ABSTRACT
Rickettsiae are obligate intracellular bacteria that can cause life-threatening illnesses and are among the oldest known vector-borne pathogens. Members of this genus are extraordinarily diverse and exhibit a broad host range. To establish intracellular infection, Rickettsia species undergo complex, multistep life cycles that are encoded by heavily streamlined genomes. As a result of reductive genome evolution, rickettsiae are exquisitely tailored to their host cell environment but cannot survive extracellularly. This host-cell dependence makes for a compelling system to uncover novel host–pathogen biology, but it has also hindered experimental progress. Consequently, the molecular details of rickettsial biology and pathogenesis remain poorly understood. With recent advances in molecular biology and genetics, the field is poised to start unraveling the molecular mechanisms of these host–pathogen interactions. Here, we review recent discoveries that have shed light on key aspects of rickettsial biology. These studies have revealed that rickettsiae subvert host cells using mechanisms that are distinct from other better-studied pathogens, underscoring the great potential of the Rickettsia genus for revealing novel biology. We also highlight several open questions as promising areas for future study and discuss the path toward solving the fundamental mysteries of this neglected and emerging human pathogen.
Keywords: Rickettsia, pathogenesis, microbial genetics, arthropod-borne pathogens, host–pathogen interactions, obligate intracellular bacteria
This review highlights the recent advances and insights gained into the unique biology of rickettsiae and key areas for future investigation.
INTRODUCTION
Bacteria dominate the biosphere and exhibit astounding diversity (Hug et al. 2016). Yet, we have merely scratched the surface of the bacterial world, and the vast majority of fascinating biology remains untapped. Much of what we know has come from studying model systems, like Escherichia coli and Listeria monocytogenes, for which there are powerful tools and large research communities. While these organisms still prove to be fruitful sources of discovery, we stand to make tremendous progress towards advancing our understanding of microbial life by expanding the range of bacteria we study to non-model systems (Alfred and Baldwin 2015; Blount 2015; Pizarro-Cerdá and Cossart 2019). With recent technological advances, especially in genetic manipulation, we are poised to delve into the molecular inner workings of diverse organisms and uncover novel biology (Alfred and Baldwin 2015; Matthews and Vosshall 2020).
In this review, we examine the unique biology of Rickettsia, a genus of obligate intracellular bacteria that includes several neglected and emerging human pathogens (Parola et al. 2013). Obligate intracellular bacteria are particularly remarkable and exciting for their extreme dependence on the host's intracellular environment. Over the course of evolutionary time, these bacteria evolved exquisitely tailored strategies to invade and hijack eukaryotic host cells to exploit them for their resources and for shelter from the humoral immune system. However, because they cannot survive extracellularly, obligate intracellular bacteria have historically been recalcitrant to laboratory manipulation, and progress towards understanding their biology has been stunted (Fields, Heinzen and Carabeo 2011; McClure et al. 2017). In the past decade, we have seen significant advancements in the study and genetic manipulation of rickettsiae (Burkhardt et al. 2011; McClure et al. 2017; Lamason, Kafai and Welch 2018; Kim et al. 2019; Narra et al. 2020). Here, we discuss these recent advances and how they have shed light on the unique infectious life cycles of rickettsiae. We also highlight several key unanswered questions that are likely to see progress over the next few years, thanks to advances in molecular biology, genetics and genomics.
THE RICKETTSIA GENUS: BROAD HOST RANGE AND SPECIES-LEVEL DIVERSITY
All Rickettsia species are obligate intracellular endosymbionts of eukaryotic host cells (Weinert 2015). This genus of alphaproteobacteria belongs to the order Rickettsiales, which also includes other obligate intracellular pathogen genera like Orientia, Ehrlichia and Anaplasma (Palmer and Azad 2012). The Rickettsia genus has extraordinary diversity at the species level, with members found in freshwater, marine and terrestrial ecosystems (Weinert 2015). Rickettsia species also exhibit a fairly broad host range. While most are associated with arthropods (e.g. ticks, lice, mites and fleas), some species are associated with hosts including protozoa, algae, plants and vertebrates. Notably, all known human pathogens in the Rickettsia genus are vectored by arthropods (Barrett and Stanberry 2009; Weinert et al. 2009; Weinert 2015; Biggs et al. 2016; Blanton 2019).
Arthropod-associated Rickettsia species are mainly transmitted vertically to their offspring through the infected cytoplasm of egg cells (Barrett and Stanberry 2009; Socolovschi et al. 2009; Weinert et al. 2009; Weinert 2015). As a result, some species of Rickettsia have evolved strategies to increase their persistence within host lineages by manipulating reproductive behavior and physiology, mainly to increase the number, survival and fitness of egg-laying individuals in a population. These strategies include male-killing, induction of parthenogenesis, cytoplasmic incompatibility and feminization of male hosts (Perlman, Hunter and Zchori-Fein 2006; Weinert et al. 2009; Weinert 2015). Interestingly, rickettsia–arthropod symbioses may extend beyond parasitism to commensalism and mutualism, with some evidence even suggesting an obligate mutualistic relationship (Perotti et al. 2006; Zchori-Fein, Borad and Harari 2006; Weinert 2015). The evolutionary and molecular underpinnings of these symbioses are not well understood, nor are the incidence and relative frequencies of any of these strategies among Rickettsia species (Weinert 2015). Some Rickettsia species have also evolved strategies to increase their persistence during vertical transmission in arthropod hosts by transovarial interference (Burgdorfer and Anacker 1981; Macaluso et al. 2002; Wright et al. 2015; Gall et al. 2016; Tomassone et al. 2018). In this process, certain species of Rickettsia (members of spotted fever group and ancestral group, see below) preclude secondary infection with another rickettsial endosymbiont, though the molecular details are not understood. Greater environmental sampling and molecular characterization of rickettsia–arthropod interactions will be necessary to gain a complete understanding of how Rickettsia species persist within arthropod populations, including those that are vectors for human diseases.
Due to its extensive diversity, the Rickettsia genus is classified into several phylogenetic groups, including spotted fever group (SFG), typhus group (TG), ancestral group (AG) and transitional group (TRG) (Gillespie et al. 2007; Barrett and Stanberry 2009; Palmer and Azad 2012; Weinert 2015; Blanton 2019). These groups have distinct intracellular life cycles that are presumably driven by genetic differences, but this has not been demonstrated and much of the rickettsial genome is uncharacterized (McClure et al. 2017; Lamason, Kafai and Welch 2018; Narra et al. 2020). While SFG and TG rickettsiae were historically categorized using antigen-based methods, these classifications have since been corroborated by genome sequences (Barrett and Stanberry 2009; Sahni et al. 2013; Weinert 2015). More recent work characterizing novel Rickettsia species has revealed greater species-level diversity than previously thought, and some newer classification systems divide the genus into as many as twelve groups (Weinert 2015). As the field continues to sequence more genomes from diverse rickettsiae, as well as relatives from the Rickettsiales order, we will gain important insights into evolution and diversification within the Rickettsia genus. This review's primary focus will be SFG and TG Rickettsia species, which are the best-studied groups at a molecular and cellular level due to their relevance as human pathogens.
RICKETTSIA CONTAINS GLOBAL, NEGLECTED AND EMERGING HUMAN PATHOGENS
The Rickettsia genus includes numerous important human pathogens, mainly in the SFG, TG and TRG clades (Barrett and Stanberry 2009; Parola et al. 2013; Weinert 2015; Biggs et al. 2016; Blanton 2019). Spotted fever rickettsioses are globally distributed and can have case-fatality rates as high as 55% without treatment, like in the case of Rocky Mountain spotted fever (Paddock and Alvarez-Hernández 2018), which the U.S. CDC considers the ‘most deadly tickborne disease in the world’ (CDC 2019a). Even with today's treatment options, Rocky Mountain spotted fever has case fatality rates of 5–10% (Biggs et al. 2016; Paddock and Alvarez-Hernández 2018). Several other SFG members cause life-threatening diseases, including R. conorii (Mediterranean spotted fever) and R. japonica (Japanese spotted fever; CDC 2019b; Barrett and Stanberry 2009; Parola et al. 2013; Blanton 2019). TG rickettsiae, which include R. typhi and R. prowazekii, exhibit mortality rates as high as 60% without treatment (Azad 2007; Barrett and Stanberry 2009; Angelakis, Bechah and Raoult 2016; Blanton 2019). The TRG member R. felis is a globally distributed flea-borne human pathogen that was first isolated in the U.S. and has more recently emerged as a common cause of non-fatal fever in Africa (Brown and Macaluso 2016). Improving our understanding of these neglected and emerging pathogens at the molecular and genetic levels will be crucial for controlling and treating these diseases.
The incidence of Rickettsia-associated diseases is currently on the rise. Around the world, there have been sporadic outbreaks of epidemic typhus and murine typhus, caused by R. prowazekii and R. typhi, respectively (Angelakis, Bechah and Raoult 2016; Blanton 2019; Caravedo Martinez, Ramírez-Hernández and Blanton 2021). Because typhus diseases spread via lice and fleas, they are often brought on by improper sanitary conditions and overcrowding. Consequently, it disproportionately affects homeless populations, those living in correctional facilities and isolated rural communities. Typhus diseases also often follow wars, spurred on by the conditions faced by troops and other people affected by warfare, and have influenced the outcomes of several wars throughout history (Azad 2007; Barrett and Stanberry 2009; Angelakis, Bechah and Raoult 2016). SFG rickettsioses have increased by at least an order of magnitude in the U.S. over the past two decades (Blanton 2019; CDC 2020). This increase in SFG rickettsioses has been driven by a variety of factors, including climate change and human migration, both of which have led to more interactions between humans and pathogen-associated arthropod vectors (Parola et al. 2013; Blanton 2019; CDC 2020). Even still, the prevalence of rickettsioses is underestimated because they are often undiagnosed or misdiagnosed (Parola et al. 2013; Biggs et al. 2016; Paddock and Alvarez-Hernández 2018; Blanton 2019). As discussed below, rickettsioses present with similar symptoms as other common infectious diseases, especially in tropical and subtropical regions, and we currently lack fast point-of-care diagnostic methods (Parola et al. 2013; Paddock and Alvarez-Hernández 2018; Blanton 2019). Unfortunately, delays in treating rickettsioses lead to poorer outcomes and possible fatality (Biggs et al. 2016; Paddock and Alvarez-Hernández 2018; Blanton 2019), underscoring the need for improved methods for detection and diagnosis.
Rickettsial diseases commonly present with one or more symptoms, including fever, headache, myalgia, rash and eschars (dry necrotic scabs), though symptoms vary among different species (Barrett and Stanberry 2009; Parola et al. 2013; Angelakis, Bechah and Raoult 2016; Biggs et al. 2016; Paddock and Alvarez-Hernández 2018; Blanton 2019). Arthropod-borne rickettsiae are transmitted to vertebrate hosts during a tick or mite bite, or they are deposited via fecal matter from lice or fleas and are subsequently introduced into the host by abrasive scratching of the skin (Barrett and Stanberry 2009; Blanton 2019). Rickettsia species then invade cells of the vasculature where localized rickettsia replication results in host cell death, giving rise to eschars in certain rickettsioses (Chan, Riley and Martinez 2010). Later in infection, rickettsia proliferation leads to increased vascular permeability, which presents as a rash (Barrett and Stanberry 2009; Biggs et al. 2016; Paddock and Alvarez-Hernández 2018; Blanton 2019). While their primary niche inside vertebrate hosts is endothelial cells, rickettsiae can infect a wide array of cell types, including macrophages, neutrophils, hepatocytes and neurons, some of which may play a role in spread throughout the body (Barrett and Stanberry 2009; Angelakis, Bechah and Raoult 2016; Biggs et al. 2016; Curto et al. 2016; Paddock and Alvarez-Hernández 2018; Blanton 2019; Sekeyová et al. 2019). Several groups have recently made progress in developing new animal models for rickettsial infection (Burke et al. 2020b; Bechah et al. 2010; Banajee et al. 2015), which will be crucial for fully understanding pathogenesis and for developing new treatments.
