively, of the immunogold-labeled GFP-KRASG12V calculated from 8 ≤ n ≤ 15 PM sheets selected from different cells. An EM-univariate nanoclustering analysis in isotonic Hepes buffers containing 5mM [K+] (polarized) or 100 mM [K+] (depolarized) was next conducted in BHK cells ectopically expressing GFP-KRASG12V, or a GFP-KRASG12V anchor mutant, or GFP-RAC1G12V with a wild-type geranylgeranylated or mutant farnesylated (C15) anchor. Mean Lmax (± SEM) (E) and mean gold labeling densities (± SEM) (F) were calculated from 10 ≤ n ≤ 15 PM sheets selected from different cells. The significance of differences between polarized and depolarized conditions for Lmax and gold labeling were evaluated using bootstrap tests or one-way ANOVA, respectively (*P < 0.05).
To further test the linkage between lipid binding specificity of prenylated PBD anchors and ΔVm responsiveness, we made use of another small GTPase, RAC1. We chose RAC1 because its C-terminal palmitoylated and geranylgeranylated PBD composed of a mix of lysine and arginine residues (LCPPPVKKRKRKCLLL) selectively binds phosphatidic acid (PA) and PIP3, while a farnesylated RAC1 mutant (RAC1G12V-C15 with a C-terminal sequence LCPPPVKKRKRKCVLS) selectively binds PtdSer (27). We found that PM depolarization did not alter the nanoclustering (Fig. 3E) or PM localization (Fig. 3F) of GFP-RAC1G12V. This is consistent with a lipid-mediated mechanism because the spatial distributions of PA and PIP3 are unresponsive to ΔVm (7). By contrast, GFP-RAC1G12V-C15, which binds PtdSer, became responsive to ΔVm, as evidenced by its elevated nanoclustering upon PM depolarization (Fig. 3E). Taken together, these results show that different PtdSer species selectively mediate KRAS responsiveness to Vm and more broadly that the spatiotemporal reorganization of PBD anchors in response to ΔVm is determined by their highly specific binding preferences for both lipid head groups and lipid acyl chains.
Selective Sorting of Distinct PtdSer Species Mediates Sensitivity of KRAS Nanoclustering to Cholesterol Depletion.
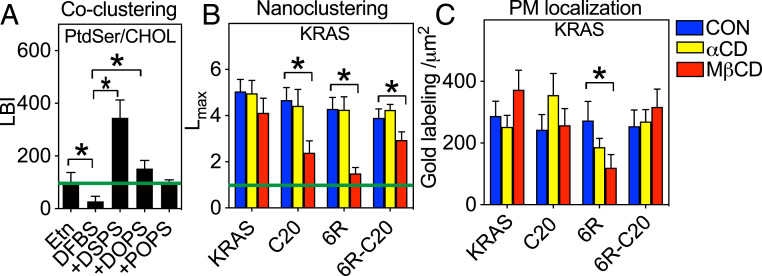
We and others have shown that the PM binding, nanoclustering, and signaling of KRAS is independent of cholesterol in cells and synthetic bilayers (8, 12, 25, 28). Fig. 2 shows that KRAS PBD anchor mutants selectively sort different PtdSer species. Phospholipids with different acyl chains may undergo differential cholesterol-dependent phase separation, such as that observed among phosphatidylcholine (PC) species (29–32). This, then, brings up an interesting possibility that the equally charged KRAS PBD anchor mutants may display distinct cholesterol preferences. To test this, we evaluated how different PtdSer species associate with cholesterol in the PM of PSA3 cells using EM-bivariate analyses to evaluate colocalization between GFP-LactC2 and mCherry-D4H, a specific cholesterol-binding domain (11, 21, 33). Fig. 4A shows that at wild-type PtdSer levels the colocalization parameter LBI for GFP-LactC2 and mCherry-D4H was at the 95% CI, suggesting that the global, heterogeneous population of ∼30 PtdSer species commonly detected in PM weakly colocalizes with cholesterol. PtdSer depletion further decreased colocalization between PtdSer and cholesterol, as evidenced by a reduction in the LBI parameter on Etn starvation (Fig. 4A). In PtdSer-depleted PSA3 cells, all three synthetic PtdSer species incorporate into the PM inner leaflet at equivalent levels (Fig. 3B). However, whereas acute addback of fully saturated DSPS significantly elevated colocalization between the PtdSer and cholesterol probes, addback of DOPS or POPS did not (Fig. 4A). The extent of POPS association with cholesterol is similar to that of the wild-type endogenous PtdSer because POPS is one of the most abundant PtdSer species in mammalian cells (11). These findings strongly suggest that PtdSer species with different acyl chains possess distinct affinities for cholesterol, similar to PC species.
