Abstract
Background
Two promising therapeutic strategies in oncology are chimeric antigen receptor-T cell (CAR-T) therapies and antibody–drug conjugates (ADCs). To be effective and safe, these immunotherapies require surface antigens to be sufficiently expressed in tumors and less or not expressed in normal tissues. To identify new targets for ADCs and CAR-T specifically targeting breast cancer (BC) molecular and pathology-based subtypes, we propose a novel in silico strategy based on multiple publicly available datasets and provide a comprehensive explanation of the workflow for a further implementation.
Methods
We carried out differential gene expression analyses on The Cancer Genome Atlas BC RNA-sequencing data to identify BC subtype-specific upregulated genes. To fully explain the proposed target-discovering methodology, as proof of concept, we selected the 200 most upregulated genes for each subtype and undertook a comprehensive analysis of their protein expression in BC and normal tissues through several publicly available databases to identify the potentially safest and viable targets.
Results
We identified 36 potentially suitable and subtype-specific tumor surface antigens (TSAs), including fibroblast growth factor receptor-4 (FGFR4), carcinoembryonic antigen-related cell adhesion molecule 6 (CEACAM6), GDNF family receptor alpha 1 (GFRA1), integrin beta-6 (ITGB6) and ectonucleotide pyrophosphatase/phosphodiesterase 1 (ENPP1). We also identified 63 potential TSA pairs that might be appropriate for co-targeting strategies. Finally, we validated subtype specificity in a cohort of our patients, multiple BC cell lines and the METABRIC database.
Conclusions
Overall, our in silico analysis provides a framework to identify novel and specific TSAs for the development of new CAR-T and antibody-based therapies in BC.
Key words: differential gene expression, breast cancer, intrinsic subtypes, CAR-T, antibody–drug conjugates, tumor surface antigens
Highlights
-
•
ADCs and CAR-T therapies target TSAs.
-
•
We propose a novel bioinformatic-based methodology to identify BC subtype-specific TSA for ADC and CAR-T.
-
•
As proof of concept we identified 36 potentially suitable and subtype-specific targets and predicted their toxicity profile.
-
•
We also identified 63 potential TSA pairs that might be appropriate for co-targeting.
-
•
The methodology was relatively easy to reproduce, less time- and resource-consuming and based on publicly available databases.
Introduction
The therapeutic scenario of breast cancer (BC) is in constant evolution and mortality rates are declining, although BC remains a major cause of death.1 Recently, two targeted immune-related therapeutic approaches are gaining substantial attention, due to impressive efficacy in advanced BC or other malignancies. The first approach is based on the use of antibody–drug conjugates (ADCs), a drug class represented by 3-8 molecules of a potent cytotoxic agent attached to a monoclonal antibody, which is directed to a specific tumor surface antigen (TSA). Examples are the already approved T-DM1 and trastuzumab deruxtecan (T-DXd),2,3 both directed to HER2, and sacituzumab govitecan directed to TROP2.4 The TSA acts as a membrane anchor that allows ADC to be internalized and release the cytotoxic agent, which eliminates the tumor cell and, in some cases, neighboring cancer cells that do not express the TSA (i.e. bystander effect; Supplementary Figure S1A, available at https://doi.org/10.1016/j.esmoop.2021.100102).5 Importantly, the activity of ADCs does not necessarily rely on the activation of the surface receptor or on the immunologic properties of the antibody component, such as its capacity to induce antibody-dependent cellular cytotoxicity.6
The second approach is chimeric antigen receptor-T cell (CAR-T) therapy, which has shown unprecedent efficacy in several hematologic malignancies.7 CAR-T therapy consists of T cells collected from autologous peripheral blood and engineered to express CARs specifically directed against the TSA of interest, without major histocompatibility complex restriction, in a non-coreceptor-dependent fashion and without depending on processing and effective presentation of target epitopes8 (Supplementary Figure S1B, available at https://doi.org/10.1016/j.esmoop.2021.100102). The impressive results obtained with CAR-T therapy in hematologic diseases justify their translation into the treatment of solid tumors. Unfortunately, the success of CAR-T therapy in treating solid tumors has been very limited so far, mostly due to (i) lack of specific TSA compared with hematologic malignancies; (ii) tumor heterogeneity and plasticity leading to tumor escape based on loss of TSA expression; (iii) immunosuppressive properties of tumor microenvironment and (iv) T-cell dysfunction driven by chronic antigen exposure.9
Intensive research efforts to overcome resistance phenomena and identify adequate targets for TSA-based immunotherapies are ongoing.10 These include, for example, combination of therapeutic strategies targeting different targets or the development of therapeutic products with the capability of co-targeting more than one molecule. Another major challenge of TSA-based immunotherapies is their potential ‘on-target, off-tumor’ effects, which can lead to severe toxicities, such as respiratory failure, multiorgan disfunction and death, as observed with the first anti-HER2 CAR-T in a patient with advanced HER2-positive (HER2+) BC.11 Thus identification of specific TSAs that are sufficiently expressed in tumor cells and less or not expressed in normal tissue cells is of utmost importance.
In addition, BC is not a single nosological entity. In fact, five major molecular subtypes [i.e. luminal A, luminal B, HER2-enriched (HER2-E), basal-like and normal-like] have been identified with differences in terms of incidence, prognosis and sensitivity to treatments.12 However, because molecular profiling of breast tumors is not readily available in most parts of the world,13 four surrogate pathology-based subgroups are broadly used to indicate tailored treatments such as anti-HER2 and endocrine therapy.14 Genomic and pathology-based classifications largely overlap but substantial discordances exist.
Here, we exploited a broad range of publicly available BC and normal tissue databases containing gene and protein expression to identify, for the first time, specific TSA in each BC subtype. Our work proposes a strategy to identify TSAs that might serve as targets for ADCs and/or CAR-T cell therapies in BC and, at the same time, identify potential TSAs for co-targeting strategies.
Methods
We downloaded several publicly available databases from the Human Protein Atlas (HPA) website, (https://www.proteinatlas.org/about/download). The original datasets names are Normal tissue data, Pathology data, RNA HPA tissue gene data and RNA Genotype-Tissue Expression (GTEx) tissue gene data. The Cancer Genome Atlas (TCGA) BC RNA-sequencing (RNA-seq) and immunohistochemical (IHC) data are available at the following website: https://www.cbioportal.org. All data were based on the HPA version 19.3 and Ensembl version 92.38.
Publicly available protein databases
The HPA normal tissue database contained expression profiles for proteins in human tissues based on IHC using tissue microarrays. The database included the Ensembl gene identifier for each gene, tissue name, cell types for each tissue, the level of expression per cell type and the gene reliability of the expression value. Protein expression was defined as either not detected–low (ND–L) or medium–high (M–H) depending on all cell types within a given tissue. If a tissue had different cell types with both ND–L and M–H, we considered the tissue group expression as M–H, in order to be more cautious.
The HPA cancer tissue database (alias ‘Pathology data’) contained expression profiles for proteins in human tumor tissues based on IHC using tissue microarrays and an assessment of expression levels and patients' survival. The database included Ensembl gene identifier, gene name, tumor name, the number of patients annotated for different staining levels (‘High’, ‘Medium’, ‘Low’ and ‘Not detected’) and log-rank P values for Kaplan–Meier analysis of correlation between messenger RNA (mRNA) expression level and patient survival. Data regarding BC were extracted from the full database.
