Abstract
In the complex process of homeostasis of phytohormones cytokinins (CKs), O-glucosylation catalyzed by specific O-glucosyltransferases represents one of important mechanisms of their reversible inactivation. The CK O-glucosyltransferases belong to a highly divergent and polyphyletic multigene superfamily of glycosyltransferases, of which subfamily 1 containing UDP-glycosyltransferases (UGTs) is the largest in the plant kingdom. It contains recently discovered O and P subfamilies present in higher plant species but not in Arabidopsis thaliana. The cis-zeatin O-glucosyltransferase (cisZOG) genes belong to the O subfamily encoding a stereo-specific O-glucosylation of cis-zeatin-type CKs. We studied different homologous genes, their domains and motifs, and performed a phylogenetic reconstruction to elucidate the plant evolution of the cisZOG gene. We found that the cisZOG homologs do not form a clear separate clade, indicating that diversification of the cisZOG gene took place after the diversification of the main angiosperm families, probably within genera or closely related groups. We confirmed that the gene(s) from group O is(are) not present in A. thaliana and is(are) also missing in the family Brassicaceae. However, cisZOG or its metabolites are found among Brassicaceae clade, indicating that remaining genes from other groups (UGT73—group D and UGT85—group G) are able, at least in part, to substitute the function of group O lost during evolution. This study is the first detailed evolutionary evaluation of relationships among different plant ZOGs within angiosperms.
Subject terms: Evolution, Molecular evolution
Introduction
Many aspects of plant growth and development are coordinated by plant hormones. Among them, cytokinins (CKs) constitute one of the major groups, playing a key role in the control of cell division and elongation as well as organogenesis and many other physiological processes in plants. Natural isoprenoid CKs include N6-(∆2-isopentenyl)adenine (iP), trans-zeatin (transZ), cis-zeatin (cisZ) and dihydrozeatin (DHZ) and their derivatives. In the complex process of CK homeostasis, N- and O-glucosylation represent important mechanisms of CK irreversible and reversible inactivation, respectively, catalyzed by specific N- and O-glucosyltransferases (for review see e.g. Refs.1,2). Products of these conjugation steps, CK N- and O-glucosides, have been frequently identified in plants3, however, their roles in CK biology still remain somewhat unclear.
The CK O-glucosyltransferases belong to the large enzyme superfamily of glycosyltransferases (GTs; EC 2.4.x.y)4. These enzymes catalyze the transfer of sugar moieties from activated donor molecules to specific acceptor molecules, forming glycosidic bonds. These enzymes can be classified into 92 families, of which family 1 glycosyltransferases, often referred to as UDP glycosyltransferases (UGTs), is the largest in the plant kingdom4. A class of UGTs is defined by the presence of a C-terminal consensus sequence and can be found both in the plants and animals5. The nomenclature of this polyphyletic multigene family has been recommended based on the evolutionary divergence6. In early studies on Arabidopsis thaliana, 107 different UGTs were recognized7, and 14 distinct groups of UGTs were found5,8. However, relationships of some individual UGTs within well-supported subgroups were not strongly resolved due to a very high similarity among closely related sequences and sequenced motifs4. Only later research revealed 16 UGT groups based on an analysis of 12 fully sequenced genomes9,10.
Biosynthesis of phytohormones CKs in plants starts with transferring an isoprenoid moiety to an adenine present either in nucleotide form (transZ- and iP-type CKs) or bound to tRNA (cisZ-types). Two distinct origins of the isoprenoid moiety have been reported—(1) the methylerythritol phosphate (MEP) pathway localized in plastids and the mevalonate (MVA) pathway in the cytosol11,12. The first CK biosynthetic products, iP nucleotides, are further specifically hydroxylated at the N6-side chain to form cisZ or transZ (for review see e.g. Ref.13), which may be subsequently conjugated to form corresponding O-glucosides. In addition, indirect evidence suggests the existence of a biosynthetic pathway for zeatins without iP intermediates14.
Another biosynthetic pathway producing cisZ-type CKs involves the release of CKs by a turnover of certain tRNAs. Although cisZ-type CKs have been reported as essential components of some tRNAs in plants15, isoprenoid CKs generally occur as structural parts of certain tRNA species of all organisms from eubacteria (but not in archaebacteria) to humans16. Considering the abundance of cisZ-types in the plant kingdom17 and early calculations of tRNA turnover rates18, tRNA degradation does not, however, seem to be a sole pathway for cisZ formation in plants.
The O-glycosylation of CKs represents a reversible step leading to rapid and efficient CK deactivation. It is catalyzed by O-glucosyltransferases that may recognize trans-zeatin (transZ), cis-zeatin (cisZ) and dihydrozeatin (DHZ), i.e. CKs having an available hydroxyl group for glucosylation. The enzymes catalyzing CK O-conjugation, O-glucosyltransferase (ZOGT, EC 2.4.1.20319) and O-xylosyltransferase (ZOXT, EC 2.4.1.20420) were first reported in the common bean (Phaseolus vulgaris) and lima bean (Phaseolus lunatus), respectively. Their biochemical characterization showed different CK substrate specificities and sugar donor recognition, and the corresponding genes were cloned by Mok’s group21,22. Later on, two genes encoding an O-glucosyltransferase specific to cisZ (cisZOG1, cisZOG2) were isolated and characterized in maize23,24, and subsequently, three cisZ-specific O-glucosyltransferases (cZOGT1, cZOGT2, and cZOGT3) were identified from rice25. In Arabidopsis, CK O-glucosylation through O-glucosyltransferases is coded by three UGTs (UGT85A1, UGT73C5 and UGT73C1) producing O-glucosides with transZ, cisZ and DHZ26. Although two of these enzymes (UGT73C5 and UGT73C1) exhibit very low activity and utilize also other substrates27,28, UGT85A1 represents zeatin O-glucosyltransferease with a preference for transZ and substantially contributes to CK homeostasis29,30.
Additionally to the findings above, researchers directed their attention to the grass family (Poaceae), namely Zea mays, Sorghum bicolor and Oryza sativa, and examined the evolutionary pattern of gene duplication of the cisZOG gene31. Having estimated duplication times for cisZOG homologs, they found cisZOG genes in tandem triplication in rice, five genes in sorghum and one maize gene.
Generally, most genes belong to larger gene families, and the analysis of gene family histories plays an important role in the study of genome evolution. The crucial point is recognition among orthologous genes referring to copies of genes that reveal the phylogeny of species and paralogous genes that evolved by duplication events. For years, knowledge on the organization of the UGTs family was limited to the model plant Arabidopsis thaliana. Analysis of this superfamily led to categorization into 54 families, including family 1, which contains UGTs8. The largest UGT1 class contains 16 subfamilies. The UDP family expanded during the transition from algae to vascular plants9. In this study based on 11 sequenced plant genomes9, five phylogenetic groups (A, D, E, G and L) have been recognized to expand more than the others during the evolution of the higher plants (8–50 members depending on plant species). Other groups were represented from 1 to 13 members. In A. thaliana group G includes only six members (about 6% of the total UGTs), group H has expanded to become the second most abundant group in this species (18%)9. Interestingly, the newly discovered phylogenetic groups O and P9 were not found in Arabidopsis thaliana. On the other hand, three sequences available in NCBI databases are named cisZOG1-3 and belong to A. thaliana.
To address this discrepancy, we directed our attention to group O, which contains unique highly conserved residues in the PSPG motif (Plant Secondary Product Glycosyltransferase; i.e., UGT Prosite consensus32) at positions 41 and 42 (His and Ser, respectively) that are not present in any other 1 UGT-glucosyltransferase phylogenetic group. The evolutionary relationships among plant ZOGs are unknown.
Here, we analyzed a large data set of publicly available amino acid sequences with emphasizing the complete genomes of cisZOGs to classify all representatives available across the plant kingdom. We studied different homologous genes belonging to the UGT1 class subfamily O within angiosperms, their domains, motifs, exon/intron organizations and their phylogenetic relationships. In the wide context of angiosperms, comparative analysis of cisZOG phylogeny and protein structural properties allowed us to identify the diversification of two main clades of monocots and eudicots. These main clades could have expanded after divergence from their common ancestor. The wide sampling of cisZOG orthologs and paralogs provides evidence of cisZOG diversification occurring after the diversification of the main angiosperm families, probably within genera or closely related groups. Additionally, we present evidence that the cisZOG gene is not present in Arabidopsis thaliana and furthermore is missing in the other members of the family Brassicaceae.
