Abstract
Coronavirus disease 2019 (COVID-19) is the latest respiratory pandemic caused by severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2). Although infection initiates in the proximal airways, severe and sometimes fatal symptoms of the disease are caused by infection of the alveolar type 2 (AT2) cells of the distal lung and associated inflammation. In this study, we develop primary human lung epithelial infection models to understand initial responses of proximal and distal lung epithelium to SARS-CoV-2 infection. Differentiated air-liquid interface (ALI) cultures of proximal airway epithelium and alveosphere cultures of distal lung AT2 cells are readily infected by SARS-CoV-2, leading to an epithelial cell-autonomous proinflammatory response with increased expression of interferon signaling genes. Studies to validate the efficacy of selected candidate COVID-19 drugs confirm that remdesivir strongly suppresses viral infection/replication. We provide a relevant platform for study of COVID-19 pathobiology and for rapid drug screening against SARS-CoV-2 and emergent respiratory pathogens.
Keywords: COVID-19, SARS-CoV-2, alveolar type 2 cells, alveoli, adult lung epithelium, primary in vitro model, remdesivir, interferon, drug discovery
Graphical abstract
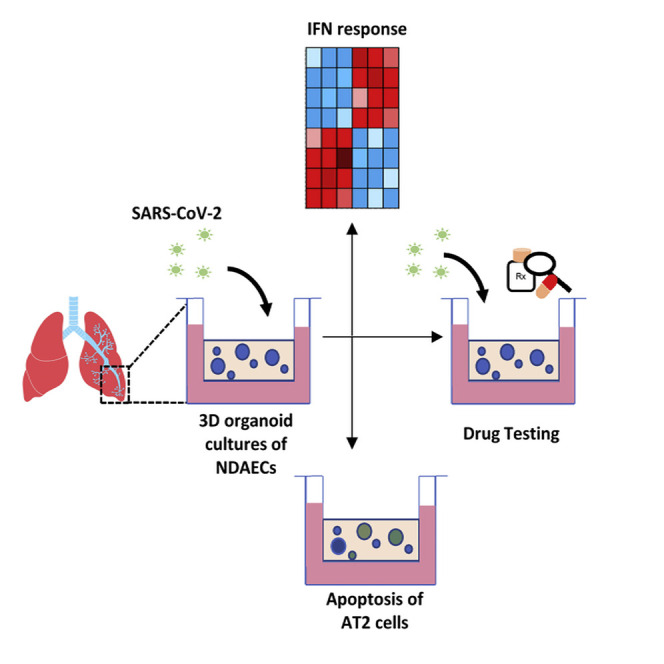
In vitro models of human lung epithelium, including diverse cell types of the proximo-distal axis, are critical for modeling infection. Mulay et al. show that alveospheres, with epithelial type 2- and type 1-like cells, are infected by SARS-CoV-2, initiating an interferon response, and serve as a platform for screening antiviral drugs.
Introduction
Coronavirus disease 2019 (COVID-19), caused by infection with the severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) virus, is the most recent of a series of severe viral infections that typically initiate in the upper respiratory tract but have the potential to cause life-threatening pneumonia as a result of infection and inflammation of the lower respiratory tract (Zhou et al., 2020; Tay et al., 2020). Unlike human coronaviruses that lead to a self-limiting upper respiratory tract infection, SARS-CoV-2 and related viruses SARS-CoV and middle eastern respiratory syndrome coronavirus (MERS-CoV) are thought to originate from bats and cause severe symptoms in their human hosts because of lack of host-pathogen adaptation (Cui et al., 2019). Similar zoonotic transmission of other respiratory pathogens, such as H1N1 influenza A virus (Vijaykrishna et al., 2010), is associated with recurrent pandemics with severe pulmonary complications seen among infected individuals caused by infection of the lower respiratory tract. Even though acute viral pneumonia appears to be a common pathological outcome of infection with these severe respiratory viruses, mechanisms leading to these adverse outcomes are poorly understood. Model systems that can closely mimic the physiology and host response in the different compartments of the lung are critical to understanding the pathogenesis of SARS-CoV-2 and for rapid drug testing against the virus. Rapidly evolving literature now suggests that SARS-CoV-2 targets various cell types of the proximal airways and the alveolar type 2 (AT2) cells of the gas exchange region of the distal lung (Hoffmann et al., 2020; Hou et al., 2020; Ziegler et al., 2020). Infection of AT2 cells drives acute respiratory distress syndrome (ARDS) in severe cases of COVID-19. We sought to develop culture models of proximal and distal human lung epithelium as a platform to study early response to SARS-CoV-2 infection and for rapid drug discovery in the setting of current and future severe respiratory viral infections.
Results
SARS-CoV-2 infects and replicates within human proximal airway epithelial cells
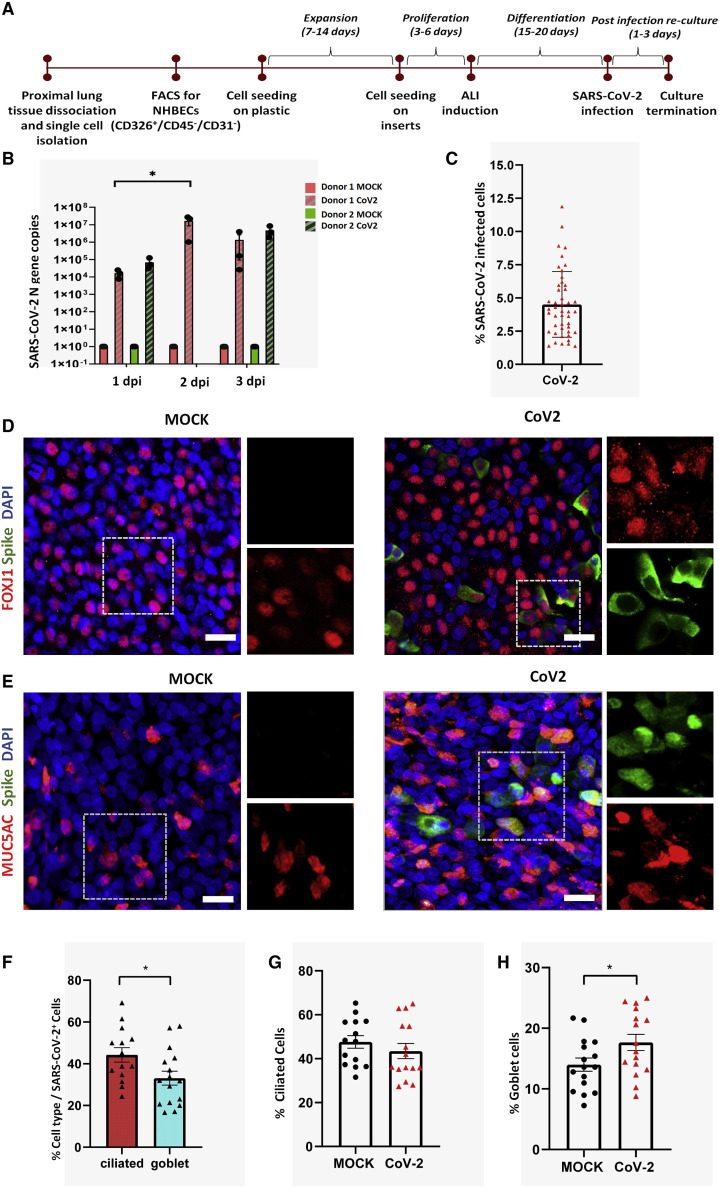
Because the upper respiratory tract represents the most likely initial site of respiratory virus infection, we initially utilized the well-established air-liquid interface (ALI) culture system to study the effect of SARS-CoV-2 infection on proximal airway epithelial cells. Normal human tracheobronchial epithelial cells (NHBECs) isolated from the trachea and upper bronchi were cultured at ALI for 16–20 days to yield a well-differentiated, pseudostratified mucociliary epithelium with approximately 1 × 105 cells per insert, of which 47% (±10.69%) were ciliated cells and 14% (±5.3%) were goblet cells. ALI cultures of proximal airway epithelial cells were infected apically with 1 × 104 median tissue culture infectious dose (TCID50) per well of SARS-CoV-2 and harvested for analysis at 1–3 days postinfection (dpi) (Figure 1 A). SARS-CoV-2 readily infected the well-differentiated proximal airway cells as indicated by SARS-CoV-2 Nucleoprotein gene (N gene) copy numbers in infected samples. N gene copy numbers for three wells of ALI cultures from two independent donor samples show that viral genome replication increased at 2 dpi compared with 1 dpi and declined at 3 dpi; however, the later decline was not statistically significant. Viral N gene was undetectable in corresponding mock cultures at all three time points (Figure 1B).
Figure 1.
SARS-CoV-2 infects normal human proximal airway cells
(A) Workflow for establishment of human proximal airway ALI cultures and their infection with SARS-CoV-2.
