Abstract
Given the limited availability of tissue, especially brain tissue, for neurological diseases and disorders research, the development of alternative biological tools for investigations of underlying molecular and genetic mechanisms is imperative. One important resource for this task is the large repositories that bank immortalized blood cells (i.e. lymphoblastoid cell lines; LCLs) from affected individuals and their unaffected family members. These repositories document demographic, phenotypic, and, in some cases, genotypic information about the donors and thus provide a ready-made sample source for hypothesis testing. Importantly, patient-specific LCLs can be used to generate induced pluripotent stem cells (iPSC) that, in turn, can be used to create specific cell types for use in mechanistic studies. To investigate this concept further, LCLs from two males (proband and sibling) were obtained from one such repository, the Autism Genetics Resource Exchange (AGRE), and iPSCs were generated by transfection with Epi5 Episomal iPSC reprogramming plasmids. Characterization of the resultant cell lines by PCR, RT-PCR, immunocytochemistry, karyotyping, and the Taqman® human pluripotent stem cell Scorecard™ Panel, was used to provide evidence of endogenous pluripotency and then to evaluate the trilineage potential of four representative clones. Results indicated that all four iPSC lines were initially pluripotent and displayed the trilineage potential predictive for successful differentiation to mesoderm, endoderm, or ectoderm-derived cell types. Compared to other published protocols, this study details a somewhat simplified approach, used here specifically for the generation and characterization of induced pluripotent stem cells from well-characterized and banked LCLs.
Keywords: Lymphoblastoid cell lines, Induced pluripotent stem cells, Reprogramming, Epstein-Barr virus
Lymphoblastoid cell lines, Induced pluripotent stem cells, Reprogramming, Epstein-Barr virus.
1. Introduction
Induced pluripotent stem cell (iPSC) technology has shown great promise for the production of large numbers of patient-specific cells that can be used in mechanistic studies and for the discovery of new therapeutic compounds (Yu et al., 2007; Takahashi et al., 2007; Mack et al., 2014). By taking advantage of the fact that the iPSC-derived cells are genetically identical to the patient from whom they are derived, patient-specific iPSCs can be differentiated along specific cell lineages, including neuronal lineages, and used to study specific neural deficits in vitro. Functional assays can be used to query molecular aspects of the specific disease phenotype, e.g. dysregulation of synapse formation, in the culture dish. This technology has been applied successfully to study a wide range of neurological diseases including Parkinson's disease (Werning et al., 2008), schizophrenia (Brennand et al., 2011), amyotrophic lateral sclerosis (Dimos et al., 2008), Rett syndrome (Marchetto et al., 2010), and Phelan-McDermid syndrome (Shcheglovitov et al., 2013).
Using autism spectrum disorder (ASD) as a neurobiological disease model, patient-derived somatic cells such as fibroblasts (Chanda et al., 2013; Brennand et al., 2012; Marchetto et al., 2011) and peripheral blood mononuclear cells (DeRosa et al., 2012) have been used to generate iPSCs. A potentially much more robust cell source for the study of, for example, ASD is available through collections such as the Autism Genetics Resource Exchange (AGRE; (Geschwind et al., 2001)) and the Simons Simplex Collection (SSC) within the Simons Foundation. The AGRE is a national repository that currently stores immortalized lymphoblastoid cell lines (LCLs) derived from individuals representing more than 2000 pedigreed simplex and multiplex families. The SSC is the core project and resource of the Simons Foundation Autism Research Initiative and contains LCLs from over 2600 simplex families. In both repositories the LCLs are accompanied by abundant phenotypic and genotypic data from the probands, siblings, and at least one parent, and all of this information is available to the research community. Applying iPSC technology to LCLs from these repositories can provide researchers with a large, well-characterized (clinically, phenotypically, and genetically) resource and an invaluable in vitro tool to test current and new hypotheses related to the biological mechanisms that underlie ASDs (Sie et al., 2009).
Methodology for reprogramming LCLs into iPSCs using feeder (Choi et al., 2011; Thomas et al., 2015; Fujimori et al., 2016) and feeder-free (Rajesh et al., 2011; Barrett et al., 2014; Kumar et al., 2016) cell culture conditions has been published previously. One group has described the differentiation of LCL-derived iPSCs into endoderm, ectoderm and mesoderm cells using a combination of pCE-hOCT3/4, pCE-hUL, pCE-mp53DD, OCT3/4, SOX2, KLF4, LMYC, LIN28 and p53 carboxy-terminal dominant-negative fragment (Kumar et al., 2016). This protocol resulted in reported LCL efficiencies ranging from 0.0102% to 0.0216%.
Here, using Epi5™ Episomal iPSC Reprogramming plasmids, we report important improvements to current LCL-specific iPSC-generating methodologies, including a more rapid reprogramming and an apparent increased efficiency. Time and cost savings were achieved by carrying out the entire reprogramming process under feeder-free conditions. Moreover, we employed a relatively new real-time PCR assay, the TaqMan® hPSC Scorecard™ Panel (Life Technologies, Inc.), to evaluate pluripotency and differentiation capacity as an alternative to using the widely accepted, but much more costly and time consuming, teratoma formation assay in immune compromised mice (Bock et al., 2011). This protocol provides a reproducible means to generate iPSCs efficiently with standardized and cost-effective reagents. IPSCs produced following this protocol can be used to generate and evaluate novel in vitro models to study a plethora of previously inaccessible cell types that underlie pathological mechanisms in neurological disease.
2. Materials and methods
2.1. Patient LCLs
Two Epstein-Barr virus immortalized lymphoblastoid cell lines (LCLs), HI2102 (Line 1) and HI2183 (Line 2), were obtained from the AGRE. HI2102 was generated from a male proband with a R300C SNP in the Shank3 gene and HI2183 is the unaffected male sibling of the proband. LCLs from these subjects were used in a previous study conducted by Kelleher et al., that reported identification of rare variants in mGluR signalling pathway genes in subjects with autism (Kelleher et al., 2012). Although using de-identified samples and data from the AGRE does not require institutional IRB approval, there is an AGRE application process to obtain the samples that requires a detailed description of how samples will be used, along with a requirement for data sharing.
2.2. Culture and maintenance of LCLs
Each LCL was cultured in T75 flasks in RPMI1640 media supplemented with 15% fetal bovine serum and 5% penicillin-streptomycin (LCL media) at 37 °C and 5% CO2 in a humidified incubator at a minimum cell density of ~500,000 cells/millilitre (mL) (all reagents were from ThermoScientific). One day prior to transfection, to ensure efficient cell proliferation, LCL density was adjusted to ~850,000 cells/mL.