Results
ZOG gene identification, conserved motif analyses and pairwise similarity approaches endorsing distinct ZOG clades
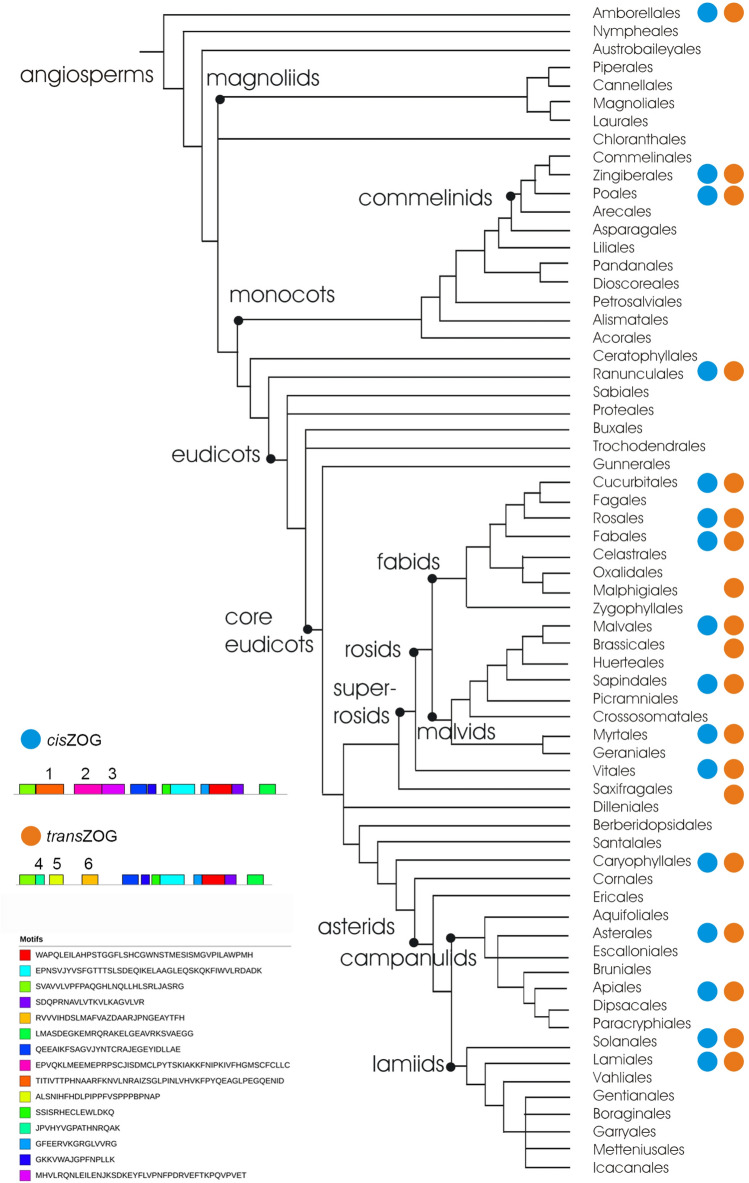
Our combined approach, BLASTP search of fully sequenced genomes via Phytozome v12 and all publicly available cis-zeatin O-glucosyltransferase homologs from databases, gave us significant results. Homology searches of taxa through databases were crucial for the accuracy of phylogenetic inference and analyses of motifs and domains. From these searches, we did not obtain sequences of Brassicaceae. Compilation of data from completely sequenced genomes allowed us to build a matrix to identify major events of gene duplication and losses in angiosperms during evolution, find trans-zeatin O-glucosyltransferase (transZOG) proteins, and explore Brassicaceae ZOGs and indicate complex outline to cis- and tranZOG distribution within angiosperms (Fig. 1). CisZOG belonging to the phylogenetic group O, which contains the PSPG motif with His and Ser at positions 41 and 42, have typical conserved motifs 1, 2 and 3 (Fig. 1). TransZOG belonging to the group D contains nine same motifs as cisZOG, but typically have motifs 4, 5 and 6 (Fig. 1).
Figure 1.
Simplified consensus phylogenetic tree of angiosperms adapted from APG IV. (2016) showing groups where cis- or transZOG were found (based on Phytozome v12) and typical sequence motifs for them. CisZOG contains conserved motifs 1,2 and 3, transZOG 4, 5 and 6.
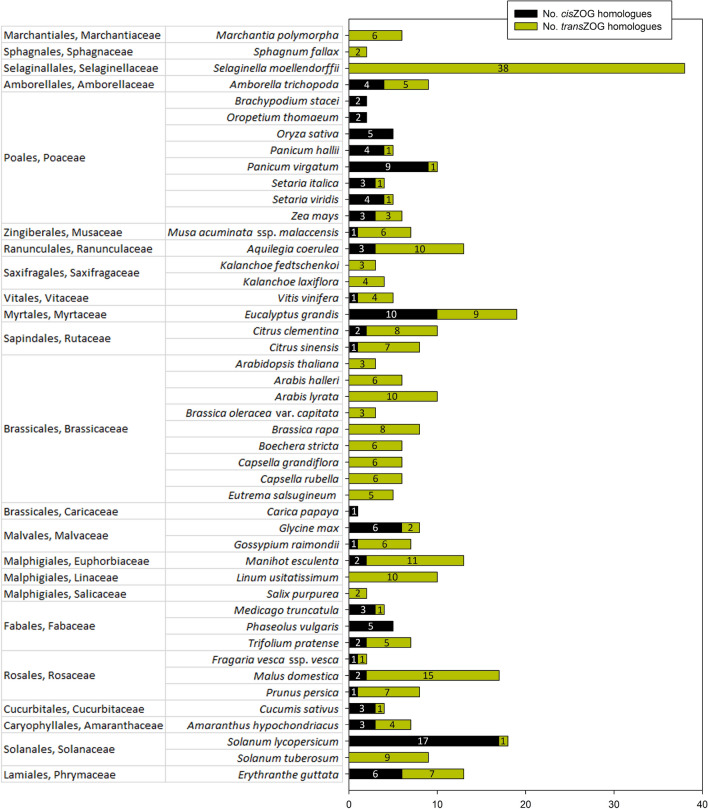
Most often, one to five cisZOG genes per plant species were revealed. However, in grasses, Panicum virgatum contained the most cisZOG homologs in monocots: nine. Four or five homologs were found in other representatives from Poales (Oryza sativa and Panicum hallii). The highest number represents plants from eudicots, Solanum lycopersicum (Solanales) with 17 cisZOG homologs and Eucalyptus grandis (Myrtales) with 10 cisZOG homologs (Fig. 2).
Figure 2.
Quantity of cis- and transZOG homologs of angiosperm species from complete genomes (Phytozome v. 12) sorted by phylogenetic system. Species from bryophytes (Marchantia polymorpha, Sphagnum phallax) and lycophytes (Selaginalla moellendorffii) are shown for comparison with angiosperms.
We screened representatives from each of the major evolutionary groups for their main motifs and domains. The cisZOG gene belonging to the O group of glycosyltransferases is characterized by a conserved protein PSPG motif (Fig. 3). This motif is specific for monocots and eudicots and has three well-characterized parts common to both groups (1: 5′-PQLEIL-3′, 2: 5′-FMSHCGWNS-3′ and 3: 5′-WPMHSDQ-3′). A detailed summary of clade/order-specific PSPG motifs is shown in Supplementary Fig. S1 (Amborellales; monocots: Asparagales-Arecales and Poales; dicots: Proteales-Ranunculales-Vitales, Fabales, Rosales, Fagales-Cucurbitales, Malphigiales, Myrtales-Sapindales-Malvales, Brassicales-Caryophyllales-Gentianales, Solanales, Lamiales-Asterales-Apiales). The conserved part were the sugar donor residues, e.g., W in the position 22, D—43 and Q—44 that are positioned to form hydrogen bonds to the sugar part of the donor as described33. Lastly mentioned, glutamine (Q) conserved in group D as well, is important for the maximal catalytic efficiency of glucosyl transfer activity34,35 and is highly conserved within all UGT groups (A-P as described by Ref.9). Furthermore, within group O the sugar donor residues W/P/H/E in the position 1/3/19/27 are invariable across all studied phylogenetically diverse species.
Figure 3.
