Abstract
Microdeletions within 1q24 have been associated with growth deficiency, varying intellectual disability, and skeletal abnormalities. The candidate locus responsible for the various phenotypic features of this syndrome has previously been predicted to lie in the area of 1q24.3, but molecular evidence of the causative gene remains elusive. Here, we report two additional patients carrying the smallest reported 1q24 deletion to date. Patient 1 exhibited intrauterine growth retardation, shortening of the long bones, frontal bossing, microstomia, micrognathia, and a language acquisition delay. Her mother, Patient 2, displayed a broad forehead and nasal bridge, thick supraorbital ridges, and toe brachydactyly, along with learning disability and language acquisition delay. The microdeletion encompasses a 94 Kb region containing exon 14 and portions of the surrounding introns of the gene encoding dynamin 3 (DNM3), resulting in an in-frame loss of 38 amino acids. This microdeletion site also contains a long non-coding RNA (DNM3OS) and three microRNAs (miR-214, miR-199A2, and miR-3120). Following culture of patient-derived and control fibroblasts, molecular analyses were performed to determine expression levels of genes affected by the heterozygous deletion. Results show decreased expression of DNM3OS and miR-214–3p in patient fibroblasts cultured in an osteogenic induction medium. Overall, our data provide further evidence to support a functional role for non-coding RNAs in regulating the skeletal phenotype, and the potential of a functionally-impaired DNM3 protein causing the non-skeletal disease pathogenesis.
Keywords: 1q24.3, Dynamin 3, Intellectual disability, Microdeletion, MicroRNA, Long non-coding RNA, Skeletal defects
1. Introduction
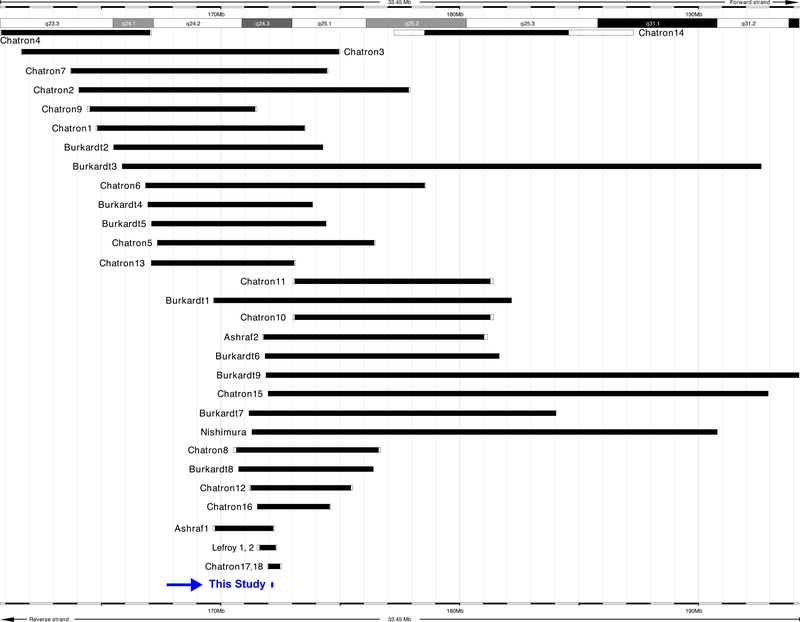
Interstitial deletions within chromosome 1q24 have been reported to confer a characteristic skeletal and intellectual phenotype. In 1982, Taysi et al. [1] described a 1q21–22→q25 deletion syndrome characterized by pre- and postnatal growth deficiency, developmental delay, intellectual disability, microcephaly, brachycephaly, and skeletal abnormalities. Several other groups later reported additional patients, more than 30 subjects in total, with microarray-confirmed deletions of varying sizes in the region of 1q24, all characterized by a distinctive skeletal phenotype and with varying levels of intellectual disability [2–5] (Fig. 1). Patients with 1q24 deletions can exhibit short stature, microcephaly, mild or profound cognitive disability, brachydactyly, clinodactyly of the fifth digit, language deficits, and autism spectrum disorder-like characteristics (Supplementary Table 1). The specifics of the underlying genotype responsible for these phenotypes, however, have remained elusive.
Fig. 1.
Alignment of the deletion in this study as compared to previously reported 1q24 deletions [2–6]. A review of published clinical phenotypes can be seen in Supplementary Table 1.
Alignment of the deletion in this study as compared to previously reported 1q24 deletions [2–6]. A review of published clinical phenotypes can be seen in Supplementary Table 1.
In the first published array comparative genomic hybridization study of 1q24q25 deletion patients, Burkardt et al. [3] hypothesized that loss of centromere protein L (CENPL, MIM# 61503) and dynamin 3 (DNM3, MIM# 611445) may be responsible for the skeletal and intellectual phenotypes, respectively. Interestingly, DNM3 is the only dynamin protein expressed in the brain, and has several reported functions in neurons and the myelin sheath, including a role in the modulation of synapse cargo trafficking by interacting with the post-synaptic Homer protein [7]. Ashraf et al. [2] characterized a distinct subset of 1q24q25 deletion patients with milder intellectual disability and corresponding smaller deletions. Importantly, CENPL was unaltered in these patients, despite the presence of the characteristic skeletal phenotype, necessitating a re-evaluation of the putative culprit gene.
Ashraf et al. [2] predicted the critical genomic interval responsible for the skeletal phenotype and short stature common to these patients to a small region of 1q24 encoding an exon of DNM3 and several non-coding RNAs. Specifically, these non-coding RNAs consist of the long non-coding RNA DNM3OS and two microRNAs (miRNAs) located within DNM3OS (miR-214 [MIM# 610721] and miR-199A2 [MIM# 610720]) which are all transcribed in the antisense direction within intron 14 under control of their own promoter. A third microRNA, miR-3120 (MIM# 614722), has also been identified within DNM3 intron 14, transcribed from the same locus as miR-214, but in the sense orientation following DNM3 transcription and mRNA processing [8]. miR-199A2 has several paralogs in the human genome (and conserved throughout the vertebrate lineage), including miR-199A1 (MIM# 610719) on chromosome 19 and miR-199B (MIM# 614791) on chromosome 9, contained within DNM2 and DNM1, respectively. No other miR-214 or miR-3120 paralogs exist in the human genome.
Prior to this study, the smallest reported deletion of 1q24 was 490 Kb, in a mother and daughter with the characteristic skeletal features but lacking profound intellectual disability [4] (Fig. 1). However, this deletion also encompasses the gene PIGC (MIM# 601730), the uncharacterized open reading frame C1orf105, and the majority of the coding sequence of DNM3 in addition to DNM3OS and the miR-214/199A2/3120 cluster.
