Significance
Osteoarthritis is a global health problem that affects load-bearing joints, causing loss of mobility and enormous healthcare costs. However, disease-modifying approaches are lacking. Here, we report a cellular mechanism of inflammatory signaling in chondrocytes, the cellular component of cartilage. We show how osteoarthritis-relevant levels of interleukin-1α reprogram articular chondrocytes so that they become more susceptible to mechanical trauma, which chondrocytes sense via Piezo1/2-mechanosensitive ion channels. We uncover that IL-1α enhances gene expression of Piezo1 in primary articular chondrocytes underlying Piezo1 enhanced function. We elucidate signaling from membrane to nucleus, including transcription factors that enhance Piezo1 expression. We also define consequences of increased expression of Piezo1, for mechanotransduction and at rest, that implicate this reprogramming mechanism in osteoarthritis pathogenesis.
Keywords: osteoarthritis, interleukin-1, PIEZO1, Piezo1 gene regulation
Abstract
Osteoarthritis (OA) is a painful and debilitating condition of synovial joints without any disease-modifying therapies [A. M. Valdes, T. D. Spector, Nat. Rev. Rheumatol. 7, 23–32 (2011)]. We previously identified mechanosensitive PIEZO channels, PIEZO1 and PIEZO2, both expressed in articular cartilage, to function in chondrocyte mechanotransduction in response to injury [W. Lee et al., Proc. Natl. Acad. Sci. U.S.A. 111, E5114–E5122 (2014); W. Lee, F. Guilak, W. Liedtke, Curr. Top. Membr. 79, 263–273 (2017)]. We therefore asked whether interleukin-1–mediated inflammatory signaling, as occurs in OA, influences Piezo gene expression and channel function, thus indicative of maladaptive reprogramming that can be rationally targeted. Primary porcine chondrocyte culture and human osteoarthritic cartilage tissue were studied. We found that interleukin-1α (IL-1α) up-regulated Piezo1 in porcine chondrocytes. Piezo1 expression was significantly increased in human osteoarthritic cartilage. Increased Piezo1 expression in chondrocytes resulted in a feed-forward pathomechanism whereby increased function of Piezo1 induced excess intracellular Ca2+ at baseline and in response to mechanical deformation. Elevated resting state Ca2+ in turn rarefied the F-actin cytoskeleton and amplified mechanically induced deformation microtrauma. As intracellular substrates of this OA-related inflammatory pathomechanism, in porcine articular chondrocytes exposed to IL-1α, we discovered that enhanced Piezo1 expression depended on p38 MAP-kinase and transcription factors HNF4 and ATF2/CREBP1. CREBP1 directly bound to the proximal PIEZO1 gene promoter. Taken together, these signaling and genetic reprogramming events represent a detrimental Ca2+-driven feed-forward mechanism that can be rationally targeted to stem the progression of OA.
Osteoarthritis (OA) is a significant global health issue with increasing population age as well as rising obesity rates (1–3). OA is characterized by progressive joint degeneration and pain, leading to significant disability and lack of mobility that further aggravates other age-associated conditions. Due to the multifactorial etiology of the disease and the lack of a full understanding of OA pathogenesis, there are no disease-modifying OA drugs (DMOADs) currently available (4, 5). However, growing evidence has documented increased levels of interleukin-1 (IL-1)–based inflammatory signaling in chondrocytes, the sole cell population in healthy articular cartilage (6–10). Articular chondrocytes express functional IL-1 receptor and respond to both isoforms of IL-1 (α and β) potently through catabolic and antianabolic activities (11). Mechanical factors, over protracted times through multiple iterative microtrauma, play a critical role in OA pathogenesis through alterations in cell-mediated mechanotransduction in cartilage (2, 12–17) and may interact with injurious loading to enhance cartilage degeneration (18). At the molecular level, we have described the presence of both mechanosensory Piezo ion channels (PIEZO1 and PIEZO2) in chondrocytes, which function synergistically in response to injurious mechanical loading (19). In the present study, we address the unanswered question whether joint inflammation, as occurs in OA, affects gene regulation and function of Piezo ion channels as a propathogenic OA mechanism. We provide affirmative and mechanistic answers on how IL-1α–mediated inflammatory signaling in articular chondrocytes up-regulates PIEZO1 gene expression and function.
Results
OA-Relevant Levels of Proinflammatory IL-1α Enhance Expression of PIEZO1.
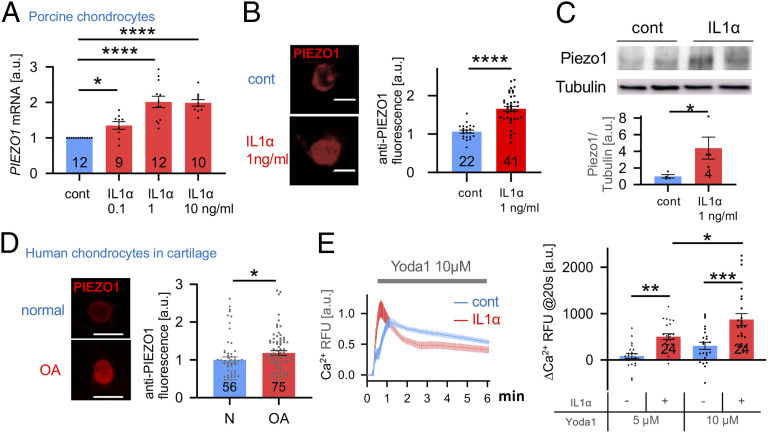
First, we found PIEZO1 messenger RNA (mRNA) was significantly increased in porcine primary articular chondrocytes in response to IL-1α over a range of physiologically and pathologically relevant concentrations (Fig. 1A). Dose dependence was clearly present between control, 0.1 ng/mL, and 1 ng/mL IL-1α, followed by a plateau of Piezo 1 mRNA levels when comparing 1 ng/mL and 10 ng/mL IL-1α. This increase also manifested as elevated PIEZO1 protein expression as shown by up-regulated PIEZO1 immunolabeling of primary porcine chondrocytes and Western blotting (Fig. 1 B and C) using a PIEZO1 antibody that we verified in a human PIEZO1−/− cell line (SI Appendix, Fig. S1). Expression of PIEZO2 was increased in response to IL-1α but not to significant degree (SI Appendix, Fig. S2). Given the importance of Piezo2 in force sensing, more focused exploration of its role in OA pathogenesis will be the subject of future studies, but here we focus on the overtly regulated Piezo1. We also probed expression of TRPV4, a well-characterized chondrocyte mechanosensor (20–23), which we found not to be regulated by IL-1α (SI Appendix, Fig. S2). We also measured PIEZO1 expression in human cartilage, where we detected significantly elevated PIEZO1 protein by immunolabeling in osteoarthritic cartilage compared to normal controls (Fig. 1D) again using the validated PIEZO1 antibody. Thus, IL-1α signaling at OA-relevant concentrations evokes significantly increased PIEZO1 mRNA expression, a finding confirmed at the protein level in articular chondrocytes and human osteoarthritic cartilage lesions. We therefore decided to test PIEZO1 function by use of Yoda-1, a specific PIEZO1 activator, and Ca2+-imaging (24). In response to Yoda-1, primary porcine chondrocytes pretreated with IL-1α exhibited enhanced Ca2+ signaling, with a robustly accelerated signal increase and a resulting vastly increased amount of Ca2+ entering the cell (Fig. 1E). We conclude that IL-1α inflammatory signaling of articular chondrocytes increases PIEZO1 expression and function in chondrocytes. Building on this finding, the concept emerges that inflammation deleteriously predisposes chondrocytes to become more sensitive to injurious levels of loading. Our finding that TRPV4, a mechanosensor that transduces physiologic levels of loading in chondrocytes, is not regulated by IL-1α suggests inflammation selectively alters chondrocyte mechanotransduction pathways. Based on this result, we conducted more in-depth Ca2+-signaling studies in response to controlled single-cell mechanical stimulation with or without IL-1α–mediated inflammation.
Fig. 1.
