Abstract
The protein molecular chaperone Hsp90 (Heat shock protein, 90 kilodalton) plays multiple roles in the biogenesis and regulation of client proteins impacting myriad aspects of cellular physiology. Amino acid alterations located throughout Saccharomyces cerevisiae Hsp90 have been shown to result in reduced client activity and temperature-sensitive growth defects. Although some Hsp90 mutants have been shown to affect activity of particular clients more than others, the mechanistic basis of client-specific effects is unknown. We found that Hsp90 mutants that disrupt the early step of Hsp70 and Sti1 interaction, or show reduced ability to adopt the ATP-bound closed conformation characterized by Sba1 and Cpr6 interaction, similarly disrupt activity of three diverse clients, Utp21, Ssl2, and v-src. In contrast, mutants that appear to alter other steps in the folding pathway had more limited effects on client activity. Protein expression profiling provided additional evidence that mutants that alter similar steps in the folding cycle cause similar in vivo consequences. Our characterization of these mutants provides new insight into how Hsp90 and cochaperones identify and interact with diverse clients, information essential for designing pharmaceutical approaches to selectively inhibit Hsp90 function.
Keywords: molecular chaperone, cochaperone, heat shock factor, tetratricopeptide repeats, Hop/Sti1
Introduction
Hsp90 is an abundant essential molecular chaperone that mediates the folding and activation of client proteins in a nucleotide-dependent cycle. Hsp90 directly or indirectly impacts the function of 10-15% of all proteins in yeast and humans (Millson et al. 2005; Zhao et al. 2005; McClellan et al. 2007; Taipale et al. 2010; Franzosa et al. 2011; Wu et al. 2012; Gopinath et al. 2014; Taipale et al. 2014). Due to its role in chaperoning oncogenic proteins and promoting cancerous growth, Hsp90 is an important drug target. However, compounds that bind the ATP binding pocket and broadly inhibit function are too toxic for widespread use (Neckers and Workman 2012; Garg et al. 2016). More selective inhibitors are clearly needed, but little is known about how to achieve precise inhibition of selective Hsp90-client protein interactions and functions.
The Hsp90 substrate folding cycle is complex. Client proteins interact with Hsp70 prior to Hsp90 and transfer of client to Hsp90 is mediated by cochaperones such as Sti1 or Cdc37. Once client binds to Hsp90, Hsp90 undergoes dramatic conformational changes and binds and hydrolyzes ATP (Li et al. 2012; Prodromou 2012; Schopf et al. 2017). Additional cochaperones interact with Hsp90 throughout the folding cycle, regulating Hsp90 ATPase activity, and/or mediating Hsp90 conformational changes. Progression of Hsp90 through a series of distinct cochaperone complexes is thought to help drive productive client folding. Increasing evidence suggests that the pattern of interacting cochaperones varies in a client-specific manner (Lorenz et al. 2014; Taipale et al. 2014; Verba et al. 2016), raising the possibility that disrupting cochaperone interactions may selectively alter Hsp90 functions. Another potential source of difference between how clients interact with Hsp90 is the location of the client binding site. Recent studies indicate that clients bind regions near the junction of the middle and carboxyl-terminal domains, although exceptions have been noted (Lorenz et al. 2014; Karagoz and Rudiger 2015; Verba et al. 2016).
Saccharomyces cerevisiae contains at least twelve Hsp90 cochaperones that form a complex network, with each likely having individual functions fine-tuning Hsp90 functions (Biebl et al. 2020). Some of the cochaperones appear to be required for most Hsp90 functions, whereas others have more restricted functions (Sahasrabudhe et al. 2017). The genetic tractability of S. cerevisiae enables dissection of the entire Hsp90 complex and it is widely used for studying the consequences of Hsp90 or cochaperone mutation on function (Borkovich et al. 1989; Nathan and Lindquist 1995; Obermann et al. 1998; Meyer et al. 2003; Johnson et al. 2007; Genest et al. 2013; Lorenz et al. 2014; Mishra et al. 2016). Prior studies suggest that some yeast Hsp90 mutations selectively affect one client more than others (Bohen 1995; Hawle et al. 2006; Flom et al. 2012; Mishra et al. 2016), but the mechanistic basis for the specific effects remain unknown. In this study, we identified a panel of yeast Hsp90 mutants that cause similar growth defects at elevated temperature, indicating they disrupt essential functions. We then sorted mutants into groups with shared complex assembly defects, such as interaction with Hsp70 or specific cochaperones, and determined whether mutants with similar defects caused similar defects in client activity. Our results indicate that some yeast Hsp90 mutants affect a broad range of Hsp90 functions, whereas others have more selective effects. 2D-DIGE protein expression profiling provided additional evidence that it is possible to group Hsp90 mutants according to overall effects on function. Further analysis of these Hsp90 mutants will provide new insights into how Hsp90 and cochaperones identify and interact with diverse clients as part of the folding cycle.
Materials and methods
Media, chemicals, antibodies, and plasmids
Standard yeast genetic methods were employed (Burke et al. 2000). Yeast were transformed by lithium acetate methods and were grown in either YPD (1% Bacto yeast extract, 2% peptone, and 2% dextrose) or defined synthetic complete media supplemented with 2% dextrose. Growth was examined by spotting 10-fold serial dilutions of yeast cultures on appropriate media, followed by incubation for two days at 30°C or 37°C. 5-fluorootic acid (5-FOA) was obtained from Toronto Research Chemicals. Galactose was obtained from Fisher. Plasmids expressing non-His-tagged or His-tagged Hsp82 have been described (Zuehlke and Johnson 2012). The plasmids pBv-src, which expresses v-src under the GAL1 promoter, and the corresponding empty vector pB656 were a gift from Frank Boschelli (Dey et al. 1996). The polyclonal antisera against Hsc82/Hsp82 recognizes the sequence STEAPVEEVPADTE, which is contained within the carboxy-terminal 20 amino acids of Hsc82, just upstream of the MEEVD sequence (Supplementary Figure S1). Antibodies against purified TIM44 and or the last 56 amino acids of Ssa1/2 were a gift from Dr. Elizabeth Craig (University of Wisconsin). The peptide antisera against Sti1 (DEAESNYKKALELDASNK; a.a. 91-108), Sba1 (KTDFDKWVDEDEQD; a.a. 118-131) and Cpr6 (GTGGESIYDEKFEDEN; a.a. 81-96) have been described and validated (Johnson et al. 2007).
Yeast strains
Strain JJ816 (hsc82::LEU2 hsp82::LEU2/YEp24-HSP82) has been described (Johnson et al. 2007). The utp21-S602F and ssl2-I690N alleles were obtained in a STI1 synthetic lethal screen (Flom et al. 2005; Tenge et al. 2014). The ssl2-I690N was previously erroneously noted as ssl2-L691I. Strain JJ712 (MATa hsp82: LEU2 hsc82::LEU2 utp21-S602F/YEp24-HSP82) expresses the utp21-S602F allele at the genomic locus. Strain 1044 (MATa hsp82: kanr hsc82::kanr ssl2::MET2/YEp24-HSP82 pRS315-ssl2-I690N) was made by standard yeast genetic techniques. The hsc82::LEU2 hsp82::LEU2/YEp24-HSP82 strain lacking CDC37 and expressing plasmid-borne cdc37-S14A (MATa JJ114) has been described (Flom et al. 2012).
Identification of novel temperature-sensitive mutations in HSC82
Plasmid pRS313-HSC82 expresses HSC82 under the endogenous promoter. For cloning purposes, additional sequences from the multiple cloning site of pet29A (Novagen) were inserted at the amino terminus (MKETAAAKFERQHMDSPDLGTLVPRGS). The additional sequences do not affect ability to rescue growth of an hsc82hsp82 strain (not shown). To construct the library, the coding sequence of HSC82 was subjected to error-prone mutagenesis using Taq polymerase and increased MgCl2 concentrations. Individual PCR products were pooled and cloned into pRS313-HSC82. Two independent libraries were constructed, with an estimated 2500 independent isolates. An estimated 5-8% of the library isolates were unable to support viability of an hsc82hsp82 strain. The library was transformed into strain JJ816. Transformants when tested for growth on 5-FOA at 30° and 37 C. Colonies that grew at 30°C but not 37°C were further analyzed. Library plasmids that produced wild-type levels of Hsc82 were rescued and sequenced fully. The indicated mutants were subsequently cloned into pRS313-His-Hsc82, which contains an amino-terminal tag with both a 6X His sequence and the Xpress epitope (Johnson et al. 2007). The growth phenotypes of the original library isolates and the His-tagged versions were indistinguishable in the assays used in this study (not shown).
Isolation of His-Hsc82 complexes and immunoblot analysis
His-Hsc82 complexes were isolated as described (26). Briefly, strain JJ816 (hsc82hsp82) expressing WT or mutant His-Hsc82 was grown overnight to an OD600 of 1.2-2.0. Cells were harvested, washed with water and resuspended in lysis buffer [20 mM Tris, pH 7.5, 100 mM KCl, 5 mM MgCl2 containing a protease inhibitor mixture (Roche Applied Science)]. Cells were disrupted in the presence of glass beads with 8 x 30 sec pulses. Hsc82 complexes were isolated by incubation with nickel resin (1 h with rocking, 4°C) followed by washes with lysis buffer plus 0.1% Tween-20 and 35 mM imidazole. Nickel resin was boiled in SDS-PAGE sample buffer and protein complexes were separated by gel electrophoresis followed by Coomassie Blue staining and/or immunoblot analysis using indicated antibodies. Coomassie Blue stained gels shown in Figure 2 were converted to grayscale. For additional immunoblot analysis, cells (0.5 O.D.600 units) were resuspended in phosphate-buffered saline containing 1 mM phenylmethylsulfonylfluoride and disrupted with glass beads in the presence of SDS and triton X-100.Chemiluminescence immunoblots were performed according to the manufacture's suggestions (Pierce, Rockford, IL, USA).
