Abstract
Aims:
The authors sought to examine associations between urinary exosomal miRNAs (exo-miRs), emerging biomarkers of renal health, and cardiorenal outcomes in early childhood.
Materials & methods:
The authors extracted exo-miRs in urine from 88 healthy Mexican children aged 4–6 years. The authors measured associations between 193 exo-miRs and cardiorenal outcomes: systolic/diastolic blood pressure, estimated glomerular filtration rate and urinary sodium and potassium levels. The authors adjusted for age, sex, BMI, socioeconomic status, indoor tobacco smoke exposure and urine specific gravity.
Results:
Multiple exo-miRs were identified meeting a false discovery rate threshold of q < 0.1. Specifically, three exo-miRs had increased expression with urinary sodium, 17 with urinary sodium-to-potassium ratio and one with decreased estimated glomerular filtration rate.
Conclusions:
These results highlight urinary exo-miRs as early-life biomarkers of children's cardiorenal health.
Keywords: : adolescence, blood pressure, childhood, exosome, extracellular vesicle, kidney, potassium, sodium
miRNAs are short, non-coding RNAs of approximately 20 nucleotides that act as post-transcriptional regulators by modifying target gene expression [1]. Urinary miRNAs are promising biomarkers of subclinical kidney damage or dysfunction because they reflect kidney signaling and histological changes at the molecular level, enabling early detection of chronic kidney disease [2] or progression of acute kidney injury [3]. Urinary miRNAs are also readily obtained in urine noninvasively and stable in stored samples.
Exosomes contain the major fraction of miRNA in urine and consequently are an ideal target to probe for molecular biomarkers of kidney diseases [4–6]. Exosomes are lipid-enclosed extracellular vesicles measuring 30–150 nm in diameter that are released by most cells in the body and play an important role in intercellular communication by carrying bioactive molecules (soluble proteins and nucleic acids such as miRNAs) to a target cell [7]. Exosomes in urine are primarily released from renal epithelial cells derived from renal tubular structures [8] and hold promise as one component of a noninvasive ‘liquid biopsy’ for detecting molecular changes in distinct nephron regions even in the absence of disease [9,10]. Thus, the study of miRNA expression in exosomes presents an opportunity in biomarker discovery for blood pressure (BP) regulation and altered renal signaling by identifying new diagnostic biomarkers and therapeutic targets. Research into exosomal miRNAs (exo-miRs) may also offer insights into the complex regulatory networks underlying kidney disease and essential hypertension [9,11].
A large portion of the world's population has or is at risk of hypertension, a chronic disease that is a risk factor for myocardial infarction, cerebrovascular disease and renal failure. Given all these connections, hypertension is a leading cause of morbidity and mortality. Although less common in the pediatric age group, hypertension affects 1–5% of children and adolescents in the US [12], and adult hypertension may have its origins in childhood. In Mexico, where the authors' study takes place, these rates are even higher, with between 6 and 11% of children and adolescents affected by hypertension [13]. Total body sodium content is a key determinant of extracellular fluid volume and thus arterial BP. Renal sodium reabsorption is regulated particularly by the thiazide-sensitive NaCl cotransporter and the epithelial sodium channel (ENaC), both of which are localized to the aldosterone-sensitive distal nephron. Sodium balance is critical to maintaining homeostatic systemic BP. BP dysregulation is linked to monogenic disorders associated with hypotension or hypertension [14].
Unraveling the role of miRNAs in sodium reabsorption and BP regulation is challenging because of the complexity of miRNA regulatory networks, which drive the phenotypic heterogeneity of hypertension. Many miRNAs are expressed with low target fidelity (i.e, a single miRNA targets multiple, even hundreds, of genes) [15,16]. This presents challenges in characterizing the miRNAs involved in BP regulation or hypertension. In addition to this apparent pleiotropy, several miRNAs are expressed in biological clusters or ‘families’. Thus, the identification of a single BP-associated miRNA can be difficult [17,18], requiring interpretations within the context of molecular networks and pathways.
The authors sought to understand the relationship between urinary exo-miR expression and children's BP and estimated glomerular filtration rate (eGFR) as well as urinary sodium and potassium levels as correlates of cardiorenal health. The authors isolated exo-miRs using differential centrifugation of spot urines and examined 193 detectable exo-miRs. The authors then used linear regression to analyze the relationship between the extracted exo-miRs and children's BP, eGFR and urine electrolyte levels. To the authors' knowledge, no previous studies have identified miRNAs associated with BP or urinary biomarkers in healthy children.
Methods
Cohort description
This study included 105 children and was nested in the longitudinal cohort study Programming Research in Obesity, Growth, Environment and Social Stress (PROGRESS) based in Mexico City. Pregnant women at 12–24 weeks of gestation and receiving care through the Mexican Social Security System were enrolled between 2007 and 2011. The women signed a letter of informed consent that included consent for future, unspecified research on chemical exposures. Institutional review boards of the participating institutions (Icahn School of Medicine at Mount Sinai and Instituto Nacional de Salud Pública) approved this study. Inclusion criteria included being 18 years or older, being free of heart or kidney disease, not using steroids or antiepileptic drugs, not consuming alcohol on a daily basis, access to a telephone and planning to reside in Mexico City for the following 3 years. Of the children born to the 948 mothers who delivered, 608 were followed up at age 4–6 years. Of these children, 594 provided urine samples. For this pilot study, 105 children who provided residual spot morning urine samples were randomly selected based on those participants who had cross-sectional measurements of lead levels in blood for kidney biomarker analyses. A letter of informed consent was obtained from the mothers of the children prior to collection of the samples. Urine samples were stored at -80°C and shipped to New York. Of the 105 samples, ten were excluded from the present analysis because of insufficient urine volume required for extracellular vesicle isolation. An additional seven samples were excluded because of a high number of undetectable probes. The authors measured sodium, potassium, specific gravity, creatinine and osmolality in the same spot urine samples. Spot urine measurements of sodium and potassium and their ratio are appropriate estimates of 24 h excretion across different populations and individual samples [19–21]. In summary, the authors analyzed 88 children's urine samples for which all covariate and outcome data were complete. The authors applied two-sample t-tests and chi-square tests to compare the demographics of this subset with the parent cohort.
BP assessment
As previously reported, children's resting BP was measured using an automated, ambulatory, oscillometric BP monitor (90207; Spacelabs Healthcare, WA, USA) [22]. After 10 minutes of rest, BP was measured using the automated Spacelabs Healthcare system with a sized cuff according to arm circumference. Two measurements were taken, with a 3–5 min rest in between measurements, as per the PROGRESS protocol at the 4–6 year study visit. The Spacelabs Healthcare system has advantages over standard sphygmomanometer measurement, including the elimination of observer bias and reduction of white coat hypertension [23,24]. To identify instances of elevated BP, the authors calculated children's diastolic BP (DBP) and systolic BP (SBP) percentiles based on sex, age and height according to the 2017 American Academy of Pediatrics guidelines [25].
