Abstract
Inflammation is an essential component of several respiratory diseases, such as chronic obstructive pulmonary disease (COPD), asthma and acute respiratory distress syndrome (ARDS). It is central to lung cancer, the leading cancer in terms of associated mortality that has affected millions of individuals worldwide. Inflammation and pulmonary manifestations are also the major causes of COVID-19 related deaths. Acute hyperinflammation plays an important role in the COVID-19 disease progression and severity, and development of protective immunity against the virus is greatly sought. Further, the severity of COVID-19 is greatly enhanced in lung cancer patients, probably due to the genes such as ACE2, TMPRSS2, PAI-1 and furin that are commonly involved in cancer progression as well as SAR-CoV-2 infection. The importance of inflammation in pulmonary manifestations, cancer and COVID-19 calls for a closer look at the underlying processes, particularly the associated increase in IL-6 and other cytokines, the dysregulation of immune cells and the coagulation pathway. Towards this end, several reports have identified epigenetic regulation of inflammation at different levels. Expression of several key inflammation-related cytokines, chemokines and other genes is affected by methylation and acetylation while non-coding RNAs, including microRNAs as well as long non-coding RNAs, also affect the overall inflammatory responses. Select miRNAs can regulate inflammation in COVID-19 infection, lung cancer as well as other inflammatory lung diseases, and can serve as epigenetic links that can be therapeutically targeted. Furthermore, epigenetic changes also mediate the environmental factors-induced inflammation. Therefore, a better understanding of epigenetic regulation of inflammation can potentially help develop novel strategies to prevent, diagnose and treat chronic pulmonary diseases, lung cancer and COVID-19.
Keywords: Inflammation, Epigenetics, COPD, ARDS, Lung cancer, COVID-19
1. Introduction
Humans are exposed to numerous infections, toxins, irritants etc. on a daily basis and inflammation is one of the ways by which a body fights them off through the involvement of immune cells, vascular system as well as other molecular mediators. The process of inflammation essentially comprises eliminating the ‘trigger’, minimizing damage, clearing of damaged cells/tissues and initiation of repair mechanisms [1]. Inflammation can either be rapid and short-lived (acute) or maybe recurring and slow and last for prolonged durations (chronic) [2], and the two subtypes markedly differ in the way different cells are mobilized, with acute inflammation involving granulocytes and the chronic inflammation involving agranulocytes. Acute inflammation can cause lung injury leading to acute respiratory distress syndrome (ARDS). Chronic inflammation can lead to (or persists in) several chronic diseases such as cancer, heart diseases, diabetes, asthma, chronic obstructive pulmonary disease (COPD), arthritis and Alzheimer’s. Thus, inflammation is an important feature that affects the cells and organ systems, and plays a critical role in both acute and chronic disease pathogenesis. Persistence of inflammation can exacerbate secondary insults and also increase susceptibilities to injuries and infections caused by microorganisms (bacteria and viruses like SARS-CoV-2).
Among the many chronic diseases linked to inflammation are several pulmonary diseases and lung cancer. Diseases such as asthma, COPD and pulmonary fibrosis are well known to be the result of persistent inflammatory processes [3]. Likewise, the role of inflammation in cancer is also well established [4,5]. Inflammation promotes tumor progression, and cancer-related inflammation affects several crucial aspects of malignancy ranging from proliferation and survival of tumor cells to angiogenesis, metastasis and response to therapies [6].
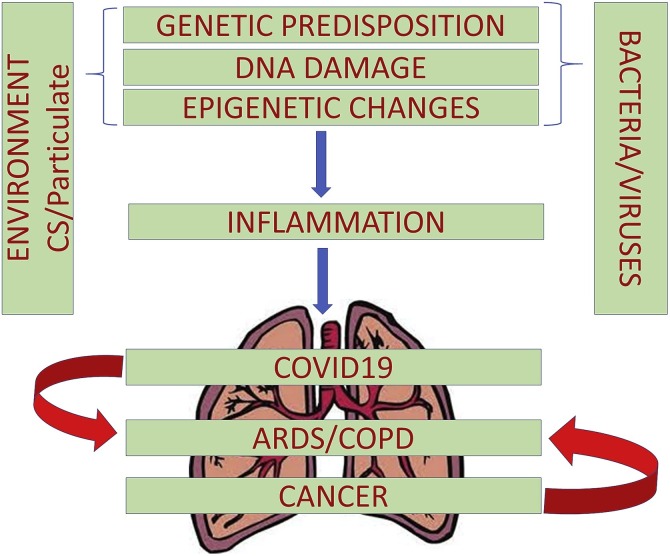
The molecular and genetic markers of inflammation have traditionally been the focus of research [[7], [8], [9]]. Recently, the role of epigenetics as the basis of inflammation is increasingly being recognized [10]. Epigenetics provides the connection between environmental and genetic factors [11]. In the absence of well understood or undruggable molecular targets in several chronic inflammatory diseases, it is possible that targeting of epigenetic changes (the changes in gene expression that are heritable but independent of changes in DNA sequence [12]), can be an effective therapeutic strategy [13]. It is becoming apparent that inflammation connects several pulmonary diseases, cancer and COVID-19 (Fig. 1 ) and there is a growing interest in understanding the epigenetic basis of inflammation. This article aims to provide a comprehensive understanding of the underlying mechanisms.
Fig. 1.
Inflammation connects COVID-19 and cancer with pulmonary diseases. In addition to genetic predisposition, several extrinsic factors such as, bacterial / viral infections and environmental agents can cause inflammation, with an emerging realization for the involvement of epigenetic regulation. Inflammation leads to pulmonary manifestations exemplified by ARDS and COPD, which are observed in COVID-19 as well as lung cancer patients.
2. Inflammation as underlying cause of pulmonary diseases
Uncontrolled inflammation is a major component of many pulmonary diseases, such as COPD, ARDS and idiopathic pulmonary fibrosis (IPF), all of which are linked to high mortality and/or morbidity rates [[14], [15], [16]]. Pulmonary inflammation is a hallmark of many acute and chronic pathophysiological inflammatory processes and there is considerable experimental and clinical evidence that pro-inflammatory cytokines, chemokines as well as adhesion molecules play a major role in its pathogenesis. The inflammatory response is complex and involves a variety of mechanisms to defend against pathogens and abnormal tissue repair [17]. In the lung, inflammation can be caused by pathogens or by exposure to toxins, pollutants, irritants, and allergens [[18], [19], [20]]. Pulmonary insults trigger a complex host response, involving an interplay of pro- and anti-inflammatory factors leading to tissue repair or organ injury and secondary pathophysiological changes [18]. Each insult triggers the release of cytokines and mediators that modify inflammatory responses in a unique way. Activation and recruitment of inflammatory cells, such as macrophages, lymphocytes, neutrophils, and eosinophils, serves as the source of different kinds of inflammatory mediators such as histamine, TNF-α, interleukins (IL-1β, IL-4, IL-5 and IL-6), prostaglandins, leukotrienes, and DAMPs (Damage Associated Molecular Patterns) that are necessary for the onset of pulmonary inflammation. The release of these inflammatory mediators correlates with the characteristics of pulmonary disease such as loss of lung function, airway hyperresponsiveness and obstruction, edema, mucus hypersecretion and lung remodeling [16].
COPD, a chronic inflammatory disease, often manifests as the combination of emphysema and chronic obstructive bronchitis, where emphysema is due to breakdown of the alveoli and the chronic obstructive bronchitis is a result of chronic inflammation of bronchial tubes. In patients with COPD, there is a characteristic pattern of inflammation with increased numbers of neutrophils in the airway lumen along with increased numbers of macrophages, T lymphocytes, and B lymphocytes [17,21]. Such inflammatory response involves both innate and adaptive mechanisms [22]. Proinflammatory cytokines, such as TNF-α, IL-1β, and IL-6, are found in increased amounts in the sputum and BALF of individuals with asthma and COPD. This is due to an amplified inflammatory response, in part through the activation of NF-κB, which leads to the increased expression of multiple inflammatory genes.
Asthma is another chronic inflammatory lung disease of the airways. Asthma involves inflammation of bronchial tubes resulting in narrower air passages and difficulty in breathing, whereas COPD involves both airways and the lung [3]. Patients with severe asthma have airway inflammation that resembles COPD, suggesting a convergence of cytokine networks [23,24]. Although over 50 cytokines have been implicated in the onset of asthma, their role in the exact pathophysiology of the disease is still poorly understood. Environmental factors are frequently associated with asthma. Given the known role of environmental factors in modulating epigenetic changes, it is conceivable that epigenetic regulation of inflammatory genes plays a critical role in the pathophysiology of this disease [25].
Acute effects of overactivated inflammation are often manifested in lung injury leading to ARDS, a devastating form of respiratory failure. Over the past several years, there have been significant advances in our understanding of ARDS pathogenesis. The risk factors leading to ARDS can be either due to direct insult to the lung like in pneumonia (bacterial, viral, fungal), inhalation injury and aspiration of gastric contents, or indirect like in sepsis, burn, and trauma, where insult to the lungs is caused by various systemic inflammatory mediators. In addition, inflammatory cell migration, fibro-proliferation and apoptosis also play important roles in the pathophysiology of ARDS [26,27]. ARDS is characterized by rapid onset of widespread inflammation in the lungs [28] with injury to the alveolar capillary barrier and extravasation of protein-rich edema fluid into the airspace [[29], [30], [31]]. The degree of alveolar epithelial injury is the major prognostic marker of ARDS. Specifically, in ARDS, pulmonary edema fluid contains high levels of pro-inflammatory cytokines including IL-1β, IL-8, IL-6, TNFα, and transforming growth factor-β1 (TGFβ1) [32]. When excessive levels of cytokines are present, they cause alveolar injury [[33], [34], [35]]. The accumulated fluid in the interstitial and air spaces of the lungs also leads to difficulty in breathing and impaired gas exchange, resulting in hypoxemia and reduced carbon dioxide excretion, which often leads to acute respiratory failure [36]. Moreover, many cell types are involved in the pathophysiology of ARDS including platelets, neutrophils, alveolar macrophages, and monocytes, as well as endothelial and alveolar epithelial cells. Microbial products or cell-injury associated endogenous molecules like DAMPs, bind to Toll-like receptors on the lung epithelium and alveolar macrophages and activate the innate immune system [37]. Mechanisms of innate immune defense, such as the formation of neutrophil extracellular traps (NETs) and histone release, can be beneficial in capturing pathogens, but may also worsen alveolar injury [38]. The immune system also generates reactive oxygen species (ROS), leukocyte proteases, chemokines, and cytokines that help neutralize pathogens, but can also adversely affect the lungs [39]. Thus, the pathophysiology of various pulmonary diseases is intricately connected with systemic inflammation and the related aberrant immune responses.
3. Pulmonary manifestations of COVID-19
No other disease has affected the global human population the way COVID-19 has. The primary target of SARS-CoV-2 infection is the respiratory tract. The nasal passage and cavity is the first to get infected and then the virus is aspirated down the trachea, bronchi and the lungs [40,41]. SARS-CoV-2 primarily utilizes the ACE2 receptor on host cell for entry. It has been proposed that ACE2 is present at low levels in the ciliated epithelial cells of the airways allowing viral entry and the antiviral response (by the host cell) to induce an increase in ACE2 expression, furthering the spread of SARS-CoV-2 infection into the respiratory mucosa and the lung alveolar epithelial cells [42,43]. It is however interesting that ACE2 expressing club cells were not found to be infected by SARS-CoV-2 [40]. Although, autopsy samples of patients that died of COVID-19 demonstrate virus positivity in these regions of the airways and lung, it is unclear as to how the viral infection causes less severe or very little effect in asymptomatic patients. Patients with severe COVID-19 progress to pneumonia, ARDS and even sepsis. The disease severity is greater in patients with existing lung diseases such as COPD. The weakened immune system may also cause superinfection with secondary bacteria or virus. Thus, survivors of a serious case of COVID-19 will have a challenging recovery as the lungs may get scarred and the lung function severely compromised.
3.1. Lung inflammation in COVID-19
Our knowledge on COVID-19 is rapidly evolving but there is a clear connection between inflammation and onset as well as progression of the disease [44]. Even in children and adolescents, the so called ‘low-risk’ categories for COVID-19, inflammation- associated aggressive form of COVID-19 has been reported [45]. Clinical resolution of inflammation can possibly impact the survival of COVID-19 patients [46]. Several studies have indicated that SARS-CoV-2 causes injury predominantly through host immune dysregulation, increase in inflammation and/or hyperinflammation leading to severity of the disease, resulting in high rate of hospitalizations and mortality. The severity of infection often depends on the intricate balance between host innate and adaptive immune response.
The SARS-CoV-2 virus enters the host cells through angiotensin-converting enzyme 2 (ACE2) binding and internalization. Loss of ACE2 function results in activation of angiotensin type 1 receptor (ATIR) and Mas receptor (MasR) along with MAPK and ERK pathways, finally resulting in the release of inflammatory mediators such as TGF-β, IL-6, MIP1α, IP10 and TNF-α. It was recently demonstrated that in severe COVID-19, SARS-CoV-2 infection augmented the release of proinflammatory cytokines and chemokines that resulted in the death of host monocyte-derived macrophages and dendritic cells [47]. Moreover, the virus processing in the host cells also results in the release of damage associated molecular patterns (DAMPs). All these events further activate the adaptive immune response leading to systemic cytokine storm and hyperinflammation and severity of infection causing multi organ injury. Targeting therapeutics against these cytokine mediators, complement systems as well as ACE2 upregulation can potentially attenuate the injury and reduce mortality [48,49]. Based on emerging evidence, it is clear that the process of inflammation is intricately involved in the SARS-CoV-2-mediated complications and a more detailed knowledge of the underlying processes can possibly help mitigate the disease.
The spectrum of lung disease from SARS-CoV-2 infection varies from lack of symptoms to mild pneumonia to severe disease associated hypoxemia, respiratory and multiorgan failure and death. Hypoxemia, unlike those of ARDS, is another atypical early disease feature of COVID-19 often referred to as “happy hypoxia” or “silent hypoxia” as it is hidden and well tolerated [50]. SARS-CoV-2-induced inflammation of the respiratory centers has been proposed as one of the causes of silent hypoxia and subsequent respiratory failure [50,51]. ARDS is the most common manifestation of severe COVID-19 disease. SARS-CoV-2 infection of the lungs causes diffused alveolar damage, apoptotic epithelial cells, interstitial inflammation, and activated T-cell responses resulting in a cytokine storm [51]. Macrophage infiltration, hyaline membrane formation, alveolar edema, and septal thickening is also observed. These features are mostly observed in the autopsies. A number of studies have been conducted to reveal the pathology of the disease [[52], [53], [54]]. In an elegant study, Ackermann et al. compared the lungs of patients who died of COVID-19 with the lungs of patient who died from H1N1 influenza [55]. Besides the common ARDS like features such as diffuse alveolar damage and perivascular T-cell infiltration that was common in both, COVID-19 patient lung had distinct features. The pulmonary vessels of COVID-19 patients had widespread thrombosis and microangiopathy. A significant increase in vascular angiogenesis via intussusceptive angiogenesis was observed. There was an increase in ACE2 expression in the endothelial cells and significant changes in endothelial morphology was observed in the lungs of COVID-19 patients. The endothelial cells were swollen and had disrupted intercellular junctions and the contact with basal membrane was lost. More interestingly, they demonstrated infection of endothelial cells with SARS-CoV-2 further supporting direct endothelial injury along with perivascular inflammation. Involvement of the endothelial system, endothelitis, endothelial dysfunction and thrombosis has been shown by others too in COVID-19 [[56], [57], [58]]. Besides viral infection, inflammatory host immune response and unrestrained complement activation plays a critical role in causing endothelitis and thrombotic events and intravascular coagulation [59].
3.2. Coagulation in COVID-19
Critically ill patients with an exacerbated inflammatory response have coagulation abnormalities [60]. Inflammation is known to initiate clotting, decrease anticoagulant mechanisms and inhibit fibrinolytic pathways. Severe infection and inflammation invariably lead to activation of the coagulation cascade and critical disseminated intravascular coagulation (DIC) which often results in multiorgan failure [61]. Thrombosis was observed in the lungs of 89 % of COVID-19 patients who died [62]. Venous thromboembolism and both proximal and distal thrombosis are a feature of COVID-19 pneumonia [63,64]. These are caused by SARS-CoV-2-induced inflammation, hypoxia and diffuse intravascular coagulation [65]. A term pulmonary intravascular coagulation (PIC) was used to describe the lung restricted vascular thrombosis [66]. Due to the close proximity of pulmonary vasculature and pneumocytes, the numerous microthrombi in the vessels cause pulmonary infarction, hemorrhage and pulmonary hypertension. Vascular endothelial cells contribute to both inflammation and coagulation via growth factor release and by their adhesion molecules. Endothelial cells themselves release and respond to cytokines by increasing fibrin formation. Endothelial cells therefore play a critical role in maintaining patent and functional capillaries and endothelial dysfunction and vascular thrombosis may contribute significantly to pulmonary disease and COVID-19 severity [67]. It has also been suggested that SARS-CoV-2 can directly or indirectly activate toll-like receptors (TLRs), particularly TLR3 and TLR4, resulting in activation of extrinsic coagulation pathway [68]. The role of TLR3 [69] and TLR4 [70], in platelet function and the overall process of coagulation is also well known and thus their activation by SARS-CoV-2 further supports a connection between coagulation and COVID-19. A recently identified consensus pattern in SARS-CoV-2 spike protein (S protein) was similar to the sequence found in proteins involved in the coagulation process [71]. This further suggested involvement of the S protein in the molecular mechanisms leading to the COVID-related coagulopathy.
4. Inflammation and Cancer connection
The connection between inflammation and cancer is rather well established with inflammation recognized as a critical component of tumor progression [4]. Inflammation predisposes to cancer and promotes all stages of tumorigenesis, namely, tumor initiation, progression and metastasis [72]. Tumor microenvironment, that plays an important role in tumor progression, is orchestrated by a number of inflammatory cells [4]. It is estimated that approximately 15–20 % of cancers are a result of infections. The infections often result in inflammation with neoplastic transformation of surrounding cells leading to cancer, thus prompting the call to target inflammation for cancer prevention and therapy [73].
As discussed earlier, inflammation is intricately connected with several pulmonary diseases. Chronic pulmonary inflammation and fibrosis, following exposure to environmental toxins, can also result in lung cancer [74]. Lung cancer is the leading cause of cancer-related deaths worldwide [75,76] and by some estimates, smokers with COPD have upto six folds increased risk of developing lung cancer, compared to smokers without COPD [77,78]. Given the inflammatory manifestations in smoking and COPD, these studies underscore the importance of inflammation in lung cancer. Further, a possible prognostic role of advanced lung cancer inflammation index (ALI) in non-small cell lung cancer (NSCLC) patients has been suggested [79]. This index, calculated using the formula ‘Body Mass Index x Serum Albumin / Neutrophil Lymphocyte Ratio’, classifies NSCLC patients into those with low (ALI > 18) or high (ALI < 18) inflammation. The patients with high inflammation (and ALI < 18) have significantly lowered overall survival, compared to patients with low inflammation (and ALI > 18). On a similar note, another index, the systemic immune-inflammation index (SII) which is calculated using the formula ‘platelet counts × neutrophil counts / lymphocyte counts’ has been proposed as an independent prognostic factor for poor survival of advanced epidermal growth factor receptor (EGFR)-mutant NSCLC patients treated with first-generation tyrosine kinase inhibitors (TKIs) [80]. EGFR-targeted therapies, involving TKIs, are relevant to NSCLC [81,82] and clearly there seems to be a role of inflammation and immune response in sensitivity to TKIs.
