Abstract
Avian genomes are small and lack some genes that are conserved in the genomes of most other vertebrates including nonavian sauropsids. One hypothesis stated that paralogs may provide biochemical or physiological compensation for certain gene losses; however, no functional evidence has been reported to date. By integrating evolutionary analysis, physiological genomics, and experimental gene interference, we clearly demonstrate functional compensation for gene loss. A large-scale phylogenetic analysis of over 1,400 SLC2 gene sequences identifies six new SLC2 genes from nonmammalian vertebrates and divides the SLC2 gene family into four classes. Vertebrates retain class III SLC2 genes but partially lack the more recent duplicates of classes I and II. Birds appear to have completely lost the SLC2A4 gene that encodes an important insulin-sensitive GLUT in mammals. We found strong evidence for positive selection, indicating that the N-termini of SLC2A4 and SLC2A12 have undergone diversifying selection in birds and mammals, and there is a significant correlation between SLC2A12 functionality and basal metabolic rates in endotherms. Physiological genomics have uncovered that SLC2A12 expression and allelic variants are associated with insulin sensitivity and blood glucose levels in wild birds. Functional tests have indicated that SLC2A12 abrogation causes hyperglycemia, insulin resistance, and high relative activity, thus increasing energy expenditures that resemble a diabetic phenotype. These analyses suggest that the SLC2A12 gene not only functionally compensates insulin response for SLC2A4 loss but also affects daily physical behavior and basal metabolic rate during bird evolution, highlighting that older genes retain a higher level of functional diversification.
Keywords: gene loss, functional compensation, new gene, positive selection, SLC2A12
Introduction
Compared with other jawed vertebrates, birds have small genomes and lack many protein encoding genes including those associated with lethality or disease phenotypes in mice or humans (Organ et al. 2007; Lovell et al. 2014). Importantly, birds avoid these disadvantages and have evolved one of the most successful and diverse vertebrate lineages. Therefore, determining why and how birds compensate biochemical or physiological defects that result from these gene losses may provide insights into the origin and evolution of birds, as well as provide a medical model for various human diseases. One hypothesis is that some conserved gene losses are related to physiological characteristics such as hyperglycemia, insulin insensitivity (SLC2A4), high metabolic rates, the loss of nonshivering thermogenesis (UCP1, SLN), and low glomerular filtration rates (KIRREL2) (Newman et al. 2013; Lovell et al. 2014; Rowland et al. 2015; Lovegrove 2017). The other hypothesis states that related gene family or different family members may supply functional compensation for gene losses (Gu et al. 2003; Liang and Li 2009; Lovell et al. 2014). To date, however, no studies provide any functional evidence in vertebrates to support these two hypotheses.
The origin of endothermy allowed birds and mammals to maintain constant body temperature through high basal metabolic rates (BMRs) in cold environments, which resulted in their elevated diversification rates and expanded global distribution (Avaria-Llautureo et al. 2019). Glucose is essential to sustains high BMRs, and the transport of glucose across the cell membranes is mediated by members of the glucose transporter (GLUT) family that are encoded by SLC2 genes (Mueckler and Thorens 2013). Hence, evolutionary changes involving SLC2 genes may be related to high BMRs in endotherms. However, the evolutionary relationship between animal SLC2 genes remains largely elusive. Insulin is an important hormone that regulates glucose uptake by stimulating SLC2A4 expression and translocation (Rose and Richter 2005; Petersen and Shulman 2018). Unlike mammals, birds are thought to have high blood glucose levels and insulin resistance due to the loss of SLC2A4, whereas evidence from chicken (Gallus gallus) and house sparrow (Passer domesticus) suggests rapid glucose uptake after exogenous insulin stimulation in birds (Sweazea and Braun 2005; Dupont et al. 2008). In mammals, 14 SLC2 genes are expressed in different tissues and possess various substrate specificities (Mueckler and Thorens 2013). Therefore, we hypothesize that paralogous genes may provide biochemical or physiological compensation for the SLC2A4 loss in birds.
To test this hypothesis, we performed phylogenetic analysis based on >1,400 putative SLC2 sequences representing ten eumetazoan phyla to reconstruct evolutionary relationships and identify instances of gene duplication and loss. We resolved the organization of the animal SLC2 gene family, and we found six new types of nonmammal SLC2 genes in vertebrates. We find that old genes have a high proportion of retention and functional diversification compared with young genes. Positive selection analyses show that N-termini of SLC2A4 and SLC2A12 evolved under diversifying selection in birds and mammals and evolutionary variations of SLC2A12 in endotherms are related to BMR.
Additionally, by combining wild bird physiological genomics with RNA interference, we confirm that SLC2A12 provides functional compensation for the loss of SLC2A4 and controls BMR in tree sparrows. In this study, we focused on tree sparrow (Passer montanus) and two snowfinch species (Onychostruthus taczanowskii and Pyrgilauda ruficollis); all species belong to the Old World sparrow family (Passeridae). Tree sparrows were introduced thousands of years ago as synanthropic colonizers of the Qinghai–Tibet Plateau, despite their altitudinal distribution from sea level to 4,400 meters above sea level (masl), whereas snowfinches are endemic natives restricted to high elevations (3,500–5,100 masl) and have resided for millions years on the Qinghai–Tibet Plateau (Qu et al. 2020). Our results revealed that genetic and expression variations of the SLC2A12 gene across elevational songbirds contribute to metabolic adaptation to highland hypoxia.
Results
New Genes, Classification, and Origin of the SLC2 Gene Family in Vertebrates
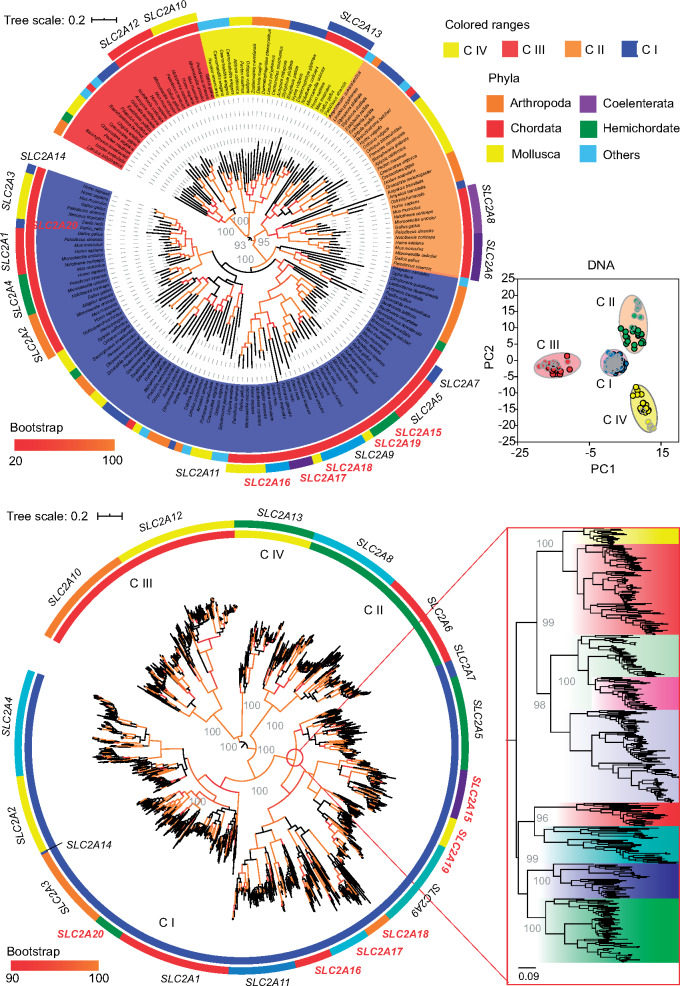
To study the evolutionary history of the SLC2 genes, we generated a maximum-likelihood tree based on a database of over 1,400 putative SLC2 sequences from ten eumetazoan phyla (Arthropoda, Brachiopoda, Chordata, Coelenterata, Hemichordata, Mollusca, Platyhelminthes, Priapuloidea, Pseudocoelomata, and Spongia). This large-scale phylogenetic analysis identified six novel SLC2 genes from nonmammal vertebrates, which we designated as SLC2A15, SLC2A16, SLC2A17, SLC2A18, SLC2A19, and SLC2A20 (fig. 1 and supplementary fig. S1 and supplementary table 1, Supplementary Material online). Of these genes, SLC2A15 is present in birds, reptiles, amphibians, and telerosts, traced to an ancestral gene (SLC2A9). SLC2A16 and SLC2A17, originated from the ancestral gene, SLC2A11, were present in three clades but respectively absent in amphibians and reptiles. However, SLC2A18, SLC2A19, and SLC2A20 were found only in telerosts and form a sister clade of gene to SLC2A11, SLC2A15, and SLC2A3. In addition, we identified two SLC2 genes, SLC2A7 and SLC2A14, which were unique genes and separately duplicated from SLC2A5 and SLC2A3 in the mammalian lineage and humans instead of other vertebrate lineages. We found total 20 types of SLC2 genes in vertebrates, 11 genes shared by all of the vertebrate lineages although 14 genes were respectively present in mammals, birds, reptiles, and amphibians and 18 genes in telerosts.
Fig. 1.
Phylogenetic analysis of SLC2 genes across ten diverse eumetazoan phyla. Maximum-likelihood tree-based 176 nucleotide sequences show the organization of all of the animal SLC2 genes (top) and identify six novel types of SLC2 gene in nonmammal vertebrate across 1,240 vertebrate SLC2 sequences (bottom). PCAs of pairwise nucleotide sequence identity of SLC2 genes also show four distinct classes. Six new genes were marked in red color.
