Abstract
Objective:
We systematically review the literature on social epigenetics, examining how empirical research to date has conceptualized and operationalized social determinants of health (SDOH).
Methods:
Using comprehensive search procedures, we identified studies that consider the impact of SDOH on DNA methylation (DNAm), the most common measure of epigenetic change in research on human adult populations. We analyzed the studies to determine: 1) which populations and environments have been investigated in the literature; 2) how SDOH are defined and operationalized; 3) which SDOH have been linked to DNAm; and 4) what lessons from the SDOH literature can be better integrated into future studies exploring the social determinants of health and epigenetic outcomes.
Results:
We identified 67 studies, with 39 to 8,397 participants. The SDOH most commonly considered were early life socioeconomic exposures and early life trauma or mental health. Our review highlights four broad challenges: a) high dependence on convenience sampling, b) limited racial/ethnic, and geographic diversity in sampling frames, c) overreliance on individual sociodemographic characteristics as proxies for broader stratification processes, and d) a focus on downstream social determinants of health and individualized experiences with social stressors.
Conclusions:
Future social epigenetics research should prioritize larger, more diverse and representative population-based samples and employ the SDOH framework to better inform the conceptualization of research questions and interpretation of findings. In particular, the simplified depiction of race/ethnicity, gender, and socioeconomic status as individual-level characteristics should be updated with an explicit acknowledgement that these characteristics are more accurately interpreted as cues used by society to differentiate subpopulations. Social epigenetics research can then more clearly elucidate the biological consequences of these social exposures for patterns of gene expression, subsequent disease etiology, and health inequities.
Keywords: Epigenetics, social determinants of health, DNA methylation, weathering, health inequities
1. Introduction
Human genome mapping has prompted massive growth in research at the intersection of the biological and social sciences, with multiple sub-fields and a rapid pace of technological innovation. Numerous publications (e.g. Bliss 2018; Landecker & Panofsky, 2013; Suzuki & von Vacano, 2018) have alerted social and behavioral scientists to the genomic revolution, urging them to become familiar with key concepts and methods in order to meaningfully contribute to this burgeoning area of study. Many researchers have eagerly answered the charge, turning social genomics and, more recently, social epigenetics into an exciting new frontier for interdisciplinary population health research (Conley & Fletcher, 2016; Mitchell et al., 2016). New biomarkers that track the influence of social and environmental determinants on health across the life course are being identified and used as both outcomes and predictors in a growing literature. Questions remain, however, about whether and how such research incorporates key concepts and methodological insights from the social sciences, and specifically the vast literature on the social determinants of health (SDOH).
We systematically review the empirical literature on social epigenetics, the subfield of genomics which asks whether and how exposures to the physical and social environment influence differential gene expression. We focus on studies linking social exposures to DNA methylation (DNAm), the most pervasive marker of epigenetic alteration in current population-based research. We analyze this literature’s definition and operationalization of the SDOH and offer suggestions for better integrating the SDOH framework into future research.
1.1. Up and Down the Stream of Causation: Genomics, Epigenetics, and Embodiment
Social epidemiologists use a running stream as a metaphor describing the causal influences flowing from distal social factors down to proximate, individual (and frequently behavioral) factors that shape health outcomes (Braveman et al., 2011; Glass & McAtee, 2006). The genomics revolution has uncovered a previously concealed portion of this stream, with social genomics and social epigenetics diverging in their depiction of the biosocial current’s direction. While social genomics explores the way in which fixed biological traits underlie socially-influenced health outcomes, social epigenetics investigates the pathways by which social factors become embodied and manifest in disparate health outcomes.
The starting point for social genomic investigations is downstream, at the level of the specific, immutable genes with which individuals are born. These studies focus on identifying gene specific nuclear polymorphisms (SNPs) and/or polygenic scores, statistically identified gene clusters that explain a portion of the variation in outcomes known to be socially patterned. Recent social genomics research incorporates upstream social, economic, and environmental factors – e.g. education (Barcellos et al., 2018; Domingue et al., 2015; Okbay et al., 2016) and neighborhood contexts (Boardman et al. 2012; Olden et al., 2015) -- via gene-environment interactions, natural experiments, and as outcomes presumed to be at least partially biologically determined. This genomic research aims to capitalize on the fixed nature of the genetic code to explore whether differences in genes and genetic polymorphisms explain physical and behavioral outcomes independently or in concert with contextual factors. To date, however, individual candidate genes and polygenic scores have accounted for a relatively small portion of observed differences in health, education, and similar outcomes (Manolio et al. 2009; Witte, 2010). Furthermore, although polygenic scores are increasingly understood as powerful statistical predictors that reflect the interactions of genes and environment (Conley et al., 2016), they are criticized for inflating the apparent causal association of genotypes with observed phenotypes while masking the mechanisms that generate social disparities (Morris et al., 2020).
The newer field of social epigenetics likewise seeks to add biological depth to our understanding of variation in health and disease, but from a different causal perspective. Taking the fundamental similarity of all human DNA profiles as a point of departure, social epigenetics asks whether and how exposures to the physical and social environment differentiate global and cell-specific gene expression. Epigenetic alterations control transcription of genetic information without changing the static underlying gene sequences (Simmons, 2008). Mechanisms include histone modification, RNA silencing, protein folding, and DNAm — the most common measure of epigenetic change in social science research to date. Methylation involves the binding of methyl groups to the cytosine nucleotide located next to a guanine nucleotide, linked by a phosphate (or CpG) site. The addition of methyl groups to the DNA regulate which genes are expressed and how, allowing cells, organs and tissues in the body to develop and differentiate over the life course from early embryogenesis through adult aging (Champagne & Curley, 2011). Epigenetics also influences inheritance of genomic changes across generations through a process called imprinting (Bartolomei, 2009). Epigenetic mechanisms are thus both a critical component of normal development and potential contributors to processes of disease and dysregulation.
While the majority of methylation marks remain stable, approximately 10–20% of CpGs do change methylation status within a 6–10 year period (Chen et al., 2016). These changes are thought to drive abnormal activation or silencing of genes which can have adverse effects on biological processes shaping health and longevity. Epigenetic changes are increasingly considered a mechanism contributing to differential aging and life expectancy (Horvath, 2013), and may be one mechanism by which deleterious social and economic exposures alter immune function, increase systemic inflammation, and influence other markers of complex chronic diseases with well-documented health disparities (Ferrucci et al., 2020; Vidrascu et al., 2019; Xia et al., 2014).
Social epigenetic research thus holds the promise of explicating a long-theorized process of embodiment, the idea that humans – as biological and social beings – biologically incorporate their social and physical environment (Krieger, 2005). The weathering hypothesis (Geronimus, 2006) was among the first and most influential attempts to explain health inequities as the biological consequences of economic hardship, discrimination, and social marginalization. Allostatic load (McEwen, 1998), telomere length (Brown et al., 2017), inflammation (McDade et al., 2006), and altered stress response have all been considered as biological indicators of weathering – measures that could demonstrate how the stresses associated with various deleterious experiences becomes embodied. Nonetheless, they have proven limited in explaining variation in health and mortality across populations (Kennedy et al., 2014). Social epigenetics represents the current frontier in bridging social science and biology, aiming to more clearly understand the pathways linking the physical, built, and social environments with differential gene expression and health disparities (Champagne & Curley, 2011; Cunliffe et al., 2016; Tung et al., 2013; Thayer & Non, 2015).
1.2. Defining Social Determinants of Health
Epigenetics’ “upstream” perspective resonates with social scientists and public health researchers who have long called attention to the contextual factors that “get under the skin” to shape population health (Syme, 1987). The World Health Organization’s Global Commission on Social Determinants of Health (CSDH, 2008) broadly defined social determinants of health (SDOH) as “the structural determinants and conditions of daily life” which are “responsible for a major part of health inequities between and within countries.” The SDOH perspective requires active recognition of “the unequal distribution of power, income, goods, and services, globally and nationally, the consequent unfairness in the immediate, visible circumstances of people’s lives – their access to health care and education, their conditions of work and leisure, their homes, communities, towns, or cities – and their chances of leading a flourishing life.”
Within the CSDH framework (Solar & Irwin, 2007), a distinction is made between “context,” “structural stratifiers,” and “intermediary determinants of health.” Context represents social, economic, and political mechanisms that generate, configure, and maintain social hierarchies (e.g., labor market, educational system). Structural stratifiers (e.g., racism, classism, heterosexism) generate social and economic divisions that shape hierarchies of power and individual socioeconomic position. In research, these stratification processes are often proxied via individual-level measures of gender, race, ethnicity, and class (among others). Context and structural stratifiers operate through intermediary material channels (e.g. housing, neighborhood infrastructure) and psychosocial pathways (e.g. experiences of adversity and discrimination, availability of social supports) to influence “downstream” behavioral factors. Taken together, context, structural stratifiers, and intermediary material and psychosocial channels are the fundamental, “upstream” social causes that underlie multiple health outcomes via multiple pathways (Braveman et al., 2011; Link & Phelan, 1995).
We also draw upon the contributions of critical race and gender scholarship (Bowleg, 2019; Nuru-Jeter et al., 2018) that has highlighted the dangers associated with relying upon sociodemographic indicators, such as race, as explanatory variables without regard for the underlying social processes (e.g., institutional discrimination) that assign privileges and disadvantages based upon social classifications. In the context of biomedical and genetics research, this atheoretical approach often leads to health inequities being interpreted as essentially rooted in individual bodies or genes, rather than in the social and economic structures that act on those bodies. We embrace the notion of upstream SDOH by examining context, structural stratifiers that are conceptually defined as social constructs, and the accompanying psychosocial factors.
Though the complex and multifaceted influence of the SDOH renders their operationalization challenging, the perspective is increasingly accepted in public health research and policy circles (Kneipp et al., 2018; RWJF Commission, 2019). A growing literature documents how pervasive social problems like racism (Williams & Mohammed, 2013), poverty (Phelan et al., 2010), and discrimination based on gender (O’Neil et al., 2018), sexual identity and orientation (Baptiste-Roberts et al., 2017), nativity (Viruell-Fuentes et al., 2012), disability status (Krahn et al., 2015), and intersections therein are linked with environmental justice and health inequities. The SDOH have been found to have greater explanatory power for observed health inequalities relative to down-stream factors. For example, a meta-analysis concluded that the number of U.S. deaths in 2000 attributable to low education, racial segregation, and low social support was comparable with the number attributable to myocardial infarction, cerebrovascular disease, and lung cancer, respectively (Galea et al., 2011). SDOH such as economic inequality also explain considerably more variation in health than individual candidate genes or polygenic scores (Bezruchka, 2012; Manolio et al., 2009; Witte, 2010). These variations suggest that SDOH have important biological consequences, with epigenetics potentially playing a role in linking individual and contextual factors with health outcomes across the life course, importantly including young through middle-adulthood (Geronimus, 2013).
We focus our systematic review on studies that have investigated the role of upstream social factors on DNAm, the most common epigenetic outcome in social science, psychology and population health research to date. We ask how empirical research on DNAm has incorporated the SDOH, consider the populations and environments that have been included in studies, and assess the ways that the SDOH have been conceptualized and operationalized in the literature. While several prior reviews have called attention to the utility of epigenetics in understanding health disparities, this is, to our knowledge, the first review to consider and evaluate the application of a SDOH framework in epigenetic research.
2. Methods and Materials
We used comprehensive search procedures to identify empirical studies examining the impact of SDOH on DNAm outcomes in adults. We analyzed the studies to determine: 1) Which populations and environments are represented in the literature; and 2) how epigenetic researchers define and operationalize SDOH across and within populations. We end by evaluating the integration of the SDOH framework into social epigenetic research and suggesting directions for future research.
2.1. Data Sources and Inclusion criteria
We defined social determinants of health to align with the upstream conceptualization and limited the scope of review to studies that assess the effects of SDOH on DNAm in adults (see figure 1). Note that while methylation was measured among adults, the SDOH indicator may have been identified at any point in the life-course. We conducted a systematic search of PubMed (1950-) and Scopus (1995-) from inception to March 2020. Searches used the following title, abstract, keyword and Medical Subject Headings (MeSH) terms: (1) social determinants of health, sociological factors, and population health and (2) DNAm, epigenetic, epigenesis, or epigenetic clock. We excluded non-English publications and papers that dealt with animal models only. Conference papers, abstracts, letters, notes, and short surveys were also excluded from the review. Systematic reviews and editorials that met our inclusion criteria of containing a focus on upstream SDOH and DNAm in adulthood were screened for relevant information to our discussion, but were not included in the final count of empirical publications covered in this systematic review.
Figure 1.
