Abstract
Sepsis, a dysregulated immune system response to infection, is among the leading causes of morbidity, mortality, and cost overruns in the Intensive Care Unit (ICU). Early prediction of sepsis can improve situational awareness amongst clinicians and facilitate timely, protective interventions. While the application of predictive analytics in ICU patients has shown early promising results, much of the work has been encumbered by high false-alarm rates and lack of trust by the end-users due to the ‘black box’ nature of these models. Here, we present DeepAISE (Deep Artificial Intelligence Sepsis Expert), a recurrent neural survival model for the early prediction of sepsis. DeepAISE automatically learns predictive features related to higher-order interactions and temporal patterns among clinical risk factors that maximize the data likelihood of observed time to septic events. A comparative study of four baseline models on data from hospitalized patients at three different healthcare systems indicates that DeepAISE produces the most accurate predictions (AUCs between 0.87 and 0.90) at the lowest false alarm rates (FARs between 0.20 and 0.25) while simultaneously producing interpretable representations of the clinical time series and risk factors.
Keywords: Deep Learning, Sepsis, Artificial Intelligence, Interpretability
1. Introduction
Sepsis is a life-threatening condition that arises when the body’s immune and inflammatory response to infection injures its own internal organs [1]. Though the condition lacks the same public notoriety as other conditions like heart attacks, 6% of all hospitalized patients in the United States carry a primary diagnosis of sepsis as compared to 2.5% for the latter [2]. When all hospital deaths are ultimately considered, nearly 35% are attributable to sepsis [2]. This condition stands in stark contrast to heart attacks which have a mortality rate of 2.7–9.6% and only cost the US $12.1 billion annually, roughly half of the cost of sepsis [3].
Starting in 2004 the Surviving Sepsis Campaign (SSC) began addressing the variations in clinical treatment regimens for sepsis and septic shock through the advocacy of evidence-based practice guidelines called ‘sepsis bundles’ [4]. The most recent recommendation from the Center for Medicare and Medicate (CMS) is a 3-hr bundle (aka, SEP-1) that in addition to obtaining diagnostic tests like cultures and lactate levels, prescribes standard treatment with broad spectrum antibiotics, IV fluid, and vasoactive drugs if necessary, all within 3 hours of a sepsis diagnosis [5]. Various investigations have demonstrated improvement in mortality amongst the septic population after timely compliance with the SEP-1 bundle [6,7]. Although there are effective protocols for treating sepsis once it has been detected, there still exists challenges in reliably identifying septic patients early in their course. As a result, there has been an increased focus on developing automated methods for reliably identifying early onset of sepsis to facilitate the timely administration of antibiotics and other life-saving interventions.
In recent years, the increased adoption of electronic health records (EHRs) in hospitals [8] has motivated the development of machine learning based surveillance tools for detection or classification [9–13] and prediction [9,14–17] of patients with sepsis or septic shock. For prediction of sepsis in particular, Desautels et al. [9] used a proprietary machine learning algorithm called InSight to achieve an Area Under the Curve (AUC) of 0.78 in predicting onset of sepsis four hours in advance. Nemati et al. [14] used a modified Weibull-Cox model on a combination of low-resolution Electronic Health Record (EHR) data and high-resolution vital signs time series data to predict onset of sepsis four hours in advance with an AUC of 0.85. Other works [15] have focused on developing models to predict septic shock, which occurs when sepsis leads to low blood pressure that persists despite treatment with intravenous fluids. However, a direct comparison of these methods is not possible for several reasons: 1) utilization of different labels for sepsis and septic shock, 2) variations in prediction horizon (finite horizon prediction vs infinite horizon prediction, 3) differences in frequency of prediction (single event classification vs sequential prediction), and 4) variations in study design and disease prevalence (case-control design vs calibrated real-world prevalence models). To date most sepsis prediction research has failed to make the transition into viable Clinical Decision Support (CDS) systems owing to the relatively low clinical tolerance for false-alarms [18], as well as the interpretability and workflow integration requirements for CDS systems [19,20]. False clinical alarms not only increase the cognitive load on clinicians but can also expose patients to unnecessary antibiotics and may contribute to emergence of antibiotic resistance pathogens [21]. Nevertheless, identifying and treating true cases of sepsis before they are clinically apparent is categorically one of the most important needs for modern medicine to address.
Consistently identifying the onset time of sepsis presents unique challenges because the condition manifests as constellation of signs and symptoms with significant variability in presentation and timing. The Third International Consensus Definitions for Sepsis (Sepsis-3) guidelines have provided two primary criteria for making a formal diagnosis of sepsis: 1) There must be a suspicion for infection (indicated by the administration of antibiotics for at least 72hrs with the concomitant collection of cultures) 2) There must be a two-point increase in the SOFA (Sequential Organ Failure Assessment) score [1]. These criteria have associated time points and from these time points, sepsis can be consistently labeled. While the Sepsis-3 criterion is considered the current standard for labeling sepsis onset time, previous consensus criteria for sepsis (based on Sepsis-1 and Sepsis-2 definitions) [22,23] remain in wide use. Additionally, there are other sepsis criteria developed by the Center for Disease Control (CDC) and Center for Medicare and Medicaid Services (CMS) for use in surveillance studies [5,24].
The primary contribution of this work is a deep learning framework for prediction of sepsis (called DeepAISE) that reduces incidents of false alarms by automatically learning predictive features related to higher-order interactions and temporal patterns among clinical risk factors for sepsis. Unlike comparable models, this algorithm maintains interpretability by tracking the top relevant features contributing to the sepsis score as a function of time, providing clinicians with rationale for alerts. Most importantly, DeepAISE is a generalizable algorithm developed using over 25,000 patient admissions to the Intensive Care Units (ICUs) at two Emory University hospitals, over 18,000 ICU admissions to the UC San Diego Health system and over 40,000 ICU admissions from the Medical Information Mart for Intensive Care-III (MIMIC-III) ICU database.
2. Materials and Methods
In this section, we describe the training and evaluation of DeepAISE and the patient cohorts utilized in this study.
2.1. Study design
We performed a retrospective study of all patients admitted to the ICUs at two hospitals within the Emory Healthcare system in Atlanta, Georgia from 2014 to 2018. This investigation was conducted according to Emory University Institutional Review Board (IRB) approved protocol #33069 and the UCSD IRB approved protocol #191098X. External evaluation of the DeepAISE algorithm was performed on two separate cohorts – a) the UCSD cohort, which consisted of all patients admitted to the ICUs at two hospitals within the UC San Diego Health system in San Diego, California from 2016 to 2019, and b) the Medical Information Mart for Intensive Care-III (MIMIC-III) ICU database [25], which is a publicly available database containing de-identified clinical data of patients admitted to the Beth Israel Deaconess Medical Center in Boston, Massachusetts from June 2001 to October 2012. Patients 18 years or older were followed throughout their ICU stay until discharge or development of sepsis, according to Sepsis-3 guidelines [1]. The purpose of this evaluation study was to show that the algorithm can be tailored to the characteristics of each local population and provide accurate predictions. During the external evaluation step, the DeepAISE model (trained on the Emory cohort) was fine-tuned and tested on the UCSD and MIMIC-III cohorts separately, and a comparison with baseline models was performed. The labels of tsuspicion, tSOFA, tsepsis-3, are used extensively throughout the work to define key time points and are clearly described in Table 1. For the purpose of defining sepsis, the order-time of antibiotics and cultures were obtained.
Table 1:
Description of defined time points utilized in the study.
| Time Point | Criteria |
|---|---|
| tsuspicion | Clinical suspicion of infection identified as the earlier timestamp of antibiotics and blood cultures within a specified duration. (If antibiotics were given first, the cultures must have been obtained within 24 hours. If cultures were obtained first, then antibiotic must have been subsequently ordered within 72 hours). Only those IV antibiotics that are commonly used for treatment of sepsis [24] which were administered for at least 72 hours were considered. |
| tSOFA | The occurrence of end organ damage as identified by a two-point deterioration in SOFA score within a 6-hour period |
| tsepsis-3 | The onset time of sepsis-3 is marked when both tsuspicion and tSOFA have happened within close proximity to each other. Specifically, tSOFA must occur 24 hours before tsuspicion or up to 12 hours after the tsuspicion (tSOFA + 24 hours > tsuspicion > tSOFA - 12 hours). The earlier of the tSOFA or tsuspicion was assigned to tsepsis-3. |
For the Emory cohort, data from the EHR (Cerner, Kansas City, MO) were extracted through a clinical data warehouse (MicroStrategy, Tysons Corner, VA). High-resolution heart rate and Mean arterial pressure time series at 2 seconds resolution were collected from select ICUs, through the BedMaster system (Excel Medical Electronics, Jupiter, FL). The BedMaster system is a third-party software connected to the hospital’s General Electric monitors for the purpose of electronic data extraction and storage of high-resolution waveforms. Patients were excluded if they developed sepsis within or prior to the first 4 hours of ICU admission (by analyzing pre-ICU IV antibiotic administration and culture acquisition) or if their length of ICU stay was less than 8 hours or more than 20 days.
