ABSTRACT
Carbon/nitrogen (C/N) balance sensing is a key requirement for the maintenance of cellular homeostasis. Therefore, cyanobacteria have evolved a sophisticated signal transduction network targeting the metabolite 2-oxoglutarate (2-OG), the carbon skeleton for nitrogen assimilation. It serves as a status reporter for the cellular C/N balance that is sensed by transcription factors NtcA and NdhR and the versatile PII-signaling protein. The PII protein acts as a multitasking signal-integrating regulator, combining the 2-OG signal with the energy state of the cell through adenyl-nucleotide binding. Depending on these integrated signals, PII orchestrates metabolic activities in response to environmental changes through binding to various targets. In addition to 2-OG, other status reporter metabolites have recently been discovered, mainly indicating the carbon status of the cells. One of them is cAMP, which is sensed by the PII-like protein SbtB. The present review focuses, with a main emphasis on unicellular model strains Synechoccus elongatus and Synechocystis sp. PCC 6803, on the physiological framework of these complex regulatory loops, the tight linkage to metabolism and the molecular mechanisms governing the signaling processes.
Keywords: nitrogen assimilation, arginine synthesis, protein-serine-phosphorylation, CO2 metabolism, energy sensing, 2-oxoglutarate
This review presents the current knowledge on the sophisticated signal transduction network evolved in cyanobacteria to maintain carbon/nitrogen homeostasis.
INTRODUCTION TO THE METABOLIC BASIS OF C/N BALANCE SENSING
Cyanobacteria are photoautotrophic organisms that build up organic molecules for cell growth by assimilating inorganic nutrients at the expense of light energy. Assimilation of carbon and nitrogen consumes the majority of the primary products of photosynthesis, ATP and reduction equivalents. Cyanobacteria maintain under balanced growth a C/N ratio of approximately 5:1 (Wolk 1973), which implies that nitrogen assimilation must constantly keep pace with CO2 fixation. Isotope labeling experiments revealed that nitrogen is assimilated in the form of ammonia via the coupled glutamine synthetase (GS)—glutamine-oxoglutarate amidotransferase (GOGAT) reaction (Meeks et al. 1977). GS is the key enzyme of nitrogen assimilation, since in this reaction, ammonia is ligated with glutamate in an ATP-dependent reaction yielding glutamine. The subsequent GOGAT reaction transfers the amido-group of glutamine to the α-C atom of 2-oxoglutarate (2-OG), resulting in two molecules of glutamate. The net yield of the GS-GOGAT cycle is therefore the conversion of 2-OG with ammonia into glutamate, consuming one ATP and two reduction equivalents. For details on the GS-GOGAT cycle and its mode of regulation, the readers are referred to comprehensive reviews on cyanobacterial ammonium assimilation (Herrero, Muro-Pastor and Flores 2001; Muro-Pastor, Reyes and Florencio 2005; Luque and Forchhammer 2008; Bolay et al. 2018). To avoid a detrimental depletion of the cellular glutamate pool by GS, its activity must be tightly adjusted to the capacity of the GOGAT reaction to refill the glutamate pool. Bacteria have evolved a plethora of mechanisms to exactly tune the activity of GS to the actual need (Leigh and Dodsworth 2007). In cyanobacteria, unlike other bacteria, GS activity is mainly regulated through small inhibitory proteins, the inactivating factors IF7 and IF17, encoded by gifA and gifB in Synechocystis strain PCC 6803 (Garcia-Dominguez, Reyes and Florencio 1999). Expression of these genes is controlled by the global nitrogen control factor NtcA, which responds to the 2-OG levels of the cell (see below) as well as by a non-coding RNA, nsiR4 (Klahn et al. 2015a) and a glutamine riboswitch in the 5′UTR of gifB. This glutamine riboswitch links regulation of the central nitrogen assimilation reaction to the glutamine status of the cells (Klahn et al. 2018). A related aptamer (termed DP aptamer) that also binds glutamine was identified by bioinformatics analysis. The structure and glutamine-binding mode of both aptamers in complex with glutamine has recently been solved (Ren et al. 2015; Huang et al. 2019).
From glutamate and glutamine, organic nitrogen is distributed to nearly all the organic nitrogen residues present in various cellular building blocks. This metabolic architecture places 2-OG, the precursor of these central amino acids, at the intersection of carbon- and nitrogen-assimilatory reactions. The metabolite 2-OG is produced from CO2 fixation products via glycolysis and the oxidative branch of the TCA cycle, the final step being isocitrate dehydrogenase. From the lack of a functional 2-OG dehydrogenase, it was assumed for a long time, that the cyanobacterial TCA cycle is incomplete and 2-OG cannot be converted to succinate (Pearce, Leach and Carr 1969). However, in the meantime, it is realized that the TCA cycle can be closed by various alternative reactions (reviewed in Esteves-Ferreira et al. 2018). The most widely distributed bypass operates through the consecutive activity of 2-OG decarboxylase and succinic semialdehyde dehydrogenase (Zhang and Bryant 2011). The sll1981 product from Synechocystis sp. PCC 6803, previously annotated as L-myo-inositol-1-phosphate synthase, was shown to function as 2-OG decarboxylase (Wang et al. 2017). A second bypass from 2-OG to succinate was proposed in Synechocystis PCC 6803 via the GABA shunt (Xiong, Brune and Vermaas 2014). In this route, glutamate is decarboxylated by glutamate decarboxylase to γ-amino butyric acid (GABA), which is then converted to succinyl-semialdehyde by a side-activity of ArgD (N-acetylornithine aminotransferase). In Synechocystis PCC 6803, which harbors the enzymes for both bypasses, the GABA shunt was proposed to be more active than the 2-OG decarboxylase route (Zhang et al. 2016). Since only small fractions of carbon seem to flow through the bypass reactions (Wan et al. 2017), the physiological significance of two alternative routes deviating the missing 2-OG dehydrogenase complex remains to be fully elucidated.
Considering the 2-OG decarboxylase reaction as a minor bypath, the flux from 2-OG to glutamate, catalyzed by GOGAT, represents the dominant metabolic route for the consumption of 2-OG among cyanobacteria. In agreement, the cellular C/N ratio of 5:1 implies that for five molecules of CO2, one molecule of ammonia has to be assimilated. This requires a high turn-over of 2-OG through GS/GOGAT activity. Thereby, despite of the recently discovered TCA cycle bypasses, changes in the level of 2-OG will mainly depend on the rate of its formation though isocitrate dehydrogenase and its consumption via the GOGAT reaction that is linked to the nitrogen assimilating GS activity. In consequence, decreasing 2-OG consumption is expected under conditions of nitrogen limitation. When ammonia assimilation via GS cannot provide enough glutamine for the subsequent GOGAT reaction, 2-OG consumption would slow down and its levels are expected to increase. This close connection of 2-OG in between carbon and nitrogen metabolism qualifies it as prime candidate to sense the cellular C/N status.
Pioneering work in Escherichia coli demonstrated that the levels of 2-OG correlate with the nitrogen supply to these bacteria (Senior 1975). The levels of 2-OG are sensed by the highly conserved PII-signaling protein (see below), which regulates various aspects of N-assimilation in many bacteria (Ninfa and Jiang 2005). The finding that the homologous Synechococcus elongatus PII protein is also a sensor of 2-OG, gave rise to the suggestion that 2-OG may also serve as an important status sensor of the cellular C/N balance in cyanobacteria (Forchhammer 1999). Indeed, measurement of 2-OG levels in Synechocystis PCC 6803 cells exposed to nitrogen starvation conditions demonstrated that 2-OG levels increased in conditions of nitrogen starvation (Muro-Pastor, Reyes and Florencio 2001b). Experiments with non-metabolizable 2-OG analogs in the filamentous, heterocyst-forming cyanobacterium Anabaena sp. PCC 7120, revealed that these compounds, which mimic 2-OG, elicit the differentiation of heterocysts, a process that is naturally triggered by nitrogen starvation (Laurent et al. 2005, reviewed by Zhang et al. 2018). Consistent with the function of 2-OG as a central status reporter metabolite, 2-OG was discovered to be not only sensed by the PII signal transduction protein but also by two transcription factors. First, the global nitrogen control factor NtcA was shown to respond to 2-OG (Tanigawa et al. 2002; Vazquez-Bermudez, Herrero and Flores 2002). Later on, NdhR, a global repressor of carbon-regulated genes was identified to use 2-OG as corepressor (Daley et al. 2012; Jiang et al. 2018). Together, these findings highlight 2-OG as a central indicator of the status of central metabolism, where carbon meets nitrogen assimilation. Intriguingly, a recent study showed that oxidative stress interferes with C/N status sensing by affecting the 2-OG levels (Robles-Rengel, Florencio and Muro-Pastor 2019).
For an efficient management of the intricate network of anabolic reactions required for cell growth, the cells have to constantly monitor the cellular C/N ration, which is apparently best achieved by the using 2-OG as a metabolic status reporter. Accordingly, they attempt to adjust the assimilation of nitrogen to the availability of fixed carbon. A drop in CO2 supply thus leads to the arrest of nitrogen assimilatory reactions, a physiological response already observed by Otto Warburg in the middle of the 20th century (reviewed by Flores et al. 2005).
Conversely, when nitrogen sources become limiting, the cells respond by increasing the capacity of nitrogen assimilation reactions: genes are induced that allow the utilization of alternative nitrogen sources, which in the case of diazotrophic cyanobacteria results in the induction of nitrogen fixation (Herrero, Muro-Pastor and Flores 2001). When non-diazotrophic cyanobacteria are completely depleted of combined nitrogen sources, they re-direct carbon metabolism towards glycogen synthesis and finally enter a dormant state in chlorosis, where they maintain viability until combined nitrogen becomes available again (Klotz et al. 2016; reviewed in Schwarz and Forchhammer 2005 and Forchhammer and Schwarz 2019).
Today, it can be assumed that the plethora of physiological responses that maintain nitrogen homeostasis and C/N balance is principally based on the sensing of status reporter metabolites, the most prominent being 2-OG. However, the deeper we dig into the molecular details, the more complex is the picture of a highly interconnected network of metabolite effector molecules, sensors and targets. This article summarizes the current state of our understanding of C/N related effector molecule sensing, with a focus on the PII signal transduction protein and recent advances in additional, PII-independent processes of C/N control.
THE PII SIGNAL TRANSDUCTION PROTEIN, A MULTITASKING 2-OXOGLUTARATE SENSOR
The family of PII signal transduction proteins consists of one of the most widely distributed signaling proteins in nature. The original designation ‘PII protein’ referred to a protein involved in post-translational regulation of GS activity in E. coli that eluted in a second peak of a gel-filtration experiment (Shapiro 1969). In general, PII proteins are involved in the regulation of various aspects of nitrogen assimilation and maintenance of carbon-nitrogen homeostasis (Fig. 1). They occur in bacteria and archaea as well as in the plastids from many Archaeplastida, descendent from the cyanobacterial endosymbiont.
Figure 1.
Metabolic basis of C/N balance sensing: schematic model of central metabolism at the intersection of CO2 fixation and nitrogen assimilation and the role of PII in controlling key reactions (indicated by red lines). Changes in the level 2-oxoglutarate, which are sensed by the PII signaling protein, report changes in the balance between CO2 fixation and ammonium assimilation: the levels are depleted by ammonium assimilation though the GS-GOGAT cycle and refilled by carbon flux originating from CO2 fixation. According to metabolic flux analysis (Wan et al. 2017), the phosphoenol pyruvate (PEP) carboxylase (PEPC) reaction plays a major role for carbon flux into the oxidative branch of the TCA cycle/2-oxoglutarate. Glutamate is the primary nitrogen donor for anabolic reactions, with arginine synthesis being of particular importance (PII controls reactions for nitrogen storage formation (N-acetyl glutamate kinase, NAGK) and fatty acid synthesis (Acetyl-CoA carboxylase, ACCase) as well as the uptake of the nitrogen sources ammonium, nitrate and urea through their respective uptake systems as indicated. The arrows width is indicative of the carbon flow.
Numerous review articles deal with various aspects of PII signaling in different organisms, describing the molecular biology as well as biochemical and physiological details of PII-signaling proteins (Merrick and Edwards 1995; Ninfa and Atkinson 2000; Arcondeguy, Jack and Merrick 2001; Forchhammer 2004, 2008; Ninfa and Jiang 2005; Leigh and Dodsworth 2007; Osanai and Tanaka 2007; Uhrig, Ng and Moorhead 2009; Huergo, Chandra and Merrick 2013; Huergo and Dixon 2015; Forchhammer and Lüddecke 2016; Forcada-Nadal et al. 2018; Bueno-Batista and Dixon 2019).
The principal function of PII proteins is conserved over all domains of life (Fig. 2). They are small trimeric signal-transducing proteins with characteristic structural and functional features (Fig. 2A). All PII proteins characterized to date are able to bind adenyl-nucleotides (ATP and ADP), and most PII proteins can in addition bind 2-OG (reviewed in Forcada-Nadal et al. 2018). Binding of the effector molecules occurs in the clefts formed at the contact sites of the subunits. Two small loops from opposing subunits (the B- and C-loop) form a clamp that coordinates the phosphates of the adenyl-nucleotides. The bound nucleotide protrudes in the cleft and extends towards the basal part of the large, surface exposed T-loop. Subtle differences in the mode of nucleotide binding result in differential contacts, leading to different conformations of the T-loop structure, depending on which nucleotide has bound (Zeth, Fokina and Forchhammer 2014). In all cases studied so far at the structural level, binding of 2-OG requires the presence of Mg2+-ATP in the binding pocket. The Mg2+ ion, partially coordinated by the phosphates of ATP, completes its hexagonal coordination sphere by interacting with the oxygen atoms from the carbonyl and carboxy moiety of 2-OG (Fokina et al. 2010; Truan et al. 2010). In addition, 2-OG makes a salt-bridge to a highly conserved Lys-residue located at the posterior end of the T-loop. The binding of 2-OG induces a significant conformational change in the T-loop structure, which becomes more tightly folded upon binding this effector. As a result of the different metabolite coordination events, the T-loop adopts various conformations, depending on the bound metabolite, and thereby, its conformation reflects the integrated information on the actual status of effector molecules. In order to transduce this information towards the signaling targets, the T-loop acts as a versatile contact module, evolved to interact with various receptor proteins. These interactions are dependent on the conformational state of the T-loop. In general, the 2-OG induced T-loop conformation abrogates most PII-protein interactions whereas different receptors may prefer the ATP- or the ADP-ligated state of PII (Zeth, Fokina and Forchhammer 2014).
Figure 2.
PII-structure and general targets of PII proteins among bacteria. (A), Cartoon representation of S. elongatus PII in complex with 2-OG-Mg2+-ATP (PDB entry 2XUL) viewed from top and side. The clefts between the subunits constitute the metabolite binding sites (for details see text). The distal part of the large, solvent exposed T-loop, is disordered and therefore, not resolved. The dotted lines connect the proximal parts of the T-loop. (B), General scheme of signal perception, integration and transduction by PII proteins, resembling the function of a central processor unit (Ninfa and Atkinson 2000). Through competitive binding of ATP or ADP and synergistic binding of 2-OG with Mg2+-ATP, PII responds to changes in the energy state and C/N balance. Moreover, in many cases, PII proteins might be modified at the tip of the T-loop by post-translational covalent modification (PTM). The various binding states and eventually covalent modifications result in differential interactions with a wide variety of receptors of the PII signal, the most prominent being transporters, key metabolic enzymes or transcription factors. Target proteins specific to cyanobacteria are written in green, universal targets in red. In grey, PII targets outside the cyanobacteria are indicated (TnrA and GlnR: Global nitrogen-transcription factors; NtrB-NtrC: Nitrogen regulatory proteins B and C; NagB: Glucosamine 6-phosphate deaminase; ATase: Adenylyl-transferase; DraT: Dinitrogenase reductase ADP-ribosyl-transferase; DraG: Dinitrogenase reductase glycohydrolase; ?: unidentified targets).
