Abstract
Background
Choroid plexus tumors (CPTs) are intraventricular brain tumors predominantly arising in children but also affecting adults. In most cases, driver mutations have not been identified, although there are reports of frequent chromosome-wide copy-number alterations and TP53 mutations, especially in choroid plexus carcinomas (CPCs).
Methods
DNA methylation profiling and RNA-sequencing was performed in a series of 47 CPTs. Samples comprised 35 choroid plexus papillomas (CPPs), 6 atypical choroid plexus papillomas (aCPPs) and 6 CPCs plus three recurrences thereof. Targeted TP53 and TERT promotor sequencing was performed in all samples. Whole exome sequencing (WES) and linked-read whole genome sequencing (WGS) was performed in 25 and 4 samples, respectively.
Results
Tumors comprised the molecular subgroups “pediatric A” (N=11), “pediatric B” (N=12) and “adult” (N=27). Copy-number alterations mainly represented whole-chromosomal alterations with subgroup-specific enrichments (gains of Chr1, 2 and 21q in “pediatric B” and gains of Chr5 and 9 and loss of Chr21q in “adult”). RNA sequencing yielded a novel CCDC47-PRKCA fusion transcript in one adult choroid plexus papilloma patient with aggressive clinical course; an underlying Chr17 inversion was demonstrated by linked-read WGS. WES and targeted sequencing showed TP53 mutations in 7/47 CPTs (15%), five of which were children. On the contrary, TERT promoter mutations were encountered in 7/28 adult patients (25%) and associated with shorter progression-free survival (log-rank test, p=0.015).
Conclusion
Pediatric CPTs lack recurrent driver alterations except for TP53, whereas CPTs in adults show TERT promoter mutations or a novel CCDC47-PRKCA gene fusion, being associated with a more unfavorable clinical course.
Keywords: choroid plexus tumor, sequencing, TP53, TERT, fusion
Key Points.
Choroid plexus tumors in children lack recurrent driver alterations except for TP53.
Choroid plexus tumors in adults show TERT promoter mutations or a novel CCDC47-PRKCA gene fusion.
Importance of the Study.
Choroid plexus tumors are rare brain tumors derived from the choroid plexus epithelium. Despite frequent numerical aneuploidy and some choroid plexus tumors harboring TP53 alterations, driver mutations have not been identified in most cases. Herein, we performed DNA methylation profiling, RNA-sequencing, whole exome sequencing and linked-read whole genome sequencing in a large cohort of choroid plexus tumors. We show that pediatric choroid plexus tumors lack recurrent driver alterations except for TP53, whereas choroid plexus tumors in adults show TERT promoter mutations and gain of chromosome 5 or, in one patient, a novel CCDC47-PRKCA gene fusion with underlying inversion on chromosome 17.
Introduction
Choroid plexus tumors are intraventricular neoplasms derived from the choroid plexus epithelium which represent 10–20% of brain tumors arising throughout the first year of life,1,2 but may also affect older children and adults. Based on histopathological features, choroid plexus tumors can be divided into choroid plexus papilloma (CPP, WHO grade I), atypical choroid plexus papilloma (aCPP, WHO grade II) and choroid plexus carcinoma (CPC, WHO grade III).3 Patients harboring CPP usually experience good long-term outcome if gross total surgical resection can be achieved, whereas aCPP is associated with an increased risk of recurrence mainly in older childen (≥ 3 years) and adults.4–7 In contrast, prognosis of CPC is poor with five-year survival of 56% and most survivors exhibiting long-term cognitive and developmental deficits.8,9 On gene expression level, unsupervised clustering segregates CPC from most CPP and aCPP,10,11 whereas epigenetic profiling using high-density DNA methylation array segregates choroid plexus tumors into three distinct DNA methylation subgroups: supratentorial pediatric low-risk choroid plexus tumors (CPP and aCPP; “pediatric A”), supratentorial pediatric high-risk choroid plexus tumors (CPP, aCPP and CPC; “pediatric B”) and infratentorial low-risk choroid plexus tumors in adults (CPP and aCPP, “adult”).12,13
Despite these recent advances, little is known about genetic alterations involved in the tumorigenesis of choroid plexus tumors. Early cytogenetic studies have demonstrated numerical aneuploidy with CPP showing hyperdiploid karyotypes14,15 and CPC mainly exhibiting widespread chromosomal losses.16,17 More recent allele-specific copy-number analyses revealed hyperdiploidy in combination with acquired uniparental disomy (aUPD) in a subset of CPC,11 but the biological relevance of these chromosomal alterations remains unclear. The majority of CPC arises in children and CPC may be associated with Li-Fraumeni syndrome, a cancer susceptibility syndrome caused by germline mutations of the TP53 tumor suppressor gene.9 Somatic TP53 mutations have been described in as many as 36–60% of CPCs10,11 and have been linked to genomic instability and poor prognosis.9 However, many CPC and the vast majority of CPP lack TP53 mutations,12,18 strongly suggesting the presence of other tumor driving events.
Next generation sequencing has been successfully employed to identify driving genetic alterations in a variety of brain tumors,19,20 but only few choroid plexus tumors have been examined so far.21,22 We thus aimed to map DNA methylation profiles and genetic events in a series of choroid plexus tumors using Infinium Methylation 450k and EPIC BeadChip arrays, RNA-sequencing, whole exome sequencing and linked-read whole genome sequencing. Here we show that pediatric choroid plexus tumors lack recurrent driver alterations except for TP53, while adult choroid plexus tumors show TERT promoter mutations or, in one patient, a not yet described CCDC47-PRKCA fusion.
Methods
Patient samples
A total of 47 choroid plexus tumors (35 CPP, 6 aCPP and 6 CPC) and 3 recurrences thereof were collected from the archives of the Institutes of Neuropathology in Münster, Berlin, Heidelberg, Aachen, Calgary, Hamburg, Regensburg, Essen, and Würzburg. Histopathology was reviewed according to 2016 WHO criteria. In addition to formalin-fixed paraffin-embedded (FFPE) samples, fresh-frozen material was available for all 50 samples. For further comparison, three non-neoplastic choroid plexus samples were kindly provided by the Netherlands Brain Bank (NBB). Investigations were approved by the Münster ethics committee (2017-707-f-S).
