Abstract
Various riboswitch classes are being discovered that precisely monitor the status of important biological processes, including metabolic pathway function, signaling for physiological adaptations, and responses to toxic agents. Biochemical components for some of these processes might make excellent targets for the development of novel antibacterial molecules, which can be broadly sought by using phenotypic drug discovery (PDD) methods. However, PDD data does not normally provide clues regarding the target for each hit compound. We have developed and validated a robust fluorescent reporter system based on a ZTP riboswitch that identifies numerous folate biosynthesis inhibitors with high sensitivity and precision. The utility of the riboswitch-based PDD strategy was evaluated using Escherichia coli bacteria by conducting a 128,310-compound high-throughput screen, which identified 78 sulfanilamide derivatives among the many initial hits. Similarly, representatives of other riboswitch classes could be employed to rapidly match antibacterial hits with the biological processes they target.
Keywords: AICAR, antibiotic, antifolate, phenotypic drug discovery, sulfonamide, sulfanilamide
Since the early 1980s, the number of novel antibacterial drugs approved for use in humans has steadily declined, reflecting the increasing scientific and commercial challenges facing those interested in pursuing antibiotic discovery and development (Gould and Bal, 2013; Ventola, 2015; Zheng et al., 2013). This reduced output for antibiotics development comes at a time when there is a critical need for new antibiotics against multi-drug resistant (MDR) bacteria, in particular for difficult to penetrate gram-negative bacterial cells (Estrada et al., 2016; Gould and Bal, 2013; Ventola, 2015). A major scientific challenge is the need to efficiently identify new chemical classes of agents that bind to well-validated cellular targets, or that operate on novel targets and pathways.
Many lead compound identification methods can be divided into phenotypic versus target-based drug discovery screening approaches (PDD and TDD, respectively) (Moffat et al., 2017). PDD screens employing model organisms gain the advantage of simultaneously pursuing multiple possible drug targets residing in their native cellular contexts. Unfortunately, when screening for inhibitors of bacterial growth, these methods usually provide little information about the specific target or mechanism of action (MOA) of the hit compound, or even the biological process that is affected. In contrast, TDD screens typically employ one known target protein or protein complex in vitro, and thus avoid the target identification problem. However, this approach does not directly resolve problems related to native cellular defenses, such as cell impermeability, drug-efflux proteins, and drug-metabolizing enzymes (Zheng et al., 2013). Although TDD methods have dominated screening campaigns in recent decades, empirical evidence suggests that PDD methods have been more effective in discovering novel drugs that were first in their class (Kotz, 2012; Swinney and Anthony, 2011).
To overcome this major drawback of PDD strategies, additional technologies have been developed to facilitate the identification of the targets of screening hits (Guiguemde et al., 2010; Kotz et al., 2012; Laggner, 2012; Payne et al., 2007). In the current report, we describe the use of a metabolite-sensing riboswitch to focus a PDD screening campaign on a specific biochemical process – folate biosynthesis and metabolism. The pathway for this universally distributed class of enzyme cofactors is one of the major targets of modern antibacterial agents (Bourne, 2014), which began with the landmark discovery and development of sulfonamide-based antibiotics (Bickel, 1988; Domagk, 1935). Therefore, the identification of different small molecules that disrupt this pathway could merit further improvement to produce new therapeutic agents.
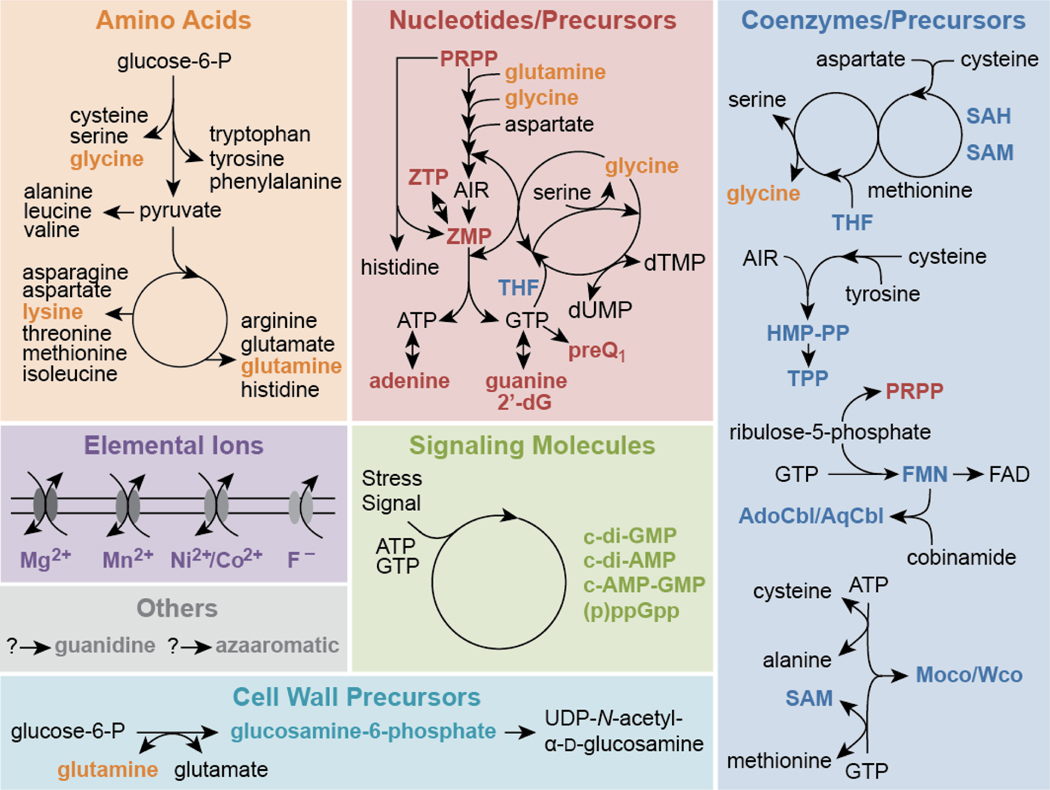
Most riboswitches are cis-acting, noncoding RNA domains embedded in particular bacterial messenger RNAs that selectively bind a natural ligand using a highly-structured ‘aptamer’ domain. Riboswitches regulate the expression of the adjoining coding region through either a transcriptional or translational ‘expression platform’ (Breaker, 2012; Sherwood and Henkin, 2016; Serganov and Nudler, 2013). More than 40 distinct riboswitch classes have been experimentally validated, and these selectively sense and respond to over 30 different ligands, such as coenzymes, nucleotide derivatives, signaling molecules, amino acids, cell wall precursors, and inorganic ions (McCown et al., 2017) (Figure 1). Many of these riboswitches are found in phylogenetically diverse bacteria, including many pathogens (Blount and Breaker, 2006; McCown et al., 2017), where they control the expression of numerous genes that are essential for viability and for virulence in certain species. These characteristics make some riboswitches well suited to serve directly as druggable targets (Blount and Breaker, 2006; Deigan and Ferré-D’Amaré, 2011; Lünse et al., 2014; Mehdizadeh Aghdam et al., 2016; Rekand and Brenk, 2017), or alternatively as sensors that report the status of key biological and biochemical responses of model bacterial species when perturbed by chemical agents (Nelson et al., 2015).
Figure 1. Biochemical Pathways and Signaling Processes Known to be Monitored by Riboswitches.
Natural ligands for riboswitches include amino acids (orange), nucleotides and their precursors or derivatives (red), coenzymes and their precursors or derivatives (dark blue), elemental ions (purple), signaling molecules (green), a cell wall precursor (light blue) and other ligands (gray). Reporter gene fusions with riboswitches for these ligands can be used to selectively monitor changes in the associated metabolic or signaling pathways. Given that many thousands of riboswitch classes are predicted to remain undiscovered, the diversity of ligands and processes monitored by natural RNA-based sensors is likely to be far greater than depicted here.
