Significance
Myosin II is the molecule that produces force in muscle contraction. Unlike the myosin head, its molecular motor, no atomic resolution structure of the ∼1000-residue–long α-helical coiled-coil tail has been reported. Here, we describe the cryo-EM atomic structure of the myosin tail within a native muscle thick filament. Three differences with crystal structures of myosin tail segments were found. The myosin head arrangement apparently alters the beginning of the tail. Striated muscle myosins have four skip residues, amino acids inserted to improve the alignment of charged residue clusters. Skips 1 and 3 agree with the crystal structures. Skip 2, which is a novel structure, and Skip 4 do not. Functional consequences are suggested by the myosin tail packing.
Keywords: cryo-electron microscopy, alpha helix coiled coil, striated muscle, invertebrate, filament
Abstract
The atomic structure of the complete myosin tail within thick filaments isolated from Lethocerus indicus flight muscle is described and compared to crystal structures of recombinant, human cardiac myosin tail segments. Overall, the agreement is good with three exceptions: the proximal S2, in which the filament has heads attached but the crystal structure doesn’t, and skip regions 2 and 4. At the head–tail junction, the tail α-helices are asymmetrically structured encompassing well-defined unfolding of 12 residues for one myosin tail, ∼4 residues of the other, and different degrees of α-helix unwinding for both tail α-helices, thereby providing an atomic resolution description of coiled-coil “uncoiling” at the head–tail junction. Asymmetry is observed in the nonhelical C termini; one C-terminal segment is intercalated between ribbons of myosin tails, the other apparently terminating at Skip 4 of another myosin tail. Between skip residues, crystal and filament structures agree well. Skips 1 and 3 also agree well and show the expected α-helix unwinding and coiled-coil untwisting in response to skip residue insertion. Skips 2 and 4 are different. Skip 2 is accommodated in an unusual manner through an increase in α-helix radius and corresponding reduction in rise/residue. Skip 4 remains helical in one chain, with the other chain unfolded, apparently influenced by the acidic myosin C terminus. The atomic model may shed some light on thick filament mechanosensing and is a step in understanding the complex roles that thick filaments of all species undergo during muscle contraction.
Striated muscles have two main filament types, thin, actin-containing and thick, myosin-containing. Actin, the building block of the thin filaments, is one of the most highly conserved proteins in nature (1). Thin filaments across muscle types are structurally very similar (2); differences are primarily found in the proteins binding F-actin such as tropomyosin, ubiquitous in striated muscle, and troponin, which if present makes the thin filament the focus of contraction regulation. The same pattern is seen in myosin filaments in the sequence level in which the nonmyosin proteins in the thick filament are not as highly conserved or absent entirely depending on species.
Conversely, thick filaments are quite variable in structure. Those from vertebrate muscle have threefold rotation symmetry, a quasi-helical arrangement of myosin heads with variable axial spacings that average 143 Å to produce a super repeat of 429 Å (3), and lengths narrowly limited to 1.6 μm governed by the giant protein titin (4). Those from invertebrates differ widely in lengths and rotational symmetries and generally have a helical arrangement of myosin heads with an axial repeat of 145 Å like that of Lethocerus flight muscle (5). Rotational symmetries as high as sevenfold are known (6), but no symmetry lower than fourfold has been reported. The protein paramyosin appears ubiquitous in thick filaments from invertebrates but absent in those of vertebrates (7). Paramyosin occupies the center of the invertebrate thick filament whereas titin binds the outer surface of the vertebrate thick filament. Such wide differences suggest that vertebrate and invertebrate thick filaments have little in common structurally other than that they contain myosin. However, myosin is highly conserved, though less so than actin, which suggests some similarity in structure, perhaps through the “curved molecular crystalline layers,” or ribbons, as the basic building block in filaments (8).
Myosin has three main domains: a folded globular domain, the molecular motor that contains the ATPase and actin-binding properties, a lever arm, which binds a pair of light chains to amplify conformation changes in the motor domain, and a long α-helical tail that spontaneously forms a parallel, two-stranded, coiled-coil dimer. At ∼1,600 Å, it is one of the longest coiled coils in nature. The myosin head has been extensively studied by many techniques, but due to its unique length, the myosin tail structure has been a challenge. The tail itself can be produced in quantity by proteolysis and forms several types of higher-ordered antiparallel aggregates (9). Myosin tail subfragments, S2 and light meromyosin, form paracrystalline arrays that have proved unsuitable for high-resolution structural studies (10, 11). A promising approach is the crystallization of tail segments stabilized using small globular domains at their ends (12, 13). However, myosin’s extreme length and systematic axial offsets within filaments (14, 15) mean that crystals of segments are unlikely to replicate the key intermolecular interactions that occur in filaments. They may, however, produce structures unaffected by external influences other than crystal contacts and thus reveal a thermodynamic “ground state” conformation.
The coiled-coil structure as a packing arrangement for α-helices was predicted by Francis Crick (16) to explain the X-ray diffraction patterns of fibrous proteins first observed by Astbury (17). Historically, the key evidence suggesting higher-order structure in muscle filaments was the presence of a 5.15 Å reflection first observed in striated muscle (17). The coiled coil explained the reflection which was subsequently verified by X-ray fiber diffraction of molluscan smooth muscle (18).
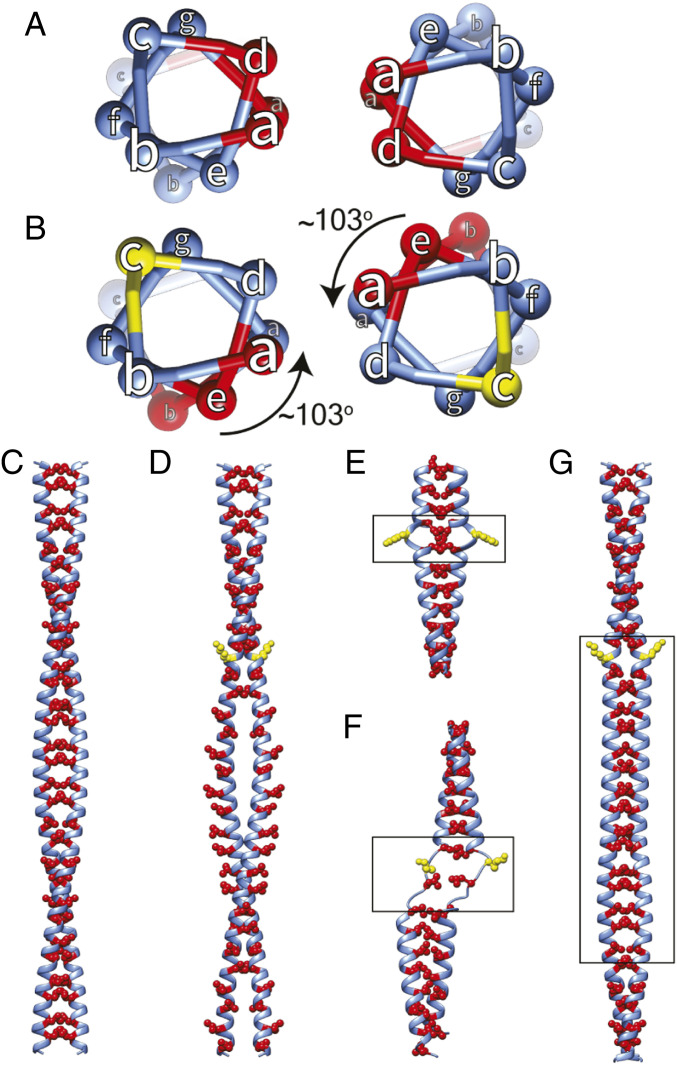
Crick’s coiled-coil model not only explained Astbury’s 5.15 Å reflection but also laid out the heptad pattern for the coiled-coil sequence, remarkably years before any coiled-coil proteins were sequenced. Confirmation for the heptad pattern came 20 y later when tropomyosin was sequenced (19). Positions in the heptad repeat are denoted as “abcdefg” with hydrophobic residues in the first and fourth positions, located in the interface of two 7/2 α-helices (Fig. 1 A and C). Interlocking residues in this “knobs-in-holes” manner play a significant part in stabilizing α-helices in a large variety of proteins (20). However, the heptad pattern is not always followed in the entirety of coiled-coil sequences.
Fig. 1.
Theoretical effect of skip residues on coiled coils. (A) Top view showing hydrophobic residues (red) in the “a” and “d” positions of the heptad repeat. (B) A skip residue insertion at position “c” (yellow) rotates the hydrophobic residue in position “d” into position “e” thus exposed to solvent. α-Helices adjust to the skip residue in different ways. (C and D) Longitudinal views of skip residue insertion. (C) Hydrophobic residues (red) buried between the paired α-helices. (D) The skip residue (yellow) shifts the “d” position by one in subsequent heptads. (E–G) Skip residue accommodation. (E) Insert a π-turn, which has an extra residue in the α-helix, (F) unfold one or both α-helices forming a loop or hinge, or (G) return the α-helix pitch to 3.6 residues/turn thereby straightening the coiled coil for about 10 α-helical turns. The box identifies the Accommodation Region where the change occurs.
The first myosin II amino acid sequence from the nematode Caenorhabditis elegans (14) revealed a very long predicted coiled-coil domain with four skip residue insertions. Skip residues, the first three of which are separated by 196 residues (7 × 28) and the last by 224 residues (8 × 28), enhance a periodic distribution of charged residues explaining the thick filament axial repeat of 145 Å. With later myosin II sequences, properties like the coiled coil, its four skip residues, and the periodic charge distributions proved to be highly conserved among species as different as nematode and human, indicative of significant functional roles. All striated muscle myosin IIs conserve these four skip residues (SI Appendix, Fig. S1). In contrast, smooth and nonmuscle myosin IIs have three skip residues, apparently missing Skip 2 (SI Appendix, Fig. S2) (21).