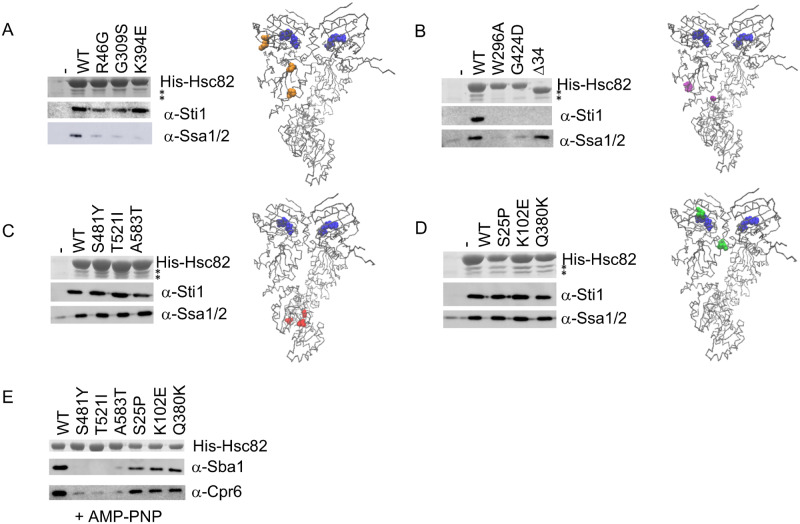
Figure 2.
Effect of mutation on Hsc82 interaction with Hsp70 and cochaperones. A-D. His-Hsc82 complexes were isolated from yeast and analyzed by SDS-PAGE. Top, Stained gel showing the predominant band corresponding to His-Hsc82. The positions of Sti1 (upper) and Ssa1/2 (lower) are indicated by asterisks. Lower, immunoblots using polyclonal antisera specific for Sti1 or Ssa1/2. Location of mutations in Hsc82 were mapped onto the closed structure of the highly homologous Hsp82 using Visual Molecular Dynamics (VMD) (http://www.ks.uiuc.edu/) [PDB 2CG9 (Ali et al. 2006)]. Bound nucleotide in each monomer is shown in blue. R46, G309 and K394 (A, orange); W296, G424 (B, purple); S481, T521, A583 (C, red); S25, K102, Q380 (D, green). (E) His-Hsc82 complexes were isolated as above except that cell lysates were incubated with 5 mM AMP-PNP for 5 min. at 30°C prior to incubation with nickel resin. Immunoblots of His-Hsc82 complexes used antisera specific for Sba1 or Cpr6 (Johnson et al. 2007).
v-src assay
Strains expressing WT or mutant HSC82 were transformed with a multicopy plasmid expressing GAL1-v-src (pBv-src) or the control plasmid (pB656) (Dey et al. 1996). Yeast cultures were grown overnight at 30° in raffinose-uracil drop-out media until mid-log phase. Twenty percent galactose was added to a final concentration of 2%. After 6 h, cultures were serially diluted 10-fold onto uracil drop out plates containing galactose. Plates were grown for 2–3 days at 30°C.
2D DIGE and mass spectrometry protein identification
Yeast cells were freshly collected and rinsed with PBS, then stored at –80 °C before sending to Applied Biomics (Hayward, CA) on dry ice for proteomics analysis. Cells were disrupted in 30 mM Tris-HCl, pH 8.8, containing 7 M urea, 2 M thiourea and 4% CHAPS using sonication. Lysate concentrations were standardized, labeled with Cy2, Cy3, and Cy5 using CyDye prior to IEF and SDS-PAGE. Image scans were carried out immediately following SDS-PAGE using Typhoon TRIO (GE Healthcare) following the protocols provided. The scanned images were then analyzed by Image QuantTL software (GE-Healthcare), and then subjected to in-gel analysis and cross-gel analysis using DeCyder software version 6.5 (GE-Healthcare). The ratio change of the protein differential expression was obtained from in-gel DeCyder software analysis. The spots of interest were picked up by Ettan Spot Picker (GE Healthcare) based on the in-gel analysis and spot picking design by DeCyder software. Trypsin-digested peptides were subject to MALDI-TOF (MS) and TOF/TOF (tandem MS/MS) performed on an Applied Biosystems 5800 mass spectrometer. The resulting peptide mass and the associated fragmentation spectra were submitted to GPS Explorer version 3.5 equipped with MASCOT search engine (Matrix science) to search the National Center for Biotechnology Information nonredundant (NCBInr) or Swiss Protein databases. Candidates with either protein score C.I.% or Ion C.I.% greater than 95 were considered significant.
Stress response pathway transcriptional activation assays
Cells expressing the plasmids pSSA3-LacZ (HSE-lacZ reporter) were grown to mid-log phase at 30°C in selective media. β-galactosidase activity was measured by adding 50 µl of cell suspension and 50 µl of Beta-Glo reagent (Promega, Madison, WI, USA) into a white 96-well plate (Lumitrac 200, Greiner) and incubating for 30 min at 30°C, followed by luminescence detection using a Synergy MX Microplate Reader (Garcia et al. 2017). Plot was prepared using Prism 8 (GraphPad Software, San Diego, CA, USA).
Data availability
Strains and plasmids are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables. Supplementary material is available at figshare: https://doi.org/10.25386/genetics.13547852.
Results
Mutations throughout Hsc82 result in temperature-sensitive growth defects when expressed in an hsc82hsp82 strain
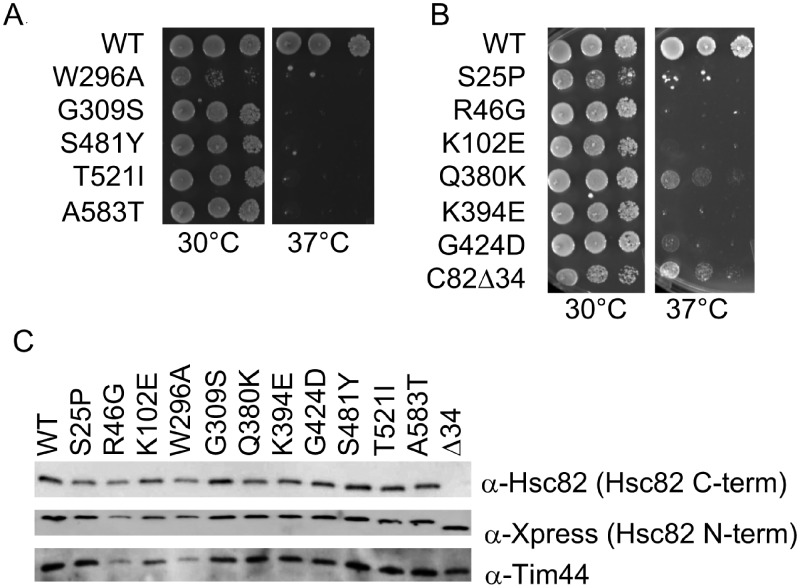
In yeast, Hsp90 is encoded by two genes, HSC82 and HSP82, which are 98% identical. Hsc82 is the more abundant isoform at optimal growth temperature (30°C), whereas Hsp82 levels are induced by elevated temperatures (37°C). Either gene is sufficient for viability of an hsc82hsp82 strain (Borkovich et al. 1989). Hsc82 contains three domains: an amino-terminal nucleotide binding domain (a.a. 1-216), a middle domain (a.a. 269-525) and a carboxyl-terminal dimerization domain (a.a. 529-705) (Ali et al. 2006). We previously analyzed a series of hsc82 mutations that result in near wild-type growth at 30°C but a strong growth defect at 37°C (W296A, G309S, S481Y, T521I and A583T) (Figure 1A) (Johnson et al. 2007). Most of those mutations are in the middle and carboxyl-terminal domains. Prior analysis suggested that the G309S, S481Y, T521I and A583T alterations caused similar in vivo effects, but that W296A had differing effects (Hawle et al. 2006; Flom et al. 2012). In order to identify novel Hsc82 mutants with potentially different allele-specific consequences, we constructed a library of hsc82 mutations using error-prone PCR. We then screened the library to identify mutations that confer temperature-sensitive growth when expressed in a hsc82hsp82 strain. We identified an additional seven mutants that result in a strong growth defect at 37°C, with little to no impacts on growth at the permissive temperature of 30˚C (See Materials and Methods for details) (Figure 1B). These mutants contain the following individual amino acid alterations in the amino-terminal domain (S25P, R46G and K102E) or middle domain (Q380K, K394E and G424D). An additional mutant obtained from the library screen is a truncation of 34 amino acids from the carboxy-terminus (Hsc82Δ34). This mutant is notable since it contains the largest known viable deletion of amino acids from the carboxy-terminus (Louvion et al. 1996). As shown in Figure 1C, all of the mutants are expressed at levels similar to WT Hsc82. The truncation mutant lacks the peptide epitope (Hsc82 C-term) located near the carboxyl-terminus used to generate anti-Hsc82 polyclonal antisera in our laboratory and previously published (Flom et al. 2006 and Supplementary Figure S1). However, when a different antibody that recognizes an epitope located in the amino-terminal tag that includes the 6X His-sequence was used [Figure 1C, anti-Xpress (Hsc82 N-term)], near wild-type levels of the truncated version of Hsc82 protein are visible. All of the mutated residues are stringently conserved from yeast to humans, and about half are conserved in the E. coli Hsp90 ortholog HtpG (Supplementary Figure S1).