To illustrate the classification and origin of these 20 genes, we collected SLC2 sequences predicted from nonvertebrate animals representing nine diverse phyla. In the maximum-likelihood tree based on nucleotide and amino acid sequences, 20 SLC2 genes were grouped into four classes and formed a sister clade to the SLC2-like sequences of invertebrates (fig. 1 and supplementary fig. S1, Supplementary Material online). Fifteen SLC2 genes were grouped into class I (SLC2A1–5, 7, 9, 11, and 14–20). Although the class I SLC2 genes formed two subclasses in vertebrates, namely, subclass I (SLC2A1–4, 14, and 20) and subclass II (SLC2A5, 7, 9, 11, and 15–19), bootstrap branch support was <95 and were derived from the ancestral genes of invertebrate class I genes. The SLC2 genes of subclass II were identified in invertebrate chordates and the acorn worm (Saccoglossus kowalevskii) but not in other invertebrates, indicating that these genes emerged after the Protostomia–Deuterostomia divergence, possibly by duplications in vertebrates calculated to occur at 356.57 Ma. Class II comprised GLUT6 and 8; class III contained SLC2A10 and 12; and SLC2A13 formed class IV as a sister clade to class III. In addition, principal component analysis (PCA) of amino acid and nucleotide identity matrixes supports four classes of SLC2 genes in terms of sequence similarity (fig. 1 and supplementary fig. S1, Supplementary Material online). Vertebrate SLC2 genes of subclass I in class I as well as other three classes formed a sister lineage to SLC2 genes from invertebrates, but SLC2-like sequences of sea squirt (Phallusia mammillata) and amphioxus (Branchiostoma belcheri) as invertebrate chordates with unique evolutionary position were grouped with the invertebrates, indicating that independent duplications of an ancestral gene generated diverse SLC2 genes in different classes after the Tunicata–Vertebrata divergence. Class I–III gene duplication events were estimated to emerge at 358.97, 457.5, and 457.5 Ma, respectively. Overall, the SLC2 genes of these four classes were independently derived from an ancestral SLC-like gene in invertebrates and those of classes I–III were duplicated during vertebrate evolution.
Gene Loss and Retention of the SLC2 Gene Family in Vertebrates
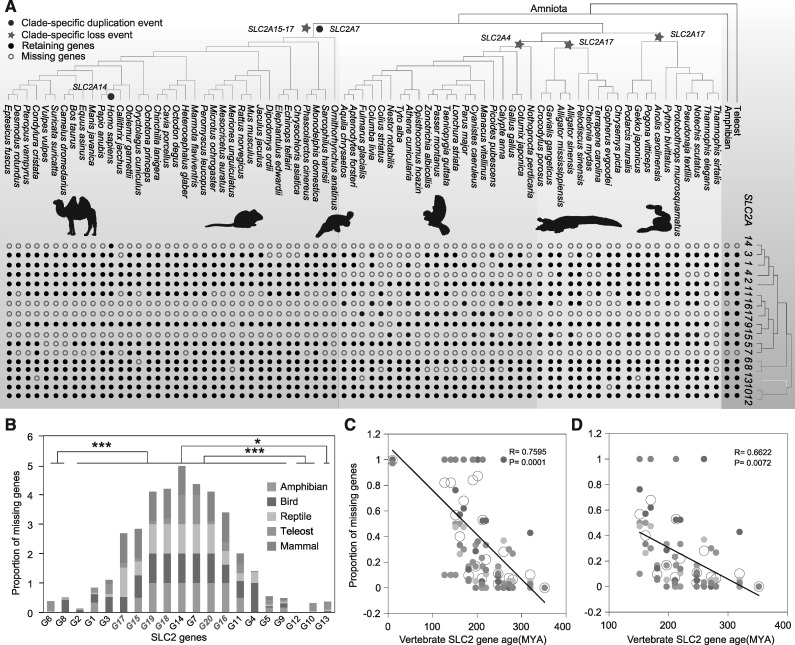
Although vertebrate SLC2 genes of classes I–III were duplicated, some vertebrate lineages did not contain certain duplicates (e.g., mammal clade lacked SLC2A15–20). We next evaluated the loss and retention all of the SLC2 genes across five vertebrate lineages, including 30 telerosts, 6 amphibians, 19 reptiles, 21 birds, and 36 mammalian species (fig. 2A and supplementary table 1, Supplementary Material online). We found that the GLUT genes of class I had a higher proportion of gene loss than the other three classes (Mann–Whitney test: P < 0.05 comparing to class IV; P value <0.0001 comparing to classes II and III) (fig. 2B). Additionally, the proportion of missing genes was significantly negatively correlated to the estimated ages (linear correlation: P = 0.0001, r = 0.7595) and relative ages (linear correlation: P < 0.0001, r = 0.8037) of all of the vertebrate SLC genes (fig. 2C and supplementary fig. 2A, Supplementary Material online). Mammals and fish independently duplicated clade-specific SLC2A7 (also SLC2A14 in human) and SLC2A18–SLC2A20, respectively (fig. 2A). This significantly negative correlation remained when these unique SLC2 genes in one lineage were excluded (linear correlation: P = 0.0072, r = 0.6622 for estimated age and P = 0.0005, r = 0.7906 for relative age) (fig. 2D and supplementary fig. 2B, Supplementary Material online). These results strongly indicate that recently duplicated genes were more likely to be lost than the older duplicates in the SLC2 gene family.
Fig. 2.
The retention and loss of SLC2 genes in different vertebrate clades. (A) SLC2 gene loss and retention in amniota species. Birds completely lost SLC2A4 appearing to result from a clade-specific gene deletion in the avian lineage, or within an archosaur organism within the dinosaurian/avian lineage ancestral to extant birds. Mammals totally lost SLC2A15–20 but independently duplicate SLC2A7 (also SLC2A14 in human). (B) Proportion of missing genes in vertebrate clades. Mann–Whitney test is used to compare differences on variant SLC2 genes. *P value <0.05, **P value <0.01, and ***P value <0.0001. (C) Significantly negative correlation between the proportion of missing genes and vertebrate SLC2 gene age (rate of evolution for nucleotide substitution). (D) Significantly negative correlation excluding certain SLC2 genes only in one lineages(SLC2A7, 14, 18, 19, and 20). Linear regression is used to test the above correlations. “Ma” represents million years ago.
The pattern of absence of these genes could be divided into two types (random loss and clade-specific loss) based on species phylogenetic relationships. Most of SLC2 genes were randomly missed across all of the vertebrate lineages. For example, SLC2A1, which encoded a basal glucose transporter, selectively lost in nonmammal vertebrates (fig. 2A). Some genes were completely missing in certain clades, such as SLC2A4 in birds, and SLC2A17 in reptiles and mammals. In particular, SLC2A4, which encoded an insulin-sensitive GLUT, is completely absent in avian lineage but was detected in other vertebrate clades including Chinese soft-shelled turtle (Pelodiscus sinensis) and Chinese alligator (Alligator sinensis) (fig. 2A). Apparently, birds lost SLC2A4 due to a clade-specific gene deletion early in the avian lineage, or within an archosaur within the dinosaurian/avian lineage ancestral to extant birds.
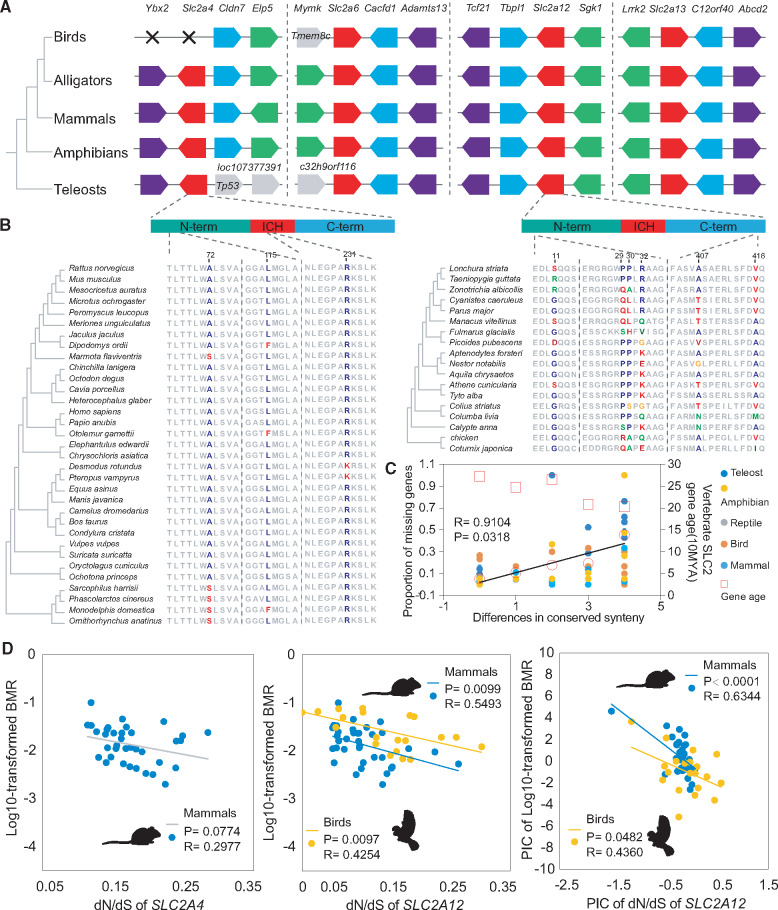
To explore why genes were lost, we examined whether lost genes among related species occurred in syntenic regions. The SLC2 genes with low proportion of missing genes, such as SLC2A2, 9, 10, 12, and 13, were always flanked within conserved syntenic loci across vertebrate clades, whereas other genes were located on chromosomes with high levels of rearrangements (fig. 3A and supplementary fig. S3A, Supplementary Material online). The differences in conserved synteny were significantly positively correlated with the proportion of missing genes (Pearson correlation: P = 0.0318, Pearson’ r = 0.9104) without the influences of gene ages (fig. 3C), indicating that chromosomal structural variations (rearrangements) may have caused these gene losses.
