Abstract
Antibiotic resistance genes (ARGs) are a relatively new type of pollutant. The rise in antibiotic resistance observed recently is closely correlated with the uncontrolled and widespread use of antibiotics in agriculture and the treatment of humans and animals. Resistant bacteria have been identified in soil, animal feces, animal housing (e.g., pens, barns, or pastures), the areas around farms, manure storage facilities, and the guts of farm animals. The selection pressure caused by the irrational use of antibiotics in animal production sectors not only promotes the survival of existing antibiotic-resistant bacteria but also the development of new resistant forms. One of the most critical hot-spots related to the development and dissemination of ARGs is livestock and poultry production. Manure is widely used as a fertilizer thanks to its rich nutrient and organic matter content. However, research indicates that its application may pose a severe threat to human and animal health by facilitating the dissemination of ARGs to arable soil and edible crops. This review examines the pathogens, potentially pathogenic microorganisms and ARGs which may be found in animal manure, and evaluates their effect on human health through their exposure to soil and plant resistomes. It takes a broader view than previous studies of this topic, discussing recent data on antibiotic use in farm animals and the effect of these practices on the composition of animal manure; it also examines how fertilization with animal manure may alter soil and crop microbiomes, and proposes the drivers of such changes and their consequences for human health.
Keywords: antibiotic resistance genes, antibiotic-resistant bacteria, antibiotic use, animal agriculture, fecal matter, manure resistome, soil resistome, plant resistome
Antibiotic Use in Animal Farms
The spread of antibiotic-resistant bacteria (ARB) is a growing problem worldwide. In 2017, the most important ARB were included in a list published by the World Health Organization (WHO). The ARB families that were believed to pose the greatest threat to human health were categorized as critical, high or medium priority, depending on the need to seek effective treatment options.
It has been estimated that antimicrobial resistance (AMR) is responsible for 25,000 deaths/year in the European Union (EU), and 700,000 worldwide. It has also been predicted that by 2050, AMR will be responsible for more deaths than cancer (European Commission, 2018). With the discovery of penicillin, in 1928, many life-threatening or even lethal diseases became curable, with clear benefits for veterinarians and animal breeders; however, since the 1960s, antibiotics have been widely applied in sub-lethal doses as growth promoters for food-producing animals (Hassan et al., 2018). Such extensive, uncontrolled use may result in the presence of low, sub-inhibitory concentrations in the tissues and guts of treated animals (Lim et al., 2020) and in the environment (Khan et al., 2013). Although the precise complete mechanisms of action remain unclear, it has been hypothesized that when applied in sub-lethal doses, antibiotics stimulate the intestinal synthesis of vitamins, lower the total amount of bacteria in the intestinal tract by reducing competition between microorganism and host for nutrients, inhibits the growth of harmful bacteria, and modifies the microbial metabolism of the rumen (Economou and Gousia, 2015).
However, the chronic application of such sub-therapeutic doses favors the selection of ARB by promoting their growth or introducing de novo mutations. One of the first reviews of the effect of sublethal doses of antibiotics on the bacterial community was published by Lorian (1975). The presence of ARB may enhance the transfer of antibiotic-resistant genes (ARGs) between enteric bacteria in the intestinal tract of the host animal, and affect quorum sensing (e.g., azithromycin in Pseudomonas aeruginosa, Staphylococcus aureus, or Streptococcus pneumoniae). In addition, the presence of antibiotics can stimulate biofilm formation and horizontal gene transfer (HGT) in some bacteria; for example, the transfer of azithromycin, ciprofloxacin or tigecycline resistance has been observed in Enterococcus faecalis and P. aeruginosa. It can also increase the rate of recombination and selected gene expression in the bacterial community, such as ciprofloxacin, norfloxacin, rifampicin, gentamycin, and tetracycline in Escherichia coli, P. aeruginosa, or S. aureus (Lorian, 1975). The mechanisms of action of sublethal levels of antibiotics on bacteria are described in detail by Andersson and Hughes (2014). In food-producing animals, antibiotic use influences the functions of enteric bacteria and can temporarily increase antibiotic resistance in the fecal microbiome (Chee-Sanford et al., 2009; Broom, 2017).
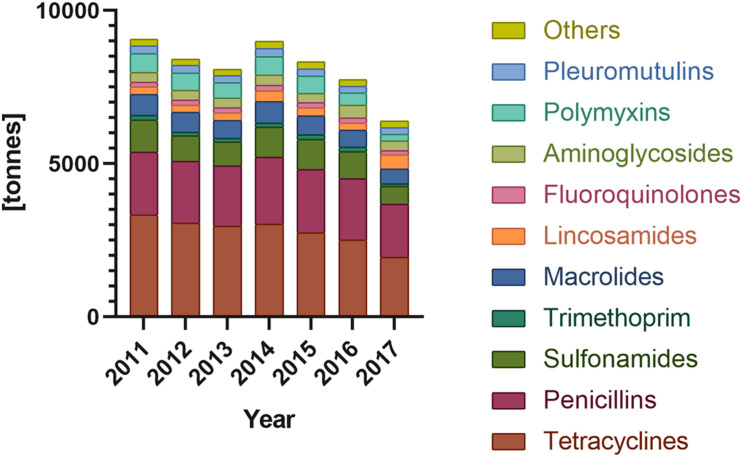
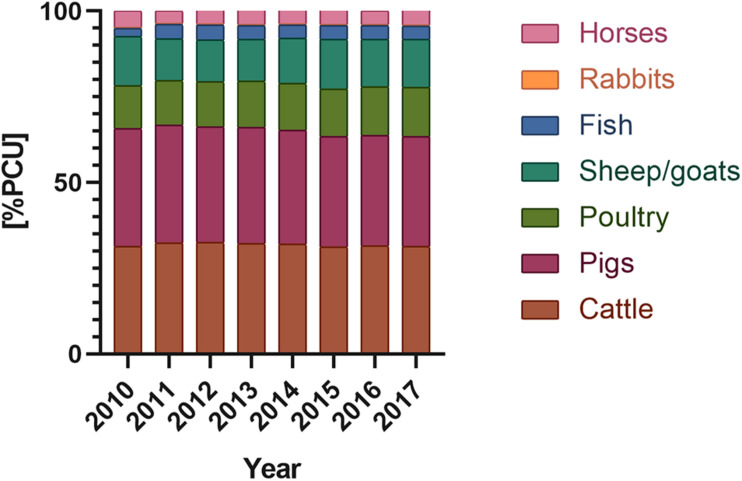
The majority of ARG transmission occurs via HGT, i.e., whereby mobile genetic elements (MGE), such as plasmids or transposons coding for ARGs, are exchanged between bacterial species, even those that are not closely related (Redondo-Salvo et al., 2020). The dynamics of transfer depend not only on the presence of positive selection, i.e., antibiotic concentrations in the environment ranging from values much higher than minimal inhibitory concentrations to those several hundred-fold lower, but also the spatial structure of the community and the presence of predators (Cairns et al., 2018). The WHO, World Organization for Animal Health (OIE), and the Food and Agriculture Organization (FAO) have officially stated that the non-human use of different types of antimicrobials may have harmful consequences for human health (So et al., 2015). Although some countries have officially restricted the use of antimicrobials in livestock to only medical purposes (e.g., the EU in 2006 in accordance to 1831/2003/EC legislation), they are still overused on some regions characterized by highly-intensive livestock production, such as the United States, Russia, India, China, and South Africa. In the US, antimicrobial treatment in food-producing animals has been estimated to account for approximately 80% of total annual use; the vast majority of these antimicrobials are believed to include essential human medicines used for the treatment of common infections, or which are necessary for performing surgeries, organ transplantations or chemotherapy in humans (Van Boeckel et al., 2015) (Figures 1, 2).
FIGURE 1.
Antibiotic sales for food-producing animals in Europe sort by antibiotic classes (according to ECDC/EFSA/EMA second joint report on the integrated analysis of the consumption of antimicrobial agents and occurrence of antimicrobial resistance in bacteria from humans and food-producing animals, 2017).
FIGURE 2.
Antimicrobials used in domestic animals (according to according to ECDC/EFSA/EMA second joint report on the integrated analysis of the consumption of antimicrobial agents and occurrence of antimicrobial resistance in bacteria from humans and food-producing animals, 2017) [PCU] – population corrected unit; PCU a technical unit of measurement, used only to estimate sales corrected by the animal population in the individual countries; 1 PCU = 1 kg of different categories of livestock and slaughtered animals.
A study of antimicrobial use in Southeast Asia indicated that, excluding feed, farmers in this region administered 46 mg of different antimicrobial compounds per kg of live pigs and 52–276 mg per kilogram of live chickens per year (Nhung et al., 2016). In addition, a 2014 study in the EU found the average intake of antimicrobials to be 123.7 mg/kg in humans and 151.5 mg/kg in animals (European Centre for Disease Prevention, and Control [ECDC], European Food Safety Authority [EFSA], and European Medicines Agency [EMA], 2017).
The use of antibiotics for non-medical purposes, or for treating entire extensive animal production facilities when a single animal falls ill, has resulted in a growth in ARB in the animal production chain. It has been estimated that 84% of the antimicrobial compounds administered in chicken farms in the Mekong Delta region were given only for prophylactic purposes; of these, the most common were penicillins, lincosamides, quinolones, and combinations of sulfonamides with trimethoprim (Nhung et al., 2016). Elsewhere, the most common antibiotics found to be present in pig, beef and turkey manure were tetracyclines (oxytetracycline and chlortetracycline), tylosin, sulfamethazine, amprolium, monensin, virginiamycin, penicillin, and nicarbazin (De Liguoro et al., 2003; Kumar et al., 2005).
The Food and Drug Administration (FDA) has approved 18 classes of antimicrobials for use in food-producing animals (Medicine, 2019). However, the WHO reports that 57% of all antimicrobials used in animal production are essential for human medicine, including amoxicillin, colistin, tetracyclines, neomycin, lincomycin, and bacitracin. Of the FDA classes, the most widely used in human medicine worldwide are the penicillins, macrolides, and fluoroquinolones, while the tetracyclines, penicillins, and sulfonamides were the highest selling for food-producing animals.
In the EU/EEA (the European Union/European Economic Area) carbapenems and monobactams are not approved for use in food-producing animals, and pleuromutilins are not authorized for use in humans. In addition, higher total consumption of the penicillins, cephalosporins (all generations) and fluoroquinolones was demonstrated by humans than food-producing animals (European Centre for Disease Prevention, and Control [ECDC], European Food Safety Authority [EFSA], and European Medicines Agency [EMA], 2017). A complete list of antibiotics allowed for use in food production animals, divided according to veterinary importance (Critically Important, Highly Important and Important) was first prepared in 2007 by the OIE in consultation with the FAO and WHO, and is constantly being revised; the most recent update was released in May 2018 by the World Assembly of OIE Delegates (International Office of Epizootics, 2015; Góchez et al., 2019).
Although environmental factors have long been known to affect human health, the relationship between the spread of AMR and antibiotic use in agriculture was first recorded in the 1969 Swann report (Soulsby, 2007). Even though antibiotics and ARGs/ARB are ancient and naturally-occurring substances, they are nevertheless considered emerging pollutants associated closely with human-impacted environments (He et al., 2019; Zhang et al., 2019). The global debate on the scale of the threat posed by antimicrobial use in livestock was intensified by the recent finding of a plasmid-mediated colistin resistance gene (mcr-1) in commensal E. coli from pigs, pork products and humans in China (Liu et al., 2016).
In 2008, the FAO, WHO, OIE, the United Nations Children’s Fund (UNICEF), United Nations System Influenza Coordination, and the World Bank developed a holistic approach entitled “Contributing to One World, One Health-A Strategic Framework for Reducing Risks of Infectious Diseases at the Animal-Human-Ecosystems Interface” including a recommendation for a One Health action plan for global health. In 2017, the European Commission adopted the WHO One Health framework, naming AMR as one of the main concerns. The proposed approach holistically regards the health of humans, animals, and different environmental compartments, such as water or soil, as a single network, where one part inseparably depends on another. This framework has three major aims: (1) making the EU a region with best practice in AMR management, (2) boosting research, development, and innovation in AMR, and (3) shaping the global approach to addressing AMR. Each aim has been divided into smaller tasks that address existing gaps in knowledge concerning AMR in the environment, such as the role of the environment in ARGs/ARB transmission, the routes of ARGs/ARB transmission, the most critical hot spots, human health risk assessment, and the establishment of tracking and detection systems.
The current limitations on antimicrobial use in livestock are explained in the document “Council conclusions on the next steps under a One Health approach to combat AMR” (2016). In addition, a good review of the existing global policies on antibiotics use in livestock is provided by Walia et al. (2019).
The Use Of Antibiotics In The Poultry Industry And Its Effect On The Poultry Manure Resistome
One of the most significant sectors of meat and egg production worldwide is that of poultry production. In 2018, global poultry population (laying hens and broilers) had reached 23.7 billion, growing from 22.85 billion in 2017 and 14.38 billion in 2000. The largest chicken producer in 2018 was China, with 5274.48 million animals, followed by Indonesia with 2384.15 million, and the US with 1973.38 million. China is also the largest egg producer globally, with 529 billion eggs, followed by the US with 106.7 billion. Moreover, in 2018, in the US alone, 22.6 million tons of broiler meat was produced. It has been predicted that in 2020, the global export of poultry meat will increase by 3.7% to reach 13.8 million tones. Thailand, Brazil, Turkey, Ukraine, the US, and the EU are the leading poultry exporters, from whom almost 80% of exported poultry meat originates (FAOSTAT, 2020).
Since the discovery of antibiotics, strict biosecurity guidelines and prevention techniques have been employed by the poultry and meat production industry to not only increase its production efficiency and eliminate the risk of zoonotic diseases, but enhance the growth and weight of the broilers (Collett et al., 2019). Although the use of antibiotics is strongly regulated in food-producing animals in the EU and the US (European Commission, 2005; World Health Organization and WHO Advisory Group on Integrated Surveillance of Antimicrobial Resistance (AGISAR), 2017), they are often used for disease prevention in many poultry exporting countries, such as Brazil and China (Roth et al., 2019). A few classes of antibiotics are approved as additions to animal fodder in Canada, such as bambermycin, bacitracin, salinomycin, penicillin, virginiamycin, and chlortetracycline (Diarra and Malouin, 2014).
In addition, in a 2017 study of eight farms in Thailand, where 14,000 3 kg broilers were raised, it was estimate that 303 mg of antibiotics were used per chicken as routine prophylaxis; this mixture included amoxicillin, colistin, doxycycline, oxytetracycline, and tilmicosin. According to the WHO, amoxicillin and colistin are critically important for human medicine, with colistin being considered the primary therapeutic option to treat diseases caused by resistant gram-negative bacteria, particularly enterobacteria (Wongsuvan et al., 2018).
In survey-based studies in Ghana in 2016, farmers from 400 poultry farms reported using antibiotics for prophylactic purposes, including penicillins (26.22%), tetracyclines (24.13%), and aminoglycosides (20.51%) (Andoh et al., 2016).
Besides growth promotion and disease prevention, antibiotics are also prescribed by veterinarians for treating illnesses. In 2015, a total of 8361 tons of antimicrobial agents were sold for veterinary use in the EU (European Medicines Agency, 2017).
Tetracyclines, with their broad spectrum of action, are one of the most frequently-used antimicrobials in the poultry industry, not only for prophylaxis or antibacterial therapy but also as growth promoters (Ljubojević et al., 2017). Interestingly, while tetracyclines constitute two-thirds of the antibiotics applied to animals in the US (Gonzalez Ronquillo and Angeles Hernandez, 2017), they accounted for less than 40% in the EU between 2015 and 2017 (OIE, 2018). However, the tetracyclines are poorly absorbed by the animal, and their active residues can concentrate in meat, urine, and feces. A study of broiler farms in Egypt, found high levels of chlortetracycline and oxytetracycline in chicken litter and droppings, these values being, respectively, 6.05 and 2.47 μg/g for chlortetracycline and 5.9 and 1.33 μg/g for oxytetracycline. Slightly lower amounts of tetracycline and doxycycline were found: 1.9 μg/g in litter and 0.46 μg/g in droppings for tetracycline, and 0.87 μg/g and 0.02 μg/g for doxycycline. More importantly, the researchers report the presence of three tetracycline resistance genes (tetM, tetW, tetQ) in the intestinal contents: tetW was observed in all analyzed samples, tetQ in 42% of them, and tetM in only 17% (Mahmoud and Abdel-Mohsein, 2019).
Despite being banned as growth promoters in the early 1970s (EC directive 70/524), the total consumption of tetracyclines in food-producing animals in Europe was still estimated at 3138 tons in 2012. Carballo et al. (2016) report that oxytetracycline was present in 37% of tested manure samples, with a maximum concentration of 0.88 mg/kg, and doxycycline in one sample, with a concentration of 0.53 mg/kg. The existence of antibiotic residues in the environment creates on-site selection pressure, may result in the presence of ARB, which are often isolated from manure samples. In Nigeria, five potential human pathogens were isolated from chicken manure, such as Salmonella typhi, E. coli, Shigella dysenteriae, S. aureus, and Aeromonas hydrophila, all of which were resistant to tetracycline (Omojowo and Omojasola, 2013).
