Graphical abstract

Keywords: Gram-negative bacteria, Prediction, Secreted protein, Transmembrane protein, Protein secretion system
Abstract
Gram-negative bacteria harness multiple protein secretion systems and secrete a large proportion of the proteome. Proteins can be exported to periplasmic space, integrated into membrane, transported into extracellular milieu, or translocated into cytoplasm of contacting cells. It is important for accurate, genome-wide annotation of the secreted proteins and their secretion pathways. In this review, we systematically classified the secreted proteins according to the types of secretion systems in Gram-negative bacteria, summarized the known features of these proteins, and reviewed the algorithms and tools for their prediction.
1. Introduction
Gram-negative (or diderm) bacteria contain two phospholipid membranes. The outer membrane encloses individual cells and separates them from extracellular environment, while the inner membrane separates bacterial cytoplasm and the periplasm, a space between the two cell membranes. Bacterial cells also have some constitutive protrusions inserted in or attached to the cell surface.
More than one third bacterial proteins undergo an extracellular translocation process from cytoplasm where they have been synthesized [1]. According to the destined location of substrate proteins, the translocation process can be classified as three major types: exportation, secretion and membrane-retention [2]. Exportation only involves the process of passing through the inner membrane actively, secretion means crossing over the outer membrane or two cell membranes completely, and membrane-retention refers in particular to the trans-membrane process after which the substrate protein is inserted in the membrane. Therefore, with a strict definition, a secreted protein should have gone through an active translocation process from cytoplasm to extracellular environment. However, in a broad sense, the proteins undergoing any type of the translocation processes described above are called secreted proteins. There are also proteins called ‘effectors’, which specifically refer to the ones translocated from bacterial cytoplasm to other cells (eukaryotic or other bacterial cells) directly via some transmembrane device contacting other cells at the distal pole. In this review, we used the broad definition, and secreted proteins include the strictly secreted proteins, transmembrane proteins, surface-associated proteins or subunit parts of surface appendages, periplasmic proteins and translocated effectors (Fig. 1).
Fig. 1.
Subcellular localization of Gram-negative bacterial proteins. The dashed arrow showed the translocation process of the proteins.
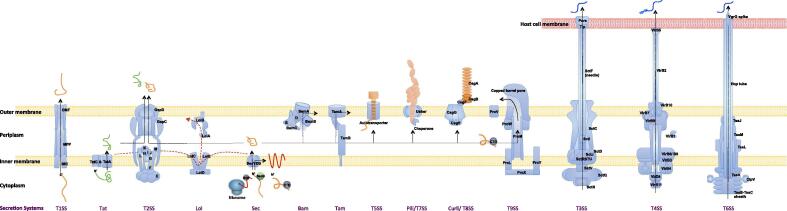
Bacteria employ multiple means to secrete proteins (Fig. 2; Table 1). The mechanisms of protein secretion in Gram-negative bacteria could be summarized as three categories: (1) one-step, two-membrane spanning secretion, (2) two-step, two-membrane spanning secretion, and (3) inner membrane spanning export. Accordingly, the protein secretion systems are divided into three major types – two-membrane spanning secretion systems, inner membrane spanning exporters and outer membrane spanning secretion systems. Based on the destiny of the secreted proteins, the two-membrane spanning secretion systems are further classified into two sub-classes, trans-membrane secretion systems and trans-membrane translocation systems. Trans-membrane secretion systems only secrete substrate proteins outside the bacterial cells, including the well-known Type I secretion Systems (T1SSs), T2SSs and T9SSs (Bacteroidetes PorSS), while trans-membrane translocation systems deliver bacterial substrate proteins into contacting cells. T3SSs, T4SSs and T6SSs are all trans-membrane translocation systems. Inner membrane spanning exporters transport proteins through Sec or Tat pathway. Outer membrane spanning secretion systems are exemplified by T5SSs, T7SSs (Chaperone-Usher pilus secretion), T8SSs (curli secretion), etc.
Fig. 2.
Secreted proteins and their transport pathways. The secretion machines are multi-protein complex, with different protein components. The protein transport processes were also indicated, with Sec and Tat pathways secreting the proteins from bacterial cytoplasm to periplasm or inner membrane, Lol pathways transporting the protein within the periplasm side of inner membrane into the periplasm side of outer membrane, Bam and Tam systems transporting periplasmic protein into outer membrane, T1SSs transporting proteins from bacterial cytoplasm to extracellular space, T2SSs and T9SSs transporting periplasmic proteins to extracellular space, and T3SSs, T4SSs and T6SSs translocating proteins from cytoplasm to host cellular cytoplasm directly. T5SSs are autotransporters that transport themselves extracellularly. The pili and curli proteins are transported out of bacterial outer membrane through T7SSs and T8SSs, respectively. The protein names or component types were shown for each secretion systems. OMF, Out Membrane Factor; MFP, Membrane Fusion Protein; IMC, Inner Membrane Component; SRP, Signal Recognition Particle.
Table 1.
Overview of protein secretion systems and the substrate features in Gram-negative bacteria.
| Secretion system | Secretion step(s) | Membrane spanning | Secretion signal | Substrate state |
|---|---|---|---|---|
| Sec | 1 | Inner | N-terminus | Unfolded |
| Tat | 1 | Inner | N-terminus | Folded |
| T1SS | 1 | Inner + Outer | C-terminus | Unfolded |
| T2SS | 2 (Sec/Tat) | Inner + Outer | N-terminus | Folded |
| T3SS1 | 1 or 2 (Sec) | Inner + Outer (+Host) | N-terminus | Unfolded |
| T4SS2 | 1 | Inner + Outer (+Host) | C-terminus | Unfolded |
| T6SS | 1 | Inner + Outer + Host | N-terminus? | Folded |
| T5SS | 2 (Sec) | Outer | N-terminus | Unfolded |
| Pili/ T7SS | 2 (Sec) | Outer | N-terminus | Folded |
| Curli/ T8SS | 2 (Sec) | Outer | N-terminus | Unfolded |
| T9SS | 2 (Sec) | Inner + Outer | C-terminus | Folded |
Notes: 1 T3SSs include non-flagella T3SSs and flagella T3SSs. Non-flagella T3SSs are translocation systems delivering substrates into host cells in one step, while flagella T3SSs involve two steps to secrete substrates extracellularly. 2 T4SSs translocate substrate proteins into host cells like T3SSs, or transport the proteins into extracellular milieu.
Effective recognition of the proteins secreted through different systems is important, which could facilitate the annotation of bacterial genomes, mechanism exploration of bacterial life processes, and prevention and control of bacterial infections and associated diseases. In recent decades, bioinformatic algorithms and methods have been introduced into the field and developed explosively, promoting the identification of proteins secreted through different systems in a large variety of bacteria. The review summarized our current knowledge on the features of different secreted proteins, and the main progress of bioinformatic applications in prediction of proteins secreted by different mechanisms in Gram-negative bacteria. At present, there are a dozen of protein secretion systems that have been reported in Gram-negative bacteria, including two inner membrane spanning export systems (Sec and Tat pathways), type I-IX secretion systems (T1SSs ~ T9SSs), and new ones. Sec and Tat pathways represent the main mechanisms mediating protein transport from cytoplasm to periplasm [3]. The periplasmic proteins can be further transported to extra-cellular matrix by certain secretion systems such as T2SSs, T5SSs and T9SSs, be secreted onto the bacterial surface such as pili secreted by T7SSs and curli secreted by T8SSs, or stay in the periplasmic space. T1SSs, T3SSs, T4SSs and T6SSs represent the major one-step two-membrane secretion systems. We will review the exporters and the tools predicting proteins exported via inner membrane first, followed by the secretion systems spanning outer membrane or both membranes and the substrate prediction methods. Transmembrane protein prediction algorithms are also summarized.
2. Inner membrane spanning exporters
In Gram-negative bacteria, there are two classical (Sec and Tat) and some non-classical inner membrane-spanning protein-exporting pathways.
2.1. The general secretion (Sec) pathway
2.1.1. Brief summary
Sec pathway is a universal protein export mechanism employed by Archeobacteria, Eubacteria and Eukaryota [3]. The Sec system is composed of a central component, SecYEG, which forms a protein-conducting channel and mediates the translocation of proteins in unfolded state into or across the plasma membrane. In Gram-negative bacteria, most periplasmic proteins and inner membrane proteins (IMPs) are exported through the SecYEG translocon, vectorially or laterally [3], [4]. The IMPs and periplasmic proteins take different targeting mechanisms, i.e., co-translational and post-translational mode, respectively [1]. For IMPs, the export involves a co-translational targeting process mediated by both signal recognition particle (SRP) and its membrane receptor FtsY. SRP binds to the N-terminal transmembrane helix (TMH) of the exported protein, forms a ribosome-nascent chain-SRP-FtsY complex, and targets the protein into the SecYEG channel. SecYEG can mediate export and insertion of the targeted protein into the inner membrane independently or in cooperation with a membrane protein insertase YidC [1], [4]. For the proteins translocated into the periplasm, a post-translational mode of export is adopted, by which an essential ATPase motor SecA recognizes the exported proteins with high affinity and empowers the transmembrane export. Other proteins could also participate in the processes of sorting, targeting and translocation, e.g., the chaperones aiding pre-protein targeting (trigger factor or SecB), the auxiliary components enhancing translocation efficiency (SecDF–YajC), etc [1], [3]. For most proteins, the two export modes are exclusive, but it is not absolutely. Some IMPs, e.g. RodZ, were found to take the co-translational mode but targeted by SecA rather than SRP [5], [6], [7].
2.1.2. Molecular features of the proteins secreted through Sec pathway
The N-terminal signal peptides (SPs) of proteins exported via Sec pathway are important for targeting, show some atypical sequential patterns, and have been explored for prediction of such types of secreted proteins [1]. A typical SP is comprised of 5–30 amino acids, which can be divided into 3 parts: a positively charged amino-terminus (N-region), a hydrophobic function domain (H-region) and a negatively charged polar carboxyl-terminus (C-region) where the cleavage site is located (Fig. 3A). There are two types of SPases that can cleave SPs, SPase I and SPase II, which can recognize different cleavage sites. SPase I cleaves the classical SPs while SPase II cleaves the SPs of lipoproteins. Positions −1 and −3 of the SPase cleavage site are often occupied by non-bulky polar amino acids (AXA pattern). Lipoprotein substrates show a pattern L[AS][GA]C at the −3 to +1 positions. The motif is recognized and cleaved by a SPase II, and the cysteine at the +1 position is lipid modified following translocation [3]. The positively charged N-region and the hydrophobic helical H-region interact with phospholipids, and are recognized by SRP, SecA or trigger factor. The signal sequences with higher hydrophobicity of the H-region show increased binding affinity for SRP [1], [3].
Fig. 3.
Sequence features of Sec/Tat SPs. (A) Sec-dependent SPs. There are two types of Sec-SPs, classical (top) and lipoprotein ones (bottom). Both of them are composed of a N-terminus region (blue), a hydrophobic region (dark blue) and a C-terminus region (grey). ‘+’ represents the region positively charged. The residue composition patterns of the C-terminal cleavage sites and corresponding SPases are shown. (B) Tat-dependent SPs. SPs targeted to Tat pathway have the sequential features similar to Sec-SPs, but generally have longer N-terminal regions which often contain a conserved motif with two consecutive arginine residues. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
It should be noted that some secretory proteins, e.g., autotransporters, have N-terminal extensions (N-AT) of varying length preceding the SPs [1]. A protein could be targeted to other pathways (e.g., Tat pathway) despite the presence of a similar SP in the N-terminus [3]. Some unfolded proteins without N-terminal SPs have been identified, which are also exported by Sec pathway [8], [9] or Sec-related non-classical pathways similar to the SecA2 pathway [10].
2.1.3. Algorithms and tools predicting proteins secreted through Sec pathway
More than a dozen of software tools have been developed to predict SPs of proteins secreted through Sec pathway (Table 2). SignalP is the most widely used program to identify the N-terminal SPs [11]. Since the first version of SignalP (SignalP 1.0) was proposed in 1997 [12], four new versions (SignalP 1.0 ~ 5.0) have been updated [13], [14]. Despite the popularity of SignalP tools and their large success in application for Sec substrate identification, other tools also have merits under certain circumstances. For example, till SignalP5.0, the other versions of SignalP can only predict Sec substrates cleaved by SPase I (Sec/SPI) but not those cleaved by SPase II (Sec/SPII) [13], [14]. For the SignalP models of Gram-negative bacteria, due to the bias of the training datasets enriched with E. coli and other γ-proteobacteria sequences, the predictive performance could be compromised for other species [11]. Some secreted proteins contained uncleaved SPs, for which SignalP cannot predict accurately [11].
Table 2.
Representative software tools predicting Sec substrates in Gram-negative bacteria.
| Tool | Algorithms | Target | URL or reference |
|---|---|---|---|
| SignalP4 | Artificial Neural Network (ANN) | Sec/SPI; Cleavage site | https://services.healthtech.dtu.dk/service.php?SignalP-4.1; [13] |
| SignalP5 | Deep Neural Network (DNN) | Sec/SPI; Sec/SPII; Tat/SPI; Cleavage site | https://services.healthtech.dtu.dk/service.php?SignalP-5.0; [14] |
| Signal-BLAST | BLASTP | Sec/SPI | http://sigpep.services.came.sbg.ac.at/signalblast.html; [15] |
| Signal-3L 2.0 | Hierarchical Mixture Model | Sec/SPI; TMH; Cleavage site | http://www.csbio.sjtu.edu.cn/bioinf/Signal-3L; [16] |
| PrediSi | Position Weight Matrix (PWM) | Sec/SPI | http://www.predisi.de; [17] |
| Signal-CF | Pseudo Amino Acid Composition;K Nearest Neighbor Classifier | Sec/SPI; Cleavage site | http://www.csbio.sjtu.edu.cn/bioinf/Signal-CF; [18] |
| LipoP | HMM | Sec/SPI; Sec/SPII; TMH | https://services.healthtech.dtu.dk/service.php?LipoP; [19] |
| SPEPlip | ANN; Regular Expression Search | Sec/SPI; Lipoprotein; Cleavage site | http://gpcr.biocomp.unibo.it/cgi/predictors/spep/pred_spepcgi.cgi; [20] |
| Phobius/ PolyPhobius | Hidden Markov Model (HMM) | Sec/SPI; Full-protein TM topology | http://phobius.sbc.su.se; [21], [22] |
| Philius | Dynamic Bayesian Network (DBN) | Sec/SPI; Full-protein TM topology; Protein type | http://www.yeastrc.org/philius; [23] |
| TOPCONS | Consensus prediction | Sec/SPI; Full-protein TM topology; Protein type | http://topcons.net; [24] |
| SPOCTOPUS | ANN and HMM | Sec/SPI; TMH | http://octopus.cbr.su.se; [25] |
| MEMSAT3/ MEMSAT-SVM | ANN; Support Vector Machine (SVM) | Sec/SPI; TMH; Re-entrant helix; Protein type | http://bioinf.cs.ucl.ac.uk/psipred; [26], [27] |
| DeepSig | Deep Convolutional Neural Network(DCNN); Grammar-Restrained Hidden Conditional Random Field | Sec/SPI; Cleavage site | https://deepsig.biocomp.unibo.it/deepsig; [28] |
| SigUNet | Convolutional Neural Network (CNN) | Sec/SPI | https://github.com/mbilab/SigUNet; [295] |
| Signal-3L 3.0 | Attention Deep Learning; Window-Based Scoring | Sec/SPI; Cleavage site | http://www.csbio.sjtu.edu.cn/bioinf/Signal-3L; [296] |
With an independent benchmarking dataset, SignalP 5.0 showed the best performance in prediction of both Sec/SPIs and Sec/SPIIs among the tools except for Signal-BLAST [14]. Signal-BLAST uses BLAST to find the sequences homologous to known SPs, and therefore shows high accuracy [15]. It is affected by the size of curated databases and similarities between query proteins and the databases. However, for a strain whose genome is newly sequenced, the homology-based methods can be applied in parallel with or before SignalP 5.0, since the former can pick out the verified or most likely proteins with SPs most precisely.
Other tools can also predict Sec/SPIIs, e.g., LipoP [19], but with performance not comparable to SignalP5.0 [14]. However, not like SignalP5.0 that predicts a protein to be Sec/SPI, Sec/SPII, Tat/SPI and others, LipoP classifies a protein as Sec/SPI, Sec/SPII, a protein with a TMH and a cytoplasmic protein [19]. Therefore, LipoP shows additional application or advantages in distinguishing TMHs from SPs or other proteins without N-terminal TMHs or SPs. There are also other software tools that can particularly distinguish TMHs from Sec/SPI SPs, e.g., Signal-3L 2.0 [11], Phobius [21], Philius [23], TOPCONS [24], SPOCTOPUS [25], etc. These tools can also have other useful application, such as full-protein TM topology prediction, protein classification (e.g., TM proteins with SP, TM proteins without SP, globular proteins with SP and globular proteins without SP) and others, in spite that they cannot predict Sec/SPII SPs, and they cannot or only poorly predict the SP cleavage sites.
Savojardo et al recently also proposed a deep learning based SP prediction tool, namely DeepSig, which can both predict Sec/SPI SPs and find the cleavage sites effectively [28]. DeepSig showed better performance than SignalP 4.1 and other tools in prediction of SP cleavage sites [28]. Although SignalP 5.0 was reported to show better performance than DeepSig in recognition of SPs, the prediction accuracy of the cleavage sites has not been compared between the two tools, and therefore DeepSig could still have the advantages in cleavage site prediction [14]. Another deep learning tool, namely SigUNet, was recently developed, which could also predict Sec/SPI SPs of gram-negative bacteria but did not show better performance than SignalP4.0 or DeepSig [295]. Signal-3L 3.0, a model using a 3-layer hybrid method of integrating deep learning algorithms and window-based scoring, showed better performance in prediction of Sec/SPI SPs of gram-negative bacteria but poorer performance in cleavage site prediction compared to SignalP 5.0 [296].
In summary, despite a batch of algorithms or tools that have been developed to predict Sec substrates, at present, SignalP 5.0 appears to have the performance superior to others and could be the first choice. However, other tools remain useful for specific purposes, e.g., cleavage site prediction, TMH / TM topology prediction, additional protein annotation, etc. Novel algorithms and tools are also required, to further improve the precision, to make taxon-specific prediction, and to distinguish the uncleaved SPs and/or other types of SPs.
2.2. The twin arginine translocation (Tat) pathway
2.2.1. Brief summary
Tat protein export system is also present in the inner membrane of many archaea, bacteria, chloroplasts, and plant mitochondria. Different from Sec pathway, Tat pathway exports folded proteins of varied size [29]. A typical Tat system is composed by subunits TatA, TatB and TatC (TatABC) or only TatA and TatC (TatAC) [30]. TatA, TatB and TatC are integral membrane proteins. TatA and TatB are homologous to each other, evolved from a common ancestor but have derived different function [29]. The TatA component of the TatAC system shows the function of both TatA and TatB of the TatABC system [29]. In Gram-negative bacterium, TatABC is the only known Tat system. The mechanism of protein translocation through Tat pathway remains largely unclear. In E. coli, TatB and TatC bind the twin arginine containing SPs of substrate proteins, followed by TatA recruitment, translocase channel formation and substrate translocation through the channel [31]. The Tat substrate export process is energized by the transmembrane proton motive force (PMF) [29]. After translocation, the substrate is released to periplasm after the SP is removed by a signal peptidase. However, not all the SPs of Tat substrates are cleaved. For example, bacterial Rieske iron–sulphur proteins have uncleaved SPs, which serve as signal anchors and are released laterally from the transporter into the membrane bilayer [29]. Some proteins with uncleaved Tat SPs can also be destined to the bacterial outer membrane with an unknown mechanism [29].
The proteins targeted to Tat pathway are much fewer than Sec substrates. In some bacteria, there is no Tat pathway [29]. However, the Tat substrates participate in various cellular processes, such as anaerobic metabolism, cell envelope biogenesis, metal acquisition and detoxification, and virulence [32]. In many important pathogens, Tat pathway is essential and closely related with the pathogenicity. Given the lack in mammals, Tat pathway and its substrates serve as ideal targets of new anti-bacterial drugs [30]. Therefore, it appears promising to predict Tat substrates, explore the mechanisms of the Tat exporting pathway, and apply the findings in drug research and development.
2.2.2. Molecular features of the proteins secreted through Tat pathway
Similar to Sec substrates, most Tat substrates also have N-terminal SPs that can be cleaved by SPase I or SPase II. The SPs of Tat substrates (Tat-SPs) also showed similar sequential features with those of Sec substrates, in spite that Tat-SPs often contain a conserved motif with two consecutive arginine residues (Fig. 3B). Tat-SPs are often longer than Sec-SPs, mainly due to their frequently longer N-regions [3]. Most computational models predict Tat substrates by recognition of Tat-SPs specifically.