Cyanobacterial PII homologues exhibit about 50%–65% amino acid sequence identity to PII paralogues from proteobacteria (Forchhammer 2004). Most cyanobacterial genomes harbor only one gene encoding a PII protein (designated as glnB gene). Some strains, however, encode highly similar PII homologues, whose physiological function needs still to be clarified (Laichoubi et al. 2011). The first cyanobacterial PII protein was identified in the non-diazotrophic strain S. elongatus PCC 6301 (Harrison et al. 1990) and the gene cloned from S. elongatus PCC 7942 (Tsinoremas et al. 1991). The biochemical properties of recombinant S. elongatus PII protein revealed high similarities to that of E. coli PII proteins GlnB and GlnK (Forchhammer and Tandeau de Marsac 1994; Forchhammer and Hedler 1997). Binding affinities were originally determined by equilibrium dialysis and ultrafiltration experiments with radio-labelled effector molecules. These measurements resulted in average binding constants, with a stoichiometry of three binding sites per trimeric PII protein. Binding of the effector molecules ATP and 2-OG was demonstrated to be mutually synergetic (Forchhammer and Hedler 1997). The synergy is explained by a tight interaction network between the γ-phosphate of ATP and the carboxyl and oxo group of 2-OG, chelating a bridging Mg2+ ion, which strengthens the affinity of both effectors to PII (Fokina et al. 2010). ADP competes with ATP for the same binding sites but ADP does not support 2-OG binding (Fokina et al. 2010a; Fokina, Herrmann and Forchhammer 2011). Intriguingly, 2-OG negatively affects ADP binding (Lapina et al. 2018). Therefore, the effector 2-OG strongly affects the competition between ATP and ADP, such that elevated levels of 2-OG guarantee the formation of PII:Mg2+-ATP:2-OG complex (Jiang and Ninfa 2009; da Rocha et al. 2013).
In addition to the mutual influence of the effectors in their binding pocket, the three sites of PII from S. elongatus were shown to be occupied in a sequential manner, through anti-cooperative interactions of the subunits (Fokina et al. 2010; Fokina et al. 2010a; Zeth, Fokina and Forchhammer 2014; Lapina et al. 2018). For each effector molecule, the first site is bound with highest affinity, then site 2 can be occupied, followed by site 3. Molecular dynamics simulation revealed the underlying molecular basis: Binding of the first effector molecule to the S1 site induces, via intramolecular signal transduction, subtle asymmetric alterations in sites S2 and S3 (Ma et al. 2014). As a result, various binding states of PII proteins are possible, which span a wide range of effector molecule concentrations (da Rocha et al. 2013). The resulting multiple conformational states, reflecting different metabolic situations, allow the differential interaction with various receptors of PII signaling. The known PII targets in cyanobacteria will be summarized in the following sections, however, preliminary results indicate that PII could regulate more targets such as phosphoenolpyruvate carboxylase (PEPC) and hypothetical proteins like Sll0944 (Fig. 2B) (Hauf 2016; Spät 2017; Watzer et al. 2019). This highly organized effector molecule binding mode is a key requirement for the versatility of PII signaling and regulatory functions. Furthermore, complex formation of PII with its receptors can change the affinities to the various effectors, as demonstrated for PII-PipX interaction (Fokina et al. 2011, Zeth, Fokina and Forchhammer 2014). This allows PII to simultaneously regulate multiple targets with different sensitivities towards effector molecules ADP, ATP and 2-OG.
Using Förster-resonance energy transfer (FRET) technology, cyanobacterial PII proteins from S. elongatus PCC 7942 and Anabaena PCC 7120, were used to develop 2-OG biosensors for in vivo quantification and live monitoring of intracellular fluctuation of 2-OG levels (Lüddecke et al. 2017; Chen et al. 2018). Those PII-based 2-OG biosensors showed to be functionally active in vitro and in vivo for Anabaena sp. and in living human cell lines, with high sensitivity in wide dynamic ranges.
As a further prerequisite to fulfill the task of a multitasking regulatory protein, PII is expressed well in excess above the level of the various PII receptors (Guerreiro et al. 2016; Forcada-Nadal et al. 2018), which will be discussed in the following sections.
THE PII PHOSPHORYLATION CYCLE OF CYANOBACTERIAL PII PROTEINS
Covalent modification of PII proteins has been described for PII proteins from Proteobacteria and Actinobacteria, where PII was found to be uridylylated or adenylylated, through covalent linkage to tyrosyl residue 51 (Merrick 2015). In these cases, the UMPylation (or AMPylation) is catalyzed by bifunctional transferases/hydrolases, whose activity responds to the cellular glutamine level: low glutamine levels favor modification of PII, high glutamine levels de-modification. These modifications alter PII target-interactions, and thus, the status of glutamine is signaled by PII.
Originally, the PII protein in S. elongatus was identified in the course of P32 labeling experiments (Harrison et al. 1990) as one of the most prominent labeled proteins in cell extracts. Subsequent biochemical analysis revealed that, in contrast to the hitherto characterized proteobacterial PII homologues, S.elongatus PII is phosphorylated at a seryl-residue (Forchhammer and Tandeau de Marsac 1994, 1995, 1995a). The phosphorylation site was identified as S49, close to Y51 (the site, which is uridylylated in E. coli PII), a position, located at the tip of the large flexible T-loop, which promotes the contacts to most PII receptors. The PII trimer can be phosphorylated on one, two or three subunits, giving rise to four different modification states of PII (a non-phosphorylated and three different phosphorylated states).
The PII phosphorylation state depends on the growth conditions: In the presence of ammonia, essentially only non-phosphorylated PII occurs. By contrast, in nitrate-grown cells, intermediate levels of PII phosphorylation were observed, with the degree of phosphorylation varying with the CO2 supply to the cells: with high inorganic carbon (Ci) supply PII was considerably more phosphorylated than under low Ci conditions. The highest degree of PII phosphorylation was observed under conditions of nitrogen starvation, or when nitrogen assimilation was inhibited by the addition of inhibitors of the GS-GOGAT cycle (Forchhammer and Tandeau de Marsac 1995; Forchhammer 2004; Kloft, Rasch and Forchhammer 2005). However, it is important to state, that PII S49 phosphorylation seems not to be a general property of all cyanobacteria. Besides S. elongatus PII phosphorylation has only been rigorously proven in Synechocystis PCC 6803 (Hisbergues et al. 1999; Spät, Macek and Forchhammer 2015). In the marine picocyanobacterium Prochlorococcus marinus PCC 9511 as well as in some Nostocales, PII phosphorylation could not be detected (Hanson et al. 1998; Palinska et al. 2002). Therefore, despite that the gene for PII is universally conserved in cyanobacteria, phosphorylation of PII may not be universal. No other cyanobacteria, particularly no strains from Subsection II, III and V have yet been investigated with respect to PII phosphorylation. Another post-translational modification was described in the filamentous cyanobacterium Anabaena PCC 7120, namely, nitration at Tyr51 (Zhang et al. 2007), even though the physiological significance of this modification remains elusive so far.
Although a PII kinase activity could be characterized in cell-free extracts of S. elongatus, purification of the enzyme failed (Forchhammer and Tandeau de Marsac 1995a; Irmler et al. 1997), nor could a kinase be identified among putative kinase mutants. A more recent phosphoproteome study indicated that PII phosphorylation may be accomplished by multiple serine/threonine protein kinases, substituting each other (Spät 2017). By contrast, a PII-specific phosphatase (termed PphA) had been identified by knock-out mutagenesis of protein phosphatase M (PPM) homologues in Synechocystis PCC 6803 (Irmler and Forchhammer 2001). In vitro, dephosphorylation of phosphorylated PII (PII-P) by PphA is affected by the ligand binding status of PII: fastest dephosphorylation occurs in the absence of any effector molecules, ATP slightly decelerates the reaction and ADP does so even more strongly. Almost complete inhibition of PII-dephosphorylation could be achieved by high concentrations of 2-OG in combination with ATP (Ruppert et al. 2002), whereas other compounds had no effect on PII-P dephosphorylation (Forchhammer et al. 2004). Together, all in vitro data suggested a model, according to which the PII modifying enzymes phosphorylate or dephosphorylate PII in response to the ligand-binding status of PII, with the ATP-2-OG bound form resulting in maximal phosphorylation of PII. Accordingly, the phosphorylation state of PII would mirror the ATP/2-OG level of the cells.
In a PphA-deficient mutant, phosphorylated forms of PII are maintained at high levels under conditions, in which rapid dephosphorylation of PII-P occurs in the wild-type. This finding indicates that other cellular phosphatases have only low reactivity towards PII-P (Kloft, Rasch and Forchhammer 2005). Apparently, PphA is able to specifically recognize phosphorylated PII in the 2-OG free state. In fact, the crystal structure of the PphA homologue from Thermosynechococcus elongatus (tPphA) revealed the presence of a flexible subdomain (termed FLAP domain) near the active center that controls the access to the catalytic site (Schlicker et al. 2008). More detailed analysis, including mutational analysis and cross-linking studies, showed that in addition to the FLAP subdomain, regions around the active site are required to ensure substrate-specific dephosphorylation. Only when a perfect fit between substrate and enzyme occurs, is the enzyme activated with Mg2+ as the catalytic metal (Su, Schlicker and Forchhammer 2011; Su and Forchhammer 2013). Furthermore, it could be shown that a third Mg2+ ion, hitherto not recognized in PPM enzymes, is required for its catalytic activity, an observation, that has general relevance for PPM phosphatases (Su, Schlicker and Forchhammer 2011).
In vivo characterization of PphA in Synechocystis sp. showed that the expression level of PphA is tuned by the nitrogen source of the growth medium. The presence of nitrate, and even stronger, the presence of nitrite, stimulated high expression levels of PphA (Kloft, Rasch and Forchhammer 2005). This suggests a particular role for PphA, presumably rapid PII-P dephosphorylation, when nitrate or nitrite are used as N-source and agrees with the fine-tuned response of the PII phosphorylation state to subtle changes in the growth conditions, when nitrate is the nitrogen source (Kloft and Forchhammer 2005).
As will be developed in the following sections, the phosphorylation state of PII has different consequences for different PII targets. As a general rule, it appears that phosphorylation abrogates the interaction with most PII targets. The interactions that are abrogated by PII phosphorylation are also negatively affected by the binding of ATP-2-OG to non-phosphorylated PII. Since in vitro phosphorylation of PII only responded to 2-OG, phosphorylation seems to be redundant to the allosteric 2-OG signal. However, it should be kept in mind that in contrast to fast signaling through allosteric 2-OG binding, phosphorylation on S49 maintains PII for prolonged periods in a non-interacting state: This is because dephosphorylation of PII-P takes more time than the dissociation of the PII-P-2-OG-ATP complex, delaying the response. Moreover, PII phosphorylation may be fine-tuned under certain metabolic situations according to hitherto unknown signals, to allow for an even more elaborated integration of metabolic signals.
PII AND THE CONTROL OF NITROGEN COMPOUND TRANSPORTERS
Cyanobacteria can use various sources of combined nitrogen with different preference. When ammonium is present, it prevents the use of other nitrogen sources by repressing the corresponding metabolic genes and inhibiting the uptake of these compounds, a phenomenon known as global nitrogen control (reviewed by Herrero, Muro-Pastor and Flores 2001; Forchhammer 2004; Esteves-Ferreira et al. 2018). At the level of gene expression, nitrogen control operates via the global regulator NtcA (see below), whereas the mechanism of uptake inhibition remained elusive. A recent analysis of the interactome of FLAG-tagged PII provided direct evidence that PII plays a central role in the control of the various nitrogen-related transporters (Figs 1 and 2B) (Watzer et al. 2019).
In many heterotrophic bacteria and in archaea, PII proteins of the GlnK subfamily directly interact with ammonium transport (Amt) proteins to regulate their activities. Typically, amt and glnK genes form an operon and GlnK specifically interacts with Amt under nitrogen excess conditions to prevent an over-accumulation of intracellular ammonium to keep the GS-GOGAT cycle in balance (Arcondeguy, Jack and Merrick 2001). In the GlnK-Amt complex, the GlnK effector molecule sites are occupied by ADP to stabilize the specific T-loop conformation for proper Amt interaction (Conroy et al. 2007; Gruswitz, O'Connell and Stroud 2007). Under nitrogen excess conditions, low 2-OG levels allow ADP to replace ATP in the GlnK complex, promoting ADP-GlnK-Amt complex formation and thereby closing the NH3 channels (Radchenko, Thornton and Merrick 2010). A PII-deficient mutant of S. elongatus showed aberrant uptake activity for methylammonium, which is used as an ammonium analogue for AMT transporters (Forchhammer and Tandeau de Marsac 1995). Furthermore, PII mutants were sensitive towards elevated ambient ammonium concentrations (Watzer et al. 2015, 2019), suggesting that cyanobacterial PII controls ammonium transport. Indeed, in the immunoprecipitate of FLAG-tagged PII from Synechocystis PCC 6803, the major ammonium permease Amt1 was strongly enriched, and bacterial-two hybrid (BACTH) analysis confirmed direct PII-Amt1 interaction (Watzer et al. 2019). The BACTH assays with different PII variants suggested that the PII-Amt1 interaction is similar to the well-known GlnK-AmtB interaction. In contrast to heterotrophic bacteria, control of ammonium uptake has an additional purpose than to adjust the supply of GS with ammonium. Excessive ammonium uptake in cyanobacteria must be prevented because high intracellular ammonium concentrations are toxic due a photosensitizing effect of ammonia on photosystem II (PSII). In combination with high-light stress, ammonia leads to detrimental photo-destruction of PSII (Drath et al. 2008; Dai, Qiu and Forchhammer 2014).
For the uptake of nitrate, one of the most abundant sources of combined nitrogen, cyanobacteria may employ two different types of transport systems: many fresh-water cyanobacteria employ an ATP-binding cassette (ABC) transport complex (NrtABCD) whereas in marine and terrestrial strains, a nitrate/nitrite permease (NrtP) of the major facilitator superfamily (MFS) has been identified (Sakamoto, Inoue-Sakamoto and Bryant 1999; reviewed in Ohashi et al. 2011). Subsequent to its uptake, nitrate is reduced to nitrite via assimilatory nitrate reductase (NarB) and further reduced to ammonia through nitrite reductase (NirA) (reviewed in Flores et al. 2005).
Uptake of nitrate is not only subjected to short-term ammonium inhibition but likewise, obstruction of CO2 fixation immediately depresses nitrate transport. Both types of short term-responses are absent in PII-deficient mutants of strains S. elongatus and Synechocystis PCC 6803 (Forchhammer and Tandeau de Marsac 1995; Lee et al. 1998; Kobayashi et al. 2005; Hisbergues et al. 1999). Phosphorylation of S49 seems to play a pervasive role in this response: replacement of the PII wild-type version by the non-phosphorylatable S49A variant creates a phenotype that shows constitutive low nitrate uptake and grows poorly on nitrate supplemented medium (Lee et al. 2000). Conversely, mutants that lack the PII-specific phosphatase PphA and thereby accumulate PII in a highly phosphorylated state are impaired in proper control of nitrate utilization. Like PII null mutants, they excrete nitrite when grown in the presence of nitrate, especially under low light conditions, indicating an imbalance between nitrate uptake and its subsequent reduction to nitrite and ammonium due to limiting reduction equivalents (Kloft and Forchhammer 2005). However, in contrast to PII-deficient mutant, the PphA mutant and PII phosphomimetic variant strains (PII variants replacing Ser 49 by Asp or Glu residues) were still able to perform short-term inhibition of nitrate uptake (Lee et al. 2000; Kobayashi et al. 2005; Kloft and Forchhammer 2005), indicating that PII might mediate short-term ammonium inhibition independent of its phosphorylation state.
