Abstract
Vemurafenib is a BRAF kinase inhibitor (BRAFi) that is used to treat melanoma patients harboring the constitutively active BRAF-V600E mutation. However, after a few months of treatment patients often develop resistance to vemurafenib leading to disease progression. Sequence analysis of drug-resistant tumor cells and functional genomic screens has identified several genes that regulate vemurafenib resistance. Reactivation of mitogen-activated protein kinase (MAPK) pathway is a recurrent feature of cells that develop resistance to vemurafenib. We performed a genome-scale CRISPR-based knockout screen to identify modulators of vemurafenib resistance in melanoma cells with a highly improved CRISPR sgRNA library called Brunello. We identified 33 genes that regulate resistance to vemurafenib out of which 14 genes have not been reported before. Gene ontology enrichment analysis showed that the hit genes regulate histone modification, transcription and cell cycle. We discuss how inactivation of hit genes might confer resistance to vemurafenib and provide a framework for follow-up investigations.
Keywords: CRISPR screen, Vemurafenib, BRAF-V600E, Melanoma, Brunello library, A375
Introduction
Vemurafenib, also known as PLX4032, is one of the first Food and Drug Administration (FDA)-approved small molecule inhibitors for the treatment of metastatic melanoma patients, specifically for patients harboring the BRAF-V600E mutation. The BRAF-V600E mutation is present in approximately 50% of melanoma cases and causes constitutive activation of BRAF and the MAPK (ERK) signaling pathway leading to uncontrolled cell proliferation (Ascierto et al. 2012). Vemurafenib binds specifically to the adenosine triphosphate (ATP) binding pocket of activated BRAF-V600E, blocks ERK1/2 activation, and induces cell cycle arrest and apoptosis (Torres-Collado et al. 2017). Although there is short-term tumor regression and enhancement of patient survival, resistance to vemurafenib frequently develops (Ascierto et al. 2012). Therefore, an understanding of mechanisms underlying vemurafenib resistance is of paramount importance to develop novel melanoma therapeutics.
Exposure of melanoma cell lines to incremental concentrations of vemurafenib resulted in drug resistance (Zecena et al. 2018). In contrast, intermittent use of vemurafenib was observed to delay acquisition of resistance. Possibly, the continuous administration of vemurafenib provides a selective pressure for drug-resistant cells to thrive (Thakur et al. 2013). Initial inhibition of BRAF-V600E by vemurafenib severely depletes MAPK output and is therefore considered to be a dormant period for resistant cells to accumulate before resulting in their over proliferation. On the other hand, excessive MAPK output causes toxicity (Morris et al. 2013; Thakur et al. 2013). Hence, the need to re-establish dynamic equilibrium with interrupted scheduled dosing of vemurafenib was emphasized (Mackiewicz-Wysocka et al. 2014).
Basal phosphorylation levels of both MEK and ERK were enhanced in vemurafenib-resistant A375 cells while AKT phosphorylation levels remained relatively unchanged (Yadav et al. 2012). Phosphorylation of downstream effectors of mTOR (decreased phospho-S6 and hyperphosphorylation of 4E-BP1) has also been shown to contribute to vemurafenib resistance (Zhan et al. 2015). Reactivation of MAPK pathway and/or activation of PI3K/AKT pathways confer resistance to BRAFi in melanoma cells (Luebker and Koepsell, 2019). MAPK activation could occur by increased expression of their upstream activators namely the receptor tyrosine kinases (RTKs). Alternatively, activating mutations within the RAS/RAF/MEK/Erk signaling pathway could recover the MAPK pathway (Luebker and Koepsell 2019). Sequence analysis of vemurafenib-resistant cancer cells has identified mutations in BRAF-V600E (amplification, truncation, alternative splicing, and fusions), activating mutations in RAS genes (NRAS, KRAS, and HRAS), RAC1, MAP2K1, and AKT and loss-of-function mutations in GNAQ/GNA11, CDKN2A, PTEN, PIK3R2, and DUSP4. However, the molecular basis of about 30–40% cases of BRAFi resistance remains unknown.
Functional genetic screens have been gaining traction as a powerful approach to identify novel cellular pathways involved in acquisition of drug-resistance. Whittaker et al. (2013) did a genome-scale RNA interference screen with a library size of 90,000 shRNAs targeting approximately 16,600 genes expressed in A375 cells (Whittaker et al. 2013). Seminal work on the genome-wide CRISPR KnockOut (GeCKO) screen (Shalem et al. 2014) for regulators for vemurafenib resistance was performed by the Zhang group using the GeCKOv1 library that covered 18,080 human genes with about 3–4 target sgRNAs per gene (Shalem et al. 2014). Only 3 genes were common among the hits from CRISPR and shRNA screens. This is consistent with observations that the correlation between the results from shRNA and CRISPR screens is poor (Morgens et al. 2016).
An improved GeCKOv2 library with minimal off-target effects and covering 19,050 human genes with 6 sgRNAs per gene (Sanjana et al. 2014) was used for a positive selection screen with vemurafenib. By determining the targeting efficiencies of 1841 sgRNAs directed at 6 mouse and 3 human genes, Doench et al. (2016) identified sequence features associated with high targeting efficiency. These features helped to formulate rules which were used to design the Avana sgRNA library that was used for a positive selection screen with vemurafenib (Doench et al. 2016). Among the top 100 hits in screens with GeCKOv1, GeCKOv2 and Avana libraries, only 19 genes were common indicating a high degree of variability in the results obtained from the three screens. By measuring the off-target activities of thousands of sgRNAs, Sanson et al. (2018) identified a metric to minimize the off-target effects of sgRNA. This metric was combined with the rules to maximize targeting efficiency (Doench et al. 2016) to design the Brunello library (Sanson et al. 2018). GeCKOv2, Avana and Brunello libraries were compared in their abilities to target the gold-standard gene sets of 1580 essential and 927 nonessential genes (Hart et al. 2014). Efficiency of the libraries was determined by the ability of sgRNAs to distinguish between essential and nonessential genes. Brunello library exhibited greater depletion of sgRNA targeting essential genes compared to the GeCKOv2 and Avana libraries but sgRNA targeting nonessential genes were unaffected (Sanson et al. 2018). Although the number of target genes in the three sgRNA libraries are comparable (Table 1), we reasoned that increased targeting efficiency coupled with minimal off-target effects of the Brunello library might identify novel modulators of vemurafenib resistance.
Table 1.
Features of different human sgRNA libraries
| Name of the library | Nature of the library | No. of genes targeted | No. of sgRNAs per target gene | Total number of sgRNAs | Reference |
|---|---|---|---|---|---|
| Geckov1 | Knockout | 18,080 | 3–4 | 64,751 | Shalem et al. (2014) |
| Geckov2 | Knockout | 19,050 | 6 | 123,441 | Sanjana et al. (2014) |
| Avana | Knockout | 18,547 | 4 | 73,782 | Doench et al. (2016) |
| Brunello | Knockout | 19,114 | 4 | 76,441 | Doench et al. (2016) |
In this study, we performed a CRISPR-Cas9 mediated genome-wide knockout screen in human melanoma cell line A375 using the Brunello library to identify novel genes that regulate vemurafenib resistance. Gene ontology (GO) enrichment analysis of the top hits suggests that alterations to MAPK pathway, epigenome and cell cycle facilitate acquisition of resistance to vemurafenib in melanoma cells.
