Summary
Autophagy is a quality control, metabolic and innate immunity process. Normative autophagy affects many cell types, including hematopoietic as well as non-hematopoietic, and promotes health in model organisms and humans. When autophagy is perturbed, this has repercussions on diseases with inflammatory components, including infections, autoimmunity and cancer, metabolic disorders, neurodegeneration, and cardiovascular and liver diseases. As a cytoplasmic degradative pathway, autophagy protects from exogenous hazards including infection and from endogenous sources of inflammation including molecular aggregates and damaged organelles. The focus of this review is on the role of autophagy in inflammation, including type I interferon responses and inflammasome outputs, from molecules to immune cells. A special emphasis is given to the intersections of autophagy with innate immunity, immunometabolism, and functions of organelles such as mitochondria and lysosomes that act as innate immunity and immunometabolic signaling platfroms.
Introduction
Mammalian autophagy is a fundamental biological process contributing to cytoplasmic quality control (Morishita and Mizushima, 2019), cellular metabolism (Herzig and Shaw, 2018; Liu and Sabatini, 2020; Marino et al., 2014; Morishita and Mizushima, 2019) and innate and adaptive immunity (Clarke and Simon, 2019; Deretic et al., 2013; Levine et al., 2011; Ma et al., 2013). The very nature of mammalian autophagy is to serve all three ‘missions’ - defense, metabolic, and quality control – and they present themselves interlinked in the context of immunity. A failure in autophagy functions is often manifested as dysregulated inflammation in animal models and human diseases (Deretic and Levine, 2018).
One may come across the word ‘autophagy’ in the literature as a catch-all term for a collection of lysosomal processes contributing to intracellular homeostasis. This includes turn-over of proteins, membranes, whole organelles, and generation of metabolic precursors at times of starvation through lysosomal degradation (Herzig and Shaw, 2018; Lin and Hardie, 2018; Liu and Sabatini, 2020; Morishita and Mizushima, 2019). Collectively, these processes contribute to cell fitness, functionality and survival. The most studied form of autophagy is macroautophagy, referred henceforth simply as autophagy. The sensu stricto autophagy pathway, whereby double membrane autophagosomes form in the cytoplasm, capture cargo, and deliver it to lysosomes for digestion (Figure 1A). In principle, there are several protein kinase, lipid kinase, and a protein-lipid conjugation system that come together to drive the formation and maturation of autophagosomes (Figure 1A) as detailed in recent reviews (Melia et al., 2020; Morishita and Mizushima, 2019). In this review we expand on the connections between autophagy and inflammation and cover aspects of autophagy in inflammation from immune molecular systems to immune cells and immunometabolism (Mills et al., 2017).
Figure 1. Autophagy and autophagy-related pathways as guardians against excessive inflammation.
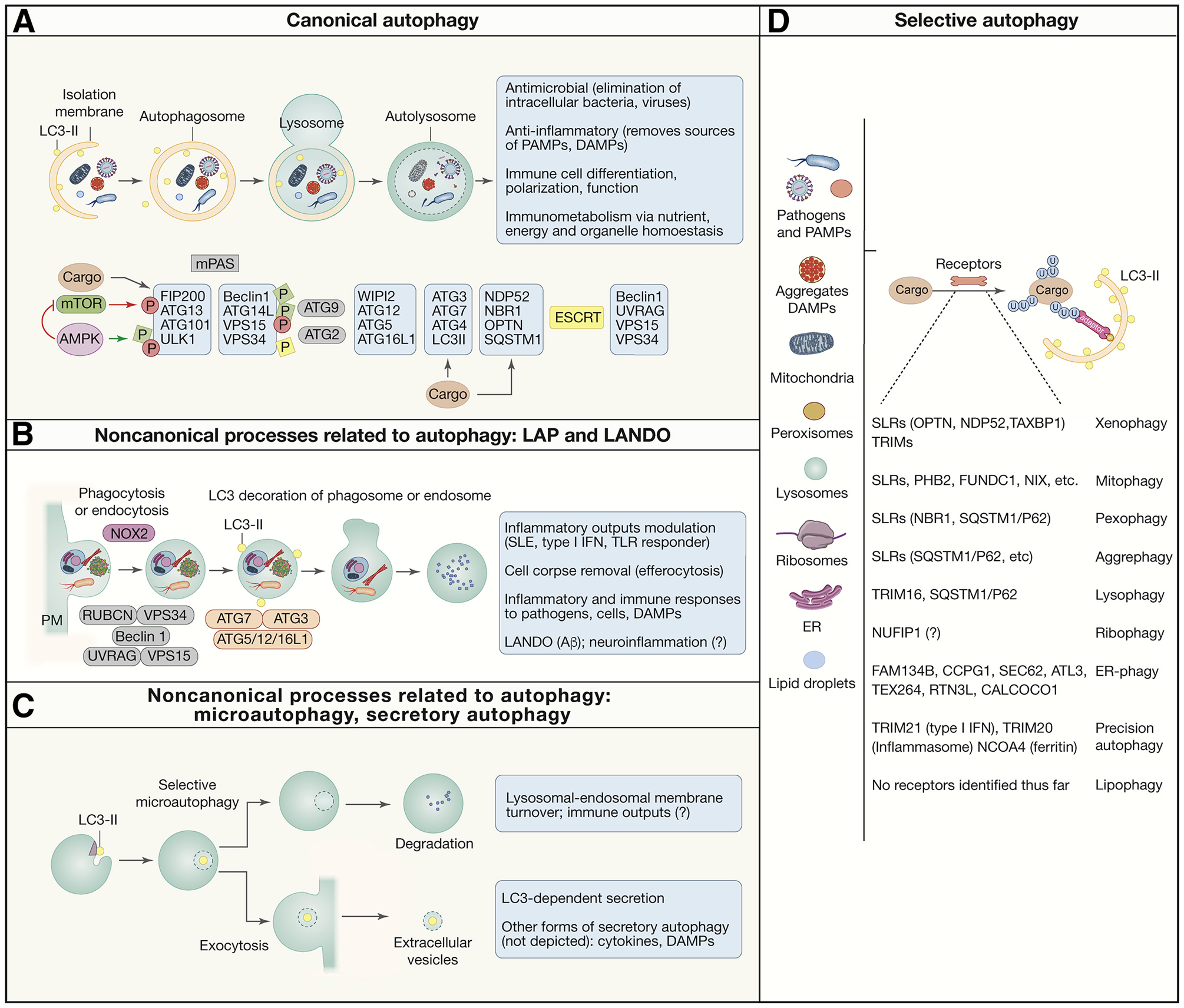
A. Simplified morphological stages of canonical autophagy. Autophagosomes have double membrane, sequester diverse cytoplasmic cargo, and typically deliver the cargo to lysosomes for degradation. Principal protein complexes controlling autophagy are shown below in approximate order of activities along the autophagy pathway. Red circles, inhibitory phosphorylation by mTOR. Green squares, activating phosphorylation by AMPK B. LC3-assocate phagocytosis (LAP) and LC-3-associated endocytosis (LANDO): noncanonical autophagy-related processes that originate starting with invaginations at the plasma membrane leading to formation of phagosomes and endosomes decorated with lipidated LC3 (LC3-II). They are single membranes that use only a subset of ATG proteins plus NOX2 (LAP) and Rubicon and are independent of the FIP200 complex. C. Other noncanonical autophagy-related processes. Selective microautophagy: LC3-II on lysosomes and endosomes contributes to formation of intralumenal vesicles that are either digested or exocytosed. Other processes (not depicted) are mentioned in the box. D. List of autophagy cargo, receptors and terminology for various types of autophagy, defined by the targeted cargo. Anti-inflammatory and related immune functions are listed in gray boxes.
Overview of autophagy pathway and its broad relationship to inflammation
Canonical autophagy (Figure 1A), as opposed to noncanonical forms of autophagy (Figure 1B,C) can capture diverse cargo (Fig. 1D). This includes invading microbes along with their pathogen associated molecular patterns (PAMPs) (Deretic et al., 2013; Gomes and Dikic, 2014; Randow and Youle, 2014). Autophagy clears cytosolic protein aggregates and condensates (Agudo-Canalejo et al., 2020; Noda et al., 2020) or ingested extracellular debris, many of which act as danger associated molecular patterns (DAMPs) (Rubinsztein et al., 2015). The cargo often defines the subtype of autophagy (Morishita and Mizushima, 2019): xenophagy (direct elimination of intracellular microbes), aggrephagy (removal of macromolecular aggregates or condensates), mitophagy (removal of mitochondria), endoplasmic reticulum (ER)-phagy (also known as reticulophagy), pexophagy (autophagy of peroxisomes), lysophagy (removal of damaged lysosomes), etc. Bulk autophagy refers to a wholesale recycling of nutrients, which sometimes happens even during so-called selective autophagy through a bystander effect (An and Harper, 2018).
The above processes affect inflammation. Given that microbes, damaged organelles, aggregates and organic or inorganic crystals are sources of inflammatory signals (PAMPs and DAMPs), the cytoplasmic clean-up function of autophagy is by default anti-inflammatory in any type of cell capable of activating cell-autonomous inflammatory response (Deretic and Levine, 2018; Deretic et al., 2013). Complementing this, autophagy sculpts the interior of immune cells by aligning mitochondrial and ER content with immune cell functions (e.g. mature naïve T cells trim down their mitochondrial content; plasma cells need to maintain their ER) (Clarke and Simon, 2019), whereas autophagy-dependent metabolic adjustments contribute to immunometabolic states affecting macrophage and T cell polarization (Riffelmacher et al., 2018), all of which impact inflammatory outputs and resolution (Mills et al., 2017; O’Neill et al., 2016).
Autophagy receptors capture intracellular cargo promoting inflammation
To remove sources of inflammation such as bacteria and endogenous pro-inflammatory sources, the autophagic cargo is often opsonized by tags such as ubiquitin, recognized by broad spectrum selective autophagy receptors termed SLRs (sequestosome-like receptors: p62 (SQSTM1), NBR1, OPTN, NDP52, TAXBP1, etc.) (Birgisdottir et al., 2013; Deretic et al., 2013) which patrol the cytosol accessing diverse targets. For example, SLRs in various combinations participate in diverse forms of anti-inflammatory autophagy: aggrephagy (Bjorkoy et al., 2005; Kirkin et al., 2009), xenophagy (Ponpuak et al., 2010; Ravenhill et al., 2019; Thurston et al., 2012; Wild et al., 2011) and mitophagy (removing sources of mitoDNA and ROS that act as DAMPs) (Lazarou et al., 2015; Vargas et al., 2019).
Intracellular organelles, even when functional, if not adjusted for total cellular content can be sources of immune and other cells’ dysfunction and systemic inflammation. This adjustment for total cellular content occurs through dedicated cognate receptors that are integral components of the target membrane (Morishita and Mizushima, 2019) with extensive repertoire per single organelle type (See Figure 1D for details). Rightsizing cell’s organellar content by autophagy is key to proper development, differentiation and function of immune cells (Clarke and Simon, 2019).
Typically, selective autophagy receptors interact with autophagosomal isolation membranes via specialized motifs (e.g. LC3 interacting region; LIR) (Johansen and Lamark, 2011; Marshall et al., 2019) that bind to mammalian Atg8 paralogs (mAtg8s: LC3A,B,B2,C and GABARAP, -L1, -L2), with LC3B being the prototypical marker for autophagic membranes (Figure 1A,D). Surprisingly, the ability of autophagic receptors to bind mAtg8s is not always essential and the initial stages of mitophagy take place even in the absence of all mAtg8s (Vargas et al., 2019). This indicates that there are other emerging possibilities to attach a receptor to autophagosomal membranes (Ravenhill et al., 2019; Smith et al., 2018; Turco et al., 2019; Vargas et al., 2019) (Figure 1A).
In summary, diverse (soluble and integral to organellar membranes) autophagy receptors orchestrate removal of exogenous and endogenous sources of inflammation by linking earmarked cargo to nascent autophagsosomes.
Autophagy’s antimicrobial and anti-inflammatory functions are interlinked
The notion that autophagy acts as cell-autonomous antimicrobial defense mechanism was long anticipated by a study showing that autophagy acts as part of antiviral defenses, and is antagonized by the herpes simplex virus protein ICP34.5 (Talloczy et al., 2002). Autophagy has been shown to be induced by eIF2α kinase PKR (EIF2AK2), which recognizes dsRNA in the context of infection by a variety of DNA and RNA viruses, as a part of innate response (Talloczy et al., 2002). Anti-viral autophagy When autophagy had been recognized in a systematic fashion as a bona fide immune process (Deretic, 2005; Levine, 2005), this was to a large extent prompted by findings that autophagy can eliminate a variety of intracellular bacteria such as Mycobacterium tuberculosis (Mtb) (Gutierrez et al., 2004). and streptococci (Nakagawa et al., 2004). This was soon followed by studies recognizing autophagy’s impact on Shigella (Ogawa et al., 2005), Salmonella (Birmingham et al., 2006), Crohn’s disease-associated adherent-invasive E. coli (Brest et al., 2011), and others. The role of autophagy as a cell-autonomous defense against intracellular microbes has been extended by recognizing that successful intracellular bacteria almost invariably encode or mount specific defenses against autophagy, which underscores antimicrobial and immunological significance of autophagy (Keller et al., 2020b). The net outcome of layers of host-pathogen measures and countermeasures often leads to distorted manifestations of autophagy’s role, as recently reviewed (Keller et al., 2020b). Since then, there has been explosive growth of the role of autophagy in immunity (Deretic and Levine, 2018), with emphasis on autophagy as an antimicrobial defense (Gomes and Dikic, 2014; Randow and Youle, 2014), immune cell development and differentiation (Clarke and Simon, 2019), polarization (Castillo et al., 2012), and inflammation (Deretic and Levine, 2018).
Autophagic capture and elimination of intracellular microbes in its purest form is referred to as xenophagy (Levine, 2005). Xenophagy rests upon recognition of pathogens entering the cells by selective autophagy receptors (Figure 1D). These receptors detect opsonized pathogens or vacuolar compartments harboring them. The opsonins can be ubiquitin (Perrin et al., 2004) recognized by SLRs or galectin 8 (Gal8) recognized by NDP52 (Montespan et al., 2017; Staring et al., 2017; Thurston et al., 2012). Autophagy can in priniciple access antibody-opsonized microbes when they penetrate the cytosol, contributing to humoral immunity against Ehrlichia chaffeensis (Velayutham et al., 2019). This occurs via the ‘cytosolic Fc receptor’ TRIM21 (McEwan et al., 2013), which acts as an adaptor for autophagy (Kimura et al., 2015). Microbes can also be directly recognized, e.g. the HIV capsid entering the cytosol is directly bound by TRIM5α (Mandell et al., 2014; Ribeiro et al., 2016). However, in that particular case, the virus has mutated away from proper recognition, and instead the autophagy machinery associated with TRIM5α contributes to inflammatory signaling through the TAK1 kinase, and transcription factors NFκB and AP1, contributing to general antiviral state (Saha et al., 2020). A complementary cooperative action of autophagy and inflammatory signaling is also seen with bacterial pathogens, as in the case of Salmonella (Noad et al., 2017).
In summary, under normal circumstances, systems supporting xenophagy and protective inflammation act as balanced, additive mechanisms. When xenophagy fails, inflammatory responses become exaggerated and, while being protective, this comes at a cost causing tissue damage and inflammatory pathology.
Autophagy and lysosomes converge upon immunometabolism
Lysosomes are intimately associated with both the initiation and completion of autophagy. Autophagosomes merge with lysosomal compartments in a process referred to as autophagosomal maturation or autophagosome-lysosome fusion (Morishita and Mizushima, 2019). This way autophagosomes deliver their cargo to be digested (Fig. 1A).