Most previous research efforts related to riboswitches and antibacterial agents involved the direct targeting of these RNAs with small molecules to disrupt normal gene regulation, thereby inhibiting bacterial cell growth. Efforts to intentionally manipulate gene expression by riboswitch-targeting agents have yielded novel compounds that appear to affect biosynthetic pathway regulation by purine (Kim et al., 2009; Mulhbacher et al., 2010) and flavin mononucleotide (FMN) (Blount et al., 2015; Howe et al., 2015) riboswitches. Similarly, the natural product and antibacterial agent roseoflavin has been shown to target FMN riboswitches (Lee et. al., 2009; Ott et al., 2009). Moreover, various other metabolite analogs (e.g., Blount et al., 2007; Sudarsan et al., 2005) and novel compounds (e.g., Chen et al. 2010; Connelly et al., 2019; Yan et al., 2018) have been found that are bound by other riboswitch classes.
In addition to these studies, we have also sought to employ riboswitches as tools to detect the effects of chemical agents that target the biological or biochemical pathways that are monitored by known riboswitch classes. For example, a genetic construct carrying a reporter gene regulated by a fluoride riboswitch was used to identify compounds that enhance the toxicity of fluoride to bacterial cells (Nelson et al., 2015). Specifically, a high-throughput screen (HTS) was conducted, using an Escherichia coli reporter strain grown in the presence of non-toxic levels (1 mM) of fluoride, to successfully identify compounds that promote fluoride uptake and/or retention. Preparation for this screen was expedited because fluoride riboswitches, like most other riboswitch classes, are modular, self-contained gene control systems that are easy to manipulate and implement. These same characteristics can be exploited to harness the function of numerous other riboswitch classes (McCown et al., 2017) to create reporter systems that monitor the effects of small molecules on biological pathways or processes in the context of PDD screens.
To demonstrate an approach for discovering antibacterial agents, herein we employ a construct based on the fusion of a reporter gene with a representative riboswitch that naturally senses the bacterial signaling molecule ZTP (5-aminoimidazole-4-carboxamide riboside 5ˊ-triphosphate) (Kim et al., 2015). ZTP is proposed to function as an ‘alarmone’ that triggers a stress response when cells are starved for various essential folate derivatives (Bochner and Ames, 1982). Specifically, the purine biosynthetic intermediate AICAR (also called ZMP) accumulates first in bacterial cells when the enzyme cofactor 10-formyltetrahydrofolate (10f-THF) is at a concentration insufficient to supply the formyl group required to close the six-membered ring of the purine base during the de novo biosynthesis of adenosine and guanosine nucleotides. ZTP is subsequently formed by the addition of two more phosphate groups to ZMP. Representatives of this riboswitch class respond to both the ZMP and ZTP forms of the alarmone (Kim et al., 2015; Jones and Ferré-D’Amaré, 2015; Trauch et al., 2015). Therefore, similar ZTP riboswitch reporter systems should permit the sensitive detection of folate distress caused by compounds examined in a cell-based HTS campaign. Indeed, several types of known antifolate compounds (trimethoprim, sulfathiazole, and methotrexate) were previously observed to activate a ZTP riboswitch reporter construct (Kim et al., 2015), whereas various other antibiotics that target biological processes different than folate biosynthesis and interconversion do not trigger a response indicative of an increase in ZTP concentration.
In the current study, we employ a ZTP riboswitch regulated reporter system in E. coli to conduct a HTS search of a small-molecule chemical library for compounds that enter a gram-negative bacterial species and disrupt purine biosynthesis, presumably via the inhibition of folate biosynthesis. The ZTP-responsive assay was first validated by testing a subset of known antifolate compounds, and then was used for screening a library of 128,310 small molecules from various compound collections. The final list of hits was formed after removing false-positive compounds that broadly trigger the riboswitch reporter construct, such as the removal of autofluorescent compounds and compounds that alter reporter gene expression even when the riboswitch carries an inactivating mutation. A final list of 80 hits were recovered, and all except two of the validated compounds carry a core structure that is characteristic of sulfonamide antibiotics that target the folate biosynthetic protein dihydropteroate synthase (DHPS). The remaining two compounds are known to inhibit folate biosynthesis using different chemical structures. These findings demonstrate the utility of employing riboswitch sensors to monitor the effects of compounds on certain key pathways in bacterial cells subjected to PDD screens.
RESULTS AND DISCUSSION
Design and Validation of a ZTP Riboswitch Reporter System for the Discovery of Folate Biosynthesis Inhibitors.
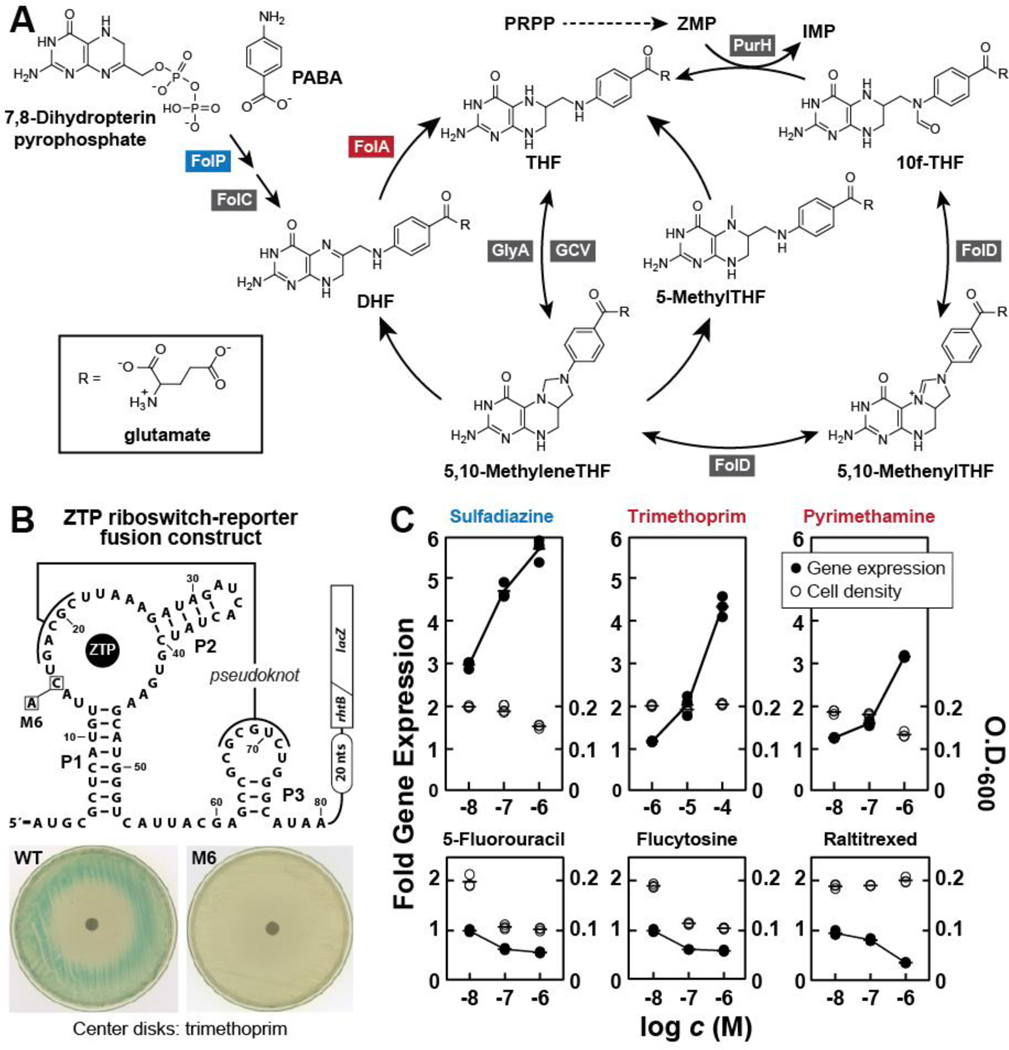
The folate biosynthesis and interconversion pathways in E. coli (Figure 2A) involve the action of several enzymes whose inhibition could ultimately lead to ZTP accumulation and riboswitch reporter activation. To synthesize inosine, bacteria need 10f-THF for a transformylase reaction with ZMP. Therefore, any disruption of 10f-THF production will cause ZMP to accumulate, and subsequently its triphosphorylated form, ZTP, will be produced (Bochner and Ames, 1982). A build-up of these ‘alarmone’ molecules triggers ZTP riboswitch mediated upregulation of certain essential folate metabolism and purine biosynthesis genes. This response is elicited by commonly used classes of antifolates that are known to inhibit folate biosynthesis or the interconversion of various natural folate derivatives that carry single carbon units.