2.3. Reprogramming LCLs
Reprogramming of LCLs (approximately 1 × 106 cells) with 2 μg Epi5™ Episomal Reprogramming plasmids (Life Technologies Inc., Carlsbad, CA) was performed via electroporation using the Amaxa Nucleofector Device II, Cell Line Kit V, and program T-020, following the Cell Line Kit V protocol (Lonza, Basel, Switzerland). The Epi5™ plasmids contain reprogramming factors Oct3/4, Sox2, Klf4, Lin28, and L-myc and two additional factors, mp53DD and EBNA, to aide in cell cycle stabilization. After transfection, each sample was transferred directly into 1 well of a 6 well plate coated with hESC-qualified Matrigel (1:60; BD Biosciences, San Jose, CA) with LCL media and incubated overnight. Twelve hours post-transfection the Epi5™ Episomal Reprogramming (“Reprogramming CD34+ Cells (Feeder-free culture)”) protocol was followed starting at Day 1 and continuing until mature iPSC colonies were apparent (at Day 15–20). Once iPSC colonies were present, it was necessary to perform an Essential 8 media (Life Technologies, Inc.) change every day to prevent differentiation. At passage 5, all clonal lines were transitioned to mTeSR-1 medium (StemCell Technologies, Vancouver, BC).
2.4. Immunocytochemistry
2.4.1. Live staining
Tra-1-60 live staining was performed using 10 μg of Tra-1-60 primary antibody (R&D Systems, Minneapolis, MN) per well of a 6-well plate in DMEM/F12 for 1 h at 37 °C and 5% CO2 in a humidified incubator. Each well was washed three times with DMEM/F12 and incubated with 1:100 dilution of secondary antibody AF488 for 1 h under the same conditions. After cells were washed three times, positive staining was visualized with a fluorescence microscope. We have also successfully live-stained with the Tra-1-60 Alexa Fluor® 488 Conjugate Kit (Life Technologies, Inc.), which provides results in less than 45 min when the manufacturer's manual is followed.
2.4.2. Fixed staining
Mature iPSC colonies in a 24 well plate were stained with Tra-1-60 (10 μg/well; R&D systems), SSEA4 (6 μg/well; StemCell Technologies), and Nanog (1:200; Genetex, Irvine, CA) primary antibodies. Cells were fixed in 4% paraformaldehyde for 15 min and washed in 0.1% Triton X-100 in phosphate buffered saline (PBS-T) three times for 5 min at room temperature. Cells were simultaneously blocked and permeabilized in a solution of 50% PBS, 45% sterile-deionized water, 0.15% Triton X-100, and 5% serum (based on secondary antibody) for 1 h at room temperature. Primary antibodies were added to 200 μl of permeabilization/blocking solution per well and incubated overnight at 4 °C. After three washes (5 min each) with PBS-T, cells were incubated in 200 μl of 1% BSA in PBS with secondary antibodies (1:250) for 30 min at room temperature. Cells were then washed, incubated with DAPI (1:500) in PBS-T, washed 3 more times, and visualized using a fluorescence microscope.
2.5. Genomic/episomal DNA PCR
Total DNA was isolated from clonal iPSCs using the DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany) according to manufacturer instructions and used to measure, by PCR, residual episomal plasmid DNA present in cells following passaging (primer sequences can be found in Table 1). PCR was performed using 10ng DNA per reaction, for 35 cycles of: 94°, 15s; 58°, 15s; 72°, 30s. PCR products were separated on a 2% agarose gel run at 50 mV for 40 min and visualized with a Kodak Gel Logic 200 imaging system (Kodak, Rochester, New York).
Table 1.
Primers used for PCR to assess pluripotency in clonal iPSC lines.
| Primer Set | Forward (5′-3′) | Reverse (5′-3′) | Size (bp) |
|---|---|---|---|
| EBNA-1 | ATCGTCAAAGCTGCACACAG | CCCAGGAGGTCCCAGTAGTCA | 665 |
| Β-actin | CCCAGGCACCAGGGCGTGAT | TCAAACATGATCTGGGTCAT | 178 |
| Oct3/4 | CGTGAAGCTGGAGAAGGAGAAGCT | CAAGGGCCGCAGCTCACACATGTTC | 250 |
| Nanog | CAGCCCCGATTCTTCCACCAGTCCC | CAGCCCCGATTCTTCCACCAGTCCC | 400 |
| DMNT3B | TGCTGCTCACAGGGCCCGATACTTC | TCCTTTCGAGCTCAGTGCACCACAAAAC | 242 |
| PODXL | TCCAGCCCCACAGCAGXATCAACTACC | CCGGGTTGAAGGTGGCTTTGACTGCTC | 226 |
2.6. RT-PCR
Total RNA was isolated from clonal iPSCs using the RNeasy Blood and Tissue Kit (Qiagen) according to manufacturer instructions and cDNAs were generated using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) in a 20μl reaction volume. Primers for EBNA-2, BZLF-1, and LMP-2 were used to measure levels of viral gene expression (derived from the LCL immortalization process with EBV) as previously reported (Rajesh et al., 2011). With these primers, PCR was run using 5μl cDNA per reaction at: 94° for 15s, 58° for 15s, and 72° for 30s, for 35 cycles.
Primers for Oct3/4, Nanog, DMNT3B, and PODXL (Table 1) were used to assess pluripotency. PCR assays were run with 5μl cDNA per reaction for 35 cycles at 94° for 15s, 62–66° for 15s and, 72° for 30s. PCR products were separated on a 2% agarose gel run at 50 mV for 40 min and visualized with a Kodak Gel Logic 200 imaging system (Kodak, Rochester, New York).
2.7. Embryoid body formation
Cells from one 10 cm2 dish of iPSCs (one per clonal line) at passage 20 were collected in single cell suspension using Accutase (StemCell Technologies), washed twice with DMEM/F12, and resuspended in EB medium supplemented with 10 μM ROCK-inhibitor (StemCell Technologies). EB medium is composed of: 79% DMEM/F12, 20% Knockout Serum Replacement, 1% non-essential amino acids, and 0.1% β-mercaptoethanol (all reagents from Gibco, Life Technologies). The cell suspension, collected from a confluent 10-cm2 dish, was applied to two wells of an AggreWell™ 400 plate (StemCell Technologies) prepared according to manufacturer instructions. At 24 h, the newly forming embryoid bodies were harvested from the AggreWell™ 400 plate as directed in the manufacturer's manual. EBs from two wells of AggreWell400 were then transferred to 3 wells of a non-adherent 6-well plate at a density of <1000 EBs per well with 5 ml EB medium. EBs were incubated at 37 °C and 5% CO2 in a humidified incubator for 7 days. Media changes occurred every other day following transfer to 6-well plates and the EBs were harvested at day 7 and 14, for qRT-PCR analysis. For media change, EBs were collected in a 15 ml conical tube and allowed to settle by gravity for 15 min. Used media was aspirated, followed by resuspension of the EBs in fresh media.