Here, we describe a 4-year-old girl and her mother, both with a heterozygous 94 Kb interstitial deletion in band 1q24 that results in the loss of DNM3OS and, hence, the miR-214/199A2/3120 cluster as well as a single exon of DNM3 which yields an in-frame amino acid deletion. We present their clinical features, compare their phenotypes with previously reported cases, and conduct molecular studies to further characterize the critical genes associated with the observed skeletal phenotype.
2. Materials and methods
2.1. Chromosomal microarray analysis
The parents signed a consent form approved by the Washington University School of Medicine Institutional Review Board. Genomic DNA from both patients was analyzed by clinical chromosomal microarray analysis (CMA) using an Affymetrix/ThermoFisher Scientific CytoScan HD Array and copy number analysis (at 50 kb/25 marker resolution) was performed using the Chromosome Analysis Suite v2.0.0195/r5758 (Affymetrix/Thermo Fisher Scientific, Waltham, MA) and BioDiscovery Nexus Discovery Edition v7.0 software. The genomic linear positions are given relative to NCBI build GRCh38.
2.2. Fibroblast isolation and culture
Full-thickness skin biopsies (3 mm diameter) were collected from both patients. Specimens were cut into small pieces (~1 mm diameter) and dispersed in 0.5 mL growth medium (DMEM containing 10% FBS, 100 U/mL penicillin, 100 μg/mL streptomycin) in one well of a sterile 6-well tissue culture dish. After a 2 h incubation period (37 °C, 5% CO2), additional growth medium was added to a final volume of 2 mL. Growth medium was changed every 3 days. The migration of fibroblasts from the skin explants was monitored. Cells were passaged when they reached 90% confluency. Non-diseased primary adult human fibroblasts (ATCC PCS-201–012) were cultured in growth medium and used as a control. In addition to culturing in baseline growth medium, fibroblast cell lines were also cultured in the presence of an osteogenic induction medium to determine if such a stimulus would alter expression of DNM3, DNM3OS, and the clustered miRNAs. Cells were first seeded into 12-well plates (2 × 105 cells/well) and cultured in growth medium overnight. After 24 h, osteogenic induction medium (αMEM containing 10% FBS, 10 mM β-glycerol phosphate, 50 μM ascorbic acid, 10 nM dexamethasone, 2 mM L-glutamine, 100 U/mL penicillin, 100 μg/mL streptomycin) was added for 3 days. Cultures were then processed for RNA extraction.
2.3. RNA and protein isolation
Total RNA, including small miRNAs, was extracted from fibroblasts (following culture in growth medium or osteogenic induction for 3 days) using the Total RNA Purification Kit (Norgen Biotek Corp). Resulting RNA was quantified by spectrophotometry (NanoDrop 1000). Protein lysates were prepared using a Cell Lysis Buffer (Cell Signaling Technology 9803S) containing protease inhibitors (Sigma P8340). Protein concentration was determined using the Bio-Rad Protein Assay Dye Reagent (Bio-Rad 5000006) and BioTek Cytation plate reader.
2.4. qPCR to determine miRNA, lncRNA, and mRNA expression
Human miRNAs (miR-199a-5p and miR-214–3p) were reverse-transcribed and quantified with the appropriate TaqMan primer/probe sets (ThermoFisher Scientific, Supplementary Table 2), reverse transcription kit and TaqMan Universal Master Mix II no UNG (ThermoFisher Scientific 4440040). Expression of mature functional miRNA strands were normalized to RNU44. LncRNA and mRNA expression were examined by quantitative reverse transcription (RT) PCR using Superscript II Reverse Transcriptase (ThermoFisher Scientific 18064014) and PowerUp SYBR Green Master Mix (ThermoFisher Scientific A25742). PPIA was used to normalize DNM3OS and DNM3 expression. Additional primers were designed to specifically amplify the region of DNM3 containing exon 14 in both control and patient RNA samples. Here, RNA was reverse-transcribed and cDNA amplified by PCR. RT-PCR products were electrophoresed on a 2% agarose gel, stained with SYBR Safe DNA gel stain (ThermoFisher) and imaged on a ChemiDoc XRS+ Molecular Imager (Bio-Rad). All PCR primer sequences are shown in Supplementary Table 2.
2.5. Western blot
Cell lysates containing protein (10 μg) were resolved by SDS-PAGE and transferred onto PVDF membranes. Immunoblotting was carried out using a rabbit polyclonal DNM3 antibody (LSBio LS-C409118) or mouse β-actin antibody (Abcam ab6267). Primary antibodies were used at 1:1000 dilution. Secondary antibodies (IRDye 800CW-labeled anti-rabbit; IRdye 680RD-labeled anti-mouse; LI-COR Biosciences) were used at 1:10,000 dilution following the manufacturer’s recommendation. Immunoblot images were generated using the LI-COR Odyssey near-infrared imager. Protein band density was quantified using LI-COR software.
3. Results
3.1. Clinical reports
3.1.1. Patient 1
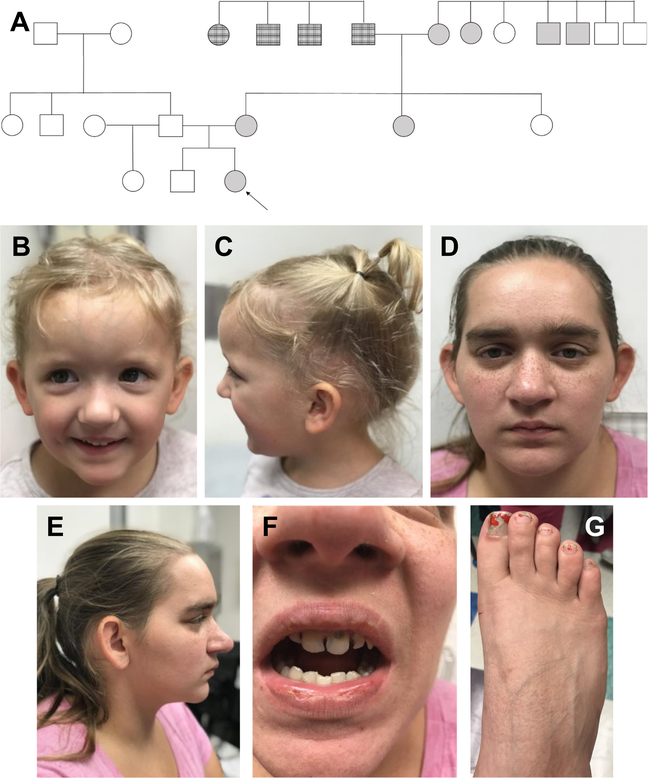
Patient 1 is a 4-year-old Caucasian female born at 37 weeks to a 20-year-old G2P1 mother (Patient 2) and 24-year-old father. Her 6-year-old brother and 2-year-old paternal half sister are of normal height and weight, and her father is of normal stature (Fig. 2A).