Lysosomes have critical signaling and immunological functions (Ballabio and Bonifacino, 2020; Herzig and Shaw, 2018; Lin and Hardie, 2018; Liu and Sabatini, 2020). Much of the signaling by AMPK and mTOR, the master regulators of cellular metabolism, and control of their activity occurs on the lysosomal membrane (Gonzalez et al., 2020; Herzig and Shaw, 2018; Jia et al., 2018; Jia et al., 2020a; Lin and Hardie, 2018; Liu and Sabatini, 2020). AMPK and mTOR work as the yin and the yang of cellular metabolism, and act in opposing ways to control both autophagy (Herzig and Shaw, 2018; Liu and Sabatini, 2020) and immunometabolism, profoundly affecting immune cell function (O’Neill and Hardie, 2013; O’Neill et al., 2016; Weichhart et al., 2015).
Lysosomal membranes can be damaged by DAMPs, PAMPs, and microbes, or permeabilized in a programmed manner, e.g. through TRAIL (TNF-related apoptosis-inducing ligand) signaling. Autophagy protects against such damage as an important part of a multi-staged lysosome-homeostatic response termed MERiT (membrane repair, removal and replacement) (Fig. 2A) (Jia et al., 2020b, c). Most stages of MERiT are driven and coordinated by galectins (Gal3, Gal8 and Gal9), a family of cytosolic lectins that recognize exposed lumenal glycans upon membrane damage and act as effectors or recruiters or effectors. The first stage of MERiT is galectin-dependent (Gal3) (Jia et al., 2020b), briefly preceded by a Gal3-independent (possibly Ca2+-dependent) step (Radulovic et al., 2018; Skowyra et al., 2018). Gal3 binds to glycans exposed to the cytosol during lysosomal membrane damage, and recruits ESCRTs to repair the lysosomal membrane (Jia et al., 2020b). The next stage of MERiT is the removal of damaged lysosomes through autophagy (‘lysophagy’) (Maejima et al., 2013), which kicks in when ESCRTs fail to repair lysosomes (Chauhan et al., 2016; Jia et al., 2020b; Papadopoulos et al., 2017). Sustained lysosomal damage inhibits mTOR (Jia et al., 2018) and activates AMPK (Jia et al., 2020a). The final stage of MERiT is lysosomal replacement. This happens through activation of TFEB, a key transcriptional factor driving expression of lysosomal genes (Napolitano and Ballabio, 2016) with multifaceted effects on inflammation (Brady et al., 2018). Protecting lysosomes by repairing them, removing them when they are damaged, and replacing them when necessary, impacts immunometabolism (Figure 2A,B) and autophagy in its anti-inflammatory functions, (Figure 1A, top box) including the removal of DAMPs, inflammasome components, and type I interferon (IFN) regulators (Figure 1D).
Figure 2. Lysosomes, mitochondria, and TBK1 in inflammation, immunometabolism and immune responses.
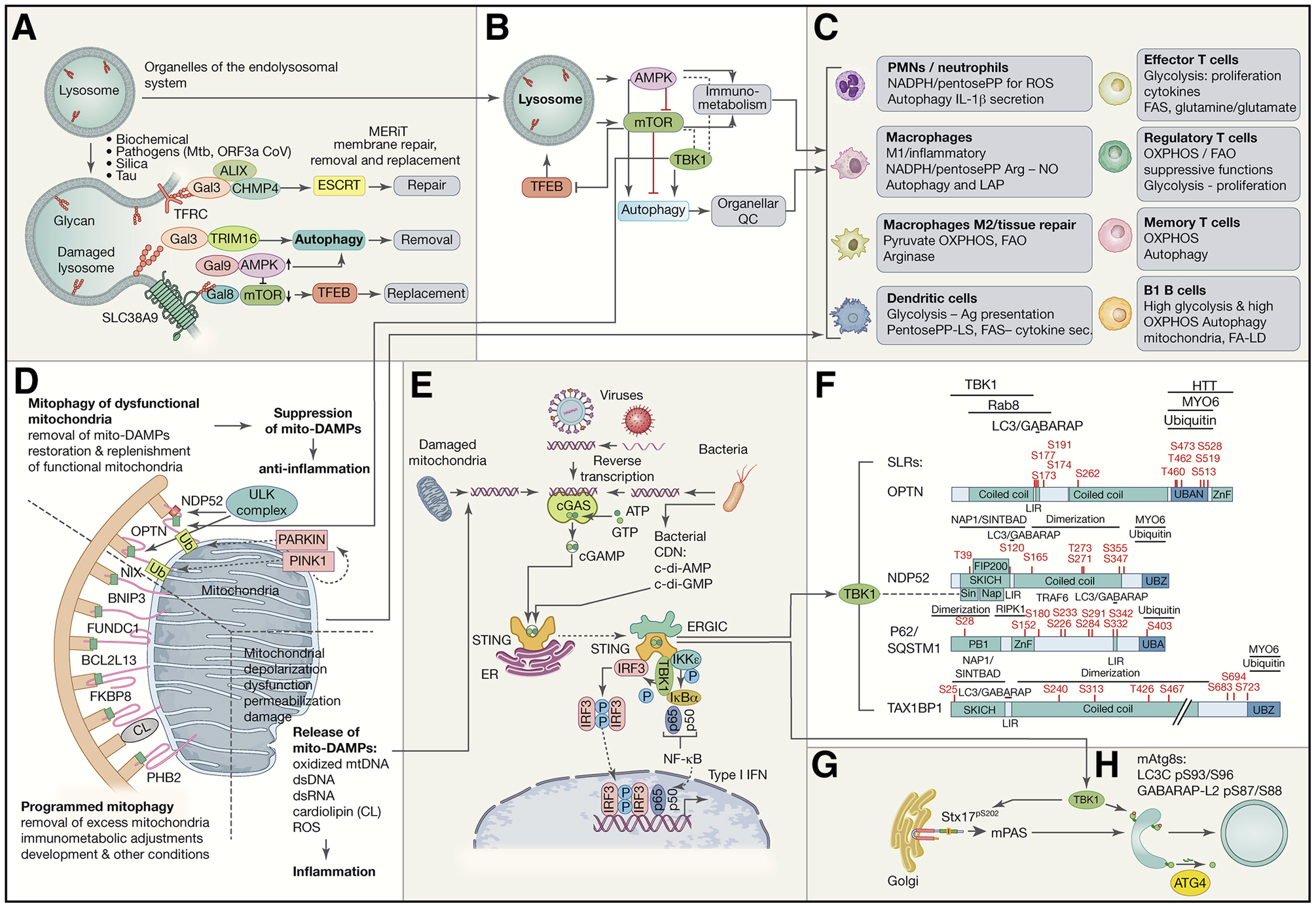
A. Lysosomal integrity is maintained by a system termed MERiT (membrane repair, removal and replacement). Galectins (Gal3, Gal8 and Gal9) recognize β-galactoside glycans exposed on cognate lysosomal membrane proteins: TFRC (transferrin receptor), SLC38A9, and LAMP1 and LAMP2 (not depicted) when lysosomal membrane is breached. They stimulate ESCRTs to repair lysosomes and AMPK to activate autophagy, whereas mTOR is inactivated. TRIM16 is one of lysophagy receptors. TFEB starts transcriptional program to replace lysosomes. B. Lysosome as the hub for AMPK, mTOR and TFEB, the three master regulators of autophagy, lysosomal system, metabolism and immunometabolism. Inhibitory and activating relationships are indicated and include TBK1 (which has positive and negative regulatory connections with AMPK and mTOR). C. Effects of signaling in B in different immune cells. D. Mitophagy, mitophagy receptors, and mitophagic suppression of inflammation caused by mitochondrial DAMPs (‘mito-DAMPs’). Dashed lines separate different types of mitophagy and proinflammatory signalng from damaged mitochiondria. Top, Pink1-Parkin system and receptors that participate in removal of dysfunctional mitochondria. Pink1 kinase and Parkin E3 ligase (phosphorylated by Pink1 to activate latent Parkin activity and recruit it to mitochondria) lead to ubiquitination and generation of phospho-ubiquitin which further amplifies Parkin activity leading to recognition of depolarized or damaged mitochondria by SLRs, NDP52 and OPTN, which bind ubiquitin. Bottom left, other mitophagy receptors act as integral membrane proteins of mitochondria and participate in mitophagy under developmental, differentiation or stress conditions. (E) Molecular machinery involved in STING-TBK1 activation in response to ectopic dsDNA, e.g. mitochondrial DNA that leaks into the cytosol (see panel D), DNA from pathogens (viruses and bacteria), etc. dsDNA binds to and stimulates cGAS that enzymatically generates cGAMP. STING stimulated by cGAMP (and other cyclic dinucleotides that may come from bacteria) activates TBK1. (F) TBK1 phosphorylates SLRs (F) modulating their ability to bind ubiquitin and mAtg8s such as LC3B, and possibly affecting other interactions. (G) TBK1 phosphorylates Syntaxin 17 (Stx17) involved in mPAS (mammalian pre-autophagosomal structure) and subsequent stages such as maturation. (H) TBK1 phosphorylates two members of the mAtg8 family rendering them resistant to delipidation by ATG4.
TFEB’s nuclear translocation from the cytoplasm to the nucleus, where it activates genes in support of lysosomal biogenesis (Napolitano and Ballabio, 2016; Settembre et al., 2011), is normally executed through inhibition of mTOR (Napolitano et al., 2020) and activation of a calcineurin phosphatase PPP3CB (Medina et al., 2015). The net result is dephosphorylation of TFEB and its translocation to and retention in the nucleus. In addition to galectins, mAtg8s (the GABARAP subset), play an unanticipated role, outside of the classical scope of association with autophagic membranes and cargo receptors, and control TFEB activation (Kumar et al., 2020; Nakamura et al., 2020). The GABARAP subset of mAtg8s are responsible for this activity (Kumar et al., 2020; Nakamura et al., 2020)
AMPK, in principle, gets synergistically co-induced with autophagy whereas mTOR is inactivated at times of metabolically (starvation) induced autophagy, This has repercussions on immunometabolism (Figure 2C) (O’Neill and Hardie, 2013; O’Neill et al., 2016; Weichhart et al., 2015), whereby active mTOR is associated with inflammatory states and robust immune responses including exit of T cells from quiescent state (Chapman et al., 2020), effector T cells, and classically activated macrophages (Weichhart et al., 2015). AMPK is often associated with immunometabolic states compatible with anti-inflammatory activities and quiescence (O’Neill and Hardie, 2013; O’Neill et al., 2016; Saravia et al., 2020). However, AMPK’s ability to stimulate catabolic pathways, of which autophagy is one component, also provides energy in support of expansion of CD4+ T helper-1 (Th1) and Th17 cells and primary T cell responses in infections (Blagih et al., 2015). AMPK helps T cell differentiation and regulatory T (Treg) cell function, supports T cell survival in glucose limiting conditions, supports T cell quiescence via fatty acid oxidation (FAO), and stimulates FAO in memory CD8+ T cells (Saravia et al., 2020). AMPK agonist metformin restores generation of memory CD8+ T cells whereas AMPKα1 is essential for their recall responses (Pearce et al., 2009).
In summary, by maintaining healthy lysosomes (Figure 2A) autophagy is important not only as a catabolic output downstream of AMPK but also as an upstream quality control of lysosomes in their role as a platfrom for mTOR’s and AMPK’s signaling and systemic and immunometabolic functions (Figure 2C) of relevance for inflammation (Chapman et al., 2020; O’Neill and Hardie, 2013; O’Neill et al., 2016; Weichhart et al., 2015).
A spectrum of non-canonical autophagy processes affect inflammation
The lysosome and the endosomal system are involved in ‘non-canonical’ autophagy-related processes that utilize portions of the autophagy machinery and affect inflammation (Figure 1B,C) (Galluzzi and Green, 2019). The prototypical noncanonical autophagy (Fugure 1B) are LC3-associated phagocytosis (LAP) and its variations such as LC3-associated endocytosis (LANDO) (Sanjuan et al., 2007), which use only portion of the autophagy factors and operate on single membrane vesicles derived from plasma membrane, i.e., phagosomes and endosomes (Figure 1B). LAP and LANDO, and several other variations can be viewed as related processes (Galluzzi and Green, 2019) (Fletcher et al., 2018; Guo et al., 2017; Heckmann et al., 2019; Lee et al., 2020; Leidal et al., 2020). They involve lipidation of LC3 (and perhaps other mAtg8s) occurring on single membrane bilayers independently of a subset of canonical autophagy factors required for double membrane autophagosomes (Galluzzi and Green, 2019; Heckmann and Green, 2019; Leidal et al., 2020; Lystad et al., 2019). Another noncanonical autophagy process referred to as microautophagy consist of direct lysosomal uptake (bypassing the need for double membrane autophagosomes) of portions of the cytoplasm or specific cytosolic targets (De Duve and Wattiaux, 1966; Schuck, 2020). Topologically, this occurs through invaginations of the lysosomal or endosomal membranes, inward pinching off of such invaginations into the lysosomal or endosomal lumen, and delivery of the captured cargo, possibly sequestered via SLRs (Mejlvang et al., 2018) or by other means. A selective form of microautophagy depends on lipidation of LC3 (Lee et al., 2020). Activation of a subset of autophagy factors on endosomes protects against viral infection (Dong et al., 2020). Finally, a process of individual protein import across lysosomal membrane for digestion, referred to as chaperone-mediated autophagy (Kaushik and Cuervo, 2018), affects immune cells (Macian, 2019; Valdor et al., 2014).
The above processes perform a variety of functions, many with inflammatory outputs (Figure 1B,C): (i) LAP-reliant dying cell elimination by efferocytosis (Boada-Romero et al., 2020) that protects against excess inflammatory cytokine production and autoimmunity such as in systemic lupus erythematosus (SLE) (Martinez et al., 2016). LAP-directed toll-like receptor-9 trafficking into a specialized type I interferon signaling compartment in plasmocytoid dendritic cells (pDC) (Henault et al., 2012). LAP’s antibacterial and anti-inflammatory action in pneumococcus-infected macrophages (Inomata et al., 2020) and anti-inflammatory and anti-fiibrogenic activity in monocytes responding to liver injury (Wan et al., 2020). (ii) Sustained β-amyloid (Aβ) aggregate removal by LANDO affecting recycling of the Aβ receptor (Heckmann et al., 2020) and preventing inflammation and memory loss in a murine model of Alzheimer’s disease (Heckmann et al., 2019). (iii) Lysosomal membrane turnover during starvation or repair of osmotically stressed lysosomal membranes (Lee et al., 2020) (iv) LC3-dependent secretion (Leidal et al., 2020). (v) Secretory autophagy (Ponpuak et al., 2015), an alternative pathway involved in secretion of inflammatory molecules such as HMGB1 and IL-1β (Dupont et al., 2011; Karmakar et al., 2020; Kimura et al., 2017) before GSDMD-pores and pyroptosis start to dominate in their release (Broz et al., 2020; Kayagaki et al., 2015; Shi et al., 2015), which appears to be a preferred way for IL-1β release by neutrophils (Karmakar et al., 2020).
Of note, secretory autophagy (Ponpuak et al., 2015) is an emerging field that includes variations such as LC3-dependent secretion of cytosolic cargo via EVs (Leidal et al., 2020), release of EVs containing decoy targets for toxins to neutralize their cytotoxic activities (Keller et al., 2020a), unconventional secretion of cytosolic proteins without signal sequences (Dupont et al., 2011; Karmakar et al., 2020; Kimura et al., 2017; Zhang et al., 2015), and nonlytic release from cells of viruses (Chen et al., 2015) and bacteria (Gerstenmaier et al., 2015) as a mode for their infectious spread. This is also an alternative solution for cells to eliminate pathogens by employing secretory autophagy rather than the degradative autophagy (Miao et al., 2015).