2.8. TaqMan® human pluripotent stem cell Scorecard™ PanelT
Total RNA was isolated from EBs derived from each of the four iPSC clonal lines at passage 20. Reverse transcription of total RNA was conducted using a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). For the Scorecard™ protocol, 8 wells of 450 μl total volume (225 μl 2X RT master mix +225 μl water containing 1μg RNA) were required for each clone. Following the Scorecard™ protocol outlined in the user manual, (with the exception of substituting RNase-free water for RNase Inhibitor (20U/μl)) the RT reactions were performed by incubating: 25 °C for 10m, 37 °C for 120m, and 85 °C for 5m, followed by a 4 °C hold. Each cDNA was applied to a TaqMan® hPSC Scorecard™ Panel 96w FAST plate using TaqMan® Gene Expression Master Mix and run on a StepOne Plus Real-Time PCR System. The experimental template file specific to the Scorecard™ Panel was downloaded from www.lifetechnologies.com/scorecardinstrument for the StepOne Plus Real-Time PCR system. In addition to the template file, PCR conditions are dependent on the specific plate used and were entered manually. For the 96w FAST plate, the ramp rate is “fast” and PCR conditions are: hold at 50 °C for 20s, then 40 cycles of 95 °C for 1s and 60 °C for 20s. Gene expression data were uploaded and analysed using the web-based hPSC Scorecard™ Analysis Software, available at www.lifetechnologies.com/scorecarddata.
2.9. Karyotyping
At passage 23 the iPSC clones were cultured in T-25 flasks and sent to Cell Line Genetics, Inc. (Madison, Wisconsin) two days prior to being 90% confluent (based on colony morphology). For each of the four iPSC line clones, cytogenetic analysis was performed on 20 G-banded metaphase cells.
3. Results
3.1. Generation of iPSCs from LCLs
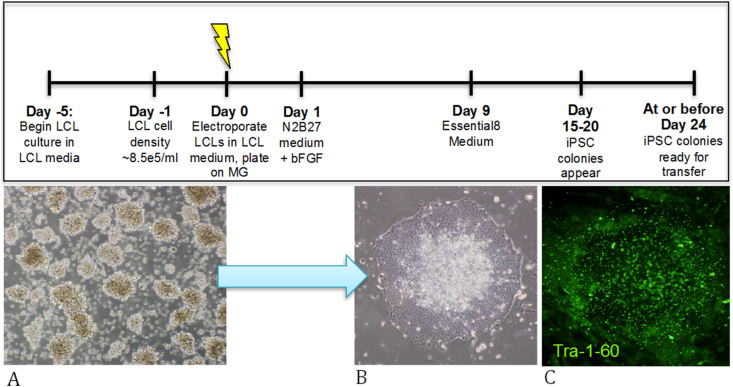
LCLs (Figure 1A) were reprogrammed to iPSCs following a single transfection with Epi5™ Episomal Reprogramming plasmids under feeder-free conditions. Post-transfection, cells were maintained in N2B27 medium followed by a transition to E8 medium on Day 9 post-transfection until iPSC colonies were ready for passaging (Figure 1B). IPSCs were first visible at day 16 (estimated to be between 13-23 clones per transfection, ~ 0.001–0.002% transfection efficiency) and mature enough for propagation and cloning at day 24 (Timeline shown in Figure 1; Top panel). Initial pluripotency was assessed via live staining with Tra-1-60 (Figure 1C). At day 24, greater than 50% of cells stained positive for Tra-1-60 and 48 iPSC colonies derived from each LCL were clonally passaged to allow for future characterization. Since the initial reprogramming carried out in our lab to now, this protocol has shown reliable reprogramming success of 100% (8/8) in AGRE lines and LCLs from the NIH Repository.
Figure 1.
Timeline for reprogramming LCLs to iPSCs. (A) Morphology of LCLs before electroporation, (B) an iPSC colony ready for propagation and, (C) subsequent Tra-1-60 live staining of iPSC colony. Images were captured using original magnification at 10X.
3.2. Characterization of clonal lines
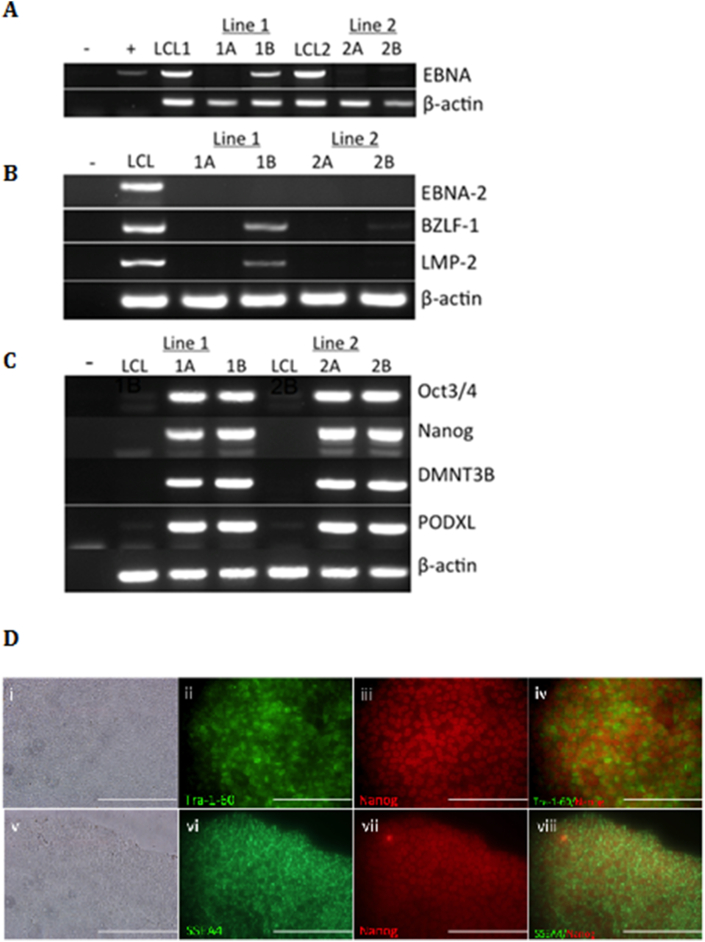
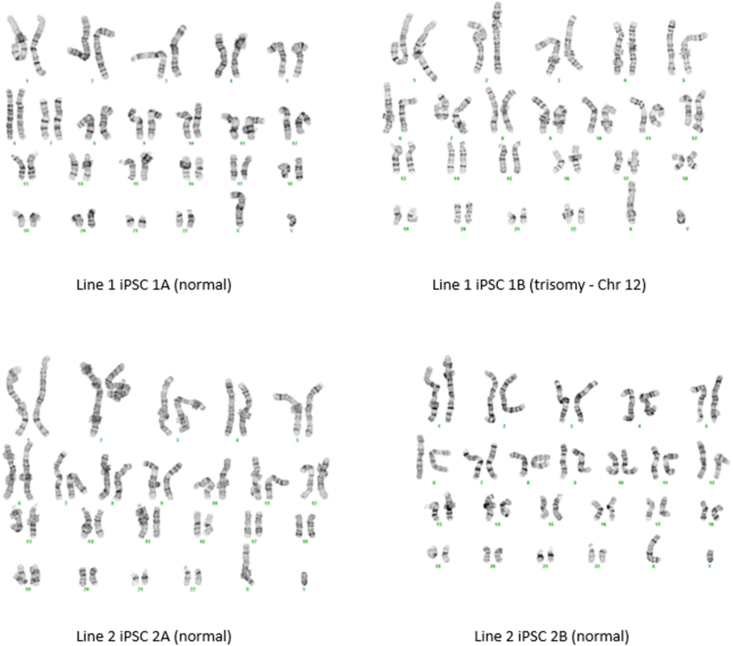