One notable but poorly understood feature of epidemic typhus is a recrudescent infection, referred to as Brill–Zinsser disease (Brill 1910; Zinsser and Castaneda 1933; Barrett and Stanberry 2009; Angelakis, Bechah and Raoult 2016). In this disease, R. prowazekii enters a dormant state that can persist asymptomatically for more than 40 years after acute infection (Barrett and Stanberry 2009; Bechah et al. 2010; Faucher et al. 2012; Angelakis, Bechah and Raoult 2016). The reactivated pathogen causes a milder form of the disease relative to the initial infection, and the trigger of reactivation is unclear (Barrett and Stanberry 2009; Bechah et al. 2010; Angelakis, Bechah and Raoult 2016). The mechanism of dormancy and identity of the host tissues in which these bacteria reside are also not well understood, though work using a recently developed mouse model identified adipose tissue as a potential reservoir (Bechah et al. 2010).
Given the status of some Rickettsia species as emerging and global health threats, as well as the classification of R. prowazekii as a select agent by the U.S. government (Azad 2007), effective therapeutics will be essential for preventing morbidity and mortality in all patients. Today, rickettsial diseases are primarily treated with tetracycline antibiotics, typically doxycycline. Chloramphenicol is the only alternative treatment option for patients with sensitivities to tetracyclines, but chloramphenicol has been associated with higher case fatality rates, and effective forms are often unavailable in hospitals in the U.S. (Barrett and Stanberry 2009; Biggs et al. 2016; Paddock and Alvarez-Hernández 2018; Blanton 2019). Many antibiotics, including β-lactams, macrolides, aminoglycosides and sulfonamides, are ineffective against rickettsiae (Biggs et al. 2016). There has been a worrying increase in antibiotic resistance among bacteria in general (Berendonk et al. 2015), and antibiotic resistance in rickettsiae is readily evolved in laboratory culture and has been found in natural isolates (Rachek et al. 1998; Troyer et al. 1998; Drancourt and Raoult 1999). Further, it is not clear how the widespread use of antibiotics and pesticides in agriculture (Berendonk et al. 2015; Antwis et al. 2017) might affect the evolution or distribution of Rickettsia species across the world. Taken together with the increased prevalence of rickettsial diseases, there is a clear need to improve our understanding of the fundamental biology and pathogenesis of rickettsiae, and to develop new strategies to diagnose, treat and prevent rickettsial infections.
THE STATE OF GENETIC TOOLS IN RICKETTSIA
Most obligate intracellular bacteria are difficult to study because they cannot be cultured axenically and must be grown within eukaryotic cell culture systems. Perhaps most impactfully, the difficulties associated with culturing and manipulating these bacteria have stunted the development of genetic tools (McClure et al. 2017). For example, even though genetic transformation has been possible for 50 years in E. coli (Mandel and Higa 1970), the first transformation in Rickettsia was only performed in 1998 (Rachek et al. 1998). While these hurdles have impeded progress towards understanding the biology of obligate intracellular bacteria (Palmer and Azad 2012; McClure et al. 2017), we have seen significant advances in rickettsial genetics over the past two decades that have revealed unique molecular mechanisms underlying the infectious life cycles of rickettsiae.
In 2009, the first published example of targeted mutagenesis was performed in R. prowazekii, in which the pld (phospholipase D) gene was knocked out by allelic exchange using a linear DNA substrate (Driskell et al. 2009). This strategy has not been widely adopted though, likely due to the poor efficiency of transformation and allelic exchange in rickettsiae. The next attempts at targeted genetic manipulation came 6 years later, with two papers describing two different methods. In R. rickettsii, the ompA gene was knocked out using group II intron retrohoming (TargeTron; Noriea, Clark and Hackstadt 2015), a proprietary technology that is no longer sold or supported. In R. typhi and R. montanensis, peptide nucleic acids were used to silence the expression of the ompB and rickA genes (Pelc et al. 2015). The PNA-based knockdown yielded slightly reduced expression of these abundant proteins when measured by western blot, with differences in expression only being detectable when comparing diluted input samples. To this date, these are the only reports of targeted genetic manipulation in Rickettsia, and the field still lacks the highly efficient and scalable tools for targeted genetic manipulation that exist in other bacteria.
As a complementary approach to targeted mutagenesis, methods for forward-genetic screening in rickettsiae have also seen significant progress. A mariner-based transposon system for random mutagenesis was developed for R. prowazekii in 2007 (Liu et al. 2007) and applied to R. rickettsii in 2010 (Kleba et al. 2010). In contrast to the targeted mutagenesis techniques described above, transposon mutagenesis allows for more efficient generation of a relatively large number of mutants from a single transformation. By improving the efficiency of the original mariner-based system from 2007, the first large-scale rickettsial mutant collection was published in 2018 using R. parkeri (Lamason, Kafai and Welch 2018). This screen isolated over 100 mutants with defects in mammalian cell infection, and the resulting collection has helped reveal the essential functions of several genes during the intracellular life cycle, including ompB (Engström et al. 2019), rickA (Reed et al. 2014; Harris et al. 2018), sca2 (Burke et al. 2020b; Reed et al. 2014; Harris et al. 2018) and sca4 (Lamason et al. 2016) (described below). These mutants have also enabled functional-genetic studies, including the first genetic complementation of a rickettsial mutant with a native promoter in 2016 (Lamason et al. 2016), which was accomplished using a shuttle vector that was developed for rickettsiae in 2011 (Burkhardt et al. 2011). A similar transposon screen using a Tn5 transposon-based system was recently conducted in R. conorii (Kim et al. 2019). A total of 53 mutant strains were isolated, including two transposon mutants that were used to demonstrate the role of O antigen in the production of Weil–Felix antibodies, which are associated with anti-rickettsial immunity and detected in serology-based diagnostics. Forward-genetic screening using transposon-based systems represents a promising avenue for identifying the roles of the many genes of unknown function in the rickettsial genome.
Other forward genetic methods, including signature-tagged mutagenesis and Tn-seq, have allowed researchers to determine gene essentiality in a wide array of bacteria in different environmental contexts (Hensel et al. 1995; Hutchison III 1999; Knuth et al. 2004; Goodman et al. 2009; van Opijnen, Bodi and Camilli 2009). These methods rely on negative selection of strains with deleterious transposon insertions, so they require high numbers of randomly generated transposon mutants. However, in rickettsiae, reaching the number of mutants needed to call gene essentiality by saturation is not currently feasible given the extremely low transformation efficiency (Clark et al. 2011; Palmer and Azad 2012). The development of more efficient methods for transformation and transposon mutagenesis will be necessary to propel Rickettsia into the realm of genome-wide screens. The introduction of more efficient and scalable methods for targeted gene knockouts or knockdowns will also be crucial for the rickettsial toolkit. The advent of tools like CRISPR-based genome editing, which has been employed in diverse bacterial species including those that are difficult to culture and manipulate (reviewed in Vigouroux and Bikard 2020), provides a promising avenue for future tool development in Rickettsia species. With more powerful tools, researchers will be able to dissect the genetic and molecular mechanisms underlying the unique rickettsial infectious life cycle.
RICKETTSIAE EMPLOY COMPLEX AND UNIQUE INTRACELLULAR LIFE CYCLES TO ESTABLISH INFECTION
Rickettsiae are entirely dependent on eukaryotic host cells for survival and have evolved complex life cycles that allow them to establish intracellular niches (Fig. 1 and Table 1; Palmer and Azad 2012; Lamason and Welch 2017; Narra et al. 2020). The rickettsial life cycle begins with adherence to the host cell, followed by uptake into a phagocytic vacuole. After escaping the phagosome, rickettsiae reside freely within the host cytosol, and some species can achieve motility by hijacking host cell actin. After replicating within the host cytosol, rickettsiae can disseminate to neighboring cells, either through lysis of the host cell or by moving directly into neighboring cells by traversing cell–cell junctions (Palmer and Azad 2012; Lamason and Welch 2017; Narra et al. 2020). In this section, we review key insights into the molecular details of this infectious life cycle.
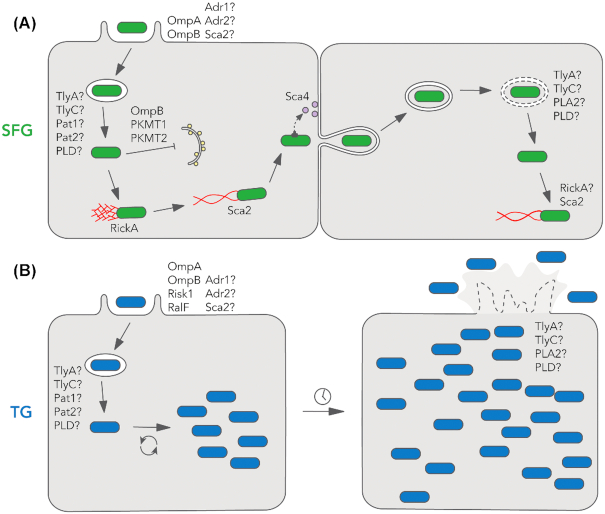
Figure 1.
The intracellular life cycles of SFG and TG rickettsiae. (A) The SFG life cycle begins with host cell invasion, during which rickettsiae enter the host cell in a phagocytic vacuole. After escaping the phagosome, SFG rickettsiae initiate two phases of actin-based motility. SFG rickettsiae avoid host autophagy through lysine methylation of their surface proteins. Starting around 8 h after invasion, SFG rickettsiae begin replicating in the host cytosol. Subsequently, they undergo cell-to-cell spread, a process in which they directly traverse the cell–cell junction and enter the neighboring cell in a double-membrane vacuole. Upon escape, they can reinitiate the life cycle. (B) The TG life cycle begins similarly to SFG with invasion followed by rapid escape from the phagocytic vacuole. After replicating to a high density, the host cell lyses and TG rickettsiae can escape and spread to neighboring cells. (A and B) Rickettsial proteins that have been implicated in the life cycle are noted. Proteins that are speculated to be involved in a process are denoted with a question mark.
Table 1.
