Abstract
Thermoregulation of virulence genes in bacterial pathogens is essential for environment-to-host transition. However, the mechanisms governing cold adaptation when outside the host remain poorly understood. Here, we found that the production of cold shock proteins CspB and CspC from Staphylococcus aureus is controlled by two paralogous RNA thermoswitches. Through in silico prediction, enzymatic probing and site-directed mutagenesis, we demonstrated that cspB and cspC 5′UTRs adopt alternative RNA structures that shift from one another upon temperature shifts. The open (O) conformation that facilitates mRNA translation is favoured at ambient temperatures (22°C). Conversely, the alternative locked (L) conformation, where the ribosome binding site (RBS) is sequestered in a double-stranded RNA structure, is folded at host-related temperatures (37°C). These structural rearrangements depend on a long RNA hairpin found in the O conformation that sequesters the anti-RBS sequence. Notably, the remaining S. aureus CSP, CspA, may interact with a UUUGUUU motif located in the loop of this long hairpin and favour the folding of the L conformation. This folding represses CspB and CspC production at 37°C. Simultaneous deletion of the cspB/cspC genes or their RNA thermoswitches significantly decreases S. aureus growth rate at ambient temperatures, highlighting the importance of CspB/CspC thermoregulation when S. aureus transitions from the host to the environment.
INTRODUCTION
The ability of pathogens to adapt to the ever-changing environmental conditions and survive outside the host under different settings is crucial for their dissemination and transmission. Their persistence on inanimate surfaces increases the risk of new host colonization and outbreaks if no regular preventive measures are taken (1). Most pathogenic bacteria, such as Enterococcus spp., Staphylococcus aureus, Escherichia coli or Streptococcus pyogenes, can survive on dry surfaces for months (2–5).
In recent years, environmental contamination by S. aureus has been recognized as a potential threat to humans (6). Approximately 20–30% of the human population are S. aureus carriers, who are responsible for its spread and colonization of the surface of a wide variety of items. Some of these include bathroom sinks, remote controls, toilet seats, computers, towels and mobile phones. The ability of S. aureus to survive for long periods of time in such diverse environments has been linked to reinfections and new host colonizations (7,8). The adaptability and opportunistic nature of this bacterium allow it to cause both hospital- and community-acquired infections, which range from superficial skin abscesses or catheter-associated infections to more serious and life-threatening events such as surgical infections, osteomyelitis, septicemia, endocarditis or pneumonia. Moreover, its capacity to resist almost all available antibiotics makes it one of the most clinically relevant pathogens worldwide (9,10).
One of the main variables that bacteria need to face when leaving the host is the shift in temperature. In general, bacterial physiology is reprogrammed through the activation of adaptive systems in response to new temperatures, which may be sensed through conformational changes of protein, DNA and RNA molecules (11–13). Exposure to low temperatures leads to the induction of members of the cold shock protein (CSP) family, which modulate changes in cell size, wall thickness and metabolic homeostasis (14–16). The cold shock adaptation process has been largely investigated in the Gram-negative bacterial model E. coli, which encodes nine CSPs (17–20). When simultaneously lacking four out of these nine variants, E. coli shows a growth defect at cold temperatures (19). In Gram-positive bacteria such as Listeria monocytogenes, csp genes are significantly induced in cold-adapted cells and bacterial growth is completely abolished in a cspA mutant at 4°C (21). A similar phenomenon has also been reported for Bacillus subtilis, Lactococcus lactis and S. aureus, when lacking one or specific combinations of the csp genes (22–26).
It is believed that post-transcriptional mechanisms drive the cold shock response in bacteria (27–29). When E. coli cells are exposed to cold, protein translation is severely decreased due to mRNA structural reorganizations that prevent optimal ribosome progression; hence, bacterial growth is arrested for about 6 h (20,30). Importantly, in contrast to other proteins, CspA production is drastically increased shortly after the temperature drops. This occurs due to rearrangements in its highly structured 5′UTR that result in a more favourably translated conformation. Once CspA reaches a sufficient concentration inside the cell, it destabilizes RNA structures that would otherwise impair translation at low temperatures (20,28,30,31). The presence of alternative structures in the E. coli cspA 5′UTR at 37°C and at cold temperatures (10 and 20°C) has been demonstrated through enzymatic and chemical probing as well as site-specific mutagenesis studies (20,28). This supports the idea of the cspA mRNA acting as an RNA thermosensor that adopts a less compact and more actively translated conformation at ambient temperatures, as opposed to the classical RNA thermometers, which generally favour translation at higher host-related temperatures (20,28,32). Interestingly, a recent report showed the presence of a similar RNA thermoswitch in the 5′UTR of the L. monocytogenes cspA mRNA that controls the expression of CspA through temperature-dependent alternative RNA structures (33). In addition, the increased translation of other CSPs at low temperatures is known to be modulated via their 5′UTRs; however, the molecular mechanisms responsible for it remain poorly understood (18,20,31,34,35).
The S. aureus genome encodes three CSP paralogues (CspA, CspB and CspC) with a protein identity >70% between them (36). It has been shown that the cspB mRNA levels are induced after cold shock (25,26), while the cspA mRNA levels remain unaltered by shifts in the temperature (37). Nevertheless, as it is the case with most bacteria, there is no knowledge of how CSP expression is coordinated at the protein level and whether they are required for S. aureus adaptation to environmental changes. In this study, we demonstrate that the production of two out of the three S. aureus CSPs is activated through a drop in temperature. This occurs when the pathogen transitions from host-related (37°C) to ambient temperatures (e.g. 22 and 28°C). The thermoregulation of CspB and CspC occurs at the post-transcriptional level through the action of two paralogous thermosensitive RNA elements located at the 5′UTR of the cspB and cspC mRNAs, respectively. We also show that the temperature-dependent repression of CspB and CspC requires the action of CspA. In addition, we predict that similar RNA thermoswitches may be involved in the control of CSP expression in several bacterial species.
MATERIALS AND METHODS
Strains, plasmids, oligonucleotides and growth conditions
The bacterial strains, plasmids and oligonucleotides used in this study are listed in Supplementary Tables S1–S3, respectively. Staphylococcus aureus strains were grown in Mueller–Hinton (MH) broth (Sigma-Aldrich), whereas E. coli was grown in LB broth (Pronadisa). The B2 (casein hydrolysate, 10 g/l; yeast extract, 25 g/l; NaCl, 25 g/l; K2HPO4, 1 g/l; glucose, 5 g/l; pH 7.5) and SuperBroth (tryptone, 30 g/l; yeast extract, 20 g/l; MOPS, 10 g/l; pH 7) media were used to prepare S. aureus and E. coli competent cells, respectively. For selective growth, media were supplemented with the appropriate antibiotics at the following concentrations: erythromycin, 1.5 μg/ml or 10 μg/ml; ampicillin, 100 μg/ml.
RNA structure predictions
To address the alternative conformations of the 5′UTRs, RNA structure predictions were carried out using both the last and the previous 2.3 mfold versions found at http://mfold.rna.albany.edu/?q=mfold/RNA-Folding-Form (38). Different 5′UTR lengths were used as queries and the default parameters were applied. All structures predicted by mfold were inspected and compared. Vienna format files from prediction results of interest were downloaded and used to draw the putative RNA structures using the VARNA software (39). Note that during the revision of this manuscript, the mfold web server was discontinued. If this situation remains unresolved over time, researchers aiming to predict alternative structures in their CSP of interest and/or reproduce our results should use RNAstructure web server (https://rna.urmc.rochester.edu/RNAstructureWeb/Servers/Predict1/Predict1.html) instead. We have obtained similar results with both web servers.
Plasmid constructions
The plasmids used in this study (Supplementary Table S2) were engineered as previously described (40). Briefly, the PCR fragments were amplified from chromosomic or plasmidic DNA with the DreamTaq DNA polymerase (Thermo Scientific) using the appropriate oligonucleotides listed in Supplementary Table S3. The PCR products were purified from agarose gels using the Macherey-Nagel NucleoSpin Gel and PCR Clean-up Kit (Thermo Scientific). The purified products were then ligated into the cloning vector pJET 1.2 (Thermo Scientific) and used to transform the E. coli XL1-blue or IM01B cells. DNA fragments were excised using the corresponding FastDigest restriction enzymes (Thermo Scientific) and ligated into the final vector with the Rapid DNA Ligation Kit (Thermo Scientific). All constructs were verified by Sanger sequencing and electroporated into S. aureus competent cells (41).
The pMAD plasmids (pMAD_cspB3xF, pMAD_cspC3xF, pMAD_Δ24cspB, pMAD_Δ24cspC, pMAD_Δ24cspB3xF, pMAD_Δ24cspC3xF) were used for chromosomal mutations, either to insert the 3xFLAG tag or to delete the first 24 nucleotides from the cspB and cspC mRNAs, respectively. Briefly, the flanking regions (AB and CD) from the target genes were amplified using specific oligos A/B and C/D (Supplementary Table S3). The flanking regions were amplified by an overlapping PCR using primers A and D and ligated into pJET. The resulting vectors were digested and the fragments corresponding to the flanking regions were inserted into a previously excised pMAD vector.
The pHRG plasmid expressed the gfp gene under the control of the Phyper promoter (42), whose sequence was adapted from the promoter that drives transcription of early genes in the SP01 B. subtilis bacteriophage to include an EcoRI site upstream of the native transcriptional start site (43). This plasmid was constructed by PCR amplification using the pCN57 plasmid (44) as a template and oligonucleotides phyper-RBSicaR-GFP and GFPend-AscI (Supplementary Table S3). The amplified product was ligated into pJET. The resulting vector was digested using SphI and AscI and the fragments of interest were inserted into the pCN47 plasmid (44). Plasmids p5′UTRcspB-gfp and p5′UTRcspC-gfp were generated by amplifying the 5′UTR sequences from the pCspB3xF and pCspC3xF plasmids (36) with oligonucleotide pairs 5UTR_CspB_FW_EcoRI/5UTR_CspB_RV_SpeI and 5′UTR_CspC_FW_EcoRI/5′UTR_CspC_RV_SpeI (Supplementary Table S3), respectively. The amplified PCR products were then digested using the EcoRI and SpeI enzymes and ligated into pHRG. Similarly, the plasmids for mutating 5′UTRs were constructed by PCR amplification from p5′UTRcspB-gfp or p5′UTRcspC-gfp using the corresponding 5′UTR mutated forward oligonucleotides (Supplementary Table S3) and 5UTR_CspB_RV_SpeI or 5′UTR_CspC_RV_SpeI (Supplementary Table S3). The amplified fragments were then digested with EcoRI/SpeI and ligated into the pHRG plasmid. The resulting vectors were named p5′UTRcspBΔ24-gfp, p5′UTRcspCΔ24-gfp, p5′UTRcspBUAU47AA-gfp, p5′UTRcspBC50G-gfp, p5′UTRcspBUU55AA-gfp, p5′UTRcspBUU55AA+UU26AA-gfp, p5′UTRcspCUU48A-gfp and p5′UTRcspBU38C+U41C-gfp (Supplementary Table S2).
Generation of csp mutants by homologous recombination
The csp mutant strains were obtained by marker-less homologous recombination, using the pMAD plasmid system (45) as previously described (36). Briefly, the cspB3xF, cspC3xF, cspB3xFΔcspA, cspC3xFΔcspA, ΔcspBC, Δ24cspB, Δ24cspC, Δ24cspB3xF, Δ24cspC3xF and Δ24cspBC strains were generated by a two-step procedure that inserts or replaces a gene of interest by a mutant allele contained within the pMAD plasmids (Supplementary Table S2) (46). The resulting mutant strains were confirmed by PCR using the corresponding oligonucleotides E and F (Supplementary Table S3). In addition, the PCR fragments were verified by Sanger sequencing.
Bacterial cultures for total protein extraction and western blotting
Staphylococcus aureus cultures were grown in Erlenmeyer flasks containing MH broth supplemented with 10 μg/ml erythromycin. Media were inoculated (1:250) with normalized bacterial aliquots, which were previously grown at 37°C and 200 rpm overnight. Bacterial cultures were incubated at different temperatures (22, 28 and 37°C) and 200 rpm until cells reached an early exponential phase (OD600nm = 0.4). Cultures were harvested by centrifugation for 10 min at 4400 × g and 4°C. Bacterial pellets were stored at −80°C until required. Pellets were thawed and washed with 700 μl of 1× phosphate-buffered saline (PBS). Next, bacteria were placed in Lysing Matrix B tubes containing acid-washed 100 μm glass beads (Sigma) and lysed for 45 s and speed 6 using a FastPrep-24 instrument (MP Biomedicals). This step was repeated once and the tubes were centrifuged for 10 min at 21 000 × g and 4°C. The supernatants containing the total protein extracts were transferred to Eppendorf tubes. Total proteins were quantified using a Bradford protein assay (Bio-Rad) and samples were prepared at the desired concentration in Laemmli buffer. Samples were stored at −20°C until needed.