In summary, ‘non-canonical’ forms of autophagy, i.e. a diverse collection of processes borrowing components of the autophagy pathway (Galluzzi and Green, 2019), impact tissue homeostasis and inflammation mirroring actions of ‘canonical autophagy’.
Mitophagy modulates inflammation
Mitophagy (Figure 2D) and other homeostatic systems maintain mitochondrial health and abundance (Youle, 2019), ensuring that mitochondria provide healthy signaling platforms for proper activation of innate immunity in response to viral and bacterial PAMPs. Mitophagy and autophagy of bacteria (xenophagy) might have co-evolved through host-microbe interactions resulting in elimination (normal outcome), endosymbiosis (mitochondria) or pathogenesis (successful intracellular parasites such as Mtb) with vestigial but powerful effects on innate immunity and inflammation (Deretic, 2010; Youle, 2019). Mitochondria are at the heart of immune responses serving as platforms for proper signaling of several innate immunity platfroms such as MAVS, whereas under pathological conditions mitochondira can themselves be sources of DAMPs (e.g. mitochondrial dsDNA; Figure 2D,E) when mitochondria are not properly maintained through mitophagy (Youle, 2019). They are also the principal contributors to immunometabolism (Figure 2C) (Mills et al., 2017) whereby switching between oxidative phosphorylation (oxphos) and glycolysis or their realignment parallels activation states of macrophages, dendritic cells, T cells and other cells, e.g. glycolysis dominating in proinflammatory classically activated macrophages and oxphos being predominant in alternatively activated macrophages involved in tissue repair. For example, the anti-inflammatory cytokine IL-10 inhibits macrophage-intrinsic mTOR and glycolysis, promotes oxphos, and induces mitophagy to cull dysfunctional mitochondria and reduce inflammatory stimulation by mitochondrial DAMPs (Ip et al., 2017).
Mitochondria’s general health, dependent upon mitophagy and related processes, is essential for productive inflammation that can clear infection or cancer and maintain tissue homeostasis. Mitochondria, when damaged (Figure 2D), become a source of endogenous DAMPs (‘mito-DAMPs’: oxidized mtDNA, dsDNA, dsRNA, cardiolipin, ROS; Figure 2D)), and these in turn stimulate a variety of pattern recognition receptors (PRRs), which lead to degenerative and other diseases (Youle, 2019). When mitophagy fails ‘mito-DAMPs’ contribute to excessive inflammation causing pathology. Defective mitophagy has been linked to such inflammation in sepsis, carcinogenesis and neurodegeneration (Li et al., 2019; Nakahira et al., 2011; Sliter et al., 2018). Curiously, this is also an aspect of the less appreciated ability of NF-κB, primarily known as a key activator of inflammation, to restrain excessive NLRP3-inflammasome activation in macrophages (Zhong et al., 2016). NF-κB prepares cells for autophagy, reflected in SQSTM1 amounts, and facilitates mitophagy to pre-empt excessive IL-1β inflammation and macrophage cell death (Zhong et al., 2016).
In summary, proper mitophagy is critical to balance innate immune responses to endogenous and exogenous sources of inflammation, unkempt mitochondria are a pathology-inducing source of endogenous ‘mito-DAMPs’ and excessive inflammation (Figure 2D,E), whereas functional mitochondria and their abundance controlled by mitophagy (Figure 2D) are important for immunometabolism (Figure 2C).
Autophagy of other organelles affects inflammation
The majority of the current literature of relevance for inflammation focuses on mitochondria. However, other organelles can be important sources or modulators of inflammation. As discussed above, lysosomes and endosomes play an important role, whereas the endoplasmic reticulum (ER) and peroxisomes are an emerging organelle of interest. For example, cyclic-di-nucleotides (CDNs) released by intracellular Gram-positive bacteria infecting phagocytes bind to the DNA sensor STING in the ER (Figure 2E), and induce ER stress (involving PERK, IRE1α, CHOP and BIP) eliciting cell death and inflammation, which is countered by ER-phagy and canonical autophagy controlled by mTOR (Moretti et al., 2017). The CDN-induced ER-phagy sequesters PERK and IRE1α (Moretti et al., 2017).
Peroxisomes, subject to selective autophagy termed pexophagy (Figure 1D), are often relegated to ‘poor cousin’ status relative to mitochondria. Nevertheless, the two represent an important continuum in fatty acid metabolism, with peroxisomes degrading very long fatty acids via β-oxidation and handing over shortened acyl-CoA via the carnitine shuttle to mitochondria for further β-oxidation and coupling to ATP synthesis. Peroxisome degrade lipid mediators of inflammation such as prostaglandins and leukotrienes, with roles in asthma, allergies and potentially in Alzheimer’s disease and related dementias, as well as support the biogenesis of precursor polyunsaturated fatty acids for specialized pro-resolving mediators that limit inflammation. Peroxisomes are also an alternative location for MAVS (in a continuum with mitochondrial location of MAVS) specializing in rapid anti-viral innate immunity responses (Bender et al., 2015; Dixit et al., 2010; Ferreira et al., 2016). The role of peroxisomes in inflammation and immunity deserves attention (Di Cara et al., 2019), and how pexophagy (Figure 1D) affects inflammation, immune cells, and immunometabolism is an unexplored and potentially interesting area of study.
Is there any hierarchy or order of importance of different organelles in the inflammatory processes impacted by autophagy? This is premature to answer and in principle difficult given the physiological and often physical interconnectedness of various organelles. Nevertheless, degradative autophagy cannot be completed without lysosomes, lysosomes house mTOR and AMPK, the very upstream regulators of autophagy and immunometabolism, and should have a special place in any hierarchy that may emerge.
Autophagy plays multiple roles in immune cells
Autophagy is one of the effector outputs (e.g. antimicrobial defense) in immune cells. It also works as a nutritional backup and immunometabolic modulator, provides quality control, and executes programmed changes in numbers and volume of intracellular organelles. The effects of autophagy and the ATG genes in specific immune cells have been extensively reviewed (Clarke and Simon, 2019; Deretic et al., 2013; Macian, 2019; Riffelmacher et al., 2018).
In brief, autophagy supports self-renewal and quiescence in stem-like cells, and remodels cellular interiors during differentiation (Riffelmacher et al., 2018). In general, different branches of the hematopoietic lineage are affected to a different extent by ATG deficiencies. Early deletion of ATGs seems to primarily suppress the lymphoid lineage and results in myeloproliferation, whereas myeloid cells maintain normal functions albeit showing hyperinflammatory phenotype, while dendritic cells perform many of their roles during immune responses even in the absence of ATGs (Clarke and Simon, 2019; Deretic et al., 2013; Macian, 2019; Riffelmacher et al., 2018). Autophagy is needed for self-renewal of B1a cells (Clarke et al., 2018; Miller et al., 2008) and for neutrophils (Riffelmacher et al., 2017). It is important for invariant NKT cell development (Salio et al., 2014). IL-15 activates AMPK and autophagy (in the face of concomitant activation of mTOR) and is necessary for survival of NKT cells (Zhu et al., 2018). In naïve T cells, autophagy is induced following stimulation (Jia et al., 2011) and is required for peripheral T cell survival and function (Li et al., 2006; Pua et al., 2009), and for survival of effector and formation of CD8+ memory T cells (Puleston et al., 2014; Xu et al., 2014). T cells upregulate autophagy upon T cell receptor (TCR) stimulation, paradoxically with concomitant mTOR activation (Botbol and Macian, 2015) that supports proliferative responses. Autophagy helps avoid T cell anergy (Mocholi et al., 2018). It appears to be a cell-autonomous modulator of CD8+ T cell metabolism with effects on anti-tumor immunity (DeVorkin et al., 2019). Recently, liver-specific tissue resident memory (Trm) CD8+ cells and unconventional T cells known as mucosal-associated invariant T cells (MAIT cells; CD161hi Va7.2+ long term residents in the liver) have been found to display high autophagy levels (Swadling et al., 2020). Enhanced autophagy could be imprinted by liver stellate cells (as well as IL-15) and is required for mitochondrial homeostasis in Trm cells (Swadling et al., 2020).The liver is of particular interest to understand T-cell tolerizing mechanisms in the face of hepatic circulation that continuously brings microbial products. Thus, the latest studies addressing autophagy in Trm cells are instructive given the need to balance clearance of virus-infected and cancerous cells while avoiding excessive inflammation and pathology in tissues. Finally, diminished autophagy in CD8+ T cells during aging contributes to vaccine efficacy decline and can be countered ex vivo by stimulating autophagy and TFEB (Alsaleh et al., 2020).
In conclusion, autophagy and autophagy factors contribute to various extents, dependent upon the cell type, to the development, differentiation, polarization, function, and immunometabolism of diverse immune cell subsets (Figure 2C).
Autophagy inhibits inflammatory signaling complexes
Genetic associations between several autophagy loci and chronic inflammatory disorders and autoimmune diseases have been reviewed recently (Deretic and Levine, 2018). Among others, these include Crohn’s disease (CD); systemic lupus erythematosus, asthma, rheumatoid arthritis, Vici syndrome, celiac disease, type 1 diabetes mellitus, familial Mediterranean fever, multiple sclerosis and other neurological disorders especially in patients with stronger neuroinflammatory components including amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD).
Genome-wide association studies (GWAS) connecting genetic polymorphisms and predisposition to diseases in humans (Consortium, 2007) have revealed connections between CD and polymorphisms in genes encoding ATG16L1 and IRGM, proteins that interact with NOD2, a familial risk factor in CD and an important innate immunity signaling hub responding to PAMPs derived from bacterial cell walls. In murine in vivo and ex vivo models, Atg16L1 regulates inflammasome activation (Saitoh et al., 2008) and prevents necroptosis and loss of Paneth cells (Matsuzawa-Ishimoto et al., 2017). ATG16L1, NOD2 and IRGM form a complex to control autophagy (Chauhan et al., 2015), whereas IRGM functionally connects with ASC, a component of canonical inflammasomes (Daussy et al., 2021). IRGM promotes xenophagy (Singh et al., 2006) and controls TFEB in reponse to viral and bacterial stimul (Kumar et al., 2020). It mediates SQSTM1-dependent autophagic degradation of cGAS, RIG-I, and TLR3 (Jena et al., 2020) and helps degrade inflammasome components (Mehto et al., 2019).
Inflammasomes are cytosolic inflammatory signaling complexes activated by PAMPs and DAMPs. They proteolytically activate IL-1β and lead to its secretion along with additional pro-inflammatory cytokines (Lamkanfi and Dixit, 2014). A canonical inflammasome consists of pro-caspase 1, ASC adaptor, and sensors such as NLRP1, NLRP3, NLRC4, which detect microbial products or sterile endogenous agonists, or AIM2 and IFI16, which sense cytosolic DNA (Lamkanfi and Dixit, 2014). Once inflammasome components assemble, activated caspase 1 processes cytosolic pro-IL-1β into mature IL-1β ready to be secreted from the cells (Lamkanfi and Dixit, 2014). Non-canonical inflammasomes do not depend on ASC and NLRPs, and instead, the agonists, such as cytosolic LPS, directly activate caspase-11 (mouse) or caspase-4 and −5 (human) resulting in proteolytic processing of gasdermin D causing an inflammatory type of cell death termed pyroptosis (Broz et al., 2020). There are crossovers between canonical and non-canonical inflammasomes as caspase-1 can also proteolytically activate gasdermin D (Kayagaki et al., 2015). Autophagy can indirectly suppress inflammasome activation by removing sources of mitochondrial DAMPs and ROS (Nakahira et al., 2011; Sumpter et al., 2016; Zhou et al., 2011). Individual inflammasome components are substrates for autophagic degradation: AIM2 (Liu et al., 2016; Shi et al., 2012), NLRP3 and ASC (Mehto et al., 2019). PYRIN (TRIM20) targets pro-Caspase-1, NLRP1, and NLRP3 for autophagic degradation (Kimura et al., 2015). Of note, anti-inflammatory cytokines, such as IL-10, suppress inflammasome activation through mitophagy lowering dysfunctional mitochondrial burden and associated NLRP3 activation in macrophages in colitis (Ip et al., 2017).
Whether non-canonical inflammasomes and pyroptosis-mediated cell death are targeted by autophagy is beginning to be explored. Autophagy factors, such as GABARAPL2, one of the seven mAtg8s, negatively control the non-canonical (murine caspase-11) inflammasome (Eren et al., 2020). GABARAPL2 tapers caspase-11 activation by LPS (Eren et al., 2020). This may relate to the findings that GABARAPs affect integrity and function of endolysosomal organelles (Kumar et al., 2020; Nakamura et al., 2020). Human IRGM, discussed above, does not affect this process (although it affects canonical inflammasome (Mehto et al., 2019)) but in mice its paralog Irgm2 does (Eren et al., 2020; Finethy et al., 2020). Atg5 or Atg14, components of both canonical and non-canonical autophagy pathways, counter non-canonical inflammasome activation and pyroptosis (Finethy et al., 2020). Forms of inflammatory cell death other then pyroptosis, e.g. necroptosis, are influenced by autophagy factors in intestinal inflammation, cancer, and other relevant models (Aden et al., 2018; Lim et al., 2019; Matsuzawa-Ishimoto et al., 2020; Matsuzawa-Ishimoto et al., 2017).
Too much of type I IFN (or for that matter any inflammatory response) may lead to pathology. Protein complexes that stimulate type I IFN production can be suppressed by autophagy factors or autophagy itself. The ATG5–ATG12 complex, independently of autophagy, inhibits RIG-I (Jounai et al., 2007). Similarly, ATG9A negatively controls trafficking of the ER-associated STING sensor and inhibits type I IFN activation (Saitoh et al., 2009). Absence of autophagy enhances RIG-I-like signaling via MAVS (Tal et al., 2009). TRIM21 targets both IKKβ (Niida et al., 2010) and IRF3 (Kimura et al., 2015) for autophagic degradation. cGAS is a substrate for selective autophagy (Chen et al., 2016) along with RIG-I, and TLR3 (Jena et al., 2020). Autophagy factors ATG16L1 and SLRs, TAX1BP1 and SQSTM1, suppress TRIF (Samie et al., 2018; Yang et al., 2017), an adaptor that activates type I IFN downstream of TLR3 and TLR4 via IRF3 (Liu et al., 2015). Autophagy furthermore suppresses type I IFN in response to intestinal microbiota (Martin et al., 2018).
Of note, one has to be mindful of cell types and type of inflammation affected by canonical and non-canonical autophagy in each specific study. Certain types of hyperinflammation such as inflammasome-driven processes observed upon genetic ablation of autophagy or LAP seem to originate, in experimental systems, predominantly but not exclusively from myeloid cells including macrophages and monocytes. Most cells in the body can produce type I IFN using various systems, of which cGAS-STING and TLR-TRIF, RIG-I-MAVS targeted by autophagy as discussed above, are some of the key pathways (McNab et al., 2015). For example, RIG-I induces type I IFN after infection with RNA viruses in fibroblasts and conventional dendritic cells, whereas plasmacytoid dendritic cells (pDCs) preferentially use TLRs (Kato et al., 2005). Although pDCs have been considered as dominant responders, recent studies indicate that in vivo a variety of other cell types are important sources of type I IFN (Ali et al., 2019), and this needs to be taken into account.
In conclusion, autophagy or individual autophagy factors target key intracellular pro-inflammatory signaling assemblies and platforms for inhibition or degradation to limit inflammation.
