Abstract
Simple Summary
The introduction of immunotherapy modified the cancer treatment landscape, especially for non-small cell lung cancer (NSCLC). Unfortunately, only a subgroup of patients benefits from this therapy. Currently, the only validated companion diagnostic test for first-line immunotherapy in metastatic NSCLC patients is testing for programmed death ligand 1 (PD-L1) expression in tumor tissues. However, obtaining tumor tissue can be challenging and it puts the patient at risk. Liquid biopsy offers an alternative, less invasive approach to select NSCLC patients who would benefit from immunotherapy and to monitor patients during their disease course. Liquid biopsy allows repetitive sampling, which makes it a useful tool in clinical practice. In this review, we discuss the challenges and opportunities of several liquid biopsy-based prognostic and predictive biomarkers in NSCLC patients receiving immunotherapy.
Abstract
In the last decade, immunotherapy has been one of the most important advances in the non-small cell lung cancer (NSCLC) treatment landscape. Nevertheless, only a subset of NSCLC patients benefits from it. Currently, the only Food and Drug Administration (FDA) approved diagnostic test for first-line immunotherapy in metastatic NSCLC patients uses tissue biopsies to determine the programmed death ligand 1 (PD-L1) status. However, obtaining tumor tissue is not always feasible and puts the patient at risk. Liquid biopsy, which refers to the tumor-derived material present in body fluids, offers an alternative approach. This less invasive technique gives real-time information on the tumor characteristics. This review addresses different promising liquid biopsy based biomarkers in NSCLC patients that enable the selection of patients who benefit from immunotherapy and the monitoring of patients during this therapy. The challenges and the opportunities of blood-based biomarkers such as cell-free DNA (cfDNA), circulating tumor cells (CTCs), exosomes, epigenetic signatures, microRNAs (miRNAs) and the T cell repertoire will be addressed. This review also focuses on the less-studied feces-based and breath-based biomarkers.
Keywords: liquid biopsy, non-small cell lung cancer, immunotherapy, molecular pathology
1. Introduction
With an estimated incidence of more than 2 million cases and approximately 1.8 million deaths annually, lung cancer is the leading cause of cancer-related death, worldwide [1]. Non-small cell lung cancer (NSCLC) accounts for approximately 85% of lung cancers [2]. At the time of diagnosis, locoregionally advanced or metastatic disease is present in more than 60% of the patients.
In the last decade, the treatment landscape and the prognosis of NSCLC patients have changed, resulting in a clinically meaningful improvement in survival and quality of life [3,4]. More specifically, targeted therapies gained a profound role in the management of NSCLC. Targeted therapy is a type of precision medicine, since cancer cells are targeted preferentially, resulting in less side effects than with traditional chemotherapy. However, only a subgroup of the NSCLC patients—harboring specific genetic alterations—benefits from this type of therapy [5]. More recently, immune checkpoint inhibitor agents targeting programmed cell death protein 1 (PD-1) or programmed cell death ligand 1 (PD-L1) have been established as a new treatment for advanced NSCLC patients. Of note is that a minority of NSCLC patients (< 20%) respond to this expensive therapy when administrated in monotherapy [6,7,8,9]. Immunotherapy is often given in combination with chemotherapy since several chemotherapeutical compounds appear to have the capacity to upregulate PD-L1 expression on cancer cells and to promote antitumor immunogenicity [10]. In 2018, the Food and Drug Administration (FDA) approved the combination of anti-PD-1 (pembrolizumab) with chemotherapy drugs (pemetrexed and carboplatin) to treat metastatic NSCLC [6].
Hence, it is crucial to identify predictive biomarkers for NSCLC patients on immunotherapy to enable the selection of patients that will benefit from this therapy. For the early detection of resistance to anti-PD-1/PD-L1 therapy, the identification of biomarkers which allow monitoring of the NSCLC patients during therapy is important [11].
Within this context, the interest in the liquid biopsy field started to grow. Liquid biopsy refers to tumor-derived material that is present in body fluids. Recently, liquid biopsy has become an attractive approach since it is a less invasive, cost-effective technique which gives real-time information on the tumor characteristics [12]. In this light, in 2016 the Cobas EGFR Mutation Test v2 (Roche) became the first liquid biopsy assay that obtained the approval of the FDA for the identification of NSCLC patients eligible for epidermal growth factor receptor (EGFR)-based targeted therapy [13].
In this review we will: (i) discuss which components of a liquid biopsy sample can be used as a predictive or prognostic biomarker in NSCLC patients on immunotherapy, and (ii) highlight the challenges and clinical applicability of each biomarker.
2. Liquid Biopsy
The treatment strategy for metastatic NSCLC patients is largely based on two parameters: the presence of specific molecular or genomic aberrations within the tumor tissue, and imaging [14]. Tissue analysis, however, faces some limitations. The procedure to obtain a tissue biopsy can put the patient at risk, sometimes it is not possible to obtain adequate tumor tissue due to the location of the tumor and studying tumor heterogeneity requires multiple biopsies which is difficult due to ethical and practical considerations [15,16,17].
Evaluating treatment response by imaging can be difficult, particularly when dealing with immunotherapy. Pseudoprogression is characterized by radiologic enlargement of the tumor mass. It is caused by the infiltration of leukocytes and associated with favorable a long-term survival. Pseudoprogression is reported in up to 6% of metastatic NSCLC treated with anti-PD-1/PD-L1 therapy. In 2009, the Response Evaluation Criteria in Solid Tumors (RECIST) scoring was adapted for the radiographic monitoring of immunotherapy receiving patients (iRECIST) in an attempt to overcome this issue [18,19].
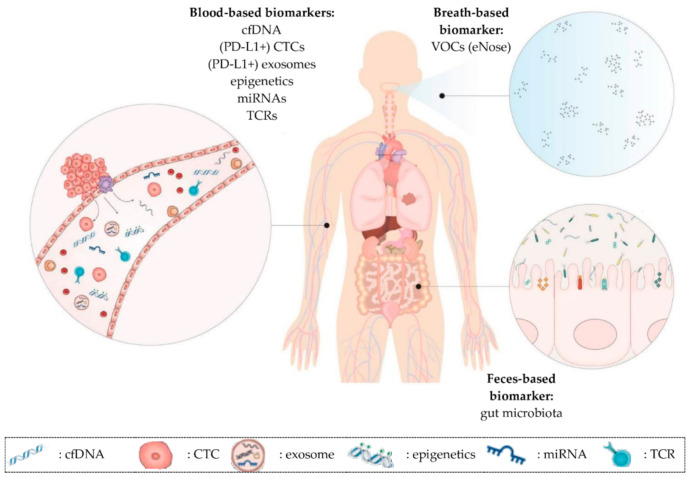
Liquid biopsies might help to solve the issues regarding the selection and monitoring of NSCLC patients since it enables repetitive sampling. It allows the analysis of cancer-associated biomarkers in biological fluids such as blood, urine or saliva. Interestingly, even in feces and breath potential biomarkers can be detected. A liquid biopsy sample consists of different (circulating) components derived from cancer as well as from healthy tissue. In this review, the characteristics of the following components will be discussed: circulating cell-free DNA (cfDNA), circulating tumor cells (CTCs), extracellular vesicles (EVs), epigenetic signatures, microRNA (miRNA), volatile organic compounds (VOCs) and the gut microbiota (Table 1, Figure 1) [20,21].
Table 1.
Summary of characteristics of different blood-based biomarkers. The different advantages and disadvantages of the blood-based biomarkers, that favor or disfavor their use in clinical settings, are shown in the table. BEAMing: beads, emulsions, amplification and magnetics; cfDNA: circulating cell-free DNA; bTMB: blood tumor mutational burden; EpCAM: epithelial cell adhesion molecule; miRNA: microRNA; NGS: next-generation sequencing; NSCLC: non-small cell lung cancer; PD-L1: programmed death-ligand 1; TCR: T cell repertoire.
| Type | Isolation Technique | Advantages | Disadvantages | Promising Biomarker | References |
|---|---|---|---|---|---|
| Cell-free DNA | Magnetic beads or spin column based | - Easy and well-established isolation procedures - Allows real-time monitoring |
- Short half-life: 16 min–2.5 h - Not stable in circulation - Long turn-around time NGS based tests - Sensitive detection methods required |
bTMB and levels of cfDNA for cancer diagnosis and treatment evaluation | [17,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53] |
| Circulating tumor cells | Antigen-dependent (e.g., EpCAM) or size and deformability based method | - Provides transcriptomic, genomic and proteomic information | - Short half-life: 1–2.4 h - Not stable in circulation |
PD-L1 analysis status to predict response on treatment | [15,25,54,55,56,57,58,59,60,61,62,63,64] |
| - Intact viable cells | - Low abundance in NSCLC | ||||
| - Isolation procedures requires high expertise and dedicated equipment | |||||
| Exosomes | Based on physical or biological properties of exosomes, immune-mediated isolation, sucrose gradient method, ultra-centrifugation | - Provides information about tumor’s biologic profile, growth rate, metastatic capacity and drug resistance - Abundant |
- Isolation can be time-consuming and can alter exosome structure - Difficult to detect due to small size |
Exosomal PD-L1 status to predict response on therapy | [25,65,66,67,68,69,70,71,72,73,74] |
| Epigenetics and miRNA | cfDNA isolation followed by methylation specific PCR or BEAMing | - Methylation makes cfDNA more stable - Allows real-time monitoring |
- Sensitive detection methods required | Methylation status of genes (signature) for cancer diagnosis and treatment evaluation | [75,76,77,78,79,80,81,82,83,84,85,86,87] |
| T cell repertoire | Density centrifugation followed by flowcytometry | - Easy isolation - Intact viable cells |
- Clustering of functionally different clones causing false positive results | The frequency, diversity and clonality of TCRs for cancer diagnosis and treatment evaluation | [41,57,88,89,90,91,92] |
Figure 1.
Overview of promising prognostic and predictive minimally invasive biomarkers in NSCLC patients on immunotherapy discussed in this review. (1) Blood-based biomarkers such as cfDNA, CTCs, exosomes, epigenetic signatures, miRNAs and TCR repertoire. These biomarkers can also be present in other liquids than blood (e.g., urine). (2) Breath-based biomarkers such as VOCs. (3) Feces-based biomarkers such as the gut microbiota. cfDNA: cell-free DNA; CTCs: circulating tumor cells; miRNAs: microRNAs; TCR: T cell receptor; VOCs: volatile organic compounds.
