Abstract
Heart regeneration occurs by dedifferentiation and proliferation of pre-existing cardiomyocytes (CMs). However, the signaling mechanisms by which injury induces CM renewal remain incompletely understood. Here, we find that cardiac injury in zebrafish induces expression of the secreted Wnt inhibitors, including Dickkopf 1 (Dkk1), Dkk3, secreted Frizzled-related protein 1 (sFrp1), and sFrp2, in cardiac tissue adjacent to injury sites. Experimental blocking of Wnt activity via Dkk1 overexpression enhances CM proliferation and heart regeneration, whereas ectopic activation of Wnt8 signaling blunts injury-induced CM dedifferentiation and proliferation. Although Wnt signaling is dampened upon injury, the cytoplasmic β-catenin is unexpectedly increased at disarrayed CM sarcomeres in myocardial wound edges. Our analyses indicated that p21-activated kinase 2 (Pak2) is induced at regenerating CMs, where it phosphorylates cytoplasmic β-catenin at Ser 675 and increases its stability at disassembled sarcomeres. Myocardial-specific induction of the phospho-mimetic β-catenin (S675E) enhances CM dedifferentiation and sarcomere disassembly in response to injury. Conversely, inactivation of Pak2 kinase activity reduces the Ser 675-phosphorylated β-catenin (pS675-β-catenin) and attenuates CM sarcomere disorganization and dedifferentiation. Taken together, these findings demonstrate that coordination of Wnt signaling inhibition and Pak2/pS675-β-catenin signaling enhances zebrafish heart regeneration by supporting CM dedifferentiation and proliferation.
Keywords: heart regeneration, Wnt signaling, PAK2 kinase, cardiomyocyte proliferation, cardiomyocyte dedifferentiation, zebrafish
Introduction
The limited regenerative response in humans is insufficient for the complete healing and recovery of cardiac function after myocardial infarction, a deficiency that is thought to contribute to the high rate of heart failure (Xin et al., 2013; Tzahor and Poss, 2017). By contrast, zebrafish heart regeneration occurs throughout life with minimal scarring, by dedifferentiation and proliferation of pre-existing cardiomyocytes (CMs) (Jopling et al., 2010; Kikuchi et al., 2010). Hearts of mice can regenerate if injured in the first week after birth, which coincides with the transient capacity for CM proliferation (Porrello et al., 2011). Developmental pathways such as Hedgehog, PDGF, Notch, Hippo, and Neuregulin1/ERBB2 signaling are necessary for heart regeneration, via diverse mechanisms involving myocardial, vascular, and epicardial tissues (Kim et al., 2010; Heallen et al., 2013; Lin et al., 2014; Zhao et al., 2014; D'Uva et al., 2015; Gemberling et al., 2015; Wang et al., 2015; Marin-Juez et al., 2016; Wu et al., 2016; Mohamed et al., 2018; Singh et al., 2018). Despite advances, there remains much to understand regarding the cellular determinants and signaling mechanisms controlling CM renewal and heart regeneration.
Wnt signaling has received considerable attention in stem cell maintenance, organogenesis, tissue homeostasis, and regeneration (Logan and Nusse, 2004; Zimmerman et al., 2012). Canonical Wnt signaling is mediated by binding of Wnt proteins to the Frizzled and LRP5/6 co-receptors, resulting in translocation of cytoplasmic β-catenin into the nucleus to activate gene transcription (Logan and Nusse, 2004). In the absence of Wnt ligands, GSK3β phosphorylates β-catenin at N-terminal residues Ser 33/Ser 37/Tyr 41 and promotes its degradation in the cytosol (Zimmerman et al., 2012). The C-terminal residues of β-catenin can also serve as phosphorylation sites, which mediates its distinct stability that is normally independent of GSK3β activity (Nelson and Nusse, 2004; Verheyen and Gottardi, 2010). For example, phosphorylation of Ser 675 of β-catenin at the C-terminus by p21-activated kinase (PAK) or protein kinase A (PKA) enhances its cytoplasmic and nuclear stability (Hino et al., 2005; Zhu et al., 2012). β-catenin localizes in various cellular compartments and executes differential cellular functions or processes, such as cell adhesion, migration, and protrusions (Nelson and Nusse, 2004; Faux et al., 2010). In adult CMs, the majority of β-catenin localizes in intercalated disks and sarcomeres and participates in CM communication, morphogenesis, cytokinesis, and cardiac function (Haq et al., 2003; Hirschy et al., 2010; Vite and Radice, 2014). Many endogenous Wnt inhibitors limit the range of Wnt signaling, including Dickkopfs (Dkks), secreted Frizzled-related proteins (sFrps), Wnt inhibitory factor (Wif), and Sclerostin (Leyns et al., 1997; Hsieh et al., 1999; Li et al., 2005; Cheng et al., 2011). Spatiotemporal control of Wnt inhibitors or activators can have potent effects in many biological contexts. During embryonic cardiogenesis, Wnt activators and inhibitors act sequentially in a positive and then negative fashion (Naito et al., 2006; Ueno et al., 2007; Ni et al., 2011). Recent pharmacological studies implicate that Wnt signaling inhibition and reduction of nuclear β-catenin are beneficial to heart regeneration in zebrafish (Zhao et al., 2019; Xie et al., 2020). However, whether β-catenin in various cellular compartments is regulated following injury and how Wnt signaling is inhibited during heart regeneration require elucidation.
PAKs are serine and threonine protein kinases that play pivotal roles in cardiovascular development and physiology, cell proliferation, polarity, and cytoskeletal rearrangements (Bokoch, 2003; Kelly et al., 2013). PAK1 participates in cardiac excitation, contractility, and vascular development (Ai et al., 2011). Mutations in PAK2 are associated with cerebral hemorrhage and cause embryonic lethal in zebrafish and mice (Buchner et al., 2007; Kelly and Chernoff, 2012). PAK1 normally mediates Wnt signaling in cancer progression or some settings of development (Zhu et al., 2012; Choe and Crump, 2015), which is achieved by cooperation with GTPases Rac1 and Cdc42 (Schlessinger et al., 2009). On the contrary, in Caenorhabditis elegans, PAK can antagonize canonical Wnt signaling in establishing the anterior/posterior polarity of the vulval lineage (Goh et al., 2012). Although our recent studies implicate the roles of Pak2 in zebrafish heart regeneration using pharmacological approach (Peng et al., 2016), whether and how Paks coordinate with Wnt signaling during heart regeneration are not understood.
Here, we provide evidence for interactions between Wnt and Pak2 signaling in the control of CM renewal during heart regeneration. Cardiac injury induces expression of multiple sets of secreted Wnt inhibitors, including epi/endocardial sFrp1/Dkk3, myocardial sFrp2/Dkk1, and endocardial Notum1b/Wif1 (Zhao et al., 2019), which dampens Wnt signaling to enable CM dedifferentiation and proliferation. During regeneration, Pak2 kinase is induced at injured heart tissues, where it increases β-catenin Ser 675 phosphorylation and stability, enhancing CM dedifferentiation and proliferation. Our findings demonstrate that the natural regenerative response to cardiac injury is to block Wnt signaling and suggest new roles and mechanisms for coordination of Pak2 kinase and Wnt signaling inhibitors during heart regeneration.
Results
Mutiple Wnt antagonists Dkks and sFrps are induced by cardiac injury
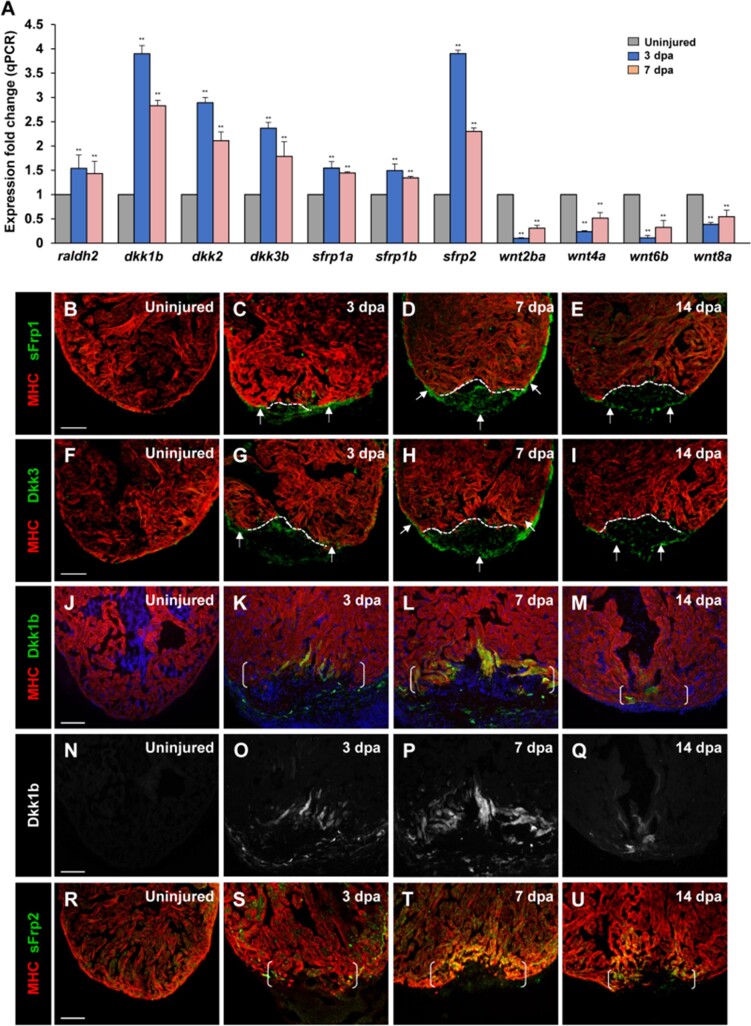
Previous studies reported upregulation of Wnt antagonists Wif1 and Notum1b in the zebrafish adult heart after cardiac injury (Zhao et al., 2019). It remains unknown whether expression of other Wnt antagonists and Wnt ligands is altered during heart regeneration. To understand the roles and mechanisms of Wnt signaling in zebrafish regenerating heart, we examined expression of Wnt antagonists and Wnt ligands following ventricular apex resection on animals at ∼6 months of age. We uncovered induction of multiple Wnt inhibitor genes, including dkk1b, dkk2, and dkk3b, as well as sfrp1a, sfrp1b, and sfrp2, upon ventricular damage (Figure 1A; Supplementary Figure S1A). By contrast, expression levels of wnt ligand genes, including wnt2ba, wnt4a, wnt6b, and wnt8a, were reduced during regeneration (Figure 1A; Supplementary Figure S1B), while the majority of genes we examined were not expressed during regeneration (Supplementary Figure S1B). We next performed immunostaining analyses and found that both sFrp1 and Dkk3 were induced in the regenerating epicardial tissue (Figure 1B–I). At 3 days post amputation (dpa), sFrp1 and Dkk3 expression were detectable in epicardial tissue adjacent to the injury site (Figure 1C and G), whereas uninjured hearts had no sFrp1 or Dkk3 expression in either the epicardium or the myocardium (Figure 1B and F). By 7 dpa, sFrp1 and Dkk3 were detectable in epicardial cells surrounding the ventricle and the wound area (Figure 1D and H). Immunostaining analyses for sFrp1 or Dkk3 in Tg(tcf21:nEGFP) epicardial reporter line validated that sFrp1 or Dkk3 were induced in the tcf21:nEGFP-marked epicardium (Supplementary Figure S2B, B′, D, and D′) compared to uninjured hearts (Supplementary Figure S2A, A′, C, and C′). At 14 dpa, sFrp1 and Dkk3 were mainly confined to epicardial cells that enclosed the regenerate (Figure 1E and I). In situ hybridization (ISH) analyses revealed similar expression patterns of dkk3 and sfrp1 transcripts during heart regeneration (Supplementary Figure S3A‒D; data not shown). In addition to the epicardial induction, we found by fluorescence in situ hybridization (FISH) analyses (Supplementary Figure S4A) that expression of dkk3b or sfrp1 was increased in endocardial cells near the injury site at 7 dpa (Supplementary Figure S4Cand E), while a small number of sfrp1 or dkk3b transcripts were detectable in endocardial cells in uninjured hearts (Supplementary Figure S4Band D). These data indicated that sFrp1 and Dkk3 are epi/endocardial secretory factors induced during heart regeneration. Analyses of a transgenic dkk1b:EGFP reporter line (Kang et al., 2013) indicated dkk1b:EGFP induction at the wounded myocardial cell edge and in nearby non-muscle cells by 3 dpa (Figure 1K and O), when compared to uninjured hearts (Figure 1J and N). dkk1b:EGFP induction peaked at 7 dpa (Figure 1L and P) and was gradually reduced by 14 dpa (Figure 1M and Q). Similarly, sFrp2 expression was enhanced at the apical edge cells of the injured myocardium at 3 dpa (Figure 1S) and peaked at 7 dpa (Figure 1T), compared with uninjured hearts (Figure 1R). By 14 dpa, sFrp2 was mainly restricted to a small number of CMs within the regenerate (Figure 1U).
