Abstract
Hypoxia induces a series of cellular adaptive responses that enable promotion of inflammation and cancer development. Hypoxia-inducible factor-1α (HIF-1α) is involved in the hypoxia response and cancer promotion, and it accumulates in hypoxia and is degraded under normoxic conditions. Here we identify prostate cancer associated transcript-1 (PCAT-1) as a hypoxia-inducible long non-coding RNA (lncRNA) that regulates HIF-1α stability, crucial for cancer progression. Extensive analyses of clinical data indicate that PCAT-1 is elevated in breast cancer patients and is associated with pathological grade, tumor size, and poor clinical outcomes. Through gain- and loss-of-function experiments, we find that PCAT-1 promotes hypoxia-associated breast cancer progression including growth, migration, invasion, colony formation, and metabolic regulation. Mechanistically, PCAT-1 directly interacts with the receptor of activated protein C kinase-1 (RACK1) protein and prevents RACK1 from binding to HIF-1α, thus protecting HIF-1α from RACK1-induced oxygen-independent degradation. These findings provide new insight into lncRNA-mediated mechanisms for HIF-1α stability and suggest a novel role of PCAT-1 as a potential therapeutic target for breast cancer.
Keywords: breast cancer, HIF-1α, hypoxia, PCAT-1, RACK1
Graphical abstract

Hypoxia induces a series of cellular adaptive responses that enable promotion of inflammation and cancer development. Liu and colleagues reveal that PCAT-1 is critical for maintaining the stability of HIF-1α in hypoxic breast cancer cells, which is related to advanced disease progression and poor prognosis.
Introduction
Breast cancer is a major public health problem, and it is the most commonly diagnosed cancer among women. The implementation of breast cancer early diagnosis, together with local surgery and radiotherapy and systemic pharmacological treatments, has resulted in substantial improvement in the therapeutic effects on breast cancer.1,2 However, cancer progression and the spread of cancer cells to other organs are the cause of death in the vast majority of patients, and the processes are complicated and involve many complex epigenetic changes.3,4 Since the pathogenic mechanisms of cancer progression are not fully understood, more studies are needed to discover and develop effective molecules and targets for diagnosis and treatment.
Hypoxia in the tumor microenvironment is associated with inflammation, tumorigenesis, and therapeutic resistance.5, 6, 7 Although hypoxia occurs at different stages of breast cancer development, it is unclear how hypoxia affects the cells during transformation. Hypoxia-inducible factor-1 (HIF-1) is a key regulator of the hypoxia response and cancer promotion.8,9 It is essential for the cellular response to hypoxia, including angiogenesis, energy metabolism, cell survival, invasion and metastasis.10,11 HIF-1 is a heterodimer composed of HIF-1α and HIF-1β subunits. HIF-1β is maintained constitutively, whereas HIF-1α accumulates rapidly in hypoxia and is degraded under normoxic conditions. In normoxia, HIF-1α is degraded by oxygen-dependent prolyl hydroxylation, ubiquitination, and proteasomal degradation.11 Under hypoxic conditions, HIF-1α evades the oxygen-dependent degradation, allowing it to translocate into the cell nucleus to form the transcriptional HIF-1α complex.10 Several studies have shown that there are oxygen-independent mechanisms of HIF-1α degradation under hypoxic conditions, including heat shock protein 90 (HSP90) and receptor of activated protein C kinase-1 (RACK1).12, 13, 14 HSP90 binds to the HIF-1α PAS domain to prevent its degradation. RACK1 competes with HSP90 for binding to HIF-1α, links HIF-1α to Elongin-C, and promotes HIF-1α degradation in an oxygen-independent manner.12,15 Thus, equilibrium between HIF-1α/RACK1 and HIF-1α/HSP90 may play a key role in maintaining HIF-1α stability, as well as cancer progression.
Long non-coding RNAs (lncRNAs) have emerged as important molecules in carcinogenesis, with various biological effects, such as genome modification, transcriptional activation, and transcriptional interference.16, 17, 18 They have been implicated as prospective biomarkers for early cancer diagnosis, with the potential to serve as new targets for cancer treatment.19,20 Previous studies have identified that lots of hypoxic-reactive lncRNAs are functionally characterized and have crucial effects on cancer progression.21 Prostate cancer associated transcript-1 (PCAT-1) is identified as a prostate cancer-overexpressed lncRNA, and it contributes to prostate cancer progression through regulation of target genes.22 The high expression of PCAT-1 has been observed in several types of cancers, including colorectal cancer,23 esophageal squamous cell carcinoma,24 hilar cholangiocarcinoma,25 osteosarcoma,26 and hepatocellular carcinoma.27 It has been shown to be involved in impairment of double-stranded DNA break repair, cancer cell proliferation, epithelial-mesenchymal transitions, migration, and invasion.28, 29, 30, 31 A recent report shows that PCAT-1 overexpression is also observed in breast cancer tissues.32 However, there is no previous report of PCAT-1 regulating hypoxia-related cancers. Currently, the clinical significance of PCAT-1 in the context of breast cancer and the molecular mechanisms by which it regulates remain largely unclear.
In this study, through the investigation of lncRNA in breast cancer, we identify that PCAT-1 is one of the significantly upregulated lncRNAs under hypoxic conditions. We reveal that PCAT-1 is critical for maintaining the stability of HIF-1α in hypoxic breast cancer cells, which is related to advanced disease progression and poor prognosis. We elucidate that PCAT-1 interacts with RACK1, which mediates oxygen-independent HIF-1α stability, thereby establishing PCAT-1 as an important lncRNA in breast cancer.
Results
The expression of PCAT-1 is induced after hypoxia treatment
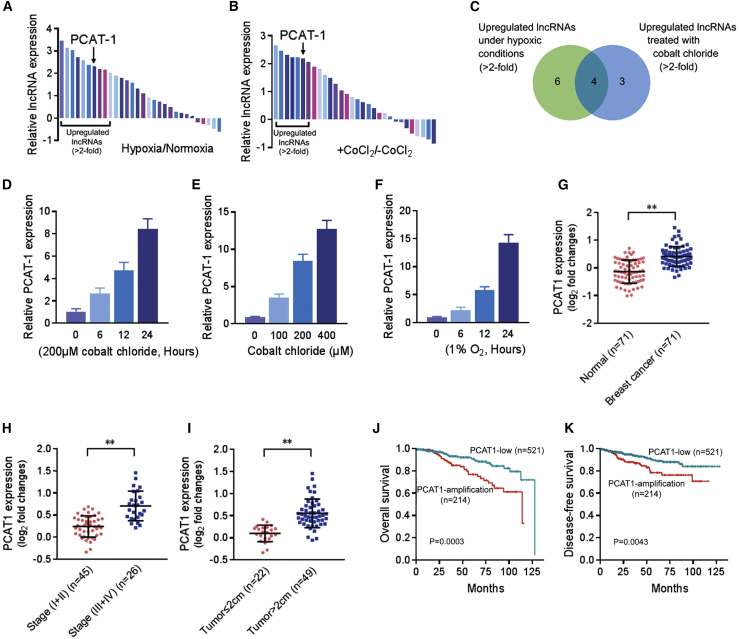
In order to determine the involvement of lncRNAs in hypoxia-related cancer progression, we selected 30 cancer-associated lncRNAs (listed in Table S1) according to previous studies and analyzed their expression in MDA-MB-231 breast cancer cells in hypoxia and normoxia.20,21 Compared with normoxic conditions, 10 lncRNAs with >2-fold expression changes were identified in hypoxia (Figure 1A). Moreover, we checked the expression of these lncRNAs and found 7 of them upregulated by >2-fold in MDA-MB-231 cells treated with cobalt chloride (CoCl2) (Figure 1B), which was used to create a pseudohypoxic environment. These lncRNAs include 4 of those induced in physical hypoxia (Figure 1C). PCAT-1 was one of the most abundantly expressed lncRNAs in response to CoCl2 treatment and displayed a time- and concentration-dependent induction (Figures 1D and 1E; Figures S1A–S1C). Similarly, a time-dependent increase of PCAT-1 was observed in MDA-MB-231 cells under hypoxic conditions (Figure 1F), confirming that PCAT-1 is a hypoxia-responsive lncRNA and is probably related to the HIF-1α signaling pathway.