Additional characterization was conducted for two clonal iPSC lines derived from Line 1 (Line 1-iPSC1A, Line 1-iPSC1B) and two clonal iPSC lines derived from Line 2 (Line 2-iPSC2A, Line 2-iPSC2B). PCR results confirmed the absence of Epi5™ plasmids in three of the four clones by passage 16 (Figure 2A) and absence of EBV gene expression by passage 18 (Figure 2B). Next, RT-PCR and immunocytochemistry validation assays were conducted to confirm pluripotency. RT-PCR revealed expression of pluripotency genes Oct3/4, Nanog, DMNT3B, and PODXL in iPSCs, but not in LCLs (Figure 2C). Since Line 1-iPSC1B still contains a substantial amount of the Epi5 plasmids, the Oct3/4 expression observed for this line may partially be due to expression of the exogenous plasmid. However, this line was also expressing Nanog, DMNT3B and PODXL, which are pluripotency genes not included in the reprogramming plasmids, indicating endogenous gene expression as well. Each clone also stained ≥90% positive for Tra-1-60, SSEA1, and Nanog expression (Figure 2D). Karyotype analyses were conducted on each of the four clonal iPSCs lines and all karyotypes were normal except for Line 1-iPSC1B, which had trisomy at chromosome 12 in four of the 20 G-banded metaphase cells tested (Figure 3).
Figure 2.
Characterization of LCL-derived iPSCs. (A) PCR-based detection of episomal plasmids in clonal iPSC lines derived from EBV-LCLs at passage 16 (P16). Total DNA was isolated from two clonal iPSC lines derived from each of two parent LCLs (Line 1 & Line 2) at multiple passages using the DNeasy Blood & Tissue kit. A 665bp PCR product is present in the agarose gel if any of the plasmids are still present in the cells. Plasmid DNA from the reprogramming kit was used as the positive control (+); the negative control had nuclease-free water in the place of DNA (-). β-actin was used as a positive loading control for each sample. (B) RT-PCR analysis of one LCL and two representative LCL-iPSC clones from each LCL at passage 18 for expression of EBV genes: EBNA-2, BZLF-1, and LMP-2. β-actin was used as a positive loading control for each sample. Negative control for each primer set consisted of using nuclease-free water in place of cDNA. Results from a single LCL is displayed, however both LCLs tested positive for all EBV genes and B-actin. (C) Validation of iPSC pluripotency by PCR. RT-PCR analysis of original LCLs and two representative LCL-iPSC clones from each LCL at passage 18 for expression of pluripotency genes: Oct3/4, Nanog, DMNT3B and PODXL. β-actin was used as a positive loading control for each sample. Negative control for each primer set consisted of using nuclease-free water in place of cDNA (-). (D) Validation of iPSC pluripotency by immunocytochemistry on passage 23 LCL-derived IPSCs. ICC was conducted on LCL-derived iPSCs following reprogramming with Epi5™ episomal reprogramming plasmids and propagation to passage 23 (i & v, brightfield). Extracellular markers for Tra-1-60 (ii) and SSEA4 (vi) appear green. Nanog was used for intracellular staining and appear red (iii, vii). Pairings of Tra-1-60/Nanog and SSEA4/Nanog indicate proper cell localization of these markers (iv, viii). All images were taken at 40X. Scale bars are 200μm.
Figure 3.
Representative karyotype from iPSC colonies. Cytogenetic analysis was performed on twenty G-banded metaphase cells from all four human iPSC lines (P23) and all twenty cells demonstrated an apparently normal male karyotype for three of the four iPSC lines.