Materials and methods
Library amplification
One hundred nanograms of human sgRNA library Brunello in lentiCRISPRv2 (Addgene, #73179) was transformed into electrocompetent Endura cells (#60242, Lucigen) in quadruplicates. Cells were transferred into a chilled electroporation cuvette and pulsed (BioRad Micropulser, #165–1200) at 10 µF, 600 Ω, 1800 V. Within 10 s of the pulse, 1975 µL of the Recovery medium was added to the cells. Cells were then incubated in a shaker incubator at 250 rpm for 1 h at 37°C before being selected on LB-Agar containing 100 μg/mL ampicillin in 245 mm Square BioAssay Dishes (Corning) at 32°C for 14 h. Transformants were pooled by rinsing the bioassay plates with 20 mL of LB twice using a cell scraper and used for plasmid DNA extraction with Endotoxin-free plasmid DNA purification (#740424-10, Macherey Nagel).
Lentivirus generation and harvesting
Lipofectamine transfection was performed on HEK293FT cells 24 h after the cells were seeded into 100 mm dishes in DMEM (Dulbecco's Modified Eagle's Medium media) supplemented with 10% Fetal Bovine Serum (FBS) (#SH30071.03, Hyclone), 2 mM L-glutamine and 1 mM sodium pyruvate. Briefly, 612.5 µL of Opti-MEM containing P3000 (14 µL), DNA mixture of lentivirus packing plasmid (2 µg PLP1, 2 µg PLP2, and 1 µg pVSVG) and Brunello library (2 µg) were added into Lipofectamine mixture (612.5 µL of Opti-MEM and 14 µL of Lipofectamine 3000). This mixture was incubated at room temperature for 15 min. Afterwards, the transfection mixture was added dropwise to the HEK293FT cells and incubated in the 37°C incubator with 5% CO2 (w/v). After 24 h, media was supplemented with 1 mM sodium butyrate to increase virus production. For the next 2 consecutive days, the viruses were harvested by centrifugation at 16,500 rpm for 90 min at 4°C. Virus pellets were re-suspended in PBS and kept at −80°C. To determine multiplicity of infection (MOI) of Brunello virus, a serial dilution of the virus at 1:10, 1:50, 1:100, 1:1000 was added into 1 106 A375 cells in a 12-well plate. Polybrene was then added at a final concentration of 5 µg/mL and then centrifuged at 3000 rpm for 90 min at 30°C. After 1 day of incubation, we equally split each virus transduced cells into two sets of medium with or without 1 µg/mL puromycin. After 3 days of incubation, cell numbers (n) were determined per condition and the percentage transduction was calculated as followed:
A MOI of 0.4 was used for large-scale screening.
Determination of IC50 for Vemurafenib in A375
IC50 was determined on days 3 and 4 after vemurafenib treatment using Real-time-Glo MT Cell Viability Assay according to the manufacturer's protocol (Promega, #G9711). Using a 96-well white plate, viability assays of 2000 A375 cells were performed in triplicates. Doubling time of DMSO-treated A375 cells was found to be about 18 h. IC50 for vemurafenib was determined with a dose-response curve.
Vemurafenib resistance screen with the Brunello library
9.6 × 107 A375 cells were transduced with 1 × 106 cells plated per transduction well (12-well plate). One hundred and ten microliters of the concentrated Brunello library (MOI = 0.4) was applied into each well containing A375 cells. Puromycin (1 µg/mL) was added to the cells 24 h post transduction and maintained for 7 days. On day 7, cells were split into DMSO or 2 µM vemurafenib conditions in duplicates with 3 × 107 cells per replicate and an additional 3.3 × 107 cells were frozen down for Brunello library genomic DNA (gDNA) analysis. DMSO-treated cells were passaged every 3–4 days. Fresh medium with 2 µM vemurafenib was added to drug-treated cells every 3–4 days. Cell pellets were taken at 7 and 14 days after vemurafenib treatment.
Genomic DNA extraction and amplicon sequencing
Cell pellets were thawed for gDNA extraction using Blood and Cell Culture DNA Midi/Maxi Kit (Qiagen). PCR was performed with Q5 Hot Start High-Fidelity 2 × Master Mix (#M0494L, New England Biolabs) with the amount of input gDNA for each samples was 15 µg. For each sample, we performed 10 separate 50 µL reactions with 1.5 µg gDNA, in each reaction using 10 forward primers with 1–10 bp staggered region to increase the diversity of the library and 1 reverse primer containing the unique barcode to differentiate the samples (Supplementary Table S1). Thermal cycling conditions included an initial denaturation step at 98°C for 3 min followed by 28 cycles of amplification (10 s at 98°C, 30 s at 60°C, and 25 s at 72°C) and a final extension step at 72°C for 5 min. Minimum number of PCR cycles to amplify the gDNA was determined by performing small-scale PCRs with varying number of cycles. All 10 PCRs of a single sample were pooled and purified using QIAquick PCR Purification Kit (Qiagen). The purified PCR product was then separated using 2% agarose gel and gel extracted with the Qiagen Gel Extraction Kit. These gel purified PCR products were stored at −20°C before being submitted for next-generation sequencing (NGS) by NovogeneAIT Genomics (Singapore) using HiSeq-SE150 platform.
Data analyses
The raw FASTQ files (.fastq.gz) were demultiplexed by NovogeneAIT Genomics (Singapore) and then uploaded to CRISPRAnalyzer (http://crispr-analyzer.dkfz.de) (Winter et al. 2017) for analysis of phenotypes against the reference Brunello library. CRISPRAnalyzer performs the alignment of the unique sgRNA sequence with the reference library to obtain the sgRNA read counts. After extraction of read count files (.txt), these files were uploaded again to CRISPRAnalyzer. To determine the log2 fold changes of gRNA read counts between treatment and control conditions, read counts with less than 20 sgRNAs were removed for analysis. Model-based Analysis of Genome-wide CRISPR/Cas9 Knockout (MAGeCK) was chosen for our analysis which scores whether each gene is enriched or depleted and generates a gene ranking list (Li et al. 2014). Default settings were used in our analysis. Genes with P-values less than 0.05 were chosen for further analysis (Yu and Yusa 2019). Shortlisted genes were analyzed using the Search Tool for the Retrieval of Interacting Genes/Proteins database (STRING) (Szklarczyk et al. 2019) for functional association. The text mining option was disabled during PPI data analysis to ensure higher confidence. GO term enrichment analysis was performed using DAVID (Database for Annotation, Visualization, and Integrated Discovery) (Huang et al. 2009a, 2009b). A list of 33 genes identified in our screen was used as input data for the DAVID Functional Annotation Tool (Huang et al. 2009a, 2009b) with default timings to identify enriched GO Terms.