Western blot analyses were performed as previously described (36). The chromosomally 3xFLAG-tagged CSPs were developed with anti-FLAG antibodies (Sigma) and the GFP samples with monoclonal anti-GFP antibodies 1:5000 (Living Colors, Clontech) and peroxidase-conjugated goat anti-mouse immunoglobulin G and M antibodies 1:2500 (Pierce-Thermo Scientific). Membranes were developed in a ChemiDoc Imaging System using the SuperSignal West Pico Chemiluminescent Substrate Kit (Thermo Scientific). Protein levels of three independent replicates were quantified by measuring band intensity with ImageJ (https://imagej.nih.gov/ij/).
RNA extraction and northern blotting
Bacteria were cultured as mentioned in the previous section. Cultures were centrifuged for 3 min at 4400 × g and 4°C. Pellets were then frozen in liquid nitrogen and stored at −80°C until needed. RNA extractions were performed as previously described (47). An appropriate amount of RNA was mixed with formaldehyde loading dye (Ambion), denatured for 15 min at 65°C and run in 1.25% agarose gels with Midori Green (Teknovas). Gels were checked for RNA integrity and appropriate loading under UV light exposure. RNAs were then transferred to 0.2 μm pore size Nitran N membranes (GE Healthcare Life Sciences) by capillarity using the NorthernMax Transfer Buffer (Ambion), for 1.5 h at room temperature. The transferred RNAs were cross-linked to the membranes by exposing them to UV light with a UV Stralinker 1800 (Stratagene). Membranes were then prehybridized using ULTRAHyb solution (Ambion) for at least 30 min at 40°C in a rotating oven. After the prehybridization step, the corresponding oligonucleotide probes (anti_3xFLAG_probe or anti-GFP probe) (Supplementary Table S3) were radioactively labelled with [γ-32P]-ATP (Perkin Elmer) following the manufacturer’s recommendation. Probes were then purified with Illustra MicroSpin G-50 columns (GE Healthcare) and incubated with membranes at 40°C overnight. Membranes were washed three times for 5 min with preheated 2× SCC, 0.1% SDS at 40°C followed by at least two washing steps of 0.1× SCC, 0.1% SDS at room temperature until the background signal was non-detectable. Membranes were developed by autoradiography for different time periods. The mRNA levels from three independent replicates were measured by densitometry of northern blot autoradiographies using ImageJ (https://imagej.nih.gov/ij/).
Molecular beacon assay
The molecular beacon (MB) assay was performed as previously described (48) with the following modifications. Briefly, the cspB and cspC ssDNA oligonucleotides (Supplementary Table S3) labelled with the 6-FAM and quencher Iowa Black FQ (IBkFQ) molecules at the 5′ and 3′ ends, respectively, were purchased from Integrated DNA Technologies. The cspA ssDNA oligonucleotide (40) was used as a control. Two picomoles of ssDNA oligonucleotides were mixed with 2.5 μl of 10× reaction buffer (100 mM Tris–HCl, pH 7.5, 300 mM KCl, 200 mM NH4Cl, 15 mM DTT, 50 mM MgCl2) and Milli-Q water up to a final volume of 25 μl. The FAM fluorescence was then measured using the Bio-Rad CFX96 Touch Real-Time PCR Detection System and the following incubation periods: 13°C, 10 min; 18 cycles of 5 min each starting at 16°C and with a 3°C increase per cycle; 5 min at 65°C; and 18 cycles of 5 min each with a 3°C decrease per cycle. FAM emission was registered throughout the incubation steps. Three independent replicates were performed.
In vitro transcription
PCR fragments containing the T7 promoter were amplified using the oligonucleotides listed in Supplementary Table S3. In vitro transcription was performed at 37°C using the Megascript T7 transcription kit (Ambion) following the manufacturer’s recommendation. RNAs were loaded into a 6% polyacrylamide gel and visualized with UV light after Midori Green (Teknovas) staining. Bands of the appropriate size were cut and the RNA was extracted using 0.5 M ammonium acetate (pH 5.5) at 15°C for 2 h. Next, 1 volume of acid phenol (pH 4.5) was added and samples were incubated at 15°C overnight. After centrifugation for 5 min at 21 000 × g and 4°C, RNAs were purified using phenol–chloroform and precipitated by adding 3 volumes of 96% ethanol and 1/10 volume of 3 M sodium acetate (pH 5.5). The mixture was incubated at −80°C for at least 1 h. Samples were then centrifuged for 30 min at 21 000 × g and 4°C. Pellets were washed with 70% ethanol, air dried and resuspended in DEPC water (Ambion). The RNA integrity was checked through agarose gel visualization and quantified by the Nanodrop system (Agilent Technologies).
5′-end labelling of RNA
Before labelling, RNAs were dephosphorylated using the FastAP enzyme (Thermo Scientific) at 37°C for 15 min. Inactivation of the enzyme was performed by heat shock at 75°C for 5 min followed by ethanol precipitation supplemented with 1 μl of GlycoBlue™ (Thermo Scientific) coprecipitant. Pellets were resuspended in 20 μl of DEPC water. Dephosphorylated nucleic acids were incubated with [γ-32P]-ATP (Perkin Elmer) and 0.4 U of PNK (Thermo Scientific) for 1 h. Labelled RNAs for enzymatic probing were purified by electrophoresis on a 6% polyacrylamide denaturing gel (8 M urea). The bands of interest were cut from the gel and the RNAs eluted with 400 μl of elution buffer (500 mM AcNH4, 1 mM EDTA and 0.1% SDS) at 4°C overnight. RNAs were then precipitated as previously described and resuspended in 80 μl DEPC water.
Enzymatic probing
The enzymatic probing experiments with RNase T1 and S1 (Thermo Scientific) were performed to determine the in vitro cspB and cspC 5′UTR structures. Labelled RNAs were first denatured (1 min at 90°C followed by 1 min in ice) and then renatured at 22 or 37°C for 15 min. Two picomoles of labelled RNA was incubated in T1 (RNA structure buffer) or S1 buffer (Thermo Scientific) supplemented with 20 μg yeast tRNA in the presence of different enzyme dilutions (T1: 1 × 10−3, 2 × 10−2 and 5 × 10–2 U/μl; S1: 5 × 10−5, 2 × 10−5 and 1 × 10−5 U/μl) for 5 min at 22 or 37°C. In parallel, alkaline hydrolysis and T1 controls were performed to assign the gel bands. Alkaline hydrolysis was accomplished by mixing 2 pmol of labelled RNA with 20 μg of yeast tRNA and 1× alkaline hydrolysis buffer (Thermo Scientific). The mix was then incubated at 95°C for 10 min. For the denaturing T1 reaction, 2 pmol of labelled RNA was preheated with 20 μg yeast tRNA and 1× RNA structure buffer for 3 min at 65°C. Samples were then incubated with 2.5 × 10−2 U/μl of RNase T1 for 1 min 10 s. All reactions were stopped by addition of 40 μl of the Stop Mix solution (0.6 M NaOAc, 4 mM EDTA, 0.1 mg/ml yeast tRNA and 1 μg GlycoBlue) and precipitated with ethanol. The pellets were washed twice with 70% ethanol, air dried and resuspended in 12 μl of loading buffer II (Thermo Scientific). Samples were loaded into a 12% polyacrylamide denaturing gel.
Bacterial growth in TSA plates
For the phenotypic growth experiments in trypticase soy agar (TSA) plates, bacteria were grown in 5 ml of MH broth at 37°C and 200 rpm overnight. The bacterial preinocula were then normalized to an OD600nm of 1. Serial dilutions were plated on TSA plates and incubated at 37°C for 24 h or at 22°C for 72 h.
Survival experiment in synthetic nasal medium
The synthetic nasal medium (SNM3) was prepared as previously described (49) and used for survival experiments of different S. aureus strains. Bacteria were grown in 5 ml of MH broth at 37°C and 200 rpm overnight. One millilitre of the preinoculum was centrifuged, washed with 1× PBS and resuspended in 1 ml of SNM3. Bacteria were then diluted 1:250 in 10 ml of SNM3 and grown at 32°C and 200 rpm for 2 days. Next, bacteria were inoculated in fresh SNM3 and grown at 22°C in static conditions in a refrigerated Infors HT Ecotron incubator. Samples were taken at different time points and serial dilutions were plated in TSA. Colony forming units (CFUs) were counted from three independent biological replicates.
Statistical analyses
Statistical significance was determined with the Mann–Whitney U test by using GraphPad Prism software. Results were considered significant if the P-value was <0.05, which was indicated as an asterisk in the corresponding plots.
RESULTS
Ambient temperatures induce the production of S. aureus CspB and CspC
In order to monitor the expression patterns of CSPs when S. aureus grows at different temperatures, we chromosomally tagged the csp genes by fusing a 3xFLAG epitope to the C-terminal end. We then incubated the tagged strains in MH medium at 37°C, which is the host-related temperature, and at 22 and 28°C, two different ambient temperatures that S. aureus may face when outside the host. Once the bacterial cultures reached the exponential phase (OD600nm = 0.4), we extracted their total RNAs and proteins to perform northern and western blot experiments. The results showed that the mRNA expression levels of all three csp genes were not significantly affected by changes in the growth temperature (Figure 1A). In agreement with this, when performing western blots, we observed that CspA3xF levels remained invariable, regardless of the growth temperature (Figure 1B). In contrast, CspB3xF and CspC3xF levels were significantly reduced at 37°C when compared to those produced at 22 and 28°C. Note that the highest CspB3xF and CspC3xF protein yields were found at 22°C (Figure 1B). The lack of correlation between the mRNA and protein levels of S. aureus CspB and CspC when grown at different temperatures, indicated the presence of putative post-transcriptional regulatory mechanisms controlling the production of both proteins.
Figure 1.
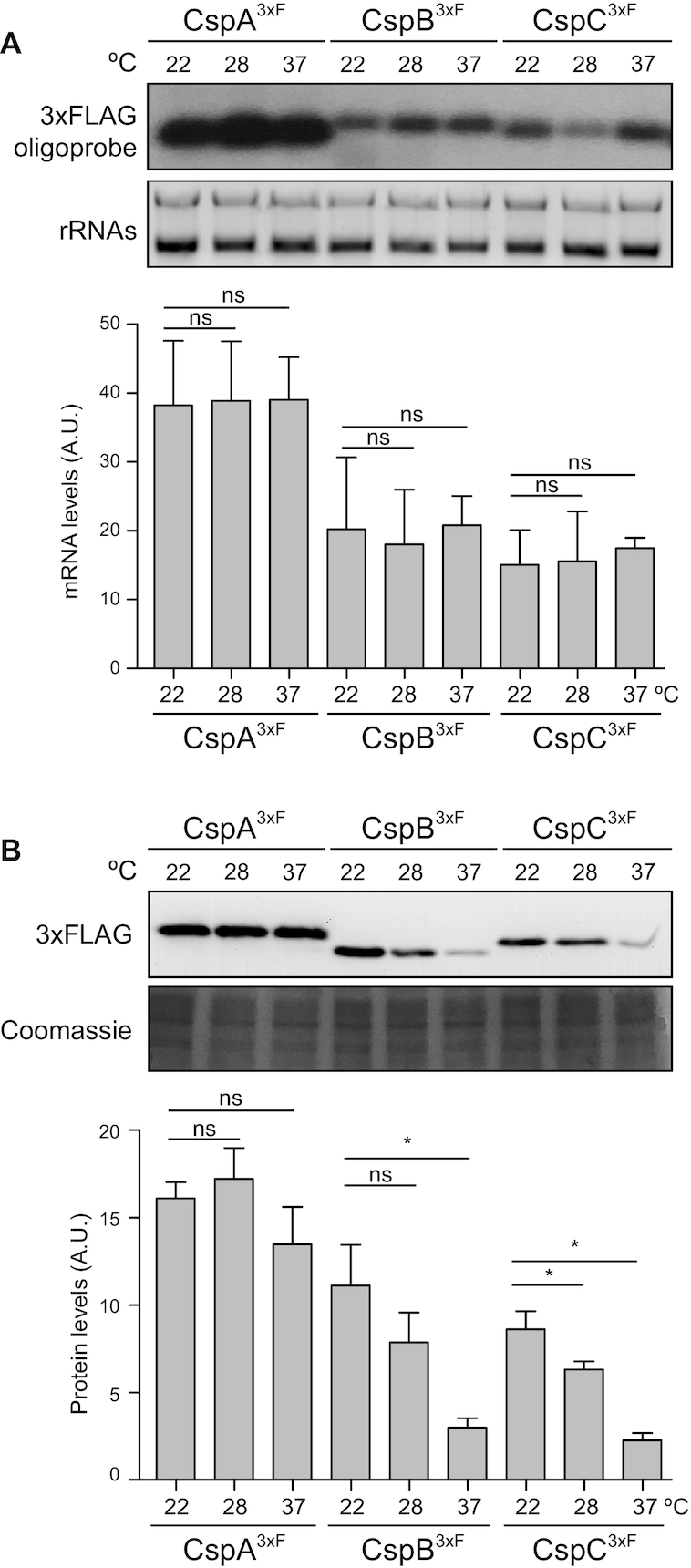
The CspB and CspC protein levels are differentially regulated in response to environmental temperatures. (A) Northern blot showing the csp mRNA levels, which were detected using a radiolabelled antisense 3xFLAG oligonucleotide probe. Midori Green-stained ribosomal RNAs are included as loading controls. (B) Western blot showing the levels of chromosomally 3xFLAG-tagged CSPs, which were developed using peroxidase-conjugated anti-FLAG antibodies and a bioluminescence kit. A Coomassie stained protein gel portion is included as a loading control. Strains were grown until exponential phase (OD600nm = 0.4) in MH broth at environmental (22 and 28°C) and host-related (37°C) temperatures. Bar plots represent the mean and standard deviation of the mRNA and protein levels from three independent biological replicates, which were determined by densitometry of mRNA and protein bands using ImageJ (https://imagej.nih.gov/ij/). Asterisks represent statistical significance (P < 0.05, Mann–Whitney U test); ns, not significant. Representative images from the triplicates are shown.