Despite the potential benefits, major hurdles need to be overcome. Liquid biopsy-based biomarkers often lack sensitivity and specificity, especially in patients with localized tumors [22]. Furthermore, workflows need to be standardized and protocols need to be harmonized. Another hurdle is the lack of reimbursement for liquid biopsy-based tests in some countries [23,24]. Ongoing clinical trials in NSCLC patients on immunotherapy address these challenges and will give us more insight on the future of this technique (Table 2).
Table 2.
Overview of ongoing clinical trials in which promising predictive and prognostic liquid biopsy-based biomarkers are investigated in NSCLC patients. Ongoing clinical trials were found at the website of https://www.Clinicaltrials.gov (accessed on 27 November 2020). An advanced search was performed with the words: “(NSCLC OR non-small cell lung cancer) AND (immunotherapy OR PD-L1 or PD-1) in combination with: (cfDNA OR cell-free DNA); (CTC or circulating tumor cell); exosomes; epigenetics; miRNA; (T cell repertoire OR TCR); gut microbiota or (eNose OR breath)”. Results of the completed trials are available, the other trials have not provided their study results on ClinicalTrials.gov yet. BC: breast cancer; CRC: colorectal cancer; H&NC: head and neck cancer; KC: kidney cancer; MC: melanoma cancer; NSCLC: non-small cell lung cancer; PC: pancreas cancer; RC: renal cancer; UC: urothelial carcinoma.
| Trial | Cancer Type | Therapy | # Patients | Markers | End Date |
|---|---|---|---|---|---|
| NCT02511288 | NSCLC | Immunotherapy, targeted therapy | 900 | cfDNA, miRNA | 2026 |
| NCT04107168 | MC, NSCLC, RC | Immunotherapy | 1.800 | microbiome | 2025 |
| NCT04146064 | H&NC, KC, MC, NSCLC, UC | Immunotherapy | 425 | eNose | 2024 |
| NCT04638751 | BC, CRC, NSCLC, PC | Chemotherapy, immunotherapy | 4.000 | microbiome | 2024 |
| NCT03926260 | NSCLC | Chemotherapy, immunotherapy, targeted therapy | 100 | cfDNA | 2023 |
| NCT04629027 | NSCLC | Immunotherapy | 80 | CTC, TCR | 2023 |
| NCT04636047 | NSCLC | Immunotherapy | 450 | cfDNA (bTMB), TCR | 2023 |
| NCT04636775 | NSCLC | Immunotherapy | 46 | microbiome | 2022 |
| NCT04427475 | NSCLC | Immunotherapy | 900 | cfDNA, miRNA | 2022 |
| NCT03512847 | NSCLC | Chemotherapy, immunotherapy | 150 | cfDNA | 2021 |
| NCT03481101 | NSCLC | Chemotherapy, immunotherapy | 60 | cfDNA | 2021 |
| NCT04291755 | CRC, NSCLC | Immunotherapy | 100 | microbiome | 2021 |
| NCT03178552 | NSCLC | Immunotherapy, targeted therapy | 660 | cfDNA (bTMB) | 2021 |
| NCT02827344 | NSCLC | Immunotherapy | 200 | CTC | 2021 |
| NCT03892096 | BC, CRC, NSCLC | Chemotherapy, immunotherapy, targeted therapy | 750 | cfDNA | 2021 |
| NCT03576937 | NSCLC | Chemotherapy, immunotherapy, targeted therapy | 210 | cfDNA | 2020 |
| NCT03986463 | NSCLC | Chemotherapy, radiotherapy | 30 | cfDNA | 2020 |
| NCT03373955 | NSCLC | Chemotherapy, immunotherapy, targeted therapy | 60 | cfDNA, TCR | 2020 |
| NCT02551211 | NSCLC | Immunotherapy | 58 | cfDNA, TCR | 2019 |
| NCT02890849 | NSCLC | Immunotherapy, radiotherapy | 60 | Exosomes, mRNA | 2019 |
| NCT01903993 | NSCLC | Chemotherapy, immunotherapy | 287 | cfDNA (bTMB) | Completed |
| NCT02008227 | NSCLC | Chemotherapy, immunotherapy | 1.225 | cfDNA (bTMB) | Completed |
3. Cell-Free DNA
cfDNA is currently the material with the greatest potential in clinical practice [25]. The term cfDNA refers to both the encapsulated DNA (in circulating vesicles) as well as the non-encapsulated free DNA. cfDNA is mostly studied in blood samples, but is also present in other body fluids such as urine, saliva and cerebrospinal fluid. Its half-life ranges from 16 min to 2.5 h, due to rapid clearance by the circulation through the kidneys, liver and spleen.
cfDNA can be derived from healthy cells, as well as from tumor cells. The fraction originated from tumor cells is named circulating cell-free tumor DNA (cf tumor DNA). Cf tumor DNA is released into the peripheral blood by three main mechanisms: apoptosis, necrosis and active secretion from EVs and CTCs. The fraction of cfDNA contributed from the tumor varies greatly, between 0.01% and more than 90% [17]. The levels of cf tumor DNA are influenced by tumor burden and other factors such as tumor location, vascularity and cellular turnover [26]. Fortunately, sensitive detection platforms like beads, emulsions, amplification and magnetics (BEAMing), digital droplet PCR (ddPCR), MassARRAY and specialized next-generation sequencing (NGS) techniques make it possible to detect low concentrations of cf tumor DNA [27].
cfDNA fragments derived from healthy cells—often referred to as genomic DNA (gDNA) contamination—stem from apoptotic or necrotic cells, particularly white blood cells which have a limited survival time. The concentration of cfDNA can be influenced by certain conditions such as inflammation, infection and even exercise [25]. For qualitative analysis of cf tumor DNA, it is important to reduce the concentration of gDNA contamination to an absolute minimum. In this context, the use of an appropriate cfDNA extraction kit which favors the isolation of cf tumor DNA fragments is crucial [28].
3.1. Blood Tumor Mutational Burden
Tumor mutational burden (TMB) can be defined as the total number of somatic non-synonymous mutations per mega base (Mb) of the genome examined. The presence of these mutations can lead to the formation of neoantigens which are recognized by the immune system as non-self, resulting in the activation of the antitumor immune response. This hypothesis suggests that NSCLC patients with a high TMB could potentially benefit from immunotherapy [29]. Notably, a high mutational load does not mean that a high number of neoantigens will be expressed on the cancer cell surface [30]. There is also evidence that the quality of neoantigens (= an elevated immune-recognition potential) is more important than the quantity of neoantigens [31,32]. The number of somatic mutations varies between different cancer types. NSCLC has one of the highest mutation frequencies (0.1 to 100 mutations/Mb), particularly in smokers [33,34]. TMB in tumors is usually evaluated using NGS-based approaches (including whole exome sequencing (WES) and comprehensive genomic profiling (CGP)) [35,36] and seems to correlate with gender, since it is higher in men than in women [37].
Herbst R.S. et al. retrospectively investigated the association between tissue TMB (tTMB) and the clinical benefit with pembrolizumab monotherapy observed in previously treated (KEYNOTE-010, NCT01905657) or treatment naïve (KEYNOTE-042, NCT02220894) PD-L1+ NSCLC patients. In both trials, improvements in overall survival (OS) as well as in progression-free survival (PFS) were generally observed for patients treated with pembrolizumab with high tTMB (≥ 175) [38]. Several research groups studied the possibility to use blood (cf tumor DNA) instead of tumor tissue to determine TMB. The group of Wang et al.—who enrolled 48 patients with advanced NSCLC treated with anti PD-(L)1 therapy—observed that blood TMB (bTMB) correlated well with tTMB calculated by WES [39]. Similar results were obtained in larger study cohorts. The randomized and retrospective POPLAR (NCT01903993, n = 287) and OAK (NCT02008227, n = 1.225) clinical trials both compared atezolizumab with the standard of care, docetaxel, in NSCLC patients. In these clinical trials, tTMB and bTMB obtained from pre-treatment plasma from the same patients were compared. A positive correlation between tTMB and bTMB in NSCLC patients was observed (Spearman rank correlation: 0.64; 95% confidence interval (CI): 0.56–0.71) [40].
In this context, determining a cut-off point for bTMB was essential. The POPLAR trial demonstrated that bTMB ≥ 16 is a clinically meaningful and technically robust cut-off point in NSCLC patients. This observation was confirmed by the OAK trial [40,41]. Interestingly, NSCLC patients on immunotherapy with high bTMB were more likely to respond to therapy. Patients from the OAK trial with bTMB ≥ 16 obtained a significant PFS benefit (hazard ratio (HR): 0.65 (95% CI: 0.47–0.92); p = 0.013) when treated with atezolizumab versus docetaxel [40]. Similar results were observed by Wang et al. [39]. Another clinical trial (B-F1RST, NCT02848651, n = 152) also showed a correlation between a high bTMB and atezolizumab response in NSCLC patients, using the same cut-off for bTMB as in the POPLAR and OAK studies [41].
These data suggest that bTMB might be a promising prognostic biomarker for NSCLC patients on immunotherapy. In this light, the FDA approved two plasma-based NGS assays for the measurement of bTMB, namely the Guardant Health (GH) Omni (500 genes, 2.1 Mb) and the Foundation Medicine (FMI) bTMB (394 genes, 1.14 Mb) panels in 2020 [42].
However, these promising results are in contrast with the preliminary data of the Checkmake 227 clinical trial (NCT02477826, n = 679). The results of this randomized trial showed a similar degree of OS benefit in patients who received nivolumab plus ipilimumab, regardless of whether they had a high or low bTMB (≥10 vs. <10 mutations per Mb, respectively) [43]. In addition, Paz-Ares et al. studied patients with non-squamous NSCLC (KEYNOTE-021 trial (NCT02039674) and KEYNOTE-189 trial (NCT02578680)) and patients with squamous NSCLC (KEYNOTE-407 trial (NCT02775435)). They found that tTMB was not significantly associated with the efficacy of first-line pembrolizumab plus platinum-based chemotherapy or with chemotherapy alone in NSCLC [44].
