Abstract
Background
Chronic tobacco smoke exposure results in a broad range of lung pathologies including emphysema, airway disease and parenchymal fibrosis as well as a multitude of extra-pulmonary comorbidities. Prior work using CT imaging has identified several clinically relevant subgroups of smoking related lung disease, but these investigations have generally lacked organ specific molecular correlates.
Research Question
Can CT imaging be used to identify clinical phenotypes of smoking related lung disease that have specific bronchial epithelial gene expression patterns to better understand disease pathogenesis?
Study Design and Methods
Using K-means clustering, we clustered participants from the COPDGene study (n = 5,273) based on CT imaging characteristics and then evaluated their clinical phenotypes. These clusters were replicated in the Detection of Early Lung Cancer Among Military Personnel (DECAMP) cohort (n = 360), and were further characterized using bronchial epithelial gene expression.
Results
Three clusters (preserved, interstitial predominant and emphysema predominant) were identified. Compared to the preserved cluster, the interstitial and emphysema clusters had worse lung function, exercise capacity and quality of life. In longitudinal follow-up, individuals from the emphysema group had greater declines in exercise capacity and lung function, more emphysema, more exacerbations, and higher mortality. Similarly, genes involved in inflammatory pathways (tumor necrosis factor-α, interferon-β) are more highly expressed in bronchial epithelial cells from individuals in the emphysema cluster, while genes associated with T-cell related biology are decreased in these samples. Samples from individuals in the interstitial cluster generally had intermediate levels of expression of these genes.
Interpretation
Using quantitative CT imaging, we identified three groups of individuals in older ever-smokers that replicate in two cohorts. Airway gene expression differences between the three groups suggests increased levels of inflammation in the most severe clinical phenotype, possibly mediated by the tumor necrosis factor-α and interferon-β pathways.
Clinical Trial Registration
COPDGene (NCT00608764), DECAMP-1 (NCT01785342), DECAMP-2 (NCT02504697)
Key Words: airway gene expression, COPD, diagnostic imaging, gene expression, imaging, interferon
Abbreviations: DECAMP, Detection of Early Lung Cancer Among Military Personnel study; EMT, epithelial mesenchymal transition; TNF-α, tumor necrosis factor-α
Chronic tobacco smoke exposure results in a broad range of lung diseases that includes emphysema, airway disease, parenchymal fibrosis, and a multitude of extrapulmonary comorbidities.1, 2, 3 Identification of more homogenous subsets of disease with the use of tools such as CT imaging may better enable clinical, epidemiologic, and genetic investigation. Prior work in this area has identified several clinically relevant subgroups,4, 5, 6, 7, 8 but these investigations generally have lacked organ-specific molecular correlates.
We sought to leverage clinical and imaging data from a large research cohort, the COPDGene Study, combined with clinical and bronchial epithelial gene expression data from the Detection of Early Lung Cancer Among Military Personnel (DECAMP) Study to identify smoking-related lung disease subgroups and to begin to determine their biologic differences.
Methods
Patients and Study Design
The COPDGene Study (NCT00608764) cohort has been described in detail previously.9 Briefly, it is a multicenter longitudinal observational investigation of smokers that is focused on the epidemiologic and genetic factors associated with COPD.10, 11, 12, 13, 14 The baseline enrollment of 10,306 COPDGene participants occurred between October 2006 and January 2011. All participants were invited to return for 5- and 10-year follow-up visits. They are also followed longitudinally through the longitudinal follow-up program. For this study, analyses were limited to those individuals who had completed both baseline and 5-year follow-up visits.
The DECAMP Study is a multicenter consortium comprised of 15 military treatment facilities, Veterans Affairs hospitals, and academic centers across the United States. Participants were recruited into one of two study protocols, designated as DECAMP-1 (NCT01785342) and DECAMP-2 (NCT02504697).15 Study participants of DECAMP-1 were adults aged ≥45 years with indeterminate pulmonary nodules and a heavy smoking history. Study participants of DECAMP-2 were aged 50 to 79 years with a heavy smoking history and a family history of lung cancer or a personal history of COPD.
Additional details regarding the study design, institutional review board approval (e-Tables 1, 2), biospecimen collection, and CT image acquisition protocols are available in the Online Supplement.
Quantitative CT Analysis
The objective imaging measurements used for cluster definition in both cohorts were obtained by previously defined methods. The breadth of possible quantitative imaging measures that could be used to define clusters of individuals with cigarette smoking-related lung diseases is beyond the scope of this study. Based on prior experience and expertise in this area, we selected a parsimonious list of imaging features to attempt to represent the breadth of both pulmonary and extrapulmonary quantitative CT metrics of lung disease.16, 17, 18, 19 These included (1) the objective characterization of interstitial features and emphysema-like tissue with the use of a local histogram-based technique, (2) the measurement of pectoralis muscle area (expressed in square centimeters) that is performed on a single axial image above the level of the aortic arch, and (3) airway wall thickness as defined by the mean thickness of 6 segmental airways from each subject.20, 21, 22, 23, 24, 25, 26, 27, 28, 29 Further details and supplemental analyses with an expanded list of quantitative measures are available in the Online Supplement.
Cluster Derivation and Statistical Analysis
Changes in the airways and lung parenchyma and extrapulmonary tissues, such as the thoracic musculature, have been shown to be related both directly to cigarette smoke exposure and, when measured using quantitative CT imaging, to clinical outcomes and disease pathophysiologic condition.16 Cluster analysis was performed with the use of a parsimonious set of variables that were selected to represent the breadth of the airway, lung parenchyma, and extrapulmonary processes that are evident in smokers. The imaging features were log-transformed and standardized as needed to address distribution skewness and range of each variable and differences between cohorts related to technical differences in image acquisition and reconstruction. K-means clustering was then applied to these variables to group the subjects into clusters. The optimum number of clusters was determined with a combination of the Silhouette and Elbow methods (e-Fig 1). Further details regarding methods of both the clustering, including variable selection, and the statistical analyses that compare the clusters are available in the Online Supplement.
Details regarding RNA isolation, sequencing, data pre-processing, and gene expression analysis are available in the online supplement.
Results
Study Population
From the COPDGene cohort, a total of 5,273 subjects completed both the baseline and 5-year follow-up visits and had CT imaging data available. These were used for the derivation of imaging clusters. Of these, 5,067 subjects had complete clinical data, and 4,954 subjects had complete mortality data. From the DECAMP study, a total of 360 subjects (169 from DECAMP-1 and 191 from DECAMP-2) had imaging data available and were used to replicate the imaging clusters. Of these, 146 subjects had bronchial epithelial samples available for bulk-RNA sequencing. Detailed demographic data on the subjects from both cohorts are presented in Table 1.
Table 1.
