Significance
In Cys-loop receptors, the affinity for neurotransmitter and the conductive/nonconductive state of the transmembrane pore depend tightly on each other. It has been previously hypothesized that wild-type side-chain–side-chain interactions straddling the interface between the extracellular and transmembrane modules are crucial for this conformational coupling. Here, we challenged this notion, and to the extent that our mutagenesis succeeded in eliminating all native interactions between side chains located across the interface, we conclude that such interactions are not required for function. Instead, function in these receptor channels seems to depend on more general properties of the extracellular–transmembrane-domain interface such as the shapes and relative positioning of the two apposed sides.
Keywords: alpha-7 nicotinic acetylcholine receptor, glutamate-gated chloride channel, ion-channel gating, chimeric constructs, pentameric ligand-gated ion channels
Abstract
One of the most fundamental questions in the field of Cys-loop receptors (pentameric ligand-gated ion channels, pLGICs) is how the affinity for neurotransmitters and the conductive/nonconductive state of the transmembrane pore are correlated despite the ∼60-Å distance between the corresponding domains. Proposed mechanisms differ, but they all converge into the idea that interactions between wild-type side chains across the extracellular–transmembrane-domain (ECD–TMD) interface are crucial for this phenomenon. Indeed, the successful design of fully functional chimeras that combine intact ECD and TMD modules from different wild-type pLGICs has commonly been ascribed to the residual conservation of sequence that exists at the level of the interfacial loops even between evolutionarily distant parent channels. Here, using mutagenesis, patch-clamp electrophysiology, and radiolabeled-ligand binding experiments, we studied the effect of eliminating this residual conservation of sequence on ion-channel function and cell-surface expression. From our results, we conclude that proper state interconversion (“gating”) does not require conservation of sequence—or even physicochemical properties—across the ECD–TMD interface. Wild-type ECD and TMD side chains undoubtedly interact with their surroundings, but the interactions between them—straddling the interface—do not seem to be more important for gating than those occurring elsewhere in the protein. We propose that gating of pLGICs requires, instead, that the overall structure of the interfacial loops be conserved, and that their relative orientation and distance be the appropriate ones for changes in one side to result in changes in the other, in a phenomenon akin to the nonspecific “bumping” of closely apposed domains.
Ion channels interconvert (“gate”) among ion-permeable and ion-impermeable conformations, and in so doing not only does the transmembrane pore alternately constrict and expand, but also, cavities throughout the protein appear, collapse, or change shape. The state-dependent binding of ligands to these cavities offers any ion channel a means to bias its conformational equilibria. In the particular case of neurotransmitter-gated ion channels, the state-dependent binding of endogenous ligands (neurotransmitters) to a subset of these cavities (the “orthosteric binding sites”) constitutes the physiological mechanism of channel activation.
Central to this mechanism is the idea that binding-site affinities and the conductive/nonconductive state of the pore are correlated (1). For example, if the pore adopts the closed-type nonconductive conformation, neurotransmitters bind with low affinity, and if the pore adopts the open, ion-conductive conformation, neurotransmitters bind with high affinity. The conformations of the neurotransmitter-binding sites and the transmembrane pore are, thus, interdependent; they are said to be “coupled.” In the superfamily of pentameric ligand-gated ion channels (pLGICs, also known as “Cys-loop” receptors), the orthosteric neurotransmitter-binding sites and the transmembrane pore map to different modules of the protein—the extracellular domain (ECD) and the transmembrane domain (TMD), respectively—and are separated by a distance of ∼60 Å (2, 3). The occurrence (in invertebrates) of soluble acetylcholine (ACh)-binding proteins (4) with high homology to the ECD of pLGICs (5) and the finding that fully functional chimeric constructs can often be generated by combining the ECD and TMD of different members of the superfamily (e.g., refs. 6–14) make the tightly coupled behavior of these two seemingly independent modules all the more remarkable.
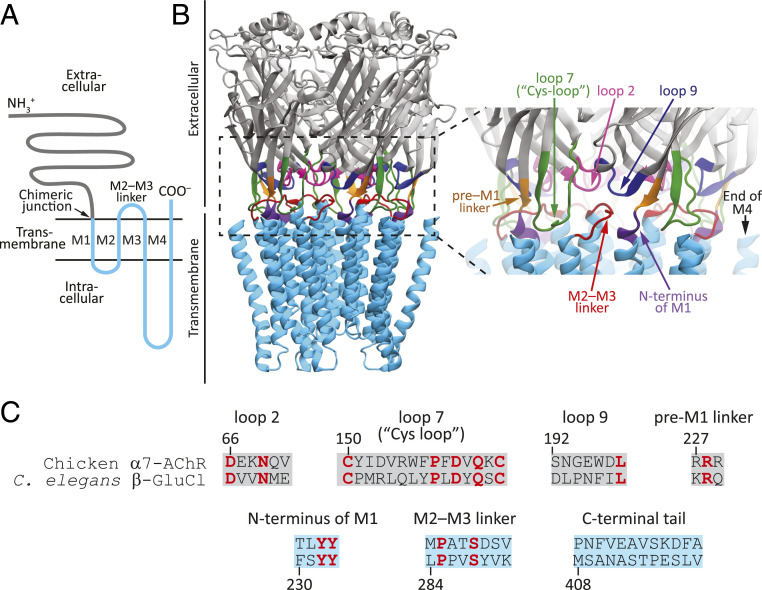
It has been suggested that interactions between wild-type side chains across the ECD–TMD interface are required for this transdomain communication to occur (e.g., refs. 3, 7, 9, 11, 13–20), a notion that is consistent with the observation that the most conserved residues in the superfamily are clustered in this very region (21). The ECD and the TMD meet at the extracellular surface of the membrane, where three loops between ECD β-strands (loops 2, 7, and 9) and the extracellular projection of the M1 α-helix (the pre-M1 linker and the first few residues of M1) closely approach a loop between two TMD α-helices (the M2–M3 linker) and the extracellular projection of the M4 α-helix (the C-terminal tail) (3) (Fig. 1). The particular aspects that need to be conserved in these structural elements to ensure a proper extracellular–intracellular signal transduction are unknown, and proposals (past and recent) as to their nature have covered the entire range of possibilities: from intact stretches of amino acids to just a few residues, and from packing, “lock-and-key” type of considerations to electrostatic ones (3, 9, 13–20, 24).
Fig. 1.
The ECD–TMD interface of pLGICs. (A) Membrane-threading pattern common to all pLGIC subunits. (B) Structural elements at the ECD–TMD interface mapped onto an X-ray crystal-structure model of α1-GluCl from C. elegans (PDB ID code 3RHW; ref. 22). The residues forming the C-terminal tail, at the end of M4, were not built in this atomic model. Furthermore, the M3–M4 linker of the crystallized construct was shortened to facilitate crystal formation. In all the constructs studied in this work, the M3–M4 linker retained its full-length, wild-type sequence. Molecular images were made with Visual Molecular Dynamics (VMD; ref. 23) using ribbon representation. (C) Sequence alignment of the structural elements at the ECD–TMD-interface of the α7-AChR from chicken and β-GluCl from C. elegans. Loops 2, 7, 9, and the pre-M1 linker map to the ECD, whereas the N terminus of M1, the M2–M3 linker, and the C-terminal tail map to the TMD. Identical residues are indicated in red.
Here, we set out to determine where along this wide range of possibilities pLGICs lie. To this end, we studied the effect of extensive mutagenesis to the ECD–TMD interfacial residues on ion-channel function and cell-surface expression. From our results, we conclude that proper state interconversion does not require conservation of sequence or physicochemical properties across the ECD–TMD interface. At variance with current models, we propose that gating of pLGICs (defined broadly, here, as all closed ⇌ open ⇌ desensitized state interconversions) only requires that the overall structure of the interfacial loops be conserved, and that their relative orientation and distance be the appropriate ones for changes in one side to result in changes in the other, in a phenomenon that could be described as the nonspecific “bumping” of closely apposed domains. Not every sequence at the interface satisfied these seemingly loose requirements, however, and we found the relationship between amino acid sequence and expression/function in this region to be far from straightforward. We discuss these findings in the context of evolutionary considerations for proteins whose signal-sensing and effector domains are covalently fused.
Results
Using a “Cut-and-Splice” Chimeric Approach.
Of all pLGICs, we decided to start our study with the α7-AChR. Besides its well-known physiological relevance and intriguing functional properties, the reasons for this choice were as follows. First, α-bungarotoxin (α-BgTx) binds to the ECD of these channels, forming a high-affinity, slowly dissociating complex that allows the quantitative estimation of channel-expression levels on the plasma membrane. This is an important consideration when poor expression, rather than inadequate function, could underlie the observed lack of channel activity of a mutant. Second, α7-AChR subunits form homomeric channels. Homomers ensure a well-defined subunit composition no matter how detrimental to channel expression a mutation may be. This is not necessarily the case for heteromeric pLGICs, the subunit composition of which may depend on the availability of correctly folded subunits. In addition, homomeric channels greatly simplify both the mutagenesis and the interpretation of the results. For all constructs studied here, mutations were introduced in all five subunits of homomeric channels. To increase the likelihood of obtaining electrophysiological recordings from poorly expressing mutants, currents were recorded in the whole-cell configuration.