TMB studies the somatic non-synonymous mutations present in a Mb. Interestingly, some studies revealed that single point mutations can also be used to monitor NSCLC patients treated with immunotherapy. Guibert et al. showed that the presence of a phosphatase and tensin homolog (PTEN) or a serine/threonine kinase 11 (STK11) mutation was correlated with early progression in NSCLC patients receiving anti PD-1 immunotherapy. In contrast, transversion mutations (substitution of a purine by a pyrimidine or vice versa) in the Ki-ras2 Kirsten rat sarcoma viral oncogene homolog (KRAS) gene and tumor suppressor gene TP53 (also known as p53) alone predicted better outcomes [45].
However, another study reported that NSCLC patients on anti PD-1 immunotherapy who harbored co-mutations with STK11 and KRAS (n = 36) had longer OS in comparison to patients who harbored STK11 mutations alone (13.6 ± 3.4 months, p = 0.049, n = 37). They further investigated the population-specific factors that influenced the survival of the cohort with STK11/KRAS mutations. The group revealed that NSCLC patients with both mutations often were: (i) older at diagnosis, (ii) more likely to have received nivolumab (as compared to pembrolizumab and atezolizumab), (iii) more likely to have longer smoking histories and (iv) harboring more targetable mutations [46]. Hence, caution is recommended, since controversial results are published regarding the presence of mutations in these genes vs. the response to immunotherapy (mutations in KRAS or STK11 gene alone vs. mutations in both genes).
Interestingly, Sun et al.—who enrolled 240 patients—observed that NSCLC patients who harbored AT-rich interacting domain-containing protein 1A gene (ARID1A) mutations or AT-rich interacting domain-containing protein 1B gene (ARID1B) mutations had a beneficial response to anti PD-(L)1 immunotherapy and a prolonged PFS [47]. Other mutations also showed promising results. The OAK and POPLAR clinical trials showed that mutations in the kelch-like ECH-associated protein 1 (KEAP1) and nuclear factor erythroid-2-related factor-2 (NFE2L2) genes were associated with poorer OS and PFS (OS: HR = 1.7, p < 0.001; PFS: HR = 1.4, p < 0.001) in NSCLC patients receiving immunotherapy [48].
We found conflicting results regarding TMB, which is not a perfect biomarker to monitor NSCLC patients on immunotherapy. Furthermore, NGS is expensive and sequencing results can vary between laboratories as a function of the use of the cut-off for TMB-high, panel size, composition and bioinformatics pipelines. NGS also has a long turn-around time. Based on the challenges and the contradictory results concerning bTMB as a biomarker, further understanding is warranted before the integration of this factor into clinical practice [43,49,50].
3.2. Levels of cfDNA
Interestingly, the levels of cfDNA in the blood circulation can also be used to monitor NSCLC patients on immunotherapy. Cabel et al. investigated the role of the concentration of cfDNA in plasma samples of patients with NSCLC, melanoma or colorectal cancer, (CRC) treated with nivolumab or pembrolizumab monotherapy. They observed a significant correlation between synchronous changes in cf tumor DNA levels and tumor size, eight weeks after the first administration of immunotherapy. They reported that patients with undetectable cf tumor DNA levels at week 8 had significantly better PFS and OS than patients with persistently detectable cf tumor DNA [51]. Other research groups confirmed these findings. Giroux Leprieur et al. studied advanced NSCLC patients during nivolumab treatment. A high cf tumor DNA concentration at two months (first tumor evaluation) and an increase in concentration compared to the baseline were associated with a poor response and no long-term clinical benefit. Low cf tumor DNA concentrations at two months were associated with a long-term benefit of nivolumab [52]. Goldberg et al. reported that a drop in cf tumor DNA level was an early marker of therapeutic efficacy and a predictor for a prolonged survival in NSCLC patients treated with immune checkpoint inhibitors [53]. It is important to note that only a small number of participants (≤28 participants) were included in all three studies. Despite the fact that these data need to be confirmed in larger study cohorts, measuring the levels of cfDNA is a potential prognostic biomarker to monitor NSCLC patients on immunotherapy.
4. CTCs
CTCs are cancer cells that are detached from the primary tumor or metastatic tumors. They are part of the metastasis process [54]. By passive shedding and/or intravasation (with epithelial–mesenchymal transition), CTCs get into the bloodstream [15]. CTCs can provide transcriptomic, genomic and proteomic information. They can be observed non-clustered as a single cell, or as a cluster of multiple CTCs. These aggregates which are comprised of a minimum of two CTCs are characterized by a significantly enhanced metastatic potential in comparison to single CTCs. The presence of CTC clusters in the blood circulation is associated with a poor prognosis in NSCLC patients [55]. CTCs are less stable than cfDNA and they have short half-life (1–2.4 h). Since the proportion of CTCs in the bloodstream is very low in NSCLC patients, dedicated equipment and high expertise is needed [56]. Currently, there are several methods to detect and isolate CTCs that take different aspects into account. The CellSearch® system (Menarini), the Epic platform (Epic sciences) and the GILUPI CellCollector (GILUPI) are based on the presence of specific antigens (e.g., epithelial cell adhesion molecule (EpCAM)) on the surface of the CTCs. In addition, antigen-independent methods have also been developed to isolate CTCs based on their size and deformability, such as Parsortix (Angle), Isolation by SizE of Tumor cells or in short ISET (Rarecells Diagnostics), Vortex VTX-1 (Vortex Bioscience) and the ClearCell FX device (Biolidics) [25,57]. Interestingly, CTCs can be frequently detected in metastatic breast cancer (BC) patients. Approximately 70% of metastatic BC patients exhibit no less than one CTC per 7.5 mL of blood and 50% exhibit no less than five CTCs per 7.5 mL [58]. The FDA even approved the CellSearch® for CTC enumeration in BC patients [59]. CTCs are considered to have been derived from more than one tumor site, resulting in a better global representation of PD-L1 expression than tissue samples [57].
The studies evaluating the concordance between PD-L1 expression by CTCs and PD-L1 expression in tissue showed controversial results. Ilie et al. compared the PD-L1 status of CTCs, using the ISET method, with the PD-L1 status in tissue in NSCLC patients. In this study, PD-L1 expression on CTCs and matched tissue biopsies were well correlated (93%) [60]. Guibert et al. studied 96 NSCLC patients treated with chemotherapy followed by immunotherapy. In contrast, they found that CTCs were more frequently PD-L1-positive than tissue (83% vs. 41%). Consequently, no correlation between tissue and CTC PD-L1 expression was observed. The ISET platform was used for the isolation of CTCs. Other research groups reported similar results as Guibert and colleagues. However, different PD-L1 analyzing methods and antibodies were used [61,62,63].
Interestingly, PD-L1-positive CTCs are shown to be a promising predictive biomarker in NSCLC patients receiving immunotherapy. Guibert et al. also presented that a higher baseline PD-L1+ CTC number (≥1%) was observed in NSCLC patients who did not respond to therapy (PFS < 6 months). Importantly, the presence of pre-treatment PD-L1+ CTCs was not significantly correlated with the clinical outcomes [61]. Another study of 24 metastatic NSCLC patients on nivolumab displayed that the presence of CTCs and the expression of PD-L1 on their surface at baseline and at 3 months of treatment were associated with poor patient outcome [64].
Due to the lack of standardized methods to detect and analyze CTCs, the results of these studies must be interpreted with caution. Furthermore, issues regarding the contradictory results about the use of CTCs as a biomarker need to be addressed. Fortunately, different clinical trials are ongoing at the moment which address these challenges. The use of CTC to determine the PD-L1 status is a potential biomarker in NSCLC patients to predict their response to immunotherapy.
5. Tumor-Derived Exosomes
EVs are lipid-bound vesicles secreted by cells into the extracellular space. EVs have a half-life from approximately a number of minutes to 6 h [65]. They have a small size (30–1.000 nm), therefore isolation of the vesicles is difficult. However, several techniques allow the extraction of EVs such as: (sucrose density gradient) ultracentrifugation, microfiltration, gel filtration, density gradient centrifugation (OptiPrep) and size-exclusive chromatography. Importantly, the isolation of EVs can be time-consuming and can alter the structure of the vesicles [66]. The three main subtypes of EVs, which differ in their biogenesis, release pathways, size, content and function, are: micro vesicles, exosomes and apoptotic bodies [67]. The most studied subtype is exosomes.
Exosomes are abundant nanosized particles with lipid bilayer membranes which have a size between 50 and 150 nm. They are secreted by several cell types, including tumor, immune and lymphoid cells, or they can be derived from the stroma [68]. Exosomes released from stromal cells are able to stimulate nearby tumor cells to metastasize. They also promote tumor cell proliferation and inhibit their apoptosis. Tumor cells, as well as immune cells, secrete immunologically active exosomes that affect the antitumor activities of other immune cells, causing a favorable environment for the tumor [67].
In this review, we will focus on tumor-derived exosomes (TEX). TEX—which only account for a small number of the total amount of exosomes—are involved in the development of cancer, the formation of metastasis and disease progression. They play an important role in cell–cell communication, and it is known that TEX are associated with the development of resistance to chemotherapy. TEX also play a role in the emergence of the radiation-induced bystander effect, which is a phenomenon wherein non-targeted cells exhibit the effects of radiation [69,70]. Furthermore, TEX have the ability to inhibit immune cell proliferation. They can also induce apoptosis (or suppression) of immune cells such as CD8+ T cells. Hence, TEX influence the sensitivity of tumor cells to immunotherapy [25,67,71].
In this context, a few research groups investigated the role that TEX might play in the selection of NSCLC patients who might benefit from immunotherapy. A study of 24 patients with lung cancer before surgery presented a correlation between the number of plasma-derived PD-L1+ exosomes and the PD-L1 expression level in the tumor tissue [72]. In contrast, Li et al., who enrolled 85 NSCLC patients, did not find a correlation between the PD-L1 status in exosomes versus in tumor tissue [73]. Gunasekaran et al. presented that PD-L1-positive TEX can be used as a predictive biomarker for NSCLC patients who are treated with immunotherapy. They observed a trend towards PD-L1 reduction in patients who responded to immunotherapy. However, this difference was not statistically significant. Interestingly, they suggested that the dynamic changes of exosomal PD-L1 (pre-treatment vs. 8 weeks of treatment) predicted the clinical outcome in terms of both PFS and OS in patients treated with immunotherapy. Again, here, a limitation of the study was that only 25 NSCLC patients were included [74].
Several challenges should be overcome to make the implementation of EVs in the clinical practice feasible. The first and foremost challenge consists of the standardization of the methodologies for EV isolation and purification [71]. Nevertheless, evaluating the PD-L1 status based on exosomes is a promising biomarker for the selection of NSCLC patients that may benefit from immunotherapy.