The Emory cohort contained a total of 25,820 patients, 1,445 of whom met the Sepsis-3 criterion four hours or later after ICU admission (see Table S2 for patient characteristics). Those who developed sepsis tended to have a slightly higher percentage of male patients compared to non-septic patients (55.2% vs. 53.2%) and had more comorbidities (Charlson Comorbidity Index [CCI] 3 vs. 2). Septic patients had longer median lengths of ICU stay (5.9 vs. 1.9 days), higher median SOFA scores (5.0 vs. 1.7), and higher hospital mortality (15.2% vs. 3.5%). The median [interquartile range (IQR)] time from ICU admission to tsepsis-3 in the Emory cohort was 24 [9, 63] hours. Out of the 25,820 patients, 70% of them were used for developing the model (training set), 10% were used for hyper-parameter optimization (validation set), and the remaining 20% formed the testing set (see Table S6 for a description of the various holdout datasets that have been used for analysis in this paper). The Emory training set contained a total of 18,074 patients out of which 1,003 patients met the Sepsis-3 criterion, and the Emory testing set contained a total of 5,165 patients out of which 287 patients met the Sepsis-3 criterion during their stay in the ICU. The UCSD and MIMIC-III cohorts were split in a similar fashion, and more details can be found in the Appendices E and F of Supplementary Material. Specifically, a) the UCSD cohort consisted of a total of 18,752 patients, 1,073 of whom met the sepsis-3 criterion four hours or later after ICU admission, and b) the MIMIC-III cohort consisted of a total of 40,474 patients, 2,276 of whom met the sepsis-3 criterion four hours or later after ICU admission.
The complete set of patient features (65 in total, see Appendix C of Supplementary Material for more details) was grouped into three categories: clinical features (e.g. heart rate, mean arterial pressure, etc.), laboratory test results (e.g. hemoglobin, creatinine, etc.) and demographic/history/context features (e.g. age, care unit type, etc.). Some of the clinical or laboratory features that were unavailable in the UCSD and MIMIC-III cohorts were treated as ‘missing’ features during fine-tuning of DeepAISE. We refer the reader to Table S1 of Supplementary material for a list of features that are present or absent in each of the three patient cohorts considered in this study.
2.2. Development of the DeepAISE model
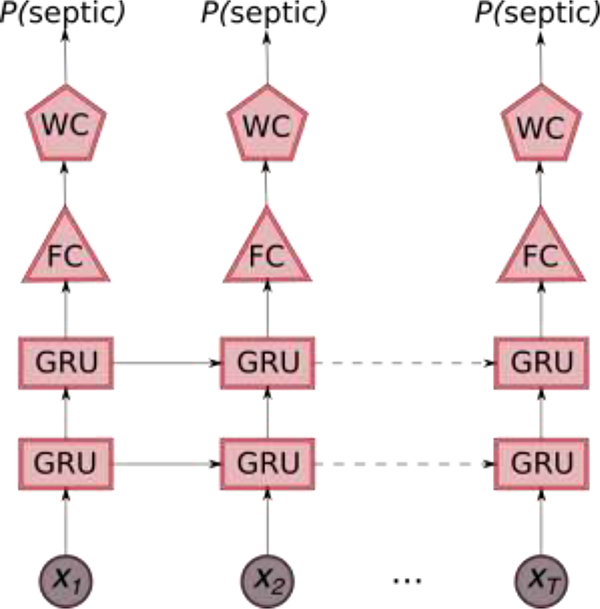
DeepAISE began producing scores four hours after ICU admission, and it was designed to predict (on an hourly basis) the probability of onset of sepsis within the next 2, 4, 6, 8, 10 and 12 hours. The two distinct characteristics of the model were a) utilization of a class of deep learning algorithms for multivariate time series data known as the Gated Recurrent Unit (GRU) [26] that allows for modeling the clinical trajectory of a patient over time, and b) deployment of a parametric survival model called the Weibull Cox proportional hazards (WCPH), which casts the problem of sepsis prediction to a time-to-event prediction framework and allows for handling of right censored outcomes [27]. The parametric survival model allows for efficient end-to-end learning of the GRU and the WCPH parameters using standard deep learning optimization techniques [28].
In preparation for time series modeling, the longitudinal data of patients were binned into consecutive windows of 1-hour duration, with the survival data for each of the time bins comprising of three elements: a set of input features x, the time to sepsis event τ and sepsis event indicator e. If a sepsis event occurred within the prediction horizon, the time interval τ would correspond to the duration of time between the time at which sepsis event occurred and the time of collection of features x, with the sepsis event indicator e set to 1. If sepsis did not occur within the prediction horizon, the time interval τ would correspond to one hour more than the duration of the prediction horizon, with the sepsis event indicator e set to 0 (i.e., a right-censored event). Instead of feeding the features of a patient directly to a WCPH model, we first fed the longitudinal data of patients into a GRU model followed by a feedforward neural network (FFNN) to learn a representation of patient’s trajectory at the current time-step, which was then fed to the WCPH model for time-to-event analysis. The WCPH model, a parametric counterpart to the familiar Cox proportional hazards model [27], represents the instantaneous risk of sepsis H(t) as a product of a baseline hazard function, , and the patient-specific sepsis risk which depends on the output of the GRU-FFNN module (or representations of a patient’s clinical trajectory at time t):
| (2) |
In the above equation, f(xt) denotes the output from the GRU-FFNN module, β represents the patient-specific hazard parameters, λ>0 is a scale parameter and ν>0 is a shape parameter for the Weibull distribution. The DeepAISE model (schematic diagram shown in Fig. 1) employed a combination of 2-layer stacked GRU framework and a modified WCPH model to predict the onset of sepsis at a regular interval of 1 hour. The parameters of the DeepAISE model, including the WCPH weight vector β, the scale and shape parameters of the Weibull distribution, and parameters of the GRU-FFNN were then learnt end-to-end by employing a mini-batch stochastic gradient descent approach that minimized the negative-log likelihood of the data. More technical details of the DeepAISE model and learning the model parameters can be found in Appendix B of Supplementary Materials. To make predictions on new patients, we used the trained model parameters and passed the longitudinal patient data sequentially through the GRU-FFNN, and the WCPH model to compute the survival scores, which provided the probability of not getting sepsis over the prediction horizon. The predicted probability of the sepsis event occurring within the prediction horizon was then defined as one minus the survival score.
Fig. 1: Schematic diagram of the Deep Artificial Intelligence Sepsis Expert (DeepAISE) model.
The 65 features that are measure/computed every hour are fed sequentially into a 2 layer stacked GRU framework, the output from the stacked GRU layer is then fed into a fully connected layer, and a modified Weibull Cox Proportional Hazards Model (WCPH) is employed to compute the probability of occurrence of sepsis within the proceeding 4 hours.
2.3. Data processing, model evaluation and statistical analysis
First, features in the Emory training set were normalized by subtracting the mean and dividing by the standard deviation (both of which were computed on the Emory training set). Next, all the remaining datasets were normalized using the mean and standard deviation computed from the Emory training set. For handling missing data, we used a simple sample-and-hold approach in all the datasets [14].
For all continuous variables, we have reported median ([25th - 75th percentile]). For binary variables, we have reported percentages. The area under receiver operating characteristic (AUC) curves statistics, specificity (1-false alarm rate) and accuracy at a fixed 85% sensitivity level were calculated to measure the performance of the models.