Rickettsial proteins with known or predicted roles during infection. Summarized details of key rickettsial proteins discussed in this review. The presence of a protein in a specific rickettsial group is defined by a full or partial homolog found in the genome of at least one species. Asterisks (*) denote studies that directly characterized the protein in the native Rickettsia species by studying a mutant strain of rickettsia. SFG, spotted fever group; TG, typhus group; TRG, transitional group; AG, ancestral group.
| Rickettsial Protein | Known host protein interactions | Function | Present | References | |
|---|---|---|---|---|---|
| Secreted | TlyA | Unknown | Predicted hemolysin with possible role in vacuole escape and/or host cell lysis |
SFG, TG, TRG, AG | R. prowazekii (Whitworth et al. 2005) |
| TlyC | Unknown | Predicted hemolysin with possible role in vacuole escape and/or host cell lysis |
SFG, TG, TRG, AG | R. prowazekii (Whitworth et al. 2005) | |
| Pat1 | Unknown | Phospholipase with possible role in vacuole escape and/or host cell lysis |
SFG, TG, TRG, AG | R. rickettsii (Walker et al. 1983; Silverman et al. 1992), R. typhi (Rahman et al. 2010, 2013) | |
| Pat2 | Unknown | Phospholipase with possible role in vacuole escape and/or host cell lysis |
SFG, TG, TRG, AG | R. rickettsii (Walker et al. 1983; Silverman et al. 1992),R. typhi (Rahman et al. 2010, 2013) | |
| PLD | Unknown | Predicted phospholipase with possible role in vacuole escape and/or host cell lysis |
SFG, TG, TRG, AG | R. prowazekii (Whitworth et al. 2005, Driskell et al. 2009*) | |
| Sca4 | Vinculin | Disrupts interaction between vinculin and α-catenin promote cell-to-cell spread |
SFG, TG, TRG, AG | R. rickettsii (Park et al. 2011), R. parkeri (Lamason et al. 2016*) | |
| Risk1 | Beclin-1 | Phosphatidylinositol 3-kinase activity promotes host cell colonization |
SFG, TG, TRG, AG | R. typhi (Voss et al. 2020) | |
| RalF | Arf6 | Guanine nucleotide exchange factor of Arf6 that promotes invasion | TG, TRG, AG | R. typhi (Rennoll-Bankert et al. 2015, 2016) | |
| RARP-2 | Unknown | Cysteine protease that contributes to trans-Golgi fragmentation |
SFG, TG, TRG, AG | R. rickettsii (Lehman et al. 2018; Aistleitner et al. 2020) | |
| Surface or membrane | Tlc1 | Unknown | ATP/ADP symporter that imports ATP from host cells |
SFG, TG, TRG, AG | R. prowazekii (Winkler 1976; Audia and Winkler 2006; Krause, Winkler and Wood 1985) |
| OmpA | α2β1 integrin | Rickettsial surface protein involved in invasion |
SFG, TRG, AG | R. conorii (Hillman, Baktash and Martinez 2013) | |
| OmpB | Ku70 | Rickettsial surface protein involved in invasion and autophagy avoidance |
SFG, TG, TRG, AG | R. conorii (Martinez et al. 2005), R. parkeri (Engström et al. 2019*) | |
| PKMT1/2 | Unknown | Lysine methylation of rickettsial surface proteins to avoid autophagy |
SFG, TG, TRG, AG | R. parkeri (Engström et al. 2020*), R. prowazekii (Abeykoon et al. 2012, 2014, 2016), R. typhi (Abeykoon et al. 2014, 2016) | |
| RickA | Arp2/3 | Mimics WASP to activate Arp2/3 to drive actin-based motility |
SFG, TRG, AG | R. conorii (Gouin et al. 2004), R. rickettsii (Jeng et al. 2004), R. parkeri (Reed et al.2014*) | |
| Sca2 | Actin, profilin | Formin-like activity that directly nucleates actin to promote motility |
SFG, TG, TRG, AG | R. rickettsii (Kleba et al. 2010*), R. parkeri (Haglund et al. 2010, Reed et al. 2014*), R. conorii (Madasu et al. 2013) |
Intracellular bacterial pathogens invade eukaryotic host cells using either the zipper or trigger mechanisms (Cossart 2004). In the zipper mechanism, the invading bacterium binds to host cell receptors, which triggers signaling cascades that cause phagocytosis of the bacterium. In rickettsiae, bacterial surface proteins bind host cell receptors to promote adherence and zipper-mediated invasion (Chan, Riley and Martinez 2010; Palmer and Azad 2012). The host proteins Ku70 and α2β1 integrin promote adherence and invasion by rickettsiae (Martinez et al. 2005; Reed, Serio and Welch 2011; Hillman, Baktash and Martinez 2013); however, an essential host cell receptor has not yet been identified. The R. conorii proteins OmpB (outer membrane protein B) and OmpA (outer membrane protein A) interact with Ku70 (Martinez et al. 2005) and α2β1 integrin (Hillman, Baktash and Martinez 2013), respectively. Other rickettsial surface proteins have also been suggested to play a role in host cell adherence and invasion, including Sca2 (Cardwell and Martinez 2009), Adr1 (Balraj, Renesto and Raoult 2009) and Adr2 (Vellaiswamy et al. 2011). The binding of rickettsiae to host cell receptors triggers a signaling cascade beginning with the activation of host tyrosine kinases and phosphoinositide 3-kinase (Martinez and Cossart 2004; Reed, Serio and Welch 2011). These tyrosine kinases then activate Rho-family GTPases, including Cdc42 (Martinez and Cossart 2004; Reed, Serio and Welch 2011) and Rac1 (Reed, Serio and Welch 2011), which in turn activate WAVE proteins (Reed, Serio and Welch 2011). These signaling events ultimately promote the activation of Arp2/3, which is the primary driver of actin polymerization during the uptake of rickettsiae by the host cell (Martinez and Cossart 2004; Reed, Serio and Welch 2011).
In contrast to the zipper mechanism, bacteria that invade host cells by the trigger mechanism use their secretion machinery to inject bacterial effectors into the host cell to activate cytosolic activators of phagocytosis directly (Cossart 2004). Recent work has suggested that rickettsial invasion might also involve elements of a trigger mechanism, possibly blurring the line between these two invasion mechanisms. It has been speculated that RickA, which nucleates host actin during actin-based motility (see below), might also be involved in activating Arp2/3 during invasion (Reed, Serio and Welch 2011; Rennoll-Bankert et al. 2015), though this has not been demonstrated experimentally. The R. typhi protein RalF is predicted to be secreted into host cells during invasion in a trigger-like mechanism (Rennoll-Bankert et al. 2015, 2016). RalF recruits the host protein Arf6 to manipulate phosphoinositol metabolism at the host plasma membrane and promote engulfment of R. typhi (Rennoll-Bankert et al. 2015, 2016). Additionally, Risk1, a phosphatidylinositol 3-kinase effector from R. typhi, has been proposed to be secreted and act on the host cell membrane during invasion (Voss et al. 2020). However, without the ability to easily generate targeted knockouts in Rickettsia, we lack direct evidence for translocation of RalF or Risk1 into host cells during invasion. Furthermore, RalF has been lost or pseudogenized in other clades, including SFG (Rennoll-Bankert et al. 2015), suggesting that rickettsiae may have evolved diverse mechanisms for host cell invasion. Indeed, the prevailing model is that Rickettsia species have evolved multiple, redundant mechanisms to adhere to and invade host cells, which could contribute to their ability to infect numerous cell types in a range of host organisms (Reed, Serio and Welch 2011; Palmer and Azad 2012; Rennoll-Bankert et al. 2016). As more powerful genetic tools are developed for rickettsiae, we will be able to broaden our understanding of invasion by directly and comprehensively studying rickettsial adhesins and invasins.
Upon successful uptake into the host cell, rickettsiae quickly escape the phagocytic vacuole and enter the host cytosol. In SFG and TG rickettsiae, cytosolic bacteria are detectable as early as 3 min post-invasion, and the majority of internalized bacteria are in the cytosol by 20 min (Teysseire, Boudier and Raoult 1995; Palmer and Azad 2012). These vacuole escape kinetics are faster relative to other cytosolic pathogens, including L. monocytogenes, Shigella flexneri and Francisella tularensis, which escape into the cytosol on the order of 15–60 min (Palmer and Azad 2012; Fredlund and Enninga 2014). While the vacuoles of these other intracellular bacteria associate with lysosomal markers (Fredlund and Enninga 2014), it is not known if rickettsia-containing vacuoles associate with lysosomal markers or if the faster escape kinetics allow them to avoid endosomal maturation. The rapid kinetics of vacuole escape suggest that rickettsiae may use mechanisms distinct from those employed by other cytosolic bacteria. The rickettsial genome encodes several enzymes that are suspected of breaking down the vacuolar membrane, including phospholipase D (PLD) (Whitworth et al. 2005; Driskell et al. 2009), the patatin-like phospholipase A2 enzymes Pat1 and Pat2 (Walker et al. 1983; Silverman et al. 1992; Blanc, Renesto and Raoult 2005; Rahman et al. 2010, 2013) and the hemolysins TlyA and TlyC (Whitworth et al. 2005). However, these genes have mostly been studied either indirectly (Walker et al. 1983; Silverman et al. 1992; Rahman et al. 2010, 2013) or by heterologous expression in bacteria like Salmonella enterica (Whitworth et al. 2005), and none have been ascribed specific roles in vacuole escape or infection dynamics. Directly probing the role of each gene has been difficult due to the lack of genetic tools, although a phospholipase D (pld) mutant was the first targeted genetic knockout created in Rickettsia (Driskell et al. 2009). Surprisingly, when pld was knocked out in R. prowazekii, no defects were observed in vacuole escape kinetics during in vitro infection of macrophages. In contrast, this pld mutant strain was unable to induce fever or weight changes in a low inoculum guinea pig infection model, suggesting that phospholipase D does contribute to pathogenesis in vivo (Driskell et al. 2009). Given that rickettsiae encode multiple membranolytic enzymes, other genes might provide functional redundancy for pld. It is also possible that phospholipase D acts during another phase of the rickettsial infectious life cycle. For instance, it could contribute to host cell lysis or lipid metabolism, or it could play a role in different cell types that were not tested, like endothelial cells. It will be interesting to see how each of the rickettsial membranolytic enzymes promotes niche establishment and pathogenesis.
Rickettsiae avoid and suppress cellular immunity to promote successful infection. For example, R. rickettsii inhibits apoptosis in a manner dependent on NF-κB (Clifton et al. 1998; Joshi et al. 2004). Pathogenic SFG rickettsiae also avoid detection by host autophagy machinery by shielding their surface proteins with lysine methylation (Abeykoon et al. 2012, 2014, 2016; Uchiyama, Kishi and Ogawa 2012; Engström et al. 2019). Indeed, transposon mutagenesis of the genes encoding two protein-lysine methyltransferases PKMT1 and PKMT2 and OmpB (Lamason, Kafai and Welch 2018) rendered these R. parkeri mutants vulnerable to autophagy-mediated clearance during infection (Engström et al. 2019, 2020). Intriguingly, R. parkeri was recently shown to exhibit more virus-like interactions with the innate immune response relative to facultative intracellular bacteria like L. monocytogenes, in that it is sensitive to IFN-I and exploits the inflammasome to avoid IFN-I (Burke et al. 2020a). The full repertoire of rickettsial factors that mediate interactions with host immunity remains to be elucidated, but future work focused on this is sure to yield valuable insights into how different cytosolic bacteria hijack their host cells.