Cigarette smoking is a major risk factor for lung cancer and the inflammatory as well as mutagenic effects of cigarette smoke promote a pro-cancer immune response [83]. The lungs of smokers are marked by activated macrophages, dendritic cells, lymphocytes and granulocytes, all resulting in an environment conducive to growth and proliferation of malignantly transformed cells [83]. However, smoking is not the only cause of lung cancer and lung cancer develops even in non-smokers potentially resulting from possible pulmonary infections, occupational dust exposure and IPF. Inflammation contributes towards lung cancer pathogenesis even in these patients [83]. A number of cellular and molecular factors mediate the inflammatory responses in lung cancer pathogenesis. The pro-inflammatory cyclooxygenase-2 (COX-2) [84] drives lung cancer progression and is a potential target for therapy [85]. Tumor associated macrophages (TAMs) are recruited within the lung cancer microenvironment wherein they induce production of several pro-inflammatory cytokines resulting in increased invasion, angiogenesis and metastasis of tumor cells. Even the tumor associated neutrophils (TANs) aid lung cancer tumorigenesis through increased expression of chemokine receptors such as CXCR3/4, CCR5/7 along with secretion of pro-inflammatory factors such as IL-6, IL-8 and MCP-1 [86]. IL-6 is produced at inflammation sites [[87], [88], [89]] and it promotes lung cancer metastasis [90]. High circulating levels of IL-6 associate with poor prognosis of lung cancer patients [91] thus making case for IL-6 blockage as an effective anticancer therapy [92]. Thus, similar to several pulmonary diseases discussed in the preceding section, inflammation is a major player in lung cancer tumorigenesis.
5. Lung cancer and COVID-19: the molecular link
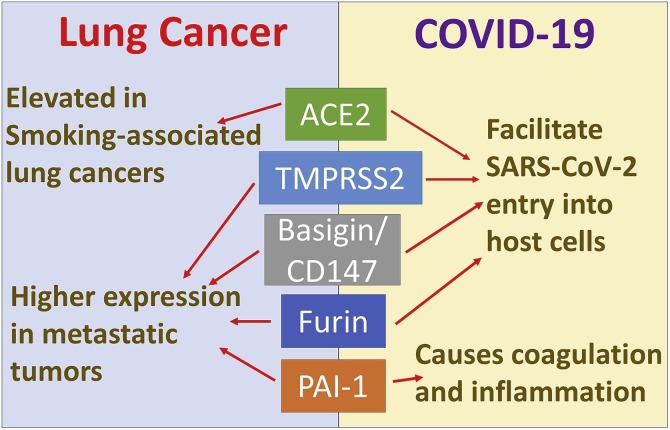
A connection between cancer and COVID-19 has been suggested [93] (Fig. 2 ). Altered expressions of ACE2, TMPRSS2 and plasminogen activator inhibitor-1 (PAI-1), and elevated levels of various cytokines may particularly be relevant with regards to an association of cancer with COVID-19. The common symptoms associated with severe COVID-19 include flu-like symptoms and ARDS. Lung cancer patients are a high-risk group for SARS-CoV-2 infections and related complications [94,95]. In fact, it has been reported that lung cancer patients in each age group, subtype, and pathological stage are much more susceptible to SARS-CoV-2 infection [96]. Based on the emerging statistics that support high rate of hospitalization, onset of ARDS, and high mortality rate, lung cancer patients represent a vulnerable population for COVID-19 [97]. Further, SARS-CoV-2 has even been detected in the pleural fluid of a lung cancer patient [98] suggesting a connection between COVID-19 and lung cancer that is not yet completely understood. In the subsections to follow, we discuss a few important genes that are common to SARS-CoV-2 infection and lung cancer, and can help explain the possible association between the two diseases.
Fig. 2.
Key molecular factors connecting lung cancer and COVID-19. A number of key cell surface proteins and enzymes play parallel roles in lung cancer progression and SARS-CoV-2 infection. A majority of them seem to play a role in the entry of SARS-CoV-2 into host cells while also being reported to be elevated in metastatic lung cancers.
5.1. ACE2 and TMPRSS2 in COVID-19 and lung cancer
ACE2 is a cellular receptor for the viral entry of SARS-COV-2. Viral S protein utilizes ACE2 receptor to fuse into the host cell membrane or enter via endocytosis and this process is regulated by type II transmembrane serine proteases (TTSPs) such as the TMPRSS2 [99]. Lung is one of the major organs with elevated ACE2 levels and ACE2 expression is significantly elevated in lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC), compared to normal tissues [100,101]. ACE2 is a human interferon-stimulated gene in airway epithelial cells [102] and ACE2 and TMPRSS2 are co-expressed in lung epithelial cells [103]. Emerging data suggests that, compared to normal individuals, those with a history of smoking and lung cancer have increased risk for SARS-CoV-2 infection due to a higher expression of ACE2 and TMPRSS2 in their lung cells [104].
TMPRSS2 is actively involved in priming the S protein of SARS-CoV-2 and its proteolytic cleavage for infectigng the host cells [105]. TMPRSS2 protein is encoded by the TMPRSS2 gene, which is widely conserved, and TMPRSS2 and ERG (ETS related oncogene) fusions are predominantly formed in tumors [106]. TMPRSS2 undergoes a developmental regulation and its expression is in a linear relationship with age in human beings [107]. Several reports have been made of the role of these proteins in causing infection and severity in case of COVID-19. The risk of this viral infection is enhanced in cancer patients as compared to non-cancer patients, owing to immunosuppression caused by malignancy or chemotherapy [108,109]. Since the primary site of infection of this virus is in the lungs, the worst outcomes are observed in the lungs [110]. Hence, lung cancer patients are believed to be at a higher risk of SARS-CoV-2 infection. Also, significant ACE2 expression level upregulation has been reported through immunohistochemistry in patients with lung cancer, more specifically in the lower airways [111]. These studies suggest an increased susceptibility of lung cancer patients to infection by SARS-CoV-2.
In a study that compared the gene expression of ACE2 and TMPRSS2 in different stages and subtypes of LUAD and LUSC with that of normal tissues [96], it was found that changes in expression levels of ACE2 and TMPRSS2 throughout different stages of lung cancer had no statistical significance except for the downregulation of TMPRSS2 in case of LUAD and that all stages of lung cancers are susceptible to the COVID-19 infection. It was speculated that this downregulation in LUAD and LUSC of TMPRSS2 could be due to its cancer suppressing properties. Comparison of ACE2 expression in different cancer tissues versus the corresponding normal tissues was also carried out in another independent study revealing a significant increase in ACE2 mRNA expression in LUAD and LUSC [112]. Another recent study [113] reported that ACE2 upregulation could actually have a protective effect in case of several different cancers including LUAD and LUSC, leading to better survival. This work suggested that several immune related pathways like T-cell receptor signaling and viral myocarditis were enhanced with high expression levels of ACE2. Further, ACE2 deficiency has been linked to a disruption of the renin-angiotensin system (RAS) that could result in an impaired immune response to virus infection more so in patients with prior underlying conditions [114]. Based on these reports, it is clear that our knowledge on the association between ACE2/TMPRSS2 expression and lung cancer is far from being conclusive, although there seems to be some trend supporting dysregulated ACE2 and TMPRSS2 expression in LUSC and LUAD patients, leading to increased susceptibility to SARS-COV-2 infection for each stage of these lung cancer subtypes. Several studies have reported DNA methylation, histone modifications and glycosylation as underlying mechanisms responsible for the increased expression of ACE2 in lung cancers [100,115,116]. Further, vorinostat (SAHA), an HDAC inhibitor, upregulates ACE2 [117]. Thus, both methylation and acetylation seem to be involved in the epigenetic regulation of ACE2 levels.
5.2. PAI-1 in lung cancer and COVID-19
Hemostatic imbalance such as increased coagulation and fibrinolysis was reported in patients with freshly diagnosed carcinoma of the lungs [118]. Dense plasma fibrin networks indicative of increased coagulation activity was observed in a study of patients with advanced lung cancer [119]. Interestingly, the plasma clot phenotype was found to be driven by smoking as plasma cotinine levels correlated with fibrin content [119]. Venous thromboembolism (VTE) is a frequent complication of advanced-stage lung cancer and its treatment. Due to these reasons, thromboprophylaxis is emerging as a common clinical practice for lung cancer patients [120]. More recently occurrence of D-Dimer along with thrombotic risk factors guides clinicians to provide preventive anticoagulant therapy to patients that have freshly undergone lung cancer surgery to reduce the incidence of VTE. Although plasma of lung cancer patients is enriched for coagulation factors, however, such a state of hypercoagulability is independent of lung cancer histology [121]. Plasma plasminogen activator inhibitor 1 (PAI) that participates in thrombotic, fibrinolytic, inflammatory and metabolic pathways is emerging as an important biomarker for non-small cell lung cancer [122].
Hypofibrinolytic state and high thrombin generation are also important in COVID-19 related thrombosis [123]. Plasma tissue plasminogen activator (tPA) and PAI-1 are highly elevated in hospitalized severe COVID-19 patients [124,125]. Therefore, PAI-1 is another factor that is common to lung cancer and COVID-19 (Fig. 2). It has been shown to promote lung cancer tumorigenesis [126]. Such pro-tumorigenic role of PAI-1 is rather paradoxical given that it is an endogenous inhibitor of oncogenic uPA/uPAR system [127]. However, PAI-1 is now regarded as a putative target for cancer therapy [128]. With regards to COVID-19, it has recently been proposed that SARS-CoV-2 gene products induce STAT-1 dysfunction which results in activation of STAT-3 as a compensatory mechanism [129]. While STAT-3 signaling is itself deregulated in lung cancer [130], in the SARS-CoV-2-infected cells, there is evidence for a positive feedback loop between STAT-3 and PAI-1 [129]. The activated PAI-1 leads to coagulopathy and secretion of pro-inflammatory chemokines and cytokines. Further, autopsies of COVID-19 patients exhibit diffuse alveolar damage and increased hyaluronan production, both of which induce PAI-1 levels [129]. This provides further evidence for enhanced PAI-1 levels in patients hospitalized with COVID-19 [125].
5.3. Other SARS-CoV-2-related genes in cancer
A number of other SARS-CoV-2 related genes, such as, basigin and paired basic amino acid cleaving enzyme (FURIN/PCSK3) have also been investigated in lung cancer [131] (Fig. 2). Basigin, also known as extracellular matrix metalloproteinase inducer (EMMPRIN) or cluster of differentiation 147 (CD147), is up-regulated in lung cancer and correlates with metastasis and poor prognosis/overall survival [[132], [133], [134]]. It is a cell surface glycoprotein on tumor cells that stimulates the production of matrix metalloproteinases and vascular endothelial growth factor [135], all of which are known to be oncogenic and conducive to cancer metastasis. It is therefore not surprising that basigin family has been implicated in bone metastasis of lung cancers [136]. Basigin expressed on host cells is understood to interact with spike protein of SARS-CoV-2 facilitating the viral infection [137] leading to basigin being regarded as a potential target for COVID-19 treatment [138]. However, a recent report contradicting this theory has suggested no evidence of interaction between basigin and SAR-CoV-2 spike protein [139]. The knowledge in the field is rapidly emerging and more studies are needed to fully understand the role of basigin in SARS-CoV-2 infection.
The role of another SARS-CoV-2-related gene, furin, on the other hand, is relatively more established. Furin is essential for the proteolytic activation and entry of SARS-CoV-2 into the human airway cells [140]. The cleavage site for furin in the spike protein is critical for SARS-CoV-2 infection and pathogenesis [141]. This furin cleavage site has a unique insertion enabling cleavage of spike protein of just the SARS-CoV-2, and not the related SARS-CoV-1 or the Middle East Respiratory Syndrome coronavirus MERS-CoV [142]. This information has led to the identification of furin inhibition as a strategy to limit SARS-CoV-2 infection [143,144]. Furin has also been investigated in lung cancer and was reported to be expressed in all the tumors, with particularly elevated levels in NSCLC, the more common lung cancer subtype, relative to the small cell lung carcinomas [145]. Furin levels positively correlate with aggressive, metastatic cancers [146] and similar to inhibition of SARS-CoV-2 infection by furin inhibitors [147], furin inhibitors can inhibit the growth, proliferation and metastasis of lung cancer cells [148,149].
6. Epigenetic regulation of inflammation
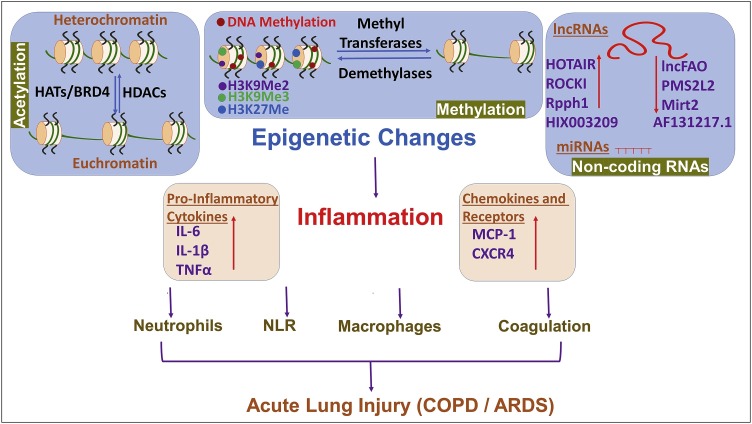
Chromatin modifying epigenetic mechanisms such as histone methylation and acetylation are involved in pathogenic injury (such as those caused by environmental insults, oxidative stress and viruses and bacteria) and remodeling of pulmonary tissues that often lead to chronic diseases such as asthma and COPD. Moreover, as stated above, it is clear that inflammation is associated with a number of pulmonary diseases, cancer and COVID-19 and, there has been considerable interest in understanding the mechanism(s) of inflammation, including the epigenetic basis of inflammation [10] (Fig. 3 ). This section focuses on methylation, acetylation and regulation through non-coding RNAs, the major investigated epigenetic pathways, in an attempt to summarize the current information on the topic.
Fig. 3.
Epigenetic regulation of inflammation. A number of epigenetic events can regulate inflammation. Modulators such as methylation and acetylation maintain a balance between heterochromatin and euchromatin with the relaxed euchromatin favoring transcription of genes. Histone acetyltransferases (HATs) and other proteins with similar activity such as bromodomain-containing protein 4 (BRD4) acetylate histones while histone deacetylases (HDACs) remove acetyl groups from histones. Methylation of DNA and histones is carried out by methyltransferases while demethylases are responsible for demethylating action. Finally, regulation through non-coding RNAs (both long non-coding RNAs ‘lncRNAs’ and microRNAs ‘miRNAs’) also comprises epigenetic regulation of inflammation. The overall result of such epigenetic events is either induction or resolution of inflammation with induction of inflammation manifested by release of pro-inflammatory cytokines, chemokines and chemokine receptors along with mobilization of neutrophils/ macrophages, elevated neutrophils-to-lymphocytes ratio (NLR) and coagulation. All these manifestations finally result in lung injury and pulmonary diseases characterized by chronic obstructive pulmonary disease (COPD) and/or acute respiratory distress syndrome (ARDS).
6.1. Role of methylation in inflammation
Epigenetic modulation of genes has been identified to regulate pro-inflammatory gene expression during the development of asthma, COPD and lung carcinogenesis [150]. Epigenetic processes are broadly carried out by DNA methylation, methylation/acetylation of histones, and non-coding RNAs. DNA methylation is the covalent binding of a methyl group to the 5′-carbon of cytosine (5mC) in cytosine-phosphate-guanine (CpG) dinucleotide sequence, mostly found in promoter/transcriptional/regulatory sites of genes [151]. DNA methyltransferases (DNMT) regulate DNA methylation and usually DNA methylation is associated with transcriptional repression while demethylation is associated with transcriptional expression of genes. Change in the status of DNA (e.g. in CpG-rich regions near transcriptional sites) and histone methylations has been identified to induce or suppress various oncogenes involved in lung cancer pathogenesis [152]. A study comparing DNA methylation and expression of inflammatory markers (IL-1β, IL-6 and IL-8) showed an inverse relationship between DNA methylation and transcriptional activation of these inflammatory genes in cancerous tissue as well as lung cancer cell lines [153]. Gene ontology analysis carried out on DNA extracted from peripheral blood of 1454 subjects identified altered methylation in 330 out of 349 CpG regions that influenced various genes, especially alpha-1-antitrypsin (AAT), involved in immune and inflammatory pathways contributing to pathogenesis of COPD [154]. Similarly, Bermingham et al., using 850 K Illumina EPIC array, demonstrated that, out of twenty eight differentially methylated sites, two were associated with development of COPD [155]. Human lung epithelium the most environmentally exposed tissue is largely affected by methylation [156]. Methylation status varies and depends on cell types affected. For instance, most hypermethylated CPGs are present in the promoter regions of bronchial epithelial cells and hypomethylated ones are present in the fibroblasts of asthmatics [157]. DNA methylation profiles of peripheral blood lymphocytes from asthma patients have been very useful to identify the epigenetic change of immune status [158]. Since DNA CPG methylation changes are more reversible than mutation, many asthma treatment strategies including dietary manipulations have generated interest [159,160]. Thus, methylation of genes affects expression of inflammation-related genes and methylation seems to be relevant in cancer as well as chronic pulmonary diseases such as asthma and COPD.
Other than the methylation of cytosine in CpG islands, DNA methylation has been reported in other sequences of DNA too which are gaining more attention in recent years. One such example is the methylation in cell-free DNA (cfDNA). It is well established that inflammation can drive necrosis and apoptosis of cells in COPD and lung cancer which results in release of cell-free DNA in blood [[161], [162], [163]]. Interestingly, methylation of cfDNA has been proposed as a diagnostic marker to differentiate between COPD, lung cancer and interstitial lung diseases [164]. Similarly, a comparative analysis between COPD patients and non-smokers revealed significant hyper-methylation in CpG regions of ANGPT1, PLD1, NFASC, ACTN4, RGS12, CAV1, PRKAG2, JARID2, HIPK2, and MECOM genes [165]. Caveolin-1 (Cav1) is known to regulate chronic inflammatory lung diseases and loss of CAV1 causes imbalance in the Th17/Treg cell in COPD patients [166]. A comparative study between ARDS patients and healthy individuals showed 30 DNA methylation sites were responsible for regulation of 10 genes that play critical role in inflammation, endothelial-epithelial barrier integrity and coagulation [167]. These studies are further corroborated by in vivo and translational studies. Mice intranasally exposed to inflammatory agent lipopolysaccharide (LPS) for two weeks reduced the global levels of cytosine hydroxymethylation (5hmc) in lung tissue [168]. Six weeks exposure to LPS increased cytosine methylation (5mc) in CDH13 (cell adhesion molecule), Ahrr, Tet1 and Rassf1 (tumor suppressors) and DAPK1 (pro-oncogene) which together highlight involvement of complex inflammatory stimulus involving methylation-based regulation of inflammation as well as tumor suppressors and oncogenes in progression of lung cancer. Further, lung cancer cells, A549, treated with IL-1β, an inflammatory cytokine, resulted in methylation of promoter region of CDH1 and histones (H3K9Me2 and H3K9Me3 and H3K27Me) resulting in epithelial mesenchymal transition (EMT) of these cells [169], thus connecting epigenetics and inflammation with pro-tumorigenic event EMT. Similarly, chronic exposure to TNF-α and TNF-β exposure also led to increased DNA methylation in CDH-1 promoter regions [169]. Collectively, there is strong evidence suggesting that DNA methylation is a mechanism that plays critical role in inflammation and resulting pulmonary diseases and cancer.
