Abstract
Background
International travel is a risk factor for incident colonization with extended spectrum beta-lactamase (ESBL)-producing organisms. These and other multidrug-resistant (MDR) bacteria are major pathogens in combat casualties. We evaluated risk factors for colonization with MDR bacteria in US military personnel travelling internationally for official duty.
Methods
TravMil is a prospective observational study enrolling subjects presenting to military travel clinics. We analysed surveys, antimicrobial use data, and pre- and post-travel perirectal swabs in military travellers to regions outside the continental USA, Canada, Western or Northern Europe, or New Zealand, presenting to one clinic from 12/2015 to 12/2017. Recovered Gram-negative isolates underwent identification and susceptibility testing (BD Phoenix). Characteristics of trip and traveller were analysed to determine risk factors for MDR organism colonization.
Results
110 trips were planned by 99 travellers (74% male, median age 38 years [IQR 31, 47.25]); 72 trips with returned pre- and post-travel swabs were completed by 64 travellers. Median duration was 21 days (IQR 12.75, 79.5). 17% travelled to Mexico/Caribbean/Central America, 15% to Asia, 57% to Africa and 10% to South America; 56% stayed in hotels and 50% in dormitories/barracks. Travellers used doxycycline (15%) for malaria prophylaxis, 11% took an antibiotic for travellers’ diarrhoea (TD) treatment (fluoroquinolone 7%, azithromycin 4%). Incident MDR organism colonization occurred in 8 travellers (incidence density 3.5/1000 travel days; cumulative incidence 11% of trips [95% CI: 4–19%]), all ESBL-producing Escherichia coli. A higher incidence of ESBL-producing E. coli acquisition was associated with travel to Asia (36% vs 7%, P = 0.02) but not with travel to other regions, TD or use of antimicrobials. No relationship was seen between fluoroquinolone or doxycycline exposure and resistance to those antimicrobials.
Conclusions
Incident colonization with MDR organisms occurs at a lower rate in this military population compared with civilian travellers, with no identified modifiable risk factors, with highest incidence of ESBL acquisition observed after South Asia travel.
Keywords: ESBL-producing Enterobacteriaceae, Eschericia coli, Perirectal swab, Colonization, Whole genome sequencing, CTX-M, Antimicrobials
Background
Multidrug-resistant (MDR) bacteria are a concern globally for international travellers in both civilian and military literature.1 MDR infections in combat casualties were noted in Iraq and Afghanistan during Operations Iraqi and Enduring Freedom (OIF/OEF) with Acinetobacter baumannii-calcoaceticus (ABC) more prevalent in casualties coming from Iraq and extended spectrum beta-lactamase (ESBL)-producing Enterobacteriaceae in casualties out of Afghanistan.2–4 In the few studies that looked at incident colonization of MDR bacteria including ESBL-producing Enterobacteriaceae in US military personnel and beneficiaries travelling internationally, rates ranged from 9 to 14%.5–7 In civilian literature, international travel has shown post-travel rates of ESBL-producing Enterobacteriaceae incident colonization ranging from 24 to 41%, with rates greater than 50% in travellers returning from South Asia.8–14 The majority of literature has described travel associated ESBL-producing Enterobacteriaceae acquisition with primarily CTX-M-producing Enterobacteriaceae. KPC-, VIM-, OXA-48- and NDM-producing Enterobacteriaceae infections have also been described following hospitalization in endemic areas.15
Prior to entering conflicts in Iraq and Afghanistan, there were few studies to predict MDR ABC colonization and infection in combat casualties. MDR bacteria have been shown to be community-associated pathogens in local, host nation personnel treated in Iraq and Afghanistan combat support hospitals; however, US military personnel have different potential exposures and typically live in a controlled environment with food and water provided from sources approved by preventive medicine.16,17 Extensive studies were conducted which demonstrated that the predominant source of ABC infection was nosocomial as little to no colonization was demonstrated in US troops prior to or at the time of injury.18–20 The source of ESBL-producing Enterobacteriaceae colonization and subsequent infection is less well established. A prospective study of US military personnel and beneficiaries travelling internationally did identify acquisition of ESBL-producing Escherichia coli, but was unable to identify clearly associated risk factors.5 No previous study has prospectively evaluated risk factors or incidence of acquisition for ESBL-producing Enterobacteriaceae colonization in travellers on military orders with a variety of destinations and missions.