Figure 1.
Hsc82 mutants used in this study. Plasmids expressing the indicated mutant form of Hsc82 were transformed into JJ816 (hsc82hsp82/HSP82). Transformants were grown in the presence of 5-FOA to counter-select for the URA3-HSP82 plasmid. Cells were then grown overnight at 30°C, serially diluted 10-fold, plated on rich media and grown for two days at the indicated temperature or harvested for immunoblot analysis. (A) Previously described hsc82 mutations. (B) Novel hsc82 mutations obtained by screening mutant library. (C) Cell lysates were analyzed by SDS-PAGE and immunoblot analysis using polyclonal antibodies specific for Hsc82/Hsp82 or the mitochondrial protein Tim44 as a loading control. Hsc82Δ34 is not recognized by the anti-Hsc82 antiserum due to the absence of the epitope near the carboxy-terminus (see Supplementary Figure S1). The Xpress antibody recognizes sequences near the amino-terminal 6XHis tag.
Hsc82 mutants have varied effects on interaction with Hsp70 and cochaperones
Clients are transferred from Hsp70 to Hsp90 in a process mediated by the cochaperone Sti1, although Hsp70 and Hsp90 also directly interact (Alvira et al. 2014; Genest et al. 2019). We determined whether mutations affect the ability of Hsc82 to interact with Hsp70 (the Ssa isoform) or Sti1. Isolation of His-tagged Hsc82 from yeast with immobilized metal affinity chromatography results in a predominant Hsc82 band in a stained gel. Copurifying Sti1 and Ssa are evident as two bands of similar intensity (each marked with an asterisk*) under His-Hsc82. We additionally performed immunoblots to more specifically resolve both proteins as well as other cochaperones. Hsc82 mutants were placed into groups that showed similar effects on Hsp70 or Sti1 interaction. The R46G, G309S and K394E mutants resulted in reduced interaction with Hsp70 but did not disrupt Sti1 interaction (Figure 2A). G309 and K394 map to a similar region within the middle domain of Hsp90 that directly contacts Hsp70. Although the levels of His-K394E in this experiment were somewhat low, this result is consistent with a prior study that showed that alterations in the Hsp70-Hsp90 binding site that includes G309 and K394 result in reduced interaction with Hsp70, but not Sti1, in vivo and in vitro (Kravats et al. 2018). R46 is located in the amino terminus (Figure 2A, right), in a recently identified second contact site between Hsp70 and Hsp90 (Wang et al. 2020).
Three mutants, W296A, G424D and Hsc82Δ34 resulted in reduced interaction with Sti1. W296A and G424D, which alter residues in the middle domain (Meyer et al. 2003), also resulted in reduced Hsp70 interaction (Figure 2B). Hsc82Δ34 migrated faster than full-length Hsc82, was isolated at wild-type levels and did not show an obvious Hsp70 interaction defect. As expected it failed to interact with Sti1 due to deletion of sequences responsible for interaction with the tetratricopeptide repeat (TPR) domain of cochaperones such as Sti1 (Scheufler et al. 2000; Lee et al. 2012).
Hsc82-S481Y, T521I and A583T showed wild-type interactions with Hsp70 and Sti1 (Figure 2C). These mutated residues cluster to a region near the junction of the middle and carboxyl-terminal domain (Karagoz and Rudiger 2015; Verba et al. 2016; Schopf et al. 2017). Hsc82-S25P, K102E and Q380K also showed wild-type interactions with Hsp70 and Sti1. These mutations alter residues located in the vicinity of ATP-binding pocket (Figure 2D).
Another well-established step in the Hsp90 pathway is nucleotide-induced dimerization of the amino-termini of Hsp90, resulting in the closed conformation (Ali et al. 2006; Hessling et al. 2009; Zierer et al. 2016). Sba1 binds the amino terminus, whereas Cpr6, which contains a TPR domain, binds the carboxyl-terminus. The presence of AMP-PNP promotes formation of the closed conformation, resulting in the interaction of Sba1 and Cpr6 with WT His-Hsc82. As we reported previously, the S481Y, T521I and A583T alterations disrupt interaction with Sba1 and Cpr6 in the presence of AMP-PNP (Johnson et al. 2007), suggesting those mutants have reduced ability to adopt the closed conformation (Figure 2E). However, S25P, K102E and Q380K maintained the ability to interact with Sba1 and Crp6.
Impact of HSC82 mutation on client activity
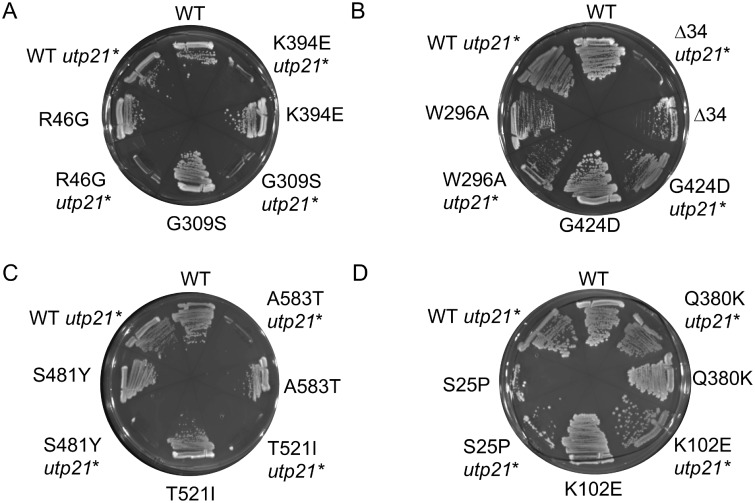
The pulldown assays indicated that the suite of Hsp90 mutants we analyzed had variable levels of interaction with Hsp70 or other cochaperones. We next determined whether the mutations also have differing effects on client activity by examining three diverse clients. Two are endogenous essential proteins: Utp21, which is involved in ribosome biogenesis, and the DNA helicase Ssl2. We also included heterologously expressed v-src kinase, a well-studied Hsp90 client (Nathan and Lindquist 1995; Meyer et al. 2003). Utp21 is an essential component of a large complex required for biogenesis of the small ribosomal subunit (Bernstein et al. 2004). Utp21 interacts with Hsp90 and mutation or inhibition of Hsp90 destabilizes the protein (Tenge et al. 2014). The utp21-S602F mutation causes a mild growth defect in otherwise wild-type cells. However, combination of the utp21-S602F allele with certain mutant forms of Hsc82, including G309S and T521I, resulted in lethality, indicating that reduced Hsc82 function further decreased the essential functions of Utp21 (Tenge et al. 2014). We compared the ability of Hsc82 mutants to support viability in either an hsc82 hsp82 strain or an hsc82 hsp82 utp21-S602F strain (marked by an asterisk *). Each strain initially contained a plasmid expressing WT HSP82 along with the indicated form of HSC82. Growth in the presence of 5-FOA counter-selects for the plasmid expressing WT HSP82, allowing analysis of the indicated mutation. As shown in Figure 3A, cells expressing WT Hsc82 were able to grow in either strain background. However, cells expressing R46G, G309S or K394E were inviable when combined with utp21-S602F. S481Y, T521I and A583T were also inviable (Figure 3C). In contrast, other than Hsc82Δ34, the remaining mutations were viable, (Figure 3, B and D). To further analyze how hsc82 mutations affect Utp21 function, the growth of each in the presence of WT UTP21 or utp21-S602F was compared. In the presence of WT HSC82, utp21-S602F results in a very slight growth defect at 37°C (Supplementary Figures S2 and S3A). For each of these hsc82 mutations, there was little or no additional growth defect in the presence of utp21-S602F, indicating that they do not significantly reduce the function of utp21-S602F.
Figure 3.
Effect of combination of hsc82 mutation and the utp21-S602F mutation on growth. Plasmids expressing WT or mutant HSC82 were transformed into strains JJ816 (hsc82hsp82/HSP82) and JJ712 (hsc82 hsp82 utp21-S602F/HSP82). Transformants were assayed for growth after 2-3 d at 30°C in the presence of 5-FOA, which counter-selects for the plasmid expressing WT HSP82.