6.2. Role of acetylation in inflammation
Acetylation is another well-established epigenetic modulator of gene expression. Addition of acetyl group to lysine (K) residues in histones as well as non-histone proteins directly influences the regulation of genes. Acetylation of proteins is carried out at two sites - Nα-terminal acetylation or ε-amino group of lysines [170,171]. Although the former is a co-translational modification and is more common, the latter is post-translational and less common but has diverse effects as it can modify protein function, and affect half-life, in addition to bringing about profound effects on the inflammatory process [171]. While addition of acetyl group is controlled by histone acetyltransferases (HAT), their removal is catalyzed by histone deacetylases (HDACs); thus, referred to as acetylation writers and erasers. The bromodomain and extra-terminal (BET) proteins categorized as readers of acetylation as they can recognize acetylated histones, are increasingly being recognized as mediators of inflammation. BETs have been shown to play crucial role in regulation of various genes involved in inflammation along with cell proliferation and apoptosis. The BET proteins namely BRD2, BRD3 and BRD4 mediate assembly of histone acetylation-dependent chromatin complexes that regulate expression of inflammatory genes [172]. BRD4 in this BET family not only identifies histone acetylation but also has HAT property, which is inducible by RelA activation [173]. BRD4 plays an important role in regulating inflammatory diseases through its direct interaction with NF-κB [172]. Moreover, BRD4 has been shown to be upregulated in airway inflammation which correlates with increased expression of IL-8 [174]. It has also been shown to play crucial role in IL-1β-induced inflammation in human airway epithelial cells and TLR3 mediated airway remodeling and virus induced lung inflammation [173,175,176]. Interestingly, treatment of lung epithelial cells with IL-1β or TNF-α results in acetylation of H4K which increases expression of granulocyte colony stimulating factor (GM-CSF) signifying role of HATs in inducing an inflammatory phenotype [177]. Similarly, pharmacologic inhibition of HDACs by Trichostatin A (TSA) leads to activation of various inflammatory cytokines while drugs like theophylline which restore histone deacetylase activity have opposing effects [178,179]. In inflammatory lung disease, such as asthma, increased expression of HATs and decreased expression of HDACs in the lung tissues has been observed [180]. Similarly, analyses carried out on lung inflammatory cells of asthmatics showed decreased HDAC activity [181]. In COPD patients, disease severity correlated with HDAC activity, which was shown to be reduced by >95 % in severely sick patients [182]. While HATs are increased in asthma, they are not much affected in COPD [182,183]. Rats exposed to cigarette smoke showed reduced HDACs activity which resulted in increased NF-κB activation [184]. Similarly, smoking in otherwise healthy individuals did not alter HAT expression but greatly reduced activity of HDACs in alveolar macrophages [185]. The importance of acetylation in the pathogenesis of inflammatory lung diseases is underscored by the fact that corticosteroids, which are known to reverse histone acetylations, are frequently used for treatment of such conditions. Lung inflammatory diseases are multifactorial and involve activation of various transcriptional factors like NF-κB, AP-1, signal transducer and activator of transcription (STAT), and nuclear factor of activated T-cells (NF-AT). Activation of these factors leads to histone acetylation and subsequently inflammation. However, it is through these transcriptional factors that corticosteroids prevent acetylation of histones and consequently suppress inflammation by switching off induced inflammatory genes [183]. Acetylation/deacetylation of glucocorticoid receptors is linked to secretory leukocyte protease inhibitor expression which is found in upper airways and regulates neutrophil elastase activity [186]. In COVID-19, hypomethylation of ACE2, frequently observed in lupus patients, has been suggested to increase SARS-CoV-2 entry [187]. Based on the collective evidence presented here, it is clear that the dynamic processes of acetylation and deacetylation regulate the expression of genes that connect inflammation with various pulmonary diseases.
6.3. Role of non-coding RNAs in inflammation
In recent years there has been considerable interest in the regulation of signaling pathways by non-coding RNAs in various human diseases [188]. Targeting non-coding RNAs, such as long non-coding RNAs (lncRNAS) [[189], [190], [191]] and miRNAs [192,193] can modulate inflammation and the associated immune responses [191]. Non-coding RNAs are a large class of RNA molecules with new members being identified regularly. They are extremely diverse in their functionality and individual non-coding RNAs modulate multiple signaling pathways. For example, lncRNAs, the non-coding RNAs longer than 200 nuceotides, can enhance or suppress transcription of inflammatory responses [194]. lncRNA HIX003209 [195], ROCKI [196], Rpph1 [197], HOTAIR [198] are some of the lncRNAs that promote inflammation while lncFAO [190], Mirt2 [199], PMS2L2 [200], AF131217.1 [201] are some of the lncRNAs that inhibit inflammation. These lncRNAs bring about their physiological effects by interacting with multiple molecular targets. This includes sponging of miRNAs and transcriptional as well as post-transcriptional regulation of target genes.
MicroRNAs, the non-coding RNAs that are much shorter and about 20–24 nucleotides long, similarly regulate inflammation through the down-regulation of their target genes. The available literature on the regulation of inflammation and the related signaling mechanisms by miRNAs is rather vast and beyond the scope of discussion here and reviewed in detail elsewhere [[202], [203], [204], [205], [206], [207], [208]]. However, similar to the regulation by lncRNAs, miRNAs have also been reported to regulate inflammation. miR-20a [209], miR-15b [210], miR-106a [211], miR-155 [212], miR-421 [213] and miR-1307 [214] are some of the miRNAs that promote inflammation while miR-10a [215], miR-24 [216], miR-124 [217], miR-146a [218], miR-181 [219] and miR-223 [220] are some of the miRNAs that inhibit inflammation. Based on such information, it is clear that non-coding RNAs have the potential to effectively regulate inflammation and related processes. There is enormous data on such activity by miRNAs and data on lncRNAs is emerging. Targeting these non-coding RNA molecules can provide an opportunity to tweak the overall process of inflammation and its impact on affected diseases.
6.4. Epigenetic regulation of inflammatory cytokines
6.4.1. Interleukin-6
Proinflammatory cytokines are produced predominantly by activated immune cells which trigger inflammatory reactions. Based on the earlier reports and our own findings [89,221], IL-6 is clearly an important pro-inflammatory cytokine in pulmonary and non-pulmonary diseases, which can be regulated epigenetically. IL-6 in turn, can epigenetically regulate other molecular factors. Its inhibition of methyltransferases DNMT1 [222] and DNMT3B [223] can possibly impact expression and activity of multiple genes. Not only a direct but even some indirect epigenetic regulation of IL-6 has been suggested. For example, in cancer, methylation and resulting repression of suppressor of cytokine signaling-1 (SOCS-1) can activate IL-6 as SOCS-1 is involved in the negative regulation of IL-6 [224].
IL-6, which is epigenetically regulated by miRNAs, can itself regulate miRNAs as well. For instance, the miRNAs, let-7 [225] and miR-149 [226] have been shown to target and inhibit IL-6. In turn, IL-6 can down-regulate miR-370 when ectopically overexpressed [227]. IL-6-mediated regulation of miR-370 possibly involves methylation, as the effect was decreased by the methylation inhibitor 5-aza-2′-deoxycytidine [227]. Similarly, IL-6 methylates and inhibits miR-142 [228]. In addition to regulation of miRNAs, IL-6 has been reported to affect methylation of some genes as well. IL-6 can elicit the loss of methylation at the promoters of cancer stem cell markers CD133 and CD44 [229]. Interestingly, such epigenetic action by IL-6 is possible only after the loss of tumor suppressor p53 that results in demethylation at IL-6 promoter and its consequent expression [229].
In dendritic cells that play important role in inflammation-induced ARDS [230], IL-6 is activated by the transcription factor Kruppel-like factor 4 (KLF4). However, instead of methylation which seems to be the most common mechanism of IL-6 activation, the effect of KLF4 involves acetylation [231]. The role of acetylation in IL-6 activation is further supported by the observation that the activation is inhibited by inhibition of acetyltransferase activity [232]. Further, there is evidence that IL-6 is demethylated and thus epigenetically activated in response to influenza virus infection [233]. Also, herpesvirus encoded viral IL-6 can induce STAT3 activity through phosphorylation as well as acetylation [234] thus supporting a robust role of IL-6-mediated epigenetic regulation in viral infections. In particular, IL-6 needs to be better scrutinized in the context of COVID-19 pandemic because a number of recent reports correlate elevated levels of IL-6 with the severity and mortality of COVID-19 [[235], [236], [237], [238]].
6.4.2. Other inflammatory cytokines
A plethora of studies have established the involvement of proinflammatory cytokines in lung disease pathogenesis. The differentiation of Th1/Th2 cells mediate inflammatory/immune responses in pulmonary diseases through action of cytokines like IL-4, IL-13, IL-5 and IFN-γ. IFN-γ is suppressed by the methyltransferase DNMT3a, which methylates CpG-53 [239]. Similarly, CpG demethylation at DNase I hypersensitive sites (DHS) in the promoter regions of IL-4 and IL-13 regulate Th2 differentiation [240]. In paraquat induced pulmonary fibrosis model, imbalance in HATs and HDAC has been shown to regulate IL-6 expression [232]. DNA methylation in promoter region has been shown to regulate STAT-6 and respective cytokines [241]. The interaction between CXCR4 and CXCL12 regulates cancer progression in various organs including lungs and it has been shown that demethylation of CXCR4 and hypermethylation of CXCL12 promoter regions is responsible for their activation [153]. A similar report published on granulomatous lung diseases shows involvement of methylation in the regulation of cytokine-cytokine receptor interaction, chemokine signaling and JAK-STAT pathway and cellular immunity [242]. Methylation of CpG islands in promoter regions of IL-1β, IL-6, and IL-8 genes have been shown to regulate the pathogenesis of lung cancer [243].
Dysregulation of histone acetylation and deacetylation also influences transcription of inflammatory cytokines, subsequently affecting the expression of inflammatory genes [244]. CXCL-10, an anti-inflammatory cytokine is possibly reduced in IPF and other inflammatory lung diseases due to acetylation on H3 and H4 histones [245]. In COPD, HDAC reduction is responsible for over expression of GM-CSF, IL-8 and TNF [185]. Other cytokines like interleukin-23 (IL-23) or receptors of some cytokines e.g. that of IL-12 are regulated by histone acetylation as well as methylation. IL-12 receptors are regulated in NSCLC cells and COPD by both histone acetylation and DNA methylation [246]. In experimental asthma DNA methylation has been shown to be involved in effector T-cell polarization causing differential secretion of TH1 and TH2 cytokines [247]. Here increase in DNA methylation at the interferon gamma promoter correlated with interferon gamma cytokine production [247,248]. In SARS-CoV-2 infected patients, cytokine storm has been reported in which there are massive increases in inflammatory cytokines, such as, interleukins, TNF, colony stimulating factors (CSFs), and growth factors (GFs). Emerging evidence suggests the involvement of epigenetic modifications in these increases [249,250]. Variable response of individuals infected with SARS-CoV-2 could possibly be influenced by epigenetic regulators of cytokines, as has been reported in lupus patients [187]. Thus, there is convincing evidence to support epigenetic regulation of inflammatory cytokines that are involved in onset/progression of lung cancer, pulmonary diseases and COVID-19.
7. MicroRNAs as epigenetic regulators connecting inflammation, lung cancer and COVID-19
As discussed above, there are numerous reports supporting a role of miRNAs in the regulation of inflammation. Given the connections between inflammation, lung cancer and COVID-19, we surveyed the literature to evaluate possible epigenetic links. The roles of miRNAs in inflammation [[202], [203], [204], [205], [206], [207], [208]] and lung cancer [[251], [252], [253], [254]] are well established, and recent literature suggests role of miRNAs in COVID-19 infections as well. For example, it has been suggested that immune responses to various viral respiratory infections, including COVID-19, are characterized by ectopic expression of miRNAs [255]. Also, some recent studies have listed the potential miRNAs that can target COVID-19-related host receptors, ACE2 or TMPRSS2, in SARS-CoV-2-infectable liver cancer cells Huh7 and lung cancer cells Calu3 [256], kidneys [257], cardiomyocytes [258] as well as in the hypothalamus [259]. A possible role of deregulated miRNAs has also been suggested in the relatively higher COVID-19 related mortality in elderly patients [260]. This section discusses some miRNAs that have been predicted/reported to be relevant to COVID-19, lung cancer and inflammation (Table 1 ).
Table 1.
miRNAs connecting COVID-19, lung cancer and inflammation.
| miRNA | SARS-CoV-2 target | Role in Lung Cancer | Role in Inflammation |
|---|---|---|---|
| miR-15b | Differentially expressed in Hamster lungs after SARS-C0V-2 infection [284] | Promotes lung cancer growth and invasion [286] | Expression positively correlates with inflammation [210] |
| miR-21 | Predicted to bind to human coronavirus RNA [268] | Oncogenic miRNA in lung cancer [269] | Regulator of inflammatory response [270] |
| miR-29 family | Has 11 binding sites on SARS-CoV-2 genome [263] | Tumor suppressor with role in therapy resistance [264,265] | Anti-inflammatory in cancer and other diseases [266,267] |
| miR-98 | Targets TMPRSS2 in lung endothelial cells [281] | Inhibits lung cancer proliferation and metastasis [283] | Expression negatively correlates with inflammatory cytokines [282] |
| miR-195 | Differentially expressed in Hamster lungs after SARS-C0V-2 infection [284] | Tumor suppressor that associates with improved survival [287] | Promotes resolution of inflammation [285] |
| miR-200 family | miR-200c is predicted to regulate ACE2 in respiratory cells [274] | Tumor suppressor and negative regulators of EMT [81,273] | Members of this family have been reported to be pro-inflammatory [275,276] as well as anti-inflammatory [277,278], |
| miR-421 | Regulates ACE2 [279] | Overexpressed in lung cancer and associates with poor prognosis [280] | Aggravates inflammatory response in lung tissues [213] |
| miR-1207 | Targeted directly by SARS-CoV-2 RNA [288] | Tumor suppressor with inhibitory effect on metastasis [289] | De-repression of its target CSF1 results in acute inflammatory response in COVID-19 [288] |
| miR-1307 | Predicted to have highest affinity for SARS-CoV-2 genome among 1872 miRNAs [261] | Promotes lung cancer growth and proliferation [262] | Promotes inflammatory responses [214] |
Numbers in parenthesis are corresponding citations. CSF1: Colony Stimulating Factor 1, EMT: Epithelial-to-Mesenchymal Transition.
A few studies have looked at the miRNAs that can potentially target SARS-CoV-2 genome. One such study reported miR-1307 as the miRNA with highest affinity for the SARS-CoV-2 genome [261]. This oncogenic miRNA associates with poor survival of lung cancer patients [262] and has been reported to promote inflammatory response [214]. In an in silico study, miR-29 family of miRNAs were found to have the most binding sites (11 sites) on SARS-Cov-2 genome [263]. Members of this family have been reported to exert tumor suppressive effects in lung cancer with role in sensitivity to cisplatin [264,265], and also found to antagonize inflammatory mechanisms in multiple diseases [266,267]. A computational approach, that evaluated interactions between miRNAs and seven human coronavirus RNAs, predicted miR-21 to be the most likely miRNA that can bind to human coronavirus RNAs resulting in potential significantly upregulated expression in human lungs after COVID-19 infection [268]. miR-21 is an oncogenic miRNA [269] whose important role in the regulation of inflammatory responses has previously been described [270].
Mir-200 family has been well characterized in cancer with numerous reports on its tumor suppressive action in multiple human cancers [271,272], including in lung cancer [81,273]. Members of this family can regulate ACE2, with miR-200c possibly the strongest candidate miRNA that can target ACE2 in respiratory cells [274]. Additionally, members of miR-200 family have been reported to regulate inflammation. They have multifaceted effects on inflammation ranging from pro-inflammatory [275,276] to anti-inflammatory [277], which might be tissue/cell-specific and may also depend on the stimulus. Interestingly, miR-200b was found to be down-regulated in lungs of COPD model mice and it was proposed as a potential therapy to treat COPD because of its ability to attenuate inflammatory responses [278]. miR-200 family is however not the only one that can regulate ACE2. Regulation of ACE2 has also been shown with miR-421 [279]. This miRNA is upregulated in lung cancer and associates with poor prognosis of lung cancer patients [280]. With regards to inflammation, ectopic up-regulation of miR-421 has been found to aggravate inflammation in lung tissues [213].
Micro RNAs can target other SARS-CoV-2 infection-related genes as well. For example, miR-98 has been shown to target TMPRSS2 [281]. This study was conducted using lung endothelial cells and such validation of TMPRSS2 targeted by miR-98 in this model system is critical, given the importance of endothelial dysfunction in the pathogenesis of COVID-19. Over-expression of miR-98 has been shown to result in diminished levels of pro-inflammatory cytokines [282] and reduced proliferation and metastasis in lung cancer [283]. Kim et al. tested the in vivo significance of miRNAs identified by bioinformatic tools and predicted to bind to the RNA of host viral entry proteins such as ACE2, TMPRSS2, and ADAM17. It was reported that miR-15b and miR-195 are significantly differentially expressed in Hamster lung tissues after infection with SARS-CoV-2 [284]. There is evidence to suggest a pro-inflammatory function of miR-15b [210] but, conversely, an anti-inflammatory function of miR-195 [285]. Consequently, while miR-15b promotes growth and invasion of lung cancer [286], miR-195 represses lung cancer and associates with improved recurrence-free and overall survival of lung cancer patients [287].
In a study that assessed role of miRNAs in COVID-19-induced acute inflammatory response, an essential role of miR-1207 in the process was identified [288]. It was revealed that miR-1207, which suppresses lung cancer progression and metastasis through the targeting of colony stimulating factor-1 (CSF1) [289], is itself targeted by SARS-CoV-2 RNA. The competitive binding of SARS-CoV-2 RNA to miR-1207 de-represses CSF1 which then induces the characteristic inflammatory response associated with COVID-19 infection [288]. This fits well with the narrative that pathogenic human coronaviruses, including SARS-CoV-2, can regulate host miRNA levels by acting as miRNA sponges [290].
Given the growing evidence for the role of miRNAs in COVID-19 infection, their putative use in COVID-19 therapy, with possibility of employing extracellular vesicles-based delivery approach, has been advocated [291,292]. The progress in the therapeutic targeting of miRNAs for the benefit of lung cancer patients in clinical settings has been extremely slow. Hopefully, the scale of current COVID-19 global pandemic can incentivize researchers to accelerate studies that can help realize clinical potential of miRNAs.
