Abstract
Clustered Regularly Interspaced Short Palindromic Repeats (CRISPRs) together with CRISPR-associated (Cas) proteins have catalysed a revolution in genetic engineering. Native CRISPR-Cas systems exist in many bacteria and archaea where they provide an adaptive immune response through sequence-specific degradation of an invading pathogen’s genome. This system has been reconfigured for use in genome editing, drug development, gene expression regulation, diagnostics, the prevention and treatment of cancers, and the treatment of genetic and infectious diseases. In recent years, CRISPR-Cas systems have been used in the diagnosis and control of viral diseases, for example, CRISPR-Cas12/13 coupled with new amplification techniques to improve the specificity of sequence-specific fluorescent probe detection. Importantly, CRISPR applications are both sensitive and specific and usually only require commonly available lab equipment. Unlike the canonical Cas9 which is guided to double-stranded DNA sites of interest, Cas13 systems target RNA sequences and thus can be employed in strategies directed against RNA viruses or for transcriptional silencing. Many challenges remain for these approach, including issues with specificity and the requirement for better mammalian delivery systems. In this review, we summarize the applications of CRISPR-Cas systems in controlling mammalian viral infections. Following necessary improvements, it is expected that CRISPR-Cas systems will be used effectively for such applications in the future.
Keywords: CRISPR, Cas13 protein, Viral disease, Detection kit, Viral RNA
Introduction
Numerous intracellular regulatory and defense mechanisms rely on complementary base-pairing between nucleic acid species for function. In addition to eukaryotic RNA interference (RNAi) and prokaryotic Argonaute-based systems, Clustered Regularly Interspaced Short Palindromic Repeats-CRISPR associated proteins (CRISPR-Cas) systems utilize nucleic acid base-pairing to suppress or destroy genetic targets (Koonin et al. 2017). These systems are adaptable and retain the memory of sequence targets derived from previous exposure to bacteriophages (Ishino et al. 1987; Mojica et al. 2005; Pourcel et al. 2005). Targeted sequences are retained in the form of spacers of 30–40 base-pairs (bp) each, separated by a series of short palindromic repeats (25–35 bp each). The cas genes themselves along with these repeats and spacers form the ‘CRISPR array’. In addition, the activity of CRISPR-Cas-mediated defenses requires proteins essential for Cas nuclease activity, the recognition of foreign nucleic acid and their uptake into the CRISPR array, as well as proteins critical for the recognition of the invading mobile genetic elements (MGEs).
CRISPR-Cas systems are divided into two distinct classes based on the structure of their effector complexes; Class 1 systems implement multiple Cas proteins for nucleic acid degradation while Class 2 systems use only one. These classes are further divided into six types and multiple subtypes based on differential Cas protein participation and CRISPR loci architectures: Class 1 systems include Cas types I, III, and IV while class 2 systems comprise types II, V, and VI (Makarova et al. 2018) (Table 1).
Table 1.
Characteristics of CRISPR-Cas systems
| Class | Type/Subtype | Effector | Nuclease domains | Target | Cut structure | tracrRNA requirement | PAM/PFS | Application |
|---|---|---|---|---|---|---|---|---|
| 1 | I | Cascade | HD nuclease domain of Cas3 | DNA | Single-strand cut (200-300nt) | NO | PAM | Genome editing, antimicrobials, gene regulation in bacteria and archaea |
| III | Csm/Cmr complex |
Cas10 PALM domain Cas7 Csm/Cmr complex |
DNA RNA |
Multiple sites | NO | Independent of PAM | Genome engineering and gene silencing | |
| IV | Complex | HD nuclease domain | DNA | Double strand | NO | PAM | Controlling plasmid propagation | |
| 2 | II | Cas9 | RuvC, HNH |
dsDNA RNA |
Blunt | Yes | 3′ GC-rich PAM |
Elimination of repetitive sequences specific Gene editing RNA knockdown RNA isolation (dCas 9) RNA imaging and tracking (dCas 9) Resistance against RNA viruses (Fn Cas9) Regulation of gene expression |
| V-A | Cas12a (Cpf1) | RuvC, NUC | dsDNA | Staggered, 5′-overhangs(7nt) | No | 5′ AT-rich PAM |
Gene editing Nucleic acid detection |
|
| V-B | Cas12b (C2c1) | RuvC | dsDNA | Staggered, 5′overhangs (5nt) | Yes | 5′ AT-rich PAM | Nucleic acid detection | |
| VI-A | Cas13a (C2c2) | 2xHEPN domain | ssRNA | Guide-dependent RNA cuts + collateral RNA cleavage | No | 3′ PFS: non-G |
RNA knockdown RNA imaging and tracking (dCas13a) Nucleic acid detection Resistance against RNA viruses |
|
| VI-B |
Cas13b (C2c6) |
2xHEPN Domain |
ssRNA | Guide-dependent RNA cuts + collateral RNA cleavage | No |
5′ PFS: non-C 3′ PFS: NANA/NNA |
RNA knockdown RNA editing Regulation of gene expression Nucleic acid detection |
dsDNA, double-stranded DNA; ssRNA, single-stranded RNA; PAM, protospacer adjacent motif; PFS, protospacer-flanking sequence
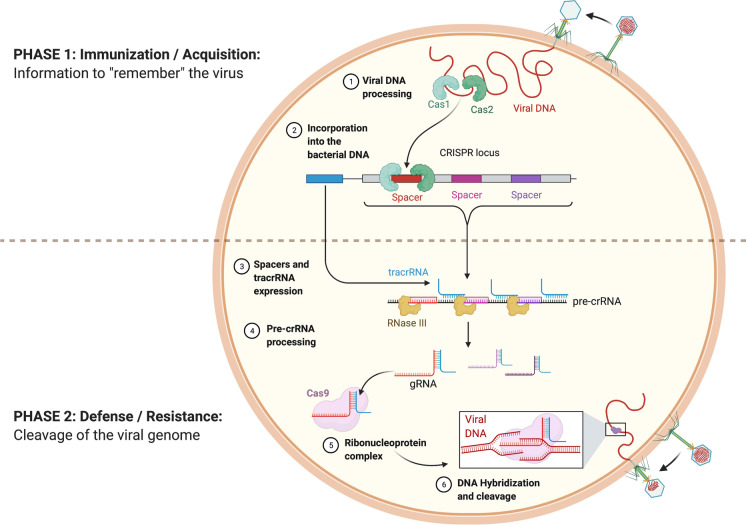
The molecular mechanisms of CRISPR-based prokaryotic adaptive immunity can be divided into two distinct processes: (i) immunization or acquistion, (ii) defense or resistance (Fig. 1). A new spacer sequence known as a protospacer is acquired from the foreign genetic elements (Yosef et al. 2012). To be acquired, the protospacer sequence for many CRISPR-Cas system types needs to be located next to a short motif called the protospacer-adjacent motif (PAM). Recognition of this motif serves as a reliable way to avoid the destruction of the host CRISPR array by its own CRISPR-Cas machinery. Cas1 and Cas2 protein activity is crucial for the adaptation stage (Yosef et al. 2012). At the crRNA biogenesis stage, precursor crRNAs (pre-crRNA) are produced by transcription of CRISPR array elements and processed into mature crRNA (Fig. 1). This maturation is performed by Cas proteins and/or cellular ribonucleases depending on the given CRISPR-Cas type (Deltcheva et al. 2011; Reeks et al. 2013). Mature crRNA is incorporated into the Cas protein effector complex. This complex uses the crRNA to interrogate foreign nucleic acids for sequence complementarity. If the requirements for both complementarity and a PAM site are satisfied, the Cas protein nuclease is activated, and the foreign genetic element is specifically targeted and degraded.
Fig. 1.
Bacterial CRISPR-Cas9 systems behave as an adaptive immune response against invading bacteriophages. Following infection (Phase 1), Cas1 and Cas2 mediate incorporation of short sequences of the viral genome as spacers within the bacterial CRISPR locus. At re-exposure (Phase 2), the CRISPR locus gets expressed as pre-crRNA, along with tracrRNA. The pre-crRNA is processed to yield guide RNAs (gRNAs) which bind the ribonucleoprotein Cas9 and target this complex to complementary sequences of the infiltrating bacteriophage genome, prompting its Cas9-mediated cleavage. Reprinted from “CRISPR-Cas9 Adaptive Immune System of Streptococcus pyogenes Against Bacteriophages”, by BioRender.com (2021). Retrieved from https://app.biorender.com/biorender-templates
Due to its high efficiency, specificity and modular nature, the CRISPR-Cas system has emerged as a revolutionary genetic engineering technology. This technology has accelerated our capacity for genome editing, drug development, control of gene expression, as well as in the diagnostics, prevention and treatment of a broad spectrum of diseases. In this review, we focus on the application of CRISPR-Cas systems to control viral diseases and the challenges faced in that field.
CRISPR-Cas9
Cas9 is the canonical class 2 type-II system nuclease. Since the first successful use of Streptococcus pyogenes Cas9 (SpCas9) for eukaryotic genome engineering, the CRISPR-Cas9 system remains the most widely adopted RNA-guided DNA targeting platform for genome editing (Cong et al. 2013; Jinek et al. 2012; Mali et al. 2013b). Cas9 contains two nuclease domains termed RuvC and HNH that function in DNA cleavage, which cut the target and non-target strands of DNA, respectively. Cas9 is guided by a trans-activating-crRNA (tracrRNAs)/crRNA complex (dual RNA hybrid), or an engineered chimeric form of this complex known as a single guide RNA (sgRNA) (Jinek et al. 2012). Upon recognition of the target DNA, Cas9 generates a double-strand break (DSB) upstream of the 5′-NGG PAM site (Deltcheva et al. 2011; Jiang and Doudna 2017; Strich and Chertow 2019). The programmability of the CRISPR-Cas9 system to target many sites for cleavage is considered its main advantage over zinc-finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs) for genome editing, technologies which were based on protein–DNA recognition (Doetschman and Georgieva 2017; Jiang and Doudna 2017). Repair of the targeted DSB and edits to the target gene are mediated by either the error-prone nonhomologous end-joining (NHEJ) or precise homology-directed repair (HDR) pathways, forming the basis of most CRISPR-Cas9 gene-editing strategies (Kim et al. 2011; Leenay et al. 2019).