Figure 1.
PCAT-1 is involved in the response to hypoxia and related to poor prognosis for breast cancer
(A and B) Expression of 30 lncRNAs in MDA-MB-231 cells under hypoxia (1% O2, 6 h) (A) or treated with cobalt chloride (200 μM, 6 h) (B) was quantified by quantitative real-time PCR and normalized to GAPDH expression. (C) The Venn diagram illustrates the number of lncRNAs elevated in hypoxia or treated with cobalt chloride. (D and E) Expression of PCAT-1 in MDA-MB-231 cells treated with cobalt chloride (200 μM) for different times (D) or with different concentrations of cobalt chloride for 24 h (E). (F) Expression of PCAT-1 in MDA-MB-231 cells under hypoxic conditions. (G) PCAT-1 expression in 71 pairs of clinical specimens containing breast cancer tissues and the normal adjacent tissues was detected by quantitative real-time PCR and normalized to GAPDH expression. (H and I) PCAT-1 expression in breast cancer tissues (n = 71) assessed by quantitative real-time PCR with tumor stage (H) and tumor size (I). (J and K) Kaplan-Meier curve for OS (J) and DFS (K) in breast cancer patients with and without genetic amplification of PCAT-1. Patients were grouped into PCAT-1-low or PCAT-1-amplification based on the PCAT log2 copy number value. The cutoff point is set as the value yielding maximum sum of sensitivity and specificity for DFS analysis.
PCAT-1 expression is associated with poor breast cancer prognosis
To assess the pathological and clinical value of PCAT-1 in breast cancer, we measured PCAT-1 expression in a panel of 71 matched pairs of clinical specimens containing breast cancer tissues and the normal adjacent tissues. This showed that PCAT-1 was significantly upregulated in the cancer tissues compared with the normal adjacent tissues (p < 0.001; Figure 1G). Moreover, statistical analysis showed that PCAT-1 overexpression was strongly correlated with advanced tumor stage (p < 0.001; Figure 1H) and tumor size (p < 0.001; Figure 1I).
To further validate the data from our studies, we surveyed public data of The Cancer Genome Atlas (TCGA) breast cancer study using the cBioPortal platform and explored the correlation between PCAT-1 gene alteration and the clinicopathological characteristics of 735 breast cancer patients (Table S2). The analyses revealed that PCAT-1 amplification was correlated with advanced tumor stage (p = 0.008) and tumor size (p = 0.002; Table 1), whereas no significant relationship with any other clinical characteristics was observed. Moreover, PCAT-1 expression was higher in Luminal B than in other subtypes of breast tumors (Figure S1D). We evaluated PCAT-1 gene alteration in relation to overall survival (OS) and disease-free survival (DFS) data on the patients (Table S2). Kaplan-Meier survival analysis was performed to compare the outcomes of patients dichotomized by PCAT log2 copy number value. Patients with PCAT-1-amplification copy number had a significantly worse OS (p < 0.001) (Figure 1J) and DFS (p = 0.004) (Figure 1K), and similar results were also observed in patients with different tumor stages (Figures S1E and S1F). In addition, similar to the prognostic values of tumor stage (I + II versus III + IV), tumor size (≤2 cm versus >2 cm), and lymph node metastasis (N0 versus N1–3), the univariate Cox proportional hazards showed that PCAT-1 amplification was another strong prognostic predictor for poor OS (hazard ratio [HR] = 2.364, p < 0.001) and DFS (HR = 2.049, p = 0.005; Table 2). Furthermore, multivariate analysis was conducted to determine risk assessment related to OS and DFS. PCAT-1 amplification was identified as an independent prognostic factor for OS (HR = 1.948, p = 0.015, multivariate analysis) and DFS (HR = 1.829, p = 0.025, multivariate analysis; Table 2). These results indicated that PCAT-1 could be used as an independent indicator of prognosis in breast cancer patients.
Table 1.
The relationship between PCAT-1 gene alteration and clinicopathological characteristics in breast cancer patients
| Clinicopathologic characteristics | PCAT-1 gene alteration |
Number of cases | p value | |
|---|---|---|---|---|
| Amplification (%) | Low (%) | |||
| Age (years) | ||||
| ≤60 | 122 (29) | 305 (71) | 427 | 0.742 |
| >60 | 92 (30) | 216 (70) | 308 | |
| Tumor stage | ||||
| I + II | 61 (38) | 100 (62) | 161 | 0.008∗∗ |
| III + IV | 153 (27) | 421 (73) | 574 | |
| Tumor size | ||||
| ≤2 cm | 41 (21) | 157 (79) | 198 | 0.002∗∗ |
| >2 cm | 173 (32) | 364 (68) | 537 | |
| Lymph node metastasis | ||||
| N0 | 97 (26) | 277 (74) | 374 | 0.062 |
| N1–3 | 117 (32) | 244 (68) | 361 | |
| Race | ||||
| Other | 50 (34) | 95 (66) | 145 | 0.098 |
| White | 144 (27) | 385 (73) | 529 | |
| Radiation | ||||
| No | 93 (29) | 225 (71) | 318 | 0.934 |
| Yes | 111 (29) | 273 (71) | 384 | |
Fisher’s exact test was used to determine the correlation between PCAT-1 gene alteration and the clinicopathological characteristics. The cutoff point is set as the value yielding maximum sum of sensitivity and specificity for disease-free survival analysis. ∗∗p < 0.01.
Table 2.
Univariate and multivariate Cox regression analyses of overall survival and disease-free survival in breast cancer patients
| Covariate |
Univariate analysis |
Multivariate analysis |
||||
|---|---|---|---|---|---|---|
| HR | 95% CI | p value | HR | 95% CI | p value | |
| Overall survival | ||||||
| Age (≤60 versus >60) | 1.826 | 1.122–2.971 | 0.015∗ | 2.097 | 1.209–3.637 | 0.008∗∗ |
| Tumor stage (I + II versus III + IV) | 2.777 | 1.697–4.545 | 0.000∗∗ | 3.587 | 1.713–7.512 | 0.001∗∗ |
| Tumor size (≤2 cm versus >2 cm) | 2.327 | 1.217–4.448 | 0.011∗ | 1.478 | 0.733–2.980 | 0.275 |
| Lymph node metastasis (N0 versus N1–3) | 2.132 | 1.284–3.537 | 0.003∗∗ | 1.280 | 0.615–2.661 | 0.509 |
| Race (Other/White) | 0.639 | 0.361–1.130 | 0.124 | 0.577 | 0.319–1.045 | 0.070 |
| Radiation (No/Yes) | 0.572 | 0.343–0.953 | 0.032∗ | 0.366 | 0.203–0.660 | 0.001∗∗ |
| PCAT-1 gene alteration (low versus amplification) | 2.364 | 1.458–3.833 | 0.000∗∗ | 1.948 | 1.135–3.341 | 0.015∗ |
| Disease-free survival | ||||||
| Age (≤60 versus >60) | 0.817 | 0.478–1.396 | 0.459 | 0.912 | 0.516–1.615 | 0.753 |
| Tumor stage (I + II versus III + IV) | 3.109 | 1.870–5.166 | 0.000∗∗ | 3.576 | 1.731–7.386 | 0.001∗∗ |
| Tumor size (≤2 cm versus >2 cm) | 2.089 | 1.086–4.017 | 0.027∗ | 1.331 | 0.669–2.644 | 0.415 |
| Lymph node metastasis (N0 versus N1–3) | 1.707 | 1.021–2.854 | 0.042∗ | 0.965 | 0.464–2.006 | 0.924 |
| Race (Other/White) | 0.525 | 0.297–0.927 | 0.026∗ | 0.553 | 0.311–0.981 | 0.043∗ |
| Radiation (No/Yes) | 0.923 | 0.553–1.538 | 0.757 | 0.599 | 0.338–1.061 | 0.079 |
| PCAT-1 gene alteration (low versus amplification) | 2.049 | 1.238–3.392 | 0.005∗∗ | 1.829 | 1.078–3.103 | 0.025∗ |
HR, hazard ratio; 95% CI, 95% confidence interval. ∗p < 0.05 and ∗∗p < 0.01.