8. Conclusions and perspectives
The relationship of inflammation with several pulmonary ailments and lung cancer is irrefutable as is the connection between pulmonary diseases and lung cancer, both in terms of pre-disposition as well as disease etiology. Over the past several months, emergence of COVID-19 has led to a lot of resources been diverted to better understand this disease. Even though the mortality rate associated with COVID-19 is low and not even comparable to many other diseases, its high infection rate has resulted in worldwide cases exceeding 68 million, resulting in more than 1.5 million deaths as of this writing during the first week of December, 2020. All the available data points to significant overlap between COVID-19 and pulmonary manifestations, and COVID-19 patients often succumb to the disease as a result of pulmonary complications. There is also a definite connection of COVID-19 with lung cancer, as lung cancer patients not only represent a more vulnerable group for SARS-CoV-2 infections but also correlate with higher mortality rates. As discussed in this article, a number of genes crucial for SARS-CoV-2 infection have also been implicated in tumorigenesis, thus explaining the COVID-19-lung cancer connection. As a mechanism, we have provided evidence connecting inflammation with pulmonary diseases, lung cancer and COVID-19, and focused on the epigenetic regulation of inflammation, including dysregulated miRNA, as this might represent novel opportunity to therapeutically target inflammation.
One of the limitations with the studies that have emerged lately during the COVID-19 pandemic is that several of these reports have been conducted on a relatively small-scale, and, additionally, a large amount of such emerging data has also been made available before the traditional peer-review. A more robust review, validation and confirmation by independent groups is needed before reaching any definitive conclusions. Regardless, it is quite apparent that inflammation, particularly the various epigenetic targets as well as modifiers of inflammation need to be further evaluated in relation to their ability to regulate pulmonary diseases, thus impacting both lung cancer and COVID-19. As an example, a number of mechanistic studies have identified the critical role of cytokines in inflammation-mediated disease etiology, with definite evidence connecting epigenetic regulation of cytokines with their expression and function. This supports the notion that a combined cytokine panel can improve the accuracy of predictive value for adverse outcome beyond standard clinical data alone [293]. This also provides opportunity for the identification of specific cytokines to stratify patients towards trials of specific immunomodulatory treatments to improve outcomes in COVID-19. Clearly, such personalized therapy involving cytokines can be beneficial for lung cancer patients as well, given the overwhelming data on cytokines in lung cancer progression. With the focus on inflammation and the resulting pulmonary manifestation, it is possible to fine tune our current understanding of lung cancer and COVID-19 with possible implications on how these diseases can be managed and treated.
Funding
This work is funded by grants from the CounterACT Program, National Institutes of Health Office of the Director (NIH OD), the National Institute of Environmental Health Sciences (NIEHS) Grants, U01ES025069 (Aftab Ahmad), U01ES028182 (SA) and the National Heart Lung and Blood Institute (NHLBI) grant numbers R01HL114933 (Aftab Ahmad).
Declaration of Competing Interest
None of the authors have any potential conflict of interest to report.
References
- 1.Rock K.L., Kono H. The inflammatory response to cell death. Annu. Rev. Pathol. 2008;3:99–126. doi: 10.1146/annurev.pathmechdis.3.121806.151456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pahwa R., Goyal A., Bansal P., Jialal I. StatPearls; Treasure Island (FL): 2020. Chronic Inflammation. [Google Scholar]
- 3.Cukic V., Lovre V., Dragisic D., Ustamujic A. Asthma and chronic obstructive pulmonary disease (COPD) - differences and similarities. Mater. Sociomed. 2012;24(2):100–105. doi: 10.5455/msm.2012.24.100-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coussens L.M., Werb Z. Inflammation and cancer. Nature. 2002;420(6917):860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balkwill F., Mantovani A. Inflammation and cancer: back to Virchow? Lancet. 2001;357(9255):539–545. doi: 10.1016/S0140-6736(00)04046-0. [DOI] [PubMed] [Google Scholar]
- 6.Gomes M., Teixeira A.L., Coelho A., Araujo A., Medeiros R. The role of inflammation in lung cancer. Adv. Exp. Med. Biol. 2014;816:1–23. doi: 10.1007/978-3-0348-0837-8_1. [DOI] [PubMed] [Google Scholar]
- 7.Marklova E. Inflammation and genes. Acta Medica Hradec Kralove (Hradec Kralove) 2007;50(1):17–21. [PubMed] [Google Scholar]
- 8.Zhao Y., Forst C.V., Sayegh C.E., Wang I.M., Yang X., Zhang B. Molecular and genetic inflammation networks in major human diseases. Mol. Biosyst. 2016;12(8):2318–2341. doi: 10.1039/c6mb00240d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Newcombe E.A., Camats-Perna J., Silva M.L., Valmas N., Huat T.J., Medeiros R. Inflammation: the link between comorbidities, genetics, and Alzheimer’s disease. J. Neuroinflammation. 2018;15(1):276. doi: 10.1186/s12974-018-1313-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bayarsaihan D. Epigenetic mechanisms in inflammation. J. Dent. Res. 2011;90(1):9–17. doi: 10.1177/0022034510378683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raghuraman S., Donkin I., Versteyhe S., Barres R., Simar D. The emerging role of epigenetics in inflammation and immunometabolism. Trends Endocrinol. Metab. 2016;27(11):782–795. doi: 10.1016/j.tem.2016.06.008. [DOI] [PubMed] [Google Scholar]
- 12.Ahmad A., Li Y., Bao B., Kong D., Sarkar F.H. Epigenetic regulation of miRNA-cancer stem cells nexus by nutraceuticals. Mol. Nutr. Food Res. 2014;58(1):79–86. doi: 10.1002/mnfr.201300528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stylianou E. Epigenetics of chronic inflammatory diseases. J. Inflamm. Res. 2019;12:1–14. doi: 10.2147/JIR.S129027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bateman E.D., Hurd S.S., Barnes P.J., Bousquet J., Drazen J.M., FitzGerald J.M., Gibson P., Ohta K., O’Byrne P., Pedersen S.E., Pizzichini E., Sullivan S.D., Wenzel S.E., Zar H.J. "Global strategy for asthma management and prevention: GINA executive summary.". Eur. Respir. J. 2008;31:143–178. doi: 10.1183/09031936.00138707. Eur Respir J 51(2) (2018). [DOI] [PubMed] [Google Scholar]
- 15.Ware L.B., Matthay M.A. The acute respiratory distress syndrome. N. Engl. J. Med. 2000;342(18):1334–1349. doi: 10.1056/NEJM200005043421806. [DOI] [PubMed] [Google Scholar]
- 16.Barnes P.J. Chronic obstructive pulmonary disease. N. Engl. J. Med. 2000;343(4):269–280. doi: 10.1056/NEJM200007273430407. [DOI] [PubMed] [Google Scholar]
- 17.Barnes P.J. Immunology of asthma and chronic obstructive pulmonary disease. Nat. Rev. Immunol. 2008;8(3):183–192. doi: 10.1038/nri2254. [DOI] [PubMed] [Google Scholar]
- 18.Moldoveanu B., Otmishi P., Jani P., Walker J., Sarmiento X., Guardiola J., Saad M., Yu J. Inflammatory mechanisms in the lung. J. Inflamm. Res. 2009;2:1–11. [PMC free article] [PubMed] [Google Scholar]
- 19.Robb C.T., Regan K.H., Dorward D.A., Rossi A.G. Key mechanisms governing resolution of lung inflammation. Semin. Immunopathol. 2016;38(4):425–448. doi: 10.1007/s00281-016-0560-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aghasafari P., George U., Pidaparti R. A review of inflammatory mechanism in airway diseases. Inflamm. Res. 2019;68(1):59–74. doi: 10.1007/s00011-018-1191-2. [DOI] [PubMed] [Google Scholar]
- 21.Brusselle G.G., Joos G.F., Bracke K.R. New insights into the immunology of chronic obstructive pulmonary disease. Lancet. 2011;378(9795):1015–1026. doi: 10.1016/S0140-6736(11)60988-4. [DOI] [PubMed] [Google Scholar]
- 22.Givi M.E., Redegeld F.A., Folkerts G., Mortaz E. Dendritic cells in pathogenesis of COPD. Curr. Pharm. Des. 2012;18(16):2329–2335. doi: 10.2174/138161212800166068. [DOI] [PubMed] [Google Scholar]
- 23.Wouters E.F. Local and systemic inflammation in chronic obstructive pulmonary disease. Proc. Am. Thorac. Soc. 2005;2(1):26–33. doi: 10.1513/pats.200408-039MS. [DOI] [PubMed] [Google Scholar]
- 24.Barnes P.J. Mediators of chronic obstructive pulmonary disease. Pharmacol. Rev. 2004;56(4):515–548. doi: 10.1124/pr.56.4.2. [DOI] [PubMed] [Google Scholar]
- 25.DeVries A., Vercelli D. Epigenetic mechanisms in asthma. Ann. Am. Thorac. Soc. 2016;13(Suppl 1):S48–50. doi: 10.1513/AnnalsATS.201507-420MG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matthay M.A., Uchida T., Fang X. Clinical acute lung injury and acute respiratory distress syndrome. Curr. Treat. Options Cardiovasc. Med. 2002;4(2):139–149. doi: 10.1007/s11936-002-0034-0. [DOI] [PubMed] [Google Scholar]
- 27.Bhandary Y.P., Shetty S.K., Marudamuthu A.S., Ji H.L., Neuenschwander P.F., Boggaram V., Morris G.F., Fu J., Idell S., Shetty S. Regulation of lung injury and fibrosis by p53-mediated changes in urokinase and plasminogen activator inhibitor-1. Am. J. Pathol. 2013;183(1):131–143. doi: 10.1016/j.ajpath.2013.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matuschak G.M., Lechner A.J. Acute lung injury and the acute respiratory distress syndrome: pathophysiology and treatment. Med. 2010;107(4):252–258. [PMC free article] [PubMed] [Google Scholar]
- 29.Han S., Mallampalli R.K. The acute respiratory distress syndrome: from mechanism to translation. J. Immunol. 2015;194(3):855–860. doi: 10.4049/jimmunol.1402513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Finigan J.H. The coagulation system and pulmonary endothelial function in acute lung injury. Microvasc. Res. 2009;77(1):35–38. doi: 10.1016/j.mvr.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 31.Gando S., Kameue T., Matsuda N., Sawamura A., Hayakawa M., Kato H. Systemic inflammation and disseminated intravascular coagulation in early stage of ALI and ARDS: role of neutrophil and endothelial activation. Inflammation. 2004;28(4):237–244. doi: 10.1023/b:ifla.0000049049.81688.fe. [DOI] [PubMed] [Google Scholar]
- 32.Olman M.A., White K.E., Ware L.B., Simmons W.L., Benveniste E.N., Zhu S., Pugin J., Matthay M.A. Pulmonary edema fluid from patients with early lung injury stimulates fibroblast proliferation through IL-1 beta-induced IL-6 expression. J. Immunol. 2004;172(4):2668–2677. doi: 10.4049/jimmunol.172.4.2668. [DOI] [PubMed] [Google Scholar]
- 33.Fukuda N., Jayr C., Lazrak A., Wang Y., Lucas R., Matalon S., Matthay M.A. Mechanisms of TNF-alpha stimulation of amiloride-sensitive sodium transport across alveolar epithelium. Am. J. Physiol. Lung Cell Mol. Physiol. 2001;280(6):L1258–65. doi: 10.1152/ajplung.2001.280.6.L1258. [DOI] [PubMed] [Google Scholar]
- 34.Elia N., Tapponnier M., Matthay M.A., Hamacher J., Pache J.C., Brundler M.A., Totsch M., De Baetselier P., Fransen L., Fukuda N., Morel D.R., Lucas R. Functional identification of the alveolar edema reabsorption activity of murine tumor necrosis factor-alpha. Am. J. Respir. Crit. Care Med. 2003;168(9):1043–1050. doi: 10.1164/rccm.200206-618OC. [DOI] [PubMed] [Google Scholar]
- 35.Dagenais A., Frechette R., Yamagata Y., Yamagata T., Carmel J.F., Clermont M.E., Brochiero E., Masse C., Berthiaume Y. Downregulation of ENaC activity and expression by TNF-alpha in alveolar epithelial cells. Am. J. Physiol. Lung Cell Mol. Physiol. 2004;286(2):L301–11. doi: 10.1152/ajplung.00326.2002. [DOI] [PubMed] [Google Scholar]
- 36.Huppert L.A., Matthay M.A., Ware L.B. Pathogenesis of acute respiratory distress syndrome. Semin. Respir. Crit. Care Med. 2019;40(1):31–39. doi: 10.1055/s-0039-1683996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Opitz B., van Laak V., Eitel J., Suttorp N. Innate immune recognition in infectious and noninfectious diseases of the lung. Am. J. Respir. Crit. Care Med. 2010;181(12):1294–1309. doi: 10.1164/rccm.200909-1427SO. [DOI] [PubMed] [Google Scholar]
- 38.Mantovani A., Cassatella M.A., Costantini C., Jaillon S. Neutrophils in the activation and regulation of innate and adaptive immunity. Nat. Rev. Immunol. 2011;11(8):519–531. doi: 10.1038/nri3024. [DOI] [PubMed] [Google Scholar]
- 39.Imai Y., Kuba K., Neely G.G., Yaghubian-Malhami R., Perkmann T., van Loo G., Ermolaeva M., Veldhuizen R., Leung Y.H., Wang H., Liu H., Sun Y., Pasparakis M., Kopf M., Mech C., Bavari S., Peiris J.S., Slutsky A.S., Akira S., Hultqvist M., Holmdahl R., Nicholls J., Jiang C., Binder C.J., Penninger J.M. Identification of oxidative stress and Toll-like receptor 4 signaling as a key pathway of acute lung injury. Cell. 2008;133(2):235–249. doi: 10.1016/j.cell.2008.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hou Y.J., Okuda K., Edwards C.E., Martinez D.R., Asakura T., Dinnon K.H., 3rd, Kato T., Lee R.E., Yount B.L., Mascenik T.M., Chen G., Olivier K.N., Ghio A., Tse L.V., Leist S.R., Gralinski L.E., Schafer A., Dang H., Gilmore R., Nakano S., Sun L., Fulcher M.L., Livraghi-Butrico A., Nicely N.I., Cameron M., Cameron C., Kelvin D.J., de Silva A., Margolis D.M., Markmann A., Bartelt L., Zumwalt R., Martinez F.J., Salvatore S.P., Borczuk A., Tata P.R., Sontake V., Kimple A., Jaspers I., O’Neal W.K., Randell S.H., Boucher R.C., Baric R.S. SARS-CoV-2 reverse genetics reveals a variable infection gradient in the respiratory tract. Cell. 2020;182(2) doi: 10.1016/j.cell.2020.05.042. 429-446 e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wolfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Muller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., Hoelscher M., Bleicker T., Brunink S., Schneider J., Ehmann R., Zwirglmaier K., Drosten C., Wendtner C. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581(7809):465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 42.Nawijn M.C., Timens W. Can ACE2 expression explain SARS-CoV-2 infection of the respiratory epithelia in COVID-19? Mol. Syst. Biol. 2020;16(7):e9841. doi: 10.15252/msb.20209841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang H., Rostami M.R., Leopold P.L., Mezey J.G., O’Beirne S.L., Strulovici-Barel Y., Crystal R.G. Expression of the SARS-CoV-2 ACE2 receptor in the human airway epithelium. Am. J. Respir. Crit. Care Med. 2020;202(2):219–229. doi: 10.1164/rccm.202003-0541OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lazzerini P.E., Boutjdir M., Capecchi P.L. COVID-19, arrhythmic risk, and inflammation: mind the gap! Circulation. 2020;142(1):7–9. doi: 10.1161/CIRCULATIONAHA.120.047293. [DOI] [PubMed] [Google Scholar]
- 45.Godfred-Cato S., Bryant B., Leung J., Oster M.E., Conklin L., Abrams J., Roguski K., Wallace B., Prezzato E., Koumans E.H., Lee E.H., Geevarughese A., Lash M.K., Reilly K.H., Pulver W.P., Thomas D., Feder K.A., Hsu K.K., Plipat N., Richardson G., Reid H., Lim S., Schmitz A., Pierce T., Hrapcak S., Datta D., Morris S.B., Clarke K., Belay E. California MIS-C response Team COVID-19-Associated multisystem inflammatory syndrome in children - United States, march-july 2020. MMWR Morb. Mortal. Wkly. Rep. 2020;69(32):1074–1080. doi: 10.15585/mmwr.mm6932e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qian S., An J., Qi F., Ye L., Chen Q., Liu X., Xie L., Li G. Tocilizumab exerts anti-inflammatory activity in six critically ill COVID-19 patients: a retrospective analysis. Ann. Transl. Med. 2020;8(14):881. doi: 10.21037/atm-20-5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zheng J., Wang Y., Li K., Meyerholz D.K., Allamargot C., Perlman S. SARS-CoV-2-induced immune activation and death of monocyte-derived human macrophages and dendritic cells. J. Infect. Dis. 2020 doi: 10.1093/infdis/jiaa753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tay M.Z., Poh C.M., Renia L., MacAry P.A., Ng L.F.P. The trinity of COVID-19: immunity, inflammation and intervention. Nat. Rev. Immunol. 2020;20(6):363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mahmudpour M., Roozbeh J., Keshavarz M., Farrokhi S., Nabipour I. COVID-19 cytokine storm: the anger of inflammation. Cytokine. 2020;133 doi: 10.1016/j.cyto.2020.155151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.U R.A., Verma K. Happy hypoxemia in COVID-19-A neural hypothesis. ACS Chem. Neurosci. 2020;11(13):1865–1867. doi: 10.1021/acschemneuro.0c00318. [DOI] [PubMed] [Google Scholar]
- 51.Stukas S., Hoiland R.L., Cooper J., Thiara S., Griesdale D.E., Thomas A.D., Orde M.M., English J.C., Chen L.Y.C., Foster D., Mitra A.R., Romano K., Sweet D.D., Ronco J.J., Kanji H.D., Chen Y.R., Wong S.L., Wellington C.L., Sekhon M.S. The association of inflammatory cytokines in the pulmonary pathophysiology of respiratory failure in critically ill patients with coronavirus disease 2019. Crit Care Explor. 2020;2(9):e0203. doi: 10.1097/CCE.0000000000000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chmielik E., Jazowiecka-Rakus J., Dyduch G., Nasierowska-Guttmejer A., Michalowski L., Sochanik A., Ulatowska-Bialas M. COVID-19 Autopsies: A Case Series from Poland. Pathobiology. 2020:1–10. doi: 10.1159/000512768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Batah S.S., Fabro A.T. Pulmonary pathology of ARDS in COVID-19: a pathological review for clinicians. Respir. Med. 2020;176 doi: 10.1016/j.rmed.2020.106239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zuk J., Zaborowska D. Alkaline elution of yeast DNA: a simple method of detection of DNA single-strand breaks. Mutagenesis. 1987;2(3):229–234. doi: 10.1093/mutage/2.3.229. [DOI] [PubMed] [Google Scholar]
