Abstract
Chemosensory neurons translate perception of external chemical cues, including odorants, tastants, and pheromones, into information that drives attraction or avoidance motor programs. In the laboratory, robust behavioral assays, coupled with powerful genetic, molecular and optical tools, have made Caenorhabditis elegans an ideal experimental system in which to dissect the contributions of individual genes and neurons to ethologically relevant chemosensory behaviors. Here, we review current knowledge of the neurons, signal transduction molecules and regulatory mechanisms that underlie the response of C. elegans to chemicals, including pheromones. The majority of identified molecules and pathways share remarkable homology with sensory mechanisms in other organisms. With the development of new tools and technologies, we anticipate that continued study of chemosensory signal transduction and processing in C. elegans will yield additional new insights into the mechanisms by which this animal is able to detect and discriminate among thousands of chemical cues with a limited sensory neuron repertoire.
Keywords: WormBook, chemosensation, signal transduction, taste, odorant, pheromone, C. elegans, olfaction, GPCR, sensory, signaling, gustation
Introduction
The first comprehensive review of C. elegans chemosensation was published in WormBook in 2006 (Bargmann 2006). This review summarized our understanding of chemosensation in the nematode at that time, beginning with work initiated in the 1970s when C. elegans was first being developed as a laboratory model system. In the 15 years since its publication, the number of labs studying chemosensation has grown considerably, along with our understanding of C. elegans nervous system function.
In this study, we focus specifically on behavioral responses of C. elegans to attractants and repellents, chemosensory neuron physiology, and chemosensory signal transduction molecules and pathways. We also briefly discuss behavioral plasticity, but only in the context of intracellular regulation of signaling cascades. By necessity, several salient topics have been omitted, including gas sensation, neuromodulation, and the mechanisms by which chemical information is processed and relayed to other neurons within sensory circuits (e.g., downstream interneurons). The ability of several C. elegans sensory neurons to detect multiple classes of stimuli (polymodality) also is not explicitly covered, but this ability suggests that, given a limited number of neurons, polymodality may be necessary to achieve maximum functionality. Other nonchemosensory functions of a subset of these neurons are described elsewhere (Goodman and Sengupta 2019).
Attractants and repellents
To effectively utilize C. elegans as a model system to study sensory neurobiological principles, systematic screens of worm responses to individual chemicals have been conducted over the years, beginning in the 1970s (Dusenbery 1973, 1974, 1975; Ward 1973). Table 1 provides a nonexhaustive list of water-soluble and volatile compounds that have been demonstrated to attract or repel wild-type animals (defined here as the Bristol N2 strain) in the laboratory. Furthermore, C. elegans can discriminate between many of these compounds (Chou et al. 1996; L'Etoile and Bargmann 2000). As in other animals, the behavioral responses of C. elegans to a specific chemical can depend on its concentration. For instance, a subset of the chemical cues that are attractive at low concentrations can elicit avoidance responses at high concentrations (Table 1).
Table 1.
A nonexhaustive list of compounds that attract or repel wild-type animals in the laboratory and the neurons demonstrated to detect them
| Chemical stimulus | Neuron(s) | Soluble (S) or Volatile (V) | Reference(s) |
|---|---|---|---|
| Attractants | |||
|
Cyclic nucleotides cAMP cGMP |
ASE (ADF, ASG, ASI) | S | |
|
Cations Na+ K+ |
ASEL (ADF, ASG, ASI) ASER (ASEL) |
S | |
|
Anions Cl− |
ASER (ADF, ASG, ASI) | S | |
| Basic pH | ASEL | S | |
|
Amino acids Lysine Histidine Cysteine Methionine |
ASE (ASG, ASI, ASK) | S | |
| Biotin | ASE (ADF, ASG, ASI) | S | (Bargmann and Horvitz 1991) |
| Pyrazine | AWA | V | (Bargmann et al. 1993) |
| Diacetyl (low) | AWA | V | (Bargmann et al. 1993) |
| Diacetyl (intermediate)a | AWA, AWC | V | (Chou et al. 2001) |
| 2,4,5-Trimethylthiazole (low) | AWA, AWC | V | (Bargmann et al. 1993) |
| Butyric acidb | AWA (AWC ?) | V | (Choi et al. 2018) |
| Isobutyric acid | AWA (AWC ?) | V | (Choi et al. 2018) |
| Benzyl proprionate | AWA, AWC | V | (Choi et al. 2018) |
| Benzaldehyde (low) | AWC (AWA) | V | |
| Isoamyl alcohol (low) | AWC (AWA) | V | (Bargmann et al. 1993) |
| 2-Butanone | AWCON | V | |
| Acetone | AWCON | V | |
| Dimethylthiazole | AWC | V | |
| 1-Methylpyrrole | AWC | V | (Choi et al. 2018) |
| 1-Pentanol | AWC | V | |
| 2-Cyclohexylethanol | AWC | V | (Choi et al. 2018) |
| 2-Ethoxythiazole | AWC | V | |
| 2-Isobutylthiazole | AWC (AWA ?) | V | |
| 2-Methylpyrazine | AWC (AWA ?) | V | (Choi et al. 2018) |
| 4-Chlorobenzyl mercaptan | AWC (AWA ?) | V | (Choi et al. 2018) |
| Benzyl mercaptan | AWC (AWA ?) | V | (Choi et al. 2018) |
| 2-Heptanone | AWCON | V | |
| 2,3-Pentanedione (low) | AWCOFF | V | |
| 2,3-Pentanedione (intermediate) c | AWA, AWC | V | (Chou et al. 2001) |
| Repellents (avoidance) | |||
| Acidic pH | ASH, ADF, ASK, ASE | S | |
| Basic pH (>10.5) | ASH | S | (Sassa et al. 2013) |
| Copper | ASH, ADL, ASE | S | |
| Cadmium | ASH, ADL, ASE | S | (Sambongi et al. 1999) |
| SDS |
ASH (ASK, ASI, ASJ) PHA, PHB (antagonistic) |
S | |
| Bitters quinine | ASH (ASK) | S | (Hilliard et al. 2004) |
| Diacetyl (high) | ASH | V | |
| 2,4,5-Trimethylthiazole (high) | V | ||
| Benzaldehyde (high) | ASH (AWB) | V | |
| Isoamyl alcohol (high) | ASH (ADL, AWB) | V | |
|
Alcohols 1-Octanol (100%) 1-Octanol (30%) |
ASH (ADL, AWB—off food) ASH |
V | |
|
Ketones 2-Nonanone |
AWB (ASH) | V | |
| Serrawettin W2 | AWB | S | (Pradel et al. 2007) |
| Phenazine-1-carboxamide | ASJ | S | (Meisel et al. 2014) |
| Pyochelin | ASJ | S | (Meisel et al. 2014) |
| Dodecanoic acid |
ASH (ADL ?, ADF ?) PHA PHB |
S | (Tran et al. 2017) |
The references include those first reporting behavioral response to the chemicals, as well as those demonstrating the neurons involved in the response. The roles of most neurons were shown by cell ablation, although some were revealed via genetic mutation or calcium imaging. Neurons with a more minor role are indicated by a smaller font. Question marks indicate neurons with a possible role in detecting a stimulus.
Chou et al. (2001) refers to 1:10 dilutions of diacetyl and 2,3-pentanedione as “high” concentration. We have indicated them here as “intermediate” to distinguish it from undiluted diacetyl and 2,3-pentanedione, which animals avoid (Yoshida et al. 2012).
Butyric acid was previously reported to be a neutral compound (Bargmann et al. 1993).
J. Thomas unpublished, cited in Bargmann et al. (1990).
Beyond having a catalog of the compounds that C. elegans can respond to, an understanding of what each compound might represent to the nematode in the wild provides context for its neuroanatomy, physiology, and sensory integration. In its natural habitat, C. elegans is typically associated with microbe-rich organic matter such as rotting fruit and vegetable matter (and also slugs) (Frézal and Félix 2015). A wide variety of bacterial strains have been found along with C. elegans in the wild, and several nonpathogenic nutritious bacterial strains (Alcaligenes sp. JUb4, Providenica sp. JUb5, Providencia sp. JUb39, and Flavobacteria sp. JUb43) release the “fruity” smelling attractive volatiles isoamyl alcohol, ethyl isobutyrate, and ethyl isovalerate (Samuel et al. 2016; Schulenburg and Félix 2017; Worthy et al. 2018a). The well-studied attractant diacetyl is also released from a Lactobacillus species that was found in rotting citrus (yazu) fruit that also contained C. elegans (Choi et al. 2016). While the natural prey of C. elegans have not been definitively identified (Schulenburg and Félix 2017), the volatile chemicals emitted by these bacteria likely provide long-range attractive cues for seeking food.
Not all soil microbes are beneficial for C. elegans, and there are nematocidal fungi and bacteria that exude chemical cues that C. elegans avoids. For example, the pathogenic bacteria Serratia marcescens releases the cyclic lipodepsipentapeptide compound serrawettin W2 (Pradel et al. 2007), the pathogen Pseudomonas aeruginosa emits phenazine-1-carboxamide (PCN) and the siderophore pyochelin (Meisel et al. 2014), and the nematocidal bacteria Streptomyces secretes dodecanoic acid (Tran et al. 2017)—all of which repel C. elegans. Aversive odorants such as 1-octanol and 2-nonanone may also indicate the presence of fungi or pathogenic bacteria (Kaminski et al. 1974; Sharpell 1985). C. elegans may also use chemical cues to be alerted to the presence of hungry nematode predators such as Pristionchus pacificus that release soluble repellent sulfolipids when they are starved (Liu et al. 2018b).
Interestingly, some nematocidal predators exploit innate attractive responses of C. elegans to specific compounds by releasing attractive chemicals. For example, at least one nematode-trapping fungus (Arthrobotrys oligospora) appears to lure its prey by releasing attractive volatile compounds that might mimic food and pheromone cues (Hsueh et al. 2017). In addition, the pathogenic bacterium B. nematocida B16 secretes an attractive odor bouquet that includes benzaldehyde and 2-heptanone (among others) that lures nematodes to their death via a “Trojan horse” mechanism (Niu et al. 2010).
Innate responses of C. elegans to chemicals can be modified by experience. The attractive chemicals butanone and acetone are emitted by the pathogenic bacteria S. marcescens and P. aeruginosa (Worthy et al. 2018b), and inexperienced worms seek out these odors. However, following pathogenic infection, animals learn to avoid these odors (Zhang et al. 2005). This plasticity provides a model for learning and vertical transmission of pathogenic bacterial memory (Moore et al. 2019). C. elegans is also able to associate chemicals with food or starvation and exhibit attraction or repulsion, respectively, to these conditioned chemicals (for examples, see Colbert and Bargmann 1997; Torayama et al. 2007; Kunitomo et al. 2013; Luo et al. 2014).
In addition to compounds produced by potentially pathogenic organisms or predators, C. elegans also avoids many compounds that are generally considered harmful at high concentrations (Table 1). These include heavy metals (e.g., copper, and cadmium), and plant alkaloids or derivatives (e.g., quinine) that are perceived as bitter by humans and are toxic for most animals (Sambongi et al. 1999; Hilliard et al. 2004). Taken together, the complex natural environment of C. elegans necessitates that these animals be able to sense and respond robustly and sensitively to a range of chemical cues for optimal survival and reproduction.
Assessing behavioral and neuronal responses
Behavioral strategies underlying C. elegans chemotaxis have been identified by studying animal movement in controlled spatial and temporal chemical gradients (Pierce-Shimomura et al. 1999, 2005; Iino and Yoshida 2009; Broekmans et al. 2016). The behavioral strategies used by C. elegans to migrate toward or away from favorable (attraction) and noxious (avoidance) chemical cues, respectively, are described in the Appendix. Here, we briefly outline the most common tools and paradigms for assessing behavioral responses, and refer the reader to the Behavior methods chapter (Hart 2006) for more detailed descriptions of chemosensory assays. Interested readers may also wish to consult these reviews for additional relevant information: (de Bono and Maricq 2005; Bargmann 2006; Bergamasco and Bazzicalupo 2006; Sengupta 2007; Hart and Chao 2010; Lockery 2011; Hobert 2013; Walker et al. 2017; Metaxakis et al. 2018).
Population assays provide good platforms to rapidly screen for mutations that disrupt sensory function, as well as to catalog chemicals that elicit behavioral responses. Typically, population assays are performed on agar-filled Petri dishes, with a gradient emanating from a point source of a stimulus (Bargmann and Horvitz 1991a; Bargmann et al. 1993). Uniform concentrations of soluble and/or volatile chemicals within quadrants of a Petri dish are also used to assess preferences (Wicks et al. 2000; Frøkjær-Jensen et al. 2008; Lee et al. 2009). These approaches can be high throughput, and allow the assessment of responses of tens to hundreds of animals in a single assay. The output behavior is either scored as an endpoint assay (often reported as a chemotaxis index) or tracked and assessed while the behavior is ongoing, thereby allowing a description of how an animal alters its locomotor behavioral strategies to respond to a stimulus over time (Brown et al. 2013; Husson et al. 2013; Tanimoto et al. 2017). The responses of single animals can also be assessed and have been used to quantitate avoidance behaviors. These assays typically measure the time for an individual animal to reverse from the aversive stimulus or report the percentage of animals that respond by reversing within a given timeframe (Troemel et al. 1995; Hart et al. 1999; Hilliard et al. 2002).
Changes in intracellular calcium levels are generally accepted as a useful readout for sensory neuron activity and are the most accessible surrogate for electrophysiological experiments in C. elegans. However, when interpreting calcium imaging data, as described below, it is important to note that there may be scenarios in which calcium signaling does not directly correlate with neuronal depolarization (Zahratka et al. 2015). To report changes in calcium, calmodulin-based fluorescent proteins have been used, including FRET-based “cameleon” (Miyawaki et al. 1997; Kerr et al. 2000; Suzuki et al. 2003; Fukuto et al. 2004; Hilliard et al. 2005) and single emission circularly permutated GFP proteins (GCaMP and its variants) (Romoser et al. 1997; Dana et al. 2019). An inverse-type reporter was also recently developed to more reliably quantify a drop in calcium following stimulation (Hara-Kuge et al. 2018). Importantly, one needs to be aware that if the reporter sequesters calcium, neurotransmission can be disrupted (Ferkey et al. 2007). Other readouts for neuronal activity/regulation include cyclic nucleotides, and cGMP levels can also be recorded (Couto et al. 2013; Shidara et al. 2017; Woldemariam et al. 2019). However, there may be subcellular differences in calcium or cyclic nucleotides, including plasma membrane versus the cell body, as well as differences in the cilia, dendrite, cell body, and axon to be considered (S. Woldemariam and N. L'Etoile, unpublished observations) (Shidara et al. 2017). Strains that express GCaMP in the nuclei of each neuron have been used to image the entire neural network in real time (Kato et al. 2015).
In addition to changes in calcium levels, opening of other nonspecific cation channels may contribute to membrane depolarization, and this needs to be considered. Thus, electrophysiological recordings provide the highest absolute and time-resolved insights into neuronal activity. Although technically difficult, this method has been used to provide high time resolution insights (Goodman et al. 1998, 2012) that include the finding that RMD (Mellem et al. 2008), AWA (Liu et al. 2018a), ASEL (Shindou et al. 2019), and other neurons (Faumont et al. 2012) fire action potentials and/or exhibit regenerative plateau potentials. To fill the gap between calcium imaging and electrophysiological recordings, genetically encoded voltage sensory hold promise and porting such sensors as the ASAP3 from mice could pave the way (Villette et al. 2019).
Microfluidics-based assays have been very useful for simultaneous recording of behavior and neuronal activity in real time (Albrecht and Bargmann 2011; Larsch et al. 2013). Briefly, animals are placed into a microfluidic device made of PDMS bonded to a coverslip and shaped into an arena within which the animals’ behavior can be observed. Within the arena, PDMS posts are arranged to provide an artificial “dirt” substrate that the animals can push against as they swim (Lockery et al. 2008). Ports flow buffer and stimulus such that they produce a laminar stream, allowing different spatiotemporal stimulus presentations. Using two cameras, one with a low and the other a high magnification objective, both locomotion and neuronal activity (e.g., calcium transients) can be monitored simultaneously (Larsch et al. 2013; Levy and Bargmann 2020).
Neurons and their contributions to chemosensation
There are 32 presumed chemosensory neurons in the hermaphrodite C. elegans nervous system. They are housed within the head amphid and inner labial organs, as well as the tail phasmid organs, and are directly or indirectly exposed to the environment (Ward et al. 1975; Ware et al. 1975; Perkins et al. 1986; White et al. 1986; Bargmann 2006; Inglis et al. 2006). An additional pair of amphid neurons (AFD) is thermosensory (Goodman and Sengupta 2019). Male-specific chemosensory neurons are described elsewhere (Barr et al. 2018). The functions of the eleven pairs of amphid and two pairs of phasmid neurons have been extensively characterized in the context of chemosensation, and are the focus here. The ADL, ADF, ASE, ASG, ASH, ASI, ASJ, and ASK neurons have simple, rod-like ciliated sensory endings that terminate within a channel formed by glial cells associated with the amphid sensilla. These neurons primarily detect soluble ligands, although ASH and ADL can also detect volatile ligands (Table 1). The AWA, AWB, and AWC amphid neurons embedded within the sheath glial cells also have ciliated sensory endings that are more complex, and these neurons appear to detect primarily volatile chemicals (Table 1). For a high-resolution ultrastructural analysis of the anterior endings of sensory neurons (and glia) see Doroquez et al. (2014) and Figure 1. The PHA and PHB nociceptive neurons in the phasmid sensilla have ciliated endings that terminate in the animal’s tail.
Figure 1.
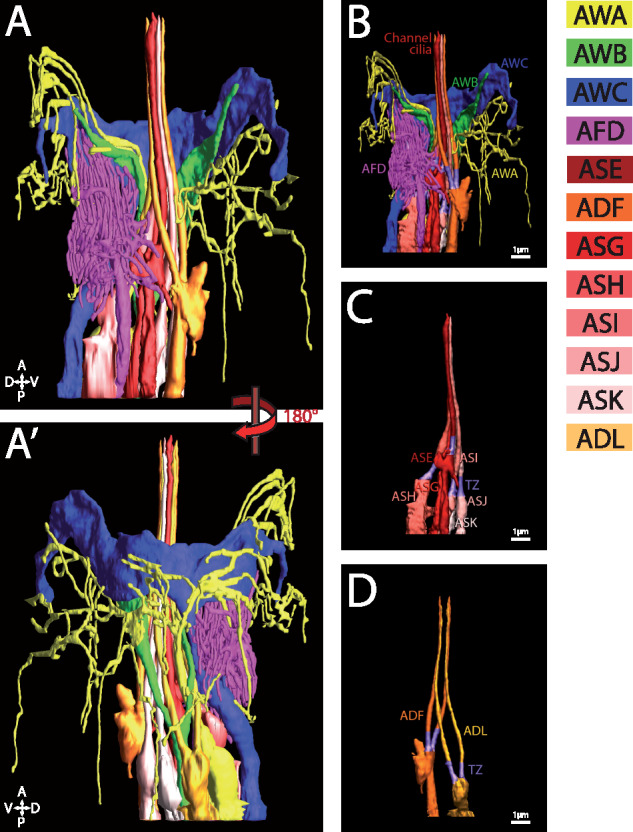
Cilia of amphid sensory neurons. (A and A′) 3 D reconstruction model of the sensory endings of 12 amphid neuronal cilia on the right side. Complex sensory endings of the winged cilia of AWA, AWB, and AWC and microvilli of the AFD neurons are shown in (B). Single (ASH, ASG, ASE, ASI, ASJ, and ASK) and double rod-shaped (ADF and ADL) channel cilia are shown in (C) and (D), respectively. Individual amphid neurons are color coded as indicated. Scale bar: 1 µm. Adapted from Doroquez et al. (2014).
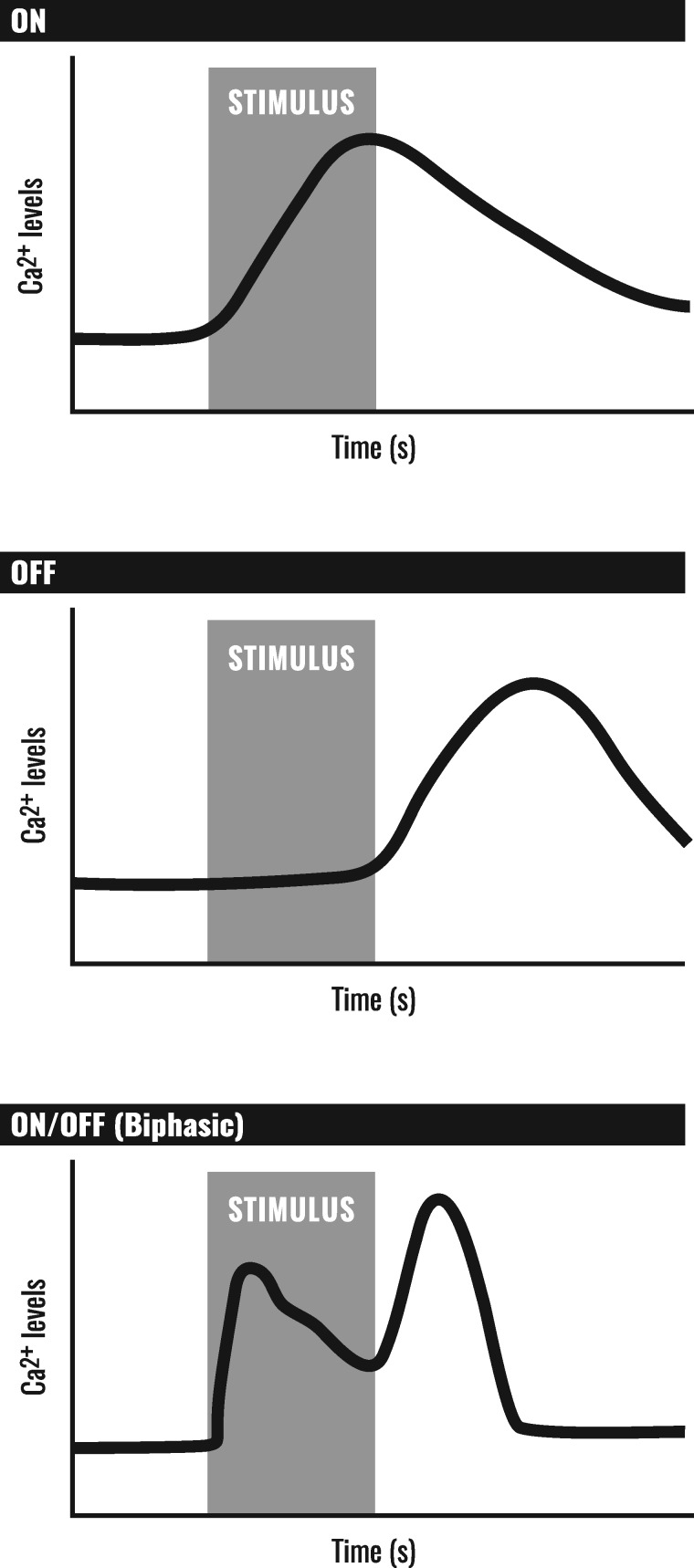
The majority of examined chemosensory neurons exhibit one of three distinct modes of response to chemical cues: (1) ON responses are increases in cytoplasmic calcium presumably due to depolarization that occurs when the concentration of the chemical cue increases; (2) OFF responses are increases in cytoplasmic calcium that occur when the concentration of the chemical cue decreases; (3) ON/OFF (biphasic) responses are increases in cytoplasmic calcium that occur in response to both the onset and offset (presentation and removal) of the chemical cue (Figure 2). In this section, we briefly discuss the response physiology, including calcium responses and electrophysiological potentials when known, of the amphid and phasmid chemosensory neurons. A detailed description of signal transduction molecules follows below.
Figure 2.
Calcium responses in sensory neurons. Sensory neurons can show a phasic increase in calcium levels upon presentation of stimulus (ON response), an increase in calcium upon removal of stimulus (OFF response), or an increase in calcium upon both the application and again with the subsequent removal of stimulus (ON/OFF or biphasic response).
ASH
The ASH sensory neurons are the main nociceptors in C. elegans. These neurons are considered to be “polymodal” because they detect a wide range of aversive stimuli, including both chemical and mechanical cues, similar to nociceptors in systems ranging from other invertebrates such as Drosophila (Tracey et al. 2003; Zhong et al. 2010; Im and Galko 2012; Johnson and Carder 2012) to vertebrates (Besson and Chaouch 1987; Treede 1999; Lee et al. 2005). Examples of ASH-detected chemical stimuli are included in (Table 1), and include high concentrations of several odorants that are normally attractive at lower concentrations.
The ASH sensory neurons exhibit a phasic ON response when presented with aversive chemical stimuli; for examples see (Fukuto et al. 2004; Hilliard et al. 2005; Mills et al. 2012; Tanimoto et al. 2017; Liu et al. 2018b). However, although the ON response appears to be the general rule for ASH, there are also experimental paradigms where an OFF (Thiele et al. 2009) or biphasic (ON and OFF) (Chronis et al. 2007; Kato et al. 2014; Wang et al. 2015) response has been observed. Analysis of ASH temporal filter properties suggests that these nociceptors integrate noxious cues over seconds to rapidly reach the activation threshold for avoidance behavior (Kato et al. 2014). ASH calcium signaling in response to chemosensory stimuli, and the effects of genetic mutations on it, are discussed extensively in the signal transduction section below. See also (Mirzakhalili et al. 2018) for additional computational modeling of ASH signaling.
While neuronal calcium flux is widely considered an indirect measure of neuronal activity, calcium transient amplitudes within the soma may not always be predictive of neuronal depolarization and synaptic signaling. For example, exposure to 1-octanol leads to ASH depolarization (Zahratka et al. 2015). But, surprisingly, while the neuromodulator serotonin (5-HT) potentiates ASH depolarization and ASH-mediated avoidance of 1-octanol, it actually decreases 1-octanol-evoked ASH calcium responses (Zahratka et al. 2015; Williams et al. 2018). These data have been interpreted to indicate that 5-HT enhances ASH excitability by suppressing a calcium-dependent inhibitory feedback loop (Williams et al. 2018). Thus, calcium signals and depolarization may not always be directly correlated.
ADL
In addition to their major role in pheromone detection (Pheromone), the ADL neurons play a minor role in chemical avoidance such that their contribution to chemical detection is often revealed only when they are ablated in combination with other sensory neurons. Single and multineuron ablation experiments have revealed a role for ADL in detecting several aversive stimuli (Table 1). In addition, ADL displays an ON response to repellent P. pacificus predator cue (Liu et al. 2018b). However, although neuronal ablation studies implicate ADL in 1-octanol avoidance (Troemel et al. 1995, 1997; Chao et al. 2004), ADL does not shows a change in calcium levels following 1-octanol exposure (Mills et al. 2012). It is possible that ADL does not respond directly to 1-octanol, or perhaps ablation of ASH causes compensatory changes in ADL and AWB (see below) responsiveness (Mills et al. 2012).
AWB
The AWB neurons detect volatile aversive chemicals. They are the primary mediators of 2-nonanone avoidance (Troemel et al. 1997) and play a minor role in the avoidance response to several other odorants (Table 1). Calcium imaging experiments revealed that AWB can respond to distinct stimuli in a variety of ways. For example, while they showed an ON response when presented with 50 mM NaCl (Zaslaver et al. 2015), these neurons are activated upon removal of 2-nonanone (OFF response) (Ha et al. 2010; Tanimoto et al. 2017). Similarly, AWB showed an OFF response upon removal of high-isoamyl alcohol (Yoshida et al. 2012) or removal of an Escherichia coli supernatant (Zaslaver et al. 2015). They also showed an unexpected ON/OFF biphasic response to a low concentration of isoamyl alcohol (10−4), which may be related to their possible (very minor) contribution to chemotaxis toward this odorant (Yoshida et al. 2012). Similar to ADL (above), 1-octanol exposure/removal did not elicit AWB calcium transients (Mills et al. 2012), although ablation studies suggest a minor role for AWB in 1-octanol avoidance (Troemel et al. 1997; Chao et al. 2004).
ASK
The ASK neuron pair was first shown to play a minor role in chemotaxis toward the amino acid lysine (Bargmann and Horvitz 1991a). Although it is unusual for a C. elegans sensory neuron to detect both attractive and aversive stimuli, ASK also contributes to the avoidance of several soluble stimuli, including SDS (Table 1). Interestingly, ASK showed an OFF response to lysine, but an ON response to SDS (Wakabayashi et al. 2009). Because ASK activation promotes reversals (Wakabayashi et al. 2004; Gray et al. 2005), suppression of calcium signaling by a chemoattractant and activation by a chemorepellent could both contribute to appropriate behavioral responses and locomotion strategies in complex chemosensory environments. For example, calcium imaging revealed that inhibition of ASK by the addition of diacetyl contributes to the disinhibition of the downstream interneuron AIA, allowing AIA to more reliably respond to diacetyl-evoked depolarization of AWA (Dobosiewicz et al. 2019). While the application of E. coli supernatant decreases ASK calcium levels, an elevation (OFF response) was seen upon its removal (Zaslaver et al. 2015). Similarly, an OFF response was also observed with removal of large (but not small) concentrations of suspended bacteria (Calhoun et al. 2015). ASK also contributes to pheromone detection (Pheromone).
AWA
The AWA olfactory neuron pair senses bacterially produced volatile cues to direct animals toward potential food sources (Bargmann et al. 1993; Larsch et al. 2013; Choi et al. 2016; Worthy et al. 2018a; Dobosiewicz et al. 2019; Table 1). However, there are sex differences in attraction to some odorants, including diacetyl (Lee and Portman 2007; White et al. 2007; Ryan et al. 2014; Barr et al. 2018).
AWA is an ON neuron that shows an elevated calcium levels in response to increases in diacetyl, pyrazine, 2-methylpyrazine, 2,4,5-trimethylthiazole and hexyl acetate (Shinkai et al. 2011; Larsch et al. 2013, 2015; Zaslaver et al. 2015; Itskovits et al. 2018; Liu et al. 2018a; Dobosiewicz et al. 2019). This neuron pair also shows an increase in calcium in response to the addition of E. coli supernatant, and a decrease in calcium upon its removal (Zaslaver et al. 2015). Activated AWA neurons signal to first order interneurons such as AIA that reduce turning probability, thereby elongating runs when an animal heads up the concentration gradient of an attractive chemical (Larsch et al. 2015).
