Abstract
Forkhead transcription factors bind a canonical consensus DNA motif, RYAAAYA (R = A/G, Y = C/T), as a monomer. However, the molecular mechanisms by which forkhead transcription factors bind DNA as a dimer are not well understood. In this study, we show that FOXO1 recognizes a palindromic DNA element DIV2, and mediates transcriptional regulation. The crystal structure of FOXO1/DIV2 reveals that the FOXO1 DNA binding domain (DBD) binds the DIV2 site as a homodimer. The wing1 region of FOXO1 mediates the dimerization, which enhances FOXO1 DNA binding affinity and complex stability. Further biochemical assays show that FOXO3, FOXM1 and FOXI1 also bind the DIV2 site as homodimer, while FOXC2 can only bind this site as a monomer. Our structural, biochemical and bioinformatics analyses not only provide a novel mechanism by which FOXO1 binds DNA as a homodimer, but also shed light on the target selection of forkhead transcription factors.
INTRODUCTION
The Forkhead box (FOX) proteins are a family of transcription factors that play important regulatory roles in a wide variety of biological functions, including cell proliferation, immunity, apoptosis and metabolism (1,2). In the human genome, 50 FOX members have been identified and can be grouped into 19 subfamilies (FOXA to FOXS) based on sequence similarity (3–5). Among the 19 subgroups, FOXOs (consisting of FOXO1, FOXO3, FOXO4 and FOXO6) are characterized by their critical regulatory roles in oxidative stress resistance and cell longevity (6–8). Emerging evidence suggests that FOXOs are tumor suppressors (9–11). It has been reported that FOXOs can interact with other transcription factors during gene regulation. For example, FOXO4 and p53 form a complex and act as a pivot in senescent cell viability (12); FOXO1 and FOXA1 or FOXA2 can form a complex on DNA and cooperate to open chromatin at insulin-regulated genes (13).
FOX transcription factors share a conserved forkhead domain, or winged-helix DNA binding domain (DBD), which recognizes and binds the consensus forkhead binding motif, RYAAAYA (R = A/G, Y = C/T) (14–17). Structurally, the FOX DBD adopts a winged-helix architecture consisting of three α-helices, three β-strands and two less conserved winged loops (wing1 and wing2) (16–19). Upon DNA binding, the third helix (H3) inserts into the major groove of the DNA in an orientation nearly perpendicular to the DNA duplex and contributes to the majority of base-specific interactions. In addition, the N-terminus and the two wing loops of DBD together play divergent roles in DNA binding by interacting with DNA phosphate groups. Although the interaction between the FOX DBD and the consensus DNA motif has been well characterized, how forkhead proteins distinguish their specific gene targets remains unclear.
Due to their functional importance, structures of multiple FOX/DNA complexes have been reported. These studies have revealed that forkhead transcription factors bind the consensus DNA motif as a monomer (16–21). Interestingly, FOXP3 and FOXP2 can bind two separate DNA sites and form domain-swapped homodimers. This unusual binding mode has implications in chromatin looping and long-range gene regulation (22–24). In addition, forkhead dimer motifs have also been identified in open chromatin using computational methods (25). A compact element with partially overlapping bases (AAATATTT, named as DIV0 site) was subsequently shown to be bound by FOXA1 in a highly cooperative fashion (26).
High-throughput SELEX (HT-SELEX) identified a possible FOXO dimeric site containing inverted repeats of two forkhead recognition sites, which can be named as DIV2 motif (termed ‘DIV’ for its diverging half-sites) according to a previous study (26,27). However, whether FOXO1 protein indeed binds DIV2 site, the biological relevance of FOXO1 binding DIV2 site, the mechanism by which FOXO1 protein recognizes and binds DIV2 site, do other forkhead transcription factors bind DIV2 site, remain unclear. In this study, we aim to address these unanswered questions.
MATERIALS AND METHODS
Protein expression and purification
The gene fragment encoding human FOXO1–DBD (UniProt ID: Q12778, residues 151–266) was cloned into pET28a expression vector. The PreScission protease cleavage site (LEVLFQGP) was inserted between the 6xHis tag and FOXO1–DBD. Recombinant protein was overexpressed in Escherichia coli Rosetta BL21 (DE3). Cells were grown at 37°C in LB culture medium, induced with 0.5 mM IPTG when the culture OD600 reached 1.0 and further cultured at 21°C for 8 h. Cells were harvested, lysed by sonication in the buffer containing 50 mM HEPES pH 7.5, 500 mM NaCl, 5 mM β-mercaptoethanol, then clarified by centrifugation. The supernatant was loaded onto a nickel-affinity chromatography column and subsequently treated with PreScission protease to remove the 6xHis fusion tag, followed by ion exchange and size exclusion chromatography for further purification. The purified protein was stored in a buffer containing 10 mM HEPES (pH 7.5), 150 mM NaCl and 1 mM Tris(2-carboxyethyl)phosphine (TCEP). Then, the samples were concentrated to 30 mg/ml and stored at −80°C.
The gene sequences encoding FOXO3-DBD (residues 158–253), FOXM1-DBD (residues 222–360), FOXC2-DBD (residues 72–171) and FOXI1-DBD (residues 119–223) were also cloned into pET28a, respectively. The recombinant proteins were expressed and purified as described above for FOXO1–DBD. FOXO1 mutants were generated by site-directed mutagenesis.
DNA oligo preparation
All forward-strand DNA oligonucleotides and their corresponding complementary strands were purchased from GenScript (Suzhou, China). Duplex DNAs were generated by annealing as previously described (28). The FKH_26 (26 indicates the length of DNA) DNA duplex contained the strands 5′-gacgaaGTAAACAcggcaggttcgtc-3′ and 5′- gacgaacctggcgTGTTTACttcgtc-3′. The palindromic DNA duplex DIV2_26 (26 indicates the length of DNA) comprised 5′-gacgaaGTAAACATGTTTACttcgtc-3′, which is a reverse complement to itself. All DNAs used for electrophoretic mobility shift assays (EMSAs) and Isothermal titration calorimetry assays were annealed to 45 and 900 μM, respectively. The DNA sequence for crystallization is 5′-ACCGTAAACATGTTTACGGT-3′, which is a reverse complement to itself.
Qualitative electrophoretic mobility shift assay (EMSA)
Qualitative EMSAs were performed as previously described (14). In brief, the binding reactions were performed in a total mixture volume of 8 μl. The EMSA buffer used here contained 20 mM HEPES, pH 7.5, 200 mM NaCl, and 10 mM MgCl2. Unlabeled DNA was incubated with protein for 20 min on ice. DNA concentration in the reaction was 6 μM. The reaction mixture was loaded onto 8% (w/v) native polyacrylamide gel using 0.5× TBE as running buffer.
