ABSTRACT
Coxiella burnetii, the etiological agent of the zoonosis Q fever, replicates inside host cells within a large vacuole displaying autolysosomal characteristics. The development of this compartment is mediated by bacterial effectors, which interfere with a number of host membrane trafficking pathways. By screening a Coxiella transposon mutant library, we observed that transposon insertions in cbu0626 led to intracellular replication and vacuole biogenesis defects. Here, we demonstrate that CBU0626 is a novel member of the Coxiella vacuolar protein (Cvp) family of effector proteins, which is translocated by the Dot/Icm secretion system and localizes to vesicles with autolysosomal features as well as Coxiella-containing vacuoles (CCVs). We thus renamed this effector CvpF for Coxiella vacuolar protein F. CvpF specifically interacts with the host small GTPase RAB26, leading to the recruitment of the autophagosomal marker MAP1LC3B/LC3B (microtubule associated protein 1 light chain 3 beta) to CCVs. Importantly, cvpF::Tn mutants were highly attenuated compared to wild-type bacteria in the SCID mouse model of infection, highlighting the importance of CvpF for Coxiella virulence. These results suggest that CvpF manipulates endosomal trafficking and macroautophagy/autophagy induction for optimal C. burnetii vacuole biogenesis.
Abbreviations: ACCM: acidified citrate cystein medium; AP: adaptor related protein complex; CCV: Coxiella-containing vacuole; Cvp: Coxiella vacuolar protein; GDI: guanosine nucleotide dissociation inhibitor; GDF: GDI dissociation factor; GEF: guanine exchange factor; LAMP1: lysosomal associated membrane protein 1; MAP1LC3B/LC3B: microtubule associated protein 1 light chain 3 beta; MTORC1: mechanistic target of rapamycin kinase MTOR complex 1; PBS: phosphate-buffered saline; PMA: phorbol myristate acetate; SQSTM1/p62: sequestosome 1; WT: wild-type.
KEYWORDS: Autophagy, Coxiella burnetii, effector protein, LC3B, RAB GTPase
Introduction
Coxiella burnetii is the causative agent of animal coxiellosis and human Q fever, considered as one of the most relevant reemerging zoonosis in Europe [1]. The symptoms of Q fever range from fatigue, long-lasting fever, and hepatitis in the acute form of the disease, to severe endocarditis in its chronic form [2]. Upon internalization by phagocytic and non-phagocytic cells, Coxiella remains in endosomes that progress in the endocytic pathway. Endosomal acidification triggers the activation of a defect in organelle trafficking genes/intracellular multiplication (Dot/Icm) type 4 secretion system (T4SS) and the translocation of bacterial effectors, which are essential for the biogenesis of the Coxiella-containing vacuole (CCV) [3–5]. Bioinformatics analysis and secretion assays using either Legionella [6] or Coxiella [7] have identified approximately 143 candidate Coxiella effectors [8]. However, very few host cell partners have been identified [9,10], and the role of Coxiella effectors in CCV biogenesis remains poorly characterized. Coxiella vacuolar proteins (Cvps) constitute a class of effectors that localize to endosomal compartments and CCVs in cells infected with wild type Coxiella. These effectors play an important role in Coxiella vacuole biogenesis, as the mutation of their corresponding genes leads to smaller CCVs [11]. CvpA interacts with adaptor related protein complex 2 (AP2) and clathrin [10]. CvpB (or Cig2) interacts with cellular lipids phosphatidylinositol 3-phosphate (PtdIns3P) and phosphatidylserine (PS) and interferes with PIKFYVE (phosphoinositide kinase, FYVE-type zinc finger containing) activity to promote autophagy-mediated homotypic fusion of CCVs [12]. The functions of CvpC, CvpD, and CvpE remain undetermined. Besides CvpB, additional effector proteins manipulate the autophagy machinery for optimal vacuole biogenesis: effectors CpeB and CpeL colocalize with the autophagosomal marker LC3B [13,14] and mutants of the effector proteins CBU0513, and Cig57 display decreased presence of LC3B at their respective vacuoles [15,16].
Macroautophagy (hereafter termed autophagy) is a highly conserved eukaryotic process used by cells to recycle and degrade cargos, such as damaged organelles and misfolded proteins, in order to acquire nutrients during periods of deprivation. It can also act as a cell defense mechanism capable of targeting invading pathogens for degradation. The mechanistic target of rapamycin kinase complex 1 (MTORC1) mediates the nutrient-sensing and regulatory functions of lysosomes. Under normal conditions, phosphorylated MTOR (mechanistic target of rapamycin kinase) localizes to the lysosome surface and downregulates autophagy. Upon starvation or treatment with the MTOR inhibitor torin1, MTOR is inactivated and delocalizes to the cytosol, which triggers a signaling cascade leading to autophagy. RAB GTPases, the key regulators of vesicular sorting and trafficking in the endocytic and secretory pathways, could regulate autophagy membrane dynamics. While RAB1, RAB5, RAB7, RAB9A, RAB11, RAB23, RAB32 and RAB33B act at different stages of autophagosome formation, RAB7, RAB8B and RAB24 participate in autophagosome maturation [17]. Recently, additional RAB GTPases, such as RAB26 and RAB37, have also been shown to participate in the autophagy process [18–21]. RAB GTPases shuttle between a cytosolic inactive GDP-bound state and a membrane-anchored active GTP-bound state. Once inserted into the membranes, these proteins can interact with other partner proteins to mediate vesicular trafficking and maturation events. Their central role in membrane dynamics and immunity made them ideal targets for intracellular bacterial pathogens, which divert their function for the development of optimal intracellular niches [22–24]. In the context of Coxiella infections, inhibiting the function or expression of RAB1, RAB5, RAB7, and RAB24 affects the vacuole biogenesis and impairs intracellular replication [12,25–29].
Multi-parametric phenotypic analysis of a Coxiella transposon library revealed that transposon insertions in the gene cbu0626 affect Coxiella vacuole biogenesis and intracellular replication [30]. Here, we demonstrate that CBU0626 is a new Coxiella vacuolar effector protein interacting with RAB26 to trigger LC3B recruitment at CCVs. Decreased LC3B recruitment at CCVs in cells challenged with cbu0626 mutants is associated with altered replication in vitro and in vivo, suggesting that diversion of autophagy by this effector is crucial for Coxiella virulence.
Results
CvpF is a Dot/Icm vacuolar effector protein required for LC3B recruitment to CCVs and for Coxiella virulence in vivo
Our phenotypic screen of a Coxiella transposon mutant library identified a significant number of genes encoding candidate Coxiella effector proteins with a role in CCV biogenesis [30]. Among these, the mutant Tn248 with a transposon insertion in the gene cbu0626 displayed a severe replication defect in epithelial cells [30]. Bioinformatics analysis indicated that cbu0626 is likely part of a PmrA-regulated operon, together with cbu0625 (also known as cig17) [31], and possesses a C-terminal E-Block motif found in type 4-secreted effectors [32] (Figure S1A). Southern blot analysis confirmed that transposon insertion in the genome of Tn248 is unique and occurs in the gene cbu0626 (Figure S1B). Complementation of the cbu0626::Tn mutation in Tn248 by the introduction of the full cbu0625-cbu0626 operon under the control of the endogenous PmrA promoter (Tn248 Comp.) restored the expression of the full cbu0626 mRNA (Figure S1C and D). We validated CBU0626 translocation by transforming either a control Coxiella transposon mutant (WT) or a T4SS-defective dotA::Tn mutant with a β-lactamase-cbu0626 fusion construct and comparing the secretion of CBU0626 with that of the previously reported effector protein CvpB [12], using the β-lactamase secretion assay. We verified the expression of all β-lactamase-tagged constructs by western blot (Figure S1E). CBU0626 secretion was comparable with that of CvpB after 24, 48, and 72 h of infection by the control Coxiella strain (Figure S1F). The absence of detectable secretion of either protein by the T4SS-defective dotA::Tn mutant indicated that CBU0626 is indeed a Dot/Icm secreted protein. Of note, T4SS-dependent translocation was still functional in the Tn248 mutant strain (Figure S1F).
To determine the subcellular localization and targets of CBU0626, we transfected U2OS cells infected with WT Coxiella Tn1832 for 4 d with a plasmid coding for mCherry-tagged CBU0626 (mCh-CBU0626, Figure 1A). The Coxiella effector colocalized with the lysosomal marker LAMP1 at CCVs, indicating that CBU0626 is a newly identified member of the Cvp family of Coxiella effector proteins. We, thus, renamed this protein CvpF for Coxiella vacuolar protein F. Next, we used our implemented image analysis algorithm [33] to validate the replication phenotypes previously observed with the cvpF::Tn mutant in our transposon library [30]. U2OS and PMA-treated THP-1 cells infected for 6 d with either the control Coxiella mutant Tn1832 (WT). We fixed and processed the cvpF::Tn transposon mutant or the complemented cvpF transposon mutant (cvpF::Tn Comp.) for immunofluorescence and quantitative PCR. Automated image analysis indicated that cvpF transposon mutant generated smaller CCVs and colonies in U2OS and THP1 cells, respectively (Figure 1B,D), and replicated less efficiently than WT bacterium (Figure 1C,E). Interestingly, in ACCM-2 axenic media, replication of the cvpF::Tn mutant was similar to WT Coxiella, the T4SS-defective dotA::Tn mutant and the cvpF::Tn complemented strain (Figure 1F), indicating that transposon insertion in cvpF only affects intracellular development of the bacterium.
Figure 1.