Figure 2. The Folate Pathway is Inhibited by Antifolates that Trigger ZTP Riboswitch-Mediated Gene Expression.
(A) Schematic representation of the metabolites of folate biosynthesis and recycling in E. coli. Enzymes listed (boxed and named for their genes) are those whose inhibition might trigger ZTP riboswitch-mediated reporter gene activation by causing a reduction in 10f-THF concentration, and a subsequent increase in ZTP concentration. Note that FolA (red, dihydrofolate reductase) is inhibited by various antibiotics such as trimethoprim and methotrexate, and FolP (blue, dihydropteroate synthase) is inhibited by sulfonamide-based antibiotics such as sulfadiazine.
(B) The design and function of a ZTP riboswitch-reporter fusion construct. (Top) Sequence and secondary structure model for a ZTP riboswitch aptamer derived from the rhtB gene of the bacterial species Pectobacterium carotovorum, depicted fused to a β-galactosidase reporter gene from E. coli as adapted from the construct created previously (Kim et al., 2015). The gene control mechanism of this riboswitch is presumed to operate by regulating ribosome access to the ribosome binding site (Shine-Dalgarno sequence). A mutant version of the ribowitch carrying a single C15A change (M6) that is known (Kim et al., 2015) to prevent ZTP binding and thus cannot activate gene expression was also used in this study. (Bottom) Agar diffusion assays depicting the effects of the addition of 10 μL of a 10 mM solution of trimethoprim (TMP) on filter disks. Plates are uniformly innoculated with E. coli cells carrying the wild-type (WT) or mutant (M6) reporter construct in LB media containing x-gal.
(C) Plots of the relative levels of reporter gene expression (β-galactosidase activity levels, left y-axis, filled circles) and bacterial cell density (O.D.600, right y-axis, open circles) versus the logarithm of the concentration (c) of various test compounds (x-axis) when tested in M9 minimal media. Gene expression values are normalized to cells grown in culture media without test compound added. Top row depicts data gathered for known antifolates (sulfadiazine, trimethoprim, and pyrimethamine) expected to affect ZTP levels. The antifolate compound names are colored to reflect their enzyme targets depicted in A. Bottom row depicts similar data for other drug compounds (5-fluorouracil, flucytosine, raltitrexed) that are believed to target certain folate-dependent enzymes whose inhibition is not expected to affect ZTP levels.
The products of the genes folP (dihydropteroate synthase) and folA (dihydrofolate reductase, DHFR) are well-established targets for widely used antibiotics (Anderson and Wright, 2014), and the inhibition of these enzymes preclude the formation of 10f-THF. The inhibition of additional enzymes in the pathways leading to 10f-THF formation could also conceivably cause ZTP accumulation. Finally, it seems possible that compounds blocking the function of PurH (AICAR transformylase/IMP cyclohydrolase) could also cause ZTP to accumulate without disrupting 10f-THF production. Thus, there are several enzymes that could serve as targets for HTS compounds to trigger ZTP production in bacterial cells (Figure 2A).
Engineered ZTP riboswitch reporter systems can be used to indirectly evaluate changes in folate metabolism by monitoring the ZTP alarmone response in cells exposed to these antifolates. Specifically, the fusion of a ZTP riboswitch to the open reading frame of a reporter gene such as β-galactosidase (lacZ gene) yields a riboswitch reporter system that reports on changes in ZTP alarmone concentrations (Kim et al., 2015). To efficiently screen chemical libraries for small-molecule inhibitors of folate biosynthesis in the current study, we employed a pair of riboswitch-reporter fusion constructs based on a ZTP riboswitch from the bacterium Pectobacterium carotovorum (Kim et al., 2015). A wild-type (WT) reporter construct carries the unaltered riboswitch sequence grafted upstream of a lacZ (β-galactosidase) reporter gene (Figure 2B, top). This construct is expected to activate reporter gene expression when ZTP accumulates as host E. coli cells are exposed to a folate biosynthesis inhibitor. To identify false positives, we also employed a second version of this riboswitch-reporter construct, called M6, which carries a disruptive mutation in the conserved ZTP aptamer that prevents ZTP binding. This disabled construct provides a means to determine whether a hit compound triggers reporter gene expression exclusively in a ZTP riboswitch-dependent manner.
The utility of the riboswitch reporter constructs was established by conducting agar-diffusion assays on bacterial plates containing M9 minimal media prepared with 5-bromo-4-chloro-3-indolyl-D-galactopyranoside (X-gal), which generates a blue color when cleaved by the β-galactosidase enzyme that is encoded by the lacZ reporter gene. As expected, the addition of trimethoprim to the center filter disk of plates inoculated with E. coli cells carrying the WT riboswitch reporter construct yields a dark blue halo as the concentration of the drug decreases (Figure 2B, bottom left). Trimethoprim prevents E. coli growth in the area closest to the disk, whereas cells that are able to replicate under extreme folate distress exhibit the highest level of reporter gene expression. In contrast, cells carrying the M6 riboswitch reporter construct fail to exhibit a change in gene expression at the cell growth/inhibition boundary (Figure 2B, bottom right). Thus, reporter gene expression requires the WT sequence of the ZTP riboswitch aptamer to function in a ZTP-dependent manner.
Furthermore, we examined the performance of the WT reporter strain by testing several antifolate compounds known to inhibit either folate biosynthesis (e.g. sulfadiazine) or interconversion (e.g. trimethoprim, pyrimethamine). These compounds all trigger a concentration-dependent increase in reporter gene expression when quantified by measuring the level of fluorescence produced by β-galactosidase activity on 4-methylumbelliferyl-β-D-glucuronide (4-MUG) in liquid M9 media cultures (Figure 2C, top row). Thus, as expected, these known antifolate compounds all increase ZTP concentration in cells. Furthermore, the riboswitch reporter assay was able to detect the effects of these antifolate compounds when present at sub-lethal concentrations. This sensitivity should make riboswitch reporter systems effective tools for high-throughput screening where costs render impractical the testing of several concentrations, where low amounts of compound are available, or hits with only weak activity might be sought.
It is important to note that some well-known antifolate compounds are not detected by the riboswitch reporter strains based on E. coli. For example, cycloguanil did not induce reporter gene expression, even at the highest concentration tested (100 μM) (Supplementary File 1). It has previously been shown that cycloguanil binds to the E. coli DHFR in vitro (Srinivasan et al., 2015). However, we also did not observe any evidence of cell growth inhibition, and so it seems possible that this compound does not efficiently cross the E. coli cell membrane. If true, this demonstrates that the ZTP riboswitch reporter system only reveals compounds that both enter cells and inhibit key enzymes responsible for maintaining adequate 10f-THF concentrations.