Cohort Characteristics for the COPDGene and Detection of Early Lung Cancer Among Military Personnel Studies
| Study | Preserved | Interstitial Predominant | Emphysema Predominant | Pa |
|---|---|---|---|---|
| COPDGene | ||||
| No. | 2,623 | 1,910 | 740 | … |
| Clinical characteristics | ||||
| Age, mean (SD), y | 60.22 (8.64) | 59.97 (9.10) | 65.57 (7.70) | <.001 |
| Male, No. (%) | 987 (37.6) | 1,364 (71.4) | 431 (58.2) | <.001 |
| Black, No. (%) | 592 (22.6) | 675 (35.3) | 120 (16.2) | <.001 |
| Current smoker, No. (%) | 1,144 (43.6) | 1,087 (56.9) | 153 (20.7) | <.001 |
| Smoking exposure, mean (SD), pack-y | 39.11 (21.09) | 48.35 (28.14) | 55.57 (26.81) | <.001 |
| BMI, mean (SD), kg/m2 | 27.92 (5.56) | 30.97 (6.22) | 25.47 (5.00) | <.001 |
| FEV1, mean (SD), % predicted | 86.58 (20.07) | 73.14 (22.08) | 40.62 (19.91) | <.001 |
| Longitudinal follow up | ||||
| Time between phase 1 and phase 2 visits, mean (SD), y | 5.35 (0.52) | 5.34 (0.56) | 5.31 (0.48) | .409 |
| Total duration of follow up, mean (SD), y | 6.85 (1.83) | 6.18 (2.18) | 5.67 (2.43) | <.001 |
| Died, No. (%) | 232 (9.3) | 323 (18.4) | 299 (42.5) | <.001 |
| Radiologic measures | ||||
| Interstitial features, mean (SD), % lung | 4.55 (2.66) | 7.89 (5.37) | 5.34 (3.27) | <.001 |
| Emphysema, mean (SD), % lung | 3.66 (5.42) | 4.97 (6.63) | 48.65 (15.22) | <.001 |
| Pectoralis muscle area, mean (SD), cm2 | 36.26 (12.38) | 48.64 (16.66) | 30.94 (10.63) | <.001 |
| Airway wall thickness, mean (SD), mm | 0.91 (0.13) | 1.25 (0.19) | 1.08 (0.22) | <.001 |
| Detection of Early Lung Cancer Among Military Personnel | ||||
| No. | 141 | 153 | 66 | |
| Clinical characteristics | ||||
| Age, mean (SD), y | 63.91 (8.11) | 66.14 (7.63) | 68.11 (6.35) | .001 |
| Male, No. (%) | 97 (68.8) | 131 (85.6) | 58 (87.9) | <.001 |
| Black, No. (%) | 13 (10.7) | 32 (22.4) | 12 (19.7) | .041 |
| Current smoker, No. (%) | 68 (51.5) | 64 (45.1) | 24 (38.7) | .227 |
| Smoking exposure, mean (SD), pack-y | 47.01 (25.89) | 49.08 (25.51) | 52.49 (26.56) | .381 |
| BMI, mean (SD), kg/m2 | 27.49 (6.01) | 28.52 (6.13) | 24.40 (5.40) | <.001 |
| FEV1, mean (SD), % predicted | 80.13 (17.27) | 73.49 (18.12) | 54.61 (19.61) | <.001 |
| Radiologic measures | ||||
| Interstitial features, mean (SD), % lung | 7.06 (4.50) | 12.62 (9.31) | 6.81 (8.18) | <.001 |
| Emphysema, mean (SD), % lung | 2.81 (2.77) | 13.08 (8.32) | 52.18 (16.60) | <.001 |
| Pectoralis muscle area, mean (SD), cm2 | 43.04 (13.62) | 47.60 (12.87) | 39.37 (10.08) | <.001 |
| Airway wall thickness, mean (SD), mm | 2.10 (0.35) | 2.22 (0.35) | 2.01 (0.36) | <.001 |
Comparisons of the absolute values of imaging variables between cohorts are limited due to technical differences in image acquisition and reconstruction.
Based on analysis of variance for continuous variables and chi-square test for categoric variables.
Characterization of Imaging Clusters From COPDGene
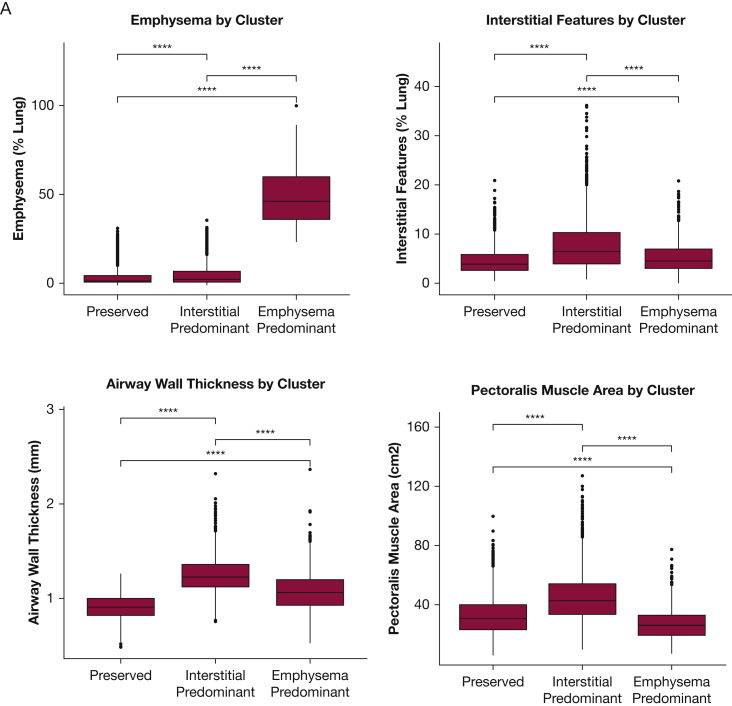
Using quantitative imaging features, we identified three distinct clusters of COPDGene participants that were labeled based on their parenchymal phenotype: preserved, interstitial predominant, and emphysema predominant (Figs 1, 2, 3A). From a CT imaging standpoint, the individuals in the preserved cluster generally had the lowest amount of parenchymal abnormalities (emphysema and interstitial features) and had normal airway wall-thickness. Those individuals in the emphysema cluster demonstrated the highest emphysema scores and mildly thickened airway walls, and those individuals in the interstitial predominant cluster had the highest amount of interstitial changes and highest airway wall-thickness.
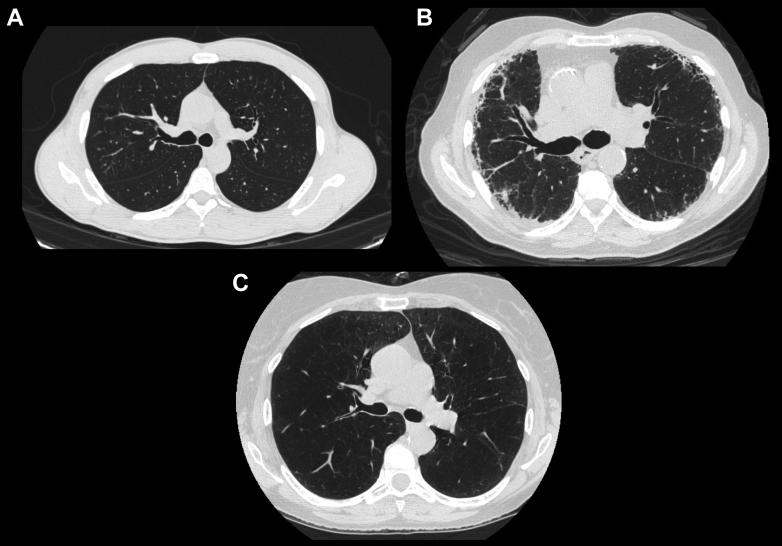
Figure 1.
A-C, Representative CT images from each of the three cluster phenotypes: A, Preserved. B, Interstitial predominent. C, Emphysema.
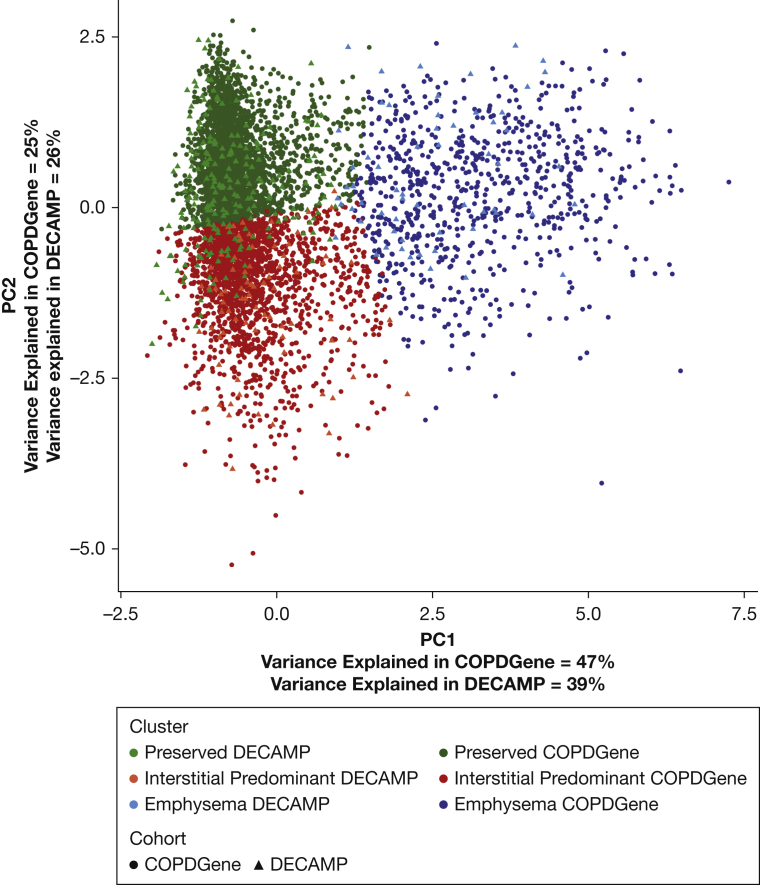
Figure 2.