We have reported the DeepAISE performance results of four hours ahead prediction on the training and testing sets of the Emory cohort. External evaluation of DeepAISE was performed on the UCSD and MIMIC-III cohorts separately. During the external evaluation step, the DeepAISE model was fine-tuned on the training set of each external cohort and was evaluated on the corresponding test set. The MIMIC-III and UCSD cohorts were split into training and testing cohorts in the ratio 80%/20%. The Emory cohort was split into training and testing cohorts in the ratio 70%/20%. The remaining 10% was used for hyper-parameter optimization (validation set). During the hyper-parameter tuning phase, the optimal hyper-parameters were chosen based on the performance of the model on the validation set. Additionally, we have also reported the performance results for 2, 4, 6, 8, 10 and 12 hours ahead prediction of onset of sepsis. Statistical comparison of all AUC curves was performed using the method of DeLong et al. [29]
2.4. Parameter Optimization
We trained each model for a total of 200 epochs using the Adam optimizer [28], with a learning rate fixed at 1e-2. The mini-batch size was fixed at a total of 1,000 patients (90% control patients, 10% septic patients), with data randomly sampled (with replacement) in every epoch. To minimize overfitting and to improve generalizability of the model, L1-L2 regularization was used with L2 regularization parameter set to 1e-3 and L1 regularization parameter set to 1e-5. Our final model consisted of 2 GRU layers stacked on top of each other (with the size of hidden state being 100 per layer), followed by 1 fully connected layer (with the size of the hidden state being 100), and the output of which was fed into a modified Weibull-Cox proportional hazards model. All of the hyper-parameters of the model: Number of GRU layers, the size of hidden state in each of GRU layers, Number of fully connected layers, the size of hidden state in each of the fully connected layers, learning rate, mini-batch size, L1 regularization parameter, and L2 regularization parameter were optimized using Bayesian optimization [30]. All pre-processing of the data was performed using Numpy [31], with the rest of the pipeline implemented using TensorFlow [32].
2.5. Testing the validity of the relevance scores and model interpretability
The contribution of individual input features to the risk score was calculated using the associated relevance scores. The relevance scores were obtained by calculating the gradient of the sepsis risk score with respect to all input features and element-wise multiplication by the corresponding input features. The relevance scores, were then z-scored and the top 10 features (i.e., most frequently observed features across patients and across time) with a z-score of larger than 1.96 (corresponding to a 95% confidence interval) were reported for analysis of the overall importance of input risk factors (or ‘global interpretability’). To test the hypothesis that individual relevance scores capture meaningful information regarding contribution of each input feature to the risk score, we performed a series of studies in which we systematically masked (or treated as missing data): 1) the top 10 features with the largest positive and negative relevance scores in a global sense (global feature replacement analysis), 2) the top 10 locally important features for each individual risk scores (local feature replacement analysis) and 3) a random set of 10 features at each point in time (repeated 100 times). Once masking was done, the resulting risk scores produced by DeepAISE and the corresponding AUCs were computed. This analysis allowed us to systematically compare the contribution of the locally important features against the globally important predictors and against the 95-percentile of a randomly selected set of features.
3. Results
3.1. DeepAISE prediction performance for sepsis onset
DeepAISE made hourly predictions, starting four hours after ICU admission, and considered a total of 65 features that were commonly available in the EHR. The Emory training and testing sets contained a total of approximately 500,000 and 125,000 hourly prediction windows, respectively. A complete list of all the input features is provided in Appendix C of Supplementary Material.
The DeepAISE model reliably predicted tsepsis-3 four hours in advance with an AUC of 0.90 (specificity of 0.80 at sensitivity of 0.85) on the Emory testing set. Slightly lower performances were observed for the UCSD and MIMIC testing sets, with the fine-tuned models achieving an AUC of 0.88 and 0.87 respectively for predicting tsepsis-3 four hours in advance (see Table 2 for more details). Additionally, the performance of DeepAISE on UCSD cohort under different levels of missingness of input features was evaluated. We observed that when the UCSD cohort was divided into three sub-groups according to percentiles of missingness (Group 1: [0–33] percentile; Group 2: [33–66] percentile; and Group 3: [66–100] percentile), Group 3 had the highest AUC and Group 1 had the highest Area Under the Precision Recall Curve (AUCpr) (see Appendix K, Table S10, Fig. S14 and Fig. S15 for more details); thus, establishing an inverse relationship between AUC and AUCpr for these groups.
Table 2: Summary of DeepAISE performance.
The Area Under the Curve (AUC) and Specificity achieved by DeepAISE on the testing (and training) sets across the three centers for various prediction horizons have been tabulated.
| Performance Metric: testing set (training set) | 4 hours | 6 hours | 12 hours |
|---|---|---|---|
| Emory cohort | |||
| AUC* | 0.90 (0.94) | 0.89 (0.90) | 0.88 (0.89) |
| Specificity | 0.80 (0.89) | 0.78 (0.84) | 0.73 (0.78) |
| UCSD cohort+ | |||
| AUC* | 0.88 (0.90) | 0.87 (0.89) | 0.85 (0.86) |
| Specificity | 0.77 (0.78) | 0.74 (0.77) | 0.68 (0.70) |
| MIMIC-III cohort# | |||
| AUC* | 0.87 (0.90) | 0.86 (0.87) | 0.83 (0.86) |
| Specificity | 0.75 (0.78) | 0.72 (0.75) | 0.69 (0.73) |
AUC = Area under the Curve; Sensitivity was fixed at 0.85
DeepAISE model (trained on the Emory cohort) fine-tuned to the UCSD cohort
DeepAISE model (trained on the Emory cohort) fine-tuned to the MIMIC-III cohort
To assess the impact of changes in institutional practices and patient populations over time (‘temporal validation’) we performed an experiment in which a model trained on Emory year-based training set (patients in Emory cohort from the year 2014 through 2017) was applied to a holdout test set collected from 2017 to 2018 (Emory year-based holdout set). The DeepAISE model achieved an AUC of 0.88 (Specificity of 0.75 at sensitivity of 0.85) on the Emory year-based holdout set (see Fig. S4 for more details), which was comparable to the model performance on external evaluation cohorts.
3.2. DeepAISE performance against other baselines
We assessed the utility of DeepAISE’s model architecture by comparing its performance against four different baseline models: 1) a Logistic Regression (LR) model, 2) a Weibull-Cox proportional hazards (WCPH) model, 3) a Feedforward Neural Network (FFNN) with two layers of 100 hidden units and a final WCPH layer for prediction of onset of sepsis, and 4) GB-Vital: This is a replication of the sepsis detection model as proposed by Mao et al. [33,34]. The model corresponds to a gradient-boosted classifier of decision trees built using six vital signs measurements: systolic blood pressure, diastolic blood pressure, heart rate, respiratory rate, oxygen saturation and temperature. A more comprehensive comparison of DeepAISE with other baseline models can be found in Supplementary Material (See Fig. S3).
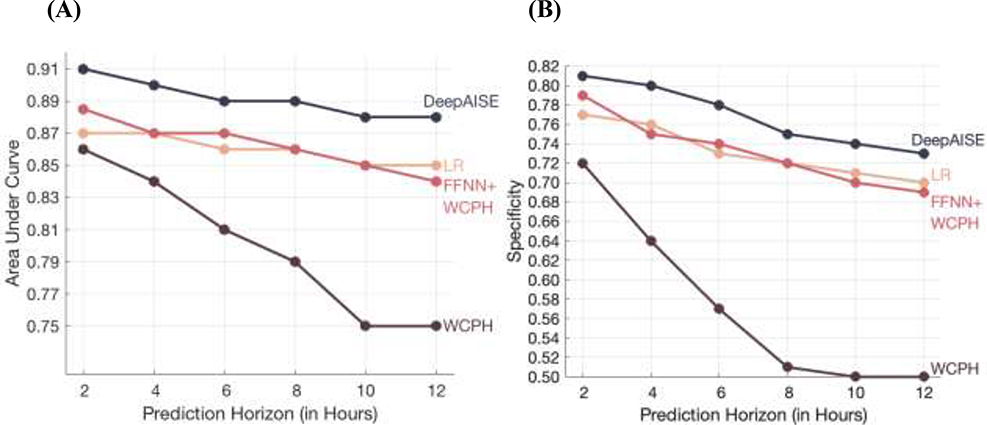
Across all prediction windows, DeepAISE consistently outperformed all other baseline classifiers (p<0.001; when AUC of DeepAISE was compared with AUC of other baseline methods) for prediction of tsepsis-3 (See Fig. 2, Fig. S3 and Table S3) and across all prediction windows, indicating that capturing temporal trends and interactions among the risk factors is important for accurate prediction of sepsis. Additionally, DeepAISE outperformed the GB-Vital baseline model which achieved AUCs of 0.711, 0.700 and 0.701 on the Emory, UCSD and MIMIC-III cohorts, respectively for prediction of onset of sepsis four hours in advance. The performance of all the models decreased with the increase in prediction horizon. For DeepAISE, the AUC on the Emory testing set decreased from 0.90 at 4-hour prediction horizon to 0.88 at 12-hour prediction horizon. We also observed that these findings were consistent with the performance of the fine-tuned model on the UCSD and MIMIC testing sets (Fig. S9 and Fig. S10).