Other types of compounds that inhibit enzymes known to use certain folate compounds as enzyme cofactors (5-fluorouracil, flucytosine, raltitrexed) (Anderson and Wright, 2014) also do not increase reporter gene expression under the concentrations tested (Figure 2C, bottom row). Therefore, as expected, the reporter construct is triggered predominantly by compounds that affect the concentration of 10f-THF, rather than by compounds that affect how folates are used by cells. One exception to this targeting expectation might be PurH (AICAR transformylase/ IMP cyclohydrolase), whose inhibition could also cause ZTP accumulation (Figure 2A). However, because the ZTP riboswitch does not recognize phosphate groups, compounds that might inhibit kinases or phosphatases that interconvert ZMP and ZTP would not be detected.
We also considered the possibility of encountering compounds that mimic the function of ZTP and directly activate gene expression by targeting the riboswitch. If the hit compound is bound by the riboswitch aptamer and activates reporter gene expression, then the output of the reporter construct will appear the same as for a compound that inhibits folate metabolism. Such riboswitch-targeting screens have been described previously (Blount et al., 2006; Mayer and Famulok, 2006; Hickey and Hammond, 2014; Howe et al., 2015; Kirchner et al. 2017). In addition, the M6 riboswitch variant would likely remain in the OFF state when exposed to such compounds, thereby confirming the hit. To distinguish hits that directly activate the reporter construct from those that affect folate metabolism, RNA-ligand binding assays such as in-line probing (Soukup and Breaker, 1999) could be performed to determine if the hit is directly bound by the riboswitch. However, no such compounds were identified for any of these alternative targets in our HTS, as further described below.
Discovery and Validation of Folate Inhibitors from a Small Molecule Library.
HTS assays were performed using the WT ZTP riboswitch reporter construct in E. coli cells grown in liquid M9 minimal media. These growth conditions are necessary to induce expression of the de novo folate and purine biosynthesis pathways (Kim et al., 2015). Under these conditions, it is expected that ZMP is actively being produced as a purine biosynthetic intermediate at the same time that folate biosynthesis is occurring. Thus, ZTP alarmone biosynthesis can be enhanced by compounds that target folate biosynthesis enzymes when cells need this pathway to be active. In a positive control assay included in each 384-well microtiter plate, 1 μM of trimethoprim was present in the culture, which yielded reporter gene expression values that were on average 3.9-fold higher than the value measured in the absence of a test compound. These control assays were reproducibly run on multiple plates over multiple weeks as the HTS campaign was pursued.
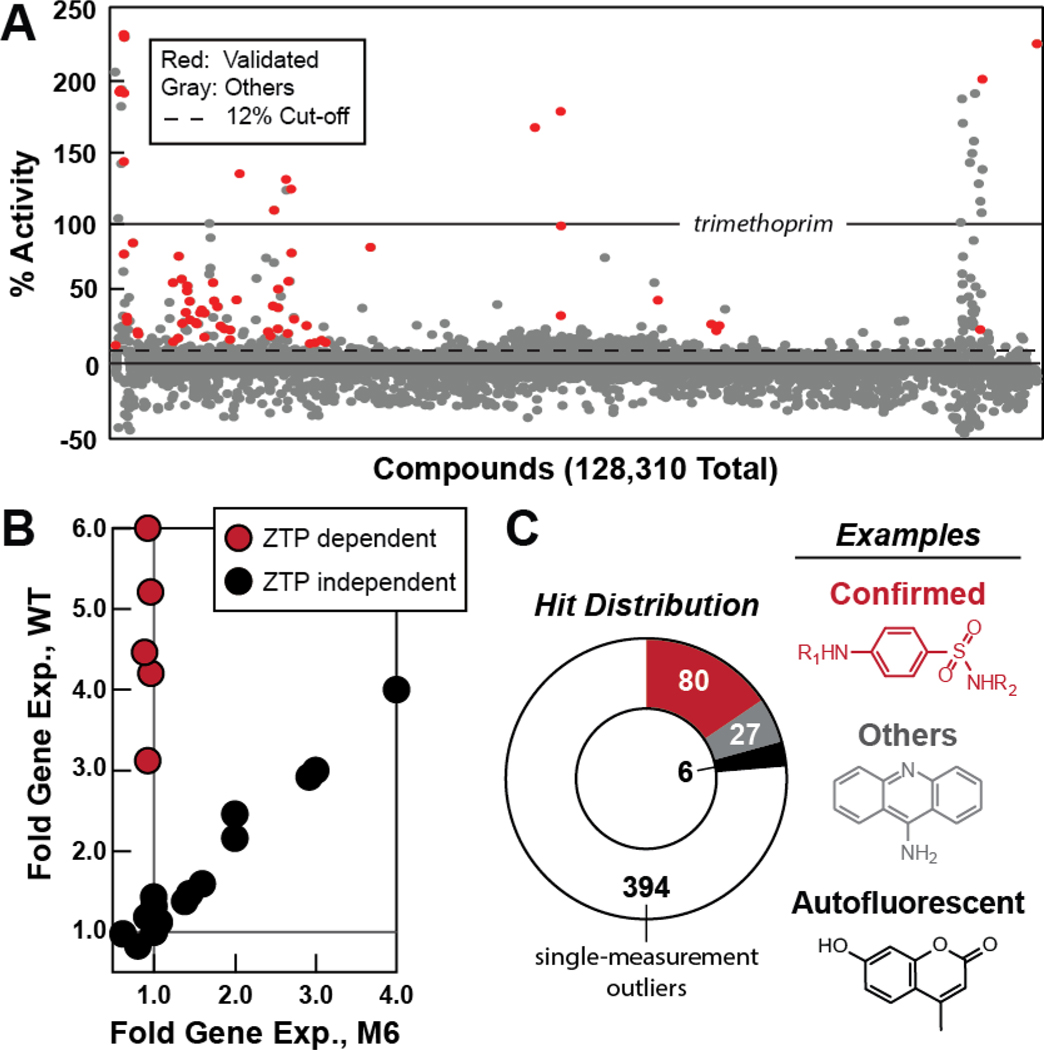
The HTS effort evaluated a total of 128,310 compounds from the small-molecule library maintained by the Yale Center for Molecular Discovery. The fluorescence signal was measured for each assay using 10 μM of test compound, and then normalized relative to the signal produced by the trimethoprim and no compound control assays on its corresponding plate. A percent activity value was subsequently determined for each compound (Figure 3A). Specifically, 0% represents the mean fluorescence of cells cultured with no added test compound, and 100% represents the mean fluorescence resulting from incubation in the presence of 1 μM trimethoprim. For each data collection run, Zˊ values ranged between 0.5 and 0.9 with an average of 0.8 (Zhang et. al., 1999), reflecting the robustness of the screening assay.
Figure 3. Validation of High-throughput Screening Hits that Trigger ZTP Dependent Flourescence Activity.
(A) Plot of the percent activity of each compound in a 128,310 small-molecule library (Yale Center for Molecular Discovery) compared to 1 μM trimethoprim (100%) for the reporter strain in M9 media. 0% activity is set as the fluoresence of cells cultured in the absense of test compound. The 12% cut-off represents the threshold for choosing hits for additional analyses. Red circles represent true-positive hits as validated through subsequent analyses.
(B) Representative data for hit compounds are plotted based on two parameters: (i) fold gene expression with the WT reporter strain versus, (ii) fold gene expression with the M6 reporter strain. Hits were considered validated if they exhibit high value for fold reporter gene expression with the WT strain, but exhibit substantially lower fold expression with the M6 strain. The data points on the diagonal distribution are false positives. Compounds were tested at 10 μM or 100 μM for each compound examined, as described in Supplementary File 1.
(C) The distribution of hits (see examples) after additional analyses reflects the observation that most hits rise above the 12% threshold due to autofluorescence (black), or as single-measurement outliers (white), or as hits that trigger reporter signal by a mechanism that does not require a functional ZTP riboswitch (gray). A total of 80 true-positive hits (red) can be classified as sulfanilamide derivatives (78), or are Dapsone and PAS.