As a food sensor, AWA's ability to detect volatiles in gradients that span large concentration ranges is likely to be important for an animal's survival. Indeed, the response properties of AWA enable animals to respond to odorants over a 100,000-fold range of concentrations (e.g., from as low as 11 nM up to 115 mM diacetyl) (Bargmann et al. 1993; Larsch et al. 2013). Calcium imaging showed that the AWA neurons themselves respond reliably over the same wide span of concentrations (Larsch et al. 2013, 2015), with oscillatory responses whose maxima remained constant and did not scale with the concentration of the odor the worm was exposed to (Larsch et al. 2015; Itskovits et al. 2018). Responses of these neurons to diacetyl sensitize rapidly at high concentrations, thereby allowing the AWA neurons to retain response sensitivity over a wide dynamic range. AWA responses also adapt to the rate of change in concentration rather than to the absolute concentration, which allows the animal to seek out odor concentrations that change most rapidly, thus allowing them to progress along the shortest route to an odor source (Itskovits et al. 2018). Interestingly, the oscillations of the left and right AWA neurons were anti-correlated, but between the two they exhibited calcium transients at each upstep of odor (Itskovits et al. 2018).
Electrophysiological recordings provided additional insights into how AWA may respond to odors over a broad dynamic range. These studies indicated that AWA fires bursts of 5–20 spikes in about 15% of trials, and these have some of the hallmarks of an action potential (Liu et al. 2018a); they are self-limiting, rising sharply then falling to a steady baseline, and they regenerate to recur as a train of spikes (Bean 2007). By imaging GCaMP while injecting current, an algorithm was trained to use the electrophysiological recording to detect spikes within the GCaMP traces. Applying this algorithm to GCaMP traces obtained when the AWA neurons were responding to intermediate concentrations of diacetyl uncovered spiking calcium signals; changes in diacetyl concentration elicited a similar spiking regime as seen with current injections (Liu et al. 2018a).
Electrophysiological investigations of AWA also revealed aspects of their responses that indicate how the neurons allow animals to ignore noise, either in the environment or generated by the animal's movement. The time threshold for AWA activation was long, about 300 ms, such that only stimuli that lasted for longer than a third of a second were able to trigger spiking (Liu et al. 2018a). This time lag was also sufficient to filter out changes in concentration that would be generated by the typical frequency of head swings generated by self-movement. This ability to filter out noise could be attributed to as yet unidentified potassium channels that increase the resistance of the AWA membrane and keep small fluctuating stimuli from depolarizing the cell (Liu et al. 2018a).
The calcium spikes generated by the AWA neurons adapt to the magnitude of the change in odor concentration over time (Liu et al. 2018a). Thus, turns should decrease as a function of an increase in odor concentration. However, because AWA activity is discontinuous, rather than directing uninterrupted runs, a decrease in AWA activity is predicted to allow turns to emerge even as an animal climbs a gradient (Itskovits et al. 2018). Thus, to model robust climbing of a gradient at higher odor concentrations, the spiking ON neuron pair had to be complemented with OFF neurons that had graded responses (Itskovits et al. 2018). The AWC neurons, with their response to intermediate concentrations of diacetyl, may fulfill this role (Dobosiewicz et al. 2019).
AWC
Many attractive odors are sensed by the paired AWC neurons (Table 1), which along with the AWA neurons are the main olfactory neurons in C. elegans (Bargmann et al. 1993). The two AWC neurons are not symmetric, as they express different G protein-coupled receptors (GPCRs) (Troemel et al. 1997; Bauer Huang et al. 2007; Vidal et al. 2018) and respond to different odorants (Table 1). Odorant bouquets from nutritive bacteria have been found to include known AWC-detected attractive volatiles (Worthy et al. 2018a). Some attractive chemicals are also released by nematophagus fungi (Hsueh et al. 2017) and pathogenic bacteria (Worthy et al. 2018b), which may coopt AWC-mediated attraction to lure C. elegans (Zhang et al. 2016). These normally attractive odors can become repulsive when worms are sickened or starved in their presence (Tsunozaki et al. 2008; Jin et al. 2016; Kaletsky et al. 2018). The AWC neurons still sense these chemicals under these conditions, but they instead direct repulsion (Tsunozaki et al. 2008; Jin et al. 2016).
Calcium imaging showed that both AWC neurons are OFF neurons (Chalasani et al. 2007, 2010). They are tonically active in buffer, showing low but constant activity that is silenced upon odor addition. Conversely, when odor (or E. coli supernatant) is withdrawn, both neurons show a sharp rise in calcium (Chalasani et al. 2007, 2010; Kato et al. 2014; Calhoun et al. 2015; Zaslaver et al. 2015; Cho et al. 2016; Hsueh et al. 2017; Hara-Kuge et al. 2018). The AWC neurons induce turns when they are active and forward runs when they are silent (Gray et al. 2005; Larsch et al. 2013; Gordus et al. 2015; Itskovits et al. 2018; Dobosiewicz et al. 2019), thereby directing runs up an attractive odor gradient.
The AWC calcium response to both odor exposure and removal is rapid (less than a second), robust and reproducible (Kato et al. 2014). Modeling showed that the speed of the response is sufficiently rapid, relative to head swings, to allow animals to track an odor gradient using the klinotaxis strategy (Izquierdo and Lockery 2010) (see Appendix), and this was experimentally verified using sensory signal transduction mutants (Kato et al. 2014). In addition, the response to a decrease in odor is graded such that it scales with both the amount of odor prior to the decrease and to the change in odor concentration (Cho et al. 2016). That is, the odor concentration is integrated over time to set the neuron's response threshold such that odor decreases that fall below the set point (Levy and Bargmann 2020) enable reliable gradient tracking.
The AWC neurons respond to some of the same odors as the AWA neurons, including diacetyl and isoamyl alcohol (Chou et al. 2001; Larsch et al. 2015; Itskovits et al. 2018; Worthy et al. 2018a). Interestingly, although AWA shows an oscillatory response to gradients of these odors, AWC responds with graded responses such that the AWC calcium signal is directly proportional to the change in stimulating odor concentration (Cho et al. 2016; Itskovits et al. 2018; Dobosiewicz et al. 2019; Levy and Bargmann 2020). When the responses of AWC and AWA are modeled together, they predict that animals are able to climb less continuous gradients more efficiently (Itskovits et al. 2018; Dobosiewicz et al. 2019). Furthermore, in contrast to a salt gradient, animals in an odor gradient (isoamyl alcohol) run faster up than down the gradient (Albrecht and Bargmann 2011). This also biases their movement toward the peak of the odor stimulus.
Levels of calcium and cGMP, the primary second messenger in AWC sensory signaling (see below), both initially decrease in the cilia and dendrites in response to onset of odor presentation. But, in the cell bodies, although calcium decreases, cGMP increases with odor onset (Shidara et al. 2017; Figure 3). How the cGMP sign is inverted between the cilia and the cell body is unclear, as is the physiological purpose of this inversion.
Figure 3.
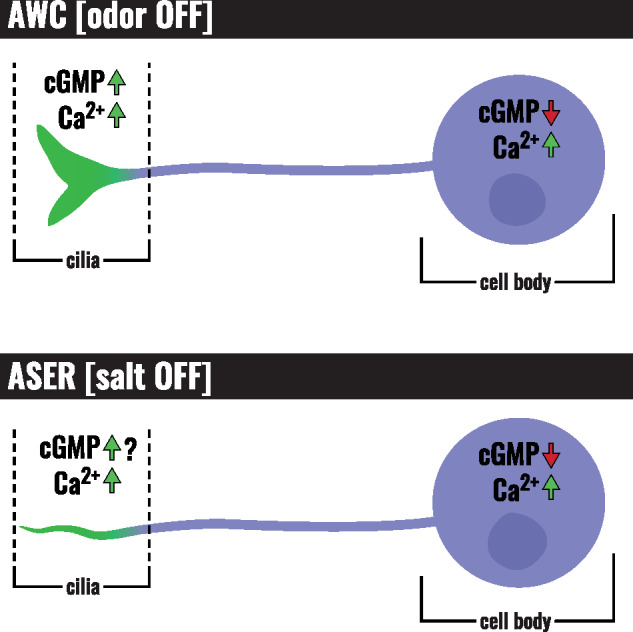
Second messenger levels in cilia versus soma. When odorant is removed from AWC, calcium levels increase in the cilia and the cell body, while cGMP levels increase slightly in the cilia but fall in the cell body. Likewise, when salt is removed from ASER, calcium levels increase in the cilia and cell body, and cGMP levels fall in the cell body. Preliminary data indicate that cGMP levels rise in the ASER cilia when salt is removed (S. Woldemariam and N. L’Etoile, personal communication).
ASI
The ASI sensory neurons play an important role in inhibiting entry into the alternative stress-resistant dauer stage under nondauer-inducing conditions (Bargmann and Horvitz 1991b; Schackwitz et al. 1996), and are the only source of DAF-7/TGF-β in C. elegans grown under standard conditions (Ren et al. 1996) (Pheromone). The ASIs also play a minor role in chemotaxis to water-soluble stimuli (Table 1), but their contribution is only revealed when ASE (major) and other sensory neurons (minor) are ablated (Bargmann and Horvitz 1991a; Kaufman et al. 2005). Ablation studies also showed a role for the ASI neurons in avoidance of worm extract (Zhou et al. 2017), SDS and P. pacificus predator cue (Liu et al. 2018b). They also promote P. aeruginosa avoidance, although it is not clear whether this is via direct detection of pathogen-released chemical cues (Cao et al. 2017). Calcium imaging experiments revealed that the ASI displays an ON response to HB101 E. coli bacteria (Gallagher et al. 2013), OP50 E. coli bacteria (Calhoun et al. 2015) and supernatant (Zaslaver et al. 2015), and Luria Broth (LB) (Gallagher et al. 2013; Davis et al. 2018), suggesting a role in food sensation. The activation of ASI by external nutrients promotes satiety quiescence (You et al. 2008; Gallagher et al. 2013). In addition, the aversive stimulus CuSO4 elicits an OFF response in ASI that allows them to modulate copper nociception in a reciprocal inhibition circuit with the primary copper detectors, the ASH neurons (Guo et al. 2015). P. pacificus predator cue also elicits an OFF calcium response in ASI (Liu et al. 2018b).
ADF
The ADF neurons are the only serotonergic sensory neurons in the hermaphrodite (Sze et al. 2000) and appear to be tonically active (Thiele et al. 2009). Thus, they are uniquely positioned to respond to environmental cues and modulate chemosensory behavioral responses. In the larva, they inhibit entry into the dauer stage under nondauer-inducing conditions (Bargmann and Horvitz 1991b; Schackwitz et al. 1996) (Pheromone). In adults, under “normoxic” conditions the ADF neurons (along with ASG and ASI) also play a minor role in chemotaxis to water-soluble stimuli (Table 1), but their contribution is only revealed when ASE (major) and other sensory neurons (minor) are ablated (Bargmann and Horvitz 1991a). However, under hypoxic conditions (e.g., those created by high bacterial metabolism in enclosed spaces) the role of ADF (and ASG) in salt chemotaxis may be enhanced due to the upregulation of 5-HT in these neurons (Pocock and Hobert 2010). Calcium imaging experiments have revealed that the ADF neurons show an ON response to E. coli supernatant (Zaslaver et al. 2015), and they respond directly to repellent levels (1/100) of isoamyl alcohol and indirectly to copper (Shao et al. 2019). ADF activation by these stimuli in turn inhibits the ASH nociceptors to modulate aversive chemosensory responses (Shao et al. 2019). The ADF neurons also show a calcium ON response to NaCl upsteps, although their activation may not be the result of direct stimulation in this context; ADF may be postsynaptic to a salt-sensitive neuron(s) (Thiele et al. 2009).
ASG
The ASGs play a minor role in inhibiting entry into the dauer stage under nondauer-inducing conditions (Bargmann and Horvitz 1991b; Schackwitz et al. 1996). In addition, under ambient (“normoxic”) oxygen conditions, the ASG neurons (along with ADF and ASI) play a minor role in chemotaxis to water-soluble stimuli (Table 1), but their contribution is only revealed when ASE (major) and other sensory neurons (minor) are ablated (Bargmann and Horvitz 1991a). However, under hypoxic conditions the role of ASG (and ADF) in salt chemotaxis may be enhanced due to the upregulation of 5-HT biosynthesis in these neurons (Pocock and Hobert 2010). Surprisingly, in contrast to the cell ablation results, calcium imaging (under normoxic conditions) did not reveal ASG calcium transients in response to either NaCl upsteps or downsteps (Thiele et al. 2009; Jang et al. 2019). However, the ASG neurons do show spontaneous calcium fluxes independent of salt stimulation, and both the frequency and average size of the activity peaks were higher after salt conditioning under starvation conditions (Jang et al. 2019). Thus, via their contribution to switching an animal’s navigation direction relative to a salt gradient, ASG activity may help animals to avoid salt concentrations associated with starvation (Jang et al. 2019).
ASJ
The major role of the ASJ neurons is to regulate dauer entry and exit (Pheromone). ASJ promotes dauer formation, such that killing these neurons significantly impaired the ability of wild-type animals to form dauers when exposed to dauer pheromone (Schackwitz et al. 1996). ASJ also promotes dauer recovery, and when the ASJ neurons are ablated animals permanently arrest in the dauer stage (Bargmann and Horvitz 1991b). In addition to these roles in the regulation of the dauer state, the ASJ neurons mediate avoidance of P. aeruginosa, most likely by detecting both secondary metabolites (Meisel et al. 2014) and nitric oxide (Hao et al. 2018) produced by these bacteria (Table 1). They also contribute to the avoidance of SDS and P. pacificus predator cue (Liu et al. 2018b). Calcium imaging experiments revealed that the application of the P. aeruginosa secondary metabolite PCN led to an increase in ASJ calcium levels (Meisel et al. 2014), as did presentation of 50 mM NaCl, pH 5 or E. coli supernatant (Zaslaver et al. 2015). Alternatively, an OFF response was seen upon removal of P. pacificus predator cue (Liu et al. 2018b). ASJ may also play a very minor role in chemotaxis to some water-soluble stimuli (Bargmann and Horvitz 1991a; Kaufman et al. 2005).
ASE
The left and right ASE neurons signal to both shared and distinct interneurons (Cook et al. 2019) (see also http://wormwiring.org) and they respond to different chemicals (Table 1). The left and right ASE neurons also express different genes, including receptor guanylyl cyclases (rGCs) that may be tuned to detect these distinct stimuli (Chang et al. 2003; Ortiz et al. 2009; Smith et al. 2013). In addition to this profound difference in sensory function, the two neurons differ in size (subtly) and electrophysiological properties (Pierce-Shimomura et al. 2001; Goldsmith et al. 2010).
The left and right ASE neurons also differ in their contribution to the locomotor strategies utilized during salt chemotaxis. ASEL responds to an increase in cations and its activity correlates with runs up the gradient, while ASER responds to decreases in anions by initiating pirouettes and decreasing run length (Figure A1). Calcium imaging studies (Pierce-Shimomura et al. 2001; Suzuki et al. 2008; Kunitomo et al. 2013; Luo et al. 2014; Wang et al. 2017; Lim et al. 2018; Shindou et al. 2019) and electrophysiology (Shindou et al. 2019) corroborate the finding that ASEL is an ON cell that depolarizes and increases intracellular calcium in response to increases in salt concentration (upsteps), while ASER is an OFF cell that depolarizes and increases intracellular calcium with decreases in salt concentration (downsteps). ASEL and ASER respond to changes in salt with a transient influx of calcium that marks the onset of the change (salt up or down, respectively) (Suzuki et al. 2008; Oda et al. 2011; Luo et al. 2014; Lim et al. 2018; Shindou et al. 2019). This combination of ON and OFF sensory cells underlies the ability of animals to reliably track a smooth gradient, composed of dissolved ion pairs, to its source (Pierce-Shimomura et al. 1999, 2001; Suzuki et al. 2008; Iino and Yoshida 2009; Izquierdo et al. 2015).
Electrical responses to current injection reveal that ASEL and ASER signal in a nonlinear regenerative manner (Goodman et al. 1998; Shindou et al. 2019) generating plateau potentials (Lockery and Goodman 2009). Responses to current injection depend on extracellular sodium and calcium in concert, but are robust to removal of either alone (Shindou et al. 2019). This observation suggests that voltage- and/or calcium-dependent channels underpin nonlinear regenerative signaling. Salt upsteps also evoke plateau potentials in ASEL and the probability of triggering this response is proportional to the change in salt concentration (Shindou et al. 2019), providing a mechanism by which ASEL detects and signals the changes in external salt concentration that drive chemotaxis. Additional channels are likely to act in concert with the voltage-gated calcium channel (VGCC) EGL-19 to allow triggering of neurotransmission.
Within ASEL, the salt upstep signal is seen as an influx of calcium in sensory cilium, dendrites, soma and axons (Lim et al. et al. 2018; Shindou et al. 2019). As described further below, cGMP is the primary second messenger in salt sensory transduction. The cGMP signal at the sensory cilia is translated into changes in intracellular calcium dynamics and further amplified via VGCCs (Shindou et al. 2019). Interestingly, although calcium levels increase in the ASEL soma as a result of a salt upstep, cGMP levels decrease (Woldemariam et al. 2019). Similarly, ASER somal calcium rises and cGMP falls in response to a salt downstep (Woldemariam et al. 2019; Figure 3). However, the mechanism underlying the opposite calcium and cGMP changes in the soma of these neurons is currently unclear.
The ASE neurons also allow an animal to tune its response to salt such that it will become attracted to the salt concentration associated with food experience (Kunitomo et al. 2013; Luo et al. 2014). Imaging ASEL and ASER calcium levels as the animal is exposed to abrupt downsteps (Kunitomo et al. 2013) or is traversing a more natural gradient (Luo et al. 2014) revealed that ASER changes the dynamics of its responses to decreases and increases in salt as a function of the salt concentration at cultivation. ASER is most active in response to decreases in salt when the animal is below this set point, driving the animal to higher salt by increasing turning (Kunitomo et al. 2013). But, when the animal is at or above the set point and tracks to a lower salt concentration, similar downsteps in salt evoke smaller (Kunitomo et al. 2013) and more complex (Luo et al. 2014) calcium transients.
PHA/PHB
The PHA and PHB neurons are located in the phasmid sensory organs of the tail of C. elegans, and their role in chemosensation was first shown in 2002 (Hilliard et al. 2002). Although ablation of PHA and PHB did not affect SDS avoidance, their ablation in combination with ASH (or ASH and ASK) leads to a stronger avoidance response than ablation of ASH alone (or ASH and ASK) (Hilliard et al. 2002). This suggested that PHA/PHB antagonize SDS avoidance that is mediated by the amphid neurons (Table 1), and that the decision to initiate backward locomotion (reversal) is based on the integration of sensory information from the head and the tail (Hilliard et al. 2002; Oren-Suissa et al. 2016). Shared connections with command interneurons in hermaphrodites further support this model (White et al. 1986) (and wormwiring.org). In addition, PHA and PHB also mediate avoidance of dodecanoic acid presented to the tail (Tran et al. 2017).
Calcium imaging experiments have shown that PHA and PHB act as polymodal nociceptors, with an ON response to SDS, aversive odors (1-octanol), high isoamyl alcohol, alkaline pH (12), high osmolarity and harsh touch (Zou et al. 2017). For each of these stimuli, the responses of PHA and PHB were similar (Zou et al. 2017). cGMP imaging of PHB also indicated that SDS triggers an increase in cGMP (Woldemariam et al. 2019), which could drive the opening of cyclic nucleotide-gated (CNG) channels that function in the phasmids (Hilliard et al. 2002). In contrast, the application of copper decreased calcium levels in PHA/PHB, while copper removal led to an increase in calcium levels (OFF response) (Zou et al. 2017). However, while the decrease in calcium signaling appears to be cell autonomous, the OFF response was abolished in unc-31 mutant animals lacking neuropeptidergic signaling, suggesting that PHA/PHB may be, in part, postsynaptically activated by copper removal via neuropeptides (Zou et al. 2017). No calcium transients were observed in response to quinine or acidic pH (Zou et al. 2017).
Chemosensory signal transduction molecules
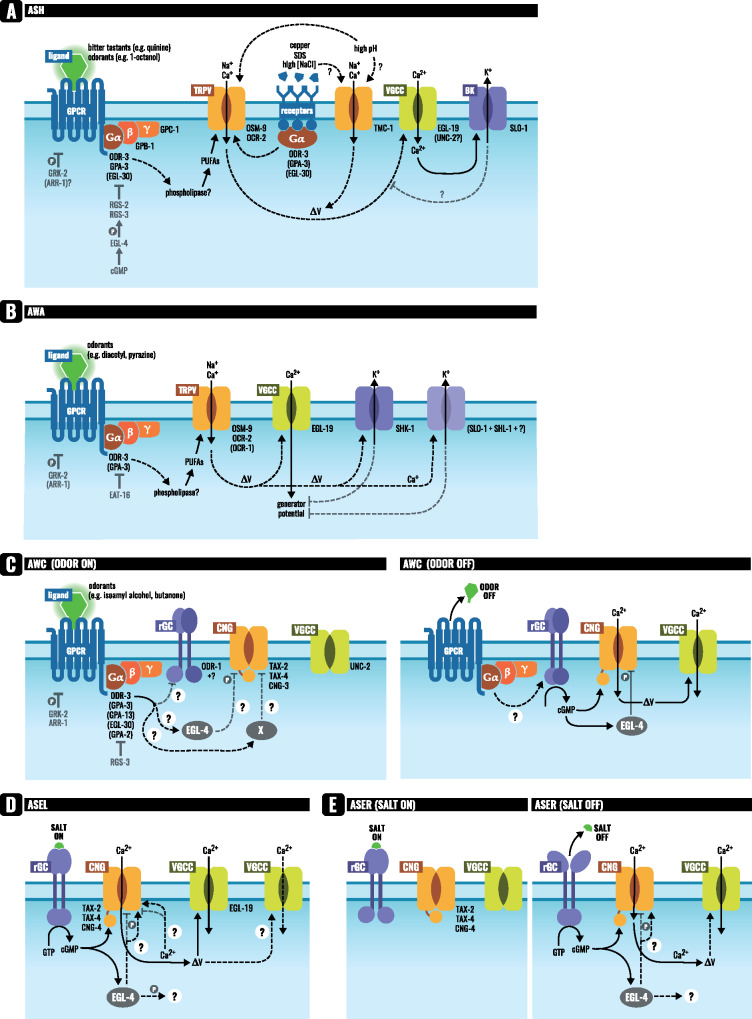
Below we describe current knowledge about the signaling molecules that transduce chemosensory information within the sensory neurons. We also refer the reader to Hobert (2013) for a broader description of the gene families that function in the C. elegans nervous system. While many gene families with neuronal functions appear to be expanded in C. elegans, a notable exception is the absence of voltage-gated sodium channels (Bargmann 1998; Hobert 2013). See Figure 4 for a summary of the signal transduction pathways that function specifically within the ASH, AWA, AWC, and ASE neurons.
Figure 4.
Signal transduction pathways in the ASH, AWA, AWC, and ASE sensory neurons. Simplified models of the potential signal transduction pathways for these representative neurons are shown. See text within the Signal Transduction section for additional details. (A) ASH: Odorant or tastant binding to a GPCR initiates G protein-coupled signaling that likely leads to the generation of PUFAs that activate TRPV channels. Stimuli may also activate other classes of receptors or channels directly. The resulting membrane depolarization activates voltage-gated calcium channels (VGCCs). In a regulatory feedback loop, ASH excitability may be dampened by a calcium-activated potassium channel. Signaling can also be downregulated at the level of GPCRs (via phosphorylation by GRK-2) or at the level of G proteins (by RGS proteins). (B) AWA: AWA signaling is initiated by odorant binding to a GPCR that initiates G protein-coupled signaling that likely leads to the generation of PUFAs that activate TRPV channels. The resulting membrane depolarization can trigger an all or none feed-forward action potential that is generated by opening of the VGCC EGL-19. The amplified voltage change opens voltage-gated potassium channels that subsequently dampen signaling. Signaling is also downregulated by GRK-2 and arrestin, and by an RGS protein. (C) AWC: In the presence of odorant, AWC is silenced. Odorant binding to a GPCR might activate a Gα that inhibits cGMP formation by guanylyl cyclases. The CNG channels may also be inhibited by EGL-4 and possibly by an unidentified protein “X.” Once odor is removed, opening of the CNG channels leads to membrane depolarization that activates VGCCs. Negative regulation of the AWC response occurs via GRK-2 and arrestin, and by an RGS protein. The cGMP-dependent protein kinase EGL-4 likely phosphorylates CNG channels during the adaptation response. (D) ASEL: Signaling is initiated when salt binds to the extracellular domain of the rGC and the intracellular cyclase domains dimerize to cyclize GTP into cGMP. The cGMP produced binds to and opens CNG channels. Membrane depolarization activates VGCCs. EGL-4 is required for calcium signals in response to salt, but its targets (besides TAX-2), and role are unknown. (E) ASER: Salt binding to the extracellular domain of the rGC inhibits cyclase activity and signaling is silenced. Signaling is initiated when salt is removed and the rGC cyclase domains dimerize to cyclize GTP into cGMP, which opens the CNG channel. Membrane depolarization activates a VGCC. Via an unknown mechanism, EGL-4 is required for the calcium flux in ASER.
G protein-coupled receptors (GPCRs)
The first expression analysis of putative C. elegans chemosensory GPCRs was undertaken over 20 years ago (Troemel et al. 1995). This foundational study, utilizing the partial genome sequence available, initially identified 41 potential C. elegans chemoreceptor genes that fell into six families (sra, srb, srg, srd, sre, and sro) based on sequence similarity with one another (Troemel et al. 1995). As completion of the full-genome sequence, a total of approximately 1,300 genes and 400 pseudogenes have been identified, and they are now classified into 19 families (15 of these comprise three major superfamilies: sra, str, srg) (Robertson and Thomas 2006; Thomas and Robertson 2008). Chemosensory GPCR genes are now known to be the largest gene family in C. elegans, comprising ∼8.5% of all its genes (Thomas and Robertson 2008). We refer the reader to the primary literature for a more thorough analysis of these gene families and their evolution (Troemel et al. 1995; Robertson 1998, 2000, 2001; Chen et al. 2005; Thomas et al. 2005; Thomas and Robertson 2008; Nagarathnam et al. 2012; Krishnan et al. 2014).
GFP-based expression analysis of a subset of the first identified putative receptor genes revealed that many were expressed in only a small subset of chemosensory neurons (Troemel et al. 1995). In addition, this work established that a single type of chemosensory neuron can express multiple chemoreceptor genes (Troemel et al. 1995). This observation has been corroborated multiple times, through studies of individual receptors and sensory neurons, and more recently by a large-scale study that examined the expression pattern of 244 rhodopsin-like (class A) C. elegans chemoreceptors (Vidal et al. 2018). A small number of C. elegans chemosensory GPCRs show left/right asymmetric gene expression, but this asymmetry has so far only been observed for the AWC sensory neuron pair (Troemel et al. 1999; Bauer Huang et al. 2007; Vidal et al. 2018). Consistent with the original findings (Troemel et al. 1995), some of the putative chemoreceptors were also found to be expressed in interneurons and motor neurons, and sometimes even in nonneuronal cells (Vidal et al. 2018). Thus, it is possible that some receptors may sense internal cues in addition to environmental stimuli. Complementing GFP-based studies with single cell transcriptional profiling (Hammarlund et al. 2018) should provide additional insights into the receptor code of individual cells.
In 1996, as the result of behavioral screens for C. elegans mutants with specific olfactory defects (odorant-response mutants), ODR-10 became the first odorant receptor in any organism to be paired with its chemical ligand, diacetyl (Sengupta et al. 1996). Consistent with a role in detecting environmental stimuli, ODR-10 is localized to the AWA sensory cilia (Sengupta et al. 1996), and ODR-10 expression conferred diacetyl responsiveness to other nondiacetyl-sensing neurons and to human HEK293 cells in culture (Zhang et al. 1997). Over the years, many groups have attempted to pair additional putative C. elegans chemoreceptors with their relevant ligands. However, these efforts have yielded only limited success. This may be due to redundancy among the chemoreceptor genes that sense a particular stimulus, or could suggest that GPCR heteromers are the primary receptors for most chemical stimuli sensed by C. elegans. The large size of the C. elegans chemoreceptor gene family also makes large-scale candidate gene approaches to de-orphanizing receptors challenging. To date, only six C. elegans (nonpheromone) chemosensory receptors have been paired with a chemical ligand (Table 2). Some GPCRs have also been characterized to be pheromone receptors, and these are described separately below (Pheromone).
Table 2.
GPCR and odorant pairings
| GPCR | Chemical Ligand | Neurons functioning in | Behavior | Reference(s) |
|---|---|---|---|---|
| ODR-10 | Diacetyl (low) | AWA | Attraction | |
| STR-2 | 2-Heptanone | AWCON | Attraction | (Zhang et al. 2016) |
| DCAR-1 |
Dihydrocaffeic acid Benzaldehyde ? (undiluted) |
ASH | Avoidance | (Aoki et al. 2011) |
| SRI-14 | Diacetyl (high) | ASH | Avoidance | (Taniguchi et al. 2014) |
| SRB-6 |
Dodecanoic acid Decanoic acid? |
ASH, ADL, ADF (head)a PHA, PHB (tail) |
Avoidance | (Tran et al. 2017) |
| STR-217 | DEET | ADL | “Confusant” | (Dennis et al. 2018) |
The limited number of C. elegans chemosensory GPCRs that have been paired with odorant ligands are shown.
SRB-6 rescued anterior response when expressed in these three head neurons, but promoters with more restrictive expression patterns were not used (Tran et al. 2017).
