Abstract
Gastric cancer is the leading cause of cancer-related mortality worldwide. Given the importance of gastric cancer in public health, identifying biomarkers associated with disease onset is an important part of precision medicine. The hedgehog signaling pathway is considered as one of the most significant widespread pathways of intracellular signaling in the early events of embryonic development. This pathway contributes also to the maintenance of pluripotency of cancer stem cells pluripotency. In this study, we analyzed the expression levels of sonic hedgehog (Shh) signaling pathway genes IHH, BOC, RAB23a and their regulatory miRNAs including MIR-195-5p, MIR-509-3-5p, MIR-6738-3p in gastric cancer patients. In addition, the impact of infection status on the expression level of those genes and their regulatory miRNAs was investigated. One hundred samples taken from 50 gastric cancer patients (50 tumoral tissues and their adjacent non-tumoral counterparts) were included in this study. There was a significant difference in all studied genes and miRNAs in tumoral tissues in comparison with their adjacent non-tumoral counterparts. The lower expression of IHH, BOC, RAB23, miR-195-5p, and miR-6738-3p was significantly associated with more advanced cancer stage. Additionally, IHH upregulation was significantly associated with CMV infection (P < 0.001). Also, receiver operating characteristic (ROC) curve analysis indicated that mir-195 was significantly related to several clinicopathological features including tumor stage, grade, age, gender, and infection status of gastric cancer and can be considered as a potential diagnostic biomarker for gastric cancer. This study confirms the important role of Shh signaling pathway genes in gastric cancer tumorigenesis and their potential as novel molecular biomarkers and therapeutic targets.
Subject terms: Gastric cancer, Biomarkers
Introduction
Gastric cancer is among the most frequently diagnosed neoplasia in both sexes and the third leading cause of death worldwide, causing about 1,000,000 new cases and 783,000 deaths in 20181. Remarkably, it has been shown that gastric cancer, similar to other types of cancer, develops upon accumulation of genetic and epigenetic abnormalities2–4. These alterations can be triggered by a wide variety of external factors such as diet, life style, and infections5–8. Relatively, genes that play important roles in various cellular processes via cell signaling pathways could be affected9,10. Inappropriate activation of one or several signaling pathways may contribute to tumor initiation and progression11,12. The sonic hedgehog (Shh) is considered as one of the most significant intracellular signaling cascades in the early events of embryonic development that regulate a wide range of biological functions, such as cell growth, differentiation, tissue patterning, and vascularisation13,14. Moreover, emerging evidence has suggested that aberrant activation of hedgehog (Hh) is associated with a variety of human cancers such as brain, gastrointestinal, lung, breast and prostate cancers as well as the maintenance of cancer stem cells15–20. Two Shh signaling pathways have been characterized: a canonical pathway that is ligand-dependent, and a non-canonical ligand-independent pathway. In the canonical Shh signaling pathway, the Hh proteins, sonic (SHH), Indian (IHH), and desert (DHH) hedgehogs bind to the receptor patched (PTCH1), which inhibits SMO, allowing SMO accumulation and translocation of activated forms of GLI family zinc finger 1 (GLI) into the nucleus, where it mediates the transcription of Hh target genes21. Furthermore, Hh co-receptors cell-adhesion-molecule-related CDON) and biregional CDON -binding protein (BOC), which encode cell surface bound members of the Ig/fibronectin-domain-superfamily, can also promote Shh signaling22,23. Also, Shh signaling can control through pathway negative regulators including Hh-interacting protein1 (HHIP) and RAB23, member RAS oncogene family (RAB23)24–26. In addition, post-transcriptional and/or translational regulations of Shh genes through micro RNAs (miRNAs) have been reported27,28. Furthermore, viral infections were shown to play an important role in Shh signaling pathway activation29–31. In a previous study, we unveiled important signaling pathways involved in the progression of gastric cancer using RNA-seq technology. The aim of this study was to evaluate the expression levels of genes associated with Shh signaling pathway that were significantly modulated and showed at least fourfold expression change, using real time PCR in gastric cancerous and non-cancerous tissues. In addition, for those dysregulated genes, we further evaluated their associations with expression levels of regulatory miRNAs and infection status.
Materials and methods
Patients
Tissue specimens used in the present study, including 50 tumoral and their adjacent non-tumoral tissues were collected during September 2015 to June 2018 at Mazandaran province, Iran hospitals. All samples were cut into approximately 0.5 cm3 pieces and individually snap-frozen in liquid nitrogen immediately after surgery, and thereafter stored at − 80 °C without thawing for further analyzes. Diagnosis of all cases was histologically confirmed by a pathologist using hematoxylin and eosin (H&E) staining, and infections including H. pylori, cytomegalovirus (CMV), human herpesvirus 6 (HHV6), and Epstein-Barr virus (EBV) were evaluated by PCR and Real-time PCR, as previously described32. Informed consent was obtained from all patients, and all procedures were approved and carried out in accordance with the guidelines of the Ethics Committee of Babol University of Medical Sciences, and were performed in compliance with Helsinki declaration.
RNA extraction
Tumoral and their adjacent non-tumoral tissues were separately crushed and homogenized with liquid nitrogen. RNX-plus reagent (Cinagen, Iran) was used to extract total RNA from samples according to the standard protocol with small modifications. Briefly, tumors were homogenized using 1 ml of RNX-plus per 10 mg of tissue. 200 μl of chloroform were added per 1 mL of RNX-plus for each sample. Samples were incubated for 10 min at room temperature and centrifuged for 15 min at 12,000×g at 4 °C. The aqueous phase containing the RNA was transferred into a new tube. Then 500 μl isopropanol was added, and the solution was incubated for 1 h and centrifuged for 40 min at 12,000×g at 4 °C. The supernatant was then discarded, and the pellet containing the RNA was washed with 500 μl 75% ethanol, left to dry for 10 min, and resuspended in 40 μl RNase-free water. The purified RNA samples were visualized after electrophoresis on 1.8% agarose gel to monitor the RNA integrity.
Prediction of miRNAs targeting BOC, IHH, and RAB23
Twelve online sequence-based target prediction algorithms have been used to predict miRNAs targeting BOC, IHH, and RAB23 mRNAs. MiRNAs that were predicted in at least seven out of twelve databases were selected for further analysis. MiR-6738-3p, miR-195-5p and miR-509-3-5p targeting the 3′′UTR of the BOC, IHH, and RAB23 mRNAs, respectively.
Quantitative real-time PCR analysis (QPCR)
Reverse transcription was performed using total RNA with gene specific stem-loop primer33,34 and SuperScript II reverse transcriptase (Invitrogen, USA). QRT-PCR was performed with the gene-specific primers listed in Table 1 using HotStar-Taq DNA Polymerase kit (Qiagen, Germany) in triplicate on the StepOne Plus real time instrument (Applied Biosystems, USA) according to the manufacturer’s instructions, to detect the expression levels of the target mRNAs and Micro-RNAs. The amplification conditions were 15 min at 95 °C, 40 cycles of 20 s at 95 °C followed by 60 s at 60 °C. The expression levels of mRNAs and miRNAs were normalized to Glyceraldehyde-3-phosphate dehydrogenase (GAPDH and RNA U6 small nuclear 1 (RNU6), respectively. The sequence of forward and reverse primers along with universal Taqman probe is presented in Table 1. The sequence of Taqman probe was FAM 5′ TGGATGTGTCTGCGGCGTTTTATCAT 3′ BHQ-1 and the sequence of reverse primer was 5′ GTATCCAGTGCTGCGACCGT 3′.
