Abstract
Background
Induction chemotherapy (IC) followed by concurrent chemoradiotherapy is the mainstay treatment for patients with locoregionally advanced nasopharyngeal carcinoma. However, some patients obtain little benefit and experience unnecessary toxicities from IC. We intended to develop a gene-expression signature that can identify beneficiaries of IC.
Methods
We screened chemosensitivity-related genes by comparing gene-expression profiles of patients with short-term tumor response or nonresponse to IC (n = 95) using microarray analysis. Chemosensitivity-related genes were quantified by digital expression profiling in a training cohort (n = 342) to obtain a gene signature. We then validated this gene signature in the clinical trial cohort (n = 187) and an external independent cohort (n = 240). Tests of statistical significance are 2-sided.
Results
We identified 43 chemosensitivity-related genes associated with the short-term tumor response to IC. In the training cohort, a 6-gene signature was developed that was highly accurate at predicting the short-term tumor response to IC (area under the curve [AUC] = 0.87, sensitivity = 87.5%, specificity = 75.6%). We further found that IC conferred failure-free survival benefits only in patients in the benefit group (hazard ratio [HR] = 0.54, 95% confidence interval [CI] = 0.34 to 0.87; P = .01) and not on those in the no-benefit group (HR = 1.25, 95% CI = 0.62 to 2.51; P = .53). In the clinical trial cohort, the 6-gene signature was also highly accurate at predicting the tumor response (AUC = 0.82, sensitivity = 87.5%, specificity = 71.8%) and indicated failure-free survival benefits. In the external independent cohort, similar results were observed.
Conclusions
The 6-gene signature can help select beneficiaries of IC and lay a foundation for a more individualized therapeutic strategy for locoregionally advanced nasopharyngeal carcinoma patients.
Nasopharyngeal carcinoma (NPC) is a unique head and neck cancer, which is highly prevalent in South China, Southeast Asia, and North Africa. Of the 130 000 newly diagnosed cases reported worldwide in 2018, more than 70% were diagnosed as locoregionally advanced NPC (LA-NPC) at initial presentation (1,2). Recently, several trials strongly supported the survival benefit of induction chemotherapy (IC) followed by concurrent chemoradiotherapy (CCRT), and it has become a new standard of care for LA-NPC (3-6). However, the short-term tumor response to IC is different among individuals, and a subset of patients with nonresponse to IC have poor survival and benefit little from IC (7,8). Conversely, IC brings increasing toxicities, treatment duration, and economic burden. Therefore, it is crucial to identify the beneficiaries of IC and thus avoid ineffective treatment.
Global gene-expression patterns can reflect individual biological features and guide personalized therapy in various tumors (9-11). Dysregulation of gene-expression results in tumor resistance to chemotherapeutic drugs and reflects tumor chemosensitivity of individuals (12-14). In NPC, several studies have reported that specific gene-expression profiles can be used for disease diagnosis, subtype classification, and prediction of prognosis (15-17). Nevertheless, no studies have been reported to effectively predict the efficacy of IC before treatment based on large-scale gene-expression profiling in NPC.
Therefore, we sought to develop and independently validate a gene-expression signature for forecasting the efficacy of IC in LA-NPC patients, which would help clinicians select beneficiaries of IC and provide individual advice for tailoring treatment decisions, paving the way toward biomarker-driven treatment strategies.
Materials and Methods
Clinical Specimens and Study Design
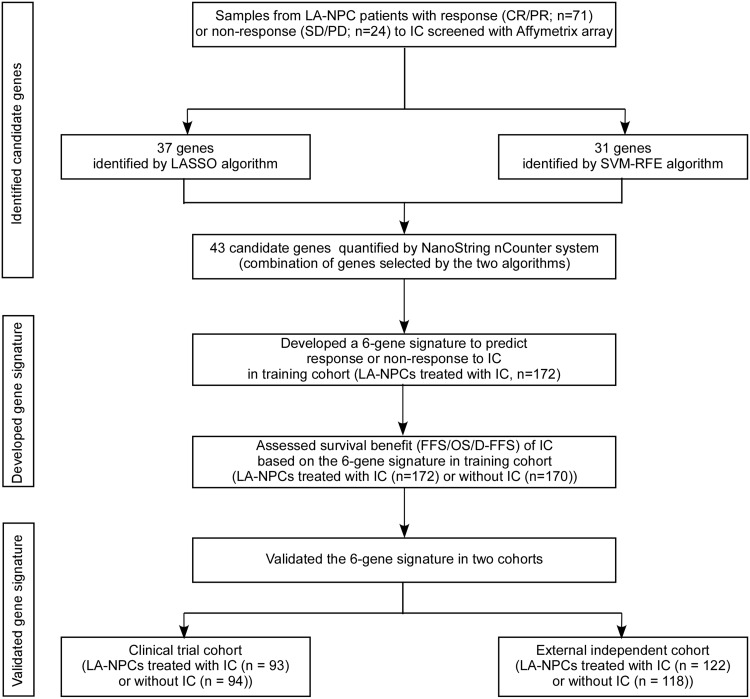
We obtained 769 pretreatment paraffin-embedded biopsy LA-NPC tissue samples. Two pathologists reevaluated all samples to confirm their eligibility with more than 70% tumor cells. In the training cohort, samples were obtained from 342 patients treated between August 21, 2009, and September 23, 2016, at the Sun Yat-sen University Cancer Center (SYSUCC; Guangzhou, China). In the validation phase, we used a phase III clinical trial (NCT01245959) as a validation cohort from which 187 samples were successfully obtained at SYSUCC between March 1, 2011, and August 22, 2013. An additional 240 samples obtained from the Affiliated Hospital of Guilin Medical College (Guilin, China) between May 17, 2010, and September 18, 2016, were used as an external independent cohort.