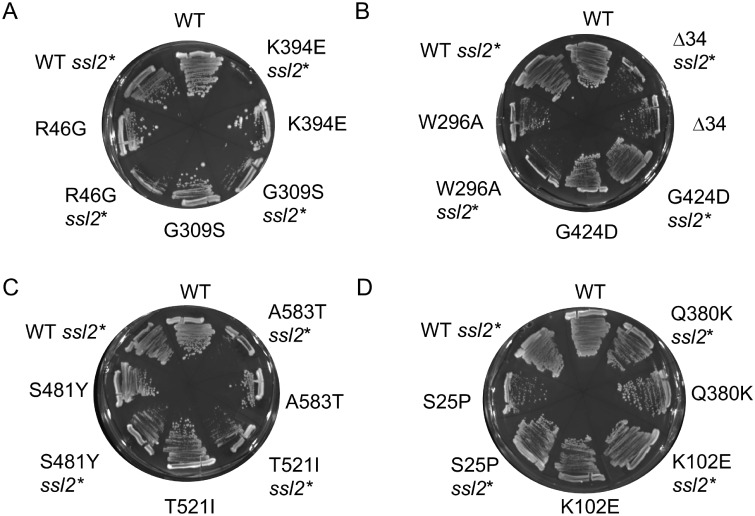
Ssl2 is an essential DNA helicase that functions in transcription and DNA repair (Lee et al. 2000). The ssl2-I690N mutation results in a mild growth defects in otherwise wild-type cells, but enhanced growth defects when combined with the hsc82-G309S mutation (Flom et al. 2005). A comparison of the growth defects of hsc82hsp82ssl2-I690N, hsc82hsp82 utp21-S602F and hsc82hsp82 cells expressing WT HSC82 is shown in Supplementary Figure S3A. The growth of each hsc82 mutant was assessed in a strain expressing the ssl2-I690N allele using an assay similar to that shown in Figure 3. The most prominent effects were observed in cells expressing R46G, G309S and K394E. Each of these mutant alleles showed a strong growth defect when combined with ssl2-I690N (marked by an asterisk *) (Figure 4A). Growth of cells expressing S481Y, T521I and A583T was moderately affected (Figure 4C). The remaining mutants were unaffected or only mildly affected (Figure 4, B and D). The growth of each mutant in the presence of WT SSL2 or ssl2-I690N was compared (Supplementary Figure S3B). R46G, G309S and K394E showed a strong growth defect at 30°C when combined with ssl2-I690N, whereas S481Y, T521I, and A583T showed a moderate growth defect at 30°C. In contrast, cells expressing the remainder of the mutants showed little enhancement of growth defects when combined with the ssl2 mutation.
Figure 4.
Effect of combination of hsc82 mutation and the ssl2-I690N mutation on growth. Plasmids expressing WT or mutant HSC82 were transformed into strains JJ816 (hsc82hsp82/HSP82) and JJ1044 (hsc82 hsp82 ssl2/ssl2-I690N HSP82). Transformants were assayed for growth after 3-4 d in at 30°C the presence of 5-FOA, which counter-selects for the plasmid expressing WT HSP82.
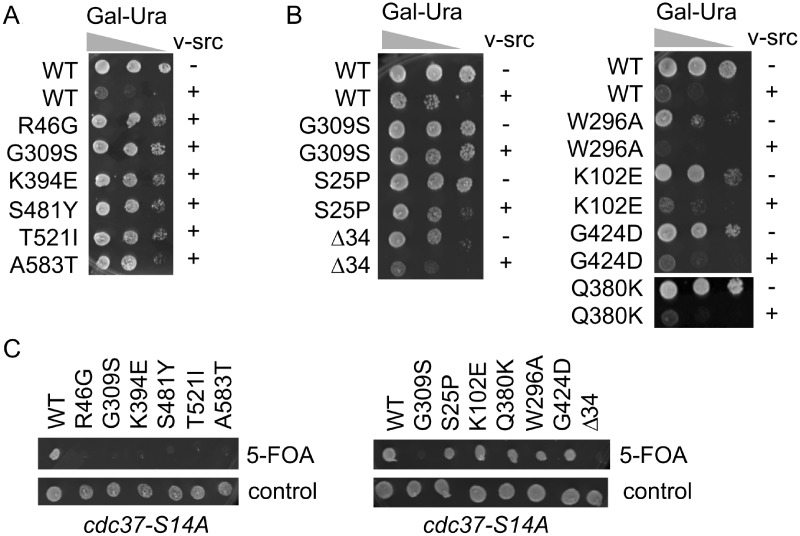
Although not native to yeast, v-src kinase is one of the most widely studied Hsp90 clients (Boczek et al. 2015; Luo et al. 2017). Overexpression of v-src kinase is toxic to yeast due to abnormal phosphotyrosine activity. Mutation of Hsp90 or cochaperones results in reduced v-src activity and toxicity (Nathan and Lindquist 1995; Dey et al. 1996). Prior reports showed that Hsp82-G313S, S485Y, T525I, and A587T, which contain substitutions in homologous residues of the other isoform of yeast Hsp90, resulted in reduced v-src activity (Supplementary Figure S1 and Nathan and Lindquist 1995; Meyer et al. 2003; Hawle et al. 2006). As shown in Figure 5A, induction of GAL-v-src production via growth on galactose-based media resulted in reduced growth of cells expressing WT Hsc82 compared to cells expressing an empty vector. However, the growth of cells expressing R46G, G309S, K394E, S481Y, T521I, or A583T was not affected, indicating reduced v-src activity. To assess v-src activity in the remainder of the mutants, we compared growth of cells expressing either empty vector or GAL-v-src (Figure 5B). The growth of cells expressing S25P was slightly inhibited. In contrast, growth of cells expressing the remainder of the mutants was sharply reduced in the presence of v-src, indicating that these mutants do not significantly affect function.
Figure 5.
Effect of HSC82 mutation of v-src kinase function. A and B. Growth of cells expressing WT or mutant HSC82 in the presence of GAL-v-src (+) empty vector (−). Cells were serially diluted 10-fold and grown in the presence of galactose for 3 days. C. Plasmids expressing WT or mutant HSC82 were transformed into strain JJ114 (hsc82 hsp82 cdc37-S14A/YEp24-HSP82). Cells were grown overnight, spotted onto cells containing 5-FOA or nonselective media, and grown for 2–3 d at 30°C.
As an independent test to determine whether some mutants have minimal effect on activity of protein kinases, hsc82 mutations were tested for synthetic lethality with a mutant allele of CDC37, cdc37-S14A. The cochaperone Cdc37 directly interacts with both Hsp90 and kinases, and previous work showed global destabilization of the yeast kinome in cells expressing cdc37-S14A (Mandal et al. 2007). Consistent with the results in Figure 5A, R46G, G309S, K394E, S481Y, T521I, and A583T were inviable when combined with the cdc37-S14A mutation (Figure 5C). In contrast, K102E, Q380K, W296A, and G424D were viable when combined with cdc37-S14A, providing additional evidence that these mutations do not dramatically alter activity of protein kinases. S25P appears to have intermediate effects on v-src activity, since it showed mild growth reduction upon v-src expression but was viable when combined with the cdc37-S14A mutation. Conversely, Δ34 showed growth reduction upon v-src expression but was inviable when combined with the cdc37-S14A mutation.
Protein expression profiling of Hsc82 mutants
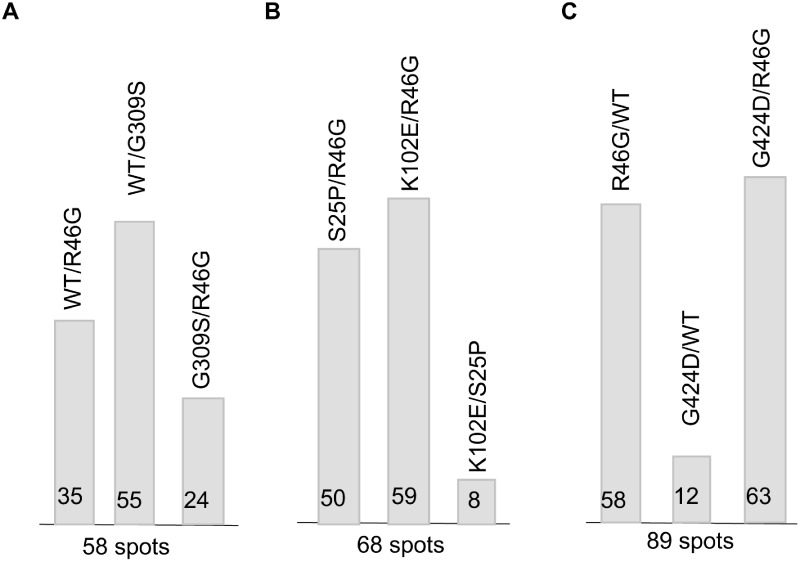
Hsp90 is known to directly or indirectly impact the function of 10-15% of all proteins in yeast and humans (Millson et al. 2005; Zhao et al. 2005; McClellan et al. 2007; Echeverria et al. 2011; Franzosa et al. 2011; Gopinath et al. 2014). Reduced Hsp90 activity results in destabilization and degradation of client proteins. However, altered activity of Hsp90-dependent transcription factors and protein kinases also results in many indirect effects due to alteration of mRNA levels. To provide more evidence that Hsc82 mutants with shared interaction defects have in vivo effects that vary from mutants with other interaction defects, we predicted that a proteomic approach might capture changes in protein levels due to both direct and indirect effects of Hsc82 mutation. We therefore employed two-dimensional fluorescence difference gel electrophoresis (2D-DIGE) to provide a protein profile of cellular changes for mutants relative to wild type cells or other mutants. Protein extracts were made from cells expressing different versions of Hsc82, then independently labeled with the dyes Cy2, Cy3, or Cy5. The samples were then mixed and resolved on the same 2-D gels to identify proteins that showed at least a 1.5-fold difference in steady-state expression levels. As an initial test, we compared the overall number of identified spots that differed in three different sets (Supplementary Table S1 and Figure S4). A comparison of cells expressing WT Hsc82, R46G or G309S, identified 58 spots with different expression (Figure 6A). The most variation was observed between WT and G309S alleles (55/58 spots). However, a comparison of R46G and G309S showed that only 23/58 spots differed. This supports our hypothesis that cells expressing R46G or G309S are more similar to each other than to cells expressing WT Hsc82. In an independent experiment, 59/68 spots differed between K102E and R46G (Figure 6B). However, only 8/68 spots varied 1.5-fold between S25P and K102E, providing additional corroborating evidence that these mutations result in similar in vivo effects. Finally, we compared cells expressing WT, R46G and G424D (Figure 6C). In this set, 58/89 spots varied between cells expressing WT Hsc82 or R46G, and only 12 spots varied between WT and G424D. The limited number of differences between cells expressing G424D and WT, combined with the large number of differences between G424D and R46G, further confirms that the G424D mutation has limited effects that differ from those of R46G.