Several Rickettsia species undergo actin-based motility, a process in which they polymerize host actin filaments at one pole of the bacterial cell surface to propel themselves throughout the host cytoplasm (Palmer and Azad 2012; Choe and Welch 2016; Lamason and Welch 2017). Unlike most cytosolic bacterial pathogens, some rickettsiae encode two distinct bacterial effector proteins for actin-based motility (Gouin et al. 2004; Jeng et al. 2004; Haglund et al. 2010; Kleba et al. 2010). The first, RickA, is found in nearly all SFG species (except R. peacockii) as well as members of AG and TRG (Jeng et al. 2004; Haglund et al. 2010; Choe and Welch 2016; Lamason and Welch 2017). RickA produces short, curved tails by mimicking WASP to activate Arp2/3 at the bacterial pole, which polymerizes branched actin networks capable of propelling the bacterium forward (Gouin et al. 2004; Jeng et al. 2004; Reed et al. 2014). This mechanism is similar to ActA in L. monocytogenes and BimA in Burkholderia thailandensis (Gouin et al. 2004; Jeng et al. 2004; Lamason and Welch 2017). The second rickettsial factor involved in actin-based motility is Sca2, which is found in SFG, AG and TRG species, as well as the TG member R. typhi (Haglund et al. 2010; Kleba et al. 2010). The best-characterized variant is the SFG Sca2, which polymerizes long, straight actin tails using a formin-like mechanism that is distinct from its eukaryotic counterpart (Haglund et al. 2010; Madasu et al. 2013; Reed et al. 2014). Unlike eukaryotic formins, which act as dimers and use formin homology domains, Sca2 exists as a monomer and uses N- and C-terminal repeat domains to nucleate and elongate actin filaments (Madasu et al. 2013). SFG Sca2-mediated motility is also dependent on several host factors, including fimbrin/T-plastin, profilin, capping protein and cofilin (Serio et al. 2010). Interestingly, members of TG and AG, like R. typhi and R. bellii, encode variants of Sca2 that lack the formin-like domains (Haglund et al. 2010; Madasu et al. 2013; Choe and Welch 2016). These orthologs still contain WH2 motifs, which might facilitate a distinct form of actin-based motility, although this has not been demonstrated. It will be intriguing to see how the differences among Sca2 orthologs impact the intracellular life cycle and pathogenesis.
Notably, transposon insertions in rickA and sca2 were isolated in R. parkeri, revealing two distinct phases of actin-based motility during its infectious life cycle (Reed et al. 2014; Lamason, Kafai and Welch 2018). Within the first 2 h of infection, RickA mediates early actin-based motility. Then, starting around 8 h post-infection, late motility is mediated by Sca2 (Reed et al. 2014). Members of SFG Rickettsia are unique among known cytosolic bacterial pathogens to undergo two phases of actin-based motility mediated by distinct effectors (Reed et al. 2014; Lamason and Welch 2017), and we are only beginning to learn how these two forms of actin-based motility shape the infectious life cycle of SFG rickettsiae and affect their pathogenesis. In mammalian cell culture, dissemination of SFG rickettsiae through the monolayer (i.e. cell-to-cell spread) is almost completely abolished in the sca2::tn mutant but only minorly impaired in the rickA::tn mutant (Reed et al. 2014; Harris et al. 2018). Given that the sca2::tn mutant is replication-competent (Reed et al. 2014; Harris et al. 2018), this suggests a model in which Sca2 is responsible for correctly positioning the bacteria within the host cell to initiate the process of cell-to-cell spread in mammalian tissues. R. parkeri Sca2 is required for efficient dissemination to host organs and lethality in an intradermal infection mouse model (Burke et al. 2020b), and R. rickettsii Sca2 has been linked to fever induction in a guinea pig infection model (Kleba et al. 2010). The primary function of RickA-mediated actin-based motility is less clear. For instance, we do not know whether it plays a small but direct role in cell-to-cell spread or if its modest phenotype in spread assays is an indirect effect stemming from other functions it has during the intracellular life cycle. RickA-mediated actin-based motility could potentially play a role in avoiding cellular immunity, similarly to ActA in L. monocytogenes (Yoshikawa et al. 2009; Cheng et al. 2018), though this has not been tested. Sca2 and RickA also mediate actin-based motility in tick cell culture (Harris et al. 2018), but they are not essential for cell-to-cell spread in this context. The rickA::tn mutant has a minor cell-to-cell spread defect in tick cells (Reed et al. 2014; Harris et al. 2018), but the sca2::tn mutant spreads similarly to wild-type R. parkeri in tick cells (Harris et al. 2018). In live ticks, both Sca2 and RickA promote early dissemination events, but the transposon mutants did not have an overall impact on dissemination at later timepoints of infection (Harris et al. 2018). Altogether, these data suggest that the roles of actin-based motility and the genetic determinants of cell-to-cell spread may vary across different host organisms. More detailed characterization of actin-based motility in diverse Rickettsia species and various host cell types will be needed to elucidate the various roles of actin-based motility during infection.
SFG rickettsiae undergo cell-to-cell spread, a process in which they directly move into neighboring cells by traversing cell–cell junctions, preserving access to the host cytoplasm and protection from the humoral immune system (Palmer and Azad 2012; Lamason and Welch 2017; Dowd, Mortuza and Ireton 2020). Cell-to-cell spread has also been observed in L. monocytogenes and S. flexneri, which use the force of actin-based motility to propel themselves into the cell–cell junction creating a protrusion into the neighboring cell (Kuehl et al. 2015; Lamason and Welch 2017; Dowd, Mortuza and Ireton 2020). This protrusion is then engulfed by the recipient cell, and the bacterium is taken up in a double-membrane vacuole. After vacuole escape, the life cycle can be reinitiated. In stark contrast to this canonical mode of cell-to-cell spread, it was recently demonstrated that R. parkeri undergoes a dramatically different process of cell-to-cell spread (Lamason et al. 2016; Lamason and Welch 2017). Despite undergoing two distinct forms of actin-based motility (Reed et al. 2014), R. parkeri loses its actin tail before protrusion initiation (Lamason et al. 2016). Consequently, these protrusions are significantly shorter in length and lifetime compared with those of L. monocytogenes (Lamason et al. 2016). How SFG rickettsiae navigate to cell–cell junctions after losing actin tails and how they manipulate the host membrane in the absence of actin-based motility is not well understood. One clue has come from work examining the function of the R. parkeri secreted effector Sca4 (Lamason et al. 2016), which contains two vinculin-binding sites (Park et al. 2011; Lamason et al. 2016). This effector blocks the association between host cell adhesion proteins vinculin and α-catenin, which normally maintains intercellular tension at adherens junctions. Sca4-mediated inhibition of vinculin decreases intercellular tension and promotes protrusion engulfment, allowing for more efficient R. parkeri cell-to-cell spread. Notably, the sca4::tn mutant is not completely defective in spread, suggesting that SFG rickettsiae rely on a suite of to-be-identified bacterial proteins to alter cell-cell junctions and drive cell-to-cell spread (Lamason et al. 2016; Lamason, Kafai and Welch 2018). Further work will be needed to determine how conserved cell-to-cell spread is within the Rickettsia genus.
In contrast to SFG rickettsiae, TG rickettsiae have more limited actin-based motility and have not been observed to undergo direct cell-to-cell spread (Teysseire, Chiche-Portiche and Raoult 1992; Haglund et al. 2010; Palmer and Azad 2012; Choe and Welch 2016). As mentioned above, this is likely due to the loss or truncation of sca2 in TG rickettsiae. Consequently, TG rickettsiae replicate in their host cells until reaching high densities (Wisseman and Waddell 1975; Palmer and Azad 2012), after which the host cells lyse and release large numbers of infectious bacteria that invade new host cells (Fig. 2). For this reason, the growth of TG rickettsiae within individual cells has been likened to the growth of bacteria in test tubes, with a brief lag phase followed by exponential growth (Palmer and Azad 2012). At high bacterial cell densities, TG rickettsiae display altered sizes and morphology (Wisseman and Waddell 1975; Silverman, Wisseman and Waddell 1980), although whether they reach a stationary phase of growth is not clear. In contrast, the growth of SFG rickettsiae has been compared to growth in a chemostat (Palmer and Azad 2012). Because SFG rickettsiae maintain low densities within individual host cells by initiating cell-to-cell spread early during their life cycle, they exhibit consistent replication rates throughout the course of infection (Fig. 2; Wisseman et al. 1976).
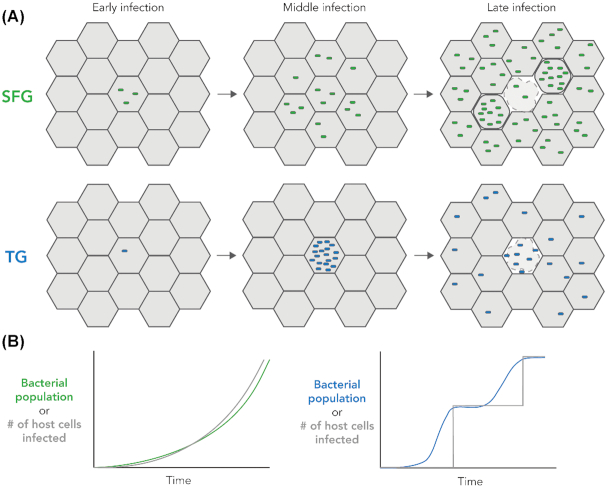
Figure 2.
Growth dynamics of SFG and TG rickettsiae. (A, top) Shortly after entry into the host cell, SFG rickettsiae begin replicating. They maintain a constant growth rate and low bacterial densities within individual cells by initiating cell-to-cell spread early in infection. Late in infection, host cell lysis occurs releasing infectious bacteria into the extracellular space. (A, bottom) After a brief initial lag phase, TG rickettsiae replicate to high densities within individual infected cells. Once the infected cell becomes saturated with bacteria, the host cell lyses and releases large quantities of infectious bacteria that subsequently invade other host cells. (B) The dynamics of bacterial growth and host cell infection are represented graphically.
Somewhat counterintuitively, TG rickettsiae do not induce significant morphological changes to their host cell during infection until lysis (Silverman, Wisseman and Waddell 1980), while SFG rickettsiae cause substantial changes to their host cells (Walker et al. 1977; Silverman and Wisseman 1979; Walker and Cain 1980; Palmer and Azad 2012). Starting between 48–72 h post-infection with SFG rickettsiae, host cells display dilation of the nuclear envelope and the rough ER, dissociation of ribosomes from the ER, loss of Golgi apparatus morphology and swelling of mitochondria. Recent work showed that the secreted effector RARP-2 contributes to the fragmentation of the trans-Golgi network during R. rickettsii infection (Lehman et al. 2018; Aistleitner et al. 2020); however, no other bacterial factors have been implicated in altering host organelle morphology. Further, it is unclear how structural changes to host organelles impact SFG rickettsial infection dynamics. It is also unknown what factors mediate host cell lysis in SFG or TG rickettsiae. More powerful tools will be essential for deciphering the genetic basis of how rickettsiae alter their host cell environment and the differences between TG and SFG rickettsiae.
In addition to the cytosolic niche, several SFG and AG Rickettsia species have been observed inside the nucleus of their host cells (Wolbach 1919; Pinkerton and Hass 1932; Philip et al. 1983; Ogata et al. 2006). Interestingly, intranuclear rickettsiae display high frequencies of actin tails with distinct dynamics relative to cytoplasmic actin tails (Heinzen et al. 1999; Ogata et al. 2006), which might be indicative of a specialized nuclear life cycle or a response to unique properties of the nucleoplasm. How these bacteria enter or escape the nucleus is not known. It is possible that rickettsiae become entrapped inside the nuclear envelope during host cell division or that they have evolved specialized mechanisms to actively invade the nucleus. However, it is not clear why other Rickettsia species, like TG members, are not observed in the nucleus. Further study of intranuclear rickettsiae will be important to determine their role in infection and pathogenesis. Moreover, the distinct behavior of intranuclear rickettsiae could provide a useful tool for studying the molecular constituents of the nucleoplasm.