2.2.3. Algorithms and tools predicting proteins secreted through Tat pathway
Not like the Sec-SP predictors, only a handful of Tat-SP predictors have been developed, including TATFIND, TatP, PRED-TAT and SignalP 5.0 (Table 3). TATFIND uses regular expression pattern matching approach and performs hydrophobicity analysis to identify Tat substrates [33], [34]. A Tat substrate was predicted by TATFIND originally if (1) there was a motif (X-1) R0 R+1 (X+2) (X+3) (X+4) in the N-terminal 35 amino acids where X represented a permitted residue from a pre-defined set, and (2) there was an uncharged peptide fragment with no fewer than 13 amino acids at the downstream of R0 R+1 [30]. TATFIND version 1.2 expanded the rules, allowed methionine at X−1 and glutamine at X+4, and provided a full list of predicted Tat substrates in 84 microorganisms [34]. TATFIND can also distinguish Tat-SPs and Sec-SPs to some extent, but cannot predict the cleave sites of Tat-SPs [34]. Bendtsen et al proposed a new method, TatP, combining pattern-matching for filtering and ANN for classification, which could classify Tat-SPs, Sec-SPs and cytoplasmic proteins with similar motifs high-accurately [35]. TatP can also predict the underlying cleavage sites of Tat-SPs [35]. The comparison between TatP and TATFIND with different independent testing datasets demonstrated that TatP showed a decreased false positive rate but an increased false negative rate [35]. PRED-TAT is an HMM-based method, which could classify Tat-SPs and Sec-SPs, predict the cleavage sites, and show higher accuracy than TATFIND and TatP [36]. Besides these tools, as mentioned above, the most recently developed SignalP 5.0 can also predict the Tat/SPI SPs and shows the best prediction performance [14].
Table 3.
Representative software tools predicting Tat substrates in Gram-negative bacteria.
| Tool | Method | Target | URL or reference |
|---|---|---|---|
| TATFIND 1.4 | Regular expression pattern; Hydrophobicity analysis | Tat/SPI; Sec/SPI | http://signalfind.org/tatfind.html; [33], [34] |
| TatP 1.0 | Regular expression pattern; ANN | Tat/SPI; Sec/SPI; Cleavage site | https://services.healthtech.dtu.dk/service.php?TatP-1.0; [35] |
| PRED-TAT | HMM | Tat/SPI; Sec/SPI; Cleavage site | http://www.compgen.org/tools/PRED-TAT; [36] |
| SignalP5 | DNN | Tat/SPI; Sec/SPI; Sec/SPII; Cleavage site | https://services.healthtech.dtu.dk/service.php?SignalP-5.0; [14] |
Generally, the tools predicting Tat substrates are limited, and currently SignalP 5.0 is the first choice. However, it should be noted that none of the tools (including SignalP 5.0) could recognize the Tat lipoprotein substrates cleaved by SPaseII. Besides the proteins with SPs, there are also Tat substrates that do not contain any targeting sequences. These proteins take a hitchhiker mechanism to be exported by Tat pathway, by forming a complex with partner proteins containing Tat-SPs and being targeted with assistance of the partners sharing the SPs [37]. E. coli hydrogenase 2 subunit, HybC, is an example of such type of Tat substrates [37]. However, the exporting mechanism is still unclear and the substrates remain largely unidentified, and consequently, corresponding prediction tools are still at a lack to date.
2.3. Non-classical exporters
The proteins hitchhiking to pass through the inner membrane via Tat pathway take a kind of non-classical secretion mechanism. In gram-negative bacteria, there are also other non-classical pathways, by which proteins without putative Sec-SPs or Tat-SPs can enter periplasm. SodA is a well-known example. SodA proteins in Helicobacter pylori, Aeromonas hydrophila, Rhizobium leguminosarum bv. viciae 3841, Rhodobacter sphaeroides and Paracoccus denitrificans all lack Sec or Tat signal peptides but are secreted into periplasm [38], [39]. Secretion of the proteins via the inner membrane is Tat independent but requires SecA and N-terminal sequences [39]. No SecA2 pathway has been found in Gram-negative bacteria and therefore the proteins could be secreted through Sec pathway in a non-classical manner like the maltose-binding protein and alkaline phosphatase in E. coli [8], [9], or there could be other similar, not-yet-identified, non-classical pathways for secretion of these proteins. There are also other similar proteins secreted by such non-classical pathways, e.g., LuxS and TtsA in Salmonella [40], [41], ChiC in Serratia marcescens [42], etc. Currently, the knowledge about these non-classical pathways and the property of substrates is quite limited, and no method has been developed to predict such secreted proteins.
3. Outer membrane and two-membrane spanning secretion systems
There are multiple secretion systems identified that span only the outer membrane (e.g., T5SSs, T7SSs and T8SSs) or both inner and outer membranes (e.g., T1SSs, T2SSs, T3SSs, T4SSs, T6SSs and T9SSs). Here, we will review the substrate proteins of each secretion system according to the naming order of the systems, which also reflects the time order for their first identification in Gram-negative bacteria.
3.1. T1SS
3.1.1. Brief summary
T1SSs have been reported in a large variety of Gram-negative bacteria, including plant and animal pathogens. They can transport the unfolded substrates outside cells through inner and outer membranes in one step [43]. A T1SS is composed by three elementary components - an ATP-binding cassette (ABC) transporter located in inner membrane, an outer membrane factor (OMF), and a membrane fusion protein (MFP) connecting the ABC transporter and OMF (Fig. 2) [43], [44]. Most OMFs belong to the multi-functional TolC family. T1SSs have a structure similar to that of resistance-nodulation-division (RND) pumps in Gram-negative bacteria which can transport small molecules such as antibiotics [45].
The T1SS substrates, also called Type 1 Secreted Effectors (T1SEs), have various biological function, such as virulence-related HlyA [46], Salmonella non-fimbrial giant adhesin SiiE [47], Legionella pneumophila RtxA [48] and Acinetobacter RTX-serralysin-like toxins [49], biofilm formation related RTX adhesin LapA [50], [51], digestion enzymes TliA and PrtA [52], etc. It appears promising to engineer T1SSs in biomedical applications owing to simple structure of the system and the frequent T1SE C-terminal signal sequences that are convenient for genetic manipulations [53].
3.1.2. Molecular features of the T1SS substrates
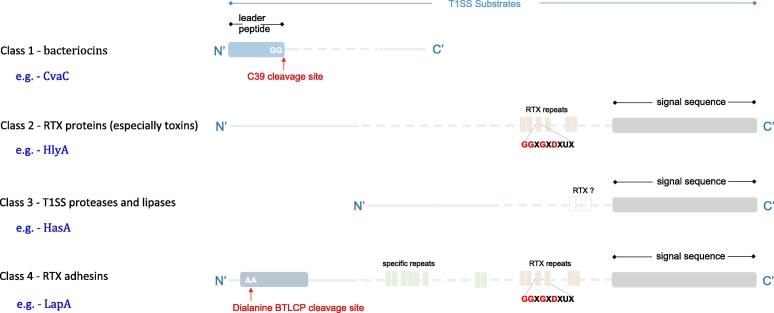
The ABC transporters of T1SSs often show high specificity in binding the substrates. According to ABC transporter types, typical T1SEs can be divided into 3 classes (Classes 1 ~ 3) (Fig. 4). Class 1 T1SEs are targeted to C39-containing ABC transporters with hydrolase activity. These T1SEs normally contain N-terminal leader peptides. The C-termini of the leader peptides contain a canonical double glycine (‘GG’) motif, which can be recognized and cleaved by the C39 domains of corresponding ABC transporters before the mature proteins being secreted through T1SSs (Fig. 4) [54]. The Class1 T1SEs are the known smallest T1SS substrates, including the small bacteriocins or microcins.
Fig. 4.
Sequence features of T1SS substrates. T1SSs can be divided into 4 groups. The substrates of Class 1 T1SSs typically contain N-terminal leader peptides (blue), while Classes 2–4 have secretion signal sequences in the C-termini (grey). Consensus sequence motifs are shown for the RTX repeats (light green and pink). RTX repeats are not necessarily present in Class 3 T1SEs. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Class 2 T1SEs are targeted to C39-like domain (CLD)-containing ABC transporters. These T1SEs have specific repeats-in-toxin (RTX) domains and are also called RTX proteins. The glycine-rich nanopeptide repeats in RTX domains show a ‘GGxGxDxUx’ consensus sequence motif where ‘x’ is any amino acid and ‘U’ represents a large or hydrophobic amino acid. Different from the Class 1 T1SEs, the RTX proteins have a large molecular mass. The CLDs of corresponding ABC transporters have a structure similar with C39 peptidase domains but do not show any hydrolase activity. The RTX proteins do not contain N-terminal leader peptides or ‘GG’ motifs as seen in Class 1 T1SEs either [54]. The Class 2 RTX T1SEs have secretion signal sequences in the C-termini, but the signal patterns and function mechanisms have not been clarified (Fig. 4) [54]. It is also unclear how the CLD-containing ABC transporters interact with the substrates.
Class 3 T1SEs are targeted to a type of ABC transporters without any additional N-terminal domain. The substrates do not necessarily contain RTX repeat sequences, but have a C-terminal secretion signal sequence as in RTX proteins (Fig. 4). They do not contain N-terminal leader peptides either. The T1SEs of this class normally have a size much smaller than RTX proteins and have hydrolytic activity. Various proteases, lipases and the iron scavenger protein HasA belong to this group [54].
Recently, a fourth class of T1SEs has been reported, exemplified by RTX adhesins [55]. Different from Classes 1–3, the RTX adhesins are transported from cytoplasm to extracellular space by two steps, and therefore considered as non-classical [55]. The class of T1SS machinery is often linked with a bacterial transglutaminase-like cysteine proteinase (BTLCP) [56]. The RTX adhesion proteins have dialanine BTLCP cleavage sites in the N-terminal retention module that can be recognized and cleaved by the machinery-coupled BTLCP in periplasm before the cross-outer membrane transport [57], [58], [59]. The currently known RTX adhesins also have specific repeats that are important for their function, RTX repeats and signal sequences in the C-termini (Fig. 4).
3.1.3. Algorithms and tools predicting T1SS substrate proteins
Despite the functional importance and the large number of T1SEs, there are very few software tools developed to predict them (Table 4). Linhartova et al combined pattern searching, HMM profiles and RPS-BLAST, to predict 1024 candidate RTX proteins from 840 bacterial genomes [60]. In 2015, Luo et al made the first try to design a machine-learning model to predict RTX proteins [61]. Luo’s method combined sequence-derived features and the random forest (RF) algorithm, and considered both the full-length T1SE sequences and the newly identified C-terminal signals to improve the prediction precision [61]. To date, algorithms or tools have not been reported for prediction of other classes of T1SEs.
Table 4.
Representative software tools predicting substrates of T1 ~ 9SSs.
3.2. T2SS
3.2.1. Brief summary
T2SSs are conserved in Gram-negative bacteria. They transport folded substrate proteins from periplasm through the outer membrane. The substrates could either be anchored in outer membrane or secreted into extracellular milieu completely. T2SS is a complicated apparatus comprised of 40–70 proteins belonging to 12–15 different families. The apparatus consists of four sub-assemblies (Fig. 2): an inner membrane platform, an outer membrane complex, a secretion ATPase and a pseudopilus located in periplasm but connecting with the inner membrane platform [62]. The secretion of T2SS substrates involves a two-step process, while the proteins must be exported into periplasm through Sec or Tat pathway before secretion [62]. If exported through Sec pathway, the protein must fold in periplasm before T2SS secretion. Structural components of the T2SS apparatus cooperate to recruit and facilitate the substrates to enter the secretin channel formed by the outer membrane complex. The inner membrane platform connects the sub-assemblies and coordinates substrate transportation. The secretion process is energized by the ATPase located in cytoplasm, while the pseudopilus pushes substrates forward to pass through the channel in a piston-like manner. The pseudopilus shows similarity to type IV pili phylogenetically and structurally [63]. The inner membrane proteins, outer membrane proteins and ATPases of T2SSs also show homology to the type IV pili system (T4P) and the tight-adherent pili system (Tad). Therefore, both T4P and Tad have been classified as subtypes of T2SSs [2]. Consequently, T2SSs can be divided into 3 classes, i.e., T2aSSs (classical secretin-dependent T2SSs), T2bSSs (T4P) and T2cSSs (Tad) [2]. More details were given for T4P and Tad systems in Section 3.7.
T2SS substrates are mainly comprised of enzymes, including proteases, lipases, phosphatases and others, which can facilitate bacteria to adapt to the environment and survive [62]. Some T2SS substrates can destroy host defenses, provide nutrients for bacteria, and facilitate bacterial colonization and diseases [64]. For example, the Acinetobacter lipases LipA and LipH as well as the protease CpaA exert important function in bacterial colonization and spread [65]. Enterohemorrhagic E. coli (EHEC) secretes YodA through T2SS to facilitate its adhesion and colonization [66]. Many pathogens also use T2SSs to secrete toxins and cause diseases. For example, the cholera toxin of Vibrio cholerae is secreted through T2SS and causes severe watery diarrhea [67]. The exotoxin A plays an important role in the lethal infection of Pseudomonas aeruginosa [68].
3.2.2. Molecular features, computational algorithms and tools of the T2SS substrate proteins
It remains an enigma how a T2SS recognizes and transports the widely distributed substrate proteins [62]. Structural studies indicated that T2SS substrate proteins contained relatively abundantβ-strands, and yet a common secretion signal has not been identified that could be specifically recognized by a T2SS [62], [69]. There could be some spatial secretion motifs comprised by the residues from different regions of a protein and formed only after protein folding or assembly [62], [70].
The algorithms and tools remain at a lack to predict the T2SS substrates, mainly because of the difficulty in seeking for common features among the molecules. The structure resolution, analysis and comparison of more T2SS substrate proteins may lend breakthrough features and lay the foundation for accurate prediction of new important T2SS substrates in future.
3.3. T3SS
3.3.1. Brief summary
T3SS is a syringe-like apparatus spanning both inner and outer membrane, with the tip of needle piercing the membrane of host cells and mediating the translocation of substrate proteins from bacterial cytoplasm into the host cytoplasm in one step [71]. Being only identified from Gram-negative bacteria, including many important animal and plant pathogens, T3SSs play important roles in bacterial interaction with host cells and the pathogenicity [72], [73]. A T3SS apparatus is comprised of ~30 structural and accessory proteins, which form multiple sub-assemblies, including a cytosolic sorting platform (SctO/L/K for the unified nomenclature for conserved components of T3SS) with an ATPase (SctN), a cytoplasmic ring (SctQ), an inner membrane export apparatus (SctR/S/T/U/V), an inner membrane ring (SctJ and SctD), an outer membrane ring (SctC), an needle assembly also called inner rod, a needle or pilus, and a translocon tip complex that is in the host cell membrane (Fig. 2) [74], [75], [76], [77], [78].
Bacterial flagella transport systems have high structural similarity to T3SSs and component proteins homologous with T3SS proteins, and they are likely to have the most recent common ancestor evolutionarily [78], [79], [80]. Therefore, the flagellar protein export system has been considered as a sub-class of T3SSs (T3bSS) [2]. The effector-translocation non-flagella T3SSs are called T3aSSs correspondingly. Not like the T3aSS needles (or pili) that mainly serve as protein translocation machine components, the homologous counterpart in T3bSS, i.e., flagella, can participate in chemotaxis, adhesion, biofilm formation, effector secretion and immune system regulation [81]. A complete T3bSS is composed by around 30 unique structure proteins with several to 10,000 s of copies [82]. A typical flagella export system contains three structural parts: the basal body which contains the reversible motor that anchors the structure to the membrane, the hook which extends out from the top of the basal body and acts as a universal joint, and the filament which extends many cell body lengths from the hook and, when rotated, forms the helical propeller [81]. Like the membrane rings and inner membrane export apparatus in T3aSSs, the basal body proteins in T3bSSs are exported through Sec pathway. Once the core T3SS is assembled, the subunit proteins of flagella (e.g., the flagellar hook FlgE and the hook-capping protein FlgD) and T3aSS needle pili are transported through respective conduit [80]. Some studies demonstrated that flagella and T3aSS pili proteins contain common secretion signals so that they can be secreted through the other injectosome [83], [84], [85].
3.3.2. Molecular features of the T3SS substrates
Like the fimbriae systems, in most cases, the flagella export systems only secrete the flagella subunit proteins, which can easily be recognized by homology searching. Therefore, here we focus on the substrates of non-flagella T3SSs.
The non-flagella T3SSs deliver a list of substrates into host cells, which often exert important function and facilitate bacterial colonization, invasion, infection and survival. These substrates are also called T3SS secreted effectors (T3SEs). A classical T3SE contains a secretion signal bearing N-terminus, C-terminal effector domain(s), and a chaperone-binding domain (CBD) connecting the termini (Fig. 5) [86], [87]. Both the N-terminal signal sequence and the CBD domain are essential for T3SS recognition, recruiting and secretion [88], [89], [90]. The length of signal sequences varied from ~5 to ~100 amino acids, and no common motif has been identified from a majority of the effectors though atypical amino acid composition bias profiles (e.g., serine/threonine/proline being enriched) were observed [91], [92]. Specific chaperones bind to T3SEs at the CBDs and unfold the T3SEs, the latter of which could only be translocated through the T3SS conduit at an unfolded status [90], [93]. The chaperones often pair with effectors, the genes co-localize in genome and they co-evolve [93], [94]. A common structural motif was identified in CBDs of a list of T3SEs from a variety of bacteria [95]. The effector domains are diverse and generally un-conserved among different species. However, there are still several large families identified from a number of bacteria of a broad diversity, e.g., YopM, YopJ, etc [73].Fig. 6B.
Fig. 5.
Sequence features of the substrates of type 3/4/6 secretion systems. (A) A classical T3SE contains a secretion signal bearing N-terminus, a C-terminal effector domain, and a CBD connecting the termini. (B) Classical T4SEs (T4aSE/T4bSE) show amino acid preference patterns in the C-terminal regions. Some of the effectors also contain essential translocation-guiding signals in the N-termini. Different from T4aSS and T4bSS effectors, T4xSS effector contains a conserved C-terminal domain termed ‘XVIPCD’. (C) Some of T6SEs contain MIX (marker for type six effectors) motif in the N-termini as the T6S signal potentially. There could be also other putative catalytic motifs as shown in example proteins (green). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 6B.
T5SSs and the features their substrates. (A) Substrate export of different types of T5SSs. (B) Sequence features of the substrates of different types of T5SSs. The pre-proteins of all these substrates also belong to Sec substrates and therefore contain SPs in N-termini. However, autotransporters (T5aSSs), TpsA exoproteins of the two-partner systems (T5bSSs) and trimeric autotransporters (T5cSSs) have extended signal peptide regions specifically (top). A T5aSS contains a passenger domain and a β-barrel translocator domain. Cleavage occurs between the two-asparagine residues located between the two domains (red arrow). A T5bSS is composed by two polypeptides, substrate TpsA and transporter TpsB. There is a conserved ‘NPNL’ motif in TpsA that is essential for its secretion. The TpsB and T5dSEs both contain POTRA (polypeptide transport-associated) motifs preceding the putative 16-stranded beta-barrel domains in the C-temini. T5cSS is composed by three polypeptides while T5eSS is inverted with an additional small periplasmic domain in the N-termini. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
There are still debates about the secretion signals of T3SEs, since some effectors appeared to have the signals contained in mRNA rather than protein level [96], [97], [98]. From DNA level, there could be conserved signatures buried in the promoter regions of effector genes or the operons that they belong to [73]. The effector genes are scattered in genome but often transcriptionally co-regulated by the pivotal T3SS regulators. The expression co-regulation requires the conservation of cis-acting elements. For many bacteria, especially plant pathogens, e.g., Pseudomonas syringae and Ralstonia spp., the motif features have been well defined in the regulon promoters of the key T3SS regulators, e.g., HrpL, HrpB, etc [99], [100].
3.3.3. Algorithms and tools predicting T3SS substrate proteins
Computational prediction of bacterial T3SEs has received a lot of research enthusiasm since the first T3SSs were identified. >20 algorithms and software tools have been developed (Table 4). Generally, the methods can be classified into 3 types: (1) methods based on sequence pattern recognition and homology searching, (2) machine-learning or statistic models mostly based on features buried in signal sequences, and (3) simple or ensemble models based on integrated features.
Homology searching of known T3SEs was the main strategy to predict new effector and achieved a large success in the early stage [101], [102]. However, a big limitation surfaced soon since the T3SEs show a large diversity in sequences and the number of experimentally verified novel effectors was very small. It was difficult to find more homologous ones based on a limited dataset of known effectors. As the number and diversity of validated effectors increased, however, the pattern-based or homology-based strategy remains an important choice for identification of partially new T3SEs [103], [72], [73]. Using homology-searching strategy and with a list of 519 non-redundant manually curated verified effectors, Hu et al recently identified 8740 T3SEs from hundreds of bacterial genomes with T3SS(s) [73]. Besides sequence patterns or homology in protein level, the promoter sequences were also studied and applied in prediction for the T3SE genes regulated by the key T3SS regulators [104], [99], [100]. However, this kind of features and effector prediction is frequently in species- or genus- specific manner.