Figure 1.
Multiple secreted Wnt inhibitors sFrps and Dkks are induced following cardiac injury. (A) Expression levels of wnt inhibitors (dkk1b, dkk2, dkk3b, sfrp1a, sfrp1b, and sfrp2) and wnt ligands (wnt2ba, wnt4a, wnt6b, and wnt8a) during cardiac regeneration. raldh2 was used as a positive control (Kikuchi et al., 2011). Data are mean ± SEM from three biological replicates and three technical replicates. Student’s t-test, **P < 0.01. (B‒U) Confocal images displaying sFrp1, Dkk3, Dkk1b, and sFrp2 expression in uninjured and injured hearts at 3, 7, and 14 dpa. Dotted line indicates amputation line. Brackets indicate amputation plane. Scale bar, 100 µm. (B‒E) Small numbers of epicardial cells adjacent to the injury site induce sFrp1 expression (arrows) at 3 dpa, whereas sFrp1 expression is little detectable in uninjured ventricles. At 7 dpa, sFrp1 expression is expanded throughout the ventricular epicardium and detectable within the regenerate (arrows). By 14 dpa, sFrp1 expression is largely restricted to the epicardial sheet surrounding the regenerate (arrows). (F‒I) Dkk3 is little detectable in uninjured ventricles. At 3 dpa, Dkk3 is expressed in epicardial cells adjacent to the injury region. By 7 dpa, Dkk3 expression is enhanced and expanded in the epicardium enclosing the ventricle and the wound (arrows). Dkk3 expression remains in the epicardial sheet surrounding the regenerate (arrows) by 14 dpa. (J–Q) Whereas Dkk1b expression is not detectable in the uninjured ventricle in Tg(dkk1b:egfp) animals (J and N), some CMs express Dkk1b at the apical edge of the wound at 3 dpa (K and O). Enhanced Dkk1b expression is detectable in the apical edge cells of the regenerating myocardium at 7 dpa (L and P). Dkk1b expression remains in a limited number of CMs within the regenerate at 14 dpa (M and Q). (R‒U) sFrp2 expression is detectable in the wounded heart at 3 dpa, enhanced at the apical cell edge cells of the injured myocardium at 7 dpa, and gradually reduced by 14 dpa. Faint expression of sFrp2 expression is detected in uninjured hearts (R).
We next examined expression of Wnt receptor genes lrp5, lrp6, and kremen1. Reverse transcriptase polymerase chain reaction (RT-PCR) and ISH analyses revealed expression of lrp5, lrp6, or kremen1 in the myocardium before ventricular resection, and their expression was unchanged during regeneration (Supplementary Figures S1A and S3E‒H; data not shown). Among expressed wnt ligand genes, ISH analyses revealed expression of the non-canonical wnt5b in the junctional region between the outflow tract and ventricle, and its expression was apparently unaltered by cardiac injury (Supplementary Figure S3I‒L). Collectively, our findings, along with others, indicate that cardiac injury causes induction and secretion of multiple Wnt antagonists, including Dkk3/sFrp1 from the epicardium/endocardium and Dkk1/sFrp2 in the myocardium, as well as elevation of Notum1b/Wif1 in the endocardium (Zhao et al., 2019), suggesting that Wnt signaling needs to be restrained to enable innate heart regeneration in zebrafish.
Suppression of Wnt signaling enhances injury-induced CM proliferation
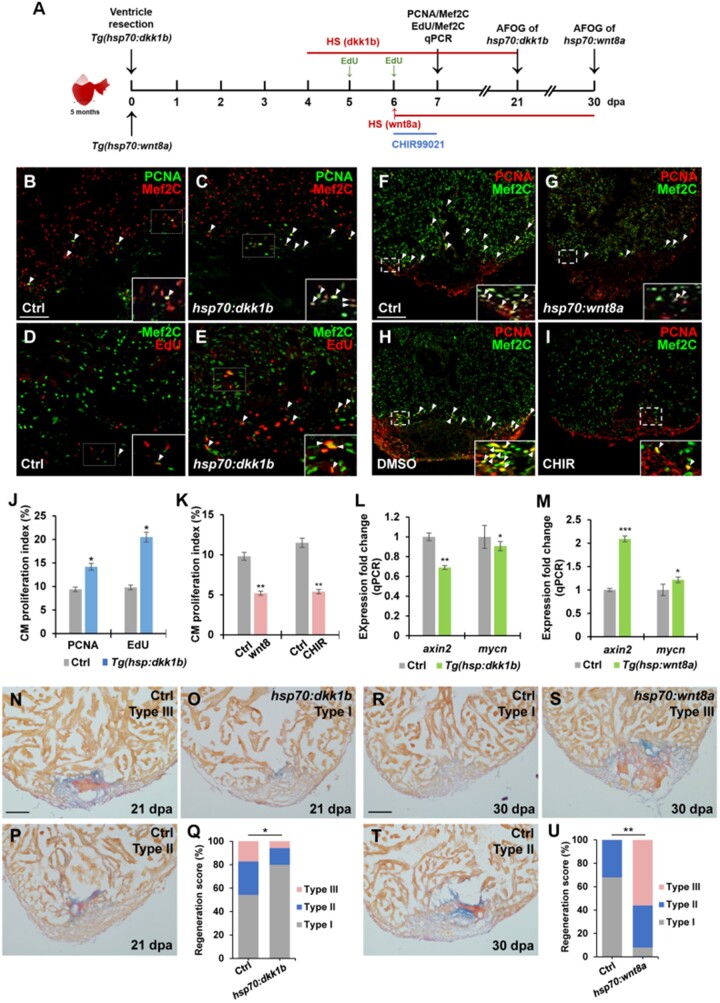
Because of induction of multiple Wnt antagonists throughout the heart following damage, we reasoned that inducible dkk1 overexpression might accelerate CM proliferation and heart regeneration through global suppressing of Wnt signaling. We assessed CM proliferation in Tg(hsp70:dkk1b) animals that enable induced expression of dkk1b by heat shock during heart regeneration (Ueno et al., 2007). We performed ventricular apex resection on control and Tg(hsp70:dkk1b) animals, and exposed them to daily heat shocks from 4 to 6 dpa at the stages when CMs are highly regenerative (Figure 2A). Injured, heat-shocked hearts were collected at 7 dpa, sectioned, and immunostained with antibodies of proliferating cell nuclear antigen (PCNA) and a CM marker Mef2C (Figure 2A). Notably, the CM proliferation index (PCNA+Mef2C+/Mef2C+) in inducible dkk1b-overexpressing hearts was increased by 74.4% over that of control hearts (Figure 2B, C, and J), consistent with previous experiments using small-molecule inhibitors IWR1 and CDMG1 (Zhao et al., 2019; Xie et al., 2020). As an independent approach to assess CM proliferation, we measured nuclear incorporation of 5-ethynyl-2-deoxyuridine (EdU), a marker of DNA synthesis. The percentage of EdU incorporation (EdU+Mef2c+/Mef2c+) in dkk1b-overexpressing hearts was 107% higher than that of control hearts (Figure 2D, E, and J). Moreover, acid fuchsin‒orange G (AFOG) staining revealed that dkk1b-overexpressing injury site contained most cardiac muscle with minimal collagen/scar tissues at 21 dpa (Type I; Figure 2O), while heat-shocked control wounds retained large patches of fibrin and collagen/scar tissues (Type III and Type II; Figure 2N and P). Quantification analyses indicated that higher percentage of hsp:dkk1b hearts displayed contiguous cardiac muscle with minimal collagen/scar tissues (Type I) than that of injured wild-type (WT) sibling hearts in wound regions (Figure 2Q), suggesting the reduction of fibrotic scars and increased cardiac muscle production in dkk1b-overexpressing hearts.
Figure 2.
Suppression of Wnt signaling enhances heart regeneration by increasing CM proliferation. (A) Schematic of heat shock experiments and Wnt agonist treatment for CM proliferation, qPCR, and AFOG analyses. Heat shock (HS): 37°C for 2 h for control and Tg(hsp70:dkk1b) animals at 4, 5, and 6 dpa for PCNA/Mef2C assay or over the time period from 4 to 21 dpa for AFOG analysis; 37°C for 4 h for control and Tg(hsp70:wnt8a) animals at 6 dpa for PCNA/Mef2C assay or over the time period from 6 to 30 dpa for AFOG analysis. EdU: injection at 5 and 6 dpa. CHIR99021: 20 µM (in DMSO) from 6 to 7 dpa. Control: 0.1% DMSO from 6 to 7 dpa. (B‒I) Confocal microscopy analyses for CM proliferation. Insets show high-magnification images in rectangles. Scale bar, 100 µm. Data are mean ± SEM from five hearts each. Student’s t-test, *P < 0.05, **P < 0.01. (B‒E and J) Confocal images of PCNA+Mef2C+ (B and C) and EdU+Mef2C+ (D and E) cells (arrowheads) in heat-shocked WT (Ctrl) and Tg(hsp70:dkk1b) hearts following resection and PCNA- or EdU-labeled CM proliferation index (J). (F‒I and K) Confocal images of PCNA+Mef2C+ cells (arrowheads) in heat-shocked Ctrl (F) and Tg(hsp70:wnt8a) (G) hearts after amputation or 0.1% DMSO-treated (H) and CHIR99021-treated (I) WT hearts following resection and PCNA-labeled CM proliferation index (K). (L and M) qPCR analysis for relative expression levels of axin2 and mycn in wounded ventricles of Ctrl and Tg(hsp70:dkk1b) (L) or Tg(hsp70:wnt8a) (M) animals. β-actin expression was used for normalization. Data are mean ± SEM from three biological replicates and three technical replicates. Student’s t-test, *P < 0.05, **P < 0.01, ***P < 0.001. (N‒U) Representative images of AFOG-stained ventricle sections and regeneration scores. (N‒P and Q) Heat-shocked Ctrl and Tg(hsp70:dkk1b) animals at 21 dpa. (R‒T and U) Heat-shocked Ctrl and Tg(hsp70:wnt8a) animals at 30 dpa. Orange: muscle; blue: collagen; red: fibrin. Scale bar, 100 µm. Fisher’s exact test, *P < 0.05, **P < 0.01.