Fig. 2: Comparison of DeepAISE performance on Emory testing set for prediction horizons of 2, 4, 6, 8, 10 and 12 hours.
The Area Under the Curve (AUC) and Specificity (at 85% Sensitivity) are shown in Panels (A) and (B), respectively.
DeepAISE = Deep Artificial Intelligence Sepsis Expert
LR = Logistic Regression
FFNN = Feedforward Neural Network
WCPH = Weibull Cox Proportional Hazard layer
In addition, a FFNN trained to predict tsepsis-3 with delta change in SOFA score as input achieved 0.54 AUC on Emory testing set, and a FFNN trained to predict tsepsis-3 with delta change in SOFA score and static covariates (such as age, gender, weight etc.) as inputs achieved 0.68 AUC on the Emory testing set. The above results show that DeepAISE scores were not simply recapitulations of the SOFA scores.
3.3. Clinical interpretation of DeepAISE predictions
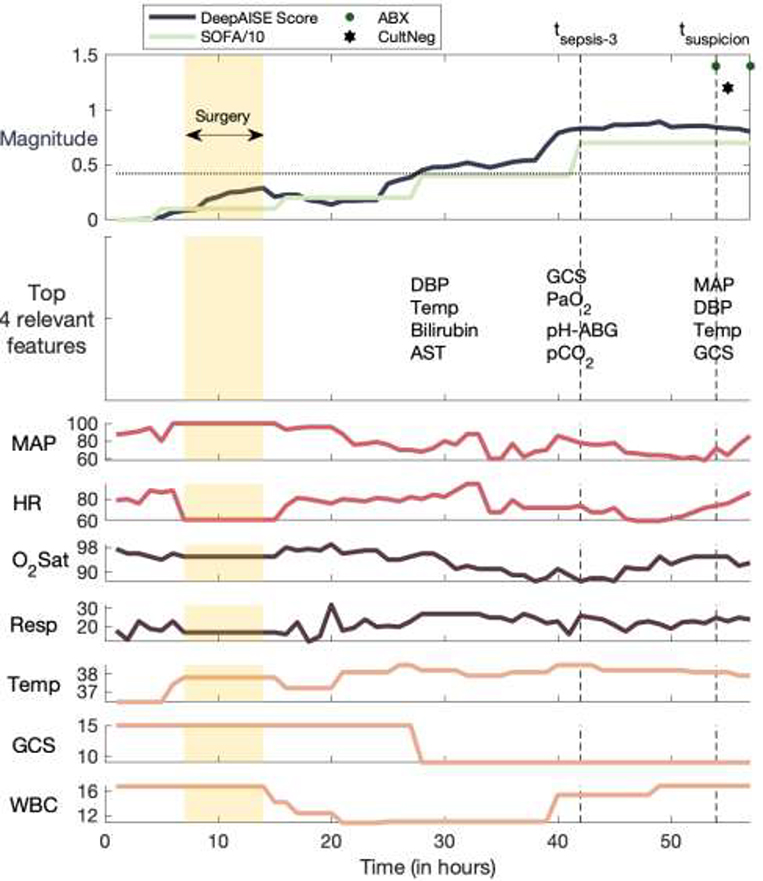
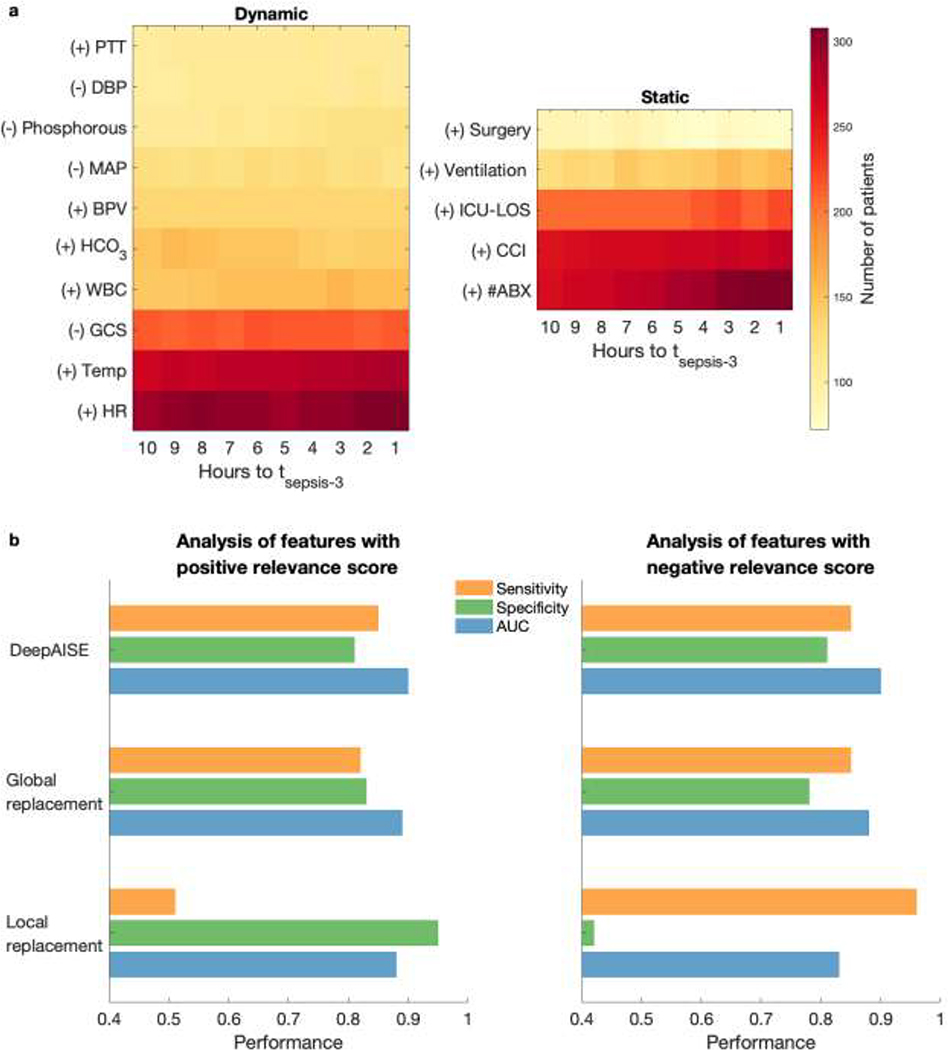
While performance characteristics of machine learning algorithms are important, providing interpretable data to the bedside clinicians that can guide diagnosis and therapeutic interventions is a critical requirement of CDS systems. To date many sepsis models have failed to demonstrate which physiologic aberrations contributed to the model’s prediction, compelling many to refer to them as “black boxes”. DeepAISE allayed these concerns by continually revealing the top patient features contributing to its predicted risk scores. Unlike many other algorithms, DeepAISE is uniquely interpretable as evidenced in Fig. 3 in which the trajectory of a septic patient who developed ventilator associated pneumonia in the ICU is displayed. In this visualization, the sepsis risk score predicted by the model is shown along with vital sign trends, and most notably, the most relevant features contributing to the risk score. In this example, early deterioration of the patient’s respiratory status was detected by the model. The model identified aberrations in PaO2, PaCO2, blood pH and Glasgow coma score (GCS) as some of the top features relevant to its prediction. The importance of each feature was calculated using the magnitude and sign of the associated relevance scores, in a fashion similar to saliency maps for convolutional neural networks [35]. To validate the clinical interpretability of the DeepAISE model, analysis of the most relevant features starting 10 hours prior to and ending at tsepsis-3 was conducted (see Fig. 5). This investigation revealed that DeepAISE ascribed importance to several features that have already been identified as risk factors for sepsis such as recent surgery, length of ICU stay, heart rate, GCS, white blood cell count, and temperature, and some less appreciated but known factors such as Phosphorus (or hypophosphatemia) [36]. This analysis provides a global view of model interpretability, whereas the individual relevance scores provide a local view of interpretability by listing the top features contributing to the risk scores for each hourly prediction window. Perturbation analysis revealed that the globally important features may not provide an accurate view of the top contributing factors to the individual risk scores. We observed that treating the top locally important features as missing values yielded a significantly lower AUC compared with a similar analysis replacing the globally most important features. (See Fig. 5; Additionally, refer to Appendix H of Supplementary Material for a more thorough analysis of global model interpretability vs local model interpretability).