(B) ALI cultures of proximal airway epithelial cells are susceptible to SARS-CoV-2 infection, indicated by viral N gene copy numbers, which peaked at 2 days postinfection (dpi); n = 3–6 cultures from 2 independent donors. Red and green colors indicate cultures from separate donors. Data were analyzed using one-way ANOVA with Tukey’s post hoc correction and represented N gene copy numbers for individual cultures ± SEM. ∗p < 0.05.
(C) Percentage of cells from proximal airway ALI cultures infected with SARS-CoV-2 at 2 dpi. n = 14–15 fields from two biological replicates.
(D and E) FOXJ1-positive ciliated cells (red) (D) and MUC5AC-positive goblet cells (red) (E) infected by SARS-CoV-2 (green). Scale bar, 20 μm. Image magnification = 2.3× with a 20× objective.
(F) Percentage of infected cells that are either ciliated cells or goblet cells.
(G and H) Change in the percentage of (G) ciliated and (H) goblet cells after SARS-CoV-2 infection. (F–H) n = 7–8 fields from two biological replicates. Data are presented as mean ± SEM (significance is determined by two-tailed t test). ∗p < 0.05.
Further analysis of proximal airway ALI cultures at 2 dpi revealed that, on average, 4.5% of total cells were infected by SARS-CoV-2 (Figure 1C). The infection was heterogenous; both ciliated and goblet cells were infected, evidenced by colocalization of SARS-CoV-2 viral capsid “spike” protein with the ciliated cell marker, FOXJ1 (Figure 1D), and the goblet cell marker, MUC5AC (Figure 1E). Ciliated cells were infected at a significantly higher rate than goblet cells. Of the total infected cells, 44.2% were ciliated and 33.1% were goblet cells (Figure 1F). We further assessed whether SARS-CoV-2 infection triggered a change in the proportions of the two cell types. We found no change in the percentage of ciliated cells after SARS-CoV-2 infection (Figure 1G). However, the proportion of MUC5AC-producing goblet cells significantly increased postinfection (Figure 1H).
SARS-CoV-2 infects and replicates within human AT2 cells
Although infection initiates in the proximal airways, a number of recent studies have shown that in severe cases of COVID-19, ARDS results because of viral infection of AT2 cells in the distal lung epithelium (Hou et al., 2020; Ziegler et al., 2020). As such, physiologically relevant alveolar infection models, including both AT1 and AT2 cells, are needed to study the response of alveolar epithelial cells to SARS-CoV-2.
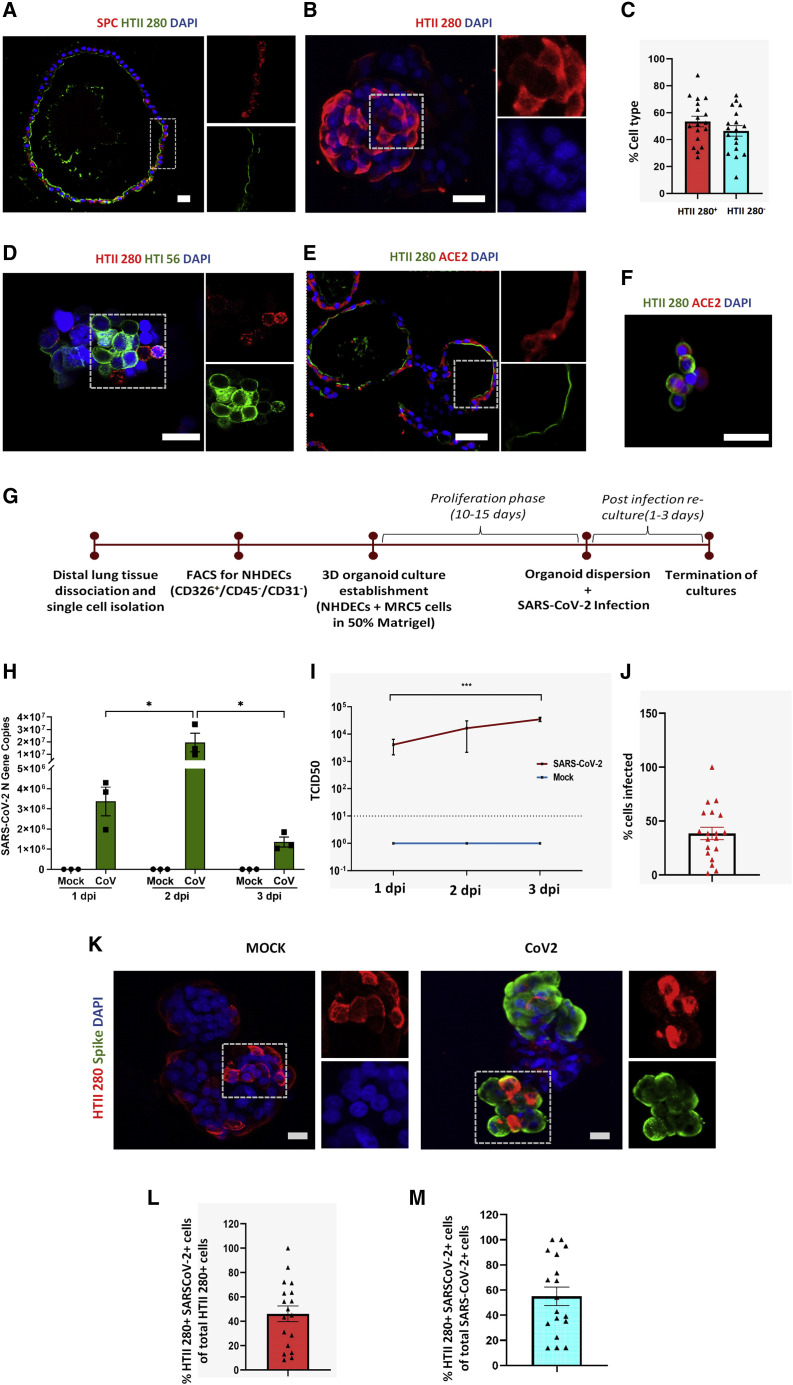
We previously published the development of the first 3D spheroid culture model of the human alveolar epithelium by co-culturing primary epithelial cells isolated from normal human distal lung tissue in the presence of stromal support cells, embedded in Matrigel (Barkauskas et al., 2013). Alveolar spheroid models were further refined to allow culture of AT2 cells in serum-free, chemically defined conditions (Konda et al., 2020). Our method enabled culture of three-dimensional alveospheres composed of self-renewing AT2 cells that express the well-established AT2 cell markers, HTII-280 and surfactant protein C (SPC) (Figures 2A and 2B). In addition, our cultures also showed the presence of some HTII-280-negative cells, which on further analysis were found to express the AT1 cell marker HTI 56 (Figures 2C and 2D), indicating AT2-to-AT1 cell differentiation capacity. We further assessed expression of the SARS-CoV-2 entry receptor, angiotensin-converting enzyme 2 (ACE2), in our alveosphere cultures and found that ACE2 staining localized to HTII-280+ AT2 cells (Figures 2E and 2F). This observation is consistent with previous reports of single-cell transcriptomic and immunofluorescence analysis of ACE2 expression in human lung (Hou et al., 2020; Sungnak et al., 2020; Ziegler et al., 2020; Muus et al., 2020).
Figure 2.
SARS-CoV-2 infects normal human distal AT2 cells
(A) Sections of Matrigel-embedded alveospheres showing colocalization of AT2 markers HTII-280 and SPC.
(B) Whole-mount staining of alveospheres dispersed from Matrigel for AT2 marker HTII-280. Scale bar = 20 μm.
(C) Percentage of HTII-280-positive and HTII-280-negative cells in alveospheres in n = 3–5 fields from three biological replicates (two-tailed t test).
(D) Whole-mount alveospheres stained for the AT2 marker HTII-280 and AT1 cell marker HTI 56.
(E and F) ACE2 staining in sectioned alveospheres (E) and in AT2 cells (F) dissociated from alveospheres. Scale bar = 20 μm.
(G) Workflow for establishment of human distal alveolar cultures and their infection with SARS-CoV-2.
(H) Absolute N gene copy numbers in SARS-CoV-2-infected cultures peaked at 2 dpi; n = 3 independent cultures. Data were analyzed using two-way ANOVA with Sidak’s post hoc correction and represented as N gene copy number for individual cultures ± SEM. ∗p <0.05.
(I) Viral load in supernatant increased from 1 to 3 dpi. ∗∗∗p < 0.001.
(J) Percentage of total cells infected (n = 3).
(K) Infection was assessed at 2 dpi by antibody against SARS-CoV-2 “spike” protein (green) and colocalized with viral AT2 cell marker HTII-280 (red). Scale bar, 20 μm.
(L) Percentage of HTII-280-positive cells infected by SARS-CoV-2; n = 3–5 fields from each of three biological replicates.
(M) Percentage of total infected cells that are HTII-280-positive AT2 cells; n = 3–5 fields from each of three biological replicates.
