Abstract
The genus Limonium, commonly known as Sea Lavenders, is one of the most species-rich genera of the family Plumbaginaceae. In this study, two new plastomes for the genus Limonium, viz. L. tetragonum and L. bicolor, were sequenced and compared to available Limonium plastomes, viz. L. aureum and L. tenellum, to understand the gene content and structural variations within the family. The loss of the rpl16 intron and pseudogenisation of rpl23 was observed. This study reports, for the first time, expansion of the IRs to include the ycf1 gene in Limonium plastomes, incongruent with previous studies. Two positively selected genes, viz. ndhF and ycf2, were identified. Furthermore, putative barcodes are proposed for the genus, based on the nucleotide diversity of four Limonium plastomes.
Keywords: Intron loss, IR expansion, positive selection, pseudogenisation, ycf1
Introduction
The family Plumbaginaceae of the order Caryophyllales is highly diverse, rich in species and displays a cosmopolitan distribution with its maximum diversity in the temperate areas of the northern hemisphere (Kubitzki 1993). It is sister to Polygonaceae (Lledó et al. 1998; Chase et al. 2016) and further classified into two subfamilies: Limonioideae (formerly Staticoideae) and Plumbaginoideae (Lledó et al. 1998, 2001; Hernández-Ledesma et al. 2015). Limonioideae is further divided into two tribes, Limonieae (consisting of 24 genera) and the monotypic Aegialitideae, whereas Plumbaginoideae consists of four genera. Limonioideae is a sub-cosmopolitan group distributed mostly in the Mediterranean and Indo-Turanian regions, but a few genera have also diversified in the Southern Hemisphere. Limonium Mill., Acantholimon Boiss. and Armeria (DC.) Willd., all belonging to Limonioideae, are the most species-rich genera, comprising 80–90% of the species in Plumbaginaceae (Koutroumpa et al. 2018).
The genus Limonium Mill., popularly known as sea lavenders, belongs to the subfamily Limonioideae and tribe Limonieae (Kubitzki 1993; Lledó et al. 2005; Malekmohammadi et al. 2017). The genus is represented by ca. 600 species and is the sole genus of Plumbaginaceae exhibiting a sub-cosmopolitan distribution (Koutroumpa et al. 2018). The genus comprises several ornamental and medicinally-important species (Malekmohammadi et al. 2017). Limonium are herbs or shrubs growing in saline or metal-rich soils, mostly in coastal areas (Kubitzki 1993). Variation of reproductive systems (sexual and apomixis), as well as events like hybridisation and polyploidy, complicate the delimitation of most of the species of Limonium (Kubitzki 1993; Cowan et al. 1998; Palacios et al. 2000; Akhani et al. 2013; Róis et al. 2016).
Certain phylogenetic studies tried to resolve the relationships within Limonium at a global scale (Lledó et al. 2005); however, many others were confined to either a specific geographic area or some specific sections of Limonium (Palacios et al. 2000; Lledó et al. 2005; Lledó et al. 2011; Akhani et al. 2013; Róis et al. 2016). More recently, Malekmohammadi et al. (2017) tried to resolve the phylogenetic relationships within Limonium including 76 species of the Mediterranean region. They found two well-supported clades for subgenera Limonium and Pteroclados (Boiss.) Pignatti, which confirmed the earlier findings by Lledó et al. (2005, 2011). Their study was based on one nuclear and several plastid loci, but lacked comprehensive sampling. Koutroumpa et al. (2018) carried out a phylogenetic study of Plumbaginaceae which included 23 genera of the family with an emphasis on the genus Limonium (201 spp.), based on three chloroplast and one nuclear marker. The study again confirmed most of the previous molecular phylogenies and led to the proposal of new sections and altering some of the existing sections. However, taxonomic difficulties, diversity in reproductive modes and sub-cosmopolitan distribution of Limonium necessitate further studies on the genus.
With the advent of sequencing technologies, the availability of large genome-scale data has made it easier to understand phylogeny and detect polyploidy events (Gompert and Mock 2017; Thomas et al. 2017; McKain et al. 2018). Five plastomes have been sequenced for Plumbaginaceae thus far, viz. Limonium sinense (Girard) Kuntze (Li et al. 2020), L. aureum (L.) Chaz. (Zhang et al. 2020), L. tenellum (Turcz.) Kuntze (Yao et al. 2019) and one each of Ceratostigma Bunge and Plumbago L. The recent phylogenomic study to understand the evolution of Caryophyllales incorporated plastome sequences of L. tenellum, Plumbago auriculata Lam. and Ceratostigma willmottianum Stapf (Yao et al. 2019). The study reported that all Plumbaginaceae members, except Limonium, exhibit expanded IR to accommodate ycf1. However, no studies have been carried out to understand and compare the structure, composition and evolution of the plastome within Plumbaginaceae and Limonium in particular.
The present study reports the plastome sequences of two Asian Limonium species, viz. L. tetragonum (Thunb.) Bullock and L. bicolor (Bunge) Kuntze and compares the structure, composition and diversity within the genus by combining them with other available plastomes. L. tetragonum is a biennial species characterised by a spicate inflorescence, yellow corolla, acute calyx with pink at the base, white in upper parts and distributed in Japan, Korea, New Caledonia and Primorye (Ohwi 1965; POWO 2021). Limonium bicolor is a perennial species, characterised by a paniculate inflorescence, yellow corolla, somewhat rounded calyx, pink to purplish at base, white in upper parts and distributed in China and Mongolia (Wu and Raven 1996; POWO 2021). The former is known for its anti-cancerous properties (Kong et al. 2008; Bae et al. 2016), while the latter is most studied for its salt glands (Li et al. 2019; Lu et al. 2020). An attempt has also been made to unravel the structural variations within Plumbaginaceae plastomes and propose molecular markers for easier discrimination of Limonium species.
Material and methods
Sampling and sequencing
Leaf samples of Limonium bicolor and L. tetragonum were collected from Meneng steppe of Dornod Province of Mongolia (Voucher No. KRIB 0070251) in June 2015 and from the coastal area of Ulsan City of the Republic of Korea (Voucher No. KRIB 0086343) in April 2018, respectively. The samples were deposited at the Herbarium of Korea Research Institute of Bioscience and Biotechnology (KRIB). DNA extraction was carried out from dried leaves using the DNeasy Plant Mini Kit (QIAGEN, Cat. No. 69104) according to the manufacturer’s protocol. For both the plastomes, a 550 bp DNA TruSeq Illumina (Illumina, San Diego, CA, USA) sequencing library was constructed. After the library preparation, the DNA samples were run in a single lane of an Illumina HiSeq 10X with a read length of 151 bp.