Fig. 2.
A. Pedigree with shaded individuals with reported short stature; note the extensive maternal family history. Arrow indicates Patient 1. B, C. Physical findings of patient 1; note mild frontal bossing, bilateral epicanthal folds, downslanting palpebral fissures, microstomia, and micrognathia. D–G. Physical Findings of patient 2; note the broad forehead and nasal bridge, thick supraorbital ridges, and toe brachydactyly.
The patient’s mother (Patient 2) was evaluated by maternal-fetal medicine and received prenatal counseling regarding inherited causes of short stature and underwent an ultrasonographic anatomy scan at 32 weeks. Femurs, humeri, ulnas, and tibias measured below the 5th percentile for gestational age. All other long bones measured at the lower limit of normal. Estimated fetal weight measured at the lower limit of normal. No other structural malformations were identified. The patient was delivered by uncomplicated spontaneous vaginal delivery under regional anesthesia following cervical ripening with oxytocin augmentation for severe maternal pre-eclampsia.
At 2.5 years of age, a skeletal survey identified frontal bossing and mild flaring of the iliac wings bilaterally but suggested no evidence of a specific skeletal dysplasia. Physical exam at age 3 years 11 months confirmed macrocephaly with mild frontal bossing, and revealed bilateral epicanthal folds and downslanting palpebral fissures, with microstomia and micrognathia (Fig. 2B, C). Rhizomelia of the bilateral upper and lower extremities was noted, along with mild lumbar lordosis. Height was 93.5 cm (4th percentile, Z = −1.75), weight was 16.2 kg (57th percentile, Z = 0.18) and occipitofrontal circumference (OFC) was 52.7 cm (98th percentile, Z = 2.05).
Upon re-evaluation at age 5 years 5 months, the height of Patient 1 was 104 cm (7th percentile, Z = −1.45), weight was 19 kg (50th percentile, Z = 0.01) and OFC was 54 cm (>98th percentile, Z > 2.05). She continued to exhibit the physical findings seen at 2 years of age and is currently receiving speech therapy for delay in speech acquisition.
The bone mineral density (BMD) of L1-L4 for patient 1 was assessed by dual-energy X-ray absorptiometry (DEXA). The average BMD within this region was 0.443 g/cm2. This is 0.5 standard deviations below the mean of the average BMD for age- and gender-matched subjects. The average BMD of femoral neck and total hip was 0.445 g/cm2 and 0.437 g/cm2, respectively (no reference range is available for these regions in children).
Bone biomarkers were measured for Patient 1, including serum alkaline phosphatase, osteocalcin, PTH, and 25-OH Vitamin D; and urine calcium and phosphorus, all of which were reported within normal reference ranges.
3.1.2. Patient 2
Patient 2 is 27 years old and 5′ tall with a speech delay requiring therapy in childhood, mild intellectual disability, and education through the 10th grade. She receives assistance for independent activities of daily living, including finances. Short stature has also been reported in the maternal aunt, both maternal grandparents, two maternal great uncles, two paternal great aunts, and one paternal great uncle. However, these individuals were not available for our clinical assessment.
Physical exam of Patient 2 revealed a broad forehead and nasal bridge, as well as thick supraorbital ridges. Brachydactyly was noted in bilateral lower extremity digits (Fig. 2E–G).
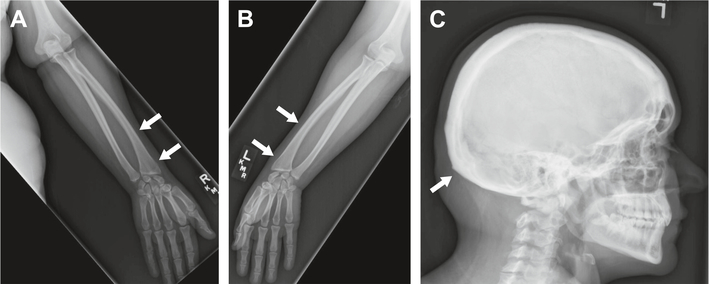
Patient 2 underwent two radiographic bone surveys: one following delivery of the proband and another four years later. The initial bone survey identified straightening of the entire spine, a likely left unilateral pars interarticularis defect at the L5 level, endplate degenerative spurring at the L1–L2 level, and intervertebral disc height loss at the L3–L4 level. The follow-up bone survey further noted skull thickening and underdevelopment of the distal radius medially with a V-shaped deformity of the carpal bones and prominent ulna; the latter findings were suggestive for Madelung-like deformity (Fig. 3). BMD of the left hip total was found to be 1.073 g/cm2. This corresponds to a Z-score of 1.1. Femoral neck is 0.969 g/cm2 with a Z-score of 1.1.
Fig. 3.
Radiographic findings of patient 2; arrows in A (right upper extremity) and B (left upper extremity) indicate Madelung-like deformity; arrow in C (skull) indicates thickening.
Bone biomarkers were measured for Patient 2 as for Patient 1, all of which were reported within normal reference ranges. Urine NTX-telopeptide was slightly elevated at 98 nmol/mmol (nmol Bone Collagen Equivalents/nmol Creatinine; normal for premenopausal women: 17–94). However, serum cross linked N-telopeptide was normal at 11.4 nM BCE (normal: 6.2–19). These measurements were normal for Patient 1.