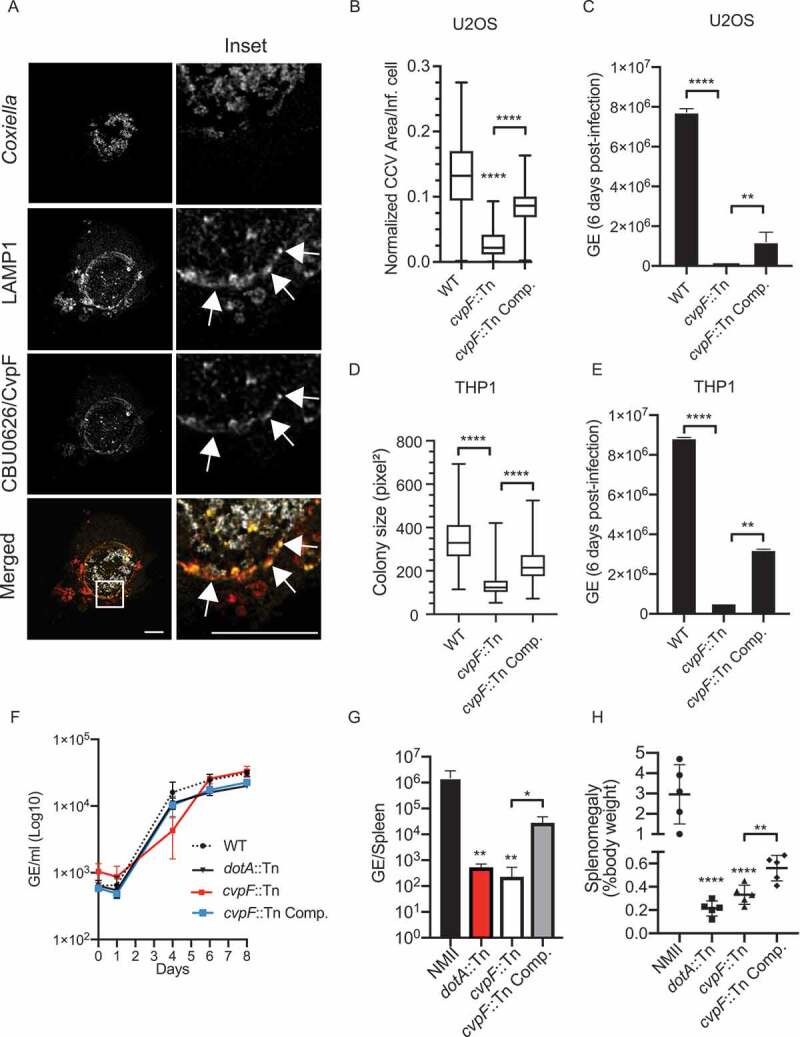
CvpF is a Dot/Icm T4BSS vacuolar effector protein important for Coxiella intracellular growth, CCV biogenesis, and virulence. (A) U2OS cells were infected with Coxiella WT GFP (gray) and transfected with pLVX-mCherry-CBU0626/CvpF (red) 4 d post-infection. We fixed cells 12 h post-transfection and labeled with an anti-LAMP1 antibody (green). White arrows indicate discrete regions of the Coxiella vacuole where CvpF and LAMP1 colocalize. (B) U2OS cells were challenged for 6 d either with Coxiella WT GFP (WT), cvpF::Tn mutant, or the complemented cvpF::Tn strain (cvpF::Tn Comp.). CCV area for each strain was determined using the Cell Profiler software. (C) U2OS cells were challenged as in B, and Genome Equivalents (GE) were determined by quantitative PCR (D) PMA-treated THP1 cells were challenged for 6 d either with Coxiella WT GFP (WT), cvpF::Tn mutant, or the complemented cvpF::Tn strain (cvpF::Tn Comp.) The colony area for each strain was determined using the Cell Profiler software. (E) THP1 cells were challenged as in D and Genome Equivalents (GE) were determined by quantitative PCR. (F) Coxiella WT GFP (WT), dotA::Tn mutant, cvpF::Tn mutant and the complemented cvpF::Tn strain (cvpF::Tn Comp.) were grown for 8 d in ACCM-2 and Genome Equivalents (GE)/ml were determined by Pico Green assay. (G) Genome Equivalents (GE) calculated using TaqMan real-time PCR with DNA purified from infected spleens of 5 SCID mice per group on day 14 after challenge with 1 × 106 GE equivalents of the strains shown. (H) Splenomegaly calculated as spleen weight as a percentage of total body weight at the time of necropsy on day 14 after infection with 1 × 106 GE equivalents of the strains listed in the figure legend. Values are mean ± SD from 3 independent experiments (n.s. = non-significant, **** = P < 0.0001, ** = P < 0.0021, * = P < 0.033, one-way ANOVA, Sidak’s multiple comparison test). Scale bars: 10 μm
We, then, investigated the in vivo relevance of CvpF using the recently developed SCID mice model [34]. Mice challenged via the IP route either with the control Coxiella strain NMII, the cvpF::Tn mutant, the complemented cvpF::Tn strain and the dotA::Tn mutant, were culled at 14 d post-infection and we assessed bacterial genome equivalents (GE) in the spleen, as well as splenomegaly (Figure 1G,H). In agreement with the observations on cultured cells, the number of cvpF::Tn mutant GE in the spleen of infected animals was significantly lower than those found for the WT bacterium and complementation of the cvpF::Tn mutant partially restored this replication defect (Figure 1G). The splenomegaly induced by the cvpF::Tn mutant was also much lower than the one provoked by WT bacterium (Figure 1H), strongly suggesting that the virulence of the cvpF::Tn mutant is severely attenuated in vivo and could be partially restored using the cvpF::Tn-complemented strain.
As CvpF appears to be important for CCV biogenesis, we investigated whether the absence of CvpF affects typical CCVs markers. More than 80% of vacuoles generated by WT Coxiella, cvpF::Tn and cvpF::Tn Comp. strains were positive for the acidification marker LysoTracker and the lysosomal/late endosomal protein LAMP1 (Figure 2A,B). However, 15.77 ± 6.8% of CCVs generated by the cvpF::Tn mutant was positive for LC3B (Figure 2C,D), while 93.56 ± 3.4% of CCVs generated by WT Coxiella and 70.13 ± 10.2% of CCVs generated by the cvpF::Tn-complemented strain were positive for LC3B (Figure 2C,D). Colocalization analysis between LAMP1 and LC3B on CCVs generated by WT, cvpF::Tn, and cvpF::Tn complemented strains (Figure 2E) showed that significantly less LC3B is found on CCVs generated by the cvpF::Tn mutant, strongly suggesting that CvpF plays a role in the recruitment of this autophagosomal marker on CCVs.
Figure 2.
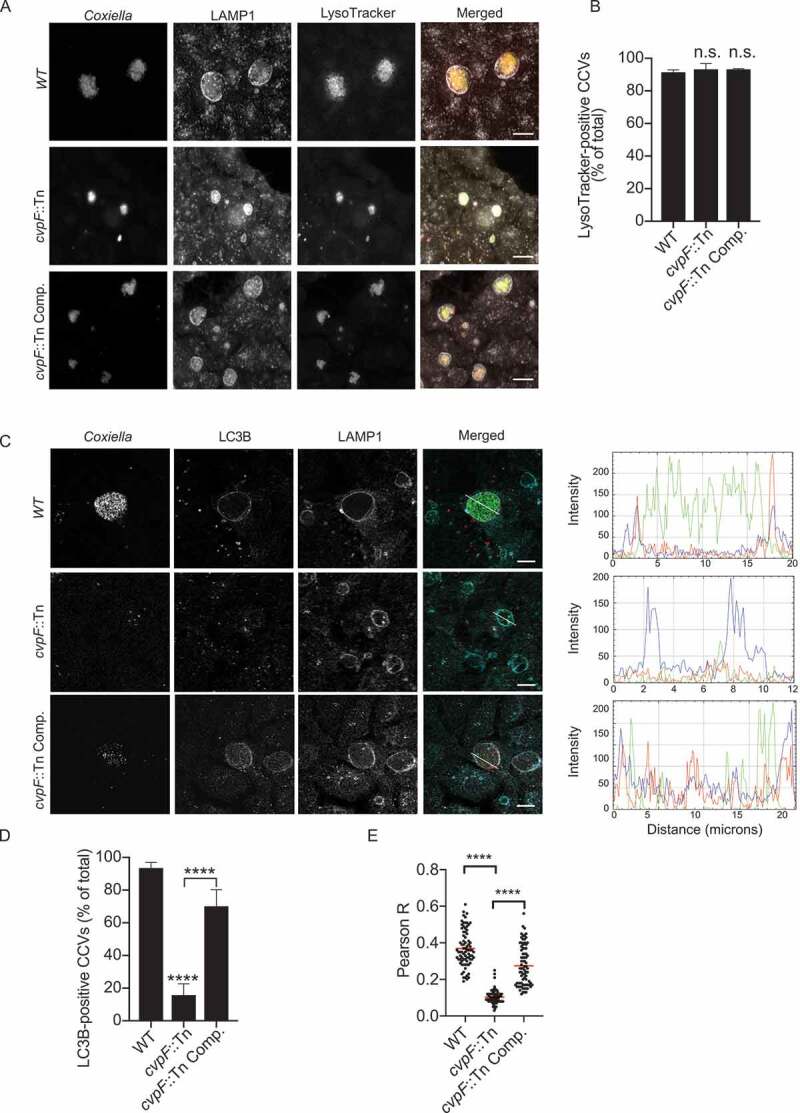
CvpF participates in LC3B recruitment to CCVs but not in their acidification. (A) U2OS cells were infected with either Coxiella WT GFP (top panels), cvpF::Tn (middle panels) or the complemented cvpF::Tn strain (cvpF::Tn Comp., bottom panels) (green) for 6 d, fixed and stained with anti-LAMP1 (pseudo-colored gray) and LysoTracker (red). (B) U2OS cells were infected as in A, and the presence of the LysoTracker probe in CCVs was scored for at least 80 cells. (C) U2OS cells were infected with either Coxiella WT GFP (top panels), cvpF::Tn (middle panels), or the complemented cvpF::Tn strain (cvpF::Tn Comp., bottom panels) for 6 d, fixed and stained with anti-LAMP1 (pseudo-colored cyan) and anti-LC3B (red). The white line in the merged image indicates the position of the profile line used for the analysis of intensity distribution. Fluorescence intensity plotting of GFP (green), LC3B (red), and LAMP1 (blue) signals are shown on the right for each strain. (D) U2OS cells were infected as in C and the presence of LC3B on CCVs was scored for at least 80 cells. Values are mean ± SD from 3 independent experiments (n.s. = non-significant, **** = P < 0.0001, one-way ANOVA, Dunnett’s multiple comparison test). Scale bars: 10 μm. (E) Pearson’s correlation coefficient between LC3B and LAMP1 signals in images acquired in C (**** = P < 0.0001, one-way ANOVA, Sidak’s multiple comparison test)
Ectopically expressed CvpF localizes to vesicular structures with autolysosomal features
The defective vacuole biogenesis phenotype observed with the cvpF transposon mutants, accompanied by the loss of LC3B at CCVs, suggested a role for this bacterial effector in re-routing autophagy components to the forming CCV. We, thus, further investigated CvpF localization in cells overexpressing either mCherry- or HA-tagged versions of the effector protein. CvpF was excluded from the Golgi complex either labeled by an anti-GM130 antibody or by expression of GFP-tagged M6PR (mannose-6-phosphate receptor, cation dependent) (Figure S2), whereas we observed partial colocalization of CvpF with markers of the endosomal sorting complex required for transport (ESCRT) complex (GFP-TSG101, GFP-VPS4A and FLAG-CHMP4B) (Figure S2). Conversely, CvpF signal overlapped with the lysosomal marker LAMP1, but also with the early endosomal and autophagosomal markers phosphatidylinositol 3-phosphate (PtdIns3P) probe 2xFYVE-GFP and LC3B (Figure 3A). Importantly, all compartments labeled by CvpF displayed a clustered perinuclear localization (Figure 3A), as opposed to cells expressing either the HA or the mCherry tags alone (Figure S3).
Figure 3.