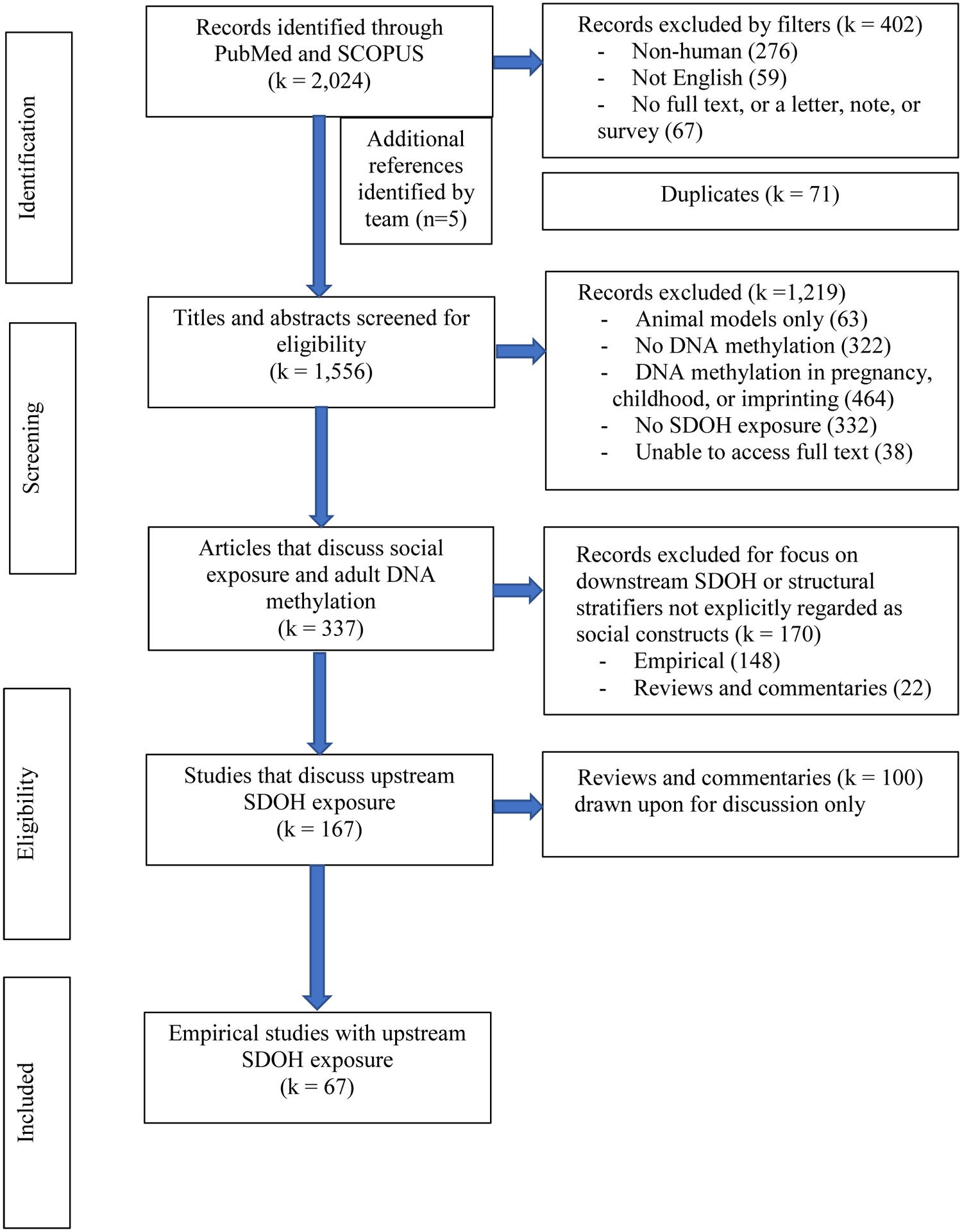
Systematic review flow chart
After combining search results in Endnote X8 and removing duplicates, a total of 1,556 articles were screened for inclusion by sequentially applying the criteria listed in figure 1. An article was tagged for exclusion based upon the first criteria that was not met. To test the review criteria and standardization of the review process across the three-member review team, each reviewer independently screened a subset (approximately 20%) of the publications to assess eligibility. Inter-reviewer disagreement was discussed and resolved by consensus or via guidance from a senior author. Following this initial step, the remainder of the studies were split between the three-member review team and screened against the inclusion criteria. Subsequent checks were periodically made by one of the principal investigators throughout this second stage of the review process. After full-text review, 67 empirical articles met the inclusion criteria. See figure 2 for full details on the selection process.
Figure 2.

Review inclusion criteria
2.2. Data Abstraction and Analysis
For each of the 67 studies, we extracted information on study design, sample size, age-range, racial/ethnic composition, SDOH exposure, study methods (e.g. epigenome-wide association studies (EWAS) vs. targeted CpG analyses), DNAm and other health outcomes assessed. Data were entered into a standardized extraction template and summarized. A formal meta-analysis was not conducted due to the heterogeneity of the studies with regard to exposure type, as well as the measurement of DNAm, and research design. Standard procedures to assess study quality were likewise not applicable.
We analyzed the studies to determine which populations and environments are represented in the literature and how they inform the investigation of upstream stratification processes and contexts. Next, we differentiated between articles that employed an explicit social frame and those that used individual-level demographic and social variables for classification of respondents. We excluded studies that listed race or ancestry as a covariate without noting what exposure it was meant to represent, leaving many readers to interpret it as a biological trait. While ancestry is indeed associated with the presence or absence of specific genes or polymorphisms, it does not clearly map on to our contemporary socially-constructed discrete enthnoracial categories (Fujimura et al., 2014; Zuberi et al., 2015). Consequently, we considered articles to employ a social frame only if they discussed race/ethnicity in the context of lived experiences. Finally, focusing on the articles written using a social frame, we examined how the SDOH were defined and operationalized in the analysis, and how they were engaged in the discussion. Below, we describe our findings and discuss the incomplete integration of the SDOH perspective into the existing social epigenetic literature.
3. Results
3.1. Study characteristics
Table 1 presents aggregate statistics on the 67 included empirical studies, published between 2000 and 2020 (see appendix A for full details on study design). Only one study examining methylation of specific loci associated with PTSD was published prior to 2010. The pace of publication accelerated after 2012 with 8–10 per year published between 2017 and 2019. A majority of studies were conducted in North America (61% n=41), predominantly within the U.S. (n=38), followed by the U.K. (n=9), other countries in Western Europe (n=7), Asia (n=4), Central and South America (n=2), and the Middle East (n=1) and Africa (n=1). Three additional studies were multinational. Half of the selected studies were drawn from sample sizes of 300 or less; the other half was partitioned between sample sizes of 301–600 (n=13), 601–1000 (n=7), and over 1000 (n=12).
Table 1.
Characteristics of empirical studies on social determinants of DNA methylation
| k | % | ||
|---|---|---|---|
| Initial Study DesignA | Case-control | 5 | 7% |
| Cohort | 60 | 90% | |
| Mixed design in multi-national study | 2 | 3% | |
| Original survey design - longitudinal | 43 | 64% | |
| Epigenetic subsample examined longitudinally | 2 | 5% | |
| Initial Sampling FrameA | Convenience | 27 | 40% |
| Probability-based | 37 | 55% | |
| Representativeness maintained in epigenetic subsample | 14 | 38% | |
| Mixed sampling frames in multi-national studies | 3 | 4% | |
| Region of Study | North America | 41 | 61% |
| United Kingdom | 9 | 13% | |
| Other Western European nations | 6 | 9% | |
| Asia | 4 | 6% | |
| Central and South America | 2 | 3% | |
| Middle East | 1 | 1% | |
| Africa | 1 | 1% | |
| Multi-national | 3 | 4% | |
| Sample Size | <100 | 15 | 22% |
| 100–300 | 20 | 30% | |
| 301–600 | 13 | 19% | |
| 601–1000 | 7 | 10% | |
| >1000 | 12 | 18% | |
| Ages includedB | 18 – 34 years | 26 | 39% |
| 35 – 60 years | 45 | 67% | |
| 61+ years | 15 | 22% | |
| Sex Composition | Female only | 13 | 19% |
| Both Sexes | 48 | 72% | |
| Racial/Ethnic Composition | North American Samples (k=41): | ||
| White only | 3 | 7% | |
| Black only | 12 | 29% | |
| Latinx only | 1 | 2% | |
| Diverse | 21 | 51% | |
| Unspecified | 4 | 10% | |
| European Samples (k=15): | |||
| White only | 4 | 20% | |
| Unspecified | 11 | 67% | |
| Other regions and multinational samples (k= 11): | |||
| Mono-ethnic | 2 | 18% | |
| Diversity across multinational sample | 2 | 18% | |
| Unspecified | 7 | 64% |
Initial design and sampling frame refers to the larger study from which the epigenetic sample was drawn. See sub-bullets for specifics on epigenetic subsamples drawn from larger cohort designs.
Categories are not mutually exclusive, and therefore will not sum to 100%.
Ninety percent of the epigenetic study designs were ancillary studies using random or purposely selected sub-populations from existing cohorts (n=60), 7% were case-control designs, and 3% (n=2) were multinational studies comprised of a mix of cohort and case-control study designs. A majority of the ancillary studies were drawn from longitudinal cohorts. Only two of the included cohort studies utilized repeated measures of DNAm to examine changes over time (Lawn et al., 2018; Sipahi et al. 2014).
Forty percent of the epigenetic studies were conducted from convenience samples (n=27). Recruitment sites for convenience samples included blood drives and community clinics (Barcelona et al., 2018; Campesi et al., 2013; Janusek et al., 2017; Santos et al., 2018), workplaces (Adams et al., 2017; Algeria-Torres et al., 2013; Goodrich et al., 2013), and ads in local media (Austin et al., 2018). In one case, a respondent-driven sampling design was employed to approximate random sampling (Kogan et al., 2018). Although many of the cohort studies were initially constructed as probability-based samples, very few of the ancillary studies (n=14) explicitly stated, in text, an effort to maintain the complex sample structure when collecting and analyzing DNA for methylation (e.g., Beach et al., 2014, 2016; Brody et al., 2016; Chen et al., 2016; Chi et al., 2016; Lei et al., 2015; Needham et al., 2015; Simons et al., 2016, 2017). Instead, epigenetic subsamples that piggybacked on population-based surveys or epidemiologic cohorts frequently relied upon voluntary response sampling (e.g., Bustamante et al., 2016, 2018; Huang et al., 2016) and employed eligibility criteria that may have compromised the original sampling frame and representativeness. For example, participation in all waves of data (e.g., Hughes et al., 2018), non-immigrant status, and white ethnicity (Borghol et al., 2012; Hughes et al., 2018; Suderman et al., 2014) are a few examples of instituted selection criteria.
3.2. Study participants
We report the age-range of study participants based upon the age at the time of DNAm measurement. Most studies were conducted with middle-aged adults (67%), age 35 to 60 years. Thirty-nine percent of the studies (n=26) included young adults under age 35 and 22% (n=15) included older adults, age 61 years or older. Only 7% (n=5) of studies included participants that spanned across young, middle, and older adult ages. Roughly three quarters of the selected studies (n=48) included men and women. Among the studies that included a single sex, a larger share were derived from pregnancy-birth cohorts and therefore focused on females only (n=19).
In reviewing the racial/ethnic composition of studies, we defined a study as predominantly monoracial if the study sample was comprised of 85% or greater of a single race. Among the 41 studies conducted in North America, 39% were monoracial – Black/African-American only (29% n=12), White only (7% n=3), or Latinx only (2% n=1). Another 21 of the North American studies had some diversity in the sample and the remainder did not explicitly identify the racial and ethnic composition of the sample.
Among the European studies, 20% (3 from the UK and 1 from Germany) specified monoracial White, non-immigrant samples. The remaining 73% of European studies did not discuss the racial and ethnic composition of their samples. Based upon information published elsewhere on the larger cohorts from which the epigenetic samples were drawn (Boyd et al., 2012; Fraser et al., 2012; Milne et al., 2017; Wadsworth et al., 2006), we infer that these samples are predominantly white. Two of the 8 studies conducted outside of North America and Europe explicitly identified the ethnic/ethnoreligious composition of their samples – Tutsi ethnicity (Perroud et al., 2014) and Jewish ancestry (Yehuda et al., 2016). One of the 3 multinational studies had diversity present across the U.S., Latin American, and Central African subsamples (Horvath et al., 2016).
3.3. Measures of DNA methylation
Thirty three percent of studies measured CpG-specific methylation on or in promotor regions of targeted genes only (n=19), 7% examined global methylation only (n=4), and another 7% combined global and CpG-specific methylation measures (n=4). The other half of studies (n=29) employed epigenome-wide methylation measures, using an exploratory approach to identify specific changes in DNAm. Roughly half of these EWAS-analyses led to investigating candidate genes or regions specific to a particular health condition of interest. Thirteen studies quantified methylation patterns across the genome to produce a composite measure of accelerated aging, 31% based on the Horvath clock (n=4), 15% on the Hannum clock (n=2), or a combination of the two (54% n=7).