The institutional ethical review committees of 2 hospitals approved this study, which analyzed anonymous data, and waived the requirement for informed consent. Two radiologists separately assessed all magnetic resonance imaging and computed tomography scans for tumor staging and response evaluation, and any disagreements were settled by consensus. All patients were staged using the 7th edition of the American Joint Committee on Cancer staging system. All patients underwent platinum-based CCRT, and IC was administered to only 387 (50.3%) of 769 patients. For IC, 60 mg/m2 docetaxel, 60 mg/m2 cisplatin, and 600 mg/m2 fluorouracil (TPF regimen) were given once every 3 weeks for 2-4 cycles. For CCRT, 80-100 mg/m2 cisplatin was administered every 3 weeks for 2-3 cycles during intensity-modulated radiotherapy. We assessed the tumor response to IC using the response evaluation criteria in solid tumors (18). Patients with complete response (CR) or partial response (PR) were designated as response group, and those with stable disease (SD) or progressive disease (PD) were defined as nonresponse group (no patients with PD).
Based on the design of screening candidate genes using high-throughput assays in small subsets of patients and validating them using low-throughput methods in a large population (17), we first profiled gene expression in 95 patients with response (CR or PR) or nonresponse (SD) to IC using Affymetrix HTA 2.0 microarray (Supplementary Table 1, available online). The 95 patients included all patients with CR (n = 32) and SD (n = 24) from the 2 SYSUCC cohorts, and 39 PR patients matched with CR/SD patients by their clinical features (age, sex, T stage, and N stage). We then detected chemosensitivity-related genes with digital expression profiling (NanoString nCounter System, NanoString Technologies, Seattle, WA) and developed a gene signature for predicting tumor response in patients who received IC in the training cohort, which was validated in the clinical trial and external independent cohorts. Because short-term tumor response can indicate long-term survival benefit (7), we further explored whether the gene signature could discriminate patients’ long-term survival benefit in patients who received IC and those who did not.
RNA Extraction and Microarray Analysis
Detailed description of RNA extraction, microarray procedure, and analysis are provided in the Supplementary Methods (available online). The data have been deposited in National Center for Biotechnology Information Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo/) with the accession number GSE132112. Briefly, after normalization and batch adjustment, we used empirical Bayes (eBayes) statistics to identify statistically significantly differentially expressed probes (eBayes P < .05) between patients with response and nonresponse to IC and then used a custom Fisher’s exact test to screen out differentially expressed transcripts with probes that were statistically significantly enriched (Fisher P < .05), as previously described (17). Meanwhile, we also performed eBayes statistics to identify differentially expressed transcripts (empirical fold-change ≥1.5 and eBayes P < .05) using the median probe values for each transcript. At last, 185 transcripts (empirical fold-change ≥1.5, eBayes P < .05, and Fisher P < .05), representing 85 unique genes, were identified.
To narrow down the number of the 85 genes for further analysis, least absolute shrinkage and selector operation (LASSO) and support vector machine-recursive feature elimination (SVM-RFE) were performed as previously described (19-21). LASSO was performed with penalization parameter λ selected by a 10-fold cross-validation approach and a minimum mean cross-validated error rule. SVM-RFE with leave-one-out cross-validation was used to identify the number of best-ranked genes. Finally, we combined genes from either the LASSO or SVM-RFE algorithms and identified 43 chemosensitivity-related genes. Detailed procedures are provided in the Supplementary Methods (available online).
Gene-Expression Signature Discovery
We then detected the expression of the 43 genes and 5 housekeeping genes using the NanoString nCounter system (NanoString Technologies, Seattle, WA). To develop a gene signature in the training cohort, we performed bootstrap LASSO logistic regression and collapsed all genes obtained in each turn, then ranked the genes by their frequencies as previously described (17,22,23). We used the decision curve analysis method to identify an optimal set of 6 genes to construct a gene signature (24). Finally, we calculated a score based on the gene-expression values weighted by the coefficients from the logistic regression model. Details are provided in the Supplementary Methods (available online).
Statistical Analysis
The primary endpoint was short-term tumor response to IC. The secondary endpoints included failure-free survival (FFS), overall survival (OS), and distant failure-free survival (D-FFS). FFS was defined as the time from the start of treatment to documented distant metastasis, locoregional recurrence, or death from any cause, whichever occurred first. OS and D-FFS were calculated from the date of treatment to the date of death or documented distant metastasis, respectively. We used receiver operating characteristic (ROC) curve to determine the accuracy of the gene signature for predicting the tumor response and comparing it with other clinical parameters. We performed multivariable logistic regression analysis to test whether the gene signature is an independent predictor. We selected the optimal cutoff point using ROC analysis in the training cohort by maximizing the sum of sensitivity and specificity (25,26) and applied it directly in another 2 cohorts. Survival rates were calculated using Kaplan-Meier curves and compared with log-rank test. The hazard ratios (HRs) and 95% confidence intervals (CIs) were calculated by univariable Cox regression analysis. We did univariable and multivariable Cox regression analyses to test the interaction between the gene signature and the IC effect. We performed the statistical analysis with SPSS software (version 22.0) and R software (version 3.4.2). A 2-sided P value less than .05 was deemed statistically significant.
Results
Patient Characteristics
We collected 769 pretreatment, nondistant metastatic LA-NPC samples for this study (Figure 1). Table 1 shows patients’ baseline characteristics in the training (n = 342), clinical trial (n = 187), and external independent cohorts (n = 240). All patients underwent platinum-based CCRT, and TPF IC was administered to 172 (50.3%) of 342 patients in the training cohort, 93 (49.7%) of 187 patients in the clinical trial cohort, and 122 (50.8%) of 240 patients in the external independent cohort. The median follow-up was 62.2 months (interquartile range [IQR] = 48.8-77.5), 69.8 months (IQR = 60.3-77.2), and 41.8 months (IQR = 34.7-50.4) for patients in the training, clinical trial, and external independent cohorts, respectively.
Figure 1.
Study design. CR = complete response; D-FFS = distant failure-free survival; FFS = failure-free survival; IC = induction chemotherapy; LA-NPC = locoregionally advanced nasopharyngeal carcinoma; LASSO = least absolute shrinkage and selector operation; OS = overall survival; PD = progressive disease; PR = partial response; SD = stable disease; SVM-RFE = support vector machine-recursive feature elimination.
Table 1.