PCAT-1 promotes breast cancer progression
To ascertain the potential role of PCAT-1 in breast cancer, we detected the expression of PCAT-1 in breast cancer cells MCF-7, MDA-MB-231, MDA-MB-468, and T47D and normal breast epithelial cell MCF10A. Compared with MCF-7, MDA-MB-468, and T47D cells with poor invasive capacity, the highly invasive MDA-MB-231 cells had higher PCAT-1 expression (Figure 2A), while MCF-7, MDA-MB-468, T47D, and MCF10A cells had a low level of PCAT-1, suggesting that PCAT-1 might be related to cell invasion in breast cancer. Similar to MDA-MB-231 cells, PCAT-1 expression was also induced in MCF-7 cells by CoCl2 under pseudohypoxia or under hypoxic conditions (Figure 2B).
Figure 2.
PCAT-1 facilitates the progression of breast cancer
(A) Expression of PCAT-1 in MCF-7, MDA-MB-231, MDA-MB-468, T47D, and MCF10A cells was analyzed by quantitative real-time PCR and normalized to GAPDH expression. (B) Expression of PCAT-1 in MCF-7 cells treated with different concentrations of cobalt chloride for 24 h or under hypoxic conditions. MDA-MB-231 cells were cultured in normoxia or hypoxia. (C–F) 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay showing growth (C), wound healing assay showing migration (D), and Transwell assay showing invasion (E) and colony formation (F) were performed in MDA-MB-231 cells transfected with PCAT-1 siRNA. (G and H) Extracellular acid ratio (ECAR) level (G) and lactate production (H) in the culture media were determined after knockdown of PCAT-1 in MDA-MB-231 cells. 2-DG, 2-deoxyglucose. The data are shown as fold difference compared with the level of negative control (NC) siRNA-transfected cells in normoxia. Data are shown as the mean ± SD. ∗p < 0.05 and ∗∗p < 0.01 compared with control.
The cell’s response to pathological hypoxia is associated with many critical aspects of cancer. To further investigate the biological significance of PCAT-1 in breast cancer, we knocked down the expression of PCAT-1 with two independent small interfering RNAs (siRNAs). Moreover, lentiviral PCAT-1 small hairpin RNA (shRNA) (shRNA-PCAT-1) and empty vector (shRNA-Control) were also stably transfected in MDA-MB-231 cells, and the related phenotypes in the cells were examined. The functional assays revealed that PCAT-1 knockdown significantly impaired cell growth, migration, invasion, and colony formation in hypoxia (Figures 2C–2F), whereas PCAT-1 overexpression had the opposite effects (Figure S2). In addition, with normalization to cell number, we found that the hypoxia-induced extracellular acidification rate (ECAR) (Figure 2G) and lactate production (Figure 2H) were significantly reduced in the PCAT-1-knockdown MDA-MB-231 cells under hypoxic conditions, indicating that PCAT-1 is essential for glycolytic metabolism in the cells.
PCAT-1 contributes to HIF-1α stability
HIF-1α is a core regulator of cellular adaptation to hypoxia, which could regulate lncRNAs at the transcriptional level via hypoxia response elements (HREs). In the present study, no HREs were found in the PCAT-1 promoter in silico prediction. Consistent with this, inhibition of either HIF-1α or HIF-2α did not significantly affect PCAT-1 induction in hypoxia (Figure S3), suggesting that PCAT-1 is not a direct HIF target lncRNA. To determine the roles of PCAT-1 in the HIF-1α signaling pathway, PCAT-1 was overexpressed in MCF-7 cells. We found that HIF-1α protein expression was increased in normoxia as well as in hypoxia (Figure 3A). However, HIF-1α mRNA expression was not changed. In addition, we investigated whether PCAT-1 regulates HIF-1α mRNA stability. We treated the cells with actinomycin D, which blocks de novo mRNA synthesis, and the HIF-1α mRNA stabilization was not affected by PCAT-1 overexpression (Figure 3B), suggesting that HIF-1α mRNA transcription and degradation are not regulated by PCAT-1. Additionally, the cells were exposed to hypoxia and then recovered in normoxia, and we found that the half-lives of HIF-1α protein were increased from 2.5 ± 0.3 to 6.5 ± 0.7 min as a consequence of PCAT-1 overexpression, indicating that PCAT-1 overexpression yielded a significant delay of HIF-1α protein degradation (Figure 3C). These results revealed that PCAT-1 overexpression increased HIF-1α expression by enhancing the stability of HIF-1α protein. In addition, PCAT-1 was depleted in MDA-MB-231 cells. HIF-1α protein expression was obviously decreased in the cells after knockdown of PCAT-1 in a hypoxic environment (Figure 3D), while HIF-1α mRNA expression and stabilization were not changed (Figure 3E). We also found that HIF-1α protein degraded more quickly during reoxygenation in the PCAT-1-knockdown cells (Figure 3F), and the half-lives of HIF-1α protein were decreased from 7.2 ± 1.1 to 3.8 ± 0.6 min, suggesting that depletion of PCAT-1 decreased the HIF-1α protein stability.
Figure 3.
Effects of PCAT-1 on HIF-1α stability
(A) Effects of PCAT-1 overexpression on HIF-1α protein and mRNA expression in MCF-7 cells. (B) Effects of PCAT-1 overexpression on HIF-1α mRNA stability. MCF-7 cells were cultured in normoxia or hypoxia; then the cells were incubated with actinomycin D (5 μg/mL) for 3 h, and the mRNA of HIF-1α was examined at the indicated times by quantitative real-time PCR. (C) Effects of PCAT-1 overexpression on HIF-1α protein stability in the cells during reoxygenation. MCF-7 cells were cultured in hypoxia (1% O2, 6 h) and incubated for 1 h in the presence of the proteasome inhibitor MG132 (10 μmol/L) and cycloheximide (50 μg/ mL); then the cells were exposed to normoxia for the indicated times. (D) Effects of PCAT-1 knockdown on HIF-1α protein expression in MDA-MB-231 cells under hypoxic conditions. (E) Effects of PCAT-1 knockdown on HIF-1α mRNA expression and stability in MDA-MB-231 cells. (F) Effects of PCAT-1 knockdown on HIF-1α protein stability in MDA-MB-231 cells during reoxygenation. Cells were cultured in hypoxia (1% O2, 6 h) and incubated for 1 h in the presence of the proteasome inhibitor MG132 (10 μmol/L) and cycloheximide (50 μg/ mL); then the cells were exposed to normoxia for the indicated times. (G) Expression of HIF-1α target genes in MDA-MB-231 cells transfected with PCAT-1 siRNA was measured by quantitative real-time PCR. (H) Expression of HIF-1α target genes in MCF-7 cells overexpressing PCAT-1 was measured by quantitative real-time PCR and normalized to RPLP0 level under hypoxic conditions.
Furthermore, to determine whether PCAT-1 modulated HIF-1α activity, we investigated the expression of HIF-1α target genes such as FLT1, VEGFA, ANGPTL4, CCNG2, SOX9, PDK1, PLOD2, LDHA, JMJD1A, GLUT1, and BNIP3 in the cells. PCAT-1 knockdown attenuated the expression of these genes in MDA-MB-231 cells in hypoxia, without a significant change of the gene expression in normoxia (Figure 3G). Reciprocally, PCAT-1 overexpression in MCF-7 cells induced the expression of HIF-1α target genes in hypoxia (Figure 3H). In addition, we found that HIF-1α transfection significantly abrogated the effects of PCAT-1 depletion on hypoxia-induced cell phenotypes in PCAT-1-silenced MDA-MB-231 cells (Figure S4), indicating that HIF-1α was a functional target of PCAT-1 in breast cancer cells.