Among the PII-interacting proteins from Synechocystis PCC 6803, the NRT complex was identified by a mass spectrometry-based pulldown analysis. Subsequent bacterial-two hybrid analysis confirmed direct interaction of PII with both ATPase subunits NrtC and NrtD (Watzer et al. 2019). Regulation of NRT by PII appears to be more complex than the simple negative regulation of AMT by PII. The non-phosphorylated form of PII has higher affinity towards NrtC and NrtD than the phosphomimetic variant PII-S49E, which is still able to mediate the short-term ammonium inhibition of AMT. Moreover, the subunit NrtC possesses a C-terminal regulatory domain with a nitrate-binding pocket (Koropatkin, Pakrasi and Smith 2006). Truncation of this domain renders the NRT complex ammonium insensitive (Kobayashi et al. 1997). Together, these facts suggest that both ATPase subunits of NRT, and in particular the regulatory domain of NtrC, act in concert with different states of PII to inhibit nitrate transport. In agreement with these data, following scenario is conceivable: The non-phosphorylated form of PII binds to NRT and adjusts the balance between nitrate uptake and its subsequent reduction. Under conditions of limiting photosystem (PS)I-reduced ferredoxin (e.g. low light), nitrite reductase, due to its higher electron demand, lags behind nitrate reductase, resulting in intermediate nitrite accumulation; as a consequence, the cells need to tune down nitrate uptake, which is achieved by non-phosphorylated PII. Inhibition of CO2 fixation results also in PII dephosphorylation (Forchhammer and Tandeau de Marsac 1995), which could explain the block of nitrate uptake under these conditions. On the other hand, phosphorylated forms of PII can also inhibit NRT, but only in response to ammonium exposure. For this response, the ammonium-prompted change of the effector molecule binding state of PII is likely the driving force. Understanding the mechanistic details, how PII and the regulatory domain of NrtC communicate and together control nitrate uptake in response to various environmental cues requires further investigation.
Analysis of the PII interactome in Synechocystis PCC 6803 finally identified the ABC-type urea transporter (UrtABCDE) as a novel PII target (Watzer et al. 2019). Urea uptake experiments confirmed that a PII-deficient mutant lost control over urea transport. The ATP-binding UrtE subunit was identified as direct interaction partner of PII. Surprisingly, BACTH assays showed that some of PII variants that were unable to interact with Amt1 or NrtC and NrtD were not negatively affected in UrtE interaction. This implies a distinct mechanism of PII-URT interaction as compared to Amt or NRT interaction. Further biochemical studies are necessary for a more detailed understanding of the control of URT and NRT transporters by PII.
PII-NAGK INTERACTION AND NITROGEN STORAGE CONTROL
The first target of cyanobacterial PII signaling, the argB product, was identified by yeast-two hybrid screenings (Burillo et al. 2004; Heinrich et al. 2004). The argB gene encodes the enzyme N-acetyl-N-glutamate kinase (NAGK), which is the controlling enzyme of the ornithine/arginine pathway. NAGK catalyzes the conversion of N-acetyl-L-glutamate to N-acetyl-L-glutamyl-phosphate, which is further converted to ornithine, from where arginine and polyamines are derived. The reaction is feed-back inhibited by the end-product of the pathway, arginine. The purpose of PII-NAGK interaction is to subject this pathway to global C/N control (Fig. 1). Simplified, it can be stated that PII, when signaling energy- and nitrogen-rich conditions (or a surplus of nitrogen relative to carbon) due to binding of Mg2+-ATP but not 2-OG, activates NAGK and reliefs it from arginine feedback inhibition, thereby enhancing the metabolite flow in this pathway (Fig. 3). As explained below, this allows the cells to store excess nitrogen under such conditions.
Figure 3.
PII controls the arginine pathway by NAGK interaction. The arginine synthesis pathway is controlled by PII-dependent activation of the rate-limiting step, the N-acetyl glutamate kinase (NAGK) reaction. Under nitrogen sufficient, low 2-OG conditions, two PII trimers (in ATP complex) symmetrically associate with the hexameric NAGK (PDB entry 2V5H) to activate the reaction and to tune down arginine feedback inhibition. This allows increased flux towards arginine, which then may be used also to store excess nitrogen in form of cyanopyhcin (for details see text). Under nitrogen-limiting conditions, the increasing 2-OG levels lead to dissociation of the complex. Free NAGK is highly sensitive towards arginine-feedback inhibition. Subsequent phosphorylation of PII robustly prevents further NAGK interaction, unless PII becomes dephosphorylated again in response to sustained nitrogen excess conditions.
The interaction between PII and NAGK was studied in great detail: a high-resolution structure of the complex was solved (Llácer et al. 2007) and the dynamic behavior of complex formation was initially analyzed by surface plasmon resonance spectroscopy (Maheswaran, Urbanke and Forchhammer 2004; Beez et al. 2009; Fokina et al. 2010a). Even more detailed insights could be achieved by setting up a FRET assay with PII and NAGK proteins fused to suitable fluorescent proteins (Lüddecke and Forchhammer 2013). The key findings of these studies can be summed up as follows: PII in its non-phosphorylated state binds to NAGK with high affinity. The complex forms in the absence of low-molecular weight effectors and is equally stable in presence of ATP. However, PII complexed with ADP, or PII in complex with 2-OG-Mg2+-ATP is unable to bind NAGK. When 2-OG is titrated to the PII-ATP-NAGK complex, dissociation of the complex occurs with an IC50 for 2-OG that resembles the dissociation constant of the third (the lowest affinity) binding site, which is in the range of 0.1 mM. This suggests that all three 2-OG sites have to be occupied to obstruct PII-NAGK interaction. This 2-OG concentration is in the range of physiologically relevant concentration (Huergo and Dixon 2015), indicating a physiologically meaningful effect. Accordingly, elevated cellular levels of 2-OG (high carbon – low nitrogen conditions) would negatively affect PII-NAGK interaction and thereby prevent NAGK activation. Intriguingly, FRET analysis revealed that even under excess concentrations of 2-OG a residual FRET signal could be observed, indicating a weak PII-NAGK interaction. It seems possible, that after dissociation of the tight complex, NAGK stays weakly associated to PII, possibly via a loose encounter complex. Formation of a transient PII-NAGK encounter complex involving a salt bridge between R233 (of NAGK) and E85 (of PII) was also postulated from the analysis of a hyperactive NAGK-binding variant of PII (PII-I86N) (Fokina et al. 2010a).
The negative effect of ADP on PII-NAGK complex formation raised the possibility, that NAGK activation by PII may be modulated by the energy status of the cells. However, careful analysis of the ADP effect revealed that in presence of ATP, a high excess of ADP over ATP is required to affect catalytic activity of NAGK in presence of PII and inhibitory concentrations of arginine, which seems to be physiologically less relevant (Fokina, Herrmann and Forchhammer 2011; Lüddecke and Forchhammer 2013). A reasonable interpretation is that NAGK activation by PII is mainly tuned by the cellular 2-OG levels. Furthermore, the phosphorylation state of PII is definitely crucial for NAGK activation: the phosphorylation site S49 organizes a hydrogen-bonding network with NAGK residues that stabilizes the complex (Fordaca-Nadal et al. 2018). Any changes at position S49 (either by phosphorylation or mutation), therefore, prevent efficient NAGK-PII interaction (Heinrich et al. 2004; Lüddecke and Forchhammer 2013). The hydrogen-bonding network, organized by S49, is the key for catalytic activation of NAGK through tightening the catalytic center (Llácer et al. 2007).
When in vivo PII dissociates from NAGK following an increase in 2-OG levels (e.g. in response to nitrogen-limited conditions), it is available as a substrate of PII kinase and can be phosphorylated. Phosphorylation itself leads to an inhibition of complex formation that persists until PII is dephosphorylated again. Therefore, when nitrogen becomes available after a period of nitrogen starvation, phosphorylated PII must first be dephosphorylated through phosphatase activity, before NAGK can be activated. This leads to a delay in activation of arginine biosynthesis, ensuring that this pathway is only activated when sufficient nitrogen is available.
The ultimate rationale for the sophisticated control of NAGK by PII appears to be the need to tightly control arginine synthesis in response to the central metabolic state of the cells, since the synthesis of arginine demands high energy and nitrogen supply. In addition to its role as an essential amino acid for protein synthesis and precursor for polyamine synthesis, arginine plays an important role as a combined nitrogen buffer and for the synthesis of nitrogen storage compounds (Llácer, Fita and Rubio 2008). A recent metabolome flux analysis demonstrated that in Synechocystis PCC 6803, arginine is an intermediate in a metabolic cycle (the ornithine-ammonia cycle), where the newly synthesized arginine is converted by arginine dihydrolase into ornithine, ammonia and CO2 (Zhang et al. 2018a). This cycle seems to optimize nitrogen acquisition under conditions of fluctuating nitrogen supply and is widespread in cyanobacteria except the Prochlorococcus and Synechococcus clade (Flores, Arevalo and Burnat 2019).
Many cyanobacteria are able to synthesize the biopolymer cyanophycin, which consists of a poly-L-aspartic acid backbone with each carboxyl group linked via isopeptide –bonds to an arginine residue (multi-[L-arginyl-poly-L-aspartic acid]) (Allen and Smith 1969; Simon 1971; Allen and Weathers 1980). Initially, cyanophycin was identified as a constituent of granular inclusions in cyanobacteria (Allen and Weathers 1980), and therefore was referred to as cyanophycine granule polypeptide (CGP). CGP is synthesized by a single enzyme, cyanophycin synthetase, CphA1, in a two-step reaction from L-aspartate and L-arginine, using one ATP per amino acid. In principle, two major functions of cyanophycin have been described. In filamentous cyanobacteria, large cyanophycin granules form in the polar neck region of heterocysts. This heterocystous CGP seems to be intimately involved in transfer of fixed nitrogen from the heterocysts to the photosynthetically active vegetative cells (Burnat, Herrero and Flores 2014). By contrast, in non-heterocyst forming cyanobacteria, CGP accumulates in most cases only transiently during unbalanced growth. When growth is impaired for various reasons but nitrogen is available, the cells are still able to store the excess nitrogen in the form of cyanophycin (Watzer and Forchhammer ).
Under balanced growth, the unicellular cyanobacterium Synechocystis PCC 6803 does not accumulate cyanophycin. The Km of CphA1 for arginine is well above the cellular concentration of this amino acid, and therefore, no synthesis occurs. However, when, as a consequence of unbalanced growth, the cellular level of arginine increases, cyanopycin synthesis can occur. Therefore, the cellular level of arginine appears to be the trigger for cyanophycin accumulation (Maheswaran et al. 2006; Watzer et al. 2015) and this response is mediated by PII: In the PII-NAGK complex, the flux into the arginine pathway increases due to NAGK activation (Maheswaran et al. 2006). When nitrogen-starved Synechocystis cells that have arrested cell growth are suddenly exposed to excess ammonia, cells of the wild-type strain increase the arginine levels, leading to transient cyanophycin accumulation. Under the same conditions, a PII-deficient mutant was unable to accumulate cyanophycin and to increase arginine levels due to a permanently low NAGK activity, due to lacking PII activation. In accord with the need of non-phosphorylated PII for NAGK activation, a PII phosphatase deficient mutant was also unable to accumulate cyanophycin: PII remained in a highly phosphorylated state and consequently, NAGK could not be activated and no cyanophycin was produced (Maheswaran et al. 2006). A recent study clarified that PII-controlled cyanophycin synthesis provides Synechocystis wild-type cells with a clear fitness advantage over cyanophycin-deficient mutant cells in environments with fluctuating nitrogen supply. Trapping assimilated nitrogen in form of cyanophycin enables Synechocystis PCC 6803 to optimize nitrogen assimilation under nitrogen-poor conditions, in particular during fluctuating nitrogen supply and day/night cycles (Watzer and Forchhammer 2018).
The PII variant I86N was identified to be a hyperactive NAGK binder (Fokina et al. 2010a). Replacing the glnB wild-type allele in Synechocystis PCC 6803 by a mutant version encoding PII-I86N, the resulting strain massively overproduces cyanophycin (Watzer et al. 2015). NAGK is permanently activated by PII-I86N, and in consequence, the cells overproduce arginine and accumulate cyanopyhcin already during balanced growth. Under optimal production conditions (phosphate-limited medium) this strain can accumulate up to 40%–50% of the cell dry mass as cyanophycin (Watzer et al. 2015; Trautmann et al. 2016). The use of PII-I86N engineered strain in the ultrahigh-density cultivation system yielded approximately a 4-fold increase in the total cyanopyhcin yield over the previously reported maximal yield in 4 days than 12 days (Lippi et al. 2018).
Whereas the role of PII-NAGK complex formation for the control of cyanophycin is very obvious, this cannot be the only explanation. Cyanophycin is only produced by a subset of cyanobacteria but strains such as S. elongatus do not synthesize it. Nevertheless, the interaction with NAGK seems to be universal among all oxygenic phototrophs that have a PII signaling system (Beez et al. 2009; Chellamuthu, Alva and Forchhammer 2013; Lapina et al. 2018; Selim et al. 2019). The PII-NAGK interaction has been conserved during the evolution from the endosymbiotic cyanobacterial ancestor to the chloroplasts of green plants (Chellamuthu, Alva and Forchhammer 2013). The need to tightly regulate the ornithine/arginine pathway in response to the C/N state of the cells appears, therefore, to be of fundamental importance. In red algae, PII-NAGK interaction perfectly resembles the situation in cyanobacteria (Lapina et al. 2018). In the further evolution of plant chloroplasts, PII has acquired an additional sensory input: a short C-terminal extension acts a low affinity glutamine-binding site and makes NAGK activation by PII glutamine-dependent. Thereby, only at elevated glutamine levels, corresponding to nitrogen-rich conditions, can PII activate NAGK (Chellamuthu et al. 2014; Selim et al. 2019).
PII-ACCase INTERACTION AND FATTY ACID ACCUMULATION
In a search for interaction partners of plant PII proteins, the biotin carboxyl carrier protein subunit (BCCP) of chloroplast acetyl-CoA carboxylase (ACCase) was identified as a plant PII interacting protein (Feria Bourrellier et al. 2010). ACCase catalyzes the committed step in fatty acid biosynthesis by carboxylating acetyl-CoA in an ATP-dependent reaction. The product of the reaction, malonyl-CoA, is then the starting building block for fatty acid synthesis. Recombinant Arabidopsis PII exerted an inhibitory effect on the activity of chloroplast ACCase, and the inhibitory effect could be reversed by 2-OG. This discovery initially suggested that in plants, PII might have acquired novel functions and might be involved in the regulation of lipid synthesis, which are a major plant carbon-storage compound that is made inside the chloroplasts.
Subsequent proteomic studies with bacterial PII proteins revealed that this property is not plant-specific but instead, PII proteins from the proteobacteria Azospirillum brasiliense and Escherichia coli also interacted with the respective biotinylated BCCP proteins (Rodrigues et al. 2014). Using a reconstituted ACCase complex, biochemical analyzes showed that the GlnB paralogues of the PII proteins from A. brasiliense and E. coli, but not the GlnK paralogues, formed a stable ternary complex with biotin carboxylase and the BCCP. Kinetic analyses demonstrated that PII proteins decrease the turnover rate of ACCase but do not affect the Km for acetyl-CoA (Gerhardt et al. 2015). Like in plants, binding of GlnB to the BCCP subunit and inhibition of ACCase activity was prevented by 2-OG. This suggested that the conformational switch in the T-loop of PII, caused by 2-OG binding, precludes interaction with biotinylated BCCP. Thus, the accumulation of 2-OG under carbon excess conditions stimulates the synthesis of the carbon storage as lipids, whereas under nitrogen-rich conditions with lowered 2-OG contents the lipid biosynthesis becomes decreased. In agreement with the assumption that the T-loop is involved in BCCP interaction, GlnB uridylylation turned out to prevent its interaction with the ACCase complex. The striking correspondence between plant and bacterial PII proteins suggested that ACCase regulation might be a highly conserved property of PII proteins. In agreement, deletion of PII encoding genes in E. coli strongly enhanced the fatty acids biosynthesis up to 80% due to the absence of the negative control of PII on the ACCase activity, while overexpression of PII reduced the malonyl-CoA levels, the product of ACCase, by 30% (Rodrigues et al. 2019).