Publicly available RNA databases
The RNA HPA tissue gene database contained the transcript expression levels summarized per gene in 37 tissues based on RNA-seq. The RNA GTEx tissue gene database contained transcript expression levels summarized per gene in 36 tissues based on RNA-seq. Both datasets included the Ensembl gene identifier, the analyzed tissue sample, the transcripts per million (TPM), the protein-TPM and the normalized expression. Within these two databases we had to define the RNA levels of expressions of each gene in each tissue. As elsewhere reported,15 we considered RNA expression as ND–L if below a log2(TPM + 1) of 4; otherwise it was defined as M–H.
TCGA database and classifications of breast cancer subtypes
All patients with tumors showing estrogen receptor (ER) and/or progesterone receptor ≥1% during the IHC analysis were considered hormone receptor-positive (HR+); otherwise they were considered HR–, according to international guidelines.14 At the same time, breast tumors were defined as HER2+ if an IHC score of 3+ was reported and/or the HER2 gene (ERBB2) was amplified using in situ hybridization-based techniques, following American Society of Clinical Oncology/College of American Pathologists (ASCO/CAP) guidelines.16 For some patients the in situ hybridization result was reported as positive or negative, whereas for some others the HER2/CEP17 ratio and average ERBB2 copy number were reported. In this case, tumors were considered HER2+ if the ratio was ≥2.0 and the average ERBB2copy number was ≥4.0 signals/cell, according to the last ASCO/CAP guidelines.16
We regrouped patients into four IHC-simplified subtypes, namely, HR+/HER2–, HR+/HER2+, HR-negative/HER2+ and triple-negative BC, if HR-negative/HER2–. PAM50 intrinsic subtypes considered for the analysis were luminal A, luminal B, HER2-E and basal-like. All tumors with IHC unknown subtypes and/or a PAM50 normal-like subtype (which often represent inadequate tumor cellularity17) were removed from the final database before carrying out genomic analysis.
Gene expression values in the TCGA dataset were represented as RNA-seq by expectation-maximization data and normalized within sample to the upper quartile of total reads. To determine intrinsic subtypes, the TCGA RNA-seq data were first adjusted to the median gene expression of an ER-balanced subset of samples. Then, the PAM50 predictor was applied as elsewhere described.18
Surface antigen check
Two reviewers (FS and PB) independently checked whether the potential candidate genes codified for proteins with extracellular domains, to assess their potential targetability with ADC and CAR-T. A third reviewer (SG) took the final decision in case of controversy. Gene products' characteristics were checked in the websites www.genecards.org, www.proteinatlas.org and www.uniprot.org (further details on Supplementary Methods, available at https://doi.org/10.1016/j.esmoop.2021.100102).
Final targets selection and subtype specificity
Genes codifying for the identified surface antigens were rechecked for expression levels in normal tissues by using the HPA RNA and GTEx RNA databases, to reassess potential risk of toxicity. Genes with ND–L expression in ≥75% of tissues only in the HPA protein database were considered as having moderate risk, while in case of ND–L expression in ≥75% of tissues in at least two or three databases, genes were considered as having low risk for toxicity. We considered as potential candidate genes all the ones with moderate/low risk for toxicity and M–H protein levels in BC (as per HPA cancer tissue database), or the ones with low toxicity risk and low protein levels in BC. Subtype specificity was assessed based on the differential gene expression analysis carried out as starting point with the TCGA BC data. In addition, Pearson's correlations among the final targets based on RNA-seq gene expression data values were carried out. If strong, good or moderate uphill significant correlations were found between two genes and a gene of the pair was found to be upregulated and within the first quartile of expression of the subtype for which the other gene was already considered a suitable target, an indication for such subtype was given. For these cases, a co-targeting strategy was also considered worth testing.
Validation of the targets' subtype specificity
Finally, we aimed at validating our potential therapeutic indications by comparing the agreement of the target genes' subtype specificity with their mRNA levels in IHC-based and molecular subtypes. Gene expression data were retrieved from the publicly available METABRIC clinicopathological and genomic data (available at https://www.cbioportal.org) and from 470 patients with BC treated at the Hospital Clinic of Barcelona. An investigational PAM50 test was also applied to a panel of BC cell lines. The validation process is fully explained in the Supplementary Methods, available at https://doi.org/10.1016/j.esmoop.2021.100102.
Statistical analyses
Two separated multiclass significance analysis of microarray (SAM) were conducted to compare gene expression profiles between PAM50 intrinsic subtypes and IHC subtypes groups, respectively. Differences were considered significant at a false discovery rate <5%.19
We assessed genes' respective protein's expression levels in normal tissues and BC tissue, along with their RNA expression levels in normal tissues, to get the final list of potential targets, following the procedure previously described.
The mRNA levels of final target genes were analyzed with Pearson's correlation tests to evaluate potential co-targets. The direction of the correlation (uphill/direct or downhill/indirect) was given by the positivity or negativity of the coefficients. Cohen's kappa was adopted in the validation process (Supplementary Methods, available at https://doi.org/10.1016/j.esmoop.2021.100102).
Statistical significance was set at P < 0.05 and all tests were two sided. All analyses were carried out with R version 3.6.1 for Mac OS X (https://rstudio.com). Hierarchical clustering was obtained with Cluster 3.0 (http://bonsai.hgc.jp/∼mdehoon/software/cluster/software.htm#ctv) and Java TreeView (http://jtreeview.sourceforge.net).
Results
Target selection process
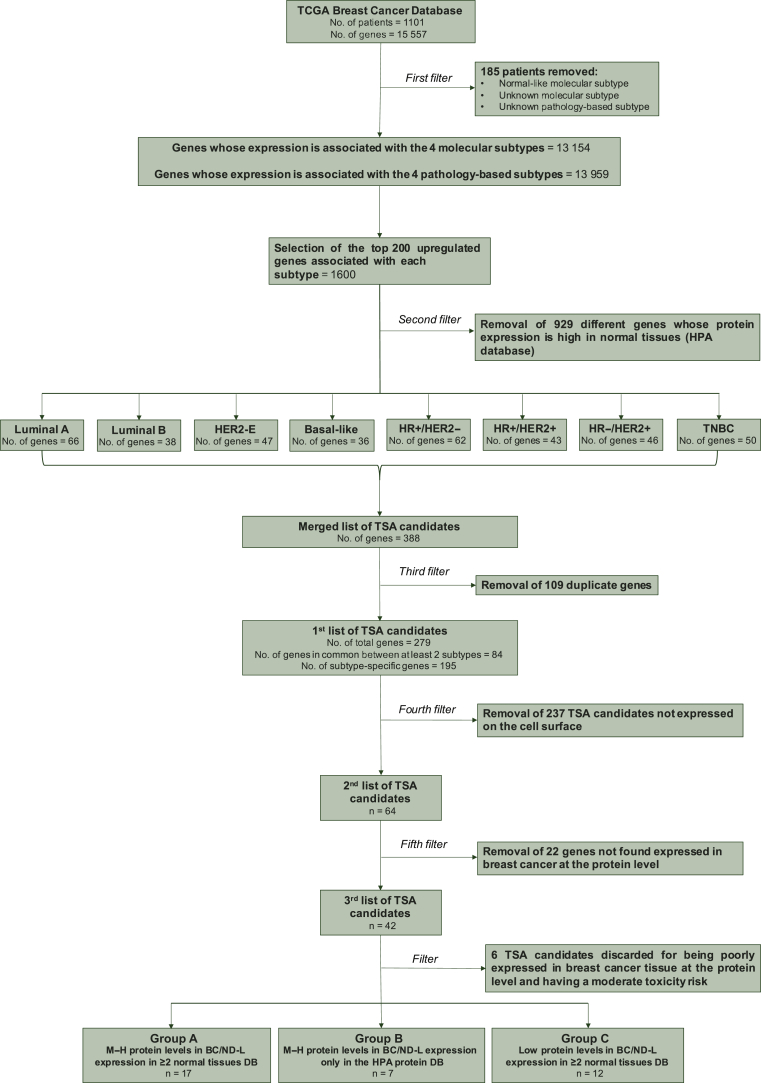
To start our target identification process (Figure 1), we interrogated The TCGA BC database, which includes 1101 patients (pts) and the expression of 15 557 genes from RNA-seq data. We removed 185 patients with an unknown subtype or with a normal-like subtype. In the final dataset of 916 patients, we identified the top expressed genes in the four BC molecular subtypes and four IHC-based testing subgroups. Overall, 13 959 and 13 154 genes were found significantly differentially expressed across the molecular and the pathology-based subtypes, respectively (false discovery rate <5%; Supplementary Figure S2 and Supplementary Table S1, available at https://doi.org/10.1016/j.esmoop.2021.100102).