Fig. 4.
KRAS PBD mutant anchors display distinct cholesterol dependence. PSA3 cells coexpressing GFP-LactC2 and the cholesterol probe RFP-D4H were grown in 10 μM Etn to maintain wild-type level of PtdSer, or depleted of endogenous PtdSer without (DFBS), or with acute addback of defined PtdSer species exactly as described in Fig. 2. Intact PM sheets prepared from the cells were coimmunolabeled with anti-GFP coupled to 6-nm gold and anti-RFP coupled to 2-nm gold and colocalization between the PtdSer and cholesterol probes assessed using bivariate K-functions quantified as mean LBI values (± SEM) from 13 ≤ n ≤ 15 PM sheets selected from different cells (A). BHK cells ectopically expressing GFP-KRASG12V, or a GFP-KRASG12V anchor mutant, were untreated (control) or treated with 2% αCD or 2% MβCD for 30 min before preparation of PM sheets, anti-GFP immunogold labeling, and univariate K-function analysis. Mean Lmax values (± SEM) (B) and gold labeling densities (C) (± SEM) calculated from 11 ≤ n ≤ 15 PM sheets selected from different cells. The significance of differences between control and treated conditions for Lmax and gold labeling were evaluated using bootstrap tests or one-way ANOVA, respectively (*P < 0.05).
We next evaluated the cholesterol dependence of different KRAS PBD anchor mutants. Baby hamster kidney (BHK) cells were incubated with 2% α-cyclodextrin (αCD) or 2% methyl-β-cyclodextrin (MβCD) for 30 min before EM-univariate analysis. MβCD acutely depletes cholesterol from the PM, whereas αCD possesses an amphipathic structure similar to MβCD but does not deplete cholesterol. The clustering of GFP-KRASG12V quantified as Lmax was unaffected by cholesterol depletion (MβCD), consistent with previous studies (8, 12, 25). By contrast, MβCD treatment strongly and significantly decreased the Lmax of GFP-KRASG12V-C20 and GFP-KRASG12V-6R (Fig. 4B) and also mislocalized GFP-KRASG12V-6R from the PM (Fig. 4C). MβCD treatment also decreased the Lmax of GFP-KRASG12V-6R-C20, albeit with less potency. The nanoclustering and PM localization of KRAS PBD anchor mutants was not affected by αCD in any of the experiments (Fig. 4 B and C). Taken together, these data show that the equally charged PBD anchors possess distinct cholesterol dependencies as governed by the cholesterol binding capacity of the PtdSer species with which they preferentially interact.
Selective Sorting of Distinct PtdSer Species Mediates PM Curvature Sensing of KRAS.
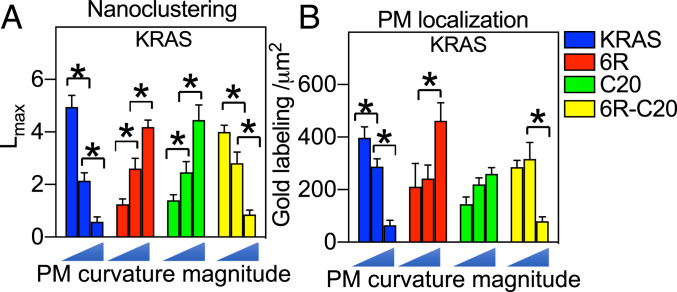
KRAS spatiotemporal organization is sensitive to membrane curvature (21), whereby flatter membranes with low curvatures enhance nanoclustering and membrane binding of full-length KRASG12V as well as the minimal membrane anchor tK-WT (21). The membrane curvature dependence of KRAS is mediated by the selectivity of the KRAS PBD anchor for mixed-chain POPS, over DSPS or DOPS (21). This follows because flattening of PM curvature enhances the PM binding and the nanoclustering of POPS but disrupts PM binding of DSPS and DOPS (21). PtdSer depletion effectively abolishes the response of KRAS to changing PM curvature (21). Acute addback of POPS, but not DSPS or DOPS, selectively restores the PM curvature sensing of KRAS (21). We therefore asked if different PBD anchors given their preferential binding of different PtdSer species would respond to changing membrane curvature in distinct manners. To do this, we manipulated the PM curvature of BHK cells by ectopically expressing the Bin-Amphiphysin-Rvs (BAR) domain of Amphiphysin 2 (BARamph2) to induce positive curvature of the PM (34, 35) or a BAR domain from the planar intestinal- and kidney-specific BAR protein (pinkBAR) to flatten the PM (36, 37). This generates three levels of PM curvature: low curvature (pinkBAR), medium curvature (unperturbed cells), and high curvature (BARamph2). As shown in Fig. 5 A and B, elevating PM curvature dose-dependently disrupted the nanoclustering and reduced the PM localization of GFP-KRASG12V, consistent with previous findings (21). By contrast, GFP-KRASG12V-6R or GFP-KRASG12V-C20 showed a diametrically opposite response, with a dose-dependent increase in nanoclustering upon elevating PM curvature (Fig. 5A). GFP-KRASG12V-6R-C20 responded to the changing PM curvature in a manner similar to KRASG12V (Fig. 5A). The PM localization of each of the KRAS PBD anchor mutants showed a trend similar to the nanoclustering response, although the changes were not as pronounced (Fig. 5B). These results show that the acyl chain composition of the PtdSer species sorted by each equally charged PBD anchor can endow differential responsiveness of the attached GTPase to mechanical perturbations of the PM.