Here, we report an atomic structure of the myosin tail from the large waterbug, Lethocerus indicus, obtained by cryoelectron microscopy (cryo-EM) of native filaments. Myosin’s unusual coiled-coil tail length in its natural environment provided an opportunity to examine its parameters in detail. Several unexpected results were observed including differences in coiled-coil structure in the vicinity of the skip residues when compared to previously reported crystal structures. Molecular dynamics (MD) simulations suggest these differences arise from interactions with other myosin tails.
Results
L. indicus Thick Filament.
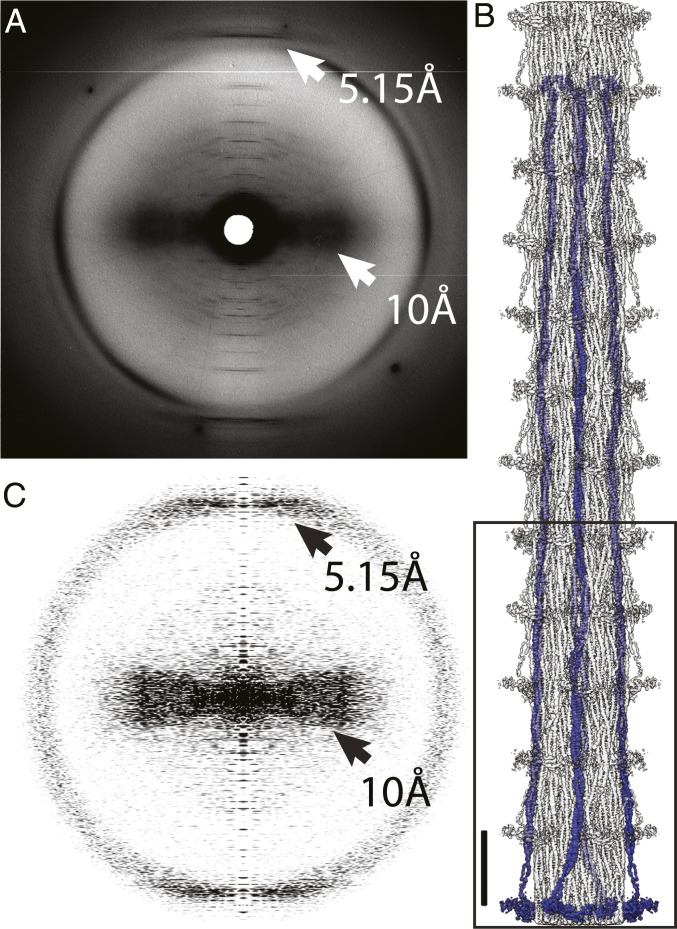
A key marker for the presence of a two-stranded α-helical coiled coil is a 5.15 Å near-meridional reflection in an X-ray fiber diagram (18). Wide-angle X-ray fiber diagrams of L. indicus flight muscle reveal the distinctive 5.15 Å layer line (Fig. 2A). After extending the reconstruction (Fig. 2B), the resulting power spectrum also reveals the characteristic 5.15 Å peak of the coiled coil, numerous meridional layer lines at increasing orders of 145 Å, and equatorial or near-equatorial intensities at 10 Å due to the α-helix packing within the coiled coil (Fig. 2C).
Fig. 2.
High-angle X-ray diffraction from Lethocerus myofibrils. (A) The 5.15 Å reflection and the broad ∼10 Å near-equatorial maximum characteristic of coiled coils (white arrows). (B) Reconstruction extended to 12 crowns using helical symmetry. Orientation has the M-line (C terminus) at the top and Z-disk (N terminus) at the bottom. The box defines the original reconstruction. (Scale bar = 145 Å.) Globular domains projecting from the backbone surface are the myosin regulatory light chains. The rest of the head is disordered at this resolution. The bend to the left at the C terminus is the nonhelical myosin C terminus. (C) Power spectrum computed from the projection of the extended reconstruction shows the same characteristics as the X-ray diagram (black arrows). The inner layer lines, too closely bunched at this magnification to be individually identified, can be seen in the supplemental material of ref. 15. Note that exact correspondence between A and C is not expected because the X-ray diagram comes from an intact muscle fiber with coiled-coil proteins such as tropomyosin and paramyosin, whereas the reconstruction shows only those features following the myosin helical repeat.
Individual myosin tails are arranged as ribbons in the filament backbone (8). Myosin heads in relaxed striated muscles are conformed into a structure known as the interacting heads motif which has an unusual orientation in Lethocerus flight muscle with the so-called “free head” binding the thick filament backbone azimuthally and the “blocked head” appearing to pin the “free head” against the backbone. No interaction occurs between the blocked-head motor domain and the coiled coil (15) as occurs in other thick filaments (3, 22). Low-pass filtering to ∼20 Å resolution reveals the heads, but at 4.25 Å resolution, only the regulatory light chains are seen (Fig. 2B).
Atomic Model Validation.
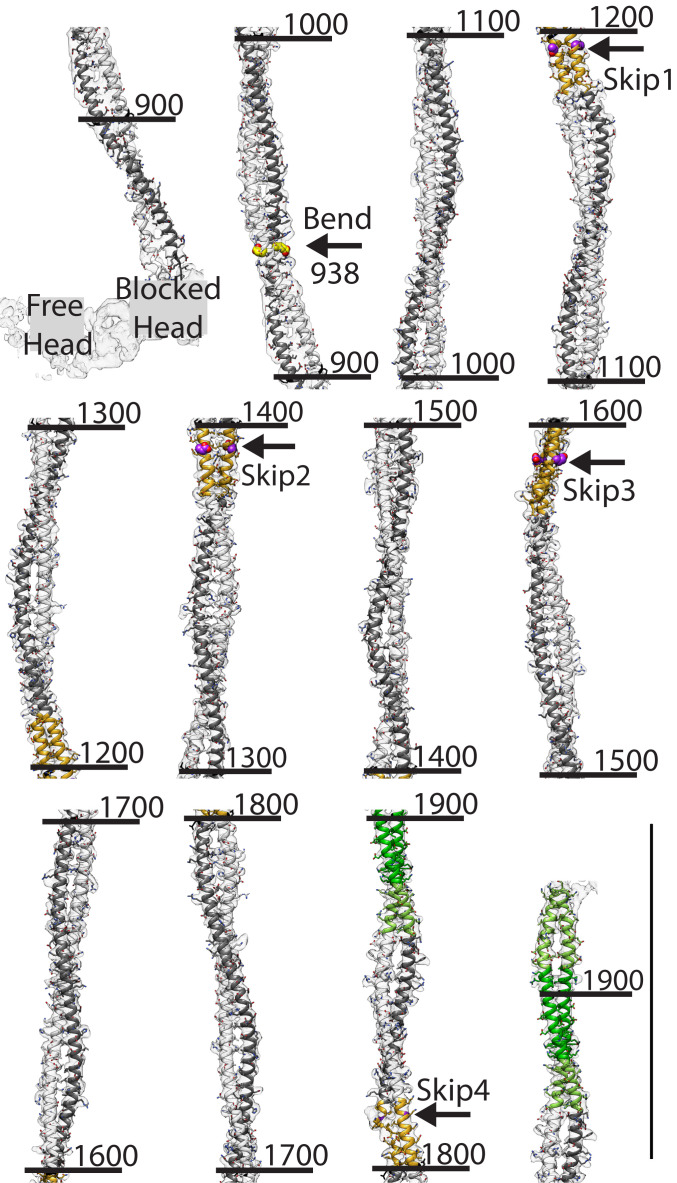
The resolution of the reconstruction is relatively low for de novo atomic model building but was facilitated by the nearly 100% α-helix content of the structure (Fig. 3). Larger side chains like histidine, lysine, and arginine are used as landmarks to minimize errors that can propagate for considerable distance at our resolution. The atomic model was built as α-helix starting from blocked head residue P836, in which the beginning of the “hook” occurs (but see Fig. 4D for an alternative). The free-head α-helix begins later at residue E856 following unfolding of the preceding 12 residues into an extended chain. On reaching Skip 4, in which the chain B helix (blocked head) is broken by the pair of glycine residues (12), the model was in phase with the break. The coiled-coil prediction score declines after residue T1930 which corresponds to the end of the coiled-coil density (SI Appendix, Fig. S3).
Fig. 3.
Coiled-coil reconstruction and fitted atomic model. View is from the outside toward the center to illustrate the azimuthal bending. Skip residues are marked with arrows. The coloring scheme has heavy chains of free head, white and blocked head, dark gray; Accommodation regions, orange; Assembly Competence Domain, green with the extended domain, light green; side chains of Skip residues 1 through 3, purple spheres; and side chain of residue 938 at start of S2 bending, yellow spheres. (Scale bar = 145 Å.) Numbers are residue numbers.