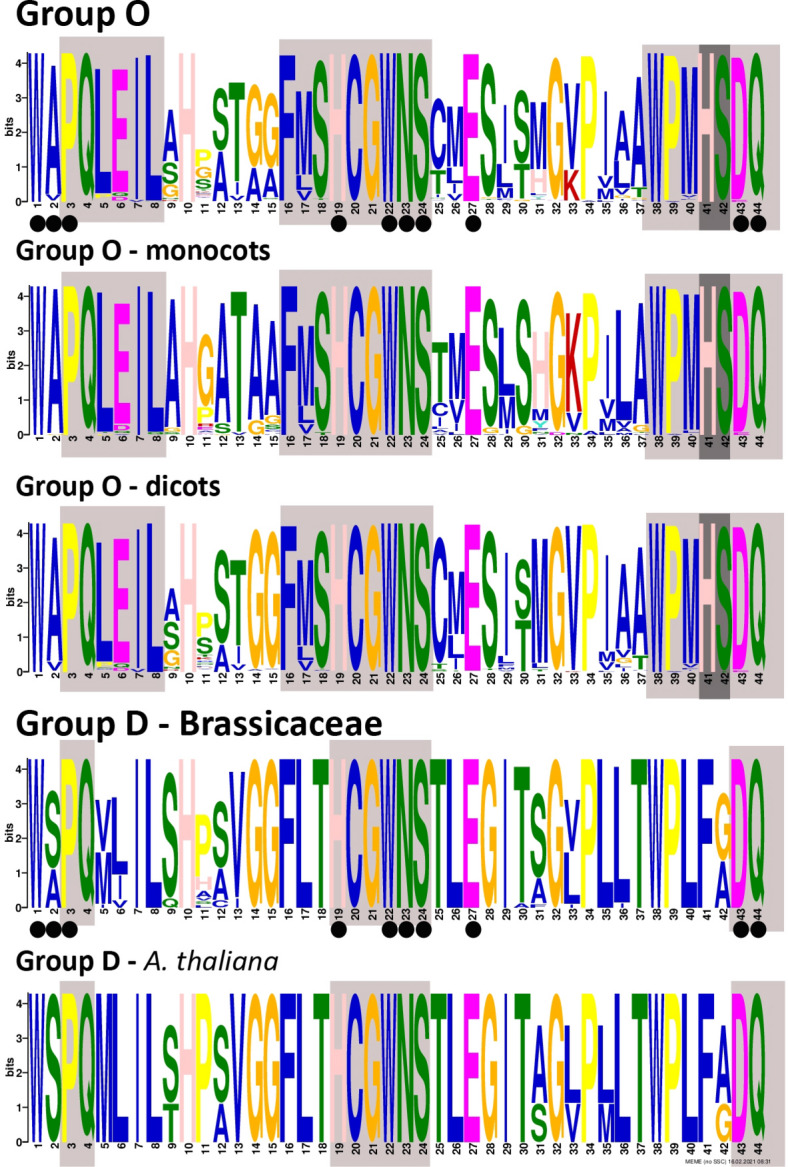
The conserved plant secondary product glycosyltransferase (PSPG) motif in group O in all taxa analyzed, monocots and eudicots in comparison with group D represented by Brassicaceae family. Highly conserved amino acids are shown in boxes (Group O: 1: 5′-PQLEIL-3′, 2: 5′-FMSHCGWNS-3′ and 3: 5′-WPMHSDQ-3′; group D: 1: 5′-PQ-3′, 2: 5′-HCGWNS-3′ and 3: 5′-DQ-3′). Group O—cisZOG is characterized by 5′-HS-3′ in the position 41 and 42 of PSPG. Circles indicate the residues interacting directly with the UDP-sugar based on available crystal structures according to Ref.9.
TransZOG, belonging to the group D have similar PSPG motif (1: 5′-PQ-3′, 2: 5′-HCGWNS-3′ and 3: 5′-DQ-3′), but differ in position 41 and 42, where they have instead of 5′-HS-3′ a 5′-FG-3′ or 5′-FA-3′ (Fig. 3).
Group H (UGT76) known to catalyze N7 and N9 glycosylation of cytokinins differ from group O (cisZOG) in position 5, 7, 11–13, 18, 20, 25, 26, 30, 31, 35–38, 40–42 of PSPG motif. Sugar donor residues are stable.
We identified 15 different conserved motifs shared among related proteins in the whole proteins (Supplementary Fig. S1), and most of the cisZOG genes had a similar intron-phasing distribution.
Furthermore, we searched the Pfam Motif Library and the NCBI Domain Architecture Retrieval Tool for unique motifs characteristic of the ZOG protein and found the most common glycosyltransferase family 28 C-terminal domain (Pfam PF04101; Supplementary Table S1) across angiosperms. Cerato-platanin (Pfam PF07249) was found in four families: Fabaceae, Malvaceae, Amaranthaceae and Solanaceae. Family Solanaceae contains one specific motif, the choline kinase N-terminus (Pfam PF04428), and similarly, members of Fabaceae contain bacterial toxin 8 (Pfam PF15545). Interestingly, Coffea arabica (Rubiaceae) contains a putative bacterial lipoprotein (DUF799, Pfam PF05643). Moreover, the sequence similarity is higher within cisZOG genes in monocots than in dicots. The cisZOG protein identity varies from 52 to 96.3% in Zea mays or 40.1–96% in Oryza sativa cv. Japonica. In general, cisZOG sequence identity varies from 23% (Amborella trichopoda and Sorghum bicolor) to 99.2% (Solanum penelli and Solanum lycopersicum). There were also differences within one genus: Nicotiana attenuata ZOG genes showed 49.2–61.6% sequence identity, N. sylvestris 51.4–78%, N. tabacum 62.2–78.8 and N. tomentosiformis 49.2–77.8%.
Phylogenetic reconstruction
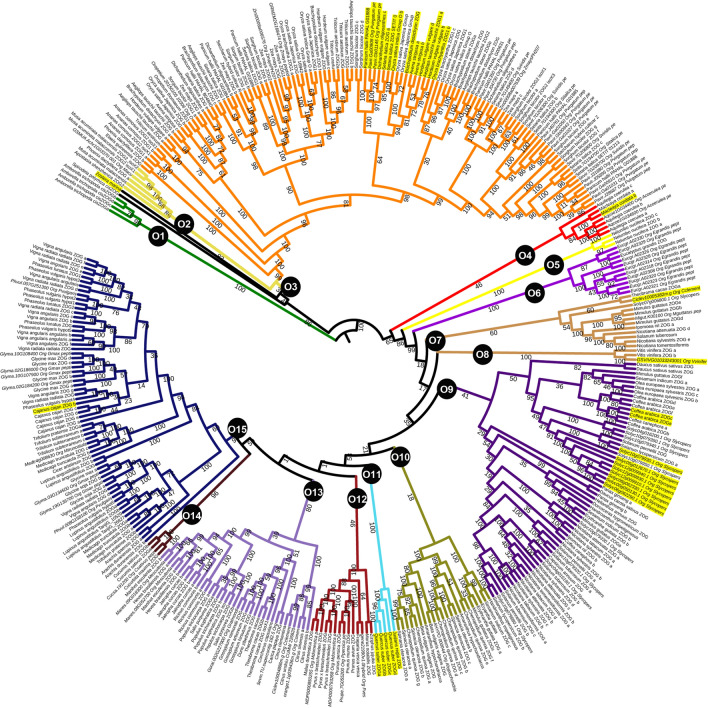
The evolutionary relationships among the cisZOG proteins were determined using maximum likelihood (ML; Fig. 4, simplified Supplementary Fig. S2) and maximum parsimony (MP; Supplementary Fig. S2a,b) analyses based on multiple alignment, producing a phylogenetic tree depicting the relationships among all currently accessible cisZOG sequences. A member of the basal angiosperm Amborella trichopoda (Amborellaceae, Amborellales), sister to the rest of the tree in our phylogeny, was used as the outgroup. The first analysis contained 116 sequences of 42 plant species with a total amino acid alignment of 1608 positions in the final data set (not shown). The second analysis with included data from homology searches via NCBI contained 376 sequences for 96 plants and 1719 positions in the aligned matrix (Fig. 4). Three hundred seventy-six sequences were analyzed for the presence of the PSPG motif, and three hundred forty-three contained the motif, i.e., only thirty-three sequences did not contain His and Ser at positions 41 and 42 (Fig. 4). Analyses with full sampling of cisZOG proteins (Fig. 4), selection of fully sequenced genomes only and selection to species with PSPG motif only resulted in the same phylogenetic results. ML and MP analyses revealed 15 distinct groups (O1–O15), MP analysis resulted in more clades (e.g., O4 is divided in 3 clades, O12 to 2 clades), however this finding should to be result of long-branch phenomenon and we can take in mind that these analyses are based on rather artificial selection of available sequence information.
Figure 4.