Methods
Study design and definitions
The Military Travel MDR Surveillance Study is a subset of the larger TravMil study: Deployment and Travel Related Infectious Disease Risk Assessment, Outcomes, and Prevention Strategies Among Department of Defense (DoD) Beneficiaries, which is approved by the Infectious Disease Institutional Review Board of the Uniformed Services University of the Health Sciences (Bethesda, MD). TravMil is a prospective, observational cohort study of DoD beneficiaries travelling outside of the USA for ≤6.5 months. Participants enrolling into TravMil at single site (Brooke Army Medical Center, TX [BAMC]) were approached about optional participation in the Military Travel MDR Surveillance Study. There were 110 travellers enrolled who were travelling for official military duties and deployments from December 2015 to December 2017. Exclusion criteria included planned travel limited to Western or Northern Europe, Canada or New Zealand; completed travel time > 6.5 months; and inability to follow-up between 3 days and 8 weeks after travel to collect a post-travel perirectal swab. Participants completed a pre-travel survey, a travel diary, a post-travel survey and an extended follow-up survey at 3 and 6 months. All demographic, itinerary, malaria prophylaxis use, contact with freshwater, detailed food and drink exposures, illnesses during and post-travel, antibiotics used during travel, and contact with medical care during travel collected as part of TravMil were analysed for risk factors for ESBL-producing Enterobacteriaceae acquisition. Antibiotic exposure was defined as taking at least one dose of an antibiotic.
Bacterial susceptibility
At the time of enrollment and again 10 days after return from travel +/− 7 days, patients were provided with BD CultureSwab™ MaxV(+) (Becton, Dickinson and Company, Franklin Lakes, NJ) for self-collection of perirectal specimens. Participants were educated on self-collection of a perirectal swab, which has been used as a valid collection method compared with stool samples.21 As the second TravMil study visit was not required until 8 weeks after return, participants were given the options to either return the swabs by paid postal package or present to BAMC clinic to drop off a self-collected swab.22 Swabs collected were plated onto sheep blood agar and MacConkey agar. Colonies demonstrating morphology consistent with Gram-negative bacterial colonies were sub-cultured onto sheep blood agar in order to assure culture purity. All isolates were frozen at −80°C in Trypticase Soy Broth with 15% glycerol. Isolates underwent automated testing (BD Phoenix™ Automated Microbiology System-Becton Dickinson, Franklin Lakes, NJ) for identification, anti-microbial susceptibility testing, and identification of ESBL-producing Enterobacteriaceae.
Molecular testing
In participants with ESBL-producing Enterobacteriaceae isolated either pre- or post-travel, all pre- and post-travel isolates of E. coli from those participants were forwarded to the multidrug-resistant organism repository and surveillance network (MRSN) for whole genome sequencing (WGS). WGS of all 25 isolates was performed on an Illumina MiSeq® (Illumina, San Diego, CA) and the overall relationship between the isolates determined using a pan-genome sequence analysis program (Panseq).
Statistical analysis
Risk factor analysis was evaluated by univariate analysis with Pearson’s χ2 and Fisher’s exact test for categorical variables and Mann–Whitney U for continuous variables. Two-tailed P values of <0.05 were considered statistically significant. Statistical analyses were performed using SPSS software (IBM® SPSS® Statistics Version 22, Chicago, Illinois). Agreement between phenotypic and genotypic antibiotic resistance profiles was measured using Cohen’s kappa coefficient.
Results
Demographics
Of 72 trips by 64 participants with returned pre- and post-travel swabs, 74% were male and median age was 38 (IQR 31, 47.25) (Table 1). The primary regions of travel were Mexico, the Caribbean, and Central America, Asia, Africa, and South America with a median duration of travel of 21 days (IQR 12.75, 79.5).
Table 1.
Baseline characteristics of participants
| Characteristics | Participant number (%) |
|---|---|
| Participants enrolled | N = 110 |
| Planned trips | N = 99 |
| Participants who returned swabs | N = 64 |
| Trips with returned swabs | N = 72 |
| Gender, Male | 81 (74%) |
| Median age (years) | 38 (IQR 31,47.25) |
| Median duration of travel (days) | 21 (IQR 12.75, 79.5) |
| Region of travel | N = 72 |
| Mexico, Caribbean and Central America | 12 (17%) |
| Asia | 11 (15%) |
| Africa | 41 (57%) |
| South America | 7 (10%) |
| Living conditions | |
| Hotel with air conditioning | 32 (56%) |
| Hotel without air conditioning | 8 (11%) |
| Military accommodations | 4 (6%) |
| Dormitories/Barracks | 36 (50%) |
| Field water exposure* | 5 (7%) |
| Local water ingestion | |
| Untreated water | 1 (1%) |
| Ice in beverage | 31 (43%) |
| Antimicrobial exposure since enrollment | 58 (81%) |
| Malaria prophylaxis | 57 (79%) |
| Chloroquine | 2 (3%) |
| Mefloquine | 0 (0%) |
| Atovaquone–proguanil | 45 (63%) |
| Doxycycline | 11 (15%) |
| Primaquine | 9 (13%) |
| Antibiotics for TD | 8 (11%) |
| Ciprofloxacin | 4 (6%) |
| Azithromycin | 3 (4%) |
| Levofloxacin | 1 (1%) |
| Systemic antibiotics for other reasons | 3 (4%) |
| Amoxicillin/clavulanate | 1 (1%) |
| Levofloxacin | 1 (1%) |
| Trimethoprim/sulfamethoxazole | 1 (1%) |
| Illness since enrollment | 19 (27%) |
| Fever | 0 (0%) |
| Influenza like illness | 9 (13%) |
| TD | 10 (14%) |
*Defined as contact with fresh water from lakes, streams or rivers (wading/bathing/swimming/etc.).