Compounds that yielded an effect greater than 12% (3 standard deviations from the mean) were chosen for further analysis, which included 507 compounds (after removing certain types of gyrase inhibitors and β-lactams that trigger expression from this and other riboswitch reporters; see Concluding Remarks). This modest requirement permits the identification of weakly-active compounds that produce only small increases in ZTP, but whose function might be improved by chemical modification through future work. Also, the low requirement permits the identification of strongly-antibacterial compounds that produce large amounts of ZTP per cell, but that reduce the total number of cells in the culture. Encouragingly, among the initial hits were numerous compounds that carry a sulfonamide linkage in the context of a sulfanilamide group. This chemical structure is a distinguishing feature of ‘sulfa’ drugs that are known to target dihydropteroate synthase (FolP) enzymes (Achari, et al., 1997). Several hits with this chemical architecture exhibit % activity values near the 12% cut off (Figure 3A, Figure S1).
The 507 hits were further examined in triplicate to remove molecules that gave consistently low percent activity values, yielding 113 that exhibited an average of greater than 12% activity. These remaining compounds were tested for autofluorescence, wherein the same assay used for the initial HTS was conducted either without the reporter substrate 4-MUG, or without inoculation with the reporter strain. By using this method, 6 of the hits were found to be autofluorescent. Some of the remaining 107 compounds carry chemical substructures that resemble previously published pan-assay interference compounds (PAINS), which are known to cause false positives in previous HTS studies (Baell and Nissink, 2017; Thorne et al., 2010). Therefore, we employed another validation step using the M6 variant of the ZTP riboswitch reporter construct (Figure 2B). This mutant riboswitch should not be activated by compounds that increase ZTP concentration because the aptamer is defective. A characteristic of certain false-positive hits will be that they exhibit fluorescence readings that are the similar regardless of whether the WT or the M6 reporter strains are used. Upon examining all commercially available hits, we classified the compounds into two general categories: those that exhibit high reporter gene expression only with the WT reporter strain, and those that exhibit similar reporter gene expression with the WT and M6 strains (Figure 3B).
Of the 107 non-autofluorescent hits, 78 compounds include a sulfanilamide group (Figure 3C, Figure S1). All hits of this chemical class that were examined trigger expression only with the WT reporter strain, suggesting that they cause an increase in ZTP concentration most likely by inhibiting FolP. Furthermore, our reporter system rediscovered Dapsone and para-Aminosalicylic acid (PAS), which are more commonly used to inhibit the growth of malaria and mycobacterium species. The abundance of active hits in the compound collections screened is partly due to the common use of the sulfonamide linkage during chemical preparation, but is also due to the fact that antibiotics targeting folate biosynthesis are well represented in the collection of FDA-approved drugs in the Yale collection, as highlighted by the recovery of Dapsone and PAS.
All 27 of the remaining compounds are formed by chemical structures that are distinct from the large collection of sulfanilamide hits (Figure 3C). Of these, 6 were not further examined because they were not commercially available, were close derivatives of other compounds tested, or were already known as nucleic acid intercalators (Jalali and Rasaee, 2015; Lerman, 1963; Lown and Hanstock, 2012). An additional 10 compounds failed to cause an increase in reporter signal when examined in assays conducted manually (apart from the automated HTS process). We speculate that some of these hits, which originally produced only a weak signal during HTS with library compounds, might have chemical changes compared to the repurchased compound. The final 11 compounds produced a concentration-dependent, fluorescence signal increase in both the WT and M6 strain assays. Thus it is unlikely that these 11 compounds function by directly targeting the riboswitch, as determined by the fact that the M6 riboswitch reporter construct yields similar results. These false positives might be due to disruption of the normal function of transcription or translation machinery, specifically in the cases of DNA/RNA-like intercalators (Figure S2).
PAINS in small-molecule libraries can be a costly nuisance for HTS studies (Baell and Nissink, 2017; Payne et al., 2007; Thorne et al., 2010). Therefore, our simple method for quickly removing these compounds improves the utility of antibiotic screening via PDD assays. The mutant riboswitch construct could also have been used to filter out false positives caused by autofluorescence, or any other signal that is not produced via ZTP accumulation, which could further reduce the steps required to create the final list of functional hit compounds. This feature, together with other attributes of riboswitches discussed above, makes HTS with riboswitch-based reporters an attractive route for antibacterial agent discovery with PDD strategies.
Structure-Activity Relationships of Sulfanilamide Derivatives.
Although no novel pharmacophores that target the folate pathway were discovered by our HTS campaign, sulfanilamide antibiotics and various derivatives of these compounds were identified by the ZTP riboswitch reporter system. Distinct members of this well-validated antibiotics class, including those with only weak activity, can be identified and quickly analyzed by using the ZTP riboswitch reporter system. By maximizing the number of initial hit compounds, valuable structure-activity relationship (SAR) data can be collected. Chemical changes can then be made to improve potency or to address other pharmacologically desirable characteristics. For example, it is known that side-effects such as renal stones can be reduced by sulfanilamide modifications that increase solubility (Bjorkman and Phillips-Howard, 1991; Perazella, 1999). Sulfanilamides with various chemical modifications can be quickly evaluated with the WT and M6 reporter strains to identify modifications that retain antifolate activity, but that might allow improvements to be made to avoid side-effects or to overcome bacterial resistance.
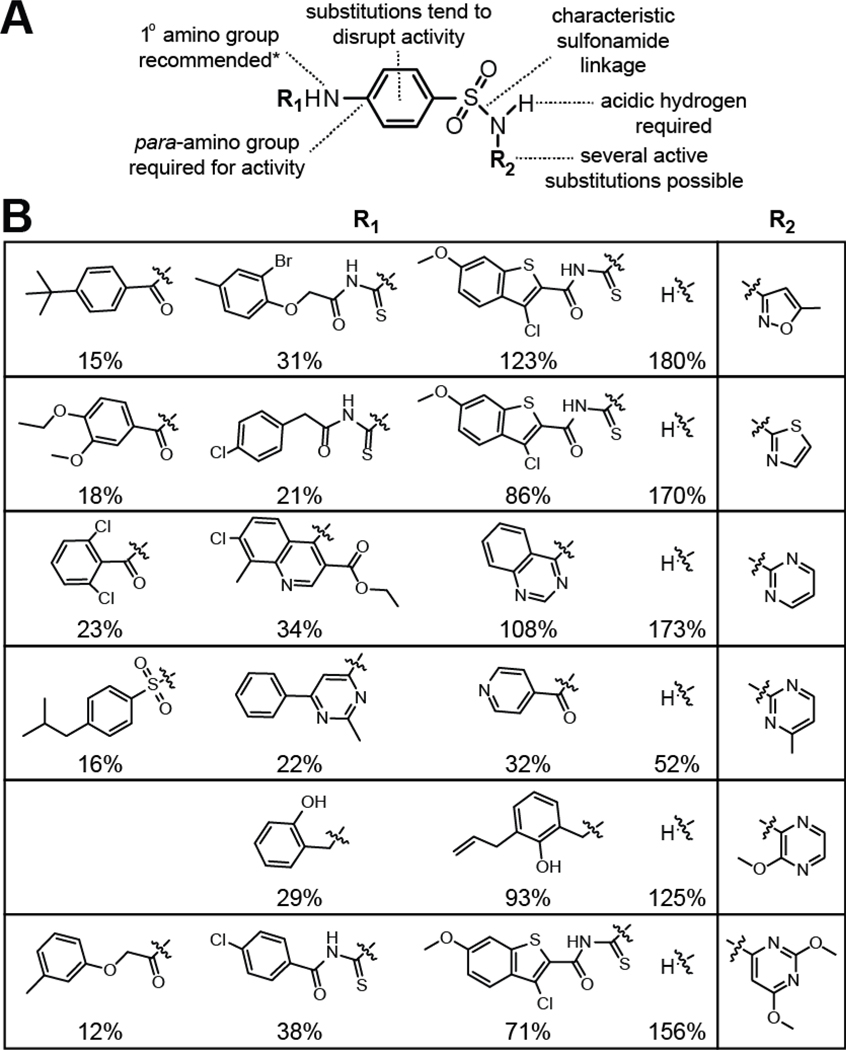
A total of 78 compounds identified in the current study (Figure 3C, Figure S1), including some known antibacterial agents, carry a core sulfanilamide structure and are likely to inhibit dihydropteroate synthase (FolP). The sulfanilamide core has two amine positions available for modification, and alterations at these sites are known to tune the effectiveness of sulfanilamide-based antibacterial agents (Hong et al. 1995) (Figure 4A). Most commonly, chemical groups have been added to the nitrogen atom of the sulfonamide linkage. Modifications at this position (R2) have been made to improve solubility, enhance membrane penetration and reduce toxicity in humans.
