Abstract
Methylglyoxal (MGO) is a reactive dicarbonyl metabolite that modifies histones in vivo and induces changes in chromatin structure and function. Here we report the synthesis and application of a chemical probe for investigating MGO-glycation. A two-step synthesis of a Cu-click compatible alkynyl oxoaldehyde probe (AlkMGO) via sequential Dess–Martin and Riley oxidations is presented. This synthesis elevates the accessibility and utility of an important tool for tracking, enriching, and studying MGO-glycation to aid in understanding its underlying biochemical functions.
Graphical Abstract
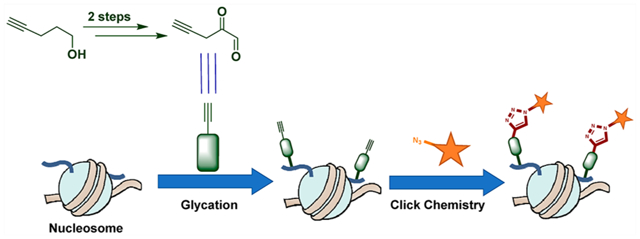
Post-translational modifications (PTMs) on amino acid side chains regulate protein properties including subcellular localization, enzymatic activity, interactions, and turnover.1 The dynamic nature of PTM regulation drives the maintenance of cellular homeostasis and has broad impacts on cellular physiology.1 Among the post-translationally modified eukaryotic proteins, histones bound in chromatin are of major importance due to their intimate relationship with DNA and subsequent effect on DNA-templated processes.2 Histone modifications can directly affect chromatin compaction as well as overall architecture by disrupting the electrostatic interactions between histones and DNA. In addition, they can recruit dedicated effector proteins that function on the chromatin template, thus altering the accessibility of a genomic region to transcription machinery and ultimately impacting cell fate.2 While most well-characterized histone tail modifications are added enzymatically,2 the same sites are capable of reacting nonenzymatically with electrophilic molecules (e.g., environmental toxins and reactive endogenous metabolites) present in their immediate cellular microenvironment.3
Recently, a new type of histone nonenzymatic covalent modification (NECM) induced by a reactive metabolite, methylglyoxal (MGO), was reported.4 MGO is an endogenous byproduct of anaerobic glycolysis, possessing a characteristically electrophilic dicarbonyl moiety reactive to amino acids such as lysines and arginines that are highly enriched in histone tails and are targets of key regulatory PTMs.5 Further oxidations and rearrangements, as well as cross-links, result in a class of species collectively known as advanced glycation end products (AGEs). This process is physiologically and pathologically important as MGO is abundant in disease states such as diabetes and cancer.6 Although MGO levels, which are a reflection of cellular glycolysis rates as well as blood sugar concentrations, are abated by scavenger enzymes such as glyoxalases I and II (GLO1 and GLO2),7 MGO-induced histone damage has been shown to accumulate in human disease.4a
In previous studies, we determined that the MGO-glycation of histones affects the cellular transcriptional program by driving changes in chromatin architecture as well as the epigenetic landscape.4a It does so by competing with enzymatic PTMs but also by changing the biophysical properties of chromatin. As MGO reacts with the positively charged side chains of lysines and arginines, we found that exposure to low concentrations of MGO disrupts the electrostatic interactions of histones with the negatively charged phosphodiester backbone of DNA, thus relaxing chromatin. However, exposure to higher concentrations of MGO induced cross-links between DNA and the histone tails, which ultimately makes chromatin less accessible.4a Finally, we identified an enzymatic regulatory mechanism for these pathophysiological adducts in the form of two different types of deglycases: DJ-1 (PARK7), which protects histones and erases the glycation mark,4 and PAD4 (PADI4), which rewrites MGO-glycation into citrullination.8 Both of these enzymes are reported as oncoproteins,9 which overexpress in breast cancer tumors,4a,8 making them potential druggable anticancer targets.9
While commercial antibodies that target MGO modifications exist, it remains a challenge to specifically track or isolate MGO-modified macromolecules due to the complexity of glycation reactions, which yield a mixture of species. As an alternative, chemical biology approaches have been developed using structurally similar derivatives of reactive metabolites, with trackable moieties, possessing temporal and pharmacokinetic attributes while maintaining high specificity. For example, a bioorthogonal handle-containing MGO mimic (2a) was synthesized (Scheme 1) and recently applied in studying proteome glycation of erythrocytes, which do not contain nuclei.10,11 Here, we design and perform an alternative two-step synthesis of the reported alkynyl methylglyoxal (AlkMGO) probe as well as a new analogue with a shorter carbon chain (2a and 2b, respectively) in gram scales (Scheme 1). We also demonstrate their reactivity and utility by characterizing histone AlkMGO-glycation (Scheme 2) in vitro and in cellulo.
Scheme 1.