Cluster Assignment Using Principal Component Analysis. Overlap of the Detection of Early Lung Cancer Among Military Personnel imaging clusters projected onto the first two principal components of the COPDGene imaging features. DECAMP = Detection of Early Lung Cancer Among Military Personnel; PC1 = principal component 1; PC2 = principal component 2.
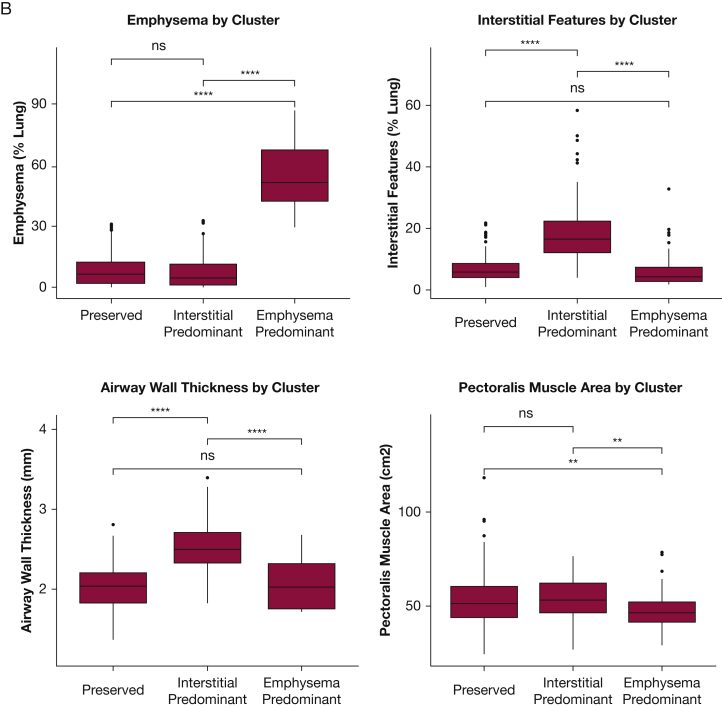
Figure 3.
A-B, Comparison of Imaging Characteristics of the Clusters in the COPDGene and Detection of Early Lung Cancer Among Military Personnel Cohorts. A, The imaging features used in the identification of the patient clusters in COPDGene were compared between patients assigned to each of the three clusters. B, The same imaging clusters were compared between DECAMP patients assigned to each of the three clusters. Global differences for each imaging feature among the three clusters were assessed using analysis of variance and found to be statistically significantly different (P < .001). Pairwise differences were assessed with the use of t-tests. Two asterisks indicate P ≤ .01; four asterisks indicate P ≤ .0001. ns = not significant at P > .05.
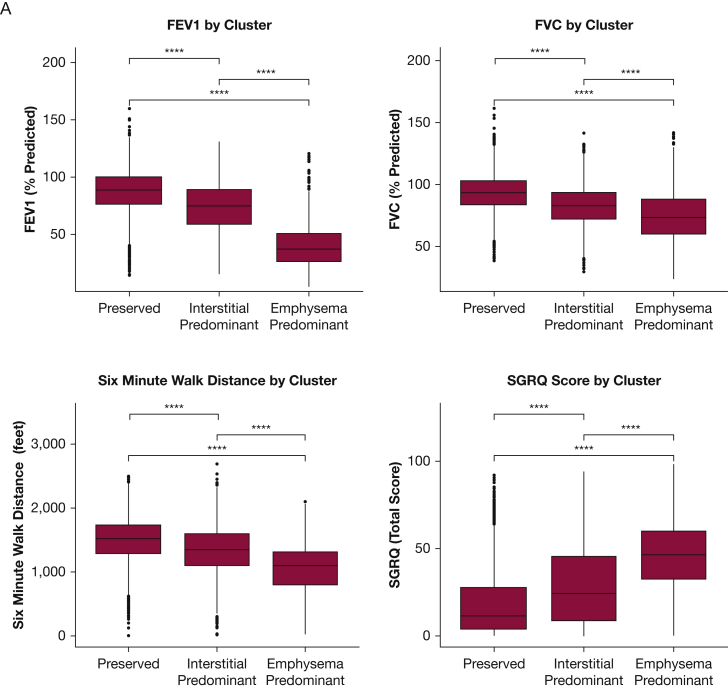
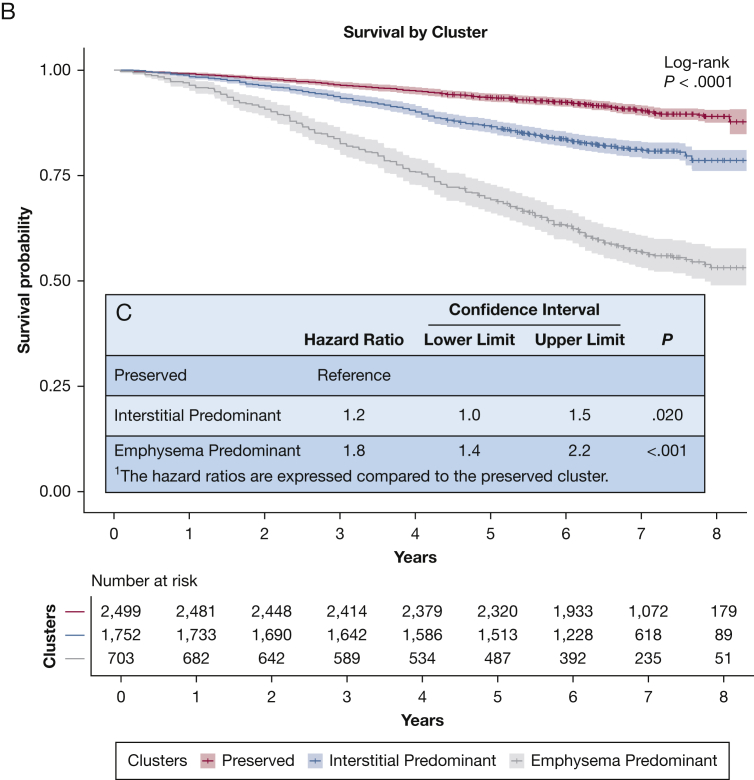
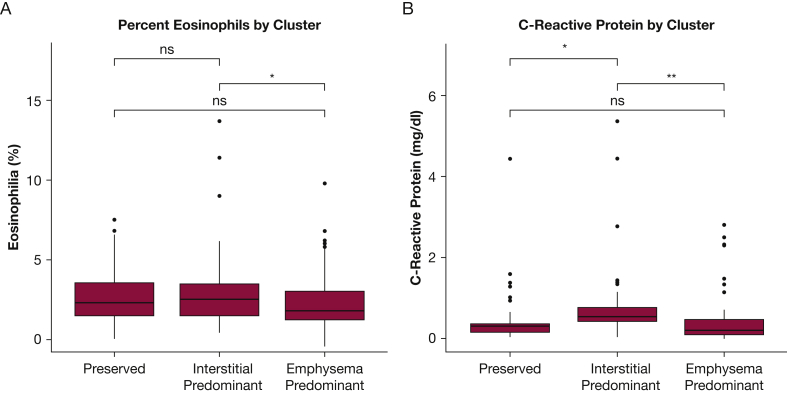
With regards to clinical characteristics (Fig 4A), individuals in the preserved cluster group tended to have normal spirometry, normal 6-minute walk distance, and preserved respiratory health quality of life. Individuals in the emphysema cluster group tended to have expiratory airflow obstruction, reduced 6-minute walk distance, and the lowest respiratory health quality of life, with the exception that the peripheral measures of inflammation the baseline clinical disease severity of those individuals in the interstitial cluster were generally intermediate between the preserved and emphysema clusters (Figs 4A, 5). In addition, the emphysema cluster had the highest mortality rate followed by the interstitial cluster and then the preserved cluster (Fig 4B, C). Similar findings were present when an expanded set of imaging features was used to define the clusters (e-Figs 2, 3) and the majority of individuals (75.5%) were assigned to the same feature-based cluster using when clustered using the expanded imaging variables as they were with the primary imaging variables (e-Table 2).
