Significance
Direct imaging and counting single molecules captured on magnetic microparticles enables diagnostic assays with ultrahigh sensitivities and wide linear detection ranges. The method offers simple assay protocols, short incubation times, minimal reagent consumption, and does not require enzymatic signal amplification. The ratiometric readout, linking the signal and binding area, eliminates the need for strict control over the number of microparticles. Direct imaging provides the sensitivity, flexibility, and simplicity required by rigorous clinical standards. Other benefits of the technique include a seamless transition between digital and analog data analysis modes, scalability, basic consumables, and the reliability to achieve robust implementation in high-throughput automated instruments. HDIA is a versatile tool that can easily be adopted for all receptor–ligand binding assays.
Keywords: single-molecule imaging, immunoassays, microparticles, fluorescence, diagnostics
Abstract
Every year, over 100 million units of donated blood undergo mandatory screening for HIV, hepatitis B, hepatitis C, and syphilis worldwide. Often, donated blood is also screened for human T cell leukemia–lymphoma virus, Chagas, dengue, Babesia, cytomegalovirus, malaria, and other infections. Several billion diagnostic tests are performed annually around the world to measure more than 400 biomarkers for cardiac, cancer, infectious, and other diseases. Considering such volumes, every improvement in assay performance and/or throughput has a major impact. Here, we show that medically relevant assay sensitivities and specificities can be fundamentally improved by direct single-molecule imaging using regular epifluorescence microscopes. In current microparticle-based assays, an ensemble of bound signal-generating molecules is measured as a whole. By contrast, we acquire intensity profiles to identify and then count individual fluorescent complexes bound to targets on antibody-coated microparticles. This increases the signal-to-noise ratio and provides better discrimination over nonspecific effects. It brings the detection sensitivity down to the attomolar (10−18 M) for model assay systems and to the low femtomolar (10−16 M) for measuring analyte in human plasma. Transitioning from counting single-molecule peaks to averaging pixel intensities at higher analyte concentrations enables a continuous linear response from 10−18 to 10−5 M. Additionally, our assays are insensitive to microparticle number and volume variations during the binding reaction, eliminating the main source of uncertainties in standard assays. Altogether, these features allow for increased assay sensitivity, wide linear detection ranges, shorter incubation times, simpler assay protocols, and minimal reagent consumption.
Billions of diagnostic and blood screening assays are performed each year, providing information critical to people’s health across the globe. Given the purpose and the volumes involved, every improvement in assay performance becomes immensely impactful. Such improvements include increasing assay sensitivity and specificity; eliminating sample predilutions by widening the detection range; shortening incubation times, thus increasing the throughput; simplifying assay protocols; and minimizing reagent consumption.
The concept of immunoassays, as a novel methodology for quantitative biological measurements, was introduced in the early 1960s (1–4). Widespread acceptance and commercialization of the technology for clinical diagnostics was drastically accelerated by the development of highly automated testing platforms in the 1980s (5–8). These instruments, reagents, and assay protocols have been further optimized, networked, and integrated into multi-instrument systems over past decades (9, 10).
However, the fundamentals of immunoassays or, in general terms, the basic principles of receptor–ligand binding interactions remain unchanged in today’s applications. The target analyte must be captured with one binding agent and be recognized by, or outcompete, another binding agent containing a label. The binding kinetics, affinities, and specificities are driven by the structure-function properties of the reagents. Hence, searching for better binding molecules is an ongoing task. Antibodies are natural robust binders, and a variety of advances have been made in the engineering of new and improved antibody constructs (11–14), but further improvement along these lines mostly requires screening increasingly larger molecular libraries, a trend that quickly reaches physical limits of the binding reaction or becomes impractical.
An alternative perspective toward better assay performance focuses on improving the measurement output after the binding reactions have taken place. Major approaches include enhancing detection sensitivity and finding new ways to reduce background signal. Before addressing these approaches, it is important to define the appropriate metrics. In terms of assay performance, functional assay sensitivity refers to the lowest concentration of analyte that can be identified and correlated with the measured signal in a statistically significant manner (15). This, of course, depends on assay background, which is the collection of all measured signals that do not scale with analyte concentration. Assay background is the main factor that limits assay sensitivity.
Signal detection, as distinct from functional assay sensitivity, refers strictly to the amplitude of the observable signal, without interpretation thereof. Clearly, better signal detection permits the identification of lower levels of a given analyte. However, enhancing the signal does not always improve assay sensitivity. In the case of high nonspecific background—when the signal-generating label interacts with reaction components other than the target analyte—much of the detected signal is not related to the assay target, and so an increased signal amplitude does not provide new information. On the other hand, stronger signal detection allows for additional options to improve signal-to-noise. For instance, nonspecific background can be greatly reduced by decreasing the concentration of labeled conjugate.
In the pursuit of enhancing signal detection, single-molecule counting is the definitive goal. A variety of single-molecule detection techniques have been developed to obtain detailed mechanistic information about biophysical reactions and to identify the properties of subpopulations, otherwise averaged out in ensemble measurements (16–22). From a diagnostics perspective, if each signal-generating molecule brought to the detector can be registered, then it can be counted (23–26), leading to the ultimate signal detection.
The Noji group was the first to demonstrate that confining enzymes in isolated, femtoliter substrate volumes allows a single enzyme molecule to quickly generate a fluorophore concentration detectable by standard microscopy (27–29). Extending the use of confined enzyme reactions, Walt and collaborators developed the single-molecule enzyme-linked immunosorbent assay, termed digital ELISA (30–32). This technique requires incubating samples with antibody-coated microparticles and complementary antibody–enzyme conjugates, loading microparticles into femtoliter-volume well arrays, sealing wells with membranes/oil, substrate incubation, multiple image acquisitions to threshold the enzymatic rates, and, finally, counting the positive wells (30, 33). In more recent work, the well arrays were replaced with droplets and nondiffusible labels (34, 35). Using enzymatic signal amplification represents a typical biochemical approach to achieve ultrasensitive detection. However, digital ELISA suffers inherent limitations at high sample concentrations because all the wells become positive and the substrate becomes exhausted, eliminating digital information and eventually saturating the intensity signal. Also, the quantitation of small molecule analytes usually requires a competitive assay format, which complicates the design and application of digital ELISA (36).
