Abstract
Much of the visual diversity of angiosperms is due to the frequent evolution of novel pigmentation patterns in flowers. The gene network responsible for anthocyanin pigmentation, in particular, has become a model for investigating how genetic changes give rise to phenotypic innovation. In the monkeyflower genus Mimulus, an evolutionarily recent gain of petal lobe anthocyanin pigmentation in M. luteus var. variegatus was previously mapped to genomic region pla2. Here, we use sequence and expression analysis, followed by transgenic manipulation of gene expression, to identify MYB5a—orthologous to the NEGAN transcriptional activator from M. lewisii—as the gene responsible for the transition to anthocyanin-pigmented petals in M. l. variegatus. In other monkeyflower taxa, MYB5a/NEGAN is part of a reaction-diffusion network that produces semi-repeating spotting patterns, such as the array of spots in the nectar guides of both M. lewisii and M. guttatus. Its co-option for the evolution of an apparently non-patterned trait—the solid petal lobe pigmentation of M. l. variegatus—illustrates how reaction-diffusion can contribute to evolutionary novelty in non-obvious ways. Transcriptome sequencing of a MYB5a RNAi line of M. l. variegatus reveals that this genetically simple change, which we hypothesize to be a regulatory mutation in cis to MYB5a, has cascading effects on gene expression, not only on the enzyme-encoding genes traditionally thought of as the targets of MYB5a but also on all of its known partners in the anthocyanin regulatory network.
Keywords: anthocyanin, color patterning, differential expression, reaction-diffusion, Turing network, feedback loops, monkeyflower, Mendelian trait, evolution of novelty
Introduction
Anthocyanins, the red and purple pigments that color many plant tissues, are largely responsible for the tremendous visual diversity of floral pigmentation in angiosperms. The evolutionary lability of anthocyanin pigmentation, combined with its ecological and agricultural importance, has helped anthocyanin biosynthesis and regulation become a model for investigating the genetic mechanisms for trait evolution.
In dicots, the anthocyanin biosynthetic pathway is functionally divided into the early biosynthetic genes (EBGs) and the late biosynthetic genes (LBGs) (Dubos et al. 2010). The LBGs are coordinately activated by an MBW complex consisting of three kinds of proteins: a subgroup-6 R2R3 MYB protein (M) that directly binds the promoters of the enzyme-encoding genes, and bHLH (B) and WD40 (W) co-factors that physically interact with the MYB (Martin and Gerats 1993; Lepiniec et al. 2006; Petroni and Tonelli 2011; Xu et al. 2014; Zhu et al. 2015; Lloyd et al. 2017). An R3 MYB protein represses anthocyanin biosynthesis by sequestering the bHLH proteins and preventing them from taking part in MBW formation (Zhu et al. 2009; Yuan et al. 2013; Albert et al. 2014).
A growing body of empirical work has demonstrated that the anthocyanin regulatory network is highly interconnected. For example, the MBW complex triggers increased transcription of its own bHLH component, as well as increasing the expression of the R3 MYB repressor (Xu et al. 2015). In some cases, the MBW complex further upregulates the R2R3 MYB activator gene (Yuan et al. 2014). This positive feedback loop is essential to generating spatial patterns via a self-organizing series of molecular interactions described as a reaction-diffusion system.
In the 1950s, Alan Turing proposed such a reaction-diffusion model to explain the developmental origins of semi-regularly repeating patterns in biological organisms, such as the spots on a cheetah or the stripes on a zebra (Turing 1952), and the idea has been further developed by others (Gierer and Meinhardt 1972; Meinhardt and Gierer 2000; Meinhardt 2012; Ball 2015). In contrast, traits that lack a repeating element are generally explained in terms of positional specification, in which overlapping zones of gene expression dictate the location of a physical feature (Nijhout 1991; Gompel et al. 2005; Werner et al. 2010; Martins et al. 2017).
Within the monkeyflower genus Mimulus (synonym Erythranthe; Barker et al. 2012; Lowry et al. 2019), floral anthocyanin traits have evolved repeatedly (Grossenbacher and Whittall 2011) and both types of patterning appear to be involved. The developmental genetics of floral anthocyanins have been studied in the greatest detail in M. lewisii and M. guttatus. In both of these species, the R2R3 MYB activator gene NEGAN/MYB5 is part of a reaction-diffusion system that creates spots of red anthocyanin pigment in the nectar guide region (Ding et al. 2020). A paralagous R2R3 MYB gene, PELAN, similarly operates as part of an MBW complex in the petal lobe region of M. lewisii, with one important difference: PELAN lacks autocatalytic ability, and thus cannot initiate reaction-diffusion dynamics. Consistent with this finding, PELAN generates a continuous field of pigment within the positionally specified petal lobe region, rather than a repeated spotting pattern, and RNAi against PELAN causes a uniform lightening of pigmentation rather than the altered spatial pattern expected under a reaction-diffusion model (Yuan et al. 2014).
At first glance, the solid purple pigmentation of Mimulus luteus var. variegatus (Figure 1A) would appear to be a classic case of positional specification. This evolutionarily recent, Mendelian trait shows no spotting, striping, or any other kind of the repetition that is thought to be the hallmark of reaction-diffusion traits. Surprisingly, when M. l. variegatus is crossed to its yellow-petaled conspecific, M. l. luteus, the offspring of the cross feature multiple spots in their petal lobes (Figure 1B). This altered spatial pattern suggests that the gain of petal lobe anthocyanin in M. l. variegatus is in fact a cryptic reaction-diffusion trait. And indeed, while solid petal color across the genus Mimulus maps to the genomic region containing the non-autocatalytic gene PELAN (pla1 in Figure 1A), petal lobe pigmentation in M. l. variegatus maps to the region containing the reaction-diffusion activator gene NEGAN (pla2 in Figure 1A).
Figure 1.
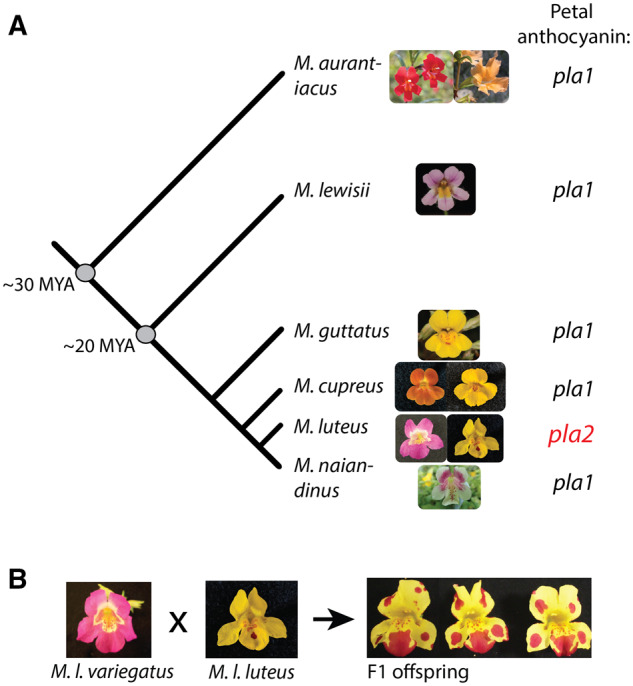
Petal anthocyanin pigmentation is evolutionarily labile within and between species of Mimulus. (A) Shifts in petal color in Mimulus have primarily been tracked to genomic region pla1. The causal gene in the pla1 genomic region has been identified as the R2R3 MYB gene MaMYB2 in M. aurantiacus (Streisfeld et al. 2013); its ortholog PELAN in M. lewisii (Yuan et al. 2014); and one or more R2R3 MYB genes at the orthologous genomic region in M. guttatus (Lowry et al. 2012), M. cupreus (Cooley et al. 2011), and M. naiandinus (A. Cooley, unpublished data). Genomic region pla2 contains MaMYB1 in M. aurantiacus; its ortholog NEGAN in M. lewisii; and MYB5 in M. guttatus, M. cupreus, M. l. variegatus, and M. naiandinus. The two varieties of M.luteus are the purple-flowered M. l.variegatus (left) and the yellow-flowered M. l. luteus (right). Photo credits: M. aurantiacus courtesy of Matt Streisfeld; M. naiandinus courtesy of Melia Matthews. Divergence times not to scale. (B) Crosses between the two varieties of M. luteus reveal a spotting mechanism underlying the apparently solid purple pigmentation of M. l. variegatus, consistent with Turing-style reaction-diffusion.
In this work, we explore the gain of an apparently unpatterned trait via a reaction-diffusion activator gene that, in other Mimulus species, creates spotting patterns. We find that MYB5a, the ortholog of NEGAN, has been co-opted for petal lobe anthocyanin pigmentation while also retaining its ancestral function of activating anthocyanin spot patterns in the nectar guide. We report the first evidence of a positive correlation in expression between the R2R3 MYB activator and its WD40 co-factor, suggesting a possible new feedback loop within the MBW transcription-factor complex. Using transcriptome data in combination with mathematical modeling, we model the interaction between the R2R3 MYB activator, MYB5a, and the R3 MYB repressor, RTO.
The results highlight how a reaction-diffusion network, and its two resulting phenotypes of nectar guide and petal lobe pigmentation, can respond in a dramatic and coordinated fashion to a relatively modest change in the expression of a single component part. The estimates of interaction strengths will help to inform and constrain future mathematical models of the evolutionary and developmental systems that give rise to the color pattern diversity of flowering plants. Finally, we note that petal lobe anthocyanin in M. l. variegatus, which looks to all appearances like a traditional positionally specified feature, is in fact controlled by a reaction-diffusion network. The existence of such “cryptic” reaction-diffusion traits suggests that the importance of this mechanism to evolutionary and developmental biology may be substantially greater than previously supposed.