PCAT-1 protects HIF-1α from RACK1-mediated oxygen-independent degradation
To determine whether the effects of PCAT-1 on the stability of HIF-1α protein are related to prolyl hydroxylase, HA-HIF-1α (P402A/P564A), which contained the proline-to-alanine substitutions and resisted hydroxylation,15 was transfected in MDA-MB-231 cells. This showed that the protein level of mutant HIF-1α could hardly be detected in PCAT-1-knockdown cells under hypoxic conditions, but it accumulated rapidly after treatment with MG132 (Figure 4A). Additionally, PCAT-1 overexpression reduced the ubiquitination of exogenous HIF-1α and increased HIF-1α stability in MCF-7 cells cotransfected with HA-HIF-1α (P402A/P564A) and PCAT-1 under hypoxic conditions (Figure 4B). Then MDA-MB-231 cells were cotransfected with PCAT-1 siRNA and RACK1 siRNA, and we found that RACK1 siRNA increased HIF-1α protein levels by 2.3-fold and blocked the degradation of HIF-1α in PCAT-1-knockdown cells (Figure 4C). Moreover, we found that RACK1 knockdown notably improved HIF-1α protein stability in MCF-7 cells cotransfected with PCAT-1 and HA-HIF-1α (P402A/P564A) (Figure 4D). In addition, PCAT-1 was overexpressed in MCF-7 cells and then the cells were treated with the HSP90 inhibitor 17-AAG, and the results showed that PCAT-1-facilitated HIF-1α protein stability was abolished after 17-AAG treatment (Figure 4E). Moreover, PCAT-1 knockdown in MDA-MB-231 cells accelerated HIF-1α degradation after 17-AAG treatment (Figure 4F).
Figure 4.
Effect of PCAT-1 on oxygen-independent HIF-1α degradation
(A and B) Stability of HA-HIF-1α (P402A/P564A) in MDA-MB-231 cells (A) transfected with PCAT-1 siRNA in the presence of MG132 (10 μmol/L) or in MCF-7 cells (B) transfected with PCAT-1 under hypoxic conditions. (C) Effect of RACK1 knockdown on HIF-1α protein stability in MDA-MB-231 cells transfected with PCAT-1 siRNA in hypoxia. (D) Effect of RACK1 knockdown on HA-HIF-1α (P402A/P564A) stability in MCF-7 cells transfected with PCAT-1 in hypoxia. (E and F) Cells were cultured in hypoxia, and then the effect of 17-AAG (1 μmol/L, 24 h) on HIF-1α protein stability in MCF-7 cells (E) transfected with PCAT-1 or in MDA-MB-231 cells (F) transfected with PCAT-1 siRNA was examined.
PCAT-1 interacts with RACK1 that mediates HIF-1α stability
RACK1 could compete with HSP90 for binding to HIF-1α and be involved in HIF-1α degradation.12 Given the established interaction of RACK1 with HIF-1α, we sought to analyze the mechanistic role of PCAT-1 in HIF-1α stability by focusing on the interaction between PCAT-1 and RACK1. We first explored the possible interaction between PCAT-1 and RACK1 through bioinformatic analysis (catRAPID), which could predict the protein-RNA associations by estimating the interaction propensity between amino acids and nucleotides. Results from catRAPID fragments analysis showed that the 1093–1172 and 1288–1367 nucleotide positions of the PCAT-1 sequence may bind to the 217–268 amino acid residues of the RACK1 protein with high affinity (Figure 5A).
Figure 5.
Bioinformatics prediction and biochemical identification of the interaction between PCAT-1 and RACK1
(A) CatRAPID fragment module prediction of the binding sites between PCAT-1 and RACK1. (B) The interaction between PCAT-1 and RACK1 was performed by RNA pull-down assay with a biotinylated PCAT-1 probe in MDA-MB-231 cells under hypoxic conditions. (C) RIP analysis of the interaction between PCAT-1 and RACK1 was performed using the anti-RACK1 antibody in MDA-MB-231 cells, and the level of PCAT-1 was determined by quantitative real-time PCR. (D) The truncated PCAT-1 was used in the RNA pull-down assay to identify the core regions of PCAT-1 for the physical interaction with RACK1. (E and F) HIF-1α stability (E) and the expression of HIF-1α target genes (F) were examined in MCF-7 cells overexpressing full-length PCAT-1 (nucleotides 1–1992) or truncated PCAT-1 (nucleotides 1–800 and 1201–1992) under hypoxic conditions. Endogenous PCAT-1 was depleted with siRNA in MDA-MB-231 cells, and the truncated PCAT-1 (nucleotides 801–1992) was overexpressed in the cells. (G–J) Growth (G), migration (H), invasion (I), and colony formation (J) were analyzed in hypoxia. ∗∗p < 0.01 compared with PCAT-1 knockdown group. (K) Schematic diagram of RACK1 functional domains is shown at the top. GST-tagged full-length or deletion mutants of RACK1 were transfected into MDA-MB-231 cells, and the potential interactions between PCAT-1 and the deletion mutants were detected by quantitative real-time PCR under hypoxic conditions. Coomassie blue staining of the GST fusion proteins is shown at the bottom.
To validate the physical interaction between PCAT-1 and RACK1, we performed RNA pull-down followed by immunoblot using RACK1 antibody in MDA-MB-231 cells. The results indicated that PCAT-1 was able to interact with RACK1 (Figure 5B). Consistently, RNA immunoprecipitation (RIP) assays further confirmed that PCAT-1 directly interacted with RACK1 (Figure 5C). Moreover, a series of PCAT-1 truncated fragment analyses demonstrated that the 3′ terminal (nucleotides 801–1200) region of the PCAT-1 was required for direct interaction with RACK1, which was confirmed by RNA pull-down assays (Figure 5D). However, RNA pull-down and RIP assays were conducted, and the results showed that PCAT-1 could not interact with HSP90 (Figure S5).
To further clarify the functional relevance of the PCAT-1/RACK1 interaction, full-length and truncated PCAT-1 (nucleotides 1–800 and 1201–1992) were overexpressed in MCF-7 cells. The results showed that, compared with the full-length PCAT-1, the ability of RACK1 binding-deficient mutant PCAT-1 to maintain HIF-1α stability and induce HIF-1α target genes was significantly impaired (Figures 5E and 5F). Moreover, overexpression of the truncated PCAT-1 (nucleotides 801–1200) in PCAT-1-silenced MDA-MB-231 cells restored the hypoxia-associated cancer phenotypes in the cells (Figures 5G–5J). Full-length RACK1 consists of seven tandemly repeated WD40 domains. To map which WD40 domain on Rack1 is required for PCAT-1 binding, we generated several deletion mutants of RACK1 consisting of WD2–7, WD3–7, WD4–7, WD5–7, WD6–7, and WD7. The RIP assays revealed that constructs containing the sixth WD40 domain could bind to PCAT-1 (Figure 5K), suggesting that the sixth WD40 domain was responsible for the PCAT-1/RACK1 interaction.
PCAT-1 reduces the binding between RACK1 and HIF-1α
To evaluate the effect of PCAT-1 on RACK1 binding to HIF-1α, MDA-MB-231 cells transfected with shRNA-PCAT-1 were treated with MG132. Immunoprecipitation results showed that PCAT-1 knockdown reduced HIF-1α binding to HSP90 but significantly increased the binding efficiency between HIF-1α and RACK1 (Figure 6A). Moreover, MCF-7 cells were cotransfected with PCAT-1 and HA-HIF-1α (P402A/P564A) under hypoxic conditions, and this showed that PCAT-1 overexpression significantly suppressed HA-HIF-1α binding to RACK1 (Figure 6B). Moreover, HSP90 inhibition by 17-AAG increased the binding efficiency between RACK1 and HIF-1α, while it can be abolished by PCAT-1 overexpression. We further investigated the effect of PCAT-1 on RACK1 dimerization. which is necessary in HIF-1α activation. FLAG-RACK1 was transfected in HEK293 cells, and then it was separated by anti-FLAG antibody immobilized on agarose resin. We further used the resin to pull down endogenous RACK1 from the MDA-MB-231 cells transfected with shRNA-PCAT-1, and the results showed that PCAT-1 knockdown increased RACK1 dimerization under hypoxic conditions (Figure 6C). Consistent with the previous study showing that serine phosphorylation of RACK1 was required for its dimerization and HIF-1α degradation,33 we found that PCAT-1 knockdown also enhanced RACK1 phosphorylation (Figure 6D).
Figure 6.