- 55.Ackermann M., Verleden S.E., Kuehnel M., Haverich A., Welte T., Laenger F., Vanstapel A., Werlein C., Stark H., Tzankov A., Li W.W., Li V.W., Mentzer S.J., Jonigk D. Pulmonary vascular endothelialitis, thrombosis, and angiogenesis in Covid-19. N. Engl. J. Med. 2020;383(2):120–128. doi: 10.1056/NEJMoa2015432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jung F., Kruger-Genge A., Franke R.P., Hufert F., Kupper J.H. COVID-19 and the endothelium. Clin. Hemorheol. Microcirc. 2020;75(1):7–11. doi: 10.3233/CH-209007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pons S., Fodil S., Azoulay E., Zafrani L. The vascular endothelium: the cornerstone of organ dysfunction in severe SARS-CoV-2 infection. Crit Care. 2020;24(1):353. doi: 10.1186/s13054-020-03062-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Varga Z., Flammer A.J., Steiger P., Haberecker M., Andermatt R., Zinkernagel A.S., Mehra M.R., Schuepbach R.A., Ruschitzka F., Moch H. Endothelial cell infection and endotheliitis in COVID-19. Lancet. 2020;395(10234):1417–1418. doi: 10.1016/S0140-6736(20)30937-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Noris M., Benigni A., Remuzzi G. The case of complement activation in COVID-19 multiorgan impact. Kidney Int. 2020;98(2):314–322. doi: 10.1016/j.kint.2020.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Winer L.K., Salyer C., Beckmann N., Caldwell C.C., Nomellini V. Enigmatic role of coagulopathy among sepsis survivors: a review of coagulation abnormalities and their possible link to chronic critical illness. Trauma Surg Acute Care Open. 2020;5(1) doi: 10.1136/tsaco-2020-000462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Levi M., Keller T.T. E. Van Gorp, H. Ten Cate, Infection and inflammation and the coagulation system. Cardiovasc. Res. 2003;60(1):26–39. doi: 10.1016/s0008-6363(02)00857-x. [DOI] [PubMed] [Google Scholar]
- 62.Hanley B., Naresh K.N., Roufosse C., Nicholson A.G., Weir J., Cooke G.S., Thursz M., Manousou P., Corbett R., Goldin R., Al-Sarraj S., Abdolrasouli A., Swann O.C., Baillon L., Penn R., Barclay W.S., Viola P., Osborn M. Histopathological findings and viral tropism in UK patients with severe fatal COVID-19: a post-mortem study. Lancet Microbe. 2020;1(6):e245–e253. doi: 10.1016/S2666-5247(20)30115-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bompard F., Monnier H., Saab I., Tordjman M., Abdoul H., Fournier L., Sanchez O., Lorut C., Chassagnon G., Revel M.P. Pulmonary embolism in patients with COVID-19 pneumonia. Eur. Respir. J. 2020;56(1) doi: 10.1183/13993003.01365-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Price L.C., McCabe C., Garfield B., Wort S.J. Thrombosis and COVID-19 pneumonia: the clot thickens! Eur. Respir. J. 2020;56(1) doi: 10.1183/13993003.01608-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Klok F.A., Kruip M., van der Meer N.J.M., Arbous M.S., Gommers D., Kant K.M., Kaptein F.H.J., van Paassen J., Stals M.A.M., Huisman M.V., Endeman H. Incidence of thrombotic complications in critically ill ICU patients with COVID-19. Thromb. Res. 2020;191:145–147. doi: 10.1016/j.thromres.2020.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McGonagle D., O’Donnell J.S., Sharif K., Emery P., Bridgewood C. Immune mechanisms of pulmonary intravascular coagulopathy in COVID-19 pneumonia. Lancet Rheumatol. 2020;2(7):e437–e445. doi: 10.1016/S2665-9913(20)30121-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rajendran P., Rengarajan T., Thangavel J., Nishigaki Y., Sakthisekaran D., Sethi G., Nishigaki I. The vascular endothelium and human diseases. Int. J. Biol. Sci. 2013;9(10):1057–1069. doi: 10.7150/ijbs.7502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Biswas I., Khan G.A. Coagulation disorders in COVID-19: role of toll-like receptors. J. Inflamm. Res. 2020;13:6. doi: 10.2147/JIR.S271768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shibamiya A., Hersemeyer K., Schmidt Woll T., Sedding D., Daniel J.M., Bauer S., Koyama T., Preissner K.T., Kanse S.M. A key role for Toll-like receptor-3 in disrupting the hemostasis balance on endothelial cells. Blood. 2009;113(3):714–722. doi: 10.1182/blood-2008-02-137901. [DOI] [PubMed] [Google Scholar]
- 70.Ding N., Chen G., Hoffman R., Loughran P.A., Sodhi C.P., Hackam D.J., Billiar T.R., Neal M.D. Toll-like receptor 4 regulates platelet function and contributes to coagulation abnormality and organ injury in hemorrhagic shock and resuscitation. Circ. Cardiovasc. Genet. 2014;7(5):615–624. doi: 10.1161/CIRCGENETICS.113.000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Buonvino S., Melino S. New Consensus pattern in Spike CoV-2: potential implications in coagulation process and cell-cell fusion. Cell Death Discov. 2020;6:134. doi: 10.1038/s41420-020-00372-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Greten F.R., Grivennikov S.I. Inflammation and Cancer: triggers, mechanisms, and consequences. Immunity. 2019;51(1):27–41. doi: 10.1016/j.immuni.2019.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Todoric J., Antonucci L., Karin M. Targeting inflammation in Cancer prevention and therapy. Cancer Prev. Res. Phila. (Phila) 2016;9(12):895–905. doi: 10.1158/1940-6207.CAPR-16-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kamp D.W., Shacter E., Weitzman S.A. Chronic inflammation and cancer: the role of the mitochondria. Oncology (Williston Park, N.Y.) 2011;25(5) 400-10, 413. [PubMed] [Google Scholar]
- 75.Ahmad A. Epigenetics in personalized management of lung Cancer. Adv. Exp. Med. Biol. 2016;890:111–122. doi: 10.1007/978-3-319-24932-2_6. [DOI] [PubMed] [Google Scholar]
- 76.Bade B.C., Dela Cruz C.S. lung Cancer 2020: epidemiology, etiology, and prevention. Clin. Chest Med. 2020;41(1):1–24. doi: 10.1016/j.ccm.2019.10.001. [DOI] [PubMed] [Google Scholar]
- 77.Yao H., Rahman I. Current concepts on the role of inflammation in COPD and lung cancer. Curr. Opin. Pharmacol. 2009;9(4):375–383. doi: 10.1016/j.coph.2009.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Young R.P., Hopkins R.J., Christmas T., Black P.N., Metcalf P., Gamble G.D. COPD prevalence is increased in lung cancer, independent of age, sex and smoking history. Eur. Respir. J. 2009;34(2):380–386. doi: 10.1183/09031936.00144208. [DOI] [PubMed] [Google Scholar]
- 79.Bacha S., Mejdoub S., Sghaier A., Sonia H., Racil H., Cheikhrouhou S., Chaouch N., Chabbou A., Megdiche M.L. Advanced lung cancer inflammation index (ALI): a new pronostic score in patients with non small cell lung cancer. Eur. Respir. J. 2016;48(suppl 60) PA518. [Google Scholar]
- 80.Deng C., Zhang N., Wang Y., Jiang S., Lu M., Huang Y., Ma J., Hu C., Hou T. High systemic immune-inflammation index predicts poor prognosis in advanced lung adenocarcinoma patients treated with EGFR-TKIs. Medicine (Baltimore) 2019;98(33) doi: 10.1097/MD.0000000000016875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ahmad A., Maitah M.Y., Ginnebaugh K.R., Li Y., Bao B., Gadgeel S.M., Sarkar F.H. Inhibition of Hedgehog signaling sensitizes NSCLC cells to standard therapies through modulation of EMT-regulating miRNAs. J. Hematol. Oncol. 2013;6(1):77. doi: 10.1186/1756-8722-6-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tang J., Salama R., Gadgeel S.M., Sarkar F.H., Ahmad A. Erlotinib resistance in lung cancer: current progress and future perspectives. Front. Pharmacol. 2013;4:15. doi: 10.3389/fphar.2013.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.O’Callaghan D.S., O’Donnell D., O’Connell F., O’Byrne K.J. The role of inflammation in the pathogenesis of non-small cell lung cancer. J. Thorac. Oncol. 2010;5(12):2024–2036. doi: 10.1097/jto.0b013e3181f387e4. [DOI] [PubMed] [Google Scholar]
- 84.Simon L.S. Role and regulation of cyclooxygenase-2 during inflammation. Am. J. Med. 1999;106(5B):37S–42S. doi: 10.1016/s0002-9343(99)00115-1. [DOI] [PubMed] [Google Scholar]
- 85.Castelao J.E., Bart R.D., 3rd, DiPerna C.A., Sievers E.M., Bremner R.M. Lung cancer and cyclooxygenase-2. Ann. Thorac. Surg. 2003;76(4):1327–1335. doi: 10.1016/s0003-4975(03)00334-5. [DOI] [PubMed] [Google Scholar]
- 86.Eruslanov E.B., Bhojnagarwala P.S., Quatromoni J.G., Stephen T.L., Ranganathan A., Deshpande C., Akimova T., Vachani A., Litzky L., Hancock W.W., Conejo-Garcia J.R., Feldman M., Albelda S.M., Singhal S. Tumor-associated neutrophils stimulate T cell responses in early-stage human lung cancer. J. Clin. Invest. 2014;124(12):5466–5480. doi: 10.1172/JCI77053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gabay C. Interleukin-6 and chronic inflammation. Arthritis Res. Ther. 2006;8(Suppl 2):S3. doi: 10.1186/ar1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ahmad S., Ahmad A. Emerging targets for treating sulfur mustard-induced injuries. Ann. N. Y. Acad. Sci. 2016;1374(1):123–131. doi: 10.1111/nyas.13095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rana T., Ahmad A., Zafar I., Mariappan N., Chandrashekar D.S., Hamid T., Husain M., Varambally S., Ahmad S., Ahmad A. MicroRNA-mediated inflammation and coagulation effects in rats exposed to an inhaled analog of sulfur mustard. Ann. N. Y. Acad. Sci. 2020 doi: 10.1111/nyas.14416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shang G.S., Liu L., Qin Y.W. IL-6 and TNF-alpha promote metastasis of lung cancer by inducing epithelial-mesenchymal transition. Oncol. Lett. 2017;13(6):4657–4660. doi: 10.3892/ol.2017.6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rice S.J., Liu X., Zhang J., Jia B., Zheng H., Belani C.P. Advanced NSCLC patients with high IL-6 levels have altered peripheral T cell population and signaling. Lung Cancer. 2019;131:58–61. doi: 10.1016/j.lungcan.2019.03.014. [DOI] [PubMed] [Google Scholar]
- 92.Ogawa H., Koyanagi-Aoi M., Otani K., Zen Y., Maniwa Y., Aoi T. Interleukin-6 blockade attenuates lung cancer tissue construction integrated by cancer stem cells. Sci. Rep. 2017;7(1):12317. doi: 10.1038/s41598-017-12017-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.van Dam P.A., Huizing M., Mestach G., Dierckxsens S., Tjalma W., Trinh X.B., Papadimitriou K., Altintas S., Vermorken J., Vulsteke C., Janssens A., Berneman Z., Prenen H., Meuris L., Vanden Berghe W., Smits E., Peeters M. SARS-CoV-2 and cancer: are they really partners in crime? Cancer Treat. Rev. 2020;89 doi: 10.1016/j.ctrv.2020.102068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dai M.Y., Chen Z., Leng Y., Wu M., Liu Y., Zhou F., Ming C., Shao N., Liu M., Cai H. Patients with lung Cancer Have high susceptibility of COVID-19: a retrospective study in Wuhan, China. Cancer Control. 2020;27(1) doi: 10.1177/1073274820960467. 1073274820960467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gupta I., Rizeq B., Elkord E., Vranic S., Al Moustafa A.E. SARS-CoV-2 Infection and Lung Cancer: Potential Therapeutic Modalities. Cancers (Basel) 2020;12(8) doi: 10.3390/cancers12082186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kong Q., Xiang Z., Wu Y., Gu Y., Guo J., Geng F. Analysis of the susceptibility of lung cancer patients to SARS-CoV-2 infection. Mol. Cancer. 2020;19(1):80. doi: 10.1186/s12943-020-01209-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Calles A., Aparicio M.I., Alva M., Bringas M., Gutierrez N., Soto J., Arregui M., Tirado V.C., Alvarez E.L., Del Monte-Millan M., Massarrah T., Galera M., Alvarez R., Martin M. Outcomes of COVID-19 in patients with lung Cancer Treated in a tertiary hospital in Madrid. Front. Oncol. 2020;10:1777. doi: 10.3389/fonc.2020.01777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Baek M.S., Kim W.Y., Lee K.J., Noh C.S. Detection of severe acute respiratory syndrome coronavirus 2 in the pleural fluid. Infect. Chemother. 2020 doi: 10.3947/ic.2020.0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Thunders M., Delahunt B. Gene of the month: TMPRSS2 (transmembrane serine protease 2) J. Clin. Pathol. 2020 doi: 10.1136/jclinpath-2020-206987. [DOI] [PubMed] [Google Scholar]
- 100.Zhang H., Quek K., Chen R., Chen J., Chen B. Expression of the SAR2-Cov-2 receptor ACE2 reveals the susceptibility of COVID-19 in non-small cell lung cancer. J. Cancer. 2020;11(18):5289–5292. doi: 10.7150/jca.49462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Li Y., Xu Q., Ma L., Wu D., Gao J., Chen G., Li H. Systematic profiling of ACE2 expression in diverse physiological and pathological conditions for COVID-19/SARS-CoV-2. J. Cell. Mol. Med. 2020 doi: 10.1111/jcmm.15607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ziegler C.G.K., Allon S.J., Nyquist S.K., Mbano I.M., Miao V.N., Tzouanas C.N., Cao Y., Yousif A.S., Bals J., Hauser B.M., Feldman J., Muus C., Wadsworth M.H., 2nd, Kazer S.W., Hughes T.K., Doran B., Gatter G.J., Vukovic M., Taliaferro F., Mead B.E., Guo Z., Wang J.P., Gras D., Plaisant M., Ansari M., Angelidis I., Adler H., Sucre J.M.S., Taylor C.J., Lin B., Waghray A., Mitsialis V., Dwyer D.F., Buchheit K.M., Boyce J.A., Barrett N.A., Laidlaw T.M., Carroll S.L., Colonna L., Tkachev V., Peterson C.W., Yu A., Zheng H.B., Gideon H.P., Winchell C.G., Lin P.L., Bingle C.D., Snapper S.B., Kropski J.A., Theis F.J., Schiller H.B., Zaragosi L.E., Barbry P., Leslie A., Kiem H.P., Flynn J.L., Fortune S.M., Berger B., Finberg R.W., Kean L.S., Garber M., Schmidt A.G., Lingwood D., Shalek A.K., Ordovas-Montanes J. H.C.A.L.B.N.E.a. lung-network@humancellatlas.org, H.C.A.L.B. network, SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell. 2020;181(5) doi: 10.1016/j.cell.2020.04.035. 1016-1035 e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Radzikowska U., Ding M., Tan G., Zhakparov D., Peng Y., Wawrzyniak P., Wang M., Li S., Morita H., Altunbulakli C., Reiger M., Neumann A.U., Lunjani N., Traidl-Hoffmann C., Nadeau K., O’Mahony L., Akdis C.A., Sokolowska M. Distribution of ACE2, CD147, CD26 and other SARS-CoV-2 associated molecules in tissues and immune cells in health and in asthma, COPD, obesity, hypertension, and COVID-19 risk factors. Allergy. 2020 doi: 10.1111/all.14429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhang Q., Yue Y., Tan H., Liu Y., Zeng Y., Xiao L. Single cell RNA-seq data analysis reveals the potential risk of SARS-CoV-2 infection among different respiratory system conditions. Front. Genet. 2020;11:942. doi: 10.3389/fgene.2020.00942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Muller M.A., Drosten C., Pohlmann S. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181(2) doi: 10.1016/j.cell.2020.02.052. 271-280 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Graff R.E., Judson G., Ahearn T.U., Fiorentino M., Loda M., Giovannucci E.L., Mucci L.A., Pettersson A. Circulating antioxidant levels and risk of prostate Cancer by TMPRSS2:ERG. Prostate. 2017;77(6):647–653. doi: 10.1002/pros.23312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Schuler B.A., Habermann A.C., Plosa E.J., Taylor C.J., Jetter C., Kapp M.E., Benjamin J.T., Gulleman P., Nichols D.S., Braunstein L.Z., Koval M., Guttentag S.H., Blackwell T.S., Vanderbilt C.-C.C., Webber S.A., Banovich N.E., Kropski J.A., Sucre J.M.S., Network H.C.A.L.B. Age-related expression of SARS-CoV-2 priming protease TMPRSS2 in the developing lung. bioRxiv. 2020 [Google Scholar]
- 108.Kamboj M., Sepkowitz K.A. Nosocomial infections in patients with cancer. Lancet Oncol. 2009;10(6):589–597. doi: 10.1016/S1470-2045(09)70069-5. [DOI] [PubMed] [Google Scholar]
- 109.Liang W., Guan W., Chen R., Wang W., Li J., Xu K., Li C., Ai Q., Lu W., Liang H., Li S., He J. Cancer patients in SARS-CoV-2 infection: a nationwide analysis in China. Lancet Oncol. 2020;21(3):335–337. doi: 10.1016/S1470-2045(20)30096-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Shereen M.A., Khan S., Kazmi A., Bashir N., Siddique R. COVID-19 infection: origin, transmission, and characteristics of human coronaviruses. J. Adv. Res. 2020;24:91–98. doi: 10.1016/j.jare.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Song J., Zeng M., Wang H., Qin C., Hou H.Y., Sun Z.Y., Xu S.P., Wang G.P., Guo C.L., Deng Y.K., Wang Z.C., Ma J., Pan L., Liao B., Du Z.H., Feng Q.M., Liu Y., Xie J.G., Liu Z. Distinct effects of asthma and COPD comorbidity on disease expression and outcome in patients with COVID-19. Allergy. 2020 doi: 10.1111/all.14517. [DOI] [PubMed] [Google Scholar]
- 112.Zhang L., Han X., Shi Y. Comparative analysis of SARS-CoV-2 receptor ACE2 expression in multiple solid tumors and matched non-diseased tissues. Infect. Genet. Evol. 2020;85 doi: 10.1016/j.meegid.2020.104428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhang Z., Li L., Li M., Wang X. The SARS-CoV-2 host cell receptor ACE2 correlates positively with immunotherapy response and is a potential protective factor for cancer progression. Comput. Struct. Biotechnol. J. 2020;18:2438–2444. doi: 10.1016/j.csbj.2020.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Verdecchia P., Cavallini C., Spanevello A., Angeli F. The pivotal link between ACE2 deficiency and SARS-CoV-2 infection. Eur. J. Intern. Med. 2020;76:14–20. doi: 10.1016/j.ejim.2020.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tikoo K., Patel G., Kumar S., Karpe P.A., Sanghavi M., Malek V., Srinivasan K. Tissue specific up regulation of ACE2 in rabbit model of atherosclerosis by atorvastatin: role of epigenetic histone modifications. Biochem. Pharmacol. 2015;93(3):343–351. doi: 10.1016/j.bcp.2014.11.013. [DOI] [PubMed] [Google Scholar]