Materials and methods
Plant sources and growth conditions
Seeds were originally collected from natural populations in central Chile in 2004, and were inbred for multiple generations prior to the work described here (Table 1). Seeds were sown in 2-in pots onto wet Black Gold potting soil (SunGro Horticulture, Agawam, MA, USA) or Miracle-Gro potting soil (Scotts Miracle-Gro Co., Marysville, OH, USA), and were misted daily until germination. For transgenic experiments, seedlings were transplanted into 6″ pots to promote large size. Plants were maintained in the Whitman College greenhouse under 16-hour days and were watered daily by an automatic misting system. After expansion of the first true leaves, fertilizer (Grow Big, FoxFarm Soil & Fertilizer Co., Arcata, CA, USA, N:P:K = 6:4:4; Open Sesame, FoxFarm Soil & Fertilizer Co., Arcata, CA, USA, N:P:K = 5:45:19; and/or Miracle-Gro Bloom Booster, Scotts Miracle-Gro Co., Marysville, OH, USA, N:P:K = 1:3:2) was applied two to three times weekly to promote growth and flowering.
Table 1.
Seed sources
| Taxon | Line ID | Generations inbred | Population | Location |
|---|---|---|---|---|
| M. luteus var. luteus | Mll-EY7 | 12 | El Yeso | 33.4°S, 70.0°W (2600 m) |
| M. luteus var. variegatus | Mlv-RC6 | 11 | Río Cipreses | 34.2°S, 70.3°W (1200 m) |
Locations from which seeds were collected are given as latitude, longitude (meters above sea level).
Expression analyses of candidate anthocyanin-activating genes
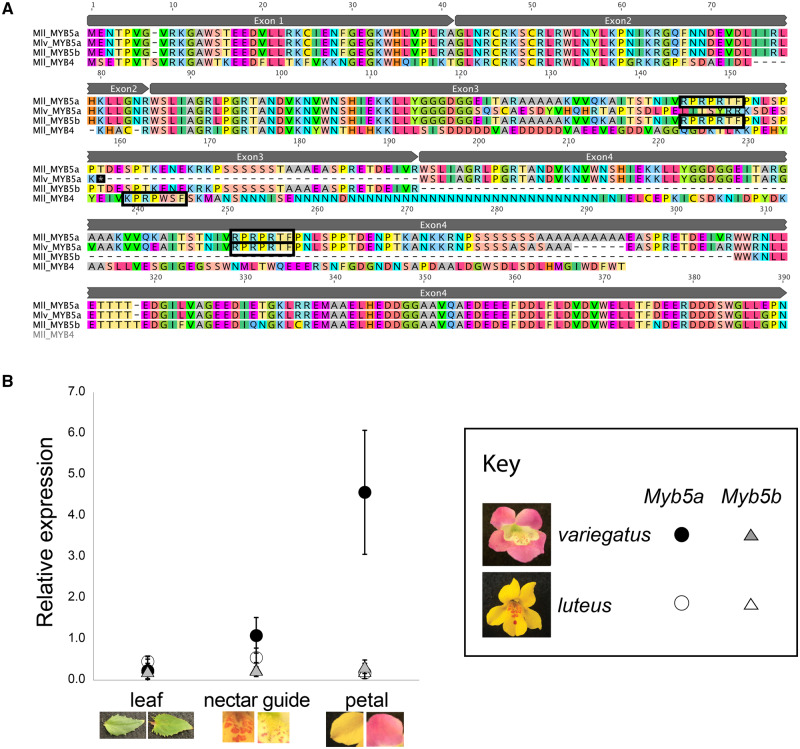
Two candidate anthocyanin-activating genes, MYB4 and MYB5, had been previously identified in M. l. variegatus by genetic mapping and sequence characterization (Cooley et al. 2011). Because members of the luteus complex are allotetraploids (Mukherjee and Vickery 1962; Vallejo-Marín et al. 2015; Edger et al. 2017), many genes have a homeologous copy located on a different chromosome, which could inflate expression estimates for a target gene if care is not taken to distinguish between the copies. In the process of investigating MYB5 in the sequenced M. l. luteus genome, we discovered its apparent homeolog, MYB5b (Figure 2A). MYB5 will therefore be referred to in this work as MYB5a, to distinguish it from its homeolog. In the M. l. luteus reference genome, the three exons that make up MYB5b are identical in their coding sequence to the first three exons of MYB5a. However, substantial divergence in the 5′-UTR (as well as downstream of the third exon) enabled the development of copy-specific primers, allowing us to disentangle the expression of MYB5a from its homeolog. We were not able to identify any homeolog of MYB4 in the M. l. luteus genome.
Figure 2.
Sequence and expression of MYB5a/NEGAN. (A) Alignment of MYB5a, MYB5b, and MYB4. Amino acid sequences were inferred by translating DNA sequences, obtained from the M. l. luteus genome and transcriptome (Edger et al. 2017; Stanton et al. 2017) and from PCR and Sanger sequencing of M. l. variegatus. Black rectangles identify putative Subgroup 6 motifs. Note that this motif, which strongly predicts ability to activate anthocyanin production, is missing from M. l. variegatus exon 3. Gray boxes denote exon boundaries for MYB5a and MYB5b. Created in Geneious 9.1 by Biomatters (www.geneious.com). (B) qRT-PCR estimates of MYB5a/NEGAN and MYB5b expression in M. l. variegatus and M. l. luteus. Each point indicates the mean of three technical replicates, ±95% CI, relative to the reference gene Actin and a reference pool of cDNA.
The gene responsible for activating anthocyanin biosynthesis in M. l. variegatus petal lobes is expected to be expressed in developing floral buds, during or just prior to the accumulation of visible anthocyanin pigment. Developmentally, this occurs approximately 72 hours before anthesis (Cooley et al. 2011), before the bud has emerged from the calyx. Endpoint and quantitative RT-PCR were used to investigate patterns of expression for each candidate gene, as well as for MYB5b, in developing floral buds and also young leaves of the purple-lobed M. l. variegatus compared to the yellow-lobed M. l. luteus. Primers were designed based on the sequenced genome of M. l. luteus (Supplementary Table S1), and their functionality in M. l. variegatus was confirmed by PCR prior to their use in expression analyses.
Although M. l. variegatus and M. l. luteus have striking differences in petal lobe pigmentation, with abundant anthocyanin production in the former and none in the latter, they share the trait of anthocyanin spotting in the nectar guide region of the corolla. Petals of developing flower buds were therefore dissected into lobe vs nectar guide components and the two tissue types were analyzed separately.
RNA was extracted using the Agilent Plant RNA Isolation Kit (Santa Clara, CA, USA). cDNA was synthesized using the ProtoScript First Strand cDNA Synthesis Kit from New England BioLabs, Inc. (Ipswich, MA, USA). Endpoint (qualitative) RT-PCR was performed on cDNA, with gDNA template as a control for primer efficacy. In addition to the MYB primers, either Actin or GAPDH primers were included as a control for successful cDNA synthesis (Supplementary Table S1). PCRs were performed with 0.2 mM dNTPs, 0.2 μM of each primer, 0.2 μL of G-Biosciences Taq DNA Polymerase per 25-μL reaction, and 1× G-Biosciences buffer. Amplification was performed with temperature settings of 95°C for 3 min, followed by 30 cycles of (95°C for 30 s, variable temperature for 30 s, 72°C for 1 min per kb of product), and then 72°C for 10 min. The annealing temperature was set to 2°C lower than the lowest melting temperature of the set of primers used in the PCR, and the minimum extension time used was 30 s.
To test for expression patterns consistent with the activation of petal lobe anthocyanin synthesis, quantitative RT-PCR was performed on MYB5a and MYB5b using SYBR-Green Brilliant III Ultra-Fast reagents (Agilent Technologies, USA), in optical 96-well plates (Greiner Bio-One, Belgium), on an MxPro3000p analyzer (Stratagene, USA). Four biological replicates were used for each taxon and tissue type, collected from different plants of the same inbred line. Each biological replicate was run in triplicate and the average of the three technical replicates was used for statistical analyses. Samples were amplified for 40 cycles of 95°C for 10 s and 51°C for 20 s. Melt curves were obtained by heating from 51°C to 95°C with a ramp speed of 0.01°C per second. The Actin gene was used as a normalizer. Raw qPCR fluorescence data were collected and analyzed by the default settings of the MxPro software v.4.10 (Agilent Technologies, USA). Amplification efficiencies for each primer pair were determined using the Ct (threshold) values obtained from a 1/4 dilution series (1:4, 1:16, 1:64, 1:256, and 1:1024).
Cloning and sequencing MYB5a splice variants
Two splice variants of M. l. variegatus MYB5a, containing exons 1-2-3 and 1-2-4 respectively, were PCR-amplified using primers MYB5a-10F and MYB5a-53R (Supplementary Table S1). PCR products were purified using a Genomic DNA Clean and Concentrate Kit (Zymo Research, Irvine, CA, USA), then cloned using the pGEM T-Easy kit and manufacturer’s protocol (Promega, Madison, WI, USA). pGEM vector containing the desired PCR product was transformed into competent JM109 E. coli cells (Promega, Madison, WI, USA). Colonies were evaluated using blue-white screening and PCR with M13 universal primers. Inserts of the expected length were sent to Eton Bioscience (San Diego, CA, USA) for Sanger sequencing.
Transgene construction
A previously built overexpression construct (Yuan et al. 2014), containing the complete NEGAN coding sequence from M. lewisii, driven by a 35S promoter, was used to test the sufficiency of MYB5a for activating petal lobe anthocyanin biosynthesis in M. l. luteus, a variety that normally lacks petal lobe anthocyanin pigmentation (Figure 1A).