Table 1.
Sequences of primers used for evaluation of SHH signaling genes and their regulatory microRNAs.
| Accession Number | Gene Name | Primers 5′ → 3′ |
|---|---|---|
| NM_002046.7 | GAPDH | Specific forward primer: |
| TGGAGTCCACTGGCGTCTTCAC | ||
| RT-PCR primer | ||
| GTCGTATCCAGTGCTGCGACCGTATGGATGTGTCTGCGGCGTTTTATCATGCACTGGATACGACAGGCATTGCTGA | ||
| NM_002181.4 | IHH | Specific forward primer: |
| CCCGTCGTGGTGTAGTCATAGAG | ||
| RT-PCR primer | ||
| GTCGTATCCAGTGCTGCGACCGTATGGATGTGTCTGCGGCGTTTTATCATGCACTGGATACGACAGGCTGAGTTGG | ||
| NM_001301861.2 | BOC | Specific forward primer: |
| CACTGGCTTGCCTCCTCCTA | ||
| RT-PCR primer | ||
| GTCGTATCCAGTGCTGCGACCGTATGGATGTGTCTGCGGCGTTTTATCATGCACTGGATACGACGCATCATCCGAG | ||
| NM_016277.5 | RAB23 | Specific forward primer |
| GCTTGTGTGCTCGTGTTCTCTAC | ||
| RT-PCR primer | ||
| GTCGTATCCAGTGCTGCGACCGTATGGATGTGTCTGCGGCGTTTTATCATGCACTGGATACGACACTTCGGCTACTAC | ||
| NR_106796.1 | MIR6738 | Specific forward primer |
| CGACCTTCTGCCTGCATTCTA | ||
| RT-PCR primer | ||
| GTCGTATCCAGTGCTGCGACCGTATGGATGTGTCTGCGGCGTTTTATCATGCACTGGATACGACCTGGGAG | ||
| NR_029712.1 | MIR195 | Specific forward primer |
| GCGGCTAGCAGCACAGAAA | ||
| RT-PCR primer | ||
| GTCGTATCCAGTGCTGCGACCGTATGGATGTGTCTGCGGCGTTTTATCATGCACTGGATACGACGCCAATA | ||
| NR_030629.1 | MIR509-3 | Specific forward primer |
| CGCATACTGCAGACGTGG | ||
| RT-PCR primer | ||
| GTCGTATCCAGTGCTGCGACCGTATGGATGTGTCTGCGGCGTTTTATCATGCACTGGATACGACCATGATTG | ||
| NR_004394.1 | RNU6-1 | Specific forward primer |
| CTCGCTTCGGCAGCACATATAC | ||
| RT-PCR primer | ||
| GTCGTATCCAGTGCTGCGACCGTATGGATGTGTCTGCGGCGTTTTATCATGCACTGGATACGACGTGTCATCCTTGC |
The target specific portion of mRNA and micro-RNA is showed in red bold. Stem-loop sequence is underlined. Tm of the micro-RNA forward primer was increased by adding a tail (green sequences) to the 5′-end of the sequences.
Statistical analysis
The relative expression of each gene was calculated by StepOne software v2.3 and the 2−ΔΔCt method. Comparisons of results between groups were performed by paired Student’s t test. To measure the diagnostic accuracy of Shh signaling genes and regulatory miRNAs in gastric cancer, ROC curve analyses were performed using MedCalc statistical software. A value of P < 0.05 was considered as statistically significant.
Ethics approval
This study was approved by the Ethics Committee of Babol University of Medical Sciences and all procedures were performed in compliance with Helsinki declaration.
Results
Clinicopathological features
The clinicopathological features of the gastric cancer patients are summarized in Table 2. Fifty patients with gastric cancer including 38 (76%) male and 12 (24%) female were enrolled in this study. The patients’ mean age and standard deviation at the time of diagnosis was 59.81 ± 14.18 years (34 to 76 years) in females and 67.65 ± 9.5 years (42 to 85 years) in males. There was a significant association between sex and age of gastric cancer patients (P = 0.0325). Tumors were mainly located at proximal position of the stomach (cardia, fundus, and body). Overall, 45.5% of tumors were located in the cardia region, and body and antrum regions of the stomach were the second and third most common sites, with 29.5% and 22.7% abundance, respectively. Overall, 74% of tumors were in stages I/II, and 26% in stages III/IV.
Table 2.
Clinicopathological characteristics of gastric cancer patients.
| Characteristics | % | |
|---|---|---|
| Gender | Male | 76 |
| Female | 24 | |
| Age | < 65 years | 42 |
| ≥ 65 years | 58 | |
| Differentiation | Well differentiated | 27.1 |
| Moderately/poorly differentiated | 72.9 | |
| Stage | I | 31.9 |
| II | 42.55 | |
| III | 23.45 | |
| IV | 2.1 | |
| Lymph node metastasis | Yes | 56.8 |
| No | 42.4 |
Expression levels of Shh signaling pathway genes and their regulatory miRNAs
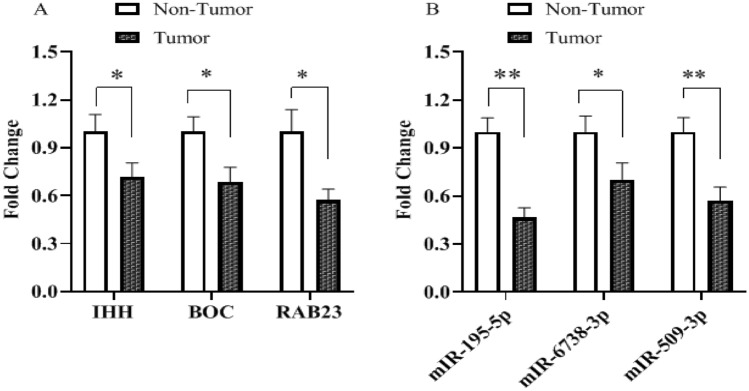
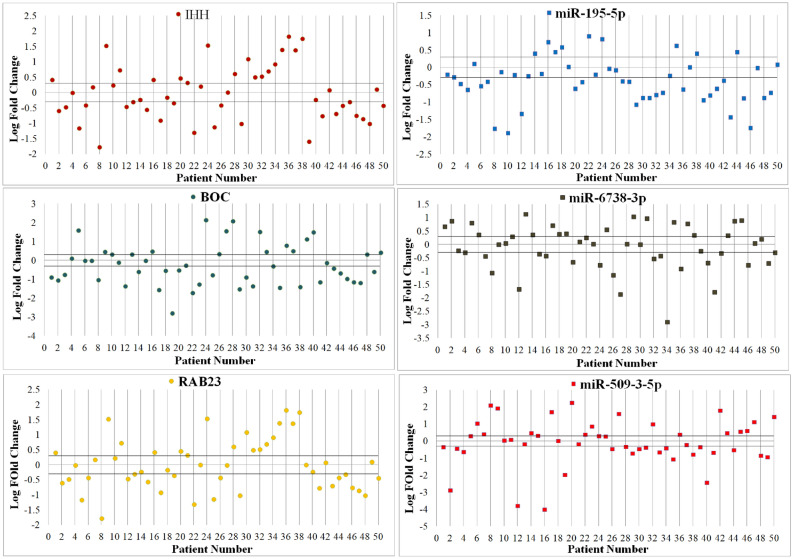
Expression levels of Shh signaling pathway genes (IHH, BOC, and RAB23) and their regulatory miRNAs (miR-195-5p, miR-6738-3p, and miR-509-3-5p) were evaluated in 50 gastric cancer patients using comparative relative real time PCR, and by comparing the expression in tumor tissues with their paired normal counterpart tissues. The results indicated that IHH, BOC, and RAB23 mRNA expression were significantly downregulated (Fig. 1). Similarly, miR-195-5p, miR-6738-3p, and miR-509-3-5p expression were also decreased significantly in gastric cancer tissues. The mean tumoral tissues expression levels for IHH, BOC, and RAB23 were 0.71, 0.68, and 0.57 respectively. Also, the expression levels for miR-195-5p, miR-6738-3p, and miR-509-3-5p in tumoral tissues were 0.46, 0.7, and 0.57 respectively. Scatter plot analysis indicated that IHH, BOC, and RAB23 were significantly down-regulated in 52%, 58%, and 50% of tumoral tissues in comparison with their adjacent non-tumoral counterparts, respectively. Also, out of 50 GC patients, 70%, 54%, and 58% showed statistically significant downregulation of miR-195-5p, miR-6738-3p, and miR-509-3-5p in tumoral tissues in comparison with their adjacent non-tumoral counterparts, respectively (Fig. 2).