3.2. Chromosomal microarray results
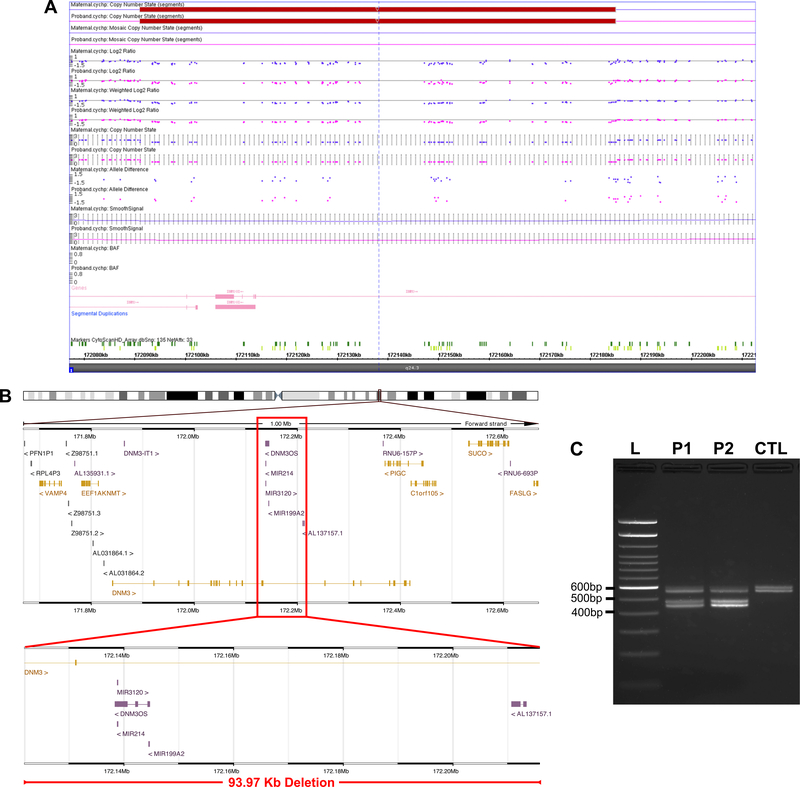
CMA in the two patients were performed as clinical tests and revealed an interstitial deletion on chromosome 1 at band q24.3 (arr [hg38] 1q24.3 (minimum: 172,121,918–172,215,889; maximum: 172,121,883–172,216,223) ×1), spanning approximately 94 Kb in length (minimum: 93.97 Kb; maximum: 94.34 Kb) and consisting of 104 markers/probes (Fig. 4A). This deletion spans a region containing a portion of introns 13 and 15 and the entirety of exon 14 of DNM3 (relative to the canonical transcript isoform, NCBI# NM_015569.5), the lncRNA DNM3OS, and three miRNAs – miR-214, miR-199A2, and miR-3120 (Fig. 4B).
Fig. 4.
A. SNP Array results for patients 1 and 2, displaying copy number loss in the indicated region. B. Genomic context of the deletion seen in these patients, indicating the chromosomal position (top), regional context (middle), and deletion (bottom). Genes are from the GENCODE Release 33 annotation visualized on the Ensembl genome browser. C. RT-PCR analysis showing the absence of exon 14 in a proportion of DNM3 transcripts generated in cells of heterozygous Patient 1 (P1) and Patient 2 (P2). The predicted size of transcripts devoid of exon 14 is 459–489 bp while transcripts containing exon 14 (114 bp) are 573–603 bp. Transcripts lacking exon 14 were not detected in cells from the control (CTL) sample. Note the presence of distinct pairs of bands in each sample that differ in size by roughly 30 bp. This is likely due to some DNM3 splice variants containing (or not) an additional 30 bp exon located between exon 13 and exon 14 in the canonical isoform.
To further demonstrate that the patient deletion mutation contains exon 14, reverse-transcription PCR was carried out using a primer pair designed to amplify a region from exon 10 to exon 16 of the DNM3 gene. As expected, Fig. 4C shows the presence of transcripts with or without exon 14 in both patient samples. Transcripts devoid of exon 14 were not found in the control sample. Interestingly, for both the exon 14-containing and exon 14-deleted bands, an additional band was visualized differing in size by roughly 30 bp. This is likely due to the fact that some DNM3 splice variants (e.g. NCBI# N_M001350204.2) have been reported to contain an additional 30 bp exon between exon 13 and exon 14 in the canonical isoform (which is not encompassed by the deletion in these patients).
3.3. Gene and protein expression analyses in patient and control fibroblasts
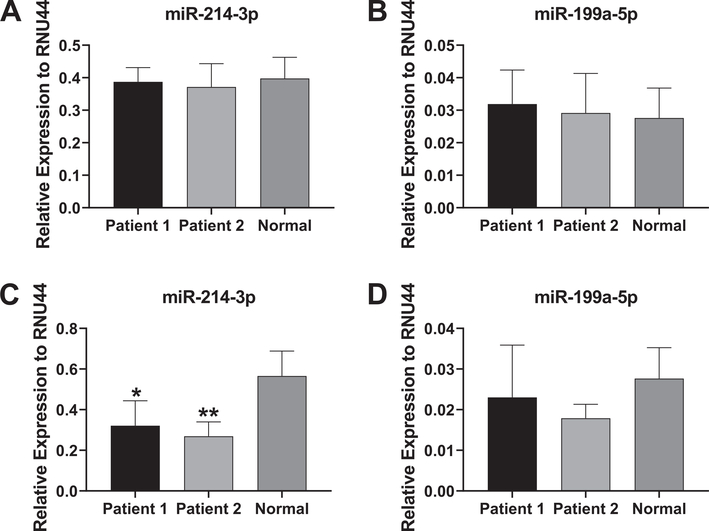
At baseline, expression levels of miR-214–3p and miR-199a-5p were similar between patient and control samples (Fig. 5A, B). However, a significant decrease in expression levels of miR-214–3p was found in patient fibroblasts relative to control fibroblasts following osteogenic induction for 3 days (Fig. 5C). Decreases in miR-199a-5p expression were seen in patient samples following osteogenic induction (Fig. 5D), but were not significant. This may be explained by the fact that expression of miR-199a-5p cannot be distinguished from transcription of the paralogous gene MIR199A1 on chromosome 19. Note that we did not include expression analysis results for miR-3120 since overall expression levels detected by qPCR were extremely low in all samples.
Fig. 5.
Expression of miR-214 and miR-199a in patient and control fibroblasts. A, B. Relative levels of the functional miR-214–3p and miR-199a-5p strands in the two patient sample fibroblasts and control, non-diseased fibroblasts at baseline. C, D. Relative levels of miR-214–3p and miR-199a-5p in patient or control fibroblasts following 3 days of osteogenic induction. miRNA levels are expressed as relative abundance to RNU44. Experiments were carried out four times and a Student’s t-test was used to determine statistical significance in miRNA expression between the control sample and each of the patient sample (*p < 0.05; **p < 0.01).