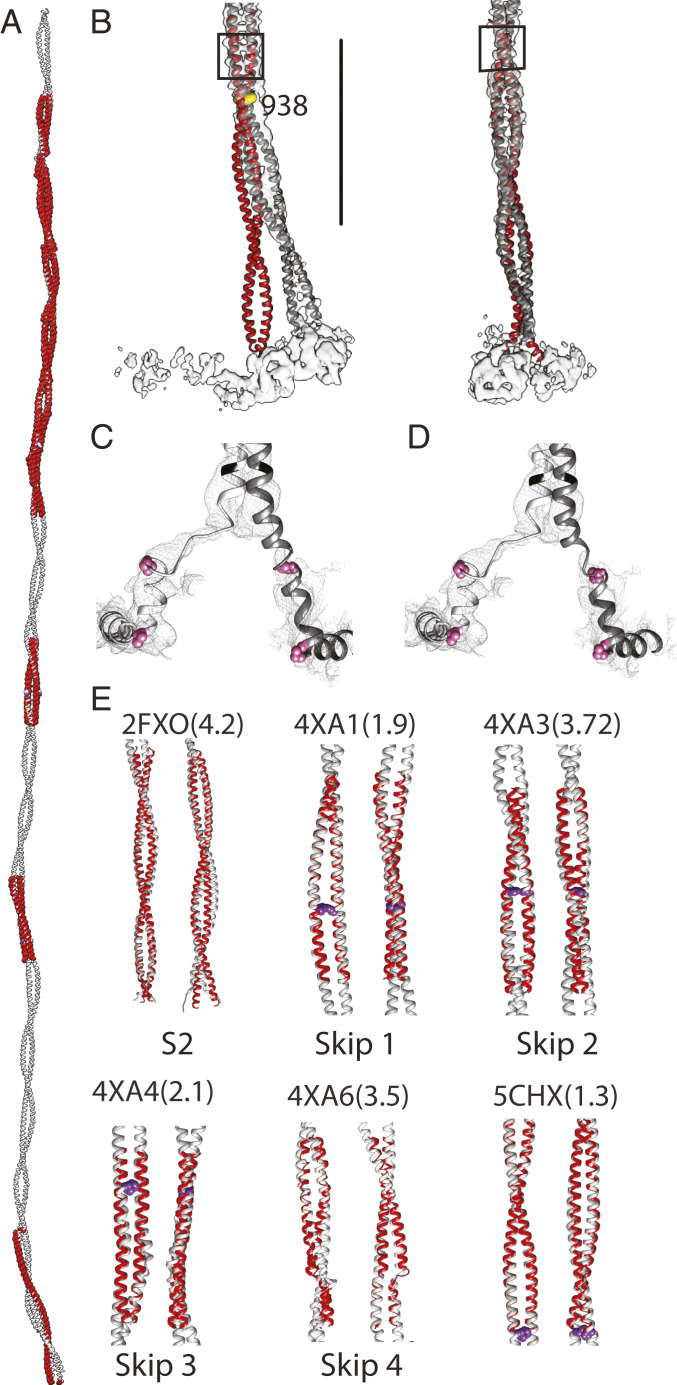
Fig. 4.
Comparison between crystal structures of human cardiac tail segments and the Lethocerus atomic model. (A) All crystallized segments (red) overlapped on the Lethocerus atomic model (gray). (B) Two orthogonal views showing the proximal S2 structures aligned using residues 945 through 963 (shown in the black box). Beyond 960, the fit is also very good. Pronounced bending starts around residue 938 (yellow sphere). (C and D) Views of the reconstruction and atomic model near the head–tail junction. Pink highlighted residues are P836 at the beginning of the “hook” and the invariant proline P846. (C) Fit imposing α-helical constraints. (D) Fit without helical constraints; three to five residues in the blocked head heavy chain (845 to 848) are unfolded. The free head heavy chain requires unfolding of 12 residues (843 to 855) to fit. (E) Cardiac myosin tail crystal structures compared to Lethocerus. Skip residues are shown using purple spheres; numbers in parenthesis are the Cα RMSDs in Ångstroms. Segments with Skip residues 1 and 2 have a higher RMSD.
The Lethocerus Coiled Coil.
The myosin coiled-coil atomic model extends from residue L860 near the head–tail junction to residue T1930 at the end of the coiled coil (Fig. 3). The myosin tail is not straight but is characterized by multiple bends, the first and most pronounced of which occurs in the proximal S2 near residue E938 in which the myosin tail enters the filament backbone (15). Notably, no hinge or break in the coiled coil is visible at this location or at the more conventional location near the cleavage site that produces heavy meromyosin (the two-headed subfragment) in which sharp bends in isolated molecules have been observed (21); the coiled coil is continuous throughout the conventional S2 region.
The tail continues axially after this bend until the next coiled-coil crossover after which it begins reversing by bending slightly rightwards. At the crossover just before Skip 1, approximately residue 1150, the tail bends to the left and the Skip 1 Accommodation Region is positioned with the two helices lying side by side on the thick filament surface. Just after the Skip 1 Accommodation Region, an azimuthal bend toward the right occurs to straighten the path of the coiled coil toward axial which it follows through the Skip 2 Accommodation Region. After Skip 2 at approximately residue 1510, another slight bend to the left occurs followed by a bend to the right just before Skip 3. The two helices at Skip 1 and Skip 2 lie side by side with respect to the radial vector, whereas they lie one on top of the other at Skip 3. The trajectory is followed to about residue 1710 where a bend to the left occurs after which the coiled coil continues through Skip 4. After the crossover immediately following Skip 4, the coiled coil bends to the right followed by a bend to the left just before the Assembly Competence Domain. The view in Fig. 3 is along a radial direction which emphases the larger azimuthal bending that the tail undergoes. Bending also occurs in the radial direction with the tail having an overall 1.5° tilt outwards (15). Notably a sharp radial bend occurs after Skip 2 at residue 1460 where the radius increases before again decreasing. After Skip 4, the radius again increases into the Assembly Competence Domain before decreasing steadily to the end of the coiled coil. These bends are quantified in SI Appendix, Fig. S4. Coiled-coil bending is greatest about an axis connecting the two chains and least about the axis perpendicular to it.
Comparison with Crystal Structures of Cardiac Myosin Tail Segments.
Thirteen crystal structures of human cardiac myosin tail segments are available in the Protein Data Bank (PDB), including the four skip residues (12) with four additional segments ranging from 1,534 to 1,697 around Skip 3 (13) and one proximal S2 crystal structure (PDB: 2FXM, 2FXO) (23). Crystal structures of coiled-coil segments cover less than half of the coiled-coil length (Fig. 4A). Coverage is most continuous between Skips 3 and 4.
The cardiac proximal S2 is a nearly straight, classical coiled coil (23), distinctly different from the proximal S2 of Lethocerus flight muscle, with its pronounced 17° bend at residue E938 (Fig. 4B). Alignment of the cardiac proximal S2 structure to residues 945 through 963 of the Lethocerus proximal S2 emphasizes the distinctive bend observed in situ (Fig. 4B) which results in a 25 Å separation between the two structures at the start of the coiled coil. α-Helix unfolding is observed in the proximal S2 at the head–tail junction (Fig. 4 C and D). If the α-helix is enforced, a continuous helix can be built from the beginning of the coiled coil through the blocked head–tail junction to the distinctive 90° bend that produces the so-called “hook” (Fig. 4C). If not enforced, the density is not sufficiently defined to maintain a continuous helix and perhaps three to five residues are unfolded (Fig. 4D). For the free head, 12 residues must be unfolded to fit the well-defined density before the coiled-coil start.
Comparison of all 13 crystal structures with our atomic model reveals good agreement for all except for the proximal S2 (PDB 2FXM, 2FXO), Skip 2 (PDB 4XA3), and Skip 4 (PDB 4XA6) regions (Fig. 4E and SI Appendix, Fig. S5). Almost all other crystal structures fit our atomic model with RMSD of <2 Å when calculated for Cα atoms. Given the differences in packing interactions in crystals and filaments, remarkable agreement in bending of the coiled coil is observed in most other regions, suggesting that these departures (bends) from an idealized, straight coiled coil are built into the structure by the amino acid sequence.
Coiled Coil Analysis.
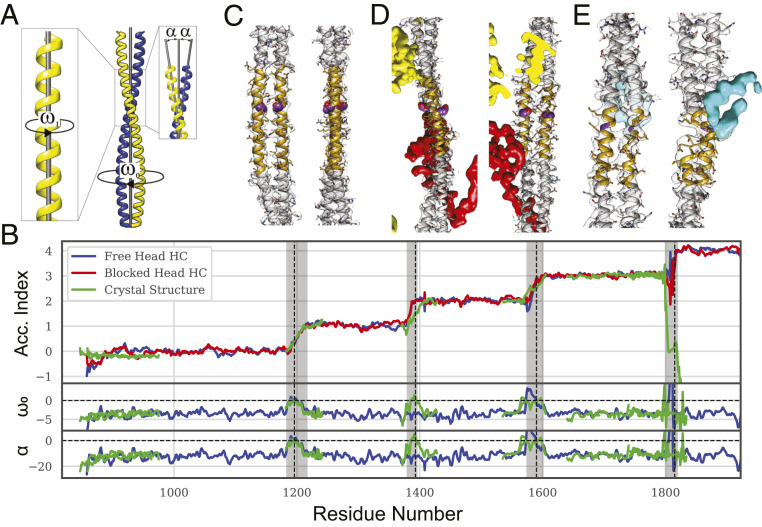
Crick’s model for two-stranded, parallel coiled coils describes a seven-residue-per-two-turns helix with general equations involving parameters R0, ω0, and φ0 which are the superhelical (coiled coil) radius, twisting frequency, and phase and R1, ω1, φ1, and α, which are the α-helical radius, frequency, phase, and pitch angle (Fig. 5A), all of which were calculated using Crick’s Coiled-Coil Parameterization (CCCP) server (24). In an ideal coiled coil, the α-helices are winding 102.8° per residue. Insertion of a skip residue alters the coiled-coil parameters, providing an objective way to identify their presence from the structure itself, in particular the winding of the α-helices, ω1, the twist of the coiled coil, ω0, and the α-helix pitch angle, α (Fig. 5B). With four skip residues, the myosin II atomic model must deviate from an ideal coiled-coil four times. Departures from ideality are tracked using a parameter called the Accommodation Index, which is the cumulative under-winding of the α-helices compared to a canonical coiled coil divided by 102.8°. The Accommodation Index is calculated using a seven-residue moving window over the length of the coiled coil and is expected to increase by one after each skip. All four skips were identified in their predicted regions by Accommodation Index increases, suggesting errors in the atomic model are small and local.
Fig. 5.
Skip residue analysis. (A) Definition of α-helix and coiled-coil parameters. (B) Coiled-coil structure analysis. Values for the Lethocerus atomic model are shown in red and blue and the cardiac crystal structure segments (green). Shaded areas indicate the Accommodation Regions, which have variable length. Accommodation Index, which reflects α-helix winding (ω1), is calculated for each chain separately; other parameters determined by the CCCP server such as the twist (ω0) and the pitch angle (α) require a coiled coil. At Skip 4, one helix formed a hinge thereby breaking the coiled coil. (C–E) Skip residue regions for Skips 1, 3, and 4. Skip residues are colored violet. (C) Skip 1. (D) Skip 3; red, flightin and yellow, myofilin. (E) Skip 4; cyan is an extra density of uncertain origin, possibly the nonhelical myosin C terminus.