ESBL-producing Enterobacteriaceae pre- and post-travel
Eleven bacterial isolates from pre- and post-travel perirectal swab isolates during 72 trips by participants were ESBL-producing Enterobacteriaceae. All ESBL-producing Enterobacteriaceae were E. coli. Multiple other Gram-negative isolates were recovered but were not MDR or ESBL-producing to include genus Acinetobacter, Aeromonas, Citrobacter, Enterobacter, Escherichia, Klebsiella, Kluyvera, Morganella, Pantoea, Proteus, Pseudomonas and Stenotrophomonas.
Eight of eleven ESBL-producing E. coli represented new acquisition (Table 2). One participant had ESBL-producing E. coli pre-travel, but was no longer colonized post-travel. Another participant had ESBL-producing E. coli both pre- and post-travel. There was a higher rate of new acquisition of ESBL-producing E. coli with travel to South Asia (India and Afghanistan) (P = 0.02). For this incident colonization, incidence density was 3.5/1000 travel days and cumulative incidence was 11% of trips (95% CI: 4–19%). No other risk factors such as living conditions, field water exposure, local water ingestion, antimicrobial exposure since enrollment or illness since enrollment was significantly associated with new acquisition of ESBL-producing E. coli for military work trips.
Table 2.
Characteristics associated with acquisition of ESBL-producing Enterobacteriaceae
| Characteristic | ESBL (n = 8) | No ESBL (n = 64) | P-value |
|---|---|---|---|
| Gender | |||
| Male | 6 (75 [34.9–96.8%]) | 51 (80 [67.2–88.7%]) | 0.67 |
| Female | 2 (25 [3.2–65.1%]) | 13 (20 [11.3–32.8%]) | 0.67 |
| Median age (years) | 39.5 (IQR 32.25, 46.0) | 41.5 (IQR 34.0, 49.0) | 0.53 |
| Median duration of travel (days) | 17.0 (IQR 7.5, 154.0) | 17.0 (IQR 8.0, 24.0) | 0.75 |
| Region of travel | |||
| Mexico, Caribbean and Central America | 0 (0 [0–36.9%]) | 12 (18.8 [11.1–30.5%]) | 0.34 |
| Asia* | 4 (50 [15.7–84.3%]) | 7 (10.9 [4.9–21.2%]) | 0.02 |
| Africa | 3 (37.5 [8.5–75.5%]) | 38 (59.4 [46.4–71.5%]) | 0.28 |
| South America | 1 (12.5 [0.3–52.7%]) | 6 (9.4 [3.5–19.3%]) | 0.58 |
| Living conditions | |||
| Hotel with air conditioning | 4 (50 [15.7–84.3%]) | 28 (43.8 [31.4–56.7%]) | 1.00 |
| Hotel without air conditioning | 1 (12.5 [0.3–52.7%]) | 7 (10.9 [4.5–21.2%]) | 1.00 |
| Military accommodations | 0 (0 [0–36.9%]) | 4 (6.3 [1.7–15.2%]) | 1.00 |
| Dormitories/Barracks | 3 (37.5 [8.5–75.5%]) | 33 (51.6 [38.7–64.2%]) | 0.71 |
| Food exposures | |||
| Food from street vendors | 2 (25 [3.2–65.1%]) | 13 (20 [11.3–32.8%]) | 0.67 |
| Meals prepared at local homes | 1 (12.5 [0.3–52.7%]) | 15 (23.4 [13.8–35.7%]) | 0.67 |
| Unpasteurized dairy products | 0 (0 [0–36.9%]) | 2 (3.1 [0.4–10.8%]) | 1.00 |
| Poorly cooked or raw meat/seafood | 1 (12.5 [0.3–52.7%]) | 6 (9.4 [3.5–19.3%]) | 0.58 |
| Poorly cooked or raw vegetables/salad | 2 (25 [3.2–65.1%]) | 7 (10.9 [4.9–21.2%]) | 0.26 |
| Field water exposure | 0 (0 [0–36.9%]) | 5 (7 [2.6–17.3%]) | 1.00 |
| Local water ingestion | |||