Figure 4. Summary of the Key Features of Sulfanilamide Antifolate Compounds.
(A) Schematic representation of the pharmacophore for sulfanilamide antibacterial agents (Hong et al., 1995; Maren, 1976). Note that the prefered R1 group is a single hydrogen, but this position can tolerate substitutions in the form of prodrugs, where removal of the R1 group to create a primary amine best mimics the PABA moiety to yield the active agent.
(B) SAR for representative sulfanilamide derivatives from the final list of validated HTS hits. Percentages reflect the average reporter gene expression values for various hits compared to trimethoprim (100%), and were established with the WT reporter strain using assays conducted in triplicate with 10 μM of each compound. The chemical moiety in the R2 column was held constant for the R1 moieties presented in each row.
Modifications on the PABA amine (R1) have been less-commonly exploited. Compounds with additions at this nitrogen have been exploited as prodrugs, wherein removal of the group is required to activate the agent. For example, the very first sulfanilamide derivative, Prontosil, functions as a prodrug that is ineffective in vitro because it needs to be metabolized into its active form in vivo (Bickel, 1988). From our data, it is apparent that large R1 moieties consistently reduce gene expression values in riboswitch reporter assays compared to the presence of a hydrogen (Figure 4B). However, some compounds with large R1 groups remain surprisingly active. Given the lack of space available in the FolP active site (Achari et al. 1997), these compounds must have the bulky R1 group removed by cells, have lost this group on storage, or (perhaps most unlikely) inhibit another enzyme in the folate pathway. Regardless, similar compounds have also emerged as hits in previous screens (Hong et al., 1995).
CONCLUDING REMARKS
Although the vast majority of compounds uncovered in the current HTS are sulfanilamide derivatives, this is most likely due to the relative abundance of this pharmacophore in the chemical library screened. The ZTP riboswitch reporter system also signaled the antifolate activity of Dapsone and PAS, thereby demonstrating the broad utility of a riboswitch that responds to the general perturbation of a metabolic pathway. The existing system should permit the rapid and efficient screening of larger and more diverse chemical libraries, and also of chemical libraries focused on a single pharmacophore to quickly evaluate the bioavailability and biochemical function of hit or lead compound derivatives.
It is important to note that some compounds in the library screened in the current study were so toxic to E. coli that little or no fluorescence signal could be measured. Among these antibacterial agents were compounds that carry a sulfonamide linkage, and therefore at least some of these are likely to kill bacteria by inhibiting the FolP enzyme. Therefore, without further action, some of the best hits would be lost in the first HTS pass. To capture these undetected hits, re-screening at a lower concentration should permit cell growth and ZTP riboswitch reporter signaling. Indeed, one notable member of the original compound library that completely inhibited cell growth in the HTS was trimethoprim. This known FolA enzyme inhibitor, like all other compounds from the library, was assayed at 10 μM. However, this concentration is lethal to E. coli, which is why we used only 1 μM trimethoprim in the control assays. Therefore, this FolA inhibitor from the original chemical library would have been detected had we conducted a second screening at a lower concentration for compounds that prevented cell growth in the original HTS.
A more direct folate monitoring system could be employed that makes use of known riboswitches for tetrahydrofolate. One widespread THF riboswitch class (Ames et al., 2010) is known to bind two THF molecules (Trausch et al., 2011) and suppress the expression of folate biosynthesis genes. A second, distinct riboswitch class has been discovered, which appears to bind only one THF molecule or a closely related natural derivative (Chen et al., 2019). These riboswitches could be used to more directly monitor folate biosynthesis, and thereby reveal compounds that inhibit genes involved in the synthesis of compounds such as DHF and THF. However, these riboswitches would likely fail to respond to compounds that disrupt the interconversion of the single carbon units carried by other natural folate derivatives. The ZTP riboswitch reporter system used in our study is expected to alert when an inhibitor of any process that disrupts 10f-THF is present. Regardless, it might be possible in the future to combine riboswitches that monitor different stages of the same biochemical pathway to define the specific step that is targeted by the hit compound. Additionally, reporter constructs based on other riboswitch classes could be used as controls to identify PAINS or that directly target specific riboswitches, which might not be identified by control constructs that carry only modest disruptive mutations.
In summary, our findings with the ZTP riboswitch reporter demonstrate that HTS on live bacterial cells can rapidly yield information both on the antibacterial function of compounds and on the pathways they target. As the number of known riboswitch classes and the ligands they sense expand with new discoveries, a far broader range of biological and biochemical processes can be monitored. Already, riboswitches exist for essential coenzymes like folates as described herein, major signaling molecules such as c-di-GMP (Lee et al., 2010; Sudarsan et al., 2008) and ppGpp (Sherlock et al., 2018), and other fundamental metabolites (McCown et al., 2017). Many bacterial species rely on these riboswitches to sense essential ligands and make crucial gene control decisions, and researchers can exploit these systems to monitor these same processes in the ongoing effort to identify novel compounds that disrupt bacterial cell growth.
METHOD DETAILS
Chemicals, DNA oligonucleotides, and bacterial strains
Chemical compounds were purchased from ChemBridge, ChemDiv, LabNetwork, and Sigma-Aldrich. DNA oligonucleotides and double stranded DNA sequences (Supplemental Table S1) were purchased from Sigma-Aldrich or Integrated DNA Technologies. E. coli BW25113 strain was obtained from the Coli Genetic Stock Center (CGSC) at Yale University.
Design of riboswitch reporter gene constructs
The WT and mutated ZTP riboswitch reporter constructs in pRS414 plasmids transformed into E. coli are the same as reported previously (Kim et al., 2015) with some modifications.
Agar diffusion assays
E. coli cultures carrying riboswitch reporter gene constructs were grown overnight ~16 h in defined minimal media (M9) liquid medium. Cultures were then spread on M9 agar plates containing X-gal (80 μg mL−1) and carbenicillin (100 μg mL−1). Autoclaved 6 mm diameter paper discs prepared from 0.35 mm thick pure cellulose chromatography paper (Fisher Scientific) were soaked with 10 μL of compound at specific concentrations and transferred to prepared agar plates. The plates were incubated overnight at 37°C prior to analysis.
Liquid-based β-galactosidase assays
Liquid-based β-galactosidase assays were performed as previously described (Nelson et al. 2015) with some modifications. In short, bacterial cell cultures were grown overnight (~16 h) in M9 with appropriate antibiotics at 37°C with shaking. The next day, 8 μL of overnight culture were then added to each well in Costar black 96-well clear-bottom plates containing 71 μL of fresh M9 and 1 μL of compound, Trimethoprim (TMP) or DMSO control. The plates were sealed with parafilm, covered with wet paper for humidity, and grown for ~18 hours at 37°C with shaking. Following incubation, the absorbance was taken at 595 nm using a Tecan Infinite M200 PRO microplate reader. Subsequently, 80 μL of Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4) and 40 μL of 1 mg mL−1 4-methylumbelliferyl-β-D-galactopyranoside (4-MUG) (dissolved in 50:50 v/v DMSO:deionized H2O) were added to each well. The mixture was mildly agitated for mixing and allowed to incubate at room temperature for 15 min. The reaction was stopped using 40 μL of 1 M Na2CO3. Excitation and emission values were measured at 360 nm and 460 nm, respectively, using a Tecan Infinite M200 PRO microplate reader. Fluorescence units were calculated as fluorescence intensity divided by the multiplication of total cell density (OD595) and the incubation time in minutes.