Assembly and annotation
The raw reads obtained after Illumina sequencing were analysed using FastQC V0.11.7 (Andrews 2010) software to ensure the quality of the reads and Phred score. The assembly was carried out using NOVOPlasty V4.2 (Dierckxsens et al. 2017). Forward and reverse reads with a read length of 150 bp and insert size of 300 bp, as well as seed sequence (rbcL of L. tetragonum), were used as input. De novo, as well as reference-based assembly, were carried out. The plastome sequence of L. aureum (MN623109) was used as a reference for the assembly of both the plastomes. The assembly and orientation of the Inverted Repeats (IRs), Large Single Copy (LSC) and Small Single Copy (SSC) regions were confirmed by NCBI blast and graphic view using Geneious prime 2020.2.2 (https://www.geneious.com). The assembled plastomes were annotated using Geneious prime 2020.2.2 (https://www.geneious.com) and Geseq – Annotation of Organellar Genomes (Tillich et al. 2017). The transfer RNAs (tRNAs) were identified with tRNA-scan-SE (Lowe and Chan 2016). Graphical maps of both plastomes were visualised using OGDRAW (Lohse et al. 2013).
Comparative plastome analysis
All Plumbaginaceae plastomes available so far were included for comparison. Four plastomes of Limonium, viz. L. aureum (MN623109), L. tenellum (MK397871), L. tetragonum, L. bicolor and Ceratostigma willmottianum (MK397862), as well as Plumbago auriculata (MH286308), were included in the analysis. The plastome of L. sinense was not included in any of the analyses due to ambiguities observed in the assembly. The selected six plastomes were aligned using a Geneious prime 2020.2.2 plugin MAFFT v.7.450 (Katoh and Standley 2013). The contraction and expansion of IRs at the junction sites were examined and plotted using IRscope (Amiryousefi et al. 2018). Percentage sequence identity for all six plastomes was plotted using MultiPIPMaker (http://pipmaker.bx.psu.edu/pipmaker/) considering L. tetragonum as a reference (Schwartz et al. 2003).
Identification of Simple Sequence Repeats (SSRs) and repeats
Simple Sequence Repeats across the four Limonium plastomes were detected using MISA online server (Beier et al. 2017). The minimum repeat units for mono-, di-, tri-, tetra-, penta- and hexanucleotide repeats were set as 10, 5, 4, 3, 3 and 3, respectively, whereas forward, reverse, palindromic and complementary repeats were identified using the programme REPuter (Kurtz et al. 2001). The threshold for repeat length was set to 30 with more than 90% similarity and the hamming distance was set to 3.
Nucleotide diversity
The four Limonium plastomes were aligned using Geneious prime 2020.2.2 plugin MAFFT v.7.450 (Katoh and Standley 2013). The aligned sequences were curated manually and, due to the presence of ambiguous ycf1 region in L. tenellum at IRb/ SSC junction, the sequences were trimmed for all four taxa before the analysis. Nucleotide diversity and sliding window analysis were performed using the software DnaSP v.6.12.01 (Rozas et al. 2017).
Codon usage
The percentage codon usage of protein-coding regions of all four Limonium plastomes was calculated using Geneious prime 2020.2.2. To examine the frequency and uniformity of Synonymous codon and codon biases, the Relative Synonymous Codon Usage (RSCU) was also determined in DnaSP v.6.12.01 software (Rozas et al. 2017).
Positive selection analysis
In order to detect protein-coding genes under selection in the genus Limonium, sequences of each gene were aligned using the MAFFT v.7.450 plugin of Geneious prime 2020.2.2. The aligned sequences were again manually checked for an end-to-end alignment. The phylogenetic tree for each protein-coding gene was constructed using the FastTree plugin (Price et al. 2010) of Geneious prime 2020.2.2. The site-specific model was performed using CODEML algorithms (Yang 1998) implemented in EasyCodeML software (Gao et al. 2019). Seven codon substitution models (M0, M1a, M2a, M3, M7, M8 and M8a) were investigated and compared to identify positively selected sites, based on likelihood ratio tests.
Phylogenomic analysis
A total of 39 plastome sequences, including six from Plumbaginaceae were considered as ingroups for phylogenomic analysis. The outgroup was composed of members of Amaranthaceae. All the plastome sequences were aligned using the MAFFT v.7.450 plugin of Geneious prime 2020.2.2. Maximum Likelihood analysis, based on the best fit model GTR+F+R5, was performed using IQtree 1.6.12-MacOSX (Nguyen et al. 2015). The best likelihood score for the consensus tree was -979751.011.
Results
General features of the plastomes
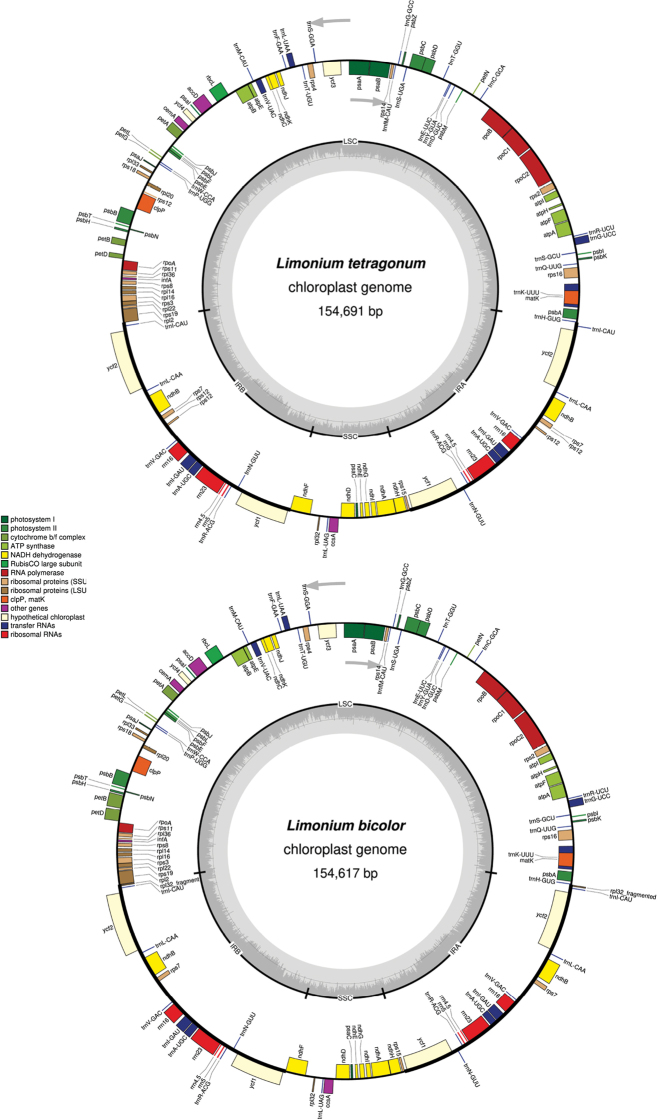
The average organelle coverage for the plastomes of L. tetragonum and L. bicolor was 1014X and 1009X, respectively. Plastomes of L. tetragonum and L. bicolor exhibited a typical quadripartite structure (Fig. 1). Total plastome lengths of L. tetragonum and L. bicolor were 154,691 bp and 154,617 bp with LSC (84,568 bp and 84,541 bp), SSC (12,997 bp and 12,964 bp) and a pair of IRs (28,563 bp and 28,556 bp), respectively (Table 1). The GC content of both plastomes was 37%. Both the plastomes exhibited 83 protein coding genes, 37 tRNA genes and four rRNA genes (duplicated in IR region) (Tables 1, 2). The assembled and annotated plastomes of L. tetragonum and L. bicolor were deposited to the NCBI database with accession numbers MW085088 and MW085089, respectively.