Quantitative electrophoretic mobility shift assay (EMSA)
5′ 6-FAM-labeled forward strand DNA oligo and their unlabeled reverse complementary DNA strand were purchased form GenScript (Suzhou, China), and annealed. 100 nM of dsDNA was incubated with varying concentrations of protein. The bands were detected with a ChemiDoc XRS scanner (Bio-Rad, USA) using 492/518 nm excitation/emission wavelengths and band intensities were quantified with ImageJ. Cooperativity factors were calculated as previously described (29).
Transient transfection assay
293T Cells were cultured in DMEM contained 10% fetal bovine serum (FBS). 293T cells were co-transfected with full-length human FOXO1-encoding plasmid pcDNA3.1-FOXO1 or the empty pcDNA3.1 plasmid, and pGL3-promoter plasmid, pGL3-3x-FKH (containing 3x FKH motif) or pGL3-3x-DIV2 (containing 3x DIV2 motif) plasmid, using Lipofectamine 2000 (Invitrogen). The transfected cells were cultured for 48 h. Luciferase activities were determined using the Firefly Luciferase Reporter Gene Assay Kit (Beyotime). The results shown are the means and standard deviations of triplicate points and are representative of at least three independent experiments.
Isothermal titration calorimetry (ITC) assays
ITC analyses were performed at 25°C using NANO ITC instrument (TA Instruments). The purified protein was placed in ITC buffer (20 mM HEPES, pH7.5, 250 mM NaCl, 10 mM MgCl2 and 1 mM TCEP) at the concentrations of 25–40 μM. DNA duplex was dissolved in the same buffer to the concentrations of 90–180 μM. The DNA was titrated in duplicate into FOX-DBDs using 2 μl injections with 300 s intervals. Data were processed using the launch NanoAnalyze software.
Fluorescence polarization (FP) assay
FP assays were performed on an Enision multilabel reader (Perkin Elmer) using 96-well optiplates (Corning). The binding reaction is carried out in 0.5× PBS with serial dilutions of FOXO1–DBD (from 1 nM to 350 nM) and 2 nM 6-FAM labeled DNA. After 30 minutes of incubation at room temperature, the dilutions were measured with λex = 480 nm and λem = 535 nm. All assays were performed in triplicate and Kd values were determined from a non-linear dose-response curve in GraphPad Prism 7.0.
Differential scanning fluorimetry (DSF) assay
DSF assays were performed using a LightCycler 480 real-time PCR device (Roche, Switzerland) as described previously (30). Protein (at a final concentration of 2.0 mg/ml) and SYPRO Orange (Sigma, USA, at a final concentration of 5.0 mg/ml) were mixed and placed into the instrument at a heating rate of 1°C/min. The fluorescence intensity versus temperature (melting curve) was measured, and a melting temperature (Tm) was calculated from the maximum value of the first derivative of the melt curve. The data were analyzed using the LightCycler 480 software, and graphics were produced using the program Origin 8.0.
Crystallization
FOX-DBD/DNA complexes were prepared by mixing the purified DBD and DIV2 at a 5:3 molar ratio with a final protein concentration of 10 mg/ml. Crystals were screened by hanging drop vapor diffusion at 4°C. The crystal of the FOXO1/DIV2 complex was grown under the conditions of 50 mM NaAc, pH 4.7, 12–18% PEG4000 (w/v), 200 mM NaCl, 10 mM MgCl2 and 1 mM TCEP. The crystal of the FOXC2/DIV2 complex was obtained in the reservoir containing 50 mM HEPES, pH 7.5, 8–12% PEG4000 (w/v), 250 mM NaCl, 10 mM MgCl2 and 1 mM TCEP. All crystals were treated with cryoprotectant containing the corresponding crystallization reservoir plus 20% glycerol, followed by flash freezing and storage in liquid nitrogen for further data collection.
Data collection and structure determination
Crystal data were collected at the BL17U1 and BL19U1 beamlines of Shanghai Synchrotron Radiation Facility (SSRF), and the data were processed by HKL2000 (31–33). Phases were determined by molecular replacement (MR) using Phenix.phaser (34). For the FOXO1/DIV2 structure, a previously determined FOXO1 structure (PDB ID: 3C06) was used as the search model (17). The initial model was manually built using the program Coot (35). Refinement was carried out with Phenix.refine (36). To solve the FOXC2/DIV2 structure, our previously determined FOXC2/DBE2 structure (PDB ID, 6AKO) was used as a search model (16).
Bioinformatics analysis
The ChIP-seq data of FOXO3 binding in human DLD1 cells was downloaded from the publicly available repository (GSE35486) (37). The raw dataset was processed with the following pipeline: reads were mapped to the reference genome (hg19 assembly, NCBI build 37) using the BWA package (38). Next, peaks were called using Cisgenome software package (42), and the peak sequences were extracted. The nearest transcription start sites to these peaks were reported, and the sequences of these peaks were searched for matches to the DIV2 motif using R package TFBSTools (39). The motif was searched with the function ‘searchSeq’ and the parameter ‘min.score = 80%’ in R package TFBSTools using the PWMatrix of the motif. The percentage depends on the minimum score for the hit(‘min.score’). The width of sampled peak regions is 200–850 bp (median: 250 bp) for FOXO3 ChIP-seq data. The width of sampled peak regions is 218–13649 bp (median: 658 bp) for FOXM1 ChIP-seq data. Both strands were searched. The number of peaks (rather than the number of motif occurrence) was counted; if a peak contained the motif, it was counted as one hit, even if the motif occurred more than once in this peak. The data of gene expression regulation of FOXO3 were from the micro-array analysis of a previous study (40).
RESULTS
FOXO1 binds the palindromic DNA motif DIV2 and mediates gene transcription
Forkhead transcription factors have been reported to recognize the canonical forkhead motif, 5′-RYAAAYA-3′, and an alternate motif, 5′-GACGC-3′ (14–16). Recently, a palindromic DNA site, DIV2, has been identified as a potential FOXO binding site from high-throughput SELEX study (27). DIV2 site consists of two canonical forkhead motifs arranged in an inverted repeat orientation, without spacer between the two motifs (Figure 1A). As two forkhead recognition motifs are present in the DIV2 site, we speculated that this DNA motif could be bound by two FOXO proteins. To determine the binding characteristics of the FOXO1 to the DIV2 site, we carried out qualitative electrophoretic mobility shift assay (EMSA) using unlabeled DNA. Two sequences were tested: FKH containing a single GTAAACA core sequence, and DIV2 containing two palindromic GTAAACA sites. As shown in Figure 1B, FOXO1–DBD formed one complex band with FKH. In contrast, FOXO1 formed two complex bands with DIV2. The lower band migrated to the similar position as the FOXO1 monomer bound to FKH, while the upper band presumably represented the dimeric complex. When the amount of FOXO1 protein increased, FOXO1–DBD formed an enhanced dimeric complex with DIV2. On the other hand, FOXO1–DBD mainly bound as a monomer on FKH even at the highest protein concentration, although a weak dimer forming band could be observed (Figure 1B). In addition, we estimated the cooperativity value (ω) after quantifying dimer, monomer and free DNA fractions from quantitative EMSAs using 6-FAM-labeled DNA. As shown in Supplementary Figure S1, FOXO1 binds DIV2 DNA with positive cooperativity (ω = 4.5), indicating that two FOXO1 molecules bind DIV site in a cooperative manner.