Fig. 5.
KRAS PBD mutant anchors respond to changing PM curvature in distinct manners. BHK cells ectopically coexpressing GFP-KRASG12V, or a GFP-KRASG12V PBD mutant anchor, together with RFP-pinkBAR, an empty vector pC1, or RFP-BARamph2 to induce low curvature, normal, or high curvature, respectively, were grown to ∼80% confluency. PM sheets were prepared on EM grids and immunogold labeling using anti-GFP coupled to 4.5-nm gold. Univariate K-functions were used to analyze the spatial distributions of each anchor. Mean Lmax values (± SEM) for each KRAS anchor (A) calculated from 12 ≤ n ≤ 16 PM sheets selected from different cells. Significant differences between Lmax values were assessed using bootstrap tests (*P < 0.05). (B) Mean number of gold particles per μm2 (± SEM) calculated for the same set of PM sheets. Significant differences between values were assessed using one-way ANOVA (*P < 0.05).
Discussion
The KRAS C-terminal membrane anchor exhibits an exquisite binding preference for monovalent PtdSer over multivalent PIP2 that extends to a preferential interaction with PtdSer species that contain mixed acyl chains (7, 11, 14, 21). In consequence, KRAS nanoclusters on the PM are enriched in mixed-chain PtdSer with one saturated acyl chain and one unsaturated acyl chain, and not with symmetric PtdSer species with two identical acyl chains (11, 21). These observations suggest that the PBD-prenyl anchors, which decorate many small GTPases, encode lipid-binding specificities that cannot be explained in terms of simple electrostatics. Indeed, MD simulations predict that the KRAS PBD anchor adopts well-defined conformational states on membrane, with only select lysine residues and the farnesyl anchor interacting extensively with the bilayer core (11). To formally and rigorously explore the generality of these observations and predictions, we engineered and compared four equally charged KRAS anchors with different combinations of arginine and lysine residues in the PBD in the context of farnesyl or geranylgeranyl groups. We show that each anchor favors distinct conformational orientations and selectively interacts with PtdSer species with different acyl chain combinations, which in turn results in distinct responses to perturbations of PM properties.
The farnesylated hexa-lysine tK anchor samples D (64%), I (29%), and O (6%) states. The dominating structures of the peptide backbone insert into the bilayer and interact with lipid acyl chains to preferentially cluster POPS rather than DSPS around the anchor. Although not directly tested here, more extensive H-bonding between poly-arginine of tK-6R and PtdSer headgroups (Fig. 1B) may lead to more preferential interactions of the saturated chains of DSPS than poly-lysine. Indeed, poly-arginine peptides penetrate membranes more effectively than poly-lysines (38). Consistent with this hypothesis, on intact cell PM farnesylated GFP-KRASG12V-6R preferentially interacted with DSPS, unlike farnesylated GFP-KRASG12V (6K) that selectively interacted with mixed-chain POPS. This preference for different PtdSer acyl chains resulted in distinct responses to cholesterol depletion. Nanoclustering of GFP-KRASG12V has consistently been shown to be insensitive to cholesterol depletion (8, 25, 28), whereas by contrast nanoclustering of GFP-KRASG12V-6R was disrupted by cholesterol depletion. This is consistent with our additional finding that DSPS, but not POPS, associates extensively with cholesterol on the PM. Similarly, the behavior of the two farnesylated anchors to changes in membrane curvature faithfully reflected the differential behavior of the respective preferred PtdSer ligand, DSPS or POPS.