Many recent studies have aimed to improve the efficiency and specificity of CRISPR-Cas9 genome editing as clinical applications of the current systems are impossible if on-target editing efficiencies are too low and non-specific off-target editing rates are too high. For one, the SpCas9 PAM requirements limit the number of targetable sites. Various strategies have been employed to expand Cas9 targeting scope, including the evolution of Cas9-derived variants with altered PAM requirements and the identification of Cas9 orthologs from other species (Chen et al. 2017; Hsu et al. 2014; Karvelis et al. 2017; Nishimasu et al. 2018).
Another strategy utilizes paired Cas9 nickases to improve site-specificity and decrease off-target DSB induction (Mali et al. 2013a; Ran et al. 2013). In this strategy, two Cas9 nickases (Cas9n) variants (D10A and H840A, respectively) are delivered together and guided to offset targets by individual sgRNAs, producing site-specific nicks on opposing strands of the DNA helix and inducing the NHEJ repair pathway. This design is proposed to reduce off-target editing activity as the repair of single-strand nicks are predominantly regulated by the high-fidelity base excision repair pathway.
The first SpCas9 mutants reported to target altered PAM sites with improved activity were SpCas9-EQR, SpCas9-VQR and, SpCas9-VRER (Kleinstiver et al. 2015). The recognition mechanisms of the altered PAM sites in these new systems were not determined, however. High-resolution crystal structures of the three SpCas9 variants in complex with a sgRNA and their target DNA identified an unexpected change in the phosphodiester backbone of the PAM duplex (induced by multiple mutations), which was proposed to be responsible for their altered PAM specificities (Hirano et al. 2016). In addition, several enhanced fidelity variants of SpCas9 such as eSpCas9 (1.1), SpCas9-HF1, and HeFSpCas9 were produced by rational design to increase on-target editing activity and improve intrinsic SpCas9 specificity (Chen et al. 2017; Kleinstiver et al. 2016; Kulcsár et al. 2017). SpCas9-NG and xCas9 are further Cas9 variants with the potential of broadening CRISPR–Cas9 PAM compatibility (Kim et al. 2020).
CRISPR–Cas12
The defining feature of Cas12, the prototypical type V Cas protein, is its capacity to cleave double-stranded DNA (dsDNA) in a staggered fashion near a 5′ PAM (TTTV) site generating 5- to 7-nucleotide overhangs. Target DNA cleavage is mediated by the single RuvC-like nuclease domain of Cas12. Unlike Cas9, Cas12a (also referred to as Cpf1) mediates pre-crRNA processing through auto-processing of its own crRNA array (Kleinstiver et al. 2019; Li et al. 2018a; Shmakov et al. 2015; Zetsche et al. 2015). This unique feature of the CRISPR–Cas12a platform is exploited in the design of simplified systems for simultaneous editing of multiple genes in vivo (Zetsche et al. 2017). Cas12a orthologs such as Cas12f have been shown to predominantly target ssDNA molecules while others such as Cas12i mainly cleave dsDNA targets (Harrington et al. 2018; Shmakov et al. 2015).
Beyond Cas9 and Cas12a effectors, Cas12b is another system potentially useful for genome editing in mammalian cells (Strecker et al. 2019; Teng et al. 2018). The native Alicyclobacillus acidoterrestris-derived Cas12b (AacCas12b) is considered unsuitable for use in mammalian genome editing applications because of the high temperatures required for nuclease activity (Shmakov et al. 2015). An engineered variant of AacCas12b that possesses nuclease activity across a wide temperature range, however, overcomes this limitation; directed by a chimeric sgRNA, this variant was demonstrated to successfully achieve genome editing in both a mammalian cell line and mice (Teng et al. 2018). The wild-type Bacillus hisashii-derived Cas12b (BhCas12b), though active at lower temperatures, cannot cleave target strands and in its native architecture cannot be utilized as an effective tool for genome editing. An engineered form of BhCas12b could be a promising addition to the toolkit for efficient genome editing in human cells (Strecker et al. 2019).
CRISPR–Cas13
Cas13 proteins (comprising subtypes A, B, C and D) are class 2 type VI nucleases. Type IV nucleases are programmable single-effector RNA-guided nucleases that specifically target and cleave RNA molecules. Precise RNA cleavage of single-stranded RNA (ssRNA) by Cas13 effectors is mediated by the function of dual higher eukaryotes and prokaryotes nucleotide-binding domains (HEPNs). These domains require recognition of a protospacer flanking sequence (PFS) at the 3′ end of the crRNA binding site for Cas13a and on both 5′ and 3′ ends for Cas13b.
The programmability of Cas13 effectors is promising for the development of a precise and flexible RNA editing platform (Abudayyeh et al. 2016; Cox et al. 2017; Gootenberg et al. 2017; Shmakov et al. 2015, 2017). For instance, the Cas13b ortholog from Prevotella sp. P5-125 (PspCas13b) was identified as an efficient tool for RNA knockdown activity in mammalian cells (Cox et al. 2017). The fusion of PspCas13b to a variant of the adenosine deaminase acting on RNA (ADAR) protein family was able to facilitate editing of transcripts, providing a platform for efficient and specific RNA editing and repair of disease-associated mutations in the human cell. The RNA targetability of other Cas13 variants (e.g. Cas 13a and Cas13d) has been successfully applied as a mammalian knockdown platform with improved specificity over RNAi (Abudayyeh et al. 2017; Konermann et al. 2018). The functional diversity among Cas13 subtypes and the low potential of off-target edits anticipates the expansion of RNA-targeting CRISPR-Cas systems into other fields, including their clinical translation to applications when DNA-targeting platforms are inappropriate.
Control of viral diseases using CRISPR-Cas systems
Cas9 systems
There are two ways in which CRISPR-Cas9 systems can be applied to the control of viral disease. Editing of mammalian genes relevant to the disease response is one approach, though many studies describe the capacity of CRISPR-Cas effectors to defend mammalian cells against DNA or RNA virus infection directly. Cas9 has demonstrated promising results in targeting both dsDNA viruses and ssRNA viruses with DNA intermediates (Liu et al. 2018; Ophinni et al. 2018; Wang et al. 2018b).
Human diseases
Human immunodeficiency virus (HIV)
HIV is an ssRNA retrovirus that infects human CD4+ T-cells, macrophages and dendritic cells. Active infection results in a profound reduction of CD4+ T-cells which can lead to a loss of cell-mediated immunity and increased risk of opportunistic infections (Johnson et al. 2013). Though antiretroviral treatment is successful in reducing the viral burden to undetectable levels, life-long treatment must be maintained. Following primary infection, the HIV genome is permanently integrated into the human genome as a DNA provirus. This provirus can be actively transcribed to produce progeny virus or remain dormant in a latent state. The latently infected cell can serve as a permanent viral reservoir which can reactivate upon cessation of antiretroviral treatment (Johnson et al. 2013).
CRISPR–Cas9 technology is a promising approach for inactivation of the integrated provirus or for preventing primary infection of naïve cells. The HIV long terminal repeats (LTRs), present at both 5′ and 3′ ends of the proviral DNA and serves as the binding site(s) for transcription factors, are attractive targets for CRISPR-Cas9-based therapeutic approaches. Using such a strategy, Hu et al. induced either complete excision of the integrated viral sequence or a fragment between two target sites within the same LTR in a latently infected cell line (Hu et al. 2014). Subsequent studies have detailed the efficiency of the CRISPR-Cas9 system in targeted cleavage of various regions of the proviral DNA (Kaminski et al. 2016b; Lebbink et al. 2017; Liao et al. 2015; Wang et al. 2016). In a study by Ophinni et al., lentiviral delivery of CRISPR-Cas9 constructs targeting the HIV-1 regulatory genes tat and rev suppressed their expression and successfully inhibited HIV-1 replication in both persistently and latently infected human CD4+ T-cell lines (Ophinni et al. 2018). Moreover, delivery of a recombinant adeno-associated viral (AAV) vector carrying Staphylococcus aureus Cas9 (SaCas9) and gRNAs targeting both LTR and gag sequences achieved effective cleavage of HIV-1 proviral DNA in transgenic mice and rats (Kaminski et al. 2016a). Further, the efficacy and function of CRISPR-Cas technologies have also been evaluated in humanized mouse models (Bella et al. 2018; Dash et al. 2019; Yin et al. 2017). Bella et al. demonstrated that treatment with lentiviral vectors expressing Cas9 and multiple HIV-targeted gRNAs abolished HIV proviral DNA from infected human peripheral blood mononuclear cells (PBMC) in humanized mice (Bella et al. 2018).
CRISPR-Cas9 can also target host genes critical for the HIV life-cycle. Dufour et al. used the CRISPR-Cas9 system to edit the human restriction factor TRIM5 to recognize the HIV capsid and inhibit active infection (Dufour et al. 2018). In another study, D366N mutation of the PSIP1 gene encoding host cell protein Lens Epithelial Derived Growth Factor p75 (LEDGF/p75) disrupted the integration of the HIV provirus into the host chromosome but preserved the cellular function of LEDGF/p75 (Lampi et al. 2019). In addition, the application of SpCas9 and U-modified tracrRNA in the ribonucleoprotein complex (RNP) delivery format was found to improve indel formation in LTRs and could reduce CCR5 surface receptor expression in primary CD4+ T-cells (Scott et al. 2019; Xu et al. 2019)Together, these results highlight the possibilities for CRISPR-Cas9 editing systems in achieving the long-sought-after functional HIV cure.
Hepatitis B virus (HBV)
Of the Hepadnaviridae family, HBV causes acute and chronic liver infections and though an effective HBV vaccine exists, curing chronic infections remains a challenge. The circular HBV DNA genome is partially double-stranded, with a short sense strand and a longer anti-sense strand. The episomal covalently closed circular DNA (cccDNA) form of the HBV genome functions as an important factor in the maintenance of HBV persistence and the inability to clear the virus (Arzumanyan et al. 2013; Dandri and Petersen 2016). Multiple in vitro and in vivo studies have shown that achieving a complete HBV cure may be possible via CRISPR-Cas gene editing (Li et al. 2017; Liu et al. 2018; Scott et al. 2017; Wang et al. 2015). Application of SpCas9 with sgRNAs targeting the HBV reverse transcriptase (RT), surface antigen or core protein successfully induced the degradation of HBV DNA and depleted cccDNA levels in chronic and de novo HBV in vitro cell models (HepAD38 and HepaRG cells, respectively) (Kennedy et al. 2015). CRISPR-Cas9 HBV-targeting in liver cell lines Huh-7 (Lin et al. 2014), HepG2 (Zhen et al. 2015), HepG2.2.15 (Karimova et al. 2015; Zhen et al. 2015), HepG2-H1.3 (Karimova et al. 2015), and HepG2.A64 (Li et al. 2017) all elicited a reduction in viral replication and disruption in cccDNA.