Thermoregulation of CspB and CspC is conducted through their 5′UTRs
The existence of post-transcriptional mechanisms that control S. aureus CSPs was supported by previous results from our group that showed that CSPs are differentially produced when expressed from the same heterologous promoter (36). Interestingly, our previous transcriptomic maps showed that the cspB and cspC mRNAs carried long 5′UTRs of 112 and 113 nucleotides, respectively (36,50). Since the 5′UTRs of CSPs in other bacterial models have been reported to undergo thermoregulation (20,23,24,28,33), we tested whether this was the case for the cspB and cspC 5′UTRs in S. aureus. Therefore, we fused both 5′UTRs plus the first five codons of the cspB and cspC mRNAs to the ATG-less gfp gene of the pHRG plasmid, generating the translational reporter plasmids p5′UTRcspB-gfp and p5′UTRcspC-gfp, respectively. Note that both reporter gene fusions were under the control of the constitutive promoter Phyper (42), whose sequence was adapted to include an EcoRI site upstream of the native transcriptional start site. We used these plasmids to transform the S. aureus wild-type (WT) strain and monitored both the chimeric mRNA levels and GFP protein production. Northen blots results shown in Figure 2A indicated that the chimeric 5′UTRcspB-gfp and 5′UTRcspC-gfp mRNA levels did not significantly change, regardless of the tested temperature (Figure 2A). In contrast, western blots evidenced higher GFP levels at 22 and 28°C when compared to 37°C (Figure 2B). These results confirmed that translation of both CspB and CspC proteins was affected by the environmental temperature and that their thermoregulation was dependent on their 5′UTRs.
Figure 2.
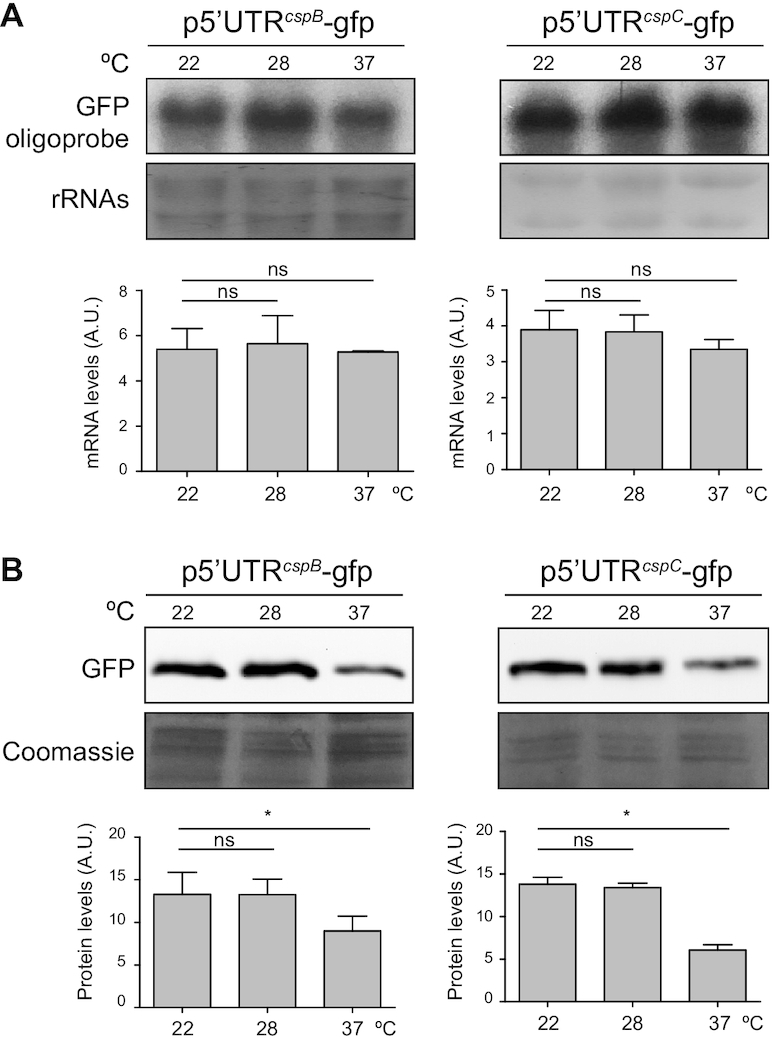
Thermoregulation of the CspB and CspC production is restricted to the cspB and cspC 5′UTRs, respectively. (A) Northern blots showing the 5′UTR-gfp chimeric mRNA levels expressed from the corresponding translational reporter strains. Bacteria were grown in MH broth supplemented with the appropriate antibiotic and incubated at 22, 28 and 37°C until exponential phase was reached. Midori Green-stained ribosomal RNAs are included as loading controls. (B) Western blots showing the GFP protein levels from the strains expressing the cspB and cspC 5′UTR-GFP reporter fusions. Membranes were developed using monoclonal anti-GFP, peroxidase-conjugated goat anti-mouse antibodies and a bioluminescent kit. Coomassie stain gel portions are included as loading controls. Bar plots represent the mean and standard deviation of mRNA and protein levels from three independent biological replicates, which were determined by densitometry of mRNA and protein bands using ImageJ (https://imagej.nih.gov/ij/). Asterisks represent statistical significance (P < 0.05, Mann–Whitney U test); ns, not significant. Representative images from the triplicates are shown.
The cspB and cspC 5′UTRs adopt mutually exclusive alternative RNA structures
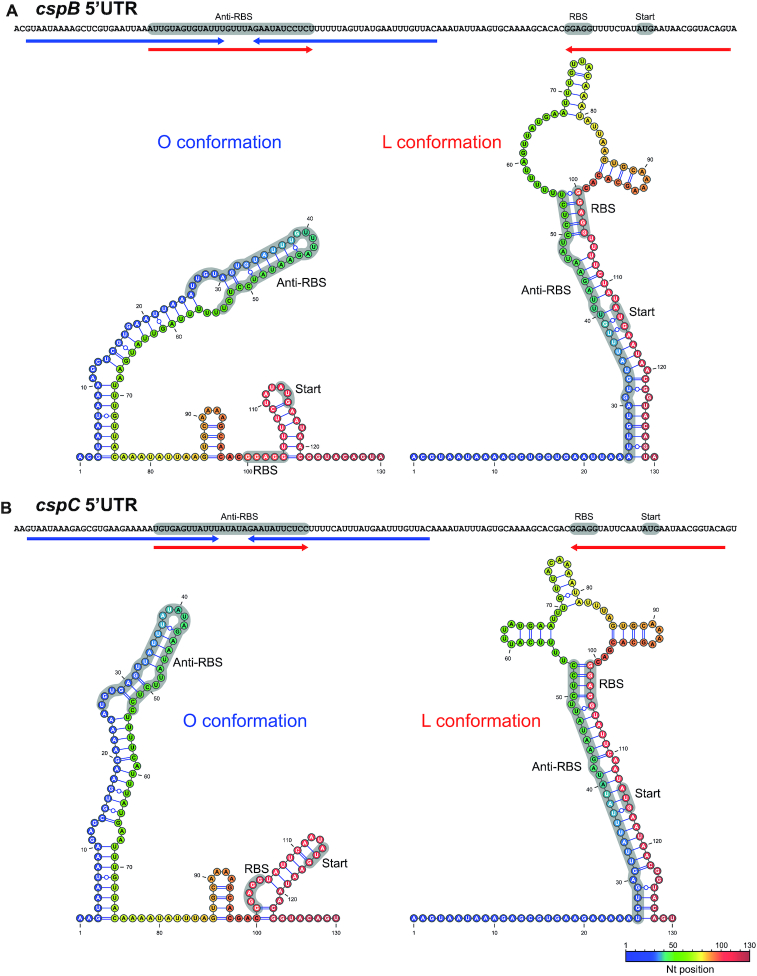
Recently, it was shown that the cspA 5′UTRs of E. coli and L. monocytogenes contain thermoresponsive secondary structure elements, which act as thermosensors that promote CspA expression at low temperatures (20,33). Using the mfold web server (38), we predicted that the cspB and cspC 5′UTRs (the first 130 nucleotides) in S. aureus can fold into two major alternative conformations. Despite minor differences, the cspB and cspC 5′UTRs displayed extremely similar alternative conformations (Figure 3). One of such conformations included a long and imperfect hairpin that comprised nucleotides ∼3–76 while leaving the ribosome binding site (RBS) exposed. We defined this folding as the ‘open’ (O) conformation. In contrast, the alternative predicted configuration formed a double-stranded structure through base pairing of nucleotides ∼25–54 with nucleotides ∼100–129 (nucleotide stretch that includes the RBS region). This nucleotide interaction prevented the utilization of the RBS by the translation machinery. For this reason, we named this secondary structure ‘locked’ (L) conformation (Figure 3). Both alternative conformations were orthologous to the one recently described for the cspA 5′UTR of L. monocytogenes, despite the lack of sequence similarity between both species (33).
Figure 3.
Putative alternative structures adopted by the cspB and cspC 5′UTRs. RNA structures were predicted using the mfold web server tool (38) and visualized and drawn with the VARNA software (39). Colours represent nucleotide positions within the RNA. Blue and red arrows below the 5′UTR sequences indicate the interacting nucleotide regions in the O and L conformations, respectively. The RBS sequence, the start codon and the anti-RBS region appear as grey shaded areas.
Considering that the 5′UTRs of cspB and cspC seemed affected by temperature (Figure 2B), we reasoned that such effect might be explained by shifts between the two alternative conformations (O and L) upon temperature changes, as previously described (20,33). Hypothetically, the O conformation would be favoured at lower temperatures, promoting CspB and CspC translation, while the L conformation would be more dominant during S. aureus growth at 37°C. To validate this idea, we performed in vitro enzymatic structural probing of the cspB and cspC 5′UTRs at 22 and 37°C using RNases T1 and S1, which cleave unpaired G and A > U > C, respectively (Supplementary Figures S1 and S2). The enzymatic cleavage patterns of the cspC 5′UTR illustrated the differences between both structures more clearly than those of the cspB 5′UTR. The cspC 5′UTR showed an increased sensitivity to RNase S1 and T1 reactivities between nucleotides 7–11, 19–24, 65 and 72 at 37°C while only few RNase cleavages (∼42–43) could be observed at 22°C, indicating that the region comprising the first ∼60–70 nucleotides adopted a double-stranded configuration at such temperature (Supplementary Figure S2). Regarding the cspB 5′UTR probing, RNAse S1 cleavage patterns were similar to those found in cspC, suggesting that both 5′UTRs might be analogously reorganized in function of temperature changes (Supplementary Figure S1). Note that although several of the RNase T1 processing correlated with the expected structure at the corresponding temperature, there were some minor cleavages that suggested that both structures might coexist at some point in vitro. The existence of two alternative structures in each of the cspB and cspC 5′UTRs indicated that they could also work as thermosensitive RNA elements to control protein expression.