Clinical characteristics of patients in the training, clinical trial, and external independent cohortsa
| Training cohort (n = 342) |
Clinical trial cohort (n = 187) |
External independent cohort (n = 240) |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Characteristic | No. of patients | Benefit group, No. (%) | No-benefit group, No. (%) | No. of patients | Benefit group, No. (%) | No-benefit group, No. (%) | No. of patients | Benefit group, No. (%) | No-benefit group, No. (%) |
| Age, y | |||||||||
| <45 | 173 | 122 (50.2) | 51 (51.5) | 115 | 79 (60.8) | 36 (63.2) | 83 | 59 (34.7) | 24 (34.3) |
| ≥45 | 169 | 121 (49.8) | 48 (48.5) | 72 | 51 (39.2) | 21 (36.8) | 157 | 111 (65.3) | 46 (65.7) |
| Sex | |||||||||
| Male | 238 | 161 (66.3) | 77 (77.8) | 155 | 104 (80.0) | 51 (89.5) | 179 | 129 (75.9) | 50 (71.4) |
| Female | 104 | 82 (33.7) | 22 (22.2) | 32 | 26 (20.0) | 6 (10.5) | 61 | 41 (24.1) | 20 (28.6) |
| WHO pathological type | |||||||||
| Undifferentiated non-keratinizing | 337 | 240 (98.8) | 97 (98.0) | 187 | 130 (100.0) | 57 (100.0) | 237 | 167 (98.2) | 70 (100.0) |
| Differentiated non-keratinizing | 5 | 3 (1.2) | 2 (2.0) | 0 | 0 (0.0) | 0 (0.0) | 3 | 3 (1.8) | 0 (0.0) |
| T stage | |||||||||
| T1 | 4 | 3 (1.2) | 1 (1.0) | 5 | 3 (2.3) | 2 (3.5) | 10 | 4 (2.4) | 6(8.6) |
| T2 | 28 | 21 (8.6) | 7 (7.1) | 11 | 9 (6.9) | 2 (3.5) | 17 | 14 (8.2) | 3 (4.3) |
| T3 | 198 | 139 (57.2) | 59 (59.6) | 111 | 74 (56.9) | 37 (64.9) | 134 | 94 (55.3) | 40 (57.1) |
| T4 | 112 | 80 (32.9) | 32 (32.3) | 60 | 44 (33.8) | 16 (28.1) | 79 | 58 (34.1) | 21 (30.0) |
| N stage | |||||||||
| N0 | 19 | 14 (5.8) | 5 (5.1) | 0 | 0 (0.0) | 0 (0.0) | 5 | 3 (1.8) | 2 (2.9) |
| N1 | 144 | 96 (39.5) | 48 (48.5) | 100 | 65 (50.0) | 35 (61.4) | 94 | 67 (39.4) | 27 (38.6) |
| N2 | 137 | 101 (41.6) | 36 (36.4) | 72 | 52 (40.0) | 20 (35.1) | 108 | 77 (45.3) | 31 (44.3) |
| N3 | 42 | 32 (13.2) | 10 (10.1) | 15 | 13 (10.0) | 2 (3.5) | 33 | 23 (13.5) | 10 (14.3) |
| TNM stage | |||||||||
| III | 201 | 140 (57.6) | 61 (61.6) | 117 | 78 (60.0) | 39 (68.4) | 143 | 100 (58.8) | 43 (61.4) |
| IV | 141 | 103 (42.4) | 38 (38.4) | 70 | 52 (40.0) | 18 (31.6) | 97 | 70 (41.2) | 27 (38.6) |
| EBV DNA | |||||||||
| <2000 | 127 | 93 (38.3) | 34 (34.3) | 60 | 43 (33.1) | 17 (29.8) | — | — | — |
| ≥2000 | 204 | 145 (59.7) | 59 (59.6) | 114 | 80 (61.5) | 34 (59.6) | — | — | — |
| NA | 11 | 5 (2.1) | 6 (6.1) | 13 | 7 (5.4) | 6 (10.5) | — | — | — |
| Treatment | |||||||||
| IC + CCRT | 172 | 120 (49.4) | 52 (52.5) | 93 | 62 (47.7) | 31 (54.4) | 122 | 90 (52.9) | 32 (45.7) |
| CCRT | 170 | 123 (50.6) | 47 (47.5) | 94 | 68 (52.3) | 26 (45.6) | 118 | 80 (47.1) | 38 (54.3) |
CCRT = concurrent chemoradiotherapy; EBV = Epstein-Barr virus; IC = induction chemotherapy; TNM = tumor–node–metastasis.
Development of a 6-Gene Signature for Predicting Short-Term Tumor Response to IC
Based on gene-expression profiling analysis, we identified 23 024 differentially expressed probes between patients with response and nonresponse to IC using eBayes statistics and then screened out 6343 differentially expressed transcripts using Fisher’s exact test. Meanwhile, we identified 385 differentially expressed transcripts using eBayes statistics based on the median probe values for each transcript. We identified 185 statistically significantly differentially expressed transcripts (empirical fold-change ≥ 1.5, eBayes P < .05, and Fisher P < .05), representing a list of 85 unique genes. We then identified 37 and 31 candidate genes that were most strongly related to the efficacy of IC by LASSO and SVM-RFE algorithms, respectively. Finally, 43 genes were obtained by combining the genes selected by these 2 algorithms (Supplementary Figure 1 and Table 2, available online). We detected the expression of these 43 genes by the digital expression profiling and then performed LASSO logistic regression analysis and decision curve analysis to select an optimal set of 6 genes to construct a 6-gene signature for predicting the tumor response only in patients who received IC (n = 172) in the training cohort (Supplementary Tables 3 and 4 and Supplementary Figure 2, available online). We generated a formula to calculate a score based on the expression values of these 6 genes weighted by their logistic regression coefficients.