3.3. Formation and characterization of embryoid bodies
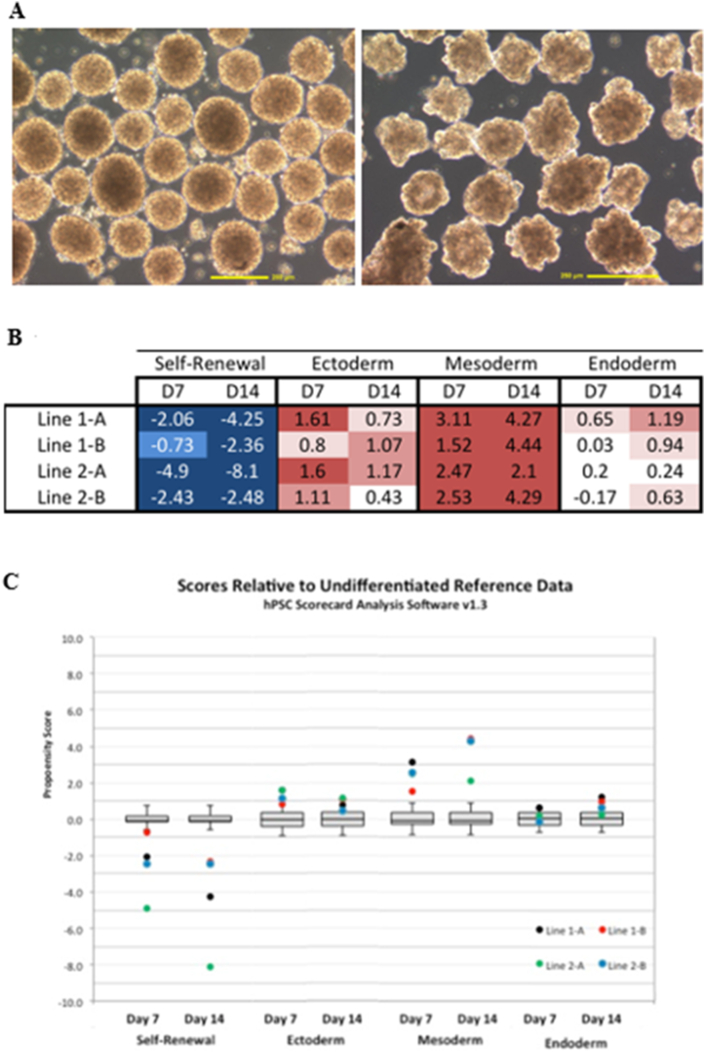
At passage 20 the reprogramming plasmid was no longer detectable and the viral RNAs were also absent from three of the four LCL-derived iPSC clones. All four clones at this time were spontaneously differentiated into embryoid bodies (EBs) for 7 and 14 days. The TaqMan® hPSC Scorecard™ Panel was used to evaluate EBs derived from the four LCL-iPSCs. Scores provided by this panel revealed pluripotency gene down-regulation upon differentiation and the potential for trilineage differentiation in one of the four clones (Figure 4). All lines had downregulation of pluripotency markers at both time points. Ectoderm and mesoderm gene expression were upregulated (red) in all cell line at days 7 and 14. Endoderm gene expression was upregulated in all cell lines by day 14. Taken together, these data suggest that all four iPSC lines have trilineage potential and may be used for differentiation protocols to mesoderm, endoderm, or ectoderm-derived cell lineages. The major milestones that patient-derived iPSCs must achieve before being considered adequate for differentiation protocols and the criteria fulfilled by the iPSC clonal lines generated in this study are summarized in Table 2.
Figure 4.
Taqman® hPSC Scorecard™ Panel analysis based on embryoid bodies (EBs) spontaneously differentiated from clonal LCL-iPSC lines. Four clonal LCLs were spontaneously differentiated for 7 and 14 days into EBs. Total RNA was isolated from each EB using the RNeasy Blood and Tissue Kit (Qiagen). cDNAs were made using a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) and applied to the 96-well Scorecard Panel. The panel contains probes to detect and report on markers of pluripotency and on trilineage potential (to ectoderm, mesoderm, and endoderm). (A) representative images (10x) of embryoid bodies at Day 2 (left image) and Day 3 (right image). Results of the day 7 and 14 time points are depicted in two formats: (B) Table of Scorecard™ values. All lines had downregulation of pluripotency markers (blue). Ectoderm and mesoderm gene expression were upregulated (red) in all cell lines at day 7 and 14. Endoderm gene expression was upregulated in all cell lines by day 14. (C) Box plot of Taqman® hPSC scores relative to reference data from undifferentiated cells.
Table 2.
Basic Criteria necessary for patient-derived iPSCs to be considered adequate for further differentiation protocols. Three out of four LCL-derived iPSC clonal lines (Line 1-iPSC1A, Line 2-iPSCA2A, Line 2-iPSC2B) generated using the protocol described in this study meet all of these criteria and could be used for ectoderm-lineage differentiation. “X” denotes the iPSC line met the criterion in each column.
| LCL-Derived iPSC Lines | iPSC Morphology | Early Tra-1-60 Positive Staining | Efficiency of Passaging | Endogenous Gene Expression of Pluripotency Markers | Normal Karyotype | Spontaneous Differentiation |
|---|---|---|---|---|---|---|
| Line 1 iPSC 1A | X | X | X | X | X | X |
| Line 1 iPSC 1B | X | X | X | |||
| Line 2 iPSC 2A | X | X | X | X | X | X |
| Line 2 iPSC 2B | X | X | X | X | X | X |
4. Discussion
It is imperative to develop tools that can be used to provide a more complete understanding of biological mechanisms that underlie complex disease so that rational therapeutic and prevention strategies can be designed. The primary goal of this study was to leverage the abundant LCL resources and, using commercially available reagents, to streamline the methodology necessary to generate patient-specific iPSCs for use in the creation of in vitro models for mechanistic studies in complex disease. Using two LCLs obtained from the AGRE and the protocols described in this manuscript, four clonal lines of iPSCs were generated in a simplified and efficient manner. These resultant iPSCs can be used for differentiation into multiple cell types, including neurons, to aide in the understanding of molecular mechanisms that underlie neurological disorders (Marchetto et al., 2011).
This is not the first report of successful reprogramming of immortalized B-lymphocytes. In 2011 two groups demonstrated successful reprogramming of LCLs (obtained from Coriell Repositories) into iPSCs with subsequent trilineage differentiation of LCL-derived iPSCs (Choi et al., 2011; Rajesh et al., 2011). These studies also provided the first evidence for the loss of EBV expression following episomal reprogramming of LCLs. This was confirmed by using RT-PCR to show the absence of expression of viral genes LMP-2 and BZLF-1 and by using real-time PCR to demonstrate the exponential decay of three EBV genome segments (EBNA-1, EBER1, and BamH1W).
Although successful reprogramming of LCLs into iPSCs has been reported previously, there are specific aspects of these published approaches that rendered replication either not desirable or not possible. First, Choi et al. performed reprogramming on feeder layers (irradiated mouse embryonic fibroblasts) which consist of cells that secrete a cocktail of non-human growth factors to influence the developing iPSCs (Choi et al., 2011). This approach was used to produce the first human derived iPSCs from fibroblasts (Takahashi et al., 2007). However the use of feeder cells for iPSC production is no longer recommended if the iPSCs may, at a later time, be transferred to a Current Good Manufacturing Practice (cGMP) environment for transplantation into patients (Ludwig et al., 2006). In the other relevant study published in 2011, Rajesh et al. used cGMP-compliant feeder-free methods throughout their reprogramming, allowing for more control over growth factors used to induce reprogramming (Rajesh et al., 2011).