Figure 1.
FOXO1 binds a palindromic DNA motif DIV2. (A) MEME analysis of the FKH and DIV2 motifs. FKH, canonical forkhead binding motif; DIV2, diverging half-sites. The position weight matrix (PWM) for both sites were obtained from FootprintDB (http://floresta.eead.csic.es/footprintdb/index.php). 2049 sites were used for FKH site, and 553 sites were used for DIV2 site. (B) DNA binding features of FOXO1–DBD were measured using qualitative EMSA using unlabeled DNA. The DNA sequences of FKH_26 and DIV2_26 are listed, and the core binding sequences are underlined. Band intensities are quantified and normalized with values shown underneath relevant lanes.
To study the biological relevance of the dimeric FOXO interaction on DIV2, we investigated the occurrence of the DIV2 motif in a FOXO3 ChIP-seq dataset (GSE35486) (37). The position weight matrix (PWM) in Figure 1A were used for the motif count analysis. A total of 242 unique peaks with DIV2 motif were identified, accounting for about a quarter of the total FOXO3 binding sites (Figure 2A). DIV2 motif was found to be centralized within these peaks (Figure 2B). Furthermore, DIV2 motif was found to be located in the promoter regions of genes such as SORL1 and TMEM140 (Figure 2C). We then analyzed the expression level of these genes upon FOXO3 activation using a previously published microarray dataset (40), and found that the expression level of SORL1 and TMEM140 were unregulated by 2.9- and 4.8-fold, respectively (Figure 2C).
Figure 2.
FOXO3 recognizes DIV2 motif in cells and promotes gene transcription. (A) Ratio of DIV2 motif in FOXO3 ChIP-seq data. (B) Composite profile of DIV2 motif located within the ChIP-seq peaks of FOXO3. The density estimation of the data is shown in black. (C) The snap view of two promoter regions that have peaks containing DIV2 motif. The positions of DIV2 sites are shown with red bars. The expression level of these genes upon FOXO3 overexpression was derived from a previously published microarray dataset. (D) 293T cells were co-transfected with FOXO1 expressing plasmid and pGL3-promoter luciferase reporter vector (empty), pGL3–3x-FKH, or pGL3–3x-DIV2. Results shown are means ± SD from triplicate samples, and are representative of three independent experiments.
In addition, we investigated the transcriptional activity of FOXO1 binding FKH site and DIV2 site. 293T cells were co-transfected with pcDNA3.1 expression (pcDNA-empty and pCDNA-FOXO1) and pGL3-promoter (pGL3-empty, pGL3–3x-FKH and pGL3–3x-DIV2) vectors. The results show that pGL3–3x-FKH increases the luciferase activity by 3.8-fold compared to pGL3-empty vector, and pGL3–3x-DIV2 shows 5.4-fold higher activity compared to pGL3–3x-FKH (Figure 2D). These observations suggest that DIV2 site may have stronger regulatory activity than that of FKH site.
Overall structure of the FOXO1/DIV2 complex
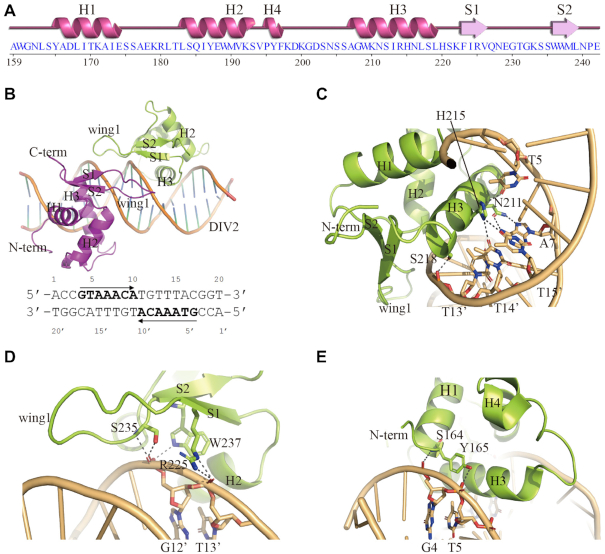
To better characterize the binding of FOXO1 to the DIV2 site, we determined the crystal structure of FOXO1–DBD in complex with a 20-bp palindromic DNA duplex containing the DIV2 motif. Secondary structure plot for FOXO1–DBD is shown in Figure 3A. The complex crystal diffracted to a resolution of 3.05 Å and belonged to the space group p212121 (Supplementary Table S1). The solved structure contains three FOXO1–DBD/DIV2 complexes in the asymmetric unit (ASU) (Supplementary Figure S2). The three complexes share similar overall structures, with RMSDs for Cα atoms of about 0.55, 0.57 and 0.65 Å, respectively. Each FOXO1–DBD/DIV2 complex is formed by one molecule of the DIV2 DNA motif and two copies of the FOXO1–DBD that bind to the DNA in a head-to-head orientation (Figure 3B). The determined crystal structure is consistent with the EMSA results showing that FOXO1–DBD forms a homodimer at the DIV2 site.
Figure 3.
Structure of the FOXO1–DBD/DIV2 ternary complex. (A) Secondary structure plot for FOXO1–DBD. (B) Structure of the FOXO1–DBD/DIV2 ternary complex. The two FOXO1–DBD molecules are colored in magenta and lime, respectively. The DNA is colored in orange. The sequences of the palindromic DIV2 DNA used for the crystallization are listed below, and the core motifs are highlighted. (C) DNA recognition by the FOXO1–DBD H3 helix. (D) DNA recognition by the FOXO1–DBD wing1. (E) DNA recognition by the FOXO1–DBD N-terminus. Hydrogen bonds are presented as dashed lines.
In the complex, FOXO1–DBD adopts the typical winged-helix fold, similar to other FOX family members (Figure 3C and Supplementary Figure S3). Overall, FOXO1–DBD comprises three stacking α-helices (H1, H2 and H3) and is capped at one end by a β-sheet composed of two antiparallel β-strands (S1 and S2) and a loop (wing 1) between S1 and S2 (Figure 3C and Supplementary Figure S3). The turn between H2 and H3 contains a 310 helix (H4), as observed in other FOX structures (15,16). The C-terminal wing2 (residues 246–265) exhibits poor electron density, suggesting that this region is flexible upon binding to DNA, consistent with previous structural studies (17).