Figure 1.
The expression level of Shh genes and their associated miRNAs. (A): IHH, BOC, and RAB23; (B) miR-195-5p, miR-6738-3p, and miR-509-3-5p in tumoral tissues in comparison with their adjacent non-tumoral counterparts. * and ** indicate significant difference P < 0.05 and P < 0.01, respectively.
Figure 2.
Scatter plot expression of Shh signaling pathway genes and their regulatory miRNAs in tumoral and adjacent non-tumoral tissues. Gene expression variations were considered in 50 gastric adenocarcinoma patients. Each plot represents a person’s gene expression change in tumor tissue in comparison with adjacent normal tissue. Horizontal lines represent cut-off values corresponding to log two-fold changes in expression. The upper and lower parts of the horizontal lines indicate upregulation, and downregulation in the tumoral tissues in comparison with non-tumoral tissues, respectively.
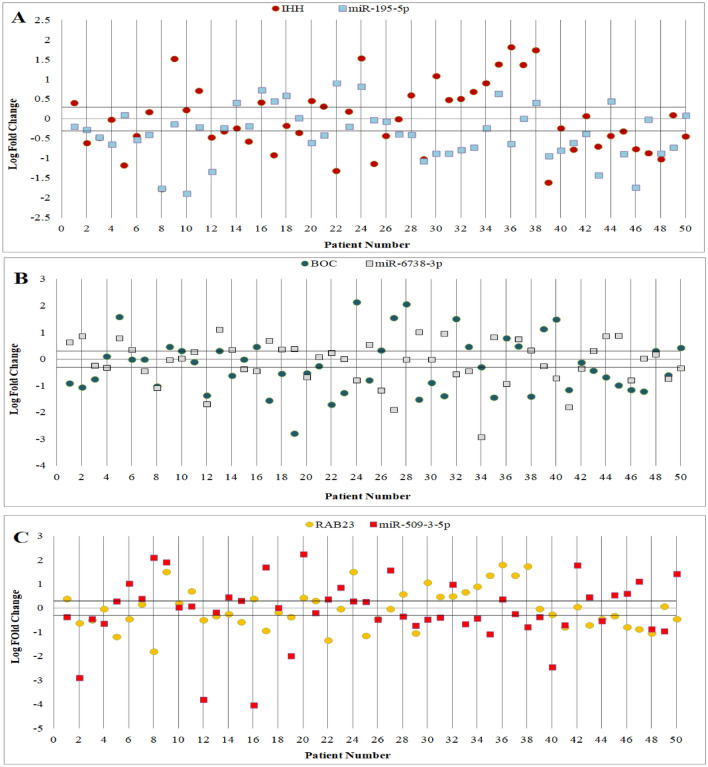
Correlation of Shh signaling pathway genes and their regulatory miRNAs
As shown in Fig. 3, analysis of relative expression levels indicated that IHH, BOC, and RAB23 were decreased in 48%, 54%, and 42% of gastric cancer tissues while 32%, 30%, and 34% of cases showed IHH, BOC, and RAB23 overexpression, respectively. Mir-195-5p was downregulated in 52% of cases. Also, 40% and 44% of cases showed miR-6738-3p and miR-509-3-5p down-regulation, respectively. 23 out of 50 (46%) gastric cancer patients indicated opposite expression pattern for IHH and its regulatory miRNA (miR-195-5p). Similarly, BOC and RAB23 and their regulatory miRNAs (miR-6738-3p and miR-509-3-5p) showed opposite expression in 56% and 50% of gastric cancer patients, respectively. Additionally, the miRNA-mRNA interaction between logarithm of fold change (Log FC) of Shh signaling pathway genes and their regulatory miRNA in early stage and advanced stage was calculated using the Pearson correlation test. A weak negative correlation was observed between Shh signaling pathway genes and their regulatory miRNA in different stages.
Figure 3.
Scatter plot of gene expression in tumoral and adjacent non-tumoral tissues of 50 gastric adenocarcinoma patients. Each plot represents a person’s gene expression change in tumor tissue compared with adjacent normal tissue. Horizontal black lines represent cut‐off values (log2 fold changes in expression). Cases above the upper line demonstrated overexpression in tumoral in comparison with non-tumoral tissues, and those below the lower line demonstrated lower expression levels (differences in expression ≥ 2; P < 0.05).