Data availability
Supplementary Figure S1 depicts the cumulative distribution of the number of reads per sgRNA in an individual genome scale CRISPR screen. Supplementary Figure S2 compares the hits obtained from CRISPR-based screens for vemurafenib resistance. Supplementary Figure S3 illustrates the proposed mechanisms for vemurafenib resistance caused by deletion of MAPK signaling pathway related hits in the screen. Supplementary Table S1 lists the sequences of primers used for amplifying sgRNA sequences from the Brunello library and for NGS. Supplementary Table S2 contains the Read count file of the 76,441 sgRNA in the Brunello library targeting 19,114 genes. Supplementary Table S3 contains two tabs. The first tab shows the ranked P-values according to MAGeCK for all the 19,114 genes after 14 days of vemurafenib treatment. The second tab only shows the values for genes that have the ranked P-value less than 0.05. Supplementary Table S4 compares the top 33 hits with top 100 enriched genes obtained in previous screens with Gecko v1, Gecko v2, and Avana sgRNA libraries. Supplementary Table S5 contains the results of STRING analysis of top 33 genes that regulate vemurafenib resistance. Supplementary Table S6 contains the results from GO analysis of the top 33 genes that regulate vemurafenib resistance using the online tools DAVID and REVIGO. Supplemental material is available at figshare: https://doi.org/10.25387/g3.13413830.
Results and discussion
Functional interrogation of genes in modulation of vemurafenib resistance via CRISPR
We sought to identify novel regulators of vemurafenib resistance by performing a genome scale CRISPR screen (Figure 1) with the highly optimized Brunello CRISPR sgRNA library. We chose A375 melanoma cells which harbor the gain-of-function BRAF-V600E mutation. Treatment of A375 cells with vemurafenib resulted in a growth arrest with an IC50 of 248.3 nM (Figure 2A).
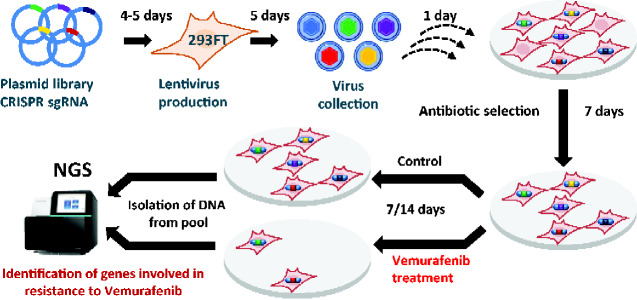
Figure 1.
Schematic representation of the workflow for the genome-wide CRISPR/Cas9 screen. HEK293FT cells were transfected with lentiviral CRISPR-Cas9 plasmid library (Brunello library with genome wide coverage and 4 sgRNAs per gene) for lentivirus production. A375 cells were then transduced with lentiviruses generated from the Brunello plasmid library. Cells were selected for successful transduction using puromycin selection for 7 days. After that, cells were either treated with 2 µM vemurafenib or DMSO. Samples were collected at days 7 and 14. gDNA was prepared from the cells and the relative sgRNA abundance was determined by PCR-mediated amplification of sgRNA sequences from gDNA followed by NGS.
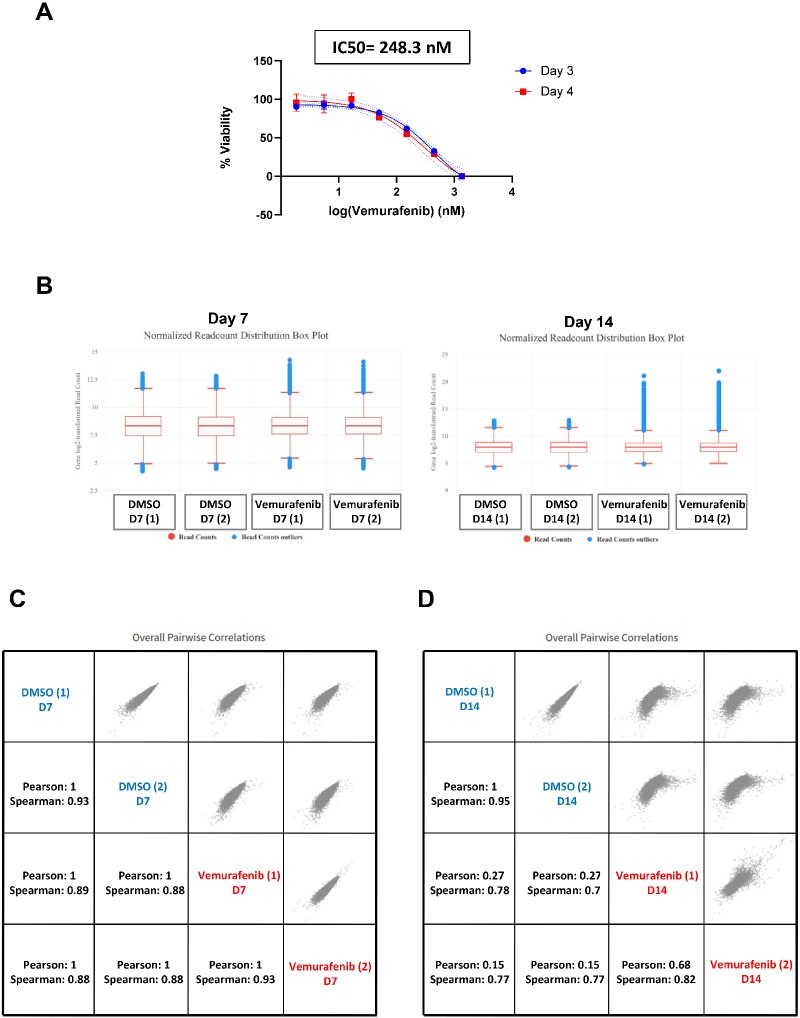
Figure 2.
NGS data analysis of the genome wide screen using CRISPRAnalyzeR. (A) Cell viability of A375 cells after treatment with various concentration of vemurafenib was determined using Real-time-Glo MT Cell Viability Assay. Percentage viability was plotted against various concentration of vemurafenib. Three days after vemurafenib treatment samples are indicated by circle shape while the 4 days after vemurafenib treatment samples are indicated by square shape. (B) Boxplot showing the read count distribution from individual sgRNAs for the DMSO-treated and vemurafenib-treated cells for days 7 and 14. For day 14, there is an increase in the number of reads for the most abundant sgRNAs in the vemurafenib- treated cells as compared to the DMSO treated cells. (C) Pairwise correlation plot of the day 7 samples with the Pearson and Spearman correlation coefficients. (D) Pairwise correlation plot of the day 14 samples with the Pearson and Spearman correlation coefficients.
To select for vemurafenib-resistant A375 cells in our genome wide CRISPR screen, we used vemurafenib at 2 µM, which is about 10-fold higher than its IC50 value and used previously in a positive selection screen (Doench et al. 2016). We transduced A375 cells with the Brunello lentiviral sgRNA library at a MOI of 0.4 and selected for transductants in the presence of puromycin for 7 days. We then harvested the puromycin-resistant cells and treated them with either DMSO or 2 µM vemurafenib for 7 and 14 days in duplicates (Figure 1). We isolated the gDNA from cells collected at day 0, 7 (DMSO or vemurafenib-treated), and day 14 (DMSO or vemurafenib-treated), amplified the sgRNA sequences from gDNA by PCR and sequenced the PCR products by deep sequencing methods.