Cα atoms in the coiled-coil backbone follow a specific geometry, which provides a means of detecting alterations propagating through the structure caused by either sequence or environment. Numerous bends of the myosin coiled coil in the filament described above and previously (15) as well as other alterations are reflected in an overall RMSD of 12 Å when fitted against an idealized, straight coiled coil (for Cα atoms) using the CCCP server (25). When the calculation is done using a moving window of seven residues, the RMSD falls to 0.34 ± 0.14 Å (Fig. 5B), indicating that our α-helices follow a coiled-coil geometry without imposing coiled-coil constraints during atomic model building. However, RMSD increases are observed in some regions, especially over Skips 2 to 4.
The proximal S2 behaves like a conventional coiled coil in crystals. In the filament, the Accommodation Index increases in the part of S2 most proximal to the head–tail junction, indicating unwinding of the individual α-helices similar to that typically produced by skip residues (Fig. 5B). Both free-head and blocked-head heavy chains indicate α-helix unwinding but by different amounts. The free-head heavy chain undergoes almost 100° of unwinding, which is an amount similar to that caused by a skip residue, though no skip residue has ever been suggested or predicted for this location. The blocked head undergoes about half as much unwinding. In parallel with the helix unwinding is an increase in the rise/residue especially near the beginning of the coiled coil (Fig. 6C). Although resolution is lower in the proximal S2 than within the filament backbone, the change in helix winding after the coiled coil start is quite striking.
Fig. 6.
Coiled coil behavior at Skip 2. (A) Plot of Accommodation Index and α-helix radius. The α-helical radius is smoothed with a seven-residue moving average. Yellow triangles indicate interactions between myosin and myofilin. The vertical dashed line is positioned on the conventional Skip 2. (B) Reconstruction in the Skip 2 region. Myofilin, yellow, binds the coiled coil at opposite ends of the Skip 2 Accommodation Region. (C) Rise per residue for the entire coiled coil. The rise varies throughout the entire coiled coil but shows a significant reduction only at Skip 2 and a significant increase at Skip 4 and the proximal S2. The rise per residue values for the MD simulation at Skip 4 are meaningless because both chains are unfolded.
Asymmetry in the coiled coil is observed all along the filament (Fig. 3). Quantifying these changes shows that near the head–tail junction, head-induced changes are relatively high (SI Appendix, Fig. S6). Both backbone and side chain atoms show high asymmetry at the start of the coiled coil while decreasing as the proximal S2 enters the backbone. In the proximal S2 crystal structure, the side chains show high asymmetry but not the backbone atoms. Asymmetry also rises around residue 1,460, at the beginning of the Skip 3 Accommodation Region, at Skip 3 itself, is particularly high at Skip 4, and rises just before the end of the coiled coil. Asymmetry at Skip 3 was also high in the crystal structure for the side chains consistent with a special role for that skip (12). Overall in the filament, asymmetry in the backbone atoms is relatively small, <1 Å RMS, whereas the asymmetry in the side chains is relatively large, between 2 and 3 Å RMS. These values are generally higher than the corresponding crystal structures, partly due to resolution differences but also due to the influence of neighboring myosin tails.
By convention, the four skip residues in the Lethocerus myosin tail sequence are located at residues 1,196, 1,393, 1,590, and 1,815, which are the c positions at the end of 28 residue–long charged regions (14).* Skip residue positions predicted from the amino acid sequence are separated by 196, 196, and 224 amino acids, close to multiples of the axial repeat in myosin filaments (∼98 residues). However, the residue assignment is conventional, and the precise skip residue is difficult to locate by the coiled-coil structure, whereas its presence is easily detected by the α-helix unwinding. Based on reported cardiac myosin tail crystal structures (12), Skips 1 through 3 in the myosin tail are expected to follow Option 3 (Fig. 1G) having parallel α-helices; Skip 4 is expected to follow Option 2 (Fig. 1F) and form a pair of loops. Each of the four skip regions exhibit varying levels of departure from an ideal coiled coil as depicted in Movies S1–S4.
Skip 1 (Fig. 5C) behaves as predicted with α-helix unwinding, detected by the increase in Accommodation Index and coiled-coil untwisting, detected by ω0 and α approaching 0.0 (Fig. 5B), and is most similar to the cardiac Skip 1 structure (12). Indeed, the plots of Accommodation Index versus residue number for the crystal structure (PDB 4XA1) and the reconstruction nearly overlap. Coiled-coil asymmetry is low at Skip 1 in both filament and crystal structure (SI Appendix, Fig. S6).
At Skip 2, neither ω0 nor α approach zero as predicted, and the coiled-coil twist remains within normal parameters. Nevertheless, the Accommodation Index increases by one. In the crystal structure, both twist and pitch angle approach zero. At 15 residues, Skip 2’s Accommodation Region is about half that predicted for Option 3.
At Skip 3 (Fig. 5D), the α-helices are under wound, the coiled coil under twisted, and the in situ structure agrees well with the crystal structure. However, in the filament both ω0, the twisting frequency, and α, the pitch angle, unpredictably become positive typical of a right-handed coiled coil. In addition, one helix becomes bent. These alterations are possibly caused by interaction with the nonmyosin protein flightin (Fig. 5D), which passes between coiled coils within this ribbon at Skip 3 (15).
Skip 4 (Fig. 5E) differs from the crystal structure in that only one α-helix incorporates a loop, chain B, the tail portion of the blocked head (15, 26). Chain A, from the free head, remains a continuous α-helix and rises one unit in Accommodation Index but not particularly smoothly as the Accommodation Index oscillates somewhat. Skip 4 is accommodated over a short range of 18 residues. Where both α-helices break forming loop, as occurs in the crystal structures, the Accommodation Index calculation fails. In the filament, the Accommodation Index increases by one as predicted, but the other coiled-coil parameters are meaningless because of the loop in chain B. At Skip 4, the increase in coiled-coil length/residue is greater than the average for the coiled coil (and the rest of the atomic model) so that fewer residues are needed to produce the same increase in length (Fig. 6C). Moreover, an unidentified nonmyosin density is observed at the Skip 4 loop, implying that α-helix unwinding is instigated by an additional interaction not present in the crystal structure.
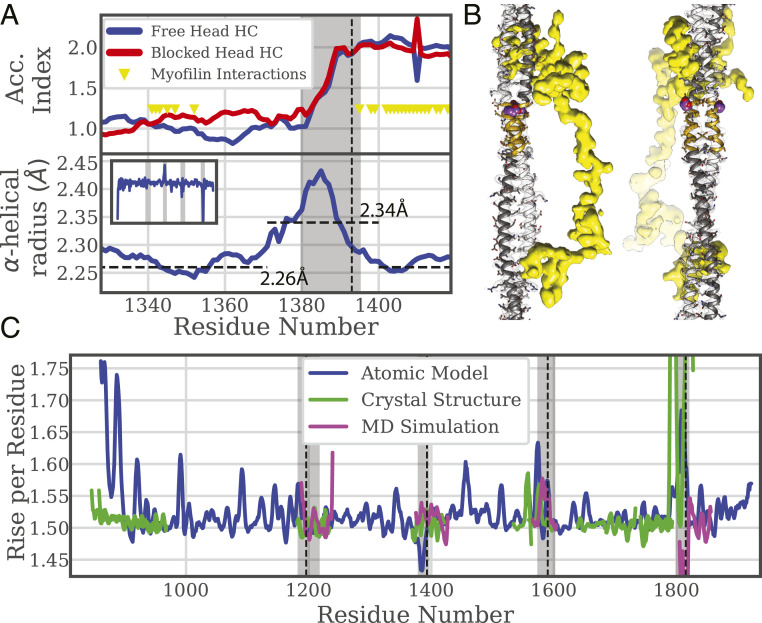
Unusual Structure of Skip 2.
As depicted in Fig. 1, the Accommodation Region of a skip residue theoretically follows the skip and lasts for ∼30 residues. Skip 2 at Lethocerus myosin residue 1,393 behaves in an unpredicted way: the α-helices are unwound as predicted, but the coiled coil is not untwisted, and the pitch angle is typical for a coiled coil. Skip 2’s 15-residue Accommodation Region precedes the conventional skip position. Higher RMSD in this region reveals the structure does not correspond to Crick’s coiled-coil model as well as elsewhere. The Ramachandran plot (SI Appendix, Fig. S7 B and C) shows that the residues in this region, including G1387, fall within the allowed α-helical region. How is Skip 2 accommodated?
A look at the α-helical radius, the radius that the backbone atoms follow in the individual helices, reveals something unusual. The radius increases before and within the Skip 2 Accommodation Region (Fig. 6A), which increases the length of the path that backbone atoms follow thus decreasing the winding frequency; one residue will wind the α-helix less at a larger radius. The average radius is 2.34 Å for residues 1,371 through 1,400 and 2.26 Å elsewhere. A 3.5% average increase over 29 residues increases the helical track by an amount equivalent to 1.02 amino acid residues, exactly what is needed to fit the extra residue and raise the Accommodation Index by one. The rise per residue of the coiled coil in this region is below average for the rest of the tail (Fig. 6C). Both features, reduced rise/residue and increased α-helix radius, are consistent with the skip residue being added with no change in length of the coiled coil.
Several factors could contribute to the unexpected changes in the Skip 2 Accommodation Region. The most obvious would be the presence of the nonmyosin protein myofilin which appears to bind the coiled coil thereby sandwiching Skip 2 and its Accommodation Region (Fig. 6B), including most of the region of increased α-helical radius and reduced added length. A possibility not visible in the segmented coiled coil is the presence of salt bridges between myosin tails.
MD Simulations.