Two RNAi constructs were built, targeting M. l. variegatus MYB5a-specific sequences in the 5′-UTR and fourth exon respectively, using the Gateway method (Earley et al. 2006; Katzen 2007). In brief, each fragment of interest was PCR-amplified using a forward primer with a 5′-CACC tag. The tag permitted the fragment to be directionally cloned into a pENTR entry vector (Thermo Fisher Scientific, Waltham, MA, USA); attL/attR recombination was used to transfer the correctly oriented fragment into the Gateway-compatible destination vector pB7GWIWG2(I) (VIB, Ghent, Belgium). M. l. variegatus genomic DNA for PCR was extracted using the E.Z.N.A. SP Plant DNA Kit (Omega Bio-Tek, Norcross, GA, USA). PCR amplification was performed using Phusion High Fidelity DNA Polymerase (New England Biolabs). The inserts and primers used for RNAi transgene construction are summarized in Table 2.
Table 2.
Construction of MYB5a/NEGAN RNAi transgenes for use in M. l. variegatus
| Insert | Insert length (bp) | Primers |
|---|---|---|
| M. l. variegatus 5′-UTR fragment | 307 | 24 F: caccCAAATTTTGGTTTTTGCCATTT |
| 37 R: TCGGTCAAATTAAATGCACA | ||
| M. l. variegatus exon 4 fragment | 377 | 60 F: caccGAAGAACGGCCAACGAC |
| 56 R: GCAGCTTCCCCGTTTCA |
Both inserts were cloned into the pB7GWIWG2(I) RNAi vector. Primers are labeled with a number and F for Forward or R for Reverse. Their sequences are shown 5′ to 3′. Nucleotides added to primers for purposes of directional cloning or restriction enzyme digests are underlined. bp, base pairs. Lowercase cacc- represents nucleotides added to primers to enable cloning into the pENTR entry vector.
Stable transformation
Agrobacterium-mediated plant transformation was performed using floral spray and vacuum infiltration as described in Yuan et al. (2013). Each experiment utilized approximately 12 robustly budding 2- to 6-month-old plants growing in 6″ pots. The commercial Agrobacterium tumefaciens strain LBA4404 (ThermoFisher Scientific) produced only two transformed seedlings from the infiltration of 67 plants (K. Om, senior thesis 2017). Subsequent experiments were performed using strain GV3101, and this yielded higher rates of transformed seedlings.
The concentration of Silwet-L77 (Lehle Seeds, Round Rock, TX, USA) used by Yuan et al. (2013) was 0.1%, or 1 mL per L of Agrobacterium culture. This unusually high concentration of Silwet was previously found to increase transformation success in Mimulus (Y.-W. Yuan, unpublished data). Acetosyringone was added to a final concentration of 0.1 M as in Yuan et al. (2013).
The predominantly outcrossing taxa M. l. luteus and M. l. variegatus were manually self-pollinated for 2 weeks after the first post-infiltration flower opened. Seeds were collected and densely sown in 25 cm × 50 cm flats. As soon as germination was observed, flats were sprayed daily with 1:1000 Finale (Farnam Companies, Inc., Phoenix, AZ, USA) to eliminate non-transgenic plants. Transgenic seedlings were grown to flowering and photographed. gDNA was extracted and the presence of the transgene was verified by PCR, using a forward primer targeted to the transgene and a reverse primer targeted to the attR2 region of the transgenic plasmid (Table 2 and Supplementary Table S1).
RNA extractions, cDNA synthesis, and endpoint RT-PCR were performed as described in the Expression Analyses section, to evaluate the expression of the target gene relative to an untransformed control plant. Two different primer pairs targeting MYB5a were used, 33 F-40R and 44 F-45R, along with primers to reference gene GAPDH (Stanton et al. 2017) as a positive control (Supplementary Table S1).
RNA extraction and library preparation for RNAseq
Transcriptome analysis was used to confirm the specific knockdown of target gene MYB5a by RNAi, and to explore the effects of this knockdown on other anthocyanin-related genes. Petal tissue for transcriptome sequencing was collected separately from three individuals of our highly inbred wild-type line of M. l. variegatus, RC6, and from three white-flowered offspring (Vrnai1.1, 1.3, and 1.5; Supplementary Figure S2) of the white-flowered RNAi transformant Vrnai1. For comparative purposes, one sample from M. naiandinus (Figure 1A) developing petals was also included.
Because the expression of anthocyanin-producing genes in Mimulus flowers is highest early in bud development, just before and after the first appearance of visible anthocyanin pigment (Cooley et al. 2011; Yuan et al. 2014), we used young buds that had not yet emerged from the calyx. Anthers were removed and the remaining petal tissue (including both lobe and nectar guide regions) was snap frozen in liquid nitrogen. RNA was extracted from each of the six samples using the Agilent Plant RNA Isolation Kit (Wilmington, DE, USA). A stranded RNA-Seq plus Ribo-Zero library preparation, followed by one lane of Illumina HiSeq 4000 50-bp single-read sequencing, was performed by the Duke University sequencing core facility (Durham, NC, USA). The FASTQ format RNA libraries were deposited in the NCBI Sequence Read Archive (SRA) repository under accession number PRJNA629107.
Transcriptome alignment
With a published M. l. luteus genome and gene feature annotation file available (Edger et al. 2017), we chose the genome splice-unaware mapping approach to assemble the M. l. variegatus transcriptomes to the M. l. luteus genome (Stein 2011; Williams et al. 2014). Sequencing quality control was performed by plotting the sequence nucleotide distribution and sequencing quality scores for all samples. Transcriptome libraries were aligned using Bowtie2(Version 2.3.5.1) under -very-sensitive-local mode (Langmead and Salzberg 2012) for best results in distinguishing homeolog expression, given that the M. luteus-group species are putative tetraploids (Mukherjee and Vickery 1962; Vickery 1995). Overall alignment rates were greater than 90% for all 7 samples. From the sequence alignment maps we counted reads per gene for all samples using the exon coordinates included in the published luteus GFF (general feature format) file. Read count was performed with software HT-seq (Version 0.11.2) (Anders et al. 2014) in Python. For detailed alignment documents, see the Supplementary Data. Pipeline code, alignment documents, and results are accessible through the GitHub repository (https://github.com/cici-xingyu-zheng/Luteus-RNA-seq) and for raw read count, see Supplementary Table S2.
Transcriptome analysis and functional annotation
Analyses of differential gene expression were conducted in DESeq2 (Version 1.26.0) in the R/Bioconductor environment (Love et al. 2014) (R Version 3.6.2; Bioconductor Release 3.10). After normalizing each gene by sequencing depth, we performed a principal component analysis plot of the seven transcriptome samples (three wild-type M. l. variegatus, three M. l. variegatus from RNAi line Vrnai1, and M. naiandinus as an outgroup). As expected, the samples clustered by treatment with the outgroup being an outlier (Supplementary Figure S2).
After performing a shrinkage estimation for dispersion to address the inaccuracy introduced by the small sample size and reduce the false positive rate (Love et al. 2014), the logarithmic fold change between RNAi treatment and control samples was used to evaluate differential expression. Following a false discovery rate control using the Benjamini–Hochberg Correction method, transcripts that were log-2-fold up- or down-regulated with a P-value <0.05 were considered to be significantly differentially expressed. Transcript expression profiles were normalized to reads per kilobase per million (RPKM) for further analysis and for plotting (Supplementary Table S3).
To annotate differentially expressed genes, all gene sequences from the M. l. luteus reference genome were translated to protein sequences using EMBOSS(6.5.7) “transeq” command and searched against Arabidopsis thaliana (TAIR) protein database (www.arabidopsis.org) using the “BLASTp” query with a e-value cut-off <10−6. For each coding sequence from M. l. luteus, the best-hit A. thaliana gene was used to annotate the transcript with a gene name and Gene Ontology (GO) annotation terms.
These annotations were then applied to the M. l. variegatus transcriptome data. GO enrichment analysis for was conducted, for genes that were differentially expressed between wild-type M. l. variegatus and the MYB5a RNAi line Vrnai1, using topGO (Version 2.36.0) with the “org. At.tair.db” database in R. Pathway enrichment tests were done using the KEGG (the Kyoto Encyclopedia of Genes and Genomes, https://www.genome.jp/kegg/;Release 93.0) pathway assignments for Arabadopsis with the KEGGREST Bioconductor package (Kanehisa et al., 2016).