- 116.Towler P., Staker B., Prasad S.G., Menon S., Tang J., Parsons T., Ryan D., Fisher M., Williams D., Dales N.A., Patane M.A., Pantoliano M.W. ACE2 X-ray structures reveal a large hinge-bending motion important for inhibitor binding and catalysis. J. Biol. Chem. 2004;279(17):17996–18007. doi: 10.1074/jbc.M311191200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sinha S., Cheng K., Schaffer A.A., Aldape K., Schiff E., Ruppin E. In vitro and in vivo identification of clinically approved drugs that modify ACE2 expression. Mol. Syst. Biol. 2020;16(7):e9628. doi: 10.15252/msb.20209628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.van Wersch J.W., Tjwa M.K. Coagulation/fibrinolysis balance and lung cancer. Haemostasis. 1991;21(2):117–123. doi: 10.1159/000216214. [DOI] [PubMed] [Google Scholar]
- 119.Zabczyk M., Krolczyk G., Czyzewicz G., Plens K., Prior S., Butenas S., Undas A. Altered fibrin clot properties in advanced lung cancer: strong impact of cigarette smoking. Med. Oncol. 2019;36(4):37. doi: 10.1007/s12032-019-1262-4. [DOI] [PubMed] [Google Scholar]
- 120.Cavaliere L. Thromboprophylaxis in ambulatory lung cancer treatment. Clin. J. Oncol. Nurs. 2013;17(1):74–79. doi: 10.1188/13.CJON.74-79. [DOI] [PubMed] [Google Scholar]
- 121.Hammouda A., Souilah S., Ferhat-Hamida M.Y., Amir Z.C., Aouichat-Bouguerra S., Hariti G. Activation of coagulation in patients with lung cancer. Ann Biol Clin (Paris) 2019;77(3):272–280. doi: 10.1684/abc.2019.1445. [DOI] [PubMed] [Google Scholar]
- 122.Sotiropoulos G.P., Kotopouli M., Karampela I., Christodoulatos G.S., Antonakos G., Marinou I., Vogiatzakis E., Lekka A., Papavassiliou A.G., Dalamaga M. Circulating plasminogen activator inhibitor-1 activity: a biomarker for resectable non-small cell lung cancer? J. BUON. 2019;24(3):943–954. [PubMed] [Google Scholar]
- 123.Nougier C., Benoit R., Simon M., Desmurs-Clavel H., Marcotte G., Argaud L., David J.S., Bonnet A., Negrier C., Dargaud Y. Hypofibrinolytic state and high thrombin generation may play a major role in SARS-COV2 associated thrombosis. J. Thromb. Haemost. 2020 doi: 10.1111/jth.15016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kang S., Tanaka T., Inoue H., Ono C., Hashimoto S., Kioi Y., Matsumoto H., Matsuura H., Matsubara T., Shimizu K., Ogura H., Matsuura Y., Kishimoto T. IL-6 trans-signaling induces plasminogen activator inhibitor-1 from vascular endothelial cells in cytokine release syndrome. Proc Natl Acad Sci U S A. 2020;117(36):22351–22356. doi: 10.1073/pnas.2010229117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zuo Y., Warnock M., Harbaugh A., Yalavarthi S., Gockman K., Zuo M., Madison J.A., Knight J.S., Kanthi Y., Lawrence D.A. Plasma tissue plasminogen activator and plasminogen activator inhibitor-1 in hospitalized COVID-19 patients. medRxiv. 2020 doi: 10.1038/s41598-020-80010-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zhu C., Shen H., Zhu L., Zhao F., Shu Y. Plasminogen activator inhibitor 1 promotes immunosuppression in human non-small cell lung cancers by enhancing TGF-Beta1 expression in macrophage. Cell. Physiol. Biochem. 2017;44(6):2201–2211. doi: 10.1159/000486025. [DOI] [PubMed] [Google Scholar]
- 127.Dass K., Ahmad A., Azmi A.S., Sarkar S.H., Sarkar F.H. Evolving role of uPA/uPAR system in human cancers. Cancer Treat. Rev. 2008;34(2):122–136. doi: 10.1016/j.ctrv.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 128.Li S., Wei X., He J., Tian X., Yuan S., Sun L. Plasminogen activator inhibitor-1 in cancer research. Biomed. Pharmacother. 2018;105:83–94. doi: 10.1016/j.biopha.2018.05.119. [DOI] [PubMed] [Google Scholar]
- 129.Matsuyama T., Kubli S.P., Yoshinaga S.K., Pfeffer K., Mak T.W. An aberrant STAT pathway is central to COVID-19. Cell Death Differ. 2020 doi: 10.1038/s41418-020-00633-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Dutta P., Sabri N., Li J., Li W.X. Role of STAT3 in lung cancer. JAKSTAT. 2014;3(4) doi: 10.1080/21623996.2014.999503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ilikci Sagkan R., Akin-Bali D.F. Structural variations and expression profiles of the SARS-CoV-2 host invasion genes in lung cancer. J. Med. Virol. 2020 doi: 10.1002/jmv.26107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Huang W.T., Yang X., He R.Q., Ma J., Hu X.H., Mo W.J., Chen G. Overexpressed BSG related to the progression of lung adenocarcinoma with high-throughput data-mining, immunohistochemistry, in vitro validation and in silico investigation. Am. J. Transl. Res. 2019;11(8):4835–4850. [PMC free article] [PubMed] [Google Scholar]
- 133.Matsumoto T., Nagashio R., Ryuge S., Igawa S., Kobayashi M., Fukuda E., Goshima N., Ichinoe M., Jiang S.X., Satoh Y., Masuda N., Murakumo Y., Saegusa M., Sato Y. Basigin expression as a prognostic indicator in stage I pulmonary adenocarcinoma. Pathol. Int. 2018;68(4):232–240. doi: 10.1111/pin.12646. [DOI] [PubMed] [Google Scholar]
- 134.Zhang X., Tian T., Zhang X., Liu C., Fang X. Elevated CD147 expression is associated with shorter overall survival in non-small cell lung cancer. Oncotarget. 2017;8(23):37673–37680. doi: 10.18632/oncotarget.16948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Belton R.J., Jr, Chen L., Mesquita F.S., Nowak R.A. Basigin-2 is a cell surface receptor for soluble basigin ligand. J. Biol. Chem. 2008;283(26):17805–17814. doi: 10.1074/jbc.M801876200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Liao C.G., Yao L., Xie W., Liu L., Wu S.D., Lu N., Huang J.G., Kong L.M., Zhang H.L. Basigin-2 upregulated by receptor activator of NF-kappaB ligand enhances lung cancer-induced osteolytic lesions. Cancer Cell Int. 2016;16:28. doi: 10.1186/s12935-016-0302-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Xia P., Dubrovska A. Tumor markers as an entry for SARS-CoV-2 infection? FEBS J. 2020 doi: 10.1111/febs.15499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ulrich H., Pillat M.M. CD147 as a target for COVID-19 treatment: suggested effects of azithromycin and stem cell engagement. Stem Cell Rev Rep. 2020;16(3):434–440. doi: 10.1007/s12015-020-09976-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Shilts J., Wright G.J. No evidence for basigin/CD147 as a direct SARS-CoV-2 spike binding receptor. bioRxiv. 2020 doi: 10.1038/s41598-020-80464-1. 2020.07.25.221036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Bestle D., Heindl M.R., Limburg H., Van Lam van T., Pilgram O., Moulton H., Stein D.A., Hardes K., Eickmann M., Dolnik O., Rohde C., Klenk H.D., Garten W., Steinmetzer T., Bottcher-Friebertshauser E. TMPRSS2 and furin are both essential for proteolytic activation of SARS-CoV-2 in human airway cells. Life Sci Alliance. 2020;3(9) doi: 10.26508/lsa.202000786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Johnson B.A., Xie X., Kalveram B., Lokugamage K.G., Muruato A., Zou J., Zhang X., Juelich T., Smith J.K., Zhang L., Bopp N., Schindewolf C., Vu M., Vanderheiden A., Swetnam D., Plante J.A., Aguilar P., Plante K.S., Lee B., Weaver S.C., Suthar M.S., Routh A.L., Ren P., Ku Z., An Z., Debbink K., Shi P.Y., Freiberg A.N., Menachery V.D. Furin cleavage site is key to SARS-CoV-2 pathogenesis. bioRxiv. 2020 doi: 10.1038/s41586-021-03237-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ord M., Faustova I., Loog M. The sequence at Spike S1/S2 site enables cleavage by furin and phospho-regulation in SARS-CoV2 but not in SARS-CoV1 or MERS-CoV. Sci. Rep. 2020;10(1):16944. doi: 10.1038/s41598-020-74101-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Cheng Y.W., Chao T.L., Li C.L., Chiu M.F., Kao H.C., Wang S.H., Pang Y.H., Lin C.H., Tsai Y.M., Lee W.H., Tao M.H., Ho T.C., Wu P.Y., Jang L.T., Chen P.J., Chang S.Y., Yeh S.H. Furin inhibitors block SARS-CoV-2 spike protein cleavage to suppress virus production and cytopathic effects. Cell Rep. 2020;33(2) doi: 10.1016/j.celrep.2020.108254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kaur U., Chakrabarti S.S., Ojha B., Pathak B.K., Singh A., Saso L., Chakrabarti S. Targeting host cell proteases to prevent SARS-CoV-2 invasion. Curr. Drug Targets. 2020 doi: 10.2174/1389450121666200924113243. [DOI] [PubMed] [Google Scholar]
- 145.Mbikay M., Sirois F., Yao J., Seidah N.G., Chretien M. Comparative analysis of expression of the proprotein convertases furin, PACE4, PC1 and PC2 in human lung tumours. Br. J. Cancer. 1997;75(10):1509–1514. doi: 10.1038/bjc.1997.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Bassi D.E., Mahloogi H., Al-Saleem L., Lopez De Cicco R., Ridge J.A., Klein-Szanto A.J. Elevated furin expression in aggressive human head and neck tumors and tumor cell lines. Mol. Carcinog. 2001;31(4):224–232. doi: 10.1002/mc.1057. [DOI] [PubMed] [Google Scholar]
- 147.Wu C., Zheng M., Yang Y., Gu X., Yang K., Li M., Liu Y., Zhang Q., Zhang P., Wang Y., Wang Q., Xu Y., Zhou Y., Zhang Y., Chen L., Li H. Furin: A Potential Therapeutic Target for COVID-19. iScience. 2020;23(10) doi: 10.1016/j.isci.2020.101642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Bassi D.E., Zhang J., Renner C., Klein-Szanto A.J. Targeting proprotein convertases in furin-rich lung cancer cells results in decreased in vitro and in vivo growth. Mol. Carcinog. 2017;56(3):1182–1188. doi: 10.1002/mc.22550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Ma Y.C., Fan W.J., Rao S.M., Gao L., Bei Z.Y., Xu S.T. Effect of Furin inhibitor on lung adenocarcinoma cell growth and metastasis. Cancer Cell Int. 2014;14:43. doi: 10.1186/1475-2867-14-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Lahousse L. Epigenetic targets for lung diseases. EBioMedicine. 2019;43:24–25. doi: 10.1016/j.ebiom.2019.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Hussain S.P., Harris C.C. Inflammation and cancer: an ancient link with novel potentials. Int. J. Cancer. 2007;121(11):2373–2380. doi: 10.1002/ijc.23173. [DOI] [PubMed] [Google Scholar]
- 152.Selamat S.A., Chung B.S., Girard L., Zhang W., Zhang Y., Campan M., Siegmund K.D., Koss M.N., Hagen J.A., Lam W.L., Lam S., Gazdar A.F., Laird-Offringa I.A. Genome-scale analysis of DNA methylation in lung adenocarcinoma and integration with mRNA expression. Genome Res. 2012;22(7) doi: 10.1101/gr.132662.111. 1197-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Tekpli X., Landvik N.E., Anmarkud K.H., Skaug V., Haugen A., Zienolddiny S. DNA methylation at promoter regions of interleukin 1B, interleukin 6, and interleukin 8 in non-small cell lung cancer. Cancer Immunol. Immunother. 2013;62(2):337–345. doi: 10.1007/s00262-012-1340-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Qiu W., Baccarelli A., Carey V.J., Boutaoui N., Bacherman H., Klanderman B., Rennard S., Agusti A., Anderson W., Lomas D.A., DeMeo D.L. Variable DNA methylation is associated with chronic obstructive pulmonary disease and lung function. Am. J. Respir. Crit. Care Med. 2012;185(4):373–381. doi: 10.1164/rccm.201108-1382OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Bermingham M.L., Walker R.M., Marioni R.E., Morris S.W., Rawlik K., Zeng Y., Campbell A., Redmond P., Whalley H.C., Adams M.J., Hayward C., Deary I.J., Porteous D.J., McIntosh A.M., Evans K.L. Identification of novel differentially methylated sites with potential as clinical predictors of impaired respiratory function and COPD. EBioMedicine. 2019;43:576–586. doi: 10.1016/j.ebiom.2019.03.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Um S.W., Kim Y., Lee B.B., Kim D., Lee K.J., Kim H.K., Han J., Kim H., Shim Y.M., Kim D.H. Genome-wide analysis of DNA methylation in bronchial washings. Clin. Epigenetics. 2018;10:65. doi: 10.1186/s13148-018-0498-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Yang Y., Haitchi H.M., Cakebread J., Sammut D., Harvey A., Powell R.M., Holloway J.W., Howarth P., Holgate S.T., Davies D.E. Epigenetic mechanisms silence a disintegrin and metalloprotease 33 expression in bronchial epithelial cells. J. Allergy Clin. Immunol. 2008;121(6) doi: 10.1016/j.jaci.2008.02.031. 1393-9, 1399 e1-14. [DOI] [PubMed] [Google Scholar]
- 158.Bae D.J., Jun J.A., Chang H.S., Park J.S., Park C.S. Epigenetic changes in asthma: role of DNA CpG methylation. Tuberc. Respir. Dis. (Seoul) 2020;83(1):1–13. doi: 10.4046/trd.2018.0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Montrose L., Ward T.J., Semmens E.O., Cho Y.H., Brown B., Noonan C.W. Dietary intake is associated with respiratory health outcomes and DNA methylation in children with asthma. Allergy Asthma Clin. Immunol. 2017;13:12. doi: 10.1186/s13223-017-0187-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Sharma S., Litonjua A. Asthma, allergy, and responses to methyl donor supplements and nutrients. J. Allergy Clin. Immunol. 2014;133(5):1246–1254. doi: 10.1016/j.jaci.2013.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Szpechcinski A., Chorostowska-Wynimko J., Struniawski R., Kupis W., Rudzinski P., Langfort R., Puscinska E., Bielen P., Sliwinski P., Orlowski T. Cell-free DNA levels in plasma of patients with non-small-cell lung cancer and inflammatory lung disease. Br. J. Cancer. 2015;113(3):476–483. doi: 10.1038/bjc.2015.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Avriel A., Rozenberg D., Raviv Y., Heimer D., Bar-Shai A., Gavish R., Sheynin J., Douvdevani A. Prognostic utility of admission cell-free DNA levels in patients with chronic obstructive pulmonary disease exacerbations. Int. J. Chron. Obstruct. Pulmon. Dis. 2016;11:3153–3161. doi: 10.2147/COPD.S113256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Durham A.L., Adcock I.M. The relationship between COPD and lung cancer. Lung Cancer. 2015;90(2):121–127. doi: 10.1016/j.lungcan.2015.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Wielscher M., Vierlinger K., Kegler U., Ziesche R., Gsur A., Weinhausel A. Diagnostic performance of plasma DNA methylation profiles in lung Cancer, Pulmonary fibrosis and COPD. EBioMedicine. 2015;2(8):929–936. doi: 10.1016/j.ebiom.2015.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Sundar I.K., Yin Q., Baier B.S., Yan L., Mazur W., Li D., Susiarjo M., Rahman I. DNA methylation profiling in peripheral lung tissues of smokers and patients with COPD. Clin. Epigenetics. 2017;9:38. doi: 10.1186/s13148-017-0335-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Sun N., Wei X., Wang J., Cheng Z., Sun W. Caveolin-1 promotes the imbalance of Th17/Treg in patients with chronic obstructive pulmonary disease. Inflammation. 2016;39(6):2008–2015. doi: 10.1007/s10753-016-0436-x. [DOI] [PubMed] [Google Scholar]
- 167.Zhang S., Wu Z., Xie J., Yang Y., Wang L., Qiu H. DNA methylation exploration for ARDS: a multi-omics and multi-microarray interrelated analysis. J. Transl. Med. 2019;17(1):345. doi: 10.1186/s12967-019-2090-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Seiler C.L., Song J.U.M., Kotandeniya D., Chen J., Kono T.J.Y., Han Q., Colwell M., Auch B., Sarver A.L., Upadhyaya P., Ren Y., Faulk C., De Flora S., La Maestra S., Chen Y., Kassie F., Tretyakova N.Y. Inhalation exposure to cigarette smoke and inflammatory agents induces epigenetic changes in the lung. Sci. Rep. 2020;10(1):11290. doi: 10.1038/s41598-020-67502-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Li R., Ong S.L., Tran L.M., Jing Z., Liu B., Park S.J., Huang Z.L., Walser T.C., Heinrich E.L., Lee G., Salehi-Rad R., Crosson W.P., Pagano P.C., Paul M.K., Xu S., Herschman H., Krysan K., Dubinett S. Chronic IL-1beta-induced inflammation regulates epithelial-to-mesenchymal transition memory phenotypes via epigenetic modifications in non-small cell lung cancer. Sci. Rep. 2020;10(1):377. doi: 10.1038/s41598-019-57285-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Glozak M.A., Sengupta N., Zhang X., Seto E. Acetylation and deacetylation of non-histone proteins. Gene. 2005;363:15–23. doi: 10.1016/j.gene.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 171.Ito K., Charron C.E., Adcock I.M. Impact of protein acetylation in inflammatory lung diseases. Pharmacol. Ther. 2007;116(2):249–265. doi: 10.1016/j.pharmthera.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 172.Nicodeme E., Jeffrey K.L., Schaefer U., Beinke S., Dewell S., Chung C.W., Chandwani R., Marazzi I., Wilson P., Coste H., White J., Kirilovsky J., Rice C.M., Lora J.M., Prinjha R.K., Lee K., Tarakhovsky A. Suppression of inflammation by a synthetic histone mimic. Nature. 2010;468(7327):1119–1123. doi: 10.1038/nature09589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Tian B., Liu Z., Litvinov J., Maroto R., Jamaluddin M., Rytting E., Patrikeev I., Ochoa L., Vargas G., Motamedi M., Ameredes B.T., Zhou J., Brasier A.R. Efficacy of novel highly specific bromodomain-containing protein 4 inhibitors in innate inflammation-driven airway remodeling. Am. J. Respir. Cell Mol. Biol. 2019;60(1):68–83. doi: 10.1165/rcmb.2017-0445OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Devaiah B.N., Gegonne A., Singer D.S. Bromodomain 4: a cellular Swiss army knife. J. Leukoc. Biol. 2016;100(4):679–686. doi: 10.1189/jlb.2RI0616-250R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Khan Y.M., Kirkham P., Barnes P.J., Adcock I.M. Brd4 is essential for IL-1beta-induced inflammation in human airway epithelial cells. PLoS One. 2014;9(4) doi: 10.1371/journal.pone.0095051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Tian B., Yang J., Zhao Y., Ivanciuc T., Sun H., Garofalo R.P., Brasier A.R. BRD4 couples NF-kappaB/RelA with airway inflammation and the IRF-RIG-I amplification loop in respiratory syncytial virus infection. J. Virol. 2017;91(6) doi: 10.1128/JVI.00007-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Ito K., Barnes P.J., Adcock I.M. Glucocorticoid receptor recruitment of histone deacetylase 2 inhibits interleukin-1beta-induced histone H4 acetylation on lysines 8 and 12. Mol. Cell. Biol. 2000;20(18):6891–6903. doi: 10.1128/mcb.20.18.6891-6903.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Yoshida M., Matsuyama A., Komatsu Y., Nishino N. From discovery to the coming generation of histone deacetylase inhibitors. Curr. Med. Chem. 2003;10(22):2351–2358. doi: 10.2174/0929867033456602. [DOI] [PubMed] [Google Scholar]