DNA recognition in the FOXO1/DIV2 complex
Due to the palindromic nature of DIV2 DNA used in crystalizing, the detailed protein–DNA interactions between the two FOXO1–DBD molecules and the DIV2 site are almost identical. FOXO1–DBD binds to the 5′-GTAAACA-3′ core site of the DIV2 motif. As observed in other FOX-DBD/DNA structures, the H3 helix of FOXO1–DBD docks into the major groove of DIV2 and makes the majority of the base-specific contacts. In detail, Asn211 forms bidentate hydrogen contacts with Ade7 through its side chain, and the His215 imidazole ring forms hydrogen bonds with both Thy13′ and Thy14′ (Figure 3C). In addition to base-specific recognition, Ser218 of the H3 helix binds the phosphate backbone of T13′ (Figure 3C). Overall, the three conserved residues Asn211, His215 and Ser218 contact with DIV2 motif in the DNA major groove by forming hydrogen interactions.
The wing1 of FOXO1–DBD also participates in DNA recognition through hydrogen interactions with the DNA backbone in the minor groove. The conserved Arg225 and Ser235 residues interact with the phosphate groups of Thy13′ and Gua12′, respectively (Figure 3D). In addition, the N-terminal residues of H1 and S2 contribute to DNA recognition by forming hydrogen bonds with DNA phosphate groups (Figure 3E). In addition to these hydrogen bond interactions, a large number of van der Waals contacts are observed in the structure and further stabilize the protein-DNA interaction (Supplementary Figure S4).
FOXO1–DBD dimer interface
An interesting feature of our FOXO1–DBD/DIV2 structure is that intermolecular protein-protein interactions are present, with clearly defined density (Supplementary Figure S5A). The dimerization is mediated by the FOXO1–DBD wing1 loop, with a small buried surface area of 386 Å2. There are four hydrogen bonds formed between the Arg225–Gln227 residues pairs of both chains (Figure 4A). In addition to these hydrogen bond interactions, several van de Waals contacts were also observed and further contribute to the stability of the dimer interface (Figure 4A).
Figure 4.
FOXO1–DBD dimer interface. (A) Protein–protein interactions in the dimer interface. Hydrogen bonds are presented as black dashed lines, and van de Waals contacts are showed as orange dashed lines. (B) The DNA binding properties of the wild type, R225E and Q227R mutants of FOXO1–DBD were measured using qualitative EMSA.
To investigate whether the interaction observed in the FOXO1 dimerization interface was important for FOXO1 DNA binding, we constructed two FOXO1 mutants (R225E, Q227R) and performed EMSA to assess their binding to DIV2. As shown in Figure 4B, both mutations did not affect the DNA binding of FOXO1 upon the FKH DNA, which bound as a monomer. However, both mutants lost the ability to form a dimer, and bound the DIV2 site only as a monomer (Figure 4B). In addition, we constructed FOXO1 R225A and Q227A mutants and assessed their binding to DIV2 site. FOXO1 R225A and Q227A mutants were able to bind DIV2 site as a dimer, however their binding affinity was weaker than that of the wild-type FOXO1 (Supplementary Figure S5B). These results suggest that the dimerization interface is important for FOXO1 binding to the DIV2 site.
DNA binding affinity and stability
We then measured the binding affinity of FOXO1–DBD to the DIV2 site as well as its single site counterpart, FKH_26, using isothermal titration calorimetry (ITC). The representative binding isotherm of DIV2_26 is presented (Figure 5A, top panel). The Kd value of FOXO1–DBD bound to FKH_26 was estimated to be 1.38 μM, while the Kd value of FOXO1–DBD toward DIV2_26 was estimated to be 0.15 μM (Figure 5A, bottom panel). In addition, we performed FP assays to measure the binding affinities. The Kd of the DIV2 site was 73 nM, while the Kd of the FKH site was 185 nM (Supplementary Figure S6). Both ITC and FP assays show that FOXO1 binds with higher affinity to the DIV2 site than the FKH site.
Figure 5.
The DIV2 site binding affinity and stability analysis of wild type FOXO1–DBD and the mutants. (A) DNA binding affinities of FOXO1–DBD were measured by ITC. The graph represents the curve of FOXO1–DBD titrated by DIV2_26 DNA (top panel). Thermodynamic parameters are summarized in the table (bottom panel). Values represent mean and standard deviation of three independent experiments (B) Melting curves of FOXO1–DBD in the absence or presence of different DNA duplexes. The corresponding Tm values are listed.
We also performed differential scanning fluorimetry (DSF) experiments to analyze the protein stability. The melting curves of FOXO1–DBD with or without DNA are shown in Figure 5B. Without DNA, the melting temperature (Tm) of FOXO1–DBD was 51.5°C. When bound to the FKH_26 DNA, the Tm increased to 57.5°C. Upon binding to the DIV2_26 DNA, an even higher Tm of 61.4°C was obtained. In addition, the DSF assays showed that the Tm of DIV2-bound R225E and Q227R deceased to 58.5 and 56.1°C, respectively (Supplementary Figure S7). These results suggest that FOXO1 binds the palindromic DIV2 site with higher thermal stability than that of FKH site.
Different forkhead transcription factors bind DIV2 site differently
In order to test whether other forkhead transcription factors can also bind DIV2 sites, we studied the DIV2 binding features of four other forkhead proteins. Similar to FOXO1–DBD, FOXO3–DBD, FOXM1–DBD and FOXI1–DBD all bind DIV2 in a dimeric manner (Figure 6A-C). Among these three proteins, FOXM1 seems to have stronger dimeric binding to the DIV2 DNA site than FOXI1 and FOXO3 (Figure 6A-C). Unexpectedly, FOXC2–DBD can only bind DIV2 sites as a monomer even at high protein concentrations (Figure 6D). As FOXM1 shows a strong tendency to form dimer on DIV2, we investigated the occurrence of the DIV2 motif in a FOXM1 ChIP-seq dataset (GSE40767) (41). A total of 21 844 unique peaks with DIV2 motif were identified, accounting for ∼46% of the total FOXM1 binding sites (Figure 6E). In addition, the intensity of ChIP-seq peaks from DIV2 sites was higher than that from FKH sites (Supplementary Figure S8).
Figure 6.