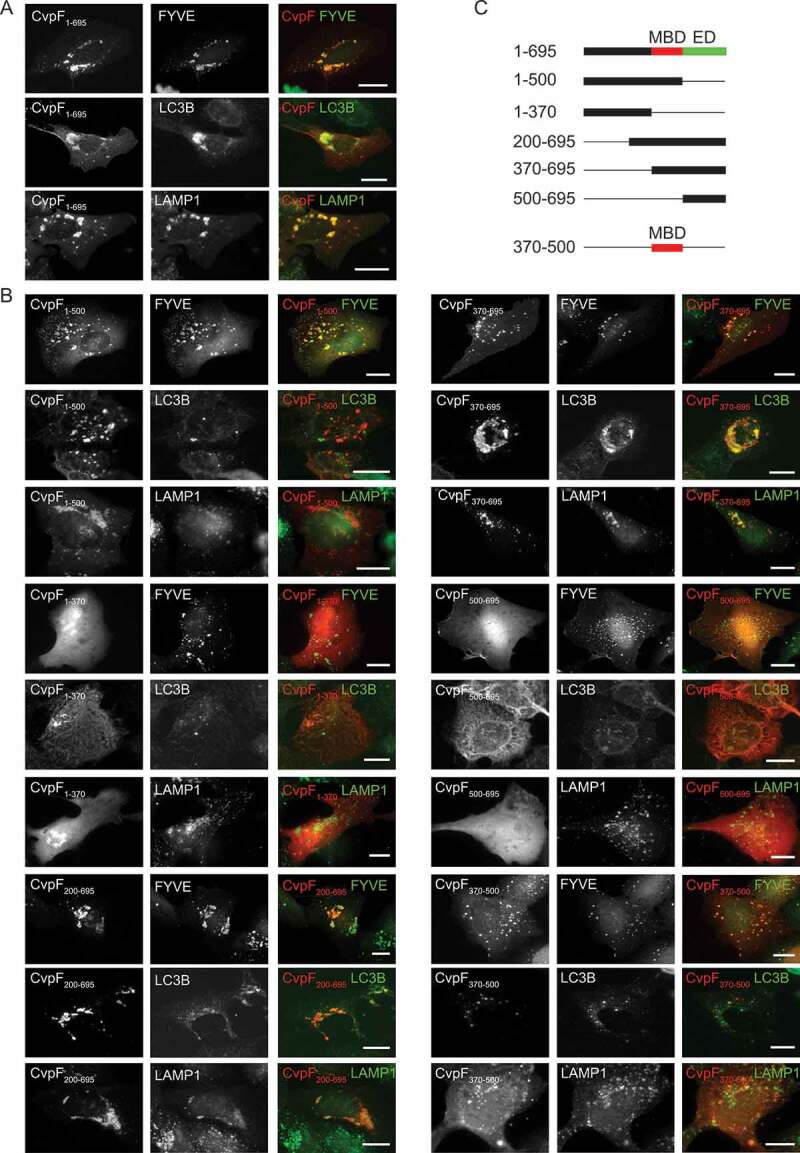
Ectopically expressed CvpF localizes to vesicles with autolysosomal features. (A) U2OS or U2OS GFP-FYVE cells were transfected with pLVX-mCherry-CvpF or pRK5-HA-CvpF (red). Cells were fixed and labeled either with anti-LC3B, or anti-LAMP1 antibodies (green). HA-CvpF was labeled with an anti-HA antibody (red). (B) U2OS or U2OS GFP-FYVE cells were transiently transfected with plasmids pLVX-mCherry or pRK5-HA fused to CvpF1-500, CvpF1-370, CvpF200-695, CvpF370-695, CvpF500-695, or CvpF370-500 (red). Cells were fixed and labeled either with anti-LC3B or anti-LAMP1 antibodies (green). HA-tagged constructs were labeled with anti-HA antibody (red). (C) Schematic representation of CvpF fragments used in A and B. MBD: membrane-binding domain, ED: effector domain. Scale bars: 10 μm
To understand how CvpF is targeted to host cell membranes, we carried out a mutational analysis of this Coxiella effector protein. We transfected U2OS cells with mCherry- or HA-tagged versions of incremental deletions either from the N-terminal or the C-terminal of CvpF, and we assessed the localization of the bacterial effector fragments, with respect to 2xFYVE-GFP, LC3B, and LAMP1 (Figure 3B,C). The deletion of the first 370 amino acids did not affect CvpF localization nor the formation of perinuclear clusters of CvpF-labeled compartments (Figure 3B). Conversely, CvpF500-695 failed to localize at cellular membranes and remained diffused in the cytoplasm (Figure 3B). Accordingly, the expression of CvpF500-695 also failed to reposition cellular compartments to the perinuclear area (Figure 3B). Deletion of the last 195 amino acids (CvpF1-500) did not affect membrane targeting of CvpF; however, the localization with LAMP1- and LC3B-positive vesicles was lost, together with the effect on the perinuclear positioning of these two markers. Of note, CvpF1-500 still localized at PtdIns3P-positive structures, which retained the perinuclear localization. Further C-terminal deletions of CvpF resulted in a loss of membrane localization and the cytoplasmic redistribution of membrane markers. This result indicated that the membrane-binding domain (MBD) of CvpF is located between amino acids 370 and 500 and that the C-terminal domain of CvpF (effector domain, ED) may play a role either in the re-routing of early endosomes to autophagosomes or the maturation of autophagosomes to autolysosomes (Figure 3C).
Bioinformatics analysis of the CvpF MBD revealed the presence of one [DERQ]XXX[LI] and 2 YXXΦ endocytic sorting motifs (Figure S4A), involved in the interaction with clathrin adaptor complexes. We, thus, generated di-leucine and tyrosine mutants to validate the functionality of these motifs. The di-leucine LLAA mutant, and the Y440A mutant, failed to affect the membrane targeting and perinuclear localization of CvpF, whereas the Y425A mutation sufficiently displaced CvpF to the cytoplasm with concomitant loss of LAMP1 and LC3B repositioning (Figure S4B and C). Accordingly, Tyr425 is required for CvpF localization at the CCV (Figure S4D). These observations suggest that Tyr425 could be part of a functional endocytic sorting motif. To date, however, we could not detect any interaction or co-localization between CvpF and adaptor complexes or clathrin (data not shown).
CvpF stimulates LC3B-II formation
Several studies have shown that Coxiella stimulates the formation of LC3B-II in a T4SS-dependent manner during infection [15,35,36]. To determine whether the secretion of CvpF participates in the formation of LC3B-II, we infected U2OS cells either with the WT GFP Coxiella strain (Tn1832), the cvpF::Tn mutant, or the complemented cvpF::Tn mutant (cvpF::Tn Comp.). Given the intracellular replication defect of the cvpF::Tn mutant Tn248, and to rule out the possibility that LC3B-II increase is dependent on the bacterial load, we also infected cells with an excess of cvpF::Tn mutant (cvpF::Tn x10) (Figure 4A). We observed an increase in LC3B-II levels for cells infected with WT Coxiella and the complemented cvpF::Tn mutant, in the presence of bafilomycin A1. We observed no LC3B-II increase for cells infected with the cvpF::Tn mutant (Figure 4A). Next, we infected the cells as in Figure 4A and starved for 3 h to determine whether the LC3B-II increase observed with WT Coxiella and the complemented cvpF::Tn mutant was not due to a blockade of the autophagy flux. We analyzed levels of SQSTM1/p62 (sequestosome 1) by western blot, which revealed that upon starvation, SQSTM1 is degraded for all conditions, indicating that CvpF does not block the starvation-induced autophagy flux but stimulates the formation of LC3B-II (Figure 4B).
Figure 4.
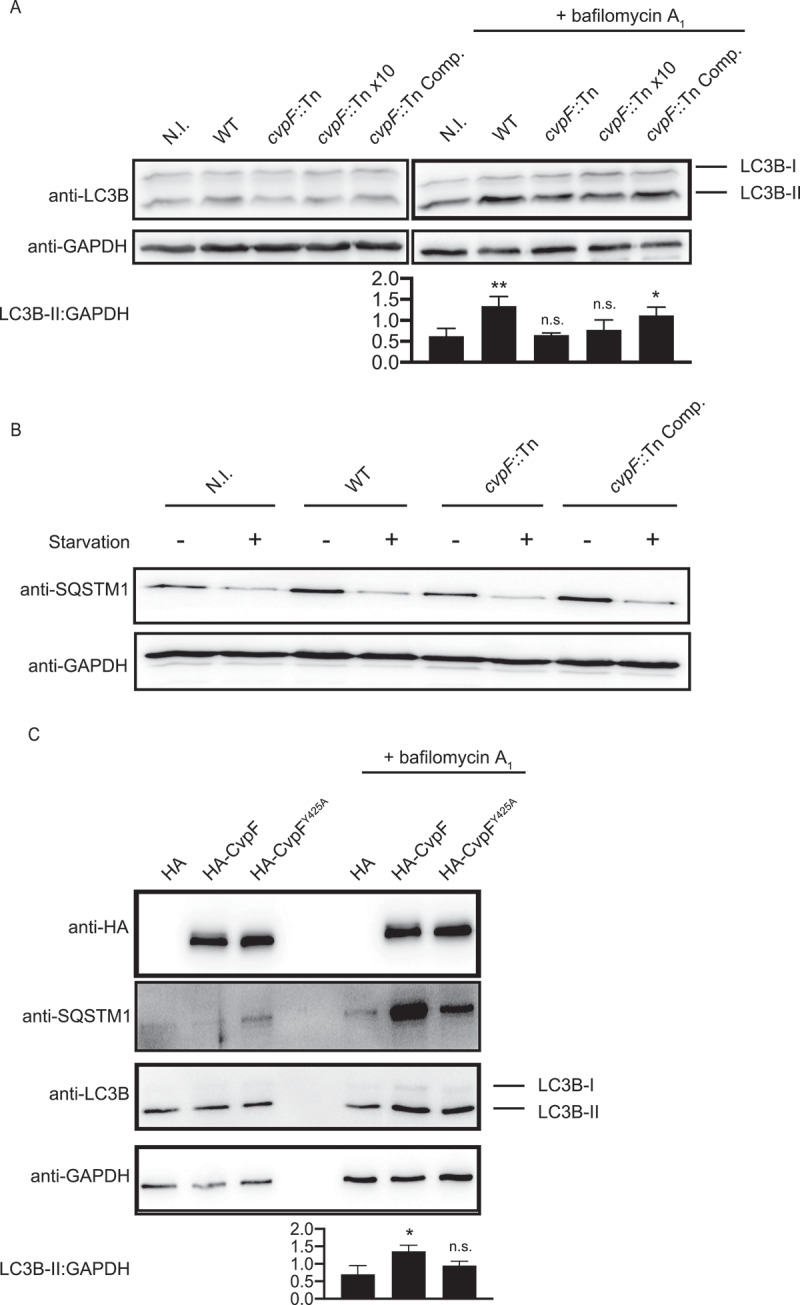
CvpF stimulates the formation of LC3B-II. (A) Immunoblot of lysates from U2OS cells left uninfected (N.I.) or infected with Coxiella WT GFP (WT), cvpF::Tn, 10 times cvpF::Tn (cvpF::Tn x10) or the complemented cvpF::Tn strain (cvpF::Tn Comp.) for 72 h in complete media. Bafilomycin A1 (25 nM) was added to the media 3 h before lysis. Immunoblots were probed with antibodies against LC3B and GAPDH. The signal ratio of LC3B-II versus GAPDH is indicated for samples treated with bafilomycin A1. (B) Immunoblot of lysates from U2OS cells left uninfected (N.I.) or infected with Coxiella WT GFP (WT), cvpF::Tn, or the complemented cvpF::Tn strain (cvpF::Tn Comp.) for 72 h in complete media. Starvation was triggered by incubation of cells in HBSS for 3 h before lysis. Immunoblots were probed with antibodies against SQSTM1 and GAPDH. (C) Immunoblot of lysates from U2OS cells transfected with pRK5-HA, pRK5-HA-CvpF or pRK5-HA-CvpFY425A mock-treated or treated with 25 nM bafilomycin A1 for 1 h. The signal ratio of LC3B-II versus GAPDH is indicated for samples treated with bafilomycin A1. Values are mean ± SD from 3 independent experiments (n.s. = non-significant, ** = P < 0.0021, * = P < 0.0033, one-way ANOVA, Dunnett’s multiple comparison test)
The vacuolar localization of CvpF, together with the decreased presence of LC3B on CCVs generated by cvpF::Tn mutants prompted us to investigate the role of CvpF in autophagosome dynamics further. We probed the individual role of CvpF on LC3B-II increase in transfected cells. We analyzed the autophagy flux by western blotting on U2OS cells expressing HA, HA-CvpF, or HA-CvpFY425A (Figure 4C). When bafilomycin A1 blocks the autophagy flux, we observed a marked increase in LC3B-II and SQSTM1 in cells expressing CvpF and HA-CvpFY425A compared to cells expressing HA, with wild type CvpF having the greatest effect on SQSTM1 levels, indicating that CvpF stimulates the autophagy flux and that its membrane localization participates in this process (Figure 4C).