G proteins
Heterotrimeric G proteins (comprised of Gα, Gβ, and Gγ subunits) transduce the signals from the transmembrane chemosensory GPCRs to different pathways in different sensory neurons [e.g., see CNG and TRP channels, below]. Briefly, in the classical G protein pathway, when ligand binds to a GPCR a conformational change in the receptor allows it to act as a guanine nucleotide exchange factor (GEF) to facilitate the exchange of GDP for GTP on Gα. Gα-GTP and Gβγ can then activate distinct effectors within the cell (McCudden et al. 2005; Weis and Kobilka 2018). The C. elegans genome encodes 21 Gα, two Gβ and two Gγ subunits. The complete family of C. elegans G proteins, and their roles in diverse processes, have been reviewed previously (Bastiani and Mendel 2006). Here, we focus specifically on the role of G proteins in chemosensory signaling, excluding pheromone responses.
Gα subunits
C. elegans has one clear ortholog of each Gα subunit family: GSA-1 (Gs), GOA-1 (Gi/o), EGL-30 (Gq), and GPA-12 (G12) (Lochrie et al. 1991; Brundage et al. 1996; Park et al. 1997; Jansen et al. 1999). The remaining 17 C. elegans Gα subunits (ODR-3, GPA-1 to GPA-11, and GPA-13 to GPA-17) are somewhat more similar to the Gi/o family, but are sufficiently divergent that they are usually referred to as nematode-specific (Roayaie et al. 1998; Jansen et al. 1999; Jovelin et al. 2003; O'Halloran et al. 2006). Consistent with a role in sensory signaling, 14 of these (ODR-3, GPA-1, GPA-2, GPA-3, GPA-4, GPA-5, GPA-6, GPA-8, GPA-9, GPA-10, GPA-11, GPA-13, GPA-14, and GPA-15) are expressed in subsets of chemosensory neurons, with individual neurons expressing multiple members of this family (Zwaal et al. 1997; Roayaie et al. 1998; Jansen et al. 1999; Lans et al. 2004). Antibody staining revealed that while some Gα subunits (ODR-3 and GPA-13) localize primarily to the sensory cilium of the neurons in which they are expressed, others (GPA-2, GPA-3, and GPA-5) localize to cilia, cell bodies and axons (Roayaie et al. 1998; Lans et al. 2004). Interestingly, GPA-6 was not found in sensory cilia, but instead was seen in cell bodies and axons (Lans et al. 2004). Thus, while some Gαs may be dedicated to transducing signals from chemosensory GPCRs that detect environmental stimuli, others may also interact with GPCRs that respond to internal signals (e.g., neurotransmitters or neuropeptides).
Consistent with localization of ODR-3 in the cilia of the AWA, AWB, AWC, ASH, and ADF head sensory neurons, odr-3 mutant animals are highly defective for response to most AWA, AWC, and ASH-detected stimuli (Bargmann et al. 1993; Roayaie et al. 1998; Yoshida et al. 2012), and partly defective for response to 2-nonanone (AWB) and quinine (ASH) (Troemel et al. 1997; Hilliard et al. 2004). The overall relative severity of the odr-3 mutants suggests that ODR-3 is the primary stimulatory Gα protein that acts downstream of chemosensory receptors in multiple sensory neurons. However, somewhat surprisingly, ODR-3 may also play an inhibitory role in AWB, affecting the time-differential property for sensory input (Tanimoto et al. 2017).
ODR-3 also transmits sensory information to influence the behavioral strategies (see Appendix) used during odor tracking. Contributing to their defect in isoamyl alcohol chemotaxis, odr-3 mutant animals were shown to be defective in klinotaxis throughout a 60-minutes chemotaxis assay using 10−2 isoamyl alcohol (Yoshida et al. 2012). A defect in klinokinesis (turning) was not observed until after 30 minutes at this concentration, suggesting that other Gα proteins might contribute to proper klinokinesis during the early time period (Yoshida et al. 2012). Although both wild-type and odr-3 animals suppress turning when moving toward isoamyl alcohol and increase turning when moving away from the odor (klinokinesis), odr-3 mutants curve in the wrong direction when moving away from the odor source (Kato et al. 2014). This may be due to altered “active sensing” during forward locomotion (Kato et al. 2014). When animals are in a spatial gradient, head swings should result in an oscillation in the odor concentration at the tip of the animal’s nose that guides steering as part of the klinotaxis strategy. However, dynamic analysis of AWC signaling in response to pulses of isoamyl alcohol showed that, in addition to being diminished, the calcium fluxes lag behind odor presentation in odr-3 mutants (Kato et al. 2014). This suggests that ODR-3 normally accelerates the AWC response to short pulses of stimulus, thereby allowing these neurons to actively sense changes in the odor gradient as the animal swings its head (Kato et al. 2014).
Because odr-3 mutant animals do retain at least a residual behavioral response to most stimuli tested, it suggests a role for additional Gα proteins in chemosensory signaling (Bargmann et al. 1993; Troemel et al. 1997; Roayaie et al. 1998; Jansen et al. 1999; Hilliard et al. 2004; Yoshida et al. 2012). Indeed, although individual mutation of most other Gα-encoding genes leads to only subtle effects on chemosensation, double and multi-mutant analyses have revealed both stimulatory and inhibitory Gα signaling roles (Jansen et al. 1999; Hilliard et al. 2004; Lans et al. 2004). For example, while ODR-3 plays a major role in AWA-mediated chemotaxis, GPA-3 also contributes, and GPA-5 plays an inhibitory role (Jansen et al. 1999; Lans et al. 2004). In the AWC neurons, ODR-3 again acts as the major transducer of chemosensory signals, along with more minor contributions from GPA-3 and GPA-13, while GPA-2 is inhibitory (Lans et al. 2004). However, GPA-2 may also contribute to butanone detection (Roayaie et al. 1998). The AWC neurons may also use GPA-3 along with EGL-30 to transduce the 2-heptanone signal from the STR-2 receptor (Zhang et al. 2016). In response to the ASH and ASK (minor) -detected stimulus quinine, GPA-3 plays a major role and ODR-3 also contributes (Hilliard et al. 2004). However, gpa-3; odr-3 double mutants are completely defective in quinine response. Interestingly, egl-30 single mutant animals are also partially defective in response to quinine, suggesting an additional role for Gq signaling (Esposito et al. 2010).
Although ODR-3 is required for ASH-mediated avoidance of high NaCl (Hukema et al. 2006), no sensory Gα has been found to play a role in NaCl chemotaxis (Roayaie et al. 1998; Hukema et al. 2006). Instead, ODR-3 and GPA-1 contribute to salt gustatory plasticity (Hukema et al. 2006).
Consistent with behavioral analyses, calcium imaging experiments showed that ASH calcium transients are significantly decreased in odr-3 mutants in response to five distinct ASH-detected stimuli: copper, glycerol, SDS, quinine (Hilliard et al. 2005), and high-isoamyl alcohol (Yoshida et al. 2012). In contrast, loss of gpa-3 alone only decreased calcium signaling in response to quinine, indicating a more repellent-specific role for GPA-3 (Hilliard et al. 2005). However, there is a complete loss of the ASH calcium flux in response to copper, glycerol, SDS and quinine in odr-3; gpa-3 double mutant animals, indicating that GPA-3 does contribute to the response to these other ASH-detected stimuli as well (Hilliard et al. 2005). Similarly, while the AWCON calcium transients of odr-3 mutants were comparable to wild-type animals, they were dramatically decreased in odr-3; gpa-3 double mutants (Yoshida et al. 2012). Animals lacking GOA-1 function fail to avoid strongly alkaline pH although the calcium influx in ASH is normal, suggesting that GOA-1 functions downstream of the OSM-9/OCR-2 TRPV channels in this context (Sassa and Maruyama 2013). GPA-11 also plays a modulatory role in ASH, acting downstream of 5-HT signaling (Chao et al. 2004).
Gβγ subunits
The two C. elegans Gβ subunits are encoded by gpb-1 and gpb-2 (van der Voorn et al. 1990; Zwaal et al. 1996; Jansen et al. 1999). GPB-1 belongs to the Gβ1–4 subtype that requires Gγ coupling for function (Smrcka 2008). gpb-1 is a ubiquitously expressed and essential gene, rendering behavioral analysis of global loss-of-function mutant animals infeasible (Zwaal et al. 1996). However, neuronally targeted RNAi experiments revealed a role for GPB-1 in chemosensory signaling (Esposito et al. 2007; Yamada et al. 2009). ASH-selective knock-down of gpb-1 leads to defective avoidance responses to quinine and high osmolarity (Esposito et al. 2007). In addition, GPB-1 acts with the Gγ subunit GPC-2 to promote AWC-mediated chemotaxis to benzaldehyde (Yamada et al. 2009). GPB-2 is most similar to the divergent vertebrate Gβ5 subunit, which can interact with the GGL domain of regulator of G protein signaling (RGS) proteins (Smrcka 2008). GPB-2 contributes to benzaldehyde olfactory adaptation (Matsuki et al. 2006; O'Halloran et al. 2009), most likely via coupling to the RGS protein EGL-10 instead of GPC-1 (Yamada et al. 2009). Targeted cell-specific knockouts of gpb-2 may aid in further characterization of its role in chemosensory signal transduction.
Animals with a loss-of-function mutation in the Gγ-encoding gene gpc-1 are defective for adaptation to the water soluble attractants (tastants) NaAc, NaCl, NH4Cl (Jansen et al. 2002), as well as gustatory plasticity in response to NaCl (Hukema et al. 2006, 2008). In the ASH nociceptors, loss of GPC-1 function leads to a partially reduced initial calcium transient in response to quinine, but not copper, glycerol, or SDS (Hilliard et al. 2005). However, consistent with the main role of GPC-1 being in adaptation, gpc-1 loss-of-function animals are also defective in sensory adaptation to all four tested ASH repellent stimuli, as assessed by calcium imaging (Hilliard et al. 2005). In addition, the Gβ subunit GPB-1 couples to GPC-1 to promote adaptation to benzaldehyde (Yamada et al. 2009). Differences in assay format may explain why the gpc-1 olfactory adaptation defect was not observed previously (Jansen et al. 2002).
Guanylyl (guanylate) cyclases (GCs)
GCs produce cGMP, the soluble messenger that regulates processes as divergent as foraging (C. elegans, D. melanogaster), learning and memory (C. elegans, R. norvegicus domesticas), vasodilation and visual signal transduction (mammals) (Osborne et al. 1997; Fujiwara et al. 2002; Sharma et al. 2016). cGMP gates the opening of CNG channels, activates cGMP-dependent protein kinases and [in most animals besides nematodes (Hobert 2013)] cyclic nucleotide-hyperpolarizing channels, and activates phosphodiesterases that ultimately degrade cGMP. GCs exist in two forms: soluble cyclases that are activated by gaseous stimuli and are not further discussed in this chapter, and the rGCs that have a transmembrane domain and can transduce gas, environmental chemical, peptide and thermal signals (Yu et al. 1997; Hallem et al. 2011; Maruyama 2017; Goodman and Sengupta 2018). rGCs can act downstream of G protein-coupled receptors via activation by Gαs and/or they can be directly regulated by ligand binding to or detachment from their extracellular domains.
C. elegans expresses 27 rGCs (Yu et al. 1997; Fitzpatrick et al. 2006; Ortiz et al. 2006) and all are found in sensory neurons, except GCY-11, which is expressed in pharyngeal muscle. Nearly half (11/27) are expressed in the gustatory ASE neurons (Ortiz et al. 2006), while the rest are expressed in other sensory neurons that also express CNG channels: ADL, AWB, AWC, ASG, ASI, ASJ, ASK, AFD, AQR, PQR, URX, PHA, and PHB, as well as a few interneurons and nonneuronal cells. Members of the large C. elegans rGC family show great heterogeneity in their extracellular ligand binding domains (Fitzpatrick et al. 2006). The expansion of this feature may reflect the evolutionary pressure this organism has experienced to sense and respond to a wide variety of ligands via guanylyl cyclase receptors, and may allow animals to respond to environmental stimuli that do not typically interact with GPCRs. Here, we focus on the role of rGCs in chemosensory signaling and also recommend this review (Maruyama 2017). See the Appendix for discussion of rGC structure and activation mechanisms, including homo- and hetero-dimer formation. Briefly, each rGC is a dimer of two polypeptides, each encoding a half-cyclase domain. Cyclase activity of the dimer requires that the half-cyclase domains come together to form an active enzyme that cyclizes cGMP from GTP. This dimerization can be regulated by ligand binding to the receptor domain, phosphorylation, or regulatory protein binding to the intracellular domains (ICDs) (Sharma et al. 2016).
rGCs and their chemosensory functions
ODR-1 and DAF-11 mRNA are co-expressed in AWC, AWB, ASI, ASJ, and ASK (Birnby et al. 2000; L'Etoile and Bargmann 2000) [http://www.cengen.org, (Hammarlund et al. 2018)]. Although an ODR-1::GFP fusion expressed from a multi-copy transgene was expressed in AWC, AWB, ASI, ASJ and ASK, a CRISPR-edited GFP-tagged ODR-1, is expressed only in AWC and AWB under standard laboratory conditions (B. Zhang, V. Paketci, C. Zuazo, B-T. Juang and N. L'Etoile, unpublished observations).
Both ODR-1 and DAF-11 are required for AWC-mediated chemotaxis to isoamyl alcohol, benzaldehyde and butanone, and for AWB-mediated repulsion from 2-nonanone (Birnby et al. 2000; L'Etoile and Bargmann 2000), and thus they were posited to act as heterodimers (Morton 2004; Ortiz et al. 2006). Indeed, DAF-11 and ODR-1 are both required downstream of the GPCR LITE-1 to mediate the response to light (Liu et al. 2010). However, there are also clues that they could act as homodimers as well as heteromers. For example, ODR-1 is exquisitely localized to the AWC cilia, while DAF-11 is expressed throughout the cell (B. Zhang, V. Paketci C. Zuazo and N. L'Etoile, unpublished observations) (Birnby et al. 2000; L'Etoile and Bargmann 2000). Loss of the cGMP-dependent protein kinase EGL-4 suppresses the benzaldehyde chemosensory defects of daf-11 mutants, but fails to suppress odr-1 defects (N. L'Etoile and C. Bargmann, unpublished results) (L'Etoile et al. 2002). Evidence from cGMP imaging also indicates that the drop in cGMP in AWC when odor is applied requires ODR-1, and only partially depends on DAF-11 (Shidara et al. 2017). The reduction in cGMP may be a consequence of negative regulation of ODR-1, perhaps by a phosphorylation of the kinase-like region, binding of a negative regulator to the hinge region, or by inhibition by a Gα protein.
In addition, although DAF-11 is required in ASJ and possibly ASK to block dauer formation (Schackwitz et al. 1996), odr-1 mutants do not show dauer phenotypes (L'Etoile and Bargmann 2000). Furthermore, although DAF-11 and GCY-27 are both required in ASJ for response to nitric oxide, they are unlikely to act as heteromers with each other in this context, as DAF-11 is required for the ON response and GCY-27 for the OFF response (Hao et al. 2018).
GCY-1, GCY-4, and GCY-22 act in ASER (Smith et al. 2013), the sensory neuron that promotes chemotaxis to the salt concentration last associated with food (Kunitomo et al. 2013; Luo et al. 2014). The GCY-22 ECD directs Cl−, I−, Br− and methionine seeking responses when appended to the ICDs of GCY-1 or GCY-4, and co-expressed in ASI in gcy-22 mutant worms (Smith et al. 2013). Surprisingly, imaging experiments in ASER showed that GCY-22 is required for both the calcium increase in response to Cl- removal, and paradoxically, the cGMP decrease in response to Cl− removal (Ortiz et al. 2009; Woldemariam et al. 2019). Furthermore, the ECD of GCY-1 is required for specific recognition of K+ ions, and the ECD of GCY-4 for I− (Smith et al. 2013).
GCY-14 is localized to the ASEL cilia and is required both for chemotaxis to Na+ and Li+ ions (Ortiz et al. 2006) and for the response of ASEL to high pH (acting as a homodimer in this case, as shown by second site suppressor mutagenesis) (Murayama et al. 2013). Mis-expression of GCY-14 (in the ASI neurons) was sufficient to confer calcium responses to alkaline pH in gcy-14 mutants, and a pH-sensitive histidine residue in its ECD was required to signal the increase in extracellular pH (although it was not required for the response of this rGC to Na+) (Murayama et al. 2013). Thus, GCY-14 is likely directly stimulated by pH upsteps (increases) to produce cGMP, with hydroxyl ions acting as the likely ligand that binds to its ECD, triggering a cascade of changes that result in dimerization and activation of the cyclase. Increased cGMP could then open CNG channels in ASEL to promote runs toward the stimulus.
GCY-27 is required in ASK, and perhaps ASH, to decrease ASH-mediated aversion of bitter tastants (Krzyzanowski et al. 2013). GCY-27 has a very short ECD, so it may only respond to intracellular ligands or, if it transduces extracellular signals, it may act as a heterodimer to do so.
GCY-12 is required to regulate an animal's body size. It is expressed in ASE and AWC, and its ECD is dispensable for body size regulation (Fujiwara et al. 2015). A possible role for this enzyme in chemosensation has yet to be described.
GCY-28 is expressed in the axons of AWC where it is required for the butanone exposure-induced switch from attraction to repulsion after prolonged starvation. It appears to act in the AWC axons, where it may affect synaptic transmission (Tsunozaki et al. 2008).
Phosphodiesterases (PDEs)
PDEs degrade cGMP and thus are crucial for regulating signaling. In vertebrate photoreceptors, rhodopsin activation by light activates PDEs that degrade cGMP, thereby decreasing the open probability of CNG channels (Fu and Yau 2007). Signaling by some chemical stimuli may similarly require rapid degradation of cGMP. C. elegans expresses six PDEs: PDE-1, PDE-2, PDE-3, PDE-4, PDE-5, and PDE-6 (Liu et al. 2010). The PDE-4 and PDE-6 proteins are homologous to human PDEs that have specificity for cAMP over cGMP (Liu et al. 2010). The remaining PDEs are most similar to those that can cleave both cAMP and cGMP (Omori and Kotera 2007). PDE-1 has a calcium regulatory domain and degrades cGMP in response to calcium increases (Couto et al. 2013). Other PDEs, such as PDE-2, are activated by cGMP and are thus capable of providing negative feedback and stabilization of cGMP levels (Couto et al. 2013; Rahi et al. 2017). Thus far, no C. elegans PDE has been shown to play a direct role in regulating chemosensory signaling, although PDE-1, -2, -3, and -5 are involved in adaptation to odor stimuli (O'Halloran et al. 2012).
Cyclic nucleotide-gated (CNG) channels
CNG cation channels, whose open probabilities are increased by the binding of cGMP or cAMP to intracellular cyclic nucleotide binding domains, play key roles as primary sensory channels in phototransduction and olfaction across species (Pifferi et al. 2006). They are localized to sensory endings where their opening/closing generates a change in membrane potential following the delivery of a chemosensory stimulus, while other voltage-gated channels that are expressed more widely in the neuron may amplify the signal (Shindou et al. 2019). Functional CNG channels are tetramers, composed of one to four A-type (alpha) and a variable number of B-type (beta) subunits arranged around a central pore (Pifferi et al. 2006). Subunit types are identified by amino acid residues within their pore domains that determine ion selectivity (Root and MacKinnon 1993; Eismann et al. 1994; Seifert et al. 1999), as well as the presence (A-type) or absence (B-type) of a leucine zipper in their C-termini (Zhong et al. 2002; Shuart et al. 2011).
The C. elegans A-type (TAX-4) (Komatsu et al. 1996, 1999) and B-type (TAX-2) (Coburn and Bargmann 1996; Coburn et al. 1998) subunits have close mammalian homologs (L’Etoile 2004; Wojtyniak et al. 2013), while the less conserved subunits (CNG-1 and CNG-3: A-types; CNG-2 and CNG-4/CHE-6: B-types) are much more diverged (Cho et al. 2004, 2005; L’Etoile 2004; Smith et al. 2013; Wojtyniak et al. 2013). In vitro experiments showed that a channel's affinity for cGMP, as well as how long it stays open once it binds cGMP, depends on which subunits comprise the channel (Komatsu et al. 1999; O'Halloran et al. 2017). For example, homomeric channels comprised of only TAX-4 (A-type) have a 10-fold higher affinity for cGMP and stay open seven times as long as a channel comprised of both TAX-4 (A-type) and TAX-2 (B-type) subunits (Komatsu et al. 1999; O'Halloran et al. 2017). Addition of other (diverged) A- or B-type subunits to the channel also changes its biophysical properties and this is important for function (O'Halloran et al. 2017). The subunit composition of each channel also dictates which subdomain of the sensory cilia the CNG channel resides within, and the subdomain each channel occupies is specific to each sensory neuron (Wojtyniak et al. 2013). Thus, the specific function each sensory neuron serves may require distinct regions of its sensory cilia to respond to cGMP with different dynamics and sensitivity.
Consistent with TAX-2 being a core component of many CNG channels, and its expression pattern (AWC, ASE, ASG, ASI, ASJ, ASK, AWB, AFD, ADE, and BAG), tax-2 mutant animals are defective for a variety of sensory responses, including chemotaxis toward the AWC-detected odorants isoamyl alcohol and benzaldehyde (Coburn and Bargmann 1996) and in AWB-mediated avoidance (Troemel et al. 1997; Yoshida et al. 2012). They are also defective in lysine chemotaxis (Coburn and Bargmann 1996). TAX-4 has a similar expression pattern (AWC, ASE, ASG, ASI, ASJ, ASK, AWB, AFD, BAG, and URX), and tax-4 mutant animals are also defective for chemotaxis toward isoamyl alcohol, benzaldehyde and 2-butanone (also detected by AWC), and partially defective for 2,4,5-trimethylthiazole (detected by AWA and AWC) (Komatsu et al. 1996). In addition, TAX-4 contributes to PHA/PHB-mediated avoidance of SDS (Hilliard et al. 2002). TAX-4 can also act in a TAX-2 independent manner, as evidenced by the finding that TAX-4, but not TAX-2, is required in ASI and ASJ to respond to sulpholipid cues secreted by the predator P. pacificus (Liu et al. 2018b).
Both tax-2 and tax-4 mutants are defective in ASE-mediated NaCl chemotaxis (Coburn and Bargmann 1996; Komatsu et al. 1996), ASEL-mediated chemotaxis toward alkaline pH (Murayama et al. 2013), and ammonium sensation (most likely mediated by AWC) (Frøkjær-Jensen et al. 2008). They are both defective in preferring the smell of P. aeruginosa PA14 over E. coli OP50 bacteria (Harris et al. 2014), S. marcescens avoidance (Pradel et al. 2007), Microbacterium nematophilum avoidance (Yook and Hodgkin 2007; Anderson and McMullan 2018) and worm extract avoidance (Zhou et al. 2017).
Imaging experiments revealed that the TAX-2 and TAX-4 subunits are required for the ASEL calcium flux in response to an NaCl upstep (Suzuki et al. 2008) and to a pH upstep (6.8 to 10) (Murayama et al. 2013). TAX-2 and TAX-4 are also required for the ASER calcium flux in response to an NaCl downstep (Suzuki et al. 2008), and TAX-4 contributes to isoamyl alcohol sensing in PHA/PHB (Zou et al. 2017).
CNG channels generate calcium fluxes that can ultimately regulate gene expression. For example, loss of either TAX-2 or TAX-4 perturbs asymmetric expression of STR-2 in AWC (Troemel et al. 1999). Both channel subunits are also necessary to transmit signals that induce daf-7 expression in the ASJ neurons and increase its expression in ASI neurons when animals are cultured on PA14 (Meisel et al. 2014). They are required for attraction to 2-heptanone, but their role in this context may be maintenance of STR-2 receptor expression in AWCON rather than in transducing the olfactory signal (Zhang et al. 2016). In addition, both TAX-2 and TAX-4 help to promote and prevent dauer formation, in different contexts, depending on the neuron they are expressed in (Pheromone) (Coburn et al. 1998). In addition to their similar expression patterns and loss-of-function phenotypes, electrophysiological data also suggest that TAX-2 and TAX-4 can form heteromeric channels (Komatsu et al. 1999; O'Halloran et al. 2017). However, complex mixtures of homomeric and heteromeric channels are likely to be expressed in sensory neurons (Wojtyniak et al. 2013).
CNG-1 is expressed in unidentified head neurons (but including ASI) and PHA/PHB in the tail (Cho et al. 2005; Wojtyniak et al. 2013). Although cng-1 mutant animals showed no defect in olfaction (AWC or AWA-mediated) or NaCl chemotaxis (Cho et al. 2005), CNG-1 is required for starvation-induced sharpening of the response to odors that are sensed by both AWCON and AWCOFF (He et al. 2016). It may also regulate sensory integration in the AIA interneurons (Shinkai et al. 2011).
CNG-2 is expressed in just a subset of the cells that express TAX-2/TAX-4: AWC, ASE, ASG, ASI, ASJ, and ASK (Wojtyniak et al. 2013). CNG-2 is required for the PA14 metabolite-induced calcium flux and daf-7 induction in ASJ (Park et al. 2020), but its function in the remaining neurons is not known. However, it may be involved in plasticity induced by cGMP and calcium signaling, as it possesses consensus cGMP-dependent protein kinase (PKG) phosphorylation and calmodulin binding sites.
CNG-3 expression is also restricted to a subset of the cells that express TAX-2/TAX-4: AWC, ASE, ASI, AWB, and AFD (Cho et al. 2004; Wojtyniak et al. 2013). However, despite being expressed in chemosensory neurons, cng-3 mutant animals showed no defect in chemotaxis to AWC (or AWA) detected odorants, or NaCl chemotaxis (Cho et al. 2004; O'Halloran et al. 2017). Instead, it plays a role in short-term (30 minutes), but not long-term (>60 minutes), adaptation of AWC to the attractive odorants benzaldehyde and 2-butanone (O'Halloran et al. 2017). Indeed, CNG-3 may be regulated by both cGMP and calcium signaling because it has a consensus PKG site at serine 20 that is required for adaptation to a 30 minutes exposure of AWC-sensed odors (O'Halloran et al. 2017) and a putative calmodulin binding site at L551-L565. Biomolecular Fluorescence Complementation (BiFC) assays suggested that CNG-3/TAX-2 and CNG-3/TAX-4 interactions likely occur in vivo (O'Halloran et al. 2017). In cell culture, the addition of CNG-3 to TAX-2/TAX-4 channels altered their gating kinetics (O'Halloran et al. 2017).
CNG-4 expression is very weak, challenging reliable identification of cells beyond AWC and ASE (Smith et al. 2013; Wojtyniak et al. 2013). Although cng-4 (also known as che-6) mutants show no defects in AWC-mediated olfaction, they are defective in the ASE-mediated response to water soluble attractants (including NaCl, cAMP and biotin) (Bargmann et al. 1993; Smith et al. 2013).
Transient receptor potential (TRP) channels
TRP channels are cation channels that are important for responses to many types of external stimuli, including light, sound, touch, temperature, and chemicals (Venkatachalam and Montell 2007; Samanta et al. 2018). The C. elegans genome encodes 23 TRP channels, including members of each of the seven TRP subfamilies (Goodman and Schwarz 2003; Hobert 2013). For an extensive review of the varied roles of these cation channels in C. elegans (see Kahn-Kirby and Bargmann 2006; Bounoutas and Chalfie 2007; Xiao and Xu 2009, 2011; Schafer 2015; Goodman and Sengupta 2019).
To date, only TRPV family members have been shown to play a role directly in chemosensory behavior in C. elegans. The TRPV channel OSM-9 was the first TRP channel shown to have a role in invertebrate chemosensation and was the first TRP channel to be functionally characterized in C. elegans (Colbert and Bargmann 1995; Colbert et al. 1997). Sequence analysis lead to the subsequent identification of four additional C. elegans TRPV family members, ocr-1, ocr-2, ocr-3, and ocr-4 (osm-9/capsaicin receptor related) (Tobin et al. 2002).
Transcriptional and translational reporters expressed from extrachromosomal arrays indicate that OSM-9 can be expressed in multiple sensory neurons, including the AWA, AWC, ASH, ADL, ASE, ADF, ASG, ASI, ASJ, and ASK head chemosensory neurons, as well as PHA and PHB in the tail (Colbert et al. 1997), while the single cell transcriptional profiling dataset [http://www.cengen.org, (Hammarlund et al. 2018)] indicates that the mRNA is most highly expressed in AWA, ADF, ADL, ASH, OLQ, PQR, PHA, and PHB. A CRISPR-edited GFP-tagged OSM-9 confirms the single cell sequencing dataset (K. Benedtti, F. Saifuddin, N. L’Etoile, personal communication). While there are a number of neurons that may express osm-9 (Colbert et al. 1997), but not any of the ocr genes (including AWC, ASE, ASG, ASI, ASJ, and ASK) (Tobin et al. 2002), each ocr gene is only expressed in a subset of cells that express OSM-9. This suggests that individual OCR channel subunits can function together with OSM-9 in distinct contexts. In particular, OCR-2 is also expressed in AWA, ASH, ADL, ADF, PHA, and PHB (Tobin et al. 2002). In cells in which OSM-9 and OCR-2 are co-expressed, they are localized to the cilia and are mutually dependent on each other for cilia localization (Tobin et al. 2002). This, combined with behavioral data (see below), suggests that these two proteins come together to function in a single channel complex to mediate primary signal transduction. An exception to this is that OCR-2 functions in an OSM-9 independent manner in ASH and ADL to generate avoidance of P. pacificus predator cue (Liu et al. 2018b). OSM-9 is not required for AWC-mediated sensory responses, and instead is required for adaptation to some AWC-sensed stimuli (Colbert and Bargmann 1995). Its exact role in adaptation is still ambiguous, as it may (Colbert et al. 1997) or may not [K. Benedtti, F. Saifuddin, N. L’Etoile, personal communication and http://www.cengen.org, (Hammarlund et al. 2018)] be expressed in AWC.osm-9 and ocr-2 mutant animals are defective in chemotaxis to odorants detected by the AWA olfactory neurons, such as diacetyl and pyrazine (Colbert et al. 1997; Tobin et al. 2002). In imaging experiments, osm-9 single and ocr-1 ocr-2 double mutant animals showed no AWA calcium flux in response to diacetyl (Larsch et al. 2015). The calcium response was also reduced 1000-fold in ocr-2 single mutant animals (Larsch et al. 2015). Thus, OSM-9 and OCR-2 are likely to be the sensory transduction channel downstream of chemical stimulation. Experiments in osm-9 mutant animals suggest that the TRPV channels also contribute to setting the threshold of AWA electrical excitability (Liu et al. 2018a).