Figure 1.
Flowchart resuming the candidate genes' selection process.
BC, breast cancer; DB, database; HER2-E, HER2-enriched; HPA, Human Protein Atlas; HR, hormone receptors; IHC, immunohistochemistry; M–H, medium–high; ND–L, not detected–low; SAM, significance analysis of microarray; TNBC, triple-negative breast cancer; +, positive; −, negative.
As a proof-of-concept, the first 200 genes from a list of all differentially expressed and upregulated genes for each BC subtype were extracted (Supplementary Table S2, available at https://doi.org/10.1016/j.esmoop.2021.100102). The whole procedure might be repeated with the subsequent subtype-specific upregulated genes in the list.
Of the 1600 identified genes, 1208 (75.5%) were unique, the others were shared between two or more subtypes (Supplementary Figure S3, available at https://doi.org/10.1016/j.esmoop.2021.100102).
TSA candidates identified using gene expression data might be expressed in normal tissues and this could have implications regarding ‘on-target, off-tumor’ effects. Thus, we checked the expression of each TSA from the top 200 subtype-specific TSA lists in normal tissues. All the TSA candidates (n = 929) with M–H expression in ≥25% of normal tissues were removed due to potential toxicity concerns (Figure 2). Finally, 279 TSA candidates were selected, of which 195 (69.9%) were subtype specific and 84 (30.1%) were common in two or more subtypes.
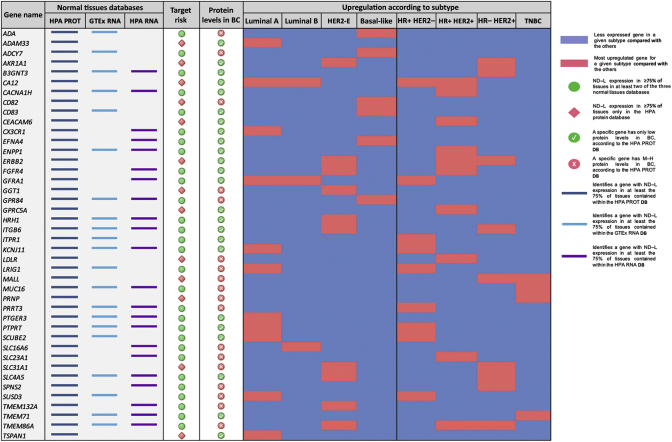
Figure 2.
Weak and strong gene candidates, with toxicity risk category and breast cancer subtype-specific upregulation.
HPA PROT, Human Protein Atlas database of protein expression in normal tissues; GTEx RNA, Genotype-Tissue Expression database of mRNA levels of human genes in normal tissues; HPA RNA, Human Protein Atlas database of mRNA levels of human genes in normal tissues. For each database column, the presence of a colored segment identifies when the corresponding gene presented a not detected and/or low (ND–L) expression in at least 75% of tissues contained within such dataset. The column ‘Target Risk’ identifies the concordance of expression levels in the three normal tissues' databases. A green circle represents an ND–L expression in ≥75% of tissues in at least two of the three databases, while a red rhombus represents an ND–L expression in ≥75% of tissues only in the HPA protein database. The column ‘Protein Levels in BC’ identifies the levels of protein expression for each gene in breast cancer tissue, as per HPA cancer tissue database. Red circles represent genes with only low protein levels, while green circles represent genes with medium/high protein levels. In the ‘Upregulation according to subtype’ section, for each breast cancer subtype the blue color identifies a less expressed gene (database TCGA, result from differential expression analysis) and the red color represents an upregulated gene for a subgroup compared with the others.
TSA-based immunotherapy requires targetable extracellular domains. Among the 279 TSA candidates, 64 (22.9%) met this criterion (Supplementary Table 3, available at https://doi.org/10.1016/j.esmoop.2021.100102). To identify which of these 64 TSA candidates were expressed at the protein level in BC, the HPA cancer tissue database was interrogated. A total of 22 (34.4%) of 64 TSA candidates had no detected protein expression in BC in the HPA dataset and were discarded. Among 42 of 64 (65.6%) TSA candidates with any protein expression in BC according to the HPA dataset, 18 (42.9%) had low protein expression and 24 (57.1%) had M–H protein expression (Figure 2). Finally, to further evaluate the expression of the 42 TSA candidates in normal tissues, two additional datasets of normal tissue based on RNA expression were explored, namely, RNA HPA and the GTEx. Of the 42 TSA candidates, 29 (69%) showed an ND–L expression in ≥75% of tissues in at least one of the other two RNA-based datasets, apart from the HPA protein database; by contrast, for 13 (31%) TSA candidates an ND–L expression was found in ≥75% of tissues only in the HPA protein database.
Based on the levels of protein expression in BC and the protein and mRNA levels in normal tissues, three groups of TSA candidates were ultimately identified (Figure 1 and Supplementary Table S4, available at https://doi.org/10.1016/j.esmoop.2021.100102): (i) strong candidates (M–H protein expression in BC and ND–L expression in ≥75% of tissues in the HPA protein database and in at least one of the two normal tissue RNA-based datasets); (ii) moderate candidates (M–H protein expression in BC and ND–L expression in ≥75% of tissues only in the HPA protein database); (iii) weak candidates (low protein expression in BC and ND–L expression in ≥75% of tissues in the HPA protein database and in at least one of the two normal tissue RNA-based datasets). Six TSA candidates (i.e. GGT1, SLC31A1, CD82, LDLR, MALL and PRNP) with low protein expression in BC and ND–L expression in ≥75% of tissues only in the HPA protein database were excluded. The final list was composed of the following 36 TSA candidates: ADA, ADAM33, ADCY7, AKR1A1, B3GNT3, CA12, CACNA1H, CD83, CEACAM6, CX3CR1, EFNA4, ENPP1, ERBB2, FGFR4, GFRA1, GRP84, GPRC5A, HRH1, ITGB6, ITPR1, KCNJ11, LRIG1, MUC16, PRRT3, PTGER3, PTPRT, SCUBE2, SLC16A6, SLC23A1, SLC4A5, SPNS2, SUSD3, TMEM71, TMEM132A, TMEM86A and TSPAN1. Full names, a description of their function, current therapeutic implications and role in cancer and association with BC are reported in the Supplementary Table S5, available at https://doi.org/10.1016/j.esmoop.2021.100102.