DNA methylation profiling
After DNA isolation from FFPE samples using the Maxwell 16 Tissue DNA Purification Kit (Promega, Fitchburg, MA, cat# AS1030), purification and bisulfite conversion were performed using standard protocols provided by the manufacturer. Samples were analyzed using the HumanMethylation450 or MethylationEPIC BeadChip array (Illumina Inc., San Diego, CA, see Supplementary Materials).
RNA-Sequencing (RNA-Seq)
Total RNA was isolated from 49 fresh-frozen tissue samples as well as three non-neoplastic choroid plexus samples from the Netherlands brain bank (NBB) using the Direct-zol-96 RNA kit (Zymo Research, Irvine, CA) according to standard protocols (Supplementary Materials).
Gene fusion detection in RNA-Sequencing data
Six different tools were used to identify gene fusions in RNA sequencing data. Arriba v1.1.0, ChimeraScan v0.4.5a, EricScript v0.5.5b, FusionCatcher v1.00 and STAR-Fusion v1.8.0 were used with default settings. InFusion v0.8 was used with the following non-default parameter: --min-fragments 3. Fusion candidates predicted by EricScript were discarded if the internal classification score (“EricScore”) was ≤ 0.5 to better discriminate between true- and false-positive calls as recommended by the authors.23 The hg38 human reference genome was used for alignment. Fusion candidates of each tools were merged into a single fusion candidate list. Details of bioinformatic analyses and validation of the CCDC47-PRKCA fusion transcript are provided in the Supplementary Materials. The whole pipeline was evaluated using RNA-Seq data of two samples with known gene fusion status (pilocytic astrocytoma with KIAA1549-BRAF fusion and ependymoma with C11orf95-RELA fusion) from the Children’s Brain Tumor Tissue Consortium (CBTTC) database (Supplementary Figure S3).
10X linked-read whole genome sequencing (WGS)
Linked-read WGS was performed on 4 CPPs. DNA was extracted using the MagAttract HMW DNA kit (Qiagen, Hilden, Germany). For details see Supplementary Materials.
Whole exome sequencing (WES)
WES was performed on 25 CPTs using the Human Core Exome Kit v1.3 + RefSeq (Twist Bioscience, San Francisco, CA, Supplementary Materials). To exclude the possibility of sample mix-up, NGSCheckMate (v1.0.0, https://github.com/parklab/NGSCheckMate) was applied to WES and RNA-Sequencing data (Supplementary Figure S2). Further details regarding WES, variant calling in WES data and subsequent filtering are provided in the Supplementary Materials.
Targeted sequencing
Sanger sequencing of the TERT promotor was performed as previously described.24 For TP53 sequencing, a custom DNA target enrichment panel (Illumina) was used for library preparation and sequencing was performed on the Illumina NextSeq 550 platform (Illumina, San Diego, CA, USA). Generated fastq files were analyzed by SeqPilot software version 5.1.0 (Modul SeqNext, JSI Medical Systems) for alignment and variant calling.
Results
Patient characteristics
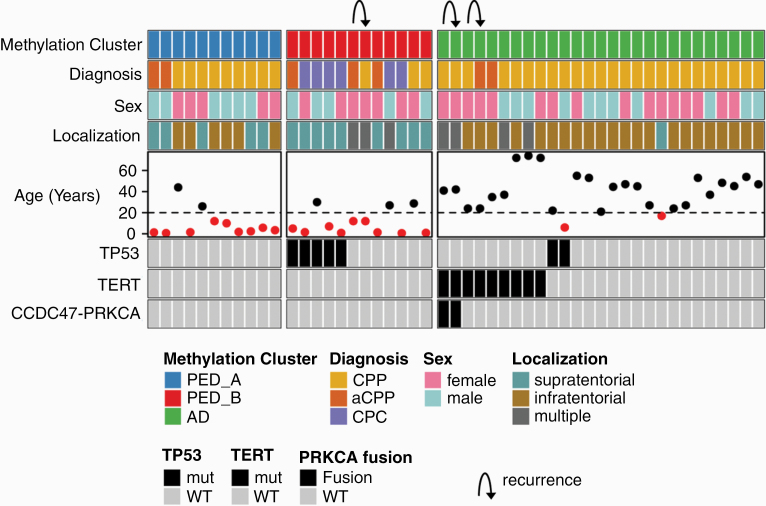
The median age of the 19 pediatric and 28 adult patients was 26 years (range, 0–74 years) (Supplementary Table S1). Fifteen tumors (32%) were located supratentorially (i.e., in the lateral ventricles), whereas 27 tumors (57%) were of fourth ventricular location and multiple locations involving supra- and infratentorial regions were observed in 5 patients (11%). Histopathologically, tumors comprised 35 CPP, 6 aCPP and 6 CPC (Figure 1). The three examined recurrences occurred 10–168 months after resection of the index samples (one pediatric aCPP and two adult CPP). Using DNA methylation-based classification (Heidelberg Brain Tumor Classifier version v11b4)13 followed by t-Distributed Stochastic Neighbor Embedding (t-SNE) analysis, all samples could be assigned to one of the three molecular choroid plexus tumor subgroups: 11 samples grouped with molecular subgroup “pediatric A” mainly comprising CPPs and aCPPs of pediatric patients and 11 choroid plexus tumors (including all 6 CPC) grouped with molecular subgroup “pediatric B,” while 25 choroid plexus tumors were classified as “adult” (Figure 1; Supplementary Figure S4). Molecular subgroup allocation was stable in the three recurrences (Supplementary Table S1).
Fig. 1.
Summary of patient characteristics and molecular alterations in choroid plexus tumors. Summary plot showing DNA methylation subgroup, diagnosis, sex, localization and age as well as TP53, TERT and PRKCA-fusion status. Age below 20 years is indicated by red dots. Arrows indicate recurrences that belong to the same individual.