We went on to utilize our alveosphere system to model SARS-CoV-2 infection of AT2 cells in vitro. In brief, isolated HTII-280+ AT2 cells were cultured in Matrigel for 10–15 days to form 3D alveospheres, which were then enzymatically released from Matrigel, followed by gently “opening” alveospheres to allow viral access to the apical membrane by pipetting three times with a P1000 tip (hereafter referred to as alveolar cultures). We then exposed alveolar cultures to SARS-CoV-2 (1 × 104 TCID50 per well) or PBS (as MOCK controls) and harvested cells for analysis 1–3 dpi (Figure 2G). Study of viral infection kinetics from 1 to 3 dpi demonstrated that absolute viral N gene copy numbers in cell lysates, which are a measure of intracellular levels of SARS-CoV-2, peaked at 2 dpi. The average N gene copy number increased from 3.3 × 106 copies at 1 dpi to 1.9 × 107 copies at 2 dpi and declined to 1.3 × 106 copies at 3 dpi. Viral N gene copy numbers were below the detection limits in corresponding mock cultures at all three time points (Figure 2H). In parallel, we also assessed the viral load present in the supernatant medium. This measure represents the amount of free virus released from the infected cells in the supernatant. Viral load increased with each day in culture, peaking at 3 dpi. Viral load in the supernatant was 10-fold higher at 3 dpi compared with 1 dpi, and this difference was statistically significant (Figure 2I). Differences in the absolute viral N gene copy numbers in cell lysates versus viral loads in supernatant medium at 3 dpi likely reflect increased cell death and virus release at the 3-dpi recovery time point (refer to Figure 4 for analysis of cell death). We used immunofluorescence confocal microscopy to verify SARS-CoV-2 infection of epithelial cells in cultures. The efficiency of infection was significantly higher in alveolar cultures compared with proximal airway ALI cultures, with SARS-CoV-2 infecting 38.6% and 4.5% of total cells in distal and proximal cultures, respectively (Figures 2J and 1C). Infection of AT2 cells was confirmed by co-localization of the viral spike protein with HTII-280+ in 2 dpi cultures (Figure 2K). Of the total HTII-280+ AT2 cells, 46% were infected by SARS-CoV-2 (Figure 2L). Of the total infected cells, 55.09% were HTII-280+ AT2 cells (Figure 2M).
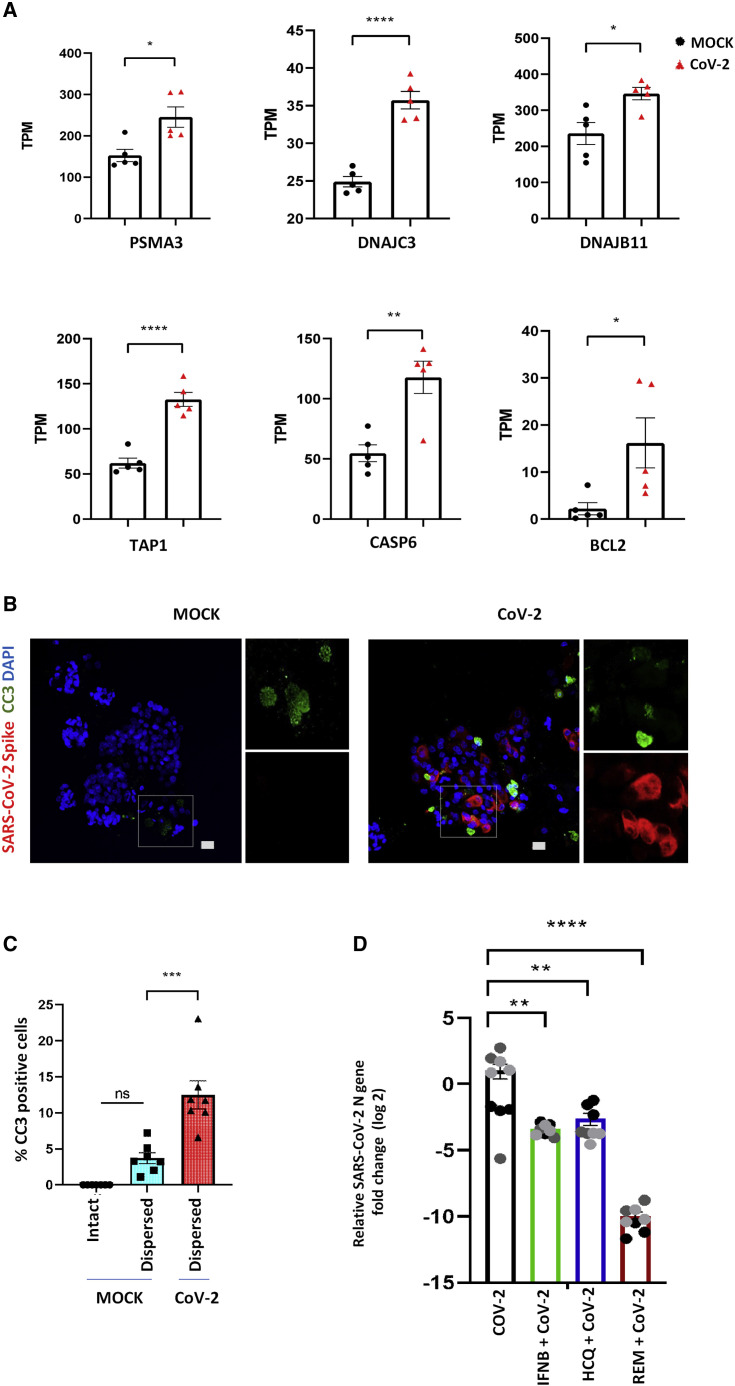
Figure 4.
SARS-CoV-2 infection triggers apoptosis of alveolar cells
(A) Quantification of normalized TPM counts for apoptosis-related genes between MOCK- and SARS-CoV-2 infected alveolar cultures (n = 5; significance is determined by two-tailed t test). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
(B) Infected AT2 cells (red) also demonstrated signs of cellular apoptosis at 3 dpi, indicated by positive staining for cleaved caspase-3 (green). Scale bar: 20 μm.
(C) Comparison of percentage of cells staining positive for CC3 in intact Matrigel-embedded MOCK alveospheres, alveolar cultures dispersed from Matrigel, and infected cultures dispersed from Matrigel. n = 7 (one-way ANOVA with Tukey’s post hoc test). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
(D) Pre-treatment of alveolar cultures with IFNB1, hydroxychloroquine, and remdesivir significantly reduced viral replication. The effect of remdesivir on viral replication was more pronounced than that of IFNB1 or hydroxychloroquine. Data are represented as log2 fold change for individual cultures ± SEM, normalized to mean infection and analyzed using one-way ANOVA with Tukey’s post hoc test. ∗∗p = 0.0012 for IFNB1, ∗∗p = 0.0044 for HCQ, and ∗∗∗∗p < 0.0001 for remdesivir. The three different colors indicate cultures from different biological replicates.