Figure 1.
Circular gene map of plastomes of L. tetragonum and L. bicolor. Genes drawn inside the circle are transcribed clockwise and those outside are counter-clockwise. Genes belonging to different functional groups are shown in different colours. The innermost circle denotes GC content across the plastome.
Table 1.
Comparison of plastome features of Plumbaginaceae members.
| Species | Limonium tetragonum | Limonium bicolor | Limonium aureum | Limonium tenellum | Ceratostigma willmottianum | Plumbago auriculata |
|---|---|---|---|---|---|---|
| Accession No. | MW085088 | MW085089 | MN623109 | MK397871 | NC041261 | NC041245 |
| Genome size (bp) | 154691 | 154617 | 154661 | 150515 | 164999 | 168765 |
| LSC length (bp) | 84568 | 84541 | 84546 | 84634 | 89454 | 91912 |
| SSC length (bp) | 12997 | 12964 | 12980 | 23755 | 13491 | 13331 |
| IR length (bp) | 28563 | 28556 | 28568 | 21063 | 31027 | 31761 |
| No. of genes duplicated in IR | 15 | 15 | 16 | 10 | 15 | 19 |
| No. of genes | 128 | 128 | 130 | 124 | 127 | 132 |
| No. of protein coding genes | 83 | 83 | 83 | 82 | 82 | 84 |
| No. of tRNA genes | 37 | 37 | 37 | 36 | 37 | 37 |
| No. of rRNA genes | 8 | 8 | 8 | 6* | 8 | 8 |
| Total GC content (%) | 37 | 37 | 37.1 | 37.1 | 37.5 | 37.2 |
*indicates annotations available as per NCBI database.
Table 2.
List of genes in the newly-sequenced plastomes of L. tetragonum and L. bicolor.
| Category | Group | Name |
|---|---|---|
| Photosynthesis-related genes | Rubisco | rbcL |
| Photosystem 1 | psaA, psaB, psaC, psaI, psaJ | |
| Photosystem 2 | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ | |
| APT synthase | atpA, atpB, atpE, atpF†, atpH, atpI | |
| Cytochrome b/f complex | petA, petB, petD, petG, petL, petN | |
| NADPH Dehydrogenase | ndhA†, ndhB†*, ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK | |
| Transcription and translation-related genes | Transcription | rpoA, rpoB, rpoC1†, rpoC2 |
| Ribosomal proteins | rps2, prs3, rps4, rps7*, rps8, rps11, rps12†*, rps14, rps15, rps16†, rps18, rps19 | |
| rpl2†, rpl14, rpl16, rpl20, rpl22, rpl23#*, rpl33, rpl36 | ||
| Translation initiation factor | infA | |
| RNA genes | Ribosomal RNA | rrn5*, rrn4.5*, rrn16*, rrn23* |
| Transfer RNA | trnA-UGC†*, trnC-GCA, trnD-GUC, trnE-UUC, trnF-GAA, trnfM-CAU, trnG-UCC, trnH-GUG, trnI-GAU†*, trnK-UUU†, trnL-CAA*, trnL-UAA, trnL-UAG†, trnM-CAU, trnN-GUU*, trnP-UGG, trnQ-UUG, trnR-ACG*, trnR-UCU, trnS-GCU, trnS-GGA, trnS-UGA, trnT-GGU, trnT-UGU, trnV-GAC*, trnW-CCA, trnY-GUA | |
| Other genes | RNA processing | matK |
| Carbon metabolism | cemA | |
| Fatty acid synthesis | accD | |
| Proteolysis | clpP† | |
| Genes of unknown function | Conserved reading frame | ycf1†*, ycf2*, ycf3†, ycf4 |
*duplicated genes, †genes with introns, #pseudogenised genes.
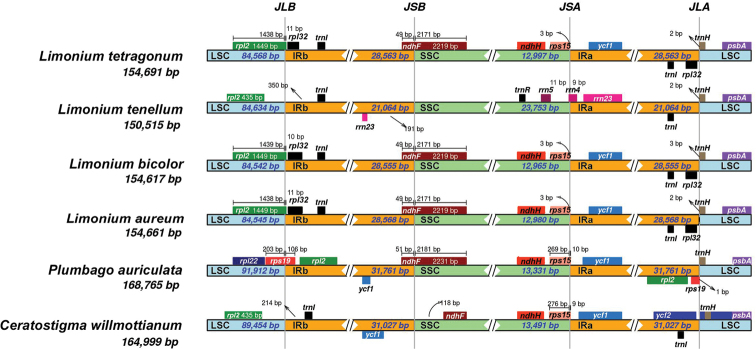
IR expansion and contraction
The IR regions of plastomes are divided by four junctions viz., IRb/LSC, IRb/SSC, IRa/SSC and IRa/LSC. All six plastomes of Plumbaginaceae (including four Limonium) were compared for their IR boundaries. The annotations available on NCBI database were used for Limonium aureum, L. tenellum, Ceratostigma willmottianum and Plumbago auriculata.
The IRb/LSC junction of three Limonium species viz. L. tetragonum, L. bicolor and L. aureum was characterised by the presence of the rpl2 gene (Fig. 2). The gene also extends in IRb with 11 bp, 10 bp and 11 bp, respectively. However, in L. tenellum and C. willmottianum, it was exclusively present in the LSC region, 937 bp and 1,041 bp away from the junction, respectively. In Plumbago, IR was expanded to include the rps19 gene at the junction, while rpl2 was duplicated in the IR region.
Figure 2.
Comparison of IRs of Plumbaginaceae plastomes.
The next junction, i.e. IRb/SSC, was characteried by the presence of the gene in L. tetragonum, L. bicolor, L. aureum and P. auriculata (49 bp of all Limonium species and 51 bp of Plumbago in IR region). In L. tenellum, the junction exhibited rrn23 (IR) and trnR-ACG (SSC), which could probably be due to wrong assembly or annotation. In Ceratostigma, ndhF appeared to be shifted to SSC, 118 bp away from IRb border.
IRa/SSC junction of all compared species was characterised by the presence of rps15 and ycf1 genes, except in L. tenellum. The IRa/LSC junction was characterised by rpl32 and trnH in L. tetragonum, L. bicolor and L. aureum, while in L. tenellum and C. willmottianum, it was characterised by ycf2 and trnH. In P. auriculata, the junction was bordered by rps19 and trnH (Fig. 2).
Plastomes of L. aureum, L. bicolor and L. tetragonum exhibited two copies of ycf1, except for L. tenellum which exhibited a single copy. All three plastomes are characterised by the ycf1 gene having a length of 5,298 bp and IR has been expanded to accommodate the ycf1 gene. Plumbago and Ceratostigma also exhibited the ycf1 gene duplicated in IRs. However, the annotation provided for L. tenellum (MK397871) exhibits a single copy of ycf1.