Arguably, a greater threat to human and animal health is posed by multidrug-resistant (MDR) bacteria, with an alarming increase being observed in the number of MDR strains isolated from the poultry meat production chain (Afridi et al., 2020). Unusually high levels of MDR bacteria, especially those of the Enterobacteriaceae, have been isolated from poultry. Portuguese studies based on livestock manure taken from pig, dairy and poultry farms and slaughterhouses found the highest level of resistance to tetracycline, trimethoprim/sulfamethoxazole, chloramphenicol, and amoxicillin/clavulanic acid in strains isolated from poultry farms. In addition, the highest prevalence of ciprofloxacin-resistant bacteria was observed in poultry, which may be related to the growing use of ciprofloxacin to treat infections. In addition, although MDR bacteria were isolated from all samples, the highest number of strains resistant to seven or more antibiotics were isolated from poultry samples. A number of chloramphenicol, quinolone, tetracycline, and sulfonamide resistance genes were identified (Amador et al., 2019).
A similar trend was also noted in Belgium. A study of AMR in commensal E. coli isolated from veal calves, young beef cattle, broiler chickens, and slaughtered pigs found the highest number of isolated MDR bacteria to be present in broiler chickens (Hanon et al., 2015).
A high prevalence of E. coli and Klebsiella pneumoniae resistant to β-lactams with ESBL (Extended-Spectrum Beta-Lactamase) and AmpC (clinically important β-lactamases; cephalosporinases) phenotypes was also identified in a study of MDR gram-negative bacteria isolated from poultry feces kept on farms in the Lebanon. The real-time PCR assay performed on 112 strains of ESBL producers detected blaSHV in 20% of strains, blaTEM in 89%, and blaCTX–M in 53%. In addition, more than half (66%) of the isolated ESBL/AmpC strains were also resistant to gentamicin, but only one AmpC β-lactamase gene (blaCMY) was found among all samples (Dandachi et al., 2018).
Resistance to extended spectrum cephalosporins (ESBL genes) has also been observed among Salmonella serovars. This could have particularly serious consequences, as these bacteria are key etiological factors in diseases among poultry, and may also cause severe illness in humans; the acquisition of antibiotic resistance may significantly limit the range of treatment options (Büdel et al., 2020). Rayamajhi et al. (2010) report the presence of ampicillin-resistant Salmonella isolates in poultry feces, the environment and egg yolks on poultry farms in South Korea (Table 1). Among all antibiotics tested, resistance was most commonly observed for sulfamethoxazole (74.7%) and nalidixic acid (63.7%), with streptomycin (38.5%), tetracycline (28.6%), and amoxicillin resistance (18.7%) being less common. Furthermore, the blaTEM–1 gene was present in all ampicillin-resistant strains and the blaDHA–1 gene in stains with lower susceptibility to cefoxitin, as indicated by β-lactamase production ability. A 2019 study from Kenya found that almost 50% of Salmonella samples isolated from manure from broilers and layers were resistant to amoxicillin. In addition, 28% of Salmonella strains were resistant to co-trimoxazole, 11%, to tetracycline and 6% to streptomycin. A search for ARGs revealed the presence of blaTEM in 46% of all isolated bacteria with the blaCTX–M gene present in 18% (Langata et al., 2019).
TABLE 1.
Antibiotic resistance bacteria and genes in poultry manure.
| Bacteria species | Phenotype | Resistance genes | Source | Country | References |
| Poultry manure | |||||
| Salmonella spp. | Extended spectrum cephalosporins | blaTEM–1, blaDHA–1 | Feces, eggshell, dead egg yolk, cloaca, liver, water, environmental dust | South Korea | Rayamajhi et al., 2010 |
| Escherichia coli | blaTEM, blaSHV, blaCTX–M | Fecal swabs | Lebanon | Dandachi et al., 2018 | |
| Klebsiella spp. | |||||
| Enterobacteriaceae | Quinolones (CIP) | qnrB, qnrC, qnrD, qnrS | Poultry manure | Portugal | Amador et al., 2019 |
| Tetracyclines (TET) | tetA, tetB, tetC, tetE, tetK, tetL, tetM, tetO | ||||
| Sulfonamides | sul1, sul2, sul3 | ||||
| Enterococcus spp. | Vancomycin (VRE) | vanA | Poultry manure | Greece | Tzavaras et al., 2012 |
| Enterococcus spp. | Macrolides (Erythromycin) | ermB, ermA, mrsC | Poultry manure | United States | Graham et al., 2009 |
| Staphylococcus spp. | Macrolides (Erythromycin) | ermA, mrsA/B | |||
| Staphylococcus spp. | MRSA | mecA; mecR1; mecI | Poultry manure | South Korea | Lee, 2006 |
Together with the high prevalence of the MDR strains among the Enterobacteriaceae family, the spread of Vancomycin-Resistant Enterococci (VRE) is widely regarded as one of the greatest threats to human health care. A key factor in the spread of vancomycin resistance is believed to involve a glycopeptide antibiotic called avoparcin, which has been applied in animal husbandry as a growth promoter. Although its use in livestock was banned in the EU early in 1997 (EC directive 97/6/EC), relatively high numbers of VRE are still observed in poultry meat production in the EU, and this can be considered a potential reservoir of vancomycin resistance. One study in Greece found that approximately 14.4% of the E. faecalis strains isolated from broilers and poultry fecal samples showed vancomycin resistance, and isolated strains demonstrated resistance to several other antibiotics, such as tetracycline (100%), erythromycin (54.4%), ampicillin (16.5%), and ciprofloxacin (30.4%). Analyses have detected the vanA gene in 14.4% of fecal samples from poultry and the vanC gene in 8.2% (Table 1) (Tzavaras et al., 2012).
Together with the emergence of VRE strains, the preservation of resistant gram-positive bacteria in stored poultry manure is also of great concern. One study by Graham et al. (2009) examined the fate of antibiotic resistant Enterococcus spp. and Staphylococcus spp. and the persistence of ARGs in litter after a 120-day storage period (Table 1). The findings revealed the presence of quinupristin-dalfopristin resistance in 11% of the isolated Staphylococcus strains and erythromycin resistance in 57%; in addition, 31% of E. faecium were resistant to both antibiotics. The most commonly identified ARGs were ermB in Enterococcus spp. and ermA in Staphylococcus spp. Moreover, the msrC gene was found in one Enterococcus isolate and this was found to be homologous with msrA found in Staphylococcus spp. It should be stressed that ARGs were found in all collected samples during a 120-day storage period or even later (Graham et al., 2009).
Staphylococcus spp. are considered typical of the chicken microflora. Although they are harmless in most cases, they are known to act as opportunistic pathogens, and can cause severe infections, particularly the bacteria that harbor ARGs. Methicillin-resistant S. aureus (MRSA) isolated from poultry may pose a considerable risk for human health, as a high chance of zoonotic transfer exists during the breeding or fattening period (Zaheer et al., 2017). MRSA bacteria were found to be present in almost 90% of fecal chicken swabs taken from small poultry farms in Nigeria, and more worryingly, in 83.3% of nasal swabs taken from personnel working on the farms (Journals and Oke, 2013). MRSA bacteria harboring mecA gene were also successfully isolated from chicken samples in South Korea (Lee, 2006) (Table 1).
Subtherapeutic doses of antibiotics used in poultry farms exert selection pressure on the bacteria of the intestinal flora of the animals, thus encouraging the spread of ARB into the environment (Suresh et al., 2018). Being located on MGEs such as plasmids, transposons and integrons, ARGs can easily be transmitted between bacteria by HGT. A good example of such spread is that of plasmid-mediated colistin resistance; this can represent a considerable risk for human health, as colistin is one of the strategic drugs used against carbapenem-resistant Enterobacteriaceae (Wang et al., 2018). Unfortunately, because of the long history of colistin usage in animals and the complete lack of surveillance until 2014 (Commission Implementing Decision 2013/652/EU), it is difficult to track colistin usage and detect resistance (Kempf et al., 2016). However, what is certain is that the first plasmid carrying the colistin resistance gene mcr-1 was isolated from Enterobacteriaceae bacteria from a pig fattening facility (Liu et al., 2016). In Brazil, of the 280 Enterobacteriaceae strains isolated from chicken fecal samples in a study from 2003 to 2015, 113 were found to be colistin resistant; the mcr-1 gene was identified in 14 E. coli strains (Fernandes et al., 2016). Elsewhere, the mcr-1 gene was detected in eight of 12 (∼66%) tested chicken manure samples (Gao et al., 2019); interestingly, the presence of detected mcr-1 genes was effectively reduced by 90% during a 30-day composting process.
Class 1 and 2 integrons are also commonly found among MDR Enterobacteriaceae isolates, which may indicate that resistance is mediated by plasmid transfer. A study of the int1 and int2 genes in clinical and livestock samples, including poultry, by Goldstein et al. (2001) found the int1 gene to be present in 79% of Salmonella isolates sampled from chickens and int2 in 2%; however, the int2 gene was present in only 14% of E. coli isolates. Furthermore, a study of 55 E. coli strains in broilers and layers in Kenya detected int1 genes in 26% of isolates, with most coming from β-lactamase positive isolates (Langata et al., 2019).
A full understanding of the development and transmission of AMR in poultry production is essential for creating effective risk management strategies for preventing the spread of resistant bacteria from animals to humans and providing safe sources of food.
The Use Of Antibiotics In The Cattle Industry And Its Effect On The Cattle Manure Resistome
Global bovine meat production reached 71.1 million tons in 2018, with the largest producers being Brazil, the US, Argentina, the EU, and Australia (FAOSTAT, 2020; Meat Market Review). In addition, there are more than 264 million dairy cows worldwide, producing nearly 600 million tons of milk every year. India has the highest number of dairy cows globally, with over 40 million cows, followed by the US, China, Pakistan, and Brazil (FAOSTAT).
The use of antibiotics in dairy cattle has the potential to stimulate the development and dissemination of ARGs among different types of enteric bacteria characteristic for ruminants, as well as potentially pathogenic species related to the fecal microbiome. Some of these bacteria may be pathogenic to humans, and hence could pose a threat to human health if they additionally acquire ARGs. Moreover, non-pathogenic bacteria from manure might transfer ARGs to pathogens existing in manure, soil, or food consumed by animals or humans (Karami et al., 2007; Brichta-Harhay et al., 2011). Furthermore, manure from dairy cows, commonly used as a soil fertilizer, may harbor diverse new variants of ARGs from the gut microbiota of cattle (Wichmann et al., 2014).
A range of antibiotics are used in the cattle production industry, including aminoglycosides, β-lactams, chloramphenicol, fluoroquinolones, glycolipids, ionophores, macrolides, quinolones, streptogramins, sulfonamides, and tetracyclines. The majority are used in dairy cow husbandry for growth promotion (when allowed), prophylaxis, and treatment of most complex diseases such as mastitis or metritis, or as dry cow therapy: the prophylactic use of antibiotics to avoid mammary gland inflammation in animals after lactation (International Office of Epizootics, 2015). In North America, the major antibiotics administered sub-therapeutically in the diet to beef and dairy cattle are chlortetracycline (16%), tylosin (10%), and sulfamethoxazole (9%). These antibiotics demonstrate quite high excretion rates, being approximately 90% for sulfamethoxazole, 65% for chlortetracycline, and, depending on the form of medication, between 50 and 100% for tylosin. Moreover, it is known that chlortetracycline strongly binds to soil particles and may accumulate in this environment, and that sulfamethoxazole persists in the environment for long periods and may be potentially washed off from the surface (Aust et al., 2008).
The most widely-used antibiotics in cattle production systems are tetracyclines, being used to treat skin, respiratory and gastrointestinal tract diseases. Furthermore, tetracycline resistance may develop rapidly because tetracycline ARGs are often located on MGE (Leclercq et al., 2016). In Europe, macrolides and lincosamides are also commonly used to treat a range of common infections in cattle such as mastitis, foot lesions, respiratory and genital infections. Many studies have examined the abundance of genes conferring resistance to MLSB (macrolides–lincosamides-streptogramin B) and tetracyclines in fecal microbial communities in beef cattle receiving antimicrobial agents, e.g., tylosin in fodder (Chen et al., 2008) (Table 2). Alexander et al. (2011) report the presence of five classes of tetracycline resistance genes in bovine fecal samples, as well as two classes of sulfonamide resistance genes and five classes of erythromycin resistance genes (Table 2).
TABLE 2.
Antibiotic-resistant bacteria and antibiotic resistance genes in cow manure.
| Bacteria species | Target antibiotics (major) | Resistance genes | Country | References |
| na | Macrolide (Erythromycin) | ermB, ermF, ermT, ermX | United States | Chen et al., 2008 |
| Tetracycline | tetA/C, tetG, and tetM, tetO, tetP, tetQ, tetS, tetT, tetW | |||
| na | Tetracycline | tetB, tetC, tetM, tetW, tetL | Canada | Alexander et al., 2011 |
| Sulfonamide | sul1, sul2 | |||
| Macrolide (Erythromycin) | ermA, ermB, ermT, ermF, ermX | |||
| na | Beta-lactam | bla2 | United States | Wichmann et al., 2014 |
| Aminoglycoside (Kanamycin) | nat, aph, aacA-aphD | |||
| Tetracycline | tetW, tetO | |||
| Chloramphenicol | cat | |||
| na | Tetracycline | tetC, tetM, tetW | Canada | Holman et al., 2019 |
| Macrolide (Erythromycin) | ermX | |||
| Sulfonamide | sul2 | |||
| Salmonella spp. | Beta Lactam | blaCMY–2, ampC | United States | Winokur et al., 2001 |
| Escherichia coli | Beta Lactam | blaCMY–2, ampC | United States | |
| Beta Lactam | blaTEM, blaCMY–2, blaCTX–M, blaSHV | Canada | Awosile et al., 2018 | |
| Polymyxin (Colistin) | mcr-1 | China | He et al., 2017b | |
| Beta Lactam | blaVIM–2, blaNDM–5 | |||
| Klebsiella spp. | Beta Lactam | blaTEM–1, blaSHV–1, blaOXA–1 | United States | Winokur et al., 2001 |
| Aminoglycosides | rmtB, aac(6)-Ib-cr | |||
| Quinoxalines | oqxAB | |||
| Quinolone | qnrS1, qnrB2 | |||
| Beta Lactam | blaNDM–5 | China | He et al., 2017a | |
| Beta Lactam | blaKPC, blaSHV, blaTEM | China | Yang et al., 2019 | |
| Multidrug | tolC | |||
| Quinolone | qnrA, qnrB | |||
| Acinetobacter spp. | Beta Lactam | blaOXA–23 | France | Poirel et al., 2012 |
| Beta Lactam | blaOXA–497 | USA | Webb et al., 2016 | |
| Beta Lactam | blaOXA–23, blaOXA–58 | Lebanon | Al Bayssari et al., 2015 | |
| Pseudomonas spp. | Beta Lactam | blaVIM–2 | Lebanon | Al Bayssari et al., 2015 |
| Enterococcus spp. | Beta Lactam | blaZ | South Africa | Tanih et al., 2017 |
| Macrolides–lincosamides-streptogramin B | ermB | |||
| Tetracycline | tetM | |||
| Glicopeptide (Vancomycin) | vanB, vanC1 | |||
| Glicopeptide (Vancomycin) | vanC, vanA | French | Haenni et al.,2009 |
na, not applicable (data obtained from metagenomic study).
The cattle industry also makes considerable use of β-lactams, first- and second-generation cephalosporins for mastitis treatment in dairy cattle. However, in the US, ceftiofur, a third-generation cephalosporin, is most commonly used for treating mastitis, respiratory disease, pododermatitis and metritis, while cefquinome, a fourth-generation cephalosporin, is applied for respiratory disease (Zalewska and Popowska, 2020).
Chambers et al. (2015) report that cows treated with ceftiofur demonstrate an increased proportion of β-lactam resistance and MDR in bacterial isolates compared to untreated controls, as well as a greater prevalence of gene sequences associated with phages, prophages, transposable elements and plasmids. These findings may suggest that treatment with this antibiotic may enhance ARG transfer. After ceftiofur treatment, additional functional shifts were noted, such as an increase in the proportion of gene sequences associated with stress response, chemotaxis and resistance to toxic compounds, and a decrease in those related to cell division, cell cycle and metabolism of aromatic compounds. In addition, measurable taxonomic shifts were observed, characterized by an increase in Bacteroidia and decrease in Actinobacteria. An increase in the number of E. coli isolates carrying blaCMY–2 was also observed during parenteral ceftiofur therapy; this can lead to a higher frequency of plasmid-mediated ARGs transfer by HGT to other enteric bacteria, including potential zoonotic pathogens. However, this would require not only the donor but also the recipient populations to be present in sufficient numbers (Stecher et al., 2012).
Although some previous studies have suggested that carbapenem-resistant bacteria are rare in livestock in the US (Webb et al., 2016; Mollenkopf et al., 2017), Vikram and Schmidt (2018) report the presence of a functional blaKPC–2 gene (K. pneumoniae carbapenemases) in the feces of beef cattle raised with antibiotics and those not; they also indicate that the blaKPC–2 gene may be mobilized. However, Agga et al. (2015) indicate that carbapenem-resistant genes are more frequently detected in effluent from US municipal wastewater treatment plant than in cattle catchment ponds for feedlot runoff or swine waste lagoons. These findings may suggest that carbapenem resistance may be more closely related to a close human environment than animal food production facilities.
Nevertheless, the dissemination of carbapenemase-producing bacteria in livestock has become a matter of global concern. Winokur et al. (2001) demonstrated the occurrence of E. coli and Salmonella spp. strains resistant to cephamycins and third-generation cephalosporins in feces samples isolated from food-producing animals. They also report the presence of carbapenemase-producing Enterobacteriaceae in dairy cow feces, e.g., K. pneumoniae harboring blaNDM–5, as well as various resistance genes including blaTEM–1, blaSHV–1, blaOXA–1, rmtB, oqxAB, qnrS1, qnrB2, aac(6)-Ib-cr (Table 2). In addition, carbapenemase-producing non-Enterobacteriaceae such as Acinetobacter spp. (related to A. lwoffii) harboring the blaOXA–23 carbapenemase gene have been isolated from dairy cattle feces, as have Acinetobacter baumannii with the blaOXA–497 gene (Bonardi and Pitino, 2019).