Switching the prenyl group on the 6R PBD from farnesyl to geranylgeranyl fundamentally changed the behavior of the hybrid anchor. The tK-6R-C20 anchor exhibited a distribution of conformational states nearly identical to the wild-type tK anchor. Concordantly, on intact cell PM, GFP-KRASG12V-6R-C20 selectively interacted with mixed-chain POPS but not the saturated DSPS, exactly matching GFP-KRASG12V with the wild-type anchor. In addition, GFP-KRASG12V-6R-C20 responded to PM perturbations, such as depolarization, cholesterol depletion, and PM curvature change in a manner identical to GFP-KRASG12V. The striking correlations between the in silico simulated conformational sampling and EM-spatial analysis of intact cell PM strongly suggest that amino acid sequences and the prenyl chains of PBD anchors synergistically encode selectivity for PtdSer acyl chains. One caveat here is that the simulations only considered the membrane anchor of KRAS. The isolated tK membrane anchor colocalizes extensively with full-length KRASG12V on the PM (25), and the isolated tK anchor and full-length KRASG12V respond to PM perturbations identically (7, 11, 12, 14, 21, 28, 39). Taken together these findings strongly suggest that the isolated tK anchor and the full-length KRASG12V interact with the same set of lipids in the PM, so that in silico predictions based on tK may be directly correlated with the in vivo interactions of KRASG12V observed by EM. However, recent work has shown that N terminus of BRAF, in addition to binding switch 1 of the G-domain, can also engage the C-terminal hypervariable region (HVR) of KRAS (40). This raises the intriguing possibility that recruiting BRAF may impact the lipid sorting specificity of KRAS, with attendant consequences for signal transmission.
The capacity for KRAS membrane-anchoring domain to discriminate between PtdSer species with different acyl chains carries important biological inferences. As the most abundant anionic phospholipid in the PM inner leaflet (comprising ∼20 mol %), PtdSer interacts with a wide variety of PM constituents, including actin, surface receptors, G proteins, and ion channels. However, many PtdSer-associating PM constituents do not colocalize. For instance, caveolae and KRAS nanoclusters are both enriched with PtdSer (14, 41), but KRAS is not a component of caveolae (14, 41, 42). Depleting PtdSer dislodges cholesterol from the PM, and vice versa (18, 43). However, PtdSer-enriched KRAS nanoclusters do not associate with cholesterol (8, 12, 22, 25, 44, 45). These data strongly imply multiple nonoverlapping PtdSer pools in the PM (14, 41, 43). These nonoverlapping PtdSer pools possess distinct properties. For example, we now show that the saturated DSPS extensively associates with cholesterol, whereas the mixed-chain POPS segregates from cholesterol. Taken together, our latest findings explain the seemingly paradoxic observations that the cholesterol-independent KRAS associates with PtdSer while cholesterol depletion mislocalizes some PtdSer from the PM. Furthermore, different PtdSer species respond to ΔVm in distinct manners. In particular, PM depolarization enhances the nanoclustering of DSPS and POPS, while not affecting the nanoclustering of the monounsaturated DOPS. This differential behavior accounts also for the different responsiveness of the KRAS mutant anchors to ΔVm.
The PM localization of KRAS mutant anchors is also sensitive to PtdSer species (Fig. 2). We have previously shown that vesicular transport of PtdSer between the PM and the recycling endosome (RE), as well as between the PM and the endoplasmic reticulum, regulate the trafficking of KRAS to the PM (15–18). Others have shown that high levels of PtdSer on the RE contribute to localization of KRAS to the RE after capture by PDEδ and release by Arl2 (46–49). Thus, to the extent that PtdSer composition of the RE broadly reflects that of the PM, the PtdSer binding preferences of the different PBD anchors will also determine their extent of binding to the RE for forward transport to the PM. In this context the extent of PM localization will reflect the capacity of the PM to retain KRAS, as well as the capacity of the RE to capture KRAS. In both locations the same PtdSer binding preferences of the anchor will operate and so together determine the fraction of KRAS maintained on the PM.