Wnt pathway activity is observed in the injured heart using T-cell factor (TCF)-mediated Wnt transgenic reporters (Stoick-Cooper et al., 2007; Zhao et al., 2019). We analyzed the endogenous Wnt signaling activity from independent approach using RNAscope analyses of a Wnt target axin2 that is also a negative regulator of Wnt signaling activity (Behrens et al., 1998). We corroborated previous reports and learned that axin2 transcript was detectable at low levels in the injured heart and slightly enriched in the wound region (Supplementary Figure S5Aand B). We next assessed expression of axin2 and another Wnt target gene mycn in heat-shocked Tg(hsp70:dkk1b) animals and noted a reduction in expression of axin2 and mycn in dkk1b-overexpressing hearts from quantitative PCR (qPCR) and RNAscope analyses (Figure 2L; Supplementary Figure S5Eand F; data not shown), indicating suppression of the Wnt signaling activity in heat-shocked Tg(hsp70:dkk1b) animals. Thus, inhibition of Wnt signaling by dkk1b overexpression at early injury stages enhances injury-induced CM proliferation, suggesting that Wnt signaling activity restrains CM proliferation during heart regeneration.
Most wnt legend genes including wnt8a were expressed at very low levels in the adult heart, some of which were even reduced following injury (Figure 1A; Supplementary Figure S1B). To assess the effects of Wnt signaling activation on CM proliferation during regeneration, we employed Tg(hsp70:wnt8a) animals, in which wnt8a is overexpressed by heat shock treatment (Ueno et al., 2007). We resected the ventricular apices of control and Tg(hsp70:wnt8a) animals, exposed them to a single heat shock at 6 dpa, and collected hearts at 7 dpa for immunostained with PCNA/Mef2C antibodies (Figure 2A). Remarkably, ectopic wnt8a expression following a single heat shock significantly reduced CM proliferation index by 47% compared to control hearts (Figure 2F, G, and K). This result is consistent with the observation that higher percentages of transgenic animal overexpressing wnt8a for 30 days after ventricle resection retained prominent fibrin and collagen/scar tissues (Type III and Type II, Figure 2S‒U), whereas control animals given heat shock replaced most fibrin and collagen/scar tissues with CMs at 30 dpa (Type I, Figure 2R and U). Next, we treated injured animals with CHIR99021, a small-molecule inhibitor of GSK-3β for specifically activating Wnt pathway (Ring et al., 2003), compared with a non-specific Wnt agonist BIO, which was utilized recently to reduce heart regeneration in zebrafish (Zhao et al., 2019). Amputated hearts exposed to 20 µM CHIR99021 for 24 h had a marked reduction of 48% in CM proliferation compared to vehicle-treated control hearts (Figure 2A, H, I, and K). RNAscope and qPCR analyses validated an increase in axin2 and mycn expression in heat-shocked Tg(hsp70:wnt8a) hearts or CHIR-treated WT fish (Figure 2M; Supplementary Figure S5Cand D; data not shown). Collectively, these findings indicate that ectopic Wnt8a signaling activation blunts the proliferative response of CMs to injury, suggesting that suppression of Wnt pathway activity is required for CM proliferation during regeneration.
Association of cytoplasmic pS675-β-catenin with disassembled sarcomeres and CM dedifferentiation following cardiac damage
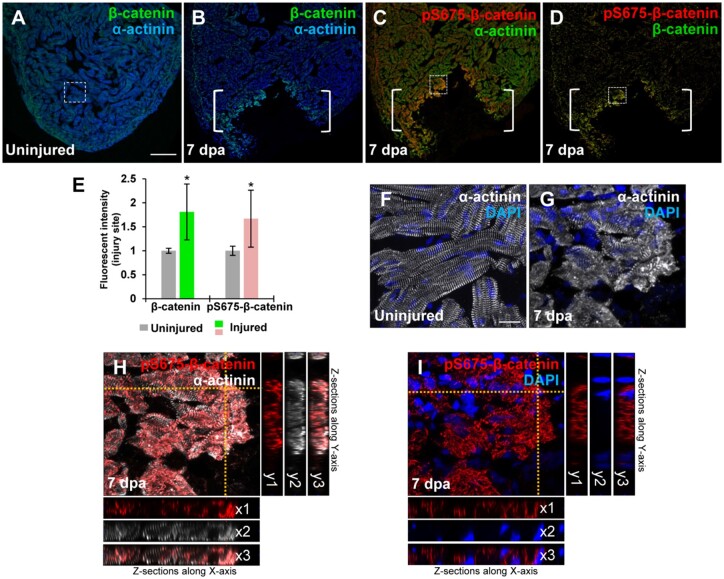
In heart regeneration contexts, proliferating CMs undergo dedifferentiation, characterized by reactivation of cardiac developmental programs and sarcomere disassembly (Jopling et al., 2010; D'Uva et al., 2015; Gemberling et al., 2015). We assessed β-catenin expression and its relationship to CM dedifferentiation in the context of Wnt inhibition following cardiac injury. β-catenin, a key downstream effector of Wnt pathway, is localized in different cellular compartments in CMs, where it executes various cellular functions (Nelson and Nusse, 2004; Faux et al., 2010). In the adult zebrafish heart, the majority of β-catenin resides in intercalated disks and cytoplasmic sarcomeres, overlapping with a Z-disk marker α-actinin (Figure 3A; Yang and Xu, 2012), which executes differential cellular functions independent of Wnt/GSK3β-mediated regulation (Valenta et al., 2012). A limited nuclear β-catenin was also detectable in adult CMs (Xie et al., 2020). Following cardiac amputation, we observed that cytoplasmic β-catenin affiliated with sarcomeres was unexpectedly increased at the injury region (Figure 3B and E). We found that the majority of elevated β-catenin was specifically recognized by Ser 675-phosphorylated β-catenin (pS675-β-catenin) antibody (Figure 3C and D), but not pS33/S37/T41-β-catenin antibody (data not shown), which reflects a stable modification form that is independent of GSK3β activity (Hino et al., 2005; Taurin et al., 2006). In uninjured hearts, myofibrils organized in regular sarcomere units exhibiting cross-striations revealed by α-actinin (Figure 3F). Following cardiac injury, myocardial sarcomeres became conspicuously disassembled in the injured edge, indicative of CM dedifferentiation (Figure 3G). We observed that pS675-β-catenin was elevated and co-localized with disassembled sarcomeres (Figure 3H) with a little nuclear detection (Figure 3I), suggestive of association of pS675-β-catenin with dedifferentiated CMs.
Figure 3.
Induction of pS675-β-catenin at disassembled sarcomeres in the injured myocardium following cardiac damage. (A) In uninjured hearts, β-catenin is detectable throughout the myocardium stained with a sarcomeric Z-disk marker α-actinin. (B‒D) Following ventricular resection, β-catenin (B) and pS675-β-catenin (C) are induced at the apical cell edge of wounded myocardia. (D) Merged panel of B and C without α-actinin. Brackets, amputation area. (E) Bar chart depicting β-catenin and pS675-β-catenin levels following ventricular resection. Fluorescent intensities were measured at the injury border zone using Image J. β-catenin or pS675-β-catenin levels in control hearts are normalized as 1. Data are mean ± SEM from five hearts for each group. Student’s t-test, *P < 0.05. (F) High-magnification image of the dash-lined area in A displaying CMs in organized sarcomeric arrays in uninjured hearts. (G‒I) High-magnification images of the dash-lined window in C exhibiting the co-localization of pS675-β-catenin (H and I) with disassembled sarcomeres (G and H) in the injured myocardial cell edge. (I) Merged panel of G and H without α-actinin. 3D analyses display co-localizations of pS675-β-catenin (red) with dissociated sarcomere components (white) along the X-axis (H, x1‒x3) and the Y-axis (H, y1‒y3), as well as the cytoplasmic localization of pS675-β-catenin in Z sections (I, x1‒x3 and y1‒y3). Scale bar, 100 µm (A‒D) and 10 µm (F‒I).
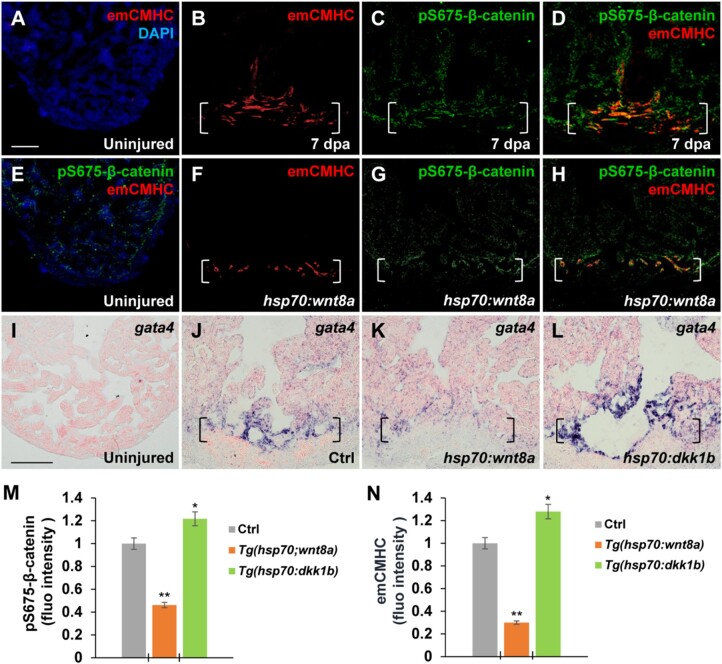
We next assessed how the elevation of cytoplasmic pS675-β-catenin relates to CM dedifferentiation and Wnt signaling inhibition. One of hallmarks of CM dedifferentiation is re-expression of cardiac embryonic or fetal genes upon injury, such as cardiac transcription factor gata4 and embryonic-specific cardiac myosin heavy chains (emCMHC) (Kikuchi et al., 2010; Kubin et al., 2011; D'Uva et al., 2015). While expression of emCMHC, labeled by N2.261 antibodies, was not detectable in uninjured hearts (Figure 4A and E; Sallin et al., 2015; Wu et al., 2016), emCMHC was induced at the apical cell edge of the wounded myocardium (Figure 4B). Notably, the induced emCMHC mostly overlapped with upregulated pS675-β-catenin at the myocardial wound edge (Figure 4C and D), consistent with the association of pS675-β-catenin with disassembled sarcomeres (Figure 3H and I). When wnt8a was experimentally induced by heat shock in Tg(hsp70:wnt8a) animals following ventricular resection, both levels of emCMHC and pS675-β-catenin were reduced (Figure 4F‒H, M, and N). In addition, expression of gata4, a key cardiac transcriptional factor (Kikuchi et al., 2010; Gupta et al., 2013), was reduced at injured myocardial cell edges in heat-shocked Tg(hsp70:wnt8a) hearts compared to control hearts (Figure 4J and K). On the contrary, inhibition of Wnt signaling via dkk1b overexpression in heat-shocked Tg(hsp70:dkk1b) animals following resection caused an increase in expression of gata4 (Figure 4J and L) as well as emCMHC and pS675-β-catenin (Figure 4M and N), suggesting that Wnt inhibition favors an immature cell state and potentially supports CM dedifferentiation following cardiac injury.
Figure 4.
Wnt signaling inhibition associates with CM dedifferentiation during heart regeneration. (A‒H) Immunofluorescence analyses showing increased emCMHC stained with N2.261 antibody and pS675-β-catenin at apical myocardial cells of the wound at 7 dpa (A‒D), which was reduced by wnt8a activation in injured Tg(hsp70:wnt8a) hearts (E‒H). N2.261 is not detectable in uninjured hearts (A and E) and overlaps mostly with induced pS675-β-catenin (D). (D and H) Merged panels of B and C and F and G, respectively. (I‒L) ISH analyses displaying gata4 expression at injured myocardial cell edges in heat-shocked Ctrl hearts (J) at 7 dpa, which was reduced in Tg(hsp:wnt8a) hearts (K) and increased in Tg(hsp:dkk1b) hearts (L) but hardly detectable in uninjured hearts (I). Brackets indicate amputation planes. Scale bar, 100 µm. (M and N) Bar charts depicting pS675-β-catenin (M) and emCMHC (N) levels in Ctrl (normalized as 1), Tg(hsp:wnt8a), and Tg(hsp:dkk1b) hearts. Fluorescent intensities were measured at the injury border zone using Image J. Data are mean ± SEM from five hearts for each group. Student’s t-test, *P < 0.05, **P < 0.01.