Effect of PCAT-1 on RACK1 binding to HIF-1α
(A) Co-immunoprecipitation of HIF-1α/RACK1 and HIF-1α/HSP90 interactions in MDA-MB-231 cells transfected with shRNA-PCAT-1. (B) Co-immunoprecipitation of HA-HIF-1α (P402A/P564A)/RACK1 and HA-HIF-1α (P402A/P564A)/HSP90 interactions in MCF-7 cells cotransfected with PCAT-1 and HA-HIF-1α (P402A/P564A) in the presence or absence of 17-AAG (0.5 mmol/L) under hypoxic conditions. (C) RACK1 dimerization was detected in MDA-MB-231 cells cotransfected with shRNA-PCAT-1 and FLAG-RACK1 in hypoxia. (D) Serine phosphorylation of RACK1 was determined in PCAT-1-knockdown MDA-MB-231 cells.
Suppression of breast cancer progression by targeting PCAT-1 in vivo
To evaluate the biological function of PCAT-1 in vivo, MDA-MB-231 cells stably transfected with shRNA-PCAT-1 or shRNA-Control lentiviral particles were inoculated into the right flank of nude mice (n = 8/group). Five weeks later, compared with shRNA-Control, the shRNA-PCAT-1 tumor growth was dramatically delayed (Figures 7A–7C).
Figure 7.
PCAT-1 is required in cancer progression and metastasis
(A) Representative images of tumors in xenograft mouse models bearing MDA-MB-231 cells transfected with shRNA-Control or shRNA-PCAT-1. Tumors were harvested at 5 weeks after implantation. (B) The volume of tumor was measured every 5 days. (C) The weights of tumors were measured after 5 weeks when mice were sacrificed. (D and E) Lung metastasis (D) and liver metastasis (E) in mice were determined by quantitative real-time PCR assays. Data are shown as mean ± SD. ∗∗p < 0.01 compared with control. (F–H) Representative H&E images and ISH detection for PCAT-1 in the xenografts (F), lungs (G), and livers (H). Scale bars, 50 μm.
We further injected the cells into the tail veins of nude mice in order to evaluate the metastatic potential of the cells. The mice were sacrificed 8 weeks later, and the lung and liver tissues were obtained for detection of human genomic DNA by quantitative real-time PCR according to the previous studies.34 We found that less human genomic DNA was present in the lungs and livers of mice injected with PCAT-1-silenced cells compared with the control group (Figures 7D and 7E), indicating that PCAT-1 knockdown suppressed circulating tumor cell colonization and prevented metastasis of breast cancer cells to the lung and liver. Moreover, we performed hematoxylin and eosin (H&E) and in situ hybridization staining (ISH) for PCAT-1 in the xenografts, livers, and lungs, and this showed that PCAT-1 expression was significantly downregulated in the shRNA-PCAT-1 group compared with the shRNA-Control group (Figure 7F–7H). Taken together, these results suggested that PCAT-1 knockdown significantly suppressed breast cancer progression and metastasis in vivo, which further confirmed the carcinogenic effects of PCAT-1 in breast cancer.
Discussion
Hypoxia is common in solid tumors, and it modulates cancer progression as well as therapeutic response. Recently, more and more observations have recognized that lncRNA can act as a regulator of cellular responses in hypoxia. Under hypoxic conditions, lncRNA plays regulatory roles through a variety of potential mechanisms such as epigenetics, lncRNA-microRNA (miRNA) interaction, and lncRNA-protein interaction.35, 36, 37 However, only a few have been confirmed in terms of the detailed mechanisms, while the roles of most others are still unclear.
Previous studies have implicated the elevated PCAT-1 expression observed in breast cancer.32 However, none of the studies has investigated the biological functions of PCAT-1 in the hypoxic environment that contributed to the malignant progression. Our present study identifies PCAT-1 as a hypoxia-inducible lncRNA and shows a distinct mechanism in breast cancer. Indeed, we find that PCAT-1 overexpression enhances HIF-1α protein stability and confers hypoxia-associated tumorigenesis including growth, migration, invasion, colony formation, and metabolic regulation. When we investigate the mechanisms of PCAT-1 promoting breast cancer under hypoxic conditions, we find that RACK1 is involved and plays a key role. As a multifunctional scaffold protein for many kinases and receptors, RACK1 binds to the PAS-A subdomain of HIF-1α and recruits an Elongin-C E3 ubiquitin ligase complex to HIF-1α, enhancing HIF-1α ubiquitination and degradation. Here, we show that PCAT-1 is capable of controlling the competition of RACK1 for binding to HIF-1α. PCAT-1 is in an important position to control the binding of RACK1 and HSP90 to HIF-1α. By binding to the sixth WD40 domain of RACK1, PCAT-1 prevents RACK1 phosphorylation and dimerization, which are required for RACK1 binding to HIF-1α. Disrupting the binding of RACK1 to HIF-1α by PCAT-1 makes it easier for HIF-1α to bind to HSP90 rather than being degraded by ubiquitin-mediated proteasomal degradation. Thus, PCAT-1 is an essential component of the survival mechanism of the cellular response to hypoxia.
As the expression of PCAT-1, RACK1, HSP90, and HIF-1α is increased in many kinds of cancers,30,38, 39, 40 the effects of PCAT-1 on the balance between HIF-1α/RACK1 and HIF-1α/HSP90 may contribute to cancer progression. It is the basis for the association of PCAT-1 with the clinicopathological prognosis of the diseases. As we have shown, PCAT-1 is almost absent or at a low level in the normal tissues, and it increased to a high level in the tumor tissues. Considering the role of PCAT-1 in the upregulation of HIF-1α, facilitation of HIF-1α stability by PCAT-1 may explain the close relation of PCAT-1 overexpression to advanced tumor stage and tumor size. In addition to hypoxia, PCAT-1 also promotes the expression of HIF-1α in normoxia. Since HIF-1α also increases in some cancer cells under normoxic conditions,41 it can be speculated that PCAT-1 also participates in the regulation of the HIF-1α signaling pathway in normoxia during cancer progression.
It is demonstrated that HIF-1α and its target genes control biological processes in cancers.40,42,43 HIF-1α and the downstream effectors have been considered as potential targets for cancer diagnosis and therapy because of their profound impacts in cancers.11,44 However, it is hard to design effective inhibitors because of the complexity of the HIF-1α signaling.43 Because lncRNA is easy to extract and detect, it has many advantages in cancer diagnosis and prognosis.19 Our work presents evidence showing that PCAT-1 is an independent risk factor for OS and DFS, suggesting the potential clinical application of PCAT-1 as a biomarker for breast cancer prognosis. Thus, it illustrates that PCAT-1 intervention through gene silencing or other strategies could present a potential new approach to anticancer therapies.
though our study supports that the expression of PCAT-1 is critical to breast cancer in normoxia and hypoxia, PCAT-1 overexpression may be the result of other modulations in cells, and it may be regulated by other factors involved in cancer progression. Our research cannot completely rule out the possibility that PCAT-1 may also enhance HIF-1α stability by regulating other factors that affect HIF-1α degradation. For example, some post-translational modifications, such as acetylation, phosphorylation, nitrosylation, and SUMOylation, have also been reported to be involved in the regulation of HIF-1α expression and activation.45 These questions should be resolved through future experiments to clarify the role and mechanism of PCAT-1 in breast cancer.
In summary, our studies highlight a previously unknown function of PCAT-1 in cancer progression under hypoxic conditions, especially its role in regulating the oxygen-independent stability of HIF-1α. The effects of PCAT-1 in HIF-1α stability not only provide new insight into the survival mechanism of cells in response to oxygen deficiency but also reveal a potential utility of PCAT-1 in prognosis and therapeutic strategy for breast cancer.
Materials and methods
Cell culture and transfection
Human breast cancer cell lines MCF-7, MDA-MB-231, MDA-MB-468, and T47D, normal breast epithelial cell MCF10A, and human embryonic kidney (HEK) 293 cells were purchased from the American Type Culture Collection (Manassas, VA, USA). The cell lines were cultured in DMEM (Gibco, USA) with 10% fetal bovine serum (FBS), 100 ng/mL streptomycin, and 100 U/mL penicillin. For normoxic conditions, mixed gas consisting of 20% O2, 5% CO2, and 75% N2 at 37°C was used. For hypoxic conditions, the cells were cultured in a gas mixture of 1% O2, 5% CO2, and 94% N2 at 37°C.