Hence, returning to the design philosophy of the original single-molecule methods, we are offering a straightforward biophysical approach for directly imaging the target molecules on surfaces or microparticles. Employing bright fluorescent conjugates helps to overcome the signal-to-background issue and allows individual analyte point sources to be observed. This type of single-molecule imaging-based detection provides a much simpler assay protocol (SI Appendix, Fig. S1) and eliminates the need for complex disposables. Replacing enzymatic signal enhancement with bright conjugates does require the use of somewhat higher quality optics and cameras than are needed for digital ELISA but does not exceed the capabilities of conventional epifluorescence microscopy. Confocal or total internal reflection fluorescence setups, as used in some other single-molecule methods (37–40), are not required. Another advantage of the direct imaging approach is that assays can be performed faster with reduced, or even zero, washing steps because the background and ambiguous peaks, or clusters, of pixels can be filtered out by image analysis algorithms. Furthermore, when employing microparticles, the spatial information is used to generate a ratiometric signal that is insensitive to microparticle number and volume variations during the binding reaction, eliminating the main source of uncertainties in standard ensemble assays. Lastly, while both the biochemical and biophysical approaches can detect single molecules at low analyte concentrations, direct biophysical methods transition seamlessly from the digital regime into analog intensity measurements at high analyte concentrations. This grants a very broad detection range that can be used to eliminate sample predilutions and the associated retesting steps.
Overall, both the biochemical and the biophysical approaches seek to gain an advantage through improved detection. Nevertheless, both approaches are built on the basic immunoassay reaction, and all antibody reactions are limited by the physical constraints of binding affinities and reaction kinetics. Therefore, it is important to note that all immunoassays merely sample the target population. Across the various capture, labeling, and detection steps, only a small fraction of the original analyte is enumerated. Single-molecule counting techniques applied to immunoassays are thus not absolute, and each assay must reference a calibration curve (i.e., a set of assay signal responses as a function of known sample concentrations). Single-molecule techniques can offer important gains in sensitivity and background discrimination, which, in turn, can be leveraged to enhance functional sensitivities of the assays as well as to reduce incubation times, reagents, and sample volumes.
In this work, we introduce an imaging approach that detects analytes with bright, dextran scaffold fluorescent complexes and incorporates digital and analog analysis modes. We demonstrate that this method, high-definition immunoassay (HDIA), enables single-molecule counting sensitivity in combination with a broad linear detection range. Lastly, we show the practical benefits of HDIA for detecting clinical biomarkers in human plasma.
Results
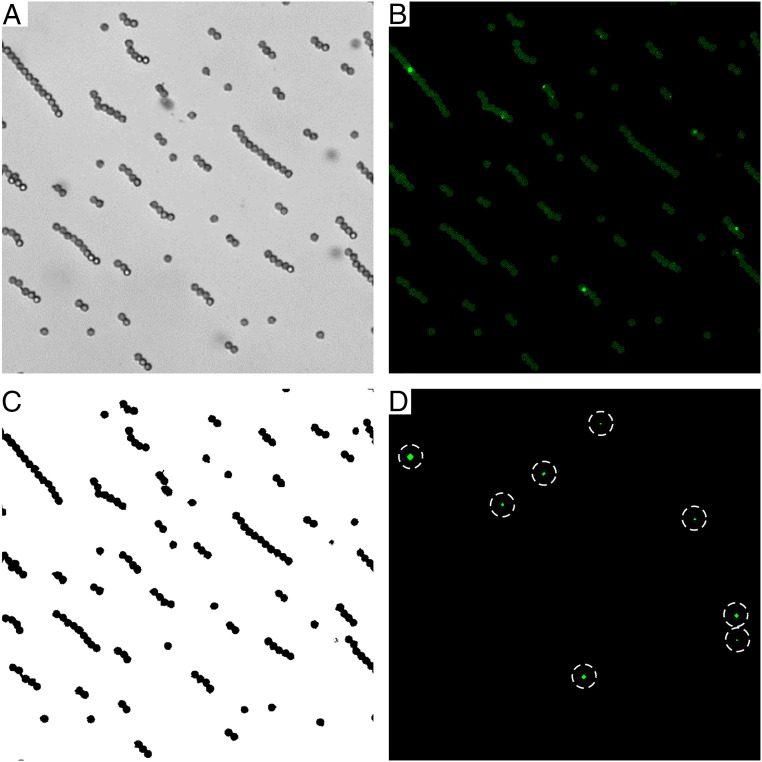
HDIA is a versatile tool that can easily be adopted for all receptor–ligand binding and screening assays. HDIA directly images antibody–analyte sandwiches. These complexes may be bound to an optical binding surface or to microparticles that have settled, or been pulled magnetically, onto a transparent support. Using magnetic microparticles as the binding solid phase simplifies manipulation, washing, and automation, which enables high-throughput diagnostics. To perform an HDIA measurement, the potential binding area must either be known a priori or, as in the case of microparticle binding surfaces, be determined from a bright-field image (Fig. 1A). The identified surface areas (microparticle pixel areas) are used to generate a binding surface mask (Fig. 1C). This mask, in turn, is applied to the fluorescence image (Fig. 1B) so that any fluorescence background not coming from a binding surface (microparticle) is excluded. We determine the average fluorescent intensity per pixel across each microparticle image and use it to calculate the median signal for all images of a given sample. Averaging over hundreds of thousands of pixels makes this a robust parameter which correlates with analyte concentration.
Fig. 1.
HDIA image acquisition and analysis. At each measurement location, two images are acquired, (A) a bright-field image to locate the microparticles and (B) a fluorescence image to record the signal. (C) A mask is made from the bright-field image to eliminate any signals not coming from a binding area/microparticle. (D) When in the digital regime, a further threshold can be applied to identify and count the bright intensity peaks (dotted circles), which indicate single antibody sandwiches.
Forming bright fluorescent antibody complexes makes it possible to resolve single analyte molecules. Therefore, at very low sample concentrations, a digital signal can be obtained. A background threshold is applied to the fluorescent image to remove microparticle autofluorescence and thereby identify the high intensity peaks of individual, fluorescently labeled antibody sandwiches (Fig. 1D). Creating this binary detection scenario—areas with peaks {1} versus areas without peaks {0}—simplifies the noise filtering, which leads to improved signal-to-noise and thus greater detection sensitivity. Moreover, HDIA processes all the images using both of the following approaches: counting peaks and totaling fluorescence intensities. These overlapping and complementary analysis regimes offer a seamless transition and a significantly expanded dynamic range.