Figure 6.
Summary of the 2D-DIGE protein expression profiling. Three independent 2D-DIGE comparisons were performed. The ratio change of the protein differential expression was obtained from in-gel DeCyder software analysis. The number of characterized protein spots that varied by at least 1.5-fold from one sample to another in each experiment is shown at the bottom of the graph. The number of spots that varied in each pair-wise comparison is shown in graphical form. A comparison of cells expressing WT Hsc82, R46G or G309S; B. A comparison of cells expressing S25P, R46G, or K102E; C. A comparison of cells expressing WT Hsc82, R46G, or G424D.
Identification of proteins that vary in abundance in an Hsc82-mutant dependent manner
A benefit of the 2D-DIGE approach is that it allows for identification of cellular proteins or processes that may be selectively affected by Hsp90 mutation. The 2D-DIGE experiments described above were conducted only once, and thus the results are viewed as preliminary findings. To help validate this approach and strengthen conclusions from the limited proteomic analysis, we first determined whether proteins identified using this method had previously been identified in other studies that examined Hsc82 and Hsp82 interactions. A limited number of spots were excised from gels, subjected to MALDI-TOF/TOF analysis and identification using the Mascot search engine and the SwissProt database, with a total of 62 different proteins identified. The complete list of identified proteins for all investigated mutants are shown in Supplementary Table S2, with proteins identified in more than one dataset shaded in gray. The mRNA or protein levels of half (31/62) of the different proteins that were identified by 2D-DIGE were previously shown to be affected by Hsp90 mutation (Echeverria et al. 2011; Flom et al. 2012; Gopinath et al. 2014) (Supplementary Table S3). Additional proteins were previously identified in genome-wide screens for Hsp90 interactors (McClellan et al. 2007; Franzosa et al. 2011; Girstmair et al. 2019). Overall, 51/62 hits from the 2D-DIGE experiment were previously shown to interact with Hsc82 or Hsp82, indicating that this approach is both robust and complementary to other successful techniques in elucidating Hsp90 partner or client proteins.
Hsc82 mutants display differences in levels of proteins regulated by the stress response
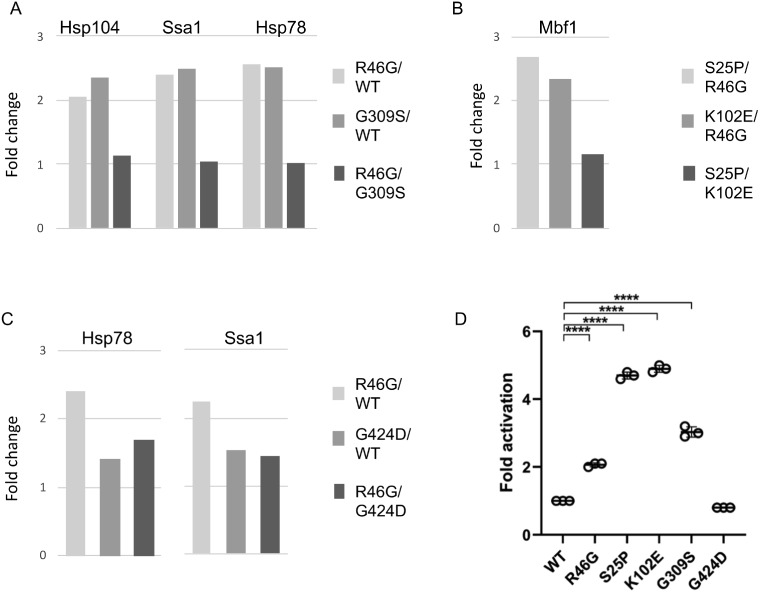
To determine the cellular processes that are affected differently by Hsc82 mutation, we used gene ontology (GO) mapping (Supplementary Table S4). Cellular processes that were most highly enriched in the subset of affected proteins were carbohydrate metabolism, response to oxidative, osmotic or heat stress, protein folding and targeting and cytosolic translation. To expand upon the 2D-DIGE studies, we focused on those proteins known to be regulated by the stress response. Mutations in the Hsp90 pathway have previously been shown to increase the basal expression of genes controlled by the heat shock transcription factor Hsf1, which can be specifically interrogated using a reporter construct bearing the Hsf1 binding sequence known as a heat shock element (HSE) (Duina et al. 1998; Liu et al. 1999). Four proteins that showed differing levels in our studies were Hsp104, Ssa1, Hsp78, and Mbf1, all of which are known targets of Hsf1 (Solis et al. 2016). Figure 7 summarizes the changes in expression levels of these proteins observed using 2D-DIGE. The Hsf1-regulated proteins were elevated in cells expressing S25P or K102E relative to R46G (Figure 7B) and elevated in cells expressing R46G or G309S relative to WT cells (Figure 7, A and C). As an independent test, we examined whether the Hsc82 mutants in this study differed in the ability to activate an HSE-lacZ reporter construct. As shown in Figure 7D, yeast expressing S25P or K102E showed a fourfold activation of the HSE-lacZ reporter construct, and R46G and G309S resulted in a twofold to threefold activation. In contrast, the G424D allele lacked detectable effects on activity of HSF1-targeted genes. These results are entirely consistent with the 2D-DIGE outcomes and verify that the observed proteomic changes reflect at least in part transcriptional potency of a known Hsp90-dependent transcription factor.
Figure 7.
Hsc82 mutation results in altered Hsf1 activity. Changes in levels of proteins known to be regulated by Hsf1 were detected in each of the three experiments as shown in Supplementary Table S1 and (A–C). (D) Cells expressing the plasmid pSSA3-LacZ (HSE-lacZ) were grown to mid-log phase at 30°C in selective media. β-galactosidase activity was measured using luminescence detection. P = 0.05*, 0.005**, 0.0005***.
The aggregate effects of the Hsc82 mutants on client activity are summarized in Table 1. The R46G, G309S and K394E mutations resulted in similar loss of Hsp70 interaction and reduced Utp21, Ssl2 and v-src activity. The S481Y, T521I and A583T mutations resulted in defects in Sba1 and Cpr6 interaction and decreased activity of all three clients tested, although the effect on Ssl2 was only moderate. The remaining mutants had selective effects. The truncation mutant Δ34 disrupted Utp21 and v-src activity but had little effect on Ssl2 function. The S25P mutation altered v-src activity but had little effect on Utp21 or Ssl2. In addition, although only some mutants were tested, S25P and K102E, showed the most pronounced effects on Hsf1 activity despite having little or no effect on Utp21, Ssl2 or v-src function.
Table 1.
Summary of Hsc82 mutant defects
| Mutation |
Domain
Altered |
Interaction defect | Utp21 | Ssl2 | Src | Hsf1 |
|---|---|---|---|---|---|---|
| R46G | N-term | Reduced Hsp70 | ↓↓ | ↓↓ | ↓↓ | ↑ |
| G309S | Middle | Reduced Hsp70 | ↓↓ | ↓↓ | ↓↓ | ↑ |
| K394E | Middle | Reduced Hsp70 | ↓↓ | ↓↓ | ↓↓ | nd |
| W296A | Middle | Reduced Hsp70, Sti1 | – | – | – | nd |
| G424D | Middle | Reduced Hsp70, Sti1 | – | – | – | – |
| Δ34 | C-term | Reduced Sti1 | ↓↓ | – | ↓ | nd |
| S481Y | Middle | Reduced Sba1, Cpr6 | ↓↓ | ↓ | ↓↓ | nd |
| T521I | C-term | Reduced Sba1, Cpr6 | ↓↓ | ↓ | ↓↓ | nd |
| A583T | C-term | Reduced Sba1, Cpr6 | ↓↓ | ↓ | ↓↓ | nd |
| S25P | N-term | None observed | – | – | ↓ | ↑↑ |
| K102E | N-term | None observed | – | – | – | ↑↑ |
| Q380K | Middle | None observed | – | – | – | nd |
nd, not determined
Discussion
Hsp90 is required for the function of hundreds of cellular proteins, including multiple oncogenic proteins in mammalian cells. Compounds that bind the ATP-binding pocket and inhibit function of Hsp90 result in reduced client activity and levels. However, the use of these compounds is limited due to unfavorable side effects (Neckers and Workman 2012; Garg et al. 2016), and alternative strategies for Hsp90 inhibition are needed. Cochaperones are known to play an important role in client maturation, (Li et al. 2012; Prodromou 2012; Schopf et al. 2017) and blocking Hsp90 interaction with cochaperones has been proposed as an alternative. However, a compound that blocks Hsp90 interaction with the kinase-specific cochaperone Cdc37 did not result in reduced client folding (Smith et al. 2015). A compound that inhibits Hsp90 interaction with the Aha1 cochaperone has been identified but the cellular effects are unknown (Stiegler et al. 2017).