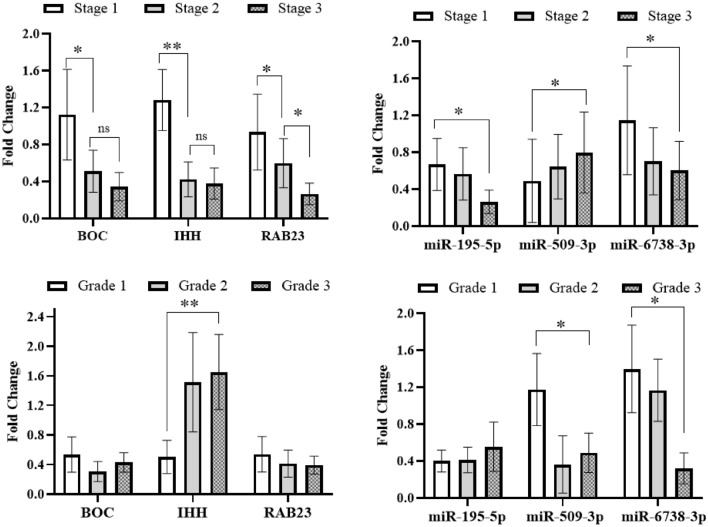
Association of clinicopathological features and Shh signaling pathway genes and their regulatory miRNAs
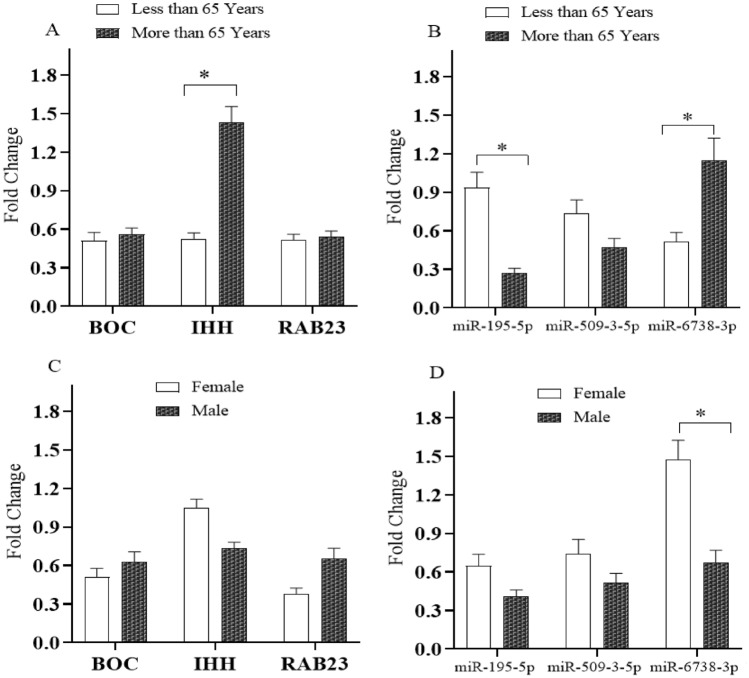
Association between the expression of Shh signaling pathway genes and their regulatory miRNAs with clinicopathological features in gastric cancer patients is illustrated in Fig. 4. A significantly associated trend was observed between the expression of studied genes and their regulatory miRNAs with TNM stage. The expression of IHH, BOC, RAB23, miR-195-5p, and miR-6738-3p decreased significantly with more advanced cancer stage, and miR-509-3-5p expression was significantly decreased during early stages in gastric cancer patients (P < 0.05). Also, IHH expression level was associated with histological type, as it was significantly lower in well differentiated gastric adenocarcinomas in comparison with moderately or poorly differentiated adenocarcinoma (P < 0.01). Furthermore, miR-6738-3p and miR-509-3-5p were significantly downregulated in poorly differentiated and moderately/poorly differentiated tumors, respectively (P < 0.05) (Fig. 4).
Figure 4.
Shh signaling pathway genes and their regulatory miRNAs fold changes in tumor gastric cancer tissues in comparison with their non-tumoral counterparts according to clinicopathological features. * and ** indicates significant difference P < 0.05 and P < 0.01, respectively.
Moreover, association study between Shh signaling pathway genes and their regulatory miRNAs according to age groups indicated that IHH and miR-6738-3p expression was significantly (P < 0.05) decreased in gastric cancer patients aged less than 65 years while miR-195-5p was significantly P < 0.05) down-regulated in gastric cancer patients older than 65 years. Moreover, a statistically significant decreased expression of miR-6738-3p was observed in male gastric cancer patients (P < 0.05) (Fig. 5).
Figure 5.
Association of the Shh signaling pathway genes and their regulatory miRNAs expression fold changes in tumoral tissues compared with their adjacent non-tumoral counterparts with age (< 65 years and ≥ 65 years) and gender. * indicates significant difference P < 0.05.
Association of the infection status and Shh signaling pathway genes and their regulatory miRNAs expression
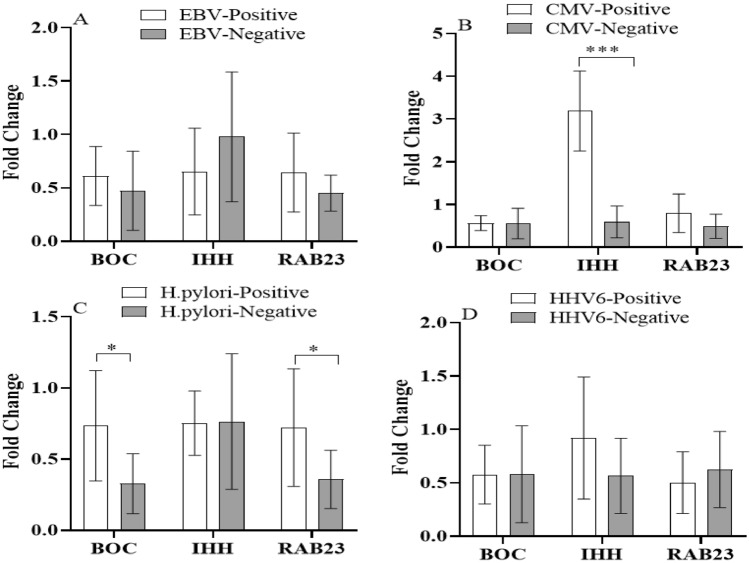
Furthermore, we investigate the effects of infections on the expression level Shh signaling pathway genes and their regulatory miRNAs in gastric cancer. As shown in Fig. 6, in HCMV positive patients, IHH expression was significantly increased (P < 0.001). Also, patients with no H. pylori infection showed lesser BOC and RAB23 expression in gastric tumoral tissues in comparison with H. pylori positive patients (P < 0.05). EBV and HHV6 infections had no significant effect on Shh signaling pathway genes and their regulatory miRNAs expression in gastric cancer tissues.
Figure 6.
Expression level of Shh signaling pathway genes in non-tumoral and tumoral gastric cancer tissues according to viral infections. * and *** indicate significant difference P < 0.05 and P < 0.001, respectively. EBV; Epstein-Barr virus, CMV; Cytomegalovirus, HHV6; Human herpesvirus 6.
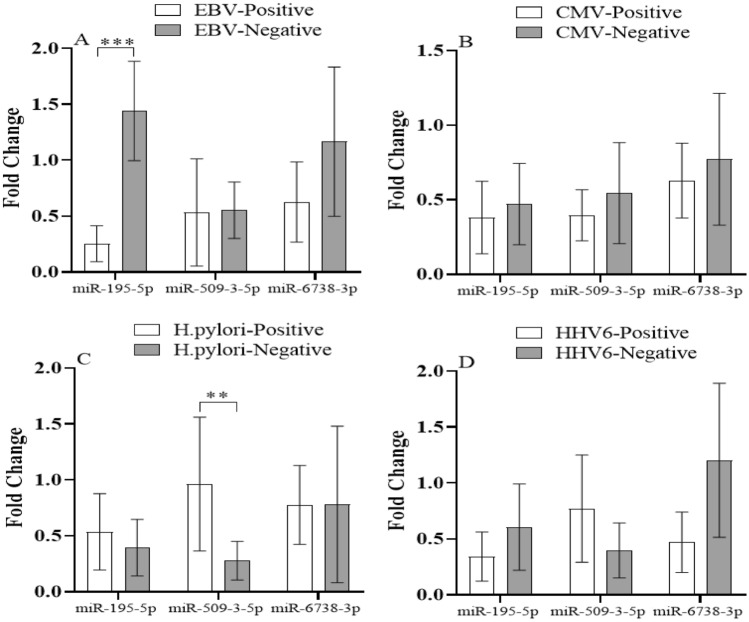
Furthermore, miR-195-5p expressions were significantly decreased in gastric cancer tissues of EBV positive patients (P < 0.001). In addition, there was a significant difference for miR-509-3-5p expression level in H. pylori positive gastric cancer patients in comparison with H. pylori negative patients (P < 0.01). HCMV infections had no significant effect on Shh signaling pathway regulatory miRNAs expression (Fig. 7).