Induction of Pak2 kinase increases β-catenin Ser 675 phosphorylation and stability during heart regeneration
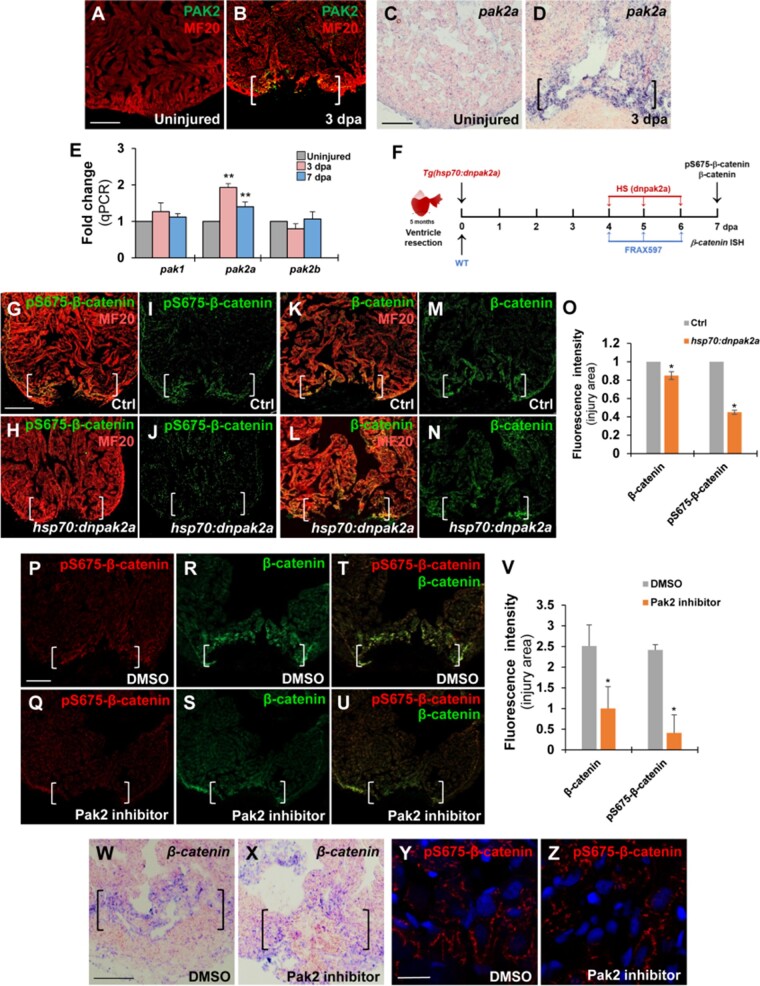
We previously reported that Pak2 plays positive roles for zebrafish heart regeneration using pharmacological approaches (Peng et al., 2016). In addition, multiple studies demonstrated interactions between PAK1 and Wnt signaling in cancer progression or during development (Goh et al., 2012; Zhu et al., 2012; Choe and Crump, 2015). In proliferative tumor cells, PAK1 phosphorylates β-catenin at the Ser 675 residue, enhancing its stability in cytosols and nuclei (Zhu et al., 2012). Thus, we assessed the possibility that Pak2 phosphorylates β-catenin in regenerating hearts. Zebrafish genome contains three pak genes, including pak1, pak2a, and pak2b. Immunostaining analyses against Pak2 indicated that Pak2 was induced at injury sites mainly in myocardial cells, some of which were also detectable in other cell types following cardiac injury (Figure 5A and B). Our ISH and qPCR analyses also revealed an increase of pak2a transcript, but not pak1 and pak2b RNAs, at the apical cell edge in injured zebrafish hearts (Figure 5C–E). To test whether Pak2 induces β-catenin Ser 675 phosphorylation following cardiac injury, we generated inducible dominant-negative Pak2a (dnPak2a) transgenic animals, in which dnPak2a mutant containing a kinase inhibitory domain and a dimerization domain was driven by the heat shock promoter (hsp70). dnPak2a acts in a dominant-negative manner by directly occupying the cleft of kinase domain that inhibits its kinase activation (Chu et al., 2004). We performed ventricular resection on control and Tg(hsp70:dnpak2a) animals, subjected them to daily heat shocks from 4 to 6 dpa, and then collected resected hearts for pS675-β-catenin and MF20 double immunostaining (Figure 5F). We found that inhibiting Pak2a activity using the dnpak2a mutant almost abolished the elevated pS675-β-catenin at the injured myocardial cell edge compared to control hearts (Figure 5G‒J and O), while total β-catenin levels were reduced to some extents (Figure 5K‒N and O), suggestive of the reduced β-catenin stability.
Figure 5.
Inhibiting Pak2 activity using dominant-negative mutation or chemical inhibitor reduces pS675-β-catenin at wound edges in the regenerating heart. (A‒D) Immunostaining and ISH analyses displaying increased Pak2 and pak2a expression, respectively, at wounded myocardial cell edge in injured hearts at 3 dpa (B and D) compared to uninjured heart (A and C). Scale bar, 100 µm. (E) qPCR analysis for expression levels of pak1, pak2a, and pak2b in uninjured and resected WT hearts at 3 or 7 dpa. β-actin expression was used for normalization. Data are mean ± SEM from three biological replicates and three technical replicates. Student’s t-test, **P < 0.01. (F) Schematic showing heat shock experiment of Tg(hsp70:dnpak2a) and FRAX597 treatment for β-catenin and pS675-β-catenin assessment. Heat shock: 37°C for 2 h daily from 4 to 6 dpa. FRAX597: 1 µM (in DMSO) from 4 to 6 dpa. Control: 0.1% DMSO from 4 to 6 dpa. (G‒V) Confocal microscopy analyses for the levels of pS675-β-catenin and total β-catenin at wounded myocardial cell edges in hearts at 7 dpa. Scale bar, 100 µm. Data are mean ± SEM from five hearts for each group. Student’s t-test, *P < 0.05. (G‒O) pS675-β-catenin was diminished whereas total β-catenin was slight reduced in Tg(hsp:dnpak2a) hearts at 7 dpa. (P‒V) pS675-β-catenin was diminished and total β-catenin was reduced in the Pak2a inhibitor FRAX597-treated hearts at 7 dpa compared to 0.1% DMSO-treated control hearts. (T and U) Merged panels of P and R and Q and S, respectively. (W and X) ISH analysis showing the same β-catenin expression level in FRAX597-treated hearts (X) compared to DMSO-treated hearts (W). (Y and Z) Immunostaining analyses revealing a reduction of cytoplasmic pS675-β-catenin in FRAX597-treated hearts (Z) compared to DMSO-treated hearts (Y) at 7 dpa. Scale bar, 10 µm.
As an independent approach to assess pS675-β-catenin, we administrated injured WT hearts using a Pak2 inhibitor FRAX597 (Figure 5F), which specifically targets the PAK ATP-binding cavity and inhibits its kinase activity (Licciulli et al., 2013). In agreement with our dnPak2a experiments, FRAX597 treatment caused a diminishment of pS675-β-catenin (Figure 5P, Q, T, U, and V) but also a reduction of total β-catenin at the wound myocardial cell edge (Figure 5R‒V). In contrast, β-catenin mRNAs were not altered in the myocardial wound edge in FRAX597-treated hearts compared to DMSO controls (Figure 5W and X). Importantly, we observed that the majority of pS675-β-catenin remained in cytosols without going to nuclei in FRAX597- or vehicle-treated hearts at 7 dpa (Figure 5Y and Z). However, human pS675-β-catenin controlled by PAK1 goes to the nucleus in colon cancer cells (Zhu et al., 2012), suggesting the distinct mechanisms underlying differentiated CMs from proliferative tumor cells. Taken together, these findings demonstrate that Pak2a kinase increases β-catenin Ser 675 phosphorylation and stability at myocardial wounds during regeneration.
Myocardial-specific induction of a constitutively active β-catenin (S675E) promotes sarcomere disassembly and CM dedifferentiation
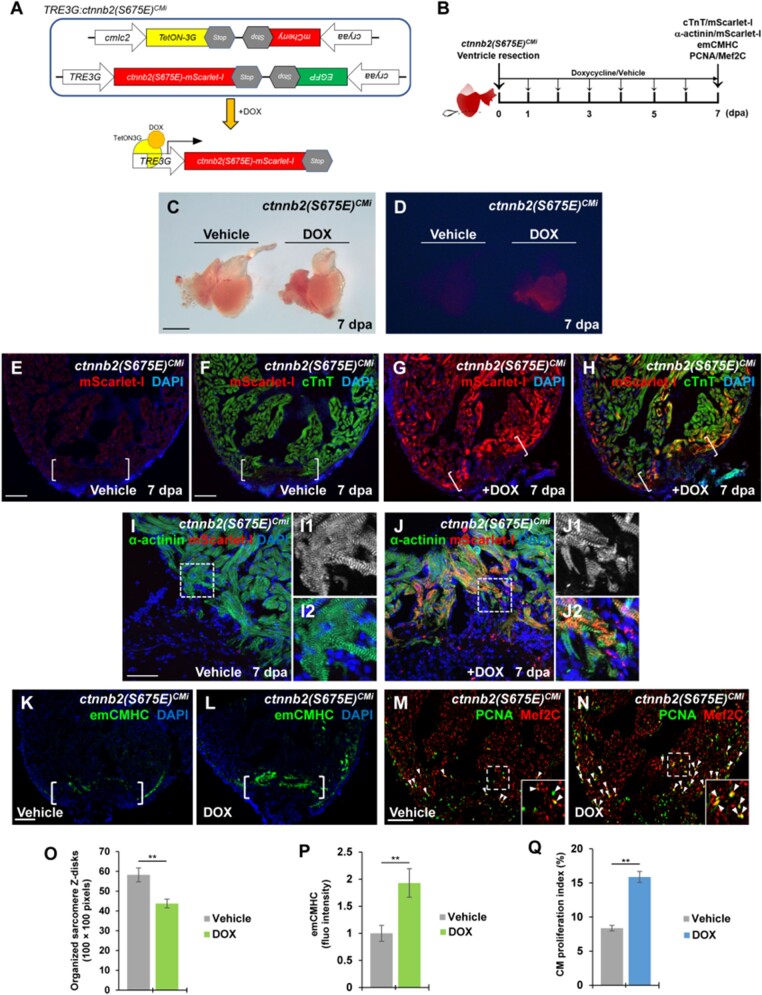
To test whether induction of β-catenin Ser 675 phosphorylation promotes CM dedifferentiation and proliferation, we converted Ser 675 to a phospho-mimetic Glu 675 (E) in Ctnnb2 (β-catenin gene) that enables constitutive phosphorylation at the residue 675. Inducible transgenic zebrafish, Tg(TRE3G:ctnnb2(S675E)-mScarlet-I, cryaa:EGFP) bearing a phospho-mimetic form of β-catenin (S675E) fused with mScarlet-I, were constructed based on a doxycycline (DOX)-inducible TetON3G system (Figure 6A). By crossing with myocardial activator line Tg(cmlc2:TetON-3G, cryaa:mCherry), we generated a double transgenic line, Tg(cmlc2:TetON-3G, cryaa:mCherry; TRE3G:ctnnb2(S675E)-mScarlet-I, cryaa:EGFP), referred to as TRE3G:ctnnb2(S675E)CMi, which enables DOX-inducible expression of β-catenin (S675E) in CMs (Figure 6A). In TRE3G:ctnnb2(S675E)CMi transgenic line, mCherry and EGFP were co-expressed in lens under the cryaa promoter and used as markers for identifying transgene in our initial screen (Supplementary Figure S6A‒D). To test whether the inducible system works, we administrated DOX or vehicle for 7 days following ventricular resection (Figure 6B). Injured hearts were isolated and subjected to immunostaining of mScarlet-I antibody that labels Ctnnb2 (S675E) and cTnT antibody that marks CMs (Figure 6B). We found that DOX-treated TRE3G:ctnnb2(S675E)CMi animals displayed red fluorescence in isolated hearts (Figure 6C and D) as well as Ctnnb2-mScarlet-I expression overlapping with cTnT adjacent to cardiac injury regions (Figure 6G and H), whereas vehicle-treated TRE3G:ctnnb2(S675E)CMi zebrafish showed absence of red fluorescence in isolated hearts (Figure 6C and D) or Ctnnb2-mScarlet-I expression in cTnT-marked hearts (Figure 6E and F).