The hairpin of the O conformation acts as a thermoresponsive element
The long hairpin found in the O conformation appeared to be critical for the reorganization of the cspB and cspC 5′UTR structures. Since it sequestered the anti-RBS region, this would imply that the hairpin would need to unfold itself to release the anti-RBS and block CspB/CspC translation (Figure 3). Seeing that such mechanism would apparently respond to temperature changes, it seemed possible for the hairpin itself to behave as a thermoresponsive element. To explore this further, we used a molecular beacon (MB) system, as previously described (40,48). The MB consisted of ∼75-mer ssDNA oligonucleotides that were designed to mimic the long hairpin found in the O conformation of the cspB and cspC 5′UTRs. Additionally, the 5′ and 3′ ends of the oligonucleotides carried the carboxyfluorescein fluorescent dye (FAM) and the IBkFQ quencher, respectively (Figure 4A). If the MB folded at low temperatures as expected, both molecules would fall in close proximity to each other, preventing fluorescence emission (48). We began by incubating the cspB and cspC MBs at 16°C for 5 min and then increased the temperature in intervals of 3°C every 5 min until 65°C were reached. After 5 min of incubation at 65°C, we gradually decreased the temperature using the same intervals. We monitored the fluorescence emission during the entire process and included the MB that mimicked the cspA 5′UTR hairpin as a control (40), since it was not dependent on temperature changes (Figure 1B). The results showed that fluorescence emission could not be registered at 16°C for the cspB and cspC MBs, indicating that at this temperature they were folded as expected. When reaching temperatures above ∼31°C, FAM fluorescence was registered, with a maximum peak at ∼43°C (Figure 4B). Correspondingly, as the temperature decreased, so did the fluorescence emission and once below 30°C no signal was detected, indicating that the hairpin structures had been restored (Figure 4B). All in all, these results showed that the cspB and cspC hairpins of the O conformation unfolded at host-related temperatures, which range from 30–33°C (human nose temperature) to 37–41°C (human infection temperature) (10,51). In contrast, the cspB and cspC MBs were completely folded (no fluorescence emission was detected) when exposed to ambient temperatures (below 30°C). Regarding the cspA MB control, it melted and emitted fluorescence when incubated above 43°C, which does not correlate with optimal growth conditions for S. aureus. Altogether, these data strongly suggested that the O conformation folds in a temperature-dependent manner, compatible with a model in which the RBSs of cspB and cspC are sequestered at host temperatures while remaining free at ambient (lower) temperatures.
Figure 4.
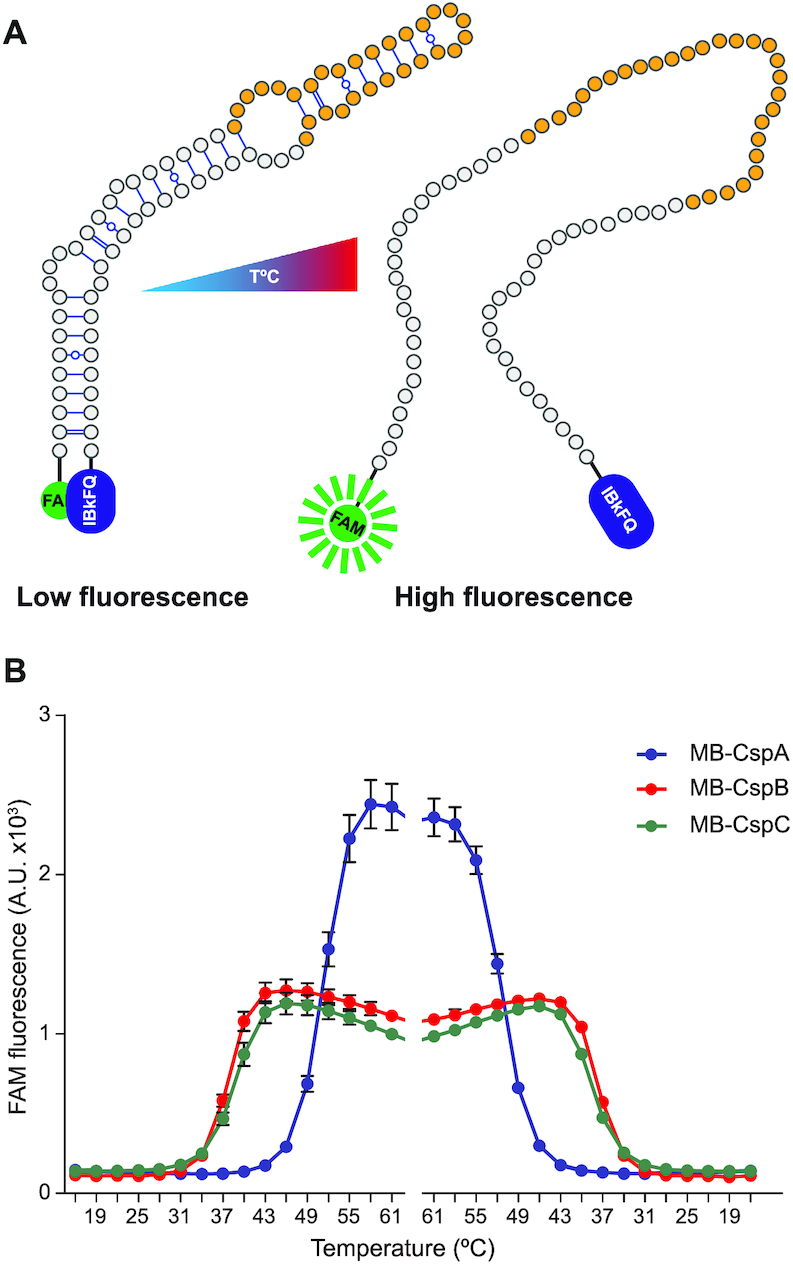
The cspB and cspC hairpins from the O conformation could work as thermoresponsive RNA elements. (A) Schematic representation of the MB design that mimics the stem loop of the O conformation from the cspB and cspC 5′UTRs. The 5′ and 3′ ends harbour the FAM and IBkFQ molecules, respectively. Fluorescence emission occurs when the FAM fluorophore is separated from the IBkFQ quencher. (B) Detection of the FAM fluorescence from MB-CspB (red), MB-CspC (green) and MB-CspA (blue) in function of the temperature. The MBs were equilibrated at 13°C for 10 min and then the temperature was increased 3°C every 5 min. After 5 min of incubation at 65°C, the temperature was gradually decreased following the inverse pattern. Fluorescence was registered for the duration of the experiment. Means and standard deviations from three independent replicates are shown.
Mutations in the cspB and cspC 5′UTRs confirm thermosensitive 5′UTR structural reorganizations in vivo
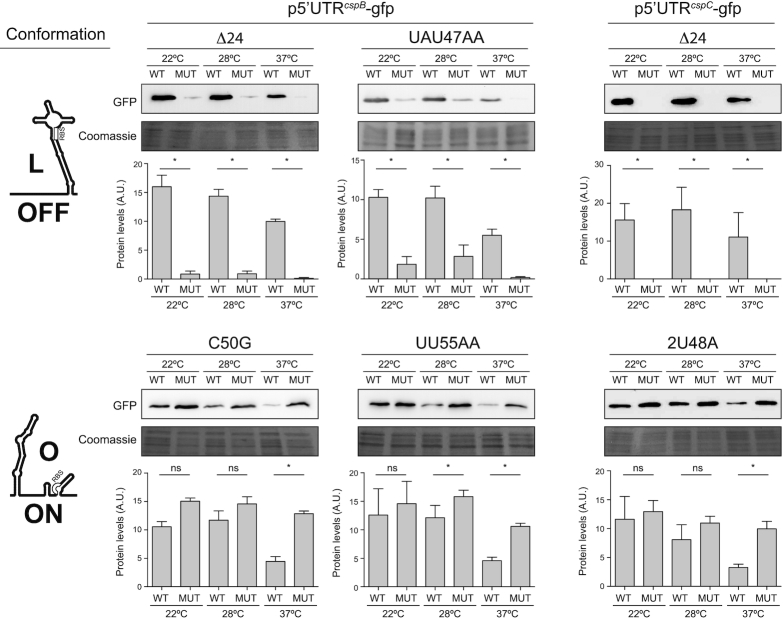
To further validate the predicted alternative 5′UTR structures in vivo, we constructed the p5′UTRcspB-gfp- and p5′UTRcspC-gfp-derived plasmids that included several mutations in the cspB and cspC 5′UTRs (Supplementary Figure S3). On the one hand, we generated the 5′UTRcspBΔ24-gfp and 5′UTRcspCΔ24-gfp plasmids, which carried a deletion of the first 24 nucleotides from both 5′UTRs (Supplementary Figure S3). Such nucleotides would be essential to form the long thermoresponsive hairpin of the O conformation (Figure 3). At the same time, this deletion would not hinder the L conformation but rather favour it (Figure 3 and Supplementary Figure S3). In agreement with this, western blot analyses revealed that the GFP expression was drastically reduced in strains expressing the 5′UTRcspBΔ24-gfp and 5′UTRcspCΔ24-gfp mRNAs (Figure 5). We next performed site-directed mutagenesis in the cspB 5′UTR of the p5′UTRcspB-gfp plasmid, by replacing nucleotides 47-UAU-49 with AA. Since nucleotides 47-UAU-49 would base pair with nucleotides 33-GUA-35 to form the hairpin structure in the O conformation, such substitution would cause a destabilization and, hence, favour the alternative L conformation (Figure 3 and Supplementary Figure S3). As anticipated, western blot results showed lower GFP levels for the strain carrying the mutated plasmids regardless of the temperature (Figure 5). On the other hand, we substituted 50-C for a G and 55-UU-56 for AA in the cspB 5′UTR of p5′UTRcspB-gfp and 48-UU-49 by an A in the cspC 5′UTR of p5′UTRcspC-gfp (Supplementary Figure S3). These mutations would favour the O conformation over the L conformation (Figure 3 and Supplementary Figure S3). Western blot results showed that these cspB and cspC 5′UTR substitutions led to similar GFP production levels (at all the tested temperatures) to the ones found in the WT p5′UTRcspB-gfp and p5′UTRcspC-gfp versions at 22°C (Figure 5). This indicated that such nucleotide substitutions made the 5′UTRs unresponsive to temperature changes while highlighting the relevance of alternative cspB and cspC 5′UTR structures in vivo for blocking or facilitating translation in response to temperature shifts.
Figure 5.
The production of CspB and CspC is controlled by alternative 5′UTR conformations in vivo. Western blots showing the effect of different mutations in the 5′UTR of the p5′UTRcspB-gfp and p5′UTRcspC-gfp plasmids on the GFP levels. Such mutations favoured one of the two 5′UTR alternative conformations. The expected conformation for each mutation, O (translation ON) or L (translation OFF), is indicated on the left hand side of the figure. Coomassie stained gel portions are included as loading controls. Bar plots represent the mean and standard deviation of protein levels from three independent biological replicates, which were determined by densitometry of protein bands using ImageJ (https://imagej.nih.gov/ij/). Asterisks represent statistical significance (P < 0.05, Mann–Whitney U test); ns, not significant. Representative images from the triplicates are shown.