| Untreated water | 0 (0 [0–36.9%]) | 1 (1.6 [0–8.4%]) | 1.00 |
| Ice in beverage | 3 (37.5 [8.5–75.5%]) | 28 (43.8 [31.4–56.7%]) | 1.00 |
| None of the above food or water exposures | 1 (12.5 [0.3–52.7%]) | 19 (29.7 [18.9–42.4%]) | 0.43 |
| Unable to wash hands regularly before eating | 1 (12.5 [0.3–52.7%]) | 11 (17.2 [8.9–28.7%]) | 1.00 |
| Antimicrobial exposure since enrollment | 6 (75 [34.9–96.8%]) | 52 (81.2 [69.5–89.9%) | 0.65 |
| Malaria prophylaxis | 6 (75 [34.9–96.8%]) | 51 (80 [67.2–88.7%]) | 0.67 |
| Chloroquine | 0 (0 [0–36.9%]) | 2 (3.1 [0.4–10.8%]) | 1.00 |
| Mefloquine | 0 (0 [0–36.9%]) | 0 (0 [0–5.6%]) | n/a |
| Atovaquone–proguanil | 4 (50 [15.7–84.3%]) | 41 (64.1 [51.5–75.7%]) | 0.46 |
| Doxycycline | 2 (25 [3.2–65.1%]) | 9 (14.1 [6.6–25.0%]) | 0.60 |
| Primaquine | 3 (37.5 [8.5–75.5%]) | 6 (9.4 [3.5–19.3%]) | 0.06 |
| Antibiotics for TD | 2 (25 [3.2–65.1%]) | 6 (9.4 [3.5–19.3%]) | 0.22 |
| Ciprofloxacin | 1 (12.5 [0.3–52.7%]) | 3 (4.7 [1–13.1%]) | 0.33 |
| Azithromycin | 1 (12.5 [0.3–52.7%]) | 2 (3.1 [0.4–10.8%]) | 0.20 |
| Levofloxacin | 0 (0 [0–36.9%]) | 1 (1.6 [0–8.4%]) | n/a |
| Systemic antibiotics for other reason | 0 (0 [0–36.9%]) | 3 (4.7 [1.0–13.1%]) | 1.00 |
| Amoxicillin/clavulanate | 0 (0 [0–36.9%]) | 1 (1.6 [0–8.4%]) | n/a |
| Levofloxacin | 0 (0 [0–36.9%]) | 1 (1.6 [0–8.4%]) | n/a |
| Sulfamethoxazole/trimethoprim | 0 (0% [0–36.9]) | 1 (1.6 [0–8.4%]) | n/a |
| Illness since enrollment | 3 (37.5 [8.5–75.5%]) | 16 (25 [15.0–37.4%]) | 0.43 |
| Fever | 0 (0 [0–36.9%]) | 0 (0–5.6%) | n/a |
| Influenza-like illness | 1 (12.5 [0.3–52.7%]) | 8 (12.5 [0.3–52.7%]) | 1.00 |
| TD | 2 (25 [3.2–65.1%]) | 8 (12.5 [5.6–23.2%]) | 0.31 |
*Isolates were from India and Afghanistan.
ESBL, extended spectrum beta-lactamase; all ESBL-producing Enterobacteriaceae were E. coli.
A total of 25 E. coli isolates were sent for WGS (Figure 1). These included 11 ESBL-producing E. coli with an additional 14 non-ESBL-producing E. coli isolates from 10 participants with ESBL-producing E. coli. All ESBL-producing E. coli isolates were unrelated based on the large separation in number of SNPs. Two patients had serial isolation of non-ESBL-producing E. coli isolates from pre- and post-travel cultures which were separated by 4–9 SNPs. Three groups of isolates formed close clusters on high-resolution SNP-based analysis. Group 1 isolates were separated by 241 SNPs. Group 2 isolates included two colonizing isolates as well as an isolate separated from the colonizing isolates by 1163 and 1165 SNPs. Group 3 isolates represented two colonizing isolates. On multi-locus sequence typing (MLST), two novel sequence types were identified in post-travel ESBL E. coli isolates.
Figure 1.
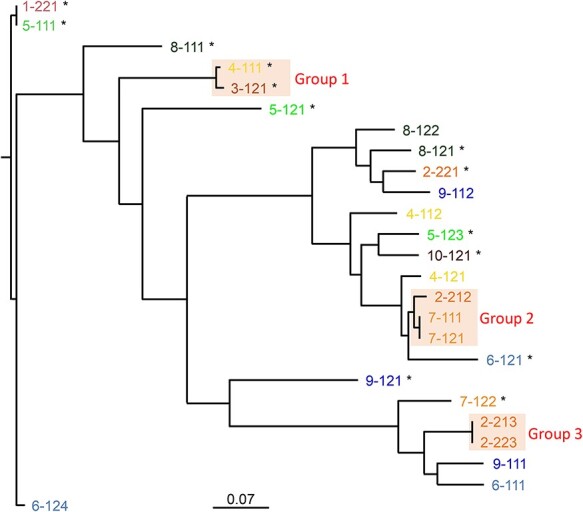
Whole genome phylogeny of the 25 E. coli. Branch lengths are indicative of relatedness and the scale bar represent approximate number of SNPs per 1000 bases. Phenotypic ESBL-producing E. coli are marked with an asterisk.