Our approach was to determine whether a panel of yeast Hsp90 mutations affect the same, or different clients, and whether it was possible to isolate groups of Hsp90 mutants with shared in vivo effects. One set of mutants that caused broad inhibition of Hsp90 functions disrupts a very early step in the pathway (Figure 8). Evidence indicates that Hsp90 clients such as the glucocorticoid receptor first bind Hsp70 and are then transferred to Hsp90 (Alvira et al. 2014; Kirschke et al. 2014; Lorenz et al. 2014). Three mutations, R46G, G309S and K394E, similarly affect this early step in the pathway. The G309S and K394E alterations were previously shown to alter a contact site between Hsp70 and Hsp90 (Kravats et al. 2018; Genest et al. 2019). Hsp82-G313S is one of the classic mutants characterized by the Lindquist lab, and Hsc82-G309S and Hsp82-G313S are substitutions in homologous residues in the two yeast Hsp90 isoforms. The two mutants result in similar temperature-sensitive growth defects and defects in v-src activity. Both mutations also disrupt Hsp90-Hsp70 interaction, suggesting they will similarly affect other functions (Nathan and Lindquist 1995; Kravats et al. 2018). Recent structural data that captures the loading complex of Hsp70, Hsp90, Hop/Sti1 and the glucocorticoid receptor indicates that Hsc82-R46 is part of a second contact site between Hsp70 and Hsp90 (Wang et al. 2020). Our data indicates that disruption of either contact site results in similar defects in vivo.
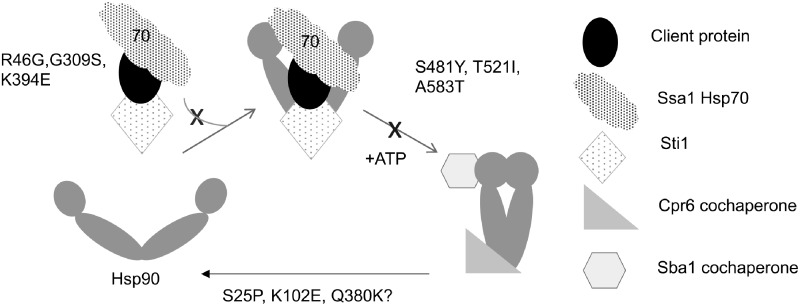
Figure 8.
Model of the effect of Hsc82 mutation on the Hsp90 folding pathway. In an accepted model of the Hsp90 folding pathway, Hsp70 and Sti1 target a client to the open conformation of Hsp90. The R46G, G309S, and K394E mutations appear to disrupt this step. Nucleotide induces formation of the closed conformation that bind Sba1 and Cpr6. S481Y, T521I, and A583T appear to disrupt this step. The proximity of S25P, K102E, and Q380K to bound nucleotide suggests they alter steps involved in regulation of nucleotide hydrolysis and/or release.
The slow, rate-determining step in the Hsp90 conformational cycle is nucleotide-induced dimerization of the amino-termini of Hsp90. This results in the closed dimer conformation that binds Sba1 and Cpr6 (Ali et al. 2006; Johnson et al. 2007; Hessling et al. 2009; Zierer et al. 2016). The S481Y, T521I, and A583T mutants disrupt this step the pathway, resulting in reduced Cpr6 and Sba1 interaction. Another mutant that resulted in strong defects is Hsc82Δ34. This mutant lacks the established binding site for TPR-containing cochaperones such as Cpr6 and Sti1, and we showed that Hsc82Δ34 disrupts interaction with Sti1. Our result that mutations that disrupt progression of established Hsp90 complexes containing Sti1, Cpr6 and/or Sba1 disrupt activity of a wide range of clients supports prior evidence that that these cochaperones regulate critical steps in client folding (Prodromou et al. 1999; Schopf et al. 2017), and is consistent with studies from the Buchner lab showed that deletion of STI1, CPR6 or SBA1 in otherwise wild-type cells results in reduced activity of a wide range of client proteins (Sahasrabudhe et al. 2017). Two additional reports examined the role of Sti1 in regulating the Hsp90 chaperone cycle, with one providing intriguing results that loss of STI1 improves protein folding in vivo (Reidy et al. 2018; Bhattacharya et al. 2020). Some of the mutants described herein that disrupt Sti1 interaction may be useful tools to better understand how cooperation between Hsp90 and Sti1 mediate protein folding. However, the Hsc82Δ34 mutant likely has pleiotropic effects on Hsp90 function due to altered interaction with additional cochaperones. Deletion of only the last four amino acids of yeast Hsp90, EEVD, was enough to disrupt stable Hsp90-Sti1 interaction but did not result in a growth defect. A prior study showed that additional sequences upstream of the EEVD also contribute to Sti1 interaction (Scheufler et al. 2000; Lee et al. 2012). The truncation site of Hsc82Δ34 is very similar to the natural carboxyl-terminus of E. coli HtpG (Supplementary Figure S1) and thus is a deletion of the entire TPR-binding domain. Our result that truncation of the TPR domain disrupts function is similar to the result that deletion of most of the TPR domain of Hsp90 alpha caused severe functional defects in mice, and that a similar truncation of human Hsp90 alpha resulted in growth defects when expressed in yeast (Grad et al. 2010).
We also identified additional mutants that have more selective effects. The W296A and G424D mutations appear to alter both Hsp70 and Sti1 interaction but did not significantly alter activity of the clients we tested. Additional experiments with purified proteins are needed to clarify the effects of these mutants since they alter residues outside known sites of contact with Hsp70 or Sti1. We previously showed that W296A results in altered transcription of genes regulated by the cAMP pathway (Flom et al. 2012). Mutations in Hsp82-W300, which alters the homologous residue, have been shown to disrupt conformation changes that coordinate domain-domain communication and information about client interaction (Rutz et al. 2018). The G424 mutation mutates a residue in a predicted hinge region, and thus may also result in altered conformation or disrupt allosteric communication (Blacklock and Verkhivker 2014). The more selective effects of these mutants suggest that either the mutations do not fully disrupt these functions or that Hsp90 conformation plays a role in client specificity (Lorenz et al. 2014; Karagoz and Rudiger 2015; Prince et al. 2015; Daturpalli et al. 2017; Mader et al. 2020). Hsp82 mutants that result in lethality alter the timing of conformational changes, including extended dwell times in the closed state (Zierer et al. 2016), and evidence suggests that the altered timing of conformational changes influences client interaction and activity (Biebl et al. 2020).
The other mutants that had selective effects altered residues near the ATP-binding site (S25P), in a lid that closes over bound nucleotide (K102E), or in a flexible loop that regulates ATP hydrolysis (Q380K) (Meyer et al. 2003; Ali et al. 2006). The biochemical effects of the K102E and the Q380K mutation have not been analyzed, but a recent study examined Hsp82-S25P (Mercier et al. 2019). Hsp82-S25P showed near wild-type ATPase activity, but the ATPase could not be stimulated by the cochaperone Aha1, indicating altered regulation of ATPase activity. Follow-up studies are required to determine how nucleotide hydrolysis, release and associated conformational changes, including time in the closed conformation, affect client specificity. A prior study that systematically mutated amino acid residues of Hsp82 identified additional mutations in the amino-terminus that had selective effects on client activity, but found that there was no correlation between client specificity and ATPase activity (Mishra et al. 2016). A more recent comprehensive analysis of Hsp82 mutations by the same group found that the mutations that were least-tolerated under a range of environmental conditions were those that affect nucleotide interaction, mediate ATP-dependent conformational changes in the N-domain of Hsp90, or those that transmit information about client interaction, such alteration of Hsp82-W300/Hsc82-W296. Additional studies are needed to determine whether the mutations we predict affect those functions also result in altered sensitivity to conditions they used, such as high salt, nitrogen depletion, or the presence of ethanol or other stressors (Flynn et al. 2020).
The 2D-DIGE studies identified proteins affected differently by Hsc82 mutation. Most of the proteins with detected changes in abundance in this study (51/62) had previously been shown to be impacted by Hsp90 alteration and/or identified as potential clients (Zhao et al. 2005; McClellan et al. 2007; Echeverria et al. 2011; Franzosa et al. 2011; Gopinath et al. 2014; Girstmair et al. 2019). The biggest benefit of this approach is that is allows a relatively easy comparison of two different mutants to determine if they have similar or different in vivo effects. This was most evident in the 2D-DIGE set that compared R46, S25P and K102E. The expression of only 8/68 spots varied 1.5-fold or more upon comparison of cells expressing S25P and K102E, indicating that they engender very similar protein expression profiles. In that same set, 59/68 spots differed between K102E and R46G. Although this technique has limitations in the number of spots that may be detected in a single experiment and skews toward proteins that are highly expressed, the types of changes detected were consistent with both other studies and our direct assays of client functions. A potential application of this technique could be to compare the in vivo effects of two different Hsp90 inhibitors or the effect of cochaperone deletion. For example, it would be interesting to determine whether an Hsp90 inhibitor that binds the ATP-binding site, and thus also alters a step in early in the folding pathway, has the same in vivo effects as cells expressing Hsc82-G309S.