Increased Piezo1 expression in IL-1α–treated porcine chondrocytes and human osteoarthritic cartilage. (A) PIEZO1 mRNA level in control and IL-1α–treated porcine chondrocytes. IL-1α increases Piezo1 mRNA levels; the number of independent experiments (= primary chondrocytes from separate joints were generated) is indicated in bars; a.u., arbitrary units. (B, Left) Representative confocal micrographs of IL-1α–treated porcine chondrocytes, immunolabeled for PIEZO1. (Right) Significantly increased PIEZO1 protein expression is found in IL-1α–treated porcine chondrocytes versus control. Numbers in bars indicate number of cells quantified; three independent cell isolations are shown. (C) Western blot in control and IL-1α–treated porcine chondrocytes. Note increased PIEZO1 protein expression with IL-1α treatment versus control, β-tubulin not regulated. The upper panels show representative immunoblots (see SI Appendix, Fig. S1 for full blot), and the lower shows bar diagram densitometric quantitation, normalized for β-tubulin; the number of independent experiments is indicated in bars. (D, Left) Representative confocal micrographs of healthy and osteoarthritic human chondrocytes; PIEZO1-specific immunolabeling of chondrocytes in human cartilage tissue (see also SI Appendix, Fig. S2 for antibody validation using PIEZO1−/− cells) (Scale bar, 10 μm). (Right) Relative expression level PIEZO1 in healthy and OA chondrocytes. The numbers in bars indicate the number of cells quantified; five independent human cartilage preparations are shown. (E) IL-1α–treated porcine chondrocytes show different Ca2+ dynamics in response to application of the PIEZO1-selective activating small molecule, Yoda-1 (Left). In this Ca2+ dynamics over time, note accelerated onset and then plateau in the IL-1α–treated cells versus protracted influx in control chondrocytes, indicative of an enhanced PIEZO1-mediated Ca2+ signaling under inflammatory conditions. (Right) Bar diagrams illustrate quantitation of the speed of onset of the Ca2+ transient; note more rapid onset under condition of IL-1α–mediated inflammation. The increased potency of 10 µM Yoda-1 versus 5 µM Yoda-1 is apparent as well; one independent cell isolation with 24 cells per experimental group. Bars represent mean ± SEM; for group comparison: t test for B and C; one-way ANOVA, Tukey’s post hoc test for A; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 significantly different from control. (Scale bars in A and D, 10 µm.)
Piezo1 Increased Expression and Enhanced Function Elevates Baseline [Ca2+]o and Renders Chondrocytes Mechanically Hypersensitive.
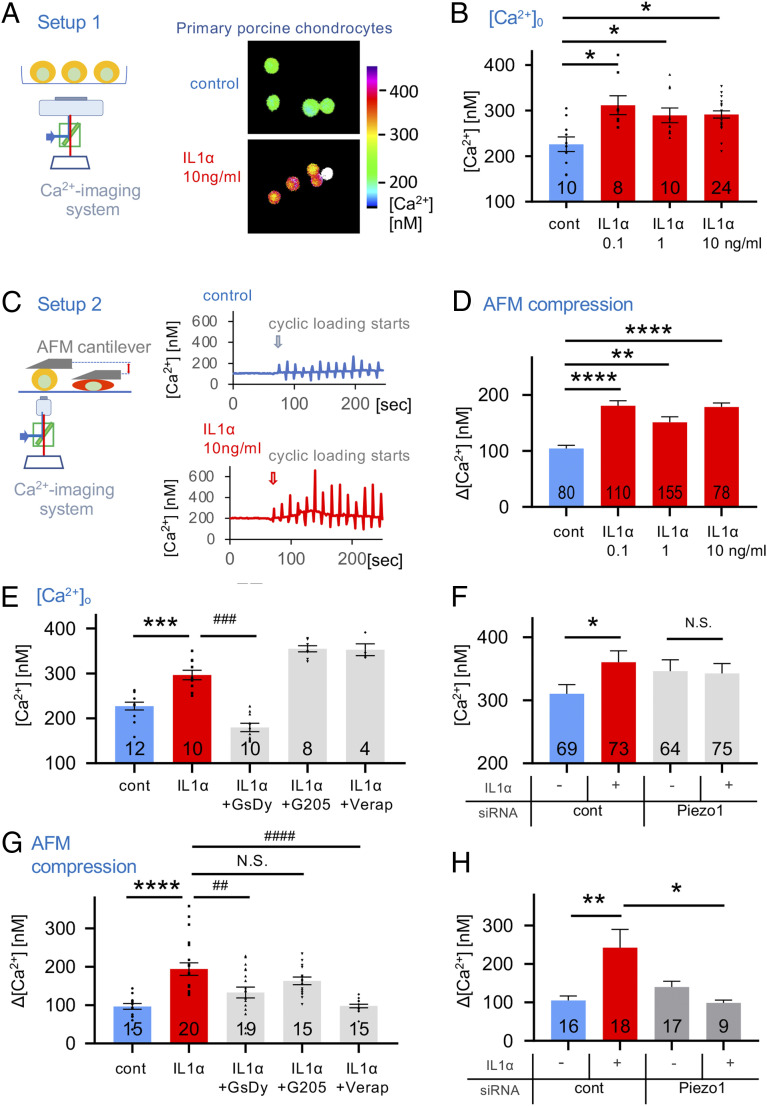
Isolated primary porcine chondrocytes were subjected to recording their steady-state intracellular calcium levels, [Ca2+]o, using ratiometric Ca2+ imaging in the presence or absence of IL-1α. Chondrocyte [Ca2+]o increased 37% in the presence of 0.1 ng/mL IL-1α. However, no further increase in [Ca2+]i was observed with higher concentration of IL-1α (Fig. 2 A and B). We conclude that IL-1α exposure, at 0.1 ng/mL, profoundly changes articular chondrocytes’ functional state by increasing basal steady-state [Ca2+]o by >30%, and that in our in vitro system, there is an exquisite sensitivity of the chondrocytes to sub-ng/ml concentrations of IL-1α, perhaps indicative of chondrocyte vulnerability in early stages of OA. We next conducted controlled compression of isolated chondrocytes, using atomic force microscopy (AFM), with a tipless cantilever to exert dynamic loading of 300 nN at a rate of 1 µm/s ramp-speed and ramps every 10 s, while simultaneously recording [Ca2+]i (Fig. 2C). The Ca2+ response to this form of mechanical load was also significantly amplified with exposure to IL-1α, again showing an appreciable response to IL-1α at 0.1 ng/mL (∼80% increase), with no further increase at 1 and 10 ng/mL (Fig. 2D) (25, 26), again an exquisite sensitivity to IL-1α in keeping with observations for basal steady-state [Ca2+]i. Thus, IL-1α as an inflammatory signal, even at subnanomolar concentrations as is likely in early stages of OA development, makes articular chondrocytes more sensitive to dynamic mechanical compression at a high but physiologically relevant force of 300 nN.
Fig. 2.
Proinflammatory IL-1α signaling alters Ca2+ dynamics in porcine articular chondrocytes. (A, Left) A schematic diagram of the setup; (Right) representative ratiometric Ca2+ images of IL-1α–treated chondrocytes. (B) Resting cytosolic Ca2+ concentrations of primary articular chondrocytes are significantly increased when exposing cells to IL-1α. The numbers in bars indicate independent experiments. (C, Left) A schematic diagram of the setup; AFM probe (flat, tipless) compresses single cells cyclically every 10 s. (Right) Ca2+ concentrations in response to cyclical compression. Loading starts at arrow-marked time point. Representative Ca2+ transients are shown: (Top) control, (Bottom) IL-1α–treated chondrocytes. (D) Mechanical-compression–induced Ca2+ transients are significantly increased when exposing primary chondrocytes to IL-1α. The numbers in bars indicate numbers of transients (increase in [Ca2+]i over resting [Ca2+]i) measured; the number of cells stimulated were 16, 23, 31, and 16 from four independent cell isolations. (E) Resting [Ca2+]i of chondrocytes that were treated with IL-1α (1 ng/mL) and inhibitors of Piezo (GsMTx4 2 µM/dynasore 5 µM), TRPV4 (GSK205 25 µM), and VGCC (verapamil 0.5 µM). Resting [Ca2+]i that is significantly elevated by exposure to IL-1α is significantly reduced below control values when inhibiting Piezo but not decreased by inhibition of TRPV4 or VGCC. The numbers in bars indicate independent experiments, with numbers of cells examined being 855, 457, 385, 387, and 153. (F) Resting [Ca2+]i of chondrocytes subjected to Piezo1 knockdown via specific siRNA. Note again the significant increase of resting [Ca2+]i when treating with IL-1α (1 ng/mL), as in E. This significant increase is absent when inhibiting Piezo1 function by transfection with Piezo1-specific siRNA (see SI Appendix, Fig. S4 for demonstration of effective knockdown of Piezo1); three independent experiments were performed with total numbers of cells indicated in bars. (G) Experimental groups as in E, but mechanical-compression–evoked Ca2+ dynamics were measured as in C and D. Note that significant IL-1α–evoked Ca2+ increase is significantly attenuated when inhibiting Piezo, but, again, there was no decrease when inhibiting TRPV4. Remarkably, there was a complete elimination of the IL-1α–evoked Ca2+ increase when inhibiting VGCC, in striking contrast to effects of VGCC on resting [Ca2+]i (E). The numbers indicate numbers of transients measured; as in D, respective number of cells was 3, 4, 4, 3, and 3. (H) Experimental setup as in G and measurement of mechanical-compression–evoked Ca2+ dynamics. We observed a significant IL-1α–mediated Ca2+ increase, which was completely absent with Piezo1 knockdown (as in F); three independent experiments were performed with the total numbers of cells indicated in bars. Bars represent mean ± SEM; for group comparison, B, D, E, and F: one-way ANOVA, Tukey’s post hoc test; *comparison IL-1α versus control, #comparison IL-1α plus treatments versus IL-1α. */#P < 0.05, **/##P < 0.01, ***/###P < 0.001, ****/####P < 0.0001, significantly different between groups. See SI Appendix, Fig. S2D for subpanels S2D, S2F, and S2H with bar diagrams and all data points.