CRISPRAnalyzer-based analysis of NGS data showed that less than 7.5% of the Brunello library sgRNAs were covered by less than 100 reads (Supplementary Figure S1, Table S2). This meant that 92.5% of the Brunello library sgRNA have a read count of more than 100, indicating a good library coverage. The sgRNA distribution in day 14 vemurafenib-treated cells was significantly different from sgRNA distribution in DMSO-treated cells (Figure 2B). Day 14 vemurafenib-treated cells showed a higher range of read counts indicating an enrichment of some sgRNA-treated cells after drug treatment (Figure 2B). Analysis of the pairwise correlation data indicated that the replicates of the day 7 treatment correlated well with a Pearson value of 1 and Spearman value of 0.93 indicating good reproducibility (Figure 2C). For the day 14 replicates, DMSO-treated samples correlated well (Pearson value = 1, Spearman = 0.95) but for the vemurafenib-treated samples, their correlation was relatively low (Pearson value = 0.68, Spearman = 0.82) (Figure 2D). This suggests that our genetic screen was not saturating and some modulators of vemurafenib resistance may have been missed out. However, the altered read count distribution of vemurafenib-resistant cells in comparison to DMSO-treated cells indicated that our screen has identified mutations that enhance resistance to vemurafenib.
Vemurafenib resistance genes
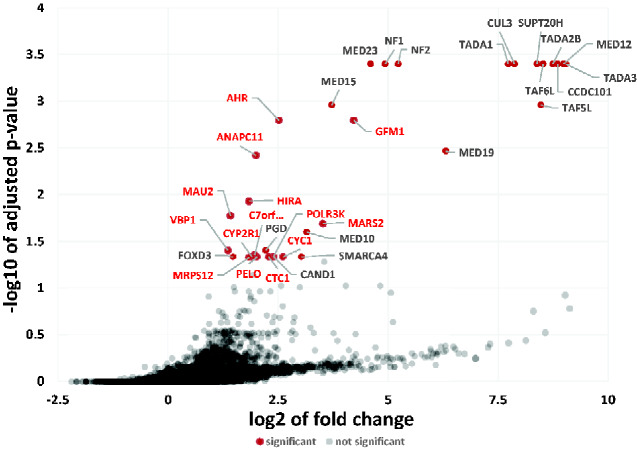
Using the CRISPRanalyzer platform to assess our NGS data, we identified 33 genes (P-value <0.05) that had significantly enriched sgRNA levels after 14 days of vemurafenib treatment (Figure 3 and Supplementary Table S3). All the top 10 hits have been reported in previous genome-wide CRISPR/Cas9 screens validating our screen for modulators of vemurafenib resistance. The top 10 enriched genes were CCDC101 (SAGA complex associated factor 29), TAF6L (TATA-box binding protein associated factor 6 like), SUPT20H (Transcription factor SPT20 homolog, SAGA complex component), TADA2B (Transcriptional Adaptor 2B), NF2 (Neurofibromin 2),
Figure 3.
Identification of vemurafenib-resistance genes using MAGeCK. The sgRNA distribution data of DMSO-treated and vemurafenib-treated cells were analyzed by MaGeCK. For each gene, the −log10P-value was plotted against its log2 fold change. Vemurafenib resistance genes were identified using a P-value threshold of 0.05. Significant hits are denoted by red dots along with gene names. Novel hit genes are in bold red font.
MED12 (Mediator complex subunit 12), TADA3 (Transcriptional adaptor 3), CUL3 (Cullin 3), TADA1 (Transcriptional adaptor 1), and MED23 (Mediator Complex Subunit 23).
Comparison with previous genetic screens
We compared our top 33 hits (P-value <0.05) with Top 100 enriched hits obtained from four previous 3 genome wide CRISPR KO screens (Li et al. 2014; Shalem et al. 2014; Doench et al. 2016). Out of our top 33 hits, only 19 were obtained in previous studies (Supplementary Table S4). As depicted in the Venn diagram (Supplementary Figure S2), only 13 hits (NF1, NF2, MED12, MED15, MED19, MED23, CUL3, TADA1, TADA2B, CCDC101, TAF5L, TAF6L, and PGD) were obtained in all the four screens. These results indicate that there is considerable variability with results obtained from independent CRISPR screens. Fourteen out of the 33 hits have not been previously reported in genome wide screens.
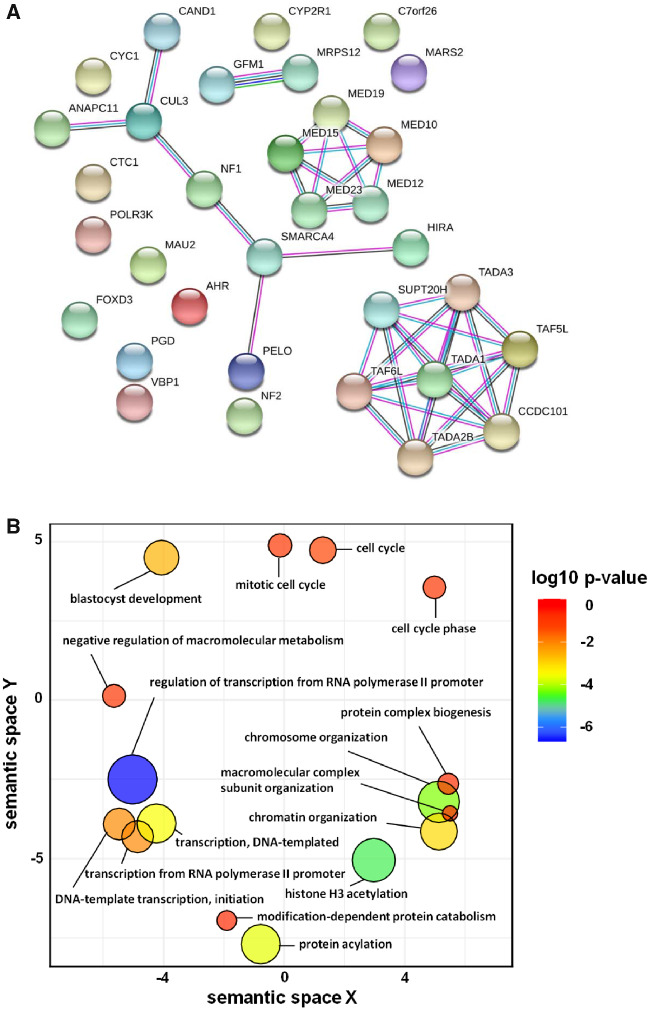
To assess the presence of functional associations between proteins that confer vemurafenib resistance, we submitted our top 33 hits to the STRING v11 database (Szklarczyk et al. 2019) to construct the protein–protein interaction (PPI) network. Proteins in the PPI network have significant overlap in their functional roles and participate in a common biological process (Szklarczyk et al. 2019). PPI network among the hits is shown in Figure 4A, which includes a total of 33 nodes and 38 edges, with the node and edge representing a target protein and PPI respectively. The average node degree is 2.3 which represents the average number of targets connected to a target. The degree of a target in a PPI reflects the strength of its role in the interaction network. Our hits interact with each other significantly with a high confidence score (average combined associated score of 0.96 and PPI enrichment P-value: < 1.0e–16) (Supplementary Table S5). Average combined associated score is an indicator of confidence of a PPI, i.e. how likely STRING judges an interaction to be true, given the available evidence. All scores rank from 0 to 1, with 1 being the highest possible confidence. MED12 interacts with MED10, MED15, MED19, and MED23 with a combined associated score of 0.999 (Figure 4A and Supplementary Table S5). The combined associated score of TADA1 with SUPT20H, TADA2B, TADA3, CCDC101, TAF5L, and TAF6L lies between 0.981 and 0.997 (Figure 4A and Supplementary Table S5). Presence of a strong functional association between the hits demonstrates the robustness of the results from the genome-scale screen.