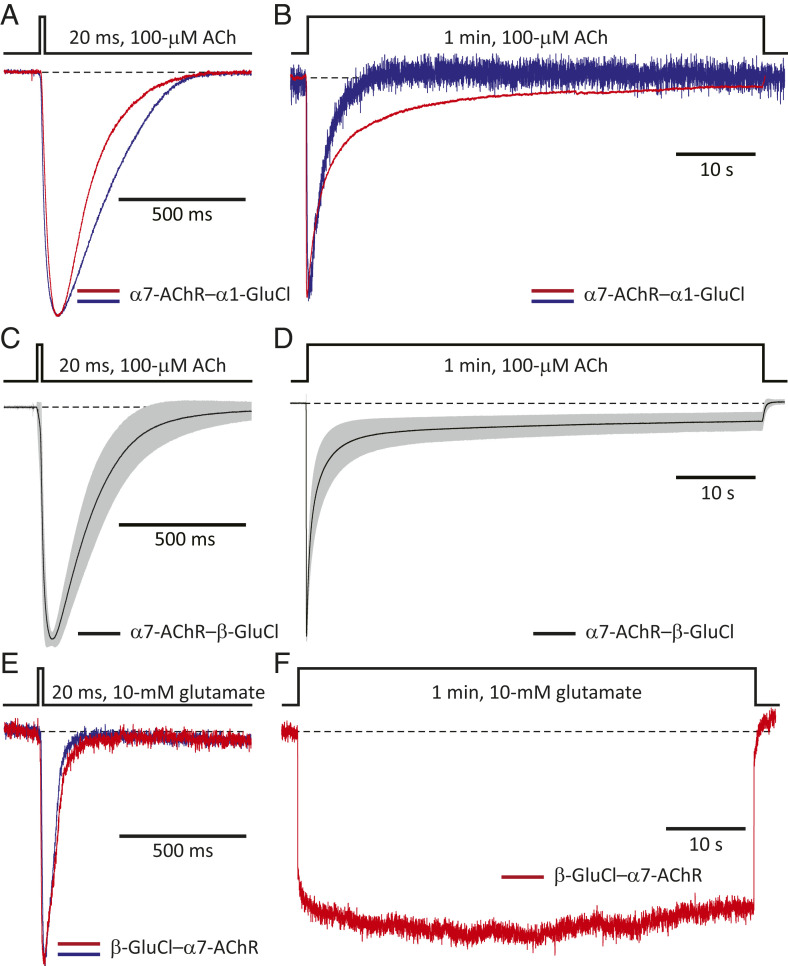
The structural elements of the ECD side of the ECD–TMD interface converge into the ion-channel pore through the M2–M3 linker. In an attempt to eliminate side-chain-specific interactions across the two sides of this interface, we replaced the M2–M3 linker of the human α7-AChR (sequence: MPATSDSV) with a string of eight alanines. This mutant, perhaps not surprisingly, expressed poorly on the surface of transiently transfected HEK-293 cells, worse than the wild-type α7-AChR by a factor of ∼15,000 [both transfections included the complementary DNAs (cDNAs) coding the chaperones RIC-3 (25) and NACHO (26) from humans]. Then, we replaced this linker with the more flexible AGTAGTAG octapeptide, but the expression of this construct was also very low (worse than the wild type by a factor of ∼40). We reasoned that not every arbitrary amino acid segment may fold properly or be tolerated in the background of a largely foreign protein. Thus, we decided to “borrow” intact sequences from other pLGICs. To minimize the probability of misfolding, we used entire protein modules in the form of ECD–TMD chimeras. Also, to minimize the probability of recapitulating across-the-interface interactions already present in the wild-type α7-AChR, we looked for parental sequences from distant orthologs. Alignment of sequences indicated little conservation between the invertebrate GluCl and the α7-AChR at the level of the ECD–TMD interface. Therefore, we combined these subunits into different chimeric constructs using the end of the β-10 strand and the first residue of the M1 α-helix as the junction site, a widely used approach (6). The chimeras between the chicken α7-AChR’s ECD and the TMD of α1-GluCl or β-GluCl from Caenorhabditis elegans both formed functional ACh-gated channels (Fig. 2 A–D), but the latter expressed better (by a factor of ∼9). The “reverse” β-GluCl–α7-AChR chimera also expressed (when the transfection included cDNA coding human RIC-3) and displayed glutamate-gated currents (Fig. 2 E and F), but the unique advantage afforded by α-BgTx–binding assays as a robust and quantitative probe of channel expression made us favor the use of the chimera with an AChR ECD. In all cases, the long linker between transmembrane α-helices M3 and M4 (∼65 residues in α1-GluCl and β-GluCl and ∼150 residues in the α7-AChR) was left intact.
Fig. 2.
Electrophysiological characterization of chimeras between the α7-AChR, and α1- and β-GluCl. Normalized inward currents recorded in the whole-cell configuration under asymmetrical KCl-concentration conditions in response to the application of 20-ms or 1-min pulses of agonist. Black dashed lines denote the zero-current baseline. (A and B) α7-AChR–α1-GluCl ECD–TMD chimera. Two representative responses to each type of pulse are shown. Each displayed response was recorded from a different cell. The membrane potential was ∼–60 mV. (C and D) α7-AChR–β-GluCl ECD–TMD (“CS”) chimera. Mean (black solid line) plus or minus one SD (gray error bars) of responses recorded from different whole-cell experiments. Data in C correspond to a total of 282 responses recorded from 35 different cells. Data in D correspond to a total of 39 responses recorded from 30 different cells. The membrane potential was ∼–60 mV. (E and F) β-GluCl– α7-AChR ECD–TMD chimera. Two representative responses to 20-ms pulses, and one to a 1-min pulse, are shown. Each displayed response was recorded from a different cell. The membrane potential was ∼–100 mV. For information about the sequences of these constructs see SI Appendix, Fig. S1 and Materials and Methods.
Our chicken–C. elegans α7-AChR–β-GluCl chimera combined two intact pLGIC modules with a single junction site (SI Appendix, Fig. S1), and thus it differed slightly from the α7-AChR–β-GluCl construct used by us in previous work (27) [first characterized by Paas and coworkers (8)]. The latter contained two mutations toward the C-terminal end of the ECD (T225I and M226I; amino acid numbering starting with the first methionine) that replaced α7-AChR’s residues with their aligned amino acids in the mouse serotonin type-3A receptor (5-HT3AR). The simpler, “cut-and-splice” chimera used here (herein referred to as the “CS” construct; we prefer not to refer to it as a “wild-type” channel) expressed well, better than the human α7-AChR (cotransfected with cDNA coding human RIC-3 and human NACHO) by a factor of ∼1.7, and better than the mouse adult-muscle AChR by a factor of ∼1.3 (accounting for the 5-to-2 ratio of α-BgTx binding sites between these two AChRs), under identical experimental conditions. Moving the chimeric-junction site to the C-terminal end of M1 reduced cell-surface expression by a factor of ∼150.
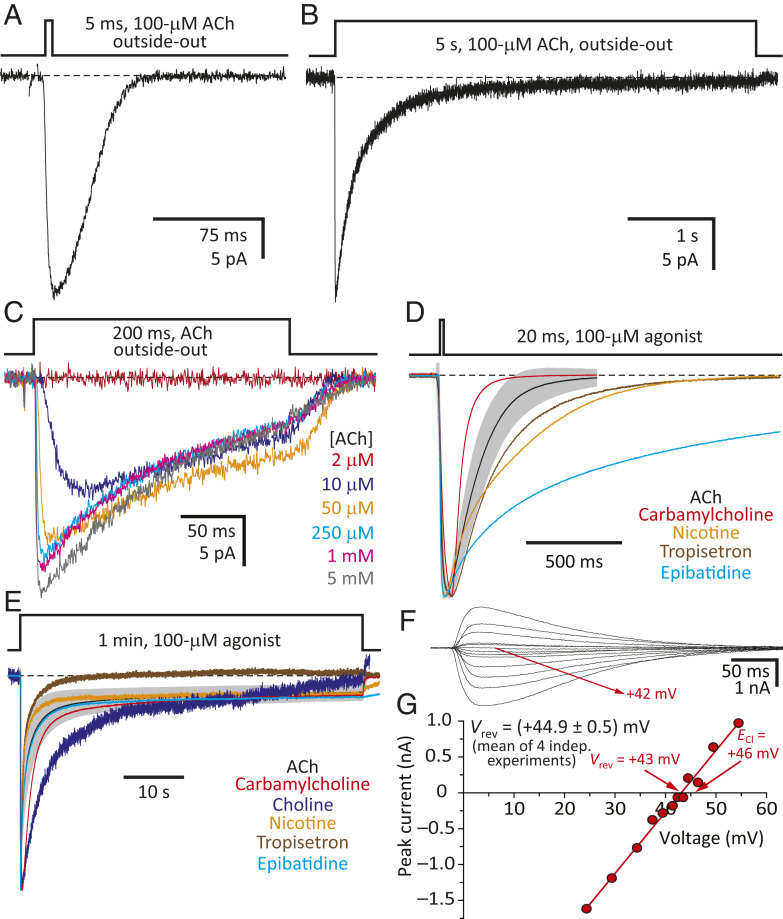
The CS channel displayed 1) the functional hallmarks common to all wild-type pLGICs, activating and desensitizing in a concentration-dependent manner upon application of agonist (single exponential time constants of ∼2 ms and 350 to 400 ms, respectively, in outside-out patches exposed to 100-μM ACh) and deactivating to a zero-current level upon agonist washout (single exponential time constant of ∼20 ms, in outside-out patches; Fig. 3 A–C); 2) activation by agonists known to open the wild-type α7-AChR (Fig. 3 D and E); 3) the high selectivity for anions expected from a channel containing a β-GluCl TMD (Fig. 3 F and G); 4) an extremely low single-channel conductance, which prevented the identification of individual channel openings (Fig. 3 A–C); and 5) kinetics of state interconversions that differed from those of the parental channels. Most conveniently in this particular regard, the CS chimera desensitized more slowly than the wild-type α7-AChR (Fig. 3B), thus allowing the recording of larger currents despite having a much lower single-channel conductance. As for the patch-to-patch differences in the kinetics of responses to agonist applications, the chimera’s range of variability seemed to be comparable (Fig. 2 C and D) to that we have observed for the human α1 glycine-receptor homomer [α1-GlyR (28)] or the bacterial pLGIC ELIC (29), for example, under similar experimental conditions. Moreover, the kinetics of the various time courses were slower in the whole-cell configuration than they were in the outside-out configuration, a finding that we attribute (at least, to some extent) to the slower perfusion of whole cells.