Antimicrobial resistance in Gram-negative organisms after travel
ESBL-producing E. coli resistance profiles demonstrated universal resistance to cefazolin. Sixty-three percent were resistant to trimethoprim–sulfamethoxazole (TMP/SMX), tetracycline and ciprofloxacin. Fifty-five percent were resistant to levofloxacin. Twenty-seven percent were resistant to tobramycin and gentamicin. Ten percent were resistant to nitrofurantoin. Isolates were universally susceptible to amikacin, ertapenem, imipenem, meropenem and piperacillin–tazobactam. On WGS, 45 different antibiotic resistance genes in 25 E. coli isolates were found. Of the 11 phenotypic ESBL-producing E. coli, 10 demonstrated genotypic resistance with presence of blaCTX-M genes (CTX-M-3 [two isolates], CTX-M-14 [six isolates], CTX-M-15 [one isolate] and CTX-M-55 [one isolate]), 9 demonstrated blaTEM genes (TEM-1 [eight isolates] and TEM-209 [one isolate]) and 8 demonstrated both blaCTX-M genes and blaTEM genes with all phenotypic ESBL-producing E. coli containing one or more ESBL genes (Table 3).
Table 3.
Genotypic and phenotypic resistance among 25 E. coli isolates
| Antibiotic resistance profile | Phenotypic resistance (%) | Genotypic resistance (%) | Concordant phenotypic and genotypic resistance (%) | Discordant phenotypic or genotypic resistance (%) | Cohen’s kappa coefficient |
|---|---|---|---|---|---|
| ESBL | 11 (44%) | 13 (52%) | 11 (44%) | 2 (8%) | 0.73 |
| Fluoroquinolone | 8 (32%) | 4 (16%) | 2 (8%) | 6 (24%) | 0.15 |
| Tetracycline | 11 (44%) | 11 (44%) | 11 44%) | 0 (0%) | 1 |
ESBL, extended spectrum beta-lactamase.
Analysis of fluoroquinolone and tetracycline resistance in all isolates from 72 trips demonstrated 6 subjects exposed to a fluoroquinolone, 11 exposed to a tetracycline and 55 exposed to neither (Table 4). Exposure to a fluoroquinolone and acquisition of both fluoroquinolone and tetracycline resistance was demonstrated (P = 0.05). Otherwise, there was no significant relationship between exposure to a fluoroquinolone or tetracycline and development of resistance. Two of six participants in our study developed fluoroquinolone resistance with exposure to levofloxacin or ciprofloxacin (P = 0.16), and 2 of 11 developed tetracycline resistance with exposure to doxycycline (P = 0.19). No participants were exposed to both a fluoroquinolone and tetracycline. Analysis of antibiotic resistance genes in 25 E. coli demonstrated eight E. coli isolates phenotypically resistant to fluoroquinolones, but only four with genotypic resistance (genes qnrB19 [three isolates] and qnrS1 [one isolate]) (Table 3). All 11 E. coli isolates with tetracycline resistance were phenotypically and genotypically concordant (genes tet(A) [four isolates], tet(B) [six isolates] and tet(C) [one isolate]).
Table 4.
Relationship between fluoroquinolone or doxycycline exposure and acquired resistance
| Characteristic | Acquired FQ resistance (n = 9) | Acquired Tet resistance (n = 27) | Acquired both FQ and Tet resistance (n = 5) | No acquired resistance (n = 41) |
|---|---|---|---|---|
| FQ exposed (n = 6) | 2 (P = 0.16) | 4 (P = 0.19) | 2 (P = 0.05) | 2 (P = 0.39) |
| Doxycycline exposed (n = 11) | 2 (P = 0.62) | 2 (P = 0.19) | 0 (P = 1.00) | 7 (P = 0.75) |
| Exposed to neither (n = 55) | 5 (P = 0.20) | 21 (P = 1.00) | 3 (P = 0.59) | 32 (P = 0.78) |
FQ, fluoroquinolone; Tet, tetracycline; *No participants were exposed to both a fluoroquinolone and doxycycline.
Discussion
In order to discover the origin of pre-injury colonization with ESBL-producing Enterobacteriaceae in a population of international travellers with official military duties or deployments, we set out to find associated risk factors through study of the prevalence of ESBL-producing Enterobacteriaceae before and after travel. This prospective study determined that there was an increased rate of incident colonization with ESBL-producing E. coli following travel to Asia especially with travellers returning from India and Afghanistan. ESBL-producing Enterobacteriaceae colonization was not pre-existing in the majority of travellers who returned with colonization of ESBL-producing E. coli. In this small study, no other individual risk factors were identified for acquisition of ESBL-producing E. coli.