CspA represses the expression of CspB and CspC in S. aureus
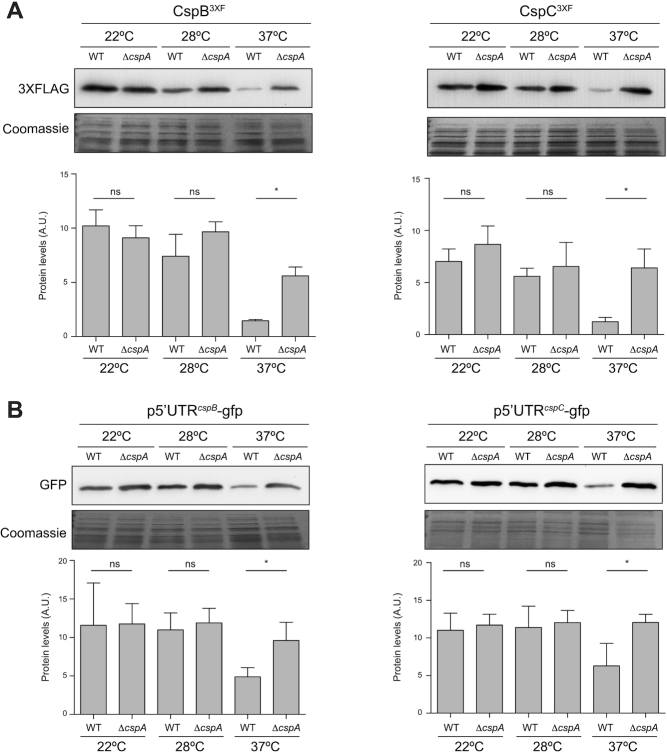
Previous results showed that the E. coli cspA and cspB thermosensors required the action of CSPs to modulate their 5′UTR structures (20). Zhang et al. reported that the structural rearrangements did not occur in a ΔcspABEG strain, which lacked the csp ORFs while retaining their 5′UTRs. In fact, these authors showed that CspA regulated its own production by binding its own mRNA and preventing the formation of a double-stranded cspA 5′UTR structure (20). To test whether CspB or CspC participated in the structural reorganizations of their own 5′UTR thermosensors and, therefore, autoregulated their production, we electroporated the translational reporter plasmids p5′UTRcspB-gfp and p5′UTRcspC-gfp into the S. aureus ΔcspB and ΔcspC strains, respectively. Western blot results showed a similar behaviour in terms of GFP expression between the WT and mutant strains for all the tested temperatures (Supplementary Figure S4). This indicated that the production of both proteins was not autoregulated, at least in the tested conditions. Previous CspA co-immunoprecipitation studies from our group showed that CspA was able to bind the cspB and cspC mRNAs in vivo. Deletion of cspA increased CspC production, indicating that the regulation of CSPs is interconnected in S. aureus (40). To corroborate these results and analyse whether CspA also modulated CspB expression, we deleted the cspA gene from the strains expressing the chromosomally tagged CspB3xF and CspC3xF proteins and performed western blots. Figure 6A shows that deletion of the cspA gene increased the CspB3xF and CspC3xF protein levels at 37°C, confirming its role in the regulation process (Figure 6A). We then analysed whether CspA might participate in the thermoregulation of CspB and CspC through their 5′UTR regions. To test this, we electroporated the translational reporter plasmids p5′UTRcspB-gfp and p5′UTRcspC-gfp in the cspA mutant and performed western blot experiments. The GFP levels in the ΔcspA mutant were higher than those in the WT strain when grown at 37°C, confirming that CspA repressed CspB and CspC through their 5′UTRs. When strains were grown at 22 and 28°C, no significant GFP differences were found (Figure 6B). Altogether, these results confirmed that CspA repressed CspB and CspC production probably by interacting with the cspB and cspC 5′UTRs.
Figure 6.
CspA participates in repressing the production of CspB and CspC at 37°C. (A) Western blot showing the chromosomal expression of CspB3xF and CspC3xF from the WT and ΔcspA strains grown in MH broth at 22, 28 and 37°C. (B) Western blot results of the GFP levels expressed from the S. aureus WT and ΔcspA strains, carrying the cspB and cspC 5′UTR-GFP translational reporter plasmids after growth in MH broth at 22, 28 and 37°C. The 3xFLAG-tagged proteins and GFP signals were developed as described in Figures 1 and 2, respectively. Coomassie stain gel portions are included as loading controls. Bar plots represent the mean and standard deviation of protein levels from three independent biological replicates, which were determined by densitometry of protein bands using ImageJ (https://imagej.nih.gov/ij/). Asterisks represent statistical significance (P < 0.05, Mann–Whitney U test); ns, not significant. Representative images from the triplicates are shown.
CspA represses CspB and CspC by favouring the L conformation at 37°C
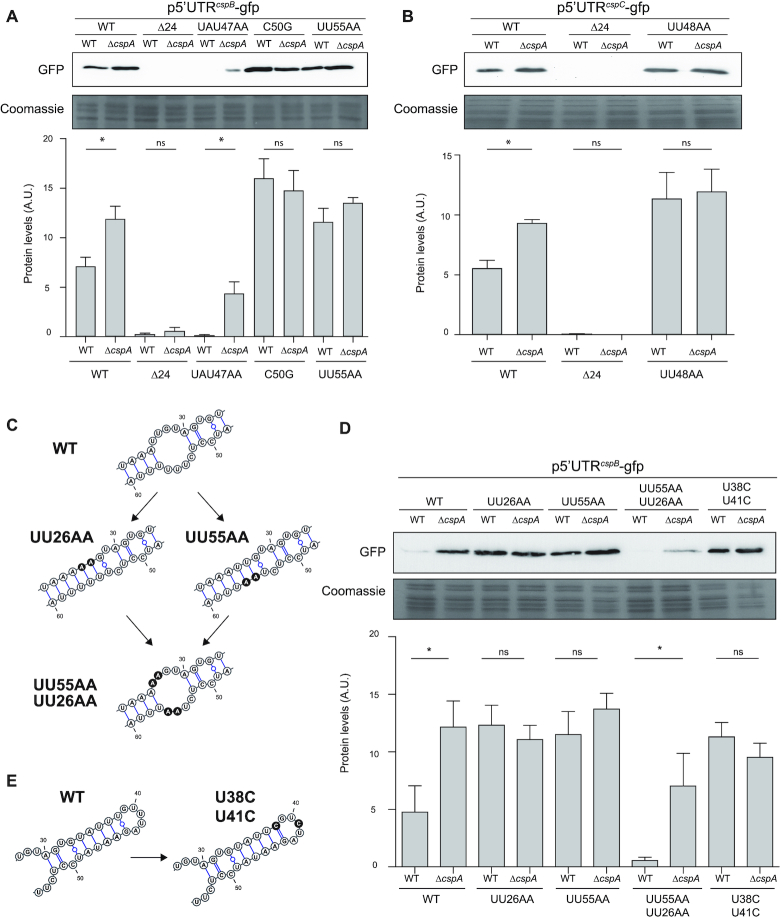
To gain further insight into how CspA regulates CspB and CspC, we considered two hypotheses: (1) CspA would directly bind the RBS and block translation, or (2) CspA would interact with the 5′UTR and favour the L conformation, repressing cspB and cspC mRNA translation. To test these hypotheses, we electroporated the cspA mutant with the translational reporter plasmids carrying all the above-mentioned 5′UTR mutations (Supplementary Figure S3). We then compared the GFP levels in the ΔcspA reporter strains grown at 37°C with those of their WT counterparts by western blot (Figure 7). We found no significant differences between the GFP expression levels in the WT and ΔcspA strains carrying the 5′UTR mutants that favour the O conformation (C50G and UU55AA in cspB 5′UTR and UU48A in cspC 5′UTR) (Figure 7A and B). This suggested that CspA could not inhibit CspB/C expression when the RBS became more accessible. Moreover, no differences were found when comparing the GFP levels of the WT and ΔcspA strains carrying the reporter plasmids that favoured the L conformation (p5′UTRcspBΔ24-gfp and p5′UTRcspCΔ24-gfp) (Supplementary Figure S3 and Figure 7A and B). This indicated that CspA was dispensable for repression when forcing the L conformation by deleting the first 24 nucleotides. We only found a slight but significant increase in the GFP expression from the ΔcspA strain carrying the cspB UAU47AA mutation (Figure 7A), which promoted the L conformation by simultaneously destabilizing the hairpin of the O conformation (Figure 3 and Supplementary Figure S3). This may indicate that destabilizing the O conformation favours the CspA repression effect. However, it may also be possible for CspA to be sensitive to 5′UTR mutations that cause changes in the nucleotide-pairing affinity. It has been proposed before that CSPs bind RNA weakly and in a non-specific way. However, in some occasions, certain RNA motifs could contribute to CSP functionality (40,52–54). For example, we showed that the autoregulation of CspA in S. aureus requires a U-rich motif located in the cspA 5′UTR (40). Notably, the cspB 5′UTR carries several U-rich motifs (Figure 3), two of which (36-UUUGUUU-42 and 54-UUUUUU-59) are involved in the folding of the O conformation. Additionally, motif 36-UUUGUUU-42 participates in the nucleotide pairing of the L conformation (Figure 3). It could be possible for CspA to target these regions at 37°C to prevent the folding of the O conformation. Note that validations in vivo turned out to be extremely difficult because the 5′UTR mutants in these potential CspA binding sites could not be constructed without affecting the folding of both alternative conformations. When comparing both U-rich regions, the second motif (54-UUUUUU-59) presented more unpaired bases in either of the two conformations than the first one. Therefore, we chose this region to examine the role of U-rich motifs in the regulation of cspB by CspA (Figure 3). To analyse whether the increased reporter expression observed in the UU55AA mutant (Figure 7A) was due to changes in the folding affinities and/or due to the inability of CspA to bind a mutated 54-UUUUUU-59 target motif, we generated an additional mutation (UU26AA; nucleotides 26-UU-27 substituted by AA) in the UU55AA mutant that reconstituted the hairpin bubble (Figures 3 and 7C). As a control, we also constructed the UU26AA mutant in the cspB 5′UTR (Figure 7C). Western blot experiments showed that CspA repressed the GFP reporter translation in the double UU55AA/UU26AA mutant (Figure 7D). This suggested that motif 54-UUUUUU-59 was dispensable for CspA to exert its repression. Next, we focused on the other U-rich motif (36-UUUGUUU-42) and generated an additional mutant (36-UUCGUCU-42) (Figure 7E). Note that these nucleotide changes were intended to avoid any significant variations in both conformations (O and L) (Figures 3 and 7E). Western blot results revealed that when we introduced the plasmid harbouring this mutation in the WT strain, the GFP levels increased and reached similar levels to the ones present in the ΔcspA strain (Figure 7D). This result suggested that CspA repressed CspB production by interacting with the cspB mRNA through the 36-UUUGUUU-42 motif. Altogether our data lead to a functional model in which CspA binds the loop of the distal stem of the O conformation and favours the L conformation that represses CspB translation at 37°C.
Figure 7.
CspA is required for promoting the L conformation at 37°C and repressing CspB and CspC production. (A, B and D). Western blots showing the GFP levels from the WT and ΔcspA strains expressing the WT and mutated cspB or cspC 5′UTR-GFP reporter plasmids. Total protein extracts were processed as described in Figure 2. Coomassie gel portions are included as loading controls. Bar plots represent the mean and standard deviation of the GFP levels from three independent biological replicates, which were determined by densitometry of protein bands using ImageJ (https://imagej.nih.gov/ij/). Asterisks represent statistical significance (P < 0.05, Mann–Whitney U test); ns, not significant. Representative images from the triplicates are shown. (C and E). Schematic representations of the mutations introduced in the cspB 5′UTR. The substituted nucleotides are shaded in black.
Thermoregulation of CSPs is required for S. aureus adaptation to ambient temperatures
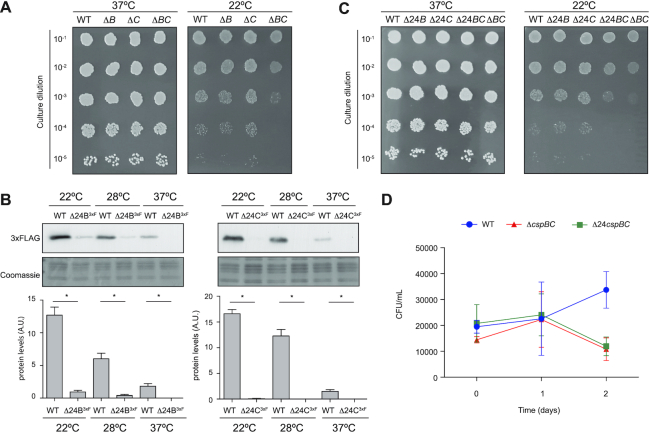
Besides its pathogenicity, the ability to colonize different niches as well as to spread rapidly by direct contact and/or through contaminated surfaces make S. aureus a life-threatening pathogen (10). The higher CspB and CspC protein expression at ambient temperatures suggested that these proteins could contribute to S. aureus adaptation when it leaves the host. To further investigate this, we plated serial dilutions of overnight cultures of the WT and isogenic ΔcspB and ΔcspC mutant strains in TSA plates and incubated them at 22 and 37°C. As shown in Figure 8A, we found no growth differences between the WT and csp mutants at the tested temperatures. Since both CSPs were induced at ambient temperatures (Figure 1B), it seemed possible for these proteins to play redundant roles, as previously described in E. coli and B. subtilis (19,24,55,56). To address this issue, we generated a double mutant strain (ΔcspBC) and analysed its growth at different temperatures. Interestingly, we observed a significant growth deficiency at 22°C when compared with the WT strain (Figure 8A). This result suggested a biological redundancy between CspB and CspC as the double deletion attenuated the ability of S. aureus to grow at low temperatures.