SECRETION SYSTEMS AND MANIPULATION OF THE HOST CELL THROUGH SECRETED EFFECTORS
Rickettsiae encode several secretion systems that foster manipulation of the host cell environment and progression through its life cycle by either secreting effector proteins into the host cell cytosol or attaching proteins to the bacterial cell surface. These include Type I, IV and V secretion systems, the Sec translocase, and the twin-arginine translocation (Tat) protein export pathway (Palmer and Azad 2012; Gillespie et al. 2014). However, the complete catalog of secreted proteins and their associations with the secretion machinery has not been elucidated.
Perhaps the most enigmatic of the rickettsial secretion machinery is the Type IV secretion system (T4SS; Fig. 3), which will be the main focus of this section (see (Gillespie et al. 2014) for a comprehensive review of rickettsial secretion systems). The rickettsial T4SS system (MPF-T class/P-T4SS) is related to the vir T4SS of the plant pathogen Agrobacterium tumefaciens, a distant relative within alphaproteobacteria, and the rickettsial T4SS genes have been renamed Rickettsiales vir homolog (rvh; Gillespie et al. 2014, 2016). T4SSs are typically comprised of 12 different subunits, although some bacteria also encode additional accessory genes, and some encode multiple complete T4SSs (Gillespie et al. 2014, 2016; Costa et al. 2015). The T4SS from the Rickettsiales order is unique because it has undergone several gene duplication events that have given rise to unprecedented complexity within a single T4SS (Gillespie et al. 2016). The rvhB4, rvhB8, rvhB9 genes each have duplicate paralogs and the rvhB6 gene has five paralogs. Each paralog is significantly divergent from one another, with as much as ∼80% divergence between paralogs and some paralogs containing additional domains, like large N- and C-terminal extensions encoded by several of the rvhB6 genes. Both sets of rvhB4, rvhB8, rvhB9 paralogs are expressed during a 24-hour window of R. typhi infection of HeLa cells (Gillespie et al. 2016). While expression of the rvhB6 paralogs has not been examined in Rickettsia, all rvhB6 paralogs are expressed during tick and human cell infection in Ehrlichia chaffeensis, a relative within the Rickettsiales order that encodes four rvhB6 genes (Bao et al. 2009). How the rickettsial T4SS uses the expanded repertoire of subunits is not clear. It will be important to determine how these paralogous subunits are incorporated into T4SS complexes. For example, different paralogs may interact to form heterogeneous T4SS complexes containing multiple variants of each paralog. Alternatively, homogenous T4SS complexes might be assembled using a single paralog at a given time, which could enable rickettsiae to control effector secretion in a manner dependent on time, life cycle stage, or host cell type. It will be interesting to see how these paralogs impact substrate specificity and secretion efficiency and, ultimately, how they impact the infectious life cycle and pathogenesis.
Figure 3.
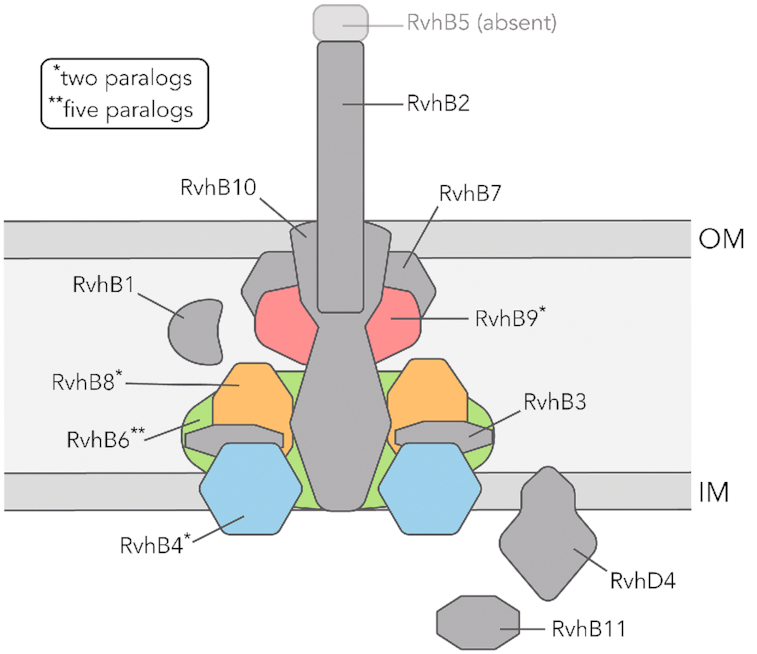
The rickettsial Type IV secretion system. The Type IV secretion system (T4SS) from the order Rickettsiales contains eleven distinct subunits termed Rickettsiales vir homologs (Rvh). Notably, the rickettsial T4SS has undergone extensive gene duplication. As a result, the RvhB4, RvhB6, RvhB8 and RvhB9 subunits have multiple paralogs (depicted in color). Also, the RvhB5 subunit is absent from the rickettsial T4SS. OM and IM indicate the rickettsial outer and inner membranes, respectively.
As Type IV substrates are difficult to predict with bioinformatic approaches, we do not know the complete catalog of Type IV effectors in rickettsiae. Still, several recent studies have identified candidate Type IV effectors. In a bacterial two-hybrid experiment, the R. typhi protein RalF was shown to interact with RvhD4, a subunit of the T4SS involved in substrate recognition, suggesting that it is a Type IV effector (Rennoll-Bankert et al. 2015, 2016), similar to its homolog in Legionella (Folly-Klan et al. 2013). Another rickettsial protein, RARP-2, coimmunoprecipitates with RvhD4, suggesting it is also a T4SS substrate (Lehman et al. 2018; Aistleitner et al. 2020). Encouraged by the identification of RalF and RARP-2, a recent investigation sought to systematically identify Type IV effectors in R. typhi using coimmunoprecipitation with RvhD4 followed by mass spectrometry (Voss et al. 2020). This approach identified seven new putative Type IV effector proteins, including Pat2, a patatin-like phospholipase and Risk1 (Voss et al. 2020). However, this screen did not identify the full repertoire of Type IV effectors, as known Type IV effectors like RARP-2 were not detected. Therefore, more sensitive or orthogonal approaches, including improved bioinformatic prediction software (Esna Ashari, Brayton and Broschat 2019), will be crucial in determining the full suite of effectors that interact with the rickettsial T4SS.
A STREAMLINED AND MINIMALIST GENOME RELIANT ON HOST CELL METABOLISM
Rickettsiae are among a small number of bacterial pathogens that primarily reside in the cytosol of eukaryotic host cells, including S. flexneri, L. monocytogenes, F. tularensis and B. pseudomallei, with many other bacterial pathogens opting for a vacuolar-replicative niche (Ray et al. 2009). It is not clear why this uneven division between cytosolic and vacuolar bacterial pathogens exists, and to what extent the cytosol is permissive to bacterial growth. The eukaryotic cytosol is known to be strongly reducing (Hwang, Sinskey and Lodish 1992; Ray et al. 2009), but its precise nutritional content has not been fully defined and may vary across different cell types (Ray et al. 2009). Some modified strains of bacteria like Bacillus subtilis (Bielecki et al. 1990) and E. coli (Monack and Theriot 2001) can replicate efficiently in eukaryotic host cells, demonstrating that the cytosol can be growth permissive for bacteria that are not strictly evolved for that niche. Additionally, recent work has revealed an increasing relevance of the cytosolic niche for bacteria that were once thought to be strictly vacuolar, including S. enterica serovar Typhimurium, Mycobacterium tuberculosis and Legionella pneumophila (reviewed in (Fredlund and Enninga 2014)). Defining the metabolic interactions and exchange between rickettsiae and their host cells will help contribute to a deeper understanding of the eukaryotic cytosol, especially as a niche for bacterial pathogens at large. Such insights will also be crucial towards the development of an axenic medium for rickettsiae, which would transform the field by massively simplifying the process of culturing and genetic manipulation, similar to what has been done for the vacuolar obligate intracellular bacterium Coxiella burnetii (Omsland et al. 2008, 2009).
Over the course of evolutionary time, rickettsiae have undergone extensive reductive genome evolution, with an average genome size of around one million base pairs (Andersson et al. 1998; Blanc et al. 2007; Palmer and Azad 2012). The cytosolic lifestyle of rickettsiae allows them to scavenge nutrients from the host cell, relieving selective pressure to maintain key biosynthetic pathways in their genomes (Blanc et al. 2007; Fuchs et al. 2012; Palmer and Azad 2012; Driscoll et al. 2017). This defined niche has also allowed rickettsiae to lose genes needed for extracellular survival. Moreover, the intracellular environment may be less conducive to horizontal gene transfer, slowing the rate of gene gain in obligate intracellular bacteria (Newton and Bordenstein 2011; Weinert and Welch 2017). Genome reduction represents a common mode of evolution throughout all kingdoms of life (Wolf and Koonin 2013) and is associated with increased virulence in some bacterial pathogens (Moran 2002; Fournier et al. 2009; Weinert and Welch 2017; Diop, Raoult and Fournier 2018). For example, the loss of genes that negatively contribute to pathogenicity (e.g. antivirulence genes; Maurelli et al. 1998; McCormick et al. 1999) and immunogenic genes (e.g. flagella; Parkhill et al. 2001, 2003; Yang 2005) have been observed in several bacterial pathogens. Further, genome size can influence factors like cell size and replication rate, with smaller genomes leading to faster growth (Koskiniemi et al. 2012; Lee and Marx 2012). In Rickettsia, the loss of regulatory genes is associated with increased pathogenicity (Fournier et al. 2009), though this has not yet been tested in vivo. Furthermore, no evidence was found for increased pathogenicity resulting from the gain of virulence genes in rickettsiae (Darby et al. 2007). Still, with a largely uncharacterized genome, more work is needed to fully understand the genetic determinants of pathogenicity in rickettsiae, as well as to provide insights into reductive genome evolution in bacterial pathogens.
Intracellular life has sculpted rickettsial genomes to become progressively more dependent on the host, while still maintaining a minimal set of genes required for infection and pathogenesis. The genomes of rickettsiae no longer encode numerous biosynthetic pathways in favor of scavenging nutrients from the host cell environment and have lost essential genes involved in glycolysis, gluconeogenesis, the pentose phosphate pathway, B vitamin synthesis and amino acid and nucleotide biosynthesis (Blanc et al. 2007; Palmer and Azad 2012; Driscoll et al. 2017). Reliance on the host cell environment for nucleotides is one potential explanation for the high AT-content of the genome (Dietel et al. 2019). Moreover, rickettsiae import energy-carrying molecules like ATP and NAD+ from the host cell, despite being able to synthesize ATP on their own. Host cell-derived energy carriers may be essential for survival since the addition of ATP or NAD+ partially restores the infectivity of extracellular rickettsiae (Bovarnick, Allen and Pagan 1953; Bovarnick and Schneider 1960). Although the NAD+ transporter has not been identified, heterologous expression studies carried out in E. coli identified the nucleotide translocase Tlc1, an ATP/ADP symporter, as the mechanism of ATP import in rickettsiae (Winkler 1976; Krause, Winkler and Wood 1985; Audia and Winkler 2006). Additionally, the genomes of rickettsiae encode biosynthetic pathways that utilize isoprenoids but lack the genes needed for isoprenoid synthesis (Driscoll et al. 2017; Ahyong et al. 2019). Recent evidence has suggested that rickettsiae import isoprenoid precursors from the host cell (Ahyong et al. 2019). This study found that statins, FDA-approved drugs that inhibit the host cell isoprenoid synthesis, impaired R. parkeri infection in vitro, providing a promising route for future drug development. Continued characterization of the metabolic interactions between rickettsiae and their mammalian and arthropod hosts will reveal new avenues for therapeutic interventions, which will be crucial given the limited options available for treating rickettsial diseases.