It was, therefore, not surprising to find that the ACCase reaction in cyanobacteria is regulated by PII as well. To identify the hitherto unclear physiological consequences of PII-ACCase interaction, cyanobacterial PII proteins were analyzed with respect to binding to BCCP (Hauf et al. 2016). Both, S. elongatus and Synechocoystis PII proteins interacted with biotinylated BCCP. The interaction only took place in the presence of ATP, implying that only the ATP-ligated state of PII is able to interact with BCCP. Titration experiments with ATP suggested that all three binding sites of PII have to be occupied by ATP in order to promote interaction with BCCP. Conversely, 2-OG inhibited the interaction of PII with BCCP in a dose-dependent manner, such that occupation of all three sites is likely necessary to antagonize the interaction with BCCP. Analysis with mutant variants showed that the phosphomimetic PII variant S49E is unable to interact with BCCP, suggesting that the phosphorylated form of PII would also not interact with BCCP. The cyanobacterial PII proteins, like their proteobacterial counterparts, tuned down the activity of ACCase. These results suggest a scenario, where high carbon/low nitrogen conditions (PII mainly in the 2-OG-Mg2+-ATP complex and furthermore phosphorylated at Ser49) would prevent the inhibitory interaction of PII with ACCase, allowing maximal ACCase activity. On the other hand, when PII is mainly in the ATP-complexed state (corresponding to high-nitrogen/low-carbon conditions under energy rich, illuminated, conditions), it would bind and tune down ACCase activity.
To clarify the physiological consequences of PII—ACCase interaction, fatty acid and acetyl-CoA levels have been compared between wild-type and PII-deficient Synechocystis cells grown under different carbon and nitrogen regimes. A small to moderate increase in lipid bodies formation and fatty acid levels were observed in PII deficient cells. With about 20%, the difference was most strongly pronounced in ammonia-grown cells, consistent with the fact that in ammonia-grown cells, PII is not phosphorylated and the 2-OG levels are expected to be low, conditions under which inhibition of ACCase by PII should be maximal. By contrast, in nitrate-grown cells, less than 10% difference was observed between wild-type and PII-deficient cells, in agreement with PII being present in partially phosphorylated forms that do not interact with ACCase. In agreement with the study from Synechocystis PCC 6803, investigation of a PII-deficient mutant of S. elongatus reported increased levels of ACCase and total lipid content (by about 29% and 34%, respectively) while the levels of acetyl-CoA were reduced by about 11% (Verma et al. 2018).
Apparently, due to the lack of PII-inhibition on ACCase activity, the steady-state pool of acetyl-CoA may be depleted by an over-active ACCase under certain conditions. Since the levels of acetyl-CoA fall well below the Km for ACCase, the net flux into fatty acid pools is not as strongly increased as the elevated activity of ACCase would intuitively suggest. In the absence of PII-regulation, the rate-limiting factor for the turn-over of ACCase reaction is the low steady-state level of acetyl-CoA, whereas in the presence of PII, ACCase-reaction is slowed down by PII-control. Since acetyl-CoA also enters into the citric acid cycle and is used for several other biochemical reactions, PII, by controlling ACCase, could control the distribution of the carbon flux towards either the TCA and GS-GOGAT cycle or towards lipid biosynthesis. In Fig. 1, a regulatory feedback loop operated by PII is proposed. Increasing 2-OG levels would enhance ACCase activity, lowering the steady state levels of acetyl-CoA and thereby decreasing the flux into the TCA cycle. Conversely, dropping 2-OG levels would cause inhibition of ACCase activity, thereby increasing the acetyl-CoA levels and promoting the flux into the TCA cycle to synthesize 2-OG as precursor for nitrogen assimilation via the GS/GOGAT cycle. In accord with this model, metabolome analysis of PII-deficient cells revealed strongly reduced levels of 2-OG and glutamate in PII-deficient mutants (Schwarz et al. 2011 and 2014).
The identification of PII control over ACCase activity could have biotechnological applications. It has already been shown that PII deficient mutants accumulate some triacylglycerols. Similarly, PII knock-down lines in green algae have been shown to accumulate lipids (Zalutskaya et al. 2015).
The PII-PipX-NtcA NETWORK AND NITROGEN-DEPENDENT TRANSCRIPTIONAL CONTROL
PipX was identified as the PII-interacting protein X (PipX), a small protein of unknown function, in yeast-two hybrid screening using S. elongatus PII as bait (Burillo et al. 2004). The pipX gene encodes a protein of 89 amino acids that showed no similarity to any protein of known function. However, PipX homologues are found in all cyanobacterial genomes. Further yeast-two hybrid analysis revealed that PipX also interacted with the global transcription factor NtcA (Espinosa et al. 2006; Espinosa, Forchhammer and Contreras 2007). Since the PII-PipX-NtcA regulatory network has been comprehensively reviewed recently (Forcada-Nadal et al. 2018; Zhang et al. 2018), only a brief summary of key points will be given in the following paragraph.
NtcA is the master regulator of nitrogen control at the transcriptional level in cyanobacteria
In the search for a factor, that is required for the expression of genes encoding enzymes for the assimilation of nitrogen sources other than ammonium, mutants have been found that can only grow in the presence of ammonium. The corresponding mutants are deficient in transcription factor NtcA, identified as a global transcription factor in S. elongatus (Luque, Flores and Herrero 1994) as well as in Anabaena PCC 7120 (Wei, Ramasubramanian and Golden 1994) (where the gene was initially termed bifA). Today, it is generally accepted that the expression of genes necessary for the assimilation of nitrogen sources alternative to ammonium, such as nitrate and nitrite, depends on the activity of NtcA. NtcA appears to be the key regulator of nitrogen-controlled gene expression in all cyanobacteria (Herrero, Muro-Pastor and Flores 2001; Herrero et al. 2004; Muro-Pastor, Reyes and Florencio 2005; Luque and Forchhammer 2008; Domínguez-Martín et al. 2018). NtcA belongs to the CRP/FNR family of transcriptional regulators and binds to a palindromic sequence with the consensus sequence GTA(N8)TAC in the promoter region of the respective genes.
The entire NtcA regulon was recently unraveled in Synechocystis PCC 6803 using a combination of RNA-seq analysis and chromatin immunoprecipitation using NtcA-specific antibodies and sequencing the retrieved DNA fragments. When starved for nitrogen, 79 genes were directly regulated by NtcA, of which 51 genes were activated and 28 genes were repressed by NtcA (Giner-Lamia et al. 2017). This study provides a comprehensive overview of the genes, whose expression depends on NtcA. In addition to genes directly related to nitrogen metabolism, several NtcA dependent genes play roles in C-metabolism and in general central cellular processes. Most of the NtcA-activated genes are mainly involved in nitrogen assimilation, encoding transporters for nitrogen scavenging such as ammonia-, nitrate/nitrite-, amino acid- and urea-transporters, N-assimilating enzymes such as GS and GOGAT subunits, nitrate and nitrite reductase subunits. Moreover, NtcA controls a variety of genes for regulatory factors, such as PII, the response regulator Rre37 and some other two component systems and NtcA itself. Furthermore, genes for oxidative stress response and genes encoding glycogen phosphorylase (sll1356) for sugar catabolism are activated by NtcA, whereas the gene for transketolase (sll1070) involved in carbon metabolism and others encoding mostly proteins of unknown function are repressed. The most strongly repressed genes are the GS inhibitor proteins IF7 and IF17 in Synechocystis PCC 6803. In addition, NtcA also controls several non-coding RNAs, for example, NsiR4 involved in the downregulation of IF7 (Klahn et al. 2015a), as well as 26 hypothetical or unknown proteins (Giner-Lamia et al. 2017).
As in other cases, the global N-regulatory transcriptional factor NtcA can positively or negatively affect gene expression. When NtcA acts as a repressor, the NtcA binding site overlaps the RNA-polymerase binding site, whereas in genes activated by NtcA, its binding site lies upstream of the RNA polymerase binding site, such that a positive interaction is possible (reviewed in Luque and Forchhammer 2008). The in vitro DNA binding affinity of NtcA is directly modulated by 2-OG (Tanigawa et al. 2002; Vazquez-Bermudez, Herrero and Flores 2002), which directly links NtcA to the cellular nitrogen status. However, 2-OG binding by NtcA is not sufficient to explain the nitrogen responsive activity of NtcA since PII deficient mutants were shown to be partially impaired in the expression of NtcA-dependent genes (Aldehni et al. 2003; Paz-Yepes, Flores and Herrero 2003). Here, the PipX protein comes into play, which directly connects the PII and NtcA regulation.
PipX is a transcriptional co-activator of NtcA. This initial suggestion (Espinosa et al. 2006 and Espinosa, Forchhammer and Contreras 2007) is now supported by a solid body of evidence. In complex with PipX, NtcA resides in the most active state for DNA binding (Camargo et al. 2014; Forcada-Nadal et al. 2014). PipX mutually exclusively binds to either NtcA or PII (Espinosa et al. 2006; Llácer et al. 2010). The partner-swapping by PipX is controlled by the 2-OG levels. In the Mg2+-ATP-2-OG complex, PII is unable to interact with PipX, whereas 2-OG-NtcA has high affinity towards PipX, forming a ternary 2-OG-NtcA-PipX complex needed for promoter recognition. Therefore, at high 2-OG levels, PipX associates to NtcA. By contrast, at low levels of 2-OG, PII is free of 2-OG and now binds avidly to PipX, whereas at the same time, NtcA loses 2-OG and concomitantly the ability to bind PipX (Fig. 4). The mechanistic details of complex formation have been elucidated by solving the structures of the PipX-PII and PipX-NtcA-2-OG complex (Llácer et al 2010; Zhao et al. 2010). The S49 phosphorylation site of the PII protein does not interfere with the PipX interaction surface, explaining the ability of phosphomimic variants of PII S49D to interact with PipX (Espinosa et al. 2006; Llácer et al 2010). PII-PipX interaction is exclusively tuned by the effector molecule binding state of PII. In the ADP-complexed state, PII has higher affinity towards PipX than in the ATP state (Zeth, Fokina and Forchhammer 2014), and conversely, PII in complex with PipX has increased affinity for ADP. Therefore, in vivo the formation of the PipX-PII complex seems to be favored when the ADP levels increase, corresponding to energy limited conditions, and this most probably represents a regulatory link between the energy and C/N status of the cells. In agreement, a study in S. elongatus revealed distinguishable localization patterns of PipX- and PII-fusions to fluorescent proteins during diurnal cycles, where they co-localized into a small-foci at the poles of S. elongatus during night phase (i.e. low ATP/ADP ratio) (Espinosa et al. 2018). A model was proposed according to which PII would trap PipX into these foci during the dark period to prevent the activation of NtcA at low ATP/ADP ratios.
Figure 4.
Indirect regulation of transcription factor NtcA by PII. Conditions of nitrogen sufficiency (low 2-OG levels) or low energy state (high ADP to ATP ratio) favor formation of the PII-PipX complex (PDB entry 3N5B), where PipX does not associate with NtcA. When 2-OG and ATP levels increase, NtcA successfully competes with PII for PipX. The NtcA-PipX complex (PDB entry 2XKO) has increased affinity to the NtcA-specific DNA binding sites and turns on transcription of NtcA-dependent genes.
The role of PipX could be even more complicated than initially anticipated, since PipX appears to be involved in other regulatory networks beyond NtcA (Espinosa et al. 2014). RNAseq analysis of PipX-deficient cells suggested the presence of at least three clusters of genes related to photosynthesis, metabolism and translation that operate in an apparently NtcA-independent manner. However, the factors to which PipX may bind to achieve this regulation remain to be characterized. PII is required to counterbalance potentially lethal processes triggered by PipX. Genetic studies in S. elongatus showed that knockout mutation glnB (encoding PII) is only successful, when the expression of PipX is abolished by spontaneous second-site mutations (Espinosa et al. 2010; Chang et al. 2013, 2014). In Nostoc punctiforme, knockout of the glnB gene was unsuccessful (Hanson et al. 1998), but so far, no attempt was made to knock out glnB in a PipX-deficient background. A glnB mutant in Anabaena PCC 7120 failed to activate NctA upon nitrogen step-down, implying that also this mutant may have acquired a second-site pipX mutation (Zhang et al. 2007).
The toxicity of PipX in absence of PII was attributed to yet unknown PipX interactions, which motivated a search for additional PipX-interacting factors in S. elongatus. Advanced methods of interaction screenings led to the discovery of a GntR-like regulator (termed PlmA) that interacts with the PII-PipX complex, while PlmA was unable to interact with PII or PipX alone (Labella et al. 2016). In complex with PII, three PipX monomers are arranged in a structure that allows interaction with PlmA. However, the physiological relevance of such novel ternary PII-PipX-PlmA complex remains elusive so far. PlmA is a putative transcription factor of yet unknown function and is universally conserved in all so-far sequenced cyanobacterial genomes and restricted to this phylum. The gene appears to be essential in S. elongatus (Rubin et al. 2015; Labella et al. 2016), but not in Synechocystis PCC 6803 or in Anabaena PCC 7120, where it seems to be involved in the regulation of photosystem stoichiometry and plasmid maintenance functions (Lee et al. 2003; Fujimori et al. 2005). A further protein, termed PipY, has been implicated in the cyanobacterial nitrogen regulatory network (Labella et al. 2017) The pipY gene is located in a conserved operon with pipX. Based on this genetic linkage, a functional relation between PipX and PipY was postulated, which was supported by phenotypic properties of respective mutants. PipY is a member of the conserved COG0325 family of PLP-binding proteins with unknown function. However, physical interaction with neither PipX nor PII was shown and it remains unclear whether PipY is involved in N-homeostasis regulation. Recently, PipX was proposed to exert a regulatory function on PipY expression in cis to maintain appropriate PipX-PipY stoichiometry (Cantos et al. 2019).
THE PII RECEPTOR PamA AND NrrA/SigE REGULATION
PamA is a poorly characterized PII target, a transmembrane protein belonging to the family of small conductance mechanosensitive channels (MscS), identified as a PII interaction partner (PII-associated membrane protein A) in a yeast-two hybrid screening (Osanai et al. 2005b). The interaction of the cytoplasmic domain of PamA with PII was shown to be negatively regulated by Mg2+-ATP-2-OG. This finding indicates that conformational changes of the T-loop of PII modulate binding to PamA, implying that this interaction is related to C/N control and is physiologically relevant. In agreement with this assumption, analysis of a PamA-deficient mutant indicated that a part of the NtcA-regulated genes were poorly expressed (nblA, ntr-operon), suggesting that PamA interaction is related to the PII-PipX-NtcA network. Remarkably, other genes of the NtcA regulon (e.g. glnA, glnB, amt1) were not affected. However, the gene for the group 2 sigma factor SigE was poorly expressed in the PamA mutant. SigE is involved in the activation of sugar catabolic genes (Osanai et al. 2005a) and its expression is induced under nitrogen-limiting conditions in an NtcA-dependent manner (Muro-Pastor, Herrero and Flores 2001a), however, it was not found to be part of the NtcA-regulon as defined by Giner-Lamia et al. (2017). In addition, the expression of sigE requires the response regulator Rre37 (NrrA for nitrogen regulated response regulator A). Both factors act in parallel to control the expression of central carbon metabolic genes (Azuma et al. 2011; Joseph et al. 2014).