Animals lacking OSM-9 or OCR-2 function display diminished responses to a broad range of ASH-detected chemical stimuli, including high benzaldehyde, high pH, 1-octanol, 2-octanone, copper, SDS and bitter tastants (including quinine) (Colbert and Bargmann 1997; Tobin et al. 2002; Hilliard et al. 2004; Ezak et al. 2010; Sassa et al. 2013; Wang et al. 2015, 2016). OSM-9 and OCR-2 may also contribute to ASH-mediated avoidance of high NaCl (Hukema et al. 2006). OSM-9 and OCR-2 also appear to contribute to social feeding by functioning in the ASH and ADL neurons that detect noxious chemicals (de Bono et al. 2002). We note that while OSM-9/OCR-2 are often referred to as being required for all ASH-mediated behaviors, in many cases the avoidance response of the presumed null mutants is reduced but not eliminated. For example, osm-9 and ocr-2 mutant animals retain substantial response to quinine (Hilliard et al. 2004; Ezak et al. 2010; Mehle et al. 2020), and even osm-9; ocr-2 double mutants show only a partial decrement in behavioral response to bitter compounds, 1-octanol, SDS and copper (Ezak et al. 2010; Mehle et al. 2020). Underscoring the likelihood that other channels contribute to ASH-mediated responses, mechanosensory stimulation of osm-9 and ocr-2 mutants (alone or in combination) evoked similar electrophysiological currents as wild-type animals (Geffeney et al. 2011). Thus, we suggest that it would be more accurate to instead consider these channels as contributing to all examined ASH-mediated behaviors. The identity of the additional channel(s) that might underlie the remaining responses to chemosensory stimuli remains unknown.
Calcium imaging experiments have confirmed a role for OSM-9 and OCR-2 in ASH chemosensory signaling. The ASH calcium flux in response to 10 mM quinine is apparently eliminated in osm-9 animals (Hilliard et al. 2005). Calcium signaling in response to 10 mM copper was also strongly reduced (Hilliard et al. 2005). Although the ASH neurons only show an ON response when presented with 1 mM CuSO4, a biphasic (ON and OFF) response was observed in response to higher (10 and 50 mM) concentrations when presented for extended durations (Wang et al. 2015). In these cases, the OFF-response was completely dependent on OSM-9, while trpa-1 (TRPA channel mutant) animals also showed a dramatic decrease in the OFF-response (Wang et al. 2015). osm-9 and ocr-2 mutants also show a diminished ASH calcium flux in response to high pH (11.2) (Sassa et al. 2013), and OSM-9 contributes to both acid- and alkali-activated electrical currents in ASH (Wang et al. 2016).
As discussed below, OSM-9 and OCR-2 are co-expressed in the ADL sensory neurons, where they mediate the avoidance of high concentrations of the dauer pheromone component ascr#3 (Jang et al. 2012). Not much is known about the sensory signaling role of OSM-9/OCR-2 in the ADF neurons, but OSM-9 does contribute to PHA/PHB calcium dynamics in response to nociceptive chemosensory stimuli (Zou et al. 2017).
Genetic and behavioral analyses have indicated that OSM-9/OCR-2 signaling depends on and can be activated by specific polyunsaturated fatty acids (PUFAs), although the lipid-mobilizing enzyme(s) that act downstream of sensory G proteins in AWA and ASH are not yet known (Kahn-Kirby et al. 2004). PUFAs are required for both AWA- and ASH-mediated chemosensory behaviors, but each sensory cell may rely on different PUFAs. While the ASH neurons may use a broad set of 20-carbon PUFAs, the AWA neurons appear more selective to EPA (eicosopentanoic acid) and AA (arachidonic acid) (Kahn-Kirby et al. 2004). The application of exogenous PUFAs can elicit behavioral avoidance (reminiscent of ASH activation) in wild-type but not osm-9 mutant animals, and was also sufficient to elicit ASH calcium fluxes that are dependent upon the OSM-9/OCR-2 channels (Kahn-Kirby et al. 2004). Together, these data suggest that the OSM-9/OCR2 TRPV channels may be directly modulated by PUFAs generated in sensory neurons in response to stimuli.
In addition to their direct role in primary signal transduction, C. elegans TRPV channels also regulate the transcription of sensory genes (Tobin et al. 2002; Zhang et al. 2004; Gruner et al. 2014). For example, osm-9 and ocr-2 mutant animals show decreased expression of the ODR-10 diacetyl receptor in the AWA olfactory neurons (Tobin et al. 2002). Although ocr-1 single mutants show no change in odr-10p::gfp expression, and it is only slightly reduced in ocr-2 animals, ocr-1; ocr-2 double mutants show little to no odr-10p::gfp expression (Tobin et al. 2002). OCR-2 (and OSM-9, weakly) promotes srh-234 chemoreceptor gene expression in ADL (Gruner et al. 2014). OSM-9 and OCR-2 are also co-expressed in the ADF, where they regulate the expression of the 5-HT biosynthetic enzyme gene tph-1 (Zhang et al. 2004). While the mechanism by which these channels control gene expression is not known, it has been proposed that the TRPV channels function in activity-dependent gene expression pathways (Tobin et al. 2002; Zhang et al. 2004). In addition, OCR-2 contains a functional nuclear localization sequence in its carboxy-terminal tail (Ezak and Ferkey 2011) and OCR-2 has been proposed to regulate gene expression in ASH (Ezak et al. 2010).
Voltage-gated calcium channels (VGCCs)
VGCCs are activated by membrane depolarization to mediate calcium influx. The channel is formed by a pore-forming α1 subunit, with its 24 transmembrane domains, that can associate with distinct auxiliary subunit combinations in different physiological contexts (Catterall 2011; Zamponi et al. 2015). The mammalian α1 subunits are classified as three types: Cav1 (L-type), Cav2 (non L-type; P/Q, N, and R), and Cav3 (T-type) (Catterall et al. 2005). The C. elegans genome encodes one of each of these main types (Hobert 2013). EGL-19 is the sole L-type (Lee et al. 1997), UNC-2 is a P/Q-type (Schafer and Kenyon 1995), and CCA-1 is the only T-type (Shtonda and Avery 2005; Steger et al. 2005). C. elegans also has two distantly related α1 subunits, NCA-1 and NCA-2 (α1 U-type), as well as two α2δ (UNC-36, TAG-180) and two β auxiliary subunits (CCB-1 and CCB-2) (Hobert 2013). To date, only mutations in egl-19 and unc-2 have been shown to affect C. elegans chemosensory signaling.
In chemosensory neurons, EGL-19 is thought to act downstream of stimulus-evoked depolarization mediated by CNG or TRPV channels, and upstream of calcium release from internal stores (Hilliard et al. 2005; Kato et al. 2014; Larsch et al. 2015; Zahratka et al. 2015; Tanimoto et al. 2017). UNC-2 may act downstream of the TAX-2/TAX-4 CNG channels in the AWC olfactory neurons (Hirotsu et al. 2000), and also stimulates tph-1 expression in ADF (Estevez et al. 2004). For additional perspective, we refer the reader to articles that model calcium signaling in C. elegans sensory neurons (Kuramochi and Iwasaki 2010; Mirzakhalili et al. 2018; Nicoletti et al. 2019).
In the AWA olfactory neurons, diacetyl-induced calcium responses are strongly reduced in egl-19 (reduction-of-function) mutant animals (Larsch et al. 2015). In addition, recent work has revealed for the first time that AWA can fire calcium-mediated action potentials, and that these are initiated by EGL-19 (Liu et al. 2018a). The action potentials are likely terminated by Shaker-type potassium channels encoded by shk-1, together with calcium-dependent processes such as calcium channel inactivation (Liu et al. 2018a).
VGCCs also function in C. elegans nociceptors. In response to 1-octanol, the ASH somal calcium flux of egl-19 mutant animals is strongly reduced, but unaffected in unc-2 mutants (Zahratka et al. 2015). However, both EGL-19 and UNC-2 are important for octanol-induced calcium signaling in ASH axons (Zahratka et al. 2015). EGL-19 is also responsible for the slow time-integral component of calcium signaling in the ASH neurons following 2-nonanone exposure (Tanimoto et al. 2017). In a regulatory feedback loop, calcium entry through EGL-19 may inhibit ASH excitability by activating the BK-type calcium-activated potassium channel SLO-1 (Williams et al. 2018). Decreased EGL-19 function also affects the AWB response to 2-nonanone, such that less calcium accumulates in the AWB cell bodies of egl-19 mutants following odorant removal (“odor down phase”) (Tanimoto et al. 2017).
ASEL responds to increases in NaCl concentration with an increase in calcium (dependent on TAX-2/TAX-4 in the cilium), and the resulting cilium membrane depolarization likely opens EGL-19 channels in ASEL dendrites, which may amplify the electrical signal (Shindou et al. 2019). Consistent with these findings, ASEL-specific RNAi knock-down of EGL-19 decreased chemotaxis toward NaCl (Shindou et al. 2019). Supporting a selective role for EGL-19 in ASEL, application of the EGL-19 antagonist nemadapine-A (Kwok et al. 2006) blocked NaCl-induced depolarization of ASEL, but not of ASER (Shindou et al. 2019). ASER expresses a voltage-dependent calcium current, although the identity of the channel involved is not known (Goodman et al. 1998).
Other channels
TMC-1
The novel family of transmembrane channel-like (TMC) proteins is conserved from worms to humans (Keresztes et al. 2003; Kurima et al. 2003). Little is known about their function in mammals, beyond their role in hearing (Kawashima et al. 2015; Yue et al. 2019). C. elegans tmc-1 encodes the transmembrane channel-like protein 1 (TMC-1) that is expressed in several sensory neurons (ASH, ADF, ASE, ADL, AQR, PQR, URX, and PHA) (Chatzigeorgiou et al. 2013). TMC-1 is required for sodium and lithium cation-induced attraction behaviors (Dao et al. 2020). TMC-1 was also shown to be required in the ASH nociceptors to mediate avoidance of high-NaCl concentrations (Chatzigeorgiou et al. 2013). Consistent with behavioral studies (Hukema et al. 2006), high concentrations of NaCl evoke a large calcium flux in the ASH neurons of wild-type animals, but this was severely diminished in tmc-1 mutant animals (Chatzigeorgiou et al. 2013). As TMC-1 selectively responds to sodium (not chloride) ions and has a high-sodium permeability, it suggests that TMC-1 may itself be an ASH nociceptive salt sensor (Chatzigeorgiou et al. 2013). However, these results were not repeated in subsequent studies (Wang et al. 2016; Dao et al. 2020), and it is not clear what differences in assay format might be contributing factors.
Alkaline pH activates an inward current in ASH, and this excitation occurs independently of G protein signaling (Wang et al. 2016). However, TMC-1 (along with a minor contribution from OSM-9) contributes to alkali-activated currents in ASH (Wang et al. 2016). In contrast, TMC-1 function is not required for acid sensation (Wang et al. 2016). Ectopic expression of TMC-1 was also able to confer alkaline sensitivity (assessed via calcium imaging and evoked current) to the normally alkaline-insensitive ASI sensory neurons (Wang et al. 2016). Combined, these results reveal a critical role for TMC-1 in sensing noxious alkaline environments, while behavioral responses to several other ASH-detected stimuli (nose touch, CuCl2, high osmolarity) are unaffected in tmc-1 mutant animals (Chatzigeorgiou et al. 2013).
DEG/ENaC sodium channels
Degenerin/epithelial Na+ channels (DEG/ENaC channels) are voltage-independent Na+ (or Na+/Ca2+) homotrimeric channels (Jasti et al. 2007; Ben-Shahar 2011). The C. elegans DEG-1 channel (Chalfie and Wolinsky 1990), most likely acting in ASK, contributes to acid avoidance behavior along with ACD-1 in glial cells (Wang et al. 2008). deg-1 and acd-1 mutant animals are also less attracted than wild-type animals to lysine, although the site of DEG-1 and ACD-1 function in this behavior has not been determined (Wang et al. 2008).
Plasticity
When chemical stimuli interact with sensory receptors, they initiate two processes: a behavioral response and adaptation to that response. Sensory adaptation is a form of plasticity that leads to a decreased response to a sensory stimulus following prolonged exposure. It allows animals to respond to new or changing stimuli in their environment while ignoring persistent signals. The change in responsiveness that takes place in peripheral cells (e.g., olfactory neurons) where sensory signal transduction occurs typically results in short-term physiological changes in these cells.
Sensory adaptation occurs over at least two timescales: milliseconds and seconds to tens of minutes. The initial feedback may be considered part of the sensory response and is required for taxis to a stimulus. In its absence, the animals may not be able to discern a gradient. Thus, chemotaxis behavioral studies need to be coupled with physiological analyses of the second messengers within a sensory transduction pathway in order to understand how a given signaling molecule contributes to both initial response and subsequent adaptation.
Regulation of G protein-coupled signaling (GPCRs, G Proteins)
In one form of sensory adaptation, desensitization, GPCR signaling is inhibited at the level of the receptors by a family of serine/threonine kinases (G protein-coupled receptor kinases, GRKs) that specifically recognize and phosphorylate the activated (agonist bound) conformation of receptors. Arrestin proteins then recognize and bind to the phosphorylated receptor, “uncouple” it from G proteins, and block its reactivation. Arrestin binding can also target the activated receptor for internalization and recycling back to the cell membrane (re-sensitization). Desensitization of GPCRs by GRKs and arrestin proteins is an important means of protecting against receptor overstimulation, and it allows cells to integrate information from multiple signaling inputs and to respond to new stimuli (Pitcher et al. 1998; Bunemann and Hosey 1999; Ferguson 2001; Pierce and Lefkowitz 2001; Komolov and Benovic 2018).
The C. elegans genome encodes two GRKs (GRK-1 and GRK-2) and one arrestin (ARR-1) (Bargmann 1998; Fukuto et al. 2004; Palmitessa et al. 2005). For a review of the varied roles of the C. elegans GRKs, see (Wood and Ferkey 2016). To date, a role for C. elegans GRK-1 in regulating chemosensory GPCRs has not been identified. However, loss of GRK-2 broadly disrupts chemoattraction and chemical avoidance in C. elegans (Fukuto et al. 2004; Ezak et al. 2010). ASH quinine-evoked calcium fluxes are also absent in grk-2 mutants (Fukuto et al. 2004). Overall, the grk-2 phenotype (chemosensory defective) is contrary to what would be expected for loss of a negative regulator of signaling (e.g., hypersensitivity). While it is possible that GRK-2 plays a positive role in chemosensory signaling, it has been proposed that there may instead be a compensatory downregulation of G protein signal transduction to protect neurons from overstimulation in the absence of GRK-2 function (Fukuto et al. 2004). In addition, consistent with the classical function of GRKs in desensitization, grk-2 mutants show excessive ASH-mediated avoidance of NaCl, which counterbalances ASE-mediated chemoattraction (Hukema et al. 2006). Furthermore, in wild-type animals GRK-2 protein levels oscillate in a circadian manner with cyclical entrainment, and GRK-2 protein levels and sensitivity to 1-octanol are inversely related (Olmedo et al. 2012).
In contrast to the broad chemosensory defects of grk-2 mutants, loss of the sole C. elegans β-arrestin (ARR-1) leads to the more expected adaptation phenotype (Palmitessa et al. 2005). Loss of ARR-1 function also leads to defective gustatory plasticity, such that arr-1 mutants do not avoid NaCl following pre-exposure as wild-type animals do (Hukema et al. 2006). However, the site of ARR-1 function in this process has not been determined.
Just downstream of receptor activation, RGS GTPase activating proteins can dampen Gα signaling by binding to Gα subunits and stabilizing the transition state for GTP hydrolysis, thus accelerating their intrinsic GTPase activity (Ross and Wilkie 2000; Hollinger and Hepler 2002; Willars 2006). This leads to the termination of downstream signaling by both the Gα and the Gβγ subunits as they re-associate. Emerging studies have also identified a role for RGS proteins in the modulation of GPCR and G protein signaling in synapses (Gerber et al. 2016).
The C. elegans genome encodes 21 proteins with RGS domains, including the two GRKs (Hobert 2013). Of these, 13 genes encode canonical RGS proteins most likely to directly regulate heterotrimeric G proteins in the manner described above. However, the in vivo role for many of the C. elegans RGS proteins remains unknown, likely due to extensive functional redundancy and/or subtle roles in regulating signaling. For an extensive review of C. elegans RGS proteins, please see Porter and Koelle (2009).
The expression pattern of RGS-3 was key to uncovering its subtle chemosensory phenotype (Ferkey et al. 2007). RGS-3 is expressed in a subset of sensory neurons (ASH, ADL, AWB, AWC, ASI, ASJ, ASK, PHA, and PHB). rgs-3 mutant animals are defective in their response to strong ASH- and AWC-detected chemosensory stimuli, but respond normally when their concentrations are decreased (Ferkey et al. 2007). Interestingly, the defective behavioral responses of rgs-3 animals to ASH-detected stimuli likely result from aberrantly elevated ODR-3 and/or GPA-3 activity and increased calcium levels that lead to decreased synaptic transmission (Ferkey et al. 2007). However, as changes in feeding status and biogenic amine levels modulate signaling levels and sensory response, after a slightly extended time off food (30 minutes) signaling was brought into the range where the hypersensitivity of rgs-3 mutant animals could be seen (as would be expected for loss of a negative regulator of signaling) (Krzyzanowski et al. 2013). rgs-2 mutants are also hypersensitive to dilute quinine at this time-point, and ASH-selective knock-down of either rgs-3 or rgs-2 leads to quinine hypersensitivity (Krzyzanowski et al. 2013). Consistent with a role in dampening G protein signaling, overexpression of either in ASH was sufficient to decrease behavioral response to quinine (Krzyzanowski et al. 2013).
Animals lacking function of the RGS protein EGL-10 are also defective in their response to ASH-detected chemosensory stimuli (including copper, quinine and 1-octanol), but in this case EGL-10 acts downstream of the TRPV channel OSM-9, perhaps by modulating ASH synaptic transmission (Esposito et al. 2010). The avoidance defects of egl-10 animals are suppressed by mutation of the RGS-encoding gene eat-16, suggesting that the two RGS proteins act in antagonistic modulatory pathways to regulate ASH sensitivity (Esposito et al. 2010). Similarly, the two may oppose each other in olfactory adaptation (Matsuki et al. 2006).
cGMP-dependent protein kinases (PKGs)
PKGs are serine/threonine kinases that are activated by cGMP binding (Lincoln et al. 2001; Hofmann 2005). The C. elegans genome encodes two PKGs, EGL-4/PKG-1, and PKG-2 (L'Etoile et al. 2002; Manning 2005). These kinases have two tandem cGMP binding domains that block access to the kinase domain in the absence of cGMP binding. cGMP binding releases this inhibition, allowing phosphorylation of target protein (Kim et al. 2016). EGL-4 is widely expressed throughout the animal (Fujiwara et al. 2002) and plays varied roles in several different sensory neurons (see below). PKG-2 may have minor roles in the animal's sensory physiology (Manning 2005), but these will not be discussed further.
EGL-4/cGMP in ASEL/R
EGL-4 may act as a regulator of the primary sensory response in the ASEL and ASER salt sensing neurons (Suzuki et al. 2008). As discussed above (see ASE), ASEL and ASER are ON and OFF neurons whose sensory responses are mediated by ligand binding and removal, respectively (Suzuki et al. 2008; Murayama et al. 2013; Smith et al. 2013; Woldemariam et al. 2019). Suzuki et al. (2008) found there is no calcium influx in either ASEL or ASER in egl-4(n479) mutants in response to salt upsteps or downsteps. Why EGL-4 might be required in both ASEL and ASER to open the tetrameric CNG channels comprised of TAX-2, TAX-4, and CNG-4/CHE-6 (Smith et al. 2013), that presumably could be opened directly by the increase in cGMP produced by guanylyl cyclase activation, is unclear. It is known from CNG channel expression studies in mammalian tissue culture that the subunits that make up the heterotetrameric channel dictate the channel's affinity for cGMP and its open probability once bound (Komatsu et al. 1999; Matulef and Zagotta 2003; O'Halloran et al. 2017). Thus, the heterotetramer that includes CNG-4/CHE-6 might require PKG phosphorylation to modulate its cGMP affinity and subsequent opening. Alternatively, or in addition to directly increasing CNG channel open probability via phosphorylation, EGL-4 may promote ASE signaling either by potentiating other parts of the signal transduction cascade to augment signaling, or by inhibiting an adaptive (negative feedback) response. EGL-4 could also act as a negative regulator of signaling such that excessive calcium signaling in egl-4 mutant animals could stimulate a calcium-dependent negative feedback loop that ultimately inhibits ASE signaling. Such a regulatory feedback mechanism would be reminiscent of loss-of-function mutations in grk-2 (Fukuto et al. 2004) and rgs-3 (Ferkey et al. 2007) (see above).
cGMP/EGL-4 in AWC
Sensory signals adapt over multiple timescales. Adaptation on the order of seconds allows the sensory neuron to respond to small increases in stimulus concentrations so that animals can climb a gradient. Failure of adaptation at this timescale partially mimics the behavioral defects of sensory signaling mutants. Sensory neurons also adapt to longer odor exposures. Wild-type animals will switch their behavioral response from being attracted to AWC-sensed odors to ignoring them if they are exposed to the odors for longer than 30 minutes in the absence of food (Bargmann et al. 1993; Colbert and Bargmann 1995). The period of decreased responsiveness scales with the length of odor exposure, such that odor adaptation is quickly reversible if odor exposures are for less than 60 minutes, but it becomes long lasting (hours) if the exposure lasts more than 60–80 minutes (L'Etoile et al. 2002; Lee et al. 2010). The change from attraction to indifference requires EGL-4 at each time scale, beginning as early as tens of seconds, and extending through minutes and hours (L'Etoile et al. 2002; Lee et al. 2010; Juang et al. 2013; O'Halloran et al. 2017; Levy and Bargmann 2020). The hours-long decrease in responsiveness may represent associative conditioning, as it requires pairing with starvation and is blocked by food (Torayama et al. 2007; Kauffman et al. 2011; Cho et al. 2016).
At very short time scales (on the order of seconds), loss of egl-4 and failure to adapt actually leads to the inhibition of chemotaxis to butanone (Daniels et al. 2000; L'Etoile et al. 2002). In egl-4 loss-of-function mutants, AWC exhibits increased calcium influx in response to changes in butanone concentration, relative to wild-type animals (Levy and Bargmann 2020). Conversely, egl-4 gain-of-function mutants show reduced calcium influx (Levy and Bargmann 2020). Thus, EGL-4 sets the threshold for calcium responsiveness in the timeframe needed for odor sensation and adaptation in a gradient (Levy and Bargmann 2020).
At slightly longer time scales (tens of minutes), it is likely that EGL-4 phosphorylates cytoplasmic targets. Consistent with this possibility, the EGL-4 consensus site on TAX-2 is required for odor adaptation (L'Etoile et al. 2002). After exposures of longer than 60–80 minutes, EGL-4 enters the AWC nucleus (O'Halloran et al. 2009; Lee et al. 2010; Cho et al. 2016), and this translocation requires cGMP binding to EGL-4 and G-protein signaling (O'Halloran et al. 2009; Lee et al. 2010). However, aberrantly high levels of cGMP (due to loss of phosphodiesterases or application of membrane permeable cGMP) blocks nuclear translocation of EGL-4, even when animals are exposed to odor (O'Halloran et al. 2012). ODR-1 function and cilia integrity are required to keep EGL-4 in the cytoplasm until worms are exposed to odor (O'Halloran et al. 2012). The residues of EGL-4, the subcellular distribution of cGMP and the cofactors that regulate EGL-4's localization within the cell remain to be fully elucidated.
Once in the nucleus, EGL-4 phosphorylates the heterochromatin binding factor HPL-2 in a small RNA- and nuclear RNAi-dependent fashion (Juang et al. 2013). One transcriptional target downregulated by HPL-2 binding is the odr-1 guanylyl cyclase-encoding gene (Juang et al. 2013). Another target, saeg-2, was predicted from gene expression studies using a constitutively active EGL-4 allele (Hao et al. 2011). SAEG-2 was recently shown to be downregulated by inherited small endogenous RNA species (Posner et al. 2019). Thus, in addition to phosphorylating cytoplasmic targets to modulate signaling within seconds to tens of minutes, EGL-4 may also modify gene expression in a heritable manner to affect AWC-mediated behaviors across generations.
cGMP/EGL-4 in ASH
EGL-4 negatively regulates response of C. elegans to select nociceptive stimuli (Krzyzanowski et al. 2013). In the ASH neurons, cGMP binding to EGL-4 likely stimulates it to phosphorylate and activate RGS-2 and RGS-3, which in turn downregulate Gα (ODR-3 and/or GPA-3) signaling (Krzyzanowski et al. 2013). Thus, similar to loss of RGS-2 or RGS-3 function (at longer time points off food), egl-4 mutant animals respond better than wild-type animals to several ASH-detected chemical stimuli (including quinine and 1-octanol) (Krzyzanowski et al. 2013). Surprisingly, the source of cGMP in this modulatory pathway appears to be other sensory neurons that are indirectly connected to ASH via a gap junction neuronal circuit (Krzyzanowski et al. 2016). Thus, diverse sets of environmental information may be integrated, via cGMP generation and movement through a neural gap junction network, to regulate nociceptive sensitivity.
OSM-9 in AWC
Beyond its role in primary signal transduction in other sensory neurons, the TRPV channel OSM-9 is also required for adaptation to the AWC-sensed odorants butanone and isoamyl alcohol (Colbert and Bargmann 1995). It is still unclear how OSM-9 promotes adaptation to AWC-sensed odorants, but it acts in both cGMP-mediated and calcium-mediated plasticity pathways. First, OSM-9 acts downstream of nuclear EGL-4; adding an additional nuclear localization signal onto EGL-4 is sufficient to drive odor adaptation and causes animals to ignore all AWC-sensed odors, unless osm-9 is also mutated (Lee et al. 2010). Second, downstream of calcium signaling, reduced TAX-6 calcineurin function results in failure to respond to isoamyl alcohol due to AWC being constitutively adapted; chemotaxis is restored to tax-6 mutants upon loss of osm-9 (Kuhara et al. 2002).
Ras/MAPK (mitogen-activated protein kinase) pathway
The levels of Ras/MAPK pathway activity are important for C. elegans olfactory responses. For example, the most loss-of-function mutations in genes of the Ras/MAPK pathway result in mild-chemotaxis defects to diacetyl (AWA), isoamyl alcohol (AWC) and 2,4,5-trimethylthiazole (AWA and AWC) (Hirotsu et al. 2000). Loss of RGEF-1b function, a putative RasGRP (activating Ras GTP exchange factor), also disrupted chemotaxis to AWA- and AWC-sensed odorants (Chen et al. 2011). Conversely, loss of the RasGAPs GAP-1 and GAP-3 (presumed negative regulators of Ras activity) also leads to mild chemotaxis defects (Gyurkó et al. 2015). GAP proteins also act, in part, through LET-60 Ras to regulate learning and memory in C. elegans (Gyurkó et al. 2015), although their site of action in this context is not known.
Recruitment of the Ras/MAPK pathway downstream of primary sensory signaling can modulate behavioral sensitivity. Application of isoamyl alcohol led to activation of Ras itself within seconds, dependent upon the function of the upstream olfactory signaling components ODR-3 and TAX-2 (Uozumi et al. 2012). Within 10 seconds of isoamyl alcohol addition, MAP kinase is activated and this is dependent upon TAX-2/TAX-4 and UNC-2 (Hirotsu et al. 2000). In addition, upregulation of pathway activity (via over active LET-60/Ras or loss of MPK-1) leads to strong chemosensory defects, particularly in response to isoamyl alcohol (Hirotsu et al. 2000; Uozumi et al. 2012). This suggests that activation of the Ras/MAPK pathway following odor exposure may be an additional mechanism to downregulate the AWC-mediated olfactory response, at least in part by modulating interneuron activity (Uozumi et al. 2012). However, the exact mechanism by which the MAPK pathway does so is unknown. A negative feedback loop also quickly inactivates Ras, and the dynamics of Ras activation may help to tune an animal’s response to changes in odor concentration during klinotaxis (Uozumi et al. 2012).
Sensing and transducing pheromone signals
C. elegans employs a remarkably rich chemical language comprised of hundreds of small molecules to communicate with conspecifics (McGrath and Ruvinsky 2019). Although the presence of molecules with biological activity in conditioned medium from C. elegans cultures has been known since the early 1980s (Golden and Riddle 1982, 1984c), the identities of a subset of these compounds were first described only several decades later (Jeong et al. 2005; Butcher et al. 2007; Srinivasan et al. 2008; Pungaliya et al. 2009). The major class of molecules that acts as pheromones in C. elegans is a structurally related family of chemicals derived from the sugar ascarylose and containing fatty acid side chains (ascarosides) (Ludewig and Schroeder 2013; Butcher 2017a; Figure 5). The specific ascarosides produced, and their relative concentrations, are regulated by the animal’s sex, developmental stage, reproductive and metabolic status, as well as experience (Butcher 2017a; Park et al. 2019b). Not surprisingly, individual ascarosides and ascaroside mixtures elicit diverse effects in responding animals, ranging from acute behavioral responses (“releaser effect”) to altered development and physiology via modulation of neuroendocrine signaling (“primer effect”) (Wyatt 2003). There is evidence that molecules other than ascarosides also act as pheromones, although their chemical identities have not been comprehensively elucidated (see below for further discussion and references). This section describes the current knowledge regarding the sensory neurons and molecules required for pheromone-mediated regulation of physiology and behavior. We refer the reader to several excellent recent reviews for information on pheromone biosynthesis and composition in C. elegans (Ludewig and Schroeder 2013; Chute and Srinivasan 2014; Butcher 2017a, b, 2019; Park et al. 2019b).
Figure 5.
Summarized overview of the chemical structure of ascaroside pheromones. The ascarylose sugar moiety (black) is attached to a fatty acid of different chain lengths (blue). Subsets of pheromone molecules also contain additional components (indicated as R1, R2, red, and green). Reproduced from McGrath and Ruvinsky (2019).