To predict potential organ-specific side-effects of the 36 TSAs, we re-interrogated the three normal tissue datasets (protein HPA, RNA HPA and GTEx). A total of 68 tissues/organs/cells were evaluated and grouped into seven main categories: gastrointestinal (GI) tract, genitourinary (GU) tract, immune system, respiratory tract, endocrine system, central nervous system (CNS) and others. Others included breast, smooth/skeletal muscle, heart, soft tissues, eye/retina, adipose tissues and hair. The most frequent potential toxicities were from the GU tract (91.7% of TSA candidates), GI tract (80.6%), CNS (55.6%), endocrine system (55.5%), immune system (50.0%), respiratory tract (50.0%), skin (30.6%), heart (25.0%) and skeletal muscle (22.2%). No potential toxicity in eye/retinal tissue was identified. A detailed prediction of organ-related side-effects can be extrapolated from Table 1.
Table 1.
Prediction of potential organ-related side-effects
| Gene ID | Gastrointestinal tract | Genitourinary tissues and organs | CNS | Respiratory tract | Endocrine tissues | Immune tissues and cells | Other tissues/organs |
|---|---|---|---|---|---|---|---|
| ADA | Appendix, duodenum, small intestine, stomach | − | − | − | − | Lymph nodes, dendritic cells, monocytes, NK cells, spleen, T cells, tonsil | Adipose tissue |
| ADAM33 | Appendix, colon, esophagus, liver, small intestine, stomach | Kidney, endometrium, epididymis, fallopian tube, gall-bladder, ovary, prostate, seminal vesicle, testis, urinary bladder, uterine cervix, vagina | Cerebral cortex | − | Adrenal gland | Spleen, tonsil | Adipose tissue, breast, placenta, smooth muscle |
| ADCY7 | Appendix, esophagus, oral mucosa, pancreas | Kidney, gall-bladder, ovary, seminal vesicle, urinary bladder, uterine cervix, vagina | Cerebellum | Lung | − | Spleen, tonsil | Heart muscle, placenta, skin |
| AKR1A1 | Appendix, colon, duodenum, esophagus, liver, pancreas, rectum, small intestine, stomach, salivary gland | Kidney, endometrium, epididymis, fallopian tube, gall-bladder, ovary, prostate, seminal vesicle, testis, urinary bladder, uterine cervix, vagina | Amygdala, basal ganglia, cerebellum, cerebral cortex, hippocampus, hypothalamus, midbrain, spinal cord | Bronchus, lung, nasopharynx | Adrenal gland, parathyroid gland, pituitary gland, thyroid gland | Bone marrow, b-cells, dendritic cells, granulocytes, lymph node, monocytes, NK cells, spleen, T cells, tonsil | Adipose tissue, breast, heart muscle, placenta, skeletal muscle, skin, smooth muscle |
| B3GNT3 | Colon, duodenum, esophagus, rectum, small intestine, stomach, salivary gland | Kidney, gall-bladder, urinary bladder | − | − | − | − | Placenta |
| CA12 | Appendix, colon, esophagus, oral mucosa, pancreas, rectum, salivary gland | Kidney, endometrium, fallopian tube, urinary bladder, uterine cervix, vagina | Basal ganglia | − | − | − | Breast, skin, smooth muscle |
| CACNA1H | Colon, esophagus | Endometrium, fallopian tube, gall-bladder, ovary, prostate, seminal vesicle, testis, urinary bladder, uterine cervix | − | Lung, nasopharynx | Adrenal gland, pituitary gland, thyroid gland | − | Skeletal muscle, smooth muscle, soft tissue |
| CD83 | Appendix | Kidney, gall-bladder, ovary, seminal vesicle, uterine cervix, vagina | Cerebellum, cerebral cortex | Lung | Adrenal gland | Bone marrow, B-cells, lymph node, monocytes, spleen, tonsil | − |
| CEACAM6 | Appendix, colon, duodenum, esophagus, rectum, small intestine, salivary gland | Gall-bladder, urinary bladder, uterine cervix, vagina | − | Lung | − | Bone marrow, tonsil | − |
| CX3CR1 | Appendix, colon, duodenum, rectum, small intestine | Kidney, gall-bladder, seminal vesicle, testis | Caudate, cerebral cortex | Lung | Adrenal gland | Dendritic cells, tonsil | Breast, placenta, skin, soft tissue |
| EFNA4 | − | − | − | − | − | B cells, dendritic cells | Skin |
| ENPP1 | Liver, pancreas, stomach | Kidney, endometrium, gall-bladder, testis | − | − | Parathyroid gland, thyroid gland | − | Placenta, skin, smooth muscle |
| ERBB2 | Appendix, colon, duodenum, esophagus, liver, pancreas, rectum, small intestine, stomach, salivary gland | Kidney, endometrium, epididymis, fallopian tube, gall-bladder, ovary, prostate, seminal vesicle, testis, urinary bladder, uterine cervix, vagina | − | Bronchus, lung, nasopharynx | Parathyroid gland, pituitary gland, thyroid gland | Tonsil | Breast, heart muscle, placenta, skeletal muscle, skin, smooth muscle |
| FGFR4 | Appendix, colon, duodenum, liver, pancreas, rectum, small intestine, stomach | Kidney, gall-bladder, ovary, seminal vesicle, testis, urinary bladder | − | Lung, nasopharynx | Adrenal gland | Spleen | − |
| GFRA1 | Appendix, colon, duodenum, liver, rectum, small intestine | Kidney, endometrium, epididymis, fallopian tube, gall-bladder, prostate, testis | − | − | − | Tonsil | Breast, heart muscle, placenta, skeletal muscle, smooth muscle |
| GPR84 | Appendix | − | Caudate, cerebellum | − | − | Bone marrow | − |
| GPRC5A | Colon, duodenum, esophagus, rectum, small intestine, stomach, salivary gland | Kidney, gall-bladder, ovary, urinary bladder, uterine cervix, vagina | Caudate, hippocampus | Lung | Thyroid gland | − | Adipose tissue, placenta |
| HRH1 | Colon, rectum | Kidney, gall-bladder | − | − | − | − | Smooth muscle |
| ITGB6 | Appendix, colon, duodenum, rectum, small intestine, stomach | Kidney, gall-bladder, urinary bladder | − | Lung | Adrenal gland, parathyroid gland | Tonsil | Breast, heart muscle, skeletal muscle |
| ITPR1 | − | Endometrium, epididymis, fallopian tube, gall-bladder, ovary, prostate, seminal vesicle | Caudate, cerebellum, cerebral cortex, hippocampus | − | Adrenal gland, parathyroid gland, thyroid gland | − | Breast, smooth muscle |
| KCNJ11 | Liver, pancreas | Prostate, testis | Cerebellum | − | − | − | Skeletal muscle, soft tissue |