In digital mode, the peaks per area (PPA) signal is measured as the ratio of the number of intensity peaks divided by the total imaged pixel area of the microparticle binding surfaces. An analyte peak is defined as a set of contiguous pixels for which each pixel is included on the microparticle mask and has an intensity value above the threshold. The PPA is expressed in units of peaks/kilopixel to make the numerical values more legible; thus
Digital analysis offers improved signal-to-noise at very low analyte concentrations, but the signal begins to saturate at higher concentrations as the peaks merge together and can no longer be resolved. At this point, the results are determined by analog analysis (i.e., total fluorescence intensity per binding area) and are reported in units of counts per pixel (CPP). At sufficiently high sample concentrations, the fluorescence intensity can saturate the camera, but a reduction in excitation power or shortened image acquisition (as used here) easily overcomes this obstacle. The overall HDIA measurement then consists of the following three components: digital, long acquisition analog, and short acquisition analog. These three analyses have overlapping regions of equivalent, valid results, making it possible to convert the signals into a single universal scale or composite signal.
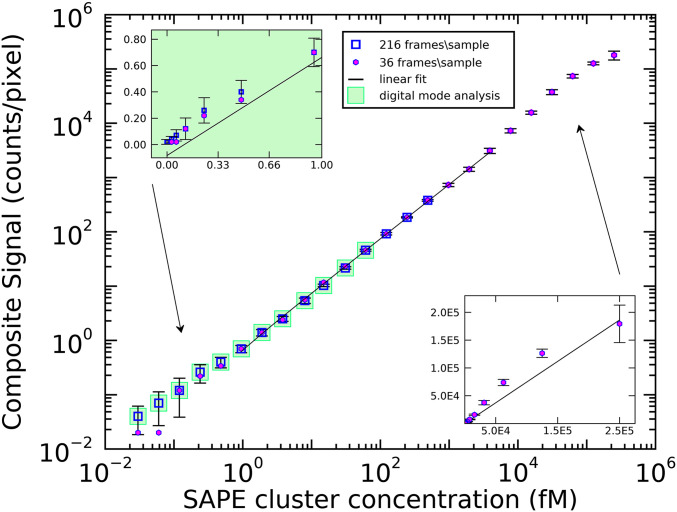
HDIA performance was characterized using a simplified model system (Fig. 2). Biotinylated magnetic microparticles were employed to directly capture fluorescent targets. In this experiment, we used very bright streptavidin phycoerythrin (SAPE) multimers (∼1 SA:12 PE). As described in Materials and Methods, a series of 24 SAPE multimer samples (0.25 nM to 30 aM) were reacted with microparticles in a glass-bottom 96-well plate and then imaged. A bright-field image (100 ms) and two fluorescence images (10 and 100 ms) were acquired at nine positions within each well. The additional 10 ms image was taken to avoid camera saturation for high concentration samples and therefore was only analyzed in analog mode. Both digital and analog modes were used to analyze the 100 ms fluorescent images. Fig. 2 shows the results of two experiments performed using the same stock solutions. In the first experiment (magenta circles), covering the full sample titration range, 36 images (4 wells) were measured for each of the 24 SAPE multimer dilutions (0.25 nM to 30 aM). In the second experiment replicating low concentration samples (blue squares), 216 images (24 wells) were acquired for each of the lowest 15 SAPE multimer concentrations and also for a buffer-only control. The full titration data (magenta circles) were converted from digital PPA values to analog CPP at the 61 fM concentration point where both modes are valid. Performing a similar rescaling from 10 ms acquisition analog values to 100 ms acquisition values at concentration 7.8 pM allows the composite signal for the entire sample range—from 30 aM to 0.25 nM—to be plotted on a single graph. The main log–log plot of Fig. 2 displays a linear fit overlaying the central region of the data. The insets (linear axes) show closeups of the low and high concentration data compared with the same fit line from the main graph. Note that at 36 frames per sample (magenta circles), the lowest two concentrations appear to bottom out. This is because, at very low analyte concentrations, only a small total number of peaks are detected (∼20) and larger sampling is required to obtain meaningful statistics. At the highest analyte concentration, 0.25 nM, the analog signal begins to saturate, approaching the limit of available binding sites on the microparticles. Nevertheless, HDIA offers a dynamic range, spanning ∼6 orders of magnitude, depending on the assay.
Fig. 2.
Model assay results. SAPE multimers are titrated onto biotinylated microparticles. Two sets of measurements were acquired and plotted on a log–log scale, one with low image density (36 frames/sample, magenta circles) and another with high image density (216 frames/sample, blue squares). The results show a good dose–response covering ∼6 orders of magnitude. A linear fit is plotted over the central section of the data, while the linear-scale inserts show the same fit plotted against the low concentration (Left Inset) and high concentration data (Right Inset). Error bars: mean ± SD.
When characterizing the sensitivity of HDIA, it is natural to assume that at very low concentrations of analyte, an intensity peak indicates only one analyte–antibody complex, not two or more clustered together. Similarly, at slightly higher concentrations, neighboring but noncontiguous peaks are treated as separate analyte complexes rather than a thresholding artifact. This should also be a valid assumption given that, with a resolution of 0.5 μm/pixel, even a large macromolecule (3 MDa ∼20 nm) would still be considered a point source. To validate these assumptions and to show that HDIA has single-molecule sensitivity, we examine the performance of HDIA compared with Poisson statistics. Poisson statistics describe the probability of a given number of discrete events, k, occurring within an interval,
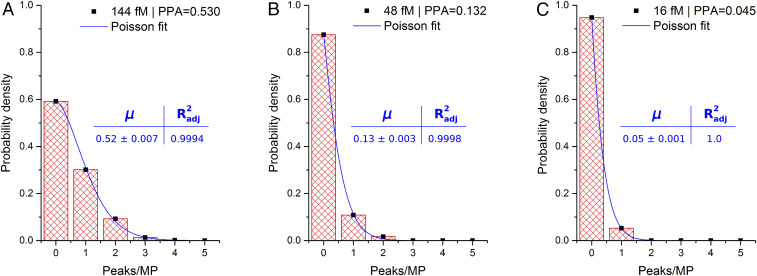
where μ is the expected event rate. Therefore, we calculate the HDIA digital signal as the number of peaks per microparticle by modifying the algorithm with a size-exclusion image filter to eliminate any objects larger than a single microparticle. Then, each individual microparticle area is sequentially mapped to the fluorescence image so that each of the detected peaks can be referenced to the microparticle it occupies. Fig. 3 shows data from three concentrations (144, 48, and 16 fM) of the SAPE multimers shown in Fig. 2. After having been reanalyzed with the modified algorithm, these data were plotted as probability density functions (PDFs) of the number of peaks per microparticle. The PDFs show an excellent fit to the Poisson model, and the Poisson fit event rates are in good agreement with the full image data PPAs (magnetic microparticle chains included). Interestingly, additional data changing the threshold instead of the concentration (SI Appendix, Fig. S2) reveal that HDIA results begin undershooting the Poisson fit at around five peaks/microparticle, indicating that the present HDIA setup can resolve up to four targets per microparticle before overlapping peaks require the switch to analog analysis mode. Overall, the alignment with Poisson statistics is strong evidence that HDIA is accurately detecting and counting single analyte molecules.