Previous studies showed that lowering Hsp90 activity altered levels of ∼900 yeast proteins and/or the mRNA encoding those proteins (Zhao et al. 2005; Echeverria et al. 2011; Gopinath et al. 2014). Since it is not practical to test the effect of a large panel of mutants on a large number of individual clients, one goal of this study was to identify endogenous yeast clients that are differently affected by Hsp90 mutation in order to expand mechanistic analysis of Hsp90 function. For example, our analysis showed that the Hsc82 mutants resulted in varied Hsf1 activity. Further analysis of the S25P and K102E mutants may help elucidate the mechanism of Hsp90 regulation of the heat shock response. The 2D-DIGE studies also identified changes in multiple proteins involved in cytosolic translation, including changes in the level of multiple ribosomal proteins. A prior study also identified a connection between Hsp90 and many ribosomal proteins and showed that Hsp90 mutation altered polysome stability (Franzosa et al. 2011). Additional proteins required for regulation of translation, translation initiation and elongation were also affected. Proteins in this category that were identified in multiple datasets include Tif6, a component of a pre-ribosomal particle; the RACK1 ortholog Asc1, which inhibits translation; Sbp1, another protein involved in repressing translation, and Eft1, elongation factor 2 (EF2). A recent study showed that Hsp90 and the cochaperone Cns1 interact with EF2 and are required for EF2 stability (Schopf et al. 2019). Other Hsp90 cochaperones have also been found to interact with the intact ribosome (Tenge et al. 2015). Additional studies are needed to determine whether our panel of Hsc82 mutants have differing effects on translation and/or EF2 stability.
In summary, our results show that Hsc82 mutants that cause similar growth defects do not have the same in vivo effects. Two sets of mutants that disrupt well characterized steps in the Hsp90 pathway likely affect a broad range of functions. However, mutation or residues presumed to be involved in regulation of nucleotide hydrolysis or release resulted in more limited in vivo effects, suggesting that selective inhibition of Hsp90 functions is possible. Our future studies will examine whether alteration of cochaperone levels exacerbates or suppresses the defects of these Hsc82 mutants. None of the mutants we used in these studies show strong dominant negative effects in an hsc82hsp82 strain expressing WT HSP82 (not shown), but future experiments will determine whether alteration of cochaperone levels unmasks dominant effects. Additional analysis of 2D-DIGE hits is also needed to identify authentic endogenous yeast clients that are differentially affected by Hsc82 mutation in order to provide new insights into Hsp90-client interaction and how client folding changes as it progresses through the pathway.
Acknowledgments
We thank Marissa Dean, Braunwyn Haverfield, Tyler Faling, and other prior lab members for assistance with isolation and characterization of these mutants. We also thank Dr. Paul LaPointe (University of Alberta) for helpful comments on the manuscript.
Funding
This research was supported by the National Institute of General Medical Sciences under Award Numbers R01GM127675 (JLJ) and R01GM127287 (KAM). JLJ obtained additional support from the College of Science at the University of Idaho and NIH grants 5P30GM103324 and P20GM104420, which fund the Institute for Modeling Complex Interactions and the Institute for Bioinformatics and Evolutionary Studies at the University of Idaho. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflicts of interest
None declared.
Literature cited
- Ali MM, Roe SM, Vaughan CK, Meyer P, Panaretou B, et al. 2006. Crystal structure of an Hsp90-nucleotide-p23/Sba1 closed chaperone complex. Nature. 440:1013–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvira S, Cuellar J, Rohl A, Yamamoto S, Itoh H, et al. 2014. Structural characterization of the substrate transfer mechanism in Hsp70/Hsp90 folding machinery mediated by Hop. Nat Commun. 5:5484. [DOI] [PubMed] [Google Scholar]
- Bernstein KA, Gallagher JE, Mitchell BM, Granneman S, Baserga SJ.. 2004. The small-subunit processome is a ribosome assembly intermediate. Eukaryot Cell. 3:1619–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya K, Weidenauer L, Luengo TM, Pieters EC, Echeverria PC, et al. 2020. The Hsp70-Hsp90 co-chaperone Hop/Stip1 shifts the proteostatic balance from folding towards degradation. Nat Commun. 11:5975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biebl MM, Riedl M, Buchner J.. 2020. Hsp90 co-chaperones form plastic genetic networks adapted to client maturation. Cell Rep. 32:108063. [DOI] [PubMed] [Google Scholar]
- Blacklock K, Verkhivker GM.. 2014. Computational modeling of allosteric regulation in the hsp90 chaperones: a statistical ensemble analysis of protein structure networks and allosteric communications. PLoS Comput Biol. 10:e1003679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boczek EE, Reefschlager LG, Dehling M, Struller TJ, Hausler E, et al. 2015. Conformational processing of oncogenic v-Src kinase by the molecular chaperone Hsp90. Proc Natl Acad Sci U S A. 112:E3189–E3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohen SP. 1995. Hsp90 mutants disrupt glucocorticoid receptor ligand binding and destabilize aporeceptor complexes. J Biol Chem. 270:29433–29438. [DOI] [PubMed] [Google Scholar]
- Borkovich KA, Farrelly FW, Finkelstein DB, Taulien J, Lindquist S.. 1989. hsp82 is an essential protein that is required in higher concentrations for growth of cells at higher temperatures. Mol Cell Biol. 9:3919–3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke D, Dawson D, Stearns T.. 2000. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. Woodbury, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Daturpalli S, Knieß RA, Lee C-T, Mayer MP.. 2017. Large rotation of the N-terminal domain of Hsp90 Is important for interaction with some but not all client proteins. J Mol Biol. 429:1406–1423. [DOI] [PubMed] [Google Scholar]
- Dey B, Caplan AJ, Boschelli F.. 1996. The Ydj1 molecular chaperone facilitates formation of active p60v-src in yeast. Mol Biol Cell. 7:91–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duina AA, Kalton HM, Gaber RF.. 1998. Requirement for Hsp90 and a CyP-40-type cyclophilin in negative regulation of the heat shock response. J Biol Chem. 273:18974–18978. [DOI] [PubMed] [Google Scholar]
- Echeverria PC, Forafonov F, Pandey DP, Muhlebach G, Picard D.. 2011. Detection of changes in gene regulatory patterns, elicited by perturbations of the Hsp90 molecular chaperone complex, by visualizing multiple experiments with an animation. BioData Min. 4:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flom G, Weekes J, Johnson JL.. 2005. Novel interaction of the Hsp90 chaperone machine with Ssl2, an essential DNA helicase in Saccharomyces cerevisiae. Curr Genet. 47:368–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flom G, Weekes J, Williams JJ, Johnson JL.. 2006. Effect of mutation of the tetratricopeptide repeat and aspartate-proline 2 domains of Sti1 on Hsp90 signaling and interaction in Saccharomyces cerevisiae. Genetics. 172:41–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flom GA, Langner E, Johnson JL.. 2012. Identification of an Hsp90 mutation that selectively disrupts cAMP/PKA signaling in Saccharomyces cerevisiae. Curr Genet. 58:149–163. [DOI] [PubMed] [Google Scholar]
- Flynn JM, Rossouw A, Cote-Hammarlof P, Fragata I, Mavor D, et al. 2020. Comprehensive fitness maps of Hsp90 show widespread environmental dependence. Elife. 9:e53810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzosa EA, Albanese V, Frydman J, Xia Y, McClellan AJ.. 2011. Heterozygous yeast deletion collection screens reveal essential targets of Hsp90. PLoS One. 6:e28211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia VM, Nillegoda NB, Bukau B, Morano KA.. 2017. Substrate binding by the yeast Hsp110 nucleotide exchange factor and molecular chaperone Sse1 is not obligate for its biological activities. Mol Biol Cell. 28:2066–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg G, Khandelwal A, Blagg BS.. 2016. Anticancer inhibitors of Hsp90 function: beyond the usual suspects. Adv Cancer Res. 129:51–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genest O, Reidy M, Street TO, Hoskins JR, Camberg JL, et al. 2013. Uncovering a region of heat shock protein 90 important for client binding in E. coli and chaperone function in yeast. Mol Cell. 49:464–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genest O, Wickner S, Doyle SM.. 2019. Hsp90 and Hsp70 chaperones: collaborators in protein remodeling. J Biol Chem. 294:2109–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girstmair H, Tippel F, Lopez A, Tych K, Stein F, et al. 2019. The Hsp90 isoforms from S. cerevisiae differ in structure, function and client range. Nat Commun. 10:3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopinath RK, You ST, Chien KY, Swamy KB, Yu JS, et al. 2014. The Hsp90-dependent proteome is conserved and enriched for hub proteins with high levels of protein-protein connectivity. Genome Biol Evol. 6:2851–2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grad I, Cederroth CR, Walicki J, Grey C, Barluenga S, et al. 2010. The molecular chaperone Hsp90alpha is required for meiotic progression of spermatocytes beyond pachytene in the mouse. PLoS One. 5:e15770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawle P, Siepmann M, Harst A, Siderius M, Reusch PH, et al. 2006. The middle domain of Hsp90 acts as a discriminator between different types of client proteins. Mol Cell Biol. 26:8385–8395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hessling M, Richter K, Buchner J.. 2009. Dissection of the ATP-induced conformational cycle of the molecular chaperone Hsp90. Nat Struct Mol Biol. 16:287–293. [DOI] [PubMed] [Google Scholar]