Maximum likelihood phylogenetic tree of cis-zeatin O-glucosyltransferase based on combined data from Phytozome v12 and GenBank. Twenty-seven species were included in 376 sequences with 1719 positions in the final data set. The ML log likelihood is − 112,690.187027. CisZOG homologs of Amborella trichopoda were used as outgroups. The tree is divided into fourteen main groups: O1. Amborellaceae, O2. Musaceae, O3. Arecaceae-Bromeliaceae-Musaceae-Orchideaceae-Poaceae, O4. Ranunclulaceae, O5. Myrtaceae, O6. Asteraceae-Apiaceae-Araliaceae-Oleaceae-Phrymaceae-Rubiaceae-Solanaceae, O7. Amaranthaceae, O8. Rutaceae-Phrymaceae, O9. Vitaceae, O10. Moraceae-Rhamnaceae-Rosaceae, O11. Cucurbitaceae, O12. Malvaceae-Euphorbiaceae-Rutaceae, O13. Fabaceae, O14. Nelumbonaceae, O15. Fagaceae. Numbers below branches indicate bootstrap support values > 50%. Yellow boxes refer to sequences without a two highly conserved residues, histidine and serine, at positions 41 and 42 in PSPGD domain, italics indicate sequences from Phytozome.
The phylogeny of single cisZOG proteins in an angiosperm lineage is mostly reflected by their taxonomy. Surprisingly, we did not find the cisZOG protein in Arabidopsis thaliana or in other representatives of the family Brassicaceae and, in general, in most of the order Brassicales except for Carica papaya. However, we also analyzed a data set of fully annotated sequences of 48 species that contained 148 cisZOG sequences (1607 bp), including representatives from Brassicaceae (Supplementary Fig. S3). We revealed that Brassicaceae species clustered together with very short branch lengths and showed different domains in comparison with the rest of the sequences (Supplementary Fig. S3). We performed an additional BLASTP search and found sequence homology with UGT73C. Our BLAST and phylogenetic analyses of annotated cisZOG in Arabidopsis thaliana (cisZOG1: AY573820.1, cisZOG2: AY573821 and cisZOG3: AY573822) revealed identity with UDP glucosyltransferase 73C (UGT73C1, UGT73C6 a UGT73C5) belonging to group D but not to group O as demonstrated by other ZOG genes.
The evolutionary history of the cisZOG gene supported independent expansions in monocots and dicots. We recognized two main groups based on clade support values: monocot clade and eudicot clade with 100% (99%) and 62% (65%) BS, respectively. Within monocots (Fig. 1, branch O3), three main basal clades contain Orchideaceae, Musaceae and Arecaceae (BS 83%), Arecaceae (BS 100%) and Bromeliaceae (BS 100%). These clades are followed by seven branches of Poaceae (BS 81–100%). In eudicots, 12 main clades were recognized (Fig. 1, branch O4–O15) with bootstrap support from 80–100%, except for three cases, where BS was lower (branch O10: Amaranthaceae, BS 18%, F: Asteraceae-Apiaceae-Araliaceae-Oleaceae-Phrymaceae-Rubiaceae-Solanaceae, BS 41%, branch O12: Moraceae-Rhamnaceae-Rosaceae, BS46%).
Discussion
Phylogenetic diversification of group O UGTs in angiosperms
Glycosyltransferases represent a highly divergent multigene family6, where the activities of some subgroups are highly conserved among different plant species, while in others, the substrate specificity shifts with relative ease5. After their expansion in vascular plant lineages, UGTs independently acquired their ability to recognize specific compounds as substrates4. UGT genes undergo rapid evolution and changes in copy number, making it difficult to identify orthologs and paralogs36.
Although CKs have been detected within the plant kingdom ranging from algae to land plants, cisZOG genes have been found only in angiosperms so far (for review see e.g. Refs.2,37). In general, ZOG belongs to group O of family 1 UGTs. It contains a low number of proteins (2–9), together with groups B (1–9), F (1–6) and N (1–13), in angiosperms9. The cisZOG and transZOG genes apparently originated and diversified during the evolution of angiosperms, which represents the most recent evolutionary explosion of embryophytes, a lineage that occupied land at least 470 million years ago38.
Identification of major protein changes and diversification to two main clades predate eudicot and monocot divergence (Fig. 3, Supplementary Fig. S2). These main clades could have expanded after divergence from their common ancestor. Our phylogeny reconstruction of group O suggests that the cisZOG phylogeny corresponds with a phylogenetic view given by genes reflecting the evolution of angiosperms in general. It is also consistent with9 who also showed that the sequences from the different plant species within each phylogenetic group generally have a tendency to cluster together, albeit in some cases they appear to be scattered. On the other hand, the cisZOG homologs do not form a clear separate clade, as described in other protein families39,40. This indicates that diversification of the cisZOG gene took place after the diversification of the main angiosperm families, probably within genera or closely related groups.
From one to five cisZOG genes per plant species were revealed. The reconstructed history of cisZOG gene duplication identified by Ref.24 presumed that the most common ancestor of grasses contained four copies of the cisZOG gene, and diversification is the result of speciation. During evolution, different numbers of cis- and transZOG genes were apparently duplicated and lost. Duplication of genes is common in plants, where whole genome duplication (WGD) or polyploidy often takes place (polyploid plants range from 30 to 70% in angiosperms). Duplication also played a role in the evolutionary history of angiosperms in determining species richness and diversification. Most of the WGD occurred during periods of environmental instability41. These events result in the retention of multiple gene paralogs that may lead to their subfunctionalization, neofunctionalization or redundancy42. Although gene duplication is followed by the loss of one of the gene copies, sometimes both copies are retained. In such case, they will initially be redundant, providing an opportunity for one of the paralogs to change function. Existing protein families did not evolve from one common ancestor but under a multiple-birth model43. A higher number of paralogs enables avoidance of purifying selection because the activity of some of them is not needed by the cell44. Moreover, analyses of WGD within Brassicales showed that gene duplication and loss rates vary across land plants, and different gene families have different probabilities of being retained following a WGD45. As shown in Tarenaya hassleriana (Brassicales, Cleomaceae), the example of evolutionary fate and functional consequences of a transposition event at the base of Brassicales resulted in the duplication of the floral regulator PISTILLATA46. It was previously pointed that gene duplication itself tends to promote divergence of gene expression, most likely just because of their redundancy44.
There is a high number of cisZOG isoforms in plants. It corresponds to the results of Ref.47, who found conserved alternative splicing events in monocots, particularly across grass species. Isoforms, as the most common result of alternative splicing, are common in plants. Genome-wide transcriptome mapping has revealed the extent of alternative splicing in plants ranging from 42 to 61%48,49. Recent data suggest that posttranscriptional regulation, especially alternative splicing, is necessary as a regulatory mechanism for plants to adapt to environmental changes50, i.e., similar to paralogs.
Missing group O UGTs within the family Brassicaceae
The glycosyltransferase superfamily contains 107 UGT genes and 10 UGT pseudogenes in Arabidopsis thaliana5. Our BLAST and phylogenetic analyses of ZOG genes revealed identity with UDP glucosyltransferase 73C (UGT73C1, UGT73C6 and UGT73C5) belonging to group D6 in Arabidopsis thaliana. The main difference between the cisZOG and UGT73C1 genes is that cisZOG utilizes UDP-glucose as a sugar donor and catalyzes the formation of O-β-D-glucosyl-cis-zeatin from cisZ24, whereas UGT73C1 is primarily involved in the O-glucosylation of trans-zeatin and DHZ26.
This agreed with the phylogenetic reconstruction of the multigene family 1 UDP glycosyltransferases by Ref.9, which revealed two phylogenetic groups, called O and P that are not present in Arabidopsis thaliana. To support this finding, the PSPG motif does not contain highly conserved residues9. The C-terminal region of UGTs contains a 44-amino acid consensus sequence. The PSPG motif. in AY573820.1 is 5′-WSPQMLILTHPAVGGFLTHCGWNSTLEGITSGVPLLTWPLFGDQ-3′ (position 41 contains Phe instead of His, and 42 Gly instead of Ser, marked in bold; Fig. 3). The PSPG consensus was originally defined as a signature motif for a plant UGT involved in glycosylation of secondary metabolites51. This motif is well conserved and is involved in binding of UDP sugar donors to the enzyme21,52. Similar to other UGTs containing the PSPG motif, we found that the cisZOG protein is characterized by only a single or very few introns. This is in contrast to the UGT80 and UGT81 gene families involved in the glycosylation of lipids and sterols and having from 5 to 13 introns32. UGTs with the PSPG motif are monophyletic32. According to Ref.9, they were lost at some stage during the plant evolution, which might be due to the massive genome reduction in this species. However, we found that the O group is missing not only in A. thaliana but also in the whole family Brassicaceae (Figs. 1, 2) as confirmed for: Arabis halleri, A. lyrata, Brassica oleracea var. capitata, B. rapa, Boechera stricta, Capsella grandiflora, C. rubella and Eutrema salsugineum. Additionally, we did not find it in any member of other 15 families belonging to Brassicales, except for Carica papaya from family Caricaceae.