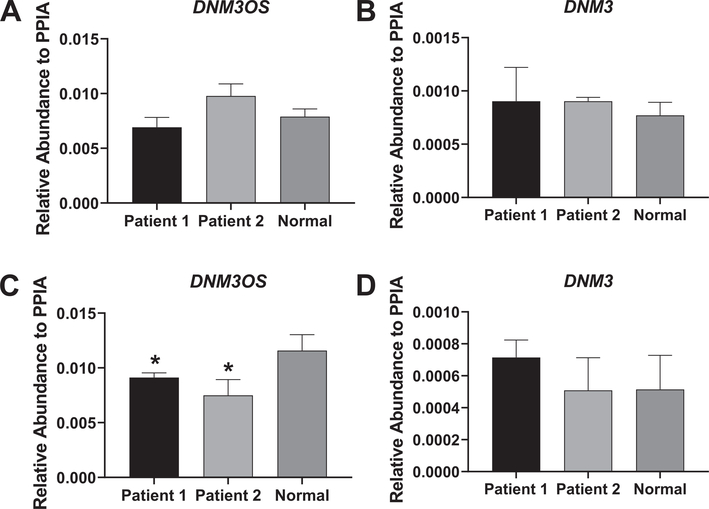
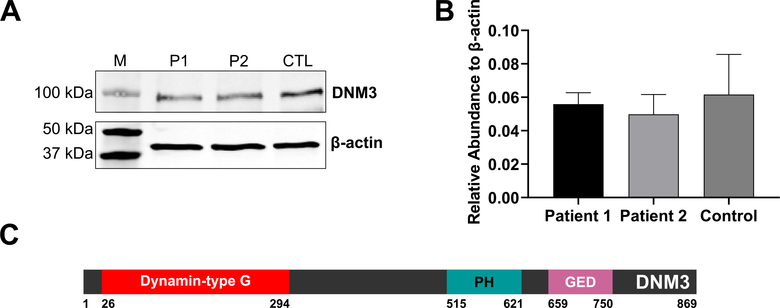
With respect to DNM3OS and DNM3 expression, no apparent differences in relative abundance levels were noted between patient and control samples. (Fig. 6A, B). However, a significant decrease in DNM3OS expression levels was seen in the patient samples following osteogenic induction compared to control (Fig. 6C). No significant differences in DNM3 gene expression following osteogenic induction were noted between patient and control samples (Fig. 6D). In agreement with the mRNA data, Fig. 7A and B show that DNM3 protein is detectable in patient and control samples and that no significant differences in expression levels between samples were found.
Fig. 6.
Expression of DNM3OS and DNM3 in patient and control fibroblasts. A, B. Relative expression of DNM3OS and DNM3 in the two patient sample fibroblasts and control, non-diseased fibroblasts at baseline. C, D. Relative expression of DNM3OS and DNM3 in patient and control fibroblasts following 3 days of osteogenic induction. Gene levels were expressed as relative abundance to PPIA. Experiments were carried out in triplicate and a Student’s t-test was used to determine statistical significance in mRNA expression between the control sample and each of the patient samples (*p < 0.05).
Fig. 7.
A. Expression of DNM3 protein in patient and control fibroblasts. Image shows a representative Western blot image to detect the presence of DNM3 or β-actin protein in lysates extracted from Patient 1 (P1), Patient 2 (P2) or control (CTL) fibroblasts. Molecular weight protein markers (M) are shown in the left lane. B. Quantification of DNM3 expression relative to β-actin from three independent experiments. C. Protein domains of DNM3 (UniProt# Q9UQ16). “Dynamin-type G”: Dynamin-type guanine nucleotide-binding (G) domain, “PH”: Pleckstrin homology domain, “GED”: GTPase effector domain [9].
4. Discussion
We describe two patients who presented with short stature and mild intellectual disability associated with the 1q24 microdeletion. We report the smallest published deletion at 1q24.3 and conclusively narrow the putative candidate region and responsible genes for the skeletal phenotype observed in this disorder. We also present molecular expression data from cells derived from patients with this 1q24.3 microdeletion.
The heterozygous deletion in the proband and her mother, identified by CMA and spanning approximately 94 Kb, results in the in-frame loss of exon 14 of DNM3 along with total loss of the long non-coding RNA gene DNM3OS and the miRNA cluster, miR-214/199A2/3120, present within DNM3OS. Despite the considerable overlap (Fig. 1), a review of concordant clinical phenotypes (Supplementary Table 1) amongst published 1q24 microdeletion syndrome patients reveals a constellation of physical findings. Some characteristics are common to all patients, even with deletions that do not encompass the region deleted in the patients presented here, but even amongst only those patients with overlapping deletions the clinical presentation is variable.
Expression analyses did not show apparent differences (i.e. decreased expression) of DNM3OS, miR-214–3p or miR-199a-5p at baseline in patient skin fibroblasts compared to non-diseased control skin fibroblasts. Data related to miR-199a-5p levels may be explained by the fact that other miR-199a-5p paralogs derived from the MIR199A1 gene exist and would therefore also be detected by qPCR. Baseline data related to expression levels of DNM3OS and miR-214–3p may be due to a lack of activation/regulation of the promoter or other enhancer elements controlling antisense transcription and expression of these non-coding RNAs in skin fibroblasts. As such, expression levels may be low in general and so any changes in levels due to a heterozygous deletion mutation may be modest or undetectable. However, it is possible that differences in baseline expression levels would be detectable in cells derived from cartilage or bone tissue, or even bone marrow-derived mesenchymal stem/stromal cells that are commonly used to study chondrogenesis and osteogenesis in vitro.
Another explanation for the lack of dose-dependent decrease in expression of these non-coding RNA genes could be due to the phenomenon known as allelic imbalance, whereby a gene/gene locus may be expressed more efficiently on one allele compared to the other [10,11]. Therefore, if the deletion mutation is in the allele that is less regulated (i.e. resulting in low expression of the gene locus), then overall modest-to-undetectable changes in gene expression would be expected when comparing cells from patients with this heterozygous mutation compared to control cells.
Given some published reports suggesting a functional role for DNM3OS, miR-214–3p and miR-199a-5p in regulating skeletal development or skeletal cell differentiation [12–19], we predicted that a medium known to induce progenitor cell differentiation toward the osteoblast lineage may regulate the promoter or other enhancer regions controlling transcription of this gene locus. As such, this treatment could potentially reveal expression differences when comparing patient and control samples. Indeed, we found significant decreases in expression levels of DNM3OS and miR-214–3p in patient samples. As already mentioned, lower levels of miR-199a-5p were not detected most likely due to the presence of other miR-199a-5p paralogs derived from the MIR199A1 gene. Overall, this data provides further evidence to suggest that deletion of the gene locus containing a long non-coding RNA and clustered miRNAs may be the major causative factor resulting in the observed skeletal phenotypes.