Exosome isolation/visualization & miRNA extraction
Urinary exosomes were isolated as previously described [26] with minor modifications. A figure highlighting the steps of this process is shown in Supplementary Figure 1. Samples were defrosted on ice and vortexed vigorously for 90 sec. Each 10 ml aliquot was spun at 300 × g (Sorvall RC-5B; Thermo Fisher Scientific, MA, USA) for 15 min at 4°C. The supernatant was carefully transferred and spun at 17,000 × g for 30 min at 4°C to remove any remaining large cellular debris. Next, the supernatant was transferred to an ultracentrifuge tube and spun at 200,000 × g for 65 min at 4°C using an SW 41 Ti rotor with swinging buckets (Optima LE-80K ultracentrifuge; Beckman, CA, USA) to obtain the low-density membrane pellet. The supernatant was carefully removed without disturbing the pellet. Fresh isolation solution was added to wash the pellet and spun for an additional 65 min. The supernatant was discarded upon completion of the spin. The pellet was resuspended in 50 μl of fresh isolation solution prior to miRNA isolation and 12 μl prior to electron microscopy visualization. MiRNA isolation was performed using an miRNeasy kit with QIAzol reagent and an RNeasy MinElute cleanup kit following the manufacturer's (Qiagen, Hilden, Germany) instructions. A 3 μl aliquot from each sample processed was analyzed using a Bioanalyzer pico chip assay (Agilent, CA, USA) to assess concentration and miRNA purity. At least 0.4 μg of miRNA from 12 μl of pellet was required for subsequent quantitative PCR (qPCR) analysis. Four randomly selected exosome fractions were selected for transmission electron microscopy to confirm exosome presence, assessed by vesicle diameter. Briefly, the material was spread on paraformaldehyde-coated copper grids and negatively stained with 1% uranyl acetate. Images were taken using an HT-7700 electron microscope (Hitachi, Tokyo, Japan) equipped with a digital camera (4 × 4 K; AMT Imaging, MA, USA), and vesicle diameter calculation was performed at the Microscopy Core at the Icahn School of Medicine at Mount Sinai.
qPCR analysis for assessment of miRNA abundance
From the 12 μl urinary exosome isolation, the authors assessed miRNA abundance for 754 miRNAs using TaqMan® OpenArray® qPCR technology (Thermo Fisher Scientific). Briefly, reverse transcription and preamplification of all RNA samples were performed using Megaplex reverse transcription primers (Thermo Fisher Scientific), Human Pool A v2.1 (4399966; Thermo Fisher Scientific) and Human Pool B v3.0 (4444281; Thermo Fisher Scientific) and Megaplex preamp primers (Thermo Fisher Scientific), Human Pool A v2.1 (4399233; Thermo Fisher Scientific) and Human Pool B v3.0 (4444748; Thermo Fisher Scientific) following the manufacturer's protocol. The qPCR was performed using QuantStudio 12 K flex (Thermo Fisher Scientific) and relative quantification cycle (Cq) values were calculated using QuantStudio software (Thermo Fisher Scientific).
qPCR data processing & normalization
To process and normalize the raw qPCR data (Cq values), the Cq values were exported from the Thermo Fisher Scientific Connect platform and the authors applied the following steps. First, the authors excluded failed wells using ROX passive reference signal (Invitrogen, CA, USA), wherein raw ROX levels below 1,000 or above 6,000 were considered technical error. Second, Cq and AmpScore thresholds were set based on replicated internal controls (RNU44, RNU48, U6). Third, nonexpressed targets were defined as those producing no detectable signal across all miRNAs in ≥70% of samples (563 targets were excluded). Fourth, because of the overall low level of expression of many probes in the samples, the authors did not apply normalization using global mean approach. For this, expression data were normalized using the deltaCq method in the NormqPCR R package [27]. The U6 RNA was chosen as an appropriate normalization control because of its stable expression across all samples (coefficient of variation: 0.069) and best unsupervised clustering of technical replicates when using this probe. The final normalized dataset included 88 samples, excluding duplicates, and 193 miRNA probes. These data were then transformed to linear space using 2−ΔCq; therefore, lower values correspond to lower expression and vice versa. Finally, the authors imputed an expression level value for ten individual data points that were missing values because of technical errors based on ROX signal (Invitrogen) values. For this step, the authors imputed the mean level of the specific probe across the other 192 measured values to preserve the maximum number of samples included in the analysis. In each of these cases, the mean value was considered an appropriate imputation of ‘average’ probe expression and to limit subsequent influence on regressions.
Electrolyte & osmolality analysis
Spot urine sodium and potassium concentrations were measured using an IL 943 flame photometer (Instrumentation Laboratory, MA, USA). The instrument was calibrated using urine standard 100/100 or 140/5 standard (Instrumentation Laboratory). Each 20 μl sample was diluted 1:100 in a cesium internal standard and analyzed in duplicate. Urine osmolality, a measure of total dissolved solutes [28], was measured using a Vapro 5520 vapor pressure osmometer (Wescor, UT, USA). The instrument was calibrated using Optimol (Wescor), 290 mmol/l standard. Each 10 μl sample was analyzed in duplicate.
Urinary creatinine & specific gravity
Measurement of the urinary creatinine concentration was performed at the Mount Sinai Clinical Chemistry Laboratory using an Architect c16000 clinical chemistry analyzer combined with Architect system software (Abbott, IL, USA). The quality control procedure was performed using a liquid unassayed Multiqual control combined with Unity real-time software (Bio-Rad Laboratories, CA, USA). Concentration conversions were performed using Architect system software (Abbott). Urine specific gravity was measured using a J157HA+ automatic refractometer (Rudolph Research Analytical, NJ, USA).
Serum cystatin C & eGFR
Measurement of serum cystatin C was performed using Quantikine® human cystatin C immunoassay (R&D Systems, MN, USA). From the serum cystatin C measurements, eGFR values were calculated using the equation eGFR = 70.69 × (cystatin C)−0.931, where cystatin C is in mg/l [29]. As serum collection was not performed when the participants were 4–6 years of age, eGFR reported herein was assessed at the subsequent study visit, when participants were 8–10 years of age.
Single miRNA regression analyses
The authors computed descriptive statistics and Spearman's correlations to characterize the miRNA expression data. The authors then used linear regression to assess associations between miRNAs, as the predictors, and multiple cardiorenal indicators, as the outcomes. The authors transformed the miRNAs to a log2 scale; thus, regression parameters can be interpreted as the change in the outcome per doubling of miRNA concentration. The cardiorenal outcomes considered for this work were SBP, DBP, urinary sodium concentration, urinary potassium concentration and ratio of urinary sodium-to-potassium concentration. The authors' regression analyses were adjusted for age, sex, BMI, socioeconomic status (SES), indoor tobacco smoke exposure and specific gravity. The reference category for child sex was male. Exposure to indoor tobacco smoke was categorized as 1 if any member of the mother's household smoked inside the home during the pregnancy and 0 otherwise. The SES categories were determined using questions related to characteristics of the household and grouped into three categories to represent a ‘low’, ‘medium’ and ‘high’ SES relative to the distribution of the study population [30]. The reference category for SES was ‘high’. The authors used the following thresholds to classify the BMI status of the participants: underweight (-2 <BMI z-score <-1), normal (-1 <BMI z-score <1), overweight (1 <BMI z-score <2) and obese (2 <BMI z-score) [31].
The authors were also interested in how the miRNAs measured at 4–6 years of age were predictive of later eGFR. Therefore, the authors constructed linear models where the outcome was eGFR (assessed at 8–10 years of age), with miRNAs as the predictors. These analyses were adjusted for age at eGFR measurement, age at miRNA measurement, sex, BMI, SES, indoor tobacco smoke exposure and specific gravity.
In supplemental analyses, the authors used multiple regressions to explore how miRNAs were affected by the covariates in the previous models. Therefore, the authors constructed linear models where the outcomes were the miRNA levels and the predictors were age, sex, BMI, SES, indoor tobacco smoke exposure and specific gravity. The authors then reported the miRNAs that were associated with age, sex and BMI (similar results were observed using BMI z-scores).
A difficulty in studying chemical concentrations in urine is the effect of hydration status. Therefore, in this study, the authors also explored how the expression levels of the miRNAs were affected by three urinary dilution measures, including specific gravity, osmolality and urinary creatinine. In these exploratory models, the authors specified the urinary dilution measures as the predictors and the miRNAs as the outcome. All of these models were adjusted for age, sex, BMI, SES and indoor tobacco smoke exposure.
Functional analysis
The authors performed downstream target prediction and functional enrichment analysis using the Ingenuity Pathway Analysis tool (Qiagen, Hilden, Germany). The experimentally measured miRNAs were overlaid onto the Ingenuity Pathway Analysis global molecular network, and putative miRNA–mRNA relationships were identified using the Ingenuity Pathway Analysis microRNA target filter. The mRNA targets were identified based on a knowledge base of highly predicted and experimentally observed relationships. Functional analysis of the target mRNAs included physiological system function enrichment and molecular network mapping. Statistical significance of each was calculated using Fisher's exact test with an alpha set at 0.05.