Since the first two back-to-back reports published ten years ago [91], [105], more and more machine-learning algorithms have been introduced in T3SE prediction. Most algorithms focused on the sequence features in T3SE signal regions. For example, EffectiveT3 mainly learned the sequential composition features of physicochemical property-binned amino acids and oligopeptides in N-termini of known T3SEs using a Naïve Bayes (NB) model [91], while SIEVE also trained sequential features in both N-termini and full length of effectors as well as in gene sequences [105]. The position-specific amino acid composition (Aac) preference of T3SE signal sequences was learned for the first time by an ANN model [106], and further observed, refined, and trained in a Bi-Profile Bayesian (BPB)-SVM model (BPBAac) [92]. Other sequence-derived features, such as codon usage and instability, constraint of neighbor Aac, etc., were also observed and learned in new models [107], [108], [109], [110]. To predict T3SEs more specifically and precisely, Hobbs et al suggested to subgroup the training datasets and to develop species-specific models (GenSET) based on the N-terminal sequence features for better prediction accuracy [111]. New algorithms such as deep learning have also been applied to predict T3SEs. For example, DeepT3 is a deep convolution neural network (CNN) model that was trained most recently to learn the sequential features of known T3SEs within the N-terminal 100 amino acids [112]. Besides the sequential features, Arnold et al, Yang et al and Wang et al also observed the property of secondary structure (SSE) and water accessibility (ACC) in the N-termini of T3SEs [113], [91], [92]. Two models, SSE-ACC and T3SEpre, were trained to learn the SSE and ACC composition features and to predict new T3SEs [113], [114]. Common tertiary structures were also found in the signal regions of T3SEs [114]. Other T3SE features were also studied extensively and applied independently for effector prediction. For example, the chaperone-effector pairing features were explored for Bordetella T3SE identification [94], while the phylogenetic profiles were also observed and used for T3SE prediction [115].
Both single models and prediction models based on single types of features were found to be less effective when independent datasets were tested. Most intuitive examples are the homology-based methods and ab intio machine-learning models. The homology-based strategies can find a lot of true ‘new’ effectors. However, they are in fact not real new ones because they showed high sequence similarity with known effectors. Such approaches depend severely on the scale of validated effectors, and cannot find the real novel effectors. The machine-learning models often over-fit the local features and provide false positive predictions despite the ability in prediction of novel effectors. To overcome the drawbacks of each strategy, several T3SE predictors integrated them as different arms to make more accurate prediction. For example, BEAN2.0 initiate a web-based T3SE prediction platform, with both homology-searching module and machine-learning models [116]. Similarly, pEffect combined homology-based prediction (PSI-BLAST) and ab intio SVM models to make comprehensive prediction of T3SEs [117]. The ensemble of individual machine-learning models was also found to achieve better performance in T3SE prediction [298], [118], [119]. A recently developed tool, Bastion3, which is an ensemble model integrating multiple types of T3SE features, was reported to achieve much better performance compared to commonly used methods [120]. An integrated prediction method, T3SEpp, was also published most recently, which takes into account of multiple-aspect features, considers both homology-searching and machine-learning techniques, and forms a hierarchical ensembler to make more precise T3SE prediction with an apparently lowered false positive rate [121].
Although different algorithms and methods have their own merits in prediction of T3SEs, the integrated prediction strategies making both homology searching and machine-learning prediction, such as pEffect, BEAN2.0 and T3SEpp, achieved better performance in average, by evaluation with different bench-marking datasets. Methods considering multiple-aspect features and hierarchically integrating multiple models, e.g., T3SEpp and Bastion3 are also recommended.
3.4. T4SS
3.4.1. Brief summary
T4SS is also a multi-component complex expressed by versatile bacterial species [122], [123]. It can mediate the transfer of DNA or protein substrates into a large range of eukaryotic and bacterial cells. Based on the substrate type, T4SSs can be divided into two major families, conjugation systems and effector translocators [122], [123]. The conjugation T4SSs are distributed in both Gram-positive and Gram-negative bacterial species, and mediate the transfer of mobile genetic elements (MGEs). The effector-translocation T4SSs are mainly found in Gram-negative bacteria, for which the substrates could be proteins, single-strand and double-strand DNA molecules [122]. There are also a few other T4SSs that can secrete DNA or protein substrates into extracellular milieu [122], [123]. Phylogenetic analysis based on the conserved T4SS components suggested that the conjugation T4SSs emerged in Gram-negative bacteria first and were expanded to Gram-positive species, followed by the most recent diversification into the dedicated effector translocation systems and others [124], [125], [126].
The effector-translocation T4SSs were classified into two broad phylogenetic groups designated as types IVA (T4aSS) and IVB (T4bSS) respectively. T4aSS is represented by the Agrobacterium tumefaciens VirB/VirD4 T4SS encoded by the R388 plasmid and the Helicobacter pylori Cag T4SS [127], [128]. The T4SSs are composed of 12 core subunits, VirB1-11 and VirD4, each with multiple copies, forming four structural sub-assemblies (Fig. 2): (1) cytoplasmic ATPases (VirB4, VirB11, VirD4), (2) inner membrane platform (VirB3, VirB6, VirB8), (3) outer membrane core complex (VirB7, VirB9, VirB10) and (4) pilus (VirB1 transglycosylase, VirB2 pilin, VirB5 pilus-tip protein). T4bSS also involves multiple (>25) proteins for assembly, among which a few are similar to VirB/VirD4 subunits and others (>20) are T4bSS specific. The Legionella pneumophila Dot/Icm T4SS is a typical example of T4bSS, which also contains the four major sub-assemblies similar to T4aSS [129], [130]. Recently, Xanthomonas citri T4SSs have been recognized as a new group, namely X-T4SSs (T4xSS), which is similar to T4aSS, but contains an uncharacteristically large VirB7 lipoprotein subunit whose C-terminal N0 domain decorates the periphery of the outer membrane layer of the core complex [131], [132], [133]. Another important feature of T4xSS is its ability to mediate the translocation of effectors into and kill competitor bacteria [134], [135].
3.4.2. Molecular features of the T4SS substrates
Like T3SSs, the protein-translocation T4SSs also show preference for the substrate effectors by specific recognition of the secretion signals (Fig. 5). Motifs or amino acid preference patterns were disclosed within the C-terminal regions of T4SS effectors (T4SEs), e.g., two positively charged amino acids separated by three or four amino acids among which at least one is negatively charged [136], frequently tiny and polar amino acids [137] and significant enrichment of glutamic acid and serine [138]. More flexible secondary structure and higher hydrophilicity were also found for the C-terminal signal regions of T4SEs [138]. Some of the effectors also contain essential translocation- guiding signals in the N-termini [139], [140]. Different from T4aSS and T4bSS effectors, T4xSS effectors interact with the effector-coupling protein VirD4 by a conserved C-terminal domain termed XVIPCD (Xanthomonas VirD4-interacting protein-conserved domain) [133], [134], [135].
The sequence features described above have been used for T4SE prediction frequently (Table 4). There are also other atypical or species-specific sequence-based features, such as the GC content, gene regulatory patterns, eukaryote-like domains, etc, which have also been used for effector identification [141], [142], [143].
3.4.3. Algorithms and tools predicting T4SS substrate proteins
The earliest T4SE prediction algorithms and tools were all species specific, e.g., the ones predicting L. pneunophila and Coxiella burnetii protein substrates [141], [142]. Burstein et al applied machine-learning algorithms in prediction of L. pneunophila T4SS effectors for the first time [141]. SVM, NN, NB and Bayesian network (BM) based models were trained to learn the genomic, evolutionary, regulatory and other specific features of L. pneunophila effectors. A voting-based strategy was adopted to combine the prediction results of different models. Moreover, the model performance improved through an iterative process of model training, prediction, validation and inclusion of newly validated effectors [141]. Chen et al combined gene selection and bioinformatic sequence feature analysis, and proposed a method to infer the T4SEs in C. burnetii [142]. Although these methods achieved ideal effect in prediction of effectors from L. pneunophila or C. burnetii, the species-specific features such as regulatory attributes limited their general application in other species.
After the L. pneunophila specific models were developed [141], the same group also trained a hidden semi-Markov model (HSMM) to represent the common Aac in effector secretion signals of Legionella and Coxiella T4SSs [144]. The model could make cross-species effector prediction, but mainly for IVB T4SSs [144]. T4EffPred and T4SEpre represent two real general T4SE predictors developed in an earlier time [138], [145]. Both T4EffPred and T4SEpre are SVM-based models and take protein sequences as input. T4EffPred takes the full-length protein sequences for feature analysis, and can classify the effectors of two types - IVA and IVB - of T4SSs [145]. However, because of the possible common features between T3SEs and T4SEs, there could be false positives of T3SEs in the T4EffPred results [143]. Wang et al manually annotated a complete list of experimentally validated T4SEs from different bacteria, observed the possible common motifs, sequential and position specific Aac, secondary structure and solvent accessibility features in C-termini of the effectors, and developed three models, i.e., T4SEpre_psAac, T4SEpre_bpbAac and T4SEpre_Joint [138]. Despite the good general and cross-species performance, T4SEpre also showed its main drawback, which was caused by the features constrained in only the C-terminal 100 amino acids of the candidate proteins [138]. In fact, at least some T4SEs also contain secretion signals at the N-termini [139], [140]. Another model was thereafter trained to overcome this limitation, with features extracted from both N-terminal 50 and C-terminal 100 amino acids of the subject proteins [146]. Similar for T3SE prediction, recently, several hierarchical ensemble models with multi-aspect features, e.g., PredT4SE-Stack and Bastion4, have been trained to improve the prediction performance for T4SEs [147], [148]. CNN-T4SE integrated three Convolutional Neural Network models training the amino acid composition, solvent accessibility and secondary structure of full-length T4SEs, achieving better performance than other tools and lower false positive predictions [298]. Other groups adopted an alternative strategy, by selecting the best optimized features, and/or training and identifying the best machine learning models, to improve the prediction performance [299], [149], [150], [151]. Some of the models have been well applied in identification of T4SEs in L. pneumophila [151] and Anaplasma phagocytophilum (OPT4e; [150]). Besides the machine-learning based methods, homology searching was also used in T4SE prediction. For example, S4TE integrated 13 sequence homology based features, including homology to known effectors, homology to eukaryotic domains, presence of subcellular localization signals, secretion signals, etc., and developed a scoring scheme to predict T4SEs mainly from α- and γ- proteobacteria [152].
T4SSs and the substrates are most complicated. A T4SS can deliver proteins, double-strand DNAs or single-strand DNAs into extracellular milieu, eukaryotic cytoplasm or competitor bacterial cytoplasm [122]. Currently, it remains unclear about the accurate clustering, distribution and substrate recognition mechanisms of T4SSs. None of the algorithms or tools can identify the possible common features in the protein and DNA substrates of the single or similar T4SSs. The machine-learning models are also unable to predict the effectors of T4SSs targeted to competitor bacterial cells. Generally, for the model species with a large number of T4SEs being validated, such as L. pneumophila, C. burnetii, A. tumefaciens, and A. phagocytophilum the species-specific models are suggested. For the species phylogenetically close to these species, homology-based screening strategies are recommended. For other species with a functional T4SS, as for T3SE prediction, the tools considering both homology and machine-learning ensemblers with multi-aspect features appeared to have better performance and therefore are recommended.
3.5. T5SS
3.5.1. Brief summary
T5SS is a special group of protein secretion system widely distributed in Gram-negative bacteria. A classical T5SS is only composed of a unique protein, which transports itself and is also called autotransporter [153]. The protein contains a β-barrel domain, which inserts into the bacterial outer membrane, forms a translocation channel and mediates the transport of the remaining protein fragments (the passenger domain) [63]. The autotransporters are secreted through the inner membrane via Sec pathway before being integrated into the outer membrane. There are also two- or multi-component T5SSs, but the conduits only span outer membrane, and the substrates need get into periplasm through Sec pathway at an unfolded state in the first place. Therefore, the pre-proteins of the T5SS proteins contain N-terminal Sec signal peptides, which are cleaved after export into periplasm [63]. Despite the simplicity in the protein composition compared to other protein secretion systems, T5SSs also show a large diversity of categories and function [153]. At present, the known T5SSs can be divided into 5 classes, namely T5aSS through T5eSS (Fig. 6A-B) [154]. T5aSS represents a classical one-component autotransporter. T5bSS is also called two-partner secretion (TPS) system, which is composed by two polypeptides, including a secreted substrate collectively designated as TpsA and a transporter protein TpsB spanning the outer membrane [154], [155]. Both TpsA and TpsB pre-proteins contain an N-terminal signal peptide that is recognized by Sec pathway. There is also a conserved TPS domain located at the N-ternimus of TpsA, which is targeted to the outer membrane protein TpsB. TpsB protein contains two periplasmic polypeptide transport-associated (POTRA) domains [154]. T5bSSs mainly transport some toxins with large volumes, such as the filamentous haemagglutinin of Bordetella pertussis [156], and the adhesins HMW1 and HMW2 of Haemophilus influenza [157]. T5cSSs, also named trimeric autotransporters, could be the most complicated autotransporters [154]. The passenger domains of T5cSSs show a large diversity while the translocation domains are highly conserved [154]. Most T5cSS substrates are adhesins, and they are also called trimeric autotransporter adhesins (TAAs). TAAs are important virulence factors in Gram-negative bacteria, e.g., the YadA proteins of enteropathogenic Yersinia spp. [158], [159]. T5dSS is the fused two-partner system, which is also composed by a single protein and has a structure similar to a T5aSS, with a C-terminal translocation domain and an N-terminal passenger domain. The prototype of T5dSSs is a patatin-like protein from Pseudomonas aeruginosa, PlpD [160]. It appears that the passenger domain of PlpD fuses with the β-barrel domain by the POTRA domain. T5dSS also contains a periplasmic domain, which is homologous to the periplasmic domains of T5bSSs [154]. The proteins secreted through T5dSSs are mostly distributed in environmental, avirulent bacterial species [154]. T5eSSs are a group of inverted autotransporters, with domains organized in an oppose direction, i.e., passenger domains formed by the C-termini and transport channels formed by the N-termini [161]. The passenger domains of T5eSSs are mainly Ig-like and lectin-like domains not found in other groups of T5SSs [162]. In addition, there is a small periplasmic domain at the N-terminal region, which shows no homology to those of T5bSSs and T5dSSs [161].
3.5.2. Molecular features, computational algorithms and tools of the T5SS substrate proteins
The T5SS substrate proteins often show high sequence conservation from the same class for the local domains, and homology searching is the most frequently adopted approach to recognize these proteins [154], [158], [163]. BLAST (blastp) is the routine tool to find the autotransporters from genome and protein databases. More sensitive, Position-Specific Iterated (PSI)-BLAST can also be used to find the T5SS proteins with lower sequence similarity with known ones [153]. Celik et al built HMM profiles to identify 1523 autotransporter proteins from numerous Chlamydiales and Fusobacteria species as well as all classes of Proteobacteria [163]. Analysis on these proteins disclosed a diversity of passenger domains besides the known proteases, adhesins and esterases [163]. Based on the conserved motifs within the β-barrel domains, the T5aSSs were clustered into 14 sequence families [163]. Zude et al further identified new T5aSS substrates in 111 publically available E. coli genomes with homology and profile based methods, and expanded the number of sequence families to 18 [164]. With the same strategies, Vo et al identified 728 autotransporter proteins of the T5aSS AIDA-I group [165]. Most recently, Goh et al used the similar sequence alignment based strategy to identify four new inverse autotransporters (IATs, T5eSS substrates) from 126 finished genomes of E. coli [166].
3.6. T6SS
3.6.1. Brief summary
T6SS is also a multi-protein complex, with a phage tail-like structure but in an opposite orientation from phage infection [167]. A typical T6SS involves ~15 proteins, assembling a two-membrane spanning nanomachine that can translocate the substrate proteins into eukaryotic or competitor bacterial cells (Fig. 2) [167]. The T6SSs are related with both bacterial pathogenicity and competition with non-self microorganisms [168], [169], [170], [171].
The known T6SSs are only distributed in Gram-negative bacteria, mostly Proteobacteria and Bacteroidetes [172], [173], [174]. Phylogenetic analysis of the T6SS core genes classified the T6SSs into three major classes: (1) group i (T6aSS) predominated in proteobacteria, (2) group ii (T6bSS) represented by the Francisella Pathogenicity Island (FPI) T6SSs and (3) group iii (T6cSS) comprised of Bacteroidetes T6SSs [173], [175], [176].
3.6.2. Molecular features, computational algorithms and tools of the T6SS substrate proteins
Till now, only a few T6SS effectors (T6SEs) have been identified experimentally. Many of them are specialized effectors, which are VgrG and Hcp proteins fused with C-terminal effector domains [177], [178], [179]. Strategies based on sequence alignment or motif pattern searching identified a list of VgrG or Hcp C-terminal extended T6SEs, and also disclosed various effector domain containing protein families, which are called ‘cargo’ effectors [180], [181], [182], [183], [184]. The cargo effectors can bind the inner surface of the Hcp tube or interact with VgrG spike or PAAR repeat-containing proteins [185], [186]. Salmon et al identified a conserved MIX (marker for type six effectors) motif in the N-termini of a group of effector-domain containing independent T6SEs (Fig. 5) [187]. By searching the motifs, a number of new potential T6SEs were identified [187], [188]. However, experiments have not been performed to examine the function of the motif. There are also a lot of non-MIX effectors [188], [189].
Bastion6 is the first machine-learning based T6SE predictor [190]. It extracted a large number of features from a very limited number of homology-filtered T6SEs, including sequence profile, evolutionary information and physicochemical property, and trained the two-layer hierarchical model [190]. Bastion6 is also restricted to process less than 500 sequences per job with amino acid count between 50 and 5000, and to overcome this inconvenience, Sen et al proposed a new tool PyPredT6 [191]. PyPredT6 also used a broadened positive training dataset, considered both the amino acid and nucleotide based sequence features, and adopted 5 different machine learning algorithms to find the consensus predictions [191]. There are also other comprehensive tools or algorithms, which can predict T3SEs, T4SEs and T6SEs simultaneously. For example, Tbooster contains three ensemble models integrating the different machine learning methods or algorithms developed by others to predict T3SEs, T4SEs and T6SEs respectively [118], while Orgsissec encodes and uses the phylogenetic profile features to predict T3SEs, T4SEs and T6SEs [115].
Generally, we have very limited knowledge about the sequence features of T6SEs, the number of validated effectors is also limited, and there are no many software tool choices for T6SE prediction at present. Experiments, thorough feature analysis and new algorithms are all urgently required to facilitate identification of more T6SEs.
3.7. T7SS - Chaperone-Usher (CU) fimbriae, T8SS – curli, and other pili secretion systems
T7SSs have been widely recognized as the ESAT-6 secretion systems (ESXs) distributed in Gram-positive bacteria, especially Mycobacteria [192], [193], [194], [195]. However, because the numerical categorization was originally used for the protein secretion systems in Gram-negative bacteria, the Chaperone-Usher (CU) pathway was suggested to be named T7SS, which was considered as an independent protein secretion system [2], [196]. In this research, we continued to use the naming scheme suggested by Desvaux et al. Pili, or named fimbriae, are a family of extracellular polymers attached at the bacterial outer membrane as non-flagella protein accessories. They have multiple functions such as adhesion, invasion, motility, biofilm formation and transmembrane transport of DNA and proteins [197], [198]. These protein accessories can be divided into 5 major classes according to their biosynthesis pathways: (1) Chaperone-usher (CU) pili (both the P and type 1 pili), and the alternative chaperone (AC) pili (such as the CS1 fimbriae and CFA/I fimbriae), (2) curli, (3) T4P, (4) the type III secretion needle pili (T3SP), and (5) type IV secretion pili (T4SP) including F-pili and T-pili [197]. Functionally, CU and AC pili, curli and T4P can help pathogens recognize, adhere and even invade target cells, but seldom transport substrates except for pilin proteins themselves, while the T3SP only serves as transporter device components and the T4SP can function in both ways [197].