Figure 8.
Simultaneous mutations in CspB and CspC RNA thermoswitches affect S. aureus growth at ambient temperatures. (A and C) Growth assay at 37 and 22°C of the different csp mutants. Bacteria were grown at 37°C and 200 rpm overnight and diluted to an OD600nm of 1. Ten-fold dilutions were plated in TSA plates and incubated at 37 and 22°C for 24 and 72 h. A representative image from three independent biological replicates is shown. (B) Western blot showing the expression of the 3xFLAG-tagged CspB and CspC proteins from the Δ24B and Δ24C mutants, which were grown at the indicated temperatures. Protein extraction and western blots were performed as previously described in Figure 1. Coomassie gel portions are included as loading controls. Bar plots show the mean and standard deviation of protein levels from three independent biological replicates, which were determined by densitometry of protein bands using ImageJ (https://imagej.nih.gov/ij/). Asterisks represent statistical significance (P < 0.05, Mann–Whitney U test); ns, not significant. Representative images from the triplicates are shown. (D) Assay mimicking the conditions that S. aureus might face when away from the host. A preinoculum was grown in TSB at 37°C and 200 rpm overnight. Bacteria were then washed, diluted 1:250 and regrown at 32°C and 200 rpm for 2 days in SNM3, resembling the nasal state. To imitate fomite contamination, bacteria were transferred to fresh SNM3 and incubated at 22°C. CFUs were counted 0, 1 and 2 days after incubation at 22°C. The plot represents the results from at least three independent replicates.
In order to evaluate the biological relevance of the thermoregulation mechanism controlling CspB and CspC expression, we constructed chromosomal mutants that modified the structural organization of the 5′UTR to prevent cspB and cspC mRNA translation. Out of the different mutations shown in Supplementary Figure S3, we chose to delete the first 24 nucleotides from both cspB and cspC mRNAs since these mutations produced the strongest GFP repression in our previous constructs (Figure 5). First, to control that these mutations produced the same effect in the chromosome as seen for the reporter plasmids, the corresponding 24 nt were deleted from the tagged cspB3xF and cspC3xF strains. We performed western blot analyses and confirmed that the chromosomal Δ24B3xF and Δ24C3xF mutant strains were not able to produce CspB3xF and CspC3xF (Figure 8B). Analogously, we studied cspB and cspC mRNA levels through northern blots (Supplementary Figure S5A). The Δ24 deletion produced no significant variations on the cspBΔ243xF mRNA levels when compared to the WT mRNA. On the contrary, the cspCΔ243xF mRNA levels were significantly lower than those of the cspC3xF WT mRNA. A similar behaviour was observed for the WT and Δ24 cspB and cspC 5′UTRs-gfp translational reporter fusion mRNAs (Supplementary Figure S5B). On the one hand, this result suggested that the decreased cspC mRNA expression observed in the Δ24 deletion was not caused by a putative cspC promoter activity disruption. Note that the reporter fusions were expressed from a constitutive heterologous promoter. On the other hand, the results indicated that the Δ24 deletion produced different outputs for the cspB and cspC mRNA levels. Whether this was a consequence of forcing the L conformation or because of a change in the mRNA stability without any functional meaning remains unanswered. Interestingly, this leaves room for speculation on how the subtle differences between such apparently similar orthologous regulatory 5′UTRs may lead to different outcomes and, perhaps, explain the need for both genes to coexist in the same genome. Regardless, the Δ24 deletions produced the expected decrease in the production of both CspB and CspC proteins and led us to engage into phenotypic analysis. Therefore, the same mutations were performed in the WT strain to create the single mutants Δ24B and Δ24C and the double mutant Δ24B–Δ24C. Overnight cultures of these strains were diluted and plated on TSA plates. As observed for the single cspB and cspC gene mutations, the Δ24B and Δ24C 5′UTR deletions did not affect S. aureus growth at ambient temperatures. In contrast, the double Δ24B–Δ24C mutant strain presented a growth defect comparable to the ΔcspBC mutant when incubated at 22°C (Figure 8C). This result confirmed that the cspB and cspC 5′UTR thermoregulation was required for CspB and CspC production and, therefore, critical for S. aureus efficient growth at ambient temperatures.
Considering that one of the major reservoirs of S. aureus in the human body is the nose and that S. aureus is often disseminated through human nasal secretions that contaminate diverse fomites (9), we recreated such scenario by growing the WT, Δ24B–Δ24C and ΔcspBC strains in Erlenmeyer flasks containing SNM3 for 2 days at 32°C and 220 rpm. SNM3 reproduces the composition of nasal secretions (49), 32°C is the temperature found in the nose (51) and agitation (220 rpm) favours aeration. After this incubation period, no growth differences were observed among the strains. Then, the bacterial cultures were normalized to an OD600nm of 0.05 using fresh SNM3 and transferred to new sterile flasks. The WT and mutant strains were incubated for 48 h at 22°C without shaking to mimic the transition between the nasal and the fomite state through mucus secretion. The number of viable S. aureus cells was then estimated by counting the CFUs per millilitre on TSA plates at different time points. As shown in Figure 8D, whereas the WT strain replicated in SNM3 at 22°C, the Δ24B–Δ24C and ΔcspBC mutant strains showed a significant growth defect after 48 h of incubation. Altogether, these data demonstrated that CspB and CspC thermoregulation is required for S. aureus adaptation and survival at ambient conditions, a scenario that this pathogen may face when it leaves its natural host.
The existence of alternative 5′UTR csp structures may apply to other bacteria
In order to analyse whether the above mentioned thermoswitch mechanism might be conserved among other bacterial species, we performed blastn analyses using the first 127 nucleotides of the cspB mRNA as a query in the Microbes NCBI website (https://blast.ncbi.nlm.nih.gov/Blast.cgi). The results showed that >50 csp genes from 43 different Staphylococcus species carried an orthologous cspB 5′UTR. The sequences were highly conserved, with an identity >74% (Supplementary Table S4). The number of CSPs among the analysed staphylococcal species was variable, ranging from 1 to 3 (36). In most of the Staphylococcus genomes containing three csp genes, two of them harboured 5′UTRs similar to S. aureus cspB/C 5′UTRs. However, among the genomes with two csp genes, only one of them carried a putative thermoswitch (Supplementary Table S4). In the case of Staphylococcus pettenkoferi and Staphylococcus kloosii, whose genomes contained just one csp gene, the 5′UTR sequence was significantly different from the S. aureus cspB 5′UTR. This might be explained by the fact that their CSP showed a high degree of identity with S. aureus CspA protein (36).
Multiple sequence alignments also revealed that the nucleotides required for adopting the alternative conformations were highly conserved among staphylococcal species (Supplementary Figures S6 and S7). Although the cspB/C 5′UTR sequence conservation was not preserved beyond the Staphylococcus genus, the fact that orthologous thermoswitch mechanisms had been previously found in E. coli and L. monocytogenes (20,28,33) indicated that the thermoregulation of CSPs based on mutually exclusive alternative 5′UTR conformations could be a common regulatory mechanism in bacteria. Since bacterial genomes often encode several CSP paralogues with a high protein identity, it might be difficult to predict which CSPs are involved in cold adaptation without further experimentation. Although the csp 5′UTRs might not share a high sequence identity with the RNA thermoswitches described so far, the existance of alternative structures could be an indicator of such function. To validate this idea, we looked for putative alternative conformations in the 5′UTRs of all the csp genes from representative bacteria such as Enterococcus faecalis, B. subtilis, Clostridium perfringens, Salmonella enterica sv Typhimurium and Pseudomonas aeruginosa (Supplementary Table S5). Since the already identified thermoswitches were located within 5′UTRs with lengths ranging from 110 to 160 nt, query sequences including at least 120 nt upstream of the start codon of each CSP were selected to predict putative secondary structures using the mfold web server (38). According to preliminary predictions, query sequences were enlarged or shortened to look for alternative folding options. Thus, we identified several putative csp 5′UTRs that displayed alternative structures resembling the ones previously described (Supplementary Table S5 and Figure 9). For example, we predicted putative RNA thermoswitches in two out of the six csp genes from E. faecalis, S. enterica sv Typhimurium and P. aeruginosa, whereas one was predicted in C. perfringens and B. subtilis (Supplementary Table S5). On the one hand, these results indicated that the alternative conformations of csp 5′UTRs may be a conserved regulatory feature in bacteria despite lacking significant sequence similarity. On the other hand, structural predictions could help identifying CSPs whose expression may be post-transcriptionally controlled by alternative RNA conformations.
Figure 9.
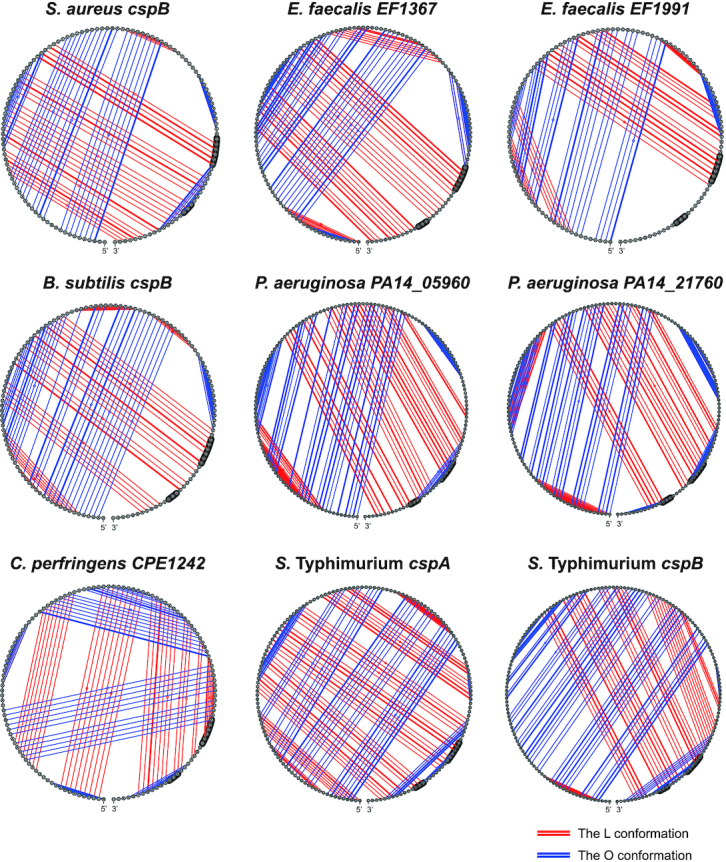
RNA alternative conformations are widely spread in csp genes of several bacteria. Circular structural representation of alternative RNA secondary structures found in the 5′UTRs of the csp mRNAs from E. faecalis, B. subtilis, C. perfringens, S. enterica sv Typhimurium and P. aeruginosa. The 5′UTR secondary structure of cspB from S. aureus is included as a control. Blue and red lines represent the pairings between nucleotides in the O and L conformations, respectively.
DISCUSSION
RNA-mediated thermoregulation is an essential process that promotes changes in gene expression and facilitates the adaptability of pathogenic bacteria when they transition from the host to the environment and vice versa. Several RNA thermosensors have been found to induce the expression of main regulators required for the activation of virulent genes in bacteria while infecting humans. Throughout this process, the zipper-like structures located at the 5′UTR of genes required for survival inside the host sense an increase in the temperature and unfold, which in turn releases the otherwise blocked RBSs. In this manner, they become accessible to the ribosomes for translation initiation (57–63). Conversely, a few RNA thermosensors have been identified as capable of activating gene expression upon drops in the temperature (20,28,33). In comparison with regular zipper-like RNA thermosensors, activation of translation at lower temperatures involves much more complex RNA structural reorganizations.
In this study, we demonstrated that S. aureus adapts to low temperatures by modulating CspB and CspC expression. The CspB and CspC thermoregulation occurred at the post-transcriptional level through the action of two thermosensitive RNA elements located at the 5′UTR of the cspB and cspC mRNAs, respectively (Figure 3). These thermosensors induced CspB and CspC translation at ambient temperatures while repressing it at host-related temperatures (Figure 2). By using conditions that mimicked the S. aureus nasal and fomite contamination, we demonstrated the importance of RNA-mediated thermoregulation for the survival of such bacterium in the environment. Chromosomal deletions of the first 24 nucleotides of the cspB and cspC mRNAs (Δ24BC strains) eliminated the transitions between the two alternative RNA conformations by forcing the folding of the 5′UTR into the L variant. This impaired S. aureus growth at 22°C (Figure 8), a similar growth defect to the one observed for the double cspB and cspC mutant.