6. Epigenetics and microRNAs
The interest in using epigenetic-based markers and miRNAs to monitor NSCLC patients on immunotherapy has recently started to grow. Epigenetic components such as DNA methylation, histone modifications, nucleosome positioning and non-coding RNAs, specifically miRNAs, largely influence tumor cells’ functioning [75].
Hypermethylation (of tumor suppressor genes) was shown to contribute to carcinogenesis. One of the main advantages of DNA methylation alterations, compared to other potential diagnostic biomarkers, is that they are remarkably stable, and generally occur early during carcinogenesis. To obtain methylated DNA, first, cfDNA isolation needs to be performed, followed by methylation-specific PCR or BEAMing [76].
Different research groups analyzed the possibility of using methylation signatures or patterns as biomarkers to monitor NSCLC patients treated with immunotherapy. Druisseaux et al. established an epigenomic profile based on a microarray DNA methylation signature (EPIMMUNE) in a set of tissue biopsies from IV stage NSCLC patients treated with nivolumab or pembrolizumab. They showed that the EPIMMUNE signature was associated with improved PFS and OS. However, no association was found between the EPIMMUNE signature and PD-L1 expression [77]. Cho et al. confirmed that methylation patterns could be used for the monitoring of NSCLC patients during therapy. They studied differentially methylated regions overlapping promoters (pDMRs) or enhancers (eDMRs) in tissue biopsies between responders and non-responders to immunotherapy. Interestingly, they identified 1.007 pDMRs and 607 eDMRs which were associated with the anti-PD-1 response [78,79].
In this context, we will highlight specific genes that might be a promising biomarker to monitor NSCLC patients treated with immunotherapy. First, the methylation status of the transcription factor T cell-related forkhead box P1 (FOXP1) was significantly associated with improved PFS and OS in NSCLC patients following immunotherapy [77]. Furthermore, hypomethylated pDMRs of the Cytohesin 1 Interacting Protein (CYTIP) and the TNF superfamily member 8 (TNFSF8) could predict response in NSCLC patients on anti-PD-1 therapy. In fact, the authors even suggested that their ability to predict the clinical outcome was superior to that of the commonly used biomarker PD-L1 [78,79].
miRNAs are small, single-stranded non-coding RNA sequences of approximately 18–22 nucleotides that are regulators of gene expression, that can be derived from either cancer cells or immune cells [80,81]. They are known to have important roles at post-transcriptional and translational levels [82]. Analysis of circulating miRNAs can be accomplished with quantitative real time-PCR (RT-PRCR), microarray or deep sequencing after initial ultracentrifugation [83].
Studies revealed that miRNAs play an important role in the regulation of PD-L1. Cortez et al. found that PD-L1 was regulated by p53 via miR-34 which regulated (in)directly the expression of several immune checkpoints [84]. Furthermore, miRNAs were associated with the clinical outcome of NSCLC patients who were treated with immunotherapy. Fan et al. presented that NSCLC patients who responded to nivolumab (n = 17) had long-term increased expression levels of miR-93, −138-5p, −200, −27a, −424, −34a, −28, −106b, −193a-3p and −181a from pre-treatment to post-treatment in their serum versus non-responders (n = 17). They reported that the high expression of these ten miRNA patterns was significantly correlated with an improvement in OS and PFS [82]. Another study of Boeri et al. displayed that a miRNA signature classifier (MSC) composed of 24 miRNAs could distinguish NSCLC patients (n = 140) that would benefit from anti PD-L1 immunotherapy from patients who would not. The MSC risk level was associated with the overall response rate (ORR (p = 0.0009), PFS (multivariate HR = 0.31; 95% CI: 0.17–0.56; p = 0.0001) and OS (multivariate HR = 0.33; 95% CI: 0.18–0.59; p = 0.0002)) [85]. In this light, Epigenomics AG developed two liquid biopsy-based tests to evaluate the methylation status of certain genes for early detection of CRC or lung cancer. The Epi proLung assay can be used to detect lung cancer by the determination of the methylation status of the short stature homeobox 2 (SHOX2) and the prostaglandin E receptor 4 (PTGER4) gene [86]. The Epi proColon assay, which detects the methylation patterns of the gene septin 9 (SEPT9) was approved by the FDA for CRC screening [87].
Despite the fact that a few technical issues need to be resolved to increase the sensitivity of this potential biomarker, the use of epigenetics and miRNAs is a promising tool to monitor NSCLC patients following immunotherapy treatment. Fortunately, several sensitive detection platforms are already commercially available.
7. T Cell Receptor Repertoire
T cell receptors (TCRs) are antigen-specific receptors that are present at the cell surface of T lymphocytes and play an important role in the immune response [41]. Isolation of T cells from patients’ blood can easily be done by density centrifugation. The biggest advantage of this technique is that T cells are still in a fully functional state after isolation. Afterwards, flow cytometry is often performed to select CD4+ and CD8+ cells. In the majority of T cells, the TCR consists of one alpha and one beta chain. Each chain contains three complementarity-determining regions (CDRs) which are hypervariable regions. The CDR3 region variability is generated by genetic recombination and is consequently unique to each TCR. Sequencing this specific region allows the determination of the TCR repertoire which contains a diversity of certain T cell clones that are responsible for anti-tumor immunity [57,88,89]. Characterization of these specific T cell clones may lead to future studies which may expand these clones in a personalized approach to potentially lengthen the duration of the sustained response to immunotherapy in the patient [90].
The possibility to use the clonality and the diversity of the TCR repertoire as a prognostic biomarker for NSCLC patients on immunotherapy was studied by several research groups. Han et al. collected blood samples from 40 NSCLC patients receiving anti PD-(L)1 immunotherapy. They observed that NSCLC patients with a high PD-1-positive CD8+ TCR diversity before treatment had a better clinical outcome in comparison to patients with a low diversity (6.4 versus 2.5 months). In addition, patients with an increased PD-1-positive CD8+ TCR clonality after immunotherapy showed longer PFS than patients with decreased clonality (7.3 versus 2.6 months [91]. In 2020, Yamauchi et al. focused on the role of the CX3C chemokine receptor 1 (CX3CR1)—which is a marker of T cell differentiation—in predicting the response to immunotherapy. They presented that the TCR frequency and clonality of the peripheral CX3CR1-positive CD8+ T cell subset (which included an enriched repertoire of tumor-specific and tumor-infiltrating CD8+ T cells) was increased in tumor-bearing mice who responded to immunotherapy. Furthermore, they observed a correlation between a positive clinical outcome and an increase in the frequency of the CX3CR1-positive subset in circulating CD8+ T cells based on the analysis of peripheral blood mononuclear cell (PBMC) samples from 36 NSCLC patients treated with nivolumab or pembrolizumab [92].
However, a few hurdles need to be resolved before the implementation in clinic is feasible. Challenges regarding the high rate of false positives, caused by the clustering of functionally different clones or the presence of artificial clones, need to be addressed. Nevertheless, TCR diversity and clonality might serve as a biomarker to monitor the response of NSCLC patients on immunotherapy [91].
8. Gut Metabolism
Feces are gaining a more profound role in the liquid biopsy field. Specifically, the interest in using the gut microbiota as a biomarker to monitor NSCLC patients treated with immunotherapy has started to grow. Even though it can be questioned whether feces are a body “liquid”, we will consider it as a liquid biopsy in concurrence with other publications [93,94]. Feces can be used to study the gut microbiota, which is composed of bacteria, protists, fungi and viruses. Interestingly, it appears that metabolic changes occurring in the gut microbiota metabolome are associated with the response to immunotherapy in NSCLC [95].
Botticelli et al. characterized the metabolomic profiling of the gut microbiota of eleven NSCLC patients receiving nivolumab. They reported that some gut microbiota, such as 2-pentanone (ketone) and tridecane (alkane), were significantly associated with early progression. In contrast, short-chain fatty acids (i.e., propionate, butyrate), lysine and nicotinic acid were significantly associated with a long-term beneficial outcome [96]. Jin et al. revealed that the diversity of the microbiome also affected the clinical outcome on immunotherapy. The research group enrolled 37 Chinese patients with advanced NSCLC treated with nivolumab. They showed that patients with a high microbiome diversity had a significantly prolonged PFS in comparison to patients with a low diversity. Furthermore, they displayed that patients with high microbiome diversity exhibited enhanced T cell memory and natural killer cell signatures in response to anti–PD-1 therapy [97]. The gut microbiota even might play a role in the development of resistance to immunotherapy. The study of Routy et al. discovered an association between a beneficial clinical response on immunotherapy and the relative abundance of Akkermansia muciniphila. These results were based on metagenomic analyses of patient stool samples that were taken at diagnosis [98]. Akkermansia muciniphila is a well-studied anaerobic bacterium which is specialized in mucus degradation and is associated with human health [99]. To validate these results, fecal microbiota transplantations (FTMs) were performed in mice. The fecal microbiota from cancer patients who developed resistance to immunotherapy was transplanted into germ-free or antibiotic-treated mice. After the FTM, mice were treated with an oral supplementation of Akkermansia muciniphila, re-establishing the response to PD-1 based immunotherapy. We also want to highlight that antibiotics can inhibit the clinical benefit of immunotherapy in advanced cancer patients [98]. Researchers treated patients (n = 121 renal cell carcinoma (RCC) and 238 NSCLC patients) with antibiotics 30 or 60 days before the start of immunotherapy. In RCC patients, the use of antibiotics was associated with an increased risk of primary progressive disease (PD) (75 versus 22%, p < 0.01), shorter PFS (median 1.9 versus 7.4 months) and shorter OS (median 17.3 versus 30.6 months) versus no use of antibiotics. In NSCLC patients, the use of antibiotics was also associated with primary PD (52% versus 43%), but decreased PFS (median 1.9 versus 3.8 months) and OS (median 7.9 versus 24.6 months). The data suggest that the modulation of antibiotic-related gut microbiota composition may be a strategy to improve clinical outcomes with immunotherapy [100].
However, confirmation of these promising data in larger study cohorts, as well as more insight and knowledge to overcome some (technical) challenges, are needed [101]. Despite the fact that the study of the microbiota is still in its infancy, it is a biomarker with great potential to monitor NSCLC patients receiving immunotherapy.