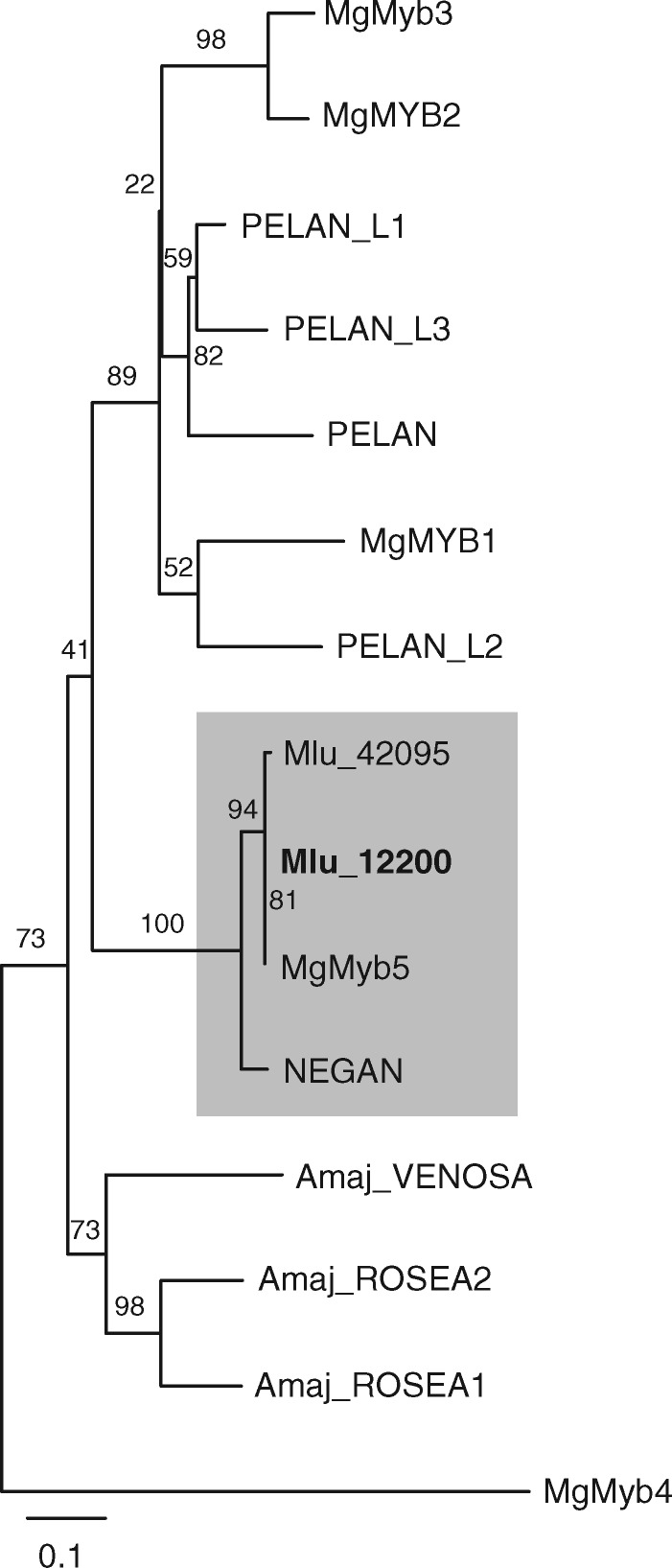
Tree construction
A gene tree was constructed to determine the relationship of Mlu_12200, the candidate MYB5a gene, to previously studied Mimulus MYBs. Sequences used to construct the gene tree in Figure 2 of Yuan et al. (2014) were downloaded from GenBank. A full list of GenBank sequence IDs are available in the Supplementary Table S4. Amino acid sequences were aligned with Clustal Omega and an ML gene tree was constructed using PhyML using a WAG substitution model and 100 bootstrap replicates. After initial tree construction, a subset of sequences that formed the core clade of MYBs of interest was realigned using Clustal Omega. The alignment cleaned manually to remove poorly aligned regions, and a tree was constructed using PhyML. The full gene tree from this analysis is available in the Supplementary Figure S3.
Regulatory and pathway analysis
In order to test for an effect of MYB5a down-regulation on the anthocyanin biosynthetic pathway, we examined the expression of genes corresponding to six core pathway enzymes that produce the cyanidin pigment found in M. l. variegatus (Cooley et al. 2008). These are: chalcone synthase (CHS), chalcone isomerase (CHI), flavonoid-3-hydroxylase (F3H), dihydroflavonol-4-reductase (DFR), anthocyanidin synthase (ANS), and UDP-flavonoid-3-glucosyl-transferase (UF3GT) (Holton and Cornish 1995; Tanaka et al. 2008). A list of M. l. variegatus transcripts with RPKM >1 and annotated with descriptions matching these enzymatic activities was obtained. Genes were considered truly orthogonal if they were the best hit in a reciprocal BLAST or had the same high BLAST score as a putative homeolog.
In addition to the enzyme-encoding genes, we were also interested in what other regulatory factors might be affected when we knock down the MYB5a transcriptional activator. In all species that have been examined, the regulatory complexes of the anthocyanin pathway include members of the MYB, basic helix-loop-helix (bHLH), and WD40 repeat families (Dooner et al. 1991; Koes et al. 2005). Therefore, we subset from the differentially expressed genes a list of genes annotated as members of these three transcriptional regulator families based on comparison to A. thaliana. Within this subset, we used reciprocal best 267 BLAST hits to identify orthologs to the genes that reported to be involved in 268 anthocyanin regulation in M. guttatus and M. lewisii.
Data availability
Seeds, bacterial strains, and plasmids are available upon request. Individual MYB4, MYB5a, and MYB5b coding sequences are located in GenBank. Whole-genome sequence for M. l. luteus was released by Edger et al. (2017) along with the draft genome and are located at https://datadryad.org/stash/dataset/doi:10.5061/dryad.d4vr0. Raw transcriptome data for M. l. luteus are published in Stanton et al. (2017) and are located at http://dx.doi.org/10.5061/dryad.84655. The M. l. variegatus transcriptome raw reads are available on the Sequence Read Archive (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA629107/). Transcriptome analysis code and alignment documents are accessible through the GitHub repository (https://github.com/cici-xingyu-zheng/Luteus-RNA-seq).
Supplementary material is available at figshare DOI: https://doi.org/10.25386/genetics.13350800.
Results
Expression of an alternately spliced R2R3 MYB, MYB5a/NEGAN, covaries with petal anthocyanin
In M. l. variegatus, only two anthocyanin-related genes were found within the genetically mapped petal anthocyanin locus pla2 (Cooley et al. 2011). The two genes, MYB4 and MYB5a, show phylogenetic similarity to the anthocyanin-activating Subgroup 6 of the R2R3 MYB gene family (Stracke et al. 2001; Cooley et al. 2011). Reciprocal BLAST searches of the M. lewisii genome indicate that MYB5a is orthologous to NEGAN, a gene shown to be responsible for nectar guide anthocyanin spots in M. lewisii (Yuan et al. 2014). NEGAN has previously been shown to be orthologous to MYB5a from M. guttatus (Yuan et al. 2014).
MYB4, while somewhat similar in sequence to MYB5a/NEGAN, has a nonconservative amino acid substitution in the functionally critical DNA-binding motif that is a hallmark of Subgroup 6 transcription factors (Figure 2A) and has a top BLAST hit on NCBI to M. guttatus LOC105953416, which encodes a GL1-like trichome differentiation protein. Additionally, unsuccessful efforts to amplify MYB4 out of M. l. variegatus floral bud cDNA, using primers that successfully amplify the gene from gDNA, indicate that this gene is not detectably expressed in developing M. l. variegatus buds (Supplementary Figure S1). Consistent with its similarity to a trichome differentiation gene, we did detect expression of MYB4 in leaf tissue (Supplementary Figure S1).
In contrast, quantitative RT-PCR revealed that MYB5a/NEGAN is strongly and specifically expressed in the anthocyanin-pigmented petal lobes of M. l. variegatus, while showing little to no expression in the non-anthocyanin-pigmented petal lobes of M. l. luteus (Figure 2B). Expression of MYB5a/NEGAN was modest in the anthocyanin-pigmented nectar guide tissue of both taxa, and undetectable in their leaf tissue (Figure 2B). The homeologous gene copy, MYB5b, showed consistently low expression across both taxa and all tissue types, at levels less than 10% of that observed in M. l. variegatus petal lobes (Figure 2B).
The anthocyanin-activating subgroup 6 R2R3 MYBs generally have three exons. The diagnostic Subgroup 6 amino acid motif is KPRPR[S/T]F in Arabidopsis, and it is located in the third exon (Stracke et al. 2001). In M. l. luteus and M. l. variegatus, however, MYB5a genomic sequence data revealed the presence of a fourth exon that is extremely similar in sequence to the third exon. In both taxa, this fourth exon appears to be intact and potentially functional, including the Subgroup 6 motif. The fourth exon differs between M. l. luteus and M. l. variegatus by 7 amino acid substitutions and one in-frame indel (Figure 2A). The third exon, although apparently intact in M. l. luteus, has a 26-bp frameshift-inducing deletion in M. l. variegatus that eliminates the critical Subgroup 6 domain, alters 35 amino acids, and creates a premature stop codon (Figure 2A). Exons one and two are identical between the taxa.
RT-PCR, followed by cloning and Sanger sequencing, revealed that exons 3 and 4 represent two alternate splice variants of MYB5a. An “exon 1-2-3” transcript was occasionally isolated from developing M. l. variegatus petals, but was never found in M. l. luteus, even in the anthocyanin-pigmented nectar guides. An “exon 1-2-4” transcript was reliably isolated from M. l. variegatus petal lobes and from the spotted nectar guide tissue of the corolla throat. In M. l. luteus, the exon 1-2-4 transcript was recovered from nectar guides but not from the (anthocyanin-free) petal lobe tissue. Thus, the exon 1-2-4 transcript of MYB5a appears to be consistently expressed in all anthocyanin-pigmented floral tissue, and absent from tissue that lacks anthocyanin.