CONCLUSION AND OUTLOOK—CURRENT CHALLENGES AND THE PATH FORWARD
Rickettsia is a genus of diverse obligate intracellular bacteria with fascinating and unique biology but remains poorly understood at a molecular and genetic level. With several global, neglected and emerging human pathogens in the genus, it will be critical to gain a deeper understanding of their biology and virulence strategies. Since their discovery over 100 years ago, rickettsiae have proven challenging to study due to their obligate intracellular nature. One of the most significant roadblocks towards understanding the enigmatic biology of rickettsiae has been the lack of genetic tools. Still, this obstacle has started to diminish with recent developments in targeted knockout and transposon mutagenesis.
In this review, we highlighted several key exciting areas of unique biology in rickettsiae and the many fundamental questions that remain. We propose that future efforts in three fundamental and complementary approaches will pave the way towards unlocking these mysteries. First, more comprehensive sampling and increased genome sequencing of Rickettsia species from diverse hosts in a variety of ecosystems will be critical for gaining a clearer picture of evolution and diversification within this genus. Second, further development of methods for profiling gene expression, metabolism and protein secretion will provide key insights into the molecular details of the intracellular life cycles of rickettsiae. Finally, new methods for genetic manipulation will be instrumental in determining the genetic basis of the intracellular life cycles and pathogenesis of rickettsiae. Altogether, the time is ripe for making fundamental advances in our understanding of rickettsiae, which will provide invaluable insights into the biology and evolution of obligate intracellular bacteria, host–pathogen interactions and eukaryotic cell biology.
ACKNOWLEDGEMENTS
We thank Allen Sanderlin, Patrick Woida and Allison Scott for critical reading of our manuscript.
Contributor Information
Jon McGinn, Department of Biology, Massachusetts Institute of Technology, 77 Massachusetts Avenue, Cambridge, MA 02139, United States.
Rebecca L Lamason, Department of Biology, Massachusetts Institute of Technology, 77 Massachusetts Avenue, Cambridge, MA 02139, United States.
FUNDING
J.M. is a Damon Runyon Fellow supported by the Damon Runyon Cancer Research Foundation (DRG-2396–20). We are also grateful for prior and current support for R.L.L. from an NIGMS K99/R00 Pathway to Independence Award (GM115765), the James H. Ferry Jr. Fund for Innovation in Research Education and the Surdna Junior Faculty Research Fund.
Conflicts of interest
None declared.
REFERENCES
- Abeykoon A, Wang G, Chao C-Cet al. Multimethylation of Rickettsia OmpB catalyzed by lysine methyltransferases. J Biol Chem. 2014;289:7691–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abeykoon AH, Chao C-C, Wang Get al. Two protein lysine methyltransferases methylate outer membrane protein B from Rickettsia. J Bacteriol. 2012;194:6410–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abeykoon AH, Noinaj N, Choi B-Eet al. Structural insights into substrate recognition and catalysis in outer membrane protein B (OmpB) by protein-lysine methyltransferases from Rickettsia. J Biol Chem. 2016;291:19962–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahyong V, Berdan CA, Burke TPet al. A metabolic dependency for host isoprenoids in the obligate intracellular pathogen Rickettsia parkeri underlies a sensitivity to the statin class of host-targeted therapeutics. mSphere. 2019;4:e00536–19.. /msphere/4/6/mSphere536-19.atom. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aistleitner K, Clark T, Dooley Cet al. Selective fragmentation of the trans-Golgi apparatus by Rickettsia rickettsii. PLoS Pathog. 2020;16:e1008582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfred J, Baldwin IT. New opportunities at the wild frontier. eLife. 2015;4:e06956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson SGE, Zomorodipour A, Andersson JOet al. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature. 1998;396:133–40. [DOI] [PubMed] [Google Scholar]
- Angelakis E, Bechah Y, Raoult D. The history of epidemic typhus. In: Drancourt R (eds). Paleomicrobiology of Humans. American Society of Microbiology, 2016, 81–92. [DOI] [PubMed] [Google Scholar]
- Antwis RE, Griffiths SM, Harrison XAet al. Fifty important research questions in microbial ecology. FEMS Microbiol Ecol. 2017;93. DOI: 10.1093/femsec/fix044. [DOI] [PubMed] [Google Scholar]
- Audia JP, Winkler HH. Study of the five Rickettsia prowazekii proteins annotated as ATP/ADP translocases (Tlc): only Tlc1 transports ATP/ADP, while Tlc4 and Tlc5 transport other ribonucleotides. JB. 2006;188:6261–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azad AF. Pathogenic rickettsiae as bioterrorism agents. Clin Infect Dis. 2007;45:S52–5. [DOI] [PubMed] [Google Scholar]
- Balraj P, Renesto P, Raoult D. Advances in Rickettsia Pathogenicity. Ann N Y Acad Sci. 2009;1166:94–105. [DOI] [PubMed] [Google Scholar]
- Banajee KH, Embers ME, Langohr IMet al. Amblyomma maculatum feeding augments Rickettsia parkeri infection in a rhesus macaque model: a pilot study. PLoS One. 2015;10:e0135175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao W, Kumagai Y, Niu Het al. Four VirB6 paralogs and VirB9 are expressed and interact in Ehrlichia chaffeensis-containing vacuoles. J Bacteriol. 2009;191:278–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett ADT, Stanberry LR. Vaccines for Biodefense and Emerging and Neglected Diseases. London: Academic, 2009. [Google Scholar]
- Bechah Y, Paddock CD, Capo Cet al. Adipose tissue serves as a reservoir for recrudescent Rickettsia prowazekii infection in a mouse model. PLoS One. 2010;5:e8547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendonk TU, Manaia CM, Merlin Cet al. Tackling antibiotic resistance: the environmental framework. Nat Rev Microbiol. 2015;13:310–7. [DOI] [PubMed] [Google Scholar]
- Bielecki J, Youngman P, Connelly Pet al. Bacillus subtilis expressing a haemolysin gene from Listeria monocytogenes can grow in mammalian cells. Nature. 1990;345:175–6. [DOI] [PubMed] [Google Scholar]
- Biggs HM, Behravesh CB, Bradley KKet al. Diagnosis and management of tickborne rickettsial diseases: rocky mountain spotted fever and other spotted fever group rickettsioses, ehrlichioses, and anaplasmosis — United States: a practical guide for health care and public health professionals. MMWR Recomm Rep. 2016;65:1–44. [DOI] [PubMed] [Google Scholar]
- Blanc G, Ogata H, Robert Cet al. Reductive genome evolution from the mother of Rickettsia. PLos Genet. 2007;3:e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G, Renesto P, Raoult D. Phylogenic analysis of rickettsial patatin-like protein with conserved phospholipase A2 active sites. Ann N Y Acad Sci. 2005;1063:83–6. [DOI] [PubMed] [Google Scholar]
- Blanton LS. The rickettsioses. Infect Dis Clin North Am. 2019;33:213–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blount ZD. The unexhausted potential of E. coli. eLife. 2015;4:e05826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bovarnick MR, Schneider L. Role of adenosine triphosphate in the hemolysis of sheep erythrocytes by typhus rickettsiae. J Bacteriol. 1960;80:344–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bovarnick NR, Allen EC, Pagan G. The influence of diphosphopyridine nucleotide on the stability of typhus rickettsiae. J Bacteriol. 1953;66:671–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brill NE. An acute infectious disease of unknown origin.: a clinical study based on 221 cases. Typhoid. Unknown infection. Am J Med Sci (1827-1924). 1910;139:484. [DOI] [PubMed] [Google Scholar]
- Brown LD, Macaluso KR. Rickettsia felis, an emerging flea-borne rickettsiosis. Curr Trop Med Rep. 2016;3:27–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgdorfer W, Anacker RLeds. Rickettsiae and Rickettsial Diseases. New York: Academic Press, 1981. [Google Scholar]
- Burke TP, Engström P, Chavez RAet al. Inflammasome-mediated antagonism of type I interferon enhances Rickettsia pathogenesis. Nat Microbiol. 2020a;5:688–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke TP, Tran CJ, Engström Pet al. Rickettsia parkeri Sca2 promotes dissemination in an intradermal infection mouse model. bioRxiv, 2020b. [Google Scholar]
- Burkhardt NY, Baldridge GD, Williamson PCet al. Development of shuttle vectors for transformation of diverse Rickettsia species. PLoS One. 2011;6:e29511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caravedo Martinez MA, Ramírez-Hernández A, Blanton LS. Manifestations and management of flea-borne rickettsioses. Res Rep Trop Med. 2021;12:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardwell MM, Martinez JJ. The Sca2 autotransporter protein from Rickettsia conorii is sufficient to mediate adherence to and invasion of cultured mammalian cells. IAI. 2009;77:5272–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (CDC) . Epidemiology and Statistics | Rocky Mountain Spotted Fever (RMSF). 2020, https://www.cdc.gov/rmsf/stats/index.html, (16 November 2020, date last accessed). [Google Scholar]
- Centers for Disease Control and Prevention (CDC) . Imported Spotted Fevers | Other Spotted Fevers. 2019b, https://www.cdc.gov/otherspottedfever/imported/index.html, (16 November 2020, date last accessed). [Google Scholar]
- Centers for Disease Control and Prevention (CDC) . RMSF: deadly, but preventable. 2019a, https://www.cdc.gov/ncezid/dvbd/media/rmsf.html (16 November 2020, date last accessed). [Google Scholar]
- Chan YG-Y, Riley SP, Martinez JJ. Adherence to and invasion of host cells by spotted fever group Rickettsia species. Front Microbiol. 2010;1:139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng MI, Chen C, Engström Pet al. Actin-based motility allows Listeria monocytogenes to avoid autophagy in the macrophage cytosol. Cell Microbiol. 2018;20:e12854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe JE, Welch MD. Actin-based motility of bacterial pathogens: mechanistic diversity and its impact on virulence. Pathog Dis. 2016;74:ftw099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark TR, Lackey AM, Kleba Bet al. Transformation frequency of a mariner-based transposon in Rickettsia rickettsii. J Bacteriol. 2011;193:4993–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifton DR, Goss RA, Sahni SKet al. NF- B-dependent inhibition of apoptosis is essential for host cell survival during Rickettsia rickettsii infection. Proc Natl Acad Sci. 1998;95:4646–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossart P. Bacterial invasion: the paradigms of enteroinvasive pathogens. Science. 2004;304:242–8. [DOI] [PubMed] [Google Scholar]
- Costa TRD, Felisberto-Rodrigues C, Meir Aet al. Secretion systems in Gram-negative bacteria: structural and mechanistic insights. Nat Rev Microbiol. 2015;13:343–59. [DOI] [PubMed] [Google Scholar]