| LRIG1 | Colon, duodenum, pancreas, stomach | Kidney, endometrium, fallopian tube, gall-bladder, prostate, testis, uterine cervix | Basal ganglia, cerebellum, cerebral cortex, hippocampus | Bronchus | − | − | Adipose tissue, beast, placenta, smooth muscle, soft tissue |
| MUC16 | − | Endometrium, fallopian tube, uterine cervix | − | Bronchus, nasopharynx | − | − | − |
| PRRT3 | − | Kidney | Caudate, cerebellum, cerebral cortex | − | Adrenal gland, parathyroid gland | Tonsil | − |
| PTGER3 | − | Kidney, endometrium, uterine cervix | − | − | − | Granulocytes | Adipose tissue, breast, smooth muscle |
| PTPRT | Duodenum, oral mucosa, rectum, stomach | Fallopian tube, gall-bladder, seminal vesicle, testis | Caudate, cerebellum, cerebral cortex, hippocampus | − | − | − | Soft tissue |
| SCUBE2 | Appendix, duodenum, liver, small intestine, stomach | Kidney, endometrium, epididymis, fallopian tube, gall-bladder, prostate, testis, urinary bladder, uterine cervix | Caudate | − | Parathyroid gland | Tonsil | Adipose tissue, breast, placenta, skin, smooth muscle |
| SLC16A6 | Colon, liver, rectum, small intestine, salivary gland | Kidney, epididymis, fallopian tube, ovary, testis | Cerebellum | − | Adrenal gland | − | Heart muscle |
| SLC23A1 | Colon, duodenum, small intestine | Kidney, fallopian tube | − | − | − | − | − |
| SLC4A5 | Appendix, colon, duodenum, rectum, small intestine, stomach | Kidney, epididymis, fallopian tube, gall-bladder, seminal vesicle, testis, urinary bladder | − | − | Thyroid gland | − | Breast |
| SPNS2 | Duodenum, esophagus, small intestine, stomach | Kidney, gall-bladder, ovary, uterine cervix, vagina | Cerebral cortex, spinal cord | Lung | − | Bone marrow | Adipose tissue, skin |
| SUSD3 | Appendix, small intestine | Kidney, endometrium, fallopian tube, ovary, testis, uterine cervix | Caudate, cerebellum, cerebral cortex, hippocampus | Nasopharynx | Adrenal gland | Bone marrow, B cells, dendritic cells, granulocytes, lymph node, spleen, T cells, tonsil | Adipose tissue, breast, heart muscle, skin, soft tissue |
| TMEM132A | Appendix, esophagus, stomach, salivary gland | Endometrium, ovary, uterine cervix | Amygdala, basal ganglia, cerebellum, cerebral cortex, hippocampus, hypothalamus, midbrain, spinal cord | Bronchus | Adrenal gland, pituitary gland | − | Placenta, skeletal muscle |
| TMEM71 | Appendix, colon, duodenum, pancreas, pancreas, rectum, small intestine, stomach, salivary gland | Kidney, endometrium, gall-bladder, testis | Caudate | Bronchus | Adrenal gland, thyroid gland | Bone marrow, dendritic cells, granulocytes, lymph node, monocytes, NK cells, spleen, T cells | Hear muscle, soft tissue |
| TMEM86A | Colon, duodenum, rectum, small intestine | Seminal vesicle, testis | Cerebral cortex | Lung | Adrenal gland, thyroid gland | − | Heart muscle, skeletal muscle, skin |
| TSPAN1 | Appendix, colon, duodenum, rectum, small intestine, stomach, salivary gland | Kidney, endometrium, epididymis, fallopian tube, gall-bladder, prostate, seminal vesicle, testis, urinary bladder, uterine cervix | − | Lung, nasopharynx | Parathyroid gland, pituitary gland, thyroid gland | − | Smooth muscle |
CNS, central nervous system.
Correlations among the final candidates and co-targeting strategies
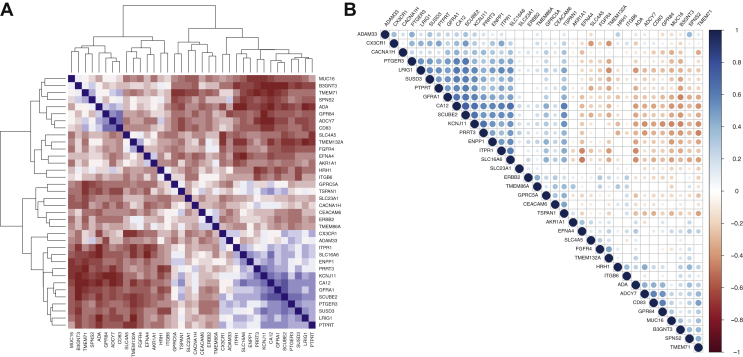
To further support subtype-specific indication, we carried out multiple Pearson's correlations among all TSA candidates (Figure 3). A strong positive correlation was found only between CA12 and SCUBE2 (r = 0.73, P < 0.001). Moderate positive correlations were found between 45 gene pairs. Detailed correlation coefficients for each gene pair are shown in Supplementary Table S6, available at https://doi.org/10.1016/j.esmoop.2021.100102. Based on the methodology that we previously explained, targets' subtype-specific indications were refined. Hence, 15 suitable targets for luminal A, 7 for luminal B, 11 for HER2-E, 6 for basal-like, 15 for HR+/HER2−, 15 for HR+/HER2+, 11 for HR−/HER2+ and 6 for triple-negative BC were ultimately identified (Table 2).
Figure 3.
Correlation matrix and heatmap of gene correlations.
(A) Heatmap representing the correlations between each gene pair and also a hierarchical clustering. Downhill/indirect correlations are shown in brown, while uphill/direct correlations are shown in blue. (B) Correlation matrix showing the significance and direction of correlation between gene pairs. Positive correlations are displayed in blue and negative correlations in brown color. Color intensity and circles' size are proportional to the correlation coefficients. Blank spaces represent nonstatistical significance of Pearson's correlation test.
Table 2.
List of final targets, as per SAM subtype specificity, gene pair correlations, levels of protein expression in breast cancer tissue and toxicity risk
| Target groups | Luminal A | Luminal B | HER2-E | Basal-like | HR+/HER2– | HR+/HER2+ | HR–/HER2+ | TNBC |
|---|---|---|---|---|---|---|---|---|
| Group A | CX3CR1 | GFRA1 | FGFR4 | EFNA4 | GFRA1 | CACNA1H | ITGB6 | TMEM71 |
| GFRA1 | SLC16A6 | HRH1 | ITPR1 | ENPP1 | SLC4A5 | B3GNT3 | ||
| KCNJ11 | ENPP1 | ITGB6 | KCNJ11 | FGFR4 | B3GNT3 | |||
| PTGER3 | KCNJ11 | SLC4A5 | PTPRT | ITGB6 | FGFR4 | |||
| PTPRT | CACNA1H | SCUBE2 | SLC16A6 | HRH1 | ||||
| SCUBE2 | ENPP1 | KCNJ11 | ||||||
| SLC16A6 | SLC16A6 | |||||||
| ITPR1 | CACNA1H | |||||||
| ENPP1 | PTGER3 | |||||||
| CX3CR1 | ||||||||
| Group B | ADAM33 | CA12 | AKR1A1 | CA12 | CA12 | AKR1A1 | ||
| CA12 | ERBB2 | TSPAN1 | CEACAM6 | ERRB2 | ||||
| TSPAN1 | CEACAM6 | ERRB2 | CEACAM6 | |||||
| GPRC5A | ||||||||
| TSPAN1 | ||||||||
| Group C | LRIG1 | PRRT3 | TMEM132A | ADA | LRIG1 | SLC23A1 | SPNS2 | MUC16 |
| SUSD3 | TMEM86A | ADCY7 | PRRT3 | TMEM86A | TMEM86A | ADCY7 | ||
| PRRT3 | SPNS2 | CD83 | SUSD3 | PRRT3 | TMEM132A | CD83 | ||
| GPR84 | TMEM132A | GPR84 | ||||||
| SPNS2 |
Given in bold are the genes added because of significant correlation and co-presence in the first quartile of expression.