In essence, nitrogen depletion activates the expression of sugar catabolic genes, including genes like zwf (encoding the key enzyme glucose-6-P dehydrogenase of the oxidative-pentose-phosphate cycle) as well as glycogen phosphorylase glgP (Ehira et al. 2017). The up-regulation of glycogen catabolizing enzymes during nitrogen depletion and the down-regulation of gluconeogenetic functions seems to be contradictory at a first glance. When cells are depleted for nitrogen, they accumulate glycogen but the genes for glycogen catabolism and glycolytic reactions are tuned on concomitantly. A physiological explanation for this paradox was provided by studying the recovery of nitrogen-starved cells. It turned out that these cells, even after having arrested metabolism and reside in a dormant state, are able to rapidly turn on glycogen catabolism, to provide energy for the recovery process (Burnap, Hagemann and Kaplan 2015; Klotz et al. 2016; Doello et al. 2018). To achieve this task, it seems necessary that the cells act anticipatory and already build up the machinery for glycogen degradation during the accumulation of this polymer. It is likely that the activity of these enzymes is mainly regulated at the post-translational level.
SigE may be only the tip of an iceberg of an even more complicated transcriptional network underlying the precisely tuned modulation of transcription in response to different nitrogen regimes and C/N states. It appears that a whole battery of transcription factors, including several group 1 and group 2 sigma factors (Imamura et al. 2006), together with NtcA, PipX, pathway-specific transcription factors like NrrA and non-coding regulatory RNAs such as NsiR4 (Klahn et al. 2015a) control gene expression under different nitrogen regimes. How the putative membrane sensor PamA and its interaction with PII contributes to fine-tuning this complex regulatory network remains to be elucidated. In any case, it represents an interesting regulatory link between carbon and nitrogen metabolism that deserves further investigation.
ROLE OF 2-OG IN THE CYANOBACTERIAL CO2 CONCENTRATING MECHANISM AND CROSS-TALK WITH OTHER EFFECTOR MOLECULES
In cyanobacteria, carbon acquisition takes place mainly via CO2 fixation reactions. RubisCO (ribulose-1,5-bisphosphate carboxylase/oxygenase) is the central CO2 fixation enzyme and uses both CO2 (carboxylase activity) and O2 (oxygenase activity) as substrates. The RubisCO carboxylation reaction generates two molecules of 3-phosphoglycerate (3-PGA) from ribulose-1,5-bisphosphate (RuBP), which is further used in the Calvin–Benson cycle, while the RubisCO oxygenase activity releases one molecule of 3-PGA and one molecule of 2-phosphoglycolate (2-PG) (reviewed in: Hohmann-Marriott and Blankenship 2011; Burnap, Hagemann and Kaplan 2015). The 2-PG is toxic to cyanobacteria and has a huge negative effect on RubisCO and the Calvin-Benson cycle. Detoxification of 2-PG can be achieved by wasteful photorespiration reactions, converting 2-PG into 3-PGA (Eisenhut et al. 2008, Hagemann et al. 2010). The selection pressure to lower the rate of photorespiration resulted in evolution of the cyanobacterial carbon-concentrating mechanism (CCM). This mechanism elevates the Ci levels around RubisCO carboxylating sites (Burnap, Hagemann and Kaplan 2015) to ensure efficient CO2 fixation even in the presence of limiting amounts of CO2/HCO3−.
Cyanobacterial CCM is composed of several uptake systems for active CO2 and HCO3− transport, carbonic anhydrase (CA) for rapid conversion of HCO3− and CO2 and a special microcompartment called carboxysome, where RubisCO and CA enzymes are present (summarized in Fig. 5) (reviewed in Burnap, Hagemann and Kaplan 2015; Long et al. 2016). The CCM is expressed at a basal level under CO2-rich conditions but under Ci limitation, CCM expression and activity is strongly enhanced (Wang, Postier and Burnap 2004; Rae et al. 2013; Burnap, Hagemann and Kaplan 2015).
Figure 5.
Schematic representation of cyanobacterial CCM. In Synechocystis PCC 6803, the carbon concentrating mechanism CCM is composed of five Ci uptake systems (among them three HCO3− transporters and two CO2 uptake systems), a Na+/H+ gradient restoring system and the carboxysome, where HCO3− is dehydrated to CO2 by carbonic anhydrase (CA) enzyme and then CO2 is fixed by RubisCO (Burnap et al. 2015; Long et al. 2016). The cyanobacterial HCO3− uptake systems consist of: a high affinity ATP-dependent ABC-type transporter (BCT1 complex), which consists of CmpA/B/C/D subunits (CmpA PDB entry 2I4B) encoded by the cmpABCD operon and two sodium-dependent bicarbonate single-subunit transporters: A low Ci inducible/high affinity/low flux transporter SbtA and a constitutive medium affinity/high flux transporter BicA. The PII-like SbtB protein (PDB entry 5O3R) is a regulatory component of the CCM. The cyanobacterial CO2 uptake systems are organized into two thylakoid-bound systems: (A), the high-affinity/low-flux/low Ci-inducible redox driven CO2 uptake NDH-I3 complex consisting of NdhB/NdhF3/NdhD3/CupA subunits and (B), the low affinity/high flux/constitutive redox driven CO2 uptake NDH-I4 complex consisting of NdhB/NdhF4/NdhD4/CupB subunits. The CO2-NDH-I3/4 complexes possess CA-like activity using the recently discovered CA (EcaB), which is interacting with CupA and CupB subunits to convert CO2 into HCO3− by hydrating CO2 and creating a proton gradient (Han et al. 2017; Sun et al. 2019).
A key to CCM is the regulated activity of multiple CO2/HCO3− uptake systems coupled with Na+/H+ gradient systems (reviewed by Burnap, Hagemann and Kaplan 2015). In Synechocystis PCC 6803, three transcriptional factors, namely, NdhR (CcmR), CmpR and CyAbrB2 have been identified to be responsible for the transcriptional regulation of the CCM components (Figge et al. 2001; Takahashi, Yamaguchi and Omata 2004; Wang, Postier and Burnap 2004; Price et al. 2008; Kaniya et al. 2013; Klahn et al. 2015; Orf et al. 2016).
The transcription factors NdhR and CmpR belong to the LysR family and are regulated by small molecular mass metabolites. CmpR is proposed to act as a transcriptional activator of the cmp operon, which encodes an ABC-type BCT1 bicarbonate transporter. Its DNA binding affinity was shown to increase with the addition of RuBP or 2-PG (Fig. 6) (Nishimura et al. 2008; Daley et al. 2012), both metabolites, whose level is expected to increase under low CO2 conditions. CmpR was found to repress expression of its own gene and to affect the phycocyanin content by controlling the gene for phycocyanine alpha-subunit phycocyanobilin lyase (Pan et al. 2016). The crystal structure of CmpR from S. elongatus was solved in a complex with RuBP, which gave a solid evidence for conformational changes induced by RuBP binding to eventually induce the cmp operon expression (Mahounga, Sun and Jiang 2018).
Figure 6.

Regulation of cyanobacterial CCM by transcription by transcription factors (TFs) NdhR and CmpR and their modulation by effector metabolites 2-OG, NADP+, 2-PG and RuBP. Dashed lines indicate regulatory interactions between the TFs to components of CCM, binding of effector metabolites to target proteins is indicated by straight arrows, with the effect on the respective target indicated by (+) for activation and (−) for inactivation.
In contrast to CmpR, transcription factor NdhR acts as a repressor of genes encoding high affinity Ci uptake systems (such as sbtA, bicA or ndhF3) and of its own gene. In vitro studies suggested that 2-OG and NADP+ act as co-repressors of NdhR (Fig. 6) (Daley et al. 2012). NADP+ is assumed to accumulate under CO2 sufficient conditions, since an efficient Calvin Cycle constantly depletes the NADPH pool. On the other hand, 2-OG is used as status reporter of the C/N balance, such that under low nitrogen conditions (high 2-OG levels), the CCM would be tuned down, which is in agreement with the diminished rate of CO2 fixation under nitrogen-deprived conditions. Recently, mechanistic insights by structure function analysis of NdhR were reported (Jiang et al. 2018). NdhR binds as a tetramer to the promoter regions thereby blocking transcriptional activation. Each subunit consists of a DNA-binding domain, connected via a linker helix to a double regulatory domain. Binding of 2-OG occurs at the interfaces between two adjacent regulatory domains thereby stabilizing the tetrameric conformation that has high DNA binding affinity. The 2-PG antagonizes the effect of 2-OG, by binding within the regulatory domains of NdhR in such a manner, that the tetramer adopts a conformation incompatible with DNA binding. As a result, NdhR dissociates from its target repressing sites. Of note, binding of 2-OG or 2-PG to NdhR is mutually exclusive. Taking into consideration the in vivo concentrations of the effectors 2-OG and 2-PG, it appears reasonable to assume that the ratio of the levels of 2-OG to 2-PG determines, whether NdhR acts as a repressor. When 2-OG levels increase due to nitrogen deficiency or excess of carbon, the CCM activity is repressed to its basal level. By contrast, at elevated levels of 2-PG, indicating the need for CO2 concentration due to increased RubisCO oxygenation reactions, the expression of CCM-related genes is de-repressed resulting in high CCM activity.
The CyAbrB2 (sll0822) factor was found to be complementary to CmpR and NdhR transcription factors, through controlling several CCM-related genes positively such as the cmpABCB, sbtAB and NDH-I3 (ndhF3/ndhD3/cupA) operons under low carbon conditions (Orf et al. 2016). Intriguingly, CyAbrB2, in addition to its involvement in CCM regulation in Synechocystis PCC 6803 (Yamauchi et al. 2011, Orf et al. 2016) was also shown to positively regulate several genes of the NtcA-regulon, including urtA, amt1, glnB, sigE and the nrt operon (Kaniya et al. 2013). Consistent with the functional role of CyAbrB in nitrogen-related gene expression, a recent study pointed towards an involvement of CyAbrB in heterocyst formation regulation in Anabaena PCC 7120 (Higo et al. 2019). These finding indicate that CyAbrB2 could integrate C- and N-metabolic pathways at the transcriptional level.
cAMP AS CO2-RELATED SECOND MESSENGER NUCLEOTIDE
In addition to the previously described status reporter metabolites of the cellular carbon supply, 2-OG and 2-PG, cyclic-AMP (cAMP), generated by soluble adenylyl cyclase (sAC) activity was shown in Synechocystis PCC 6803 and Anabaena PCC 7120 (Hammer, Hodgson and Cann 2006) to respond to varying bicarbonate concentrations (Chen et al. 2000; Steegborn et al. 2005). Sensing the cellular HCO3− level by sAC enzymes seems to be universal and evolutionary conserved across different phyla separated by millions of years (Chen et al. 2000). However, the physiological significance remained undetermined. Hypothetically, sAC could play a role as an evolutionarily conserved HCO3− sensor in prokaryotes and eukaryotes to detect the fluctuation in Ci and accordingly modulates the level of the second messenger cAMP (Chen et al. 2000; Raven 2006). Intriguingly, cAMP is one of the most prevalent second messengers in nature, playing fundamental roles in regulating many crucial physiological processes related to carbon metabolism, which also includes the well-understood response of enterobacteria to glucose (Cann et al. 2003; Steegborn et al. 2005; Gancedo 2013). Among cyanobacteria, cAMP has been shown to be involved in respiration, light sensing and cell motility (Agostoni and Montgomery 2014).
Recently, one of the components of cyanobacterial CCM, the products of the sbtAB operon (slr1512-slr1513), were shown to link cAMP sensing with carbon acquisition (Selim et al. 2018). The gene for the sodium-dependent HCO3− transporter SbtA is located next to a putative regulatory protein, termed SbtB (Srivastava, Pisareva and Norling 2005, Du et al. 2014, Burnap, Hagemann and Kaplan 2015, Long et al. 2016). The sbtB gene product belongs to the widespread PII superfamily, including PII-like proteins that do not represent canonical PII proteins (Forchhammer and Lüddecke 2016). Transcriptional analysis of Synechocystis PCC 6803 and S. elongatus revealed that SbtB is co-expressed with SbtA: highly expressed under Ci limitation and strongly repressed under high CO2 conditions (Wang, Postier and Burnap 2004, Schwarz et al. 2011). A phosphoproteome study of Synechocystis PCC 6803 implied that SbtA and SbtB are a phosphoproteins, whose phosphorylation level oppositely responds to nitrogen limitation: Phosphorylation of SbtA is down regulated while SbtB protein is highly phosphorylated on Thr53 (i.e. resembling posttranslational phosphorylation of the canonical PII protein) under nitrogen starvation conditions (Spät, Macek and Forchhammer 2015). Heterologous expression of SbtB and SbtA in E. coli suggested that SbtB inhibited the HCO3− uptake activity of SbtA, implying that SbtB is likely a regulator of SbtA (Du et al. 2014).
Biochemical and structural analysis of SbtB protein showed that, despite of its low sequence conservation with canonical PII proteins, the SbtB structure perfectly matches the PII architecture. Although the effector molecule binding site of PII proteins is poorly conserved, SbtB binds the adenyl nucleotides ATP, ADP, AMP in an anti-cooperative manner, very similar to the binding properties of canonical PII. However, in contrast to any PII family member studied so far, SbtB binds cAMP and the affinity to this second messenger nucleotide was higher than that for the metabolic adenyl nucleotides (Selim et al. 2018). The structure of SbtB in complex with AMP or cAMP revealed that the bound nucleotides perfectly match the structure of bound adenyl nucleotides in canonical PII proteins.
To substantiate a link between cAMP sensing and Ci signaling by SbtB, the levels of adenyl nucleotides under various Ci conditions were analyzed. Under low carbon conditions, the intracellular cAMP levels dropped while AMP levels increased, and vice versa, cAMP levels increased with ambient Ci levels while AMP levels dropped, implying that cAMP represents a high carbon signal and AMP a low carbon signal. Addition of AMP to cell-free extracts supported the membrane bound state of SbtB, which was also observed in vivo under low Ci conditions. The membrane-localized state of SbtB depends on the presence of SbtA and is therefore, an indicator of SbtA-SbtB interaction. By contrast, cAMP did not support the membrane-localized state of SbtB. In agreement with cAMP controlling the localization of SbtB, an the SbtB protein was constitutively membrane associated in an adenylyl cyclase deficient mutant (∆Cya1). With purified SbtB, cAMP successfully competed with AMP for SbtB binding under any tested condition. These results suggest that SbtB monitors the Ci state via cAMP binding in conjunction with energy state sensing through the competitive binding of AMP.
Physiological analysis of an SbtB-deficient mutant in Synechocystis PCC 6803 suggested that SbtB is required for efficient Ci acclimatization and growth at low Ci conditions. Also, the expression of bicA was disturbed in the sbtB-deficient mutant, implying that SbtB might play a more general role than just controlling the SbtA transporter, in analogy to the versatile role of canonical PII proteins in nitrogen control.
Altogether, SbtB appears an additional component of the cyanobacterial CCM responding to the intracellular cAMP/AMP levels, which depend on the Ci supply of the cells (Fig. 7). These findings highlight an evolutionary conserved role of cAMP as an indicator of the cellular carbon status across different domains of the life (Chen et al. 2000; Steegborn et al. 2005; Agostoni and Montgomery 2014; Burnap, Hagemann and Kaplan 2015; Hennon et al. 2015).
Figure 7.