Next, we co-transfected HA-CvpF and HA-CvpFY425A with the tandem-fluorescent probe TF-LC3. This probe consists of an LC3B linked to GFP and RFP. Since acidic conditions quench GFP fluorescence, this probe labels autophagosomes in green and red, while autolysosomes appear only red. In transfected cells, HA-CvpF localizes to clustered vesicles, only displaying red fluorescence corresponding to autolysosomes (Figure S5A). Conversely, CvpFY425A displayed a diffuse localization, and transfected cells displayed non-acidified autophagosomes similarly to HA-transfected cells (Figure S5A). Upon addition of bafilomycin A1, CvpF was still found present on clustered vesicles, but these were GFP- and RFP-positive (Figure S5B). In bafilomycin A1-treated cells, HA and HA-CvpFY425A – transfected cells displayed non-acidified autophagosomes similarly to the mock-treated condition (Figure S5B). CvpF-dependent clustering of vesicles rendered the scoring of LC3B-positive puncta impossible. Of note, vesicular swelling and clustering triggered by overexpression of LAMP1-Flag did not affect the TF-LC3 probe signal in the presence or absence of bafilomycin A1, indicating that CvpF alone can stimulate the formation of autophagosomes and autolysosomes. Furthermore, its membrane localization seems critical to act on this endosomal system.
Finally, to rule out the possibility that CvpF stimulates autophagy by inhibiting MTORC1, we stained U2OS cells transfected with HA, HA-CvpF, or HA-CvpFY425A for MTOR and we labeled lysosomes with LysoTracker (Figure S6A). The expression of CvpF or CvpFY425A did not affect the localization of MTOR on lysosomes, indicating that CvpF does not alter MTORC1 activity to stimulate autophagy.
CvpF interacts with the autophagy-related RAB GTPase RAB26
Yeast two-hybrid screening identified the small GTPase RAB26 as a candidate interactor of CvpF. Of note, RAB26 has been previously involved in lysosomal positioning [37], autophagosome maturation [18,19], and vesicle-mediated secretion of adrenergic receptors [38]. We, thus, validated the interaction using a co-immunoprecipitation approach on U2OS cells co-expressing HA-CvpF and GFP-RAB26 (Figure 5A). The lack of interaction in cells co-expressing HA-CvpF together with GFP-tagged versions of RAB5, RAB7, RAB9, RAB11, or RAB37 (the closest homolog of RAB26 with 54% amino acid identity), indicated that the CvpF-RAB26 interaction is specific (Figure 5A). In agreement with the interactomics analysis, fluorescence microscopy indicated that CvpF and RAB26 localize at the same cellular compartments, and that expression of CvpF increases membrane targeting of RAB26 (Figure 5B). Of note, we still observed interactions in cells co-expressing CvpFY425A and RAB26, indicating that the membrane targeting of CvpF is not required for the interaction with RAB26 (Fig. S6B). However, ectopically expressed CvpFY425A does not alter GFP-RAB26 distribution in cells, indicating that the interaction of CvpF with membranes is required to relocalize RAB26 (Figure S6C). Further analysis using the CvpF truncations revealed that the domain 370–695 of CvpF is sufficient to relocalize RAB26 onto vesicles (Figure S7).
Figure 5.
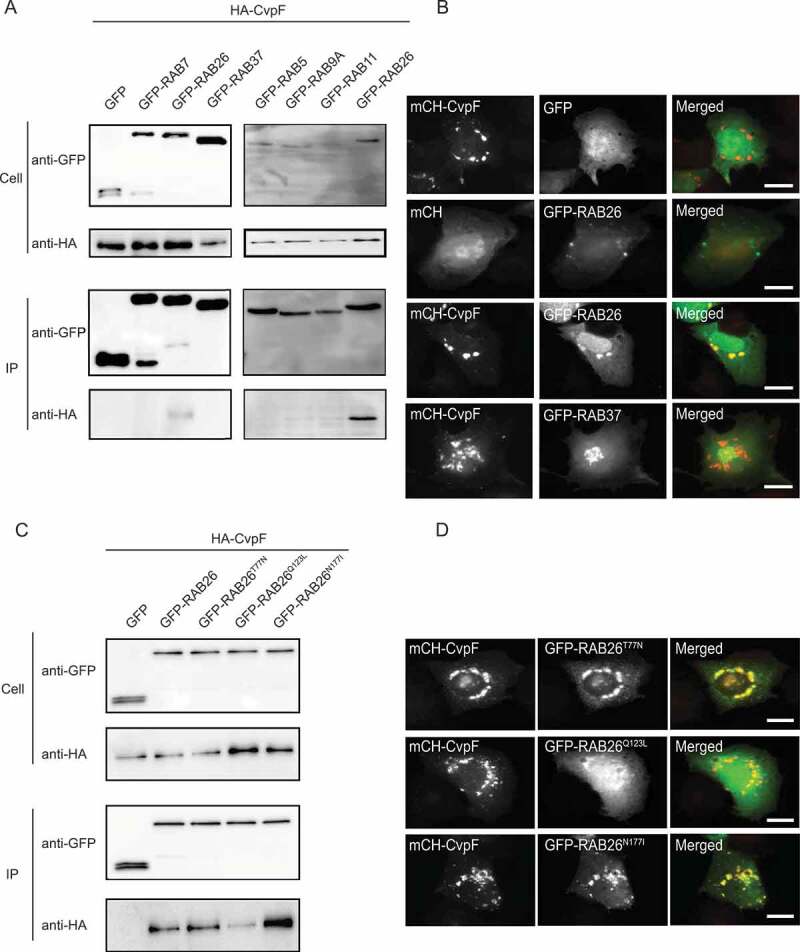
CvpF interacts specifically with RAB26. (A) A co-immunoprecipitation assay using U2OS cells co-transfected with pRK5-HA-CvpF and pLVX-GFP, pLVX-GFP-RAB5, peGFP-RAB7, peGFP-RAB9, pLVX-GFP-RAB11, pLVX-GFP-RAB26, pLVX-GFP-RAB37. RAB GTPases and CvpF were detected using anti-GFP and anti-HA antibodies, respectively. (B) U2OS cells were transfected with pLVX-mCherry or pLVX-mCherry-CvpF (red) and pLVX-GFP, pLVX-GFP-RAB26 or pLVX-GFP-RAB37 (green). (C) A co-immunoprecipitation assay using U2OS cells co-transfected with pRK5-HA-CvpF and pLVX-GFP, pLVX-GFP-RAB26, pLVX-GFP-RAB26T77N, pLVX-GFP-RAB26Q123L, pLVX-GFP-RAB26N177I. The RAB GTPases and CvpF were detected using anti-GFP and anti-HA antibodies, respectively. (D) U2OS cells were transiently transfected with pLVX-mCherry-CvpF (red) and pLVX-GFP-RAB26T77N, pLVX-GFP-RAB26Q123L or pLVX-GFP-RAB26N177I (green). Scale bars: 10 μm
To investigate whether CvpF binds preferentially to active or inactive forms of RAB26, we generated GFP-tagged dominant-negative (GFP-RAB26T77N), dominant-positive (GFP-RAB26Q123L) and guanosine-free (GFP-RAB26N177I) versions of RAB26. We co-expressed these constructs in U2OS cells, in combination with HA-CvpF for the co-immunoprecipitation assay or mCherry-CvpF, to investigate the respective intracellular localization of the two proteins. Interactomics analysis indicated that CvpF preferentially binds to inactive RAB26 (Figure 5C), which we confirmed by fluorescence microscopy. Indeed, co-expression of either GFP-RAB26T77N or GFP-RAB26N177I with mCherry-CvpF resulted in the membrane targeting of the small GTPase, conversely, we observed little or no overlapping between mCherry-CvpF and GFP-RAB26Q123L (Figure 5D). Together, these observations suggest that CvpF might act as a guanosine exchange factor (GEF) or GDI displacement factor (GDF), anchoring and/or activating RAB26 on either early endosomal or pre-autophagosomal structures.
CvpF triggers the recruitment of RAB26 to CCVs
CvpF localization at CCVs (Figure 1A) and its interaction with RAB26 (Figure 5A) suggested that the small GTPase might be recruited at CCVs and be involved in vacuole biogenesis. To test whether RAB26 recruitment at CCVs is CvpF-dependent, we infected U2OS cells with WT Coxiella, cvpF::Tn or cvpF::Tn Comp. and transfected with a plasmid expressing GFP-RAB26 (Figure 6A). Indeed, GFP-RAB26 became recruited on 75.4 ± 2.72% of CCVs generated by WT Coxiella and 66.9 ± 6.12% of CCVs generated by cvpF::Tn Comp. strains but only 23.23 ± 3.78% of CCVs generated by cvpF::Tn mutants were positive for RAB26 (Figure 6B). Colocalization analysis between LAMP1 and RAB26 on CCVs generated by WT, cvpF::Tn and cvpF::Tn complemented strains (Figure 6C) indicated that CvpF stimulates the presence of RAB26 at the CCV.