Binding properties of other forkhead transcription factors to DIV2 site using qualitative EMSA. (A) FOXO3–DBD. (B) FOXM1–DBD. (C). FOXI1–DBD. (D) FOXC2–DBD. (E) Ratio of DIV2 motif in FOXM1 ChIP-seq data. (F) Multiple sequence alignment of forkhead DBD wing1. (G) DNA binding properties of chimeric forkhead proteins. FOXO1-WT: FOXO1–DBD wildtype; FOXO1c2w: FOXO1–DBD protein with its wing1 region substituted with FOXC2 wing1; FOXC2o1w: FOXC2–DBD protein with its wing1 region substituted with FOXO1 wing1. (H) Structure of FOXC2–DBD/DIV2 complex.
The wing1 region is less conserved among forkhead DBDs (Figure 6F), and our results suggest that the wing1 of FOXO1 plays an important role in the dimeric binding of FOXO1 to the DIV2 site. In order to test the importance of the wing1 region, we constructed two chimera proteins: FOXO1c2w (FOXO1–DBD protein with its wing1 region substituted with FOXC2 wing1) and FOXC2o1w (FOXC2–DBD protein with its wing1 region substituted with FOXO1 wing1). When FOXO1 wing1 is replaced with FOXC2 wing1, FOXO1c2w loses its ability to bind DIV2 site as a homodimer, and binds DIV2 site as a monomer; however, when FOXC2 wing1 is replaced with FOXO1 wing1, FOXC2o1w still binds DIV2 site as a monomer, and fails to gain the ability to bind DIV2 site as a homodimer (Figure 6G). These results suggest that the wing1 region of FOXO1 is important for its binding to DIV2 site as a homodimer, however, there may be other factors that also contribute to the mechanism by which FOXC2 can only bind DIV2 site as a monomer.
Structure of FOXC2 in complex with the DIV2 DNA site
To study the mechanism by which FOXC2–DBD can only bind DIV2 site as a monomer, we carried out crystallographic studies of FOXC2–DBD in complex with the DIV2 motif. The structure of the FOXC2/DIV2 complex was determined at 2.85 Å (Supplementary Table S1). In the FOXC2/DIV2 structure, FOXC2–DBD binds DIV2 at a 1:1 molar ratio, supporting our biochemical observation (Figure 6H). In the experiment, we used the same 20-bp DIV2 DNA containing two forkhead binding sites. However, only one binding site was bound by FOXC2-DBD. It is worth noting that the end of the unbound site has very poor electron density, therefore part of the DNA is missing in the deposited pdb (6LBM).
We compared the structural variations between FOXO1/DIV2 and FOXC2/DIV2 complexes by superimposing them. The two DBDs share a similar overall winged-helix fold. The superposition of FOXC2-DBD on FOXO1–DBD gave 0.47 Å root mean square deviations (RMSD) for all aligned Cα atoms. The structural variations are mainly located in the wing1 and wing2 regions (Supplementary Figure S9A). FOXC2 exhibits similar major groove recognition to the DIV2 site as FOXO1 (Supplementary Figure S9B). FOXC2 wing1 region is also involved in DNA recognition in the corresponding structure, however its conformation is different from that of FOXO1 (Supplementary Figure S9C). FOXO1 wing1 is arranged in an orientation nearly parallel to the DNA, while the wing1 of FOXC2 points outward, away from the DNA (Supplementary Figure S9A). In the FOXC2/DIV2 structure, the FOXC2 wing2 is involved in DNA recognition by interacting with DNA backbone phosphate groups (Supplementary Figure S9D).
To elucidate the mechanism by which FOXC2 can only bind DIV2 site as a monomer, we made a computational dimeric binding model of FOXC2–DBD on the DIV2 site. To our surprise, no obvious clash was present between the two FOXC2–DBD molecules in our model (Supplementary Figure S10). This observation, together with the fact that FOXC2o1w chimeric protein cannot gain the ability to bind DIV2 site as a dimer, suggest that there might be some unknown mechanisms preventing FOXC2 from dimerizing over the DIV2 site.
We then performed qualitative EMSA experiments to test how altered half-site spacing affects dimeric binding of FOXO1. Of the five tested (DIV0 to DIV4), FOXO1 had the strongest tendency to dimerize on DIV0, followed by DIV2; FOXO1 mainly bound DIV1, DIV3 and DIV4 as monomer, although a weak dimer forming band could be observed at high protein concentration (Supplementary Figure S11). We further tested the binding cooperativity of FOXO1 on the DIV0 site using quantitative EMSA experiments using 6-FAM-labeled DNA, and found that FOXO1 bound the DIV0 site with high cooperativity, with a cooperativity factor of 54 (Supplementary Figure S12).
These results suggest that altered half-site spacing between half-sites affects dimeric binding of FOXO1.
Comparison with published FOX/DNA structures
In order to provide more structural insights into the molecular mechanism by which FOX proteins bind DNA, we compared our structure with previously published FOX/DNA structures (Supplementary Figure S13). FOXO1 and FOXA2 bind FKH site as monomer (17,42), while two FOXP3 molecules form a domain-swapped dimer and bind two separate DNA sites. In the FOXM1/DNA complex structure, a 19-bp duplex contained two FOXM1 recognition sites in a palindromic orientation (atTGTTTA-TAAACAgcccg) is bound by two FOXM1 molecules, however structural, functional and bioinformatics data showed no preference of FOXM1 for this tandem recognition sequences (19). In a recently published FOXO1/DNA structure, one FOXO1 molecule binds FKH site, while the other FOXO1 molecule binds the pseudo-continuous DNA helices in the crystal packing (43). Interestingly, the crystallographically stacked DNA helices resemble the DIV0 configuration, which were bound by two FOXO1 molecules from opposite sides (Supplementary Figure S14A). In our FOXO1/DIV2 complex, two FOXO1 molecules bind DIV2 DNA from nearly vertical orientations and formed intermolecular interaction mediated by the wing1 regions of both FOXO1 molecules (Supplementary Figure S14B).
DISCUSSION
To achieve specificity in gene regulation, a common mechanism adopted by many transcription factors is cooperative binding to the same DNA site (29,44,45). Previous studies showed that most forkhead transcription factors bind the consensus DNA motif as a monomer (Figure 1) (16–19). In this study, we showed that FOXO1 binds DIV2 site as a homodimer, with intermolecular protein-protein interaction mediated by the wing1 region of FOXO1. Forming a dimer on a DNA site has at least two impacts on DNA recognition. First, it may lead to more specific DNA binding, since DIV2 sites are much less abundant than FKH sites. Second, it may enhance protein/DNA binding affinity. FOXO1 binds DIV2 with a higher affinity than that of its single-site counterpart, FKH.