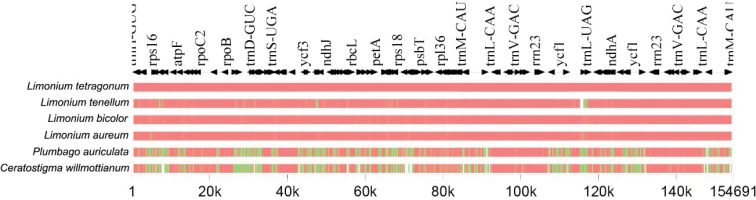
Structural comparison
Plastomes of Limonium tetragonum, L. tenellum, L. bicolor and L. aureum were compared with two Plumbaginaceae plastomes, keeping L. tetragonum as a reference. Sequence divergence amongst the four compared Limonium plastomes was similar as compared to Plumbago and Ceratostigma. Limonium tenellum exhibited partial deletion at the IRa/LSC junction in the ycf1 gene (Fig. 3). Our results suggest that the gene rpl23 has been pseudogenised in all the studied plastomes of the three genera of Plumbaginaceae. The length of rpl23 in all four Limonium plastomes was observed to be 171 bp, 270 bp in Plumbago and 50 bp in Ceratostigma. A similar loss of intron was observed in L. aureum, L. bicolor and L. tetragonum in our study. In all four compared species of Limonium, the length of rpl16 was 408 bp without any intron. However, the two other genera of Plumbaginaceae, i.e. Plumbago and Ceratostigma, exhibited the presence of intron. The length of the gene was 1,449 bp in the former, while in the latter, it was 1,469 bp.
Figure 3.
Plastome alignment of Plumbaginaceae members.
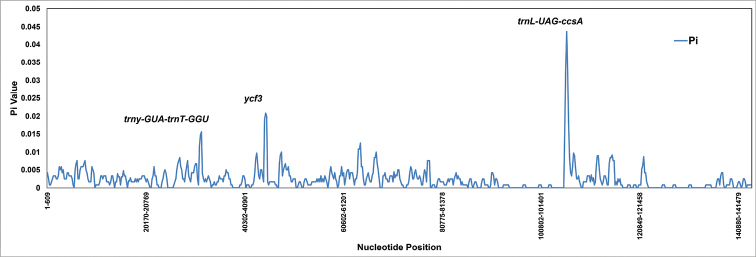
Nucleotide diversity
The nucleotide diversity (Pi) of four Limonium plastomes was analysed, except for the ycf1 region, which was removed due to ambiguous alignment. Sliding window analysis yielded some regions with higher Pi values. High nucleotide diversity was found in two spacer regions viz. trnY-GUA-trnT-GGU, trnL-UAG-ccsA and one gene ycf3 with Pi values 0.015, 0.043 and 0.02, respectively (Fig. 4).
Figure 4.
Nucleotide diversity and hotspot regions between Limonium plastomes. The X-axis represents the nucleotide position and Y-axis represents nucleotide diversity (Pi).
SSR and repeat analyses
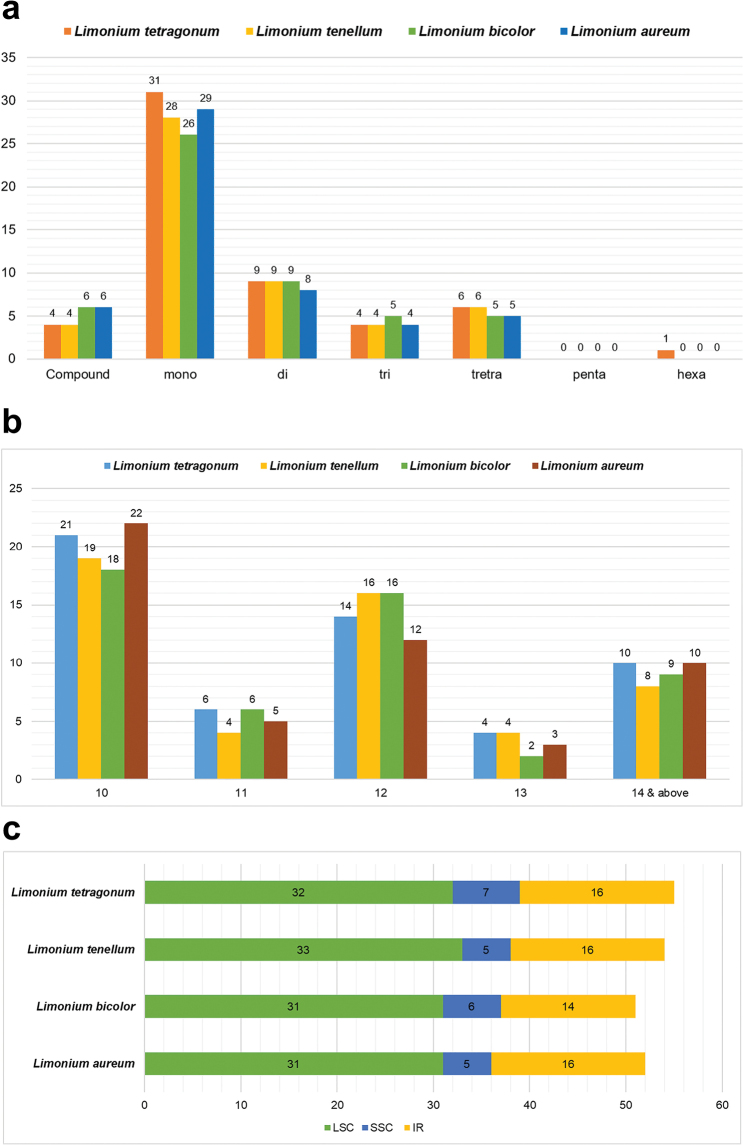
The plastomes of L. bicolor exhibited 6 compound, 26 mono-, 9 di-, 5 tri- and 5 tetranucleotide repeats, while L. tetragonum exhibited 4 compound, 31 mono-, 9 di-, 4 tri-, 6 tetra- and 1 hexanucleotide repeats (Fig. 5a). The size of repeats ranged from 10 to 168 in L. bicolor and 10 to 109 in L. tetragonum (Fig. 5b). Repeats in both the plastomes were found to be AT-rich. The highest number of SSRs was found in the LSC region, followed by IR and SSC regions in both the plastomes (Fig. 5c).
Figure 5.
Repeat analysis of four Limonium plastomes. a. Types of repeats, b. Size of repeats, c. Position of repeats.
All four species of Limonium exhibited only forward and palindrome type in REPuter analysis (Suppl. material 1).