A 2015 study found that a combination of SpCas9 and eight gRNAs targeting the conserved regions of different HBV genotypes significantly inhibited HBV replication both in vitro and in vivo (Liu et al. 2015). Liu et al. have reported efficient inhibition of HBV replication by AAV9-delivered SaCas9/gRNA in a mouse model of chronic HBV infection (Liu et al. 2018). In a similar study, recombinant AAV8-packaged CRISPR-SaCas9 was used to inhibit HBV expression in HBV transgenic mice: HBV surface antigen (HBsAg) and HBV antigen (HBeAg) serum levels significantly decreased following treatment (Li et al. 2018b). Moreover, base editing, which uses the SpCas9-BE (SpCas9-base editor) to introduce nonsense point mutations, could successfully inhibit viral gene expression in an in vitro HBV infection model (Yang et al. 2020).
Though these in vivo and in vitro studies have shown the potential of CRISPR-Cas9 in the elimination of HBV infection, further studies are needed to improve our understanding of cccDNA biology and HBV replication in order to develop effective editing systems that could be used to eradicate HBV infection.
Herpesviruses (HSV-1, EBV, HCMV)
Viruses of the Herpesviridae family (to which Herpes simplex virus type-1 (HSV-1), Epstein-Barr virus (EBV), and human cytomegalovirus (HCMV) belong) are dsDNA viruses characterised by their ability to establish life-long infections in the host. HSV-1 is the prototypical human Alphaherpesvirus and is the causative agent of herpes labialis, herpes keratitis and more severe sequelae such as viral encephalitis and neonatal herpes. Current antiviral drugs are ineffective against latent infection of HSV-1, especially in the trigeminal ganglion (TG) neurons. Many recent studies have approached this difficult virus through CRISPR-Cas9 gene editing approaches. Chen et al. attempted to inhibit HSV-1 replication in African green monkey Vero cells and cultured primary TG neurons using SpCas9 and SaCas9 systems targeting two essential HSV-1 genes (ICP0 and ICP4); demonstrating significant impairment of viral replication in both cell types (Chen et al. 2020). A study by Xu et al. reported CRISPR-Cas9-mediated knockout of the viral UL7 gene diminished genome replication, attenuated neurovirulence and decreased pathologies in mice (Xu et al. 2016). The inhibitory effect of CRISPR-Cas9 systems in HSV-1 infection by targeting different HSV-1 loci has also been reported in other studies (Li et al. 2018c; Roehm et al. 2016; Russell et al. 2015; van Diemen et al. 2016).
EBV is responsible for several human malignancies such as Burkitt’s lymphoma and nasopharyngeal carcinoma. During the latent stage of viral infection, the EBV genome is maintained in an episomal form in host B cells and epithelial cells; here, only a limited number of proteins are expressed, namely those involved in the maintenance of the episomal genome (e.g. EBV nuclear antigens-1 (EBNAs-1) and proteins involved in cell transformation and induction of invasion) (Raab-Traub 2012; Yuen et al. 2018b). CRISPR-Cas9 targeting of EBV in Raji cells (a Burkitt’s lymphoma cell line) derived from patients with latent EBV infection significantly reduced EBV replication and apoptotic events (Wang and Quake 2014). Similarly, complete clearance of EBV in latently infected lymphoma cells has been achieved by Cas9-targeting of viral EBNA-1 (van Diemen et al. 2016). Yuen et al. successfully deleted 558 bp in the promoter region of BamHI-A rightward transcripts (BART) with CRISPR, which led to decreased levels of miR-Bart3 and reduced viral yields in latently infected EBV models (Yuen et al. 2015). Later application of multiple EBV-targeting gRNAs demonstrated down regulation of EBV DNA loads and lytic replication in latently infected nasopharyngeal carcinoma cells (Yuen et al. 2018a).
Human cytomegalovirus (HCMV) causes severe disease in neonates and immunocompromised individuals. Like all herpesviruses, HCMV can establish latent infection within the host, and reactivation is considered the main cause of HCMV-induced morbidity and mortality in immunocompromised individuals (Damato and Winnen 2002; Stern et al. 2019). Van Diemen et al. report the efficient impairment of HCMV replication by CRISPR-Cas9 gene editing in an in vitro cell model (van Diemen et al. 2016). Further, multiplexed CRISPR-Cas9 targeting of HCMV essential genes UL122/123 in human cells demonstrated successful abrogation of virus replication (Gergen et al. 2018).
While these findings highlight the potential for CRISPR-Cas9 systems in eradicating herpesviruses from the body, again additional in vitro and in vivo studies are needed to prove the capability of the system.
Human Papillomaviruses (HPVs)
HPVs are considered the etiological agents of almost all cervical carcinomas and several other cancers. E6 and E7 antigens are responsible for inducing and maintaining the oncogenic properties of HPV by disrupting the host cell tumor suppressors p53 and retinoblastoma protein (Rb), respectively (Mirabello et al. 2017). Though no effective therapy has been developed for HPV-associated carcinogenesis, CRISPR-Cas9 systems have the potential to provide effective treatment in HPV-associated cancer (Kennedy et al. 2014; Zhen and Li 2017). Cas9 targeting of the E6 or E7 oncogenes in HPV-induced cancer cell lines resulted in cleavage of the HPV genome. Successful inactivation of the oncogenes and increased p53 or pRb expression successfully induced cancer cell death (Kennedy et al. 2014). In a study by Zhen et al., in vitro and in vivo targeting of E6/E7 by Cas9 in combination with the anti-cancer drug cisplatin provided promising results for the treatment of cervical cancer. Additionally, the potential of CRISPR-Cas9 as a highly specific alternative treatment for patients with cervical cancer was demonstrated by Yoshiba et al.: in in vitro and in vivo animal models of high-risk HPV-positive cervical cancer, transduction of an AAV carrying E6-targeting gRNA (AAV-sgE6) and Cas9 suppressed the growth of cell lines and tumor growth in a mouse model following intratumoral administration of AAV-sgE6 (Yoshiba et al. 2019). Future in vivo studies is needed to confirm the efficiency of CRISPR-Cas9 based approaches in the treatment of HPV-associated malignancy.
Human Polyomavirus 2 (or John Cunningham Virus, JC Virus (JCV))
JCV is a causative agent of progressive multifocal leukoencephalopathy (PML) and belongs to the human polyomavirus family. The small circular dsDNA genome of JCV comprises three regions: the early coding region, which encodes the regulatory T antigens; the late coding region, which encodes the viral capsid proteins (VP1-3); and the non-coding control region (NCCR), which contains the viral promoters and origin of replication (Jiang et al. 2009). Some of the currently available treatments are ineffective against the debilitating disease (Tavazzi et al. 2012). The CRISPR-Cas system has therefore been proposed as an alternative approach. Wollebo et al. introduced mutations in the JCV genome using Cas9 to specifically target the viral DNA sequences encoding the T-antigen, strongly inhibiting viral replication in primary human fetal glial cells (Wollebo et al. 2015). In another study, inhibition of JCV replication by Cas9 targeting of the late coding region and NCCR of the JCV genome was demonstrated in vitro. While in vitro studies are suggestive of the potential of CRISPR-Cas9 applications in the treatment of JCV-mediated PML, in vivo studies are still needed to confirm clinical translatability.
Though these examples demonstrate the power of CRISPR systems to tackle viral diseases of humans, there remain several hurdles on the road to widespread clinical application. Perhaps the most significant of these is reagent delivery to a large enough proportion of target cells in situ to benefit clinical outcome. While ethical considerations surrounding human germline editing can restrict delivery options, similar constraints do not apply when considering CRISPR approaches to viral disease in other species.
Livestock diseases
Genome editing studies of host genes are possible in livestock, where ethical concerns are lesser and a precedent for cloned organisms has been established.
Classical swine fever virus (CSFV)
Classical swine fever, caused by the classical swine fever virus (CSFV), is one of the most detrimental diseases in the swine livestock industry, responsible for significant economic losses (Xie et al. 2018). A member of the Flaviviridae family, CSFV is an enveloped virus that possesses a single-stranded positive-sense RNA genome (Shi et al. 2016). CSFV can be spread both by horizontal and vertical transmission (De Smit 2000). Xie et al. combined CRISPR-Cas9 and RNAi technologies to generate anti-CSFV transgenic pigs with a knock-in of a defined antiviral small hairpin RNA (shRNA) at the porcine ROSA26 (pROSA26) locus. These transgenic pigs were found to effectively limit CSFV replication upon viral challenge and reduce CSFV-associated clinical signs and mortality. Importantly, disease resistance could be stably transmitted to the F1-generation (Xie et al. 2018). A more recent study using a knock-in of the antiviral porcine RSAD2 gene (pRSAD2) at the pROSA26 locus produced a pRSAD2-KI pig that constitutively overexpressed pRSAD2 and reduced CSFV infection (Xie et al. 2020).
Porcine reproductive and respiratory syndrome virus (PRRSV)
Like CSFV, PRRSV causes severe economic losses for the global swine industry. Highly pathogenic PRRSV (HP-PRRSV), originating from a genotype 2 PRRSV, is more virulent than classical PRRSV and further exacerbates the economic impact (Yang et al. 2018). PRRSV exclusively infects monocyte/macrophage cell lines and the macrophage-specific CD163 and CD169 proteins were identified as co-receptors for the virus: CD169 is expressed on the cell surface and CD163 localizes within the internalizing transport vesicles (Calvert et al. 2007; Van Gorp et al. 2008). Using CRISPR-Cas9 gene targeting and somatic cell nuclear transfer (SCNT) technologies, some studies generated CD163 knockout pigs with CRISPR-Cas9 systems that were completely resistant to HP-PRRSV infection (Whitworth et al. 2014, 2016) and Yang et al. generating CD163-knockout pig that achieved similar results (Yang et al. 2018). Other studies generating CD163 knockout pigs with CRISPR-Cas9 systems achieved similar results (Whitworth et al. 2014, 2016). Given that CD163 has a range of important biological functions (Onofre et al. 2009), CRISPR editing strategies to minimize the whole organism effects of CD163 knockout were designed to remove only domain 5 of CD163. These animals were completely resistant to PRRSV infection and retained the biological activities associated with the remaining CD163 functional domains (Burkard et al. 2017, 2018).