TBK1 contributes to control of autophagy
TBK1 plays a role in autophagy and inflammation in a special way (Figure 2E–H). TBK1 is best known to immunologists in the context of type I IFN responses (Fitzgerald et al., 2003; Liu et al., 2015) downstream of PAMPs and DAMPs (Figure 2E). TBK1 activates type I IFN (Fitzgerald et al., 2003; Liu et al., 2015) via IRF3 and, together with IKKε (Balka et al., 2020), activates NFκB (Figure 2E). TBK1 influences inflammatory processes and diseases in neuroinflammation (Ahmad et al., 2016; Gerbino et al., 2020), autoimmunity (Hasan and Yan, 2016), cancer (Zhu et al., 2019), and viral infections. For example, severe COVID-19 is associated with genetic polymorphisms in TBK1 and other parts of the type I IFN activation pathways (Zhang et al., 2020a). TBK1 works as a kinase activating type I IFN response downstream of RIG-I-MAVS, MDA5-MAVS, cGAS-STING and TLR-TRIF pathways responding to cytosolic dsRNA, dsDNA, and other PAMPs (Liu et al., 2015) (Figure 2E). cGAS recognizes dsDNA, generates a second messenger cyclic-AMP-GMP (cGAMP) which in turn activates STING and TBK1 to induce phosphorylation of IRF3 and activate type I IFN expression (Li et al., 2013; Liu et al., 2015; Zhang et al., 2019).
In addition to type I IFN activation, TBK1 is important for autophagy (Kumar et al., 2019; Pilli et al., 2012; Ravenhill et al., 2019; Richter et al., 2016; Vargas et al., 2019; Wild et al., 2011; Zachari et al., 2019) (Figure 2E,F). TBK1 works in autophagosome formation (Kumar et al., 2019; Zachari et al., 2019) and interacts with and phosphorylates autophagic receptors (Pilli et al., 2012; Ravenhill et al., 2019; Richter et al., 2016; Vargas et al., 2019; Wild et al., 2011) (Figure 2F–G). Additionally, TBK1 has direct connections with immunometabolism regulators, mTOR (Antonia et al., 2019; Zhu et al., 2019) and AMPK (Zhao et al., 2018) (Figure 2B).
Multiple SLRs (OPTN1, NDP52, SQSTM1, TAX1BP1) depend on TBK1 for optimal receptor function in their antibacterial and antiviral autophagic roles (Pilli et al., 2012; Sparrer et al., 2017; Thurston et al., 2009; Wild et al., 2011). TBK1 extensively phosphorylates multiple regions in SLRs (Richter et al., 2016), including those that recognize LC3 on autophagosomal membranes (Wild et al., 2011) and ubiquitin on the cargo (Pilli et al., 2012) (Figure 2F).
TBK1 intersects with TRIMs, a group of proteins that participate in autophagy as regulators and selective autophagy receptors (Di Rienzo et al., 2020; Kimura et al., 2015; Kimura et al., 2016; Mandell et al., 2014; Versteeg et al., 2013). TBK1 associates with TRIM23 in response to permissive HSV-1 infection, phosphorylates S403 of the UBA domain of SQSTM1, and the tripartite complex TRIM23-TBK1-SQSTSM1 (Sparrer et al., 2017) augments SQSTM1’s ability to recognize ubiquitin-opsonized targets. SQSTSM1 also plays a role in feedback inhibition of type I IFN output by curbing STING-TBK1 activation (Prabakaran et al., 2018). SQSTM1, phosphorylated by TBK1 at Ser403 within its ubiquitin binding domain (Pilli et al., 2012), efficiently binds to K63-ubiquitinated STING to promote its selective autophagic degradation thus attenuating type I IFN response to cytosolic dsDNA and other stimuli (Prabakaran et al., 2018).
In addition to autophagic receptors, TBK1 is involved in autophagosome formation (Kumar et al., 2019) (Figure 2G) and stabilizes a subset of mAtg8s in their lipidated state (Herhaus et al., 2020) (Figure 2H). In these functions, TBK1 is involved in anti-bacterial autophagy (Pilli et al., 2012; Thurston et al., 2009; Wild et al., 2011), anti-viral autophagy (Sparrer et al., 2017; Yamashiro et al., 2020), mitophagy (Vargas et al., 2019), and even in ER-phagy when it is provoked by bacterial PAMPs (Moretti et al., 2017).
Whereas type I IFN is a pillar of antiviral responses, in bacterial infections it has been generally considered as a promotor of bacterial survival and pathogenesis (Ahn and Barber, 2019). However, cGAS-STING-TBK1 induces autophagy, and it can eliminate intracellular bacteria. For example, the activation of the STING-TBK1 system by bacterial CDNs (e.g. c-di-AMP, c-di-GMP), which are mimetics of cGAMP, or bacterial dsDNA released into the cytosol, represents a PAMP-PRR combination that activates antibacterial autophagy in a TBK1-dependent fashion (Figure 2E–G). The simultaneous type I IFN activation may be incidental to antibacterial action of TBK1 through autophagy. A mixed-function model emerges with autophagy and type I IFN as two parallel cGAS-STING-TBK1 outputs, that contribute to antiviral actions (Gui et al., 2019; Yamashiro et al., 2020). In bacterial infections however, the antimicrobial effects are attributable to autophagy whereas type I IFN activation may be an epiphenomenon that is neutral or with immunological consequence by suppressing inflammasome-driven inflammation (Burke et al., 2020; Guarda et al., 2011; Labzin et al., 2016; Ludigs et al., 2012; Reboldi et al., 2014) of relevance in diseases such as tuberculosis (Berry et al., 2010; Novikov et al., 2011).
In summary, TBK1 is a multifaceted regulator of autophagy pathway, including initiation, cargo capture, and preparation of autophagosomes for maturation. It is also at a special crossroad between type I IFN responses and autophagy.
Autophagy affects a spectrum of disease through inflammation
From infections to neurodegeneration, autophagy plays a role in human diseases (Levine and Kroemer, 2019; Mizushima and Levine, 2020). Here, we provide vignettes, rather than a comprehensive coverage, emphasising autophagy’s ability to modulate inflammation.
Neurodegeneration:
Parkinson’s disease (PD) has been associated with inflammation, including elevated IL-1β, TNFα, IFNγ, IL-6, IL-4, IL-10, IL-15, etc., detected in patient serum and CSF (Dzamko et al., 2015). Some of the predisposition factors such as α-synuclein, which can act as a DAMP to stimulate PRRs, and LRRK2 (a member of the RIPK family) have been associated with autophagy (Dzamko et al., 2015). Exogenously endocytosed α-synuclein can cause lysosomal damage and induce autophagy (Hoffmann et al., 2019), which can also be caused by protopathic tau (Jia et al., 2020b) associated with Alzheimer’s disease. Of particular interest is the system consisting of PRKN (parkin, an E3 ubiquitin ligase) and PINK1 (a ubiquitin kinase) (Figure 2D) capable of driving particularly efficient form of mitophagy (Youle, 2019). In mouse models, PINK1 and PRKN have been difficult to connect with manifestations of parkinsonism. However, when Prkn−/− or Pink1−/− mice are subjected to mitochondrial stress (mutations in mitochondrial DNA, extreme exercise), two important phenotypes co-emerged – inflammation and symptoms of PD (Sliter et al., 2018). Both phenotypes could be rescued by eliminating STING in these mice, connecting the dots between mitophagy failure, mitochondrial DAMP release, and the cytosolic DNA sensing system working though STING to induce inflammation (Sliter et al., 2018).
Cancer:
The role of autophagy in cancer has been extensively studied from tumor cell biology to clinical trials (Towers et al., 2020). In principle, cell-autonomous effects of autophagy are tumor-suppressive during oncogenesis and tumor-promoting in established tumors. FUNDC1 is one of the many selective mitophagy receptors embedded in the outer mitochondrial membrane (Figure 2D), removing defunct mitochondria through autophagy under hypoxic conditions and stress. Mechanistically, FUNDC1’s LIR is tonically phosphorylated by Src under normoxic conditions, whereas hypoxia induces its dephosphorylation thus allowing it to bind to LC3 and promote selective mitophagy (Liu et al., 2012). FUNDC1 accumulates in human hepatocellular carcinomas (Li et al., 2019), the most common primary liver cancer in adults associated with chronic liver inflammation. Genetic ablation of Fundc1 in hepatocytes promotes hepatocarcinogenesis associated with dysfunctional mitochondria, inflammasome activation, release of IL-1β, and hepatocyte proliferation (Li et al., 2019). Later on, FUNDC1 supports primary hepatic tumor growth through mitochondrial homeostasis and metabolic impact on cancer cells.
With cancer, the role of cancer cell-extrinsic factors, e.g. tumor microenvironment and even more importantly immune surveillance, is just as critical. Reprograming tumor-associated macrophages in murine models of hepatocarcinoma and melanoma from alternatively activated to tumor-suppressing classically activated phenotypes through inhibition of lysosomal function and activation of TFEB (see MERiT above as a potential context) plus switching to glycolysis, can decrease Treg cells and enhance antitumor activity of infiltrating IFNγ-producing CD8+ T (Chen et al., 2018). Metabolomics and other measures indicate that CD8+ T cells switch more readily to glycolysis in tamoxifen-inducible whole body Atg14l−/−, Atg5−/−, or Atg6l1−/− mice to reject mammary breast tumor implants (DeVorkin et al., 2019). Along those lines, autophagy contributes to T cell tolerizing processes in the liver that normally counter continuous stimulation with microbial products brought in by hepatic circulation (Swadling et al., 2020). This leads to tumor immunotolerance, attributable to enablement of regulatory T cells (Poillet-Perez et al., 2020).
Recent studies indicate that pancreatic ductal adenocarcinoma cells (PDAC) downregulate MHC-I glycoproteins on their surface, important for recalcitrance of PDAC to immunotherapy with check point inhibitors (Yamamoto et al., 2020). This occurs through lysosomal degradation of MHC I in PDAC cells potentially via a conventional autophagy mechanism (based on ATG14L, ATG13 or ULK1 dependence) and is not related to non-canonical process of LAP or LANDO, ruled out in Rbcn (RUBICON) downregulation experiments (Yamamoto et al., 2020). Nevertheless, LAP by tumor-associated macrophages appears to be immunosuppressive (Cunha et al., 2018). In a model with engrafted murine B16F10 melanoma, reduced tumor volume and lung micrometastases have been observed in mice with myeloid cell lacking Rbcn (a distinguishing feature of LAP; Figure 1C) as well as Becn1, Atg5, but not Fip200 or Atg14l (necessary for canonical autophagy). In these mice, better pro-inflammatory expression of STING-mediated type I IFN responses spur anti-tumor effects of tumor infiltrating T cells (Cunha et al., 2018). Another recent study has shown that radiation treatment causes stronger ‘abscopal’ (remote site) benefit in a syngeneic model with mouse mammary carcinoma TS/A cells when Atg5−/− or Atg7−/− TS/A clones were used or upon chloroquine treatment (chloroquine inhibits autophagy maturation and lysosomal function) (Yamazaki et al., 2020). This anti-tumor activity correlates with increased type I IFN secretion and depends on mitochondrial DNA release, STING, and cGAS (Yamazaki et al., 2020). Chloroquine (its derivative hydroxychloroquine is being tested in cancer clinical trials), is most likely inhibiting mitophagy thus enhancing radiation treatment-induced immunogenicity through mitoDNA-STING-cGAS and type I IFN response. In another study, the interplay of autophagy factors and STING was of significance in a high tumor mutational burden model (Poillet-Perez et al., 2020). Thus, given that type I IFN plays an important anti-cancer role in general (Zitvogel et al., 2015), and that cGAS synergizes with immune check point blockade by anti-PDL-1 antibody (Wang et al., 2017), the role of autophagy (or LAP) together with type I IFN responses connected through STING-TBK1 is an area of clinical promise in cancer.
Cardiovascular disease:
Certain aspects of heart failure, atherogenesis, and cardiovascular diseases have been associated with autophagy, inflammation, and immunometabolism and the role of macrophages in atherosclerotic plaques (Razani et al., 2012; Zhang et al., 2020b). In mouse models, defective autophagy in macrophages is associated with proatherogenic inflammasome activation IL-1β secretion and inflammation, attributed to increased burden of cholesterol crystals in macrophages, a hallmark of atheromatous plaques, (Razani et al., 2012). Of note, cholesterol crystals, if undissolved, remain trapped in lysosomal compartments and can cause lysosomal damage and downstream NLRP3 inflammasome activation (Duewell et al., 2010). Curiously, high protein diet has been recently linked to mTOR activation in macrophages, suppressing mitophagy and autophagy thus contributing to plaque progression and instability (Zhang et al., 2020b).
Infectious diseases:
In infection models, the role of autophagy (or individual ATG genes) reveals enormous complexity. Xenophagy, when it occurs in vivo, is accompanied by an equal or more important action of autophagy in controlling inflammation and tissue damage. This is evident from mouse models of Mtb infection, with xenophagy being less remarkable and best detected synergistically with antibiotics (Choi et al., 2018; Pahari et al., 2020), whereby absence of autophagy factors is dominated by inflammation. Increased lung inflammation is observed in Mtb-infected mice with defective Atg5 in the myeloid lineage (Castillo et al., 2012; Kimmey et al., 2015; Watson et al., 2012) and whole-body Smurf1−/− mice (Franco et al., 2017). Mice lacking Gal3 or Gal8 are more susceptible to Mtb infections, suggesting components of MERiT being at play (Jia et al., 2018; Jia et al., 2020b). The hyperinflammatory state observed in mice upon inactivation of certain Atg genes can be protective in some experimental situations: myeloid-specific inactivation of Fip200, Atg5, Atg7, or Atg14l, etc., elevates basal respiratory tract inflammation and confers resistance to influenza (Lu et al., 2016) or reactivation of murine herpesvirus (Park et al., 2016).
With regards to COVID-19, there are connections between coronavirus infections and autophagy. Both coronaviruses and autophagy involve formation of intracellular double membranes, albeit they differ (Cottam et al., 2011; Cottam et al., 2014; Fung and Liu, 2019; Gassen, 2020; Prentice et al., 2004; Reggiori et al., 2010). The features and the basis for COVID-19 morbidity and mortality are complex and not understood. They vary from asymptomatic infection to respiratory failure, and can involve multiple organs and systems (Berlin et al., 2020; Gandhi et al., 2020; van de Veerdonk et al., 2020; Zhang et al., 2020c). At the immunological level, this has been proposed to corelate with pre-existing T cells that are cross-reactive with the spike protein of SARS-CoV-2 (Braun et al., 2020) or with the proposed three immunotypes (Mathew et al., 2020). Recent analyses have uncovered, sometimes cryptic during other infections, genetic errors in type I IFN pathways including TLR3 and TBK1 (Zhang et al., 2020a) and autoantibodies against type I IFN (Bastard et al., 2020) in patients with severe COVID-19. Of note, TLR3 and dsRNA ligands induce autophagy (Delgado et al., 2008), whereas TBK1 plays a role in autophagy (Kumar et al., 2019; Pilli et al., 2012; Sparrer et al., 2017; Thurston et al., 2009; Wild et al., 2011). Clearly, autophagy and type I IFN systems overlap as discussed in sections above. Given an important role of type I IFN in COVID-19, and the interplay between autophagy and type I IFN as well as the fact that autophagy affects other systems that drive or suppress inflammatory responses during SARS-CoV-2 infection, these relationships are worth investigating.
Concluding remarks
The field of autophagy in immunity and inflammation continues to evolve in both fundamental and translational aspects. In principle, most human diseases have an inflammatory component and this in turn creates a window of opportunity and a challenge to develop autophagy-based therapeutics. To achieve this safely, we must continue studies aimed at understanding autophagy’s fundamental biology. We also must keep in mind two things at all times: that autophagy is a primordial immune mechanism and that it is hardwired into quality control processes and metabolism. This may appear difficult to disentangle. An encouragement to embrace this complexity can be found in Albert Einstein’s quote: “In the middle of every difficulty lies opportunity”. Autophagy provides both.