Codon usage
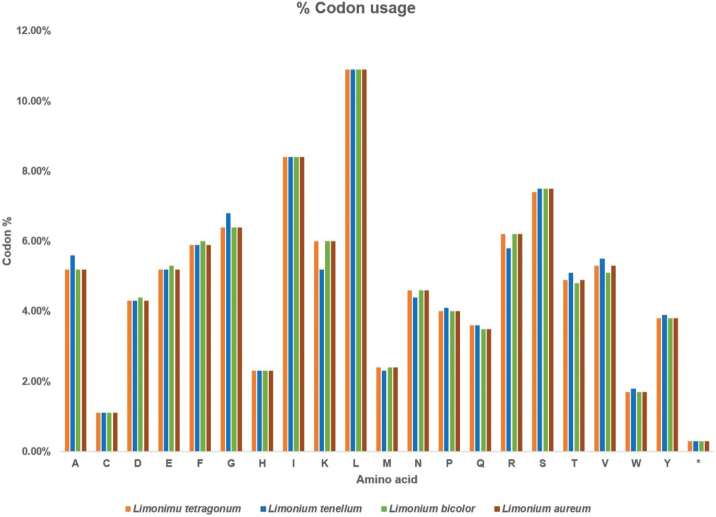
The four Limonium plastomes were compared for their codon usage. The plastomes of L. tetragonum, L. tenellum, L. bicolor and L. aureum exhibited 27,290, 24,093, 26,682 and 27,308 codons, respectively. Leucine was the most abundant while Cysteine was the least abundant amino acid in all the compared plastomes (Fig. 6). Codon usage was biased towards A and T in all the compared plastomes. The highest codon preference was 1.88, 1.89, 1.82 and 1.91 while the lowest was 0.33, 0.36, 0.37 and 0.36, respectively, in L. tetragonum, L. tenellum, L. bicolor and L. aureum (Suppl. material 2). Codon usage was biased towards A and T in all the compared plastomes.
Figure 6.
Codon usage of four Limonium species. X-axis: Amino acid, Y-axis: codon usage in percentage. * indicates the stop codon.
Positive selection analysis
A total of 79 consensus protein-coding genes of four Limonium species were evaluated with respect to selective pressure. Two genes were found to have undergone positive selection viz. ndhF and ycf2 with ω values 2.2278 and 19.657, respectively (Suppl. material 3).
Phylogenomic analysis
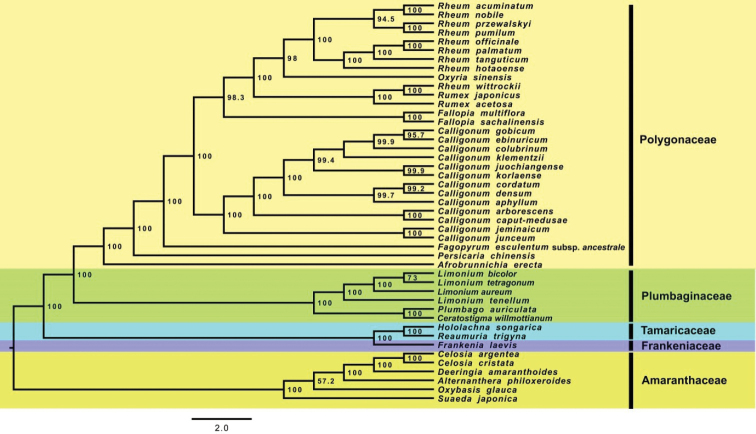
In the phylogenomic analysis, the representatives of Limonium formed a strongly-supported monophyletic group (BS = 100), in which L. bicolor was recovered as sister to L. tetragonum (BS = 73), with L. aureum and L. tenellum being successive sisters (BS = 100) to the clade of L. bicolor and L. tetragonum. Plumbago auriculata and Ceratostigma willmottianum also formed a monophyletic group (BS = 100), sister to the Limonium clade (BS = 100). All these made Plumbaginaceae a strongly-supported (BS = 100) monophyletic group (Fig. 7).
Figure 7.
Phylogenomic tree from the Maximum Likelihood analysis showing the placement of the studied four Limonium species. Nodal support is represented by bootstrap percentages.
Discussion
In this study, two Limonium plastomes were assembled and the structure and composition of four Limonium plastomes were compared. The plastomes were conserved in terms of size and structure ranging from 154,617 to 154,691 bp, except for L. tenellum with 150,515 bp. Expansion and contraction of IRs/SSC account for huge variation, evolutionary events and also affect the plastome sizes (Zhu et al. 2016; Darshetkar et al. 2019; Maurya et al. 2020). Yao et al. (2019) reported the expansion of IRs in Polygonaceae and Plumbaginaceae members to accommodate ycf1, except for the genus Limonium. However, the study included a single plastome sequence of Limonium tenellum. In all the Limonium plastomes, except L. tenellum, IR has been expanded to accommodate ycf1, unlike that reported by Yao et al. (2019). We assume the single copy of ycf1 in L. tenellum could be a result of erroneous assembly and annotation. Sequencing more plastomes for the genus will help to better understand the arrangement and position of ycf1 in Limonium.
Ribosomal Protein L23 is a protein component of the 60s large ribosomal subunit. The comprehensive study of plastomes of Caryophyllales (Yao et al. 2019) reported that the gene has undergone pseudogenisation at least 11 times in the order. Pseudogenisation has also been reported in the family Polygonaceae, a sister family of Plumbaginaceae (Logacheva et al. 2008). The results of our study corroborate these earlier studies. Transfer of the functional copy of the gene to the nucleus has been reported for many plastid genes in angiosperms (Millen et al. 2001; Jansen et al. 2011; Daniell et al. 2016). The functional copy of rpl23 might have been transferred to the nucleus, even in the members of Plumbaginaceae as predicted by Yao et al. (2019). Ribosomal Protein L16 codes for a protein component of the 50s ribosomal subunit. The loss of rpl16 intron was reported in Limonium gmelinii Kuntze and Limonium latifolium Kuntze (Campagna and Downie 1998). Recently, Yao et al. (2019) reported the loss of rpl16 intron in L. tenellum plastome. Hence, our study confirms the loss of rpl16 intron on the branch leading to Limonium as reported by Yao et al. (2019).
The value of the ratio of synonymous and nonsynonymous substitutions (Ka/Ks or ω) above 1 indicates that the corresponding genes experience positive selection, however, ω values ranging from 0.5 to 1 indicate relaxed selection (Tomoko 1995). The ndhF gene was found to be positively selected in the genus Rheum L. of Polygonaceae. The study reported that the higher expression levels of ndhF were observed in Rheum under environmental stress conditions (Li et al. 2016). Most of the Limonium species also grow in saline soils, which could be the reason behind the adaptive evolution of the gene. Positive selection of the ycf2 gene has already been reported in several angiosperms (Jiang et al. 2018; Zhong et al. 2019).
The sampling for the phylogenomic analysis followed the studies of Walker et al. (2018) and Yao et al. (2019). The ingroup consisted of plastomes belonging to the FTPP clade, i.e. Frankeniaceae, Tamaricaceae, Plumbaginaceae and Polygonaceae (they were all recovered as strongly-supported monophyletic groups), while the outgroup consisted of species belonging to Amaranthaceae. The position of the four Limonium species studied fits with the earlier studies (Yao et al. 2019). In the present study, phylogenetic position of four Limonium species belonging to subgenus Limonium (Koutroumpa et al. 2018) was studied. A study based on rbcL, trnL intron and trnL-F intergenic spacer included two species viz. L. tenellum and L. tetragonum, which were resolved in the same clade, but were placed into two different subsections, according to Boissier (1848), Rhodanthae and Chrysanthae, respectively. However, the study treated them under subgenus Limonium (Lledo et al. 2005). Later, Malekmohammadi et al. (2017) studied the phylogeny for the genus, based on several plastid and single nuclear (ITS) markers. The study included L. tetragonum and L. aureum and both the species were placed in the “L. aureum clade”. Recently, Koutroumpa et al. (2018) carried out phylogenetic studies of the genus, based on three chloroplast (trnL-F, matK and rbcL) and one nuclear marker (ITS). Three species, L. tetragonum, L. aureum and L. tenellum were included in the study. These three species were all resolved in the same clade. These species were earlier placed under sect. Plathymenium, L. tetragonum and L. aureum belonging to subsect. Chrysanthae and L. tenellum under subsect. Rhodanthae (Boissier 1848). Earlier studies reported plastome data dealing with single Limonium species and placed them as sister to Plumbago species (Yao et al. 2019; Li et al. 2020; Zhang et al. 2020). The present phylogenetic study, for the first time, included L. bicolor and analysed four Limonium plastomes together. However, inclusion of more Limonium plastomes would help in understanding the intrageneric relationships.