Figure 4.
A-C, Comparison of Clinical Characteristics and Mortality Rates of the Clusters in the COPDGene Cohort. A, The clinical characteristics identified in COPDGene were compared among the three clusters. Global differences for each clinical characteristic among the three clusters were assessed with the use of analysis of variance and found to be statistically significantly different (P < .001). Pairwise differences were assessed with the use of t-tests without adjustment for multiple comparisons. Four asterisks indicate P ≤ .0001. B, The survival rate of the three clusters identified in COPDGene is demonstrated in this Kaplan-Meier curve. Individuals in the emphysema-predominant cluster had the lowest 5-year survival rate; individuals in the preserved cluster had the highest 5-year survival rate. C, The inset table shows the results of multivariable Cox regression analyses that compared the interstitial predominant cluster and emphysema cluster with the preserved cluster. Note that these analyses were adjusted for age, sex, race, smoking status, and FEV1 at baseline. SGRQ = St. George's Respiratory Questionnaire.
Figure 5.
A-B, Peripheral Eosinophilia and Inflammation in the COPDGene Cohort. A, The percent of peripheral WBCs that are eosinophils by imaging cluster. B, C-reactive protein by imaging cluster. Global differences for each biomarker among the three clusters were assessed with the use of analysis of variance and found to be statistically significantly different for C-reactive protein (P = .003) and not significant for eosinophilia (P = .05). Pairwise differences were assessed with the use of t-tests without adjustment for multiple comparisons. One asterisk indicates P ≤ .05; two asterisks indicate P ≤ .01. ns = not significant at P > .05.
The COPDGene patients in the emphysema cluster had the greatest evidence of disease activity and disease progression during follow up, followed by those in the interstitial group. For example, relative to the preserved group, those in the emphysema group had greater declines in both FEV1 and 6-minute walk distance, gained more emphysema, and had a higher rate of exacerbations (Tables 2, 3). Those patients in the interstitial group did not have a significantly higher rate of lung function decline than those in the preserved group, but they did have a higher rate of decline in exercise capacity, gained more emphysema, and had a higher rate of exacerbations than those in the preserved group (Tables 2, 3). Finally, over 5 years of follow-up, the majority of individuals (79.4%) remained in their original cluster (e-Table 3).
Table 2.
Rate of Acute Respiratory Disease Events by Cluster
| Variable | Incidence Rate Ratioa | 95% CI |
P | |
|---|---|---|---|---|
| Lower Limit | Upper Limit | |||
| Preserved | Reference | |||
| Interstitial predominant | 1.14 | 1.00 | 1.30 | .043 |
| Emphysema predominant | 1.32 | 1.12 | 1.56 | <.001 |
Expressed compared with the preserved cluster (eg, those individuals in the emphysema predominant cluster had 32% more acute respiratory disease events over the course of follow up than those in the preserved cluster).
Table 3.
Longitudinal Changes in Clinical Measures by Cluster
| Variable | Difference in Annual Changea | 95% CI |
P | |
|---|---|---|---|---|
| Lower Limit | Upper Limit | |||
| FEV1, % predicted | ||||
| Preserved | Reference | |||
| Interstitial predominant | 0.02 | −0.10 | 0.15 | .703 |
| Emphysema predominant | −0.66 | −0.87 | −0.44 | <.001 |
| 6-Minute walk distance, ft/y | ||||
| Preserved | Reference | |||
| Interstitial predominant | −6.2 | −10.6 | −1.7 | .007 |
| Emphysema predominant | −15.9 | −23.4 | −8.5 | <.001 |
| Emphysema, % lung volume/y | ||||
| Preserved | Reference | |||
| Interstitial predominant | 0.13 | 0.03 | 0.23 | .011 |
| Emphysema predominant | 0.30 | 0.13 | 0.47 | <.001 |
Expressed as the annualized absolute difference in the change of that cluster compared to the preserved cluster (eg, those individuals in the emphysema predominant cluster lost an average of 15.9 feet more per year over the course of follow up than those in the preserved cluster.
Replication of Imaging Clusters in External Cohort (the DECAMP Study)
After clustering, the first two principal components of the analytic variables from COPDGene were plotted against each other for a geometric interpretation of the grouped data points. The participants from the DECAMP study were clustered independently based on the same imaging variables from COPDGene and projected onto the same principal component plot (Fig 2) and by the similarity in the distribution of the imaging characteristics within the DECAMP clusters (Fig 3B). Similar to COPDGene, in the DECAMP cohort, those in the preserved cluster and those in the emphysema cluster had the least severe and most severe clinical phenotypes, respectively, and the interstitial cluster had an intermediate clinical phenotype (Table 1).
Gene Expression Profiling of Imaging Clusters
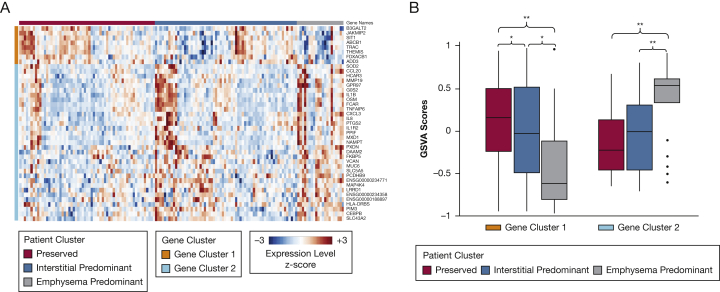
We analyzed the bronchial epithelial gene expression associated with imaging cluster membership using a subset of individuals from the DECAMP study. Although no genes were differentially expressed when we compared the interstitial group with either the emphysema or preserved predominant groups, we identified 41 genes that were expressed differentially between the preserved and emphysema clusters (false discovery rate, < 0.25) (Fig 6A). Eight of these were expressed at lower levels in individuals from the emphysema cluster, including those involved in T-cell biology (T-cell receptor alpha constant, T-cell receptor antigen, thymocyte-expressed molecule involved in selection, which have a regulatory role in both positive and negative T-cell selection during late thymocyte development, and signaling threshold regulating transmembrane adaptor 1, which negatively regulates T-cell antigen receptor-mediated signaling in T cells). Among the 33 genes that were expressed at higher levels in individuals from the emphysema cluster are genes that are related to acute and chronic inflammation (chemokine ligand 20, IL1B, IL8, and IL1R2), tumor necrosis factor-alpha (TNF-α) signaling pathway (TNFA1P6, CXCL3), and mucus production (MUC6) (e-Table 4).
Figure 6.