Recent advances find methylation across the genome can vary by cell composition and tissue site. A majority of studies (81% n=54) relied solely on peripheral blood as the source of DNA collection with 21 specifying use of peripheral blood mononuclear cells and another 14 specifying use of blood leucocytes. Among these studies, 30 used venipuncture blood draw, 3 combined venipuncture and blood spots, and the remaining did not provide enough detail to discern blood collection approach. Seven studies used a buccal cheek swab or spit saliva. Two studies derived DNA from tumor tissue and another 4 studies combined blood with saliva, brain, or adipose tissue. The most common approach to quantifying DNAm was to use microarray-based methods (n=32), followed by pyrosequencing (n=18).
Of the 67 studies reviewed, 43% (n=29) tested DNAm as a potential mediator to one or more health outcomes. A total of 12 health domains were examined across these 29 studies, with mood disorders and internalizing behaviors (n=9) and inflammatory markers or cortisol response (n=7) being the most commonly studied. Other health outcomes included cardiometabolic risks, sleep phenotypes, cancers, stress, having a low birthweight baby, externalizing behaviors and substance-use, Parkinson’s disease, and self-rated health.
3.4. Defining and measuring SDOH
Table 2 categorizes studies based upon the primary SDOH investigated and provides information on the indicators used to define the SDOH. We excluded 148 empirical papers for investigating a.) downstream social determinants of health only (e.g., health behaviors such as diet, physical activity) or b.) racial/ethnic and gender differences without explicit discussion on the social meaning of these variables. One hundred and sixteen of the 148 excluded empirical papers included a measure of race/ethnicity or gender, yet treated these factors as biological traits, essentialized variables, or allowed for ambiguity in interpretation by not defining these variables or elaborating upon in the discussion. As such, only 7 studies were included in this review for examining race/ethnicity, gender, or geography (Barfield et al., 2019; Campesi et al., 2013; Guerrero-Preston et al., 2019; Horvath et al., 2016; Subramanyam et al., 2013; Tajuddin et al., 2019; Toyooka et al., 2003) as a primary SDOH while also employing a social frame. Examples of qualifying language for inclusion can be found in Subramanyam et al. (2013) who described race/ethnicity as “a social factor because of evidence showing strong patterning of various social exposures by race/ethnicity in the United States” or Campesi et al. (2013) who stated that the term sex-gender reflects “both physiological distinctions between sex and environmental and social influences, such as social status.”
Table 2.
Social determinants and health outcomes examined in epigenetic studies on DNA methylation
| Social Determinant of HealthA | Operationalization of the SDOHB |
|---|---|
| Individual-level stratifiers (k=28): | |
| Childhood SES only (5) | Family income or poverty status (4); TANF (3); financial strain (3); parental education (3); parental occupation (2); parental unemployment (2); single parent household (1). |
| Adulthood SES only (6) | Family income (3); financial strain (2); education (3); occupation (1); housing tenure (1); and area-level deprivation (3). |
| SES across the lifecourse (11) | Childhood: occupation (5); parental education (3); family income (1); single-parent household (1). Adulthood: education (6), occupation (4), family income (4); wealth (1); employment status (1); financial strain (1). Trajectories: occup.-->occup. (3), educ./assets-->educ. (2). |
| Sociodemographics as stratification proxy (7) | Race/ethnicity (5); sex/gender (5); geography (1). |
| Traumas (k=26): | |
| Unspecified (2) | As assessed in PTSD screening (2). |
| Genocide (2) | Holocaust survivor or offspring of survivor (1); Rwanda genocide survivor (1). |
| Discrimination (3) | Perceived racism (2); perceived discrimination (2); Moderator: social support (1). |
| Early life adversity: ACEs/SES (19) | Physical, sexual, emotional abuse, or maltreatment (15); parental illness (6); parental separation (4); and included childhood SES in measure of adversity (5). Moderators: parenting repetoire (3); social support (1). |
| Workplace exposures (k=6): | |
| Physical environment/toxicants (3) | PAHs in brickmaking (1); carbon nanotubes in engineering (1); mercury in dentistry (1). |
| Social environment (3) | Shiftwork (2); job strain (1). |
| Neighborhood exposures (k=15): | |
| Physical toxicants (n=6) | Air pollution (3); microbial environment (1); proximity to heavy metal mining (1) or PCB manufacturing (1). |
| Social and economic environment (9) | Neighborhood deprivation (5); community crime (2) or violence (1); worse social environment [aesthetics, safety, and cohesion] (1). |
the number of papers that included that SDOH domain as a primary focus in the epigenetic analysis.
the number of times the SDOH was captured via that particular measure.
Within our framework, socioeconomic status (SES) then became the most commonly studied SDOH (33% n=22 as the primary focus). Five papers focused on low-SES exposure in childhood, 6 in adulthood, and another 11 investigated the combined influence of childhood and adulthood SES or trajectories (e.g., upwardly/downwardly mobile, stable). Measures of SES were most commonly constructed at the individual-level, based upon indicators of the parental or respondent’s own income, education and occupation. In fewer cases, assessments of household income, wealth, poverty status, TANF receipt, home ownership, self-reported unmet needs, and family composition (e.g. single-parent household) were included in composite measures of SES.
Early-life adversity was the next most commonly investigated SDOH in epigenetic studies (28% n=19), with adverse childhood experiences (ACEs) centered in analyses. ACE scores tallied experiences with violence, abuse, household presence of substance use or mental health problems, and in a few instances, parental separation or death. In 5 of these studies, abuse-related trauma was investigated in conjunction with childhood SES, assessed by researchers interested in the collective and long-term effect of material and psychosocial adversity in childhood. Another 2 studies investigated trauma exposure via standard PTSD assessment protocols, unconstrained to a specific lifestage (Sipahi et al., 2014; Uddin et al., 2010). Other studies explored the trauma of discrimination, measured at societal and interpersonal levels. For example, one study examined state-sponsored persecution inflicted upon Jewish Holocaust survivors and intergenerational trauma carried forward to adult offspring (Yehuda et al., 2016) and another investigated direct exposure to the Rwandan genocide among women of Tutsi ethnicity (Perroud et al., 2014). Three other studies examined reports of interpersonal experiences of racism and discrimination among African-American women (Barcelona et al., 2018), Latina women during the perinatal period (Santos et al. 2018), and African-American young adults (Brody, 2016a).
A third of included studies focused on place-based social exposures external to the household setting (n=21). While the examination of physical toxins is prevalent in epigenetic studies, we included only 9 in our review based upon a discussion of policies, institutions, or places as sources of differential distributions in risk exposures. Workplace physical exposures included toxins present in brickmaking (Algeria-Torres et al., 2013), carbon nanotubes at a production facility (Ghosh et al., 2017), and mercury in the dental occupation (Goodrich et al. 2013). Neighborhood physical toxins included air pollution (n=3) (Chi et al., 2012; Gondalia et al., 2019; Peluso et al., 2012), and living in a mining community (Castillo et al., 2017) or proximally close to a PCB manufacturing plant (Pittman et al., 2020). Another 12 place-based studies focused on psychosocial exposures, such as shiftwork or job strain in the workplace (Adams et al., 2017; Bhatti et al., 2015) or social and economic features of a neighborhood, including neighborhood-level deprivation (n=5), community crime and violence (n=3), aesthetics and cohesion (n=1) (Guerrero-Preston et al., 2019; Janusek et al., 2017; Hsu et al., 2019; Lei et al., 2015; McCartney et al., 2017; McClelland et al., 2016; McGuiness et al., 2012; Simons et al., 2017; Smith et al., 2017).
4. Discussion
Social epigenetics has incorporated lessons from the SDOH literature by extending inquiry into the role of economic deprivation, childhood adversities, discrimination, and place-based stressors on DNAm in adulthood. However, our review highlights gaps that constrain knowledge production on how human bodies biologically respond to their social environments. We identified four broad challenges: a.) limited racial/ethnic, and geographic diversity in sampling frames, b.) high dependence on convenience sampling, c.) overreliance on sociodemographic characteristics as proxies for stratification processes, and d.) a focus on downstream SDOH and individualized experiences with social stressors. Each of these points are discussed in further detail below.
4.1. Representativeness
Echoing patterns observed in genomics research (Popejoy & Fullerton, 2016), a majority of the epigenetic studies reviewed were conducted in the Global North, among monoracial populations. Over 80% of reviewed studies originated from North America and Western Europe. Within North American studies, 40% focused on a singular race group, often African Americans. In European studies, a majority were conducted with predominantly white samples – a determination we made from sample descriptions, or in unspecified circumstances, information extracted from external documentation. The lack of racial/ethnic diversity in genomics studies stems from the field’s pattern of sampling from established cohorts and its conception of race as solely a biological marker of ancestry rather than as a system of social classification with social, economic, and biological consequences for groups (Bentley et al., 2017; Matthew, 2019). We find similar practices in epigenetics research.
Particularly in the European context, the lack of diversity is partly dictated by cohort sampling frames established in the mid 1990s, when less attention was directed toward the role of racism and xenophobia in class disparities. Also, a significant proportion (36% n=24) of studies neglected to specify the racial/ethnic composition of their sample, requiring readers to search elsewhere for this information or assume that countries are a proxy for their dominant racial/ethnic group. These practices make it impossible to know if observed findings hold across groups or global regions absent from epigenetic samples. The omission matters not because race is presumed to have biological relevance, but rather because the social conditions in which people live differ by race/ethnicity and geography due to long-standing power structures. These social factors differentiate exposure to SDOH and drive variation in gene expression and disease.
Slightly under half of all studies relied upon small, convenience samples. Recruitment of populations readily available at community clinics, blood drives, or ads in local media risk underrepresentation of subgroups that are harder to reach yet inherently important to fully answering questions about the SDOH. Likely underrepresented are both extremes on the spectrum of social privilege: the most oppressed due to issues of access (e.g., undocumented status, lack of health insurance, distal proximity to clinics) and the most privileged due to economic resources that afford buying out of public services. Consequently, variation decreases in the sample and researchers are less able to make inference to these particular subpopulations and to detect differences across groups.
Even in ancillary studies attached to population-based cohorts, sampling bias has likely been introduced via attrition and selection criteria employed in determining epigenetic subsamples. For example, in some of the longitudinal surveys, attrition due to death, migration, and incarceration has limited representativeness of epigenetic subsamples drawn from latter waves of data (e.g., Beach et al., 2010, 2011; Loucks et al., 2016). Reliance on voluntary response sampling has also confined epigenetic subsamples to the most accessible part of populations (e.g., Bustamante et al., 2016, 2018; Huang et al., 2016). In other cases, eligibility criteria applied to epigenetic subsamples, such as participation in all waves of data (e.g., Hughes et al., 2018) or non-immigrant and white ethnicity (e.g., Borghol et al., 2012; Hughes et al., 2018; Suderman et al., 2014) further restricted analyses to less-representative subpopulations. Eligibility criteria employed at the outset of some population-based surveys (e.g., singleton births within marriage in the MRC National Survey on Health and Development, English-speakers in the pSoBid cohort, and absence of clinical cardiovascular disease and baseline weight less than 300 pounds in the MESA cohort) may raise further questions about the representativeness of even the larger, more diverse cohort studies. Future epigenetic studies derived from longitudinal cohorts must openly discuss the limitations imposed by the original sample design.
To fully understand the role of epigenetics in population-based health inequities, we recommend that contemporary cohort studies integrate the SDOH as key exposures at the onset of study design, rather than retroactively. Doing so means prioritizing data collection from diverse samples that can demonstrate whether and how racial dynamics and distinctive contexts impact epigenetic changes and disease outcomes. While recruitment of diverse samples will require broader outreach and additional resources, the importance of investment in such efforts is increasingly acknowledged by funding agencies (i.e., NIH All of Us Research Program) who have begun to direct more resources towards them.
4.2. Heterogeneity in measurement of DNA methylation and associations
Our review highlights the rapid technological advances in the use of DNAm as a marker of biological weathering. Early studies focused on global methylation, measuring the overall percent hyper or hypo-methylation in human tissue or blood as indicators of genomic stability. The advent of array-based assays supported a move towards genome-wide and epigenome-wide approaches. The newest EPIC Chip Array includes over 850 thousand CpG sites that can be mapped to specific regions of DNA, supporting the identification of candidate genes and the testing of more specific hypotheses about SDOH exposures and outcomes such as altered methylation in stress and neuroendocrine pathways. Array-based assays have led to DNAm measures of accelerated aging that rely upon a varying number of CpGs and statical “clock” algorithms, including the most prominent Horvath and Hannum clocks. However, heterogeneity and rapid changes in analytic methods used to characterize DNAm renders quantitative comparisons across studies difficult.