MYB5a/NEGAN is sufficient and necessary for activating petal anthocyanin in M. luteus
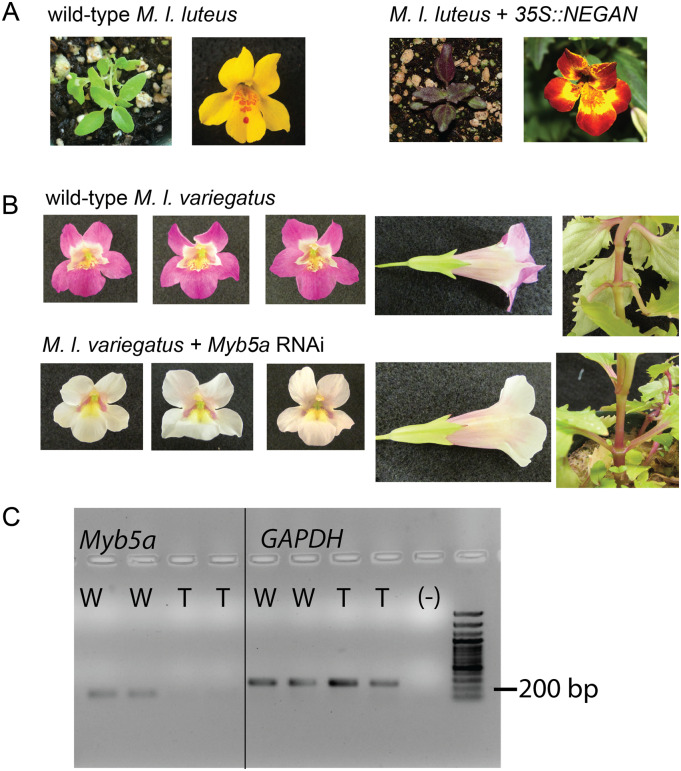
A pilot experiment showed that NEGAN from M. lewisii, when overexpressed with the 35S promoter, was sufficient to activate anthocyanin production in the petal lobe, as well as the nectar guide region and leaf tissue, of the normally yellow-petaled M. l. luteus (Figure 3A). Encouraged by this result, we used a 35S-driven RNAi transgene to test the hypothesis that the exon 1-2-4 transcript of MYB5a is necessary for petal lobe anthocyanin pigmentation in the purple-petaled M. l. variegatus. In the strongest RNAi line, “Vrnai1,” anthocyanin pigmentation was completely abolished in both the petal lobe and the nectar guide (Figure 3B). Anthocyanin pigmentation in the stems, the leaves, and the adaxial side of the corolla appeared to be unaffected, indicating that the effect of MYB5a knockdown is spatially specific. Successful knockdown of MYB5a in line Vrnai1 was demonstrated via qualitative RT-PCR (Figure 3C), and confirmed through transcriptome analysis (next section).
Figure 3.
MYB5a/NEGAN is sufficient and necessary for activation of petal lobe anthocyanins. (A) Overexpression of the coding sequence of MYB5a/NEGAN from M. lewisii in the normally yellow-petaled M. l. luteus activates anthocyanin biosynthesis in both leaf and petal tissue. (B) RNAi targeting MYB5a exon 4 in the normally purple-petaled M. l. variegatus eliminates anthocyanin biosynthesis in the petal lobes and nectar guides, but not elsewhere in the plant. The strongest of 8 RNAi lines, Vrnai1, is shown here. See Supplementary Data for images of the other RNAi lines. (C) Qualitative (end-point) RT-PCR on cDNA from developing petal lobes of wild type (W) and transgenic (T) M. l. variegatus reveals a reduction in MYB5a expression in the Vrnai1 transgenic line. Reference gene GAPDH was used as a positive control.
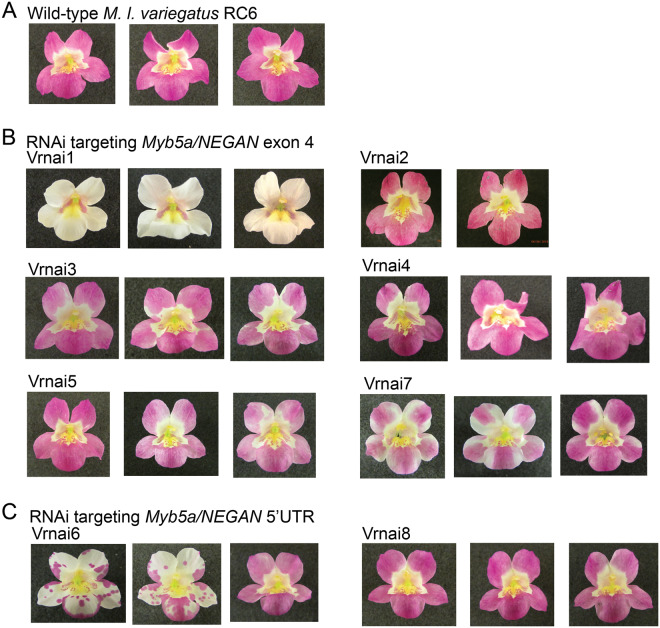
Considerable variation in the severity of the RNAi phenotype was observed across different transgenic lines, as is typical for this technique (Figure 4). From 12 wild-type M. l. variegatus transformed with RNAi targeting exon 4 of MYB5a, six putatively transgenic seedlings were obtained. Four of these (Vrnai1, Vrnai3, Vrnai5, Vrnai7) had a visible reduction in petal lobe anthocyanin pigmentation. From an additional 12 wild-type M. l. variegatus transformed with RNAi targeting the 5′-UTR of MYB5a, two putatively transgenic seedlings were obtained. One of these (Vrnai6) had a visible reduction in petal lobe anthocyanin pigmentation. Genotyping with a MYB5a forward primer and the attR2 transgene-specific reverse primer confirmed the presence of the RNAi transgene in all eight lines. In line Vrnai6, the RNAi phenotype declined over the lifetime of the plant (Figure 4). This phenomenon has been shown to be caused by methylation and epigenetic silencing, particularly of the 35S promoter, in both A. thaliana and the tobacco species Nicotiana attenuata (Daxinger et al. 2008; Mlotshwa et al. 2010; Weinhold et al. 2013).
Figure 4.
Transgene effects vary between and within insertion lines. (A) Wild-type M. l. variegatus. (B) Stable transgenic lines, for which the presence of an RNAi transgene targeting MYB5a/NEGAN exon 4 was confirmed by genotyping. (C) Stable transgenic lines, for which the presence of an RNAi transgene targeting MYB5a/NEGAN 5′-UTR was confirmed by genotyping.
The Vrnai1 plant was self-fertilized, and eight of the resulting seeds were planted and grown to flowering. As expected for a single-copy (heterozygous) insertion of a dominantly acting transgene in the Vrnai1 plant, we observed a 3:1 ratio of six white-flowered and two wild-type plants among the offspring (Supplementary Figure S2). The six white-flowered offspring exhibited slight anthocyanin mottling on some flowers (Supplementary Figure S2).
Transcriptomic differences between wild type and MYB5a RNAi lines of M. l. variegatus
We found a total of 632 genes that were significantly differentially expressed between wild-type and Vrnai1 lines of M. l. variegatus, with 346 genes down-regulated and 290 up-regulated in the Vrnai1 line (Supplementary Table S5). The differentially expressed genes are enriched in a variety of functions including response to UV-B, anthocyanin-containing compound biosynthesis, pollen exine formation, and phenylpropanoid biosynthetic process (P-value <0.001) (Supplementary Table S6). A pathway enrichment analysis identified two significantly enriched pathways (Supplementary Table S7): 1. cutin, suberine and wax biosynthesis, and 2. flavonoid biosynthesis, which includes the anthocyanin biosynthetic pathway (Holton and Cornish 1995).
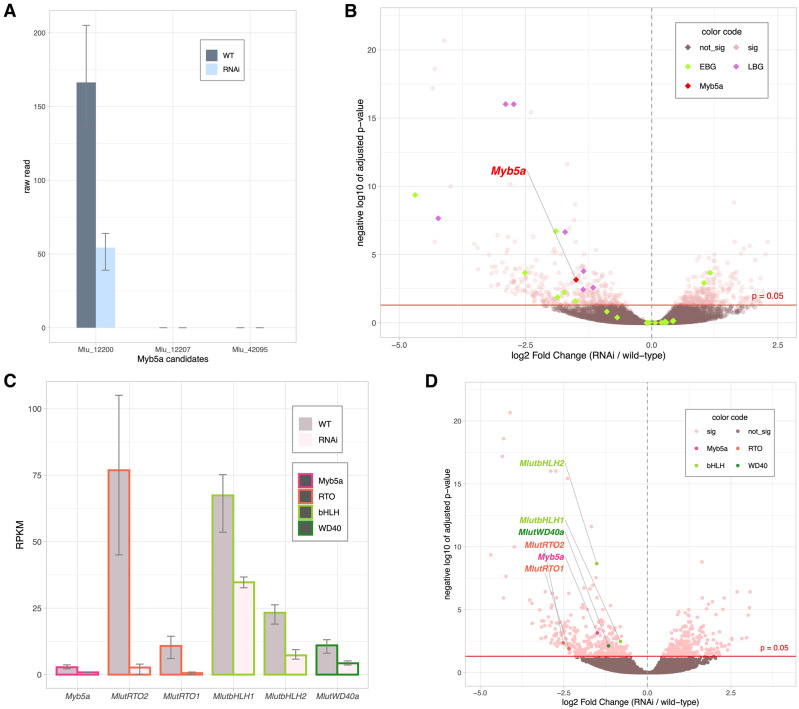
The M. l. variegatus MYB5a DNA sequence, which had previously been determined by PCR and Sanger sequencing, had best hits to three coding sequences in the M. l. luteus reference genome: Mlu 12200, Mlu 12207, and Mlu 42095. In the M. l. variegatus transcriptomes, the latter two transcripts were not detectably expressed in developing petal tissue, in either wild-type or RNAi lines of M. l. variegatus, eliminating them from further consideration as the petal anthocyanin activator. In contrast, Mlu 12200 showed expression patterns consistent with our expectation for MYB5a: it was robustly expressed in all six libraries, with threefold higher expression in the wild-type compared to Vrnai1 (Figure 5A). Both the Mlu 12200 and the Mlu 42095 transcripts from M. l. variegatus grouped phylogenetically with M. guttatus MYB5 and M. lewisii NEGAN (Figure 6).
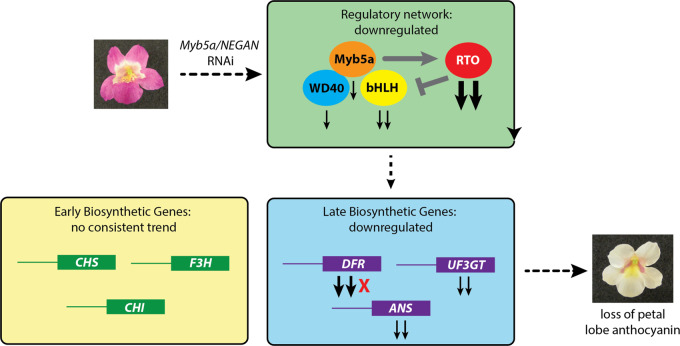
Figure 5.