- Curto P, Simões I, Riley SPet al. Differences in intracellular fate of two spotted fever group Rickettsia in macrophage-like cells. Front Cell Infect Microbiol. 2016;6. DOI: 10.3389/fcimb.2016.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darby AC, Cho N-H, Fuxelius H-Het al. Intracellular pathogens go extreme: genome evolution in the Rickettsiales. Trends Genet. 2007;23:511–20. [DOI] [PubMed] [Google Scholar]
- Dietel A-K, Merker H, Kaltenpoth Met al. Selective advantages favour high genomic AT-contents in intracellular elements. PLos Genet. 2019;15:e1007778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diop A, Raoult D, Fournier P-E. Rickettsial genomics and the paradigm of genome reduction associated with increased virulence. Microbes Infect. 2018;20:401–9. [DOI] [PubMed] [Google Scholar]
- Dowd GC, Mortuza R, Ireton K. Molecular mechanisms of intercellular dissemination of bacterial pathogens. Trends Microbiol. 2020:S0966842X20301876. DOI: 10.1016/j.tim.2020.06.008.. [DOI] [PubMed] [Google Scholar]
- Drancourt M, Raoult D. Characterization of mutations in the rpoB gene in naturally rifampin-resistant Rickettsia species. Antimicrob Agents Chemother. 1999;43:2400–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll TP, Verhoeve VI, Guillotte MLet al. Wholly Rickettsia! reconstructed metabolic profile of the quintessential bacterial parasite of eukaryotic cells. mBio. 2017;8:mBio.00859-17, e00859-17. DOI: 10.1128/mBio.00859-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driskell LO, Yu X, Zhang Let al. Directed mutagenesis of the Rickettsia prowazekii pld gene encoding Phospholipase D. IAI. 2009;77:3244–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engström P, Burke TP, Iavarone ATet al. Lysine methylation shields an intracellular pathogen from ubiquitylation. bioRxiv. 2020;11.20.392290. 10.1101/2020.11.20.392290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engström P, Burke TP, Mitchell Get al. Evasion of autophagy mediated by Rickettsia surface protein OmpB is critical for virulence. Nat Microbiol. 2019;4:2538–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esna Ashari Z, Brayton KA, Broschat SL. Prediction of T4SS effector proteins for Anaplasma phagocytophilum using OPT4e, a new software tool. Front Microbiol. 2019;10:1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faucher J-F, Socolovschi C, Aubry Cet al. Brill-Zinsser disease in Moroccan man, France, 2011. Emerg Infect Dis. 2012;18:171–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields KA, Heinzen RA, Carabeo R. The obligate intracellular lifestyle. Front Microbio. 2011;2. DOI: 10.3389/fmicb.2011.00099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folly-Klan M, Alix E, Stalder Det al. A novel membrane sensor controls the localization and ArfGEF activity of bacterial RalF. PLoS Pathog. 2013;9:e1003747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier P-E, El Karkouri K, Leroy Qet al. Analysis of the Rickettsia africae genome reveals that virulence acquisition in Rickettsia species may be explained by genome reduction. BMC Genomics. 2009;10:166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredlund J, Enninga J. Cytoplasmic access by intracellular bacterial pathogens. Trends Microbiol. 2014;22:128–37. [DOI] [PubMed] [Google Scholar]
- Fuchs TM, Eisenreich W, Heesemann Jet al. Metabolic adaptation of human pathogenic and related nonpathogenic bacteria to extra- and intracellular habitats. FEMS Microbiol Rev. 2012;36:435–62. [DOI] [PubMed] [Google Scholar]
- Gall CA, Reif KE, Scoles GAet al. The bacterial microbiome of Dermacentor andersoni ticks influences pathogen susceptibility. ISME J. 2016;10:1846–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie JJ, Beier MS, Rahman MSet al. Plasmids and rickettsial evolution: insight from Rickettsia felis. PLoS One. 2007;2:e266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie JJ, Kaur SJ, Rahman MSet al. Secretome of obligate intracellular Rickettsia. FEMS Microbiol Rev. 2014. DOI: 10.1111/1574-6976.12084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie JJ, Phan IQH, Driscoll TPet al. The Rickettsia type IV secretion system: unrealized complexity mired by gene family expansion. Pathog Dis. 2016;74:ftw058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman AL, McNulty NP, Zhao Yet al. Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell Host Microbe. 2009;6:279–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouin E, Egile C, Dehoux Pet al. The RickA protein of Rickettsia conorii activates the Arp2/3 complex. Nature. 2004;427:457–61. [DOI] [PubMed] [Google Scholar]
- Haglund CM, Choe JE, Skau CTet al. Rickettsia Sca2 is a bacterial formin-like mediator of actin-based motility. Nat Cell Biol. 2010;12:1057–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris EK, Jirakanwisal K, Verhoeve VIet al. Role of Sca2 and RickA in the dissemination of Rickettsia parkeri in Amblyomma maculatum. Infect Immun. 2018;86:e00123–18., /iai/86/6/e00123-18.atom. DOI: 10.1128/IAI.00123-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinzen RA, Grieshaber SS, Van Kirk LSet al. Dynamics of actin-based movement by Rickettsia rickettsii in vero cells. Infect Immun. 1999;67:4201–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensel M, Shea J, Gleeson Cet al. Simultaneous identification of bacterial virulence genes by negative selection. Science. 1995;269:400–3. [DOI] [PubMed] [Google Scholar]
- Hillman RD, Baktash YM, Martinez JJ. OmpA-mediated rickettsial adherence to and invasion of human endothelial cells is dependent upon interaction with α2β1 integrin: ompA-α2β1 integrin mediated rickettsial invasion. Cell Microbiol. 2013;15:727–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hug LA, Baker BJ, Anantharaman Ket al. A new view of the tree of life. Nat Microbiol. 2016;1:16048. [DOI] [PubMed] [Google Scholar]
- Hutchison CA III. Global transposon mutagenesis and a minimal Mycoplasma genome. Science. 1999;286:2165–9. [DOI] [PubMed] [Google Scholar]
- Hwang C, Sinskey AJ, Lodish HF. Oxidized redox state of glutathione in the endoplasmic reticulum. Science. 1992;257:1496–502. [DOI] [PubMed] [Google Scholar]
- Jeng RL, Goley ED, D'Alessio JAet al. A Rickettsia WASP-like protein activates the Arp2/3 complex and mediates actin-based motility. Cell Microbiol. 2004;6:761–9. [DOI] [PubMed] [Google Scholar]
- Joshi SG, Francis CW, Silverman DJet al. NF-κB activation suppresses host cell apoptosis during Rickettsia rickettsii infection via regulatory effects on intracellular localization or levels of apoptogenic and anti-apoptotic proteins. FEMS Microbiol Lett. 2004;234:333–41. [DOI] [PubMed] [Google Scholar]
- Kim HK, Premaratna R, Missiakas DMet al. Rickettsia conorii O antigen is the target of bactericidal Weil–Felix antibodies. Proc Natl Acad Sci USA. 2019;116:19659–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleba B, Clark TR, Lutter EIet al. Disruption of the Rickettsia rickettsii Sca2 autotransporter inhibits actin-based motility. IAI. 2010;78:2240–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knuth K, Niesalla H, Hueck CJet al. Large-scale identification of essential Salmonella genes by trapping lethal insertions: essential Salmonella genes. Mol Microbiol. 2004;51:1729–44. [DOI] [PubMed] [Google Scholar]
- Koskiniemi S, Sun S, Berg OGet al. Selection-driven gene loss in bacteria. PLos Genet. 2012;8:e1002787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause DC, Winkler HH, Wood DO. Cloning and expression of the Rickettsia prowazekii ADP/ATP translocator in Escherichia coli. Proc Natl Acad Sci. 1985;82:3015–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehl CJ, Dragoi A-M, Talman Aet al. Bacterial spread from cell to cell: beyond actin-based motility. Trends Microbiol. 2015;23:558–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamason RL, Bastounis E, Kafai NMet al. Rickettsia Sca4 reduces vinculin-mediated intercellular tension to promote spread. Cell. 2016;167:670–83.. e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamason RL, Kafai NM, Welch MD. A streamlined method for transposon mutagenesis of Rickettsia parkeri yields numerous mutations that impact infection. PLoS One. 2018;13:e0197012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamason RL, Welch MD. Actin-based motility and cell-to-cell spread of bacterial pathogens. Curr Opin Microbiol. 2017;35:48–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M-C, Marx CJ. Repeated, selection-driven genome reduction of accessory genes in experimental populations. PLos Genet. 2012;8:e1002651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman SS, Noriea NF, Aistleitner Ket al. The rickettsial ankyrin repeat protein 2 is a Type IV secreted effector that associates with the endoplasmic reticulum. mBio. 2018;9:e00975–18., /mbio/9/3/mBio.00975-18.atom. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z-M, Tucker AM, Driskell LOet al. mariner-based transposon mutagenesis of Rickettsia prowazekii. Appl Environ Microbiol. 2007;73:6644–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macaluso KR, Sonenshine DE, Ceraul SMet al. Rickettsial infection in Dermacentor variabilis (Acari: ixodidae) inhibits transovarial transmission of a second Rickettsia. J Med Entomol. 2002;39:809–13. [DOI] [PubMed] [Google Scholar]
- Madasu Y, Suarez C, Kast DJet al. Rickettsia Sca2 has evolved formin-like activity through a different molecular mechanism. Proc Natl Acad Sci. 2013;110:E2677–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel M, Higa A. Calcium-dependent bacteriophage DNA infection. J Mol Biol. 1970;53:159–62. [DOI] [PubMed] [Google Scholar]
- Martinez JJ, Cossart P. Early signaling events involved in the entry of Rickettsia conorii into mammalian cells. J Cell Sci. 2004;117:5097–106. [DOI] [PubMed] [Google Scholar]
- Martinez JJ, Seveau S, Veiga Eet al. Ku70, a component of DNA-dependent protein kinase, is a mammalian receptor for Rickettsia conorii. Cell. 2005;123:1013–23. [DOI] [PubMed] [Google Scholar]
- Matthews BJ, Vosshall LB. How to turn an organism into a model organism in 10 ‘easy’ steps. J Exp Biol. 2020;223:jeb218198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurelli AT, Fernández RE, Bloch CAet al. “Black holes” and bacterial pathogenicity: a large genomic deletion that enhances the virulence of Shigella spp. and enteroinvasive Escherichia coli. Proc Natl Acad Sci USA. 1998;95:3943–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure EE, Chávez ASO, Shaw DKet al. Engineering of obligate intracellular bacteria: progress, challenges and paradigms. Nat Rev Microbiol. 2017;15:544–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick BA, Fernandez MI, Siber AMet al. Inhibition of Shigella flexneri-induced transepithelial migration of polymorphonuclear leucocytes by cadaverine. Cell Microbiol. 1999;1:143–55. [DOI] [PubMed] [Google Scholar]
- Monack DM, Theriot JA. Actin-based motility is sufficient for bacterial membrane protrusion formation and host cell uptake. Cell Microbiol. 2001;3:633–47. [DOI] [PubMed] [Google Scholar]
- Moran NA. Microbial minimalism. Cell. 2002;108:583–6. [DOI] [PubMed] [Google Scholar]