Figure 3. Summary of autophagy relationship with inflammation, innate immunity, adaptive immunity, and diseases.
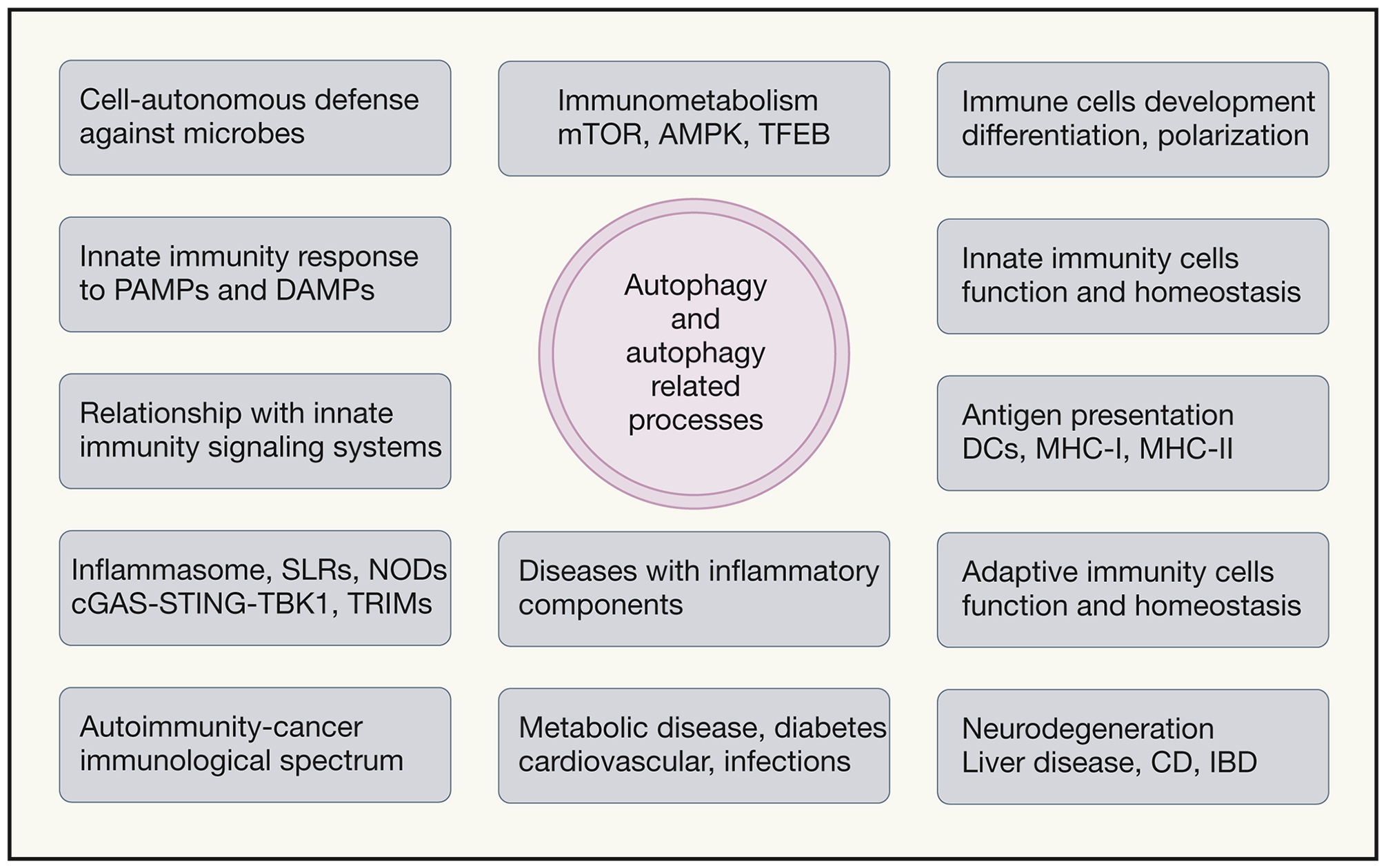
Autophagy and autophagy related processes (LAP, LANDO, microautophagy, CMA) affect immunometabolism and both innate and adaptive immune cells and immune responses. Left, special functions that autophagy plays: defense against intracellular microbes (xenophagy), response to pathogen associated molecular patterns, relationships with various innate immune response signaling platforms such as inflammasomes, NLRs including NODs, STING-TBK1, and TRIMs. Center top, immunometabolism and connections to AMPK, mTORC1, TFEB and autophagy. Right, contributions of autophagy (through immunometabolism, organelle homeostasis and degradation of specific protein complexes) to differentiation, polarization and function of macrophages, DCs, B1 B cells, plasma cells, T cells, and in antigen presentation. Bottom, examples of diseases with inflammatory components affected by autophagy.
Acknowledgments
The author dedicates this work to the memory of Dr. Beth Levine. VD thanks Drs. Judy Cannon and Andrew Thorburn for comments. This work was supported by NIH grants R37AI042999 and R01AI111935 and center grant P20GM121176.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The author declares no competing interests.
References
- Aden K, Tran F, Ito G, Sheibani-Tezerji R, Lipinski S, Kuiper JW, Tschurtschenthaler M, Saveljeva S, Bhattacharyya J, Hasler R, et al. (2018) ATG16L1 orchestrates interleukin-22 signaling in the intestinal epithelium via cGAS-STING. J Exp Med 215, 2868–2886.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agudo-Canalejo J, Schultz SW, Chino H, Migliano SM, Saito C, Koyama-Honda I, Stenmark H, Brech A, May AI, Mizushima N, et al. (2020). Wetting regulates autophagy of phase-separated compartments and the cytosol. Nature. [DOI] [PubMed] [Google Scholar]
- Ahmad L, Zhang SY, Casanova JL, and Sancho-Shimizu V (2016). Human TBK1: A Gatekeeper of Neuroinflammation. Trends Mol Med 22, 511–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn J, and Barber GN (2019). STING signaling and host defense against microbial infection. Exp Mol Med 51, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali S, Mann-Nuttel R, Schulze A, Richter L, Alferink J, and Scheu S (2019). Sources of Type I Interferons in Infectious Immunity: Plasmacytoid Dendritic Cells Not Always in the Driver’s Seat. Front Immunol 10, 778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsaleh G, Panse I, Swadling L, Zhang H, Richter FC, Meyer A, Lord J, Barnes E, Klenerman P, Green C, et al. (2020). Autophagy in T cells from aged donors is maintained by spermidine and correlates with function and vaccine responses. Elife 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An H, and Harper JW (2018). Systematic analysis of ribophagy in human cells reveals bystander flux during selective autophagy. Nat Cell Biol 20, 135–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonia RJ, Castillo J, Herring LE, Serafin DS, Liu P, Graves LM, Baldwin AS, and Hagan RS (2019). TBK1 Limits mTORC1 by Promoting Phosphorylation of Raptor Ser877. Sci Rep 9, 13470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balka KR, Louis C, Saunders TL, Smith AM, Calleja DJ, D’Silva DB, Moghaddas F, Tailler M, Lawlor KE, Zhan Y, et al. (2020). TBK1 and IKKepsilon Act Redundantly to Mediate STING-Induced NF-kappaB Responses in Myeloid Cells. Cell Rep 31, 107492. [DOI] [PubMed] [Google Scholar]
- Ballabio A, and Bonifacino JS (2020). Lysosomes as dynamic regulators of cell and organismal homeostasis. Nat Rev Mol Cell Biol 21, 101–118. [DOI] [PubMed] [Google Scholar]
- Bastard P, Rosen LB, Zhang Q, Michailidis E, Hoffmann HH, Zhang Y, Dorgham K, Philippot Q, Rosain J, Beziat V, et al. (2020). Auto-antibodies against type I IFNs in patients with life-threatening COVID-19. Science. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender S, Reuter A, Eberle F, Einhorn E, Binder M, and Bartenschlager R (2015). Activation of Type I and III Interferon Response by Mitochondrial and Peroxisomal MAVS and Inhibition by Hepatitis C Virus. PLoS Pathog 11, e1005264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlin DA, Gulick RM, and Martinez FJ (2020). Severe Covid-19. N Engl J Med. [DOI] [PubMed] [Google Scholar]
- Berry MP, Graham CM, McNab FW, Xu Z, Bloch SA, Oni T, Wilkinson KA, Banchereau R, Skinner J, Wilkinson RJ, et al. (2010). An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature 466, 973–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birgisdottir AB, Lamark T, and Johansen T (2013). The LIR motif - crucial for selective autophagy. Journal of cell science 126, 3237–3247. [DOI] [PubMed] [Google Scholar]
- Birmingham CL, Smith AC, Bakowski MA, Yoshimori T, and Brumell JH (2006). Autophagy controls Salmonella infection in response to damage to the Salmonella-containing vacuole. J Biol Chem 281, 11374–11383. [DOI] [PubMed] [Google Scholar]
- Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, and Johansen T (2005). p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol 171, 603–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagih J, Coulombe F, Vincent EE, Dupuy F, Galicia-Vazquez G, Yurchenko E, Raissi TC, van der Windt GJ, Viollet B, Pearce EL, et al. (2015). The energy sensor AMPK regulates T cell metabolic adaptation and effector responses in vivo. Immunity 42, 41–54. [DOI] [PubMed] [Google Scholar]
- Boada-Romero E, Martinez J, Heckmann BL, and Green DR (2020). The clearance of dead cells by efferocytosis. Nat Rev Mol Cell Biol 21, 398–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botbol Y, and Macian F (2015). Assays for Monitoring Macroautophagy Activity in T cells. Methods Mol Biol 1343, 143–153. [DOI] [PubMed] [Google Scholar]
- Brady OA, Martina JA, and Puertollano R (2018). Emerging roles for TFEB in the immune response and inflammation. Autophagy 14, 181–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun J, Loyal L, Frentsch M, Wendisch D, Georg P, Kurth F, Hippenstiel S, Dingeldey M, Kruse B, Fauchere F, et al. (2020). SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature. [DOI] [PubMed] [Google Scholar]
- Brest P, Lapaquette P, Souidi M, Lebrigand K, Cesaro A, Vouret-Craviari V, Mari B, Barbry P, Mosnier JF, Hebuterne X, et al. (2011). A synonymous variant in IRGM alters a binding site for miR-196 and causes deregulation of IRGM-dependent xenophagy in Crohn’s disease. Nat Genet 43, 242–245. [DOI] [PubMed] [Google Scholar]
- Broz P, Pelegrin P, and Shao F (2020). The gasdermins, a protein family executing cell death and inflammation. Nat Rev Immunol 20, 143–157. [DOI] [PubMed] [Google Scholar]
- Burke TP, Engstrom P, Chavez RA, Fonbuena JA, Vance RE, and Welch MD (2020). Inflammasome-mediated antagonism of type I interferon enhances Rickettsia pathogenesis. Nat Microbiol 5, 688–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo EF, Dekonenko A, Arko-Mensah J, Mandell MA, Dupont N, Jiang S, Delgado-Vargas M, Timmins GS, Bhattacharya D, Yang H, et al. (2012). Autophagy protects against active tuberculosis by suppressing bacterial burden and inflammation. Proceedings of the National Academy of Sciences of the United States of America 109, E3168–3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman NM, Boothby MR, and Chi H (2020). Metabolic coordination of T cell quiescence and activation. Nat Rev Immunol 20, 55–70. [DOI] [PubMed] [Google Scholar]
- Chauhan S, Kumar S, Jain A, Ponpuak M, Mudd MH, Kimura T, Choi SW, Peters R, Mandell M, Bruun JA, et al. (2016). TRIMs and Galectins Globally Cooperate and TRIM16 and Galectin-3 Co-direct Autophagy in Endomembrane Damage Homeostasis. Dev Cell 39, 13–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan S, Mandell MA, and Deretic V (2015). Mechanism of action of the tuberculosis and Crohn disease risk factor IRGM in autophagy. Autophagy, 0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Xie J, Fiskesund R, Dong W, Liang X, Lv J, Jin X, Liu J, Mo S, Zhang T, et al. (2018). Chloroquine modulates antitumor immune response by resetting tumor-associated macrophages toward M1 phenotype. Nat Commun 9, 873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Meng Q, Qin Y, Liang P, Tan P, He L, Zhou Y, Chen Y, Huang J, Wang RF, et al. (2016). TRIM14 Inhibits cGAS Degradation Mediated by Selective Autophagy Receptor p62 to Promote Innate Immune Responses. Mol Cell. [DOI] [PubMed] [Google Scholar]
- Chen YH, Du W, Hagemeijer MC, Takvorian PM, Pau C, Cali A, Brantner CA, Stempinski ES, Connelly PS, Ma HC, et al. (2015). Phosphatidylserine vesicles enable efficient en bloc transmission of enteroviruses. Cell 160, 619–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SW, Gu Y, Peters RS, Salgame P, Ellner JJ, Timmins GS, and Deretic V (2018). Ambroxol Induces Autophagy and Potentiates Rifampin Antimycobacterial Activity. Antimicrob Agents Chemother 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke AJ, Riffelmacher T, Braas D, Cornall RJ, and Simon AK (2018). B1a B cells require autophagy for metabolic homeostasis and self-renewal. J Exp Med 215, 399–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke AJ, and Simon AK (2019). Autophagy in the renewal, differentiation and homeostasis of immune cells. Nat Rev Immunol 19, 170–183. [DOI] [PubMed] [Google Scholar]
- Consortium (2007). Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447, 661–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottam EM, Maier HJ, Manifava M, Vaux LC, Chandra-Schoenfelder P, Gerner W, Britton P, Ktistakis NT, and Wileman T (2011). Coronavirus nsp6 proteins generate autophagosomes from the endoplasmic reticulum via an omegasome intermediate. Autophagy 7, 1335–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottam EM, Whelband MC, and Wileman T (2014). Coronavirus NSP6 restricts autophagosome expansion. Autophagy 10, 1426–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha LD, Yang M, Carter R, Guy C, Harris L, Crawford JC, Quarato G, Boada-Romero E, Kalkavan H, Johnson MDL, et al. (2018). LC3-Associated Phagocytosis in Myeloid Cells Promotes Tumor Immune Tolerance. Cell 175, 429–441 e416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daussy CF, Monard SC, Guy C, Munoz-Gonzalez S, Chazal M, Anthonsen MW, Jouvenet N, Henry T, Dreux M, Meurs EF, et al. (2021). The Inflammasome Components NLRP3 and ASC Act in Concert with IRGM To Rearrange the Golgi Apparatus during Hepatitis C Virus Infection. J Virol 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Duve C, and Wattiaux R (1966). Functions of lysosomes. Annu Rev Physiol 28, 435–492. [DOI] [PubMed] [Google Scholar]
- Delgado MA, Elmaoued RA, Davis AS, Kyei G, and Deretic V (2008). Toll-like receptors control autophagy. Embo J 27, 1110–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V (2005). Autophagy in innate and adaptive immunity. Trends Immunol 26, 523–528. [DOI] [PubMed] [Google Scholar]
- Deretic V (2010). Autophagy of intracellular microbes and mitochondria: two sides of the same coin? F1000 Biol Rep 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V, and Levine B (2018). Autophagy balances inflammation in innate immunity. Autophagy, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V, Saitoh T, and Akira S (2013). Autophagy in infection, inflammation and immunity. Nat Rev Immunol 13, 722–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVorkin L, Pavey N, Carleton G, Comber A, Ho C, Lim J, McNamara E, Huang H, Kim P, Zacharias LG, et al. (2019). Autophagy Regulation of Metabolism Is Required for CD8(+) T Cell Anti-tumor Immunity. Cell Rep 27, 502–513 e505. [DOI] [PubMed] [Google Scholar]
- Di Cara F, Andreoletti P, Trompier D, Vejux A, Bulow MH, Sellin J, Lizard G, Cherkaoui-Malki M, and Savary S (2019). Peroxisomes in Immune Response and Inflammation. Int J Mol Sci 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Rienzo M, Romagnoli A, Antonioli M, Piacentini M, and Fimia GM (2020). TRIM proteins in autophagy: selective sensors in cell damage and innate immune responses. Cell Death Differ 27, 887–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit E, Boulant S, Zhang Y, Lee AS, Odendall C, Shum B, Hacohen N, Chen ZJ, Whelan SP, Fransen M, et al. (2010). Peroxisomes are signaling platforms for antiviral innate immunity. Cell 141, 668–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Yang Y, Zou Z, Zhao Y, Ci B, Zhong L, Bhave M, Wang L, Kuo YC, Zang X, et al. (2020). Sorting nexin 5 mediates virus-induced autophagy and immunity. Nature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duewell P, Kono H, Rayner KJ, Sirois CM, Vladimer G, Bauernfeind FG, Abela GS, Franchi L, Nunez G, Schnurr M, et al. (2010). NLRP3 inflammasomes are required for atherogenesis and activated by cholesterol crystals. Nature 464, 1357–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont N, Jiang S, Pilli M, Ornatowski W, Bhattacharya D, and Deretic V (2011). Autophagy-based unconventional secretory pathway for extracellular delivery of IL-1beta. EMBO J 30, 4701–4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzamko N, Geczy CL, and Halliday GM (2015). Inflammation is genetically implicated in Parkinson’s disease. Neuroscience 302, 89–102. [DOI] [PubMed] [Google Scholar]
- Eren E, Planes R, Bagayoko S, Bordignon PJ, Chaoui K, Hessel A, Santoni K, Pinilla M, Lagrange B, Burlet-Schiltz O, et al. (2020). Irgm2 and Gate-16 cooperatively dampen Gram-negative bacteria-induced caspase-11 response. EMBO Rep 21, e50829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira AR, Magalhaes AC, Camoes F, Gouveia A, Vieira M, Kagan JC, and Ribeiro D (2016). Hepatitis C virus NS3–4A inhibits the peroxisomal MAVS-dependent antiviral signalling response. J Cell Mol Med 20, 750–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finethy R, Dockterman J, Kutsch M, Orench-Rivera N, Wallace GD, Piro AS, Luoma S, Haldar AK, Hwang S, Martinez J, et al. (2020). Dynamin-related Irgm proteins modulate LPS-induced caspase-11 activation and septic shock. EMBO Rep 21, e50830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald KA, McWhirter SM, Faia KL, Rowe DC, Latz E, Golenbock DT, Coyle AJ, Liao SM, and Maniatis T (2003). IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat Immunol 4, 491–496. [DOI] [PubMed] [Google Scholar]