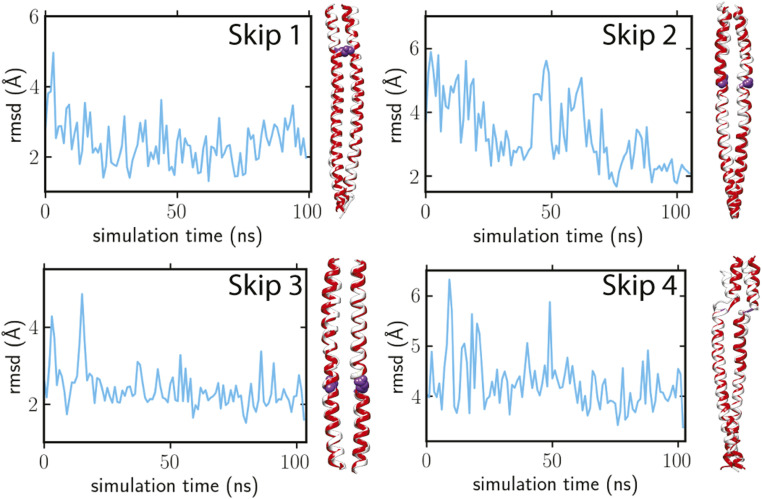
To determine if the skip region structures were a consequence of their sequences or their environment, we performed MD simulations starting from our cryo-EM atomic models for the four skip regions (SI Appendix, Methods). The simulations, each of which ran for 100 nanoseconds, showed that the structures remain steady for Skip 1, 3, and 4 or progress steadily from the in-filament arrangement toward the more conventional arrangement for Skip 2 (Movies S5–S8). The RMSD values of Skip 1 and 3 remained around 2 to 3 Å, demonstrating the intrinsic conformations of these regions are well preserved independent of the filament environment (Fig. 7).
Fig. 7.
MD simulations for the four skip regions. In each panel, the left shows the progress of the Cα RMSD between filament and crystal structures over time; the right shows the final conformation (white ribbon) compared to the crystal structure (red ribbon). The skip residue side chains are highlighted in purple spheres.
For Skip 2, the RMSD dropped to around 2 Å, during which the coiled coil untwisted from the initial conformation (Fig. 7), with ω0 approaching zero for residues 1,380 to 1,390 (SI Appendix, Fig. S8). The final structural snapshot of Skip 2 resembles Option 3 of Fig. 1, found in the crystal structure (Fig. 7). Thus, the filament Skip 2 structure seems to be imposed on it by its local environment.
The simulation for Skip 4, which started with one chain helical and the other chain unfolded, ended with both chains unfolded. Skip 4 residue G1815 unwound both helices, making the final simulated conformation close to the crystal structure (Option 2 of Fig. 1). The ω0 values near the skip residue became positive, whereas the α-helices on either side of the hinge remained in a uniform coiled-coil configuration (SI Appendix, Fig. S8). The relatively larger RMSD around 4 Å (Fig. 7) suggests conformational variability in the Accommodation Region of Skip 4, indicating a strong influence from the surrounding interactions with neighboring coiled coils and the extra density, which is possibly the myosin C terminus.
Discussion
Numerous atomic structures exist for the heads of myosin IIs from different species and in different catalytic states (27), but structures for the myosin tail have been limited to short segments (12, 13, 23). Here is a structure of a complete myosin tail in its native context at a resolution sufficient for building a de novo atomic model. Despite 545 to 490 million years of evolutionary divergence between vertebrates and invertebrates (28) and the different appearances of their thick filaments, comparison of the Lethocerus myosin tail structure with myosin tail segments from human cardiac muscle reveals a surprising degree of agreement. Their piecewise similarity suggests that when packed into a thick filament, the overall tail structure of vertebrate myosin is likely to be very similar. This further supports the proposal that curved molecular crystalline layers (ribbons) are a universal packing strategy for myosin filaments (8). Where they differ, which is primarily around two skip residues, suggests loci for changes in filament properties. These observations are consistent with filament structures from two divergent insect species, which showed nearly identical myosin tail arrangements within the ribbons despite significant differences in composition and structure (15, 29).
Asymmetry of Myosin II.
Both myosin II heavy chains are identical in sequence but in many contexts form surprisingly asymmetric structures. The most notable of these is the ATPase-inhibited interacting heads motif (26, 30). Myosin heads in a filament are in a fundamentally asymmetric environment, even if initially positioned symmetrically about the head–tail junction. Anticlockwise azimuthal movements of the blocked head toward the free head or the thin filament are unobstructed. Identical anticlockwise movements by the free head encounter the filament backbone. The reconstruction and atomic model provide evidence that head movements to achieve the interacting heads motif unwind and unfold the tail α-helices asymmetrically. In solution, the free and blocked head positions can be assumed by either polypeptide chain; once the tail is incorporated into the filament, which heavy chain becomes the free head or the blocked head is fixed. The evidence for this is seen in the agreement in the number of coiled-coil turns in the proximal S2 of filaments and crystal structures. Reversing the roles of free and blocked heads would require inserting (or removing) a half turn of the proximal S2 coiled coil, which if done randomly would reduce the proximal S2 resolution considerably. Although the asymmetric head–head interaction is best characterized, a number of others are observed here.
Many myosin II molecules have nonhelical C termini that vary in length (e.g., 39 residues in Lethocerus and Drosophila and 10 to 11 residues in vertebrate striated muscles) (SI Appendix, Fig. S9). In Lethocerus, the blocked-head C-terminal segment, which can be tracked from the end of the coiled coil to its last residue, is intercalated between ribbons within the filament backbone. The free-head nonhelical C terminus is not fully resolved, but a reasonable hypothesis is that the acidic residues at its end constitute the extra density at the blocked-head Skip 4. The KGGKK motif at Skip 4 is highly basic whereas the C terminus of Lethocerus, DLHEDMM, is acidic and may favor unfolding of the α-helix at Skip 4. If the assumption is correct, the heavy chain asymmetry at Skip 4 is not simply a consequence of insufficient C termini to affect both chains. Only one chain is freely accessible from the central core of the filament, the other is not easily reached. Thus, the two C termini, like the N termini, have nonsymmetrical roles.
Unusual Structure of the Proximal S2.
At the head–tail junction, we observed a number of changes and, for clarity, define some terminology. We use the term “unfolded” to indicate a change from α-helix to an extended polypeptide chain or a loop. A second structural change in which the α-helix is retained but with altered pitch we call α-helix “unwinding.” α-Helix unwinding is observed at all of the skip residues and near the head–tail junction. Although the pitch of the α-helix fluctuates along the entire tail, systematic α-helix overwinding is not observed. A third change, uncoiling of the coiled coil, has been described, which usually means a separation of the proximal S2 chains near the head–tail junction. At the resolution in which this has been reported, it is unclear whether the α-helix has been retained or not, but in many cases, it is clear that the chains have separated (e.g ref. 31). A fourth possibility, not generally described, is a change in the “twist” of the coiled coil. In the filament, we observe only α-helix unfolding leading to polypeptide chain separation and α-helix unwinding within the coiled coil. Although changes in the coiled-coil twist of S2 are possible, they seem to be small in filaments, because the cardiac and Lethocerus proximal S2 are in good agreement regarding coiled-coil pitch despite the fact that one is observed in a crystal with no heads attached and the other in the filament with both heads attached.
At the head–tail junction, 12 residues of the free-head α-helix are unfolded into an extended chain. Unfolding of the free-head heavy chain, in combination with the separation of the heavy chains caused by the RLC presence, prevents the coiled coil from forming until residue L860 rather than the head–tail junction defined as two residues past the invariant proline (32) between residues N849 and V850 in Lethocerus. At the blocked head–tail junction, it is unclear whether the α-helix has been maintained or has partially unfolded, the resolution being insufficient to distinguish between the two. After the free-head α-helix reforms at residue E856, it joins the blocked-head α-helix forming a coiled coil. Although technically the coiled coil has uncoiled, the more significant change is that the free-head α-helix has unfolded. The free head also binds the thick filament backbone displacing the proximal S2 azimuthally by 17° (15), producing the unusual orientation of the interacting heads motif in this muscle. Thus, uncoiling of the coiled coil in Lethocerus filaments encompasses both unwinding and unfolding of the α-helix.
One coarse-gained modeling study of the conformational change forming the interacting heads motif found that both heads underwent anticlockwise (unwinding) torsional rotations about their S2 α-helices of greater than 100° and were seen to alter the structure of the S2 (33). In that study, which did not incorporate a thick filament presence, alterations in the proximal S2 helices were about equal. Structural alterations propagated throughout the coiled coil to a degree that depended on the length of the coiled coil. In Lethocerus, we see evidence that both helices are unwound, but not equally, and the α-helix unwound the most is also unfolded for 12 residues. α-Helix unwinding is confined to the first 40 residues of the coiled coil for the free head and 20 residues for the blocked head (Fig. 5B), approximately the same number of residues that unwind following a skip residue. We have not found a quantifiable structural change indicating torsional effects propagate beyond the region of α-helix unwinding. Thus, the asymmetry in the interacting heads motif extends beyond the head–head interaction into the proximal coiled coil. Whether it extends much beyond the clearly observed effects is uncertain.
Antibodies used to separate the two helical strands at the head–tail junction found that the amount of separation could be interpreted as either α-helix unfolding or tail uncoiling (31). Crystal structures of the proximal S2 observed poor order for the first two heptads indicating coiled-coil instability at the head–tail junction (23, 34). Other studies have observed asymmetry along the coiled coil, ideally possessing C2 symmetry about the coiled-coil axis, but this asymmetry varied by isoform, being greater in scallop than in cardiac myosin (32). Motility assays show that replacement of the native sequence at the myosin head–tail junction with a leucine zipper reduces unitary displacement when compared with wild-type (35), indicating that coiled-coil instability is necessary for normal function. Loss of regulation in smooth muscle myosin, which requires a two-headed species, is also observed when a leucine zipper is substituted at the head–tail junction (36). The present work on Lethocerus thick filaments shows coiled-coil instability involves not just chain separation but α-helix unfolding.