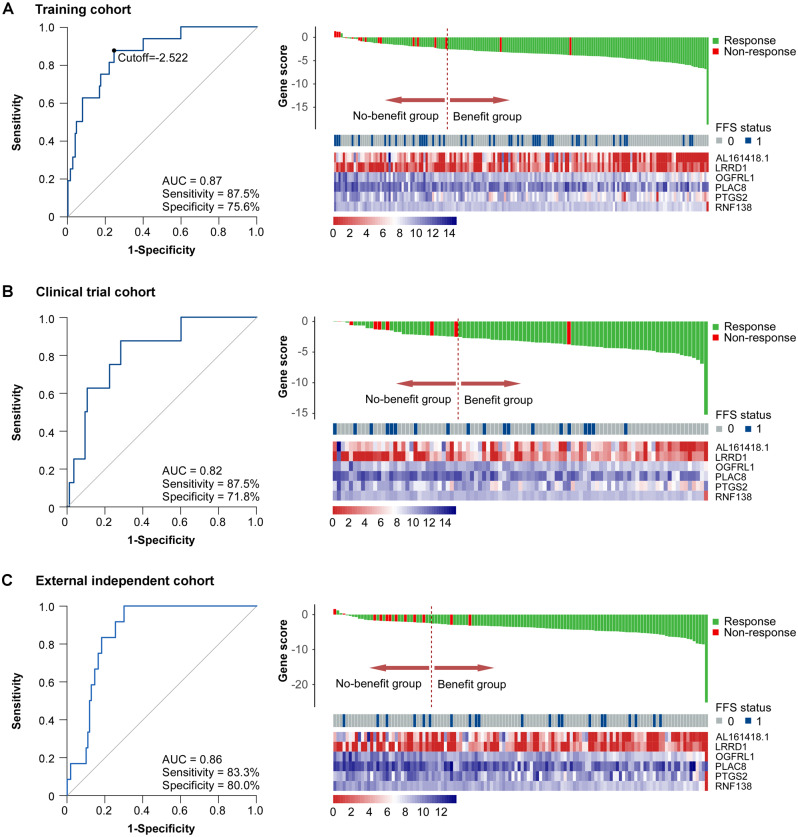
Gene score = -26.01361 + (0.36816 × expression of AL161418.1) - (0.64682 × expression of LRRD1) + (0.70213 × expression of OGFRL1) + (0.07571 × expression of PLAC8) + (0.24862 × expression of PTGS2) + (1.54186 × expression of RNF138).
Prediction of Short-Term Response to IC by the 6-Gene Signature and Clinical Parameters
We evaluated the performance of the 6-gene signature for the prediction of tumor response using ROC analysis in patients who received IC in the training cohort. The 6-gene signature had a statistically significantly higher AUC than each single gene (Supplementary Figure 3, available online; all P < .05). Moreover, the 6-gene signature showed an accuracy of 0.87 (95% CI = 0.78 to 0.94) for predicting the tumor response with a cutoff value of -2.522, with a sensitivity of 87.5% and a specificity of 75.6% (Figure 2A).
Figure 2.
Predictive performance of the 6-gene signature for the short-term response to induction chemotherapy (IC). Left panel: Receiver operating characteristic (ROC) curve analysis of the 6-gene signature for predicting the short-term tumor response to IC in patients who received IC in the training (A, n = 172), clinical trial (B, n = 93), and external independent (C, n = 122) cohorts. Right panel: Distributions of gene score, FFS status of patients, and the expression levels of the 6 genes from the signature: the dotted line represents the cutoff (-2.522) used to divide patients into the no-benefit and benefit groups in the training (A), clinical trial (B), and external independent (C) cohorts. AUC = area under ROC curve; FFS = failure-free survival.
To validate the robustness of our 6-gene signature, we performed a validation analysis in patients who received IC (n = 93) in a clinical trial cohort. Using the same formula and cutoff developed in the training cohort, the 6-gene signature showed an accuracy of 0.82 (95% CI = 0.68 to 0.94) with a sensitivity of 87.5% and a specificity of 71.8% (Figure 2B). To validate the performance of our 6-gene signature in different populations, we used patients who received IC (n = 122) from an external independent cohort and obtained a predictive accuracy of 0.86 (95% CI = 0.79 to 0.93) with a sensitivity of 83.3% and a specificity of 80.0% (Figure 2C).
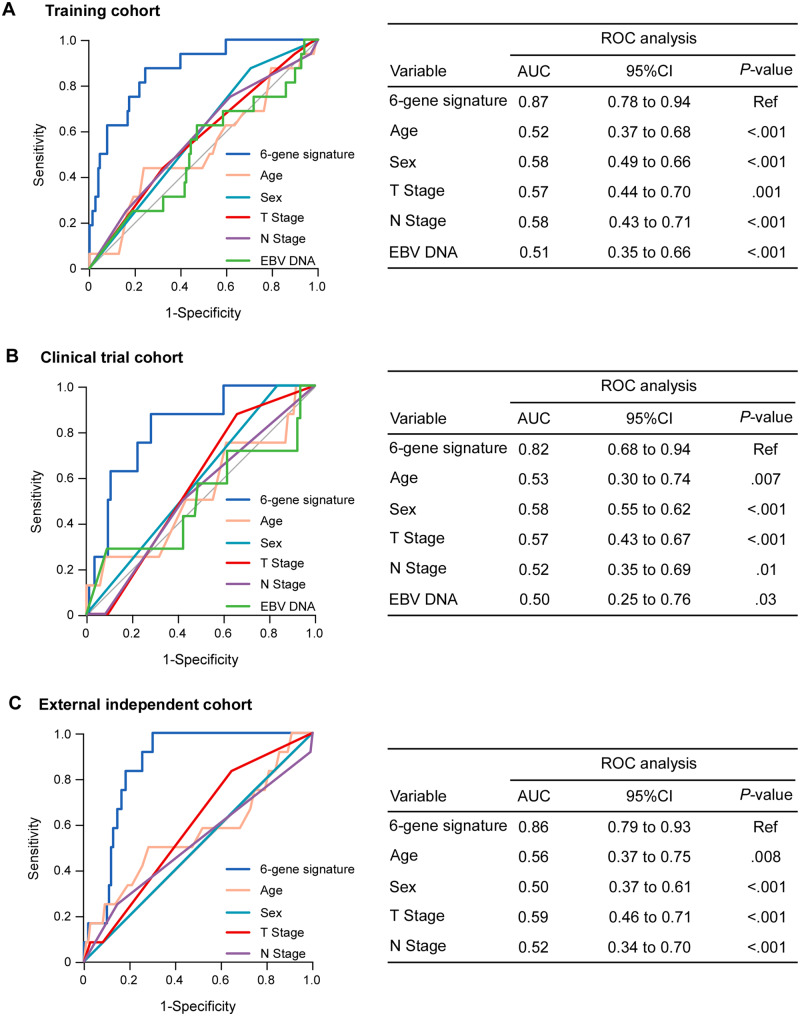
We then compared the performance of the 6-gene signature with that of other clinical parameters for the prediction of the tumor response. The 6-gene signature had a statistically significantly superior predictive capability compared with the other clinical variates in the training cohort (Figure 3A; all P < .05), which was an independent predictor for tumor response according to multivariable analysis (odds ratio [OR] = 29.69, 95% CI = 6.11 to 144.23; P < .001; Supplementary Table 5, available online). Similar results were observed in the clinical trial and external independent cohorts (Figure 3, B and C; all P < .05; Supplementary Table 5, available online).