A study of rectal samples of dairy cattle in France by Poirel et al. (2012) identified the presence of Acinetobacter spp. isolates that were resistant to penicillins, combinations of penicillins with β-lactamase inhibitors, and carbapenems, but which were fully susceptible to cefotaxime. The identified bacteria also demonstrated reduced susceptibility to ceftazidime, and were resistant to tetracycline, kanamycin, and fosfomycin. The isolates with the blaOXA–23 gene were found to express β-lactamase OXA-23, which is widespread among A. baumannii (Table 2). Furthermore, the main vehicle for the blaOXA–23 gene among Acinetobacter spp. was identified as transposon Tn2008.
Klebsiella pneumoniae harboring the blaNDM–5 gene was also isolated from samples of feces from dairy cows with mastitis in Jiangsu Province, China (Table 2). In all isolates, the blaNDM–5 gene was found on an approximately 46 kb self-transmissible IncX3 pNDM-MGR194-like plasmid (He et al., 2017a). Additionally, He et al. (2017b) isolated and identified three NDM-5-producing E. coli isolates from dairy cows, including one co-producing the transferrable colistin resistance gene mcr-1 and another co-harboring the carbapenemase gene blaVIM–2 (Table 2). The blaNDM–5- and mcr-1-harboring plasmids reduced the fitness of their bacterial hosts but maintained stability in the recipient strain. The mcr-1-carrying plasmid could be conjugated into NDM-5-positive E. coli isolates in vitro, thereby generating strains that eventually achieve pan resistance.
In addition, Al Bayssari et al. (2015) report the detection of VIM2-producing (blaVIM–2) P. aeruginosa and OXA-23-producing (blaOXA–23) A. baumannii in samples of cattle feces. They also identified the co-occurrence of blaOXA–23 and blaOXA–58 in the same isolate of A. baumannii (Table 2). The exact origins of these genes remain undefined, but they are known to have previously transferred from environmental bacteria into species with clinical relevance (Jiang et al., 2017; Waglechner and Wright, 2017). Woodford et al. (2014) note that the blaOXA–48 family of enzymes occurs naturally in Shewanella spp., a genus that inhabits lake sediments, and that OXA-23 carbapenemase originates from the environmental species Acinetobacter radioresistens, suggesting that these carbapenemase genes have probably been transferred from environmental bacteria into the animal microbiome, through the close contact between the host animal and the environment.
Importantly, commensal bacteria from animal intestines may serve as a reservoir of ARGs. E. coli is commonly used as an indicator species for monitoring AMR dynamics, especially for critical antimicrobials in veterinary and medicine, such as extended-spectrum cephalosporins (ESC) and fluoroquinolones (Reynolds et al., 2020). A number of studies have found E. coli strain O157 isolated from cattle feces from commercial and communal farms to be resistant to erythromycin, tetracycline, sulfamethoxazole, chloramphenicol, kanamycin, ampicillin and streptomycin (Ateba and Bezuidenhout, 2008; Volkova et al., 2012). Most infections caused by E. coli O157: H7 result from the consumption of food and water contaminated with the fecal matter of infected animals (Beauvais et al., 2018). Increased ESC resistance has frequently been reported in E. coli and Salmonella enterica from dairy cattle, and this was found to be caused mainly by ESBL or AmpC; many of these organisms have been classified as MDR (Sawant et al., 2007; Seiffert et al., 2013).
Kanwar et al. (2013) detected the presence of tetA, tetB, and blaCMY–2 genes in the feces of feedlot cattle; interestingly, the findings indicate that the use of chlortetracycline treatment following ceftiofur therapy could elevate the level of ceftiofur resistance. Jiang et al. (2006) report high resistance to ceftriaxone among various gram-positive and gram-negative bacteria isolated from calf feces.
Another important concern for human medicine is presented by the development of vancomycin resistance in bacterial strains with clinical significance such as Enterococcus spp. and Staphylococcus spp. Beukers et al. (2017) identified E. faecium and E. faecalis, which are potentially harmful to humans, in bovine feces; however, the Tn917 transposon conferring MLSB resistance was identified only in E. faecium and E. hirae. In addition, a study of cattle feces by Tanih et al. (2017) indicated the presence of four Enterococcus species, such as E. hirae, E. faecium, E. durans, and E. faecalis harboring tetracycline, erythromycin, penicillin, and vancomycin resistance genes (Table 2).
Jackson et al. (2011) examined AMR in ten different Enterococcus species, including E. hirae, E. faecalis, and E. faecium, in feces samples from US dairy cattle. In all species, the most prevalent resistance against an array of 17 antimicrobials was found to be against lincomycin, while all strains were resistant to at least one of the tested antimicrobials. Ten different types of AMR were observed among the E. hirae isolates, with one E. hirae isolate demonstrating MDR against seven antimicrobials. Interestingly, some E. hirae isolates were also resistant to one of the newest antimicrobials, daptomycin. In addition, E. hirae, E. faecalis, E. faecium, and another undetermined Enterococcus species demonstrated persistence to four antimicrobials (Jackson et al., 2011). Elsewhere, intrinsic vanC-mediated and acquired vanA-mediated resistance has also been reported in enterococci isolated from French cattle (Haenni et al., 2009) (Table 2).
Although pathogens resistant to only one antibiotic may cause severe infections that are very hard to treat, MDR bacteria, such as those isolated from cattle feces, are particularly dangerous. Yang et al. (2019) identified MDR in ESBL+ K. pneumoniae isolated from cow feces. The strain showed resistance to 13 antibiotics from almost all commonly-used antimicrobial classes. In addition, the following resistance genes were found: blaKPC, blaSHV, blaTEM, qnrA, qnrB, and tolC (Table 2). Most carbapenemases present in this K. pneumoniae strain belonged to the KPC group.
Studies conducted in Canada have shed light on the ARGs associated with ESC and fluoroquinolone resistance in Enterobacteriaceae in dairy calves. Awosile et al. (2018) identified E. coli isolates carrying blaTEM, blaCMY–2, blaCTXM, and blaSHV genes in the feces of dairy cattle (Table 2). In Germany, Klotz et al. (2019) isolated MDR strains from various non-fermenting bacilli, such as A. baumannii; these strains demonstrated resistance to ampicillin, amoxicillin-clavulanic acid, cefalexin, ceftiofur, nitrofurantoin, and chloramphenicol, with intermediate resistance observed for piperacillin (6%) and rifampicin (25%). Interestingly, the highest frequency of A. baumannii was observed in dairy cows, followed by beef cattle and calves.
Many studies focusing on the diversity of known and novel ARGs have been conducted. Wichmann et al. (2014) used a combination of functional metagenomics with PacBio sequencing to characterize the resistome of dairy cow manure. The prepared DNA libraries indicated the presence of genes coding resistance to β-lactams, kanamycin, tetracycline, and chloramphenicol (Table 2). In addition, the researchers identified 80 unique and functional ARGs, suggesting that the ARGs found in animal manure may originate from other genera such as Proteobacteria, Firmicutes, Bacteroidetes, and Actinobacteria. Hence, the full resistome appears to consist of phylogenetically varied organisms; in addition, many ARGs seem to be flanked by MGE such as transposases and insertion sequences, or may be created de novo like the newly-identified chloramphenicol resistance genes.
Research data indicates that even a single antimicrobial application can have a strong effect on the diversity and abundance of ARGs. Holman et al. (2019) found the fecal microbiota of beef cattle to be significantly altered on the second and fifth day after a single injection of either oxytetracycline or tulathromycin, and that they remained at this level until the 34th day after administration. Although five resistance genes (ermX, sul2, tetC, tetM, and tetW) were detected in the fecal microbiome, only tetM and tetW differed significantly between the animal groups divided according to the medicine administered: oxytetracycline application increased the prevalence of tetM in the fecal sample, while tulathromycin boosted the ratio of tetM to tetW resistance genes compared to animals receiving no antibiotic treatment. The presence of a single antibiotic in a bacteria-living niche may exert on-site selective pressure, thus facilitating the maintenance of ARGs, not only against this particular antimicrobial but also against unrelated resistance determinants through co-resistance; for example, co-transfer of ermB and tetM has been observed in the presence of erythromycin in Streptococcus pyogenes isolates (Brenciani et al., 2007). Fecal bacteria isolated from conventionally-raised cows, where antibiotics are added either as a growth promoter or for prophylaxis, tend to be more resistant to antibiotics than those from cows raised organically, i.e., where antibiotics application is forbidden or strongly supervised (Sato et al., 2005; Halbert et al., 2006); however, it seems that the observed increase in ARGs may be only temporary (Tragesser et al., 2006; Singer et al., 2008).
Interestingly, even cattle never exposed to antibiotics can also carry ARB and ARGs, such as a fluoroquinolone-, tetracycline- or β-lactam resistance genes (Durso et al., 2011). In addition, Thames et al. (2012) have detected genes coding resistance to tetracycline, sulfonamide, and macrolide in samples from newborn calves. In contrast, Roesch et al. (2006) found similar levels of antibiotic resistance between organic farms and conventional ones.
Reaching a common understanding that uncontrolled and unlimited usage of antibiotics may contribute to environmental pollution seems crucial to stopping the propagation of resistant bacteria in food animal production systems (Call et al., 2013).
The Use of Antibiotics in the Pig Industry and Its Effect on the Swine Manure Resistome
Worldwide pig production has risen steadily during the last 50 years, reaching approximately 120 million tons in 2018. The leading producers of pig meat are China, with almost 45% of global production, followed by the US, Germany, Spain, and Vietnam. These five countries account for nearly 65% of world pig meat production (FAOSTAT, 2020).
An analysis of commercial pig feed formulation available in Vietnam has revealed that it may contain up to 55.4% antimicrobials, with the most common being bacitracin (24.8%), chlortetracycline (23.9%), and florfenicol (17.4%) (Van Cuong et al., 2016). It is estimated that 286.6 mg of in-feed antimicrobials are used to raise 1 kg of live pig (Van Cuong et al., 2016). Approximately 55.5% of antimicrobials administered to pigs are classified by the WHO as important, very important, or critically important for humans. A list of the antimicrobials intended for use in animals has been published by the OIE as a “list of antimicrobial agents of veterinary importance” (International Office of Epizootics, 2015).
One group of antibiotics considered as essential for human medicine is the polymyxins. Polymyxins are not routinely applied parenterally in animals because of their toxicity; however, one polymyxin, colistin, has been used extensively in pig production outside of North America as an oral treatment for neonatal colibacillosis (Dowling, 2013). In Southeastern Asia (India, China), colistin is used as a last-resort therapeutic option to treat severe infections caused by MDR gram-negative bacteria, including P. aeruginosa and A. baumannii (Catry et al., 2015). It is widely used in pig production for prophylactic (single animal), metaphylactic (whole pen or herd), and therapeutic purposes.
In addition, mcr-1, a plasmid-mediated colistin resistance gene, firstly identified in China (Liu et al., 2016), has also been found in Vietnamese pigs (Van Cuong et al., 2016). Colistin is also widely used in Europe (Catry et al., 2015; Kempf et al., 2016), and this may play a crucial role in the spread of colistin resistance in bacteria: although mcr-1 has been identified in porcine fecal samples across Europe, it appears to be more predominant in Germany, Netherlands, Belgium, Denmark, Italy, Great Britain (Brauer et al., 2016; Guenther et al., 2017; Pulss et al., 2017), and Spain (Quesada et al., 2016), which may be due to differences in the regional regulations regarding its application. In addition, other variants of this gene have been found: mcr-2 and mcr-3 in China (Yin et al., 2017) and mcr-2 in Germany (Roschanski et al., 2017).
Just as in cattle breeding, the use of ceftiofur and cefquinome in pig farming may be related to the spread of resistance to third and fourth generation cephalosporins among Salmonella spp. Resistance to ceftiofur in pigs was first identified in 2002 (blaCMY) (Hanson et al., 2002). A similar observation was also made in 2015 on Danish pig farms, where it was found that frequent use of third and fourth generation cephalosporins appeared to be related to the occurrence of ESBL producing E. coli. This resistance was also maintained over a prolonged period in animal herds (Andersen et al., 2015).
Some of the most extensively-studied ARGs are those conferring tetracycline resistance because this antimicrobial is widely administered to pigs. Research conducted on the hog population in the US in 2000 revealed a high level (71%) of tetracycline resistance in isolated Enterococcus spp. (Haack and Andrews, 2000). The gene tetM is known to confer the widest host range of tetracycline resistance and is often associated with MGEs, enhancing its transfer rate from one bacterium to another. More importantly, a number of tetracycline-resistant genes have been found not only in fecal swabs but also near swine feedlots. The genes found in pig fattening farms may code for a range of resistance mechanisms, such as efflux pumps, ribosomal protection peptides, or enzymatic modifications (Wu et al., 2010). The same genes have also been reported previously in swine lagoons, swine manure, storage pits, and groundwater located near pig production facilities (Table 3) (Chee-Sanford et al., 2001; Mackie et al., 2006; Koike et al., 2007). A study of the resistance profile of MDR E. coli from a pig farm and its surroundings in Germany (Table 3) identified similar ARGs in samples taken from stable flies and from the feces of barn dogs, indicating the potential for AMR to spread across the surrounding environment (Guenther et al., 2017).
TABLE 3.
Antibiotic resistance genes in pig.
| Resistance genes | Strain | Source | County | References |
| mcr-1 | E. coli | Intensive pig farm | China | Liu et al., 2016 |
| na | Pig | Vietnam | Van Cuong et al., 2016 | |
| Porcine fecal samples | Germany Netherlands Belgium Denmark Italy Great Britain | Brauer et al., 2016; Guenther et al., 2017; Pulss et al., 2017 | ||
| mcr-2 | E. coli | Porcine fecal sample | China | Yin et al., 2017 |
| Pooled feces and boot swab samples | Germany | Roschanski et al., 2017 | ||
| mcr-3 | E. coli | Porcine fecal sample | China | Yin et al., 2017 |
| tetA, tetC, tetE, tetG, tetK, tetL, tetA/P, tetM, tetO, tetQ, tetS, tetT, tetW, tetB/P, tetX | na | Porcine feces swine lagoons manure storage pits groundwater near pig production facilities | Chee-Sanford et al., 2001; Mackie et al., 2006; Koike et al., 2007; Wu et al., 2010 | |
| aadA5, aph(3′)-Ic-like, blaCTX–M–1, dfrA17, strA, strB, sul2-like, tet(B), aadA1, blaTEM–1B, dfrA1, mcr-1, sul1, sul2, tetA, aadA2, cmlA1-like, mph(A), sul3, tet(A)-like | E. coli | Pig manure | Germany | Guenther et al., 2017 |
| blaTEM–1B-like, strA-like, strB-like, tetA, aadA1, aadA2, aph(3′)-Ia-like, blaTEM–1B, cmlA1-like, dfrA1, mcr-1, sul3 | Animal boot swabs | |||
| aadA1, blaTEM–1B, dfrA1, dfrA14-like, mcr-1, strA, strB, sul1, sul2, tet(A) | Stable fly | |||
| blaTEM–1B-like, dfrA1-like, mcr-1, strA-like, strB-like, tetA | Barn dog feces | |||
| blaCTX–M, ermB, mcr-1, optrA, qnrS, tet(40), aac(6′)-lm | na | Pig feces | Belgium, Bulgaria, Germany, Denmark, Spain, France, Italy, Netherlands, Poland | Munk et al., 2018 |
| mexF, oprD, aadA2, aadD, aacA/adhD, aphA1, fox5, tetPB, tetQ, tet(32), tetL, tetO, ermT, ermX, lnuB, vatE, sat4 | na | Pig feces | Zhao et al., 2018 | |
| tet36, tetM, tetQ tetX, tetP(A), tet40, ermF, mefA, sul1, sul2, aac(6)-le, aph(6)-ld, catB3, catA16, oxa9, cfxa, mexD | na | Swine fattening facility surroundings (well water, swine wastewater, soil, fishpond) | China | He et al., 2019 |
| sul1, tet(32), tetL, tetM, tetO, tetBP-03, tetQ, tetW, tetX, floR, mexF, aadA, aadA1, aadA2, aadD, aph(2′)-Id, aphA3, ermA, ermB, vatE, sat4 | na | Pig slurry | China | Pu et al., 2018 |
| blaOXA–48, blaTEM–1B, aph(3′)-Ia, aadA1, aadA2, aac(3)-IId, Inu(F), qnrS1, floR, cmlA1, sul2, sul3, tet(A), tet(M), dfrA12, aph(4)-Ia, aadA5, aac(3)-IVa, armA, dfrA17 | E. coli pathovars | Porcine fecal samples | Germany, Netherlands Belgium Denmark Italy Great Britain | Pulss et al., 2017 |
| tetM, tetO, blaTEM–1, blaSHV–2, sul2, aph(3′′)-Ib, aph(6′)-Ic, aph(6′)-Ib, ermD, mdfA, mdtH, mdtL, rosA, tet(B), bcr, adeA, armB, mdtF, mdtN, mdtO, mdtP, oprA, tolC, acrA | na | Porcine feces samples | The US | Looft et al., 2012 |
na, not applicable – dataset was obtained during metagenomic study.