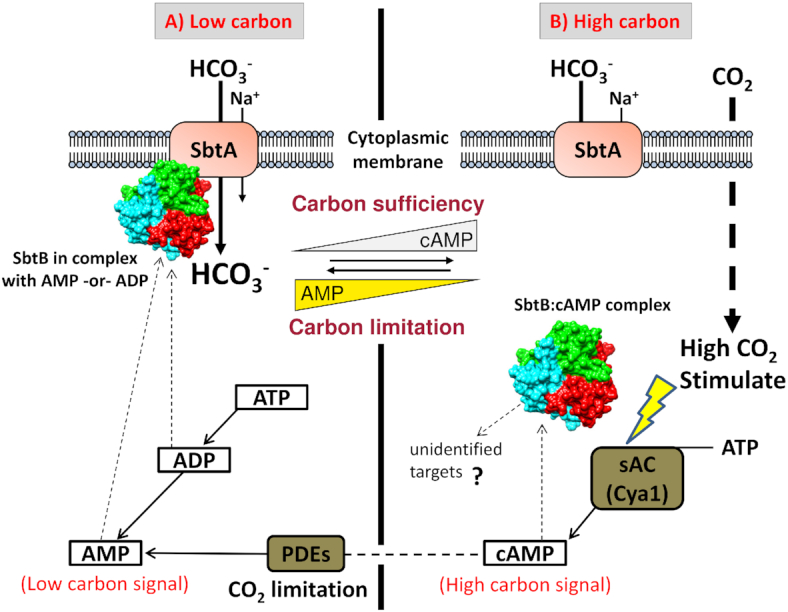
Model for the proposed regulatory mechanism of the PII-like protein SbtB. The interactions of SbtB are modulated by the formation of various SbtB—adenylnucleotide complexes. Under carbon-limiting conditions (part A), SbtA/SbtB proteins are highly expressed and form a membrane-localized complex, favored by the binding of ADP or AMP to SbtB. Under the low carbon conditions, the levels of AMP are high, presumably due to energy consuming CO2 uptake activity. When low carbon-adapted cells are shifted to high carbon conditions (part B), soluble adenylate cyclase (sAC) Cya1 becomes activated by CO2 and produces cAMP. When SbtB interacts with cAMP, it no longer stays bound to the membrane. Instead, soluble, cAMP-complexed SbtB may now be able to interact with additional targets (model according to Selim et al. 2018).
CONCLUDING REMARKS
As already pointed out by Galperin (2005), Cyanobacteria are unique within the bacterial kingdom, as they are mainly concerned about their internal homeostasis, in contrast to most other bacteria, which actively monitor their environment. This ‘introvert’ life style of cyanobacteria requires them to constantly monitor their intracellular milieu. Maintenance of C/N homeostasis is one of the most fundamental aspects of cellular homeostasis for an autotrophic organism, as it is essential for vegetative growth to tightly interconnect CO2 fixation and N-assimilation. This article summarized current knowledge on the mechanisms how cyanobacteria monitor the C/N balance and which regulatory mechanisms are effecting the adjustment of homeostasis. An extremely complex network of interwoven signal transduction pathway emerges, in which the sensing of 2-OG as a central status reporter of the cellular C/N balance plays the key role. This signal is integrated with various other signals, such as the energy state through the PII-signaling protein, or multiple signals are perceived by two component systems that converge with the NtcA regulon. Moreover, 2-OG converges with 2-PG level monitoring for CCM regulation. To a certain extent, cells are able to cope with imbalances, which they can tolerate by buffering them through the accumulation of carbon or nitrogen storage polymers. However, when the imbalance exceeds the internal buffering capacity, specific acclimation processes are employed which overlap with the homeostatic control. For example, for the induction of nitrogen chlorosis, the 2-OG signal, perceived by NtcA, is combined with redox-stress signal perceived by the Hik33/RpaB relay (Klotz et al. 2015). Only when all players and factors are identified, a mathematical model can help to predict cyanobacterial responses towards metabolic perturbations, a prerequisite for the application of cyanobacteria as photosynthetic cell factories.
ACKNOWLEDGEMENTS
We appreciate helpful comments on the manuscript by Martin Hagemann and Wolfgang Hess. We are indebted to the Deutsche Forschungsgemeinschaft for continuous funding of the work in the authors lab (Grants Fo195/9-2 and Fo195/13-1)
Contributor Information
Karl Forchhammer, Lehrstuhl für Mikrobiologie, Universität Tübingen, Auf der Morgenstelle 28, D-72076 Tübingen, Germany.
Khaled A Selim, Lehrstuhl für Mikrobiologie, Universität Tübingen, Auf der Morgenstelle 28, D-72076 Tübingen, Germany.
Conflict of interest. None declared.
REFERENCES
- Agostoni M, Montgomery BL. Survival strategies in the aquatic and terrestrial world: the impact of second messengers on cyanobacterial processes. Life (Basel). 2014;4:745–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldehni FM, Sauer J, Spielhaupter Cet al.. Signal transduction protein P(II) is required for NtcA-regulated gene expression during nitrogen deprivation in the cyanobacterium Synechococcus elongatus strain PCC 7942. J Bacteriol. 2003;185:2582–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen MM, Smith AJ. Nitrogen chlorosis in blue-green algae. Arch Microbiol. 1969;69:114–20. [DOI] [PubMed] [Google Scholar]
- Allen MM, Weathers PJ. Structure and composition of cyanophycin granules in the cyanobacterium Aphanocapsa 6308. J Bacteriol. 1980;141:959–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcondeguy T, Jack R, Merrick M. P(II) signal transduction proteins, pivotal players in microbial nitrogen control. Microbiol Mol Biol R: MMBR. 2001;65:80–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuma M, Osanai T, Hirai MYet al.. A response regulator Rre37 and an RNA polymerase sigma factor SigE represent two parallel pathways to activate sugar catabolism in a cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 2011;52:404–12. [DOI] [PubMed] [Google Scholar]
- Beez S, Fokina O, Herrmann Cet al.. N-acetyl-L-glutamate kinase (NAGK) from oxygenic phototrophs: P(II) signal transduction across domains of life reveals novel insights in NAGK control. J Mol Biol. 2009;389:748–58. [DOI] [PubMed] [Google Scholar]
- Bolay P, Muro-Pastor MI, Florencio FJet al.. The distinctive regulation of cyanobacterial glutamine synthetase. Life (Basel). 2018;8:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourrellier Feria, A.B. Valot, B. Guillotet al.. Chloroplast acetyl-CoA carboxylase activity is 2-oxoglutarate-regulated by interaction of PII with the biotin carboxyl carrier subunit. Proc Natl Acad Sci USA. 2010;107:502–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bueno-Batista M, Dixon R. Manipulating nitrogen regulation in diazotrophic bacteria for agronomic benefit. Biochem Soc Trans. 2019;47:603–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burillo S, Luque I, Fuentes Iet al.. Interactions between the nitrogen signal transduction protein PII and N-acetyl glutamate kinase in organisms that perform oxygenic photosynthesis. J Bacteriol. 2004;186:3346–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnap RL, Hagemann M, Kaplan A. Regulation of CO2 concentrating mechanism in cyanobacteria. Life (Basel). 2015;5:348–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnat M, Herrero A, Flores E. Compartmentalized cyanophycin metabolism in the diazotrophic filaments of a heterocyst-forming cyanobacterium. Proc Natl Acad Sci USA. 2014;111:3823–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camargo S, Valladares A, Forchhammer Ket al.Effects of PipX on NtcA-dependent promoters and characterization of the cox3 promoter region in the heterocyst-forming cyanobacterium Anabaena sp. PCC 7120. FEBS Lett. 2014;588:1787–94. [DOI] [PubMed] [Google Scholar]
- Cann MJ, Hammer A, Zhou Jet al.. A defined subset of adenylyl cyclases is regulated by bicarbonate ion. J Biol Chem. 2003;278:35033–8. [DOI] [PubMed] [Google Scholar]
- Cantos R, Labella JI, Espinosa Jet al.. The nitrogen regulator PipX acts in cis to prevent operon polarity. Environ Microbiol Rep. 2019;11:495–507. [DOI] [PubMed] [Google Scholar]
- Chang Y, Takatani N, Aichi Met al.. Evaluation of the effects of P(II) deficiency and the toxicity of PipX on growth characteristics of the P(II)-Less mutant of the cyanobacterium Synechococcus elongatus. Plant Cell Physiol. 2013;54:1504–14. [DOI] [PubMed] [Google Scholar]
- Chellamuthu VR, Alva V, Forchhammer K. From cyanobacteria to plants: conservation of PII functions during plastid evolution. Planta. 2013;237:451–62. [DOI] [PubMed] [Google Scholar]
- Chellamuthu VR, Ermilova E, Lapina Tet al.. A Widespread Glutamine-Sensing Mechanism in the Plant Kingdom. Cell. 2014;159:1188–99. [DOI] [PubMed] [Google Scholar]
- Chen HL, Latifi A, Zhang CCet al.. Biosensors-Based In vivo quantification of 2-oxoglutarate in cyanobacteria and proteobacteria. Life (Basel). 2018;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YQ, Cann MJ, Litvin TNet al.Soluble adenylyl cyclase as an evolutionarily conserved bicarbonate sensor. Science. 2000;289:625–8. [DOI] [PubMed] [Google Scholar]
- Conroy MJ, Durand A, Lupo Det al.. The crystal structure of the Escherichia coli AmtB-GlnK complex reveals how GlnK regulates the ammonia channel. Proc Natl Acad Sci USA. 2007;104:1213–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai GZ, Qiu BS, Forchhammer K. Ammonium tolerance in the cyanobacterium Synechocystis sp. strain PCC 6803 and the role of the psbA multigene family. Plant Cell Environment. 2014;37:840–51. [DOI] [PubMed] [Google Scholar]
- Daley SM, Kappell AD, Carrick MJet al.. Regulation of the cyanobacterial CO2-concentrating mechanism involves internal sensing of NADP+ and alpha-ketogutarate levels by transcription factor CcmR. PLoS One. 2012;7:e41286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Rocha RA, Weschenfelder TA, de Castilhos Fet al.. Mathematical model of the binding of allosteric effectors to the Escherichia coli PII signal transduction protein GlnB. Biochemistry. 2013;52:2683–93. [DOI] [PubMed] [Google Scholar]
- Doello S, Klotz A, Makowka Aet al.. A specific glycogen mobilization strategy enables rapid awakening of dormant cyanobacteria from chlorosis. Plant Physiol. 2018;177:594–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domínguez-Martín MA, López-Lozano A, Clavería-Gimeno Ret al.. Differential NtcA responsiveness to 2-oxoglutarate underlies the diversity of C/N balance Regulation in Prochlorococcus. Front Microbiol. 2018;8:2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drath M, Kloft N, Batschauer Aet al.Ammonia Triggers Photodamage of Photosystem II in the Cyanobacterium Synechocystis sp. Strain PCC 6803. Plant Physiol. 2008;147:206–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du JH, Forster B, Rourke Let al.. Characterisation of cyanobacterial bicarbonate transporters in E. coli shows that SbtA homologs are functional in this heterologous expression system. PLoS One. 2014;9:e115905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehira S, Shimmori Y, Watanabe Set al.. The nitrogen-regulated response regulator NrrA is a conserved regulator of glycogen catabolism in beta-cyanobacteria. Microbiology. 2017;163:1711–9. [DOI] [PubMed] [Google Scholar]
- Eisenhut M, Ruth W, Haimovich Met al.. The photorespiratory glycolate metabolism is essential for cyanobacteria and might have been conveyed endosymbiontically to plants. Proc Natl Acad Sci USA. 2008;105:17199–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa J, Castells MA, Laichoubi KBet al.. Effects of spontaneous mutations in PipX functions and regulatory complexes on the cyanobacterium Synechococcus elongatus strain PCC 7942. Microbiology. 2010;156:1517–26. [DOI] [PubMed] [Google Scholar]
- Espinosa J, Forchhammer K, Burillo Set al.. Interaction network in cyanobacterial nitrogen regulation: PipX, a protein that interacts in a 2-oxoglutarate dependent manner with PII and NtcA. Mol Microbiol. 2006;61:457–69. [DOI] [PubMed] [Google Scholar]
- Espinosa J, Forchhammer K, Contreras A. Role of the Synechococcus PCC 7942 nitrogen regulator protein PipX in NtcA-controlled processes. Microbiology. 2007;153:711–8. [DOI] [PubMed] [Google Scholar]
- Espinosa J, Labella JI, Cantos Ret al.. Energy drives the dynamic localization of cyanobacterial nitrogen regulators during diurnal cycles. Environ Microbiol. 2018;20:1240–52. [DOI] [PubMed] [Google Scholar]
- Espinosa J, Rodriguez-Mateos F, Salinas Pet al.. PipX, the coactivator of NtcA, is a global regulator in cyanobacteria. Proc Natl Acad Sci USA. 2014;111:E2423–2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteves-Ferreira AA, Inaba M, Fort Aet al.. Nitrogen metabolism in cyanobacteria: metabolic and molecular control, growth consequences and biotechnological applications. Crit Rev Microbiol. 2018;44:541–60. [DOI] [PubMed] [Google Scholar]
- Figge RM, Cassier-Chauvat C, Chauvat Fet al.. Characterization and analysis of an NAD(P)H dehydrogenase transcriptional regulator critical for the survival of cyanobacteria facing inorganic carbon starvation and osmotic stress. Mol Microbiol. 2001;39:455–68. [DOI] [PubMed] [Google Scholar]
- Flores E, Arevalo S, Burnat M. Cyanophycin and arginine metabolism in cyanobacteria. Algal Res. 2019;42:101577. [Google Scholar]
- Flores E, Frias JE, Rubio LMet al.. Photosynthetic nitrate assimilation in cyanobacteria. Photosynth Res. 2005;83:117–33. [DOI] [PubMed] [Google Scholar]
- Fokina O, Chellamuthu VR, Forchhammer Ket al.. Mechanism of 2-oxoglutarate signaling by the Synechococcus elongatus P-II signal transduction protein. Proc Natl Acad Sci USA. 2010;107:19760–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fokina O, Chellamuthu VR, Zeth Ket al.. A Novel signal transduction protein P-II variant from Synechococcus elongatus PCC 7942 indicates a two-step process for NAGK-P-II complex formation. J Mol Biol. 2010a;399:410–21. [DOI] [PubMed] [Google Scholar]
- Fokina O, Herrmann C, Forchhammer K. Signal-transduction protein P-II from Synechococcus elongatus PCC 7942 senses low adenylate energy charge in vitro. Biochem J. 2011;440:147–56. [DOI] [PubMed] [Google Scholar]
- Forcada-Nadal A, Forchhammer K, Rubio V. SPR analysis of promoter binding of Synechocystis PCC 6803 transcription factors NtcA and CRP suggests cross-talk and sheds light on regulation by effector molecules. FEBS Lett. 2014;588:2270–6. [DOI] [PubMed] [Google Scholar]
- Forcada-Nadal A, Llacer JL, Contreras Aet al.. The PII-NAGK-PipX-NtcA Regulatory Axis of Cyanobacteria: A Tale of Changing Partners, Allosteric Effectors and Non-covalent Interactions. Front Mol Biosci. 2018;5:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forchhammer K, Hedler A. Phosphoprotein PII from cyanobacteria–analysis of functional conservation with the PII signal-transduction protein from Escherichia coli. Eur J Biochem. 1997;244:869–75. [DOI] [PubMed] [Google Scholar]
- Forchhammer K, Irmler A, Kloft Net al.. P-II signalling in unicellular cyanobacteria: analysis of redox-signals and energy charge. Physiol Plant. 2004;120:51–56. [DOI] [PubMed] [Google Scholar]
- Forchhammer K, Lüddecke J. Sensory properties of the PII signalling protein family. FEBS J. 2016;283:425–37. [DOI] [PubMed] [Google Scholar]
- Forchhammer K, Schwarz R. Nitrogen chlorosis in unicellular cyanobacteria - a developmental program for surviving nitrogen deprivation. Environ Microbiol. 2019;21:1173–84. [DOI] [PubMed] [Google Scholar]
- Forchhammer K, Tandeau de Marsac N. Functional analysis of the phosphoprotein PII (glnB gene product) in the cyanobacterium Synechococcus sp. strain PCC 7942. J Bacteriol. 1995;177:2033–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forchhammer K, Tandeau de Marsac N. Phosphorylation of the P-II Protein (Glnb Gene-Product) in the Cyanobacterium Synechococcus sp. strain PCC-7942 - Analysis of in-Vitro Kinase-Activity. J Bacteriol. 1995a;177:5812–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forchhammer K, Tandeau de Marsac N. The PII protein in the cyanobacterium Synechococcus sp. strain PCC 7942 is modified by serine phosphorylation and signals the cellular N-status. J Bacteriol. 1994;176:84–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forchhammer K. Global carbon/nitrogen control by PII signal transduction in cyanobacteria: from signals to targets. FEMS Microbiol Rev. 2004;28:319–33. [DOI] [PubMed] [Google Scholar]
- Forchhammer K. P(II) signal transducers: novel functional and structural insights. Trends Microbiol. 2008;16:65–72. [DOI] [PubMed] [Google Scholar]
- Forchhammer K. The PII protein in Synechococcus PCC 7942 senses and signals 2-oxoglutarate under ATP-replete conditions. The Phototrophic Prokaryotes. 1999:549–53. [Google Scholar]
- Fujimori T, Higuchi M, Sato Het al.. The mutant of sll1961, which encodes a putative transcriptional regulator, has a defect in regulation of photosystem stoichiometry in the cyanobacterium Synechocystis sp PCC 6803. Plant Physiol. 2005;139:408–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin MY. A census of membrane-bound and intracellular signal transduction proteins in bacteria: bacterial IQ, extroverts and introverts. BMC Microbiol. 2005;5:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gancedo JM. Biological roles of cAMP: variations on a theme in the different kingdoms of life. Biol Rev. 2013;88:645–68. [DOI] [PubMed] [Google Scholar]
- Garcia-Dominguez M, Reyes JC, Florencio FJ. Glutamine synthetase inactivation by protein-protein interaction. Proc Natl Acad Sci USA. 1999;96:7161–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhardt ECM, Rodrigues TE, Muller-Santos Met al.. The Bacterial signal transduction protein GlnB regulates the committed step in fatty acid biosynthesis by acting as a dissociable regulatory subunit of acetyl-CoA carboxylase. Mol Microbiol. 2015;95:1025–35. [DOI] [PubMed] [Google Scholar]
- Giner-Lamia J, Robles-Rengel R, Hernandez-Prieto MAet al.. Identification of the direct regulon of NtcA during early acclimation to nitrogen starvation in the cyanobacterium Synechocystis sp. PCC 6803. Nucleic Acids Res. 2017;45:11800–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruswitz F, O'Connell J 3rd, Stroud RM. Inhibitory complex of the transmembrane ammonia channel, AmtB, and the cytosolic regulatory protein, GlnK, at 1.96 A. Proc Natl Acad Sci USA. 2007;104:42–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerreiro AC, Penning R, Raaijmakers LMet al.. Monitoring light/dark association dynamics of multi-protein complexes in cyanobacteria using size exclusion chromatography-based proteomics. J Proteomics. 2016;142:33–44. [DOI] [PubMed] [Google Scholar]
- Hagemann M, Eisenhut M, Hackenberg Cet al.. Pathway and importance of photorespiratory 2-phosphoglycolate metabolism in cyanobacteria. Adv Exp Med Biol. 2010;675:91–108. [DOI] [PubMed] [Google Scholar]
- Hammer A, Hodgson DR, Cann MJ. Regulation of prokaryotic adenylyl cyclases by CO2. Biochem J. 2006;396:215–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson TE, Forchhammer K, Tandeau de Marsac Net al.. Characterization of the glnB gene product of Nostoc punctiforme strain ATCC 29133: glnB or the PII protein may be essential. Microbiology. 1998;144:1537–47. [DOI] [PubMed] [Google Scholar]
- Han XL, Sun N, Xu Met al.. Co-ordination of NDH and Cup proteins in CO2 uptake in cyanobacterium Synechocystis sp. PCC 6803. J Exp Bot. 2017;68:3869–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison MA, Keen JN, Findlay JBet al.. Modification of a glnB-like gene product by photosynthetic electron transport in the cyanobacterium Synechococcus 6301. FEBS Lett. 1990;264:25–28. [DOI] [PubMed] [Google Scholar]
- Hauf W, Schmid K, Gerhardt ECet al.. Interaction of the Nitrogen Regulatory Protein GlnB (PII) with Biotin Carboxyl Carrier Protein (BCCP) Controls Acetyl-CoA Levels in the Cyanobacterium Synechocystis sp. PCC 6803. Front Microbiol. 2016;7:1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauf W. Regulation of carbon polymer accumulation in Synechocystis sp. PCC 6803. PhD Dissertation, Tübingen University, Germany, 2016.