Figure 3.
Prediction of short-term response to induction chemotherapy (IC) by the 6-gene signature and clinical variables in the training and 2 validation cohorts. Receiver operating characteristic (ROC) curve analysis of the 6-gene signature and clinical variables (age, sex, T stage, N stage, EBV DNA) for the prediction of short-term tumor response to IC in patients who received IC in the training (A, n = 172), clinical trial (B, n = 93), and external independent (C, n = 122) cohorts. The 95% confidence interval of the AUC and P value were estimated using the bootstrap method. AUC = area under ROC curve; CI = confidence interval; EBV = Epstein-Barr virus.
Association of the 6-Gene Signature with Long-Term Survival
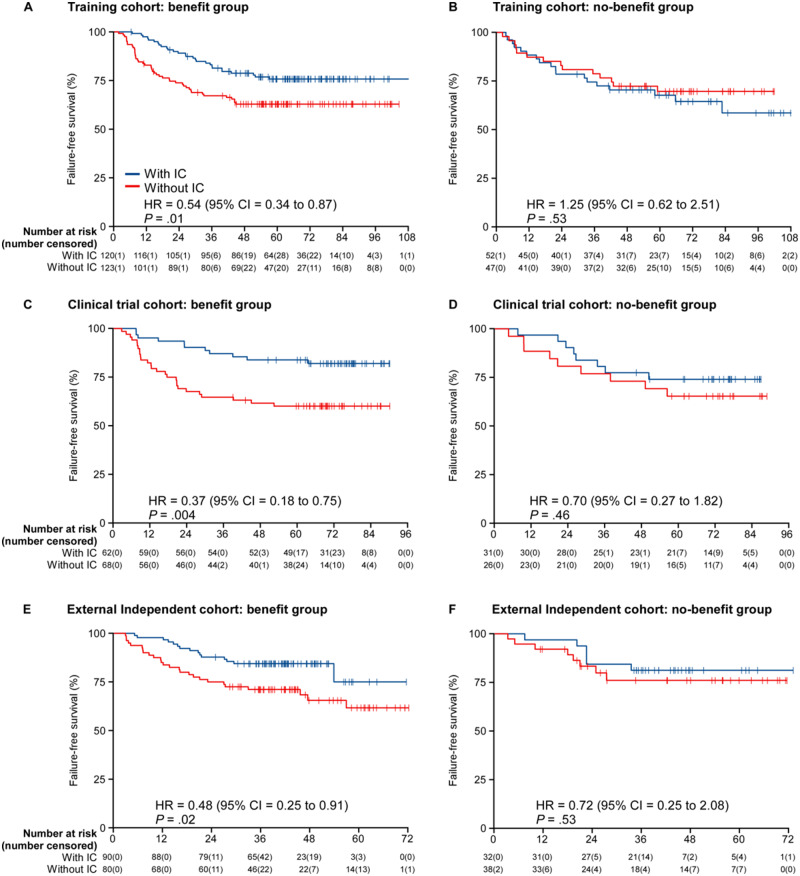
Because short-term tumor response can predict long-term survival benefit, we analyzed whether the 6-gene signature could discriminate the survival benefit of IC. All patients in the training cohort, including those who received IC and those who did not, were divided into the clinical benefit group and no-benefit group with the above cutoff (-2.522) determined by ROC analysis. Survival analysis demonstrated that patients whose gene signature predicted clinical benefit segregated IC from non-IC patients (FFS: HR = 0.54, 95% CI = 0.34 to 0.87; P = .01; OS: HR = 0.43, 95% CI = 0.24 to 0.79; P = .005; D-FFS: HR = 0.49, 95% CI = 0.28 to 0.85; P = .01), but this demonstration of IC benefit was not seen in patients whose signature predicted poor clinical benefit (FFS: HR = 1.25, 95% CI = 0.62 to 2.51; P = .53; OS: HR = 0.70, 95% CI = 0.26 to 1.89; P = .48; D-FFS: HR = 0.85, 95% CI = 0.38 to 1.93; P = .70; Figure 4, A and B; Supplementary Figures 4 and 5, available online).
Figure 4.
Kaplan-Meier curves of failure-free survival according to treatment with or without induction chemotherapy (IC) in the benefit and no-benefit groups. A) Benefit group of the training cohort (n = 243); B) no-benefit group of the training cohort (n = 99); C) benefit group of the clinical trial cohort (n = 130); D) no-benefit group of the clinical trial cohort (n = 57); E) benefit group of the external independent cohort (n = 170); F) no-benefit group of the external independent cohort (n = 70). We calculated P values with the 2-sided unadjusted log-rank test and hazard ratios (HRs) with univariable Cox regression analysis. Data in each x axis are number at risk (number censored). CI = confidence interval; HR = hazard ratio.