Figure 6.
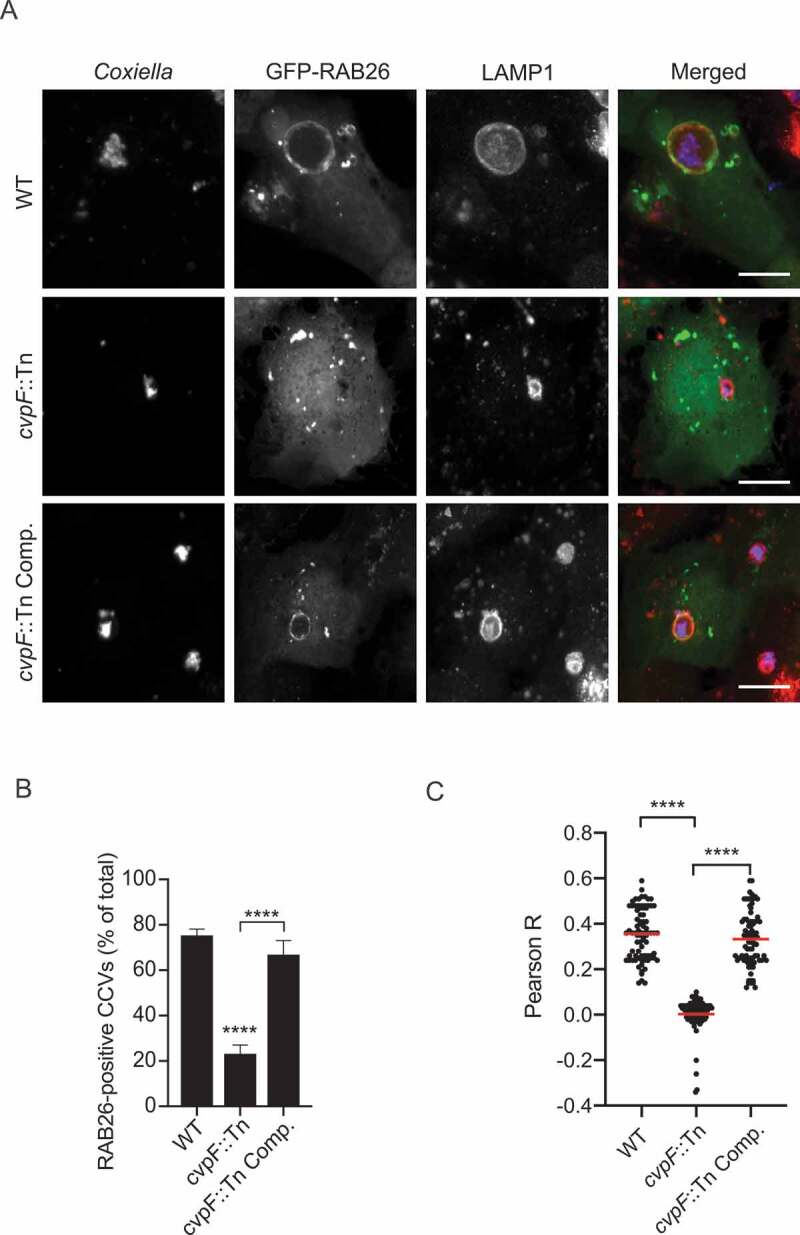
CvpF stimulates the recruitment of RAB26 to CCVs. (A) U2OS cells were infected with either Coxiella WT GFP (WT, top panels), cvpF::Tn (middle panels), or the complemented cvpF::Tn strain (cvpF::Tn Comp., bottom panels) and transfected with pLVX-GFP-RAB26 (green) 4 d post-infection. We fixed cells 12 h post-transfection and stained with anti-LAMP1 (red) and anti-Coxiella (blue) antibodies. (B) U2OS cells were infected, as in A, and the presence of RAB26 on CCVs was scored for at least 80 cells per condition. Values are mean ± SD from 3 independent experiments (n.s. = non-significant, **** = P < 0.0001, one-way ANOVA, Bonferroni’s multiple comparison test). Scale bars: 10 μm. (C) Pearson’s correlation coefficient between RAB26 and LAMP1 signals in images acquired in A (**** = P < 0.0001, one-way ANOVA, Sidak’s multiple comparison test)
Inhibition of RAB26 activity alters LC3B recruitment to CCVs and CCV development
Active RAB26 has been shown to interact with ATG16L1 [18,19], which is key for LC3B anchoring to membranes. We, thus, determined the requirement of active RAB26 for CCV biogenesis and investigated the presence of the LC3B autophagy marker on CCVs in cells expressing GFP, GFP-RAB26, GFP-RAB26T77N, GFP-RAB26Q123L, and GFP-RAB26N177I. The LC3B signal on CCVs generated by Coxiella NMII became much reduced in cells transfected with the inactive forms of RAB26 (GFP-RAB26T77N and GFP-RAB26N177I) compared to cells expressing either RAB26 wild type (GFP-RAB26) or active RAB26 (GFP-RAB26Q123L) (Figure 7A). Of note, loss of LC3B was accompanied by a reduction in the size of CCVs (Figure 7B).
Figure 7.
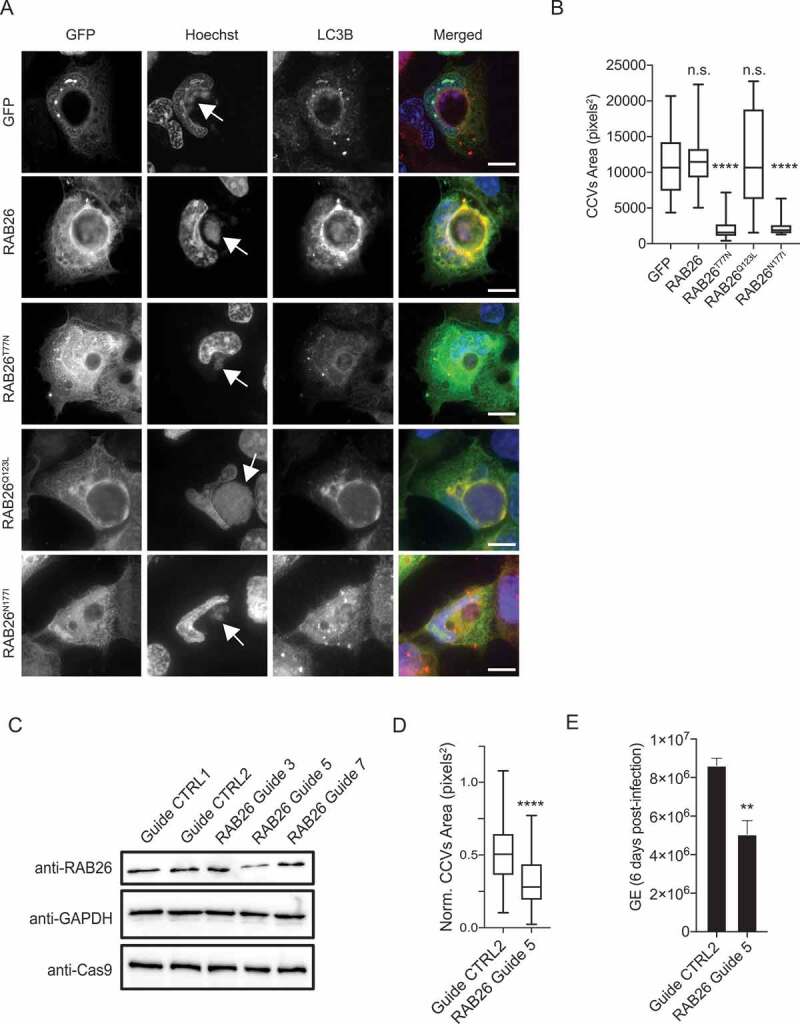
RAB26 activity is essential for CCV biogenesis. (A) U2OS cells infected with Coxiella NMII were transfected with pLVX-GFP, pLVX-GFP-RAB26, pLVX-GFP-RAB26T77N, pLVX-GFP-RAB26Q123L or pLVX-GFP-RAB26N177I (green) 2 d post-infection. We fixed cells 24 h post-transfection and stained with anti-LC3B (red) and stained DNA using Hoechst 33,258 (blue). White arrows indicate Coxiella colonies. (B) Cells were treated as in A, and the CCV area was measured for at least 100 cells per condition. Values are mean ± SD from 3 independent experiments (n.s. = non-significant, **** = P < 0.0001, one-way ANOVA, Dunnett’s multiple comparison test). (C) Immunoblot of lysates from U2OS Cas9 cells expressing non-targeting guides (Guide CTRL1 and guide CTRL2) or RAB26-targeting guides (RAB26 Guide 3, 5 and 7). Immunoblots were probed with antibodies against RAB26, GAPDH and Cas9. (D) U2OS Cas9 cells expressing the non-targeting guide CTRL2 or the RAB26-targeting guide 5 were challenged for 6 d with Coxiella WT GFP (WT). The normalized CCV area for each condition was determined using the Cell Profiler software. (E) U2OS cells were challenged as in D and Genome Equivalents (GE) were determined by quantitative PCR. Values are mean ± SD from 3 independent experiments (n.s. = non-significant, **** = P < 0.0001, ** = P < 0.001, unpaired t test). Scale bars: 10 μm
To confirm the importance of RAB26 for CCV development, we inhibited endogenous RAB26 expression using the CRISPR-Cas9 genome editing system. We used three RAB26-targeting guides, and guide 5 was the most efficient in decreasing RAB26 expression (Figure 7C). We infected U2OS cells treated with either non-targeting guides or RAB26-targeting guide 5 with WT Coxiella, and we determined the CCV area and Genome Equivalents (GE). We observed a marked decrease in CCV area (Figure 7D), as well as bacterial replication (Figure 7E) in cells deficient in RAB26 expression, indicating that RAB26-mediated autophagy is key to the development of CCVs.
Co-expression of RAB26 with CvpF stimulates the formation of LC3B-II
To determine whether the interaction between CvpF and RAB26 stimulates the recruitment of LC3B onto vesicles, we transfected U2OS cells with HA-CvpF, in combination with either GFP, GFP-RAB26, GFP-RAB26T77N, GFP-RAB26Q123L, or GFP-RAB26N177I. While the co-expression of RAB26 wild type and RAB26Q123L with CvpF led to an increase of the LC3B signal onto vesicles, the concomitant expression of the dominant-negative and guanosine-free forms of RAB26 with CvpF led to a moderate increase of the LC3B signal (Figure 8). These observations indicate that active RAB26 can stimulate the recruitment of LC3B onto vesicles and that CvpF could activate RAB26 to trigger the recruitment of LC3B onto vesicles.
Figure 8.