Fig. 3.
Poisson distributions of peaks per microparticle in model assay. The readout of HDIA is always ratiometric (signal/area), but defining an area equivalent to a single microparticle allows the data to be fit to Poisson distributions. The histograms show detected peaks per microparticle for three SAPE concentrations: (A) 144 fM, (B) 48 fM, and (C) 16 fM. The fits show excellent agreement, signifying discrete signals (i.e., single antibody sandwich detection). n = 216.
The true test of a diagnostic technology is its ability to process biological samples and generate clinically relevant results despite the chief obstacle of immunoassays: nonspecific interactions. Finding a bright conjugate that functions well in assay conditions without exacerbating the nonspecific signal was a challenge. Several types of macromolecules were investigated, including SAPE multimers, nanospheres, quantum dots, polymers, and DNA dendrimers, but none exhibited both a universally efficient dose–response and a low nonspecific background across multiple assays. Only one class of macromolecular constructs containing dextran, bovine serum albumin (BSA), biotin and antibody, in conjunction with SAPE, has shown good performance across all tested assays. Dextran-BSA-biotin (DBB) is a scaffold molecule, contains multiple biotin tags, and can also be decorated with multiple Fab antibody fragments. This creates a multivalent conjugate that can have super bright fluorescence after binding multiple SAPEs. The DBB-Fabs used in this study are megadalton macromolecules prepared by first cross-linking dextran (molecular weight 150,000 Da) with multiple BSAs and then activating with succinimidyl 4-(N-maleimidomethyl)cyclohexane-1-carboxylate linker at 10× molar excess. Next, anti-HIV recombinant Fab or anti-thyroid stimulating hormone (TSH) recombinant Fab was reacted with dextran-BSA at a 1:1 molar ratio (to the BSA). After purification, N-hydroxysuccinimidobiotin was reacted with dextran-BSA-Fab at 10× molar excess. The resultant molecules are difficult to characterize as they have a relatively broad size distribution (1 × 106 to 3 × 106 Da), and the overlapping absorbance spectra of BSA and Fab prevent full characterization by spectrophotometry. Combining spectrophotometric and fluorescent correlation spectroscopy (FCS) data suggests that there are roughly 12 to 16 BSA molecules per dextran and a similar number of Fab fragments. Reacting 1:1 SAPE (∼1 SA per PE) secondary conjugate with DBB-Fab creates a further distribution of labels. The experimentally determined, minimal peak-over-background discrimination level was obtained using a series of SAPE multimers with known PE/multimer ratios. The results suggested that five or more 1:1 SAPE labels bound to a single DBB make it detectable. In the digital mode, a random distribution of DBB brightnesses may lead to some intensity spikes falling below the threshold and being ignored. However, this cannot create a systematic bias. It might also appear that a DBB brightness distribution could be an additional source of variation in the analog mode, but after summing intensities across all the microparticles within a frame and then selecting the median signal across all the frames/sample, any such fluctuations will have been averaged out.
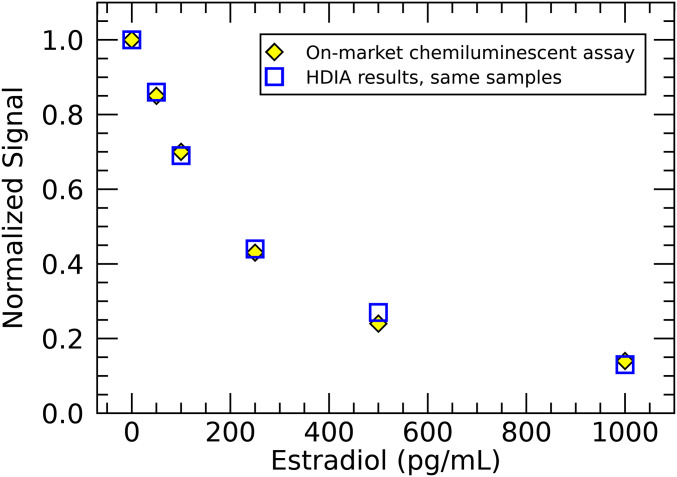
To assess the performance of the HDIA technique in genuine diagnostic assays, three Abbott assays were chosen. First, we present a competitive estradiol immunoassay to demonstrate the ease of implementing HDIA in a competitive format. Estradiol, the most common estrogen, is an important target for monitoring ovarian function. As a small hormone molecule (272 Da), it is better suited to a competitive assay in which the estradiol in patient sample competes with labeled synthetic estradiol for binding sites on an anti-estradiol antibody-coated microparticle. By simply exchanging the chemiluminescently labeled estradiol tracer with biotinylated estradiol and adding a secondary 1:1 SAPE conjugate (SI Appendix, Fig. S3), we converted a commercial diagnostic Abbott kit for use in estradiol-HDIA. This competitive HDIA estradiol assay was compared with the on-market chemiluminescence-based assay by measuring a standard set of estradiol calibrators (0, 50, 100, 250, 500, and 1,000 pg/mL). Fig. 4 shows that the results of the two assays are identical. Clearly, the estradiol-HDIA exhibits equivalent diagnostic sensitivity and spans the full clinically relevant range. It is important to underscore that this conversion was completed with minimal reagent modification and without laborious optimizations.
Fig. 4.
Competitive estradiol HDIA. The dose response of a competitive format HDIA estradiol assay is compared with that of Abbott’s on-market chemiluminescence-based test by measuring six calibrator samples. The normalized results are equivalent.