Transmissible gastroenteritis coronavirus (TGEV)
TGEV is an acute, rapidly spreading coronavirus disease which infects pigs of all ages. High morbidity and mortality have been reported for piglets less than 1 week old (Doyle and Hutchings 1946). Gene-edited pigs with a knockout of exon 2 of aminopeptidase N, a putative host cell receptor for TGEV, demonstrated resistance to TGEV (Whitworth et al. 2019).
In summary, though CRISPR-Cas9 systems for viral disease control in humans show significant promise, system delivery remains problematic. The production of genome-edited livestock with improved resistance and resilience to viral disease clearly demonstrates the power of CRISPR-Cas9 technology for a subset of viruses, but gaps remain.
Cas 13 systems
CRISPR-Cas13 systems complement Cas9 systems with their ability to target ssRNA viruses. Freije et al. describe the development of a CRISPR-Cas13 technology called CARVER (Cas13-assisted restriction of viral expression and readout), demonstrating an efficient antiviral response against three mammalian ssRNA viruses in cell culture (lymphocytic choriomeningitis virus (LCMV), influenza A virus (IAV) and vesicular stomatitis virus (VSV)) (Freije et al. 2019). This programmable diagnostic and antiviral technology could be applied to a variety of ssRNA viruses.
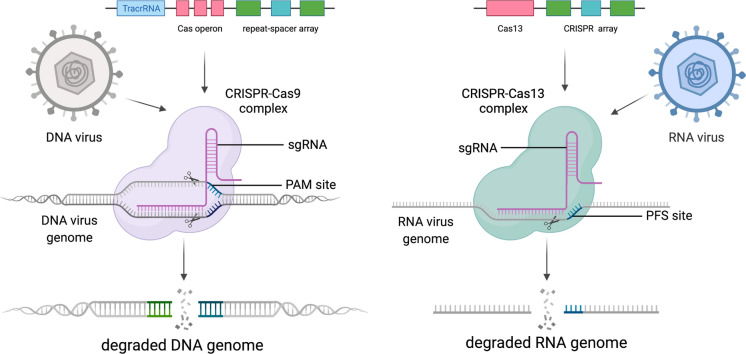
A prophylactic antiviral CRISPR-Cas13d architecture called PAC-MAN successfully reduced SARS-CoV-2 and IAV infection in human cells (Abbott et al. 2020). This system is a promising tool for the inhibition of broad coronavirus infections, relevant in light of the current COVID-19 pandemic (Abbott et al. 2020). In a recent preprint, Xu et al. describe Cas13e.1, part of a novel family of small type VI CRISPR-Cas systems, which similarly demonstrated high cleavage activity when targeted against SARS-CoV-2 and IAV genomes (Xu et al. 2020). In another study, the PRRSV genome was completely knocked down in cell culture by the use of the CRISPR-Cas13b system (Cui et al. 2020). Cas13 has also been used to inhibit HIV-1 infection by targeting and cleaving HIV-1 RNA (Yin et al. 2020). These findings provide opportunities for more effective prophylactic and therapeutic strategies against existing and emerging viral diseases. Figure 2 descript the degradation of viral genomes using CRISPR-Cas9 and Cas13.
Fig. 2.
Schematic of viral nucleic acid degradation using Cas9 and Cas13. Created with BioRender.com
Pathogen detection based on CRISPR-Cas system
Cas9 has formed the basis of the majority of DNA gene editing systems and has been modified to enable the targeting of RNAs in living cells. RCas9 has been developed by incorporation of RNA-specific PAM sequences with complimentary guide RNAs. Catalytically active RCas9 has the potential to be used for diagnostic and therapeutic purposes (Batra et al. 2017; Nelles et al. 2016; O’Connell et al. 2014) (Table 2).
Table 2.
Nucleic acid detection platforms based on CRISPR‐Cas systems
| CRISPR‐Cas system | Platform | Effector | Nucleic Acid Target | Signal Amplification Method | Fluorescent Signal | Applications | References |
|---|---|---|---|---|---|---|---|
| Cas13 | SHERLOCK | LwCas13a | DNA/RNA | RPA | FAM | Virus detection, human DNA genotyping, cancer mutations | Gootenberg et al. (2017) |
| SHERLOCKV2 | CcaCas13b PsmCas13b LwaCas13a | DNA/RNA | RPA | FAM,TEX, Cy5,HEX | Multiple analyte detection (up to four targets) | Myhrvold et al. (2018) | |
| SHERLOCK + HUDSON | LwCas13a | DNA/RNA | RPA | FAM | Identify the virus directly from body fluids | Gootenberg and Abudayyeh (2018) | |
| CARMEN | Cas13a | PCR or RPA | Fluorescence microscopy |
Multiplexed pathogen detection; more than 4,500 crRNA–target pairs on a single array |
Ackerman et al. (2020) | ||
| Cas 12 | DETECTOR | LbCas12a | DNA | RPA | FAM | SNP detection | Liang et al. (2019) |
| HOLMES | LbCas12a | DNA/RNA | PCR; RT-PCR | HEX | Virus genotyping and human | Harrington et al. (2018) | |
| HOLMESv2 | AacCas12b | DNA/RNA | LAMP; RT-LAMP; Asymmetric PCR | HEX,FAM | SNP detection | Li et al. (2019) | |
| Cas14 | DETECTOR | Cas14a | DNA/RNA | RPA | λex: 485 nm; λem: 535 nm | SNP identification | Aquino-Jarquin (2019) |
| Cas9 | CAS-EXPAR | SpCas9 | DNA/RNA | EXPAR | SYBR Green I | SNP genotype discrimination | Huang and Zhou (2018) |
| NASBACC | SpCas9 | RNA | NASBA | Luciferase | Virus detection | Pardee et al. (2016) |
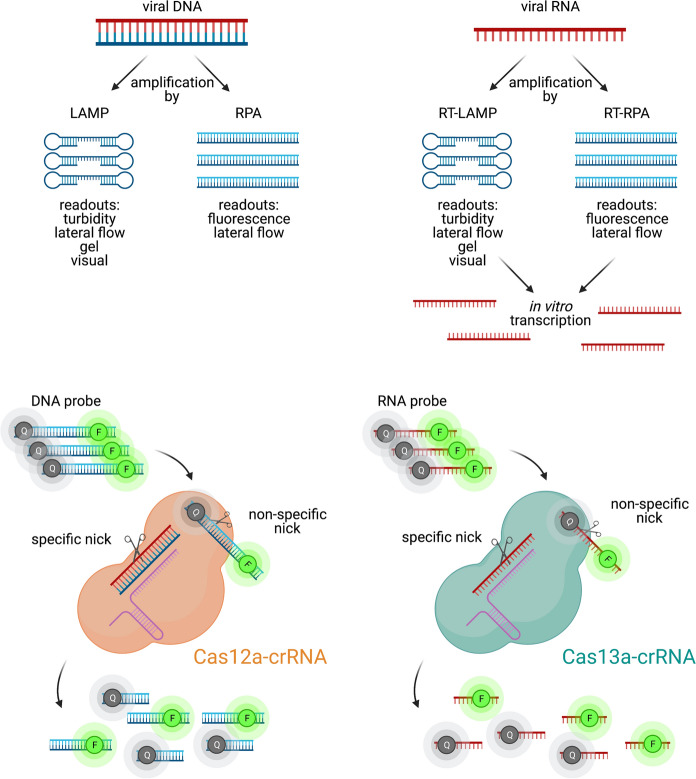
Several simple RNA detection strategies that do not require special instruments have been developed in recent years (Notomi et al. 2000; Piepenburg et al. 2006). Reverse transcription-recombinase polymerase amplification (RT–RPA) and reverse transcription-loop-mediated isothermal amplification (RT-LAMP) are highly sensitive RNA amplification methods that are further enhanced with sequence-specific detection modules like hybridization-based fluorescent-oligonucleotide probes (Becherer et al. 2020; Bhadra et al. 2020). RT-RPA or RT-LAMP methods have since been combined with CRISPR-Cas systems to take advantage of Cas-induced cleavage of bystander nucleic acid probes (Gootenberg et al. 2018) (Fig. 3). Non-specific cleavage of single-stranded fluorescent probes by Cas proteins can reverse reporter quenching and produce detectable signals (Fig. 3). Such assays can also be performed on lateral flow strips, a method much more accessible because of strip portability and a simple visual result readout (Myhrvold et al. 2018). These CRISPR diagnostic methods are both highly sensitive and specific (Gootenberg et al. 2018; Harrington et al. 2018). For example, a recent preprint by Zhang et al. described a protocol based on the Cas13-based Specific High Sensitivity Enzymatic Reporter UnLOCKing (SHERLOCK) system for detection of SARS-CoV-2, sensitive to as few as two copies of extracted SARS-CoV-2 RNA (Zhang et al. 2020). Other CRISPR-Cas systems including Cas12, Cas13 and Cas14 have also been implemented to detect nucleic acid (Table 2).
Fig. 3 .
Schematic of viral nucleic acid detection with CRISPR Cas12 and 13. Nucleic acid targets can be amplified by LAMP/RPA (DNA) and RT-LAMP/RPA (RNA). Cas12- and Cas13-crRNA complexes cleave introduced target-specific fluorescent probes (F) on DNA and RNA, respectively, removing quencher (Q) moieties and producing detectable fluorescent signals. Created with BioRender.com
These systems have been applied to the detection of SARS-CoV-2, Zika virus, HIV and HPV (Table 3).
Table 3.