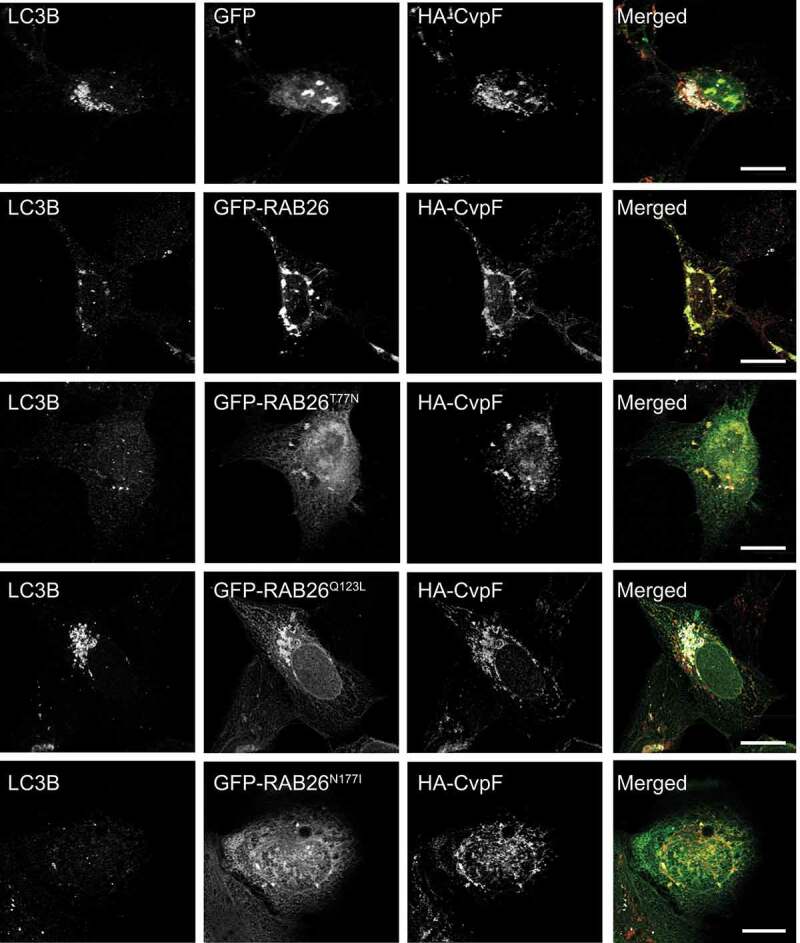
Co-expression of CvpF and RAB26 stimulates the formation of LC3B-positive endosomes. U2OS cells were transfected with pLVX-GFP, pLVX-GFP-RAB26, pLVX-GFP-RAB26T77N, pLVX-GFP-RAB26Q123L, or pLVX-GFP-RAB26N177I (green) and pRK5-HA or pRK5-HA-CvpF. Cells were fixed and stained with anti-LC3B (red) and anti-HA (blue). Scale bars: 10 μm
Discussion
Coxiella-containing vacuoles (CCVs) are unique compartments with autolysosomal features. Key to the generation of CCVs is the secretion of bacterial effectors that divert host cell mechanisms to shape an adequate environment for bacterial replication. Here, we identified a new Coxiella effector protein important for vacuole biogenesis and replication of the bacterium in epithelial and myeloid cell lines. As recent studies have highlighted, the fact that defective vacuole biogenesis impacts Coxiella virulence in insect and mammalian models of infection [12,34,39], we, thus, investigated the in vivo relevance of CvpF using the recently developed SCID mouse model. This model is capable of assessing two traits relevant to virulence, such as the ability to cause splenomegaly (a surrogate for inflammation) and the ability to replicate within the spleen (replication defect). The cvpF::Tn mutant was severely attenuated for both virulence readouts, and we found even less GE in the spleens than in the dotA::Tn mutant. We hypothesize that this is a result of the impaired vacuole synthesis, but it could be due to the ability of the cvpF::Tn mutant to secrete proteins, which may alert the innate immune system. In agreement with the in vitro observations, the complemented strain was able to partially rescue the ability of cvpF::Tn to replicate within the spleen and marginally increased the amount of splenomegaly.
Stimulation of autophagy leads to increased CCV size, suggesting that autophagy provides nutrients and membranes to the expanding CCV [28]. It has been shown that infection of cultured myeloid and epithelial cells with Coxiella wild type leads to increased lipidation of LC3B [35,40] and that this lipidation is dependent on the secretion of bacterial effectors [15,36]. Besides lipidation of LC3B, secretion of bacterial effectors via the T4SS is required for the acquisition of autophagosomes and autophagosomal markers to the CCV [12,35,39,40]. Here, we showed how the secreted effector CvpF diverts RAB26-dependent autophagy to favor the generation of Coxiella-containing vacuoles in vitro, and the virulence of the bacterium in vivo. Indeed, most of CCVs generated by the cvpF::Tn mutant are devoid of the autophagosomal marker LC3B, suggesting that CvpF could favor CCV biogenesis by manipulating autophagy. Interestingly, SQSTM1 levels are stabilized during Coxiella infections [40,41], and SQSTM1 is recruited to CCVs in a T4SS-dependent but LC3B-independent manner. We showed here an increase of steady-state levels of LC3B-II and SQSTM1 in cells infected with WT Coxiella and the cvpF::Tn complemented strains compared to cells infected with the cvpF::Tn mutant or in uninfected cells. This observation is similar to what is observed in HeLa cells infected with the cig57::Tn mutant [15]. Indeed, Coxiella effector Cig57 has been shown to influence autophagy via clathrin recruitment to CCVs, as cig57::Tn mutant CCVs display decreased levels of clathrin and LC3B [15,42]. However, contrary to CvpF, Cig57 alone was not capable of inducing LC3B lipidation [15]. This observation raises the possibility that CvpF and Cig57 could be functionally linked through the recruitment of host cell proteins RAB26 and clathrin, respectively, to CCVs.
Cells expressing HA-CvpF show decreased steady-state levels of SQSTM1 but unaltered levels of steady-state LC3B-II. The increased levels of SQSTM1 and LC3B-II in the presence of bafilomycin A1 suggest that ectopically expressed CvpF could stimulate the degradation of SQSTM1 and the lipidation of LC3B-II. However, in the context of infection, multiple effectors are translocated that could counteract some of CvpF actions (i.e., SQSTM1 degradation, stimulation of SQSTM1 transcription). The fact that cells infected with the cvpF::Tn mutant display less SQSTM1 compared to cells infected with the WT strain could be the result of other Coxiella effectors. Overall, ectopically expressed CvpF stimulates the formation of LC3B-II but also the degradation of SQSTM1, suggesting that CvpF would be involved in the formation of autolysosomes, but other effectors could stabilize SQSTM1 during infection.
Several studies have shown that mature autophagosomes are recruited to the CCV [39,40] and that another effector protein, CvpB/Cig2, is important to recruit LC3B to CCVs and to maintain the CCV in an autolysosomal stage of maturation [35,39]. This is achieved via the CvpB-dependent stabilization of phosphatidylinositol 3-phosphate at CCVs [12]. Interestingly, in cvpB/cig2::Tn mutants, LC3B lipidation, and SQSTM1 levels are increased. This raises the possibility that Coxiella effectors may act in sequence to modify the lipid and protein signature of the CCV, and to alter the physiological behavior of the endosomal network. In this case, effectors such as CvpB and Cig57 would initiate CCV maturation events, and CvpF could then stimulate the autophagy flux to provide membranes and nutrients to expand CCVs and permit optimal Coxiella replication.
Ectopic expression showed that CvpF has a strong capacity to remodel the endosomal network by clustering early and late endosomal vesicles, as well as autophagosomal structures in the perinuclear area of transfected cells. Of note, the dynein-dependent movement of autophagosomes is used in cells to promote their efficient encounter with lysosomes [43], a mechanism that could be used by CvpF to drive transport and maturation of vesicles toward an autolysosomal state. Interestingly, CvpF possesses an internal motif that corresponds to a putative eukaryotic-like endocytic sorting motif that could interact with adaptor complexes (APs), similar to the vacuolar effector CvpA [10]. Indeed, mutation of a critical tyrosine residue in this motif led to the complete relocalization of CvpF to the cytosol of transfected cells. Furthermore, this tyrosine could also be part of an extended LC3-interacting region (LIR) motif (xx[WFY]xx[LIV]) [44]. However, direct interaction of CvpF with APs and LC3B still needs to be experimentally demonstrated.
Several RAB GTPases have been shown to decorate the CCV (RAB1 [29], RAB7 [26], and RAB24 [28]), all of them being important for its biogenesis. Importantly, CvpF is the first Coxiella effector protein to interact with RAB GTPases. Specific interaction of CvpF with RAB26 recruits the small GTPase to CCVs and RAB26-depletion, or inactivation decreases LC3B recruitment at CCVs, which is accompanied by a strong reduction in CCVs size, indicating that active RAB26 and RAB26-dependent stimulation of autophagy favor the development of CCVs. Accordingly, RAB26 participates in the transport of lysosomes to perinuclear regions [37] and autophagy [18,19], as its active form is capable of interacting with ATG16L1 to stimulate LC3B lipidation.
Intracellular pathogens commonly manipulate RAB GTPases for the establishment of their replicative niche [24]. For example, Legionella pneumophila secretes at least 7 effectors targeting the RAB GTPase RAB1 for the development of its vacuole [45,46]. CvpF preferentially interacts with the inactive form of RAB26 and stimulates its presence on endosomal vesicles, suggesting that CvpF may act either as a GDI Dissociation Factor (GDF) or Guanine Exchange Factor (GEF) for RAB26. This hypothesis remains to be experimentally validated. Interestingly, CvpF still localizes with inactive RAB26 in the perinuclear area, suggesting that the CvpF-dependent endosomal relocation does not rely on RAB26 activity.
Thus, in this study, we identified the first Coxiella effector protein capable of interacting with an autophagy-related RAB GTPase to stimulate the autophagy flux. With a repertoire of more than 130 effector proteins, Coxiella could possess additional RAB-modifying proteins that could favor its development in mammalian hosts. CvpF could open the way for the characterization of novel RAB-targeting effectors.
Material and methods
Bacterial strains, cell lines, and growth conditions
Strains used in this study are listed in Supplementary Table 1. Coxiella burnetii strains were grown in ACCM-2 [47] supplemented with kanamycin (Sigma-Aldrich, K1377; 340 μg/ml) or chloramphenicol (Sigma-Aldrich, C0378; 3 μg/ml), as appropriate in a humidified atmosphere of 5% CO2 and 2.5% O2 at 37°C. U2OS (ATCC, HTB-96), U2OS mCherry, U2OS GFP-FYVE (kindly provided by Dr. Tassula Proikas-Cezanne, Eberhard Karls University Tübingen, Germany), U2OS Cas9, and HEK293T (ATCC, CRL-11268) cell lines were routinely maintained in DMEM with GlutaMAX (GIBCO, 61965–026) containing 10% fetal calf serum (FCS; Sigma-Aldrich, F7524) and THP-1 (ATCC, TIB-202) cells were routinely maintained in RPMI with GlutaMAX (GIBCO, 61,870–010) containing 10% FCS. All cells were grown in a humidified atmosphere of 5% CO2 at 37°C. THP-1 cells were differentiated into macrophages by treatment with 25 ng/ml phorbol myristate acetate (PMA, Sigma-Aldrich, P8134) for 24 h. Cell growth medium was supplemented with 600 μg/ml of geneticin G418 (Gibco, 10131035), 10 μg/ml of blasticidin (InvivoGen, ant-bl-05) and/or 1 μg/ml of puromycin (Sigma-Aldrich, P8833) as appropriate.