A study of the resistomes of non-typhoidal Salmonella strains isolated from swine fecal samples found the highest levels of AMR to be against tetracycline (80% of isolates), streptomycin (43.4%), and sulfamethoxazole (36%). The results also indicated that the microbes isolated from farms raising animals with antimicrobials tended to demonstrate greater resistance to most classes of antibiotics than antimicrobial-free farms (Gebreyes et al., 2006). Furthermore, MDR Campylobacter coli have been found within pig production systems (Thakur and Gebreyes, 2005).
More than 400 ARGs were identified in a study conducted in nine European countries (Belgium, Bulgaria, Germany, Denmark, Spain, France, Italy, Netherlands, and Poland) on 181 pig herds and 178 poultry farms. These studies indicated that fecal pig resistome [blaCTX–M, ermB, mcr-1, optrA, qnrS, tet(40), aac(6′)-lm] differs significantly between countries. This discovery appears to be closely related to the antimicrobial usage policy of each country: countries with a strict policy tend to have a lower rate of antibiotic resistance. Conversely, pig herds located in the same country or countries with a similar antimicrobial usage policy tend to demonstrate similar ARGs patterns. The newly-identified optrA gene (enterococcal linezolid resistance group), which codes resistance to both oxazolidinone and amphenicols (florfenicol), was detected in countries with the highest usage of amphenicols, such as Bulgaria, Italy, and Spain (Munk et al., 2018).
Zhao et al. (2018) identified a total number of 146 ARGs in animal feces from seven large-scale Chinese pig farms, i.e., two with high levels of medication and five with low levels, including eight transposon-transposase genes and two class 1 integron-integrase genes. These ARGs genes represented all major classes of resistance mechanisms: 44.5% conferring antibiotic deactivation, 34.3% coding for efflux pumps, and 19.2% cellular protection. These mechanisms provide resistance for all major antimicrobial classes: aminoglycoside (aadA2, aadD, aacA/adhD, aphA1 [aka kanR] – 20%), β-lactam (fox5 – 14%), tetracycline [tetPB, tetQ, tet(32), tetL, tetO -14%], MLSB (ermT, ermX, lnuB, vatE – 14%), vancomycin (8%), sulfonamide (2%), chloramphenicol (2%), and others (sat4 – 4%). The copy number of the gene ranges from 2.72 × 109 to 1.34 × 1010, divided mostly between tetracycline (4.77 × 109), MLSB (3.41 × 109), and aminoglycoside (4.02 × 109) ARGs (Zhao et al., 2018).
A wide variety of ARGs have also been found in wastewater from pig farms. Of these, tetracycline resistance genes predominated, followed by MLSB resistance and sulfonamide resistance, with aminoglycoside resistance, phenicol resistance, and β-lactam resistance being slightly less abundant (Table 3) (He et al., 2019). In addition, the initially identified genes were also identified in pig wastewater-irrigated soils; however, their type and relative abundance varied depending on the sampling depth.
Another analysis conducted in China in 2018 identified 21 ARGs and three transposons (Table 3) in pig slurry originating from a high production livestock facility. The ARGs were divided into four resistance mechanisms: cellular protection (43%), antibiotic deactivation (38%), efflux pump (14%), and others (5%). They thus conferred resistance to seven major classes of antibiotics: tetracycline (38%), florfenicol-chloramphenicol-amphenicol (29%), MLSB (14%), aminoglycoside (9%), sulfonamide (5%), and others (5%) (Pu et al., 2018).
A study conducted by Pulss et al. (2017) also detected many other ARGs in an E. coli strain carrying an OXA-181 carbapenemase (Table 3).
An analysis of the feces from animals supplemented with typical performance-enhancing antibiotics (chlortetracycline, sulfamethazine, and penicillin) in the US revealed increased abundance and diversity of ARGs in the gut microbiome compared with animals raised without antibiotics. Six types of resistance mechanisms were found to be more abundant in the treated animals: those coding for tetracycline efflux pumps, class A β-lactamases, sulfonamide resistance genes, aminoglycoside O-phosphotransferase, and two types of multidrug efflux: a major facilitator superfamily transporter, and a resistance-nodulation-cell division transporter system (Table 3). However, some genes were also detected at high frequencies in non-medicated pigs. It is possible that their presence may reflect antibiotic resistance developed by farm animals due to selection pressure exerted by over 50 years of antimicrobial treatment (Looft et al., 2012).
Additionally, heavy metals such as zinc, copper, lead, cadmium, chrome, and nickel have been commonly found in animal manure. It has been proven that high doses of copper and zinc enhance the growth of animals (Poole, 2017), and it is possible that these were supplied as mineral addition to fodder; however, others, such as lead, cadmium, chrome or nickel, may have originated in corroded metal parts of installations (Zhao et al., 2018). It is known that heavy metal tolerance is linked to AMR: it may select for resistant bacteria and facilitate the persistence of resistance genes by cross-resistance. It has also been proven that the presence of sublethal doses of heavy metals in the environment facilitates the conjugative transfer of ARGs (Zhang et al., 2018).
The use of copper and zinc in pig fodder has been found to be closely associated with the presence of MDR Salmonella spp. (Barton, 2014). Compared to copper-sensitive strains, copper-resistant strains are more likely to be resistant to ampicillin, tetracycline, chloramphenicol, or sulfonamides, as well as to penicillin, just as nickel-resistant strains are also resistant to ampicillin. Moreover, lead resistance may be linked to resistance to β-lactams, mercury tolerance to streptomycin resistance, or lead and mercury tolerance to ampicillin resistance (Sabry et al., 1997; Bass et al., 1999; Berg et al., 2010). Such cross-resistance between heavy metals and antibiotic resistance in the pig production chain has been confirmed by Hölzel et al. (2012).
An interesting example of a plasmid carrying both ARGs and metal tolerance genes was described by Fang et al. (2016). One of the most common incompatibility groups of plasmids in the Enterobacteriaceae is IncHI2. It has been identified in E. coli, S. enterica, K. pneumoniae, and Enterobacter cloacae isolated from humans, chickens, and rarely swine. The plasmid may harbor a number of metal tolerance genes, such as pcoABCDRSE (efflux systems to detoxify copper), silESRCBAP (efflux systems to detoxify silver), arsCBRH (efflux systems to detoxify arsenic), merEDACPTR (Tn1696-related mercury operon), and terZABCDEF and terY3Y2XY1W (tellurite resistance systems). It also contains a number of AGRs coding resistance to amphenicols (floR), aminoglycosides (armA, aac-Ib/aac-Ib-cr), β-lactams (blaCTX–M, blaCMY, blaSHV, blaIMP, blaVIM), quinolones (oqxAB, qnrA1, qnrS1 and qnrB2), and fosfomycin (fosA3). A plasmid harboring both ARGs and metal tolerance genes, Sal-1457, has been identified in Salmonella infantis isolates obtained from the feces and carcasses of goats, cattle and chickens in Australia; it has been found to contain aph(3″)-I, aph(6″)-ld, blaTEM–1, and dfrA5, together with an arsenic resistance operon (Wilson et al., 2019). A very detailed study found almost 17% of analyzed bacterial genomes in samples obtained from multiple environments to carry both ARGs and metal tolerance genes (Pal et al., 2015).
Changes in the Soil Resistome Occurring Due to Manure Application
The soil has been identified as a critical source of ARGs, not only because of the presence of a diverse range of bacteria able to produce natural antibiotics (Lang et al., 2010) but mainly because of the application of natural manure on crop fields, which might contain ARGs or antibiotics: only a small amount of antibiotics are absorbed or metabolized by animals, with about 75% of the administered drug being excreted into the feces or urine (Zhang and Zhang, 2011).
Soil fertilization with animal manure is a widespread agricultural practice, not only in Europe but worldwide, especially in less developed countries (Heuer et al., 2011). This kind of manure is known to be a rich source of nutrients and organic matter for fertilizing fields. The use of organic manure as fertilizer can also be a practical approach to animal waste management by lowering the cost of its disposal: a single dairy cow can produce 54 kg of wet manure per day, a pig – 6.4 kg, a sheep – 2.5 kg, and a chicken – 0.2 kg (Girotto and Cossu, 2017). Furthermore, fertilizing arable soil with manure can play an essential role in the active cycling of chemicals which are crucial for optimal crop growth and development, such as phosphorus or nitrogen. However, besides its doubtless advantages, the misuse or overuse of manure field applications may account for the excessive amount of these elements in soils and the accumulation of heavy metals added to feeders.
Antibiotics can react and accumulate in specific ways following manure application depending on physicochemical properties of soil and climate conditions. A study of columns filled with types of soil by Pan and Chu (2017) found that the leaching of antibiotics is generally higher in sandy soil than in clay and silty soil. Antibiotics such as norfloxacin and tetracycline were also found to persist longer at the soil surface, contrary to sulfamethazine and erythromycin that tend to reach deeper layers of soil and groundwater.
The full biotransformation and degradation of antimicrobials may take up to 150 days in bovine manure (De Liguoro et al., 2003). Although manure is an essential and sufficient source of nitrogen for agricultural soil, it is also a rich source of antibiotics and ARB, which may be transferred to the environment and survive there even for several months (Merchant et al., 2012). Therefore, it is extremely important to develop effective treatment strategies for manure used as natural fertilizer, with the aim of eliminating, or at least reducing, the risk of releasing antibiotics, ARGs, and ARB to the environment (Zalewska and Popowska, 2020).
Some antibiotics with high adsorption capacity, such as tetracyclines and quinolones, can actively adhere to soil particles, making them non-biodegradable. They can thus accumulate in agricultural soil amended with manure, changing the natural microbial community structure and promoting the maintenance of resistance genes (Li et al., 2011). There are several possible explanations for the increasing abundance of ARGs observed in manured soils. Firstly, it is possible that, following manure application, any ARGs present in bacteria that will survive in the environment can be simply transferred to new hosts by HGT. Alternatively, residues of active antibiotic compounds in manure can induce new mutations in soil bacteria, or enrich and sustain pre-existing ones. Lastly, animal waste rich in organic matter can enhance the growth of resistant resident bacteria in soil (Udikovic-Kolic et al., 2014; Xie et al., 2018). In addition, heavy metals present in soil act as major stress factors in the environment, and as antibiotics and heavy metals can share the same regulatory responses, their presence may also promote ARG selection via co-selection and cross-resistance mechanisms (Imran et al., 2019).
A study in Shandong Province, China examined the abundance of ARB and ARGs in farmland soil fertilized with chicken manure (Zhao et al., 2017). The results indicate the presence of a number of ARB and ten AGRs, such as tetW, tetO, tetT, tetM, tetA, tetL, tetQ, sul1, sul2, and sul3, in soil samples collected from four fields. In all samples, researchers observed significant correlations between the concentration of applied sulfonamides and the abundance of sulfonamide resistance genes.
Similar studies from Finland examined changes in ARGs and MGE abundance in the soil after fertilization with swine and cow manure. The results identified the presence of ARGs, previously detected in animal waste, in fertilized soil. However, while the abundance of ARGs associated with manure generally decreased following 2 and 6 weeks after fertilization, the levels of some ARGs, such as those conferring resistance to disinfectants, aminoglycosides, and vancomycin, remained elevated compared to unfertilized soil, even after 6 weeks (Muurinen et al., 2017). In addition, Marti et al. (2014) report greater abundance of selected genes related to AMR and gene transfer, such as sul1, ermB, strB, int1, and repA, in soils fertilized with cow and swine manure than non-manured soil.
Studies also indicate that the increase in ARB observed in soil treated with cow manure may be due to the bloom of resistant species already present in the soil, rather than resistant bacteria introduced from the manure itself (Udikovic-Kolic et al., 2014). The manure used in this study was obtained from dairy cows that had not been treated with antibiotics.
The length of time for which ARG levels remain elevated varies considerably depends on various factors, such as weather conditions or soil type. Increased levels of sul1 and sul2 genes were observed for over 4 months following swine manure application in field plots in Germany (Jechalke et al., 2014), while intI1 persisted in soil for 10 months after swine manure application in agricultural fields in the United Kingdom (Byrne-Bailey et al., 2011). Tighter regulations regarding manure application are in force in the US: manure application must be performed no less than 120 days before harvesting crops that have direct contact with manured soil and no less than 90 days for crops with no direct contact with the soil (Ferguson and Ziegler, 2004).
Changes Of The Resistome In Plants Growing On Manure-Fertilized Soil
Fruits and vegetables often harbor non-pathogenic epiphytic microflora; however, many studies have reported contamination with pathogens or different types of ARGs; such contamination may occur pre-harvest via soil and organic fertilizers such as manure, sewage sludge, and irrigation water. This is a serious consideration as any contaminated vegetables intended to be consumed raw may act as vehicles for the spread of ARB and ARGs to humans (Hölzel et al., 2018).
Sub-inhibitory concentrations of antibiotics in plant tissues have been found to be potential drivers of antibiotic resistance in endophytic bacteria, and small amounts of antibiotics, such as tetracycline, can trigger HGT between different bacteria (Redondo-Salvo et al., 2020).
Antibiotic resistance genes can also be transferred to plants from common soil bacteria via root endophytes, and these have been found to survive in the root (Bulgarelli et al., 2012, 2013; Lundberg et al., 2012). Some endophytes appear to be are closely related to human pathogens, particularly opportunistic ones (Rosenblueth and Martínez-Romero, 2006), and this similarity could pose a potential threat to human health. Solomon et al. (2002) demonstrated the transmission of E. coli O157: H7 from manure-contaminated soil and irrigation water to lettuce; it is believed that these bacteria can enter the lettuce through the root system and propagate throughout the edible portion of the plant.
The soil may well represent the main source of ARGs to the plant, as indicted by the large degree of overlap between ARGs in the plant microbiome and those in the soil resistome (Yang et al., 2018). ARB associated with soil and manure may enter the plant microbiome by colonizing the roots, which are in direct contact with soil, or the aboveground parts, potentially through air particulates or the motility of root endophytes (Zhu et al., 2017; Guron et al., 2019). In addition, plants have been found to take up antibiotic residues from manure-amended soil; this may exert long-term pressure in the plant, facilitating the acquisition of drug resistance and its spread across the plant resistome (Chen et al., 2019a).
The phyllosphere, i.e., aerial leaf surfaces, is a specific niche harboring diverse species of bacteria and other microbes (Bulgarelli et al., 2013). Many previous studies have examined the impact of manure application on the levels of ARGs in the phyllosphere of leafy vegetables (Chen et al., 2016, 2017), or changes in ARGs abundance and dissemination (Marti et al., 2013; Tien et al., 2017; Murray et al., 2019). MGE such as intI1 and genes encoding transposase have been detected in leaf endophytes, as well as in the phyllosphere of lettuce (Wang et al., 2015; Zhu et al., 2017), maize (Chen et al., 2016), Brassica chinensis L (Chen et al., 2019a), and Coriandrum sativum L (Chen et al., 2019b).
After excretion by animals, pollutants such as antibiotics, antibiotic residues, ARB and ARGs may be transported through the environment via runoff, leaching and manure application (McEachran et al., 2015). These can accumulate in soil and increase the risk of selection pressure and crop contamination with ARB and ARGs (Hu et al., 2010). The ability of plants to take up antibiotics depends on several biotic and abiotic factors, as well as the type of crop: fruit and grain demonstrate a lower ability to absorb contaminants than leafy and root vegetables (Christou et al., 2019).
Manure application has been found to increase the abundance of ARB and ARGs in soil (Udikovic-Kolic et al., 2014; McKinney et al., 2018). In addition, manure-amended soils have been associated with increased detection of ARB and ARGs on lettuce and root vegetables; however, this has not been associated with all crops or ARGs (Marti et al., 2013; Rahube et al., 2014; Wang et al., 2015). It is important to note that while fecal bacteria can survive for weeks to months in the environment, depending on species and temperature, genetic elements can persist for much longer, regardless of cell viability (Chee-Sanford et al., 2009). ARB, which are naturally found in manure, may attach to the crops grown in the amended soil and multiply in this new potentially more favorable environment. As a consequence, ARB can become dominant among the natural resident bacterial population (Baquero, 2011; Wichmann et al., 2014).
Humans can become exposed to bacteria carrying ARGs and various pathogens by consuming contaminated vegetables (Berger et al., 2010; van Hoek et al., 2015). Yang et al. (2014) examined the effect of soil amendment with chicken manure on the distribution of antibiotic resistant endophytic bacteria (AREB) and MDR endophytic bacteria in celery, pak choi and cucumber. High numbers of bacteria resistant to at least one antibiotic were identified in all vegetables; however, MDR bacteria were found to be absent from the surface of the tissue or inside the vegetables. However, Yang et al. (2016) report that the application of chicken manure or organic fertilizer increases the populations of MDR bacteria in soil and MDR endophytic bacteria in pak choi. Yang et al. (2014) found the highest amounts of cultivable endophytic ARB in celery roots. This is not surprising, as in most plants, roots generally have the highest rates of total cultivable AREB, and hence may serve as a reservoir for potentially pathogenic bacteria or ARB (Berg et al., 2005).
A study of ARB in wheat by Yang et al. (2009) found that AREB were most prevalent in the roots, followed by the leaves, with no AREB detected in the seeds. A report by Sengeløv et al. (2003) indicated that the occurrence of tetracycline-resistant bacteria in the soil was elevated after pig manure slurry treatment, but this fell to control values over the following 5 months. Furthermore, Schmitt et al. (2006) found that both pig manure and the amended soil contain a wide variety of tetracycline ARGs following fertilization. A number of other publications indicate that ARGs can infiltrate the edible parts of commercial crops and that their presence and concentrations depend upon farming practices (Manaia et al., 2018; Cerqueira et al., 2019a, b).