The phylogeny of Brassicales shows three main paleopolyploidization events53. At-γ is now recognized to be the same duplication as the paleohexapolyploidization detected in both Carica and Vitis54,55. This event is shared by all rosids and potentially all eudicots but is likely not as old as the origin of the angiosperms53. The Carica genome did not contain evidence of having undergone At-β even though both Arabidopsis and Carica belong to the same order, Brassicales. At-β duplication is a core Brassicales genome duplication. Moreover, Cleome spinosa (Brassicales, Cleomaceae) did not find evidence of the At-α event55 typical of Brassicaceae. Br-α is typical of the Brassica genome56.
Within Brassicaceae, the Carica papaya genome is unique in other features. An older WGD event in the Arabidopsis thaliana lineage (At-β) is not shared by C. papaya56 but is shared by all sequenced Brassicaceae57,58. As summarized by Ref.59, the ancestor of all Caricaceae underwent a single WGD event, and chromosome numbers and genome sizes appear stable since60–62. Brassicaceae have undergone at least three ancestral polyploidization events63; however, no data are available for any Cariaceae genome duplications. Moreover, Carica papaya has been recently shown the sole exception not only for the ZOG gene but also for the Lon evolutionary history, as the Lon gene is the only gene among land plants containing a single copy64. Papaya carries the preduplication ancestral Lon gene placed at the beginning of the model for Lon evolution in plants64. We can speculate that cisZOG gene loss in Brassicales occurred before the first WGD (At-β65) and diversification of Caricaceae-Moringaceae-Akaniaceae-Tropaeolaceae.
Also representatives of the closely related order Malvales contain the cisZOG gene (Gossypium raimondii, Glycine max; for details see Fig. 2). Both Brassicales and Malvales are descendants of the paleohexaploid genome common to all eudicots having 21 chromosome pairs resulting from triplication of ancestral n = 766. Recently, UGTs belonging to group O were found neither in Linum ussitatisimum67 nor in lower plant lineages. These results indicate an independent loss of the cisZOG gene in different plant lineages.
Methylerythritol phosphate and mevalonate biosynthesis pathways
Although zeatin O-glucosides have been first discovered in plants already in the last century (transZ O-glucoside68,69; cisZ O-glucoside70,71), knowledge of when they first appeared during plant evolution is missing. The enzymes involved in O-glycosylation of transZ and cisZ in plants and their encoding genes have been well characterized in plants21–24,26. Whereas only transZ (and partial DHZ) were the substrates of O-glucosyltransferase and O-xylosyltransferase from lima bean (Phaseolus lunatus) and common bean (Phaseolus vulgaris), respectively21,22, two O-glucosyltranferases identified in maize had strict specificity for cisZ23,24. In maize and other Poaceae representatives, cisZ O-glucoside and its riboside were reported major CK forms, mostly representing altogether > 80% of the total CKs, in our previous research17. Here, we revealed two to nine cisZOG homologs in Poaceae, three found in maize (Fig. 2). Three UGTs capable of O-glucosylation are known to occur in Arabidopsis, recognizing transZ and DHZ preferentially26. It is consistent with the absence of cisZOG gene in the Brassicaceae family reported here and the lack of cisZ O-glucosides in selected Brassicaceae species (data not shown).
Moreover, two possible biosynthesis pathways for the isoprenoid moiety of CKs, the MEP and the MVA pathways, occur in plants11,12 and play a role in cisZ and transZ biosyntheses10. The prenyl group of transZ (and iP) is mainly produced through the MEP pathway in plastids10. On the other hand, the prenylated adenine moiety of tRNA is typical for cisZ, whose formation in Arabidopsis may involve the transfer of isoprenoid precursor dimethylallyl diphosphate (DMAPP) from the MVA pathway to tRNA in the cytosol10. Distinct origins of DMAPP for transZ and cisZ biosynthesis suggest a potentially separate modulation of these CK species levels in plants. The MEP pathway is present in many bacteria and in the chloroplasts of all phototropic organisms. In contrast, the MVA pathway has been found in animals, fungi, plant cytoplasm, archaeobacteria, and some eubacteria11. We pointed out that the MEP pathway is phylogenetically old being found in bryophytes (Marchantia polymorpha and Sphagnum phallax) and lycophytes (Selaginella moellendorffii) (Fig. 2). However, the MEP pathway is present in a higher group of plants, angiosperms often have the MVA pathway, and sometimes only the MVA pathway is present (currently found in five species; Fig. 2). Surprisingly, in Brassicaceae, only transZ was found, and the MEP pathway was confirmed (Fig. 3). A similar situation was found in Saxifragales (Kalanchoe spp.), only one member of Salicaceae (Salix purpurea) and Solanaceae (Solanum tuberosum). The pattern in angiosperms is not associated with the evolutionary history of the plants. In general, it is not clear which properties are related to the abundance of the cis isomer in a particular plant species72. The possibilities include specific environmental conditions, biotic interactions and lifestyle72.
Conclusion
We identified ZOG proteins in over 376 unique accessions in 96 plant species. Our study indicates the expansion of ZOG proteins in both monocots and eudicots. We mainly identified 1–5 putative ZOG proteins in plants with fully sequenced genomes; however, there are exceptions with many more homologs. They were classified into 15 main groups. We confirmed that the cisZOG gene is not present in Arabidopsis thaliana and is also missing in the family Brassicaceae and most of the other members of the order Brassicales. Similarly, cisZOG was not found in Malphigiales or Saxifragales. However, except for Carica papaya (Brassicales, Caricaceae), only a few representatives from the plant kingdom exist fitting these criteria with results possibly affected by not-so-deeply sequenced genomes. Thus, these data provide a foundation for further detailed studies of the chromosomal locations of ZOG homologs, their secondary structures, expression patterns, as well as interaction partners. Two possible biosynthesis pathways for the isoprenoid moiety of transZ- and cisZ-type CKs, the MEP and the MVA ones, occur in plants11,12. Based on cis- and transZOG distribution, only the phylogenetically older MEP pathway was found in bryophytes and lycophytes. The MEP pathway occurs in a higher group of plants as well; in angiosperms, it is often involved together with the cisZOG gene in the MVA pathway, and sometimes only the MVA pathway is present. Surprisingly, in Brassicaceae, only transZ was found, and the MEP pathway was confirmed. To date, the pattern in angiosperms is apparently not associated with the evolutionary history of the plants.
Materials and methods
Bioinformatic identification of ZOG homologs
We combined two approaches, homology searches via BLASTP from available databases and compilation of all publicly available sequences associated with zeatin O-glucosyltransferase to date. First, the Zea mays cisZOG protein originally reported by Ref.23 was used as a query sequence for BLASTP in NCBI protein databases (http://www.ncbi.nlm.nih.gov; not shown separately). Then, we used the homology search tool BLASTP to scan sequences via Phytozome v1273 (https://phytozome.jgi.doe.gov) and find more orthologs in different plant species to build the matrix to identify major events of ZOG gene duplication and losses in angiosperms during evolution and to explore evolutionary diversification of ZOGs within the plant kingdom. We performed extensive BLASTP searches using default parameters adjusted to the lowest E-value (< 1e−10) to obtain as many sequences as possible from GenBank. Via these searches, we identified only cisZOG homologs and any representative from Brassicaceae. Second, to collect transZOG, we searched for ZOG proteins among the annotated genomic sequences from the Phytozome v12 database. Finally, to obtain a more comprehensive set of genes, the Phytozome database was also searched for genes annotated as ‘zeatin O-glucosyltransferase’, and sequences were checked by alignment. The hits obtained from the two searches were then combined, and the redundant sequences were removed. Accession numbers for all retrieved sequences used in analyses are provided in Supplementary Table S2 and alignment in Supplementary Table S3. As a result, we combined sequences representing both attempts into one matrix of cisZOG O-glucosyltransferases to unravel the main groups and show patterns of sequence conservation and evolution.
We have two reasons for this exhaustive sampling, rather than only using completely sequenced genomes, although the actual number of species with full genomes available is quite large (Phytozome). First, taxon sampling is thought to impact the accuracy of phylogenetic inference, so we aimed to assess how stable the evolutionary relationships are with the inclusion of additional taxa. Second, we used a complete data set for motif and domain analyses. We eliminated duplicates (i.e., identical sequences of approximately the same length) from all searches. Protein isoforms with the same length were also used because the differential expression patterns producing protein isoforms from various tissues suggested that isoforms could have different biological functions in vivo74. However, we also used the IsoCel program75 to select from an alternative potential isoform dataset optimized for tree reconstruction. To prevent incorrect inference of gene duplication and losses from species available only from NCBI, we employed for this purpose only sequences obtained from completely sequenced genomes.