SARS-CoV-2 infection of AT2 cells induces a robust pro-inflammatory response
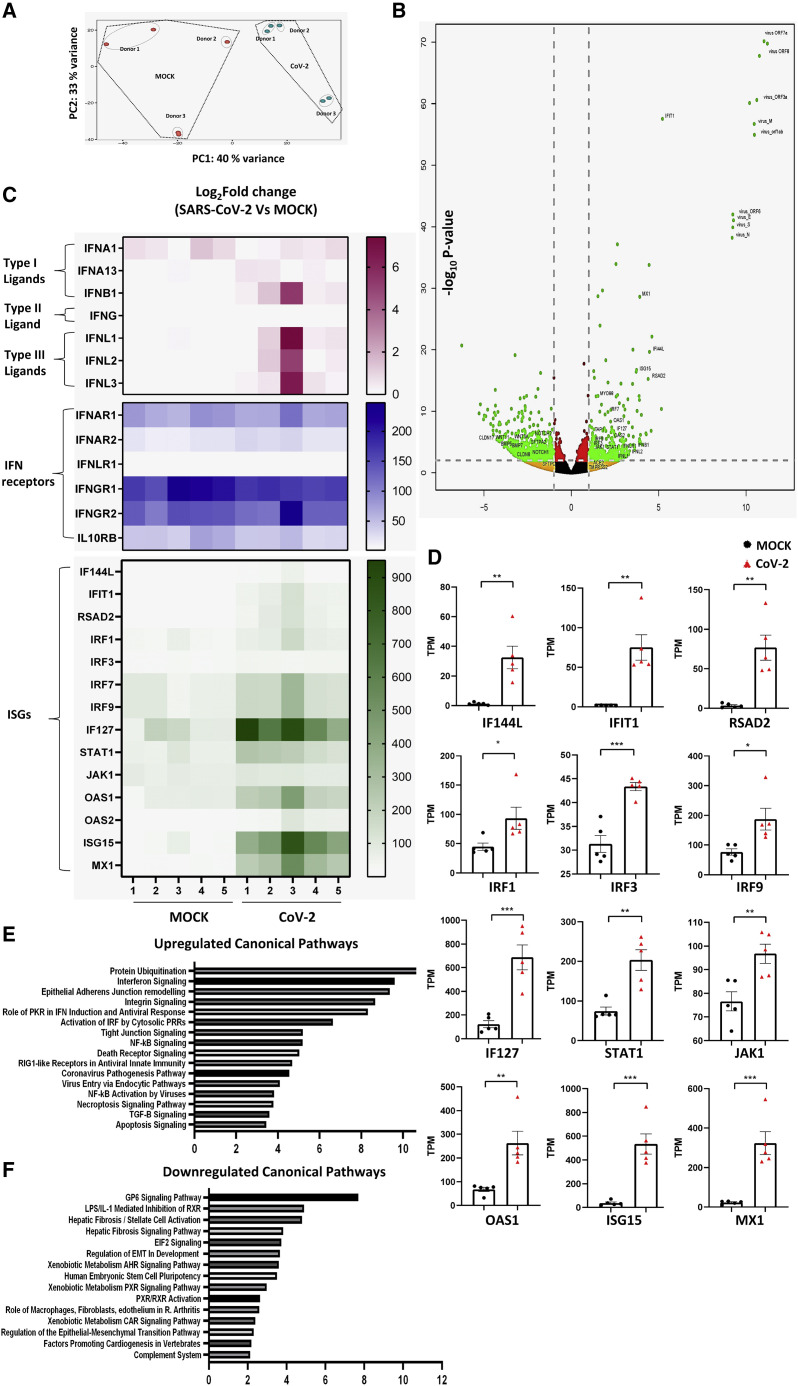
Having established a model that enables robust infection of AT2 cells by SARS-CoV-2, we further evaluated the utility of this system to study host pathogen responses. Alveolar cultures were infected with SARS-CoV-2, and cells were harvested 2 dpi for transcriptomic analysis via global RNA sequencing (RNA-seq; five samples overall, of which three donor cell preparations were represented for both MOCK- and SARS-CoV-2-infected cultures). Significant changes in transcript profiles were observed between SARS-CoV-2-infected and mock alveolar cultures (Figure 3 A). Heatmap (Figure S1A) and volcano plots (Figure 3B) of differentially expressed genes revealed high levels of SARS-CoV-2 viral RNA, such as coding sequences for Virus_N, virus_ORF1ab, virus_ORF3a, and virus_ORF7a, further confirming SARS-CoV-2 genome replication and/or gene transcription in alveolar cultures. Infected cultures showed induction of a number of viral transcripts (Figures S1A and S1B) and proinflammatory transcripts related to viral infections, including type 1 and type 3 interferon-related genes and their downstream targets. The type 1 interferon ligands IFNA13 and IFNB1 and type 3 interferon ligands IFNL1, IFNL2, and IFNL3 were upregulated in the infected cultures (Figure 3C), although their cumulative changes were not statistically significant (Figures 3C and S2A). Our alveolar cultures also expressed interferon receptor transcripts irrespective of viral infection status, with IFNAR2 and IFNLR1 being significantly upregulated in infected cultures compared with mock-infected controls (Figures 3C and S2B). Donor variability was seen in magnitude of infection between cultures, wherein a direct correlation existed with induction of IFN-related ligand and receptor gene expression. Additionally, multiple downstream interferon signaling genes (ISGs), such as IF144L, IFIT1, RSAD2 IRF1, IRF3, IRF7, IRF9 IF127, STAT1, JAK1, OAS1, OAS2 ISG15, and MX1, were significantly upregulated in infected cultures (Figures 3C and 3D). These observed increases in gene transcript levels were further validated by qRT-PCR (Figure S2C).
Figure 3.
Primary human alveolar cultures as a model to study SARS-CoV-2-induced host response
(A) Principal-component analysis of infected alveolar cultures and MOCK cultures showing variance in the global transcriptome of the two groups (n = 5 from three biological replicates for both MOCK and SARS-CoV-2-infected cultures).
(B) Volcano plot of gene expression changes in SARS-CoV-2-infected versus mock cultures defined by p value and >2-fold change.
(C) Heatmap of normalized TPM counts for various interferon ligands, receptors, and ISGs.
(D) Quantification of normalized TPM counts of various downstream ISGs between MOCK and SARS-CoV-2-infected cultures (n = 5; significance determined by two-tailed t test). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
(E and F) The most highly upregulated (E) and downregulated (F) canonical pathways in SARS-CoV-2-infected alveolar cultures.
Analysis of differentially regulated canonical pathways revealed that other proinflammatory and viral-sensing pathways, such as nuclear factor κB (NF-κB) activation, transforming growth factor β (TGF-β) signaling and retinoic acid inducible gene 1 (RIG1)-like receptor signaling, were also upregulated in virus-infected cells (Figure 3E). In contrast, various signaling pathways related to regulation of stem cell activation and fate, such as Notch, Wnt, and BMP signaling, were downregulated (Figures 3B and 3E). However, we did not observe significant changes in expression of the AT2 cell markers, including SFTPC, SFTPA1, SFTPA2, and SFTPD (Figures 3B). Furthermore, we did not observe a change in transcript levels for viral entry genes such as ACE2 and TMPRSS2 (Figure 3B). Taken together, these data indicate that SARS-CoV-2 infection induces significant upregulation of innate immune response genes in alveolar cells without the participation of recruited immune cells.
SARS-CoV-2 infection of AT2 triggers cellular apoptosis
Another key pathway that was activated in SARS-CoV2-infected alveolar cultures included the protein ubiquitination pathway. In particular, we saw a significant increase in transcripts for genes whose products act either as chaperons/co-chaperons or sensitize cells to apoptosis. For example, we saw a significant upregulation of PSMA3, DNAJC3, DNAB11, and TAP1 (Figure 4 A), all of which are typically upregulated by cellular stress and interact with key components of apoptotic pathways. Furthermore, we also observed upregulation of apoptosis-related genes CASP6 and BCL2 in SARS-CoV-2 infected cultures (Figure 4A). In order to confirm increased cellular apoptosis in SARS-CoV2-infected cultures, we performed immunofluorescent staining of MOCK and infected cultures for the apoptotic marker, cleaved caspase 3 (CC3). By 3 dpi, infected cultures exhibited significantly enhanced staining for CC3 compared with mock cultures. Interestingly, only a proportion of the apoptotic cells were infected, thus suggesting that SARS-CoV-2 infection led to direct and indirect cytopathic effects on cultured alveolar epithelial cells (Figures 4B and 4C). SARS-CoV-2-induced apoptosis of neighboring uninfected epithelial cells suggests the potential for non-cell-autonomous effects of viral infection on alveolar epithelial integrity. Together, our data suggest that SARS-CoV-2 infection triggers both cell-autonomous and non-cell-autonomous apoptosis that may contribute to alveolar injury. We conclude that cultures of primary alveolar epithelial cells serve as a robust model for studying the effect of SARS-CoV-2 infection on adult distal lung alveolar epithelium.
Alveolar cultures as a tool to screen therapeutic targets against SARS-CoV-2 infection
Finally, we utilized the distal alveolar cultures to study the effect of a selected panel of drugs against SARS-CoV-2 infection and replication, including the known anti-viral cytokine, IFNB1, and investigational drugs for COVID-19 treatment, remdesivir (Wang et al., 2020; Beigel et al., 2020) and hydroxychloroquine (Pastick et al., 2020; Liu et al., 2020). Treatment of alveolar cultures with IFNB1 leads to a statistically significant, 3.2-log reduction in viral N gene RNA compared with control-infected alveolar cultures (Figure 4D). Hydroxychloroquine led to an overall 2.4-log reduction in viral N gene expression compared with average infection in untreated alveolar cultures (Figure 4D). However, variable effects of hydroxychloroquine were observed on viral replication/gene expression that were donor epithelium dependent (Figure S2). Remdesivir showed the strongest effect on viral replication in alveolar cultures, resulting in a 9-log decrease in viral N gene expression compared with the average infection in untreated cultures (Figure 4D). This effect was consistent irrespective of donor origin of epithelial cells, confirming it as a direct-acting antiviral (DAA) agent targeting viral-specific RNA polymerase (Figure S3). Taken together, our data show that our model of primary human alveolar cultures represents a highly relevant preclinical tool to assess SARS-CoV-2 infection and replication, and serves as a sensitive platform for drug screening and validation.