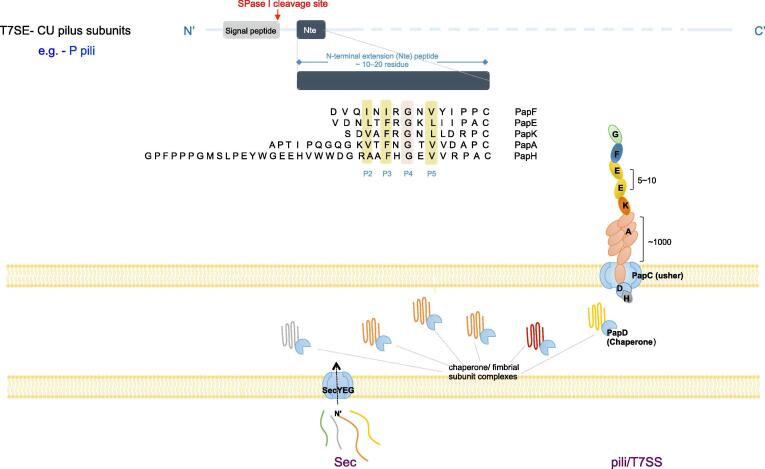
CU pathway, also named T7SS, is a ubiquitous protein accessory attached on bacterial cell surface [199]. The system is of simple structure, involving two assembly proteins: a specific periplasmic chaperone and an outer membrane assembly platform also called usher (Fig. 2; Fig. 7) [198]. The general concept of CU pathway contains the AC pili, for which the chaperone is not specific. All the structural proteins of CU pathway contain typical N-terminal signal peptides that are recognized and exported out of inner membrane through Sec pathway. The structure and protein transport mechanisms of CU pathway show similarity to T5SS, but the substrates of CU pathway fold in periplasmic space before being transported through the outer membrane [200]. CU system has 6 phylogenetic clades: α-, β-, γ- (subdivided into γ1, γ2, γ3 and γ4 sub-clades), κ-, π- and σ- fimbriae. The members from each phylogenetic clade have the common operon structure which encodes the fimbriae subunits of the similar protein domains [201]. The α clade is exemplified by CS1 fimbriae and secreted through AC pathway. The type P pili and type 1 pili belong to π and γ1 clade, respectively. The κ clade is mainly represented by the K88 (F4) fimbriae, while the σ-fimbriae refers in particular to the spore coat protein U from Myxococcus xanthus [197], [202], [203]. The β-clade fimbriae remain conceptual and are derived according to the sequences, as have not been observed for expression or assembly in any bacteria [197], [203].
Fig. 7.
Sequence features and the transport of the T7SS substrates. Pilus subunits contain SPs in the N-termini. The proteins are taken up by their cognate chaperones within periplasm, and a donor strand complementation (DSC) reaction occurs, by which a motif of four alternating hydrophobic residues (termed P1 to P4) on the chaperone G1 ftrand are inserted into a hydrophobic groove (known as the P1 to P4 pockets) of the pilus subunits so that a correct folding of the pilus subunits is catalyzed. CU pilus subunits also contain a 10–20 residue-long N-terminal extension (Nte) peptide that is sequentially conserved. During CU pilus subunit polymerization, the complementing G1 strand donated by the chaperone is replaced by the Nte on the subunit of the incoming chaperone–subunit complex. The assembly reaction is termed donor strand exchange (DSE). After DSE, the P2 to P5 pockets of the subunit groove are occupied by the hydrophobic residues (termed P2–P5) of the incoming subunit Nte. The P4 Gly residue in Nte sequences is strictly conserved.
Curli, a kind of functional amyloid fibers in nature, are the main protein compositions of the complex extracellular matrix of many enteric bacteria including E. coli and Salmonella species (Fig. 2) [204], [205]. As a type of secretion apparatus, Curli system is also called the extracellular nucleation-precipitation (ENP) pathway [206] or T8SS [2]. Curli fibers are linear, noncovalent polymers composed of the major and minor subunits CsgA and CsgB, respectively. These subunit proteins are transported through the ENP pathway at an unfolded state with the assistance of the accessory proteins CsgE, CsgF and CsgG [207], [208]. Curli are implicated in surface adhesion, cell aggregation, biofilm formation, infection and host inflammation [207].
Different from the CU pili and curli, the T4P system is independent of Sec pathway but requires the assembly of a two-membrane spanning, ATP-powering transporter apparatus. The pilin contains an unusual N-methylated amino terminus, a conserved hydrophobic N-terminal region composed of 25 residues, and a C-terminal disulphide bond [209]. T4P has long fibers (1–4 μm), strong and flexible dynamic filaments, which are formed and disassembled quickly by polymerization and depolymerization of the plilin subunits respectively [210]. T4P shows large structural similarity to T2SS [209], and was considered as a subtype of T2SS, i.e., T2bSS [2]. T4P contains two subtypes, type IVa (T4aP) and type IVb (T4bP). T4aP is distributed in various Gram-negative bacteria, while T4bP has only been reported in human intestinal bacteria [209]. Recently, McCallum et al introduced the ‘T4P-like system’ to describe the T4P and their alike systems with similar structure and transporting mechanisms, and classify them into 5 subtypes: T4aP, T2aSS, T4bP, Tad/Flp pili (T2cSS), Com pili, and archaeal flagellum (archaellum) [211].
T3SP is the core component of the T3SS apparatus. The pilus is a short, stiff filament (animal pathogens or symbionts) or a long flexible pilus (plant pathogens or symbionts). The distal polar structure allows bacteria to reach the plasma membrane of target cells [212]. The pilin subunits of T3SP are also transported by T3SSs and have the sequence features of T3SEs [212], [213]. Similar to T3SP, T4SP is the core component of the T4SS apparatus. T4SP plays dual roles by serving as the conduit of substrates other than T4SP pilin proteins, and functioning in adhesion of bacteria with target cells [214], [215]. T4SP was divided into two subtypes: IncF-like pili (conjugative pili produced by Inc-F, -H, -T and -J systems) and IncP-like pili (conjugative pili produced by Inc-P, -N, -W systems) [197], [216]. Dependent on T4aSSs and composed by VirB-like components, the IncP-like pili are short (<1 μm) rigid rods with 8–12 nm in diameter. The IncF-like pili, in contrast, are long (2–20 μm) and flexible appendages, which depend on T4bSSs and are composed by both VirB-like components and other proteins not present in IncP-like pili [197].
As protein secretion systems, except the T3SP and T4SP, the other fimbriae pathways have only been reported to transport the pilin subunit proteins themselves. Sequence alignment based strategies can identify these fimbriae systems and the pilin proteins.
3.8. T9SS – PorSS
T9SS, also known as PorSS, is a protein transport system specifically deployed by the Fibrobacteres–Chlorobi–Bacteroidetes (FCB) superphylum, which also serves as an important pathogenic factor in severe periodontal diseases [217], [218]. T9SS is a two-membrane spanning protein secretion system (Fig. 2). The protein-conducting translocon SprA (also named SOV) located in bacterial outer membrane is the core component of T9SS. SprA forms a large (36-strand) single polypeptide transmembrane β-barrel in bacterial outer membrane [218].
T9SS substrates are exported through bacterial plasma membrane via Sec pathway, folded in periplasm and then targeted to the T9SS translocon [219]. The substrates are large (100–650 kDa) multi-domain proteins, containing N-terminal Sec signal peptides and C-terminal folded domains (CTDs) composed by ~100 amino acids where T9SS targeting signals are located [218], [219], [220]. The CTDs have been proven to play essential roles in secretion, modification, and attachment of the substrates to cell surface [220]. The signal patterns of T9SS substrates have not been fully studied, and their prediction is mainly homology searching based [220]. By building HMM profiles for three conserved sequence motifs in CTDs and screening in 21 completely sequenced genomes of Bacteroidetes phylum, Veith et al predicted 663 CTD-containing proteins [221]. These proteins function as proteases, glycosidases, motility adhesins, hemagglutinins and internalins [221].
3.9. Non-classical and novel protein transporting systems
There are also some non-classical secreted proteins that do not have signal sequences and are not secreted through the known secretion systems [222]. Besides the non-classical pathways descried before by which proteins could be exported from cytoplasm to periplasm, there are also other transporting systems that can mediate the transport of proteins into extracellular space. ClyA is an example that is secreted through non-classical pathways in Gram-negative bacteria [222]. ClyA is a pore-forming protein encoded by E. coli and other intestinal bacteria, which is toxic to mammalian cells. ClyA does not bear an N-terminal signal peptide but can be released from the outer membrane of E. coli. The protein is likely to be secreted to the extracellular milieu as outer-membrane vesicles (OMV) [223].
Novel protein transporting systems remain to be disclosed [224]. Most recently, an extracellular contractile injection system (eCIS) was recognized as an independent protein translocation system [225], [226]. To be precise, the eCISs are not protein secretion system, since they only mediate the translocation of secreted proteins into host cells. Structurally, eCISs resemble headless bacteriophages and share evolutionarily related proteins such as the tail tube, sheath, and baseplate complex [225]. Three sets of eCISs were independently identified previously, including the anti-feeding prophages (AFPs) in Serratia entomophila [227], the Photorhabdus virulence cassettes (PVCs) [228] and metamorphosis-associated contractile (MAC) structure identified in Pseudoalteromonas luteoviolacea [229]. Using these three verified eCISs and the phage-like-protein translocation structures (PLTSs) screened by BLAST [230], Chen et al built the core protein HMM profiles and updated them iteratively, and detected 631 eCIS-like loci from 11,699 publicly available complete bacterial genomes [226]. The eCISs are distributed among Gram-negative, Gram-positive bacteria and archaea. They are phylogenetically diverse and form six clusters [226]. Both eCISs and T6SSs are CISs, which encode proteins homologous to the phage contractile tails, deliver effectors to mediate bacterial-host interactions. However, eCISs differ from T6SSs apparently in the mode of action - the eCIS devices are released into the extracellular space, bind to and deliver substrates into target host cells [225], [229], [231]. Only a paucity of eCIS effectors have been identified, while the possible sequence signal features and their modes of action remain unknown [232], [233], [234].
Holin-like protein secretion systems (also named Type 10 Secretion Systems, TXSSs) were also reported in Gram-negative bacteria, mediating the transport of specific proteins into extracellular milieu from periplasm [42]. The systems are different from T2SSs and the two types of secretion systems recognize different substrates. The mechanism of the substrate recognition and secretion of TXSSs remain unclear, and there are only a few substrates that have been identified to secrete through this pathway.
4. Prediction of transmembrane proteins
Transmembrane proteins (TMPs) participate in molecular recognition, signal transduction and transmembrane transport, playing roles in various diseases [235]. They are also important molecular targets for many commercial drugs [236], [237]. TMPs pass through cell membrane either with transmembrane α-helices (TMHs) exclusively or with β-barrels formed by transmembrane β-strands [235], [294]. TMH proteins constitute 20–30% of the proteome of most organisms [235], [238], [294]. β-barrel proteins are mainly distributed in gram-negative and acid-fast bacteria, chloroplasts and mitochondria [239]. In Gram-negative bacteria, TMH proteins are mainly located in the inner membrane while β-barrel proteins are distributed in outer membrane [295], [239], [240], [241].
4.1. Features and prediction of TMH proteins
TMH proteins show the fragmental bias of hydrophobicity and electric charges [242]. Peptide fragments on the cytoplasmic side of the membrane are often enriched with positively charged residues [243]. A number of software tools predicting TMHs and the TM topology have been developed based on these two features [244], [245] (Table 5). There are also other features that were observed and applied, including the length of helix, grammar constrains of cytoplasmic and non-cytoplasmic loops, etc [244]. The tools can be simply classified into physicochemical property and statistics based and machine learning methods, the latter of which depend on a certain size of validated proteins and therefore were developed later than the former. Most of the tools can predict both TMHs (or TMH proteins) and the TM topology (Table 5).
Table 5.
Representative software tools predicting TMHs.
| Tool | Method | Target | URL or reference |
|---|---|---|---|
| TOPPred2 | Physiochemical property and statistics based | TMH; TM topology | http://bioweb.pasteur.fr/seqanal/interfaces/toppred.html; [246] |
| SOSUI | Physiochemical property and statistics based | TMH | http://www.tuat.ac.jp/mitaku/sosui/; [247] |
| SCAMPI | Physiochemical property and statistics based | TMH; TM topology | http://topcons.cbr.su.se/; [248] |
| PHDhtm | ANN | TMH | [249] |
| MEMSAT3 | ANN | TMH; TM topology; Sec/SPI | http://bioinf.cs.ucl.ac.uk/memsat/; [26] |
| SPOCTOPUS | NN + HMM | TMH; TM topology; Sec/SPI | http://octopus.cbr.su.se/; [25] |
| SOMPNN | PNN | TMH | http://www.csbio.sjtu.edu.cn/bioinf/SOMPNN/; [250] |
| TMSEG | NN + RF | TMH; TM topology | www.predictprotein.org; [251] |
| TMHMM 2.0 | HMM | TMH; TM topology | https://services.healthtech.dtu.dk/service.php?TMHMM; [244] |
| HMMTOP2 | HMM | TMH; TM topology | http://www.enzim.hu/hmmtop; [252] |
| Phobius/ PolyPhobius | HMM | TMH; TM topology; Sec/SPI | http://phobius.sbc.su.se/; [22] |
| Philius | DBN | TMH; TM topology; Sec/SPI | http://www.yeastrc.org/philius/; [23] |
| MEMSAT-SVM | SVM | TMH; TM topology; Sec/SPI; Re-entrant helix | http://bioinf.cs.ucl.ac.uk/psipred/; [27] |
| MemBrain | OET-KNN | TMH; Sec/SPI | http://www.csbio.sjtu.edu.cn/bioinf/MemBrain/; [253], [254] |
TOPPred2, SOSUI and SCAMPI are representative physicochemical property based models predicting TMH proteins. TOPPred2 used a trapezoid sliding window and hydrophobicity scale to predict TM fragments, followed by seeking the best topology according to the ‘positive-inside’ charge bias rule [246]. SOSUI improved the TMH prediction performance by introduction of 4 physicochemical parameters - the hydropathy index of Kyte and Doolittle, the amphiphilicity index of polar side chains, the index of amino acid charges, and the length of each sequence - to classify TM and soluble proteins and to predict the topology of TMH proteins [247]. SCAMPI adopted a position-specific membrane-insertion propensity scale and the ‘positive-inside’ rule to predict the topology of TMHs, reaching the performance comparable to the best-performed machine learning tools at the time [248].
Both NN and HMM are most frequently used to train the machine learning models predicting TMH fragments, TMH proteins or their topology. PHPhtm [249], SPOCTOPUS [25] and MEMSAT3 [26] are the representative NN models. PHDhtm used the phylogenetic information derived from multiple sequence alignments and amino acid composition features to predict the locations of TMH fragments in the TMPs [249]. Both SPOCTOPUS and MEMSAT3 integrated a SP prediction step to better distinguish TMH fragments [25], [26]. Besides TMH recognition, they can also predict the TM topology of TMPs as well as Sec/SPIs [25], [26]. There are also other novel NN models developed recently. Yu et al proposed a SOMPNN model combining a self-organizing map (SOM) with a probabilistic neural network (PNN) model [250]. SOMPNN used SOM to learn the knowledge of helix distribution hidden in the training datasets adaptively, and predicted TMH fragments with PNN. The model showed the advantages of high computational efficiency and low requirements in the prior hypothesis of parameters [250]. Another method, TMSEG, integrated multiple models, including NN, RF and experience filters, to identify TMPs and predict TMH fragments and the topology accurately [251].
The HMM models showed more advantages in prediction of the TM and non-TM state transition and are therefore widely used for TM topology too. A list of HMM models have been developed, such as TMHMM [244], HMMTOP [252], Phobius [21] and PolypPhobius [22]. TMHMM trained models for each region of TMPs, including helix caps, middle of helix, regions close to the membrane and globular domains [244]. HMMTOP depends on the amino acid distribution difference among structural components of proteins, and the version 2 allows users to submit other location related information of the fragments to improve the prediction accuracy [252]. Both TMHMM and HMMTOP have been widely used for TMH and TM topology prediction, and yet neither of them distinguishes SPs. Phobius and PolyPhobius solved such a problem and provide the module to classify TMH fragments and SPs, as was also described in Section 2.1 [21], [22]. Compared to Phobius, PolyPhobius incorporated information from homologs, and the prediction performance was improved substantially [22].
Other machine-learning algorithms have also been used to predict TMH proteins, such as the Dynamic Bayesian Networks (DBNs) based model Philius [23], the SVM model MEMSAT-SVM [27], and the optimized evidence-theoretic K-nearest neighbor (OET-KNN) model MemBrain [253], [54], etc. Philius, MEMSAT-SVM and MemBrain can also distinguish the TMHs from SPs [23], [27], [253], [254].
4.2. Features and prediction of β-barrel proteins
It is difficult to identify β-barrel TMPs experimentally, while traditional TM prediction methods can also hardly predict β-barrel TMPs due to their smaller size of TM regions than TMHs [255], [256], [257]. However, there are still a number of software tools that have been developed to predict these proteins, using physicochemical property analysis, statistic measures or machine learning algorithms (Table 6).
Table 6.
Representative software tools predicting β-barrel OMPs.
| Tool | Method | Target | URL or reference |
|---|---|---|---|
| Neuwald’s | Motif searching | TMβ-strand | [261] |
| Gromiha’s | Physicochemical property, structure and statistics based | TM β-strand | [262] |
| BBF | Physicochemical property, structure and statistics based | β-barrel OMP | [263] |
| BOMP | Physicochemical property, structure and statistics based | β-barrel OMP | http://www.bioinfo.no/tools/bomp; [264] |
| transFold | Physicochemical property, structure and statistics based | TM β-barrel; residue side-chain orientations; inter-strand residue contact; strand inclination | http://bioinformatics.bc.edu/clotelab/transFold; [265] |
| HHomp | Sequence similarity searching | β-barrel OMP | http://toolkit.tuebingen.mpg.de/hhomp; [266] |
| Freeman-Wimley | Physicochemical property, structure and statistics based | TM β-barrel | http://www.tulane.edu/~biochem/WW/apps.html; [256] |
| OM-TOPO-PREDICT | NN | TM β-strand; OMP topology | http://strucbio.biologie.unikonstanz.de/-kay/om-topo-predict.html (Page not found); [267] |
| B2TMPRED | NN | TM β-strand; OMP topology | http://www.biocomp.unibo.it; [268] |
| TMBETA-NET | NN | TM β-strand | http://psfs.cbrc.jp/tmbeta-net/; [269] |
| TMBpro | NN | TM β-barrel; secondary structure; β-contacts; tertiary structure | http://www.igb.uci.edu/servers/psss.html; [270] |
| TBBPred | NN + SVM | TM β-barrel | http://www.imtech.res.in/raghava/tbbpred; [270] |
| TMbeta-SVM | SVM (sequential Aac + residue pairs) | β-barrel OMP | http://tmbeta-svm.cbrc.jp; [276] |
| PredβTM | SVM (position-specific Aac + residue pairs) | TM β-strand | http://transpred.ki.si/; [277] |
| BOCTOPUS/ BOCTOPUS2 | SVM; HMM | OMP topology | http://boctopus.cbr.su.se/; [284], [285] |
| HMMB2TMR | HMM | OMP topology | [273] |
| PROFtmb | HMM (beta-hairpin motifs) | β-barrel OMP; non-β-barrel OMP | http://www.rostlab.org/services/PROFtmb/; [274] |
| PRED-TMBB | HMM; | OMP; soluble protein | http://bioinformatics.biol.uoa.gr/PRED-TMBB; [275] |
| ConBBPRED | Consensus approaches | β-strand; OMP topology | http://bioinformatics.biol.uoa.gr/ConBBPRED; [286] |
| TMB-Hunt | k-NN | β-barrel TMP; non-β-barrel TMP | http://www.bioinformatics.leeds.ac.uk/betaBarrel; [278] |
| IDQD | Quadratic discriminant analysis | β-barrel TMP; TMH; global protein | [280] |
| TMBETADISC-RBF | Radial Basis Function (RBF) Networks | OMP | http://rbf.bioinfo.tw/~sachen/OMP.html; [281] |
| GRHCRF | Grammatical-Restrained Hidden Conditional Random Fields (GRHCRFs) | OMP | http://www.biocomp.unibo.it/~savojard/biocrf-0.9.tar.gz; [282] |
| BetAware | N-to-1 Extreme Learning | β-barrel TMP | http://betaware.biocomp.unibo.it/BetAware; [255] |
| BETAWARE | N-to-1 network encoding and ELM training algorithm | β-barrel TMP | http://www.biocomp.unibo.it/~savojard/betawarecl; [283] |
| MemBrain-TMB | Statistical machine learning | β-barrel TMP | www.csbio.sjtu.edu.cn/bioinf/MemBrain-TMB; [257] |
| Koehler's | NN | β-barrel TMP; TMH | [272] |
The amphipathicity of TM chains, i.e., the alternating patterns of hydrophobic-hydrophilic residues, was first used for prediction of β-barrel TMPs [257], [258], [259], [260]. Other statistic features were also adopted. For example, Neuwald et al proposed a new Gibbs-sampling algorithm to detect the repeated motif features buried in β-strands of bacteria outer membrane proteins (OMPs) [261]. Gromiha et al predicted the TM β-chains of bacterial porin family by statistical analysis of amino acid bias and the prior knowledge on protein structural properties [262]. Zhai et al used multiple statistics-based features including secondary structure, hydrophilicity, amphipathicity and N-terminal target sequence patterns to develop a program named BBF, with which β-barrel TMPs were detected from the E. coli genome [263]. BOMP based on the C-terminal motif features of β-barrel TM proteins and the typical amino acid property of TM β-strands to predict the β-barrel OMPs in Gram-negative bacterial genomes [264]. The transFold web server described all potential conformations based on multi-tape S-attribute grammars, and then used a dynamic programming algorithm to predict the structure and residue contacts of TM β-barrels [265]. HHomp based on the finding that all TM β-barrels are homologous to each other, and therefore used a database of profile HMMs to identify new β-barrel OMPs based on more sensitive profile-profile alignments [266]. Freeman and Wimley also proposed a method to predict genes encoding β-barrel TMPs from genome databases by analyzing the physicochemical properties of the proteins [256].