Pheromone signal transduction in the regulation of development and physiology
Regulation of dauer formation
C. elegans larvae develop into reproductive adults via one of two mutually exclusive developmental trajectories (Fielenbach and Antebi 2008). Under conditions of plentiful food, low temperature, and low-population density (and thus low levels of pheromone), L1 larvae progress sequentially through the subsequent L2–L4 larval stages to develop into reproductive adult hermaphrodites. However, under adverse environmental conditions, L1 larvae instead enter into the long-lived and stress-resistant dauer stage (Cassada and Russell 1975; Golden and Riddle 1982, 1984b, c). When conditions improve, dauer larvae exit the dauer stage and develop into reproductive post-dauer adults (Fielenbach and Antebi 2008). Both high concentrations of pheromone and high temperature are instructive for dauer formation (Golden and Riddle 1982, 1984a, b; Ailion and Thomas 2000), whereas food levels are generally permissive (Golden and Riddle 1984a; Neal et al. 2015; O'Donnell et al. 2018).
Of the many ascarosides that have been identified, a subset including ascr#1, ascr#2, ascr#3, ascr#5, ascr#8, and icas#9 has been shown to regulate dauer entry (Jeong et al. 2005; Butcher et al. 2007, 2008, 2009a; Srinivasan et al. 2008; Pungaliya et al. 2009). Pheromone chemoreceptors necessary for dauer formation include the GPCRs SRBC-64 and SRBC-66 expressed specifically in ASK (Kim et al. 2009), SRG-36 and SRG-37 expressed specifically in ASI (McGrath et al. 2011), and DAF-37 and DAF-38 expressed in ASI, ASK, and IL2 (Park et al. 2012). Although SRBC-64 and SRBC-66 act nonredundantly, SRG-36 and SRG-37 act partially redundantly to mediate dauer formation (Kim et al. 2009; McGrath et al. 2011; Lee et al. 2019; Table 3). DAF-37 acts in ASK to modulate dauer formation (Park et al. 2012), and has been suggested to heterodimerize with DAF-38 (Park et al. 2012; Table 3). With the exception of DAF-38 whose localization pattern has not been reported, these receptors are localized to the sensory ciliated endings of the corresponding expressing neurons (Kim et al. 2009; McGrath et al. 2011; Park et al. 2012).
Table 3.
Summary of discussed sensory neurons and signaling molecules that transduce ascaroside signals in multiple contexts
| Sensory neuron(s) | Chemoreceptors | Other signaling molecules | Context |
|---|---|---|---|
| ASK |
SRG-64, SRG-66, DAF-37, DAF-38 |
GPA-2, GPA-3, DAF-11 TAX-2, TAX-4 |
Dauer formation |
| DAF-37 | ? | Lifespan | |
| ? | TAX-4 | Attraction | |
| ASI | SRG-36, SRG-37 |
GPA-2, GPA-3 DAF-11 TAX-2, TAX-4 |
Dauer formation |
| SRX-43 | Foraging | ||
| ADL | ? | GPA-3 | Lipid metabolism |
| ? | OSM-9, OCR-2 | Reproductive physiology | |
| ? | OSM-9, OCR-2 | Avoidance | |
| ? | ? | Pathogen learning | |
| ASH | TYRA-2 | GPA-6 | Avoidance |
| ASJ, ADL | SRX-44 | ? | Foraging |
| ASJ, AWB, AWC | ? | TAX-2, TAX-4 | Reproductive physiology |
Question marks indicate neurons with a possible role in detecting a stimulus.
srbc-64 and srbc-66 mutants exhibit strong defects in dauer formation induced by low concentrations of ascr#1, ascr#2, and ascr#3, with weaker defects in ascr#5-induced dauer entry (Kim et al. 2009). In contrast, srg-36 srg-37 double mutants are specifically defective only in ascr#5-induced dauer formation (McGrath et al. 2011) and daf-37 mutants exhibit specific defects in ascr#2-induced dauer entry (Park et al. 2012). Moreover, photoaffinity-labeled ascr#2 has been shown to bind DAF-37 expressed heterologously in mammalian cells (Park et al. 2012). As the conditions used to induce dauer formation greatly influence the rate of dauer entry (Golden and Riddle 1984a), it is possible that the extent of contribution of each GPCR is distinct in different conditions (Lee et al. 2019), in part accounting for the presence of multiple receptors for each ascaroside in the regulation of dauer formation. The requirement for receptors is also likely to be distinct at different ascaroside concentrations. The receptive range and tuning breadth of these receptors remain to be comprehensively assessed either genetically or biochemically.
The signaling events downstream of the GPCRs in dauer formation are not well understood. Heterologous expression experiments suggest that ascarosides may act as inverse agonists of SRBC-64 and SRBC-66 although whether these receptors act similarly in vivo is unclear (Kim et al. 2009). The GPA-2 and GPA-3 Gα proteins have been shown to be necessary for dauer formation, and are expressed in ASK and ASI as well as in other neuron types (Zwaal et al. 1997; Jansen et al. 1999; Table 3). Mutations in the DAF-11 rGC, and TAX-2 and TAX-4 cGMP-gated channel subunits, also lead to dauer formation defects (Riddle et al. 1981; Vowels and Thomas 1994; Coburn and Bargmann 1996; Komatsu et al. 1996; Ailion and Thomas 2000; Birnby et al. 2000). However, since these genes are expressed broadly and are implicated in transducing dauer-regulatory chemosensory as well as thermosensory signals, it is not clear whether pheromones signal via regulation of intracellular cGMP concentrations. Interestingly, ascarosides do not appear to modulate intracellular calcium dynamics in either ASK or ASI via these receptors (Kim et al. 2009; McGrath et al. 2011). However, mis-expression of SRG-36 or SRG-37 in the ASH nociceptive neurons is sufficient to drive ascr#5-induced avoidance and regulate calcium dynamics in these neurons in adult animals (McGrath et al. 2011), indicating that these receptors are able to couple with calcium signaling pathways in specific contexts. Pheromone signals are integrated with food and temperature cues over hours-long timescales during development to regulate expression of neuroendocrine ligand genes such as the daf-7 TGF-β and daf-28 insulin-like peptide to drive the dauer decision; in this context these molecules act as primer pheromones (Ren et al. 1996; Schackwitz et al. 1996; Li et al. 2003; Wyatt 2003; Cornils et al. 2011; Schaedel et al. 2012; Avery 2014; Entchev et al. 2015; Neal et al. 2015; O'Donnell et al. 2018). How pheromone cues sensed by their cognate receptors in multiple sensory neurons are transduced, and how these signals are integrated with food and temperature information to regulate neuroendocrine signaling, remain open questions.
Regulation of lifespan and physiology
In addition to regulating a larval developmental decision, ascarosides also regulate C. elegans lifespan. Exposure of late stage larvae and adult hermaphrodites to ascr#2 and ascr#3 extends lifespan and increases stress resistance; ascr#2 mediates this effect via the DAF-37 GPCR in ASK (Kawano et al. 2005; Ludewig et al. 2013; Table 3). The excreted nonascaroside molecule nacq#1 N-acetylated glutamine derivative has recently been shown to accelerate reproductive development and shorten lifespan; ascr#2 and ascr#3 antagonize these effects of nacq#1 (Ludewig et al. 2017, 2019; Wharam et al. 2017). In contrast, chemicals such as ascr#10 produced by males shorten hermaphrodite lifespan and can kill other males (Maures et al. 2014; Shi et al. 2017; Ludewig et al. 2019). Although ciliated sensory neurons have been implicated in sensing these small molecules in the context of lifespan regulation, the required signal transduction pathways in these neurons are largely unknown.
Exposure to ascarosides also affects additional aspects of C. elegans physiology. A recent study has shown that pheromone can regulate body fat stores (Hussey et al. 2017). In one underlying pathway, ascr#3 acts via the GPA-3 Gα protein in ADL to downregulate intracellular cAMP levels (Table 3). cAMP signaling in turn modulates cholinergic signaling to regulate expression of the atgl-1 triglyceride lipase in intestinal cells (Hussey et al. 2017). Ascarosides also regulate reproductive physiology in C. elegans hermaphrodites. Low concentrations of ascr#3 and ascr#10 mixtures in ratios normally produced by males regulate hermaphrodite reproductive development, sperm guidance toward oocytes and aging-dependent loss of germline progenitor cells (Aprison and Ruvinsky 2015, 2016, 2017). In this context, the ascr#10 signal requires OSM-9/OCR-2 TRPV channel function in ADL in hermaphrodites; ascr#10 signaling is antagonized by ascr#3 signaling in the ASJ, AWB, and AWC neurons mediated via cGMP (Aprison and Ruvinsky 2017; Table 3). Additional ascarosides including ascr#2 and ascr#3 have been reported to also affect sperm motility in the oviduct (McKnight et al. 2014). Together, these observations indicate that different pheromone components, singly or together, have complex effects on diverse aspects of C. elegans physiology.
Pheromone signal transduction in the regulation of behavior
Attraction and aversion
In addition to modulating development and physiology, pheromones elicit acute behavioral responses. A key characteristic of responses such as attraction and avoidance of pheromone is that they are highly state-dependent. Pheromone-elicited behaviors are modulated by sex, environmental conditions, internal state, and past experience. A subset of the sensory neurons and circuits mediating these behavioral responses in adults has been identified, although little is known about the required sensory signaling molecules.
Both the attraction-promoting ASK and nociceptive ADL neurons respond to ascarosides (Macosko et al. 2009; Jang et al. 2012; Fenk and de Bono 2017; Wu et al. 2019). ASK and ADL comprise a subset of the “spokes” of a hub-and-spoke circuit motif (White et al. 1986; Macosko et al. 2009; Jang et al. 2012; Figure 6). In this circuit, the RMG inter/motor neuron is the central hub that is connected to spoke sensory neurons via gap junctions (Jang et al. 2017). In addition to being electrically coupled to RMG, each spoke sensory neuron as well as RMG itself also has chemical synapses to interneurons (White et al. 1986). Behavioral, genetic, and imaging analyses have indicated that sensory inputs into individual spokes of this circuit are integrated by RMG in a state-dependent manner, and via NPR-1 neuropeptide Y receptor-mediated signaling (de Bono and Bargmann 1998), to regulate both sensory responses in the spoke neurons, as well as synaptic outputs from the circuit, thereby modulating pheromone responses as a function of context (Macosko et al. 2009; Jang et al. 2012, 2017; Fenk and de Bono 2017). Interestingly, ASK also mediates attraction to the icas#3 ascaroside, but these behaviors are NPR-1- and RMG-independent (Srinivasan et al. 2012; Table 3).
Figure 6.
The RMG hub motor/interneurons are synaptically and electrically connected to O2-sensing, nociceptive and pheromone-sensing neurons. The RMG hub-and-spoke circuit integrates external and internal state information to modulate pheromone avoidance and attraction. Wiring based on White et al. (1986).
Conditions of high NPR-1 signaling in RMG enable robust pheromone response in ADL, and weaker response in ASK, resulting in net avoidance of ascarosides, whereas upon loss of npr-1 signaling, ASK and ADL ascaroside responses are increased and decreased, respectively, thereby driving weak attraction (Macosko et al. 2009; Jang et al. 2012; Figure 6). Pheromone responses in these sensory neurons are also further modulated by prior oxygen experience via RMG and URX, another oxygen-sensing neuronal spoke in the RMG-centered circuit (Fenk and de Bono 2017; Figure 6). Synaptic output but not sensory responses of ADL to ascr#3 are also regulated by the animal’s satiety state such that starved animals exhibit enhanced ascr#3 avoidance (Ryu et al. 2018). Moreover, early ascr#3 exposure has recently been shown to potentiate ascr#3 avoidance in adult hermaphrodites via modulation of ADL-driven synaptic activity (Hong et al. 2017). Finally, ADL but not ASK pheromone responses are sexually dimorphic (Jang et al. 2012). Males exhibit additional sexually dimorphic ascaroside responses that are mediated by both sex-shared and sex-specific sensory neurons (Srinivasan et al. 2008, 2012; Pungaliya et al. 2009; Narayan et al. 2016; Fagan et al. 2018). We refer the reader to the Wormbook chapter by Barr et al. (2018) for details on sensory neurons required for male-specific pheromone-elicted behaviors.
Transduction of pheromone signals requires the TAX-4 cGMP-gated channel and the OCR-2 and OSM-9 TRPV channels in ASK and ADL, respectively (Macosko et al. 2009; Jang et al. 2012; Table 3). The GPA-3 Gα protein has also been implicated in ascaroside avoidance behaviors (Park et al. 2017). Additional required signaling molecules including receptors in these sensory neurons are largely unknown, although DAF-37 has been implicated in ascr#2 sensation in ASK in adult animals (Park et al. 2012). Chemoreceptors mediating ascaroside responses in males have not been characterized.
Nonascaroside components also elicit avoidance and attraction in C. elegans (Zhou et al. 2017). Both male and hermaphrodite C. elegans avoid the internal fluid that is leaked from injured animals (Zhou et al. 2017). The active chemicals in this fluid are unlikely to be ascarosides but appear to be nonvolatile and require direct contact to result in repulsion (Zhou et al. 2017). Avoidance is mediated in part by cGMP signaling in the ASI and ASK sensory neurons but does not require the known ascaroside receptors that are expressed in these neuron types (Zhou et al. 2017). The presence of sperm in the hermaphrodite gonad decreases their ”sex appeal” to males via the production of nonascaroside volatile chemicals (Morsci et al. 2011; Leighton et al. 2014). Nonascaroside chemicals produced by gravid hermaphrodites also robustly attract wild-type sexually mature males at a distance but have no effect on hermaphrodite behavior (Chasnov et al. 2007; White et al. 2007). Male attraction to a subset of these ”sex pheromones” is largely mediated by the sex-shared AWA and AWC, as well as the male-specific CEM sensory neurons (Chasnov et al. 2007; White et al. 2007). Responses to volatile sex pheromones in the AWA sensory neurons have recently been shown to be mediated by the SRD-1 GPCR (Wan et al. 2019). SRD-1 is expressed in AWA, ASI, and ADF in adult but not larval males, with expression in AWA and ADF being sexually dimorphic (Troemel et al. 1995; Wan et al. 2019). Sex pheromone receptors in other sensory neurons in males are as yet unidentified.
Avoidance of osas#9
A particularly intriguing class of small molecule pheromones in C. elegans are ascarosides that are connected to byproducts of other metabolic pathways. For instance, osas molecules are comprised of ascarosides connected to succinylated octopamine, the invertebrate analog of norepinephrine. Correlated with upregulation of octopamine by nutrient deprivation (Tao et al. 2016), osas#2, osas#9, and osas#10 are produced specifically by starved animals, although production appears to be restricted to L1 larvae (Artyukhin et al. 2013). osas#9 (derived from ascr#9) elicits robust avoidance behaviors by starved larvae and adults, suggesting that this molecule acts as a signal promoting dispersal from unfavorable conditions (Artyukhin et al. 2013; Chute et al. 2019). Interestingly, consistent with the presence of an octopamine moiety on osas#9, avoidance appears to be mediated by the TYRA-2 tyramine/octopamine receptor and the GPA-6 Gα protein in the ASH nociceptive neurons (Table 3), although receptors in addition to TYRA-2 expressed in other sensory neuron types may also contribute to the response (Rex et al. 2005; Chute et al. 2019). tyra-2 expression is upregulated upon starvation (Chute et al. 2019), providing a plausible mechanism for enhanced osas#9 aversion by starved animals. These observations suggest that C. elegans has co-opted both a neurotransmitter and neurotransmitter receptor for inter-organismal signaling of environmental conditions, and raise the possibility that additional related molecules and transduction pathways communicate unique contextual cues.
Regulation of olfactory behavioral plasticity
In addition to directly eliciting behaviors, pheromone experience can modulate responses to other chemical cues in adult C. elegans in part via regulation of sensory gene expression. For instance, it has long been known that exposure to an initially attractive chemical in the absence of food subsequently abolishes attraction to that chemical (Colbert and Bargmann 1995; Hirotsu and Iino 2005). The extent of this behavioral plasticity is modulated by prior ascaroside exposure (Yamada et al. 2010) and is abolished in daf-22 mutants which fail to produce ascarosides (Golden and Riddle 1985; Butcher et al. 2009b; Yamada et al. 2010). Pheromone downregulates expression of the olfactory plasticity-antagonizing neuropeptide snet-1 in pheromone-sensing neurons such as ASK and ASI to decrease attraction (Yamada et al. 2010). Pheromones also modulate expression of a subset of GPCRs in sensory neurons, although the behavioral consequence of this regulation is currently unclear (Peckol et al. 2001; Kim et al. 2009; Park et al. 2019a). At least a subset of pheromone-mediated modulation of sensory gene expression is mediated by known pheromone receptors such as SRBC-64 and SRBC-66 (Kim et al. 2009; Park et al. 2019a).
In a well-characterized learning paradigm, C. elegans learns to avoid odors associated with the pathogenic bacteria P. aeruginosa PA14 following a period of feeding on this bacterial strain and subsequent infection (“training”) (Zhang et al. 2005). Addition of a mixture of ascr#2, ascr#3, and ascr#5 to the training plates was found to significantly decrease pathogen avoidance behavior and increase pathogen resistance in C. elegans in trained animals in part via modulation of insulin signaling from sensory neurons such as AWA and ADL (Wu et al. 2019). The ascaroside mixture was shown to increase intracellular calcium in ADL but not AWA (Wu et al. 2019). While training with PA14 decreased pheromone responses in ADL, prior pheromone exposure was sufficient to abolish this suppression in trained animals (Wu et al. 2019). These observations indicate that C. elegans integrates information about social context and population density to adaptively modulate feeding decisions. The signaling pathways mediating pheromone responses in ADL in naïve and trained animals in this context has not been examined.
Modulation of exploratory behavior
On a uniform concentration of bacterial food, C. elegans spontaneously switches between locomotory behavioral states referred to as roaming and dwelling (Fujiwara et al. 2002; Ben Arous et al. 2009). Roaming animals are active and explore the bacterial lawn, whereas dwelling animals exhibit slow speeds and restrict their movement to a small region by increasing the frequency of high-angle turns (Fujiwara et al. 2002; Ben Arous et al. 2009). In addition to satiety state (Fujiwara et al. 2002; Shtonda and Avery 2006; Ben Arous et al. 2009; Flavell et al. 2013), exposure to a subset of ascarosides including ascr#2, ascr#3, ascr#8, and icas#9 has been shown to regulate foraging behavior (Greene et al. 2016a). In the presence of relatively high concentrations of these chemicals, exploration is suppressed primarily via decreasing the fraction and duration of time animals spend in the roaming state. Analyses of natural variation in ascaroside-mediated regulation of foraging in multiple C. elegans strains identified the SRX-43 chemoreceptor in the ASI chemosensory neurons that specifically mediates icas#9-induced regulation of exploratory behavior (Greene et al. 2016a, b; Table 3). icas#9 regulates daf-7 TGF-β and daf-28 ILP expression in ASI (Greene et al. 2016a, b), indicating that as in dauer formation this ascaroside acts as a primer pheromone via SRX-43 to modulate exploratory behavior. icas#9- and SRX-43-mediated regulation of exploration is further modulated by icas#9 acting via the SRX-44 GPCR in ASJ and ADL (Greene et al. 2016b; Table 3).
Why might ascarosides suppress foraging? An interesting possibility has recently been proposed in the context of ascr#10-mediated suppression of exploration. The male-specific ascr#10 reduces exploratory behavior in hermaphrodites in part via increased serotonergic signaling (Aprison and Ruvinsky 2019a). Reduced roaming enhances mating success (Aprison and Ruvinsky 2019a), suggesting a plausible physiological relevance for the ascaroside-mediated suppression of foraging. Providing an intriguing example of how pheromones can additionally coordinate physiology and behavior, ascr#10-induced upregulation of serotonergic signaling also promotes egg-laying which in turn is permissive for ascr#10-mediated suppression of foraging (Aprison and Ruvinsky 2019b). Although ascr#10 has been shown to act via ADL and the OSM-9 TRPV channel to modulate reproductive physiology (Aprison and Ruvinsky 2017), whether this pathway is also involved in modulation of foraging behavior has not been established.
Conclusions
Chemical stimuli, including odorants and tastants, provide information about individuals of the same species, food availability, food quality, and environmental threats. From the studies described here, unifying themes have emerged, as well as new questions. For example, while each C. elegans sensory neuron expresses multiple GPCRs, it is not yet clear how a neuron might discriminate between stimuli that couple to multiple G proteins and activate shared downstream signal transduction components. Continued receptor de-orphanization efforts may also shed light on whether C. elegans chemosensory GPCRs are dedicated to select chemicals, or can respond (alone or in combination) to a range of stimuli. We can now watch, in real time, how both calcium and cyclic nucleotide messages traverse the sensory neuron, which will allow analysis of the role subcellular localization of these signals may play in sensory transmission, integration (or partitioning), and regulation. Whether the dynamics of nuclear translocation of signaling and transcription factors play a role in shaping the output signals from sensory neurons is another area left to explore. Furthermore, how narrow or broad is the role for left/right neuronal asymmetry? And, while the sensory neuron itself is the site of extensive plasticity, how are signals from multiple sensory neurons integrated within the nervous system to sharpen context- and experience-dependent behavioral responses? With the continued development of tools for single-cell and circuit-level analyses, as well as computational approaches to analyze complex data sets, future work will continue the quest begun five decades ago to understand sensory signaling and behavior in the “simple” model organism, C. elegans.
Acknowledgments
We are grateful to Miriam Goodman, Jonathan Pierce, and Patrick McGrath for valuable discussions and comments on this work, to Miriam Goodman for contributions to the behavior strategies section, and to Joseph Hill for creating the illustrations. We also thank members of the Ferkey, L’Etoile, and Sengupta labs for feedback.
Funding
This work was supported by the National Institutes of Health (R01DC015758 to D.M.F, R01DC005991 and R01NS087544 to N.D.L), and the National Science Foundation (NSF IOS 1655118 to P.S.).
Conflicts of interest
None declared.
Appendix
Behavioral strategies used to track toward or avoid chemical cues
C. elegans navigate, exploit and survive shifting environments by responding to volatile and dissolved chemical cues that may be encountered as nonuniform concentration gradients. Moreover, these signals can travel in waves or plumes from distant sources. Thus, the sensory neurons that detect these stimuli need to respond to stimuli that vary over orders of magnitude. To accurately locate the source of an attractive stimulus, the sensory neuron must also be able to adapt to the local concentration to enable response to further concentration changes and, thus, allow the animal to ascend the stimulus gradient. In theory, these neurons may also be able to integrate stimulus concentration over time in order to track discontinuous stimuli (e.g., waves of odor) (Kato et al. 2014; Levy and Bargmann 2020).
Worms use multiple motor strategies to locate the source of attractive chemical cues and to avoid noxious ones. Location of an attractive source appears to be primarily mediated via klinotaxis and klinokinesis on a gradient. When removed from food, animals initially exhibit a brief period of area-restricted search characterized by frequent high-angle turns (Wakabayashi et al. 2004; Gray et al. 2005), which is followed by increased periods of roaming (Tsalik and Hobert 2003; Hills et al. 2004; Wakabayashi et al. 2004; Gray et al. 2005; Roberts et al. 2016). Klinotaxing animals run smoothly up a chemical gradient, gradually reorienting by biasing head swings such that they angle up the gradient (Figure A1) (Iino and Yoshida 2009; Izquierdo et al. 2015). This weathervane-like steering depends on chemosensory signals that modulate ongoing sinusoidal head sweeps (Iino and Yoshida 2009; Kato et al. 2014; Satoh et al. 2014; Izquierdo et al. 2015). Although animals maintain a constant crawling speed of 0.12 mm/s as they run directly up a salt gradient (Iino and Yoshida 2009), their speed in an olfactory gradient can be altered by odor concentration (Albrecht and Bargmann 2011).
The major contributor to chemotaxis, klinokinesis, is also termed a biased random walk and pirouette strategy (Pierce-Shimomura et al. 1999, 2005; Wakabayashi et al. 2004; Gray et al. 2005; Luo et al. 2014) (Figure A1). A pirouette is a bout of one or more coupled reversals and omega turns, during which an animal bends head to tail into the shape of the Greek letter Ω (Croll 1975b; Pierce-Shimomura et al. 1999; Broekmans et al. 2016). In general, when moving up the gradient of an attractive chemical, animals suppress the frequency of reversals and turns and increase forward run duration, whereas when moving down the gradient, animals increase reversal frequency and terminate runs. The converse strategy is used when avoiding a repellent (Tanimoto et al. 2017) or high-salt concentration (Kunitomo et al. 2013). When exiting the pirouette, the probability that the animal’s head will be oriented up the chemical gradient for an attractive chemical is higher than chance alone (Pierce-Shimomura et al. 1999). Each strategy depends upon the animal’s ability to monitor the change in chemical concentration as a function of time as the worm crawls through a spatial chemical gradient.
Combinations of klinotaxis and klinokinesis may be employed in response to all attractive odors and soluble cues (Iino and Yoshida 2009). Interestingly, feedback from motor neurons and the substrate animals are crawling on or swimming through may shape the strategy that animals use to locate an attractive chemical (Hendricks et al. 2012; Hendricks and Zhang 2013). Animals have also been observed to pause, perhaps to integrate stimuli over time (Roberts et al. 2016; Steuer Costa et al. 2019).
Worms also display acute avoidance responses when confronted with a noxious chemical cue. When presented at the nose, the worm halts forward locomotion and initiates backing within seconds. Such reversals are often coupled to an omega turn, enabling the animal to continue moving in a different direction (Croll 1975a, b). Animals exposed to noxious chemicals applied to their tails accelerate forward (Hilliard et al. 2002). In the wild, additional strategies, including three dimensional maneuvers, are likely to be employed (Bilbao et al. 2018).
rGC structure and activation
rGCs can be both activated and inactivated by extracellular ligands as well as by intracellular calcium binding factors, phosphorylation and ATP binding, and by effectors of G protein-coupled signaling (Sharma et al. 2016; Maruyama 2017). Extracellular ligands activate the single pass transmembrane rGCs by binding to their amino-terminal extracellular domain (ECD). Mammalian rGCs are obligate homodimers comprised of identical alpha and beta subunits whose activities are regulated by conformational changes upon ligand binding to the ECD (Sharma et al. 2016). The ECD is followed by a transmembrane domain and an intracellular kinase homology domain (KHD), then an ∼50 residue hinge region and a carboxy-terminal cyclase domain that is inactive until it dimerizes (Sharma et al. 2016; Maruyama 2017).
C. elegans chemosensory responses to soluble ligands are likely to be transduced by direct binding of ligand to the ECD of rGCs (Figure 4, D and E): sodium and hydroxyl ions to GCY-14 (ASEL) (Murayama et al. 2013), K+ to GCY-1 (ASER), Br− and I− to GCY-4 (ASER), and Cl−, Br−, and I− to GCY-22 (ASER) (Smith et al. 2013). When each ECD was appended to a different intracellular rGC domain, and the fusion protein was mis-expressed in a nonnative cell type, appropriate ligand responsiveness (based on the ECD) was restored to animals that lacked the original respective rGC.
Although many rGCs may be activated by ligand binding, some, such as the human retinal rGC GUCY2D (Duda et al. 1996), have residues in their KHDs that allow dimerization in the absence of a ligand. This raises the possibility that some rGCs may be tonically dimerized and active, and ligand binding disrupts the dimerization to inactivate the cyclase. Consistent with this possibility, GCY-8 (which is normally expressed in the thermosensory AFD cells) was constitutively active when expressed in mammalian tissue culture cells and was inactivated by chloride binding to its ECD (Singhvi et al. 2016). The conserved D(F/H/Y)G motif in the KHD of GCY-8, as well as many other C. elegans rGCs, is thought to indicate whether a rGC has basal activity.
Constitutive basal rGC dimerization and activity, that is inactivated by ligand binding and restored by ligand release, could explain how stimulus application would silence (and removal trigger) calcium influx in a sensory neuron that expresses CNG channels and exhibits an OFF calcium response. Most C. elegans rGCs have residues in the hinge region that are consistent with the possibility that they are tonically active. By contrast, in vertebrate vision cGMP decreases are executed by GPCR-activated phosphodiesterases (Sharma and Duda 2014).
Intracellular regulators can also directly activate rGCs via mechanisms that are independent of ligand binding, but also result in moving the catalytic subunits together (for cGMP cyclization) or apart (for inhibition of this reaction) (Sharma et al. 2016; Maruyama 2017). One such proposed activator class is the Gα subunits released from seven transmembrane G protein-coupled receptors in response to their stimulation (Winger et al. 2008; Maruyama 2017). For example, ODR-1 (L'Etoile and Bargmann 2000) and GCY-12 (Fujiwara et al. 2015) may be primarily regulated through their ICDs. When the crystal structure of the cyclase domain of the soluble green algae guanylyl cyclase was solved and compared to that of an adenylyl cyclase, shared features indicated that the rGCs, like the adenylyl cyclases, could possibly be regulated (activated or inactivated) by G alpha binding to the hinge region (Winger et al. 2008). Olfactory signaling may depend on this type of intracellular regulation; ODR-1 is required for chemotaxis to all AWC-sensed odors typically tested, but removal of its ECD did not affect odor taxis (L'Etoile and Bargmann 2000). This indicates that ODR-1 may be directly regulated by GPCR signaling, possibly via a Gα (Figure 4C). Likewise, the ECD of GCY-12 was not required for its role in body size regulation (Fujiwara et al. 2015), suggesting that it also is likely activated by intracellular regulators that are downstream of other receptor pathways. Alternatively, ODR-1 and GCY-12 may heterodimerize with another rGC that is activated (or inactivated) by an extracellular ligand.
In the mammalian retina, rGCs are activated by calcium-dependent guanylyl cyclase activator proteins (GCAPs), which bind to the hinge region of rGCs at the conserved residues DIVGFTALSAESTPMQVV. By binding to this region, they promote association of the catalytic domains of the alpha and beta subunits into an active cyclase, but this activation is independent of ligand binding. The calcium that activates the retinal GCs is supplied by the opening of CNG cation channels in the dark, and this promotes the dark current (Sharma and Duda 2014). All C. elegans rGCs contain portions of this hinge sequence and may thus be activated by C. elegans calcium-activated GC regulators such as NCS-1 and others (Gomez et al. 2001; Wang et al. 2013).