- Johnson JL, Halas A, Flom G.. 2007. Nucleotide-dependent interaction of Saccharomyces cerevisiae Hsp90 with the Cochaperone proteins Sti1, Cpr6, and Sba1. Mol Cell Biol. 27:768–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karagoz GE, Rudiger SG.. 2015. Hsp90 interaction with clients. Trends Biochem Sci. 40:117–125. [DOI] [PubMed] [Google Scholar]
- Kirschke E, Goswami D, Southworth D, Griffin PR, Agard DA.. 2014. Glucocorticoid receptor function regulated by coordinated action of the Hsp90 and Hsp70 chaperone cycles. Cell. 157:1685–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kravats AN, Hoskins JR, Reidy M, Johnson JL, Doyle SM, et al. 2018. Functional and physical interaction between yeast Hsp90 and Hsp70. Proc Natl Acad Sci U S A. 115:E2210–E2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BS, Bi L, Garfinkel DJ, Bailis AM.. 2000. Nucleotide excision repair/TFIIH helicases RAD3 and SSL2 inhibit short-sequence recombination and Ty1 retrotransposition by similar mechanisms. Mol Cell Biol. 20:2436–2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C-T, Graf C, Mayer F J, Richter S M, Mayer M P. 2012. Dynamics of the regulation of Hsp90 by the co-chaperone Sti1. EMBO J. 31:1518–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Soroka J, Buchner J.. 2012. The Hsp90 chaperone machinery: Conformational dynamics and regulation by co-chaperones. Biochim Biophys Acta. 1823:624–635. [DOI] [PubMed] [Google Scholar]
- Liu XD, Morano KA, Thiele DJ.. 1999. The yeast Hsp110 family member, Sse1, is an Hsp90 cochaperone. J Biol Chem. 274:26654–26660. [DOI] [PubMed] [Google Scholar]
- Lorenz OR, Freiburger L, Rutz DA, Krause M, Zierer BK, et al. 2014. Modulation of the hsp90 chaperone cycle by a stringent client protein. Mol Cell. 53:941–953. [DOI] [PubMed] [Google Scholar]
- Louvion JF, Warth R, Picard D.. 1996. Two eukaryote-specific regions of Hsp82 are dispensable for its viability and signal transduction functions in yeast. Proc Natl Acad Sci U S A. 93:13937–13942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Q, Boczek EE, Wang Q, Buchner J, Kaila VR.. 2017. Hsp90 dependence of a kinase is determined by its conformational landscape. Sci Rep. 7:43996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mader SL, Lopez A, Lawatscheck J, Luo Q, Rutz DA, et al. 2020. Conformational dynamics modulate the catalytic activity of the molecular chaperone Hsp90. Nat Commun. 11:1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal AK, Lee P, Chen JA, Nillegoda N, Heller A, et al. 2007. Cdc37 has distinct roles in protein kinase quality control that protect nascent chains from degradation and promote posttranslational maturation. J Cell Biol. 176:319–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClellan AJ, Xia Y, Deutschbauer AM, Davis RW, Gerstein M, et al. 2007. Diverse cellular functions of the hsp90 molecular chaperone uncovered using systems approaches. Cell. 131:121–135. [DOI] [PubMed] [Google Scholar]
- Mercier R, Wolmarans A, Schubert J, Neuweiler H, Johnson JL, et al. 2019. The conserved NxNNWHW motif in Aha-type co-chaperones modulates the kinetics of Hsp90 ATPase stimulation. Nat Commun. 10:1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer P, Prodromou C, Hu B, Vaughan C, Roe SM, et al. 2003. Structural and functional analysis of the middle segment of hsp90. Implications for ATP hydrolysis and client protein and cochaperone interactions. Mol Cell. 11:647–658. [DOI] [PubMed] [Google Scholar]
- Millson SH, Truman AW, King V, Prodromou C, Pearl LH, et al. 2005. A two-hybrid screen of the yeast proteome for Hsp90 interactors uncovers a novel Hsp90 chaperone requirement in the activity of a stress-activated mitogen-activated protein kinase, Slt2p (Mpk1p). Eukaryot Cell. 4:849–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra P, Flynn JM, Starr TN, Bolon DN.. 2016. Systematic mutant analyses elucidate general and client-specific aspects of Hsp90 function. Cell Rep. 15:588–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan DF, Lindquist S.. 1995. Mutational analysis of Hsp90 function: interactions with a steroid receptor and a protein kinase. Mol Cell Biol. 15:3917–3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neckers L, Workman P.. 2012. Hsp90 molecular chaperone inhibitors: are we there yet? Clin Cancer Res. 18:64–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obermann WM, Sondermann H, Russo AA, Pavletich NP, Hartl FU.. 1998. In vivo function of Hsp90 is dependent on ATP binding and ATP hydrolysis. J Cell Biol. 143:901–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince TL, Kijima T, Tatokoro M, Lee S, Tsutsumi S, et al. 2015. Client proteins and small molecule inhibitors display distinct binding preferences for constitutive and stress-induced HSP90 isoforms and their conformationally restricted mutants. PLoS One. 10:e0141786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prodromou C. 2012. The ‘active life’ of Hsp90 complexes. Biochim Biophys Acta. 1823:614–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prodromou C, Siligardi G, O'Brien R, Woolfson DN, Regan L, et al. 1999. Regulation of Hsp90 ATPase activity by tetratricopeptide repeat (TPR)-domain co-chaperones. EMBO J. 18:754–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reidy M, Kumar S, Anderson DE, Masison DC.. 2018. Dual roles for yeast Sti1/Hop in regulating the Hsp90 chaperone cycle. Genetics. 209:1139–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutz DA, Luo Q, Freiburger L, Madl T, Kaila VRI, et al. 2018. A switch point in the molecular chaperone Hsp90 responding to client interaction. Nat Commun. 9:1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahasrabudhe P, Rohrberg J, Biebl MM, Rutz DA, Buchner J.. 2017. The plasticity of the Hsp90 co-chaperone system. Mol Cell. 67:947–961. e5. [DOI] [PubMed] [Google Scholar]
- Scheufler C, Brinker A, Bourenkov G, Pegoraro S, Moroder L, et al. 2000. Structure of TPR domain-peptide complexes: critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell. 101:199–210. [DOI] [PubMed] [Google Scholar]
- Schopf FH, Biebl MM, Buchner J.. 2017. The HSP90 chaperone machinery. Nat Rev Mol Cell Biol. 18:345–360. [DOI] [PubMed] [Google Scholar]
- Schopf FH, Huber EM, Dodt C, Lopez A, Biebl MM, et al. 2019. The co-chaperone Cns1 and the recruiter protein Hgh1 link Hsp90 to translation elongation via chaperoning elongation factor 2. Mol Cell. 74:73–87. e8. [DOI] [PubMed] [Google Scholar]
- Smith JR, de Billy E, Hobbs S, Powers M, Prodromou C, et al. 2015. Restricting direct interaction of CDC37 with HSP90 does not compromise chaperoning of client proteins. Oncogene. 34:15–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solis EJ, Pandey JP, Zheng X, Jin DX, Gupta PB, et al. 2016. Defining the essential function of yeast Hsf1 reveals a compact transcriptional program for maintaining eukaryotic proteostasis. Mol Cell. 63:60–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiegler SC, Rubbelke M, Korotkov VS, Weiwad M, John C, et al. 2017. A chemical compound inhibiting the Aha1-Hsp90 chaperone complex. J Biol Chem. 292:17073–17083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taipale M, Jarosz DF, Lindquist S.. 2010. HSP90 at the hub of protein homeostasis: emerging mechanistic insights. Nat Rev Mol Cell Biol. 11:515–528. [DOI] [PubMed] [Google Scholar]
- Taipale M, Tucker G, Peng J, Krykbaeva I, Lin ZY, et al. 2014. A quantitative chaperone interaction network reveals the architecture of cellular protein homeostasis pathways. Cell. 158:434–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenge VR, Knowles J, Johnson JL.. 2014. The ribosomal biogenesis protein Utp21 interacts with Hsp90 and has differing requirements for Hsp90-associated proteins. PLoS One. 9:e92569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenge VR, Zuehlke AD, Shrestha N, Johnson JL.. 2015. The Hsp90 cochaperones Cpr6, Cpr7, and Cns1 interact with the intact ribosome. Eukaryot Cell. 14:55–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verba KA, Wang RY, Arakawa A, Liu Y, Shirouzu M, et al. 2016. Atomic structure of Hsp90-Cdc37-Cdk4 reveals that Hsp90 traps and stabilizes an unfolded kinase. Science. 352:1542–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RY-R, Noddings CM, Kirschke E, Myasnikov AG, Johnson JL, et al. 2020. GR chaperone cycle mechanism revealed by cryo-EM: inactivation of GR by GR:Hsp90:Hsp70:Hop client-loading complex. bioRxiv 11.05.370247.
- Wu Z, Moghaddas Gholami A, Kuster B.. 2012. Systematic identification of the HSP90 candidate regulated proteome. Mol Cell Proteomics. 11:M111 016675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R, Davey M, Hsu YC, Kaplanek P, Tong A, et al. 2005. Navigating the chaperone network: an integrative map of physical and genetic interactions mediated by the hsp90 chaperone. Cell. 120:715–727. [DOI] [PubMed] [Google Scholar]
- Zierer BK, Rubbelke M, Tippel F, Madl T, Schopf FH, et al. 2016. Importance of cycle timing for the function of the molecular chaperone Hsp90. Nat Struct Mol Biol. 23:1020–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuehlke AD, Johnson JL.. 2012. Chaperoning the chaperone: a role for the co-chaperone Cpr7 in modulating Hsp90 Function in Saccharomyces cerevisiae. Genetics. 191:805–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains and plasmids are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables. Supplementary material is available at figshare: https://doi.org/10.25386/genetics.13547852.