Figure 4.
STRING analysis and GO term enrichment analysis of the 33 hits from the CRISPR screen. (A) Shortlisted genes were analyzed using the Search Tool for the Retrieval of Interacting Genes/Proteins database (STRING v11). Network nodes represent proteins and the edges represent PPIs. Results of STRING analysis are presented in Supplementary Table S5. (B) GO term enrichment analysis was performed using DAVID. GO terms are clustered together based on semantic similarity in the 2-dimensional scatterplot. P-value which indicates the enrichment strength in the annotation category is denoted by the bubble color and the GO term frequency is denoted by the bubble size. Results of DAVID analysis are presented in Supplementary Table S6.
To gain insights into the vemurafenib resistance mechanisms in A375 cells, we performed GO enrichment analysis with the top 33 genes that regulate resistance to vemurafenib using the online functional annotation tool DAVID (Huang et al. 2009a, 2009b). Many GO terms under Biological Process were redundant under different annotation clusters (Supplementary Table S6). We removed the redundant GO terms using the online tool REVIGO (Supek et al. 2011). Visualization of GO data by REVIGO (Figure 4B) indicated that genes that modulate vemurafenib resistance are involved in histone/protein acetylation, chromosome/chromatin organization, cell cycle, protein complex biogenesis, DNA template transcription/initiation and regulation of transcription from RNA Pol II promoter. Although validation of top hits with targeted knockouts is necessary, we discuss below how the identified hit genes might regulate resistance to vemurafenib on the basis of information available in the literature.
MAPK pathway
BRAF, the target of vemurafenib, is the RAF component of the MAPK signaling pathway that is frequently hyperactivated in cancer cells (Pratilas et al. 2012), MAPK pathway consists of 1G protein RAS, and 3 kinases RAF, MEK, and ERK. Binding of a growth factor to the RTK activates the Ras protein (Supplementary Figure S3). Activated Ras phosphorylates RAF, which is then followed by sequential activation of MEK and ERK (Supplementary Figure S3). Four genes NF1, CUL3, NF2, and MED12 involved in the MAPK signaling pathway were among our top 11 hits. We discuss how deletion of these 4 genes could confer vemurafenib resistance below.
NF1 (Neurofibromin 1), a large protein consisting of over 2800 amino acid residues, contains a domain similar to the catalytic domain of GTPase Activating Protein (GAP). NF1 negatively regulates RAS by stimulating its GTPase activity (Supplementary Figure S3) (Kiuru and Busam 2017). Therefore, loss of NF1 would result in activation of Ras and MAPK signaling pathways.
CUL3 is a subunit of multiple E3 ubiquitin ligases and its loss is expected to cause stabilization of several proteins. CUL3 was recently shown to enhance MAPK signaling by stabilization and Src-dependent activation of RAC1 (Vanneste et al. 2020). Interestingly, CUL3 is also required for proteasomal degradation of NF1 (Hollstein and Cichowski 2013) but its effect on RAC1 appears to override its effect of NF1 stability in terms of MAPK signaling and vemurafenib resistance (Vanneste et al. 2020). RAC1 belongs to the Rho family of small GTP-binding proteins which transduce extracellular signals from growth factor, integrin and G-protein-coupled receptors (Wertheimer et al. 2012). Loss of CUL3 stabilizes RAC1 and therefore hyperactivates RAC1 (Supplementary Figure S3) (Vanneste et al. 2020). RAC1-GTP activates PAK1 (Supplementary Figure S3), leading to the downstream activation of MEK and bypassing the upstream BRAF inhibition (De et al. 2019).
NF2 encodes merlin which is mutated in various forms of cancer such as schwannomas, mesotheliomas, breast, prostate, colorectal, hepatic, clear cell renal cell carcinoma and melanomas (Petrilli and Fernández-Valle 2016). Loss of merlin was reported to cause vemurafenib resistance and the presence of merlin even at very low levels was sufficient to maintain vemurafenib sensitivity (Petrilli and Fernández-Valle 2016). Merlin inhibits Rac by preventing its localization to the plasma membrane (Okada et al. 2005) and by inhibiting the p21-activated kinase PAK1 (Kissil et al. 2003). So, inactivation of merlin would result in Rac activation and stimulation of the MAPK pathway. Loss of merlin activates other growth promoting pathways such as Hippo and mTOR (Petrilli and Fernández-Valle 2016) explaining the incidence of NF2 mutations in several forms of cancers.
MED12 is a part of the Transcriptional MEDIATOR complex that physically binds to TGF-βR2 and inhibits the TGF-βR2 signaling pathway. TGF-β activates Ras-MEK-ERK signaling via the nonSMAD pathway (Zhang, 2009). Binding of TGF-β activates TGFβR which activates Ras by promoting the formation of ShcA/Grb2/Sos complex (Zhang, 2009; Chapnick et al. 2011; Gui et al. 2012) (Supplementary Figure S3). MED12 suppression would result in activation of TGF-βR signaling pathway leading to increased RAS-MEK-ERK signaling. Mutations in MED12 confer resistance to a number of drugs used against colon cancer, melanoma and liver cancer (Huang et al. 2012). We also identified 4 additional subunits of the MEDIATOR complex, namely MED10, MED15, MED19, and MED23 which have all been reported in previous genetic screens (Shalem et al. 2014; Doench et al. 2016; Le Sage et al. 2017).
To summarize, loss of negative regulators of MAPK pathway (deletion of NF1), loss of degradation signals leading to stabilization of MAPK signaling (deletion of CUL3), loss of Rac inhibitors (deletion of NF2) and loss of inhibition of upstream pathways (deletion of MED12) could result in increased MAPK signaling and vemurafenib resistance. These 4 genes were obtained in previous screens in agreement with the notion that activation of MAPK signaling pathway is the main mechanism leading to vemurafenib resistance.
Epigenetic regulation
Post-translational histone modifications such as acetylation, methylation and sumoylation have significant effects on gene expression. Histone acetylation is commonly associated with activation of gene expression whereas histone methylation is linked to either activation or repression of gene expression [28]. Studies of epigenomic alterations revealed a loss of histone acetylation and histone H3 Lys 4 methylation (H3K4me2/3) on regulatory regions proximal to specific cancer-regulatory genes involved in important signaling pathways driving melanoma [14]. We identified several genes related to post-translational histone modification in our screen namely TAF6L, TAF5L, CCDC101, SUPT20H, TADA2B, TADA1, TADA3, HIRA, and SMARCA4.
Seven genes (TAF6L, TAF5L, CCDC101, SUPT20H, TADA2B, TADA1, and TADA3) encode subunits of the SPT3-TAFII31-GCN5L acetylase (STAGA complex). STAGA is a chromatin-acetylating transcription coactivator and regulates numerous cellular processes like transcription, splicing and DNA repair through coordination of multiple histone post-translational modifications (Martinez et al. 2001; Baker and Grant 2007).