- Fletcher K, Ulferts R, Jacquin E, Veith T, Gammoh N, Arasteh JM, Mayer U, Carding SR, Wileman T, Beale R, et al. (2018). The WD40 domain of ATG16L1 is required for its non-canonical role in lipidation of LC3 at single membranes. EMBO J 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco LH, Nair VR, Scharn CR, Xavier RJ, Torrealba JR, Shiloh MU, and Levine B (2017). The Ubiquitin Ligase Smurf1 Functions in Selective Autophagy of Mycobacterium tuberculosis and Anti-tuberculous Host Defense. Cell Host Microbe 21, 59–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung TS, and Liu DX (2019). Human Coronavirus: Host-Pathogen Interaction. Annu Rev Microbiol 73, 529–557. [DOI] [PubMed] [Google Scholar]
- Galluzzi L, and Green DR (2019). Autophagy-Independent Functions of the Autophagy Machinery. Cell 177, 1682–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi RT, Lynch JB, and Del Rio C (2020). Mild or Moderate Covid-19. N Engl J Med. [DOI] [PubMed] [Google Scholar]
- Gassen NC P. J; Bajaj T; Dethloff F; Emanuel J; Weckmann K; Heinz DE; Heinemann N; ennarz M; Richter A (2020). Analysis of sars-cov-2-controlled autophagy reveals spermidine, mk-2206, and niclosamide as putative antiviral therapeutics. BioRxiv. [Google Scholar]
- Gerbino V, Kaunga E, Ye J, Canzio D, O’Keeffe S, Rudnick ND, Guarnieri P, Lutz CM, and Maniatis T (2020). The Loss of TBK1 Kinase Activity in Motor Neurons or in All Cell Types Differentially Impacts ALS Disease Progression in SOD1 Mice. Neuron 106, 789–805 e785. [DOI] [PubMed] [Google Scholar]
- Gerstenmaier L, Pilla R, Herrmann L, Herrmann H, Prado M, Villafano GJ, Kolonko M, Reimer R, Soldati T, King JS, et al. (2015). The autophagic machinery ensures nonlytic transmission of mycobacteria. Proc Natl Acad Sci U S A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes LC, and Dikic I (2014). Autophagy in antimicrobial immunity. Mol Cell 54, 224–233. [DOI] [PubMed] [Google Scholar]
- Gonzalez A, Hall MN, Lin SC, and Hardie DG (2020). AMPK and TOR: The Yin and Yang of Cellular Nutrient Sensing and Growth Control. Cell Metab 31, 472–492. [DOI] [PubMed] [Google Scholar]
- Guarda G, Braun M, Staehli F, Tardivel A, Mattmann C, Forster I, Farlik M, Decker T, Du Pasquier RA, Romero P, et al. (2011). Type I interferon inhibits interleukin-1 production and inflammasome activation. Immunity 34, 213–223. [DOI] [PubMed] [Google Scholar]
- Gui X, Yang H, Li T, Tan X, Shi P, Li M, Du F, and Chen ZJ (2019). Autophagy induction via STING trafficking is a primordial function of the cGAS pathway. Nature 567, 262–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Chitiprolu M, Roncevic L, Javalet C, Hemming FJ, Trung MT, Meng L, Latreille E, Tanese de Souza C, McCulloch D, et al. (2017). Atg5 Disassociates the V1V0-ATPase to Promote Exosome Production and Tumor Metastasis Independent of Canonical Macroautophagy. Dev Cell 43, 716–730 e717. [DOI] [PubMed] [Google Scholar]
- Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, and Deretic V (2004). Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell 119, 753–766. [DOI] [PubMed] [Google Scholar]
- Hasan M, and Yan N (2016). Therapeutic potential of targeting TBK1 in autoimmune diseases and interferonopathies. Pharmacol Res 111, 336–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckmann BL, and Green DR (2019). LC3-associated phagocytosis at a glance. J Cell Sci 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckmann BL, Teubner BJW, Boada-Romero E, Tummers B, Guy C, Fitzgerald P, Mayer U, Carding S, Zakharenko SS, Wileman T, et al. (2020). Noncanonical function of an autophagy protein prevents spontaneous Alzheimer’s disease. Sci Adv 6, eabb9036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckmann BL, Teubner BJW, Tummers B, Boada-Romero E, Harris L, Yang M, Guy CS, Zakharenko SS, and Green DR (2019). LC3-Associated Endocytosis Facilitates beta-Amyloid Clearance and Mitigates Neurodegeneration in Murine Alzheimer’s Disease. Cell 178, 536–551 e514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henault J, Martinez J, Riggs JM, Tian J, Mehta P, Clarke L, Sasai M, Latz E, Brinkmann MM, Iwasaki A, et al. (2012). Noncanonical Autophagy Is Required for Type I Interferon Secretion in Response to DNA-Immune Complexes. Immunity 37, 986–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herhaus L, Bhaskara RM, Lystad AH, Gestal-Mato U, Covarrubias-Pinto A, Bonn F, Simonsen A, Hummer G, and Dikic I (2020). TBK1-mediated phosphorylation of LC3C and GABARAP-L2 controls autophagosome shedding by ATG4 protease. EMBO Rep 21, e48317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzig S, and Shaw RJ (2018). AMPK: guardian of metabolism and mitochondrial homeostasis. Nat Rev Mol Cell Biol 19, 121–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann AC, Minakaki G, Menges S, Salvi R, Savitskiy S, Kazman A, Vicente Miranda H, Mielenz D, Klucken J, Winkler J, et al. (2019). Extracellular aggregated alpha synuclein primarily triggers lysosomal dysfunction in neural cells prevented by trehalose. Sci Rep 9, 544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inomata M, Xu S, Chandra P, Meydani SN, Takemura G, Philips JA, and Leong JM (2020). Macrophage LC3-associated phagocytosis is an immune defense against Streptococcus pneumoniae that diminishes with host aging. Proceedings of the National Academy of Sciences, 202015368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip WKE, Hoshi N, Shouval DS, Snapper S, and Medzhitov R (2017). Anti-inflammatory effect of IL-10 mediated by metabolic reprogramming of macrophages. Science 356, 513–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jena KK, Mehto S, Nath P, Chauhan NR, Sahu R, Dhar K, Das SK, Kolapalli SP, Murmu KC, Jain A, et al. (2020). Autoimmunity gene IRGM suppresses cGAS-STING and RIG-I-MAVS signaling to control interferon response. EMBO Rep 21, e50051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia J, Abudu YP, Claude-Taupin A, Gu Y, Kumar S, Choi SW, Peters R, Mudd MH, Allers L, Salemi M, et al. (2018). Galectins Control mTOR in Response to Endomembrane Damage. Mol Cell 70, 120–135 e128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia J, Bissa B, Brecht L, Allers L, Choi SW, Gu Y, Zbinden M, Burge MR, Timmins G, Hallows K, et al. (2020a). AMPK, a Regulator of Metabolism and Autophagy, Is Activated by Lysosomal Damage via a Novel Galectin-Directed Ubiquitin Signal Transduction System. Mol Cell 77, 951–969 e959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia J, Claude-Taupin A, Gu Y, Choi SW, Peters R, Bissa B, Mudd MH, Allers L, Pallikkuth S, Lidke KA, et al. (2020b). Galectin-3 Coordinates a Cellular System for Lysosomal Repair and Removal. Dev Cell 52, 69–87 e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia J, Claude-Taupin A, Gu Y, Choi SW, Peters R, Bissa B, Mudd MH, Allers L, Pallikkuth S, Lidke KA, et al. (2020c). MERIT, a cellular system coordinating lysosomal repair, removal and replacement. Autophagy 16, 1539–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia W, Pua HH, Li QJ, and He YW (2011). Autophagy regulates endoplasmic reticulum homeostasis and calcium mobilization in T lymphocytes. Journal of immunology 186, 1564–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen T, and Lamark T (2011). Selective autophagy mediated by autophagic adapter proteins. Autophagy 7, 279–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jounai N, Takeshita F, Kobiyama K, Sawano A, Miyawaki A, Xin KQ, Ishii KJ, Kawai T, Akira S, Suzuki K, et al. (2007). The Atg5 Atg12 conjugate associates with innate antiviral immune responses. Proc Natl Acad Sci U S A 104, 14050–14055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karmakar M, Minns M, Greenberg EN, Diaz-Aponte J, Pestonjamasp K, Johnson JL, Rathkey JK, Abbott DW, Wang K, Shao F, et al. (2020). N-GSDMD trafficking to neutrophil organelles facilitates IL-1beta release independently of plasma membrane pores and pyroptosis. Nat Commun 11, 2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Sato S, Yoneyama M, Yamamoto M, Uematsu S, Matsui K, Tsujimura T, Takeda K, Fujita T, Takeuchi O, et al. (2005). Cell type-specific involvement of RIG-I in antiviral response. Immunity 23, 19–28. [DOI] [PubMed] [Google Scholar]
- Kaushik S, and Cuervo AM (2018). The coming of age of chaperone-mediated autophagy. Nat Rev Mol Cell Biol 19, 365–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayagaki N, Stowe IB, Lee BL, O’Rourke K, Anderson K, Warming S, Cuellar T, Haley B, Roose-Girma M, Phung QT, et al. (2015). Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature 526, 666–671. [DOI] [PubMed] [Google Scholar]
- Keller MD, Ching KL, Liang FX, Dhabaria A, Tam K, Ueberheide BM, Unutmaz D, Torres VJ, and Cadwell K (2020a). Decoy exosomes provide protection against bacterial toxins. Nature 579, 260–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller MD, Torres VJ, and Cadwell K (2020b). Autophagy and microbial pathogenesis. Cell Death Differ 27, 872–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmey JM, Huynh JP, Weiss LA, Park S, Kambal A, Debnath J, Virgin HW, and Stallings CL (2015). Unique role for ATG5 in neutrophil-mediated immunopathology during M. tuberculosis infection. Nature 528, 565–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura T, Jain A, Choi SW, Mandell MA, Schroder K, Johansen T, and Deretic V (2015). TRIM-mediated precision autophagy targets cytoplasmic regulators of innate immunity. J Cell Biol 210, 973–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura T, Jia J, Kumar S, Choi SW, Gu Y, Mudd M, Dupont N, Jiang S, Peters R, Farzam F, et al. (2017). Dedicated SNAREs and specialized TRIM cargo receptors mediate secretory autophagy. EMBO J 36, 42–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura T, Mandell M, and Deretic V (2016). Precision autophagy directed by receptor regulators - emerging examples within the TRIM family. J Cell Sci 129, 881–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkin V, Lamark T, Sou YS, Bjorkoy G, Nunn JL, Bruun JA, Shvets E, McEwan DG, Clausen TH, Wild P, et al. (2009). A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell 33, 505–516. [DOI] [PubMed] [Google Scholar]
- Kumar S, Gu Y, Abudu YP, Bruun JA, Jain A, Farzam F, Mudd M, Anonsen JH, Rusten TE, Kasof G, et al. (2019). Phosphorylation of Syntaxin 17 by TBK1 Controls Autophagy Initiation. Dev Cell 49, 130–144 e136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Jain A, Choi SW, da Silva GPD, Allers L, Mudd MH, Peters RS, Anonsen JH, Rusten TE, Lazarou M, et al. (2020). Mammalian Atg8 proteins and the autophagy factor IRGM control mTOR and TFEB at a regulatory node critical for responses to pathogens. Nat Cell Biol 22, 973–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labzin LI, Lauterbach MA, and Latz E (2016). Interferons and inflammasomes: Cooperation and counterregulation in disease. J Allergy Clin Immunol 138, 37–46. [DOI] [PubMed] [Google Scholar]
- Lamkanfi M, and Dixit VM (2014). Mechanisms and Functions of Inflammasomes. Cell 157, 1013–1022. [DOI] [PubMed] [Google Scholar]
- Lazarou M, Sliter DA, Kane LA, Sarraf SA, Wang C, Burman JL, Sideris DP, Fogel AI, and Youle RJ (2015). The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature 524, 309–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Lamech L, Johns E, and Overholtzer M (2020). Selective Lysosome Membrane Turnover Is Induced by Nutrient Starvation. Dev Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidal AM, Huang HH, Marsh T, Solvik T, Zhang D, Ye J, Kai F, Goldsmith J, Liu JY, Huang YH, et al. (2020). The LC3-conjugation machinery specifies the loading of RNA-binding proteins into extracellular vesicles. Nat Cell Biol 22, 187–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B (2005). Eating Oneself and Uninvited Guests; Autophagy-Related Pathways in Cellular Defense. Cell 120, 159–162. [DOI] [PubMed] [Google Scholar]
- Levine B, and Kroemer G (2019). Biological Functions of Autophagy Genes: A Disease Perspective. Cell 176, 11–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Mizushima N, and Virgin HW (2011). Autophagy in immunity and inflammation. Nature 469, 323–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Capan E, Zhao Y, Zhao J, Stolz D, Watkins SC, Jin S, and Lu B (2006). Autophagy is induced in CD4+ T cells and important for the growth factor-withdrawal cell death. J Immunol 177, 5163–5168. [DOI] [PubMed] [Google Scholar]
- Li W, Li Y, Siraj S, Jin H, Fan Y, Yang X, Huang X, Wang X, Wang J, Liu L, et al. (2019). FUN14 Domain-Containing 1-Mediated Mitophagy Suppresses Hepatocarcinogenesis by Inhibition of Inflammasome Activation in Mice. Hepatology 69, 604–621. [DOI] [PubMed] [Google Scholar]
- Li XD, Wu J, Gao D, Wang H, Sun L, and Chen ZJ (2013). Pivotal roles of cGAS-cGAMP signaling in antiviral defense and immune adjuvant effects. Science 341, 1390–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J, Park H, Heisler J, Maculins T, Roose-Girma M, Xu M, McKenzie B, van Lookeren Campagne M, Newton K, and Murthy A (2019). Autophagy regulates inflammatory programmed cell death via turnover of RHIM-domain proteins. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SC, and Hardie DG (2018). AMPK: Sensing Glucose as well as Cellular Energy Status. Cell Metab 27, 299–313. [DOI] [PubMed] [Google Scholar]
- Liu GY, and Sabatini DM (2020). mTOR at the nexus of nutrition, growth, ageing and disease. Nat Rev Mol Cell Biol 21, 183–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Feng D, Chen G, Chen M, Zheng Q, Song P, Ma Q, Zhu C, Wang R, Qi W, et al. (2012). Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nature cell biology 14, 177–185. [DOI] [PubMed] [Google Scholar]
- Liu S, Cai X, Wu J, Cong Q, Chen X, Li T, Du F, Ren J, Wu YT, Grishin NV, et al. (2015). Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science 347, aaa2630. [DOI] [PubMed] [Google Scholar]
- Liu T, Tang Q, Liu K, Xie W, Liu X, Wang H, Wang RF, and Cui J (2016). TRIM11 Suppresses AIM2 Inflammasome by Degrading AIM2 via p62-Dependent Selective Autophagy. Cell Rep 16, 1988–2002. [DOI] [PubMed] [Google Scholar]