Taken together, our data strongly suggest that multiple nonoverlapping PtdSer pools enriched with distinct PtdSer species contribute to the compartmentalization and specificity of cell signaling from the PM. Acyl chain-dependent lateral compartmentalization of other lipid types has been observed in various model systems. For example, different PC species display clear immiscibility in model bilayers and MD simulations and contribute to the formation of liquid-ordered and liquid-disordered domains in bilayers and lipid rafts in biomembranes (29–31, 50). Similarly, cholesterol has been shown to compartmentalize into spatially nonoverlapping pools in biological membranes (51). These parallel cholesterol pools have been shown to possess distinct properties, such as sensitivity to sphingomyelinase treatment (51). Although anionic lipids have been mostly considered for the contribution of their charged headgroups to electrostatic interactions, we now show convincing evidence that PtdSer species possess similar acyl-chain-dependent immiscibility that can contribute to the complexity of compartmentalization of prenylated GTPases on the PM which detect PtdSer acyl chain structure.
In support of this broader interpretation of our study we note that the C-terminal HVRs of RAS and RHO family GTPases that contains the membrane anchor sequence are 94 to 99% conserved across mammalian species (SI Appendix, Tables S1 and S2), suggesting important conserved biological roles. The predicted prenyl group is 100% conserved and the few amino acid changes that do occur do not involve the polybasic clusters and/or palmitoyl chains that comprise the anchor. By contrast, no HVR or anchor sequence is shared between any pair of RAS or RHO proteins. The inference is that the conserved variation may indeed reflect a complex code for lipid binding specificity that includes both headgroup and acyl chain recognition; this different lipid binding capacity may contribute to differential lateral spatial distribution and intracellular transport and hence biological function.
Materials and Methods
MD Simulations.
All-atom MD simulations were conducted on tK-6R and tK-6R-C20 bound to a POPC/POPS (20%) bilayer and on tK and tK-6R bound to a bilayer in which POPS was replaced by DSPS. Each system contained four peptides, two each in the top and bottom leaflet, and simulated for for up to 2.5 µs using the CHARMM36 force field and the NAMD program. In addition, we extended our previous simulations of tK and tK-C20 to 2 to 2.5 μs to ensure a direct comparison among the six systems. The rest of the simulation details are the same as in a previous report (11). Similarly, analyses of the trajectories in terms of peptide backbone structural properties, patterns of peptide–lipid hydrogen bond interactions, and 2D radial pair distributions were as described previously (11). The 2D MSDs of the C-alpha atoms of the prenyl group in tK or 6R were calculated based on a 300-ns segment spanning 1.3 and 1.7 µs of the respective trajectoires, dividing the data into blocks of 100 ns to estimate errors.
Cell Culture and Plasmids.
BHK cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% bovine calf serum (BCS), while PSA3 cells were maintained in F-12K medium containing 10% FBS. To manipulate endogenous PS levels, PSA3 cells grown in F-12K medium containing 10% DFBS were supplemented with 10 μM Etn for 72 h before harvesting. For transient transfection in EM-spatial analysis, 2 × 105 BHK or PSA3 cells were seeded in a 3.5-cm dish. Following overnight growth, cells were incubated with 1 mL optiMEM containing 0.8 μg of appropriate complementary DNA plasmid (GFP-KRASG12V, GFP-KRASG12V-C20, GFP-KRASG12V-6R, or GFP-KRASG12V-6R-C20) and 7 μL lipofectamine for 5 h. Cells were then switched to normal growth media (DMEM containing 10% BCS for BHK cells, F-12K containing 10% DFBS for PSA3 cells) for overnight growth before experiments.
EM-Spatial Analysis.
EM-univariate spatial analysis.
The univariate K-function analysis quantifies the lateral spatial distribution of a single population of immunolabeled gold nanoparticles on intact PM sheets (7, 11). Intact PM sheets of BHK or PSA3 cells expressing GFP-tagged proteins/peptides were attached to copper EM grids, fixed with 4% paraformaldehyde and 0.1% gluaraldehyde, immunolabeled with 4.5-nm gold nanoparticles conjugated to anti-GFP antibody, and negative-stained with uranyl acetate. TEM was used to image the gold nanoparticles on the PM at 100,000× magnification. ImageJ was then used to assign the coordinates of every gold particle. Nanoclustering of the gold particles within a 1-μm2 PM area was calculated using Ripley’s K-function. The analysis tests a null hypothesis that all points in the selected area are distributed randomly:
| [1] |
| [2] |
where K(r) designates the univariate K-function for the number of gold nanoparticles (n) in an intact PM area of A; r = length scale between 1 and 240 nm with an increment of 1 nm; || · || = Euclidean distance, where the condition of ||xi − xj|| ≤ r yields an indicator function of 1(·) = 1 and the condition of ||xi − xj|| > r yields indicator function of 1(·) = 0. To accomplish an unbiased edge correction, wij−1 characterizes the fraction of the circumference of a circle that has the center at xi and radius ||xi − xj||. K(r) is transformed into L(r) – r, which is then normalized against the 99% CI estimated via Monte Carlo simulations. L(r) − r = 0 for all the corresponding r values indicates a complete random distribution of the nanoparticles. L(r) − r > 99% CI at the corresponding values of r indicates statistically meaningful clustering at the defined length scale. For each condition, at least 15 PM sheets were imaged, analyzed, and pooled. Statistical significance between the polarized and depolarized conditions was weighed via comparing our calculated point patterns against 1,000 bootstrap samples in bootstrap tests (7, 11).