Results
Characteristics of study participants
The demographic information for the 88 participants in this study is presented in Table 1. Slightly more than half of the population were girls, and the average age was 4.7 years (range: 4.0–6.4 years). Children's BMI ranged from 12.8 to 21.9 kg/m2, with an average of 15.6 kg/m2. Based on BMI z-score, 12.5% were underweight, 64.7% were of normal weight and 14.8% were overweight and 8.0% obese [31]. The percentages of overweight and obese children are lower than those of a representative sample of Mexican preschool children [32]. The urinary specific gravity and concentrations of creatinine and sodium were comparable to a reference population of children of similar ages [33,34]. The urinary potassium concentrations in these children were statistically significantly higher than those seen in a reference population of similar age [33]. Two children in this study met the criteria for elevated BP (≥90th percentile) as defined by the Subcommittee on Screening Management of High Blood Pressure in Children and Adolescents [25]. One child in this cohort had an eGFR below 60 ml/min/1.73 m2 [35]. The eGFR values for the other participants in this study were within the normal physiological range for this age. The demographics of this group did not deviate significantly from the parent cohort (data not shown). SBP was slightly higher in this subset than in the parent cohort, but this difference did not survive a family-wise error correction.
Table 1. . Demographic characteristics of the 88 children participating in this PROGRESS subcohort at 4–6 years of age.
| Demographic | Category | n (%) |
|---|---|---|
| Sex | ||
| Female | 47 (53) | |
| Male | 41 (47) | |
| Indoor tobacco smoke exposure | ||
| Yes | 28 (32) | |
| No | 60 (68) | |
| Socioeconomic status | ||
| Low | 46 (52) | |
| Medium | 35 (40) | |
| High | 7 (8) | |
| Mean ± SD (range), n | ||
| Age, years | 4.7 ± 0.5 (4.0–6.4), 88 | |
| BMI, kg/m2 | 15.6 ± 1.7 (12.8–21.9), 88 | |
| Urine creatinine, mg/dl | 72.0 ± 34.3 (10.4–156.0), 87 | |
| Urine specific gravity, kg/m3 | 1.02 ± 0.01 (1.01–1.03), 88 | |
| Urine osmolality, mmol/kg | 653 ± 201 (272-1100), 86 | |
| Systolic blood pressure, mmHg | 84 ± 7.6 (70–120), 88 | |
| Diastolic blood pressure, mmHg | 52 ± 6.5 (35–76), 88 | |
| Urinary sodium, mmol/l | 130 ± 49 (15–268), 86 | |
| Urinary potassium, mmol/l | 74.9 ± 40.4 (15.5–207.6), 86 | |
| Urinary sodium-to-potassium ratio | 2.29 ± 1.49 (0.09–8.21), 86 | |
| Serum cystatin C, age 8–10 years, mg/l | 0.74 ± 0.19 (0.39–1.59), 68 | |
| eGFRCysC, 8–10 years, ml/min/1.73 m2 | 99.3 ± 24.3 (46.8–170), 68 | |
eGFRCysC: Estimated glomerular filtration rate using the cystatin C-based equation; SD: Standard deviation.
Characteristics of exosomes & exo-miRs
The authors confirmed the presence of exo-miRs via transmission electron microscopy in randomly selected samples. The average diameter of the exosomes was 89 nm, with an overall distribution in exosome diameter ranging from 15 to 329 nm (representative exosome size distributions are shown in Supplementary Figure 2). Detection rates and expression levels for the miRNAs are shown in Supplementary Table 1. There were 29 miRNAs detected in 100% of the samples. The exo-miRs with the highest expression levels were from the miR-30 family, including miR-30c-5p, miR-30a-5p, miR-30b-5p, miR-30a-3p and miR-30e-3p. The Spearman's correlation matrix for the miRNAs is shown in Supplementary Figure 3.
Association between exo-miRs & cardiorenal indicators: urinary electrolyte levels & BP
In adjusted models, three exo-miRs were associated with urinary sodium concentration after false discovery rate (FDR) correction (Table 2): miR-1180-3p, miR-34a-3p and miR-32-5p. A doubling of miR-1180-3p, miR-34a-3p and miR-32-5p was associated with increased urinary sodium concentration of 12.9 (95% CI: 6.2–19.6), 9.8 (95% CI: 3.9–15.7) and 19.8 mmol/l (95% CI: 7.9–31.8), respectively. The authors also identified four exo-miRs (miR-32-5p, miR-9-3p, miR-215-5p and miR-135a-5p) associated with urinary potassium concentration; however, none of these associations survived FDR correction.
Table 2. . Association between exo-miRs and urinary sodium concentration.†.
| Names | β (95% CI) | Uncorrected p-value | FDR-corrected p-value |
|---|---|---|---|
| hsa-miR-1180-3p | 12.9 (6.2–19.6) | 0.0002 | 0.03 |
| hsa-miR-34a-3p | 9.8 (3.9–15.7) | 0.0011 | 0.07 |
| hsa-miR-32-5p | 19.8 (7.9–31.8) | 0.0012 | 0.07 |
Regression coefficients reflect changes in urinary sodium concentration per doubling of exo-miR concentration.
Models were adjusted for age, sex, BMI, indoor tobacco smoke exposure, socioeconomic status and specific gravity.
exo-miRs: Exosomal miRNAs; FDR: False discovery rate.
Seventeen exo-miRs were associated with the ratio of urinary sodium-to-potassium concentrations after FDR correction (Table 3). Of these, the exo-miRs that had the strongest associations ranked by p-value were miR-32-5p, miR-378a-5p, miR-34a-3p and miR-223-3p. A doubling in expression of miR-32-5p, miR-378a-5p, miR-34a-3p and miR-223-3p was associated with an increased urinary sodium-to-potassium ratio of 0.6 (95% CI: 0.3–1.0), 0.5 (95% CI: 0.2–0.8), 0.4 (95% CI: 0.2–0.6) and 0.2 (95% CI: 0.1–0.3), respectively. The three exo-miRs that were associated with urinary sodium concentration – miR-1180-3p, miR-34a-3p and miR-32-5p – were also associated with the urinary sodium-to-potassium ratio before FDR correction.
Table 3. . Association between exo-miRs and urinary sodium-to-potassium ratio.†.
| Names | β (95% CI) | Uncorrected p-value | FDR-corrected p-value |
|---|---|---|---|
| hsa-miR-32-5p | 0.6 (0.3–1.0) | 0.0002 | 0.02 |
| hsa-miR-378a-5p | 0.5 (0.2–0.8) | 0.0003 | 0.02 |
| hsa-miR-34a-3p | 0.4 (0.2–0.6) | 0.0004 | 0.02 |
| hsa-miR-223-3p | 0.2 (0.1–0.3) | 0.0014 | 0.07 |
| hsa-miR-31-3p | 0.2 (0.1–0.3) | 0.0028 | 0.09 |
| hsa-miR-16-5p | 0.2 (0.1–0.4) | 0.0044 | 0.09 |
| hsa-miR-375 | 0.2 (0.1–0.4) | 0.0055 | 0.09 |
| hsa-miR-328-3p | 0.3 (0.1–0.4) | 0.0057 | 0.09 |
| hsa-miR-15b-5p | 0.2 (0.1–0.3) | 0.0057 | 0.09 |
| hsa-miR-128-3p | 0.3 (0.1–0.4) | 0.0061 | 0.09 |
| hsa-miR-195-5p | 0.4 (0.1–0.8) | 0.0062 | 0.09 |
| hsa-miR-15a-5p | 0.3 (0.1–0.5) | 0.0067 | 0.09 |
| hsa-miR-25-3p | 0.2 (0.1–0.4) | 0.0070 | 0.09 |
| hsa-miR-1180-3p | 0.3 (0.1–0.6) | 0.0073 | 0.09 |
| hsa-miR-148a-3p | 0.2 (0.1–0.4) | 0.0078 | 0.09 |
| hsa-miR-340-3p | 0.3 (0.1–0.5) | 0.0079 | 0.09 |
| hsa-miR-424-5p | 0.3 (0.1–0.5) | 0.0083 | 0.09 |
Regression coefficients reflect changes in the ratio per doubling of exo-miR concentration.