We next examined the mechanism underlying this Ca2+ sensitization. Steady-state [Ca2+]o was attenuated by GsMTx4 combined with dynasore (GsDy), a combination previously shown to inhibit Piezo channels in articular chondrocytes (19, 27, 28). We observed no effect on [Ca2+]i by inhibition of the chondrocyte mechanosensitive channel TRPV4 with selective inhibitor GSK205, or inhibition of voltage-gated Ca2+ channels (VGCC) with verapamil, also expressed in chondrocytes (Fig. 2E). Chemical inhibition of Piezo channels with GsMTx4 combined with dynasore can possibly have off-target effects to contribute to the observed effect. We therefore knocked down Piezo1 with small-interfering RNA (siRNA). Using a knockdown strategy that we employed previously (19), we again observed effective reduction of Piezo1 expression in articular chondrocytes with specific Piezo1-targeting siRNA versus control, both in the native state and in IL-1α–mediated inflammatory signaling (SI Appendix, Fig. S3). When using this approach to inhibit function of Piezo1, in lieu of GsMTx4 combined with dynasore, we confirmed and extended our initial findings, namely that IL-1α–mediated increase of basal steady-state [Ca2+]o in articular chondrocytes depends on Piezo1 (Fig. 2F). We then asked this same question for the mechanical-load–induced Ca2+ response. Interestingly, the mechanical-load–induced Ca2+ response was also dependent on PIEZO1, as shown by strong reduction of the increase in [Ca2+]i by GsMTx4/dynasore (Fig. 2G). Again, this finding was also confirmed and extended by our Piezo1 siRNA-mediated approach (Fig. 2H). These findings indicate that increased expression of Piezo1 in IL-1α–mediated inflammation was responsible for baseline steady-state [Ca2+]i increase as well as the mechanical-load–evoked [Ca2+]i increase. TRPV4 was not involved in either condition. We noted with great interest that VGCC were also key for the mechanical-load–evoked [Ca2+]i increase, in clear contrast to these channels’ lack of contribution to increased baseline steady-state [Ca2+]i. Therefore, the mechanical-load–evoked [Ca2+]i increase relies on both PIEZO1 and VGCC, potentially functioning as a Ca2+-amplification mechanism of directly mechanically activated PIEZO1 channels, a finding consistent with our earlier characterization of PIEZO-mediated mechanotransduction in chondrocytes (19). This is not the case without mechanical load, where increased baseline steady-state [Ca2+]o relies solely on increased expression of PIEZO1 evoked by inflammatory IL-1α signaling, consistent with studies suggesting that low levels of Ca2+ permeation through PIEZO1 are present at resting membrane tensions due in part to cytoskeletal forces (29–31). Taken together, increased Piezo1 expression evoked by IL-1α profoundly changes the basal function of articular chondrocytes by increasing the steady-state baseline [Ca2+]o by >30%. Increased Piezo1 expression in turn makes these cells hypersensitive to cyclic mechanical loading with mildly injurious stimuli, resulting in additional Ca2+ influx. Furthermore, VGCC function together with PIEZO1 only under direct deformational mechanical loading in the form of a mechanical stress amplification mechanisms, not at resting state.
Piezo1 Increased Expression and Enhanced Function Rarefies F-Actin, Makes Chondrocytes Vulnerable to Mechanical Injury.
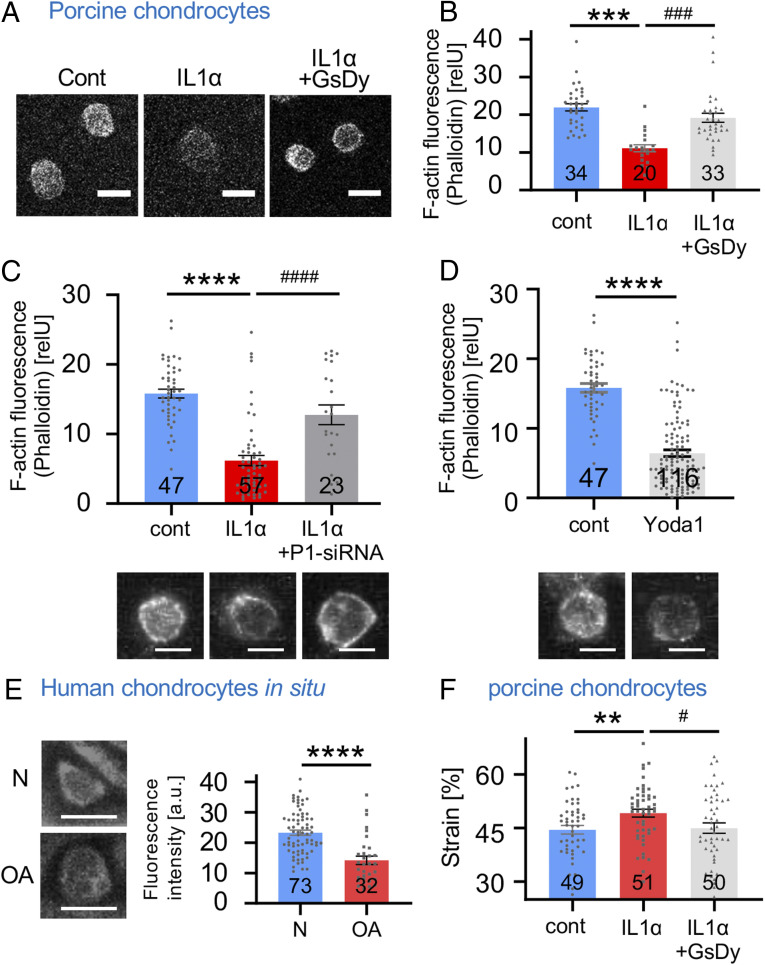
We next hypothesized that inflammatory signaling in articular chondrocytes might exert an effect on the cytoskeleton, the subcellular substrate of physical stiffness (modulus) of the cell, thus protecting or exposing chondrocytes to increased strain resulting from the thousands of cyclic mechanical loads each day that chondrocytes in weight-bearing joints undergo. We used phalloidin staining of porcine chondrocytes to visualize filamentous actin (F-actin) because force transmission through F-actin is an essential mechanotransduction process in chondrocytes (Fig. 3A) (32, 33). We found that F-actin was significantly reduced in IL-1α–exposed chondrocytes (Fig. 3B). If Piezo signaling was inhibited with GsMTx4/dynasore, F-actin was restored. Again, we observed results similar to inhibition of Piezo signaling with GsMTx4/dynasore when knocking down Piezo1 with siRNA (Fig. 3C). In view of this finding, we asked whether selective activation of Piezo1 with small molecule, Yoda-1, would also impact chondrocytes' F-actin labeling pattern. We generated affirmative results (Fig. 3D), indicating that Piezo-mediated Ca2+ influx suffices to rapidly affect cytoskeletal organization of the F-actin cytoskeleton of articular chondrocytes.
Fig. 3.