Civilian studies have also associated ESBL-producing E. coli incident colonization with international travel, but the majority of studies have been done outside of US cohorts.8,9,11,23,24 The one prior prospective civilian study conducted in an American cohort from New York City looking at pre- and post-travel colonization with ESBL-producing Enterobacteriaceae showed solely the isolation of E. coli as did this study. The pre-travel colonization rate of ESBL-producing E. coli was 1.7% while post-travel was 25% in the New York City cohort.10 These rates are similar to other studies outside the USA.8,9,23 Consistent with another study that included US military personnel and beneficiaries travelling internationally, this study showed similarly low rates of colonization with 2% pre- and 11% post-travel of ESBL-producing E. coli5. This study had a longer duration of international travel (median of 21 days compared to 12 days with the previous US military personnel and beneficiaries cohort), which was more similar to the New York City cohort (median of 21 days).5,10
This was the first study to prospectively evaluate risk factors or incidence of acquisition for ESBL-producing Enterobacteriaceae colonization in travellers on military orders with a variety of destinations and missions. Travel in the military is unique in that travellers were not visiting friends and relatives or having casual contact with locals, food and water supply is regulated, and pre-travel counselling is required. The trips included deployments and short-term travel to include support missions, so these results may not be fully generalizable to the larger population of military personnel at highest risk for combat casualties. While the sample size is relatively low, military members staying in either hotels or military accommodations had similarly low incidence of acquisition. The effect of antibiotic exposure and malaria prophylaxis was not found to have statistical significance for acquiring ESBL-producing Enterobacteriaceae. One of five participants who took a fluoroquinolone for travellers’ diarrhoea (TD) acquired ESBL-producing E. coli, which is lower than seen in Swedish cohort (three of three) and a US traveller cohort (one of two).5,8 Two of eleven participants exposed to doxycycline for malaria prophylaxis acquired ESBL-producing E. coli.
Within this cohort, E. coli isolates from participants with ESBL-producing Enterobacteriaceae isolated either pre- or post-travel were sent for WGS. Overall, the majority of isolates were separated by a greater relatedness threshold than 10 SNPs indicating that post-travel ESBL-producing Enterobacteriaceae were strains acquired while travelling rather than through horizontal acquisition of an ESBL in a colonizing E. coli.25 Consistent with other studies, the most common ESBL were variants of blaCTX-M and blaTEM genes.8–13,15 In addition to beta-lactam resistance, acquisition of fluoroquinolone and tetracycline resistance was observed compared with pre-travel isolates.13 Exposure to ciprofloxacin for TD and acquisition of both fluoroquinolone and tetracycline resistance was the only significant relationship demonstrated (P = 0.05).
The study had several limitations. Although the targeted enrollment over the proposed study period was similar to studies with acquisition rates of 24–41%, our relatively low rate of ESBL-producing Enterobacteriaceae colonization resulted in a small cohort size and made the determination of risk factors for acquisition more difficult to determine as the study was not powered to assess individual risk factors. The population is closer in age (median 38 years) to an active duty population of combat related injury and infection (median 24–25) compared with previous studies of travel-related colonization in DoD beneficiaries (median 64).3,5 There is the possibility of decreased yield of organism in screening for ESBL-producing Enterobacteriaceae colonization through the use of perirectal swabs rather than stool culture. Numerous studies have however shown adequate sensitivity of perirectal swabs (90%) compared with stool culture in isolation of resistant Gram-negative bacilli, and other studies have successfully implemented perirectal swabs for screening purposes.5,9,21 Finally, no clinical significance can be drawn relating these episodes of incident colonization with potential future infectious complications.
Findings of this study support previously observed findings in travel-related colonization studies showing acquisition of antimicrobial resistance in US military personnel travellers. Significant findings included travel to South Asia was found to have higher incident colonization with ESBL-producing Enterobacteriaceae. Compared with civilian literature, acquisition rates of resistant organisms remain comparatively low in our population of study; although, a relationship to individual risk factors for acquisition of ESBL-producing Enterobacteriaceae could not be determined. This study suggests that US military personnel travelling on official duty, particularly in South Asia, are at risk for incident colonization of ESBL-producing Enterobacteriaceae. For the purposes of pre-travel counselling and military planning to reduce post-injury infection, further prospective studies are required to better determine individual risk factors for acquisition of ESBL-producing Enterobacteriaceae.
Author Contributions
Gregory Buchek, MD: Associate investigator, preparation and review of manuscript.
Katrin Mende, PhD: Associate investigator, study development, laboratory specimen processing, summarizing laboratory results, review of manuscript.
Kalyani Telu, MS: SAS Programmer, generate data sets, review of manuscript.
Susan Kaiser, BS: Laboratory specimen processing.
Jamie Fraser, MPH: Research coordinator, review of manuscript.
Indrani Mitra, MS: Data analyst/Epidemiologist, review of manuscript.
Jason Stam, BS: Bioinformaticist, whole genome sequencing.
Tahaniyat Lalani, MBBS, MHS: Principal investigator of TravMil, review of manuscript.
David Tribble, MD, DrPH: Associate investigator, review of manuscript.
Heather C Yun, MD: Principal investigator, preparation and review of manuscript.
Funding
This work (IDCRP-037) was conducted by the Infectious Disease Clinical Research Program (IDCRP), a DoD program executed by the Uniformed Services University of the Health Sciences (USU) through a cooperative agreement with The Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc. (HJF). This project has been supported with federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health (NIH), under Inter-Agency Agreement Y1-Al-5072 and the Department of Defense Global Emerging Infections Surveillance.