Marti et al. (2013) evaluated the occurrence of ARB on the surface of vegetables that are often eaten raw, such as tomatoes, cucumbers, peppers, carrots, radishes and lettuce, and compared their presence in vegetables grown in inorganically-fertilized soil with those in soil fertilized with dairy or swine manure. The selected vegetables included a range of roots, fruits, and leafy vegetables with different degrees of presentation to key environmental factors (e.g., sun, rain, or wind). It was found that fertilization only had a significant effect on the abundance of amoxicillin-clavulanic acid, ampicillin, and cefoxitin resistance genes. Interestingly, in the case of carrots, a reduction in the abundance of ampicillin- or cefoxitin-resistant bacteria was observed compared to the control group. Across the sample as a whole, eight gene targets (sul2, tetB, tetT, ermA, ermF, qnrB, blaPSE, and blaOXA–20) were detected on at least one vegetable sample. Plants grown in dairy manure-amended soil carried sul2, ermF, qnrB, blaPSE, and blaOXA–20, while those grown on swine manure-amended soil harbored sul2, tetB, tetT, ermA, ermF, and blaOXA–20 (Marti et al., 2013).
Cerqueira et al. (2019b) examined the distribution of ARGs in soil, rhizospheric soil, roots, and leaves for tomato, lettuce, and broad beans grown in nine commercial plots. All plots had been fertilized with manure obtained from fairly unusual sources such as horse or pigeon feces. Of the ARGs, the most prevalent was blaTEM, which was detected in all analyzed samples. The data showed that ARGs loads decreased gradually from soil to fruit, resulting in a 100- to 1000-fold dilution for most studied genes; in addition, the ARGs concentration and composition of edible plant parts was strongly dependent on the initial soil ARGs content (Cerqueira et al., 2019b).
Fogler et al. (2019) found the compositions of microbiota and resistomes associated with vegetable surfaces to be strongly influenced by the amendment of soil with raw manure collected from dairy cows during antibiotic administration, especially clindamycin. Their findings indicate that the genes tetW and sul1 are ubiquitous and present in high abundance in the commensal bacteria inhabiting humans and domestic animals, and that they demonstrate a consequently high representation in waste streams, a low abundance in less-affected environments and a uniform and a highly-conserved DNA sequence; they may therefore serve as valuable markers of soil contamination with ARB, ARGs, and antibiotic residues originating from manure. In addition, lettuce grown in manure-amended soil demonstrated a more diverse bacterial composition than lettuce grown in soil supplied with chemical fertilizers. No significant differences were observed between the relative abundances of detected bacterial phyla/classes based on the type of amendment (Fogler et al., 2019).
Wang et al. (2015) identified ten tetracycline resistance genes (tetA, tetP, tetC, tetG, tetL, tetB, tetM, tetO, tetW, tetX,) and two sulfonamide resistance genes (sul1 and sul2), along with one int1 gene, in root endophytes, leaf endophytes, and phyllosphere microorganisms from vegetable samples grown in manure-amended soil. The prevalence of ARGs was generally lower in leaf endophytes than in the root or phyllosphere microorganisms. Interestingly, it was observed that, for lettuce and endive, the occurrence of ARGs, such as sul1, sul2, tetC, and tetG, in root endophytes, leaf endophytes and the phyllosphere members, was also affected by growth period and species of plant (Wang et al., 2015).
A considerable number of studies have reported the presence of significant numbers of ARB on consumer-ready vegetables, such as lettuce and spinach leaves (Abriouel et al., 2008; Bezanson et al., 2008). Zhang et al. (2020) constructed a pot experiment to determine the effect of poultry and cattle manure application on resistome shifts in the plant microbiome of harvested cherry radish. A total of 144 ARGs conferring resistance to eight major classes of antibiotics were detected across all samples. The abundance of MGE was found to be positively correlated with individual ARGs coding for resistance to aminoglycoside, fluoroquinolone, quinolone, florfenicol-chloramphenicol-amphenicol, sulfonamide and tetracycline, and MDR determinants including MLSB. The results suggest that manure application significantly increased the abundance of ARGs in the rhizosphere and phyllosphere but not in the endophytes of the root, i.e., the edible part (Zhang et al., 2020).
The abundance of two MGEs (intI and tnpA-05) also appears to significantly increase after poultry manure application, indicating that not only does manure application directly introduce ARGs to the soil, but also may increase the HGT rate for ARGs; it also suggests that the transmission of ARGs from manure/soil to the surface of vegetables may be the predominant dissemination route. However, although the ARB involved in the transfer colonize as root endophytes, they demonstrate very little transfer to plant tissues; this suggests that the risk of transmitting external ARGs to the food chain is low. Therefore, root vegetables like cherry radish might be at a lower risk of ARGs contamination than leafy vegetables like lettuce (Zhang et al., 2019, 2020).
The degree to which livestock and agricultural land act as reservoirs of antibiotic-resistant bacteria, and how these two factors interact are relatively unknown (Tyrrell et al., 2019). Many studies have attempted to trace the direction of gene transfer from the environment to manure and determine its implications for future antibiotic resistance management and microbial ecology (Cook et al., 2014; Nesme and Simonet, 2015). In addition, evidence suggests that the primary pathway of gene acquisition from different environments may be HGT, including transfer from soils to the genomes of pathogenic bacteria (Allen et al., 2009; Forsberg et al., 2012; Nesme et al., 2014). Furthermore, DNA element class 1 integrons allow bacteria to adapt and evolve by integrating foreign genes from the environment through the capture of MGEs. This phenomenon has played an essential role in spreading AMR from non-pathogenic bacteria to pathogenic bacteria in the environment (Zhu et al., 2017).
Concluding Remarks
Although many promising solutions aimed at the reduction of bacterial resistance and overuse of antibiotics in the fields of animal production have emerged (Allen et al., 2013; Cheng et al., 2014; Tabashsum and Biswas, 2019), it seems that no single approach will be able to replace all the antibiotic applications in the animal production sector. Fortunately, public awareness is growing of the harmful effect of antibiotic usage on farm animals, and with it consumer demand for food products with guaranteed quality obtained from animals raised humanely and with minimal environmental impact. However, the demand for animal-derived food products is increasing with rising global population and economic growth (Figure 3). This global trend is well illustrated in the FAOSTAT database. Hence, the combination of steadily growing consumption with lack of regulation or intervention strategies, particularly in developing and middle-income countries, will no doubt result in increasing levels of antimicrobial usage, mostly due to the shift from extensive farming to large-scale production systems (Van Boeckel et al., 2015). It is assumed that the global consumption of antimicrobials in livestock and humans will increase by 67% between 2010 (63,151 tons ± 1560) and 2030 (105,596 ± 3605), of which approximately two-thirds will be associated with the rising number of food-producing animals (Bloomer and McKee, 2018).
FIGURE 3.
Animal production worldwide (according to FAOSTAT, 2020).
The relationship between antimicrobial use in farm animals and spread of ARB/ARGs is undeniable, as is the risk of their spread to pathogens posing a threat to human health. Antibiotic resistance should be considered a global threat, because neither bacteria nor genes respect geographical or national obstacles.
The most direct way to restrain the spread of AMR is to reduce or optimize their application in animal farming practices. The first step is to improve animal keeping conditions and herd management systems: animals remain healthy when they receive good quality, well-balanced fodder, and are kept in facilities with exceptional hygiene. Moreover, a range of prevention strategies such as vaccination, or the addition of probiotics, prebiotics, or bioactive compounds (e.g., antimicrobial peptides) in fodder can be used to protect vast herds from infection, and subsequently limit antimicrobial usage. In addition, guidelines should be prepared for good practice regarding livestock waste, such as manure management or wastewater treatment strategies (Zhao et al., 2020).
Author Contributions
MZ and MP: conceptualization and writing-review and editing. MZ, AB, AC, and MP: writing – original draft preparation. MZ, AB, and AC: visualization. MP: supervision, project administration, and funding acquisition. All authors read and agreed to the published version of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This review research was supported by the National Science Centre (NCN), Poland, grants numbers UMO-2017/25/Z/NZ7/03026 under the European Horizon 2020, The Joint Programming Initiative on Antimicrobial Resistance (5th JPIAMR Joint Call): “INART – Intervention of antimicrobial resistance transfer into the food chain” (http://www.inart-project.eu/index.html) and UMO- 2019/32/Z/NZ8/00011 (NCN) within the BiodivERsA Call 2018: “ANTIVERSA – Biodiversity as an ecological barrier for the spread of clinically relevant antibiotic resistance in the environment”. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Abriouel H., Omar N. B., Molinos A. C., López R. L., Grande M. J., Martínez-Viedma P., et al. (2008). Comparative analysis of genetic diversity and incidence of virulence factors and antibiotic resistance among enterococcal populations from raw fruit and vegetable foods, water and soil, and clinical samples. Int. J. Food Microbiol. 123 38–49. 10.1016/j.ijfoodmicro.2007.11.067 [DOI] [PubMed] [Google Scholar]
- Afridi O. K., Ali J., Chang J. H. (2020). Next-generation sequencing based gut resistome profiling of broiler chickens infected with multidrug-resistant Escherichia coli. Animals 10:2350. 10.3390/ani10122350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agga G. E., Arthur T. M., Durso L. M., Harhay D. M., Schmidt J. W. (2015). Antimicrobial-resistant bacterial populations and antimicrobial resistance genes obtained from environments impacted by livestock and municipal waste. PLoS One 10:e0132586. 10.1371/journal.pone.0132586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Bayssari C., Dabboussi F., Hamze M., Rolain J.-M. (2015). Emergence of carbapenemase-producing Pseudomonas aeruginosa and Acinetobacter baumannii in livestock animals in Lebanon. J. Antimicrob. Chemother. 70 950–951. 10.1093/jac/dku469 [DOI] [PubMed] [Google Scholar]
- Alexander T. W., Yanke J. L., Reuter T., Topp E., Read R. R., Selinger B. L., et al. (2011). Longitudinal characterization of antimicrobial resistance genes in feces shed from cattle fed different subtherapeutic antibiotics. BMC Microbiol. 11:19. 10.1186/1471-2180-11-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen H. K., Levine U. Y., Looft T., Bandrick M., Casey T. A. (2013). Treatment, promotion, commotion: antibiotic alternatives in food-producing animals. Trends Microbiol. 21 114–119. 10.1016/j.tim.2012.11.001 [DOI] [PubMed] [Google Scholar]
- Allen H. K., Moe L. A., Rodbumrer J., Gaarder A., Handelsman J. (2009). Functional metagenomics reveals diverse beta-lactamases in a remote Alaskan soil. ISME J. 3 243–251. 10.1038/ismej.2008.86 [DOI] [PubMed] [Google Scholar]
- Amador P., Fernandes R., Prudêncio C., Duarte I. (2019). Prevalence of antibiotic resistance genes in multidrug-resistant Enterobacteriaceae on portuguese livestock manure. Antibiotics 8:23. 10.3390/antibiotics8010023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen V. D., Jensen V. F., Vigre H., Andreasen M., Agersø Y. (2015). The use of third and fourth generation cephalosporins affects the occurrence of extended-spectrum cephalosporinase-producing Escherichia coli in Danish pig herds. Vet. J. 204 345–350. 10.1016/j.tvjl.2015.03.014 [DOI] [PubMed] [Google Scholar]
- Andersson D. I., Hughes D. (2014). Microbiological effects of sublethal levels of antibiotics. Nat. Rev. Microbiol. 12 465–478. 10.1038/nrmicro3270 [DOI] [PubMed] [Google Scholar]
- Andoh L. A., Dalsgaard A., Obiri-Danso K., Newman M. J., Barco L., Olsen J. E. (2016). Prevalence and antimicrobial resistance of Salmonella serovars isolated from poultry in Ghana. Epidemiol. Infect. 144 3288–3299. 10.1017/S0950268816001126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ateba C. N., Bezuidenhout C. C. (2008). Characterisation of Escherichia coli O157 strains from humans, cattle and pigs in the North-West Province, South Africa. Int. J. Food Microbiol. 128 181–188. 10.1016/j.ijfoodmicro.2008.08.011 [DOI] [PubMed] [Google Scholar]
- Aust M. O., Godlinski F., Travis G. R., Hao X., McAllister T. A., Leinweber P., et al. (2008). Distribution of sulfamethazine, chlortetracycline and tylosin in manure and soil of Canadian feedlots after subtherapeutic use in cattle. Environ. Pollut. 156 1243–1251. 10.1016/j.envpol.2008.03.011 [DOI] [PubMed] [Google Scholar]
- Awosile B., McClure J., Sanchez J., Rodriguez-Lecompte J. C., Keefe G., Heider L. C. (2018). Salmonella enterica and extended-spectrum cephalosporin-resistant Escherichia coli recovered from Holstein dairy calves from 8 farms in New Brunswick. Canada. J. Dairy Sci. 101 3271–3284. 10.3168/jds.2017-13277 [DOI] [PubMed] [Google Scholar]
- Baquero F. (2011). The 2010 Garrod Lecture: the dimensions of evolution in antibiotic resistance: ex unibus plurum et ex pluribus unum. J. Antimicrob. Chemother. 66 1659–1672. 10.1093/jac/dkr214 [DOI] [PubMed] [Google Scholar]
- Barton M. D. (2014). Impact of antibiotic use in the swine industry. Curr. Opin. Microbiol. 19 9–15. 10.1016/j.mib.2014.05.017 [DOI] [PubMed] [Google Scholar]
- Bass L., Liebert C. A., Lee M. D., Summers A. O., White D. G., Thayer S. G., et al. (1999). Incidence and characterization of integrons, genetic elements mediating multiple-drug resistance, in avian Escherichia coli. Antimicrob. Agents Chemother. 43 2925–2929. 10.1128/AAC.43.12.2925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauvais W., Gart E. V., Bean M., Blanco A., Wilsey J., McWhinney K., et al. (2018). The prevalence of Escherichia coli O157: H7 fecal shedding in feedlot pens is affected by the water-to-cattle ratio: a randomized controlled trial. PLoS One 13:e0192149. 10.1371/journal.pone.0192149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg G., Eberl L., Hartmann A. (2005). The rhizosphere as a reservoir for opportunistic human pathogenic bacteria. Environ. Microbiol. 7 1673–1685. 10.1111/j.1462-2920.2005.00891.x [DOI] [PubMed] [Google Scholar]
- Berg J., Thorsen M. K., Holm P. E., Jensen J., Nybroe O., Brandt K. K. (2010). Cu Exposure under field conditions coselects for antibiotic resistance as determined by a novel cultivation-independent bacterial community tolerance assay. Environ. Sci. Technol. 44 8724–8728. 10.1021/es101798r [DOI] [PubMed] [Google Scholar]
- Berger C. N., Sodha S. V., Shaw R. K., Griffin P. M., Pink D., Hand P., et al. (2010). Fresh fruit and vegetables as vehicles for the transmission of human pathogens. Environ. Microbiol. 12 2385–2397. 10.1111/j.1462-2920.2010.02297.x [DOI] [PubMed] [Google Scholar]
- Beukers A. G., Zaheer R., Goji N., Amoako K. K., Chaves A. V., Ward M. P., et al. (2017). Comparative genomics of Enterococcus spp. isolated from bovine feces. BMC Microbiol. 17:52. 10.1186/s12866-017-0962-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezanson G. S., MacInnis R., Potter G., Hughes T. (2008). Presence and potential for horizontal transfer of antibiotic resistance in oxidase-positive bacteria populating raw salad vegetables. Int. J. Food Microbiol. 127 37–42. 10.1016/j.ijfoodmicro.2008.06.008 [DOI] [PubMed] [Google Scholar]
- Bloomer E., McKee M. (2018). Policy options for reducing antibiotics and antibiotic-resistant genes in the environment. J. Public Health Policy. 39 389–406. 10.1057/s41271-018-0144-x [DOI] [PubMed] [Google Scholar]