Previous studies have shown that FOXA1 binds the DIV0 site in a highly cooperative fashion, and addition of spacers separating the half-sites decreased the cooperativity (26). In our study, FOXO1 had the strongest tendency to dimerize on the DIV0 site, followed by the DIV2 site; while FOXO1 mainly bound the DIV1, DIV3, and DIV4 sites as monomer. These results suggest that altered half-site spacing can affects dimeric binding of forkhead proteins, which may subsequently affect the target gene selection and gene expression. In addition, our studies show that FOXO1 binds the DIV0 site with high cooperativity, suggesting that the DIV0 site may also be an important dimeric site for FOXO1.
Forkhead transcription factors possess a highly conserved DBD but have divergent wing1 regions. Previously, the wing1 region has been reported to engage in DNA recognition as well as interaction with NFAT (46,47). Here, we find that FOXO1 wing1 region plays an important role in mediating protein-protein interaction in the homodimer interface. Therefore, FOXO1 may not be able to interact with NFAT when bound to DIV2 site. In addition to FOXO1, our biochemical assays show that FOXO3, FOXM1 and FOXI1 can bind DIV2 DNA site as homodimer in vitro. Among these proteins, FOXM1 seems to have stronger dimeric binding to the DIV2 site. On the other hand, one forkhead transcription factor, FOXC2, can only bind the DIV2 site as a monomer. Sequence alignment of the wing1 regions among these proteins shows that the two amino acids (R225 and Q227 of FOXO1) are not well conserved. The corresponding residues in FOXM1 are arginine and threonine, while those in FOXC2 are lysine and Proline. The sequence variations at the wing1 regions among the forkhead transcription factors may contribute to the difference in their binding properties to DIV2 DNA site. In addition, R225C and R225H mutations have been identified in the COSMIC database (48), suggesting that these mutations may have some effects in human cancer.
Analysis of FOXO3-binding sites using ChIP-seq data showed that about a quarter of FOXO3-binding sites contain DIV2 motif. Interestingly, FOXO3 binds the DIV2 site located within the promoter regions of SORL1 and TMEM140 genes, and upregulate gene expression. Our transient transfection assays also suggest that the palindromic DIV2 site may have stronger regulatory activity than that of a single FKH site. The fact that FOXO can bind both DIV2 and FKH motifs with different binding affinities suggests a mechanism of target selection by FOXO proteins. FOXOs may preferentially bind the high-affinity palindromic DIV2 site at low expression level, while binding both DIV2 and FKH sites at high expression level. For instance, the expression level of FOXO1 is reduced in degenerated discs during aging (49); FOXO1 expression is up-regulated in some cancer cells such as esophageal cancer (50). Therefore, depending on its expression level, FOXO protein may regulate different gene sets, leading to different biological functions.
In summary, our biochemical, structural and bioinformatic studies provide evidence that FOXO1 binds a novel palindromic DIV2 site as a homodimer, with wing1 region mediating the dimerization. Our results show that FOXO1 dimerization enhances FOXO1 affinity for DNA, and promotes FOXO1-mediated transcription regulation. In addition, our studies reveal that four of the five forkhead transcription factors tested can bind the DIV2 site as homodimer, with the exception of FOXC2, which can only bind DIV2 site as a monomer. Whether DIV2 site is a common physiological binding site for most forkhead transcription factors, and the mechanism by which FOXC2 protein can only bind DIV2 site as a monomer, need to be further investigated in the future.
DATA AVAILABILITY
Atomic coordinates and structure factors for the reported crystal structures have been deposited with the Protein Data bank under accession codes 6LBI (FOXO1–DBD/DIV2) and 6LBM (FOXC2-DBD/DIV2).
Supplementary Material
ACKNOWLEDGEMENTS
We thank the staffs from BL17U and BL19U1 beamline of National Facility for Protein Science in Shanghai (NFPS) at Shanghai Synchrotron Radiation Facility, for assistance during data collection. We thank Dr Michael R. Stallcup for proofreading.
Contributor Information
Jun Li, Department of Oncology, NHC Key Laboratory of Cancer Proteomics, Laboratory of Structural Biology, National Clinical Research Center for Geriatric Disorders, Xiangya Hospital, Central South University, Changsha, Hunan 410008, China.
Shuyan Dai, Department of Oncology, NHC Key Laboratory of Cancer Proteomics, Laboratory of Structural Biology, National Clinical Research Center for Geriatric Disorders, Xiangya Hospital, Central South University, Changsha, Hunan 410008, China.
Xiaojuan Chen, Department of Oncology, NHC Key Laboratory of Cancer Proteomics, Laboratory of Structural Biology, National Clinical Research Center for Geriatric Disorders, Xiangya Hospital, Central South University, Changsha, Hunan 410008, China.
Xujun Liang, Department of Oncology, NHC Key Laboratory of Cancer Proteomics, Laboratory of Structural Biology, National Clinical Research Center for Geriatric Disorders, Xiangya Hospital, Central South University, Changsha, Hunan 410008, China.
Lingzhi Qu, Department of Oncology, NHC Key Laboratory of Cancer Proteomics, Laboratory of Structural Biology, National Clinical Research Center for Geriatric Disorders, Xiangya Hospital, Central South University, Changsha, Hunan 410008, China.
Longying Jiang, Department of Oncology, NHC Key Laboratory of Cancer Proteomics, Laboratory of Structural Biology, National Clinical Research Center for Geriatric Disorders, Xiangya Hospital, Central South University, Changsha, Hunan 410008, China.
Ming Guo, Department of Oncology, NHC Key Laboratory of Cancer Proteomics, Laboratory of Structural Biology, National Clinical Research Center for Geriatric Disorders, Xiangya Hospital, Central South University, Changsha, Hunan 410008, China.
Zhan Zhou, Department of Oncology, NHC Key Laboratory of Cancer Proteomics, Laboratory of Structural Biology, National Clinical Research Center for Geriatric Disorders, Xiangya Hospital, Central South University, Changsha, Hunan 410008, China.
Hudie Wei, Department of Oncology, NHC Key Laboratory of Cancer Proteomics, Laboratory of Structural Biology, National Clinical Research Center for Geriatric Disorders, Xiangya Hospital, Central South University, Changsha, Hunan 410008, China.
Huajun Zhang, Department of Oncology, NHC Key Laboratory of Cancer Proteomics, Laboratory of Structural Biology, National Clinical Research Center for Geriatric Disorders, Xiangya Hospital, Central South University, Changsha, Hunan 410008, China.
Zhuchu Chen, Department of Oncology, NHC Key Laboratory of Cancer Proteomics, Laboratory of Structural Biology, National Clinical Research Center for Geriatric Disorders, Xiangya Hospital, Central South University, Changsha, Hunan 410008, China.
Lin Chen, Molecular and Computational Biology Program, Department of Biological Sciences and Department of Chemistry, University of Southern California, Los Angeles, California 90089, United States.