Antibodies and reagents
Hoechst 33258 (94403), anti-mouse (12–349), and anti-rabbit (12–348) HRP-conjugated antibodies, rabbit anti-LC3B (L8918), rabbit anti-LAMP1 (L1418), mouse anti-GAPDH (G8795), mouse anti-Flag (F1804) and bafilomycin A1 (B1793) were purchased from Sigma-Aldrich. Rabbit monoclonal anti-GOLGA2/GM130 (ab52649) and rabbit polyclonal anti-RAB26 (ab187151) were purchased from AbCam. Mouse monoclonal anti-beta-lactamase (MA1-20370) was from ThermoFisher Scientific. Mouse anti-LC3B (ALX-803-080-C100) was from Enzo Life Sciences. Rabbit anti-HA (sc-7392) was from Santa Cruz Biotechnology, and mouse anti-HA (66006-1-lg) was from Proteintech. Rabbit polyclonal anti-SQSTM1/p62 (GTX100685) was from GeneTex. Phalloidin coupled to Alexa Fluor 647 (A22287) and LysoTracker Red (L7528) were purchased from Life technologies. Rabbit anti-GFP (PABG1-20) was from Chromotek. Rabbit anti-MTOR (2983S) was from Cell Signaling Technology. Mouse anti-Cas9 (844302) was from BioLegend. Rabbit anti-Coxiella NMII antibodies [30] were generated by Covalab. Mouse and rabbit IgG conjugated to Alexa Fluor 488 (A-11029 and A-11008), 555 (A-21147 and A-21429), or 647 (A-21239 and A-21245), as well as Prolong Gold antifade mounting reagent (P36930) were purchased from Invitrogen. Paraformaldehyde (15700) was provided by Electron Microscopy Sciences, PA.
Plasmids
Plasmids and primers used in this study are listed in Supplementary Tables 2 and 3, respectively. DNA sequences were amplified by PCR using Phusion polymerase (New England Biolabs, M0530L) and gene-specific primers (Sigma-Aldrich). All site-directed mutagenesis was performed by PCR overlap extension with the mentioned primer pairs.
Complementation in C. burnetii
The operon comprising cbu0625 promoter (600 nt upstream cbu0625 start codon), cbu0625, and cbu0626/cvpF was amplified from Coxiella RSA439 NMII genomic DNA using Prom-0625-NheI-Fw and CvpF-XhoI-Rv and cloned into pUCR6K-miniTn7-Kan (kindly provided by Prof. Robert Heinzen, Rocky Mountain Laboratories, MT, USA) to generate Mini-Tn7-Kan-prom625-0625-0626. Preparation of electrocompetent Coxiella cvpF::Tn was performed as described in [33] and the bacteria were electroporated with an equimolar amount of Mini-Tn7-Kan-prom625-0625-0626 (carrying the transposon prom625-0625-0626) and pTnS2 (carrying the Tn7 transposase, kindly provided by Prof. Robert Heinzen) plasmids prior to selection and amplification in ACCM-2 containing appropriate antibiotics.
Lentivirus production and stable cell line engineering
To produce lentiviral particles, HEK293T cells were cotransfected with an HIV-1 based genome coding plasmid (pLentiCas9-Blast [Addgene, 47948], pLentiGuide-Puro [Addgene, 68463], or pLVX-mCherry-C1 [Clontech, 632561]), pCMV-dR8.91 (HIV-1 GagPol, kindly provided by Dr. Antoine Gross, IRIM UMR9004 CNRS, Montpellier, France) and pMD.G (VSV-G, kindly provided by Dr. Antoine Gross, IRIM UMR9004 CNRS, Montpellier, France) at a ratio of 1:1:0.5, respectively. The medium was replaced after 6 h, and viral particles were harvested 42 h later, filtered on 0.45-μm filters (Clearline, 146561), and directly used to transduce U2OS target cells. After 4 to 6 h, the transduction medium was replaced with fresh DMEM containing 10% FCS, and the relevant antibiotics were added 48 h later to select transduced U2OS Cas9 and U2OS mCherry cells.
Reverse transcription and PCR
Coxiella strains Tn1832 (WT), Tn248, or Tn248 Comp. were grown for 7 d in 5 ml of ACCM-2 containing the appropriate antibiotics. Bacteria were centrifuged for 30 min at 21,000 × g at 4°C and total RNA was extracted using the RNA easy kit (Qiagen, 74104) or the Direct-zol RNA miniprep kit (Zymo Research, R2051), according to the manufacturer’s recommendations. DNA was further removed using DNase I (New England Biolabs, M0303S). 1 μg of total RNA was used for reverse transcription using the High Capacity cDNA reverse transcription kit (Applied Biosystems, 4368814) or the SuperScript VILO cDNA synthesis kit (Invitrogen, 11754–050) according to the manufacturer’s recommendations. Gene-specific primers (Supplementary Table 3) were used to amplify the cDNA corresponding to ompA (cbu1260), cbu0625, and cbu0626/cvpF.
Real-time PCR
Lightcycler 480 SYBR Green I master mix (Thermo Fisher Scientific, 4887352001) was used for real-time PCR and readouts was acquired on a Lightcycler480 real-time system (Roche) according to the manufacturer’s instructions. The relative levels of transcripts were calculated by the ΔΔCT threshold cycle (CT) method using dotA as the internal control. The relative levels of mRNA from the WT samples were adjusted to 1 and served as the basal control value. Each experiment was done in biological triplicate. For the calibration curve construction, aliquots of the plasmid pCR2.1::dotA (Provided by Dr. Robert Heinzen, 8 × 1010 copies/μl) in dilutions ranging from 8 × 102 to 8 × 109 copy numbers were applied. Three replicative runs of a dotA real-time PCR for all samples were performed. For the standard curves, the threshold cycle (Cq) values were calculated using the Lightcycler480 Software (Roche).
Immunofluorescence staining
Cells were either fixed in 3% paraformaldehyde in PBS (ThermoFisher Scientific, 14190–094) at room temperature for 30 min, or in methanol/acetone (1:1) at −20°C for 5 min. Samples were rinsed in PBS and incubated in BSS (0.5% BSA (Sigma-Aldrich, A2058), 50 mM NH4Cl in PBS pH 7.4) or FBS (5% fetal bovine serum [Sigma-Aldrich, F7524] in PBS pH 7.4) blocking solutions. Primary and secondary antibodies were diluted in the respective blocking solutions and used for immunofluorescence staining.
Co-immunoprecipitation
U2OS cells were transfected with equimolar amounts of plasmids expressing GFP- and HA-tagged constructs. After 24 h, cells were trypsinized, washed twice in PBS and incubated in the co-IP buffer (20 mM Tris-HCl pH 7.6, 200 mM NaCl, 0.5 mM EDTA, 1% IGEPAL CA-630 [Sigma-Aldrich, I8896], Complete protease inhibitor [Roche, 11836170001]) on ice for 30 min. Following a 10-min spin at 21,000 x g at 4°C, cell lysates were incubated with magnetic GFP-trap beads (Chromotek, gtma-20) for 1 h at 4°C on a roller. Beads were washed thrice with co-IP buffer, and proteins were collected in Laemmli buffer.
Beta-lactamase translocation assay
For C. burnetii effector translocation assays, cells were cultured in black, clear-bottomed, 96-well plates (Greiner Bio-One, 655,090), and infected with the appropriate C. burnetii strain (MOI of 100) for 24, 48, and 72 h. C. burnetii expressing β-lactamase alone was used as a negative control; C. burnetii expressing β-Lactamase-tagged CvpB was used as a positive control. Cell monolayers were loaded with the fluorescent substrate CCF4/AM (LiveBLAzer-FRET B/G loading kit; Invitrogen, K1095) in a solution containing 15 mM probenecid (Sigma-Aldrich, P36400). Cells were incubated in the dark for 1 h at room temperature and imaged using an EVOS inverted fluorescence microscope (ThermoFisher Scientific, UK). Images were acquired using a DAPI and GFP filter cubes. The image analysis software CellProfiler (Broad Institute, MA) was used to segment and identify all cells in the sample (GFP) and positive cells (DAPI) and to calculate the intensity of fluorescence in each channel. The percentage of positive cells versus the total number of cells was then calculated and used to evaluate effector translocation.
Microscopy and image analysis
Samples were imaged with a Zeiss Axio Imager Z1 epifluorescence microscope (Carl Zeiss, Germany) connected to a CoolSNAP HQ2 CCD camera (Teledyne Photometrics, Tucson, AZ). Images were acquired with 40x oil immersion objectives and processed with Metamorph (Molecular Devices, San Jose, CA). High throughput image acquisition was performed on an epifluorescence automated microscope (Cellomics, ThermoFisher Scientific, Pittsburgh, PA) equipped with a 20X objective. Image analysis was performed using the CellProfiler 3.0 software, as previously described (Martinez et al., 2015). Briefly, Hoechst 33258 (Sigma-Aldrich, 94403) staining, cytoplasmic mCherry fluorescence produced by U2OS mCherry cells and LAMP1 labeling were used to identify nucleus, cytoplasm, and lysosomal/late endosomal compartments, respectively. The GFP signal was used to identify C. burnetii colonies, and CCVs were segmented based on the LAMP1 antibody labeling and GFP fluorescence, respectively. After sorting non-infected and infected cells, a morphometrical analysis was carried out over a population of at least 200 objects (CCVs and/or C. burnetii colonies) per condition. Confocal microscopy was performed using a TCS SP8 HyVolution microscope (Leica, Germany) connected to a DFC9000 sCMOS camera or an SP5-SMD microscope (Leica, Germany). Images were acquired with 63x oil immersion objectives and processed with LAS-AF (Leica, Germany). Fluorescence was measured using Fiji analysis software (version 2.0.0) and the Multicolor line profile plot macro (University of Leicester). LC3B and RAB26 recruitment at CCVs were scored as follow: The mean ± confidence interval (CI) Pearson’s correlation coefficient between LAMP1 and the cellular marker of interest (LC3B or RAB26) was calculated using Icy analysis software from triplicate experiments representing a total of at least 80 CCVs per condition (WT, cvpF::Tn, cvpF::Tn Comp.). The mean + positive CI calculated from cells infected with the cvpF::Tn mutant was then taken as a threshold to calculate the percentage of CCVs positive for either marker.