High-throughput screening was performed l largely as described above, with some modifications. Defined minimal media with appropriate antibiotic was added via combidrop in 10 μL volumes to each well of 384-well plates. To each well, 20 nl of a 10 mM solution of each compound tested was added via pintool, followed by the addition of 10 μL of a 1:5 dilution of an overnight culture of E. coli containing the ZTP reporter. The reporter cells with TMP or DMSO were included on each plate as controls. Assays testing for autofluorescence of compounds omitted cells or 4-MUG from the plate. Bacteria were grown in a humidified incubator for ~18 hours at 37°C with shaking. Reporter gene expression was analyzed by calculating the percent activity of a compounds fluorescence compared to the DMSO control set at 0% and the TMP control set at 100%.
Supplementary Material
SIGNIFICANCE.
There is a pressing need to identify new lead compounds and additional targets that can be pursued for the development of useful antibacterial therapeutics. Indeed, it is likely that novel antibiotics will forever need to be sought, given the inevitable evolutionary emergence of resistant strains. Riboswitches that monitor the status of essential biochemical pathways in bacteria can enhance phenotypic drug development screens both by identifying agents that obstruct normal cellular function, and by linking the agent to its target pathway. In the current report, we describe the use of a ZTP riboswitch reporter system to identify compounds that primarily target the folate biosynthesis pathway to uncover numerous sulfanilamide derivatives from a large chemical library. Moreover, there are riboswitches for more than 30 fundamental ligands, including other essential enzyme cofactors, nucleotide derivatives, signaling molecules, and amino acids. It is likely that thousands of additional riboswitch classes remain to be discovered, which would greatly expand the ability to monitor diverse biochemical processes, thereby aiding the search for compounds that interfere with fundamental bacterial metabolism and physiology.
ACKNOWLEDGMENTS
We thank Sheila Umlauf and her colleagues at the Yale Center for Molecular Discovery for implementing the HTS data collection process, and Alan Sutherland and his colleagues at L2 Diagnostics LLC for their helpful suggestions. We also thank the members of the Breaker Lab for helpful conversations. R.M.A. was supported by the National Science Foundation Graduate Research Fellowship Program (DGE1122492). K.R.P was supported in part by the Medical Scientist Training Program (NIH/NIGMS T32 GM007205). This work was also supported by an NIH grant to R.R.B. (GM022778, NIAID AI136794). R.R.B. is also supported by the Howard Hughes Medical Institute.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental material is available for this article.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Achari A, Somers DO, Champness JN, Bryant PK, Rosemond J, and Stammers DK (1997). Crystal structure of the anti-bacterial sulfonamide drug target dihydropteroate synthase. Nat Struct Biol. 4, 490–497. [DOI] [PubMed] [Google Scholar]
- Ames TD, Rodionov DA, Weinberg Z, Breaker RR (2010). A eubacterial riboswitch class that senses the coenzyme tetrahydrofolate. Chem. Biol 17, 681–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson AC, and Wright DL (2014). Antifolate agents: a patent review (2010–2013). Expert Opin. Ther. Pat 24, 687–697. [DOI] [PubMed] [Google Scholar]
- Baell JB, and Nissink JWM (2017). Seven Year Itch: Pan-Assay Interference Compounds (PAINS) in 2017 Utility and Limitations. ACS Chem. Biol 13, 36–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickel MH (1988). The development of sulfonamides (1932–1938) as a focal point in the history of chemotherapy. Gesnerus 45, 67. [PubMed] [Google Scholar]
- Bjorkman A and Phillips-Howard PA (1991). Adverse reactions to sulfa drugs: implications for malaria chemotherapy. Bull World Health Organ 69, 297–304. [PMC free article] [PubMed] [Google Scholar]
- Blount KF, Breaker RR (2006). Riboswitches as antibacterial drug targets. Nat. Biotechnol 24, 1558. [DOI] [PubMed] [Google Scholar]
- Blount KF, Puskarz I, Penchovsky R, Breaker R. (2006). Development and application of a high-throughput assay for glmS riboswitch activators. RNA Biol. 3, 77–81. [DOI] [PubMed] [Google Scholar]
- Blount KF, Wang JX, Lim J, Sudarsan N, Breaker RR (2007). Antibacterial lysine analogs that target lysine riboswitches. Nat. Chem. Biol 3, 44–49. [DOI] [PubMed] [Google Scholar]
- Blount KF, Megyola C, Plummer M, Osterman D, O’Connell T, Aristoff P, Quinn C, Chrusciel RA, Poel TJ, Schostarez HJ, et al. (2015). Novel riboswitch-binding flavin analog that protects mice against Clostridium difficile infection without inhibiting cecal flora. Antimicrob. Agents Chemother 59, 5736–5746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochner BR, Ames BN (1982). ZTP (5-amino 4-imidazole carboxamide riboside 50 -triphosphate): a proposed alarmone for 10-formyl-tetrahydrofolate deficiency. Cell 29, 929–937. [DOI] [PubMed] [Google Scholar]
- Bourne CR (2014). Utility of the biosynthetic folate pathway for targets in antimicrobial discovery. Antibiotics 3, 1–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breaker RR (2012). Riboswitches and the RNA world. Cold Spring Harb. Perspect. Biol 4, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Cressina E, Leeper FJ, Smith AG, Abell C. (2010). A fragment-based approach to identifying ligands for riboswitches. ACS Chem. Biol 5, 355–358. [DOI] [PubMed] [Google Scholar]
- Connelly CM, Numata T, Boer RE, Moon MH, Sinniah RS, Barchi JJ, Ferré-D’Amaré AR, Schneekloth JS Jr. (2019) Synthetic ligands for PreQ1 riboswitches provide structural and mechanistic insights into targeting RNA tertiary structure. Nat. Commun 10, 1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deigan KE, Ferré-D’Amaré AR (2011). Riboswitches: discovery of drugs that target bacterial gene-regulatory RNAs. Acc. Chem. Res 44, 1329–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domagk G. (1935). Ein beitrag zur chemotherapie der bakteriellen infektionen. Deutsche Medizinische Wochenschrift. 61, 250–253. [Google Scholar]
- Estrada A, Wright DL, and Anderson AC (2016). Antibacterial Antifolates: From Development through Resistance to the Next Generation. Cold Spring Harb. Perspect. Med 6, a028324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould IM, and Bal AM (2013). New antibiotic agents in the pipeline and how they can overcome microbial resistance. Virulence 4, 185–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiguemde WA, Shelat AA, Bouck D, Duffy S, Crowther GJ, Davis PH, Smithson DC, Connelly M, Clark J, Zhu F, Jiménez-Díaz MB (2010). Chemical genetics of Plasmodium falciparum. Nature 465, 311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickey SF, Hammond MC (2014). Structure-guided design of fluorescent S-adenosylmethionine analogs for a high-throughput screen to target SAM-I riboswitch RNAs. Chem. Biol 21, 345–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong YL, Hossler PA, Calhoun DH, and Meshnick SR (1995). Inhibition of recombinant Pneumocystis carinii dihydropteroate synthetase by sulfa drugs. Antimicrob. Agents Chemother 39, 1756–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe JA et al. (2015). Selective small-molecule inhibition of an RNA structural element. Nature 526, 672–677. [DOI] [PubMed] [Google Scholar]