Upon further inspection, we found the thermosensitive structural shifts to be orthologous to the ones recently described for E. coli and L. monocytogenes cspA mRNAs (20,33). These type of RNA structural rearrangements, which repress translation at higher temperatures while activating it at lower temperatures, are also present in the 5′UTR of the bacteriophage cIII gene (64). Recently, regulatory 5′UTRs that activate the expression of the holin-like protein CidA at low temperatures have been described for S. aureus (65). Hussein et al. propose that the cidA gene is transcribed by two alternative promoters, generating two different 5′UTRs of 36 and 82 nt, respectively. Translational reporter fusions revealed that both 5′UTRs post-transcriptionally activated CidA translation at low temperatures, the longer cidA 5′UTR being the one responsible for the highest CidA protein expression. Other than the closed conformation identified by Hussein et al. (65), we could also predict by mfold a putative alternative conformation in the longer cidA 5′UTR that would make the RBS available to the ribosome at low temperatures (Supplementary Figure S8). This open conformation would involve a long hairpin comprising nucleotides 5–66 of the cidA 5′UTR that would sequester the anti-RBS motif (Supplementary Figure S8), resembling the mechanism found for CspB and CspC. It would be interesting to investigate which are the signals responsible for the activation of each of the cidA alternative promoters.
Altogether, these examples indicated that the thermoregulatory mechanism based on RNA conformational switches could be widely extended. In agreement with this idea, we were able to predict orthologous cspB thermosensors in all Staphylococcus species, with the exception of S. pettenkoferi and S. kloosii, whose genomes only encoded CspA. Note that CspA expression was not affected by temperature shifts in S. aureus.
Considering that multiple csp genes are present in the core genome of most bacteria, one would expect this type of RNA thermoswitch to be distributed beyond sequence conservation. In fact, we found that orthologous alternative structures could also be predicted in the 5′UTRs of csp mRNAs from several representative bacteria, including E. faecalis, B. subtilis, C. perfringens, S. enterica sv Typhimurium and P. aeruginosa (Figure 9). Moreover, examples like the cIII and cidA thermosensors indicated that these regulatory elements may not be restricted to the csp gene family (64,65).
Although the cspB and cspC 5′UTR structures significantly differ from the regular RNA thermosensors (11,32,63), the hairpin of the O conformation seems to work like the already known zipper-like structures. However, the outcome of this mechanism is rather different as instead of freeing the RBS when melting, it is the anti-RBS region that is released. Therefore, in addition to masking RBSs or anti-RBSs, it could be speculated that bacterial genomes may encode alternative thermosensitive hairpins that may also sequester other binding sites for ribonucleases, RNA-binding proteins and/or non-coding RNAs, making such motifs available or unavailable depending on the environmental temperature.
In agreement with previous results from E. coli and B. subtilis studies, we showed that besides from shifts in the temperature, S. aureus cspB and cspC genes can also be negatively regulated by a csp paralogue (20,24,55). In E. coli, CspA downregulates the expression of CspB, CspE and CspG proteins (55). Additionally, in B. subtilis the lack of one member of the CSP family produces the upregulation of the others, suggesting that CSPs control is interconnected (24). Previous RNA immunoprecipitation experiments from our group showed that the cspB and cspC mRNAs were targeted by CspA in S. aureus (40). Here, we propose CspA as an essential player that favours the L conformation over the O conformation at 37°C (Figure 7). Although CspA regulates its own expression in S. aureus by preventing the folding of a long hairpin in the cspA 5′UTR (40), such hairpin is unsensitive to physiological temperatures (Figure 1). Since CspA levels remain unaltered upon temperature changes, it is difficult to explain how CspA can repress CspB/C translation on its own (Figure 1). We reasoned that if it takes part in regulating both of its paralogues, it should work in conjunction with temperature-dependent elements. CspA seems to interact with one of the U-rich motifs located at the 5′UTRs. Specifically, mutations in the cspB 36-UUUGUUU-42 motif abolished CspA repression (Figure 7). Interestingly, this motif is located at the top of the hairpin that is unfolded when temperature increases (Figure 4). This interaction would prevent the folding of the long hairpin in the O conformation at 37°C and allow the anti-RBS to base pair with the RBS in the L conformation. In agreement with this, our in vivo experiments showed how deleting the cspA gene increased CspB and CspC expression at 37°C (Figure 6). On the contrary, when testing the effect of the presence/absence of CspA on the expression of CspB and CspC at 22°C no significant differences were found. In other words, there was not a temperature-dependent repression of CspB and CspC expression in the absence of CspA. This could be explained by the fact that the RNA secondary structures are more rigid at lower temperatures (20). The affinity between the RNA regions forming the hairpin of the O conformation at 22°C is probably higher than their affinity for CspA. Therefore, it would seem that CspA cannot disrupt the folding of such hairpin at ambient temperatures. We further confirmed this by performing site-directed mutations in the 5′UTRs that altered the affinities of the different RNA regions between them (Figure 5). In most cases, thermoregulation was eliminated and, although it cannot be excluded that some of these mutations could also modify the accessibility of CspA to their binding site, we suggest the thermoregulatory mechanism to be based on the different affinities of the distinct elements involved. These affinities would be naturally altered by temperature shifts.
It remains to be elucidated whether CspB and CspC are functionally redundant in a mechanistic way. Previously, functional redundancy was found among E. coli, Salmonella and B. subtilis CSPs (19,24,55,56). However, we recently showed that the phenotypes of a cspA mutant could not be restored by the expression of the CspB or CspC proteins despite their high protein identity, suggesting a certain functional specificity for S. aureus CSPs. The CspA specificity related to SigB-associated phenotypes was concentrated in proline 58 of the CspA protein (36). Interestingly, amino acids occupying the same position (58) in the CspB and CspC were different (note that all S. aureus CSPs are 66 amino acids long). It is possible that, despite their similarities, S. aureus CSPs have different preferences when it comes to sequence affinities, as previously described for E. coli and Salmonella CSPs (53,56). Determination of the RNA targetomes would help elucidating how CspB and CspC participate on regulating gene expression when S. aureus grows at low temperatures.
In summary, our study unveils the thermoregulatory mechanism that controls the expression of CspB and CspC upon temperature shifts. The biological relevance of this thermoregulation is highlighted by the fact that it is required for S. aureus optimal growth and survival in ambient conditions. Moreover, we propose the CSP regulation based on RNA alternative conformations to be widely spread in bacteria.
DATA AVAILABILITY
Any data or material that support the findings of this study can be made available by the corresponding author upon request.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Prof. Cristina Solano for critical reading of this manuscript, Anne-Catherine Helfer for excellent technical assistance in probing experiments and Dr Emanuelle Charpentier for pCN51 plasmid (ref. NR-46149), which was provided by the Network on Antimicrobial Resistance in Staphylococcus aureus (NARSA) for distribution by BEI Resources, NIAID, NIH. We acknowledge support of the publication fee by the CSIC Open Access Publication Support Initiative through its Unit of Information Resources for Research (URICI).
Contributor Information
Arancha Catalan-Moreno, Instituto de Agrobiotecnología, IdAB, CSIC-Gobierno de Navarra, Avda. de Pamplona 123, 31192 Mutilva, Navarra, Spain.
Marta Cela, Instituto de Agrobiotecnología, IdAB, CSIC-Gobierno de Navarra, Avda. de Pamplona 123, 31192 Mutilva, Navarra, Spain.
Pilar Menendez-Gil, Instituto de Agrobiotecnología, IdAB, CSIC-Gobierno de Navarra, Avda. de Pamplona 123, 31192 Mutilva, Navarra, Spain.
Naiara Irurzun, Instituto de Agrobiotecnología, IdAB, CSIC-Gobierno de Navarra, Avda. de Pamplona 123, 31192 Mutilva, Navarra, Spain.
Carlos J Caballero, Instituto de Agrobiotecnología, IdAB, CSIC-Gobierno de Navarra, Avda. de Pamplona 123, 31192 Mutilva, Navarra, Spain.
Isabelle Caldelari, Architecture et Réactivité de l’ARN, Université de Strasbourg, CNRS, UPR 9002, F-67000 Strasbourg, France.
Alejandro Toledo-Arana, Instituto de Agrobiotecnología, IdAB, CSIC-Gobierno de Navarra, Avda. de Pamplona 123, 31192 Mutilva, Navarra, Spain.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
A.T.-A. was supported by the European Research Council under the European Union's Horizon 2020 research and innovation program [ERC-CoG-2014-646869]; and the Spanish Ministry of Science and Innovation [PID2019-105216GB-I00] grants. I.C. was supported by Centre National de la Recherche Scientifique (CNRS) through an International Project of Scientific Cooperation between CNRS and CSIC [PICS07507]; and by the framework of the labEx Net RNA, French National Research Agency [ANR-10-LABX-0036 and IMCBio ANR-17-EURE-0023]. Funding for open access charge: CSIC Open Access Publication Support Initiative, Unit of Information Resources for Research (URICI).
Conflict of interest statement. None declared.