The coiled coil in relaxed Lethocerus thick filaments starts at residue L860 and continues comparatively straight until just before entering the backbone at about residue E938. A pronounced 17° bend breaks the run of symmetry here and defines the termination of the proximal S2. Between the proximal S2 end and the C terminus, the coiled coil bends back and forth, in and out, generally facilitating close approach between neighboring tails within the ribbon, which are separated by multiples of 3 × 145 Å. Less close approach occurs between neighboring tails of different ribbons for which the separation is by 1, 2, 4, or 5 × 145 Å (15). Notably, the axis about which the coiled coil bends in filaments is aligned with the interchain axis of the coiled coil with little bending in the perpendicular direction suggesting anisotropic coiled-coil flexibility consistent with MD simulations of the dynein coiled coil (37). Coiled-coil bending further disrupts local symmetry between the two chains. This bending seems important for ribbon formation because the bends are conserved even in the crystal structures of myosin tail segments, for which the arrangement between molecules in the crystals is unrelated to their placement in the filament. Similarly, the coiled-coil protein tropomyosin, which has a superhelical structure in thin filaments, also has some superhelical structure in crystals (38).
Comparison with Human Cardiac Muscle Myosin II.
The crystal structures of the skip residue segments of the cardiac myosin tail behave as expected theoretically, showing α-helical unwinding and coiled-coil untwisting for Skips 1 through 3 and loop formation at Skip 4. The myosin structure in filaments only partially follows expectations. Differences in the amino acid sequence are a possible effect. However, the human cardiac and Lethocerus fight muscle sequences in the tail are 52% identical overall and 73% identical in the charged amino acids. Amino acid sequence similarities are even higher. Sequence identity varies along the myosin tail, being highest just before Skip 1 and higher than average at Skips 1, 2, 4, and the ACD. Identity is lower than average at Skip 3, where the proximal S2 enters the backbone, and after the end of the coiled coil (SI Appendix, Fig. S10).
Differences in structure might be attributable to packing interactions in crystals and filaments. Myosin tail segments in filaments are never in close proximity with the same segment of another tail as a consequence of the 3 × 145 Å myosin tail offset within ribbons, exactly opposite of what occurs in a crystal. However, the cardiac myosin tail segments outside of the skip regions agree well with the corresponding tail segment in filaments, which is contrary to expectations based only on the skip regions. Myosin II molecules in solution show greater flexibility near skip residues than within the intervening regions (21). In the filament backbone, solution flexibility in the skip regions is expressed as changes relative to the crystal structures. Assuming the crystal structures represent a ground state energetically, deviations in filaments likely arise from factors such as asymmetric packing interactions, nearest-neighbor salt bridges, the acidic, nonhelical, myosin C terminus, and nonmyosin proteins in the backbone, which in Lethocerus would be flightin or myofilin. Larger changes in the tail structure at the skip residues due to external factors are consistent with their greater flexibility.
Skip 1.
The Skip 1 segment of the myosin tail has a somewhat unusual structure with distinctive bends both before and after its Accommodation Region so that the segment of parallel α-helices is angled with respect to the filament axis. However, Skip 1 is the region that agreed best with the cardiac crystal structure which also showed one of these bends in the coiled coil. Skip 1’s location on the filament surface results in fewer opportunities for structure modifications influenced by neighboring myosin tails or nonmyosin proteins. Cardiac muscle has titin and mybp-c as potential modifiers of its structure in thick filaments, though there seems to be no evidence that the Skip 1 region is involved in their binding. Lethocerus thick filaments apparently have no proteins decorating the backbone surface. Moreover, sequence conservation in the Skip 1 region is above the average for the rest of the backbone (SI Appendix, Fig. S10).
Skip 2.
Skip 2 is unusual in myosin IIs, being found in striated muscle myosin II but not smooth muscle or nonmuscle myosin II (21). At a resolution of ∼7 Å at which individual α-helices can be resolved, the Skip 2 Accommodation Region is indistinguishable from a canonical coiled coil. However, the increase in Accommodation Index and the α-helical winding frequency measured at higher resolution show a one-residue insertion being accommodated through an increase in the α-helical radius (Fig. 6A), thereby increasing the helical track by the length of one residue without increasing the length of the coiled coil. The effect is further validated when the increase in coiled-coil length for each added residue is computed; the increase is lowest in the Skip 2 Accommodation region (Fig. 6C).
Skip 2’s effect on the cardiac coiled-coil crystal structure is distinctly different from Lethocerus. In the crystal structure, Skip 2 causes untwisting of the coiled coil similar to the effects of Skips 1 and 3 and in accord with prediction (Fig. 1G). In cultured cardiac myocytes, replacement of the cardiac Skip 2 region with the correspond sequence from smooth or nonmuscle myosin II, which lack the skip residue, caused no obvious phenotype change (12). Our result shows why. In situ, Skip 2 is accommodated in an unusual way to preserve the pitch of the coiled coil, which readily accommodates a region whose predicted structure is a canonical coiled coil. If this explanation is further substantiated, it makes the remarkable prediction that the Skip 2 region in vertebrate striated muscle thick filaments is similar to that of Lethocerus flight muscle. The nearest common ancestor, which is unknown if one exists, must have appeared at least 550 Mya, before the appearance of vertebrates (28).
Skip 2’s unusual structure in filaments is likely due to either nearest neighbor myosin tails or the nonmyosin protein myofilin. However, as argued below, in Lethocerus, the nonmyosin proteins may be stabilizing the structure rather than causing it.
Skip 3 and Skip 4.
Skips 3 and 4 follow this same pattern. Skip 3 agrees well with the crystal structure, the one difference apparently due to the nonmyosin protein flightin, which penetrates the ribbon near Skip 3 (Fig. 5D). Skip 4 differs significantly from the crystal structure in that only one chain is unfolded, the other chain remaining helical with an increased rise/residue.
Relevance for Vertebrate Muscle.
Historically, the myosin filament backbone has been regarded as a passive participant in muscle contraction, performing the role of a scaffold from which myosin heads extend to interact with the thin filament and which bears the resulting tension. That concept is evolving due to investigations into one of the longest known and least understood features of vertebrate muscle contraction, how the axial spacing change from 143 Å in relaxed muscle to 145 Å on activation disorders the arrangement of myosin heads on the thick filament (39–41). Rigor vertebrate muscle also has a 145 Å axial repeat (39) with all the heads bound to actin rather than each other consistent with coupling of filament length changes to the arrangement of myosin heads. More recent studies have refined this effect into the concept of the thick filament as a tension sensor (42, 43). Perhaps some speculation on a molecular explanation would be useful even given limited experimental observations on the topic. First, we explore the question of why invertebrates maintain a 145 Å spacing.
Most invertebrate thick filaments already have a 145 Å axial spacing in the relaxed state (e.g., refs. 5, 44–47) and others. Of thick filaments studied in detail, all invertebrate filaments contain the protein paramyosin, whereas no vertebrate thick filament has this protein. The paramyosin axial repeat is 725 Å, which is 5 × 145 Å (48), so its presence could influence the myosin tail axial spacing to match, if not exactly, at least close enough to provide a vernier for filament length specification. Paramyosin is likely involved in determining the length of the invertebrate thick filament (49, 50), a role performed by the protein titin in vertebrates (4). Thick filament length specification in Drosophila asynchronous flight muscle also involves the protein flightin (51) and may involve others such as myofilin. Flightin and myofilin have domains lining the inner wall of the myosin tail annulus which interact with myosin and could interact with paramyosin. Flightin and myofilin as well as the blocked-head myosin C terminus have unfolded domains intercalated between the ribbons making multiple contacts with the myosin tails (15, 29). An adjustment of only 0.02 Å in the average rise per residue distributed over the ∼90 residues in each 145 Å repeat would be sufficient to match the paramyosin repeat. A reasonable speculation is that flightin, myofilin, the myosin C-terminal domain, and paramyosin are involved in maintaining the 145 Å crown spacing in asynchronous flight muscle; their absence would provide an opportunity for the myosin tail to assume another axial spacing or revert to a possibly more preferred 143 Å repeat.
The 143 Å crown spacing is characterized by the so-called M3 reflection (the meridional third order of the 429 Å average repeat of the myosin head spacing) and its 71.5 Å second order (42), the M6, which is largely regarded as coming from the myosin tail arrangement in the backbone. The M6 intensity is strong in relaxed vertebrate striated muscle and decreases significantly on activation and tension generation. In Lethocerus, both the M3, 145 Å spacing, and M6, 72.5 Å, equivalent reflections are strong with the M3 being especially strong due to the perpendicular orientation of the interacting heads motif (5). The Lethocerus M6 intensity is enhanced by the nonmyosin proteins which are located axially between the myosin heads (15). Applied tension produces a tiny change in the M3 spacing, a 0.25° reduction in helical angle, a small change in the M3 intensity, and no reported change in the M6 intensity (5). The latter would imply the small length change was distributed uniformly along the 10 crown spacings comprising the tail. If the myosin tail arrangement of human cardiac and Lethocerus in relaxed muscle were very similar, which seems a reasonable possibility, a uniformly distributed change in the axial spacing of the backbone from 143 Å in cardiac to 145 Å would not change the M6 intensity. A more focused change not distributed uniformly might be more consistent with the M6 intensity change. It is perhaps noteworthy that a focused change in a single myosin tail would be spaced by 429 Å along the ribbons, which is the spacing between mybp-c in vertebrate striated muscle (52).
Two places in the myosin tail, Skips 2 and 4, suggest loci for a more focused increase in the axial period propagating outward rather than a uniform increase. The unique packing of myosin tails within the ribbons would affect the propagation of a focused length change. A change triggered on one region of a myosin tail can propagate both axially and laterally because of the extensive tail overlaps and close packing. In other words, a focused length change initiated in a single myosin tail can propagate axially along the ribbon as a consequence of propagating laterally across the ribbon.