Models were adjusted for age, sex, BMI, indoor tobacco smoke exposure, socioeconomic status and specific gravity.
exo-miRs: Exosomal miRNAs; FDR: False discovery rate.
A doubling of miR-150-5p was associated with a 1.6 mmHg (95% CI: 0.2–3.0) higher SBP, whereas a doubling of miR-500a-3p, miR-15a-5p, miR-27a-5p and miR-29c-5p was associated with decreased SBP by 1.1 (95% CI: 0.1–2.1), 0.8 (95% CI: 0.1–1.6), 0.8 (95% CI: 0.0–1.6) and 1.3 mmHg (95% CI: 0.0–2.6), respectively (Supplementary Table 2). Additionally, the authors observed associations between DBP and miR-27a-5p (β [95% CI]: -1.1 [-1.9 to -0.4]) as well as miR-29b-2-5p (β [95% CI]: -1.3 [-;2.2 to -0.3]) (Supplementary Table 3). These associations were not significant after FDR correction.
Association between exo-miRs & eGFR at ages 8–10
Four exo-miRs were associated with reduced eGFR at ages 8–10 (Table 4). Specifically, a doubling in miR-520e, miR-425-3p, miR-645 and miR-140-3p was associated with decreases of 12.1 (95% CI: 5.6–18.6), 6.1 (95% CI: 1.6–10.5), 3.6 (95% CI: 0.6–6.7) and 10.8 ml/min/1.73 m2 (95% CI: 1.0–20.6), respectively. The association between eGFR and miR-520e survived an FDR correction for multiple comparisons. Representative plots highlighting selected associations between exo-miRs and cardiorenal outcomes are shown in Figure 1.
Table 4. . Association between exo-miRs and eGFR assessed at 8–10 years old.†.
| Names | β (95% CI) | Uncorrected p-value | FDR-corrected p-value |
|---|---|---|---|
| hsa-miR-520e | -12.1 (-18.6 to -5.6) | 0.0003 | 0.05 |
| hsa-miR-425-3p | -6.1 (-10.5 to -1.6) | 0.0073 | 0.71 |
| hsa-miR-645 | -3.6 (-6.7 to -0.6) | 0.0202 | 0.94 |
| hsa-miR-140-3p | -10.8 (-20.6 to -1.0) | 0.0303 | 0.94 |
Regression coefficients reflect changes in eGFR from a doubling of exo-miR concentration.
Models were adjusted for age at urine collection and eGFR measurement, sex, BMI, indoor tobacco smoke exposure, socioeconomic status and specific gravity.
exo-miRs: Exosomal miRNAs; FDR: False discovery rate.
Figure 1. . Representative plots of highlighted associations between exo-miRs and cardiorenal outcomes.
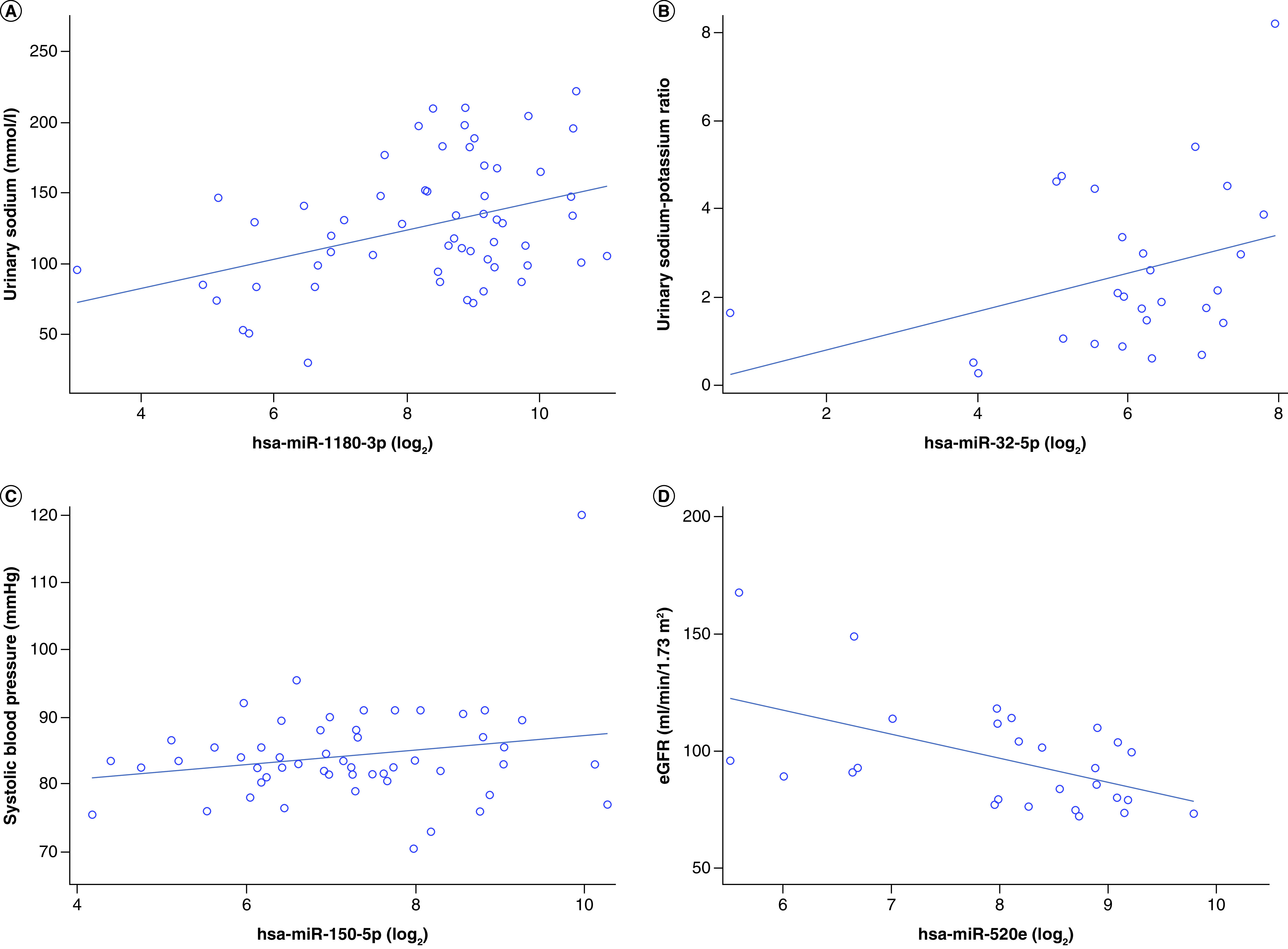
Representative plots of the top (based on lowest p-value) associations between exo-miRs and cardiorenal outcomes, including (A) urinary sodium and miR-1180 (n = 59), (B) urinary sodium-to-potassium ratio and miR-32-5p (n = 27), (C) systolic blood pressure and miR-150-5p (n = 53) and (D) eGFR and miR-520e (n = 27). The plots are unadjusted regression lines.
eGFR: Estimated glomerular filtration rate; exo-miRs: Exosomal miRNAs.
Association between exo-miRs, age, sex & BMI
Thirty-eight exo-miRs were at least marginally associated with age (Supplementary Table 4); however, only miR-184 survived an FDR correction for multiple comparisons. A 1-year increase in age was associated with a 290% (95% CI: 110–600) increase in miR-184. Additionally, the authors identified 25 exo-miRs at least marginally associated with sex (Supplementary Table 5), two of which, miR-223-3p and miR-184, survived an FDR correction. Notably, both miR-223-3p and miR-184 were higher in girls compared with boys. The authors found 87 exo-miRs associated with BMI. Of these, 85 had an FDR-corrected p < 0.1 (Supplementary Table 6), although none had an FDR-corrected p < 0.05.