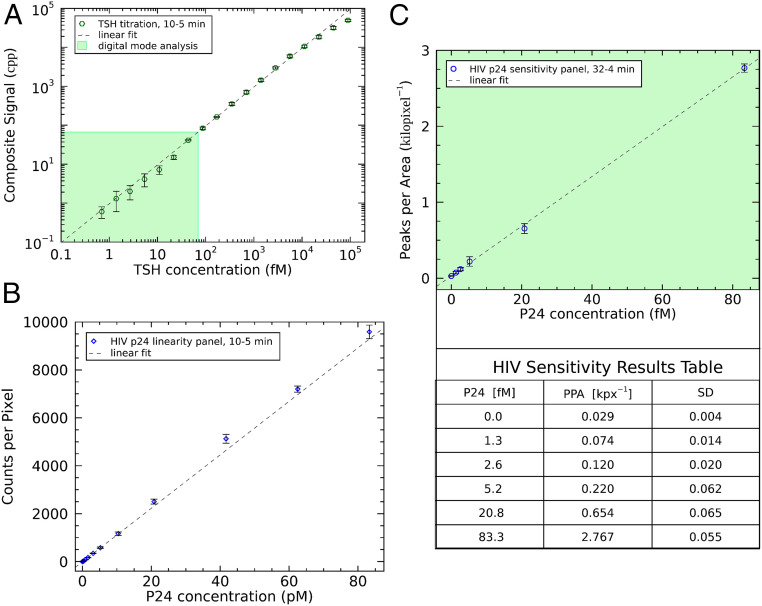
To expand the benefits of HDIA detection, two additional standard Abbott immunoassays were modified to use DBB-Fab conjugates and the following 1:1 SAPE fluorescent labels: a TSH assay and a HIV p24 antigen assay. The TSH assay demonstrates the utility of a broad dynamic range because both low and high levels of TSH—as well as the degree of change from normal—are clinically relevant. Therefore, a diagnostic TSH assay (SI Appendix, Fig. S4) must be able to quantify an extremely broad range of TSH concentrations. Twofold serial dilutions of a high concentration, recombinant TSH protein stock were made using a buffer matrix. Normal serum cannot be used because it contains endogenous TSH. We have shown that HDIA detection in rigorously TSH-depleted serum achieves equivalent background levels to the buffer matrix (SI Appendix, Table S1). In this experiment, we measured 18 TSH samples with concentrations ranging from 0.7 fM to 89 pM. Due to the broad range, a composite signal was constructed, as in the model assay (Fig. 2), and plotted on a log–log scale as a function of the starting TSH sample concentrations (Fig. 5A). By comparing the data with the linear fit (dotted line), the highest TSH concentration signals are observed to flatten due to the saturation of available binding sites. The lowest TSH concentration data point, 0.7 fM, has a signal-to-background ratio of ∼2, which indicates the assay sensitivity limit. This assay spans five orders of magnitude of TSH concentrations. In its sensitivity, range, and linearity, TSH-HDIA significantly exceeds the performance of all current commercial TSH diagnostic assays.
Fig. 5.
HDIA in diagnostic applications. (A) High-concentration recombinant TSH samples were diluted in buffer matrix and incubated for 10 min with microparticles, 5 min with DBB-Fab, and 5 min with 1:1 SAPE. The results are plotted on a log–log scale, accompanied by a linear fit (n = 3). (B) An HDIA HIV assay (10 min with microparticles and DBB; 5 min with SAPE) was performed using a high concentration HIV sample and subsequent dilutions in negative human plasma. The data were analyzed in HDIA analog mode and plotted against a linear fit (n = 4). (C) A longer assay format (32 min with microparticles and DBB; 4 min with SAPE) was used to probe HDIA sensitivity in human plasma. The digitally analyzed results are plotted with a linear fit, and the associated data table shows that the assay is sensitive down to ∼1 fM (n = 3). Error bars: mean ± SD.
Two HIV p24 assay protocols (SI Appendix, Fig. S5) were used to show the linearity and sensitivity of HDIA. To demonstrate assay linearity (Fig. 5B), human-sourced HIV-1 viral lysate was serially diluted into pooled viral-negative human plasma (0.2 to 83 pM). At these relatively high concentrations, only the analog CPPs were recorded. These CPPs are plotted in Fig. 5B as a function of p24 concentration (pM) along with a linear fit to the data. The results show an excellent linear trend that would be expected to continue up into higher picomolar concentrations before saturating the available binding sites. Unfortunately, such high concentration HIV samples were not available.
A second HIV p24 experiment (Fig. 5C) was focused on determining the sensitivity of the assay using plasma samples with femtomolar p24 concentrations (1.3 to 83 fM and a background control). The assay was performed in triplicate, and the primary incubation was 32 min. The longer reaction time increases the amount of analyte captured by microparticles. At these low concentrations, the digital analysis approach was used. In Fig. 5C, the PPA is plotted against p24 concentration (fM) along with a linear fit. The data follow a linear dose–response down to the lowest concentration measured, 1.3 fM, which has a signal-to-background ratio of 2.5 (Fig. 5C, table). Note that there is sufficient signal amplitude to extend the assay into attomolar p24 concentrations if better ways to reduce nonspecific background could be found. The simplest path would be to further decrease the detection conjugate concentration, but this results in much longer incubation times and quickly becomes impractical. Single-molecule counting can provide highly sensitive detection, but the fundamental constraints of the antibody–antigen interaction and nonspecific binding set the ultimate boundaries.
Discussion
Digital detection has gained some buzzword status in the diagnostics field, but there is nothing miraculous about digital analysis. For example, whether counting targets or averaging intensity, the analysis is performed on the same fluorescent images. The information content is exactly the same. The advantage of digital analysis is a few-fold improvement in signal-to-noise because in a binary system consisting only of “ones” and “zeroes,” differentiating signal from noise is much simpler than in contiguous distributions. Genuine binary distributions are rarely found in biological spheres such as diagnostics. Therefore, digital detection methods are constructed to generate data that approximate a binary system such that the discretization error is minimized. If this assumption is valid, then the conversion of the data to a digital model suppresses noise, as would the conversion of a noisy sinusoidal signal to a square pulse. However, it is unlikely to improve the signal-to-noise by more than an order of magnitude, and the less noisy the original analog signal, the less there is to be gained by a digital approach.
In a true binary diagnostics experiment, the dose–response a priori would be absolutely linear, but in practice, this doesn’t happen. Therefore, diagnostic assays always employ a calibration curve, constructed from known samples, so that a monotonically—or even a piecewise monotonically—increasing or decreasing dose–response can be accurately correlated with analyte concentration. Because of the complexities of the multistep assay process (probabilistic capture, detection, and labeling), as well as other losses due to surface adhesion and washing, neither an analog nor digital approach can return absolute analyte numbers. A calibration curve must always be used to translate the measured signal into an analyte concentration.