Conflict of interest
Author statements each declare that there is no conflict of interest.
Disclaimer
The contents of this publication are the sole responsibility of the author(s) and do not necessarily reflect the view, opinions or policies of Uniformed Services University of the Health Sciences (USUHS), the DoD, The Departments of the Army, Navy, or Air Force, Brooke Army Medical Center, the Henry M. Jackson Foundation for the Advancement of Military Medicine (HJF) and Walter Reed Army Institute of Research (WRAIR). Mention of trade names, commercial products or organization does not imply endorsement by the US Government. The investigators have adhered to the policies for protection of human subjects as prescribed in AR 70-25.
Author Statements
Gregory Buchek, MD: The authors have declared no conflicts of interest.
Katrin Mende, PhD: The authors have declared no conflicts of interest.
Kalyani Telu, MS: The authors have declared no conflicts of interest.
Susan Kaiser, BS: The authors have declared no conflicts of interest.
Jamie Fraser, MPH: The authors have declared no conflicts of interest.
Indrani Mitra, MS: The authors have declared no conflicts of interest.
Jason Stam, BS: The authors have declared no conflicts of interest.
Tahaniyat Lalani, MBBS, MHS: The authors have declared no conflicts of interest.
David Tribble, MD, DrPH: The authors have declared no conflicts of interest.
Heather C. Yun, MD: The authors have declared no conflicts of interest.
Contributor Information
Gregory Buchek, Brooke Army Medical Center, JBSA Fort Sam Houston, TX, USA; Uniformed Services University of the Health Sciences, Bethesda, MD, USA.
Katrin Mende, Brooke Army Medical Center, JBSA Fort Sam Houston, TX, USA; Infectious Disease Clinical Research Program, Department of Preventive Medicine and Biostatistics, Uniformed Services University of the Health Sciences, Bethesda, MD, USA; Henry M Jackson Foundation for the Advancement of Military Medicine, Bethesda, MD, USA.
Kalyani Telu, Infectious Disease Clinical Research Program, Department of Preventive Medicine and Biostatistics, Uniformed Services University of the Health Sciences, Bethesda, MD, USA; Henry M Jackson Foundation for the Advancement of Military Medicine, Bethesda, MD, USA.
Susan Kaiser, Brooke Army Medical Center, JBSA Fort Sam Houston, TX, USA; Infectious Disease Clinical Research Program, Department of Preventive Medicine and Biostatistics, Uniformed Services University of the Health Sciences, Bethesda, MD, USA; Henry M Jackson Foundation for the Advancement of Military Medicine, Bethesda, MD, USA.
Jamie Fraser, Infectious Disease Clinical Research Program, Department of Preventive Medicine and Biostatistics, Uniformed Services University of the Health Sciences, Bethesda, MD, USA; Henry M Jackson Foundation for the Advancement of Military Medicine, Bethesda, MD, USA.
Indrani Mitra, Infectious Disease Clinical Research Program, Department of Preventive Medicine and Biostatistics, Uniformed Services University of the Health Sciences, Bethesda, MD, USA; Henry M Jackson Foundation for the Advancement of Military Medicine, Bethesda, MD, USA.
Jason Stam, Walter Reed Army Institute of Research, Silver Spring, MD, USA.
Tahaniyat Lalani, Infectious Disease Clinical Research Program, Department of Preventive Medicine and Biostatistics, Uniformed Services University of the Health Sciences, Bethesda, MD, USA; Henry M Jackson Foundation for the Advancement of Military Medicine, Bethesda, MD, USA.
David Tribble, Infectious Disease Clinical Research Program, Department of Preventive Medicine and Biostatistics, Uniformed Services University of the Health Sciences, Bethesda, MD, USA.
Heather C Yun, Brooke Army Medical Center, JBSA Fort Sam Houston, TX, USA; Uniformed Services University of the Health Sciences, Bethesda, MD, USA.