A-B, Emphysema Cluster-Related Gene Expression. A, With the use of linear modeling, 41 genes were identified to be differentially expressed between the preserved and emphysema cluster (false discovery rate, <0.25). Of these, eight genes were decreased in the airway epithelial cells from individuals in the emphysema cluster relative to the preserved cluster group (gene cluster 1), and 33 genes were increased in the emphysema cluster relative to the preserved cluster group (gene cluster 2). Note: a complete list of gene names is available in the Online Supplement. B, Gene set variation analysis was used to summarize the expression of each gene cluster in each sample. Variation in these summary scores was then examined as a function of imaging clusters. For both gene clusters, there was a significant difference between the imaging clusters by analysis of variance (each, P < .001). Post-hoc Tukey’s honestly significant difference test was applied to examine the pairwise differences between groups. ∗P ≤ .05; ∗∗P ≤ .01.
To more fully characterize the biology of the gene expression differences associated with the imaging clusters, gene set enrichment analysis was performed on preranked gene lists created by the pair-wise comparison of all combinations from the three clusters to identify enrichment of pathway-related gene sets from the KEGG, Reactome, and Gene Ontology databases. Gene sets with significant enrichment (gene set enrichment analysis q, < 0.05) from individuals in the emphysema cluster relative to those in the preserved cluster included upregulated pathways involved in the acute and chronic inflammatory response, TNF-α signaling via NF-κB pathway, and epithelial mesenchymal transition (EMT) (e-Table 4, e-Fig 4), while several common pathways are upregulated in the interstitial predominant group relative to the preserved cluster that included TNF-α signaling via NF-κB pathway and EMT and WNT/β-catenin signaling (e-Table 5). We also used the Crowd Extracted Expression of Differential Signatures tool30 to search its library of manually curated signatures from the Gene Expression Ominbus for experimental perturbations that lead to a pattern of gene expression alterations that are similar to the emphysema signature. Gene expression signatures from five published datasets that involved the response to interferon-β were identified as concordant (GSE26104,31 GSE19392,32 GSE3920,33 GSE125066,34 and GSE4840035); we found that the gene set variation analysis scores from the signature of genes increased in the emphysema cluster is significantly increased in peripheral blood mononuclear cell after interferon-β treatment (GSE26104; Fig 7). We found a similar increase in the gene set variation analysis scores from the emphysema-increased signature in datasets that examined the response of bronchial epithelial cells, hepatocytes, fibroblasts, and endothelial cells to interferon-β32, 33, 34, 35 (e-Fig 5).
Figure 7.
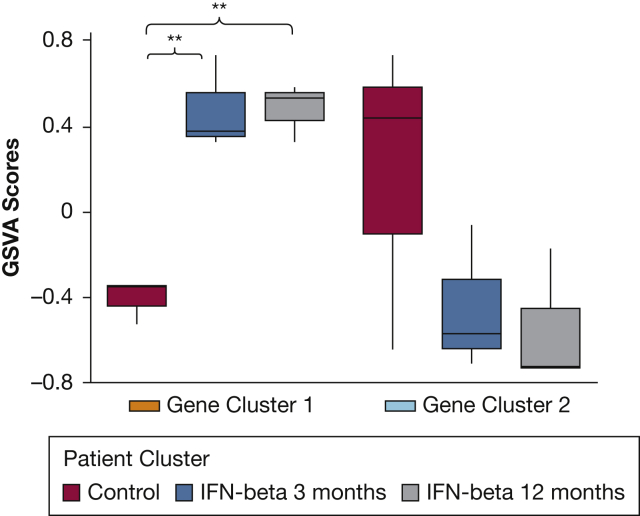
Gene Set Variation Analysis of Emphysema Signature Genes in Peripheral Blood Mononuclear Cells. Following Interferon-β Treatment. Gene set variation analysis was used to summarize the expression of each emphysema signature gene cluster in a published dataset of peripheral blood mononuclear cells from patients at baseline or after interferon-β treatment (GSE2610418). This demonstrates that the genes increased in the bronchial airway of patients in the emphysema-cluster group are induced significantly in peripheral blood mononuclear cells from patients with multiple sclerosis who are treated with interferon-β. Post hoc Tukey’s honestly significant difference test was applied to examine the pairwise differences between groups. Two asterisks indicate P ≤ .01. GSVA = gene set variation analysis.
In addition, we were able to identify emphysema cluster-associated gene expression changes in the bronchial airway that reflect COPD-associated processes in the airway from an external cohort.36 We were also able to characterize the interstitial cluster using gene set variation analysis with those genes that were expressed differentially between the emphysema and preserved clusters to summarize the expression of the emphysema-increased and emphysema-decreased genes in each of the three groups (Fig 6B). Further details of these analyses are available in the Online Supplement.
Discussion
Using quantitative CT imaging from the COPDGene cohort, we identified three subgroups of smokers that have specific clinical characteristics. Individuals classified in the emphysema cluster have worse physiologic measures, reduced exercise capacity, worse respiratory health-related quality of life, and a lower survival rate. Those who are classified in the preserved cluster have little-to-no emphysema, relatively normal physiologic measures, a normal exercise capacity and symptoms assessment, and a higher survival rate when compared with the emphysema cluster; and the interstitial cluster is composed of individuals whose level of disease is intermediate in severity. Furthermore, we identified the same imaging clusters in an independent cohort (the DECAMP study) and demonstrated that they not only had similar baseline clinical characteristics but also had specific gene expression correlates in bronchial epithelial cells. More specifically, by leveraging bronchial epithelial gene expression from patients in the DECAMP cohort, we identified differential gene expression patterns when comparing the emphysema cluster with the preserved cluster that replicated previously published COPD signatures36 and that are intermediate in their expression levels in the interstitial group.
Various studies have used spirometry, clinical symptoms, and imaging studies to identify features associated with particular disease behavior and outcomes in hopes of identifying unique patient subgroups.4,23,37, 38, 39, 40 In the COPDGene cohort for example, Castaldi et al4 identified four clusters with differing clinical and genetic associations. Although their clustering approach used both clinical and radiologic variables, in many ways the clusters they identified are similar to those found in our study and included two relatively preserved clusters, a severe emphysema cluster, and an airways disease cluster that appears clinically similar to that which we termed interstitial predominant. Our approach suggests that these groups, for the most part, can be identified with imaging features alone. In addition, our work demonstrates the presence of these specific clusters in two very different cohorts, which suggests that the radiologic variables we used for clustering may represent broadly distinct thoracic manifestations of smoking.
A central question raised by this work and other COPD phenotyping efforts is how to interpret the patient groups that were identified. One possibility is that the radiographic subtypes represent a single trajectory of disease progression (ie, individuals from the preserved group with progressive disease move into the interstitial group and then to the emphysema group). In this model, the interstitial features identified on CT imaging likely represent inflammation and edema that has been observed to precede the development of emphysema in some cases.27,41 If this hypothesis is correct, then the identification of interstitial features may enable earlier disease detection. To some extent, this is supported by the shifts seen in cluster assignments over 5 years of follow up in this study. For example, it may be that those individuals who transition from the interstitial group to the emphysema group are progressing along the pathway mentioned earlier and those that transition from emphysema to interstitial are those that are experiencing the development of concomitant fibrosis or having evidence of ongoing disease activity. Unfortunately, limited data both in terms of time points and the number of individuals who change groups prevent us from fully exploring this possibility, and additional work using multistate modeling and cohorts with multiple points of follow up is necessary to determine if this is the case. Another possibility is that these three groups represent three distinct phenotypes of response to cigarette smoke: a group relatively resistant to smoking, another group that has evidence of inflammation and experiences the development of airway and interstitial disease, and a third group that experiences the development of progressive emphysema. This mirrors the clinical observation that various lung-function trajectories lead to COPD42 and suggests that these different radiographic phenotypes may reflect distinct pathogenic mechanisms43 that all result in a common physiologic abnormality (ie, airflow limitation). In this schema, a subset of the second group, those with interstitial predominance, may have early or subtle pulmonary fibrosis and may go on to experience more advanced fibrotic disease, as has been suggested for patients with visually defined interstitial lung abnormalities.41
Although future long-term studies will be needed to fully answer this central question regarding what each cluster represents, the gene expression patterns demonstrated by the current analysis help to explain some of the issues related to disease progression. For example, our data suggest that gene expression downstream of interferon-β signaling is activated in patients with emphysema relative to patients from the preserved cluster. This may be due to a COPD-associated increase in airway epithelial production of interferon-β, because conditioned media from human bronchial epithelial cells from patients with COPD that is grown at an air-liquid interface has increased levels of cytokines and interferons relative to conditioned media from similarly grown human bronchial epithelial cells from healthy donors.44 The role of interferons is intrinsically to promote an antimicrobial state in cells that are infected as well as neighboring cells to help limit the spread of infection.45 Furthermore, they stimulate the adaptive immune system via antigen presentation and natural killer cell activation in response to infection.46 But they also play a role in noninfected cells because they are secreted constitutively in low amounts by many tissues in order for them to be primed for future responses.47,48 Interestingly, while interferon-β is increased in the airway epithelium, its production is impaired in BAL fluid49 and resected lung tissue50 from patients with COPD. Further research is necessary to determine whether these differences reflect tissue-specific differences or differences in the clinical contexts in which these studies were performed.