Figure 7.
Expression level of Shh signaling pathway regulatory miRNAs in non-tumoral and tumoral gastric cancer tissues according to viral infections. ** and *** indicates significant difference P < 0.01 and 0.001, respectively. EBV; Epstein-Barr virus, CMV; Cytomegalovirus, HHV6; Human herpesvirus 6.
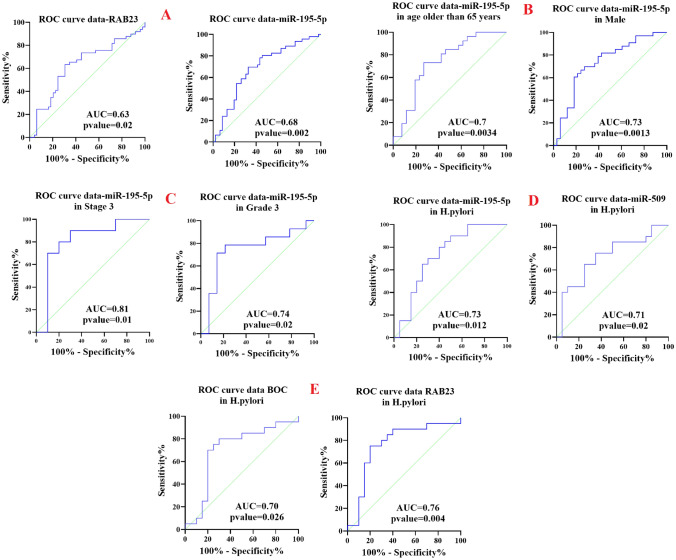
Receiver operating characteristic (ROC) curve analysis
ROC curve analysis used to reveal whether studied genes and their regulatory miRNAs can serve as diagnostic biomarkers (Fig. 8)35. As it is evident, total area under the curves (AUCs) of RAB23 (AUC = 0.63, sensitivity 71% and specificity 51%, P = 0.02) and miR-195-5p (AUC = 0.68, sensitivity 80% and specificity 54%, P = 0.002) were > 60%, suggesting that RAB23 and miR-195-5p can serve as diagnostic biomarkers for distinguishing patients with gastric cancer from healthy controls. ROC curve analysis was used to evaluate the predictive potential of Shh signaling pathway genes and their regulatory miRNAs with clinical parameters including gender, age, tumor grade, and TNM stage. The ROC analyses indicated that miR-195-5p expression levels were able to reliably distinguish late stages and grade from early stages and grade, with AUC 0.81 (sensitivity 90% and specificity 70%, P = 0.01) and 0.74 (sensitivity 78.57% and specificity 71.43%, P = 0.02), respectively. In addition, differences in the miR-195-5p expression level could successfully discriminate gastric cancer males from females (AUC = 0.73, sensitivity 81.8% and specificity 57.5%, P = 0.0013), as well as those belonging to the age group older than 65 years from younger than than 65 years (AUC = 0.73, sensitivity 80.77% and specificity 57.69%, P = 0.003). Furthermore, ROC curve analysis was applied to study the potential of Shh signaling pathway genes and their regulatory miRNAs as a biomarker in gastric cancer patients infected with EBV, CMV, HHV6, and H. pylori. ROC curve analysis showed good predictive accuracy for BOC, RAB23a, miR-195-5p, and miR-509-3-5p in discriminating H. pylori infected patients from uninfected patients (AUC of 0.703 for BOC, sensitivity 80% and specificity 70%, P = 0.02; AUC of 0.76 for RAB23a, sensitivity 90% and specificity 65%, P = 0.0045; AUC of 0.73 for miR-195-5p, sensitivity 80% and specificity 60%, , P = 0.012; AUC of 0.71 for miR-509-3-5p, sensitivity 75% and specificity 65%, P = 0.02).
Figure 8.
Receiver operating characteristics (ROC) curves for gene expression of potential biomarkers in gastric cancer. (A): The AUC of RAB-23 and miR-195-5p in gastric canccer was 0.63 and 0.68, respectively (B): The AUC of miR-195-5p in gastric cancer patients older than 65 years and male patients were 0.7 and 0.73, respectively (C): The AUC of miR-195-5p in advanced stage and grade in gastric cancer patients were 0.81 and 0.74, respectively (D): The AUC of miR-195-5p and miR-509 in H.pylori negative gastric cancer patients was 0.73 and 0.71, respectively (E): The AUC of BOC and RAB23 in H.pylori negative gastric cancer patients was 0.7 and 0.76, respectively. AUC: the area under the curve.