HER2-E, HER2-enriched; HR, hormone receptor; TNBC, triple-negative breast cancer; +, positive; –, negative; SAM, significance analysis of microarray.
The identification of molecules coexpressed within the same BC subtype might help overcoming the resistance based on the loss of expression of TSA.9 Therefore we carried out multiple Pearson's correlations among the final proposed targets within each subtype. For each gene pair with strong, good or moderate uphill correlations a co-targeting strategy was considered worth testing. Overall, 63 different gene pairs were identified (Supplementary Table S7, available at https://doi.org/10.1016/j.esmoop.2021.100102).
Check for surface targets in ongoing CAR-T trials for breast cancer and main ADC targets in breast tumors
Once identified as the best candidates, we decided to check whether our potential targets had already been exploited in ongoing/already terminated CAR-T clinical trials and which were the characteristics of already studied targets and why they were not comprised in our list. On 11 March 2020 we carried out a search in the online clinical trials databases www.clinicaltrials.gov and www.carglobaltrials.com. The literature research outcome is reported in Supplementary Results, available at https://doi.org/10.1016/j.esmoop.2021.100102. The identified targets were ERBB2, MUC1, MSLN (mesothelin), MET and CEACAM5 (CEA), GD2, KLRK1 (protein NKG2D), MS4A1 (protein CD20), EPCAM, CD274 (protein PD-L1), ROR1 and CD133/PROM1 (Supplementary Table S8, available at https://doi.org/10.1016/j.esmoop.2021.100102). We also searched in PubMed the main ADC targets in BC in current development. The most promising or already targeted in the practice appeared to be TACSD2 (protein TROP-2), SLC39A6 (protein LIV-1), ERBB2 (protein HER2) and ERBB3 (protein HER3).4
All of these targets were found to be differentially expressed in our analysis but, with the exception of ERBB2, were not included in our list, mainly because they were not among the top 200 most upregulated genes for any of the PAM50 and IHC subtypes (Supplementary Table S9, available at https://doi.org/10.1016/j.esmoop.2021.100102). Conversely, LIV-1, CD133, MSLN and GD2 were not present in our target list for different reasons (Supplementary Results, available at https://doi.org/10.1016/j.esmoop.2021.100102).
Validation of the targets' subtype specificity
We assessed the targets' subtype specificity as detailed in the Supplementary Methods, available at https://doi.org/10.1016/j.esmoop.2021.100102. The overall results suggest that there was a moderate/strong agreement with both our clinical and in silico models. Some of the most well-established BC laboratory models were also sufficiently reliable to further explore the efficacy of the targets identified with our methodology in further preclinical studies (Supplementary Results, Supplementary Table S10 and Supplementary Figures S4 and S5, available at https://doi.org/10.1016/j.esmoop.2021.100102).
Discussion
We identified 36 TSA candidates for the personalized treatment of breast tumors and 63 potential TSA pairs that might be appropriate for co-targeting strategies. Only six candidates were already in use or under investigation in breast or other cancer types.
Recent advances in molecular biology have paved the way for more personalized treatment approaches in BC.20 The identification of different molecular subtypes and their prognostic role is already changing the therapeutic decision making14; molecular subtyping, genomic and mutational profiling in tumor tissue or liquid biopsy is also driving the development of innovative clinical trials for both the early and advanced disease.21, 22, 23, 24 Within this innovative scenario, where the old ‘one-size-fits-all’ paradigm is rapidly shifting toward a patient-centered, biomarker/genetic-driven personalized therapeutic approach, the need to find new personalized treatment strategies is crucial. What makes this a hard task, among other issues, is the vast amount of available scientific information, sometimes contradictory, sometimes incomplete or very preliminary, which implies time-consuming, potentially expensive and prone-to-bias subjective evaluation of preclinical research with the objective of discovering new potential therapeutic targets and treatment strategies that could adapt to specific patient populations or diseases with specific molecular characteristics. In a recent paper by MacKay et al.15 the possibility of exploiting publicly available database of gene expression and protein-level data in tissues was proposed. In fact, we adopted the same cut-offs for defining levels of expression at both mRNA and protein levels. However, compared with other studies, our starting point was gene expression data with a special focus on BC molecular subtypes. Moreover, we carried out an in-depth review of the available evidence on our candidate genes, to include only the ones that had sufficient evidence to support their presence on the cell membrane. In addition, we identified potential subtype-specific co-targets, a strategy that might be particularly effective to overcome resistance based on antigen loss of expression or mutations that lead to antigen modifications.25
The identified candidate antigens are TSA proved to be overexpressed in several BC subtypes, but potentially expressed at lower levels also in normal organs, raising safety concerns. The normal tissue expression sites revealed that the most frequent side-effects should be expected at the GU and GI tract, with colitis, appendicitis, gastritis, renal and vesical toxicities being the most likely form of potential adverse events. Some respiratory and endocrine toxicities might also be possible, with lungs and adrenal glands being the most at-risk sites, respectively. Of note, the most relevant toxicities observed in previous trials of ADC or CAR-T therapy directed against some of our targets showed concordant toxicity profiles, indirectly suggesting the goodness of our model.11,26 For example, anti-HER2 CAR-T have shown lethal pulmonary toxicity and anti-HER2 directed drugs, such as trastuzumab, present cardiotoxicity as a typical, although not frequent, side-effect.27,28 In fact, our results showed as two potential toxicity sites for ERBB2-directed therapeutic products both lung and heart. At the same time, the first toxicity filter was passed by ERBB2, suggesting that with our methodology we may effectively detect potential toxicities which, at the same time, are not frequent and/or might be overcome with appropriate countermeasures. In addition, potentially good candidates, such as LIV-1, were not selected due to our toxicity filter. Considering that LIV-1 is currently under evaluation for ADC therapy, we can conclude that from this perspective our methodology is sufficiently conservative. In any case, strategies for preventing predictable adverse events should be pursued before starting in-human studies. This is especially relevant in the case of CAR-T therapy, where T cells work as living drugs, and can greatly proliferate in the patient.
Different strategies have been proposed to mitigate toxicity when targeting TSA with ADC or CAR-T cells. One possible strategy includes the modulation of the affinity of the scFv for its binding target. A lowered affinity might prevent the therapeutic product to significantly bind to its target on normal tissues' cells, while effectively binding to the same target present on cancer cells at higher densities.29,30 In the case of CAR-T therapy, next-generation CARs are being engineered as AND-gate circuits, so that T-cell activation is only achieved when CARs recognize a specific combination of different antigens.31, 32, 33, 34 In this regard, our subtype-specific identification of co-expressed antigens may guide the design of safer CARs that can only drive full T-cell activation when antigen A and antigen B are being co-expressed in the same BC tissue. Another approach that could be used when targeting the proposed novel antigens, that is currently under clinical investigation for CAR-T cells, includes the genetic modification of T cells to express suicide genes or safety switches, so that CAR-T cells can be selectively eliminated from the patient in the event of severe toxicity.35,36 Finally, some toxicities might be dose dependent, and infusion with reduced doses of CAR-T can prevent toxicities but still provide patients with optimal therapeutic effects.37 With respect to ADCs, such kind of modulations are not possible. However, with our model a considerable part of in-human toxicities might be predicted before even starting phase I trials. This might lead to a more accurate selection of TSAs to target with novel ADCs and/or predispose, whenever possible, preventive or neutralizing countermeasures.