Yongheng Chen, Department of Oncology, NHC Key Laboratory of Cancer Proteomics, Laboratory of Structural Biology, National Clinical Research Center for Geriatric Disorders, Xiangya Hospital, Central South University, Changsha, Hunan 410008, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [81570537 to Y.C., 81974074 to Y.C., 31900880 to H.W.]; Science and Technology Planning Project of Hunan Province [2018TP1017 to Y.C.]. Funding for open access charge: National Natural Science Foundation of China.
Conflict of interest statement. None declared.
REFERENCES
- 1. Katoh M., Igarashi M., Fukuda H., Nakagama H.. Cancer genetics and genomics of human FOX family genes. Cancer Lett. 2013; 328:198–206. [DOI] [PubMed] [Google Scholar]
- 2. Golson M.L., Kaestner K.H.. Fox transcription factors: from development to disease. Development. 2016; 143:4558–4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Myatt S.S., Lam E.W.. The emerging roles of forkhead box (Fox) proteins in cancer. Nat. Rev. Cancer. 2007; 7:847–859. [DOI] [PubMed] [Google Scholar]
- 4. Lam E.W., Brosens J.J., Gomes A.R., Koo C.Y.. Forkhead box proteins: tuning forks for transcriptional harmony. Nat. Rev. Cancer. 2013; 13:482–495. [DOI] [PubMed] [Google Scholar]
- 5. Kaestner K.H., Knochel W., Martinez D.E.. Unified nomenclature for the winged helix/forkhead transcription factors. Genes Dev. 2000; 14:142–146. [PubMed] [Google Scholar]
- 6. Martins R., Lithgow G.J., Link W.. Long live FOXO: unraveling the role of FOXO proteins in aging and longevity. Aging Cell. 2016; 15:196–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Murtaza G., Khan A.K., Rashid R., Muneer S., Hasan S.M.F., Chen J. FOXO transcriptional factors and Long-Term living. Oxid. Med. Cell. Longev. 2017; 2017:3494289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lee S., Dong H.H.. FoxO integration of insulin signaling with glucose and lipid metabolism. J. Endocrinol. 2017; 233:R67–R79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Paik J.H., Kollipara R., Chu G., Ji H., Xiao Y., Ding Z., Miao L., Tothova Z., Horner J.W., Carrasco D.R.et al.. FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell. 2007; 128:309–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Trinh D.L., Scott D.W., Morin R.D., Mendez-Lago M., An J., Jones S.J., Mungall A.J., Zhao Y., Schein J., Steidl C.et al.. Analysis of FOXO1 mutations in diffuse large B-cell lymphoma. Blood. 2013; 121:3666–3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Coomans de Brachene A., Demoulin J.B.. FOXO transcription factors in cancer development and therapy. Cell. Mol. Life Sci.: CMLS. 2016; 73:1159–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Baar M.P., Brandt R.M.C., Putavet D.A., Klein J.D.D., Derks K.W.J., Bourgeois B.R.M., Stryeck S., Rijksen Y., van Willigenburg H., Feijtel D.A.et al.. Targeted apoptosis of senescent cells restores tissue homeostasis in response to chemotoxicity and aging. Cell. 2017; 169:132–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schill D., Nord J., Cirillo L.A.. FoxO1 and FoxA1/2 form a complex on DNA and cooperate to open chromatin at insulin-regulated genes. Biochem. Cell Biol. 2019; 97:118–129. [DOI] [PubMed] [Google Scholar]
- 14. Li J., Jiang L., Liang X., Qu L., Wu D., Chen X., Guo M., Chen Z., Chen L., Chen Y.. DNA-binding properties of FOXP3 transcription factor. Acta Biochim. Biophys. Sin. (Shanghai). 2017; 49:792–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rogers J.M., Waters C.T., Seegar T.C.M., Jarrett S.M., Hallworth A.N., Blacklow S.C., Bulyk M.L. Bispecific forkhead transcription factor FoxN3 recognizes two distinct motifs with different DNA shapes. Mol. Cell. 2019; 74:245–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen X., Wei H., Li J., Liang X., Dai S., Jiang L., Guo M., Qu L., Chen Z., Chen L.et al.. Structural basis for DNA recognition by FOXC2. Nucleic Acids Res. 2019; 47:3752–3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Brent M.M., Anand R., Marmorstein R.. Structural basis for DNA recognition by FoxO1 and its regulation by posttranslational modification. Structure. 2008; 16:1407–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Clark K.L., Halay E.D., Lai E., Burley S.K.. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature. 1993; 364:412–420. [DOI] [PubMed] [Google Scholar]
- 19. Littler D.R., Alvarez-Fernandez M., Stein A., Hibbert R.G., Heidebrecht T., Aloy P., Medema R.H., Perrakis A.. Structure of the FoxM1 DNA-recognition domain bound to a promoter sequence. Nucleic Acids Res. 2010; 38:4527–4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tsai K.-L., Sun Y.-J., Huang C.-Y., Yang J.-Y., Hung M.-C., Hsiao C.-D.. Crystal structure of the human FOXO3a-DBD/DNA complex suggests the effects of post-translational modification. Nucleic Acids Res. 2007; 35:6984–6994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dai S., Li J., Zhang H., Chen X., Guo M., Chen Z., Chen Y.. Structural basis for DNA recognition by FOXG1 and the characterization of disease-causing FOXG1 mutations. J. Mol. Biol. 2020; 432:6146–6156. [DOI] [PubMed] [Google Scholar]
- 22. Stroud J.C., Wu Y., Bates D.L., Han A., Nowick K., Paabo S., Tong H., Chen L.. Structure of the forkhead domain of FOXP2 bound to DNA. Structure. 2006; 14:159–166. [DOI] [PubMed] [Google Scholar]
- 23. Chen Y., Chen C., Zhang Z., Liu C.C., Johnson M.E., Espinoza C.A., Edsall L.E., Ren B., Zhou X.J., Grant S.F.et al.. DNA binding by FOXP3 domain-swapped dimer suggests mechanisms of long-range chromosomal interactions. Nucleic Acids Res. 2015; 43:1268–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ostrow A.Z., Kalhor R., Gan Y., Villwock S.K., Linke C., Barberis M., Chen L., Aparicio O.M.. Conserved forkhead dimerization motif controls DNA replication timing and spatial organization of chromosomes in S. cerevisiae. PNAS. 2017; 114:E2411–E2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jankowski A., Szczurek E., Jauch R., Tiuryn J., Prabhakar S.. Comprehensive prediction in 78 human cell lines reveals rigidity and compactness of transcription factor dimers. Genome Res. 2013; 23:1307–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang X., Srivastava Y., Jankowski A., Malik V., Wei Y., Del Rosario R.C., Cojocaru V., Prabhakar S., Jauch R.. DNA-mediated dimerization on a compact sequence signature controls enhancer engagement and regulation by FOXA1. Nucleic Acids Res. 2018; 46:5470–5486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jolma A., Yan J., Whitington T., Toivonen J., Nitta K.R., Rastas P., Morgunova E., Enge M., Taipale M., Wei G.et al.. DNA-binding specificities of human transcription factors. Cell. 2013; 152:327–339. [DOI] [PubMed] [Google Scholar]