Fig. 3: A clinically interpretable example of DeepAISE.
The DeepAISE score is shown for a septic patient according to the Third International Consensus Definitions for Sepsis (Sepsis-3). Commonly recorded hourly vital signs of the patient, including heart rate (HR), mean arterial blood pressure (MAP), respiratory rate (RESP), temperature (TEMP), oxygen saturation (O2Sat) are shown. The most significant features contributing to the DeepAISE score are listed immediately below the DeepAISE Scores (for clarity of presentation, only selected time points are shown). The horizontal dashed line indicates the prediction threshold corresponding to a sensitivity of 0.85. Refer to Appendix C of Supplementary Material for more details on the abbreviated features.
Fig. 5: Most common features contributing to an elevated risk score.
(a) Every hour DeepAISE identifies the top features contributing to an individual septic patient’s risk score. The left subfigure demonstrates the frequency of the top ten dynamic features (ordered according to the magnitude of the relevance score) across the septic patient population (in the Emory cohort) preceding tsepsis-3 and the right subfigure demonstrates the frequency of the top five static features that are seen preceding tsepsis-3. Features with positive gradient with respect to the sepsis risk score are identified by ‘(+)’. Features with negative gradient with respect to the sepsis risk score are identified by ‘(−)’. (b) Summary of performance of DeepAISE (on the Emory testing set) when global feature replacement analysis and local feature replacement analysis were performed for features with positive relevance score (left subfigure; see Table S7) and negative relavance score (right subfigure; see Table S8). Note that the performance (AUC) of DeepAISE when a random set of 10 features at each point in time were masked (repeated 100 times) was 0.899 [0.886, 0.901]. The sensitivity and specificity values reported for global feature replacement analysis and local feature replacement analysis were measured at threshold corresponding to 0.85 sensitivity for the original model.
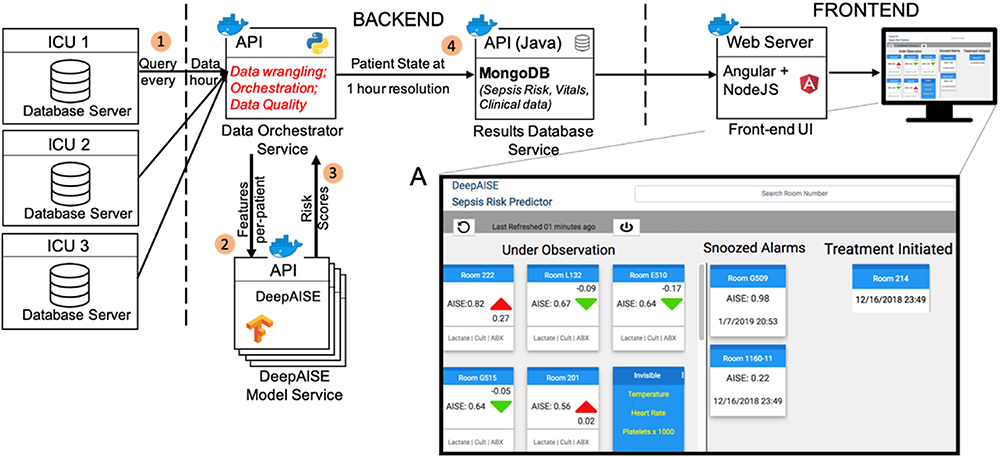
Additionally, DeepAISE was implemented in a real-time setting and a decision support user interface (UI) was designed to communicate the risk scores to the clinical team. The resultant UI shown in Fig. 6 was designed to present a list of patients sorted by DeepAISE risk score for predicting tsepsis-3 four hours in advance (See Appendix K of Supplementary Material for more details). Square cards that include the sepsis risk score as well as the change in score over the past hour are used to represent a single patient. Individual cards can be flipped via a single mouse-click to reveal the top factors contributing to the presented score. To improve individual and unit situational awareness regarding patient interventions and assessments, users can drag-and-drop patient cards into columns representing different treatment categories.
Fig. 6: Block diagram of the DeepAISE software platform.
EHR is queried for the required data elements (1). EHR data is then prepared by the Data Orchestrator Service and passed to the DeepAISE Model Service for DeepAISE score computation (2). The resulting scores are computed and returned to the Data Orchestrator Service using a predefined set of API calls (3). The scores are then managed by a time series database in the Results Database Service (4), which provides the required data for the UI implemented using client-side Javascript. The figure inset shows a live integration of DeepAISE UI in a tele-ICU workflow.
3.4. Inferring significance of individual patient trajectories
Clinicians have long appreciated that trends in patient metrics are often more telling than discrete point values. The high dimensional nature of the data used to represent a patient is challenging to represent. Display of patient trajectories as they pass from states of sickness to health provides yet another opportunity to inform the clinician about a patient’s expected clinical course.
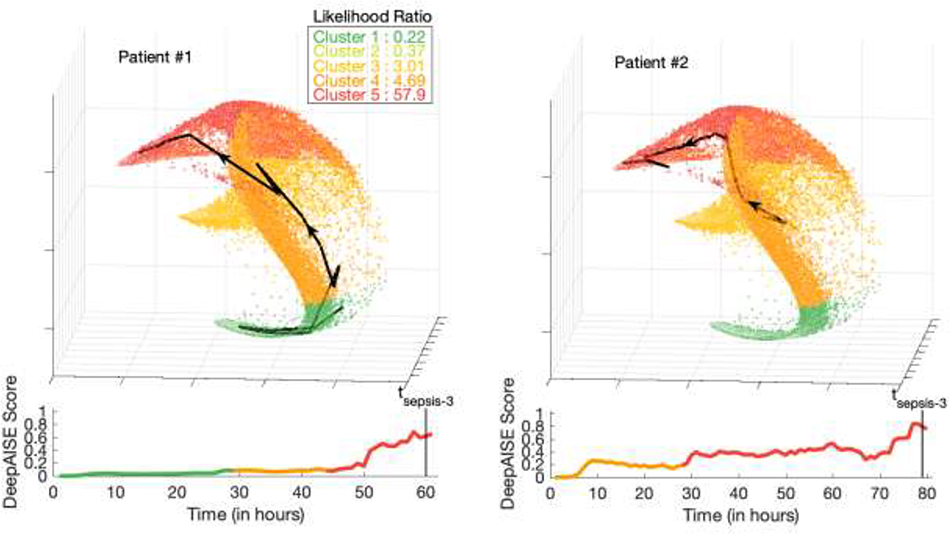
Each point on the manifold shown in Fig. 4 is a 3D representation (projection) of the patient’s 65 features, constructed via first learning a 100-dimensional representation (last layer of the DeepAISE model) followed by dimensionality reduction via Spectral clustering [37]. The individual axes in Fig. 4 denote a unique weighted combination of the learned 100-dimensional representations, designed to construct a 3D space that preserves the distance among the original data points as much as possible. Two exemplar 3D patient trajectories are presented in Fig. 4. Patient 2 was in a state of good health (specifically no suspicion for infection) prior to developing a subdural hemorrhage which prompted admission. This patient went on to be diagnosed with a ventilator associated pneumonia two days after an emergent craniectomy. In contrast Patient 1, who was several weeks status post craniectomy for stroke, was readmitted with a culture positive pneumonia present on admission. The manifold in Fig. 4 shows that trajectories for patient 1 and 2 follow similar terminal patterns; however, correctly assigns them different starting positions with patient 2 starting from a comparatively higher risk cluster. The specific trajectory of an ICU patient may be useful in categorizing infectious phenotypes and detecting anomalous physiological dynamics.
Fig. 4: Visualization of DeepAISE time series representations.
The trajectory of the DeepAISE score for two septic patients (Patient #1 and Patient #2) from ICU admission until sepsis diagnosis is displayed below a larger manifold that makes use of spectral clustering to visually display a patient’s physiologic journey through their illness (each point on the graph represents one hour of data from one patient). The colors for both patients in the plots were chosen based on the predicted sepsis risk score (green represents the lowest predicted sepsis risk score, and red represents the highest predicted sepsis risk score). A similar figure with septic patients highlighted is shown in Fig. S6 of Supplementary Material. (a) Patient #1 (P1) was a 63-year-old female admitted for a left sided subdural hemorrhage who underwent a craniectomy on hospital day zero. This patient remained intubated after surgery and began receiving treatment for a culture proven ventilator associated pneumonia the afternoon of hospital day number three. DeepAISE identified this patient as being septic nearly 24hrs before clinical treatment was implemented (See Fig. S7). (b) Patient #2 (P2) was a 70-year-old male who was admitted for altered mental status and seizures after vascular coiling of a middle cerebral artery (MCA) aneurysm. P2 was intubated on admission and began treatment for a culture proven ventilator associated pneumonia on hospital day five however DeepAISE made its sepsis prediction nearly 36 hours prior to this time, after demonstrating a steadily worsening score since admission (See Fig. S8).