Apart from the STAGA complex, factors mediating chromatin organization such as histone chaperones (HIRA) and SMARCA4 (SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4) were among our top hits. Mutations in SWI/SNF chromatin remodeling genes have been associated with invasive melanomas (Shain et al. 2015). SMARCA4 promotes chromatin accessibility around double-strand breaks (DSBs) (Rother and van Attikum 2017) and HIRA is recruited to DSBs, facilitating restoration of chromatin structure by depositing histones (Polo, 2015). Therefore, loss of HIRA and SMARCA4 is expected to cause genomic instability, a hallmark of tumor cells (Negrini et al. 2010).
Cell cycle
Dysregulation of cell cycle can boost cell division rates by overriding cell cycle arrests, inhibiting apoptosis and by promoting genetic instability. We found four genes related to cell cycle in our screen namely FOXD3, ANAPC11, PELO, and MAU2 with the last three being novel hits.
FOXD3 (Forkhead transcription factor) was previously reported to play a role in resistance toward the precursor of vemurafenib, PLX4720 (Smit et al. 2014). As a potent antagonist of melanoma proliferation (Abel and Aplin 2010), FOXD3 prevents melanoma cell migration and invasion in a Rho-associated protein kinase dependent manner (Katiyar and Aplin, 2011). Mutant BRAF signaling results in FOXD3 downregulation (Abel and Aplin 2010), which was also linked to epithelial–mesenchymal transition (EMT)-like phenotype (Chu et al. 2014). Attenuation of mutant BRAF signaling caused increased FOXD3 levels (Abel and Aplin 2010). Overexpression of FOXD3 inhibited growth of melanoma cells by causing a p53-dependent cell cycle arrest (Abel and Aplin 2010). Deletion of FOXD3 in vemurafenib-treated A375 cells could therefore overcome the cell cycle arrest and result in vemurafenib-resistance.
MAU2 interacts with NIPBL/SCC2 to form a heterodimeric complex that is required for loading of cohesin complex onto chromatin (Bermudez et al. 2012; Bot et al. 2017). This enables cohesin to tether sister chromatids together immediately after their generation during DNA replication until their separation during anaphase. ANAPC11 is the catalytic subunit of the anaphase promoting complex/cyclosome (APC/C) that regulates progression through mitosis and the G1 phase of the cell cycle. Loss of MAU2 and ANAPC11 could enhance genetic instability which could accelerate the acquisition of mutations causing vemurafenib resistance.
Translation
Misregulation of protein translation is a key mechanism that drives cellular transformation and tumor growth (Ruggero 2013). We found that 5 of the top 33 hit genes encode proteins involved in translation, with 3 of them being mitochondrial. The 5 genes encode PELO (ribosome rescue/mRNA surveillance factor), POLR3K (DNA-dependent RNA polymerase involved in 5S rRNA and tRNA synthesis), GFM1 (G elongation Factor Mitochondrial 1), MRPS12 (Mitochondrial Ribosomal Protein S12), and MARS2 (mitochondrial Methionine-tRNA synthetase 2).
PELO is an evolutionarily conserved gene (Shamsadin et al. 2002) required for ribosomal disassembly (Pisareva et al. 2011). PELO is involved in the no-go decay (NGD) surveillance mechanism which leads to mRNA degradation installed ribosomal elongation complexes (ECs) (Doma and Parker 2006). PELO knockdown has been reported to activate PI3K/AKT (Pedersen et al. 2014; Elkenani et al. 2016) and Bone Morphogenetic Protein (BMP) signaling pathways (Elkenani et al. 2016), which resulted in epidermal hyperplasia in mice (Elkenani et al. 2016). Furthermore, conditional deletion of PELO in mouse epidermal stem cells resulted in their hyperproliferation (Liakath-Ali et al. 2018).
Notably, three mitochondrial translation proteins (GFM1, MRPS12, and MARS2) were among the novel 14 hits. BRAF mutant cells have increased rates of glycolysis and reduced oxidative phosphorylation in comparison to wild type cells (Haq et al. 2013). Treatment of BRAF mutant cells with vemurafenib reduces glycolytic flux and increases mitochondrial oxidative phosphorylation resulting in severe oxidative stress (Corazao-Rozas et al. 2013). It would be informative to test whether inhibition of mitochondrial translation relieves oxidative stress in vemurafenib-treated BRAF mutant cells thereby providing a fitness advantage.
Other cellular processes
We obtained AHR (Aryl Hydrocarbon/dioxin receptor) which encodes a ligand-dependent transcription factor in our screen. In response to agonist binding, AHR translocates to the nucleus and activates the expression of xenobiotic metabolism genes like the P450 family of enzymes. Intriguingly, stable activation of AHR was reported to be required for resistance to BRAF-inhibitors in melanoma (Corre et al. 2018). Vemurafenib was shown to bind to a noncanonical substrate binding site of AHR and inhibit the canonical AHR signaling pathway (Corre et al. 2018). Our result appears to be at odds with this observation. However, AHR was shown to have either oncogenic or tumor suppressive effects depending on cellular phenotype (Contador-Troca et al. 2015). AHR knockdown promoted melanoma in mouse (Contador-Troca et al. 2015). In addition, the AHR levels in human metastatic melanomas were reduced in comparison to benign nevi (Benign nevi refer to abnormal cell growth arising from melanocytes that are noncancerous) (Contador-Troca et al. 2015).
Another novel hit Ctc1 is a component of the CST (Ctc1, Stn1, and Ten1) complex, which functions as a terminator of telomerase activity (Chen et al. 2012). Depletion of Cst1 boosted telomerase activity and telomere elongation (Chen et al. 2012). Interestingly, downregulation of the Ras pathway by inhibition of MEK/ERK kinases promoted telomere DNA damage and fragility in aggressive lung cancer and glioblastoma (GBM) mouse models (Bejarano et al. 2019). It is conceivable that increased telomere length caused by deletion of Ctc1 might provide a growth advantage in vemurafenib-treated cells.
Our list of hits included CYC1 (mitochondrial ubiquinol-cytochrome c oxidoreductase: functions in the Electron Transport Chain), CYP2R1 (Cytochrome P450 family 2 subfamily R member 1: a 25-hydroxylase involved in Vitamin D3 metabolism), CAND1 (cullin-associated neddylation-dissociated protein 1: a F-Box exchange factor), VBP1 (VHL binding protein 1: binds to E3 Ubiquitin ligase Von Hippel Lindau), PGD (phosphogluconate dehydrogenase: Enzyme in the pentose phosphate pathway) and C7orf26 (Chromosome 7 Open Reading Frame 26: Uncharacterized protein). All of these hits except CAND1 and PGD are novel.
In summary, genome-scale screen for modulators of vemurafenib resistance in A375 cells with the Brunello library identified 19 previously reported genes and 14 novel hits. Confirming and dissecting how inactivation of novel genes results in vemurafenib resistance might generate new therapeutic strategies for countering melanoma. Newly identified hits can potentially serve as melanocytic biomarkers for cancer detection.
Acknowledgments
C.J.H.G. and J.H.W. performed the CRISPR-based genetic screen. C.E.F., C.J.H.G., and J.H.W. did the analysis of the NGS data. BXT and CRC supervised the lentiviral production and transduction experiments. Y.H.L. and D.L. supervised the bioinformatics and CRISPR experiments respectively. P.A., J.H.W., and C.J.H.G. wrote the MS. P.A. conceived and supervised the entire project.
Contributor Information
Corinna Jie Hui Goh, Bioinformatics Institute (BII), A*STAR, Singapore 138671, Singapore.