Transfection of the cells with siRNAs was performed with Lipofectamine RNAiMAX (Invitrogen, USA) according to the manufacturer’s instructions. For siRNA-mediated knockdown of PCAT-1, two different siRNAs were synthesized by GenePharma (Shanghai, China) using the following sequences: si-PCAT-1#1: 5′-GCAGAAACACCAAUGGAUAUU-3′; si-PCAT-1#2: 5′-GAACCUAACUGGACUUUAAUU-3′. The specific siRNAs for RACK1, HIF-1α, and HIF-2α were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). The lentiviral PCAT-1 shRNA (shRNA-PCAT-1) was constructed by Hanbio (Shanghai, China) using the following sequence: 5′-AUACAUAAGACCAUGGAAAU-3′.28 The PCAT-1, truncated PCAT-1 (Δ1–400), truncated PCAT-1 (Δ401–800), truncated PCAT-1 (Δ801–1200), truncated PCAT-1 (Δ1201–1600), truncated PCAT-1 (Δ1601–1992), truncated PCAT-1 (Δ1–800), and truncated PCAT-1 (Δ1201–1992) overexpressed lentiviruses were constructed by Hanbio (Shanghai, China). To express and purify glutathione S-transferase (GST)-RACK1, a full length of RACK1 and its truncated RACK1 cDNAs were amplified by PCR using the appropriate primers and subcloned in frame into pGEX4T-1 to make the GST fusion protein using bacterial expression vectors. The HIF-1α mutant coding plasmid HA-HIF-1α-P402A/P564A-pcDNA3 was purchased from Addgene (Cambridge, MA, USA).
Tissue specimens
Tumors and the adjacent breast tissues used in this study were surgical specimens from patients with breast cancer. All samples were fresh frozen in liquid nitrogen and stored at −80°C until RNA extraction. The study protocol was approved by the Ethics Committee of the Second Hospital of Hebei Medical University (Hebei, China), and informed consent was obtained from all participants.
RNA extraction and quantitative real-time PCR
Total RNA was extracted by TRIzol Reagent (Invitrogen, USA). Equal amounts of RNA were reverse-transcribed with the Transcriptor First Strand cDNA Synthesis Kit (Roche), and quantitative real-time PCR was performed with the SYBR PrimeScript RT-PCR Kit (Takara, Dalian, China). Unless otherwise specified, the amplified transcript level of each specific gene was normalized against the glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mRNA level. The primers were provided by Shenggong (Shanghai, China). The primer sequences are listed in Table S3.
Bioinformatics analysis
TCGA Breast Cancer datasets were retrieved from the cBioPortal for Cancer Genomics. PCAT-1 gene log2 copy number from cBioPortal was downloaded. Patients were grouped into PCAT-1-low or PCAT-1-amplification based on PCAT-1 log2 copy number. The cutoff point of PCAT-1 gene log2 copy number calls on the cases was determined as the value yielding maximum sum of sensitivity and specificity. The clinical data (including the overall survival data and disease-free survival data) were obtained from the TCGA Pan-Cancer Clinical Data Resource.46 The patients with overall event time <130 months were used in this study. The survival curves were constructed according to the Kaplan-Meier method and compared with the log-rank test.
Cell growth assay
The cell growth was monitored by MTT assay. Briefly, cells were treated and cultured into 96-well plates. A total of 20 μL of MTT (Sigma, USA) dissolved in PBS at 5 mg/mL was added to the wells and then incubated for an additional 4 h at 37°C. A total of 100 μL of DMSO was added to dissolve the formed formazan crystals, and the optical density was measured on the microplate reader.
Colony formation assay
Cells were seeded in 6-well plates (200 cells/well) and treated for 14 days, and then the cells were fixed with methanol and stained with 1% crystal violet solution. The numbers of cell colonies from three dishes were counted.
Wound healing and invasion assay
For the wound healing assays, cells were treated and cultured in 6-well plates until almost totally confluent. Then, the artificial wounds were scratched with a sterile pipette tip, and images were captured at 0 h and 48 h under the microscope.
Cell invasion assays were performed with Transwell chambers (8 μm pores, Corning, USA) coated with Matrigel (BD Biosciences, USA). Cells were treated and suspended in medium with 1% FBS in the upper chambers. Medium containing 10% FBS was used as a chemoattractant in the bottom chamber. After incubation for 24 h, the migrated or invaded cells were fixed, stained, and counted under an inverted microscope.
Measurement of ECAR
The ECAR was measured with the Seahorse Extracellular Flux Analyzer XF96 (Seahorse Bioscience, USA) according to the manufacturer’s instructions. Briefly, the treated cells were seeded at a density of 105 per well and allowed to adhere overnight. Before the assay, the cells were washed with assay medium followed by a sequential injection of 10 mmol/L glucose, 1 μmol/L oligomycin, and 100 mmol/L 2-deoxyglucose (2-DG). ECAR is expressed as mpH/min, and three replicates were performed for each group.
Lactate production assay
Cells were seeded into 6-well plates, and the culture medium was collected for measurement of lactate concentration. The level of lactate in the culture medium was determined with the L-Lactate Assay Kit (Abcam, USA), following the manufacturer’s instructions.
Immunoblot analysis and co-immunoprecipitation (coIP)
For immunoblot analysis, radioimmunoprecipitation assay (RIPA) buffer was added and incubated for 30 min, followed by centrifugation; the supernatant contained the total protein. After determination of the protein concentrations by the Bradford method, proteins were separated by SDS-PAGE and transferred onto polyvinylidene fluoride (PVDF) membranes. The membranes were blocked with 5% fat-free milk powder in Tris-buffered saline containing Tween-20 (TBST) buffer and then exposed to primary antibodies at 4°C overnight. After incubation with secondary antibodies, signals were detected with the enhanced chemiluminescence (ECL) detection system.
For immunoprecipitation, protein extracts from the cells were determined by the Bradford method. For the experiment, 500 μg of protein was immunoprecipitated with indicated antibodies at 4°C overnight, followed by incubation with protein A Sepharose beads for 2 h. The immunoprecipitates were washed and resolved by SDS-PAGE and immunoblotted with the indicated antibodies.
For coIP, cell lysates were mixed with mouse/rabbit IgG or primary antibodies, and then the antibody-protein complex was incubated with Protein A/G PLUS-Agarose (Santa Cruz Biotechnology, USA). The agarose-antibody-protein complex was collected and then assessed by immunoblot analysis. The following antibodies were used: anti-HIF-1α (Abcam), anti-RACK1 (Cell Signaling Technology), anti-HSP90 (Abcam), anti-ubiquitin (Abcam), anti-p-Ser (Abcam), anti-HA (Abcam), anti-FLAG (Sigma), and anti-β-actin (Abcam).
RNA pull-down assay
Biotin-labeled PCAT-1 and its fragments were transcribed with Biotin RNA Labeling Mix (Roche) and T7 RNA polymerase (Invitrogen, USA), treated with RNase-free DNase I (Invitrogen, USA), and purified with the RNeasy Mini Kit (QIAGEN, Germany). Pull-downs were performed with biotinylated RNA and cell extracts. The bead-RNA-protein complexes were isolated with streptavidin magnetic beads (Sigma, USA) and subjected to standard immunoblot analysis.
RIP
The RIP assay was performed with the Magna RIP RNA Binding Protein Immunoprecipitation Kit (Millipore, USA). Briefly, cells were harvested and lysed in lysis buffer containing protease inhibitor cocktail and RNase inhibitors. Cell extracts were co-immunoprecipitated using the primary antibody, and the retrieved RNA was subjected to quantitative real-time PCR analysis to demonstrate the presence of the binding products using respective primers.
Xenograft experiments
The animal studies were performed as described previously.47,48 Female BALB/c nude mice (4–5 weeks old) were purchased from Charles River (Beijing, China). All experimental procedures were approved by the Institutional Animal Care and Use Committee of the Second Hospital of Hebei Medical University (Hebei, China). A total of 106 MDA-MB- (transfected with sh-Control or sh-PCAT-1) were injected into the right flank or the tail vein of mice (n = 8/group), respectively. The tumor volume was measured every 5 days, and tumor volume was estimated by the formula V (mm3) = 1/2 × length × width2. At the end of the experiment, the tumor tissues in the tumor growth xenograft model were removed and weighed. The lung and liver in the metastasis model were perfused with PBS and subjected to quantitative real-time PCR with primers for human HK2 gene and mouse and human 18S rRNA. The remaining tumor tissues, lung, and liver were fixed in formalin, embedded in paraffin, and analyzed by H&E staining.