Our methodology has limitations worth noting. First, the target identification is subject to some variability depending on the assumptions under which the SAM is conducted. Further, with our methodology, we do not provide targets deriving from tumor-specific splicing variants (e.g. EGFRvIII)38 or protein products derived from gene translocations or mutations (e.g. NRG1/heregulin),39 nor molecules with several subunits and thus codified by multiple genes. We also do not provide targets directly deriving from post-transcriptional modifications, such as glycosylation (e.g. GD2, Tn-Muc1),10,40 or shared among all BC subtypes, which might be limiting especially in the development process of ADCs, given the efficacy that some of these therapeutics have shown beyond subtypes.41 By contrast, the proposed strategy presents several advantages: (i) the possibility of sparing a significant amount of time, along with human and economic resources, to systematically explore the available literature, which is also ever changing and sometimes producing contradictory evidence that might lead to incorrect interpretations; (ii) the possibility of accurately predicting the most probable toxicities and adopting preventive strategies to reduce/overcome them; (iii) an easy reproducibility and (iv) an immediate application to all other differentially overexpressed genes. In fact, we considered this as a proof-of-concept study to propose a novel methodology to identify BC TSA with an expected low or acceptable and predictable toxicity profile. We also provided a validation with another publicly available dataset, along with further assessment in a cohort of patients' BC tissues and different BC cell lines, showing an overall acceptable agreement with our subtype-specific indications.
This study might pave the way for a reasoned and comprehensive approach to construct new CAR-T cells and ADCs to be further tested in the preclinical and clinical settings.
Acknowledgements
FS is the recipient of an European Society for Medical Oncology (ESMO) Fellowship. However, any views, opinions, findings, conclusions or recommendations expressed in this material are those solely of the authors and do not necessarily reflect those of ESMO.
Funding
This work was supported by the Italian Government grant Programma di Ricerca Scientifica di Rilevante Interesse Nazionale (PRIN) 2017 – 2017EKMFTN_006 (to FS), by the Fundación Científica Asociación Española Contra el Cáncer: AECC- Postdoctoral17-1062 (to FB-M), by Instituto de Salud Carlos III (PI16/00904 and PI19/01846 to AP), AACR Career Development Awards for Translational Breast Cancer Research (no grant number), the Breast Cancer Research Foundation (19-20-26-PRAT to AP), Breast Cancer Now (2018NovPCC1294 to AP), European Union's Horizon 2020 and innovation programme (H2020-SC1-BHC-2018- 2020), Fundació La Marató TV3 (201935-30 to AP), Pas a Pas (no grant number; to AP), Save the Mama (no grant number; to AP) and the Spanish Ministry of Science and Innovation under a Ramon y Cajal grant (RYC2018-024442-I to SG).
Disclosure
SDP has declared honoraria from Roche, Pfizer, Astra-Zeneca, Novartis, Celgene, Eli Lilly, Amgen and Eisai. SG is an inventor on patents related to CAR-T cell therapy, filed by the University of Pennsylvania and licensed to Novartis or Tmunity; she receives research funding from Gilead and Roche and personal honoraria from Roche, Gilead, Celgene and Bristol-Myers Squibb. AP has declared an immediate family member being employed by Novartis; personal honoraria from Pfizer, Novartis, Roche, MSD Oncology, Lilly and Daiichi Sankyo; travel, accommodations and expenses paid by Daiichi Sankyo; research funding from Roche and Novartis; consulting/advisory role for NanoString Technologies, Amgen, Roche, Novartis, Pfizer and Bristol-Myers Squibb and patent PCT/EP2016/080056. All other authors have declared no conflicts of interest.
Data sharing
The original databases are all freely available online. Databases obtained after processing the originals, as described in the ‘Methods’ section, and containing raw data for the study analyses are available from the corresponding authors upon reasonable request. The R codes used for this study are available from the corresponding authors upon reasonable request. No original codes have been specifically developed for this study.
Contributor Information
F. Schettini, Email: schettini@clinic.cat.
S. Guedan, Email: sguedan@clinic.cat.
Supplementary data
References
- 1.DeSantis C.E., Ma J., Gaudet M.M. Breast cancer statistics, 2019. CA Cancer J Clin. 2019;69(6):438–451. doi: 10.3322/caac.21583. [DOI] [PubMed] [Google Scholar]
- 2.Modi S., Saura C., Yamashita T. Trastuzumab deruxtecan in previously treated HER2-positive breast cancer. N Engl J Med. 2020;382(7):610–621. doi: 10.1056/NEJMoa1914510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Verma S., Miles D., Gianni L. Trastuzumab emtansine for HER2-positive advanced breast cancer. N Engl J Med. 2012;367(19):1783–1791. doi: 10.1056/NEJMoa1209124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bardia A. March 2020. Antibody-drug conjugates: present and future. Paper presented at the Miami Breast Cancer Conference 5-8. Miami, USA. [Google Scholar]
- 5.Ogitani Y., Hagihara K., Oitate M. Bystander killing effect of DS-8201a, a novel anti-human epidermal growth factor receptor 2 antibody-drug conjugate, in tumors with human epidermal growth factor receptor 2 heterogeneity. Cancer Sci. 2016;107(7):1039–1046. doi: 10.1111/cas.12966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chau C.H., Steeg P.S., Figg W.D. Antibody-drug conjugates for cancer. Lancet Lond Engl. 2019;394(10200):793–804. doi: 10.1016/S0140-6736(19)31774-X. [DOI] [PubMed] [Google Scholar]
- 7.Martinez M., Moon E.K. CAR T cells for solid tumors: new strategies for finding, infiltrating, and surviving in the tumor microenvironment. Front Immunol. 2019;10:128. doi: 10.3389/fimmu.2019.00128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guedan S., Ruella M., June C.H. Emerging cellular therapies for cancer. Annu Rev Immunol. 2019;37:145–171. doi: 10.1146/annurev-immunol-042718-041407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rodriguez-Garcia A., Palazon A., Noguera-Ortega E. CAR-T cells hit the tumor microenvironment: strategies to overcome tumor escape. Front Immunol. 2020;11:1109. doi: 10.3389/fimmu.2020.01109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guedan S., Calderon H., Posey A.D., Maus M.V. Engineering and design of chimeric antigen receptors. Mol Ther Methods Clin Dev. 2019;12:145–156. doi: 10.1016/j.omtm.2018.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bonifant C.L., Jackson H.J., Brentjens R.J., Curran K.J. Toxicity and management in CAR T-cell therapy. Mol Ther Oncolytics. 2016;3:16011. doi: 10.1038/mto.2016.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prat A., Carey L.A., Adamo B. Molecular features and survival outcomes of the intrinsic subtypes within HER2-positive breast cancer. J Natl Cancer Inst. 2014;106:dju152. doi: 10.1093/jnci/dju152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muftah A.A., Aleskandarany M.A., Ellis I.O., Rakha E.A. Molecular-based diagnostic, prognostic and predictive tests in breast cancer. In: Khan A., Ellis I.O., Hanby A.M., editors. Precision Molecular Pathology Breast Cancer. Springer; New York, New York: 2015. pp. 177–195. [Google Scholar]