Fig. 3.
Functional properties of the α7-AChR–β-GluCl CS chimera. (A and B) Inward currents recorded in the outside-out configuration under asymmetrical KCl-concentration conditions in response to the application of 5-ms or 5-s pulses of 100-μM ACh. In these examples obtained from the same patch, the peak-current value was ∼25 pA; discrete single-channel events could not be identified. (C) Inward currents recorded in the outside-out configuration under the same ion conditions as in A and B in response to the application of 200-ms pulses of the indicated concentrations of ACh. (D and E) Normalized inward currents recorded in the whole-cell configuration under asymmetrical KCl-concentration conditions in response to the application of 20-ms or 1-min pulses of the indicated agonist at a concentration of 100 μM. For ACh, the mean ± 1 SD values of several recordings are indicated with a black solid line and gray error bars, respectively, as in Fig. 2 C and D. In D, short, 20-ms pulses of the slowly activating agonist choline did not elicit measurable currents; longer pulses were needed. (F and G) Current–voltage (I–V) data recorded in the whole-cell configuration from a representative cell and I–V curve of its rectilinear portion. The equilibrium potentials (calculated at 22 °C using ion activities) were ∼–51 mV for K+ and ∼+46 mV for Cl–. The reversal potential (corrected for the liquid-junction potential) obtained from four independent experiments was (+44.9 ± 0.5) mV. The membrane potential was ∼–70 mV for A–C and ∼–60 mV for D and E. For A–E, black dashed lines denote the zero-current baseline.
Eliminating Conserved Residues from the M2–M3 Linker.
Alignment of both parental sequences revealed a small, but nonzero, number of conserved residues at the ECD–TMD interface (Fig. 1C and SI Appendix, Fig. S1). Thus, we wondered whether the full functionality of the chimera could be ascribed to the “survival” of parent-like interactions between these side chains in the daughter construct. In fact, all pLGIC pairs that have been reported to give rise to functional channels when cut and spliced as ECD–TMD chimeras display a larger degree of sequence identity at the interface than suggested by overall, entire-sequence identity values. Furthermore, many of the functional ECD–TMD pLGIC chimeras described in the literature contain mutations that make the sequences on both sides of the interface resemble the sequence of one or the other parent (e.g., refs. 9, 11, 14). Here, instead of aiming to preserve across-the-interface wild-type contacts, we sought to eliminate them.
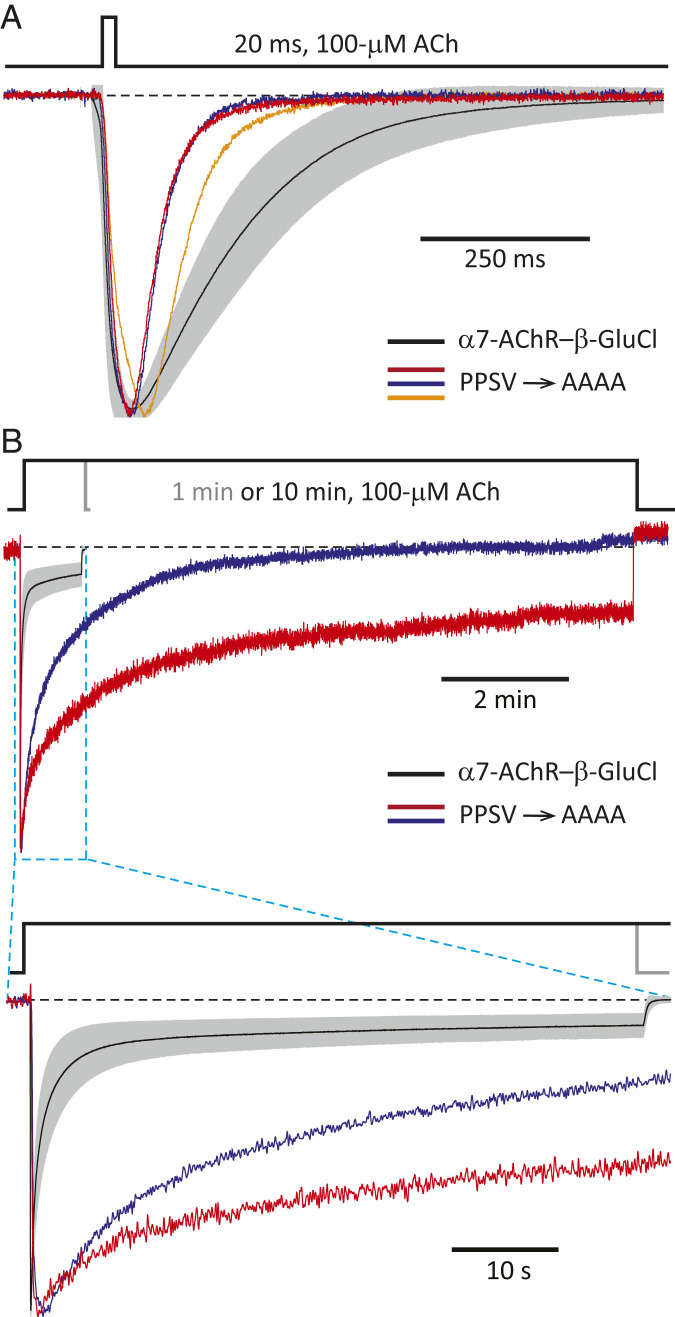
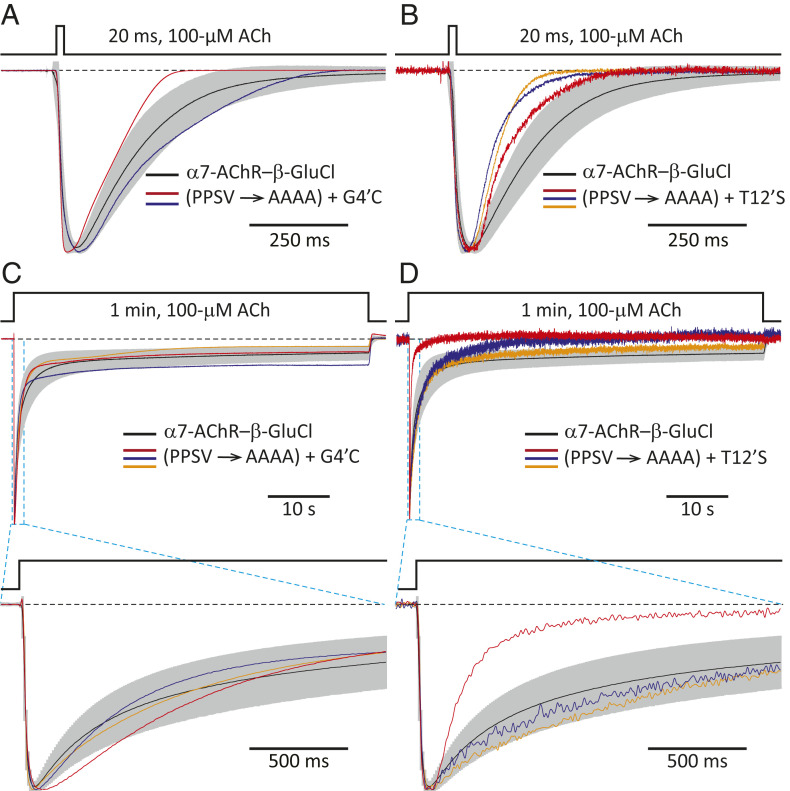
In our case, on the TMD side of the interface, there is no conservation of sequence at the level of the (extracellular) C-terminal tail between the α7-AChR and β-GluCl (Fig. 1C). The same is the case for the most extracellular residues of the M2, M3, and M4 α-helices (SI Appendix, Fig. S1), which (on the basis of atomic models of other pLGICs) are expected to protrude into the aqueous space and contribute to this side of the interface. At the level of the M2–M3 linker, however, a proline and a serine are conserved between the α7-AChR (MPATSDSV; conserved residues are indicated in italics) and β-GluCl (LPPVSYVK; Fig. 1C). Moreover, the M2–M3 linker of β-GluCl (that is, the M2–M3 linker present in the chimera) has a second proline that may also be regarded as aligned with the α7-AChR’s single proline, and a valine that is misaligned with that of the α7-AChR by only one position. Thus, we mutated these four residues (Pro-285, Pro-286, Ser-288, and Val-290) to alanines (L284PPVSYVK → L284AAVAYAK; mutated residues are underlined), and the resulting construct gave rise to a channel that expressed relatively well (mutant-to-CS ratio = 0.53; SI Appendix, Table S1) and, most remarkably, displayed the functional properties expected from a pLGIC (Fig. 4). It could be argued, however, that the time course of entry into desensitization of this multiple mutant was uncharacteristically slow for a pLGIC (Fig. 4B; note the duration of ACh application), and that this anomalous behavior could be a direct consequence of the absence of necessary interactions across the mismatched ECD–TMD interface. Nevertheless, we found that adding single-site mutations elsewhere in the protein, far from the interface—such as G266C (at position 4ʹ of the M2 α-helix) or T274S (at position 12ʹ of M2)—speeds up desensitization of this “wild-type-contactless” construct to CS-chimera levels while having little to no effect on the kinetics of activation or deactivation (Fig. 5 and SI Appendix, Fig. S2). The Cα–Cα distance between position 4ʹ and the center of the M2–M3 linker [measured in an atomic model of α1-GluCl; Protein Data Bank (PDB) ID code 3RHW (22)] is ∼30 Å, and that between position 12ʹ and the center of the M2–M3 linker is ∼20 Å. Needless to say, neither mutation to M2 can have any effect on the extent of sequence conservation at the level of the M2–M3 linker. Hence, to the extent that our mutagenesis succeeded in eliminating all native interactions between side chains located across the interface, it seems inescapable to conclude that the conservation of matching pairs of amino acids across the ECD–TMD interface is not necessary for proper ion-channel gating. It is worth noting, here, that the kinetics of activation, deactivation, and desensitization of the CS chimera in whole-cell recordings are similar to those of, for example, the (full-length) wild-type α1-GlyR recorded under the same experimental conditions (SI Appendix, Fig. S3). Therefore, as far as the kinetics of state interconversion are concerned, the contactless mutants in Fig. 5 are typical members of the pLGIC superfamily.