Another challenge is heterogeneity in tissue samples and differential cell composition. Array based assays have the advantage of using validated statistical adjustments to account for differences, while others extract monocytes and leucocytes prior to DNA extraction to account for this source of variability across blood samples. As methods continue to evolve, the lack of standardization in DNA source and extraction, processing and analytic platform, and statistical approaches render comparability across studies challenging.
In this review, many studies were derived from longitudinal cohorts, yet only 2 examined DNAm changes over time, and none examined longitudinal shifts in DNA among middle-aged or older adults. Further, the reliance on cross-sectional designs mean that epigenetics was often examined as an outcome, rather than as a predictor of health outcomes. Increased longitudinal follow-up in epigenetics studies will allow scholars to determine whether methylation may indeed be understood as early biomarkers of socially determined health inequities.
4.3. Breadth and Depth in the Measurement of SDOH
Under our search protocol for upstream SDOH we found that adverse childhood experiences and SES have received the most attention from epigenetic researchers. Initially much of this work was motivated by research on the associations between childhood adversity, adult methylation patterns, and mood and adjustment disorders. Increasingly, interest has expanded into SES over the lifecourse and the association with metabolic disorders and cardiovascular health. Across studies, measures were typically operationalized at the individual- or household-level, approximating childhood adversity through self-reported experiences with abuse and neglect, or proxying household SES from parental education, occupation, wealth, and unmet needs.
Fewer studies incorporated SES at neighborhood-levels, perhaps due to a lack of geocoded data. In the future, we recommend expanded inquiry into area-level measures, as the social science literature has documented nuanced ways that contextual measures, both geographic and socioeconomic, matter to health. For example, historical and current practices of residential segregation in the U.S. have left Black and Latinx families living in neighborhoods with higher concentrations of poverty and fewer resources and opportunities than their white counterparts with similar incomes. This contextual exposure is in turn associated with heightened stress and poorer physical and mental health (Massey & Wagner, 2018; Williams & Collins, 2001). Other studies have revealed that living in a neighborhood with a greater percentage (up to a threshold) of neighbors from the same racial/ethnic background, is protective of health for some racial/ethnic minoritized subgroups, especially at the lowest levels of socioeconomic position (Bécares et al., 2014; Linnenbringer et al., 2020). Theorized mechanisms suggest that greater in-group representation offers respite from daily microaggressions and environmental cues that bolster a sense of belonging, social support, and self-affirmation.
Epigenetic investigations that aim to interrogate the dynamic relationship between place and health could fruitfully incorporate area-level measures of SES (e.g. family income and wealth, home ownership, etc.) as well as measures of racial/ethnic composition to illuminate both deleterious and protective neighborhood features and explore their impacts on diverse subpopulations. More could also be done to incorporate existing knowledge on the health impacts of place-based institutions and policies beyond residences and workplaces. Linking information on transportation, banking, corrections, border protections and national security (Rudolph et al., 2013) to epigenetic data could open up intriguing new avenues of research.
Although sparse, the literature has expanded into traumas more characteristic of interactions external to the household context, such as experiences with discrimination. Standard perceived discrimination scales (Krieger et al., 2005; Landrine & Klonoff, 1996; Williams et al., 1997) were employed, assessing both interpersonal and major life experiences. In one instance, state-sanctioned violence was assessed in examining the Holocaust as a predictor of DNAm. The paucity of these studies, however, suggests many opportunities remain for the field of epigenetic research to delve further into this area of investigation, e.g. by incorporating the conceptualization and measurement of ethnoracism beyond the African-American experience (Ikram et al., 2015; Schunck et al., 2014), and exploring structural factors that capture intersectional and interlocking systems of oppression (Bailey et al., 2017).
Notably, numerous studies included measures of race/ethnicity or gender without adequately specifying what social exposures these variables are intended to proxy or represent. This suggests that social epigenetics research has yet to fully assimilate calls from social scientists to guard against biological or cultural essentialism in discussions of differences across socially-defined subpopulations (Zuberi et al., 2015). Addressing social disparities by race, gender, or other social identity will first require that researchers acknowledge them as proxies for hierarchies of power and make efforts to directly measure the structures and processes responsible for the differential distribution of resources across groups.
5. Limitations
Our review offers the first assessment of the integration and operationalization of a SDOH framework in epigenetic research. The epigenetic literature is growing quickly, and our review focuses particularly on studies employing DNAm -- the most pervasive measure of epigenetic change -- as a key outcome. Multiple other measures are available, and future studies could assess the extent to which studies focused on such measures engage with the SDOH. Furthermore, even among methylation studies, there is substantial variation in cell and tissue types, and in epigenetic measurements developed over time and employed for samples at different ages. This heterogeneity and rapid changes in the basic science of epigenetic processes complicates inferences about the association between various SDOH and epigenetic markers. The present study focuses on the operationalization of SDOH in social epigenetic research, and leaves a more formal quantitative assessment of the strength and direction of specific associations to subsequent research.
The complex and multifaceted influence of the SDOH renders their operationalization in quantitative research challenging. It is possible that researchers considered the multiple intersecting ways in which social and economic conditions shape health outcomes in diverse populations but were constrained by the available samples and extant measures in their datasets. We highlight the limited way in which social determinants are operationalized as individual characteristics rather than systemic ones that impact populations with the goal of spurring conceptual innovation and technical progress on the social side of the epigenetic frontier.
6. Conclusion and Recommendations
Calls for social scientists to learn the terminology, concepts, methods, and insights of epigenetic research abound (Bliss, 2018; Suzuki & von Vacano, 2018). To date, however, the notion that researchers specializing in epigenetics would benefit from incorporating key concepts and methodological insights from the substantial literature on the SDOH has had less traction. There are significant upstream social processes that shape lived experiences and can contribute to biological weathering that are not currently considered as part of the literature, but have potential for expanding the impact of social epigenetic research. Our review reveals the incomplete integration of the SDOH perspective into the literature and points to potential interventions that can substantially bolster the production of knowledge in this area.
First, disregard for the ways in which social stratification by race and geography may impact epigenetics has limited racial/ethnic and geographic diversity in the populations being studied. Epigenetic studies feature slightly more diverse populations than their genomic counterparts which rely almost exclusively on data from white respondents of European ancestry (Martin et al., 2019; Popejoy & Fullerton, 2016). Still, our review highlights the need for epigenetic studies in larger representative, population-based samples that will better support analyses about the connections between various SDOH and DNAm patterns. Statistically well-powered samples will also allow researchers to go beyond documenting differences in methylation across subpopulations to testing more nuanced hypotheses about particular exposures that put communities at risk of poorer health.
Second, the epigenetics literature frequently operationalized the social determinants of health as individual sociodemographic traits like parental education, occupation income, race/ethnicity or individualized experiences with social stressors such as poverty, discrimination, or abuse. While this research has generated important insights into the biosocial mechanisms that shape health outcomes, relying on individual characteristics as proxies for stratification processes has limited the ability of research to point to upstream, systemic factors as key determinants of population health. To more accurately portray the way that systems of social stratification produce real differences in gene expression, biological functioning, and health, social epigenetics research should eschew a simplified depiction of race/ethnicity, gender, and socioeconomic status as individual-level characteristics and explicitly acknowledge that these characteristics are more accurately interpreted as cues used by society to differentiate people. The SDOH framework teaches us that social cues are used to distinguish and order groups, allocating power to some and excluding others. These dynamics differentiate social and economic opportunities as well as in biomarkers and health outcomes (Chowkwanyun et al., 2018). Future research in epigenetics could employ this framework to better inform the conceptualization of research questions and interpretation of findings on a population level.
Finally, epigenetic research must guard against the tendency for biological reductionism and determinism that have entangled prior biosocial research enterprises (Frank, 2015; Morris et al., 2020). Epigenetics is at the frontier of bridging social science and biology, and its causal logic is highly compatible with the SDOH framework. While much progress remains to be made on understanding the biological mechanisms that underly epigenetic changes, maintaining the SDOH perspective is crucial if the field is to realize its promise of explicating how context, structural stratifiers, and intermediary mechanisms get under our skins and shape population health.
Research Highlights.
Often, SDOH were operationalized as individual-level traits and experiences
Measuring upstream social processes will expand the reach of DNAm research
Larger, more diverse and representative samples are needed to support SDOH analyses
A SDOH lens more effectively elucidates biological consequences of social exposures
Employing a SDOH lens also guards against the tendency for biological reductionism
ACKNOWLEDGEMENTS
The first author also gratefully acknowledges writing time supported by an NICHD training grant (T32HD049302) to the University of Wisconsin-Madison School of Medicine and Public Health. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Appendix A.
Selected Studies and Study Design
| Study | Study demographics | SDOH of primary interest | Epigenetics (tissue source, m. measure) | Health outcomes |
|---|---|---|---|---|
|
Adams et al. 2017 USA |
Seattle Female and Male Shift Worker Study n=328 39.9% male Age range: 20–55 yrs 77% White, 22% Non-White |
Workplace exposure: shiftwork | Peripheral blood Genome-wide m. |
|
|
Alegria.-Torres et al. 2013 Mexico |
Traditional brick producers n=39 100% male Mean age 42.5 yrs (SD 15.84) No mention of race/ethnicity, or nativity |
Workplace exposure: toxins (PAHs) in brickmaking | Peripheral blood Global (Alu) and local m. of TNF-α, IFN- γ, IL-6, IL-12 |
|
|
Austin et al. 2018 Canada |
Vancouver cohort stratified by SES n=335 45% male Age range: 15 to 55 yrs 73% White |
Life-course SES trajectory | Peripheral blood Horvath epigenetic clock |
|
|
Barcelona et al. 2018 USA |
Intergen. Impact of Genetic & Psych Factors on BP n=159 0% male Age range: 21 to 49 yrs 100% Black/AA |
Perceived racism & discrimination | Saliva Genome-wide m. |
|
|
Barfield et al. 2019 USA |
Multi-Ethnic Study of Atherosclerosis (MESA) and the Cardiovascular Health Study (CHS) n=619 in MESA and n=483 in CHS 53% female in MESA and 63% female in CHS Mean age 68 yrs in MESA and 74 yrs in CHS 21% Black/AA, 33% Latinx, 46% White in MESA 49% Black/AA and 51% White in CHS |