We initially attempted to generate iPSCs using the protocol described by Rajesh et al. however, despite multiple attempts, were unsuccessful. One trivial reason for our initial lack of success may be due to our inability to replicate the specific transfection parameters of the Lonza 96-well Shuttle™ system used by the Rajesh group - an instrument that we did not have available in our laboratory. However, while we were able to successfully generate iPSCs using a different transfection system (Amaxa Nucleofector Device) we acknowledge that this step may still present a limitation for replication of the transfection success reported here. Therefore, without direct comparison, we concluded that the protocol described by Rajesh et al., and the protocol described here, are both valid approaches for generating iPSCs from LCLs but may not be easy to replicate without the exact instrument and settings that were reported. Recently, Barrett et al. described an alternative protocol to generate iPSCs from LCLs, citing the unreliability of previously published protocols. However, the Barrett approach requires generating individual stocks of endo-free plasmids and full validation of vector expression prior to reprogramming, as opposed to removing the high risk of plasmid variability by using commercial reagents (Barrett et al., 2014).
The method outlined in the current study, in addition to using only commercially-available reagents, has the added advantage of producing LCL-derived iPSCs in as little as 21 days, as opposed to 32–35 days or more using the Rajesh or Barrett protocol. In addition to using an alternative transfection system and parameters, we utilized standardized and updated materials that are: (1) more optimal for hESC and iPSC cultures, (2) commonly used in stem cell laboratories and, (3) more efficient for somatic cell reprogramming. First, the N2B27 reprogramming media applied to LCLs post-transfection includes 7 components essential for reprogramming, while previous papers used up to 13 components, including 6 costly small molecule inhibitors. Furthermore, only one of the 6 small molecule inhibitors (βFGF) is used in the N2B27 media, which successfully yielded iPSCs in this study. Following culture in N2B27 media, reprogramming LCLs were transitioned to Essential 8™ media on Day 9. This media, generated by Chen and colleagues, was produced solely to simplify the chemical components of feeder-free cell culture mediums and to offer a more cost-effective approach to pluripotent stem cell maintenance (Chen et al., 2011).
The episomal plasmids used for reprogramming in Rajesh et al., following testing with multiple combinations of Oct3/4, Sox2, Nanog, Lin 28, Klf4, c-Myc, L-Myc, and SV40-T, resulted in the observation that a minimum combination of Oct3/4, Sox2, Nanog, and SV40-T antigen was sufficient to reprogram the LCLs. The commercially available Epi5™ Episomal Reprogramming kit used in this study contains plasmids expressing five reprogramming factors Oct3/4, Sox2, Klf4, Lin28, and L-myc. These plasmids are smaller in size, ranging from 6-11kb (compared to 12–17.5 kb plasmids used by Rajesh et al.) which renders them potentially more efficient in entering the LCLs through openings in cell membranes during transfection. While both systems use OriP/EBNA1 mediated nuclear transport vectors to increase transfection efficiency, the Epi5™ Episomal Reprogramming plasmids contain additional components, mp53DD and additional EBNA, to increase transfection efficiency and survivability of future iPSCs (Hong et al., 2009; Spike and Wahl, 2011).
The Epi5™ Episomal iPSC Reprogramming manual projects an efficiency of 0.04%–0.3%, producing at most 3000 iPSCs from one million fibroblasts or CD34+ cells, but does not describe a protocol for immortalized LCLs. Although (to avoid cell death) we did not directly count the number of individual iPSCs, we were able to generate and clone approximately 48 mature iPSC colonies from each of the two original LCLs (one million LCLs per reaction). From those 48 clones, 8 were propagated until passage 10, and 2 clones were taken to passage 21 and used for subsequent characterization.
Traditionally iPSC pluripotentiality is evaluated by injecting iPSCs under the kidney capsule of immune-compromised mice and then assaying the subsequent teratoma growth for cell types derived from all three germ layers. Because it involves the use of mice, this assay is very expensive and typically takes months to complete, making it both time- and cost-inefficient. Recently, Bock et al. described the development of a PCR-based assay that can be used to measure pluripotency and predict germ layer predisposition in hPSCs (Bock et al., 2011). From this early work, the TaqMan® hPSC Scorecard™ Panel from Life Technologies was developed and released in late 2013. This assay provides robust characterization of pluripotent cells and has shown to be consistent with in vitro differentiation and teratoma results (Fergus et al., 2014a, 2014b; Goh et al., 2013). The characterization of the four iPSC clones described herein corroborates previous data generated using the TaqMan® hPSC Scorecard™ Panel. All four clones spontaneously differentiated into EBs at passage 20 for 7 and 14 days and were tested using a Scorecard™ panel. When harvesting EBs on Day 7, one of the four clones was without lineage bias (Line-iPSC1a) and was recommended, based on the “score”, for future differentiation protocols. The other three clones, Line1-iPSC1B, Line2-iPSC2A and Line2-iPSC2B, did not show up-regulation of endoderm genes during EB formation, suggesting that these three iPSC clones are not truly pluripotent, if left to differentiate without lineage-specific guidance. However, based on data shown by Fergus et al. at ISSCR (Fergus et al., 2014a, Fergus et al., 2014b), endoderm lineage “scores” do not reach those of ectoderm or mesoderm until after 14 days of spontaneous differentiation (Fergus et al., 2014a, 2014b). Since manufacturer protocols recommending 7 days of differentiation were followed, this may help to explain why the three iPS cell lines do not show positive endoderm expression in 7 day-old EBs. In EBs harvested at day 14, endoderm-specific gene expression was apparent in all 4 clones. These data are consistent with endoderm differentiation protocols (Spence et al., 2011) and LTI data suggesting induction of endoderm differentiation takes longer than ectoderm or mesoderm (Fergus et al., 2014a, 2014b). Overall, analysis of the Scorecard™ data resulted in a finding that our clonal iPSCs were deemed suitable for use in differentiation protocols for all three germ cell lineages.