Discussion
Our study provides an in vitro model to investigate SARS-CoV-2 infection along the proximal-distal axis of the human lung epithelium. Most of the current knowledge of SARS-CoV-2 infection of the lung comes from experiments in non-physiological cell lines that express ACE2, the receptor for SARS-CoV-2 entry and attachment, such as Vero E6 cells (Chu et al., 2020) or lung cell lines with or without exogenous expression of ACE2 (Hoffmann et al., 2020; Cagno, 2020). However, these models do not replicate the complex physiology of the different polarized lung epithelial cell types. Moreover, commonly used animal models for preclinical drug screening, such as mice, are not natural hosts to SARS-CoV-2 infection. Therefore, there is an urgent need for development of model systems that can mimic the physiology and host response of the human lung and provide a relevant platform for screening potential therapeutic agents that target SARS-CoV-2 infection and replication.
More recently, some studies have utilized primary epithelial cultures of nasal and proximal airway epithelium and organ on chip methods to study SARS-CoV-2 infection of the upper airways, the initial site of entry and infection (Blanco-Melo et al., 2020; Si et al., 2020; Hou et al., 2020; Ravindra et al., 2020). We utilized the well-differentiated ALI culture system to study SARS-CoV-2 infection of proximal airway epithelial cells and found that SARS-CoV-2 predominantly infected ciliated cells in addition to a sub-population of goblet cells. This observation is congruent with previous reports of infection of ciliated cells by SARS-CoV (Jia et al., 2005) and recent reports of infection of ciliated cells by SARS-CoV-2, in in vitro cultures of proximal airways (Hou et al., 2020; Lamers et al., 2020), as well as COVID-19 patient biopsies (Hou et al., 2020). A number of studies employing single-cell RNA-seq analysis have described the expression of ACE2 in the respiratory tract (Sungnak et al., 2020; Ziegler et al., 2020; Hou et al., 2020; Qi et al., 2020) and in the nasal passages and large airways, and the highest levels of ACE2 expression have been reported in ciliated cells (Lee et al., 2020; Hou et al., 2020; Ziegler et al., 2020).
Although proximal airways are the initial site of SARS-CoV-2 attachment and infection, it is infection and inflammation of the distal lung that drive the severe and fatal symptoms in COVID-19 among highly susceptible patients. Recent single-cell transcriptome studies have shown that ACE2 is predominantly expressed by AT2 cells (Hou et al., 2020; Qi et al., 2020). AT2 cells are bifunctional cells that serve as facultative progenitors, contributing to epithelial maintenance in addition to fulfilling specialized functions, such as surfactant production (Barkauskas et al., 2013; Rackley and Stripp, 2012). In this study, we describe the development of an in vitro model of primary AT2 cells isolated from the adult human lung. In our model, AT2 cells were readily infected by SARS-CoV-2 and demonstrated a significant upregulation of heat shock proteins and chaperon-inducing cellular apoptosis. Moreover, our model demonstrated cell-autonomous and non-cell-autonomous apoptosis induced by viral infection that may contribute to alveolar injury seen in COVID-19.
Infection of AT2 cells was accompanied by a cell-intrinsic proinflammatory response and upregulation of the interferon signaling pathway, characterized by upregulation of type 1 and type 3 interferon genes, as well as several ISGs. Recent studies have reported that interferon response is diminished or delayed in severe cases of COVID-19, similar to that seen in severe cases of MERS and SARS-CoV (Blanco-Melo et al., 2020; Channappanavar et al., 2016, 2019). However, we observed a moderate interferon response in our model cells after SARS-CoV-2 infection. A recently published report of SARS-CoV-2 infection of pluripotent stem cell-derived AT2 cells also demonstrated an induction of interferon signaling response similar to our model of primary adult AT2 cells (Huang et al., 2020). The presence of an IFN response in cells derived from normal donors may explain the lack of a severe phenotype in a large proportion of COVID-19 patients. It will be of interest to evaluate the host response to SARS-CoV-2 infection in primary AT2 cells derived from donors with pre-existing lung conditions in the future. It is also important to note that our model system does not consist of an immune cell component. We speculate that the epithelial innate immune response may provide activating signals leading to global activation of the host immune response and provide therapeutic targets for mitigation of uncontrolled lung inflammation and adverse patient outcomes. To better understand host immune response to SARS-CoV-2 infection, co-culture systems of infected AT2 cells with immune cells can be developed in the future.
Further, we tested the efficacy of selected potential therapeutic agents against SARS-CoV-2 infection of AT2 cells and found that viral infection and/or replication was strongly suppressed by the candidate drug, remdesivir. Remdesivir is currently the most promising candidate drug and has been granted emergency use authorization for treatment of hospitalized COVID-19 patients by the US Food and Drug Administration (FDA) (Wang et al., 2020; Beigel et al., 2020). Type 1 interferons are known antiviral agents, and IFNB1 has been suggested to reduce SARS-CoV-2 infection in Vero cells (Clementi et al., 2020; Mantlo et al., 2020) and more recently in patients with severe COVID-19 in an ongoing clinical trial (ClinicalTrials.gov: NCT04385095). Consistent with these findings, we also observed significant suppression of viral infection upon treatment of alveolar cultures with IFNB1. Thus, our platform provides a relevant model to effectively screen and validate drugs targets against SARS-CoV-2.
Limitations
Our human alveolar model cultures provide a relevant resource to study the host epithelial responses to SARS-CoV-2 infection and a preclinical tool for rapid screening of drugs against COVID-19 and future emergent respiratory pathogens. Recently, two similar studies have reported the use of primary lung alveolar organoids for the study of SARS-CoV-2 infections (Katsura et al., 2020; Youk et al., 2020). Both studies use long-term feeder-free cultures to study the effect of SARS-CoV-2 infection of AT2 cells and demonstrated an induction of interferon signaling response similar to our model. However, our model utilizes a feeder-based system of AT2-derived organoids cultured in the presence of human lung fibroblasts. Moreover, we have also demonstrated differentiation of AT2 cells into AT1 cells in our model. Although our current study focuses on SARS-CoV-2 infection of AT2 cells, the presence of the different cell types of the distal lung in our model allows for the potential to dissect cell-type-specific responses to SARS-CoV-2 infection.
Currently available models also lack the immune and endothelial components, which are key drivers of adverse outcomes in COVID-19 patients. Perhaps adapting our alveolar model to an organ of the chip system that facilitates co-culture of distal lung epithelial cells with fibroblasts, endothelial cells, and immune cells may help to develop a more cohesive model of the distal lung and help to mitigate this limitation in future studies.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse anti- FOXJ1 | Thermo Fisher Scientific | Cat # 14-9965-82; RRID: AB_1548835 |
| Mouse anti- Mucin 5AC, 45M1 | Thermo Fisher Scientific | Cat # MA5-12178; RRID: AB_10978001 |
| Mouse anti- HTII-280 | Terrace Biotech | Cat # TB-27AHT2-280; RRID: AB_2832931 |
| Mouse anti- HTI-56 | Terrace Biotech | Cat # TB-29AHT1-56; RRID: AB_2847898 |
| Guinea Pig anti- SARS-CoV-2 spike (S) protein | BEI Resources | NR-10361 |
| Rabbit anti- Cleaved caspase-3 | Cell Signaling Technology | Cat # 9661; RRID: AB_2341188 |
| Mouse anti- CD31 | BioLegend | Cat # 303104; RRID: AB_314330 |
| Mouse anti- CD45 | BioLegend | Cat # 304054; RRID: AB_2564154 |
| Mouse anti- CD326 | BioLegend | Cat # 324208; RRID: AB_756082 |
| Bacterial and virus strains | ||
| SARS-CoV-2, isolate USA-WA1/2020 GenBank: MT020880) | BEI Resources | (NR-52281) |
| Biological samples | ||
| Normal Lung Tissue | International Institute for the Advancement of Medicine (IIAM) | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Hydroxychloroquine | Selleck Chemicals | Cat # S4430 |
| Remdesivir | Selleck Chemicals | Cat # S8932 |
| IFNB1 stock | Provided by Dr Jay Kolls | N/A |
| Liberase | Millipore Sigma | Cat # 5401127001 |
| Critical commercial assays | ||
| CD31 MicroBead Kit, human | Miltenyi Biotec | Cat # 130-091-935 |
| CD45 MicroBead Kit, human | Miltenyi Biotec 130- | Cat # 130-045-801 |
| Deposited data | ||
| Raw and analyzed data | This paper | GEO:GSE160435 |
| Experimental models: cell lines | ||
| MRC5 human lung fibroblast cells | ATCC | CCL-171 |
| Oligonucleotides | ||
| Primers for SARS-CoV-2 N1 gene (See Table S1) | CDC’s resources for research labs | N/A |
| Primers for IFN Stimulated genes (See Table S1) | This paper | N/A |
| Software and algorithms | ||
| ImageJ | N/A | https://imagej.nih.gov/ij/ |
| GraphPad Prism 8 | GraphPad | RRID:SCR_002798 |
| Other | ||
| Pneumacult Ex media | Stem Cell Technologies | Cat #05008 |
| Pneumacult ALI media | Stem Cell Technologies | Cat # 05001 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Barry Stripp (barry.stripp@cshs.org).
Materials availability
AT2 cell cultures or primary proximal and distal epithelial cells will be made available upon installment of a material transfer agreement (MTA).