The remarkable single-molecule sensitivity of HDIA depends on the ability to detect and count single intensity peaks, which on the surface, sounds trivial. Microparticles provide an enormous advantage in terms of capture capacity, ease of washing, and automation. However, most polymer microparticles exhibit autofluorescence so that the binding surface itself has an inherent fluorescent background. To date, we have not found a completely nonfluorescent magnetic microparticle that also maintains good population homogeneity and robust surface coating properties. Therefore, the fluorescent label must be bright enough to be resolved from the autofluorescent background. A single fluorophore dye does not meet this criterion, nor even a very bright fluorescent protein with multiple chromophores such as phycoerythrin. The solution then is a macromolecular conjugate that contains antibodies, or antibody fragments, as well as a number of bright fluorophores. DBB-Fab is, thus far, the only conjugate family which exhibits robust dose–responses and minimal nonspecific binding across a range of diagnostic assays.
Theoretically speaking, since HDIA can detect single molecules, its potential detection ability approaches infinity. Of course, that ignores the issue of bringing said molecules within the detector’s purview and also discounts the background. The next level of pragmatism in comparing fact to theory is to consider a clean system, as described in Fig. 2, involving merely the capture and signal-generating constructs in a simple buffer. The minimum sensitivity is then limited by signal-to-background. From the Fig. 2 data, a typical control sample (nine images) was observed to contain ∼2 peaks. As the nine images cover only 12% of the available 96-well surface area, this should be multiplied by 8 to get a total of 16 peaks of background in a 100 µL sample. Requiring a signal-to-noise ratio of 3, we calculate the minimum clean system detection sensitivity to be 48 molecules/100 µL or 800 zeptomolar. This is an impressive potential detection limit, but even in the dramatically simplified model system, the detection limit was 30 aM (Fig. 2), 40-times higher than the calculated limit. First and foremost, this is a consequence of binding efficiencies. We never capture all of the available analyte in a given sample. In fact, between binding kinetics, capture efficiency, conjugate binding efficiency, and detection efficiency, less than 5% of the available analyte contributes to the final results. In summary, physical chemistry, the practical constraints of sample handling, and the difficulty of collecting scarce target molecules cannot be eliminated by a detection method, be it ever so sensitive and flexible. HDIA and other single-molecule detection approaches offer improvements but stand upon the fundamental strengths and weaknesses of immunoassays.
HDIA offers simplicity, fewer steps, and greater flexibility than digital ELISA (SI Appendix, Fig. S1). Dynamic range is critical in many diagnostic assays, and HDIA’s advantage here deserves reiteration, as digital ELISA has inherent constraints limiting measurements at high concentrations. HDIA can also be straightforwardly implemented in a competitive format, used to quantify small molecules such as steroid hormones, thyroxines, and various drugs. Moreover, HDIA uses many of the same reagents as standard immunoassays, eliminating much of the accompanying development and optimization work, as would be necessary for competitive digital ELISA (36). Nevertheless, for both single-molecule techniques, the chief weakness—and most continuous fight in designing assays—remains the presence of nonspecific binding events, which stems from assay conjugate/reagent interactions and clouds the detection signal. Because nonspecific interactions strongly depend on the specific reagents involved, they vary from assay to assay. Therefore, thorough experimental tinkering remains an essential part of the art of assay development.
HDIA can trade against the advantages of single-molecule sensitivity to enhance other aspects of assay performance. For example, the simplest way of confronting nonspecific binding is to reduce the concentration of labeled conjugate. However, this also slows the reaction kinetics. Similarly, increased washing and detergents reduce background but usually decrease genuine signal. A more sensitive detection technique provides an edge in low signal situations, which can be used to the optimize signal-to-noise, shorten reaction times, or minimize sample volumes, as fewer targets arriving at the detector will still produce meaningful results.
In summary, HDIA combines the advantages of magnetic microparticle-based immunoassays with direct single-molecule counting. It covers a wide dynamic range of concentrations while simultaneously achieving single-molecule sensitivity. The flexibility of the imaging approach simplifies background discrimination and multiplexing. All this can be accomplished with simple assay protocols and reduced sample and reagent volumes. These advantages, combined with a ratiometric readout, make HDIA an excellent technology for improved signal detection and implementation in the highly automated, high-throughput world of clinical immunoassay diagnostics.
Materials and Methods
Microscope Instrumentation.
An inverted IX83 microscope (Olympus) was used to measure samples with bright-field illumination from a pE 100 white light-emitting diode (LED) (CoolLED) and excitation light produced by an X-Cite Xylis LED illumination system (Excelitas) in conjunction with a Cy3 Olympus filter cube set (Edmund Optics). The detection setup employs a 20× air objective (UPlanSApo, numerical aperture = 0.75, Olympus) and a PCO.panda camera (PCO). Microparticles in 96-well optical-bottom plates (Thermo Fisher Nunc) were pulled down by 1 to 2 s of resting on neodymium magnets (DynaMag-96 bottom, Thermo Fisher). MetaMorph software (Molecular Devices) coordinated imaging measurements, directing a ProScan III xy-stage (Prior Scientific Instruments) to acquire one bright-field image (100 ms, 1.3% LED power) and one (or two) fluorescence image(s) (10 and 100 ms, 25% LED power) at nine positions within each 96-well plate. Olympus ZeroDrift IX3-ZDC2 maintained focus throughout the measurement. Image processing algorithms—written in Metamorph, Matlab, and R—removed very large or very bright aggregates, determined total pixel area of the microparticles, and evaluated total microparticle fluorescence intensity and/or numbers of peaks above a threshold. The ratio of the number of PPA of interest as well as the intensity CPP area were recorded for each image, and the median PPA and median total intensity values from the nine positions were reported for each sample. Median values are used because it is difficult to remove all fluorescent artifacts during image processing without compromising genuine data; the median therefore returns much more robust results than the mean. When seeking to measure very low analyte concentrations, the median is replaced with a modified robust mean. For high concentration samples, these return virtually identical results, but at very low concentrations, the median and a standard robust mean will almost always return a value of zero since most images contain no peaks. Under these circumstances, when the median equals zero, the modified robust mean has been programmed to reintroduce rejected images which contained either one or two peaks and then recalculate the mean.
Conjugate Characterization.