References
- 1. Holubar M. Antimicrobial resistance: a global public health emergency further exacerbated by international travel. J Travel Med 2020; 27:1–2. [DOI] [PubMed] [Google Scholar]
- 2. Murray CK, Blyth DM. Acquisition of multidrug-resistant gram-negative organisms during travel. Mil Med 2017; 182:26–33. [DOI] [PubMed] [Google Scholar]
- 3. Tribble DR, Conger NG, Fraser S et al. Infection-associated clinical outcomes in hospitalized medical evacuees after traumatic injury: trauma infectious disease outcome study. J Trauma 2011; 71:S33–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hospenthal DR, Crouch HK, English JF et al. Multidrug-resistant bacterial colonization of combat-injured personnel at admission to medical centers after evacuation from Afghanistan and Iraq. J Trauma 2011; 71:S52–7. [DOI] [PubMed] [Google Scholar]
- 5. Blyth DM, Mende K, Maranich AM et al. Antimicrobial resistance acquisition after international travel in U.S. Travelers. Trop Dis Travel Med Vaccines 2016; 2:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vento TJ, Cole DW, Mende K et al. Multidrug-resistant gram-negative bacteria colonization of healthy us military personnel in the us and Afghanistan. BMC Infect Dis 2013; 13:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gilbert LJ, Li P, Murray CK et al. Multidrug-resistant gram-negative bacilli colonization risk factors among trauma patients. Diagn Microbiol Infect Dis 2016; 84:358–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tangden T, Cars O, Melhus A, Lowdin E. Foreign travel is a major risk factor for colonization with escherichia coli producing ctx-m-type extended-spectrum beta-lactamases: a prospective study with Swedish volunteers. Antimicrob Agents Chemother 2010; 54:3564–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Paltansing S, Vlot JA, Kraakman ME et al. Extended-spectrum beta-lactamase-producing enterobacteriaceae among travelers from the Netherlands. Emerg Infect Dis 2013; 19:1206–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Weisenberg SA, Mediavilla JR, Chen L et al. Extended spectrum beta-lactamase-producing enterobacteriaceae in international travelers and non-travelers in New York city. PLoS One 2012; 7:e45141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ostholm-Balkhed A, Tarnberg M, Nilsson M et al. Travel-associated faecal colonization with ESBL-producing enterobacteriaceae: incidence and risk factors. J Antimicrob Chemother 2013; 68:2144–53. [DOI] [PubMed] [Google Scholar]
- 12. von Wintersdorff CJ, Penders J, Stobberingh EE et al. High rates of antimicrobial drug resistance gene acquisition after international travel, the Netherlands. Emerg Infect Dis 2014; 20:649–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kennedy K, Collignon P. Colonisation with escherichia coli resistant to "critically important" antibiotics: a high risk for international travellers. Eur J Clin Microbiol Infect Dis 2010; 29:1501–6. [DOI] [PubMed] [Google Scholar]
- 14. Furuya-Kanamori L, Stone J, Yakob L et al. Risk factors for acquisition of multidrug-resistant enterobacterales among international travellers: a synthesis of cumulative evidence. J Travel Med 2020; 27:1–10. [DOI] [PubMed] [Google Scholar]
- 15. van der Bij AK, Pitout JD. The role of international travel in the worldwide spread of multiresistant enterobacteriaceae. J Antimicrob Chemother 2012; 67:2090–100. [DOI] [PubMed] [Google Scholar]
- 16. Sutter DE, Bradshaw LU, Simkins LH et al. High incidence of multidrug-resistant gram-negative bacteria recovered from afghan patients at a deployed US military hospital. Infect Control Hosp Epidemiol 2011; 32:854–60. [DOI] [PubMed] [Google Scholar]
- 17. Ake J, Scott P, Wortmann G et al. Gram-negative multidrug-resistant organism colonization in a us military healthcare facility in Iraq. Infect Control Hosp Epidemiol 2011; 32:545–52. [DOI] [PubMed] [Google Scholar]
- 18. Griffith ME, Ceremuga JM, Ellis MW et al. Acinetobacter skin colonization of us army soldiers. Infect Control Hosp Epidemiol 2006; 27:659–61. [DOI] [PubMed] [Google Scholar]
- 19. Scott P, Deye G, Srinivasan A et al. An outbreak of multidrug-resistant Acinetobacter Baumannii-Calcoaceticus complex infection in the us military health care system associated with military operations in Iraq. Clin Infect Dis 2007; 44:1577–84. [DOI] [PubMed] [Google Scholar]
- 20. Murray CK, Roop SA, Hospenthal DR et al. Bacteriology of war wounds at the time of injury. Mil Med 2006; 171:826–9. [DOI] [PubMed] [Google Scholar]
- 21. Lautenbach E, Harris AD, Perencevich EN et al. Test characteristics of perirectal and rectal swab compared to stool sample for detection of fluoroquinolone-resistant Escherichia coli in the gastrointestinal tract. Antimicrob Agents Chemother 2005; 49:798–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mende K, Beckius ML, Hospenthal DR. Recovery of multidrug-resistant bacteria from swabs stored for durations of 1 and 4 weeks under conditions mimicking long-distance-shipping conditions. J Clin Microbiol 2014; 52:1798–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Peirano G, Laupland KB, Gregson DB, Pitout JD. Colonization of returning travelers with ctx-m-producing Escherichia coli. J Travel Med 2011; 18:299–303. [DOI] [PubMed] [Google Scholar]
- 24. OstholmBalkhed A, Tarnberg M, Nilsson M et al. Duration of travel-associated faecal colonisation with esbl-producing enterobacteriaceae - a one year follow-up study. PLoS One 2018; 13:e0205504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schurch AC, Arredondo-Alonso S, Willems RJL, Goering RV. Whole genome sequencing options for bacterial strain typing and epidemiologic analysis based on single nucleotide polymorphism versus gene-by-gene-based approaches. Clin Microbiol Infect 2018; 24:350–4. [DOI] [PubMed] [Google Scholar]