Fig. 4.
Wild-type side-chain–side-chain contacts across the ECD–TMD interface of pLGICs are not required for gating. Normalized inward currents recorded from the P285A + P286A + S288A + V290A M2–M3-linker mutant (PPSV → AAAA) of the α7-AChR– β-GluCl CS chimera in the whole-cell configuration under asymmetrical KCl-concentration conditions in response to the application of 20-ms or 10-min pulses of 100-μM ACh. The membrane potential was ∼–60 mV. Black dashed lines denote the zero-current baseline. (A and B) Three representative responses to 20-ms pulses and two to 10-min pulses are shown. Each displayed response was recorded from a different cell. For comparison, the averaged responses (mean ± 1 SD) of the CS chimera (without any additional mutation) to 20-ms or 1-min pulses of 100-μM ACh are also shown, as in Fig. 2 C and D (mean: black solid line; SD: gray error bars).
Fig. 5.
Mutations in M2 restore CS-like kinetics of desensitization to the PPSV → AAAA mutant. Normalized inward currents recorded in the whole-cell configuration under asymmetrical KCl-concentration conditions in response to the application of 20-ms or 1-min pulses of 100-μM ACh. The membrane potential was ∼–60 mV. Black dashed lines denote the zero-current baseline. (A and B) Responses to 20-ms pulses. (C and D) Responses to 1-min pulses. Two or three representative responses from each of the quintuple mutants to each type of pulse are shown. Each displayed response was recorded from a different cell. For comparison, the averaged responses (mean ± 1 SD) of the CS chimera (without any additional mutation) to 20-ms or 1-min pulses of 100-μM ACh are also shown, as in Fig. 2 C and D (mean: black solid line; SD: gray error bars).
We also replaced this chimera’s M2–M3 linker with that of the α7-AChR with the goal of contrasting the behavior of a channel having a completely mismatched interface with that of a construct in which the ECD’s interfacial segments face the same M2–M3 linker as they do in the wild-type α7-AChR. Although this chimera expressed robustly (ratio = 1.5) we could not record any currents. Because the α7-AChR’s M2–M3 linker contains an aspartate that could create an unfavorable environment for the anions passing through the β-GluCl pore, we mutated this acidic residue to lysine (M284PATSDSV → M284PATSKSV). Once again, however, although this mutant channel expressed very well (ratio = 2.0), no currents could be recorded. It is likely that a slightly different (shorter, longer, or shifted) stretch of α7-AChR M2–M3-linker amino acids was required to obtain a functional chimera (see, for example, ref. 30). If this were the case, this result would highlight the idea that these loops need to be correctly oriented relative to each other in order to function properly. Surface expression, on the other hand, was not compromised at all in these two mutants.
We then took a second look at the highly variable (and therefore, difficult-to-align) C-terminal tail. Although our alignment suggested that no residues are conserved between the parental sequences (Fig. 1C), both tails contain asparagines (one each), prolines (one each), and ionizable residues (three in the α7-AChR and one in β-GluCl). Hence, we mutated the chimera’s Asn-411, Pro-415, and Glu-416 to alanine, both individually and as a triple mutant. We also shortened the chimera’s tail by removing the last 9 out of ∼12 residues (M408SANASTPESLV → M408SA; “N411stop”), thereby eliminating the stretch containing Asn-411, Pro-415, and Glu-416. Finally, we replaced the chimera’s β-GluCl C-terminal tail with that of the α7-AChR (M408SANASTPESLV → P408NFVEAVSKDFA; “α7-AChR C-terminal tail”). Notably, these six mutants expressed well (ratios = 1.2, 1.4, 1.0, 1.1, 0.74, and 0.27, respectively), and currents elicited from them in response to short and long applications of ACh were characteristic of fully functional pLGICs (SI Appendix, Fig. S4). All in all, these results indicate that side-chain–side-chain interactions between the C-terminal tails and the ECD are not required for proper gating in this superfamily.
Eliminating Conserved Residues from Loops 2, 7, and 9.
The results obtained with the M2–M3 linker (Figs. 4 and 5) and the C-terminal tail (SI Appendix, Fig. S4) were compelling regarding the lack of a requirement for the conservation of matching sequences (or, even, of physicochemical properties) across the ECD–TMD interface for function to be retained. Indeed, it suffices to mutate only one of the partners of a pairwise interaction (either one) to eliminate it. Consider a disulfide bond, for example: Mutating either cysteine eliminates the interaction—there is no need to mutate both cysteines individually, let alone at the same time, as a double mutant. However, for the sake of completeness, and because so much has been written about the role of conserved residues in the extracellular “half” of the ECD–TMD interface in pLGIC gating, we extended the mutagenesis to loops 2, 7, and 9, and the extracellular projection of M1. We did this with the understanding that the eventual finding of total-loss-of-function mutations on this side of the interface could not be ascribed to the elimination of ECD–TMD side-chain–side-chain interactions that are essential for gating (because their existence was already ruled out; Figs. 4 and 5 and SI Appendix, Fig. S4), but rather, to the disruption of other phenomena that are required to keep the ECD as a functional module, such as intradomain interactions.