Rarefied F-actin cytoskeleton in IL-1α–exposed porcine articular chondrocytes and human OA cartilage. (A) Representative confocal fluorescent micrographs (confocal slice) of porcine articular chondrocytes; cytoskeleton with F-actin is visualized. (B) Quantitation of fluorescence intensity (arbitrary units, a.u.) of F-actin (phalloidin labeling); also see Materials and Methods and SI Appendix, Fig. S1. IL-1α exposure significantly diminished F-actin, and Piezo inhibition with GsMTx4/dynasore completely rescued the rarefication of the F-actin network. The numbers indicate the number of cells analyzed, with three independent isolations of primary chondrocytes for each group. (C) Quantitation of fluorescence intensity of F-actin as in B. IL-1α exposure significantly diminished F-actin, and Piezo1-targeting siRNA significantly attenuated the rarefication of the F-actin network. The numbers indicate the number of cells analyzed, with four independent isolations of primary chondrocytes for each group. Representative confocoal fluorescent micrograph (z-stack) is shown below. (D) Quantitation of fluorescence intensity of F-actin as in B. Short-term treatment with Piezo1-activating compound Yoda-1 (10 µM, 30 min) significantly diminished F-actin. The numbers indicate the number of cells analyzed, with four independent isolations of primary chondrocytes for each group. Representative confocoal fluorescent micrograph (z-stack) is shown below. (E) Representative confocal fluorescent micrographs of human cartilage (confocal slice), healthy (normal) versus osteoarthritic (OA); cytoskeleton with F-actin is visualized. F-actin was significantly decreased in OA. The numbers indicate number of cells analyzed. (F) Strain of control, IL-1α–treated, and IL-1α/GsMTx4/dynasore–treated porcine chondrocytes. IL-1α–treated cells (1 ng/mL) have significantly higher strain than vehicle-treated cells. Note complete restitution to lower strain when inhibiting Piezo with GsMTx4/dynasore in IL-1α–treated chondrocytes. The numbers indicate number of cells analyzed from three independent isolations of primary chondrocytes for each group. The bars represent mean ± SEM; for group comparison, B, C, and E: one-way ANOVA, Tukey’s post hoc test; *comparison IL-1α versus control, #comparison IL-1α plus GsMtx4/Dyn versus IL-1α. */#P < 0.05, **/##P < 0.01, and ***/###P < 0.001, significantly different between groups. Group comparison D: t test; *P < 0.05, ****P < 0.0001, significantly different between groups. (Scale bars in A, C, D, and E, 10 µm.)
These results indicate that Ca2+ influx through up-regulated PIEZO1 leads to readily apparent changes of the chondrocytic cytoskeleton, namely a rarefication of F-actin. We did not find similar regulation of β-tubulin (SI Appendix, Fig. S4), nor did we see β-actin mRNA or total chondrocytic β-actin protein down-regulated in response to inflammatory IL-1α signaling (SI Appendix, Fig. S5). Thus, the F-actin rarefication in response to increased expression of PIEZO1 occurs at the level of intracellular protein processing and aggregation, not gene expression of β-actin. With its subplasmalemmal localization, F-actin provides structure and mechanical stability to the spherical chondrocytes upon compression.
We next wanted to verify this concept in human OA cartilage and found that F-actin was also significantly reduced in OA articular chondrocytes, with F-actin rarefication appearing very similar to that observed in IL-1α–treated porcine articular chondrocytes (Fig. 3E).
We therefore investigated the influence of IL-1α on the mechanical properties and force-deformation behavior of porcine chondrocytes. Exposure to IL-1α led to significantly increased cellular deformation in response to the same magnitude of mechanical loading, a result of a decreased cellular modulus (Fig. 3F). This phenomenon translates to increased cellular mechanical strain and potentially microtrauma upon compression. When inhibiting Piezo signaling with GsMTx4/dynasore, the cellular modulus and force-deformation properties reverted to control levels absent IL-1α. This finding, together with the PIEZO1-regulated F-actin polymerization, indicates that excess Ca2+ entering the chondrocyte via over-expressed PIEZO1, upon mechanical load, decreases cellular mechanical stiffness of chondrocytes due to rarefied F-actin in the cortical region of the cell. Decreased cellular rigidity will facilitate cellular mechanical microtrauma and thus increase likelihood of healthy chondrocytes transitioning into a degenerated OA phenotype. Importantly, both F-actin rarefication and decreased cellular rigidity in response to inflammatory IL-1α signaling were completely reversible when inhibiting Piezo signaling.
IL-1α Signaling in Chondrocytes Causing Piezo1 Increased Expression and Enhanced Function: from Membrane to Nucleus.
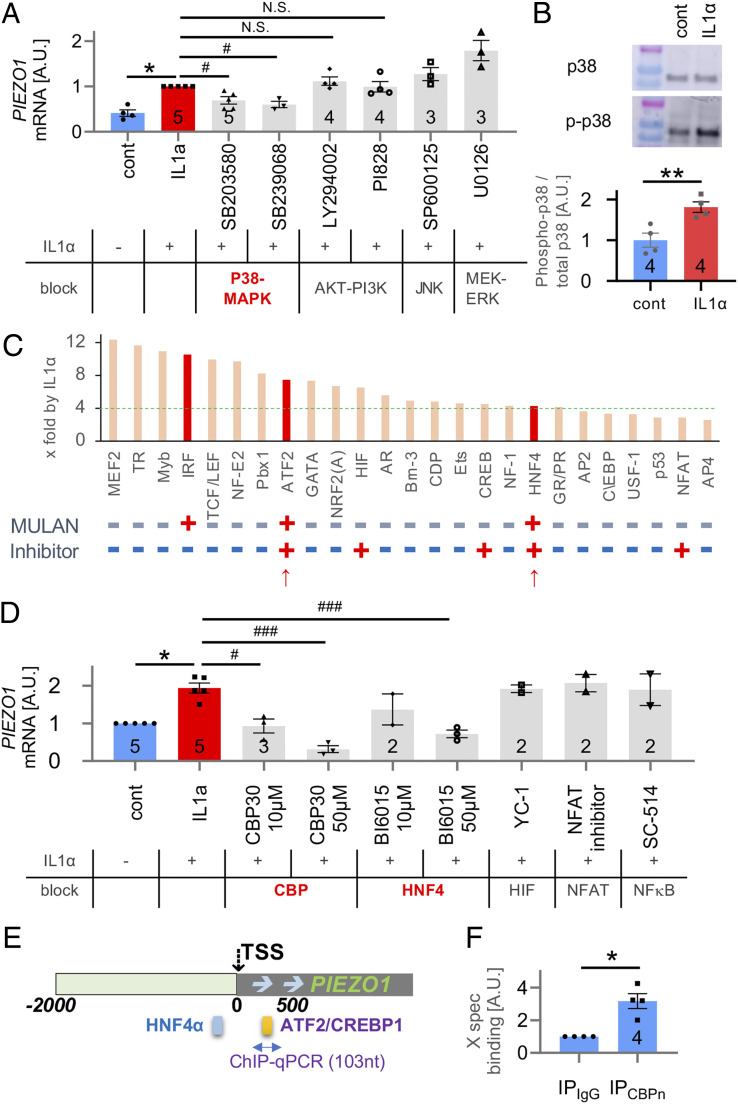
Based on these findings, we next investigated signal transduction from membrane-bound IL-1 receptor (IL1R) via intracellular cytoplasmic signaling to nuclear transcriptional mechanisms that regulate the PIEZO1 gene in articular chondrocytes in response to IL-1α. Since IL-1α will engage the IL-1 receptor type 1 (IL-1RI), known to be expressed by chondrocytes (6, 11), we assessed cellular signal transduction that up-regulates expression of PIEZO1 by using a candidate approach. We tested MAP-kinases, known to be downstream of IL-1RI and downstream kinase, MKK3/6, namely MEK-ERK, JUN, p38, and AKT-PI3K. Of these, only chemical inhibitors of p38-MAP kinase attenuated PIEZO1 mRNA expression in porcine articular chondrocytes when exposed to IL-1α (Fig. 4A). Inhibiting the other kinases had no effect. In support, the phosphorylated isoform of p38 was significantly increased in response to IL-1α stimulation, whereas its total protein abundance was unchanged (Fig. 4B). Phosphorylated p38 is known to traffic to the nucleus to regulate gene expression (34, 35). To identify transcription factors (TFs) that can up-regulate PIEZO1 in articular chondrocytes, we conducted a delimited TF screening experiment. We found 19 TFs to be up-regulated in porcine articular chondrocytes in response to IL-1α out of a total of 96 assessed TFs (Fig. 4C). We next analyzed PIEZO1 regulatory DNA sequences from 2,000 base pairs (bp) upstream of the transcriptional start site to 500 bp downstream of the transcriptional start site (TSS) in human, cow, and chimpanzee. Using the MULAN DNA sequence analysis program, we generated predicted binding sites present in all three species tested (SI Appendix, Fig. S6). We further interrogated ATF2/CREBP1 and HNF4 because they identified from the 96-TF screen and were ≥4× up-regulated, they were present in all species’ proximal PIEZO1 promoter in MULAN, and they can be reliably inhibited with well-established selective small molecule inhibitors (Fig. 4C and SI Appendix, Fig. S6). We selected HIF, NFAT, and NF-κB as controls, expected not to regulate PIEZO1 in response to IL-1α because they were not picked up by MULAN or the 96-TF screen. Of these TFs, in porcine articular chondrocytes, inhibition of ATF2/CREBP1 and HNF4 significantly and dose dependently attenuated the increase of PIEZO1 expression in response to IL-1α (Fig. 4D). As expected, inhibition of HIF, NFAT, and NFκB had no effect on PIEZO1 expression. Of the TFs that we identified to enhance expression of PIEZO1, ATF2 and CREBP1 are family members known to bind to a consensus DNA binding site and to form protein–protein complexes that also include p38 (36, 37). HNF4 has also been demonstrated to be part of such multiprotein complexes by binding to p38 and CREB (38, 39). Based on the MULAN bioinformatics analysis (Fig. 4E and SI Appendix, Fig. S6), we predicted direct binding of CREBP/ATF2 and HNF4 to the proximal PIEZO1 promoter to regulate the PIEZO1 gene rather than indirect regulation through other transcriptional enhancer or derepression mechanisms. We examined CREBP1 binding to the PIEZO1 promoter by chromatin immunoprecipitation (ChIP)-qPCR using DNA extracted from porcine articular chondrocytes after subjecting these cells to immunoprecipitation for CREBP1 (Fig. 4F). We amplified sequences from the predicted ATF2/CREBP1 binding site in the vicinity of the PIEZO1 TSS. When immunoprecipitating with anti-CREBP1, we detected a robust ∼3× increase of PIEZO1-promoter–amplified DNA than with control antibody, indicating direct binding of CREBP1 to the probed site. Given the known interaction between CREBP1, ATF2, and HNF4, we regard it as highly likely that ATF2 and HNF4 also participate in a multiprotein enhancer complex that directly binds to the proximal PIEZO1 promoter and up-regulates PIEZO1 gene expression.