Discussion
Gastric cancer is among the five most frequently diagnosed cancers, and is highly heterogeneous. Accumulating evidence strongly indicate that aberrant activation of multiple signaling pathways can contribute to gastric cancer development. Consequently, cancer stem cells (CSCs) are key driving cells for growth and metastasis of this tumor type. It has been demonstrated that Shh signaling pathway is implicated in maintaining the pluripotency of CSCs, and aberrant activation of this pathway is associated with the development and progression of various types of cancer. In this study, we investigated the clinicopathological features of gastric carcinoma as well as expression levels of Shh signaling pathway genes and their regulatory miRNAs in gastric cancer patients. Although gastric cancer is common in both sexes, its incidence is higher in males, and is more frequently observed in younger female patients36–38. Our demographic findings are consistent with these reports. We also investigated the expression level of Shh signaling genes including IHH, BOC, and RAB23 in gastric cancer patients. Remarkably, we observed that IHH expression was decreased in tumoral tissues in comparison with adjacent non-tumoral tissues. Also, IHH expression strongly correlated with the stage and grade of malignancy as well as with CMV infection. IHH is one of the three protein ligands in the mammalian hedgehog signaling pathway, and plays an essential role in bone growth and differentiation. The expression level of IHH was found to be upregulated in certain tumors such as basal cell carcinoma, pancreatic cancer, and medulloblastomas39. Also, immunohistochemical study indicated that IHH expression was increased in pancreatic ductal adenocarcinoma in comparison with paracancer tissue and benign lesions, and this expression was associated with tumor grade, lymph node metastasis, tumor invasion, and poor overall survival40. In contrast, loss of IHH expression can promote the development of dysplasia in colon carcinogenesis via Wnt signaling pathway41. Also, epidermal deletion of IHH can promote squamous skin tumor formation and increased malignant tumor progression and metastasis as well as prolonged loss of IHH expression leads to inflammation and mucosal damage42. In addition, stromal activation of Hh signaling pathways by IHH suppresses tumor growth and metastases through angiogenesis and reduction of reactive oxygen species (ROS) activity. However, the tumor suppressor or oncogenic role of IHH in cancer is controversial. Relatively, our data demonstrated a decrease in BOC mRNA levels in tumoral tissues, and this expression was associated with the tumor’s stage and H.pylori infection. BOC is a co-receptor in Shh signaling pathway and a component of cell-surface receptor complex that mediates cell–cell interactions. However, its relative contribution to cancer risk is currently unknown. The hedgehog co-receptors including GAS1, CDON, and BOC modulate the levels of HH responsiveness in pancreatic fibroblasts, and loss of BOC and GAS1 was shown to reduce HH activity while promoting pancreatic tumor growth through the induction of angiogenic factors43. In addition, BOC may induce DNA damage and promote progression of early medulloblastoma to advanced tumors via increasing the incidence of loss of heterozygosity (LOH) of its coreceptor PTCH144. Moreover, our results indicated that RAB23 was downregulated in 42% of gastric cancer tissues while it was overexpressed in 34%of cases. Also, the expression of RAB23 was significantly associated with more advanced cancer stage and H. pylori infection. RAB23 is recognized as a negative regulator of the Shh signaling pathway, and also as the target of many proteins involved in cancer development. However, it remains controversial whether RAB23 acts as an oncogene or tumor suppressor. Although more and more studies have introduced RAB23 as an oncogene in variety of human cancers, there is some evidence indicating that RAB23 plays a tumor suppressive role during carcinogenesis. RAB23 through interaction with SUFU can inhibit GLI transcriptional activities and its nuclear localization. In addition, overexpression of RAB23 inhibits breast cancer cells viability and proliferation, and induces cell apoptosis45. Moreover, transient overexpression of miR-367 in medulloblastoma cells caused decreased RAB23 expression resulting in increased medulloblastoma cell proliferation46.
It is now established that both genetic and epigenetic alterations contribute to gastric cancer development. Changes in miRNAs expression, as epigenetic modulators, via regulation of cancer-related genes have a primary role in cancer onset and progression. We studied the Shh signaling pathway regulatory miRNAs by in silico analysis, and experimentally validated the expression levels of these miRNAs in gastric cancer patients. Through in silico analysis, we identified three miRNAs, including miR-195-5p, miR-6738-3p, and miR-509-3-5p, which could bind to the 3′ UTRs of Shh signaling genes and modulate the hedgehog pathway. While there is no evidence supporting a role for miR-6738-3p in cancer development, our findings showed that miR-6738-3p expression was decreased in tumoral tissues in comparison with adjacent non-tumoral tissues. Also, its expression was associated with the clinicopathological features of gastric cancer patients including stage, grade, gender, and age. Our in silico analysis predicted that mir-195-5p binding site was located in the 3′ UTR of IHH, and experimental data indicated that mir-195-5p was significantly down-regulated in tumor samples in comparison with their adjacent non-tumoral tissues. Also, mir-195-5p expression was strongly associated with the advanced cancer stage and age of gastric cancer patients, and correlated with EBV infection. Mir-195-5p is one of the well-studied miRNA that is strongly connected with various types of cancer including those of digestive system, respiratory system, urinary system, reproductive system, bone, brain, head and neck, skin, and endocrine cancer. Nevertheless, despite its strong tumor suppressive effects, there is evidence that miR-195 has an oncogenic role in some cancers. Therefore, whether mir-195-5p functions as a tumor suppressor or oncogene is still under debate. It has been reported that miR-195-5p had significant effect on oncogenicity in various types of cancer through binding to complementary sequences in crucial genes of signaling pathways. In stomach cancer, evidence had indicated that mir-195-5p is able to sensitize chemotherapeutic resistant gastric cancer cells to 5-fluorouracil and cisplatin by directly targeting ZNF139 and AKT3, respectively47,48. Also, Epstein–Barr virus-encoded miRNA BART1 through regulating the abundance of cellular tumor suppressive miRNAs such as miR-150, miR-152, and miR-195 was shown to induce tumor metastasis in nasopharyngeal carcinoma. This result further strengthens our observation that miR-195 expression was significantly decreased in gastric cancer tissues of EBV positive patients49.
Mir-509 is one of the anti-oncogene miRNAs that was reported to be downregulated in many prevalent human cancers. Mir-509 functions as a tumor suppressor in pancreatic and breast cancer via targeting MDM2 proto-oncogene (MDM2) and superoxide dismutase 2 (SOD2)50,51. Also, miR-509 as epithelial-mesenchymal transition miRNA was shown to induce the expression of E-cadherin, and inhibit cell motility and invasion52. In addition, miR-509 can inhibit cell motility and invasion through targeting tribbles pseudokinase 2 (TRIB2) in osteosarcoma53. Our findings showed that miR-509 was significantly down-regulated in tumor tissues in comparison with their adjacent non-tumoral tissues, and lower miR-509 expression in tumoral tissues was associated with high tumor grade, stage, and H. pylori infection. In line with our results, a similar study demonstrated that lower miR-509-3-5P expression was associated with advanced tumor stage and poor differentiation in gastric cancer tissues. Additionally, this lower expression promoted the migration and invasion abilities of gastric cancer cells by targeting podocalyxin like (PODXL) gene54. However, our finding is in disagreement with another study indicated miR-509 upregulation in H. pylori-negative gastric cancer55.
Gastric cancer can be considered a complex disease that is influenced by multiple genes, miRNAs, and environmental factors. Finding valuable biomarkers for the diagnosis and prognosis of gastric cancer is difficult. Therefore, using deep convolutional neural network learning method is a very promising way to predict disease risk based on biomarkers56. Our ROC curve analysis indicated that mir-195 can be considered a good biomarker as it was significantly related to several clinicopathological features of gastric cancer including stage, grade, age, gender, and infection status. In conclusion, the present study demonstrated that the expression level of Shh signaling pathway genes and their regulatory miRNAs were significantly associated with gastric cancer. Also, the expression level of these genes was significantly associated with clinicopathological features including tumor stage, grade, age, gender, and infection status in gastric cancer patients. Although Hh signaling inhibitors have been developed, and two inhibitors, vismodegib and sonidegib, have been approved by the U.S. Food and Drug Administration (FDA) to treat basal cell carcinoma and medulloblastoma, none of them have been approved for gastric cancer. Thus, more researches to elucidate factors regulating Shh signaling pathway as well as their detailed mechanism of action in gastric cancer are necessary. Also, it is well known that immune regulations may affect gastric cancer stage development, and patients with common variable immunodeficiency (CVID) have a high risk of gastric cancer57. Recently, in vivo study indicated that immunodeficiency can promote adaptive alterations of host gut or tissue-based microbiome58. Therefore, additional studies on alterations of gastric mucosal immunity and microbiota and effects on signaling pathways are needed to better understand the gastric carcinogenesis.
Author contributions
S.F.: Methodology, Formal analysis, Investigation, Writing—Original Draft. N.N.: Resources, Visualization. M.R.: Investigation, Visualization. D.S.: Formal analysis, Visualization. H.A.N.: Conceptualization, Writing—Review & Editing, Supervision, Project administration.