Jin Huei Wong, Bioinformatics Institute (BII), A*STAR, Singapore 138671, Singapore.
Chadi El Farran, Epigenetics and Cell Fates Laboratory, A*STAR Institute of Molecular and Cell Biology, Singapore 138673, Singapore.
Ban Xiong Tan, Experimental Drug Development Centre, A*STAR, Singapore 138670, Singapore.
Cynthia R Coffill, p53Lab, A*STAR, Singapore 138648, Singapore.
Yuin-Hain Loh, Epigenetics and Cell Fates Laboratory, A*STAR Institute of Molecular and Cell Biology, Singapore 138673, Singapore.
David Lane, p53Lab, A*STAR, Singapore 138648, Singapore.
Prakash Arumugam, Bioinformatics Institute (BII), A*STAR, Singapore 138671, Singapore; Singapore Institute for Food and Biotechnology Innovation, Singapore 138632, Singapore.
Funding
This research was supported by the Agency for Science, Technology and Research (A*STAR) under its Industry Alignment Fund—Pre-Positioning Programme (IAF-PP) grant number H18/01/a0/B14 as part of the A*STAR Innovations in Food and Chemical Safety (IFCS) Programme.
Conflicts of interest: None declared.
Literature cited
- Abel EV, Aplin AE.. 2010. FOXD3 is a mutant B-RAF-regulated inhibitor of G1-S progression in melanoma cells. Cancer Res. 70: 2891–2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ascierto PA, Kirkwood JM, Grob JJ, Simeone E, Grimaldi AM, et al. 2012. The role of BRAF V600 mutation in melanoma. J Transl Med. 10:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker SP, Grant PA.. 2007. The SAGA continues: expanding the cellular role of a transcriptional co-activator complex. Oncogene. 26:5329–5340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejarano L, Bosso G, Louzame J, Serrano R, Gómez‐Casero E, et al. 2019. Multiple cancer pathways regulate telomere protection. EMBO Mol Med. 11:1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermudez VP, Farina A, Higashi TL, Du F, Tappin I, et al. 2012. In vitro loading of human cohesin on DNA by the human Scc2-Scc4 loader complex. Proc Natl Acad Sci USA. 109:9366–9371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bot C, Pfeiffer A, Giordano F, Manjeera DE, Dantuma NP, et al. 2017. Independent mechanisms recruit the cohesin loader protein NIPBL to sites of DNA damage. J Cell Sci. 130:1134–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapnick DA, Warner L, Bernet J, Rao T, Liu X.. 2011. Partners in crime: the TGFβ and MAPK pathways in cancer progression. Cell Biosci. 1:42–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LY, Redon S, Lingner J.. 2012. The human CST complex is a terminator of telomerase activity. Nature. 488:540–544. [DOI] [PubMed] [Google Scholar]
- Chu TL, Zhao HM, Li Y, Chen AX, Sun X, et al. 2014. FoxD3 deficiency promotes breast cancer progression by induction of epithelial-mesenchymal transition. Biochem Biophys Res Commun. 446:580–584. [DOI] [PubMed] [Google Scholar]
- Contador-Troca M, Alvarez-Barrientos A, Merino JM, Morales-Hernández A, Rodríguez MI, et al. 2015. Dioxin receptor regulates aldehyde dehydrogenase to block melanoma tumorigenesis and metastasis. Mol Cancer 14:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corazao-Rozas P, Guerreschi P, Jendoubi M, André F, Jonneaux A, et al. 2013. Mitochondrial oxidative stress is the Achille’s heel of melanoma cells resistant to BRAF-mutant inhibitor. Oncotarget. 4:1986–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corre S, Tardif N, Mouchet N, Leclair HM, Boussemart L, et al. 2018. Sustained activation of the Aryl hydrocarbon receptor transcription factor promotes resistance to BRAF-inhibitors in melanoma. Nat Commun. 9:4775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De P, Aske JC, Dey N.. 2019. RAC1 takes the lead in solid tumors. Cells. 8:382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, et al. 2016. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol. 34:184–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doma MK, Parker R.. 2006. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature. 440:561–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkenani M, Nyamsuren G, Raju P, Liakath-Ali K, Hamdaoui A, et al. 2016. Pelota regulates epidermal differentiation by modulating BMP and PI3K/AKT signaling pathways. J Invest Dermatol. 136:1664–1671. [DOI] [PubMed] [Google Scholar]
- Gui T, Sun Y, Shimokado A, Muragaki Y.. 2012. The roles of mitogen-activated protein kinase pathways in TGF- β -induced epithelial-mesenchymal transition. J Signal Transduct. 2012:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haq R, Shoag J, Andreu-Perez P, Yokoyama S, Edelman H, et al. 2013. Oncogenic BRAF regulates oxidative metabolism via PGC1α and MITF. Cancer Cell. 23:302–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart T, Brown KR, Sircoulomb F, Rottapel R, Moffat J.. 2014. Measuring error rates in genomic perturbation screens: gold standards for human functional genomics. Mol Syst Biol. 10:733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollstein PE, Cichowski K.. 2013. Identifying the ubiquitin ligase complex that regulates the NF1 tumor suppressor and Ras. Cancer Discov. 3:880–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA.. 2009a. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA.. 2009b. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 4:44–57. [DOI] [PubMed] [Google Scholar]
- Huang S, Hölzel M, Knijnenburg T, Schlicker A, Roepman P, et al. 2012. MED12 controls the response to multiple cancer drugs through regulation of TGF-β receptor signaling. Cell. 151:937–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katiyar P, Aplin AE.. 2011. FOXD3 regulates migration properties and RND3 expression in melanoma cells. Mol Cancer Res. 9:545–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissil JL, Wilker EW, Johnson KC, Eckman MS, Yaffe MB, et al. 2003. Merlin, the product of the Nf2 tumor suppressor gene, is an inhibitor of the p21-activated kinase, Pak1. Mol Cell 12:841–849. [DOI] [PubMed] [Google Scholar]
- Kiuru M, Busam KJ.. 2017. The NF1 gene in tumor syndromes and melanoma. Lab Invest. 97:146–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Sage C, Lawo S, Panicker P, Scales TME, Rahman SA, et al. 2017. Dual direction CRISPR transcriptional regulation screening uncovers gene networks driving drug resistance. Sci Rep. 7:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Xu H, Xiao T, Cong L, Love MI, et al. 2014. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol. 15:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liakath-Ali K, Mills EW, Sequeira I, Lichtenberger BM, Pisco AO, et al. 2018. An evolutionarily conserved ribosome-rescue pathway maintains epidermal homeostasis. Nature. 556:376–380. [DOI] [PubMed] [Google Scholar]
- Luebker SA, Koepsell SA.. 2019. Diverse mechanisms of BRAF inhibitor resistance in melanoma identified in clinical and preclinical studies. Front Oncol. 9:268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackiewicz-Wysocka M, Krokowicz Ł, Kocur J, Mackiewicz J.. 2014. Resistance to vemurafenib can be reversible after treatment interruption. Medicine (Baltimore) 93:e157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez E, Palhan VB, Tjernberg A, Lymar ES, Gamper AM, et al. 2001. Human STAGA complex is a chromatin-acetylating transcription coactivator that interacts with pre-mRNA splicing and DNA damage-binding factors in vivo. Mol Cell Biol. 21:6782–6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgens DW, Deans RM, Li A, Bassik MC.. 2016. Systematic comparison of CRISPR/Cas9 and RNAi screens for essential genes. Nat Biotechnol. 34:634–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris EJ, Jha S, Restaino CR, Dayananth P, Zhu H, et al. 2013. Discovery of a novel ERK inhibitor with activity in models of acquired resistance to BRAF and MEK inhibitors. Cancer Discov. 3:742–750. [DOI] [PubMed] [Google Scholar]