Long non-coding RNAs (lncRNAs) are defined as transcripts of 200 nucleotides or longer that do not encode protein. They can exist in the nucleus or cytoplasm to regulate a number of cellular processes including cell growth and differentiation [20–22]. Common mechanisms reported for lncRNAs include binding to chromatin, RNA or proteins to induce their functional effects. A number of published reports suggest that specific lncRNAs can regulate skeletal development and homeostasis [23,24]. Of relevance to this current study, deletion of Dnm3os in mice results in short stature, craniofacial hypoplasia, neural arch defects, vertebral abnormalities, and osteopenia [19]. Importantly, expression of Dnm3os does not appear to be correlated with that of Dnm3 since its deletion in the Dnm3os knock-out mouse model did not affect Dnm3 expression levels. These findings strongly suggest that Dnm3os and/or the miR-214/miR-199a-2 cluster regulate aspects of skeletal development and support the hypothesis that the heterozygous deletion of these non-coding RNAs in the patients reported in this study is responsible for the observed skeletal phenotypes. The Watanabe et al. study and others [25] suggest that the main function of Dnm3os is to act as a precursor for generation of functional miRNA strands. However, other lncRNA roles for Dnm3os have been proposed including one study reporting its regulation of proliferation and apoptosis in a chondrocyte cell line by sponging miR-126 and modulating IGF1 signaling [26]. Therefore, it is not altogether clear if the skeletal defects observed in the 1q24.3 patients is due to altered miRNA levels and function and/or altered Dnm3os function independent of its role in serving as a miRNA precursor.
Since their discovery in C. elegans in 1993, miRNAs are now recognized as critical epigenetic regulators of gene expression [27]. The major function of miRNAs is to suppress protein expression via binding to specific sites in the 3′-UTR of target mRNAs [28]. Tens to hundreds of mRNAs can potentially be targeted by a specific miRNA in a given cell type [29]. Subsequently, cellular pathways and networks may be affected due to altered miRNA levels, thereby resulting in changes in a number of cell processes such as differentiation, metabolism, proliferation and apoptosis. A large body of published studies exist suggesting that miRNAs play an important role in regulating skeletal development and homeostasis [30]. With respect to the miR-199a/214 cluster, it has been shown to be evolutionarily conserved across vertebrate lineages [31]. This study, and others, found expression of this cluster within and around many skeletal regions during development [19,32]. Importantly, expression patterns of Dnm3 in zebrafish was found in the developing nervous system, particularly the neural tube, brain and eyes and did not overlap with miR-199/214 [31]. This further supports the notion that the miRNA cluster regulates skeletal tissue development and homeostasis while the host gene, Dnm3, may regulate neural function.
Many studies have reported functional effects of miR-199a and miR-214 in bone and cartilage development. For example, both miRNAs have been shown to be regulated by Twist 1, a transcription factor implicated in osteoblast differentiation [33]. In addition, miR-199a-5p has been shown to enhance osteogenesis by either targeting TET2 [14] or regulating the HIF-1A pathway [12]. Another report showed that miR-199a-3p can be regulated by BMP-2 to affect Smad1 activity and inhibit chondrogenesis [34]. On the other hand, miR-214 has been shown to inhibit osteoblast differentiation by targeting the transcription factor Osterix [35] or ATF4 [36] and has also been reported to promote osteoclast differentiation [37]. Studies have also shown that miR-214 antagomirs promote matrix mineralization and osteoblast activity in an osteoporosis mouse model [38–40]. Neither over- nor under-mineralization of bone was measured by DEXA scan of patients 1 and 2, and the reported phenotype of the 1q24.3 microdeletion syndrome does not suggest an increased bone density phenotype. However, it should be highlighted that the majority of functional studies on miR-199a or miR-214 have involved analyzing their individual function and not their effects as a cluster. The consequences of over-expressing or inhibiting one of these miRNAs in vitro or in vivo does not provide appropriate insight into the functional effects of a heterozygous deletion of this miRNA cluster, and indeed the entire Dnm3os region. A mouse model containing the same microdeletion as the patients described in this study would be more informative.
No changes in DNM3 mRNA or protein levels were observed between patient and control samples (Fig. 7A, B). This is not surprising given that the microdeletion results in an in-frame loss of exon 14. This exon encodes 38 amino acids within the DNM3 pleckstrin homology (PH) domain (Fig. 7C), including several residues annotated to act as homodimer interfaces [9]. PH domains are thought to play a role in recruiting and localizing proteins to cellular membranes and may play a role in protein-protein interactions [41]. Intriguingly, replacement of the PH domain in DNM1, a close relative of DNM3, with a polyhistidine linker resulted in a functionally active protein in vitro, albeit with impaired scaffold formation [42]. We previously reported two patients with a missense mutation in the DNM1 PH domain who exhibited a mild phenotype distinct from that traditionally seen in patients with mutations in other DNM1 functional domains [43]. The specific role of the PH domain in DNM3 function remains uncharacterized and the functional significance of its partial loss is not understood. Therefore, while protein (a mix of wild type and mutant protein devoid of exon 14) was detected in heterozygous patient cells by Western blotting, it has yet to be determined if altered protein function is a major cause of the brain-related issues in these patients involving intellectual disability. An additional possibility is the role of miR-3120 in regulating neural/brain function. There are very few reports of miR-3120 in the published literature. However, one study showed that miR-3120 regulates endocytic processes in neurons, similar to the function of its host gene DNM3 [8]. Therefore, additional effects of miR-3120 deletion in these patients may also contribute to the neurological and cognitive phenotypes observed.
Others groups, including Burkardt et al. [3] and Ashraf et al. [2], have speculated that the miR-214/199A2 cluster may be responsible for the skeletal phenotype characteristic of the 1q24.3 microdeletion syndrome. The clinical and molecular data of the two patients presented in this study, with the smallest 1q24.3 microdeletion reported to date, lends support to this claim. Taken together, our findings, as well as those previously reported, suggest it is likely that the skeletal defects are due to deletion of DNM3OS and the miR-199/214 cluster. Given the expression patterns and reported functions for DNM3 and miR-3120, we speculate that altered DNM3 protein function and possibly also reduced miR-3120 expression may be responsible for the cognitive phenotypes observed in both patients. We acknowledge that restricting our studies to quantitative expression analyses cannot provide definitive conclusions related to the functional and mechanistic outcomes of the heterozygous patient deletion mutation reported in this study. Future work involving testing patient and control RNA samples by RNA-Seq and pathway analyses would likely provide further insights into what genes, cellular pathways and networks are altered as a result of the deletion mutation in question.
Supplementary Material
Acknowledgments
This work was supported by R01 AR064191 (to A.M.). J.L.S. was supported by T32 GM07200 (to the Washington University in St. Louis Medical Scientist Training Program).
Footnotes
Declaration of competing interest
None.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bone.2020.115705.