Competing interests
The authors declare no competing interests.
Footnotes
The original online version of this Article was revised: In the original version of this Article Haleh Akhavan-Niaki was incorrectly affiliated with ‘Department of Surgery, Rouhani Hospital Babol University of Medical Sciences, Babol, Iran’. The correct affiliations are ‘Cellular and Molecular Biology Research Center, Health Research Institute, Babol University of Medical Sciences, Babol, Iran’ and ‘Department of Genetics, Faculty of Medicine, Babol University of Medical Sciences, Babol, Iran’.
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
8/2/2021
A Correction to this paper has been published: 10.1038/s41598-021-95379-8
References
- 1.Bray F, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Herceg Z, Hainaut P. Genetic and epigenetic alterations as biomarkers for cancer detection, diagnosis and prognosis. Mol. Oncol. 2007;1(1):26–41. doi: 10.1016/j.molonc.2007.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.You JS, Jones PA. Cancer genetics and epigenetics: Two sides of the same coin? Cancer Cell. 2012;22(1):9–20. doi: 10.1016/j.ccr.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yamashita K, Sakuramoto S, Watanabe M. Genomic and epigenetic profiles of gastric cancer: Potential diagnostic and therapeutic applications. Surg Today. 2011;41(1):24–38. doi: 10.1007/s00595-010-4370-5. [DOI] [PubMed] [Google Scholar]
- 5.Mathers JC, Strathdee G, Relton CL. Induction of epigenetic alterations by dietary and other environmental factors. Adv Genet. 2010;71:3–39. doi: 10.1016/B978-0-12-380864-6.00001-8. [DOI] [PubMed] [Google Scholar]
- 6.Alegria-Torres JA, Baccarelli A, Bollati V. Epigenetics and lifestyle. Epigenomics. 2011;3(3):267–277. doi: 10.2217/epi.11.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fattahi S, et al. Infection-associated epigenetic alterations in gastric cancer: New insight in cancer therapy. J. Cell. Physiol. 2018;233(12):9261–9270. doi: 10.1002/jcp.27030. [DOI] [PubMed] [Google Scholar]
- 8.Morimoto K. Life-Style and Genetic Factors that Determine the Susceptibility to the Production of Chromosome Damage. In: Obe G, Natarajan AT, editors. Chromosomal Aberrations: Basic and Applied Aspects. Berlin: Springer; 1990. pp. 287–301. [Google Scholar]
- 9.Baudot A, de la Torre V, Valencia A. Mutated genes, pathways and processes in tumours. EMBO Rep. 2010;11(10):805–810. doi: 10.1038/embor.2010.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sever, R. and J.S. Brugge, Signal transduction in cancer. Cold Spring Harb Perspect Med, 2015. 5(4). [DOI] [PMC free article] [PubMed]
- 11.Deng R. Therapeutic effects of guggul and its constituent guggulsterone: Cardiovascular benefits. Cardiovasc. Drug Rev. 2007;25(4):375–390. doi: 10.1111/j.1527-3466.2007.00023.x. [DOI] [PubMed] [Google Scholar]
- 12.Nisar S, et al. Exploring dysregulated signaling pathways in cancer. Curr. Pharm. Des. 2020;26(4):429–445. doi: 10.2174/1381612826666200115095937. [DOI] [PubMed] [Google Scholar]
- 13.Armas-Lopez L, et al. The Hedgehog-GLI pathway in embryonic development and cancer: Implications for pulmonary oncology therapy. Oncotarget. 2017;8(36):60684–60703. doi: 10.18632/oncotarget.19527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heussler HS, Suri M. Sonic hedgehog. Mol. Pathol. 2003;56(3):129–131. doi: 10.1136/mp.56.3.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu Q, et al. Hedgehog signaling regulates brain tumor-initiating cell proliferation and portends shorter survival for patients with PTEN-coexpressing glioblastomas. Stem Cells. 2008;26(12):3018–3026. doi: 10.1634/stemcells.2008-0459. [DOI] [PubMed] [Google Scholar]
- 16.Saqui-Salces M, Merchant JL. Hedgehog signaling and gastrointestinal cancer. Biochem. Biophys. Acta. 2010;1803(7):786–795. doi: 10.1016/j.bbamcr.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Savani M, et al. Sonic hedgehog pathway expression in non-small cell lung cancer. Ther. Adv. Med. Oncol. 2012;4(5):225–233. doi: 10.1177/1758834012450362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Riobo-Del Galdo NA, Lara Montero Á, Wertheimer EV. Role of hedgehog signaling in breast cancer: Pathogenesis and therapeutics. Cells. 2019;8(4):375. doi: 10.3390/cells8040375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gonnissen A, Isebaert S, Haustermans K. Hedgehog signaling in prostate cancer and its therapeutic implication. Int. J. Mol. Sci. 2013;14(7):13979–14007. doi: 10.3390/ijms140713979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cochrane CR, et al. Hedgehog signaling in the maintenance of cancer stem cells. Cancers (Basel) 2015;7(3):1554–1585. doi: 10.3390/cancers7030851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carballo GB, et al. A highlight on Sonic hedgehog pathway. Cell Commun. Signal. 2018;16(1):11. doi: 10.1186/s12964-018-0220-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Song JY, et al. Distinct structural requirements for CDON and BOC in the promotion of Hedgehog signaling. Dev. Biol. 2015;402(2):239–252. doi: 10.1016/j.ydbio.2015.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Izzi L, et al. Boc and Gas1 each form distinct Shh receptor complexes with Ptch1 and are required for Shh-mediated cell proliferation. Dev. Cell. 2011;20(6):788–801. doi: 10.1016/j.devcel.2011.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chuang P-T, Kawcak TN, McMahon AP. Feedback control of mammalian Hedgehog signaling by the Hedgehog-binding protein, Hip1, modulates Fgf signaling during branching morphogenesis of the lung. Genes Dev. 2003;17(3):342–347. doi: 10.1101/gad.1026303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Evans TM, et al. Methods in Enzymology. Cambridge: Academic Press; 2005. Characterization of Rab23, a Negative Regulator of Sonic Hedgehog Signaling; pp. 759–777. [DOI] [PubMed] [Google Scholar]
- 26.Eggenschwiler JT, Espinoza E, Anderson KV. Rab23 is an essential negative regulator of the mouse Sonic hedgehog signalling pathway. Nature. 2001;412(6843):194–198. doi: 10.1038/35084089. [DOI] [PubMed] [Google Scholar]
- 27.Akhtar N, Makki MS, Haqqi TM. MicroRNA-602 and microRNA-608 regulate sonic hedgehog expression via target sites in the coding region in human chondrocytes. Arthritis Rheumatol. 2015;67(2):423–434. doi: 10.1002/art.38952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang Z, et al. miR-326 is downstream of Sonic hedgehog signaling and regulates the expression of Gli2 and smoothened. Am. J. Respir. Cell Mol. Biol. 2014;51(2):273–283. doi: 10.1165/rcmb.2013-0127OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fattahi S, Pilehchian Langroudi M, Akhavan-Niaki H. Hedgehog signaling pathway: Epigenetic regulation and role in disease and cancer development. J Cell Physiol. 2018;233(8):5726–5735. doi: 10.1002/jcp.26506. [DOI] [PubMed] [Google Scholar]
