Abstract
Purpose
We report on the first identified cluster of the B.1.1.7 variant of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infections in the northeast of Italy.
Methods
The cluster was recognized in January 2021 with an epidemiological started from the hospitalization of a 68-year-old man suffering from coronavirus disease 2019 (COVID-19) related pneumonia and we surprisingly found three families involved in the same cluster.
Results
We retrospectively rebuilt the pathway of infection and performed a virological analysis.
Conclusion
This allow us to make clear the very high attack rate and the great infective capacity of this B.1.1.7 variant of SARS-CoV-2.
Keywords: COVID-19, UK variant, Sars-COV-2
Background
A new emergent variant of SARS-CoV-2, called B.1.1.7 and characterized by an unusually large number of mutations, was first identified in the UK in Sep 2020 [1–5]. It has quickly disseminated and has become highly prevalent in London and southeast England, besides being detected in numerous other countries around the world, including the Netherlands, Denmark, and the United States [6, 7]. In Italy, the first case of the B.1.1.7 variant was identified in December 2020 in a traveler who had arrived from the UK [8]. Since then, the variant has been reported in different Italian regions [9].
Case
On Jan 7, 2021, a 68-year-old patient with hypertension presented to the Infectious Diseases Unit of Padua with a six-day history of fever and dry cough. Eight days before, he had a positive SARS-CoV-2 real-time polymerase chain reaction (RT-PCR) nasopharyngeal swab (cycle threshold [Ct] of 15.3 [10]). At admission the physical examination was notable for a body temperature of 39.0 °C, a respiratory rate of 28/min, and an oxygen saturation of 68 mmHg. The laboratory workup revealed a low white blood cell count (3400 cells/µL), a mild lymphopenia (970 cells/µL), a significant increase of C-Reactive Protein (109 mg/L; n.v < 6), and a respiratory alkalosis (pH = 7.49). A new nasopharyngeal swab confirmed the SARS-CoV-2 infection (Ct of 19.7) and a viral whole genome sequencing (WGS) of the respiratory tract sample was performed on the grounds of epidemiological suspicion that the B.1.1.7 variant was involved. The chest computed tomography showed multifocal bilateral and peripheral ground-glass pulmonary opacities (Fig. 1). The patient was treated with Remdesivir [200 mg intravenous (i.v.) loading dose at day 1 and 100 mg at days 2–5], Dexamethasone (6 mg i.v. per day), convalescent plasma and low-molecular weight heparin (4000 international units per day). He completely recovered in nine days. At discharge, he continued isolation at home being the nasopharyngeal swab still positive (Ct of 33.9). The WGS result was available two days after the discharge and showed the presence of all the mutations typical of the B.1.1.7 lineage.
Fig. 1.
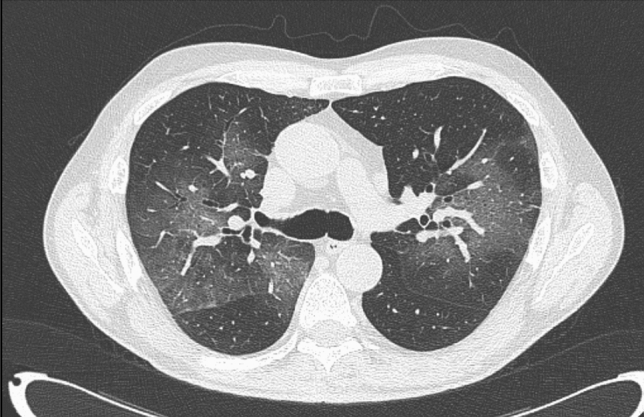
Multifocal bilateral and peripheral ground-glass pulmonary opacities
Epidemiological investigation
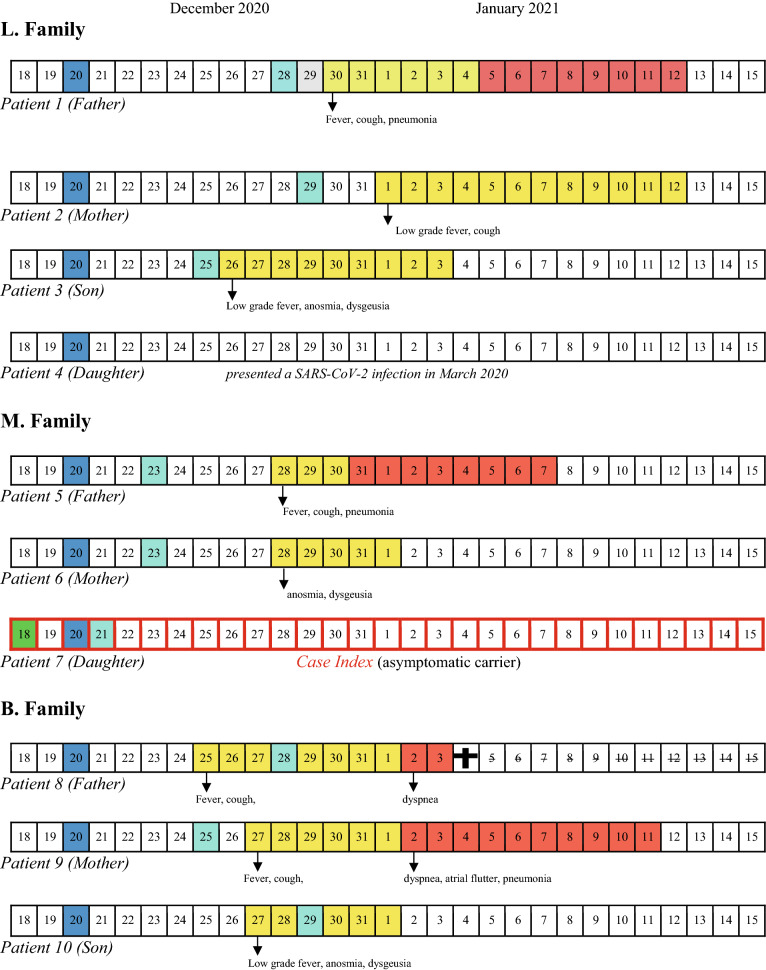