ISH
ISH was used to detect the presence of PCAT-1 with digoxigenin-labeled probes designed and synthesized by Exiqon (Shanghai, China). ISH was performed with the ISH Kit (Boster Bio-Engineering, Wuhan, China), and the stains were then scored using the following criteria as described previously:49 0 = negative; 1 = weak staining; 2 = moderate staining; and 3 = strong staining.
Statistical analysis
The overall and disease-free survival curves were calculated by the Kaplan-Meier method, and the differences were assessed with the log-rank test. The nonparametric Mann-Whitney U test served to assess the differences of PCAT-1 expression in clinical specimens. Differences between two independent groups were tested with two-tailed Student’s t test. Comparison of multiple groups was made with one- or two-way ANOVA tests. Univariate and multivariate Cox proportional hazards regression models were performed to explore independent prognostic factors. Data are expressed as mean ± SD. Difference was considered statistically significant at p <0.05.
Acknowledgments
This work was supported by National Natural Science Foundation of China (no. 81402510, 11672332, 11932013), National Key R&D Program of China (no. 2016YFC1101500) and Natural Science Foundation of Tianjin (no. 18JCQNJC09600).
Author contributions
B.L. and J.G. designed the study. J.W., X.C., H.H., Y.S., and N.Z. performed the experiments. J.W., X.X., and X.C. collected clinical samples. M.Y., N.Z., and J.G. analyzed the data. J.G. and B.L. wrote the paper. All authors read and approved the final manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2021.02.034.
Contributor Information
Jianzhao Gao, Email: gaojz@nankai.edu.cn.
Bin Liu, Email: liubin810103@yahoo.com.
Supplemental information
References
- 1.Harbeck N., Penault-Llorca F., Cortes J., Gnant M., Houssami N., Poortmans P., Ruddy K., Tsang J., Cardoso F. Breast cancer. Nat. Rev. Dis. Primers. 2019;5:66. doi: 10.1038/s41572-019-0111-2. [DOI] [PubMed] [Google Scholar]
- 2.Waks A.G., Winer E.P. Breast Cancer Treatment: A Review. JAMA. 2019;321:288–300. doi: 10.1001/jama.2018.19323. [DOI] [PubMed] [Google Scholar]
- 3.Redig A.J., McAllister S.S. Breast cancer as a systemic disease: a view of metastasis. J. Intern. Med. 2013;274:113–126. doi: 10.1111/joim.12084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Esteva F.J., Hubbard-Lucey V.M., Tang J., Pusztai L. Immunotherapy and targeted therapy combinations in metastatic breast cancer. Lancet Oncol. 2019;20:e175–e186. doi: 10.1016/S1470-2045(19)30026-9. [DOI] [PubMed] [Google Scholar]
- 5.Wouters B.G., Koritzinsky M. Hypoxia signalling through mTOR and the unfolded protein response in cancer. Nat. Rev. Cancer. 2008;8:851–864. doi: 10.1038/nrc2501. [DOI] [PubMed] [Google Scholar]
- 6.Nobre A.R., Entenberg D., Wang Y., Condeelis J., Aguirre-Ghiso J.A. The Different Routes to Metastasis via Hypoxia-Regulated Programs. Trends Cell Biol. 2018;28:941–956. doi: 10.1016/j.tcb.2018.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu J.N., Bu W., Shi J. Chemical Design and Synthesis of Functionalized Probes for Imaging and Treating Tumor Hypoxia. Chem. Rev. 2017;117:6160–6224. doi: 10.1021/acs.chemrev.6b00525. [DOI] [PubMed] [Google Scholar]
- 8.Palazon A., Goldrath A.W., Nizet V., Johnson R.S. HIF transcription factors, inflammation, and immunity. Immunity. 2014;41:518–528. doi: 10.1016/j.immuni.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choudhry H., Harris A.L. Advances in Hypoxia-Inducible Factor Biology. Cell Metab. 2018;27:281–298. doi: 10.1016/j.cmet.2017.10.005. [DOI] [PubMed] [Google Scholar]
- 10.Rankin E.B., Giaccia A.J. Hypoxic control of metastasis. Science. 2016;352:175–180. doi: 10.1126/science.aaf4405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Semenza G.L. Targeting HIF-1 for cancer therapy. Nat. Rev. Cancer. 2003;3:721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 12.Liu Y.V., Semenza G.L. RACK1 vs. HSP90: competition for HIF-1 alpha degradation vs. stabilization. Cell Cycle. 2007;6:656–659. doi: 10.4161/cc.6.6.3981. [DOI] [PubMed] [Google Scholar]
- 13.Zhou Z., Liu F., Zhang Z.S., Shu F., Zheng Y., Fu L., Li L.Y. Human rhomboid family-1 suppresses oxygen-independent degradation of hypoxia-inducible factor-1α in breast cancer. Cancer Res. 2014;74:2719–2730. doi: 10.1158/0008-5472.CAN-13-1027. [DOI] [PubMed] [Google Scholar]
- 14.Yang S.J., Park Y.S., Cho J.H., Moon B., An H.J., Lee J.Y., Xie Z., Wang Y., Pocalyko D., Lee D.C. Regulation of hypoxia responses by flavin adenine dinucleotide-dependent modulation of HIF-1α protein stability. EMBO J. 2017;36:1011–1028. doi: 10.15252/embj.201694408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Y.V., Baek J.H., Zhang H., Diez R., Cole R.N., Semenza G.L. RACK1 competes with HSP90 for binding to HIF-1alpha and is required for O(2)-independent and HSP90 inhibitor-induced degradation of HIF-1alpha. Mol. Cell. 2007;25:207–217. doi: 10.1016/j.molcel.2007.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liz J., Esteller M. lncRNAs and microRNAs with a role in cancer development. Biochim. Biophys. Acta. 2016;1859:169–176. doi: 10.1016/j.bbagrm.2015.06.015. [DOI] [PubMed] [Google Scholar]
- 17.Ransohoff J.D., Wei Y., Khavari P.A. The functions and unique features of long intergenic non-coding RNA. Nat. Rev. Mol. Cell Biol. 2018;19:143–157. doi: 10.1038/nrm.2017.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Engreitz J.M., Ollikainen N., Guttman M. Long non-coding RNAs: spatial amplifiers that control nuclear structure and gene expression. Nat. Rev. Mol. Cell Biol. 2016;17:756–770. doi: 10.1038/nrm.2016.126. [DOI] [PubMed] [Google Scholar]
- 19.Huarte M. The emerging role of lncRNAs in cancer. Nat. Med. 2015;21:1253–1261. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 20.Prensner J.R., Chinnaiyan A.M. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1:391–407. doi: 10.1158/2159-8290.CD-11-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang Y.N., Zhang K., Hu Z.M., Qi H.X., Shi Z.M., Han X.H., Han Y.W., Hong W. Hypoxia-regulated lncRNAs in cancer. Gene. 2016;575:1–8. doi: 10.1016/j.gene.2015.08.049. [DOI] [PubMed] [Google Scholar]
- 22.Prensner J.R., Iyer M.K., Balbin O.A., Dhanasekaran S.M., Cao Q., Brenner J.C., Laxman B., Asangani I.A., Grasso C.S., Kominsky H.D. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat. Biotechnol. 2011;29:742–749. doi: 10.1038/nbt.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ge X., Chen Y., Liao X., Liu D., Li F., Ruan H., Jia W. Overexpression of long noncoding RNA PCAT-1 is a novel biomarker of poor prognosis in patients with colorectal cancer. Med. Oncol. 2013;30:588. doi: 10.1007/s12032-013-0588-6. [DOI] [PubMed] [Google Scholar]