Another pathway of interest that was identified in our study is TNF-α signaling via NF-κB pathway that is upregulated in the emphysema cluster. Although this pathway has been implicated in many different processes in the body, it has been shown specifically to play a central role in airway inflammation in asthma and COPD. Many NF-κB-mediated processes are insensitive to the actions of steroids, and some investigators have proposed targeting NF-κB signaling as a potential intervention for steroid-refractory airway disease.51, 52, 53 Our current work suggests that such an intervention may be of particular importance in those patients with emphysema predominant disease. Regarding other salient findings from the genetic analyses, we found that genes involved in the EMT are upregulated in airway epithelium from subjects in the emphysema-cluster group relative to the preserved-cluster group. In addition to its role in promoting metastasis, EMT has been implicated in peribronchiolar fibrosis, which is observed in the small airways of patients with COPD and contributes to airway obstruction.54 Genes involved in WNT/β-catenin signaling are also elevated in patients in the emphysema cluster. This pathway has been shown to play a role in the pathogenesis of chronic pulmonary diseases such as COPD and pulmonary fibrosis by regulating airway inflammation, remodeling, and EMT.55 Finally, increased expression of the imaging cluster-associated gene expression signature in bronchial epithelial cells from individuals in the interstitial predominant cluster lends credence to the hypothesis that interstitial features may represent imaging indices of early disease processes within COPD, such as tissue destruction, inflammation, and parenchymal remodeling stemming from smoke exposure.23,29,37,56 Combined, these findings suggest that, not only is there an underlying molecular basis for the three patient clusters, but also these biologic signals impact gene expression in airway epithelial cells that might either reflect or contribute to the appearance of the underlying imaging phenotype.
One of the strengths of our study is the ability to identify imaging clusters that replicate across two separate cohorts, despite differences in patient population and image processing. Another strength is the ability to associate imaging cluster membership with gene expression differences in the bronchial airway epithelium, because these gene expression associations argue for an underlying biologic cause for the imaging-based subgroups and begin to suggest the molecular processes that differentiate the groups. Our study did have several limitations as well. The two cohorts are not identical and have differences in inclusion and exclusion criteria. For instance, COPDGene excluded patients with interstitial lung disease, which potentially could limit the ability to draw accurate conclusions regarding interstitial features. No such exclusion criterion was included in the DECAMP study. Also, and notably, the demographics of the two cohorts are quite different, as are several of the imaging variables, with the latter differences likely related both to demographics and differences in image acquisition techniques. The replication of the clusters in the DECAMP study suggests that they may be robust to these differences; however, additional work is needed in other cohorts to determine whether this is the case. Another potential limitation is the small sample size from the DECAMP study, which further restricted the size of the three radiographic subgroups. Of the 360 individuals in the DECAMP study whose imaging was available for analysis, only 146 samples were available for RNA sequencing. These sample size limitations might limit the power to define the molecular differences fully among the three subgroups. Finally, we selected imaging variables based on prior knowledge and experience and on available data in the DECAMP cohort in particular. Although the majority of individuals were classified into the same cluster with the use of an expanded set of imaging variables in the supplemental COPDGene analyses and the clinical characteristics of clusters based on the expanded imaging variables were similar to those in the primary analysis, there were some differences between the interstitial and preserved groups in particular, which should be explored in additional work in other replication cohorts. Similarly, from an exposure standpoint, although we focused on tobacco smoke exposure for this work, future studies with more robust environmental and occupational exposure data, as well as longer term follow-up data, preferably starting in childhood, are needed to determine how these clusters relate to other potential etiologies of COPD.57, 58, 59
In summary, in two distinct cohorts, clustering smokers with the use of quantitative CT imaging-based measures enabled the identification of three subgroups of disease that have organ-specific molecular correlates. Further work is needed to better understand the significance of these clusters, the pathophysiologic and molecular differences between them, and their potential usefulness for defining disease prognosis and management.
Acknowledgments
Author contributions: E. B. and S. Y. A. contributed equally to this work (co-first author); M. E. L. and G. R. W. contributed equally to this work (co-senior author). E. B. is the guarantor of this paper and takes full responsibility for the integrity of the work as a whole. E. B., S. Y. A., F. D., K. X., J. R., and H. M. analyzed data and interpreted results; E. B., S. Y. A., and K. X. prepared the figures; E. B. and S. Y. A. drafted manuscript; E. B., S. Y. A., K. X., D. W., C. S., M. E. L., and G. W. edited and revised manuscript; all the others approved the final version of manuscript
Financial/nonfinancial disclosures: The authors have reported to CHEST the following: M. K. H. reports personal fees from GlaxoSmithKline, Boehringer Ingelheim, AstraZeneca, Sunovion, and Novartis not related to this manuscript. R. P. B. served on the advisory boards for GlaxoSmithKline, Mylan Pharmaceuticals, Boehringer Ingelheim and received research grants from GlaxoSmithKline, and Mylan Pharmaceuticals not related to this manuscript. T. J. D. reports a research grant from BMS and is involved in a clinical trial funded by Genentech. C. S. and D. W. are employees of Johnson and Johnson Services, Inc. D. A. reports grants from American College of Radiology Imaging Network (ACRIN) and personal fees from Veracyte sitting on an Advisory Board. A. S. is an employee of Johnson and Johnson Services, Inc, and has received personal fees from Veracyte Inc outside the submitted work. Ra. S. J. E. reports personal fees from Toshiba, Boehringer Ingelheim, and Eolo Medical; he is also a founder and co-owner of Quantitative Imaging Solutions. M. E. L. reports grants and personal fees from Johnson and Johnson and is a shareholder in Metera Pharmaceuticals; he also has a patent US PTO 9,677,138 issued. G. R. W. is a consultant and sits on an advisory board for Boehringer Ingelheim, PulmonX, Janssen Pharmaceuticals, and GlaxoSmithKline and is a founder and co-owner of Quantitative Imaging Solutions. S. Y. A. is co-owner of Quantitative Imaging Solutions. None declared (E. B., F. D., K. X., J. R., H. M., E. M., E. A. R., S. E. M., Ru. S. J. E., I. O. R., J. C. R., X. X., H. L., G. L., G. S., M. W., C. D.)
Role of sponsors: Boehringer Ingelheim Pharmaceuticals, Inc. (BIPI) and Johnson and Johnson Services, Inc. (JJSI) had no role in the design, analysis or interpretation of the results in this study; BIPI and JJSI were given the opportunity to review the manuscript for medical and scientific accuracy as it relates to BIPI and JJSI substances, as well as intellectual property considerations. The sponsor had no role in the design of the study, the collection and analysis of the data, or the preparation of the manuscript.
Additional information: The e-Appendix, e-Figures, and e-Tables can be found in the Supplemental Materials section of the online article.