Our patient (Patient 1) was presumably exposed to SARS-CoV-2 on Dec 20, 2020, during a Christmas gathering lunch attended by ten relatives from three different families (L., M., and B. family). The chronology of SARS-CoV-2 tests and symptom’s evolution of all the familie’s members is shown in Table 1. The niece of Patient 1 (Patient 7: index case) attended the lunch the day after her return from London. She had performed a SARS-CoV-2 RT-PCR test on December 18, resulted negative. However, a new molecular test performed for traveller’s surveillance purposes was found positive on Dec 21 (Ct of 19.5). As a consequence, all the other nine individuals who attended the lunch were immediately isolated in their home and did not meet each other anymore. By the time of the family lunch, neither the index case nor the other family members had symptoms of SARS-CoV-2 infection. Those who acquired the infection started to get sick five–ten days later, whereas the index case remained asymptomatic [11]. Overall, a secondary transmission occurred in eight persons, of which four had mild symptoms and four were hospitalized. Among the latter, there was a 70-year old man (Patient 8) with a past-medical history of cardiovascular disease and advanced chronic kidney disease. He had rapidly progressed to an acute respiratory distress syndrome and died after two days of hospitalization.
Table 1.
Chronology of tests, symptom’s onset and evolution of the three families cluster
Blue square Christmas lunch, Light blue squares SARS-CoV-2 RT-PCR-positive test, Green square SARS-CoV-2 RT-PCR-negative test, Yellow squares presence of symptoms, Red squares hospitalization, Black cross day of death
Overall, the median duration of nasopharyngeal swab PCR positivity was 17 days (range 11–25), with no important differences between hospitalized and not hospitalized patients. The presence of the B.1.1.7 variant was confirmed by WGS not only for Patient 1 but also for Patient 2. Unfortunately, the WGS was not performed on the Index Case because of the unavailability of the methodic in the laboratory where the positive PCR test was carried out.
The daughter of Patient 1 (Patient 4) was the only one who did not acquire the infection. She reported having had a previous paucisymptomatic SARS-CoV-2 infection, occurred in Mar 2020. Her serum contained indeed high titers of SARS-CoV-2-specific antibodies [IgM = 1182 kAU/L], IgG 4346 kAU/L (Positive: > 1100 kAU/L).
Virological analysis
The detection of SARS-CoV-2 N2 gene was performed using RT PCR in accordance with CDC guidelines. The diagnostic procedures are accredited by Accredia since Oct 30, 2020 [12].
Complete genome sequencing was performed on the RNA isolated from the nasopharyngeal swab of Patient 1 using an Illumina MiSeq platform (Illumina, San Diego, CA, USA) and an in-house protocol for target amplification. After trimming and filtering for quality, reads were aligned against the reference genome (GenBank: NC_045512.2) using BWA-mem [13]. The sequence reads were able to cover 99.6% of the reference genome with a mean coverage depth of 2292. The sequence was deposited in GISAID under accession number EPI_ISL_875558.
According to the PANGOLIN application [13, 14], the virus was assigned to the lineage B.1.1.7 (variant VOC 202012/01) and possessed all the 14 mutations and the three deletions defining this lineage [14].