9. The Electronic Nose
Recently, studying air as a non-invasive biomarker in cancer patients has gained more attention. Again, here, consistent with other liquid biopsy-related publications, exhaled air will be considered as a “liquid” biopsy. Exhaled breath consists of thousands of volatile organic compounds (VOCs). VOCs were identified in 1970 and since then, breath analysis has transformed from a relatively unknown area to a high-throughput breath omics research field [102]. Nowadays, more than 3000 different VOCs in human breath have been identified [102,103,104]. These volatile compounds are produced by several metabolic processes within the human body. Since these processes can be induced by or altered due to disease, it is believed that VOCs cause a specific “breath print” for different diseases. The electronic nose (eNose) was developed, aiming to detect those different breath prints or VOC patterns [104,105].
In 2019, de Vries et al. studied the role of breath to distinguish non-responders from responders to nivolumab or pembrolizumab in NSCLC patients. They collected the exhaled breath data of 143 NSCLC patients at baseline. Samples were taken by a metal oxide semiconductor eNose. They reported that the eNose contributed significantly at baseline in differentiating between patients who responded and who did not at 3 months of anti PD-1 treatment. This study revealed that the eNose could be used to predict individual patient response to immunotherapy [106]. Furthermore, a clinical trial (NCT04146064) is ongoing which addresses breath print analysis as a potential predicting factor for response to immunotherapy. In this trial, 425 participants (with NSCLC, melanoma kidney cancer, urothelial carcinoma (UC) and head and neck cancer (H&NC)) will be included.
Currently, some challenges prevent this technique from being widespread in clinical practice, such as the stability of the VOCs. Furthermore, the lack of standardized sampling and analysis methods needs to be addressed to implement this technique in clinic. After resolving these challenges, it seems that eNose will find its way to routine practice [102].
10. Conclusions and Future Perspectives
Recently, the introduction of immune checkpoint inhibitor agents targeting PD-1 or PD-L1 has radically modified lung cancer care. Since immunotherapy preferentially targets cancer cells it causes less side effects than traditional chemotherapy. However, only ~20% of the NSCLC patients benefit from it. Therefore, it is crucial to identify: (i) predictive biomarkers in order to select patients who will benefit from immunotherapy and (ii) prognostic biomarkers to monitor patients during therapy, which enables the rapid detection of treatment resistance. Currently, PD-L1 tissue testing is used for the selection of responders to immunotherapy, and imaging is used to monitor patients during their disease course. Unfortunately, obtaining a tissue biopsy is not always feasible due to, e.g., the location of the tumor, and the amount of CT scans per patient each year is limited.
Liquid biopsy, or the tumor-derived material present in body fluids, may overcome these difficulties. The biggest advantage of this technique in comparison to imaging is its minimally invasive character, which allows repetitive sampling. Furthermore, a liquid biopsy sample can be obtained at almost any moment (regardless of, e.g., the location of the tumor or the health status of the patient) which is an advantage compared to the gold standard PD-L1.
In this review we discussed liquid biopsy-based biomarkers that can be used to select NSCLC patients who will benefit from immunotherapy, such as PD-L1-positive CTCs and exomes. Next, we also highlighted minimally invasive prognostic biomarkers, such as: cf tumor DNA (bTMB and cf tumor DNA levels), methylation signatures, miRNAs, the TCR repertoire, the gut microbiota and VOCs. They are likely to eventually be translated to the clinic in the future. Nevertheless, despite its potential, liquid biopsy is still hampered by some limitations. The biggest bottleneck in most studies is the small sample size. Promising biomarkers should be validated in larger patient cohorts. Secondly, liquid biopsy is limited by the lack of standardization and the absence of broadly accepted standard operating procedures. Lastly, detection problems that arise due to the low abundance of most liquid biopsy compounds need to be resolved. Ongoing clinical trials in NSCLC patients on immunotherapy address these challenges and will give us more insight on the future of liquid biopsy as a biomarker. In summary, nowadays, liquid biopsy is a great additional tool for NSCLC patients on immunotherapy, next to the existing techniques (imaging and PD-L1 tissue testing). However, we hope that one day liquid biopsy can replace the current techniques in order to improve the patients comfort and quality of life.
Acknowledgments
The authors would like to thank Lauren Augustus to create the illustration of this manuscript (Figure 1).
Abbreviations
| (b)TMB | (blood) tumor mutational burden |
| NFE2L2 | nuclear factor erythroid-2-related factor-2 |
| ARID1A | AT-rich interacting domain-containing protein 1A gene |
| ARID1B | AT-rich interacting domain-containing protein 1B gene |
| BC | breast cancer |
| BEAMing | beads, emulsions, amplification and magnetics |
| CDRs | complementarity-determining regions |
| cf tumor DNA | circulating cell-free tumor DNA |
| cfDNA | circulating cell-free DNA |
| CGP | comprehensive genomic profiling |
| CI | confidence interval |
| CRC | colorectal cancer |
| CTCs | circulating tumor cells |
| CX3CR1 | CX3C chemokine receptor 1 |
| CYTIP | Cytohesin 1 Interacting Protein |
| ddPCR | digital droplet PCR |
| eDMRs | differentially methylated regions overlapping enhancers |
| EGFR | epidermal growth factor receptor |
| eNose | electronic nose |
| EpCAM | epithelial cell adhesion molecule |
| EVs | extracellular vesicles |
| FDA | food and drug administration |
| FMTs | fecal microbiota transplantations |
| FOXP1 | forkhead box P1 |
| gDNA | genomic DNA |
| H&NC | head and neck cancer |
| HR | hazard ratio |
| ISET | Isolation by SizE of Tumor cells |
| KEAP1 | kelch-like ECH-associated protein 1 |
| KRAS | Ki-ras2 Kirsten rat sarcoma viral oncogene homolog |
| Mb | mega base |
| miRNA | microRNA |
| MSC | miRNA signature classifier |
| NGS | next-generation sequencing |
| NSCLC | non-small cell lung cancer |
| ORR | overall response rate |
| OS | overall survival |
| PBMC | peripheral blood mononuclear cell |
| PD | progressive disease |
| PD-1 | programmed cell death protein 1 |
| PD-L1 | programmed cell death ligand 1 |
| pDMRs | differentially methylated regions overlapping promoters |
| PFS | progression-free survival |
| PTEN | phosphatase and TENsin homolog |
| PTGER4 | prostaglandin E receptor 4 |
| RCC | renal cell carcinoma |
| RECIST | response evaluation criteria in solid tumors |
| RT-PCR | real time-PCR |
| SEPT9 | septin 9 |
| SHOX2 | short stature homeobox 2 |
| STK11 | serine/threonine kinase 11 |
| TCR | T cell receptor |
| TEX | tumor-derived exosomes |
| tTMB | tissue tumor mutational burden |
| TNFSF8 | TNF superfamily member 8 |
| UC | urotherial carcininoma |
| VOCs | volatile organic compounds |
| WES | whole exome sequencing |
Author Contributions
Conceptualization, E.A., K.Z. and P.P.; methodology, E.A. and K.Z.; software, E.A. and K.Z.; validation, E.A. and K.Z.; formal analysis, E.A. and K.Z.; investigation, E.A. and K.Z.; resources, E.A. and K.Z.; data curation, E.A. and K.Z.; writing—original draft preparation, E.A.; writing—review and editing, E.A., K.Z., V.S., J.R., S.J., G.B., G.R., M.P. and P.P.; visualization, E.A.; supervision, K.Z. and P.P.; project administration, E.A. and K.Z.; funding acquisition, K.Z., L.S. and P.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Stand up To Cancer (Kom Op Tegen Kanker), the Flemish Cancer Society (Grant Number: 00000000116000000177). P.P. received research grants and/or consulting fees from AstraZeneca, Boehringer-Ingelheim, BMS, MSD, Pfizer and Roche.
Data Availability Statement
The data presented in this review are openly available, references are listed below.