EM-bivariate coclustering analysis.
Colocalization between two populations of gold immunolabeling GFP-tagged and RFP-tagged proteins/peptides is quantified using the bivariate K-function colocalization analysis (7, 11). Intact apical PM sheets of cells coexpressing GFP- and RFP-tagged proteins/peptides were attached and fixed to EM grids. The PM sheets were immunolabeled with 2-nm gold conjugated to anti-RFP antibody and 6-nm gold linked to anti-GFP antibody, respectively. X/Y coordinates of each gold nanoparticle were assigned in ImageJ and the colocalization between the two gold populations was calculated using a bivariate K-function. The analysis is designed to test the null hypothesis that the two point populations spatially segregate from each other (Eqs. 3–6):
| [3] |
| [4] |
| [5] |
| [6] |
where Kbiv(r) is the bivariate estimator composed of two separate bivariate K-functions: Kbs(r) describes the distribution of all the big 6-nm gold particles (b = big gold) with respect to each 2-nm small gold particle (s = small gold) and Ksb(r) describes the distribution of all the small gold particles with respect to each big gold particle. The value of nb is the number of 6-nm big gold particles and the value of ns is the number of 2-nm small gold particles within a PM area of A. Other notations follow the same description as explained in Eqs. 1 and 2. Kbiv(r) is then linearly transformed into Lbiv(r)-r, which was normalized against the 95% CI. An Lbiv(r)-r value of 0 indicates spatial segregation between the two populations of gold particles, whereas an Lbiv(r)-r value above the 95% CI of 1 at the corresponding distance of r indicates yields statistically significant colocalization at certain distance yields. Area under the curve (from the x axis) for each Lbiv(r)-r curve was calculated within a fixed range 10 < r < 110 nm and was termed bivariate Lbiv(r)-r integrated (or LBI):
| [7] |
For each condition, > 15 apical PM sheets were imaged, analyzed and pooled, shown as mean of LBI values ± SEM. Statistical significance between conditions was evaluated via comparing against 1,000 bootstrap samples as described (7, 11).
Supplementary Material
Acknowledgments
This work was supported by a grant from the NIH (National Institute of General Medical Sciences) to A.A.G. and J.F.H. (R01 GM124233).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2014605118/-/DCSupplemental.
Data Availability.
All study data are included in the article and/or SI Appendix.
Change History
July 9, 2021: The supporting information has been updated.