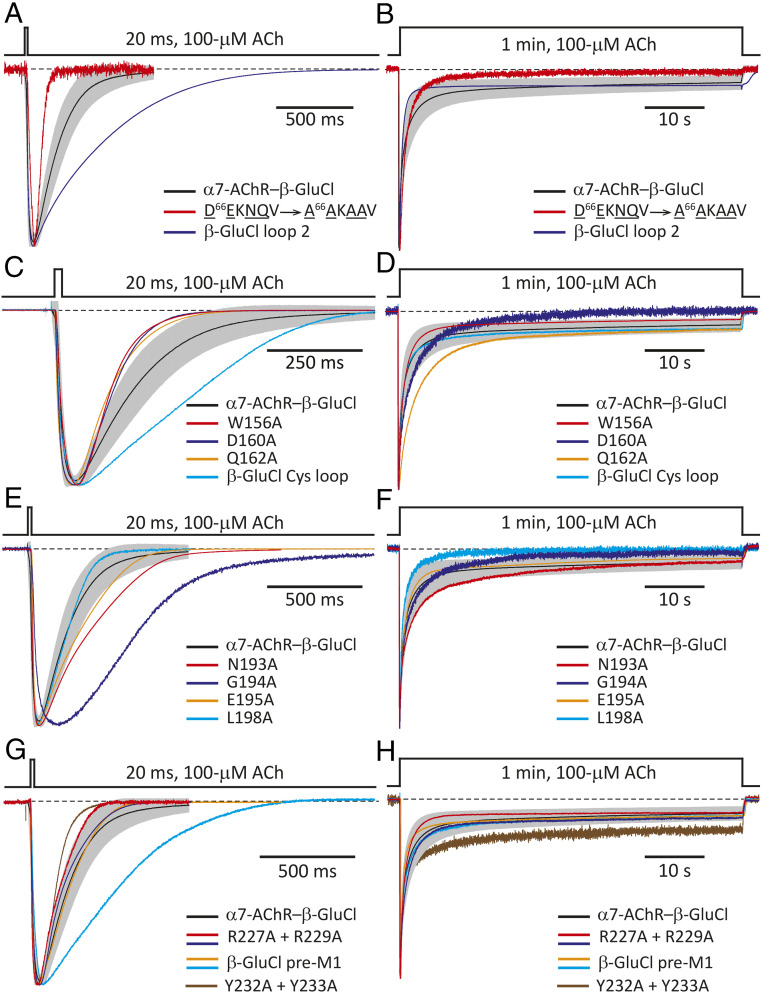
In loop 2, an aspartate and an asparagine are conserved between the α7-AChR (DEKNQV) and β-GluCl (DVVNME; Fig. 1C). Moreover, loop 2 of the α7-AChR has a glutamate and a glutamine that are misaligned by only one position with the aspartate and asparagine of β-GluCl, and thus, may also be considered to be conserved. Therefore, we mutated loop 2’s Asp-66, Glu-67, Asn-69, and Gln-70 to alanines (D66EKNQV → A66AKAAV). Although this multiple mutant expressed poorly (ratio = 0.011), well-expressing cells could be found. The channel activated more slowly and deactivated faster than the CS chimera, but kinetic details aside the channel’s behavior was characteristic of pLGICs (Fig. 6 A and B). Furthermore, replacing the entire α7-AChR loop 2 with that of β-GluCl (“β-GluCl loop 2”) gave rise to a channel that expressed well (ratio = 1.3) and displayed unremarkable functional properties (Fig. 6 A and B).
Fig. 6.
Eliminating conserved residues from the ECD side of the interface and the N terminus of M1. Normalized inward currents recorded from the indicated CS-chimera mutants in the whole-cell configuration under asymmetrical KCl-concentration conditions in response to the application of 20-ms or 1-min pulses of 100-μM ACh. The membrane potential was ∼–60 mV. Black dashed lines denote the zero-current baseline. (A and B) Loop-2 mutants. (C and D) Cys-loop (loop-7) mutants. (E and F) Loop-9 mutants. (G and H) Pre-M1-linker and N terminus-of-M1 mutants. One or two representative responses from each mutant to each type of pulse are shown. Each displayed response was recorded from a different cell. For comparison, the averaged responses (mean ± 1 SD) of the CS chimera (without any additional mutation) to 20-ms or 1-min pulses of 100-μM ACh are also shown, as in Fig. 2 C and D (mean: black solid line; SD: gray error bars).
In loop 7 (that is, the “Cys-loop”), five residues are conserved between the α7-AChR (CYIDVRWFPFDVQKC) and β-GluCl (CPMRLQLYPLDYQSC; Fig. 1C): the flanking cysteines, an aspartate, a glutamine, and a universally conserved (21) proline. Hence, we proceeded to mutate Pro-158, Asp-160, and Gln-162, individually, to alanines; the flanking, disulfide-bonded cysteines were not mutated, here, because they have been known to be essential for the expression of functional receptors (31). Mutation of the conserved Pro-158 to alanine or glutamine (C150YIDVRWFPFDVQKC → C150YIDVRWFA/QFDVQKC) nearly abolished expression, and although the glycine mutant expressed better (ratio = 0.010), its expression was still very low. Alanine mutations were better tolerated at positions 160 (ratio = 0.047) and 162 (ratio = 0.71), and both single-site mutants displayed CS-like functional properties (Fig. 6 C and D); the double mutant, however, expressed very poorly (ratio = 0.0096). We also mutated Phe-157 (which aligns with a tyrosine in β-GluCl) and the preceding (also aromatic) Trp-156, individually, to alanine. Both single mutants expressed relatively well (ratios = 0.35 and 0.69, respectively), but whereas the W156A mutant’s behavior was CS-like (Fig. 6 C and D), the F157A mutant displayed no detectable currents. As was the case for loop 2, replacing the entire α7-AChR Cys-loop with that of β-GluCl (“β-GluCl Cys-loop”) gave rise to a channel that, despite its modest expression levels (ratio = 0.15), displayed all the hallmarks of an ACh-gated pLGIC (Fig. 6 C and D).
In loop 9, only a leucine is conserved between the α7-AChR (SNGEWDL) and β-GluCl (DLPNFIL; Fig. 1C), and although the L198A mutant (S192NGEWDL → S192NGEWDA) expressed poorly (ratio = 0.053), CS-like currents could be recorded from cells that expressed well (Fig. 6 E and F). The central motif of this loop in the α7-AChR (GEW) resembles that of β-GluCl (PNF), thus suggesting that the conservation of some physicochemical properties at the level of this loop may be required for the functionality of the extracellular side of the interface. Although single-site mutants G194A and E195A expressed well (ratio = 1.2 and 0.33, respectively) and gave rise to characteristic ACh-gated currents (Fig. 6 E and F), the W196A mutant expressed poorly (ratio = 0.0025), and currents could not be recorded. Asn-193, in the α7-AChR side of the chimera, may be regarded as aligned with the central asparagine of β-GluCl’s PNF motif, and thus we mutated it to alanine. The N193A mutant expressed well (ratio = 0.54), and its behavior was CS-like (Fig. 6 E and F). Finally, upon replacing the three central residues of the α7-AChR’s loop 9 with those of β-GluCl (S192NGEWDL → S192NPNFDL), the expression was reduced (ratio = 0.084), and currents could not be recorded.
In the pre-M1 linker, an arginine is conserved between the α7-AChR (RRR) and β-GluCl (KRQ; Fig. 1C). Mutation of this nearly universally conserved arginine (21) to alanine (R227RR → R227AR) decreased expression to a large extent (ratio = 0.0023), and currents could not be detected. Mutation of the flanking arginines, instead, to alanines (R227RR → A227RA) or to their corresponding residues in β-GluCl (R227RR → K227RQ; “β-GluCl pre-M1”) decreased expression less markedly (ratio = 0.018 and 0.17, respectively) and ACh-gated currents could be recorded (Fig. 6 G and H). Upon insertion of a fourth arginine, however, the channel failed to display detectable currents despite expressing well (ratio = 0.66).
In the CS chimera (as well as in many other pLGIC ECD–TMD chimeric constructs), the pre-M1 linker marks the end of the ECD module; the residues that follow (F230SYYLV···) correspond to the N terminus of the M1 α-helix of the other parent sequence (SI Appendix, Fig. S1). An inspection of atomic models (e.g., refs. 18, 19, 22) reveals that the first few residues of M1 protrude into the extracellular side and contribute to the interface. Here, two tyrosines are conserved between the α7-AChR (TLYY) and β-GluCl (FSYY; Fig. 1C), and a tyrosine or a phenylalanine occupy either or both of these positions in most (if not all) functionally characterized pLGIC-forming subunit sequences. Sitting right at the junction between modules, the decision as to whether these residues contribute to one or the other side of the interface seems somewhat arbitrary, but the design of our chimera assumed that the N terminus of M1 belongs to the transmembrane side (and hence, has a β-GluCl sequence). Despite the high conservation of aromatic residues at these two positions, the alanine double mutant (F230SYY → F230SAA) expressed very well (ratio = 0.78), and the corresponding currents displayed CS-like time courses (Fig. 6 G and H). This result adds experimental evidence to the notion that wild-type–like interactions between side chains located across the interface are not required for function.
Discussion
Membrane-embedded receptors transmit signals between aqueous compartments of a cell. A signal-receiving module binds ligand from one side of the membrane, and a signal-transducing module translates this binding event into the catalysis of a chemical reaction on the other side or a transmembrane ion flow. In heterotrimeric G protein/arrestin-dependent metabotropic receptors, for example, these two modules map to separate proteins (the G protein-coupled receptor [GPCR] and the G protein/arrestin), whereas in neurotransmitter-gated ion channels, receptor tyrosine kinases/phosphatases, and receptor guanylyl cyclases, these two modules are covalently fused. Because GPCRs bind to certain heterotrimeric G proteins but not to others, these two types of protein must have coevolved under the pressure of recognizing each other among a large number of other similar proteins in the membrane. It does not seem surprising, then, that defined stretches of amino acids on both the GPCR and the G protein have been found to be critical determinants of this selective interaction (32–37). It is in this precise context that the reported requirement for the conservation of matching sequences (7, 11, 13, 14, 16) or even packing (3, 9) or electrostatic properties (15, 20, 24) across the ECD–TMD interface of pLGICs seemed intriguing to us. Certainly, in contrast to the situation in GPCR signaling (in which case, physiologically aberrant GPCR–G protein pairs need to be avoided) the two modules of pLGICs are part of the same protein, and thus the “correct” combination is inevitable. The only requirement for the ECD–TMD interface of pLGICs is to ensure the proper conversion of a ligand affinity-changing conformational change in the ECD into the gating of a transmembrane ion flow.