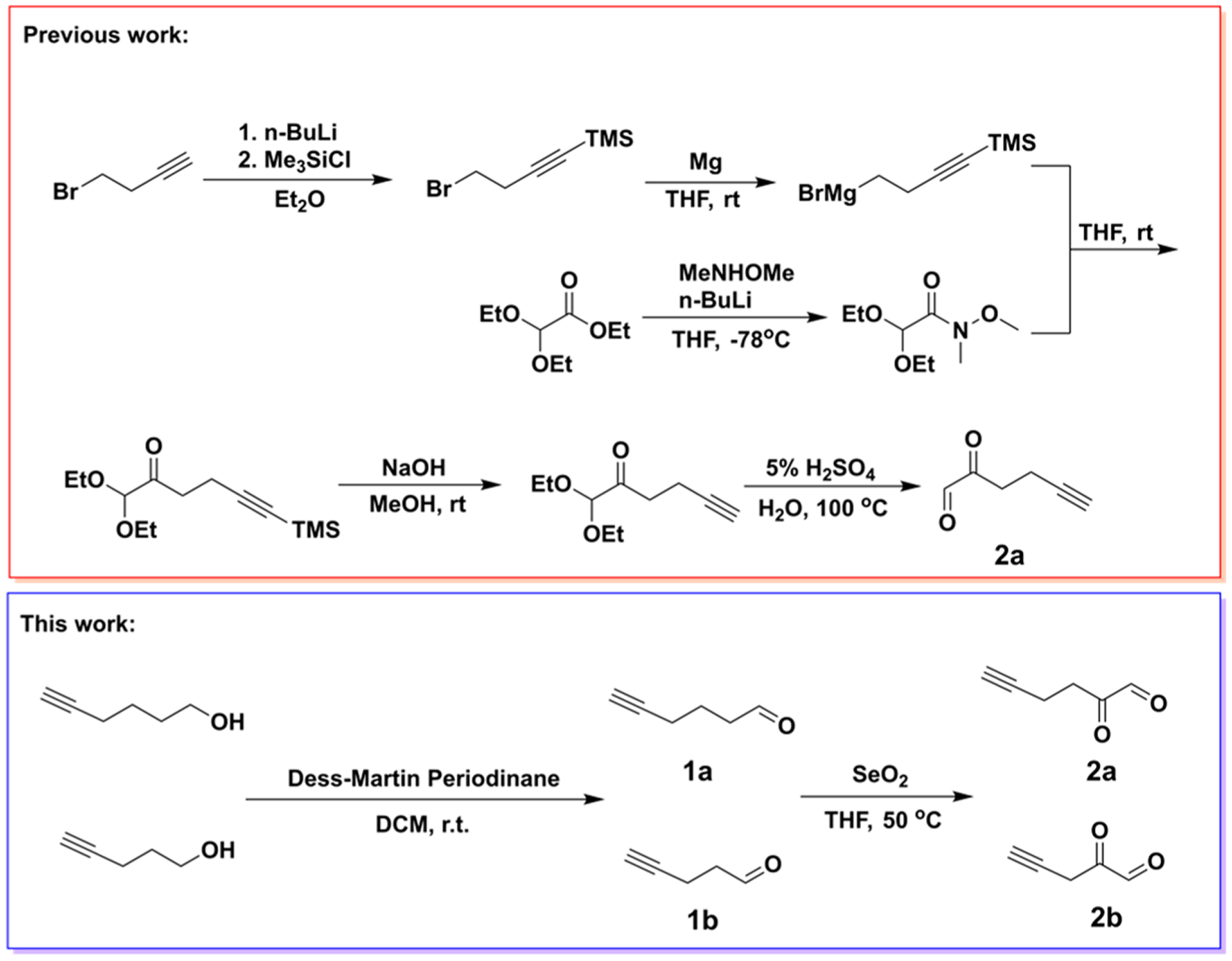
Synthetic Route of AlkMGO Probes 2a and 2b
Scheme 2.
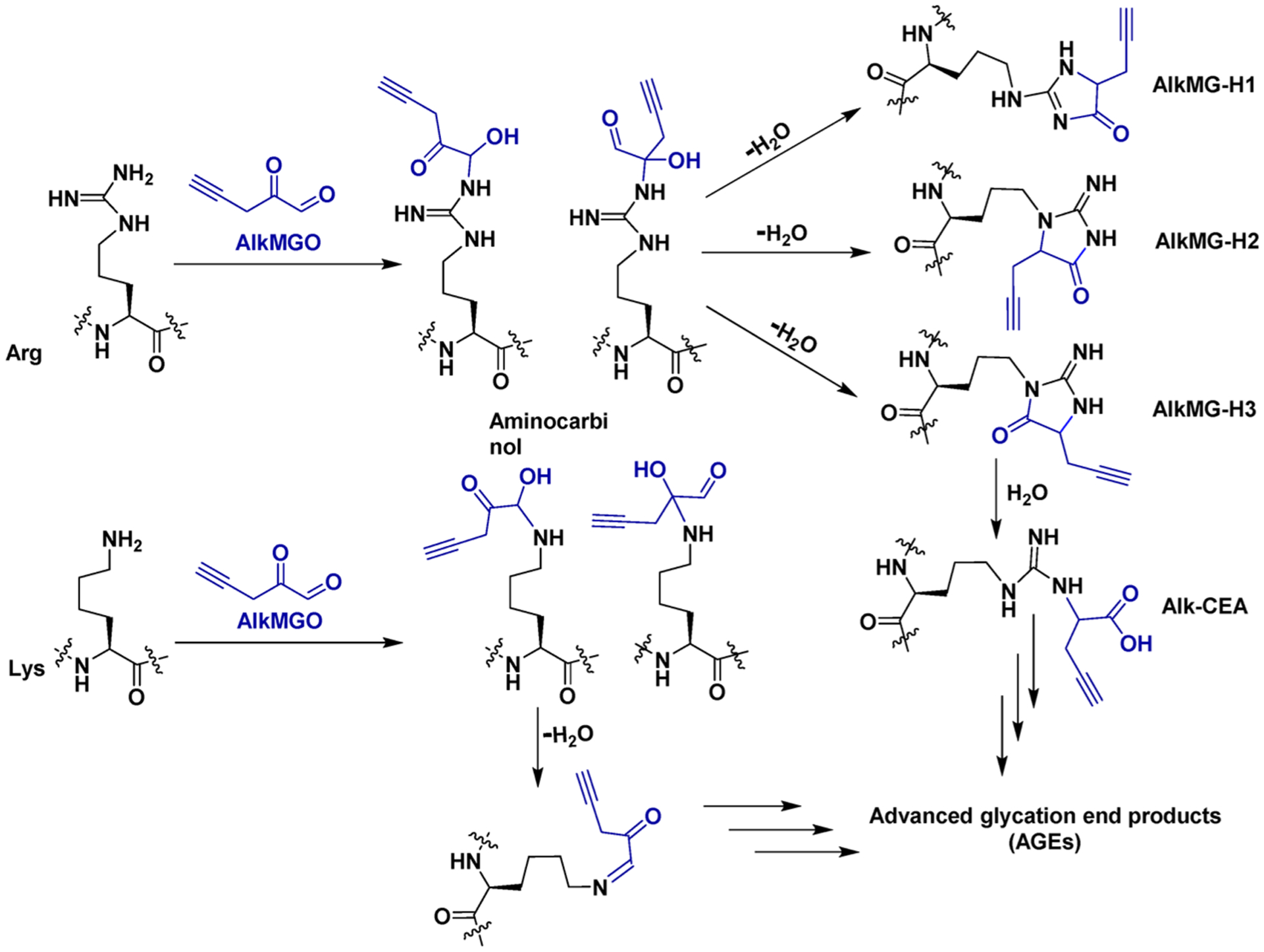
Complex Pathways of AlkMGO (2b)-Mediated Glycation on Lysine (Lys) and Arginine (Arg) Histone Residues
To accomplish the synthesis of these AlkMGO probes, the hydroxyl groups of commercially available 5-hexyn-1-ol or 4-pentyn-1-ol were first converted to their respective aldehydes (1a and 1b) through the Dess–Martin oxidation. The reactions were performed in dichloromethane (DCM) at room temperature, and the products were purified by column chromatography. Subsequent SeO2-catalyzed Riley oxidation of the purified aldehydes was conducted in tetrahydrofuran (THF) at 50 °C to yield the alkynyl α-oxoaldehydes 2a and 2b (Scheme 1). The final products were purified by column chromatography (or vacuum distillation), and the structures were confirmed by HRMS (direct injection and using o-phenylenediamine for derivatization), 1H NMR, and 13C NMR analyses. Although compound 2a has a longer carbon chain than both MGO and 2b, it has been shown to have a similar reactivity as MGO against the scavenger enzymes GLO1 and GLO2.11 Thus, a commercial MGO probing assay was used to examine whether the newly synthesized AlkMGO probe 2b also has a similar chemical reactivity as unmodified MGO. The assay consists of a set of engineered enzymes and a