SAPE and DBB macromolecules were characterized by FCS. Six SAPE multimers from Agilent—PJ39S, PJRS27, PJRS25, PJRS34, PJRS20, and PJRS301—were diluted 50-fold in a commercial zwitterionic sulfonic acid buffer system (HBS-EP, GE Healthcare). Then, 50 µL of each SAPE multimer along with 50 µL of monomeric phycoerythrin were loaded into a 384-microwell plate and measured on an FCS instrument at 780 nm, 2.5 mW laser power after the objective and 50 kHz sampling frequency, acquiring 10 million data points per sample (SI Appendix, Fig. S6). The data were subdivided into 5,000-point segments (0.1 s) analyzed using G(1) analysis code written in IDL software (Harris Geospatial). This results in brightness histograms (2,000 occurrences) for each sample, showing the distribution of brightness species within in SAPE multimer sample (SI Appendix, Fig. S7).
To characterize the synthesis of dextran-BSA, dextran-BSA was labeled with Alexa-488, purified with a Zeba column (Thermo Fisher), and measured on a Cary 4000 spectrophotometer (Agilent) to determine BSA concentration (BSA-ε280 = 43,824 cm−1 ⋅ M−1, Dextran-ε280 = 0). DB-AF488 was further diluted and the number of molecules determined by FCS, yielding a ratio with the previously determined BSA concentration to be an average of ∼15 BSA molecules per dextran scaffold.
Model Assay.
Magnetic microparticles, 4.7 µm diameter, as described below, were biotinylated to allow direct conjugation with SAPE multimers (∼12 PE per multimer, PJS34, Agilent). SAPE multimer stock concentration was determined by a combination of absorbance and FCS measurements. SAPE multimers were diluted to 250 pM in Architect wash buffer (phosphate buffer, Abbott Laboratories). The same 250 pM stock of SAPE multimers was used to generate 1) 24 samples—1 mL, twofold dilutions from 250 pM to 0.03 fM; and 2) 16 samples—10 mL, twofold dilutions from 488 fM to 30 aM as well as a zero control. Each sample (100 µL) was placed in an optical-bottom 96-well plate (Nunc, Thermo Fisher) along with microparticles (5 µL, 0.1% solids) and incubated, shaking at 37 °C for 60 min. After incubation, all samples were washed on a microplate washer (Biotek) and measured on the fluorescence microscope.
TSH Assay.
Magnetic microparticles, 4.7 µm diameter (PL6604-0090AB, Agilent), were coated with anti-TSH antibody (Abbott) for antigen capture. DBBs labeled with anti-TSH Fab (Abbott) were used as the primary conjugate, with 1:1 PE:SA labels as the secondary conjugate. The 100 µL samples from twofold serial dilutions of TSH calibrator (89 pM, Abbott, recombinant TSH in Tris buffer) diluted in Architect Calibrator 1 buffer matrix (Abbott, Tris solution) and 2.5 µL microparticles (0.1% solids) were pipetted into a 96-well plate along with a buffer matrix only control. Following a 10 min incubation, there was a 1 min wash in Abbott blocking buffer, a second 5 min conjugate binding step with 50 µL DBB-Fab in blocking buffer, a detergent wash step, a final 5 min binding step with 50 µL 12 nM SAPE conjugate, and 3 × 100 µL washes in detergent buffer (Architect wash buffer, Abbott). The sample plate was processed on a KingFisher magnetic microparticle processor maintained at 37 °C. The completed assay microparticles were transferred to an optical-bottom 96-well plate and imaged.
HIV p24 Assay.
Magnetic microparticles, 4.7 µm diameter (PL6604-0090AB, Agilent), were coated with anti-p24 antibody (Abbott) for antigen capture. DBBs labeled with anti-p24 Fab (Abbott) were used as the primary conjugate, with SAPE (∼1:1 PE:SA, PJ39S, Agilent) binding to the DBB-Fab as the secondary conjugate/signal-generating molecule. Human-sourced, purified HIV-1 viral lysate with high HIV p24 levels was serially diluted into pooled, recalcified viral-negative human plasma as described in the HIV Ab/Ag Combo assay package insert (http://www.corelaboratory.abbott/us/en/offerings/segments/infectious-disease). The p24 levels were determined using the Abbott HIV Ab/Ag Combo assay, generating a concentration panel ranging from 83 pM to 200 fM, along with a plasma-only background control. Using a protocol similar to the TSH assay above, 100 µL p24 sample, 5 µL microparticles (0.05% solids), and 50 µL DBB-Fab diluted in a blocking buffer (Abbott) were incubated at 37 °C for 10 (or 32) min, washed 5 (or 4) min with SAPE, washed three more times, and imaged.
Supplementary Material
Acknowledgments
We would like to convey our everlasting gratitude to all our Abbott colleagues without whom this work could not have been completed. Mark Pope, Brenda Calfin, Felicia Bogdan, and Zhen Lin prepared dextran-based conjugates. Quinn Best synthesized biotinylated estradiol. We express our appreciation to Xiaoxing Qiu and Clinton Huizenga for advice on optimizing HIV assays. Special thanks go to Frank Holas for the sheep monoclonal antibody-coated TSH microparticles and the high concentration TSH calibrator. He and George Manderino shared their valuable TSH assay experience. We are also very thankful to Christine Rudolph and Steve Dahlen for providing HIV and TSH sample panels. Many reagents described in Materials and Methods were taken from assay kits manufactured by our Operations organization.