Figure 6.
Induction of phospho-mimetic β-catenin (S675E) in adult CMs enhances sarcomere disassembly and CM dedifferentiation. (A) Schematic diagram of double transgenes of TRE3G:ctnnb2(S675E)CMi. DOX treatment allows binding of TetON-3G to the TRE3G promoter, enabling the tissue-specific transcription of ctnnb2(S675E) in CMs. (B) Experimental strategy to induce myocardial ctnnb2(S675E) overexpression during the course of heart regeneration. Ctnnb2(S675E) induction (mScarlet-I/cTnT), sarcomere disassembly (α-actinin/mScarlet-1), CM dedifferentiation (emCMHC), and proliferation (PCNA/Mef2C) were examined at 7 dpa in vehicle-treated and TOX-treated TRE3G:ctnnb2(S675E)CMi animals. (C and D) Brightfield (C) or red fluorescence (D) images of whole-mount hearts. mScarlet-I signal was only detected in myocardium of DOX-treated hearts. Scale bar, 500 µm. (E‒H) Representative confocal fluorescence images of cardiac sections immunostained for cTnT (green) and mScarlet-I (red). Scale bar, 100 µm. (I, J, and O) Myocardial wound edge regions in heart sections were co-stained for α-actinin (green) and mScarlet-I (red). Scale bar, 50 µm. (I1, I2, J1, and J2) Magnified panels show high-magnification images of the boxed regions in I and J. (O) Quantification of organized sarcomere Z-disks in α-actinin-marked CMs (100 × 100 pixels). (K‒N, P, and Q) Confocal image analyses of emCMHC expression (K and L) and PCNA+Mef2C+ cells (arrowheads, M and N). Brackets indicate amputation planes. Insets show high-magnification images of PCNA+Mef2C+ cells in boxed regions. (P) Fluorescent intensities depicting emCMHC levels were measured at the injury border zone using Image J. emCMHC levels in vehicle-treated hearts are normalized as 1. (Q) PCNA-labeled CM proliferation indices. Scale bar, 100 µm. For bar chart analyses in O‒Q, data are mean ± SEM from five hearts for each group. Student’s t-test, **P < 0.01.
Next, we assessed CM dedifferentiation status in DOX-treated and vehicle-treated TRE3G:ctnnb2(S675E)CMi animals following ventricle resection. Injured hearts were immunostained with mScarlet-I antibody and the sarcomeric Z-disk marker α-actinin antibody for sarcomere disarray analyses or the dedifferentiation marker emCMHC antibody for embryonic/fetal gene analyses (Figure 6B). DOX-treated TRE3G:ctnnb2(S675E)CMi CMs in the wound edge showed higher degree of sarcomere structure disruption (Figure 6I, I1, I2, J, J1, J2, and O). Ctnnb2 (S675E) expression was detected to overlap with α-actinin on sarcomeres (Figure 6J and J2), consistent with previous experiments (Figure 3H and I). We found that emCMHC was markedly induced in DOX-treated TRE3G:ctnnb2(S675E)CMi injured hearts at 7 dpa compared to vehicle-treated hearts (Figure 6K, L, and P), ndicating the enhancement of sarcomere disassembly and CM dedifferentiation by the phospho-mimetic β-catenin (S675E). Lastly, we tested whether induction of constitutive β-catenin (S675E) phosphorylation in adult CMs promotes CM proliferation using PCNA/Mef2 immunostaining analysis (Figure 6B). Consistent with the dedifferentiation effect of β-catenin (S675E), CM proliferation indices (PCNA+Mef2+/Mef2+) in DOX-treated TRE3G:ctnnb2(S675E)CMi zebrafish were increased by 89.6% over that of vehicle-treated animals (Figure 6M, N, and Q). Taken together, these findings indicate that myocardial-specific induction of constitutive β-catenin (S675E) phosphorylation is sufficient to enhance CM dedifferentiation, which in turn accelerates CM proliferation in response to cardiac damage.
Pak2 kinase is required for heart regeneration by supporting CM dedifferentiation and proliferation
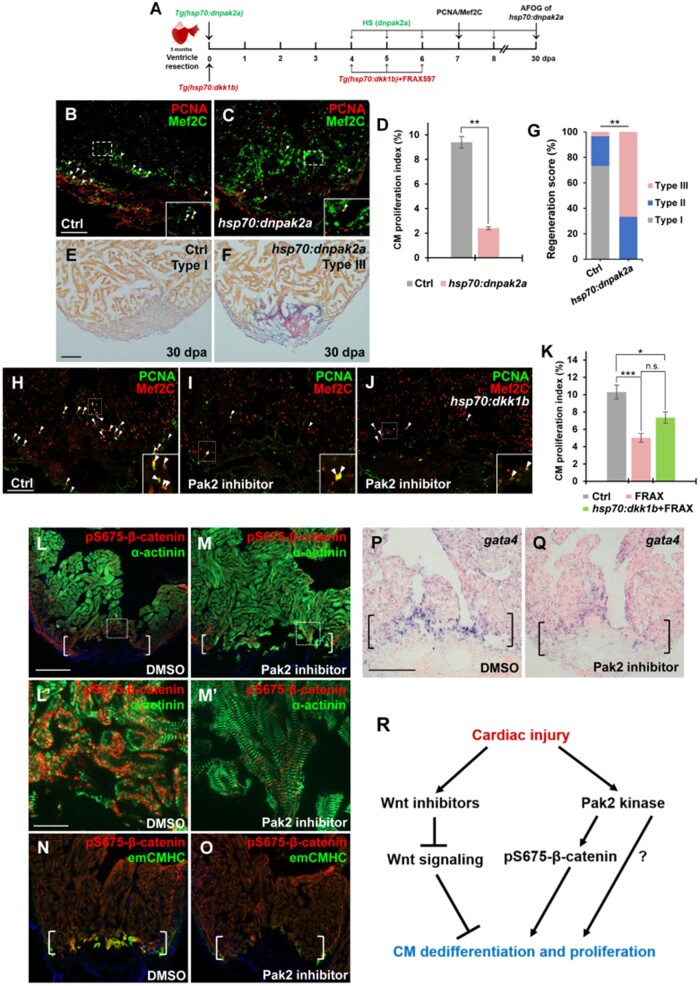
To determine whether a loss of Pak2 kinase activity impairs CM proliferation following cardiac damage, control and Tg(hsp70:dnpak2a) animals were subjected to ventricular apex resection and daily heat shocks from 4 to 6 dpa. Cardiac sections were immunostained with PCNA/Mef2 antibodies for CM proliferation analyses at 7 dpa (Figure 7A). We found that inhibiting Pak2 activity in dnpak2a mutants caused a reduction of CM proliferation by 74.5% compared to control hearts (Figure 7B‒D). We next conducted AFOG analyses and found that higher percentages of dnPak2a hearts displayed prominent fibrin and collagen/scar tissues (Type III) after ventricular resection at 30 dpa (Figure 7E‒G). These data together indicate that Pak2 kinase activity is required for cardiac muscle regeneration and wound healing. We next tested whether inhibiting Pak2 kinase activity attenuated CM proliferation in Wnt antagonist dkk1b-overexpressing hearts after injury. Injured control and Tg(hsp70:dkk1b) animals were treated with FRAX597 from 4 to 6 dpa and wound edge CM proliferation was analyzed at 7 dpa (Figure 7A). We found that suppression of Pak2 activity can markedly reduce CM proliferation in dkk1b-overexpressing hearts to the same extent as FRAX-treated WT hearts (Figure 7H‒K), suggesting that Pak2 acts synergistically with Wnt antagonists to regulate myocardial regeneration.
Figure 7.
Inhibition of Pak2 activity impairs CM dedifferentiation and proliferation. (A) Schematic of heat shock experiments and FRAX597 treatment for PCNA/Mef2C analyses. Heat shock: 37°C for 2 h daily from 4 to 6 dpa for PCNA/Mef2C assay or over the time period from 4 to 30 dpa for AFOG analysis; treatment: 1 µM FRAX597 or 0.1% DMSO from 4 to 6 dpa. (B‒D) Confocal image analyses displaying PCNA+Mef2C+ cells (arrowheads, B and C) and PCNA-labeled CM proliferation index (D) in Ctrl and Tg(hsp:dnpak2a) hearts at 7 dpa. Insets indicate high-magnification images in dash-lined areas. Data are mean ± SEM from five hearts for each group. Student’s t-test, **P < 0.01. (E‒G) Representative images (E and F) and quantification of regeneration scores (G) of ventricle sections from heat-shocked Ctrl and Tg(hsp70:dnpak2a) animals at 30 dpa and stained with AFOG. Orange: muscle; blue: collagen; red: fibrin. Fisher’s exact test. **P < 0.01. (H‒K) Confocal image analyses displaying PCNA+Mef2C+ cells (arrowheads) and quantification of CM proliferation indices (K) in DMSO-treated (H, Ctrl), Pak2 inhibitor (FRAX597)-treated (I), and FRAX597-treated Tg(hsp:dkk1b) (J) hearts at 7 dpa. Insets indicate high-magnification images of dash-lined area. Data are represented as mean ± SEM from five hearts for each group. Statistical significance was calculated using one-way ANOVA followed by Tukey’s test. n.s., none significance; *P < 0.05, ***P < 0.001. (L‒O) Immunostaining analyses displaying reduction of pS675-β-catenin (L and M) and emCMHC (N and O) at apical myocardial cells of the wounded edge in FRAX597-treated hearts compared to DMSO-treated hearts at 7 dpa. (L′ and M′) High-magnification images of dash-lined windows in L and M, respectively, exhibiting disassembled sarcomeres in DMSO-treated hearts (L′) and relatively normal striated sarcomeres in FRAX597-treated hearts (M′). (P and Q) ISH analysis displaying a reduction of gata4 at the wounded myocardial cell edge in FRAX597-treated hearts compared to DMSO-treated hearts at 7 dpa. Brackets indicate amputation planes. Scale bar, 100 µm (except 10 µm in L′ and M′). (R) A working model for Wnt/Pak2/pS675-β-catenin signaling events regulating heart regeneration. Cardiac injury induces multiple Wnt antagonists that restrain Wnt signaling throughout the wounded heart, enabling CM dedifferentiation and proliferation. In the context of Wnt signaling inhibition, Pak2 is upregulated in cardiac wounds, where it phosphorylates cytoplasmic β-catenin at the Ser 675 residue and increases its stability in disassembled sarcomeres, enhancing CM renewal and heart regeneration.
Because Pak2 regulates pS675-β-catenin during regeneration, we assessed whether zebrafish Pak2 directly phosphorylates β-catenin at Ser 675 in vitro. We created the zebrafish β-catenin-S675A (serine to alanine) mutant, which mimics unphosphorylated forms. Pak2a tagged by HA was immunoprecipitated and subjected to kinase assays using purified proteins of glutathione S-transferase (GST)-tagged β-catenin-WT or GST-tagged β-catenin-S675A. In the presence of ATP, co-incubation of β-catenin-WT with Pak2a caused a marked increase in Ser 675 phosphorylation, whereas this phosphorylation was completely abrogated by β-catenin S675A substitution (Supplementary Figure S7), consistent with the specific kinase activity in human PAK1 (Zhu et al., 2012). Importantly, treatment of a specific Pak2 inhibitor FRAX597 or FRAX486 caused a reduction of β-catenin Ser 675 phosphorylation (Supplementary Figure S7). We noticed that pS675-β-catenin rather than unphosphorylated β-catenin can be recognized using a monoclonal antibody (D2F1; Supplementary Figure S7), indicating that D2F1 antibody specifically recognizes zebrafish pS675-β-catenin. Thus, the in vitro findings that zebrafish Pak2a phosphorylates β-catenin at the Ser 675 residue are consistent with our in vivo results in the setting of heart regeneration.