Sociodemographics: race/ethnicity | Peripheral blood Genome-wide m. and ID of differentially methylated regions mapped to specific genes |
Sleep phenotypes |
|
Beach et al. 2010 USA |
Iowa Adoptee Study n=192 50% male Mean age 48 (range 35 to 69) No mention of race/ethnicity, or nativity |
Adverse childhood experience: childhood physical/sexual abuse | Peripheral blood Local m. of serotonin transporter promoter region SLC6A4 |
|
|
Beach et al. 2011 USA |
Iowa Adoptee Study n=155 0% male Middle-aged, but range unknown No mention of race/ethnicity, or nativity |
Adverse childhood experience: childhood sexual abuse | Peripheral blood Local m. of serotonin transporter protein 5HTT |
Antisocial personality |
|
Beach et al. 2014 USA |
Strong African American Healthy Adult Project n=388 50% male Mean age 19.2 yrs 100% Black/AA |
Childhood SES and financial strain | Saliva Genome-wide and local m. of the 5-HTTL-linked polymorphic region |
|
|
Beach et al. 2016 USA |
Strong African American Healthy Adult Project n=398 50% male Mean age 19.2 yrs 100% Black/AA |
Childhood SES and financial strain Protective parenting style |
Peripheral blood Genome-wide m. |
Self-reported health |
|
Bhatti et al. 2015 USA |
Seattle-based health care workers n=139 41% male Age range: 22 to 40 yrs 77% White, 15% Asian, 4% Black/AA, 4% Other |
Workplace exposure: shiftwork status | Peripheral blood Genome-wide m. and local m. of circadian genes |
|
|
Borghol et al. 2012 UK |
1958 British Birth Cohort Study n=40 100% male Age: 45 yrs 100% White, non-immigrant |
Childhood and adulthood SES Adulthood financial strain |
Peripheral blood Genome-wide m. and differentially m. regions by SEP |
|
|
Brody et al. 2016 (a) USA |
Strong African American Healthy Adult Project and Adults in the Making Project n=616 40% male Age: 20 yrs (SHAPE) and 22 yrs (AIM) 100% Black/AA |
Perceived racial discrimination Family social support |
Peripheral blood Genome-wide m., Hannum epigenetic clock |
|
|
Brody et al. 2016 (b) USA |
Strong African American Healthy Adult Project n=399 45.4% male Age: 20 yrs 100% Black/AA |
Adverse childhood experience: parental depression, harsh parenting | Peripheral blood Genome-wide m., Horvath epigenetic clock |
|
|
Bustamante et al. 2016 USA |
Detroit Neighborhood Health Study n=147 38.2% male Mean age 49.6 yrs 17% White, 77% Black/AA, 6% Other |
Adverse childhood experiences: childhood maltreatment Major Depressive Disorder |
Peripheral blood Local m. of glucocorticoid receptor NR3C1 |
|
|
Bustamante et al. 2018 USA |
Detroit Neighborhood Health Study n=112 44.6% male Mean age 50.74 yrs (SD 12.96) 16% White, 77% Black/AA, 7% Other |
Adverse childhood experiences: childhood maltreatment | Peripheral blood Local m. of FKBP5 gene, involved in negative feedback loop of HPA-axis |
Depressive symptomology, MDD |
|
Campesi et al. 2013 Italy |
Blood drive n=168 49.4% male Age range: 18 to 40 yrs No mention of race/ethnicity, or nativity |
Sociodemographics: gender as a marker of social status | Peripheral blood Global m. |
|
|
Castillo et al. 2017 Chile |
Residents of mining and control community n=208 46.6% male Age range: 52 to 85 yrs No mention of race/ethnicity, or nativity |
Neighborhood exposure: living in a mining community | Peripheral blood Global m. |
Parkinson’s disease |
|
Chen et al. 2016 USA |
Strong African American Healthy Adult Project n=330 % male unknown Age: 19 yrs 100% Black/AA |
Childhood SES and financial strain | Peripheral blood Genome-wide m., Hannum and Horvath epigenetic clock |
|
|
Chi et al. 2016 USA |
Multi-Ethnic Study of Atherosclerosis n=1207 48.4% male Mean age 69.6 yrs (SD 9.4) 47.2% White, 21.2% Black/AA, 31.6% Hisp./Latinx |
Neighborhood exposure: air pollution | Peripheral blood Global m. (Alu and LINE-1) and local m. of candidate genes |
|
|
Fiorito, et al. 2017 Multinational Italy, Australia, Ireland |
EPIC Italy, Melbourne Coll Cohort Study, TILDA n=5111 Male: 38% EPIC, 61% MCSS, 50% TILDA Mean age 53.3 EPIC, 59 MCCS, 62.1 yrs TILDA No mention of race/ethnicity, or nativity |
Adulthood SES Life-course SES trajectory |
Peripheral blood Genome-wide m., Hannum and Horvath epigenetic clock |
|
|
Ghosh et al. 2017 Netherlands |
Workplace sample of factory workers n=67 77.6% male Mean age 35 yrs No mention of race/ethnicity, or nativity |
Workplace exposure: toxin (carbon nanotubes) | Peripheral blood Global (LINE-1) and local m. of promoter region of candidate genes (DNMT1, HDAC4, NPAT/ATM, TGF-β, and SKI) |
|
|
Gondalia et al. 2019 USA |
12 subsamples from the Women’s Health Initiative and Atherosclerosis Risk in Communities Study n=8397 in total 83% female across all subsamples Mean age 61.3 yrs (SD 7.4) 45.8% Black/AA, 8.4% Hisp./Latinx, 45.8% White |
Neighborhood exposure: air pollution | Peripheral blood Genome-wide m. and ID of differentially m. regions mapped to specific genes |
|
|
Goodrich et al. 2013 USA |
Workplace sample of dental professionals n=131 49.6% male Mean age 55.8 yrs No mention of race/ethnicity, or nativity |
Workplace exposure: toxin exposure (mercury) via dental amalgams | Buccal swab Global m. (LINE-1) and candidate genes related to mercury toxicity (DNMT1, SEPW1, SEPP1) |
|
|
Gouin et al. 2017 Canada |
l’Etude longitudinale des enfants de maternelle au Quebec n=46 50% male Age: 27 yrs 100% White |
Childhood SES Adverse childhood experiences: childhood abuse |
Peripheral blood Local m. of candidate gene OXTR, involved in social behaviors |
Anxiousness, disruptiveness |
| Guerrero-Preston et al. 2019 USA | Patients treated for head and neck cancers n=496 75% male Mean age 58 yrs 50% Black/AA, 50% White |
Sociodemographics: race/ethnicity Neighborhood exposure: zip code level median household income; home ownership and vacancy; insurance coverage rates |
Tumor tissue Local m. of the promoter regions of EDNRB, NID2, PAX5, and PAX1 |
Head and neck cancers |
|
Horvath et al. 2016 Multinational |
Women’s Health Initiative; Bogalusa; Parkinson’s Disease, Envir. and Genes; and 9 small datasets n=5162 Age range across studies: 0 to 93 yrs 57% White, 27% African ancestry (AA and Central African), 13% Hispanic, 2.5% East Asian, 1% Tsimane Amerindian |
Sociodemographics: race/ethnicity, gender | Peripheral blood, saliva, and brain tissue Genome-wide m., and Horvath epigenetic clock |
Coronary heart disease |
|
Houtepen et al. 2018 UK |
AVON longitudinal study and MRC NSHD n=1332 across both cohorts 0% male Mean age 47.14 ALSPAC, 53.45 MRC No mention of race/ethnicity, or nativity |
Adverse childhood experiences: childhood maltreatment, abuse, and illness; parental death/separation, physical or mental illness; suboptimal maternal bonding | Peripheral blood, buccal swab Genome-wide m. and differentially m. regions by ACEs, and candidate gene probes |
|
|
Hsu et al. 2019 USA |
Arkansas Rural Community Health Study n=39 100% female Mean age 48 yrs (SD 12.0) 51% Black/AA, 49% White |
Neighborhood exposure: county poverty rate | Saliva Genome-wide m. and ID of differentially m. regions |
Telomere length |
|
Huang et al. 2016 Israel |
Jerusalem Perinatal Study Family Follow-up n=613 0% male Mean age 32 yrs No mention of race/ethnicity, or nativity |
Childhood SES | Peripheral blood Local m. of 5 cardiometabolic and stress- response genes ABCA1, INS-IGF2, HSD11B2, and NR3C1 |
Cardiometabolic risk, LBW baby |
|
Hughes et al. 2018 UK |
UK Household Longitudinal Study n=1099 42.4% male Mean age 58.4 yrs (SD 14.9) 100% White |
Childhood and adulthood SES | Peripheral blood Genome-wide m., Hannum and Horvath epigenetic clock |
|
|
Janusek et al. 2017 USA |
Low-SES Chicago neighborhood cohort n=34 100% male Mean age 20.2 yrs (range 18 to 25) 100% Black/AA |
Childhood SES Adverse childhood experiences: maltreatment, abuse, parental separation and/or mental illness, HH dysfunction, community violence |
Peripheral blood Local m. of IL-6 promoter |
Salivary IL-6 response |
|
Klinger-König et al. 2019 Germany |
Study of Health in Pomerania (SHIP-2, TREND-0) n=3965 52% female Mean age 54.2 yrs (SD 14.4) 100% White |
Adverse childhood experiences: childhood maltreatment and depression | Peripheral blood Local m. of FKBP5, involved in negative feedback loop of HPA-axis function |
Depressive symptomology |
|
Kogan et al. 2018 USA |
African American men in rural South Georgia n=358 100% male Mean age 21.85 yrs (SD 1.27) 100% Black/AA |
Adverse childhood experiences: childhood trauma, substance use Social support: prosocial ties |
Saliva Local m. of promoter region of OXTR gene |
Substance use |
|
Lam et al. 2012 Canada |
Vancouver cohort stratified by SES n=92 38% male Mean age 33.04 yrs (range 24 to 45; SD 5.03) 68% White, 32% Non-White |
Sociodemographics: race, sex Childhood and adulthood SES |
Peripheral blood Genome-wide m. and ID of differentially m. regions |
Perceived stress, cortisol output |
|
Lawn et al. 2018 UK |
AVON longitudinal study and MRC NSHD n=1762 0% male Mean age 29 yrs at time1 and 47 yrs at time 2 No mention of race/ethnicity, or nativity |
Childhood and adulthood SES Adverse childhood experiences: maltreatment, abuse, and parental separation and/or mental illness |
Peripheral blood, buccal swab Genome-wide m., Horvath epigenetic clock |
|
|
Lei et al. 2015 USA |
Family and Community Health Study n=99 0% male Mean age 48.33 yrs 100% Black/AA |
Neighborhood exposure: high crime | Peripheral blood Local m. of serotonin transporter gene 5HTT |
Depressive symptomology |
|
Loucks et al. 2016 USA |
New England Family Study n=141 47.5% male Mean age 47 yrs 66% White, 34% non-White |
Childhood SES | Peripheral blood and subcutaneous adipose tissue Genome-wide m. and ID of differentially m. regions |
Body mass index |
|
McCartney et al. 2018 Scotland |
Generation Scotland: Scottish Family Health Study n=5100 38% male Mean age 48.51 yrs No mention of race/ethnicity, or nativity |
Adulthood SES Neighborhood exposure: neighborhood deprivation index |
Peripheral Blood Genome-wide m., Hannum and Horvath epigenetic clock |
|
|
McClelland et al. 2016 Scotland |
Psych., Soc., and Bio. Determinants of Ill Health n=666 49% male Age range 35 – 64 years No mention of race/ethnicity, or nativity |
Adulthood SES Neighborhood exposure: neighborhood SES |
Peripheral blood Global m. |
|
|
McCrory et al. 2019 Ireland |
Irish Longitudinal Study on Ageing (TILDA) n=490 50% female Mean age 62.2 yrs (SD 8.3) No mention of race/ethnicity, or nativity |
Childhood and adulthood SES Life-course SES trajectory |
Peripheral blood Genome-wide m. Horvath, Hannum, and Levine epigenetic clocks |
Timed up and go measure |
|
McDade et al. 2017 Philippines |
Cebu Longitudinal Health and Nutrition Survey n=494 20% male Age range: 20 to 22 years No mention of race/ethnicity, or nativity |
Childhood SES Adverse childhood experiences: parental separation, microbial envi. |
Peripheral blood Genome-wide m. and ID of differentially m. regions relevant to inflammation |
Inflammatory markers (CRP, IL-6, TNFα, IL-10, IFNγ, IL-1β, and IL-8) |
| McDade et al. 2019 Philippines |
Cebu Longitudinal Health and Nutrition Survey n=489 20% male Mean age 20.9 yrs No mention of race/ethnicity, or nativity |
Life-course SES trajectory | Peripheral blood Genome-wide m. and ID of differentially m. regions |
|
|
McGuinness et al. 2012 Scotland |
Psych., Soc., and Bio. Determinants of Ill Health n=239 49% male Age range: 35 to 64 yrs No mention of race/ethnicity, or nativity |
Adulthood SES | Peripheral blood Global m. |
Cardiovascular biomarkers-fibrinogen and IL-6 |
|
Miller et al. 2015 USA |
Adults in the Making Study n=292 36.3% male Age: 22 yrs 100% Black/AA |
Childhood SES and financial strain and resilience (self-control) | Peripheral blood Genome-wide m., Hannum and Horvath epigenetic clock |
Depressive symptomology, substance use, aggressive and internalizing behavior |
|
Moser et al. 2015 Switzerland |
Mothers tested with stress/non-stress child stimuli n=46 100% female Mean age 34.3 yrs No mention of race/ethnicity, or nativity |
Adverse experiences: interpersonal violence exposure | Buccal swab Local m. of candidate gene (BDNF) |
Neural activity and generalized anxiety |
|
Needham et al. 2015 USA |
Multi-Ethnic Study of Atherosclerosis n=1231 41% male Mean age: 69.55 yrs (range 55 to 94, SD 9.35) 47.2% White, 21.2% Black/AA, 31.6% Hisp./Latinx |
Childhood and adulthood SES Life-course SES trajectory |
Peripheral blood Local m. of candidate genes in stress response and inflammation (AVP, FKBP5, F8, CCL1, OXTR, CD1D, KLRG1, NLRP12, PYDC1,…) |
|
|
Peluso et al. 2012 Thailand |
Ma Ta Phut industrial state and rural area n=177 87% male Mean age: 31.2 yrs No mention of race/ethnicity, or nativity |
Neighborhood exposure: air pollution | Peripheral blood Global m. (LINE-1 and Alu) candidate genes p53, p16, and IL-6) |
|
|
Peng et al. 2018 USA |
The Twins Heart Study and the Mood and Methylation Study n=238 (119 monozygotic twin pairs) 71% male Mean age 55 yrs for male pairs, 36 yrs female pairs No mention of race/ethnicity, or nativity |
Adverse childhood experiences: physical, emotional, or sexual abuse other traumas (e.g., natural disaster, family mental illness or separation of parents) | Peripheral blood Genome-wide and local m. of candidate genes involved in stress response (BDNF, NR3C1, SLC6A4, MAOA, MAOB) |