Copy-number variations
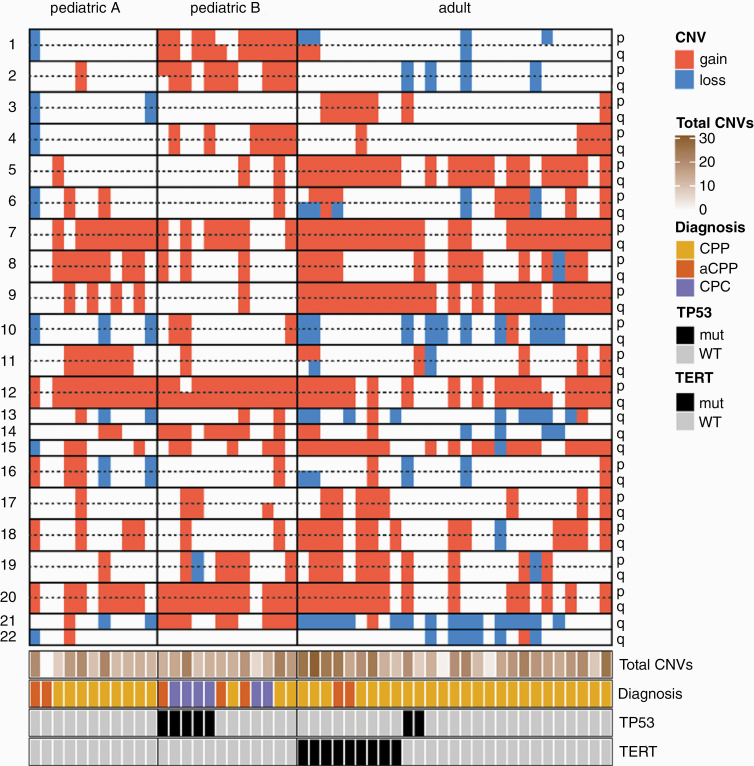
Copy-number alterations mainly comprised whole-chromosomal or chromosomal-arm alterations (Figure 2). The most frequently observed copy-number alterations were gains of chr12q (77%), chr12p (72%) and of whole chromosome 7 (72%). Most frequent chromosomal losses were chr21q (34%), whole chromosome 10 (23%) and chr13q (19%). However, in 2 of 3 recurrences, additional focal alterations were observed (Supplementary Figure S5). Only one choroid plexus tumor showed a balanced CNV profile. Copy-number profiles suggesting chromothripsis was not present in any sample. The total number of chromosomal arms being affected by gains or losses did not differ between the three molecular subgroups (pediatric A: 13 ± 6, pediatric B: 15 ± 5, adult: 15 ± 6, mean ± SD, P=N.S., One-Way ANOVA). Gains of chromosome 1, 2 and 21q were significantly enriched in “pediatric B” tumors (Fisher’s exact test, p < 1x10-3), while gains of whole chromosome 5 and 9 as well as losses of chromosome 21q are enriched in tumors of the “adult” subgroup (Fisher’s exact test, p < 1x10-3).
Fig. 2.
Copy-number alterations in choroid plexus tumors. Summary plot showing chromosomal imbalances according to the molecular subgroups. Total CNV count per sample (total number of chromosomal arms being affected by gains or losses) did not differ between the three molecular subgroups.
Gene fusion detection
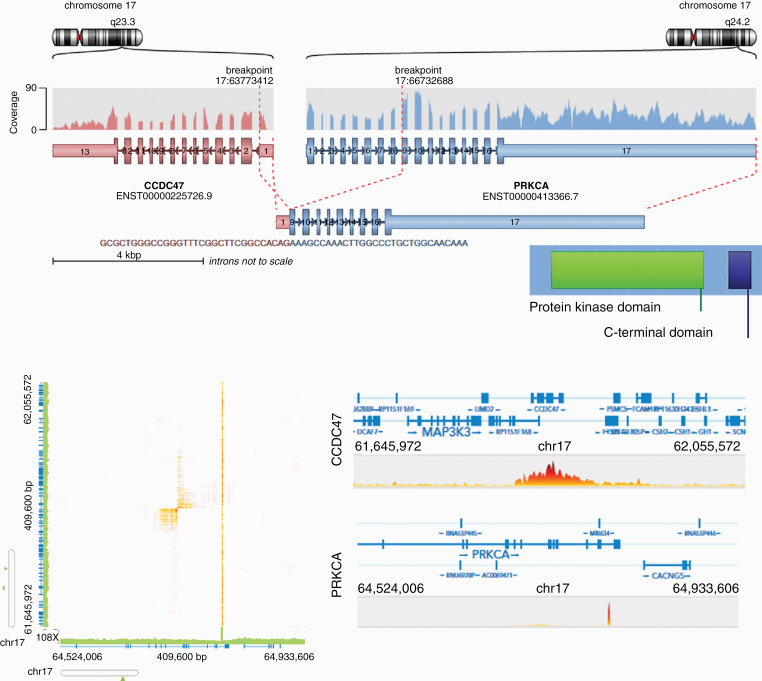
RNA sequencing of 49 samples was achieved with an average total number of 33.6 million paired-end reads (range, 23–45 million reads). After alignment against the hg38 reference genome, a total of 6 different fusion calling tools were used resulting in a total of 10491 gene fusion candidates. Overlap of fusion candidates predicted by each tool was poor with 9604/1049 (92%) fusion candidates uniquely called by either ericscript or chimerascan (Supplementary Figure S6A). The following filter criteria were applied: (i) fusion candidates were called by ≥ 3 tools, (ii) read support > 10, (iii) remove read-through fusion transcripts, (iv) remove fusions that were detected in non-neoplastic choroid plexus tissue or in the fusion hub database (Supplementary Figure S6B). Application of these filter criteria resulted in a final list of three fusion candidates (Table 1). While PTPRN2-WDR60 and UBR5-TMEM67 were each found in only 1 sample, CCDC47-PRKCA was detected in two recurrent tumors of the same patient (Figure 1; Supplementary Table S1). After initial resection, the infratentorial CPP of this patient had shown an aggressive biological behavior and a total of four recurrences including spinal (involving cervical, thoracic, lumbar and sacral segments) and supratentorial metastases occurred. The tumor was treated using a multimodal approach (etoposide, carboplatin, vincristine, cyclophosphamide, bevacizumab combined with radiotherapy). Treatment response was moderate with slight progression of spinal manifestations and currently, the patient is stable under continued bevacizumab treatment. The overall survival was 86 months since first tumor resection. Using the oncofuse algorithm25 to predict the oncogenic relevance of a fusion, only the CCDC47-PRKCA fusion (but not PTPRN2-WDR60 and UBR5-TMEM67) received a high driver probability score (0.99) and a low score for being a passenger event (0.09). The distance between the breakpoints is 2.9 Mb and the fusion is predicted to connect exon 1 (5’ UTR) of CCDC47 to exon 9 of PRKCA (Figure 3A). The fusion was subsequently validated by Sanger sequencing of cDNA using primers that span the breakpoint (Supplementary Figure S7). Using the orffinder tool (https://www.ncbi.nlm.nih.gov/orffinder/), the chimeric sequence was predicted to contain an open reading frame including the full-length protein kinase domain and the C-terminal domain of PRKCA. As CCDC47 is located on the minus-strand and PRKCA is located on the plus-strand of chromosome 17, the fusion most likely occurred due to an inversion event. This assumption was further supported by the fact that some fusion calling tools (including chimerascan, arriba, fusioncatcher and infusion) also predicted a reverse fusion with PRKCA as 5’ fusion partner and CCDC47 as 3’ fusion partner. Analysis of available RNA-sequencing datasets from two different platforms (17 CPPs and 4 CPCs from the CBTTC platform, 24 CPCs from the St. Jude platform) did not reveal any other case with a CCDC47-PRKCA fusion using our fusion calling pipeline and the Linux “grep” command for the fusion breakpoint sequence in various string lengths (data not shown).