- Bonardi S., Pitino R. (2019). Carbapenemase-producing bacteria in food-producing animals, wildlife and environment: a challenge for human health. Ital. J. Food Saf. 8:7956. 10.4081/ijfs.2019.7956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brauer A., Telling K., Laht M., Kalmus P., Lutsar I., Remm M., et al. (2016). Plasmid with colistin resistance gene mcr-1 in extended-spectrum-β-lactamase-producing Escherichia coli strains isolated from pig slurry in Estonia. Antimicrob. Agents Chemother. 60 6933–6936. 10.1128/AAC.00443-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenciani A., Bacciaglia A., Vecchi M., Vitali L. A., Varaldo P. E., Giovanetti E. (2007). Genetic elements carrying erm(B) in Streptococcus pyogenes and association with tet(M) tetracycline resistance gene. Antimicrob. Agents Chemother. 51 1209–1216. 10.1128/AAC.01484-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brichta-Harhay D. M., Arthur T. M., Bosilevac J. M., Kalchayanand N., Shackelford S. D., Wheeler T. L., et al. (2011). Diversity of multidrug-resistant salmonella enterica strains associated with cattle at harvest in the United States. Appl. Environ. Microbiol. 77 1783–1796. 10.1128/AEM.01885-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broom L. J. (2017). The sub-inhibitory theory for antibiotic growth promoters. Poult. Sci. 96 3104–3108. 10.3382/ps/pex114 [DOI] [PubMed] [Google Scholar]
- Büdel T., Kuenzli E., Campos-Madueno E. I., Mohammed A. H., Hassan N. K., Zinsstag J., et al. (2020). On the island of Zanzibar people in the community are frequently colonized with the same MDR Enterobacterales found in poultry and retailed chicken meat. J. Antimicrob. Chemother. 75 2432–2441. 10.1093/jac/dkaa198 [DOI] [PubMed] [Google Scholar]
- Bulgarelli D., Rott M., Schlaeppi K., Ver Loren van Themaat E., Ahmadinejad N., Assenza F., et al. (2012). Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 488 91–95. 10.1038/nature11336 [DOI] [PubMed] [Google Scholar]
- Bulgarelli D., Schlaeppi K., Spaepen S., Ver Loren van Themaat E., Schulze-Lefert P. (2013). Structure and functions of the bacterial microbiota of plants. Annu. Rev. Plant Biol. 64 807–838. 10.1146/annurev-arplant-050312-120106 [DOI] [PubMed] [Google Scholar]
- Byrne-Bailey K. G., Gaze W. H., Zhang L., Kay P., Boxall A., Hawkey P. M., et al. (2011). Integron prevalence and diversity in manured soil. Appl. Environ. Microbiol. 77 684–687. 10.1128/AEM.01425-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns J., Ruokolainen L., Hultman J., Tamminen M., Virta M., Hiltunen T. (2018). Ecology determines how low antibiotic concentration impacts community composition and horizontal transfer of resistance genes. Commun. Biol. 1:35. 10.1038/s42003-018-0041-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Call D. R., Matthews L., Subbiah M., Liu J. (2013). Do antibiotic residues in soils play a role in amplification and transmission of antibiotic resistant bacteria in cattle populations? Front. Microbiol. 4:193. 10.3389/fmicb.2013.00193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carballo M., Aguayo S., González M., Esperon F., de la Torre A. (2016). Environmental assessment of tetracycline’s residues detected in pig slurry and poultry manure. J. Environ. Prot. 7 82–92. 10.4236/jep.2016.71008 [DOI] [Google Scholar]
- Catry B., Cavaleri M., Baptiste K., Grave K., Grein K., Holm A., et al. (2015). Use of colistin-containing products within the European Union and European Economic Area (EU/EEA): development of resistance in animals and possible impact on human and animal health. Int. J. Antimicrob. Agents 46 297–306. 10.1016/j.ijantimicag.2015.06.005 [DOI] [PubMed] [Google Scholar]
- Cerqueira F., Matamoros V., Bayona J., Elsinga G., Hornstra L. M., Piña B. (2019a). Distribution of antibiotic resistance genes in soils and crops. A field study in legume plants (Vicia faba L.) grown under different watering regimes. Environ. Res. 170 16–25. 10.1016/j.envres.2018.12.007 [DOI] [PubMed] [Google Scholar]
- Cerqueira F., Matamoros V., Bayona J. M., Berendonk T. U., Elsinga G., Hornstra L. M., et al. (2019b). Antibiotic resistance gene distribution in agricultural fields and crops. A soil-to-food analysis. Environ. Res. 177:108608. 10.1016/j.envres.2019.108608 [DOI] [PubMed] [Google Scholar]
- Chambers L., Yang Y., Littier H., Ray P., Zhang T., Pruden A., et al. (2015). Metagenomic analysis of antibiotic resistance genes in dairy cow feces following therapeutic administration of third generation cephalosporin. PLoS One 10:e0133764. 10.1371/journal.pone.0133764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chee-Sanford J. C., Aminov R. I., Krapac I. J., Garrigues-Jeanjean N., Mackie R. I. (2001). Occurrence and diversity of tetracycline resistance genes in lagoons and groundwater underlying two swine production facilities. Appl. Environ. Microbiol. 67 1494–1502. 10.1128/AEM.67.4.1494-1502.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chee-Sanford J. C., Mackie R. I., Koike S., Krapac I. G., Lin Y.-F., Yannarell A. C., et al. (2009). Fate and transport of antibiotic residues and antibiotic resistance genes following land application of manure waste. J. Environ. Qual. 38 1086–1108. 10.2134/jeq2008.0128 [DOI] [PubMed] [Google Scholar]
- Chen J., Fluharty F. L., St-Pierre N., Morrison M., Yu Z. (2008). Technical note: occurrence in fecal microbiota of genes conferring resistance to both macrolide-lincosamide-streptogramin B and tetracyclines concomitant with feeding of beef cattle with tylosin. J. Anim. Sci. 86 2385–2391. 10.2527/jas.2007-0705 [DOI] [PubMed] [Google Scholar]
- Chen Q., An X., Li H., Su J., Ma Y., Zhu Y.-G. (2016). Long-term field application of sewage sludge increases the abundance of antibiotic resistance genes in soil. Environ. Int. 92–93 1–10. 10.1016/j.envint.2016.03.026 [DOI] [PubMed] [Google Scholar]
- Chen Q.-L., An X.-L., Zhu Y.-G., Su J.-Q., Gillings M. R., Ye Z.-L., et al. (2017). Application of struvite alters the antibiotic resistome in soil, rhizosphere, and phyllosphere. Environ. Sci. Technol. 51 8149–8157. 10.1021/acs.est.7b01420 [DOI] [PubMed] [Google Scholar]
- Chen Q.-L., Cui H.-L., Su J.-Q., Penuelas J., Zhu Y.-G. (2019a). Antibiotic resistomes in plant microbiomes. Trends Plant Sci. 24 530–541. 10.1016/j.tplants.2019.02.010 [DOI] [PubMed] [Google Scholar]
- Chen Q.-L., Zhu D., An X.-L., Ding J., Zhu Y.-G., Cui L. (2019b). Does nano silver promote the selection of antibiotic resistance genes in soil and plant? Environ. Int. 128 399–406. 10.1016/j.envint.2019.04.061 [DOI] [PubMed] [Google Scholar]
- Cheng G., Hao H., Xie S., Wang X., Dai M., Huang L., et al. (2014). Antibiotic alternatives: the substitution of antibiotics in animal husbandry? Front. Microbiol. 5:217. 10.3389/fmicb.2014.00217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christou A., Papadavid G., Dalias P., Fotopoulos V., Michael C., Bayona J. M., et al. (2019). Ranking of crop plants according to their potential to uptake and accumulate contaminants of emerging concern. Environ. Res. 170 422–432. 10.1016/j.envres.2018.12.048 [DOI] [PubMed] [Google Scholar]
- Collett S. R., Smith J. A., Boulianne M., Owen R. L., Gingerich E., Singer R. S., et al. (2019). “Principles of disease prevention, diagnosis, and control,” in Diseases of Poultry, eds Swayne D. E., et al. (Hoboken, NJ: John Wiley & Sons, Ltd; ), 1–78. 10.1002/9781119371199.ch1 [DOI] [Google Scholar]
- Cook K. L., Netthisinghe A. M. P., Gilfillen R. A. (2014). Detection of pathogens, indicators, and antibiotic resistance genes after land application of poultry litter. J. Environ. Qual. 43:1546. 10.2134/jeq2013.10.0432 [DOI] [PubMed] [Google Scholar]
- Dandachi I., Sokhn E. S., Dahdouh E. A., Azar E., El-Bazzal B., Rolain J.-M., et al. (2018). Prevalence and characterization of multi-drug-resistant gram-negative bacilli isolated from lebanese poultry: a nationwide study. Front. Microbiol. 9:550. 10.3389/fmicb.2018.00550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Liguoro M., Cibin V., Capolongo F., Halling-Sørensen B., Montesissa C. (2003). Use of oxytetracycline and tylosin in intensive calf farming: evaluation of transfer to manure and soil. Chemosphere 52 203–212. 10.1016/S0045-6535(03)00284-4 [DOI] [PubMed] [Google Scholar]
- Diarra M. S., Malouin F. (2014). Antibiotics in Canadian poultry productions and anticipated alternatives. Front. Microbiol. 5:282. 10.3389/fmicb.2014.00282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowling P. M. (2013). “Peptide antibiotics,” in Antimicrobial Therapy in Veterinary Medicine, eds Giguère S., Prescott J. F., Dowling P. M. (Hoboken, NJ: John Wiley & Sons, Ltd; ), 10.1002/9781118675014.ch11 [DOI] [Google Scholar]
- Durso L. M., Harhay G. P., Bono J. L., Smith T. P. L. (2011). Virulence-associated and antibiotic resistance genes of microbial populations in cattle feces analyzed using a metagenomic approach. J. Microbiol. Methods 84 278–282. 10.1016/j.mimet.2010.12.008 [DOI] [PubMed] [Google Scholar]
- Economou V., Gousia P. (2015). Agriculture and food animals as a source of antimicrobial-resistant bacteria. Infect. Drug Resist. 8 49–61. 10.2147/IDR.S55778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Centre for Disease Prevention, and Control [ECDC], European Food Safety Authority [EFSA], and European Medicines Agency [EMA] (2017). ECDC/EFSA/EMA second joint report on the integrated analysis of the consumption of antimicrobial agents and occurrence of antimicrobial resistance in bacteria from humans and food-producing animals. EFSA J. 15:e04872. 10.2903/j.efsa.2017.4872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Commission (2005). Regulation 1831/2003/EC on Additives for Use in Animal Nutrition, Replacing Directive 70/524/EEC on Additives in Feeding-stuffs. [Google Scholar]
- European Commision (2018). A European One Health Action Plan Against Antimicrobial Resistance (AMR); 2017/C 212/01. Available online at: https://ec.europa.eu/health/antimicrobial-resistance/eu-action-on-antimicrobial-resistance_en [Google Scholar]
- European Medicines Agency (2017). ESVAC Report – Ninth ESVAC Report: Sales of Veterinary Antimicrobial Agents in 31 European Countries in 2017. Available online at: https://www.ema.europa.eu/en/documents/report/sales-veterinary-antimicrobial-agents-31-european-countries-2017_en.pdf [Google Scholar]
- Fang L., Li X., Li L., Li S., Liao X., Sun J., et al. (2016). Co-spread of metal and antibiotic resistance within ST3-IncHI2 plasmids from E. coli isolates of food-producing animals. Sci. Rep. 6:25312. 10.1038/srep25312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAOSTAT (2020). Available online at: http://www.fao.org/faostat/en/#home [Google Scholar]
- Ferguson J. J., Ziegler M. R. (2004). Guidelines for Purchase and Application of Poultry Manure for Organic Crop Production. EDIS 2004. Available online at: https://journals.flvc.org/edis/article/view/111168 (accessed March 12, 2020). [Google Scholar]
- Fernandes M. R., Moura Q., Sartori L., Silva K. C., Cunha M. P., Esposito F., et al. (2016). Silent dissemination of colistin-resistant Escherichia coli in South America could contribute to the global spread of the mcr-1 gene. Eurosurveillance 21:30214. 10.2807/1560-7917.ES.2016.21.17.30214 [DOI] [PubMed] [Google Scholar]
- Fogler K., Guron G. K. P., Wind L. L., Keenum I. M., Hession W. C., Krometis L.-A., et al. (2019). Microbiota and antibiotic resistome of lettuce leaves and radishes grown in soils receiving manure-based amendments derived from antibiotic-treated cows. Front. Sustain. Food Syst. 3:22. 10.3389/fsufs.2019.00022 [DOI] [Google Scholar]
- Forsberg K. J., Reyes A., Wang B., Selleck E. M., Sommer M. O. A., Dantas G. (2012). The shared antibiotic resistome of soil bacteria and human pathogens. Science 337 1107–1111. 10.1126/science.1220761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Lu C., Shen D., Liu J., Ma Z., Yang B., et al. (2019). Elimination of the risks of colistin resistance gene (mcr-1) in livestock manure during composting. Environ. Int. 126 61–68. 10.1016/j.envint.2019.02.015 [DOI] [PubMed] [Google Scholar]
- Gebreyes W. A., Thakur S., Morgan Morrow W. E. (2006). Comparison of prevalence, antimicrobial resistance, and occurrence of multidrug-resistant Salmonella in antimicrobial-free and conventional pig production. J. Food Prot. 69 743–748. 10.4315/0362-028X-69.4.743 [DOI] [PubMed] [Google Scholar]
- Girotto F., Cossu R. (2017). “Animal waste: opportunities and challenges,” in Sustainable Agriculture Reviews Sustainable Agriculture Reviews, ed. Lichtfouse E. (Cham: Springer International Publishing; ), 1–13. 10.1007/978-3-319-48006-0_1 [DOI] [Google Scholar]
- Góchez D., Raicek M., Pinto Ferreira J., Jeannin M., Moulin G., Erlacher-Vindel E. (2019). OIE annual report on antimicrobial agents intended for use in animals: methods used. Front. Vet. Sci. 6:317. 10.3389/fvets.2019.00317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein C., Lee M. D., Sanchez S., Hudson C., Phillips B., Register B., et al. (2001). Incidence of class 1 and 2 integrases in clinical and commensal bacteria from livestock, companion animals, and exotics. Antimicrob. Agents Chemother. 45 723–726. 10.1128/AAC.45.3.723-726.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez Ronquillo M., Angeles Hernandez J. C. (2017). Antibiotic and synthetic growth promoters in animal diets: review of impact and analytical methods. Food Control 72 255–267. 10.1016/j.foodcont.2016.03.001 [DOI] [Google Scholar]
- Graham J. P., Evans S. L., Price L. B., Silbergeld E. K. (2009). Fate of antimicrobial-resistant enterococci and staphylococci and resistance determinants in stored poultry litter. Environ. Res. 109 682–689. 10.1016/j.envres.2009.05.005 [DOI] [PubMed] [Google Scholar]
- Guenther S., Falgenhauer L., Semmler T., Imirzalioglu C., Chakraborty T., Roesler U., et al. (2017). Environmental emission of multiresistant Escherichia coli carrying the colistin resistance gene mcr-1 from German swine farms. J. Antimicrob. Chemother. 72 1289–1292. 10.1093/jac/dkw585 [DOI] [PubMed] [Google Scholar]
- Guron G. K. P., Arango-Argoty G., Zhang L., Pruden A., Ponder M. A. (2019). Effects of dairy manure-based amendments and soil texture on lettuce- and radish-associated microbiota and resistomes. mSphere 4:e239-19. 10.1128/mSphere.00239-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haack B. J., Andrews R. E., Jr. (2000). Isolation of Tn916-like conjugal elements from swine lot effluent. Can. J. Microbiol. 46 542–549. [DOI] [PubMed] [Google Scholar]
- Haenni M., Saras E., Châtre P., Meunier D., Martin S., Lepage G., et al. (2009). vanA in Enterococcus faecium, Enterococcus faecalis, and Enterococcus casseliflavus detected in French cattle. Foodborne Pathog. Dis. 6 1107–1111. 10.1089/fpd.2009.0303 [DOI] [PubMed] [Google Scholar]
- Halbert L. W., Kaneene J. B., Ruegg P. L., Warnick L. D., Wells S. J., Mansfield L. S., et al. (2006). Evaluation of antimicrobial susceptibility patterns in Campylobacter spp isolated from dairy cattle and farms managed organically and conventionally in the midwestern and northeastern United States. J. Am. Vet. Med. Assoc. 228 1074–1081. 10.2460/javma.228.7.1074 [DOI] [PubMed] [Google Scholar]
- Hanon J.-B., Jaspers S., Butaye P., Wattiau P., Méroc E., Aerts M., et al. (2015). A trend analysis of antimicrobial resistance in commensal Escherichia coli from several livestock species in Belgium (2011–2014). Prev. Vet. Med. 122 443–452. 10.1016/j.prevetmed.2015.09.001 [DOI] [PubMed] [Google Scholar]
- Hanson R., Kaneene J. B., Padungtod P., Hirokawa K., Zeno C. (2002). Prevalence of Salmonella and E. coli, and their resistance to antimicrobial agents, in farming communities in northern Thailand. Se. Asian. J. Trop. Med. 33 120–126. [PubMed] [Google Scholar]
- Hassan Y. I., Lahaye L., Gong M. M., Peng J., Gong J., Liu S., et al. (2018). Innovative drugs, chemicals, and enzymes within the animal production chain. Vet. Res. 49:71. 10.1186/s13567-018-0559-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L.-Y., He L.-K., Liu Y.-S., Zhang M., Zhao J.-L., Zhang Q.-Q., et al. (2019). Microbial diversity and antibiotic resistome in swine farm environments. Sci. Total Environ. 685 197–207. 10.1016/j.scitotenv.2019.05.369 [DOI] [PubMed] [Google Scholar]