4. Discussion
In this work, a deep learning model was used to automatically learn complex features, including temporal trends and higher-order interactions among the risk factors, to accurately predict the likelihood of sepsis in patients admitted to the ICU up to 12 hours in advance. DeepAISE was developed to predict tsepsis-3 (Sepsis-3 criterion) in this study. We observed that the four hours ahead prediction AUC of DeepAISE (on Emory testing set) was 0.90 for tsepsis-3. Additionally, we have included varying prediction windows to illustrate how DeepAISE performs at various time frames to illustrate potential uses (Fig. 2, Fig. S3, S9 and S10 in Supplementary Material). For a 12 hour ahead prediction horizon, specificity of 0.73 was achieved (on the Emory cohort) at 0.85 sensitivity versus specificity of 0.80 for 4 hours ahead prediction. A model with lower specificity would have a higher false positive rate, which could potentially lead to additional cognitive burden on the clinical team who are ultimately responsible for evaluating patients for initiation of SEP-1 bundle. All our findings were externally reproduced with the UCSD and MIMIC-III patient cohorts, providing supporting evidence that the DeepAISE algorithm can be tailored and applied to a geographically diverse patient population.
Another advantage of the proposed deep learning approach is in its ability to provide the top factors contributing to the risk score for every point in time for each patient (i.e., local interpretability). The distinction between global and local notions of interpretability (i.e., what features are contributing to the sepsis risk score for the cohort at large versus an individual patient’s hourly prediction window) is most notable when dealing with models capable of capturing higher order interactions and temporal trends in the data. As a result, the degree of risk associated with a factor (e.g., temperature) is a function of other factors in a multiplicative sense (e.g., hypothermia and old age are together a greater risk factor than either by itself). Similarly, the temporal context of a risk factor can alter its contribution to a given risk score calculation (e.g., leukocytosis immediately after surgery may not be unexpected and may contribute differently to the risk for sepsis). These multiplicative and temporal factors (which are captured by the DeepAISE model) result in variations in the importance of risk factors when viewed from a local, hourly prediction perspective for each patient. Note that traditional logistic regression models and decision trees are not capable of making such inferences unless the relevant features are hand-crafted by the experts and included in the model.
A major barrier to wide adoption of modern machine learning based CDS tools in clinical practice has been their “black box” nature [38]. While it is important to design deep learning models with high performance, it is imperative to build models that provide interpretable data to bedside clinicians that can augment their understanding of the disease process and can contribute to the selection and initiation of appropriate treatments. DeepAISE was designed to be transparent by: 1) continually revealing the top causes contributing to the sepsis risk score (see Fig. 3), 2) providing a lower dimensional view of the patients’ trajectory (see Fig. 4), and 3) providing a prioritized list of patients at risk for sepsis (see Fig. 6). These three attributes allow the bedside clinician to identify pathologic deviations from expected physiology early and in real-time throughout the duration of patients’ hospital admission. Further longitudinal usability studies are required to validate the utility of this feature to improve situational awareness and assist clinicians with independent evaluation of patient’s risk for sepsis prior to initiation of interventions.
We have shown that the top causes can be broken down into two categories of positively and negatively contributing factors to the risk score (see Appendix H of supplementary material). Notably, this analysis shed insight on the input features contributing significantly to the sensitivity (positive contributors) and specificity (negative contributors) of DeepAISE (see Fig. 5). Since one of the key limitations of using EHR data is the intermittent nature of laboratory measurements, we hypothesize that one may use the knowledge of the top contributing factors to protocolize the ordering of laboratory tests, to ensure specific updated measurements of these factors are available to the algorithm, thus improving model sensitivity and specificity. Our results indicate that the degree of data missingness was inversely correlated with the prevalence of sepsis within a given subgroup of patients (see Appendix K, Table S10). DeepAISE had the highest AUC on the subgroup with the highest level of missingness and the lowest prevalence of sepsis; since data missingness pushes the risk score to lower values which translates to more specific predictions at the cost of reduced sensitivity and positive predictive value. The net effect was a higher AUC due to the higher prevalence of negative labels within this subgroup. Further work is required to assess the performance of DeepAISE under varying degrees of missingness of the top contributing factors to the risk score. This is particularly important as one extends such algorithms to non-ICU units where patients are not as frequently monitored.
In recent years, several machine learning (ML) based models for early prediction of sepsis and septic shock have been proposed and have been summarized in detail by Moor et al. [16] and Fleuren et al. [17]; although variations in experimental design and definitions of sepsis make a direct comparison of these methods impractical. Desautels et al. [9] proposed a proprietary machine learning model called InSight to predict sepsis in ICU patients. Their model used a combination of vital signs, pulse oximetry, GCS, and age as input features. An earlier version of this algorithm relied on the Sepsis-3 definition (specifically tSOFA) to train its model and was able to reliably identify (detect) patients at the time they had met the Sepsis-3 criteria, with a 4-hours ahead prediction AUC of 0.74, which is comparable to performance reported by Amland et al.[39]. Following the Sepsis-3 definition, Nemati et al. [14] achieved an AUC of 0.85 for 4 hours ahead prediction of sepsis, by combining 65 data points including low-resolution data from the EHR and high-resolution vital sign time series features from the bedside monitors. In comparison to existing works, some of the novel contributions of this study are as follows. First, this study is the amongst the first that has been validated on data from three different healthcare systems. Second, majority of the existing works have approached prediction of sepsis as a classification problem while we have approached it as a survival analysis problem (within a Weibull-Cox survival analysis framework), which enables us to model right-censored outcomes (e.g., patients who were transferred or died prior to observation of severe sepsis). Third, while other works have explored interpretability methods, this is the first work that has utilized sensitivity analysis (see section 3.3; Fig. 5) to validate relevance score as a metric for identifying the top features contributing to the predicted risk score. Finally, this work is among the very few that has explored clinical implementation of a deep-learning based sepsis prediction algorithm (Fig. 6). In particular, the DeepAISE User interface has been designed to communicate both the predicted risk score and the most relevant features contributing to risk score to the clinical care team.
Experimental design can have a pronounced effect on the reported AUC of machine learning algorithms. A commonly utilized method known as the ‘case-control’ design (which includes the majority of studies involving biomarkers) significantly overestimates the true prevalence of positive labels and can result in highly optimistic reported performances in the literature when compared to a ‘sequential prediction’ design [14]. Assuming a sepsis prevalence of 8% in the ICU population (after excluding all cases of sepsis developed before ICU admission), a median time-to-sepsis of 23 hours, and a 6-hours ahead prediction window, typically only 1–2% of the observed windows include a positive label for sepsis. The case-control study design assumes the timing of sepsis is known a priori and seeks to show that certain physiological or biomarker signatures preceding this time are significantly different than that of the non-septic control patients. The resulting algorithms, which are tuned to a 50% prevalence of positive labels, tend to produce high rates of false alarms when deployed prospectively.
In general, statistical evaluation methods (such as the AUC) have a limited applicability when evaluating the clinical utility of such algorithms, although they can provide quantitative metrics for the comparison of various algorithms. In practice, performance metrics are only meaningful when coupled with appropriate clinical protocols that describe the course of action in response to the associated risk alerts. Simple clinical actions (such as ‘snoozing’ the alarm for X hours if the patient did not meet the clinical threshold to initiate therapy) can significantly alter the false-alarm rate (defined as 1-Specificity) and the associated AUC of an algorithm in practice.