We then validated the discriminative performance of the 6-gene signature for the survival benefit of IC in another 2 cohorts. In the clinical trial cohort, patients in the benefit group benefited from IC, with an improved FFS (HR = 0.37, 95% CI = 0.18 to 0.75; P = .004), OS (HR = 0.37, 95% CI = 0.17 to 0.85; P = .01), and D-FFS (HR = 0.41, 95% CI = 0.18 to 0.93; P = .03), whereas patients in the no-benefit group did not benefit from IC (FFS: HR = 0.70, 95% CI = 0.27 to 1.82; P = .46; OS: HR = 1.00, 95% CI = 0.27 to 3.74; P = .99; D-FFS: HR = 0.62, 95% CI = 0.14 to 2.76; P = .53; Figure 4, C and D; Supplementary Figures 4 and 5, available online). Similar results (benefit group: FFS: HR = 0.48, 95% CI = 0.25 to 0.91; P = .02; OS: HR = 0.43, 95% CI = 0.21 to 0.88; P = .02; D-FFS: HR = 0.43, 95% CI = 0.21 to 0.90; P = .02; no-benefit group: FFS: HR = 0.72, 95% CI = 0.25 to 2.08; P = .53; OS: HR = 0.69, 95% CI = 0.19 to 2.46; P = .57; D-FFS: HR = 1.25, 95% CI = 0.33 to 4.66; P = .75) were obtained in the external independent cohort (Figure 4, E and F; Supplementary Figures 4 and 5, available online).
We further pooled all patients to test the interaction between the 6-gene signature and the IC treatment. Univariable analysis showed that the 6-gene signature had a statistically significant interaction with IC treatment (Pinteraction = .02). After multivariable adjustment by clinical variables, the 6-gene signature remained independently associated with IC treatment (Pinteraction = .01).
Discussion
In this multicenter cohort study, we identified a 6-gene signature with high predictive performance for forecasting the efficacy of IC in LA-NPC patients. It could aid clinicians to select IC beneficiaries and lay a foundation for a more individualized therapeutic strategy for LA-NPC patients.
Currently, IC plus CCRT is recommended for LA-NPC patients, because it can eradicate micrometastasis early and is well tolerated by patients. Clinical trials indicate that TPF IC can statistically significantly improve survival (3-5,27-29). However, a clinical trial from our group shows that IC only yields an absolute survival benefit of 8% at the expense of increasing toxicities, and a subset of patients do not benefit from IC (3,4). Moreover, patients with detectable plasma Epstein-Barr virus DNA after IC have inferior survival (30). Additionally, the tumor response differs among individuals, and patients with nonresponse to IC have poor survival and benefit little from IC (7,8). However, both the plasma Epstein-Barr virus DNA and tumor response cannot predict IC efficacy and guide therapeutic choices before treatment. Thus, it is urgent to identify novel pretreatment biomarkers to tailor therapy regimens and avoid ineffective treatments.
The prediction of IC efficacy is the key determinant for appropriate treatment decisions in LA-NPC patients. Multigene biomarkers for predicting the efficacy of chemotherapy have been reported in many cancers (12-14). In this study, we developed a 6-gene signature that could accurately differentiate patients with nonresponse to IC from those with response, whereas other widely used clinical variables could not. These results indicated that the 6-gene signature could reflect the biological characteristics of LA-NPC and provide information different from the conventional clinical characteristics, aiding clinicians to identify beneficiaries of IC before treatment and thus avoid unnecessary ineffective IC therapy.
Moreover, short-term tumor response can indicate long-term survival benefit (7,8). Here, we found that patients in the benefit group benefited from IC, whereas those in the no-benefit group did not. Long-term survival is influenced by many other factors, such as tumor response to radiotherapy and CCRT. Indeed, there is a subset of patients with poor response to IC who have good clinical outcomes, and there are patients with good response to IC who nevertheless have poor clinical outcome. Therefore, we recommend that patients with the benefit gene signature receive IC. For patients with the no-benefit gene signature, there was no clinical benefit to IC. Our 6-gene signature could screen out these IC nonresponse patients to directly receive effectual CCRT, avoiding unnecessary IC-related toxicities, expense, and a prolonged waiting period before CCRT. In addition, in patients with the benefit gene signature, TPF IC yielded an absolute overall survival benefit of 13%, 19%, and 22% in the training, clinical trial, and external validation cohort, respectively. However, TPF IC only yielded an overall benefit of 8% in our randomised controlled trial. Thus, whereas in the original TPF trial, 12 patients had to receive TPF IC for 1 to obtain clinical benefit, using our gene signature as a selector, the number to treat for one to benefit would be reduced to 5 patients.
Gene dysregulation is a hallmark of cancer and plays important roles in tumorigenesis, aggressiveness, and therapeutic resistance. Some genes in our signature have been reported to be associated with resistance to chemotherapeutic drugs involved in the TPF regimen. PTGS2, also known as COX-2, is widely reported to be associated with the chemoresistance to multiple drugs. In NPC, PTGS2 promotes chemoresistance to 5-fluorouracil by recruiting mitochondrial translocation of p53, increasing the activity of Drp1 and inducing mitochondrial fission (31,32). PLAC8 can regulate the expression of inflammation-associated genes, leading to the malignant progression and cisplatin resistance (33,34). RNF138, a ubiquitin E3 ligase, which is recruited to DNA damage sites, mediates Ku80 ubiquitylation and promotes homologous recombination, thereby contributing to cisplatin resistance (35,36). However, because of limited research on OGFRL1, LRRD1, and AL161418.1, the functions and mechanisms of these genes remain to be elucidated. These findings support that our 6-gene classifier can effectively reflect chemosensitivity and guide individual IC treatment for LA-NPC patients.
To our knowledge, this is the first large-scale study to investigate global gene-expression profiles and establish a signature for predicting IC efficacy. Our results demonstrated that the 6-gene signature could effectively predict the efficacy of IC, and it had been well validated in a clinical trial cohort and an external independent cohort. The 6-gene signature can aid clinicians in identifying IC beneficiaries and tailoring therapeutic regimens according to individual tumor biology. It should be noted that this signature is specific for TPF regimen, and its predictive value in other regimens such as gemcitabine-cisplatin requires further investigation. We also acknowledge that this signature needs to be confirmed in large-scale, multinational prospective studies before it can be used in the clinics.
Funding
This work was supported by grants from the Key-Area Research and Development Program of Guangdong Province (2019B020230002), the National Natural Science Foundation of China (81930072; 81922057), the Natural Science Foundation of Guangdong Province (2017A030312003; 2018B030306045; 2017A030310264), the Guangdong Special Support Program (2017TQ04R754); the Health and Medical Collaborative Innovation Project of Guangzhou City, China (201803040003); the Innovation Team Development Plan of the Ministry of Education (IRT_17R110); and the Overseas Expertise Introduction Project for Discipline Innovation (111 Project, B14035).