RNAi knockdown of MYB5a (transcript Mlu 12200) coordinately down-regulates Late Biosynthetic Genes and the entire anthocyanin regulatory network. (A) Transcript Mlu 12200 corresponds to Myb5a/NEGAN. Bars show average read-count (n = 3) of wild-type and RNAi lines respectively, for candidate transcripts Mlu 12200, Mlu 12207, and Mlu 42095. Error bars mark the highest and lowest read counts; only Mlu 12200 has any mapped reads. (B) Differentially expressed anthocyanin biosynthesis genes. Green, EBGs (Early Biosynthetic Genes). Purple, LBGs (Late Biosynthetic Genes). Log-2-fold change (RNAi vs wild-type) is shown on the x axis, and −log 10 of adjusted P-value on the y axis. (C) Transcript level fold change of anthocyanin regulators. Normalized expression fold change is shown in RPKM (per million mapped reads). Error bars represent maximum and minimum expression levels among the samples (n = 3). (D) The six regulatory genes of interest (Myb5a, MlutbHLH1, MlutbHLH2, MlutRTO1, MlutRTO2, and MlutWD40a) are highlighted in the gene expression volcano plot.
Figure 6.
Maximum likelihood tree of anthocyanin-related R2R3 MYBs. Mg, Mimulus guttatus; Mlu, Mimulus luteus; Amaj, Antirrhinum majus. Mlu_12200 was identified as the anthocyanin activator MYB5a on the basis of its grouping with M. guttatus MYB5 and M. lewisii NEGAN, and its patterns of expression in developing petal tissue from wild-type vs MYB5a RNAi lines of M. luteus var. variegatus. Its paralog, Mlu_42095, is not detectably expressed in the developing anthocyanin-pigmented petals of M. l. variegatus. Scale bar indicates substitutions per site. See Supplementary Figure S3 for an untrimmed tree containing additional taxa and genes.
To check for off-target effects of the RNAi transgene, we asked whether any other Subgroup 6 R2R3 MYB genes were significantly down-regulated in the RNAi line. Five MYBs were identified among the down-regulated genes (Mlu 24690, Mlu 05348, Mlu 27563, Mlu 17841, and Mlu 00921), but none of them contained a Subgroup 6 motif. We conclude that the loss-of-pigment phenotype observed in line Vrnai1 is due to the reduction in MYB5a expression alone, and was not caused by incidental down-regulation of another anthocyanin-activating MYB gene.
RNAi knockdown of MYB5a reduces transcription of the late anthocyanin biosynthesis genes
We identified a total of 30 genes that are annotated to have enzymatic functions corresponding to the six core enzymes in the cyanidin pathway (Holton and Cornish 1995) (Supplementary Table S8). The EBGs (CHS, CHI, and F3H) are particularly enriched in copy number, each having more than the two homeologous copies expected in a tetraploid.
In eudicots, the anthocyanin biosynthetic genes are usually divided into two groups, the early (EBGs) and late (LBGs) biosynthetic genes, with the latter being tightly regulated by an MBW complex (Martin and Gerats 1993; Lepiniec et al. 2006; Dubos et al. 2010; Petroni and Tonelli 2011; Xu et al. 2014; Lloyd et al. 2017). In Arabidopsis, CHS, CHI, and F3H belong to the EBGs, producing the precursors of not only flavonoid pigments but other flavonol compounds. DFR, ANS, and UF3GT comprise the LBGs. Conserved as the enzymatic pathway is, though, the break point of regulation between early and late genes can vary across species (Streisfeld and Rausher 2009). We utilized our transcriptomic data to determine where the breakpoint occurs in M. l. variegatus.
Consistent with expectations from the literature, the anthocyanin biosynthetic genes in M. l. variegatus show a dichotomous pattern of response to MYB5a down-regulation. The genes earlier in the pathway—CHS, CHI, and F3H—had no consistent pattern of expression change (Figure 5B and Supplementary Figure S4), while the genes later in the pathway—DFR, ANS, and UF3GT—were consistently down-regulated in Vrnai1 (Figure 5B and Supplementary Figure S5). All copies of the LBGs had statistically significant change in the same direction (lower expression in the MYB5a RNAi line relative to wild-type), while the multiple copies of the EBGs showed a mix of expression increase, decrease, and no change in the RNAi line. This breakpoint is consistent with observations in the congeneric M. aurantiacus (Streisfeld et al. 2013). In summary, the results suggest that MYB5a controls anthocyanin production by regulating the LBGs, which include multiple copies of DFR, ANS, and UF3GT.
RNAi knockdown of MYB5a reduces transcription of other anthocyanin regulators
Within the list of differentially expressed genes, we identified 20 genes that are annotated to be transcription factors with MYB or helix-loop-helix (HLH) domains, or that encode a WD40 protein that could potentially belong to a MYB–bHLH–WD40 regulatory complex (Supplementary Table S9). Since different MBW genes regulate a variety of traits besides anthocyanin synthesis and gene functions might have diverged between Mimulus and Arabidopsis, we further compared this subset of genes to the MBW regulatory genes within M. lewisii, the Mimulus species with the most thoroughly characterized anthocyanin regulatory network. One WD40 gene (labeled MlutWD40a) was the best hit to M. lewisii MlWD40a, a gene that co-activates anthocyanin expression in the corolla. Two bHLH genes (MlutbHLH1 and MlutbHLH2) have the highest hit to the same bHLH gene in M. lewisii, ANbHLH3, indicating they are duplicates of each other. Interestingly, in M. lewisii, ANbHLH3 is not detectably expressed in the petal lobes, suggesting a functional diversification in the bHLH co-factors as well as in MYB5a/NEGAN (Yuan et al. 2014).
Recent studies show that single-repeat R3 MYBs are activated by the R2R3 MYB member of the MBW complex, and inhibit anthocyanin biosynthesis by directly interacting with the bHLH component of the same complex. In two Mimulus species, M. lewisii and M. guttus, the same R3 MYB (RTO) is reported to down-regulate floral anthocyanin production. We found that two R3 MYB genes, Mlu 13044 (MlutR3MYB1) and Mlu 33990 (MlutR3MYB2), both have the highest BLAST hit to this RTO gene and are strongly and significantly down-regulated in the MYB5a RNAi line, Vrnai1. Their down-regulation in Vrnai1 is consistent with the model that the R2R3 MYB, MYB5a, is an activator of its own inhibitor, although the impact of MYB5a on two apparently homeologous gene copies is unique.
The activating properties of the anthocyanin-activating R2R3 MYB on itself, its R3 MYB inhibitor, and its bHLH cofactor, have been previously shown in other taxa (Yuan et al. 2014; Xu et al. 2015). We hypothesized that in M. l. variegatus RNAi lines, the bHLH and RTO candidate anthocyanin regulators that we identified would show the same directional expression change as MYB5a (Li 2014). In fact, our expression results showed that down-regulating MYB5a in the RNAi line of M. l. variegatus led to a down-regulation in all the putative regulatory genes (Supplementary Table S10), surprisingly including the WD40 genes (Figure 5, C and D). The involvement of the WD40 genes in a positive feedback loop has not, to our knowledge, been reported previously. The activators of anthocyanin biosynthesis—including R2R3 MYB, bHLH, and WD40 transcription factors—were all two- to threefold down-regulated in the RNAi line. The two R3 MYB inhibitors showed much more dramatic effects: they were 19- and 29-fold down-regulated (Figure 5, C and D).
Discussion
Here we demonstrate that MYB5a is both sufficient and necessary for the evolutionarily recent, single-locus gain of petal lobe anthocyanin within M. luteus. We identify an unusual four-exon structure to this Subgroup 6 R2R3 MYB, along with evidence for alternative splicing. While the role of coding vs cis-regulatory evolution (Hoekstra and Coyne 2007; Stern and Orgogozo 2008) was not explicitly examined in this paper, we found that patterns of expression of MYB5a corresponded to pigmentation: the gene was strongly expressed in the heavily anthocyanin-pigmented petal lobes of M. l. variegatus; not detectably expressed in the petal lobes of M. l. luteus, a conspecific which lacks petal lobe anthocyanin; and modestly expressed in the partly pigmented nectar guides of both taxa. Further supporting the importance of expression change rather than coding change is that the orthologous coding sequence from the evolutionarily distant M. lewisii, which is approximately 20 MY divergent from M. l. variegatus (Nie et al. 2006), was highly effective at activating anthocyanin in the normally yellow-flowered M. l. luteus. We therefore predict that the causal mutation is either cis to MYB5a, or located in a trans-regulator of MYB5a that is closely linked to MYB5a within the mapped pla2 genomic region. Additional transgenic experiments, such as coding or promoter swaps, will help to clarify the extent and nature of MYB5a functional divergence in M. l. variegatus.
The adaptive significance of this evolutionary change is not yet fully understood. Flower color differences in Mimulus can have dramatic effects on pollinator behavior (Schemske and Bradshaw 1999). However, in wild populations in Chile, both M. l. luteus and M. l. variegatus are pollinated by the native Chilean bumblebee Bombus dahlbomii, which did not exhibit either collective or individual flower color preferences in a mixed-phenotype swarm of Mimulus hybrids (Cooley et al. 2008). Anthocyanins also protect plants against a variety of environmental stressors including extremes of heat, cold, drought, and UV radiation (Chalker-Scott 1999). The closely related M. cupreus, which recently gained petal lobe anthocyanins independently of M. l. variegatus (Figure 1A), does show greater drought tolerance than the yellow-flowered M. l. luteus (Stanton et al. 2016), but this work has not yet been extended to M. l. variegatus.