While we have strictly adhered to the Sepsis-3 criteria for defining septic labels in our study, it has been noted that this criterion is too stringent and the sensitivity of early detection is lost to an increased specificity [40,41]. The Sepsis-3 criterion utilized in this study is an acausal clinical construct for demarcating the onset time of sepsis, and as such cannot be used in a clinical setting for early detection of sepsis; however, a predictive analytic risk score when trained to predict the associated onset-time can combine the specificity advantages of Sepsis-3 with the benefits of early recognition. Moreover, it is critical to appreciate that making a clinical diagnosis of sepsis carries much greater value than simply identifying ‘poor health’ or general decompensation. True cases of sepsis can be positively impacted by the rapid administration of broad-spectrum antibiotics, IV fluids, and vasopressors if indicated [42,43] where as “decompensated” patients still need further assessment to ascertain the etiology of the deterioration and to determine appropriate intervention.
A potential use case of DeepAISE is to facilitate compliance with standardized care bundles such as SEP-1 [5], which advocates for obtaining blood cultures, administering broad spectrum antibiotics, measuring lactate, and starting appropriate fluid resuscitation if clinically indicated, all within 3 hours of clinical recognition of sepsis. We anticipate that a likely clinical workflow may include 1) flagging of a patient by the DeepAISE risk score with a prediction horizon of 4 hours, 2) independent evaluation of the patient by a bedside caregiver (this may include ordering of additional labs such as lactate), 3) followed by ordering of cultures prior to ordering of antibiotics, and 4) completion of the SEP-1 bundle components. The predictive ability of DeepAISE and enumeration of top causes contributing to the risk score is remarkable because clinicians will be able to independently evaluate the algorithm’s rationale for flagging a patient, and when clinically appropriate begin implementing components of the sepsis bundle much earlier. In fact, a recent study provided critical evidence [44] that longer intervals from antibiotic order to infusion are associated with higher mortality rates in septic and septic shock patients, thus emphasizing the importance of improving workflow related factors to the care of this patient population.
Sepsis survivors often suffer from high rates of readmission [45] and many survivors of sepsis face life-long, debilitating sequelae as a result of the disease [46]. Future extensions of this work will involve performing prospective clinical trials to validate DeepAISE’s real-time predictions in a clinical setting; however, our findings provide significant clinical evidence for a radical change to the sepsis treatment paradigm that has real-time high-dimensional data analysis and model transparency at its center.
Supplementary Material
Highlights.
Sepsis is among the leading causes of morbidity, mortality, and cost overruns in the Intensive Care Unit (ICU).
Early prediction of sepsis can improve situational awareness amongst clinicians and facilitate timely, protective interventions.
DeepAISE (Deep Artificial Intelligence Sepsis Expert), a recurrent neural survival model for the early prediction of sepsis.
DeepAISE maintains interpretability by tracking the top relevant features contributing to the sepsis score as a function of time, providing clinicians with rationale for alerts.
Acknowledgments:
We would like to acknowledge our funding sources. Additionally, we would like to thank Drs. Andre Holder and Russell Jeter for their constructive comments on the manuscript, and the members of our tele-ICU team especially Dr. Timothy G. Buchman and Cheryl Hiddleson for feedback regarding the user interface, Chad Robichaux for help with data extraction and curation from the Emory Clinical Data Warehouse, and Fatemeh Amrollahi for her contribution to development of the software pipeline.
Funding: Dr. Nemati is funded by the National Institutes of Health (NIH), award #K01ES025445. Dr. Josef is funded by the Surgical Critical Care Initiative (SC2i), funded by the Department of Defense Defense Health Program Joint Program Committee 6 / Combat Casualty Care (USUHS HT9404-13-1-0032 and HU0001-15-2-0001). The opinions or assertions contained herein are the private ones of the author and are not to be construed as official or reflecting the views of the Department of Defense, the Uniformed Services University of the Health Sciences, the NIH or any other agency of the US Government.
Footnotes
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: MIMIC-III is a de-identified critical care dataset and is publicly available. Access to de-identified data of Emory cohort has been made available as a part of the PhysioNet Challenge 2019. The UCSD data is not publicly available. Access to the computer code used in this research is available upon request to the corresponding author.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Singer M, Deutschman CS, Seymour CW, Shankar-Hari M, Annane D, Bauer M, Bellomo R, Bernard GR, Chiche J-D, Coopersmith CM, Hotchkiss RS, Levy MM, Marshall JC, Martin GS, Opal SM, Rubenfeld GD, van der Poll T, Vincent J-L, Angus DC, The third international consensus definitions for sepsis and septic shock (Sepsis-3), JAMA. 315 (2016) 801–810. 10.1001/jama.2016.0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Rhee C, Dantes R, Epstein L, Murphy DJ, Seymour CW, Iwashyna TJ, Kadri SS, Angus DC, Danner RL, Fiore AE, Jernigan JA, Martin GS, Septimus E, Warren DK, Karcz A, Chan C, Menchaca JT, Wang R, Gruber S, Klompas M, Incidence and trends of sepsis in US hospitals using clinical vs claims data, 2009–2014, JAMA. 318 (2017) 1241–1249. 10.1001/jama.2017.13836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Torio CM, Moore BJ, National Inpatient Hospital Costs: The Most Expensive Conditions by Payer, 2013: Statistical Brief #204, in: Healthcare Cost and Utilization Project (HCUP) Statistical Briefs, Agency for Healthcare Research and Quality (US), Rockville (MD), 2006. http://www.ncbi.nlm.nih.gov/books/NBK368492/ (accessed September 3, 2018). [PubMed] [Google Scholar]
- [4].Dellinger RP, Carlet JM, Masur H, Gerlach H, Calandra T, Cohen J, Gea-Banacloche J, Keh D, Marshall JC, Parker MM, Ramsay G, Zimmerman JL, Vincent J-L, Levy MM, Surviving Sepsis Campaign guidelines for management of severe sepsis and septic shock, Intensive Care Medicine. 30 (2004) 536–555. 10.1007/s00134-004-2210-z. [DOI] [PubMed] [Google Scholar]
- [5].Centers for Medicare & Medicaid Services, QualityNet—inpatient hospitals specifications manual. Quality website. https://www.qualitynet.org/inpatient/specifications-manuals. Accessed August, 2020, (n.d.).
- [6].Rhee C, Filbin MR, Massaro AF, Bulger AL, McEachern D, Tobin KA, Kitch BT, Thurlo-Walsh B, Kadar A, Koffman A, Pande A, Hamad Y, Warren DK, Jones TM, O’Brien C, Anderson DJ, Wang R, Klompas M, Compliance With the National SEP-1 Quality Measure and Association With Sepsis Outcomes: A Multicenter Retrospective Cohort Study*, Critical Care Medicine. 46 (2018) 1585–1591. 10.1097/CCM.0000000000003261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Han X, Edelson DP, Snyder A, Pettit N, Sokol S, Barc C, Howell MD, Churpek MM, Implications of centers for medicare & medicaid services severe sepsis and septic shock early management bundle and initial lactate measurement on the management of sepsis, Chest. 154 (2018) 302–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].DesRoches CM, Campbell EG, Rao SR, Donelan K, Ferris TG, Jha A, Kaushal R, Levy DE, Rosenbaum S, Shields AE, Electronic health records in ambulatory care—a national survey of physicians, New England Journal of Medicine. 359 (2008) 50–60. [DOI] [PubMed] [Google Scholar]