Notes
Role of the funder: The funding sponsors had no role in the study design; data collection, analysis, and interpretation; writing of the report; or the decision to submit this manuscript for publication. The corresponding authors had full access to the data and had final responsibility for the decision to submit this manuscript for publication.
Disclosures: The authors have no actual or potential conflicts of interest to declare.
Author contributions: YL: Data collection; Data analysis and interpretation; Writing of the manuscript; Statistical analysis. Y-QL: Data collection; Data analysis and interpretation; Revision of the manuscript. WJ: Data collection; Revision of the manuscript. X-HH: Data collection; Data analysis and interpretation; Revision of the manuscript. W-XG: Data analysis and interpretation; Data collection; Revision of the manuscript; Statistical analysis. YZ: Data collection; Revision of the manuscript. W-HH: Data collection; Revision of the manuscript. Y-QW: Data collection; Revision of the manuscript. Y-LL: Data collection; Revision of the manuscript. J-YL: Data collection; Revision of the manuscript. WCSC: Data analysis and interpretation; Revision of the manuscript. J-PY: Revision of the manuscript. JZ: Data collection; Revision of the manuscript. J-WC: Data collection. L-ZL: Data collection; Revision of the manuscript. LL: Data collection; Revision of the manuscript. LC: Data collection; Revision of the manuscript. F-YX: Data collection; Revision of the manuscript. W-FL: Data collection; Revision of the manuscript. Y-PM: Data collection; Revision of the manuscript. XL: Data collection; Revision of the manuscript. Y-PC: Data collection; Revision of the manuscript. L-LT: Study design; Data collection; Revision of the manuscript. YS: Data analysis and interpretation; Study design; Writing of the manuscript. NL: Study design; Data collection; Data analysis and interpretation; Writing of the manuscript; Statistical analysis. JM: Study design; Data analysis and interpretation; Writing of the manuscript.
Acknowledgments: We thank Hanqi Yin and Ganlin Xu (Guangzhou Longsee-MicroV Biotechnology Corporation, China) for statistical consultation.
Data availability statement
The microarray data have been deposited in www.ncbi.nlm.nih.gov/geo/ with the accession number GSE132112, and the key raw data have been deposited at Research Data Deposit public platform (www.researchdata.org.cn), with an approval number of RDDB2020000883.
Supplementary Material
References
- 1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A.. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. [DOI] [PubMed] [Google Scholar]
- 2. Ren Y, Qiu H, Yuan Y, et al. Evaluation of 7th edition of AJCC staging system for nasopharyngeal carcinoma. J Cancer. 2017;8(9):1665–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sun Y, Li W, Chen N, et al. Induction chemotherapy plus concurrent chemoradiotherapy versus concurrent chemoradiotherapy alone in locoregionally advanced nasopharyngeal carcinoma: a phase 3, multicentre, randomised controlled trial. Lancet Oncol. 2016;17(11):1509–1520. [DOI] [PubMed] [Google Scholar]
- 4. Li WF, Chen NY, Zhang N, et al. Concurrent chemotherapy with/without induction chemotherapy in locoregionally advanced nasopharyngeal carcinoma: long-term results of phase 3 randomized controlled trial. Int J Cancer. 2019;145(1):295–305. [DOI] [PubMed] [Google Scholar]
- 5. Frikha M, Auperin A, Tao Y, et al. A randomized trial of induction docetaxel-cisplatin-5FU followed by concomitant cisplatin-RT versus concomitant cisplatin-RT in nasopharyngeal carcinoma (GORTEC 2006-02). Ann Oncol. 2018;29(3):731–736. [DOI] [PubMed] [Google Scholar]
- 6. Zhang Y, Chen L, Hu GQ, et al. Gemcitabine and cisplatin induction chemotherapy in nasopharyngeal carcinoma. N Engl J Med. 2019;381(12):1124–1135. [DOI] [PubMed] [Google Scholar]
- 7. Peng H, Chen L, Li W, et al. Tumor response to neoadjuvant chemotherapy predicts long-term survival outcomes in patients with locoregionally advanced nasopharyngeal carcinoma: a secondary analysis of a randomized phase 3 clinical trial. Cancer. 2017;123(9):1643–1652. [DOI] [PubMed] [Google Scholar]
- 8. Liu LT, Tang LQ, Chen QY, et al. The prognostic value of plasma Epstein-Barr viral DNA and tumor response to neoadjuvant chemotherapy in advanced-stage nasopharyngeal carcinoma. Int J Radiat Oncol Biol Phys. 2015;93(4):862–869. [DOI] [PubMed] [Google Scholar]
- 9. Sparano JA, Gray RJ, Makower DF, et al. Adjuvant chemotherapy guided by a 21-gene expression assay in breast cancer. N Engl J Med. 2018;379(2):111–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sparano JA, Gray RJ, Ravdin PM, et al. Clinical and genomic risk to guide the use of adjuvant therapy for breast cancer. N Engl J Med. 2019;380(25):2395–2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cheong J, Yang H, Kim H, et al. Predictive test for chemotherapy response in resectable gastric cancer: a multi-cohort, retrospective analysis. Lancet Oncol. 2018;19(5):629–638. [DOI] [PubMed] [Google Scholar]
- 12. Paik S, Tang G, Shak S, et al. Gene expression and benefit of chemotherapy in women with node-negative, estrogen receptor-positive breast cancer. J Clin Oncol. 2006;24(23):3726–3734. [DOI] [PubMed] [Google Scholar]
- 13. Ulloa-Montoya F, Louahed J, Dizier B, et al. Predictive gene signature in MAGE-A3 antigen-specific cancer immunotherapy. J Clin Oncol. 2013;31(19):2388–2395. [DOI] [PubMed] [Google Scholar]