Table 1.
Filtered gene fusion detection results
| 5“ Gene | 5“ Location | 3“ Gene | 3“ Location | Supporting reads | Samples |
|---|---|---|---|---|---|
| CCDC47 | 17:63773412 | PRKCA | 17:66732688 | 32 | CPP-10, CPP-23 |
| PTPRN2 | 7:158166931 | WDR60 | 7:158956583 | 14 | CPP-8 |
| UBR5 | 8:102272529 | TMEM67 | 8:93785222 | 85 | aCPP-6 |
Fig. 3.
Molecular features of the CCDC47-PRKCA fusion transcript. (A) The CCDC47-PRKCA fusion transcript juxtaposes the exon 1 of CCDC47 to exon 9 of PRKCA. Retained protein domains include the full-length protein kinase domain and the C terminal domain of PRKCA. (B) Linked-read whole genome sequencing showing an inversion event on chromosome 17q juxtaposing CCDC47 to PRKCA.
10X linked-read whole genome sequencing
To further explore structural variants (SVs), 10X linked-read whole genome sequencing (WGS) of four CPP samples (including one sample with a CCDC47-PRKCA fusion, Supplementary Table S1) was performed. The libraries were prepared on the Chromium platform resulting in a mean sequencing coverage of 36.6X producing approximately 900 million reads per samples with mean insert size of 360bp and an average molecule length between 21kb-59kb. First, a targeted analysis of the sample with known CCDC47-PRKCA fusion was performed. Using WGS data, we were able to identify the 2.9Mb inversion on chromosome 17 leading to the fusion between PRKCA and CCDC47 (Figure 3B). The inversion was called by both SvABA and NAIBR.
The entire dataset was then run through our in-house analysis pipeline. After germline calls from our database were removed and low-quality calls filtered, we were left with 60 unique SV calls which were manually validated. No SV calls expected to be somatic were recurrent across tumors and we did not find any recurrently affected genes.
Whole exome sequencing of choroid plexus tumors
Exome sequencing was performed in 25 choroid plexus tumors of 9 children and 15 adults of the DNA methylation subgroups “pediatric A” (n=6), “pediatric B” (n=7) and “adult” (n=12) (Supplementary Table S1). On average, 106 million reads (range, 75–151 million reads) were generated, resulting in coverage of targeted exome regions to a mean depth of 196X per sample (range, 133X – 282X). The median percentage of targeted bases which covered ≥50X across samples was 99%. Variant calling with appreci8, a recently developed pipeline combining and filtering the output of 8 variant callers,26 resulted in 18148 variants (Supplementary Figure S8). The overlap of the variants was high with >99.99% of the variants being called by ≥2 variant callers. According to our filter criteria (Supplementary Figure S9), a total of 526 non-synonymous single nucleotide variants (SNVs) and small-scale somatic insertion/deletions (indels) in coding regions were identified (Supplementary Table S2). The range of (likely) pathogenic variants and variants of uncertain significance per tumor was 11 to 44, with a median and standard deviation (SD) of 20 ± 8 mutations per tumor. Nine variants occurring in TP53, SF3B1, ERBB4, ERCC2, MSH6, TSC2 and SMARCA4 were classified as likely pathogenic or pathogenic according to the joint AMP-ASCO-CAP 2017 guidelines for cancer variant interpretation.27 As TP53 was the only recurrently mutated gene, targeted sequencing was performed for the remaining cases. As shown in Table 2, TP53 mutations were observed in 7/47 samples (4 CPC, 2 CPP and 1 aCPP) of five children and two adults and comprised missense mutations (R110L, R158C, K164fs*, C229G, R267L, A347T) and a small inframe deletion involving the DNA binding domain (N235_Y236). Variant allele frequencies (VAF) of TP53 mutations were above 70% in five cases, suggesting biallelic alterations in these tumors. Allele specific copy-number analyses from all four cases with available WES data revealed copy-neutral loss of heterozygosity (CN-LOH) of chromosome 17 in all three pediatric TP53-mutated cases (CPC-1, aCPP-1 and CPP-9) (Supplementary Figure S10A) and on RNA-sequencing, allelic imbalances of nearby single nucleotide polymorphisms (SNPs) were observed in all seven samples (Supplementary Figure S10B). Tumors harboring TP53 mutations did not show more chromosomal aberrations as compared to TP53-wildtype tumors (14 ± 5 vs. 16 ± 6, mean ± SD, P=N.S., Student´s t-test, Figure 2). A TSC2 mutation