Viral disease detection based on CRISPR‐Cas systems
| Virus | Method | Cas Effector | Targeted Substrate | References |
|---|---|---|---|---|
| HPV | CtPCR | SpCas9 | dsDNA | Wang et al. (2018a) |
| CARP (ctPCR2.0) | SpCas9 | dsDNA | Zhang et al. (2018a) | |
| ctPCR3.0 | SpCas9 | dsDNA | Zhang et al. (2018b) | |
| DETECTOR | LbCas12a | dsDNA | Chen et al. (2018) | |
| ZIKV | NASBACC | SpCas9 | RNA | Pardee et al. (2016) |
| LwCas13a | SHERLOCK | dsDNA/RNA | Bhattacharya et al. (2020) | |
| CcaCas13b | SHERLOCK v2 | dsDNA/RNA | Gootenberg and Abudayyeh (2018) | |
| SARS COV-2 | Cas13 | SHINE | DNA/RNA | Arizti-Sanz et al. (2020) |
| AapCas12b | STOPCovid | DNA/RNA | Joung et al. (2020) | |
| Cas12 | iSCANSARSCOV-2 | DNA/RNA | Ali et al. (2020) | |
| HIV-1 (as integrated DNA) | AsCas12a | SCAN | DNA | Nouri et al. (2020) |
Conclusion
CRISPR-Cas systems have shown themselves to be incredibly powerful tools for gene editing, viral knockout, and pathogen detection. Concerns remain, however, for clinical applications of such technologies, requiring off-target edit rates to be dramatically reduced and effective methods of delivery confirmed for in vivo use. CRISPR-Cas9 has been developed predominantly for DNA editing applications. CRISPR-Cas12 is most commonly utilized in sequence detection kits. Cas13 is useful in editing RNA targets, including RNA viruses of clinical relevance. Because of their sequence programmability, much research has focused on the use of CRISPR-Cas technologies for improved accuracy in the development of disease treatment and diagnostic platforms. Despite the potential risk of non-specific RNA targeting effects, we believe that the CRISPR-Cas13 system presents a higher level of specificity over the available RNA-targeting approaches. Continued development of Cas variants with increased specificities and activities is critical for the application of these technologies to viral infection control.
Declarations
Conflict of interest
The authors declare that they have no conflict of interest. The research reported here did not involve experimentation with human participants or animals.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Abbott TR, Dhamdhere G, Liu Y, Lin X, Goudy L, Zeng L, Chemparathy A, Chmura S, Heaton NS, Debs R. Development of CRISPR as an antiviral strategy to combat SARS-CoV-2 and influenza. Cell. 2020;181(4):865–876. doi: 10.1016/j.cell.2020.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abudayyeh OO, Gootenberg JS, Konermann S, Joung J, Slaymaker IM, Cox DB, Shmakov S, Makarova KS, Semenova E, Minakhin L. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 2016;353:6299. doi: 10.1126/science.aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abudayyeh OO, Gootenberg JS, Essletzbichler P, Han S, Joung J, Belanto JJ, Verdine V, Cox DBT, Kellner MJ, Regev A, Lander ES, Voytas DF, Ting AY, Zhang F. RNA targeting with CRISPR-Cas13. Nature. 2017;550:280–284. doi: 10.1038/nature24049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackerman CM, Myhrvold C, Thakku SG, Freije CA, Metsky HC, Yang DK, Simon HY, Boehm CK, Kosoko-Thoroddsen T-SF, Kehe J. Massively multiplexed nucleic acid detection with Cas13. Nature. 2020;582(7811):277–282. doi: 10.1038/s41586-020-2279-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali Z, Aman R, Mahas A, Rao GS, Tehseen M, Marsic T, Salunke R, Subudhi AK, Hala SM, Hamdan SM, Pain A, Alofi FS, Alsomali A, Hashem AM, Khogeer A, Almontashiri NAM, Abedalthagafi M, Hassan N, Mahfouz MM. iSCAN: An RT-LAMP-coupled CRISPR-Cas12 module for rapid, sensitive detection of SARS-CoV-2. Virus Res. 2020;288:198129. doi: 10.1016/j.virusres.2020.198129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aquino-Jarquin G. CRISPR-Cas14 is now part of the artillery for gene editing and molecular diagnostic. Nanomedicine. 2019;18:428–431. doi: 10.1016/j.nano.2019.03.006. [DOI] [PubMed] [Google Scholar]
- Arizti-Sanz J, Freije CA, Stanton AC, Boehm CK, Petros BA, Siddiqui S, Shaw BM, Adams G, Kosoko-Thoroddsen TF, Kemball ME, Gross R, Wronka L, Caviness K, Hensley LE, Bergman NH, MacInnis BL, Lemieux JE, Sabeti PC, Myhrvold C (2020) Integrated sample inactivation, amplification, and Cas13-based detection of SARS-CoV-2. bioRxiv [DOI] [PMC free article] [PubMed]
- Arzumanyan A, Reis HM, Feitelson MA. Pathogenic mechanisms in HBV- and HCV-associated hepatocellular carcinoma. Nat Rev Cancer. 2013;13:123–135. doi: 10.1038/nrc3449. [DOI] [PubMed] [Google Scholar]
- Batra R, Nelles DA, Pirie E, Blue SM, Marina RJ, Wang H, Chaim IA, Thomas JD, Zhang N, Nguyen V. Elimination of toxic microsatellite repeat expansion RNA by RNA-targeting Cas9. Cell. 2017;170(899–912):e810. doi: 10.1016/j.cell.2017.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becherer L, Borst N, Bakheit M, Frischmann S, Zengerle R, von Stetten F. Loop-mediated isothermal amplification (LAMP)–review and classification of methods for sequence-specific detection. Anal Methods. 2020;12:717–746. doi: 10.1039/C9AY02246E. [DOI] [Google Scholar]
- Bella R, Kaminski R, Mancuso P, Young WB, Chen C, Sariyer R, Fischer T, Amini S, Ferrante P, Jacobson JM, Kashanchi F, Khalili K. Removal of HIV DNA by CRISPR from patient blood engrafts in humanized mice. Mol Ther Nucleic Acids. 2018;12:275–282. doi: 10.1016/j.omtn.2018.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhadra S, Riedel TE, Lakhotia S, Tran ND, Ellington AD (2020) High-surety isothermal amplification and detection of SARS-CoV-2, including with crude enzymes. bioRxiv [DOI] [PMC free article] [PubMed]
- Bhattacharya M, Sharma AR, Patra P, Ghosh P, Sharma G, Patra BC, Lee SS, Chakraborty C. Development of epitope-based peptide vaccine against novel coronavirus 2019 (SARS-COV-2): immunoinformatics approach. J Med Virol. 2020;92:618–631. doi: 10.1002/jmv.25736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkard C, Lillico SG, Reid E, Jackson B, Mileham AJ, Ait-Ali T, Whitelaw CBA, Archibald AL. Precision engineering for PRRSV resistance in pigs: macrophages from genome edited pigs lacking CD163 SRCR5 domain are fully resistant to both PRRSV genotypes while maintaining biological function. PLoS Pathog. 2017;13:e1006206. doi: 10.1371/journal.ppat.1006206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkard C, Opriessnig T, Mileham AJ, Stadejek T, Ait-Ali T, Lillico SG, Whitelaw CBA, Archibald AL (2018) Pigs lacking the scavenger receptor cysteine-rich domain 5 of CD163 are resistant to porcine reproductive and respiratory syndrome virus 1 infection. Journal of virology 92 [DOI] [PMC free article] [PubMed]
- Calvert JG, Slade DE, Shields SL, Jolie R, Mannan RM, Ankenbauer RG, Welch S-KW. CD163 expression confers susceptibility to porcine reproductive and respiratory syndrome viruses. J Virol. 2007;81:7371–7379. doi: 10.1128/JVI.00513-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JS, Dagdas YS, Kleinstiver BP, Welch MM, Sousa AA, Harrington LB, Sternberg SH, Joung JK, Yildiz A, Doudna JA. Enhanced proofreading governs CRISPR–Cas9 targeting accuracy. Nature. 2017;550:407–410. doi: 10.1038/nature24268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JS, Ma E, Harrington LB, Da Costa M (2018) CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. 360:436–439 [DOI] [PMC free article] [PubMed]
- Chen Y, Zhi S, Liang P, Zheng Q, Liu M, Zhao Q, Ren J, Cui J, Huang J, Liu Y, Songyang Z. Single AAV-mediated CRISPR-SaCas9 inhibits HSV-1 replication by editing ICP4 in trigeminal ganglion neurons. Mol Ther Methods Clin Dev. 2020;18:33–43. doi: 10.1016/j.omtm.2020.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DB, Gootenberg JS, Abudayyeh OO, Franklin B, Kellner MJ, Joung J, Zhang F. RNA editing with CRISPR-Cas13. Science. 2017;358:1019–1027. doi: 10.1126/science.aaq0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J, Techakriengkrai N, Nedumpun T, Suradhat S. Abrogation of PRRSV infectivity by CRISPR-Cas13b-mediated viral RNA cleavage in mammalian cells. Sci Rep. 2020;10:9617. doi: 10.1038/s41598-020-66775-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damato EG, Winnen CW. Cytomegalovirus infection: perinatal implications. J Obstet Gynecol Neonatal Nurs. 2002;31:86–92. doi: 10.1111/j.1552-6909.2002.tb00026.x. [DOI] [PubMed] [Google Scholar]
- Dandri M, Petersen J. Mechanism of hepatitis B virus persistence in hepatocytes and its carcinogenic potential. Clin Infect Dis. 2016;62(Suppl 4):S281–288. doi: 10.1093/cid/ciw023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dash PK, Kaminski R, Bella R, Su H, Mathews S, Ahooyi TM, Chen C, Mancuso P, Sariyer R, Ferrante P, Donadoni M, Robinson JA, Sillman B, Lin Z, Hilaire JR, Banoub M, Elango M, Gautam N, Mosley RL, Poluektova LY, McMillan J, Bade AN, Gorantla S, Sariyer IK, Burdo TH, Young W-B, Amini S, Gordon J, Jacobson JM, Edagwa B, Khalili K, Gendelman HE. Sequential LASER ART and CRISPR treatments eliminate HIV-1 in a subset of infected humanized mice. Nat Commun. 2019;10:2753. doi: 10.1038/s41467-019-10366-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smit A. Laboratory diagnosis, epizootiology, and efficacy of marker vaccines in classical swine fever: a review. Vet Q. 2000;22:182–188. doi: 10.1080/01652176.2000.9695054. [DOI] [PubMed] [Google Scholar]
- Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doetschman T, Georgieva T. Gene editing with CRISPR/Cas9 RNA-directed nuclease. Circ Res. 2017;120:876–894. doi: 10.1161/CIRCRESAHA.116.309727. [DOI] [PubMed] [Google Scholar]
- Doyle L, Hutchings L. A transmissible gastroenteritis in pigs. J Am Vet Med Assoc. 1946;108:257–259. [PubMed] [Google Scholar]
- Dufour C, Claudel A, Joubarne N, Merindol N, Maisonnet T, Masroori N, Plourde MB, Berthoux L. Editing of the human TRIM5 gene to introduce mutations with the potential to inhibit HIV-1. PLoS ONE. 2018;13:e0191709. doi: 10.1371/journal.pone.0191709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freije CA, Myhrvold C, Boehm CK, Lin AE, Welch NL, Carter A, Metsky HC, Luo CY, Abudayyeh OO, Gootenberg JS. Programmable inhibition and detection of RNA viruses using Cas13. Mol Cell. 2019;76(826–837):e811. doi: 10.1016/j.molcel.2019.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gergen J, Coulon F, Creneguy A, Elain-Duret N, Gutierrez A, Pinkenburg O, Verhoeyen E, Anegon I, Nguyen TH, Halary FA. Multiplex CRISPR/Cas9 system impairs HCMV replication by excising an essential viral gene. PLoS ONE. 2018;13:e0192602. doi: 10.1371/journal.pone.0192602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gootenberg JS, Abudayyeh OO (2018) Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. 360:439–444 [DOI] [PMC free article] [PubMed]
- Gootenberg JS, Abudayyeh OO, Lee JW, Essletzbichler P, Dy AJ, Joung J, Verdine V, Donghia N, Daringer NM, Freije CA. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017;356:438–442. doi: 10.1126/science.aam9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gootenberg JS, Abudayyeh OO, Kellner MJ, Joung J, Collins JJ, Zhang F. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science. 2018;360:439–444. doi: 10.1126/science.aaq0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington LB, Burstein D, Chen JS, Paez-Espino D, Ma E, Witte IP, Cofsky JC, Kyrpides NC, Banfield JF, Doudna JA. Programmed DNA destruction by miniature CRISPR-Cas14 enzymes. Science. 2018;362:839–842. doi: 10.1126/science.aav4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano S, Nishimasu H, Ishitani R, Nureki O. Structural basis for the altered PAM specificities of engineered CRISPR-Cas9. Mol Cell. 2016;61:886–894. doi: 10.1016/j.molcel.2016.02.018. [DOI] [PubMed] [Google Scholar]
- Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Kaminski R, Yang F, Zhang Y, Cosentino L, Li F, Luo B, Alvarez-Carbonell D, Garcia-Mesa Y, Karn J. RNA-directed gene editing specifically eradicates latent and prevents new HIV-1 infection. Proc Natl Acad Sci. 2014;111:11461–11466. doi: 10.1073/pnas.1405186111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M, Zhou X (2018) Clustered regularly interspaced short palindromic repeats/Cas9 triggered isothermal amplification for site-specific nucleic acid detection. 90:2193–2200 [DOI] [PubMed]
- Ishino Y, Shinagawa H, Makino K, Amemura M, Nakata A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J Bacteriol. 1987;169:5429–5433. doi: 10.1128/jb.169.12.5429-5433.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F, Doudna JA. CRISPR–Cas9 structures and mechanisms. Annu Rev Biophys. 2017;46:505–529. doi: 10.1146/annurev-biophys-062215-010822. [DOI] [PubMed] [Google Scholar]
- Jiang M, Abend JR, Johnson SF, Imperiale MJ. The role of polyomaviruses in human disease. Virology. 2009;384:266–273. doi: 10.1016/j.virol.2008.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson TP, Patel K, Johnson KR, Maric D, Calabresi PA, Hasbun R, Nath A. Induction of IL-17 and nonclassical T-cell activation by HIV-Tat protein. Proc Natl Acad Sci U S A. 2013;110:13588–13593. doi: 10.1073/pnas.1308673110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung J, Ladha A, Saito M, Segel M, Bruneau R, Huang M-lW, Kim N-G, Yu X, Li J, Walker BD (2020) Point-of-care testing for COVID-19 using SHERLOCK diagnostics. medRxiv
- Kaminski R, Bella R, Yin C, Otte J, Ferrante P, Gendelman HE, Li H, Booze R, Gordon J, Hu W, Khalili K. Excision of HIV-1 DNA by gene editing: a proof-of-concept in vivo study. Gene Ther. 2016;23:690–695. doi: 10.1038/gt.2016.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminski R, Chen Y, Fischer T, Tedaldi E, Napoli A, Zhang Y, Karn J, Hu W, Khalili K. Elimination of HIV-1 genomes from human T-lymphoid cells by CRISPR/Cas9 gene editing. Sci Rep. 2016;6:1–15. doi: 10.1038/srep22555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimova M, Beschorner N, Dammermann W, Chemnitz J, Indenbirken D, Bockmann JH, Grundhoff A, Lüth S, Buchholz F, Schulze zur Wiesch J, Hauber J. CRISPR/Cas9 nickase-mediated disruption of hepatitis B virus open reading frame S and X. Sci Rep. 2015;5:13734. doi: 10.1038/srep13734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karvelis T, Gasiunas G, Siksnys V. Harnessing the natural diversity and in vitro evolution of Cas9 to expand the genome editing toolbox. Curr Opin Microbiol. 2017;37:88–94. doi: 10.1016/j.mib.2017.05.009. [DOI] [PubMed] [Google Scholar]
- Kennedy EM, Kornepati AVR, Goldstein M, Bogerd HP, Poling BC, Whisnant AW, Kastan MB, Cullen BR. Inactivation of the human papillomavirus E6 or E7 gene in cervical carcinoma cells by using a bacterial CRISPR/Cas RNA-guided endonuclease. J Virol. 2014;88:11965–11972. doi: 10.1128/JVI.01879-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy EM, Bassit LC, Mueller H, Kornepati AV, Bogerd HP, Nie T, Chatterjee P, Javanbakht H, Schinazi RF, Cullen BR. Suppression of hepatitis B virus DNA accumulation in chronically infected cells using a bacterial CRISPR/Cas RNA-guided DNA endonuclease. Virology. 2015;476:196–205. doi: 10.1016/j.virol.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Um E, Cho S-R, Jung C, Kim H, Kim J-S. Surrogate reporters for enrichment of cells with nuclease-induced mutations. Nat Methods. 2011;8:941. doi: 10.1038/nmeth.1733. [DOI] [PubMed] [Google Scholar]
- Kim HK, Lee S, Kim Y, Park J, Min S, Choi JW, Huang TP, Yoon S, Liu DR, Kim HH. High-throughput analysis of the activities of xCas9, SpCas9-NG and SpCas9 at matched and mismatched target sequences in human cells. Nat Biomed Eng. 2020;4:111–124. doi: 10.1038/s41551-019-0505-1. [DOI] [PubMed] [Google Scholar]
- Kleinstiver BP, Prew MS, Tsai SQ, Topkar VV, Nguyen NT, Zheng Z, Gonzales APW, Li Z, Peterson RT, Yeh J-RJ, Aryee MJ, Joung JK. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015;523:481–485. doi: 10.1038/nature14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen NT, Zheng Z, Joung JK. High-fidelity CRISPR–Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529:490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver BP, Sousa AA, Walton RT, Tak YE, Hsu JY, Clement K, Welch MM, Horng JE, Malagon-Lopez J, Scarfò I. Engineered CRISPR–Cas12a variants with increased activities and improved targeting ranges for gene, epigenetic and base editing. Nat Biotechnol. 2019;37:276–282. doi: 10.1038/s41587-018-0011-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konermann S, Lotfy P, Brideau NJ, Oki J, Shokhirev MN, Hsu PD. Transcriptome engineering with RNA-targeting type VI-D CRISPR effectors. Cell. 2018;173:665–676.e614. doi: 10.1016/j.cell.2018.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Makarova KS, Zhang F. Diversity, classification and evolution of CRISPR-Cas systems. Curr Opin Microbiol. 2017;37:67–78. doi: 10.1016/j.mib.2017.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulcsár PI, Tálas A, Huszár K, Ligeti Z, Tóth E, Weinhardt N, Fodor E, Welker E. Crossing enhanced and high fidelity SpCas9 nucleases to optimize specificity and cleavage. Genome Biol. 2017;18:1–17. doi: 10.1186/s13059-017-1318-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampi Y, Van Looveren D, Vranckx LS, Thiry I, Bornschein S, Debyser Z, Gijsbers R. Targeted editing of the PSIP1 gene encoding LEDGF/p75 protects cells against HIV infection. Sci Rep. 2019;9:1–15. doi: 10.1038/s41598-019-38718-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebbink RJ, De Jong DC, Wolters F, Kruse EM, Van Ham PM, Wiertz EJ, Nijhuis M. A combinational CRISPR/Cas9 gene-editing approach can halt HIV replication and prevent viral escape. Sci Rep. 2017;7:41968. doi: 10.1038/srep41968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leenay RT, Aghazadeh A, Hiatt J, Tse D, Roth TL, Apathy R, Shifrut E, Hultquist JF, Krogan N, Wu Z. Large dataset enables prediction of repair after CRISPR–Cas9 editing in primary T cells. Nat Biotechnol. 2019;37:1034–1037. doi: 10.1038/s41587-019-0203-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Sheng C, Wang S, Yang L, Liang Y, Huang Y, Liu H, Li P, Yang C, Yang X, Jia L, Xie J, Wang L, Hao R, Du X, Xu D, Zhou J, Li M, Sun Y, Tong Y, Li Q, Qiu S, Song H. Removal of integrated hepatitis B virus DNA using CRISPR-Cas9. Front Cell Infect Microbiol. 2017;7:91. doi: 10.3389/fcimb.2017.00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Zeng C, Dong Y. Design and assessment of engineered CRISPR–Cpf1 and its use for genome editing. Nat Protoc. 2018;13:899. doi: 10.1038/nprot.2018.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Sheng C, Liu H, Wang S, Zhao J, Yang L, Jia L, Li P, Wang L, Xie J, Xu D, Sun Y, Qiu S, Song H. Inhibition of HBV expression in HBV transgenic mice using AAV-delivered CRISPR-SaCas9. Front Immunol. 2018;9:2080. doi: 10.3389/fimmu.2018.02080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Bi Y, Xiao H, Sun L, Ren Y, Li Y, Chen C, Cun W. CRISPR-Cas9 system-driven site-specific selection pressure on herpes simplex virus genomes. Virus Res. 2018;244:286–295. doi: 10.1016/j.virusres.2017.03.010. [DOI] [PubMed] [Google Scholar]