Sequence alignment
Amino acid sequences were aligned using the Clustal Omega algorithm76 in the Mobyle platform77, with homology detection by HMM–HMM comparisons78. We screened data after alignment in the BioEdit program79.
Conserved motifs and domains were analyzed in Geneious 11.0.3 (https://www.geneious.com) and MEME 4.11.2 (http://meme-suite.org)80. The MEME search was set to identify a maximum of 50 motifs for each protein with a wide sequence motif range from 2 to 50 and a total number of sites ranging from 2 to 600. The Pfam Motif Library81 and NCBI Domain Architecture Retrieval Tool82 were used to analyze the conserved motifs. The number and arrangement of introns and exons were analyzed using Gene Structure Display Server version 2.083 by aligning the coding sequences with the genomic sequences.
First, we analyzed all sequences independent of their annotations, with no prior assumptions. Later, cisZOG homologs based on sequence identity were checked to determine if they contained the PSPG motif. Because group O contains only two small changes in the PSPG motif and homologs lacking such mutations might still be evolutionarily related to other cisZOGs, we included these sequences in the analyses but marked them to show which do not have PSPG motifs His and Ser at positions 41 and 42. In total, we identified 376 unique accessions in 96 plant species (Supplementary Table S2).
Phylogenetic and comparative analyses
Maximum likelihood (ML) topology searches were performed in RAxML 8.2.484 to examine differences in optimality between alternative topologies. The analysis involved 376 amino acid sequences and a total of 1711 positions in the final dataset. 1000 replications were run for bootstrap values. To confirm and compare results we used maximum parsimony method (MP). The MP tree was obtained using the Tree-Bisection-Regrafting (TBR) algorithm85 with search level 1 in which the initial trees were obtained by the random addition of sequences (10 replicates). Evolutionary analyses were conducted in MEGA786. The bootstrap consensus tree inferred from 500 replicates87. Phylogenetic trees were constructed and modified with iTOL v3.488.
Supplementary Information
Author contributions
L.Z.D. designed the study, designed and performed phylogenetic analyses and wrote the manuscript with V.M. and invaluable support from D.H. All authors discussed the results and contributed to the final manuscript.
Funding
The authors gratefully acknowledge the financial support from the Czech Science Foundation (Grants no. 19-02699S and 19-12262S) and the Ministry of Education, Youth and Sports from European Regional Development Fund-Project "Centre for Experimental Plant Biology" (No. CZ.02.1.01/0.0/0.0/16_019/0000738). The access to the computing and storage facilities owned by parties and projects contributing to the National Grid Infrastructure MetaCentrum provided under the program “Projects of Large Infrastructure for Research, Development, and Innovations” (LM2010005) was highly appreciated, as was the access to the CERIT-SC computing and storage facilities provided under the program “Center CERIT Scientific Cloud”, part of the Operational Program Research and Development for Innovations, reg. no. CZ.1.05/3.2.00/08.0144. LZD is grateful to Dr. Guy Perrière for his kindness in introducing the IsoSel program and selecting protein isoforms.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Lenka Záveská Drábková, Email: lenka.zaveska.drabkova@gmail.com.
Václav Motyka, Email: vmotyka@ueb.cas.cz.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-87047-8.
References
- 1.Spíchal L. Cytokinins—Recent news and views of evolutionally old molecules. Funct. Plant Biol. 2012;39:267–284. doi: 10.1071/FP11276. [DOI] [PubMed] [Google Scholar]
- 2.Hluska T, Hlusková L, Emery RJN. The Hulks and the Deadpools of the cytokinin universe: A dual strategy for cytokinin production, translocation, and signal transduction. Biomolecules. 2021;11:209. doi: 10.3390/biom11020209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pokorná E, Hluska T, Galuszka P, Hallmark HT, Dobrev PI, Záveská Drábková L, Filipi T, Holubová K, Plíhal O, Rashotte AM, Filepová R, Malbeck J, Novák O, Spíchal L, Brzobohatý B, Mazura P, Zahajská L, Motyka V. Cytokinin N-glucosides: Occurrence, metabolism and biological activities in plants. Biomolecules. 2021;11:24. doi: 10.3390/biom11010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yonekura-Sakakibara K, Hanada K. An evolutionary view of functional diversity in family 1 glycosyltransferases. Plant J. 2011;66:182–193. doi: 10.1111/j.1365-313X.2011.04493.x. [DOI] [PubMed] [Google Scholar]
- 5.Li Y, Baldauf S, Lim E-K, Bowles DJ. Phylogenetic analysis of the UDP-glycosyltransferase multigene family of Arabidopsis thaliana. J. Biol. Chem. 2001;276:4338–4343. doi: 10.1074/jbc.M007447200. [DOI] [PubMed] [Google Scholar]
- 6.Mackenzie PI, Owens IS, Burchel B, Bock KW, Bairoch A, Bélanger A, et al. The UDP glucosyltransferase gene family: Recommended nomenclature updated based on evolutionary divergence. Pharmacogenetics. 1997;7:255–269. doi: 10.1097/00008571-199708000-00001. [DOI] [PubMed] [Google Scholar]
- 7.Bowles D, Isayenkova J, Lim EK, Poppenberger B. Glycosyltransferases: Managers of small molecules. Curr. Opin. Plant Biol. 2005;8:254–263. doi: 10.1016/j.pbi.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 8.Ross J, Li Y, Lim E-K, Bowles DJ. Higher plant glycosyltransferases. Genome Biol. 2001;2:reviews3004. doi: 10.1186/gb-2001-2-2-reviews3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caputi L, Malnoy M, Goremykin V, Nikiforova S, Materns S. A genome-wide phylogenetic reconstruction of family 1 UDP-glycosyltransferases revealed the expansion of the family during the adaptation of plants to life on land. Plant J. 2012;69:1030–1042. doi: 10.1111/j.1365-313X.2011.04853.x. [DOI] [PubMed] [Google Scholar]
- 10.Huang J, Pang C, Fan C, Song M, Yu J, Wei H, et al. Genome-wide analysis of the family 1 glycosyltransferases in cotton. Mol. Genet. Genom. 2015;290:1805–1818. doi: 10.1007/s00438-015-1040-8. [DOI] [PubMed] [Google Scholar]
- 11.Rohmer M. Mevalonate-independent methylerythritol phosphate pathway for isoprenoid biosynthesis. Elucidation and distribution. Pure Appl. Chem. 2003;75:375–387. doi: 10.1351/pac200375020375. [DOI] [Google Scholar]
- 12.Kasahara H, Takei K, Ueda N, Hishiyama S, Yamaya T, Kamiyam Y, et al. Distinct isoprenoid origins of cis and trans-zeatin biosyntheses in Arabidopsis. J. Biol. Chem. 2004;279:14049–14054. doi: 10.1074/jbc.M314195200. [DOI] [PubMed] [Google Scholar]
- 13.Sakakibara H. Cytokinins: Activity, biosynthesis, and translocation. Annu. Rev. Plant Biol. 2006;57:431–449. doi: 10.1146/annurev.arplant.57.032905.105231. [DOI] [PubMed] [Google Scholar]
- 14.Åstot C, Doležal K, Nordström A, Wang Q, Kunkel T, Moritz T, et al. An alternative cytokinin biosynthesis pathway. PNAS. 2000;97:14778–14783. doi: 10.1073/pnas.260504097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kamínek M. Evolution of tRNA and origin of the two positional isomers of zeatin. J. Theor. Biol. 1974;48:489–492. doi: 10.1016/S0022-5193(74)80018-4. [DOI] [PubMed] [Google Scholar]
- 16.Persson BC, Esberg B, Ólafsson Ó, Björk GR. Synthesis and function of isopentenyl adenosine derivatives in tRNA. Biochimie. 1994;76:1152–1160. doi: 10.1016/0300-9084(94)90044-2. [DOI] [PubMed] [Google Scholar]
- 17.Gajdošová S, Spíchal L, Kamínek M, Hoyerová K, Novák O, Dobrev PI, Galuszka P, Klíma P, Gaudinová A, Žižková E, Hanuš J, Dančák M, Trávníček B, Pešek B, Krupička M, Vaňková R, Strnad M, Motyka V. Distribution, biological activities, metabolism, and the conceivable function of cis-zeatin-type cytokinins in plants. J. Exp. Bot. 2011;62:2827–2840. doi: 10.1093/jxb/erq457. [DOI] [PubMed] [Google Scholar]
- 18.Klämbt D. The biogenesis of cytokinins in higher plants: Our present knowledge. In Physiology and Biochemistry of Cytokinins in Plants. (eds. Kamínek, M., Mok, D. W. S. & Zažímalová, E.) 25–27 (SPB Academic, 1992).