chromophore whose reduction produces a stable signal that is directly proportional to the amount of MGO in the measured samples. Our analysis demonstrates that the synthetic AlkMGO probe 2b has a comparable reactivity to MGO in this assay (Figure 1A). These data highlight that, although the alkyne moiety modifies the three-dimensional structure of MGO, it does not affect the reactivity of the dicarbonyl nor the ability of MGO to be recognized by scavenger enzymes.
Figure 1.
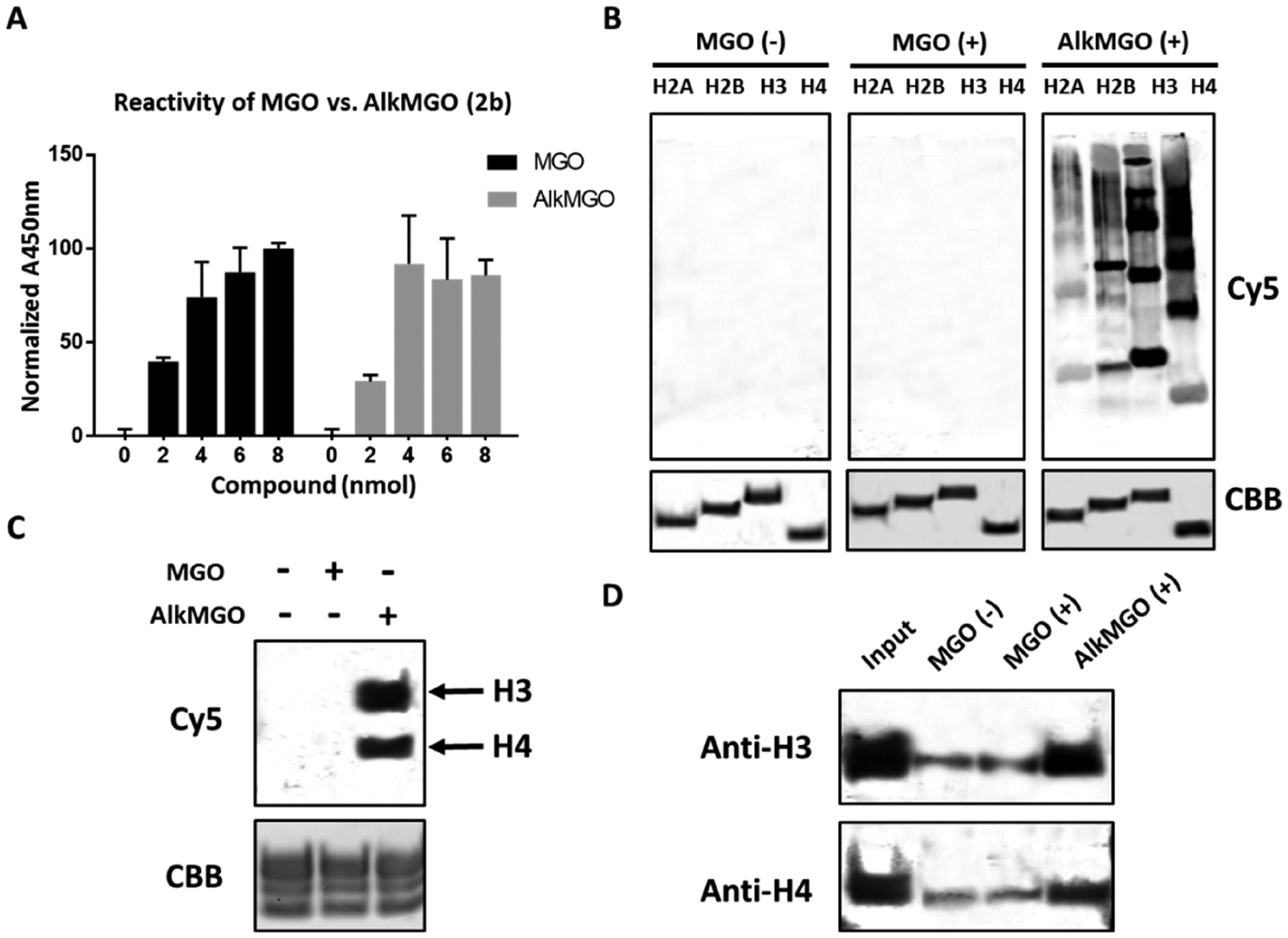
AlkMGO has a similar reactivity as MGO in vitro. (A) A commercial assay for MGO activity was used to evaluate the recognition of AlkMGO by a set of engineered enzymes. (B) Recombinant core histones or (C) assembled nucleosome core particles (NCPs) were incubated with buffer, MGO, or AlkMGO (2b) followed by a standard click reaction with Cy5 azide as described in the Experimental Section. Samples were separated on SDS-PAGE and imaged by in-gel fluorescence. Loading controls were taken at time zero, loaded on SDS-PAGE, and stained by Coomassie Brilliant Blue (CBB). (D) Glycated NCPs from panel C were derivatized with biotin azide and then enriched using magnetic avidin beads as described in the Experimental Section. The beads were washed, and the enriched proteins were separated on SDS-PAGE and analyzed by western blot analysis with the indicated antibodies.