Association between exo-miRs, specific gravity, osmolality & urinary creatinine concentration
The authors assessed and compared urine specific gravity, osmolality and creatinine to correct for differences in hydration status. The authors found 16, 9 and 22 exo-miRs that showed at least a marginal association with specific gravity, osmolality and urinary creatinine, respectively (p < 0.1) (Supplementary Tables 7–9). Of these, 7, 5 and 11, respectively, had p < 0.05. Of note, miR-99a-3p, miR-664a-3p and miR-101-3p were the exo-miRs that had the strongest associations with with specific gravity, osmolality and urinary creatinine concentration. However, none of these associations survived an FDR correction.
Functional network & pathway analysis of miRNAs & targeted genes
The authors analyzed the three exo-miRs associated with urinary sodium concentration for downstream target prediction and functional enrichment analysis. The authors identified 343 mRNAs that were experimentally observed or highly predicted targets of the three exo-miRs. Notably, miR-32-5p, which had the largest effect estimate of the upregulated exo-miRs, had 207 targets (the authors note that miR-32-5p also has a seed miRNA sequence identical to miR-92a-3p). Additionally, miR-1180-3p and miR-34a-5p were associated with increased urinary sodium concentration and had 68 and 72 mRNA targets, respectively. To understand the kidney-related biological functions of the mRNA targets, the authors performed enrichment analysis on the 343 mRNA targets, including physiological system function enrichment and molecular network mapping. The top nephrotoxic pathways included renal necrosis/cell death, glomerular injury, renal proliferation and renal hyperplasia/hyperproliferation. The top identified regulator network was enriched for diseases and functions, including ‘cell death of kidney cell lines’, ‘mineralization of bone’, ‘development of connective tissue’, ‘hypoplasia of organ’, ‘quantity of IgM’ and ‘thoracic hypoplasia’. Further, targets of miR-92a-3p were identified in both insulin (PTEN, RAP1B, PIK3R3) and aldosterone (DNAJB9, DNAJC30, PIK3R3) receptor signaling pathways.
Discussion
In a sample of 88 Mexican children free of clinically apparent kidney or cardiac disease, the authors identified a series of exo-miRs associated with cross-sectional measures of cardiorenal health, including BP and urine electrolytes. The authors identified multiple exo-miRs associated with electrolyte biomarkers measured in the same urine sample; specifically, 3 exo-miRs showed increased expression with urinary sodium concentration and 17 showed increased expression with urinary sodium-to-potassium ratio. The authors also identified a number of exo-miRs putatively associated with BP, one of which, miR-27a-5p, was associated with lower SBP and DBP. The authors also found that miR-520e was associated with decreased eGFR assessed approximately 4 years later in childhood. The authors' findings, combined with prior studies, are encouraging for future studies of urinary biomarker discovery. However, as with all observational studies, replication in other cohorts and populations will be necessary to assess the reproducibility and generalizability of the authors' results.
The authors' results align with prior work demonstrating that short, non-coding RNAs, specifically miRNAs, are potential biomarkers in cardiovascular, renal and diabetic disease [36–38]. As summarized earlier, excess sodium reabsorption by NaCl cotransporter and ENaC in the aldosterone-sensitive distal nephron is associated with elevated BP and hypertension [39], whereas a high-potassium diet has been reported to lower BP in both human and animal models, especially if there is salt sensitivity [40]. The urinary sodium-to-potassium ratio is an indicator of risk of cardiovascular disease [41] and dietary pattern [19,41]. Furthermore, cross-sectional studies have shown that urinary sodium-to-potassium ratio and SBP are correlated, following similar trends to that of sodium and SBP [19,42–44]. Indeed, the urinary sodium-to-potassium ratio has been suggested to have a stronger impact on BP levels than salt levels alone [44]. The authors' study found that more miRNAs were associated with the ratio of urinary sodium-to-potassium concentrations than either urinary sodium or potassium concentration alone.
There are multiple pathways by which changes in the identified exo-miRs are related to cardiorenal health, including disrupted ion transport and endothelial injury/inflammation. Recent evidence suggests that miRNAs play an integral role as intermediates of kidney hormone signaling – specifically, aldosterone and insulin – which alter ion transport (i.e., ENaC activity) [45–48]. miRNA expression may directly and indirectly regulate ENaC in the lung and kidney [45]. For example, miR-16, which was associated with a higher urinary sodium-to-potassium ratio in the current study, binds to the beta subunit of ENaC, thereby reducing expression in lung epithelial cells [49]. Additionally, among the identified electrolyte-associated miRNAs, the authors' findings are in line with prior work. Exposure to a combination of exo-miRs, including miR-148a-3p, has been found to reduce levels of the renal outer medullary potassium channel [50]. It is thus biologically plausible that this would result in reduced potassium reabsorption. MiR-32-5p, which has the same seed sequence as miR-92a-3p, is involved in the endothelial injury and inflammatory response, acting as a possible diagnostic biomarker for acute myocardial infarction [51], coronary heart disease [52], worse renal injury-associated atherosclerosis [53]. Additionally, increased levels of serum miR-92a-3p are associated with decreased eGFR [54].
Although the authors' study is the first to identify urinary exo-miRs associated with BP in healthy children, the findings align with observations in adults and animal studies. For example, serum levels of miR-29c-5p and miR-27a-5p are associated with essential and pulmonary hypertension in adults [36,37,55]. In a mouse model, miR-27 increased ENaC activity, even in the absence of aldosterone, in cortical collecting duct cells [48]. Another BP-associated exo-miR from the current study, miR-150-5p, has been found in diabetic rat models to be associated with renal fibrosis, atrial fibrillation and coronary artery disease as well as inflammatory response and atrial injury [37,38,56–58]. Additionally, although the authors are the first to identify the association between miR-520e and subsequent decreases in eGFR, a number of exo-miRs in the same ‘family’ as miR-520e are increased in individuals with preeclampsia, the pathophysiology of which is characterized by glomerular dysfunction [59]. There is some overlap between the miRNAs identified in this study and those identified in pediatric populations with clinically relevant preexisting conditions. For example, miR-520 was also found to correlate with pulmonary vascular resistance index, a measure of cardiac stress, in a population of children with pediatric pulmonary arterial hypertension [60]. Previous research has also shown miR-29c to be predictive of fetal congenital heart defects [61].
The authors' study has multiple strengths. The PROGRESS participants are free of clinical cardiovascular or renal disease. Further, the ongoing PROGRESS study has carefully collected covariate data from the participants, enabling the study of whether exo-miRs may serve as preclinical biomarkers of cardiorenal health. The authors' exosome isolation methodology largely followed the recommendations of the International Society for Extracellular Vesicles with only a few differences. The authors used differential centrifugation, considered a ‘gold standard’ in exosome isolation. The International Society for Extracellular Vesicles recommends the use of western blot, high-resolution flow cytometry and electron microscopy to assess exosomes [62]. In this study, the authors employed transmission electron microscopy to help characterize the vesicles. Future studies will also include flow cytometry and quantify the presence of isolated exosomes via specific immunolabeling of protein markers. In this work, the authors used the OpenArray real-time qPCR platform (Thermo Fisher Scientific), which uses real-time PCR for miRNA expression analyses. The platform shows superior accuracy and sensitivity compared with sequencing technologies and does not require validation using another platform [63]. The authors detected and analyzed 193 exo-miRs as potential biomarkers of cardiorenal health. Thus, the authors did not make a priori assumptions about which exo-miRs would serve as more informative biomarkers. To the authors' knowledge, this is the first study to evaluate associations between exo-miRs and BP as well as urinary biomarkers in healthy children. Moreover, the authors assessed and compared three different measures of urinary dilution – urinary creatinine concentration, osmolality and specific gravity – in the analyses. Concentrations of urinary creatinine show intra- and inter-individual variability, specifically by age, sex, BMI, physical activity, diet and time of specimen collection [64,65]. Specific gravity and osmolality are less affected by participant demographics and show greater reproducibility and less systematic variation [66–69]. Although the authors' results did not vary significantly, specific gravity was selected to account for hydration status in this study since it was measured in all participants and analyses identified that it correlated with a moderate number of exo-miRs. The authors were able to measure electrolyte concentrations in the same spot urines as the exo-miRs, enabling a more nuanced exploration of cross-sectional associations between exo-miRs and cardiorenal parameters.