Conclusions
The present study has made an effort to understand the structural changes in the plastomes of Plumbaginaceae by including two newly-generated Limonium plastome sequences. The study also confirms the loss of rpl16 intron in the genus Limonium and pseudogenisation of the rpl23 gene in Plumbaginaceae. Our results also revealed, for the first time, the expansion of the IRs to accommodate the ycf1 gene in Limonium as in other Plumbaginaceae members. The annotation available for L. tenellum exhibits ycf1 in the SSC region. Hence, the sequencing of more plastomes would aid in identifying the exact position of ycf1. Two positively-selected genes were identified, viz. ndhF and ycf2. The positive selection of these genes could be linked to the evolution of ndhF to adapt to extreme environmental conditions, such as salt stress. It would be interesting to identify the adaptive sites in the ndhF amino acid by adding more ndhF sequences of Limonium species, while the expansion of IRs and accommodation of ycf2 genes could be related to the re-arrangement of the plastome. The function of ycf2 is still not clear, but it would be interesting to study the ycf2 evolution and re-arrangement in the whole order. High nucleotide diversity was observed in two spacer regions trnY-GUA--trnT-GGU, trnL-UAG--ccsA and one gene ycf3, which could be used as potential DNA barcodes for the genus. Future studies will focus on identifying adaptive codon sites in positively-selected genes and correlating those with the habitats and environmental conditions and validation of the proposed barcodes by including more Limonium species.
Acknowledgements
AMD, SM and RKC are grateful to the Director, Agharkar Research Institute for providing facilities. AMD is thankful to CSIR for Senior Research Fellowship. This research was supported by the grant from the KRIBB Initiative Program of the Republic of Korea and the Bio and Medical Technology Development Program of the National Research Foundation (NRF) and funded by the Korean Government (MSIT) (NRF-2016K1A1A8A01939075).
Citation
Darshetkar AM, Maurya S, Lee C, Bazarragchaa B, Batdelger G, Janchiv A, Jeong EJ, Choi S, Choudhary RK, Kim S-Y (2021) Plastome analysis unveils Inverted Repeat (IR) expansion and positive selection in Sea Lavenders (Limonium, Plumbaginaceae, Limonioideae, Limonieae). PhytoKeys 175: 121–139. https://doi.org/10.3897/phytokeys.175.61054
Funding Statement
This research was supported by the grant from the KRIBB Initiative Program of the Republic of Korea and the Bio and Medical Technology Development Program of the National Research Foundation (NRF) and funded by the Korean government (MSIT) (NRF-2016K1A1A8A01939075)
Contributor Information
Sangho Choi, Email: decoy0@kribb.re.kr.
Ritesh Kumar Choudhary, Email: rkchoudhary@aripune.org.
Soo-Yong Kim, Email: soodole@kribb.re.kr.
Supplementary materials
The output of the repeat analysis of four Limonium plastomes
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Ashwini M. Darshetkar, Satish Maurya, Changyoung Lee, Badamtsetseg Bazarragchaa, Gantuya Batdelger, Agiimaa Janchiv, Eun Ju Jeong, Sangho Choi, Ritesh Kumar Choudhary, Soo-Yong Kim
Data type
molecular analysis
Relative Synonymous Codon Usage
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Ashwini M. Darshetkar, Satish Maurya, Changyoung Lee, Badamtsetseg Bazarragchaa, Gantuya Batdelger, Agiimaa Janchiv, Eun Ju Jeong, Sangho Choi, Ritesh Kumar Choudhary, Soo-Yong Kim
Data type
molecular data
List of Positively selected genes
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Ashwini M. Darshetkar, Satish Maurya, Changyoung Lee, Badamtsetseg Bazarragchaa, Gantuya Batdelger, Agiimaa Janchiv, Eun Ju Jeong, Sangho Choi, Ritesh Kumar Choudhary, Soo-Yong Kim
Data type
molecular data
References
- Akhani H, Malekmohammadi M, Mahdavi P, Gharibiyan A, Chase MW. (2013) Phylogenetics of the Irano-Turanian taxa of Limonium (Plumbaginaceae) based on ITS nrDNA sequences and leaf anatomy provides evidence for species delimitation and relationships of lineages. Botanical Journal of the Linnean Society 171(3): 519–550. 10.1111/boj.12015 [DOI] [Google Scholar]
- Amiryousefi A, Hyvönen J, Poczai P. (2018) IRscope: An online program to visualize the junction sites of chloroplast genomes. Bioinformatics 34(17): 3030–3031. 10.1093/bioinformatics/bty220 [DOI] [PubMed] [Google Scholar]
- Andrews S. (2010) Babraham bioinformatics-FastQC a quality control tool for high throughput sequence data. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- Bae M-J, Karadeniz F, Lee S-G, Seo Y, Kong C-S. (2016) Inhibition of MMP-2 and MMP-9 Activities by Limonium tetragonum Extract. Preventive Nutrition and Food Science 21(1): 38–43. 10.3746/pnf.2016.21.1.38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beier S, Thiel T, Münch T, Scholz U, Mascher M. (2017) MISA-web: A web server for microsatellite prediction. Bioinformatics 33(16): 2583–2585. 10.1093/bioinformatics/btx198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boissier E. (1848) Plumbaginales. In: de Candolle AP (Ed. ) Prodromussystematis naturalis regni vegetabilis, Treuttel et Wurz, Paris, France 12: 617–696. [Google Scholar]
- Campagna ML, Downie SR. (1998) The Intron in Chloroplast Gene rpl16 is missing from the flowering plant families Geraniaceae, Goodeniaceae, and Plumbaginaceae. Transactions of the Illinois State Academy of Science. Illinois State Academy of Science 91: 1–11. [Google Scholar]
- Chase MW, Christenhusz MJM, Fay MF, Byng JW, Judd WS, Soltis DE, Mabberley DJ, Sennikov AN, Soltis PS, Stevens PF. (2016) An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Botanical Journal of the Linnean Society 181(1): 1–20. 10.1111/boj.12385 [DOI] [Google Scholar]
- Cowan RMJ, Ingrouille M, Lledó MD. (1998) The taxonomic treatment of agamosperms in the genus Limonium Mill. (Plumbaginaceae). Folia Geobotanica 33(3): 353–366. 10.1007/BF03216212 [DOI] [Google Scholar]
- Daniell H, Lin C-S, Yu M, Chang W-J. (2016) Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genome Biology 17(1): e134. 10.1186/s13059-016-1004-2 [DOI] [PMC free article] [PubMed]