Sequencing of the Spike protein of Patient 2 was also performed and confirmed all the mutations typical of the lineage B.1.1.7 namely del 69–70, del 144, N501Y, A570D, D614G, P681H, T716I, S982A, and D1118H.
Discussion
We described a cluster of the B.1.1.7 variant of SARS-CoV-2 infections. To the best of our knowledge, this is the first household-associated transmission reported in the northeast of Italy. The epidemiological investigation suggested that the family cluster, encompassing eight secondary cases, was initiated by a person returning from London and that the transmission occurred during a Christmas lunch. The incubation periods for all the eight persons with confirmed SARS-CoV-2 infection matched well with the time of the lunch and no elements emerged which suggest that these people may have been exposed to the virus before or after that lunch. Moreover, the genomic sequencing of two out of nine infected individuals confirmed the B.1.1.7 variant.
This family cluster shows some remarkable features. First, the index case was an asymptomatic carrier of SARS-CoV-2 [11, 15]. However, the attack rate was very high, resulting in all susceptible persons acquiring the infection. This confirms the ability of the new variant to spread more easily and quickly than other variants, as it has already been described [16]. We lack detailed information on movements, seat allocations, toilets use or crockery sharing among the guests during the lunch. However, such festive family gathering are traditionally very long and guests are normally used to move around. Therefore, it is likely that the prolonged and unprotected close contact may have favored the viral transmission through both the respiratory and the contact routes [17].
Second, not only the outbreak could have been avoided if containment measures were respected, but it is also unlikely that the B.1.1.7 variant cluster would have been detected without testing the index case immediately after her return to Italy. At that time, a Decree-Law banned all private gatherings involving more than two persons from different families [18]. Moreover, the rules adopted by the regional authorities provided that all travelers from UK should perform a mandatory test after the arrival on the Italian soil in addition to the requirement of a negative test performed within 48 h before the arrival. It is worth also mentioning that the prompt contact tracing further avoided the transmission of the infection in the community, confirming that preventive measures of isolation are additional effective public health interventions, especially in the case of new variants [19].
Third, the presence of SARS-CoV2 B.1.1.7 variant in Patient 1 was unknown at the beginning, and the clinical management was similar to all other COVID-19 cases. As already described, the new strain does not seem to be associated with a more severe clinical disease [3, 20, 21]. However, in this current cluster, four out of nine patients were hospitalized with moderate/severe diseases and one died. On the other side, the deceased patient presented several underlying comorbidities, which are widely known to be associated with a high mortality risk in COVID-19 patients [22]. Furthermore, the median duration of PCR positivity in our family cluster resulted to be similar to that commonly observed in COVID-19 patients and the severity of the disease of individual patients did not seem to correlate with the duration of viral shedding.
Finally, it is worth noting that Patient 4, even if effectively exposed to the virus, did not acquire the infection. It is reasonable to assume that the previous infection, confirmed by the presence of high anti-SARS-CoV-2 IgG titer, had a protective role. This finding may be an indirect proof that SARS-CoV-2 wild-type antibodies are cross-protective, and that the B.1.1.7 variant might not be able to evade the vaccination or the natural immune response.
Declarations
Conflict of interest
All the authors declare that they have no conflict of interest.
Human rights statements and informed consent
All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1964 and its later amendments. Informed consent was obtained from all patients for being included in the study.
References
- 1.Rambaut A, Loman N, Pybus O, Barclay W, Barrett J, Carabelli A, et al. Preliminary genomic characterization of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations. 2020 Dec. Available from https://virological.org/t/preliminary-genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-the-uk-defined-by-a-novel-set-of-spike-mutations/563.
- 2.Mahase E. Covid-19: what have we learnt about the new variant in the UK? BMJ. 2020;371:m4944. doi: 10.1136/bmj.m4944. [DOI] [PubMed] [Google Scholar]
- 3.Wise J. Covid-19: new coronavirus variant is identified in UK. BMJ. 2020;371:m4857. doi: 10.1136/bmj.m4857. [DOI] [PubMed] [Google Scholar]
- 4.World Health Organization. SARS-CoV-2 variant—United Kingdom. 21 Dec 2020. Available from https://www.who.int/csr/don/21-december-2020-sars-cov2-variant-united-kingdom/en.
- 5.Salzberger B, Buder F, Lampl B, Ehrenstein B, Hitzenbichler F, Holzmann T, Schmidt B, Hanses F. Epidemiology of SARS-CoV-2. Infection. 2020;8:1–7. doi: 10.1007/s15010-020-01531-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.European Centre for Disease Prevention and Control . Rapid increase of a SARS-CoV-2 variant with multiple spike protein mutations observed in the United Kingdom–20 December 2020. Stockholm: ECDC; 2020. [Google Scholar]
- 7.Center for Disease Control and Prevention, Available from https://www.cdc.gov/coronavirus/2019-ncov/more/science-and-research/scientific-brief-emerging-variants.html.