Our RNAi results indicate that MYB5a is responsible for anthocyanin pigmentation in two distinct regions of the M. l. variegatus corolla: the nectar guide and the petal lobe. The role of MYB5a/NEGAN in nectar guide spotting appears to be conserved across much of the genus. Its deployment to the petal lobes is unique: in diverse species including M. lewisii, M. aurantiacus, M. cupreus, and M. naiandinus (Cooley and Willis 2009; Cooley et al. 2011; Streisfeld et al. 2013; Yuan et al. 2014), petal lobe anthocyanin pigmentation is controlled by ancient paralogs of MYB5a/NEGAN, located at the functionally similar but genetically unlinked genomic region pla1 (Figure 1).
The R2R3 MYB gene PELAN, which is responsible for petal lobe pigmentation in M. lewisii, differs fundamentally from MYB5a/NEGAN in that it does not appear to function as part of a reaction-diffusion network (Yuan et al. 2014). Several observations indicate that MYB5a in M. l. variegatus is operating via reaction-diffusion, despite the apparently unpatterned nature of its petal lobe anthocyanin phenotype. (1) Crossing M. l. variegatus to the conspecific variety M. l. luteus reveals a pattern of anthocyanin spots in the petal lobes of the F1 hybrids, rather than a spatially uniform reduction in pigment intensity. (2) Knocking down MYB5a in M. l. variegatus eliminates the nectar guide spots, indicating that MYB5a has retained this ancestral ability which relies on reaction-diffusion (Ding et al. 2020). Our transcriptome data show that no other Subgroup 6 R2R3 MYB was significantly down-regulated in the MYB5a RNAi line, confirming that the loss of nectar guide spots is unlikely to be caused by an off-target effect of the RNAi transgene silencing a paralogous MYB gene. (3) The RNAi lines with weaker phenotypes did not show a spatially uniform reduction in pigment intensity, as has been observed for RNAi against the non-autocatalytic R2R3 MYB PELAN (Yuan et al. 2014). Rather, pigment was completely lost in portions of the petal, and the remaining pigment was sometimes expressed in a spotted pattern (i.e., line Vrnai6; Figure 4).
The genomic region harboring PELAN appears to have been used for petal pigmentation across Mimulus more often than the genomic region harboring MYB5a/NEGAN (Figure 1). More work is required to determine whether the inability of PELAN to self-activate is a recently derived trait within M. lewisii, or a conserved and evolutionarily ancient feature. It is interesting to note that the orthologous (pla1) genomic region in M. guttatus has been found to create semi-stochastically distributed red dots of anthocyanin pigment on the calyx (Lowry et al. 2012), suggesting that the causal gene in this instance is capable of reaction-diffusion-type patterning. While circumstantial, this discovery suggests that the lack of self-activation in PELAN in M. lewisii may be the exception rather than the rule for anthocyanin regulatory networks in Mimulus.
The repeated gains and losses of anthocyanin pigmentation across Mimulus (Grossenbacher and Whittall 2011), combined with the genomic and molecular resources that characterize the genus (Sobel and Streisfeld 2013; Twyford et al. 2015; Yuan 2019), create fertile ground for investigating how two major developmental approaches to trait formation—reaction-diffusion and positional specification—are utilized in the evolution of biological diversity. One challenge in studying anthocyanin pigmentation by either mechanism is that the number of interactions in the regulatory network makes it difficult to precisely predict the effects of perturbing any one component. Yet, the ability to make such predictions is central to generating and testing mechanistic hypotheses about how the system functions.
Mathematical modeling provides a potentially powerful solution to this problem. Yet, empirical estimates for many of the necessary parameters—such as the strength of the interactions among the different regulatory proteins—are lacking. Discovering the values of such parameters is essential in order to constrain a wide universe of possible models to a narrower range of biologically plausible ones.
One striking result from our transcriptome data was that an RNAi knockdown of MYB5a/NEGAN had a much stronger effect on the RTO repressor than on MYB5a itself (Figure 7). To further explore this result, we used mathematical modeling to identify conditions that could give rise to the observed patterns of gene expression change between wild-type and Vrnai1 lines of M. l. variegatus. We found that the experimental observation that a knock-down of MYB5a/NEGAN decreases the RTo concentration is consistent with activator-inhibitor systems that serve as minimal models of the MBW regulatory network (see Supplementary File S1). Working with such a minimal model, we found it straightforward to identify parameter values that demonstrate how a knock-down of MYB5a/NEGAN can result in a relatively larger fold change of the inhibitor concentration. Additionally, by changing parameters, we are able to demonstrate a wide variety of theoretical behaviors consistent with previous experimental results (Ding et al. 2020). According to this model, a knock-down of activator will always cause a decrease in inhibitor concentration; the relative size of the inhibitor change will vary depending on the parameter values.
Figure 7.
Solid black arrows indicate the direction of gene expression change, following RNAi knockdown of MYB5a/NEGAN in M. l. variegatus. The number of black arrows corresponds to the number of gene copies identified in the transcriptome. Red X, gene expression near zero in the RNAi line; thin black arrow, approximately 3-fold downregulation in the RNAi line; medium black arrow, approximately 9-fold downregulation; thick black arrow, more than 18-fold downregulation. Grey symbols denote positive and negative regulatory interactions. RTO is an R3 MYB protein that inhibits anthocyanin biosynthesis by sequestering bHLH proteins away from the MBW complex.
Overall, our transcriptome analyses and models highlight the inter-relatedness of expression patterns across the network of anthocyanin regulatory and biosynthetic genes (Figure 7). These results illustrate how the network can respond dynamically to expression changes of a single network component, creating an avenue for a relatively modest expression change—such as that caused by a single cis-regulatory mutation—to have a major effect on the transcriptome as well as on the ultimate phenotype.
Acknowledgments
The authors gratefully acknowledge assistance with molecular work from Baoqing Ding, Lauren Stanley, Rachel Eguia, Johanna Au, Jonah Rodewald, and Jeanette Schwensen. We thank Larry North for construction of a vacuum infiltration apparatus, Carter Cooke and Bella Rivera for greenhouse assistance, and Amy LaFountain and Qiaoshan Lin for assistance in obtaining transgenic seeds.
Funding
This material is based upon work supported by the Murdock College Research Program for Life Sciences Award 2015270, National Science Foundation microMORPH training grant, and National Science Foundation - Division of Environmental Biology award 1655311 to A.M.C.; and National Science Foundation - Division of Mathematical Sciences award 1951646 and National Institutes of Health - National Center for Complementary and Integrative Health award R01AT01816 to G.D.C.S.
Conflicts of interest
None declared.
Literature cited
- Albert NW, Davies KM, Lewis DH, Zhang H, Montefiori M, et al. 2014. A conserved network of transcriptional activators and repressors regulates anthocyanin pigmentation in eudicots. Plant Cell. 26:962–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Theodor PT, Huber WH. 2015. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball P. 2015. Forging patterns and making waves from biology to geology: a commentary on Turing (1952) ‘the chemical basis of morphogenesis’. Philos Trans R Soc B. 370:20140218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker W, Nesom G, Beardsley PM, Fraga NS. 2012. A taxonomic conspectus of Phrymaceae: a narrowed circumscription for Mimulus, new and resurrected genera, and new names and combinations. Phytoneuron 39:1–60. [Google Scholar]
- Chalker-Scott L. 1999. Environmental significance of anthocyanins in plant stress responses. Photochem Photobiol. 70:1–9. [Google Scholar]
- Cooley A, Carvallo G, Willis J. 2008. Is floral diversification associated with pollinator divergence? Flower shape, flower colour and pollinator preference in Chilean Mimulus. Ann Bot. 101:641–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooley AM, Modliszewski JL, Rommel ML, Willis JH. 2011. Gene duplication in Mimulus underlies parallel floral evolution via independent trans-regulatory changes. Curr Biol. 21:700–704. [DOI] [PubMed] [Google Scholar]
- Cooley AM, Willis JH. 2009. Genetic divergence causes parallel evolution of flower color in Chilean Mimulus. New Phytol. 183:729–739. [DOI] [PubMed] [Google Scholar]
- Daxinger L, Hunter B, Sheikh M, Jauvion V, Gasciolli V, et al. 2008. Unexpected silencing effects from t-DNA tags in Arabidopsis. Trends Plant Sci. 13:4–6. [DOI] [PubMed] [Google Scholar]
- Ding B, Patterson EL, Holalu SV, Li J, Johnson GA, et al. 2020. Two MYB proteins in a self-organizing activator-inhibitor system produce spotted pigmentation patterns. Curr Biol. 30:802–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooner HK, Robbins TP, Jorgensen RA. 1991. Genetic and developmental control of anthocyanin biosynthesis. Annu Rev Genet. 25:173–199. [DOI] [PubMed] [Google Scholar]
- Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, et al. 2010. MYB transcription factors in Arabidopsis. Trends Plant Sci. 15:573–581. [DOI] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, et al. 2006. Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45:616–629. [DOI] [PubMed] [Google Scholar]