References
- 1.Cox A. D., Fesik S. W., Kimmelman A. C., Luo J., Der C. J., Drugging the undruggable RAS: Mission possible? Nat. Rev. Drug Discov. 13, 828–851 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Downward J., Targeting RAS signalling pathways in cancer therapy. Nat. Rev. Cancer 3, 11–22 (2003). [DOI] [PubMed] [Google Scholar]
- 3.Cox A. D., Der C. J., Philips M. R., Targeting RAS membrane association: Back to the future for anti-RAS drug discovery? Clin. Cancer Res. 21, 1819–1827 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou Y., Hancock J. F., Ras nanoclusters: Versatile lipid-based signaling platforms. Biochim. Biophys. Acta 1853, 841–849 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Zhou Y., Hancock J. F., Deciphering lipid codes: K-ras as a paradigm. Traffic 19, 157–165 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tian T., et al., Plasma membrane nanoswitches generate high-fidelity Ras signal transduction. Nat. Cell Biol. 9, 905–914 (2007). [DOI] [PubMed] [Google Scholar]
- 7.Zhou Y., et al., Signal transduction. Membrane potential modulates plasma membrane phospholipid dynamics and K-Ras signaling. Science 349, 873–876 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Plowman S. J., Muncke C., Parton R. G., Hancock J. F., H-ras, K-ras, and inner plasma membrane raft proteins operate in nanoclusters with differential dependence on the actin cytoskeleton. Proc. Natl. Acad. Sci. U.S.A. 102, 15500–15505 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghosh S., Strum J. C., Sciorra V. A., Daniel L., Bell R. M., Raf-1 kinase possesses distinct binding domains for phosphatidylserine and phosphatidic acid. Phosphatidic acid regulates the translocation of Raf-1 in 12-O-tetradecanoylphorbol-13-acetate-stimulated Madin-Darby canine kidney cells. J. Biol. Chem. 271, 8472–8480 (1996). [DOI] [PubMed] [Google Scholar]
- 10.Inder K., et al., Activation of the MAPK module from different spatial locations generates distinct system outputs. Mol. Biol. Cell 19, 4776–4784 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou Y., et al., Lipid-sorting specificity encoded in K-ras membrane anchor regulates signal output. Cell 168, 239–251.e16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weise K., et al., Membrane-mediated induction and sorting of K-Ras microdomain signaling platforms. J. Am. Chem. Soc. 133, 880–887 (2011). [DOI] [PubMed] [Google Scholar]
- 13.Lin W. C., et al., H-Ras forms dimers on membrane surfaces via a protein-protein interface. Proc. Natl. Acad. Sci. U.S.A. 111, 2996–3001 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou Y., et al., Signal integration by lipid-mediated spatial cross talk between Ras nanoclusters. Mol. Cell. Biol. 34, 862–876 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cho K. J., et al., Staurosporines disrupt phosphatidylserine trafficking and mislocalize Ras proteins. J. Biol. Chem. 287, 43573–43584 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cho K. J., et al., Inhibition of acid sphingomyelinase depletes cellular phosphatidylserine and mislocalizes K-ras from the plasma membrane. Mol. Cell. Biol. 36, 363–374 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van der Hoeven D., et al., Fendiline inhibits K-Ras plasma membrane localization and blocks K-Ras signal transmission. Mol. Cell. Biol. 33, 237–251 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van der Hoeven D., et al., Sphingomyelin metabolism is a regulator of KRAS function. Mol. Cell. Biol. 38, e00373-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kattan W. E., et al., Targeting plasma membrane phosphatidylserine content to inhibit oncogenic KRAS function. Life Sci. Alliance 2, e201900431 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tan L., et al., Acylpeptide hydrolase is a novel regulator of KRAS plasma membrane localization and function. J. Cell Sci. 132, jcs232132 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liang H., et al., Membrane curvature sensing of the lipid-anchored K-Ras small GTPase. Life Sci. Alliance 2, e201900343 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Janosi L., Gorfe A. A., Segregation of negatively charged phospholipids by the polycationic and farnesylated membrane anchor of Kras. Biophys. J. 99, 3666–3674 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee S., et al., Impaired retrograde membrane traffic through endosomes in a mutant CHO cell defective in phosphatidylserine synthesis. Genes Cells 17, 728–736 (2012). [DOI] [PubMed] [Google Scholar]