- He T., Wang Y., Sun L., Pang M., Zhang L., Wang R. (2017a). Occurrence and characterization of blaNDM-5-positive Klebsiella pneumoniae isolates from dairy cows in Jiangsu. China. J. Antimicrob. Chemother. 72 90–94. 10.1093/jac/dkw357 [DOI] [PubMed] [Google Scholar]
- He T., Wei R., Zhang L., Sun L., Pang M., Wang R., et al. (2017b). Characterization of NDM-5-positive extensively resistant Escherichia coli isolates from dairy cows. Vet. Microbiol. 207 153–158. 10.1016/j.vetmic.2017.06.010 [DOI] [PubMed] [Google Scholar]
- Heuer H., Schmitt H., Smalla K. (2011). Antibiotic resistance gene spread due to manure application on agricultural fields. Curr. Opin. Microbiol. 14 236–243. 10.1016/j.mib.2011.04.009 [DOI] [PubMed] [Google Scholar]
- Holman D. B., Yang W., Alexander T. W. (2019). Antibiotic treatment in feedlot cattle: a longitudinal study of the effect of oxytetracycline and tulathromycin on the fecal and nasopharyngeal microbiota. Microbiome 7:86. 10.1186/s40168-019-0696-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hölzel C. S., Müller C., Harms K. S., Mikolajewski S., Schäfer S., Schwaiger K., et al. (2012). Heavy metals in liquid pig manure in light of bacterial antimicrobial resistance. Environ. Res. 113 21–27. 10.1016/j.envres.2012.01.002 [DOI] [PubMed] [Google Scholar]
- Hölzel C. S., Tetens J. L., Schwaiger K. (2018). Unraveling the role of vegetables in spreading antimicrobial-resistant bacteria: a need for quantitative risk assessment. Foodborne Pathog. Dis. 15 671–688. 10.1089/fpd.2018.2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X., Zhou Q., Luo Y. (2010). Occurrence and source analysis of typical veterinary antibiotics in manure, soil, vegetables and groundwater from organic vegetable bases, northern China. Environ. Pollut. 2992–2998. 10.1016/j.envpol.2010.05.023 [DOI] [PubMed] [Google Scholar]
- Imran M., Das K. R., Naik M. M. (2019). Co-selection of multi-antibiotic resistance in bacterial pathogens in metal and microplastic contaminated environments: an emerging health threat. Chemosphere 215 846–857. 10.1016/j.chemosphere.2018.10.114 [DOI] [PubMed] [Google Scholar]
- International Office of Epizootics (ed.) (2015). OIE standards, Gidelines and Resolution on Antimicrobial Resistance and the Use of Antimicrobial Agents. Paris: OIE. [Google Scholar]
- Jackson C. R., Lombard J. E., Dargatz D. A., Fedorka-Cray P. J. (2011). Prevalence, species distribution and antimicrobial resistance of enterococci isolated from US dairy cattle. Lett. Appl. Microbiol. 52 41–48. 10.1111/j.1472-765X.2010.02964.x [DOI] [PubMed] [Google Scholar]
- Jechalke S., Heuer H., Siemens J., Amelung W., Smalla K. (2014). Fate and effects of veterinary antibiotics in soil. Trends Microbiol. 22 536–545. 10.1016/j.tim.2014.05.005 [DOI] [PubMed] [Google Scholar]
- Jiang X., Ellabaan M., Charusanti P., Munck C., Blin K., Tong Y., et al. (2017). Dissemination of antibiotic resistance genes from antibiotic producers to pathogens. Nat. Commun. 8:15784. 10.1038/ncomms15784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X., Yang H., Dettman B., Doyle M. P. (2006). Analysis of fecal microbial flora for antibiotic resistance in ceftiofur-treated calves. Foodborne Pathog. Dis. 3 355–365. 10.1089/fpd.2006.3.355 [DOI] [PubMed] [Google Scholar]
- Journals I., Oke A. A. (2013). Incidence of methicillin-resistant Staphylococcus aureus (MRSA) in a small poultry in south West, Nigeria. IOSR J. Agric. Vet. Sci. 5 53–55. 10.9790/2380-0535355 [DOI] [Google Scholar]
- Kanwar N., Scott H. M., Norby B., Loneragan G. H., Vinasco J., McGowan M., et al. (2013). Effects of ceftiofur and chlortetracycline treatment strategies on antimicrobial susceptibility and on tet(A), tet(B), and bla CMY-2 resistance genes among E. coli isolated from the feces of feedlot cattle. PLoS One 8:e80575. 10.1371/journal.pone.0080575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karami N., Martner A., Enne V. I., Swerkersson S., Adlerberth I., Wold A. E. (2007). Transfer of an ampicillin resistance gene between two Escherichia coli strains in the bowel microbiota of an infant treated with antibiotics. J. Antimicrob. Chemother. 60 1142–1145. 10.1093/jac/dkm327 [DOI] [PubMed] [Google Scholar]
- Kempf I., Jouy E., Chauvin C. (2016). Colistin use and colistin resistance in bacteria from animals. Int. J. Antimicrob. Agents 48 598–606. 10.1016/j.ijantimicag.2016.09.016 [DOI] [PubMed] [Google Scholar]
- Khan G. A., Berglund B., Khan K. M., Lindgren P.-E., Fick J. (2013). Occurrence and abundance of antibiotics and resistance genes in rivers, canal and near drug formulation facilities – a study in Pakistan. PLoS One 8:e62712. 10.1371/journal.pone.0062712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klotz P., Higgins P. G., Schaubmar A. R., Failing K., Leidner U., Seifert H., et al. (2019). Seasonal occurrence and carbapenem susceptibility of bovine Acinetobacter baumannii in Germany. Front. Microbiol. 10:272. 10.3389/fmicb.2019.00272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike S., Krapac I. G., Oliver H. D., Yannarell A. C., Chee-Sanford J. C., Aminov R. I., et al. (2007). Monitoring and source tracking of tetracycline resistance genes in lagoons and groundwater adjacent to swine production facilities over a 3-year period. Appl. Environ. Microbiol. 73 4813–4823. 10.1128/AEM.00665-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar K., C. Gupta S., Chander Y., Singh A. K. (2005). Antibiotic use in agriculture and its impact on the terrestrial environment. Adv. Agron. 87 1–54. 10.1016/S0065-2113(05)87001-4 [DOI] [Google Scholar]
- Lang K. S., Anderson J. M., Schwarz S., Williamson L., Handelsman J., Singer R. S. (2010). Novel florfenicol and chloramphenicol resistance gene discovered in alaskan soil by using functional metagenomics. Appl. Environ. Microbiol. 76 5321–5326. 10.1128/AEM.00323-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langata L. M., Maingi J. M., Musonye H. A., Kiiru J., Nyamache A. K. (2019). Antimicrobial resistance genes in Salmonella and Escherichia coli isolates from chicken droppings in Nairobi, Kenya. BMC Res. Notes 12:22. 10.1186/s13104-019-4068-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclercq S. O., Wang C., Zhu Y., Wu H., Du X., Liu Z., et al. (2016). Diversity of the tetracycline mobilome within a Chinese pig manure sample. Appl. Environ. Microbiol. 82 6454–6462. 10.1128/AEM.01754-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. H. (2006). Occurrence of methicillin-resistant Staphylococcus aureus strains from cattle and chicken, and analyses of their mecA, mecR1 and mecI genes. Vet. Microbiol. 114 155–159. 10.1016/j.vetmic.2005.10.024 [DOI] [PubMed] [Google Scholar]
- Li Y.-W., Wu X.-L., Mo C.-H., Tai Y.-P., Huang X.-P., Xiang L. (2011). Investigation of sulfonamide, tetracycline, and quinolone antibiotics in vegetable farmland soil in the Pearl River Delta Area, Southern China. J. Agric. Food Chem. 59 7268–7276. 10.1021/jf1047578 [DOI] [PubMed] [Google Scholar]
- Lim S.-K., Kim D., Moon D.-C., Cho Y., Rho M. (2020). Antibiotic resistomes discovered in the gut microbiomes of Korean swine and cattle. GigaScience 9:giaa043. 10.1093/gigascience/giaa043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.-Y., Wang Y., Walsh T. R., Yi L.-X., Zhang R., Spencer J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16 161–168. 10.1016/S1473-3099(15)00424-7 [DOI] [PubMed] [Google Scholar]
- Ljubojević D., Pelić M., Puvača N., Milanov D. (2017). Resistance totetracycline in Escherichia coli isolates from poultry meat: epidemiology, policy and perspective. Worlds Poult. Sci. J. 73 409–417. 10.1017/S0043933917000216 [DOI] [Google Scholar]
- Looft T., Johnson T. A., Allen H. K., Bayles D. O., Alt D. P., Stedtfeld R. D., et al. (2012). In-feed antibiotic effects on the swine intestinal microbiome. Proc. Natl. Acad. Sci. U. S. A. 109 1691–1696. 10.1073/pnas.1120238109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorian V. (1975). Some effects of subinhibitory concentrations of antibiotics on bacteria. Bull. N. Y. Acad. Med. 51 1046–1055. [PMC free article] [PubMed] [Google Scholar]
- Lundberg D. S., Lebeis S. L., Paredes S. H., Yourstone S., Gehring J., Malfatti S., et al. (2012). Defining the core Arabidopsis thaliana root microbiome. Nature 488 86–90. 10.1038/nature11237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackie R. I., Koike S., Krapac I., Chee-Sanford J., Maxwell S., Aminov R. I. (2006). Tetracycline residues and tetracycline resistance genes in groundwater impacted by swine production facilities. Anim. Biotechnol. 17 157–176. 10.1080/10495390600956953 [DOI] [PubMed] [Google Scholar]
- Mahmoud M. A. M., Abdel-Mohsein H. S. (2019). Hysterical tetracycline in intensive poultry farms accountable for substantial gene resistance, health and ecological risk in Egypt- manure and fish. Environ. Pollut. 255:113039. 10.1016/j.envpol.2019.113039 [DOI] [PubMed] [Google Scholar]
- Manaia C. M., Rocha J., Scaccia N., Marano R., Radu E., Biancullo F., et al. (2018). Antibiotic resistance in wastewater treatment plants: tackling the black box. Environ. Int. 115 312–324. 10.1016/j.envint.2018.03.044 [DOI] [PubMed] [Google Scholar]
- Marti R., Scott A., Tien Y.-C., Murray R., Sabourin L., Zhang Y., et al. (2013). Impact of manure fertilization on the abundance of antibiotic-resistant bacteria and frequency of detection of antibiotic resistance genes in soil and on vegetables at harvest. Appl. Environ. Microbiol. 79 5701–5709. 10.1128/AEM.01682-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marti R., Tien Y.-C., Murray R., Scott A., Sabourin L., Topp E. (2014). Safely coupling livestock and crop production systems: how rapidly do antibiotic resistance genes dissipate in soil following a commercial application of swine or dairy manure? Appl. Environ. Microbiol. 80 3258–3265. 10.1128/AEM.00231-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEachran A. D., Blackwell B. R., Hanson J. D., Wooten K. J., Mayer G. D., Cox S. B., et al. (2015). Antibiotics, bacteria, and antibiotic resistance genes: aerial transport from cattle feed yards via particulate matter. Environ. Health Perspect. 123 337–343. 10.1289/ehp.1408555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney C. W., Dungan R. S., Moore A., Leytem A. B. (2018). Occurrence and abundance of antibiotic resistance genes in agricultural soil receiving dairy manure. FEMS Microbiol. Ecol. 94:fiy010. 10.1093/femsec/fiy010 [DOI] [PubMed] [Google Scholar]
- Medicine (2019). Committee on Human Health Risk Assessment of Using Subtherapeutic Antibiotics in Animal Feeds. Washington, DC: The National Academies Press. [Google Scholar]
- Merchant L. E., Rempel H., Forge T., Kannangara T., Bittman S., Delaquis P., et al. (2012). Characterization of antibiotic-resistant and potentially pathogenic Escherichia coli from soil fertilized with litter of broiler chickens fed antimicrobial-supplemented diets. Can. J. Microbiol. 58 1084–1098. 10.1139/w2012-082 [DOI] [PubMed] [Google Scholar]
- Mollenkopf D. F., Stull J. W., Mathys D. A., Bowman A. S., Feicht S. M., Grooters S. V., et al. (2017). Carbapenemase-producing Enterobacteriaceae recovered from the environment of a swine farrow-to-finish operation in the United States. Antimicrob. Agents Chemother. 61:e01298-16. 10.1128/AAC.01298-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munk P., Knudsen B. E., Lukjancenko O., Duarte A. S. R., Van Gompel L., Luiken R. E. C., et al. (2018). Abundance and diversity of the faecal resistome in slaughter pigs and broilers in nine European countries. Nat. Microbiol. 3 898–908. 10.1038/s41564-018-0192-9 [DOI] [PubMed] [Google Scholar]
- Murray R., Tien Y.-C., Scott A., Topp E. (2019). The impact of municipal sewage sludge stabilization processes on the abundance, field persistence, and transmission of antibiotic resistant bacteria and antibiotic resistance genes to vegetables at harvest. Sci. Total Environ. 651 1680–1687. 10.1016/j.scitotenv.2018.10.030 [DOI] [PubMed] [Google Scholar]
- Muurinen J., Stedtfeld R., Karkman A., Pärnänen K., Tiedje J., Virta M. (2017). Influence of manure application on the environmental resistome under finnish agricultural practice with restricted antibiotic use. Environ. Sci. Technol. 51 5989–5999. 10.1021/acs.est.7b00551 [DOI] [PubMed] [Google Scholar]
- Nesme J., Cécillon S., Delmont T. O., Monier J.-M., Vogel T. M., Simonet P. (2014). Large-scale metagenomic-based study of antibiotic resistance in the environment. Curr. Biol. 24 1096–1100. 10.1016/j.cub.2014.03.036 [DOI] [PubMed] [Google Scholar]
- Nesme J., Simonet P. (2015). The soil resistome: a critical review on antibiotic resistance origins, ecology and dissemination potential in telluric bacteria. Environ. Microbiol. 17 913–930. 10.1111/1462-2920.12631 [DOI] [PubMed] [Google Scholar]
- Nhung N. T., Cuong N. V., Thwaites G., Carrique-Mas J. (2016). Antimicrobial usage and antimicrobial resistance in animal production in southeast asia: a review. Antibiotics 5:37. 10.3390/antibiotics5040037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- OIE (2018). OIE Annual Report on Antimicrobial Agents Intended for Use in Animals – Better Understanding of the Global Situation. Third Report. Available online at: https://rr-africa.oie.int/wp-content/uploads/2019/09/annual_report_amr_3.pdf [Google Scholar]
- Omojowo F., Omojasola F. (2013). Antibiotic resistance pattern of bacterial pathogens isolated from poultry manure used to fertilize fish ponds in new Bussa. Nigeria. Albanian J. Agric. Sci. 12 81–85. [Google Scholar]
- Pal C., Bengtsson-Palme J., Kristiansson E., Larsson D. G. J. (2015). Co-occurrence of resistance genes to antibiotics, biocides and metals reveals novel insights into their co-selection potential. BMC Genomics 16:964. 10.1186/s12864-015-2153-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan M., Chu L. M. (2017). Transfer of antibiotics from wastewater or animal manure to soil and edible crops. Environ. Pollut. 231 829–836. 10.1016/j.envpol.2017.08.051 [DOI] [PubMed] [Google Scholar]
- Poirel L., Berçot B., Millemann Y., Bonnin R. A., Pannaux G., Nordmann P. (2012). Carbapenemase-producing Acinetobacter spp. in Cattle. France. Emerg. Infect. Dis. 18 523–525. 10.3201/eid1803.111330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole K. (2017). At the nexus of antibiotics and metals: the impact of Cu and Zn on antibiotic activity and resistance. Trends Microbiol. 25 820–832. 10.1016/j.tim.2017.04.010 [DOI] [PubMed] [Google Scholar]
- Pu C., Liu H., Ding G., Sun Y., Yu X., Chen J., et al. (2018). Impact of direct application of biogas slurry and residue in fields: in situ analysis of antibiotic resistance genes from pig manure to fields. J. Hazard. Mater. 344 441–449. 10.1016/j.jhazmat.2017.10.031 [DOI] [PubMed] [Google Scholar]
- Pulss S., Semmler T., Prenger-Berninghoff E., Bauerfeind R., Ewers C. (2017). First report of an Escherichia coli strain from swine carrying an OXA-181 carbapenemase and the colistin resistance determinant MCR-1. Int. J. Antimicrob. Agents 50 232–236. 10.1016/j.ijantimicag.2017.03.014 [DOI] [PubMed] [Google Scholar]
- Quesada A., Ugarte-Ruiz M., Iglesias M. R., Porrero M. C., Martínez R., Florez-Cuadrado D., et al. (2016). Detection of plasmid mediated colistin resistance (MCR-1) in Escherichia coli and Salmonella enterica isolated from poultry and swine in Spain. Res. Vet. Sci. 105 134–135. 10.1016/j.rvsc.2016.02.003 [DOI] [PubMed] [Google Scholar]
- Rahube T. O., Marti R., Scott A., Tien Y.-C., Murray R., Sabourin L., et al. (2014). Impact of fertilizing with raw or anaerobically digested sewage sludge on the abundance of antibiotic-resistant coliforms, antibiotic resistance genes, and pathogenic bacteria in soil and on vegetables at harvest. Appl. Environ. Microbiol. 80 6898–6907. 10.1128/AEM.02389-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayamajhi N., Jung B. Y., Cha S. B., Shin M. K., Kim A., Kang M. S., et al. (2010). Antibiotic resistance patterns and detection of blaDHA-1 in Salmonella species isolates from chicken farms in South Korea. Appl. Environ. Microbiol. 76 4760–4764. 10.1128/AEM.02536-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redondo-Salvo S., Fernández-López R., Ruiz R., Vielva L., de Toro M., Rocha E. P. C., et al. (2020). Pathways for horizontal gene transfer in bacteria revealed by a global map of their plasmids. Nat. Commun. 11:3602. 10.1038/s41467-020-17278-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds L. J., Sala-Comorera L., Martin N. A., Nolan T. M., Stephens J. H., Gitto A., et al. (2020). Correlation between antimicrobial resistance and faecal contamination in small urban streams and bathing waters. Sci. Total Environ. 739:140242. 10.1016/j.scitotenv.2020.140242 [DOI] [PubMed] [Google Scholar]