With the new probe 2b in hand, we next aimed to test its reactivity toward histones (Scheme 2), which we have shown by immunoassays to be the primary substrates of MGO in cells. The recombinant core histones H2A, H2B, H3, and H4 were incubated with AlkMGO and subsequently conjugated to the Cy5 fluorophore label using Cy5 azide via established copper-assisted click chemistry.11 The fluorophore-conjugated proteins were visualized by in-gel fluorescence, and as shown in Figure 1B, no background is detected in control samples. In line with our results using antibody-based immunoblot assays, the most reactive histones in this assay are H3 and H4. Cross-linking products are also observed, as exhibited by the higher molecular weight bands in the AlkMGO-treated lane of the in-gel fluorescence channel. This protocol was extended to a more native substrate in the form of nucleosome core particles (NCPs), which represent the fundamental unit of eukaryotic chromatin. NCPs are composed of a histone octamer containing two copies of each core histone (H2A, H2B, H3, and H4) wrapped by 147 base pairs of DNA with a strong nucleosome positioning sequence.4a Results from the reaction of our probe with NCPs indicate a similar reactivity as MGO, with histones H3 and H4 once again the primary targets of glycation (Figure 1C). To further test the utility of the probe for enrichment, following AlkMGO-treatment of NCPs, biotin azide was similarly conjugated via click chemistry to adducts containing the alkyne handle. Next, magnetic avidin beads were used to pull down glycated substrates, and the presence of histones H3 and H4 was determined by western blot analysis. The results, presented in Figure 1D, show that both H3 and H4 are highly enriched, demonstrating again the similar reactivity of AlkMGO to MGO and its suitability for pull-down assays. All of the aforementioned data suggest that the new AlkMGO probe represents a close mimic of MGO and can be utilized for a number of applications, including enrichment of glycated histones and as an alternative method to western blot analysis.
To test the capacity of cells to internalize the AlkMGO probe as well as evaluate its intracellular reactivity, the compound was added to the media of 293T human embryonic kidney cells, which were incubated with the probe for 12 h. Thereafter, cells were lysed and fractionated to soluble proteins and histone fractions. Click labeling of the histone fraction using Cy5 azide followed by in-gel fluorescence was performed to visualize the glycation of histones, which indeed occurs in a dose-dependent manner (Figure 2A). A low background in samples lacking AlkMGO confirms the specificity of the click reaction and analysis procedure. Importantly, similar to our in vitro analyses, the primary glycation substrates are histones H3 and H4, and increased cross-linking is observed at high concentration treatments (Figure 2A). Comparing these results to those generated using an Anti-MGO antibody in an immunoassay4a reveals similar cross-linking aggregation patterns, indicating that both methods illustrate the same species and act as complementary detection approaches. A competition assay (Figure S1) and the detection of enzymatic histone PTMs depletion following treatment (Figure S2) further demonstrate that the new AlkMGO probe has the same reactivity and selectivity against histones as MGO.
Figure 2.
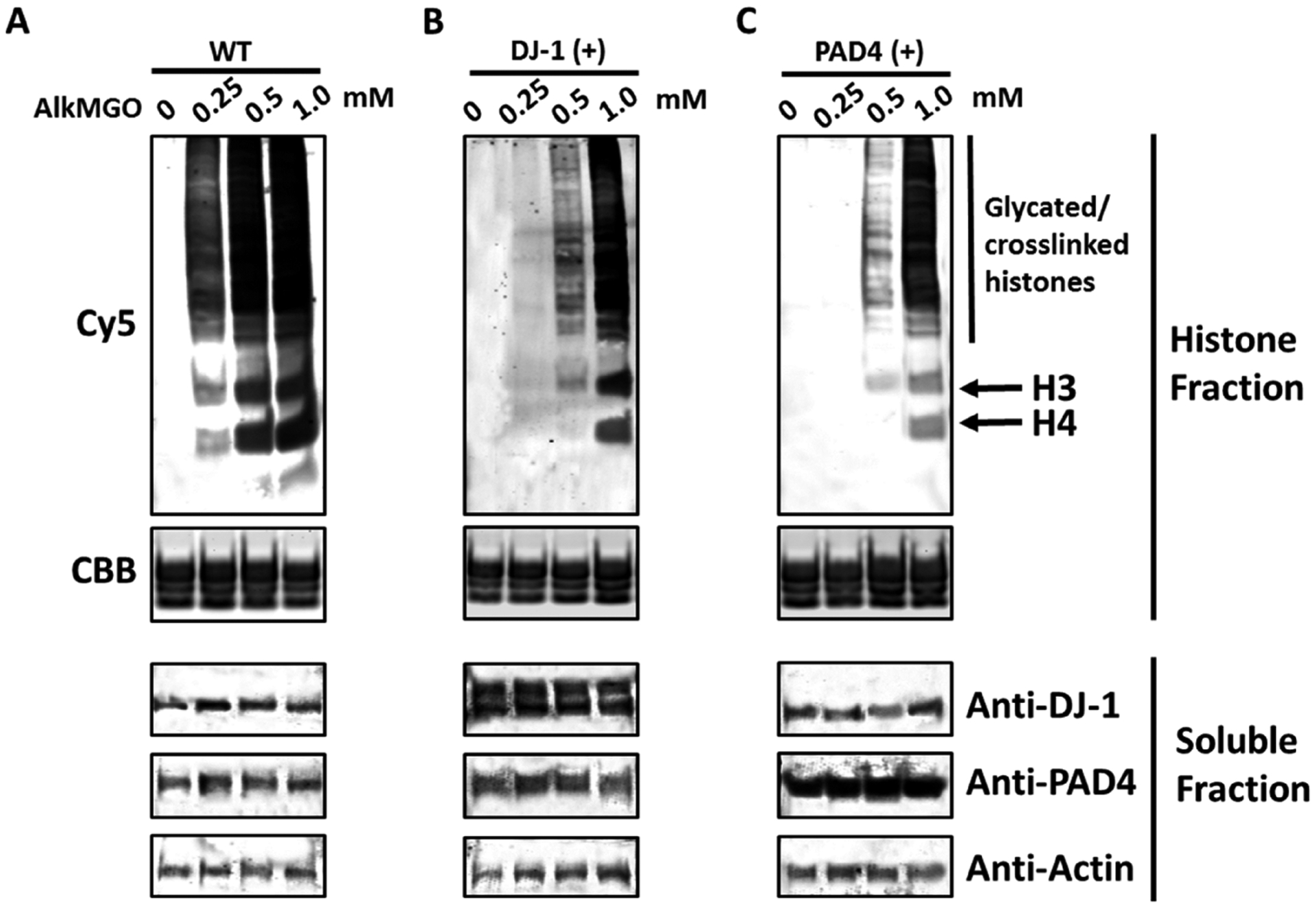
AlkMGO is internalized by mammalian cells, and histone AlkMGO-glycation is subjected to enzymatic regulation. HEK 293T cells were treated with increasing amounts of AlkMGO for 12 h. Following 2 h of recovery, cells were transiently transfected with (A) empty vectors or constructs to overexpress either (B) DJ-1 or (C) PAD4 for 24 h. Next, cells were harvested and fractionated to histone and soluble protein fractions. The extracted histones were derivatized with Cy5-azide as described in the Experimental Section. Samples were separated by SDS-PAGE, and gels were imaged by in gel-fluorescence followed by Coomassie Brilliant Blue staining for loading control. The soluble fractions were analyzed by western blot analysis with the indicated antibodies.