within the tuberin domain (E805K) was identified in aCPP-2 and allele specific copy-number analyses revealed CN-LOH of chromosome 16 in this tumor (Supplementary Figure S11). Similarly, a ERCC2 mutation (R965C) and CN-LOH of chromosome 19 were encountered in CPC-3 (Supplementary Figure S12). In contrast, missense mutations of SMARCA4 (S132P), SF3B1 (P1171L), MSH6 (K295R) and ERBB4 (R842W) were detected at VAFs around 50% in both whole exome and RNA sequencing data (Table 2) and the SMARCA4-missense-mutated case showed retained (normal) BRG1/SMARCA4 protein expression (Supplementary Figure S13). Manual inspection of the TERT promoter including the cases examined with linked-read whole genome sequencing as well as Sanger sequencing of the remaining cases revealed c.-124C>T (frequently called C228T) mutations in 8/50 samples (16%) and 1/50 samples (2%) with c.-146C>T (frequently called C250T) mutation (Supplementary Table S1; Figure 1). Mutations exclusively occurred in tumors of adults assigned the molecular subgroup “adult” (Figure 1) and were associated with significantly higher expression levels of the TERT gene (38.8 vs. 1.3 normalized counts; Kruskal-Wallis test, p= 1.3x10-6; Supplementary Figure S14A). Of note, there was no association of TERT expression and gain of chromosome 5 (Supplementary Figure S14B). Only one tumor (CPC-2) showed increased TERT expression without underlying promotor mutation (or gain of chromosome 5) (Supplementary Table S1). Additionally, CPP-7 showed a coding mutation of uncertain significance in TERT (A1014T), which was, however, not associated with increased TERT expression (Supplementary Table S1, Supplementary Table S2). Next, variants classified as “uncertain significance” according to the AMP-ASCO-CAP 2017 guidelines were also analyzed. A total of 42 genes were mutated in >1 sample (Supplementary Table S2). Manual curation and inspection revealed 2 JAK2 missense mutations (V414I in CPP-14 and M38I in CPC-3) as other promising candidates with CPC-3 showing evidence of allelic imbalance in RNA sequencing data favoring the mutated allele (Supplementary Figure S15).
Table 2.
Relevant coding mutations
| #ID | Gene | Location | Reference | Nucleotide change | Amino acid change | VAF WES (%) | VAF RNA-Seq (%) |
|---|---|---|---|---|---|---|---|
| CPC-1 | TP53 | chr17:7674254 | NM_001126113 | c.704_709del | p.N235_Y236del | 71 | 81 |
| CPC-2 | TP53 | chr17:7675140 | NM_001126113 | c.C472T | p.R158C | 59 | 78 |
| aCPP-1 | TP53 | chr17:7673820 | NM_001126113 | c.G800T | p.R267L | 91 | 86 |
| CPP-9 | TP53 | chr17:7676040 | NM_001126113 | c.G329T | p.R110L | 82 | 80 |
| CPP-30 | TP53 | chr17:7577596 | NM_001126113 | c.T685G | p.C229G | 64 | 89 |
| CPC-4 | TP53 | chr17:7578440 | NM_001126113 | c.491delA | p.K164Sfs*6 | 84 | 100 |
| CPC-6 | TP53 | chr17:7573988 | NM_001126112 | c.G1039A | p.A347T | 94 | 93 |
| CPP-5 | SMARCA4 | chr19:11098416 | NM_001128845 | c.T934C | p.S312P | 44 | 62 |
| aCPP-4 | SF3B1 | chr2:198260807 | NM_012433 | c.C3512T | p.P1171L | 50 | 52 |
| CPP-14 | ERBB4 | chr2:212295789 | NM_001042599 | c.C2524T | p.R842W | 51 | 70 |
| aCPP-2 | TSC2 | chr16:2129290 | NM_001318831 | c.G2413A | p.E805K | 93 | 100 |
| CPC-3 | ERCC2 | chr19:45855574 | NM_000400 | c.C2083T | p.R965C | 84 | 100 |
| CPP-8 | MSH6 | chr2:48026006 | NM_000179 | c.A884G | p.K295R | 50 | 53 |
| CPP-14 | JAK2 | chr9:5072537 | NM_001322204 | c.G1240A | p.V414I | 52 | 45 |
| CPC-3 | JAK2 | chr9:5050778 | NM_001322204 | c.G114A | p.M38I | 81 | 77 |
Patient outcome
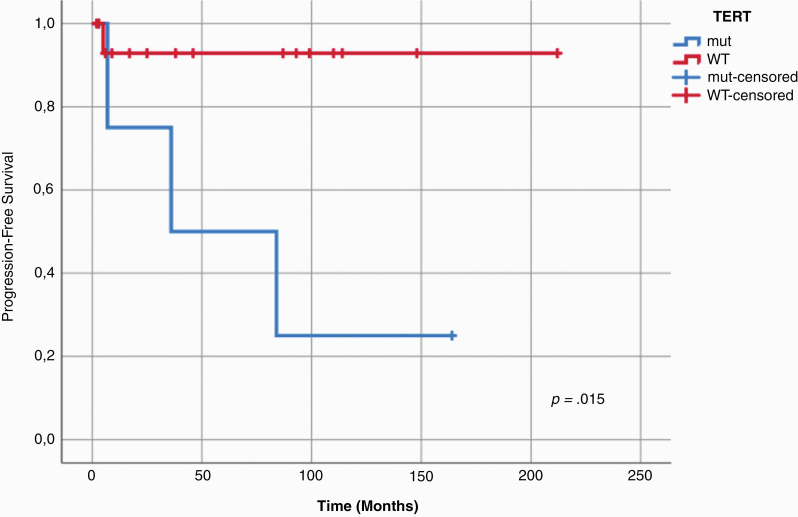
Follow-up information could be retrieved for 39/47 patients (83%) with a mean follow-up time of 74 months (Supplementary Table S1). Throughout the entire cohort, TP53 status was significantly associated with both shorter progression-free survival (log-rank test, p=0.004) and overall survival (log-rank test, p=0.024), and TERT status was associated with shorter progression-free survival (log-rank test, p=0.046) (Supplementary Figure S16). As TERT promotor mutations exclusively occurred in the “adult” subgroup, progression-free survival was next examined in 21 patients with tumors of this subgroup (16 TERT-wt and 5 TERT-mut) showing that TERT promotor mutation status may predict shorter progression-free survival in this subgroup (log-rank test, p=0.015; Figure 4).