Side chains interact with their microenvironment, and interactions bridging the ECD–TMD interface of pLGICs are likely to be only a fraction of the several interactions an interfacial side chain establishes. Clearly, interactions may also take place between side chains on the same side of the interface, within each domain. As a result, the finding that a mutation to an interfacial residue abolishes function cannot be uniquely ascribed to the elimination of across-the-interface interactions; it may well be that it is the intradomain interactions established by this residue that are essential. The only result that could be interpreted unambiguously, then, would be the absence of a detrimental effect of a mutation on function, in which case one would have to conclude that interactions across the interface (as well as those within the corresponding domain) involving the mutated residue are not required for gating. The mutations to the M2–M3 linker (Figs. 4 and 5), the C-terminal tail (SI Appendix, Fig. S4), and the N terminus of M1 (Fig. 6 G and H) described in this work are expected to have eliminated all possible native-like interactions between side chains across the ECD–TMD interface, and thus the finding that these mutants function well has a most unequivocal mechanistic interpretation: Wild-type–like side-chain–side-chain interactions straddling the interface are not required for gating. By no means should this result be taken to imply, however, that the intra- or interdomain interactions established by the highly conserved side chains at the interface are not important. Indeed, mutations to these residues affected cell-surface expression and function (SI Appendix, Table S1), and although some mutants expressed well and remained functional, these parameters may have been altered beyond what would be compatible with normal physiology. Even small changes in expression and/or the kinetics of gating may have marked effects on the in vivo response of pLGIC-expressing cells to neurotransmitters.
Our results suggest that any chimeric construct assembled from intact (that is, wild-type) ECD and TMD modules would be functional; neither sequence conservation nor side-chain packing, electrostatic, or hydrogen-bond donor–acceptor complementarity between the two “halves” seems to be required. Thus, as long as the individual modules are functional in the context of their respective full ion channels—and that the domains are fused without adding or deleting intervening residues—so would the newly generated chimera. The eventual inability to record currents from a particular combination of wild-type ECD and TMD would likely reflect an exceedingly low unliganded-gating equilibrium constant of the new channel (in such a way that opening cannot be elicited by known agonists), a situation that could be offset by gain-of-function mutations engineered elsewhere in the protein. A lower-than-usual value for the unliganded-gating equilibrium constant would be as unremarkable as an unusually high value, and it would simply be one of the possible outcomes for a protein assembled from two halves that have not evolved together under the pressure of communicating in a physiologically relevant manner. In fact, the chimera between the ECD of α1-GluCl and the TMD of β-GluCl (both from C. elegans), for example, was found to display a high constitutive activity (higher than that of either parental channel; ref. 38), an observation that could be accounted for by a higher-than-usual unliganded-gating equilibrium constant.
Much has been written about the importance of side-chain–side-chain interactions across the ECD–TMD interface of pLGICs, including meticulous analyses of interatomic distances and detailed descriptions of the kinds of noncovalent bond involved (e.g., refs. 18, 19). In many cases, the emerging picture is one of exquisite complementarity between the two sides. However, using site-directed mutagenesis to eliminate the residual conservation of sequence present in a chimeric construct, we found that such specific side-chain interactions are not required for proper pLGIC operation. The pairwise interactions between ECD and TMD side chains inferred from interatomic distances in structural models evidently exist, but they do not seem to be essential for gating. Thus, although mutations to conserved interfacial residues could render a pLGIC inactive (as was the case for the F157A mutation in loop 7, the insertion of a fourth arginine in the pre-M1 linker, and the β-GluCl → α7-AChR conversion of the M2–M3 linker’s sequence), the underlying mechanism would not be the disruption of a specific interaction between side chains bridging the two sides of the interface, but rather, an adverse effect on some intradomain interaction or an unfavorable change in the relative positioning of the two domains. We hypothesize that this conclusion extends to the entire superfamily. Certainly, it seems unlikely that such a fundamental aspect of ion-channel gating could hold true for only some, but not all, pLGICs. After all, biochemical and electrophysiological evidence—and more recently, atomic models of several members of the superfamily in closed, open, and desensitized states—have indicated that the structural aspects of gating are essentially the same for the different pLGICs from animals. We are, thus, left with a picture that is consistent with conformational changes in either side of the ECD–TMD interface being transmitted to the other side as a result of their close apposition. Almost undoubtedly, chemically well-defined side-chain–side-chain interactions across the ECD–TMD interface contribute to the stability of the protein and the energetics of gating, but so do pairwise interactions in many other regions of the protein (e.g., refs. 39–44).
Although the ideas presented above may suggest a simple, nonsophisticated type of operation for the system of ECD–TMD interfacial loops, the relationship between amino acid sequence (on the one hand) and cell-surface expression and function (on the other) seemed to be complex. For example, the chimera in which four out of the eight residues of the β-GluCl M2–M3 linker (including two prolines) were mutated to alanine expressed and worked well, but the chimera in which only Pro-158 (in loop 7) was mutated did not express, and that in which only Phe-157 (also in loop 7) was mutated expressed but did not work. Furthermore, the construct in which the entire β-GluCl M2–M3 linker was replaced by the aligned residues in the α7-AChR (in such a way that loops 2, 7, and 9, the pre-M1 linker, and the M2–M3 linker come from the same channel) did not give rise to detectable currents despite expressing even better than the background construct.
More remains to be learned about how the ECD–TMD interfacial loops couple conformational changes in the neurotransmitter-binding sites to those in the transmembrane pore (upon channel opening) and vice versa (upon closing; ref. 45). We anticipate that exploration of the role of ECD–TMD interactions involving backbone atoms as well as the development and application of nonelectrophysiological approaches to probe function in electrically silent mutants will be critical components of this endeavor.
Materials and Methods
cDNA Clones, Mutagenesis, and Heterologous Expression.
cDNA coding the chimeric construct consisting of the ECD of α7-AChR from chicken (accession number: P22770)—containing the T225I and M226I mutations (amino acid numbering starting with the first methionine)—and the TMD of β-GluCl from C. elegans (accession number: Q17328) in pMT3 (8) was provided by Y. Paas, Bar-Ilan University, Ramat Gan, Israel. We mutated this construct (I225T and I226M) so as to revert the sequence of its ECD to that of the wild-type α7-AChR from chicken; the sequence is shown in SI Appendix, Fig. S1. Throughout this paper, we refer to this chimera as the CS construct. These and all other mutations were engineered using the QuikChange kit (Agilent Technologies), and the sequences of the resulting cDNAs were verified by dideoxy sequencing of the entire coding region (ACGT). cDNA coding the chimera consisting of the ECD of β-GluCl from C. elegans and the TMD of α7-AChR from chicken in pcDNA3.1 was synthesized by Genscript and was subcloned into the pMT3 vector; the sequence is shown in SI Appendix, Fig. S1. cDNA coding the chimera consisting of the ECD of the chicken α7-AChR (containing the T225I and M226I mutations) and the TMD of C. elegans α1-GluCl (accession number: G5EBR3) in pcDNA3.1 was synthesized by Genscript and was subcloned into the pMT3 vector. The chimeric-junction site was analogous to that of the CS construct, that is, the C-terminal end of the α7-AChR portion was the RRR sequence of the pre-M1 linker, and the N-terminal end of the α1-GluCl portion was the FSFY sequence at the beginning of M1. cDNA coding the human α7-AChR (accession number: P36544) in pcDNA3.1 was purchased from addgene (62276); that coding isoform 1 of human RIC-3 (accession number: Q7Z5B4; ref. 25) in pcDNA3.1 was provided by W. N. Green, University of Chicago, Chicago, IL, and that coding human NACHO (TMEM35A; accession number: Q53FP2; ref. 26) in pCMV6-XL5 was purchased from OriGene Technologies Inc. (SC112910). cDNAs coding the mouse α1, β1, δ, and ε subunits of the (muscle) AChR (accession numbers: P04756, P09690, P02716, and P20782, respectively) in pRBG4 were provided by S. M. Sine, Mayo Clinic, Rochester, MN, whereas that coding the rat α1-GlyR (accession number: P07727-2) in pcDNA3.1 was provided by M. M. Slaughter, University at Buffalo, Buffalo, NY. Wild-type and mutant channels were heterologously expressed in transiently transfected adherent HEK-293 cells (calcium-phosphate method using 187.5 ng DNA/cm2) grown at 37 °C and 5% CO2. Transfections proceeded for 16 to 18 h, after which the cell-culture medium (Dulbecco’s modified Eagle’s medium; Gibco) containing the DNA precipitate was replaced by fresh medium. Electrophysiological recordings and cell surface-expression assays started ∼24 h later.