- 24.Raghupathy R., et al., Transbilayer lipid interactions mediate nanoclustering of lipid-anchored proteins. Cell 161, 581–594 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prior I. A., Muncke C., Parton R. G., Hancock J. F., Direct visualization of Ras proteins in spatially distinct cell surface microdomains. J. Cell Biol. 160, 165–170 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee Y., et al., High-throughput single-particle tracking reveals nested membrane domains that dictate KRas(G12D) diffusion and trafficking. eLife 8, e46393 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maxwell K. N., Zhou Y., Hancock J. F., Rac1 nanoscale organization on the plasma membrane is driven by lipid binding specificity encoded in the membrane anchor. Mol. Cell. Biol. 38, e00186-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou Y., Cho K. J., Plowman S. J., Hancock J. F., Nonsteroidal anti-inflammatory drugs alter the spatiotemporal organization of Ras proteins on the plasma membrane. J. Biol. Chem. 287, 16586–16595 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Veatch S. L., Keller S. L., Organization in lipid membranes containing cholesterol. Phys. Rev. Lett. 89, 268101 (2002). [DOI] [PubMed] [Google Scholar]
- 30.Veatch S. L., Keller S. L., Separation of liquid phases in giant vesicles of ternary mixtures of phospholipids and cholesterol. Biophys. J. 85, 3074–3083 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baumgart T., Hess S. T., Webb W. W., Imaging coexisting fluid domains in biomembrane models coupling curvature and line tension. Nature 425, 821–824 (2003). [DOI] [PubMed] [Google Scholar]
- 32.Simons K., Gerl M. J., Revitalizing membrane rafts: New tools and insights. Nat. Rev. Mol. Cell Biol. 11, 688–699 (2010). [DOI] [PubMed] [Google Scholar]
- 33.Maekawa M., Fairn G. D., Complementary probes reveal that phosphatidylserine is required for the proper transbilayer distribution of cholesterol. J. Cell Sci. 128, 1422–1433 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Frost A., Unger V. M., De Camilli P., The BAR domain superfamily: Membrane-molding macromolecules. Cell 137, 191–196 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peter B. J., et al., BAR domains as sensors of membrane curvature: The amphiphysin BAR structure. Science 303, 495–499 (2004). [DOI] [PubMed] [Google Scholar]
- 36.Pykäläinen A., et al., Pinkbar is an epithelial-specific BAR domain protein that generates planar membrane structures. Nat. Struct. Mol. Biol. 18, 902–907 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao H., Pykäläinen A., Lappalainen P., I-BAR domain proteins: Linking actin and plasma membrane dynamics. Curr. Opin. Cell Biol. 23, 14–21 (2011). [DOI] [PubMed] [Google Scholar]
- 38.Kardani K., Milani A., H Shabani S., Bolhassani A., Cell penetrating peptides: The potent multi-cargo intracellular carriers. Expert Opin. Drug Deliv. 16, 1227–1258 (2019). [DOI] [PubMed] [Google Scholar]
- 39.Lakshman B., et al., Quantitative biophysical analysis defines key components modulating recruitment of the GTPase KRAS to the plasma membrane. J. Biol. Chem. 294, 2193–2207 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Terrell E. M., et al., Distinct binding preferences between ras and raf family members and the impact on oncogenic ras signaling. Mol. Cell 76, 872–884.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ariotti N., et al., Caveolae regulate the nanoscale organization of the plasma membrane to remotely control Ras signaling. J. Cell Biol. 204, 777–792 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Collins B. M., Davis M. J., Hancock J. F., Parton R. G., Structure-based reassessment of the caveolin signaling model: Do caveolae regulate signaling through caveolin-protein interactions? Dev. Cell 23, 11–20 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kay J. G., Koivusalo M., Ma X., Wohland T., Grinstein S., Phosphatidylserine dynamics in cellular membranes. Mol. Biol. Cell 23, 2198–2212 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kapoor S., et al., The role of G-domain orientation and nucleotide state on the Ras isoform-specific membrane interaction. Eur. Biophys. J. 41, 801–813 (2012). [DOI] [PubMed] [Google Scholar]
- 45.Erwin N., Patra S., Winter R., Probing conformational and functional substates of calmodulin by high pressure FTIR spectroscopy: Influence of Ca2+ binding and the hypervariable region of K-Ras4B. Phys. Chem. Chem. Phys. 18, 30020–30028 (2016). [DOI] [PubMed] [Google Scholar]
- 46.Chandra A., et al., The GDI-like solubilizing factor PDEδ sustains the spatial organization and signalling of Ras family proteins. Nat. Cell Biol. 14, 148–158 (2011). [DOI] [PubMed] [Google Scholar]
- 47.Ismail S. A., et al., Arl2-GTP and Arl3-GTP regulate a GDI-like transport system for farnesylated cargo. Nat. Chem. Biol. 7, 942–949 (2011). [DOI] [PubMed] [Google Scholar]
- 48.Schmick M., et al., KRas localizes to the plasma membrane by spatial cycles of solubilization, trapping and vesicular transport. Cell 157, 459–471 (2014). [DOI] [PubMed] [Google Scholar]
- 49.Zimmermann G., et al., Small molecule inhibition of the KRAS-PDEδ interaction impairs oncogenic KRAS signalling. Nature 497, 638–642 (2013). [DOI] [PubMed] [Google Scholar]
- 50.Veatch S. L., Keller S. L., A closer look at the canonical ‘Raft Mixture’ in model membrane studies. Biophys. J. 84, 725–726 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Das A., Brown M. S., Anderson D. D., Goldstein J. L., Radhakrishnan A., Three pools of plasma membrane cholesterol and their relation to cholesterol homeostasis. eLife 3, e02882 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.