Depressive symptomology |
|
Perroud et al. 2014 Rwanda |
Mothers of Tutsi ethnicity and their children n=50 100% female Mean age of mothers 45 yrs 100% Tutsi ethnicity |
Exposure to Rwandan genocide | Peripheral blood Local m. of candidate genes (NR3C1, NR3C2) |
PTSD, depressive symptomology, cortisol, GR and mineralcorticoid receptor levels |
|
Pittman et al. 2020 USA |
Anniston Community Health Survey I and II n=518 in ACHS-I and n=299 in ACHS-II 71% female in ACHS-I and 75% female in ACHS-II Mean age 54.6 yrs @ ACHS-I, 62.8 yrs @ ACHS-II 40% Black/AA, 60% White in ACHS-I 52% Black/AA, 48% White in ACHS-II |
Living in community with PCB manufacturing facility | Stored blood clot in ACHS-I and whole blood in ACHS-II | |
|
Santos et al. 2018 USA |
Pregnant Latina women in North Carolina n=147 0% male Mean age 27.6 yrs 100% Latina |
Perceived discrimination & ethnic-specific discrimination | Peripheral blood Local methylation of NR3CI, BDNF, FKBP5 genes |
|
|
Schür et al. 2018 Netherlands |
Adults with a mental health disorder, a family history of mental disorder, or healthy controls n=241 60% male Mean age 37.4 yrs No mention of race/ethnicity, or nativity |
Adverse childhood experiences: childhood trauma; emotional, physical, and sexual abuse; and emotional and physical neglect | Peripheral blood Local m. of the GR-1F region |
Salivary cortisol response |
|
Simons et al. 2016 USA |
Family and Community Health Study - caregivers n=100 0% male Mean age: 48.5 yrs 100% Black/AA |
Adulthood SES Adulthood financial strain |
Peripheral blood Genome-wide m., Hannum epigenetic clock |
|
|
Simons et al. 2017 USA |
Family and Community Health Study - caregivers n=100 0% male Mean age: 48.5 yrs (SD 9.30) 100% Black/AA |
Adulthood financial strain Neighborhood exposure: high crime |
Peripheral blood Genome-wide m. and local m. of oxytocin gene (OXTR) |
Depressive symptomology, negative schema (pessimism, distrust) |
|
Sipahi et al. 2014 USA |
Detroit Neighborhood Health Study n=60 23.3% male Mean age 55.1 yrs 77% Black/AA, 20% White, 3% Other |
Trauma exposure, PTSD | Peripheral blood or blood spot Local m. of candidate genes (DNMT1, DNMT3A, DNMT3L, and DNMT3B) |
Severity of trauma response |
|
Smith et al. 2017 USA |
Multi-Ethnic Study of Atherosclerosis n=1226 52% male Mean age 69.6 yrs 47% White, 22% Black/AA, 31% Hispanic/Latinx |
Neighborhood exposure: neighborhood SES and social environment (aesthetic, safety, and social cohesion) | Peripheral blood Local m. of candidate response and inflammation (AVP, FKBP5, OXTR, F8, CD1D, KLRG1,…) |
|
| Song et al. 2013 Japan |
Occupational Study of Social Class and Health n=360 91% male Age range: mid 30s to mid 50s No mention of race/ethnicity, or nativity |
Workplace exposure: job strain | Saliva Local m. of the BDNF gene involved in function of the adult brain |
|
|
Stringhini et al. 2015 Italy |
European Prospective Investigation into Cancer and Nutrition Cohort (EPIC) n=857 42.8% male Age range mid 40s to mid 60s No mention of race/ethnicity, or nativity |
Childhood and adulthood SES Life-course SES trajectory |
Peripheral blood Genome-wide and local m. of candidate genes involved in inflammation (ADM,GPR132, IL1A, CCL20, CXCL2…) |
|
| Subramanyam et al. 2013 USA |
Multi-Ethnic Study of Atherosclerosis n=1002 48% male Mean age 61 yrs (range 45 to 84 yrs) White, Black/AA, Hispanic/Latinx |
Sociodemographics: race/ethnicity Childhood and adulthood SES |
Peripheral blood Global m. (Alu and LINE-1) |
|
|
Suderman et al. 2014 UK |
1958 British Birth Cohort Study n=40 100% male Age: 45 yrs 100% White |
Adverse childhood exposures: physical, emotional and sexual abuse Adulthood SES |
Peripheral blood Genome-wide m. and ID of differentially m. regions |
|
|
Tajuddin et al. 2019 USA |
Healthy Aging in Neighborhoods of Diversity across the Life Span (HANDLS) Study n=487 51% male Mean age 48.7 yrs 50.4% Black/AA, 49.6% White |
Sociodemographics: Race, sex, and poverty status | Peripheral blood Genome-wide m. Horvath and Hannum epigenetic clock |
|
|
Tehranifar et al. 2013 USA |
New York Women’s Birth Cohort n=89 0% male Mean age 43 yrs 31% Black/AA, 26% White, 43% Hispanic/Latinx |
Childhood and adulthood SES | Peripheral blood Global m. (Sat2, Alu, LINE-1) |
|
|
Toyooka et al. 2003 Multinational USA, Taiwan, Japan, and Australia |
Patients with primary non-small cell lung cancers n=598 69% male Mean age 63 yrs No mention of race/ethnicity, or nativity |
Sociodemographics: gender, geography | Lung tissue Local m. of 7 genes (p16, RASSFIA, APC, RARβ, CDH13, MGMT, GSTP1) involved in lung cancers |
Lung cancers |
|
Uddin et al. 2010 USA |
Detroit Neighborhood Health Study n=100 40% male Mean age 45.8 yrs 79% Black/AA, 14% White, 7% Other |
Trauma exposure as assessed via PTSD | Peripheral blood Genome-wide m. and ID of differentially-methylated regions by PTSD status |
|
|
Uddin et al. 2013 USA |
Detroit Neighborhood Health Study n=100 40% male Mean age 45.8 yrs 79% Black/AA, 14% White, 7% Other |
Adulthood SES (as a moderator) | Peripheral blood Genome-wide m. and ID of differentially-m. regions by PTSD status |
PTSD, postraumatic symptom severity |
|
Yehuda et al. 2014 USA |
Adult children of Holocaust survivors and controls n=95 100% male Mean age 56.6 yrs |
Adverse childhood experiences: maternal or paternal PTSD | Peripheral blood Local m. of the GR-1F promoter |
|
|
Yehuda et al. 2016 USA |
Holocaust survivors, children of survivors, controls n=71 Survivors: 60% male; Children: 23% male Age range: early 40s to mid 80s 100% Jewish |
Trauma exposure: Holocaust Adverse childhood experiences: maternal or paternal PTSD, physical or sexual abuse |
Peripheral blood Local m. of FKBP5 intron 7 binding protein 5 |
Cortisol |
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams CD, Jordahl KM, Copeland W, Mirick DK, Song X, Sather CL,…Bhatti P (2017). Nightshift work, chronotype, and genome-wide DNA methylation in blood. Epigenetics, 12(10), 833–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Algería-Torres JA, Barretta F, Bares-Esquivel LE, Carrizales-Yáñez L, Pérez-Maldonado IN, Baccarelli A, & Bertazzi PA (2013). Epigenetic markers of exposure to polycyclic aromatic hydrocarbons in Mexican brickmakers: A pilot study. Chemosphere, 91, 475–480. [DOI] [PubMed] [Google Scholar]
- Austin MK, Chen E, Moss KM, McEwen L, Maclsaac JL, Kobor MS, & Miller GE (2018). Early-life socioeconomic disadvantage, not current, predicts accelerated epigenetic aging of monocytes. Psychoneuroendocrinology, 97, 131–134. [DOI] [PubMed] [Google Scholar]
- Bailey ZD, Krieger N, Agénor M, Graves J, Linos N, & Bassett MT (2017). Structural racism and health inequities in the USA: Evidence and intervention. Lancet 389, 1453–1463. [DOI] [PubMed] [Google Scholar]
- Baptiste-Roberts K, Oranuba E, Werts N, & Edwards LV (2017). Addressing health care disparities among sexual minorities. Obstetrics & Gynecology 44(1): 71–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barcellos SH, Carvalho LS, & Turley P (2018). Education can reduce health differences related to genetic risk of obesity. Proceedings of the National Academy of Sciences 115 (42), E9765–E9772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barcelona de Mendoza V, Huang Y, Crusto C, Sun YV, & Taylor JY (2018). Perceived racial discrimination and DNA methylation among African American Women in the InterGEN study. Biological Research for Nursing, 20(2), 145–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barfield R, Wang H, Liu Y, Brody JA, Swenson B, Li R,…Sofer T (2019). Epigenome-wide association analysis of daytime sleepiness in the Multi-Ethnic Study of Atherosclerosis reveals African-American-specific associations. Sleep, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolomei MS (2009) Genomic imprinting: Employing and avoiding epigenetic processes. Genes Dev 23:2124–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beach SRH, Brody GH, Kim S, & Cui J (2014). Is serotonin transporter genotype associated with epigenetic susceptibility or vulnerability? Examination of the impact of SES risk on African American youth. Dev Psychopathol, 26(2), 289–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beach SRH, Brody GH, Todorov AA, Gunter TD, & Phlibert RA (2010). Methylation at SLC6A4 is linked to family history of child abuse: An examination of the Iowa adoptee sample. Am J Med Genet B Neuropsychiatr Genet, 153B(2), 710–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beach SRH, Brody GH, Todorov AA Gunter TD, & Philbert RA (2011). Methylation at 5HTT mediates the impact of child sex abuse on women’s antisocial behavior: An examination of the Iowa adoptee sample. Psychosom Med. 73(1), 83–87L [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beach SR, Lei MK, Brody GH, Kim S, Barton AW, Dogan MV, & Philibert RA (2016). Parenting, socioeconomic status risk, and later young adult health: Exploration of opposing indirect effects via DNA methylation, Child Dev, 87, 111–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bécares L, Nazroo J, & Jackson J (2014). Ethnic density and depressive symptoms among African Americans: Threshold and differential effects across social and demographic subgroups. Am J Public Health 104, 2334–2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley AR, Callier S, Rotimi CN (2017). Diversity and inclusion in genomic research: Why the uneven progress? J Community Genet 8, 255–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezruchka S (2012). The hurrider I go the behinder I get: The deteriorating international ranking of U.S. health status. Annual Review of Public Health 33, 157–173. [DOI] [PubMed] [Google Scholar]
- Bhatti P, Zhang Y, Song X, Makar KW, Sather CL, Kelsey KT,…Wang P 2015). Nightshift work and genome-wide DNA methylation. Chronobiology International, 32(1), 103–112. [DOI] [PubMed] [Google Scholar]
- Bliss C (2018). Social by Nature: The Promise and Peril of Sociogenomics. Stanford, CA: Stanford University Press. [Google Scholar]
- Boardman JD, Barnes LL, Wilson RS, Evans DA, & Mendes de Leon CF (2012). Social disorder, APOE-E4 genotype, and change in cognitive function among older adults living in Chicago. Social Science & Medicine, 74(10), 1584–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghol N, Suderman M, McArdle W, Racine A, Hallett M, Pembrey M,…Szyf M (2012). Associations with early-life socio-economic position in adult DNA methylation. Epigenetic Epidemiology 41, 62–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowleg L (2019). Perspectives from the social sciences: Critically engage public health. AJPH, 109(1), 15–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd A, Golding J, Macleod J, Lawlor D, Fraser A, Henderson J,…Davey Smith G (2012). Cohort profile: The ‘children of the 90s’ – the index offspring of the Avon Longitudinal Study of Parents and Children. International Journal of Epidemiology, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braveman P, Egerter S, & Williams DR (2011). The social determinants of health: Coming of age.” Annual Review Public Health, 32, 381–398. [DOI] [PubMed] [Google Scholar]
- Brody GH, Miller GE, Yu T, Beach SR, & Chen E (2016a). Supportive family environments ameliorate the link between racial discrimination and epigenetic aging: A replication across two longitudinal cohorts, Psychol Sci, 27, 530–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brody GH, Yu T, Chen E, Beach SR, & Miller GM (2016b). Family-centered prevention ameliorates the longitudinal association between risky family processes and epigenetic aging, J Child Psychol Psychiatry, 57, 566–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown L, Needham B, & Ailshire J (2016). Telomere length among older U.S. adults: Differences by race/ethnicity, gender, and age. Journal of Aging and Health 29(8), 1350–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante AC, Aiello AE Galea S, Ratanatharathorn A, Noronha C, Wildman DE, & Uddin M (2016). Glucocorticoid receptor DNA methylation, childhood maltreatment and major depression. J Affect Disord. 206, 181–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante AC, Aiello AE, Guffanti G, Galea S, Wildman DE, & Uddin M (2018). FKBP5 DNA methylation does not mediate the association between childhood maltreatment and depression symptom severity in the Detroit Neighborhood Health Study. Journal of Psychiatric Research 96, 39–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campesi I, Carru C, Zinelu A, Occhioni S, Sanna M, Palermo M,…Franconi F (2013). Regular cigarette smoking influences the transsulfuration pathway, endothelial function, and inflammation biomarkers in sex-gender specific manner in healthy young humans. Am J Transl Res 5(5), 497–509. [PMC free article] [PubMed] [Google Scholar]
- Castillo S, Muñoz P, Behrens MI, Diaz-Grez F, Segura-Aguilar J (2017). On the role of mining exposure in epigenetic effects in Parkinson’s disease. Neurotox Res 32, 172–174. [DOI] [PubMed] [Google Scholar]