To test whether the new AlkMGO probe can be utilized in studying the enzymatic regulations of histone glycation, we turned to our previously established pulse-chase assay in mammalian cells.4a,8 In this case, 293T cells were first treated with increasing amounts of AlkMGO for 12 h, and after a media change, followed by 2 h of recovery, cells were transiently transfected with either the regulatory proteins DJ-1 (histone glycation eraser) or PAD4 (histone glycation rewriter) for an additional 24 h. Thereafter, the cells were harvested, histones were extracted and analyzed through the clicking of a Cy5 fluorophore as described above. As shown in Figure 2B,C, the overexpression of either regulatory enzyme after treatment with the AlkMGO probe led to a reduction in the fluorescent output signal. These results suggest that both deglycases, DJ-1, and PAD4, are able to reverse AlkMGO-induced histone glycation. Moreover, it illustrates the versatility of DJ-1 and PAD4 in terms of substrate tolerance and outlines the key roles they play in cellular mechanisms surrounding histone glycation regulation. Finally, these results indicate that the exogenous treatment of AlkMGO (2b) in cultured cell lines can induce histone glycation that is regulated by the deglycases DJ-1 and PAD4, mimicking the dynamic nature of glycation and deglycation in metabolically compromised cells.4a,8
MGO-induced histone glycation is a newly identified pathophysiological damage that links metabolism and epigenetic regulation.4,12 However, the complexity of MGO-adducts on both proteins (Scheme 2) and nucleic acids presents a challenge in detecting and studying these events on chromatin both in vitro and in vivo. Over the past decades, chemical biology strategies have shown increasing applicability in characterizing small molecule-mediated protein PTMs,13,14 particularly in glycol-adducts,14 providing a number of key tools and advancing our understanding of their functions. The synthetic route performed in this study can be used for producing not only the previously reported AlkMGO probe 2a but also a newly developed probe 2b that is a closer mimic of MGO. The in vitro and in cellulo evaluations of its biochemical reactivity demonstrate that 2b can be used for detecting and enriching glycated proteins.
In summary, here we have described an efficient two-step synthetic route to generate alkynyl-modified methylglyoxal derivatives used to monitor histone glycation in vitro and in cells. The AlkMGO probe was also applied to enrich modified histones and study the biochemical mechanisms of two distinct histone deglycases, DJ-1 and PAD4, making it well-suited for future applications for both high-throughput-based approaches as well as in vivo fluorescence imaging assays.
EXPERIMENTAL SECTION
Materials and Methods.
All solvents and reagents for the synthesis were purchased from Sigma-Aldrich Corporation or Fisher Scientific and used without further purification. NMR spectra were recorded on Bruker UltraShield Plus 500 MHz instruments at 24 °C in the designated solvents. Chemical shifts are expressed in ppm relative to TMS (1H, 0 ppm) or solvent signals: CD2 Cl2 (1H, 5.32 ppm; 13C, 53.5 ppm) or THF-d8 (1H, 3.58 ppm; 13C, 67.21 ppm); coupling constants are expressed in hertz (Hz). High performance liquid chromatography electrospray ionization mass spectrometry (HPLC-ESI-MS) analysis was performed on an Agilent 6120 Quadrupole LC/MS spectrometer (Agilent Technologies). Biochemical reagents and cell culture media were purchased from Fisher Scientific or Sigma-Aldrich Corporation unless otherwise stated. Western blots and gels were imaged on an Odyssey CLx Imaging System (Li-Cor).
General Procedure for the Synthesis of 1.
A mixture of the alkyne alcohol (1 equiv, 35.66 mmol) and Dess–Martin Periodinane (1.02 equiv, 36.54 mmol) in 100 mL of dichloromethane was vigorously stirred at room temperature for 12 h. Thereafter, the reaction was cooled in an ice–water bath for 10 min before a white precipitate was filtered off using celite. The filtrate was quenched using 250 mL of saturated sodium bicarbonate containing 2.5% sodium thiosulfate solution and stirred for 1 h at room temperature. The two layers were separated, and the aqueous phase was extracted three times using dichloromethane. The combined organic fractions were dried over anhydrous sodium sulfate, filtered over cotton, and then concentrated under reduced pressure, producing a yellow oil. The crude was purified on a silica column using 5–20% ether in pentane. The product was tracked using thin-layer chromatography (TLC) in 2:5 ethyl acetate/hexane eluent and visualized using potassium permanganate stain. The pure fractions were combined and concentrated under reduced pressure to yield a colorless oil.