Conflicts of Interest
The authors declare no potential conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Tun A.M., Thein K.Z., Thein W.L., Guevara E. Checkpoint inhibitors plus chemotherapy for first-line treatment of advanced non-small cell lung cancer: A systematic review and meta-analysis of randomized controlled trials. Future Sci. OA. 2019;5:FSO421. doi: 10.2144/fsoa-2019-0081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rijavec E., Coco S., Genova C., Rossi G., Longo L., Grossi F. Liquid Biopsy in Non-Small Cell Lung Cancer: Highlights and Challenges. Cancers. 2019;12:17. doi: 10.3390/cancers12010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doroshow D.B., Herbst R.S. Treatment of Advanced Non-Small Cell Lung Cancer in 2018. JAMA Oncol. 2018;4:569–570. doi: 10.1001/jamaoncol.2017.5190. [DOI] [PubMed] [Google Scholar]
- 5.Yuan M., Huang L.L., Chen J.H., Wu J., Xu Q. The emerging treatment landscape of targeted therapy in non-small-cell lung cancer. Signal. Transduct. Target. Ther. 2019;4:61. doi: 10.1038/s41392-019-0099-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu M., Guo F. Recent updates on cancer immunotherapy. Precis. Clin. Med. 2018;1:65–74. doi: 10.1093/pcmedi/pby011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aguiar P.N., Jr., Perry L.A., Penny-Dimri J., Babiker H., Tadokoro H., de Mello R.A., Lopes G.L., Jr. The effect of PD-L1 testing on the cost-effectiveness and economic impact of immune checkpoint inhibitors for the second-line treatment of NSCLC. Ann. Oncol. 2018;29:1078. doi: 10.1093/annonc/mdx478. [DOI] [PubMed] [Google Scholar]
- 8.Kim J.Y., Choi J.K., Jung H. Genome-wide methylation patterns predict clinical benefit of immunotherapy in lung cancer. Clin. Epigenet. 2020;12:119. doi: 10.1186/s13148-020-00907-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duffy M.J., Crown J. Biomarkers for Predicting Response to Immunotherapy with Immune Checkpoint Inhibitors in Cancer Patients. Clin. Chem. 2019;65:1228–1238. doi: 10.1373/clinchem.2019.303644. [DOI] [PubMed] [Google Scholar]
- 10.Bailly C. Combined cytotoxic chemotherapy and immunotherapy of cancer: Modern times. NAR Cancer. 2020;2:zcaa002. doi: 10.1093/narcan/zcaa002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sui H., Ma N., Wang Y., Li H., Liu X., Su Y., Yang J. Anti-PD-1/PD-L1 Therapy for Non-Small-Cell Lung Cancer: Toward Personalized Medicine and Combination Strategies. J. Immunol. Res. 2018;2018:6984948. doi: 10.1155/2018/6984948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muinelo-Romay L., Garcia-Gonzalez J., Leon-Mateos L. Lung Cancer and Liquid Biopsy: Realities and Challenges in Routine Clinical Practice. Arch. Bronconeumol. 2019;55:289–290. doi: 10.1016/j.arbres.2018.11.011. [DOI] [PubMed] [Google Scholar]
- 13.Kwapisz D. The first liquid biopsy test approved. Is it a new era of mutation testing for non-small cell lung cancer? Ann. Transl. Med. 2017;5:46. doi: 10.21037/atm.2017.01.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pakkala S., Ramalingam S.S. Personalized therapy for lung cancer: Striking a moving target. JCI Insight. 2018;3 doi: 10.1172/jci.insight.120858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Rubis G., Rajeev Krishnan S., Bebawy M. Liquid Biopsies in Cancer Diagnosis, Monitoring, and Prognosis. Trends Pharmacol. Sci. 2019;40:172–186. doi: 10.1016/j.tips.2019.01.006. [DOI] [PubMed] [Google Scholar]
- 16.Overman M.J., Modak J., Kopetz S., Murthy R., Yao J.C., Hicks M.E., Abbruzzese J.L., Tam A.L. Use of research biopsies in clinical trials: Are risks and benefits adequately discussed? J. Clin. Oncol. 2013;31:17–22. doi: 10.1200/JCO.2012.43.1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Diaz L.A., Jr., Bardelli A. Liquid biopsies: Genotyping circulating tumor DNA. J. Clin. Oncol. 2014;32:579–586. doi: 10.1200/JCO.2012.45.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herbreteau G., Vallee A., Charpentier S., Normanno N., Hofman P., Denis M.G. Circulating free tumor DNA in non-small cell lung cancer (NSCLC): Clinical application and future perspectives. J. Thorac. Dis. 2019;11:S113–S126. doi: 10.21037/jtd.2018.12.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vrankar M., Unk M. Immune RECIST criteria and symptomatic pseudoprogression in non-small cell lung cancer patients treated with immunotherapy. Radiol. Oncol. 2018;52:365–369. doi: 10.2478/raon-2018-0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Delft F., Koffijberg H., Retel V., Heuvel M.V.D., IJzerman M. The Validity and Predictive Value of Blood-Based Biomarkers in Prediction of Response in the Treatment of Metastatic Non-Small Cell Lung Cancer: A Systematic Review. Cancers. 2020;12:1120. doi: 10.3390/cancers12051120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Augustus E., Van Casteren K., Sorber L., van Dam P., Roeyen G., Peeters M., Vorsters A., Wouters A., Raskin J., Rolfo C., et al. The art of obtaining a high yield of cell-free DNA from urine. PLoS ONE. 2020;15:e0231058. doi: 10.1371/journal.pone.0231058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Babayan A., Pantel K. Advances in liquid biopsy approaches for early detection and monitoring of cancer. Genome Med. 2018;10:21. doi: 10.1186/s13073-018-0533-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Castro-Giner F., Gkountela S., Donato C., Alborelli I., Quagliata L., Ng C.K.Y., Piscuoglio S., Aceto N. Cancer Diagnosis Using a Liquid Biopsy: Challenges and Expectations. Diagnostics. 2018;8:31. doi: 10.3390/diagnostics8020031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grolz D., Hauch S., Schlumpberger M., Guenther K., Voss T., Sprenger-Haussels M., Oelmuller U. Liquid Biopsy Preservation Solutions for Standardized Pre-Analytical Workflows-Venous Whole Blood and Plasma. Curr. Pathobiol. Rep. 2018;6:275–286. doi: 10.1007/s40139-018-0180-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brozos-Vazquez E.M., Diaz-Pena R., Garcia-Gonzalez J., Leon-Mateos L., Mondelo-Macia P., Pena-Chilet M., Lopez-Lopez R. Immunotherapy in nonsmall-cell lung cancer: Current status and future prospects for liquid biopsy. Cancer Immunol. Immunother. 2020 doi: 10.1007/s00262-020-02752-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Siravegna G., Mussolin B., Venesio T., Marsoni S., Seoane J., Dive C., Papadopoulos N., Kopetz S., Corcoran R.B., Siu L.L., et al. How liquid biopsies can change clinical practice in oncology. Ann. Oncol. 2019;30:1580–1590. doi: 10.1093/annonc/mdz227. [DOI] [PubMed] [Google Scholar]
- 27.Elazezy M., Joosse S.A. Techniques of using circulating tumor DNA as a liquid biopsy component in cancer management. Comput. Struct. Biotechnol. J. 2018;16:370–378. doi: 10.1016/j.csbj.2018.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Diefenbach R.J., Lee J.H., Kefford R.F., Rizos H. Evaluation of commercial kits for purification of circulating free DNA. Cancer Genet. 2018;228:21–27. doi: 10.1016/j.cancergen.2018.08.005. [DOI] [PubMed] [Google Scholar]
- 29.Heeke S., Hofman P. Tumor mutational burden assessment as a predictive biomarker for immunotherapy in lung cancer patients: Getting ready for prime-time or not? Transl. Lung Cancer Res. 2018;7:631–638. doi: 10.21037/tlcr.2018.08.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Galuppini F., Dal Pozzo C.A., Deckert J., Loupakis F., Fassan M., Baffa R. Tumor mutation burden: From comprehensive mutational screening to the clinic. Cancer Cell Int. 2019;19:209. doi: 10.1186/s12935-019-0929-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Romero D. Immunotherapy: Relying on quality over quantity. Nat. Rev. Clin. Oncol. 2018;15:6–7. doi: 10.1038/nrclinonc.2017.189. [DOI] [PubMed] [Google Scholar]
- 32.Dolgin E. Neoantigen Quality Predicts Immune Response, Survival. Cancer Discov. 2018;8:6. doi: 10.1158/2159-8290.CD-NB2017-165. [DOI] [PubMed] [Google Scholar]
- 33.Lawrence M.S., Stojanov P., Polak P., Kryukov G.V., Cibulskis K., Sivachenko A., Carter S.L., Stewart C., Mermel C.H., Roberts S.A., et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alexandrov L.B., Ju Y.S., Haase K., Van Loo P., Martincorena I., Nik-Zainal S., Totoki Y., Fujimoto A., Nakagawa H., Shibata T., et al. Mutational signatures associated with tobacco smoking in human cancer. Science. 2016;354:618–622. doi: 10.1126/science.aag0299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berland L., Heeke S., Humbert O., Macocco A., Long-Mira E., Lassalle S., Lespinet-Fabre V., Lalvee S., Bordone O., Cohen C., et al. Current views on tumor mutational burden in patients with non-small cell lung cancer treated by immune checkpoint inhibitors. J. Thorac. Dis. 2019;11:S71–S80. doi: 10.21037/jtd.2018.11.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fang W., Ma Y., Yin J.C., Hong S., Zhou H., Wang A., Wang F., Bao H., Wu X., Yang Y., et al. Comprehensive Genomic Profiling Identifies Novel Genetic Predictors of Response to Anti-PD-(L)1 Therapies in Non-Small Cell Lung Cancer. Clin. Cancer Res. 2019;25:5015–5026. doi: 10.1158/1078-0432.CCR-19-0585. [DOI] [PubMed] [Google Scholar]
- 37.Xiao D., Pan H., Li F., Wu K., Zhang X., He J. Analysis of ultra-deep targeted sequencing reveals mutation burden is associated with gender and clinical outcome in lung adenocarcinoma. Oncotarget. 2016;7:22857–22864. doi: 10.18632/oncotarget.8213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Herbst R.S. Association between tissue TMB (tTMB) and clinical outcomes with pembrolizumab monotherapy (pembro) in PD-L1-positive advanced NSCLC in the KEYNOTE-010 and -042 trials. Ann. Oncol. 2019;30:v916–v917. doi: 10.1093/annonc/mdz394.077. [DOI] [Google Scholar]