As induction of the phospho-mimetic β-catenin (S675E) promotes CM dedifferentiation during regeneration (Figure 6), we reasoned that Pak2-mediated pS675-β-catenin upregulation might be involved in CM dedifferentiation. To test this idea, we administrated FRAX597 to animals with resected hearts and collected tissue for immunostaining with pS675-β-catenin and α-actinin antibodies. Consistent with previous experiments (Figure 3H and I), injured hearts exhibited an increase of pS675-β-catenin (Figure 7L) at disassembled sarcomeres (Figure 7L′; Supplementary Figure S8A) in the myocardial wound edge, whereas inhibition of Pak2a kinase activity by FRAX597 treatment caused a marked reduction of pS675-β-catenin in this tissue with reduced sarcomere disassembly (Figure 7M and M′; Supplementary Figure S8B). emCMHC+pS675-β-catenin+ heart muscle at the injury site was also conspicuously reduced in FRAX597-treated hearts, compared to control hearts (Figure 7N and O; Supplementary Figure S8C‒F). Accordingly, we observed the reduced expression of the transcriptional progenitor marker gata4 and other cardiac fetal gene markers α-smooth muscle actin (α-SMA) and smooth muscle actinin (actn1) (Kubin et al., 2011; D'Uva et al., 2015) at injured myocardial edges in FRAX597-treated hearts, compared to control hearts (Figure 7P and Q; Supplementary Figure S8G‒J). These findings indicate that Pak2-mediated pS675-β-catenin upregulation contributes to CM dedifferentiation.
Discussion
In this study, we provide evidence that coordination of Wnt signaling inhibition and Pak2/pS675-β-catenin signaling enhances zebrafish heart regeneration. Based on our findings and others, we propose a model for Wnt/Pak2 signaling events in regulating heart regeneration (Figure 7R). Cardiac injury triggers induction of multiple secreted Wnt antagonists at cardiac wounds, including epi/endocardial Dkk3/sFrp1, myocardial Dkk1/sFrp2, and endocardial Wif1/Notum1b (Zhao et al., 2019). This type of multimodal repressive mechanism indicates that Wnt signaling must be dampened to enable natural heart regeneration. In the context of Wnt signaling inhibition, Pak2 kinase is induced in injured heart tissues, where it phosphorylates sarcomere β-catenin at Ser 675 and increases its stability in disassembled sarcomeres, promoting CM dedifferentiation and proliferation. How Pak2 is induced in the context of Wnt signaling inhibition remains unknown. It would be likely that non-canonical Wnt signaling might be upregulated after injury to influence Pak2 expression. Nonetheless, these findings demonstrate that zebrafish heart regeneration requires Wnt signaling inhibition and Pak2-mediated pS675-β-catenin signaling, both of which function coordinately to CM dedifferentiation and proliferation.
The influence of Wnt signaling on organ regeneration appears to be context dependent. While studies demonstrate Wnt signaling as a positive regulator of regeneration in myriad vertebrate tissues and organs, including teleost fish fins, mammalian blood, and intestinal epithelium (Kawakami et al., 2006; Barker et al., 2007; Stoick-Cooper et al., 2007; Congdon et al., 2008; Choi et al., 2013; Kang et al., 2013), our findings uncover that regulated inhibition of Wnt signaling contributes to regeneration of zebrafish hearts. Heart regeneration occurs via stem cell-independent mechanisms (Jopling et al., 2010; Kikuchi et al., 2010; Senyo et al., 2013; D'Uva et al., 2015). Inhibition of Wnt signaling favors dedifferentiation of pre-existing CMs followed by proliferation and redifferentiation during heart regeneration. In this study, multiple Wnt antagonists, including Dkk1, Dkk3, sFrp1, and sFrp2, were found to upregulate following injury. We focused on Dkk1 rather than other Wnt inhibitors, because Dkk1 is the typical antagonist of canonical Wnt signaling, which exerts inhibitory functions by binding to LRP6 of the receptor complex (Stoick-Cooper et al., 2007; Moro et al., 2012). Moreover, Dkk1 is induced only in the injured myocardium, whereas other Wnt antagonists are upregulated in the epicardium and endocardium, indicating the cell-autonomous role of Dkk1 for cardiac muscle regeneration. We have demonstrated that dkk1b overexpression in Tg(hsp70:dkk1b) zebrafish enhances CM dedifferentiation and proliferation through inhibiting canonical Wnt target genes axin2 and mycn (Figure 2L; Supplementary Figure S5Eand F). It will be important to test whether and how other Wnt antagonists such as Dkk3/sFrp1 modulate Wnt signaling during heart regeneration and to assess how Dkk3/sFrp1 secreted from epi/endocardial cells influence CM renewal and wound healing.
We find that pS675-β-catenin regulated by Pak2 is markedly increased in the injured myocardium during cardiac regeneration. The majority of upregulated pS675-β-catenin overlaps with emCMHC and co-localizes with disassembled sarcomeres, in association with CM dedifferentiation. Our findings about conditional expression of phospho-mimetic β-catenin (S675E) in adult CMs support the direct role of Ser 675 phosphorylation in sarcomere disassembly and CM dedifferentiation in injured hearts (Figure 6). This is in agreement with the function of β-catenin in enhancing CM dedifferentiation in the context of active Neuregulin-1/ERBB2 signaling, examined in cultured murine CMs (D'Uva et al., 2015). Moreover, inactivation of Pak2 activity reduces pS675-β-catenin at cardiac wounds and attenuates CM dedifferentiation (Figure 7L‒Q). Considering that β-catenin is the specific substrate of Pak2 kinase (Supplementary Figure S7), these findings together indicate that PAK2-mediated pS675-β-catenin induction is most likely to contribute to CM dedifferentiation and proliferation (Figure 7R). It is also possible that Pak2 regulates CM regeneration via other factors or substrates (Figure 7R). As dnpak2a mutant or FRAX597 inhibitor specifically inhibits the Pak2 kinase activity (Licciulli et al., 2013), it is unlikely that any Pak2-non-kinase activity associates with CM dedifferentiation/proliferation processes. It will be important to test whether Pak2/pS675-β-catenin interacts with Neuregulin-1/ERBB2 signaling during cardiac regeneration, to test by which mechanisms Pak2/pS675-β-catenin signaling regulates the dedifferentiation process, and to determine whether there are other unknown substrates of Pak2 involved in heart regeneration in coordinating with Wnt signaling inhibition.
Studies in our and other labs have uncovered an injury-dependent Wnt inhibitory role for zebrafish heart regeneration, and this is likely to be conserved in mammals. However, some groups observed the confounding results in heart regeneration using heat-shocked Wnt modifier transgenic zebrafish after cryoinjury (Ozhan and Weidinger, 2015). This discrepancy might be the result of different injury contexts. In murine hearts after myocardial infarction, inhibiting Wnt signaling by overexpression of sFrp1, Dkk3, or Igfb4 (Barandon et al., 2003; Bao et al., 2015; Wo et al., 2016) ameliorates myocardial cell apoptosis, reduces adverse remodeling, and inhibits functional deterioration (Bergmann, 2010). Administration of Porcupine inhibitor WNT974 or chemical inhibitor CDMG1 not only reduces cardiac fibrosis but also stimulates CM proliferation in the infarcted border zone (Moon et al., 2017; Yang et al., 2017; Xie et al., 2020). Hence, our findings along with these indicate that regulation of Wnt/Pak2 signaling may be instrumental in strategies to improve survival, and even regeneration, after myocardial infarction, highlighting a potential inroad for therapeutic intervention.
Materials and methods
Zebrafish strains and chemical inhibitors
Transgenic lines used in this study include Tg(cmlc2:EGFP) (Huang et al., 2003), Tg(cmlc2:nDsRed) (Mably et al., 2003), Tg(hsp70:wnt8a) (Ueno et al., 2007), Tg(hsp70:dkk1b) (Ueno et al., 2007), Tg(dkk1b:EGFP) (Kang et al., 2013), Tg(tcf21:nEGFP) (Wang et al., 2011), Tg(fli1a:EGFP) (Lawson and Weinstein, 2002), and Tg(cmlc2:TetON-3G, cryaa:mCherry). Experimental procedures involving animals were advised and approved by the Institutional Animal Care and Use Committee at Fudan University. CDMG1, FRAX597, and FRAX486 (Selleck) were dissolved in DMSO to a 10 mM stock solution. BIO (Sigma) and CHIR99021 (Sigma) were dissolved in DMSO to 1 and 5 mM stock solutions, respectively. Adult transgenic TRE3G:ctnnb2(S675E)CMi fish were daily treated with 50 mg/L DOX (Sigma) over the window of heart regeneration.
Generation of Tg(hsp70:dnpak2a) animals
The dominant-negative pak2a (pak283‒149) was generated by deleting CRIB motif, which retains the dimerization domain, the kinase inhibitory domain, and the inhibitory switch domain (Chu et al., 2004). MultiSite Gateway Kit (Invitrogen) was used to clone the hsp70 promoter and dnpak2a fragment into the pDESTol2pA2 vector. The hsp70–dnpak2a construct was co-injected with Tol2 transposase mRNA into one-cell stage embryos to generate transgenic lines.
Generation of Tg(TRE3G:ctnnb2(S675E)-mScarlet-I, cryaa:EGFP) animals
Full-length coding sequences of ctnnb2 were generated from zebrafish heart cDNA and subcloned into TOPO blunt cloning vector (Invitrogen). S675E phospho-mimetic mutation (Arias-Romero et al., 2013) was introduced using Q5 site-directed mutagenesis kit (NEB). Amplified ctnnb2(S675E) fragment and mScarlet-I fragment were subcloned into Tol2-mediated TRE3G empty vector containing cryaa:EGFP using HIFI assembly kit (NEB). The purified TRE3G:ctnnb2(S675E)-mScarlet-I, cryaa:EGFP construct was co-injected with Tol2 transposase mRNA into one-cell stage embryos to generate transgenic lines. Stable transgenic lines were screened under a stereo-fluorescence microscope.