- Darshetkar AM, Datar MN, Tamhankar S, Li P, Choudhary RK. (2019) Understanding evolution in Poales: Insights from Eriocaulaceae plastome. PLoS ONE 14(8): e0221423. 10.1371/journal.pone.0221423 [DOI] [PMC free article] [PubMed]
- Dierckxsens N, Mardulyn P, Smits G. (2017) NOVOPlasty: De novo assembly of organelle genomes from whole genome data. Nucleic Acids Research 45: e18–e18. 10.1093/nar/gkw955 [DOI] [PMC free article] [PubMed]
- Gao F, Chen C, Arab DA, Du Z, He Y, Ho SYW. (2019) EasyCodeML: A visual tool for analysis of selection using CodeML. Ecology and Evolution 9(7): 3891–3898. 10.1002/ece3.5015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gompert Z, Mock KE. (2017) Detection of individual ploidy levels with genotyping-by-sequencing (GBS) analysis. Molecular Ecology Resources 17(6): 1156–1167. 10.1111/1755-0998.12657 [DOI] [PubMed] [Google Scholar]
- Hernández-Ledesma P, Berendsohn WG, Borsch T, Von Mering S, Akhani H, Arias S, Castañeda-Noa I, Eggli U, Eriksson R, Flores-Olvera H. (2015) A taxonomic backbone for the global synthesis of species diversity in the angiosperm order Caryophyllales. Willdenowia 45(3): 281–383. 10.3372/wi.45.45301 [DOI] [Google Scholar]
- Jansen RK, Saski C, Lee S-B, Hansen AK, Daniell H. (2011) Complete Plastid Genome Sequences of Three Rosids (Castanea, Prunus, Theobroma): Evidence for at least two independent transfers of rpl22 to the nucleus. Molecular Biology and Evolution 28(1): 835–847. 10.1093/molbev/msq261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang P, Shi F-X, Li M-R, Liu B, Wen J, Xiao H-X, Li L-F. (2018) Positive selection driving cytoplasmic genome evolution of the medicinally important Ginseng plant genus Panax. Frontiers of Plant Science 9: e359. 10.3389/fpls.2018.00359 [DOI] [PMC free article] [PubMed]
- Katoh K, Standley DM. (2013) MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Molecular Biology and Evolution 30(4): 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong C-S, Um Y-R, Lee J-I, Kim Y-A, Lee J-S, Seo Y-W. (2008) Inhibition effects of extracts and its solvent fractions isolated from Limonium tetragonum on growth of human cancer cells. KSBB Journal 23: 177–182. [Google Scholar]
- Koutroumpa K, Theodoridis S, Warren BH, Jiménez A, Celep F, Doğan M, Romeiras MM, Santos‐Guerra A, Fernández‐Palacios JM, Caujapé‐Castells J, Moura M, de Sequeira MM, Conti E. (2018) An expanded molecular phylogeny of Plumbaginaceae, with emphasis on Limonium (sea lavenders): Taxonomic implications and biogeographic considerations. Ecology and Evolution 8(24): 12397–12424. 10.1002/ece3.4553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubitzki K. (1993) The Families and Genera of Vascular Plants: Flowering Plants: Dicotyledons, Magnoliid, Hamamelid and Caryophyllid Families. Springer, Berlin, Heidelberg & New York, 653 pp. [Google Scholar]
- Kurtz S, Choudhuri JV, Ohlebusch E, Schleiermacher C, Stoye J, Giegerich R. (2001) REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Research 29(22): 4633–4642. 10.1093/nar/29.22.4633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Liu H, Mao S. (2016) Adaptive evolution of ndhF gene in the genus Rheum (Polygonaceae). Guihaia 36: 101–106. [Google Scholar]
- Li J, Zhao C, Zhang M, Yuan F, Chen M. (2019) Exogenous melatonin improves seed germination in Limonium bicolor under salt stress. Plant Signaling & Behavior 14(11): 1659705. 10.1080/15592324.2019.1659705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Xu B, Yang Q, Wang T, Zhu Q, Lin Y, Liu Z-L. (2020) The complete chloroplast genome sequence of Limonium sinense (Plumbaginaceae). Mitochondrial DNA. Part B, Resources 5(1): 556–557. 10.1080/23802359.2019.1710286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lledó MD, Crespo MB, Cameron KM, Fay MF, Chase MW. (1998) Systematics of Plumbaginaceae based upon cladistic analysis of rbcL sequence data. Systematic Botany 23(1): 21–29. 10.2307/2419571 [DOI] [Google Scholar]
- Lledó MD, Karis PO, Crespo MB, Cameron KM, Fay MF, Chase MW. (2001) Phylogenetic position and taxonomic status of the genus Aegialitis and subfamilies Staticoideae and Plumbaginoideae (Plumbaginaceae): Evidence from plastid DNA sequences and morphology. Plant Systematics and Evolution 229(1–2): 107–124. 10.1007/s006060170021 [DOI] [Google Scholar]
- Lledó MD, Crespo MB, Fay MF, Chase MW. (2005) Molecular phylogenetics of Limonium and related genera (Plumbaginaceae): Biogeographical and systematic implications. American Journal of Botany 92(7): 1189–1198. 10.3732/ajb.92.7.1189 [DOI] [PubMed] [Google Scholar]
- Lledó MD, Karis PO, Crespo MB, Fay MF, Chase MW. (2011) Endemism and evolution in Macaronesian and Mediterranean Limonium. In: Bramwell D, Caujapé-Castells J. (Eds) The Biology of Island Floras.Cambridge University Press, Cambridge, 325–337. 10.1017/CBO9780511844270.014 [DOI]
- Logacheva MD, Samigullin TH, Dhingra A, Penin AA. (2008) Comparative chloroplast genomics and phylogenetics of Fagopyrum esculentum ssp. ancestrale – A wild ancestor of cultivated buckwheat. BMC Plant Biology 8(1): e59. 10.1186/1471-2229-8-59 [DOI] [PMC free article] [PubMed]
- Lohse M, Drechsel O, Kahlau S, Bock R. (2013) OrganellarGenomeDRAW – A suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Research 41(W1): W575–W581. 10.1093/nar/gkt289 [DOI] [PMC free article] [PubMed]
- Lowe TM, Chan PP. (2016) tRNAscan-SE On-line: Integrating search and context for analysis of transfer RNA genes. Nucleic Acids Research 44(W1): W54–W57. 10.1093/nar/gkw413 [DOI] [PMC free article] [PubMed]
- Lu C, Feng Z, Yuan F, Han G, Guo J, Chen M, Wang B. (2020) The SNARE protein LbSYP61 participates in salt secretion in Limonium bicolor. Environmental and Experimental Botany 176: e104076. 10.1016/j.envexpbot.2020.104076 [DOI]
- Malekmohammadi M, Akhani H, Borsch T. (2017) Phylogenetic relationships of Limonium (Plumbaginaceae) inferred from multiple chloroplast and nuclear loci. Taxon 66(5): 1128–1146. 10.12705/665.8 [DOI] [Google Scholar]
- Maurya S, Darshetkar AM, Datar MN, Tamhankar S, Li P, Choudhary RK. (2020) Plastome data provide insights into intra and interspecific diversity and ndh gene loss in Capparis (Capparaceae). Phytotaxa 432(2): 206–220. 10.11646/phytotaxa.432.2.10 [DOI] [Google Scholar]