Fig. 4.
Prognostic role of TERT promoter mutation status in adult CPT patients. Kaplan-Meier estimates of progression-free survival examined in 21 patients with CPP of the molecular subgroup “adult” suggest a prognostic role of TERT promoter mutation status (log-rank test, p = 0.015).
Discussion
Using a whole-transcriptome, whole-exome, and linked-read whole genome sequencing approach in a large cohort of epigenetically well characterized choroid plexus tumors, the genetic landscape of choroid plexus tumors was further refined. In line with previous observations,9–12 large-scale chromosomal imbalances were frequent and on whole-exome sequencing TP53 mutations were only present in a fraction of tumors. While the majority of TP53-mutant choroid plexus tumors were encountered in the molecular subgroup “pediatric B,” TERT promoter mutations resulting in new binding sites for members of the E-twenty-six (ETS) transcription factor family28 were exclusively detected in choroid plexus papillomas (CPP and aCPP) from adult patients. This finding is novel but well in line with the fact that TERT promoter mutations are associated with older age in other cancer types.29,30 Moreover, TERT promoter mutations were associated with shorter progression-free survival and might thus serve as a molecular marker to identify adult patients with CPP/aCPP at increased risk for recurrence, although this finding needs to be validated in larger cohorts. Recent large-scale analyses from the Pan-Cancer Analysis of Whole Genomes (PCAWG) consortium revealed that TERT promoter mutations are among the most common non-coding driver mutations across all cancers.31,32TP53 mutations in turn represents the most recurrent driver alteration in coding regions across adult and pediatric cancers.19,32 Although likely pathogenic variants of SF3B1, ERBB4, ERCC2, MSH6, TSC2 and SMARCA4 were found in CPP and aCPP samples, VAFs of the missense mutations (except for TSC2) was around 50% and a potential role in the pathogenesis of choroid plexus tumors needs to be determined by functional assays in the context of germ line testing, which was not available for the cases of the present retrospective series.
Ependymomas also show frequent DNA copy-number alterations33,34 and no recurrent SNVs or indels,33,35 but large-scale transcriptomic studies have revealed C11orf95–RELA fusion transcripts in a substantial proportion of supratentorial ependymomas.33 While RNA sequencing has become a precise and efficient standard for genome-wide screening of fusion transcripts, bioinformatic tools for fusion detection may show poor precision and recall metrics. It is therefore recommended to use an ensemble approach integrating the results of different programs.36 Using a total of six fusion callers and only considering candidate fusions detected by at least 3 tools with a read support of > 10 that have not been detected in non-neoplastic tissues, we found a total of 3 fusion candidates with only one fusion being predicted as an oncogenic driver.25 The CCDC47-PRKCA fusion in this CPP patient with aggressive clinical course leads to a chimeric transcript involving CCDC47 as the 5’ fusion partner and PRKCA, a protein kinase that has been previously detected in other tumors,37,38 as the 3’ fusion partner. The alpha isoform of protein kinase C is encoded by PRKCA and represents a family member of calcium and phospholipid-dependent serine/threonine kinases which can be activated by diacylglycerol and phorbol esters being involved in tumor formation and progression.39 Fusions involving PRKCA have been previously described in papillary glioneuronal tumor (PGNT) and a melanocytic skin neoplasm.37,38 Although the 5’ fusion partners were different (SLC44A1 in PGNT and ATP2B4 in the melanocytic skin neoplasm) in these tumors, the 3’ breakpoint in exon 9 of PRKCA was identical to our CCDC47-PRKCA fusion, thus preserving the catalytic kinase domain and cropping the 5’ regulatory domain from the resulting chimeric transcript. CCDC47 is located on the minus strand of chromosome 17q23.3, whereas the location of PRKCA is on the plus strand of chromosome 17q24.2 suggesting that this fusion arises due to an inversion on chromosome 17. Recently, 10X linked-read sequencing has been shown to combine low per-base error with long-range information for improved structural variation (SV) detection.40 Using this technology, we were able to confirm an inversion on chromosome 17q in this tumor. Of note, the CCDC47-PRKCA fusion has been detected in two recurrences of an adult patient with CPP. The clinical course of this patient was remarkable for an adult patient with infratentorial CPP including a total of four recurrences and supratentorial and spinal metastases despite an aggressive treatment regime including chemo- and radiotherapy. Although this fusion was not detected in any other sample in our cohort, the presence of fusion transcripts might play a role in a small subset of choroid plexus tumors. In retrospective clinical studies, adult patients with CPP and aCPP were shown to have a higher tendency of recurrence5 as compared to their pediatric counterparts7 and might warrant further investigation. Recently, a recurrent p.D463H mutation within the PRKCA kinase domain has been described in chordoid gliomas,41 but, however, manual inspection did not reveal any non-synonymous variant within the coding region of PRKCA in our samples.
One limitation of our study is that annotation and prioritization were biased towards known cancer mutations using the AMP/ASCO/CAP 201727 classification scheme that gives higher weight to mutations occurring in genes that already have been associated to other cancer types. Another drawback of this retrospective study is the fact that no matched blood samples or normal tissues were available for comparison. Further prospective analyses with matched control samples to unambiguously identify somatic mutations are desirable. This also includes more detailed examinations for mutations in non-coding regions that were beyond the scope of the present study. However, publicly available data derived from whole-genome sequencing of tumors and matched control samples from 20 TP53- and TERT-wildtype choroid plexus tumors comprising 16 CPPs and 4 CPCs of the Children’s Brain Tumor Tissue Consortium (CBTTC) lacked recurrent SNVs, indels and focal CNVs (https://pedcbioportal.org, accessed: 20.09.2020), further supporting the hypothesis that most choroid plexus tumors do not show recurrent driver mutations on the DNA level that are detectable with available methods.