The Scorecard™ Panel assay may ultimately provide a cost-effective screening tool for ESCs and iPSCs that would facilitate more rapid advancements in stem cell research (Barrett et al., 2014; Chen et al., 2011). Results from this assay have the potential to save countless hours currently being spent attempting to differentiate iPSC clones into cell types that they are not able to produce. These results also highlight the utility and necessity of propagating multiple clonal lines from each parent LCL when making iPSCs. Although clonal lines are by definition genetically identical to the parent cell line, they can differ epigenetically from other clones derived from the same parent line. For this study we chose to propagate and characterize two clonal lines for each parent line. Although this is a modest number relative to some other reports, we have shown 3 iPSC lines, Line1-iPSC1A, Line1-iPSC1B, and Line2-iPSC2B, to be fully pluripotent and therefore suitable for differentiation protocols - a 75% success rate of clonal iPSC lines. One line, Line2-iPSC2A, did not meet all requirements for optimal downstream use. Another cell line, Line1-iPSC1B, was consistently different from the other three lines and likely needs to be passaged further to become plasmid- and EBV-independent (Figure 2). Based on these findings, we recommend propagation and characterization of at least two clonal lines from each parent LCL.
Generation of iPSCs from patient-derived cells enables the development of disease-specific cellular models, platforms for drug screening (Guan et al., 2014), and primary sources for cell replacement therapy (Wernig et al., 2008; Rauch et al., 2006; Zhang et al., 2010). The ability to use state-of-the-art technology to generate iPSCs from LCLs elevates repositories, such as the AGRE and SSC, as well as dozens of other disease-specific LCL repositories available through the Coriell Institute for Medical Research, to even more important status for accelerating research into neurodevelopmental disorders like autism. If this technology were to be broadly applied by, for example, the autism research community, results using LCL-derived iPSCs will become integrative and inter-comparable. As a potential added benefit, this cell culture model system can be readily scaled up to produce a relatively low-cost, high-throughput screening tool for pharmaceuticals (Dolmetsch and Geschwind, 2011).
5. Conclusion
This methods paper describes an efficient and reproducible protocol for the generation and characterization of patient-specific induced pluripotent stem cells from lymphoblastoid cell lines. Characterization of these lines showed that they are suitable for use in the generation of multiple cell types that can be used to study neurobiological aspects that underlie disease. Adoption of this strategy would significantly enhance the value of available cell banking repositories (e.g. Coriell Institute for Medical Research; coriell.org) for the study of mechanisms that underlie disease.
Declarations
Author contribution statement
Stephen J. Walker, David L Mack: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Ashley L Wagoner: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Dana Leavitt: Analyzed and interpreted the data; Wrote the paper.
Funding statement
This work was supported by the 2013 John H. Tietze Stem Cell Scientist Award at the University of Washington Institute for Stem Cell and Regenerative Medicine (DLM) and an award from the Jane Botsford Johnson Foundation (SJW).
Data availability statement
Data will be made available on request.
Declaration of interests statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
We gratefully acknowledge the resources provided by the Autism Genetic Resource Exchange (AGRE) Consortium∗ and the participating AGRE families. The Autism Genetic Resource Exchange is a program of Autism Speaks and is supported, in part, by grant 1U24MH081810 from the National Institute of Mental Health to Clara M. Lajonchere (PI).
References
- Barrett R., Ornelas L., Yeager N., Mandefro B., Sahabian A., Lenaeus L., Targan S.R., Svendsen C.N., Sareen D. Reliable generation of induced pluripotent stem cells from human lymphoblastoid cell lines. Stem Cell Trans. Med. 2014;3:1429–1434. doi: 10.5966/sctm.2014-0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock C., Kiskinis E., Verstappen G., Gu H., Boulting G., Smith Z.D., Ziller M., Croft G.F., Amoroso M.W., Oakley D.H., Gnirke A., Eggan K., Meissner A. Reference maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell. 2011;144:439–452. doi: 10.1016/j.cell.2010.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennand K.J., Simone A., Jou J., Gelboin-Burkhart C., Tran N., Sangar S., Li Y., Mu Y., Chen G., Yu D., McCarthy S., Sebat J., Gage F.H. Modelling schizophrenia using human induced pluripotent stem cells. Nature. 2011;473:221–225. doi: 10.1038/nature09915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennand K.J., Simone A., Tran N., Gage F.H. Modeling psychiatric disorders at the cellular and network levels. Mol. Psychiatr. 2012;17:1239–1253. doi: 10.1038/mp.2012.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanda S., Marro S., Wernig M., Südhof T.C. Neurons generated by direct conversion of fibroblasts reproduce synaptic phenotype caused by autism-associated neuroligin-3 mutation. Proc. Natl. Acad. Sci. U.S.A. 2013;110:16622–16627. doi: 10.1073/pnas.1316240110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., Gulbranson D.R., Hou Z., Bolin J.M., Ruotti V., Probasco M.D., Smuga-Otto K., Howden S.E., Diol N.R., Propson N.E., Wagner R., Lee G.O., Antosiewicz-Bourget J., Teng J.M., Thomson J.A. Chemically defined conditions for human iPSC derivation and culture. Nat. Methods. 2011;8:424–429. doi: 10.1038/nmeth.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S.M., Liu H., Chaudhari P., Kim Y., Cheng L., Feng J., Sharkis S., Ye Z., Jang Y.-Y. Reprogramming of EBV-immortalized B-lymphocyte cell lines into induced pluripotent stem cells. Blood. 2011;118:1801–1805. doi: 10.1182/blood-2011-03-340620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRosa B.A., Van Baaren J.M., Dubey G.K., Lee J.M., Cuccaro M.L., Vance J.M., Pericak-Vance M.A., Dykxhoorn D.M. Derivation of autism spectrum disorder-specific induced pluripotent stem cells from peripheral blood mononuclear cells. Neurosci. Lett. 2012;516:9–14. doi: 10.1016/j.neulet.2012.02.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimos J.T., Rodolfa K.T., Niakan K.K., Weisenthal L.M., Mitsumoto H., Chung W., Croft G.F., Saphier G., Leibel R., Goland R., Wichterle H., Henderson C.E., Eggan K. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science. 2008;321:1218–1221. doi: 10.1126/science.1158799. [DOI] [PubMed] [Google Scholar]