- 19.Dixon SC, Martin RC, Mok MC, Shaw G, Mok DWS. Zeatin glycosylation enzymes in Phaseolus. Isolation of O-glucosyltransferase from P. lunatus and comparison to O-xylosyltransferase from P. vulgaris. Plant Physiol. 1989;90:1316–1321. doi: 10.1104/pp.90.4.1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Turner JE, Mok DWS, Mok MC, Shaw G. Isolation and partial purification of an enzyme catalyzing the formation of O-xylosylzeatin in Phaseolus vulgaris embryos. Proc. Natl. Acad. Sci. U. S. A. 1987;84:3714–3717. doi: 10.1073/pnas.84.11.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin RC, Mok MC, Mok DWS. Isolation of a cytokinin gene, ZOG1, encoding zeatin O-glucosyltransferase from Phaseolus lunatus. Proc. Natl. Acad. Sci. U. S. A. 1999;96:284–289. doi: 10.1073/pnas.96.1.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin RC, Mok MC, Mok DW. A gene encoding the cytokinin enzyme zeatin O-xylosyltransferase of Phaseolus vulgaris. Plant Physiol. 1999;120:553–558. doi: 10.1104/pp.120.2.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martin RC, Mok MC, Habben JE, Mok DW. A maize cytokinin gene encoding an O-glucosyltransferase specific to cis-zeatin. PNAS. 2001;98:5922–5926. doi: 10.1073/pnas.101128798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Veach YK, Martin RC, Mok DWS, Malbeck J, Vaňková R, Mok MC. O-Glucosylation of cis-zeatin in maize characterization of genes, enzymes, and endogenous cytokinins. Plant Phys. 2003;131:1374–1380. doi: 10.1104/pp.017210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kudo T, Makita N, Kojima M, Tokunaga H, Sakakibara H. Cytokinin Activity of cis-zeatin and phenotypic alterations induced by overexpression of putative cis-zeatin-O-glucosyltransferase in rice. Plant Physiol. 2012;160:319–331. doi: 10.1104/pp.112.196733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hou B, Lim E-K, Higgins GS, Bowles DJ. N-glucosylation of cytokinins by glycosyltransferases of Arabidopsis thaliana. J. Biol. Chem. 2004;279:47822–47832. doi: 10.1074/jbc.M409569200. [DOI] [PubMed] [Google Scholar]
- 27.Poppenberger B, Fujioka S, Soeno K, George GL, Vaistij FE, Hiranuma S, Seto H, Takatsuto S, Adam G, Yoshida S, Bowles D. The UGT73C5 of Arabidopsis thaliana glucosylates brassinosteroids. Proc. Natl. Acad. Sci. U. S. A. 2005;102:15253–15258. doi: 10.1073/pnas.0504279102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gandia-Herrero F, Lorenz A, Larson T, Graham IA, Bowles DJ, Rylott EL, Bruce NC. Detoxification of the explosive 2,4,6-trinitrotoluene in Arabidopsis: Discovery of bifunctional O- and C-glucosyltransferases. Plant J. 2008;56:963–974. doi: 10.1111/j.1365-313X.2008.03653.x. [DOI] [PubMed] [Google Scholar]
- 29.Jin S-H, Ma X-M, Kojima M, Sakakibara H, Wang Y-W, Hou B-K. Overexpression of glucosyltransferase UGT85A1 influences trans-zeatin homeostasis and trans-zeatin responses likely through O-glucosylation. Planta. 2013;237:991–999. doi: 10.1007/s00425-012-1818-4. [DOI] [PubMed] [Google Scholar]
- 30.Šmehilová M, Dobrůšková J, Novák O, Takáč T, Galuszka P. Cytokinin-specific glycosyltransferases possess different roles in cytokinin homeostasis maintenance. Front. Plant Sci. 2016;7:1264. doi: 10.3389/fpls.2016.01264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Swigoňová Z, Bennetzen JL, Messig J. Structure and evolution of the r/b chromosomal regions in rice, maize and sorgum. Genetics. 2005;169:891–906. doi: 10.1534/genetics.104.034629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paquette S, Lindberg Møller B, Bak S. On the origin of family 1 plant glycosyltransferases. Phytochemistry. 2003;62:399–413. doi: 10.1016/S0031-9422(02)00558-7. [DOI] [PubMed] [Google Scholar]
- 33.Osmani SA, Bak S, Imberty A, Olsen CE, Møller BL. Catalytic key amino acids and UDP-sugar donor specificity of a plant glucuronosyltransferase UGT94B1: Molecular modeling substantiated by site-specific mutagenesis and biochemical analyses. Plant Physiol. 2008;48:1295–1308. doi: 10.1104/pp.108.128256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kubo A, Arai Y, Nagashima S, Yoshikawa T. Alteration of sugar donor specificities of plant glycosyltransferases by a single point mutation. Arch. Biochem. Biophys. 2004;429:198–203. doi: 10.1016/j.abb.2004.06.021. [DOI] [PubMed] [Google Scholar]
- 35.Ono E, Homma Y, Horikawa M, Kunikane-Doi S, Imai H, Takahashi S, Kawai Y, Ishiguro M, Fukui Y, Nakayama T. Functional differentiation of the glycosyltransferases that contribute to the chemical diversity of bioactive flavonol glycosides in grapevines (Vitis vinifera) Plant Cell. 2010;22:2856–2871. doi: 10.1105/tpc.110.074625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schweiger W, Pasquet J-C, Nussbaumer T, Kovalsky Paris MP, Wiesenberger GW, et al. Functional characterization of two clusters of Brachypodium distachyon UDP-glycosyltransferases encoding putative deoxynivalenol detoxification genes. MPMI. 2013;26:781–792. doi: 10.1094/MPMI-08-12-0205-R. [DOI] [PubMed] [Google Scholar]
- 37.Spíchal L. Cytokinins—Recent news of old molecules. Funct. Plant Biol. 2012;39:267–284. doi: 10.1071/FP11276. [DOI] [PubMed] [Google Scholar]
- 38.Rubinstein CV, Gerriene P, de la Puente GS, Astini RA, Steemans P. Early Middle Ordovician evidence for land plants in Argentina (eastern Gondwana) New Phytol. 2010;188:365–369. doi: 10.1111/j.1469-8137.2010.03433.x. [DOI] [PubMed] [Google Scholar]
- 39.Záveská Drábková L, Honys D. Evolutionary history of callose synthases in terrestrial plants with emphasis on proteins involved in male gametophyte development. PLoS ONE. 2017;13:e0187331. doi: 10.1371/journal.pone.0187331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Little A, Schwerdt JG, Shirley NJ, Khor SF, Neumann K, O´Donovan LA, et al. Revised phylogeny of cellulose synthase gene superfamily: Insights into cell wall evolution. Plant. Phys. 2018;117:1124–1141. doi: 10.1104/pp.17.01718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Landis JB, Soltis DE, Li Z, Marx HE, Barker MS, Tank DC, Soltis PS. Impact of whole-genome duplication events on diversification rates in angiosperms. Am. J. Bot. 2018;105:348–363. doi: 10.1002/ajb2.1060. [DOI] [PubMed] [Google Scholar]
- 42.Freeling M. Bias in plant gene content following different sorts of duplication: Tandem, whole-genome, segmental, or by transposition. Ann. Rev. Plant. Biol. 2009;60:433–453. doi: 10.1146/annurev.arplant.043008.092122. [DOI] [PubMed] [Google Scholar]
- 43.Choi IG, Kim SH. Evolution of protein structural classes and protein sequence families. PNAS. 2006;3:14056–14061. doi: 10.1073/pnas.0606239103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sikosek, T. & Bornberg-Bauer, E. Evolution before and after gene duplication? In Evolution After Gene Duplication (eds. Dittmar, K. & Liberles, D.) 106–131 (Wiley-Blackwell, 2010).