Hex-5-ynal (1a).
Following the general procedure, 3 g of 4-pentyn-1-ol was used as the starting material and 1a was prepared as a colorless oil (2.1 g, 72%).
1H NMR (500 MHz, CD2Cl2): δ 9.75 (t, J = 1.3 Hz, 1H), 2.55 (td, J = 7.3, 1.3 Hz, 3H), 2.24 (td, J = 7.0, 2.7 Hz, 3H), 2.00 (t, J = 2.7 Hz, 1H), 1.81 (p, J = 7.1 Hz, 3H). 13C{1H} NMR (151 MHz, CD2Cl2): δ 202.0, 83.7, 69.3, 42.9, 21.3, 18.1.
Pent-4-ynal (1b).
Following the general procedure, 3.5 g of 5-hexyn-1-ol was used as the starting material, and 1b was prepared as a colorless oil (2.6 g, 76%).
1H NMR (500 MHz, CD2Cl2): δ 9.73 (s, 1H), 2.65 (t, J = 7.1 Hz,2H), 2.47 (dd, J = 7.1, 2.6 Hz, 2H), 2.00 (t, J = 2.5 Hz, 1H). 13C{1H}NMR (151 MHz, CD2Cl2): δ 200.5, 82.9, 69.2, 42.7, 11.9.
General Procedure for the Synthesis of 2.
To a mixture of the alkyne aldehyde (1 equiv, 6.1 mmol) in 10 mL of tetrahydrofuran, selenium dioxide powder (1.02 equiv, 6.2 mmol) was added. The reaction was stirred at 50 °C for 2.5 h and monitored by TLC in 2:5 ethyl acetate/hexane eluent for visualizing the disappearance of the starting material. The reaction was next filtered through celite before direct vacuum distillation or being quenched with a saturated sodium bicarbonate solution (20 mL). The organic solvent was removed using a vacuum, and the aqueous phase was extracted three times using diethyl ether. The combined organic fractions were washed with brine and dried over anhydrous sodium sulfate, and the solvent was removed under reduced pressure to produce a crude red oil. The oil was purified using a silica column with 0–20% ether in pentane. Fractions were analyzed by TLC, combined, and concentrated under reduced pressure to yield a colorless liquid.
Synthesis of 2-Oxohex-5-ynal (2a).
Following the general procedure, 2a was prepared as a colorless oil (200 mg, 34%).
1H NMR (500 MHz, THF-d8): δ 9.53 (s, 1H), 2.19 (t, J = 7.3 Hz, 1H), 2.09−2.00 (m, 1H), 1.57 (p, J = 7.2 Hz, 1H). 13C{1H} NMR(151 MHz, THF-d8): δ 201.5, 174.8, 83.8, 69.9, 32.8, 18.1. HRMS (ESI): calcd for C6H6O2 (2a) [M + H]+ 110.0368, found 110.0370; calcd for C12H10N2 (2a + o-phenylenediamine) [M + H]+ 183.0922, found 183.0920.
Synthesis of 2-Oxopent-4-ynal (2b).
Following the general procedure, 2b was prepared as a colorless oil (250 mg, 43.1%).
1H NMR (500 MHz, THF-d8): δ 9.50 (s, 1H), 2.44 (t, J = 7.2 Hz, 2H), 2.26 (dd, J = 7.5, 3.4 Hz, 3H), 2.07 (t, J = 2.7 Hz, 1H), 2.04 (t, J = 2.5 Hz, 1H). 13C{1H} NMR (151 MHz, THF-d8): δ 200.0, 173.2,83.2, 69.9, 33.5. HRMS (ESI): calcd for C5H4O2 (2b) [M + H]+96.0211, found 96.0214; calcd for C11H8N2 (2b + o-phenylenediamine) [M + H]+ 169.0766, found 169.0768.
Methylglyoxal Activity Assay.
A commercially available methylglyoxal assay kit (Abcam ab241006) was used as per the manufacturer’s instructions to verify the presence of the dicarbonyl. Absorbance measurements were taken at 450 nm for each concentration of the MGO standard or AlkMGO probe in triplicate using a SPECTRAmax M series microplate reader. The corresponding data were analyzed using SoftMax Pro software.
Recombinant Histone Expression and Purification.
Recombinant human histones H2A, H2B, H3.2, and H4 were expressed in E. coli BL21 (DE3) or E. coli C41 (DE3), extracted from inclusion bodies by guanidine hydrochloride, and purified by flash reverse chromatography or semipreparative HPLC as previously described.4a The purity of the histones was confirmed by SDS-PAGE (Figure 1B and Figure S4) and RP-LC-ESI-MS (Figure S3).
Histone Octamer and “601” DNA Preparation.
Histone octamers were prepared as previously described.4a Briefly, recombinant histones were dissolved in unfolding buffer (20 mM Tris-HCl, 6 M GdmCl, 0.5 mM DTT, pH 7.5), and combined with the following stoichiometry: 1.1 equiv of H2A, 1.1 equiv of H2B, 1 equiv of H3.2, 1 equiv of H4. The combined histone solution was adjusted to 1 mg/mL concentration and transferred to a dialysis cassette with a 7000 Da molecular cutoff. Octamers were assembled by dialysis at 4 °C against 3 × 1L of octamer refolding buffer (10 mM Tris-HCl, 2 M NaCl, 0.5 mM EDTA, 1 mM DTT, pH 7.5) and subsequently purified by size exclusion chromatography on a Superdex S200 10/300 column. Fractions containing octamers were combined, concentrated, diluted with glycerol to a final 50% v/v, and stored at −20 °C. The 147-bp 601 DNA fragment was prepared by digestion from a plasmid containing 30 copies of the desired sequence (flanked by blunt EcoRV sites on either side) and purified by PEG-6000 precipitation as previously described.4a
Preparation of Nucleosome Core Particles.