A sequence alignment of 351 striated muscle myosin II sequences, 167 invertebrate and 184 vertebrate species, showed that all observed myosins had C-terminal extensions with highly acidic termini of average length 16 for vertebrates and 37 for invertebrates (SI Appendix, Fig. S9). It may be possible that the longer C-terminal length found in general for invertebrate myosin II is an adaption to the presence of paramyosin, as mentioned above. The KGGKK sequence motif at Skip 4 is also highly conserved among striated muscle myosin IIs. Smooth and nonmuscle myosin IIs lack both the KGGKK motif and acidic C terminus and have filaments built throughout by antiparallel tail packing (8, 53, 54), with one known exception (55): vertebrates switch from antiparallel packing in the bare zone to parallel tail packing in the A-band. It would seem reasonable that the KGGKK motif and the acidic C terminus are involved in parallel packing of myosin tails in the striated muscle A-band. Because the KGGKK motif can unfold, it could provide a focus for a tension-induced increase in axial spacing. However, Skip 4 is located inside the filament backbone making it difficult to see how a change here could affect the conformation of the myosin heads.
At Skip 2, the added amino acid residue is accommodated by an increase in α-helix radius and a reduction in rise/residue with no change in length of the myosin tail. Skip 2 would therefor seem also well poised for a focused length change. The increase in α-helix radius and reduced rise/residue comes with an apparent energy cost. Outside of the filament context, the Skip 2 region in the crystal structure and in MD simulations shown here follow Option 3 (Fig. 1G) and untwist into parallel helices (Fig. 7 and Movie S6). We hypothesize that the Skip 2 structure is maintained in flight muscle by the nonmyosin protein myofilin because it sandwiches the Skip 2 Accommodation Region. What is puzzling is why flight muscle has not made use of a structural change in Skip 2 as part of its activation. Perhaps for Lethocerus, the slight change in helical angle resulting from a stretch, which provides better alignment between myosin origins and targets on actin for myosin binding (5) and has been hypothesized to activate the blocked head (56), has greater importance than an axial spacing change.
Vertebrate striated muscle lacks the long pitch helical tracks present in Lethocerus and Drosophila thick filaments and instead has an axial period matching the 3 × 143 Å myosin tail offset in the ribbons. Skip 2 is also located close to the filament surface but could also impact segments of tails on the thick filament surface through lateral interactions. Structural changes in Skips 2 and 4 could through lateral interactions alter the structure of six of the 10 crown spacings in a myosin tail. If the others are unchanged, then the crown segments of the myosin tail would become heterogeneous possibly accounting for the loss in M6 intensity.
Arguing against a conformational change at Skip 2 in vertebrates is its replacement by a smooth muscle sequence lacking Skip 2, which produced no observable phenotype in cultured cardiomyocytes (12). Possibly, a phenotype too small to be observed in a cardiomyocyte could be amplified in both space and time to an observable phenotype in the live animal. Perhaps cultured myocytes are contracting without any stretch activation as occurs in heart muscle.
Materials and Methods
Thick filament suspensions from glycerinated L. indicus flight muscle were prepared and grids frozen as previously described (15). Images were recorded on a DE-64 direct electron detector operated in integration mode. Defocus ranged primarily between 1.3 and 3 μm (SI Appendix, Fig. S11). Filaments were picked manually (SI Appendix, Fig. S12). cisTEM was used for two-dimensional classification (SI Appendix, Fig. S13) and three-dimensional refinement (SI Appendix, Fig. S14) (57). The Fourier Shell Correlation resolution varied depending on the mask used on the half-maps (SI Appendix, Fig. S15A). MonoRes, a program for determining spatially variable resolution (58), suggests the resolution of the heads of ∼15 Å (SI Appendix, Fig. S15 B and C), regulatory light chain 5 to 6 Å, and the backbone uniformly at 4.2 Å (SI Appendix, Fig. S15C). Helical parameters calculated using relion_helix_toolbox (59) yielded a rise of 149.56 Å and a helical twist of 34.08°, higher than previously observed (15) and 0.33° higher than the 33.75° reported by X-ray fiber diffraction (5). The pixel size was thus reset from 1.009 to 0.978 to scale the helical rise to the known value of 145 Å (5). Sharpening was done using local Deblur (60) (SI Appendix, Fig. S15 E–G). Using relion_helix_toolbox, we extended the structure into a 2,0483 box sufficient to contain a full myosin molecule, which was subsequently segmented out by the Segger algorithm in chimera (61). The complete myosin tail ∼1600 Å long was used to build the atomic model in a single piece. Larger side chains were well resolved and used as guides while making the model, in later corrections and refinements (SI Appendix, Fig. S15G), and to differentiate alternatively spliced regions in the tail (SI Appendix, Fig. S16) (62). Constraints were unnecessary to build the atomic model de novo in COOT (63) and to refine the model in Phenix (64). For a structure at our resolution, the PDB validation statistics are very good (SI Appendix, Fig. S7A). Molecular dynamics simulations were performed on all four Skip regions using the graphics processing unit-accelerated version of Amber18 using the ff14SB force field (65, 66). Further details are given in SI Appendix.
Supplementary Material
Acknowledgments
This research was funded by grants from the National Institute of General Medical Science of the NIH including R01 GM030598 to K.A.T., R01-GM031749 to JAM, P41 GM103622 to T.C.I., National Institute of Arthritis, Musculoskeletal and Skin Diseases Grant R01 AR014317 to R.J.E., and an American Heart Association Predoctoral Fellowship 20PRE35120273 to H.R. The Titan Krios was partially funded by NIH Grant S10 RR25081 to K.A.T. The DE-64 was purchased using funds from NIH Grant U24 GM116788 to the Southeastern Consortium for Microscopy of MacroMolecular Machines. This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. We thank Prof. Donald L. D. Caspar for his encouragement, insights, and comments and Drs. Belinda Bullard and Ivan Rayment for comments on an earlier version. We dedicate this paper to the late Prof. Michael K. Reedy who inspired our work on Lethocerus flight muscle and provided inspiration throughout the 38 years of our collaboration, and to the recently deceased Prof. John Squire, whose insight into the myosin tail arrangement in thick filaments was four decades ahead of its time but ultimately proved correct.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
* Equivalent residues in human cardiac β-myosin II are obtained by subtracting 8 from the Lethocerus residue number (i.e., 1188, 1385, 1582, and 1807).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2024151118/-/DCSupplemental.
Data Availability
Atomic model, segmented density map, and raw image data have been deposited in PDB, EMDB, and EMPIAR (PDB entry ID 7KOG (67) EMDB entry ID EMD-22975 (68). The raw data link is through the EMDB entry ID EMD-22975 (69).
References
- 1.Sheterline P., Clayton J., Sparrow J., Actin (Oxford University Press, U.S.A., 1995), pp. 1–103. [Google Scholar]
- 2.Galkin V. E., VanLoock M. S., Orlova A., Egelman E. H., A new internal mode in F-actin helps explain the remarkable evolutionary conservation of actin’s sequence and structure. Curr. Biol. 12, 570–575 (2002). [DOI] [PubMed] [Google Scholar]
- 3.Al-Khayat H. A., Kensler R. W., Squire J. M., Marston S. B., Morris E. P., Atomic model of the human cardiac muscle myosin filament. Proc. Natl. Acad. Sci. U.S.A. 110, 318–323 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Whiting A., Wardale J., Trinick J., Does titin regulate the length of muscle thick filaments? J. Mol. Biol. 205, 263–268 (1989). [DOI] [PubMed] [Google Scholar]
- 5.Perz-Edwards R. J., et al., X-ray diffraction evidence for myosin-troponin connections and tropomyosin movement during stretch activation of insect flight muscle. Proc. Natl. Acad. Sci. U.S.A. 108, 120–125 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vibert P., Craig R., Electron microscopy and image analysis of myosin filaments from scallop striated muscle. J. Mol. Biol. 165, 303–320 (1983). [DOI] [PubMed] [Google Scholar]
- 7.Levine R. J., Elfvin M., Dewey M. M., Walcott B., Paramyosin in invertebrate muscles. II. Content in relation to structure and function. J. Cell Biol. 71, 273–279 (1976). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Squire J. M., General model of myosin filament structure. 3. Molecular packing arrangements in myosin filaments. J. Mol. Biol. 77, 291–323 (1973). [DOI] [PubMed] [Google Scholar]
- 9.Harrison R. G., Lowey S., Cohen C., Assembly of myosin. J. Mol. Biol. 59, 531–535 (1971). [DOI] [PubMed] [Google Scholar]
- 10.Lowey S., Goldstein L., Cohen C., Luck S. M., Proteolytic degradation of myosin and the meromyosins by a water-insoluble polyanionic derivative of trypsin: Properties of a helical subunit isolated from heavy meromyosin. J. Mol. Biol. 23, 287–304 (1967). [DOI] [PubMed] [Google Scholar]
- 11.Szent-Gyorgyi A. G., Cohen C., Philpott D. E., Light meromyosin fraction I: A helical molecule from myosin. J. Mol. Biol. 2, 133–142 (1960). [Google Scholar]
- 12.Taylor K. C., et al., Skip residues modulate the structural properties of the myosin rod and guide thick filament assembly. Proc. Natl. Acad. Sci. U.S.A. 112, E3806–E3815 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Korkmaz E. N., et al., A composite approach towards a complete model of the myosin rod. Proteins 84, 172–189 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McLachlan A. D., Karn J., Periodic charge distributions in the myosin rod amino acid sequence match cross-bridge spacings in muscle. Nature 299, 226–231 (1982). [DOI] [PubMed] [Google Scholar]
- 15.Hu Z., Taylor D. W., Reedy M. K., Edwards R. J., Taylor K. A., Structure of myosin filaments from relaxed Lethocerus flight muscle by cryo-EM at 6 Å resolution. Sci. Adv. 2, e1600058 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crick F. H. C., The packing of α-helices: Simple coiled coils. Acta Crystallogr. 6, 689–697 (1953). [Google Scholar]