- Roesch M., Perreten V., Doherr M. G., Schaeren W., Schällibaum M., Blum J. W. (2006). Comparison of antibiotic resistance of udder pathogens in dairy cows kept on organic and on conventional farms. J. Dairy Sci. 89 989–997. 10.3168/jds.S0022-0302(06)72164-6 [DOI] [PubMed] [Google Scholar]
- Roschanski N., Falgenhauer L., Grobbel M., Guenther S., Kreienbrock L., Imirzalioglu C., et al. (2017). Retrospective survey of mcr-1 and mcr-2 in German pig-fattening farms, 2011–2012. Int. J. Antimicrob. Agents 50 266–271. 10.1016/j.ijantimicag.2017.03.007 [DOI] [PubMed] [Google Scholar]
- Rosenblueth M., Martínez-Romero E. (2006). Bacterial endophytes and their interactions with hosts. Mol. Plant Microbe Interact. 19 827–837. 10.1094/MPMI-19-0827 [DOI] [PubMed] [Google Scholar]
- Roth N., Käsbohrer A., Mayrhofer S., Zitz U., Hofacre C., Domig K. J. (2019). The application of antibiotics in broiler production and the resulting antibiotic resistance in Escherichia coli: a global overview. Poult. Sci. 98 1791–1804. 10.3382/ps/pey539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabry S. A., Ghozlan H. A., Abou-Zeid D.-M. (1997). Metal tolerance and antibiotic resistance patterns of a bacterial population isolated from sea water. J. Appl. Microbiol. 82 245–252. 10.1111/j.1365-2672.1997.tb03580.x [DOI] [PubMed] [Google Scholar]
- Sato K., Bartlett P. C., Saeed M. A. (2005). Antimicrobial susceptibility of Escherichia coli isolates from dairy farms using organic versus conventional production methods. J. Am. Vet. Med. Assoc. 226 589–594. 10.2460/javma.2005.226.589 [DOI] [PubMed] [Google Scholar]
- Sawant A. A., Hegde N. V., Straley B. A., Donaldson S. C., Love B. C., Knabel S. J., et al. (2007). Antimicrobial-resistant enteric bacteria from dairy cattle. Appl. Environ. Microbiol. 73 156–163. 10.1128/AEM.01551-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt H., Stoob K., Hamscher G., Smit E., Seinen W. (2006). Tetracyclines and tetracycline resistance in agricultural soils: microcosm and field studies. Microb. Ecol. 51 267–276. 10.1007/s00248-006-9035-y [DOI] [PubMed] [Google Scholar]
- Seiffert S. N., Hilty M., Perreten V., Endimiani A. (2013). Extended-spectrum cephalosporin-resistant Gram-negative organisms in livestock: an emerging problem for human health? Drug. Resist. Updat. 16 22–45. 10.1016/j.drup.2012.12.001 [DOI] [PubMed] [Google Scholar]
- Sengeløv G., Agersø Y., Halling-Sørensen B., Baloda S. B., Andersen J. S., Jensen L. B. (2003). Bacterial antibiotic resistance levels in Danish farmland as a result of treatment with pig manure slurry. Environ. Int. 28 587–595. 10.1016/s0160-4120(02)00084-3 [DOI] [PubMed] [Google Scholar]
- Singer R. S., Patterson S. K., Wallace R. L. (2008). Effects of therapeutic ceftiofur administration to dairy cattle on Escherichia coli dynamics in the intestinal tract. Appl. Environ. Microbiol. 74 6956–6962. 10.1128/AEM.01241-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- So A. D., Shah T. A., Roach S., Chee Y. L., Nachman K. E. (2015). An integrated systems approach is needed to ensure the sustainability of antibiotic effectiveness for both humans and animals. J. Law. Med. Ethics 43 38–45. 10.1111/jlme.12273 [DOI] [PubMed] [Google Scholar]
- Solomon E. B., Yaron S., Matthews K. R. (2002). Transmission of Escherichia coli O157:H7 from contaminated manure and irrigation water to lettuce plant tissue and its subsequent internalization. Appl. Environ. Microbiol. 68 397–400. 10.1128/aem.68.1.397-400.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soulsby L. (2007). Antimicrobials and animal health: a fascinating nexus. J. Antimicrob. Chemother. 60 i77–i78. 10.1093/jac/dkm164 [DOI] [PubMed] [Google Scholar]
- Stecher B., Denzler R., Maier L., Bernet F., Sanders M. J., Pickard D. J., et al. (2012). Gut inflammation can boost horizontal gene transfer between pathogenic and commensal Enterobacteriaceae. Proc. Natl. Acad. Sci. U. S. A. 109 1269–1274. 10.1073/pnas.1113246109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suresh G., Das R. K., Brar S. K., Rouissi T., Ramirez A. A., Chorfi Y., et al. (2018). Alternatives to antibiotics in poultry feed: molecular perspectives. Crit. Rev. Microbiol. 44 318–335. 10.1080/1040841X.2017.1373062 [DOI] [PubMed] [Google Scholar]
- Tabashsum Z., Biswas D. (2019). “Chapter 17 - acceptable alternative growth promoters for organic farm animal production,” in Safety and Practice for Organic Food, eds Biswas D., Micallef S. A. (Cambridge, MA: Academic Press; ), 345–363. 10.1016/B978-0-12-812060-6.00017-9 [DOI] [Google Scholar]
- Tanih G. N., Okoh A. I., Ndip R. N., Nwodo U. U., Green E. (2017). Multidrug resistance in Enterococcus species of faecal origin from commercial dairy lactating cattle: public health concern. Asian Pac. J. Trop. Dis. 7 665–670. 10.12980/apjtd.7.2017D7-154 [DOI] [Google Scholar]
- Thakur S., Gebreyes W. A. (2005). Prevalence and antimicrobial resistance of Campylobacter in antimicrobial-free and conventional pig production systems. J. Food Prot. 68 2402–2410. 10.4315/0362-028X-68.11.2402 [DOI] [PubMed] [Google Scholar]
- Thames C. H., Pruden A., James R. E., Ray P. P., Knowlton K. F. (2012). Excretion of antibiotic resistance genes by dairy calves fed milk replacers with varying doses of antibiotics. Front. Microbiol. 3:139. 10.3389/fmicb.2012.00139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tien Y.-C., Li B., Zhang T., Scott A., Murray R., Sabourin L., et al. (2017). Impact of dairy manure pre-application treatment on manure composition, soil dynamics of antibiotic resistance genes, and abundance of antibiotic-resistance genes on vegetables at harvest. Sci. Total Environ. 581–582 32–39. 10.1016/j.scitotenv.2016.12.138 [DOI] [PubMed] [Google Scholar]
- Tragesser L. A., Wittum T. E., Funk J. A., Winokur P. L., Rajala-Schultz P. J. (2006). Association between ceftiofur use and isolation of Escherichia coli with reduced susceptibility to ceftriaxone from fecal samples of dairy cows. Am. J. Vet. Res. 67 1696–1700. 10.2460/ajvr.67.10.1696 [DOI] [PubMed] [Google Scholar]
- Tyrrell C., Burgess C. M., Brennan F. P., Walsh F. (2019). Antibiotic resistance in grass and soil. Biochem. Soc. Trans. 47 477–486. 10.1042/BST20180552 [DOI] [PubMed] [Google Scholar]
- Tzavaras I., Siarkou V. I., Zdragas A., Kotzamanidis C., Vafeas G., Bourtzi-Hatzopoulou E., et al. (2012). Diversity of vanA-type vancomycin-resistant Enterococcus faecium isolated from broilers, poultry slaughterers and hospitalized humans in Greece. J. Antimicrob. Chemother. 67 1811–1818. 10.1093/jac/dks166 [DOI] [PubMed] [Google Scholar]
- Udikovic-Kolic N., Wichmann F., Broderick N. A., Handelsman J. (2014). Bloom of resident antibiotic-resistant bacteria in soil following manure fertilization. Proc. Natl. Acad. Sci. U.S.A. 111 15202–15207. 10.1073/pnas.1409836111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Boeckel T. P., Brower C., Gilbert M., Grenfell B. T., Levin S. A., Robinson T. P., et al. (2015). Global trends in antimicrobial use in food animals. Proc. Natl. Acad. Sci. U.S.A. 112 5649–5654. 10.1073/pnas.1503141112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Cuong N., Nhung N. T., Nghia N. H., Mai Hoa N. T., Trung N. V., Thwaites G., et al. (2016). Antimicrobial consumption in medicated feeds in vietnamese pig and poultry production. EcoHealth 13 490–498. 10.1007/s10393-016-1130-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vikram A., Schmidt J. W. (2018). Functional blaKPC-2 sequences are present in U.S. beef cattle feces regardless of antibiotic use. Foodborne Pathog. Dis. 15 444–448. 10.1089/fpd.2017.2406 [DOI] [PubMed] [Google Scholar]
- Volkova V. V., Lanzas C., Lu Z., Gröhn Y. T. (2012). Mathematical model of plasmid-mediated resistance to ceftiofur in commensal enteric Escherichia coli of cattle. PLoS One 7:e36738. 10.1371/journal.pone.0036738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waglechner N., Wright G. D. (2017). Antibiotic resistance: it’s bad, but why isn’t it worse? BMC Biol. 15:84. 10.1186/s12915-017-0423-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoek A. H. A. M., Veenman C., van Overbeek W. M., Lynch G., de Roda Husman A. M., Blaak H. (2015). Prevalence and characterization of ESBL- and AmpC-producing Enterobacteriaceae on retail vegetables. Int. J. Food Microbiol. 204 1–8. 10.1016/j.ijfoodmicro.2015.03.014 [DOI] [PubMed] [Google Scholar]
- Walia K., Sharma M., Vijay S., Shome B. R. (2019). Understanding policy dilemmas around antibiotic use in food animals & offering potential solutions. Indian J. Med. Res. 149 107–118. 10.4103/ijmr.IJMR_2_18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F.-H., Qiao M., Chen Z., Su J.-Q., Zhu Y.-G. (2015). Antibiotic resistance genes in manure-amended soil and vegetables at harvest. J. Hazard. Mater. 299 215–221. 10.1016/j.jhazmat.2015.05.028 [DOI] [PubMed] [Google Scholar]
- Wang R., Dorp L., van, Shaw L. P., Bradley P., Wang Q., et al. (2018). The global distribution and spread of the mobilized colistin resistance gene mcr-1. Nat. Commun. 9:1179. 10.1038/s41467-018-03205-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb H. E., Bugarel M., den Bakker H. C., Nightingale K. K., Granier S. A., Scott H. M., et al. (2016). Carbapenem-resistant bacteria recovered from faeces of dairy cattle in the high plains region of the USA. PLoS One 11:e0147363. 10.1371/journal.pone.0147363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichmann F., Udikovic-Kolic N., Andrew S., Handelsman J. (2014). Diverse antibiotic resistance genes in dairy cow manure. mBio 5:e01017. 10.1128/mBio.01017-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson A., Fox E. M., Fegan N., Kurtböke D. Í. (2019). Comparative genomics and phenotypic investigations into antibiotic, heavy metal, and disinfectant susceptibilities of Salmonella enterica strains isolated in Australia. Front. Microbiol. 10:1620. 10.3389/fmicb.2019.01620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winokur P. L., Vonstein D. L., Hoffman L. J., Uhlenhopp E. K., Doern G. V. (2001). Evidence for transfer of CMY-2 AmpC beta-lactamase plasmids between Escherichia coli and Salmonella isolates from food animals and humans. Antimicrob. Agents Chemother. 45 2716–2722. 10.1128/AAC.45.10.2716-2722.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wongsuvan G., Wuthiekanun V., Hinjoy S., Day N. P., Limmathurotsakul D. (2018). Antibiotic use in poultry: a survey of eight farms in Thailand. Bull. World Health Organ. 96 94–100. 10.2471/BLT.17.195834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodford N., Wareham D. W., Guerra B., Teale C. (2014). Carbapenemase-producing Enterobacteriaceae and non-Enterobacteriaceae from animals and the environment: an emerging public health risk of our own making? J. Antimicrob. Chemother. 69 287–291. 10.1093/jac/dkt392 [DOI] [PubMed] [Google Scholar]
- World Health Organization and WHO Advisory Group on Integrated Surveillance of Antimicrobial Resistance (AGISAR) (2017). Critically Important Antimicrobials for Human Medicine: Ranking of Antimicrobial Agents for Risk Management of Antimicrobial Resistance Due to Non-Human Use, 5th Revision. Avialable online at: https://apps.who.int/iris/handle/10665/255027 [Google Scholar]
- Wu N., Qiao M., Zhang B., Cheng W.-D., Zhu Y.-G. (2010). Abundance and diversity of tetracycline resistance genes in soils adjacent to representative swine feedlots in China. Environ. Sci. Technol. 44 6933–6939. 10.1021/es1007802 [DOI] [PubMed] [Google Scholar]
- Xie W.-Y., Shen Q., Zhao F. J. (2018). Antibiotics and antibiotic resistance from animal manures to soil: a review. Eur. J. Soil Sci. 69 181–195. 10.1111/ejss.12494 [DOI] [Google Scholar]
- Yang F., Deng B., Liao W., Wang P., Chen P., Wei J. (2019). High rate of multiresistant Klebsiella pneumoniae from human and animal origin. Infect. Drug Resist. 12 2729–2737. 10.2147/IDR.S219155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Liu W., Zhu D., Hou J., Ma T., Wu L., et al. (2018). Application of biosolids drives the diversity of antibiotic resistance genes in soil and lettuce at harvest. Soil Biol. Biochem. 122 131–140. 10.1016/j.soilbio.2018.04.017 [DOI] [Google Scholar]
- Yang Q., Ren S., Niu T., Guo Y., Qi S., Han X., et al. (2014). Distribution of antibiotic-resistant bacteria in chicken manure and manure-fertilized vegetables. Environ. Sci. Pollut. Res. Int. 21 1231–1241. 10.1007/s11356-013-1994-1 [DOI] [PubMed] [Google Scholar]
- Yang Q., Zhang H., Guo Y., Tian T. (2016). Influence of chicken manure fertilization on antibiotic-resistant bacteria in soil and the endophytic bacteria of pakchoi. Int J Environ Res Public Health 13:662. 10.3390/ijerph13070662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q., Zhang J., Zhu K., Zhang H. (2009). Influence of oxytetracycline on the structure and activity of microbial community in wheat rhizosphere soil. J. Environ. Sci. 21 954–959. 10.1016/S1001-0742(08)62367-0 [DOI] [PubMed] [Google Scholar]
- Yin W., Li H., Shen Y., Liu Z., Wang S., Shen Z., et al. (2017). Novel plasmid-mediated colistin resistance gene mcr-3 in Escherichia coli. mBio 8:e543-17. 10.1128/mBio.00543-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaheer Z., Rahman S. U., Zaheer I., Abbas G., Younas T. (2017). Methicillin-resistant Staphylococcus aureus in poultry- an emerging concern related to future epidemic. Matrix Sci. Medica 1 15–18. 10.26480/msm.01.2017.15.18 [DOI] [Google Scholar]
- Zalewska M., Popowska M. (2020). “Antimicrobial/antibiotic resistance genes due to manure and agricultural waste applications,” in Antibiotics and Antimicrobial Resistance Genes: Environmental Occurrence and Treatment Technologies Emerging Contaminants and Associated Treatment Technologies, ed. Hashmi M. Z. (Cham: Springer International Publishing; ), 139–161. 10.1007/978-3-030-40422-2_6 [DOI] [Google Scholar]
- Zhang X.-X., Zhang T. (2011). Occurrence, abundance, and diversity of tetracycline resistance genes in 15 sewage treatment plants across China and other global locations. Environ. Sci. Technol. 45 2598–2604. 10.1021/es103672x [DOI] [PubMed] [Google Scholar]
- Zhang Y., Gu A. Z., Cen T., Li X., He M., Li D., et al. (2018). Sub-inhibitory concentrations of heavy metals facilitate the horizontal transfer of plasmid-mediated antibiotic resistance genes in water environment. Environ. Pollut. 237 74–82. 10.1016/j.envpol.2018.01.032 [DOI] [PubMed] [Google Scholar]
- Zhang Y.-J., Hu H.-W., Chen Q.-L., Singh B. K., Yan H., Chen D., et al. (2019). Transfer of antibiotic resistance from manure-amended soils to vegetable microbiomes. Environ. Int. 130:104912. 10.1016/j.envint.2019.104912 [DOI] [PubMed] [Google Scholar]
- Zhang Y.-J., Hu H.-W., Chen Q.-L., Yan H., Wang J.-T., Chen D., et al. (2020). Manure application did not enrich antibiotic resistance genes in root endophytic bacterial microbiota of cherry radish plants. Appl. Environ. Microbiol. 86:e2106-19. 10.1128/AEM.02106-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Wang J., Zhu L., Ge W., Wang J. (2017). Environmental analysis of typical antibiotic-resistant bacteria and ARGs in farmland soil chronically fertilized with chicken manure. Sci. Total Environ. 593–594 10–17. 10.1016/j.scitotenv.2017.03.062 [DOI] [PubMed] [Google Scholar]
- Zhao Y., Su J.-Q., An X.-L., Huang F.-Y., Rensing C., Brandt K. K., et al. (2018). Feed additives shift gut microbiota and enrich antibiotic resistance in swine gut. Sci. Total Environ. 621 1224–1232. 10.1016/j.scitotenv.2017.10.106 [DOI] [PubMed] [Google Scholar]
- Zhao Y., Yang Q. E., Zhou X., Wang F.-H., Muurinen J., Virta M. P., et al. (2020). Antibiotic resistome in the livestock and aquaculture industries: status and solutions. Crit. Rev. Environ. Sci. Technol. 1–38. 10.1080/10643389.2020.1777815 [DOI] [Google Scholar]
- Zhu B., Chen Q., Chen S., Zhu Y.-G. (2017). Does organically produced lettuce harbor higher abundance of antibiotic resistance genes than conventionally produced? Environ. Int. 98 152–159. 10.1016/j.envint.2016.11.001 [DOI] [PubMed] [Google Scholar]