- Narra HP, Sahni A, Walker DHet al. Recent research milestones in the pathogenesis of human rickettsioses and opportunities ahead. Fut Microbiol. 2020;15:753–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton ILG, Bordenstein SR. Correlations between bacterial ecology and mobile DNA. Curr Microbiol. 2011;62:198–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noriea NF, Clark TR, Hackstadt T. Targeted knockout of the Rickettsia rickettsii OmpA surface antigen does not diminish virulence in a mammalian model system. mBio. 2015;6:e00323–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata H, La Scola B, Audic Set al. Genome sequence of Rickettsia bellii illuminates the role of amoebae in gene exchanges between intracellular pathogens. PLos Genet. 2006;2:e76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omsland A, Cockrell DC, Fischer ERet al. Sustained axenic metabolic activity by the obligate intracellular bacterium Coxiella burnetii. JB. 2008;190:3203–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omsland A, Cockrell DC, Howe Det al. Host cell-free growth of the Q fever bacterium Coxiella burnetii. Proc Natl Acad Sci. 2009;106:4430–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paddock CD, Alvarez-Hernández G. Rickettsia rickettsii (Rocky Mountain Spotted Fever). Principles and Practice of Pediatric Infectious Diseases. Elsevier, 2018, 952–7.. e2. [Google Scholar]
- Palmer GH, Azad AFeds. Intracellular Pathogens II: Rickettsiales. Washington, DC: ASM Press, 2012. [Google Scholar]
- Park H, Lee JH, Gouin Eet al. The Rickettsia surface cell antigen 4 applies mimicry to bind to and activate vinculin. J Biol Chem. 2011;286:35096–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhill J, Sebaihia M, Preston Aet al. Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Nat Genet. 2003;35:32–40. [DOI] [PubMed] [Google Scholar]
- Parkhill J, Wren BW, Thomson NRet al. Genome sequence of Yersinia pestis, the causative agent of plague. Nature. 2001;413:523–7. [DOI] [PubMed] [Google Scholar]
- Parola P, Paddock CD, Socolovschi Cet al. Update on tick-borne rickettsioses around the world: a geographic approach. Clin Microbiol Rev. 2013;26:657–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelc RS, McClure JC, Kaur SJet al. Disrupting protein expression with peptide nucleic acids reduces infection by obligate intracellular Rickettsia. PLoS One. 2015;10:e0119283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman SJ, Hunter MS, Zchori-Fein E. The emerging diversity of Rickettsia. Proc R Soc B. 2006;273:2097–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perotti bacteria MA, Clarke HK, Turner BDet al. Rickettsia as obligate and mycetomic. FASEB J. 2006;20:2372–4. [DOI] [PubMed] [Google Scholar]
- Philip RN, Casper EA, Anacker RLet al. Rickettsia bellii sp. nov.: a tick-borne Rickettsia, widely distributed in the United States, that is distinct from the spotted fever and typhus biogroups. Int J Syst Bacteriol. 1983;33:94–106. [Google Scholar]
- Pinkerton H, Hass GM. Spotted fever : I. Intranuclear rickettsiae in spotted fever studied in tissue culture. J Exp Med. 1932;56:151–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizarro-Cerdá J, Cossart P. Microbe profile: listeria monocytogenes: a paradigm among intracellular bacterial pathogens. Microbiology. 2019;165:719–21. [DOI] [PubMed] [Google Scholar]
- Rachek LI, Tucker AM, Winkler HHet al. Transformation of Rickettsia prowazekii to rifampin resistance. J Bacteriol. 1998;180:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman MS, Ammerman NC, Sears KTet al. Functional characterization of a phospholipase A(2) homolog from Rickettsia typhi. J Bacteriol. 2010;192:3294–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman MS, Gillespie JJ, Kaur SJet al. Rickettsia typhi possesses phospholipase A2 enzymes that are involved in infection of host cells. PLoS Pathog. 2013;9:e1003399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray K, Marteyn B, Sansonetti PJet al. Life on the inside: the intracellular lifestyle of cytosolic bacteria. Nat Rev Microbiol. 2009;7:333–40. [DOI] [PubMed] [Google Scholar]
- Reed SCO, Lamason RL, Risca VIet al. Rickettsia actin-based motility occurs in distinct phases mediated by different actin nucleators. Curr Biol. 2014;24:98–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed SCO, Serio AW, Welch MD. Rickettsia parkeri invasion of diverse host cells involves an Arp2/3 complex, WAVE complex and Rhofamily GTPasedependent pathway. Cell Microbiol. 2011:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rennoll-Bankert KE, Rahman MS, Gillespie JJet al. Which way in? The RalF Arf-GEF orchestrates Rickettsia host cell invasion. PLoS Pathog. 2015;11:e1005115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rennoll-Bankert KE, Rahman MS, Guillotte MLet al. RalF-mediated activation of Arf6 controls Rickettsia typhi invasion by co-opting phosphoinositol metabolism. Infect Immun. 2016;84:3496–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahni SK, Narra HP, Sahni Aet al. Recent molecular insights into rickettsial pathogenesis and immunity. Fut Microbiol. 2013;8:1265–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekeyová Z, Danchenko M, Filipčík Pet al. Rickettsial infections of the central nervous system. PLoS Negl Trop Dis. 2019;13:e0007469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serio AW, Jeng RL, Haglund CMet al. Defining a core set of actin cytoskeletal proteins critical for actin-based motility of Rickettsia. Cell Host Microbe. 2010;7:388–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman DJ, Santucci LA, Meyers Net al. Penetration of host cells by Rickettsia rickettsii appears to be mediated by a phospholipase of rickettsial origin. Infect Immun. 1992;60:2733–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman DJ, Wisseman CL, Waddell A. In vitro studies of rickettsia-host cell interactions: ultrastructural study of Rickettsia prowazekii-infected chicken embryo fibroblasts. Infect Immun. 1980;29:778–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman DJ, Wisseman CL. In vitro studies of rickettsia-host cell interactions: ultrastructural changes induced by Rickettsia rickettsii infection of chicken embryo fibroblasts. Infect Immun. 1979;26:714–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Socolovschi C, Mediannikov O, Raoult Det al. The relationship between spotted fever group Rickettsiae and Ixodid ticks. Vet Res. 2009;40:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teysseire N, Boudier JA, Raoult D. Rickettsia conorii entry into Vero cells. Infect Immun. 1995;63:366–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teysseire N, Chiche-Portiche C, Raoult D. Intracellular movements of Rickettsia conorii and R. typhi based on actin polymerization. Res Microbiol. 1992;143:821–9. [DOI] [PubMed] [Google Scholar]
- Tomassone L, Portillo A, Nováková Met al. Neglected aspects of tick-borne rickettsioses. Parasites Vectors. 2018;11:263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troyer JM, Radulovic S, Andersson SGet al. Detection of point mutations in rpoB gene of rifampin-resistant Rickettsia typhi. Antimicrob Agents Chemother. 1998;42:1845–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiyama T, Kishi M, Ogawa M. Restriction of the growth of a nonpathogenic spotted fever group Rickettsia. FEMS Immunol Med Microbiol. 2012;64:42–7. [DOI] [PubMed] [Google Scholar]
- van Opijnen T, Bodi KL, Camilli A. Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat Methods. 2009;6:767–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vellaiswamy M, Kowalczewska M, Merhej Vet al. Characterization of rickettsial adhesin Adr2 belonging to a new group of adhesins in α-proteobacteria. Microb Pathog. 2011;50:233–42. [DOI] [PubMed] [Google Scholar]
- Vigouroux A, Bikard D. CRISPR tools to control gene expression in bacteria. Microbiol Mol Biol Rev. 2020;84, DOI: 10.1128/MMBR.00077-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss OH, Gillespie JJ, Lehman SSet al. Risk1, a phosphatidylinositol 3-kinase effector, promotes Rickettsia typhi intracellular survival. mBio. 2020;11:e00820–20., /mbio/11/3/mBio.00820-20.atom. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker DH, Cain BG. The rickettsial plaque. Evidence for direct cytopathic effect of Rickettsia rickettsii. Lab Invest. 1980;43:388–96. [PubMed] [Google Scholar]
- Walker DH, Firth WT, Ballard JGet al. Role of phospholipase-associated penetration mechanism in cell injury by Rickettsia rickettsii. Infect Immun. 1983;40:840–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker DH, Harrison A, Henderson Fet al. Identification of Rickettsia rickettsii in a guinea pig model by immunofluorescent and electron microscopic techniques. Am J Pathol. 1977;86:343–58. [PMC free article] [PubMed] [Google Scholar]
- Weinert LA, Welch JJ. Why might bacterial pathogens have small genomes?. Trends Ecol Evol. 2017;32:936–47. [DOI] [PubMed] [Google Scholar]
- Weinert LA, Werren JH, Aebi Aet al. Evolution and diversity of Rickettsia bacteria. BMC Biol. 2009;7:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert LA. The diversity and phylogeny of Rickettsia. In: Morand S, Krasnov BR, Littlewood DTJ (eds). Parasite Diversity and Diversification. Cambridge: Cambridge University Press, 2015, 150–81. [Google Scholar]
- Whitworth T, Popov VL, Yu X-Jet al. Expression of the Rickettsia prowazekii pld or tlyC gene in Salmonella enterica serovar Typhimurium mediates phagosomal escape. Infect Immun. 2005;73:6668–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler HH. Rickettsial permeability. An ADP-ATP transport system. J Biol Chem. 1976;251:389–96. [PubMed] [Google Scholar]
- Wisseman CL, Edlinger EA, Waddell ADet al. Infection cycle of Rickettsia rickettsii in chicken embryo and L-929 cells in culture. Infect Immun. 1976;14:1052–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisseman CL, Waddell AD. In vitro studies on rickettsia-host cell interactions: intracellular growth cycle of virulent and attenuated Rickettsia prowazeki in chicken embryo cells in slide chamber cultures. Infect Immun. 1975;11:1391–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolbach SB. Studies on rocky mountain spotted fever. J Med Res. 1919;41:1–198. [PMC free article] [PubMed] [Google Scholar]
- Wolf YI, Koonin EV. Genome reduction as the dominant mode of evolution. Bioessays. 2013;35:829–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright CL, Sonenshine DE, Gaff HDet al. Rickettsia parkeri transmission to Amblyomma americanum by cofeeding with Amblyomma maculatum (Acari: ixodidae) and potential for spillover. J Med Entomol. 2015;52:1090–5. [DOI] [PubMed] [Google Scholar]
- Yang F. Genome dynamics and diversity of Shigella species, the etiologic agents of bacillary dysentery. Nucleic Acids Res. 2005;33:6445–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa Y, Ogawa M, Hain Tet al. Listeria monocytogenes ActA-mediated escape from autophagic recognition. Nat Cell Biol. 2009;11:1233–40. [DOI] [PubMed] [Google Scholar]
- Zchori-Fein E, Borad C, Harari AR. Oogenesis in the date stone beetle, Coccotrypes dactyliperda, depends on symbiotic bacteria. Physiol Entomol. 2006;31:164–9. [Google Scholar]
- Zinsser H, Castaneda MR. On the isolation from a case of Brill's disease of a typhus strain resembling the European type. N Engl J Med. 1933;209:815–9. [Google Scholar]