- Li L, Li S, Wu N, Wu J, Wang G, Zhao G, Wang J. HOLMESv2: a CRISPR-Cas12b-assisted platform for nucleic acid detection and DNA methylation quantitation. ACS Synth Biol. 2019;8:2228–2237. doi: 10.1021/acssynbio.9b00209. [DOI] [PubMed] [Google Scholar]
- Liang M, Li Z, Wang W, Liu J, Liu L, Zhu G, Karthik L, Wang M, Wang K-F, Wang Z. A CRISPR-Cas12a-derived biosensing platform for the highly sensitive detection of diverse small molecules. Nat Commun. 2019;10:1–9. doi: 10.1038/s41467-019-11648-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao H-K, Gu Y, Diaz A, Marlett J, Takahashi Y, Li M, Suzuki K, Xu R, Hishida T, Chang C-J. Use of the CRISPR/Cas9 system as an intracellular defense against HIV-1 infection in human cells. Nat Commun. 2015;6:1–10. doi: 10.1038/ncomms7413. [DOI] [PubMed] [Google Scholar]
- Lin S-R, Yang H-C, Kuo Y-T, Liu C-J, Yang T-Y, Sung K-C, Lin Y-Y, Wang H-Y, Wang C-C, Shen Y-C, Wu F-Y, Kao J-H, Chen D-S, Chen P-J. The CRISPR/Cas9 system facilitates clearance of the intrahepatic HBV templates in vivo. Mol Ther Nucleic Acids. 2014;3:e186. doi: 10.1038/mtna.2014.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Hao R, Chen S, Guo D, Chen Y. Inhibition of hepatitis B virus by the CRISPR/Cas9 system via targeting the conserved regions of the viral genome. J Gen Virol. 2015;96:2252–2261. doi: 10.1099/vir.0.000159. [DOI] [PubMed] [Google Scholar]
- Liu Y, Zhao M, Gong M, Xu Y, Xie C, Deng H, Li X, Wu H, Wang Z. Inhibition of hepatitis B virus replication via HBV DNA cleavage by Cas9 from Staphylococcus aureus. Antiviral Res. 2018;152:58–67. doi: 10.1016/j.antiviral.2018.02.011. [DOI] [PubMed] [Google Scholar]
- Makarova KS, Wolf YI, Koonin EV. Classification and nomenclature of CRISPR-Cas systems: where from here? CRISPR J. 2018;1:325–336. doi: 10.1089/crispr.2018.0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, Kosuri S, Yang L, Church GM. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol. 2013;31:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirabello L, Yeager M, Yu K, Clifford GM, Xiao Y, Zhu B, Cullen M, Boland JF, Wentzensen N, Nelson CW, Raine-Bennett T, Chen Z, Bass S, Song L, Yang Q, Steinberg M, Burdett L, Dean M, Roberson D, Mitchell J, Lorey T, Franceschi S, Castle PE, Walker J, Zuna R, Kreimer AR, Beachler DC, Hildesheim A, Gonzalez P, Porras C, Burk RD, Schiffman M. HPV16 E7 genetic conservation is critical to carcinogenesis. Cell. 2017;170:1164–1174.e1166. doi: 10.1016/j.cell.2017.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojica FJ, García-Martínez J, Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol. 2005;60:174–182. doi: 10.1007/s00239-004-0046-3. [DOI] [PubMed] [Google Scholar]
- Myhrvold C, Freije CA, Gootenberg JS, Abudayyeh OO, Metsky HC, Durbin AF, Kellner MJ, Tan AL, Paul LM, Parham LA. Field-deployable viral diagnostics using CRISPR-Cas13. Science. 2018;360:444–448. doi: 10.1126/science.aas8836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelles DA, Fang MY, O’Connell MR, Xu JL, Markmiller SJ, Doudna JA, Yeo GW. Programmable RNA tracking in live cells with CRISPR/Cas9. Cell. 2016;165:488–496. doi: 10.1016/j.cell.2016.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimasu H, Shi X, Ishiguro S, Gao L, Hirano S, Okazaki S, Noda T, Abudayyeh OO, Gootenberg JS, Mori H. Engineered CRISPR-Cas9 nuclease with expanded targeting space. Science. 2018;361:1259–1262. doi: 10.1126/science.aas9129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:e63–e63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nouri R, Jiang Y, Lian XL, Guan W (2020) Sequence-specific recognition of HIV-1 DNA with solid-state CRISPR-Cas12a-assisted nanopores (SCAN). 5:1273–1280 [DOI] [PubMed]
- O’Connell MR, Oakes BL, Sternberg SH, East-Seletsky A, Kaplan M, Doudna JA. Programmable RNA recognition and cleavage by CRISPR/Cas9. Nature. 2014;516:263–266. doi: 10.1038/nature13769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onofre G, Kolácková M, Jankovicová K, Krejsek J. Scavenger receptor CD163 and its biological functions. Acta Medica (Hradec Kralove) 2009;52:57–61. doi: 10.14712/18059694.2016.105. [DOI] [PubMed] [Google Scholar]
- Ophinni Y, Inoue M, Kotaki T, Kameoka M. CRISPR/Cas9 system targeting regulatory genes of HIV-1 inhibits viral replication in infected T-cell cultures. Sci Rep. 2018;8:7784. doi: 10.1038/s41598-018-26190-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardee K, Green AA, Takahashi MK, Braff D, Lambert G, Lee JW, Ferrante T, Ma D, Donghia N, Fan M, Daringer NM, Bosch I, Dudley DM, O'Connor DH, Gehrke L, Collins JJ. Rapid, low-cost detection of zika virus using programmable biomolecular components. Cell. 2016;165:1255–1266. doi: 10.1016/j.cell.2016.04.059. [DOI] [PubMed] [Google Scholar]
- Piepenburg O, Williams CH, Stemple DL, Armes NA. DNA detection using recombination proteins. PLoS Biol. 2006;4:e204. doi: 10.1371/journal.pbio.0040204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourcel C, Salvignol G, Vergnaud G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology (Reading, England) 2005;151:653–663. doi: 10.1099/mic.0.27437-0. [DOI] [PubMed] [Google Scholar]
- Raab-Traub N. Novel mechanisms of EBV-induced oncogenesis. Curr Opin Virol. 2012;2:453–458. doi: 10.1016/j.coviro.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Lin C-Y, Gootenberg JS, Konermann S, Trevino AE, Scott DA, Inoue A, Matoba S, Zhang Y. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeks J, Naismith JH, White MF. CRISPR interference: a structural perspective. Biochem J. 2013;453:155–166. doi: 10.1042/BJ20130316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roehm PC, Shekarabi M, Wollebo HS, Bellizzi A, He L, Salkind J, Khalili K. Inhibition of HSV-1 replication by gene editing strategy. Sci Rep. 2016;6:23146. doi: 10.1038/srep23146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell TA, Stefanovic T, Tscharke DC. Engineering herpes simplex viruses by infection–transfection methods including recombination site targeting by CRISPR/Cas9 nucleases. J Virol Methods. 2015;213:18–25. doi: 10.1016/j.jviromet.2014.11.009. [DOI] [PubMed] [Google Scholar]
- Scott T, Moyo B, Nicholson S, Maepa MB, Watashi K, Ely A, Weinberg MS, Arbuthnot P. ssAAVs containing cassettes encoding SaCas9 and guides targeting hepatitis B virus inactivate replication of the virus in cultured cells. Sci Rep. 2017;7:1–11. doi: 10.1038/s41598-017-07642-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott T, Urak R, Soemardy C, Morris KV. Improved Cas9 activity by specific modifications of the tracrRNA. Sci Rep. 2019;9:1–11. doi: 10.1038/s41598-019-52616-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi B-J, Liu C-C, Zhou J, Wang S-Q, Gao Z-C, Zhang X-M, Zhou B, Chen P-Y. Entry of classical swine fever virus into PK-15 cells via a pH-, dynamin-, and cholesterol-dependent, clathrin-mediated endocytic pathway that requires Rab5 and Rab7. J Virol. 2016;90:9194–9208. doi: 10.1128/JVI.00688-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmakov S, Abudayyeh OO, Makarova KS, Wolf YI, Gootenberg JS, Semenova E, Minakhin L, Joung J, Konermann S, Severinov K. Discovery and functional characterization of diverse class 2 CRISPR-Cas systems. Mol Cell. 2015;60:385–397. doi: 10.1016/j.molcel.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmakov S, Smargon A, Scott D, Cox D, Pyzocha N, Yan W, Abudayyeh OO, Gootenberg JS, Makarova KS, Wolf YI. Diversity and evolution of class 2 CRISPR–Cas systems. Nat Rev Microbiol. 2017;15:169–182. doi: 10.1038/nrmicro.2016.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern L, Withers B, Avdic S, Gottlieb D, Abendroth A, Blyth E, Slobedman B. Human cytomegalovirus latency and reactivation in allogeneic hematopoietic stem cell transplant recipients. Front Microbiol. 2019;10:1186. doi: 10.3389/fmicb.2019.01186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strecker J, Jones S, Koopal B, Schmid-Burgk J, Zetsche B, Gao L, Makarova KS, Koonin EV, Zhang F. Engineering of CRISPR-Cas12b for human genome editing. Nat Commun. 2019;10:1–8. doi: 10.1038/s41467-018-08224-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strich JR, Chertow DS (2019) CRISPR-Cas biology and its application to infectious diseases. J Clin Microbiol 57 [DOI] [PMC free article] [PubMed]