Machine-learning algorithms have also been widely applied in prediction of β-barrel TMPs (Table 6). NN and HMM are most frequently adopted though other models are also used such as SVM, k-NN, etc. As one of the earliest applications, Diederichs et al trained an NN model to predict the topology of β-chain OMPs [267]. B2TMPRED [268], TMBETA-NET [269], TBBPred [270] and TMBpro [270] are also NN-based methods. B2TMPRED considered phylogenetic information [268], TMBETA-NET introduced the concept of “residue probability” [269], while TBBPred trained both NN and SVM models and combined them to predict the β-barrel regions in proteins [270]. The TMBpro suite includes three modules, which can predict the secondary structure, β-contacts and tertiary structure of β-barrel TMPs with TMBpro-SS, TMBpro-CON and TMBpro-3D module respectively [271]. Koehler et al proposed a ANN based method, which can predict TM β-strands and TMHs simultaneously [272].
The HMM models are represented by HMMB2TMR [273], PROFtmb [274], PRED-TMBB [275]. There are also tools based on other algorithms, e.g., SVM based TMbeta-SVM [276] and PredβTM [277], k-NN based TMB-Hunt [278] and OMP-kNN [279], and others like IDQD [280], TMBETADISC-RBF [281], GRHCRFs [282], BetAware [255], BETAWARE [283] and MemBrain-TMB [257]. The links or references for these tools and the brief description were shown in Table 6. It is noteworthy that some tools combined multiple models to improve the prediction performance of β-barrel TM proteins, e.g., TMBETA-NET described above [270], BOCTOPUS [284], BOCTOPUS2 [285] and ConBBPRED [286]. BOCTOPUS and BOCTOPUS2 trained SVM models to predict the location of each residue and to detect the likely TM β-strands, followed by building HMM models to analyze the global topology of the β-barrel OMPs [284], [285]. ConBBPRED is a consensus prediction method integrating the results of 9 NN, HMM or SVM models, which can predict the β-strands and the topology of β-barrel OMPs with higher accuracy than individual models [286].
5. Integrated prediction pipelines and other applications
There are also some tools designed to predict protein subcellular localization, e.g., PSORTb, PSSM-S and FUEL-mLoc, which can also predict the extracellular (secreted) proteins and TMPs, but without specific secretion pathway information [287], [288], [293]. PREFFECTOR is another representative of tools predicting proteins secreted through non-specific pathways [289]. It combined effector proteins secreted though T1 ~ 6SSs of Gram-negative bacteria and trained models to classify general effectors from non-effectors regardless of the secretion system knowledge [289]. Because specific predictors require the prior knowledge about the specific secretion pathways or secreted proteins, PREFFECTOR would show the advantage in finding novel effectors secreted by unknown mechanisms. Other integrated prediction pipelines for secreted proteins include the ones predicting T3SEs, T4SEs and T6SEs (e.g., Tbooster and Orgsissec) and those predicting SPs/TMH fragments (e.g., Phobius, Philius and SPOCTOPUS) or TMHs/TMBs (e.g., Koehler’s method) simultaneously, as described before.
Besides the secreted proteins, tools have also been developed to predict the secretion devices. For example, SSPred can recognize T1-4SSs and Sec pathways [290]. T346Hunter can find T3SS, T4SS and T6SS gene clusters from bacterial genomes [291]. TXSScan can predict T1-6SSs, T9SSs, flagella T3SSs, T4P and Tad fimbriae systems [292]. There are also tools predicting individual secretion systems, which will not be discussed in this review.
6. Summary and perspectives
In this review, we summarized protein secretion systems and the bioinformatic tools predicting these secreted proteins in Gram-negative bacteria. Precise prediction and classification of the secreted proteins is important for both bacterial genome annotation and molecular mechanism exploration of bacterial virulence, drug resistance and other important biological phonotypes. A large number of algorithms and tools have been developed. However, there remains a long way till our ultimate destination.
First of all, there is often a gap between computational scientists and experimental biologists. Despite the high accuracy of software tools demonstrated by the developers, the non-homology based effector predictors (especially for T3SEs, T4SEs and T6SEs) have yet seldom been successfully applied to identify novel effectors by wet-lab researchers. More enthusiasms have been put in new algorithms rather than the biological side, e.g., new features. Most effector prediction tools are general and no specific biological prior information is considered, such as species, secretion system subtypes and regulation conduit specificity. For the software tools themselves, few groups collected, annotated and filtered the training proteins manually and carefully. The size and distribution of negative protein dataset was not optimized either. However, these aspects are really important for the development of a practically useful prediction model. Secondly, our current knowledge on protein secretion systems and the secretion mechanisms remains limited. There are new protein secretion systems that remain to be identified. Except for few pathways, the secretion mechanisms are not fully clear for a majority of the known protein secretion systems. Only a paucity of secreted proteins is experimentally validated for many newly disclosed secretion systems, and it is very difficult to identify common features stably.
There are still challenges and improvement requirements for the current algorithms and tools. For example, too many tools have been developed for some protein secretion systems, e.g., T3SS and T4SS. It is difficult for experimental biologists to select the most appropriate tool. For other systems, there is still a lack of tools, e.g., T1SS and T2SS. An integrated pipeline is also desired urgently for comprehensive annotation of different types of secreted proteins. Moreover, most of the current software tools were designed for individual bacterial strains and the individual genome-derived proteome. New algorithms, databases and tools are useful and desired to facilitate evaluation of the secretome of microbiota with the metagenomic, metatranscriptomic or metaproteomic data [300], [301], [302].
All the authors certify that they have seen and approved the final version of the manuscript being submitted. They warrant that the article is the authors' original work, hasn't received prior publication and isn't under consideration for publication elsewhere.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
Y.W. was supported by Natural Science Funds of Shenzhen (JCYJ201607115221141; JCYJ20190808165205582) and a fund for Youth Innovation Team of Shenzhen University (no. 406/0000080805). Z.C. and X.H. were supported by Funds for the Cultivation of Guangdong College Students’ Scientific and Technological Innovation, Climbing Program (pdjh2021b0432 and pdjha0427). Z.C. was supported by a National Undergraduate Training Program of China for Innovation and Entrepreneurship (no. 201910590003).
References
- 1.Tsirigotaki A., De Geyter J., Sostaric N., Economou A., Karamanou S. Protein export through the bacterial Sec pathway. Nat Rev Microbiol. 2017;15:21–36. doi: 10.1038/nrmicro.2016.161. [DOI] [PubMed] [Google Scholar]
- 2.Desvaux M., Hebraud M., Talon R., Henderson I.R. Secretion and subcellular localizations of bacterial proteins: a semantic awareness issue. Trends Microbiol. 2009;17:139–145. doi: 10.1016/j.tim.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 3.Natale P., Bruser T., Driessen A.J. Sec- and Tat-mediated protein secretion across the bacterial cytoplasmic membrane–distinct translocases and mechanisms. Biochim Biophys Acta. 2008;1778:1735–1756. doi: 10.1016/j.bbamem.2007.07.015. [DOI] [PubMed] [Google Scholar]
- 4.Luirink J., von Heijne G., Houben E., de Gier J.W. Biogenesis of inner membrane proteins in Escherichia coli. Annu Rev Microbiol. 2005;59:329–355. doi: 10.1146/annurev.micro.59.030804.121246. [DOI] [PubMed] [Google Scholar]
- 5.Zhou Y., Ueda T., Müller M. Signal recognition particle and SecA cooperate during export of secretory proteins with highly hydrophobic signal sequences. PLoS ONE. 2014;9 doi: 10.1371/journal.pone.0092994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang S., Yang C.I., Shan S.O. SecA mediates cotranslational targeting and translocation of an inner membrane protein. J Cell Biol. 2017;216:3639–3653. doi: 10.1083/jcb.201704036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang S., Jomaa A., Jaskolowski M., Yang C.I., Ban N., Shan S.O. The molecular mechanism of cotranslational membrane protein recognition and targeting by SecA. Nat Struct Mol Biol. 2019;26:919–929. doi: 10.1038/s41594-019-0297-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Derman A.I., Puziss J.W., Bassford P.J., Jr, Beckwith J. A signal sequence is not required for protein export in prlA mutants of Escherichia coli. EMBO J. 1993;12:879–888. doi: 10.1002/j.1460-2075.1993.tb05728.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gouridis G., Karamanou S., Gelis I., Kalodimos C.G., Economou A. Signal peptides are allosteric activators of the protein translocase. Nature. 2009;462:363–367. doi: 10.1038/nature08559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feltcher M.E., Braunstein M. Emerging themes in SecA2-mediated protein export. Nat Rev Microbiol. 2012;10:779–789. doi: 10.1038/nrmicro2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nielsen H. Predicting Secretory Proteins with SignalP. Methods Mol Biol. 2017;1611:59–73. doi: 10.1007/978-1-4939-7015-5_6. [DOI] [PubMed] [Google Scholar]
- 12.Nielsen H., Engelbrecht J., Brunak S., von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 13.Petersen T.N., Brunak S., von Heijne G., Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 14.Almagro Armenteros J.J., Tsirigos K.D., Sonderby C.K., Petersen T.N., Winther O., Brunak S. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat Biotechnol. 2019;37:420–423. doi: 10.1038/s41587-019-0036-z. [DOI] [PubMed] [Google Scholar]
- 15.Frank K., Sippl M.J. High-performance signal peptide prediction based on sequence alignment techniques. Bioinformatics. 2008;24:2172–2176. doi: 10.1093/bioinformatics/btn422. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Y.Z., Shen H.B. Signal-3L 2.0: A Hierarchical Mixture Model for Enhancing Protein Signal Peptide Prediction by Incorporating Residue-Domain Cross-Level Features. J Chem Inf Model. 2017;57:988–999. doi: 10.1021/acs.jcim.6b00484. [DOI] [PubMed] [Google Scholar]
- 17.Hiller K., Grote A., Scheer M., Munch R., Jahn D. PrediSi: prediction of signal peptides and their cleavage positions. Nucleic Acids Res. 2004;32:W375–W379. doi: 10.1093/nar/gkh378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chou K.C., Shen H.B. Signal-CF: a subsite-coupled and window-fusing approach for predicting signal peptides. Biochem Biophys Res Commun. 2007;357:633–640. doi: 10.1016/j.bbrc.2007.03.162. [DOI] [PubMed] [Google Scholar]
- 19.Juncker A.S., Willenbrock H., Von Heijne G., Brunak S., Nielsen H., Krogh A. Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci. 2003;12:1652–1662. doi: 10.1110/ps.0303703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fariselli P., Finocchiaro G., Casadio R. SPEPlip: the detection of signal peptide and lipoprotein cleavage sites. Bioinformatics. 2003;19:2498–2499. doi: 10.1093/bioinformatics/btg360. [DOI] [PubMed] [Google Scholar]
- 21.Kall L., Krogh A., Sonnhammer E.L. A combined transmembrane topology and signal peptide prediction method. J Mol Biol. 2004;338:1027–1036. doi: 10.1016/j.jmb.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 22.Kall L., Krogh A., Sonnhammer E.L. An HMM posterior decoder for sequence feature prediction that includes homology information. Bioinformatics. 2005;21(Suppl 1):i251–i257. doi: 10.1093/bioinformatics/bti1014. [DOI] [PubMed] [Google Scholar]
- 23.Reynolds S.M., Kall L., Riffle M.E., Bilmes J.A., Noble W.S. Transmembrane topology and signal peptide prediction using dynamic bayesian networks. PLoS Comput Biol. 2008;4 doi: 10.1371/journal.pcbi.1000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsirigos K.D., Peters C., Shu N., Kall L., Elofsson A. The TOPCONS web server for consensus prediction of membrane protein topology and signal peptides. Nucleic Acids Res. 2015;43:W401–W407. doi: 10.1093/nar/gkv485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Viklund H., Bernsel A., Skwark M., Elofsson A. SPOCTOPUS: a combined predictor of signal peptides and membrane protein topology. Bioinformatics. 2008;24:2928–2929. doi: 10.1093/bioinformatics/btn550. [DOI] [PubMed] [Google Scholar]
- 26.Jones D.T. Improving the accuracy of transmembrane protein topology prediction using evolutionary information. Bioinformatics. 2007;23:538–544. doi: 10.1093/bioinformatics/btl677. [DOI] [PubMed] [Google Scholar]
- 27.Nugent T., Jones D.T. Transmembrane protein topology prediction using support vector machines. BMC Bioinf. 2009;10:159. doi: 10.1186/1471-2105-10-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Savojardo C., Martelli P.L., Fariselli P., Casadio R. DeepSig: deep learning improves signal peptide detection in proteins. Bioinformatics. 2018;34:1690–1696. doi: 10.1093/bioinformatics/btx818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Palmer T., Berks B.C. The twin-arginine translocation (Tat) protein export pathway. Nat Rev Microbiol. 2012;10:483–496. doi: 10.1038/nrmicro2814. [DOI] [PubMed] [Google Scholar]
- 30.De Buck E., Lammertyn E., Anne J. The importance of the twin-arginine translocation pathway for bacterial virulence. Trends Microbiol. 2008;16:442–453. doi: 10.1016/j.tim.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 31.Muller M. Twin-arginine-specific protein export in Escherichia coli. Res Microbiol. 2005;156:131–136. doi: 10.1016/j.resmic.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 32.Lee P.A., Tullman-Ercek D., Georgiou G. The bacterial twin-arginine translocation pathway. Annu Rev Microbiol. 2006;60:373–395. doi: 10.1146/annurev.micro.60.080805.142212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rose R.W., Bruser T., Kissinger J.C., Pohlschroder M. Adaptation of protein secretion to extremely high-salt conditions by extensive use of the twin-arginine translocation pathway. Mol Microbiol. 2002;45:943–950. doi: 10.1046/j.1365-2958.2002.03090.x. [DOI] [PubMed] [Google Scholar]
- 34.Dilks K., Rose R.W., Hartmann E., Pohlschroder M. Prokaryotic utilization of the twin-arginine translocation pathway: a genomic survey. J Bacteriol. 2003;185:1478–1483. doi: 10.1128/JB.185.4.1478-1483.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bendtsen J.D., Nielsen H., Widdick D., Palmer T., Brunak S. Prediction of twin-arginine signal peptides. BMC Bioinf. 2005;6:167. doi: 10.1186/1471-2105-6-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bagos P.G., Nikolaou E.P., Liakopoulos T.D., Tsirigos K.D. Combined prediction of Tat and Sec signal peptides with hidden Markov models. Bioinformatics. 2010;26:2811–2817. doi: 10.1093/bioinformatics/btq530. [DOI] [PubMed] [Google Scholar]
- 37.Rodrigue A., Chanal A., Beck K., Muller M., Wu L.F. Co-translocation of a periplasmic enzyme complex by a hitchhiker mechanism through the bacterial tat pathway. J Biol Chem. 1999;274:13223–13228. doi: 10.1074/jbc.274.19.13223. [DOI] [PubMed] [Google Scholar]
- 38.Leclere V., Bechet M., Blondeau R. Functional significance of a periplasmic Mn-superoxide dismutase from Aeromonas hydrophila. J Appl Microbiol. 2004;96:828–833. doi: 10.1111/j.1365-2672.2004.02231.x. [DOI] [PubMed] [Google Scholar]
- 39.Krehenbrink M., Edwards A., Downie J.A. The superoxide dismutase SodA is targeted to the periplasm in a SecA-dependent manner by a novel mechanism. Mol Microbiol. 2011;82:164–179. doi: 10.1111/j.1365-2958.2011.07803.x. [DOI] [PubMed] [Google Scholar]
- 40.Kint G., Sonck K.A., Schoofs G., De Coster D., Vanderley-den J., De Keersmaecker S.C. 2D proteome analysis initiates new insights on the Salmonella typhimu- rium LuxS protein. BMC Microbiol. 2009;9:198. doi: 10.1186/1471-2180-9-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fowler C.C., Chang S.J., Gao X., Geiger T., Stack G., Galán J.E. Emerging insights into the biology of typhoid toxin. Curr Opin Microbiol. 2017;35:70–77. doi: 10.1016/j.mib.2017.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hamilton J.J., Marlow V.L., Owen R.A., Costa Mde A., Guo M., Buchanan G. A holin and an endopeptidase are essential for chitinolytic protein secretion in Serratia marcescens. J Cell Biol. 2014;207:615–626. doi: 10.1083/jcb.201404127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spitz O., Erenburg I.N., Beer T., Kanonenberg K., Holland I.B., Schmitt L. Type I Secretion Systems-One Mechanism for All? Microbiol Spectr. 2019;7(2) doi: 10.1128/microbiolspec.psib-0003-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Du D., Wang Z., James N.R., Voss J.E., Klimont E., Ohene-Agyei T. Structure of the AcrAB-TolC multidrug efflux pump. Nature. 2014;509:512–515. doi: 10.1038/nature13205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blair J.M., Richmond G.E., Piddock L.J. Multidrug efflux pumps in Gram-negative bacteria and their role in antibiotic resistance. Future Microbiol. 2014;9:1165–1177. doi: 10.2217/fmb.14.66. [DOI] [PubMed] [Google Scholar]
- 46.Felmlee T., Pellett S., Welch R.A. Nucleotide sequence of an Escherichia coli chromosomal hemolysin. J Bacteriol. 1985;163:94–105. doi: 10.1128/jb.163.1.94-105.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barlag B., Hensel M. The giant adhesin SiiE of Salmonella enterica. Molecules. 2015;20:1134–1150. doi: 10.3390/molecules20011134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fuche F., Vianney A., Andrea C., Doublet P., Gilbert C. Functional type 1 secretion system involved in Legionella pneumophila virulence. J Bacteriol. 2015;197:563–571. doi: 10.1128/JB.02164-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harding C.M., Pulido M.R., Di Venanzio G., Kinsella R.L., Webb A.I., Scott N.E. Pathogenic Acinetobacter species have a functional type I secretion system and contact-dependent inhibition systems. J Biol Chem. 2017;292:9075–9087. doi: 10.1074/jbc.M117.781575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.El-Kirat-Chatel S., Boyd C.D., O'Toole G.A., Dufrene Y.F. Single-molecule analysis of Pseudomonas fluorescens footprints. ACS Nano. 2014;8:1690–1698. doi: 10.1021/nn4060489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guo S., Vance T.D.R., Stevens C.A., Voets I., Davies P.L. RTX Adhesins are Key Bacterial Surface Megaproteins in the Formation of Biofilms. Trends Microbiol. 2019;27:453–467. doi: 10.1016/j.tim.2018.12.003. [DOI] [PubMed] [Google Scholar]
- 52.Son M., Moon Y., Oh M.J., Han S.B., Park K.H., Kim J.G. Lipase and protease double-deletion mutant of Pseudomonas fluorescens suitable for extracellular protein production. Appl Environ Microbiol. 2012;78:8454–8462. doi: 10.1128/AEM.02476-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ryu J., Lee U., Park J., Yoo D.H., Ahn J.H. A vector system for ABC transporter-mediated secretion and purification of recombinant proteins in Pseudomonas species. Appl Environ Microbiol. 2015;81:1744–1753. doi: 10.1128/AEM.03514-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kanonenberg K., Schwarz C.K., Schmitt L. Type I secretion systems - a story of appendices. Res Microbiol. 2013;164:596–604. doi: 10.1016/j.resmic.2013.03.011. [DOI] [PubMed] [Google Scholar]