Footnotes
Competing interest statement: All authors are employed by Abbott Laboratories, which funded the research.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2025033118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Miles L. E. M., Hales C. N., Labelled antibodies and immunological assay systems. Nature 219, 186–189 (1968). [DOI] [PubMed] [Google Scholar]
- 2.Yalow R. S., Berson S. A., Assay of plasma insulin in human subjects by immunological methods. Nature 184 (suppl. 21), 1648–1649 (1959). [DOI] [PubMed] [Google Scholar]
- 3.Yalow R. S., Berson S. A., Immunoassay of endogenous plasma insulin in man. J. Clin. Invest. 39, 1157–1175 (1960). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wide L., Porath J., Radioimmunoassay of proteins with the use of Sephadex-coupled antibodies. Biochim. Biophys. Acta, Gen. Subj. 130, 257–260 (1966). [Google Scholar]
- 5.Lequin R. M., Enzyme immunoassay (EIA)/enzyme-linked immunosorbent assay (ELISA). Clin. Chem. 51, 2415–2418 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Alexander T. S., Human immunodeficiency virus diagnostic testing: 30 years of evolution. Clin. Vaccine Immunol. 23, 249–253 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rao M., Kapila K., Gupta R. M., Enzyme linked immunosorbent assays revisited. Med. J. Armed Forces India 53, 45–51 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andreotti P. E., et al., Immunoassay of infectious agents. Biotechniques 35, 850–859 (2003). [DOI] [PubMed] [Google Scholar]
- 9.Wheeler M. J., Automated immunoassay analysers. Ann. Clin. Biochem. 38, 217–229 (2001). [DOI] [PubMed] [Google Scholar]
- 10.Boyd J. C., Hawker C. D., “Automation in the clinical laboratory” in Tietz Textbook of Clinical Chemistry and Molecular Diagnostics, Burtis C. A., Ashwood E. R., Bruns D. E., Eds. (Elsevier Saunders, St. Louis, MO, 2012), pp. 478–483. [Google Scholar]
- 11.Tiller K. E., Tessier P. M., Advances in antibody design. Annu. Rev. Biomed. Eng. 17, 191–216 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baran D., et al., Principles for computational design of binding antibodies. Proc. Natl. Acad. Sci. U.S.A. 114, 10900–10905 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Birch J. R., Racher A. J., Antibody production. Adv. Drug Deliv. Rev. 58, 671–685 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Liu H. F., Ma J., Winter C., Bayer R., Recovery and purification process development for monoclonal antibody production. MAbs 2, 480–499 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Armbruster D. A., Pry T., Limit of blank, limit of detection and limit of quantitation. Clin. Biochem. Rev. 29 (suppl. 1), S49–S52 (2008). [PMC free article] [PubMed] [Google Scholar]
- 16.Hirschfeld T., Optical microscopic observation of single small molecules. Appl. Opt. 15, 2965–2966 (1976). [DOI] [PubMed] [Google Scholar]
- 17.Moerner W. E., Kador L., Optical detection and spectroscopy of single molecules in a solid. Phys. Rev. Lett. 62, 2535–2538 (1989). [DOI] [PubMed] [Google Scholar]
- 18.Orrit M., Bernard J., Single pentacene molecules detected by fluorescence excitation in a p-terphenyl crystal. Phys. Rev. Lett. 65, 2716–2719 (1990). [DOI] [PubMed] [Google Scholar]
- 19.Betzig E., Chichester R. J., Single molecules observed by near-field scanning optical microscopy. Science 262, 1422–1425 (1993). [DOI] [PubMed] [Google Scholar]
- 20.Funatsu T., Harada Y., Tokunaga M., Saito K., Yanagida T., Imaging of single fluorescent molecules and individual ATP turnovers by single myosin molecules in aqueous solution. Nature 374, 555–559 (1995). [DOI] [PubMed] [Google Scholar]
- 21.Ha T., et al., Probing the interaction between two single molecules: Fluorescence resonance energy transfer between a single donor and a single acceptor. Proc. Natl. Acad. Sci. U.S.A. 93, 6264–6268 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Noji H., Yasuda R., Yoshida M., Kinosita K. Jr, Direct observation of the rotation of F1-ATPase. Nature 386, 299–302 (1997). [DOI] [PubMed] [Google Scholar]
- 23.Skinner J. P., Tetin S. Y., Rapid single-molecule imaging in cyclic olefin copolymer channels. Microsc. Res. Tech. 78, 309–316 (2015). [DOI] [PubMed] [Google Scholar]
- 24.Smith L., Kohli M., Smith A. M., Expanding the dynamic range of fluorescence assays through single-molecule counting and intensity calibration. J. Am. Chem. Soc. 140, 13904–13912 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Macdonald P. J., Ruan Q., Tetin S. Y., Direct single-molecule counting for immunoassay applications. Anal. Biochem. 566, 139–145 (2019). [DOI] [PubMed] [Google Scholar]
- 26.Mickert M. J., et al., Measurement of sub-femtomolar concentrations of prostate-specific antigen through single-molecule counting with an upconversion-linked immunosorbent assay. Anal. Chem. 91, 9435–9441 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Rondelez Y., et al., Microfabricated arrays of femtoliter chambers allow single molecule enzymology. Nat. Biotechnol. 23, 361–365 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Sakakihara S., Araki S., Iino R., Noji H., A single-molecule enzymatic assay in a directly accessible femtoliter droplet array. Lab Chip 10, 3355–3362 (2010). [DOI] [PubMed] [Google Scholar]
- 29.Kim S. H., et al., Large-scale femtoliter droplet array for digital counting of single biomolecules. Lab Chip 12, 4986–4991 (2012). [DOI] [PubMed] [Google Scholar]
- 30.Rissin D. M., et al., Single-molecule enzyme-linked immunosorbent assay detects serum proteins at subfemtomolar concentrations. Nat. Biotechnol. 28, 595–599 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rissin D. M., Walt D. R., Digital readout of target binding with attomole detection limits via enzyme amplification in femtoliter arrays. J. Am. Chem. Soc. 128, 6286–6287 (2006). [DOI] [PubMed] [Google Scholar]
- 32.Walt D. R., Optical methods for single molecule detection and analysis. Anal. Chem. 85, 1258–1263 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilson D. H., et al., The simoa hd-1 analyzer: A novel fully automated digital immunoassay analyzer with single-molecule sensitivity and multiplexing. J. Lab. Autom. 21, 533–547 (2016). [DOI] [PubMed] [Google Scholar]
- 34.Cohen L., et al., Single molecule protein detection with attomolar sensitivity using droplet digital enzyme-linked immunosorbent assay. ACS Nano 14, 9491–9501 (2020). [DOI] [PubMed] [Google Scholar]
- 35.Wu C., Garden P. M., Walt D. R., Ultrasensitive detection of attomolar protein concentrations by dropcast single molecule assays. J. Am. Chem. Soc. 142, 12314–12323 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang X., Cohen L., Wang J., Walt D. R., Competitive immunoassays for the detection of small molecules using single molecule arrays. J. Am. Chem. Soc. 140, 18132–18139 (2018). [DOI] [PubMed] [Google Scholar]
- 37.Jain A., et al., Probing cellular protein complexes using single-molecule pull-down. Nature 473, 484–488 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jain A., Liu R., Xiang Y. K., Ha T., Single-molecule pull-down for studying protein interactions. Nat. Protoc. 7, 445–452 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Todd J., et al., Ultrasensitive flow-based immunoassays using single-molecule counting. Clin. Chem. 53, 1990–1995 (2007). [DOI] [PubMed] [Google Scholar]
- 40.Wu A. H., Fukushima N., Puskas R., Todd J., Goix P., Development and preliminary clinical validation of a high sensitivity assay for cardiac troponin using a capillary flow (single molecule) fluorescence detector. Clin. Chem. 52, 2157–2159 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.