Fig. 4.
Identification of IL-1α–induced signal transduction that results in increased PIEZO1 gene expression by delimited screening in porcine articular chondrocytes. (A) Inhibitors of p38 MAP-kinase significantly attenuated IL-1α–induced PIEZO1-mRNA increase in primary chondrocytes (IL-1α 1 ng/mL for 3 d, for all panels) but not inhibitors of MAP-kinases, JNK, MEK-ERK, and PI3K. (B) Western blot analysis of p38 phosphorylation indicates a significant increase of phospho-p38; total p38 does not change. (C) Starting with 96 TFs, their activation was assessed in response to IL-1α treatment; increased x-fold over baseline, ranked, and top 25 are shown here. Note the green dotted line at 4×, with 19 of the top 25 TFs ≥4. Below the bar diagram, labeled “MULAN”, note binary score (+/−) indicating whether the respective TF was found to have a predicted binding site in the proximal PIEZO1 promoter (see E, also SI Appendix, Fig. S6) using MULAN TF-binding prediction program. Indicated below, labeled “Inhibitor,” is whether the respective TF can be inhibited with a well-established selective small-molecule inhibitor compound. The two red arrows point toward the two TFs that fulfill all criteria, ATF2 and HNF4. (D) TFs that impact IL-1α–induced PIEZO1-mRNA increase were identified using selective compounds. HNF4 and ATF2/CREBP (the latter having the same DNA binding site as ATF2, known to form TF complexes) were confirmed as relevant; HIF, NFAT, and NFkB were not found to be involved. Inhibitors of ATF2/CREBP1 and HNF4 significantly attenuated PIEZO1-mRNA increase. The dose–response relationship of ATF2/CREBP1 inhibitor CBP30 showed a Pearson correlation coefficient = −0.85, P = 0.0008, indicating a significant correlation. The respective metrics for HNF4 inhibitor BI6015 were Pearson correlation coefficient = −0.86, P = 0.0014, also indicative of a signification correlation. (E) A schematic of predicted binding sites of HNF4 and ATF2/CREBP1 TFs in the proximal PIEZO1 promoter, as revealed by the bioinformatics platform MULAN. Please see also SI Appendix, Fig. S6. (F) Direct CREBP1 binding to the PIEZO1 promoter was assessed by chromatin immunoprecipitation followed by qPCR. Note fivefold increased abundance with IL-1α signaling versus control, indicative of direct CREBP1 binding to the PIEZO1 proximal promoter site indicated in E (yellow bar, between 0 and +500 PIEZO1 promoter). For A, B, D, and F, the numbers in bars indicate number of independent isolations of primary chondrocytes. The bars represent mean ± SEM; for group comparison B and F, t test, *P < 0.05; for group comparisons A and D, one-way ANOVA, Tukey’s post hoc test; *comparison IL-1α versus control, #comparison IL-1α plus treatment versus IL-1α. */#P < 0.05.
Discussion
Taken together, our findings demonstrate a signaling mechanism from membrane to nucleus, from the IL-1α–IL-1RI complex, and then via MKK3/6 to p38 MAP-kinase (as previously established), phospho-p38 to nuclear signaling of TF CREBP1, which binds PIEZO1 regulatory DNA sequences together with TFs HNF4 and ATF2 (Fig. 5). These TFs then enhance PIEZO1 gene expression. This signaling cascade leads to overexpression of Piezo1 in articular chondrocytes in response to inflammatory signaling. Overexpressed PIEZO1 channels in turn cause increased resting [Ca2+]i and increased mechanical-stress–evoked Ca2+ increase, indicative of mechanical hypersensitivity, “hypermechanotransduction” (40, 41). Increased resting [Ca2+]o, via PIEZO1, leads to rarefication of F-actin which decreases the stiffness of the chondrocytes. This results in increased cellular deformation in response to mechanical loading. This phenomenon represents a detrimental feed-forward mechanism, as increased deformation elevates the opening probability of mechanosensitive PIEZO channels, amplified by PIEZO1 being overexpressed. We speculate that the rarefied F-actin cytoskeleton enhances cytoplasmic availability of p38 for phosphorylation, which happens after activation of IL-1RI, based on previous demonstration of the F-actin cytoskeleton influencing gene expression (42, 43).
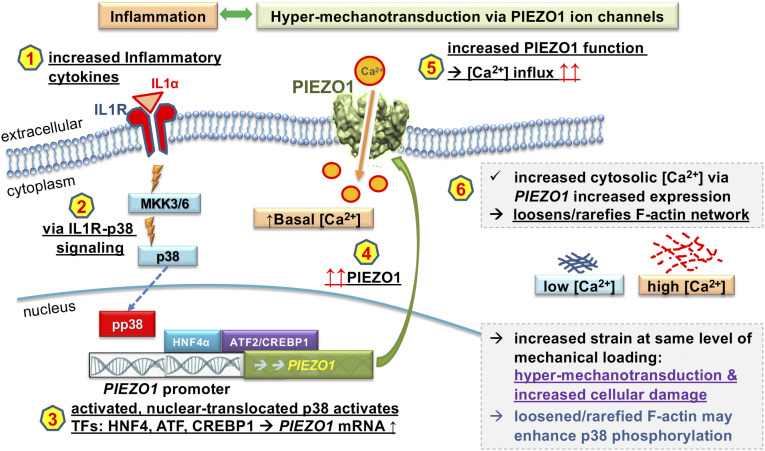
Fig. 5.
Feed-forward pathogenesis of OA relying on chondrocytic inflammatory signaling, which results in Piezo1 increased function. A schematic of our findings and proposed OA pathogenetic mechanism. Signaling hubs and their consequences, upon activation as we demonstrate, are shown as a sequence 1 to 6. Shown in black letters, 1 to 6 is supported by our findings and known background. We interpret the result as “hypermechanotransduction & increased cellular damage,” in purple and speculate, in blue, that a loosened/rarefied F-actin may enhance p38 phosphorylation by making p38 more available for kinases.
PIEZO1 gain-of-function mutations have been reported in humans, typically associated with an erythrocyte disorder, xerocytosis, an anemia phenotype, and also a resistance to malaria in Africans (44, 45). Increased or premature OA has not been reported in these cases. We believe that a genetically encoded gain-of-function of PIEZO1 can set up altered homeostatic regulation in Piezo1-expressing cells because the impact of the gain-of-function channel is present throughout all stages of development. Joint-inflammation–evoked increased gene expression and function of Piezo1 in articular chondrocytes forces an adaptation on the cell that might well be maladaptive, whereas in the case of PIEZO1 gain-of-function mutations, there is no adaptation to it.
Another interesting spotlight is cast on our study by the previous investigation of Blythe et al., who reported that Piezo1 activation leads to production of inflammatory cytokine IL-6 in cardiac fibroblasts via p38 MAP-kinase signaling, thus perhaps involved in cardiac remodeling in response to mechanical stimuli (46). This paper and ours report feed-forward mechanisms; however, conceptually, this study (46) has a starting point of Piezo1 activation by mechanical stimuli, whereas we are looking at inflammatory signaling by IL-1α as starting point, downstream of it increased expression of Piezo1 which then facilitates hyper mechanotransduction and subsequent cellular damage via repeat microtrauma (Fig. 5) at the heart of the feed-forward mechanism that we describe here.