Fig. 3.
Conserved synteny and positive selection of mammal SLC2A4 and bird SLC2A12. (A) Schematic of syntenic region encoding SLC2 genes from four distinct classes in vertebrates. “X” symbols denote absence of genes. (B) PAML identifying two (codons 72 and 115) and four (codons 11, 29, 30, and 32) positively selected sites on the N-terminal domains of mammal SLC2A4 and bird SLC2A12. (C) Significantly positive correlation between the proportion of missing genes and differences of conserved synteny using Pearson correlation analysis. (D) Selective constraint of SLC2A4 and SLC2A12 showing relationship between evolutionary changes of SLC2A12 and the BMRs in endotherms using linear regression analysis. PIC represents the result for phylogenetic independent contrast.
Functional Divergence and Strong Signatures of Positive Selection
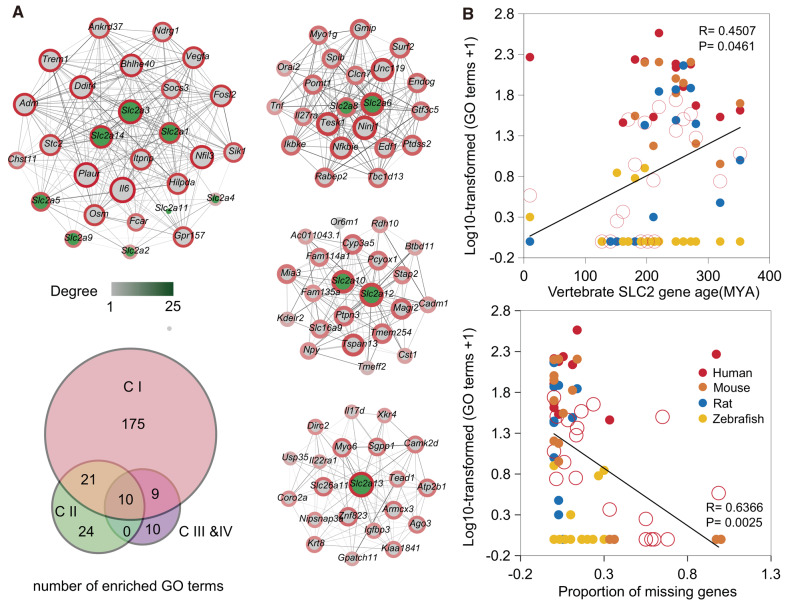
To determine the functional conservation and divergence among these genes of four classes, we performed gene coexpression and Gene Ontology (GO) analyses using transcriptomic data from human, mouse, rat, and zebrafish. There were nonoverlapping coexpressed genes among the four networks, but these gene networks shared some GO terms which had a statistically significant extent (fig. 4A). The class I gene network had more GO terms (215 vs. 55 and 29) than the other three classes for more SLC2 genes (7 vs. 2 and 3). However, ten significantly enriched GO terms were shared among the four networks (fig. 4A), indicating functional conservation and divergence through different coexpressed genes in each class. When we considered each SLC2 gene, we found that the number of GO terms was associated with the estimated SLC2 gene ages (linear correlation: P = 0.0461, r = 0.4507), relative gene ages (linear correlation: P = 0.0048, r = 0.6035), and the proportion of missing genes (linear correlation: P = 0.0025, r = 0.6366) (fig. 4B and supplementary fig. 2C, Supplementary Material online). These results revealed that the early duplicates may have more functional diversification than the recent duplicates which have lost more genes in the SLC2 gene family.
Fig. 4.
Functional conservation and divergence among these four distinct classes of SLC2 genes. (A) In human, nonoverlapping coexpressed gene networks among four class SLC2 genes, but existence of shared statistically significant enrichment. (B) In single gene network, the number of GO terms showing association with vertebrate SLC2 gene age and proportion of missing genes using linear regression analysis.
Glucose is an essential molecule that provides energy to sustain high BMRs, so we hypothesized that SLC2 genes may play an important role in metabolism during vertebrate evolution, particularly the evolution of endotherms. To qualify the difference in dN/dS ratios of endotherms with various metabolic rates, we calculated the dN/dS values of different genes within the same species clade and of different species lineages within the same gene. SLC2A12 in bird clade showed a higher average pairwise dN/dS (Analysis of variance: mean value = 0.1851, P < 0.05) and highest dN/dS for the M0 test (0.2418) compared with the other SLC2 genes (<0.24) which were found in all of the vertebrate lineages, implying that this gene evolved under relaxed selection or diversifying selection in birds (supplementary fig. S3B and supplementary table 2, Supplementary Material online, and table 1). To test these probabilities, we used site models in Phylogenetic Analysis by Maximum Likelihood (PAML) to test these genes for evidence of positive selection in five vertebrate clades and these models included four pair of models (m1a vs. m2a, m2a vs. m2a_fixed, m7 vs. m8, and m8a vs. m8). We found strong positive selection of the SLC2A12 gene in birds in all of the PAML tests, whereas we failed to find evidence of positive selection in any two tests on other bird SLC2 genes, including SLC2A10, which is a duplication of SLC2A12. PAML analyses identified six sites were highlighted with a high posterior probability of having evolved under positive selection (Bayes empirical Bayes (BEB) ≥ 85%). Four (codons 11, 29, 30, and 32) and two (codons 407 and 416) of these sites separately clustered within the N-termini and C-termini of the gene and showed extensive diversification in amino acid properties across birds (table 1 and fig. 3B). These results indicated that the SLC2A12 gene underwent strong positive selection relative to the other avian SLC2 genes.
Table 1.
Results of Selection Tests across Different Vertebrate Lineages.
| Lineage | nb. | m0 | m1a versus m2a | m2a versus m2a_fixed | m7 versus m8 | m8a versus m8 | Sites |
|---|---|---|---|---|---|---|---|
| Species | dN/dS | BEB ≥ 85% | |||||
| SLC2A4 | |||||||
| Mammal | 33 | 0.0577 | 0.999 | 1 | 9.85e-05 | 1.20e-04 | 72A, 115L, 231R |
| Reptile | 12 | 0.0610 | 1 | 1 | 0.810 | 0.230 | N/A |
| Amphibian | 6 | 0.0630 | 1 | 1 | 0.095 | 0.583 | N/A |
| Teleost | 29 | 0.0806 | 1 | 1 | 0.247 | 1.09e-04 | N/A |
| SLC2A12 | |||||||
| Mammal | 36 | 0.1430 | 1 | 1 | 0.999 | 0.655 | N/A |
| Bird | 18 | 0.2418 | 9.77e-04 | 1.97e-04 | 9.73e-05 | 5.09e-05 | 11G, 29P, 30P, 32K, 407A, 416A |
| Reptile | 19 | 0.2099 | 1 | 1 | 0.250 | 0.177 | N/A |
| Amphibian | 6 | 0.1339 | 1 | 1 | 0.527 | 0.802 | N/A |
| Teleost | 30 | 0.0957 | 1 | 1 | 0.996 | 0.093 | N/A |
Italic values highlight P-values below 0.05.
Birds are descendants of therapod dinosaurs and known to have higher metabolic rates to sustain constant body temperature similar to mammals. We therefore hypothesized that mammalian species also have SLC2 genes subjected to similar selective pressures as avian SLC2A12. By performing likelihood ratio tests, we found strong support for positive selection acting on the SLC2A4 gene in less conservative m7 versus m8 and m8a versus m8 tests compared with the more conservative m1a versus m2a and m2a versus m2a_fixed tests (table 1). In contrast, no positive selection was detected in mammalian SLC2A12 or other SLC2 genes in any two tests (supplementary table 2, Supplementary Material online). PAML analyses identified three substitutions as likely under diversifying selection with a high posterior probability (BEB ≥ 85%). Similar to our findings in avian SLC2A12, two of these sites (codons 72 and 115) were located in the N-terminal domain of mammalian SLC2A4 (table 1 and fig. 3C). In other vertebrate clades, we found no evidence for diversifying selection acting on either SLC2A12 or SLC2A4, although other SLC2 genes were positively selected (supplementary table 2, Supplementary Material online). These findings serve as evidence of positive selection on SLC2A12 or SLC2A4 genes separately belonging to SLC2 gene classes in birds and mammals, representing two clades of endotherms, implying that functional convergence resulted from the N-terminal domain of SLC2 genes and resulting in physiological adaptation.
To further determine the relationships among SLC2A12 and SLC2A4 genes and energy metabolism, we conducted linear regression analysis by comparing dN/dS values to BMRs for 36 mammals and 21 avian species. The log 10-transformed BMR was negatively correlated to the dN/dS value of SLC2A4 (linear correlation: r = 0.2977) in mammals, although these correlations were only marginally near significant (P = 0.0774) (fig. 3D). Importantly, the dN/dS ratio of SLC2A12 gene was significantly negatively correlated to BMRs (P values 0.0097, r = 0.4254) in mammals (fig. 3D). In birds, we also observed a significantly negative correlation between the dN/dS ratio for SLC2A12 and BMRs (linear correlation: P = 0.0099, r = 0.5493) (fig. 3D). Thus, in endotherms (mammals and birds), there was a tendency that the dN/dS ratio of SLC2A12 decreased with the BMR (linear correlation: P = 0.0158, r = 0.3184). To exclude phylogenetic influence, we performed phylogenetic independent contrast (PIC) between the dN/dS value and BMRs. After restricting the analysis to independent contrasts, significant negative relationships between dN/dS ratios and BMR were observed in both birds (PIC: P = 0.0482, r = 0.4360) and mammals (P < 0.0001, r = 0.6344), which suggests that evolution of SLC2A12 is related to the BMR in endotherms.