- Edger PP, Smith R, McKain MR, Cooley AM, Vallejo-Marin M, et al. 2017. Subgenome dominance in an interspecific hybrid, synthetic allopolyploid, and a 140-year-old naturally established neo-allopolyploid monkeyflower. Plant Cell. 29:2150–2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gierer A, Meinhardt H. 1972. A theory of biological pattern formation. Kybernetik. 12:30–39. [DOI] [PubMed] [Google Scholar]
- Gompel N, Prud’homme B, Wittkopp PJ, Kassner VA, Carroll SB. 2005. Chance caught on the wing: cis-regulatory evolution and the origin of pigment patterns in Drosophila. Nature. 433:481–487. [DOI] [PubMed] [Google Scholar]
- Grossenbacher DL, Whittall JB. 2011. Increased floral divergence in sympatric monkeyflowers. Evolution. 65:2712–2718. [DOI] [PubMed] [Google Scholar]
- Hoekstra HE, Coyne JA. 2007. The locus of evolution: evo devo and the genetics of adaptation. Evolution. 61:995–1016. [DOI] [PubMed] [Google Scholar]
- Holton TA, Cornish EC. 1995. Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell. 7:1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. 2016. Kegg as a reference resource for gene and protein. Nucleic Acids Res. 44: D457–D462. [DOI] [PMC free article] [PubMed]
- Katzen F. 2007. Gateway® recombinational cloning: a biological operating system. Exp Opin Drug Discov. 2:571–589. [DOI] [PubMed] [Google Scholar]
- Koes R, Verweij W, Quattrocchio F. 2005. Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci. 10:236–242. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepiniec L, Debeaujon I, Routaboul J-M, Baudry A, Pourcel L, et al. 2006. Genetics and biochemistry of seed flavonoids. Annu Rev Plant Biol. 57:405–430. [DOI] [PubMed] [Google Scholar]
- Li S. 2014. Transcriptional control of flavonoid biosynthesis: fine-tuning of the MYB-bHLH-WD40 (MBW) complex. Plant Signal Behav. 9:e27522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd A, Brockman A, Aguirre L, Campbell A, Bean A, et al. 2017. Advances in the MYB–bHLH–WD repeat (MBW) pigment regulatory model: addition of a wrky factor and co-option of an anthocyanin MYB for betalain regulation. Plant Cell Physiol. 58:1431–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry DB, Sheng CC, Lasky JR, Willis JH. 2012. Five anthocyanin polymorphisms are associated with an R2R3-MYB cluster in Mimulus guttatus (Phrymaceae). Am J Bot. 99:82–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry DB, Sobel JM, Angert AL, Ashman T-L, Baker RL, et al. 2019. The case for the continued use of the genus name Mimulus for all monkeyflowers. Taxon. 68:617–623. [Google Scholar]
- Martin C, Gerats T. 1993. Control of pigment biosynthesis genes during petal development. Plant Cell. 5:1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins TR, Jiang P, Rausher MD. 2017. How petals change their spots: cis-regulatory re-wiring in Clarkia (Onagraceae). New Phytol. 216:510–518. [DOI] [PubMed] [Google Scholar]
- Meinhardt H. 2012. Turing’s theory of morphogenesis of 1952 and the subsequent discovery of the crucial role of local self-enhancement and long-range inhibition. Interface Focus. 2:407–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinhardt H, Gierer A. 2000. Pattern formation by local self-activation and lateral inhibition. Bioessays. 22:753–760. [DOI] [PubMed] [Google Scholar]
- Mlotshwa S, Pruss GJ, Gao Z, Mgutshini NL, Li J, et al. 2010. Transcriptional silencing induced by Arabidopsis t-DNA mutants is associated with 35s promoter siRNAs and requires genes involved in siRNA-mediated chromatin silencing. Plant J. 64:699–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee BB, Vickery RK. 1962. Chromosome counts in the section simiolus of the genus Mimulus (Scrophulariaceae). v. the chromosomal homologies of M. guttatus and its allied species and varieties. Madrono. 16:141–155. [Google Scholar]
- Nie Z-L, Sun H, Beardsley PM, Olmstead RG, Wen J. 2006. Evolution of biogeographic disjunction between Eastern Asia and Eastern North America in Phryma (Phrymaceae). Am J Bot. 93:1343–1356. [DOI] [PubMed] [Google Scholar]
- Nijhout H. 1991. Pattern formation in biological systems. In: Lectures in Complex Systems, SFI Studies in the Sciences of Complexity. Vol. IV. Nadel L, Stein D, editors. p. 159–187. Addison Wesley Publishing Company. [Google Scholar]
- Petroni K, Tonelli C. 2011. Recent advances on the regulation of anthocyanin synthesis in reproductive organs. Plant Sci. 181:219–229. [DOI] [PubMed] [Google Scholar]
- Schemske DW, Bradshaw H. 1999. Pollinator preference and the evolution of floral traits in monkeyflowers (Mimulus). Proc Natl Acad Sci U S A. 96:11910–11915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobel JM, Streisfeld MA. 2013. Flower color as a model system for studies of plant evo-devo. Front Plant Sci. 4:321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanton K, Valentin CM, Wijnen ME, Stutstman S, Palacios JJ, et al. 2016. Absence of postmating barriers between a selfing vs. outcrossing Chilean Mimulus species pair. Am J Bot. 103:1030–1040. [DOI] [PubMed] [Google Scholar]
- Stanton KA, Edger PP, Puzey JR, Kinser T, Cheng P, et al. 2017. A whole-transcriptome approach to evaluating reference genes for quantitative gene expression studies: a case study in Mimulus. G3 (Bethesda). 7:1085–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein LD. 2011. An introduction to the informatics of “next-generation” sequencing. Curr Protoc Bioinformatics. 36:11. [DOI] [PubMed] [Google Scholar]
- Stern DL, Orgogozo V. 2008. The loci of evolution: how predictable is genetic evolution? Evolution. 62:2155–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke R, Werber M, Weisshaar B. 2001. The R2R3-MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol. 4:447–456. [DOI] [PubMed] [Google Scholar]
- Streisfeld MA, Rausher MD. 2009. Altered trans-regulatory control of gene expression in multiple anthocyanin genes contributes to adaptive flower color evolution in Mimulus aurantiacus. Mol Biol Evol. 26:433–444. [DOI] [PubMed] [Google Scholar]
- Streisfeld MA, Young WN, Sobel JM. 2013. Divergent selection drives genetic differentiation in an R2R3-MYB transcription factor that contributes to incipient speciation in Mimulus aurantiacus. PLoS Genet. 9:e1003385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Sasaki N, Ohmiya A. 2008. Biosynthesis of plant pigments: anthocyanins, betalains and carotenoids. Plant J. 54:733–749. [DOI] [PubMed] [Google Scholar]
- Turing A. 1952. The chemical theory of morphogenesis. Philos Trans R. Soc. 13:5–72. [Google Scholar]
- Twyford AD, Streisfeld MA, Lowry DB, Friedman J. 2015. Genomic studies on the nature of species: adaptation and speciation in Mimulus. Mol Ecol. 24:2601–2609. [DOI] [PubMed] [Google Scholar]
- Vallejo-Marín M, Buggs RJ, Cooley AM, Puzey JR. 2015. Speciation by genome duplication: repeated origins and genomic composition of the recently formed allopolyploid species Mimulus peregrinus. Evolution. 69:1487–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickery RK. 1995. Speciation by aneuploidy and polyploidy in Mimulus (Scrophulariaceae). Great Basin Nat. 55:174–176. [Google Scholar]
- Weinhold A, Kallenbach M, Baldwin IT. 2013. Progressive 35s promoter methylation increases rapidly during vegetative development in transgenic Nicotiana attenuata plants. BMC Plant Biol. 13:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T, Koshikawa S, Williams TM, Carroll SB. 2010. Generation of a novel wing colour pattern by the Wingless morphogen. Nature. 464:1143–1148. [DOI] [PubMed] [Google Scholar]
- Williams AG, Thomas S, Wyman SK, Holloway AK. 2014. RNA-seq data: challenges in and recommendations for experimental design and analysis. Curr Protoc Hum Genet. 83:11.13.1–11.13.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Dubos C, Lepiniec L. 2015. Transcriptional control of flavonoid biosynthesis by MYB–bHLH–WDR complexes. Trends Plant Sci. 20:176–185. [DOI] [PubMed] [Google Scholar]
- Xu W, Grain D, Bobet S, Gourrierec JL, Thévenin J, et al. 2014. Complexity and robustness of the flavonoid transcriptional regulatory network revealed by comprehensive analyses of MYB–bHLH–WDR complexes and their targets in Arabidopsis seed. New Phytol. 202:132–144. [DOI] [PubMed] [Google Scholar]
- Yuan Y-W. 2019. Monkeyflowers (Mimulus): new model for plant developmental genetics and evo-devo. New Phytol. 222:694–700. [DOI] [PubMed] [Google Scholar]
- Yuan Y-W, Sagawa JM, Frost L, Vela JP, Bradshaw HD Jr. 2014. Transcriptional control of floral anthocyanin pigmentation in monkeyflowers (Mimulus). New Phytol. 204:1013–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y-W, Sagawa JM, Young RC, Christensen BJ, Bradshaw HD. 2013. Genetic dissection of a major anthocyanin qtl contributing to pollinator-mediated reproductive isolation between sister species of Mimulus. Genetics. 194:255–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H-F, Fitzsimmons K, Khandelwal A, Kranz RG. 2009. Cpc, a single-repeat R3 MYB, is a negative regulator of anthocyanin biosynthesis in Arabidopsis. Mol Plant. 2:790–802. [DOI] [PubMed] [Google Scholar]
- Zhu Z, Wang H, Wang Y, Guan S, Wang F, et al. 2015. Characterization of the cis elements in the proximal promoter regions of the anthocyanin pathway genes reveals a common regulatory logic that governs pathway regulation. J Exp Bot. 66:3775–3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Seeds, bacterial strains, and plasmids are available upon request. Individual MYB4, MYB5a, and MYB5b coding sequences are located in GenBank. Whole-genome sequence for M. l. luteus was released by Edger et al. (2017) along with the draft genome and are located at https://datadryad.org/stash/dataset/doi:10.5061/dryad.d4vr0. Raw transcriptome data for M. l. luteus are published in Stanton et al. (2017) and are located at http://dx.doi.org/10.5061/dryad.84655. The M. l. variegatus transcriptome raw reads are available on the Sequence Read Archive (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA629107/). Transcriptome analysis code and alignment documents are accessible through the GitHub repository (https://github.com/cici-xingyu-zheng/Luteus-RNA-seq).
Supplementary material is available at figshare DOI: https://doi.org/10.25386/genetics.13350800.