Footnotes
FUNDING/SUPPORT: The COPDGene Study (NCT00608764) is supported by NHLBI R01 HL089897 and R01 HL089856 and by the COPD Foundation through contributions made to an Industry Advisory Board comprised of AstraZeneca, Boehringer-Ingelheim, GlaxoSmithKline, Novartis, Pfizer, Siemens and Sunovion. The DECAMP study is supported by funds from the Department of Defense (W81XWH-11-2-0161), the National Cancer Institute (U01CA196408), and Johnson and Johnson Services, Inc (JJSI). Additional funding for this work includes National Institutes of Health grants: K08-HL145118 (S. Y. A.), R01-HL107246 (G. R. W.; Raúl S. J. E.), R01-HL116933 (G. R. W.; Raúl S. J. E.), R21-HL140422 (Raúl S. J. E.), P01-HL114501 (G. R. W.), R01-HL089856 (G. R. W.; Raúl S. J. E.), K23-HL119558 (T. J. D.) and T32-HL007633 (S. E. M.). As well as from DOD Grant PR171782 (I. O. R. and G. R. W.), Boehringer-Ingelheim Pharmaceuticals, Inc. (G. R. W.), and the Pulmonary Fibrosis Foundation (S. Y. A.).
Supplementary Data
References
- 1.Leopold J.G., Gough J. The centrilobular form of hypertrophic emphysema and its relation to chronic bronchitis. Thorax. 1957;12(3):219–235. doi: 10.1136/thx.12.3.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hernandez J.A., Anderson A.E., Holmes W.L., Foraker A.G. Pulmonary parenchymal defects in dogs following prolonged cigarette smoke exposure. Am Rev Respir Dis. 1966;93(1):78–83. doi: 10.1164/arrd.1966.93.1.78. [DOI] [PubMed] [Google Scholar]
- 3.Divo M.J., Casanova C., Marin J.M. COPD comorbidities network. Eur Respir J. 2015;46(3):640–650. doi: 10.1183/09031936.00171614. [DOI] [PubMed] [Google Scholar]
- 4.Castaldi P.J., Dy J., Ross J. Cluster analysis in the COPDGene study identifies subtypes of smokers with distinct patterns of airway disease and emphysema. Thorax. 2014;69(5):415–422. doi: 10.1136/thoraxjnl-2013-203601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garcia-Aymerich J., Gómez F.P., Benet M. Identification and prospective validation of clinically relevant chronic obstructive pulmonary disease (COPD) subtypes. Thorax. 2011;66(5):430–437. doi: 10.1136/thx.2010.154484. [DOI] [PubMed] [Google Scholar]
- 6.Rennard S.I. Chronic obstructive pulmonary disease: linking outcomes and pathobiology of disease modification. Proc Am Thorac Soc. 2006;3(3):276–280. doi: 10.1513/pats.200512-129SF. [DOI] [PubMed] [Google Scholar]
- 7.Rennard S.I., Locantore N., Delafont B. Identification of five chronic obstructive pulmonary disease subgroups with different prognoses in the ECLIPSE cohort using cluster analysis. Ann Am Thorac Soc. 2015;12(3):303–312. doi: 10.1513/AnnalsATS.201403-125OC. [DOI] [PubMed] [Google Scholar]
- 8.Sieren J.P., Newell J.D., Barr R.G. SPIROMICS protocol for multicenter quantitative computed tomography to phenotype the lungs. Am J Respir Crit Care Med. 2016;194(7):794–806. doi: 10.1164/rccm.201506-1208PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Regan E.A., Hokanson J.E., Murphy J.R. Genetic epidemiology of COPD (COPDGene) study design. COPD. 2010;7(1):32–43. doi: 10.3109/15412550903499522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.American Thoracic Society Standardization of spirometry, 1994 Update. Am J Respir Crit Care Med. 1995;152(3):1107–1136. doi: 10.1164/ajrccm.152.3.7663792. [DOI] [PubMed] [Google Scholar]
- 11.ATS Committee on Proficiency Standards for Clinical Pulmonary Function Laboratories ATS statement: guidelines for the six-minute walk test. Am J Respir Crit Care Med. 2002;166(1):111–117. doi: 10.1164/ajrccm.166.1.at1102. [DOI] [PubMed] [Google Scholar]
- 12.Kim D.K., Jacobson F.L., Washko G.R. Clinical and radiographic correlates of hypoxemia and oxygen therapy in the COPDGene Study. Respir Med. 2011;105(8):1211–1221. doi: 10.1016/j.rmed.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mahler D.A., Wells C.K. Evaluation of clinical methods for rating dyspnea. Chest. 1988;93(3):580–586. doi: 10.1378/chest.93.3.580. [DOI] [PubMed] [Google Scholar]
- 14.Jones P.W., Quirk F.H., Baveystock C.M. The St. George’s Respiratory Questionnaire. Respir Med. 1991;85(suppl B):25–31. doi: 10.1016/s0954-6111(06)80166-6. [DOI] [PubMed] [Google Scholar]
- 15.Billatos E., Duan F., Moses E. Detection of early lung cancer among military personnel (DECAMP) consortium: study protocols. BMC Pulm Med. 2019;19(1):59. doi: 10.1186/s12890-019-0825-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanders K.J.C., Ash S.Y., Washko G.R., Mottaghy F.M., Schols A.M.W.J. Imaging approaches to understand disease complexity: chronic obstructive pulmonary disease as a clinical model. J Appl Physiol. 2017;124(2):512–520. doi: 10.1152/japplphysiol.00143.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Albert R.K., Connett J., Bailey W.C. Azithromycin for prevention of exacerbations of COPD. N Engl J Med. 2011;365(8):689–698. doi: 10.1056/NEJMoa1104623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anzueto A, Heijdra Y, Hurst JR. Controversies in COPD. European Respiratory Society; 2015.
- 19.Washko G.R. Diagnostic imaging in COPD. Semin Respir Crit Care Med. 2010;31(3):276–285. doi: 10.1055/s-0030-1254068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Diaz A.A., Zhou L., Young T.P. Chest CT measures of muscle and adipose tissue in COPD: gender-based differences in content and in relationships with blood biomarkers. Acad Radiol. 2014;21(10):1255–1261. doi: 10.1016/j.acra.2014.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McDonald M.-L.N., Diaz A.A., Ross J.C. Quantitative computed tomography measures of pectoralis muscle area and disease severity in chronic obstructive pulmonary disease: s cross-sectional study. Ann Am Thorac Soc. 2014;11(3):326–334. doi: 10.1513/AnnalsATS.201307-229OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kinsey C.M., Estépar R.S.J., van der Velden J., Cole B.F., Christiani D.C., Washko G.R. Lower pectoralis muscle area is associated with a worse overall survival in non-small cell lung cancer. Cancer Epidemiol Prev Biomark. 2017;26(1):38–43. doi: 10.1158/1055-9965.EPI-15-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ash S.Y., Harmouche R., Putman R.K. Clinical and genetic associations of objectively identified interstitial changes in smokers. Chest. 2017;152(4):780–791. doi: 10.1016/j.chest.2017.04.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ash S.Y., Harmouche R., Ross J.C. The objective identification and quantification of interstitial lung abnormalities in smokers. Acad Radiol. 2017;24(8):941–946. doi: 10.1016/j.acra.2016.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ash S.Y., Harmouche R., Ross J.C. Interstitial features at chest CT enhance the deleterious effects of emphysema in the COPDGene Cohort. Radiology. 2018;288(2):600–609. doi: 10.1148/radiol.2018172688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim V., Desai P., Newell J.D. Airway wall thickness is increased in COPD patients with bronchodilator responsiveness. Respir Res. 2014;15(1):84. doi: 10.1186/s12931-014-0084-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Putman R.K., Hatabu H., Araki T. Association between interstitial lung abnormalities and all-cause mortality. JAMA. 2016;315(7):672–681. doi: 10.1001/jama.2016.0518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hunninghake G.M., Hatabu H., Okajima Y. MUC5B promoter polymorphism and interstitial lung abnormalities. N Engl J Med. 2013;368(23):2192–2200. doi: 10.1056/NEJMoa1216076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Washko G.R., Lynch D.A., Matsuoka S. Identification of early interstitial lung disease in smokers from the COPDGene Study. Acad Radiol. 2010;17(1):48–53. doi: 10.1016/j.acra.2009.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Z, Monteiro CD, Jagodnik KM, et al. Extraction and analysis of signatures from the Gene Expression Omnibus by the crowd. Nat Commun [Internet] 2016 [cited 2019 Nov 18];7. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5052684/. Accessed March 1, 2020. [DOI] [PMC free article] [PubMed]