Physiological and Genomic Evidence Indicates Vital Function of SLC2A12 in Wild Songbirds
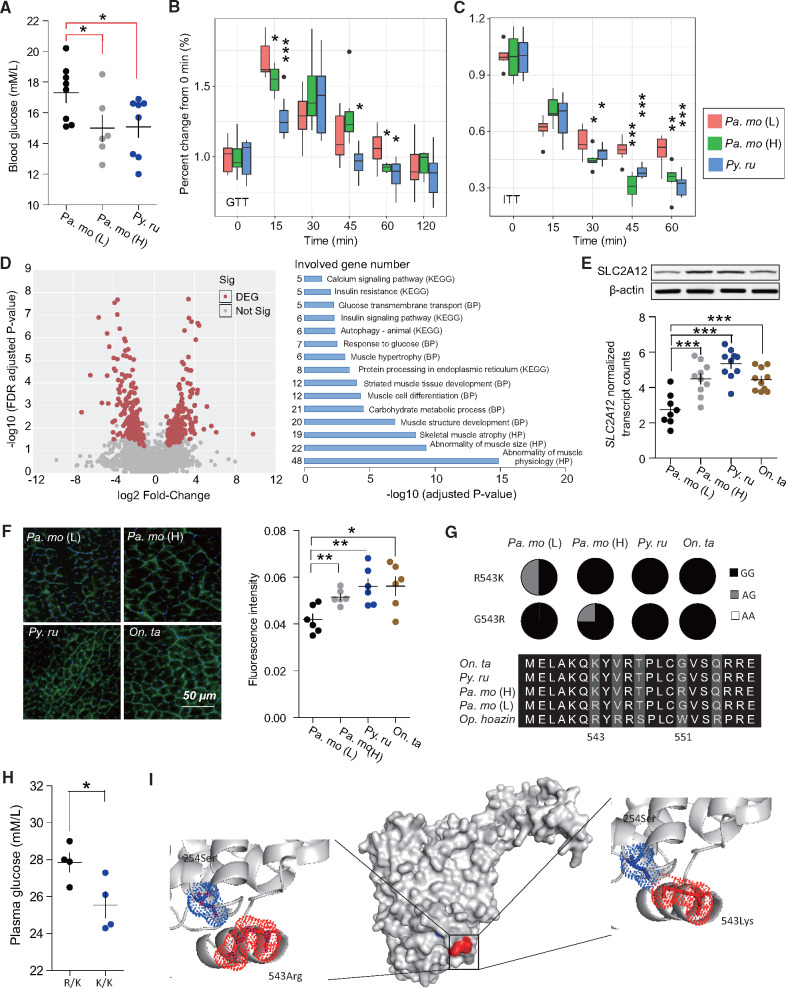
We next examined high-altitude birds to provide additional evidence for the function of SLC2A12. High-altitude hypoxia forces the increase of glucose utilization on small birds and mammals (Schippers et al. 2012), but the mechanism remains unknown. Insulin plays an important role in glucose metabolism and we recently found that high-altitude songbirds usually had lower blood glucose and similar insulin content compared with lowland birds (fig. 5A), so we speculated that highland birds might have improved insulin sensitivity relative to their lowland partners. To test this, we performed glucose and insulin tolerance tests (GTT and ITT) across altitudinal songbirds. Highland birds had more rapid normalization of blood glucose levels at 15 and 60 min (also at 45 min in snow fiches) post an oral average of 3 mg/g body weight d-glucose (P values <0.05) (fig. 5B). Next, to investigate whether highland songbirds improved insulin sensitivity, we measured blood glucose levels at 15, 30, 45, and 60 min after injection of 1.5 mU/g body weight human insulin. In highland songbirds, insulin administration caused significant decrease in blood glucose levels at 30 min and continued to 60 min after insulin injection (t-test: P < 0.05) (fig. 5C), in contrast to low-altitude birds. Altogether, these data indicated that high-altitude songbirds indeed increased insulin sensitivity to improve glucose uptake.
Fig. 5.
Physiological and genomic evidence showed vital function of SLC2A12 in wild songbirds. (A) Blood glucose levels are lower in highland songbirds in contrast to lowland birds (n = 8 for Pa. mo (L) and snowfinch, and 6 for Pa. mo (H)). (B, C) Highland birds increase glucose tolerance and insulin sensitivity. Two tailed t-test is used to perform significant analysis. *P value <0.05, **P value <0.01, and ***P value <0.0001. D) RNA-seq analysis identifies 407 DEGs shared by all highlanders, and these genes enrich in insulin resistance and insulin signaling pathway functions (n = 8 for Pa. mo (L), and 10 for highland birds). (E) Protein and gene abundance of SLC2A12 are overexpressed in highlander muscles (n = 8 for Pa. mo (L), and 10 for snowfinches and Pa. mo (H)). (F) Immunofluorescence staining shows more SLC2A12 translocated to the myolemma in highland birds. Green and blue indicate SLC2A12 protein and nuclei, respectively. (G) Genotypes of the lowlander and highlanders at SLC2A12 1703 SNP. The putatively advantageous homozygote is shown in black, and heterozygote was shown in gray. (H) SLC2A12-G alleles at site 1073 is associated with low blood glucose level in lowland populations (n = 4 each). (I) Modeling of the structural changes in SLC2A12 caused by the arginine–lysine dimorphism at residue 543 predicted a van der Waals interaction is formed between 543Lys and 254Ser. Latin name abbreviation is presented as follow. Pa. mo (L) for lowland tree sparrow (Passer montanus); Pa. mo (H) for highland tree sparrow (Passer montanus); Py. ru for rufous-necked snowfinch (Pyrgilauda ruficollis); On. ta for white-rumped snowfinch (Onychostruthus taczanowskii); Op. hoazin for hoatzin (Opisthocomus hoazin).
Previous studies have suggested that birds have high blood glucose levels and low sensitivity to insulin due to the absence of GLUT4 (SLC2A4) (Braun and Sweazea 2008; Polakof et al. 2011). Alternatively, another hypothesis suggested high blood glucose connected with high activities (e.g., flight capacity) (Braun and Sweazea 2008). Our above findings confirm that birds can respond to insulin stimulation despite the lack of GLUT4. Additionally, we found that lowland songbirds showed incapacity of movement and even convulsion when blood glucose levels dropped from 22.81 to 5.95 mM/l, whereas similar responses were observed only at lower levels (meantree_sparrow = from 14.77 to 3.73 mM/l and meansnowfinch from 18.43 to 4.38 mM/l), indicating that high glycemia is important for maintaining daily movement.
Why do birds remain sensitivity to insulin stimulation despite GLUT4 loss? To investigate the molecular mechanisms underlying insulin sensitivity, we performed comparative transcriptome analyses using pectoralis muscles of 38 highland and lowland songbirds (again same species). We identified 407 differentially expressed genes (DEGs) shared by the pectoralis muscles of all highland birds, and functional annotation indicated that these enriched the categories of “insulin resistance (KEGG)” and “insulin signaling pathway (KEGG)” using human functional enrichment profiling from G: PROFILER (fig. 5D). We did not find the expression of orthologs to gene encoding GLUT4, whereas expression of SLC2A12 gene encoding another glucose transporter protein 12 (GLUT12) was specifically upregulated in highland birds’ pectoralis muscles. Western blot confirmed that protein levels of GLUT12 were also overexpressed (fig. 5E). Immunofluorescence staining of skeletal muscles confirmed more SLC2A12 translocation to the myolemma in highland birds (fig. 5F). Additionally, we also detected the expression of seven other transporters in pectoral muscle, but no expression difference of these genes was found between highland and lowland birds (supplementary fig. S4, Supplementary Material online). Thus, SLC2A12 correlates with insulin sensitivity and glucose metabolism in birds.
To assess whether variation in the SLC2A12 gene was the result of natural selection, we investigated genetic variation in 44 birds of the three species and identified alleles resulting from nonsynonymous substitutions for the gene. We identified two nonsynonymous sites between high- and low-elevation populations, whereas only one nonsynonymous site had significantly different allele frequencies (Fisher’s exact test: P = 0.0219). In contrast to SLC2A12, no missense substitutions were identified on other SLC2 genes between populations from different elevations. The SLC2A12 gene showed an A-to-G transition at site 1703, causing an arginine-to-lysine nonsynonymous substitution at site 543, with a higher A frequency in highland songbirds (all 100% at site 1703 versus 75%; fig. 5G), suggesting that this single-nucleotide polymorphism (SNP) was a standing variant (in lowland populations) and is fixed in highland birds. Additionally, in low-elevation birds, the plasma glucose content of individuals that are heterozygous at this site was higher than homozygous birds (27.85 vs. 25.55 mM/l, t-test: P = 0.0392) (fig. 5H). To address the possibility that the R543K substitution may affect protein function, we performed a homology model of the two SLC2A12 allotypes based on the crystal structure of SLC2A3 protein (PDB 4ZWC) using SWISS-MODEL. Modeling showed that there is no atomic contact between 543Arg and 254Ser, whereas 543Lys had an additional carbon atom relative to Ser, and a van der Waals interaction is formed between 543Lys and 254Ser (fig. 5I). These results indicate that SLC2A12 experiences more natural selection than other SLC2 genes, which is related with 543K variant influencing blood glucose concentration.