REFERENCES
- 1. Uhlemann A.C., Knox J., Miller M., Hafer C., Vasquez G., Ryan M., Vavagiakis P., Shi Q., Lowy F.D.. The environment as an unrecognized reservoir for community-associated methicillin resistant Staphylococcus aureus USA300: a case-control study. PLoS One. 2011; 6:e22407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Scott E., Bloomfield S.F.. The survival and transfer of microbial contamination via cloths, hands and utensils. J. Appl. Bacteriol. 1990; 68:271–278. [DOI] [PubMed] [Google Scholar]
- 3. Kramer A., Schwebke I., Kampf G.. How long do nosocomial pathogens persist on inanimate surfaces? A systematic review. BMC Infect. Dis. 2006; 6:130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Neely A.N., Maley M.P.. Survival of enterococci and staphylococci on hospital fabrics and plastic. J. Clin. Microbiol. 2000; 38:724–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Walther B.A., Ewald P.W.. Pathogen survival in the external environment and the evolution of virulence. Biol. Rev. Camb. Philos. Soc. 2004; 79:849–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Knox J., Uhlemann A.C., Lowy F.D.. Staphylococcus aureus infections: transmission within households and the community. Trends Microbiol. 2015; 23:437–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. David M.Z., Daum R.S.. Community-associated methicillin-resistant Staphylococcus aureus: epidemiology and clinical consequences of an emerging epidemic. Clin. Microbiol. Rev. 2010; 23:616–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Davis M.F., Iverson S.A., Baron P., Vasse A., Silbergeld E.K., Lautenbach E., Morris D.O.. Household transmission of meticillin-resistant Staphylococcus aureus and other staphylococci. Lancet Infect. Dis. 2012; 12:703–716. [DOI] [PubMed] [Google Scholar]
- 9. Wertheim H.F.L., Melles D.C., Vos M.C., van Leeuwen W., van Belkum A., Verbrugh H.A., Nouwen J.L.. The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect. Dis. 2005; 5:751–762. [DOI] [PubMed] [Google Scholar]
- 10. Tong S.Y.C., Davis J.S., Eichenberger E., Holland T.L., Fowler V.G.. Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 2015; 28:603–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Klinkert B., Narberhaus F.. Microbial thermosensors. Cell. Mol. Life Sci. 2009; 66:2661–2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schumann W. Thermosensor systems in eubacteria. Adv. Exp. Med. Biol. 2012; 739:1–16. [DOI] [PubMed] [Google Scholar]
- 13. Lam O., Wheeler J., Tang C.M.. Thermal control of virulence factors in bacteria: a hot topic. Virulence. 2014; 5:852–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Onyango L.A., Hugh Dunstan R., Roberts T.K., Macdonald M.M., Gottfries J.. Phenotypic variants of staphylococci and their underlying population distributions following exposure to stress. PLoS One. 2013; 8:e77614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Barria C., Malecki M., Arraiano C.M.. Bacterial adaptation to cold. Microbiology. 2013; 159:2437–2443. [DOI] [PubMed] [Google Scholar]
- 16. Alreshidi M.M., Dunstan R.H., Macdonald M.M., Smith N.D., Gottfries J., Roberts T.K.. Metabolomic and proteomic responses of Staphylococcus aureus to prolonged cold stress. J. Proteomics. 2015; 121:44–55. [DOI] [PubMed] [Google Scholar]
- 17. Fang L., Jiang W., Bae W., Inouye M.. Promoter-independent cold-shock induction of cspA and its derepression at 37°C by mRNA stabilization. Mol. Microbiol. 1997; 23:355–364. [DOI] [PubMed] [Google Scholar]
- 18. Uppal S., Akkipeddi V.S.N.R., Jawali N.. Posttranscriptional regulation of cspE in Escherichia coli: involvement of the short 5′-untranslated region. FEMS Microbiol. Lett. 2008; 279:83–91. [DOI] [PubMed] [Google Scholar]
- 19. Xia B., Ke H., Inouye M.. Acquirement of cold sensitivity by quadruple deletion of the cspA family and its suppression by PNPase S1 domain in Escherichia coli. Mol. Microbiol. 2001; 40:179–188. [DOI] [PubMed] [Google Scholar]
- 20. Zhang Y., Burkhardt D.H., Rouskin S., Li G.-W., Weissman J.S., Gross C.A.. A stress response that monitors and regulates mRNA structure is central to cold shock adaptation. Mol. Cell. 2018; 70:274–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schmid B., Klumpp J., Raimann E., Loessner M.J., Stephan R., Tasara T.. Role of cold shock proteins in growth of Listeria monocytogenes under cold and osmotic stress conditions. Appl. Environ. Microbiol. 2009; 75:1621–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chapot-Chartier M.P., Schouler C., Lepeuple A.S., Gripon J.C., Chopin C.. Characterization of cspB, a cold-shock-inducible gene from Lactococcus lactis, and evidence for a family of genes homologous to the Escherichia coli cspA major cold shock gene. J. Bacteriol. 1997; 179:5589–5593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Willimsky G., Bang H., Fischer G., Marahiel M.A.. Characterization of cspB, a Bacillus subtilis inducible cold shock gene affecting cell viability at low temperatures. J. Bacteriol. 1992; 174:6326–6335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Graumann P., Wendrich T.M., Weber M.H., Schröder K., Marahiel M.A.. A family of cold shock proteins in Bacillus subtilis is essential for cellular growth and for efficient protein synthesis at optimal and low temperatures. Mol. Microbiol. 1997; 25:741–756. [DOI] [PubMed] [Google Scholar]
- 25. Duval B.D., Mathew A., Satola S.W., Shafer W.M.. Altered growth, pigmentation, and antimicrobial susceptibility properties of Staphylococcus aureus due to loss of the major cold shock gene cspB. Antimicrob. Agents Chemother. 2010; 54:2283–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Anderson K.L., Roberts C., Disz T., Vonstein V., Hwang K., Overbeek R., Olson P.D., Projan S.J., Dunman P.M.. Characterization of the Staphylococcus aureus heat shock, cold shock, stringent, and SOS responses and their effects on log-phase mRNA turnover. J. Bacteriol. 2006; 188:6739–6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Brandi A. Massive presence of the Escherichia coli ‘major cold-shock protein’ CspA under non-stress conditions. EMBO J. 1999; 18:1653–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Giuliodori A.M., Di Pietro F., Marzi S., Masquida B., Wagner R., Romby P., Gualerzi C.O., Pon C.L.. The cspA mRNA is a thermosensor that modulates translation of the cold-shock protein CspA. Mol. Cell. 2010; 37:21–33. [DOI] [PubMed] [Google Scholar]
- 29. Gualerzi C.O., Maria Giuliodori A., Pon C.L.. Transcriptional and post-transcriptional control of cold-shock genes. J. Mol. Biol. 2003; 331:527–539. [DOI] [PubMed] [Google Scholar]
- 30. Goldstein J., Pollitt N.S., Inouye M.. Major cold shock protein of Escherichia coli. Proc. Natl Acad. Sci. U.S.A. 1990; 87:283–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yamanaka K., Mitta M., Inouye M.. Mutation analysis of the 5′ untranslated region of the cold shock cspA mRNA of Escherichia coli. J. Bacteriol. 1999; 181:6284–6291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Narberhaus F., Waldminghaus T., Chowdhury S.. RNA thermometers. FEMS Microbiol. Rev. 2006; 30:3–16. [DOI] [PubMed] [Google Scholar]
- 33. Ignatov D., Vaitkevicius K., Durand S., Cahoon L., Sandberg S.S., Liu X., Kallipolitis B.H., Rydén P., Freitag N., Condon C.et al.. An mRNA–mRNA interaction couples expression of a virulence factor and its chaperone in Listeria monocytogenes. Cell Rep. 2020; 30:4027–4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Phadtare S., Alsina J., Inouye M.. Cold-shock response and cold-shock proteins. Curr. Opin. Microbiol. 1999; 2:175–180. [DOI] [PubMed] [Google Scholar]
- 35. Wang N., Yamanaka K., Inouye M.. CspI, the ninth member of the CspA family of Escherichia coli, is induced upon cold shock. J. Bacteriol. 1999; 181:1603–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Catalan-Moreno A., Caballero C.J., Irurzun N., Cuesta S., López Sagaseta J., Toledo-Arana A.. One evolutionarily selected amino acid variation is sufficient to provide functional specificity in the cold shock protein paralogs of Staphylococcus aureus. Mol. Microbiol. 2020; 113:826–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lioliou E., Sharma C.M., Caldelari I., Helfer A.C., Fechter P., Vandenesch F., Vogel J., Romby P.. Global regulatory functions of the Staphylococcus aureus endoribonuclease III in gene expression. PLoS Genet. 2012; 8:e1002782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003; 31:3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Darty K., Denise A., Ponty Y.. VARNA: interactive drawing and editing of the RNA secondary structure. Bioinformatics. 2009; 25:1974–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Caballero C.J., Menendez-Gil P., Catalan-Moreno A., Vergara-Irigaray M., García B., Segura V., Irurzun N., Villanueva M., Ruiz de Los Mozos I., Solano C.et al.. The regulon of the RNA chaperone CspA and its auto-regulation in Staphylococcus aureus. Nucleic Acids Res. 2018; 46:1345–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lee J.C. Electrotransformation of staphylococci. Methods Mol. Biol. 1995; 47:209–216. [DOI] [PubMed] [Google Scholar]
- 42. Balestrino D., Hamon M.A., Dortet L., Nahori M.-A., Pizarro-Cerda J., Alignani D., Dussurget O., Cossart P., Toledo-Arana A.. Single-cell techniques using chromosomally tagged fluorescent bacteria to study Listeria monocytogenes infection processes. Appl. Environ. Microbiol. 2010; 76:3625–3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lee G., Talkington C., Pero J.. Nucleotide sequence of a promoter recognized by Bacillus subtilis RNA polymerase. Mol. Gen. Genet. 1980; 180:57–65. [DOI] [PubMed] [Google Scholar]
- 44. Charpentier E., Anton A.I., Barry P., Alfonso B., Fang Y., Novick R.P.. Novel cassette-based shuttle vector system for Gram-positive bacteria. Appl. Environ. Microbiol. 2004; 70:6076–6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Arnaud M., Chastanet A., Debarbouille M.. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, Gram-positive bacteria. Appl. Environ. Microbiol. 2004; 70:6887–6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Valle J., Toledo-Arana A., Berasain C., Ghigo J.-M., Amorena B., Penadés J.R., Lasa I.. SarA and not sigma B is essential for biofilm development by Staphylococcus aureus. Mol. Microbiol. 2003; 48:1075–1087. [DOI] [PubMed] [Google Scholar]
- 47. Toledo-Arana A., Dussurget O., Nikitas G., Sesto N., Guet-Revillet H., Balestrino D., Loh E., Gripenland J., Tiensuu T., Vaitkevicius K.et al.. The Listeria transcriptional landscape from saprophytism to virulence. Nature. 2009; 459:950–956. [DOI] [PubMed] [Google Scholar]
- 48. Menendez-Gil P., Caballero C.J., Solano C., Toledo-Arana A.. Fluorescent molecular beacons mimicking RNA secondary structures to study RNA chaperone activity. Methods Mol. Biol. 2020; 2106:41–58. [DOI] [PubMed] [Google Scholar]
- 49. Krismer B., Liebeke M., Janek D., Nega M., Rautenberg M., Hornig G., Unger C., Weidenmaier C., Lalk M., Peschel A.. Nutrient limitation governs Staphylococcus aureus metabolism and niche adaptation in the human nose. PLoS Pathog. 2014; 10:e1003862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lasa I., Toledo-Arana A., Dobin A., Villanueva M., de los Mozos I.R., Vergara-Irigaray M., Segura V., Fagegaltier D., Penadés J.R., Valle J.et al.. Genome-wide antisense transcription drives mRNA processing in bacteria. Proc. Natl Acad. Sci. U.S.A. 2011; 108:20172–20177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lindemann J., Leiacker R., Rettinger G., Keck T.. Nasal mucosal temperature during respiration. Clin. Otolaryngol. Allied Sci. 2002; 27:135–139. [DOI] [PubMed] [Google Scholar]
- 52. Jiang W., Hou Y., Inouye M.. CspA, the major cold-shock protein of Escherichia coli, is an RNA chaperone. J. Biol. Chem. 1997; 272:196–202. [DOI] [PubMed] [Google Scholar]
- 53. Phadtare S., Inouye M.. Sequence-selective interactions with RNA by CspB, CspC and CspE, members of the CspA family of Escherichia coli. Mol. Microbiol. 1999; 33:1004–1014. [DOI] [PubMed] [Google Scholar]
- 54. Sachs R., Max K.E.A., Heinemann U., Balbach J.. RNA single strands bind to a conserved surface of the major cold shock protein in crystals and solution. RNA. 2012; 18:65–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Shenhar Y., Biran D., Ron E.Z.. Resistance to environmental stress requires the RNA chaperones CspC and CspE. Environ. Microbiol. Rep. 2012; 4:532–539. [DOI] [PubMed] [Google Scholar]
- 56. Michaux C., Holmqvist E., Vasicek E., Sharan M., Barquist L., Westermann A.J., Gunn J.S., Vogel J.. RNA target profiles direct the discovery of virulence functions for the cold-shock proteins CspC and CspE. Proc. Natl Acad. Sci. U.S.A. 2017; 114:6824–6829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Johansson J., Mandin P., Renzoni A., Chiaruttini C., Springer M., Cossart P.. An RNA thermosensor controls expression of virulence genes in Listeria monocytogenes. Cell. 2002; 110:551–561. [DOI] [PubMed] [Google Scholar]
- 58. Waldminghaus T., Heidrich N., Brantl S., Narberhaus F.. FourU: a novel type of RNA thermometer in Salmonella. Mol. Microbiol. 2007; 65:413–424. [DOI] [PubMed] [Google Scholar]
- 59. Loh E., Kugelberg E., Tracy A., Zhang Q., Gollan B., Ewles H., Chalmers R., Pelicic V., Tang C.M.. Temperature triggers immune evasion by Neisseria meningitidis. Nature. 2013; 502:237–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Weber G.G., Kortmann J., Narberhaus F., Klose K.E.. RNA thermometer controls temperature-dependent virulence factor expression in Vibrio cholerae. Proc. Natl Acad. Sci. U.S.A. 2014; 111:14241–14246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Righetti F., Nuss A.M., Twittenhoff C., Beele S., Urban K., Will S., Bernhart S.H., Stadler P.F., Dersch P., Narberhaus F.. Temperature-responsive in vitro RNA structurome of Yersinia pseudotuberculosis. Proc. Natl Acad. Sci. U.S.A. 2016; 113:7237–7242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Twittenhoff C., Heroven A.K., Mühlen S., Dersch P., Narberhaus F.. An RNA thermometer dictates production of a secreted bacterial toxin. PLoS Pathog. 2020; 16:e1008184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Loh E., Righetti F., Eichner H., Twittenhoff C., Narberhaus F.. RNA thermometers in bacterial pathogens. Microbiol. Spectrum. 2018; 6:RWR-0012-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Altuvia S., Kornitzer D., Teff D., Oppenheim A.B.. Alternative mRNA structures of the cIII gene of bacteriophage lambda determine the rate of its translation initiation. J. Mol. Biol. 1989; 210:265–280. [DOI] [PubMed] [Google Scholar]
- 65. Hussein H., Fris M.E., Salem A.H., Wiemels R.E., Bastock R.A., Righetti F., Burke C.A., Narberhaus F., Carroll R.K., Hassan N.S.et al.. An unconventional RNA-based thermosensor within the 5′UTR of Staphylococcus aureus cidA. PLoS One. 2019; 14:e0214521. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Any data or material that support the findings of this study can be made available by the corresponding author upon request.