The study also has limitations. The sample size of 88 was limited because of the temporal and financial constraints of exo-miR isolation and quantification. Therefore, the authors' analyses suffered from a lack of power to detect associations that were robust to an FDR correction. However, despite the small sample size, the authors identified 3 exo-miRs that were significantly associated with urinary sodium concentration after FDR correction and 17 exo-miRs that were associated with the urinary sodium-to-potassium ratio after FDR correction. The authors did not collect information about the exact time of urine collection at this visit and so were unable to account for this in the analyses; however, the urine samples were the first or second urine of the day. Subsequent PROGRESS study visits have recorded the time of urine collection. The authors' exosome isolation methods differed from the recommendations of the International Society for Extracellular Vesicles in two ways. First, the authors did not employ a particle filtration step to correct for contamination from particles during centrifugation because it can cause deformation of vesicles as well as the breakup of larger particles, resulting in contaminating fragments passing through. Second, the authors did not employ a Tamm–Horsfall protein disruption step because of evidence that it does not significantly improve exosome recovery [70]. This pilot study assessed electrolyte concentrations in previously collected spot urines rather than 24 h excretion. However, 24 h excretion levels as well as associated urinary dilution measures can be hard to obtain in large population-based studies because of burdensome participant collection and errors associated with under- or over-collection [19,20]. Additionally, spot urines are reliable estimates of 24 h excretion of sodium and potassium for associations with BP [21] and can be used to estimate 24 h sodium and potassium intake [21,71]. Finally, urine contains several elements, including free-floating nucleic acids, exosomes and various cell types [8,72–74]. These cell types may be released from kidney intrinsic cells or cells facing the urinary tract [8,73]; however, the authors' methods did not distinguish the cell source.
Conclusion
In this exploratory study, the authors identified a set of exo-miRs associated with eGFR, cross-sectional BP and urinary electrolyte biomarkers measured in a single spot urine sample, encouraging future studies of urinary biomarker discovery. To the author's knowledge, no previous studies have identified exo-miRs associated with BP, eGFR and urinary biomarkers in healthy children. Additional longitudinal studies expanding on these findings are needed to determine whether these changes lead to clinically apparent kidney or cardiovascular dysfunction as well as the efficacy of exo-miRs as noninvasive biomarkers for predicting long-term cardiorenal health.
Future perspective
A large number of children are at risk of developing cardiorenal problems that can impact health throughout the life span. Assessment of miRNA expression in urine exosomes presents the opportunity to develop novel, noninvasive biomarkers of cardiorenal health. Further study of miRNAs as biomarkers of cardiorenal health will lead to discovery of a subset that are most predictive of clinically apparent kidney or cardiovascular dysfunction in adolescence and adulthood. Different combinations of miRNAs may even inform diagnoses of multiple conditions.
Summary points.
Urinary miRNAs are promising biomarkers for subclinical kidney damage or dysfunction.
In this exploratory study, the authors sought to understand the relationship between urinary miRNA expression and children's blood pressure and estimated glomerular filtration rate as well as urine sodium and potassium levels as correlates of cardiorenal health.
Using linear regressions, the authors measured associations between 193 miRNAs and cardiorenal outcomes, including blood pressure, estimated glomerular filtration rate and urinary sodium and potassium levels.
The authors identified a number of miRNAs putatively associated with blood pressure and estimated glomerular filtration rate. The authors also identified multiple miRNAs associated with electrolyte biomarkers measured in the same urine sample–specifically, urine sodium concentration and urinary sodium-to-potassium ratio.
The authors' findings, combined with prior studies, are encouraging for future studies of urinary biomarker discovery.
Additional longitudinal studies expanding on these findings are needed to determine whether these changes lead to clinically apparent kidney or cardiovascular dysfunction as well as the efficacy of miRNAs as noninvasive biomarkers of long-term cardiorenal health.
Acknowledgments
The authors thank J Ashton and K Schooping at the University of Rochester Genomics Core for their assistance with OpenArray quantitative PCR and D Benson, W Janssen and A Sowa at the Microscopy Core at the Icahn School of Medicine at Mount Sinai for visualization and morphological characterization of extracellular vesicles. The authors also thank the ABC Medical Center in Mexico City, Mexico, for providing the research facilities necessary to complete this work. Finally, the authors thank N McRae for assistance with data curation and G M Hair for preliminary data analyses and drafting of the work.
Footnotes
Author contributions
Data analysis, data interpretation and drafting of the work: Y Levin-Schwartz. Data interpretation and critical revision of the work for important intellectual content: P Curtin. Data acquisition, data interpretation and drafting of the work: D Flores. Data acquisition, data analysis, drafting of the work and critical revision of the work for important intellectual content: V N Aushev. Data acquisition and critical revision of the work for important intellectual content: M Tamayo-Ortiz. Data interpretation and critical revision of the work for important intellectual content: K Svensson. Data acquisition, data interpretation and critical revision of the work for important intellectual content: I Pantic. Data acquisition and critical revision of the work for important intellectual content: G Estrada-Gutierrez. Data acquisition and critical revision of the work for important intellectual content: M L Pizano-Zárate. Data interpretation and critical revision of the work for important intellectual content: C Gennings. Data interpretation, drafting of the work and critical revision of the work for important intellectual content: L M Satlin. Data interpretation and critical revision of the work for important intellectual content: A A Baccarelli. Data interpretation and critical revision of the work for important intellectual content: M M Tellez-Rojo. Data interpretation and critical revision of the work for important intellectual content: R O Wright. Conception of the work, data acquisition, data interpretation, drafting of the work and critical revision of the work for important intellectual content: A P Sanders.
Financial & competing interests disclosure
This work was supported in part by funding from the Mount Sinai Children's Center Foundation and the NIH (T32HD049311, K99ES027508, R00ES027508, P30ES023515, R01ES013744, R01ES020268, P30DK079307, R24ES028522 and R01ES021357). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Ethical conduct of research
The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved. Institutional review boards of the participating institutions (Icahn School of Medicine at Mount Sinai and Instituto Nacional de Salud Pública) approved this study. The women who were eligible for the PROGRESS cohort and agreed to participate signed a letter of informed consent.
Data sharing statement
Access to the data is limited because of a data sharing agreement approved by the institutional review board at Mount Sinai. However, the data from this study are accessible to qualified researchers upon reasonable request pursuant to the following restrictions to ensure the privacy of human subjects. Researchers interested in accessing PROGRESS data must send their resume or curriculum vitae and Collaborative Institutional Training Initiative training certificate to the institutional review board chair, I Wilets (ilene.wilets@mssm.edu). They must also send a data analysis plan to the principal investigators for PROGRESS, RO Wright (robert.wright@mssm.edu), M Tellez-Rojo (mmtellez@insp.mx) and AA Baccarelli (andrea.baccarelli@columbia.edu). When this process is completed and the request is approved, the PROGRESS data analyst, N McRae (nia.x.mcrae@mssm.edu), will send a de-identified dataset via the secure data sharing platform box.
Open access
This work is licensed under the Attribution-NonCommercial-NoDerivatives 4.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
References
Papers of special note have been highlighted as: • of interest; •• of considerable interest
- 1.Catalanotto C, Cogoni C, Zardo G. MicroRNA in control of gene expression: an overview of nuclear functions. Int. J. Mol. Sci. 17(10), 1712 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Papadopoulos T, Belliere J, Bascands JL, Neau E, Klein J, Schanstra JP. miRNAs in urine: a mirror image of kidney disease? Expert Rev. Mol. Diagn. 15(3), 361–374 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Sonoda H, Lee BR, Park KH et al. miRNA profiling of urinary exosomes to assess the progression of acute kidney injury. Sci. Rep. 9(1), 4692 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Demonstrated using a rat renal ischemia-reperfusion injury to model acute kidney injury that the dynamics of exosomal miRNAs (exo-miRs) in urine mirror the progression of acute kidney injury.