- 8.“A lab in Italy reported a cluster of the U.K. coronavirus variant. But that wasn’t enough to stop the spread.” Available from https://www.google.it/amp/s/www.washingtonpost.com/world/europe/italy-covid-uk-variant/2021/01/16/0732bd24-544e-11eb acc592d2819a1ccb_story.html%3foutputType=amp.
- 9.“Caso italiano è una 007, 'alta viralità'.” Ansa 2020 Dec. Available from https://www.ansa.it/sito/notizie/cronaca/2020/12/21/covid-tre-positivi-e-due-dubbi-su-volo-londra-palermo_555903dc-0265-4c54-8958-b5433fa20e65.html.
- 10.Lavezzo E, Franchin E, Ciavarella C, Cuomo-Dannenburg G, Barzon N, Del Vecchio C, et al. Suppression of a SARS-CoV-2 outbreak in the Italian municipality of Vo’. Nature. 2020;584(7821):425–429. doi: 10.1038/s41586-020-2488-1. [DOI] [PubMed] [Google Scholar]
- 11.Hasanoglu I, Korukluoglu G, Asilturk D, Cosgun Y, Kalem AK, Altas AB, Kayaaslan B, Eser F, Kuzucu EA, Guner R. Higher viral loads in asymptomatic COVID-19 patients might be the invisible part of the iceberg. Infection. 2021;49(1):117–126. doi: 10.1007/s15010-020-01548-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.US Centers for Disease Control and Prevention. 2019-novel coronavirus (2019-nCoV) real-time rRT-PCR panel primers and probes. Washington (DC): Department of Health and Human Services; 2020 [cited 2020 Sep 29].
- 13.Available from https://pangolin.cog-uk.io.
- 14.Houtgast EJ, Sima VM, Bertels K, Al-Ars Z. Hardware acceleration of BWA-MEM genomic short read mapping for longer read lengths. Comput Biol Chem. 2018;75:54–64. doi: 10.1016/j.compbiolchem.2018.03.024. [DOI] [PubMed] [Google Scholar]
- 15.Huff HV, Singh A. Asymptomatic transmission during the coronavirus disease 2019 pandemic and implications for public health strategies. Clin Infect Dis. 2020;71:2752–2756. doi: 10.1093/cid/ciaa654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rambaut A, Holmes EC, O'Toole Á, Hill V, McCrone JT, Ruis C, et al. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol. 2020;5:1403–1407. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chu D, Akl E, Duda S, Solo K, Yaacoub S, Schünemann H, et al. Physical distancing, face masks, and eye protection to prevent person-to-person transmission of SARS-CoV-2 and COVID-19: a systematic review and meta-analysis. Lancet. 2020;395:1973–1987. doi: 10.1016/S0140-6736(20)31142-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Decreto Natale 18 Dicembre 2020, no 172, Ulteriori disposizioni urgenti per fronteggiare i rischi sanitari connessi alla diffusione del virus COVID-19; Gazzetta Ufficiale della Repubblica Italiana, 19 Dec 2020.
- 19.New and Emerging Respiratory Virus Threats Advisory Group. NERVTAG meeting on SARS-CoV-2 variant under investigation VUI-202012/01. 18 Dec 2020. Available from https://khub.net/documents/135939561/338928724/SARS-CoV2+variant+under+investigation%2C+meeting+minutes.pdf/962e866b-161f-2fd5–1030–32b6ab467896?t=1608491166921.
- 20.Calza L, Bon I, Tadolini M, Borderi M, Colangeli V, Badia L, Verucchi G, Rossini G, Vocale C, Gaibani P, Viale P, Attard L. COVID-19 in patients with HIV-1 infection: a single-centre experience in northern Italy. Infection. 2020;3:1–5. doi: 10.1007/s15010-020-01492-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grasselli G, Greco M, Zanella A, Albano G, Antonelli M, Bellani G, et al. Risk factors associated with mortality among patients with COVID-19 in intensive care units in Lombardy, Italy. JAMA Intern Med. 2020;180:1345–1355. doi: 10.1001/jamainternmed.2020.3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leung K, Shum MH, Leung GM, Lam TT, Wu JT. Early transmissibility assessment of the N501Y mutant strains of SARS-CoV-2 in the United Kingdom, October to November 2020. Euro Surveill. 2021;26(1):1–7. doi: 10.2807/1560-7917.ES.2020.26.1.2002106. [DOI] [PMC free article] [PubMed] [Google Scholar]