- 45.Tiley GP, Ané C, Burleigh G. Evaluating and characterizing ancient whole-genome duplications in plants with gene count data. Genome Biol. Evol. 2015;8(4):1023–1037. doi: 10.1093/gbe/evw058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de Bruijn S, Zhao T, Muiño JM, Schranz EM, Angenent GC, Kaufmann K. PISTILLATA paralogs in Tarenaya hassleriana have diverged in interaction specificity. BMC Plant Biol. 2018;18:368. doi: 10.1186/s12870-018-1574-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mei W, Boatwright L, Feng G, Schnable JC, Barbazuk WB. Evolutionarily conserved alternative splicing across monocots. Genetics. 2017;207:465–480. doi: 10.1534/genetics.117.300189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marquez Y, Brown JWS, Simpson C, Barta A, Kalyna M. Transcriptome survey reveals increased complexity of the alternative splicing landscape in Arabidopsis. Genome Res. 2012;22:1184–1195. doi: 10.1101/gr.134106.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reddy ASN, Marquez Y, Kalyna M, Barta A. Complexity of the alternative splicing landscape in plants. Plant Cell. 2013;25:3657–3683. doi: 10.1105/tpc.113.117523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shang X, Cao Y, Ma L. Alternative splicing in plant genes: A means of regulating the environmental fitness of plants. Int. J. Mol. Sci. 2017;18:432. doi: 10.3390/ijms18020432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hughes J, Hughes MA. Multiple secondary plant product UDP-glucose glucosyltransferase genes expressed in cassava. DNA Seq. 1994;5:41–49. doi: 10.3109/10425179409039703. [DOI] [PubMed] [Google Scholar]
- 52.Ostrowski M, Jakubovska A. UDP-glycosyltransferases of plant hormones. Adv. Cell Biol. 2014;4:43–60. doi: 10.2478/acb-2014-0003. [DOI] [Google Scholar]
- 53.Baker MS, Vogel H, Schranz ME. Paleopolyploidy in the Brassicales: Analyses of the cleome transcriptome elucidate the history of genome duplications in Arabidopsis and other Brassicales. Genome Biol. Evol. 2009;1:391–399. doi: 10.1093/gbe/evp040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lyons E, Pedersen B, Kane J, Alam M, Ming R, Tang H, et al. Finding and comparing syntenic regions among Arabidopsis and the outgroups papaya, poplar, and grape: CoGe with rosids. Plant Physiol. 2008;148:1772–1781. doi: 10.1104/pp.108.124867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schranz ME, Mitchell-Olds T. Independent ancient polyploidy events in the sister families Brassicaceae and Cleomaceae. Plant Cell. 2006;18:1152–1165. doi: 10.1105/tpc.106.041111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ming R, Hou S, Feng Y, Yu Q, Dionne-Laporte A, Saw JH, et al. The draft genome of the transgenic tropical fruit tree papaya (Carica papaya L.) Nature. 2008;452:991–996. doi: 10.1038/nature06856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.The Brassica rapa Genome Sequencing Consortium The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 2011;43:1035–1039. doi: 10.1038/ng.919. [DOI] [PubMed] [Google Scholar]
- 58.Dassanayake M, Oh D-H, Haas JS, Hernandez A, Hong H, Ali S, et al. The genome of the extremophile crucifer Thellungiella parvula. Nat. Genet. 2011;43:913–918. doi: 10.1038/ng.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rockinger A, Souza A, Carvalho FA, Renner SS. Chromosome number reduction in the sister clade of Carica papaya with concomitant genome size doubling. Am. J. Bot. 2016;103:1082–1088. doi: 10.3732/ajb.1600134. [DOI] [PubMed] [Google Scholar]
- 60.Tang H, Bowers JE, Wang X, Ming R, Alam M, Paterson AH. Synteny and collinearity in plant genomes. Science. 2008;320:486–488. doi: 10.1126/science.1153917. [DOI] [PubMed] [Google Scholar]
- 61.Rice A, Glick L, Abadi S, Einhorn M, Kopelman NM, Salman-Minkov A, et al. The chromosome counts database (CCDB) A community resource of plant chromosome numbers. New Phytol. 2015;206:19–26. doi: 10.1111/nph.13191. [DOI] [PubMed] [Google Scholar]
- 62.Gschwend AR, Wai CM, Zee F, Arumuganathan AK, Ming R. Genome size variation among sex types in dioecious and trioecious Caricaceae species. Euphytica. 2013;189:461–469. doi: 10.1007/s10681-012-0815-9. [DOI] [Google Scholar]
- 63.Tang HB, Wang XY, Bowers JE, Ming R, Alam M, Paterson AH, et al. Unraveling ancient hexaploidy through multiply-aligned angiosperm gene maps. Genome Res. 2008;18:1944–1954. doi: 10.1101/gr.080978.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tsitsekian D, Daras G, Alatzas A, Templalexis D, Hatzopoulos P, Rigas S. Comprehensive analysis of Lon proteases in plants highlights independent gene duplication events. J. Exp. Bot. 2019;70:2185–2197. doi: 10.1093/jxb/ery440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Edger PP, Heidel-Fischer HM, Bekaert M, Rota J, Glöckner G, Platts AE, et al. The butterfly plant arms-race escalated by gene and genome duplications. PNAS. 2015;112:8362–8366. doi: 10.1073/pnas.1503926112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Salse J. Ancestors of modern plant crops. Curr. Opin. Plant. Biol. 2016;30:134–142. doi: 10.1016/j.pbi.2016.02.005. [DOI] [PubMed] [Google Scholar]
- 67.Barvkar VT, Pardeshi VC, Kale SM, Kadoo NY, Gupta VS. Phylogenomic analysis of UDP glycosyltransferase 1 multigene family in Linum usitatissimum identified genes with varied expression patterns. BMC Genom. 2012;13:175. doi: 10.1186/1471-2164-13-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Horgan R. A new cytokinin metabolite. Biochem. Biophys. Res. Commun. 1975;65:358–363. doi: 10.1016/S0006-291X(75)80101-X. [DOI] [PubMed] [Google Scholar]
- 69.Parker CW, Letham DS, Wilson MM, Jenkins ID, MacLeod JK, Summons RE. The identity of two new cytokinin metabolites. Ann. Bot. 1975;39:375–376. doi: 10.1093/oxfordjournals.aob.a084951. [DOI] [Google Scholar]
- 70.Takagi M, Yokota T, Murofushi N, Saka H, Takahashi N. Quantitative changes of free-base, riboside, ribotide and glucoside cytokinins in developing rice grains. Plant Growth Regul. 1989;8:349–364. doi: 10.1007/BF00024665. [DOI] [Google Scholar]
- 71.Wagner BM, Beck E. Cytokinins in the perennial herb Urtica dioica L. as influenced by its nitrogen status. Planta. 1993;190:511–518. doi: 10.1007/BF00224790. [DOI] [Google Scholar]
- 72.Schäfer M, Brütting C, Canales IM, Großkinsky DK, Vaňková R, Baldwin IT, Meldau S. The role of cis-zeatin-type cytokinins in plant growth regulation and mediating responses to environmental interactions. J. Exp. Bot. 2015;66:4873–4884. doi: 10.1093/jxb/erv214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012;40:1178–1186. doi: 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen Y, Zou M, Cao Y. Transcriptome analysis of the Arabidopsis semi-in vivo pollen tube guidance system uncovers a distinct gene expression profile. J. Plant. Biol. 2014;57:93–105. doi: 10.1007/s12374-013-0272-6. [DOI] [Google Scholar]
- 75.Philippon H, Souvane A, Brochier-Armanet C, Perrière G. IsoSel: Protein Isoform Selector for phylogenetic reconstructions. PLoS ONE. 2017;12:e0174250. doi: 10.1371/journal.pone.0174250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Néron B, Ménager H, Maufrais C, Joly N, Maupetit J, Letort S, et al. Mobyle: A new full web bioinformatics framework. Bioinformatics. 2009;25:3005–3011. doi: 10.1093/bioinformatics/btp493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Söding J. Protein homology detection by HMM–HMM comparison. Bioinformatics. 2005;21:951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- 79.Hall TA. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids Symp. 1999;41:95–98. [Google Scholar]
- 80.Bailey TL, Bodén M, Buske FA, Frith M, Grant CE, Clementi L, et al. MEME SUITE: Tools for motif discovery and searching. Nucl. Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bateman A, Birney E, Cerruti L, Durbin R, Etwiller L, Eddy SR, et al. The Pfam protein families database. Nucl. Acids Res. 2002;30(1):276–280. doi: 10.1093/nar/30.1.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Geer LY, Domarchev M, Lipman DJ, Bryant SH. CDART: Protein homology by domain architecture. Genome Res. 2002;12(10):1619–1623. doi: 10.1101/gr.278202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hu B, Jin J, Guo A-Y, Zhang H, Luo J, Gao G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics. 2015;31:1296–1297. doi: 10.1093/bioinformatics/btu817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nei M, Kumar S. Molecular Evolution and Phylogenetics. Oxford University Press; 2000. [Google Scholar]
- 86.Kumar S, Stecher G, Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 88.Letunic I, Bork P. Interactive Tree Of Life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;8:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.