Animal experiments
SCID (C.B-17/LcrHsd-Prkdcscid) mice were handled and infected, as previously described [34]. All animal procedures were done in compliance with Texas A&M University IACUC (AUP#2016-0370). DNA was purified from infected organs, and primers and probe for IS1111 were used to determine Genome Equivalents (GE) using TaqMan real-time PCR.
Statistical analysis
Statistical analyses of data were performed using Prism software (GraphPad, San Diego, CA). For experiments requiring statistical analysis, an adapted statistical test was performed, as described in the corresponding figure legends.
Supplementary Material
Acknowledgments
This work was supported by the French National Research Agency (ANR; ANR-14-CE14-0012-01, project AttaQ) and by the ERA-NET Infect-ERA (ANR-13-IFEC-0003, project EUGENPATH). FAS is the recipient of a fellowship from the Agence Nationale Bourses Gabon (ANBG). We acknowledge the imaging facility MRI, member of the national infrastructure France-BioImaging supported by the French National Research Agency (ANR-10-INBS-04, «Investments for the future»). We thank Dr. Martine Biard-Piechaczyk, Dr. Lucile Espert, and Dr. Fabien Blanchet (IRIM CNRS UMR9004 Montpellier, France) for scientific advice and sharing material.
Funding Statement
This work was supported by the Agence Nationale de la Recherche [ANR-13-IFEC-0003];Agence Nationale de la Recherche [ANR-14-CE14-0012-01];Agence Nationale de la Recherche [ANR-10-INBS-04].
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
References
- [1].Quaglio GL, Demotes-Mainard J, Loddenkemper R.. Emerging and re-emerging infectious diseases: a continuous challenge for Europe. Eur Respir J. 2012;40:1312–1314. [DOI] [PubMed] [Google Scholar]
- [2].Waag DM, Fritz DL. Q fever. Biodefense Res Methodol Anim Model. Second Ed. 2012; 12:179–196. [Google Scholar]
- [3].Moffatt JH, Newton P, Newton HJ. Coxiella burnetii: turning hostility into a home. Cell Microbiol. 2015;17:621–631. [DOI] [PubMed] [Google Scholar]
- [4].Lührmann A, Newton HJ, Bonazzi M. Beginning to understand the role of the type IV secretion system effector proteins in Coxiella burnetii pathogenesis. Curr Top Microbiol Immunol. 2017;413:243–268. [DOI] [PubMed] [Google Scholar]
- [5].Voth DE, Heinzen RA. Lounging in a lysosome: the intracellular lifestyle of Coxiella burnetii. Cell Microbiol. 2007;9:829–840. [DOI] [PubMed] [Google Scholar]
- [6].Carey KL, Newton HJ, Lührmann A, et al. The Coxiella burnetii Dot/Icm system delivers a unique repertoire of type IV effectors into host cells and is required for intracellular replication. PLoS Pathog. 2011;7(5):e1002056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Newton HJ, McDonough JA, Roy CR. Effector protein translocation by the Coxiella burnetii Dot/Icm type iv secretion system requires endocytic maturation of the pathogen-occupied vacuole. PLoS One. 2013;8:e54566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Larson CL, Martinez E, Beare PA, et al. Right on Q: genetics begin to unravel Coxiella burnetii host cell interactions. Future Microbiol. 2016;11:919–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Lührmann A, Nogueira CV, Carey KL, et al. Inhibition of pathogen-induced apoptosis by a Coxiella burnetii type IV effector protein. Proc Natl Acad Sci U S A. 2010;107:18997–19001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Larson CL, Beare PA, Howe D, et al. Coxiella burnetii effector protein subverts clathrin mediated vesicular trafficking for pathogen vacuole biogenesis. Proc Natl Acad Sci U S A. 2013;110:E4770–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Larson CL, Beare PA, Voth DE, et al. Coxiella burnetii effector proteins that localize to the parasitophorous vacuole membrane promote intracellular replication. Infect Immun. 2015;83:661–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Martinez E, Allombert J, Cantet F, et al. Coxiella burnetii effector CvpB modulates phosphoinositide metabolism for optimal vacuole development. Proc Natl Acad Sci U S A. 2016;113:E3260–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Voth DE, Beare PA, Howe D, et al. The Coxiella burnetii cryptic plasmid is enriched in genes encoding type IV secretion system substrates. J Bacteriol. 2011;193:1493–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Maturana P, Graham JG, Sharma UM, et al. Refining the plasmid-encoded type IV secretion system substrate repertoire of Coxiella burnetii. J Bacteriol. 2013;195:3269–3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Latomanski EA, Newton HJ. Interaction between autophagic vesicles and the Coxiella-containing vacuole requires CLTC (clathrin heavy chain). Autophagy. 2018;14:1710–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Crabill E, Schofield WB, Newton HJ, et al. Dot/Icm-translocated proteins important for biogenesis of the Coxiella burnetii-containing vacuole identified by screening of an effector mutant sublibrary. Infect Immun. 2018;86:e00758–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ao X, Zou L, Wu Y. Regulation of autophagy by the Rab GTPase network. Cell Death Differ. 2014;21:348–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Binotti B, Pavlos NJ, Riedel D, et al. The GTPase RaB26 links synaptic vesicles to the autophagy pathway. Elife. 2014;2015:1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Dong W, He B, Qian H, et al. RAB26-dependent autophagy protects adherens junctional integrity in acute lung injury. Autophagy. 2018;14:1677–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Song Y, Shang D, Cheng H, et al. The small GTPase RAB37 functions as an organizer for autophagosome biogenesis. Autophagy. 2018;14:727–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Sheng Y, Song Y, Li Z, et al. RAB37 interacts directly with ATG5 and promotes autophagosome formation via regulating ATG5-12-16 complex assembly. Cell Death Differ. 2018;25:918–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Spanò S, Galán JE. Taking control: hijacking of RAB GTPases by intracellular bacterial pathogens. Small GTPases. 2018;9:182–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Stein MP, Müller MP, Wandinger-Ness A. Bacterial pathogens commandeer RAB GTPases to establish intracellular niches. Traffic. 2012;13:1565–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Martinez E, Siadous FA, Bonazzi M. Tiny architects: biogenesis of intracellular replicative niches by bacterial pathogens. FEMS Microbiol Rev. 2018;42:425–447. [DOI] [PubMed] [Google Scholar]
- [25].McDonough JAJ, Newton HJHHJ, Klum S, et al. Host pathways important for Coxiella burnetii infection revealed by genome-wide RNA interference screening. MBio. 2013;4:e00606–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Berón W, Gutierrez MG, Rabinovitch M, et al. Coxiella burnetii localizes in a Rab7-labeled compartment with autophagic characteristics. Infect Immun. 2002;70:5816–5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Romano PS, Gutierrez MG, Berón W, et al. The autophagic pathway is actively modulated by phase II Coxiella burnetii to efficiently replicate in the host cell. Cell Microbiol. 2007;9:891–909. [DOI] [PubMed] [Google Scholar]
- [28].Gutierrez MG, Vázquez CL, Munafó DB, et al. Autophagy induction favours the generation and maturation of the Coxiella-replicative vacuoles. Cell Microbiol. 2005;7:981–993. [DOI] [PubMed] [Google Scholar]
- [29].Campoy EM, Martín Zoppino FC, Colombo MI. The early secretory pathway contributes to the growth of the Coxiella-replicative niche. Infect Immun. 2011;79:402–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Martinez E, Cantet F, Fava L, et al. Identification of OmpA, a Coxiella burnetii protein involved in host cell invasion, by multi-phenotypic high-content screening. PLoS Pathog. 2014;10:e1004013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Zusman T, Aloni G, Halperin E, et al. The response regulator PmrA is a major regulator of the icm/dot type IV secretion system in Legionella pneumophila and Coxiella burnetii. Mol Microbiol. 2007;63:1508–1523. [DOI] [PubMed] [Google Scholar]
- [32].Lifshitz Z, Burstein D, Peeri M, et al. Computational modeling and experimental validation of the Legionella and Coxiella virulence-related type-IVB secretion signal. Proc Natl Acad Sci U S A. 2013;110:E707–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Martinez E, Cantet F, Bonazzi M. Generation and multi-phenotypic high-content screening of Coxiella burnetii transposon mutants. J Vis Exp. 2015;2015:4–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].van Schaik EJ, Case ED, Martinez E, et al. The SCID mouse model for identifying virulence determinants in Coxiella burnetii. Front Cell Infect Microbiol. 2017;7:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Newton HJ, Kohler LJ, McDonough JA, et al. A screen of Coxiella burnetii mutants reveals important roles for Dot/Icm effectors and host autophagy in vacuole biogenesis. PLoS Pathog. 2014;10(7):e1004286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Larson CL, Sandoz KM, Cockrell DC, et al. Noncanonical Inhibition of mTORC1 by Coxiella burnetii promotes replication within a phagolysosome-like vacuole. MBio. 2019;10(1):e02816–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Jin RU, Mills JC. RAB26 coordinates lysosome traffic and mitochondrial localization. J Cell Sci. 2014;127:1018–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Lambert NA, Lan T-H, Li C, et al. Rab26 modulates the cell surface transport of α 2 -adrenergic receptors from the golgi. J Biol Chem. 2012;287:42784–42794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Kohler LJ, Reed SR, Sarraf SA, et al. Effector protein cig2 decreases host tolerance of infection by directing constitutive fusion of autophagosomes with the Coxiella-containing vacuole. MBio. 2016;7:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Winchell CG, Graham JG, Kurten RC, et al. Coxiella burnetii type IV secretion-dependent recruitment of macrophage autophagosomes. Infect Immun. 2014;82:2229–2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Winchell CG, Dragan AL, Brann KR, et al. Coxiella burnetii subverts p62/sequestosome 1 and activates Nrf2 signaling in human macrophages. Infect Immun. 2018;86:e00608–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Latomanski EA, Newton P, Khoo CA, et al. The effector Cig57 hijacks FCHO-mediated vesicular trafficking to facilitate intracellular replication of Coxiella burnetii. PLoS Pathog. 2016;12(12):e1006101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Kimura S, Noda T, Yoshimori T. Dynein-dependent movement of autophagosomes mediates efficient encounters with lysosomes. Cell Struct Funct. 2008;33:109–122. [DOI] [PubMed] [Google Scholar]
- [44].Kalvari I, Tsompanis S, Mulakkal NC, et al. iLIR: a web resource for prediction of Atg8-family interacting proteins. Autophagy. 2014;10:913–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Goody RS, Itzen A. Modulation of small Gtpases by Legionella. Curr Top Microbiol Immunol. 2013;376:117–133. [DOI] [PubMed] [Google Scholar]
- [46].Wang Z, McCloskey A, Cheng S, et al. Regulation of the small GTPase Rab1 function by a bacterial glucosyltransferase. Cell Discov. 2018;4:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Omsland A, Beare PA, Hill J, et al. Isolation from animal tissue and genetic transformation of Coxiella burnetii are facilitated by an improved axenic growth medium. Appl Environ Microbiol. 2011;77:3720–3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