- 17.Astbury W. T., On the structure of biological fibres and the problem of muscle. Proc. R. Soc. Lond. B Biol. Sci. 134, 303–328 (1947). [DOI] [PubMed] [Google Scholar]
- 18.Cohen C., Holmes K. C., X-ray diffraction evidence for alpha-helical coiled-coils in native muscle. J. Mol. Biol. 6, 423–432 (1963). [DOI] [PubMed] [Google Scholar]
- 19.Sodek J., Hodges R. S., Smillie L. B., Jurasek L., Amino-acid sequence of rabbit skeletal tropomyosin and its coiled-coil structure. Proc. Natl. Acad. Sci. U.S.A. 69, 3800–3804 (1972). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lupas A., Coiled coils: New structures and new functions. Trends Biochem. Sci. 21, 375–382 (1996). [PubMed] [Google Scholar]
- 21.Offer G., Skip residues correlate with bends in the myosin tail. J. Mol. Biol. 216, 213–218 (1990). [DOI] [PubMed] [Google Scholar]
- 22.Woodhead J. L., et al., Atomic model of a myosin filament in the relaxed state. Nature 436, 1195–1199 (2005). [DOI] [PubMed] [Google Scholar]
- 23.Blankenfeldt W., Thomä N. H., Wray J. S., Gautel M., Schlichting I., Crystal structures of human cardiac beta-myosin II S2-Delta provide insight into the functional role of the S2 subfragment. Proc. Natl. Acad. Sci. U.S.A. 103, 17713–17717 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmidt N. W., Grigoryan G., DeGrado W. F., The accommodation index measures the perturbation associated with insertions and deletions in coiled-coils: Application to understand signaling in histidine kinases. Protein Sci. 26, 414–435 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grigoryan G., Degrado W. F., Probing designability via a generalized model of helical bundle geometry. J. Mol. Biol. 405, 1079–1100 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wendt T., Taylor D., Trybus K. M., Taylor K., Three-dimensional image reconstruction of dephosphorylated smooth muscle heavy meromyosin reveals asymmetry in the interaction between myosin heads and placement of subfragment 2. Proc. Natl. Acad. Sci. U.S.A. 98, 4361–4366 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holmes K. C., Geeves M. A., The structural basis of muscle contraction. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355, 419–431 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shu D. G., et al., Lower Cambrian vertebrates from south China. Nature 402, 42–46 (1999). [Google Scholar]
- 29.Daneshparvar N., et al., CryoEM structure of Drosophila flight muscle thick filaments at 7 Å resolution. Life Sci. Alliance 3, e202000823 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burgess S. A., et al., Structures of smooth muscle myosin and heavy meromyosin in the folded, shutdown state. J. Mol. Biol. 372, 1165–1178 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Knight P. J., Dynamic behaviour of the head-tail junction of myosin. J. Mol. Biol. 255, 269–274 (1996). [DOI] [PubMed] [Google Scholar]
- 32.Brown J. H., et al., An unstable head-rod junction may promote folding into the compact off-state conformation of regulated myosins. J. Mol. Biol. 375, 1434–1443 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tama F., Feig M., Liu J., Brooks C. L. III, Taylor K. A., The requirement for mechanical coupling between head and S2 domains in smooth muscle myosin ATPase regulation and its implications for dimeric motor function. J. Mol. Biol. 345, 837–854 (2005). [DOI] [PubMed] [Google Scholar]
- 34.Li Y., et al., Visualization of an unstable coiled coil from the scallop myosin rod. Nature 424, 341–345 (2003). [DOI] [PubMed] [Google Scholar]
- 35.Lauzon A. M., Fagnant P. M., Warshaw D. M., Trybus K. M., Coiled-coil unwinding at the smooth muscle myosin head-rod junction is required for optimal mechanical performance. Biophys. J. 80, 1900–1904 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trybus K. M., Freyzon Y., Faust L. Z., Sweeney H. L., Spare the rod, spoil the regulation: Necessity for a myosin rod. Proc. Natl. Acad. Sci. U.S.A. 94, 48–52 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lippert L. G., et al., Angular measurements of the dynein ring reveal a stepping mechanism dependent on a flexible stalk. Proc. Natl. Acad. Sci. U.S.A. 114, E4564–E4573 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stewart M., Crystalline sheets of tropomyosin. J. Mol. Biol. 174, 231–238 (1984). [DOI] [PubMed] [Google Scholar]
- 39.Haselgrove J. C., X-ray evidence for conformational changes in the myosin filaments of vertebrate striated muscle. J. Mol. Biol. 92, 113–143 (1975). [DOI] [PubMed] [Google Scholar]
- 40.Huxley H. E., Stewart A., Sosa H., Irving T., X-ray diffraction measurements of the extensibility of actin and myosin filaments in contracting muscle. Biophys. J. 67, 2411–2421 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wakabayashi K., et al., X-ray diffraction evidence for the extensibility of actin and myosin filaments during muscle contraction. Biophys. J. 67, 2422–2435 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Linari M., et al., Force generation by skeletal muscle is controlled by mechanosensing in myosin filaments. Nature 528, 276–279 (2015). [DOI] [PubMed] [Google Scholar]
- 43.Irving M., Regulation of contraction by the thick filaments in Skeletal muscle. Biophys. J. 113, 2579–2594 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Millman B. M., Bennett P. M., Bennett P. M., Structure of the cross-striated adductor muscle of the scallop. J. Mol. Biol. 103, 439–467 (1976). [DOI] [PubMed] [Google Scholar]
- 45.Wray J. S., Structure of the backbone in myosin filaments of muscle. Nature 277, 37–40 (1979). [DOI] [PubMed] [Google Scholar]
- 46.Irving T. C., Maughan D. W., In vivo x-ray diffraction of indirect flight muscle from Drosophila melanogaster. Biophys. J. 78, 2511–2515 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Padrón R., et al., The myosin interacting-heads motif present in live tarantula muscle explains tetanic and posttetanic phosphorylation mechanisms. Proc. Natl. Acad. Sci. U.S.A. 117, 11865–11874 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bear R. S., Selby C. C., The structure of paramyosin fibrils according to x-ray diffraction. J. Biophys. Biochem. Cytol. 2, 55–69 (1956). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bullard B., Luke B., Winkelman L., The paramyosin of insect flight muscle. J. Mol. Biol. 75, 359–367 (1973). [DOI] [PubMed] [Google Scholar]
- 50.Mackenzie J. M. Jr., Epstein H. F., Paramyosin is necessary for determination of nematode thick filament length in vivo. Cell 22, 747–755 (1980). [DOI] [PubMed] [Google Scholar]
- 51.Reedy M. C., Bullard B., Vigoreaux J. O., Flightin is essential for thick filament assembly and sarcomere stability in Drosophila flight muscles. J. Cell Biol. 151, 1483–1500 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Craig R., Offer G., The location of C-protein in rabbit skeletal muscle. Proc. R. Soc. Lond. B Biol. Sci. 192, 451–461 (1976). [DOI] [PubMed] [Google Scholar]
- 53.Craig R., Megerman J., Assembly of smooth muscle myosin into side-polar filaments. J. Cell Biol. 75, 990–996 (1977). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu J. Q., Harder B. A., Uman P., Craig R., Myosin filament structure in vertebrate smooth muscle. J. Cell Biol. 134, 53–66 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sulbarán G., et al., An invertebrate smooth muscle with striated muscle myosin filaments. Proc. Natl. Acad. Sci. U.S.A. 112, E5660–E5668 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hu Z., Taylor D. W., Edwards R. J., Taylor K. A., Coupling between myosin head conformation and the thick filament backbone structure. J. Struct. Biol. 200, 334–342 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Grant T., Rohou A., Grigorieff N., cisTEM, user-friendly software for single-particle image processing. eLife 7, e35383 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vilas J. L., et al., MonoRes: Automatic and accurate estimation of local resolution for electron microscopy maps. Structure 26, 337–344.e4 (2018). [DOI] [PubMed] [Google Scholar]
- 59.He S., Scheres S. H. W., Helical reconstruction in RELION. J. Struct. Biol. 198, 163–176 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ramírez-Aportela E., et al., Automatic local resolution-based sharpening of cryo-EM maps. Bioinformatics 36, 765–772 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pintilie G. D., Zhang J., Goddard T. D., Chiu W., Gossard D. C., Quantitative analysis of cryo-EM density map segmentation by watershed and scale-space filtering, and fitting of structures by alignment to regions. J. Struct. Biol. 170, 427–438 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fee L., Lin W., Qiu F., Edwards R. J., Myosin II sequences for Lethocerus indicus. J. Muscle Res. Cell Motil. 38, 193–200 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 64.Adams P. D., et al., PHENIX: A comprehensive python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Salomon-Ferrer R., Götz A. W., Poole D., Le Grand S., Walker R. C., Routine microsecond molecular dynamics simulations with AMBER on GPUs. 2. Explicit solvent particle mesh ewald. J. Chem. Theory Comput. 9, 3878–3888 (2013). [DOI] [PubMed] [Google Scholar]
- 66.Maier J. A., et al., ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 11, 3696–3713 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rahmani H., Hu Z., Daneshparvar N., Taylor D., Taylor K. A.. Lethocerus Myosin II complete coiled-coil domain resolved in its native environment. Protein Data Bank. 10.2210/pdb7KOG/pdb. Deposited 9 November 2020. [DOI]
- 68.Rahmani H., Hu Z., Daneshparvar N., Taylor D., Taylor K. A.. Lethocerus Myosin II complete coiled-coil domain resolved in its native environment. Protein Data Bank. https://www.ebi.ac.uk/pdbe/entry/emdb/EMD-22975. Deposited 9 November 2020.
- 69.Rahmani H., Hu Z., Daneshparvar N., Taylor D., Taylor K. A.. Lethocerus Myosin II complete coiled-coil domain resolved in its native environment. Protein Data Bank. https://www.emdataresource.org/EMD-22975. Deposited 9 November 2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic model, segmented density map, and raw image data have been deposited in PDB, EMDB, and EMPIAR (PDB entry ID 7KOG (67) EMDB entry ID EMD-22975 (68). The raw data link is through the EMDB entry ID EMD-22975 (69).