Electrophysiology.
Our preliminary experiments showed that the single-channel conductance of the CS channel was unmeasurably low, and that several mutations to the ECD–TMD interface reduced protein expression. Thus, in an attempt to increase the number of channels contributing to the observed currents, electrophysiological recordings from mutant constructs were performed in the whole-cell patch-clamp configuration. The higher probability of recording data from poorly expressing mutants amply justified the slower and more variable perfusion of whole cells compared to that of excised patches. The outside-out configuration was only used in the case of the CS chimeric construct to estimate the kinetics of activation, deactivation, and entry into desensitization with more precision. Furthermore, to increase the amplitude of the recorded currents, the distribution of ions across the membrane was such that the reversal potential for Cl– was positive, and the holding potential was negative. Currents were recorded at ∼22 °C with an effective bandwidth of 5 kHz using an Axopatch 200B amplifier (Molecular Devices), digitized at 1 to 100 kHz, and analyzed using pCLAMP 11.1 software (Molecular Devices). Series-resistance compensation was used and set to ∼80%; throughout the paper, the given values of the membrane potential assumed that this compensation zeroed the series-resistance error. The reference Ag/AgCl wire was connected to the extracellular solution through an agar bridge containing 200 mM KCl. Agonist-concentration jumps were applied to whole cells or outside-out patches of membrane using a piece of double-barreled “θ-tubing” (Siskiyou; ref. 46). The flow of solution through the θ-tube was controlled using a gravity-fed system (ALA BPS-8; ALA Scientific Instruments), and the movement of the θ-tube was achieved using a piezoelectric arm (Burleigh-LSS-3100; discontinued) controlled by pCLAMP 11.1 software (Molecular Devices). The latter signals were low-pass-filtered (900C; Frequency Devices) at a cutoff frequency of 15 to 25 Hz prior to their arrival at the piezoelectric arm to reduce ringing in the θ-tube motion. In the whole-cell configuration, cells remained attached to a piece of poly-l-lysine–coated glass or glycol-modified polyethylene terephthalate (PETG; Thermo Fisher) coverslip, placed at the bottom of the recording chamber, throughout the experiment. In this configuration, the perfusion system achieved a solution-exchange time of ∼4 ms for the t10 to 90% and ∼10 ms for the t90 to 10%, as estimated from changes in the liquid-junction current measured with an open-tip patch pipette. In the outside-out configuration, these values were ∼0.4 ms for the t10 to 90% and ∼0.8 ms for the t90 to 10%. Although slower than the pressure-driven perfusion that we have used previously (46–48), the gravity-fed system was favored here in an attempt to increase the stability of patches; the collection of abundant data was deemed more important, in this particular case, than the highly accurate estimation of kinetic parameters from any construct. Charge selectivity was inferred from reversal potentials estimated under near KCl-dilution conditions (27). Whole-cell current–voltage (I–V) curves were generated by plotting the peak values of current transients elicited by brief (20-ms) applications of 100 μM ACh at membrane potentials straddling the reversal potential. Reversal potentials were estimated as the x-axis intercept of linear fits to only the linear portion of the recorded I–V curves. In these experiments, the pipette solution was 110 mM KCl, 40 mM KF, and 5 mM Hepes/KOH, pH 7.4 (measured osmolarity: ∼280 mOsm; Wescor), and the extracellular solution (flowing through the two barrels of a piece of θ-tubing) was 15 mM KCl, 5 mM Hepes/KOH, pH 7.4, and enough mannitol (∼230 mM) to reach an osmolarity of ∼270 mOsm, with or without 100 μM ACh. We included F– in the pipette solution because the stability of seals seemed to increase when this anion was present on the intracellular side. With these intra- and extracellular solutions, the equilibrium potentials were ∼–51 mV for K+ and ∼+46 mV for Cl–. Following work by Fatima-Shad and Barry on anion-selective pLGICs (49), we assumed a negligible permeability to F–. The liquid-junction potential between the pipette and bath solutions (calculated with the JPCalc module in pCLAMP 11.1 software to be ∼–1.3 mV at a temperature of 22 °C) was offset, and the membrane-potential values were corrected accordingly. In all other whole-cell experiments, the pipette solution was the same as that used in reversal-potential experiments, and the extracellular solution was 5 mM KCl, 5 mM Hepes/KOH, pH 7.4, and enough mannitol (∼250 mM) to reach an osmolarity of ∼275 mOsm, with or without agonist. With these intra- and extracellular solutions, the equilibrium potential for Cl– was ∼+73 mV. The liquid-junction potential (calculated to be ∼–1.6 mV) was offset. α7-AChR agonists were applied at a concentration of 100 μM, whereas glutamate and glycine were applied at concentrations of 10 mM and 1 mM, respectively. In outside-out experiments, the pipette solution was 100 mM KF, 40 mM KCl, 1 mM CaCl2, 11 mM EGTA, and 10-mM Hepes/KOH, pH 7.4 (∼290 mOsm), and the extracellular solution was 5 mM KCl, 5 mM Hepes/KOH, pH 7.4, and enough mannitol (∼250 mM) to reach an osmolarity of ∼275 mOsm, with or without 100 μM ACh. With these intra- and extracellular solutions, the equilibrium potential for Cl– was ∼+50 mV. The liquid-junction potential (calculated to be ∼–10 mV) was offset. In all cases, transfected cells were bathed in the corresponding extracellular solution without agonist.
Cell-Surface Expression.
The number of receptors on the plasma membrane of transfected HEK-293 cells was estimated from the amount of [125I]α-BgTx (PerkinElmer) bound upon incubation with ∼30 nM toxin for 4 h at 4 °C in suspension. This concentration of α-BgTx is larger than the dissociation equilibrium constant of the toxin from α7-AChR binding sites (50) by a factor >10. None of the mutations reported in this study occurred at positions predicted to affect the affinity of the α7-AChR for α-BgTx (51). The low temperature during the incubation was intended to minimize the uptake of toxin through endocytosis. Once the incubation was completed, the cells were pelleted by centrifugation, and the pellets were resuspended and washed with ice-cold phosphate-buffered saline solution. The pelleting–washing steps were repeated a second time, and the final (third) pellet was resuspended and solubilized in a solution consisting of 0.1 N NaOH and 1% (wt/vol) sodium dodecyl sulfate at 65 °C for 30 min. The radioactivity associated with each cell pellet was counted in a γ-counter for 1 min, and the amount of protein was estimated using the bicinchoninic-acid assay (Thermo Fisher). To account for the nonspecific binding of toxin, controls consisting of HEK-293 cells transfected with cDNA coding pLGIC subunits that do not form α-BgTx-binding sites (we used a mixture of β1, δ, and ε AChR subunits from mouse) were run in parallel. Throughout the paper, the expression levels of mutant chimeric constructs were indicated as mutant-to-CS ratios, and they are presented together in SI Appendix, Table S1.