- Lu Q, Yokoyama CC, Williams JW, Baldridge MT, Jin X, DesRochers B, Bricker T, Wilen CB, Bagaitkar J, Loginicheva E, et al. (2016). Homeostatic Control of Innate Lung Inflammation by Vici Syndrome Gene Epg5 and Additional Autophagy Genes Promotes Influenza Pathogenesis. Cell Host Microbe 19, 102–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludigs K, Parfenov V, Du Pasquier RA, and Guarda G (2012). Type I IFN-mediated regulation of IL-1 production in inflammatory disorders. Cellular and molecular life sciences : CMLS 69, 3395–3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lystad AH, Carlsson SR, de la Ballina LR, Kauffman KJ, Nag S, Yoshimori T, Melia TJ, and Simonsen A (2019). Distinct functions of ATG16L1 isoforms in membrane binding and LC3B lipidation in autophagy-related processes. Nat Cell Biol 21, 372–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Galluzzi L, Zitvogel L, and Kroemer G (2013). Autophagy and cellular immune responses. Immunity 39, 211–227. [DOI] [PubMed] [Google Scholar]
- Macian F (2019). Autophagy in T Cell Function and Aging. Front Cell Dev Biol 7, 213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maejima I, Takahashi A, Omori H, Kimura T, Takabatake Y, Saitoh T, Yamamoto A, Hamasaki M, Noda T, Isaka Y, et al. (2013). Autophagy sequesters damaged lysosomes to control lysosomal biogenesis and kidney injury. EMBO J 32, 2336–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandell MA, Jain A, Arko-Mensah J, Chauhan S, Kimura T, Dinkins C, Silvestri G, Munch J, Kirchhoff F, Simonsen A, et al. (2014). TRIM proteins regulate autophagy and can target autophagic substrates by direct recognition. Dev Cell 30, 394–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino G, Pietrocola F, Eisenberg T, Kong Y, Malik SA, Andryushkova A, Schroeder S, Pendl T, Harger A, Niso-Santano M, et al. (2014). Regulation of autophagy by cytosolic acetyl-coenzyme A. Mol Cell 53, 710–725. [DOI] [PubMed] [Google Scholar]
- Marshall RS, Hua Z, Mali S, McLoughlin F, and Vierstra RD (2019). ATG8-Binding UIM Proteins Define a New Class of Autophagy Adaptors and Receptors. Cell 177, 766–781 e724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin PK, Marchiando A, Xu R, Rudensky E, Yeung F, Schuster SL, Kernbauer E, and Cadwell K (2018). Autophagy proteins suppress protective type I interferon signalling in response to the murine gut microbiota. Nat Microbiol 3, 1131–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J, Cunha LD, Park S, Yang M, Lu Q, Orchard R, Li QZ, Yan M, Janke L, Guy C, et al. (2016). Noncanonical autophagy inhibits the autoinflammatory, lupus-like response to dying cells. Nature 533, 115–119. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Mathew D, Giles JR, Baxter AE, Oldridge DA, Greenplate AR, Wu JE, Alanio C, Kuri-Cervantes L, Pampena MB, D’Andrea K, et al. (2020). Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science 369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzawa-Ishimoto Y, Hine A, Shono Y, Rudensky E, Lazrak A, Yeung F, Neil JA, Yao X, Chen YH, Heaney T, et al. (2020). An intestinal organoid-based platform that recreates susceptibility to T-cell-mediated tissue injury. Blood 135, 2388–2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzawa-Ishimoto Y, Shono Y, Gomez LE, Hubbard-Lucey VM, Cammer M, Neil J, Dewan MZ, Lieberman SR, Lazrak A, Marinis JM, et al. (2017). Autophagy protein ATG16L1 prevents necroptosis in the intestinal epithelium. J Exp Med 214, 3687–3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwan WA, Tam JC, Watkinson RE, Bidgood SR, Mallery DL, and James LC (2013). Intracellular antibody-bound pathogens stimulate immune signaling via the Fc receptor TRIM21. Nat Immunol 14, 327–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNab F, Mayer-Barber K, Sher A, Wack A, and O’Garra A (2015). Type I interferons in infectious disease. Nat Rev Immunol 15, 87–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina DL, Di Paola S, Peluso I, Armani A, De Stefani D, Venditti R, Montefusco S, Scotto-Rosato A, Prezioso C, Forrester A, et al. (2015). Lysosomal calcium signalling regulates autophagy through calcineurin and TFEB. Nat Cell Biol 17, 288–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehto S, Jena KK, Nath P, Chauhan S, Kolapalli SP, Das SK, Sahoo PK, Jain A, Taylor GA, and Chauhan S (2019). The Crohn’s Disease Risk Factor IRGM Limits NLRP3 Inflammasome Activation by Impeding Its Assembly and by Mediating Its Selective Autophagy. Mol Cell 73, 429–445 e427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mejlvang J, Olsvik H, Svenning S, Bruun JA, Abudu YP, Larsen KB, Brech A, Hansen TE, Brenne H, Hansen T, et al. (2018). Starvation induces rapid degradation of selective autophagy receptors by endosomal microautophagy. J Cell Biol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melia TJ, Lystad AH, and Simonsen A (2020). Autophagosome biogenesis: From membrane growth to closure. J Cell Biol 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y, Li G, Zhang X, Xu H, and Abraham SN (2015). A TRP Channel Senses Lysosome Neutralization by Pathogens to Trigger Their Expulsion. Cell 161, 1306–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller BC, Zhao Z, Stephenson LM, Cadwell K, Pua HH, Lee HK, Mizushima NN, Iwasaki A, He YW, Swat W, et al. (2008). The autophagy gene ATG5 plays an essential role in B lymphocyte development. Autophagy 4, 309–314. [DOI] [PubMed] [Google Scholar]
- Mills EL, Kelly B, and O’Neill LAJ (2017). Mitochondria are the powerhouses of immunity. Nat Immunol 18, 488–498. [DOI] [PubMed] [Google Scholar]
- Mizushima N, and Levine B (2020). Autophagy in Human Diseases. N Engl J Med 383, 1564–1576. [DOI] [PubMed] [Google Scholar]
- Mocholi E, Dowling SD, Botbol Y, Gruber RC, Ray AK, Vastert S, Shafit-Zagardo B, Coffer PJ, and Macian F (2018). Autophagy Is a Tolerance-Avoidance Mechanism that Modulates TCR-Mediated Signaling and Cell Metabolism to Prevent Induction of T Cell Anergy. Cell Rep 24, 1136–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montespan C, Marvin SA, Austin S, Burrage AM, Roger B, Rayne F, Faure M, Campell EM, Schneider C, Reimer R, et al. (2017). Multi-layered control of Galectin-8 mediated autophagy during adenovirus cell entry through a conserved PPxY motif in the viral capsid. PLoS Pathog 13, e1006217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti J, Roy S, Bozec D, Martinez J, Chapman JR, Ueberheide B, Lamming DW, Chen ZJ, Horng T, Yeretssian G, et al. (2017). STING Senses Microbial Viability to Orchestrate Stress-Mediated Autophagy of the Endoplasmic Reticulum. Cell 171, 809–823 e813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morishita H, and Mizushima N (2019). Diverse Cellular Roles of Autophagy. Annu Rev Cell Dev Biol 35, 453–475. [DOI] [PubMed] [Google Scholar]
- Nakagawa I, Amano A, Mizushima N, Yamamoto A, Yamaguchi H, Kamimoto T, Nara A, Funao J, Nakata M, Tsuda K, et al. (2004). Autophagy defends cells against invading group A Streptococcus. Science 306, 1037–1040. [DOI] [PubMed] [Google Scholar]
- Nakahira K, Haspel JA, Rathinam VA, Lee SJ, Dolinay T, Lam HC, Englert JA, Rabinovitch M, Cernadas M, Kim HP, et al. (2011). Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol 12, 222–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura S, Shigeyama S, Minami S, Shima T, Akayama S, Matsuda T, Esposito A, Napolitano G, Kuma A, Namba-Hamano T, et al. (2020). LC3 lipidation is essential for TFEB activation during the lysosomal damage response to kidney injury. Nat Cell Biol. [DOI] [PubMed] [Google Scholar]
- Napolitano G, and Ballabio A (2016). TFEB at a glance. Journal of Cell Science In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napolitano G, Di Malta C, Esposito A, de Araujo MEG, Pece S, Bertalot G, Matarese M, Benedetti V, Zampelli A, Stasyk T, et al. (2020). A substrate-specific mTORC1 pathway underlies Birt-Hogg-Dube syndrome. Nature 585, 597–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niida M, Tanaka M, and Kamitani T (2010). Downregulation of active IKK beta by Ro52-mediated autophagy. Molecular immunology 47, 2378–2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noad J, von der Malsburg A, Pathe C, Michel MA, Komander D, and Randow F (2017). LUBAC-synthesized linear ubiquitin chains restrict cytosol-invading bacteria by activating autophagy and NF-kappaB. Nat Microbiol 2, 17063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda NN, Wang Z, and Zhang H (2020). Liquid-liquid phase separation in autophagy. J Cell Biol 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novikov A, Cardone M, Thompson R, Shenderov K, Kirschman KD, Mayer-Barber KD, Myers TG, Rabin RL, Trinchieri G, Sher A, et al. (2011). Mycobacterium tuberculosis triggers host type I IFN signaling to regulate IL-1beta production in human macrophages. Journal of immunology 187, 2540–2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill LA, and Hardie DG (2013). Metabolism of inflammation limited by AMPK and pseudo-starvation. Nature 493, 346–355. [DOI] [PubMed] [Google Scholar]
- O’Neill LA, Kishton RJ, and Rathmell J (2016). A guide to immunometabolism for immunologists. Nat Rev Immunol 16, 553–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M, Yoshimori T, Suzuki T, Sagara H, Mizushima N, and Sasakawa C (2005). Escape of intracellular Shigella from autophagy. Science 307, 727–731. [DOI] [PubMed] [Google Scholar]
- Pahari S, Negi S, Aqdas M, Arnett E, Schlesinger LS, and Agrewala JN (2020). Induction of autophagy through CLEC4E in combination with TLR4: an innovative strategy to restrict the survival of Mycobacterium tuberculosis. Autophagy 16, 1021–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos C, Kirchner P, Bug M, Grum D, Koerver L, Schulze N, Poehler R, Dressler A, Fengler S, Arhzaouy K, et al. (2017). VCP/p97 cooperates with YOD1, UBXD1 and PLAA to drive clearance of ruptured lysosomes by autophagy. EMBO J 36, 135–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Buck MD, Desai C, Zhang X, Loginicheva E, Martinez J, Freeman ML, Saitoh T, Akira S, Guan JL, et al. (2016). Autophagy Genes Enhance Murine Gammaherpesvirus 68 Reactivation from Latency by Preventing Virus-Induced Systemic Inflammation. Cell Host Microbe 19, 91–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce EL, Walsh MC, Cejas PJ, Harms GM, Shen H, Wang LS, Jones RG, and Choi Y (2009). Enhancing CD8 T-cell memory by modulating fatty acid metabolism. Nature 460, 103–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin AJ, Jiang X, Birmingham CL, So NS, and Brumell JH (2004). Recognition of bacteria in the cytosol of Mammalian cells by the ubiquitin system. Curr Biol 14, 806–811. [DOI] [PubMed] [Google Scholar]
- Pilli M, Arko-Mensah J, Ponpuak M, Roberts E, Master S, Mandell MA, Dupont N, Ornatowski W, Jiang S, Bradfute SB, et al. (2012). TBK-1 Promotes Autophagy-Mediated Antimicrobial Defense by Controlling Autophagosome Maturation. Immunity 37, 223–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poillet-Perez L, Sharp DW, Yang Y, Laddha SV, Ibrahim M, Bommareddy PK, Hu ZS, Vieth J, Haas M, Bosenberg MW, et al. (2020). Autophagy promotes growth of tumors with high mutational burden by inhibiting a T-cell immune response. Nature Cancer 1, 923–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponpuak M, Davis AS, Roberts EA, Delgado MA, Dinkins C, Zhao Z, Virgin H.W.t., Kyei GB, Johansen T, Vergne I, et al. (2010). Delivery of cytosolic components by autophagic adaptor protein p62 endows autophagosomes with unique antimicrobial properties. Immunity 32, 329–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponpuak M, Mandell MA, Kimura T, Chauhan S, Cleyrat C, and Deretic V (2015). Secretory autophagy. Curr Opin Cell Biol 35, 106–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabakaran T, Bodda C, Krapp C, Zhang BC, Christensen MH, Sun C, Reinert L, Cai Y, Jensen SB, Skouboe MK, et al. (2018). Attenuation of cGAS-STING signaling is mediated by a p62/SQSTM1-dependent autophagy pathway activated by TBK1. EMBO J 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentice E, Jerome WG, Yoshimori T, Mizushima N, and Denison MR (2004). Coronavirus replication complex formation utilizes components of cellular autophagy. J Biol Chem 279, 10136–10141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pua HH, Guo J, Komatsu M, and He YW (2009). Autophagy is essential for mitochondrial clearance in mature T lymphocytes. J Immunol 182, 4046–4055. [DOI] [PubMed] [Google Scholar]
- Puleston DJ, Zhang H, Powell TJ, Lipina E, Sims S, Panse I, Watson AS, Cerundolo V, Townsend AR, Klenerman P, et al. (2014). Autophagy is a critical regulator of memory CD8(+) T cell formation. Elife 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radulovic M, Schink KO, Wenzel EM, Nahse V, Bongiovanni A, Lafont F, and Stenmark H (2018). ESCRT-mediated lysosome repair precedes lysophagy and promotes cell survival. EMBO J 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randow F, and Youle RJ (2014). Self and nonself: how autophagy targets mitochondria and bacteria. Cell Host Microbe 15, 403–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravenhill BJ, Boyle KB, von Muhlinen N, Ellison CJ, Masson GR, Otten EG, Foeglein A, Williams R, and Randow F (2019). The Cargo Receptor NDP52 Initiates Selective Autophagy by Recruiting the ULK Complex to Cytosol-Invading Bacteria. Mol Cell 74, 320–329 e326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razani B, Feng C, Coleman T, Emanuel R, Wen H, Hwang S, Ting JP, Virgin HW, Kastan MB, and Semenkovich CF (2012). Autophagy links inflammasomes to atherosclerotic progression. Cell Metab 15, 534–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reboldi A, Dang EV, McDonald JG, Liang G, Russell DW, and Cyster JG (2014). Inflammation. 25-Hydroxycholesterol suppresses interleukin-1-driven inflammation downstream of type I interferon. Science 345, 679–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F, Monastyrska I, Verheije MH, Cali T, Ulasli M, Bianchi S, Bernasconi R, de Haan CA, and Molinari M (2010). Coronaviruses Hijack the LC3-I-positive EDEMosomes, ER-derived vesicles exporting short-lived ERAD regulators, for replication. Cell Host Microbe 7, 500–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro CM, Sarrami-Forooshani R, Setiawan LC, Zijlstra-Willems EM, van Hamme JL, Tigchelaar W, van der Wel NN, Kootstra NA, Gringhuis SI, and Geijtenbeek TB (2016). Receptor usage dictates HIV-1 restriction by human TRIM5alpha in dendritic cell subsets. Nature. [DOI] [PubMed] [Google Scholar]