- 14.Cardoso F., Kyriakides S., Ohno S. Early breast cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2019;30(10):1674. doi: 10.1093/annonc/mdz189. [DOI] [PubMed] [Google Scholar]
- 15.MacKay M., Afshinnekoo E., Rub J. The therapeutic landscape for cells engineered with chimeric antigen receptors. Nat Biotechnol. 2020;38(2):233–244. doi: 10.1038/s41587-019-0329-2. [DOI] [PubMed] [Google Scholar]
- 16.Wolff A.C., Hammond M.E.H., Allison K.H. Human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists Clinical Practice Guideline Focused Update. J Clin Oncol. 2018;36(20):2105–2122. doi: 10.1200/JCO.2018.77.8738. [DOI] [PubMed] [Google Scholar]
- 17.Prat A., Perou C.M. Deconstructing the molecular portraits of breast cancer. Mol Oncol. 2011;5(1):5–23. doi: 10.1016/j.molonc.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ciriello G., Gatza M.L., Beck A.H. Comprehensive molecular portraits of invasive lobular breast cancer. Cell. 2015;163(2):506–519. doi: 10.1016/j.cell.2015.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tusher V.G., Tibshirani R., Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98(9):5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buono G., Schettini F., Perri F. The impact of translational research in breast cancer care: can we improve the therapeutic scenario? Anticancer Agents Med Chem. 2018;18(6):832–836. doi: 10.2174/1871520617666171103105247. [DOI] [PubMed] [Google Scholar]
- 21.André F., Bachelot T., Commo F. Comparative genomic hybridisation array and DNA sequencing to direct treatment of metastatic breast cancer: a multicentre, prospective trial (SAFIR01/UNICANCER) Lancet Oncol. 2014;15(3):267–274. doi: 10.1016/S1470-2045(13)70611-9. [DOI] [PubMed] [Google Scholar]
- 22.Llombart-Cussac A., Cortés J., Paré L. HER2-enriched subtype as a predictor of pathological complete response following trastuzumab and lapatinib without chemotherapy in early-stage HER2-positive breast cancer (PAMELA): an open-label, single-group, multicentre, phase 2 trial. Lancet Oncol. 2017;18(4):545–554. doi: 10.1016/S1470-2045(17)30021-9. [DOI] [PubMed] [Google Scholar]
- 23.Prat A., Saura C., Pascual T. Ribociclib plus letrozole versus chemotherapy for postmenopausal women with hormone receptor-positive, HER2-negative, luminal B breast cancer (CORALLEEN): an open-label, multicentre, randomised, phase 2 trial. Lancet Oncol. 2020;21(1):33–43. doi: 10.1016/S1470-2045(19)30786-7. [DOI] [PubMed] [Google Scholar]
- 24.Bidard F.-C., Pistilli B., Dalenc F. Abstract PD2-06: circulating ESR1 mutation detection rate and early decrease under first line aromatase inhibitor and palbociclib in the PADA-1 trial (UCBG-GINECO) Cancer Res. 2019;79(suppl 4):PD2-06. [Google Scholar]
- 25.Dotti G., Gottschalk S., Savoldo B., Brenner M.K. Design and development of therapies using chimeric antigen receptor-expressing T cells. Immunol Rev. 2014;257(1):107–126. doi: 10.1111/imr.12131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Q., Ping J., Huang Z. CAR-T cell therapy in cancer: tribulations and road ahead. J Immunol Res. 2020;2020:1924379. doi: 10.1155/2020/1924379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morgan R.A., Yang J.C., Kitano M. Case report of a serious adverse event following the administration of T cells transduced with a chimeric antigen receptor recognizing ERBB2. Mol Ther J Am Soc Gene Ther. 2010;18(4):843–851. doi: 10.1038/mt.2010.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Slamon D.J., Leyland-Jones B., Shak S. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001;344(11):783–792. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- 29.Mazor Y., Sachsenmeier K.F., Yang C. Enhanced tumor-targeting selectivity by modulating bispecific antibody binding affinity and format valence. Sci Rep. 2017;7(1):40098. doi: 10.1038/srep40098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu X., Jiang S., Fang C. Affinity-tuned ErbB2 or EGFR chimeric antigen receptor T cells exhibit an increased therapeutic index against tumors in mice. Cancer Res. 2015;75:3596–3607. doi: 10.1158/0008-5472.CAN-15-0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lim W.A., June C.H. The principles of engineering immune cells to treat cancer. Cell. 2017;168(4):724–740. doi: 10.1016/j.cell.2017.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roybal K., Rupp L., Morsut L. Precision tumor recognition by T cells with combinatorial antigen-sensing circuits. Cell. 2016;164:770–779. doi: 10.1016/j.cell.2016.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu C.-Y., Roybal K.T., Puchner E.M. Remote control of therapeutic T cells through a small molecule-gated chimeric receptor. Science. 2015;350(6258):aab4077. doi: 10.1126/science.aab4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kloss C.C., Condomines M., Cartellieri M. Combinatorial antigen recognition with balanced signaling promotes selective tumor eradication by engineered T cells. Nat Biotechnol. 2013;31(1):71–75. doi: 10.1038/nbt.2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Di Stasi A., Tey S.-K., Dotti G. Inducible apoptosis as a safety switch for adoptive cell therapy. N Engl J Med. 2011;365(18):1673–1683. doi: 10.1056/NEJMoa1106152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang X., Chang W.-C., Wong C.W. A transgene-encoded cell surface polypeptide for selection, in vivo tracking, and ablation of engineered cells. Blood. 2011;118(5):1255–1263. doi: 10.1182/blood-2011-02-337360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hegde M., Joseph S.K., Pashankar F. Tumor response and endogenous immune reactivity after administration of HER2 CAR T cells in a child with metastatic rhabdomyosarcoma. Nat Commun. 2020;11(1):3549. doi: 10.1038/s41467-020-17175-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O'Rourke D.M., Nasrallah M.P., Desai A. A single dose of peripherally infused EGFRvIII-directed CAR T cells mediates antigen loss and induces adaptive resistance in patients with recurrent glioblastoma. Sci Transl Med. 2017;9(399):eaaa0984. doi: 10.1126/scitranslmed.aaa0984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang H.-E., Chin S.-F., Ginestier C. A recurrent chromosome breakpoint in breast cancer at the NRG1/neuregulin 1/heregulin gene. Cancer Res. 2004;64(19):6840–6844. doi: 10.1158/0008-5472.CAN-04-1762. [DOI] [PubMed] [Google Scholar]
- 40.Posey A.D., Schwab R.D., Boesteanu A.C. Engineered CAR T cells targeting the cancer-associated Tn-Glycoform of the membrane mucin MUC1 control adenocarcinoma. Immunity. 2016;44(6):1444–1454. doi: 10.1016/j.immuni.2016.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Criscitiello C., Morganti S., Curigliano G. Antibody-drug conjugates in solid tumors: a look into novel targets. J Hematol Oncol. 2021;14(1):20. doi: 10.1186/s13045-021-01035-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.