- 39.Wang Z., Duan J., Cai S., Han M., Dong H., Zhao J., Zhu B., Wang S., Zhuo M., Sun J., et al. Assessment of Blood Tumor Mutational Burden as a Potential Biomarker for Immunotherapy in Patients With Non-Small Cell Lung Cancer With Use of a Next-Generation Sequencing Cancer Gene Panel. JAMA Oncol. 2019;5:696–702. doi: 10.1001/jamaoncol.2018.7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gandara D.R., Paul S.M., Kowanetz M., Schleifman E., Zou W., Li Y., Rittmeyer A., Fehrenbacher L., Otto G., Malboeuf C., et al. Blood-based tumor mutational burden as a predictor of clinical benefit in non-small-cell lung cancer patients treated with atezolizumab. Nat. Med. 2018;24:1441–1448. doi: 10.1038/s41591-018-0134-3. [DOI] [PubMed] [Google Scholar]
- 41.Sesma A., Pardo J., Cruellas M., Galvez E.M., Gascon M., Isla D., Martinez-Lostao L., Ocariz M., Pano J.R., Quilez E., et al. From Tumor Mutational Burden to Blood T Cell Receptor: Looking for the Best Predictive Biomarker in Lung Cancer Treated with Immunotherapy. Cancers. 2020;12:2974. doi: 10.3390/cancers12102974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qiu P., Poehlein C.H., Marton M.J., Laterza O.F., Levitan D. Measuring Tumor Mutational Burden (TMB) in Plasma from mCRPC Patients Using Two Commercial NGS Assays. Sci. Rep. 2019;9:114. doi: 10.1038/s41598-018-37128-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hellmann M.D., Paz-Ares L., Bernabe Caro R., Zurawski B., Kim S.W., Carcereny Costa E., Park K., Alexandru A., Lupinacci L., de la Mora Jimenez E., et al. Nivolumab plus Ipilimumab in Advanced Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2019;381:2020–2031. doi: 10.1056/NEJMoa1910231. [DOI] [PubMed] [Google Scholar]
- 44.Paz-Ares L. Pembrolizumab (pembro) plus platinum-based chemotherapy (chemo) for metastatic NSCLC: Tissue TMB (tTMB) and outcomes in KEYNOTE-021, 189, and 407. Ann. Oncol. 2019;30:v917–v918. doi: 10.1093/annonc/mdz394.078. [DOI] [Google Scholar]
- 45.Guibert N., Jones G., Beeler J.F., Plagnol V., Morris C., Mourlanette J., Delaunay M., Keller L., Rouquette I., Favre G., et al. Targeted sequencing of plasma cell-free DNA to predict response to PD1 inhibitors in advanced non-small cell lung cancer. Lung Cancer. 2019;137:1–6. doi: 10.1016/j.lungcan.2019.09.005. [DOI] [PubMed] [Google Scholar]
- 46.Basher F., Saravia D., Fanfan D., Cotta J.A., Lopes G. Impact of STK11 and KRAS co-mutations on outcomes with immunotherapy in non-small cell lung cancer. J. Clin. Oncol. 2020;38 doi: 10.1200/JCO.2020.38.15_suppl.e15135. [DOI] [Google Scholar]
- 47.Sun D., Tian L., Zhu Y., Wo Y., Liu Q., Liu S., Li H., Hou H. Subunits of ARID1 serve as novel biomarkers for the sensitivity to immune checkpoint inhibitors and prognosis of advanced non-small cell lung cancer. Mol. Med. 2020;26:78. doi: 10.1186/s10020-020-00208-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu H., Yu Y., Zheng Y., Xu B., Zheng S., Zeng F., Xie W., Huang L., Li F., Lin W., et al. KEAP1/NFE2L2 as a prognostic biomarker on immunotherapy and correlation with immune infiltrates in non-small cell lung cancer (NSCLC) J. Clin. Oncol. 2020;38 doi: 10.1200/JCO.2020.38.15_suppl.e21551. [DOI] [Google Scholar]
- 49.Sholl L.M., Hirsch F.R., Hwang D., Botling J., Lopez-Rios F., Bubendorf L., Mino-Kenudson M., Roden A.C., Beasley M.B., Borczuk A., et al. The Promises and Challenges of Tumor Mutation Burden as an Immunotherapy Biomarker: A Perspective from the International Association for the Study of Lung Cancer Pathology Committee. J. Thorac. Oncol. 2020;15:1409–1424. doi: 10.1016/j.jtho.2020.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fancello L., Gandini S., Pelicci P.G., Mazzarella L. Tumor mutational burden quantification from targeted gene panels: Major advancements and challenges. J. Immunother. Cancer. 2019;7:183. doi: 10.1186/s40425-019-0647-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cabel L., Riva F., Servois V., Livartowski A., Daniel C., Rampanou A., Lantz O., Romano E., Milder M., Buecher B., et al. Circulating tumor DNA changes for early monitoring of anti-PD1 immunotherapy: A proof-of-concept study. Ann. Oncol. 2017;28:1996–2001. doi: 10.1093/annonc/mdx212. [DOI] [PubMed] [Google Scholar]
- 52.Giroux Leprieur E., Herbretau G., Dumenil C., Julie C., Giraud V., Labrune S., Dumoulin J., Tisserand J., Emile J.F., Blons H., et al. Circulating tumor DNA evaluated by Next-Generation Sequencing is predictive of tumor response and prolonged clinical benefit with nivolumab in advanced non-small cell lung cancer. Oncoimmunology. 2018;7:e1424675. doi: 10.1080/2162402X.2018.1424675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Goldberg S.B., Narayan A., Kole A.J., Decker R.H., Teysir J., Carriero N.J., Lee A., Nemati R., Nath S.K., Mane S.M., et al. Early Assessment of Lung Cancer Immunotherapy Response via Circulating Tumor DNA. Clin. Cancer Res. 2018;24:1872–1880. doi: 10.1158/1078-0432.CCR-17-1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shen Z., Wu A., Chen X. Current detection technologies for circulating tumor cells. Chem. Soc. Rev. 2017;46:2038–2056. doi: 10.1039/C6CS00803H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Manjunath Y., Upparahalli S.V., Suvilesh K.N., Avella D.M., Kimchi E.T., Staveley-O’Carroll K.F., Li G., Kaifi J.T. Circulating tumor cell clusters are a potential biomarker for detection of non-small cell lung cancer. Lung Cancer. 2019;134:147–150. doi: 10.1016/j.lungcan.2019.06.016. [DOI] [PubMed] [Google Scholar]
- 56.Kapeleris J., Kulasinghe A., Warkiani M.E., Vela I., Kenny L., O’Byrne K., Punyadeera C. The Prognostic Role of Circulating Tumor Cells (CTCs) in Lung Cancer. Front. Oncol. 2018;8:311. doi: 10.3389/fonc.2018.00311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hofman P., Heeke S., Alix-Panabieres C., Pantel K. Liquid biopsy in the era of immuno-oncology: Is it ready for prime-time use for cancer patients? Ann. Oncol. 2019;30:1448–1459. doi: 10.1093/annonc/mdz196. [DOI] [PubMed] [Google Scholar]
- 58.Thery L., Meddis A., Cabel L., Proudhon C., Latouche A., Pierga J.Y., Bidard F.C. Circulating Tumor Cells in Early Breast Cancer. JNCI Cancer Spectr. 2019;3:pkz026. doi: 10.1093/jncics/pkz026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Moussavi-Harami S.F., Wisinski K.B., Beebe D.J. Circulating Tumor Cells in Metastatic Breast Cancer: A Prognostic and Predictive Marker. J. Patient Cent. Res. Rev. 2014;1:85–92. doi: 10.17294/2330-0698.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ilie M., Szafer-Glusman E., Hofman V., Chamorey E., Lalvee S., Selva E., Leroy S., Marquette C.H., Kowanetz M., Hedge P., et al. Detection of PD-L1 in circulating tumor cells and white blood cells from patients with advanced non-small-cell lung cancer. Ann. Oncol. 2018;29:193–199. doi: 10.1093/annonc/mdx636. [DOI] [PubMed] [Google Scholar]
- 61.Guibert N., Delaunay M., Lusque A., Boubekeur N., Rouquette I., Clermont E., Mourlanette J., Gouin S., Dormoy I., Favre G., et al. PD-L1 expression in circulating tumor cells of advanced non-small cell lung cancer patients treated with nivolumab. Lung Cancer. 2018;120:108–112. doi: 10.1016/j.lungcan.2018.04.001. [DOI] [PubMed] [Google Scholar]
- 62.Janning M., Kobus F., Babayan A., Wikman H., Velthaus J.L., Bergmann S., Schatz S., Falk M., Berger L.A., Bottcher L.M., et al. Determination of PD-L1 Expression in Circulating Tumor Cells of NSCLC Patients and Correlation with Response to PD-1/PD-L1 Inhibitors. Cancers. 2019;11:835. doi: 10.3390/cancers11060835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Koh Y., Yagi S., Akamatsu H., Kanai K., Hayata A., Tokudome N., Akamatsu K., Higuchi M., Kanbara H., Nakanishi M., et al. Heterogeneous Expression of Programmed Death Receptor-ligand 1 on Circulating Tumor Cells in Patients With Lung Cancer. Clin. Lung Cancer. 2019;20:270–277. doi: 10.1016/j.cllc.2019.03.004. [DOI] [PubMed] [Google Scholar]
- 64.Nicolazzo C., Raimondi C., Mancini M., Caponnetto S., Gradilone A., Gandini O., Mastromartino M., Del Bene G., Prete A., Longo F., et al. Monitoring PD-L1 positive circulating tumor cells in non-small cell lung cancer patients treated with the PD-1 inhibitor Nivolumab. Sci. Rep. 2016;6:31726. doi: 10.1038/srep31726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gangadaran P., Hong C.M., Ahn B.C. Current Perspectives on In Vivo Noninvasive Tracking of Extracellular Vesicles with Molecular Imaging. Biomed. Res. Int. 2017;2017:9158319. doi: 10.1155/2017/9158319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Konoshenko M.Y., Lekchnov E.A., Vlassov A.V., Laktionov P.P. Isolation of Extracellular Vesicles: General Methodologies and Latest Trends. Biomed. Res. Int. 2018;2018:8545347. doi: 10.1155/2018/8545347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Olejarz W., Dominiak A., Zolnierzak A., Kubiak-Tomaszewska G., Lorenc T. Tumor-Derived Exosomes in Immunosuppression and Immunotherapy. J. Immunol. Res. 2020;2020:6272498. doi: 10.1155/2020/6272498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Maia J., Caja S., Strano Moraes M.C., Couto N., Costa-Silva B. Exosome-Based Cell-Cell Communication in the Tumor Microenvironment. Front. Cell Dev. Biol. 2018;6:18. doi: 10.3389/fcell.2018.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jella K.K., Rani S., O’Driscoll L., McClean B., Byrne H.J., Lyng F.M. Exosomes are involved in mediating radiation induced bystander signaling in human keratinocyte cells. Radiat. Res. 2014;181:138–145. doi: 10.1667/RR13337.1. [DOI] [PubMed] [Google Scholar]
- 70.Jabalee J., Towle R., Garnis C. The Role of Extracellular Vesicles in Cancer: Cargo, Function, and Therapeutic Implications. Cells. 2018;7:93. doi: 10.3390/cells7080093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pasini L., Ulivi P. Extracellular Vesicles in Non-Small-Cell Lung Cancer: Functional Role and Involvement in Resistance to Targeted Treatment and Immunotherapy. Cancers. 2019;12:40. doi: 10.3390/cancers12010040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kim D.H., Kim H., Choi Y.J., Kim S.Y., Lee J.E., Sung K.J., Sung Y.H., Pack C.G., Jung M.K., Han B., et al. Exosomal PD-L1 promotes tumor growth through immune escape in non-small cell lung cancer. Exp. Mol. Med. 2019;51:1–13. doi: 10.1038/s12276-019-0295-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li C., Li C., Zhi C., Liang W., Wang X., Chen X., Lv T., Shen Q., Song Y., Lin D., et al. Clinical significance of PD-L1 expression in serum-derived exosomes in NSCLC patients. J. Transl. Med. 2019;17:355. doi: 10.1186/s12967-019-2101-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gunasekaran M. Exosomal PD-L1 expression as non-invasive biomarker for immune checkpoint inhibitors in non-small cell lung cancer. J. Immunol. 2020;204 [Google Scholar]