- 179.Cosio B.G., Tsaprouni L., Ito K., Jazrawi E., Adcock I.M., Barnes P.J. Theophylline restores histone deacetylase activity and steroid responses in COPD macrophages. J. Exp. Med. 2004;200(5):689–695. doi: 10.1084/jem.20040416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Ito K., Caramori G., Lim S., Oates T., Chung K.F., Barnes P.J., Adcock I.M. Expression and activity of histone deacetylases in human asthmatic airways. Am. J. Respir. Crit. Care Med. 2002;166(3):392–396. doi: 10.1164/rccm.2110060. [DOI] [PubMed] [Google Scholar]
- 181.Cosio B.G., Mann B., Ito K., Jazrawi E., Barnes P.J., Chung K.F., Adcock I.M. Histone acetylase and deacetylase activity in alveolar macrophages and blood mononocytes in asthma. Am. J. Respir. Crit. Care Med. 2004;170(2):141–147. doi: 10.1164/rccm.200305-659OC. [DOI] [PubMed] [Google Scholar]
- 182.Hogg J.C., Chu F., Utokaparch S., Woods R., Elliott W.M., Buzatu L., Cherniack R.M., Rogers R.M., Sciurba F.C., Coxson H.O., Pare P.D. The nature of small-airway obstruction in chronic obstructive pulmonary disease. N. Engl. J. Med. 2004;350(26):2645–2653. doi: 10.1056/NEJMoa032158. [DOI] [PubMed] [Google Scholar]
- 183.Barnes P.J., Adcock I.M., Ito K. Histone acetylation and deacetylation: importance in inflammatory lung diseases. Eur. Respir. J. 2005;25(3):552–563. doi: 10.1183/09031936.05.00117504. [DOI] [PubMed] [Google Scholar]
- 184.Marwick J.A., Kirkham P.A., Stevenson C.S., Danahay H., Giddings J., Butler K., Donaldson K., Macnee W., Rahman I. Cigarette smoke alters chromatin remodeling and induces proinflammatory genes in rat lungs. Am. J. Respir. Cell Mol. Biol. 2004;31(6):633–642. doi: 10.1165/rcmb.2004-0006OC. [DOI] [PubMed] [Google Scholar]
- 185.Ito K., Lim S., Caramori G., Chung K.F., Barnes P.J., Adcock I.M. Cigarette smoking reduces histone deacetylase 2 expression, enhances cytokine expression, and inhibits glucocorticoid actions in alveolar macrophages. FASEB J. 2001;15(6):1110–1112. [PubMed] [Google Scholar]
- 186.Rimstad E., Poppe T., Evensen O., Hyllseth B. Inoculation of infectious pancreatic necrosis virus serotype Sp did not cause pancreas disease in Atlantic salmon (Salmo salar L.) Acta Vet. Scand. 1991;32(4):503–510. doi: 10.1186/BF03546951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Sawalha A.H., Zhao M., Coit P., Lu Q. Epigenetic dysregulation of ACE2 and interferon-regulated genes might suggest increased COVID-19 susceptibility and severity in lupus patients. Clin. Immunol. 2020;215 doi: 10.1016/j.clim.2020.108410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Ahmad A. Non-coding RNAs: A tale of junk turning into treasure. Noncoding RNA Res. 2016;1(1):1–2. doi: 10.1016/j.ncrna.2016.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Chew C.L., Conos S.A., Unal B., Tergaonkar V. Noncoding RNAs: Master Regulators of Inflammatory Signaling. Trends Mol. Med. 2018;24(1):66–84. doi: 10.1016/j.molmed.2017.11.003. [DOI] [PubMed] [Google Scholar]
- 190.Nakayama Y., Fujiu K., Yuki R., Oishi Y., Morioka M.S., Isagawa T., Matsuda J., Oshima T., Matsubara T., Sugita J., Kudo F., Kaneda A., Endo Y., Nakayama T., Nagai R., Komuro I., Manabe I. A long noncoding RNA regulates inflammation resolution by mouse macrophages through fatty acid oxidation activation. Proc Natl Acad Sci U S A. 2020;117(25):14365–14375. doi: 10.1073/pnas.2005924117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Chen J., Ao L., Yang J. Long non-coding RNAs in diseases related to inflammation and immunity. Ann. Transl. Med. 2019;7(18):494. doi: 10.21037/atm.2019.08.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Tahamtan A., Teymoori-Rad M., Nakstad B., Salimi V. Anti-inflammatory MicroRNAs and their potential for inflammatory diseases treatment. Front. Immunol. 2018;9:1377. doi: 10.3389/fimmu.2018.01377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Bime C., Gurguis C.I., Hecker L., Desai A.A., Wang T., Garcia J.G.N. In: TranSlating MicroRNAs to the Clinic. Laurence J., editor. Academic Press; Boston: 2017. Chapter 6 - MicroRNAs in inflammatory lung disease; pp. 135–177. [Google Scholar]
- 194.Mathy N.W., Chen X.M. Long non-coding RNAs (lncRNAs) and their transcriptional control of inflammatory responses. J. Biol. Chem. 2017;292(30):12375–12382. doi: 10.1074/jbc.R116.760884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Yan S., Wang P., Wang J., Yang J., Lu H., Jin C., Cheng M., Xu D. Long non-coding RNA HIX003209 promotes inflammation by sponging miR-6089 via TLR4/NF-kappaB signaling pathway in rheumatoid arthritis. Front. Immunol. 2019;10:2218. doi: 10.3389/fimmu.2019.02218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Zhang Q., Chao T.C., Patil V.S., Qin Y., Tiwari S.K., Chiou J., Dobin A., Tsai C.M., Li Z., Dang J., Gupta S., Urdahl K., Nizet V., Gingeras T.R., Gaulton K.J., Rana T.M. The long noncoding RNA ROCKI regulates inflammatory gene expression. EMBO J. 2019;38(8) doi: 10.15252/embj.2018100041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Zhang P., Sun Y., Peng R., Chen W., Fu X., Zhang L., Peng H., Zhang Z. Long non-coding RNA Rpph1 promotes inflammation and proliferation of mesangial cells in diabetic nephropathy via an interaction with Gal-3. Cell Death Dis. 2019;10(7):526. doi: 10.1038/s41419-019-1765-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Obaid M., Udden S.M.N., Deb P., Shihabeddin N., Zaki M.H., Mandal S.S. LncRNA HOTAIR regulates lipopolysaccharide-induced cytokine expression and inflammatory response in macrophages. Sci. Rep. 2018;8(1):15670. doi: 10.1038/s41598-018-33722-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Du M., Yuan L., Tan X., Huang D., Wang X., Zheng Z., Mao X., Li X., Yang L., Huang K., Zhang F., Wang Y., Luo X., Huang D., Huang K. The LPS-inducible lncRNA Mirt2 is a negative regulator of inflammation. Nat. Commun. 2017;8(1):2049. doi: 10.1038/s41467-017-02229-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Li X., Yu M., Chen L., Sun T., Wang H., Zhao L., Zhao Q. LncRNA PMS2L2 protects ATDC5 chondrocytes against lipopolysaccharide-induced inflammatory injury by sponging miR-203. Life Sci. 2019;217:283–292. doi: 10.1016/j.lfs.2018.12.020. [DOI] [PubMed] [Google Scholar]
- 201.Lu Q., Meng Q., Qi M., Li F., Liu B. Shear-sensitive lncRNA AF131217.1 inhibits inflammation in HUVECs via regulation of KLF4. Hypertension. 2019;73(5):e25–e34. doi: 10.1161/HYPERTENSIONAHA.118.12476. [DOI] [PubMed] [Google Scholar]
- 202.Ma Y., Han W., Yang L., He L., Wang H. The regulation of miRNAs in inflammation-related carcinogenesis. Curr. Pharm. Des. 2015;21(21):3023–3031. doi: 10.2174/1381612821666150514105606. [DOI] [PubMed] [Google Scholar]
- 203.Nejad C., Stunden H.J., Gantier M.P. A guide to miRNAs in inflammation and innate immune responses. FEBS J. 2018;285(20):3695–3716. doi: 10.1111/febs.14482. [DOI] [PubMed] [Google Scholar]
- 204.Hirschberger S., Hinske L.C., Kreth S. MiRNAs: dynamic regulators of immune cell functions in inflammation and cancer. Cancer Lett. 2018;431:11–21. doi: 10.1016/j.canlet.2018.05.020. [DOI] [PubMed] [Google Scholar]
- 205.Sethi A., Kulkarni N., Sonar S., Lal G. Role of miRNAs in CD4 T cell plasticity during inflammation and tolerance. Front. Genet. 2013;4:8. doi: 10.3389/fgene.2013.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Aguilera-Aguirre L., Hao W., Pan L., Li X., Saavedra-Molina A., Bacsi A., Radak Z., Sur S., Brasier A.R., Ba X., Boldogh I. Pollen-induced oxidative DNA damage response regulates miRNAs controlling allergic inflammation. Am. J. Physiol. Lung Cell Mol. Physiol. 2017;313(6):L1058–L1068. doi: 10.1152/ajplung.00141.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Chakraborty C., Sharma A.R., Sharma G., Lee S.S. The interplay among miRNAs, major cytokines, and cancer-related inflammation. Mol. Ther. Nucleic Acids. 2020;20:606–620. doi: 10.1016/j.omtn.2020.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Markopoulos G.S., Roupakia E., Tokamani M., Alabasi G., Sandaltzopoulos R., Marcu K.B., Kolettas E. Roles of NF-kappaB signaling in the regulation of miRNAs impacting on inflammation in Cancer. Biomedicines. 2018;6(2) doi: 10.3390/biomedicines6020040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Z. Liu, H. Yu, Q. Guo, MicroRNA‑20a promotes inflammation via the nuclear factor‑κB signaling pathway in pediatric pneumonia, (1791-3004 (Electronic)). [DOI] [PubMed]
- 210.Qianru C., Xueyuan H., Bing Z., Qing Z., Kaixin Z., Shu L. Regulation of H2S-induced necroptosis and inflammation in broiler bursa of Fabricius by the miR-15b-5p/TGFBR3 axis and the involvement of oxidative stress in this process. J. Hazard. Mater. 2020 doi: 10.1016/j.jhazmat.2020.124682. [DOI] [PubMed] [Google Scholar]
- 211.Sanctuary M.R., Huang R.H., Jones A.A., Luck M.E., Aherne C.M., Jedlicka P., de Zoeten E.F., Collins C.B. 2019. miR-106a Deficiency Attenuates Inflammation in Murine IBD Models, (1935-3456 (Electronic)) p. 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.O’Connell R.M., Taganov K.D., Boldin M.P., Cheng G., Baltimore D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci U S A. 2007;104(5):1604–1609. doi: 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Yuan H.S., Xiong D.Q., Huang F., Cui J., Luo H. MicroRNA-421 inhibition alleviates bronchopulmonary dysplasia in a mouse model via targeting Fgf10. J. Cell. Biochem. 2019;120(10):16876–16887. doi: 10.1002/jcb.28945. [DOI] [PubMed] [Google Scholar]
- 214.Pasquali L. 2017. MiR-1307 Is Upregulated in Psoriasis Keratinocytes and Promotes Keratinocyte Inflammatory Response. [Google Scholar]
- 215.Fang Y., Shi C., Manduchi E., Civelek M., Davies P.F. MicroRNA-10a regulation of proinflammatory phenotype in athero-susceptible endothelium in vivo and in vitro. Proc Natl Acad Sci U S A. 2010;107(30):13450–13455. doi: 10.1073/pnas.1002120107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Fordham J.B., Naqvi A.R., Nares S. miR-24 regulates macrophage polarization and plasticity. J. Clin. Cell. Immunol. 2015;6(5) doi: 10.4172/2155-9899.1000362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Xiao Y.T., Wang J., Lu W., Cao Y., Cai W. Downregulated expression of microRNA-124 in pediatric intestinal failure patients modulates macrophages activation by inhibiting STAT3 and AChE. Cell Death Dis. 2016;7(12):e2521. doi: 10.1038/cddis.2016.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Lambert K.A., Roff A.N., Panganiban R.P., Douglas S., Ishmael F.T. MicroRNA-146a is induced by inflammatory stimuli in airway epithelial cells and augments the anti-inflammatory effects of glucocorticoids. PLoS One. 2018;13(10) doi: 10.1371/journal.pone.0205434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Hutchison E.R., Kawamoto E.M., Taub D.D., Lal A., Abdelmohsen K., Zhang Y., Wood W.H., 3rd, Lehrmann E., Camandola S., Becker K.G., Gorospe M., Mattson M.P. Evidence for miR-181 involvement in neuroinflammatory responses of astrocytes. Glia. 2013;61(7):1018–1028. doi: 10.1002/glia.22483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Yan Y., Lu K., Ye T., Zhang Z. MicroRNA223 attenuates LPSinduced inflammation in an acute lung injury model via the NLRP3 inflammasome and TLR4/NFkappaB signaling pathway via RHOB. Int. J. Mol. Med. 2019;43(3):1467–1477. doi: 10.3892/ijmm.2019.4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Mariappan N., Husain M., Zafar I., Singh V., Smithson K.G., Crowe D.R., Pittet J.F., Ahmad S., Ahmad A. Extracellular nucleic acid scavenging rescues rats from sulfur mustard analog-induced lung injury and mortality. Arch. Toxicol. 2020;94(4):1321–1334. doi: 10.1007/s00204-020-02699-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Hegde M., Guruprasad K.P., Ramachandra L., Satyamoorthy K., Joshi M.B. Interleukin-6-mediated epigenetic control of the VEGFR2 gene induces disorganized angiogenesis in human breast tumors. J. Biol. Chem. 2020;295(34):12086–12098. doi: 10.1074/jbc.RA120.012590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.So J.Y., Skrypek N., Yang H.H., Merchant A.S., Nelson G.W., Chen W.D., Ishii H., Chen J.M., Hu G., Achyut B.R., Yoon E.C., Han L., Huang C., Cam M.C., Zhao K., Lee M.P., Yang L. Induction of DNMT3B by PGE2 and IL6 at distant metastatic sites promotes epigenetic modification and breast Cancer colonization. Cancer Res. 2020;80(12):2612–2627. doi: 10.1158/0008-5472.CAN-19-3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Lee T.L., Yeh J., Van Waes C., Chen Z. Epigenetic modification of SOCS-1 differentially regulates STAT3 activation in response to interleukin-6 receptor and epidermal growth factor receptor signaling through JAK and/or MEK in head and neck squamous cell carcinomas. Mol. Cancer Ther. 2006;5(1):8–19. doi: 10.1158/1535-7163.MCT-05-0069. [DOI] [PubMed] [Google Scholar]
- 225.Iliopoulos D., Hirsch H.A., Struhl K. An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell. 2009;139(4):693–706. doi: 10.1016/j.cell.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Li P., Shan J.X., Chen X.H., Zhang D., Su L.P., Huang X.Y., Yu B.Q., Zhi Q.M., Li C.L., Wang Y.Q., Tomei S., Cai Q., Ji J., Li J.F., Chouchane L., Yu Y.Y., Sun F.Z., Xu Z.H., Liu B.Y., Zhu Z.G. Epigenetic silencing of microRNA-149 in cancer-associated fibroblasts mediates prostaglandin E2/interleukin-6 signaling in the tumor microenvironment. Cell Res. 2015;25(5):588–603. doi: 10.1038/cr.2015.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Meng F., Wehbe-Janek H., Henson R., Smith H., Patel T. Epigenetic regulation of microRNA-370 by interleukin-6 in malignant human cholangiocytes. Oncogene. 2008;27(3):378–386. doi: 10.1038/sj.onc.1210648. [DOI] [PubMed] [Google Scholar]
- 228.Chiou G.Y., Chien C.S., Wang M.L., Chen M.T., Yang Y.P., Yu Y.L., Chien Y., Chang Y.C., Shen C.C., Chio C.C., Lu K.H., Ma H.I., Chen K.H., Liu D.M., Miller S.A., Chen Y.W., Huang P.I., Shih Y.H., Hung M.C., Chiou S.H. Epigenetic regulation of the miR142-3p/interleukin-6 circuit in glioblastoma. Mol. Cell. 2013;52(5):693–706. doi: 10.1016/j.molcel.2013.11.009. [DOI] [PubMed] [Google Scholar]
- 229.D’Anello L., Sansone P., Storci G., Mitrugno V., D’Uva G., Chieco P., Bonafé M. Epigenetic control of the basal-like gene expression profile via Interleukin-6 in breast cancer cells. Mol. Cancer. 2010;9:300. doi: 10.1186/1476-4598-9-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Li L., Dong L., Zhao D., Gao F., Yan J. Classical dendritic cells regulate acute lung inflammation and injury in mice with lipopolysaccharideinduced acute respiratory distress syndrome. Int. J. Mol. Med. 2019;44(2):617–629. doi: 10.3892/ijmm.2019.4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Rosenzweig J.M., Glenn J.D., Calabresi P.A., Whartenby K.A. KLF4 modulates expression of IL-6 in dendritic cells via both promoter activation and epigenetic modification. J. Biol. Chem. 2013;288(33):23868–23874. doi: 10.1074/jbc.M113.479576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Hu L., Yu Y., Huang H., Fan H., Hu L., Yin C., Li K., Fulton D.J., Chen F. Epigenetic regulation of interleukin 6 by histone acetylation in macrophages and its role in paraquat-induced pulmonary fibrosis. Front. Immunol. 2016;7:696. doi: 10.3389/fimmu.2016.00696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Tang B., Zhao R., Sun Y., Zhu Y., Zhong J., Zhao G., Zhu N. Interleukin-6 expression was regulated by epigenetic mechanisms in response to influenza virus infection or dsRNA treatment. Mol. Immunol. 2011;48(8):1001–1008. doi: 10.1016/j.molimm.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 234.Li W., Wang Q., Qi X., Guo Y., Lu H., Chen Y., Lu Z., Yan Q., Zhu X., Jung J.U., Tosato G., Gao S.J., Lu C. Viral interleukin-6 encoded by an oncogenic virus promotes angiogenesis and cellular transformation by enhancing STAT3-mediated epigenetic silencing of caveolin 1. Oncogene. 2020;39(23):4603–4618. doi: 10.1038/s41388-020-1317-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Notz Q., Schmalzing M., Wedekink F., Schlesinger T., Gernert M., Herrmann J., Sorger L., Weismann D., Schmid B., Sitter M., Schlegel N., Kranke P., Wischhusen J., Meybohm P., Lotz C. Pro- and anti-inflammatory responses in severe COVID-19-Induced acute respiratory distress syndrome-an observational pilot study. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.581338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Zhang J., Hao Y., Ou W., Ming F., Liang G., Qian Y., Cai Q., Dong S., Hu S., Wang W., Wei S. Serum interleukin-6 is an indicator for severity in 901 patients with SARS-CoV-2 infection: a cohort study. J. Transl. Med. 2020;18(1):406. doi: 10.1186/s12967-020-02571-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Kwon J.S., Kim J.Y., Kim M.C., Park S.Y., Kim B.N., Bae S., Cha H.H., Jung J., Kim M.J., Lee M.J., Choi S.H., Chung J.W., Shin E.C., Kim S.H. Factors of severity in patients with COVID-19: Cytokine/Chemokine concentrations, viral load, and antibody responses. Am. J. Trop. Med. Hyg. 2020 doi: 10.4269/ajtmh.20-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Udomsinprasert W., Jittikoon J., Sangroongruangsri S., Chaikledkaew U. Circulating levels of Interleukin-6 and Interleukin-10, but not tumor necrosis factor-alpha, as potential biomarkers of severity and mortality for COVID-19: systematic review with meta-analysis. J. Clin. Immunol. 2020 doi: 10.1007/s10875-020-00899-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Jones B., Chen J. Inhibition of IFN-gamma transcription by site-specific methylation during T helper cell development. EMBO J. 2006;25(11):2443–2452. doi: 10.1038/sj.emboj.7601148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Lee D.U., Agarwal S., Rao A. Th2 lineage commitment and efficient IL-4 production involves extended demethylation of the IL-4 gene. Immunity. 2002;16(5):649–660. doi: 10.1016/s1074-7613(02)00314-x. [DOI] [PubMed] [Google Scholar]