- 28. Chen Y., Bates D.L., Dey R., Chen P.H., Machado A.C., Laird-Offringa I.A., Rohs R., Chen L.. DNA binding by GATA transcription factor suggests mechanisms of DNA looping and long-range gene regulation. Cell Rep. 2012; 2:1197–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. BabuRajendran N., Palasingam P., Narasimhan K., Sun W., Prabhakar S., Jauch R., Kolatkar P.R.. Structure of Smad1 MH1/DNA complex reveals distinctive rearrangements of BMP and TGF-beta effectors. Nucleic Acids Res. 2010; 38:3477–3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dai S., Sun C., Tan K., Ye S., Zhang R.. Structure of thrombospondin type 3 repeats in bacterial outer membrane protein A reveals its intra-repeat disulfide bond-dependent calcium-binding capability. Cell Calcium. 2017; 66:78–89. [DOI] [PubMed] [Google Scholar]
- 31. Minor W., Cymborowski M., Otwinowski Z., Chruszcz M.. HKL-3000: the integration of data reduction and structure solution–from diffraction images to an initial model in minutes. Acta Crystallogr. D, Biol. Crystallogr. 2006; 62:859–866. [DOI] [PubMed] [Google Scholar]
- 32. Wang Q.-S., Zhang K.-H., Cui Y., Wang Z.-J., Pan Q.-Y., Liu K., Sun B., Zhou H., Li M.-J., Xu Q.et al.. Upgrade of macromolecular crystallography beamline BL17U1 at SSRF. Nucl. Sci. Tech. 2018; 29:68. [Google Scholar]
- 33. Zhang W.-Z., Tang J.-C., Wang S.-S., Wang Z.-J., Qin W.-M., He J.-H.. The protein complex crystallography beamline (BL19U1) at the Shanghai Synchrotron Radiation Facility. Nucl. Sci. Tech. 2019; 30:170. [Google Scholar]
- 34. McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J.. Phaser crystallographic software. J. Appl. Crystallogr. 2007; 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Emsley P., Cowtan K.. Coot: model-building tools for molecular graphics. Acta Crystallogr. D, Biol. Crystallogr. 2004; 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 36. Afonine P.V., Grosse-Kunstleve R.W., Echols N., Headd J.J., Moriarty N.W., Mustyakimov M., Terwilliger T.C., Urzhumtsev A., Zwart P.H., Adams P.D.. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D, Biol. Crystallogr. 2012; 68:352–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Eijkelenboom A., Mokry M., de Wit E., Smits L.M., Polderman P.E., van Triest M.H., van Boxtel R., Schulze A., de Laat W., Cuppen E.et al.. Genome-wide analysis of FOXO3 mediated transcription regulation through RNA polymerase II profiling. Mol. Syst. Biol. 2013; 9:638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Li H., Durbin R.. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010; 26:589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tan G., Lenhard B.. TFBSTools: an R/bioconductor package for transcription factor binding site analysis. Bioinformatics. 2016; 32:1555–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ferber E.C., Peck B., Delpuech O., Bell G.P., East P., Schulze A.. FOXO3a regulates reactive oxygen metabolism by inhibiting mitochondrial gene expression. Cell Death Differ. 2012; 19:968–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sanders D.A., Ross-Innes C.S., Beraldi D., Carroll J.S., Balasubramanian S.. Genome-wide mapping of FOXM1 binding reveals co-binding with estrogen receptor alpha in breast cancer cells. Genome Biol. 2013; 14:R6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Li J., Dantas Machado A.C., Guo M., Sagendorf J.M., Zhou Z., Jiang L., Chen X., Wu D., Qu L., Chen Z.et al.. Structure of the forkhead domain of FOXA2 bound to a complete DNA consensus Site. Biochemistry. 2017; 56:3745–3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Singh P., Han E.H., Endrizzi J.A., O’Brien R.M., Chi Y.I.. Crystal structures reveal a new and novel FoxO1 binding site within the human glucose-6-phosphatase catalytic subunit 1 gene promoter. J. Struct. Biol. 2017; 198:54–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Babayeva N.D., Wilder P.J., Shiina M., Mino K., Desler M., Ogata K., Rizzino A., Tahirov T.H.. Structural basis of Ets1 cooperative binding to palindromic sequences on stromelysin-1 promoter DNA. Cell Cycle. 2010; 9:3054–3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Baburajendran N., Jauch R., Tan C.Y., Narasimhan K., Kolatkar P.R.. Structural basis for the cooperative DNA recognition by Smad4 MH1 dimers. Nucleic Acids Res. 2011; 39:8213–8222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bandukwala H.S., Wu Y., Feuerer M., Chen Y., Barboza B., Ghosh S., Stroud J.C., Benoist C., Mathis D., Rao A.et al.. Structure of a domain-swapped FOXP3 dimer on DNA and its function in regulatory T cells. Immunity. 2011; 34:479–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wu Y., Borde M., Heissmeyer V., Feuerer M., Lapan A.D., Stroud J.C., Bates D.L., Guo L., Han A., Ziegler S.F.et al.. FOXP3 controls regulatory T cell function through cooperation with NFAT. Cell. 2006; 126:375–387. [DOI] [PubMed] [Google Scholar]
- 48. Tate J.G., Bamford S., Jubb H.C., Sondka Z., Beare D.M., Bindal N., Boutselakis H., Cole C.G., Creatore C., Dawson E.et al.. COSMIC: the catalogue of somatic mutations in cancer. Nucleic Acids Res. 2019; 47:D941–D947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Alvarez-Garcia O., Matsuzaki T., Olmer M., Masuda K., Lotz M.K.. Age-related reduction in the expression of FOXO transcription factors and correlations with intervertebral disc degeneration. J. Orthop. Res. 2017; 35:2682–2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Grupp K., Uzunoglu F.G., Melling N., Hofmann B., El Gammal A.T., Grotelüschen R., Heumann A., Bellon E., Reeh M., Wolters-Eisfeld G.et al.. FOXO1 overexpression and loss of pSerine256-FOXO1 expression predicts clinical outcome in esophageal adenocarcinomas. Sci. Rep. 2018; 8:17370. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structure factors for the reported crystal structures have been deposited with the Protein Data bank under accession codes 6LBI (FOXO1–DBD/DIV2) and 6LBM (FOXC2-DBD/DIV2).