SLC2A12 Displays Functional Compensation on Insulin Response and Glucose Metabolism in Birds
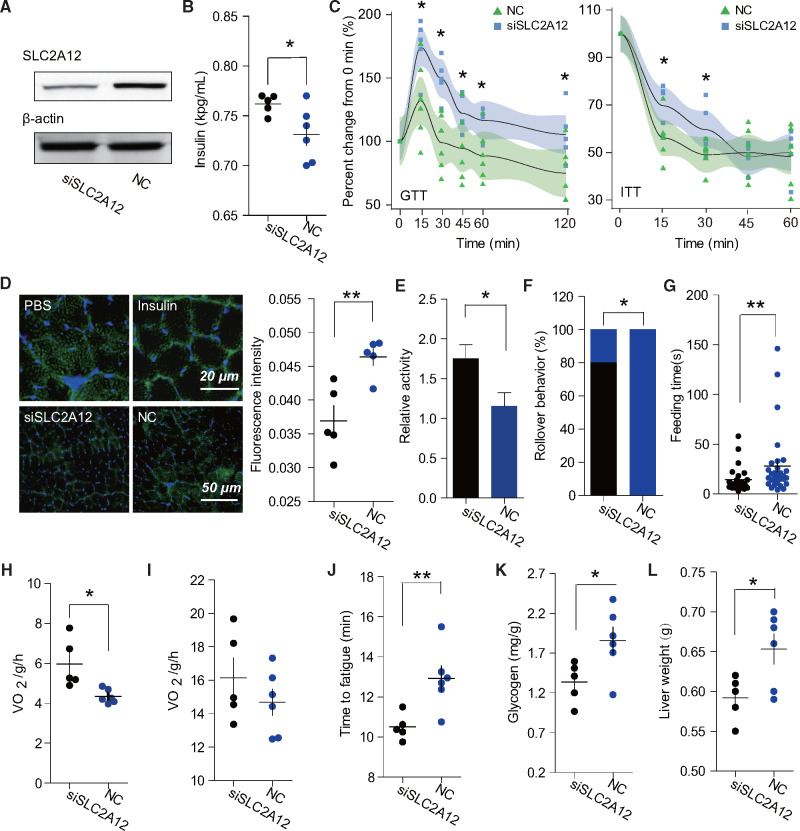
To further test the importance of SLC2A12 in insulin response and glucose metabolism, we depleted SLC2A12 expression using in vivo RNA interference in tree sparrow skeletal muscles. Western blot assessed that SLC2A12 protein level normalized to β-actin was significantly decreased in flight muscle of SLC2A12-silenced sparrows than in the muscles of control group tree sparrows (fig. 6A). Blood glucose was different between SLC2A12 abrogation and control group tree sparrows after 3 h fasting (24.06 vs. 21.85 mM/l; t-test: P = 0.0432) but not 6 h fasting (17.28 vs. 17.20 mM/l; t-test: P = 0.8194). Furthermore, insulin contents were increased in SLC2A12-silenced birds (0.7620 vs. 0.7314 kpg/ml; P value 0.0420) (fig. 6B). SLC2A12-silenced sparrows exhibited glucose intolerance and decreased insulin sensitivity during GTT and ITT (fig. 6C). In addition, functional experimentation confirmed that the SLC2A12 protein appears to be located in intracellular compartments in noninsulin injection state (Phosphate Buffer Saline (PBS) injection); after insulin injection, however, more SLC2A12 protein appeared at or near the plasma membrane of the muscle fiber (fig. 6D). A reduced portion of SLC2A12 from the intracellular to the plasma membrane component was observed in SLC2A12 abrogation birds after insulin injection (fig. 6D). These finding showed that SLC2A12-silencing causes hyperglycemia and insulin resistance, resembling a diabetic phenotype, and thus SLC2A12 gene functionally compensates for insulin response due to SLC2A4 gene loss in birds.
Fig. 6.
SLC2A12 abrogation affected insulin response and physiological metabolism. (A) Reduction of GLUT12 protein in SLC2A12 gene knockdown. (B) SLC2A12 abrogation birds have high blood insulin levels (n = 5 and 6 for abrogation and control birds). (C) SLC2A12 abrogation birds decrease glucose tolerance and insulin sensitivity. (D) Immunofluorescence staining shows a general cellular distribution of GLUT12 protein under PBS and insulin-stimulated conditions in control birds (top), and weaker GLUT12 fluorescence intensity in the myolemma after insulin stimulation in SLC2A12 abrogation birds compared with control birds (n = 5 each). Green and blue indicate GLUT12 protein and nuclei, respectively. (E) Knockdown of SLC2A12 in birds increases relative activity (n = 5 and 6). (F) Rollover behavior is usually observed in silenced birds after feeding. (G) Gene-silenced birds decrease feeding time in contrast to control birds. Knockdown of SLC2A12 increases BMRs (H) but not exercise metabolic rates (I). (J) Control birds have more fatigue resistance than abrogation birds. Abrogation birds reduce glycogen storage in skeletal muscle (K) and have light liver weight (L).
Notably, SLC2A12-silenced tree sparrows showed more corporeal activity and fidgeting compared with control birds (fig. 6E and supplementary video 1, Supplementary Material online). SLC2A12 abrogation birds frequently showed obvious and frequent rollover behavior from top to bottom in the cage, which was not usually observed in control birds (fig. 6F, χ2 test: P = 0.03426). This behavior usually occurred after birds were feed and it was well known a dramatic increase in blood glucose when feeding, which indicated a physical way to drop blood glucose when disruption of SLC2A12 expression (supplementary videos 2 and 3, Supplementary Material online). Additionally, SLC2A12-silenced sparrows had shorter feeding time than control birds (14.28 vs. 28.03 s, Mann–Whitney test P value 0.0075) (fig. 6G and supplementary videos 2 and 3, Supplementary Material online). Hence, SLC2A12 knockdown in tree sparrows results in modifications of physical behavior and feeding strategy to maintain glucose homeostasis.
To determine if the decrease in SLC2A12 was associated with metabolism, we measured energy expenditure of tree sparrow at rest and during exercise. SLC2A12-silenced sparrows showed a significant increase in O2 consumption (5.968 vs. 4.344 ml/g/h, t-test: P = 0.0125) at the resting state compared with control tree sparrows (fig. 6H); however, there was no difference at exercise state (16.15 vs. 14.68 ml/g/h, t-test: P = 0.3176) (fig. 6I). Birds with low expression of SLC2A12 were more sensitive to fatigue and were 23.1% lower than control birds at a constant speed of 15 m/min (10.50 vs. 12.92 min, t-test: P = 0.0097) (fig. 6J). Additionally, glycogen quantification detected by biochemical assays found that the skeletal muscle of SLC2A12 abrogation sparrows had lower glycogen content (1.337 vs. 1.857 mg/g, t-test: P = 0.0365) (fig. 6K). SLC2A12-silenced sparrows also displayed a dramatic reduction in liver weight compared with control birds (0.5920 vs. 0.6533 g, t-test: P = 0.0313) (fig. 6L). These findings suggest that lower expression of SLC2A12 reduces glucose uptake and glycogen storage in flight muscle, and thus decreases fatigue and increases BMRs in songbirds.
Discussion
A total of 14 vertebrate SLC2 genes were grouped into three or five distinct classes based on sequence similarities of limited species (Wilson-O’Brien et al. 2010; Mueckler and Thorens 2013; Jia et al. 2019). Our large-scale phylogenetic analysis across ten diverse eumetazoan phyla revealed that six new types of SLC2 genes exist in nonmammal vertebrates and all these 20 genes fall into four distinct types in the phylogenetic tree based on nucleotide and amino acid sequences. Although the variable number of SLC2 genes varies in invertebrate species likely resulting from poor annotation, phylogenetic reconstruction shows that duplication events in different SLC2 classes independently occurred between 350 and 560 Ma after Tunicata–Vertebrata divergence, which probably relates to whole-genome duplications at the invertebrate–vertebrate transition and tissue specificity for glucose transportation in vertebrates (Putnam et al. 2008; Mueckler and Thorens 2013). Class I comprises most of the SLC2 paralogs (15 genes), and three novel genes were identified only in telerosts, possibly reflecting two or more rounds of whole-genome duplications in telerosts (Putnam et al. 2008). The SLC2 genes of subclass II in class I were recently derived from the split of Protostomia–Deuterostome and then also diversified in vertebrates, which is likely related to the transportation of specific metabolites in complex circulatory system such as regulation on blood fructose (SLC2A5) and urate (SLC2A9) (Mueckler and Thorens 2013).
Additionally, each SLC2 gene shows selection or even was totally missing in different vertebrate lineages, and recently duplicated genes were more probably lost, which may result from chromosome structure variations. SLC2A1 and SLC2A4, two key players in global body glucose transport and insulin-stimulating blood glucose clearance in mammals, are selectively or completely missing in other animals. Homozygous deletion of SLC2A1 affects the survival of the preimplantation embryos, and mice die beyond embryonic day 14 (Wang et al. 2006). In humans, mutations in the SLC2A1 gene cause GLUT1 deficiency syndrome, wherein patients exhibit development delay and neurobehavioral symptoms due to lack of glucose supplement in the nervous systems (Darryl et al. 2002). Muscle specific knockout of the SLC2A4 gene disrupts sustain growth, normal cellular glucose metabolism and causes insulin resistance and glucose intolerance at an early age (Katz et al. 1995; Zisman et al. 2000). Additionally, SLC2 gene deficient mouse models show that other GLUT deletion in mammals cannot cause insulin resistance or diabetic phenotype except for SLC2A4 (Zisman et al. 2000; Darryl et al. 2002; Mueckler and Thorens 2013). Duplicated genes often overlap in function and expression, indicating that duplicate genes play a remarkable role in functional compensation (Gu et al. 2003; Liao and Zhang 2007; Liang and Li 2009). However, loss of function can be also compensated by alternative metabolic pathways and regulatory networks but not the other duplicated genes (Gu et al. 2003). Despite the importance of these two genes in mammals, gene coexpression analyses of each SLC2 gene show that old duplicates may indeed have more functional diversification compared with young ones, indicating potential functional compensation for the losses of these genes.