Thus, there are at least two independent ways that C. elegans rGC activity can be regulated: one driven by ligand binding to or detachment from the extracellular domain, which triggers a cascade of structural changes that orient the cyclase domains into their active conformation, and the other by intracellular factors binding to the cyclase and hinge region. These two mechanisms can act on the same rGC at the same time.
rGC homodimers and heterodimers
Each individual mammalian rGC cyclase domain contains all the residues needed to coordinate magnesium and bind to GTP, but they nonetheless require dimerization to cyclize GTP into cGMP (Morton 2004). Although mammalian rGCs and C. elegans GCY-14 act as obligate homodimers, a number of C. elegans rGCs have been hypothesized to act as obligate heterodimers because they lack key GTP-binding and Mg++-coordinating residues that would only be provided by a complementary heterodimer (Morton 2004). Experimental support for this hypothesis comes from studies of the ICDs of the AWC-expressed rGCs ODR-1 and DAF-11, which are both required for chemotaxis to all AWC-sensed odors (Birnby et al. 2000; L'Etoile and Bargmann 2000). In order to determine if the ECD of ASEL-expressed GCY-14 directly responds to alkaline pH, the ECD of GCY-14 was appended to the ICD of either ODR-1 or DAF-11 (Murayama et al. 2013). The alkaline seeking and calcium influx (ON response) in response to a pH increase was only restored in gcy-14 mutants upon expression of both, but not single, fusion proteins in ASEL. This indicates that the ICDs of ODR-1 and DAF-11 can heterodimerize in ASEL, and that the ECD can direct them to increase production of cGMP in response to extracellular ligand binding. Formal proof that ODR-1 and DAF-11 heterodimerize in their native cell types awaits additional experiments.
GCY-1, GCY-4, and GCY-22 may also act as either homodimers (based on sequence analysis) or heterodimers. In support of the latter possibility, mis-expression of GCY-4 in ASI restored iodide responsiveness to animals lacking ASE, but only when co-expressed with GCY-22 (Smith et al. 2013). Confirmation of heterodimer formation for these rGCs awaits biochemical or tissue culture experiments. The ability to heterodimerize could provide unique cGMP output patterns—possibly allowing different kinetics and downstream regulation, as well as combinatorial ligand binding. For example, if the six rGCs known to be expressed in ASER can homo- and hetero-dimerize, this would give rise to 21 distinct receptors, for a similarly wide array of possible ligands. Such diversity may make sense for an animal that relies on chemosensory cues to navigate its world.
Figure A1.
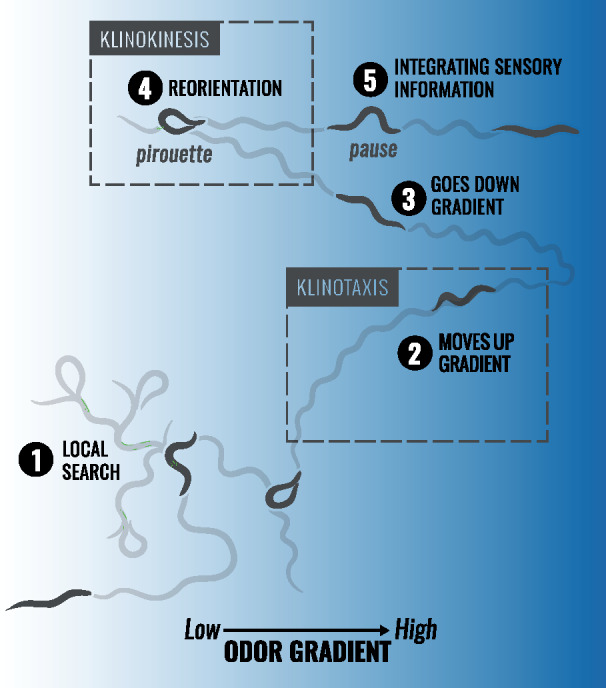
C. elegans movement in a gradient. When initially removed from food, the animal executes a search for food in its local vicinity by making short forward movements, backing and making omega turns (1). If the animal senses an attractive chemical gradient (blue) it can run up the concentration gradient and slightly curve into the higher concentration in a movement pattern called klinotaxis (2). If it veers from the gradient and travels to lower concentrations (3), it can reorient by executing a series of reversals and omega turns (termed a pirouette) from which the head is more likely to be reoriented such that it is directed up the gradient (4). Klinokinesis is also known as a biased random walk in which animals run up a gradient, and if they leave the gradient, reorient with turns. This is the major strategy used to travel up the gradient. Animals have been shown to pause (5), which may allow for sensory integration before resuming the search for the source of the attractive chemical cue.
Literature cited
- Ailion M, Thomas JH.. 2000. Dauer formation induced by high temperatures in Caenorhabditis elegans. Genetics 156:1047–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht DR, Bargmann CI.. 2011. High-content behavioral analysis of Caenorhabditis elegans in precise spatiotemporal chemical environments. Nat Methods 8:599–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson A, McMullan R.. 2018. Neuronal and non-neuronal signals regulate Caernorhabditis elegans avoidance of contaminated food. Philos Trans R Soc Lond B Biol Sci. 373:20170255. [10.1098/rstb.2017.0255] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki R, Yagami T, Sasakura H, Ogura K, Kajihara Y, et al. 2011. A seven-transmembrane receptor that mediates avoidance response to dihydrocaffeic acid, a water-soluble repellent in Caenorhabditis elegans. J Neurosci. 31:16603–16610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aprison EZ, Ruvinsky I.. 2015. Sex pheromones of C. elegans males prime the female reproductive system and ameliorate the effects of heat stress. PLoS Genet. 11:e1005729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aprison EZ, Ruvinsky I.. 2016. Sexually antagonistic male signals manipulate germline and soma of C. elegans hermaphrodites. Curr Biol. 26:2827–2833. [DOI] [PubMed] [Google Scholar]
- Aprison EZ, Ruvinsky I.. 2017. Counteracting ascarosides act through distinct neurons to determine the sexual identity of C. elegans pheromones. Curr Biol. 27:2589–2599.e2583. [DOI] [PubMed] [Google Scholar]
- Aprison EZ, Ruvinsky I.. 2019a. Coordinated behavioral and physiological responses to a social signal are regulated by a shared neuronal circuit. Curr Biol. 29:4108–4115.ae4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aprison EZ, Ruvinsky I.. 2019b. Dynamic regulation of adult-specific functions of the nervous system by signaling from the reproductive system. Curr Biol. 29:4116–4123.be4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artyukhin AB, Yim JJ, Srinivasan J, Izrayelit Y, Bose N, et al. 2013. Succinylated octopamine ascarosides and a new pathway of biogenic amine metabolism in Caenorhabditis elegans. J Biol Chem. 288:18778–18783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avery L. 2014. A model of the effect of uncertainty on the C. elegans L2/L2d decision. PLoS ONE 9:e100580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargmann CI. 1998. Neurobiology of the Caenorhabditis elegans genome. Science 282:2028–2033. [DOI] [PubMed] [Google Scholar]
- Bargmann CI. 2006. Chemosensation in C. elegans. In: The C. elegans Research Community WormBook, editor. WormBook.
- Bargmann CI, Hartwieg E, Horvitz HR.. 1993. Odorant-selective genes and neurons mediate olfaction in C. elegans. Cell 74:515–527. [DOI] [PubMed] [Google Scholar]
- Bargmann CI, Horvitz HR.. 1991a. Chemosensory neurons with overlapping functions direct chemotaxis to multiple chemicals in C. elegans. Neuron 7:729–742. [DOI] [PubMed] [Google Scholar]
- Bargmann CI, Horvitz HR.. 1991b. Control of larval development by chemosensory neurons in Caenorhabditis elegans. Science 251:1243–1246. [DOI] [PubMed] [Google Scholar]
- Bargmann CI, Thomas JH, Horvitz HR.. 1990. Chemosensory cell function in the behavior and development of Caenorhabditis elegans. Cold Spring Harb Symp Quant Biol. 55:529–538. [DOI] [PubMed] [Google Scholar]
- Barr MM, Garcia LR, Portman DS.. 2018. Sexual dimorphism and sex differences in Caenorhabditis elegans neuronal development and behavior. Genetics 208:909–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastiani C, Mendel J.. 2006. Heterotrimeric G proteins in C. elegans In: The C. elegans Research Community WormBook, editor. WormBook. [DOI] [PMC free article] [PubMed]
- Bauer Huang SL, Saheki Y, VanHoven MK, Torayama I, Ishihara T, et al. 2007. Left-right olfactory asymmetry results from antagonistic functions of voltage-activated calcium channels and the raw repeat protein OLRN-1 in C. elegans. Neural Dev. 2:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bean BP. 2007. The action potential in mammalian central neurons. Nat Rev Neurosci. 8:451–465. [DOI] [PubMed] [Google Scholar]
- Ben Arous J, Laffont S, Chatenay D.. 2009. Molecular and sensory basis of a food related two-state behavior in C. elegans. PLoS One 4:e7584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shahar Y. 2011. Sensory functions for degenerin/epithelial sodium channels (DEG/ENaC). Adv Genet. 76:1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergamasco C, Bazzicalupo P.. 2006. Chemical sensitivity in Caenorhabditis elegans. Cell Mol Life Sci. 63:1510–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besson JM, Chaouch A.. 1987. Peripheral and spinal mechanisms of nociception. Physiol Rev. 67:67–186. [DOI] [PubMed] [Google Scholar]
- Bilbao A, Patel AK, Rahman M, Vanapalli SA, Blawzdziewicz J.. 2018. Roll maneuvers are essential for active reorientation of Caenorhabditis elegans in 3D media. Proc Natl Acad Sci USA. 115:E3616–E3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnby DA, Link EM, Vowels JJ, Tian H, Colacurcio PL, et al. 2000. A transmembrane guanylyl cyclase (DAF-11) and Hsp90 (DAF-21) regulate a common set of chemosensory behaviors in Caenorhabditis elegans. Genetics 155:85–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bounoutas A, Chalfie M.. 2007. Touch sensitivity in Caenorhabditis elegans. Pflugers Arch. 454:691–702. [DOI] [PubMed] [Google Scholar]
- Broekmans OD, Rodgers JB, Ryu WS, Stephens GJ.. 2016. Resolving coiled shapes reveals new reorientation behaviors in C. elegans. Elife 5:e17227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AE, Yemini EI, Grundy LJ, Jucikas T, Schafer WR.. 2013. A dictionary of behavioral motifs reveals clusters of genes affecting Caenorhabditis elegans locomotion. Proc Natl Acad Sci USA. 110:791–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brundage L, Avery L, Katz A, Kim UJ, Mendel JE, et al. 1996. Mutations in a C. elegans Gqα gene disrupt movement, egg laying, and viability. Neuron 16:999–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunemann M, Hosey MM.. 1999. G-protein coupled receptor kinases as modulators of G-protein signalling. J Physiol. 517:5–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butcher RA. 2017a. Decoding chemical communication in nematodes. Nat Prod Rep. 34:472–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butcher RA. 2017b. Small-molecule pheromones and hormones controlling nematode development. Nat Chem Biol. 13:577–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butcher RA. 2019. Natural products as chemical tools to dissect complex biology in C. elegans. Curr Opin Chem Biol. 50:138–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butcher RA, Fujita M, Schroeder FC, Clardy J.. 2007. Small-molecule pheromones that control dauer development in Caenorhabditis elegans. Nat Chem Biol. 3:420–422. [DOI] [PubMed] [Google Scholar]
- Butcher RA, Ragains JR, Clardy J.. 2009. An indole-containing dauer pheromone component with unusual dauer inhibitory activity at higher concentrations. Org Lett. 11:3100–3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butcher RA, Ragains JR, Kim E, Clardy J.. 2008. A potent dauer pheromone component in Caenorhabditis elegans that acts synergistically with other components. Proc Natl Acad Sci USA. 105:14288–14292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butcher RA, Ragains JR, Li W, Ruvkun G, Clardy J, et al. 2009. Biosynthesis of the Caenorhabditis elegans dauer pheromone. Proc Natl Acad Sci USA. 106:1875–1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun AJ, Tong A, Pokala N, Fitzpatrick JA, Sharpee TO, et al. 2015. Neural mechanisms for evaluating environmental variability in Caenorhabditis elegans. Neuron 86:428–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Kajino-Sakamoto R, Doss A, Aballay A.. 2017. Distinct roles of sensory neurons in mediating pathogen avoidance and neuropeptide-dependent immune regulation. Cell Rep. 21:1442–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassada RC, Russell RL.. 1975. The dauer larva, a post-embryonic developmental variant of the nematode Caenorhabditis elegans. Dev Biol. 46:326–342. [DOI] [PubMed] [Google Scholar]
- Catterall WA. 2011. Voltage-gated calcium channels. Cold Spring Harb Perspect Biol. 3:a003947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catterall WA, Perez-Reyes E, Snutch TP, Striessnig J.. 2005. International union of pharmacology. XLVIII. Nomenclature and structure-function relationships of voltage-gated calcium channels. Pharmacol Rev. 57:411–425. [DOI] [PubMed] [Google Scholar]
- Chalasani SH, Chronis N, Tsunozaki M, Gray JM, Ramot D, et al. 2007. Dissecting a circuit for olfactory behaviour in Caenorhabditis elegans. Nature 450:63–70. [DOI] [PubMed] [Google Scholar]
- Chalasani SH, Kato S, Albrecht DR, Nakagawa T, Abbott LF, et al. 2010. Neuropeptide feedback modifies odor-evoked dynamics in Caenorhabditis elegans olfactory neurons. Nat Neurosci. 13:615–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalfie M, Wolinsky E.. 1990. The identification and suppression of inherited neurodegeneration in Caenorhabditis elegans. Nature 345:410–416. [DOI] [PubMed] [Google Scholar]
- Chang S, Johnston RJ Jr., Hobert O.. 2003. A transcriptional regulatory cascade that controls left/right asymmetry in chemosensory neurons of C. elegans. Genes Dev. 17:2123–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao MY, Komatsu H, Fukuto HS, Dionne HM, Hart AC.. 2004. Feeding status and serotonin rapidly and reversibly modulate a Caenorhabditis elegans chemosensory circuit. Proc Natl Acad Sci USA. 101:15512–15517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chasnov JR, So WK, Chan CM, Chow KL.. 2007. The species, sex, and stage specificity of a Caenorhabditis sex pheromone. Proc Natl Acad Sci USA. 104:6730–6735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatzigeorgiou M, Bang S, Hwang SW, Schafer WR.. 2013. tmc-1 encodes a sodium-sensitive channel required for salt chemosensation in C. elegans. Nature 494:95–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Fu Y, Ren M, Xiao B, Rubin CS.. 2011. A RasGRP, C. elegans RGEF-1b, couples external stimuli to behavior by activating LET-60 (Ras) in sensory neurons. Neuron 70:51–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N, Pai S, Zhao Z, Mah A, Newbury R, et al. 2005. Identification of a nematode chemosensory gene family. Proc Natl Acad Sci USA. 102:146–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho CE, Brueggemann C, L'Etoile ND, Bargmann CI.. 2016. Parallel encoding of sensory history and behavioral preference during Caenorhabditis elegans olfactory learning. Elife 5:e14000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SW, Cho JH, Song HO, Park CS.. 2005. Identification and characterization of a putative cyclic nucleotide-gated channel, CNG-1, in C. elegans. Mol Cells 19:149–154. [PubMed] [Google Scholar]
- Cho SW, Choi KY, Park CS.. 2004. A new putative cyclic nucleotide-gated channel gene, cng-3, is critical for thermotolerance in Caenorhabditis elegans. Biochem Biophys Res Commun. 325:525–531. [DOI] [PubMed] [Google Scholar]
- Choi JI, Lee HK, Kim HS, Park SY, Lee TY, et al. 2018. Odor-dependent temporal dynamics in Caenorhabitis elegans adaptation and aversive learning behavior. PeerJ. 6:e4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JI, Yoon KH, Subbammal Kalichamy S, Yoon SS, Il Lee J.. 2016. A natural odor attraction between lactic acid bacteria and the nematode Caenorhabditis elegans. ISME J. 10:558–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou JH, Bargmann CI, Sengupta P.. 2001. The Caenorhabditis elegans odr-2 gene encodes a novel Ly-6-related protein required for olfaction. Genetics 157:211–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou JH, Troemel ER, Sengupta P, Colbert HA, Tong L, et al. 1996. Olfactory recognition and discrimination in Caenorhabditis elegans. Cold Spring Harb Symp Quant Biol. 61:157–164. [PubMed] [Google Scholar]
- Chronis N, Zimmer M, Bargmann CI.. 2007. Microfluidics for in vivo imaging of neuronal and behavioral activity in Caenorhabditis elegans. Nat Methods 4:727–731. [DOI] [PubMed] [Google Scholar]
- Chute CD, DiLoreto EM, Zhang YK, Reilly DK, Rayes D, et al. 2019. Co-option of neurotransmitter signaling for inter-organismal communication in C. elegans. Nat Commun. 10:3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chute CD, Srinivasan J.. 2014. Chemical mating cues in C. elegans. Semin Cell Dev Biol. 33:18–24. [DOI] [PubMed] [Google Scholar]
- Coburn CM, Bargmann CI.. 1996. A putative cyclic nucleotide-gated channel is required for sensory development and function in C. elegans. Neuron 17:695–706. [DOI] [PubMed] [Google Scholar]
- Coburn CM, Mori I, Ohshima Y, Bargmann CI.. 1998. A cyclic nucleotide-gated channel inhibits sensory axon outgrowth in larval and adult Caenorhabditis elegans: a distinct pathway for maintenance of sensory axon structure. Development 125:249–258. [DOI] [PubMed] [Google Scholar]
- Colbert HA, Bargmann CI.. 1995. Odorant-specific adaptation pathways generate olfactory plasticity in C. elegans. Neuron 14:803–812. [DOI] [PubMed] [Google Scholar]
- Colbert HA, Bargmann CI.. 1997. Environmental signals modulate olfactory acuity, discrimination, and memory in Caenorhabditis elegans. Learn Mem. 4:179–191. [DOI] [PubMed] [Google Scholar]
- Colbert HA, Smith TL, Bargmann CI.. 1997. OSM-9, a novel protein with structural similarity to channels, is required for olfaction, mechanosensation, and olfactory adaptation in Caenorhabditis elegans. J Neurosci. 17:8259–8269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook SJ, Jarrell TA, Brittin CA, Wang Y, Bloniarz AE, et al. 2019. Whole-animal connectomes of both Caenorhabditis elegans sexes. Nature 571:63–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornils A, Gloeck M, Chen Z, Zhang Y, Alcedo J.. 2011. Specific insulin-like peptides encode sensory information to regulate distinct developmental processes. Development 138:1183–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couto A, Oda S, Nikolaev VO, Soltesz Z, de Bono M.. 2013. In vivo genetic dissection of O2-evoked cGMP dynamics in a Caenorhabditis elegans gas sensor. Proc Natl Acad Sci USA. 110:E3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croll NA. 1975a. Behavioural analysis of nematode movement. Adv Parasitol. 13:71–122. [DOI] [PubMed] [Google Scholar]
- Croll NA. 1975b. Components and patterns in behavior of nematode Caenorhabditis elegans. J Zool. 176:159–176. [Google Scholar]
- Dana H, Sun Y, Mohar B, Hulse BK, Kerlin AM, et al. 2019. High-performance calcium sensors for imaging activity in neuronal populations and microcompartments. Nat Methods 16:649–657. [DOI] [PubMed] [Google Scholar]
- Daniels SA, Ailion M, Thomas JH, Sengupta P.. 2000. egl-4 acts through a transforming growth factor-β/SMAD pathway in Caenorhabditis elegans to regulate multiple neuronal circuits in response to sensory cues. Genetics 156:123–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dao J, Lee A, Drecksel DK, Bittlingmaier NM, Nelson TM.. 2020. Characterization of TMC-1 in C. elegans sodium chemotaxis and sodium conditioned aversion. BMC Genet. 21:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis KC, Choi YI, Kim J, You YJ.. 2018. Satiety behavior is regulated by ASI/ASH reciprocal antagonism. Sci Rep. 8:6918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bono M, Bargmann CI.. 1998. Natural variation in a neuropeptide Y receptor homolog modifies social behavior and food response in C. elegans. Cell 94:679–689. [DOI] [PubMed] [Google Scholar]
- de Bono M, Maricq AV.. 2005. Neuronal substrates of complex behaviors in C. elegans. Annu Rev Neurosci. 28:451–501. [DOI] [PubMed] [Google Scholar]
- de Bono M, Tobin DM, Davis MW, Avery L, Bargmann CI.. 2002. Social feeding in Caenorhabditis elegans is induced by neurons that detect aversive stimuli. Nature 419:899–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis EJ, Dobosiewicz M, Jin X, Duvall LB, Hartman PS, et al. 2018. A natural variant and engineered mutation in a GPCR promote DEET resistance in C. elegans. Nature 562:119–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobosiewicz M, Liu Q, Bargmann CI.. 2019. Reliability of an interneuron response depends on an integrated sensory state. Elife 8:e50566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doroquez DB, Berciu C, Anderson JR, Sengupta P, Nicastro D.. 2014. A high-resolution morphological and ultrastructural map of anterior sensory cilia and glia in Caenorhabditis elegans. Elife 3:e01948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duda T, Goraczniak R, Surgucheva I, Rudnicka-Nawrot M, Gorczyca WA, et al. 1996. Calcium modulation of bovine photoreceptor guanylate cyclase. Biochemistry 35:8478–8482. [DOI] [PubMed] [Google Scholar]
- Dusenbery DB. 1973. Countercurrent separation: a new method for studying behavior of small aquatic organisms. Proc Natl Acad Sci USA. 70:1349–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dusenbery DB. 1974. Analysis of chemotaxis in the nematode Caenorhabditis elegans by countercurrent separation. J Exp Zool. 188:41–47. [DOI] [PubMed] [Google Scholar]
- Dusenbery DB. 1975. The avoidance of D-tryptophan by the nematode Caenorhabditis elegans. J Exp Zool. 193:413–418. [DOI] [PubMed] [Google Scholar]
- Eismann E, Muller F, Heinemann SH, Kaupp UB.. 1994. A single negative charge within the pore region of a cGMP-gated channel controls rectification, Ca2+ blockage, and ionic selectivity. Proc Natl Acad Sci USA. 91:1109–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entchev EV, Patel DS, Zhan M, Steele AJ, Lu H, et al. 2015. A gene-expression-based neural code for food abundance that modulates lifespan. Elife 4:e06259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito G, Amoroso MR, Bergamasco C, Schiavi ED, Bazzicalupo P.. 2010. The G protein regulators EGL-10 and EAT-16, the Giα GOA-1 and the G(q)α EGL-30 modulate the response of the C. elegans ASH polymodal nociceptive sensory neurons to repellents. BMC Biol. 8:138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito G, Schiavi ED, Bergamasco C, Bazzicalupo P.. 2007. Efficient and cell specific knock-down of gene function in targeted C. elegans neurons. Gene 395:170–176. [DOI] [PubMed] [Google Scholar]
- Estevez M, Estevez AO, Cowie RH, Gardner KL.. 2003. The voltage-gated calcium channel UNC-2 is involved in stress-mediated regulation of tryptophan hydroxylase. J Neurochem. 88:102–113. [DOI] [PubMed] [Google Scholar]
- Ezak MJ, Ferkey DM.. 2011. A functional nuclear localization sequence in the C. elegans TRPV channel OCR-2. PLoS One 6:e25047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezak MJ, Hong E, Chaparro-Garcia A, Ferkey DM.. 2010. Caenorhabditis elegans TRPV channels function in a modality-specific pathway to regulate response to aberrant sensory signaling. Genetics 185:233–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagan KA, Luo J, Lagoy RC, Schroeder FC, Albrecht DR, et al. 2018. A single-neuron chemosensory switch determines the valence of a sexually dimorphic sensory behavior. Curr Biol. 28:902–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faumont S, Lindsay TH, Lockery SR.. 2012. Neuronal microcircuits for decision making in C. elegans. Curr Opin Neurobiol. 22:580–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenk LA, de Bono M.. 2017. Memory of recent oxygen experience switches pheromone valence in Caenorhabditis elegans. Proc Natl Acad Sci USA. 114:4195–4200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson SS. 2001. Evolving concepts in G protein-coupled receptor endocytosis: the role in receptor desensitization and signaling. Pharmacol Rev. 53:1–24. [PubMed] [Google Scholar]
- Ferkey DM, Hyde R, Haspel G, Dionne HM, Hess HA, et al. 2007. C. elegans G protein regulator RGS-3 controls sensitivity to sensory stimuli. Neuron 53:39–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fielenbach N, Antebi A.. 2008. C. elegans dauer formation and the molecular basis of plasticity. Genes Dev. 22:2149–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick DA, O'Halloran DM, Burnell AM.. 2006. Multiple lineage specific expansions within the guanylyl cyclase gene family. BMC Evol Biol. 6:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell SW, Pokala N, Macosko EZ, Albrecht DR, Larsch J, et al. 2013. Serotonin and the neuropeptide PDF initiate and extend opposing behavioral states in C. elegans. Cell 154:1023–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frézal L, Félix MA.. 2015. C. elegans outside the Petri dish. Elife 4:e05849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjær-Jensen C, Ailion M, Lockery SR.. 2008. Ammonium-acetate is sensed by gustatory and olfactory neurons in Caenorhabditis elegans. PLoS ONE 3:e2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Yau KW.. 2007. Phototransduction in mouse rods and cones. Pflugers Arch. 454:805–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara M, Hino T, Miyamoto R, Inada H, Mori I, et al. 2015. The importance of cGMP signaling in sensory cilia for body size regulation in Caenorhabditis elegans. Genetics 201:1497–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara M, Sengupta P, McIntire SL.. 2002. Regulation of body size and behavioral state of C. elegans by sensory perception and the EGL-4 cGMP-dependent protein kinase. Neuron 36:1091–1102. [DOI] [PubMed] [Google Scholar]
- Fukuto HS, Ferkey DM, Apicella AJ, Lans H, Sharmeen T, et al. 2004. G protein-coupled receptor kinase function is essential for chemosensation in C. elegans. Neuron 42:581–593., [DOI] [PubMed] [Google Scholar]
- Gallagher T, Kim J, Oldenbroek M, Kerr R, You YJ.. 2013. ASI regulates satiety quiescence in C. elegans. J Neurosci. 33:9716–9724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geffeney SL, Cueva JG, Glauser DA, Doll JC, Lee TH, et al. 2011. DEG/ENaC but not TRP channels are the major mechanoelectrical transduction channels in a C. elegans nociceptor. Neuron 71:845–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber KJ, Squires KE, Hepler JR.. 2016. Roles for regulator of G protein signaling proteins in synaptic signaling and plasticity. Mol Pharmacol. 89:273–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden JW, Riddle DL.. 1982. A pheromone influences larval development in the nematode Caenorhabditis elegans. Science 218:578–580. [DOI] [PubMed] [Google Scholar]
- Golden JW, Riddle DL.. 1984a. The Caenorhabditis elegans dauer larva: developmental effects of pheromone, food, and temperature. Dev Biol. 102:368–378. [DOI] [PubMed] [Google Scholar]
- Golden JW, Riddle DL.. 1984b. A Caenorhabditis elegans dauer-inducing pheromone and an antagonistic component of the food supply. J Chem Ecol. 10:1265–1280. [DOI] [PubMed] [Google Scholar]
- Golden JW, Riddle DL.. 1984c. A pheromone-induced developmental switch in Caenorhabditis elegans: temperature-sensitive mutants reveal a wild-type temperature-dependent process. Proc Natl Acad Sci USA. 81:819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden JW, Riddle DL.. 1985. A gene affecting production of the Caenorhabditis elegans dauer-inducing pheromone. Mol Gen Genet. 198:534–536. [DOI] [PubMed] [Google Scholar]
- Goldsmith AD, Sarin S, Lockery S, Hobert O.. 2010. Developmental control of lateralized neuron size in the nematode Caenorhabditis elegans. Neural Dev. 5:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez M, De Castro E, Guarin E, Sasakura H, Kuhara A, et al. 2001. Ca2+ signaling via the neuronal calcium sensor-1 regulates associative learning and memory in C. elegans. Neuron 30:241–248. [DOI] [PubMed] [Google Scholar]
- Goodman MB, Hall DH, Avery L, Lockery SR.. 1998. Active currents regulate sensitivity and dynamic range in C. elegans neurons. Neuron 20:763–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman MB, Lindsay TH, Lockery SR, Richmond JE.. 2012. Electrophysiological methods for Caenorhabditis elegans neurobiology. Methods Cell Biol. 107:409–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman MB, Schwarz EM.. 2003. Transducing touch in Caenorhabditis elegans. Annu Rev Physiol. 65:429–452. [DOI] [PubMed] [Google Scholar]
- Goodman MB, Sengupta P.. 2018. The extraordinary AFD thermosensor of C. elegans. Pflugers Arch. 470:839–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman MB, Sengupta P.. 2019. How Caenorhabditis elegans senses mechanical stress, temperature, and other physical stimuli. Genetics 212:25–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordus A, Pokala N, Levy S, Flavell SW, Bargmann CI.. 2015. Feedback from network states generates variability in a probabilistic olfactory circuit. Cell 161:215–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray JM, Hill JJ, Bargmann CI.. 2005. A circuit for navigation in Caenorhabditis elegans. Proc Natl Acad Sci USA. 102:3184–3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene JS, Brown M, Dobosiewicz M, Ishida IG, Macosko EZ, et al. 2016a. Balancing selection shapes density-dependent foraging behaviour. Nature 539:254–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene JS, Dobosiewicz M, Butcher RA, McGrath PT, Bargmann CI.. 2016b. Regulatory changes in two chemoreceptor genes contribute to a Caenorhabditis elegans QTL for foraging behavior. Elife 5:e21454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruner M, Nelson D, Winbush A, Hintz R, Ryu L, et al. 2014. Feeding state, insulin and NPR-1 modulate chemoreceptor gene expression via integration of sensory and circuit inputs. PLoS Genet. 10:e1004707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M, Wu TH, Song YX, Ge MH, Su CM, et al. 2015. Reciprocal inhibition between sensory ASH and ASI neurons modulates nociception and avoidance in Caenorhabditis elegans. Nat Commun. 6:5655. [DOI] [PubMed] [Google Scholar]
- Gyurkó MD, Csermely P, Sőti C, Steták A.. 2015. Distinct roles of the RasGAP family proteins in C. elegans associative learning and memory. Sci Rep. 5:15084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha HI, Hendricks M, Shen Y, Gabel CV, Fang-Yen C, et al. 2010. Functional organization of a neural network for aversive olfactory learning in Caenorhabditis elegans. Neuron 68:1173–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallem EA, Spencer WC, McWhirter RD, Zeller G, Henz SR, et al. 2011. Receptor-type guanylate cyclase is required for carbon dioxide sensation by Caenorhabditis elegans. Proc Natl Acad Sci USA. 108:254–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammarlund M, Hobert O, Miller DM, Sestan N.. 2018. The CeNGEN project: the complete gene expression map of an entire nervous system. Neuron 99:430–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y, Xu N, Box AC, Schaefer L, Kannan K, et al. 2011. Nuclear cGMP-dependent kinase regulates gene expression via activity-dependent recruitment of a conserved histone deacetylase complex. PLoS Genet. 7:e1002065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y, Yang W, Ren J, Hall Q, Zhang Y, et al. 2018. Thioredoxin shapes the C. elegans sensory response to Pseudomonas produced nitric oxide. Elife 7:e36833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara-Kuge S, Nishihara T, Matsuda T, Kitazono T, Teramoto T, et al. 2018. An improved inverse-type Ca2+ indicator can detect putative neuronal inhibition in Caenorhabditis elegans by increasing signal intensity upon Ca2+ decrease. PLoS ONE 13:e0194707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris G, Shen Y, Ha H, Donato A, Wallis S, et al. 2014. Dissecting the signaling mechanisms underlying recognition and preference of food odors. J Neurosci. 34:9389–9403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart AC. 2006. Behavior. In: The C. elegans Research Community WormBook, editor. WormBook, doi/10.1895/wormbook.1.87.1.