Also of note, our study focuses on inflammatory reprogramming mechanisms of articular chondrocytes. We document increased gene expression and function of Piezo1 after 72 h of exposure to IL-1α. Early changes in response to IL-1α might differ how they can impact the F-actin cytoskeleton (47). However, we wish to emphasize that long-term reprogramming likely has increased disease relevance for OA, a long-term chronic process. Our findings are interesting when viewed against cytoskeletal sequalae observed with Piezo1 loss-of-function in nonskeletal cells (48, 49). In these elegant studies, though, there was no inflammatory injury.
In summary, we have identified a pathogenic feed-forward signaling and gene-regulatory mechanism in weight-bearing articular chondrocytes. This mechanism appears relevant for the pathogenesis of OA because it is a molecular mechanism linking inflammatory signaling with resulting hyper mechanotransduction via up-regulated PIEZO1 expression and function. As one result of PIEZO1 overexpression and resulting excess steady-state Ca2+, we found cytoskeletal changes of F-actin that facilitate cellular mechanotrauma, predisposing and contributing to chondrocyte dedifferentiation and degeneration as the foundation of the irreversible cartilage damage in “degenerative” OA. Breaking through this feed-forward mechanism therapeutically provides a rational target in the quest for OA disease-modifying therapies.
Materials and Methods
Porcine Chondrocyte Isolation and IL-1α Treatment.
Primary articular chondrocytes were harvested from articular cartilage of the femoral condyles of skeletally mature (2 to 3 y old) female pigs as described in ref. 19. The isolated chondrocytes were seeded on 12 mm coverslips and tissue culture plates. The next day, chondrocytes were exposed to the proinflammatory cytokine, IL-1α (R&D Biosystems), at 0, 0.1, 1, and 10 ng/mL for 2 to 3 d (25, 26). Ca2+ imaging, mechanical stimulation experiments, and RT-qPCR were performed approximately 60 to 72 h after IL-1α treatment. Culture medium was changed every 2 d.
Measurement of Steady-State Intracellular Ca2+ Concentration [Ca2+]o.
To measure cytosolic [Ca2+] level, chondrocytes on coverslips were loaded with Ca2+-sensitive Fura-2-AM dye (2 μM for 40 min; Invitrogen) and imaged using a ratiometric Ca2+ imaging platform (Intracellular Imaging, Setup 1 in Fig. 2A) as described in ref. 19. Cytosolic [Ca2+] was determined using Incytim-2 software (Intracellular Imaging), based on 340:380 ratio and calibration of standard solutions, measuring [Ca2+]i in individual chondrocytes.
Measurement of Compression-Induced Ca2+ Transients.
Mechanically induced Ca2+ influx of individual chondrocytes was measured using a custom-built AFM/Ca2+ setup consisting of the atomic force microscope (Bioscope; Veeco or MFP-3D; Asylum) and a ratiometric Ca2+ imaging microscope (Intracellular Imaging; Fig. 2C) as described in ref. 19. Briefly, AFM probe (flat, tipless) compresses single cells cyclically with trigger force of 300∼500 nN every 10 s for 2 min, and the cytosolic [Ca2+] transients in response to cyclical compression mechanics were measured during the loading (1 ∼2 frames). Spring constants of the tipless cantilever were 0.5 to 14 N/m (Novasan or Bruker Probes), the compression rate was 1 to 2 μm/s, and the experiments were conducted at 37 °C. The five maximum Ca2+ influx of each chondrocyte were determined ([Ca2+]max), and influx was determined as ∆[Ca2+] = [Ca2+]max − [Ca2+]o.
Measurement of Intracellular Ca2+ Concentration Using Fluorescent Imaging Plate Reader Assay.
Our approach followed previously reported methods (50, 51). Isolated chondrocytes were plated in a 96-well plate at 30,000 cells per well for 12 h. Media were then supplemented with either 0 ng/mL or 10 ng/mL porcine IL-1α and incubated for 72 h. Cellular response in each well was measured via fluo-4 intracellular calcium dye using the fluo-4 NW calcium assay kit (Molecular Probes) according to manufacturer’s directions. Intracellular Ca2+ using a fluorescent imaging plate reader. PIEZO1 stimulation was performed by supplementing Yoda-1 at 5 µM or 10 µM. A vehicle (dimethyl sulfoxide (DMSO), 0 µM Yoda-1) control group was also run to measure the baseline intracellular Ca2+ levels.
Treatment with Compounds and GsMTx4.
Chondrocytes were treated with compounds for 1∼2 h during Fura-2-AM loading process: GsMTx4 peptide (2 μM, provided by Philip Gottlieb, State University of New York at Buffalo), dynasore (5 μM, Tocris), and verapamil (0.5 μM, Sigma). To identify signal transduction mechanisms and transcription factors that regulate IL-1α–induced Piezo1 expression, inhibitors were coincubated with IL-1α (1 ng/mL) for 3 d. Inhibitors used were SGCCBP30 (10 to 50 μM, Cayman Chemical), BI6015 (10 to 50 μM, Cayman), YC-1 (10 μM, Cayman), nuclear factor of activated T cells (NFAT) inhibitor (10 μM, Cayman), SB203580 (10 μM, VWR), SB239063 (10 μM, Sigma Aldrich), LY294002 (10 μM, VWR), PI828 (10 μM, Fisher Scientific), SP600125 (10 μM, VWR), and U0126 (10 μM, VWR).
mRNA Expression in Isolated Chondrocytes by RT-qPCR.
After treatments, chondrocytes’ total RNA was extracted using TRIzol for RT-qPCR. PCR specificity was confirmed by gel electrophoresis and dissociation curve analysis. GAPDH or 18S were used as a housekeeping gene for normalization, and the gene-under-study mRNA level was quantified using the 2-ΔΔCT method, as in refs. 52 and 53, with normalization to control as in refs. 54 and 55.
Immunocytochemistry.
Porcine articular chondrocytes were cultured on glass coverslips, fixed with 4% paraformaldehyde at 4 °C for 15 min, permeabilized with 2% Triton X-100, and blocked with 5% donkey or goat serum for 30 min. Then, cells were exposed to primary antibodies at 4 °C overnight and then to fluorescent secondary antibody. Immunolabeling was visualized using a Zeiss 780 confocal microscope. Anti-PIEZO1 (NBP1-78537, Novus) was used. Chondrocytes were also labeled with fluorescent Phalloidin (phalloidin-CF568, Biotium) to visualize F-actin or with BT7R-dylight488 (Thermo Fisher) to visualize β-tubulin and then imaged. A Zeiss 780 confocal microscope (Plan Apochromat 40×/1.4 NA oil immersion objective) or a BX61 Olympus upright microscope (X-Apochromat 40×/1.4 NA oil immersion objective) equipped with high-resolution charge-coupled device (CCD) camera and ISEE software (ISEE Imaging Systems) were used. For confocal laser scanning microscopy, confocal slices or z-stacks were acquired following refs. 56 and 57.
Circular chondrocytes were selected as region-of-interest (ROI), and their ROI integrated density was determined using ImageJ, subtracted for noncellular background in immunocytochemistry applications, yielding individual metric data points per cell, see also SI Appendix, Fig. S1.
Immunohistochemistry.
Paraffin-embedded tissue sections (15 µm) of human articular cartilage from healthy (n = 5) and osteoarthritic (n = 6) anonymized donors were used (Articular Engineering). Sections were immunolabeled with Piezo1-antibody (NBP1-78537) followed by fluorescent secondary antibody (Alexa Fluor 594) and imaged. For F-actin staining, sections were labeled with fluorescent Phalloidin (phalloidin-CF350, Biotium) and imaged under a Zeiss 780 confocal microscopy platform (Plan Apochromat 40×/1.4 NA oil immersion objective), again acquiring z-stacks as described above.
Western Blot.
We followed standard methods (56, 58). Briefly, cells were lysed with CHAPS buffer and separated on a 10% sodium dodecyl sulfate (SDS) polyacrylamide gel and then transferred to polyvinylidene difluoride (PVDF) membrane filters (0.45 mm pore; Millipore). Membranes were blocked with 5% dry milk, protein levels were detected by using primary antibodies of PIEZO1 (Proteintech, 15939-1-AP), p38 (Cell Signaling, #8690), phospho-p38 (Cell Signaling, #4511) secondary antibody (Jackson Immuno Research, anti-rabbit peroxidase-conjugated); and chemoluminescence substrate (ECL-Advance, GE Healthcare). Abundance was quantified using ImagePro Plus software.
siRNA Transfection to Knock-Down Piezo1.