Birds and mammals are two parallel clades that have evolved endothermy with constant body temperature. However, birds lack some functional genes involving in physiological metabolism in contrast to mammals. It is reported that SLC2A12 similar to SLC2A4 exhibited a high affinity for d-glucose over other hexoses (e.g., d-galactose, d-fructose, and l-glucose) in Xenopus laevis oocytes, and that SLC2A12 gene deficiency impacts glucose uptake and the development of the embryonic heart in zebrafish which lacks a functional SLC2A4 gene (Rogers et al. 2003; Jiménez-Amilburu et al. 2015). Additionally, the evolutionary rate of SLC2A12 genes is negatively associated with the high BMR across 36 mammalian and 21 bird species without phylogenetic influence despite undergoing negative selection in mammals, which is beneficial to endothermy during vertebrate evolution. A recent study revealed that SLC2A12 gene was an urate transporter and regulated the blood urate level in mice (Toyoda et al. 2020). These evidences might highlight the functional diversification and essentiality of old genes, and functional compensation owing to similar positive selection on N-terminal domain.
Genetic diversity and expression levels of SLC2A12 in high- versus low-elevation songbirds further support our hypothesis that SLC2A12 has been under continuous selection for the adaptation of birds to high-altitude environments. After comparing high and low elevation, we find one nonsynonymous allele with remarkable difference in allelic frequency and function between highland and lowland birds. Structural modeling predicts that the single missense substitution in Lys543 results in a new molecular interaction between 543Lys and 254Ser. The Lys543 variant results in lower blood glucose levels and is a heterozygote advantage to lowland populations. High- and low-elevation tree sparrows were diverged thousands of years ago (Qu et al. 2020), and all highland birds harbor the Lys543 variant, whereas this variant occurs in only 70% of lowland birds. The upregulation of SLC2A12 in highland birds also increases glucose uptake and improves glycolytic metabolism thereby allowing hypoxia survival. An alternative mechanism is that SLC2A12 overexpression improves insulin sensitivity and prevents hyperglycemia because highland birds indeed exhibit glucose tolerance and insulin sensitivity. Previous studies have suggested that SLC2A12 functions as a basal and insulin-independent glucose transporter, although it could be a secondary insulin-sensitive glucose transporter (Purcell et al. 2011; Waller et al. 2013). Our study provides robust evidence for translocation of SLC2A12 from intracellular compartments to plasma membrane under insulin injection in contrast to noninsulin stimulation. In vivo gene interference in tree sparrows indicates that knockdown of SLC2A12 in skeletal muscle as example of the most important glucose metabolic tissue causes glucose intolerance and insulin resistance, resembling a diabetes phenotype. Therefore, the SLC2A12 gene functionally compensates for the loss of SLC2A4 in avian evolution and has been undergoing natural selection for extreme environmental adaptation.
Conclusion
In this study, we analyzed the phylogeny and physiological genomics of SLC2 genes to understand gene selective loss and functional compensation in relation to its evolutionary history. Here, we provided a totally new classification of SLC2 gene family in animals and identified six novel genes in nonmammal vertebrates. These SLC2 genes in each subclass and class independently derived from the splits of Protostomia–Deuterostome or Tunicata–Vertebrata, and then were duplicated in vertebrate. We showed that younger SLC2 genes were more likely easy to lost compared with older duplicates, which was probably caused by chromosome structure variation. We found that SLC2A12 in birds paralleling SLC2A4 in mammals suffered from strong positive selection and it was related with BMRs in endotherms, indicating that the SLC2A12 gene likely provided functional compensation for the loss of SLC2A4 in birds. By combining physiological genomics with experimental gene interference, we confirmed that SLC2A12 provided some compensatory functions and contributed to metabolic adaptation to hypoxia in highland birds, which tested our hypothesis that paralogous genes may provide biochemical or physiological compensation for the SLC2A4 loss in birds.
Materials and Methods
Phylogenetic Analysis
Five vertebrate genomes (Homo sapiens, Gallus gallus, Pelodiscus sinensis, Microcaecilia unicolor, and Notothenia coriiceps) and genomes of Drosophila melanogaster and Caenorhabditis elegans were downloaded from Ensembl (Zerbino et al. 2018). All of the potential orthologs for the human SLC2A1–14 genes were retrieved from these genomes through BLAST analysis. An e-value of <105 was used in reciprocal BLAST searches with additional constraint that at least 60% of sequences are covered by the alignment (Altschul et al. 1997). To expand the taxa sampled, we used BlastN to search other 148 animal species across ten different phyla including Arthropoda (n = 16), Brachiopoda (n = 1), Chordata (n = 109), Coelenterata (n = 7), Hemichordata (n = 1), Mollusca (n = 9), Platyhelminthes (n = 1), Priapuloidea (n = 1), Pseudocoelomata (n = 2), and Spongia (n = 1) (supplementary table 1, Supplementary Material online). A total of 1,496 SLC2-like sequence across 155 species were identified. To resolve the evolutionary relationship of the animal SLC2 genes, multiple sequence alignment of 176 sequences across ten animal phyla was performed using the ClustalX program (Larkin et al. 2007). To assess the organization of vertebrate SLC2 genes, 1,240 of 1,406 SLC2 sequences were selected for phylogenetic reconstruction after excluding 166 SLC2 sequences for short length. Phylogenetic trees were constructed with PhyML (Guindon et al. 2010) using maximum-likelihood analysis based on both nucleotide and amino acid sequences, and bootstrap tests were separately carried out with 1,000 iterations and 500 iterations, respectively. The nucleotide and amino acid analyses separately used the codon-based GTR + G + I mode of substitution and the JTT substitution model. Substitution models were estimated using the program ModelFinder (Kalyaanamoorthy et al. 2017). Phylogenetic trees were visualized in iTOL (Letunic and Bork 2016) and FigTree v1.4.3 (http://tree.bio.ed.ac.uk/software/figtree/). A pairwise identity matrix for nucleotide and amino acid sequences was computed and then PCA was performed using Jalview (Waterhouse et al. 2009) to distinguish SLC2 genes.
Divergence Time Estimations
The relative age of each SLC2 gene was presented as the rate of evolution for nucleotide substitution at each ancestral node of each SLC2 gene across vertebrates based on the phylogenetic tree of SLC2 genes of ten eumetazoan phyla. Because classes I–III gene duplications emerged after Tunicata–Vertebrata split, we also estimated divergence time of each gene class using MEGA X clocks (Kumar et al. 2018). The root of the ingroup tree was set 457.5–636.1 Ma corresponding to the Tunicata–Vertebrata separation (https://fossilcalibrations.org/). Two internal nodes were constrained as follows: Mammalia split (164.9–201.5 Ma) and Chimpanzee–Human split (6.5–10 Ma), corresponding to clade-specific duplications of SLC2A7 and SLC2A14, respectively. For certain SLC2 gene duplications in vertebrates and SLC2A4 gene loss in birds, duplication or loss nodes were constrained and estimated by ancestral and descendant speciation events.
Gene Loss and Synteny Analysis
To further confirm the gene losses in different vertebrate lineages, we grouped 1,406 SLC2 sequences into the above organization and calculated the proportion of SLC2 gene retention and loss in 30 teleosts, 6 amphibians, 19 reptiles, 21 birds, and 36 mammals. The proportion of missing genes was calculated using the ratio of species without certain SLC2 genes to all detected species. Differences in synteny blocks encoding SLC2 genes were identified using NCBI (http://www.ncbi.nlm.nih.gov/gene/). Syntenic regions comprised neighboring four genes and we defined these syntenic regions as five classes according to gene order and name across five diverse vertebrate clades.
Gene Coexpression Networks
We generated human gene coexpression networks for four distinct classes of SLC2 genes using transcriptomic data from GeneMANIA (Warde-Farley et al. 2010) which we visualized in Cytoscape. We identified all of the coexpressed genes for each SLC2 genes in human, mouse, rat and zebrafish according to gene expression data from GeneMANIA (http://genemania.org). We performed functional enrichment analysis using G: PROFILER (Reimand et al. 2007) on coexpressed genes and calculated the number of enriched genes for each SLC2 gene after applying a false discovery rate (FDR) correction. Correlation was tested between log-transformed the number of GO terms and SLC2 gene age as well as proportion of missing genes using linear regression. To construct gene coexpression networks of single SLC2 genes, transcriptomic data of human, mouse, rat, and zebrafish were used for functional enrichment.
Selection Analyses
To calculate evolutionary rates, we generated phylogenetic trees corresponding to these SLC2 genes across vertebrate species using the method described above. The overall ratio of nonsynonymous substitution rate to synonymous substitution rate (dN/dS) for each gene over the entire phylogeny was inferred based on the codon alignments within PAML (Yang 2007) using the site model Model0. We tested for positive selection using four pair of models including m1a versus m2a, m2a versus m2a_fixed, m7 versus m8, and m8a versus m8 with free of any gaps and stop codons. Sites under positive selection were defined as those with elevated dN/dS compared with the expectation under neutral evolution (dN/dS = 1). We calculated P values based on a χ2 test with two, and one degree of freedom respectively for the above four pairs of models. Genes with P values <0.05 of any two model tests were defined to be under positive selection. To explore the relationship between evolutionary changes in the SLC2A12 gene and BMRs in endotherms, we used Model1 of CodeML in the PAML package to compute the dN/dS ratios of this gene along with each branch of the tree for various lineages. Model1 fits the overall substitution rate and the dN/dS ratio with branch-specific values. Liner regression and PIC were performed using R packages. The pairwise dN/dS ratios for codon alignments were calculated using SNAP v2.1.1 website (https://www.hiv.lanl.gov/content/sequence/SNAP/SNAP.html). The dN/dS of all pairs of each SLC2 gene (was stained in all vertebrates) was estimated in each lineage and the average dN/dS is presented in the text.
Collection of BMRs
We obtained body mass and BMRs values for the above species from previous studies (McNab 2009; Clarke et al. 2010). For mammals and birds, final BMRs were corrected by the body masses (mass-specific metabolic rate).