- 24.Qin H.D., Liao X.Y., Chen Y.B., Huang S.Y., Xue W.Q., Li F.F., Ge X.S., Liu D.Q., Cai Q., Long J. Genomic Characterization of Esophageal Squamous Cell Carcinoma Reveals Critical Genes Underlying Tumorigenesis and Poor Prognosis. Am. J. Hum. Genet. 2016;98:709–727. doi: 10.1016/j.ajhg.2016.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi J., Li X., Zhang F., Kong L., Zhang X., Cheng Y., Guan Q., Cao X., Zhu W., Ou K. The Plasma LncRNA Acting as Fingerprint in Hilar Cholangiocarcinoma. Cell. Physiol. Biochem. 2018;49:1694–1702. doi: 10.1159/000493613. [DOI] [PubMed] [Google Scholar]
- 26.Zhang X., Zhang Y., Mao Y., Ma X. The lncRNA PCAT1 is correlated with poor prognosis and promotes cell proliferation, invasion, migration and EMT in osteosarcoma. OncoTargets Ther. 2018;11:629–638. doi: 10.2147/OTT.S152063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wen J., Xu J., Sun Q., Xing C., Yin W. Upregulation of long non coding RNA PCAT-1 contributes to cell proliferation, migration and apoptosis in hepatocellular carcinoma. Mol. Med. Rep. 2016;13:4481–4486. doi: 10.3892/mmr.2016.5075. [DOI] [PubMed] [Google Scholar]
- 28.Prensner J.R., Chen W., Iyer M.K., Cao Q., Ma T., Han S., Sahu A., Malik R., Wilder-Romans K., Navone N. PCAT-1, a long noncoding RNA, regulates BRCA2 and controls homologous recombination in cancer. Cancer Res. 2014;74:1651–1660. doi: 10.1158/0008-5472.CAN-13-3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo H., Ahmed M., Zhang F., Yao C.Q., Li S., Liang Y., Hua J., Soares F., Sun Y., Langstein J. Modulation of long noncoding RNAs by risk SNPs underlying genetic predispositions to prostate cancer. Nat. Genet. 2016;48:1142–1150. doi: 10.1038/ng.3637. [DOI] [PubMed] [Google Scholar]
- 30.Shang Z., Yu J., Sun L., Tian J., Zhu S., Zhang B., Dong Q., Jiang N., Flores-Morales A., Chang C., Niu Y. LncRNA PCAT1 activates AKT and NF-κB signaling in castration-resistant prostate cancer by regulating the PHLPP/FKBP51/IKKα complex. Nucleic Acids Res. 2019;47:4211–4225. doi: 10.1093/nar/gkz108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang L., Wang Y., Chen J., Wang Y., Zhao Y., Wang Y., Ma Y., Chen X., Liu W., Li Z. Long noncoding RNA PCAT1, a novel serum-based biomarker, enhances cell growth by sponging miR-326 in oesophageal squamous cell carcinoma. Cell Death Dis. 2019;10:513. doi: 10.1038/s41419-019-1745-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sarrafzadeh S., Geranpayeh L., Ghafouri-Fard S. Expression Analysis of Long Non-Coding PCAT-1in Breast Cancer. Int. J. Hematol. Oncol. Stem Cell Res. 2017;11:185–191. [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Y.V., Hubbi M.E., Pan F., McDonald K.R., Mansharamani M., Cole R.N., Liu J.O., Semenza G.L. Calcineurin promotes hypoxia-inducible factor 1alpha expression by dephosphorylating RACK1 and blocking RACK1 dimerization. J. Biol. Chem. 2007;282:37064–37073. doi: 10.1074/jbc.M705015200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen Y., Zhang B., Bao L., Jin L., Yang M., Peng Y., Kumar A., Wang J.E., Wang C., Zou X. ZMYND8 acetylation mediates HIF-dependent breast cancer progression and metastasis. J. Clin. Invest. 2018;128:1937–1955. doi: 10.1172/JCI95089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zeng Z., Xu F.Y., Zheng H., Cheng P., Chen Q.Y., Ye Z., Zhong J.X., Deng S.J., Liu M.L., Huang K. LncRNA-MTA2TR functions as a promoter in pancreatic cancer via driving deacetylation-dependent accumulation of HIF-1α. Theranostics. 2019;9:5298–5314. doi: 10.7150/thno.34559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Y., Chen W., Lian J., Zhang H., Yu B., Zhang M., Wei F., Wu J., Jiang J., Jia Y. The lncRNA PVT1 regulates nasopharyngeal carcinoma cell proliferation via activating the KAT2A acetyltransferase and stabilizing HIF-1α. Cell Death Differ. 2020;27:695–710. doi: 10.1038/s41418-019-0381-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shih J.W., Chiang W.F., Wu A.T.H., Wu M.H., Wang L.Y., Yu Y.L., Hung Y.W., Wang W.C., Chu C.Y., Hung C.L. Long noncoding RNA LncHIFCAR/MIR31HG is a HIF-1α co-activator driving oral cancer progression. Nat. Commun. 2017;8:15874. doi: 10.1038/ncomms15874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Duff D., Long A. Roles for RACK1 in cancer cell migration and invasion. Cell. Signal. 2017;35:250–255. doi: 10.1016/j.cellsig.2017.03.005. [DOI] [PubMed] [Google Scholar]
- 39.Wu J., Liu T., Rios Z., Mei Q., Lin X., Cao S. Heat Shock Proteins and Cancer. Trends Pharmacol. Sci. 2017;38:226–256. doi: 10.1016/j.tips.2016.11.009. [DOI] [PubMed] [Google Scholar]
- 40.Schito L., Semenza G.L. Hypoxia-Inducible Factors: Master Regulators of Cancer Progression. Trends Cancer. 2016;2:758–770. doi: 10.1016/j.trecan.2016.10.016. [DOI] [PubMed] [Google Scholar]
- 41.Lee Y.H., Bae H.C., Noh K.H., Song K.H., Ye S.K., Mao C.P., Lee K.M., Wu T.C., Kim T.W. Gain of HIF-1α under normoxia in cancer mediates immune adaptation through the AKT/ERK and VEGFA axes. Clin. Cancer Res. 2015;21:1438–1446. doi: 10.1158/1078-0432.CCR-14-1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Keith B., Johnson R.S., Simon M.C. HIF1α and HIF2α: sibling rivalry in hypoxic tumour growth and progression. Nat. Rev. Cancer. 2011;12:9–22. doi: 10.1038/nrc3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nordgren I.K., Tavassoli A. Targeting tumour angiogenesis with small molecule inhibitors of hypoxia inducible factor. Chem. Soc. Rev. 2011;40:4307–4317. doi: 10.1039/c1cs15032d. [DOI] [PubMed] [Google Scholar]
- 44.Yee Koh M., Spivak-Kroizman T.R., Powis G. HIF-1 regulation: not so easy come, easy go. Trends Biochem. Sci. 2008;33:526–534. doi: 10.1016/j.tibs.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 45.Kuschel A., Simon P., Tug S. Functional regulation of HIF-1α under normoxia--is there more than post-translational regulation? J. Cell. Physiol. 2012;227:514–524. doi: 10.1002/jcp.22798. [DOI] [PubMed] [Google Scholar]
- 46.Liu J., Lichtenberg T., Hoadley K.A., Poisson L.M., Lazar A.J., Cherniack A.D., Kovatich A.J., Benz C.C., Levine D.A., Lee A.V., Cancer Genome Atlas Research Network An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics. Cell. 2018;173:400–416.e11. doi: 10.1016/j.cell.2018.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu B., Wen J.K., Li B.H., Fang X.M., Wang J.J., Zhang Y.P., Shi C.J., Zhang D.Q., Han M. Celecoxib and acetylbritannilactone interact synergistically to suppress breast cancer cell growth via COX-2-dependent and -independent mechanisms. Cell Death Dis. 2011;2:e185. doi: 10.1038/cddis.2011.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu B., Han M., Sun R.H., Wang J.J., Zhang Y.P., Zhang D.Q., Wen J.K. ABL-N-induced apoptosis in human breast cancer cells is partially mediated by c-Jun NH2-terminal kinase activation. Breast Cancer Res. 2010;12:R9. doi: 10.1186/bcr2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang A., Zhao J.C., Kim J., Fong K.W., Yang Y.A., Chakravarti D., Mo Y.Y., Yu J. LncRNA HOTAIR Enhances the Androgen-Receptor-Mediated Transcriptional Program and Drives Castration-Resistant Prostate Cancer. Cell Rep. 2015;13:209–221. doi: 10.1016/j.celrep.2015.08.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.