- Champagne FA, & Curley JP (2011). Epigenetic Influence of the Social Environment. In Petronic A & Mill J (Eds.), Brain, Behavior and Epigenetics: Epigenetics and Human Health. Springer-Verlag Berlin Heidelberg. [Google Scholar]
- Chen BH, Marioni RE, Colicino E, Peters MJ, Ward-Caviness CK, Tsai P,…Horvath S (2016). DNA methylation-based measures of biological age: meta-analysis predicting time to death. Aging 8(9), 1844–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E, Miller GE, Yu T, & Brody GH (2016). The great recession and health risks in African American youth. Brain Behav Immun 53, 234–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi G Liu CY, MacDonald JW, Barr RG, Donohue KM, Hensley MD,…Kaufman JD (2016). Long-term outdoor air pollution and DNA methylation in circulating monocytes: Results from the Multi-Ethnic Study of Atherosclerosis (MESA), Environmental Health: A Global Access Science Source, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowkwanyun M, Bayer R, & Galea S (2018). “ Precision” Public Health-Between Novelty and Hype. The New England Journal of Medicine, 379(15), 1398. [DOI] [PubMed] [Google Scholar]
- Commission on the Social Determinants of Health (2008). Closing the gap in a generation: health equity through action on the social determinants of health. Final Report of the Commission on Social Determinants of Health. Geneva, World Health Organization. [Google Scholar]
- Conley D, & Fletcher J (2018). The genome factor: What the social genomics revolution reveals about ourselves, our history, and the future. Princeton University Press. [Google Scholar]
- Conley D, Laidley TM, Boardman JD, & Domingue BW (2016). Changing Polygenic Penetrance on Phenotypes in the 20th Century among Adults in the US Population. Scientific reports, 6, 30348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunliffe VT (2016). The epigenetic impacts of social stress: How does social adversity become biologically embedded? Epigenomics 8(12), 1653–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingue BW, Belsky DW, Conley D, Mullan Harris K, & Boardman JD (2015). Polygenic influence on educational attainment: New evidence from the National Longitudinal Study of Adolescent to Adult Health. AERA Open 1(3), 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrucci L, Gonzalez-Freire M, Fabbri E, Simonsick E, Tanaka T, Moore Z, Salimi S, Sierra F, de Cabo R. (2020). Measuring biological aging in humans: A quest. Aging Cell 19, e13080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorito G, Polidoro S, Dugue P, Kivimaki M, Ponzi E,…Vineis P (2017). Social adversity and epigenetic aging: A multi-cohort study on socioeconomic differences in peripheral blood DNA methylation. Scientific Reports 7, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank R (2015). Back to the future? The emergence of a geneticized conceptualization of race in sociology. The ANNALS of the American Academy of Political and Social Science, 661(1), 51–64. [Google Scholar]
- Fraser A, Macdonald-Wallis C Tilling K, Boyd A, Golding J, Davey Smith G,…Lawlor DA (2012). Cohort profile: The AVON Longitudinal Study of Parents and Children: ALSPAC mothers cohort. International Journal of Epidemiology, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimura J, Bolnick DA, Rajagopalan R, Kaufman JS, Lewontin RC, Duster T,…Marks J (2014). Clines without classes: How to make sense of human variation. Sociological Theory 32(3), 208–227. [Google Scholar]
- Galea S, Tracy M, Hoggatt KJ, Dimaggio C, & Karpati A (2011). Estimated deaths attributable to social factors in the United States. American Journal of Public Health 101, 1456–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geronimus AT (2013). Deep integration: Letting the epigenome out of the bottle without losing sight of the structural origins of population health. Am J Public Health, 103(103): S56–S63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geronimus AT, Hicken M, Keene D, & Bound J (2006). “Weathering” and age patterns of allostatic load scores among blacks and whites in the United States. American Journal of Public Health, 96(5), 826–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh M, Öner D, Poels K, Tabish AM, Vlaanderen J, Pronk A,…Godderis L (2017). Changes in DNA methylation induced by multi-walled carbon nanotube exposure in the workplace. Nanotoxicology, 11(9–10), 1195–1210. [DOI] [PubMed] [Google Scholar]
- Gondalia R, Baldassari A, Holliday KM, Justice AE, Méndez-Giráldez R, Stewart JD, & Whitsel EA (2019). Methylome-wide association study provides evidence of particulate matter air pollution-associated DNA methylation. Environmental International 132, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass TA, & McAtee MJ (2006). Behavioral science at the crossroads in public health: Extending horizons, envisioning the future. Social Science & Medicine 62, 1650–1671. [DOI] [PubMed] [Google Scholar]
- Goodrich JM, Basu N, Franzblau A, & Dolinoy DC (2013). Mercury biomarkers and DNA methylation among Michigan dental professionals. Environmental and Molecular Mutagenesis 54, 195–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouin JP, Zhou QQ Boivin M, Coté, Hebert M, Ouellet-Morin I,…Vitaro F (2017). Associations among oxytocin receptor gene (OXTR) DNA methylation in adulthood, exposure to early life adversity, and childhood trajectories of anxiousness. Scientific Reports, 7, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero-Preston R, Lawson F, Rodriguez-Torres S, Noordhuis MG, Pirini F, Manuel L,…Sidransky D (2019). JAK3 variant, immune signatures, DNA methylation, and social determinants linked to survival racial disparities in head and neck cancer patients. Cancer Prevention Research 12(4), 255–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S (2013). DNA methylation age of human tissues and cell types. Genome Biology, 14, R115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S, Gurven M, Levine ME, Trumble BC, Kaplan H, Allayee H,…Assimes T (2016). An epigenetic clock analysis of race/ethnicity, sex, and coronary heart disease. Genome Biology 17(171), 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtepen LC, Hardy R, Maddock J Kuh D, Anderson EL, Relton CL,…Howe LD (2018). Childhood adversity and DNA methylation in two population-based cohorts. Translational Psychiatry, 8(266), 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu P, Kadlubar S, Su LJ, Acheampong D, Rogers LJ, Runnells G,…Schootman M (2019). County poverty levels influence genome-wide DNA methylation profiles in African American and European American women. Transl Cancer Res 8(2), 683–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang JY, Gavin AR, Richardson TS, Rowhani-Rahbar A, Siscovick DS, Hochner H,…Enquobahrie DA (2016). Accounting for life-course exposures in epigenetic biomarker association studies: Early life socioeconomic position, candidate gene DNA methylation, and adult cardiometabolic risk. American J of Epidemiol, 184(7), 520–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes A, Smart M, Gorrie-Stone T, Hannon E, Mill J, Bao Y,L,…Kumari M (2018). Socioeconomic position and DNA methylation age acceleration across the life course. Am J Epidemiol. 187(11), 2346–2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikram UZ, Snijder MB, Fassaert TJL, Schene AH Kunst AE, & Stronks K (2015). The contribution of perceived ethnic discrimination to the prevalence of depression. The European Journal of Public Health, 25(2), 243–248. [DOI] [PubMed] [Google Scholar]
- Janusek LW, Tell D, Gaylord-Harden N, & Mathews HL (2017). Relationship of childhood adversity and neighborhood violence to a proinflammatory phenotype in emerging adult African American men: An epigenetic link. Brain, Behavior, and Immunity 60, 126–135. [DOI] [PubMed] [Google Scholar]
- Kennedy BK, Berger SL, Brunet A, Campisi J, Cuervo A, Epel ES,…Sierra F (2014). Geroscience: Linking aging to chronic disease. Cell, 159(4), 709–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinger-König Hertel, J., Van der Auwera S, Frenzel S, Pfeiffer L, Waldenberger M,…Grabe HJ (2019). Methylation of the FKBP5 gene in association with FKBP5 genotypes, childhood maltreatment and depression. Neuropsychoparmacology 44, 930–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kneipp SM, Schwartz TA, Drevdahl DJ, Canales MK, Santacroce S, Santos HP Jr, & Anderson R (2018). Trends in health disparities, health inequity, and social determinants of health research: A 17-year analysis of NINR, NHLBI, and NIMHD funding. Nursing Research 67(3), 231–241. [DOI] [PubMed] [Google Scholar]
- Kogan SM, Cho J, Beach SRH, Smith AK, & Nishitani S (2018). Oxytocin receptor gene methylation and substance use problems among young African American men. Drug and Alcohol Dependence 192, 309–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krahn GL, Klein Walker D, & Correa-De-Araujo R (2015). Persons with disabilities as an unrecognized health disparity population. American Journal of Public Health 105(S2), S198–S206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger N (2005). Embodiment: A conceptual glossary for epidemiology. Journal of Epidemiology and Community Health 59, 350–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger N, Smith K, Naishadham D, Hartman C, & Barbeau E (2005). Experiences of discrimination: validity and reliability of a self-report measure for population health research on racism and health. Social Science & Medicine, 61(7),1576–1596. [DOI] [PubMed] [Google Scholar]
- Lam LL, Emberly E, Fraser HB, Neumann SM, Chen E, Miller GE, & Kobor MS (2012). Factors underlying variable DNA methylation in a human community cohort. PNAS, 109(2), 17253–17260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landecker H, & Panofsky A (2013). From social structure to gene regulation, and back: A critical introduction to environmental epigenetics for sociology. Annual Review of Sociology 39, 333–357. [Google Scholar]
- Landrine H & Klonoff EA (1996). The schedule of racist events: A measure of racial discrimination and a study of its negative physical and mental health consequences. Journal of Black Psychology 22, 144–168. [Google Scholar]
- Lawn RB, Anderson EL, Suderman M, Simpkin AJ, Gaunt TR, Teschendorff AE,…Howe LD (2018). Psychosocial adversity and socioeconomic position during childhood and epigenetic age: Analysis of two prospective cohort studies, Human Molecular Genetics, 27, 1301–08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei MK, Beach SRH, Simons RL, & Philibert RA (2015). Neighborhood crime and depressive symptoms among African American women: Genetic moderation and epigenetic mediation of effects, Social Science and Medicine, 146, 120–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link BG, & Phelan JC (1995). Social conditions as fundamental causes of disease. Journal of Health and Social Behavior, Extra Issue: Forty Years of Medical Sociology: The State of the Art and Directions for the Future, 80–94. [PubMed] [Google Scholar]
- Linnenbringer E, Geronimus AT, Davis KL, Bound J, Ellis L, & Gomez SL (2020). Associations between breast cancer subtype and neighborhood socioeconomic and racial composition among Black and White women. Breast Cancer Research and Treatment, 180: 437–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loucks EB, Huang Y, Agha G, Eaton CB, Gilman SE, Buka SL, & Kelsey KT (2016). Epigenetic mediators between childhood socioeconomic disadvantage and mid-life body mass index: The New England Family Study. Psychosomatic Medicine, 78, 1053–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ,…Visscher PM (2009). Finding the missing heritability of complex diseases. Nature 461(8), 747–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin AR, Kanai M, Kamatani Y, Okada Y, Neale BM, & Daly MJ (2019). Clinical Use of Current Polygenic Risk Scores May Exacerbate Health Disparities. Nature Genetics 51(4),584–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthew DB (2019). Two threats to precision medicine equity. Ethn Dis 29(3): 629–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massey DS, & Wagner B (2018). Segregation, Stigma, and Stratification: A Biosocial Model. In Major B, Dovidio JF, & Link BG (Eds.), the Oxford Handbook of Stigma, Discrimination, and Health (pp. 142–162). Oxford University Press. [Google Scholar]
- McCartney DL, Stevenson AJ, Walker RM, Gibson J, Morris SW, Campbell A,…Marioni RE (2018). Investigating the relationship between DNA methylation age acceleration and risk factors for Alzheimer’s disease. [DOI] [PMC free article] [PubMed]
- McClelland R, Christensen K, Mohammed S, McGuinness D, Cooney J, Bakshi A,…Shiels PG (2016). Accelerated ageing and renal dysfunction links lower socioeconomic status and dietary phosphate intake. Aging, 8(5), 1135–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCrory C, Fiorito G, Cheallaigh CN, Polidoro S, Karisola P, Alenius H,…& Kenny RA (2019). How does socio-economic position (SEP) get biologically embedded? A comparison of allostatic load and the epigenetic clock(s). Psychoneuroendocrinology 104, 64–73. [DOI] [PubMed] [Google Scholar]