As described in ref. 19, isolated chondrocytes were immediately nucleofected with siRNA to target the Sus scofra Piezo1 mRNA (pool of siRNAs with the following sense strands: 5′-CAGCGAGAUCUCGCACUCCAUCUU-3′, 5′-UACGACCUGCUGCAGCUCCUGUU-3′, and 5′-ACCCGCUGGCCAUGCAGUUCUUUU-3′ all synthesized from Dharmacon) or a nontargeting construct siRNA (siNTC, Invitrogen). Nucleofection was done with the Lonza 4D-Nucleofector according to manufacturer’s guidelines using the ER-100 protocol. Nucleofected cells were plated on glass coverslips overnight in fetal bovine serum (FBS)–containing feed media (without Pen-strep) before changing media to contain either 0 or 1 ng/mL porcine IL-1α. Cells were left incubating for 48 h. PIEZO1 mRNA levels were quantified by RT-qPCR.
Chondrocyte Strain Measurement.
To measure the cellular strain applied with AFM, cells were plated on coverslip glass and supplemented with IL-1α, IL-1α+GsMTx4+dynasore, or vehicle control as described above. To measure deformation and strain of cells under each treatment, we used an AFM to apply a 25 nN load to each tested cell. After loading each cell, the coverslip adjacent to the cell was also loaded to measure the baseline position. Load curves for each cell and coverslip load consist of the z-position (height) and AFM cantilever deflection. AFM cantilever deflection was used to estimate the applied load using the cantilever stiffness. A custom-written MATLAB code, based on the work of Chang et al. (59), was used to determine the contact point of the cantilever with the cell (zcp), the contact point of the cantilever with the coverslip (zcs), and the point at which the cell deformed under the 25 nN load (zdef). While cells are generally considered to be complex materials and modeled under mixture material frameworks consisting of viscoelastic and elastic components, this code approximates the cellular material response closely surrounding the contact point as an elastic material. Using a Hertzian-contact–type framework, the contact point is approximated based on this assumption. The nominal cellular strain percentage, e, was computed as deformation of the cell to the load divided by the height of the cell, e = (zcp − zdef)/(zcp − zcs)×100. Our MATLAB script (adapted from ref. 59) is shared as freeware.
Discovery of Transcription Factors Regulating the IL-1α–Induced Piezo1 Expression by Transcription Factor Array.
Nuclear extracts of porcine chondrocytes were prepared using Nuclear Extraction Kit (SK-0001, Signosis), and then the transcription factor (TF) Activation Profiling Plate Array II (FA-1002, Signosis) was used to monitor 96 TFs simultaneously, according to manufacturer protocol.
Briefly, the nuclear extracts were incubated with biotin-labeled probes which were designed based on the consensus sequences of TF binding sites. The TF-probe complexes were purified, and then the bound probes were separated from the complex. The detached probes were hybridized in 96-well plates where each well is specifically coated with complimentary sequences of the probes. The bound DNA probes are mixed with horseradish peroxidase (HRP)-Streptavidin conjugates, and the luminescence was read in microplate luminometer. The luminescence of control and the IL-1α–treated chondrocytes were compared.
TF Binding Site Analysis by Bioinformatics.
TF binding sites within the PIEZO1 promoter were analyzed and predicted using the comprehensive, web-based Evolutionary Conserved Region Browser (http://ecrbrowser.dcode.org). The promotor regions of PIEZO1 [−2,000 nt to +500 nt, transcription start site (TSS) defined as zero] in three different species were aligned, using high-quality genome sequences with a defined PIEZO1 TSS: human (hg19 Chromosome 16: 88853372-88850872), cow (bostau6 Chromosome 18: 14041407-14039769), and chimpanzee (pantro3 Chromosome 16: 88511045-88508550). The MULAN TF-binding prediction algorithm, within ecrbrowser, was run on these three species to identify candidate TF binding sites.
ChIP Assay.
ChIP assays were performed using primary articular porcine chondrocyte nuclear extract, and cell isolation from 4 individual pigs was conducted. The Magnify ChIP kit (Thermo Fisher) was used according to manufacturer's instructions and as described previously (52, 60).
Anti-CRBP1–immunoprecipitated DNA was purified to perform qPCR to determine the relative abundance of PIEZO1 promoter DNA fragments. Conserved sequences of cow (bostau6 Chromosome 18: 14041407-14039769, AC_000175.1) and pig (NC_010448.3) were aligned to generate primers for ChIP-qPCR. A 103-nucleotide amplicon was generated from the pig PIEZO1 promoter: (CTCCGGATTAAACAGCTCCAGGCAGGAAGC-CCGCCTTCTCCCAGATTGGTCAGGAAGTCCTGATGCAAGTTTGCGCGTTTTCTTTCTCTCTCTCTCTCTCTTT; CREBP1 binding site is underlined). The sequences of the forward and backward primers are as follows: For-CTCCGGATTAAACAGCTCCA and Rev-AAAGAGAGAGAGAGAGAGAAAGAAA.
Statistical Analysis.
Data are presented as mean ± SEM. Student’s t test or one-way ANOVA, Tukey’s post hoc test for group-to-group comparisons were used to determine the statistical significance. For correlation, Pearson’s correlation coefficient and its P value were calculated. All statistical analysis was conducted using GraphPad Prism 8.4.
Supplementary Material
Acknowledgments
We thank Dr. Michelle Yeo (Duke University) for assistance with TF array tests and Dr. Jorg Grandl (Duke University) for donating the PIEZO1−/− HEK293t cell line, for discussion of PIEZO channel activity, and for careful reading of the manuscript. Dr. Andrey Bortsov (Duke University) provided advice with statistical testing. This work was supported by NIH Grants AR072999, AR074240, AR073221, AR065653, AG15768, AG46927, DE027454, P30 AR069655, P30 AR074992, and P30 AR073752; the Arthritis Foundation; and the Michael Ross Haffner Foundation.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2001611118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Zhang Y., Jordan J. M., Epidemiology of osteoarthritis. Clin. Geriatr. Med. 26, 355–369 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guilak F., Biomechanical factors in osteoarthritis. Best Pract. Res. Clin. Rheumatol. 25, 815–823 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deveza L. A., Loeser R. F., Is osteoarthritis one disease or a collection of many? Rheumatology (Oxford) 57 (suppl. 4), iv34–iv42 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van Spil W. E., Kubassova O., Boesen M., Bay-Jensen A. C., Mobasheri A., Osteoarthritis phenotypes and novel therapeutic targets. Biochem. Pharmacol. 165, 41–48 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Karsdal M. A., et al., Disease-modifying treatments for osteoarthritis (DMOADs) of the knee and hip: Lessons learned from failures and opportunities for the future. Osteoarthritis Cartilage 24, 2013–2021 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Blom A. B., van der Kraan P. M., van den Berg W. B., Cytokine targeting in osteoarthritis. Curr. Drug Targets 8, 283–292 (2007). [DOI] [PubMed] [Google Scholar]
- 7.Kraus V. B., et al., Effects of intraarticular IL1-Ra for acute anterior cruciate ligament knee injury: A randomized controlled pilot trial (NCT00332254). Osteoarthritis Cartilage 20, 271–278 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Olson S. A., et al., The role of cytokines in posttraumatic arthritis. J. Am. Acad. Orthop. Surg. 22, 29–37 (2014). [DOI] [PubMed] [Google Scholar]
- 9.Loeser R. F., Collins J. A., Diekman B. O., Ageing and the pathogenesis of osteoarthritis. Nat. Rev. Rheumatol. 12, 412–420 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chevalier X., Eymard F., Richette P., Biologic agents in osteoarthritis: Hopes and disappointments. Nat. Rev. Rheumatol. 9, 400–410 (2013). [DOI] [PubMed] [Google Scholar]
- 11.McNulty A. L., Rothfusz N. E., Leddy H. A., Guilak F., Synovial fluid concentrations and relative potency of interleukin-1 alpha and beta in cartilage and meniscus degradation. J. Orthop. Res. 31, 1039–1045 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sanchez-Adams J., Leddy H. A., McNulty A. L., O’Conor C. J., Guilak F., The mechanobiology of articular cartilage: Bearing the burden of osteoarthritis. Curr. Rheumatol. Rep. 16, 451 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Griffin T. M., Guilak F., The role of mechanical loading in the onset and progression of osteoarthritis. Exerc. Sport Sci. Rev. 33, 195–200 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Anderson D. D., et al., Post-traumatic osteoarthritis: Improved understanding and opportunities for early intervention. J. Orthop. Res. 29, 802–809 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diekman B. O., et al., Intra-articular delivery of purified mesenchymal stem cells from C57BL/6 or MRL/MpJ superhealer mice prevents post-traumatic arthritis. Cell Transplant. 22, 1395–1408 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chubinskaya S., et al., Articular cartilage injury and potential remedies. J. Orthop. Trauma 29 (suppl. 12), S47–S52 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Furman B. D., et al., Targeting pro-inflammatory cytokines following joint injury: acute intra-articular inhibition of interleukin-1 following knee injury prevents post-traumatic arthritis. Arthritis Res. Therapy 16, 10.1186/ar4591 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patwari P., et al., Proteoglycan degradation after injurious compression of bovine and human articular cartilage in vitro: Interaction with exogenous cytokines. Arthritis Rheum. 48, 1292–1301 (2003). [DOI] [PubMed] [Google Scholar]