- Hart AC, Chao MY.. 2010. From Odors to Behaviors in Caenorhabditis elegans in the Neurobiology of Olfaction. Boca Raton, FL: CRC Press. [PubMed] [Google Scholar]
- Hart AC, Kass J, Shapiro JE, Kaplan JM.. 1999. Distinct signaling pathways mediate touch and osmosensory responses in a polymodal sensory neuron. J Neurosci. 19:1952–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Altshuler-Keylin S, Daniel D, L'Etoile ND, O'Halloran D.. 2016. The cyclic nucleotide gated channel subunit CNG-1 instructs behavioral outputs in Caenorhabditis elegans by coincidence detection of nutritional status and olfactory input. Neurosci Lett. 632:71–78. [DOI] [PubMed] [Google Scholar]
- Hendricks M, Ha H, Maffey N, Zhang Y.. 2012. Compartmentalized calcium dynamics in a C. elegans interneuron encode head movement. Nature 487:99–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendricks M, Zhang Y.. 2013. Complex RIA calcium dynamics and its function in navigational behavior. Worm 2:e25546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilliard MA, Apicella AJ, Kerr R, Suzuki H, Bazzicalupo P, et al. 2005. In vivo imaging of C. elegans ASH neurons: cellular response and adaptation to chemical repellents. Embo J. 24:63–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilliard MA, Bargmann CI, Bazzicalupo P.. 2002. C. elegans responds to chemical repellents by integrating sensory inputs from the head and the tail. Curr Biol. 12:730–734. [DOI] [PubMed] [Google Scholar]
- Hilliard MA, Bergamasco C, Arbucci S, Plasterk RH, Bazzicalupo P.. 2004. Worms taste bitter: ASH neurons, QUI-1, GPA-3 and ODR-3 mediate quinine avoidance in Caenorhabditis elegans. Embo J. 23:1101–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hills T, Brockie PJ, Maricq AV.. 2004. Dopamine and glutamate control area-restricted search behavior in Caenorhabditis elegans. J Neurosci. 24:1217–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirotsu T, Iino Y.. 2005. Neural circuit-dependent odor adaptation in C. elegans is regulated by the Ras-MAPK pathway. Genes Cells 10:517–530. [DOI] [PubMed] [Google Scholar]
- Hirotsu T, Saeki S, Yamamoto M, Iino Y.. 2000. The Ras-MAPK pathway is important for olfaction in Caenorhabditis elegans. Nature 404:289–293. [DOI] [PubMed] [Google Scholar]
- Hobert O. 2013. The neuronal genome of Caenorhabditis elegans. WormBook 1–106, doi: 10.1895/wormbook.1.161.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann F. 2005. The biology of cyclic GMP-dependent protein kinases. J Biol Chem. 280:1–4. [DOI] [PubMed] [Google Scholar]
- Hollinger S, Hepler JR.. 2002. Cellular regulation of RGS proteins: modulators and integrators of G protein signaling. Pharmacol Rev. 54:527–559. [DOI] [PubMed] [Google Scholar]
- Hong M, Ryu L, Ow MC, Kim J, Je AR, et al. 2017. Early Pheromone Experience Modifies a Synaptic Activity to Influence Adult Pheromone Responses of C. elegans. Curr Biol. 27:3168–3177.e3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsueh YP, Gronquist MR, Schwarz EM, Nath RD, Lee CH, et al. 2017. Nematophagous fungus Arthrobotrys oligospora mimics olfactory cues of sex and food to lure its nematode prey. Elife 6:e20023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hukema RK, Rademakers S, Dekkers MP, Burghoorn J, Jansen G.. 2006. Antagonistic sensory cues generate gustatory plasticity in Caenorhabditis elegans. Embo J. 25:312–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hukema RK, Rademakers S, Jansen G.. 2008. Gustatory plasticity in C. elegans involves integration of negative cues and NaCl taste mediated by serotonin, dopamine, and glutamate. Learn Mem. 15:829–836. [DOI] [PubMed] [Google Scholar]
- Hussey R, Stieglitz J, Mesgarzadeh J, Locke TT, Zhang YK, et al. 2017. Pheromone-sensing neurons regulate peripheral lipid metabolism in Caenorhabditis elegans. PLoS Genet. 13:e1006806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husson SJ, Costa WS, Schmitt C, Gottschalk A.. 2013. Keeping track of worm trackers. WormBook 1–17, doi: 10.1895/wormbook.1.156.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iino Y, Yoshida K.. 2009. Parallel use of two behavioral mechanisms for chemotaxis in Caenorhabditis elegans. J Neurosci. 29:5370–5380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im SH, Galko MJ.. 2012. Pokes, sunburn, and hot sauce: Drosophila as an emerging model for the biology of nociception. Dev Dyn. 241:16–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglis PN, Ou G, Leroux MR, Scholey JM.. 2006. The sensory cilia of Caenorhabditis elegans. In: The C. elegans Research Community WormBook, editor. WormBook. [DOI] [PMC free article] [PubMed]
- Itskovits E, Ruach R, Kazakov A, Zaslaver A.. 2018. Concerted pulsatile and graded neural dynamics enables efficient chemotaxis in C. elegans. Nat Commun. 9:2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izquierdo EJ, Lockery SR.. 2010. Evolution and analysis of minimal neural circuits for klinotaxis in Caenorhabditis elegans. J Neurosci. 30:12908–12917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izquierdo EJ, Williams PL, Beer RD.. 2015. Information flow through a model of the C. elegans klinotaxis circuit. PLoS ONE 10:e0140397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang H, Kim K, Neal SJ, Macosko E, Kim D, et al. 2012. Neuromodulatory state and sex specify alternative behaviors through antagonistic synaptic pathways in C. elegans. Neuron 75:585–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang H, Levy S, Flavell SW, Mende F, Latham R, et al. 2017. Dissection of neuronal gap junction circuits that regulate social behavior in Caenorhabditis elegans. Proc Natl Acad Sci USA. 114:E1263–E1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang MS, Toyoshima Y, Tomioka M, Kunitomo H, Iino Y.. 2019. Multiple sensory neurons mediate starvation-dependent aversive navigation in Caenorhabditis elegans. Proc Natl Acad Sci USA. 116:18673–18683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen G, Thijssen KL, Werner P, van der Horst M, Hazendonk E, et al. 1999. The complete family of genes encoding G proteins of Caenorhabditis elegans. Nat Genet. 21:414–419. [DOI] [PubMed] [Google Scholar]
- Jansen G, Weinkove D, Plasterk RH.. 2002. The G protein gamma subunit gpc-1 of the nematode C. elegans is involved in taste adaptation. EMBO J. 21:986–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasti J, Furukawa H, Gonzales EB, Gouaux E.. 2007. Structure of acid-sensing ion channel 1 at 1.9 A resolution and low pH. Nature 449:316–323. [DOI] [PubMed] [Google Scholar]
- Jeong PY, Jung M, Yim YH, Kim H, Park M, et al. 2005. Chemical structure and biological activity of the Caenorhabditis elegans dauer-inducing pheromone. Nature 433:541–545. [DOI] [PubMed] [Google Scholar]
- Jin X, Pokala N, Bargmann CI.. 2016. Distinct circuits for the formation and retrieval of an imprinted olfactory memory. Cell 164:632–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson WA, Carder JW.. 2012. Drosophila nociceptors mediate larval aversion to dry surface environments utilizing both the painless TRP channel and the DEG/ENaC subunit, PPK1. PLoS ONE 7:e32878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovelin R, Ajie BC, Phillips PC.. 2003. Molecular evolution and quantitative variation for chemosensory behaviour in the nematode genus Caenorhabditis. Mol Ecol. 12:1325–1337. [DOI] [PubMed] [Google Scholar]
- Juang BT, Gu C, Starnes L, Palladino F, Goga A, et al. 2013. Endogenous nuclear RNAi mediates behavioral adaptation to odor. Cell 154:1010–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn-Kirby AH, Bargmann CI.. 2006. TRP channels in C. elegans. Annu Rev Physiol. 68:719–736. [DOI] [PubMed] [Google Scholar]
- Kahn-Kirby AH, Dantzker JL, Apicella AJ, Schafer WR, Browse J, et al. 2004. Specific polyunsaturated fatty acids drive TRPV-dependent sensory signaling in vivo. Cell 119:889–900. [DOI] [PubMed] [Google Scholar]
- Kaletsky R, Yao V, Williams A, Runnels AM, Tadych A, et al. 2018. Transcriptome analysis of adult Caenorhabditis elegans cells reveals tissue-specific gene and isoform expression. PLoS Genet. 14:e1007559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminski E, Stawicki S, Wasowicz E.. 1974. Volatile flavor compounds produced by molds of aspergillus, penicillium, and fungi imperfecti. Appl Microbiol. 27:1001–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato S, Kaplan HS, Schrodel T, Skora S, Lindsay TH, et al. 2015. Global brain dynamics embed the motor command sequence of Caenorhabditis elegans. Cell 163:656–669. [DOI] [PubMed] [Google Scholar]
- Kato S, Xu Y, Cho CE, Abbott LF, Bargmann CI.. 2014. Temporal responses of C. elegans chemosensory neurons are preserved in behavioral dynamics. Neuron 81:616–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffman A, Parsons L, Stein G, Wills A, Kaletsky R, et al. 2011. C. elegans positive butanone learning, short-term, and long-term associative memory assays. J Vis Exp. 2490, doi: 10.3791/2490. [DOI] [PMC free article] [PubMed]
- Kaufman A, Keinan A, Meilijson I, Kupiec M, Ruppin E.. 2005. Quantitative analysis of genetic and neuronal multi-perturbation experiments. PLoS Comput Biol. 1:e64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano T, Kataoka N, Abe S, Ohtani M, Honda Y, et al. 2005. Lifespan extending activity of substances secreted by the nematode Caenorhabditis elegans that include the dauer-inducing pheromone. Biosci Biotechnol Biochem. 69:2479–2481. [DOI] [PubMed] [Google Scholar]
- Kawashima Y, Kurima K, Pan B, Griffith AJ, Holt JR.. 2015. Transmembrane channel-like (TMC) genes are required for auditory and vestibular mechanosensation. Pflugers Arch. 467:85–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keresztes G, Mutai H, Heller S.. 2003. TMC and EVER genes belong to a larger novel family, the TMC gene family encoding transmembrane proteins. BMC Genomics 4:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr R, Lev-Ram V, Baird G, Vincent P, Tsien RY, et al. 2000. Optical imaging of calcium transients in neurons and pharyngeal muscle of C. elegans. Neuron 26:583–594. [DOI] [PubMed] [Google Scholar]
- Kim JJ, Lorenz R, Arold ST, Reger AS, Sankaran B, et al. 2016. Crystal structure of PKG I:cGMP complex reveals a cGMP-mediated dimeric interface that facilitates cGMP-induced activation. Structure 24:710–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Sato K, Shibuya M, Zeiger DM, Butcher RA, et al. 2009. Two chemoreceptors mediate developmental effects of dauer pheromone in C. elegans. Science 326:994–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu H, Jin YH, L'Etoile N, Mori I, Bargmann CI, et al. 1999. Functional reconstitution of a heteromeric cyclic nucleotide-gated channel of Caenorhabditis elegans in cultured cells. Brain Res. 821:160–168. [DOI] [PubMed] [Google Scholar]
- Komatsu H, Mori I, Rhee JS, Akaike N, Ohshima Y.. 1996. Mutations in a cyclic nucleotide-gated channel lead to abnormal thermosensation and chemosensation in C. elegans. Neuron 17:707–718. [DOI] [PubMed] [Google Scholar]
- Komolov KE, Benovic JL.. 2018. G protein-coupled receptor kinases: past, present and future. Cell Signal. 41:17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan A, Almen MS, Fredriksson R, Schioth HB.. 2014. Insights into the origin of nematode chemosensory GPCRs: putative orthologs of the SRW family are found across several phyla of protostomes. PLoS ONE 9:e93048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzyzanowski MC, Brueggemann C, Ezak MJ, Wood JF, Michaels KL, et al. 2013. The C. elegans cGMP-dependent protein kinase EGL-4 regulates nociceptive behavioral sensitivity. PLoS Genet. 9:e1003619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzyzanowski MC, Woldemariam S, Wood JF, Chaubey AH, Brueggemann C, et al. 2016. Aversive behavior in the nematode C. elegans is modulated by cGMP and a neuronal gap junction network. PLoS Genet. 12:e1006153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhara A, Inada H, Katsura I, Mori I.. 2002. Negative regulation and gain control of sensory neurons by the C. elegans calcineurin TAX-6. Neuron 33:751–763. [DOI] [PubMed] [Google Scholar]
- Kunitomo H, Sato H, Iwata R, Satoh Y, Ohno H, et al. 2013. Concentration memory-dependent synaptic plasticity of a taste circuit regulates salt concentration chemotaxis in Caenorhabditis elegans. Nat Commun. 4:2210. [DOI] [PubMed] [Google Scholar]
- Kuramochi M, Iwasaki Y.. 2010. Quantitative Modeling of Neuronal Dynamics inC. elegans in Neural Information Processing Theory and AlgorithmsBerlin, Heidelberg: Springer. [Google Scholar]
- Kurima K, Yang Y, Sorber K, Griffith AJ.. 2003. Characterization of the transmembrane channel-like (TMC) gene family: functional clues from hearing loss and epidermodysplasia verruciformis. Genomics 82:300–308. [DOI] [PubMed] [Google Scholar]
- Kwok TC, Ricker N, Fraser R, Chan AW, Burns A, et al. 2006. A small-molecule screen in C. elegans yields a new calcium channel antagonist. Nature 441:91–95. [DOI] [PubMed] [Google Scholar]
- L’Etoile ND. 2004. Chemosensory Transduction in Caenorhabditis elegans. In: Frings S, Bradley J, editors. Transduction Channels in Sensory Cells. John Wiley & Sons, Inc. p. 73–97. [Google Scholar]
- L'Etoile ND, Bargmann CI.. 2000. Olfaction and odor discrimination are mediated by the C. elegans guanylyl cyclase ODR-1. Neuron 25:575–586. [DOI] [PubMed] [Google Scholar]
- L'Etoile ND, Coburn CM, Eastham J, Kistler A, Gallegos G, et al. 2002. The cyclic GMP-dependent protein kinase EGL-4 regulates olfactory adaptation in C. elegans. Neuron 36:1079–1089. [DOI] [PubMed] [Google Scholar]
- Lans H, Rademakers S, Jansen G.. 2004. A network of stimulatory and inhibitory Gα-subunits regulates olfaction in Caenorhabditis elegans. Genetics 167:1677–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsch J, Flavell SW, Liu Q, Gordus A, Albrecht DR, et al. 2015. A circuit for gradient climbing in C. elegans Chemotaxis . Cell Rep. 12:1748–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsch J, Ventimiglia D, Bargmann CI, Albrecht DR.. 2013. High-throughput imaging of neuronal activity in Caenorhabditis elegans. Proc Natl Acad Sci USA. 110:E4266–E4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D, Zdraljevic S, Cook DE, Frezal L, Hsu JC, et al. 2019. Selection and gene flow shape niche-associated variation in pheromone response. Nat Ecol Evol. 3:1455–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Jee C, McIntire SL.. 2009. Ethanol preference in C. elegans. Genes Brain Behav. 8:578–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JI, O'Halloran DM, Eastham-Anderson J, Juang BT, Kaye JA, et al. 2010. Nuclear entry of a cGMP-dependent kinase converts transient into long-lasting olfactory adaptation. Proc Natl Acad Sci USA. 107:6016–6021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Portman DS.. 2007. Neural sex modifies the function of a C. elegans sensory circuit. Curr Biol. 17:1858–1863. [DOI] [PubMed] [Google Scholar]
- Lee RY, Lobel L, Hengartner M, Horvitz HR, Avery L.. 1997. Mutations in the α1 subunit of an L-type voltage-activated Ca2+ channel cause myotonia in Caenorhabditis elegans. EMBO J. 16:6066–6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Lee CH, Oh U.. 2005. Painful channels in sensory neurons. Mol Cells 20:315–324. [PubMed] [Google Scholar]
- Leighton DH, Choe A, Wu SY, Sternberg PW.. 2014. Communication between oocytes and somatic cells regulates volatile pheromone production in Caenorhabditis elegans. Proc Natl Acad Sci USA. 111:17905–17910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leinwand SG, Yang CJ, Bazopoulou D, Chronis N, Srinivasan J, et al. 2015. Circuit mechanisms encoding odors and driving aging-associated behavioral declines in Caenorhabditis elegans. Elife 4:e10181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy S, Bargmann CI.. 2020. An adaptive-threshold mechanism for odor sensation and animal navigation. Neuron 105:534–548.e513. [DOI] [PubMed] [Google Scholar]
- Li W, Kennedy SG, Ruvkun G.. 2003. daf-28 encodes a C. elegans insulin superfamily member that is regulated by environmental cues and acts in the DAF-2 signaling pathway. Genes Dev. 17:844–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim JP, Fehlauer H, Das A, Saro G, Glauser DA, et al. 2018. Loss of CaMKI function disrupts salt aversive learning in C. elegans. J Neurosci. 38:6114–6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln TM, Dey N, Sellak H.. 2001. Invited review: cGMP-dependent protein kinase signaling mechanisms in smooth muscle: from the regulation of tone to gene expression. J Appl Physiol. 91:1421–1430. [DOI] [PubMed] [Google Scholar]
- Liu J, Ward A, Gao J, Dong Y, Nishio N, et al. 2010. C. elegans phototransduction requires a G protein-dependent cGMP pathway and a taste receptor homolog. Nat Neurosci. 13:715–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Kidd PB, Dobosiewicz M, Bargmann CI.. 2018a. C. elegans AWA olfactory neurons fire calcium-mediated all-or-none action potentials. Cell 175:57–70.e17. [DOI] [PubMed] [Google Scholar]
- Liu Z, Kariya MJ, Chute CD, Pribadi AK, Leinwand SG, et al. 2018b. Predator-secreted sulfolipids induce defensive responses in C. elegans. Nat Commun. 9:1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lochrie MA, Mendel JE, Sternberg PW, Simon MI.. 1991. Homologous and unique G protein a subunits in the nematode Caenorhabditis elegans. Cell Regul. 2:135–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockery SR. 2011. The computational worm: spatial orientation and its neuronal basis in C. elegans. Curr Opin Neurobiol. 21:782–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockery SR, Goodman MB.. 2009. The quest for action potentials in C. elegans neurons hits a plateau. Nat Neurosci. 12:377–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockery SR, Lawton KJ, Doll JC, Faumont S, Coulthard SM, et al. 2008. Artificial dirt: microfluidic substrates for nematode neurobiology and behavior. J Neurophysiol. 99:3136–3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludewig AH, Artyukhin AB, Aprison EZ, Rodrigues PR, Pulido DC, et al. 2019. An excreted small molecule promotes C. elegans reproductive development and aging. Nat Chem Biol. 15:838–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludewig AH, Gimond C, Judkins JC, Thornton S, Pulido DC, et al. 2017. Larval crowding accelerates C. elegans development and reduces lifespan. PLoS Genet. 13:e1006717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludewig AH, Izrayelit Y, Park D, Malik RU, Zimmermann A, et al. 2013. Pheromone sensing regulates Caenorhabditis elegans lifespan and stress resistance via the deacetylase SIR-2.1. Proc Natl Acad Sci USA. 110:5522–5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludewig AH, Schroeder FC.. 2013. Ascaroside signaling in C. elegans. WormBook 1–22, doi: 10.1895/wormbook.1.155.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L, Gabel CV, Ha HI, Zhang Y, Samuel AD.. 2008. Olfactory behavior of swimming C. elegans analyzed by measuring motile responses to temporal variations of odorants. J Neurophysiol. 99:2617–2625. [DOI] [PubMed] [Google Scholar]
- Luo L, Wen Q, Ren J, Hendricks M, Gershow M, et al. 2014. Dynamic encoding of perception, memory, and movement in a C. elegans chemotaxis circuit. Neuron 82:1115–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macosko EZ, Pokala N, Feinberg EH, Chalasani SH, Butcher RA, et al. 2009. A hub-and-spoke circuit drives pheromone attraction and social behaviour in C. elegans. Nature 458:1171–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning G. 2005. Genomic overview of protein kinases. In: The C. elegans Research Community WormBook, editor. WormBook. [DOI] [PMC free article] [PubMed]
- Maruyama IN. 2016. Receptor guanylyl cyclases in sensory processing. Front Endocrinol. 7:173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuki M, Kunitomo H, Iino Y.. 2006. Goα regulates olfactory adaptation by antagonizing Gqα-DAG signaling in Caenorhabditis elegans. Proc Natl Acad Sci USA. 103:1112–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matulef K, Zagotta WN.. 2003. Cyclic nucleotide-gated ion channels. Annu Rev Cell Dev Biol. 19:23–44. [DOI] [PubMed] [Google Scholar]
- Maures TJ, Booth LN, Benayoun BA, Izrayelit Y, Schroeder FC, et al. 2014. Males shorten the life span of C. elegans hermaphrodites via secreted compounds. Science 343:541–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCudden CR, Hains MD, Kimple RJ, Siderovski DP, Willard FS.. 2005. G-protein signaling: back to the future. Cell Mol Life Sci. 62:551–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath PT, Ruvinsky I.. 2019. A primer on pheromone signaling in Caenorhabditis elegans for systems biologists. Curr Opin Syst Biol. 13:23–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath PT, Xu Y, Ailion M, Garrison JL, Butcher RA, et al. 2011. Parallel evolution of domesticated Caenorhabditis species targets pheromone receptor genes. Nature 477:321–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnight K, Hoang HD, Prasain JK, Brown N, Vibbert J, et al. 2014. Neurosensory perception of environmental cues modulates sperm motility critical for fertilization. Science 344:754–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehle EA, Sojka SE, Medha KC, Zel RM, Reese J.. et al. 2020. The C. elegans TRPV channel proteins OSM-9 and OCR-2 contribute to aversive chemical sensitivity. MicroPubl Biol. 2020. [10.17912/micropub.biology.000277] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisel JD, Panda O, Mahanti P, Schroeder FC, Kim DH.. 2014. Chemosensation of bacterial secondary metabolites modulates neuroendocrine signaling and behavior of C. elegans. Cell 159:267–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellem JE, Brockie PJ, Madsen DM, Maricq AV.. 2008. Action potentials contribute to neuronal signaling in C. elegans. Nat Neurosci. 11:865–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metaxakis A, Petratou D, Tavernarakis N.. 2018. Multimodal sensory processing in Caenorhabditis elegans. Open Biol. 8:180049. [10.1098/rsob.180049] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills H, Wragg R, Hapiak V, Castelletto M, Zahratka J, et al. 2012. Monoamines and neuropeptides interact to inhibit aversive behaviour in Caenorhabditis elegans. EMBO J. 31:667–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzakhalili E, Epureanu BI, Gourgou E.. 2018. A mathematical and computational model of the calcium dynamics in Caenorhabditis elegans ASH sensory neuron. PLoS ONE 13:e0201302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyawaki A, Llopis J, Heim R, McCaffery JM, Adams JA, et al. 1997. Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature 388:882–887. [DOI] [PubMed] [Google Scholar]
- Moore RS, Kaletsky R, Murphy CT.. 2019. Piwi/PRG-1 argonaute and TGF-b mediate transgenerational learned pathogenic avoidance. Cell 177:1827–1841 e1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morsci NS, Haas LA, Barr MM.. 2011. Sperm status regulates sexual attraction in Caenorhabditis elegans. Genetics 189:1341–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton DB. 2004. Atypical soluble guanylyl cyclases in Drosophila can function as molecular oxygen sensors. J Biol Chem. 279:50651–50653. [DOI] [PubMed] [Google Scholar]
- Murayama T, Takayama J, Fujiwara M, Maruyama IN.. 2013. Environmental alkalinity sensing mediated by the transmembrane guanylyl cyclase GCY-14 in C. elegans. Curr Biol. 23:1007–1012. [DOI] [PubMed] [Google Scholar]
- Nagarathnam B, Kalaimathy S, Balakrishnan V, Sowdhamini R.. 2012. Cross-genome clustering of human and C. elegans G-protein coupled receptors. Evol Bioinform Online 8:229–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayan A, Venkatachalam V, Durak O, Reilly DK, Bose N, et al. 2016. Contrasting responses within a single neuron class enable sex-specific attraction in Caenorhabditis elegans. Proc Natl Acad Sci USA. 113:E1392–E1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neal SJ, Takeishi A, O'Donnell MP, Park J, Hong M, et al. 2015. Feeding state-dependent regulation of developmental plasticity via CaMKI and neuroendocrine signaling. Elife 4:e10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicoletti M, Loppini A, Chiodo L, Folli V, Ruocco G, et al. 2019. Biophysical modeling of C. elegans neurons: Single ion currents and whole-cell dynamics of AWCon and RMD. PLoS ONE 14:e0218738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Q, Huang X, Zhang L, Xu J, Yang D, et al. 2010. A Trojan horse mechanism of bacterial pathogenesis against nematodes. Proc Natl Acad Sci USA. 107:16631–16636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell MP, Chao P-H, Kammenga JE, Sengupta P.. 2018. Rictor/TORC2 mediates gut-to-brain signaling in the regulation of phenotypic plasticity in C. elegans. PLoS Genet. 14:e1007213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Halloran DM, Altshuler-Keylin S, Lee JI, L'Etoile ND.. 2009. Regulators of AWC-mediated olfactory plasticity in Caenorhabditis elegans. PLoS Genet. 5:e1000761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Halloran DM, Altshuler-Keylin S, Zhang X-D, He C, Morales-Phan C, et al. 2017. Contribution of the cyclic nucleotide gated channel subunit, CNG-3, to olfactory plasticity in Caenorhabditis elegans. Sci Rep. 7:169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Halloran DM, Fitzpatrick DA, McCormack GP, McInerney JO, Burnell AM.. 2006. The molecular phylogeny of a nematode-specific clade of heterotrimeric G-protein a-subunit genes. J Mol Evol. 63:87–94. [DOI] [PubMed] [Google Scholar]
- O'Halloran DM, Hamilton OS, Lee JI, Gallegos M, L'Etoile ND.. 2012. Changes in cGMP levels affect the localization of EGL-4 in AWC in Caenorhabditis elegans. PLoS ONE 7:e31614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda S, Tomioka M, Iino Y.. 2011. Neuronal plasticity regulated by the insulin-like signaling pathway underlies salt chemotaxis learning in Caenorhabditis elegans. J Neurophysiol. 106:301–308. [DOI] [PubMed] [Google Scholar]
- Olmedo M, O'Neill JS, Edgar RS, Valekunja UK, Reddy AB, et al. 2012. Circadian regulation of olfaction and an evolutionarily conserved, nontranscriptional marker in Caenorhabditis elegans. Proc Natl Acad Sci USA. 109:20479–20484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omori K, Kotera J.. 2007. Overview of PDEs and their regulation. Circ Res. 100:309–327. [DOI] [PubMed] [Google Scholar]
- Oren-Suissa M, Bayer EA, Hobert O.. 2016. Sex-specific pruning of neuronal synapses in Caenorhabditis elegans. Nature 533:206–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz CO, Etchberger JF, Posy SL, Frøkjær-Jensen C, Lockery S, et al. 2006. Searching for neuronal left/right asymmetry: genomewide analysis of nematode receptor-type guanylyl cyclases. Genetics 173:131–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz CO, Faumont S, Takayama J, Ahmed HK, Goldsmith AD, et al. 2009. Lateralized gustatory behavior of C. elegans is controlled by specific receptor-type guanylyl cyclases. Curr Biol. 19:996–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne KA, Robichon A, Burgess E, Butland S, Shaw RA, et al. 1997. Natural behavior polymorphism due to a cGMP-dependent protein kinase of Drosophila. Science 277:834–836. [DOI] [PubMed] [Google Scholar]
- Palmitessa A, Hess HA, Bany IA, Kim YM, Koelle MR, et al. 2005. Caenorhabditus elegans arrestin regulates neural G protein signaling and olfactory adaptation and recovery. J Biol Chem. 280:24649–24662. [DOI] [PubMed] [Google Scholar]
- Park D, Hahm JH, Park S, Ha G, Chang GE, et al. 2017. A conserved neuronal DAF-16/FoxO plays an important role in conveying pheromone signals to elicit repulsion behavior in Caenorhabditis elegans. Sci Rep. 7:7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park D, O'Doherty I, Somvanshi RK, Bethke A, Schroeder FC, et al. 2012. Interaction of structure-specific and promiscuous G-protein-coupled receptors mediates small-molecule signaling in Caenorhabditis elegans. Proc Natl Acad Sci USA. 109:9917–9922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Choi W, Dar AR, Butcher RA, Kim K.. 2019a. Neuropeptide signaling regulates pheromone-mediated gene expression of a chemoreceptor gene in C. elegans. Mol Cells 42:28–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Meisel JD, Kim DH.. 2020. Immediate activation of chemosensory neuron gene expression by bacterial metabolites is selectively induced by distinct cyclic GMP-dependent pathways in Caenorhabditis elegans. PLoS Genet. 16:e1008505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Ohshima S, Tani T, Ohshima Y.. 1997. Structure and expression of the gsa-1 gene encoding a G protein a (s) subunit in C. elegans. Gene 194:183–190. [DOI] [PubMed] [Google Scholar]