- 55.Smith T.J., Sondermann H., O'Toole G.A. Type 1 does the two-step: type 1 secretion substrates with a functional periplasmic intermediate. J Bacteriol. 2018;200:e00168–e218. doi: 10.1128/JB.00168-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ginalski K., Kinch L., Rychlewski L., Grishin N. BTLCP proteins: a novel family of bacterial transglutaminase-like cysteine proteinases. Trends Biochem Sci. 2004;29:392–395. doi: 10.1016/j.tibs.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 57.Smith T.J., Font M.E., Kelly C.M., Sondermann H., O'Toole G.A. An N-terminal retention module anchors the giant adhesin LapA of Pseudomonas fluorescens at the cell surface: a novel sub-family of type I secretion systems. J Bacteriol. 2018;200:e00734–e817. doi: 10.1128/JB.00734-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.D'Auria G., Jiménez N., Peris-Bondia F., Pelaz C., Latorre A., Moya A. Virulence factor Rtx in Legionella pneumophila, evidence suggesting it is a modular multifunctional protein. BMC Genomics. 2008;9:14. doi: 10.1186/1471-2164-9-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Boyd C.D., Smith T.J., El-Kirat-Chatel S., Newell P.D., Dufrêne Y.F., O'Toole G.A. Structural features of the Pseudomonas fluorescens biofilm adhesin LapA required for LapG-dependent cleavage, biofilm formation, and cell surface localization. J Bacteriol. 2014;196:2775–2788. doi: 10.1128/JB.01629-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Linhartova I., Bumba L., Masin J., Basler M., Osicka R., Kamanova J. RTX proteins: a highly diverse family secreted by a common mechanism. FEMS Microbiol Rev. 2010;34:1076–1112. doi: 10.1111/j.1574-6976.2010.00231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Luo J., Li W., Liu Z., Guo Y., Pu X., Li M. A sequence-based two-level method for the prediction of type I secreted RTX proteins. Analyst. 2015;140:3048–3056. doi: 10.1039/c5an00311c. [DOI] [PubMed] [Google Scholar]
- 62.Korotkov K.V., Sandkvist M., Hol W.G. The type II secretion system: biogenesis, molecular architecture and mechanism. Nat Rev Microbiol. 2012;10:336–351. doi: 10.1038/nrmicro2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Green E.R., Mecsas J. Bacterial Secretion Systems: An Overview. Microbiol Spectr. 2016;4 doi: 10.1128/microbiolspec.VMBF-0012-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sandkvist M. Type II secretion and pathogenesis. Infect Immun. 2001;69:3523–3535. doi: 10.1128/IAI.69.6.3523-3535.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Harding C.M., Kinsella R.L., Palmer L.D., Skaar E.P., Feldman M.F. Medically Relevant Acinetobacter Species Require a Type II Secretion System and Specific Membrane-Associated Chaperones for the Export of Multiple Substrates and Full Virulence. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ho T.D., Davis B.M., Ritchie J.M., Waldor M.K. Type 2 secretion promotes enterohemorrhagic Escherichia coli adherence and intestinal colonization. Infect Immun. 2008;76:1858–1865. doi: 10.1128/IAI.01688-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sandkvist M., Michel L.O., Hough L.P., Morales V.M., Bagdasarian M., Koomey M. General secretion pathway (eps) genes required for toxin secretion and outer membrane biogenesis in Vibrio cholerae. J Bacteriol. 1997;179:6994–7003. doi: 10.1128/jb.179.22.6994-7003.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jyot J., Balloy V., Jouvion G., Verma A., Touqui L., Huerre M. Type II secretion system of Pseudomonas aeruginosa: in vivo evidence of a significant role in death due to lung infection. J Infect Dis. 2011;203:1369–1377. doi: 10.1093/infdis/jir045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pineau C., Guschinskaya N., Robert X., Gouet P., Ballut L., Shevchik V.E. Substrate recognition by the bacterial type II secretion system: more than a simple interaction. Mol Microbiol. 2014;94:126–140. doi: 10.1111/mmi.12744. [DOI] [PubMed] [Google Scholar]
- 70.Korotkov K.V., Sandkvist M. Architecture, Function, and Substrates of the Type II Secretion System. EcoSal Plus. 2019;8 doi: 10.1128/ecosalplus.esp-0034-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hueck C.J. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol Mol Biol Rev. 1998;62:379–433. doi: 10.1128/mmbr.62.2.379-433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang Y., Huang H., Sun M., Zhang Q., Guo D. T3DB: an integrated database for bacterial type III secretion system. BMC Bioinf. 2012;13:66. doi: 10.1186/1471-2105-13-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hu Y., Huang H., Cheng X., Shu X., White A.P., Stavrinides J. A global survey of bacterial type III secretion systems and their effectors. Environ Microbiol. 2017;19:3879–3895. doi: 10.1111/1462-2920.13755. [DOI] [PubMed] [Google Scholar]
- 74.Kubori T., Matsushima Y., Nakamura D., Uralil J., Lara-Tejero M., Sukhan A. Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science. 1998;280:602–605. doi: 10.1126/science.280.5363.602. [DOI] [PubMed] [Google Scholar]
- 75.Schraidt O., Marlovits T.E. Three-dimensional model of Salmonella’s needle complex at subnanometer resolution. Science. 2011;331:1192–1195. doi: 10.1126/science.1199358. [DOI] [PubMed] [Google Scholar]
- 76.Hu B., Morado D.R., Margolin W., Rohde J.R., Arizmendi O., Picking W.L. Visualization of the type III secretion sorting platform of Shigella flexneri. Proc Natl Acad Sci U S A. 2015;112(4):1047–1052. doi: 10.1073/pnas.1411610112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Worrall L.J., Hong C., Vuckovic M., Deng W., Bergeron J.R.C., Majewski D.D. Near-atomic-resolution cryo-EM analysis of the Salmonella T3S injectisome basal body. Nature. 2016;540:597–601. doi: 10.1038/nature20576. [DOI] [PubMed] [Google Scholar]
- 78.Deng W., Marshall N.C., Rowland J.L., McCoy J.M., Worrall L.J., Santos A.S. Assembly, structure, function and regulation of type III secretion systems. Nat Rev Microbiol. 2017;15:323–337. doi: 10.1038/nrmicro.2017.20. [DOI] [PubMed] [Google Scholar]
- 79.Abby S.S., Rocha E.P. The non-flagellar type III secretion system evolved from the bacterial flagellum and diversified into host-cell adapted systems. PLoS Genet. 2012;8 doi: 10.1371/journal.pgen.1002983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Diepold A., Armitage J.P. Type III secretion systems: the bacterial flagellum and the injectisome. Philos Trans R Soc Lond B Biol Sci. 2015;370 doi: 10.1098/rstb.2015.0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chaban B., Hughes H.V., Beeby M. The flagellum in bacterial pathogens: For motility and a whole lot more. Semin Cell Dev Biol. 2015;46:91–103. doi: 10.1016/j.semcdb.2015.10.032. [DOI] [PubMed] [Google Scholar]
- 82.Morimoto Y.V., Minamino T. Structure and function of the bi-directional bacterial flagellar motor. Biomolecules. 2014;4:217–234. doi: 10.3390/biom4010217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Young B.M., Young G.M. YplA is exported by the Ysc, Ysa, and flagellar type III secretion systems of Yersinia enterocolitica. J Bacteriol. 2002;184:1324–1334. doi: 10.1128/JB.184.5.1324-1334.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Warren S.M., Young G.M. An amino-terminal secretion signal is required for YplA export by the Ysa, Ysc, and flagellar type III secretion systems of Yersinia enterocolitica biovar 1B. J Bacteriol. 2005;187:6075–6083. doi: 10.1128/JB.187.17.6075-6083.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ince D., Sutterwala F.S., Yahr T.L. Secretion of Flagellar Proteins by the Pseudomonas aeruginosa Type III Secretion-Injectisome System. J Bacteriol. 2015;197:2003–2011. doi: 10.1128/JB.00030-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Izore T., Job V., Dessen A. Biogenesis, regulation, and targeting of the type III secretion system. Structure. 2011;19:603–612. doi: 10.1016/j.str.2011.03.015. [DOI] [PubMed] [Google Scholar]
- 87.Büttner D. Protein export according to schedule: architecture, assembly, and regulation of type III secretion systems from plant- and animal-pathogenic bacteria. Microbiol Mol Biol Rev. 2012;76:262–310. doi: 10.1128/MMBR.05017-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lloyd S.A., Norman M., Rosqvist R., Wolf-Watz H. Yersinia YopE is targeted for type III secretion by N-terminal, not mRNA, signals. Mol Microbiol. 2001;39:520–531. doi: 10.1046/j.1365-2958.2001.02271.x. [DOI] [PubMed] [Google Scholar]
- 89.Lee S.H., Galan J.E. Salmonella type III secretion- associated chaperones confer secretion-pathway specificity. Mol Microbiol. 2004;51:483–495. doi: 10.1046/j.1365-2958.2003.03840.x. [DOI] [PubMed] [Google Scholar]
- 90.Stebbins C.E., Galan J.E. Maintenance of an unfolded polypeptide by a cognate chaperone in bacterial type III secretion. Nature. 2001;414:77–81. doi: 10.1038/35102073. [DOI] [PubMed] [Google Scholar]
- 91.Arnold R., Brandmaier S., Kleine F., Tischler P., Heinz E., Behrens S. Sequence-based prediction of type III secreted proteins. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang Y., Zhang Q., Sun M.A., Guo D. High-accuracy prediction of bacterial type III secreted effectors based on position-specific amino acid composition profiles. Bioinformatics. 2011;27:777–784. doi: 10.1093/bioinformatics/btr021. [DOI] [PubMed] [Google Scholar]
- 93.Thomas N.A., Ma I., Prasad M.E., Rafuse C. Expanded roles for multicargo and class 1B effector chaperones in type III secretion. J Bacteriol. 2012;194:3767–3773. doi: 10.1128/JB.00406-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Panina E.M., Mattoo S., Griffith N., Kozak N.A., Yuk M.H., Miller J.F. A genome-wide screen identifies a Bordetella type III secretion effector and candidate effectors in other species. Mol Microbiol. 2005;58:267–279. doi: 10.1111/j.1365-2958.2005.04823.x. [DOI] [PubMed] [Google Scholar]
- 95.Lilic M., Vujanac M., Stebbins C.E. A common structural motif in the binding of virulence factors to bacterial secretion chaperones. Mol Cell. 2006;21:653–664. doi: 10.1016/j.molcel.2006.01.026. [DOI] [PubMed] [Google Scholar]
- 96.Anderson D.M., Schneewind O.A. mRNA signal for the type III secretion of Yop proteins by Yersinia enterocolitica. Science. 1997;278:1140–1143. doi: 10.1126/science.278.5340.1140. [DOI] [PubMed] [Google Scholar]
- 97.Anderson D.M., Fouts D.E., Collmer A., Schneewind O. Reciprocal secretion of proteins by the bacterial type III machines of plant and animal pathogens suggests universal recognition of mRNA targeting signals. Proc Natl Acad Sci USA. 1999;96:12839–12843. doi: 10.1073/pnas.96.22.12839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Niemann G.S., Brown R.N., Mushamiri I.T., Nguyen N.T., Taiwo R., Stufkens A. RNA type III secretion signals that require Hfq. J Bacteriol. 2013;195:2119–2125. doi: 10.1128/JB.00024-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fouts D.E., Abramovitch R.B., Alfano J.R., Baldo A.M., Buell C.R., Cartinhour S. Genomewide identification of Pseudomonas syringae pv. tomato DC3000 promoters controlled by the HrpL alternative sigma factor. Proc Natl Acad Sci U S A. 2002;99:2275–2280. doi: 10.1073/pnas.032514099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cunnac S., Occhialini A., Barberis P., Boucher C., Genin S. Inventory and functional analysis of the large Hrp regulon in Ralstonia solanacearum: identification of novel effector proteins translocated to plant host cells through the type III secretion system. Mol Microbiol. 2004;53:115–128. doi: 10.1111/j.1365-2958.2004.04118.x. [DOI] [PubMed] [Google Scholar]
- 101.Petnicki-Ocwieja T., Schneider D.J., Tam V.C., Chancey S.T., Shan L., Jamir Y. Genomewide identification of proteins secreted by the Hrp type III protein secretion system of Pseudomonas syringae pv. tomato DC3000. Proc Natl Acad Sci U S A. 2002;99:7652–7657. doi: 10.1073/pnas.112183899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Tobe T., Beatson S.A., Taniguchi H., Abe H., Bailey C.M., Fivian A. An extensive repertoire of type III secretion effectors in Escherichia coli O157 and the role of lambdoid phages in their dissemination. Proc Natl Acad Sci U S A. 2006;103:14941–14946. doi: 10.1073/pnas.0604891103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Guo Z., Cheng X., Hui X., Shu X., White A.P., Hu Y. Curr Bioinform. 2018;13:280–289. [Google Scholar]
- 104.Vencato M., Tian R., Alfano J.R., Buell C.R., Cartinhour S., DeClerck G.A. Bioinformatics-enabled identification of the HrpL regulon and type III secretion system effector proteins of Pseudomonas syringae pv. phaseolicola 1448A. Mol Plant Microbe Interact. 2006;19:1193–1206. doi: 10.1094/MPMI-19-1193. [DOI] [PubMed] [Google Scholar]
- 105.Samudrala R., Heffron F., McDermott J.E. Accurate prediction of secreted substrates and identification of a conserved putative secretion signal for type III secretion systems. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lower M., Schneider G. Prediction of type III secretion signals in genomes of gram-negative bacteria. PLoS ONE. 2009;4 doi: 10.1371/journal.pone.0005917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sato Y., Takaya A., Yamamoto T. Meta-analytic approach to the accurate prediction of secreted virulence effectors in gram-negative bacteria. BMC Bioinf. 2011;12:442. doi: 10.1186/1471-2105-12-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Schechter L.M., Valenta J.C., Schneider D.J., Collmer A., Sakk E. Functional and computational analysis of amino acid patterns predictive of type III secretion system substrates in Pseudomonas syringae. PLoS ONE. 2012;7 doi: 10.1371/journal.pone.0036038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wang Y., Sun M., Bao H., White A.P. T3_MM: a Markov model effectively classifies bacterial type III secretion signals. PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0058173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Dong X., Zhang Y.J., Zhang Z. Using weakly conserved motifs hidden in secretion signals to identify type-III effectors from bacterial pathogen genomes. PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0056632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hobbs C.K., Porter V.L., Stow M.L., Siame B.A., Tsang H.H., Leung K.Y. Computational approach to predict species-specific type III secretion system (T3SS) effectors using single and multiple genomes. BMC Genomics. 2016;17:1048. doi: 10.1186/s12864-016-3363-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Xue L., Tang B., Chen W., Luo J. DeepT3: deep convolutional neural networks accurately identify Gram-negative bacterial type III secreted effectors using the N-terminal sequence. Bioinformatics. 2019;35:2051–2057. doi: 10.1093/bioinformatics/bty931. [DOI] [PubMed] [Google Scholar]
- 113.Yang Y., Zhao J., Morgan R.L., Ma W., Jiang T. Computational prediction of type III secreted proteins from gram-negative bacteria. BMC Bioinf. 2010;11(Suppl 1):S47. doi: 10.1186/1471-2105-11-S1-S47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wang Y., Sun M., Bao H., Zhang Q., Guo D. Effective identification of bacterial type III secretion signals using joint element features. PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0059754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zalguizuri A., Caetano-Anolles G., Lepek V.C. Phylogenetic profiling, an untapped resource for the prediction of secreted proteins and its complementation with sequence-based classifiers in bacterial type III, IV and VI secretion systems. Brief Bioinform. 2019;20:1395–1402. doi: 10.1093/bib/bby009. [DOI] [PubMed] [Google Scholar]
- 116.Dong X., Lu X., Zhang Z. BEAN 2.0: an integrated web resource for the identification and functional analysis of type III secreted effectors. Database (Oxford) 2015;2015:bav064. doi: 10.1093/database/bav064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Goldberg T., Rost B., Bromberg Y. Computational prediction shines light on type III secretion origins. Sci Rep. 2016;6:34516. doi: 10.1038/srep34516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.An Y., Wang J., Li C., Leier A., Marquez-Lago T., Wilksch J. Comprehensive assessment and performance improvement of effector protein predictors for bacterial secretion systems III, IV and VI. Brief Bioinform. 2018;19:148–161. doi: 10.1093/bib/bbw100. [DOI] [PubMed] [Google Scholar]
- 119.Zeng C., Zou L. An account of in silico identification tools of secreted effector proteins in bacteria and future challenges. Brief Bioinform. 2019;20:110–129. doi: 10.1093/bib/bbx078. [DOI] [PubMed] [Google Scholar]
- 120.Wang J., Li J., Yang B., Xie R., Marquez-Lago T.T., Leier A. Bastion3: a two-layer ensemble predictor of type III secreted effectors. Bioinformatics. 2019;35:2017–2028. doi: 10.1093/bioinformatics/bty914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hui X., Chen Z., Lin M., Zhang J., Hu Y., Zeng Y. T3SEpp: an integrated prediction pipeline for bacterial type III secreted effectors. mSystems. 2020;5:e00288–e320. doi: 10.1128/mSystems.00288-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Grohmann E., Christie P.J., Waksman G., Backert S. Type IV secretion in Gram-negative and Gram-positive bacteria. Mol Microbiol. 2018;107:455–471. doi: 10.1111/mmi.13896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Li Y.G., Hu B., Christie P.J. Biological and Structural Diversity of Type IV Secretion Systems. Microbiol Spectr. 2019;7(2) doi: 10.1128/microbiolspec.psib-0012-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bhatty M., Laverde Gomez J.A., Christie P.J. The expanding bacterial type IV secretion lexicon. Res Microbiol. 2013;164:620–639. doi: 10.1016/j.resmic.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Guglielmini J., de la Cruz F., Rocha E.P. Evolution of conjugation and type IV secretion systems. Mol Biol Evol. 2013;30:315–331. doi: 10.1093/molbev/mss221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Guglielmini J., Neron B., Abby S.S., Garcillan-Barcia M.P., de la Cruz F., Rocha E.P. Key components of the eight classes of type IV secretion systems involved in bacterial conjugation or protein secretion. Nucleic Acids Res. 2014;42:5715–5727. doi: 10.1093/nar/gku194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Chandran Darbari V., Waksman G. Structural Biology of Bacterial Type IV Secretion Systems. Annu Rev Biochem. 2015;84:603–629. doi: 10.1146/annurev-biochem-062911-102821. [DOI] [PubMed] [Google Scholar]
- 128.Christie P.J. The Mosaic type IV secretion systems. EcoSalPlus. 2016;7 doi: 10.1128/ecosalplus.esp-0020-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Nagai H., Kubori T. Type IVB Secretion Systems of Legionella and Other Gram-Negative Bacteria. Front Microbiol. 2011;2:136. doi: 10.3389/fmicb.2011.00136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kwak M.J., Kim J.D., Kim H., Kim C., Bowman J.W., Kim S. Architecture of the type IV coupling protein complex of Legionella pneumophila. Nat Microbiol. 2017;2:17114. doi: 10.1038/nmicrobiol.2017.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Souza D.P., Andrade M.O., Alvarez-Martinez C.E., Arantes G.M., Farah C.S., Salinas R.K. A component of the Xanthomonadaceae type IV secretion system combines a VirB7 motif with a N0 domain found in outer membrane transport proteins. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1002031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Sgro G.G., Costa T.R.D., Cenens W., Souza D.P., Cassago A., Coutinho de Oliveira L. Cryo-EM structure of the bacteria-killing type IV secretion system core complex from Xanthomonas citri. Nat Microbiol. 2018;3:1429–1440. doi: 10.1038/s41564-018-0262-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sgro G.G., Oka G.U., Souza D.P., Cenens W., Bayer-Santos E., Matsuyama B.Y. Bacteria-Killing Type IV Secretion Systems. Front Microbiol. 2019;10:1078. doi: 10.3389/fmicb.2019.01078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Souza D.P., Oka G.U., Alvarez-Martinez C.E., Bisson-Filho A.W., Dunger G., Hobeika L. Bacterial killing via a type IV secretion system. Nat Commun. 2015;6:6453. doi: 10.1038/ncomms7453. [DOI] [PubMed] [Google Scholar]
- 135.Bayer-Santos E., Cenens W., Matsuyama B.Y., Oka G.U., Di Sessa G., Mininel I.D.V. The opportunistic pathogen Stenotrophomonas maltophilia utilizes a type IV secretion system for interbacterial killing. PLoS Pathog. 2019;15(9) doi: 10.1371/journal.ppat.1007651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hohlfeld S., Pattis I., Puls J., Plano G.V., Haas R. A C-terminal translocation signal is necessary, but not sufficient for type IV secretion of the Helicobacter pylori CagA protein. Mol Microbiol. 2006;59:1624–1637. doi: 10.1111/j.1365-2958.2006.05050.x. [DOI] [PubMed] [Google Scholar]
- 137.Kubori T., Hyakutake A., Nagai H. Legionella translocates an E3 ubiquitin ligase that has multiple U-boxes with distinct functions. Mol Microbiol. 2008;67:1307–1319. doi: 10.1111/j.1365-2958.2008.06124.x. [DOI] [PubMed] [Google Scholar]