- Negrini S, Gorgoulis VG, Halazonetis TD.. 2010. Genomic instability an evolving hallmark of cancer. Nat Rev Mol Cell Biol. 11:220–228. [DOI] [PubMed] [Google Scholar]
- Okada T, Lopez-Lago M, Giancotti FG.. 2005. Merlin/NF-2 mediates contact inhibition of growth by suppressing recruitment of Rac to the plasma membrane. J Cell Biol. 171:361–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen K, Canals F, Prat A, Tabernero J, Arribas J.. 2014. PELO negatively regulates HER receptor signalling and metastasis. Oncogene. 33:1190–1197. [DOI] [PubMed] [Google Scholar]
- Petrilli AM, Fernández-Valle C.. 2016. Role of Merlin/NF2 inactivation in tumor biology. Oncogene. 35:537–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisareva VP, Skabkin MA, Hellen CUT, Pestova TV, Pisarev AV.. 2011. Dissociation by Pelota, Hbs1 and ABCE1 of mammalian vacant 80S ribosomes and stalled elongation complexes. EMBO J. 30:1804–1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo SE. 2015. Reshaping chromatin after DNA damage: the choreography of histone proteins. J Mol Biol. 427:626–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratilas CA, Xing F, Solit DB.. 2012. Targeting oncogenic BRAF in human cancer. Curr Top Microbiol Immunol. 355:83–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rother MB, van Attikum H.. 2017. DNA repair goes hip-hop: SMARCA and CHD chromatin remodellers join the break dance. Phil Trans R Soc B Biol Sci. 372:20160285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggero D. 2013. Translational control in cancer etiology. Cold Spring Harb Perspect Biol. 5:a012336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana NE, Shalem O, Zhang F.. 2014. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 11:783–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanson KR, Hanna RE, Hegde M, Donovan KF, Strand C, et al. 2018. Optimized libraries for CRISPR-Cas9 genetic screens with multiple modalities. Nat Commun. 9:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shain AH, Yeh I, Kovalyshyn I, Sriharan A, Talevich E, et al. 2015. The genetic evolution of melanoma from precursor lesions. N Engl J Med. 373:1926–1936. [DOI] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, et al. 2014. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 343:84–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamsadin R, Adham IM, Engel W.. 2002. Mouse pelota gene(Pelo): cDNA cloning, genomic structure, and chromosomal localization. Cytogenet Genome Res. 97:95–99. [DOI] [PubMed] [Google Scholar]
- Smit MA, Maddalo G, Greig K, Raaijmakers LM, Possik PA, et al. 2014. ROCK 1 is a potential combinatorial drug target for BRAF mutant melanoma. Mol Syst Biol. 10:772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supek F, Bošnjak M, Škunca N, Šmuc T.. 2011. Revigo summarizes and visualizes long lists of gene ontology terms. PLoS One. 6:e21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, et al. 2019. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47:D607–D613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur MD, Salangsang F, Landman AS, Sellers WR, Pryer NK, et al. 2013. Reveals a strategy to forestall drug resistance. Nature. 494:251–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Collado AX, Nazarian R, Jazirehi AR.. 2017. Rescue of cell cycle progression in BRAFV600E inhibitor–resistant human melanoma by a chromatin modifier. Tumour Biol. 39:101042831772162. [DOI] [PubMed] [Google Scholar]
- Vanneste M, Feddersen CR, Varzavand A, Zhu EY, Foley T, et al. 2020. Functional genomic screening independently identifies CUL3 as a mediator of vemurafenib resistance via Src-Rac1 signaling axis. Front Oncol. 10:442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertheimer E, Gutierrez-Uzquiza A, Rosemblit C, Lopez-Haber C, Sosa MS, et al. 2012. Rac signaling in breast cancer: a tale of GEFs and GAPs. Cell Signal. 24:353–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker SR, Theurillat J-P, Van Allen E, Wagle N, Hsiao J, et al. 2013. A genome-scale RNA interference screen implicates NF1 loss in resistance to RAF inhibition. Cancer Discov. 3:350–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter J, Schwering M, Pelz O, Rauscher B, Zhan T, et al. 2017. CRISPRAnalyzeR: Interactive analysis, annotation and documentation of pooled CRISPR screens. BioRxiv. 1–11. doi:10.1101/109967. [Google Scholar]
- Yadav V, Zhang X, Liu J, Estrem S, Li S, et al. 2012. Reactivation of Mitogen-activated Protein Kinase (MAPK) pathway by FGF Receptor 3 (FGFR3)/Ras mediates resistance to vemurafenib in human B-RAF V600E mutant melanoma. J Biol Chem. 287:28087–28098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JSL, Yusa K.. 2019. Genome-wide CRISPR-Cas9 screening in mammalian cells. Methods. 164-165:29–35. [DOI] [PubMed] [Google Scholar]
- Zecena H, Tveit D, Wang Z, Farhat A, Panchal P, et al. 2018. Systems biology analysis of mitogen activated protein kinase inhibitor resistance in malignant melanoma. BMC Syst Biol. 12:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan Y, Dahabieh MS, Rajakumar A, Dobocan MC, M’Boutchou MN, et al. 2015. The role of eIF4E in response and acquired resistance to vemurafenib in melanoma. J Invest Dermatol. 135:1368–1376. [DOI] [PubMed] [Google Scholar]
- Zhang YE. 2009. Non-Smad pathways in TGF-β signaling. Cell Res. 19:128–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Supplementary Figure S1 depicts the cumulative distribution of the number of reads per sgRNA in an individual genome scale CRISPR screen. Supplementary Figure S2 compares the hits obtained from CRISPR-based screens for vemurafenib resistance. Supplementary Figure S3 illustrates the proposed mechanisms for vemurafenib resistance caused by deletion of MAPK signaling pathway related hits in the screen. Supplementary Table S1 lists the sequences of primers used for amplifying sgRNA sequences from the Brunello library and for NGS. Supplementary Table S2 contains the Read count file of the 76,441 sgRNA in the Brunello library targeting 19,114 genes. Supplementary Table S3 contains two tabs. The first tab shows the ranked P-values according to MAGeCK for all the 19,114 genes after 14 days of vemurafenib treatment. The second tab only shows the values for genes that have the ranked P-value less than 0.05. Supplementary Table S4 compares the top 33 hits with top 100 enriched genes obtained in previous screens with Gecko v1, Gecko v2, and Avana sgRNA libraries. Supplementary Table S5 contains the results of STRING analysis of top 33 genes that regulate vemurafenib resistance. Supplementary Table S6 contains the results from GO analysis of the top 33 genes that regulate vemurafenib resistance using the online tools DAVID and REVIGO. Supplemental material is available at figshare: https://doi.org/10.25387/g3.13413830.