References
- [1].Taysi K, Sekhon GS, Hillman RE, A new syndrome of proximal deletion of the long arm of chromosome 1: 1q21–23 leads to 1q25, Am. J. Med. Genet 13 (1982) 423–430, 10.1002/ajmg.1320130411. [DOI] [PubMed] [Google Scholar]
- [2].Ashraf T, Collinson MN, Fairhurst J, Wang R, Wilson LC, Foulds N, Two further patients with the 1q24 deletion syndrome expand the phenotype: a possible role for the miR199–214 cluster in the skeletal features of the condition, Am. J. Med. Genet. A 167 (2015) 3153–3160, 10.1002/ajmg.a.37336. [DOI] [PubMed] [Google Scholar]
- [3].Burkardt DD, Rosenfeld JA, Helgeson ML, Angle B, Banks V, Smith WE, Gripp KW, Moline J, Moran RT, Niyazov DM, et al. , Distinctive phenotype in 9 patients with deletion of chromosome 1q24-q25, Am. J. Med. Genet. A 155 (2011) 1336–1351, 10.1002/ajmg.a.34049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Chatron N, Haddad V, Andrieux J, Désir J, Boute O, Dieux A, Baumann C, Drunat S, Gérard M, Bonnet C, et al. , Refinement of genotype-phenotype correlation in 18 patients carrying a 1q24q25 deletion, Am. J. Med. Genet. A 167 (2015) 1008–1017, 10.1002/ajmg.a.36856. [DOI] [PubMed] [Google Scholar]
- [5].Lefroy H, Fox O, Javaid MK, Makaya T, Shears DJ, 1q24 deletion syndrome. Two cases and new insights into genotype-phenotype correlations, Am. J. Med. Genet. A 176 (2018) 2004–2008, 10.1002/ajmg.a.40426. [DOI] [PubMed] [Google Scholar]
- [6].Nishimura A, Hiraki Y, Shimoda H, Nishimura G, Tadaki H, Tsurusaki Y, Miyake N, Saitsu H, Matsumoto N, De novo deletion of 1q24.3-q31.2 in a patient with severe growth retardation, Am. J. Med. Genet. A 152A (2010) 1322–1325, 10.1002/ajmg.a.33371. [DOI] [PubMed] [Google Scholar]
- [7].Lu J, Helton TD, Blanpied TA, Rácz B, Newpher TM, Weinberg RJ, Ehlers MD, Postsynaptic positioning of endocytic zones and AMPA receptor cycling by physical coupling of Dynamin-3 to Homer, Neuron 55 (2007) 874–889, 10.1016/j.neuron.2007.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Scott H, Howarth J, Lee YB, Wong LF, Bantounas I, Phylactou L, Verkade P, Uney JB, MiR-3120 is a mirror microRNA that targets heat shock cognate protein 70 and auxilin messenger RNAs and regulates clathrin vesicle uncoating, J. Biol. Chem 287 (2012) 14726–14733, 10.1074/jbc.M111.326041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Consortium UniProt, Uniprot: a worldwide hub of protein knowledge, Nucleic Acids Res. 47 (2019) D506–D515, 10.1093/nar/gky1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Palacios R, Gazave E, Goñi J, Piedrafita G, Fernando O, Navarro A, Villoslada P, Allele-specific gene expression is widespread across the genome and biological processes, PLoS One 4 (2009), e4150, 10.1371/journal.pone.0004150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Veitia RA, Bottani S, Birchler JA, Gene dosage effects: nonlinearities, genetic interactions, and dosage compensation, Trends Genet. 29 (2013) 385–393, 10.1016/j.tig.2013.04.004. [DOI] [PubMed] [Google Scholar]
- [12].Chen X, Gu S, Chen BF, Shen WL, Yin Z, Xu GW, Hu JJ, Zhu T, Li G, Wan C, Ouyang HW, Lee TL, Chan WY, Nanoparticle delivery of stable miR-199a-5p agomir improves the osteogenesis of human mesenchymal stem cells via the HIF1a pathway, Biomaterials 53 (2015) 239–250, 10.1016/j.biomaterials.2015.02.071. [DOI] [PubMed] [Google Scholar]
- [13].Li D, Liu J, Guo B, Liang C, Dang L, Lu C, He X, Cheung HYS, Xu L, Lu C, He B, Liu B, Shaikh AB, Li F, Wang L, Yang Z, Au DWT, Peng S, Zhang Z, Zhang BT, Pan X, Qian A, Shang P, Xiao L, Jiang B, Wong CKC, Xu J, Bian Z, Liang Z, Guo DA, Zhu H, Tan W, Lu A, Zhang G, Osteoclast-derived exosomal mir-214–3p inhibits osteoblastic bone formation, Nat. Commun 7 (2016) 10872, 10.1038/ncomms10872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Qi XB, Jia B, Wang W, Xu GH, Guo JC, Li X, Liu JN, Role of miR-199a-5p in osteoblast differentiation by targeting TET2, Gene 726 (2020) 144193, 10.1016/j.gene.2019.144193. [DOI] [PubMed] [Google Scholar]
- [15].Roberto VP, Gavaia P, Nunes MJ, Rodrigues E, Cancela ML, Tiago DM, Evidences for a new role of mir-214 in chondrogenesis, Sci. Rep 8 (2018) 3704, 10.1038/s41598-018-21735-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Tang J, Yu H, Wang Y, Duan G, Wang B, Li W, Zhu Z, Microrna-199a counteracts glucocorticoid inhibition of bone marrow mesenchymal stem cell osteogenic differentiation through regulation of klotho expression in vitro, Cell Biol. Int (2020), 10.1002/cbin.11460. [DOI] [PubMed] [Google Scholar]
- [17].Teng JW, Ji PF, Zhao ZG, Mir-214–3p inhibits β-catenin signaling pathway leading to delayed fracture healing, Eur. Rev. Med. Pharmacol. Sci 22 (2018) 17–24, 10.26355/eurrev_201801_14095. [DOI] [PubMed] [Google Scholar]
- [18].Wang X, Guo B, Li Q, Peng J, Yang Z, Wang A, Li D, Hou Z, Lv K, Kan G, Cao H, Wu H, Song J, Pan X, Sun Q, Ling S, Li Y, Zhu M, Zhang P, Peng S, Xie X, Tang T, Hong A, Bian Z, Bai Y, Lu A, Li Y, He F, Zhang G, Li Y, mir-214 targets atf4 to inhibit bone formation, Nat. Med 19 (2013) 93–100, 10.1038/nm.3026. [DOI] [PubMed] [Google Scholar]
- [19].Watanabe T, Sato T, Amano T, Kawamura Y, Kawamura N, Kawaguchi H, Yamashita N, Kurihara H, Nakaoka T, Dnm3os, a non-coding RNA, is required for normal growth and skeletal development in mice, Dev. Dyn 237 (2008) 3738–3748, 10.1002/dvdy.21787. [DOI] [PubMed] [Google Scholar]
- [20].Akhade VS, Pal D, Kanduri C, Long noncoding RNA: genome organization and mechanism of action, Adv. Exp. Med. Biol 1008 (2017) 47–74, 10.1007/978-981-10-5203-3_2. [DOI] [PubMed] [Google Scholar]