- Richter B, Sliter DA, Herhaus L, Stolz A, Wang C, Beli P, Zaffagnini G, Wild P, Martens S, Wagner SA, et al. (2016). Phosphorylation of OPTN by TBK1 enhances its binding to Ub chains and promotes selective autophagy of damaged mitochondria. Proc Natl Acad Sci U S A 113, 4039–4044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riffelmacher T, Clarke A, Richter FC, Stranks A, Pandey S, Danielli S, Hublitz P, Yu Z, Johnson E, Schwerd T, et al. (2017). Autophagy-Dependent Generation of Free Fatty Acids Is Critical for Normal Neutrophil Differentiation. Immunity 47, 466–480 e465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riffelmacher T, Richter FC, and Simon AK (2018). Autophagy dictates metabolism and differentiation of inflammatory immune cells. Autophagy 14, 199–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinsztein DC, Bento CF, and Deretic V (2015). Therapeutic targeting of autophagy in neurodegenerative and infectious diseases. J Exp Med 212, 979–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha B, Chisholm D, Kell AM, and Mandell MA (2020). A non-canonical role for the autophagy machinery in anti-retroviral signaling mediated by TRIM5alpha. PLoS Pathog 16, e1009017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh T, Fujita N, Hayashi T, Takahara K, Satoh T, Lee H, Matsunaga K, Kageyama S, Omori H, Noda T, et al. (2009). Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc Natl Acad Sci U S A 106, 20842–20846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T, Omori H, Noda T, Yamamoto N, Komatsu M, et al. (2008). Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature 456, 264–268. [DOI] [PubMed] [Google Scholar]
- Salio M, Puleston DJ, Mathan TS, Shepherd D, Stranks AJ, Adamopoulou E, Veerapen N, Besra GS, Hollander GA, Simon AK, et al. (2014). Essential role for autophagy during invariant NKT cell development. Proc Natl Acad Sci U S A 111, E5678–5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samie M, Lim J, Verschueren E, Baughman JM, Peng I, Wong A, Kwon Y, Senbabaoglu Y, Hackney JA, Keir M, et al. (2018). Selective autophagy of the adaptor TRIF regulates innate inflammatory signaling. Nat Immunol 19, 246–254. [DOI] [PubMed] [Google Scholar]
- Sanjuan MA, Dillon CP, Tait SW, Moshiach S, Dorsey F, Connell S, Komatsu M, Tanaka K, Cleveland JL, Withoff S, et al. (2007). Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature 450, 1253–1257. [DOI] [PubMed] [Google Scholar]
- Saravia J, Raynor JL, Chapman NM, Lim SA, and Chi H (2020). Signaling networks in immunometabolism. Cell Res 30, 328–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck S (2020). Microautophagy - distinct molecular mechanisms handle cargoes of many sizes. J Cell Sci 133. [DOI] [PubMed] [Google Scholar]
- Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, Erdin SU, Huynh T, Medina D, Colella P, et al. (2011). TFEB links autophagy to lysosomal biogenesis. Science 332, 1429–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi CS, Shenderov K, Huang NN, Kabat J, Abu-Asab M, Fitzgerald KA, Sher A, and Kehrl JH (2012). Activation of autophagy by inflammatory signals limits IL-1beta production by targeting ubiquitinated inflammasomes for destruction. Nat Immunol 13, 255–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Zhao Y, Wang K, Shi X, Wang Y, Huang H, Zhuang Y, Cai T, Wang F, and Shao F (2015). Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature 526, 660–665. [DOI] [PubMed] [Google Scholar]
- Singh SB, Davis AS, Taylor GA, and Deretic V (2006). Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science 313, 1438–1441. [DOI] [PubMed] [Google Scholar]
- Skowyra ML, Schlesinger PH, Naismith TV, and Hanson PI (2018). Triggered recruitment of ESCRT machinery promotes endolysosomal repair. Science 360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sliter DA, Martinez J, Hao L, Chen X, Sun N, Fischer TD, Burman JL, Li Y, Zhang Z, Narendra DP, et al. (2018). Parkin and PINK1 mitigate STING-induced inflammation. Nature 561, 258–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MD, Harley ME, Kemp AJ, Wills J, Lee M, Arends M, von Kriegsheim A, Behrends C, and Wilkinson S (2018). CCPG1 Is a Non-canonical Autophagy Cargo Receptor Essential for ER-Phagy and Pancreatic ER Proteostasis. Dev Cell 44, 217–232 e211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparrer KMJ, Gableske S, Zurenski MA, Parker ZM, Full F, Baumgart GJ, Kato J, Pacheco-Rodriguez G, Liang C, Pornillos O, et al. (2017). TRIM23 mediates virus-induced autophagy via activation of TBK1. Nat Microbiol 2, 1543–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staring J, von Castelmur E, Blomen VA, van den Hengel LG, Brockmann M, Baggen J, Thibaut HJ, Nieuwenhuis J, Janssen H, van Kuppeveld FJ, et al. (2017). PLA2G16 represents a switch between entry and clearance of Picornaviridae. Nature 541, 412–416. [DOI] [PubMed] [Google Scholar]
- Sumpter R Jr., Sirasanagandla S, Fernandez AF, Wei Y, Dong X, Franco L, Zou Z, Marchal C, Lee MY, Clapp DW, et al. (2016). Fanconi Anemia Proteins Function in Mitophagy and Immunity. Cell 165, 867–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swadling L, Pallett LJ, Diniz MO, Baker JM, Amin OE, Stegmann KA, Burton AR, Schmidt NM, Jeffery-Smith A, Zakeri N, et al. (2020). Human Liver Memory CD8(+) T Cells Use Autophagy for Tissue Residence. Cell Rep 30, 687–698 e686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tal MC, Sasai M, Lee HK, Yordy B, Shadel GS, and Iwasaki A (2009). Absence of autophagy results in reactive oxygen species-dependent amplification of RLR signaling. Proc Natl Acad Sci U S A 106, 2770–2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talloczy Z, Jiang W, Virgin H.W.t., Leib DA, Scheuner D, Kaufman RJ, Eskelinen EL, and Levine B (2002). Regulation of starvation- and virus-induced autophagy by the eIF2alpha kinase signaling pathway. Proc Natl Acad Sci U S A 99, 190–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurston TL, Ryzhakov G, Bloor S, von Muhlinen N, and Randow F (2009). The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat Immunol 10, 1215–1221. [DOI] [PubMed] [Google Scholar]
- Thurston TL, Wandel MP, von Muhlinen N, Foeglein A, and Randow F (2012). Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature 482, 414–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towers CG, Wodetzki D, and Thorburn A (2020). Autophagy and cancer: Modulation of cell death pathways and cancer cell adaptations. J Cell Biol 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turco E, Witt M, Abert C, Bock-Bierbaum T, Su MY, Trapannone R, Sztacho M, Danieli A, Shi X, Zaffagnini G, et al. (2019). FIP200 Claw Domain Binding to p62 Promotes Autophagosome Formation at Ubiquitin Condensates. Mol Cell 74, 330–346 e311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdor R, Mocholi E, Botbol Y, Guerrero-Ros I, Chandra D, Koga H, Gravekamp C, Cuervo AM, and Macian F (2014). Chaperone-mediated autophagy regulates T cell responses through targeted degradation of negative regulators of T cell activation. Nat Immunol 15, 1046–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Veerdonk FL, Netea MG, van Deuren M, van der Meer JW, de Mast Q, Bruggemann RJ, and van der Hoeven H (2020). Kallikrein-kinin blockade in patients with COVID-19 to prevent acute respiratory distress syndrome. Elife 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas JNS, Wang C, Bunker E, Hao L, Maric D, Schiavo G, Randow F, and Youle RJ (2019). Spatiotemporal Control of ULK1 Activation by NDP52 and TBK1 during Selective Autophagy. Mol Cell 74, 347–362 e346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velayutham TS, Kumar S, Zhang X, Kose N, Walker DH, Winslow G, Crowe JE Jr., and McBride JW (2019). Ehrlichia chaffeensis Outer Membrane Protein 1-Specific Human Antibody-Mediated Immunity Is Defined by Intracellular TRIM21-Dependent Innate Immune Activation and Extracellular Neutralization. Infect Immun 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versteeg Gijs A., Rajsbaum R, Sánchez-Aparicio Maria T., Maestre Ana M., Valdiviezo J, Shi M, Inn K-S, Fernandez-Sesma A, Jung J, and García-Sastre A (2013). The E3-Ligase TRIM Family of Proteins Regulates Signaling Pathways Triggered by Innate Immune Pattern-Recognition Receptors. Immunity 38, 384–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J, Weiss E, Ben Mkaddem S, Mabire M, Choinier P-M, Picq O, Thibault-Sogorb T, Hegde P, Pishvaie D, Bens M, et al. (2020). LC3-associated phagocytosis protects against inflammation and liver fibrosis via immunoreceptor inhibitory signaling. Science Translational Medicine 12, eaaw8523. [DOI] [PubMed] [Google Scholar]
- Wang H, Hu S, Chen X, Shi H, Chen C, Sun L, and Chen ZJ (2017). cGAS is essential for the antitumor effect of immune checkpoint blockade. Proc Natl Acad Sci U S A 114, 1637–1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson RO, Manzanillo PS, and Cox JS (2012). Extracellular M. tuberculosis DNA Targets Bacteria for Autophagy by Activating the Host DNA-Sensing Pathway. Cell 150, 803–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weichhart T, Hengstschlager M, and Linke M (2015). Regulation of innate immune cell function by mTOR. Nat Rev Immunol 15, 599–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild P, Farhan H, McEwan DG, Wagner S, Rogov VV, Brady NR, Richter B, Korac J, Waidmann O, Choudhary C, et al. (2011). Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science 333, 228–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Araki K, Li S, Han JH, Ye L, Tan WG, Konieczny BT, Bruinsma MW, Martinez J, Pearce EL, et al. (2014). Autophagy is essential for effector CD8(+) T cell survival and memory formation. Nat Immunol 15, 1152–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K, Venida A, Yano J, Biancur DE, Kakiuchi M, Gupta S, Sohn ASW, Mukhopadhyay S, Lin EY, Parker SJ, et al. (2020). Autophagy promotes immune evasion of pancreatic cancer by degrading MHC-I. Nature 581, 100–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashiro LH, Wilson SC, Morrison HM, Karalis V, Chung JJ, Chen KJ, Bateup HS, Szpara ML, Lee AY, Cox JS, et al. (2020). Interferon-independent STING signaling promotes resistance to HSV-1 in vivo. Nat Commun 11, 3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki T, Kirchmair A, Sato A, Buque A, Rybstein M, Petroni G, Bloy N, Finotello F, Stafford L, Navarro Manzano E, et al. (2020). Mitochondrial DNA drives abscopal responses to radiation that are inhibited by autophagy. Nat Immunol 21, 1160–1171. [DOI] [PubMed] [Google Scholar]
- Yang Q, Liu TT, Lin H, Zhang M, Wei J, Luo WW, Hu YH, Zhong B, Hu MM, and Shu HB (2017). TRIM32-TAX1BP1-dependent selective autophagic degradation of TRIF negatively regulates TLR3/4-mediated innate immune responses. PLoS Pathog 13, e1006600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youle RJ (2019). Mitochondria-Striking a balance between host and endosymbiont. Science 365. [DOI] [PubMed] [Google Scholar]
- Zachari M, Gudmundsson SR, Li Z, Manifava M, Cugliandolo F, Shah R, Smith M, Stronge J, Karanasios E, Piunti C, et al. (2019). Selective Autophagy of Mitochondria on a Ubiquitin-Endoplasmic-Reticulum Platform. Dev Cell 50, 627–643 e625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Shang G, Gui X, Zhang X, Bai XC, and Chen ZJ (2019). Structural basis of STING binding with and phosphorylation by TBK1. Nature 567, 394–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Kenny S, Ge L, Xu K, and Schekman R (2015). Translocation of interleukin-1beta into a vesicle intermediate in autophagy-mediated secretion. Elife 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Bastard P, Liu Z, Le Pen J, Moncada-Velez M, Chen J, Ogishi M, Sabli IKD, Hodeib S, Korol C, et al. (2020a). Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science 370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Sergin I, Evans TD, Jeong SJ, Rodriguez-Velez A, Kapoor D, Chen S, Song E, Holloway KB, Crowley JR, et al. (2020b). High-protein diets increase cardiovascular risk by activating macrophage mTOR to suppress mitophagy. Nat Metab 2, 110–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Tan Y, Ling Y, Lu G, Liu F, Yi Z, Jia X, Wu M, Shi B, Xu S, et al. (2020c). Viral and host factors related to the clinical outcome of COVID-19. Nature 583, 437–440. [DOI] [PubMed] [Google Scholar]
- Zhao P, Wong KI, Sun X, Reilly SM, Uhm M, Liao Z, Skorobogatko Y, and Saltiel AR (2018). TBK1 at the Crossroads of Inflammation and Energy Homeostasis in Adipose Tissue. Cell 172, 731–743 e712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Z, Umemura A, Sanchez-Lopez E, Liang S, Shalapour S, Wong J, He F, Boassa D, Perkins G, Ali SR, et al. (2016). NF-kappaB Restricts Inflammasome Activation via Elimination of Damaged Mitochondria. Cell 164, 896–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Yazdi AS, Menu P, and Tschopp J (2011). A role for mitochondria in NLRP3 inflammasome activation. Nature 469, 221–225. [DOI] [PubMed] [Google Scholar]
- Zhu L, Li Y, Xie X, Zhou X, Gu M, Jie Z, Ko CJ, Gao T, Hernandez BE, Cheng X, et al. (2019). TBKBP1 and TBK1 form a growth factor signalling axis mediating immunosuppression and tumourigenesis. Nat Cell Biol 21, 1604–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Xie X, Zhang L, Wang H, Jie Z, Zhou X, Shi J, Zhao S, Zhang B, Cheng X, et al. (2018). TBK-binding protein 1 regulates IL-15-induced autophagy and NKT cell survival. Nat Commun 9, 2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zitvogel L, Galluzzi L, Kepp O, Smyth MJ, and Kroemer G (2015). Type I interferons in anticancer immunity. Nat Rev Immunol 15, 405–414. [DOI] [PubMed] [Google Scholar]