- McDade TW, Hawkley L, Cacioppo JT (2006). Psychosocial and behavioral predictors of inflammation in middle-aged and older adults: The Chicago Health, Aging, and Social Relations Study. Psychosomatic Medicine, 68(3), 376–381. [DOI] [PubMed] [Google Scholar]
- McDade TW, Ryan C, Jones MJ, Maclsaac JL, Morin AM, Meyer JM,…& Kuzawa C (2017). Social and physical environments early in development predict DNA methylation of inflammatory genes in young adulthood. PNAS, 114(20), 7611–7616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen BS (1998). Protective and damaging effects of stress mediators. New England Journal of Medicine, 338(3), 171–179. [DOI] [PubMed] [Google Scholar]
- McGuinness D, McGlynn LM, Johnson PCD, MacIntyre A, Batty GD, Burns H,…& Shiels PG (2012). Socio-economic status is associated with epigenetic differences in the pSoBid cohort. Intl J. of Epi 41, 151–160. [DOI] [PubMed] [Google Scholar]
- Miller GE, Yu T, Chen E, & Brody GH (2015). Self-control forecasts better psychosocial outcomes but faster epigenetic aging in low-SES youth. PNAS, 112(33), 10325–10330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne RL, Fletcher AS, MacInnis RJ,…Giles GG (2017). Cohort profile: The Melbourne Collaborative Cohort Study (Health 2020). International Journal of Epidemiology, 1757–1757i. [DOI] [PubMed] [Google Scholar]
- Mitchell C, Schneper LM, & Notterman DA (2016). DNA methylation, early life environment, and health outcomes. Pediatric Research, 79(1), 212–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris TT, Davies NM, Hemani G, & Smith GD (2020). Population phenomena inflate genetic associations of complex social traits. Science Advances, 6(16), eaay0328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser DA, Paoloni-Giacobino A, Stenz L, Adouan W, Manini A, Suardi F,… Schechter S (2015). BDNF methylation and maternal brain activity in a violence-related sample. PLoS ONE, 10(12), e0143427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needham BL, Smith JA, Zhao W, Wang X, Mukherjee B, Kardia SL,…Diez Roux AV (2015). Life course socioeconomic status and DNA methylation in genes related to stress reactivity and inflammation: The multi-ethnic study of atherosclerosis, Epigenetics, 10, 958–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Institutes of Health All of Us Research Program (2020, December 23). Why is diversity important to the All of Us research program? https://allofus.nih.gov/about/diversity-and-inclusion
- Nuru-Jeter AM, Michaels EK, Thomas MD, Reeves AN, Thorpe RJ Jr., & LaVeist TA (2018). Relative roles of race versus socioeconomic position in studies of health inequalities: A matter of interpretation. Annual Review of Public Health 39, 169–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okbay A, Beauchamp JP, Fontana MA Lee JJ, Pers TH, Rietveld CA,…Benjamin DJ (2016). Genome-wide association study identifies 74 loci associated with educational attainment. Nature 533, 539–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olden K, Olden HA, & Lin Y (2015). The role of the epigenome in translating neighborhood disadvantage into health disparities. Current Environmental Health Reports 2, 163–170. [DOI] [PubMed] [Google Scholar]
- O’Neil A, Scovelle AJ, Milner AJ, & Kavanagh A (2018). Gender/sex as a social determinant of cardiovascular risk. Circulation 137, 854–864. [DOI] [PubMed] [Google Scholar]
- Peluso M, Bollati V, Munnia A, Srivatanakul P, Jedpiyawongse A, Sangrajrang S,…Baccarelli AA (2012). DNA methylation differences in exposed workers and nearby residents of the Ma Ta Phut industrial estate, Rayong, Thailand. Intl. J. of Epi, 41, 1753–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H, Zhu Y, Strachan E, Fowler E, Bacarus T, Roy-Byrne P,…Zhao J (2018). Childhood trauma, DNA methylation of stress-related genes, and depression: Findings from two monozygotic twin studies. Psychosom Med., 80(7), 599–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perroud N, Rutembesa E, Paoloni-Giacobino A, Mutubaruka J, Mutesa L, Stenz L,…Karege F (2014). The Tutsi genocide and transgenerational transmission of maternal stress: Epigenetics and biology of the HPA axis. the World Journal of Biological Psych. 15, 334–345. [DOI] [PubMed] [Google Scholar]
- Phelan JC, Link BG, & Tehranifar P (2010). Social conditions as fundamental causes of health inequalities: Theory, evidence, and policy implications. Journal of Health and Social Behavior 51(S), S28–S40. [DOI] [PubMed] [Google Scholar]
- Pittman GS, Wang X, Campbell MR, Coulter SJ, Olson JR, Pavuk M,…Bell DA (2020). Polychlorinated biphenyl exposure and DNA methylation in the Anniston Community Health Survey. Epigenetics, 15(4), 337–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popejoy AB, & Fullerton SM (2016). Genomics is failing on diversity. Nature 13(538): 161–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert Wood Johnson Foundation (2019, December 12) Our Focus Areas. https://www.rwjf.org/en/our-focus-areas.html
- Rudolph L, Caplan J, Ben-Moshe K, & Dillon L (2013). Health in all policies: a guide for state and local governments. American Public Health Association. [Google Scholar]
- Santos HP, Nephew BC, Bhattacharya A, Tan X, Smith L, Alyamani RAS,…Murgatroyd. (2018). Discrimination exposure and DNA methylation of stress-related genes in Latina mothers. Psychoneuroendocrinology, 98, 131–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schunck R, Reiss K, & Razum O (2015). Pathways between perceived discrimination and health among immigrants: evidence from a large national panel survey in Germany. Ethnicity & health, 20(5), 493–510. [DOI] [PubMed] [Google Scholar]
- Schür RR, van Leeuwen JMC, Houtepen LC, Joëls M, Kahn RS, Boks MP, & Vinkers CH (2018). Glucocorticoid receptor exon 1F methylation and the cortisol stress response in health and disease. Psychoneuroendocrinology, 97182–189. [DOI] [PubMed] [Google Scholar]
- Simmons D (2008) Epigenetic influence and disease. Nature Education 1(1), 6. [Google Scholar]
- Simons RL, Lei MK, Beach SRH, Cutrona CE, & Philibert RA (2017). Methylation of the oxytocin receptor gene mediates the effect of adversity on negative schemas and depression, Dev Psychopathol, 29, 725–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons RL, Lei MK, Beach SR, Philibert RA, Cutrona CE, Gibbons FX, & Barr A (2016). Economic hardship and biological weathering: The epigenetics of aging in a U.S. sample of black women, Soc Sci Med, 150, 192–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipahi L, Wildman DE, Aiello AE, Koenen KC, Galea S, Abbas A, & Uddin M (2014). Longitudinal epigenetic variation of DNA methyltransferase genes is associated with vulnerability to post-traumatic stress disorder, Psychol Med, 44, 3165–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JA, Zhao W, Wang X, Ratliff SM, Mukherjee B, Kardia SLR,…Needham BL (2017). Neighborhood characteristics influence DNA methylation of genes involved in stress response and inflammation: The Multi-Ethnic Study of Atherosclerosis, Epigenetics, 12, 662–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solar O, & Irwin A (2010) A conceptual framework for action on the social determinants of health. Social Determinants of Health Discussion Paper 2 (Policy and Practice). WHO. https://www.who.int/sdhconference/resources/ConceptualframeworkforactiononSDH_eng.pdf [Google Scholar]
- Song Y, Miyaki K, Suzuki T, Sasaki Y, Tsutsumi, Kawakami N,…Shimbo T (2014). Altered DNA methylation status of human brain derived neurotrophis factor gene could be useful as biomarker of depression. Am J Med Genet, 165B, 357–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, & von Vacano DA (2018). Reconsidering race: Social science perspectives on racial categories in the age of genomics. Oxford University Press. [Google Scholar]
- Stringhini S, Polidoro S, Sacerdote C, Kelly RS, van Veldhoven K, Agnoli C,…&Vineis P (2015). Life-course socioeconomic status and DNA methylation of genes regulating inflammation. Intl J of Epi, 44(4), 1320–1330. [DOI] [PubMed] [Google Scholar]
- Suderman M, Borghol N, Pappas JJ, Pereira SMP, Pembrey M, Hertzman C, & Szyf M (2014). Childhood abuse is associated with methylation of multiple loci in adult DNA. BMC Medical Genomics 7(13), 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syme SL (1987). Social determinants of disease. Annals of Clinical Research, 19(2), 44–52. [PubMed] [Google Scholar]
- Tajuddin S, Hernandez DG, Chen BH, Hooten NN, Mode N, Nails MA,…& Evans MK (2019). Novel age-associated DNA methylation changes and epigenetic age acceleration in middle-aged African Americans and whites. Clinical Epigenetics, 11(119): 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tehranifar P, Wu H, Fan X, Flom JD, Ferris JS, Cho YH,…& Terry MB (2013). Early life socioeconomic and genomic DNA methylation in mid-life. Epigenetics 8(1), 23–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thayer Z & Non AL (2015). Anthropology meets epigenetics: Current and future directions. American Anthropologist 117(4), 722–735. [Google Scholar]
- Toyooka S, Maruyama R, Toyooka KO, McLerran D, Feng Z, Fukuyama Y,…Gazdar AF (2003). Smoke exposure, histologic type and geography-related differences in the methylation profiles of non-small cell lung cancer. Int. J. Cancer, 103, 153–160. [DOI] [PubMed] [Google Scholar]
- Tung J,& Gilad Y (2013). Social environmental effects on gene regulation. Cellular and Molecular Life Sciences 70, 4323–4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin M, Aiello AE, Wildman DE Koenen KC, Pawelec G, de los Santos R,…& Galea S (2010). Epigenetic and immune function profiles associated with posttraumatic stress disorder. PNAS, 107(20), 9470–9475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin M, Galea S, Chang SC, Koenen KC, Goldmann E, Wildman DE, & Aiello AE (2013). Epigenetic signatures may explain the relationship between socioeconomic position and risk of mental illness: Preliminary findings from an urban community based sample. Biodemography Soc Biol, 59(1), 68–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velupillai YN, Packard CJ, Batty GD, Bezlyak V, Burns H, Cavanagh J…& Tannahill C (2008). Psychological, social and biological determinants of ill health (pSoBid): Study protocol of a population-based study. MC Public Health, 8(126), 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidrascu EM, Bashore AC, Howard TD, & Moore JB (2019). Effects of early- and mid-life stress on DNA methylation of genes associated with subclinical cardiovascular disease and cognitive impairment: a systematic review. BMC Medical Genetics 20(39), 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viruell-Fuentes E, Miranda PY, & Abdulrahim S (2012). More than culture: Structural racism, intersectionality theory, and immigrant health. Social Science & Medicine 75, 2099–2106. [DOI] [PubMed] [Google Scholar]
- Wadsworth M, Kuh D, Richards M, & Hardy R (2006). Cohort profile: The 1946 National Birth Cohort (MRC National Survey of Health and Development). International Journal of Epidemiology, 35, 49–54. [DOI] [PubMed] [Google Scholar]
- Williams DR, & Collins C (2001). Racial Residential Segregation: A Fundamental Cause of Racial Disparities in Health. Public Health Reports 116(5), 404–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams David R., & Mohammed SA (2013). Racism and Health I: Pathways and Scientific Evidence. American Behavioral Scientist, 57(8): 1152–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams DR,Y, Jackson J, & Anderson NB. 1997. Racial differences in physical and mental health: socioeconomic status, stress, and discrimination. Journal of Health Psychology, 2(3), 335–351. [DOI] [PubMed] [Google Scholar]
- World Health Organization. (2020, September. 1). Social Determinants of Health. https://www.who.int/social_determinants/en/
- Witte JS (2010). Genome-wide association studies and beyond. Annual Review of Public Health 31: 9–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y, Ding Y, Liu X, Chen X, Cheng S, Li L,…Wang Y (2014). Racial/ethnic disparities in human DNA methylation. Biochimica et Biophysica Acta 1846, 258–262. [DOI] [PubMed] [Google Scholar]
- Yehuda R, Daskalakis NP, Bierer LM, Bader HN, Klengel T, Holsboer F, & Binder EB (2016). Holocaust exposure induced intergenerational effects on FKBP5 methylation. Biological Psychiatry, 80, 372–380. [DOI] [PubMed] [Google Scholar]
- Yehuda R, Daskalakis N, Lehrner A, Desarnaud F, Bader HN, Makotkine I,…& Meaney MJ (2014). Influences of maternal and paternal PTSD on epigenetic regulation of the glucocorticoid receptor gene in Holocaust survivor offspring. Am J Psychiatry, 171(8), 872–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuberi T, Patterson EJ, & Stewart QT (2015). Race, Methodology and Social Construction in the Genomic Era. The ANNALS of the American Academy of Political and Social Science, 661, 109–127. [Google Scholar]