- Park JY, Joo HJ, Park S, Paik YK.. 2019b. Ascaroside pheromones: chemical biology and pleiotropic neuronal functions. Int J Mol Sci. 20:3898. [10.3390/ijms20163898] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peckol EL, Troemel ER, Bargmann CI.. 2001. Sensory experience and sensory activity regulate chemosensory receptor gene expression in Caenorhabditis elegans. Proc Natl Acad Sci USA. 98:11032–11038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins LA, Hedgecock EM, Thomson JN, Culotti JG.. 1986. Mutant sensory cilia in the nematode Caenorhabditis elegans. Dev Biol. 117:456–487. [DOI] [PubMed] [Google Scholar]
- Pierce KL, Lefkowitz RJ.. 2001. Classical and new roles of β-arrestins in the regulation of G-protein-coupled receptors. Nat Rev Neurosci. 2:727–733. [DOI] [PubMed] [Google Scholar]
- Pierce-Shimomura JT, Dores M, Lockery SR.. 2005. Analysis of the effects of turning bias on chemotaxis in C. elegans. J Exp Biol. 208:4727–4733. [DOI] [PubMed] [Google Scholar]
- Pierce-Shimomura JT, Faumont S, Gaston MR, Pearson BJ, Lockery SR.. 2001. The homeobox gene lim-6 is required for distinct chemosensory representations in C. elegans. Nature 410:694–698. [DOI] [PubMed] [Google Scholar]
- Pierce-Shimomura JT, Morse TM, Lockery SR.. 1999. The fundamental role of pirouettes in Caenorhabditis elegans chemotaxis. J Neurosci. 19:9557–9569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pifferi S, Boccaccio A, Menini A.. 2006. Cyclic nucleotide-gated ion channels in sensory transduction. FEBS Lett. 580:2853–2859. [DOI] [PubMed] [Google Scholar]
- Pitcher JA, Freedman NJ, Lefkowitz RJ.. 1998. G protein-coupled receptor kinases. Annu Rev Biochem. 67:653–692. [DOI] [PubMed] [Google Scholar]
- Pocock R, Hobert O.. 2010. Hypoxia activates a latent circuit for processing gustatory information in C. elegans. Nat Neurosci. 13:610–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter MY, Koelle MR.. 2009. Insights into RGS protein function from studies in Caenorhabditis elegans. Prog Mol Biol Transl Sci. 86:15–47. [DOI] [PubMed] [Google Scholar]
- Posner R, Toker IA, Antonova O, Star E, Anava S, et al. 2019. Neuronal small RNAs control behavior transgenerationally. Cell 177:1814–1826.e1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradel E, Zhang Y, Pujol N, Matsuyama T, Bargmann CI, et al. 2007. Detection and avoidance of a natural product from the pathogenic bacterium Serratia marcescens by Caenorhabditis elegans. Proc Natl Acad Sci USA. 104:2295–2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pungaliya C, Srinivasan J, Fox BW, Malik RU, Ludewig AH, et al. 2009. A shortcut to identifying small molecule signals that regulate behavior and development in Caenorhabditis elegans. Proc Natl Acad Sci USA. 106:7708–7713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahi SJ, Larsch J, Pecani K, Katsov AY, Mansouri N, et al. 2017. Oscillatory stimuli differentiate adapting circuit topologies. Nat Methods 14:1010–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren P, Lim CS, Johnsen R, Albert PS, Pilgrim D, et al. 1996. Control of C. elegans larval development by neuronal expression of a TGF-beta homolog. Science 274:1389–1391. [DOI] [PubMed] [Google Scholar]
- Rex E, Hapiak V, Hobson R, Smith K, Xiao H, et al. 2005. TYRA-2 (F01E11.5): a Caenorhabditis elegans tyramine receptor expressed in the MC and NSM pharyngeal neurons. J Neurochem. 94:181–191. [DOI] [PubMed] [Google Scholar]
- Riddle DL, Swanson MM, Albert PS.. 1981. Interacting genes in nematode dauer larva formation. Nature 290:668–671. [DOI] [PubMed] [Google Scholar]
- Roayaie K, Crump JG, Sagasti A, Bargmann CI.. 1998. The Gα protein ODR-3 mediates olfactory and nociceptive function and controls cilium morphogenesis in C. elegans olfactory neurons. Neuron 20:55–67. [DOI] [PubMed] [Google Scholar]
- Roberts WM, Augustine SB, Lawton KJ, Lindsay TH, Thiele TR, et al. 2016. A stochastic neuronal model predicts random search behaviors at multiple spatial scales in C. elegans. Elife 5:e12572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson HM. 1998. Two large families of chemoreceptor genes in the nematodes Caenorhabditis elegans and Caenorhabditis briggsae reveal extensive gene duplication, diversification, movement, and intron loss. Genome Res. 8:449–463. [DOI] [PubMed] [Google Scholar]
- Robertson HM. 2000. The large srh family of chemoreceptor genes in Caenorhabditis nematodes reveals processes of genome evolution involving large duplications and deletions and intron gains and losses. Genome Res. 10:192–203. [DOI] [PubMed] [Google Scholar]
- Robertson HM. 2001. Updating the str and srj (stl) families of chemoreceptors in Caenorhabditis nematodes reveals frequent gene movement within and between chromosomes. Chem Senses 26:151–159. [DOI] [PubMed] [Google Scholar]
- Robertson HM, Thomas JH.. 2006. The putative chemoreceptor families of C. elegans. In: The C. elegans Research Community WormBook, editor. WormBook. [DOI] [PMC free article] [PubMed]
- Romoser VA, Hinkle PM, Persechini A.. 1997. Detection in living cells of Ca2+-dependent changes in the fluorescence emission of an indicator composed of two green fluorescent protein variants linked by a calmodulin-binding sequence. A new class of fluorescent indicators. J Biol Chem. 272:13270–13274. [DOI] [PubMed] [Google Scholar]
- Root MJ, MacKinnon R.. 1993. Identification of an external divalent cation-binding site in the pore of a cGMP-activated channel. Neuron 11:459–466. [DOI] [PubMed] [Google Scholar]
- Ross EM, Wilkie TM.. 2000. GTPase-activating proteins for heterotrimeric G proteins: regulators of G protein signaling (RGS) and RGS-like proteins. Annu Rev Biochem. 69:795–827. [DOI] [PubMed] [Google Scholar]
- Ryan DA, Miller RM, Lee K, Neal SJ, Fagan KA, et al. 2014. Sex, age, and hunger regulate behavioral prioritization through dynamic modulation of chemoreceptor expression. Curr Biol. 24:2509–2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu L, Cheon Y, Huh YH, Pyo S, Chinta S, et al. 2018. Feeding state regulates pheromone-mediated avoidance behavior via the insulin signaling pathway in Caenorhabditis elegans. EMBO J. 37:e98402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samanta A, Hughes TET, Moiseenkova-Bell VY.. 2018. Transient receptor potential (TRP) channels. Subcell Biochem. 87:141–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambongi Y, Nagae T, Liu Y, Yoshimizu T, Takeda K, et al. 1999. Sensing of cadmium and copper ions by externally exposed ADL, ASE, and ASH neurons elicits avoidance response in Caenorhabditis elegans. Neuroreport 10:753–757. [DOI] [PubMed] [Google Scholar]
- Sambongi Y, Takeda K, Wakabayashi T, Ueda I, Wada Y, et al. 2000. Caenorhabditis elegans senses protons through amphid chemosensory neurons: proton signals elicit avoidance behavior. Neuroreport 11:2229–2232. [DOI] [PubMed] [Google Scholar]
- Samuel BS, Rowedder H, Braendle C, Felix MA, Ruvkun G.. 2016. Caenorhabditis elegans responses to bacteria from its natural habitats. Proc Natl Acad Sci USA. 113:E3941–E3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassa T, Maruyama IN.. 2013. A G-protein alpha subunit, GOA-1, plays a role in C. elegans avoidance behavior of strongly alkaline pH. Commun Integr Biol. 6:e26668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassa T, Murayama T, Maruyama IN.. 2013. Strongly alkaline pH avoidance mediated by ASH sensory neurons in C. elegans. Neurosci Lett. 555:248–252. [DOI] [PubMed] [Google Scholar]
- Satoh Y, Sato H, Kunitomo H, Fei X, Hashimoto K, et al. 2014. Regulation of experience-dependent bidirectional chemotaxis by a neural circuit switch in Caenorhabditis elegans. J Neurosci. 34:15631–15637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schackwitz WS, Inoue T, Thomas JH.. 1996. Chemosensory neurons function in parallel to mediate a pheromone response in C. elegans. Neuron 17:719–728. [DOI] [PubMed] [Google Scholar]
- Schaedel ON, Gerisch B, Antebi A, Sternberg PW.. 2012. Hormonal signal amplification mediates environmental conditions during development and controls an irreversible commitment to adulthood. PLoS Biol. 10:e1001306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer WR. 2015. Mechanosensory molecules and circuits in C. elegans. Pflugers Arch. 467:39–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer WR, Kenyon CJ.. 1995. A calcium-channel homologue required for adaptation to dopamine and serotonin in Caenorhabditis elegans. Nature 375:73–78. [DOI] [PubMed] [Google Scholar]
- Schulenburg H, Félix MA.. 2017. The natural biotic environment of Caenorhabditis elegans. Genetics 206:55–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert R, Eismann E, Ludwig J, Baumann A, Kaupp UB.. 1999. Molecular determinants of a Ca2+-binding site in the pore of cyclic nucleotide-gated channels: S5/S6 segments control affinity of intrapore glutamates. EMBO J. 18:119–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta P. 2007. Generation and modulation of chemosensory behaviors in C. elegans. Pflugers Arch. 454:721–734. [DOI] [PubMed] [Google Scholar]
- Sengupta P, Chou JH, Bargmann CI.. 1996. odr-10 encodes a seven transmembrane domain olfactory receptor required for responses to the odorant diacetyl. Cell 84:899–909. [DOI] [PubMed] [Google Scholar]
- Shao J, Zhang X, Cheng H, Yue X, Zou W, et al. 2019. Serotonergic neuron ADF modulates avoidance behaviors by inhibiting sensory neurons in C. elegans. Pflugers Arch. 471:357–363. [DOI] [PubMed] [Google Scholar]
- Sharma RK, Duda T.. 2014. Membrane guanylate cyclase, a multimodal transduction machine: history, present, and future directions. Front Mol Neurosci. 7:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma RK, Duda T, Makino CL.. 2016. Integrative signaling networks of membrane guanylate cyclases: Biochemistry and Physiology . Front Mol Neurosci. 9:83. [10.3389/fnmol.2016.00083] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpell FH. 1985. Microbial Flavors and Fragrances, in Comprehensive Biotechnology: The Principles, Applications, and Regulations of Biotechnology in Industry, Agriculture, and Medicine. Oxford: Pergamon Press; [Oxfordshire]; New York. [Google Scholar]
- Shi C, Runnels AM, Murphy CT.. 2017. Mating and male pheromone kill Caenorhabditis males through distinct mechanisms. Elife 6:e23493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shidara H, Hotta K, Oka K.. 2017. Compartmentalized cGMP responses of olfactory sensory neurons in Caenorhabditis elegans. J Neurosci. 37:3753–3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindou T, Ochi-Shindou M, Murayama T, Saita EI, Momohara Y, et al. 2019. Active propagation of dendritic electrical signals in. Sci Rep. 9:3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinkai Y, Yamamoto Y, Fujiwara M, Tabata T, Murayama T, et al. 2011. Behavioral choice between conflicting alternatives is regulated by a receptor guanylyl cyclase, GCY-28, and a receptor tyrosine kinase, SCD-2, in AIA interneurons of Caenorhabditis elegans. J Neurosci. 31:3007–3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shtonda BB, Avery L.. 2005. CCA-1, EGL-19 and EXP-2 currents shape action potentials in the Caenorhabditis elegans pharynx. J Exp Biol. 208:2177–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shtonda BB, Avery L.. 2006. Dietary choice behavior in Caenorhabditis elegans. J Exp Biol. 209:89–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuart NG, Haitin Y, Camp SS, Black KD, Zagotta WN.. 2011. Molecular mechanism for 3:1 subunit stoichiometry of rod cyclic nucleotide-gated ion channels. Nat Commun. 2:457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhvi A, Liu B, Friedman CJ, Fong J, Lu Y, et al. 2016. A glial K/Cl transporter controls neuronal receptive ending shape by chloride inhibition of an rGC. Cell 165:936–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith HK, Luo L, O’Halloran D, Guo D, Huang X-Y, et al. 2013. Defining specificity determinants of cGMP mediated gustatory sensory transduction in Caenorhabditis elegans. Genetics 194:885–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smrcka AV. 2008. G protein βγ subunits: central mediators of G protein-coupled receptor signaling. Cell Mol Life Sci. 65:2191–2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan J, Kaplan F, Ajredini R, Zachariah C, Alborn HT, et al. 2008. A blend of small molecules regulates both mating and development in Caenorhabditis elegans. Nature 454:1115–1118., [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan J, von Reuss SH, Bose N, Zaslaver A, Mahanti P, et al. 2012. A modular library of small molecule signals regulates social behaviors in Caenorhabditis elegans. PLoS Biol. 10:e1001237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steger KA, Shtonda BB, Thacker C, Snutch TP, Avery L.. 2005. The C. elegans T-type calcium channel CCA-1 boosts neuromuscular transmission. J Exp Biol. 208:2191–2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steuer Costa W, Van der Auwera P, Glock C, Liewald JF, Bach M, et al. 2019. A GABAergic and peptidergic sleep neuron as a locomotion stop neuron with compartmentalized Ca2+ dynamics. Nat Commun. 10:4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Kerr R, Bianchi L, Frøkjær-Jensen C, Slone D, et al. 2003. In vivo imaging of C. elegans mechanosensory neurons demonstrates a specific role for the MEC-4 channel in the process of gentle touch sensation. Neuron 39:1005–1017. [DOI] [PubMed] [Google Scholar]
- Suzuki H, Thiele TR, Faumont S, Ezcurra M, Lockery SR, et al. 2008. Functional asymmetry in Caenorhabditis elegans taste neurons and its computational role in chemotaxis. Nature 454:114–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sze JY, Victor M, Loer C, Shi Y, Ruvkun G.. 2000. Food and metabolic signalling defects in a Caenorhabditis elegans serotonin-synthesis mutant. Nature 403:560–564. [DOI] [PubMed] [Google Scholar]
- Taniguchi G, Uozumi T, Kiriyama K, Kamizaki T, Hirotsu T.. 2014. Screening of odor-receptor pairs in Caenorhabditis elegans reveals different receptors for high and low odor concentrations. Sci Signal. 7:ra39. [DOI] [PubMed] [Google Scholar]
- Tanimoto Y, Yamazoe-Umemoto A, Fujita K, Kawazoe Y, Miyanishi Y, et al. 2017. Calcium dynamics regulating the timing of decision-making in C. elegans. Elife 6:e21629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao J, Ma YC, Yang ZS, Zou CG, Zhang KQ.. 2016. Octopamine connects nutrient cues to lipid metabolism upon nutrient deprivation. Sci Adv. 2:e1501372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiele TR, Faumont S, Lockery SR.. 2009. The neural network for chemotaxis to tastants in Caenorhabditis elegans is specialized for temporal differentiation. J Neurosci. 29:11904–11911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JH, Kelley JL, Robertson HM, Ly K, Swanson WJ.. 2005. Adaptive evolution in the SRZ chemoreceptor families of Caenorhabditis elegans and Caenorhabditis briggsae. Proc Natl Acad Sci USA. 102:4476–4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JH, Robertson HM.. 2008. The Caenorhabditis chemoreceptor gene families. BMC Biol. 6:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobin D, Madsen D, Kahn-Kirby A, Peckol E, Moulder G, et al. 2002. Combinatorial expression of TRPV channel proteins defines their sensory functions and subcellular localization in C. elegans neurons. Neuron 35:307–318. [DOI] [PubMed] [Google Scholar]
- Torayama I, Ishihara T, Katsura I.. 2007. Caenorhabditis elegans integrates the signals of butanone and food to enhance chemotaxis to butanone. J Neurosci. 27:741–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tracey WD Jr., Wilson RI, Laurent G, Benzer S.. 2003. painless, a Drosophila gene essential for nociception. Cell 113:261–273. [DOI] [PubMed] [Google Scholar]
- Tran A, Tang A, O'Loughlin CT, Balistreri A, Chang E, et al. 2017. C. elegans avoids toxin-producing Streptomyces using a seven transmembrane domain chemosensory receptor. Elife 6:e23770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treede RD. 1999. Transduction and transmission properties of primary nociceptive afferents. Ross Fiziol Zh Im I M Sechenova 85:205–211. [PubMed] [Google Scholar]
- Troemel ER, Chou JH, Dwyer ND, Colbert HA, Bargmann CI.. 1995. Divergent seven transmembrane receptors are candidate chemosensory receptors in C. elegans. Cell 83:207–218. [DOI] [PubMed] [Google Scholar]
- Troemel ER, Kimmel BE, Bargmann CI.. 1997. Reprogramming chemotaxis responses: sensory neurons define olfactory preferences in C. elegans. Cell 91:161–169. [DOI] [PubMed] [Google Scholar]
- Troemel ER, Sagasti A, Bargmann CI.. 1999. Lateral signaling mediated by axon contact and calcium entry regulates asymmetric odorant receptor expression in C. elegans. Cell 99:387–398. [DOI] [PubMed] [Google Scholar]
- Tsalik EL, Hobert O.. 2003. Functional mapping of neurons that control locomotory behavior in Caenorhabditis elegans. J Neurobiol. 56:178–197. [DOI] [PubMed] [Google Scholar]
- Tsunozaki M, Chalasani SH, Bargmann CI.. 2008. A behavioral switch: cGMP and PKC signaling in olfactory neurons reverses odor preference in C. elegans. Neuron 59:959–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uozumi T, Hirotsu T, Yoshida K, Yamada R, Suzuki A, et al. 2012. Temporally-regulated quick activation and inactivation of Ras is important for olfactory behaviour. Sci Rep. 2:500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Voorn L, Gebbink M, Plasterk RH, Ploegh HL.. 1990. Characterization of a G protein β-subunit gene from the nematode Caenorhabditis elegans. J Mol Biol. 213:17–26. [DOI] [PubMed] [Google Scholar]
- Venkatachalam K, Montell C.. 2007. TRP channels. Annu Rev Biochem. 76:387–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal B, Aghayeva U, Sun H, Wang C, Glenwinkel L, et al. 2018. An atlas of Caenorhabditis elegans chemoreceptor expression. PLoS Biol. 16:e2004218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villette V, Chavarha M, Dimov IK, Bradley J, Pradhan L, et al. 2019. Ultrafast two-photon imaging of a high-gain voltage indicator in awake behaving mice. Cell 179:1590–1608 e1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vowels JJ, Thomas JH.. 1994. Multiple chemosensory defects in daf-11 and daf-21 mutants of Caenorhabditis elegans. Genetics 138:303–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi T, Kimura Y, Ohba Y, Adachi R, Satoh Y, et al. 2009. In vivo calcium imaging of OFF-responding ASK chemosensory neurons in C. elegans. Biochim Biophys Acta 1790:765–769. [DOI] [PubMed] [Google Scholar]
- Wakabayashi T, Kitagawa I, Shingai R.. 2004. Neurons regulating the duration of forward locomotion in Caenorhabditis elegans. Neurosci Res. 50:103–111. [DOI] [PubMed] [Google Scholar]
- Walker DS, Chew YL, Schafer WR.. 2017. Genetics of Behavior in C. elegans in the Oxford Handbook of Invertebrate Neurobiology. Oxford University Press. [Google Scholar]
- Wan X, Zhou Y, Chan CM, Yang H, Yeung C, et al. 2019. SRD-1 in AWA neurons is the receptor for female volatile sex pheromones in C. elegans males. EMBO Rep. 20:e46288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, O’Halloran D, Goodman MB.. 2013. GCY-8, PDE-2, and NCS-1 are critical elements of the cGMP-dependent thermotransduction cascade in the AFD neurons responsible for C. elegans thermotaxis. J Gen Physiol. 142:437–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Sato H, Satoh Y, Tomioka M, Kunitomo H, et al. 2017. A gustatory neural circuit of Caenorhabditis elegans generates memory-dependent behaviors in Na+ chemotaxis. J Neurosci. 37:2097–2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Xu ZJ, Wu YQ, Qin LW, Li ZY, et al. 2015. Off-response in ASH neurons evoked by CuSO4 requires the TRP channel OSM-9 in Caenorhabditis elegans. Biochem Biophys Res Commun. 461:463–468. [DOI] [PubMed] [Google Scholar]
- Wang X, Li G, Liu J, Liu J, Xu XZ.. 2016. TMC-1 mediates alkaline sensation in C. elegans through nociceptive neurons. Neuron 91:146–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Apicella A Jr., Lee SK, Ezcurra M, Slone RD, et al. 2008. A glial DEG/ENaC channel functions with neuronal channel DEG-1 to mediate specific sensory functions in C. elegans. EMBO J. 27:2388–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward S. 1973. Chemotaxis by the nematode Caenorhabditis elegans: identification of attractants and analysis of the response by use of mutants. Proc Natl Acad Sci USA. 70:817–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward S, Thomson N, White JG, Brenner S.. 1975. Electron microscopical reconstitution of the anterior sensory anatomy of the nematode Caenorhabditis elegans. J Comp Neurol. 160:313–337. [DOI] [PubMed] [Google Scholar]
- Ware RW, Clark D, Crossland K, Russell RL.. 1975. The nerve ring of the nematode Caenorhabditis elegans: sensory input and motor output. J Comp Neurol. 162:71–110. [Google Scholar]
- Weis WI, Kobilka BK.. 2018. The molecular basis of G protein-coupled receptor activation. Annu Rev Biochem. 87:897–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wes PD, Bargmann CI.. 2001. C. elegans odour discrimination requires asymmetric diversity in olfactory neurons. Nature 410:698–701. [DOI] [PubMed] [Google Scholar]
- Wharam B, Weldon L, Viney M.. 2017. Pheromone modulates two phenotypically plastic traits - adult reproduction and larval diapause - in the nematode Caenorhabditis elegans. BMC Evol Biol. 17:197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JG, Southgate E, Thomson JN, Brenner S.. 1986. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci. 314:1–340. [DOI] [PubMed] [Google Scholar]
- White JQ, Nicholas TJ, Gritton J, Truong L, Davidson ER, et al. 2007. The sensory circuitry for sexual attraction in C. elegans males. Curr Biol. 17:1847–1857. [DOI] [PubMed] [Google Scholar]
- Wicks SR, de Vries CJ, van Luenen HG, Plasterk RH.. 2000. CHE-3, a cytosolic dynein heavy chain, is required for sensory cilia structure and function in Caenorhabditis elegans. Dev Biol. 221:295–307. [DOI] [PubMed] [Google Scholar]
- Willars GB. 2006. Mammalian RGS proteins: multifunctional regulators of cellular signalling. Semin Cell Dev Biol. 17:363–376. [DOI] [PubMed] [Google Scholar]
- Williams PDE, Zahratka JA, Rodenbeck M, Wanamaker J, Linzie H, et al. 2018. Serotonin disinhibits a Caenorhabditis elegans sensory neuron by suppressing Ca2+-dependent negative feedback. J Neurosci. 38:2069–2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winger JA, Derbyshire ER, Lamers MH, Marletta MA, Kuriyan J.. 2008. The crystal structure of the catalytic domain of a eukaryotic guanylate cyclase. BMC Struct Biol. 8:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtyniak M, Brear AG, O'Halloran DM, Sengupta P.. 2013. Cell- and subunit-specific mechanisms of CNG channel ciliary trafficking and localization in C. elegans. J Cell Sci. 126:4381–4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woldemariam S, Nagpal J, Hill T, Li J, Schneider MW, et al. 2019. Using a robust and sensitive GFP-based cGMP sensor for real-time imaging in intact Caenorhabditis elegans. Genetics 213:59–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood JF, Ferkey DM.. 2016. GRK roles in C. elegans. In: Gurevich VV, Gurevich EV, Tesmer JJG, editors. G Protein-Coupled Receptor Kinases. Humana Press. p. 283–299. [Google Scholar]
- Worthy SE, Haynes L, Chambers M, Bethune D, Kan E, et al. 2018a. Identification of attractive odorants released by preferred bacterial food found in the natural habitats of C. elegans. PLoS ONE 13:e0201158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worthy SE, Rojas GL, Taylor CJ, Glater EE.. 2018b. Identification of odor blend used by Caenorhabditis elegans for pathogen recognition. Chem Senses 43:169–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T, Duan F, Yang W, Liu H, Caballero A, et al. 2019. Pheromones modulate learning by regulating the balanced signals of two insulin-like peptides. Neuron 104:1095–1109 e1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt TD. 2003. Pheromones and Animal Behavior: Communication by Smell and Taste. Cambridge: Cambridge University Press. [Google Scholar]
- Xiao R, Xu XZ.. 2009. Function and regulation of TRP family channels in C. elegans. Pflugers Arch. 458:851–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao R, Xu XZ.. 2011. C. elegans TRP channels. Adv Exp Med Biol. 704:323–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Hirotsu T, Matsuki M, Butcher RA, Tomioka M, et al. 2010. Olfactory plasticity is regulated by pheromonal signaling in Caenorhabditis elegans. Science 329:1647–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Hirotsu T, Matsuki M, Kunitomo H, Iino Y.. 2009. GPC-1, a G protein γ-subunit, regulates olfactory adaptation in Caenorhabditis elegans. Genetics 181:1347–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yook K, Hodgkin J.. 2007. Mos1 mutagenesis reveals a diversity of mechanisms affecting response of Caenorhabditis elegans to the bacterial pathogen Microbacterium nematophilum. Genetics 175:681–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Hirotsu T, Tagawa T, Oda S, Wakabayashi T, et al. 2012. Odour concentration-dependent olfactory preference change in C. elegans. Nat Commun. 3:739.,. [DOI] [PubMed] [Google Scholar]
- You YJ, Kim J, Raizen DM, Avery L.. 2008. Insulin, cGMP, and TGF-b signals regulate food intake and quiescence in C. elegans: a model for satiety. Cell Metab. 7:249–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S, Avery L, Baude E, Garbers DL.. 1997. Guanylyl cyclase expression in specific sensory neurons: a new family of chemosensory receptors. Proc Natl Acad Sci USA. 94:3384–3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue X, Sheng Y, Kang L, Xiao R.. 2019. Distinct functions of TMC channels: a comparative overview. Cell Mol Life Sci. 76:4221–4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahratka JA, Williams PD, Summers PJ, Komuniecki RW, Bamber BA.. 2015. Serotonin differentially modulates Ca2+ transients and depolarization in a C. elegans nociceptor. J Neurophysiol. 113:1041–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamponi GW, Striessnig J, Koschak A, Dolphin AC.. 2015. The physiology, pathology, and pharmacology of voltage-gated calcium channels and their future therapeutic potential. Pharmacol Rev. 67:821–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaslaver A, Liani I, Shtangel O, Ginzburg S, Yee L, et al. 2015. Hierarchical sparse coding in the sensory system of Caenorhabditis elegans. Proc Natl Acad Sci USA. 112:1185–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Zhao N, Chen Y, Zhang D, Yan J, et al. 2016. The signaling pathway of Caenorhabditis elegans mediates chemotaxis response to the attractant 2-heptanone in a trojan horse-like pathogenesis. J Biol Chem. 291:23618–23627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Sokolchik I, Blanco G, Sze JY.. 2004. Caenorhabditis elegans TRPV ion channel regulates 5HT biosynthesis in chemosensory neurons. Development 131:1629–1638. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Chou JH, Bradley J, Bargmann CI, Zinn K.. 1997. The Caenorhabditis elegans seven-transmembrane protein ODR-10 functions as an odorant receptor in mammalian cells. Proc Natl Acad Sci USA. 94:12162–12167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Lu H, Bargmann CI.. 2005. Pathogenic bacteria induce aversive olfactory learning in Caenorhabditis elegans. Nature 438:179–184. [DOI] [PubMed] [Google Scholar]
- Zhong H, Molday LL, Molday RS, Yau KW.. 2002. The heteromeric cyclic nucleotide-gated channel adopts a 3A:1B stoichiometry. Nature 420:193–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong L, Hwang RY, Tracey WD.. 2010. Pickpocket is a DEG/ENaC protein required for mechanical nociception in Drosophila larvae. Curr Biol. 20:429–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Loeza-Cabrera M, Liu Z, Aleman-Meza B, Nguyen JK, et al. 2017. Potential nematode alarm pheromone induces acute avoidance in Caenorhabditis elegans. Genetics 206:1469–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou W, Cheng H, Li S, Yue X, Xue Y, et al. 2017. Polymodal responses in C. elegans phasmid neurons rely on multiple intracellular and intercellular signaling pathways. Sci Rep. 7:42295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwaal RR, Ahringer J, van Luenen HG, Rushforth A, Anderson P, et al. 1996. G proteins are required for spatial orientation of early cell cleavages in C. elegans embryos. Cell 86:619–629. [DOI] [PubMed] [Google Scholar]
- Zwaal RR, Mendel JE, Sternberg PW, Plasterk RH.. 1997. Two neuronal G proteins are involved in chemosensation of the Caenorhabditis elegans Dauer-inducing pheromone. Genetics 145:715–727. [DOI] [PMC free article] [PubMed] [Google Scholar]