RNA Sequencing and Differential Expression Analysis
Total RNA was extracted from flight muscle using TRIzol RNA isolation reagents (Invitrogen Corp., Carlsbad, CA). RNA integrity was assessed using the RNA Nano 6000 Assay Kit and the Agilent Bioanalyzer 2100 system (Agilent Technologies, CA, USA). A total amount of 3 µg RNA per sample was used as input material for RNA sample preparation. Sequencing libraries were generated using NEBNext UltraTM RNA Library Prep Kit for Illumina (NEB, USA) following manufacturer’s recommendations and index codes were added to attribute sequences to each sample.
Trimmomatic (Bolger et al. 2014) was used to filter reads containing adapter, reads containing ploy-N, and low quality reads based on read quality checked with FASTQC (Andrews 2016). The parameters used were as follows: sliding window = 4-bp; Phred33 quality scores = 20; min read length = 50. Adapter sequences, when detected, were removed. Clean data with high quality were mapped from each species to respective genomes (Qu et al. 2020) using STAR with default parameters (Dobin et al. 2013). We used the reciprocal best-hit method to generate tree sparrow–rufous-necked snowfinch and tree sparrow–white-rumped snowfinch orthologs, respectively. The orthologs shared by three species were obtained by intersecting the lists of the above two orthologs. After the reads were mapped to the reference genomes, expression quantification of the genes and transcripts was performed using RSEM (Li and Dewey 2011). Expression levels for genes with one-to-one orthologs in all three bird species (n = 12,951) were normalized with the relative log expression method across muscle samples (Anders and Huber 2010; Maza 2016).
To identify genes related with insulin sensitivity across altitudinal songbirds, we performed differential expression analysis. DEGs were calculated based on the negative binomial distribution and independent filtering was enabled in the program Desq2 with a FDR < 0.05 based on Benjamini–Hochberg method to account for type I error in identifying significantly DEGs (Love et al. 2014). The cut off values for log2-fold change were set at 0.59 and −0.59. Only genes with count number > 10 in at least four samples were included in differential gene expression analysis. We then implemented GO categories within modules positively and negatively correlated with muscle traits using G: PROFILER (FDR < 0.05) (Reimand et al. 2007).
Genetic Variation Analysis
Cleaned RNA sequencing reads from high and low-altitude populations (12 vs. 12 individuals) were aligned to Tree Sparrow CDS reference using BWA (Li and Durbin 2010) with default parameters. Only reads that were mapped uniquely to the reference were retained. After high-quality filtering with custom scripts, we called SNPs on all 24 individuals with Samtools. Local realignment over indel positions was performed using GATK (McKenna et al. 2010). SNPs with missing data in two or more individuals in elevation populations were excluded. Fisher’s exact test was used to determine the statistical significance of the allele frequency differences between highland and lowland populations at exonic SNP positions.
Structural Modeling
Structural modeling was performed on the SWISS-MODEL server (Biasini et al. 2014), using the crystal structure of SLC2A3 protein (PDB 4ZWC) as template. Structure visualization and analysis of residue interactions at the mutation site K543 were performed using the PyMol.
Immunostaining
Dissected muscle tissues were fixed in 4% paraformaldehyde overnight at 4 °C, embedded in Optimal cutting temperature compound (OCT), and then cut into sections of 8-μm thickness. The sections were washed twice in 0.1 M PBS. Serial sections were incubated in primary antibody diluted 1:300 at 4 °C overnight (rabbit anti-GLUT12 polyclonal antibody; bs-2540R; Bioss, Beijing, China). Following overnight incubation, the samples were washed in PBS. Presence of the primary antibody was visualized by incubating sections for 30 min in the dark at room temperature with a fluorescent secondary antibody at a 1:300 dilution (goat anti-rabbit antibodies; Bioss). 4',6-diamidino-2-phenylindole (DAPI) was used for the nuclear staining.
Western Blot
Western blots were performed on samples of pectoral major muscle, as previously described (Frolova et al. 2009). In total, 50 mg of muscle were grinded with liquid nitrogen and then the powder was lysed in 1 ml lysis buffer, 50 mM Tris HCl (pH 7.5), 150 mM NaCl, 5 mM “ethylenediaminetetraacetic acid, 0.5% Nonidet P-40, and completeMini protease inhibitor cocktail (BC3640; Solarbio). The samples were placed on ice for 4 min and then centrifugated at 12,000 rpm for 15 min at 4 °C. In total, 350 μl of the supernatant were transferred to a 2-ml auto-sampler vial and were diluted by 3-fold of loading buffer. Protein concentration was quantified using the BCA Protein Assay Kit (PC0020; Solarbio). Supernatant was then separated using Sodium dodecylsulphate-polyacrylamide gel electrophoresis (SDS-PAGE) for 1 h at 140 V and transferred to PVDF for 2 h at 90 V. Nonspecific antibody bind in was blocked in 5% nonfat dry milk powder in 1 Tris-buffered saline for 1 h. Blots were incubated in 1% nonfat dry milk powder in Tris-buffered saline with 0.05% Tween 20 overnight at 4 °C with the following antibodies: rabbit anti-GLUT12 (bs-2540R; 1:1,000; Bioss), and mouse anti-β-actin (bsm-33036M; 1:1,000; Bioss). Blots were incubated with goat anti-rabbit (bs-0295G; 1:3,000; Bioss) for 1 h at room temperature. Blots were detected using protein detection reagents (AY0371; Solarbio).
In Vivo Experiments and RNA Interference
The experimental birds were transported to the laboratory at the Institute of Zoology, Chinese Academy of Sciences after capture. Birds were weighed and housed individually in screen cages to avoid bird injury (35 cm × 35 cm × 35 cm) at a constant temperature (25 ± 1 °C) and 12 h:12 h L:D photoperiod. After acclimation to laboratory for about 14 days, 11 individuals with similar weights were randomly separated into five groups for insulin sensitivity and hypoxia experiments. For insulin sensitivity trials, one group of birds (n = 5) was administered in flight muscle with 1 nmol of a mixed double-stranded SLC2A12 siRNA (GACTGTAACTGAGCTCATT; CCAGGAAACTTCACTGTCA; CATACGAGATACTCATTGT; Ribobio, Guangzhou, China) specifically targeting Eurasian tree sparrow SLC2A12 mRNA. Control birds (n = 6) were administered with 1 nmol of a double-stranded scramble control siRNA (Ribobio) that has the same GC ratio as the SLC2A12 siRNA. Birds were injected with target siRNA every 2 days during cage cleaning for a brief period (<1 h) and were provided with water and mixed seed ad libitum. Control group was injected with equal normal saline. siRNA was vertically injected to 2 mm deep of the pectoralis major using microsyringe. The site of injection was closed to bird carina about 0.5 cm. Experimental period was 8 days.
Behavioral Measures
In the laboratory, general activity patterns were measured using video recordings, and each treatment was recorded for at least 4 h in daylight. The behaviors were divided into two parts, namely, eating and sedentary. Then, we calculated the time when birds had been actively eating and sedentary.
Respirometry
We measured the rates of oxygen consumption VO2 by open flow-through respirometry (Foxbox). For resting metabolic rate, birds were maintained in a 1.0-l transparent metabolic chamber with an air flow rate of 500 ml/min and O2 and CO2 concentration within the chamber were recorded for over 15 min. The lowest 1-min average of oxygen consumption was taken as the resting metabolic rate. For exercise oxygen consumption, we used a metabolic flight wheel as previously described (Chappell et al. 1999). An incurrent flow rate of 5 l/min and a subsampling rate of 100 ml/min were used. We exercised the birds using an initial speed of wheel rotation (∼0.3 m/s) and increased the rotation speed by ∼0.1 m/s every 2 min until birds could no longer keep position and VO2 no longer increased with increasing speed. The tests lasted for 20 min.
Glucose and Insulin Tolerance Tests
For the GTT, an oral gavage of 50% d-glucose (3 mg/g body wt; Sigma) was performed after 16 h fasting. Blood glucose was measured at 0, 15, 30, 45, 60, and 120 min via the medial metatarsal vein. For the ITT, human insulin (1.5 mU/g body wt; Novonordisk) was intraperitoneally injected after the birds underwent 3 h of fasting. Blood glucose was measured at 0, 15, 30, 45, and 60 min via the medial metatarsal vein. Glucose and insulin were measured using an Accu-Check blood glucose meter (Roche Diagnostics) and an insulin ELISA kit (Cloud-Clone), respectively.
Statistical Analyses
Graphpad Prism software was used to perform statistical analyses and graphs were performed using this software and R packages. Statistical test used, number of independent experiments, and P values were listed in individual figure legends.
Animal Experiment Statement
All procedures performed on animal were approved by the Animal Care Committee of Institute of Zoology, Chinese Academy of Sciences. The approval number was IOZ20160049.
Data Availability
RNA sequencing data have been deposited at the National Center for Biotechnology Information short read archive under bioproject PRJNA523449. All other data are available from the corresponding authors upon reasonable request.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
Many thanks to Zuohua Yin, Yongbin Chang, and Dan Zhu for fieldwork and laboratory space. Thanks to Matthew Miller for constructed edition and comments and to Yong Zhang for many technique comments. Thanks also to the editor and two anonymous reviewers for constructive comments and suggestions for improving the manuscript. This work was funded by National Natural Science Foundation of China (No. 31630069 to F.L.), the Strategic Priority Research Program of the Chinese Academy of Sciences (Grant Nos. XDA19050202 and XDB13020300 to F.L.), the Second Tibetan Plateau Scientific Expedition and Research (STEP) program (Grant No. 2019QZKK0501 to F.L.), and the Third Xinjiang Pre-Scientific Expedition and Research Program (2019FY100204 to F.L.).
Author Contributions
F.L. and Y.X. designed and performed the experiments and wrote the manuscript.