In a recently published study from the Pan-Cancer Analysis of Whole Genomes (PCAWG) consortium, two tumor entities had a surprisingly low fraction of patients with identifiable driver mutations: chromophobe renal cell carcinoma and pancreatic neuroendocrine cancers.32 As in choroid plexus tumors, a remarkable feature of both tumor entities is a high degree of numerical aneuploidy, raising the possibility that certain combinations of whole chromosomal gains and losses may be sufficient to drive tumorigenesis in the absence of small-scale events such as SNVs, indels or genes fusions. Epimutations, i.e. specific heritable changes in epigenetic modification of DNA, as well as loss of imprinting represent other possible mechanisms to initiate tumorigenesis42,43 and warrant closer investigation in future studies using genome-wide comparison of CPT and non-neoplastic choroid plexus tissue.
In conclusion, our data further confirm that pediatric choroid plexus tumors lack recurrent driver alterations except for TP53. In addition, we find that adult choroid plexus tumors may show TERT promoter mutations or rarely a CCDC47-PRKCA fusion transcript, which was associated with an aggressive clinical course.
Funding
This study was funded by Deutsche Forschungsgemeinschaft (TH 2345/1-1) and further supported by a grant from La Fundación Bancaria ”la Caixa” (ID 100010434, code LCF/BQ/PI19/11690009) as well as a Young Investigator Award of the Alex´s Lemonade Stand Foundation to BR. LS is a fellow of the BIH-Charité Clinical Scientist Program and supported by a DKTK Young Investigator grant. US was supported by the Fördergemeinschaft Kinderkrebszentrum Hamburg. Further support was provided by the Canada Foundation for Innovation (CFI), Compute Canada Resource Allocation Project (WST-164-AB) and Genome Innovation Node (244819) to JR. MH is supported by IZKF Münster (Ha3/017/20).
Conflict of interest statement. The authors declare no potential conflicts of interest.
Authorship statement. Conceptualization: CT, BR, WP, MH; Investigation: CT, PS, MZ, SO, LS, AK, JN, US, FS, LR, KK, MP, MJR, JS, CR, OG, CMM, KL; Analysis and interpretation of the data: CT, PS, MZ, RS, JR, BR, MH. Funding acquisition: CT, BR, JR. All authors were involved in the writing of the manuscript and have read and approved the final version.
Supplementary Material
Acknowledgment
We thank patients and their families for their support of the study. The fusion calling pipeline was evaluated using data made available by The Children’s Brain Tumor Tissue Consortium (CBTTC). The Netherlands Brain Bank kindly provided non-neoplastic choroid plexus tissue samples. We also acknowledge the excellent technical support Core Facility Genomics of the Medical Faculty Münster. We thank Dr. Kathrin Oehl-Huber and Dr. Hannah Rabenstein (Institute of Human Genetics Ulm) for performing targeted TP53 sequencing. Susanne Peetz-Dienhart provided expert technical assistance for TERT sequencing. Carolin Walter and Sarah Sandmann (Institute of Medical Informatics, University Hospital Münster, Münster, Germany) provided expert bioinformatic advice.
References
- 1. Ostrom QT, Gittleman H, Liao P, et al. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2007–2011. Neuro Oncol. 2014;16(November):iv1–iv63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rickert CH, Paulus W. Tumors of the choroid plexus. Microsc Res Tech. 2001;52(1):104–111. [DOI] [PubMed] [Google Scholar]
- 3. Louis DN, Perry A, Reifenberger G, et al. World Health Organization Histological Classification of Tumours of the Central Nervous System. France: International Agency for Research on Cancer; 2016. [Google Scholar]
- 4. Siegfried A, Morin S, Munzer C, et al. A French retrospective study on clinical outcome in 102 choroid plexus tumors in children. J Neurooncol. 2017;135(1):151–160. [DOI] [PubMed] [Google Scholar]
- 5. Safaee M, Oh MC, Sughrue ME, et al. The relative patient benefit of gross total resection in adult choroid plexus papillomas. J Clin Neurosci. 2013;20(6):808–812. [DOI] [PubMed] [Google Scholar]
- 6. Jeibmann A, Hasselblatt M, Gerss J, et al. Prognostic implications of atypical histologic features in choroid plexus papilloma. J Neuropathol Exp Neurol. 2006;65(11):1069–1073. [DOI] [PubMed] [Google Scholar]
- 7. Thomas C, Ruland V, Kordes U, et al. Pediatric atypical choroid plexus papilloma reconsidered: increased mitotic activity is prognostic only in older children. Acta Neuropathol. 2015;129(6):925–927. [DOI] [PubMed] [Google Scholar]
- 8. Wrede B, Hasselblatt M, Peters O, et al. Atypical choroid plexus papilloma: clinical experience in the CPT-SIOP-2000 study. J Neurooncol. 2009;95(3):383–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tabori U, Shlien A, Baskin B, et al. TP53 alterations determine clinical subgroups and survival of patients with choroid plexus tumors. J Clin Oncol. 2010;28(12):1995–2001. [DOI] [PubMed] [Google Scholar]
- 10. Japp AS, Gessi M, Messing-Jünger M, et al. High-resolution genomic analysis does not qualify atypical plexus papilloma as a separate entity among choroid plexus tumors. J Neuropathol Exp Neurol. 2015;74(2):110–120. [DOI] [PubMed] [Google Scholar]
- 11. Merino DM, Shlien A, Villani A, et al. Molecular characterization of choroid plexus tumors reveals novel clinically relevant subgroups. Clin Cancer Res. 2015;21(1):184–192. [DOI] [PubMed] [Google Scholar]
- 12. Thomas C, Sill M, Ruland V, et al. Methylation profiling of choroid plexus tumors reveals 3 clinically distinct subgroups. Neuro Oncol. 2016;18(6):790–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Capper D, Jones DTW, Sill M, et al. DNA methylation-based classification of central nervous system tumours. Nature. 2018;555(7697): 469–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Donovan MJ, Yunis EJ, DeGirolami U, Fletcher JA, Schofield DE. Chromosome aberrations in choroid plexus papillomas. Genes Chromosomes Cancer. 1994;11(4):267–270. [DOI] [PubMed] [Google Scholar]
- 15. Roland B, Pinto A. Hyperdiploid karyotype in a choroid plexus papilloma. Cancer Genet Cytogenet. 1996;90(2):130–131. [DOI] [PubMed] [Google Scholar]