- 4.Lyu LL, Feng Y, Liu BC. Urinary biomarkers for chronic kidney disease with a focus on gene transcript. Chin. Med. J. (Engl.) 130(18), 2251–2256 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Merchant ML, Rood IM, Deegens JKJ, Klein JB. Isolation and characterization of urinary extracellular vesicles: implications for biomarker discovery. Nat. Rev. Nephrol. 13(12), 731–749 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]; • Focuses primarily on human studies of exo-miRs and explores their role as mediators of renal pathophysiology.
- 6.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 9(6), 654–659 (2007). [DOI] [PubMed] [Google Scholar]
- 7.Jeppesen DK, Fenix AM, Franklin JL et al. Reassessment of exosome composition. Cell 177(2), 428–445.e18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cavanaugh C, Perazella MA. Urine sediment examination in the diagnosis and management of kidney disease: core curriculum 2019. Am. J. Kidney Dis. 73(2), 258–272 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Harrill AH, Sanders AP. Urinary microRNAs in environmental health: biomarkers of emergent kidney injury and disease. Curr. Environ. Health Rep. 7(2), 101–108 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Provides an overview of the utility of miRNAs and exo-miRs in biomarker discovery for kidney diseases in the field of environmental health.
- 10.Salih M, Fenton RA, Zietse R, Hoorn EJ. Urinary extracellular vesicles as markers to assess kidney sodium transport. Curr. Opin. Nephrol. Hypertens. 25(2), 67–72 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Kriegel AJ, Mladinov D, Liang M. Translational study of microRNAs and its application in kidney disease and hypertension research. Clin. Sci. 122(10), 439–447 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Provides an overview of multiple translational studies of miRNAs detectable in body fluids and highlights their potential value as diagnostic or prognostic biomarkers.
- 12.Moyer VA, US Preventive Services Task Force. Screening for primary hypertension in children and adolescents: U.S. Preventive Services Task Force recommendation statement. Ann. Intern. Med. 159(9), 613–619 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Guerrero-Romero F, Rodriguez-Moran M, Hernandez-Ronquillo G et al. Low serum magnesium levels and its association with high blood pressure in children. J. Pediatr. 168, 93–98.e1 (2016). [DOI] [PubMed] [Google Scholar]
- 14.O'shaughnessy KM, Karet FE. Salt handling and hypertension. J. Clin. Invest. 113(8), 1075–1081 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ho J, Kreidberg JA. MicroRNAs in renal development. Pediatr. Nephrol. 28(2), 219–225 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marques FZ, Booth SA, Charchar FJ. The emerging role of non-coding RNA in essential hypertension and blood pressure regulation. J. Hum. Hypertens. 29(8), 459–467 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Li JY, Yong TY, Michael MZ, Gleadle JM. Review: the role of microRNAs in kidney disease. Nephrology (Carlton) 15(6), 599–608 (2010). [DOI] [PubMed] [Google Scholar]
- 18.Kabekkodu SP, Shukla V, Varghese VK, D’ Souza J, Chakrabarty S, Satyamoorthy K. Clustered miRNAs and their role in biological functions and diseases. Biol. Rev. 93(4), 1955–1986 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Iwahori T, Miura K, Ueshima H. Time to consider use of the sodium-to-potassium ratio for practical sodium reduction and potassium increase. Nutrients 9(7), 700 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iwahori T, Miura K, Ueshima H et al. Estimating 24-h urinary sodium/potassium ratio from casual (‘spot’) urinary sodium/potassium ratio: the INTERSALT study. Int. J. Epidemiol. 46(5), 1564–1572 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mente A, O'donnell MJ, Rangarajan S et al. Association of urinary sodium and potassium excretion with blood pressure. N. Engl. J. Med. 371(7), 601–611 (2014). [DOI] [PubMed] [Google Scholar]
- 22.Sanders AP, Svensson K, Gennings C et al. Prenatal lead exposure modifies the effect of shorter gestation on increased blood pressure in children. Environ. Int. 120, 464–471 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gillman MW, Cook NR. Blood pressure measurement in childhood epidemiological studies. Circulation 92(4), 1049–1057 (1995). [DOI] [PubMed] [Google Scholar]
- 24.Mattu GS, Perry TL Jr, Wright JM. Comparison of the oscillometric blood pressure monitor (BPM-100 (Beta)) with the auscultatory mercury sphygmomanometer. Blood Press. Monit. 6(3), 153–159 (2001). [DOI] [PubMed] [Google Scholar]
- 25.Flynn JT, Kaelber DC, Baker-Smith CM et al. Clinical practice guideline for screening and management of high blood pressure in children and adolescents. Pediatrics 140(3), e20171904 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Pisitkun T, Shen RF, Knepper MA. Identification and proteomic profiling of exosomes in human urine. Proc. Natl Acad. Sci. U. S. A. 101(36), 13368–13373 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perkins JR, Dawes JM, Mcmahon SB, Bennett DL, Orengo C, Kohl M. ReadqPCR and NormqPCR: R packages for the reading, quality checking and normalisation of RT-qPCR quantification cycle (Cq) data. BMC Genomics 13(1), 296 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chadha V, Garg U, Alon US. Measurement of urinary concentration: a critical appraisal of methodologies. Pediatr. Nephrol. 16(4), 374–382 (2001). [DOI] [PubMed] [Google Scholar]
- 29.Ng DK, Schwartz GJ, Schneider MF, Furth SL, Warady BA. Combination of pediatric and adult formulas yield valid glomerular filtration rate estimates in young adults with a history of pediatric chronic kidney disease. Kidney Int. 94(1), 170–177 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Svensson K, Just AC, Fleisch AF et al. Prenatal salivary sex hormone levels and birth-weight-for-gestational age. J. Perinatol. 39(7), 941–948 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.The WHO child growth standards 2007. www.who.int/childgrowth/en/
- 32.Hernandez-Cordero S, Cuevas-Nasu L, Moran-Ruan MC, Mendez-Gomez Humaran I, Avila-Arcos MA, Rivera-Dommarco JA. Overweight and obesity in Mexican children and adolescents during the last 25 years. Nutr. Diabetes 7(3), e247 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kristbjornsdottir OK, Halldorsson TI, Thorsdottir I, Gunnarsdottir I. Association between 24-hour urine sodium and potassium excretion and diet quality in six-year-old children: a cross sectional study. Nutr. J. 11(1), 94 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barr DB, Wilder LC, Caudill SP, Gonzalez AJ, Needham LL, Pirkle JL. Urinary creatinine concentrations in the U.S. population: implications for urinary biologic monitoring measurements. Environ. Health Perspect. 113(2), 192–200 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Levin A, Stevens PE, Bilous RW et al. Kidney Disease: Improving Global Outcomes (KDIGO) CKD work group. KDIGO 2012 clinical practice guideline for the evaluation and management of chronic kidney disease. Kidney Int. Supp. 3(1), 1–150 (2013). [Google Scholar]
- 36.Jusic A, Devaux Y, Action EU-CC. Noncoding RNAs in hypertension. Hypertension 74(3), 477–492 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu G, Jose PA, Zeng C. Noncoding RNAs in the regulatory network of hypertension. Hypertension 72(5), 1047–1059 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xie Y, Jia Y, Cuihua X, Hu F, Xue M, Xue Y. Urinary exosomal microRNA profiling in incipient type 2 diabetic kidney disease. J. Diabet. Res. 2017, 1–10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Coffman TM. The inextricable role of the kidney in hypertension. J. Clin. Invest. 124(6), 2341–2347 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Staruschenko A. Beneficial effects of high potassium. Hypertension 71(6), 1015–1022 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yi SS, Curtis CJ, Angell SY, Anderson CA, Jung M, Kansagra SM. Highlighting the ratio of sodium to potassium in population-level dietary assessments: cross-sectional data from New York City, USA. Public Health Nutr. 17(11), 2484–2488 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stamler J, Elliott P, Chan Q, For The IRG. INTERMAP appendix tables, tables of contents (tables A). J. Hum. Hypertens. 17(9), 665–758 (2003).14504623 [Google Scholar]
- 43.Rose GSJ, Stamler R, Elliott P et al. Intersalt: an international study of electrolyte excretion and blood pressure. Results for 24 hour urinary sodium and potassium excretion. Intersalt Cooperative Research Group. BMJ 297(6644), 319–328 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Valentino G, Hernandez C, Tagle R et al. Urinary sodium-to-potassium ratio and body mass index in relation to high blood pressure in a national health survey in Chile. J. Clin. Hypertens. 22(6), 1041–1049 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ding Y, Zhao R, Zhao X, Matthay MA, Nie HG, Ji HL. ENaCs as both effectors and regulators of MiRNAs in lung epithelial development and regeneration. Cell. Physiol. Biochem. 44(3), 1120–1132 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jacobs ME, Kathpalia PP, Chen Y, Thomas SV, Noonan EJ, Pao AC. SGK1 regulation by miR-466g in cortical collecting duct cells. Am. J. Physiol. Renal Physiol. 310(11), F1251–F1257 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blass G, Klemens CA, Brands MW, Palygin O, Staruschenko A. Postprandial effects on ENaC-mediated sodium absorption. Sci. Rep. 9(1), 4296 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]; • Uses a fasting rat model to demonstrate that epithelial sodium channel (ENaC) transport is affected by insulin levels.