- 19.Lee W., et al., Synergy between Piezo1 and Piezo2 channels confers high-strain mechanosensitivity to articular cartilage. Proc. Natl. Acad. Sci. U.S.A. 111, E5114–E5122 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O’Conor C. J., Leddy H. A., Benefield H. C., Liedtke W. B., Guilak F., TRPV4-mediated mechanotransduction regulates the metabolic response of chondrocytes to dynamic loading. Proc. Natl. Acad. Sci. U.S.A. 111, 1316–1321 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Servin-Vences M. R., Richardson J., Lewin G. R., Poole K., Mechanoelectrical transduction in chondrocytes. Clin. Exp. Pharmacol. Physiol. 45, 481–488 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Servin-Vences M. R., Moroni M., Lewin G. R., Poole K., Direct measurement of TRPV4 and PIEZO1 activity reveals multiple mechanotransduction pathways in chondrocytes. eLife 6, e21074 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barrett-Jolley R., Lewis R., Fallman R., Mobasheri A., The emerging chondrocyte channelome. Front. Physiol. 1, 135 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Syeda R., et al., Chemical activation of the mechanotransduction channel Piezo1. eLife 4, e07369 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stevens A. L., Wishnok J. S., White F. M., Grodzinsky A. J., Tannenbaum S. R., Mechanical injury and cytokines cause loss of cartilage integrity and upregulate proteins associated with catabolism, immunity, inflammation, and repair. Mol. Cell. Proteomics 8, 1475–1489 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McNulty A. L., Estes B. T., Wilusz R. E., Weinberg J. B., Guilak F., Dynamic loading enhances integrative meniscal repair in the presence of interleukin-1. Osteoarthritis Cartilage 18, 830–838 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee W., Guilak F., Liedtke W., Role of Piezo channels in joint health and injury. Curr. Top. Membr. 79, 263–273 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bae C., Sachs F., Gottlieb P. A., The mechanosensitive ion channel Piezo1 is inhibited by the peptide GsMTx4. Biochemistry 50, 6295–6300 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lewis A. H., Grandl J., Mechanical sensitivity of Piezo1 ion channels can be tuned by cellular membrane tension. eLife 4, e12088 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pathak M. M., et al., Stretch-activated ion channel Piezo1 directs lineage choice in human neural stem cells. Proc. Natl. Acad. Sci. U.S.A. 111, 16148–16153 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ellefsen K. L., et al., Myosin-II mediated traction forces evoke localized Piezo1-dependent Ca2+ flickers. Commun. Biol. 2, 298 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Erickson G. R., Northrup D. L., Guilak F., Hypo-osmotic stress induces calcium-dependent actin reorganization in articular chondrocytes. Osteoarthritis Cartilage 11, 187–197 (2003). [DOI] [PubMed] [Google Scholar]
- 33.Chao P. H., West A. C., Hung C. T., Chondrocyte intracellular calcium, cytoskeletal organization, and gene expression responses to dynamic osmotic loading. Am. J. Physiol. Cell Physiol. 291, C718–C725 (2006). [DOI] [PubMed] [Google Scholar]
- 34.Chuderland D., Seger R., Protein-protein interactions in the regulation of the extracellular signal-regulated kinase. Mol. Biotechnol. 29, 57–74 (2005). [DOI] [PubMed] [Google Scholar]
- 35.Wei S., Siegal G. P., Mechanisms modulating inflammatory osteolysis: A review with insights into therapeutic targets. Pathol. Res. Pract. 204, 695–706 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lau E., Ronai Z. A., ATF2 - at the crossroad of nuclear and cytosolic functions. J. Cell Sci. 125, 2815–2824 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lopez-Bergami P., Lau E., Ronai Z., Emerging roles of ATF2 and the dynamic AP1 network in cancer. Nat. Rev. Cancer 10, 65–76 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guo H., Gao C., Mi Z., Zhang J., Kuo P. C., Characterization of the PC4 binding domain and its interactions with HNF4alpha. J. Biochem. 141, 635–640 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Dell H., Hadzopoulou-Cladaras M., CREB-binding protein is a transcriptional coactivator for hepatocyte nuclear factor-4 and enhances apolipoprotein gene expression. J. Biol. Chem. 274, 9013–9021 (1999). [DOI] [PubMed] [Google Scholar]
- 40.Parpaite T., Coste B., Piezo channels. Curr. Biol. 27, R250–R252 (2017). [DOI] [PubMed] [Google Scholar]
- 41.Gnanasambandam R., Gottlieb P. A., Sachs F., The kinetics and the permeation properties of Piezo channels. Curr. Top. Membr. 79, 275–307 (2017). [DOI] [PubMed] [Google Scholar]
- 42.Haudenschild D. R., Nguyen B., Chen J., D’Lima D. D., Lotz M. K., Rho kinase-dependent CCL20 induced by dynamic compression of human chondrocytes. Arthritis Rheum. 58, 2735–2742 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li S., Jia X., Duance V. C., Blain E. J., The effects of cyclic tensile strain on the organisation and expression of cytoskeletal elements in bovine intervertebral disc cells: An in vitro study. Eur. Cell. Mater. 21, 508–522 (2011). [DOI] [PubMed] [Google Scholar]
- 44.Ma S., et al., Common PIEZO1 allele in african populations causes RBC dehydration and attenuates plasmodium infection. Cell 173, 443–455.e12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cahalan S. M., et al., Piezo1 links mechanical forces to red blood cell volume. eLife 4, e07370 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Blythe N. M., et al., Mechanically activated Piezo1 channels of cardiac fibroblasts stimulate p38 mitogen-activated protein kinase activity and interleukin-6 secretion. J. Biol. Chem. 294, 17395–17408 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pritchard S., Guilak F., Effects of interleukin-1 on calcium signaling and the increase of filamentous actin in isolated and in situ articular chondrocytes. Arthritis Rheum. 54, 2164–2174 (2006). [DOI] [PubMed] [Google Scholar]
- 48.Nonomura K., et al., Mechanically activated ion channel PIEZO1 is required for lymphatic valve formation. Proc. Natl. Acad. Sci. U.S.A. 115, 12817–12822 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McHugh B. J., Murdoch A., Haslett C., Sethi T., Loss of the integrin-activating transmembrane protein Fam38A (Piezo1) promotes a switch to a reduced integrin-dependent mode of cell migration. PLoS One 7, e40346 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kanju P., et al., Small molecule dual-inhibitors of TRPV4 and TRPA1 for attenuation of inflammation and pain. Sci. Rep. 6, 26894, 10.1038/srep26894 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nims R. J., et al., A synthetic mechanogenetic gene circuit for autonomous drug delivery in engineered tissues. Sci. Adv. 7, eabd9858 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yeo M., Berglund K., Augustine G., Liedtke W., Novel repression of Kcc2 transcription by REST-RE-1 controls developmental switch in neuronal chloride. J. Neurosci. 29, 14652–14662 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Livak K. J., Schmittgen T. D., Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
- 54.Pires B. R. B., et al., NF-kappaB regulates redox status in breast cancer subtypes. Genes (Basel) 9, 320 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nejadnik H., et al., MR imaging features of gadofluorine-labeled matrix-associated stem cell implants in cartilage defects. PLoS One 7, e49971 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Phan M. N., et al., Functional characterization of TRPV4 as an osmotically sensitive ion channel in porcine articular chondrocytes. Arthritis Rheum. 60, 3028–3037 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sasazaki Y., Seedhom B. B., Shore R., Morphology of the bovine chondrocyte and of its cytoskeleton in isolation and in situ: Are chondrocytes ubiquitously paired through the entire layer of articular cartilage? Rheumatology (Oxford) 47, 1641–1646 (2008). [DOI] [PubMed] [Google Scholar]
- 58.Leddy H. A., et al., Follistatin in chondrocytes: The link between TRPV4 channelopathies and skeletal malformations. FASEB J. 28, 2525–2537 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chang Y. R., et al., Automated AFM force curve analysis for determining elastic modulus of biomaterials and biological samples. J. Mech. Behav. Biomed. Mater. 37, 209–218 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yeo M., et al., Bisphenol A delays the perinatal chloride shift in cortical neurons by epigenetic effects on the Kcc2 promoter. Proc. Natl. Acad. Sci. U.S.A. 110, 4315–4320 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.