- 138.Wang Y., Wei X., Bao H., Liu S.L. Prediction of bacterial type IV secreted effectors by C-terminal features. BMC Genomics. 2014;15:50. doi: 10.1186/1471-2164-15-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Marchesini M.I., Herrmann C.K., Salcedo S.P., Gorvel J.P., Comerci D.J. In search of Brucella abortus type IV secretion substrates: screening and identification of four proteins translocated into host cells through VirB system. Cell Microbiol. 2011;13:1261–1274. doi: 10.1111/j.1462-5822.2011.01618.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Myeni S., Child R., Ng T.W., Kupko J.J., 3rd, Wehrly T.D., Porcella S.F. Brucella modulates secretory trafficking via multiple type IV secretion effector proteins. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Burstein D., Zusman T., Degtyar E., Viner R., Segal G., Pupko T. Genome-scale identification of Legionella pneumophila effectors using a machine learning approach. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Chen C., Banga S., Mertens K., Weber M.M., Gorbaslieva I., Tan Y. Large-scale identification and translocation of type IV secretion substrates by Coxiella burnetii. Proc Natl Acad Sci U S A. 2010;107:21755–21760. doi: 10.1073/pnas.1010485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Xu S., Zhang C., Miao Y., Gao J., Xu D. Effector prediction in host-pathogen interaction based on a Markov model of a ubiquitous EPIYA motif. BMC Genomics. 2010;11(Suppl 3):S1. doi: 10.1186/1471-2164-11-S3-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Lifshitz Z., Burstein D., Peeri M., Zusman T., Schwartz K., Shuman H.A. Computational modeling and experimental validation of the Legionella and Coxiella virulence-related type-IVB secretion signal. Proc Natl Acad Sci U S A. 2013;110:E707–E715. doi: 10.1073/pnas.1215278110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zou L., Nan C., Hu F. Accurate prediction of bacterial type IV secreted effectors using amino acid composition and PSSM profiles. Bioinformatics. 2013;29:3135–3142. doi: 10.1093/bioinformatics/btt554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Wang Y., Guo Y., Pu X., Li M. Effective prediction of bacterial type IV secreted effectors by combined features of both C-termini and N-termini. J Comput Aided Mol Des. 2017;31:1029–1038. doi: 10.1007/s10822-017-0080-z. [DOI] [PubMed] [Google Scholar]
- 147.Xiong Y., Wang Q., Yang J., Zhu X., Wei D.Q. PredT4SE-Stack: Prediction of Bacterial Type IV Secreted Effectors From Protein Sequences Using a Stacked Ensemble Method. Front Microbiol. 2018;9:2571. doi: 10.3389/fmicb.2018.02571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Wang J., Yang B., An Y., Marquez-Lago T., Leier A., Wilksch J. Systematic analysis and prediction of type IV secreted effector proteins by machine learning approaches. Brief Bioinform. 2019;20:931–951. doi: 10.1093/bib/bbx164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Esna Ashari Z., Dasgupta N., Brayton K.A., Broschat S.L. An optimal set of features for predicting type IV secretion system effector proteins for a subset of species based on a multi-level feature selection approach. PLoS ONE. 2018;13 doi: 10.1371/journal.pone.0197041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Esna Ashari Z., Brayton K.A., Broschat S.L. Prediction of T4SS Effector Proteins for Anaplasma phagocytophilum Using OPT4e. A New Software Tool. Front Microbiol. 2019;10:1391. doi: 10.3389/fmicb.2019.01391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Esna Ashari Z., Brayton K.A., Broschat S.L. Using an optimal set of features with a machine learning-based approach to predict effector proteins for Legionella pneumophila. PLoS ONE. 2019;14 doi: 10.1371/journal.pone.0202312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Meyer D.F., Noroy C., Moumene A., Raffaele S., Albina E., Vachiery N. Searching algorithm for type IV secretion system effectors 1.0: a tool for predicting type IV effectors and exploring their genomic context. Nucleic Acids Res. 2013;41:9218–9229. doi: 10.1093/nar/gkt718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Fan E., Chauhan N., Udatha D.B., Leo J.C., Linke D. Type V Secretion Systems in Bacteria. Microbiol Spectr. 2016;4 doi: 10.1128/microbiolspec.VMBF-0009-2015. [DOI] [PubMed] [Google Scholar]
- 154.Leo J.C., Grin I., Linke D. Type V secretion: mechanism(s) of autotransport through the bacterial outer membrane. Philos Trans R Soc Lond B Biol Sci. 2012;367:1088–1101. doi: 10.1098/rstb.2011.0208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Jacob-Dubuisson F., Locht C., Antoine R. Two-partner secretion in Gram-negative bacteria: a thrifty, specific pathway for large virulence proteins. Mol Microbiol. 2001;40:306–313. doi: 10.1046/j.1365-2958.2001.02278.x. [DOI] [PubMed] [Google Scholar]
- 156.Lambert-Buisine C., Willery E., Locht C., Jacob-Dubuisson F. N-terminal characterization of the Bordetella pertussis filamentous haemagglutinin. Mol Microbiol. 1998;28:1283–1293. doi: 10.1046/j.1365-2958.1998.00892.x. [DOI] [PubMed] [Google Scholar]
- 157.St Geme J.W., 3rd, Yeo H.J. A prototype two-partner secretion pathway: the Haemophilus influenzae HMW1 and HMW2 adhesin systems. Trends Microbiol. 2009;17:355–360. doi: 10.1016/j.tim.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 158.Linke D., Riess T., Autenrieth I.B., Lupas A., Kempf V.A. Trimeric autotransporter adhesins: variable structure, common function. Trends Microbiol. 2006;14:264–270. doi: 10.1016/j.tim.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 159.Tamm A., Tarkkanen A.M., Korhonen T.K., Kuusela P., Toivanen P., Skurnik M. Hydrophobic domains affect the collagen-binding specificity and surface polymerization as well as the virulence potential of the YadA protein of Yersinia enterocolitica. Mol Microbiol. 1993;10:995–1011. doi: 10.1111/j.1365-2958.1993.tb00971.x. [DOI] [PubMed] [Google Scholar]
- 160.Salacha R., Kovacic F., Brochier-Armanet C., Wilhelm S., Tommassen J., Filloux A. The Pseudomonas aeruginosa patatin-like protein PlpD is the archetype of a novel Type V secretion system. Environ Microbiol. 2010;12:1498–1512. doi: 10.1111/j.1462-2920.2010.02174.x. [DOI] [PubMed] [Google Scholar]
- 161.Oberhettinger P., Schutz M., Leo J.C., Heinz N., Berger J., Autenrieth I.B. Intimin and invasin export their C-terminus to the bacterial cell surface using an inverse mechanism compared to classical autotransport. PLoS ONE. 2012;7 doi: 10.1371/journal.pone.0047069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Bodelon G., Palomino C., Fernandez L.A. Immunoglobulin domains in Escherichia coli and other enterobacteria: from pathogenesis to applications in antibody technologies. FEMS Microbiol Rev. 2013;37:204–250. doi: 10.1111/j.1574-6976.2012.00347.x. [DOI] [PubMed] [Google Scholar]
- 163.Celik N., Webb C.T., Leyton D.L., Holt K.E., Heinz E., Gorrell R. A bioinformatic strategy for the detection, classification and analysis of bacterial autotransporters. PLoS ONE. 2012;7 doi: 10.1371/journal.pone.0043245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Zude I., Leimbach A., Dobrindt U. Prevalence of autotransporters in Escherichia coli: what is the impact of phylogeny and pathotype? Int J Med Microbiol. 2014;304:243–256. doi: 10.1016/j.ijmm.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 165.Vo J.L., Martinez Ortiz G.C., Subedi P., Keerthikumar S., Mathivanan S., Paxman J.J. Autotransporter Adhesins in Escherichia coli Pathogenesis. Proteomics. 2017;17 doi: 10.1002/pmic.201600431. [DOI] [PubMed] [Google Scholar]
- 166.Goh K.G.K., Moriel D.G., Hancock S.J., Phan M.D., Schembri M.A. Bioinformatic and Molecular Analysis of Inverse Autotransporters from Escherichia coli. mSphere. 2019:4. doi: 10.1128/mSphere.00572-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Leiman P.G., Basler M., Ramagopal U.A., Bonanno J.B., Sauder J.M., Pukatzki S. Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc Natl Acad Sci U S A. 2009;106:4154–4159. doi: 10.1073/pnas.0813360106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Chow J., Mazmanian S.K. A pathobiont of the microbiota balances host colonization and intestinal inflammation. Cell Host Microbe. 2010;7:265–276. doi: 10.1016/j.chom.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Hood R.D., Singh P., Hsu F., Güvener T., Carl M.A., Trinidad R.R.S. A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe. 2010;7:25–37. doi: 10.1016/j.chom.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Alteri C.J., Himpsl S.D., Pickens S.R., Lindner J.R., Zora J.S., Miller J.E. Multicellular bacteria deploy the type VI secretion system to preemptively strike neighboring cells. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Trunk K., Peltier J., Liu Y.C., Dill B.D., Walker L., Gow N.A.R. The type VI secretion system deploys antifungal effectors against microbial competitors. Nat Microbiol. 2018;3(8):920–931. doi: 10.1038/s41564-018-0191-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Boyer F., Fichant G., Berthod J., Vandenbrouck Y., Attree I. Dissecting the bacterial type VI secretion system by a genome wide in silico analysis: What can be learned from available microbial genomic resources? BMC Genomics. 2009;10:104. doi: 10.1186/1471-2164-10-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Li J., Yao Y., Xu H.H., Hao L., Deng Z., Rajakumar K. SecReT6: a web-based resource for type VI secretion systems found in bacteria. Environ Microbiol. 2015;17:2196–2202. doi: 10.1111/1462-2920.12794. [DOI] [PubMed] [Google Scholar]
- 174.Coyne M.J., Roelofs K.G., Comstock L.E. Type VI secretion systems of human gut Bacteroidales segregate into three genetic architectures, two of which are contained on mobile genetic elements. BMC Genomics. 2016;17:58. doi: 10.1186/s12864-016-2377-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Barret M., Egan F., O'Gara F. Distribution and diversity of bacterial secretion systems across metagenomic datasets. Environ Microbiol Rep. 2013;5(1):117–126. doi: 10.1111/j.1758-2229.2012.00394.x. [DOI] [PubMed] [Google Scholar]
- 176.Russell A.B., Wexler A.G., Harding B.N., Whitney J.C., Bohn A.J., Goo Y.A. A type VI secretion-related pathway in Bacteroidetes mediates interbacterial antagonism. Cell Host Microbe. 2014;16:227–236. doi: 10.1016/j.chom.2014.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Pukatzki S., Ma A.T., Revel A.T., Sturtevant D., Mekalanos J.J. Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proc Natl Acad Sci USA. 2007;104:15508–15513. doi: 10.1073/pnas.0706532104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Ma J., Pan Z., Huang J., Sun M., Lu C., Yao H. The Hcp proteins fused with diverse extended-toxin domains represent a novel pattern of antibacterial effectors in type VI secretion systems. Virulence. 2017;8:1189–1202. doi: 10.1080/21505594.2017.1279374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Lien Y.W., Lai E.M. Type VI Secretion Effectors: Methodologies and Biology. Front Cell Infect Microbiol. 2017;7:254. doi: 10.3389/fcimb.2017.00254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Russell A.B., Singh P., Brittnacher M., Bui N.K., Hood R.D., Carl M.A. A widespread bacterial type VI secretion effector superfamily identified using a heuristic approach. Cell Host Microbe. 2012;11(5):538–549. doi: 10.1016/j.chom.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Shneider M.M., Buth S.A., Ho B.T., Basler M., Mekalanos J.J., Leiman P.G. PAAR-repeat proteins sharpen and diversify the type VI secretion system spike. Nature. 2013;500:350–353. doi: 10.1038/nature12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Russell A.B., LeRoux M., Hathazi K., Agnello D.M., Ishikawa T., Wiggins P.A. Diverse type VI secretion phospholipases are functionally plastic antibacterial effectors. Nature. 2013;496:508–512. doi: 10.1038/nature12074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Koskiniemi S., Lamoureux J.G., Nikolakakis K.C., t'Kint de Roodenbeke C., Kaplan M.D., Low D.A. Rhs proteins from diverse bacteria mediate intercellular competition. Proc Natl Acad Sci USA. 2013;110:7032–7037. doi: 10.1073/pnas.1300627110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Ma L.S., Hachani A., Lin J.S., Filloux A., Lai E.M. Agrobacterium tumefaciens deploys a superfamily of type VI secretion dnase effectors as weapons for interbacterial competition in planta. Cell Host Microbe. 2014;16:94–104. doi: 10.1016/j.chom.2014.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Ho B.T., Dong T.G., Mekalanos J.J. A view to a kill: The bacterial type VI secretion system. Cell Host Microbe. 2014;15:9–21. doi: 10.1016/j.chom.2013.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Flaugnatti N., Le T.T., Canaan S., Aschtgen M.S., Nguyen V.S., Blangy S. A phospholipase A1 antibacterial Type VI secretion effector interacts directly with the C-terminal domain of the VgrG spike protein for delivery. Mol Microbiol. 2016;99:1099–1118. doi: 10.1111/mmi.13292. [DOI] [PubMed] [Google Scholar]
- 187.Salomon D., Kinch L.N., Trudgian D.C., Guo X., Klimko J.A., Grishin N.V. Marker for type VI secretion system effectors. Proc Natl Acad Sci U S A. 2014;111:9271–9276. doi: 10.1073/pnas.1406110111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Dar Y., Salomon D., Bosis E. The Antibacterial and Anti-Eukaryotic Type VI Secretion System MIX-Effector Repertoire in Vibrionaceae. Mar Drugs. 2018;16:433. doi: 10.3390/md16110433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Jana B., Fridman C.M., Bosis E., Salomon D. A modular effector with a DNase domain and a marker for T6SS substrates. Nat Commun. 2019;10:3595. doi: 10.1038/s41467-019-11546-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Wang J., Yang B., Leier A., Marquez-Lago T.T., Hayashida M., Rocker A. Bastion6: a bioinformatics approach for accurate prediction of type VI secreted effectors. Bioinformatics. 2018;34:2546–2555. doi: 10.1093/bioinformatics/bty155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Sen R., Nayak L., De R.K. PyPredT6: A python-based prediction tool for identification of Type VI effector proteins. J Bioinform Comput Biol. 2019;17:1950019. doi: 10.1142/S0219720019500197. [DOI] [PubMed] [Google Scholar]
- 192.Abdallah A.M., Gey van Pittius N.C., Champion P.A., Cox J., Luirink J., Vandenbroucke-Grauls C.M. Type VII secretion–mycobacteria show the way. Nat Rev Microbiol. 2007;5:883–891. doi: 10.1038/nrmicro1773. [DOI] [PubMed] [Google Scholar]
- 193.Warne B., Harkins C.P., Harris S.R., Vatsiou A., Stanley-Wall N., Parkhill J. The Ess/Type VII secretion system of Staphylococcus aureus shows unexpected genetic diversity. BMC Genomics. 2016;17:222. doi: 10.1186/s12864-016-2426-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Bottai D., Groschel M.I., Brosch R. Type VII Secretion Systems in Gram-Positive Bacteria. Curr Top Microbiol Immunol. 2017;404:235–265. doi: 10.1007/82_2015_5015. [DOI] [PubMed] [Google Scholar]
- 195.Vaziri F., Brosch R. ESX/Type VII Secretion Systems-An Important Way Out for Mycobacterial Proteins. Microbiol Spectr. 2019;7 doi: 10.1128/microbiolspec.psib-0029-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Desvaux M., Hebraud M., Talon R., Henderson I.R. Outer membrane translocation: numerical protein secretion nomenclature in question in mycobacteria. Trends Microbiol. 2009;17:338–340. doi: 10.1016/j.tim.2009.05.008. [DOI] [PubMed] [Google Scholar]
- 197.Fronzes R., Remaut H., Waksman G. Architectures and biogenesis of non-flagellar protein appendages in Gram-negative bacteria. EMBO J. 2008;27:2271–2280. doi: 10.1038/emboj.2008.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Sauer F.G., Remaut H., Hultgren S.J., Waksman G. Fiber assembly by the chaperone-usher pathway. Biochim Biophys Acta. 2004;1694:259–267. doi: 10.1016/j.bbamcr.2004.02.010. [DOI] [PubMed] [Google Scholar]
- 199.Waksman G., Hultgren S.J. Structural biology of the chaperone-usher pathway of pilus biogenesis. Nat Rev Microbiol. 2009;7:765–774. doi: 10.1038/nrmicro2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Thanassi D.G., Stathopoulos C., Karkal A., Li H. Protein secretion in the absence of ATP: the autotransporter, two-partner secretion and chaperone/usher pathways of gram-negative bacteria (review) Mol Membr Biol. 2005;22:63–72. doi: 10.1080/09687860500063290. [DOI] [PubMed] [Google Scholar]
- 201.Nuccio S.P., Baumler A.J. Evolution of the chaperone/usher assembly pathway: fimbrial classification goes Greek. Microbiol Mol Biol Rev. 2007;71:551–575. doi: 10.1128/MMBR.00014-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Galkin V.E., Kolappan S., Ng D., Zong Z., Li J., Yu X. The structure of the CS1 pilus of enterotoxigenic Escherichia coli reveals structural polymorphism. J Bacteriol. 2013;195:1360–1370. doi: 10.1128/JB.01989-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Busch A., Waksman G. Chaperone-usher pathways: diversity and pilus assembly mechanism. Philos Trans R Soc Lond B Biol Sci. 2012;367:1112–1122. doi: 10.1098/rstb.2011.0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Van Gerven N., Klein R.D., Hultgren S.J., Remaut H. Bacterial amyloid formation: structural insights into curli biogensis. Trends Microbiol. 2015;23:693–706. doi: 10.1016/j.tim.2015.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Evans M.L., Chapman M.R. Curli biogenesis: order out of disorder. Biochim Biophys Acta. 2014;1843:1551–1558. doi: 10.1016/j.bbamcr.2013.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Soto G.E., Hultgren S.J. Bacterial adhesins: common themes and variations in architecture and assembly. J Bacteriol. 1999;181:1059–1071. doi: 10.1128/jb.181.4.1059-1071.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Barnhart M.M., Chapman M.R. Curli biogenesis and function. Annu Rev Microbiol. 2006;60:131–147. doi: 10.1146/annurev.micro.60.080805.142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Bhoite S., van Gerven N., Chapman M.R., Remaut H. Curli Biogenesis: Bacterial Amyloid Assembly by the Type VIII Secretion Pathway. EcoSal Plus. 2019;8 doi: 10.1128/ecosalplus.esp-0037-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Craig L., Pique M.E., Tainer J.A. Type IV pilus structure and bacterial pathogenicity. Nat Rev Microbiol. 2004;2:363–378. doi: 10.1038/nrmicro885. [DOI] [PubMed] [Google Scholar]
- 210.Merz A.J., So M., Sheetz M.P. Pilus retraction powers bacterial twitching motility. Nature. 2000;407:98–102. doi: 10.1038/35024105. [DOI] [PubMed] [Google Scholar]
- 211.McCallum M., Burrows L.L., Howell P.L. The Dynamic Structures of the Type IV Pilus. Microbiol Spectr. 2019;7 doi: 10.1128/microbiolspec.psib-0006-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Cornelis G.R. The type III secretion injectisome. Nat Rev Microbiol. 2006;4:811–825. doi: 10.1038/nrmicro1526. [DOI] [PubMed] [Google Scholar]
- 213.Chatterjee S., Chaudhury S., McShan A.C., Kaur K., De Guzman R.N. Structure and biophysics of type III secretion in bacteria. Biochemistry. 2013;52:2508–2517. doi: 10.1021/bi400160a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Babic A., Lindner A.B., Vulic M., Stewart E.J., Radman M. Direct visualization of horizontal gene transfer. Science. 2008;319:1533–1536. doi: 10.1126/science.1153498. [DOI] [PubMed] [Google Scholar]
- 215.Schroder G., Lanka E. The mating pair formation system of conjugative plasmids-A versatile secretion machinery for transfer of proteins and DNA. Plasmid. 2005;54:1–25. doi: 10.1016/j.plasmid.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 216.Lawley T.D., Klimke W.A., Gubbins M.J., Frost L.S. F factor conjugation is a true type IV secretion system. FEMS Microbiol Lett. 2003;224:1–15. doi: 10.1016/S0378-1097(03)00430-0. [DOI] [PubMed] [Google Scholar]