- 75.Baylin S.B., Jones P.A. Epigenetic Determinants of Cancer. Cold Spring Harb. Perspect. Biol. 2016;8 doi: 10.1101/cshperspect.a019505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Revelo A.E., Martin A., Velasquez R., Kulandaisamy P.C., Bustamante J., Keshishyan S., Otterson G. Liquid biopsy for lung cancers: An update on recent developments. Ann. Transl. Med. 2019;7:349. doi: 10.21037/atm.2019.03.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Duruisseaux M., Martinez-Cardus A., Calleja-Cervantes M.E., Moran S., Castro de Moura M., Davalos V., Pineyro D., Sanchez-Cespedes M., Girard N., Brevet M., et al. Epigenetic prediction of response to anti-PD-1 treatment in non-small-cell lung cancer: A multicentre, retrospective analysis. Lancet Respir. Med. 2018;6:771–781. doi: 10.1016/S2213-2600(18)30284-4. [DOI] [PubMed] [Google Scholar]
- 78.Cho J.W., Hong M.H., Ha S.J., Kim Y.J., Cho B.C., Lee I., Kim H.R. Genome-wide identification of differentially methylated promoters and enhancers associated with response to anti-PD-1 therapy in non-small cell lung cancer. Exp. Mol. Med. 2020;52:1550–1563. doi: 10.1038/s12276-020-00493-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Perrier A., Didelot A., Laurent-Puig P., Blons H., Garinet S. Epigenetic Mechanisms of Resistance to Immune Checkpoint Inhibitors. Biomolecules. 2020;10:1061. doi: 10.3390/biom10071061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Niveditha D., Jasoria M., Narayan J., Majumder S., Mukherjee S., Chowdhury R., Chowdhury S. Common and Unique microRNAs in Multiple Carcinomas Regulate Similar Network of Pathways to Mediate Cancer Progression. Sci. Rep. 2020;10:2331. doi: 10.1038/s41598-020-59142-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jia Y., Wei Y. Modulators of MicroRNA Function in the Immune System. Int. J. Mol. Sci. 2020;21:2357. doi: 10.3390/ijms21072357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fan J., Yin Z., Xu J., Wu F., Huang Q., Yang L., Jin Y., Yang G. Circulating microRNAs predict the response to anti-PD-1 therapy in non-small cell lung cancer. Genomics. 2020;112:2063–2071. doi: 10.1016/j.ygeno.2019.11.019. [DOI] [PubMed] [Google Scholar]
- 83.Lu Y.T., Delijani K., Mecum A., Goldkorn A. Current status of liquid biopsies for the detection and management of prostate cancer. Cancer Manag. Res. 2019;11:5271–5291. doi: 10.2147/CMAR.S170380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cortez M.A., Ivan C., Valdecanas D., Wang X., Peltier H.J., Ye Y., Araujo L., Carbone D.P., Shilo K., Giri D.K., et al. PDL1 Regulation by p53 via miR-34. J. Natl. Cancer Inst. 2016;108 doi: 10.1093/jnci/djv303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Boeri M., Milione M., Proto C., Signorelli D., Lo Russo G., Galeone C., Verri C., Mensah M., Centonze G., Martinetti A., et al. Circulating miRNAs and PD-L1 Tumor Expression Are Associated with Survival in Advanced NSCLC Patients Treated with Immunotherapy: A Prospective Study. Clin. Cancer Res. 2019;25:2166–2173. doi: 10.1158/1078-0432.CCR-18-1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Constancio V., Nunes S.P., Henrique R., Jeronimo C. DNA Methylation-Based Testing in Liquid Biopsies as Detection and Prognostic Biomarkers for the Four Major Cancer Types. Cells. 2020;9:624. doi: 10.3390/cells9030624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dong L., Ren H. Blood-based DNA Methylation Biomarkers for Early Detection of Colorectal Cancer. J. Proteom. Bioinform. 2018;11:120–126. doi: 10.4172/jpb.1000477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Burtrum D.B., Kim S., Dudley E.C., Hayday A.C., Petrie H.T. TCR gene recombination and alpha beta-gamma delta lineage divergence: Productive TCR-beta rearrangement is neither exclusive nor preclusive of gamma delta cell development. J. Immunol. 1996;157:4293–4296. [PubMed] [Google Scholar]
- 89.Rosati E., Dowds C.M., Liaskou E., Henriksen E.K.K., Karlsen T.H., Franke A. Overview of methodologies for T-cell receptor repertoire analysis. BMC Biotechnol. 2017;17:61. doi: 10.1186/s12896-017-0379-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Qin A., Street L., Cease K., Viglianti B.L., Warren E.H., Zhao L., Ramnath N. Clinical Determinants of Durable Clinical Benefit of Pembrolizumab in Veterans With Advanced Non-Small-Cell Lung Cancer. Clin. Lung Cancer. 2017;18:559–564. doi: 10.1016/j.cllc.2017.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Han J., Duan J., Bai H., Wang Y., Wan R., Wang X., Chen S., Tian Y., Wang D., Fei K., et al. TCR Repertoire Diversity of Peripheral PD-1(+)CD8(+) T Cells Predicts Clinical Outcomes after Immunotherapy in Patients with Non-Small Cell Lung Cancer. Cancer Immunol. Res. 2020;8:146–154. doi: 10.1158/2326-6066.CIR-19-0398. [DOI] [PubMed] [Google Scholar]
- 92.Yamauchi T. A circulating T-cell differentiation marker to predict response to immune checkpoint inhibitors. BioRxiv. 2020 doi: 10.1101/2020.06.13.095844. [DOI] [Google Scholar]
- 93.Amelio I., Bertolo R., Bove P., Buonomo O.C., Candi E., Chiocchi M., Cipriani C., Di Daniele N., Ganini C., Juhl H., et al. Liquid biopsies and cancer omics. Cell Death Discov. 2020;6:131. doi: 10.1038/s41420-020-00373-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chen D., Xu T., Wang S., Chang H., Yu T., Zhu Y., Chen J. Liquid Biopsy Applications in the Clinic. Mol. Diagn. Ther. 2020;24:125–132. doi: 10.1007/s40291-019-00444-8. [DOI] [PubMed] [Google Scholar]
- 95.Ramirez-Labrada A.G., Isla D., Artal A., Arias M., Rezusta A., Pardo J., Galvez E.M. The Influence of Lung Microbiota on Lung Carcinogenesis, Immunity, and Immunotherapy. Trends Cancer. 2020;6:86–97. doi: 10.1016/j.trecan.2019.12.007. [DOI] [PubMed] [Google Scholar]
- 96.Botticelli A., Vernocchi P., Marini F., Quagliariello A., Cerbelli B., Reddel S., Del Chierico F., Di Pietro F., Giusti R., Tomassini A., et al. Gut metabolomics profiling of non-small cell lung cancer (NSCLC) patients under immunotherapy treatment. J. Transl. Med. 2020;18:49. doi: 10.1186/s12967-020-02231-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jin Y., Dong H., Xia L., Yang Y., Zhu Y., Shen Y., Zheng H., Yao C., Wang Y., Lu S. The Diversity of Gut Microbiome is Associated With Favorable Responses to Anti-Programmed Death 1 Immunotherapy in Chinese Patients With NSCLC. J. Thorac. Oncol. 2019;14:1378–1389. doi: 10.1016/j.jtho.2019.04.007. [DOI] [PubMed] [Google Scholar]
- 98.Routy B., Le Chatelier E., Derosa L., Duong C.P.M., Alou M.T., Daillere R., Fluckiger A., Messaoudene M., Rauber C., Roberti M.P., et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. 2018;359:91–97. doi: 10.1126/science.aan3706. [DOI] [PubMed] [Google Scholar]
- 99.Kostopoulos I., Elzinga J., Ottman N., Klievink J.T., Blijenberg B., Aalvink S., Boeren S., Mank M., Knol J., de Vos W.M., et al. Akkermansia muciniphila uses human milk oligosaccharides to thrive in the early life conditions in vitro. Sci. Rep. 2020;10:14330. doi: 10.1038/s41598-020-71113-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Derosa L., Hellmann M.D., Spaziano M., Halpenny D., Fidelle M., Rizvi H., Long N., Plodkowski A.J., Arbour K.C., Chaft J.E., et al. Negative association of antibiotics on clinical activity of immune checkpoint inhibitors in patients with advanced renal cell and non-small-cell lung cancer. Ann. Oncol. 2018;29:1437–1444. doi: 10.1093/annonc/mdy103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ocariz-Diez M., Cruellas M., Gascon M., Lastra R., Martinez-Lostao L., Ramirez-Labrada A., Pano J.R., Sesma A., Torres I., Yubero A., et al. Microbiota and Lung Cancer. Opportunities and Challenges for Improving Immunotherapy Efficacy. Front. Oncol. 2020;10:568939. doi: 10.3389/fonc.2020.568939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Van Geffen W.H., Lamote K., Costantini A., Hendriks L.E.L., Rahman N.M., Blum T.G., van Meerbeeck J. The electronic nose: Emerging biomarkers in lung cancer diagnostics. Breathe. 2019;15:e135–e141. doi: 10.1183/20734735.0309-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rocco G., Pennazza G., Santonico M., Longo F., Rocco R., Crucitti P., Antonelli Incalzi R. Breathprinting and Early Diagnosis of Lung Cancer. J. Thorac. Oncol. 2018;13:883–894. doi: 10.1016/j.jtho.2018.02.026. [DOI] [PubMed] [Google Scholar]
- 104.Behera B., Joshi R., Anil Vishnu G.K., Bhalerao S., Pandya H.J. Electronic nose: A non-invasive technology for breath analysis of diabetes and lung cancer patients. J. Breath Res. 2019;13:024001. doi: 10.1088/1752-7163/aafc77. [DOI] [PubMed] [Google Scholar]
- 105.De Vries R., Brinkman P., van der Schee M.P., Fens N., Dijkers E., Bootsma S.K., de Jongh F.H., Sterk P.J. Integration of electronic nose technology with spirometry: Validation of a new approach for exhaled breath analysis. J. Breath Res. 2015;9:046001. doi: 10.1088/1752-7155/9/4/046001. [DOI] [PubMed] [Google Scholar]
- 106.De Vries R., Muller M., van der Noort V., Theelen W., Schouten R.D., Hummelink K., Muller S.H., Wolf-Lansdorf M., Dagelet J.W.F., Monkhorst K., et al. Prediction of response to anti-PD-1 therapy in patients with non-small-cell lung cancer by electronic nose analysis of exhaled breath. Ann. Oncol. 2019;30:1660–1666. doi: 10.1093/annonc/mdz279. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data presented in this review are openly available, references are listed below.