- 30.Pereira TDA, et al. Viral factors induce Hedgehog pathway activation in humans with viral hepatitis, cirrhosis, and hepatocellular carcinoma. Lab. Invest. J. Tech. Methods Pathol. 2010;90(12):1690–1703. doi: 10.1038/labinvest.2010.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Port RJ, et al. Epstein-Barr virus induction of the Hedgehog signalling pathway imposes a stem cell phenotype on human epithelial cells. J. Pathol. 2013;231(3):367–377. doi: 10.1002/path.4245. [DOI] [PubMed] [Google Scholar]
- 32.Fattahi S, et al. Prevalence of multiple infections and the risk of gastric adenocarcinoma development at earlier age. Diagn. Microbiol. Infect. Dis. 2018;92(1):62–68. doi: 10.1016/j.diagmicrobio.2018.04.015. [DOI] [PubMed] [Google Scholar]
- 33.Fattahi S, et al. Application of unique sequence index (USI) barcode to gene expression profiling in gastric adenocarcinoma. J. Cell. Commun. Signal. 2017;11(1):97–104. doi: 10.1007/s12079-017-0376-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fattahi S, et al. Development of a universal taqman probe for mRNA gene expression analysis. Iran. J. Sci. Technol. Trans. A Sci. 2018;42(2):363–370. [Google Scholar]
- 35.Yu H, et al. LEPR hypomethylation is significantly associated with gastric cancer in males. Exp. Mol. Pathol. 2020;116:104493. doi: 10.1016/j.yexmp.2020.104493. [DOI] [PubMed] [Google Scholar]
- 36.Rawla P, Barsouk A. Epidemiology of gastric cancer: Global trends, risk factors and prevention. Przeglad Gastroenterol. 2019;14(1):26–38. doi: 10.5114/pg.2018.80001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zaręba KP, et al. Stomach cancer in young people—a diagnostic and therapeutic problem. Przeglad Gastroenterol. 2019;14(4):283–285. doi: 10.5114/pg.2019.90254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cormedi MCV, et al. Survival and prognosis of young adults with gastric cancer. Clinics. 2018;73(suppl 1):e651s. doi: 10.6061/clinics/2018/e651s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kayed H, et al. Indian hedgehog signaling pathway: Expression and regulation in pancreatic cancer. Int. J. Cancer. 2004;110(5):668–676. doi: 10.1002/ijc.20194. [DOI] [PubMed] [Google Scholar]
- 40.Zou Q, et al. Association of chloride intracellular channel 4 and Indian hedgehog proteins with survival of patients with pancreatic ductal adenocarcinoma. Int. J. Exp. Pathol. 2016;97(6):422–429. doi: 10.1111/iep.12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van den Brink GR, et al. Indian Hedgehog is an antagonist of Wnt signaling in colonic epithelial cell differentiation. Nat. Genet. 2004;36(3):277–282. doi: 10.1038/ng1304. [DOI] [PubMed] [Google Scholar]
- 42.Kakanj P, et al. Indian Hedgehog controls proliferation and differentiation in skin tumorigenesis and protects against malignant progression. Cell. Rep. 2013;4(2):340–351. doi: 10.1016/j.celrep.2013.06.037. [DOI] [PubMed] [Google Scholar]
- 43.Mathew E, et al. Dosage-dependent regulation of pancreatic cancer growth and angiogenesis by hedgehog signaling. Cell. Rep. 2014;9(2):484–494. doi: 10.1016/j.celrep.2014.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mille F, et al. The Shh receptor Boc promotes progression of early medulloblastoma to advanced tumors. Dev. Cell. 2014;31(1):34–47. doi: 10.1016/j.devcel.2014.08.010. [DOI] [PubMed] [Google Scholar]
- 45.Liu Y, et al. Effect of Rab23 on the proliferation and apoptosis in breast cancer. Dev. Cell. 2015;34(4):1835–44. doi: 10.3892/or.2015.4152. [DOI] [PubMed] [Google Scholar]
- 46.Kaid C, et al. miR-367 promotes proliferation and stem-like traits in medulloblastoma cells. Cancer Sci. 2015;106(9):1188–1195. doi: 10.1111/cas.12733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ye R, et al. Expression of miR-195 is associated with chemotherapy sensitivity of cisplatin and clinical prognosis in gastric cancer. Oncotarget. 2017;8(57):97260–97272. doi: 10.18632/oncotarget.21919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nie H, et al. miR-195-5p regulates multi-drug resistance of gastric cancer cells via targeting ZNF139. Oncol. Rep. 2018;40(3):1370–1378. doi: 10.3892/or.2018.6524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cai L, et al. Epstein-Barr virus-encoded microRNA BART1 induces tumour metastasis by regulating PTEN-dependent pathways in nasopharyngeal carcinoma. Nat. Commun. 2015;6(1):7353. doi: 10.1038/ncomms8353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li X, et al. miR-509-5p inhibits cellular proliferation and migration via targeting MDM2 in pancreatic cancer cells. Onco Targets Ther. 2017;10:4455–4464. doi: 10.2147/OTT.S130378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Song YH, et al. MicroRNA-509-5p functions as an anti-oncogene in breast cancer via targeting SOD2. Eur. Rev. Med. Pharmacol. Sci. 2017;21(16):3617–3625. [PubMed] [Google Scholar]
- 52.Hiramoto H, et al. miR-509-5p and miR-1243 increase the sensitivity to gemcitabine by inhibiting epithelial-mesenchymal transition in pancreatic cancer. Sci. Rep. 2017;7(1):4002. doi: 10.1038/s41598-017-04191-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guo J, et al. miR-509-5p inhibits the proliferation and invasion of osteosarcoma by targeting TRIB2. Biomed. Res. Int. 2019;2019:2523032. doi: 10.1155/2019/2523032. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 54.Zhang J, et al. miR-509-3-5P inhibits the invasion and lymphatic metastasis by targeting PODXL and serves as a novel prognostic indicator for gastric cancer. Oncotarget. 2017;8(21):34867–34883. doi: 10.18632/oncotarget.16802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chang H, et al. Different microRNA expression levels in gastric cancer depending on Helicobacter pylori infection. Gut Liver. 2015;9(2):188–196. doi: 10.5009/gnl13371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu M, et al. A multi-model deep convolutional neural network for automatic hippocampus segmentation and classification in Alzheimer’s disease. Neuroimage. 2020;208:116459. doi: 10.1016/j.neuroimage.2019.116459. [DOI] [PubMed] [Google Scholar]
- 57.Leone P, et al. Common variable immunodeficiency and gastric malignancies. Int. J. Mol. Sci. 2018;19(2):451–463. doi: 10.3390/ijms19020451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zheng S, et al. Immunodeficiency promotes adaptive alterations of host gut microbiome: An observational metagenomic study in mice. Front Microbiol. 2019;10:2415. doi: 10.3389/fmicb.2019.02415. [DOI] [PMC free article] [PubMed] [Google Scholar]