- 31.Malhotra S., Bustamante M.F., Pérez-Miralles F. Search for specific biomarkers of IFNβ bioactivity in patients with multiple sclerosis. PLoS One. 2011;6(8) doi: 10.1371/journal.pone.0023634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shapira S.D., Gat-Viks I., Shum B.O.V. A physical and regulatory map of host-influenza interactions reveals pathways in H1N1 infection. Cell. 2009;139(7):1255–1267. doi: 10.1016/j.cell.2009.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Indraccolo S., Pfeffer U., Minuzzo S. Identification of genes selectively regulated by IFNs in endothelial cells. J Immunol. 2007;178(2):1122–1135. doi: 10.4049/jimmunol.178.2.1122. [DOI] [PubMed] [Google Scholar]
- 34.Matta S.K., Olias P., Huang Z. Toxoplasma gondii effector TgIST blocks type I interferon signaling to promote infection. Proc Natl Acad Sci U S A. 2019;116(35):17480–17491. doi: 10.1073/pnas.1904637116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bolen C.R., Ding S., Robek M.D., Kleinstein S.H. Dynamic expression profiling of type I and type III interferon-stimulated hepatocytes reveals a stable hierarchy of gene expression. Hepatology. 2014;59(4):1262–1272. doi: 10.1002/hep.26657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Steiling K., van den Berge M., Hijazi K. A dynamic bronchial airway gene expression signature of chronic obstructive pulmonary disease and lung function impairment. Am J Respir Crit Care Med. 2013;187(9):933–942. doi: 10.1164/rccm.201208-1449OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Castaldi P.J., San José Estépar R., Mendoza C.S. Distinct quantitative computed tomography emphysema patterns are associated with physiology and function in smokers. Am J Respir Crit Care Med. 2013;188(9):1083–1090. doi: 10.1164/rccm.201305-0873OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schroeder J.D., McKenzie A.S., Zach J.A. Relationships between airflow obstruction and quantitative CT measurements of emphysema, air trapping, and airways in subjects with and without chronic obstructive pulmonary disease. AJR Am J Roentgenol. 2013;201(3):W460–W470. doi: 10.2214/AJR.12.10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Han M.K., Kazerooni E.A., Lynch D.A. Chronic obstructive pulmonary disease exacerbations in the COPDGene study: associated radiologic phenotypes. Radiology. 2011;261(1):274–282. doi: 10.1148/radiol.11110173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Han M.K., Agusti A., Calverley P.M. Chronic obstructive pulmonary disease phenotypes. Am J Respir Crit Care Med. 2010;182(5):598–604. doi: 10.1164/rccm.200912-1843CC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ash S.Y., Washko G.R. Interstitial lung abnormalities: risk and opportunity. Lancet Respir Med. 2017;5(2):95–96. doi: 10.1016/S2213-2600(17)30006-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Agusti A., Faner R. Lung function trajectories in health and disease. Lancet Respir Med. 2019;7(4):358–364. doi: 10.1016/S2213-2600(18)30529-0. [DOI] [PubMed] [Google Scholar]
- 43.Agustí A., Hogg J.C. Update on the pathogenesis of chronic obstructive pulmonary disease. N Engl J Med. 2019;381(13):1248–1256. doi: 10.1056/NEJMra1900475. [DOI] [PubMed] [Google Scholar]
- 44.Schneider D., Ganesan S., Comstock A.T. Increased cytokine response of rhinovirus-infected airway epithelial cells in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2010;182(3):332–340. doi: 10.1164/rccm.200911-1673OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ivashkiv L.B., Donlin L.T. Regulation of type I interferon responses. Nat Rev Immunol. 2014;14(1):36–49. doi: 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Singanayagam A., Loo S.-L., Calderazzo M.A. Anti-viral immunity is impaired in COPD patients with frequent exacerbations. Am J Physiol Lung Cell Mol Physiol. 2019;317(6):L893–L903. doi: 10.1152/ajplung.00253.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gough D.J., Messina N.L., Clarke C.J.P., Johnstone R.W., Levy D.E. Constitutive type I interferon modulates homeostatic balance through tonic signaling. Immunity. 2012;36(2):166–174. doi: 10.1016/j.immuni.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Crotta S., Davidson S., Mahlakoiv T. Type I and type III interferons drive redundant amplification loops to induce a transcriptional signature in influenza-infected airway epithelia. PLoS Pathog. 2013;9(11) doi: 10.1371/journal.ppat.1003773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mallia P., Message S.D., Gielen V. Experimental rhinovirus infection as a human model of chronic obstructive pulmonary disease exacerbation. Am J Respir Crit Care Med. 2011;183(6):734–742. doi: 10.1164/rccm.201006-0833OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.García-Valero J, Olloquequi J, Montes JF, et al. Deficient pulmonary IFN-β expression in COPD patients. PLoS ONE [Internet] 2019 [cited 2019 Oct 9];14(6). https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6553750/. Accessed March 1, 2020.
- 51.Schuliga M. NF-kappaB signaling in chronic inflammatory airway disease. Biomolecules. 2015;5(3):1266–1283. doi: 10.3390/biom5031266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Edwards M.R., Bartlett N.W., Clarke D., Birrell M., Belvisi M., Johnston S.L. Targeting the NF-kappaB pathway in asthma and chronic obstructive pulmonary disease. Pharmacol Ther. 2009;121(1):1–13. doi: 10.1016/j.pharmthera.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zaynagetdinov R., Sherrill T.P., Gleaves L.A. Chronic NF-κB activation links COPD and lung cancer through generation of an immunosuppressive microenvironment in the lungs. Oncotarget. 2015;7(5):5470–5482. doi: 10.18632/oncotarget.6562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Milara J., Peiró T., Serrano A., Cortijo J. Epithelial to mesenchymal transition is increased in patients with COPD and induced by cigarette smoke. Thorax. 2013;68(5):410–420. doi: 10.1136/thoraxjnl-2012-201761. [DOI] [PubMed] [Google Scholar]
- 55.Shi J, Li F, Luo M, Wei J, Liu X. Distinct Roles of Wnt/β-Catenin Signaling in the Pathogenesis of Chronic Obstructive Pulmonary Disease and Idiopathic Pulmonary Fibrosis. Mediators Inflamm [Internet] 2017 [cited 2018 Nov 17];2017. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5447271/. Accessed March 1, 2020. [DOI] [PMC free article] [PubMed]
- 56.Ostridge K, Williams N, Kim V, et al. Distinct emphysema subtypes defined by quantitative CT analysis are associated with specific pulmonary matrix metalloproteinases. Respir Res [Internet] 2016 [cited 2018 Nov 13];17. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4962504/. Accessed March 1, 2020. [DOI] [PMC free article] [PubMed]
- 57.Lange P., Celli B., Agustí A. Lung-function trajectories leading to chronic obstructive pulmonary disease. N Engl J Med. 2015;373(2):111–122. doi: 10.1056/NEJMoa1411532. [DOI] [PubMed] [Google Scholar]
- 58.Rice M.B., Ljungman P.L., Wilker E.H. Long-term exposure to traffic emissions and fine particulate matter and lung function decline in the Framingham heart study. Am J Respir Crit Care Med. 2015;191(6):656–664. doi: 10.1164/rccm.201410-1875OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Porter K.L., Green F.H.Y., Harley R.A. Evaluation of the pulmonary toxicity of ambient particulate matter from Camp Victory, Iraq. J Toxicol Environ Health A. 2015;78(23-24):1385–1408. doi: 10.1080/15287394.2015.1072611. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.