- Heinrich A, Maheswaran M, Ruppert Uet al.. The Synechococcus elongatus P signal transduction protein controls arginine synthesis by complex formation with N-acetyl-L-glutamate kinase. Mol Microbiol. 2004;52:1303–14. [DOI] [PubMed] [Google Scholar]
- Hennon GMM, Ashworth J, Groussman RDet al.. Diatom acclimation to elevated CO2 via cAMP signalling and coordinated gene expression. Nat Clim Change. 2015;5:761–U179. [Google Scholar]
- Herrero A, Muro-Pastor AM, Flores E. Nitrogen control in cyanobacteria. J Bacteriol. 2001;183:411–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero A, Muro-Pastor AM, Valladares Aet al.. Cellular differentiation and the NtcA transcription factor in filamentous cyanobacteria. FEMS Microbiol Rev. 2004;28:469–87. [DOI] [PubMed] [Google Scholar]
- Higo A, Nishiyama E, Nakamura Ket al.. cyAbrB transcriptional regulators as safety devices to inhibit heterocyst differentiation in Anabaena. J Bacteriol. 2019;201:e00244–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hisbergues M, Jeanjean R, Joset Fet al.. Protein PII regulates both inorganic carbon and nitrate uptake and is modified by a redox signal in synechocystis PCC 6803. FEBS Lett. 1999;463:216–20. [DOI] [PubMed] [Google Scholar]
- Hohmann-Marriott MF, Blankenship RE. Evolution of photosynthesis. Annu Rev Plant Biol. 2011;62:515–48. [DOI] [PubMed] [Google Scholar]
- Huang L, Wang J, Watkins AMet al.. Structure and ligand binding of the glutamine-II riboswitch. Nucleic Acids Res. 2019;47:7666–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huergo LF, Chandra G, Merrick M. PII signal transduction proteins: nitrogen regulation and beyond. FEMS Microbiol Rev. 2013;37:251. [DOI] [PubMed] [Google Scholar]
- Huergo LF, Dixon R. The Emergence of 2-Oxoglutarate as a Master Regulator Metabolite. Microbiol Mol Biol Rev. 2015;79:419–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura S, Tanaka K, Shirai Met al.. Growth phase-dependent activation of nitrogen-related genes by a control network of group 1 and group 2 sigma factors in a cyanobacterium. J Biol Chem. 2006;281:2668–75. [DOI] [PubMed] [Google Scholar]
- Irmler A, Forchhammer K. A PP2C-type phosphatase dephosphorylates the PII signaling protein in the cyanobacterium Synechocystis PCC 6803. Proc Natl Acad Sci USA. 2001;98:12978–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irmler A, Sanner S, Dierks Het al.. Dephosphorylation of the phosphoprotein P-II in Synechococcus PCC 7942: identification of an ATP and 2-oxoglutarate-regulated phosphatase activity. Mol Microbiol. 1997;26:81–90. [DOI] [PubMed] [Google Scholar]
- Jiang P, Ninfa AJ. Sensation and signaling of alpha-ketoglutarate and adenylylate energy charge by the Escherichia coli PII signal transduction protein require cooperation of the three ligand-binding sites within the PII trimer. Biochemistry. 2009;48:11522–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang YL, Wang XP, Sun Het al.. Coordinating carbon and nitrogen metabolic signaling through the cyanobacterial global repressor NdhR. Proc Natl Acad Sci USA. 2018;115:403–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph A, Aikawa S, Sasaki Ket al.. Rre37 stimulates accumulation of 2-oxoglutarate and glycogen under nitrogen starvation in Synechocystis sp. PCC 6803. FEBS Lett. 2014;588:466–71. [DOI] [PubMed] [Google Scholar]
- Kaniya Y, Kizawa A, Miyagi Aet al.. Deletion of the transcriptional regulator cyAbrB2 deregulates primary carbon metabolism in Synechocystis sp. PCC 6803. Plant Physiol. 2013;162:1153–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klahn S, Bolay P, Wright PRet al.. A glutamine riboswitch is a key element for the regulation of glutamine synthetase in cyanobacteria. Nucleic Acids Res. 2018;46:10082–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klahn S, Orf I, Schwarz Det al.. Integrated Transcriptomic and Metabolomic Characterization of the Low-Carbon Response Using an ndhR Mutant of Synechocystis sp. PCC 6803. Plant Physiol. 2015;169:1540–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klahn S, Schaal C, Georg Jet al.. The sRNA NsiR4 is involved in nitrogen assimilation control in cyanobacteria by targeting glutamine synthetase inactivating factor IF7. Proc Natl Acad Sci USA. 2015a;112:E6243–6252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloft N, Forchhammer K. Signal transduction protein P-II phosphatase PphA is required for light-dependent control of nitrate utilization in Synechocystis sp strain PCC 6803. J Bacteriol. 2005;187:6683–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloft N, Rasch G, Forchhammer K. Protein phosphatase PphA from Synechocystis sp. PCC 6803: the physiological framework of PII-P dephosphorylation. Microbiology. 2005;151:1275–83. [DOI] [PubMed] [Google Scholar]
- Klotz A, Georg J, Bucinska Let al.. Awakening of a dormant cyanobacterium from nitrogen chlorosis reveals a genetically determined program. Curr Biol. 2016;26:2862–72. [DOI] [PubMed] [Google Scholar]
- Klotz A, Reinhold E, Doello Set al.. Nitrogen starvation acclimation in Synechococcus elongatus: Redox-Control and the role of nitrate reduction as an electron sink. Life. 2015;5:888–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M, Rodriguez R, Lara Cet al.. Involvement of the C-terminal domain of an ATP-binding subunit in the regulation of the ABC-type nitrate/nitrite transporter of the Cyanobacterium Synechococcus sp. strain PCC 7942. J Biol Chem. 1997;272:27197–201. [DOI] [PubMed] [Google Scholar]
- Kobayashi M, Takatani N, Tanigawa Met al.. Posttranslational regulation of nitrate assimilation in the cyanobacterium Synechocystis sp. strain PCC 6803. J Bacteriol. 2005;187:498–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koropatkin NM, Pakrasi HB, Smith TJ. Atomic structure of a nitrate-binding protein crucial for photosynthetic productivity. Proc Natl Acad Sci USA. 2006;103:9820–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labella JI, Cantos R, Espinosa Jet al.. PipY, a Member of the Conserved COG0325 Family of PLP-Binding Proteins, Expands the Cyanobacterial Nitrogen Regulatory Network. Front Microbiol. 2017;8:1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labella JI, Obrebska A, Espinosa Jet al.. Expanding the Cyanobacterial Nitrogen Regulatory Network: The GntR-Like Regulator PlmA Interacts with the PII-PipX Complex. Front Microbiol. 2016;7:1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laichoubi KB, Beez S, Espinosa Jet al.. The nitrogen interaction network in Synechococcus WH5701, a cyanobacterium with two PipX and two P(II)-like proteins. Microbiology. 2011;157:1220–8. [DOI] [PubMed] [Google Scholar]
- Lapina T, Selim KA, Forchhammer Ket al.. The PII signaling protein from red algae represents an evolutionary link between cyanobacterial and Chloroplastida PII proteins. Sci Rep. 2018;8:790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent S, Chen H, Bedu Set al.. Nonmetabolizable analogue of 2-oxoglutarate elicits heterocyst differentiation under repressive conditions in Anabaena sp. PCC 7120. Proc Natl Acad Sci USA. 2005;102:9907–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HM, Flores E, Forchhammer Ket al.. Phosphorylation of the signal transducer PII protein and an additional effector are required for the PII-mediated regulation of nitrate and nitrite uptake in the Cyanobacterium Synechococcus sp. PCC 7942. Eur J Biochem. 2000;267:591–600. [DOI] [PubMed] [Google Scholar]
- Lee HM, Flores E, Herrero Aet al.. A role for the signal transduction protein PII in the control of nitrate/nitrite uptake in a cyanobacterium. FEBS Lett. 1998;427:291–5. [DOI] [PubMed] [Google Scholar]
- Lee MH, Scherer M, Rigali Set al.. PlmA, a new member of the GntR family, has plasmid maintenance functions in Anabaena sp. strain PCC 7120. J Bacteriol. 2003;185:4315–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh JA, Dodsworth JA. Nitrogen regulation in bacteria and archaea. Annu Rev Microbiol. 2007;61:349–77. [DOI] [PubMed] [Google Scholar]
- Lippi L, Bahr L, Wustenberg Aet al., 2018. Exploring the potential of high-density cultivation of cyanobacteria for the production of cyanophycin. Algal Res. 31:363–6. [Google Scholar]
- Llácer JL, Contreras A, Forchhammer Ket al.. The crystal structure of the complex of PII and acetylglutamate kinase reveals how PII controls the storage of nitrogen as arginine. Proc Natl Acad Sci USA. 2007;104:17644–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llácer JL, Espinosa J, Castells MAet al.. Structural basis for the regulation of NtcA-dependent transcription by proteins PipX and PII. Proc Natl Acad Sci USA. 2010;107:15397–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llácer JL, Fita I, Rubio V. Arginine and nitrogen storage. Curr Opin Struct Biol. 2008;18:673–81. [DOI] [PubMed] [Google Scholar]
- Long BM, Rae BD, Rolland Vet al.. Cyanobacterial CO2-concentrating mechanism components: function and prospects for plant metabolic engineering. Curr Opin Plant Biol. 2016;31:1–8. [DOI] [PubMed] [Google Scholar]
- Luque I, Flores E, Herrero A. Molecular mechanism for the operation of nitrogen control in cyanobacteria. Embo J. 1994;13:2862–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luque I, Forchhammer K. Nitrogen assimilation and C/N balance sensing. In: Herrero A., Flores E., (eds). The Cyanobacteria: Molecular Biology, Genomics and Evolution. Norfolk, UK: Caister Academic Press, 2008. [Google Scholar]
- Lüddecke J, Forchhammer K. From PII signaling to metabolite sensing: a novel 2-oxoglutarate sensor that details PII - NAGK complex formation. PLoS One. 2013;8:e83181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüddecke J, Francois L, Spat Pet al.. PII protein-derived FRET sensors for quantification and live-cell imaging of 2-oxoglutarate. Sci Rep. 2017;7:1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma CW, Lüddecke J, Forchhammer Ket al.. Population shift of binding pocket size and dynamic correlation analysis shed new light on the anticooperative mechanism of PII protein. Proteins. 2014;82:1048–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maheswaran M, Urbanke C, Forchhammer K. Complex formation and catalytic activation by the PII signaling protein of N-acetyl-L-glutamate kinase from Synechococcus elongatus strain PCC 7942. J Biol Chem. 2004;279:55202–10. [DOI] [PubMed] [Google Scholar]
- Maheswaran M, Ziegler K, Lockau Wet al.. P-II-regulated arginine synthesis controls accumulation of cyanophycin in Synechocystis sp strain PCC 6803. J Bacteriol. 2006;188:2730–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahounga DM, Sun H, Jiang YL. Crystal structure of the effector-binding domain of Synechococcus elongatus CmpR in complex with ribulose 1,5-bisphosphate. Acta Crystallogr F Struct Biol Commun. 2018;74:506–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeks JC, Wolk CP, Thomas Jet al.. The pathways of assimilation of 13NH4+ by the cyanobacterium, Anabaena cylindrica. J Biol Chem. 1977;252:7894–900. [PubMed] [Google Scholar]
- Merrick M. Post-translational modification of P-II signal transduction proteins. Front Microbiol. 2015;5:763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrick MJ, Edwards RA. Nitrogen Control in Bacteria. Microbiol Rev. 1995;59:604–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muro-Pastor AM, Herrero A, Flores E. Nitrogen-regulated group 2 sigma factor from Synechocystis sp. strain PCC 6803 involved in survival under nitrogen stress. J Bacteriol. 2001a;183:1090–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muro-Pastor MI, Reyes JC, Florencio FJ. Ammonium assimilation in cyanobacteria. Photosynth Res. 2005;83:135–50. [DOI] [PubMed] [Google Scholar]
- Muro-Pastor MI, Reyes JC, Florencio FJ. Cyanobacteria perceive nitrogen status by sensing intracellular 2-oxoglutarate levels. J Biol Chem. 2001b;276:38320–8. [DOI] [PubMed] [Google Scholar]
- Ninfa AJ, Atkinson MR. PII signal transduction proteins. Trends Microbiol. 2000;8:172–9. [DOI] [PubMed] [Google Scholar]
- Ninfa AJ, Jiang P. PII signal transduction proteins: sensors of alpha-ketoglutarate that regulate nitrogen metabolism. Curr Opin Microbiol. 2005;8:168–73. [DOI] [PubMed] [Google Scholar]
- Nishimura T, Takahashi Y, Yamaguchi Oet al.. Mechanism of low CO2-induced activation of the cmp bicarbonate transporter operon by a LysR family protein in the cyanobacterium Synechococcus elongatus strain PCC 7942. Mol Microbiol. 2008;68:98–109. [DOI] [PubMed] [Google Scholar]
- Ohashi Y, Shi W, Takatani Net al.. Regulation of nitrate assimilation in cyanobacteria. J Exp Bot. 2011;62:1411–24. [DOI] [PubMed] [Google Scholar]
- Orf I, Schwarz D, Kaplan Aet al.. CyAbrB2 Contributes to the Transcriptional Regulation of Low CO2 Acclimation in Synechocystis sp. PCC 6803. Plant Cell Physiol. 2016;57:2232–43. [DOI] [PubMed] [Google Scholar]
- Osanai T, Kanesaki Y, Nakano Tet al.. Positive regulation of sugar catabolic pathways in the cyanobacterium Synechocystis sp. PCC 6803 by the group 2 sigma factor sigE. J Biol Chem. 2005a;280:30653–9. [DOI] [PubMed] [Google Scholar]