- [9].Desautels T, Calvert J, Hoffman J, Jay M, Kerem Y, Shieh L, Shimabukuro D, Chettipally U, Feldman MD, Barton C, Wales DJ, Das R, Prediction of sepsis in the intensive care unit with minimal electronic health record data: a machine learning approach, JMIR Medical Informatics. 4 (2016) e28. 10.2196/medinform.5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Horng S, Sontag DA, Halpern Y, Jernite Y, Shapiro NI, Nathanson LA, Creating an automated trigger for sepsis clinical decision support at emergency department triage using machine learning, PloS One. 12 (2017) e0174708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Brown SM, Jones J, Kuttler KG, Keddington RK, Allen TL, Haug P, Prospective evaluation of an automated method to identify patients with severe sepsis or septic shock in the emergency department, BMC Emergency Medicine. 16 (2016) 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Giuliano KK, Physiological monitoring for critically ill patients: testing a predictive model for the early detection of sepsis, AJCC. 16 (2007) 122–130; quiz 131. [PubMed] [Google Scholar]
- [13].Shashikumar SP, Li Q, Clifford GD, Nemati S, Multiscale network representation of physiological time series for early prediction of sepsis, Physiological Measurement. 38 (2017) 2235–2248. 10.1088/1361-6579/aa9772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Nemati S, Holder A, Razmi F, Stanley MD, Clifford GD, Buchman TG, An interpretable machine learning model for accurate prediction of sepsis in the ICU, Crit Care Med. 46 (2018) 547–553. 10.1097/CCM.0000000000002936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Henry KE, Hager DN, Pronovost PJ, Saria S, A targeted real-time early warning score (TREWScore) for septic shock, Science Translational Medicine. 7 (2015) 299ra122–299ra122. 10.1126/scitranslmed.aab3719. [DOI] [PubMed] [Google Scholar]
- [16].Moor M, Rieck B, Horn M, Jutzeler C, Borgwardt K, Early Prediction of Sepsis in the ICU using Machine Learning: A Systematic Review., MedRxiv (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Fleuren LM, Klausch TLT, Zwager CL, Schoonmade LJ, Guo T, Roggeveen LF, Swart EL, Girbes ARJ, Thoral P, Ercole A, Hoogendoorn M, Elbers PWG, Machine learning for the prediction of sepsis: a systematic review and meta-analysis of diagnostic test accuracy, Intensive Care Medicine. (2020). 10.1007/s00134-019-05872-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Awry ABTG, Sepsis early warning scoring systems: The ideal tool remains elusive!, J Crit Care. (2018). [DOI] [PubMed] [Google Scholar]
- [19].Shortliffe EH, Sepúlveda MJ, Clinical decision support in the era of artificial intelligence, JAMA. 320 (2018) 2199–2200. [DOI] [PubMed] [Google Scholar]
- [20].Norrie J, The challenge of implementing AI models in the ICU, The Lancet Respiratory Medicine. 6 (2018) 886–888. 10.1016/S2213-2600(18)30412-0. [DOI] [PubMed] [Google Scholar]
- [21].Ventola CL, The antibiotic resistance crisis: part 1: causes and threats, Pharmacy and Therapeutics. 40 (2015) 277. [PMC free article] [PubMed] [Google Scholar]
- [22].Dellinger RP, Levy MM, Rhodes A, Annane D, Gerlach H, Opal SM, Sevransky JE, Sprung CL, Douglas IS, Jaeschke R, Osborn TM, Nunnally ME, Townsend SR, Reinhart K, Kleinpell RM, Angus DC, Deutschman CS, Machado FR, Rubenfeld GD, Webb SA, Beale RJ, Vincent J-L, Moreno R, Surviving Sepsis Campaign Guidelines Committee including the Pediatric Subgroup, Surviving sepsis campaign: international guidelines for management of severe sepsis and septic shock: 2012, Critical Care Medicine. 41 (2013) 580–637. 10.1097/CCM.0b013e31827e83af. [DOI] [PubMed] [Google Scholar]
- [23].Bone RC, Balk RA, Cerra FB, Dellinger RP, Fein AM, Knaus WA, Schein RM, Sibbald WJ, Definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. The ACCP/SCCM Consensus Conference Committee. American College of Chest Physicians/Society of Critical Care Medicine, Chest. 101 (1992) 1644–1655. [DOI] [PubMed] [Google Scholar]
- [24].Center for Disease Control and Prevention, Hospital toolkit for adult sepsis surveillance, US Department of Health and Human Services. (2018). https://www.cdc.gov/sepsis/pdfs/Sepsis-Surveillance-Toolkit-Mar-2018_508.pdf. [Google Scholar]
- [25].Johnson AE, Pollard TJ, Shen L, Li-wei HL, Feng M, Ghassemi M, Moody B, Szolovits P, Celi LA, Mark RG, MIMIC-III, a freely accessible critical care database, Scientific Data. 3 (2016) 160035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Cho K, Van Merriënboer B, Gulcehre C, Bahdanau D, Bougares F, Schwenk H, Bengio Y, Learning phrase representations using RNN encoder-decoder for statistical machine translation, ArXiv Preprint ArXiv:1406.1078. (2014). [Google Scholar]
- [27].Cox DR, Regression models and life-tables, in: Breakthroughs in Statistics, Springer, 1992: pp. 527–541. [Google Scholar]
- [28].Kingma DP, Ba J, Adam: A method for stochastic optimization, ArXiv Preprint ArXiv:1412.6980. (2014). [Google Scholar]
- [29].DeLong ER, DeLong DM, Clarke-Pearson DL, Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach, Biometrics. (1988) 837–845. [PubMed] [Google Scholar]
- [30].Snoek J, Larochelle H, Adams RP, Practical bayesian optimization of machine learning algorithms, Advances in Neural Information Processing Systems. (2012) 2951–2959. [Google Scholar]
- [31].Oliphant TE, A guide to NumPy, Trelgol Publishing USA, 2006. [Google Scholar]
- [32].Abadi M, Barham P, Chen J, Chen Z, Davis A, Dean J, Devin M, Ghemawat S, Irving G, Isard M, Tensorflow: a system for large-scale machine learning., OSDI. 16 (2016) 265–283. [Google Scholar]
- [33].Mao Q, Jay M, Hoffman JL, Calvert J, Barton C, Shimabukuro D, Shieh L, Chettipally U, Fletcher G, Kerem Y, Zhou Y, Das R, Multicentre validation of a sepsis prediction algorithm using only vital sign data in the emergency department, general ward and ICU, BMJ Open. 8 (2018) e017833. 10.1136/bmjopen-2017-017833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Lauritsen SM, Kristensen M, Olsen MV, Larsen MS, Lauritsen KM, Jørgensen MJ, Lange J, Thiesson B, Explainable artificial intelligence model to predict acute critical illness from electronic health records, Nature Communications. 11 (2020) 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Simonyan K, Vedaldi A, Zisserman A, Deep inside convolutional networks: Visualising image classification models and saliency maps, ArXiv Preprint ArXiv:1312.6034. (2013). [Google Scholar]
- [36].Barak V, Schwartz A, Kalickman I, Nisman B, Gurman G, Shoenfeld Y, Prevalence of hypophosphatemia in sepsis and infection: the role of cytokines, The American Journal of Medicine. 104 (1998) 40–47. [DOI] [PubMed] [Google Scholar]
- [37].Chen W-Y, Song Y, Bai H, Lin C-J, Chang EY, Parallel spectral clustering in distributed systems, IEEE Transactions on Pattern Analysis and Machine Intelligence. 33 (2011) 568–586. [DOI] [PubMed] [Google Scholar]
- [38].Castelvecchi D, Can we open the black box of AI?, Nature News. 538 (2016) 20. [DOI] [PubMed] [Google Scholar]
- [39].Amland RC, Sutariya BB, Quick Sequential [Sepsis-Related] Organ Failure Assessment (qSOFA) and St. John Sepsis Surveillance Agent to Detect Patients at Risk of Sepsis: An Observational Cohort Study, Am J Med Qual. 33 (2018) 50–57. 10.1177/1062860617692034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Cortés-Puch I, Hartog CS, Opening the Debate on the New Sepsis Definition Change Is Not Necessarily Progress: Revision of the Sepsis Definition Should Be Based on New Scientific Insights, Am. J. Respir. Crit. Care Med 194 (2016) 16–18. 10.1164/rccm.201604-0734ED. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Carneiro AH, Póvoa P, Gomes JA, Dear Sepsis-3, we are sorry to say that we don’t like you, Rev Bras Ter Intensiva. 29 (2017) 4–8. 10.5935/0103-507X.20170002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Sterling SA, Miller WR, Pryor J, Puskarich MA, Jones AE, The Impact of Timing of Antibiotics on Outcomes in Severe Sepsis and Septic Shock: A Systematic Review and Meta-analysis, Critical Care Medicine. 43 (2015) 1907–1915. 10.1097/CCM.0000000000001142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Levy MM, Rhodes A, The Surviving Sepsis Campaign Bundle: 2018 Update, Critical Care Medicine. 46 (2018) 4. [DOI] [PubMed] [Google Scholar]
- [44].Kashiouris MG, Zemore Z, Kimball Z, Stefanou C, Fisher B, de Wit M, Pedram S, Sessler CN, Supply Chain Delays in Antimicrobial Administration After the Initial Clinician Order and Mortality in Patients With Sepsis, Critical Care Medicine. 47 (2019) 1388–1395. [DOI] [PubMed] [Google Scholar]
- [45].Gadre SK, Shah M, Mireles-Cabodevila E, Patel B, Duggal A, Epidemiology and Predictors of 30-Day Readmission in Patients With Sepsis, Chest. 155 (2019) 483–490. [DOI] [PubMed] [Google Scholar]
- [46].Shankar-Hari M, Rubenfeld GD, Understanding long-term outcomes following sepsis: implications and challenges, Current Infectious Disease Reports. 18 (2016) 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.