Mononucleosome assembly was performed as previously described using salt dilution.4a Briefly, purified histone octamers were mixed together with “601” DNA (1:1 molar ratio) in 2 M salt solution (10 mM Tris pH 7.5, 2 M NaCl, 1 mM EDTA, 1 mM DTT) in 10 μL. After incubation at 37 °C for 15 min, the mixture was gradually diluted (9 × 15 min) at 30 °C by dilution buffer (10 mM Tris pH 7.5, 10 mM NaCl, 1 mM EDTA, 1 mM DTT) to 200 μL. The assembled mononucleosomes were concentrated and then characterized by native gel electrophoresis (5% acrylamide gel, 0.5 × TBE, 120 V, 40 min) and ethidium bromide (EtBr) staining.
In Vitro Glycation Studies and in-Gel Imaging.
The free histone glycation assay (100 μL) was prepared on ice and contained 20 μM of the designated histone (H2A, H2B, H3, or H4) in 1× PBS buffer (pH 7.4) and 0.5 mM of either MGO or AlkMGO (2b). The reactions were incubated at 37 °C for 12 h and quenched by α-aminoguanidine. Protein samples were added to a premixed solution containing 3 μL of 10 mM Cy5 azide (Sigma-Aldrich, 777323), 10 μL of a 3:7 mixture of 50 mM CuSO4, and 100 mM THPTA, and vortexed. Thereafter, 5 μL of 100 mM freshly made TCEP was added to initiate the click reaction followed by incubation (1 h) at room temperature. Then, 10 μL of 0.5 M EDTA was added to quench the reactions. Excess reagents were removed by MeOH/CHCl3 protein precipitation followed by centrifugation. Pellets were washed in 500 μL of MeOH/H2O (9:1) prior to the second centrifugation. The air-dried protein samples were then analyzed by SDS-PAGE, followed by in-gel imaging using the Odyssey CLx Imaging System (wavelength 680 nm). The gel was stained using standard Coomassie staining to evaluate total protein loading. Nucleosome glycation assays were similarly prepared using 0.1 μM NCPs and 2 mM of either MGO or AlkMGO (2b). The reactions were prepared on ice, incubated at 37 °C for 72 h, and quenched by α-aminoguanidine. The glycated NCPs were labeled via a similar click reaction and analyzed by SDS-PAGE, followed by in-gel fluorescence.
Enrichment of AlkMGO-Modified Histones.
Fifty μL of NCPs treated with AlkMGO (2b) was added to a premixed solution containing 3 μL of 10 mM biotin azide (Sigma-Aldrich, 762024), 10 μL of a 3:7 mixture of 50 mM CuSO4, and 100 mM THPTA and vortexed. Thereafter, 5 μL of 100 mM freshly made TCEP was added to initiate the click reaction followed by incubation (1 h) at room temperature. Then, 10 μL of 0.5 M EDTA was added to quench the reactions. Excess reagents were removed by MeOH/CHCl3 protein precipitation or concentration–dilution using a 0.5 mL Centrifugal Filter (3K, Millipore). The modified histone proteins were enriched by BSA-blocked Streptavidin Magnetic Beads (Thermo Scientific). Next, beads were washed three times with 1× PBS buffer (pH 7.4), boiled, separated on SDS-PAGE, and analyzed by western blot with anti-H3 (Abcam, ab10799) and anti-H4 (CST, 2592S) antibodies.
In Cellulo Deglycation Assays.
HEK 293T cells were treated with a gradient of AlkMGO 2b (0, 0.25, 0.5, and 1.0 mM) for 12 h, after which the medium was changed to AlkMGO-free DMEM. Cells were cultured for an additional 2 h, after which they were transfected with the indicated plasmid using the Lipofectamine 2000 Transfection Reagent (Thermo Fisher Scientific) according to the manufacturer’s protocol. The pGw1-Myc-DJ1 plasmid was a kind gift from Mark Cookson (Addgene plasmid no. 29347), and the pCMV-HA-PAD4 plasmid was reported in our previous study.8 After overnight incubation, cells were harvested and fractionated to cytosolic and histone fractions as described before.4a,8 After dialysis, the histone fraction was analyzed by the aforementioned click labeling and in-gel imaging, while DJ-1 and PAD4 overexpression was detected by western blot analysis using anti-DJ-1 (CST, 5933S) and anti-PAD4 (Abcam, ab50332) antibodies. Anti-Actin was used as a loading control for soluble fractions.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Jian Li at the Scripps Research Institute and Dr. Yao Xu and Dr. Yu Zong at MSKCC for their insightful suggestions and generous help in the synthesis presented in this study. Work in the David lab is supported by R21 DA044767, CCSG core grant P30 CA008748, SPORE P50-CA192937 from the National Institutes of Health. In addition, work in the lab is supported by the Tri-institutional Therapeutic Discovery Institute, the Mr. William H. Goodwin and Mrs. Alice Goodwin and the Commonwealth Foundation for Cancer Research and the Center for Experimental Therapeutics at MSKCC, the Pershing Square Sohn Cancer Alliance and Cycle for Survival. Y.D. is a Josie Robertson Young Investigator.
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.joc.9b02504.
Competition treatment of the AlkMGO probe and MGO to 293T cells, impacts of AlkMGO-glycation on other histone PTMs, LC-ESI-MS analysis of recombinant histones, full gels and uncropped immunoblots, HRMS analysis of compounds 2a and 2b, and NMR spectra of the synthesized compounds in this study (PDF)
The authors declare no competing financial interest.