- Osanai T, Sato S, Tabata Set al.. Identification of PamA as a PII-binding membrane protein important in nitrogen-related and sugar-catabolic gene expression in Synechocystis sp. PCC 6803. J Biol Chem. 2005b;280:34684–90. [DOI] [PubMed] [Google Scholar]
- Osanai T, Tanaka K. Keeping in touch with PII: PII-interacting proteins in unicellular cyanobacteria. Plant Cell Physiol. 2007;48:908–14. [DOI] [PubMed] [Google Scholar]
- Palinska KA, Laloui W, Bedu Set al.. The signal transducer P(II) and bicarbonate acquisition in Prochlorococcus marinus PCC 9511, a marine cyanobacterium naturally deficient in nitrate and nitrite assimilation. Microbiology. 2002;148:2405–12. [DOI] [PubMed] [Google Scholar]
- Pan L-L, Onai K, Uesaka Ket al.. Transcriptional regulation of CmpR, the LysR family protein involved in CO2-responsive gene regulation in the cyanobacterium Synechococcus elongatus. Biomed Genet Genomics. 2016;1:1–6. [Google Scholar]
- Paz-Yepes J, Flores E, Herrero A. Transcriptional effects of the signal transduction protein P(II) (glnB gene product) on NtcA-dependent genes in Synechococcus sp. PCC 7942. FEBS Lett. 2003;543:42–46. [DOI] [PubMed] [Google Scholar]
- Pearce J, Leach CK, Carr NG. The incomplete tricarboxylic acid cycle in the blue-green alga Anabaena variabilis. J Gen Microbiol. 1969;55:371–8. [DOI] [PubMed] [Google Scholar]
- Price GD, Badger MR, Woodger FJet al.. Advances in understanding the cyanobacterial CO2-concentrating-mechanism (CCM): functional components, Ci transporters, diversity, genetic regulation and prospects for engineering into plants. J Exp Bot. 2008;59:1441–61. [DOI] [PubMed] [Google Scholar]
- Radchenko MV, Thornton J, Merrick M. Control of AmtB-GlnK complex formation by intracellular levels of ATP, ADP, and 2-oxoglutarate. J Biol Chem. 2010;285:31037–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rae BD, Long BM, Badger MRet al.. Functions, compositions, and evolution of the two types of Carboxysomes: Polyhedral microcompartments that facilitate CO2 fixation in cyanobacteria and some proteobacteria. Microbiol Mol Biol R. 2013;77:357–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raven JA. Sensing inorganic carbon: CO2 and HCO3. Biochem J. 2006;396:e5–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren A, Xue Y, Peselis Aet al.. Structural and dynamic basis for Low-Affinity, High-Selectivity binding of L-Glutamine by the Glutamine Riboswitch. Cell Rep. 2015;13:1800–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robles-Rengel R, Florencio FJ, Muro-Pastor MI. Redox interference in nitrogen status via oxidative stress is mediated by 2-oxoglutarate in cyanobacteria. New Phytol. 2019;224:216–28. [DOI] [PubMed] [Google Scholar]
- Rodrigues TE, Gerhardt EC, Oliveira MAet al.. Search for novel targets of the PII signal transduction protein in Bacteria identifies the BCCP component of acetyl-CoA carboxylase as a PII binding partner. Mol Microbiol. 2014;91:751–61. [DOI] [PubMed] [Google Scholar]
- Rodrigues TE, Sassaki GL, Valdameri Get al.. Fatty acid biosynthesis is enhanced in Escherichia coli strains with deletion in genes encoding the PII signaling proteins. Arch Microbiol. 2019;201:209–14. [DOI] [PubMed] [Google Scholar]
- Rubin BE, Wetmore KM, Price MNet al.. The essential gene set of a photosynthetic organism. Proc Natl Acad Sci USA. 2015;112:E6634–6643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruppert U, Irmler A, Kloft Net al.. The novel protein phosphatase PphA from Synechocystis PCC 6803 controls dephosphorylation of the signalling protein P-II. Mol Microbiol. 2002;44:855–64. [DOI] [PubMed] [Google Scholar]
- Sakamoto T, Inoue-Sakamoto K, Bryant DA. A novel nitrate/nitrite permease in the marine Cyanobacterium Synechococcus sp. strain PCC 7002. J Bacteriol. 1999;181:7363–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlicker C, Fokina O, Kloft Net al.. Structural analysis of the PP2C phosphatase tPphA from Thermosynechococcus elongatus: a flexible flap subdomain controls access to the catalytic site. J Mol Biol. 2008;376:570–81. [DOI] [PubMed] [Google Scholar]
- Schwarz D, Nodop A, Huge Jet al.. Metabolic and Transcriptomic Phenotyping of Inorganic Carbon Acclimation in the Cyanobacterium Synechococcus elongatus PCC 7942. Plant Physiol. 2011;155:1640–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz D, Orf I, Kopka Jet al.. Effects of inorganic carbon limitation on the metabolome of the Synechocystis sp. PCC 6803 mutant defective in glnB encoding the central regulator PII of cyanobacterial C/N acclimation. Metabolites. 2014;4:232–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz R, Forchhammer K. Acclimation of unicellular cyanobacteria to macronutrient deficiency: emergence of a complex network of cellular responses. Microbiology. 2005;151:2503–14. [DOI] [PubMed] [Google Scholar]
- Selim KA, Haase F, Hartmann MDet al.. PII-like signaling protein SbtB links cAMP sensing with cyanobacterial inorganic carbon response. Proc Natl Acad Sci USA. 2018;115:E4861–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selim KA, Lapina T, Forchhammer Ket al.. Interaction of N-acetyl-l-glutamate kinase with the PII signal transducer in the non-photosynthetic alga Polytomella parva: Co-evolution towards a hetero-oligomeric enzyme. FEBS J. 2019. doi:10.1111/febs.14989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senior PJ. Regulation of nitrogen metabolism in Escherichia coli and Klebsiella aerogenes: studies with the continuous-culture technique. J Bacteriol. 1975;123:407–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro BM. The glutamine synthetase deadenylylating enzyme system from Escherichia coli. Resolution into two components, specific nucleotide stimulation, and cofactor requirements. Biochemistry. 1969;8:659–70. [DOI] [PubMed] [Google Scholar]
- Simon RD. Cyanophycin Granules from the Blue-Green Alga Anabaena cylindrica: A Reserve material consisting of copolymers of aspartic acid and arginine. Proc Natl Acad Sci USA. 1971;68:265–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spät P, Macek B, Forchhammer K. Phosphoproteome of the cyanobacterium Synechocystis sp. PCC 6803 and its dynamics during nitrogen starvation. Front Microbiol. 2015;6:248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spät P. Establishment of quantitative proteomics in cyanobacteria reveals O-phosphorylation as fundamental process for stress-acclimation to nitrogen deficiency. PhD Dissertation, Tübingen University, Germany, 2017.
- Srivastava R, Pisareva T, Norling B. Proteomic studies of the thylakoid membrane of Synechocystis sp. PCC 6803. Proteomics. 2005;5:4905–16. [DOI] [PubMed] [Google Scholar]
- Steegborn C, Litvin TN, Levin LRet al.. Bicarbonate activation of adenylyl cyclase via promotion of catalytic active site closure and metal recruitment. Nat Struct Mol Biol. 2005;12:32–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su JY, Forchhammer K. Determinants for substrate specificity of the bacterial PP2C protein phosphatase tPphA from Thermosynechococcus elongatus. FEBS J. 2013;280:694–707. [DOI] [PubMed] [Google Scholar]
- Su JY, Schlicker C, Forchhammer K. A Third Metal Is Required for Catalytic Activity of the Signal-transducing Protein Phosphatase M tPphA. J Biol Chem. 2011;286:13481–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun N, Han XL, Xu Met al.. A thylakoid-located carbonic anhydrase regulates CO2 uptake in the cyanobacterium Synechocystis sp. PCC 6803. New Phytol. 2019;222:206–17. [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Yamaguchi O, Omata T. Roles of CmpR, a LysR family transcriptional regulator, in acclimation of the cyanobacterium Synechococcus sp. strain PCC 7942 to low-CO(2) and high-light conditions. Mol Microbiol. 2004;52:837–45. [DOI] [PubMed] [Google Scholar]
- Tanigawa R, Shirokane M, Maeda Si Set al.. Transcriptional activation of NtcA-dependent promoters of Synechococcus sp. PCC 7942 by 2-oxoglutarate in vitro. Proc Natl Acad Sci USA. 2002;99:4251–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trautmann A, Watzer B, Wilde Aet al.. Effect of phosphate availability on cyanophycin accumulation in Synechocystis sp. PCC 6803 and the production strain BW86. Algal Res. 2016;20:189–96. [Google Scholar]
- Truan D, Huergo LF, Chubatsu LSet al.. A new P(II) protein structure identifies the 2-oxoglutarate binding site. J Mol Biol2010;400:531–9. [DOI] [PubMed] [Google Scholar]
- Tsinoremas NF, Castets AM, Harrison MAet al.. Photosynthetic electron transport controls nitrogen assimilation in cyanobacteria by means of posttranslational modification of the glnB gene product. Proc Natl Acad Sci USA. 1991;88:4565–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhrig RG, Ng KK, Moorhead GB. PII in higher plants: a modern role for an ancient protein. Trends Plant Sci. 2009;14:505–11. [DOI] [PubMed] [Google Scholar]
- Vazquez-Bermudez MF, Herrero A, Flores E. 2-Oxoglutarate increases the binding affinity of the NtcA (nitrogen control) transcription factor for the Synechococcus glnA promoter. FEBS Lett. 2002;512:71–74. [DOI] [PubMed] [Google Scholar]
- Verma E, Chakraborty S, Tiwari Bet al.. Transcriptional regulation of acetyl CoA and lipid synthesis by PII protein in Synechococcus PCC 7942. J Basic Microbiol. 2018;58:187–97. [DOI] [PubMed] [Google Scholar]
- Wang HL, Postier BL, Burnap RL. Alterations in global patterns of gene expression in Synechocystis sp. PCC 6803 in response to inorganic carbon limitation and the inactivation of ndhR, a LysR family regulator. J Biol Chem. 2004;279:5739–51. [DOI] [PubMed] [Google Scholar]
- Wang X, Lei G, Wu Xet al.. Expression, purification and characterization of sll1981 protein from cyanobacterium Synechocystis sp. PCC 6803. Protein Expr Purif. 2017;139:21–28. [DOI] [PubMed] [Google Scholar]
- Wan N, DeLorenzo DM, He Let al.. Cyanobacterial carbon metabolism: Fluxome plasticity and oxygen dependence. Biotechnol Bioeng. 2017;114:1593–602. [DOI] [PubMed] [Google Scholar]
- Watzer B, Engelbrecht A, Hauf Wet al.. Metabolic pathway engineering using the central signal processor PII. Microbial Cell Factories. 2015;14:192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watzer B, Forchhammer K. Cyanophycin synthesis optimizes nitrogen utilization in the unicellular cyanobacterium Synechocystis sp. Strain PCC 6803. Appl Environ Microbiol. 2018;84:e01298–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watzer B, Spät P, Neumann Net al.. The signal transduction protein PII controls ammonium, nitrate and urea uptake in cyanobacteria. Front Microbiol. 2019;10:1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei TF, Ramasubramanian TS, Golden JW. Anabaena sp. strain PCC 7120 ntcA gene required for growth on nitrate and heterocyst development. J Bacteriol. 1994;176:4473–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolk CP. Physiology and cytological chemistry blue-green algae. Bacteriol Rev. 1973;37:32–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong W, Brune D, Vermaas WF. The gamma-aminobutyric acid shunt contributes to closing the tricarboxylic acid cycle in Synechocystis sp. PCC 6803. Mol Microbiol. 2014;93:786–96. [DOI] [PubMed] [Google Scholar]
- Yamauchi Y, Kaniya Y, Kaneko Yet al.. Physiological roles of the cyAbrB transcriptional regulator pair Sll0822 and Sll0359 in Synechocystis sp. strain PCC 6803. J Bacteriol. 2011;193:3702–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalutskaya Z, Kharatyan N, Forchhammer Ket al.. Reduction of PII signaling protein enhances lipid body production in Chlamydomonas reinhardtii. Plant Sci. 2015;240:1–9. [DOI] [PubMed] [Google Scholar]
- Zeth K, Fokina O, Forchhammer K. Structural basis and Target-Specific modulation of ADP Sensing by the Synechococcus elongatus PII signaling protein. J Biol Chem. 2014;289:8960–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CC, Zhou CZ, Burnap RLet al.. Carbon/Nitrogen Metabolic Balance: Lessons from Cyanobacteria. Trends Plant Sci. 2018;23:1116–30. [DOI] [PubMed] [Google Scholar]
- Zhang H, Liu Y, Nie Xet al.. The cyanobacterial ornithine-ammonia cycle involves an arginine dihydrolase. Nat Chem Biol. 2018a;14:575–81. [DOI] [PubMed] [Google Scholar]
- Zhang S, Bryant DA. The tricarboxylic acid cycle in cyanobacteria. Science. 2011;334:1551–3. [DOI] [PubMed] [Google Scholar]
- Zhang S, Qian X, Chang Set al.. Natural and synthetic variants of the tricarboxylic acid cycle in cyanobacteria: Introduction of the GABA Shunt into Synechococcus sp. PCC 7002. Front Microbiol. 2016;7:1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Pu H, Wang Qet al.. PII is important in regulation of nitrogen metabolism but not required for heterocyst formation in the Cyanobacterium Anabaena sp. PCC 7120. J Biol Chem. 2007;282:33641–8. [DOI] [PubMed] [Google Scholar]
- Zhao MX, Jiang YL, Xu BYet al.. Crystal structure of the cyanobacterial signal transduction protein PII in complex with PipX. J Mol Biol. 2010;402:552–9. [DOI] [PubMed] [Google Scholar]