- Dolmetsch R., Geschwind D.H. The human brain in a dish: the promise of iPSC-derived neurons. Cell. 2011;145:831–834. doi: 10.1016/j.cell.2011.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fergus J., Quintanilla R., Lakshmipathy U. Characterizing pluripotent stem cells using the TaqMan(®) hPSC Scorecard (TM) Panel. Methods Mol. Biol. 2014;1307:25–37. doi: 10.1007/7651_2014_109. [DOI] [PubMed] [Google Scholar]
- Fergus J., Quintanilla R., Lakshmipathy U. International Society of Stem Cell Research. 2014. Utility of Taqman Scorecard Assay in the assessment of functional pluripotent stem cells. [Google Scholar]
- Fujimori K., Tezuka T., Ishiura H., Mitsui J., Doi K., Yoshimura J., Tada H., Matsumoto T., Isoda M., Hashimoto R., Hattori N., Takahashi T., Morishita S., Tsuji S., Akamatsu W., Okano H. Modeling neurological diseases with induced pluripotent cells reprogrammed from immortalized lymphoblastoid cell lines. Mol. Brain. 2016;9:88. doi: 10.1186/s13041-016-0267-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geschwind D.H., Sowinski J., Lord C., Iversen P., Shestack J., Jones P., Ducat L., Spence S.J. The autism genetic resource exchange: a resource for the study of autism and related neuropsychiatric conditions. Am. J. Hum. Genet. 2001;69:463–466. doi: 10.1086/321292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh P.A., Caxaria S., Casper C., Rosales C., Warner T.T., Coffey P.J., Nathwani A.C. A systematic evaluation of integration free reprogramming methods for deriving clinically relevant patient specific induced pluripotent stem (iPS) cells. PloS One. 2013;8 doi: 10.1371/journal.pone.0081622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan X., Mack D.L., Moreno C.M., Strande J.L., Mathieu J., Shi Y., Wang Z., Liu G., Lawlor M.W., Moorefield E.C., Jones T.N., Fugate J.A., Furth M.E., Murry C.E., Ruohola-Baker H., Zhang Y., Santana L.F., Childers M.K. Dystrophin-deficient cardiomyocytes derived from human urine: new biologic reagents for drug discovery. Stem Cell Res. 2014;12:467–480. doi: 10.1016/j.scr.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong H., Takahashi K., Ichisaka T., Aoi T., Kanagawa O., Nakagawa M., Okita K., Yamanaka S. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature. 2009;460:1132–1135. doi: 10.1038/nature08235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher R.J., Geigenmüller U., Hovhannisyan H., Trautman E., Pinard R., Rathmell B., Carpenter R., Margulies D. High-throughput sequencing of mGluR signaling pathway genes reveals enrichment of rare variants in autism. PloS One. 2012;7 doi: 10.1371/journal.pone.0035003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Curran J.E., Glahn D.C., Blangero J. Utility of lymphoblastoid cell lines for Induced pluripotent stem cell generation. Stem Cell. Int. 2016;2016 doi: 10.1155/2016/2349261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig T.E., Bergendahl V., Levenstein M.E., Yu J., Probasco M.D., Thomson J.A. Feeder-independent culture of human embryonic stem cells. Nat. Methods. 2006;3:637–646. doi: 10.1038/nmeth902. [DOI] [PubMed] [Google Scholar]
- Mack D.L., Guan X., Wagoner A., Walker S.J., Childers M.K. Disease-in-a-dish: the contribution of patient-specific induced pluripotent stem cell technology to regenerative rehabilitation. Am. J. Phys. Med. Rehabil. 2014;93:S155–168. doi: 10.1097/PHM.0000000000000141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetto M.C., Brennand K.J., Boyer L.F., Gage F.H. Induced pluripotent stem cells (iPSCs) and neurological disease modeling: progress and promises. Hum. Mol. Genet. 2011;20:R109–115. doi: 10.1093/hmg/ddr336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetto M.C., Carromeu C., Acab A., Yu D., Yeo G.W., Mu Y., Chen G., Gage F.H., Muotri A.R. A model for neural development and treatment of Rett syndrome using human induced pluripotent stem cells. Cell. 2010;143:527–539. doi: 10.1016/j.cell.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajesh D., Dickerson S.J., Yu J., Brown M.E., Thomson J.A., Seay N.J. Human lymphoblastoid B-cell lines reprogrammed to EBV-free induced pluripotent stem cells. Blood. 2011;118:1797–1800. doi: 10.1182/blood-2011-01-332064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch U., Hänsgen A., Hagl C., Holland-Cunz S., Schäfer K.-H. Isolation and cultivation of neuronal precursor cells from the developing human enteric nervous system as a tool for cell therapy in dysganglionosis. Int. J. Colorectal Dis. 2006;21:554–559. doi: 10.1007/s00384-005-0051-z. [DOI] [PubMed] [Google Scholar]
- Shcheglovitov A., Shcheglovitova O., Yazawa M., Portmann T., Shu R., Sebastiano V., Krawisz A., Froehlich W., Bernstein J.A., Hallmayer J.F., Dolmetsch R.E. SHANK3 and IGF1 restore synaptic deficits in neurons from 22q13 deletion syndrome patients. Nature. 2013;503:267–271. doi: 10.1038/nature12618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sie L., Loong S., Tan E.K. Utility of lymphoblastoid cell lines. J. Neurosci. Res. 2009;87:1953–1959. doi: 10.1002/jnr.22000. [DOI] [PubMed] [Google Scholar]
- Spence J.R., Mayhew C.N., Rankin S.A., Kuhar M.F., Vallance J.E., Tolle K., Hoskins E.E., Kalinichenko V.V., Wells S.I., Zorn A.M., Shroyer N.F., Wells J.M. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature. 2011;470:105–109. doi: 10.1038/nature09691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spike B.T., Wahl G.M. p53, stem cells, and reprogramming: tumor suppression beyond guarding the genome. Genes Cancer. 2011;2:404–419. doi: 10.1177/1947601911410224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Thomas S.M., Kagan C., Pavlovic B.J., Burnett J., Patterson K., Pritchard J.K., Gilad Y. Reprogramming LCLs to iPSCs results in recovery of donor-specific gene expression signature. PLoS Genet. 2015;11 doi: 10.1371/journal.pgen.1005216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernig M., Zhao J.-P., Pruszak J., Hedlund E., Fu D., Soldner F., Broccoli V., Constantine-Paton M., Isacson O., Jaenisch R. Neurons derived from reprogrammed fibroblasts functionally integrate into the fetal brain and improve symptoms of rats with Parkinson’s disease. Proc. Natl. Acad. Sci. U. S. A. 2008;105:5856–5861. doi: 10.1073/pnas.0801677105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Vodyanik M.A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J.L., Tian S., Nie J., Jonsdottir G.A., Ruotti V., Stewart R., Slukvin, II Thomson J.A. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Zhang D., Brinas I.M., Binder B.J., Landman K.A., Newgreen D.F. Neural crest regionalisation for enteric nervous system formation: implications for Hirschsprung’s disease and stem cell therapy. Dev. Biol. 2010;339:280–294. doi: 10.1016/j.ydbio.2009.12.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.