- Tavazzi E, White MK, Khalili K. Progressive multifocal leukoencephalopathy: clinical and molecular aspects. Rev Med Virol. 2012;22:18–32. doi: 10.1002/rmv.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng F, Cui T, Feng G, Guo L, Xu K, Gao Q, Li T, Li J, Zhou Q, Li W. Repurposing CRISPR-Cas12b for mammalian genome engineering. Cell Discovery. 2018;4:63. doi: 10.1038/s41421-018-0069-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Diemen FR, Kruse EM, Hooykaas MJ, Bruggeling CE, Schürch AC, van Ham PM, Imhof SM, Nijhuis M, Wiertz EJ, Lebbink RJ. CRISPR/Cas9-mediated genome editing of herpesviruses limits productive and latent infections. PLoS Pathog. 2016;12:e1005701. doi: 10.1371/journal.ppat.1005701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Gorp H, Van Breedam W, Delputte PL, Nauwynck HJ. Sialoadhesin and CD163 join forces during entry of the porcine reproductive and respiratory syndrome virus. J Gen Virol. 2008;89:2943–2953. doi: 10.1099/vir.0.2008/005009-0. [DOI] [PubMed] [Google Scholar]
- Wang J, Quake SR. RNA-guided endonuclease provides a therapeutic strategy to cure latent herpesviridae infection. Proc Natl Acad Sci USA. 2014;111:13157–13162. doi: 10.1073/pnas.1410785111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Xu Z-W, Liu S, Zhang R-Y, Ding S-L, Xie X-M, Long L, Chen X-M, Zhuang H, Lu F-M. Dual gRNAs guided CRISPR/Cas9 system inhibits hepatitis B virus replication. World J Gastroenterol WJG. 2015;21:9554. doi: 10.3748/wjg.v21.i32.9554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Zhao N, Berkhout B, Das AT. A combinatorial CRISPR-Cas9 attack on HIV-1 DNA extinguishes all infectious provirus in infected T cell cultures. Cell Rep. 2016;17:2819–2826. doi: 10.1016/j.celrep.2016.11.057. [DOI] [PubMed] [Google Scholar]
- Wang Q, Zhang B, Xu X, Long F, Wang J. CRISPR-typing PCR (ctPCR), a new Cas9-based DNA detection method. Sci Rep. 2018;8:1–13. doi: 10.1038/s41598-018-32329-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Wang W, Cui YC, Pan Q, Zhu W, Gendron P, Guo F, Cen S, Witcher M, Liang C. HIV-1 employs multiple mechanisms to resist Cas9/single guide RNA targeting the viral primer binding site. J Virol. 2018;92:e01135–e11118. doi: 10.1128/JVI.01135-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitworth KM, Lee K, Benne JA, Beaton BP, Spate LD, Murphy SL, Samuel MS, Mao J, O'Gorman C, Walters EM. Use of the CRISPR/Cas9 system to produce genetically engineered pigs from in vitro-derived oocytes and embryos. Biol Reprod. 2014;91(78):71–13. doi: 10.1095/biolreprod.114.121723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitworth KM, Rowland RR, Ewen CL, Trible BR, Kerrigan MA, Cino-Ozuna AG, Samuel MS, Lightner JE, McLaren DG, Mileham AJ. Gene-edited pigs are protected from porcine reproductive and respiratory syndrome virus. Nat Biotechnol. 2016;34:20–22. doi: 10.1038/nbt.3434. [DOI] [PubMed] [Google Scholar]
- Whitworth KM, Rowland RR, Petrovan V, Sheahan M, Cino-Ozuna AG, Fang Y, Hesse R, Mileham A, Samuel MS, Wells KD. Resistance to coronavirus infection in amino peptidase N-deficient pigs. Transgenic Res. 2019;28:21–32. doi: 10.1007/s11248-018-0100-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollebo HS, Bellizzi A, Kaminski R, Hu W, White MK, Khalili K. CRISPR/Cas9 system as an agent for eliminating polyomavirus JC infection. PLoS ONE. 2015;10:e0136046. doi: 10.1371/journal.pone.0136046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Pang D, Yuan H, Jiao H, Lu C, Wang K, Yang Q, Li M, Chen X, Yu T. Genetically modified pigs are protected from classical swine fever virus. PLoS Pathog. 2018;14:e1007193. doi: 10.1371/journal.ppat.1007193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Jiao H, Xiao H, Jiang Y, Liu Z, Qi C, Zhao D, Jiao S, Yu T, Tang X. Generation of pRSAD2 gene knock-in pig via CRISPR/Cas9 technology. Antiviral Res. 2020;174:104696. doi: 10.1016/j.antiviral.2019.104696. [DOI] [PubMed] [Google Scholar]
- Xu X, Fan S, Zhou J, Zhang Y, Che Y, Cai H, Wang L, Guo L, Liu L, Li Q. The mutated tegument protein UL7 attenuates the virulence of herpes simplex virus 1 by reducing the modulation of α-4 gene transcription. Virol J. 2016;13:1–12. doi: 10.1186/s12985-016-0600-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Wang J, Liu Y, Xie L, Su B, Mou D, Wang L, Liu T, Wang X, Zhang B, Zhao L, Hu L, Ning H, Zhang Y, Deng K, Liu L, Lu X, Zhang T, Xu J, Li C, Wu H, Deng H, Chen H. CRISPR-edited stem cells in a patient with HIV and acute lymphocytic leukemia. N Engl J Med. 2019;381:1240–1247. doi: 10.1056/NEJMoa1817426. [DOI] [PubMed] [Google Scholar]
- Xu C, Zhou Y, Xiao Q, He B, Geng G, Wang Z, Cao B, Wang X, Zhou D, Yuan T (2020) Novel miniature CRISPR–Cas13 systems from uncultivated microbes effective in degrading SARS-CoV-2 sequences and influenza viruses
- Yang H, Zhang J, Zhang X, Shi J, Pan Y, Zhou R, Li G, Li Z, Cai G, Wu Z. CD163 knockout pigs are fully resistant to highly pathogenic porcine reproductive and respiratory syndrome virus. Antiviral Res. 2018;151:63–70. doi: 10.1016/j.antiviral.2018.01.004. [DOI] [PubMed] [Google Scholar]
- Yang Y-C, Chen Y-H, Kao J-H, Ching C, Liu I-J, Wang C-C, Tsai C-H, Wu F-Y, Liu C-J, Chen P-J (2020) Permanent inactivation of HBV genomes by CRISPR/Cas9-mediated non-cleavage base editing. Mol Ther Nucleic Acids [DOI] [PMC free article] [PubMed]
- Yin C, Zhang T, Qu X, Zhang Y, Putatunda R, Xiao X, Li F, Xiao W, Zhao H, Dai S. In vivo excision of HIV-1 provirus by saCas9 and multiplex single-guide RNAs in animal models. Mol Ther. 2017;25:1168–1186. doi: 10.1016/j.ymthe.2017.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin L, Zhao F, Sun H, Wang Z, Huang Y, Zhu W, Xu F, Mei S, Liu X, Zhang D (2020) CRISPR/Cas13a inhibits HIV-1 infection. Mol Ther Nucl Acids [DOI] [PMC free article] [PubMed]
- Yosef I, Goren MG, Qimron U. Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res. 2012;40:5569–5576. doi: 10.1093/nar/gks216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshiba T, Saga Y, Urabe M, Uchibori R, Matsubara S, Fujiwara H, Mizukami H. CRISPR/Cas9-mediated cervical cancer treatment targeting human papillomavirus E6. Oncol Lett. 2019;17:2197–2206. doi: 10.3892/ol.2018.9815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen K-S, Chan C-P, Wong N-HM, Ho C-H, Ho T-H, Lei T, Deng W, Tsao SW, Chen H, Kok K-H. CRISPR/Cas9-mediated genome editing of Epstein–Barr virus in human cells. J Gen Virol. 2015;96:626–636. doi: 10.1099/jgv.0.000012. [DOI] [PubMed] [Google Scholar]
- Yuen K-S, Wang Z-M, Wong N-HM, Zhang Z-Q, Cheng T-F, Lui W-Y, Chan C-P, Jin D-Y. Suppression of Epstein-Barr virus DNA load in latently infected nasopharyngeal carcinoma cells by CRISPR/Cas9. Virus Res. 2018;244:296–303. doi: 10.1016/j.virusres.2017.04.019. [DOI] [PubMed] [Google Scholar]
- Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, Van Der Oost J, Regev A. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche B, Heidenreich M, Mohanraju P, Fedorova I, Kneppers J, DeGennaro EM, Winblad N, Choudhury SR, Abudayyeh OO, Gootenberg JS, Wu WY, Scott DA, Severinov K, van der Oost J, Zhang F. Multiplex gene editing by CRISPR–Cpf1 using a single crRNA array. Nat Biotechnol. 2017;35:31–34. doi: 10.1038/nbt.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Wang Q, Xu X, Xia Q, Long F, Li W, Shui Y, Xia X, Wang J. Detection of target DNA with a novel Cas9/sgRNAs-associated reverse PCR (CARP) technique. Anal Bioanal Chem. 2018;410:2889–2900. doi: 10.1007/s00216-018-0873-5. [DOI] [PubMed] [Google Scholar]
- Zhang B, Xia Q, Wang Q, Xia X, Wang J. Detecting and typing target DNA with a novel CRISPR-typing PCR (ctPCR) technique. Anal Biochem. 2018;561–562:37–46. doi: 10.1016/j.ab.2018.09.012. [DOI] [PubMed] [Google Scholar]
- Zhang F, Abudayyeh OO, Gootenberg JS (2020) A protocol for detection of COVID-19 using CRISPR diagnostics. A protocol for detection of COVID-19 using CRISPR diagnostics 8
- Zhen S, Li X. Oncogenic human papillomavirus: application of CRISPR/Cas9 therapeutic strategies for cervical cancer. Cell Physiol Biochem. 2017;44:2455–2466. doi: 10.1159/000486168. [DOI] [PubMed] [Google Scholar]
- Zhen S, Hua L, Liu YH, Gao LC, Fu J, Wan DY, Dong LH, Song HF, Gao X. Harnessing the clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated Cas9 system to disrupt the hepatitis B virus. Gene Ther. 2015;22:404–412. doi: 10.1038/gt.2015.2. [DOI] [PubMed] [Google Scholar]