Data and code availability
Original gene expression data from this study have been deposited to Gene Expression Omnibus with accession number GEO: GSE160435.
Experimental model and subject details
Tissue Procurement
Human lung tissue was obtained from ‘normal’ (without prior history of lung disease) deceased organ donors in compliance with consent procedures developed by International Institute for the Advancement of Medicine (IIAM) and approved by the Internal Review Board at Cedar-Sinai Medical Center.
Donor 1: 52-year-old Male
Donor 2: 37-year-old Male
Donor 3: 60-year-old Female
Donor 4: 78-year-old Female
Donor 5: 72-year-old Female
Donor 6: 2-year-old Male
Donor 7: 51-year-old Male
Donors 1 and 2 were used for SARS-CoV-2 infection of proximal airway cultures whereas donors 3-7 were used for SARS-CoV-2 infection of alveolar cultures. Donors 5,6 and 7 were used for RNA seq studies.
Primary cell cultures
Human proximal airway cells (tracheo-bronchial epithelial cells) and distal lung small airway epithelial cells were isolated from freshly excised normal human lungs obtained from transplant donors with lungs unsuitable for transplant, procured from IIAM under IRB-approved protocol. Proximal cells expanded in Pneumault Ex medium and differentiated in Pneumacult ALI medium as described in methods details. Distal small airway epithelial cells were cultured as 3D alveospheres in Pneumacult ALI medium as described in method details.
Virus strain
SARS-CoV-2, isolate USA-WA1/2020 (NR-52281), was sourced from the Biodefense and Emerging Infections (BEI) Resources of the National Institute of Allergy and Infectious Diseases (NIAID, GenBank: MT020880) All studies involving SARS-CoV-2 infection of proximal airway and AT2 cells were conducted in the UCLA BSL3 High-Containment Facility. SARS-CoV-2 was amplified once in Vero-E6 cells and viral stocks were stored at −80°C. Vero-E6 cells were cultured in Eagle’s minimal essential medium (MEM) (Corning) supplemented with 10% fetal bovine serum (Corning), penicillin-streptomycin, L-glutamine (GIBCO) and HEPES (GIBCO). Specifics of viral culture medium are described in method details.
Method details
Cell isolation
Human lung tissue was processed as described previously (Konda et al., 2020; Carraro et al., 2020) with the following modifications. For isolation of proximal airway cells, trachea and the first 2-3 generation of bronchi were slit vertically and enzymatically digested with Liberase (50 μg/mL) and DNase 1 (25 μg/mL) incubated at 37°C with mechanical agitation for 20 minutes, followed by gentle scraping of epithelial cells from the basement membrane. The remaining tissue was finely minced and further digested for 40 minutes at 37°C. For distal alveolar cell isolation, small airways of 2mm diameter or less and surrounding parenchymal tissue was minced finely and enzymatically digested for 40-60 minutes as described before. Total proximal or distal dissociated cells were passed through a series of cell strainers of decreasing pore sizes from 500 μm to 40 μm under vacuum pressure and depleted of immune and endothelial cells by magnetic associated cell sorting (MACS) in accordance to the manufacturing protocol (Miltenyi Biotec). Viable epithelial cells were further enriched by fluorescence associated cell sorting (FACS) using DAPI (Thermo Fisher Scientific) and antibodies against EPCAM (CD326), CD45 and CD31 (Biolegend) on a BD Influx cell sorter (Becton Dickinson).
Culture and differentiation of proximal airway epithelial cells at air liquid interface
FACS enriched proximal airway epithelial cells were expanded in T25 or T75 flasks coated with bovine type I collagen (Purecol, Advanced biomatrix) in Pneumacult Ex media (STEMCELL Technologies), supplemented with 1X Penicillin-Streptomycin-Neomycin (PSN) Antibiotic Mixture (Thermo Fisher Scientific) and 10μM Rho kinase inhibitor, Y-27632 (STEMCELL technologies). Upon confluence cells were dissociated using 0.05% Trypsin-EDTA (Thermo Fisher Scientific) and seeded onto collagen coated 0.4 μm pore size transparent cell culture inserts in a 24-well supported format (Corning) at a density of 7.5 × 104 cells per insert. Cells were initially cultured submerged with 300 μL of Pneumacult Ex media in the apical chamber and 700 μL in the basement chamber for 3-5 days. Upon confluence, cells were cultured at air liquid interface in 700 μL Pneumacult ALI media (STEMCELL Technologies) supplemented with 1X PSN and media was changed every 48 hr. Cultures were maintained at 37°C in a humidified incubator (5% CO2) and used for SARS-CoV-2 infection after 16-20 days of differentiation.
Culture of 3D alveospheres
Five thousand FACS enriched distal lung epithelial cells were mixed with 7.5 × 104 MRC5 human lung fibroblast cells (ATCC CCL-171) and resuspended in a 50:50 (v/v) ratio of ice cold Matrigel (Corning) and Pneumacult ALI medium. 100uL of the suspension was seeded onto the apical surface of a 0.4 μm pore-size cell culture insert in a 24 well supported format. After polymerization of Matrigel, 700 μL of Pneumacult ALI medium was added to the basement membrane. Media was supplemented with 50 μg per ml of Gentamycin (Sigma Aldrich) for the first 24 hr. and 10μM Rho kinase inhibitor for the first 48 hr. 2 μM of the Wnt pathway activator, CHIR-99021 (STEMCELL technologies) was added to the media at 48hrs and maintained for the entire duration of culture. Media was changed every 48 hr. Cultures were maintained at 37°C in a humidified incubator (5% CO2) and used for SARS-CoV-2 infection after 15-20 days.
SARS-CoV-2 infection of proximal ALI and alveolar type 2 cultures
All studies involving SARS-CoV-2 infection of proximal airway and AT2 cells were conducted in the UCLA BSL3 High-Containment Facility. SARS-CoV-2, isolate USA-WA1/2020, was sourced from the Biodefense and Emerging Infections (BEI) Resources of the National Institute of Allergy and Infectious Diseases (NIAID). SARS-CoV-2 was amplified once in Vero-E6 cells and viral stocks were stored at −80°C. Vero-E6 cells were cultured in Eagle’s minimal essential medium (MEM) (Corning) supplemented with 10% fetal bovine serum (Corning), penicillin-streptomycin (100 units/ml, GIBCO), 2 mM L-glutamine (GIBCO) and 10 mM HEPES (GIBCO). Cells were incubated at 37°C in a humidified incubator (5% CO2).
Prior to infection of alveolar cultures, Matrigel was dissolved by adding 500 μL of Dispase (500 μg/ml, GIBCO) to the apical and basement chambers of inserts and incubating for 1 hour at 37°C. Alveospheres were harvested, washed, gently dispersed with a P1000 tip by pipetting up and down 3 times such that the alveospheres were ‘popped open’, exposing the apical surface of the cells. Hereafter the cultures were referred to as alveolar cultures. SARS-CoV-2 inoculum (1x10ˆ4 TCID50 per well) was added to the alveolar cultures in a 2 mL conical tube and incubated for 2 hours at 37°C (5% CO2). Every 15 minutes, tubes were gently mixed to facilitate virus adsorption on to the cells. Subsequently, inoculum was replaced with fresh Pneumacult ALI medium and alveolar cultures were transferred to the apical chamber of the insert in 100 μL volume with 500 μL of the medium in the basement chamber. Cultures were incubated at 37°C (5% CO2) and harvested at indicated time points for sample collection.
For infection of proximal airway ALI cultures, 100 μl of SARS-CoV-2 virus inoculum (1x10ˆ4 TCID50 per well) was added on to the apical chamber of inserts. Cells were incubated at 37°C (5% CO2) for 2 hours for virus adsorption. Subsequently, the cells were washed and fresh Pneumacult ALI media (500 μl in the base chamber) was added. Cells were incubated at 37°C (5% CO2 and harvested for analysis at indicated time points for sample collection.
Viral titers
Proximal ALI and alveolar cultures were infected with 1x10ˆ4 TCID50 per well of SARS-CoV-2 as described before. For AT2 cells, at each time point (1,2 and 3dpi), media from the basal chamber from mock and SARS-CoV-2 infected wells were collected and stored at −80°C. For ALI cultures, apical washes at each time point were collected and stored at 80°C. Viral production by infected cultures was measured by quantifying TCID50 (Median Tissue Culture Infectious Dose) as described (Gauger and Vincent, 2014). Briefly, Vero-E6 cells were plated in 96-well plates at a density of 5 x103 cells/well. The following day, samples collected from the cultures at the different time points were subjected to 10-fold serial dilutions (10−1 to 10−6) and inoculated onto Vero-E6 cells. The cells were incubated at 37°C with 5% CO2. After 72 hours, each inoculated well was examined for presence or absence of viral CPE and percent infected dilutions immediately above and immediately below 50% were determined. TCID50 was calculated based on the method of Reed and Muench.