Ventricular resection, real-time PCR, and ISH analyses
Adult zebrafish (4‒12 months of age) were used for ventricular resection surgery as previously described (Poss et al., 2002). Briefly, 20% of ventricular muscle was removed at the apex with iridectomy scissors. Total RNAs were extracted from uninjured, 3 dpa, and 7 dpa hearts and used for cDNA synthesis using PrimeScript™ II 1st Strand cDNA Synthesis Kit (TaKaRa). Primers for dkk1a, dkk1b, dkk2, dkk3a, dkk3b, sfrp1a, sfrp1b, sfrp2, sfrp3, lrp5, lrp6, kremen1, wnt1, wnt2, wnt2ba, wnt2bb, wnt3, wnt3a, wnt4a, wnt4b, wnt5b, wnt6b, wnt7ba, wnt7bb, wnt8a, wnt8b, wnt9a, wnt10a, wnt10b, wnt 11, wnt11r, and wnt 16 were designed and used for semi RT-PCR analyses. Specific primers for dkk1b, dkk2, dkk3b, sfrp1a, sfrp1b, sfrp2, wnt2ba, wnt2bb, wnt4a, wnt6b, wnt8a, axin2, mycn, pak1, pak2a, and pak2b were designed and used for qPCR analyses. Real-time PCR was performed with SYBR Green (TOYOBO) using Roche LightCycler® 480 II PCR system. β-actin expression was used for normalization and target gene expression levels were analyzed using 2−ΔΔCt method (Livak and Schmittgen, 2001). For each biological sample, 3‒5 biological replicates and 3‒5 technical replicates were used for qPCR. ISH analyses were performed on cryosections of hearts as described (Lepilina et al., 2006). FISH analyses were performed on cryosections from Tg(fli1a:EGFP) hearts as described previously (Munch et al., 2017). PCR fragments of sfrp1a, dkk3b, wnt5b, lrp6, β-catenin, pak2a, gata4, α-SMA, and actn1 were subcloned into pGEM-T vector (Promega). Digoxigenin-labeled cRNA probes were transcribed using T7 and SP6 RNA polymerase (Invitrogen). In situ signals were detected using anti-digoxigenin-AP (Roche), visualized by NBT/BCIP substrates (Roche), and imaged by a Nikon Eclipse Ni microscope (Nikon) and Nikon Digital Sight DS-Ri1 camera (Nikon). Fluorescence in situ signals were detected using TSA plus fluorescence kit (PerkinElmer) and imaged by Zeiss LSM880 confocal microscope.
Histology and immunofluorescence methods
Injured hearts were extracted, fixed, and cryosectioned. Fibrin and scar analyses by AFOG staining were performed as previously described (Poss et al., 2002). Briefly, sections were hydrated and incubated in Bouin’s solution containing 5% acetic acid, 9% formaldehyde, and 0.9% picric acid (Sigma). Slides were then placed in AFOG solution, containing analine blue (Fisher), orange G (Sigma), and acid fuchsin (Sigma). Myofibril, fibrin, and collagen were visualized as orange, red, and blue, respectively. CM proliferation experiments were performed as previously described (Fang et al., 2013). Fish density (one fish/500 ml) was arranged to examine heart regeneration. Primary antibodies used in this study include anti-DsRed (Clontech #632496; 1:100), anti-Mef2C (Santa Cruz #sc-313; 1:100), anti-Dkk3 (Novus Biologicals #NBP2-37430; 1:50), anti-sFrp1 (Abcam #ab126613; 1:50), anti-sFrp2 (Abcam #ab92667; 1:50), anti-PCNA (Sigma #P8825; 1:100), anti-β-catenin (Abcam #ab32572; 1:100), anti-pS675-β-catenin (Cell Signaling Technology #4176; 1:100), anti-α-actinin (Sigma #A7732; 1:100), anti-MHC (DSHB; 1:100), anti-emCMHC (DSHB; 1:100), anti-PAK2 (Abcam #76293; 1:100), anti-GFP (Aves Labs #GFP-1010; 1:500), and anti-mScarlet-I (NanoTag Biotechnologies #N1302; 1:250). Secondary antibodies include goat anti-mouse Alexa-Fluor-488 (Invitrogen #A-11001; 1:1000), goat anti-mouse Alexa-Fluor-555 (Invitrogen #A-21422; 1:1000), goat anti-rabbit Alexa-Fluor-488 (Invitrogen #A27034; 1:1000), goat anti-rabbit Alexa-Fluor-555 (Invitrogen #A-21428; 1:1000), and goat anti-chicken Alexa-Fluor-488 (Invitrogen #A-11039; 1:1000). For each heart, Z-stacks were acquired from 10-μm-thick sections using Zeiss Axio Observer. To quantify CM proliferation, three sections from each heart containing injury border zone were chosen for Apotome imaging using a 10× objective. The numbers of Mef2c+ and PCNA+Mef2c+ cells, as well as nDsRed+ and nDsRed+EdU+ cells, in the injury border zone were counted by ImageJ2x software. Percentage of PCNA+Mef2c+/Mef2c+ or nDsRed+EdU+/nDsRed+ cells was calculated and summed to determine the CM proliferation index. To quantify regenerates shown by AFOG staining at indicated time, we score heart histological sections according to criteria: ‘Type I’ indicates complete regeneration displaying contiguous myocardial wall and little scarring; ‘Type II’ indicates partial regeneration displaying partial myocardial wall formation and moderate scarring; ‘Type III’ indicates block in regeneration displaying prominent scarring. For each biological sample, five hearts were used for AFOG analyses and CM proliferation experiments, repeated twice. Click-iT® EdU Alexa-Fluor-488 and 555 Imaging Kit (Invitrogen) was used for EdU detection. AFOG-stained images were obtained using Nikon Eclipse Ni microscope (Nikon) and Nikon Digital Sight DS-Ri1 camera (Nikon). Confocal images were obtained using a Zeiss LSM710 microscope.
Pak2 kinase assay
For PAK kinase assays, HEK293T cells were transfected with pcDNA3.1-HA-PAK2. After 48 h, cells were lysed with lysis buffer (50 mM HEPES, pH7.5, 150 mM NaCl, 1 mM EDTA, 1% Nonidet P-40, 10 mM pyrophosphate, 10 mM glycerophosphate, 50 mM NaF, 1.5 mM Na3VO4, complete protease inhibitor cocktail (Sigma), 1 mM DTT, and 1 mM PMSF) and immunoprecipitated with agarose-conjugated HA-tagged antibody (Abmart). The immunoprecipitates were washed three times with lysis buffer, once with wash buffer (40 mM HEPES and 200 mM NaCl), and once with kinase assay buffer (30 mM HEPES, 50 mM potassium acetate, and 5 mM MgCl2). The immunoprecipitated HA-PAK2 was subjected to kinase assay in the presence of 500 μM ATP and 1.5 μg of GST-tagged β-catenin-WT or β-catenin-S675A purified from Escherichia coli as substrates. PAK inhibitor FRAX597 or FRAX486 was added to the reaction mixtures by incubating for 60 min at 30°C with shaking at a concentration of 2 μM. The reactions were then terminated with SDS sample buffer and boiled before analysis by SDS PAGE with related antibodies.
RNAscope analysis
RNAscope (Advanced Cell Diagnostics, ACD) for Danio rerio axin2 was performed on injured control, Tg(hsp70:dkk1b), and Tg(hsp70:wnt8a) heart cryosections. The Dr-axin2 RNAscope probe was designed by ACD. RNAscope probe hybridization, amplification, and detection were performed according to the protocol of RNAscope Multiplex Fluorescent Reagent Kit V2 user manual (ACD). Following the final step of washing of RNAscope detection, immunostaining for CM visualization with CT3 antibody (DSHB; 1:100) was performed as described above. Images were taken using Zeiss LSM880 confocal microscope. Image analysis was performed using Zen 3.0 software (Zeiss).
Statistical analysis
Statistical tests, P-values, and sample sizes are indicated in the figure legends. All statistical values are presented as mean ± standard error of the mean (SEM). Statistical analyses were performed with GraphPad Prism 8 or Microsoft Excel. Student’s t-test was used to analyze comparisons between two groups. One-way analysis of variance (ANOVA) with Tukey’s test was used to assess the difference between three independent groups. Fisher’s exact test was used to assess the regeneration score. P < 0.05 was considered statistically significant.
Supplementary material
Supplementary material is available at Journal of Molecular Cell Biology online.
Supplementary Material
Acknowledgements
We acknowledge Guozhen Wu for invaluable assistance with fish care. We are grateful to Mark Mercola and members of TPZ laboratory for comments on the manuscript and helpful discussions.
Funding
This research was supported by grants from the Ministry of Science and Technology of China (2018YFA0801004 and 2018YFA0800103) and National Science Foundation of China (NSFC31530044 and NSFC31970780).
Conflict of interest: none declared.
References
- Ai X., Jiang A., Ke Y., et al. (2011). Enhanced activation of p21-activated kinase 1 in heart failure contributes to dephosphorylation of connexin 43. Cardiovasc. Res. 92, 106–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias-Romero L.E., Villamar-Cruz O., Huang M., et al. (2013). Pak1 kinase links ErbB2 to β-catenin in transformation of breast epithelial cells. Cancer Res. 73, 3671–3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao M.W., Cai Z., Zhang X.J., et al. (2015). Dickkopf-3 protects against cardiac dysfunction and ventricular remodelling following myocardial infarction. Basic Res. Cardiol. 110, 25. [DOI] [PubMed] [Google Scholar]
- Barandon L., Couffinhal T., Ezan J., et al. (2003). Reduction of infarct size and prevention of cardiac rupture in transgenic mice overexpressing FrzA. Circulation 108, 2282–2289. [DOI] [PubMed] [Google Scholar]
- Barker N., van Es J.H., Kuipers J., et al. (2007). Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449, 1003–1007. [DOI] [PubMed] [Google Scholar]
- Behrens J., Jerchow B.A., Wurtele M., et al. (1998). Functional interaction of an axin homolog, conductin, with β-catenin, APC, and GSK3β. Science 280, 596–599. [DOI] [PubMed] [Google Scholar]
- Bergmann M.W. (2010). WNT signaling in adult cardiac hypertrophy and remodeling: lessons learned from cardiac development. Circ. Res. 107, 1198–1208. [DOI] [PubMed] [Google Scholar]
- Bokoch G.M. (2003). Biology of the p21-activated kinases. Annu. Rev. Biochem. 72, 743–781. [DOI] [PubMed] [Google Scholar]
- Buchner D.A., Su F., Yamaoka J.S., et al. (2007). pak2a mutations cause cerebral hemorrhage in redhead zebrafish. Proc. Natl Acad. Sci. USA 104, 13996–14001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z., Biechele T., Wei Z., et al. (2011). Crystal structures of the extracellular domain of LRP6 and its complex with DKK1. Nat. Struct. Mol. Biol. 18, 1204–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe C.P., Crump J.G. (2015). Eph–Pak2a signaling regulates branching of the pharyngeal endoderm by inhibiting late-stage epithelial dynamics. Development 142, 1089–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y.S., Zhang Y., Xu M., et al. (2013). Distinct functions for Wnt/β-catenin in hair follicle stem cell proliferation and survival and interfollicular epidermal homeostasis. Cell Stem Cell 13, 720–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu P.C., Wu J., Liao X.C., et al. (2004). A novel role for p21-activated protein kinase 2 in T cell activation. J. Immunol. 172, 7324–7334. [DOI] [PubMed] [Google Scholar]
- Congdon K.L., Voermans C., Ferguson E.C., et al. (2008). Activation of Wnt signaling in hematopoietic regeneration. Stem Cells 26, 1202–1210. [DOI] [PubMed] [Google Scholar]
- D'Uva G., Aharonov A., Lauriola M., et al. (2015). ERBB2 triggers mammalian heart regeneration by promoting cardiomyocyte dedifferentiation and proliferation. Nat. Cell Biol. 17, 627–638. [DOI] [PubMed] [Google Scholar]
- Fang Y., Gupta V., Karra R., et al. (2013). Translational profiling of cardiomyocytes identifies an early Jak1/Stat3 injury response required for zebrafish heart regeneration. Proc. Natl Acad. Sci. USA 110, 13416–13421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faux M.C., Coates J.L., Kershaw N.J., et al. (2010). Independent interactions of phosphorylated β-catenin with E-cadherin at cell‒cell contacts and APC at cell protrusions. PLoS One 5, e14127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemberling M., Karra R., Dickson A.L., et al. (2015). Nrg1 is an injury-induced cardiomyocyte mitogen for the endogenous heart regeneration program in zebrafish. eLife 4, e05871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh K.Y., Ng N.W., Hagen T., et al. (2012). p21-activated kinase interacts with Wnt signaling to regulate tissue polarity and gene expression. Proc. Natl Acad. Sci. USA 109, 15853–15858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta V., Gemberling M., Karra R., et al. (2013). An injury-responsive gata4 program shapes the zebrafish cardiac ventricle. Curr. Biol. 23, 1221–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haq S., Michael A., Andreucci M., et al. (2003). Stabilization of β-catenin by a Wnt-independent mechanism regulates cardiomyocyte growth. Proc. Natl Acad. Sci. USA 100, 4610–4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heallen T., Morikawa Y., Leach J., et al. (2013). Hippo signaling impedes adult heart regeneration. Development 140, 4683–4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hino S., Tanji C., Nakayama K.I., et al. (2005). Phosphorylation of β-catenin by cyclic AMP-dependent protein kinase stabilizes β-catenin through inhibition of its ubiquitination. Mol. Cell. Biol. 25, 9063–9072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschy A., Croquelois A., Perriard E., et al. (2010). Stabilised β-catenin in postnatal ventricular myocardium leads to dilated cardiomyopathy and premature death. Basic Res. Cardiol. 105, 597–608. [DOI] [PubMed] [Google Scholar]