- Jalali F, and Rasaee G. (2015). Electrochemical, spectroscopic, and theoretical stuies on the interaction between azathioprine and DNA. Int. J. Biol. Macromol 81, 427–434. [DOI] [PubMed] [Google Scholar]
- Jones CP, Ferré-D’Amaré AR (2015). Recognition of the bacterial alarmone ZMP through long-distance association of two RNA sub-domains. Nat. Struct. Mol. Biol 22, 679–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JN, Blount KF, Puskarz I, Lim J, Breaker RR (2009). Design and antimicrobial action of purine analogues that bind guanine riboswitches. ACS Chem. Biol 4, 919–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim PB, Nelson JW, Breaker RR (2015). An ancient riboswitch class in bacteria regulates purine biosynthesis and one-carbon metabolism. Mol. Cell 57, 317–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchner M, Schorpp K, Hadian K, Schneider S. (2017). An in vitro high-throughput screening for riboswitch ligands using a reverse reporter gene system. Sci. Rep 7, 7732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotz J. (2012). Phenotypic screening, take two. Science-Business eXchange 5, 380. [Google Scholar]
- Laggner C, Kokel D, Setola V, Tolia A, Lin H, Irwin JJ, Keiser MJ, Cheung CYJ, Minor DL Jr, Roth BL, and Peterson RT (2012). Chemical informatics and target identification in a zebrafish phenotypic screen. Nat. Chem. Biol 8, 144–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee ER, Baker JL, Weinberg Z, Sudarsan N, Breaker RR (2010). An allosteric self-splicing ribozyme triggered by a bacterial second messenger. Science 329, 845–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee ER, Blount KF, Breaker RR (2009). Roseoflavin is a natural antibacterial compound that binds FMN riboswitches and regulates gene expression. RNA Biol. 6, 187–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerman LS (1963). The structure of the DNA-acridine complex. Proc. Natl. Acad. Sci. USA 49, 94–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lown JW, Hanstock CC (1985). High Field 1H-NMR analysis of the 1:1 intercalation complex of the antitumor agent mitoxantrone and the DNA duplex [d(CpGpCpG)]2. J. Biomol. Struct. Dyn 2, 1097–106. [DOI] [PubMed] [Google Scholar]
- Lünse CE, Schüller A, Mayer G. (2014). The promise of riboswitches as potential antibacterial drug targets. Int. J. Med. Microbiol 304, 79–92. [DOI] [PubMed] [Google Scholar]
- Mayer G, Famulock M. (2006). High-throughput-compatible assay for glmS riboswitch metabolite dependence. Chembiochem 7, 602–604. [DOI] [PubMed] [Google Scholar]
- McCown PJ, Corbino KA, Stav S, Sherlock ME, Breaker RR (2017). Riboswitch diversity and distribution. RNA 23, 995–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehdizadeh Aghdam E, Hejazi MS, Barzegar A. (2016). Riboswitches: from living biosensors to novel targets of antibiotics. Gene 592, 244–259. [DOI] [PubMed] [Google Scholar]
- Moffat JG, Vincent F, Lee JA, Eder J, Prunotto M. (2017). Opportunities and challenges in phenotypic drug discovery: an industry perspective. Nat. Rev. Drug. Discov 16, 531–543. [DOI] [PubMed] [Google Scholar]
- Mulhbacher J, Brouillette E, Allard M, Fortier LC, Malouin F, Lafontaine DA (2010). Novel riboswitch ligands as selective inhibitors of guanine-related metabolic pathways. PLoS Pathog. 6, e1000865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson JW, Plummer MS, Blount KF, Ames TD, Breaker RR (2015). Small molecule fluoride toxicity agonists. Chem. Biol 22, 527–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott E, Stolz J, Lehmann M, Mack M 2009. The RFN riboswitch of Bacillus subtilis is a target for the antibiotic roseoflavin produced by Streptomyces davawensis. RNA Biol. 6, 276–280. [DOI] [PubMed] [Google Scholar]
- Payne DJ, Gwynn MN, Holmes DJ, Pompliano DL (2007). Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat. Rev. Drug Discov 6, 29. [DOI] [PubMed] [Google Scholar]
- Perazella MA (1999). Crystal-induced acute renal failure. Am. J. Med 106, 459–465. [DOI] [PubMed] [Google Scholar]
- Rekand IH, Brenk R. (2017). Ligand design for riboswitches, an emerging target class for novel antibiotics. Future Med. Chem 9, 1649–1663. [DOI] [PubMed] [Google Scholar]
- Serganov A, Nudler E. (2013). A decade of riboswitches. Cell 152, 17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherlock ME, Sudarsan N, Breaker RR (2018). Riboswitches for the alarmone ppGpp expand the collection of RNA-based signaling systems. Proc. Natl. Acad. Sci. USA 115, 6052–6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood AV, Henkin TM (2016). Riboswitch-mediated gene regulation: novel RNA architectures dictate gene expression responses. Annu. Rev. Microbiol 70, 361–374. [DOI] [PubMed] [Google Scholar]
- Soukup GA, Breaker RR (1999). Relationship between internucleotide linkage geometry and the stability of RNA. RNA 5, 1308–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan B, Tonddast-Navaei S, Skolnick J. (2015). Ligand binding studies, preliminary structure–activity relationship and detailed mechanistic characterization of 1-phenyl-6, 6-dimethyl-1, 3, 5-triazine-2, 4-diamine derivatives as inhibitors of Escherichia coli dihydrofolate reductase. Eur. J. Med. Chem 103, 600–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudarsan N, Cohen-Chalamish S, Nakamura S, Emilsson GM, Breaker RR (2005). Thiamine pyrophosphate riboswitches are targets for the antimicrobial compound pyrithiamine. Chem. Biol 12, 1325–1335. [DOI] [PubMed] [Google Scholar]
- Sudarsan N, Lee ER, Weinberg Z, Moy RH, Kim JN, Link KH, Breaker RR (2008). Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science 321, 411–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinney DC, and Anthony J. (2011). How were new medicines discovered?. Nat. Rev. Drug Discov 10, 507. [DOI] [PubMed] [Google Scholar]
- Maren TH (1976), Relations between structure and biological activity of sulfonamides. Annu. Rev. Pharmacol. Toxicol 16, 309–327. [DOI] [PubMed] [Google Scholar]
- Thorne N, Auld DS, Inglese J. (2010). Apparent activity in high-throughput screening: origins of compound-dependent assay interference. Curr. Opin. Chem. Biol 14, 315–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trauch JJ, Ceres P, Reyes FE, Batey RT (2011). The structure of a tetrahydrofolate-sensing riboswitch reveals two ligand binding sites in a single aptamer. Structure 19, 1413–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trausch JJ, Marcano-Velázquez JG, Matyjasik MM, Batey RT (2015). Metal ion-mediated nucleobase recognition by the ZTP riboswitch. Chem. Biol 22, 829–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventola CL (2015). The antibiotic resistance crisis Part 1: causes and threats. P. T 40, 277–83. [PMC free article] [PubMed] [Google Scholar]
- Chen X, Arachchilage GM, Breaker RR (2019). Biochemical validation of a second class of tetrahydrofolate riboswitches in bacteria. RNA. “in press” [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan LH, Le Roux A, Boyapelly K, Lamontagne AM, Archambault MA, Picard-Jean F, Lalonde-Seguin D, St-Pierre E, Najmanovich RJ, Fortier LC, et al. (2018). Purine analogs targeting the guanine riboswitch as potential antibiotics against Clostridioides difficile. Eur. J. Med. Chem 143, 755–768. [DOI] [PubMed] [Google Scholar]
- Zhang JH, Chung TD, Oldenburg KR (1999). A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen 4, 67–73. [DOI] [PubMed] [Google Scholar]
- Zheng W, Thorne N, McKew JC (2013). Phenotypic screens as a renewed approach for drug discovery. Drug Discov. Today 18, 1067–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.