Drug validation experiments
Hydroxychloroquine (Selleck Chemicals Cat. No. S4430) and Remdesivir (Selleck Chemicals Cat. No. S8932) were dissolved in DMSO to a stock concentration of 10 mM. IFNB1 stock of 10ˆ6 units/ml was provided by Dr. Jay Kolls. To test the efficacy of the drugs against SARS-CoV-2 infection/replication in AT2 cells, cultures were suspended in media containing treated with 10 μM of Hydroxychloroquine or Remdesivir, or 100 units/ml of IFNB1 3 hours prior to SARS-CoV-2 infection. Viral infections were performed as described previously. Drugs were maintained in the media for the duration of culture post infection. alveolar cultures without drug treatment in the presence or absence (mock) of viral infections were included as controls.
Immunofluorescence staining
1-3 days after SARS-CoV-2 infection, cultures were fixed by adding 500μL of 4% paraformaldehyde in phosphate-buffered saline (PBS) for 20 minutes. Fixed samples were permeabilized and blocked for 1 hour in a ‘blocking buffer’ containing PBS, 2% bovine serum albumin, 5% goat serum, 5% donkey serum and 0.3% Triton X-100. Primary antibodies diluted in the blocking buffer were applied to samples and incubated overnight at 4°C. The following primary antibodies were used: FOXJ1 (1:300, Thermo Fisher Scientific, Cat. No. 14-9965-82); Mucin 5AC, 45M1 (1:500, Thermo Fisher Scientific, Cat. No. MA5-12178); HTII-280 (1:500, Terrace Biotech, Cat. No. TB-27AHT2-280); HTI-56 (1:100, Terrace Biotech, Cat. No. TB-29AHT1-56) SARS-CoV-2 spike (S) protein (1:100, BEI Resources Polyclonal Anti-SARS Coronavirus (antiserum, Guinea Pig), NR-10361); Cleaved caspase-3 (1:200, Cell Signaling Technology Cat. No. 9661). Samples were washed 3 times for 5 minutes each with PBS. Appropriate secondary antibodies conjugated to fluorophores (Thermo Fisher Scientific) were applied to the samples for 1 hour at room temperature. Samples were washed 3 times for 5 minutes each with PBS followed by addition of DAPI (1:5000) for 5 minutes. Insert membranes were carefully detached from their transwell support with a fine scalpel, and transferred to a glass microscopic slide. Samples were mounted using Fluoromount G (Thermo Fisher Scientific), imaged on a Zeiss LSM 780 Confocal Microscope and images were processed using Zen Blue software (Zeiss).
Quantitative real time PCR (qRT-PCR)
Total RNA was extracted from mock and SARS-CoV-2 infected proximal airway ALI and alveolar cultures lysed in Trizol (Thermo Fisher Scientific) using the chloroform-iso-propanol-ethanol method. 500ng of RNA was reversed transcribed into cDNA in a 20 μL reaction volume using iScript cDNA synthesis kit (Biorad) in accordance to manufacturer’s guidelines. qRT-PCR was performed on 10ng of CDNA per reaction in triplicates for each sample using SYBR green master mix (Thermo Fisher Scientific) on a 7500 Fast Real Time PCR system (Applied biosystems). Primers sequences for detection of SARS-CoV-2 N gene were obtained from the Center for Disease Control’s resources for research labs. Primer pairs used are described in Table S1
SARS-CoV-2 copy numbers
SARS-CoV-2 RNA transcript levels in infected samples were quantified by comparing them to a standard curve generated using serial ten-fold dilutions (101-109 copies) of a SARS-CoV-2 N gene containing plasmid.
Bulk RNA sequencing
2 days after SARS-CoV-2 infection, cells were lysed in RLT buffer and total RNA was extracted for bulk RNA sequencing using a QIAGEN RNEasy Mini Kit. RNA quality was analyzed using the 2100 Bioanalyzer (Agilent Technologies) and quantified using QubitTM (ThermoFisher Scientific). Library construction was performed by the Cedars-Sinai Applied Genomics, Computation and Translational Core using the Lexogen RiboCop rRNA Depletion kit and Swift Biosciences RNA Library Sciences. Sequencing was performed using the NovaSeq 6000 (Illumina) with single-end 75bp sequencing chemistry. On average, about 20 million reads were generated from each sample. Raw reads were aligned using Star aligner 2.6.1 (Dobin et al., 2013)/RSEM 1.2.28 (Li and Dewey, 2011) with default parameters, using a custom human GRCh38 transcriptome reference downloaded from https://www.gencodegenes.org, containing all protein coding and long non-coding RNA genes based on human GENCODE version 33 annotation with SARS-Cov2 virus genome MT246667.1 https://www.ncbi.nlm.nih.gov/nuccore/MT246667.1.
Differential gene expression was determined by DESeq2 (Love et al., 2014). Top differential genes were determined based on fold change and test statistics. Pathway analysis was performed using Ingenuity Pathway Analysis.
Quantification and statistical analysis
Statistical analysis was performed using GraphPad Prism version 8 software. Data are presented as linear fold change or log2 fold change ± SEM. SARS-CoV-2 infection time course for ALI and alveolar cultures was analyzed using Two- Way ANOVA with Sidak’s post hoc correction. Drug validation experiments were analyzed using One-way ANOVA with Tukey’s post hoc correction. N numbers and p values are indicated in corresponding figure legends. TPM counts and relative gene expression for ISGs between MOCK and infected cultures were analyzed using Two-tailed t tests. ∗p < 0.5, ∗∗p < 0.01, ∗∗∗p < 0.001,∗∗∗∗p < 0.0001.
Acknowledgments
We would like to thank the Cedars-Sinai Applied Genomics, Computation and Translational Core for their help with performing bulk RNA-seq. The following reagents were obtained through BEI Resources, NIAID, National Institutes of Health (NIH): Polyclonal Anti-SARS Coronavirus (antiserum, Guinea Pig), NR-10361; and SARS-Related Coronavirus 2, Isolate USA-WA1/2020, NR-52281. This work was supported by the NIH (5RO1HL135163-04, PO1HL108793-08, 3UG3NS105703-03S1, T32HL134637, and 1R01EY032149), the BMS/Celgene IDEAL Consortium, CIRM EDUC2-08383, Parker B. Francis Fellowship Program, UCLA DGSOM and Broad Stem Cell Research Center institutional award (OCRC #20-15), and a California Institute for Regenerative Medicine TRAN Award (TRAN1COVID19-11975).
Author contributions
A.M., G.G., and B.K. performed experiments. A.M. and B.R.S. designed studies and wrote the manuscript. G.G., S.B., J.M.V., C.K., C.S., A.P., P.P., J.S.d.A., and C.G.-d.-A. prepared samples and provided reagents. C.Y. and A.M. analyzed RNA-seq data. A.M., C.F.K., B.G., B.R.S., and V.A. interpreted results. J.K.K. and D.A.P. provided IFNB1 and guidance for drug validation experiments. B.R.S. and V.A. supervised.
Declaration of interests
The authors declare no competing interests.
Published: May 4, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.109055.