References
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ.. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25(17):3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Huber W.. 2010. Differential expression analysis for sequence count data. Nat Prec. [DOI] [PMC free article] [PubMed]
- Andrews S. 2016. FastQC: a quality control tool for high throughput sequence data. Available from: http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
- Avaria-Llautureo J, Hernández CE, Rodríguez-Serrano E, Venditti C.. 2019. The decoupled nature of basal metabolic rate and body temperature in endotherm evolution. Nature 572(7771):651–654. [DOI] [PubMed] [Google Scholar]
- Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Kiefer F, Cassarino TG, Bertoni M, Bordoli L, et al. 2014. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 42(W1):W252–W258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun EJ, Sweazea KL.. 2008. Glucose regulation in birds. Comp Biochem Physiol B Biochem Mol Biol. 151(1):1–9. [DOI] [PubMed] [Google Scholar]
- Chappell MA, Bech C, Buttemer WA.. 1999. The relationship of central and peripheral organ masses to aerobic performance variation in house sparrows. J Exp Biol. 202:2269–2279. [DOI] [PubMed] [Google Scholar]
- Clarke A, Rothery P, Isaac NJ.. 2010. Scaling of basal metabolic rate with body mass and temperature in mammals. J Anim Ecol. 79(3):610–619. [DOI] [PubMed] [Google Scholar]
- Darryl C, Wang D, Pascual JM, Ho YY.. 2002. Glucose transporter protein syndromes. Int Rev Neurobiol. 51:259–288. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR.. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29(1):15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont J, Tesseraud S, Derouet M, Collin A, Rideau N, Crochet S, Godet E, Cailleau-Audouin E, Métayer-Coustard S, Duclos MJ, et al. 2008. Insulin immuno-neutralization in chicken: effects on insulin signaling and gene expression in liver and muscle. J Endocrinol. 197(3):531–542. [DOI] [PubMed] [Google Scholar]
- Frolova A, Flessner L, Chi M, Kim ST, Foyouzi-Yousefi N, Moley KH.. 2009. Facilitative glucose transporter type 1 is differentially regulated by progesterone and estrogen in murine and human endometrial stromal cells. Endocrinology 150(3):1512–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z, Steinmetz LM, Gu X, Scharfe C, Davis RW, Li W-H.. 2003. Role of duplicate genes in genetic robustness against null mutations. Nature 421(6918):63–66. [DOI] [PubMed] [Google Scholar]
- Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O.. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59(3):307–321. [DOI] [PubMed] [Google Scholar]
- Jia B, Yuan DP, Lan WJ, Xuan YH, Jeon CO.. 2019. New insight into the classification and evolution of glucose transporters in the Metazoa. FASEB J. 33(6):7519–7528. [DOI] [PubMed] [Google Scholar]
- Jiménez-Amilburu V, Jong-Raadsen S, Bakkers J, Spaink HP, Marín-Juez R.. 2015. GLUT12 deficiency during early development results in heart failure and a diabetic phenotype in zebrafish. J Endocrinol. 224(1):1–15. [DOI] [PubMed] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS.. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz EB, Stenbit AE, Hatton K, DePinhot R, Charron MJ.. 1995. Cardiac and adipose tissue abnormalities but not diabetes in mice deficient in GLUT4. Nature 377(6545):151–155. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K.. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23(21):2947–2948. [DOI] [PubMed] [Google Scholar]
- Letunic I, Bork P.. 2016. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44(W1):W242–W245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN.. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf. 12(1):323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R.. 2010. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 26(5):589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H, Li W-H.. 2009. Functional compensation by duplicated genes in mouse. Trends Genet. 25(10):441–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao B-Y, Zhang J.. 2007. Mouse duplicate genes are as essential as singletons. Trends Genet. 23(8):378–381. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovegrove BG. 2017. A phenology of the evolution of endothermy in birds and mammals. Biol Rev. 92(2):1213–1240. [DOI] [PubMed] [Google Scholar]
- Lovell PV, Wirthlin M, Wilhelm L, Minx P, Lazar NH, Carbone L, Warren WC, Mello CV.. 2014. Conserved syntenic clusters of protein coding genes are missing in birds. Genome Biol. 15(12):565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maza E. 2016. In papyro comparison of TMM (edgeR), RLE (DESeq2), and MRN normalization methods for a simple two-conditions-without-replicates RNA-seq experimental design. Front Genet. 7:164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20(9):1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNab BK. 2009. Ecological factors affect the level and scaling of avian BMR. Comp Biochem Physiol A Mol Integr Physiol. 152(1):22–45. [DOI] [PubMed] [Google Scholar]
- Mueckler M, Thorens B.. 2013. The SLC2 (GLUT) family of membrane transporters. Mol Aspects Med. 34(2–3):121–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman SA, Mezentseva NV, Badyaev AV.. 2013. Gene loss, thermogenesis, and the origin of birds. Ann NY Acad Sci. 1289(1):36–47. [DOI] [PubMed] [Google Scholar]
- Organ CL, Shedlock AM, Meade A, Pagel M, Edwards SV.. 2007. Origin of avian genome size and structure in non-avian dinosaurs. Nature 446(7132):180–184. [DOI] [PubMed] [Google Scholar]
- Petersen MC, Shulman GI.. 2018. Mechanisms of insulin action and insulin resistance. Physiol Rev. 98(4):2133–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polakof S, Mommsen TP, Soengas JL.. 2011. Glucosensing and glucose homeostasis: from fish to mammals. Comp Biochem Physiol B Biochem Mol Biol. 160(4):123–149. [DOI] [PubMed] [Google Scholar]
- Purcell SH, Aerni-Flessner LB, Willcockson AR, Diggs-Andrews KA, Fisher SJ, Moley KH.. 2011. Improved insulin sensitivity by GLUT12 overexpression in mice. Diabetes 60(5):1478–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam NH, Butts T, Ferrier DEK, Furlong RF, Hellsten U, Kawashima T, Robinson- Rechavi M, Shoguchi E, Terry A, Yu J-K, et al. 2008. The amphioxus genome and the evolution of the chordate karyotype. Nature 453(7198):1064–1071. [DOI] [PubMed] [Google Scholar]
- Qu Y, Chen C, Xiong Y, She H, Zhang YE, Cheng Y, DuBay S, Li D, Ericson P, Hao Y, et al. 2020. Rapid phenotypic evolution with shallow genomic differentiation during early stages of high elevation adaptation in Eurasian Tree Sparrows. Natl Sci Rev. 7(1):113–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimand J, Kull M, Peterson H, Hansen J, Vilo J.. 2007. g: profiler—a web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res. 35(Suppl 2):W193–W200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers S, Chandler JD, Clarke AL, Petrou S, Best JD.. 2003. Glucose transporter GLUT12-functional characterization in Xenopus laevis oocytes. Biochem Biophys Res Commun. 308(3):422–426. [DOI] [PubMed] [Google Scholar]
- Rose AJ, Richter EA.. 2005. Skeletal muscle glucose uptake during exercise: how is it regulated? Physiology 20(4):260–270. [DOI] [PubMed] [Google Scholar]
- Rowland LA, Bal NC, Periasamy M.. 2015. The role of skeletal-muscle-based thermogenic mechanisms in vertebrate endothermy. Biol Rev. 90(4):1279–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schippers M-P, Ramirez O, Arana M, Pinedo-Bernal P, McClelland GB.. 2012. Increase in carbohydrate utilization in high-altitude Andean mice. Curr Biol. 22(24):2350–2354. [DOI] [PubMed] [Google Scholar]
- Sweazea KL, Braun EJ.. 2005. Glucose transport by English sparrow (Passer domesticus) skeletal muscle: have we been chirping up the wrong tree? J Exp Zool. 303A(2):143–153. [DOI] [PubMed] [Google Scholar]
- Toyoda Y, Takada T, Miyata H, Matsuo H, Kassai H, Nakao K, Nakatochi M, Kawamura Y, Shimizu S, Shinomiya N, et al. 2020. Identification of GLUT12/SLC2A12 as a urate transporter that regulates the blood urate level in hyperuricemia model mice. Proc Natl Acad Sci U S A. 117(31):18175–18177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waller AP, George M, Kalyanasundaram A, Kang C, Periasamy M, Hu K, Lacombe VA.. 2013. GLUT12 functions as a basal and insulin-independent glucose transporter in the heart. Biochim Biophys Acta Mol Basis Dis. 1832(1):121–127. [DOI] [PubMed] [Google Scholar]
- Wang D, Pascual JM, Yang H, Engelstad K, Mao X, Cheng J, Yoo J, Noebels JL, De Vivo DC.. 2006. A mouse model for Glut-1 haploinsufficiency. Hum Mol Genet. 15(7):1169–1179. [DOI] [PubMed] [Google Scholar]
- Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT, et al. 2010. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 38(Suppl 2):W214–W220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ.. 2009. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25(9):1189–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson-O’Brien AL, Patron N, Rogers S.. 2010. Evolutionary ancestry and novel functions of the mammalian glucose transporter (GLUT) family. BMC Evol Biol. 10(1):152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24(8):1586–1591. [DOI] [PubMed] [Google Scholar]
- Zerbino DR, Achuthan P, Akanni W, Amode MR, Barrell D, Bhai J, Billis K, Cummins C, Gall A, Girón CG, et al. 2018. Ensembl 2018. Nucleic Acids Res. 46(D1):D754–D761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zisman A, Peroni OD, Abel ED, Michael MD, Mauvais-Jarvis F, Lowell BB, Wojtaszewski JFP, Hirshman MF, Virkamaki A, Goodyear LJ, et al. 2000. Targeted disruption of the glucose transporter 4 selectively in muscle causes insulin resistance and glucose intolerance. Nat Med. 6(8):924–928. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA sequencing data have been deposited at the National Center for Biotechnology Information short read archive under bioproject PRJNA523449. All other data are available from the corresponding authors upon reasonable request.