- 217.Sato K., Naito M., Yukitake H., Hirakawa H., Shoji M., McBride M.J. A protein secretion system linked to bacteroidete gliding motility and pathogenesis. Proc Natl Acad Sci U S A. 2010;107:276–281. doi: 10.1073/pnas.0912010107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Lauber F., Deme J.C., Lea S.M., Berks B.C. Type 9 secretion system structures reveal a new protein transport mechanism. Nature. 2018;564:77–82. doi: 10.1038/s41586-018-0693-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Lasica A.M., Ksiazek M., Madej M., Potempa J. The Type IX Secretion System (T9SS): Highlights and Recent Insights into Its Structure and Function. Front Cell Infect Microbiol. 2017;7:215. doi: 10.3389/fcimb.2017.00215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Veith P.D., Glew M.D., Gorasia D.G., Reynolds E.C. Type IX secretion: the generation of bacterial cell surface coatings involved in virulence, gliding motility and the degradation of complex biopolymers. Mol Microbiol. 2017;106:35–53. doi: 10.1111/mmi.13752. [DOI] [PubMed] [Google Scholar]
- 221.Veith P.D., Nor Muhammad N.A., Dashper S.G., Likic V.A., Gorasia D.G., Chen D. Protein substrates of a novel secretion system are numerous in the Bacteroidetes phylum and have in common a cleavable C-terminal secretion signal, extensive post-translational modification, and cell-surface attachment. J Proteome Res. 2013;12:4449–4461. doi: 10.1021/pr400487b. [DOI] [PubMed] [Google Scholar]
- 222.Bendtsen J.D., Kiemer L., Fausboll A., Brunak S. Non-classical protein secretion in bacteria. BMC Microbiol. 2005;5:58. doi: 10.1186/1471-2180-5-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Wai S.N., Lindmark B., Soderblom T., Takade A., Westermark M., Oscarsson J. Vesicle-mediated export and assembly of pore-forming oligomers of the enterobacterial ClyA cytotoxin. Cell. 2003;115:25–35. doi: 10.1016/s0092-8674(03)00754-2. [DOI] [PubMed] [Google Scholar]
- 224.Christie P.J. The Rich Tapestry of Bacterial Protein Translocation Systems. Protein J. 2019;38:389–408. doi: 10.1007/s10930-019-09862-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Jiang F., Li N., Wang X., Cheng J., Huang Y., Yang Y. Cryo-EM Structure and Assembly of an Extracellular Contractile Injection System. Cell. 2019;177:370–383. doi: 10.1016/j.cell.2019.02.020. [DOI] [PubMed] [Google Scholar]
- 226.Chen L., Song N., Liu B., Zhang N., Alikhan N.F., Zhou Z. Genome-wide Identification and Characterization of a Superfamily of Bacterial Extracellular Contractile Injection Systems. Cell Rep. 2019;29:511–521. doi: 10.1016/j.celrep.2019.08.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Hurst M.R., Glare T.R., Jackson T.A. Cloning Serratia entomophila antifeeding genes–a putative defective prophage active against the grass grub Costelytra zealandica. J Bacteriol. 2004;186:5116–5128. doi: 10.1128/JB.186.15.5116-5128.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Yang G., Dowling A.J., Gerike U., ffrench-Constant R.H., Waterfield N.R. Photorhabdus virulence cassettes confer injectable insecticidal activity against the wax moth. J Bacteriol. 2006;188:2254–2261. doi: 10.1128/JB.188.6.2254-2261.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Shikuma N.J., Pilhofer M., Weiss G.L., Hadfield M.G., Jensen G.J., Newman D.K. Marine tubeworm metamorphosis induced by arrays of bacterial phage tail-like structures. Science. 2014;343:529–533. doi: 10.1126/science.1246794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Sarris P.F., Ladoukakis E.D., Panopoulos N.J., Scoulica E.V. A phage tail-derived element with wide distribution among both prokaryotic domains: a comparative genomic and phylogenetic study. Genome Biol Evol. 2014;6:1739–1747. doi: 10.1093/gbe/evu136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Leiman P.G., Shneider N.M. Contractile tail machines of bacteriophages. Adv Exp Med Biol. 2012;726:93–114. doi: 10.1007/978-1-4614-0980-9_5. [DOI] [PubMed] [Google Scholar]
- 232.Ericson C.F., Eisenstein F., Medeiros J.M., Malter K.E., Cavalcanti G.S., Zeller R.W. A contractile injection system stimulates tubeworm metamorphosis by translocating a proteinaceous effector. Elife. 2019;8 doi: 10.7554/eLife.46845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Rocchi I., Ericson C.F., Malter K.E., Zargar S., Eisenstein F., Pilhofer M. A Bacterial Phage Tail-like Structure Kills Eukaryotic Cells by Injecting a Nuclease Effector. Cell Rep. 2019;28:295–301. doi: 10.1016/j.celrep.2019.06.019. [DOI] [PubMed] [Google Scholar]
- 234.Vlisidou I., Hapeshi A., Healey J.R., Smart K., Yang G., Waterfield N.R. The Photorhabdus asymbiotica virulence cassettes deliver protein effectors directly into target eukaryotic cells. Elife. 2019;8 doi: 10.7554/eLife.46259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Reeb J., Kloppmann E., Bernhofer M., Rost B. Evaluation of transmembrane helix predictions in 2014. Proteins. 2015;83:473–484. doi: 10.1002/prot.24749. [DOI] [PubMed] [Google Scholar]
- 236.Yildirim M.A., Goh K.I., Cusick M.E., Barabasi A.L., Vidal M. Drug-target network. Nat Biotechnol. 2007;25:1119–1126. doi: 10.1038/nbt1338. [DOI] [PubMed] [Google Scholar]
- 237.Bakheet T.M., Doig A.J. Properties and identification of human protein drug targets. Bioinformatics. 2009;25:451–457. doi: 10.1093/bioinformatics/btp002. [DOI] [PubMed] [Google Scholar]
- 238.Stevens T.J., Arkin I.T. Do more complex organisms have a greater proportion of membrane proteins in their genomes? Proteins. 2000;39:417–420. doi: 10.1002/(sici)1097-0134(20000601)39:4<417::aid-prot140>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 239.Schulz G.E. Transmembrane beta-barrel proteins. Adv Protein Chem. 2003;63:47–70. doi: 10.1016/s0065-3233(03)63003-2. [DOI] [PubMed] [Google Scholar]
- 240.Elofsson A., von Heijne G. Membrane protein structure: prediction versus reality. Annu Rev Biochem. 2007;76:125–140. doi: 10.1146/annurev.biochem.76.052705.163539. [DOI] [PubMed] [Google Scholar]
- 241.Tsirigos K.D., Bagos P.G., Hamodrakas S.J. OMPdb: a database of {beta}-barrel outer membrane proteins from Gram-negative bacteria. Nucleic Acids Res. 2011;39:D324–D331. doi: 10.1093/nar/gkq863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Kennedy S.J. Structures of membrane proteins. J Membr Biol. 1978;42:265–279. doi: 10.1007/BF01870362. [DOI] [PubMed] [Google Scholar]
- 243.Heijne G. The distribution of positively charged residues in bacterial inner membrane proteins correlates with the trans-membrane topology. EMBO J. 1986;5:3021–3027. doi: 10.1002/j.1460-2075.1986.tb04601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Krogh A., Larsson B., von Heijne G., Sonnhammer E.L. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 245.von Heijne G. Membrane protein structure prediction. Hydrophobicity analysis and the positive-inside rule. J Mol Biol. 1992;225:487–494. doi: 10.1016/0022-2836(92)90934-c. [DOI] [PubMed] [Google Scholar]
- 246.Claros M.G., von Heijne G. TopPred II: an improved software for membrane protein structure predictions. Comput Appl Biosci. 1994;10:685–686. doi: 10.1093/bioinformatics/10.6.685. [DOI] [PubMed] [Google Scholar]
- 247.Hirokawa T., Boon-Chieng S., Mitaku S. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics. 1998;14:378–379. doi: 10.1093/bioinformatics/14.4.378. [DOI] [PubMed] [Google Scholar]
- 248.Bernsel A., Viklund H., Falk J., Lindahl E., von Heijne G., Elofsson A. Prediction of membrane-protein topology from first principles. Proc Natl Acad Sci U S A. 2008;105:7177–7181. doi: 10.1073/pnas.0711151105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Rost B., Casadio R., Fariselli P., Sander C. Transmembrane helices predicted at 95% accuracy. Protein Sci. 1995;4:521–533. doi: 10.1002/pro.5560040318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Yu D.J., Shen H.B., Yang J.Y. SOMPNN: an efficient non-parametric model for predicting transmembrane helices. Amino Acids. 2012;42:2195–2205. doi: 10.1007/s00726-011-0959-2. [DOI] [PubMed] [Google Scholar]
- 251.Bernhofer M., Kloppmann E., Reeb J., Rost B. TMSEG: Novel prediction of transmembrane helices. Proteins. 2016;84:1706–1716. doi: 10.1002/prot.25155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Tusnady G.E., Simon I. The HMMTOP transmembrane topology prediction server. Bioinformatics. 2001;17:849–850. doi: 10.1093/bioinformatics/17.9.849. [DOI] [PubMed] [Google Scholar]
- 253.Shen H., Chou J.J. MemBrain: improving the accuracy of predicting transmembrane helices. PLoS ONE. 2008;3 doi: 10.1371/journal.pone.0002399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Yin X., Yang J., Xiao F., Yang Y., Shen H.B. MemBrain: An Easy-to-Use Online Webserver for Transmembrane Protein Structure Prediction. Nanomicro Lett. 2018;10:2. doi: 10.1007/s40820-017-0156-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Savojardo C., Fariselli P., Casadio R. Improving the detection of transmembrane beta-barrel chains with N-to-1 extreme learning machines. Bioinformatics. 2011;27:3128–3138. doi: 10.1093/bioinformatics/btr549. [DOI] [PubMed] [Google Scholar]
- 256.Freeman T.C., Jr., Wimley W.C. A highly accurate statistical approach for the prediction of transmembrane beta-barrels. Bioinformatics. 2010;26:1965–1974. doi: 10.1093/bioinformatics/btq308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Yin X., Xu Y.Y., Shen H.B. Enhancing the prediction of transmembrane beta-barrel segments with chain learning and feature sparse representation. IEEE/ACM Trans Comput Biol Bioinform. 2016;13:1016–1026. doi: 10.1109/TCBB.2016.2528000. [DOI] [PubMed] [Google Scholar]
- 258.Jeanteur D., Lakey J.H., Pattus F. The bacterial porin superfamily: sequence alignment and structure prediction. Mol Microbiol. 1991;5:2153–2164. doi: 10.1111/j.1365-2958.1991.tb02145.x. [DOI] [PubMed] [Google Scholar]
- 259.Vogel H., Jahnig F. Models for the structure of outer-membrane proteins of Escherichia coli derived from raman spectroscopy and prediction methods. J Mol Biol. 1986;190:191–199. doi: 10.1016/0022-2836(86)90292-5. [DOI] [PubMed] [Google Scholar]
- 260.Schirmer T., Cowan S.W. Prediction of membrane-spanning beta-strands and its application to maltoporin. Protein Sci. 1993;2:1361–1363. doi: 10.1002/pro.5560020820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Neuwald A.F., Liu J.S., Lawrence C.E. Gibbs motif sampling: detection of bacterial outer membrane protein repeats. Protein Sci. 1995;4:1618–1632. doi: 10.1002/pro.5560040820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Gromiha M.M., Majumdar R., Ponnuswamy P.K. Identification of membrane spanning beta strands in bacterial porins. Protein Eng. 1997;10:497–500. doi: 10.1093/protein/10.5.497. [DOI] [PubMed] [Google Scholar]
- 263.Zhai Y., Saier M.H., Jr The beta-barrel finder (BBF) program, allowing identification of outer membrane beta-barrel proteins encoded within prokaryotic genomes. Protein Sci. 2002;11:2196–2207. doi: 10.1110/ps.0209002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Berven F.S., Flikka K., Jensen H.B., Eidhammer I. BOMP: a program to predict integral beta-barrel outer membrane proteins encoded within genomes of Gram-negative bacteria. Nucleic Acids Res. 2004;32:W394–W399. doi: 10.1093/nar/gkh351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Waldispuhl J., Berger B., Clote P., Steyaert J.M. transFold: a web server for predicting the structure and residue contacts of transmembrane beta-barrels. Nucleic Acids Res. 2006;34:W189–W193. doi: 10.1093/nar/gkl205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Remmert M., Linke D., Lupas A.N., Soding J. HHomp–prediction and classification of outer membrane proteins. Nucleic Acids Res. 2009;37:W446–W451. doi: 10.1093/nar/gkp325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Diederichs K., Freigang J., Umhau S., Zeth K., Breed J. Prediction by a neural network of outer membrane beta-strand protein topology. Protein Sci. 1998;7:2413–2420. doi: 10.1002/pro.5560071119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Jacoboni I., Martelli P.L., Fariselli P., De Pinto V., Casadio R. Prediction of the transmembrane regions of beta-barrel membrane proteins with a neural network-based predictor. Protein Sci. 2001;10:779–787. doi: 10.1110/ps.37201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Gromiha M.M., Ahmad S., Suwa M. Neural network-based prediction of transmembrane beta-strand segments in outer membrane proteins. J Comput Chem. 2004;25:762–767. doi: 10.1002/jcc.10386. [DOI] [PubMed] [Google Scholar]
- 270.Natt N.K., Kaur H., Raghava G.P. Prediction of transmembrane regions of beta-barrel proteins using ANN- and SVM-based methods. Proteins. 2004;56:11–18. doi: 10.1002/prot.20092. [DOI] [PubMed] [Google Scholar]
- 271.Randall A., Cheng J., Sweredoski M., Baldi P. TMBpro: secondary structure, beta-contact and tertiary structure prediction of transmembrane beta-barrel proteins. Bioinformatics. 2008;24:513–520. doi: 10.1093/bioinformatics/btm548. [DOI] [PubMed] [Google Scholar]
- 272.Koehler J., Mueller R., Meiler J. Improved prediction of trans-membrane spans in proteins using an Artificial Neural Network. IEEE Symp Comput Intell Bioinforma Comput Biol Proc. 2009;2009:68–74. doi: 10.1109/CIBCB.2009.4925709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Martelli P.L., Fariselli P., Krogh A., Casadio R. A sequence-profile-based HMM for predicting and discriminating beta barrel membrane proteins. Bioinformatics. 2002;18(Suppl 1):S46–S53. doi: 10.1093/bioinformatics/18.suppl_1.s46. [DOI] [PubMed] [Google Scholar]
- 274.Bigelow H.R., Petrey D.S., Liu J., Przybylski D., Rost B. Predicting transmembrane beta-barrels in proteomes. Nucleic Acids Res. 2004;32:2566–2577. doi: 10.1093/nar/gkh580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Bagos P.G., Liakopoulos T.D., Spyropoulos I.C., Hamodrakas S.J. A Hidden Markov Model method, capable of predicting and discriminating beta-barrel outer membrane proteins. BMC Bioinf. 2004;5:29. doi: 10.1186/1471-2105-5-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Park K.J., Gromiha M.M., Horton P., Suwa M. Discrimination of outer membrane proteins using support vector machines. Bioinformatics. 2005;21:4223–4229. doi: 10.1093/bioinformatics/bti697. [DOI] [PubMed] [Google Scholar]
- 277.Roy Choudhury A., Novic M. PredbetaTM: A Novel beta-Transmembrane Region Prediction Algorithm. PLoS ONE. 2015;10 doi: 10.1371/journal.pone.0145564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Garrow A.G., Agnew A., Westhead D.R. TMB-Hunt: an amino acid composition based method to screen proteomes for beta-barrel transmembrane proteins. BMC Bioinf. 2005;6:56. doi: 10.1186/1471-2105-6-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Yan C., Hu J., Wang Y. Discrimination of outer membrane proteins using a K-nearest neighbor method. Amino Acids. 2008;35:65–73. doi: 10.1007/s00726-007-0628-7. [DOI] [PubMed] [Google Scholar]
- 280.Lin H. The modified Mahalanobis Discriminant for predicting outer membrane proteins by using Chou's pseudo amino acid composition. J Theor Biol. 2008;252:350–356. doi: 10.1016/j.jtbi.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 281.Ou Y.Y., Gromiha M.M., Chen S.A., Suwa M. TMBETADISC-RBF: Discrimination of beta-barrel membrane proteins using RBF networks and PSSM profiles. Comput Biol Chem. 2008;32:227–231. doi: 10.1016/j.compbiolchem.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 282.Fariselli P., Savojardo C., Martelli P.L., Casadio R. Grammatical-Restrained Hidden Conditional Random Fields for Bioinformatics applications. Algorithms Mol Biol. 2009;4:13. doi: 10.1186/1748-7188-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Mooney C., Wang Y.H., Pollastri G. SCLpred: protein subcellular localization prediction by N-to-1 neural networks. Bioinformatics. 2011;27:2812–2819. doi: 10.1093/bioinformatics/btr494. [DOI] [PubMed] [Google Scholar]
- 284.Hayat S., Elofsson A. BOCTOPUS: improved topology prediction of transmembrane beta barrel proteins. Bioinformatics. 2012;28:516–522. doi: 10.1093/bioinformatics/btr710. [DOI] [PubMed] [Google Scholar]
- 285.Hayat S., Peters C., Shu N., Tsirigos K.D., Elofsson A. Inclusion of dyad-repeat pattern improves topology prediction of transmembrane beta-barrel proteins. Bioinformatics. 2016;32:1571–1573. doi: 10.1093/bioinformatics/btw025. [DOI] [PubMed] [Google Scholar]
- 286.Bagos P.G., Liakopoulos T.D., Hamodrakas S.J. Evaluation of methods for predicting the topology of beta-barrel outer membrane proteins and a consensus prediction method. BMC Bioinf. 2005;6:7. doi: 10.1186/1471-2105-6-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Yu N.Y., Wagner J.R., Laird M.R., Melli G., Rey S., Lo R. PSORTb 3.0: Improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics. 2010;26:1608–1615. doi: 10.1093/bioinformatics/btq249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Wan S., Mak M.W., Kung S.Y. FUEL-mLoc: feature-unified prediction and explanation of multi-localization of cellular proteins in multiple organisms. Bioinformatics. 2017;33(5):749–750. doi: 10.1093/bioinformatics/btw717. [DOI] [PubMed] [Google Scholar]
- 289.Dhroso A., Eidson S., Korkin D. Genome-wide prediction of bacterial effector candidates across six secretion system types using a feature-based statistical framework. Sci Rep. 2018;8:17209. doi: 10.1038/s41598-018-33874-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Pundhir S., Kumar A. SSPred: A prediction server based on SVM for the identification and classification of proteins involved in bacterial secretion systems. Bioinformation. 2011;6:380–382. doi: 10.6026/97320630006380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Martinez-Garcia P.M., Ramos C., Rodriguez-Palenzuela P. T346Hunter: a novel web-based tool for the prediction of type III, type IV and type VI secretion systems in bacterial genomes. PLoS ONE. 2015;10 doi: 10.1371/journal.pone.0119317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Abby S.S., Cury J., Guglielmini J., Neron B., Touchon M., Rocha E.P. Identification of protein secretion systems in bacterial genomes. Sci Rep. 2016;6:23080. doi: 10.1038/srep23080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Dehzangi A., Heffernan R., Sharma A., Lyons J., Paliwal K., Sattar A. Gram-positive and Gram-negative protein subcellular localization by incorporating evolutionary-based descriptors into Chou׳ s general PseAAC. J Theor Biol. 2015;364:284–294. doi: 10.1016/j.jtbi.2014.09.029. [DOI] [PubMed] [Google Scholar]
- 294.Sueki A., Stein F., Savitski M.M., Selkrig J., Typas A. Systematic localization of Escherichia coli membrane proteins. mSystems. 2020;5(2):e00808–e00819. doi: 10.1128/mSystems.00808-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Wu J., Liu Y.C., Chang D.T.H. SigUNet: signal peptide recognition based on semantic segmentation. BMC Bioinf. 2019;20(Suppl 24):677. doi: 10.1186/s12859-019-3245-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Zhang W.X., Pan X., Shen H.B. Signal-3L 3.0: Improving Signal Peptide Prediction through Combining Attention Deep Learning with Window-Based Scoring. J Chem Inf Model. 2020;60(7):3679–3686. doi: 10.1021/acs.jcim.0c00401. [DOI] [PubMed] [Google Scholar]
- 297.Li J., Wei L., Guo F., Zou Q. EP3: an ensemble predictor that accurately identifies type III secreted effectors. Brief Bioinform. 2020:bbaa008. doi: 10.1093/bib/bbaa008. [DOI] [PubMed] [Google Scholar]
- 298.Hong J., Luo Y., Mou M., Fu J., Zhang Y., Xue W. Convolutional neural network-based annotation of bacterial type IV secretion system effectors with enhanced accuracy and reduced false discovery. Brief Bioinform. 2020;21(5):1825–1836. doi: 10.1093/bib/bbz120. [DOI] [PubMed] [Google Scholar]
- 299.Chen T., Wang X., Chu Y., Wang Y., Jiang M., Wei D.Q. T4SE-XGB: interpretable sequence-based prediction of type IV secreted effectors using eXtreme gradient boosting algorithm. Front Microbiol. 2020;11 doi: 10.3389/fmicb.2020.580382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Almeida A., Nayfach S., Boland M., Strozzi F., Beracochea M., Shi Z.J. A unified catalog of 204,938 reference genomes from the human gut microbiome. Nat Biotechnol. 2021;39(1):105–114. doi: 10.1038/s41587-020-0603-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Liu J., Lian Q., Chen Y., Qi J. Amino acid based de Bruijn graph algorithm for identifying complete coding genes from metagenomic and metatranscriptomic short reads. Nucleic Acids Res. 2019;47(5) doi: 10.1093/nar/gkz017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Lai L.A., Tong Z., Chen R., Pan S. Metaproteomics study of the gut microbiome. Methods Mol Biol. 2019;1871:123–132. doi: 10.1007/978-1-4939-8814-3_8. [DOI] [PubMed] [Google Scholar]