Supplemental information
References
- Barkauskas C.E., Cronce M.J., Rackley C.R., Bowie E.J., Keene D.R., Stripp B.R., Randell S.H., Noble P.W., Hogan B.L. Type 2 alveolar cells are stem cells in adult lung. J. Clin. Invest. 2013;123:3025–3036. doi: 10.1172/JCI68782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beigel J.H., Tomashek K.M., Dodd L.E., Mehta A.K., Zingman B.S., Kalil A.C., Hohmann E., Chu H.Y., Luetkemeyer A., Kline S. Remdesivir for the Treatment of Covid-19—Preliminary Report. N. Engl. J. Med. 2020;383:1813–1826. doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Melo D., Nilsson-Payant B.E., Liu W.C., Uhl S., Hoagland D., Møller R., Jordan T.X., Oishi K., Panis M., Sachs D. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell. 2020;181:1036–1045.e9. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cagno V. SARS-CoV-2 cellular tropism. Lancet Microbe. 2020;1:e2–e3. doi: 10.1016/S2666-5247(20)30008-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carraro G., Mulay A., Yao C., Mizuno T., Konda B., Petrov M., Lafkas D., Arron J.R., Hogaboam C.M., Chen P. Single-Cell Reconstruction of Human Basal Cell Diversity in Normal and Idiopathic Pulmonary Fibrosis Lung. Am. J. Respir. Crit. Care Med. 2020;202:1540–1550. doi: 10.1164/rccm.201904-0792OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Channappanavar R., Fehr A.R., Vijay R., Mack M., Zhao J., Meyerholz D.K., Perlman S. Dysregulated Type I Interferon and Inflammatory Monocyte-Macrophage Responses Cause Lethal Pneumonia in SARS-CoV-Infected Mice. Cell Host Microbe. 2016;19:181–193. doi: 10.1016/j.chom.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Channappanavar R., Fehr A.R., Zheng J., Wohlford-Lenane C., Abrahante J.E., Mack M., Sompallae R., McCray P.B., Jr., Meyerholz D.K., Perlman S. IFN-I response timing relative to virus replication determines MERS coronavirus infection outcomes. J. Clin. Invest. 2019;129:3625–3639. doi: 10.1172/JCI126363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H., Chan J.F., Yuen T.T., Shuai H., Yuan S., Wang Y., Hu B., Yip C.C., Tsang J.O., Huang X. Comparative tropism, replication kinetics, and cell damage profiling of SARS-CoV-2 and SARS-CoV with implications for clinical manifestations, transmissibility, and laboratory studies of COVID-19: an observational study. Lancet Microbe. 2020;1:e14–e23. doi: 10.1016/S2666-5247(20)30004-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clementi N., Ferrarese R., Criscuolo E., Diotti R.A., Castelli M., Scagnolari C., Burioni R., Antonelli G., Clementi M., Mancini N. Interferon-β-1a Inhibition of Severe Acute Respiratory Syndrome-Coronavirus 2 In Vitro When Administered After Virus Infection. J. Infect. Dis. 2020;222:722–725. doi: 10.1093/infdis/jiaa350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauger P.C., Vincent A.L. Serum virus neutralization assay for detection and quantitation of serum-neutralizing antibodies to influenza A virus in swine. Methods Mol. Biol. 2014;1161:313–324. doi: 10.1007/978-1-4939-0758-8_26. [DOI] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y.J., Okuda K., Edwards C.E., Martinez D.R., Asakura T., Dinnon K.H., 3rd, Kato T., Lee R.E., Yount B.L., Mascenik T.M. SARS-CoV-2 Reverse Genetics Reveals a Variable Infection Gradient in the Respiratory Tract. Cell. 2020;182:429–446.e14. doi: 10.1016/j.cell.2020.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Hume A.J., Abo K.M., Werder R.B., Villacorta-Martin C., Alysandratos K.D., Beermann M.L., Simone-Roach C., Lindstrom-Vautrin J., Olejnik J. SARS-CoV-2 Infection of Pluripotent Stem Cell-Derived Human Lung Alveolar Type 2 Cells Elicits a Rapid Epithelial-Intrinsic Inflammatory Response. Cell Stem Cell. 2020;27:962–973.e7. doi: 10.1016/j.stem.2020.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia H.P., Look D.C., Shi L., Hickey M., Pewe L., Netland J., Farzan M., Wohlford-Lenane C., Perlman S., McCray P.B., Jr. ACE2 receptor expression and severe acute respiratory syndrome coronavirus infection depend on differentiation of human airway epithelia. J. Virol. 2005;79:14614–14621. doi: 10.1128/JVI.79.23.14614-14621.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsura H., Sontake V., Tata A., Kobayashi Y., Edwards C.E., Heaton B.E., Konkimalla A., Asakura T., Mikami Y., Fritch E.J. Human Lung Stem Cell-Based Alveolospheres Provide Insights into SARS-CoV-2-Mediated Interferon Responses and Pneumocyte Dysfunction. Cell Stem Cell. 2020;27:890–904.e8. doi: 10.1016/j.stem.2020.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konda B., Mulay A., Yao C., Beil S., Israely E., Stripp B.R. Isolation and Enrichment of Human Lung Epithelial Progenitor Cells for Organoid Culture. J. Vis. Exp. 2020;2020:161. doi: 10.3791/61541. [DOI] [PubMed] [Google Scholar]
- Lamers M.M., Beumer J., van der Vaart J., Knoops K., Puschhof J., Breugem T.I., Ravelli R.B.G., Paul van Schayck J., Mykytyn A.Z., Duimel H.Q. SARS-CoV-2 productively infects human gut enterocytes. Science. 2020;369:50–54. doi: 10.1126/science.abc1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I.T., Nakayama T., Wu C.-T., Goltsev Y., Jiang S., Gall P.A., Liao C.-K., Shih L.-C., Schürch C.M., McIlwain D.R. Robust ACE2 protein expression localizes to the motile cilia of the respiratory tract epithelia and is not increased by ACE inhibitors or angiotensin receptor blockers. medRxiv. 2020 doi: 10.1101/2020.05.08.20092866. [DOI] [Google Scholar]
- Li B., Dewey C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Cao R., Xu M., Wang X., Zhang H., Hu H., Li Y., Hu Z., Zhong W., Wang M. Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro. Cell Discov. 2020;6:16. doi: 10.1038/s41421-020-0156-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantlo E., Bukreyeva N., Maruyama J., Paessler S., Huang C. Antiviral activities of type I interferons to SARS-CoV-2 infection. Antiviral Res. 2020;179:104811. doi: 10.1016/j.antiviral.2020.104811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muus C., Luecken M.D., Eraslan G., Waghray A., Heimberg G., Sikkema L., Kobayashi Y., Vaishnav E.D., Subramanian A., Smilie C., Jagadeesh K. Integrated analyses of single-cell atlases reveal age, gender, and smoking status associations with cell type-specific expression of mediators of SARS-CoV-2 viral entry and highlights inflammatory programs in putative target cells. bioRxiv. 2020 doi: 10.1101/2020.04.19.049254. [DOI] [Google Scholar]
- Pastick K.A., Okafor E.C., Wang F., Lofgren S.M., Skipper C.P., Nicol M.R., Pullen M.F., Rajasingham R., McDonald E.G., Lee T.C. Review: Hydroxychloroquine and Chloroquine for Treatment of SARS-CoV-2 (COVID-19) Open Forum Infect. Dis. 2020;7:ofaa130. doi: 10.1093/ofid/ofaa130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi F., Qian S., Zhang S., Zhang Z. Single cell RNA sequencing of 13 human tissues identify cell types and receptors of human coronaviruses. Biochem. Biophys. Res. Commun. 2020;526:135–140. doi: 10.1016/j.bbrc.2020.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rackley C.R., Stripp B.R. Building and maintaining the epithelium of the lung. J. Clin. Invest. 2012;122:2724–2730. doi: 10.1172/JCI60519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravindra N.G., Alfajaro M.M., Gasque V., Habet V., Wei J., Filler R.B., Huston N.C., Wan H., Szigeti-Buck K., Wang B. Single-cell longitudinal analysis of SARS-CoV-2 infection in human bronchial epithelial cells. bioRxiv. 2020 doi: 10.1101/2020.05.06.081695. [DOI] [Google Scholar]
- Si L., Bai H., Rodas M., Cao W., Oh C.Y., Jiang A., Nurani A., Zhu D.Y., Goyal G., Gilpin S.E. Human organs-on-chips as tools for repurposing approved drugs as potential influenza and COVID19 therapeutics in viral pandemics. bioRxiv. 2020 doi: 10.1101/2020.04.13.039917. [DOI] [Google Scholar]
- Sungnak W., Huang N., Bécavin C., Berg M., Queen R., Litvinukova M., Talavera-López C., Maatz H., Reichart D., Sampaziotis F., HCA Lung Biological Network SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. 2020;26:681–687. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay M.Z., Poh C.M., Rénia L., MacAry P.A., Ng L.F.P. The trinity of COVID-19: immunity, inflammation and intervention. Nat. Rev. Immunol. 2020;20:363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijaykrishna D., Poon L.L., Zhu H.C., Ma S.K., Li O.T., Cheung C.L., Smith G.J., Peiris J.S., Guan Y. Reassortment of pandemic H1N1/2009 influenza A virus in swine. Science. 2010;328:1529. doi: 10.1126/science.1189132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zhang D., Du G., Du R., Zhao J., Jin Y., Fu S., Gao L., Cheng Z., Lu Q. Remdesivir in adults with severe COVID-19: a randomised, double-blind, placebo-controlled, multicentre trial. Lancet. 2020;395:1569–1578. doi: 10.1016/S0140-6736(20)31022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youk J., Kim T., Evans K.V., Jeong Y.I., Hur Y., Hong S.P., Kim J.H., Yi K., Kim S.Y., Na K.J. Three-Dimensional Human Alveolar Stem Cell Culture Models Reveal Infection Response to SARS-CoV-2. Cell Stem Cell. 2020;27:905–919.e10. doi: 10.1016/j.stem.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler C.G.K., Allon S.J., Nyquist S.K., Mbano I.M., Miao V.N., Tzouanas C.N., Cao Y., Yousif A.S., Bals J., Hauser B.M., HCA Lung Biological Network. Electronic address: lung-network@humancellatlas.org. HCA Lung Biological Network SARS-CoV-2 Receptor ACE2 Is an Interferon-Stimulated Gene in Human Airway Epithelial Cells and Is Detected in Specific Cell Subsets across Tissues. Cell. 2020;181:1016–1035.e19. doi: 10.1016/j.cell.2020.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Original gene expression data from this study have been deposited to Gene Expression Omnibus with accession number GEO: GSE160435.