- 241.Liu Y., Li H., Xiao T., Lu Q. Epigenetics in immune-mediated pulmonary diseases. Clin. Rev. Allergy Immunol. 2013;45(3):314–330. doi: 10.1007/s12016-013-8398-3. [DOI] [PubMed] [Google Scholar]
- 242.Yang I.V., Konigsberg I., MacPhail K., Li L., Davidson E.J., Mroz P.M., Hamzeh N., Gillespie M., Silveira L.J., Fingerlin T.E., Maier L.A. DNA methylation changes in lung immune cells are associated with granulomatous lung disease. Am. J. Respir. Cell Mol. Biol. 2019;60(1):96–105. doi: 10.1165/rcmb.2018-0177OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Baird A.M., Leonard J., Naicker K.M., Kilmartin L., O’Byrne K.J., Gray S.G. IL-23 is pro-proliferative, epigenetically regulated and modulated by chemotherapy in non-small cell lung cancer. Lung Cancer. 2013;79(1):83–90. doi: 10.1016/j.lungcan.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 244.He L.X., Tang Z.H., Huang Q.S., Li W.H. DNA Methylation: A Potential Biomarker of Chronic Obstructive Pulmonary Disease. Front. Cell Dev. Biol. 2020;8:585. doi: 10.3389/fcell.2020.00585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Bowman R.V., Wright C.M., Davidson M.R., Francis S.M., Yang I.A., Fong K.M. Epigenomic targets for the treatment of respiratory disease. Expert Opin. Ther. Targets. 2009;13(6):625–640. doi: 10.1517/14728220902926119. [DOI] [PubMed] [Google Scholar]
- 246.Suzuki M., Wada H., Yoshino M., Tian L., Shigematsu H., Suzuki H., Alaa M., Tamura H., Fujiwara T., Nagato K., Motohashi S., Moriya Y., Hoshino H., Yoshida S., Shibuya K., Hiroshima K., Nakatani Y., Yoshino I. Molecular characterization of chronic obstructive pulmonary disease-related non-small cell lung cancer through aberrant methylation and alterations of EGFR signaling. Ann. Surg. Oncol. 2010;17(3):878–888. doi: 10.1245/s10434-009-0739-3. [DOI] [PubMed] [Google Scholar]
- 247.Brand S., Kesper D.A., Teich R., Kilic-Niebergall E., Pinkenburg O., Bothur E., Lohoff M., Garn H., Pfefferle P.I., Renz H. DNA methylation of TH1/TH2 cytokine genes affects sensitization and progress of experimental asthma. J. Allergy Clin. Immunol. 2012;129(6) doi: 10.1016/j.jaci.2011.12.963. 1602-10 e6. [DOI] [PubMed] [Google Scholar]
- 248.Kabesch M., Tost J. Recent findings in the genetics and epigenetics of asthma and allergy. Semin. Immunopathol. 2020;42(1):43–60. doi: 10.1007/s00281-019-00777-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.McCoy K., Peterson A., Tian Y., Sang Y. Immunogenetic association underlying severe COVID-19. Vaccines (Basel) 2020;8(4) doi: 10.3390/vaccines8040700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Salkowska A., Karwaciak I., Karas K., Dastych J., Ratajewski M. SARS-CoV-2 proteins induce IFNG in Th1 lymphocytes generated from CD4+ cells from healthy, unexposed polish donors. Vaccines (Basel) 2020;8(4) doi: 10.3390/vaccines8040673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Ahn Y.H., Ko Y.H. Diagnostic and therapeutic implications of microRNAs in non-small cell lung Cancer. Int. J. Mol. Sci. 2020;21(22) doi: 10.3390/ijms21228782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Pozza D.H., De Mello R.A., Araujo R.L.C., Velcheti V. MicroRNAs in Lung Cancer Oncogenesis and Tumor Suppression: How it Can Improve the Clinical Practice? Curr. Genomics. 2020;21(5):372–381. doi: 10.2174/1389202921999200630144712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Tirpe A., Gulei D., Tirpe G.R., Nutu A., Irimie A., Campomenosi P., Pop L.A., Berindan-Neagoe I. Beyond Conventional: The New Horizon of Anti-Angiogenic microRNAs in Non-Small Cell Lung Cancer Therapy. Int. J. Mol. Sci. 2020;21(21) doi: 10.3390/ijms21218002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Hu C., Hui K., Jiang X. Effects of microRNA regulation on antiangiogenic therapy resistance in non-small cell lung cancer. Biomed. Pharmacother. 2020;131 doi: 10.1016/j.biopha.2020.110557. [DOI] [PubMed] [Google Scholar]
- 255.Mirzaei R., Mahdavi F., Badrzadeh F., Hosseini-Fard S.R., Heidary M., Jeda A.S., Mohammadi T., Roshani M., Yousefimashouf R., Keyvani H., Darvishmotevalli M., Sani M.Z., Karampoor S. The emerging role of microRNAs in the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection. Int. Immunopharmacol. 2020 doi: 10.1016/j.intimp.2020.107204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Pierce J.B., Simion V., Icli B., Pérez-Cremades D., Cheng H.S., Feinberg M.W. Computational analysis of targeting SARS-CoV-2, viral entry proteins ACE2 and TMPRSS2, and interferon genes by host MicroRNAs. Genes (Basel) 2020;11(11) doi: 10.3390/genes11111354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Widiasta A., Sribudiani Y., Nugrahapraja H., Hilmanto D., Sekarwana N., Rachmadi D. Potential role of ACE2-related microRNAs in COVID-19-associated nephropathy. Noncoding RNA Res. 2020;5(4):153–166. doi: 10.1016/j.ncrna.2020.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Lu D., Chatterjee S., Xiao K., Riedel I., Wang Y., Foo R., Bär C., Thum T. MicroRNAs targeting the SARS-CoV-2 entry receptor ACE2 in cardiomyocytes. J. Mol. Cell. Cardiol. 2020;148:46–49. doi: 10.1016/j.yjmcc.2020.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Mukhopadhyay D., Mussa B.M. Identification of novel hypothalamic MicroRNAs as promising therapeutics for SARS-CoV-2 by regulating ACE2 and TMPRSS2 expression: an in Silico Analysis. Brain Sci. 2020;10(10) doi: 10.3390/brainsci10100666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Fulzele S., Sahay B., Yusufu I., Lee T.J., Sharma A., Kolhe R., Isales C.M. COVID-19 virulence in aged patients might Be impacted by the host cellular MicroRNAs Abundance/Profile. Aging Dis. 2020;11(3):509–522. doi: 10.14336/AD.2020.0428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Balmeh N., Mahmoudi S., Mohammadi N., Karabedianhajiabadi A. Predicted therapeutic targets for COVID-19 disease by inhibiting SARS-CoV-2 and its related receptors. Inform Med Unlocked. 2020;20 doi: 10.1016/j.imu.2020.100407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Du X., Wang S., Liu X., He T., Lin X., Wu S., Wang D., Li J., Huang W., Yang H. MiR-1307-5p targeting TRAF3 upregulates the MAPK/NF-kappaB pathway and promotes lung adenocarcinoma proliferation. Cancer Cell Int. 2020;20:502. doi: 10.1186/s12935-020-01595-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Jafarinejad-Farsangi S., Jazi M.M., Rostamzadeh F., Hadizadeh M. High affinity of host human microRNAs to SARS-CoV-2 genome: an in silico analysis. Noncoding RNA Res. 2020;5(4):222–231. doi: 10.1016/j.ncrna.2020.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Sun D.M., Tang B.F., Li Z.X., Guo H.B., Cheng J.L., Song P.P., Zhao X. MiR-29c reduces the cisplatin resistance of non-small cell lung cancer cells by negatively regulating the PI3K/Akt pathway. Sci. Rep. 2018;8(1):8007. doi: 10.1038/s41598-018-26381-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Liu X., Lv X., Yang Q., Jin H., Zhou W., Fan Q. MicroRNA-29a functions as a tumor suppressor and increases cisplatin sensitivity by targeting NRAS in lung Cancer. Technol. Cancer Res. Treat. 2018;17 doi: 10.1177/1533033818758905. 1533033818758905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Botta C., Cuce M., Pitari M.R., Caracciolo D., Gulla A., Morelli E., Riillo C., Biamonte L., Gallo Cantafio M.E., Prabhala R., Mignogna C., Di Vito A., Altomare E., Amodio N., Di Martino M.T., Correale P., Rossi M., Giordano A., Munshi N.C., Tagliaferri P., Tassone P. MiR-29b antagonizes the pro-inflammatory tumor-promoting activity of multiple myeloma-educated dendritic cells. Leukemia. 2018;32(4):1003–1015. doi: 10.1038/leu.2017.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Eken S.M., Christersdottir T., Winski G., Sangsuwan T., Jin H., Chernogubova E., Pirault J., Sun C., Simon N., Winter H., Backlund A., Haghdoost S., Hansson G.K., Halle M., Maegdefessel L. miR-29b mediates the chronic inflammatory response in radiotherapy-induced vascular disease. JACC Basic Transl. Sci. 2019;4(1):72–82. doi: 10.1016/j.jacbts.2018.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Nersisyan S., Engibaryan N., Gorbonos A., Kirdey K., Makhonin A., Tonevitsky A. Potential role of cellular miRNAs in coronavirus-host interplay. PeerJ. 2020;8:e9994. doi: 10.7717/peerj.9994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Zheng W., Zhao J., Tao Y., Guo M., Ya Z., Chen C., Qin N., Zheng J., Luo J., Xu L. MicroRNA-21: A promising biomarker for the prognosis and diagnosis of non-small cell lung cancer. Oncol. Lett. 2018;16(3):2777–2782. doi: 10.3892/ol.2018.8972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Sheedy F.J. Turning 21: induction of miR-21 as a key switch in the inflammatory response. Front. Immunol. 2015;6:19. doi: 10.3389/fimmu.2015.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Feng X., Wang Z., Fillmore R., Xi Y. MiR-200, a new star miRNA in human cancer. Cancer Lett. 2014;344(2):166–173. doi: 10.1016/j.canlet.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Ahmad A., Aboukameel A., Kong D., Wang Z., Sethi S., Chen W., Sarkar F.H., Raz A. Phosphoglucose isomerase/autocrine motility factor mediates epithelial-mesenchymal transition regulated by miR-200 in breast cancer cells. Cancer Res. 2011;71(9):3400–3409. doi: 10.1158/0008-5472.CAN-10-0965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Liu C., Hu W., Li L.L., Wang Y.X., Zhou Q., Zhang F., Song-Yang Y.Y., Zhu W., Sun C.C., Li D.J. Roles of miR-200 family members in lung cancer: more than tumor suppressors. Future Oncol. 2018;14(27):2875–2886. doi: 10.2217/fon-2018-0155. [DOI] [PubMed] [Google Scholar]
- 274.Bozgeyik I. Therapeutic potential of miRNAs targeting SARS-CoV-2 host cell receptor ACE2. Meta Gene. 2021;27 doi: 10.1016/j.mgene.2020.100831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Yu J., Chen J., Yang H., Chen S., Wang Z. Overexpression of miR200a3p promoted inflammation in sepsisinduced brain injury through ROSinduced NLRP3. Int. J. Mol. Med. 2019;44(5):1811–1823. doi: 10.3892/ijmm.2019.4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Reddy M.A., Jin W., Villeneuve L., Wang M., Lanting L., Todorov I., Kato M., Natarajan R. Pro-inflammatory role of microrna-200 in vascular smooth muscle cells from diabetic mice. Arterioscler. Thromb. Vasc. Biol. 2012;32(3):721–729. doi: 10.1161/ATVBAHA.111.241109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Rokavec M., Wu W., Luo J.L. IL6-mediated suppression of miR-200c directs constitutive activation of inflammatory signaling circuit driving transformation and tumorigenesis. Mol. Cell. 2012;45(6):777–789. doi: 10.1016/j.molcel.2012.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Shen Z., Xuan W., Wang H., Sun F., Zhang C., Gong Q., Ge S. miR-200b regulates cellular senescence and inflammatory responses by targeting ZEB2 in pulmonary emphysema. Artif. Cells Nanomed. Biotechnol. 2020;48(1):656–663. doi: 10.1080/21691401.2020.1725029. [DOI] [PubMed] [Google Scholar]
- 279.Niu W., Wu F., Cui H., Cao W., Chao Y., Wu Z., Fan M., Liang C. Network pharmacology analysis to identify phytochemicals in traditional chinese medicines that may regulate ACE2 for the treatment of COVID-19. Evid. Complement. Alternat. Med. 2020;2020 doi: 10.1155/2020/7493281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Duan F.G., Wang M.F., Cao Y.B., Dan L., Li R.Z., Fan X.X., Khan I., Lai H.L., Zhang Y.Z., Hsiao W.W., Yao X.J., Wu Q.B., Liu L., Tang Y.J., Leung E.L. MicroRNA-421 confers paclitaxel resistance by binding to the KEAP1 3’UTR and predicts poor survival in non-small cell lung cancer. Cell Death Dis. 2019;10(11):821. doi: 10.1038/s41419-019-2031-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Matarese A., Gambardella J., Sardu C., Santulli G. miR-98 regulates TMPRSS2 expression in human endothelial cells: key implications for COVID-19. Biomedicines. 2020;8(11) doi: 10.3390/biomedicines8110462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Rom S., Dykstra H., Zuluaga-Ramirez V., Reichenbach N.L., Persidsky Y. miR-98 and let-7g* protect the blood-brain barrier under neuroinflammatory conditions. J. Cereb. Blood Flow Metab. 2015;35(12):1957–1965. doi: 10.1038/jcbfm.2015.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Jiang F., Yu Q., Chu Y., Zhu X., Lu W., Liu Q., Wang Q. MicroRNA-98-5p inhibits proliferation and metastasis in non-small cell lung cancer by targeting TGFBR1. Int. J. Oncol. 2019;54(1):128–138. doi: 10.3892/ijo.2018.4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Kim W.R., Park E.G., Kang K.W., Lee S.M., Kim B., Kim H.S. Expression analyses of MicroRNAs in Hamster lung tissues infected by SARS-CoV-2. Mol. Cells. 2020;43(11):953–963. doi: 10.14348/molcells.2020.0177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Bras J.P., Silva A.M., Calin G.A., Barbosa M.A., Santos S.G., Almeida M.I. miR-195 inhibits macrophages pro-inflammatory profile and impacts the crosstalk with smooth muscle cells. PLoS One. 2017;12(11) doi: 10.1371/journal.pone.0188530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Wang H., Zhan Y., Jin J., Zhang C., Li W. MicroRNA-15b promotes proliferation and invasion of nonsmall cell lung carcinoma cells by directly targeting TIMP2. Oncol. Rep. 2017;37(6):3305–3312. doi: 10.3892/or.2017.5604. [DOI] [PubMed] [Google Scholar]
- 287.Yu X., Zhang Y., Cavazos D., Ma X., Zhao Z., Du L., Pertsemlidis A. miR-195 targets cyclin D3 and survivin to modulate the tumorigenesis of non-small cell lung cancer. Cell Death Dis. 2018;9(2):193. doi: 10.1038/s41419-017-0219-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Bertolazzi G., Cipollina C., Benos P.V., Tumminello M., Coronnello C. miR-1207-5p can contribute to dysregulation of inflammatory response in COVID-19 via targeting SARS-CoV-2 RNA. Front. Cell. Infect. Microbiol. 2020;10 doi: 10.3389/fcimb.2020.586592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Dang W., Qin Z., Fan S., Wen Q., Lu Y., Wang J., Zhang X., Wei L., He W., Ye Q., Yan Q., Li G., Ma J. miR-1207-5p suppresses lung cancer growth and metastasis by targeting CSF1. Oncotarget. 2016;7(22):32421–32432. doi: 10.18632/oncotarget.8718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Bartoszewski R., Dabrowski M., Jakiela B., Matalon S., Harrod K.S., Sanak M., Collawn J.F. SARS-CoV-2 may regulate cellular responses through depletion of specific host miRNAs. Am. J. Physiol. Lung Cell Mol. Physiol. 2020;319(3):L444–l455. doi: 10.1152/ajplung.00252.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.El-Nabi S.H., Elhiti M., El-Sheekh M. A new approach for COVID-19 treatment by micro-RNA. Med. Hypotheses. 2020;143 doi: 10.1016/j.mehy.2020.110203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Khalaj K., Figueira R.L., Antounians L., Lauriti G., Zani A. Systematic review of extracellular vesicle-based treatments for lung injury: are EVs a potential therapy for COVID-19? J. Extracell. Vesicles. 2020;9(1) doi: 10.1080/20013078.2020.1795365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Burke H., Freeman A., Cellura D.C., Stuart B.L., Brendish N.J., Poole S., Borca F., Phan H.T.T., Sheard N., Williams S., Spalluto C.M., Staples K.J., Clark T.W., Wilkinson T.M.A. R.C. investigators, Inflammatory phenotyping predicts clinical outcome in COVID-19. Respir. Res. 2020;21(1):245. doi: 10.1186/s12931-020-01511-z. [DOI] [PMC free article] [PubMed] [Google Scholar]