REFERENCES
- (1).(a) Deribe YL; Pawson T; Dikic I Post-translational modifications in signal integration. Nat. Struct. Mol. Biol 2010, 17, 666–672. [DOI] [PubMed] [Google Scholar]; (b) Müller MM Post-translational modifications of protein backbones: Unique functions, mechanisms, and challenges. Biochemistry 2018, 57, 177–185. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Kouzarides T Chromatin modifications and their function. Cell 2007, 128, 693–705. [DOI] [PubMed] [Google Scholar]
- (2).(a) Peterson CL; Laniel MA Histones and histone modifications. Curr. Biol 2004, 14, R546–R551. [DOI] [PubMed] [Google Scholar]; (b) Strahl BD; Allis CD The language of covalent histone modifications. Nature 2000, 403, 41–45. [DOI] [PubMed] [Google Scholar]; (c) Jenuwein T; Allis CD Translating the histone code. Science 2001, 293, 1074–1080. [DOI] [PubMed] [Google Scholar]; (d) Patel DJ; Wang Z Readout of epigenetic modifications. Annu. Rev. Biochem 2013, 82, 81–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).(a) Zheng Q; Prescott NA; Maksimovic I; David Y De)Toxifying the epigenetic code. Chem. Res. Toxicol 2019, 32, 796–807. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Harmel R; Fiedler D Features and regulation of nonenzymatic posttranslational modifications. Nat. Chem. Biol 2018, 14, 244–252. [DOI] [PubMed] [Google Scholar]
- (4).(a) Zheng Q; Omans ND; Leicher R; Osunsade A; Agustinus AS; Finkin-Groner E; D’Ambrosio H; Liu B; Chandarlapaty S; Liu S; David Y Reversible histone glycation is associated with disease-related changes in chromatin architecture. Nat. Commun 2019, 10, 1289. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Galligan JJ; Wepy JA; Streeter MD; Kingsley PJ; Mitchener MM; Wauchope OR; Beavers WN; Rose KL; Wang T; Spiegel DA; Marnett LJ Methylglyoxal-derived posttranslational arginine modifications are abundant histone marks. Proc. Natl. Acad. Sci. U. S. A 2018, 115, 9228–9233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Allaman I; Bélanger M; Magistretti PJ Methylglyoxal, the dark side of glycolysis. Front. Neurosci 2015, 9, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).(a) Chaplen FW; Fahl WE; Cameron DC Evidence of high levels of methylglyoxal in cultured Chinese hamster ovary cells. Proc. Natl. Acad. Sci. U. S. A 1998, 95, 5533–5538. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Nokin MJ; Durieux F; Peixoto P; Chiavarina B; Peulen O; Blomme A; Turtoi A; Costanza B; Smargiasso N; Baiwir D; Scheijen JL; Schalkwijk CG; Leenders J; De Tullio P; Bianchi E; Thiry M; Uchida K; Spiegel DA; Cochrane JR; Hutton CA; De Pauw E; Delvenne P; Belpomme D; Castronovo V; Bellahcène A Methylglyoxal, a glycolysis side-product, induces Hsp90 glycation and YAP-mediated tumor growth and metastasis. eLife 2016, 5, e19375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).(a) Distler MG; Palmer AA Role of Glyoxalase 1 (Glo1) and methylglyoxal (MG) in behavior: recent advances and mechanistic insights. Front. Genet 2012, 3, 250. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Xu Y; Chen X Glyoxalase II, a detoxifying enzyme of glycolysis byproduct methylglyoxal and a target of p63 and p73, is a pro-survival factor of the p53 family. J. Biol. Chem 2006, 281, 26702–26713. [DOI] [PubMed] [Google Scholar]
- (8).Zheng Q; Osunsade A; David Y Protein arginine deiminase 4 antagonizes methylglyoxal-induced histone glycation. bioRxiv 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).(a) Cao J; Lou S; Ying M; Yang B DJ-1 as a human oncogene and potential therapeutic target. Biochem. Pharmacol 2015, 93, 241–250. [DOI] [PubMed] [Google Scholar]; (b) Chang X; Han J; Pang L; Zhao Y; Yang Y; Shen Z Increased PADI4 expression in blood and tissues of patients with malignant tumors. BMC Cancer 2009, 9, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Tashiro S; Caaveiro JMM; Nakakido M; Tanabe A; Nagatoishi S; Tamura Y; Matsuda N; Liu D; Hoang QQ; Tsumoto K Discovery and optimization of inhibitors of the parkinson’s disease associated protein DJ-1. ACS Chem. Biol 2018, 13, 2783–2793. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Lewis HD; Liddle J; Coote JE; Atkinson SJ; Barker MD; Bax BD; Bicker KL; Bingham RP; Campbell M; Chen YH; Chung CW; Craggs PD; Davis RP; Eberhard D; Joberty G; Lind KE; Locke K; Maller C; Martinod K; Patten C; Polyakova O; Rise CE; Rüdiger M; Sheppard RJ; Slade DJ; Thomas P; Thorpe J; Yao G; Drewes G; Wagner DD; Thompson PR; Prinjha RK; Wilson DM Inhibition of PAD4 activity is sufficient to disrupt mouse and human NET formation. Nat. Chem. Biol 2015, 11, 189–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Sibbersen C; Palmfeldt J; Hansen J; Gregersen N; Jørgensen KA; Johannsen M Development of a chemical probe for identifying protein targets of α-oxoaldehydes. Chem. Commun. (Cambridge, U. K.) 2013, 49, 4012–4014. [DOI] [PubMed] [Google Scholar]
- (11).Sibbersen C; Schou Oxvig AM; Bisgaard Olesen S; Nielsen CB; Galligan JJ; Jørgensen KA; Palmfeldt J; Johannsen M Profiling of methylglyoxal blood metabolism and advanced glycation end-product proteome using a chemical probe. ACS Chem. Biol 2018, 13, 3294–3305. [DOI] [PubMed] [Google Scholar]
- (12).Diao X Histone glycation: Linking metabolic perturbation with epigenetic misregulation in cancer. AIMS Genet. 2019, 6, 14–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).(a) Yang A; Cho K; Park HS Chemical biology approaches for studying posttranslational modifications. RNA Biol. 2018, 15, 427–440. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Chu GC; Pan M; Li J; Liu S; Zuo C; Tong ZB; Bai JS; Gong Q; Ai H; Fan J; Meng X; Huang YC; Shi J; Deng H; Tian C; Li YM; Liu L Cysteine-aminoethylation-assisted chemical ubiquitination of recombinant histones. J. Am. Chem. Soc 2019, 141, 3654–3663. [DOI] [PubMed] [Google Scholar]
- (14).Bertozzi CR; Kiessling LL Chemical glycobiology. Science 2001, 291, 2357–2364. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