- [21].Batista PJ, Chang HY, Long noncoding RNAs: cellular address codes in development and disease, Cell 152 (2013) 1298–1307, 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Flynn RA, Chang HY, Long noncoding RNAs in cell-fate programming and reprogramming, Cell Stem Cell 14 (2014) 752–761, 10.1016/j.stem.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Huynh NPT, Anderson BA, Guilak F, McAlinden A, Emerging roles for long noncoding RNAs in skeletal biology and disease, Connect. Tissue Res. 58 (2017) 116–141, 10.1080/03008207.2016.1194406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Sun H, Peng G, Ning X, Wang J, Yang H, Deng J, Emerging roles of long noncoding RNA in chondrogenesis, osteogenesis, and osteoarthritis, Am. J. Transl. Res 11 (2019) 16–30. [PMC free article] [PubMed] [Google Scholar]
- [25].Savary G, Dewaeles E, Diazzi S, Buscot M, Nottet N, Fassy J, Courcot E, Henaoui IS, Lemaire J, Martis N, Van der Hauwaert C, Pons N, Magnone V, Leroy S, Hofman V, Plantier L, Lebrigand K, Paquet A, Lino Cardenas CL, Vassaux G, Hofman P, Günther A, Crestani B, Wallaert B, Rezzonico R, Brousseau T, Glowacki F, Bellusci S, Perrais M, Broly F, Barbry P, Marquette CH, Cauffiez C, Mari B, Pottier N, The long noncoding rna DNM3OS is a reservoir of fibromirs with major functions in lung fibroblast response to TGF-β and pulmonary fibrosis, Am. J. Respir. Crit. Care Med 200 (2019) 184–198, 10.1164/rccm.201807-12370C. [DOI] [PubMed] [Google Scholar]
- [26].Ai D, Yu F, LncRNA DNM3OS promotes proliferation and inhibits apoptosis through modulating IGF1 expression by sponging MiR-126 in CHON-001 cells, Diagn. Pathol 14 (2019) 106, 10.1186/s13000-019-0877-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Lee RC, Feinbaum RL, Ambros V, The C elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14, Cell 75 (1993) 843–854, 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- [28].Bartel DP, MicroRNAs: genomics, biogenesis, mechanism, and function, Cell 116 (2004) 281–297, 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- [29].Bartel DP, MicroRNAs: target recognition and regulatory functions, Cell 136 (2009) 215–233, 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].McAlinden A, Im GI, MicroRNAs in orthopaedic research: disease associations, potential therapeutic applications, and perspectives, J. Orthop. Res 36 (2018) 33–51, 10.1002/jor.23822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Desvignes T, Contreras A, Postlethwait JH, Evolution of the miR199–214 cluster and vertebrate skeletal development, RNA Biol. 11 (2014) 281–294, 10.4161/rna.28141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Loebel DAF, Tsoi B, Wong N, Tam PPL, A conserved noncoding intronic transcript at the mouse Dnm3 locus, Genomics 85 (2005) 782–789, 10.1016/j.ygeno.2005.02.001. [DOI] [PubMed] [Google Scholar]
- [33].Lee YB, Bantounas I, Lee DY, Phylactou L, Caldwell MA, Uney JB, Twist-1 regulates the miR-199a/214 cluster during development, Nucleic Acids Res. 37 (2009) 123–128, 10.1093/nar/gkn920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Lin EA, Kong L, Bai XH, Luan Y, Liu CJ, miR-199a, a bone morphogenic protein 2-responsive microRNA, regulates chondrogenesis via direct targeting to Smad1, J. Biol. Chem 284 (2009) 11326–11335, 10.1074/jbc.M807709200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Shi K, Lu J, Zhao Y, Wang L, Li J, Qi B, Li H, Ma C, MicroRNA-214 suppresses osteogenic differentiation of C2C12 myoblast cells by targeting Osterix, Bone 55 (2013) 487–494, 10.1016/j.bone.2013.04.002. [DOI] [PubMed] [Google Scholar]
- [36].Wang X, Guo B, Li Q, Peng J, Yang Z, Wang A, Li D, Hou Z, Lv K, Kan G, et al. , miR-214 targets ATF4 to inhibit bone formation, Nat. Med 19 (2012) 93–100, 10.1038/nm.3026. [DOI] [PubMed] [Google Scholar]
- [37].Zhao C, Sun W, Zhang P, Ling S, Li Y, Zhao D, Peng J, Wang A, Li Q, Song J, Wang C, Xu X, Xu Z, Zhong G, Han B, Chang YZ, Li Y, miR-214 promotes osteoclastogenesis by targeting Pten/PI3k/Akt pathway, RNA Biol. 12 (2015) 343–353, 10.1080/15476286.2015.1017205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Cai M, Yang L, Zhang S, Liu J, Sun Y, Wang X, A bone-resorption surface-targeting nanoparticle to deliver anti-miR214 for osteoporosis therapy, Int. J. Nanomedicine 12 (2017) 7469–7482, 10.2147/IJN.S139775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Cao F, Zhan J, Chen X, Zhang K, Lai R, Feng Z, miR-214 promotes periodontal ligament stem cell osteoblastic differentiation by modulating Wnt/β-catenin signaling, Mol. Med. Rep 16 (2017) 9301–9308, 10.3892/mmr.2017.7821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Guo Y, Li L, Gao J, Chen X, Sang Q, miR-214 suppresses the osteogenic differentiation of bone marrow-derived mesenchymal stem cells and these effects are mediated through the inhibition of the JNK and p38 pathways, Int. J. Mol. Med 39 (2016) 71–80, 10.3892/ijmm.2016.2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Scheffzek K, Welti S, Pleckstrin homology (PH) like domains - versatile modules in protein-protein interaction platforms, FEBS Lett. 586 (2012) 2662–2673, 10.1016/j.febslet.2012.06.006. [DOI] [PubMed] [Google Scholar]
- [42].Dar S, Pucadyil TJ, The pleckstrin-homology domain of dynamin is dispensable for membrane constriction and fission, Mol. Biol. Cell 28 (2017) 152–160, 10.1091/mbc.E16-09-0640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Brereton E, Fassi E, Araujo GC, Dodd J, Telegrafi A, Pathak SJ, Shinawi M, Mutations in the PH domain of DNM1 are associated with a nonepileptic phenotype characterized by developmental delay and neurobehavioral abnormalities, Molecular Genetics & Genomic Medicine 6 (2018) 294–300, 10.1002/mgg3.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.