- 14. Wen J, Yang H, Liu MZ, et al. Gene expression analysis of pretreatment biopsies predicts the pathological response of esophageal squamous cell carcinomas to neo-chemoradiotherapy. Ann Oncol. 2014;25(9):1769–1774. [DOI] [PubMed] [Google Scholar]
- 15. Zeng Z, Zhou Y, Xiong W, et al. Analysis of gene expression identifies candidate molecular markers in nasopharyngeal carcinoma using microdissection and cDNA microarray. J Cancer Res Clin Oncol. 2006;133(2):71–81. [DOI] [PubMed] [Google Scholar]
- 16. Wang S, Li X, Li Z, et al. Gene expression profile changes and possible molecular subtypes in differentiated-type nonkeratinizing nasopharyngeal carcinoma. Int J Cancer. 2011;128(4):753–762. [DOI] [PubMed] [Google Scholar]
- 17. Tang X, Li Y, Liang S, et al. Development and validation of a gene expression-based signature to predict distant metastasis in locoregionally advanced nasopharyngeal carcinoma: a retrospective, multicentre, cohort study. Lancet Oncol. 2018;19(3):382–393. [DOI] [PubMed] [Google Scholar]
- 18. Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer. 2009;45(2):228–247. [DOI] [PubMed] [Google Scholar]
- 19. Tibshirani R. Regression shrinkage and selection via the Lasso. J R Stat Soc Series B. 1996;58(1):267–288. [Google Scholar]
- 20. Huang ML, Hung YH, Lee WM, Li RK, Jiang BR.. SVM-RFE based feature selection and Taguchi parameters optimization for multiclass SVM classifier. Sci World J. 2014;2014:795624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Qiu J, Peng B, Tang Y, et al. CpG methylation signature predicts recurrence in early-stage hepatocellular carcinoma: results from a multicenter study. J Clin Oncol. 2017;35(7):734–742. [DOI] [PubMed] [Google Scholar]
- 22. Xu R, Wei W, Krawczyk M, et al. Circulating tumour DNA methylation markers for diagnosis and prognosis of hepatocellular carcinoma. Nat Mater. 2017;16(11):1155–1161. [DOI] [PubMed] [Google Scholar]
- 23. Olmos D, Brewer D, Clark J, et al. Prognostic value of blood mRNA expression signatures in castration-resistant prostate cancer: a prospective, two-stage study. Lancet Oncol. 2012;13(11):1114–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Vickers AJ, Elkin EB.. Decision curve analysis: a novel method for evaluating prediction models. Med Decis Mak. 2006;26(6):565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pataer A, Kalhor N, Correa AM, et al. Neutrophil-lymphocyte and platelet-lymphocyte ratios as prognostic factors after stereotactic radiation therapy for early-stage non-small-cell lung cancer. J Thorac Oncol. 2015;10(2):280–285. [DOI] [PubMed] [Google Scholar]
- 26. Shen Q, Fan J, Yang XR, et al. Serum DKK1 as a protein biomarker for the diagnosis of hepatocellular carcinoma: a large-scale, multicentre study. Lancet Oncol. 2012;13(8):817–826. [DOI] [PubMed] [Google Scholar]
- 27. Bae WK, Hwang JE, Shim HJ, et al. Phase II study of docetaxel, cisplatin, and 5-FU induction chemotherapy followed by chemoradiotherapy in locoregionally advanced nasopharyngeal cancer. Cancer Chemother Pharmacol. 2010;65(3):589–595. [DOI] [PubMed] [Google Scholar]
- 28. Du C, Ying H, Zhou J, et al. Experience with combination of docetaxel, cisplatin plus 5-fluorouracil chemotherapy, and intensity-modulated radiotherapy for locoregionally advanced nasopharyngeal carcinoma. Int J Clin Oncol. 2013;18(3):464–471. [DOI] [PubMed] [Google Scholar]
- 29. Kong L, Zhang Y, Hu C, et al. Effects of induction docetaxel, platinum, and fluorouracil chemotherapy in patients with stage III or IVA/B nasopharyngeal cancer treated with concurrent chemoradiation therapy: final results of 2 parallel phase 2 clinical trials. Cancer. 2017;123(12):2258–2267. [DOI] [PubMed] [Google Scholar]
- 30. Huang CL, Sun ZQ, Guo R, et al. Plasma Epstein-Barr virus DNA load after induction chemotherapy predicts outcome in locoregionally advanced nasopharyngeal carcinoma. Int J Radiat Oncol Biol Phys. 2019;104(2):355–361. [DOI] [PubMed] [Google Scholar]
- 31. Zhou T, Zhang S, He C, et al. Downregulation of mitochondrial cyclooxygenase-2 inhibits the stemness of nasopharyngeal carcinoma by decreasing the activity of dynamin-related-protein 1. Theranostics. 2017;7(5):1389–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shi C, Guan Y, Zeng L, et al. High COX-2 expression contributes to a poor prognosis through the inhibition of chemotherapy-induced senescence in nasopharyngeal carcinoma. Int J Oncol. 2018;53(3):1138–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zou L, Chai J, Gao Y, Guan J, Liu Q, Du J.. Down-regulated PLAC8 promotes hepatocellular carcinoma cell proliferation by enhancing PI3K/Akt/GSK3β/Wnt/β-catenin signaling. Biomed Pharmacother. 2016;84:139–146. [DOI] [PubMed] [Google Scholar]
- 34. Shi L, Xiao L, Heng B, Mo S, Chen W, Su Z.. Overexpression of placenta specific 8 is associated with malignant progression and poor prognosis of clear cell renal cell carcinoma. Int Urol Nephrol. 2017;49(7):1165–1176. [DOI] [PubMed] [Google Scholar]
- 35. Lu Y, Han D, Liu W, et al. RNF138 confers cisplatin resistance in gastric cancer cells via activating Chk1 signaling pathway. Cancer Biol Ther. 2018;19(12):1128–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ismail IH, Gagne JP, Genois MM, et al. The RNF138 E3 ligase displaces Ku80 to promote DNA end resection and regulate DNA repair pathway choice. Nat Cell Biol. 2015;17(11):1446–1457. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The microarray data have been deposited in www.ncbi.nlm.nih.gov/geo/ with the accession number GSE132112, and the key raw data have been deposited at Research Data Deposit public platform (www.researchdata.org.cn), with an approval number of RDDB2020000883.