- 48.Liu X, Edinger RS, Klemens CA et al. A microRNA cluster miR-23-24-27 is upregulated by aldosterone in the distal kidney nephron where it alters sodium transport. J. Cell Physiol. 232(6), 1306–1317 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]; • Uses both in vivo and in vitro methods to demonstrate that clusters of miRNAs are upregulated in the kidney by aldosterone and affect ENaC-mediated Na+ transport.
- 49.Tamarapu Parthasarathy P, Galam L, Huynh B et al. MicroRNA 16 modulates epithelial sodium channel in human alveolar epithelial cells. Biochem. Biophys. Res. Commun. 426(2), 203–208 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gracia T, Wang X, Su Y et al. Urinary exosomes contain microRNAs capable of paracrine modulation of tubular transporters in kidney. Sci. Rep. 7(1), 1–11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]; • Exposed cultured renal epithelial cells to urinary exosomes, demonstrating cellular exosomal uptake and reduced levels of potassium channels.
- 51.Dai Y, Yan T, Gao Y. Silence of miR-32-5p promotes endothelial cell viability by targeting KLF2 and serves as a diagnostic biomarker of acute myocardial infarction. Diagn. Pathol. 15(1), 19–19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mari-Alexandre J, Barcelo-Molina M, Sanz-Sanchez J et al. Thickness and an altered miRNA expression in the epicardial adipose tissue is associated with coronary heart disease in sudden death victims. Rev. Esp. Cardiol. 72(1), 30–39 (2019). [DOI] [PubMed] [Google Scholar]
- 53.Wiese CB, Zhong J, Xu ZQ et al. Dual inhibition of endothelial miR-92a-3p and miR-489-3p reduces renal injury-associated atherosclerosis. Atherosclerosis 282, 121–131 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kocyigit I, Taheri S, Sener EF et al. Serum micro-rna profiles in patients with autosomal dominant polycystic kidney disease according to hypertension and renal function. BMC Nephrol. 18(1), 179 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huang Y, Tang S, Huang C et al. Circulating miRNA29 family expression levels in patients with essential hypertension as potential markers for left ventricular hypertrophy. Clin. Exp. Hypertens. 39(2), 119–125 (2017). [DOI] [PubMed] [Google Scholar]
- 56.Scrutinio D, Conserva F, Passantino A, Iacoviello M, Lagioia R, Gesualdo L. Circulating microRNA-150-5p as a novel biomarker for advanced heart failure: a genome-wide prospective study. J. Heart Lung Transplant. 36(6), 616–624 (2017). [DOI] [PubMed] [Google Scholar]
- 57.Goren Y, Meiri E, Hogan C et al. Relation of reduced expression of MiR-150 in platelets to atrial fibrillation in patients with chronic systolic heart failure. Am. J. Cardiol. 113(6), 976–981 (2014). [DOI] [PubMed] [Google Scholar]
- 58.Liu Y, Liu L-Y, Jia Y, Sun Y-Y, Ma F-Z. Role of microRNA-15a-5p in the atherosclerotic inflammatory response and arterial injury improvement of diabetic by targeting FASN. Biosci. Rep. 39(7), 1–14 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 59.Hromadnikova I, Kotlabova K, Ondrackova M et al. Circulating C19MC microRNAs in preeclampsia, gestational hypertension, and fetal growth restriction. Mediators Inflamm. 2013, 1–12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kheyfets VO, Sucharov CC, Truong U et al. Circulating miRNAs in pediatric pulmonary hypertension show promise as biomarkers of vascular function. Oxid. Med. Cell Longev. 2017, 1–11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhu S, Cao L, Zhu J et al. Identification of maternal serum microRNAs as novel non-invasive biomarkers for prenatal detection of fetal congenital heart defects. Clin. Chim. Acta 424, 66–72 (2013). [DOI] [PubMed] [Google Scholar]
- 62.Lötvall J, Hill AF, Hochberg F et al. Minimal experimental requirements for definition of extracellular vesicles and their functions: a position statement from the International Society for Extracellular Vesicles. J. Extracell. Ves. 3(1) 1–6 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mestdagh P, Hartmann N, Baeriswyl L et al. Evaluation of quantitative miRNA expression platforms in the microRNA quality control (miRQC) study. Nat. Methods 11(8), 809–815 (2014). [DOI] [PubMed] [Google Scholar]
- 64.Cocker J, Mason HJ, Warren ND, Cotton RJ. Creatinine adjustment of biological monitoring results. Occup. Med. 61(5), 349–353 (2011). [DOI] [PubMed] [Google Scholar]
- 65.Fortin MC, Carrier G, Bouchard M. Concentrations versus amounts of biomarkers in urine: a comparison of approaches to assess pyrethroid exposure. Environ. Health 7(55), 1–13 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Macpherson S, Arbuckle TE, Fisher M. Adjusting urinary chemical biomarkers for hydration status during pregnancy. J. Expo. Sci. Environ. Epidemiol. 28(5), 481–493 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Suwazono Y, Akesson A, Alfven T, Jarup L, Vahter M. Creatinine versus specific gravity-adjusted urinary cadmium concentrations. Biomarkers 10(2–3), 117–126 (2005). [DOI] [PubMed] [Google Scholar]
- 68.Sauvé J-F, Lévesque M, Huard M et al. Creatinine and specific gravity normalization in biological monitoring of occupational exposures. J. Occup. Environ. Hyg. 12(2), 123–129 (2015). [DOI] [PubMed] [Google Scholar]
- 69.Baron S, Courbebaisse M, Lepicard EM, Friedlander G. Assessment of hydration status in a large population. Br. J. Nutr. 113(1), 147–158 (2014). [DOI] [PubMed] [Google Scholar]
- 70.Cheng L, Sun X, Scicluna BJ, Coleman BM, Hill AF. Characterization and deep sequencing analysis of exosomal and non-exosomal miRNA in human urine. Kidney Int. 86(2), 433–444 (2014). [DOI] [PubMed] [Google Scholar]
- 71.Kawasaki T, Itoh K, Uezono K, Sasaki H. A simple method for estimating 24 h urinary sodium and potassium excretion from second morning voiding urine specimen in adults. Clin. Exp. Pharmacol. Physiol. 20(1), 7–14 (1993). [DOI] [PubMed] [Google Scholar]
- 72.Yasui Y, Tatsumi N, Park K, Koezuka T. Urinary sediment analyzed by flow cytometry. Cytometry 22(1), 75–79 (1995). [DOI] [PubMed] [Google Scholar]
- 73.Ringsrud KM. Cells in the urine sediment. Lab. Med. 32(3), 153–155 (2001). [Google Scholar]
- 74.Nauwelaerts SJD, Van Geel D, Delvoye M et al. Selection of a noninvasive source of human DNA envisaging genotyping assays in epidemiological studies: urine or saliva? J. Biomol. Tech. 31(1), 27–35 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]