- McKain MR, Johnson MG, Uribe‐Convers S, Eaton D, Yang Y. (2018) Practical considerations for plant phylogenomics. Applications in Plant Sciences 6(3): e1038. 10.1002/aps3.1038 [DOI] [PMC free article] [PubMed]
- Millen RS, Olmstead RG, Adams KL, Palmer JD, Lao NT, Heggie L, Kavanagh TA, Hibberd JM, Gray JC, Morden CW, Calie PJ, Jermiin LS, Wolfe KH. (2001) Many parallel losses of infA from chloroplast DNA during angiosperm evolution with multiple independent transfers to the nucleus. The PLANT Cell 13(3): 645–658. 10.1105/tpc.13.3.645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. (2015) IQ-TREE: A fast and effective stochastic algorithm for estimating Maximum-Likelihood phylogenies. Molecular Biology and Evolution 32(1): 268–274. 10.1093/molbev/msu300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohwi J. (1965) Flora of Japan (rev. ed.). Shibundo Co. Ltd., Tokyo, 344 pp. [Google Scholar]
- Palacios C, Rosselló JA, González-Candelas F. (2000) Study of the evolutionary relationships among Limonium Species (Plumbaginaceae) using nuclear and cytoplasmic molecular markers. Molecular Phylogenetics and Evolution 14(2): 232–249. 10.1006/mpev.1999.0690 [DOI] [PubMed] [Google Scholar]
- POWO (2021) Plants of the World Online. Facilitated by the Royal Botanic Gardens, Kew. http://www.plantsoftheworldonline.org/ [accessed 15 February 2021]
- Price MN, Dehal PS, Arkin AP. (2010) FastTree 2 – Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE 5(3): e9490. 10.1371/journal.pone.0009490 [DOI] [PMC free article] [PubMed]
- Róis AS, Sádio F, Paulo OS, Teixeira G, Paes AP, Espírito-Santo D, Sharbel TF, Caperta AD. (2016) Phylogeography and modes of reproduction in diploid and tetraploid halophytes of Limonium species (Plumbaginaceae): Evidence for a pattern of geographical parthenogenesis. Annals of Botany 117(1): 37–50. 10.1093/aob/mcv138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas J, Ferrer-Mata A, Sánchez-DelBarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE, Sánchez-Gracia A. (2017) DnaSP 6: DNA sequence polymorphism analysis of large data sets. Molecular Biology and Evolution 34(12): 3299–3302. 10.1093/molbev/msx248 [DOI] [PubMed] [Google Scholar]
- Schwartz S, Elnitski L, Li M, Weirauch M, Riemer C, Smit A, Program NCS, Green ED, Hardison RC, Miller W. (2003) MultiPipMaker and supporting tools: Alignments and analysis of multiple genomic DNA sequences. Nucleic Acids Research 31(13): 3518–3524. 10.1093/nar/gkg579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas GWC, Ather SH, Hahn MW. (2017) Gene-Tree Reconciliation with MUL-Trees to resolve polyploidy events. Systematic Biology 66(6): 1007–1018. 10.1093/sysbio/syx044 [DOI] [PubMed] [Google Scholar]
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. (2017) GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Research 45(W1): W6–W11. 10.1093/nar/gkx391 [DOI] [PMC free article] [PubMed]
- Tomoko O. (1995) Synonymous and nonsynonymous substitutions in mammalian genes and the nearly neutral theory. Journal of Molecular Evolution 40(1): 56–63. 10.1007/BF00166595 [DOI] [PubMed] [Google Scholar]
- Walker JF, Yang Y, Feng T, Timoneda A, Mikenas J, Hutchison V, Edwards C, Wang N, Ahluwalia S, Olivieri J, Walker‐Hale N, Majure LC, Puente R, Kadereit G, Lauterbach M, Eggli U, Flores‐Olvera H, Ochoterena H, Brockington SF, Moore MJ, Smith SA. (2018) From cacti to carnivores: Improved phylotranscriptomic sampling and hierarchical homology inference provide further insight into the evolution of Caryophyllales. American Journal of Botany 105(3): 446–462. 10.1002/ajb2.1069 [DOI] [PubMed] [Google Scholar]
- Wu Z, Raven PH. (1996) Flora of China. Science Press (Beijing) & Missouri Botanical Garden Press (St. Louis), 15: 1–387. [Google Scholar]
- Yang Z. (1998) Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Molecular Biology and Evolution 15(5): 568–573. 10.1093/oxfordjournals.molbev.a025957 [DOI] [PubMed] [Google Scholar]
- Yao G, Jin J-J, Li H-T, Yang J-B, Mandala VS, Croley M, Mostow R, Douglas NA, Chase MW, Christenhusz MJM, Soltis DE, Soltis PS, Smith SA, Brockington SF, Moore MJ, Yi T-S, Li D-Z. (2019) Plastid phylogenomic insights into the evolution of Caryophyllales. Molecular Phylogenetics and Evolution 134: 74–86. 10.1016/j.ympev.2018.12.023 [DOI] [PubMed] [Google Scholar]
- Zhang X, Xu Y, Liu X. (2020) Complete plastome sequence of Limonium aureum, a medicinal and ornamental species in China. Mitochondrial DNA. Part B, Resources 5(1): 333–334. 10.1080/23802359.2019.1703608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Q, Yang S, Sun X, Wang L, Li Y. (2019) The complete chloroplast genome of the Jerusalem artichoke (Helianthus tuberosus L.) and an adaptive evolutionary analysis of the ycf2 gene. PeerJ 7: e7596. 10.7717/peerj.7596 [DOI] [PMC free article] [PubMed]
- Zhu A, Guo W, Gupta S, Fan W, Mower JP. (2016) Evolutionary dynamics of the plastid inverted repeat: The effects of expansion, contraction, and loss on substitution rates. The New Phytologist 209(4): 1747–1756. 10.1111/nph.13743 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The output of the repeat analysis of four Limonium plastomes
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Ashwini M. Darshetkar, Satish Maurya, Changyoung Lee, Badamtsetseg Bazarragchaa, Gantuya Batdelger, Agiimaa Janchiv, Eun Ju Jeong, Sangho Choi, Ritesh Kumar Choudhary, Soo-Yong Kim
Data type
molecular analysis
Relative Synonymous Codon Usage
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Ashwini M. Darshetkar, Satish Maurya, Changyoung Lee, Badamtsetseg Bazarragchaa, Gantuya Batdelger, Agiimaa Janchiv, Eun Ju Jeong, Sangho Choi, Ritesh Kumar Choudhary, Soo-Yong Kim
Data type
molecular data
List of Positively selected genes
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Ashwini M. Darshetkar, Satish Maurya, Changyoung Lee, Badamtsetseg Bazarragchaa, Gantuya Batdelger, Agiimaa Janchiv, Eun Ju Jeong, Sangho Choi, Ritesh Kumar Choudhary, Soo-Yong Kim
Data type
molecular data