Supplementary Material
Acknowledgments
We thank N. Godellas for assistance with cell culture and molecular biology; Y. Paas (Bar-Ilan University), W. N. Green (University of Chicago), M. M. Slaughter (University at Buffalo), and S. M. Sine (Mayo Clinic College of Medicine) for cDNAs; and M. Choi, S. Gough, N. Kowalczyk, A. Metropoulos, and S. Romo for additional technical assistance. This work was supported by a grant from the NIH (R01-NS042169 to C.G.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2021016118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Jackson M. B., Perfection of a synaptic receptor: Kinetics and energetics of the acetylcholine receptor. Proc. Natl. Acad. Sci. U.S.A. 86, 2199–2203 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Unwin N., Nicotinic acetylcholine receptor at 9 A resolution. J. Mol. Biol. 229, 1101–1124 (1993). [DOI] [PubMed] [Google Scholar]
- 3.Miyazawa A., Fujiyoshi Y., Unwin N., Structure and gating mechanism of the acetylcholine receptor pore. Nature 423, 949–955 (2003). [DOI] [PubMed] [Google Scholar]
- 4.Smit A. B., et al., A glia-derived acetylcholine-binding protein that modulates synaptic transmission. Nature 411, 261–268 (2001). [DOI] [PubMed] [Google Scholar]
- 5.Brejc K., et al., Crystal structure of an ACh-binding protein reveals the ligand-binding domain of nicotinic receptors. Nature 411, 269–276 (2001). [DOI] [PubMed] [Google Scholar]
- 6.Eiselé J.-L., et al., Chimaeric nicotinic-serotonergic receptor combines distinct ligand binding and channel specificities. Nature 366, 479–483 (1993). [DOI] [PubMed] [Google Scholar]
- 7.Grutter T., et al., Molecular tuning of fast gating in pentameric ligand-gated ion channels. Proc. Natl. Acad. Sci. U.S.A. 102, 18207–18212 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sunesen M., et al., Mechanism of Cl- selection by a glutamate-gated chloride (GluCl) receptor revealed through mutations in the selectivity filter. J. Biol. Chem. 281, 14875–14881 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Duret G., et al., Functional prokaryotic-eukaryotic chimera from the pentameric ligand-gated ion channel family. Proc. Natl. Acad. Sci. U.S.A. 108, 12143–12148 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Magnus C. J., et al., Chemical and genetic engineering of selective ion channel-ligand interactions. Science 333, 1292–1296 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tillman T. S., Seyoum E., Mowrey D. D., Xu Y., Tang P., ELIC-α7 nicotinic acetylcholine receptor (α7nAChR) chimeras reveal a prominent role of the extracellular-transmembrane domain interface in allosteric modulation. J. Biol. Chem. 289, 13851–13857 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmandt N., et al., A chimeric prokaryotic pentameric ligand-gated channel reveals distinct pathways of activation. J. Gen. Physiol. 146, 323–340 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ghosh B., Tsao T.-W., Czajkowski C., A chimeric prokaryotic-eukaryotic pentameric ligand gated ion channel reveals interactions between the extracellular and transmembrane domains shape neurosteroid modulation. Neuropharmacology 125, 343–352 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Price K. L., Lummis S. C. R., Characterization of a 5-HT3–ELIC chimera revealing the sites of action of modulators. ACS Chem. Neurosci. 9, 1409–1415 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Kash T. L., Jenkins A., Kelley J. C., Trudell J. R., Harrison N. L., Coupling of agonist binding to channel gating in the GABA(A) receptor. Nature 421, 272–275 (2003). [DOI] [PubMed] [Google Scholar]
- 16.Bouzat C., et al., Coupling of agonist binding to channel gating in an ACh-binding protein linked to an ion channel. Nature 430, 896–900 (2004). [DOI] [PubMed] [Google Scholar]
- 17.Calimet N., et al., A gating mechanism of pentameric ligand-gated ion channels. Proc. Natl. Acad. Sci. U.S.A. 110, E3987–E3996 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Du J., Lü W., Wu S., Cheng Y., Gouaux E., Glycine receptor mechanism elucidated by electron cryo-microscopy. Nature 526, 224–229 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang X., Chen H., Michelsen K., Schneider S., Shaffer P. L., Crystal structure of human glycine receptor-α3 bound to antagonist strychnine. Nature 526, 277–280 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Lev B., et al., String method solution of the gating pathways for a pentameric ligand-gated ion channel. Proc. Natl. Acad. Sci. U.S.A. 114, E4158–E4167 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jaiteh M., Taly A., Hénin J., Evolution of pentameric ligand-gated ion channels: Pro-loop receptors. PLoS One 11, e0151934 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hibbs R. E., Gouaux E., Principles of activation and permeation in an anion-selective Cys-loop receptor. Nature 474, 54–60 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Humphrey W., Dalke A., Schulten K., VMD: Visual molecular dynamics. J. Mol. Graph. 14, 33–38, 27–28 (1996). [DOI] [PubMed] [Google Scholar]
- 24.Xiu X., Hanek A. P., Wang J., Lester H. A., Dougherty D. A., A unified view of the role of electrostatic interactions in modulating the gating of Cys loop receptors. J. Biol. Chem. 280, 41655–41666 (2005). [DOI] [PubMed] [Google Scholar]
- 25.Treinin M., RIC-3 and nicotinic acetylcholine receptors: Biogenesis, properties, and diversity. Biotechnol. J. 3, 1539–1547 (2008). [DOI] [PubMed] [Google Scholar]
- 26.Gu S., et al., Brain α7 nicotinic acetylcholine receptor assembly requires NACHO. Neuron 89, 948–955 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Cymes G. D., Grosman C., Identifying the elusive link between amino acid sequence and charge selectivity in pentameric ligand-gated ion channels. Proc. Natl. Acad. Sci. U.S.A. 113, E7106–E7115 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Papke D., Grosman C., The role of intracellular linkers in gating and desensitization of human pentameric ligand-gated ion channels. J. Neurosci. 34, 7238–7252 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar P., et al., Cryo-EM structures of a lipid-sensitive pentameric ligand-gated ion channel embedded in a phosphatidylcholine-only bilayer. Proc. Natl. Acad. Sci. U.S.A. 117, 1788–1798 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bertrand D., et al., Positive allosteric modulation of the α7 nicotinic acetylcholine receptor: Ligand interactions with distinct binding sites and evidence for a prominent role of the M2-M3 segment. Mol. Pharmacol. 74, 1407–1416 (2008). [DOI] [PubMed] [Google Scholar]
- 31.Mishina M., et al., Location of functional regions of acetylcholine receptor α-subunit by site-directed mutagenesis. Nature 313, 364–369 (1985). [DOI] [PubMed] [Google Scholar]
- 32.Kobilka B. K., et al., Chimeric α 2-,β 2-adrenergic receptors: Delineation of domains involved in effector coupling and ligand binding specificity. Science 240, 1310–1316 (1988). [DOI] [PubMed] [Google Scholar]
- 33.Kubo T., et al., Location of a region of the muscarinic acetylcholine receptor involved in selective effector coupling. FEBS Lett. 241, 119–125 (1988). [DOI] [PubMed] [Google Scholar]
- 34.Conklin B. R., Farfel Z., Lustig K. D., Julius D., Bourne H. R., Substitution of three amino acids switches receptor specificity of Gq α to that of Gi α. Nature 363, 274–276 (1993). [DOI] [PubMed] [Google Scholar]
- 35.Kim J.-M., et al., Light-driven activation of β 2-adrenergic receptor signaling by a chimeric rhodopsin containing the β 2-adrenergic receptor cytoplasmic loops. Biochemistry 44, 2284–2292 (2005). [DOI] [PubMed] [Google Scholar]
- 36.Flock T., et al., Selectivity determinants of GPCR-G-protein binding. Nature 545, 317–322 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.García-Nafría J., Tate C. G., Cryo-EM structures of GPCRs coupled to Gs, Gi and Go. Mol. Cell. Endocrinol. 488, 1–13 (2019). [DOI] [PubMed] [Google Scholar]
- 38.Etter A., Cully D. F., Schaeffer J. M., Liu K. K., Arena J. P., An amino acid substitution in the pore region of a glutamate-gated chloride channel enables the coupling of ligand binding to channel gating. J. Biol. Chem. 271, 16035–16039 (1996). [DOI] [PubMed] [Google Scholar]
- 39.Filatov G. N., White M. M., The role of conserved leucines in the M2 domain of the acetylcholine receptor in channel gating. Mol. Pharmacol. 48, 379–384 (1995). [PubMed] [Google Scholar]
- 40.Labarca C., et al., Channel gating governed symmetrically by conserved leucine residues in the M2 domain of nicotinic receptors. Nature 376, 514–516 (1995). [DOI] [PubMed] [Google Scholar]
- 41.Ohno K., et al., Congenital myasthenic syndrome caused by prolonged acetylcholine receptor channel openings due to a mutation in the M2 domain of the epsilon subunit. Proc. Natl. Acad. Sci. U.S.A. 92, 758–762 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grosman C., Auerbach A., Asymmetric and independent contribution of the second transmembrane segment 12′ residues to diliganded gating of acetylcholine receptor channels: A single-channel study with choline as the agonist. J. Gen. Physiol. 115, 637–651 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guzmán G. R., et al., Tryptophan scanning mutagenesis in the αM3 transmembrane domain of the Torpedo californica acetylcholine receptor: Functional and structural implications. Biochemistry 42, 12243–12250 (2003). [DOI] [PubMed] [Google Scholar]
- 44.Purohit P., Gupta S., Jadey S., Auerbach A., Functional anatomy of an allosteric protein. Nat. Commun. 4, 2984 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grosman C., Zhou M., Auerbach A., Mapping the conformational wave of acetylcholine receptor channel gating. Nature 403, 773–776 (2000). [DOI] [PubMed] [Google Scholar]
- 46.Elenes S., Ni Y., Cymes G. D., Grosman C., Desensitization contributes to the synaptic response of gain-of-function mutants of the muscle nicotinic receptor. J. Gen. Physiol. 128, 615–627 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Elenes S., Decker M., Cymes G. D., Grosman C., Decremental response to high-frequency trains of acetylcholine pulses but unaltered fractional Ca2+ currents in a panel of “slow-channel syndrome” nicotinic receptor mutants. J. Gen. Physiol. 133, 151–169 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Papke D., Gonzalez-Gutierrez G., Grosman C., Desensitization of neurotransmitter-gated ion channels during high-frequency stimulation: A comparative study of Cys-loop, AMPA and purinergic receptors. J. Physiol. 589, 1571–1585 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fatima-Shad K., Barry P. H., Anion permeation in GABA- and glycine-gated channels of mammalian cultured hippocampal neurons. Proc. Biol. Sci. 253, 69–75 (1993). [DOI] [PubMed] [Google Scholar]
- 50.Corringer P.-J., et al., Identification of a new component of the agonist binding site of the nicotinic α 7 homooligomeric receptor. J. Biol. Chem. 270, 11749–11752 (1995). [DOI] [PubMed] [Google Scholar]
- 51.Marinou M., Tzartos S. J., Identification of regions involved in the binding of α-bungarotoxin to the human α7 neuronal nicotinic acetylcholine receptor using synthetic peptides. Biochem. J. 372, 543–554 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.