- 16. Li YS, Fan YS, Armstrong RF. Endoreduplication and telomeric association in a choroid plexus carcinoma. Cancer Genet Cytogenet. 1996;87(1):7–10. [DOI] [PubMed] [Google Scholar]
- 17. Neumann E, Kalousek DK, Norman MG, Steinbok P, Cochrane DD, Goddard K. Cytogenetic analysis of 109 pediatric central nervous system tumors. Cancer Genet Cytogenet. 1993;71(1):40–49. [DOI] [PubMed] [Google Scholar]
- 18. Carlotti CG Jr, Salhia B, Weitzman S, et al. Evaluation of proliferative index and cell cycle protein expression in choroid plexus tumors in children. Acta Neuropathol. 2002;103(1):1–10. [DOI] [PubMed] [Google Scholar]
- 19. Gröbner SN, Worst BC, Weischenfeldt J, et al. ; ICGC PedBrain-Seq Project; ICGC MMML-Seq Project . The landscape of genomic alterations across childhood cancers. Nature. 2018;559(7714):E10. [DOI] [PubMed] [Google Scholar]
- 20. Northcott PA, Buchhalter I, Morrissy AS, et al. The whole-genome landscape of medulloblastoma subtypes. Nature. 2017;547(7663):311–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tong Y, Merino D, Nimmervoll B, et al. Cross-species genomics identifies TAF12, NFYC, and RAD54L as choroid plexus carcinoma oncogenes. Cancer Cell. 2015;27(5):712–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Taher MM, Hassan AA, Saeed M, et al. Next generation DNA sequencing of atypical choroid plexus papilloma of brain: Identification of novel mutations in a female patient by Ion Proton. Oncol Lett. 2019;18(5):5063–5076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Benelli M, Pescucci C, Marseglia G, Severgnini M, Torricelli F, Magi A. Discovering chimeric transcripts in paired-end RNA-seq data by using EricScript. Bioinformatics. 2012;28(24):3232–3239. [DOI] [PubMed] [Google Scholar]
- 24. Qu Y, Shi L, Wang D, et al. Low frequency of TERT promoter mutations in a large cohort of gallbladder and gastric cancers. Int J Cancer. 2014;134(12):2993–2994. [DOI] [PubMed] [Google Scholar]
- 25. Shugay M, Ortiz de Mendíbil I, Vizmanos JL, Novo FJ. Oncofuse: a computational framework for the prediction of the oncogenic potential of gene fusions. Bioinformatics. 2013;29(20):2539–2546. [DOI] [PubMed] [Google Scholar]
- 26. Sandmann S, Karimi M, de Graaf AO, et al. appreci8: a pipeline for precise variant calling integrating 8 tools. Bioinformatics. 2018;34(24):4205–4212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li MM, Datto M, Duncavage EJ, et al. Standards and guidelines for the interpretation and reporting of sequence variants in cancer: a joint consensus recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J Mol Diagn. 2017;19(1):4–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Huang FW, Hodis E, Xu MJ, Kryukov GV, Chin L, Garraway LA. Highly recurrent TERT promoter mutations in human melanoma. Science. 2013;339(6122):957–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vinagre J, Almeida A, Pópulo H, et al. Frequency of TERT promoter mutations in human cancers. Nat Commun. 2013;4:2185. [DOI] [PubMed] [Google Scholar]
- 30. Koelsche C, Sahm F, Capper D, et al. Distribution of TERT promoter mutations in pediatric and adult tumors of the nervous system. Acta Neuropathol. 2013;126(6):907–915. [DOI] [PubMed] [Google Scholar]
- 31. Rheinbay E, Nielsen MM, Abascal F, et al. ; PCAWG Drivers and Functional Interpretation Working Group; PCAWG Structural Variation Working Group; PCAWG Consortium . Analyses of non-coding somatic drivers in 2,658 cancer whole genomes. Nature. 2020;578(7793):102–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. The ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium. Pan-cancer analysis of whole genomes. Nature. 2020;578(7793):82–93. doi:10.1038/s41586-020-1969-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Parker M, Mohankumar KM, Punchihewa C, et al. C11orf95-RELA fusions drive oncogenic NF-κB signalling in ependymoma. Nature. 2014;506(7489):451–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pajtler KW, Witt H, Sill M, et al. Molecular classification of ependymal tumors across all CNS compartments, histopathological grades, and age groups. Cancer Cell. 2015;27(5):728–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mack SC, Witt H, Piro RM, et al. Epigenomic alterations define lethal CIMP-positive ependymomas of infancy. Nature. 2014;506(7489):445–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liu S, Tsai WH, Ding Y, et al. Comprehensive evaluation of fusion transcript detection algorithms and a meta-caller to combine top performing methods in paired-end RNA-seq data. Nucleic Acids Res. 2016;44(5):e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bridge JA, Liu XQ, Sumegi J, et al. Identification of a novel, recurrent SLC44A1-PRKCA fusion in papillary glioneuronal tumor. Brain Pathol. 2013;23(2):121–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bahrami A, Lee S, Wu G, et al. Pigment-synthesizing melanocytic neoplasm with protein kinase C alpha (PRKCA) fusion. JAMA Dermatol. 2016;152(3):318–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Martiny-Baron G, Fabbro D. Classical PKC isoforms in cancer. Pharmacol Res. 2007;55(6):477–486. [DOI] [PubMed] [Google Scholar]
- 40. Elyanow R, Wu HT, Raphael BJ. Identifying structural variants using linked-read sequencing data. Bioinformatics. 2018;34(2):353–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rosenberg S, Simeonova I, Bielle F, et al. A recurrent point mutation in PRKCA is a hallmark of chordoid gliomas. Nat Commun. 2018;9(1):2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Crucianelli F, Tricarico R, Turchetti D, et al. MLH1 constitutional and somatic methylation in patients with MLH1 negative tumors fulfilling the revised Bethesda criteria. Epigenetics. 2014;9(10): 1431–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Steenman MJ, Rainier S, Dobry CJ, Grundy P, Horon IL, Feinberg AP. Loss of imprinting of IGF2 is linked to reduced expression and abnormal methylation of H19 in Wilms’ tumour. Nat Genet. 1994;7(3): 433–439. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.