- Hsieh J.C., Kodjabachian L., Rebbert M.L., et al. (1999). A new secreted protein that binds to Wnt proteins and inhibits their activities. Nature 398, 431–436. [DOI] [PubMed] [Google Scholar]
- Huang C.J., Tu C.T., Hsiao C.D., et al. (2003). Germ-line transmission of a myocardium-specific GFP transgene reveals critical regulatory elements in the cardiac myosin light chain 2 promoter of zebrafish. Dev. Dyn. 228, 30–40. [DOI] [PubMed] [Google Scholar]
- Jopling C., Sleep E., Raya M., et al. (2010). Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature 464, 606–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J., Nachtrab G., Poss K.D. (2013). Local Dkk1 crosstalk from breeding ornaments impedes regeneration of injured male zebrafish fins. Dev. Cell 27, 19–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami Y., Rodriguez Esteban C., Raya M., et al. (2006). Wnt/β-catenin signaling regulates vertebrate limb regeneration. Genes Dev. 20, 3232–3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly M.L., Chernoff J. (2012). Mouse models of PAK function. Cell. Logist. 2, 84–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly M.L., Astsaturov A., Chernoff J. (2013). Role of p21-activated kinases in cardiovascular development and function. Cell. Mol. Life Sci. 70, 4223–4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi K., Holdway J.E., Major R.J., et al. (2011). Retinoic acid production by endocardium and epicardium is an injury response essential for zebrafish heart regeneration. Dev. Cell 20, 397–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi K., Holdway J.E., Werdich A.A., et al. (2010). Primary contribution to zebrafish heart regeneration by gata4+ cardiomyocytes. Nature 464, 601–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Wu Q., Zhang Y., et al. (2010). PDGF signaling is required for epicardial function and blood vessel formation in regenerating zebrafish hearts. Proc. Natl Acad. Sci. USA 107, 17206–17210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubin T., Poling J., Kostin S., et al. (2011). Oncostatin M is a major mediator of cardiomyocyte dedifferentiation and remodeling. Cell Stem Cell 9, 420–432. [DOI] [PubMed] [Google Scholar]
- Lawson N.D., Weinstein B.M. (2002). In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev. Biol. 248, 307–318. [DOI] [PubMed] [Google Scholar]
- Lepilina A., Coon A.N., Kikuchi K., et al. (2006). A dynamic epicardial injury response supports progenitor cell activity during zebrafish heart regeneration. Cell 127, 607–619. [DOI] [PubMed] [Google Scholar]
- Leyns L., Bouwmeester T., Kim S.H., et al. (1997). Frzb-1 is a secreted antagonist of Wnt signaling expressed in the Spemann organizer. Cell 88, 747–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Zhang Y., Kang H., et al. (2005). Sclerostin binds to LRP5/6 and antagonizes canonical Wnt signaling. J. Biol. Chem. 280, 19883–19887. [DOI] [PubMed] [Google Scholar]
- Licciulli S., Maksimoska J., Zhou C., et al. (2013). FRAX597, a small molecule inhibitor of the p21-activated kinases, inhibits tumorigenesis of neurofibromatosis type 2 (NF2)-associated Schwannomas. J. Biol. Chem. 288, 29105–29114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z., von Gise A., Zhou P., et al. (2014). Cardiac-specific YAP activation improves cardiac function and survival in an experimental murine MI model. Circ. Res. 115, 354–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Logan C.Y., Nusse R. (2004). The Wnt signaling pathway in development and disease. Annu. Rev. Cell Dev. Biol. 20, 781–810. [DOI] [PubMed] [Google Scholar]
- Mably J.D., Mohideen M.A., Burns C.G., et al. (2003). Heart of glass regulates the concentric growth of the heart in zebrafish. Curr. Biol. 13, 2138–2147. [DOI] [PubMed] [Google Scholar]
- Marin-Juez R., Marass M., Gauvrit S., et al. (2016). Fast revascularization of the injured area is essential to support zebrafish heart regeneration. Proc. Natl Acad. Sci. USA 113, 11237–11242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed T.M.A., Ang Y.S., Radzinsky E., et al. (2018). Regulation of cell cycle to stimulate adult cardiomyocyte proliferation and cardiac regeneration. Cell 173, 104–116.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon J., Zhou H., Zhang L.S., et al. (2017). Blockade to pathological remodeling of infarcted heart tissue using a porcupine antagonist. Proc. Natl Acad. Sci. USA 114, 1649–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moro E., Ozhan-Kizil G., Mongera A., et al. (2012). In vivo Wnt signaling tracing through a transgenic biosensor fish reveals novel activity domains. Dev. Biol. 366, 327–340. [DOI] [PubMed] [Google Scholar]
- Munch J., Grivas D., Gonzalez-Rajal A., et al. (2017). Notch signalling restricts inflammation and serpine1 expression in the dynamic endocardium of the regenerating zebrafish heart. Development 144, 1425–1440. [DOI] [PubMed] [Google Scholar]
- Naito A.T., Shiojima I., Akazawa H., et al. (2006). Developmental stage-specific biphasic roles of Wnt/β-catenin signaling in cardiomyogenesis and hematopoiesis. Proc. Natl Acad. Sci. USA 103, 19812–19817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson W.J., Nusse R. (2004). Convergence of Wnt, β-catenin, and cadherin pathways. Science 303, 1483–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni T.T., Rellinger E.J., Mukherjee A., et al. (2011). Discovering small molecules that promote cardiomyocyte generation by modulating Wnt signaling. Chem. Biol. 18, 1658–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozhan G., Weidinger G. (2015). Wnt/β-catenin signaling in heart regeneration. Cell Regen. 4, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng X., He Q., Li G., et al. (2016). Rac1‒PAK2 pathway is essential for zebrafish heart regeneration. Biochem. Biophys. Res. Commun. 472, 637–642. [DOI] [PubMed] [Google Scholar]
- Porrello E.R., Mahmoud A.I., Simpson E., et al. (2011). Transient regenerative potential of the neonatal mouse heart. Science 331, 1078–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poss K.D., Wilson L.G., Keating M.T. (2002). Heart regeneration in zebrafish. Science 298, 2188–2190. [DOI] [PubMed] [Google Scholar]
- Ring D.B., Johnson K.W., Henriksen E.J., et al. (2003). Selective glycogen synthase kinase 3 inhibitors potentiate insulin activation of glucose transport and utilization in vitro and in vivo. Diabetes 52, 588–595. [DOI] [PubMed] [Google Scholar]
- Sallin P., de Preux Charles A.S., Duruz V., et al. (2015). A dual epimorphic and compensatory mode of heart regeneration in zebrafish. Dev. Biol. 399, 27–40. [DOI] [PubMed] [Google Scholar]
- Schlessinger K., Hall A., Tolwinski N. (2009). Wnt signaling pathways meet Rho GTPases. Genes Dev. 23, 265–277. [DOI] [PubMed] [Google Scholar]
- Senyo S.E., Steinhauser M.L., Pizzimenti C.L., et al. (2013). Mammalian heart renewal by pre-existing cardiomyocytes. Nature 493, 433–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh B.N., Koyano-Nakagawa N., Gong W., et al. (2018). A conserved HH‒Gli1‒Mycn network regulates heart regeneration from newt to human. Nat. Commun. 9, 4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoick-Cooper C.L., Weidinger G., Riehle K.J., et al. (2007). Distinct Wnt signaling pathways have opposing roles in appendage regeneration. Development 134, 479–489. [DOI] [PubMed] [Google Scholar]
- Taurin S., Sandbo N., Qin Y., et al. (2006). Phosphorylation of β-catenin by cyclic AMP-dependent protein kinase. J. Biol. Chem. 281, 9971–9976. [DOI] [PubMed] [Google Scholar]
- Tzahor E., Poss K.D. (2017). Cardiac regeneration strategies: staying young at heart. Science 356, 1035–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno S., Weidinger G., Osugi T., et al. (2007). Biphasic role for Wnt/β-catenin signaling in cardiac specification in zebrafish and embryonic stem cells. Proc. Natl Acad. Sci. USA 104, 9685–9690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenta T., Hausmann G., Basler K. (2012). The many faces and functions of β-catenin. EMBO J. 31, 2714–2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheyen E.M., Gottardi C.J. (2010). Regulation of Wnt/β-catenin signaling by protein kinases. Dev. Dyn. 239, 34–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vite A., Radice G.L. (2014). N-cadherin/catenin complex as a master regulator of intercalated disc function. Cell Commun. Adhes. 21, 169–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Cao J., Dickson A.L., et al. (2015). Epicardial regeneration is guided by cardiac outflow tract and Hedgehog signalling. Nature 522, 226–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Panakova D., Kikuchi K., et al. (2011). The regenerative capacity of zebrafish reverses cardiac failure caused by genetic cardiomyocyte depletion. Development 138, 3421–3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wo D., Peng J., Ren D.N., et al. (2016). Opposing roles of Wnt inhibitors IGFBP-4 and Dkk1 in cardiac ischemia by differential targeting of LRP5/6 and β-catenin. Circulation 134, 1991–2007. [DOI] [PubMed] [Google Scholar]
- Wu C.C., Kruse F., Vasudevarao M.D., et al. (2016). Spatially resolved genome-wide transcriptional profiling identifies BMP signaling as essential regulator of zebrafish cardiomyocyte regeneration. Dev. Cell 36, 36–49. [DOI] [PubMed] [Google Scholar]
- Xie S., Fu W., Yu G., et al. (2020). Discovering small molecules as Wnt inhibitors that promote heart regeneration and injury repair. J. Mol. Cell Biol. 12, 42–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin M., Olson E.N., Bassel-Duby R. (2013). Mending broken hearts: cardiac development as a basis for adult heart regeneration and repair. Nat. Rev. Mol. Cell Biol. 14, 529–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D., Fu W., Li L., et al. (2017). Therapeutic effect of a novel Wnt pathway inhibitor on cardiac regeneration after myocardial infarction. Clin. Sci. 131, 2919–2932. [DOI] [PubMed] [Google Scholar]
- Yang J., Xu X. (2012). α-actinin2 is required for the lateral alignment of Z discs and ventricular chamber enlargement during zebrafish cardiogenesis. FASEB J. 26, 4230–4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Ben-Yair R., Burns C.E., et al. (2019). Endocardial Notch signaling promotes cardiomyocyte proliferation in the regenerating zebrafish heart through Wnt pathway antagonism. Cell Rep. 26, 546–554.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Borikova A.L., Ben-Yair R., et al. (2014). Notch signaling regulates cardiomyocyte proliferation during zebrafish heart regeneration. Proc. Natl Acad. Sci. USA 111, 1403–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G., Wang Y., Huang B., et al. (2012). A Rac1/PAK1 cascade controls β-catenin activation in colon cancer cells. Oncogene 31, 1001–1012. [DOI] [PubMed] [Google Scholar]
- Zimmerman Z.F., Moon R.T., Chien A.J. (2012). Targeting Wnt pathways in disease. Cold Spring Harb. Perspect. Biol. 4, a008086. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.